Abstract
Study Objectives
Dysregulation of sleep is associated with metabolic diseases, and metabolic rate (MR) is acutely regulated by sleep-wake behavior. In humans and rodent models, sleep loss is associated with obesity, reduced metabolic rate, and negative energy balance, yet little is known about the neural mechanisms governing interactions between sleep and metabolism.
Methods
We have developed a system to simultaneously measure sleep and MR in individual Drosophila, allowing for interrogation of neural systems governing interactions between sleep and metabolic rate.
Results
Like mammals, MR in flies is reduced during sleep and increased during sleep deprivation suggesting sleep-dependent regulation of MR is conserved across phyla. The reduction of MR during sleep is not simply a consequence of inactivity because MR is reduced ~30 minutes following the onset of sleep, raising the possibility that CO2 production provides a metric to distinguish different sleep states in the fruit fly. To examine the relationship between sleep and metabolism, we determined basal and sleep-dependent changes in MR is reduced in starved flies, suggesting that starvation inhibits normal sleep-associated effects on metabolic rate. Further, translin mutant flies that fail to suppress sleep during starvation demonstrate a lower basal metabolic rate, but this rate was further reduced in response to starvation, revealing that regulation of starvation-induced changes in MR and sleep duration are genetically distinct.
Conclusions
Therefore, this system provides the unique ability to simultaneously measure sleep and oxidative metabolism, providing novel insight into the physiological changes associated with sleep and wakefulness in the fruit fly.
Keywords: Drosophila, metabolism, respirometry, calorimetry, sleep
Statement of Significance
Metabolic disorders are associated with sleep disturbances, yet our understanding of the mechanisms underlying interactions between sleep and metabolism remains limited. Here, we describe a novel system to simultaneously record sleep and metabolic rate in single Drosophila. Our findings reveal that uninterrupted sleep bouts of 30 minutes or greater are associated with a reduction in metabolic rate providing a physiological readout of sleep. Use of this system, combined with existing genetic tools in Drosophila, will facilitate identification of novel sleep genes and neurons, with implications for understanding the relationship between sleep loss and metabolic disease.
INTRODUCTION
Dysregulation of sleep is strongly linked to metabolism-related pathologies, and reciprocal interactions between sleep and metabolism suggest these processes are integrated at the cellular and molecular levels.1,2 In mammals, metabolic rate (MR) is reduced during sleep raising the possibility that sleep provides a mechanism of energy conservation or partitioning.3 Although a reduction in MR and energy expenditure during sleep has been documented in mammalian and avian species,4,5 little is known about the genetic and neural mechanisms governing the effects of sleep on MR. The fruit fly, Drosophila melanogaster, displays all the behavioral characteristics of sleep and provides a powerful system for genetic investigation of interactions between sleep and diverse physiological processes.2,6,7 Here, we describe a novel single-fly respirometry assay in the fruit fly, designed to simultaneously measure sleep and whole-body MR that allows for genetic interrogation of the mechanisms regulating interactions between these processes.
Sleep is characterized by physiological changes in brain activity or through the behavioral correlates that accompany these changes.8 Flies, like mammals, display distinct electrophysiological patterns that correlate with wake and rest.9,10 Additionally, flies display all the behavioral hallmarks of sleep including extended periods of behavioral quiescence, rebound following deprivation, increased arousal threshold, and species-specific posture.6,7 Sleep in Drosophila is typically defined by 5 minutes of behavioral quiescence because this correlates with other behavioral characteristics used to define sleep.7 Although these behavioral metrics of sleep have been studied extensively, significantly less is known about physiological changes associated with sleep in flies.
In rodents and humans, MR is elevated in response to sleep deprivation and reduced during sleep, supporting the notion that metabolic processes are acutely regulated by sleep state.11–13 In flies and other small insects, stop-flow respirometry can be used to monitor CO2 production, a by-product of oxidative metabolism and a proxy for MR.14 Here, we describe a system to simultaneously measure sleep and MR in individual fruit flies. Our findings reveal that MR is reduced when flies sleep, and uninterrupted sleep bouts of ~30 minutes or greater are associated with an additional reduction in MR, indicating that flies exhibit sleep stages that are physiologically distinct. Further, we find that starvation inhibits sleep-associated reductions in MR, suggesting feeding state influence physiological changes associated with sleep. These findings suggest that sleep-dependent reductions in MR previously observed in mammals are conserved in the fruit fly and further support the notion that sleep provides a mechanism for energy conservation.
MethodS
Drosophila Maintenance and Fly Stocks
Flies were grown and maintained on standard food (Bloomington Recipe, Genesee Scientific). Flies were maintained in incubators (Powers Scientific; Dros52) at 25°C on a 12:12 light/dark cycle, with humidity set to 55%–65%. The wild-type line used in this manuscript is the w1118 fly strain (Bloomington Stock #5905). The trsnnull allele is an excision of the trsnEY06981 locus derived from mobilizing the EPgy2 insertion.15 This allele removes the entire coding region of the gene and represents a null mutation that has been outcrossed to the w1118 background and has previously been described as Δtrsn.15 Unless noted in the figures, all experiments are performed in 3- to 5-day-old mated female flies.
Measurement of MR and Locomotor Activity
MR was measured at 25°C through indirect calorimetry, measuring the CO2 production of individual flies with a Li-7000 CO2 analyzer (LI-COR), which was calibrated with pure CO2 before each run. A stop-flow, push-through respirometry setup was constructed using Sable Systems equipment (Sable Systems International). The experimental setup included sampling CO2 from an empty chamber to assess baseline levels, alongside five behavioral chambers, each measuring CO2 production of a single fly. The weight of flies used for analysis were not taken into account because body size and energy stores are not perturbed in trsnnull flies and do not vary significantly in w1118 flies. Further, previous work using a comparable system suggests weight will have little effect on CO2 measurements unless there is an excess of >50% differences in size between individuals.14,16 To measure CO2 output, air was flushed from each chamber for 50 seconds providing a readout of CO2 accumulation over a 5-minute period. This 5-minute interval allows the coordinate and simultaneous activity-based assessment of sleep. The first 20 minutes of recordings were not included in analyses because this time was necessary to purge the system of ambient air and residual CO2 from the closed system. Dehumidified, CO2 free air was pumped through a mass flow control valve (Side-Trak 840 Series; Sierra Instruments, Inc.) to maintain the experimental flow rate of 100 mL/min. The air was then passed through water-permeable Nafion tubing (Perma Pure, LLC, Lakewood, New Jersey, USA, #TT-070) immersed in a reservoir containing deionized H2O to rehumidify the air before reaching the behavioral chambers. Nonpermeable Bev-A line tubing (United States Plastic Corp., Lima, Ohio, USA, #56280) was used throughout the rest of the system.
Experiments were conducted by placing single flies in 70 mm × 20 mm glass tubes that fit a custom-built Drosophila Locomotor Activity Monitor (Trikinetics, Waltham, Massachusetts) with three sets of infrared (IR) beams for activity detection. The monitor was connected to a computer to record beam breaks every minute for each animal using standard Drosophila Activity Monitor (DAMS) activity software (Trikinetics, Waltham, Massachusetts) as previously described.17 These data were used to calculate sleep information by extracting immobility bouts of 5 minutes using a custom-generated python program. The total activity from all three beams was summed for each time point in order to determine overall activity. Video recordings for analysis of feeding activity were acquired using a handheld USB Digital microscope (Vivida, 2MP #eheV1-USBpro) camera at 12 fps with VirtualDub software (v.1.10.4). Each 60-minute video recording occurred between ZT01-04 to prevent circadian differences in sleep, feeding, and MR. During video recording, flies were simultaneously assayed for activity and MR, with the stop flow set to collect CO2 output every 2 minutes. Videos were manually scored for feeding activity in corresponding 2-minute intervals as a “feeding” or “nonfeeding” bin.
