 A screening of 18 compounds identified five coumarin derivatives with subnanomolar activity against the 5HT1A receptor.
A screening of 18 compounds identified five coumarin derivatives with subnanomolar activity against the 5HT1A receptor.
Abstract
A series of 18 new 5-[3-(4-aryl-1-piperazinyl)propoxy]coumarin derivatives from the corresponding bromoalkyl derivatives have been designed and synthesized by us using a microwave-assisted protocol. Radioligand binding assays of this series of compounds as well as a previously synthesized series of 17 structurally-similar compounds showed that six systems have very high affinities to the 5-HT1A receptor (0.3–1.0 nM) and good selectivity against the 5-HT2A receptor. Molecular docking, structural studies and structure–activity relationship studies were used to gain more insight into the atomistic details of ligand binding and rationalize the obtained results.
1. Introduction
Despite the wealth of known serotonin and dopamine receptor antagonists, agonists and partial agonists, there is still a lot of interest in designing and synthesizing new agents interacting with these G protein-coupled receptors (GPCRs) and potentially modulating the release of various neurotransmitters. 5-HT is a neurotransmitter that plays an important role in several brain functions and are involved in the pathogenesis of several psychiatric and neurological disorders, including anxiety, depression, schizophrenia, and Parkinson's disease.1 Among many classes of chemical compounds, those containing the N-arylpiperazine moiety are often biologically active against one or several serotonin 5-HT1 receptors.2 Linking a carbocyclic or heterocyclic system to one of the piperazine nitrogen atoms via a lipophilic chain has been shown to result in enhanced selectivity against the 5-HT1A receptor versus the α1 and D2 receptors3,4 and/or 5-HT1A/D2A receptors.5
Oxygen-containing heterocyclic compounds, including coumarins, are on the other hand an important class of natural compounds, reported to have antitumor, antifungal and antimicrobial activities.6,7 7-Hydroxycoumarin derivatives have been also shown to increase central nervous system (CNS) activity.8 For example, arylpiperazines linked to a coumarin system via a propyloxy chain, such as 4-methyl-7-(3-(4-phenylpiperazin-1-yl)propoxy)coumarin, have pronounced affinity towards both D2A and 5-HT1A receptors.9,10 7-(4-(4-(6-fluorobenzo[d]isoxazol-3-yl)-piperidin-1-yl)butoxy)-4-methyl-8-chloro-2H-chromen-2-one possessed, on the other hand, unique pharmacological features which include high affinity for dopamine D2/D3 and serotonin 5-HT1A/5-HT2A receptors. This compound has also shown low affinity for 5-HT2C and H1 receptors and hERG channels.11 A similar compound, 7-(4-(4-(6-fluorobenzo[d]isoxazol-3-yl)piperidin-1-yl)butoxy)-6-methyl-2,3-dihydrocyclopenta[c]chromen-4(1H)-one, has shown high affinity for dopamine D2/D3 receptors and serotonin 5-HT1A/5-HT2A receptors coupled with a low affinity for H1 receptors. In animal models, this compound inhibited apomorphine-induced climbing and MK-801-induced hyperactivity without observable catalepsy at the highest dose tested; interestingly, this compound was more potent than clozapine.12 Other examples of coumarin-containing derivatives with CNS activity include 7-[3-(4-aryl-1-piperazinyl)propoxy]coumarins, which showed high affinity for α1A, D2 and 5-HT2A receptors.13
In general, there is a wealth of 7-hydroxycoumarin derivatives which have been synthesized and comprehensively tested in biological screenings but the library of derivatives of 5-hydroxycoumarin is less prominent, though also potentially interesting. As a result we decide to combine two important classes of compounds commonly used in CNS disorder therapies, namely systems containing the N-arylpiperazine moiety and 5-hydroxycoumarin derivatives to design new leads. Previously we have synthesized a series of 18 derivatives of 8-acetyl-7-hydroxy-4-methylcoumarin with five compounds possessing affinities to 5HT1A receptors in the 0.8–1.3 nM range.14 In this work we have designed a series of N-arylpiperazine derivatives attached to 5-hydroxy-4,7-dimethylcoumarin via a propyloxy chain. We report the synthesis, structural analysis and molecular docking studies of a series of 18 coumarin derivatives characterized by a high affinity to 5-HT1A and good selectivity against the 5-HT2A receptors. The goal of this study was to achieve an optimum interaction with serotonin receptors for a new series of chemical systems (3b–11b, 3d–10d) and compare them with a previously synthesized, structurally similar series of compounds (3a–11a, 3c–10c) with potential cytotoxic and antimicrobial activities (Scheme 1).15 The synthesized series of arylpiperazine derivatives, prepared in this study, can be used as a starting point for further optimization of ligands against 5-HT receptors.
Scheme 1. Synthesis of aryl/heteroarylpiperazinyl derivatives of 5-hydroxy-4,7-dimethylcoumarin.
