Abstract
A deeper appreciation of the complex architecture of African genomes is critical to the global effort to understand human history, biology and differential distribution of disease by geography and ancestry. Here, we report on how the growing engagement of African populations in genome science is providing new insights into the forces that shaped human genomes before and after the Out-of-Africa migrations. As a result of this human evolutionary history, African ancestry populations have the greatest genomic diversity in the world, and this diversity has important ramifications for genomic research. In the case of pharmacogenomics, for instance, variants of consequence are not limited to those identified in other populations, and diversity within African ancestry populations precludes summarizing risk across different African ethnic groups. Exposure of Africans to fatal pathogens, such as Plasmodium falciparum, Lassa Virus and Trypanosoma brucei rhodesiense, has resulted in elevated frequencies of alleles conferring survival advantages for infectious diseases, but that are maladaptive in modern-day environments. Illustrating with cardiometabolic traits, we show that while genomic research in African ancestry populations is still in early stages, there are already many examples of novel and African ancestry-specific disease loci that have been discovered. Furthermore, the shorter haplotypes in African genomes have facilitated fine-mapping of loci discovered in other human ancestry populations. Given the insights already gained from the interrogation of African genomes, it is imperative to continue and increase our efforts to describe genomic risk in and across African ancestry populations.
Introduction
Africa is the birthplace of anatomically modern humans and the geographic origin of human expansion across the globe within the last 100 000 years (1–3). Consequently, anatomically modern humans have lived longer on the African continent than anywhere else in the world (4–6). This long evolutionary history has led to several important genetic characteristics, as revealed by the growing engagement of African people in genomic projects such as the International HapMap, the 1000 Genomes, and the African Genome Variation Projects (AGVP) (7–9). Insights provided by these projects include the existence of more haplotypes, lower levels of linkage disequilibrium (LD), more divergent patterns of LD, and more complex patterns of population substructure in Africans compared with other populations. Anthropological and epidemiological studies are providing new understanding of the cultural and social characteristics of African populations, including linguistic diversity representing approximately a third of the world’s languages, complex dietary patterns, varied farming practices and uneven economic development (5,6). These characteristics have resulted in varying levels of urbanization and differences in the distribution of non-genetic risk factors for disease across African populations.
A high degree of population subdivision across Africa has been observed, with nearly as many ancestries within Africa as in the rest of the world combined (3,5,10). On average, African populations display the highest level of within populations genetic diversity such that genetic diversity declines with distance from Africa (6). Consistent with the out-of-Africa model of human origin, the number of variant sites per genome is highest among Africans (∼5 million variants) compared with individuals of East Asian, European, or South Asian ancestry (∼4.0–4.2 million variants) (8). Genetic differentiation between African ancestries (e.g. FST is 0.054 between Khoisan and Omotic ancestries) can exceed that between pairs of non-African ancestries (e.g. 0.024 between Southern and Northern European ancestries or 0.042 between Arabian and Indian ancestries) (3,5). As a function of physical distance, LD decays faster in African populations than in non-African populations, resulting in shorter haplotypes among Africans as illustrated in Figure 1 (8). Admixture has occurred between African ethno-linguistic groups and both Eurasian and hunter-gatherer populations in regionally distinctive ways (9). Furthermore, genomes sampled from the Americas display considerable African admixture with average African proportion as high as 80% in populations such as African Americans (AA) (3,4,8). Notably, individuals with African admixture show great variability in the number of variants and the degree of variability is roughly proportional to the degree of recent African ancestry in their genomes (8).
Figure 1.
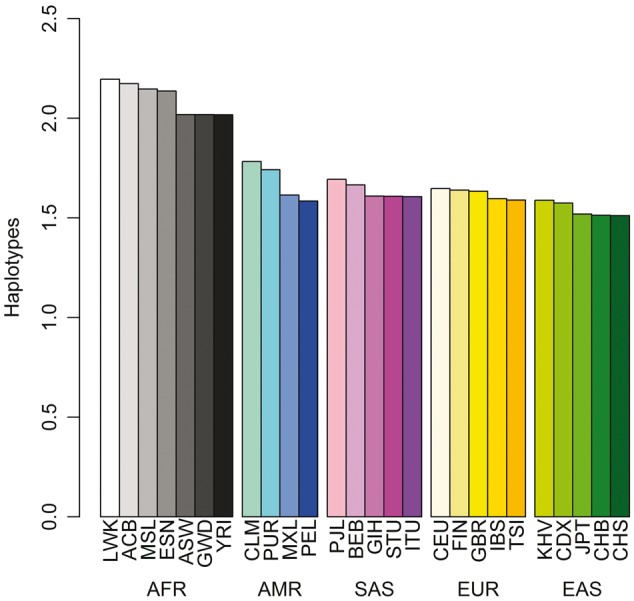
Haplotype diversity in the 1000 Genomes Project. Haplotypes were counted over windows of six consecutive diallelic SNPs, with a slide of one SNP. For each sample, all counted haplotypes have a frequency ≥1%. See Supplementary Material, Table S1 in (8) for the sample descriptors.
Recent genomic studies also show that the current generation of genome-wide association study (GWAS) arrays are inefficient for interrogating African genomes, motivating a major effort by the Human Heredity and Health in Africa (H3Africa) Consortium and Illumina to develop more efficient genome-wide arrays by interrogating whole-genome sequences of thousands of Africans sampled across the continent. Using examples from infectious and cardiometabolic disorders, this review will illustrate how African genomes and environments have shaped the distributions of diseases and variable drug response across the continent, with implications for health disparities among its global diasporan populations, including AA. In addition, implications of some of the unique characteristics of the African genomes for the performance of imputation and GWAS studies are discussed.
The Impact of Infectious Diseases on Inherited Diseases
Infectious diseases have impacted the human genome. Some mutations, while conferring protection against fatal pathogens, can be associated with costs in the form of inherited diseases. For example, seven loci in the human genome associated with protection against different forms of malaria are associated with several inherited diseases (Table 1).
Table 1.
Loci protective against malaria that are associated with inherited diseases
| Gene | Cost | References |
|---|---|---|
| HBA | α thalassemia | (134) |
| HBB | β thalassemia | (134) |
| HBB | sickle cell | (135,136) |
| G6PD | favism, hemolysis, jaundice | (137-139) |
| CD36 | impaired fatty acid metabolism, glucose intolerance, atherosclerosis, arterial hypertension, diabetes, cardiomyopathy, Alzheimer disease | (140,141) |
| ACKR1 | benign ethnic neutropenia | (142-145) |
| HBG1 | hyperuricemia, hypertension | (146,147) |
| GYPA/GYPB/GYPE | ovalocytosis, anemia | (148) |
Two genome-wide admixture mapping studies performed in AA identified a locus at chromosome 22q12.3 at which African ancestry conferred increased risk for several forms of end-stage kidney disease (11,12). Subsequent fine-mapping identified two haplotypes in APOL1 called G1 and G2 (13,14). APOL1 carrying G2 affords protection against Trypanosoma brucei rhodesiense because the G2 deletion prevents binding of a trypanosomal virulence factor called serum resistance-associated protein (15). Derived allele frequencies for G1 are highest in West-Central Africa, particularly in Nigeria and Ghana, and zero among non-African ancestry populations (16). APOL1 variants have been associated with increased risk of multiple nondiabetic nephropathies and cardiovascular disease (17–23), accounting for a large proportion of observed disparities in renal disease between individuals with and without African ancestry. However, not all individuals with two copies of the APOL1 variants develop kidney disease, suggesting the existence of other genetic and/or environmental modifying factors. Understanding the mechanism through which APOL1 influences kidney diseases is a major biomedical priority. A recent study has demonstrated that APOL1 risk alleles are pathogenic. This study overcame the fact that APOL1 appears to exist only in a subset of primate lineages by generating transgenic mice with podocyte-specific inducible expression of APOL1. This study demonstrated that APOL1 risk variants adversely affect endosomal trafficking and autophagy, leading to inflammatory-mediated podocyte death and glomerular scarring (24).
Lassa virus, the cause of Lassa fever, has origins in the reservoir of the rodent Mastomys natalensis in Nigeria (25). The main cellular receptor for Lassa virus is α-dystroglycan (DAG1) (26). Glycosylation of α-dystroglycan by the enzyme LARGE is required for binding of Old World arenaviruses including Lassa virus (27). Signals for natural selection have been detected at LARGE in populations with West African ancestry (28–30). Mutations in DAG1 and LARGE are associated with congenital muscular dystrophies (31–34), raising the possibility that an increased risk of congenital muscular dystrophies is a cost for increased resistance to arenavirus infection.
