Abstract
Structural equation modeling is commonly used to capture complex structures of relationships among multiple variables, both latent and observed. We propose a general class of structural equation models with a semiparametric component for potentially censored survival times. We consider nonparametric maximum likelihood estimation and devise a combined Expectation-Maximization and Newton-Raphson algorithm for its implementation. We establish conditions for model identifiability and prove the consistency, asymptotic normality, and semiparametric efficiency of the estimators. Finally, we demonstrate the satisfactory performance of the proposed methods through simulation studies and provide an application to a motivating cancer study that contains a variety of genomic variables. Supplementary materials for this article are available online.
Keywords: Integrative analysis, Joint modeling, Latent variables, Model identifiability, Nonparametric maximum likelihood estimation, Survival analysis
1. INTRODUCTION
Structural equation modeling (SEM) is a very general and powerful approach to capture complex relationships among multiple factors, both observed and latent (Bollen 1989). A typical SEM framework consists of a structural model that connects latent variables and a measurement model that relates latent variables to observed variables. SEM is extremely popular in the social sciences and psychology, where unmeasured quantities and psychological constructs, such as human intelligence and creativity, can be related to and investigated through observed data. The text of Bollen (1989) has been cited more than 20,000 times. Recently, SEM has gained popularity in medical and public health research (Dahly et al. 2009; Naliboff et al. 2012).
Our interest in SEM was motivated by its potential application to integrative analysis in genomic studies. Recent technological advances have made it possible to collect different types of genomic data, including DNA copy number, SNP genotype, DNA methylation level, and expression levels of mRNA, microRNA, and protein, on a large number of subjects. There is a growing interest in integrating these genomic platforms so as to understand their biological relationships and predict disease progression and death, which are considered potentially censored survival times (The Cancer Genome Atlas (TCGA); https://tcga-data.nci.nih.gov/tcga/).
SEM with discrete survival times has been studied by Rabe-Hesketh et al. (2001; 2004), Muthén and Masyn (2005), and Moustaki and Steele (2005). For continuous survival time, Larsen (2004; 2005) adopted the proportional hazards model (Cox 1972) with a single latent variable to capture the association between the survival time and other observed variables; Asparouhov et al. (2006) considered a more general formulation of the association among the latent and observed variables. SEM with the Cox proportional hazards model for the survival component has been adopted for more complex settings, such as multivariate survival times (Stoolmiller and Snyder 2006) and competing risks (Stoolmiller and Snyder 2013). A popular software program, Mplus (Muthén and Muthén 1998–2015), has implemented SEM with survival data under the proportional hazards model. The estimation of the nonparametric baseline hazard function is based on piecewise-constant splines, and no theoretical justification is available. In fact, the standard error estimator for the baseline hazard function is incorrect.
In this article, we propose a general SEM framework that includes a semiparametric component of the measurement model for potentially censored survival times. Specifically, we formulate the effects of latent and observed covariates on survival times through a broad class of semiparametric transformation models that includes the proportional hazards model as a special case. The observed covariates may include manifest variables that depend on latent variables. We study nonparametric maximum likelihood estimation (NPMLE), under which the cumulative hazard functions are estimated by step functions with jumps at observed survival times.
The proposed SEM is reminiscent of joint modeling for survival and longitudinal data (Henderson et al. 2000; Tsiatis and Davidian 2004). With the latter, the observed longitudinal variables are considered error-prone measurements of some underlying latent variables, but the measurements themselves are not causal determinants of the survival time. By contrast, our SEM framework allows latent variables to have direct effects on survival times, as well as indirect effects through other manifest variables. In addition, our framework accommodates much more complex relationships among latent variables.
A major challenge in our theoretical development is model identifiability. Even for an SEM with normally distributed variables, no single set of conditions exists that is both necessary and sufficient for model identifiability. Methods that deal with special cases of the normal SEM were proposed by Bollen (1989), Reilly and O’Brien (1996), Vicard (2000), and Bollen and Davis (2009), among others. Most of the methods are based on the fact that identifiability can be established by solving the equations relating the first two model-implied moments to the sample moments. This approach is not directly applicable to models with nonparametric components, as infinite-dimensional parameters cannot be identified through a finite number of equations. Because the proportional hazards structure results in a likelihood function that takes the form of a Laplace transform, however, we are able to develop sufficient conditions under which the identifiability of a semiparametric SEM can be established by inspecting simpler parametric models.
Another theoretical challenge is the invertibility of the information operator. For the information operator to be invertible, we require that the score statistic along any nontrivial submodel is non-zero. As in the case of model identifiability, general conditions for the invertibility of the information operator for semiparametric models do not exist. In the existing work involving latent variables for survival times (Kosorok et al. 2004; Zeng and Lin 2010), verifying the invertibility of the information operator involves inspecting the local behavior of the score statistic around the zero survival time. This approach does not make full use of the variability of the score statistic contributed by the survival times and cannot deal with the proposed general modeling framework. We show that the invertibility of the information operator can be verified by inspecting the parametric components of the SEM under some mild conditions in the survival model.
The rest of this article is structured as follows. In Section 2, we formulate the model and describe our approach to establish model identifiability. In Section 3, we discuss the numerical implementation of the NPMLE. In Section 4, we present theoretical results for model identifiability and describe the asymptotic properties of the estimators. In Section 5, we report the results from simulation studies. In Section 6, we provide an application to the TCGA data, which motivated this work. We make some concluding remarks in Section 7 and relegate theoretical proofs to the Appendix.
2. BASIC FRAMEWORK
2.1 Model and Likelihood
Let η denote a q-vector of latent variables, Y denote an r-vector of uncensored manifest variables, (T1, …, TK) denote K potentially censored survival times, and W and Z denote two vectors of observed covariates. Without loss of generality, assume that the support of the covariates includes zero. We specify the conditional distributions of η given Z, Y given Z and η, and Tk given W, Z, Y, and η as follows:
| (1) |
| (2) |
| (3) |
where Fη (· | Z, ν) denotes a q-variate normal distribution function indexed by a parameter vector ν, FY (· | Z, η; ψ) denotes an r-variate parametric distribution function indexed by a parameter vector ψ, ΛTk is the cumulative hazard function of Tk given (W,Z,Y, η), Gk is a known increasing function, Λk is an unspecified positive increasing function with Λk(0) = 0, and (ϑk, βk, αk,ϕk) are unknown regression parameters.
Model (1) is the structural model of the latent variables. Model (2) is the measurement model of Y. We assume that Y and η are independent of W given Z. Models (1) and (2) represent the existing SEM framework with Y not restricted to be normally distributed. Equation (3) includes the proportional hazards and proportional odds models as special cases with the choices of Gk (x) = x and Gk (x) = log (1 + x), respectively. The proportional hazards model has been considered in the literature.
The survival time Tk is subject to right censoring by Ck. It is assumed that (C1, …, CK) are independent of (T1, …, TK) and η conditional on Y, Z, and W. Define T̃k = min(Tk,Ck) and Δk = I(Tk ≤ Ck), where I(·) is the indicator function. For a sample of size n, the observed data consist of 𝒪i ≡ (T̃1i, … T̃Ki,Δ1i, …, ΔKi,Yi,Zi,Wi) (i = 1, …, n).
Let θ denote the collection of all Euclidean parameters, and write 𝒜 = (Λ1, …, ΛK). The likelihood function for θ and 𝒜 is proportional to
| (4) |
where f′(x) = df(x)/dx for any function f, , and . The NPMLE is defined to be the maximizer of Ln(θ,𝒜), in which Λk is treated as a step function with jumps at T̃ki with Δki = 1 (i = 1, …, n).
2.2 Model Identifiability
We describe our approach to establish model identifiability in this section and defer the technical details to Section 4. The identifiability results can be summarized by two simple rules. Suppose that we have arranged the survival times such that for some 0 ≤ K1 ≤ min(q,K), each of (T1, …, TK1) regresses on and only on one latent variable and a set of covariates that are independent of the latent variables. (We allow K1 = 0 if no survival time satisfies the given conditions, in which case Rule 1 below is vacuous.) We call an observed variable X an indicator of a latent variable η if X follows a generalized linear model with η as a covariate and is independent of all other manifest variables and survival times conditional on η. We have the following rules:
Rule 1. The latent variables attached to (T1, …, TK1) can be treated as observed if each of (T1, …, TK1) depends on at least one observed covariate.