2. Experimental
2.1. General methods
Reagents were purchased from Aldrich or Merck, of the higher grade available, and used without further purification. Solvents were used as received from commercial suppliers, and no further attempts were made to purify or dry them. Melting points were determined with a ElectroThermal 9001 Digital Melting Point apparatus (ElectroThermal, Essex, UK) and are uncorrected. A microwave oven, Plazmatronika 1000 W, equipped with a single mode cavity suitable for microscale synthesis and a microwave choked outlet connected to an external condenser set to 30% power was used. High resolution mass spectra were recorded on a Quattro LCT (TOF). 1H NMR and 13C NMR spectra in solution were recorded at 25 °C with a Varian Unity plus-300 spectrometer and standard Varian software was employed (Varian, Inc., Palo Alto, CA, USA). TLC was carried out using Kieselgel 60 F254 sheets (Merck, Darmstadt, Germany) and spots were visualized by UV – 254 and 365 nm. Kieselgel 60 was used for column chromatography.
2.2. General procedure to obtain compounds 2b and 2d
A mixture of 5-hydroxy-4,7-dimethylcoumarin (1b) (0.190 g, 1 mmol) or 6-acetyl-5-hydroxy-4,7-dimethylcoumarin (1d) (0.232 g, 1 mmol), 1,3-dibrompropane (0.34 cm3, 3 mmol), anhydrous K2CO3 (0.1 g), and a catalytic amount of KI were placed in a microwave flask. The mixture was refluxed (acetonitrile) in the temperature range 70–80 °C in the monomode microwave oven (300 W) and monitored by TLC on silica-gel plates (eluent CHCl3–MeOH 10 : 0.25). Two cycles were needed to obtain 5-(3-bromopropoxy)-4,7-dimethylcoumarin (2b) or 6-acetyl-5-(3-bromopropoxy)-4,7-dimethylcoumarin (2d). Then the solvent was removed on a rotary evaporator and the oily residue was purified by column chromatography (chloroform : hexane 5 : 3).
2.3. General procedure to obtain compounds 3b–11b and 3d–11d
A mixture of derivative 2b or 2d (1 mmol, 0.310 g or 1 mmol, 0.353 g, respectively) and the corresponding amine (2 mmol), anhydrous K2CO3 (0.3 g), and a catalytic amount of KI were placed in a microwave flask. The mixture was refluxed (acetonitrile) in the temperature range 70–80 °C in the monomode microwave oven (300 W) and monitored by TLC on silica-gel plates (eluent CHCl3–MeOH 10 : 0.25). Four cycles were needed to obtain 5-[3-(4-aryl-1-piperazinyl)propoxy]coumarins. Then the mixture was filtered off and the solvent was evaporated. The residue was purified by column chromatography (chloroform : hexane, 5 : 2). Atom numbering, 1H NMR and 13C NMR spectra of all synthesized compounds is available in the ESI.†
2.4. Pharmacology
Preparation of solutions of test and reference compounds for radioligand binding assays
1 mM stock solutions of tested compounds were prepared in DMSO. Serial dilutions of compounds were prepared in a 96-well microplate in assay buffer using an automated pipetting system, epMotion 5070 (Eppendorf). Each compound was tested in 6 concentrations from 10–5 to 10–10 M (final concentration).
5-HT2A receptor binding assay
Radioligand binding was performed using membranes from CHO-K1 cells stably transfected with the human 5-HT2A receptor (CHO-K1 cells which had undergone 10 passages were acquired from Perkin Elmer – product number ES-310-C). All assays were carried out in duplicate. 50 μl working solution of the tested compounds, 50 μl [3H]-ketanserin (final concentration 1 nM) and 150 μl diluted membranes (7 μg protein per well) prepared in assay buffer (50 mM Tris, pH 7.4, 4 mM CaCl2, 0.1% ascorbic acid) were transferred to a polypropylene 96-well microplate using a 96-well pipetting station, Rainin Liquidator (Mettler Toledo). Mianserin (10 μM) was used to define nonspecific binding. The microplate was covered with sealing tape, mixed and incubated for 60 minutes at 27 °C. The reaction was terminated by rapid filtration through a GF/B filter mate presoaked with 0.5% polyethyleneimine for 30 minutes. Ten rapid washes with 200 μl 50 mM Tris buffer (4 °C, pH 7.4) were performed using an automated harvester system, Harvester-96 MACH III FM (Tomtec). The filter mates were dried at 37 °C in a forced air fan incubator and then a solid scintillator, MeltiLex, was melted on the filter mates at 90 °C for 5 minutes. Radioactivity was counted in a MicroBeta2 scintillation counter (PerkinElmer). Data were fitted by a one-site curve-fitting equation with Prism 6 (GraphPad Software) and Ki values were estimated from the Cheng–Prusoff equation.
