Boulay et al. find that a subset of microsatellite repeats is transcriptionally active in Ewing sarcoma and that silencing individual repeats abolishes local nascent transcription and leads to markedly reduced expression of putative target genes.
Keywords: Ewing sarcoma, pediatric cancer, epigenetics, EWS-FLI1, enhancer therapy, epigenome editing
Abstract
Various types of repetitive sequences are dysregulated in cancer. In Ewing sarcoma, the oncogenic fusion protein EWS-FLI1 induces chromatin features typical of active enhancers at GGAA microsatellite repeats, but the function of these sites has not been directly demonstrated. Here, by combining nascent transcription profiling with epigenome editing, we found that a subset of GGAA microsatellite repeats is transcriptionally active in Ewing sarcoma and that silencing individual repeats abolishes local nascent transcription and leads to markedly reduced expression of putative target genes. Epigenome silencing of these repeat sites does not affect gene expression in unrelated cells, can prevent the induction of gene expression by EWS-FLI1, and, in the case of a GGAA repeat that controls SOX2 expression from a distance of 470 kb, is sufficient to impair the growth of Ewing sarcoma xenografts. Using an experimental approach that is broadly applicable to testing different types of repetitive genomic elements, our study directly demonstrates that specific repeat microsatellites can have critical gene regulation functions in cancer and thus represent tumor-specific vulnerabilities that may be exploited to develop new therapies.
Repeat elements constitute a large fraction of the human genome and for the most part have not been assigned specific biological functions (Lander et al. 2001; de Koning et al. 2011). Recent studies have shown widespread epigenetic and transcriptional dysregulation of repeat elements in various tumor types (Ting et al. 2011; Riggi et al. 2014; Rooney et al. 2015; Burns 2017), highlighting the need to elucidate their role in cellular transformation and tumor progression. However, understanding of their implication in cancer has been hampered by their repetitive nature and broad genomic distribution, which constitute major challenges toward studying specific biological contributions.
Ewing sarcoma, the second most common pediatric bone cancer, is characterized by specific chromosomal translocations that generate fusions between the EWSR1 gene and members of the ETS family of transcription factors, most commonly FLI1 (Delattre et al. 1992). The EWS-FLI1 fusion protein has been shown to operate as a major regulator of chromatin states that can either activate or repress enhancer activity, depending on the underlying DNA sequence of its binding sites (Riggi et al. 2014). EWS-FLI1 binding is most clearly associated with enhancer activation at a large set of GGAA microsatellite repeats where the fusion protein behaves as a pioneer factor to induce chromatin opening and the deposition of histone modifications associated with active enhancer elements (H3K4me1 and H3K27ac) (Gangwal et al. 2008; Guillon et al. 2009; Riggi et al. 2014; Tomazou et al. 2015; Boulay et al. 2017). Remarkably, the activation of GGAA microsatellites is highly specific to Ewing sarcoma, since these sites remain in a closed chromatin conformation in all other cell types examined (Riggi et al. 2014). Moreover, we showed recently that the ability to bind and activate GGAA repeats is a neomorphic property of EWS-FLI1 that requires the phase transition properties of the EWS prion-like domain and is not shared by wild-type FLI1 (Boulay et al. 2017).
Despite the fact that active GGAA repeat elements are a major feature of the epigenomic landscape of Ewing sarcoma, their functional implication in gene regulation and tumorigenesis has not been directly demonstrated. Assessment of the precise functional contributions of GGAA repeats is thus a key missing link toward our understanding of gene regulation by EWS-FLI1. To resolve this issue, we designed an experimental strategy for testing the function of specific repeat enhancer sites by targeting immediately adjacent unique genomic sequences with CRISPR/dCas9-KRAB (Gilbert et al. 2013, 2014; Thakore et al. 2015; Klann et al. 2017). We show that this approach leads to silencing of repeat elements by local spreading of repressive H3K9me3 marks, which in turn abolishes local nascent transcription and H3K27ac activation marks and dramatically decreases the expression of putative target genes. Moreover, silencing of a single EWS-FLI1-bound GGAA repeat enhancer 470 kb from the SOX2 locus was sufficient to impair the growth of Ewing sarcoma xenografts. Taken together, our results directly demonstrate that specific microsatellite repeats can make critical contributions to oncogenic gene expression programs that are essential for tumor maintenance.
Results
EWS-FLI1-bound GGAA microsatellites are transcriptionally active in Ewing sarcoma
EWS-FLI1-bound GGAA repeat microsatellites have been proposed to operate as distal enhancers in Ewing sarcoma based on active chromatin features. Given that evidence of activity at these sites has been observed only in Ewing sarcoma (Riggi et al. 2014) in association with tumor-specific phase transitions (Boulay et al. 2017), we sought to determine whether they exhibit other typical enhancer features. In particular, local transcription at enhancers has been shown recently to be a strong indicator of functional engagement in target gene activation (Li et al. 2016). We therefore tested for the presence of nascent noncoding enhancer RNAs (eRNAs) at GGAA repeats in Ewing sarcoma cells by performing nuclear run-on (NRO) experiments (Roberts et al. 2015) followed by high-throughput sequencing (NRO-seq).
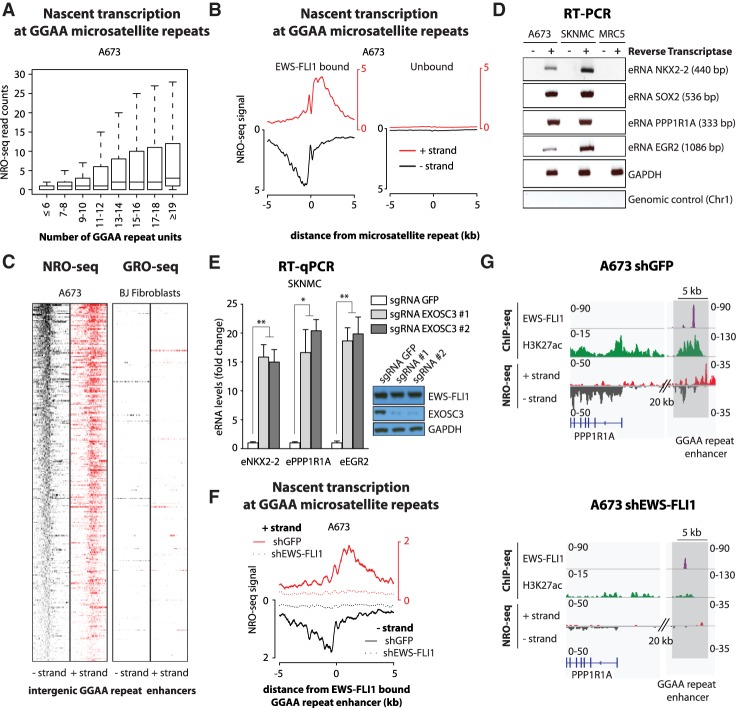
Our results show that a subset of GGAA repeats, particularly those with high numbers of repeat units (10 and above), displayed eRNA expression in Ewing sarcoma cells (Fig. 1A; Supplemental Fig. S1A). Moreover, EWS-FLI1 binding strongly predicted transcriptional activity at GGAA microsatellite repeats (Fig. 1B; Supplemental Fig. S1B), and these sites had high levels of transcriptional activity compared with other distal H3K27ac sites (Supplemental Fig. S1C). These signals were also specific, since analysis of GRO-seq (global run-on [GRO] combined with high-throughput sequencing) data from normal fibroblasts did not detect evidence of nascent transcription at GGAA repeats (Fig. 1C; Supplemental Fig. S1D), whereas BJ cells exhibited strong GRO-seq signals compared with Ewing sarcoma cells at other genomic locations (Supplemental Fig. S1E; Slobodin et al. 2017). A subset of these sites was validated by RT–PCR and gel electrophoresis in two Ewing sarcoma cell lines (Fig. 1D) as well as by RT-qPCR (real-time quantitative RT–PCR) in spheroid cultures derived from primary tumors (Supplemental Fig. S1F). Finally, CRISPR–Cas9-mediated knockout of EXOSC3, a core member of the exosome complex, strongly increased eRNA levels detected by RT-qPCR, demonstrating that the stability of eRNAs associated with GGAA repeats is regulated by the exosome complex in Ewing sarcoma, as observed for other eRNAs in different models (Fig. 1E; Pefanis et al. 2015).
