In this study, Lee et al. analyze pre-mRNA splicing patterns, RNA-binding sites, and RNA structures near these binding sites coordinately controlled by two splicing factors: the heterogeneous nuclear ribonucleoprotein hnRNPA1 and the RNA helicase DDX5. RNA-binding sites were identified in the nuclear transcriptome using cell-fractionated eCLIP, and these data sets were integrated with in vivo SHAPE chemical RNA structure probing data, which was used to map binding sites in RNA targets and often flanked regions of higher chemical reactivity, suggesting an organized nature of nuclear pre-mRNPs.
Keywords: pre-mRNA splicing, eCLIP, hnRNPA1, DDX5, RNA structure probing
Abstract
Alternative premessenger RNA (pre-mRNA) splicing is a post-transcriptional mechanism for controlling gene expression. Splicing patterns are determined by both RNA-binding proteins and nuclear pre-mRNA structure. Here, we analyzed pre-mRNA splicing patterns, RNA-binding sites, and RNA structures near these binding sites coordinately controlled by two splicing factors: the heterogeneous nuclear ribonucleoprotein hnRNPA1 and the RNA helicase DDX5. We identified thousands of alternative pre-mRNA splicing events controlled by these factors by RNA sequencing (RNA-seq) following RNAi. Enhanced cross-linking and immunoprecipitation (eCLIP) on nuclear extracts was used to identify protein–RNA-binding sites for both proteins in the nuclear transcriptome. We found a significant overlap between hnRNPA1 and DDX5 splicing targets and that they share many closely linked binding sites as determined by eCLIP analysis. In vivo SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) chemical RNA structure probing data were used to model RNA structures near several exons controlled and bound by both proteins. Both sequence motifs and in vivo UV cross-linking sites for hnRNPA1 and DDX5 were used to map binding sites in their RNA targets, and often these sites flanked regions of higher chemical reactivity, suggesting an organized nature of nuclear pre-mRNPs. This work provides a first glimpse into the possible RNA structures surrounding pre-mRNA splicing factor-binding sites.
Alternative premessenger RNA (pre-mRNA) splicing is an important mechanism that cells use to modulate gene expression and protein isoform diversity at the post-transcriptional level. Many studies have indicated that both RNA-binding proteins (Fu and Ares 2014; Lee and Rio 2015)—such as heterogeneous nuclear ribonucleoproteins (hnRNPs) (Martinez-Contreras et al. 2007; Geuens et al. 2016) and serine- and arginine-rich (SR) proteins (Howard and Sanford 2015; Jeong 2017)—and RNA structure (Warf and Berglund 2010; McManus and Graveley 2011) play roles in determining patterns of intron splicing in specific cases. In humans, >95% of genes are alternatively spliced (Barbosa-Morais et al. 2012; Merkin et al. 2012), and often these alternative pre-mRNA processing events have implications for health and disease, as disease gene mutations that affect the splicing process result in human genetic disorders (Singh and Cooper 2012; Li et al. 2016). It is also known that proteins can act to alter RNA structure by either promoting annealing of complementary RNA strands (Herschlag 1995) or disrupting base-paired regions in long RNAs (Jarmoskaite and Russell 2011; Smola et al. 2016). DEAD/H-box family RNA helicase proteins play important roles in intron removal as part of the spliceosomal machinery (Jarmoskaite and Russell 2011; Will and Luhrmann 2011). Biochemical studies using purified proteins have also confirmed that RNA-binding proteins along with their auxiliary low-complexity or intrinsically disordered regions can function as “RNA chaperones” to promote novel RNA structures and RNA-folding pathways (Herschlag 1995).
The use of high-throughput RNA sequencing (RNA-seq) methods in conjunction with either mutations in splicing factor genes or RNAi/CRISPR interference have allowed the identification of thousands of RNA splicing events that are either directly or indirectly controlled by specific RNA-binding proteins (Fu and Ares 2014; Lee and Rio 2015). UV photochemical RNA–protein cross-linking and immunoprecipitation (CLIP) followed by high-throughput RNA-seq methods—HITS-CLIP (high-throughput sequencing CLIP) (Darnell 2010), PAR-CLIP (photoactivatable ribonucleoside-enhanced CLIP) (Hafner et al. 2010), iCLIP (individual nucleotide-resolution UV CLIP) (Huppertz et al. 2014), eCLIP (enhanced CLIP) (Van Nostrand et al. 2016), and irCLIP (infrared CLIP) (Zarnegar et al. 2016)—have allowed direct mapping of RNA–protein-binding sites in vivo and, in many cases, informed on both motifs (transcripts and transcript regions) that a given RNA-binding protein or protein complex can recognize (Marchese et al. 2016; Wheeler et al. 2018).
The recent development of methods for chemical probing and cross-linking of RNA structures inside living cells has allowed a transcriptome-wide view of both local RNA secondary structure and long-range RNA–RNA interactions (Kwok et al. 2015; Smola et al. 2015, 2016; Flynn et al. 2016; Graveley 2016; Kwok 2016). These methods use either dimethyl sulfate (DMS) (Rouskin et al. 2014) or 2′OH acylation (SHAPE [selective 2′-hydroxyl acylation and profiling experiment]) (Flynn et al. 2016) reagents to probe ssRNA regions mapped by reverse transcriptase stops or mutations at modified sites and 4-hydroxymethyl trioxalen (psoralen) to cross-link RNA–RNA duplexes in vivo followed by digestion with single-strand-specific RNases and library preparation (ligation of interacting RNA [LIGR] followed by high-throughput sequencing [LIGR-seq] [Sharma et al. 2016] and psoralen analysis of RNA interactions and structures [PARIS] [Lu et al. 2016]). These are powerful methods to map protein–RNA interaction sites and RNA secondary structures across the transcriptome.
In order to investigate the relationship between pre-mRNA alternative splicing (AS) patterns, splicing factor-binding sites, and local RNA structures, we integrated data sets that identify splicing pattern alterations, RNA–protein interaction eCLIP data, and RNA structure probing data. We chose to study two RNA-binding proteins known to play roles in pre-mRNA splicing and act as RNA chaperone proteins: human hnRNPA1 and the RNA helicase DDX5. hnRNPA1 is a well-known splicing factor that binds to splicing silencer elements and can antagonize the effect of the SR family of splicing activators (Guil et al. 2003; Expert-Bezancon et al. 2004). hnRNPA1 is one member of the family of hnRNPs that possesses RNA chaperone activity, can promote RNA–RNA annealing, and can enhance hammerhead ribozyme cleavage in vitro (Herschlag et al. 1994; Portman and Dreyfuss 1994). This activity is manifest by the C-terminal RGG-rich intrinsically disordered region or prion-like domain (Harrison and Shorter 2017). In contrast to the RNA–RNA annealing activity of hnRNPA1, DDX5 is an ATP-dependent RNA helicase belonging to the DEAD/H-box helicase family. These proteins typically unwind RNA structures or can translocate along RNA and displace RNA-bound proteins using the energy of ATP hydrolysis (Jarmoskaite and Russell 2011). Of the many possible DEAD/H-box RNA helicase family members, DDX5 has been shown to affect splicing of the tau gene exon 10 (Camats et al. 2008; Kar et al. 2011) and the ras pre-mRNA (Guil et al. 2003). Furthermore, the genome-wide effects of RNA helicases on pre-mRNA splicing decisions have not been investigated. Here, we used RNAi to deplete hnRNPA1 and DDX5 mRNA and protein and performed RNA-seq to assess pre-mRNA splicing pattern changes in human K562 cells using a new software algorithm called the junction usage model (JUM) (Wang and Rio 2017). These results show that thousands of splicing pattern changes are observed upon hnRNPA1 and DDX5 knockdown, with more changes detected in the DDX5 knockdown condition. Furthermore, there is a significant overlap in hundreds of splicing targets for both proteins. We also performed eCLIP RNA-binding immunoprecipitation experiments using nuclear extracts (Van Nostrand et al. 2016) to define in vivo binding sites on target pre-mRNAs and deduced binding motifs for both proteins. hnRNPA1 eCLIP data show motifs similar to previously reported in vitro SELEX sites, and our nuclear eCLIP tags indicate differential binding of hnRNPA1 to distinct transcript regions compared with the whole-cell eCLIP tags identified by ENCODE. Analysis of DDX5 nuclear eCLIP tags led to deduction of a GC-rich binding motif, typically in regions with single-stranded character adjacent to RNA duplexes. Finally, we used an icSHAPE (in vivo click SHAPE) whole-cell chemical RNA probing data set (Spitale et al. 2015) to deduce RNA structures around hnRNPA1 and DDX5 exonic eCLIP tags found on splicing target transcripts. Using the icSHAPE probing data, we can deduce local RNA structures (comparing reactivity profiles of in vivo probed vs. in vitro deproteinized chemically treated RNA samples) on coordinately controlled hnRNAP1/DDX5 target RNAs containing closely clustered eCLIP tags and binding motifs for both proteins. These experiments show that the positions with hnRNPA1 eCLIP tag cross-linking sites and enriched binding motifs often show much less single-stranded chemical reactivity in vivo, indicating that in vivo protein binding blocks chemical probing (i.e., footprints), since hnRNPA1 is known to bind ssRNA. The DDX5 eCLIP tag cross-linking sites or enriched binding motifs typically appear to be near ssRNA or bulged RNA regions but are often located adjacent to putative dsRNA regions, suggesting that the short motifs may function to load the helicase onto the RNA at specific sites. These initial findings indicate that chemical RNA structure probing can be used in combination with protein binding and splicing pattern changes to find unique features and deduce putative RNA structures on transcripts whose pre-mRNA splicing patterns are coordinately controlled by multiple specific RNA-binding proteins.