Flies were briefly anesthetized using CO2 for sorting at least 24 hours before the start of an experiment to allow for metabolic recovery. For all experiments, flies were loaded into chambers by mouth pipette to avoid confounding effects of anesthesia and allowed to acclimate in the system with the air flowing for 12–24 hours before behavior experiments, unless otherwise specified. To control for effects of diet composition, all experimental flies were fed a consistent diet. Each chamber contained a single food vial containing 1% agar plus 5% sucrose (Sigma) with red food coloring (McCormick), which we have previously shown to result in sleep comparable to standard fly food.18 For starvation experiments, flies had access to 1% agar dissolved in dH2O and were acclimated for 12 hours during lights on with access to agar alone, with analyses beginning at ZT12 at lights off. All experimental runs included analysis of both experimental flies and relevant controls in a randomized order to account for any subtle variation between runs.
Pharmacology
Pharmacological-induced sleep was achieved through administration of gabaxodol (4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridin-3-ol hydrochloride, THIP hydrochloride; Sigma Aldrich #85118-33-8) at the dosage of 0.1 mg/mL, as previously described.19 Gaboxadol was dissolved in dH2O with 1% agar and 5% sucrose. Flies were loaded into the respirometry system with the gaboxadol 2 hours before lights off (ZT10) and were maintained on the drug throughout the duration of the experiment, as described in the text.
Sleep Deprivation
Flies were acclimated to the respirometry system during the daytime (ZT0-12). For mechanical sleep deprivation, flies were shaken every 2–3 minutes for 12 hours in the modified DAMs monitor/respirometry system throughout the nighttime (ZT12-24) while simultaneously measuring MR. The mechanical stimulus was applied using a vortexer (Fisher Scientific, MultiTube Vortexer) and a repeat cycle relay switch (Macromatic, TR63122). Sleep rebound and corresponding MR was measured the following day from ZT0-ZT12.
Sleep, Metabolic, and Statistical Analyses
Respirometry recordings were analyzed using ExpeData PRO software (Sable Systems International, v1.8.4). The CO2 lag time from the chamber to the analyzer was corrected, the baseline was subtracted from each behavioral chamber, and the absolute CO2 levels (ppm) was converted to μL/hr using the recorded air flow rate. Integrating the CO2 trace revealed the total CO2 produced, or the average MR, per fly for each recording. These data were exported to Excel, where metabolic output was matched to activity, and sleep analyses were performed using a custom python program. Since individual flies were measured for either a 12-hour or 24-hour experimental duration (described in text), our raw data included resampling of MR or beam crosses for each hour. To account for these repeated hourly measures, we determined the mean of the hourly readings for each individual fly before our statistical analyses represented in the graphs, meaning that each fly is represented once and the “N” reported in each figure specifically refers to the number of individual flies assayed in the experiment. To detect significant differences for activity (number of beam crosses), mean VCO2 (μL/hour) or total sleep (minutes), we employed a Student t-test (day vs. night; untreated control vs. gaboxadol-treated; fed vs. starved), two-way analysis of variance (ANOVA) with Sidak’s multiple comparison correction (female, day vs. night; male, day vs. night), and a two-way ANOVA with Sidak’s multiple comparison correction (w1118, fed vs. starved; trsn, fed vs. starved), when appropriate using InStat software (GraphPad Software 6.0). The two-tailed p-value used to test significance is denoted as p < .05.
To account for individual-specific differences in MR, we surveyed the MR throughout longer sleep bouts by calculating percent change in MR. This was determined by subtracting the MR during the first 5 minutes asleep from the MR during each of the subsequent 5 minutes asleep for the entire length of the sleep bout, divided by the MR during the first 5 minutes asleep, multiplied by 100 (eg, [{first 5 minutes MR} – {20 minutes MR}/{first 5 minutes MR}] × 100). We note some flies exhibited longer sleep bouts; however, this analysis was restricted to sleep bouts up to 60 minutes due to limited replicates with extended bout lengths. Moreover, a similar approach was employed as described above for analysis of MR for flies with repeated sleep bouts. If a single fly demonstrated multiple distinct sleep bouts, we determined the mean percent change in MR for each fly at each sleep bin. For these analyses, we performed a one-way ANOVA with Sidak correction comparing the initial percent change in MR (5-minute bin) to each of the subsequent sleep bins (15–60 minute bins at 5-minute intervals) using InStat software (GraphPad Software 6.0) with significance denoted as p < .05.
We applied a linear regression model to characterize the relationship between both absolute vCO2 versus activity (number of beam crossings) and percent change in MR and sleep duration using InStat software (GraphPad Software 6.0) with significance denoted as p < .05. Comparison of slopes derived from regression lines in fed versus starved states was performed using analysis of covariance (F-statistic; GraphPad Software 6.0). Before modeling, we performed pretests, including: generation of residual versus fitted plots to determine homogeneity of variance, normal Q-Q plot, Pearson correlation table and linear model assumptions (B.L.U.E.). The culmination of these tests indicated that our data were both normally distributed and appropriate for linear regression modeling.
