Abstract
The fate of mesenchymal stem cells (MSCs) in the perivascular niche, as well as factors controlling their fate, is poorly understood. Here, we study MSCs in the perivascular microenvironment of endothelial capillaries by modifying a synthetic 3D biomimetic poly(ethylene glycol) (PEG)‐hydrogel system in vitro. We show that MSCs together with endothelial cells form micro‐capillary networks specifically in soft PEG hydrogels. Transcriptome analysis of human MSCs isolated from engineered capillaries shows a prominent switch in extracellular matrix (ECM) production. We demonstrate that the ECM phenotypic switch of MSCs can be recapitulated in the absence of endothelial cells by functionalizing PEG hydrogels with the Notch‐activator Jagged1. Moreover, transient culture of MSCs in Notch‐inducing microenvironments reveals the reversibility of this ECM switch. These findings provide insight into the perivascular commitment of MSCs by use of engineered niche‐mimicking synthetic hydrogels.
Keywords: extracellular matrix, mesenchymal stem cell, Notch signaling, perivascular niche, synthetic hydrogel
Subject Categories: Signal Transduction, Stem Cells, Vascular Biology & Angiogenesis
Introduction
Blood capillaries consist of an inner endothelial cell layer and a surrounding wall of perivascular cells 1. During the formation of new blood capillaries, perivascular cells are recruited by endothelial cells and contribute to the stabilization and regulation of newly formed endothelial capillaries. Interestingly, the perivascular microenvironment of endothelial capillaries within different tissues has been proposed to potentially function as a reservoir for mesenchymal stem/progenitor cells (MSCs) 2, 3, 4, 5, 6, 7, 8. This has led to the constantly debated hypothesis that during tissue regeneration, MSCs are mobilized from blood capillaries and that MSCs and perivascular cells are closely related cell types. Although the perivascular origin of MSCs remains controversial 9, MSCs can act as perivascular support cells 10, 11, 12, 13. However, how MSCs adapt to the perivascular environment and which niche signals regulate their perivascular adaptation has not been addressed comprehensively. This knowledge gap exists mainly due to the lack of MSC‐specific markers allowing for cell tracing experiments and difficulties in specifically manipulating cells within their perivascular niche in vivo 14. Therefore, advanced in vitro models could be beneficial to systematically address the regulation of MSCs in a defined perivascular microenvironment.
The elucidation as well as the incorporation of specific niche signals into natural extracellular matrix (ECM) hydrogels remains difficult due to inherent bioactivities and therefore requires defined and tunable materials. To mimic native cell niches in vitro, 3D microenvironments with application‐specific and systematically tailored properties are required 15. Fully synthetic materials, such as poly (ethylene glycol) (PEG)‐based or hyaluronic acid (HA)‐based hydrogels 16, 17, 18, can be generated with defined physical and chemical properties and provide reproducible and tunable 3D platforms for the creation of defined cell microenvironments. These synthetic hydrogels are suited to mimic complex developmental processes in vitro, as excellently demonstrated for the promotion of neural tube formation, angiogenesis, epithelial cyst formation, and culture of small intestine organoids 19, 20, 21, 22, 23, 24, 25, 26.
To specifically control the fate of MSCs in engineered 3D microenvironments, stiffness and degradability as well as the presentation of matrix‐immobilized small molecules and growth factors have been engineered in synthetic hydrogels 27, 28, 29. However, there is still a lack of basic biological knowledge on mechanisms governing MSC function and fate within the perivascular niche and how this niche might be successfully mimicked.
Here, we engineer 3D micro‐capillary networks within PEG‐hydrogels to control and determine the fate of MSCs in a defined environment, with the aim to mimic their native niche. We show that this synthetic, ECM‐free hydrogel system becomes modified by ECM deposition from human bone marrow MSCs (BM‐MSCs), which defines the cellular microenvironment immediately after cell embedment. In co‐culture with human endothelial cells or endothelial progenitor cells, we show that MSCs behave as perivascular‐like cells and support endothelial capillaries, which occur as a function of PEG hydrogel stiffness and are dependent on MSC‐derived ECM. Unbiased transcriptome analysis of MSCs, when isolated from these engineered capillaries, demonstrated a prominent switch in the production of basement membrane ECM and Notch signaling pathway genes compared to monocultured MSCs. By functionalizing PEG hydrogels with the Notch‐activating ligand Jagged1, we recapitulated the microenvironment‐dependent induction of the perivascular MSC phenotype in the absence of endothelial cells. Moreover, transient exposure of MSCs to Jagged1‐modified microenvironments revealed a reversible Notch‐induced ECM switch. In summary, we have established a fully defined perivascular niche model in vitro and we have used it to uncover a novel Notch‐regulated and reversible ECM switch in MSCs.
Results
BM‐MSCs rapidly modify engineered microenvironments with their own ECM
3D microenvironments were engineered by enzymatically cross‐linking star‐shaped 8‐arm PEG precursor molecules that are end‐functionalized by substrate sequences for FXIIIa (Fig 1A) 30, 31. A matrix metalloproteinase (MMP)‐sensitive degradation domain in one of the precursor molecules and the simultaneously cross‐linked integrin adhesion peptide RGD render this synthetic environment bioactive. To address the contribution of BM‐MSCs to the engineered microenvironment, we embedded them in PEG hydrogels and investigated cell‐derived, secreted ECM by immunostaining without cell membrane permeabilization. Intriguingly, we found that 4 h post‐embedment BM‐MSCs were not yet spread but had already lined their encapsulation cavities with a shell of secreted fibronectin (FN) (Fig 1B). After 24 h, cells started to spread and to move out from their FN‐cavities. Based on the very early appearance of FN, its abundancy in mesenchymal cells, and its key role during cell adhesion and tissue morphogenesis, we asked whether the cell‐derived FN influences the interaction of cells with the engineered microenvironment. We knocked down FN in BM‐MSCs by siRNAs (Fig EV1A and B) and embedded FN‐depleted cells in PEG‐hydrogels (Fig 1C). Indeed, these FN‐depleted BM‐MSCs could not form FN‐networks. Additionally, FN‐depleted cells were less elongated and less spread than their control cells after 3 days of culture as determined by automated measurement of cell spreading indexes (Fig 1C). These data show that hydrogel‐embedded BM‐MSCs reshape their microenvironment by depositing ECM components, such as FN, which influences their behavior. Importantly, within 7 days BM‐MSCs weaved a fibrillar meshwork of several structural ECM proteins into their surrounding environment, as shown by immunostaining of exclusively extracellular proteins (Fig 1D). This cell‐derived ECM meshwork comprised cellular FN, heparan sulfate proteoglycan‐2 (HSPG‐2/perlecan), as well as collagen type 1 (COL1) and type 3 (COL3). In summary, PEG hydrogels provide a 3D system to study MSCs within their own ECM.
Figure 1. Engineered cell microenvironments in PEG hydrogels and their modification by BM‐MSC‐derived ECM .
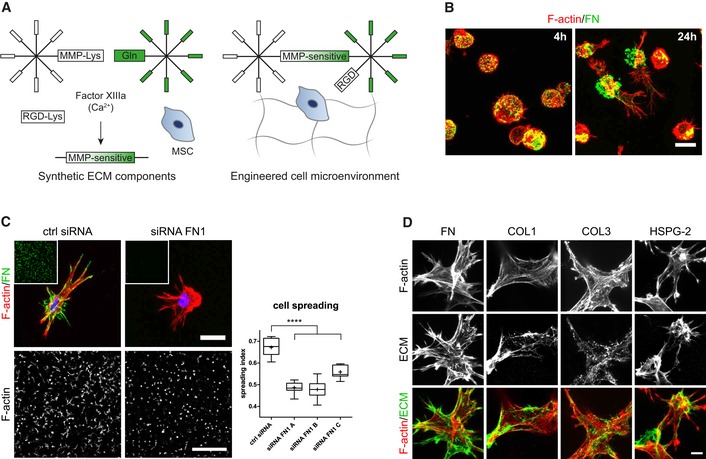
- Schematic of synthetic, FXIIIa cross‐linked PEG hydrogels containing the integrin adhesion ligand RGD and MMP‐sensitive sites.
- BM‐MSCs (F‐actin, red) secreted and deposited cell‐derived fibronectin (green) to sites of initial embedment in PEG hydrogels after 4 and 24 h of culture. Scale bar: 20 μm.
- Cellular fibronectin modulated spreading of BM‐MSCs. Representative individual cells with intact or knockdown fibronectin expression after 3 days of culture within PEG hydrogels. Scale bar: 20 μm. Insets show overview images of the fibronectin stained hydrogels (green) at the same low magnification as the representative F‐actin images used for quantification of spreading (Scale bar: 500 μm). Quantification of cell spreading: Spreading index between 0 for circular cells and 1 for elongated cells. Box plot (25th and 75th percentiles) with whiskers (5th and 95th percentiles), median (line) and mean (+), n = 9, ANOVA with Bonferroni's post hoc test ****P < 0.0001.
- Representative immunofluorescence images of BM‐MSCs (F‐actin, red) and deposited ECM components (green) after 7 days of culture within PEG hydrogels. Scale bar: 10 μm.
Figure EV1. Verification of fibronectin knockdown in BM‐MSCs by siRNA gene silencing.
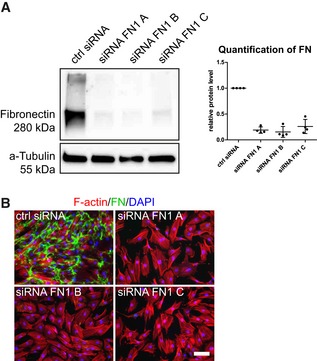
- Efficiency of siRNA‐mediated fibronectin knockdown in BM‐MSCs analyzed by Western blot analysis 48 h after knockdown initiation. Quantification shows the Western blot signal intensity of fibronectin normalized to a‐tubulin, n = 4, individual data points and mean (line) ± SD.
- Absence of cellular fibronectin networks analyzed by immunofluorescence of cell‐derived fibronectin 3 days after successful knockdown. Scale bar: 100 μm.
Source data are available online for this figure.