Rule 2. If each latent variable has a separate continuous indicator and the distributions of the latent variables and the indicators are identifiable, then the whole model is identifiable.
To illustrate the usefulness of the two identifiability rules, we present two examples.
Example 1
Consider the model depicted in Figure 1. In the model, Y1, Y2, and Y3 are conditionally independent normal manifest variables of η, and T is a survival time that follows the proportional hazards model with covariate η. Assume that the regression parameter of Y1 on η is fixed to be one, E(η) = 0, and the regression parameters of η in the models of Y2 and Y3 are non-zero.
Figure 1.
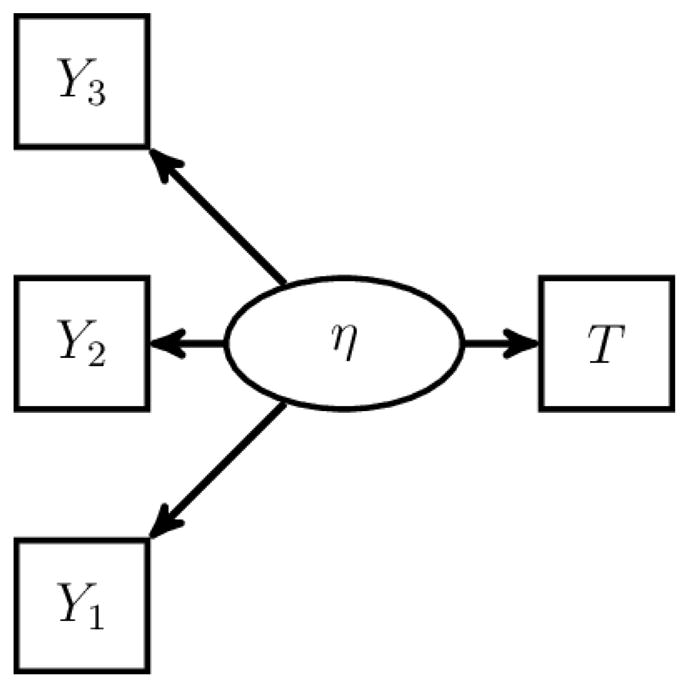
The First Example of SEM to Illustrate the Identifiability Rules. The SEM con- sists of one latent variable, one survival time, and three conditionally independent normal manifest variables.
The above model is similar to the joint model for survival and longitudinal variables. By Bollen (1989)’s three-indicator rule, the model of (Y1, Y2, Y3, η) is identifiable. With Y1 serving as an indicator of η, Rule 2 implies that the remaining parameters are identifiable. Note that Rule 1 is not applicable in this case because T does not depend on an independent covariate. In fact, the model is not identifiable without (Y1, Y2, Y3) because the scale of the baseline hazard function and the variance of η cannot be separated.
Example 2
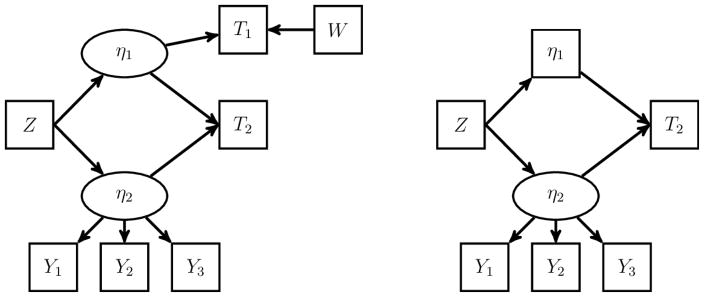
Consider the model depicted on the left-hand side of Figure 2. In the model, Y1, Y2, and Y3 are conditionally independent normal manifest variables of η2, T1 is a survival time that follows the proportional hazards model with covariates W and η1, and T2 is a survival time that follows the proportional hazards model with covariates η1 and η2. Assume that W and Z are non-constant and linearly independent, the regression parameters for the latent variables in the models of T1 and Y1 are fixed to be one, E(η1) = E(η2) = 0, and the regression parameters of W in the model of T1 and η2 in the models of Y2 and Y3 are non-zero.
Figure 2.
The Second Example of SEM to Illustrate the Identifiability Rules. The left panel is an SEM that consists of two latent variables, two observed covariates, two survival times, and three conditionally independent normal manifest variables. The right panel is an intermediate step in identifying the SEM on the left.
First, we use T1 to help identify the latent variable distributions. By Rule 1, η1 can be treated as observed when identifying the model. The problem thus reduces to identifying the model shown on the right-hand side of Figure 2. The model can then be shown identifiable by the arguments used in Example 1.
3. COMPUTATION OF THE NPMLE
In this section, we use Z to denote both W and Z with βk (k = 1, …,K) as the corresponding vector of regression parameters. Application of a transformation Gk can be viewed as inclusion of an extra latent variable log sk in the regression equation, where sk is a random variable with density gk such that . We adopt the Expectation-Maximization (EM) algorithm (Dempster et al. 1977) by treating the latent variables, including those introduced by the transformations, as missing data. We perform occasional Newton-Raphson steps to speed up the convergence.
In the combined algorithm, either an EM step or a Newton-Raphson step is performed at each iteration. To avoid confusion, we call the latter an outer Newton-Raphson step. For an EM step, note that the conditional expectation for any function φ of (ηi, si) ≡ (ηi, s1i, …, sKi) given the observed data is
where Λk {t} is the jump size of the step function Λk at t, s = (s1, …, sK), , and 𝒞 equals the above integral evaluated at φ(·, ·) = 1. We use the Gauss-Hermite quadrature to approximate the integrals. To reduce the number of abscissas, we adopt an adaptive quadrature approach (Liu and Pierce 1994). Denote the approximation of the conditional expectation as Ê(·). After taking expectation on the functions involved, we update (βk, αk,ϕk) by the one-step Newton-Raphson algorithm on
Then, we update the cumulative baseline hazard function by
where (βk, αk,ϕk) are evaluated at the current estimates. In addition, we update the remaining parameters at the maximum of
If a closed-form solution is not available, then we apply the one-step Newton-Raphson algorithm to the above expression instead. The above algorithm can be generalized to the case where components of Yi that do not appear in the model of the survival times are missing at random for some subjects. In this case, we simply drop the corresponding fY terms in the evaluation of Ê and the complete-data log-likelihood.
For an outer Newton-Raphson step, we apply the one-step Newton-Raphson algorithm directly to the logarithm of Ln(θ,𝒜) given in (4) using a similar adaptive quadrature approximation. At the current estimates, the first derivative of log Ln(θ,𝒜), i.e., the score statistic, is the same as the first derivative of the expected complete-data log-likelihood. The Hessian matrix used in the Newton-Raphson algorithm can be obtained by Louis (1982)’s formula.
To determine whether an EM step or an outer Newton-Raphson step is to be performed, we keep track of the difference in the log-likelihood at the previous iteration, either an EM or an outer Newton-Raphson step, and the difference at the previous outer Newton-Raphson step. For each iteration, if the log-likelihood difference at the previous step is too small relative to that at the previous outer Newton-Raphson step, then an outer Newton-Raphson step is performed; otherwise, an EM step is performed. Upon convergence, Louis (1982)’s formula is used to obtain the information matrix for the estimation of the standard errors.
The reason that we use a combination of the EM and Newton-Raphson algorithms instead of the Newton-Raphson algorithm alone is two-fold. First, EM steps are more stable, which is important, especially in early iterations. Second, in the estimation of the survival model under the EM algorithm, the regression parameters can be obtained by maximizing the partial-likelihood-type function, and the estimators of the baseline hazard functions take the form of the Breslow estimator. Unlike the Newton-Raphson algorithm, the EM algorithm does not involve the inversion of a high-dimensional matrix.