5-HT1A receptor binding assay
Radioligand binding was performed using membranes from CHO-K1 cells stably transfected with the human 5-HT1A receptor (PerkinElmer). All assays were carried out in duplicate. 50 μl working solution of the tested compounds, 50 μl [3H]-8-OH-DPAT (final concentration 1 nM) and 150 μl diluted membranes (10 μg protein per well) prepared in assay buffer (50 mM Tris, pH 7.4, 10 mM MgSO4, 0.5 mM EDTA, 0.1% ascorbic acid) were transferred to a polypropylene 96-well microplate using a 96-well pipetting station, Rainin Liquidator (Mettler Toledo). Serotonin (10 μM) was used to define nonspecific binding. The microplate was covered with sealing tape, mixed and incubated for 60 minutes at 27 °C. The reaction was terminated by rapid filtration through a GF/C filter mate presoaked with 0.3% polyethyleneimine for 30 minutes. Ten rapid washes with 200 μl 50 mM Tris buffer (4 °C, pH 7.4) were performed using an automated harvester system Harvester-96 MACH III FM (Tomtec). The filter mates were dried at 37 °C in a forced air fan incubator and then a solid scintillator, MeltiLex, was melted on the filter mates at 90 °C for 4 minutes. Radioactivity was counted in a MicroBeta2 scintillation counter (PerkinElmer). Data were fitted by a one-site curve-fitting equation with Prism 6 (GraphPad Software) and Ki values were estimated from the Cheng–Prusoff equation.
2.5. Molecular docking studies
The homology model for the docking part of the study has been prepared using Prime.16 BLAST homology search revealed that human dopamine receptor D3 had the most similar sequence to 5HT1A (among all templates available in Prime) and we used it to build the homology model. The obtained model was verified to have all the amino acid residues implied in the binding of various agonists and antagonists in the binding pocket as well as appropriate distances between crucial residues, in accordance with previous studies.17,18
All ligands have been prepared manually in Maestro (Maestro, Schrödinger, LLC, New York, NY, 2016). We used LigPrep software (LigPrep, Schrödinger, LLC, New York, NY, 2016.) to obtain the starting structure for docking and Epik to quickly evaluate the pKa of the nitrogen atom in the piperazine ring.19 In the next step of the investigation we used a flexible docking protocol, as implemented in Autodock 4.2.20 Atomic interaction energy grids have been calculated using probes corresponding to each atomic type found in the ligand, at standard 0.375 Å grid resolution. We have used a 48 × 52 × 40 Å box centered protein binding pocket. In the docking part we have used Autodock 4.2 with the Lamarckian Genetic Algorithm and standard options, but including 100 docking poses per compound and 5 000 000 energy evaluations per docking.21 In all docking experiments each ligand has been treated in a fully flexible manner with the Gasteiger partial charges added by AutoDockTools 4. We have separately docked all compounds in both possible protonation states (charged and neutral); in Table 2 we report the Ki values only for the protonation state giving the lower Ki value. The protein has been treated as a rigid model, also with Gasteiger partial charges, with seven residues of the binding pocket (D116, V117, W358, F361, F362, N386 and Y390) described in a fully flexible manner. Ki values reported in Table 2 were obtained by converting estimated ligand binding energies using the standard equation for Gibbs free energy. ADME analysis, presented in the ESI,† has been performed using the QikProp software.
Table 2. Computationally estimated Ki values of serotonergic receptor 5HT1A and pKa values of the basic piperazine nitrogen atom of the studied compounds.