Results
Analysis of pre-mRNA splicing targets for hnRNPA1
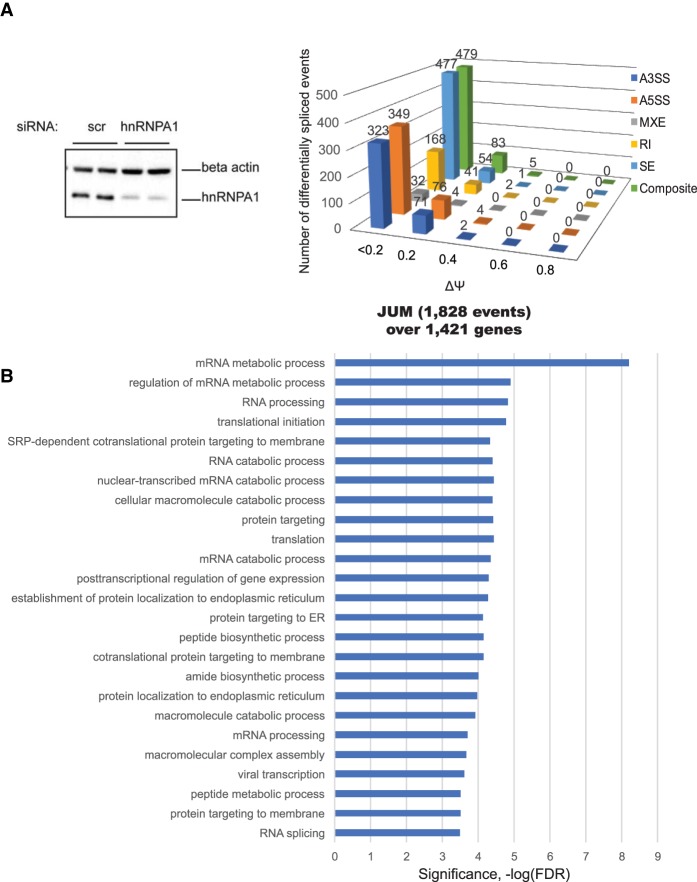
In order to identify pre-mRNA splicing target transcripts controlled by hnRNPA1 in human K562 cells, we compared AS pattern changes in hnRNPA1 knockdown cells with a nonspecific control knockdown cell sample by high-throughput RNA-seq. K562 is a lymphoblast cell line that has been extensively characterized for transcription profiles, genome-wide histone marks, and chromatin accessibility from the ENCODE project. Following RNAi knockdown with an 81% knockdown efficiency (Fig. 1A, left panel), we processed RNA from two biological replicate samples into strand-specific RNA-seq libraries and sequenced using the Illumina platform and acquired ∼40 million Illumina 100-base-pair (bp) paired-end reads per biological replicate.
Figure 1.
Quantitation of differential AS events controlled by hnRNPA1 from RNA-seq using JUM. (A, left panel) siRNA-mediated knockdown of hnRNPA1 at the protein level. K562 cells were transfected with either nonspecific control siRNA oligos (scr si) or hnRNPA1 duplex siRNA oligos. After a second round of siRNA transfection, the cells were harvested for RNA isolation or protein lysates. (Right panel) JUM is a splicing annotation-independent method for determining pre-mRNA splicing patterns from RNA-seq data. Only splice junction-spanning reads were taken into account for quantitation. This resulted in a quantitative comparison of AS events (1828) whose splicing patterns were significantly altered in the hnRNPA1 knockdown samples versus the control (false discovery rate [FDR], P < 0.05), covering 1421 genes. The expression levels of a set of annotated events was quantitated to determine distributions over “percent spliced in” (PSI or Ψ) values. (B) Gene ontology (GO) enrichment analysis of the hnRNPA1 splicing target genes from the JUM splicing analysis against 10,043 expressed genes (reads per kilobase per million mapped reads [RPKM] > 1) in K562 cells. Categories of related genes are listed at the left, and the enrichment significance (−log FDR) is indicated along the X-axis.
Recently, we developed a new splicing analysis software tool called JUM to identify differentially spliced pre-mRNA transcripts from RNA-seq data (Wang et al. 2016; Wang and Rio 2017). The JUM method does not depend on any pre-existing splice junction annotations and thus is able to detect both novel and previously known splicing events specific to the sample. JUM uses RNA-seq reads that map exclusively over splice junctions for AS quantification using a robust statistical algorithm, deduces patterns of splicing, and calculates a percent spliced in (PSI or Ψ) value (Wang et al. 2016; Wang and Rio 2017). We detected 1828 differential AS events over 1421 genes upon hnRNPA1 knockdown using JUM (Fig. 1A, right panel). The detected differential splicing events were deconvoluted into different patterns of splicing with discreet ranges of PSI (or Ψ) values, including not only the conventionally recognized alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), mutually exclusive exon (MXE), intron retention (RI), and skipped exon (SE) patterns but also a more complex “composite” class of splicing pattern. This composite classification is a novel feature of JUM where two or more combinations of the conventional patterns are observed in alternatively spliced transcripts (Fig. 1A, right panel). If a complex combination of AS events exists over a single splicing structure, JUM places the splicing structure into this separate category, allowing accurate quantitation of the standard, less complex patterns of pre-mRNA splicing events mentioned above. Indeed, upon hnRNPA1 knockdown, we detected 479 composite events or complex alternative pre-mRNA splicing events, indicating that a large percentage of differential splicing events consists of a mixture of different types and combinations of conventionally recognized AS patterns (Fig. 1A, right panel).
We also used the MISO (Mixture of Isoforms) software (Katz et al. 2010) to detect differential splicing events upon hnRNPA1 knockdown (Supplemental Fig. S1A). MISO quantitates the expression of alternatively spliced mRNAs from RNA-seq data using a Bayesian model to estimate and measure differential expression and requires the use of a preannotated library of splice events. We found a total of 3111 differential AS changes upon hnRNPA1 knockdown over 1742 genes using MISO (Supplemental Fig. S1A). Of the total 3111 detected differential AS events upon hnRNPA1 knockdown, 1052 had a ΔΨ or ΔPSI value >0.2 or 20%. When we compared the splicing target RNAs from the JUM and MISO analyses, we detected 575 overlapping differential splicing targets of hnRNPA1 (Supplemental Table 1; Supplemental Fig. S1A, right panel). We previously observed partially overlapping subsets of splicing targets comparing JUM with MISO, which may be due to differences in the underlying algorithms (Wang and Rio 2017). We also experimentally validated the ΔΨ estimates calculated by JUM and MISO for several AS events using RT–PCR assays (Supplemental Fig. S3). We selected several random targets from the JUM and MISO analyses from the SE splicing category that had a ΔPSI >0.2 and a few with a ΔPSI <0.2 (e.g., hnRNPD, ALAS2, FN1, and CD97) for validation. For most of the splicing targets tested by RT–PCR (11 out of 13), the differential splicing patterns were detected by both JUM and MISO, and the ΔΨ value calculated experimentally from the RT–PCR assays correlated well with the ΔΨ estimates calculated from the RNA-seq data (Supplemental Fig. S2, primer information listed in Supplemental Table 3). Gene ontology (GO) term analysis of JUM-identified hnRNPA1 splicing targets shows clustering of transcripts functionally enriched in cotranslational protein targeting to the endoplasmic reticulum (ER), RNA processing, and translation (Fig. 1B; Supplemental Table 4).