4. THEORETICAL PROPERTIES
4.1 Identifiability Conditions
As discussed in Section 2, we set aside K1 survival times, T1, …, TK1, that are used to identify the distribution of the underlying latent variables. We assume that span(ϕ1, …, ϕK1) = ℝK1. We can choose the K1 survival times such that each is associated with a few, preferably only one, latent variables. (K1 is allowed to be 0, in which case we rely solely on the manifest variable Y to identity the latent variable distribution.) Without loss of generality, we assume that ϕk = ek (k = 1, …, K1), where ek is a q-vector with 1 at the kth position and 0 elsewhere. This assumption can be satisfied by applying a linear transformation to the latent variables. Effectively, we fix the scale of the first K1 latent variables, as is common when establishing model identifiability for SEM. We partition η into (η1, η2), where η1 ≡ (η11, …, η1K1) consists of the first K1 components of η.
We consider the following identifiability conditions. For the “baseline” hazard functions (Λ1, …, ΛK), we only require identifiability on [0, τ], where τ denotes the study duration.
-
(C1)
If (1,WT,ZT,YT)Tc = 0 almost surely for some vector c of appropriate dimension, then c = 0. For k = 1, …,K, λk is continuous and strictly positive on [0, τ], and there exists a positive and measurable function gk such that , where mk is the Lebesgue measure or the counting measure at 1.
-
(C2)
For k = 1, …,K1, E(YTαk | Z = 0) = 0, and E(η | Z = 0) = 0. Also, for any vectors c1 and c2 of appropriate dimensions, E(eYTc1+ηTc2 | Z) is finite almost surely.
-
(C3)
For k = 1, …,K1, ϑk is non-zero, and αk and βk are zero.
-
(C4)
Consider two sets of parameters (ψ, ν) and (ψ̃, ν̃). Let fY,η1 be the density of (Y, η1) given Z. Then, fY,η1 (Y, η1 | Z; ψ, ν) = fY,η1 (Y, η1 | Z; ψ̃, ν̃) for all Z, Y, and η1 implies that ψ = ψ̃ and ν = ν̃.
-
(C5)
Let (Y+, ) be the components of (Y, η1) that appear in the regression of Tk for some k = K1 + 1, …,K, and let (Y−, ) be the remaining components. Let Fη2|Y,η1 be the distribution function of η2 given (Z,Y, η1) with (Y−, ) treated as a parameter vector. Then, η2 is complete sufficient in {Fη2|Y,η1 (· | Z,Y, η1): Z = z0,Y+ = y0, for any fixed z0, y0, and η10.
Remark 1
Condition (C1) pertains to basic requirements on the covariates, the baseline hazard functions, and the transformation functions such that the survival model with observed covariates is identifiable. If mk is a point mass at 1, then Gk is simply the identity function. Condition (C2) fixes the location parameters of the latent variables and the manifest variables that appear in the regression models of the first K1 survival times. Condition (C3) requires that the first K1 survival times depend only on their corresponding latent variable and W. The presence of a covariate besides the latent variable is necessary for distinguishing the contributions of the baseline hazard function and the latent variable to the distribution of a survival time that follows a mixture distribution. Condition (C4) requires that the model with observed (Y, η1) is identifiable. Condition (C5) requires that η2 is complete sufficient conditional on (Y, η1), where components of (Y, η1) that do not appear in the regression of Tk (k = K1 + 1, …, K) are treated as parameters, and the rest are held fixed. Conditions (C2) and (C3) are vacuous if K1 = 0, and condition (C5) is vacuous if K1 = K.
We have the following identifiability result.
Theorem 1
Under conditions (C1)–(C5), the model specified by (1)–(3) is identifiable.
Remark 2
The condition that αk and βk are zero for k = 1, …, K1 separates the first K1 survival times from the remaining observed variables that are associated with the latent variables. This condition is used to simplify the presentation of the identifiability conditions. In the proof of Theorem 1, we consider generalized versions of conditions (C3)–(C5), where αk and βk are allowed to be non-zero.
Remark 3
Theorem 1 implies that the distribution of the latent variable underlying a given survival time can be completely identified if the survival time only regresses on the latent variable and a set of independent covariates. Thus, the survival times make it easy to identify the model, as only a single survival time is enough to identify an underlying latent variable. By contrast, this property does not hold for normal random variables.
Remark 4
The derivation of model identifiability from condition (C5) utilizes the property of complete sufficient statistics. The derivation is applicable to general latent variable models; the general result is given by Lemma 1 in the Appendix. Lemma 1 allows for the establishment of model identifiability by inspecting just a part of the model. It includes Reilly and O’Brien (1996)’s side-by-side rule, which states that the loadings of an observed variable on any number of latent variables are identifiable if each of the latent variables is attached to a separate independent observed variable whose distribution is identifiable, as a special case.
4.2 Asymptotic Properties
Let d be the dimension of θ, θ0 denote the true value of θ, and Λ0k denote the true value of Λk (k = 1, …, K). We impose the following conditions.
-
(D1)
The parameter θ0 lies in the interior of a compact set Θ ⊂ ℝd, and the function Λ0k is continuously differentiable with on [0, τ] for each k = 1, …,K.
-
(D2)
With probability one, P(T̃ki = τ | W,Z) > δ0 (k = 1, …,K) for some fixed δ0 > 0.
-
(D3)Consider any fixed Z and (ψ, ν) ∈ Θψν, where Θψν consists of the (ψ, ν)-component of every θ ∈ Θ. For any constant a1 > 0 and δ = 0, 1,Also, for j = 1, 2, 3, there exists a constant a2 > 0 such thatIn addition, for some positive constants Mj and cj, Nj ∈ ℝr, and φ1 ∈ ℓ∞(ℝr),
where b = S (η) for some one-to-one linear transformation S.
-
(D4)The function Gk is four-times differentiable, Gk (0) = 0, and for some positive constants κ1k, κ2k, K1k, and K2k. Also, Gk(x)/xρk → Mk or Gk(x)/log(x) → Mk as x → ∞ for some positive constants Mk and ρk. In addition, exp {−Gk(x)} ≤ μk (1 + x)−κ3k for some μk and κ3k > κ2k + 1. Furthermore, for some rk,
where and denote the second and jth derivatives of Gk, respectively.
-
(D5)Let (Z(k),Y(k)) be the components of (Z,Y) that appear in the regression of Tk (k = 1, …,K1). For any vectors h1, h2k, and h3k of appropriate dimensions, if
is equal to 0 for all Z, Y, and η1, then h1 = 0, h2k = 0, and h3k = 0 for k = 1, …,K1, where fY,η1 is defined in condition (C4).
Remark 5
Conditions (D1)–(D4) are similar to the conditions of Zeng and Lin (2010) for joint modeling of longitudinal and survival data. Extra conditions are imposed on the transformations and the distributions of Y and η to accommodate the presence of unbounded covariate Y in the survival model. Condition (D5) is for the invertibility of the information operator. If αk = 0 and βk = 0 for k = 1, …, K1, then condition (D5) simply requires that the information matrix of the model for (Y, η1) is invertible. This is parallel to condition (C4) for identifiability.
Remark 6
The conditions for identifiability and the invertibility of the information operator (C1)–(C5), and (D5) differ significantly from the corresponding conditions (C5) and (C7) of Zeng and Lin (2010). The latter are stated under very general settings, but they are hard to verify for specific models, especially under our SEM framework. By contrast, our conditions are easier to verify and have intuitive interpretations. For the model in Example 1, (D5) simply requires that the model of (Y1, Y2, Y3, η1) given Z has a non-zero score statistic, which clearly holds.
Let 𝒜0 = (Λ01, …,Λ0K) and (θ̂, 𝒜̂) be the NPMLE of (θ, 𝒜). Also, let 𝒱 = {v ∈ ℝd, |v| ≤ 1} and 𝒬 = {h (t): ||h (t)||V [0,τ] ≤ 1} with ||·||V [0,τ] being the total variation norm on [0, τ]. We consider (θ̂ − θ0, 𝒜̂ − 𝒜0) as a random element in l∞(𝒱 × 𝒬K) with
We have the following results.