| Compound | K i (nM) | pKa |
| 3a | 3.73 | 7.55 |
| 4a | 29.07 | 7.44 |
| 5a | 2.09 | 7.55 |
| 6a | 15.56 | 7.39 |
| 7a | 10.74 | 7.27 |
| 8a | 3.37 | 7.49 |
| 9a | 5.38 | 7.39 |
| 10a | 5.16 | 7.55 |
| 11a | 11.57 | 6.86 |
| 3b | 2.46 | 7.19 |
| 4b | 26.08 | 7.08 |
| 5b | 3.38 | 7.19 |
| 6b | 54.54 | 6.98 |
| 7b | 2.77 | 6.91 |
| 8b | 1.22 | 7.13 |
| 9b | 1.99 | 7.03 |
| 10b | 10.57 | 7.19 |
| 11b | 39.97 | 6.50 |
| 3c | 21.25 | 7.55 |
| 4c | 75.84 | 7.44 |
| 5c | 11.42 | 7.55 |
| 6c | 7.33 | 7.39 |
| 7c | 33.63 | 7.27 |
| 8c | 29.65 | 7.49 |
| 9c | 5.65 | 7.39 |
| 11c | 97.67 | 6.86 |
| 3d | 2.22 | 7.19 |
| 4d | 63.60 | 7.08 |
| 5d | 4.07 | 7.19 |
| 6d | 85.07 | 6.98 |
| 7d | 11.43 | 6.91 |
| 8d | 3.24 | 7.13 |
| 9d | 1.57 | 7.03 |
| 10d | 6.86 | 7.19 |
| 11d | 16.75 | 6.50 |
3. Results and discussion
3.1. Chemistry
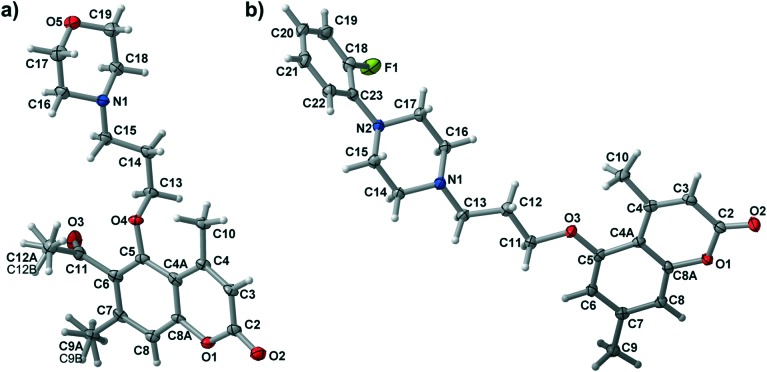
The synthesis of new aryl/heteroarylpiperazinyl derivatives of 5-hydroxy-4,7-dimethylcoumarin (3b–11b, 3d–10d) was carried out through an efficient synthetic protocol outlined in Scheme 1. The bromoalkyl derivatives of coumarins (2b, 2d) were obtained according to the procedure described earlier and used as an alkylating agent in the next step.13 Starting compounds 5-hydroxy-4,7-dimethylcoumarin (1a) and 6-acetyl-5-hydroxy-4,7-dimethylcoumarin (1b) were converted to 5-(3-bromopropoxy)-4,7-dimethylcoumarin (2b) and 6-acetyl-5-(3-bromopropoxy)-4,7-dimethylcoumarin (2d), respectively, by refluxing with 1,3-dibromopropane in the presence of KI and K2CO3 in acetonitrile using microwave irradiation. In the second step, reactions with the corresponding N-substituted piperazine in acetonitrile and in the presence of K2CO3 and KI yielded the final compounds 3b–11b and 3d–10d, respectively. All synthesized compounds were purified by column chromatography using appropriate solvents. The structures of target compounds were characterized by 1H NMR, 13C NMR and HRMS spectrometry, and the molecular structures of 3b and 6d were confirmed by X-ray crystallography (see ESI†). Numbering of atoms for both structures together with atomic displacement parameters at the 50% probability level is presented in Fig. 1.
Fig. 1. Numbering scheme and thermal ellipsoid at the 50% probability level for 6d a) and 3b b).
3.2. Pharmacology and structure–activity relationship
All studied compounds were tested for their affinity to 5-HT1A and 5-HT2A receptors and most of them showed affinities in the nanomolar range (Table 1). For 5-HT1A we can immediately notice a trend, since most of the compounds from the a group (3-carbon linker) show lower Ki values than those from group b (4-carbon linker), but interestingly compounds of the c group (3-carbon linker) are on average worse than those of group d (4-carbon linker). These results suggest that the 4-carbon linker allows the ligands to find more favorable interactions with residues in the binding pocket, but the addition of the acetyl group to the C-6 position makes the ligand too large to optimally fit into the pocket.
Table 1. Binding affinities to serotonergic receptors of the reference and tested compounds.
| Compound |
K
i (nM) ± SEM |
|
| 5-HT1A [3H]8-OH-DPAT | 5-HT2A [3H]ketanserin | |
| 3a | 1.7 ± 0.05 | 120.0 ± 9.0 |
| 4a | 6.7 ± 0.2 | 2300.0 ± 250.0 |
| 5a | 1.0 ± 0.1 | 184.0 ± 4.7 |
| 6a | 3500.0 ± 350.0 | >10 000.0 |
| 7a | 809.0 ± 80.0 | >10 000.0 |
| 8a | 60.0 ± 2.0 | 465.0 ± 34.7 |
| 9a | 8.0 ± 0.4 | 167.0 ± 15.0 |
| 10a | 1.5 ± 0.2 | 112.0 ± 4.4 |
| 11a | 389.0 ± 21.0 | >5000.0 |
| 3b | 3.0 ± 0.1 | 55.0 ± 3.7 |
| 4b | 186.0 ± 7.0 | 4565.0 ± 269.0 |
| 5b | 6.0 ± 0.8 | 106.0 ± 11.0 |