Figure 1.
EWS-FLI1-bound GGAA microsatellite repeats are transcriptionally active. (A) Box plots show NRO-seq read counts at intergenic GGAA microsatellite repeats in A673 Ewing sarcoma cells based on the number of consecutive repeats. Reads were counted in a 1-kb window centered on GGAA microsatellite repeats. (B) Composite plots show NRO-seq signals at intergenic GGAA repeats that are either EWS-FLI1-bound (n = 473; left) or unbound (n = 7232; right) in A673 Ewing sarcoma cells. The X-axis represents a 10-kb window centered on GGAA microsatellite repeats. (C) Heat maps show NRO-seq signal density in Ewing sarcoma A673 cells (left) and GRO-seq signal density in BJ fibroblasts (right) at the top 300 transcriptionally active intergenic EWS-FLI1-bound GGAA repeats in A673 cells. Ten-kilobase windows in each panel are centered on EWS-FLI1-bound GGAA repeats. (D) RT–PCR shows the detection of eRNAs at EWS-FLI1-bound GGAA repeats in Ewing sarcoma cells A673 and SKNMC. MRC5 fibroblasts were used as negative control cells. Additional negative controls lack reverse transcriptase or map to a nontranscribed region on chromosome 1. GAPDH was used as a positive control of reverse transcription. Amplicon sizes are indicated in parentheses. (E) RT-qPCR shows the increased detection of eRNAs at EWS-FLI1-bound GGAA repeats in SKNMC cells expressing Cas9 and single-guide RNAs (sgRNAs) targeting the EXOSC3 coding sequence. (Inset) EXOSC3 and EWS-FLI1 protein levels assessed by immunoblotting in the same experiments. GAPDH was used as loading control. Cells were collected 7 d after lentiviral transduction. (F) Composite plots show NRO-seq signals at intergenic EWS-FLI1-bound GGAA repeats (n = 473) in A673 cells 96 h after infection with either a control shRNA (plain line) or a shRNA targeting FLI1 (dotted line). Enhancers found in intragenic regions or <5 kb from genes expressed in Ewing sarcoma were removed from consideration. The X-axis represents a 10-kb window centered on EWS-FLI1-bound GGAA microsatellite repeats. (G) Example shows decreased nascent transcription at an EWS-FLI1-bound GGAA repeat enhancer near PPP1R1A in A673 cells infected with either a control shRNA (left) or a shRNA targeting FLI1 (right). ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) tracks of FLI1 (EWS-FLI1), H3K27ac, and NRO-seq. Regions of interest are highlighted in light gray. (*) P-value < 0.05; (**) P-value < 0.01. See also Supplemental Figures S1 and S2.
EWS-FLI1-bound GGAA repeat enhancers thus possess not only features of active chromatin but also the transcriptional activity that is characteristic of functional enhancers (see examples in Supplemental Fig. S1G). Importantly, transcriptional activity at GGAA repeat enhancers was completely dependent on EWS-FLI1 levels (Fig. 1F,G; Supplemental Fig. S2A,B, showing intergenic EWS-FLI1-bound GGAA repeats). Indeed, as observed previously for the active histone mark H3K27ac (Riggi et al. 2014), nascent transcription was abrogated at these sites upon EWS-FLI1 knockdown (Supplemental Fig. S2C–E), concomitant with decreased expression of their neighboring putative target genes. Nascent transcription can thus also serve as a powerful complementary measurement of EWS-FLI1-dependent GGAA repeat activity.
Silencing of specific EWS-FLI1-bound GGAA repeat enhancers has major effects on local chromatin states and eRNA transcription
We next sought to epigenetically silence specific GGAA repeat enhancers using a CRISPR dCas9-KRAB system capable of inducing focal chromatin-repressive states by a combination of histone deacetylation and H3K9me3 deposition (Gilbert et al. 2014; Thakore et al. 2015). To design guide RNAs targeting GGAA repeat-flanking regions, we focused on EWS-FLI1-bound GGAA repeat enhancers neighboring a set of the most responsive EWS-FLI1 putative target genes defined by integrating RNA sequencing (RNA-seq) gene expression profiles from EWS-FLI1 down-regulation in tumor cell lines (A673 and SKNMC) and EWS-FLI1 expression in mesenchymal stem cells (MSCs), a putative cell of origin for this tumor type (Supplemental Fig. S3A,B; Supplemental Table S1; Riggi et al. 2005, 2008). The majority of these genes was found within 500 kb of EWS-FLI1-bound GGAA repeat enhancers in Ewing sarcoma cells (Supplemental Fig. S3C; Supplemental Table S1) and was expressed at very low levels in MSCs (Supplemental Fig. S3D), where these genes were frequently associated with the H3K27me3 Polycomb mark (Supplemental Fig. S3E). Activation of GGAA microsatellites by introduction of EWS-FLI1 in MSCs resulted in marked activation of these putative target promoters (Fig. S3F–H).
To avoid potential nonspecific effects of CRISPR silencing, genes associated with enhancers found in intragenic regions or <5 kb from genes expressed in Ewing sarcoma were removed from consideration. This approach yielded a total of 27 high-confidence putative target genes associated with 30 EWS-FLI1-bound GGAA repeat enhancers (Supplemental Table S2), from which we selected six candidate enhancers neighboring the genes that encode the transcription factors NKX2-2, SOX2, POU3F1, and EGR2; the membrane protein NPY1R; and the inhibitor of phosphatase PPP1R1A. These enhancers were located at variable genomic distances from their putative target genes, ranging from 15 to 470 kb.