The RNA helicase DDX5 controls AS of thousands of target pre-mRNAs
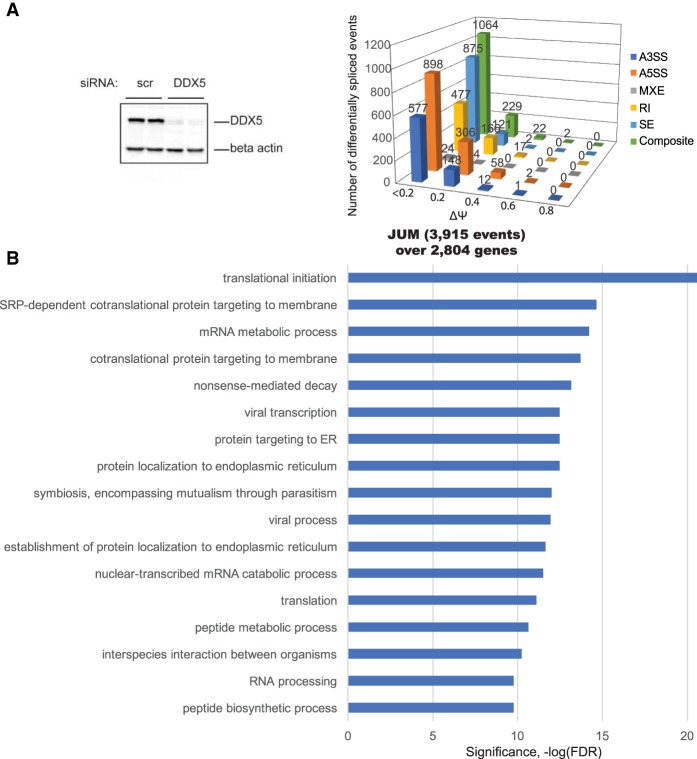
We performed RNAi knockdown of DDX5 in human K562 cells with 93% efficiency (Fig. 2A, left panel) and processed RNA from the cells into strand-specific RNA-seq libraries. We again analyzed the RNA-seq data from the DDX5 knockdown samples using JUM and MISO to detect and quantitate changes in pre-mRNA splicing patterns. This analysis revealed many more splicing changes in DDX5 knockdowns compared with the hnRNPA1 knockdowns. The JUM analysis found 3915 differential splicing events over 2704 genes upon DDX5 knockdown (Fig. 2A, right panel; Supplemental Table 2). MISO analysis uncovered 5294 differential splicing events over 2514 genes; 2415 out of 5294 differential AS events had ΔΨ/ΔPSI >0.2 or 20% (Supplemental Fig. S1B). Again, we identified some splicing targets that were unique for either JUM or MISO in the DDX5 knockdown data sets. GO term analysis of the JUM-identified DDX5 splicing targets is shown in Figure 2B and Supplemental Table 5. Interestingly, the prominent GO terms enriched in the DDX5 target transcripts were similar to those found associated with hnRNPA1; namely, cotranslational protein targeting, RNA processing, nonsense-mediated mRNA decay, and translation.
Figure 2.
The RNA helicase DDX controls alternative pre-mRNA splicing of thousands of target RNAs. (A, left panel) K562 cells were transfected with either nonspecific control siRNA oligos (scr si) or DDX5 duplex siRNA oligos. After a second round of siRNA transfection, the cells were harvested for RNA isolation or protein lysates. Protein lysates were immunoblotted with DDX5 antibody to detect the efficiency of siRNA-mediated knockdown at the protein level. (Right panel) Detection of changes in AS events upon DDX5 knockdown in human K562 cells using JUM. JUM analysis revealed 3915 AS events whose splicing patterns were significantly altered in DDX5 RNAi knockdown samples versus the control scrambled siRNA samples, covering 2804 genes (FDR, P < 0.05). (B) GO enrichment analysis of 2804 DDX5 splicing target genes from JUM splicing analysis. Categories of related genes are listed at the left, and the enrichment significance (−log FDR) is indicated along the X-axis.
Analysis of RNA-binding sites on the nuclear transcriptome for hnRNPA1 and DDX5
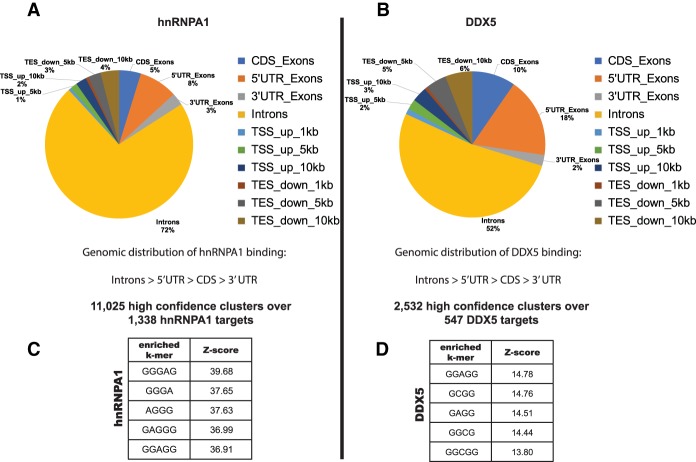
In order to determine the RNA-binding sites for hnRNPA1 and DDX5 on nuclear RNAs and correlate this information with the differentially spliced target RNAs, we performed in vivo eCLIP experiments. eCLIP has optimized many steps in the procedure, resulting in more comprehensive RNA-binding protein target identification (Van Nostrand et al. 2016). We wanted to enrich for nuclear pre-mRNA targets bound by hnRNPA1 and DDX5 and, since both proteins are found in the nucleus and the cytoplasm, prepared nuclear extracts from K562 cells as the starting material for the eCLIP assays. Initial analysis showed that the majority of the nuclear eCLIP tags for both hnRNPA1 and DDX5 mapped to intronic regions in comparison with other genomic regions but also to 5′ untranslated regions (UTRs) (Fig. 3A,B). This is an especially prominent result for the DDX5 nuclear eCLIP tags, since 18% of the uniquely mapping reads localized to 5′ UTRs, and 52% mapped to introns. This result again confirms that we are enriching for nuclear pre-mRNA and is consistent with the roles of both hnRNPA1 (Jean-Philippe et al. 2013; Lemieux et al. 2015) and DDX5 (Fuller-Pace and Ali 2008; Fuller-Pace 2013) in the nucleus.
Figure 3.
Nuclear eCLIP-seq (eCLIP combined with high-throughput sequencing) for hnRNPA1 and DDX5 reveals thousands of protein-binding sites on the human transcriptome. (A,B) Pie charts showing the differential distribution of hnRNPA1- and DDX5-binding sites within different categories of genomic regions. hnRNPA1 nuclear eCLIP generated 11,025 high-confidence clusters (FDR, P < 0.05) over 1338 targets. DDX5 nuclear eCLIP generated 2532 high-confidence clusters (FDR, P < 0.05) over 547 targets. k-mer enrichment motif analysis of the eCLIP clusters is shown for hnRNPA1 (C) and DDX5 (D). Motifs were generated by extracting k-mers from reads after comparing with control data sets with reads randomly distributed over the same genomic features.
The hnRNPA1 eCLIP data analysis using the pyCRAC package (Webb et al. 2014) resulted in 11,025 nuclear eCLIP clusters over 1338 hnRNPA1 target pre-mRNAs. Motif analysis indicated that hnRNPA1 nuclear eCLIP tags showed enrichment for the consensus high-affinity hnRNPA1-binding site UAGGGA/U (Fig. 3C), similar to the motif identified previously by in vitro SELEX experiments (Burd and Dreyfuss 1994) and also determined by iCLIP-seq (iCLIP combined with high-throughput sequencing) (Bruun et al. 2016), confirming that in vivo binding of hnRNPA1 mirrors the binding preferences observed with the purified protein. DDX5 was shown to bind to the tau pre-mRNA in the stem–loop region downstream from exon 10 (Kar et al. 2011), and, unlike hnRNPA1, there is no known binding motif for DDX5. False discovery rate (FDR) filtering (P < 0.05) resulted in 2532 DDX5 eCLIP clusters over 547 target pre-mRNAs. When we performed k-mer motif analysis to extract enriched motifs from the DDX5 eCLIP data sets, compared with randomly distributed control data sets over the same genomic features, a short GC-rich motif was found to be enriched in the DDX5-bound peaks (Fig. 3D). It is possible that GC-rich regions are present near a single-stranded region adjacent to an RNA duplex that DDX5 might be binding.
Correlation of hnRNPA1/DDX5 binding to the differentially spliced target RNAs
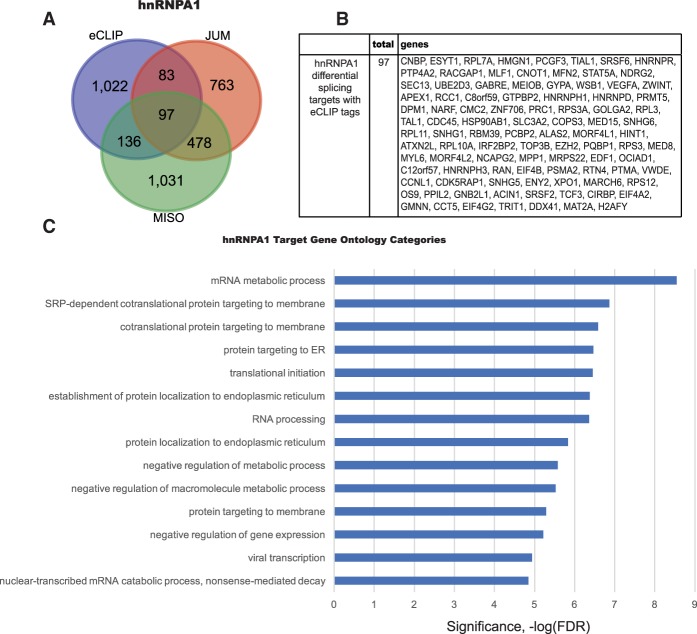
We compared hnRNPA1 nuclear eCLIP targets with the differentially spliced hnRNPA1 targets identified by the JUM and MISO RNA-seq analyses. We identified 316 differentially spliced target RNAs with nuclear hnRNPA1 eCLIP tags (Fig. 4A). Among these targets, 97 targets were shown to be differentially spliced in both the JUM and MISO RNA-seq data. These genes are listed in Figure 4B. The results indicate that only a subset of pre-mRNAs with hnRNPA1 nuclear eCLIP tags is differentially spliced (∼24%). This seems consistent with the knowledge that hnRNPA1 has other cellular functions besides its role in pre-mRNA splicing, such as in nuclear RNA export, telomere biogenesis, and microRNA processing (Guil and Caceres 2007; Chaudhury et al. 2010). However, it is also known that the RNA–protein UV cross-linking efficiency is low, and so perhaps only a subset of the hnRNPA1 protein was covalently attached to RNA and retrieved after immunoprecipitation. It is also possible that some pre-mRNA splicing changes detected upon hnRNPA1 knockdown were not a direct consequence of hnRNPA1 binding. We also performed GO term analysis on these 1338 hnRNPA1 target RNAs with eCLIP tags, and they are involved in regulation of gene expression, SRP-dependent cotranslational protein targeting to membrane, protein localization to the ER, translational initiation, and RNA processing (Fig. 4C), very similar to the GO term analysis of differentially spliced genes identified upon hnRNPA1 knockdown.