RESULTS
Long-Term Recordings of Sleep and MR
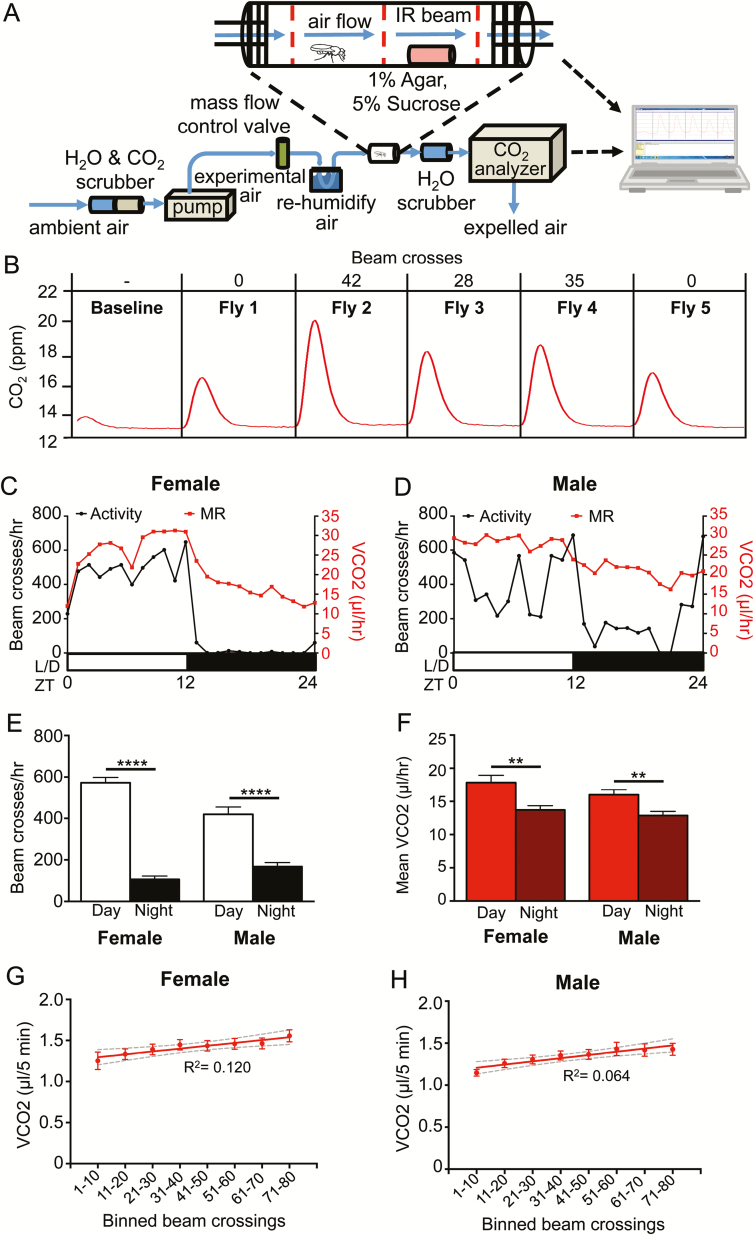
To simultaneously measure the effects of sleep on MR, we designed a stop-flow respirometry system coupled to a custom-built DAM system (Figure 1A). Each DAM chamber contained three IR beams for precise detection of locomotor activity of a single fly.20 Humidified, fully oxygenated air was passed through each chamber, preventing desiccation and allowing for long-term recordings. After exiting the chamber, air was dehumidified and passed through a CO2 analyzer. The system was set to a stop-flow configuration, where the CO2 accumulation in each chamber was measured every 5 minutes, and these were matched to the corresponding locomotor activity of individual flies within this period (Figure 1B). Flies are diurnal with elevated locomotor levels during the day compared to night, and these activity patterns were maintained in the respirometry system in both male and female flies (Figure 1C and D), indicating that the moderate airflow used in this system does not disrupt sleep-wake behavior. In both male and female flies, the mean MR was elevated during the daytime compared to the night, supporting the notion that CO2 production is associated with periods of high activity (Figure 1E and F). Examination of CO2 levels in individual flies revealed a weak correlation in both females and males between total locomotor activity and CO2 levels (Figure 1G and H). However, vCO2 was significantly elevated in females with activity of >60 beam breaks and males >50 beam breaks per 5-minute bin compared to the 1–10 beam breaks bin, suggesting MR is elevated during periods of robust activity (Figure 1G and H). Therefore, this system effectively measures locomotor activity and MR simultaneously in individual Drosophila.
Figure 1—
A system to measure MR in single flies. (A) MR was measured through indirect calorimetry. A stop-flow respirometry system measured the CO2 produced by single flies placed inside of a 70 mm long × 20 mm diameter glass tube. Each fly had access to 1% agar and 5% sucrose. Activity and sleep were measured simultaneously as MR using a Drosophila Locomotor Activity Monitor with three infrared beams running through each behavior chamber. The computer counted the number of beam breaks. (B) A representative 5-minute reading, with the activity in number of beam crosses and the amount of CO2 produced by each fly over time. (C) The MR and activity for one female fly. (D) The MR and activity for one male fly. (E) The activity of female (N = 24; p < .0001) and male (N = 35; p < .0001) flies in beam crosses per hour, over 12 hours of day and night (two-way ANOVA F(1,114) = 171.9, p < .0001). Condition-by-sex interaction is significant (two-way ANOVA F(1,114) = 15.30, p < .001). (F) The MR of female (N = 24; p < .01) and male (N = 35; p < .01) flies as CO2 produced per hour, over 12 hours of day and night (two-way ANOVA F(1,114) = 21.27, p < .0001). Condition-by-sex interaction is not significant (two-way ANOVA F(1,114) = 0.4137, p > .50). (G) Linear regression of absolute vCO2 readout versus activity of female flies (N = 24 each bin; R2 = 0.120) and (H) male flies (N = 35 each bin; R2 = 0.064). Gray dashed lines indicate 95% confidence interval. One-way ANOVA comparing the vCO2 at the 1–10 beam crossings bin to each subsequent beam crossing bin: females >60 crossings (N = 24 each bin; p < .05) and males >60 crossings (N = 35 each bin; p < .05). ANOVA = analysis of variance; IR = infrared.
MR Is Reduced in Sleeping Drosophila
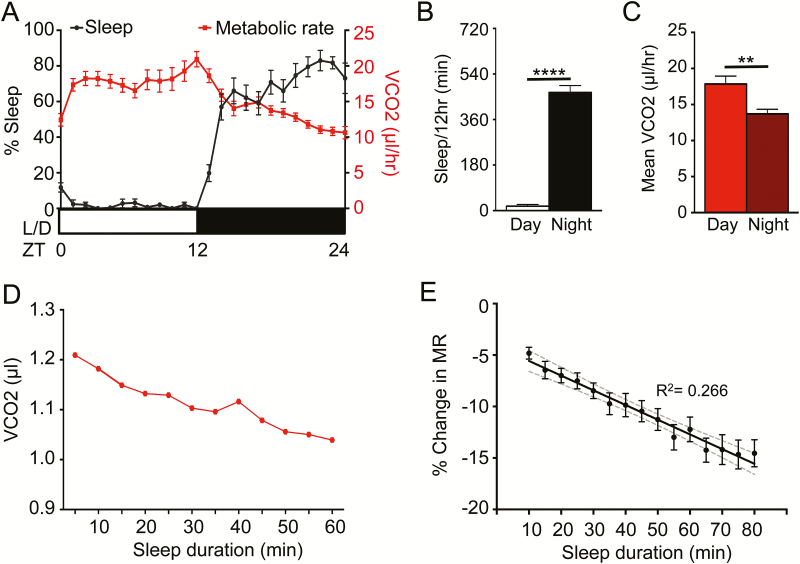
Five minutes of immobility in Drosophila associates with relevant behavioral and physiological sleep metrics, allowing for sleep duration to be inferred from periods of behavioral quiescence.7,10 To measure MR during sleep, female flies were acclimated in the locomotor chambers for 24 hours, followed by continuous measurements of sleep and MR for an additional 24 hours. Flies slept significantly more during the night (ZT12-24), which corresponded with a reduction in MR (Figure 2A–C). To determine whether changes in MR are associated with sleep bout duration, we investigated changes in CO2 production during sleep bouts. CO2 production during a single representative 60-minute sleep bout revealed a reduction in MR as sleep progressed (Figure 2D). To account for individual variation between replicates, change in MR was calculated as percent change for each 5-minute interval throughout the sleep bout compared to the first 5 minutes of sleep. To avoid confounds resulting from circadian differences in MR, analysis was limited to nighttime sleep. Regression analysis revealed a significant relationship between of vCO2 and sleep bout length (Figure 2E). Comparing the average percent change in MR during sleep for each individual bout revealed MR was significantly reduced following 35 minutes of sleep, indicating that longer periods of uninterrupted sleep are associated with reduced MR. Percent change in MR continued to decline as sleep progressed until reaching a maximum percent change in MR of ~−12% to 15% after 50 minutes of sleep. To confirm that reduction of MR during sleep is not simply due to lack of feeding activity, we compared MR during feeding and nonfeeding bins from ZT1-ZT3 and did not detect significant differences in MR between feeding and waking nonfeeding periods (Supplementary Figure S1). Moreover, we performed standard allometric analysis of body size versus MR to identify if weight variation among individual flies could function as a covariate affecting MR21,22 and determined that there is no effect of variation in body weight on MR (n = 34, R2 = 0.030). Therefore, reduced CO2 production is associated with consolidated sleep bouts, revealing that MR can be functionally separated from overall activity.