BM‐MSCs and endothelial cells co‐cultured in tailored PEG‐hydrogels form micro‐capillary networks in defined conditions
Next, to convert the MSC‐derived microenvironment presented above into a perivascular microenvironment, we aimed at tailoring PEG hydrogel properties and culture conditions for optimal in vitro vessel morphogenesis and micro‐capillary network formation. To achieve this aim, we embedded BM‐MSCs together with human umbilical vein endothelial cells (HUVEC) in a 1:1 ratio in 3D PEG‐hydrogels (Fig 2A). After 7 days of culture, we evaluated the formation of 3D micro‐capillary networks by CD31 immunostaining of endothelial cells (Fig 2B). BM‐MSCs and endothelial cells failed to assemble into micro‐capillary networks in the absence of FGF‐2. However, when co‐cultures were conducted in the presence of FGF‐2, micro‐capillary network formation occurred in a dose‐dependent manner up to 50 ng/ml FGF‐2 (Figs 2B and EV2A). We also tested the pro‐angiogenic growth factor VEGF‐A and found that micro‐capillary networks can be induced by VEGF‐A (Fig EV2B). However, in our system, VEGF‐A seems to be less potent than FGF‐2 at the tested concentrations of 50 and 200 ng/ml VEGF‐A, and we saw no significant difference between these VEGF‐A concentrations. We next examined the impact of matrix stiffness and material density on network formation by comparing hydrogels of varying PEG amounts (dry mass content 1–3% with corresponding storage moduli 74 Pa–2,157 Pa, respectively; Fig 2C) in the presence of 50 ng/ml FGF‐2 (Figs 2D and EV2C). The overall length of CD31‐positive micro‐capillaries was equally high in very soft matrices (1–1.3% PEG with 74–276 Pa, respectively), while there was a slight reduction in 1.7% PEG matrices (470 Pa). However, in matrices above 2% PEG (> 762 Pa) the length of micro‐capillaries decreased significantly and CD31‐networks were almost completely absent in matrices of 3% PEG (2,157 Pa). Taken together, a combination of FGF‐2 and very soft PEG matrices supports the robust formation of 3D micro‐capillary networks by endothelial cells (CD31‐positive) and BM‐MSCs (CD31‐negative; Fig 2E).
Figure 2. Engineering of 3D perivascular microenvironments by defining parameters for micro‐capillary formation.
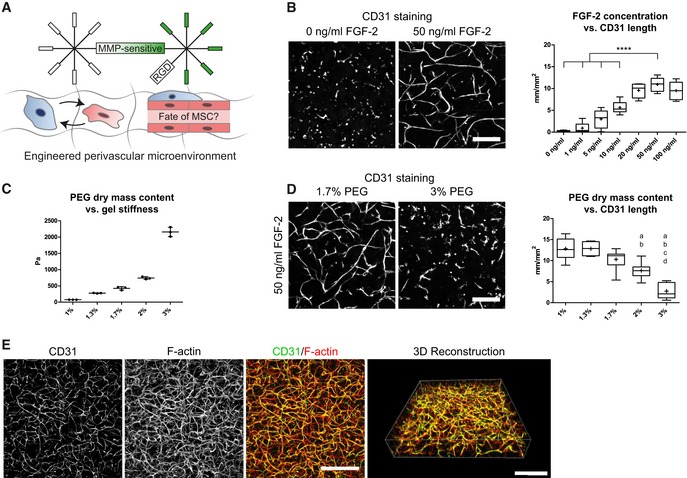
-
AEngineering of perivascular microenvironments by the co‐culture of MSCs and endothelial cells (EC), resulting in the cell‐autonomous establishment of micro‐capillaries containing perivascular localized MSCs.
-
B–D(B, D) Representative immunofluorescence images of micro‐capillary networks formed by BM‐MSCs and endothelial cells (CD31) after 7 days of 3D co‐culture in PEG hydrogels. Scale bars: 200 μm. (B) Quantitative analysis of the absolute length of CD31‐positive networks depending on FGF‐2 concentration (n = 6, ANOVA with Bonferroni's post hoc test ****P < 0.0001) and (D) physical matrix properties by PEG dry mass content [n = 8, ANOVA with Bonferroni's post hoc test shows significant differences from 1% PEG (a), 1.3% PEG (b), 1.7% PEG (c), and 2% PEG (d)]. Box plots in (B and D) show 25th and 75th percentiles with whiskers at 5th and 95th percentiles, median (line), and mean (+). (C) Correlation of PEG dry mass content and matrix stiffness assessed by rheological measurement of the corresponding storage moduli, n = 3, individual data points and mean (line) ± SD.
-
C3D micro‐capillary network formed by co‐cultures of BM‐MSCs and endothelial cells in 1.7% PEG matrices and in the presence of 50 ng/ml FGF‐2. Scale bars: 500 μm.
Source data are available online for this figure.
Figure EV2. 3D micro‐capillary network formation in PEG hydrogels depends on growth factor concentration, PEG matrix stiffness, and MMP activity.
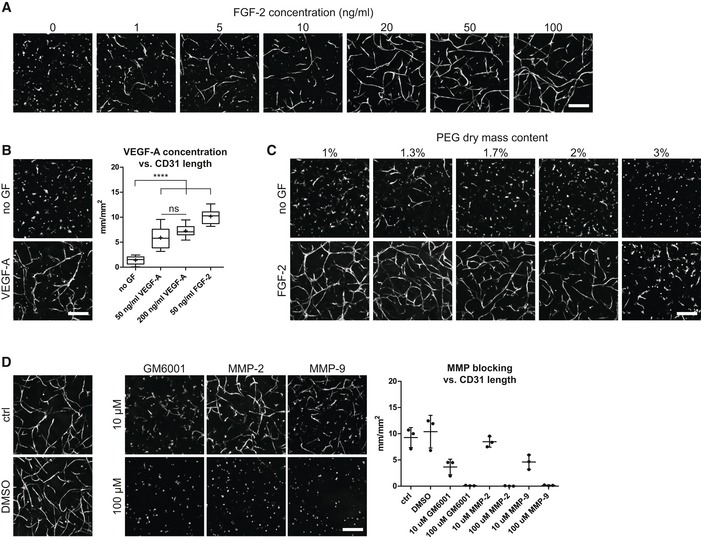
- Impact of FGF‐2 concentration in the medium.
- Impact of VEGF‐A concentration in the medium. Quantitative analysis of the absolute length of CD31‐positive networks depending on VEGF‐A concentration (n = 8). Box plot shows 25th and 75th percentiles with whiskers at 5th and 95th percentiles, median (line), and mean (+). ANOVA with Bonferroni's post hoc test (****P < 0.0001) shows significant differences from no growth factor (no GF) control. Both VEGF‐A conditions are not significantly different from each other, but each from the FGF‐2 group.
- Impact of physical matrix properties by PEG dry mass content.
- Impact of MMP activity by MMP inhibitors. Quantitative analysis of the MMP inhibition effect on the absolute length of CD31‐positive micro‐capillary networks. Individual data points and mean (line) ± SD, n = 3, ANOVA with Bonferroni's post hoc test: All MMP‐blocking conditions except 10 μM MMP‐2 are significantly reduced compared to control and DMSO control groups, with P < 0.05.
To gain insights into the structural details of these micro‐capillaries, we investigated PEG hydrogels by light and transmission electron microscopy (TEM; Fig 3). We show that BM‐MSCs reside around the endothelial capillaries and express the perivascular marker proteins PDGFR‐β/CD140b, MCAM/CD146, and SMA/ACTA2 (Fig 3A). Using TEM, we demonstrate that micro‐capillaries have lumens of several micrometers in diameter and are surrounded by a layer of cell‐derived ECM (Fig 3B). Moreover, endothelial cells were in very tight contact with perivascular localized BM‐MSCs that are enshrouded in the cell‐derived ECM layer. Ultrastructural analysis shows that the deposited ECM layer is of heterogeneous composition, including collagen microfibrils, and is associated with cellular extensions. We next investigated the composition of the perivascular ECM network by means of immunofluorescence analysis without cell membrane permeabilization (Fig 3C). We found FN, COL1, and COL3 to be present in the extracellular space around the micro‐capillaries. Notably, the perivascular ECM layer also consisted of the basement membrane components collagen type 4 (COL4), laminin, and HSPG‐2. Intriguingly, by using FN‐depleted BM‐MSCs we observed that in the absence of BM‐MSC‐derived FN, the micro‐capillary formation is completely inhibited, indicating that BM‐MSC‐deposited ECM and particularly FN play a key role in this process (Fig 3D). However, matrix remodeling in synthetic hydrogels likely is not just featured by newly deposited cellular ECM but is also dependent on the proteolytic breakdown of the synthetic matrix. To test this assumption, we blocked matrix metalloproteinase (MMP) activity using chemical inhibitors (Fig EV2D). Indeed, the broadband MMP inhibitor GM6001 completely prevented the formation of micro‐capillary networks. Furthermore, inhibitors of MMP‐2 and MMP‐9 also blocked the capillary formation at high concentrations. However, at these high concentrations, a potential inhibition of other MMPs cannot be ruled out. Taken together, favorable conditions for the culture of 3D micro‐capillaries were featured by balanced hydrogel degradation and cell‐derived ECM deposition, enabling heterocellular adhesions and morphogenesis to occur.
Figure 3. Characterization of micro‐capillaries, perivascular cells, and perivascular ECM .
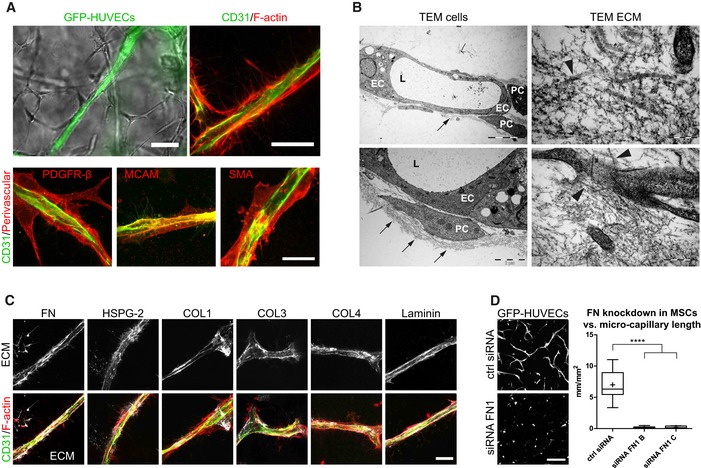
- Bright‐field and fluorescence microscopy images show endothelial capillaries (GFP, CD31) that are surrounded by perivascular‐orientated cells and are positively stained for PDGFR‐β, MCAM, and SMA (day 7). Scale bar bright‐field image: 50 μm. Scale bar fluorescence images: 20 μm.