Figure 4.
hnRNPA1 differentially spliced transcripts with nuclear eCLIP peaks in human K562 cells. (A) Venn diagram showing the overlap between the differential splicing target RNAs and nuclear hnRNPA1-binding targets in K562 cells. (B) A list of common differential splicing targets from JUM and MISO analysis that contain nuclear hnRNPA1 eCLIP targets. (C) GO enrichment of hnRNPA1 targets containing nuclear eCLIP tags revealed that these target RNAs are involved in processes such as protein localization to the ER, the mRNA metabolic process, translational initiation, and nonsense-mediated decay.
We also performed a comparison of DDX5 nuclear eCLIP tag-containing transcripts with the differentially spliced target RNAs from the JUM and MISO RNA-seq analysis upon DDX5 RNAi knockdown. This analysis identified 172 differentially spliced target RNAs with nuclear DDX5 eCLIP tags, and 58 genes out of 174 were determined to be differentially spliced target RNAs controlled by DDX5 from the JUM and MISO analyses (Supplemental Fig. S3A,B). GO term analysis of the 547 DDX5 splicing targets with eCLIP tags revealed that these DDX5-bound targets are involved in the regulation of gene expression, RNA metabolism, protein localization to the ER, and transcription (Supplemental Fig. S3C).
Coordinated regulation of many splicing targets by hnRNPA1 and DDX5
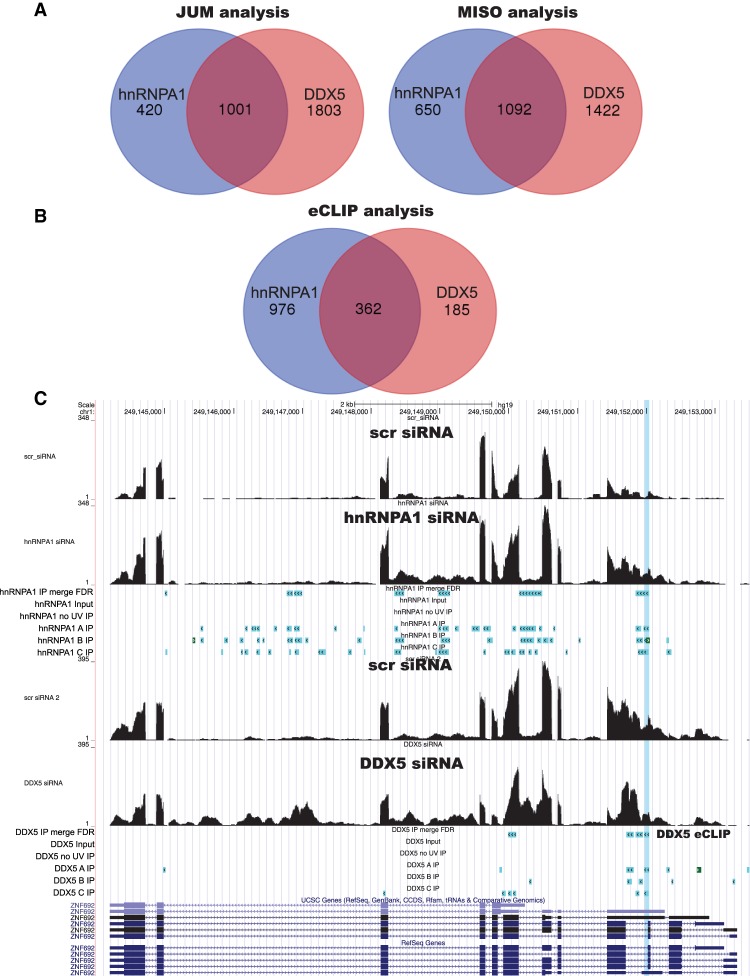
We noticed that the DDX5-binding sites determined by eCLIP analysis sometimes overlapped with hnRNPA1-binding sites. When we examined whether there is overlap between the splicing targets of hnRNPA1 and DDX5, we also found that there was a significant overlap between the splicing targets of these two RNA chaperones (Fig. 5A). Over ∼66% of hnRNPA1 and ∼40% DDX5 splicing targets are shared between hnRNPA1 and DDX5, as detected by both JUM and MISO RNA-seq analysis. When we compared the binding targets of hnRNPA1 and DDX5 as determined by eCLIP, we found that the majority of DDX5-bound RNAs is also bound by hnRNPA1 (Fig. 5B). These data suggest that there is coordinated regulation of pre-mRNA splicing between hnRNPA1 and DDX5. DDX5 was shown to play a role in the pre-mRNA splicing of the proto-oncogene c-H-Ras (HRAS), and hnRNPA1 negatively regulated the HRAS upstream intron splicing in vitro (Guil et al. 2003). Although we found DDX5-binding sites on HRAS by eCLIP, only one of the hnRNPA1 eCLIP replicates was found on HRAS RNA, and corresponding eCLIP tags did not pass the FDR testing (FDR, P < 0.05). Although hnRNPA1 eCLIP reads on the HRAS pre-mRNA did not pass our FDR testing, we found other common targets of hnRNPA1 and DDX5 binding, with a total of 1078 binding sites (Supplemental Table 6) over 362 target RNAs (FDR P < 0.05). We examined whether there was a positive or negative effect of hnRNPA1 or DDX5 on these splicing targets and determined that both hnRNPA1 and DDX5 can act as a splicing activator or repressor on these pre-mRNAs. Supplemental Table 6 lists the genomic coordinates for the shared binding sites of hnRNPA1 and DDX5.
Figure 5.
A large number of hnRNPA1 and DDX5 splicing targets are coregulated. (A) JUM and MISO analysis independently identified significant overlap between the splicing targets of hnRNPA1 and DDX5. Approximately 66% of hnRNPA1 and 40% DDX5 splicing targets are shared between hnRNPA1 and DDX5. (B) eCLIP analysis confirms that the binding targets are shared between hnRNPA1 and DDX5. (C) Genome browser shot of the ZNF692 gene region with RNA-seq data upon hnRNPA1 knockdown (top) and DDX5 knockdown (bottom), with corresponding nuclear CLIP peak cluster regions shown below. The shared hnRNPA1/DDX5-binding site is highlighted on the genome browser.
Use of in vivo RNA structure chemical probing (icSHAPE) data to model RNA structures around protein-binding sites for hRNPA1 and DDX5
RNA chaperone proteins function to assist correct RNA folding and prevent RNA from misfolding. These RNA–protein interactions can also promote or disrupt RNA–RNA base-pairing interactions to alter RNA structure. Both hnRNPA1 (Herschlag et al. 1994; Portman and Dreyfuss 1994) and the RNA helicase DDX5 can act as RNA chaperones. To examine how hnRNPA1 and DDX5 binding to the transcriptome relates to local RNA secondary structures, we used differential transcriptome-wide icSHAPE chemical RNA reactivities (Flynn et al. 2016) to generate potential RNA structures around the nuclear eCLIP exonic tag-binding sites for both proteins on several of the more abundant transcripts from our identified target genes. We used publicly available whole-cell HEK293 icSHAPE data from a previous study (Lu et al. 2016). The icSHAPE data were generated from whole-cell RNA, and so the read density for this data set is higher on exons found in more abundant transcripts. This fact limited the number and location of protein-binding sites that we could examine where the read density was high enough to generate RNA structures. Therefore, we examined several cases where eCLIP tags for hnRNPA1 and DDX5 were found on exons from abundant transcripts close to the observed AS events in order to deduce possible local RNA structures around the protein-binding sites on the splicing target RNAs. A control data set was also generated using deproteinized and refolded RNA treated with NAI-N3 in vitro (“in vitro control”) (Lu et al. 2016). In both cases, purified RNA was then reverse-transcribed where the SHAPE-modified adduct on the 2′-hydroxyl group reactive in the ssRNA regions stops reverse transcriptase, resulting in truncated cDNA products. The in vivo/in vitro icSHAPE reactivity profiles can then be incorporated into RNA structure prediction programs to generate potential RNA secondary structures using single-stranded versus double-stranded region constraints (Aviran and Pachter 2014; Matthews 2014).