Figure 2—
MR is reduced during sleep state. Female control flies (w1118) were allowed to acclimate in the system for 24 hours. (A) MR shows an inverse pattern to their sleep (N = 24). (B) Total minutes of sleep per 12 hours of day and night for B (N = 24; p < .001). (C) The MR of female flies as CO2 produced per hour, over 24 hours of day and night (N = 24; p < .002). (D) The MR throughout a single, representative sleep bout during the night. (E) Linear regression model comparing percent change in MR versus sleep duration, binned per 5 minutes (N = 24; R2 = 0.266). Gray dashed lines indicate 95% confidence interval. One-way ANOVA comparing the initial percent change in MR at the 10-minute sleep bin to each subsequent sleep bin reveals significant differences after 35 minutes asleep (N = 24 each sleep bin; p < .05). ANOVA = analysis of variance.
MR During Sleep Deprivation and Rebound Sleep
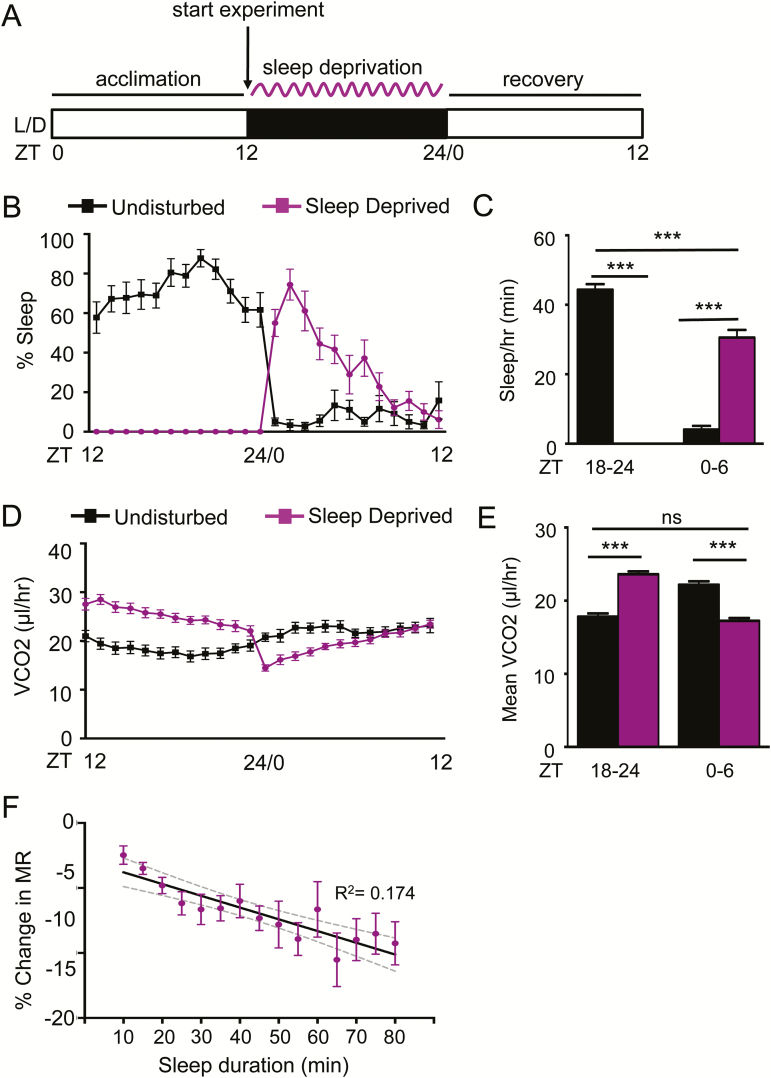
To further examine the relationship between sleep and MR, we sleep deprived flies during the night (ZT12-24) and measured vCO2 during deprivation and recovery (Figure 3A). Consistent with previous findings, sleep deprivation significantly increased sleep the following day (ZT0-6) compared to nonsleep deprived controls (Figure 3B–C). MR was elevated in sleep-deprived flies during deprivation (ZT12-24), and reduced during recovery (ZT0-6), fortifying the notion that reduced MR is associated with sleep. There was a significant correlation between MR and sleep bout duration, indicating that similar to nighttime sleep in undisturbed flies, prolonged bouts of daytime sleep are associated with reduced MR (Figure 3F). Rebound sleep demonstrated a significant reduction in metabolic as sleep duration progressed beyond 35 minutes (Figure 3F), further supporting the notion that daytime rebound recapitulates physiologically similar sleep-associated metabolic changes to nighttime sleep.
Figure 3—
MR is elevated during sleep deprivation and reduced during rebound. (A) Female control flies (w1118) were acclimated during the day (ZT0-12). Mechanical sleep deprivation was applied during the 12-hour night (ZT12-24), and recovery was assessed the following day (ZT0-12). (B) Sleep-deprived flies (N = 15; purple) sleep more during the first 6 hours of daytime following deprivation (ZT0-6) relative to undisturbed controls (N = 15; black). (C) Quantification of total sleep shows that flies were sufficiently sleep deprived during nighttime (ZT18-24; p < .0001) and demonstrated increased sleep during the recovery period (ZT0-6; p < .0001). (D) Hourly profile of MR in sleep deprived and control flies. (E) Quantification MRs demonstrates elevated MR during sleep deprivation (ZT18-24; p < .0001) and reduced MR during recovery (ZT0-6; p < .0001). MR during recovery in sleep-deprived flies is comparable to levels of control flies during normal nighttime sleep (p > .327). (F) Regression analysis comparing percent change in MR versus sleep duration, binned per 5 minutes (N = 15; R2 = 0.174). Gray dashed lines indicate 95% confidence interval. One-way ANOVA comparing the initial percent change in MR at 10-minute sleep bin to each subsequent sleep bin reveals significant differences after 35 minutes asleep (N = 15 each sleep bin; p < .05). ANOVA = analysis of variance.
MR Is Reduced During Pharmacologically Induced Sleep
Gamma-amino butyric acid (GABA) signaling promotes sleep in diverse species, and the GABA-A receptor agonist gabaxodol potently induces sleep in Drosophila.19,23–25 To determine the effects of pharmacologically induced sleep on MR, we housed flies on agar containing 0.1 mg/mL gaboxadol and 5% sucrose in the respirometry chambers and measured the effects on sleep and MR (Figure 4A). Consistent with previous studies, sleep was elevated in gaboxadol-treated flies compared to controls throughout the 12-hour daytime recording19 (Figure 4B and C). Notably, MR was reduced in gaboxadol-treated flies during the daytime compared to controls, confirming that pharmacologically induced sleep lowers MR (Figure 4D and E). These experiments were limited to analysis of daytime sleep, therefore, we could not determine percent change in MR of untreated w1118 flies across sleep bouts, since control flies sleep very little during the day in this paradigm. Sleep bout length in gaboxadol-treated flies was associated with reduced MR (Figure 4F). Moreover, comparison of the of percent change in MR of each subsequent sleep bin relative to the first change at 10 minutes shows a robust reduction in MR in gaboxadol-treated flies after 30 minutes of sleep (Figure 4F). Because the percent change in MR is comparable to the MRs of wild-type flies during night sleep, it is possible that pharmacologically induced daytime sleep is physiologically comparable to nighttime sleep.