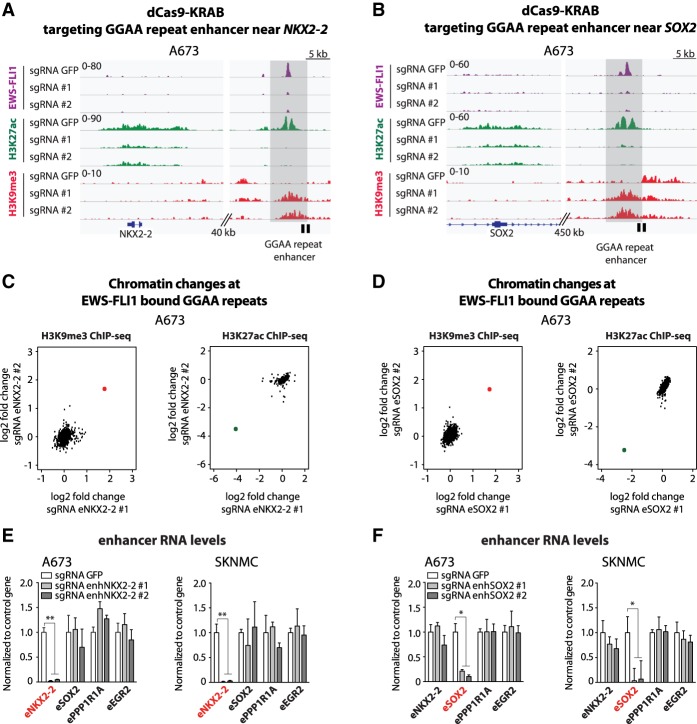
We first assessed chromatin changes mediated by CRISPR dCas9-KRAB targeting unique sites adjacent to GGAA repeats associated with the NKX2-2 and SOX2 genomic loci. Control cells exhibited strong signals for EWS-FLI1 and the active chromatin mark H3K27ac at these sites, whereas the repressive H3K9me3 mark was not detected (Fig. 2A,B). Upon lentiviral transduction of the tumor cells with CRISPR dCas9-KRAB constructs, we observed substantial changes in chromatin marks at the targeted sites. Consistent with histone deacetylase and H3K9 methylase activities of the KRAB domain, we observed the virtually complete disappearance of H3K27ac signals, which were replaced by the focal spread of H3K9me3 marks over a region of ∼3 kb adjacent to the targeted GGAA repeat (Fig. 2A,B). Remarkably, we also observed the disappearance of EWS-FLI1 signals (Fig. 2A,B), suggesting that the changes in chromatin state induced by our editing strategy were profound enough to compete with the direct binding of the oncogenic fusion protein. These results were further confirmed by ChIP-qPCR (chromatin immunoprecipitation [ChIP] combined with qPCR) at the EWS-FLI1-bound GGAA repeat associated with the SOX2 locus (Supplemental Fig. S4A).
Figure 2.
Site-specific GGAA repeat enhancer silencing using dCas9-KRAB. (A,B) ChIP-seq tracks of FLI1 (EWS-FLI1), H3K27ac, and H3K9me3 signals over the NKX2-2 (A) and SOX2 (B) loci in A673 cells expressing dCas9-KRAB and a sgRNA targeting either the GFP control sequence or GGAA repeat enhancers activated by EWS-FLI1 in Ewing sarcoma. Target regions are shown in light gray, and locations of sgRNA-binding sites are illustrated by a black box. (C,D) Scatter plots show the H3K9me3 and H3K27ac changes observed by ChIP-seq at EWS-FLI1-bound GGAA repeat enhancers (n = 812) in A673 cells lentivirally induced with dCas9-KRAB and sgRNAs targeting enhancers near NKX2-2 (C) or SOX2 (D). A red dot for H3K9me3 and a green dot for H3K27ac show the positions of the targeted enhancers in each experiment. Signals were calculated over a 5-kb window centered on EWS-FLI1-bound GGAA repeats. (E,F) RT-qPCR shows the specific decreases of eRNA levels at targeted EWS-FLI1-bound GGAA repeat enhancers near NKX2-2 (E) or SOX2 (F) in Ewing sarcoma cell lines (A673 and SKNMC). Cells were collected 7 or 8 d after lentiviral transduction. (*) P-value < 0.05; (**) P-value < 0.01. See also Supplemental Figures S3 and S4A.
ChIP-seq analysis of chromatin marks across all EWS-FLI1-bound GGAA repeat enhancers revealed a high degree of specificity for the observed changes in chromatin marks at targeted loci. Notably, whereas we identified pronounced increases in H3K9me3 and decreases in H3K27ac marks at the corresponding targeted sites, other EWS-FLI1-bound GGAA repeats remained virtually unaffected in each experiment (Fig. 2C,D).
To further assess the functional impact of targeted epigenome editing on enhancer activity, we measured changes in eRNA levels upon site-specific repeat silencing. As expected, induction of chromatin-repressive states at GGAA repeats resulted in transcriptional abrogation of the corresponding eRNAs (Fig. 2E,F), whereas eRNA expression levels remained unaltered at other EWS-FLI1-bound GGAA sites (Fig. 2E,F). In summary, our observations suggest that site-specific epigenetic silencing of genomic repeats by CRISPR dCAS9-KRAB provides a powerful and selective approach to evaluate the functional role of repetitive genomic elements in cancer.
Site-specific silencing of GGAA repeat enhancers strongly decreases the expression of putative target genes in Ewing sarcoma
Having demonstrated robust changes in activity at specifically targeted GGAA repeats through epigenome silencing, we next sought to determine the impact on their putative target gene expression. We found that NKX2-2 and SOX2 transcripts and proteins levels were strongly decreased upon targeting of their corresponding repeat elements (Fig. 3A,B, left and right, respectively), whereas expression of other GGAA repeat-associated genes remained stable (Supplemental Fig. S4B).
Figure 3.
EWS-FLI1-bound GGAA repeat enhancers directly control putative target gene expression. (A,B) NKX2-2 and SOX2 mRNA and protein levels assessed by RT-qPCR (left) and immunoblotting (right), respectively, in two Ewing sarcoma cell lines (SKNMC and A673) expressing dCas9-KRAB and a sgRNA targeting GGAA repeat enhancers activated by EWS-FLI1 near NKX2-2 or SOX2. sgRNAs against GFP were used as controls. Protein levels are quantified by immunoblots. (C–F, top) ChIP-seq tracks of FLI1 (EWS-FLI1) and H3K27ac over putative target genes and associated GGAA repeat enhancers activated by EWS-FLI1 in Ewing sarcoma. Target regions are shown in light gray, and locations of sgRNA-binding sites are illustrated by a black box. (Bottom) Expression levels of putative target genes assessed by RT-qPCR in two Ewing sarcoma cell lines (SKNMC and A673) expressing dCas9-KRAB and a sgRNA targeting either GFP or neighboring GGAA repeat enhancers activated by EWS-FLI1 in Ewing sarcoma. Cells were collected 7 or 8 d after lentiviral transduction. (*) P-value < 0.05; (**) P-value < 0.01. See also Supplemental Figure S4B.