| 6b | 1304.0 ± 50.0 | >10 000.0 |
| 7b | 1210.0 ± 120.0 | >10 000.0 |
| 8b | 161.0 ± 15.0 | 70.0 ± 6.4 |
| 9b | 4.0 ± 0.8 | 42.0 ± 1.5 |
| 10b | 3.0 ± 0.2 | 200.0 ± 17.9 |
| 11b | 450.0 ± 13.0 | >5000.0 |
| 3c | 0.7 ± 0.05 | 9.0 ± 0.3 |
| 4c | 4.8 ± 0.08 | 2500.0 ± 84.0 |
| 5c | 15.0 ± 0.5 | 520.0 ± 37.0 |
| 6c | 250.0 ± 3.0 | 4460.0 ± 160.0 |
| 7c | 33.0 ± 1.0 | 46.0 ± 0.3 |
| 8c | 7.0 ± 1.0 | 1.7 ± 0.2 |
| 9c | 250.0 ± 29.0 | 2650.0 ± 215.0 |
| 11c | 308.0 ± 14.4 | 1595.0 ± 84.0 |
| 3d | 0.3 ± 0.05 | 28.0 ± 0.5 |
| 4d | 11.0 ± 1.5 | 3000.0 ± 334.0 |
| 5d | 0.4 ± 0.02 | 48.0 ± 2.8 |
| 6d | 850.0 ± 11.0 | >10 000.0 |
| 7d | 159.0 ± 8.5 | 286.0 ± 12.0 |
| 8d | 1.2 ± 0.08 | 4.0 ± 0.5 |
| 9d | 0.4 ± 0.03 | 66.0 ± 3.0 |
| 10d | 0.3 ± 0.04 | 64.0 ± 2.3 |
| 11d | 154.0 ± 6.6 | >10 000.0 |
| Serotonin | 1.1 ± 0.1 | — |
| Mianserin | — | 1.2 ± 0.06 |
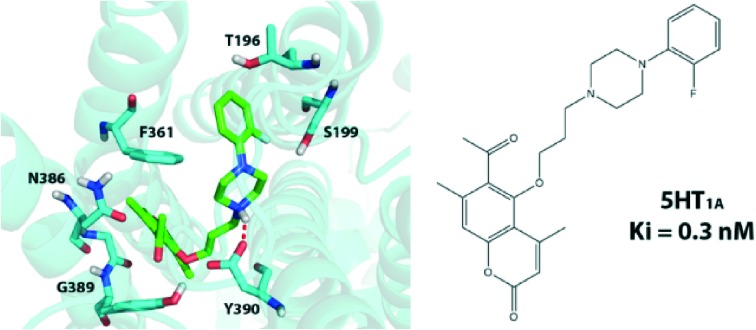
Analyzing the 5-HT1A results in depth we see that some substituents at the R2 position (3, 5, 9 and 10) give, in general, very favorable results with very low Ki values. In the group of compounds without the acetyl group (3a–11a, 3b–11b) only 4,7-dimethyl-5-{4-[4-(3-methoxyphenyl)piperazin-1-yl]butoxy}coumarin (5a) was active at the serotonin level with a Ki value of 1.0 nM (Ki for serotonin was found to be 1.1 nM), while in the group of compounds with the acetyl group (3c–10c, 3d–11d) 6-acetyl-4,7-dimethyl-5-{4-[4-(2-fluorophenyl)piperazin-1-yl]butoxy}coumarin (3c), 6-acetyl-4,7-dimethyl-5-{3-[4-(2-fluorophenyl) piperazin-1-yl]propoxy}coumarin (3d), 6-acetyl-4,7-dimethyl-5-{3-[4-(3-methoxyphenyl)piperazin-1-yl]propoxy}coumarin (5d), 6-acetyl-4,7-dimethyl-5-{3-[4-(2,3-dichlorophenyl)piperazin-1-yl]propoxy}coumarin (9d) and 6-acetyl-4,7-dimethyl-5-{3-[4-(2-cyanophenyl)piperazin-1-yl]propoxy}coumarin (10d) have all shown Ki values lower than 1.0 nM (Ki = 0.7 nM for 3c, 0.3 for 3d, 0.4 for 5d, 0.4 for 9d and 0.3 for 10d). Clearly, (2-fluorophenyl)piperazinyl (3), (3-methoxyphenyl)piperazinyl (5), (2,3-dichlorophenyl) piperazinyl (9) and (2-cyanophenyl)piperazinyl (10) moieties are able to form favorable interactions within the binding pocket of the receptor. From this data we can conclude that for the investigated arylpiperazinyl derivatives of 5-hydroxy-4,7-dimethylcoumarin introduction of fluoro-, methoxy-, dichloro- or cyano- substituents in the ortho, meta or ortho and meta positions of the phenyl group increases their affinities to 5-HT1A receptors (Ki = 0.3–1.0 nM). Replacing the piperazine with a morpholine ring displayed a dramatic decrease in affinities for 5-HT1A receptors (Ki = 250.0–3500.0 nM, see compounds 6a–d). The same is true in the case of replacement of the phenyl ring with a heterocyclic moiety (compounds 4a–d, 11a–d), which also causes a decrease in affinities (Ki = 154.0–450.0 nM). Introduction of the nitro- or chloro- substituents in the para position of the phenyl ring also decreases the activity, regardless of the alkyl chain length or the presence of an acetyl group in the coumarin core (see compounds 7a–d, 8a–d). It is worth noting that we have previously obtained similar results for arylpiperazinyl derivatives of 8-acetyl-7-hydroxy-4-methylcoumarin and the results between those two classes of compounds are very similar.14 In particular compounds with the highest affinities to the 5-HT1A receptor from both series bear either the (2-fluorophenyl)piperazinyl or (3-methoxyphenyl)piperazinyl moieties, which is also in agreement with the data reported earlier by Chen et al.11 Similarly, both sets show low affinities upon the introduction of either the morpholine ring or the (4-nitro)piperazinyl moiety.