Figure 4—
Reduced MR during pharmacologically induced sleep. (A) Female w1118 flies were loaded on sucrose or sucrose containing 0.1 mg/mL gaboxadol 2 hours before lights out (ZT10), acclimated to the system for 12 hours during the night phase and were measured for 12 hours (ZT0-12) during the following day. (B) Daytime sleep was significantly elevated in gaboxadol-treated flies (green) compared to flies fed sucrose alone (black). (C) Quantification of total sleep reveals gaboxadol-treated flies (N = 15) sleep significantly longer than untreated controls (N = 14; p < .0001). (D) MR was reduced throughout the 12-hour day. (E) Quantification of mean MR reveals a significant reduction in gaboxadol-treated flies (N = 15) compared to controls (N = 14; p < .0001). (F) Linear regression of percent change in MR versus sleep duration, binned per 5 minutes (N = 15; R2 = 0.200). Gray dashed lines indicate 95% confidence interval. One-way ANOVA comparing the initial percent change in MR at the 10-minute sleep bin to each subsequent sleep bin reveals significant differences after 30 minutes asleep. (N = 15 each sleep bin; p < .05). ANOVA = analysis of variance.
The Effects of Starvation on Sleep and MR
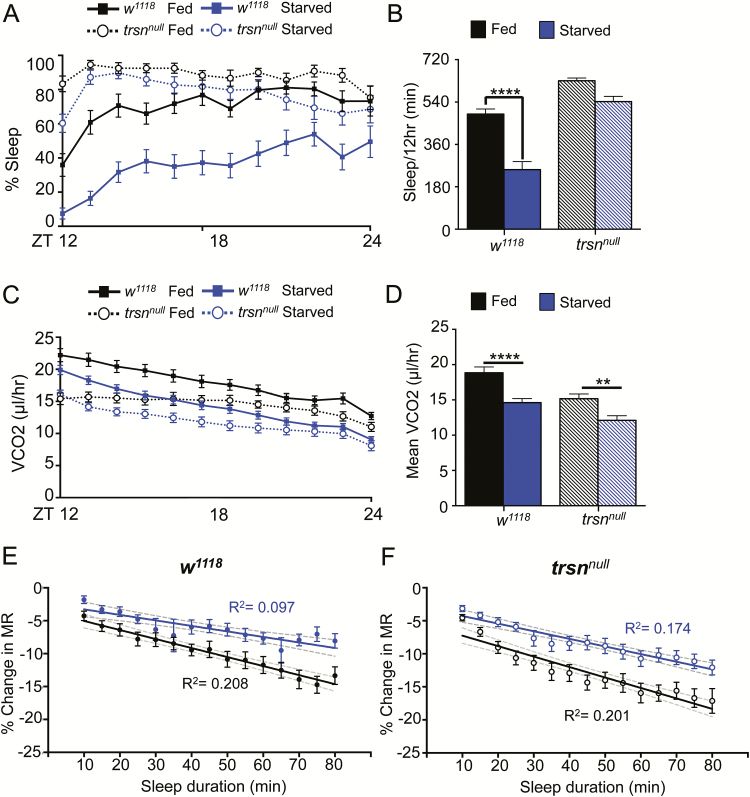
In mammals, starvation potently suppresses sleep and MR.26 Further, flies suppress sleep shortly after the onset of food deprivation, presumably to increase foraging behavior.18,27 To determine how starvation-induced sleep suppression impacts metabolic function, we compared the MR of fed and starved female w1118 flies. Flies were acclimated for 12 hours on food or agar, and MR was measured during the 12-hour night phase (ZT12-24; Figure 5A). In agreement with previous findings, sleep was reduced in starved flies throughout the 12-hour recording period28,29 (Figure 5B and C). Despite the loss of sleep in starved flies, MR was lower in starved animals compared to fed counterparts, providing further support that MR in Drosophila is modulated independently from locomotor activity (Figure 5D and E). There was a significantly stronger relationship between sleep bout length and MR in fed flies, providing evidence that starvation impairs sleep-associated physiological changes on MR (Figure 5F). To determine the effect of starvation on sleep-dependent regulation of MR, we compared the MR of each sleep bout in fed and starved animals. In fed flies, MR was reduced following 40 minutes of sleep compared to the first 5 minutes of sleep, yet when starved, MR is not significantly reduced as sleep progresses, further supporting the notion that starvation impedes sleep (Figure 5F). Taken together, these findings reveal that CO2 production is reduced in starved flies without affecting sleep-dependent changes in MR.
Figure 5—
MR and sleep are reduced in starved flies. (A) Flies were fed or starved while acclimating to the system for 12 hours during the day (ZT0-12) before measurement throughout the night (ZT12-24). (B) Flies starved on agar (blue) slept less than flies housed on 5% sucrose (black) during the 12-hour night period. (C) Quantification of total sleep over the 12-hour night period reveals a significant reduction in starved flies (N = 30) compared to fed controls (N = 29; p < .0001). (D) MR is lower throughout the 12-hour nighttime period in starved flies. (E) Quantification of mean vCO2 production over this period reveals a signification reduction in starved animals (N = 30) relative to controls (N = 29; p < .01). (F) Regression analysis comparing percent change in MR versus sleep duration, binned per 5 minutes reveals a correlation in fed flies (N = 29; R2 = 0.201), but little effect in starved flies (N = 26 each sleep bin, four flies did not have any sleep bouts when starved; R2 = 0.067). Gray dashed lines indicate 95% confidence interval of each line. Comparison of the regression lines indicate that the slopes are different between the fed versus starved state (F = 5.319; p < .05). One-way ANOVA comparing the initial percent change in MR at the 10-minute sleep bin to each subsequent sleep bin within each group reveals significant differences after 40 minutes asleep in fed flies (N = 29 each sleep bin; p < .05) and no significant differences in starved flies (N = 26 each sleep bin). ANOVA = analysis of variance.
Metabolic Changes During Sleep Are Intact in translin Mutant Flies
It is possible that shared genes regulate starvation-induced reductions in sleep duration and sleep-dependent regulation of MR. We previously identified the RNA-binding protein translin (trsn), as essential for starvation-induced sleep suppression.29 Energy stores and feeding behavior are normal in trsn deficient flies, yet they fail to suppress sleep in response to starvation, suggesting trsn is required for the integration of sleep and metabolic state.29 To determine whether trsn affects MR, we measured sleep and MR in fed and starved trsn mutant flies. Flies were loaded into the respirometry system and allowed to acclimate for 12 hours during the day. Sleep and MR were then measured for the duration of the night phase (ZT12-24). In agreement with previous findings, control flies robustly suppressed sleep when starved on agar, while there was no significant effect of starvation on sleep duration in trsnnull flies (Figure 6A and B). In both control and trsnnull flies, CO2 production was reduced during starvation, suggesting trsn is not required for modulating MR in accordance with feeding state. These findings fortify the notion that MR can be regulated independently from both sleep and locomotor activity (Figure 6C and D). Interestingly, while MR was further reduced in trsnnull flies upon starvation, the basal MR of fed trsnnull flies was lower than w1118 controls (Figure 6C and D). For both w1118 and trsnnull flies, there was a stronger relationship between MR and sleep bout duration in fed flies than starved flies (Figure 6E and F), fortifying the notion that sleep-dependent changes in MR are not disrupted in trsnnull flies. Together, these results indicate that trsn is required for starvation-induced sleep suppression but is dispensable for sleep-induced modulation of MR.