We anticipated that RNA sites bound by hnRNPA1 and DDX5 in vivo might have a negative VTD (“vivo–vitro difference,” which compares the reactivity profile across the region at each nucleotide) value because the protein-bound nucleotides would be undermodified by the icSHAPE reagent in vivo compared with the in vitro probed RNA samples. Indeed, in some cases, the positions of both the hnRNPA1- and DDX5-binding motifs determined by eCLIP tag k-mer enrichment analysis are located near the negative VTD regions of the plots (Supplemental Figs. S4, S6, S9C, S10C), delineating sites of RNA–protein interaction. However, there are also situations where the eCLIP tag cross-link sites are located in positive VTD regions, as if those sites are more accessible to the icSHAPE reagent due to protein binding. The in vivo and in vitro RNA secondary structures were generated for the transcripts containing the selected nuclear eCLIP peaks using the RNA structure program (Matthews 2014) and incorporating the icSHAPE constraints. The resulting structures were visualized using the VARNA software (Darty et al. 2009).
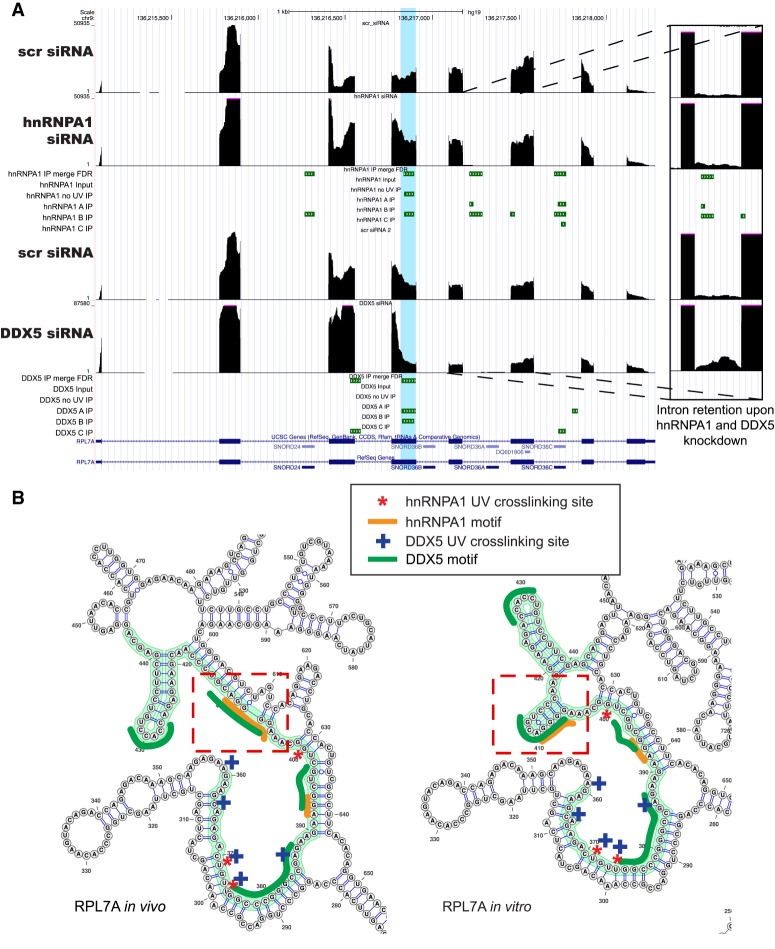
The pre-mRNA encoding ribosomal protein L7A (RPL7A), a component of the 60S subunit, is differentially spliced upon knockdown of either DDX5 or hnRNPA1. Both hnRNPA1 and DDX5 eCLIP tags are found on the transcript and share a binding site (hg19 chromosome 9: 136,215,834–13,621,896+). We also compared the in vivo versus in vitro icSHAPE reactivities over the eCLIP cluster regions and found several negative VTD regions, indicating possible in vivo protein-binding sites due to reduced SHAPE reactivity in vivo (Supplemental Fig. S4) for both hnRNPA1 and DDX5. The red dashed lines on the VTD profile in Supplemental Figure S4 indicate the UV cross-linking sites of the two proteins. This VTD profile difference was reflected in the RNA secondary structures derived from the RNA structure program with icSHAPE constraints (Fig. 6B, cf. dashed rectangle regions). We observed that one of the predicted step–loop regions was absent from the in vivo sample compared with the in vitro probed sample (Fig. 6B) proximal to an hnRNPA1 UV cross-linking site. We detected the hnRNPA1-enriched binding motif AGGGA, resembling a portion of the hnRNPA1 in vitro SELEX site, near the nuclear hnRNPA1 eCLIP clusters. We also found the GC-rich motifs for DDX5 from the eCLIP data in either the putative duplex region of the RNA near the adjacent stem–loop region or, in some cases, the predicted single-stranded region of the RNA. This observation may indicate that DDX5 binding occurs at single–double-strand junctions, consistent with these being sites of RNA helicase loading.
Figure 6.
Alternative pre-mRNA splicing of RPL7A mRNA is regulated by hnRNPA1 and DDX5. (A) Genome browser shot of the RPL7A gene region with RNA-seq data upon hnRNPA1 knockdown (top) and DDX5 knockdown (bottom), with corresponding nuclear CLIP peak cluster regions shown below. (B) icSHAPE-constrained in vivo and in vitro secondary structures for the RPL7A RNA. The nuclear eCLIP cluster/peak region is highlighted in green, the hnRNPA1 cross-link sites are marked with red asterisks, and the DDX5 cross-link sites are marked with blue plus signs. The dotted red rectangles indicate regions of the RNA with secondary structural changes between the in vivo and in vitro icSHAPE constraints. The enriched hnRNPA1 motifs are indicated by orange lines, and the enriched GC-rich motifs for DDX5 is indicated by dark-green lines.
RPS12 is a component of the 40S ribosomal subunit and was determined to be differentially spliced upon knockdown of hnRNPA1 and DDX5. hnRNPA1 and DDX5 share one protein-binding region (hg19 chromosome 6: 133,135,889–133,135,984+) on RPS12 RNA. We generated RNA secondary structures (Supplemental Fig. S5) as described above and examined the VTD profile (Supplemental Fig. S6) around the exonic binding sites for hnRNPA1 and DDX5. We detected a negative VTD region near the hnRNPA1 and DDX5 UV cross-linking sites (Supplemental Fig. S6). A GC-rich region was found within the bulged region of loop 2 in the in vivo structure, which was largely base-paired in the in vitro structure. We also observed the disappearance of loop 1 in the in vivo structure compared with the in vitro structure prediction based on SHAPE reactivities, possibly indicating hnRNPA1 and/or DDX5 binding to this region in vivo.
Prothymosin α (PTMA) is a nuclear protein associated with cell proliferation. The PTMA pre-mRNA was determined to be differentially spliced by both hnRNPA1 and DDX5. Although both hnRNPA1 and DDX5 eCLIP clusters are found on the PTMA transcripts, they bind to separate regions. We visualized the exonic region of PTMA transcript bound by hnRNPA1 and found multiple putative hnRNPA1-binding motifs within this region, with UV cross-linking sites detected throughout the negative VTD region (Supplemental Fig. S8). Unlike what was observed with predicted RNA secondary structures derived previously that encompass both hnRNPA1- and DDX5-binding sites, we observed more RNA secondary structure around this region in both the in vivo and the in vitro structures, with numerous hnRNPA1-binding motifs and cross-linking sites (Supplemental Fig. S7). Interestingly, there seemed to be two regions that actually became more open and chemically reactive in the in vivo structure (positive VTD) (indicated by red rectangles in Supplemental Fig. S7), which have numerous hnRNPA1 cross-links in these two regions (Supplemental Fig. S7).
Heat-shock protein 90α family class B member 1 (HSP90AB1) encodes the cytosolic 90-kDa heat-shock protein. This molecular chaperone protein promotes folding of proteins involved in cell cycle control and signal transduction. Splicing of the HSP90AB1 pre-mRNA is regulated by both hnRNPA1 and DDX5. We mapped five shared binding sites for hnRNPA1 and DDX5 on the HSP90AB1 transcript, with two shared binding sites directly adjacent to each other (hg19 chromosome 6: 44,215,108–44,215, 204+, chromosome 6: 44,215,474–44,215,520+, chromosome 6: 2,218,055–44,218,114+, chromosome 6: 44,218, 134–44,218,208+, and chromosome 6: 44,219,874–44, 219,938+). We visualized two different exonic regions encompassing three of the shared binding sites.
We found DDX5 UV cross-linking sites right near the negative VTD region (Supplemental Fig. S9C), followed by a putative hnRNPA1-binding site (AGGAGGGU). This negative VTD region is visualized as the loop 1 region on the RNA secondary structure (Supplemental Fig. S9B), where DDX5 was bound to a single–double-stranded junction. There is a nearby GC-rich DDX5 motif (Supplemental Fig. S9B, indicated in green) and hnRNPA1-binding motif (Supplemental Fig. S9B, indicated in orange) as well as UV cross-linking sites for both proteins (Supplemental Fig. S9B).