- Transmission electron microscopy (TEM) images show endothelial cells (EC) forming lumenized (L) micro‐capillaries that are surrounded by perivascular‐like cells (PC) and embedded in a dense network of ECM (black arrows). Ultrastructural analysis showing the heterogeneous ECM composition including collagen microfibrils (black arrowheads) and the close interaction of cellular extensions with the ECM. Scale bars clockwise starting upper left: 5 μm, 200 nm, 500 nm, and 2 μm.
- Representative immunofluorescence images of the perivascular ECM layer by exclusive labeling of extracellular deposited ECM proteins (white) and of micro‐capillaries (CD31 in green, F‐actin in red). Scale bar: 20 μm.
- Micro‐capillary formation using BM‐MSCs with intact or knockdown fibronectin expression in co‐cultures with GFP‐HUVECs. Images (GFP) and quantification after 3 days of culture within PEG hydrogels. Scale bar: 200 μm. Quantitative analysis shows the length of GFP‐positive micro‐capillary networks from co‐cultures using BM‐MSCs treated with control siRNA and two different siRNAs to FN1. Box plot (25th and 75th percentiles) with whiskers (5th and 95th percentiles), median (line) and mean (+), n = 12, ANOVA with Bonferroni's post hoc test ****P < 0.0001.
Micro‐capillaries promote the perivascular commitment of BM‐MSCs
Next, using the optimized micro‐capillary cultures (1.7% PEG, 50 ng/ml FGF‐2), we investigated the molecular changes BM‐MSCs undergo in the perivascular microenvironment by unbiased transcriptome analysis (Fig EV3A). To retrieve BM‐MSCs from either micro‐capillary networks or monocultures, we digested PEG‐hydrogels by collagenase and isolated CD31‐negative cells by FACS (Fig EV3A and B). Successful cell separation was verified by qRT–PCR for CD31/PECAM1 mRNA expression in isolated populations (Fig EV3C). In subsequently isolated and deep sequenced mRNA of BM‐MSCs, we detected 16,839 transcripts. Among this absolute number of detected transcripts, we found 551 to be strongly and significantly upregulated and 771 downregulated in micro‐capillary network‐derived BM‐MSCs (Fig EV3D). The 100 most significantly differentially expressed genes are displayed as a heatmap in Fig EV3E (see Dataset EV1 for full list). To systematically investigate the relationship of the differentially expressed genes in BM‐MSCs, we functionally enriched genes within the gene ontology (GO) domain Cellular Component (Fig 4). As a result, Cellular Component terms that are overrepresented (gene‐enriched) appear as clusters in the visualization map of Fig 4A (see Dataset EV2 for full list of labeled functional terms and genes). To gain deeper insights into the transcriptome changes that BM‐MSCs undergo in the perivascular environment, we addressed the largest clusters of the gene enrichment (Fig 4B). Strikingly, the most prominent changes in perivascular BM‐MSCs were related to components of the ECM, as the upregulation of one large ECM cluster and the downregulation of another cluster indicate that BM‐MSCs are reprogrammed to adjust their ECM contribution (Fig 4B: clusters 1,2). More specifically, the induced ECM genes included many collagenous and non‐collagenous components of the vascular basement membrane (COL4A1‐2, COL15A1, COL18A1, NID1, NID2, LAMB3, LAMA4, LAMC1, NTN4) as well as fibrillar collagens (COL1A2, COL3A1, COL5A1‐3). Conversely, rather rare collagens (COL11A2, COL17A1, COL8A2, COL28A1) and specialized basement membrane components (COL4A3, COL4A4, LAMA3) were reduced. Other clusters in the GO Cellular Component domain indicate a reorganization of the cell surface including an induction of integrins and ion/voltage‐dependent channels (Fig 4B: clusters 3–6). When alternatively enriching the differentially expressed genes in the GO domain Biological Process, we detected prominent clusters of blood vessel regulation and vasoconstriction, cell–matrix interactions, and metal and ion homeostasis, all pointing toward a contractile phenotype of perivascular BM‐MSCs (Fig EV4A, see Dataset EV3 for full list of labeled terms and genes). Interestingly, we also found many genes that are associated with perivascular cells and many target genes of the Notch signaling pathway to be induced in perivascular BM‐MSCs (Fig EV4B). Overall, our transcriptome data show that the interaction with endothelial capillaries drives BM‐MSCs to obtain a perivascular phenotype, reprogram their ECM expression, and display increased expression of Notch target genes.
Figure EV3. Genome‐wide transcriptional comparison of BM‐MSCs isolated from micro‐capillary networks with control cultured BM‐MSCs.
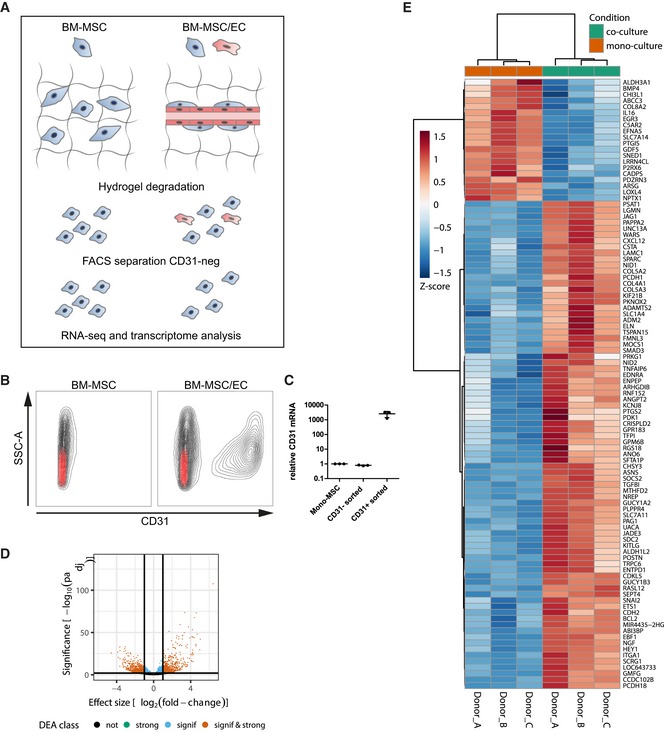
- Schematic of experimental workflow. BM‐MSCs (n = 3 donors) were cultured alone or in micro‐capillary co‐cultures with endothelial cells (EC). After 7 days, cells were retrieved from PEG hydrogels and BM‐MSCs were isolated by FACS‐sorting for non‐endothelial (CD31‐negative) cells. BM‐MSC‐isolated RNAs were analyzed by next‐generation sequencing and differential gene expression.
- FACS separation of BM‐MSCs and ECs by means of CD31‐staining. Black represents the full cell population of beforehand as single and live determined cells. Red represents the staining pattern of the isotype control antibody.
- CD31/PECAM1 mRNA expression in FACS separated cell populations after 7 days of culture in PEG hydrogels measured by qRT–PCR. n = 3, individual data points and mean (line) ± SD.
- Volcano plot of differentially expressed genes between mono‐ and co‐cultured BM‐MSCs. The horizontal line corresponds to a FDR = 0.01 and genes below this line are considered as not significant (not), while genes above are considered as significant (signif). The two vertical lines correspond to a log2‐fold change of 1 in expression and genes outside this range are considered as strongly affected (strong; signif & strong).
- Heatmap of gene expression by BM‐MSCs monocultured or cultured in the perivascular microenvironment. Depicted are the 100 most significantly changed genes due to culture in the perivascular microenvironment. Hierarchical clustering separates genes with positive log2 fold change (increased expression in perivascular BM‐MSCs, red) from negative change (reduced expression in perivascular BM‐MSCs, blue) (A full list of differentially expressed genes is displayed in Dataset EV1).
Source data are available online for this figure.
Figure 4. Functional enrichment map of differentially expressed genes (DEG) in BM‐MSCs isolated from micro‐capillary networks.
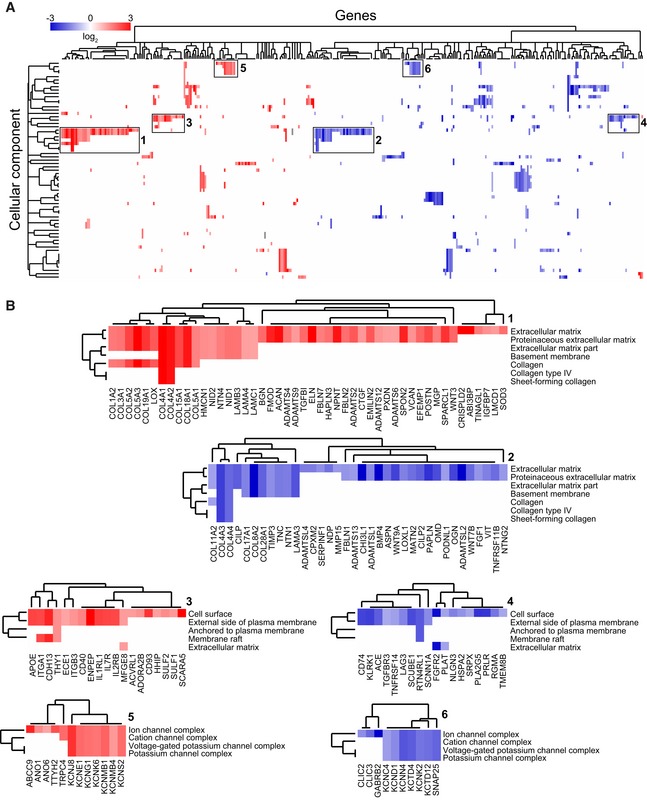
- Genes identified as DEG following transcriptome analysis are enriched and clustered by the gene ontology domain GO Cellular Component. Up‐ and downregulated genes in perivascular BM‐MSCs are displayed in red and blue, respectively (a list of differentially expressed genes with their corresponding Cellular Component terms is displayed in Dataset EV2).
- Strongly represented GO Cellular Component clusters show a strong rearrangement of the ECM including the basement membrane (1, 2), the rearrangement of matrix interaction partners and receptors on the cell surface (3, 4), and of membrane ion channel complexes (5, 6).
Figure EV4. Functional enrichment map of differentially expressed genes (DEG) and induction of Notch pathway target genes and perivascular markers in BM‐MSCs isolated from micro‐capillary networks.