Pharmacogenomics
The vast genetic diversity in African ancestry individuals (8,9) has pharmacogenomic consequences (16,35). Not only are there marked differences in allele frequencies at pharmacogenomic loci between African versus other ancestry populations, these variants also differ importantly among African ancestry populations (36,37). For example, the HLA-B*57:01 allele is strongly associated with serious adverse reactions to the HIV/AIDS drug abacavir. Among the Yoruba in Nigeria, the risk allele is absent, but it is carried by 14% of Kenyan Masai. Importantly, among the Luhya of Kenya, this variant is much less frequent (3%), showing that even the descriptor ‘Kenyan’ is inadequate to describe potential pharmacogenomic risk (38). Although practicing precision medicine may be prohibitively expensive in many parts of Africa, pharmacogenomic discoveries have already been used to improve drug safety in some countries by considering the average frequency of risk alleles in their population before selecting medicines for use (39,40). Further progress will require a more complete understanding of the genetic diversity of African ancestry individuals at pharmacogenomic loci.
Examples of Genomic Studies in African and AA Populations
Hypertension and blood pressure traits
We conducted the first GWAS of hypertension (HTN) and blood pressure (BP) traits in AA (n = 1017) (41). One of the three significant loci identified is a HTN gene (CACNA1H) that is a drug target for a class of calcium channel blockers. Another (SLC24A4) is a strong candidate for BP regulation (41). African ancestry individuals have since been included in several other published GWAS and meta-analyses of HTN and BP traits, including an international study that identified 29 significant loci in over 200 000 individuals of European ancestry (EA) with replication in over 70 000 non-European (East Asian, South Asian and African) ancestry individuals (42). The COGENT Network has conducted a GWAS of systolic blood pressure (SBP) and diastolic blood pressure (DBP) in AA and Nigerians (n = 29 378) that identified three novel loci (EVX1-HOXA, RSPO3 and PLEKHG1) in addition to two known loci (ULK4 and SOX6) (43). Another COGENT study used a novel methodological approach that systemically integrated association evidence from multiple traits to identify four loci (CHIC2, EVX1-HOXA, IGFBP1/IGFBP3 and CDH17) associated with HTN-related traits, some of which were missed or undetectable in a single-trait analysis (44). A recent investigation of BP traits in African ancestry individuals from eight cohorts (n = 15 914) (45) identified rare variants associated with SBP and DBP in ten genes, including AFF1, GAPDHS, SLC28A3, COL6A1, CRYBA2, KRBA1, SEL1L3, YOD1, CCDC13 and QSOX1. This study provided important risk information on rare variants that complemented published findings for common variants in AA and continental Africans.
The only primary GWAS for HTN/BP in Africans to date used a pathway-focused approach to study Nigerians (n = 1614). Evidence in support of biological relevance was found for the ADRA1 receptor pathway (46). A recent GWAS for metabolic syndrome (MetSyn) in Africans (n = 1427; described in more detail below) demonstrated that variants in KSR2 and MBNL1 displayed significant pleiotropic associations with BP in the single trait analysis (47). Our recent analysis of the GWAS data available through the AGVP (n = 1481) (9) identified several loci under selection in African populations, including a highly differentiated variant (rs1378940) in the CSK region which has been reported to be associated with HTN and BP in multiple GWAS (42,48–50). Although these studies provide some evidence in support of the genetic basis of BP regulation in African ancestry populations, the field awaits the publication of the first adequately powered GWAS of BP traits in Africans.
Type 2 diabetes and related traits
Type 2 diabetes (T2D) is a devastating disease that impacts quality of life and increases risk of complications and cardiovascular disease. In the USA, the prevalence of T2D is approximately twice as high among AA compared with EA individuals (51), and the prevalence of T2D is anticipated to increase by 140% in Africa by 2040 (52). The largest genome-wide study of T2D in Africa included 1775 West Africans (53), in which 106 previously reported loci across the genome were evaluated. One of these loci, the well-established TCF7L2 locus (54–56), reached genome-wide significance. There was evidence of transferability for 39% of these loci to West Africans. Although it is probable that the proportion of known T2D loci with evidence of transferability to African populations will grow as sample sizes increase, genetic diversity across world populations (particularly for African versus non-African ancestry individuals) at T2D loci has been noted (57).
Considerably more genomic research has been conducted in AA for T2D (58–63). Notable among these is the largest GWAS of T2D in AA (n = 23 827) (60) which identified several significant loci including TCF7L2, HMGA2 and KCNQ1 that were previously reported and HLA-B and INS-IGF2 that were novel findings. Interestingly, HLA-B and INS-IGF2 have been associated with type 1 diabetes; new research supports some overlap in the pathways underlying susceptibility to both type 1 diabetes and T2D (64). The signal at INS-IGF2 has since been replicated in Africans (65). Other studies of common variants have identified two novel signals: RND3/RBM43 in 1994 AA (61) and an independent association within the known locus HMGA2 in 9681 AA (62). Analyses of low frequency and rare variants have not reported novel discoveries among AA (58,63).
Studies of T2D-related traits insulin and insulin resistance in African ancestry individuals have also yielded novel findings. A trans-ethnic meta-analysis of fasting glucose and insulin in 20 209 AA and 57 292 EA individuals revealed two novel loci for fasting insulin: FAM133A and PELO (66). Similar to proportions reported for T2D loci, 43% of EA-identified loci were shared with AA (66). A GWAS of fasting insulin and insulin resistance in 927 non-T2D AA with follow-up and meta-analysis in 570 non-T2D West Africans identified SC4MOL and TCERG1L as novel loci for these traits (67).
The MetSyn is a clustering of traits that have been shown to dramatically increase risk of T2D and cardiovascular disease. A large study of MetSyn in 15 148 AA was conducted using the Metabochip genotyping array (68). Of the five MetSyn-associated loci, one was in a known glucose locus (TCF7L2) and four were in lipids loci (LPL, APOA5, CETP and APOC1/APOE/TOMM40), consistent with previous reports of the contribution of lipids and T2D loci to MetSyn risk. Only one genetic or genomic study of MetSyn in Africans has been reported. This study included participants from Ghana and Nigeria (n = 1427) with replication and meta-analysis in Kenyans (n = 174) and in AA (n = 2475). This analysis identified two African ancestry-specific variants that were associated with MetSyn, one near CA10 and one in CTNNA3. The signal at CA10 was subsequently observed to be associated with T2D in Africans (65). Additionally, two variants (near RALYL and KSR2) that are not unique to African populations were significantly associated with MetSyn. This study also replicated previous MetSyn associations with LPL and CETP that had been found in EA (47).
Obesity and inflammation
The growing global obesity epidemic is particularly devastating for African ancestry populations on the continent and in the diaspora (69–71). Although it is evident that lifestyle changes are the major drivers of the obesity epidemic, it is well established that genetic factors play a role in individual susceptibility to excess weight gain and related metabolic disorders (72). Indeed, the heritability of obesity has been estimated to be as high as 60% (73) and over 300 loci have been identified to be associated with measures of adiposity (47,73,74). Several genetic/genomics studies of obesity traits have included African ancestry populations (47,75–93). Collectively, these studies have identified or replicated variants in FTO, MC4R, MTCH2, TFAP2A, SEC16B, TMEM18, NEGR1 and POMC in African ancestry populations. Variants in the FTO gene are associated with obesity in West Africans (94) and in Black South Africans, suggesting that common genetic variants contribute to obesity risk across populations (83,95,96).
We recently published the first GWAS of BMI in Africans (n = 1570), which included replication in independent samples of Africans (n = 1411) and AA (n = 9020). This study identified an African-specific variant (rs80068415) in Semaphorin-4D (SEMA4D) that was associated with increased BMI (73). The largest meta-analysis of measures of adiposity that includes African ancestry individuals (n = 52 895) (74) identified two novel loci: TCF7L2/HABP2 (sex-combined analysis) and SPRYD7/DLEU2 (ancestry-combined analysis). Consistent with observed sexual dimorphism in the genetic determinants of anthropometric markers (97,98), five additional loci were found to be associated with adiposity in sex-stratified analysis in the African ancestry cohort: IRX4/IRX2 (women), INTS10/LPL (men), MLC1 (men), SSX2IP (women) and PDE3B (women) (74).
A number of tissues, including adipose and muscle, are involved in the pathophysiology of obesity and obesity-related traits (99). An epigenome-wide association study of BMI, BMI change, and waist circumference in 2097 AA identified 15 novel methylation sites near genes implicated in lipid metabolism, immune response, and cytokine signaling (76). An eQTL study in AA identified ∼2000 transcripts with at least one significant cis-eQTL in adipose and muscle tissues (76). Comparative transcriptomics between 14 obese AA with T2D and 6 obese AA without T2D in omental adipose tissue identified a number of canonical pathways that were overrepresented and overlaid with T2D signaling pathways including telomere extension by telomerase (HNRNPA1, TNKS2), D-myo-inositol (1,4,5)-trisphosphate biosynthesis (PIP5K1A, PIP4K2A), and regulation of actin-based motility by Rho (ARPC3). Additionally, five miRNAs (miR-320b, miR-381-3p, miR-3679-3p, miR-494-3p and miR-141-3p) were predicted as regulators of the expression changes identified in the omental adipose tissue of obese diabetics (100).