We also visualized a region on the HSP90AB1 RNA that carried shared binding sites for hnRNPA1 and DDX5 (hg19 chromosome 6: 2,218,055–44,218,114+ and chromosome 6: 44,218,134–44,218,208+). The beginning of the visualized exon was found to be enriched for hnRNPA1 and DDX5 UV cross-linking sites (Supplemental Fig. S10C) near the stem–loop region, which also contained single-stranded bulged regions. This portion of the RNA is near a negative VTD region, but we did not find much change in this region when we compared the in vivo and in vitro deduced RNA structures (Supplemental Fig. S10B), possibly due to reduced SHAPE reactivities in this region. More differences were observed between the in vivo versus in vitro RNA secondary structures encompassing mostly DDX5 eCLIP cross-linking sites, including the shared hnRNPA1- and DDX5-binding site (hg19 chromosome 6: 44,218,134–44,218,208+) (Supplemental Fig. S10B, bottom, red dotted rectangles). Taken together, these structures are one of the first examples of how transcriptome-wide chemical RNA probing data can be used in conjunction with RNA splicing pattern and protein-binding data to examine putative local RNA structures near splicing factor-binding sites.
Discussion
The nuclear transcriptome in eukaryotic cells undergoes extensive pre-mRNA processing reactions prior to the export of functional mRNA. Intron removal by spliceosomal pre-mRNA splicing is a critical step in the post-transcriptional regulation of gene expression that can greatly expand transcriptome and proteomic diversity (Lee and Rio 2015). Nearly all human gene transcripts are alternatively spliced (Barbosa-Morais et al. 2012; Merkin et al. 2012), and pre-mRNA splicing regulation involves both cis and trans-acting components, composed of short RNA regulatory sequence elements embedded in the pre-mRNA that are bound by nuclear RNA-binding proteins (Fu and Ares 2014). Positive or negative control by splicing factors involves RNA–protein interactions with splicing enhancers or silencer elements, but also RNA structure and RNA–RNA base-pairing interactions and the effects of chromatin modifications can play roles in specifying splicing patterns (Lee and Rio 2015). Here, we focused on two known splicing factors that can affect RNA structure—hnRNPA1 and DDX5—to begin to examine how in vivo chemical RNA structure probing data can be used to deduce local RNA secondary structures near splicing factor-binding sites in target transcripts for these proteins.
Using RNAi and RNA-seq, we determined that splicing of thousands of transcripts is controlled by hnRNPA1 in human K562 cells. A prior study used microarrays and RNA-seq to examine targets for several hnRNPs, including hnRNPA1 (Huelga et al. 2012). hnRNPA1 is an abundant hnRNP that has been well characterized as a splicing factor and often functions as a splicing repressor binding to splicing silencer elements (Han et al. 2010; Jean-Philippe et al. 2013). While the DEAD/H-box protein DDX5 had been implicated in AS of several specific pre-mRNAs, no comprehensive global examination of DDX5 splicing targets has been reported. Recently, DDX5 was found as a component of a large neuronal RNA–protein complex containing the RbFox protein called LASR that controls neuronal pre-mRNA splicing (Damianov et al. 2016). Here, we identified thousands of hnRNPA1 and DDX5 differential splicing targets (Figs. 1, 2; Supplemental Fig. S1). Our analysis revealed that hnRNPA1 can act as both a splicing activator and a splicing repressor, as has been shown for other hnRNPs (Huelga et al. 2012; Rossbach et al. 2014). Surprisingly, our analysis of DDX5 resulted in the identification of thousands of differentially spliced target RNAs upon RNAi knockdown. It is possible that the knockdown of DDX5, an RNA helicase, could disrupt the distribution of splicing factors on some transcripts through RNP remodeling and thus indirectly affect pre-mRNA splicing patterns. However, we found that a large fraction of hnRNPA1 and DDX5 splicing target pre-mRNAs overlaps and that both proteins can bind to numerous target-binding partners, as determined by eCLIP (Fig. 5), and share >1000 binding sites (Supplemental Table 6). Coordinate control has been observed before in Drosophila with hnRNP and SR proteins (Blanchette et al. 2005) and with human hnRNPs (Huelga et al. 2012). However, the case that we outline here involves splicing control by two distinct classes of proteins: a classical hnRNP known to act as a splicing repressor at splicing silencers (Caputi et al. 1999; Jean-Philippe et al. 2013) and that is able to alter RNA structure (Herschlag et al. 1994; Portman and Dreyfuss 1994) and a DEAD/H-box ATP-dependent RNA helicase/translocase (Fuller-Pace 2013).
We were also interested in determining where hnRNPA1 and DDX5 were bound on the nuclear transcriptome. Because hnRNPA1 and DDX5 are located in both the nucleus and cytoplasm, we performed UV cross-linking and immunoprecipitation experiments using nuclear extracts from UV-treated cells to avoid detection of cytoplasmic RNA-binding sites for these proteins. Using a modified version of iCLIP called eCLIP (Van Nostrand et al. 2016), we identified nuclear hnRNPA1- and DDX5-binding sites in K562 cells and compared these binding events with the differential splicing targets. We identified 1338 nuclear hnRNPA1-binding target RNAs, and these eCLIP targets contained a match to the consensus high-affinity in vitro SELEX-binding site, (U/A)GGG(A/U) (Fig. 3). We also identified 547 nuclear DDX5-binding target RNAs and found that these nuclear DDX5 targets were enriched in a short GC-rich motif. This result was surprising, since we expected DDX5, an RNA helicase, to bind RNA nonspecifically. Interestingly, a recent study on the role of a paralogous RNA helicase, DDX17 (Rm62 in Drosophila), showed by iCLIP that DDX17 recognized a CU/CA-rich motif, suggesting distinct RNA sequence preferences for this family of RNA helicases (Moy et al. 2014). DDX17 functions in viral immune responses in the cytoplasm and in microRNA biogenesis in the nucleus. It should be also noted that only a subset of hnRNPA1 and DDX5 nuclear eCLIP-bound RNAs was found to be differentially spliced by our RNA-seq data analysis. This result may not be surprising, since both hnRNPA1 and DDX5 perform other cellular functions in the nucleus, such as transcriptional regulation, nuclear RNA export, microRNA processing, etc.
Here, we attempted to integrate in vivo chemical RNA probing data with protein-binding sites and changes in pre-mRNA splicing patterns. icSHAPE data (Flynn et al. 2016; Lu et al. 2016) were used to deduce RNA structures that are positioned on splicing target RNAs near hnRNPA1 and DDX5 nuclear binding sites determined from eCLIP RNA–protein interaction data. The differences between in vivo and in vitro icSHAPE chemical RNA reactivity data can be used to infer sites of RNA–protein interactions (Spitale et al. 2015; Flynn et al. 2016) or changes in RNA structure, including those involved with pre-mRNA splicing. Analysis of the VTD profiles (vivo–vitro icSHAPE reactivities) showed that hnRNPA1 eCLIP peaks were often found near the negative VTD profile region (less chemically reactive compared with the in vitro probed samples), while hnRNPA1 UV cross-linked sites resided in the peak region on the VTD profile (Supplemental Figs. S4, S6, S8, S9C, S10C). Interestingly, DDX5 nuclear eCLIP peaks were found in both the positive and the negative VTD profile regions, near stem–loop regions (Supplemental Figs. S4, S6, S9C, S10C). We suspect that DDX5 will bind to both ssRNA regions and adjacent duplex RNA. We also found GC-rich motifs near the nuclear DDX5 eCLIP peaks. These results demonstrate how comparing in vivo versus in vitro icSHAPE data and using these constraints can allow deduction of putative RNA structures near the protein-binding sites and, in conjunction with eCLIP-binding data, allowed us to visualize how these sites can be accessed by splicing factors to control alternative pre-mRNA splicing. Our findings also provide a complementary approach to using chemical structure probing of pools of putative splicing regulatory sequences in vitro (Taliaferro et al. 2016).
Interestingly, our results are consistent with a broad transcriptome-wide study of three RNA-binding proteins showing that CLIP tag sites not bound in vivo tend to have RNA secondary structures associated with them but that bone fide splicing regulatory elements have less structure (Taliaferro et al. 2016). These observations are consistent with our findings that protein-binding sites for both hnRNPA1 and DDX5 on splicing target transcripts occur in less structured regions yet appear to have complex structures adjacent to the sites of RNA–protein interaction. This idea is also related to findings made about noncoding RNAs, where the RNA structure provides a scaffold for protein binding (Zappulla and Cech 2006). Thus, pre-mRNA may also contain both open less structured regions to allow binding of splicing activators or repressor proteins to enhancer or silencer elements (Lee and Rio 2015) and more complex, folded RNA structures nearby.
While the primary RNA sequence elements in pre-mRNAs that regulate AS are being identified and studied, the contribution of secondary RNA structures to AS decisions is less well understood. In summary, we used a combination of RNA-seq and in vivo RNA-binding specificities to identify how the RNA chaperones hnRNPA1 and DDX5 can coregulate thousands of splicing targets. An unexpected result was the finding that these two RNA-binding proteins share a large portion of splicing targets in common and also share more than ∼1000 protein-binding regions in vivo. We also demonstrated how in vivo eCLIP binding data can be used in conjunction with icSHAPE data to deduce putative RNA structures near the protein-binding sites and suggest how these sites can be accessed by splicing factors to control alternative pre-mRNA splicing. These data suggest that pre-mRNAs may also contain both open less structured regions to allow binding of splicing activator or repressor proteins to enhancer or silencer elements (Lee and Rio 2015), perhaps aided by RNA helicases such as DDX5, and more complex folded RNA structures nearby. Thus, local RNA structure may play a critical role in AS control by either exposing or masking protein-binding sites in cells (Taliaferro et al. 2016).