These observations were further expanded by targeting four additional GGAA repeat enhancers located near the PPP1R1A, NPY1R, POU3F1, and EGR2 gene loci. Each of these EWS-FLI1-bound repeat sites was silenced using two specific single-guide RNAs (sgRNAs) in two Ewing sarcoma cell lines (Fig. 3C–F, top panel). Similar to the results obtained with NKX2-2 and SOX2, silencing of additional GGAA repeat enhancers invariably resulted in a marked decrease in expression of their associated genes (Fig. 3C–F, bottom panel) without altering the transcriptional profile of other repeat-associated genes (Supplemental Fig. S4B). These experiments strongly support the strength and specificity of our enhancer targeting strategy and identify the major functional role played by GGAA repeat enhancers in regulating the expression of their putative target genes.
Silencing Ewing sarcoma-specific microsatellite enhancers does not affect gene expression in unrelated cells
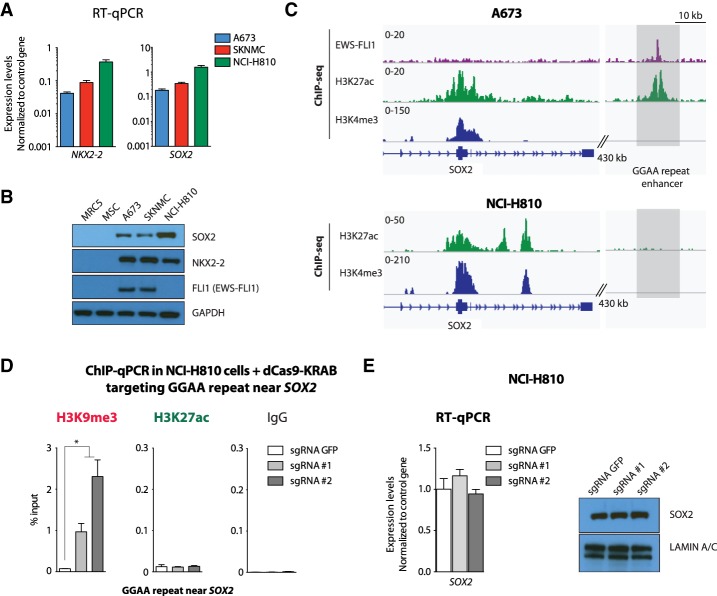
Enhancer elements represent cell type-specific distal regulatory units capable of coordinating the precise spatio–temporal distribution of gene expression required to define different cell states. EWS-FLI1-bound GGAA repeat enhancers were found to be specifically active only in Ewing sarcoma among a large collection of normal and tumor cell types (Riggi et al. 2014). Consistent with this observation, the transcriptional effects observed upon site-specific GGAA repeat silencing (Fig. 3) should be restricted to Ewing sarcoma cells and not detected in other cell types. To confirm this hypothesis, the dCas9-KRAB construct and specific sgRNAs for NKX2-2 and SOX2 repeat enhancers were transduced into a lung cancer cell line, NCI-H810, known to express high levels of both transcripts (Fig. 4A,B). As expected, since these cells do not express EWS-FLI1 (Fig. 4B), ChIP-seq analysis of their H3K27ac distribution failed to show any hallmarks of active enhancers at GGAA repeat sites near these gene loci (Fig. 4C; Supplemental Fig. S5A). Accordingly, despite the efficient deposition of the H3K9me3 marks at the same sites, as detected by ChIP-qPCR (Fig. 4D; Supplemental Fig. S5B), no effect was observed on either SOX2 or NKX2-2 mRNA and protein levels in NCI-H810 cells (Fig. 4E; Supplemental Fig. S5C), confirming the high selectivity of this targeting strategy that relies on the cell type-specific enhancer landscape.
Figure 4.
Silencing a Ewing sarcoma-specific GGAA repeat enhancer does not affect SOX2 expression levels in NCI-H810 lung cancer cells. (A,B) NKX2-2 and SOX2 are expressed at similar levels in lung carcinoma NCI-H810 cells and Ewing sarcoma cells. RT-qPCR (A) and immunoblotting (B) show similar expression levels. EWS-FLI1 is detected only in A673 and SKNMC Ewing sarcoma cells. GAPDH was used as loading control. (C) ChIP-seq for H3K4me3 and H3K27ac (A673 and NCI-H810) and FLI1 (EWS-FLI1) (A673) shows that an EWS-FLI1-bound GGAA repeat near SOX2 is specifically active in Ewing sarcoma. (D) ChIP-qPCR experiments confirm the absence of H3K27ac and the deposition of H3K9me3 at a Ewing sarcoma-specific enhancer near SOX2 in lentivirally transduced NCI-H810 cells with dCas9-KRAB and a sgRNA targeting either the GFP control sequence or the GGAA repeat site near SOX2. (E) RT-qPCR and immunoblotting show that targeting of a Ewing sarcoma-specific repeat enhancer near SOX2 in NCI-H810 cells does not affect SOX2 expression levels in the same experiments. LAMIN A/C was used as loading control. Cells were collected 8 or 10 d after lentiviral transduction. (*) P-value < 0.05. See also Supplemental Figure S5.
Silencing a Ewing sarcoma-specific microsatellite enhancer decreases de novo induction of NKX2-2 in MSCs
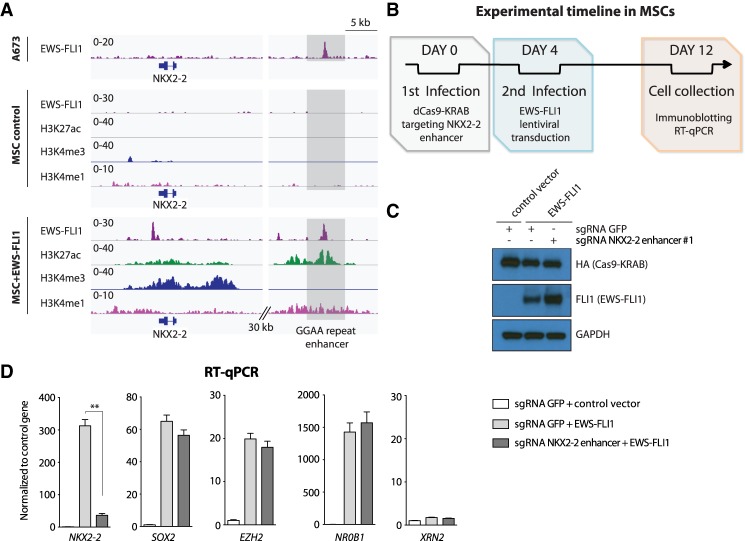
We next considered whether repression of a GGAA microsatellite repeat in non-Ewing sarcoma cells could prevent gene activation by EWS-FLI1. For these experiments, we turned to MSCs, in which NKX2-2 expression is virtually undetectable but becomes strongly induced upon lentiviral transduction of EWS-FLI1 and the concomitant activation of the associated GGAA repeat enhancer (Fig. 5A). MSCs were initially transduced with dCas9-KRAB and a specific sgRNA targeting the GGAA microsatellite repeat near the NKX2-2 locus and subsequently lentivirally infected with EWS-FLI1 (Fig. 5B,C). Consistent with our results in Ewing sarcoma cells (Fig. 3), NKX2-2 induction was strongly impaired, whereas expression of other EWS-FLI1 target genes remained unaltered (Fig. 5D).