Inflammation is a hallmark of obesity and its complications (101). An association between obesity and circulating levels of adipokines has been observed in Africans (102–104). Serum concentrations of adipokines and inflammatory markers are under genetic control (105–107). Surprisingly, except for C reactive protein levels, serum levels of interleukins IL-10, IL-1RA and IL-6 appear to be under the control of trans-QTL. Adjusting for BMI did not alter the strength of these associations, suggesting that the identified variants directly influence interleukin levels (106). Adiponectin, the only adipokine known to be decreased in obesity, has an anti-inflammatory property and is associated with obesity in African diasporan populations (103). Paradoxically, in metabolically healthy but obese individuals (MHO), adiponectin is increased compared with metabolically unhealthy individuals (108). The association between high adiponectin and MHO suggests that the absence of inflammation may be protective against the development of obesity-related co-morbidities. Furthermore, our characterization of MHO in AA using shot-gun proteomics revealed that under-expression of inflammatory markers was characteristic of MHO (109). Polymorphisms in adiponectin have been associated with obesity in populations of African ancestry (84) and this association seems to be modulated by the proportion of EA: the incidence of obesity was higher in individuals with the G allele and a higher proportion of African ancestry (84).
Serum lipids
In contrast to the increased prevalence of HTN, T2D, and obesity observed with increasing degree of westernization, there is remarkable similarity in the distribution of serum lipid parameters across African ancestry populations. In the USA, interethnic differences in serum lipid distributions are consistently observed, with AA having a generally healthier lipid profile than European Americans despite differences in lifestyle factors that would be expected to produce a worse lipid profile among AA. This observation, coupled with the relative similarity in distribution of lipids between West Africans and AA with even greater differences in lifestyle characteristics, suggests that the healthier lipid profile among AA has a genetic basis (110).
Primary GWAS of serum lipids have yet to be performed in Africans. However, GWAS conducted in AA have identified a number of novel loci for lipid parameters. Common variant association studies have identified novel loci for low-density lipoprotein in ICAM1 (111,112) and for HDL in CD36 (112) and EXOC3L1 (113). Large-scale exome sequencing projects in AA have identified associated rare/low-frequency variants in novel loci COL18A1 and PCSK7 (114), as well as known loci APOC3 (115), PCSK9 (116), CETP (114), LCAT (114), APOE (114) and ZNF259 (114).
A predominant focus in the published work on the genomics of serum lipids in African ancestry individuals, however, is not in discovery analyses, but in establishing the transferability of loci discovered primarily in EA studies (59,112,117–119). It has been estimated that 42% of lipids loci have replicated in AA (117). Despite modest statistical replication, analyses have noted a consistent direction of effect for most loci (112,120–122), suggesting that replication may be achieved with increased sample sizes. However, effect sizes may appear diluted in AA compared with EA, due to interethnic differences in LD between the tag and causal SNP (122).
An efficient strategy that has been used to address the issue of trans-ethnic replication in the presence of interethnic differences in LD patterns and minor allele frequencies is first sequencing the gene of interest in African ancestry individuals to identify variants present before conducting analyses. We sequenced five lipids loci (LPL, ABCA1, PON1, LCAT and SERPINE1) in 48 AA, with follow-up in 1694 AA. As expected, LPL was associated with serum lipids; however, the ancestral background on which this variant occurred modified this association. The associations were much stronger among those with EA at this locus, and not significant among those with only African ancestry, in whom the associations were strikingly similar to those in West Africans (123). Sequencing of known lipids genes has also been performed in studies of Africans, identifying novel associations in LDLR (124), APOB (125), CETP (126), SCARB1 (127) and LPL (128).
Examples of Fine-Mapping of Loci Using Reduced LD in African Genomes
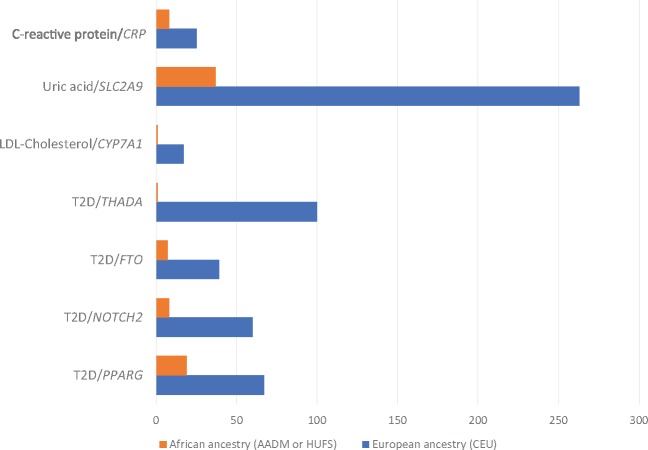
As discussed earlier, the demographic histories of African ancestry individuals have resulted in shorter segments of linked alleles compared with the relatively longer segments observed in other human populations (Fig. 1). The distinct patterns created by these relative segmental lengths can be leveraged in genomic studies for fine-mapping loci associated with disease and non-disease traits (129). One of the earliest demonstrations of this principle was for the refinement of the association between variants in TCF7L2 and T2D (55). The considerable interest and enthusiasm accompanying the discovery was tempered by the observation that the association spanned over many markers and a fairly broad region due to the strong LD in EA populations. Using the weaker LD in West Africans, we were able to refine initial association between T2D and three markers (the composite allele X of microsatellite DG10S478 and the T alleles of SNPs rs12255372 and rs7903146) which were in strong LD in EA populations (54). In the West African sample, the association was not significant for DG10S478, weaker for rs12255372 and statistically significant for rs7903146, indicating that either rs7903146 is the risk variant or its closest known correlate (55). Using sequence data on this region from the 1000 Genomes Project shows how critical the finer-grained LD in West Africans was in refining the original association (Fig. 2). Another notable example is the fine-mapping of the SLC2A9 locus originally reported to be associated with serum uric acid concentrations in EA populations. In our GWAS in AA, four variants in this locus achieved genome-wide significance for association with uric acid (130). Using an r2 cutoff of ≥0.5, the 10 top ranking SNPs fall within the 263 kb range in EA but remarkably 9 of the 10 top ranked SNPs lie within the 37 kb interval and 3 out of the 4 SNPs that achieved genome-wide significance lie in an ∼1.3 kb interval. We also found other notable examples of fine-mapping (Fig. 3) from our genome-wide studies of Africans or AA in the CYP7A1 locus for LDL-cholesterol (117) and in the CRP promoter locus for C-reactive protein (105). A recent trans-ethnic fine-mapping study of over 54 000 individuals of diverse ancestries reduce the region of interest for three known lipid loci (LIPC, PLTP and APOA5) to a single variant by taking advantage of the reduced LD in the AA included in the analyses (131). Last, an attempt to fine-map genetic association signals using expression quantitative trait loci (eQTL) in six 1000 Genomes populations (Utah residents with Northern and Western European ancestry [CEU]; Han Chinese in Beijing, China [CHB]; Gujarati Indian from Houston, Texas [GIH]; Japanese in Tokyo, Japan [JPT]; Luhya in Webuye, Kenya [LWK] and Yoruba in Ibadan, Nigeria [YRI]) demonstrated how African sampes can facilitate improved localization (8). Overall, between 17.5 and 19.5% of the top eQTL variants overlapped annotated transcription factor-binding sites (TFBSs) within each population. However, a meta-analysis approach that combined pairs of populations resulted in an increase in the proportion of top eQTL variants overlapping TFBSs to 19.2–21.6% consistent with improved localization. Notably, the inclusion of an African population facilitated the greatest increase in overlap between top variants and TFBSs (8).
Figure 2.
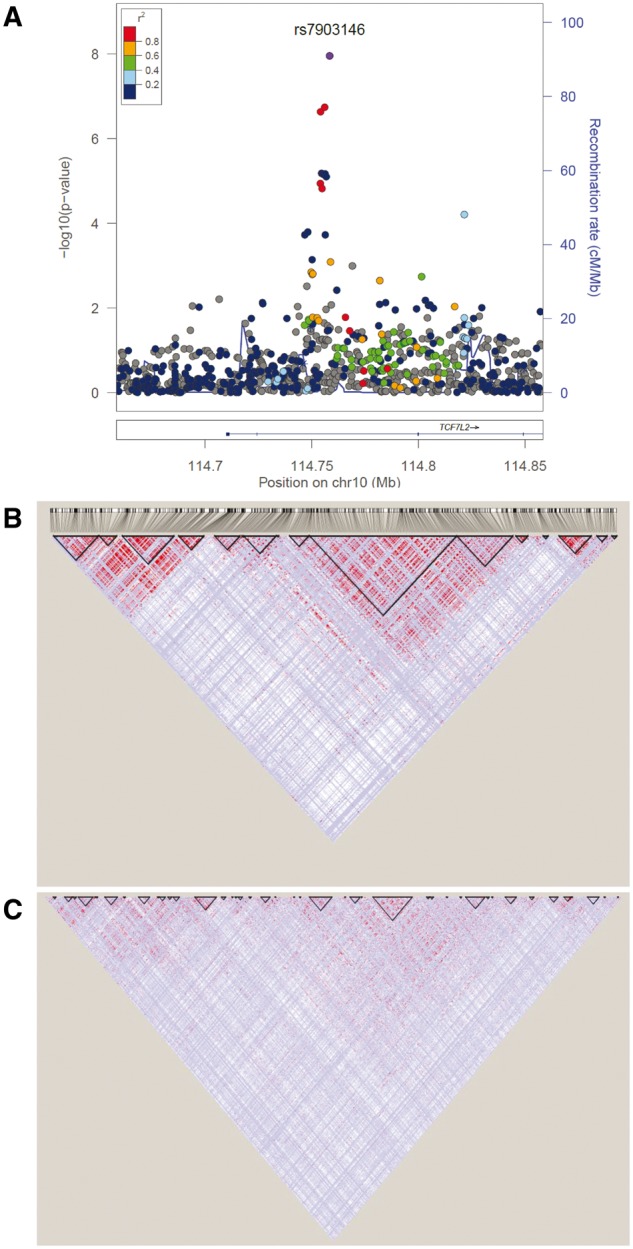
Fine-mapping of TCF7L2. (A) Regional plot of the TCF7L2 rs7903146 association among West Africans (55). (B) LD plot of the region in 1000 Genomes CEU. (C) LD plot of the region in 1000 Genomes YRI. Consistent with the original report based on genotyping a few SNPS around the locus (54), both sequence and genotype data showed a much smaller haplotype around the lead SNP in the African samples. Note the fine-grained haplotype structure across the whole region in the African ancestry compared with the EA sample.
