Abstract
Appropriate and abundant sources of bone-forming osteoblasts are essential for bone tissue engineering. Pluripotent stem cells can self-renew and thereby offer a potentially unlimited supply of osteoblasts, a significant advantage over other cell sources. We generated mouse embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) from transgenic mice expressing rat 2.3 kb type I collagen promoter-driven green fluorescent protein (Col2.3GFP), a reporter of the osteoblast lineage. We demonstrated that Col2.3GFP ESCs and iPSCs can be successfully differentiated to osteoblast lineage cells that express Col2.3GFP in vitro. We harvested GFP+ osteoblasts differentiated from ESCs. Genome wide gene expression profiles validated that ESC- and iPSC-derived osteoblasts resemble calvarial osteoblasts, and that Col2.3GFP expression serves as a marker for mature osteoblasts. Our results confirm the cell identity of ESC- and iPSC-derived osteoblasts and highlight the potential of pluripotent stem cells as a source of osteoblasts for regenerative medicine.
Keywords: Embryonic stem cells, Induced pluripotent stem cells, osteoblasts, differentiation, tissue engineering
1. Introduction
Bone is generally capable of self-repair, for example following a fracture. However, in some instances such as a critical-sized defect or tumor-mediated destruction, localized bone regeneration is impaired. In such cases a source of bone-forming osteoblasts in combination with tissue engineering could provide a regenerative option for bony defects. Because osteoblasts are found in mineralized tissues, they have been difficult to harvest in large numbers. Traditional approaches of obtaining primary osteoblasts rely upon cell outgrowth from surgically resected bone fragments, or differentiation from bone marrow mesenchymal stem cells (MSCs) [1, 2]. These approaches are limited by the need for invasive biopsies or surgical specimens, and in the case of adult bone marrow MSCs, low frequency and gradual loss of multipotency and proliferative potential [3–5].
Pluripotent stem cells can self-renew and differentiate into any tissue, and therefore represent a theoretically unlimited source of osteoblasts. Pluripotent stem cells include embryonic stem cells (ESCs) derived from the inner cell mass of the blastocyst, and induced pluripotent stem cells (iPSCs) derived from somatic cells by the introduction of four transcription factors [6]. We have recently reported that both ESCs and iPSCs can give rise to osteoblasts in vivo using a skeletal complementation model, and that these osteoblasts can further rescue a defective hematopoietic bone marrow microenvironment [7]. However, since in vivo transplantation of ESCs and iPSCs carries the risk of teratoma formation, osteoblasts derived from ESC and iPSC differentiation in vitro might be preferred for bone tissue engineering purposes. Several recent studies have focused on differentiating both ESCs and iPSCs into osteoblasts [8–10]. ESCs and iPSCs can be differentiated to the osteoblast lineage by first forming embryoid bodies (EBs), in which mesoderm lineage cells differentiate to osteoblast lineage cells under osteogenic factors including ascorbic acid, ß-glycerophosphate, and dexamethasone [11–27]. ESCs and iPSCs can also be differentiated to osteoblast lineage cells through monolayer culture without first forming suspended EBs [28–35].
Osteoblast differentiation in culture is frequently assayed by expression of osteoblast-related genes, formation of bony nodules, and mineralization of the surrounding extracellular matrix [8]. However, differentiation of pluripotent stem cells typically results in heterogeneous cellular populations, and even the presence of only a small fraction of osteoblasts can yield positive results in assays of osteoblast gene expression and mineralization. Not surprisingly, in vitro osteoblast assays do not accurately predict bone formation in vivo [29], and osteoblast gene expression and mineralization are not sufficient to demonstrate osteogenic maturation [36]. Furthermore, these assays do not allow for quantitation of osteoblast frequency within a mixed population, making it difficult to directly compare the efficiency of various differentiation protocols.
The ability to selectively enrich for live mature osteoblasts within heterogeneous pluripotent stem cell-derived cell populations would be an important first step to optimizing osteoblast differentiation from ESCs and iPSCs, but has been hindered by the lack of cell surface markers that uniquely distinguish osteoblasts. For example Stro-1, alkaline phosphatase, and Cadherin-11 antibodies have been used to isolation osteoblasts, however these proteins are not unique to osteoblasts [37, 38]. In mice fluorescent reporters are now available to distinguish early osteoprogenitors (Osterix-GFP), maturing osteoblasts (rat type I collagen-GFP in Col2.3GFP-expressing cells, GFP), and differentiated osteoblasts (Osteocalcin-GFP) [39–41]. We have further demonstrated that Col2.3GFP intensity can also be used to distinguish early and late stages of osteoblast differentiation [42]. We sought to combine these two approaches by deriving ESC and iPSC lines from Col2.3GFP reporter mice. Here we report that both ESC and iPSC lines can be derived, expanded and differentiated to yield GFP+ osteoblasts. We confirmed the osteoblast identity of ESC- and iPSC-derived osteoblasts using RNA-sequencing and hierarchical clustering, and demonstrate that GFP+ sorted cells are more similar to freshly isolated GFP+ osteoblasts from Col2.3GFP transgenic mouse bone than to unsorted cells based on whole genome expression profiling. While the efficiency of osteoblast differentiation remains low as determined by frequency of GFP+ cells, the Col2.3GFP reporter provides a means to identify and harvest an enriched population of pluripotent stem cell-derived mature osteoblasts.