Theorem 2
Under conditions (C1)–(C5) and (D1)–(D5),
; and
n1/2(θ̂ − θ0, 𝒜̂ − 𝒜0) →d 𝒢 in l∞(𝒱 × 𝒬K), where 𝒢 is a continuous zero-mean Gaussian process. Furthermore, the limiting covariance matrix of n1/2(θ̂ − θ0) attains the semiparametric efficiency bound.
Remark 7
The proof of Theorem 2 relies on the Donsker properties of certain classes of functions. It is more challenging to establish the Donsker results in our setting than in previous settings (e.g., Kosorok et al. (2004) and Zeng and Lin (2010)) because the likelihood function of the proposed model may contain the unbounded variable Y.
Remark 8
A key step in proving the asymptotic normality of the NPMLE is to show that the information operator is invertible. The result is given by Lemma 2 in the Appendix, which states that condition (D5), together with conditions (C1)–(C3), and (C5), implies that the information operator of the model is invertible. With this result, we can verify the invertibility of the information operator of the semiparametric model by inspecting the parametric part of the model that contains the observed and latent variables. For the frailty models in Kosorok et al. (2004), verification of the invertibility of the information operator involves inspection of the local behavior of the score around T = 0. However, that approach is limited to frailty distributions that are indexed by a one-dimensional parameter and is not directly applicable to cases with more complex latent variable distributions such as those in our setting.
5. SIMULATION STUDIES
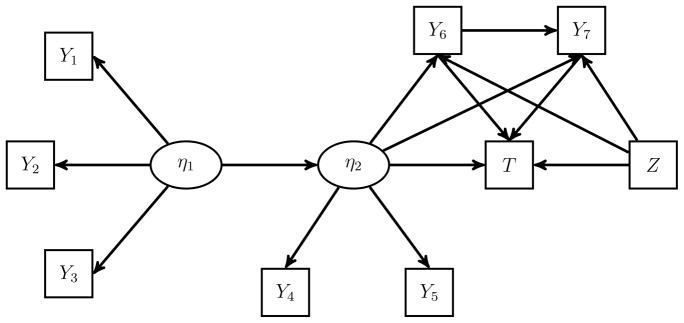
We considered a model with covariates Z = (Z1,Z2)T, two latent variables (η1, η2), observed continuous variables (Y1, …, Y5), observed binary variables (Y6, Y7), and a survival time T. Their distributions are given by
The parameters ϕY1 and ϕY4 are fixed to be one. The model is depicted in Figure 3.
Figure 3.
Model Used in Simulation Studies. The SEM consists of two latent variables, an observed covariate, seven binary or normal manifest variables, and a survival time that regresses on the latent variable, some manifest variables, and the observed covariates.
We set Z1 and Z2 to independent standard normal and Bernoulli(0.5), respectively, and Λ0(t) = t2. We considered the class of logarithmic transformations G(x) = r−1 log(1 + rx) with r = 0 or 1, which correspond to the proportional hazards and proportional odds models, respectively. We generated the censoring times from Exp(c), where c was chosen to yield approximately 30% censored observations. We set (Y1, …, Y5) to be missing completely at random for 30% of the subjects. We set the sample size to 400 and set the number of abscissa points to 20 for each Gauss-Hermite quadrature. We simulated 1,000 datasets for each setting. The results are summarized in Table 1.
Table 1.
Simulation Results for the SEM with Two Latent Variables.
| Dep | Ind | Proportional Hazards Model
|
Proportional Odds Model
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Param | Bias | SE | SEE | CP | Param | Bias | SE | SEE | CP | ||
| T | Z1 | 0.100 | 0.004 | 0.075 | 0.073 | 0.95 | 0.100 | 0.004 | 0.109 | 0.107 | 0.94 |
| Z2 | −0.200 | −0.004 | 0.180 | 0.182 | 0.95 | −0.200 | 0.008 | 0.264 | 0.270 | 0.95 | |
| Y6 | 0.100 | 0.001 | 0.145 | 0.143 | 0.95 | 0.100 | 0.000 | 0.214 | 0.208 | 0.95 | |
| Y7 | 0.200 | −0.007 | 0.167 | 0.163 | 0.95 | 0.200 | −0.007 | 0.233 | 0.236 | 0.95 | |
| η2 | 0.500 | 0.020 | 0.167 | 0.166 | 0.96 | 0.500 | 0.024 | 0.225 | 0.223 | 0.96 | |
| Λ0(t1) | 0.202 | 0.003 | 0.050 | 0.049 | 0.95 | 0.212 | 0.004 | 0.073 | 0.071 | 0.95 | |
| Λ0(t2) | 0.518 | 0.010 | 0.121 | 0.118 | 0.95 | 0.608 | 0.019 | 0.204 | 0.201 | 0.94 | |
| Λ0(t3) | 1.103 | 0.042 | 0.258 | 0.251 | 0.95 | 1.588 | 0.088 | 0.550 | 0.542 | 0.95 | |
| Y1 | Int | 0.000 | 0.000 | 0.074 | 0.073 | 0.94 | 0.000 | 0.001 | 0.074 | 0.073 | 0.94 |
| Var | 1.000 | −0.015 | 0.132 | 0.128 | 0.96 | 1.000 | −0.016 | 0.132 | 0.128 | 0.97 | |
| Y2 | Int | 0.000 | 0.002 | 0.076 | 0.073 | 0.94 | 0.000 | 0.002 | 0.076 | 0.073 | 0.94 |
| η1 | 1.000 | 0.022 | 0.220 | 0.204 | 0.94 | 1.000 | 0.021 | 0.218 | 0.204 | 0.94 | |
| Var | 1.000 | −0.013 | 0.130 | 0.129 | 0.96 | 1.000 | −0.012 | 0.129 | 0.129 | 0.96 | |
| Y3 | Int | 0.000 | 0.001 | 0.076 | 0.073 | 0.95 | 0.000 | 0.002 | 0.076 | 0.073 | 0.95 |
| η1 | 1.000 | 0.021 | 0.214 | 0.203 | 0.95 | 1.000 | 0.021 | 0.215 | 0.204 | 0.96 | |
| Var | 1.000 | −0.010 | 0.132 | 0.128 | 0.96 | 1.000 | −0.010 | 0.133 | 0.129 | 0.96 | |
| Y4 | Int | 0.000 | −0.001 | 0.077 | 0.075 | 0.95 | 0.000 | −0.001 | 0.077 | 0.076 | 0.95 |
| Var | 1.000 | −0.026 | 0.193 | 0.184 | 0.96 | 1.000 | −0.032 | 0.215 | 0.210 | 0.96 | |
| Y5 | Int | 0.000 | −0.002 | 0.078 | 0.076 | 0.94 | 0.000 | −0.001 | 0.079 | 0.076 | 0.94 |
| η2 | 1.000 | 0.048 | 0.296 | 0.276 | 0.94 | 1.000 | 0.057 | 0.339 | 0.323 | 0.95 | |
| Var | 1.000 | −0.031 | 0.195 | 0.187 | 0.96 | 1.000 | −0.036 | 0.219 | 0.214 | 0.96 | |
| Y6 | Int | 0.000 | −0.005 | 0.286 | 0.273 | 0.94 | 0.000 | −0.006 | 0.286 | 0.273 | 0.94 |
| Z1 | −0.500 | 0.001 | 0.112 | 0.113 | 0.95 | −0.500 | 0.001 | 0.112 | 0.113 | 0.95 | |
| Z2 | 0.500 | 0.021 | 0.307 | 0.297 | 0.95 | 0.500 | 0.021 | 0.307 | 0.297 | 0.95 | |
| η2 | 0.000 | 0.008 | 0.226 | 0.220 | 0.96 | 0.000 | 0.007 | 0.228 | 0.221 | 0.97 | |
| Y7 | Int | 1.000 | 0.032 | 0.338 | 0.345 | 0.96 | 1.000 | 0.030 | 0.337 | 0.345 | 0.96 |
| Z1 | 1.000 | 0.028 | 0.148 | 0.148 | 0.96 | 1.000 | 0.028 | 0.148 | 0.148 | 0.96 | |
| Z2 | 0.200 | 0.002 | 0.333 | 0.342 | 0.96 | 0.200 | 0.002 | 0.333 | 0.342 | 0.96 | |
| Y6 | −0.200 | −0.011 | 0.264 | 0.264 | 0.95 | −0.200 | −0.009 | 0.264 | 0.264 | 0.95 | |
| η2 | 0.000 | 0.006 | 0.262 | 0.253 | 0.97 | 0.000 | 0.008 | 0.266 | 0.255 | 0.97 | |
| η1 | Var | 0.500 | 0.009 | 0.139 | 0.134 | 0.95 | 0.500 | 0.009 | 0.138 | 0.134 | 0.95 |
| η2 | η1 | 0.500 | 0.008 | 0.156 | 0.147 | 0.93 | 0.500 | 0.008 | 0.160 | 0.151 | 0.94 |
| Var | 0.500 | 0.007 | 0.175 | 0.168 | 0.96 | 0.500 | 0.013 | 0.194 | 0.191 | 0.97 | |
NOTE: Each row corresponds to the regression parameter of the dependent variable “Dep” on the independent variable “Ind” or some other parameter in the model of “Dep”. “Int” and “Var” stand for the intercept and error variance, respectively. The parameters Λ0(t1), Λ0(t2), and Λ0(t3) correspond to the cumulative baseline hazard function values at the 25%, 50%, and 75% quantiles of the survival time. The true value of a parameter is given under “Param”. “Bias” is the empirical bias; “SE” is the empirical standard error; “SEE” is the empirical mean of the standard error estimator; and “CP” is the empirical coverage probability of the 95% confidence interval.