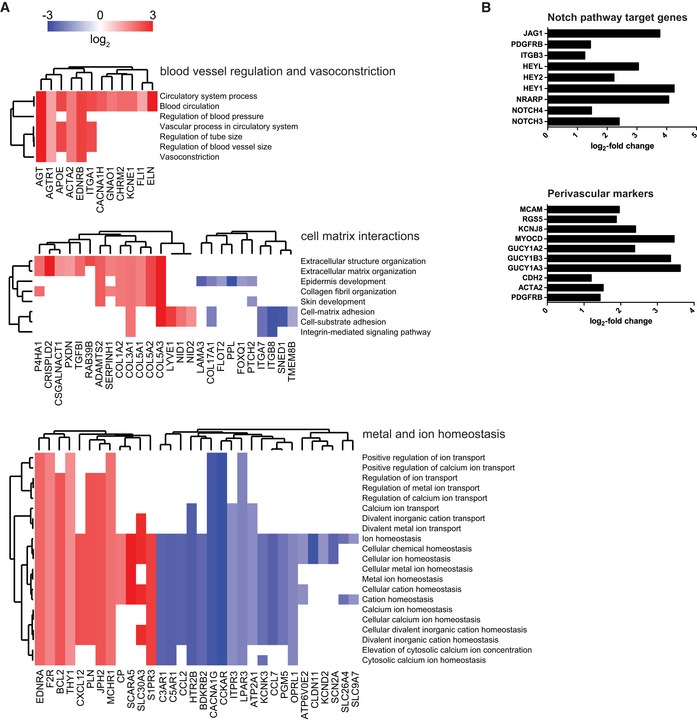
- Genes identified as DEG following transcriptome analysis are enriched and clustered by the gene ontology domain GO Biological Process. Up‐ and downregulated genes in perivascular BM‐MSCs are displayed in red and blue, respectively. GO Biological Process clusters show clusters of blood vessel regulation, cell–matrix interactions, and metal and ion homeostasis (a list of differentially expressed genes with their corresponding Biological Process terms is displayed in Dataset EV3).
- Induction of Notch pathway target genes and perivascular markers in BM‐MSCs isolated from micro‐capillary networks. Log2‐fold change of identified differentially expressed genes following transcriptome analysis in BM‐MSCs isolated from micro‐capillary networks compared to monocultured BM‐MSCs.
Source data are available online for this figure.
Perivascular microenvironment‐mediated commitment of MSCs is not cell source dependent
We examined the induction of basement membrane ECM genes, components and targets of the Notch pathway (NOTCH3 and JAG1), and a perivascular marker (PDGFRB) in BM‐MSCs isolated from micro‐capillaries by qRT–PCR, and could confirm the expression pattern seen in the RNA‐seq analysis (Fig 5A). Furthermore, we confirm this ECM reprogramming does not only occur when FGF‐2 is used as pro‐angiogenic growth factor in the system, but also when BM‐MSCs isolated from VEGF‐A‐induced micro‐capillaries were examined (Fig EV5A). We next asked whether the induced expression of basement membrane ECM and Notch components are cell source dependent. To address this question, we used mesenchymal progenitor cells from human adipose tissue, which are adipose‐derived stromal cells (ASCs), and we performed 3D co‐cultures of ASCs and HUVECs. Hence, we isolated ASCs from engineered micro‐capillary networks and compared their gene expression to monocultured ASCs by qRT–PCR (Fig 5B). Similar to what was observed for BM‐MSCs, basement membrane ECM components and Notch target genes were induced in perivascular ASCs. Specifically, COL4A1, COL4A2, COL18A1, COL5A3, NOTCH3, and JAG1 were the 6 most induced genes in both MSC types. To next investigate whether the induced expression pattern in MSCs is dependent on the endothelia cell type, we formed micro‐capillary networks with human ASCs and human endothelial progenitor cells from the umbilical cord blood, also known as endothelial colony‐forming cells (ECFCs; Fig 5C). We found that ASCs from ECFC micro‐capillaries compared to ASCs from monocultures within the investigated gene subset upregulated the same top 6 genes as BM‐MSCs and ASCs when co‐cultured with HUVECs. In summary, our data show that mesenchymal progenitor cells from bone marrow and adipose tissue upregulate components of the vascular basement membrane ECM when cultured together with endothelial cells (HUVECs or ECFCs) in PEG hydrogels. Furthermore, we found that in all three applied cell combinations the Notch pathway was affected in MSCs.
Figure 5. Jagged1‐functionalized PEG hydrogels recapitulate perivascular microenvironment promoted expression of basement membrane ECM and Notch target genes.
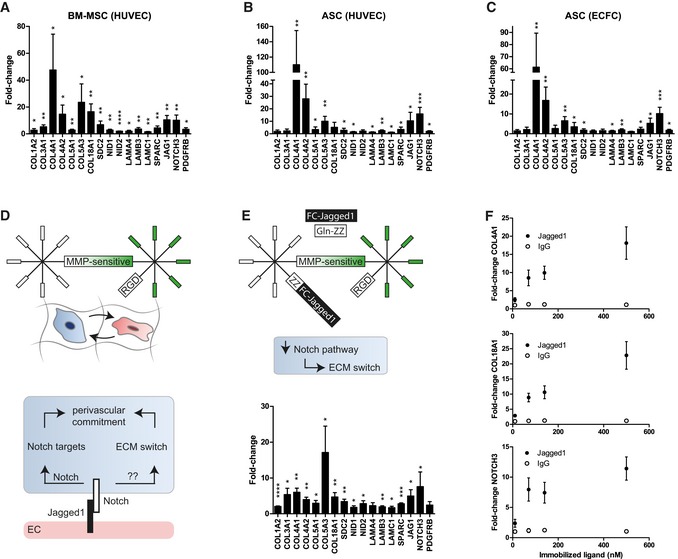
-
A–CInduction of gene expression in MSCs after 7 days of participation in 3D micro‐capillary networks versus monocultured MSCs in PEG hydrogels. (A) BM‐MSCs isolated from co‐cultures with HUVECs (n = 5). (B) ASC isolated from co‐cultures with HUVECs (n = 6). (C) ASC isolated from co‐cultures with ECFCs (n = 6).
-
DSchematic showing that in optimized synthetic hydrogel‐based co‐cultures, endothelial cells, via Jagged1‐Notch‐mediated cell–cell interaction, might promote the upregulation of ECM and Notch target genes in MSCs.
-
EConcept of endothelial cell interaction mimicking hydrogels. Modular designed PEG hydrogels contain in addition to the integrin ligand RGD and MMP‐sensitive sites immobilized Jagged1. This Jagged1 microenvironment induces the perivascular ECM switch in MSCs in the absence of endothelial cells. BM‐MSCs were cultured for 7 days in Jagged1‐functionalized PEG hydrogels (n = 4).
-
FGene induction depends on the concentration of hydrogel‐immobilized Jagged1 ligand (n = 4).
Figure EV5. ECM reprogramming in BM‐MSCs isolated from micro‐capillary co‐cultures induced by VEGF‐A (A) and reversible microenvironment‐controlled ECM switch in MSCs (B).
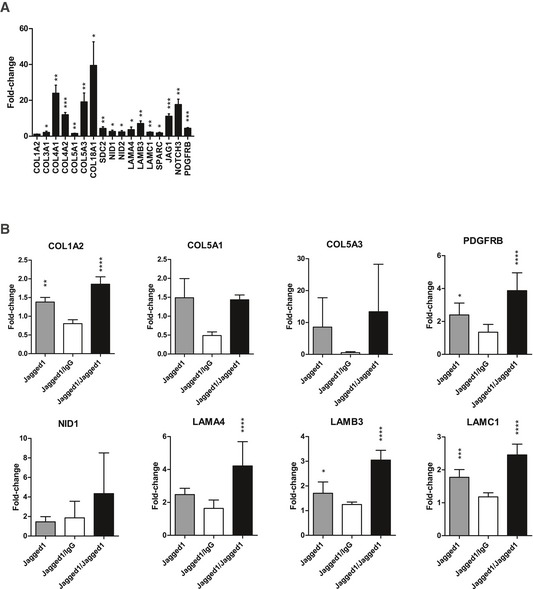
- Induction of gene expression in BM‐MSCs after 7 days of participation in 3D micro‐capillary networks versus monocultured BM‐MSCs in the presence of 200 ng/ml VEGF‐A (n = 4). Data are displayed as relative fold changes compared to MSCs monocultured. Gene expression was analyzed by qRT–PCR and normalized on three reference genes (GAPDH, YWHAZ, EEF1A1). Bars represent mean values ± SD, and one‐sample t‐tests address the significance of the induction: *P < 0.05, **P < 0.01, ***P < 0.001.
- Gene expression of BM‐MSCs after (7 days) culture in Jagged1‐functionalized 3D microenvironments and subsequent (7 days) exposure to new Jagged1‐functionalized or IgG control microenvironments. Gene expression was analyzed by qRT–PCR and normalized on three reference genes (GAPDH, YWHAZ, EEF1A1). Bars represent mean values ± SD, n = 5; ANOVA with Bonferroni's post hoc test shows significant differences from IgG control: *P < 0.05, **P < 0.01, ***P < 0.001,****P < 0.0001.
Source data are available online for this figure.
Jagged1‐modified PEG hydrogels recapitulate the perivascular ECM switch in MSCs
Jagged1 is a cell surface protein exposed on endothelial cells and is able to trigger the Notch pathway in receiving cells (Fig 5D). We therefore reasoned that in our perivascular microenvironments endothelial Jagged1 could be an important factor to control the observed expression changes in MSCs. Thus, we sought to engineer microenvironments that mimic endothelial cell‐derived cell–cell interactions by immobilized Jagged1 (Fig 5E). To incorporate Jagged1 into PEG hydrogels, we made use of their modular design and applied an earlier described protein A‐based linker (Gln‐ZZ). This linker via a ZZ‐domain binds Fc‐tagged proteins with high affinity and via a Gln‐domain is covalently tethered to PEG hydrogels 21, 32. Hence, the pre‐incubation of Gln‐ZZ with Jagged1‐Fc or IgG‐Fc leads to the formation of stable protein complexes that are immobilized within PEG hydrogels by Factor XIIIa. Next, we examined whether monocultured BM‐MSCs induce the vascular basement membrane and the Notch pathway in Jagged1‐modified 3D microenvironments compared to IgG control‐modified microenvironments by qRT–PCR (Fig 5E). In fact, hydrogel‐immobilized Jagged1 (70 nM) did induce the Notch pathway in BM‐MSCs as shown by induction of NOTCH3, JAG1, and PDGFRB. Moreover, all tested ECM genes that were induced in BM‐MSCs by endothelial cells (Fig 5A) were also induced in BM‐MSCs by the matrix‐immobilized Jagged1 (70 nM). However, since the levels of Jagged1‐mediated gene induction were lower than in BM‐MSCs isolated from co‐cultures with endothelial cells, we investigated whether the concentration of immobilized Jagged1 influences the gene induction in mono‐cultured BM‐MSCs (Fig 5F). Our data show that the Jagged1‐mediated induction of three example genes (COL4A1, COL18A1, and NOTCH3) occurs in a dose‐dependent manner and that using 70 nM is within the non‐saturated region of the induction. Intriguingly, the Jagged1‐mediated ECM switch is reversible when BM‐MSCs revert from Jagged1‐modified to control (IgG‐modified) microenvironments (Figs 6 and EV5B). Furthermore, the continuous exposure to Jagged1 maintained the MSC's perivascular phenotype over the observed culture period of 14 days. Together, these data show for the first time that Notch signaling is involved in reversibly reprogramming the ECM expression of MSCs toward a vascular basement membrane phenotype, and that Notch‐inducing hydrogels can be used to study such conditional reprogramming in the absence of endothelial cells.