All investigated compounds showed rather weak affinity to 5-HT2A receptors, usually much weaker than that to 5-HT1A (see Table 1). Approximately half of them gave Ki values in the micromolar range, rendering them completely uninteresting from a practical point of view. Only three of them (6-acetyl-4,7-dimethyl-5-{4-[4-(4-chlorophenyl)piperazin-1-yl]butoxy}coumarin (8c), 6-acetyl-4,7-dimethyl-5-{3-[4-(4-chlorophenyl) piperazin-1-yl]propoxy}coumarin (8d) and 6-acetyl-4,7-dimethyl-5-{4-[4-(2-fluorophenyl)piperazin-1-yl]butoxy}coumarin (3c)) had affinities below the 10 nM threshold, with Ki values of 1.7, 4.0 and 9.0 nM, respectively. Compounds bearing the (4-chlorophenyl)piperazine moiety (8c and 8d) and an acetyl group at the 6-position of the coumarin ring produced the best results regardless of the alkyl linker length. Nevertheless, none of the studied compounds reached the Ki value of mianserin, a known 5HT2A antagonist/inverse agonist (Ki = 1.2 nM). It is worth pointing out, though, that compounds 8b and 8c displayed higher affinity towards 5-HT2A than 5-HT1A.
3.3. Molecular docking studies
The results of the computational part of this study are presented in Table 2. We can see that the general trends of binding to 5-HT1A receptors are reproduced well, since the strongest binding to this receptor was predicted for compounds belonging to the 3, 5, 9 and 10 series. In many cases, particularly for ligands with the smallest/best Ki values, computationally estimated values are very close to the experimental ones, i.e. for 3b (experimental Ki of 3.0 ± 0.1 nM versus computational value of 2.46 nM). Additionally, all presented ligand poses exhibit the crucial interaction between the basic nitrogen atom of piperazine and D116, known to play a vital role in the stabilization of the ligands of this class in the binding pocket.17
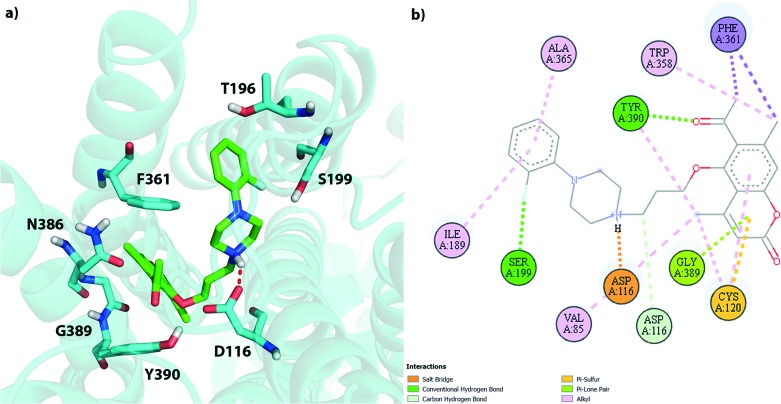
Based on the computational results we can try to rationalize the obtained experimental Ki values. Fig. 2 presents the binding pocket and all ligand-protein interactions found for 3d – the compound with the lowest experimental Ki value (0.3 ± 0.05 nM) found in this study. We can see that there are multiple favorable interactions between the ligand and the residues in the binding pocket. The most important ones are the salt bridge between the basic piperazine nitrogen atom and D116 as well as the hydrogen bonds between S199 and the fluorine atom of the phenyl group and Y390 and the acetyl group. There are also multiple van der Waals interactions which stabilize both parts of the ligand. Due to its relatively high flexibility compound 3d is able to find a number of relatively strong and favorable interactions, which translate into a very low Ki value.
Fig. 2. Schematic representation of the 3d pose in the 5HT1A binding pocket; a) 3D view of all residues within 3 Å of the ligand; b) 2D view of all residues interacting with the ligand.
Franchini et al. synthesized earlier a number of series of compounds based on 1,4-dioxa-spiro[4.5]decane, 1-oxa-4-thiaspiro- and 1,4-dithiaspiro[4.5]decane or similar scaffolds connected to either piperazine or aryloxyethylamine derivatives, with high structural similarity to our series.22–24 As expected their obtained binding poses are similar to ours. All piperazine-derived ligands are anchored by the strong interaction between the basic nitrogen atom and D116. The phenylpiperazynyl part of our ligands is positioned between helices 5 and 6, also very similarly to the previous results. All compounds belonging to the (2-fluorophenyl)piperazinyl or (3-methoxyphenyl)piperazinyl family (3 and 5) form, however, a strong interaction with S199, not described earlier, which would explain their strong binding. The coumarin part, on the other hand, is located close to helices 1, 2 and 3, similarly to the 1,4-dioxa/dithia-spiro[4.5]decane core forming hydrogen bonds with W358/Y390, as shown in Fig. 2.