Materials and methods
Cells
Human K562 cells (American Type Culture Collection) were maintained at 37°C in RPMI medium supplemented with 15% FBS, 1 mM sodium pyruvate (Gibco, Life Technologies), and 1× nonessential amino acids (Gibco, Life Technologies).
RNAi for hnRNPA1 and DDX5 in K562 cells
siRNA duplexes (Ambion, Life Technologies) were generated against the following target sequences or were commercially purchased: hnRNPA1 siRNA 1 (CAGCTGAGGAAGCTCTTCA; sequence provided by Dr. James Manley), hnRNPA1 siRNA 2 (Ambion, Life Technologies, s6710), DDX5 siRNA 1 (Ambion, Life Technologies, s4009), and DDX5 siRNA 2 (Ambion, Life Technologies, s4008). Nonspecific control siRNA duplex 1 [scrambled siRNA (scr si)] was purchased from Life Technologies (4390843). For all knockdown experiments, combinations of hnRNPA1 siRNA 1 and 2 (hnRNPA1 combo siRNA) or DDX5 siRNA 1 and 2 (DDX5 combo siRNA) were used.
For electroporation-based transfection, 4.5 × 105 K562 cells were used per reaction in a six-well plate. The cells were washed twice with 1× PBS. For each electroporation reaction, 100 μL of Nucleofector V-Kit and 10 μL of 50 µM hnRNPA1 combo siRNA, DDX5 combo siRNA, or scr si were prepared. The cell pellets were resuspended with the siRNA duplex suspension, and then cells/siRNA duplex oligo suspensions were transferred into cuvettes and electroporated using Nucleofector program (T-016). Immediately after electroporation, 400 µL of the pre-equilibrated culture medium was added to the cuvette and transferred to a six-well plate. Twenty-four hours after transfection, the medium was changed with fresh medium. Samples were harvested for protein lysates for immunoblotting or for RNA isolation 72 h after transfection.
RNA-seq library preparation and sequencing
After RNA isolation using RNeasy minikit (Qiagen, 74104) followed by 30 min of DNase treatment (Ambion, AM2238) at 37°C, poly(A)+ RNA transcript was isolated [NEBNext poly(A) mRNA magnetic isolation module; New England Biolabs, E7490] from 1 µg of total RNA for RNA library preparation and sequencing using NEBNext Ultra directional RNA library preparation kit for Illumina (New England Biolabs, E7420S) according to the manufacturer's instruction. The samples were sequenced on an Illumina HiSeq 2500 with 100-bp paired-end reads at the Vincent J. Coates Genomics Sequencing Laboratory at the University of California at Berkeley.
Analysis of pre-mRNA splicing patterns using JUM and MISO
For JUM analysis, RNA-seq reads were mapped to hg38 using STAR with the two-pass mode (Dobin et al. 2013) for junction discovery. Only unique mapped reads were considered in the downstream JUM analysis. Only splice junctions that received more than five reads in each of the replicates of the RNAi and the control knockdown samples were considered as valid junctions for downstream analysis.
JUM exclusively used RNA-seq reads that were mapped to splice junctions for AS analysis. JUM defined the basic AS quantification unit as “AS structures,” which describe any set of splice junctions that share the same 5′ or 3′ splice sites, with each splice junction in an AS structure defined as a “sub-AS junction.” JUM then calculated the “usage” of each sub-AS junction in every AS structure, defined as the relative level of the sub-AS junction compared with all of the sub-AS junctions within the same AS structure. JUM then profiled for any AS structures whose usage of sub-AS junctions significantly changed between conditions. Finally, JUM assembled all profiled AS structures into AS events that fell into the conventionally recognized categories of AS patterns as well as the “composite” category, based on the unique topological features of the splice graphs that represent each AS pattern, respectively (Wang and Rio 2017). A P-value of 0.05 was used as the statistical cutoff for differentially spliced AS events.
For MISO analysis (Katz et al. 2010), the reads from replicate samples were mapped to the hg19 genome using TopHat (--library-type fr-firststrand) (Trapnell et al. 2009) for stranded RNA-seq libraries and then merged for differential splicing analysis. The duplicate sample reads were merged and then sorted using SAMtools (Li et al. 2009) for MISO analysis. The ΔΨSJ estimate was calculated using only splice-junction and alternative exon–body reads. The minimum number of inclusion and exclusion RNA-seq reads over each detected splicing junction was set to 10, and differential splicing events were detected using the following options using MISO: filter_events.py --num-inc 1 --num-exc 1 --num-sum-inc-exc 10 --delta-psi --bayes-factor 10. The differential splicing events were filtered to contain only the AS events with Bayes factor ≥10. Differential splicing of alternative exons entailed a difference in the PSI (or Ψ) values.
Validation of MISO and JUM estimates by RT–PCR
One microgram of total RNA was reverse-transcribed according to the manufacturer's instructions (Bio-Rad, 1708891) and subjected to RT–PCR with the following conditions: 30 sec at 98°C (one cycle); 10 sec at 98°C, 30 sec at 60°C, and 30 sec at 72°C (35 cycles); and 5 min at 72°C (one cycle). The primer sets used in the RT–PCR reaction are listed in Supplemental Table 3. RT–PCR products were resolved, visualized, and quantitated by use of an Agilent Technologies Bioanalyzer.
eCLIP for hnRNPA1 and DDX5 from K562 cells
eCLIP was performed as described previously (Van Nostrand et al. 2016)—with the exception of using nuclear extract prepared from K562 cells instead of whole-cell extract—with the following antibodies: monoclonal anti-hnRNPA1 antibody, clone 9H10 (Sigma-Aldrich, R4528 RRID:AB_261962); monoclonal anti-hnRNPA1 antibody, clone 4B10 (Sigma-Aldrich, R9778 RRID:AB_477477); and polyclonal anti-DDX5 antibody (Abcam, ab21696 RRID:AB_446484).
Nuclear extract prepared from 15 × 106 cells was used for the immunoprecipitation reaction and prepared into eCLIP libraries according to a previously published method (Van Nostrand et al. 2016). As with previous CLIP/iCLIP protocols, this method used UV cross-linking of intact cells to covalently link RNA to cellular RNA-binding proteins. In our case, nuclei and nuclear extracts were prepared, followed by RNase I digestion to fragment the nuclear RNA prior to immunoprecipitation of target proteins and associated bound RNAs. The membrane-isolated RNA–protein adducts were proteolyzed, and the purified RNA was reverse-transcribed and further processed into high-throughput sequencing libraries.
eCLIP maintained the single-nucleotide resolution of iCLIP because an indexed 3′ RNA adapter was ligated to the cross-linked RNA fragments, and a 3′ ssDNA adapter was ligated to the cDNA following reverse transcription. This ssDNA adapter contained a random barcode to identify PCR duplicates and tag unique RNA fragments during later sequencing read processing steps. We performed eCLIP using nuclear extracts from K562 cells and monoclonal anti-hnRNPA1 or polyclonal anti-DDX5 antibodies. The RNA-binding protein complexes were isolated from polyacrylamide gels based on the expected molecular weight of hnRNPA1 and DDX5 plus a higher 75-kDa region, as described for the iCLIP method (Huppertz et al. 2014). The bound protein was degraded, and RNA was isolated and further processed into dsDNA sequencing libraries as described (Van Nostrand et al. 2016). The uniquely barcoded eCLIP samples were then pooled for sequencing. The final library material was quantified on the Bioanalyzer high-sensitivity DNA chip (Agilent) and sequenced on an Illumina HiSeq 4000 with 100-bp paired-end reads at the Vincent J. Coates Genomics Sequencing Laboratory at the University of California at Berkeley.
eCLIP-seq (eCLIP combined with high-throughput sequencing) analysis
eCLIP-seq reads were processed, quality-filtered, and collapsed to eliminate PCR duplicates, and then random barcodes from the adapters were removed before mapping to the hg19 genome. The reads were preprocessed prior to mapping using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit). Reads were quality-filtered based on quality score (fastq_quality_filter -q 25 -p 80), and PCR duplicates were collapsed (fastx_collapser). The adapter sequences were then trimmed using fastx_clipper. A second round of clipping was performed to ensure trimming off the 3′ adapters on read 2 as described previously (Van Nostrand et al. 2016), and random-mer sequences were trimmed. Any reads <15 nucleotides (nt) were discarded, and only read 2, the read that was enriched for termination at the cross-linking site, was considered for mapping. Only uniquely mapped reads were used for further analysis. The reads were mapped to hg19 using STAR using the following parameters: --outFilterMultimapNmax 1 --quantMode TranscriptomeSAM GeneCounts --outReadsUnmapped fastx --outSAMtype BAM SortedByCoordinate. Uniquely mapped reads were filtered using SAMtools (samtools view –bq 1) and then used for cluster identification using pyCRAC (Webb et al. 2014). eCLIP clusters were generated using at least five overlapping unique cDNA reads generated after removal of PCR duplicates (pyClusterReads.py --cic=5 --mutsfreq=20). De novo motifs for hnRNPA1 and DDX5 were determined using the pyMotif tool from the pyCRAC package with the following options: --n 100 -r 50 --k_min=4 --k_max=8. Z-scores were calculated to indicate overrepresentation of the k-mer sequence in the experimental data compared against k-mers from reads randomly distributed over the same genomic features. pyCalculateFDRs were used to filter out statistically significant clusters over the regions with a read coverage of at least 5 (--min=5) and FDR P < 0.05.