Figure 6. Reversible microenvironment‐controlled ECM switch in BM‐MSCs.
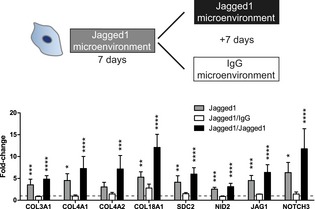
Gene expression of BM‐MSCs after (7 days) culture in Jagged1‐functionalized 3D microenvironments and subsequent (7 days) exposure to new Jagged1‐functionalized or IgG control microenvironments. Gene expression was analyzed by qRT–PCR and normalized on 3 reference genes (GAPDH, YWHAZ, EEF1A1). Bars represent mean values ± SD, n = 5, ANOVA with Bonferroni's post hoc test shows significant differences from IgG baseline control (dashed line): *P < 0.05, **P < 0.01, ***P < 0.001,****P < 0.0001.Source data are available online for this figure.
Discussion
Here, we investigated the fate of MSCs in a 3D in vitro model of the perivascular niche engineered in synthetic hydrogels. To create our model, we employed PEG hydrogels that are formed from fully defined building blocks and are free of exogenous ECM. We show that the often‐neglected reciprocal interactions of cells with the surrounding biomaterial lead to a dynamic reshaping of the initial microenvironment by the deposition of cell‐derived endogenous ECM, which influences cellular behavior and tissue morphogenesis. These findings are particularly important for MSCs, as they are ECM‐producing and tissue‐templating cells.
In addition to modulating their own microenvironment and providing ECM, MSCs have been shown to support endothelial cell functions during micro‐capillary formation within natural ECM hydrogel materials 10, 11, 12, 13, 33, 34. However, the elucidation of specific niche signals and the control over the niche regulatory processes remain very difficult in natural materials. Therefore, we engineered micro‐capillary networks in synthetic hydrogels to study the fate of MSCs in a defined in vitro model of the perivascular niche. In line with previous work, we found that capillary formation is favored at low hydrogel stiffness and material density 19, 35, 36, 37, 38. Additionally, the well‐known pro‐migratory and pro‐angiogenic factor FGF‐2 strongly induced the micro‐capillary network formation, which is dependent on MMP‐mediated matrix degradation. Hence, it is conceivable that micro‐capillary network assembly is most efficient if culture conditions best support cell migration and communication as well as matrix remodeling, all key functions required during native vascular processes.
In these native vessels mimicking micro‐capillaries, BM‐MSCs significantly induced the expression of numerous markers that are associated with perivascular cells and showed a prominent switch in the expression of ECM components. Importantly, we show that BM‐MSCs strongly induced the expression of general and capillary basement membrane‐specific components, while glomerular, corneal, or epidermal basement membrane proteins were downregulated 39, 40, 41, 42, 43. Our data significantly expand earlier findings on the expression of basement membrane components in collagen co‐cultured bovine pericytes and endothelial cells 44. Furthermore, by using endothelial cells and MSCs from different tissue sources, we provided evidence that the induced expression of vascular basement membrane ECM in perivascular MSCs upon endothelial cell contact is highly conserved.
Interestingly, we observed the triggering of the Notch pathway in perivascular BM‐MSCs, which, in the case of Notch3 signaling, has been shown to be important for perivascular cell regulation 45, 46, 47, 48, 49. Activation of the Notch pathway in perivascular cells occurs via the transmembrane protein Notch that interacts with endothelial cell surface‐presented Jagged1 49, 50, 51, 52, 53, 54. Furthermore, the transmembrane protein Jagged1 has previously been shown to contribute to cell fate decisions by cell–cell communication and either functions as agonist or antagonist for Notch signaling when presented in immobilized or soluble forms, respectively 55, 56. Therefore, our findings together with these earlier observations suggest that endothelial cells stimulate Notch target genes in perivascular MSCs in a Jagged1‐dependent manner and thereby contribute to their perivascular commitment. Indeed, we show for the first time that Notch‐inducing biomaterials induce the expression of vascular basement membrane ECM components in MSCs and thereby bypass the need for endothelial cells in promoting this perivascular ECM switch in vitro. Furthermore, the temporary stimulation of Notch signaling demonstrates that the perivascular ECM switch of MSCs is a microenvironmentally dependent and reversible process. Therefore, it is tempting to speculate that these different in vitro states of MSCs might represent a perivascular and a mobilized cell population in vivo. However, while we have shown that Jagged‐Notch signaling plays an important role for the perivascular ECM switch in vitro, the combination of many signaling cues likely influence this ECM switch and the perivascular commitment in vivo.
We envision that our Notch‐inducing hydrogels will enable to further elucidate the function of perivascular niches in tissue regeneration and disease. Furthermore, Notch is a conserved signaling pathway that is involved in the regulation of cell proliferation, cell death as well as cell fate, and differentiation in a broad range of developmental processes 57. Therefore, we believe heterogeneous cell–cell contact mimicking Notch‐inducing 3D hydrogels will be a tool to study various developmental and pathological processes under highly reproducible conditions. In future, hydrogels containing other cell–cell adhesion ligands such as cadherins, junctional adhesion molecules, or nectins can be readily designed to generate novel materials for the 3D modeling of specific development and disease‐related processes.
Materials and Methods
Cell culture
Human bone marrow‐derived mesenchymal stem cells (BM‐MSCs) were isolated as described previously 58 from bone marrow aspirates of healthy donors obtained during orthopedic surgical procedures after informed consent and in accordance with the local ethical committee (University Hospital Basel; Prof. Dr. Kummer; approval date 26/03/2007 Ref Number 78/07). Human adipose‐derived stromal cells (ASCs) were isolated as described previously 59, 60 from adipose tissue obtained during routine surgical procedures of healthy donors after informed consent and in accordance with the local ethical committee (Ethikkommission beider Basel [EKKB], Ref Number 78/07). ASCs and BM‐MSCs were maintained in MEM alpha (with nucleosides, Gibco) supplemented with FBS (10%, Gibco), penicillin (100 U/ml, Gibco), streptomycin (100 μg/ml, Gibco), and FGF‐2 (5 ng/ml, PeproTech). Human umbilical vein endothelial cells (HUVECs from PromoCell; GFP‐HUVECs from PELO Biotech) and human endothelial colony‐forming cells (ECFCs) isolated from term umbilical cord blood as described previously 61 were maintained in EGM‐2 (EBM‐2 basal medium supplemented with the EGM‐2 SingleQuots growth factor kit, both from Lonza) and FCS (10%) in collagen coated flasks (0.15 mg/ml rat‐tail collagen, Corning). All cells were cultured at 37°C in a humidified atmosphere at 5% CO2.
TG‐PEG matrix formation and culture
One milliliter of FXIIIa (200 U/ml, Fibrogammin, CSL Behring) was activated with 100 μl of thrombin (20 U/ml, Sigma‐Aldrich) for 30 min at 37°C. Small aliquots of FXIIIa were stored at –80°C for further use. Hydrogels with final dry mass contents of 1–3% w/v were prepared by stoichiometrically balanced ([Lys]/[Gln] = 1) precursor solutions of n‐PEG‐Gln and n‐PEG‐MMP‐sensitive‐Lys (previously described 30, 31) in 50 mM Tris buffer pH 7.6 containing 50 mM CaCl2. Additionally, Lys‐RGD at a final concentration of 50 μM and cells at final concentration 3 Mio/ml for co‐cultures (1.5:1.5 Mio/ml) and 1, 1.5, or 3 Mio/ml for MSC monocultures were added. Subsequently, PEG cross‐linking was initiated by addition of 10 U/ml FXIIIa and disk‐shaped matrices were prepared between hydrophobic glass slides (treated with SigmaCote, Sigma‐Aldrich) separated by 0.95 mm high spacers. Final hydrogels (volumes described in the following subsections) were cultured in medium containing MEM alpha, FCS (10%), penicillin (100 U/ml), streptomycin (100 μg/ml), and human recombinant FGF‐2 or VEGF‐A 165 (0–200 ng/ml as indicated in the results, Prepotech) at 37°C in a humidified atmosphere at 5% CO2 for time periods as indicated in the results. Medium was replaced after 4 days.
For MMP inhibition experiments, 1.7% TG‐PEG hydrogels were cultured for 7 days in full medium containing 50 ng/ml FGF‐2 and the following chemical MMP inhibitors at 10 or 100 μM: GM6001 (Calbiochem, 364206), MMP‐2 Inhibitor III (Calbiochem, 444288), MMP‐9 Inhibitor I (Calbiochem, 444278).
Characterization of hydrogel stiffness by in situ rheometry
Hydrogel gelation was analyzed on a rheometer (MCR 301, Anton Paar) equipped with 20 mm plate–plate geometry (PP20, Anton Paar) at RT and under humidified atmosphere. After adding FXIIIa to a reaction of 100 μl, containing precursors in stoichiometrically balanced amounts (8‐PEG‐Gln and 8‐PEG‐MMP‐sensitive‐Lys, 1–3% w/v), the mixture was quickly vortexed and precisely loaded onto the center of the bottom plate. The upper plate was lowered to a measuring gap size of 0.2 mm, ensuring proper loading of the space between the plates according to DIN 51810‐1 and the dynamic oscillating measurement was started. The evolution of storage and loss moduli at a constant angular frequency of 0.5 rad/s and constant shear strain of 0.1% was recorded for 30 min.