The estimators of all parameters are virtually unbiased for both the proportional hazards and proportional odds models. The standard error estimators accurately reflect the true variations, and the coverage probabilities of the confidence intervals are close to the nominal level. Standard error estimators for the parameters in the survival model are larger under the proportional odds model than under the proportional hazards model. As a result, the standard error estimators for the parameters associated with η2 are larger. The standard error estimators for the remaining parameters are very similar between the two models.
We also evaluated Mplus (Muthén and Muthén 1998–2015) under the proportional hazards model, and the results are presented in Table S.1 of the Supplementary Materials. The results for the Euclidean parameters are similar to those presented in Table 1. Mplus provides estimator for the baseline hazard function instead of the cumulative baseline hazard function. Its standard error estimator does not reflect the true variation, and the coverages of the confidence intervals are far below the nominal level.
6. REAL DATA ANALYSIS
We analyzed a dataset on patients with serous ovarian cancer from the TCGA project (The Cancer Genome Atlas Research Network 2011). Genomic variables include DNA copy number, SNP genotype, DNA methylation level, and levels of expression of mRNA, microRNA, total protein, and phosphorylated protein. Demographic and clinical variables include age at diagnosis, race, tumor stage, tumor grade, time to tumor progression, and time to death. There are a total of 586 patients. The median follow-up time was about 2.5 years, and roughly 30% of the patients were lost to follow-up before tumor progression or death. The data are available from http://gdac.broadinstitute.org/.
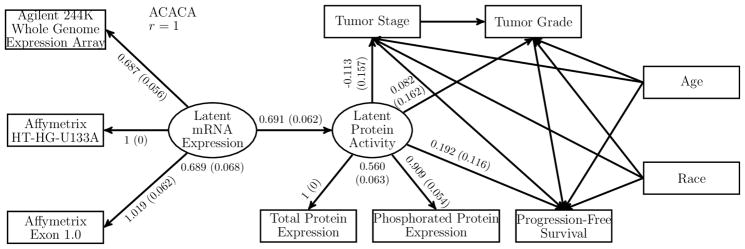
We focused on the integrative analysis of clinical outcomes and expression levels of mRNA, total protein, and phosphorylated protein. We considered mRNA expression as a latent variable that can only be observed with error through three microarray platforms, namely Agilent 244K Whole Genome Expression Array, Affymetrix HT-HG-U133A, and Affymetrix Exon 1.0. We assumed that the effects of a gene on clinical outcomes are mediated through unobserved protein activity. The latent protein activity is modified by mRNA expression and is manifest through the observed protein expression measurements, which were obtained from the reverse-phase protein arrays platform. Figure 4 depicts the SEM fit for each gene. We assumed that the observed variables follow the distributions described in Section 5, with (Y1, Y2, Y3) being the three microarray measurements, (Y4, Y5) = (Total protein expression, Phosphorylated protein expression), (Y6, Y7) = (Tumor stage, Tumor grade), (Z1, Z2) = (Age, Race), and T being progression-free survival time.
Figure 4.
Results from the SEM Analysis of the Gene ACACA. Analysis results are from 542 patients with ovarian cancer in the TCGA project. The numbers besides an arrow correspond to the point estimate and standard error estimate (in parentheses) of the regression parameter. The numbers below the latent variables correspond to the point estimate and standard error estimate (in parentheses) of the error variance.
We dichotomized tumor stage into stage II/III versus stage IV and tumor grade into grade 2 versus grade 3/4. Race was dichotomized into white and non-white. We allowed mRNA expression and protein expression data to be missing for some subjects. We excluded patients with tumor stage I or grade 1, as those patients may have a disease that is biologically different from that of patients with tumors of other stages or grades. For each gene, we fit the class of transformation models with G(x) = r−1 log(1 + rx) over a grid of r = (0, 0.1, …, 2). We selected the model with the smallest AIC or, equivalently, the largest log-likelihood value.
We present the results for the gene ACACA. The sample size is 542. About 30% of the subjects do not have protein expression data, and over 10% of the subjects miss at least one mRNA expression measurement. The best-fitting model is obtained at r = 1, which corresponds to the proportional odds model. The point estimates and standard error estimates of the parameters associated with the latent variables are shown in Figure 4. The remaining results are shown in Table S.2 of the Supplementary Materials. The latent variables have strong positive association with the measurement platforms. As expected, latent protein activity and latent mRNA expression are highly correlated. Latent protein activity is positively associated with progression-free survival time, with a p-value of 0.100. Specifically, higher latent protein activity is associated with shorter progression-free survival time, which agrees with the findings of the literature (Menendez and Lupu 2007). The association of ACACA with tumor stage or tumor grade is weak.
The results for the parameters in the non-survival models are similar between r = 0 and 1. The parameters in the survival model have different interpretations between r = 0 and 1. With r = 0, a unit increase in latent protein activity would have a multiplicative effect of exp(0.068) on the hazard function. With r = 1, a unit increase in the latent protein activity would have a multiplicative effect of exp(−0.192) on the survival odds. For this dataset, the proportional odds model provides much stronger evidence for the effect of protein activity on progression-free survival than the proportional hazards model.
For the Cox proportional hazards model, we also present the results from Mplus in Table S.2. The results from NPMLE and Mplus are similar for most parameters. There are considerable differences between the cumulative baseline hazard function estimates. The standard error estimates for the cumulative baseline hazard function are not available from Mplus.
For comparisons, we also fit a proportional odds model without latent variables for progression-free survival on the covariates and the two protein expression variables, where the subjects with missing protein expression data were discarded. The p-value of the Wald test for the joint effect of protein expression is 0.157. With r = 0, the Wald test p-value is 0.578. Therefore, analyses based on standard models fail to conclude a strong association between the protein expression and progression-free survival. The power of the proposed SEM framework stems from the appropriate handling of missing data, the dimension reduction of the observed covariates, and the flexibility of the survival model.
7. DISCUSSION
In this article, we consider semiparametric SEM for potentially right-censored survival time data. We prove the consistency, asymptotic normality, and semiparametric efficiency of the NPMLE. We propose new rules for establishing model identifiability and invertibility of the information operator. We construct an EM algorithm to compute the NPMLE and introduce occasional Newton-Raphson steps to accelerate the convergence.