Figure 6—
Sleep-dependent changes in metabolism are intact in trsnnull flies. (A) Sleep did not significantly differ between trsnnull flies housed on sucrose or starved on agar alone. (B) Quantification of total nighttime sleep (ZT12-ZT24) revealed sleep is significantly lower in w1118 control flies (N = 30) housed on agar compared to fed (N = 28; p < .0001), while there is no significant difference between trsnnull flies (N = 28) housed on 5% sucrose or agar alone (N = 25; p > .05; two-way ANOVA F(1,107) = 42.52, p < .0001). Treatment-by-genotype interaction is significant (two-way ANOVA F(1,107) = 11.24), p < .01). (C) MR is lower in control and trsnnull flies housed on agar compared to flies housed on 5% sucrose. (D) Quantification revealed MR is lower in both starved trsnnull flies and controls (w1118, p < .0001; trsnnull, p < .01; two-way ANOVA F(1,107) = 28.22, p < .0001). There is no effect of treatment-by-genotype interaction (two-way ANOVA F(1,107) = 0.7162, p > .30). (E) Applied linear regression model comparing percent change in MR versus sleep duration, binned per 5 minutes reveals a correlation in w1118 fed flies (N = 28; R2 = 0.208), but only a weak effect in w1118 starved flies (N = 25, 5 flies did not sleep on agar; R2 = 0.097). Gray dashed lines indicate 95% confidence interval of each line. Comparison of the regression lines indicate that the slopes are different between the w1118 fed versus starved state (F = 7.09725, p< .01). One-way ANOVA comparing the initial percent change in MR at the 10-minute sleep bin to each subsequent sleep bin within each group reveals significant differences after 40 minutes asleep in fed flies (N = 28 each sleep bin; p < .05) and differences in starved flies beyond 55 minutes (N = 25 each sleep bin; p < .05). (F) Regression analysis model comparing percent change in MR versus sleep duration, binned per 5 minutes reveals a correlation in trsnnull fed flies (N = 28; R2 = 0.201), but only a weak effect in trsnnull starved flies (N = 25; R2 = 0.183). Gray dashed lines indicate 95% confidence interval of each line. Comparison of the regression lines indicates that the slopes do not differ between the trsnnull fed versus starved state (F = 5.0557, p < .05). One-way ANOVA comparing the initial percent change in MR at the 10-minute sleep bin to each subsequent sleep bin within each group reveals significant differences after 25 minutes asleep in trsnnull fed flies (N = 28 each sleep bin; p < .05) and differences in trsnnull starved flies beyond 30 minutes (N = 25 each sleep bin; p < .05). ANOVA = analysis of variance.
DISCUSSION
MR is regulated in accordance with environmental changes and life history, providing a metric for whole-body metabolic function. While mammalian studies typically determine MR via O2 consumption or respiratory quotient (ratio of CO2 eliminated/O2 consumed), studies in Drosophila commonly measure CO2 production because it is directly correlated with O2 input and accurately reflects MR.14,16,30 Previous systems investigating MR in Drosophila have used single flies or populations to measure changes in CO2 in response to aging, temperature change, and dietary restriction.16,31,32 Here, we have modified a previously described single-fly respirometry system and DAM system to simultaneously measure MR and sleep. This system can measure CO2 production and locomotor activity over a 24-hour period, providing the ability to measure the relationship between sleep and metabolism, providing a system to investigate the complex relationships between diverse genetic and environmental factors with MR.
Sleep-Metabolism Interactions in Mammals and Arthropods
Regulation of sleep and metabolism is conserved at the molecular and physiological levels between Drosophila and mammals.33,34 Similar to mammals, flies modulate sleep and feeding in accordance with metabolic state, providing a system to investigate the genetic underpinnings of these behaviors.2 For instance, when starved, both flies and mammals suppress sleep presumably to forage for food.18,35,36 The finding that sleep-dependent reductions in MR are conserved in Drosophila supports the notion that an essential function of sleep is metabolic regulation. A number of previous studies suggest total sleep duration is positively correlated with basal MR, supporting the notion that sleep may be an adaptive mechanism of energy conservation.37,38 However, a meta-analysis study examining over 40 different mammalian species revealed a negative relationship between sleep and basal MR, opposing the energy conservation model of sleep.39 In humans, reduced MR during sleep accounts for as much as a 15% energy savings.40–42 Our findings reveal a similar reduction of MR during sleep in fruit flies, suggesting this may provide an evolutionarily adaptive mechanism to conserve energy.
While our study is the first to examine the relationship between sleep and MR in Drosophila, previous studies implicated shared genetic or environmental factors in the regulation of sleep and metabolic function. For example, dopamine potently suppresses sleep in Drosophila, and flies harboring a mutation in the dopamine transporter gene fumin (fmn) exhibit reduced sleep and increased CO2 production,43,44 suggesting dopamine regulates both sleep and metabolic state. Importantly, MR remained elevated in fumin mutant flies when motor neurons were genetically silenced, indicating that the elevated MR does not result from differences in locomotor activity.44 Moreover, long-term sleep deprivation in Pacific beetle cockroach, Diploptera punctate, caused significant increases in O2 consumption and elevated basal MR compared to controls, indicating that sleep loss impacts metabolism.45 These data are in agreement with our findings, where we identify increased MR during sleep deprivation and reduced MR during sleep rebound or pharmacologically induced sleep, revealing a fundamental and direct relationship between sleep and lower MR, indicating that changes in CO2 production during sleep are due to changes in basal MR, rather than reduced locomotor activity.
Environmental Factors Regulating Sleep and Metabolism
In Drosophila, sleep and MR are influenced by diet, temperature, and age.46,47 Here, we discover that starvation conditions impede the physiological changes associated with normal sleep. This simultaneous assessment of sleep and metabolic state can be applied to determine how MR and sleep are related to starvation resistance. Selection for starvation-resistant Drosophila through experimental evolution results in flies that can survive over 2 weeks without food and exhibit a host of metabolic and developmental differences, thus providing a system to examine interactions between metabolism and behavior.48,49 The starvation-resistant flies exhibit increased body size, energy stores, and reduced MR, providing numerous mechanisms for energy conservation.49–51 Previously, we reported that sleep duration is increased in starvation-resistant flies and proposed that this provides an additional mechanism for energy conservation.50,52 Application of this approach measuring sleep and MR will provide the ability to determine MR in asleep and awake flies and identify whether reduced MR in starvation-resistant flies is a consequence of increased sleep or these traits have evolved in parallel.
Evidence for Sleep-Associated Regulation of MRs in Drosophila
In birds and mammals, sleep is associated with changes in cortical activity resulting in defined stages of sleep, such as rapid eye movement and nonrapid eye movement, which differ in physiology and function.53,54 In Drosophila, sleep studies have primarily used behavioral quiescence and body postures to denote sleep, and much less is known about how sleep impacts physiology. Recording of local field potentials in tethered animals reveals distinct differences between quiescent and active states, and sleep is associated with a reduction in 15–30 Hz local-field potentials. The reduction in 15–30 Hz oscillations is at its greatest following 15 minutes of immobility, suggesting that this physiological change in neuronal activity represents a deeper form of sleep, along with coordinate increases in arousal threshold.9,10 Evaluating sleep intensity in male and female Drosophila using an arousal-testing paradigm during extended nighttime sleep bouts identified a gradual decrease in responsiveness until a second, deeper sleep state was reached after ~30 minutes.10,55 Consistent with these findings, we report that MR decreases with sleep duration, reaching a significant reduction ~30 minutes following sleep onset. Taken together, these findings suggest MR may provide a physiological indicator of sleep intensity that compliments existing electrophysiology and behavioral methods of analysis to define a deeper sleep state in flies.