Immunofluorescence and confocal laser scanning microscopy (CLSM)
For immunofluorescence staining of ECM components, primary antibodies were added directly to the medium of cultures and incubated for 4 h at growth conditions. Afterward, hydrogels (10 μl) were washed twice with PBS followed by fixation in 4% PFA for 30 min at RT. Next, gels were washed for 1 h with PBS and incubated with the corresponding secondary antibodies and with Rhodamine phalloidin (Molecular Probes 1:500) in PBS containing 1% BSA (Albumin Fraction V, AppliChem) for 4 h at RT.
For immunofluorescence staining of CD31, hydrogels (10 μl) were washed once with PBS followed by fixation in 4% PFA for 30 min at RT. Fixed gels were washed twice with PBS and permeabilized in 0.3% Triton X‐100/PBS (Sigma) containing 1% BSA (Albumin Fraction V, AppliChem) for 30 min at RT. Afterward, hydrogels were incubated with a primary antibody to CD31 in 1% BSA/PBS over night at 4°C. Next, gels were washed 1 h with PBS and incubated with the corresponding secondary antibody and with Rhodamine phalloidin (1:500) in PBS containing 1% BSA (Albumin Fraction V, AppliChem) for 4 h at RT.
For co‐stains of ECM components and CD31, primary antibodies to ECM components were added directly to the medium of cultures and incubated for 4 hours at growth conditions. Afterward, hydrogels (10 μl) were washed twice with PBS followed by fixation in 4% PFA for 30 min at RT. Fixed gels were washed twice with PBS and permeabilized in 0.3% Triton X‐100/PBS (Sigma) containing 1% BSA (Albumin Fraction V, AppliChem) for 30 min at RT. Afterward, hydrogels were incubated with a primary antibody to CD31 in 1% BSA/PBS over night at 4°C. Next, gels were washed 1 h with PBS and incubated with the corresponding secondary antibodies and with SiR‐Actin (Spirochrome 1:1,000) in PBS containing 1% BSA (Albumin Fraction V, AppliChem) for 4 h at RT. Finally, stained hydrogels were washed with PBS for 3 h prior to image acquisition.
Confocal images were acquired by an inverted laser scanning microscope (TCS SP5, Leica). Images were processed with ImageJ software (Fiji version 1.48.u, April 2014) and are depicted as projections of Z‐stacks. To quantify the overall length of CD31‐positive structures, 100 μm Z‐stacks were taken by recording 25 planes at 4 μm Z‐steps. Images were Z‐projected and measured by the AngioQuant tool 62 in Matlab (version R2013b, MathWorks Inc, USA). The overall length of CD31‐positive structures was normalized to the image area (mm/mm2).
Fibronectin knockdown experiments
BM‐MSCs were seeded in MEM alpha supplemented with FBS, penicillin, and streptomycin at 50,000 cells per well of a 12‐well plate. 24 h post‐seeding, knockdown was initiated following the standard protocol of lipofectamine RNAiMAX (Invitrogen): 20 pmol siRNA was premixed for 5 min with 3 μl lipofectamine in 100 μl OptiMEM (Invitrogen) and added to cells at a final concentration of 20 nM. The following silencer select siRNAs were used: Negative control No 1 siRNA (4390843, Ambion), FN1 siRNA: A (ID: s223585, 4427037, Ambion), B (ID: s223586, 4427037, Ambion), C (ID: s5321, 4427037, Ambion).
After 48 h of incubation, the knockdown efficiency was evaluated by Western blot analysis using a standard protocol. The fibronectin signal intensities were quantified by the gels tool of ImageJ software (Fiji version 1.48.u, April 2014) and normalized to tubulin signal intensities. For spreading experiments, knockdown and control cells were obtained after 48 h of knockdown and embedded in 1.7% TG‐PEG hydrogels (10 μl) containing 50 μM RGD at a cell concentration of 1 Mio/ml. After 3 days of culture, hydrogels were immunostained for FN and F‐actin as described above. To quantify the cellular spreading, 100 μm Z‐stacks of F‐actin stainings were taken by recording 10 planes at 10 μm Z‐steps. Images were Z‐projected, binarized, and automatically measured by the analyze particles tool of ImageJ software (Fiji version 1.48.u, April 2014). The spreading index is the inversion of cell circularity = 1−circularity (4pi(area/perimeter2)); a value of 0 indicates a perfect circle, and a value of 1 indicates an increasingly elongated shape.
For co‐culture experiments, knockdown and control BM‐MSCs were obtained after 48 h of knockdown and embedded together with GFP‐HUVECs in 1.7% TG‐PEG hydrogels (10 μl) containing 50 μM RGD. After 3 days of culture in full medium containing 50 ng/ml FGF‐2, the overall length of GFP‐positive structures was quantified as described above for CD31‐positive structures.
Transmission electron microscopy
For TEM microscopy, hydrogels (20 μl) were washed once with PBS followed by fixation in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4, 540 mOsmol/kg H2O). Hydrogels were washed 3 times in 0.1 M sodium cacodylate buffer (pH 7.4, 340 mOsmol/kg H2O) and post‐fixed in 1% OsO4 (in 0.1 M sodium cacodylate buffer pH 7.4, 340 mOsmol/kg H2O). Hydrogels were again washed 3 times in 0.1 M sodium cacodylate buffer (pH 7.4, 340 mOsmol/kg H2O) and subsequently gradually dehydrated by an increasing concentration series of ethanol as follows: 70, 80, 90, 96% for 30 min and finally twice in 99% and 100% for 15 min. Last, ethanol was replaced by 100% acetone. Finally, specimens were embedded in epoxy resin. Ultrathin sections were obtained at 70 nm thickness on a microtome (Ultracut S, Reichert), and images were obtained using an electron microscope (EM 400T, Philips) and Morada Soft Imaging System (Olympus).
FACS separation, RNA‐sequencing, and DEG Analysis
BM‐MSCs from three different donors were encapsulated in monoculture (1.5 Mio/ml) or 1:1 co‐culture with HUVECs (1.5:1.5 Mio/ml) within 1.7% TG‐PEG hydrogels (10 × 40 μl) and cultured in medium containing MEM alpha, FCS (10%), penicillin (100 U/ml), streptomycin (100 μg/ml), and human recombinant FGF‐2 (50 ng/ml) at 37°C in a humidified atmosphere at 5% CO2 for 7 days. Medium was exchanged after 4 days. To retrieve BM‐MSCs, hydrogels were pooled and degraded in 0.5 mg/ml collagenase A (Sigma, 11088793001) at 37°C for 60–90 min. Next, cells were washed once in FACS buffer (PBS, 1 mM EDTA, 2% FCS) and incubated for 2 min in trypsin‐EDTA (Gibco, 25300062) at 37°C. Cells were washed twice in FACS buffer and stained with a monoclonal PE‐labeled CD31 antibody for 45 min at 4°C. Cells were washed twice and resuspended in FACS buffer. Cell viability was assessed by SYTOX blue and red stain (Molecular Probes, 1:1,500). CD31‐positive and CD31‐negative cells were separated by the cell sorter BD FACSAria III using a nozzle size of 100 μm (BD Bioscience). CD31‐negative cells were collected, washed once in PBS, and lysed in the RLT‐buffer from RNeasy Micro Kit (Qiagen). Subsequently, total RNA was isolated from cells following the manufacturer's instructions.
Next, the quality of the RNA was assessed with the Agilent TapeStation 4200. Concentration was determined with RiboGreen (Life Technologies) and measured on the Infinite M1000 Pro plate reader (Tecan). 200 ng total RNA was used for RNA‐seq library prep with the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to manufacturer's specifications. Libraries were pooled and sequenced SR81 with the Illumina NextSeq 500 system using the High Output Kit v2 (75 cycles). Primary data analysis was performed with the Illumina RTA version 2.4.11, bcl2fastq v2.18.0.12.
Starting from the raw.fastq files the reads were mapped against the human reference genome and simultaneously soft‐clipped using the STAR aligner (version 2.4.0). Standard quality control (Picard, RSeQC, QoRTs) was run to assess the quality of the resulting alignments. Subsequently, strand‐specific (reversely stranded) read counting was performed using featureCount (subread, version 1.5.0). To avoid technical artifacts, data were filtered such that genes where the average over all samples was < 5 counts were removed. This resulted in 40,980 transcripts removed out of 57,825 in total, leaving 16,845 transcripts for further analysis. Transcripts were mapped to HGNC symbols and ENTREZID's using the org.Hs.e.g.db package version 3.4.0. Differential expression was modeled using generalized linear regression of the negative binomial family, as implemented in the DESeq2 package 63 of the R environment for statistical computing 64. A list of differentially expressed genes (DEG) was compiled with identified genes having a false discovery rate < 0.01 and fold change of more than of 2.
Functional enrichment maps were generated as described previously 65, 66. Over‐representation of gene ontology terms was calculated for the list of DEG using High‐Throughput GoMiner 67. One thousand randomizations were performed, and data were thresholded for a 0.05 false discovery rate. Over‐represented terms with ≥ 5 and ≤ 500 assigned genes were reported. Fold change values were mapped onto genes assigned to each over‐represented term, and the data matrix was subjected to hierarchical clustering analysis on the basis of uncentered Pearson correlation and a complete‐linkage matrix using Cluster 3.0 68. Clustered data were visualized using Java TreeView 69.
Quantitative real‐time PCR (qRT–PCR)
BM‐MSCs or ASCs in monoculture (1.5 Mio/ml) or 1:1 co‐culture with HUVECs or ECFCs (1.5:1.5 Mio/ml) were encapsulated in 1.7% TG‐PEG hydrogels (40 μl) and cultured in medium containing MEM alpha, FCS (10%), penicillin (100 U/ml), streptomycin (100 μg/ml), and human recombinant FGF‐2 (50 ng/ml) or VEGF‐A 165 (200 ng/ml) at 37°C in a humidified atmosphere at 5% CO2 for 7 days. Medium was exchanged after 4 days. RNA from retrieved and separated BM‐MSC or ASC was obtained as described above. For quantitative real‐time PCR (qRT–PCR), 300 ng RNA were converted into 80 μl cDNA by means of the High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT–PCR was carried out using 1.5 μl cDNA template, the TaqMan Universal PCR Master Mix (Applied Biosystems), and the Viia 7 Real‐Time PCR System (Applied Biosystems). The following TaqMan primer/probe sets were used for gene expression tests: Hs00266237_m1 (COL4A1), Hs01098873_m1 (COL4A2), Hs00181017_m1 (COL18A1), Hs00609133_m1 (COL5A1), Hs01555669_m1 (COL5A3), Hs01081432_m1 (SDC2), Hs00915875_m1 (NID1), Hs00201233_m1 (NID2), Hs01128537_m1 (NOTCH3), Hs00943809_m1 (COL3A1), Hs00169777_m1 (PECAM1), Hs00164099_m1 (COL1A2), Hs00234160_m1 (SPARC), Hs00165078_m1 (LAMB3), Hs00267056_m1 (LAMC1), Hs00935293_m1 (LAMA4), Hs01019589_m1 (PDGFRB), Hs01070032_m1 (JAG1). Data were normalized on the expression of the following genes: Hs02758991_g1 (GAPDH), Hs03044281_g1 (YWHAZ), and Hs00265885_g1 (EEF1A1). Relative gene expression was calculated by the comparative Ct method.