Figure 3.
Fine-mapping examples. Examples from studies of C-reactive protein (104), serum uric acid (129), serum lipids (116), and T2D (53) showing how smaller haplotype blocks in African ancestry populations helped refine genome-wide significant loci. Data are shown for fine-mapping analyses conducted in sub-Saharan Africans from Nigeria, Ghana and Kenya (AADM) and in AA from the Washington DC region in the United States (HUFS).
Considerations for Future Studies
In the context of the growing success stories of the genomics of African ancestry populations, we highlight some issues for consideration for the design of future initiatives. First, a careful review of the literature shows that the sampling of African ancestry populations remains a patchwork process, with efforts ranging from collecting convenience samples (i.e. samples that are easy to get or already available) to the more systematic approaches implemented by the 1000 Genomes, AGVP and the H3Africa projects, among others. In the context of sub-Saharan Africa, the 1000 Genomes Project focused on populations genetically close to the Yoruba (8). The African Genome Variation, Egypt Genome, and H3Africa Projects have contributed samples from populations in Benin, Botswana, Burkina Faso, Cameroon, Côte d’Ivoire, Democratic Republic of the Congo, Egypt, Ethiopia, The Gambia, Ghana, Guinea, Kenya, Mali, Nigeria, South Africa, Uganda and Zambia. These samples are predominantly from Niger-Congo-speaking populations, largely as a consequence of the Bantu Expansion. Many linguistic groups are poorly or not represented, including speakers of Afroasiatic, Khoisan and Nilo-Saharan languages, as well as other populations such as Central African Pygmies, Hadza, Laal and Sandawe. In the past, for reasons including ongoing wars, it has been extremely difficult to sample some regions of the African continent. Resolution of some of these human conflicts are presenting us with new opportunities for filling in these gaps. However, the achievement of this goal must include robust community engagements and involvement of local scientists as true partners and address local ethical issues that are likely to arise.
A second issue for consideration is what sample sizes are needed. The answer, of course, depends on the question being asked. For association analysis of single variants, the sample size is inversely proportional to r2 between the tag and causal variants (132). Thus, genotyping arrays that provide higher correlation between typed tag variants and untyped causal variants will be more efficient, requiring smaller sample sizes. Larger sample sizes are needed for lower frequency alleles or haplotypes and for smaller effect sizes. For diversity projects, sample sizes are inversely proportional to FST. Given the lower LD in African ancestry populations, a more efficient GWAS array, such as the H3Africa consortium array under development, will provide better opportunities to identify susceptibility loci.
A third issue is the adequacy of available reference panels for imputation in African ancestry populations. It is heartening to observe that this issue has received considerable attention from the genomic scientific community such that available reference panels are constantly improving. However, based on whole-genome sequencing, we demonstrated recently that the inclusion of more genetic diversity from the AGVP to the 1000 Genomes reference panels resulted in substantial improvement in imputation accuracy for Sotho population from South Africa (9). This result suggested poor representation of some haplotypes (e.g. Khoe-San haplotypes in Sotho) in the 1000 Genomes Project reference panel and calls for the inclusion of more diverse whole-genome sequenced samples in public genomic reference panels (9). A recent contribution to this effort was made by the southern African Human Genome Program (SAHGP), which is at the forefront of whole-genome sequencing in Africa of Africans by Africans. Although limited in scope, the SAHGP is demonstrating the feasibility of major sequencing effort led by continental scientists with local funding (133). However, larger future efforts such as GWAS based on whole-genome sequences in Africa will require a substantially higher commitment of resources from national and international funding agencies.
Conclusions
Genomic science is contributing novel insights into biology and human history, especially in EA populations. The engagement of Africans and it’s diasporan populations in genomic science is still limited but growing (Fig. 4). Some of the challenges facing a broader understanding of the genomics of African people are highlighted in Box 1. Although the ability to summarize the transferability of loci identified in other populations is a notable advance, genomic diversity among African populations offers the potential to identify novel loci that could enhance our understanding of pathophysiology and prompt new treatment strategies that could benefit at-risk Africans as well as worldwide populations. As exemplified in this review, the successes proceeding from the still limited genomic research in African ancestry populations should motivate the field to promote and support the increased efforts into this area.
Figure 4.
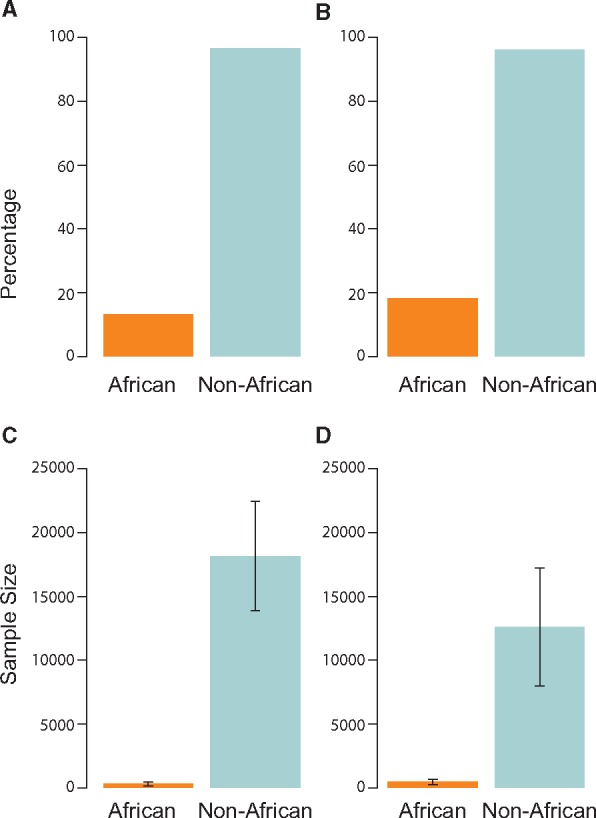
Inclusion of African ancestry individuals in GWAS. (A) Percentage of studies including individuals with African ancestry versus non-African ancestry individuals in the discovery sample. (B) Percentage of studies including individuals with African ancestry versus non-African ancestry individuals in the replication sample. (C). Average sample size of individuals with African ancestry versus non-African ancestry individuals in the discovery sample. (D) Average sample size of individuals with African ancestry versus non-African ancestry individuals in the replication sample. Whiskers indicate 95% CIs. Data compiled from the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas; date last accessed May 17, 2017) from 2015 to 2017.
Box 1.
Key challenges to the improved understanding of the genomics of African peoples
Too few African ancestry scientists in genomics
Limited funding opportunities, especially from national governments
Inadequate genomic research infrastructure
Lack of optimum genomics tools (e.g. GWAS arrays) and methodologies for the interrogation of African genomes
Need for more discovery- versus replication-focused analyses
Larger studies of common, complex traits needed
Insufficient characterization of diverse African ethnicities
Further characterization of pharmacogenomic loci across the vast genomic diversity of African ancestry populations essential for clinical care
Appropriately considering environmental background in studies of complex diseases, given vast environmental heterogeneity across African ancestry populations.
Acknowledgements
The authors would like to acknowledge the contribution of Darryl Leja, National Human Genome Research Institute, National Institutes of Health, USA, to the design of a figure for this article.
Conflict of Interest statement. None declared.
Funding
This research was supported by the Intramural Research Program of the National Human Genome Research Institute in the Center for Research in Genomics and Global Health (CRGGH, Z01HG200362). Center for Research in Genomics and Global Health is also supported by National Institute of Diabetes and Digestive and Kidney Diseases, Center for Information Technology, and the Office of the Director at the National Institutes of Health.