Figure 5.
Silencing a Ewing sarcoma-specific GGAA repeat enhancer impairs de novo induction of NKX2-2 in MSCs. (A) ChIP-seq for FLI1 (EWS-FLI1) in A673 cells and FLI1 (EWS-FLI1), H3K27ac, H3K4me3, and H3K4me1 in MSCs lentivirally transduced with a control vector or EWS-FLI1. EWS-FLI1 can activate a GGAA repeat enhancer near NKX2-2. (B) Schematic showing the experimental strategy used in our assays. MSCs were lentivirally transduced with dCas9-KRAB and either sgRNA control (GFP) or a sgRNA targeting the EWS-FLI1-bound GGAA repeat enhancer near NKX2-2 before EWS-FLI1 lentiviral induction. (C) Immunoblotting confirms EWS-FLI1 expression in MSCs upon lentiviral induction. GAPDH was used as loading control. (D) RT-qPCR shows the specific decreased mRNA levels of NKX2-2 in MSCs infected with dCas9-KRAB and a sgRNA targeting the Ewing sarcoma-specific GGAA repeat enhancer site near NKX2-2 before EWS-FLI1 induction. SOX2, EZH2, and NR0B1 are other EWS-FLI1-induced genes not affected in these experiments. XRN2 was used as a control gene. (**) P-value < 0.01.
Altogether, our results demonstrate that GGAA repeat sites can be converted by EWS-FLI1 into fully active enhancer elements (which strongly influence the regulation of target gene expression) and that the transcriptional effects resulting from site-specific epigenome editing can be tumor-specific.
Silencing of a single specific GGAA repeat enhancer can inhibit tumor growth in vivo
Finally, to measure the biological contribution of a single GGAA microsatellite repeat enhancer in Ewing sarcoma, we asked whether epigenome silencing of an intergenic GGAA site might be sufficient to impair tumor growth in vivo. Among the panel of six repeat enhancers shown to control EWS-FLI1-induced genes in our experiments (Fig. 3), we selected the repeat enhancer that regulates SOX2 expression from a distance of 470 kb, based on our previous demonstration that this target gene plays an oncogenic role in Ewing sarcoma (Riggi et al. 2010; Ren et al. 2016).
In order to test the role of this GGAA repeat enhancer on xenograft tumor growth, A673 and SKNMC Ewing sarcoma cells were transduced with dCas9-KRAB along with two sgRNAs targeting the enhancer, selected with puromycin, and injected subcutaneously into the flanks of NOD-scidγ (NSG) mice (Fig. 6A). Remarkably, we observed a marked decrease in both the weight and volume of the xenografts derived from tumor cells subjected to enhancer silencing for both A673 and SKNMC cell lines (Fig. 6B,C).
Figure 6.
Silencing of a GGAA repeat enhancer reduces Ewing sarcoma tumor growth in vivo. (A) Schematic showing the experimental strategy used in our assays. Ewing sarcoma cells (A673 and SKNMC) were lentivirally transduced with dCas9-KRAB and either sgRNA control (GFP) or a sgRNA targeting a EWS-FLI1-bound GGAA repeat enhancer neighboring SOX2 before subcutaneous injection in immunocompromised NSG mice. (B,C) Inhibition of a repeat microsatellite enhancer near SOX2 markedly reduces tumor growth in immunocompromised mice, as assessed by tumor weight (B) and volume (C) 3 wk after subcutaneous injection. Six tumors were assayed per condition. (*) P-value < 0.05; (**) P-value < 0.01.
Taken together, our results demonstrate that individual GGAA microsatellite enhancers activated by EWS-FLI1 constitute key requirements for the establishment of the Ewing sarcoma oncogenic regulatory program and can be selectively disrupted using site-specific epigenome-editing technologies.
Discussion
Active GGAA repeat elements are a major feature of the chromatin landscape induced by EWS-FLI1 in Ewing sarcoma, but their function as distal regulatory elements has not been tested. Our studies demonstrate that epigenome editing can be used to silence repeat elements and directly demonstrate that specific GGAA repeats perform critical gene regulation functions in Ewing sarcoma by operating as powerful distal enhancers. Importantly, since the activation of GGAA microsatellite repeats is a neomorphic property of EWS-FLI1 that is not shared by the wild-type transcription factor FLI1 (Boulay et al. 2017), the regulatory activity of these repeat elements represents a tumor-specific mechanism that may be exploited for the development of new therapies. Considering the increasing evidence of repeat element dysregulation in cancer, our results also raise the possibility that repeats may be associated with enhancer functions in other tumor types. As in the case of Ewing sarcoma, the activity of specific repeats may be intimately connected with oncogenic events and thus may also point to therapeutic opportunities.
Unlike many instances where distal regulatory elements are part of complex networks and make only incremental contributions to the expression of their associated target genes, our studies directly demonstrate that individual GGAA microsatellite repeat sites activated by EWS-FLI1 can be major determinants of gene expression. Indeed, the enhancers selected for testing were located near some of the genes that are most highly responsive to the presence of EWS-FLI1, and our epigenome-editing experiments showed major effects on gene activation. Moreover, the lack of expression of these genes in MSCs and their association with repressive chromatin marks in these cells suggest that their expression is induced de novo by GGAA repeat enhancers during transformation by EWS-FLI1.
The importance of the powerful regulatory effects mediated by individual GGAA repeat elements is further supported by our finding that silencing a single microsatellite located 470 kb distal to SOX2 is sufficient to impair tumor growth. Our data further show that silencing this same site in unrelated cells did not affect gene expression and thus are in keeping with the fact that GGAA repeats do not appear to have open chromatin in a diverse panel of cell lines (Riggi et al. 2014). Individual repeat enhancers thus may constitute vulnerabilities that are specific to cancer cells, and strategies aimed at targeting these sites individually or in combination could have therapeutic value and a large therapeutic window. Importantly, in the context of efforts aimed at targeting EWS-FLI1 itself, EWS-FLI1-bound GGAA repeats observed genome-wide represent a large set of additional tumor-specific targets that may be effective even in cases of resistance to EWS-FLI1-directed therapies.