Jagged1‐modified PEG hydrogels
Recombinant human Jagged1 Fc Chimera (R&D Systems, 1277‐JG‐050 FC‐IgG) or control IgG1 Fc‐fragment (R&D Systems, 110‐HG‐100 FC‐IgG) were immobilized to TG‐PEG hydrogels at a final molar concentration of 10, 70, 140, or 500 nM by a 30 min pre‐incubation step with 5 μM (final molar concentration) of Gln‐ZZ protein (previously characterized 32). ZZ is a synthetic protein A analogue with super high affinity to the Fc‐fragment of immunoglobulins. The FXIIIa‐mediated immobilization of the Gln‐ZZ‐Fc‐Jagged1 protein complex to TG‐PEG leads to the presentation of Jagged1 within hydrogels. BM‐MSCs were encapsulated at 3 Mio/ml in Jagged1‐ or IgG‐modified TG‐PEG hydrogels (1.7% w/v, 50 μM RGD) and cultured in medium containing MEM alpha, FCS (10%), penicillin (100 U/ml), streptomycin (100 μg/ml), and human recombinant FGF‐2 (50 ng/ml) at 37°C in a humidified atmosphere at 5% CO2. Medium was exchanged every 4 days. After 7 days, RNA was extracted as described above. Alternatively, after 7 days of culture BM‐MSC were retrieved from Jagged1‐ or IgG‐modified TG‐PEG hydrogels by collagenase treatment (0.5 mg/ml in PBS), washed twice in medium and again encapsulated in either Jagged1‐ or IgG‐modified TG‐PEG hydrogels for another 7 days. RNA was extracted, and qRT–PCR was conducted as described above.
Statistical analysis
All graphs and statistical tests were done using GraphPad Prism (GraphPad software version 5.04, San Diego California, USA). Mean values were compared by one‐way analysis of variance (ANOVA) followed by Bonferroni's post hoc test when more than two mean values were compared. Significance of relative gene expression in qRT–PCRs experiments was tested by one‐sample t‐tests. Statistical significance was accepted with *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Further information on the way of data presentation is found in the particular figure legends.
Antibodies
| Antibody | Application/dilution | Catalog number |
|---|---|---|
| Mouse anti‐CD31 | ICC 1:150 | BD Bioscience 555444 |
| Rabbit a‐CD31 | ICC 1:50 | Abcam ab28364 |
| Mouse a‐FN (IST‐9) | ICC 1:300 | Santa Cruz sc59826 |
| Mouse a‐COL1 | ICC 1:150 | Abcam ab90395 |
| Rabbit a‐COL3 | ICC 1:150 | Abcam ab7778 |
| Mouse a‐COL4 | ICC 1:150 | Abcam ab6311 |
| Mouse a‐HSPG‐2 | ICC 1:200 | Abcam ab23418 |
| Rabbit a‐Laminin 1+2 | ICC 1:150 | Abcam ab7463 |
| Goat a‐mouse‐488 | ICC 1:200 | Abcam ab150113 |
| Goat a‐rabbit‐488 | ICC 1:200 | Abcam ab150077 |
| Goat a‐rabbit‐568 | ICC 1:200 | Abcam ab175471 |
| Goat a‐mouse‐555 | ICC 1:200 | Biolegend 405324 |
| Mouse a‐alpha‐Tubulin | WB: 1:2,000 | Sigma T‐5168 |
| Mouse a‐FN (IST4) | WB 1:1,000 | Sigma F‐0916 |
| Goat a‐mouse‐HRP | WB 1:2,000 | DAKO P0447 |
| Mouse a‐SMA | ICC 1:100 | Sigma A‐2547 |
| Rabbit a‐PDGFR‐β | ICC 1:100 | Cell Signaling 3169 |
| Rabbit a‐MCAM | ICC 1:200 | Abcam ab75769 |
| Mouse a‐CD31‐PE | FACS 1:50 | BD Bioscience 560983 |
| Mouse IgG1, k isotype control‐PE | FACS 1:50 | BD Bioscience 556650 |
Availability of data and materials
The RNA‐sequencing data from this publication have been deposited to the Annotare database https://www.ebi.ac.uk/fg/annotare and assigned the identifier E‐MTAB‐6849.
Author contribution
UB conducted all experiments, collected, and evaluated data. QV‐M performed FACS experiment. ERH and JTE assisted with evaluation of RNA sequencing data. JG, AS, and IM isolated and characterized primary human cells. VD conducted TEM analysis. UB, VM, JGS, and ME designed the project. UB, QV‐M, ERH, JGS, VM, and ME wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expaned View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We would like to thank Marcy Zenobi‐Wong (ETH Zurich) and Philipp Fisch (ETH Zurich) for support with rheology measurements, Christian Beisel (Genomics Facility Basel, ETH Zurich) for support on RNA sequencing, Michael Prummer (NEXUS, ETH Zurich) for differential gene expression analysis, Werner Graber (University of Bern) for TEM microscopy, Philipp Lienemann and Aida Kurmanavicius (University Hospital of Zurich) for protein expression of TG‐ZZ and the Flow Cytometry Core Facility of the ETH. We thank Matthias Lütolf (EPF Lausanne) for commenting on the manuscript. This work has been supported by the European Union's Seventh Framework Programme (iTERM grant agreement No. 607868) and by the Swiss National Science Foundation (310030‐169808/1). E.R.H is supported by an EMBO long‐term fellowship (ALTF 922‐2016).
EMBO Reports (2018) 19: e45964
References
- 1. Gaengel K, Genove G, Armulik A, Betsholtz C (2009) Endothelial‐mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol 29: 630–638 [DOI] [PubMed] [Google Scholar]
- 2. Shi S, Gronthos S (2003) Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 18: 696–704 [DOI] [PubMed] [Google Scholar]
- 3. Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M et al (2007) Self‐renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 131: 324–336 [DOI] [PubMed] [Google Scholar]
- 4. Crisan M, Chen CW, Corselli M, Andriolo G, Lazzari L, Peault B (2009) Perivascular multipotent progenitor cells in human organs. Ann N Y Acad Sci 1176: 118–123 [DOI] [PubMed] [Google Scholar]
- 5. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L et al (2008) A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3: 301–313 [DOI] [PubMed] [Google Scholar]
- 6. Feng J, Mantesso A, De Bari C, Nishiyama A, Sharpe PT (2011) Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci U S A 108: 6503–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corselli M, Chen CW, Sun B, Yap S, Rubin JP, Peault B (2012) The tunica adventitia of human arteries and veins as a source of mesenchymal stem cells. Stem Cells Dev 21: 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Billaud M, Donnenberg VS, Ellis BW, Meyer EM, Donnenberg AD, Hill JC, Richards TD, Gleason TG, Phillippi JA (2017) Classification and functional characterization of vasa vasorum‐associated perivascular progenitor cells in human aorta. Stem Cell Reports 9: 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guimaraes‐Camboa N, Cattaneo P, Sun Y, Moore‐Morris T, Gu Y, Dalton ND, Rockenstein E, Masliah E, Peterson KL, Stallcup WB et al (2017) Pericytes of multiple organs do not behave as mesenchymal stem cells in vivo . Cell Stem Cell 20: 345–359.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Au P, Tam J, Fukumura D, Jain RK (2008) Bone marrow‐derived mesenchymal stem cells facilitate engineering of long‐lasting functional vasculature. Blood 111: 4551–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melero‐Martin JM, De Obaldia ME, Kang SY, Khan ZA, Yuan L, Oettgen P, Bischoff J (2008) Engineering robust and functional vascular networks in vivo with human adult and cord blood‐derived progenitor cells. Circ Res 103: 194–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Traktuev DO, Prater DN, Merfeld‐Clauss S, Sanjeevaiah AR, Saadatzadeh MR, Murphy M, Johnstone BH, Ingram DA, March KL (2009) Robust functional vascular network formation in vivo by cooperation of adipose progenitor and endothelial cells. Circ Res 104: 1410–1420 [DOI] [PubMed] [Google Scholar]
- 13. Alimperti S, Mirabella T, Bajaj V, Polacheck W, Pirone DM, Duffield J, Eyckmans J, Assoian RK, Chen CS (2017) Three‐dimensional biomimetic vascular model reveals a RhoA, Rac1, and N‐cadherin balance in mural cell‐endothelial cell‐regulated barrier function. Proc Natl Acad Sci U S A 114: 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kfoury Y, Scadden DT (2015) Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell 16: 239–253 [DOI] [PubMed] [Google Scholar]
- 15. Murphy WL, McDevitt TC, Engler AJ (2014) Materials as stem cell regulators. Nat Mater 13: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seliktar D (2012) Designing cell‐compatible hydrogels for biomedical applications. Science (New York, NY) 336: 1124–1128 [DOI] [PubMed] [Google Scholar]
- 17. Caliari SR, Burdick JA (2016) A practical guide to hydrogels for cell culture. Nat Methods 13: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cruz‐Acuna R, Garcia AJ (2017) Synthetic hydrogels mimicking basement membrane matrices to promote cell‐matrix interactions. Matrix Biol 57–58: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moon JJ, Saik JE, Poche RA, Leslie‐Barbick JE, Lee SH, Smith AA, Dickinson ME, West JL (2010) Biomimetic hydrogels with pro‐angiogenic properties. Biomaterials 31: 3840–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP (2016) Neural tube morphogenesis in synthetic 3D microenvironments. Proc Natl Acad Sci U S A 113: E6831–E6839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP (2014) 3D niche microarrays for systems‐level analyses of cell fate. Nat Commun 5: 4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Enemchukwu NO, Cruz‐Acuna R, Bongiorno T, Johnson CT, Garcia JR, Sulchek T, Garcia AJ (2016) Synthetic matrices reveal contributions of ECM biophysical and biochemical properties to epithelial morphogenesis. J Cell Biol 212: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S, Nih LR, Bachman H, Fei P, Li Y, Nam E, Dimatteo R, Carmichael ST, Barker TH, Segura T (2017) Hydrogels with precisely controlled integrin activation dictate vascular patterning and permeability. Nat Mater 16: 953–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez‐Moran P, Clevers H, Lutolf MP (2016) Designer matrices for intestinal stem cell and organoid culture. Nature 539: 560–564 [DOI] [PubMed] [Google Scholar]
- 25. Cruz‐Acuna R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, Garcia‐Hernandez V, Miller AJ, Spence JR, Nusrat A et al (2017) Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 19: 1326–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gjorevski N, Lutolf MP (2017) Synthesis and characterization of well‐defined hydrogel matrices and their application to intestinal stem cell and organoid culture. Nat Protoc 12: 2263–2274 [DOI] [PubMed] [Google Scholar]