References
- 1. Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. (2017) Tracing the peopling of the world through genomics. Nature, 541, 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shriner D., Tekola-Ayele F., Adeyemo A., Rotimi C.N. (2016) Ancient human migration after out-of-Africa. Sci. Rep, 6, 26565.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shriner D., Tekola-Ayele F., Adeyemo A., Rotimi C.N. (2014) Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci. Rep., 4, 6055.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell M.C., Hirbo J.B., Townsend J.P., Tishkoff S.A. (2014) The peopling of the African continent and the diaspora into the new world. Curr. Opin. Genet. Dev., 29, 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rotimi C.N., Tekola-Ayele F., Baker J.L., Shriner D. (2016) The African diaspora: history, adaptation and health. Curr. Opin. Genet. Dev., 41, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.M., Doumbo O.. et al. (2009) The genetic structure and history of Africans and African Americans. Science, 324, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. International HapMap C., Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F.. et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature, 467, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. The 1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudhury A.. et al. (2015) The African Genome Variation Project shapes medical genetics in Africa. Nature, 517, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker J.L., Rotimi C.N., Shriner D. (2017) Human ancestry correlates with language and reveals that race is not an objective genomic classifier. Sci. Rep., 7, 1572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kopp J.B., Smith M.W., Nelson G.W., Johnson R.C., Freedman B.I., Bowden D.W., Oleksyk T., McKenzie L.M., Kajiyama H., Ahuja T.S.. et al. (2008) MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet., 40, 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kao W.H.L., Klag M.J., Meoni L.A., Reich D., Berthier-Schaad Y., Li M., Coresh J., Patterson N., Tandon A., Powe N.R.. et al. (2008) MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet., 40, 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Uscinski Knob A.L.. et al. (2010) Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science, 329, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tzur S., Rosset S., Shemer R., Yudkovsky G., Selig S., Tarekegn A., Bekele E., Bradman N., Wasser W.G., Behar D.M.. et al. (2010) Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet., 128, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomson R., Genovese G., Canon C., Kovacsics D., Higgins M.K., Carrington M., Winkler C.A., Kopp J., Rotimi C., Adeyemo A.. et al. (2014) Evolution of the primate trypanolytic factor APOL1. Proc. Natl. Acad. Sci. U. S. A., 111, E2130–E2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baker J.L., Shriner D., Bentley A.R., Rotimi C.N. (2016) Pharmacogenomic implications of the evolutionary history of infectious diseases in Africa. Pharmacogenomics J, 17, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Genovese G., Tonna S.J., Knob A.U., Appel G.B., Katz A., Bernhardy A.J., Needham A.W., Lazarus R., Pollak M.R. (2010) A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int., 78, 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopp J.B., Nelson G.W., Sampath K., Johnson R.C., Genovese G., An P., Friedman D., Briggs W., Dart R., Korbet S.. et al. (2011) APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol., 22, 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parsa A., Kao W.H.L., Xie D., Astor B.C., Li M., Hsu C.-Y., Feldman H.I., Parekh R.S., Kusek J.W., Greene T.H.. et al. (2013) APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med., 369, 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freedman B.I., Langefeld C.D., Andringa K.K., Croker J.A., Williams A.H., Garner N.E., Birmingham D.J., Hebert L.A., Hicks P.J., Segal M.S.. et al. (2014) End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthr. Rheumatol., 66, 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ito K., Bick A.G., Flannick J., Friedman D.J., Genovese G., Parfenov M.G., DePalma S.R., Gupta N., Gabriel S.B., Taylor H.A. Jr. et al. (2014) Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ. Res., 114, 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mukamal K.J., Tremaglio J., Friedman D.J., Ix J.H., Kuller L.H., Tracy R.P., Pollak M.R. (2016) APOL1 Genotype, Kidney and Cardiovascular Disease, and Death in Older Adults. Arterioscler. Thromb. Vasc. Biol., 36, 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kasembeli A.N., Duarte R., Ramsay M., Mosiane P., Dickens C., Dix-Peek T., Limou S., Sezgin E., Nelson G.W., Fogo A.B.. et al. (2015) APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J. Am. Soc. Nephrol., 26, 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beckerman P., Bi-Karchin J., Park A.S.D., Qiu C., Dummer P.D., Soomro I., Boustany-Kari C.M., Pullen S.S., Miner J.H., Hu C.-A.A.. et al. (2017) Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat. Med., 23, 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersen K.G., Shapiro B.J., Matranga C.B., Sealfon R., Lin A.E., Moses L.M., Folarin O.A., Goba A., Odia I., Ehiane P.E.. et al. (2015) Clinical sequencing uncovers origins and evolution of lassa virus. Cell, 162, 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao W., Henry M.D., Borrow P., Yamada H., Elder J.H., Ravkov E.V., Nichol S.T., Compans R.W., Campbell K.P., Oldstone M.B.A. (1998) Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science, 282, 2079–2081. [DOI] [PubMed] [Google Scholar]
- 27. Hara Y., Kanagawa M., Kunz S., Yoshida-Moriguchi T., Satz J.S., Kobayashi Y.M., Zhu Z., Burden S.J., Oldstone M.B.A., Campbell K.P. (2011) Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc. Natl. Acad. Sci. U. S. A., 108, 17426–17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R.. et al. (2007) Genome-wide detection and characterization of positive selection in human populations. Nature, 449, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fumagalli M., Pozzoli U., Cagliani R., Comi G.P., Bresolin N., Clerici M., Sironi M. (2010) Genome-wide identification of susceptibility alleles for viral infections through a population genetics approach. PLOS Genet., 6, e1000849.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersen K.G., Shylakhter I., Tabrizi S., Grossman S.R., Happi C.T., Sabeti P.C. (2012) Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci., 367, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longman C., Brockington M., Torelli S., Jimenez-Mallebrera C., Kennedy C., Khalil N., Feng L., Saran R.K., Voit T., Merlini L.. et al. (2003) Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of α-dystroglycan. Hum. Mol. Genet., 12, 2853–2861. [DOI] [PubMed] [Google Scholar]
- 32. Clarke N.F., Maugenre S., Vandebrouck A., Urtizberea J.A., Willer T., Peat R.A., Gray F., Bouchet C., Manya H., Vuillaumier-Barrot S.. et al. (2011) Congenital muscular dystrophy type 1D (MDC1D) due to a large intragenic insertion/deletion, involving intron 10 of the LARGE gene. Eur. J. Hum. Genet., 19, 452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meilleur K.G., Zukosky K., Medne L., Fequiere P., Powell-Hamilton N., Winder T.L., Alsaman A., El-Hattab A.W., Dastgir J., Hu Y.. et al. (2014) Clinical, pathologic, and mutational spectrum of dystroglycanopathy caused by LARGE mutations. J. Neuropathol. Exp. Neurol., 73, 425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riemersma M., Mandel H., van Beusekom E., Gazzoli I., Roscioli T., Eran A., Gershoni-Baruch R., Gershoni M., Pietrokovski S., Vissers L.E.. et al. (2015) Absence of α- and β-dystroglycan is associated with Walker-Warburg syndrome. Neurology, 84, 2177–2182. [DOI] [PubMed] [Google Scholar]
- 35. Li J., Zhang L., Zhou H., Stoneking M., Tang K. (2011) Global patterns of genetic diversity and signals of natural selection for human ADME genes. Hum. Mol. Genet., 20, 528–540. [DOI] [PubMed] [Google Scholar]
- 36. Ramos E., Doumatey A., Elkahloun A.G., Shriner D., Huang H., Chen G., Zhou J., McLeod H., Adeyemo A., Rotimi C.N. (2014) Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J., 14, 217–222. [DOI] [PubMed] [Google Scholar]
- 37. Tekola-Ayele F., Adeyemo A., Aseffa A., Hailu E., Finan C., Davey G., Rotimi C.N., Newport M.J. (2015) Clinical and pharmacogenomic implications of genetic variation in a southern Ethiopian population. Pharmacogenomics J., 15, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rotimi C.N., Jorde L.B. (2010) Ancestry and disease in the age of genomic medicine. N. Engl. J. Med., 363, 1551–1558. [DOI] [PubMed] [Google Scholar]
- 39. Nordling L. (2017) How the genomics revolution could finally help Africa. Nature, 544, 20–22. [DOI] [PubMed] [Google Scholar]
- 40. Nyakutira C., Roshammar D., Chigutsa E., Chonzi P., Ashton M., Nhachi C., Masimirembwa C. (2008) High prevalence of the CYP2B6 516G–>T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur. J. Clin. Pharmacol., 64, 357–365. [DOI] [PubMed] [Google Scholar]
- 41. Adeyemo A., Gerry N., Chen G., Herbert A., Doumatey A., Huang H., Zhou J., Lashley K., Chen Y., Christman M.. et al. (2009) A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet., 5, e1000564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. International Consortium for Blood Pressure Genome-Wide Association, S., Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C.. et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Franceschini N., Fox E., Zhang Z., Edwards T.L., Nalls M.A., Sung Y.J., Tayo B.O., Sun Y.V., Gottesman O., Adeyemo A.. et al. (2013) Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am. J. Hum. Genet., 93, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu X., Feng T., Tayo B.O., Liang J., Young J.H., Franceschini N., Smith J.A., Yanek L.R., Sun Y.V., Edwards T.L.. et al. (2015) Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am. J. Hum. Genet., 96, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nandakumar P., Lee D., Richard M.A., Tekola-Ayele F., Tayo B.O., Ware E., Sung Y.J., Salako B., Ogunniyi A., Gu C.C.. et al. (2017) Rare coding variants associated with blood pressure variation in 15 914 individuals of African ancestry. J. Hypertens, 35, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reder N.P., Tayo B.O., Salako B., Ogunniyi A., Adeyemo A., Rotimi C., Cooper R.S. (2012) Adrenergic alpha-1 pathway is associated with hypertension among Nigerians in a pathway-focused analysis. PLoS One, 7, e37145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tekola-Ayele F., Doumatey A.P., Shriner D., Bentley A.R., Chen G., Zhou J., Fasanmade O., Johnson T., Oli J., Okafor G.. et al. (2015) Genome-wide association study identifies African-ancestry specific variants for metabolic syndrome. Mol. Genet. Metab., 116, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kato N., Loh M., Takeuchi F., Verweij N., Wang X., Zhang W., Kelly T.N., Saleheen D., Lehne B., Mateo Leach I.. et al. (2015) Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet., 47, 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T.. et al. (2009) Genome-wide association study of blood pressure and hypertension. Nat. Genet., 41, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S.. et al. (2009) Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet., 41, 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Menke A., Casagrande S., Geiss L., Cowie C.C. (2015) Prevalence of and trends in diabetes among adults in the United States, 1988-2012. JAMA, 314, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 52. International Diabetes Federation. (2015). IDF Diabetes Atlas, 7th edn. Brussels, Belgium: International Diabetes Federation.