Our results show that the combination of epigenome profiling and functional testing with dCas9-KRAB provides an effective strategy for identifying functional repeat elements. Epigenome profiling has shown previously that a large fraction of EWS-FLI1-binding events occurs at GGAA microsatellites, where the fusion protein induces chromatin marks typical of active enhancers (Gangwal et al. 2008; Riggi et al. 2014; Tomazou et al. 2015). Our NRO-seq profiles now show that a subset of EWS-FLI1-bound GGAA microsatellites is transcribed and produces eRNAs, which recently have been closely linked to functional enhancer activity and chromatin looping (Li et al. 2016). As demonstrated in our experiments, candidate repeat loci identified through these methodologies can be effectively tested by targeting unique sequences adjacent to microsatellites with dCas9-KRAB. Our ChIP-seq profiling and recent studies in other systems (Thakore et al. 2015) show limited local spreading of H3K9me3 marks upon introduction of dCas9-KRAB. This is sufficient for repeat silencing, and the reduction in spatial resolution compared with CRISPR–Cas9 targeting is compensated for by the ability to specifically inactivate a single genomic location containing a repeat sequence. In addition, silencing is achieved without inducing DNA damage, thereby avoiding the risk of nonspecific effects on cell viability (Aguirre et al. 2016; Munoz et al. 2016).
Interestingly, effective targeting using dCas9-KRAB also provides an opportunity to gain mechanistic insights beyond functional validation of specific enhancers. For example, the fact that dCas9-KRAB was sufficient to evict EWS-FLI1 from GGAA repeat microsatellites suggests that the removal of histone acetylation and the accumulation of H3K9me3 can induce a chromatin environment that prevents EWS-FLI1 binding to DNA. This was further supported by experiments in which dCAS9-KRAB targeting preceding the introduction of EWS-FLI1 strongly decreased the induction of NKX2-2 by the fusion protein in MSCs. The absence of H3K9me3 at GGAA microsatellite repeats thus may be a prerequisite for EWS-FLI1 pioneer activity leading to enhancer activation during the first steps of transformation and may be an important feature of the cells of origin of Ewing sarcoma.
In conclusion, our studies establish a methodology for silencing repeat elements and directly demonstrate that GGAA microsatellites activated by EWS-FLI1 operate as powerful distal regulatory elements in Ewing sarcoma. These findings also suggest that the dysregulation of repeat elements in other tumor types may have important consequences for oncogenic gene expression programs. The systematic functional characterization of tumor-specific regulatory elements through epigenome editing thus may point to vulnerabilities that could be exploited for the development of new therapies in many types of cancer.
Materials and methods
MSCs and tumor spheres
MSCs and primary tumor samples were collected with approval from the Institutional Review Board of the Centre Hospitalier Universitaire Vaudois (University of Lausanne). Samples were deidentified prior to our analysis. Primary bone marrow-derived MSCs were cultured in Iscove's modified Dulbecco's medium (IMDM) containing 10% fetal calf serum (FCS) and 10 ng/mL platelet-derived growth factor BB (PeproTech). Ewing sarcoma patient-derived tumor spheres were cultured in IMDM (Gibco) supplemented with 20% knockout serum (Gibco), 10 ng/mL human recombinant EGF and bFGF (Invitrogen), and 1% penicillin/streptomycin (Gibco) in ultralow attachment flasks (Corning), as described previously (Suva et al. 2009).
Cell lines
Cell lines were obtained from American Type Culture Collection (ATCC), and media were obtained from Life Technologies. Ewing sarcoma cell lines SKNMC and A673 were grown in RPMI. NCI-H810 and MRC5 cells were grown in RPMI and EMEM, respectively. All media were supplemented with 10% FBS, and cells were cultured at 37°C with 5% CO2. Cells were maintained and split every 2–3 d according to ATCC's recommendations.
Definition of genes induced by EWS-FLI1
Genes strongly induced by EWS-FLI1 were defined from previously generated RNA-seq data (Boulay et al. 2017) based on the following criteria: (1) greater than threefold decrease upon EWS-FLI1 knockdown in both A673 and SKNMC Ewing sarcoma cells, (2) greater than threefold increase in MSCs lentivirally induced with EWS-FLI1 (DESeq2-corrected P-value of <0.05) (Love et al. 2014), and (3) expression values of at least 1 FPKM (fragments per kilobase per million mapped fragments) in A673 and SKNMC cells prior to EWS-FLI1 knockdown and in MSCs upon lentiviral induction with EWS-FLI1. Differences in expression between Ewing sarcoma tumor samples and normal tissues were calculated for 92 induced genes present in publicly available microarray data (GSE: GSE68776) (Svoboda et al. 2014) and are displayed in a row-normalized heat map (Supplemental Fig. S3B). Distances between transcription start sites (TSSs) and EWS-FLI1-bound GGAA repeats (Supplemental Fig. S3C; Riggi et al. 2014) were compared with nonresponsive genes (minimum expression of 1 FPKM in A673 and SKNMC Ewing cell lines and log2 fold change expression of <0.3 in knockdown and up-regulation experiments).
Annotation of GGAA microsatellite repeats
GGAA repeats were defined as intervals with four or more consecutive GGAA/TTCC units in the human genome (hg19) using BedTools (Quinlan and Hall 2010). Repeat intervals within 100 bases of each other were merged prior to further processing. In Figure 1, A and B, and Supplemental Figure S1, A and B, we focused our analysis on distal sites by excluding microsatellite repeats within 5000 bases from the boundaries of RefSeq-annotated genes and sites with high H3K4me3 ChIP-seq signals (more than eight normalized units in either A673 or SKNMC) to account for unannotated gene promoters. Each category contained 4126, 637, 700, 632, 608, 412, 227, and 363 sites, respectively. EWS-FLI1-bound GGAA repeats were defined based on EWS-FLI1 ChIP-seq as before (Riggi et al. 2014). In Figure 1C and Supplemental Figure S1D, the same exclusion criteria were applied to EWS-FLI1-bound GGAA repeats. In Supplemental Figure S1C, for comparison of NRO signals at EWS-FLI1-bound GGAA repeats versus other active enhancers, we applied the same exclusion criteria as above to H3K27ac peaks in order to isolate distal peaks and identified two categories: (1) peaks intersecting EWS-FLI1-bound GGAA repeats (Riggi et al. 2014) and (2) peaks that are not bound by EWS-FLI1 and do not contain four or more GGAA repeat units.
Definition of targetable EWS-FLI1-bound GGAA repeat enhancers
The set of targetable EWS-FLI1-bound GGAA repeat enhancers (Supplemental Table S2) was obtained by identifying EWS-FLI1-bound GGAA repeat enhancers (Riggi et al. 2014) within 500 kb of the 111 EWS-FLI1-induced genes defined above. Genes whose nearest enhancer lies within 5 kb of any gene expressed (>1 FPKM) in either A673 or SKNMC cells were removed from this analysis. When several candidate enhancers were assigned to a gene, we chose to target the most active site based on H3K27ac ChIP-seq levels.
V2-dCas9-KRAB and guide RNA cloning
The V2-dCas9-KRAB construct was generated by replacing wild-type Cas9 from V2-Cas9 plasmid (Addgene, 52961) with mutant dCas9-KRAB (Addgene, 60954). Cas9 was removed from the V2-Cas9 plasmid using BamH1 and Xba1. dCas9-KRAB was extracted using Nhe1 and partial BamH1 digestion. The plasmid sequence was verified using next-generation sequencing (Massachusetts General Hospital Center for Computational and Integrative Biology [CCIB] DNA Core). sgRNAs were cloned into V2-dCas9-KRAB following standard protocols.