Interestingly, there is a relatively large discrepancy in the experimental values of Ki and their computational estimations for a number of studied compounds (though the general trend of computational estimates of Kiversus experimental values is well preserved). These discrepancies are particularly pronounced for compounds 6a, 7a, 6c and 9c, with computational estimates of Ki one to two orders of magnitude lower than the experimental values. We can offer two explanations for these results. The first possible explanation involves the pKa values of the basic nitrogen atom of the piperazine moiety. For most studied systems computational estimates of pKa values of this atom yield a slightly basic result (7.10–7.55). This is, however, not true for compounds 6b, 7b, 11b, 11c, 6d, 7d and 11d with pKa values below 7. The less basic piperazine nitrogen atoms of these systems are not able to form strong interactions with Asp116, greatly reducing their binding affinities. Since the accuracy of our pKa estimates is around 0.5–1 pH unit it is likely that other systems with large experimental Ki values, including 6a, 7a, 6c and 9c, are also affected by this feature. The second possible explanation can be attributed to the imperfection of our computational model of the 5HT1A receptor, particularly in modelling the binding site. The poor experimental affinities of the 7a–d series can be likely attributed to the size of these particular compounds, since the nitro group in the para position makes them the largest molecules in the investigated set. These results stipulate that our modelled binding site is slightly too large, since it can still incorporate these ligands and suggest strong binding, which is not in agreement with experimental data.
4. Conclusions
In this study we described the synthesis of a new series of 18 derivatives of 5-hydroxy-4,7-dimethylcoumarin (3b–11b, 3d–11d). We also performed pharmacological evaluation of the synthesized compounds as well as a series previously reported, structurally similar compounds (3a–11a, 3c–10c) as potential antipsychotic agents. Among all synthesized derivatives, compounds 3d, 5d, 9d and 10d showed very high, subnanomolar affinities to the serotonin 5-HT1A receptor and good selectivity against the 5-HT2A receptor. In this series of compounds the presence of the three-carbon alkyl linker between the coumarin and piperazine moieties, the acetyl group in the C-6 position of the coumarin ring and the substituents in the ortho, meta or ortho and meta positions of the phenyl group of piperazine all increase the affinities to the 5-HT1A receptor. On the other hand replacing the piperazine with morpholine or with a heterocyclic ring displayed a large decrease in affinities to the 5-HT1A receptors, as did the introduction of the chloro- or nitro- substituents in the para position of the phenyl ring in the piperazine moiety.
Crystals of 4,7-dimethyl-5-{3-[4-(2-fluorophenyl)piperazin-1-yl]propoxy}coumarin (3b) and 6-acetyl-4,7-dimethyl-5-[3-(morpholin-4-yl)propoxy]coumarin (6d) were investigated using a single crystal X-ray diffraction technique and provide more evidence of their structure as well as showed some interesting intermolecular bonding patterns. Computational studies revealed, on the other hand, that compound with the highest affinity to 5HT1A (3d) form a number of strong interactions with the protein's binding pocket, including a salt bridge to D116 and hydrogen bonds to S199 and Y390. Poor binding of some of investigated compounds can be attributed to a relatively low pKa value of the piperazine nitrogen atom and the change of the total charge of the ligand, which weakens the crucial salt bridge with D116.
Conflict of interest
The authors declare no competing interest.
Supplementary Material
Acknowledgments
This project was supported by the Medical University of Warsaw, Faculty of Pharmacy grant no FW24/NM1/15 and FW24/NM1/16. The X-ray structures were determined in the Advanced Crystal Engineering Laboratory (aceLAB) at the Faculty of Chemistry, University of Warsaw.