One contribution of Theorem 1 is that it reduces a semiparametric identifiability problem to a parametric one; it shows that the inclusion of the semiparametric component does not make the model less identifiable but, in some sense, makes the model more easily identifiable. With that being said, the result hinges on correct specification of the model and does not guarantee empirical identifiability in a finite sample. Therefore, care should be taken when fitting a model that is nearly non-identifiable. Another main result of ours is given by Lemma 1. This lemma is applicable to a wide range of latent variable models and allows one to deduce the identifiability of a model by inspecting just part of it.
Invertibility of the information operator has received much less attention in the literature than model identifiability. In this article, we prove a general result for invertibility of the information operator. It is evident from the proof that the invertibility of the information operator can be established using techniques similar to those used to establish model identifiability. Specifically, the key to the proof of the identifiability of the mixture Cox model is that with the presence of a covariate that is independent of the latent variable, the contributions to the likelihood from the latent variable and the baseline hazard function can be separated by considering different values of the covariate. (In a normal mixture model, however, we lack such identifiability results precisely because the random effect and error term are combined linearly and their distributions cannot be distinguished.) As a result, if two sets of parameters give rise to the same marginal survival function, then they must do so by giving rise to the same random-effect distribution. Based on the proportional hazards structure, we prove a parallel result for the invertibility of the information operator: the existence of a submodel with zero score implies that the random-effect distribution has zero score along that submodel as well. Therefore, to ensure the invertibility of the information operator of the mixture Cox model, one only has to ensure that the information matrix of the random-effect distribution is invertible.
Our work can be extended in several directions. First, one may be interested in expanding the model by inclusion of more latent and observed variables. As the number of variables increases, the number of parameters to be estimated increases as well. Then, it may be desirable to perform variable selection. Because a single variable may be associated with multiple parameters, one may prefer not to treat parameters as the basic unit of selection, as in traditional lasso methods (Tibshirani 1996). Instead, methods like group lasso (Yuan and Lin 2006) that penalize parameters associated with a variable as a group may be considered.
In our model, the distribution of the manifest variable Y is fully parametric. One can allow a nonparametric transformation on Y. A major challenge arises in extending the asymptotic results to unbounded nonparametric transformation, as the estimator of the transformation function can be unbounded (Zeng and Lin 2010).
Finally, it would be of interest to consider interval-censored data. Interval censoring results in a different likelihood function, which makes the computation of the NPMLE and the derivation of its asymptotic properties challenging, even for univariate survival time data. The asymptotic theory for interval-censored data is only available in a few simple cases; see Huang and Wellner (1997) for a review.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health grants R01GM047845, R01CA082659, and P01CA142538.
APPENDIX: TECHNICAL DETAILS
We present the following conditions, which are clearly implied by conditions (C3)–(C5):
-
(C3′)
For k = 1, …, K1, ϑk is a non-zero vector.
-
(C4′)
Consider two sets of parameters (αk, βk, ψ, ν) and (α̃k, β̃k, ψ̃, ν̃) for k = 1, …, K1, and let η̃1 = (η̃11, …, η̃1K1), where η̃1k = YT(αk−α̃k)+ZT(βk−β̃k)+η1k. Let fY, η1 be the density of (Y, η1) given Z. Then, fY, η1 (Y, η1 | Z; ψ, ν) = fY, η1 (Y, η̃1 | Z; ψ̃, ν̃) for all Z, Y, and η1 implies that αk = α̃k, βk = β̃k, ψ = ψ̃, and ν = ν̃.
-
(C5′)
For k = K1 + 1, …, K, let (Y(k), ) be the components of (Y, η1, η2) that appear in the regression of Tk, and let (Y−(k), ) be the remaining components. If is non-empty, then (Y−(k), ) is non-empty, and is complete sufficient in { } for any fixed z0, y0, and η10, where is the distribution function of given (Z,Y, η1) with (Y−(k), ) treated as a parameter vector.
We prove Theorem 1 under the generalized conditions (C1), (C2), and (C3′)-(C5′). The proof makes use of two lemmas given at the end of this appendix. We first provide an overview of the proof. For any two sets of parameters (ϑk, αk, βk, ϕk, Λk, ψ, ν) and (ϑ̃k, α̃k, β̃k, ϕ̃k, Λ̃k, ψ̃, ν̃), assume that the likelihood values at the two sets of parameters are identical almost surely. By definition, the model is identifiable if the equality of the likelihood values implies the equality of the two sets of parameters. We derive the equality of the two sets of parameters in the following steps:
By conditions (C1), (C2), and (C3′) and the identifiability of the mixture Cox model (Kortram et al. 1995), ϑk = ϑ̃k and Λk = Λ̃k for k = 1, …, K1.
With some algebraic manipulation, the likelihood function can be expressed in the form of the Laplace transform of the distribution of a function of (Y, η1). The uniqueness of the Laplace transform, together with condition (C4′), implies that (αk, βk, ψ, ν) = (α̃k, β̃k, ψ̃, ν̃) for k = 1, …, K1.
By the uniqueness of the Laplace transform and the complete sufficiency of η2 imposed by condition (C5′), the equality of the likelihood functions of (TK1+1, …, TK, Y) implies the equality of the likelihood functions of (TK1+1, …, TK, Y, η). By the identifiability of the Cox model, we conclude that (ϑk, αk, βk, ϕk) = (ϑ̃k, α̃k, β̃k, ϕ̃k) for k = K1 + 1, …, K.
Proof of Theorem 1
The likelihood is given in (4). Here, we consider a single observation and drop the subscript i. Using the arguments in Section 10.1 of Zeng and Lin (2010), we can set each survival time to be right censored at any time point within [0, τ] when establishing identifiability. Consider two sets of parameters (ϑk, αk, βk, ϕk, Λk, ψ, ν) and (ϑ̃k, α̃k, β̃k, ϕ̃k, Λ̃k, ψ̃, ν̃) such that the likelihood values for an observation with the K survival times being right censored are equal almost surely, i.e.,
| (A.1) |
for all t1, …, tK ∈ [0, τ], W, Z, and Y, where fY,η is the density of (Y, η) given Z. If mk is a point mass at one, then sk is fixed at one, gk = 1, and the integration with respect to mk(sk) can be omitted. For simplicity of description, assume that mk is the Lebesgue measure. Note that
Thus, a transformation model can be written as a random-effect proportional hazards model with known distributions (g1, …, gK) for random effects (s1, …, sK) with finite means.
First, we show that the baseline hazard functions of the first K1 survival times are identifiable. For each k = 1, …, K1, set tl → 0 for l ≠ k on both sides of (A.1). On each side of the resulting equation, integration with respect to Y results in the likelihood of a mixture Cox model with skeYTαk+η1k or skeYTα̃k+η1k as a latent variable. Let E(· | Z) and Ẽ(· | Z) be the expectations under fY,η(· | Z; ψ, ν) and fY,η(· | Z; ψ̃, ν̃), respectively. Theorem 3 of Kortram et al. (1995) implies that E(skeYTαk+η1k | Z = 0)Λk = Ẽ(skeYTα̃k+η1k | Z = 0)Λ̃k on [0, τ], ϑk = ϑ̃k, and the distribution of E(skeYTαk+η1k | Z = 0)−1skeYTαk+η1k under fY,η(· | Z;ψ, ν) is equal to that of Ẽ(skeYTα̃k+η1k | Z = 0)−1skeYTα̃k+η1k under fY,η(· |Z; ψ̃, ν̃). Because E(YTαk + η1k | Z = 0) = Ẽ(YT α̃k + η1k| Z = 0) = 0 by condition (C2), we see that Λk = Λ̃k on [0, τ].