A Role for translin in Regulating MR
The RNA-binding protein trsn is a proposed integrator of sleep and metabolic state, and flies deficient for trsn fail to suppress sleep in response to starvation.29 Notably, the defect in trsn-mutant flies is specific to regulation sleep regulation because trsn-deficient flies have normal feeding and energy stores.29 Here, we find that starvation reduces MR in trsn mutant and wild-type flies. Even though trsn mutants demonstrate starvation-induced sleep suppression, our findings indicate that MR can still be modulated in trsn mutants in a starved state. We identify a lower basal MR in fed trsn mutants compared to controls, though this may be attributable to the trend toward increased nighttime sleep in trsn mutant flies. In addition to trsn, a number of additional genes and transmitters have been identified as regulating starvation-induced sleep suppression or hyperactivity, including Octopamine, clock, and the glucagon-like adipokinetic hormone.35,56,57 Therefore, this assay provides a direct readout of metabolic response to starvation and can be used for more detailed investigation of the mechanisms underlying the integration of sleep and metabolic state.
Future Applications for Investigation of MR in Drosophila
Beyond our initial analysis of the relationship between sleep and MR in Drosophila, this system allows for genetic screens or targeted genetic manipulations to identify novel genes regulating sleep, MR, and the integration of these processes. For example, the mushroom bodies, fan-shaped body, and circadian neurons modulate sleep and wakefulness in Drosophila,23,58–61 and the effects of manipulating these systems on sleep-dependent modulation of MR could be measured using this system. Similarly, application of this system could identify novel genes, neurons or environmental factors required for changes in MR during sleep. Recent studies in flies have identified neural circuits involved in sleep homeostasis,62–64 yet little is known about the physiological changes associated with rebound sleep in flies. In addition to sleep, the circadian system regulates metabolism in flies and mammals.65,66
In addition to measuring MR, respirometry can also be used to measure specific molecules being metabolized. The simultaneous measurement of CO2 and O2 enables the ability to identify the exchange ratios of O2 consumed and CO2 produced and ultimately infer the specific energy fuels utilized.67,68 More specifically, the ratio between the CO2 produced and O2 consumed at a steady state, also known as the respirometry quotient (RQ) or the respirometry exchange ratio which quantifies the same ratio but at any time point (eg, during exercise), can be used to identify food sources metabolized including fat (RQ = ~0.7), carbohydrates (RQ = ~1.0), or protein (RQ = ~0.8–0.9).14 Respirometry measurements have been used to identify substrates metabolized in both mammals69,70 and invertebrates.16 However, the sensitivity of O2 detection is lower than CO2, thus preventing detection of O2 changes in single flies, yet recent studies indicate that sleep can be measured in group-housed Drosophila.71 Therefore, it may be feasible for future studies utilizing groups of flies to determine metabolized energy stores using this system.
CONCLUSIONS
We describe a system for simultaneously measuring sleep and MR and, further, identify dynamic regulation of MR during individual sleep bouts. This system denotes MR as a readily identifiable marker of the physiological changes associated with sleep, which can be universally applied to examine the function of novel sleep genes and neurons in Drosophila. Ultimately, this unique system can be applied to examine precise interactions between numerous aspects of life history and circadian function coordinately with MR.
SUPPLEMENTARY MATERIAL
Supplementary material is available at SLEEP online.
FUNDING
This work was supported by NIH grants 1R01NS085252 to ACK and R15NS080155 to ACK and JRD.
DISCLOSURE STATEMENT
None declared.
Supplementary Material
ACKNOWLEGMENTS
The authors are grateful to Dr. Allen Gibbs (University of Nevada, Las Vegas) for initial guidance in establishing the respirometry system, as well as Mark Spencer (Trikinetics) and Dr. Thomas Foerster (Sable Systems), for technical advice and assistance designing this system. Dr. Paul Shaw (Washington University, St. Louis) provided critical experimental suggestions and feedback.
References
- 1. Reutrakul S, Van Cauter E. Interactions between sleep, circadian function, and glucose metabolism: implications for risk and severity of diabetes. Ann N Y Acad Sci. 2014; 1311: 151–173. [DOI] [PubMed] [Google Scholar]
- 2. Yurgel M, Masek P, DiAngelo JR, Keene A. Genetic dissection of sleep-metabolism interactions in the fruit fly. J Comp Physiol a Neuroethol Sens Neural Behav Physiol. 2015;201(9): 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt MH. The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neurosci Biobehav Rev. 2014; 47: 122–153. [DOI] [PubMed] [Google Scholar]
- 4. Berger RJ, Phillips NH. Energy conservation and sleep. Behav Brain Res. 1995; 69(1-2): 65–73. [DOI] [PubMed] [Google Scholar]
- 5. Walker JM, Berger RJ. Sleep as an adaptation for energy conservation functionally related to hibernation and shallow torpor. Prog Brain Res. 1980; 53: 255–278. [DOI] [PubMed] [Google Scholar]
- 6. Hendricks JC, Finn SM, Panckeri KA, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000; 25(1): 129–138. [DOI] [PubMed] [Google Scholar]
- 7. Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000; 287(5459): 1834–1837. [DOI] [PubMed] [Google Scholar]
- 8. Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984; 8(3): 269–300. [DOI] [PubMed] [Google Scholar]
- 9. Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002; 12(22): 1934–1940. [DOI] [PubMed] [Google Scholar]
- 10. van Alphen B, Yap MH, Kirszenblat L, Kottler B, van Swinderen B. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013; 33(16): 6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caron AM, Stephenson R. Energy expenditure is affected by rate of accumulation of sleep deficit in rats. Sleep. 2010; 33(9): 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valenti G, Bonomi AG, Westerterp KR. Quality sleep is associated with overnight metabolic rate in healthy older adults. J Gerontol A Biol Sci Med Sci. 2016; 72(4): 567–571. [DOI] [PubMed] [Google Scholar]
- 13. Spaeth AM, Dinges DF, Goel N. Resting metabolic rate varies by race and by sleep duration. Obesity. 2015; 23(12): 2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lighton J.Measuring Metabolic Rates: A Manual for Scientists. Oxford University Press; 2008. [Google Scholar]
- 15. Claussen M, Koch R, Jin ZY, Suter B. Functional characterization of Drosophila Translin and Trax. Genetics. 2006; 174(3): 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Voorhies WA, Khazaeli AA, Curtsinger JW. Testing the “rate of living” model: further evidence that longevity and metabolic rate are not inversely correlated in Drosophila melanogaster. J Appl Physiol (1985). 2004; 97(5): 1915–1922. [DOI] [PubMed] [Google Scholar]
- 17. Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) system. Cold Spring Harb Protoc. 2010; 2010(11): pdb.prot5518. [DOI] [PubMed] [Google Scholar]
- 18. Keene AC, Duboué ER, McDonald DM, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010; 20(13): 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dissel S, Angadi V, Kirszenblat L, et al. Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol. 2015; 25(10): 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garbe DS, Bollinger WL, Vigderman A, et al. Context-specific comparison of sleep acquisition systems in Drosophila. Biol Open. 2015; 4(11): 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heusner AA. Energy metabolism and body size. I. Is the 0.75 mass exponent of Kleiber’s equation a statistical artifact? Respir Physiol. 1982; 48(1): 1–12. [DOI] [PubMed] [Google Scholar]
- 22. Schilman PE, Waters JS, Harrison JF, Lighton JR. Effects of temperature on responses to anoxia and oxygen reperfusion in Drosophila melanogaster. J Exp Biol. 2011; 214(Pt 8): 1271–1275. [DOI] [PubMed] [Google Scholar]