Lentiviral generation
Lentivirus was produced in 293T LentiX cells (Clontech) by LT1 (Mirus Bio) transfection with gene delivery vector and packaging vectors GAG/POL and VSV plasmids. Viral supernatants were collected 72 h after transfection and concentrated using LentiX concentrator (Clontech). Virus-containing pellets were resuspended in PBS and added dropwise on cells in the presence of medium supplemented with 6 µg/mL polybrene. Selection of lentivirally infected cells was achieved with puromycin used at 0.75 µg/mL for MSCs, 1 µg/mL for SK-N-MC, 2 µg/mL for A673, and 4 µg/mL for NCI-H810. Overexpression or knockdown efficiency was determined by Western blot analysis and RT-qPCR.
RT-qPCR
For gene expression assays, total RNA was isolated from cells using NucleoSpin RNA Plus (Clontech). cDNA was obtained using SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). One microgram of template total RNA and random hexamers were used for each reaction. Real-time PCR amplification was performed using fast SYBR Green master mix (Life Technologies) and specific PCR primers in a LightCycler 480 instrument (Roche). The oligonucleotides used are shown in Supplemental Table S3. Relative quantification of each target, normalized to an endogenous control (GAPDH or HPRT1), was performed using the comparative Ct method (Applied Biosystems). Error bars indicate the standard deviation of three technical replicates and represent at least two independent biological experiments. Statistical analyses were performed by Student's t-test. In heat maps (Supplemental Fig. S4B), log2 RT-qPCR expression values were averaged across biological replicates in each condition.
Western blot analysis
Western blotting was performed using standard protocols. Primary antibodies used for Western blotting are listed in Supplemental Table S3. Secondary antibodies were goat anti-rabbit, anti-mouse (Bio-Rad), and anti-rat (Invitrogen) immunoglobulin G-horseradish peroxidase-conjugated (1:10,000 dilution). Membranes were developed using Western Lightning Plus-ECL enhanced chemiluminescence substrate (PerkinElmer) and visualized using photographic film.
ChIP-seq
ChIP assays were carried out on A673, MSCs, and NCI-H810 cultures of ∼2 million to 5 million cells per sample and per epitope, following the procedures described previously (Mikkelsen et al. 2007). In brief, chromatin from formaldehyde-fixed cells was fragmented to a size range of 200–700 bases with a Branson 250 sonifier. Solubilized chromatin was immunoprecipitated with the indicated antibodies overnight at 4°C. Antibody–chromatin complexes were pulled down with protein G Dynabeads (Life Technologies), washed, and then eluted. After cross-link reversal and RNase A and proteinase K treatment, immunoprecipitated DNA was extracted with AMP Pure beads (Beckman Coulter). ChIP DNA was quantified with Qubit. ChIP DNA samples (1–5 ng) were used to prepare sequencing libraries, and ChIP DNA and input controls were sequenced with the NextSeq 500 Illumina genome analyzer.
ChIP-seq bioinformatic processing
Reads were aligned to hg19 using bwa (Li and Durbin 2009). Aligned reads were then filtered to exclude PCR duplicates and extended to 200 base pairs (bp) to approximate fragment sizes. Density maps were generated by counting the number of fragments overlapping each position using igvtools, and normalized to 10 million reads. Average ChIP-seq signals across intervals were calculated using bwtool (Pohl and Beato 2014). Signals shown in heat maps (100-bp windows) and composite plots (10-bp window) were calculated using bwtool (Pohl and Beato 2014). Heat map signals are in log2 scale, centered on the TSS of EWS-FLI1-induced genes, and capped at the 99th percentile.
ChIP-qPCR
ChIP was performed as described above. qPCR was performed using fast SYBR Green master mix (Life Technologies) and specific PCR primers in a LightCycler 480 instrument (Roche). The oligonucleotides used are shown in Supplemental Table S3. Relative quantification of each target, normalized to input control, was performed using the comparative Ct method (Applied Biosystems). Error bars indicate the standard deviation of three technical replicates and represent at least two independent biological experiments.
NRO assay
NRO assays were performed as described recently (Roberts et al. 2015) with minor modifications. Briefly, cells were collected by cold trypsinization and washed with cold PBS before counting, and 10 million to 15 million cells were aliquoted per reaction. Nuclei were isolated in NP-40 lysis buffer (10 mM Tris-HCl at pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) for 5 min on ice, sedimented by centrifugation at 300g for 4 min at 4°C, and resuspended in 100 µL of nucleus storage buffer (50 mM Tris-HCl at pH 8, 0.1 mM EDTA, 5 mM MgCl2, 40% glycerol). NRO transcription was performed for 30 min at 30°C after adding 150 µL of transcription buffer (20 mM Tris-HCl at pH 8, 5 mM MgCl2, 300 mM KCl) supplemented with 4 mM fresh DTT (Sigma-Aldrich), 3.33 mM ATP, 1.66 mM UTP, 3.33 mM CTP, 3.33 mM GTP, 1.66 mM BrUTP (Roche), and 100 U of RNaseOUT (Life Technologies). Nuclear RNA was isolated using NucleoSpin microRNA kit (Clontech). For each immunoprecipitation, 30 µL of protein G Dynabeads (Life Technologies) was washed twice in PBST (1× PBS, 0.1% Tween-20) and incubated with rotation for 10 min at room temperature with 2 µg of anti-BrdU monoclonal antibody (Santa Cruz Biotechnology). Next, 150 µL of blocking buffer (PBST supplemented with 0.1% polyvinylpyrrolidone [Sigma-Aldrich] and 0.1% UltraPure BSA [Life Technologies]) was added, and the reaction was incubated with rotation for 30 min longer at room temperature. Anti-BrdU-bound beads were collected using a magnet and resuspended in 100 µL of PBSTR (PBST supplemented with 8 U/mL RNaseOUT). Six micrograms of NRO-RNA was denatured for 5 min at 65°C, mixed with anti-BrdU-bound beads, and incubated with rotation for 30 min at room temperature. Immunocomplexes were collected using a magnet and washed three times with PBSTR. Finally, purified RNA was isolated using the NucleoZOL extraction kit (Clontech) and eluted in 20 µL of ultrapure water.
NRO-RT-qPCR
Ten microliters of purified NRO-RNA or 600 ng of nuclear input RNA was used for each reaction. cDNA was obtained using SuperScript VILO cDNA synthesis kit with random hexamers (Thermo Fisher Scientific). Real-time PCR amplification was performed using fast SYBR Green master mix (Life Technologies) and specific PCR primers in a LightCycler 480 instrument (Roche). The oligonucleotides used are shown in Supplemental Table S3. Relative quantification of each target, normalized to an endogenous control (GAPDH or HPRT1), was performed using the comparative Ct method (Applied Biosystems). Error bars indicate the standard deviation of three technical replicates. Statistical analyses were performed by Student's t-test.