Footnotes
†Electronic supplementary information (ESI) available: Single crystal X-ray diffraction data for 3b and 6d, NMR spectra with analysis and HR-MS spectra for all new investigated compounds. CCDC 1541420 and 1541465. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7md00281e
References
- Nichols D. E., Nichols C. D. Chem. Rev. 2008;108:1614. doi: 10.1021/cr078224o. [DOI] [PubMed] [Google Scholar]
- Martin E. G., Elgin Jr. R. J., Mathiasen J. R., Davis C. B., Kesslick J. M., Baldy W. J., Shank R. P., Di Stefano D. L., Fedde C. L., Scott M. K. J. Med. Chem. 1989;32:1052. doi: 10.1021/jm00125a020. [DOI] [PubMed] [Google Scholar]
- Ransom R. W., Asarch K. B., Shih J. C. J. Neurochem. 1986;46:68. doi: 10.1111/j.1471-4159.1986.tb12926.x. [DOI] [PubMed] [Google Scholar]
- Perrone R., Berardi F., Colabufo N. A., Leopoldo M., Tortorella V. J. J. Med. Chem. 1999;42:490. doi: 10.1021/jm980420n. [DOI] [PubMed] [Google Scholar]
- van Steen B. J., Wijngaarden I., Tulp M., Soudjin W. J. Med. Chem. 1993;36:2751. doi: 10.1021/jm00071a006. [DOI] [PubMed] [Google Scholar]
- Riveiro M. E., De Kimpe N., Moglioni A., Vázquez R., Monczor F., Shayo C., Davio C. Curr. Med. Chem. 2010;17:1325. doi: 10.2174/092986710790936284. [DOI] [PubMed] [Google Scholar]
- Mandala D., Valeru A., Pochampalli J., Vankadari S. R., Tigulla P., Gatla R., Tahmpu R. Med. Chem. Res. 2013;22:5481. [Google Scholar]
- Skalicka-Woźniak K., Orhanb I. E., Cordellc G. A., Nabavi S. M., Budzyńska B. Pharmacol. Res. 2016;103:188. doi: 10.1016/j.phrs.2015.11.023. [DOI] [PubMed] [Google Scholar]
- Santana L., Uriarte E., Fall Y., Teijeira M., Teran C., Garcia-Martinez E., Tolf B. R. Eur. J. Med. Chem. 2002;37:503. doi: 10.1016/s0223-5234(02)01357-0. [DOI] [PubMed] [Google Scholar]
- Teran C., Santana L., Uriarte E., Fall Y., Unelius L., Tolf B. R. Bioorg. Med. Chem. Lett. 1998;8:3567. doi: 10.1016/s0960-894x(98)00646-5. [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang S., Xu X., Liu X., Yu M., Zhao S., Liu S., Qiu Y., Zhang T., Bi-Feng L., Zhang G. J. Med. Chem. 2013;56:4671. doi: 10.1021/jm400408r. [DOI] [PubMed] [Google Scholar]
- Chen Y., Lan Y., Wang S., Zhang H., Xu X., Liu X., Yu M., Liu B.-F., Zhang G. Eur. J. Med. Chem. 2014;74:427. doi: 10.1016/j.ejmech.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gomez J. C., Santana L., Uriarte E., Brea J., Villazon M., Loza M. I., De Luca M., Rivas M. E., Montenegrob G. Y., Fontenlab J. A. Bioorg. Med. Chem. Lett. 2003;13:175. doi: 10.1016/s0960-894x(02)00933-2. [DOI] [PubMed] [Google Scholar]
- Ostrowska K., Młodzikowska K., Głuch-Litwin M., Gryboś A., Siwek A. Eur. J. Med. Chem. 2017;137:108. doi: 10.1016/j.ejmech.2017.05.047. [DOI] [PubMed] [Google Scholar]
- Ostrowska K., Grzeszczuk D., Maciejewska D., Młynarczuk-Biały I., Czajkowska A., Sztokfisz A., Dobrzycki Ł., Kruszewska H. Monatsh. Chem. 2016;147:1615. [Google Scholar]
- Jacobson M. P., Pincus L. D., Rapp C. S., Day T. J. F., Honig B., Shaw D. E., Friesner R. A. Proteins: Struct., Funct., Bioinf. 2004;55:351. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Liegeois J.-F., Lespagnard M., Salas E. M., Mangin F., Scuvee-Moreau J., Dilly S. ACS Med. Chem. Lett. 2014;5:358. doi: 10.1021/ml4004843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M., Kołaczkowski M., Pawłowski M., Bojarski A. J. J. Med. Chem. 2006;49:205. doi: 10.1021/jm050826h. [DOI] [PubMed] [Google Scholar]
- Greenwood J. R., Calkins D., Sullivan A. P., Shelley J. C. J. Comput.-Aided Mol. Des. 2010;24:591. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. J. Comput. Chem. 2009;16:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Xu L., Wolan D. W., Wilson I. A., Olson A. J. J. Med. Chem. 2004;47:6681. doi: 10.1021/jm049504o. [DOI] [PubMed] [Google Scholar]
- Prandi A., Franchini S., Manasieva L. I., Fossa P., Cichero E., Marucci G., Bucciono M., Cilia A., Pirona L., Brasili L. J. Med. Chem. 2012;55:23. doi: 10.1021/jm200421e. [DOI] [PubMed] [Google Scholar]
- Franchini S., Battisti U. M., Baraldi A., Prandi A., Fossa P., Cichero E., Tait A., Sorbi C., Marucci G., Cilia A., Pirona L., Brasili L. Eur. J. Med. Chem. 2014;87:248. doi: 10.1016/j.ejmech.2014.09.070. [DOI] [PubMed] [Google Scholar]
- Franchini S., Manasieva L. I., Sorbi C., Battisti U. M., Fossa P., Cichero E., Denora N., Iacobazzi R. M., Cilia A., Pirona L., Ronsisvalle S., Arico G., Brasili L. Eur. J. Med. Chem. 2017;125:435. doi: 10.1016/j.ejmech.2016.09.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.