- 53. Adeyemo A.A., Tekola-Ayele F., Doumatey A.P., Bentley A.R., Chen G., Huang H., Zhou J., Shriner D., Fasanmade O., Okafor G.. et al. (2015) Evaluation of genome wide association study associated type 2 diabetes susceptibility loci in Sub Saharan Africans. Front. Genet., 6, 335.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grant S.F.A., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A.. et al. (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet., 38, 320–323. [DOI] [PubMed] [Google Scholar]
- 55. Helgason A., Palsson S., Thorleifsson G., Grant S.F.A., Emilsson V., Gunnarsdottir S., Adeyemo A., Chen Y., Chen G., Reynisdottir I.. et al. (2007) Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat. Genet., 39, 218–225. [DOI] [PubMed] [Google Scholar]
- 56. Palmer N.D., Hester J.M., An S.S., Adeyemo A., Rotimi C., Langefeld C.D., Freedman B.I., Ng M.C., Bowden D.W. (2011) Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes, 60, 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xian G.G. A comparative analysis of genetic diversity of candidate genes associated with type 2 diabetes in worldwide populations. Yíchuán, 38, 543–559. [DOI] [PubMed] [Google Scholar]
- 58. Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J.. et al. (2016) The genetic architecture of type 2 diabetes. Nature, 536, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lettre G., Palmer C.D., Young T., Ejebe K.G., Allayee H., Benjamin E.J., Bennett F., Bowden D.W., Chakravarti A., Dreisbach A.. et al. (2011) Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project. PLoS Genet., 7, e1001300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ng M.C., Shriner D., Chen B.H., Li J., Chen W.M., Guo X., Liu J., Bielinski S.J., Yanek L.R., Nalls M.A.. et al. (2014) Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet., 10, e1004517.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Palmer N.D., McDonough C.W., Hicks P.J., Roh B.H., Wing M.R., An S.S., Hester J.M., Cooke J.N., Bostrom M.A., Rudock M.E.. et al. (2012) A genome-wide association search for type 2 diabetes genes in African Americans. PLoS One, 7, e29202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saxena R., Elbers Clara C., Guo Y., Peter I., Gaunt Tom R., Mega J.L., Lanktree Matthew B.. et al. (2012) Large-scale gene-centric meta-analysis across 39 studies identifies Type 2 diabetes loci. Am. J. Hum. Genet., 90, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wessel J., Chu A.Y., Willems S.M., Wang S., Yaghootkar H., Brody J.A., Dauriz M., Hivert M.F., Raghavan S., Lipovich L.. et al. (2015) Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat. Commun., 6, 5897.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dooley J., Tian L., Schonefeldt S., Delghingaro-Augusto V., Garcia-Perez J.E., Pasciuto E., Di Marino D., Carr E.J., Oskolkov N., Lyssenko V.. et al. (2016) Genetic predisposition for beta cell fragility underlies type 1 and type 2 diabetes. Nat. Genet., 48, 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Adeyemo A.A., Sun M., Chen J., Wheeler E., Mahajan A., Pirie F., Doumatey A., Sandhu M., Morris A., Motala A.. et al. (2017) A Meta-Analysis of Genome-Wide Association Studies for Type 2 Diabetes in Africans [abstract]. Diabetes., 66 (suppl 1), A47. [Google Scholar]
- 66. Liu C.-T., Raghavan S., Maruthur N., Kabagambe Edmond K., Hong J., Ng Maggie C., Hivert M.-F., Lu Y.. et al. (2016) Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am. J. Hum. Genet., 99, 56–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen G., Bentley A., Adeyemo A., Shriner D., Zhou J., Doumatey A., Huang H., Ramos E., Erdos M., Gerry N.. et al. (2012) Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans. Hum. Mol. Genet., 21, 4530–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carty C.L., Bhattacharjee S., Haessler J., Cheng I., Hindorff L.A., Aroda V., Carlson C.S., Hsu C.-N., Wilkens L., Liu S.. et al. (2014) Comparative analysis of metabolic syndrome components in over 15,000 African Americans identifies pleiotropic variants: results from the PAGE study. Circul. Cardiovasc. Genet., 7, 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Adeboye B., Bermano G., Rolland C. (2012) Obesity and its health impact in Africa: a systematic review. Cardiovasc. J. Afr., 23, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Flegal K.M., Kruszon-Moran D., Carroll M.D., Fryar C.D., Ogden C.L. (2016) Trends in Obesity Among Adults in the United States, 2005 to 2014. JAMA, 315, 2284–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Visscher T.L., Seidell J.C. (2001) The public health impact of obesity. Annu. Rev. Public Health, 22, 355–375. [DOI] [PubMed] [Google Scholar]
- 72. Temelkova-Kurktschiev T., Stefanov T. (2012) Lifestyle and genetics in obesity and type 2 diabetes. Exp. Clin. Endocrinol. Diabetes, 120, 1–6. [DOI] [PubMed] [Google Scholar]
- 73. Chen G., Doumatey A.P., Zhou J., Lei L., Bentley A.R., Tekola-Ayele F., Adebamowo S.N., Baker J.L., Fasanmade O., Okafor G.. et al. (2017) Genome-wide analysis identifies an african-specific variant in SEMA4D associated with body mass index. Obesity (Silver Spring), 25, 794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ng M.C.Y., Graff M., Lu Y., Justice A.E., Mudgal P., Liu C.T., Young K., Yanek L.R., Feitosa M.F., Wojczynski M.K.. et al. (2017) Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African ancestry anthropometry genetics consortium. PLoS Genet., 13, e1006719.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chu A.Y., Deng X., Fisher V.A., Drong A., Zhang Y., Feitosa M.F., Liu C.T., Weeks O., Choh A.C., Duan Q.. et al. (2017) Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Int. J. Obes. (Lond.), 49, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Demerath E.W., Guan W., Grove M.L., Aslibekyan S., Mendelson M., Zhou Y.H., Hedman A.K., Sandling J.K., Li L.A., Irvin M.R.. et al. (2015) Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet., 24, 4464–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fulford A.J., Ong K.K., Elks C.E., Prentice A.M., Hennig B.J. (2015) Progressive influence of body mass index-associated genetic markers in rural Gambians. J. Med. Genet., 52, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gardner K.R., Sapienza C., Fisher J.O. (2015) Genetic and epigenetic associations to obesity-related appetite phenotypes among African-American children. Pediatr. Obes., 10, 476–482. [DOI] [PubMed] [Google Scholar]
- 79. Graff M., North K.E., Richardson A.S., Young K.L., Mazul A.L., Highland H.M., Mohlke K.L., Lange L.A., Lange E.M., Mullan Harris K.. et al. (2017) BMI loci and longitudinal BMI from adolescence to young adulthood in an ethnically diverse cohort. Int. J. Obes. (Lond.), 41, 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Graff M., Richardson A.S., Young K.L., Mazul A.L., Highland H., North K.E., Mohlke K.L., Lange L.A., Lange E.M., Harris K.M.. et al. (2016) The interaction between physical activity and obesity gene variants in association with BMI: Does the obesogenic environment matter?. Health Place, 42, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lange L.A., Graff M., Lange E.M., Young K.L., Richardson A.S., Mohlke K.L., North K.E., Harris K.M., Gordon-Larsen P. (2016) Evidence for Association between SH2B1 Gene Variants and Glycated Hemoglobin in Nondiabetic European American Young Adults: The Add Health Study. Ann. Hum. Genet., 80, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ngwa E.N., Sobngwi E., Atogho-Tiedeu B., Noubiap J.J., Donfack O.S., Guewo-Fokeng M., Mofo E.P., Fosso P.P., Djahmeni E., Djokam-Dadjeu R.. et al. (2015) Association between the rs12255372 variant of the TCF7L2 gene and obesity in a Cameroonian population. BMC Res. Notes, 8, 717.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pillay V., Crowther N.J., Ramsay M., Smith G.D., Norris S.A., Lombard Z. (2015) Exploring genetic markers of adult obesity risk in black adolescent South Africans-the Birth to Twenty Cohort. Nutr. Diabetes, 5, e157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Riestra P., Gebreab S.Y., Xu R., Khan R.J., Bidulescu A., Correa A., Tekola-Ayele F., Davis S.K. (2015) Gender-specific associations between ADIPOQ gene polymorphisms and adiponectin levels and obesity in the Jackson Heart Study cohort. BMC Med. Genet., 16, 65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sajuthi S.P., Sharma N.K., Chou J.W., Palmer N.D., McWilliams D.R., Beal J., Comeau M.E., Ma L., Calles-Escandon J., Demons J.. et al. (2016) Mapping adipose and muscle tissue expression quantitative trait loci in African Americans to identify genes for type 2 diabetes and obesity. Hum. Genet., 135, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Samaan Z., Lee Y.K., Gerstein H.C., Engert J.C., Bosch J., Mohan V., Diaz R., Yusuf S., Anand S.S., Meyre D. (2015) Obesity genes and risk of major depressive disorder in a multiethnic population: a cross-sectional study. J. Clin. Psychiatry, 76, e1611–e1618. [DOI] [PubMed] [Google Scholar]