Determination of RNA structures around eCLIP tags in target pre-mRNAs
HEK293 in vitro icSHAPE (7165 transcripts) and in vivo icSHAPE (10,164 transcripts) data were obtained from NCBI (GSE74353) (Lu et al. 2016). The VTD profile was calculated at each nucleotide position to assess the difference between the in vivo and in vitro RNA reactivities along the transcript. RNA secondary structure was derived using RNAstructure (Mathews 2006) with icSHAPE constraints (--SHAPEintercept -0.6 --SHAPEslope 1.8). The generated structures were visualized using the VARNA software (Darty et al. 2009). The icSHAPE profiles were then correlated to the exon-overlapping eCLIP cluster (--cic=5 --mutsfreq=20, FDR>0.05) positions for the differentially spliced targets upon hnRNPA1 and DDX5 knockdown, as determined from RNA-seq analysis. For each eCLIP cluster, VTD profiles were generated across the transcript coordinate. In the plots, red dashed lines indicate potential UV cross-link sites of RNP complexes.
Data deposition
The data reported here have been deposited in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115442).
Supplementary Material
Acknowledgments
We thank members of the Center for RNA Systems Biology and Dr. Doug Black for helpful comments and suggestions. We thank Dr. Bo Li (Broad Institute of Massachusetts Institute of Technology and Harvard University), Dr. Lior Pachter (California Institute of Technology), Robert Tunney (University of California at Berkeley), and Dr. Liana Lareau (University of California at Berkeley) for their help with structural data analysis not included here. We thank Eric Van Nostrand (Dr. Geo Yeo's laboratory, University of California at San Diego) and Dr. Jernej Ule (The Francis Crick Institute) for very helpful discussions regarding eCLIP and iCLIP. We thank Dr. Ryan Flynn (Dr. H. Chang's laboratory, Stanford University) for helpful FastiCLIP and icSHAPE discussions. This work used the Vincent J. Coates Genomics Sequencing Laboratory at University of California at Berkeley, supported by National Institutes of Health (NIH) S10 Instrumentation grants S10RR025622, S10RR029668, and S10RR027303. This research used the Savio computational cluster resource provided by the Berkeley Research Computing program at the University of California at Berkeley (supported by the University of California at Berkeley Chancellor, Vice Chancellor for Research, and Chief Information Officer). This work was funded by the Arnold O. Beckman Post-doctoral Fellowship (to Q.W.), an NIH National Institute of General Medical Sciences Systems Biology Center grant (P50 GM102706; J. Cate, PI), and NIH grants R01 GM097352 and R35 GM11812.
Author contributions: D.C.R. conceived the study. Y.J.L. used the following software to generate the data and figures: Bedtools (2.22.1), FASTX-Toolkit (0.0.13), MISO (0.5.4), GOrilla, RNAstructure (6.0), SAMtools (1.3.1), STAR (2.5.2), pyCRAC (1.2.2.1), TopHat (2.0.8), and VARNA (3.93). Y.J.L performed RNA-seq analysis using MISO; performed downstream JUM analysis; generated figures for both JUM and MISO data, including GO term analysis; performed eCLIP-seq analysis; compared different data sets; and generated RNA structures and VTD profiles. Q.W. developed the JUM (1.3.7) software and used SAMtools (1.3.1), STAR (2.5.2), and JUM (1.3.7) for RNA-seq JUM analysis. Y.J.L. validated the results. Y.J.L. performed the investigation. D.C.R. acquired the resources. Y.J.L. and D.C.R. wrote the original draft. Y.J.L., D.C.R., and Q.W. reviewed and edited the manuscript. Y.J.L. visualized the study. D.C.R. supervised the study. D.C.R. was the project administrator. D.C.R. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.316034.118.
References
- Aviran S, Pachter L. 2014. Rational experiment design for sequencing-based RNA structure mapping. RNA 20: 1864–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC. 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev 19: 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun GH, Doktor TK, Borch-Jensen J, Masuda A, Krainer AR, Ohno K, Andresen BS. 2016. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol 14: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J 13: 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camats M, Guil S, Kokolo M, Bach-Elias M. 2008. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One 3: e2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J 18: 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Chander P, Howe PH. 2010. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16: 1449–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Ying Y, Lin CH, Lee JA, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, et al. 2016. Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell 165: 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. 2010. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert-Bezancon A, Sureau A, Durosay P, Salesse R, Groeneveld H, Lecaer JP, Marie J. 2004. hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of β-tropomyosin exon 6B. J Biol Chem 279: 38249–38259. [DOI] [PubMed] [Google Scholar]
- Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. 2016. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc 11: 273–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV. 2013. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta 1829: 756–763. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV, Ali S. 2008. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans 36: 609–612. [DOI] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. 2016. The hnRNP family: insights into their role in health and disease. Hum Genet 135: 851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. 2016. RNA matchmaking: finding cellular pairing partners. Mol Cell 63: 186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF. 2007. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14: 591–596. [DOI] [PubMed] [Google Scholar]
- Guil S, Gattoni R, Carrascal M, Abian J, Stevenin J, Bach-Elias M. 2003. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol 23: 2927–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010. PAR-CLIP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SP, Tang YH, Smith R. 2010. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430: 379–392. [DOI] [PubMed] [Google Scholar]
- Harrison AF, Shorter J. 2017. RNA-binding proteins with prion-like domains in health and disease. Biochem J 474: 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. 1995. RNA chaperones and the RNA folding problem. J Biol Chem 270: 20871–20874. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. 1994. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J 13: 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JM, Sanford JR. 2015. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 6: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. 2014. iCLIP: protein–RNA interactions at nucleotide resolution. Methods 65: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. 2011. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA 2: 135–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Philippe J, Paz S, Caputi M. 2013. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci 14: 18999–19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S. 2017. SR proteins: binders, regulators, and connectors of RNA. Mol Cells 40: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A, Fushimi K, Zhou X, Ray P, Shi C, Chen X, Liu Z, Chen S, Wu JY. 2011. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem–loop structure at the 5′ splice site. Mol Cell Biol 31: 1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK. 2016. Dawn of the in vivo RNA structurome and interactome. Biochem Soc Trans 44: 1395–1410. [DOI] [PubMed] [Google Scholar]
- Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. 2015. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci 40: 221–232. [DOI] [PubMed] [Google Scholar]
- Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B, Blanchette M, Monette A, Mouland AJ, Wellinger RJ, Chabot B. 2015. A function for the hnRNP A1/A2 proteins in transcription elongation. PLoS One 10: e0126654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. 2016. RNA splicing is a primary link between genetic variation and disease. Science 352: 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese D, de Groot NS, Lorenzo Gotor N, Livi CM, Tartaglia GG. 2016. Advances in the characterization of RNA-binding proteins. Wiley Interdiscip Rev RNA 7: 793–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. 2007. hnRNP proteins and splicing control. Adv Exp Med Biol 623: 123–147. [DOI] [PubMed] [Google Scholar]
- Mathews DH. 2006. RNA secondary structure analysis using RNAstructure. Curr Protoc Bioinformatics 13: 12.6.1–12.6.14. [DOI] [PubMed] [Google Scholar]
- Matthews DH. 2014. RNA secondary structure analysis using RNAstructure. Curr Protoc Bioinformatics 46: 12.6.1–12.6.25. [DOI] [PubMed] [Google Scholar]
- McManus CJ, Graveley BR. 2011. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev 21: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy RH, Cole BS, Yasunaga A, Gold B, Shankarling G, Varble A, Molleston JM, tenOever BR, Lynch KW, Cherry S. 2014. Stem–loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell 158: 764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman DS, Dreyfuss G. 1994. RNA annealing activities in HeLa nuclei. EMBO J 13: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Khrameeva E, Schreiner S, Konig J, Curk T, Zupan B, Ule J, Gelfand MS, Bindereif A. 2014. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol 11: 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O'Hanlon D, Blencowe BJ. 2016. Global mapping of human RNA–RNA interactions. Mol Cell 62: 618–626. [DOI] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. 2012. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18: 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. 2015. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry 54: 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci 113: 10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Lambert NJ, Sudmant PH, Dominguez D, Merkin JJ, Alexis MS, Bazile C, Burge CB. 2016. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol Cell 64: 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Rio D. 2017. The junction usage model (JUM): a method for comprehensive annotation-free differential analysis of tissue-specific global alternative pre-mRNA splicing patterns. bioRxiv 10.1101/116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Taliaferro JM, Klibaite U, Hilgers V, Shaevitz JW, Rio DC. 2016. The PSI-U1 snRNP interaction regulates male mating behavior in Drosophila. Proc Natl Acad Sci 113: 5269–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. 2010. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci 35: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S, Hector RD, Kudla G, Granneman S. 2014. PAR-CLIP data indicate that Nrd1–Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol 15: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Van Nostrand EL, Yeo GW. 2018. Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip Rev RNA 9: e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. 2006. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol 71: 217–224. [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. 2016. irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods 13: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.