- 27. Benoit DS, Schwartz MP, Durney AR, Anseth KS (2008) Small functional groups for controlled differentiation of hydrogel‐encapsulated human mesenchymal stem cells. Nat Mater 7: 816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caliari SR, Vega SL, Kwon M, Soulas EM, Burdick JA (2016) Dimensionality and spreading influence MSC YAP/TAZ signaling in hydrogel environments. Biomaterials 103: 314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Metzger S, Lienemann PS, Ghayor C, Weber W, Martin I, Weber FE, Ehrbar M (2015) Modular poly(ethylene glycol) matrices for the controlled 3D‐localized osteogenic differentiation of mesenchymal stem cells. Adv Healthc Mater 4: 550–558 [DOI] [PubMed] [Google Scholar]
- 30. Ehrbar M, Rizzi SC, Hlushchuk R, Djonov V, Zisch AH, Hubbell JA, Weber FE, Lutolf MP (2007) Enzymatic formation of modular cell‐instructive fibrin analogs for tissue engineering. Biomaterials 28: 3856–3866 [DOI] [PubMed] [Google Scholar]
- 31. Ehrbar M, Rizzi SC, Schoenmakers RG, Miguel BS, Hubbell JA, Weber FE, Lutolf MP (2007) Biomolecular hydrogels formed and degraded via site‐specific enzymatic reactions. Biomacromol 8: 3000–3007 [DOI] [PubMed] [Google Scholar]
- 32. Lienemann PS, Karlsson M, Sala A, Wischhusen HM, Weber FE, Zimmermann R, Weber W, Lutolf MP, Ehrbar M (2013) A versatile approach to engineering biomolecule‐presenting cellular microenvironments. Adv Healthc Mater 2: 292–296 [DOI] [PubMed] [Google Scholar]
- 33. Ghajar CM, Blevins KS, Hughes CC, George SC, Putnam AJ (2006) Mesenchymal stem cells enhance angiogenesis in mechanically viable prevascularized tissues via early matrix metalloproteinase upregulation. Tissue Eng 12: 2875–2888 [DOI] [PubMed] [Google Scholar]
- 34. Ghajar CM, Kachgal S, Kniazeva E, Mori H, Costes SV, George SC, Putnam AJ (2010) Mesenchymal cells stimulate capillary morphogenesis via distinct proteolytic mechanisms. Exp Cell Res 316: 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghajar CM, Chen X, Harris JW, Suresh V, Hughes CC, Jeon NL, Putnam AJ, George SC (2008) The effect of matrix density on the regulation of 3‐D capillary morphogenesis. Biophys J 94: 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kniazeva E, Putnam AJ (2009) Endothelial cell traction and ECM density influence both capillary morphogenesis and maintenance in 3‐D. Am J Physiol Cell Physiol 297: C179–C187 [DOI] [PubMed] [Google Scholar]
- 37. Vigen M, Ceccarelli J, Putnam AJ (2014) Protease‐sensitive PEG hydrogels regulate vascularization in vitro and in vivo . Macromol Biosci 14: 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chwalek K, Tsurkan MV, Freudenberg U, Werner C (2014) Glycosaminoglycan‐based hydrogels to modulate heterocellular communication in in vitro angiogenesis models. Sci Rep 4: 4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B et al (2001) Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet 27: 205–208 [DOI] [PubMed] [Google Scholar]
- 40. Kruegel J, Miosge N (2010) Basement membrane components are key players in specialized extracellular matrices. Cell Mol Life Sci 67: 2879–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ricard‐Blum S (2011) The collagen family. Cold Spring Harb Perspect Biol 3: a004978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yurchenco PD (2011) Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 3: a004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suh JH, Miner JH (2013) The glomerular basement membrane as a barrier to albumin. Nat Rev Nephrol 9: 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stratman AN, Malotte KM, Mahan RD, Davis MJ, Davis GE (2009) Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation. Blood 114: 5091–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morrow D, Guha S, Sweeney C, Birney Y, Walshe T, O'Brien C, Walls D, Redmond EM, Cahill PA (2008) Notch and vascular smooth muscle cell phenotype. Circ Res 103: 1370–1382 [DOI] [PubMed] [Google Scholar]
- 46. Armulik A, Genove G, Betsholtz C (2011) Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 21: 193–215 [DOI] [PubMed] [Google Scholar]
- 47. Wang Y, Pan L, Moens CB, Appel B (2014) Notch3 establishes brain vascular integrity by regulating pericyte number. Development 141: 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kofler NM, Cuervo H, Uh MK, Murtomaki A, Kitajewski J (2015) Combined deficiency of Notch1 and Notch3 causes pericyte dysfunction, models CADASIL, and results in arteriovenous malformations. Sci Rep 5: 16449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Volz KS, Jacobs AH, Chen HI, Poduri A, McKay AS, Riordan DP, Kofler N, Kitajewski J, Weissman I, Red‐Horse K (2015) Pericytes are progenitors for coronary artery smooth muscle. eLife 4: e10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA (2008) Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc Natl Acad Sci U S A 105: 1955–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jin S, Hansson EM, Tikka S, Lanner F, Sahlgren C, Farnebo F, Baumann M, Kalimo H, Lendahl U (2008) Notch signaling regulates platelet‐derived growth factor receptor‐beta expression in vascular smooth muscle cells. Circ Res 102: 1483–1491 [DOI] [PubMed] [Google Scholar]
- 52. Liu H, Kennard S, Lilly B (2009) NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial‐expressed JAGGED1. Circ Res 104: 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scheppke L, Murphy EA, Zarpellon A, Hofmann JJ, Merkulova A, Shields DJ, Weis SM, Byzova TV, Ruggeri ZM, Iruela‐Arispe ML et al (2012) Notch promotes vascular maturation by inducing integrin‐mediated smooth muscle cell adhesion to the endothelial basement membrane. Blood 119: 2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poulos MG, Guo P, Kofler NM, Pinho S, Gutkin MC, Tikhonova A, Aifantis I, Frenette PS, Kitajewski J, Rafii S et al (2013) Endothelial Jagged‐1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep 4: 1022–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Small D, Kovalenko D, Kacer D, Liaw L, Landriscina M, Di Serio C, Prudovsky I, Maciag T (2001) Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src‐dependent chord‐like phenotype. J Biol Chem 276: 32022–32030 [DOI] [PubMed] [Google Scholar]
- 56. Urs S, Roudabush A, O'Neill CF, Pinz I, Prudovsky I, Kacer D, Tang Y, Liaw L, Small D (2008) Soluble forms of the Notch ligands Delta1 and Jagged1 promote in vivo tumorigenicity in NIH3T3 fibroblasts with distinct phenotypes. Am J Pathol 173: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Papadimitropoulos A, Piccinini E, Brachat S, Braccini A, Wendt D, Barbero A, Jacobi C, Martin I (2014) Expansion of human mesenchymal stromal cells from fresh bone marrow in a 3D scaffold‐based system under direct perfusion. PLoS One 9: e102359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guven S, Mehrkens A, Saxer F, Schaefer DJ, Martinetti R, Martin I, Scherberich A (2011) Engineering of large osteogenic grafts with rapid engraftment capacity using mesenchymal and endothelial progenitors from human adipose tissue. Biomaterials 32: 5801–5809 [DOI] [PubMed] [Google Scholar]
- 60. Osinga R, Di Maggio N, Todorov A, Allafi N, Barbero A, Laurent F, Schaefer DJ, Martin I, Scherberich A (2016) Generation of a bone organ by human adipose‐derived stromal cells through endochondral ossification. Stem Cells Transl Med 5: 1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zeisberger SM, Zoller S, Riegel M, Chen S, Krenning G, Harmsen MC, Sachinidis A, Zisch AH (2010) Optimization of the culturing conditions of human umbilical cord blood‐derived endothelial colony‐forming cells under xeno‐free conditions applying a transcriptomic approach. Genes Cells 15: 671–687 [DOI] [PubMed] [Google Scholar]
- 62. Niemisto A, Dunmire V, Yli‐Harja O, Zhang W, Shmulevich I (2005) Robust quantification of in vitro angiogenesis through image analysis. IEEE Trans Med Imaging 24: 549–553 [DOI] [PubMed] [Google Scholar]
- 63. Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T et al (2015) Orchestrating high‐throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Horton ER, Byron A, Askari JA, Ng DHJ, Millon‐Fremillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D et al (2015) Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 17: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mayorca‐Guiliani AE, Madsen CD, Cox TR, Horton ER, Venning FA, Erler JT (2017) ISDoT: in situ decellularization of tissues for high‐resolution imaging and proteomic analysis of native extracellular matrix. Nat Med 23: 890–898 [DOI] [PubMed] [Google Scholar]
- 67. Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK et al (2005) High‐Throughput GoMiner, an ‘industrial‐strength’ integrative gene ontology tool for interpretation of multiple‐microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics 6: 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Hoon MJ, Makita Y, Imoto S, Kobayashi K, Ogasawara N, Nakai K, Miyano S (2004) Predicting gene regulation by sigma factors in Bacillus subtilis from genome‐wide data. Bioinformatics (Oxford, England) 20(Suppl 1): i101–i108 [DOI] [PubMed] [Google Scholar]
- 69. Saldanha AJ (2004) Java Treeview–extensible visualization of microarray data. Bioinformatics (Oxford, England) 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expaned View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
The RNA‐sequencing data from this publication have been deposited to the Annotare database https://www.ebi.ac.uk/fg/annotare and assigned the identifier E‐MTAB‐6849.