NRO-seq
For NRO-seq, NRO was performed as described above, but five to 10 immunoprecipitations were simultaneously prepared and pooled before final RNA purification. Nine microliters of purified NRO-RNA was treated with RiboGone oligonucleotides to remove ribosomal RNA (Clontech). Illumina-compatible sequencing libraries were constructed using SMARTer stranded total RNA sample preparation kit (Clontech) according to the manufacturer's instructions and sequenced with the NextSeq 500 Illumina genome analyzer.
NRO-seq and GRO-seq bioinformatic processing
Reads were aligned to GRCh37(hg19) using bwa-mem (Li 2013). PCR duplicates were removed using Picard (http://broadinstitute.github.io/picard). We generated read count totals in each interval (Fig. 1A; Supplemental Fig. S1A,C) as well as genome-wide signals (Fig. 1B,C,F,G; Supplemental Fig. S1B,D,G) using BedTools coverage. Strand-specific signals were generated from bam files obtained by separating reads that aligned to forward and reverse strands. Genome-wide smoothed signals were generated by counting reads in 150-bp sliding windows with 20-bp overlap. All signals were normalized for sequencing depth and scaled to 10 million reads. GRO-seq data for BJ cells were obtained from Gene Expression Omnibus (GEO; GSE96717) (Slobodin et al. 2017) and processed identically to NRO-seq data. In Supplemental Figure S1E, GRO-seq peaks were called in BJ cells with MACS2 using a ChIP-seq input as a background control (GEO: ENCSR000DQE/ENCFF001HAG). Intergenic sites were defined by excluding intervals that fell within 5000 bases from the boundaries of RefSeq-annotated genes (TSS − 5000 to transcription end site + 5000). In order to account for unannotated gene promoters, we also excluded intervals that had an average normalized H3K4me3 ChIP-seq signal >8 in BJ cells (GEO: ENCSR000DWS/ ENCFF001FZQ).
In vivo tumorigenic assays
For in vivo experiments, A673 and SKNMC cells were infected with lentiviral vectors expressing dCas9-KRAB and sgRNAs targeting either the GFP sequence (control) or the GGAA repeat enhancer identified near the SOX2 locus. After 48 h of puromycin selection, 1 × 106 cells for each condition were injected subcutaneously into NOD-scid IL2Rγnull mice (The Jackson Laboratory). Mice were monitored daily for tumor development and sacrificed 3 wk later, when tumor weight and volume were assessed. Experimental protocols involving mice were approved by the Veterinary Service of the Canton of Vaud, Switzerland (Etat de Vaud, Service Vétérinaire), under authorization number VD2488. Statistical analyses were performed by Student's t-test.
Quantification and statistical analysis
All statistical details of experiments are included in the figure legends or specific Materials and Methods section.
Data and software availability
The data accompanying this study have been deposited into GEO under accession number GSE106925.
Additional resources
To aid our analysis, we also used our publicly available data sets for EWS-FLI1, H3K4me1, H3K4me3, and H3K27ac ChIP-seq in A673 and SKNMC cells (GEO:GSE61953) (Riggi et al. 2014) and RNA-seq in A673, SKNMC, and MSCs (GEO: GSE94278) (Boulay et al. 2017). We also used publicly available data sets for GRO-seq in BJ cells (GEO: GSE96717) (Slobodin et al. 2017), H3K4me3 ChIP-seq in BJ cells (ENCODE: ENCSR000DWS/ENCFF001FZQ) and its matched input control (ENCODE: ENCSR000DQE/ENCFF001HAG), expression microarray in Ewing sarcoma tumor samples and normal tissues (GEO: GSE68776) (Svoboda et al. 2014), and Cancer Cell Line Encyclopedia (https://data.broadinstitute.org/ccle_legacy_data/mRNA_expression/CCLE_Expression_Entrez_2012-09-29.gct; Barretina et al. 2012).
Competing interest statement
M.N.R. receives research support from Advanced Cell Diagnostics and is a consultant for Loxo Oncology.
Supplementary Material
Acknowledgments
We thank the members of the Rivera, Bernstein, and Suva laboratories for valuable guidance and advice. A.V. is supported by the Medic Foundation, the Swiss Cancer Research Foundation, and the Botnar Foundation. I.S. is supported by Swiss National Science Foundation grant 310030_169563 and the Swiss Cancer League (KLS 3365-02-2014). N.R. is supported by a Swiss National Science Foundation professorship grant (PP00P3-157468/1), Oncosuisse (KFS-3973-08-2016), and the Medic Foundation. M.N.R. receives research support from the V Foundation for Cancer Research and the Charley Davidson Children's Cancer Fund at the MassGeneral Hospital for Children.
Author contributions: G.B., A.V., N.R., and M.N.R designed the study and wrote the manuscript. I.S. provided necessary reagents and conceptual advice and wrote the manuscript. A.V., G.B., N.R., and L.C.B. performed the experiments. S.I. and G.B. conducted bioinformatic analyses.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.315192.118.
References
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. 2016. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov 6: 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. 2017. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171: 163–178 e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17: 415–424. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359: 162–165. [DOI] [PubMed] [Google Scholar]
- Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, Boucher KM, Watkins WS, Jorde LB, Graves BJ, et al. 2008. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci 105: 10149–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. 2009. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE 4: e4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. 2017. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol 35: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997 [q-bio.GN]. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17: 207–223. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, Golji J, Ruddy DA, Yu K, McAllister G, et al. 2016. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov 6: 900–913. [DOI] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. 2015. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161: 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A, Beato M. 2014. bwtool: a tool for bigWig files. Bioinformatics 30: 1618–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Ren T, Yang K, Wang S, Bao X, Zhang F, Guo W. 2016. Inhibition of SOX2 induces cell apoptosis and G1/S arrest in Ewing's sarcoma through the PI3K/Akt pathway. J Exp Clin Cancer Res 35: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, Garcia-Echeverria C, Hoffmann F, Trumpp A, Stamenkovic I. 2005. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 65: 11459–11468. [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V, et al. 2008. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 68: 2176–2185. [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suva D, et al. 2010. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 24: 916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML, Rossetti NE, Boonseng WE, Oksuz O, Cook EB, et al. 2014. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 26: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TC, Hart JR, Kaikkonen MU, Weinberg MS, Vogt PK, Morris KV. 2015. Quantification of nascent transcription by bromouridine immunocapture nuclear run-on RT-qPCR. Nat Protoc 10: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. 2015. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160: 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink JA, Loayza-Puch F, Elkon R, Agami R. 2017. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169: 326–337 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Stehle JC, Baumer K, Tercier S, Joseph JM, Suva D, Clement V, Provero P, Cironi L, et al. 2009. Identification of cancer stem cells in Ewing's sarcoma. Cancer Res 69: 1776–1781. [DOI] [PubMed] [Google Scholar]
- Svoboda LK, Harris A, Bailey NJ, Schwentner R, Tomazou E, von Levetzow C, Magnuson B, Ljungman M, Kovar H, Lawlor ER. 2014. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics 9: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. 2015. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12: 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. 2011. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331: 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P, Kubicek S, Bock C, Kovar H. 2015. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep 10: 1082–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.