- 23. Parisky KM, Agosto J, Pulver SR, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008; 60(4): 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008; 11(3): 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wafford KA, Ebert B. Gaboxadol—a new awakening in sleep. Curr Opin Pharmacol. 2006; 6(1): 30–36. [DOI] [PubMed] [Google Scholar]
- 26. Adamantidis A, de Lecea L. Sleep and metabolism: shared circuits, new connections. Trends Endocrinol Metab. 2008; 19(10): 362–370. [DOI] [PubMed] [Google Scholar]
- 27. Linford NJ, Chan TP, Pletcher SD. Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 2012; 8(5): e1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDonald DM, Keene AC. The sleep-feeding conflict: understanding behavioral integration through genetic analysis in Drosophila. Aging (Albany NY). 2010; 2(8): 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murakami K, Yurgel ME, Stahl BA, et al. Translin is required for metabolic regulation of sleep. Curr Biol. 2016; 26(7): 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yatsenko AS, Marrone AK, Kucherenko MM, Shcherbata HR. Measurement of metabolic rate in Drosophila using respirometry. J. Vis. Exp. 2014;(88):e51681. doi:10.3791/51681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mölich AB, Förster TD, Lighton JR. Hyperthermic overdrive: oxygen delivery does not limit thermal tolerance in Drosophila melanogaster. J Insect Sci. 2012; 12: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hulbert AJ, Clancy DJ, Mair W, Braeckman BP, Gems D, Partridge L. Metabolic rate is not reduced by dietary-restriction or by lowered insulin/IGF-1 signalling and is not correlated with individual lifespan in Drosophila melanogaster. Exp Gerontol. 2004; 39(8): 1137–1143. [DOI] [PubMed] [Google Scholar]
- 33. Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008; 18(15): R670–R679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Padmanabha D, Baker KD. Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab. 2014:1–10. [DOI] [PubMed] [Google Scholar]
- 35. Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004; 167(1): 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Danguir J, Nicolaidis S. Dependence of sleep on nutrients’ availability. Physiol Behav. 1979; 22(4): 735–740. [DOI] [PubMed] [Google Scholar]
- 37. Elgar MA, Pagel MD, Harvey PH. Sleep in mammals. Anim. Behav. 1988;36(5):1407–1419. [Google Scholar]
- 38. Lesku JA, Roth TC, 2nd, Amlaner CJ, Lima SL. A phylogenetic analysis of sleep architecture in mammals: the integration of anatomy, physiology, and ecology. Am Nat. 2006; 168(4): 441–453. [DOI] [PubMed] [Google Scholar]
- 39. Capellini I, Nunn CL, McNamara P, Preston BT, Barton RA. Energetic constraints, not predation, influence the evolution of sleep patterning in mammals. Funct Ecol. 2008; 22(5): 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shapiro CM, Goll CC, Cohen GR, Oswald I. Heat production during sleep. J Appl Physiol Respir Environ Exerc Physiol. 1984; 56(3): 671–677. [DOI] [PubMed] [Google Scholar]
- 41. Garby L, Kurzer MS, Lammert O, Nielsen E. Energy expenditure during sleep in men and women: evaporative and sensible heat losses. Hum Nutr Clin Nutr. 1987; 41(3): 225–233. [PubMed] [Google Scholar]
- 42. Goldberg GR, Prentice AM, Davies HL, Murgatroyd PR Overnight and basal metabolic rates in men and women. Eur. J. Clin. Nutr. 1988;42(2):137–144. [PubMed] [Google Scholar]
- 43. Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005; 25(32): 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ueno T, Tomita J, Kume S, Kume K. Dopamine modulates metabolic rate and temperature sensitivity in Drosophila melanogaster. PLoS One 2012;7(2). doi:10.1371/journal.pone.0031513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stephenson R, Chu KM, Lee J. Prolonged deprivation of sleep-like rest raises metabolic rate in the Pacific beetle cockroach, Diploptera punctata (Eschscholtz). J Exp Biol. 2007; 210(Pt 14): 2540–2547. [DOI] [PubMed] [Google Scholar]
- 46. Yurgel ME, Masek P, DiAngelo J, Keene AC. Genetic dissection of sleep-metabolism interactions in the fruit fly. J Comp Physiol a Neuroethol Sens Neural Behav Physiol. 2015; 201(9): 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Griffith LC. Neuromodulatory control of sleep in Drosophila melanogaster: integration of competing and complementary behaviors. Curr Opin Neurobiol. 2013; 23(5): 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwasinger-Schmidt TE, Kachman SD, Harshman LG. Evolution of starvation resistance in Drosophila melanogaster: measurement of direct and correlated responses to artificial selection. J Evol Biol. 2012; 25(2): 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Masek P, Reynolds LA, Bollinger WL, et al. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J Exp Biol. 2014; 217(Pt 17): 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slocumb ME, Regalado JM, Yoshizawa M, et al. Enhanced sleep is an evolutionarily adaptive response to starvation stress in Drosophila. PLoS One. 2015; 10(7): e0131275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harshman LG, Hoffmann AA, Clark AG. Selection for starvation resistance in Drosophila melanogaster: physiological correlates, enzyme activities and multiple stress responses. J. Evol. Biol. 1999; 12:370–379. [Google Scholar]
- 52. Masek P, Reynolds L a, Bollinger WL, et al. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J Exp Biol. 2014; 217(Pt 17): 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Newman SM, Paletz EM, Rattenborg NC, Obermeyer WH, Benca RM. Sleep deprivation in the pigeon using the Disk-Over-Water method. Physiol Behav. 2008; 93(1-2): 50–58. [DOI] [PubMed] [Google Scholar]
- 54. Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005; 437(7063): 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Faville R, Kottler B, Goodhill GJ, Shaw PJ, van Swinderen B. How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci Rep. 2015; 5: 8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010; 65(5): 670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Keene AC, Duboué ER, McDonald DM, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010; 20(13): 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006; 441(7094): 757–760. [DOI] [PubMed] [Google Scholar]
- 59. Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006; 441(7094): 753–756. [DOI] [PubMed] [Google Scholar]
- 60. Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011; 332(6037): 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guo F, Yu J, Jung HJ, et al. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature. 2016; 536(7616): 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu S, Liu Q, Tabuchi M, Wu MN. Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell. 2016; 165(6): 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Seidner G, Robinson JE, Wu M, et al. Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol. 2015; 25(22): 2928–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Donlea JM, Pimentel D, Miesenböck G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014; 81(4): 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011; 13(6): 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lin JD, Liu C, Li S. Integration of energy metabolism and the mammalian clock. Cell Cycle. 2008; 7(4): 453–457. [DOI] [PubMed] [Google Scholar]
- 67. Arrese E, Soulages J. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 2010; 55: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Marron MT, Markow TA, Kain KJ, Gibbs AG. Effects of starvation and desiccation on energy metabolism in desert and mesic Drosophila. J Insect Physiol. 2003; 49(3): 261–270. [DOI] [PubMed] [Google Scholar]
- 69. Melanson EL, Ingebrigtsen JP, Bergouignan A, Ohkawara K, Kohrt WM, Lighton JR. A new approach for flow-through respirometry measurements in humans. Am J Physiol Regul Integr Comp Physiol. 2010; 298(6): R1571–R1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sinitskaya N, Gourmelen S, Schuster-Klein C, Guardiola-Lemaitre B, Pévet P, Challet E. Increasing the fat-to-carbohydrate ratio in a high-fat diet prevents the development of obesity but not a prediabetic state in rats. Clin Sci (Lond). 2007; 113(10): 417–425. [DOI] [PubMed] [Google Scholar]
- 71. Liu C, Haynes PR, Donelson NC, Aharon S, Griffith LC. Sleep in populations of Drosophila melanogaster. eNeuro 2015;2(4): doi:10.1523/ENEURO.0071-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.