Second, we show that the likelihood function takes the form of a Laplace transform and use the uniqueness of the Laplace transform to prove the identifiability of (αk, βk, ψ, ν) (k = 1, …, K1). Setting tk → 0 for k = K1 + 1, …, K and W = 0 on both sides of (A.1), we have
| (A.2) |
Let U = (U1, …, UK1), Uk = skeη1k, and fU|Y be the density function of U given Z and Y. By the uniqueness of the Laplace transform, for any continuous functions f and f̃, any open set 𝒮, and any positive real numbers c and c̃,
implies that f(t) = (c/c̃)f̃(ct/c̃) for all t > 0. Therefore, the equality of (A.2) for all t1, …, tK1, Z, and Y implies that
| (A.3) |
for all U, Z, and Y, where Ũ = (Ũ1, …, ŨK1), and Ũk = eZT(βk−β̃k)+YT(αk−α̃k)Uk. Let fη1|Y be the density of η1 given Z and Y. By the definition of U,
where ḡk(v) = gk(ev). Thus, (A.3) implies that
| (A.4) |
Consider two arbitrary continuous functions f, g: ℝ → ℝ. Note that
for any s such that the integrals are defined, where is the convolution of f and g. Therefore, (f * g)(·) = 0 implies that
which, if g is positive, implies that f(·) = 0 by the uniqueness of the bilateral Laplace 27 transform (Chareka 2007). Because ḡk(·)e(·) is positive, (A.4) implies that fη1|Y (η1 | Z,Y; ψ, ν) = fη1|Y (η̃1 | Z,Y; ψ̃, ν̃), where η̃1 is defined in condition (C4′). By condition (C4′), (αk, βk, ψ, ν) = (α̃k, β̃k, ψ̃, ν̃) for k = 1, …, K1.
It remains to identify the parameters associated with (TK1+1, …, TK). By the uniqueness of the Laplace transform, (A.1) implies that
for all tK1+1, …, tK, W, Z, Y, and η1, i.e., η1 can be treated as observed for identifying the remaining parameters. Under condition (C5′), we can use the arguments in the proof of Lemma 1 to show that the integrands in the above equality are equal at each value of η2. We conclude that (ϑk, αk, βk, ϕk, Λk) = (ϑ̃k, α̃k, β̃k, ϕ̃k, Λ̃k) for k = K1+1, …, K.
We provide an overview for the proof of Theorem 2. The consistency of the NPMLE is proved in the following steps:
By conditions (D2)–(D4), the NPMLE exists, i.e., Λ̂k(τ) < ∞.
By conditions (D3) and (D4), Λ̂k(τ) is uniformly bounded. Helly’s selection theorem then implies that every subsequence of Λ̂k has a further converging subsequence.
By the Glivenko-Cantelli properties of the log-likelihood and related functions given by Lemma S2 in the Supplementary Materials, the identifiability of the model, and the non-negativity of the Kullback-Leibler divergence, we conclude the consistency of the NPMLE.
The asymptotic normality of the NPMLE follows mainly from the arguments of van der Vaart (1998, pp. 419–424). Donsker properties of the score and related functions are given by Lemma S2 in the Supplementary Materials, and the invertibility of the information operator is given by Lemma 2.
Proof of Theorem 2
We use Z to denote both W and Z with βk (k = 1, …, K) being the corresponding vector of regression parameters. Let
Ψ̇θ(𝒪i; θ, 𝒜) be the derivative of Ψ(𝒪i; θ, 𝒜) with respect to θ, and Ψ̇k(𝒪i; θ, 𝒜)[Hk] be the derivative of Ψ(𝒪i; θ, 𝒜) along the path (Λk + εHk).
First, we prove the consistency. By condition (D4),
Thus, condition (D3) implies that
| (A.5) |
where ℱ(𝒪i; θ) is a random variable with |E{log ℱ(𝒪i; θ)}| < ∞ for any θ. By condition (D2), P(T̃ki = τ) is positive. Therefore, if Λk (τ) = ∞, then the right-hand side of (A.5) is zero for large n. We conclude that Λ̂k (τ) < ∞, such that the NPMLE exists.
We then show that lim supnΛ̂k (τ) < ∞ almost surely. From (A.5),
Let . Clearly,
The second term on the right-hand side of the above equation is Op (1). Thus,
Note that (κ3k − κ2k − 1) is positive by condition (D4). Using the partitioning argument similar to those of Murphy (1994) and Parner (1998), we can show that the right-hand side of the above inequality tends to −∞ if lim supn Λ̂k (τ) = ∞. By definition of (θ̂, 𝒜̂), the left-hand side of the inequality is bounded below by an Op(1) term. Therefore, Λ̂k (τ) is uniformly bounded.
Given the boundedness of Λ̂k (τ), Helly’s selection theorem implies that, for any subsequence of n, we can always choose a further subsequence such that Λ̂k converges pointwise to some monotone function and θ̂ converges to θ*. The desired consistency result follows if we can show that and θ* = θ0 almost surely. With an abuse of notation, let {n}1,2, … be the subsequence. Define
By Lemma S2 in the Supplementary Materials and the properties of Donsker (and therefore, Glivenko-Cantelli) classes,
uniformly on [0, τ]. Because the score function along the path Λk = Λ0k + εI (· ≥ s) with other parameters fixed at their true values has zero expectation,
Algebraic manipulation yields that the uniform limit of Λ̃k on [0, τ] is Λ0k. Note that
We have shown that the numerator of the integrand in the above equation converges uniformly. Similarly, we can show that the denominator of the integrand in the above equation converges uniformly to |E{Ψ̇k(𝒪i; θ*,𝒜*)[I (s ≤ ·)]/Ψ(𝒪i; θ*,𝒜*)}| and that the limit is bounded away from 0. Because Λ̃k converges uniformly to Λ0k, which is differentiable with respect to t, is also differentiable with respect to t. It follows that dΛ̂k/dΛ̃k converges uniformly to on [0, τ], where . As n−1 log Ln(θ̂, 𝒜̂) − n−1 log Ln(θ0, 𝒜̃) is non-negative,
By the Glivenko-Cantelli properties of the class of functions of log Ψ(𝒪i; θ, 𝒜) given by Lemma S2 and the uniform convergence of dΛ̂k/dΛ̃k, setting n → ∞ on both sides of the above inequality yields
The left-hand side of the above inequality is the negative Kullback-Leibler distance of the density indexed by (θ*,𝒜*). From the identifiability of the model implied by Theorem 1, we conclude that θ* = θ0 and . The desired consistency result follows.
To prove the asymptotic normality of the NPMLE, we adopt the arguments of van der Vaart (1998, pp. 419–424). Let ℘n be the empirical measure determined by n i.i.d. observations, and let ℘ be the true probability measure. Let ℓ̇θ(θ, 𝒜) be the derivative of log Ln(θ, 𝒜) with respect to θ, and let ℓ̇k(θ, 𝒜)[Hk] be the derivative of log Ln(θ, 𝒜) along the path (Λk + εHk). For any v ∈ ℝd and 𝒲 = (h1, …, hK) with hk ∈ BV [0, τ], where BV [0, τ] is the space of functions of bounded variation on [0, τ], we have
In addition,
Therefore,
| (A.6) |
From the Donsker properties of the classes of functions of ℓ̇θ and ℓ̇k implied by Lemma S2 and the consistency of θ̂ and 𝒜̂, we conclude that the left-hand side of (A.6) equals
This term converges to a Gaussian process in l∞(𝒱 ×𝒬K). By the Taylor series expansion, the right-hand side of (A.6) is of the form
where ℬ ≡ (B1,B21, …,B2K) is the information operator and is linear in ℝd ×BV [0, τ]K. By Lemma 2, ℬ is invertible. The rest of the proof then follows the arguments of van der Vaart (1998, pp. 419–424). Finally, because vTθ̂ is an asymptotically linear estimator of vTθ0 with the influence function lying in the space spanned by the score functions, θ̂ is an efficient estimator for θ0.
The following two lemmas are used in the proofs of Theorem 1 and Theorem 2 and are proved in Section S2 of the Supplementary Materials.