- 87. Sung Y.J., Perusse L., Sarzynski M.A., Fornage M., Sidney S., Sternfeld B., Rice T., Terry J.G., Jacobs D.R. Jr., Katzmarzyk P.. et al. (2016) Genome-wide association studies suggest sex-specific loci associated with abdominal and visceral fat. Int. J. Obes. (Lond.) 40, 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yan J., Wang X., Tao H., Yang W., Luo M., Lin F. (2015) Lack of association between leptin G-2548A polymorphisms and obesity risk: evidence based on a meta-analysis. Obes. Res. Clin. Pract., 9, 389–397. [DOI] [PubMed] [Google Scholar]
- 89. Yoneyama S., Yao J., Guo X., Fernandez-Rhodes L., Lim U., Boston J., Buzkova P., Carlson C.S., Cheng I., Cochran B.. et al. (2017) Generalization and fine mapping of European ancestry-based central adiposity variants in African ancestry populations. Int. J. Obes. (Lond.), 41, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Young K.L., Graff M., North K.E., Richardson A.S., Mohlke K.L., Lange L.A., Lange E.M., Harris K.M., Gordon-Larsen P. (2015) Interaction of smoking and obesity susceptibility loci on adolescent BMI: the National Longitudinal Study of Adolescent to Adult Health. BMC Genet., 16, 131.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhu A.Z., Cox L.S., Ahluwalia J.S., Renner C.C., Hatsukami D.K., Benowitz N.L., Tyndale R.F. (2015) Genetic and phenotypic variation in UGT2B17, a testosterone-metabolizing enzyme, is associated with BMI in males. Pharmacogenet. Genomics, 25, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ng M.C., Hester J.M., Wing M.R., Li J., Xu J., Hicks P.J., Roh B.H., Lu L., Divers J., Langefeld C.D.. et al. (2012) Genome-wide association of BMI in African Americans. Obesity (Silver Spring), 20, 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Monda K.L., Chen G.K., Taylor K.C., Palmer C., Edwards T.L., Lange L.A., Ng M.C., Adeyemo A.A., Allison M.A., Bielak L.F.. et al. (2013) A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat. Genet., 45, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Adeyemo A., Chen G., Zhou J., Shriner D., Doumatey A., Huang H., Rotimi C. (2010) FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes, 59, 1549–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yako Y.Y., Echouffo-Tcheugui J.B., Balti E.V., Matsha T.E., Sobngwi E., Erasmus R.T., Kengne A.P. (2015) Genetic association studies of obesity in Africa: a systematic review. Obes. Rev., 16, 259–272. [DOI] [PubMed] [Google Scholar]
- 96. Lombard Z., Crowther N.J., van der Merwe L., Pitamber P., Norris S.A., Ramsay M. (2012) Appetite regulation genes are associated with body mass index in black South African adolescents: a genetic association study. BMJ Open, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Magi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E.. et al. (2015) New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liu C.T., Monda K.L., Taylor K.C., Lange L., Demerath E.W., Palmas W., Wojczynski M.K., Ellis J.C., Vitolins M.Z., Liu S.. et al. (2013) Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet., 9, e1003681.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Argiles J.M., Lopez-Soriano J., Almendro V., Busquets S., Lopez-Soriano F.J. (2005) Cross-talk between skeletal muscle and adipose tissue: a link with obesity? Med. Res. Rev., 25, 49–65. [DOI] [PubMed] [Google Scholar]
- 100. Doumatey A.P., Xu H., Huang H., Trivedi N.S., Lei L., Elkahloun A., Adeyemo A., Rotimi C.N. (2015) Global Gene Expression Profiling in Omental Adipose Tissue of Morbidly Obese Diabetic African Americans. J Endocrinol. Metab., 5, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Xu H. (2013) Obesity and metabolic inflammation. Drug Discov. Today Dis. Mechan., 10, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Doumatey A.P., Lashley K.S., Huang H., Zhou J., Chen G., Amoah A., Agyenim-Boateng K., Oli J., Fasanmade O., Adebamowo C.A.. et al. (2010) Relationships among obesity, inflammation, and insulin resistance in African Americans and West Africans. Obesity (Silver Spring), 18, 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Meilleur K.G., Doumatey A., Huang H., Charles B., Chen G., Zhou J., Shriner D., Adeyemo A., Rotimi C. (2010) Circulating adiponectin is associated with obesity and serum lipids in West Africans. J. Clin. Endocrinol. Metab., 95, 3517–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Charles B.A., Doumatey A., Huang H., Zhou J., Chen G., Shriner D., Adeyemo A., Rotimi C.N. (2011) The roles of IL-6, IL-10, and IL-1RA in obesity and insulin resistance in African-Americans. J. Clin. Endocrinol. Metab., 96, E2018–E2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Doumatey A.P., Chen G., Tekola Ayele F., Zhou J., Erdos M., Shriner D., Huang H., Adeleye J., Balogun W., Fasanmade O.. et al. (2012) C-reactive protein (CRP) promoter polymorphisms influence circulating CRP levels in a genome-wide association study of African Americans. Hum. Mol. Genet., 21, 3063–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tekola Ayele F., Doumatey A., Huang H., Zhou J., Charles B., Erdos M., Adeleye J., Balogun W., Fasanmade O., Johnson T.. et al. (2012) Genome-wide associated loci influencing interleukin (IL)-10, IL-1Ra, and IL-6 levels in African Americans. Immunogenetics, 64, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schleinitz D. (2015) Genetic determination of serum levels of diabetes-associated adipokines. Rev. Diabet. Stud., 12, 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Doumatey A.P., Bentley A.R., Zhou J., Huang H., Adeyemo A., Rotimi C.N. (2012) Paradoxical hyperadiponectinemia is associated with the metabolically healthy obese (MHO) phenotype in African Americans. J Endocrinol Metab., 2, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Doumatey A.P., Zhou J., Zhou M., Prieto D., Rotimi C.N., Adeyemo A. (2016) Proinflammatory and lipid biomarkers mediate metabolically healthy obesity: A proteomics study. Obesity (Silver Spring), 24, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bentley A.R., Rotimi C.N. (2017) Interethnic differences in serum lipids: implications for cardiometabolic disease risk in African ancestry populations. Global Heart, 12, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Musunuru K., Romaine S.P.R., Lettre G., Wilson J.G., Volcik K.A., Tsai M.Y., Taylor H.A., Schreiner P.J., Rotter J.I., Rich S.S.. et al. (2012) Multi-ethnic analysis of lipid-associated loci: the NHLBI CARe project. PLoS One, 7, e36473.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Elbers C.C., Guo Y., Tragante V., van Iperen E.P.A., Lanktree M.B., Castillo B.A., Chen F., Yanek L.R., Wojczynski M.K., Li Y.R.. et al. (2012) Gene-centric meta-analysis of lipid traits in African, East Asian and Hispanic populations. PLoS One, 7, e50198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lanktree M.B., Elbers C.C., Li Y., Zhang G., Duan Q., Karczewski K.J., Guo Y., Tragante V., North K.E., Cushman M.. et al. (2015) Genetic meta-analysis of 15,901 African Americans identifies variation in EXOC3L1 is associated with HDL concentration. J. Lipid Res., 56, 1781–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Peloso Gina M., Auer Paul L., Bis Joshua C., Voorman A., Morrison Alanna C., et al. (2014) Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 Whites and Blacks. Am. J. Hum. Genet., 94, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. TG, T., HDL Working Group of the Exome Sequencing Project, N.H., Lung, and Institute, B. (2014) Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med., 371, 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Peloso G.M., Lange L.A., Varga T.V., Nickerson D.A., Smith J.D., Griswold M.E., Musani S., Polfus L.M., Mei H., Gabriel S.. et al. (2016) Association of exome sequences with cardiovascular traits among Blacks in the Jackson Heart Study. Circ. Cardiovasc. Genet., 9, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Adeyemo A., Bentley A.R., Meilleur K.G., Doumatey A.P., Chen G., Zhou J., Shriner D., Huang H., Herbert A., Gerry N.P.. et al. (2012) Transferability and Fine Mapping of genome-wide associated loci for lipids in African Americans. BMC Med. Genet., 13, 88–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dumitrescu L., Carty C.L., Taylor K., Schumacher F.R., Hindorff L.A., Ambite J.L., Anderson G., Best L.G., Brown-Gentry K., Bůžková P.. et al. (2011) Genetic determinants of lipid traits in diverse populations from the population architecture using genomics and epidemiology (PAGE) study. PLoS Genet., 7, e1002138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chang M-h., Ned R.M., Hong Y., Yesupriya A., Yang Q., Liu T., Janssens A.C.J.W., Dowling N.F. (2011) Racial/Ethnic Variation in the Association of Lipid-Related Genetic Variants With Blood Lipids in the US Adult Population. Circ. Cardiovasc. Genet, 4, 523.. [DOI] [PubMed] [Google Scholar]