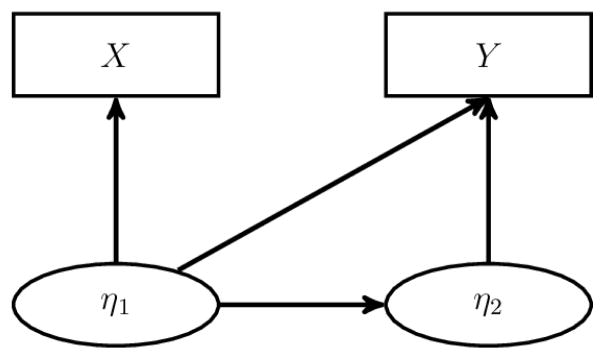
Lemma 1
Let Model A be
where (X,Y) are observed, and (η1, η2) are latent. Model A is depicted in Figure A.1. Let , and . Assume that: (a) for any density functions f̃Y |η and f̃η2|η1,
implies that (fY |η, fη2|η1) = (f̃Y |η, f̃η2|η1), i.e., the model for Y is identifiable if η1 is observed, (b) FX|η1 and Fη1 are identifiable based on (X,Y), and (c) η1 is a complete sufficient statistic in {Fη1|X(· | X) : X ∈ 𝒳}, where Fη1|X is the conditional distribution function of η1 given X, and 𝒳 is the range of X. Then, Model A is identifiable. A sufficient condition for η1 to be complete sufficient is that the density of X is of the form
where X = (X1, …,Xq), η1 ↦ (s1(η1), …, sq(η1)) is one-to-one, and bj is non-zero on some open set.
Figure A.1.
SEM Considered in Lemma 1. The SEM consists of two sets of latent variables and two sets of observed variables that may all be multivariate. The observed variable X depends only on the latent variable η1, but the observed variable Y depends on both sets of latent variables.
Lemma 2
Under conditions (C1), (C2), (C3′), (C5′), and (D5), the model given by (1)–(3) has an invertible information operator.
Footnotes
The supplementary materials contain proofs of Lemmas 1 and 2, additional theoretical results, simulation results for Mplus, and additional data analysis results.
References
- Asparouhov T, Masyn K, Muthen B. Continuous Time Survival in Latent Variable Models. ASA Section on Biometrics; Proceedings of the Joint Statistical Meeting; Seattle. August 2006; 2006. pp. 180–187. [Google Scholar]
- Bollen KA. Structural Equations With Latent Variables. New York: Wiley; 1989. [Google Scholar]
- Bollen KA, Davis WR. Two Rules of Identification for Structural Equation Models. Structural Equation Modeling. 2009;16:523–536. [Google Scholar]
- Chareka P. A Finite-Interval Uniqueness Theorem for Bilateral Laplace Transforms. International Journal of Mathematics and Mathematical Sciences. 2007;2007 [Google Scholar]
- Cox DR. Regression Models and Life-Tables. Journal of the Royal Statistical Society: Series B. 1972;34:187–220. [Google Scholar]
- Dahly D, Adair L, Bollen K. A Structural Equation Model of the Developmental Origins of Blood Pressure. International Journal of Epidemiology. 2009;38:538–548. doi: 10.1093/ije/dyn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum Likelihood From Incomplete Data via the EM Algorithm. Journal of the Royal Statistical Society: Series B. 1977;39:1–38. [Google Scholar]
- Henderson R, Diggle P, Dobson A. Joint Modelling of Longitudinal Measurements and Event Time Data. Biostatistics. 2000;1:465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]
- Huang J, Wellner JA. Interval Censored Survival Data: a Review of Recent Progress. In: Lin DY, Fleming TR, editors. Proceedings of the First Seattle Symposium in Biostatistics. New York: Springer; 1997. pp. 123–169. [Google Scholar]
- Kortram RA, van Rooij ACM, Lenstra AJ, Ridder G. Constructive Identification of the Mixed Proportional Hazards Model. Statistica Neerlandica. 1995;49:269–281. [Google Scholar]
- Kosorok MR, Lee BL, Fine JP. Robust Inference for Univariate Proportional Hazards Frailty Regression Models. The Annals of Statistics. 2004;32:1448–1491. [Google Scholar]
- Larsen K. Joint Analysis of Time-to-Event and Multiple Binary Indicators of Latent Classes. Biometrics. 2004;60:85–92. doi: 10.1111/j.0006-341X.2004.00141.x. [DOI] [PubMed] [Google Scholar]
- Larsen K. The Cox Proportional Hazards Model With a Continuous Latent Variable Measured by Multiple Binary Indicators. Biometrics. 2005;61:1049–1055. doi: 10.1111/j.1541-0420.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Pierce DA. A Note on Gauss-Hermite Quadrature. Biometrika. 1994;81:624–629. [Google Scholar]
- Louis TA. Finding the Observed Information Matrix When Using the EM Algorithm. Journal of the Royal Statistical Society: Series B. 1982;44:226–233. [Google Scholar]
- Menendez JA, Lupu R. Fatty Acid Synthase and the Lipogenic Phenotype in Cancer Pathogenesis. Nature Reviews Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Moustaki I, Steele F. Latent Variable Models for Mixed Categorical and Survival Responses, With an Application to Fertility Preferences and Family Planning in Bangladesh. Statistical Modelling. 2005;5:327–342. [Google Scholar]
- Murphy SA. Consistency in a Proportional Hazards Model Incorporating a Random Effect. The Annals of Statistics. 1994;22:712–731. [Google Scholar]
- Muthén B, Masyn K. Discrete-Time Survival Mixture Analysis. Journal of Educational and Behavioral Statistics. 2005;30:27–58. [Google Scholar]
- Muthén L, Muthén B. Mplus User’s Guide. 7. Los Angeles, CA: Muthén & Muthén; 1998–2015. [Google Scholar]
- Naliboff BD, Kim SE, Bolus R, Bernstein CN, Mayer EA, Chang L. Gastrointestinal and Psychological Mediators of Health-Related Quality of Life in IBS and IBD: a Structural Equation Modeling Analysis. The American Journal of Gastroenterology. 2012;107:451–459. doi: 10.1038/ajg.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parner E. Asymptotic Theory for the Correlated Gamma-Frailty Model. The Annals of Statistics. 1998;26:183–214. [Google Scholar]
- Rabe-Hesketh S, Skrondal A, Pickles A. Generalized Multilevel Structural Equation Modeling. Psychometrika. 2004;69:167–190. [Google Scholar]
- Rabe-Hesketh S, Yang S, Pickles A. Multilevel Models for Censored and Latent Responses. Statistical Methods in Medical Research. 2001;10:409–427. doi: 10.1177/096228020101000604. [DOI] [PubMed] [Google Scholar]
- Reilly T, O’Brien RM. Identification of Confirmatory Factor Analysis Models of Arbitrary Complexity: the Side-By-Side Rule. Sociological Methods & Research. 1996;24:473–491. [Google Scholar]
- Stoolmiller M, Snyder J. Modeling Heterogeneity in Social Interaction Processes Using Multilevel Survival Analysis. Psychological Methods. 2006;11:164–177. doi: 10.1037/1082-989X.11.2.164. [DOI] [PubMed] [Google Scholar]
- Stoolmiller M, Snyder J. Embedding Multilevel Survival Analysis of Dyadic Social Interaction in Structural Equation Models: Hazard Rates as Both Outcomes and Predictors. Journal of Pediatric Psychology. 2013:1–11. doi: 10.1093/jpepsy/jst076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society: Series B. 1996;58:267–288. [Google Scholar]
- Tsiatis AA, Davidian M. Joint Modeling of Longitudinal and Time-To- Event Data: an Overview. Statistica Sinica. 2004;14:809–834. [Google Scholar]
- van der Vaart AW. Asymptotic Statistics. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- Vicard P. On the Identification of a Single-Factor Model With Correlated Residuals. Biometrika. 2000;87:199–205. [Google Scholar]
- Yuan M, Lin Y. Model Selection and Estimation in Regression With Grouped Variables. Journal of the Royal Statistical Society: Series B. 2006;68:49–67. [Google Scholar]
- Zeng D, Lin DY. A General Asymptotic Theory for Maximum Likelihood Estimation in Semiparametric Regression Models With Censored Data. Statistica Sinica. 2010;20:871–910. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.