- 120. Coram M.A., Duan Q., Hoffmann T.J., Thornton T., Knowles J.W., Johnson N.A., Ochs-Balcom H.M., Donlon T.A., Martin L.W., Eaton C.B.. et al. (2013) Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet., 92, 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J.. et al. (2010) Biological, clinical, and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Carlson C.S., Matise T.C., North K.E., Haiman C.A., Fesinmeyer M.D., Buyske S., Schumacher F.R., Peters U., Franceschini N., Ritchie M.D.. et al. (2013) Generalization and dilution of association results from European GWAS in populations of Non-European ancestry: the PAGE study. PLoS Biol., 11, e1001661.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bentley A.R., Chen G., Shriner D., Doumatey A.P., Zhou J., Huang H., Mullikin J.C., Blakesley R.W., Hansen N.F., Bouffard G.G.. et al. (2014) Gene-based sequencing identifies lipid-influencing variants with ethnicity-specific effects in African Americans. PLoS Genet., 10, e1004190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ibe U.K.U. Analysis of mutations causing familial hypercholesterolaemia in black South African patients of different ancestr. S. Afr. Med. J., 107, 145–148. [DOI] [PubMed] [Google Scholar]
- 125. Miller S.A., Hooper A.J., Mantiri G.A., Marais D., Tanyanyiwa D.M., McKnight J., Burnett J.R. (2016) Novel APOB missense variants, A224T and V925L, in a black South African woman with marked hypocholesterolemia. J. Clin. Lipidol., 10, 604–609. [DOI] [PubMed] [Google Scholar]
- 126. Pirim D., Wang X., Niemsiri V., Radwan Z.H., Bunker C.H., Hokanson J.E., Hamman R.F., Barmada M.M., Demirci F.Y., Kamboh M.I. (2016) Resequencing of the CETP gene in American whites and African blacks: association of rare and common variants with HDL-cholesterol levels. Metabolism, 65, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Niemsiri V., Wang X., Pirim D., Radwan Z.H., Bunker C.H., Barmada M.M., Kamboh M.I., Demirci F.Y. (2015) Genetic contribution of SCARB1 variants to lipid traits in African Blacks: a candidate gene association study. BMC Med. Genet., 16, 106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pirim D., Wang X., Radwan Z.H., Niemsiri V., Bunker C.H., Barmada M.M., Kamboh M.I., Demirci F.Y. (2015) Resequencing of LPL in African Blacks and associations with lipoprotein–lipid levels. Eur. J. Hum. Genet., 23, 1244–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Charles B.A., Shriner D., Rotimi C.N. (2014) Accounting for linkage disequilibrium in association analysis of diverse populations. Genet. Epidemiol., 38, 265–273. [DOI] [PubMed] [Google Scholar]
- 130. Charles B.A., Shriner D., Doumatey A., Chen G., Zhou J., Huang H., Herbert A., Gerry N.P., Christman M.F., Adeyemo A.. et al. (2011) A genome-wide association study of serum uric acid in African Americans. BMC Med. Genomics, 4, 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zubair N., Graff M., Luis Ambite J., Bush W.S., Kichaev G., Lu Y., Manichaikul A., Sheu W.H., Absher D., Assimes T.L.. et al. (2016) Fine-mapping of lipid regions in global populations discovers ethnic-specific signals and refines previously identified lipid loci. Hum. Mol. Genet., 25, 5500–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Pritchard J.K., Przeworski M. (2001) Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet., 69, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Choudhury A., Hazelhurst S., Mulder N., Joubert F., Aron S., Gamieldien J., Botha G., Meintjes A., Chimusa E., Jalali M.. et al. (2016), Southern African Human Genome Programme: Deep whole genome sequencing provides insights into the genetic architecture of Southern Africans. 9th Meeting of the African Society of Human Genetics (AfSHG), May 16, 2016. Dakar, Senegal. [Google Scholar]
- 134. Shang X., Xu X. (2017) Update in the genetics of thalassemia: What clinicians need to know. Best Practice & Research Clinical Obstetrics & Gynaecology, 39, 3–15. [DOI] [PubMed] [Google Scholar]
- 135. Ngo Bitoungui V.J., Pule G.D., Hanchard N., Ngogang J., Wonkam A. (2015) Beta-globin gene haplotypes among cameroonians and review of the global distribution: is there a case for a single sickle mutation origin in Africa? OMICS, 19, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lindenau J.D., Wagner S.C., de Castro S.M., Hutz M.H. (2016) The effects of old and recent migration waves in the distribution of HBB*S globin gene haplotypes. Genet. Mol. Biol., 39, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Carson P.E., Flanagan C.L., Ickes C.E., Alving A.S. (1956) Enzymatic deficiency in primaquine-sensitive erythrocytes. Science, 124, 484–485. [DOI] [PubMed] [Google Scholar]
- 138. Cappellini M.D., Fiorelli G. (2008) Glucose-6-phosphate dehydrogenase deficiency. Lancet, 371, 64–74. [DOI] [PubMed] [Google Scholar]
- 139. Shah S.S., Rockett K.A., Jallow M., Sisay-Joof F., Bojang K.A., Pinder M., Jeffreys A., Craik R., Hubbart C., Wellems T.E.. et al. (2016) Heterogeneous alleles comprising G6PD deficiency trait in West Africa exert contrasting effects on two major clinical presentations of severe malaria. Malar. J., 15, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Rać M.E., Safranow K., Poncyljusz W. (2007) Molecular basis of human CD36 gene mutations. Mol. Med., 13, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Huang B.-H., Liao P.-C. (2015) Tracing evolutionary relicts of positive selection on eight malaria-related immune genes in mammals. Innate Immun., 21, 463–476. [DOI] [PubMed] [Google Scholar]
- 142. Reich D., Nalls M.A., Kao W.H.L., Akylbekova E.L., Tandon A., Patterson N., Mullikin J., Hsueh W.-C., Cheng C.-Y., Coresh J.. et al. (2009) Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLOS Genetics, 5, e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hsieh M.M., Tisdale J.F., Rodgers G.P., Young N.S., Trimble E.L., Little R.F. (2010) Neutrophil count in African Americans: lowering the target cutoff to initiate or resume chemotherapy? J. Clin. Oncol., 28, 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Thobakgale C.F., Ndung'u T. (2014) Neutrophil counts in persons of African origin. Curr. Opin. Hematol., 21, 50–57. [DOI] [PubMed] [Google Scholar]
- 145. Triska P., Soares P., Patin E., Fernandes V., Cerny V., Pereira L. (2015) Extensive Admixture and Selective Pressure Across the Sahel Belt. Genome Biol. Evol., 7, 3484–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mmbando B.P., Mgaya J., Cox S.E., Mtatiro S.N., Soka D., Rwezaula S., Meda E., Msaki E., Snow R.W., Jeffries N.. et al. (2015) Negative Epistasis between Sickle and Foetal Haemoglobin Suggests a Reduction in Protection against Malaria. PLoS One, 10, e0125929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shriner D., Kumkhaek C., Doumatey A.P., Chen G., Bentley A.R., Charles B.A., Zhou J., Adeyemo A., Rodgers G.P., Rotimi C.N. (2015) Evolutionary context for the association of gamma-globin, serum uric acid, and hypertension in African Americans. BMC Med. Genet., 16, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Network M.G.E. (2015) A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature, 526, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]