2. Materials and methods
2.1. Generation and culture of mouse Col2.3GFP ESCs and Col2.3GFP iPSCs
Mouse ESC lines were derived from Col2.3GFP transgenic mice [41]. Following timed mating, pregnant mice were sacrificed at E3.5 and blastocysts were collected. Blastocysts were plated on inactivated mouse embryonic fibroblast (MEF) feeder cells in ESC derivation medium comprised of DMEM supplemented with 15% KnockOut Serum Replacement (KOSR), 1× penicillin-streptomycin, 2 mM GlutaMAX, 1 mM sodium pyruvate, 0.1 mM MEM NEAA, 0.1 mM 2-mercaptoethanol (all from Invitrogen, Carlsbad, CA), 103 IU of Leukemia Inhibitory Factor (LIF, Millipore, Billerica, MA), 1 µM PD0325901 (Thermo Fisher Scientific, Inc., Waltham, MA) and 3 µM CHIR99021 (Sigma, St Louis, MO). PD0325901 (an inhibitor of MEK1/2 signaling) and CHIR99021 (an inhibitor of glycogen synthase kinase-3ß signaling) are widely used to maintain self-renewal and pluripotency [43]. Blastocyst outgrowth disaggregation, stem cell passage and maintenance were processed as previously described [44]. mESCs were maintained in either ESC medium on MEF feeder cells or in feeder free condition in serum free ESC medium comprised of basal medium (50 % DMEM/F12, 50 % Neurobasal medium, 0.5× N2 supplement, 0.5× B27 supplement (all from Invitrogene), 25 µg/mL BSA fraction V (Sigma), 1× penicillin-streptomycin, 2 mM GlutaMAX, 1 mM sodium pyruvate, 0.1 mM MEM NEAA, 0.1 mM 2-mercaptoethanol) supplemented with 103 IU of LIF, 1 µM PD0325901 and 3 µM CHIR99021 [43]. ESC medium was the same as derivation medium except KOSR was replaced by Fetal Bovine Serum (FBS, Invitrogen). Genotyping of ESCs was performed by PCR for the presence of GFP (5’-TCATCTGCACCACCGGCAAGC-3’; 5’-AGCAGGACCATGTGATCGCGC-3’).
4-in-1 lentiviral reprogramming vector encoding Oct4, Klf4, Sox2, and c-Myc was kindly provided by Dr. Axel Schambach (Hannover Medical School, Hannover, Germany) [45]. Human embryonic kidney line 293T cells were transducted with lentiviral vectors along with the packaging vectors psPAX2 and pMAD2.G (Addgene; Cat. No. 16620 and 16619, respectively) using Lipofectamine 2000 regent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Viral supernatants were collected after 48 hours and concentrated by using Centrifugal Filter Units (Merck Millipore Ltd, IRELAND) according to the manufacturer’s instruction to produce virus stocks with titers of 1 × 108 to 1 × 109 infectious units per milliliter. Mouse tail tip fibroblasts (TTPs) were derived from Col2.3GFP transgenic mice, and seeded at 1 × 105 cells/well in fibroblast growing medium (DMEM medium supplemented with 10 % FBS). On day 2, the medium was replaced with virus mixed with fibroblast growing medium containing 8 ng/ml polybrene. After 24 hours, the virus was washed out and cells recovered in fresh fibroblast growing medium. On day 3, the cells were trypsinized and split (1:6) to new dishes and thereafter cultured in ESC medium from day 4. Upon appearance of ESC-like colonies, single colonies were picked based on morphology and expanded on MEF feeder cells. ESC medium was replaced every other day and iPSCs were passaged every 2–4 days based on the confluency. Similarly, iPSCs were maintained in either ESC medium on MEF feeder cells or in feeder free condition in serum free ESC medium.
2.2. Differentiation of mouse Col2.3GFP ESCs and Col2.3GFP iPSCs toward osteoblasts
Osteogenic differentiation was performed through embryoid body (EB) formation, EB plating and differentiation as previously described [23]. Briefly, dissociated mouse Col2.3GFP ESCs and Col2.3GFP iPSCs were cultured as EBs in suspension in 6 well ultra-low cluster plates for 6 days with ESC medium or serum free ESC medium without LIF, PD0325901 and CHIR99021. 0.1 µM retinoic acid (RA) was added at day 3 and washed out at day 5. After 6 days, EBs were plated in osteogenesis medium (ESC medium or serum free ESC medium without LIF, PD0325901 and CHIR99021, supplemented with 50 µg/mL ascorbic acid, 10 mM β-glycerol phosphate, and 100 nM dexamethasone (all from Sigma, St. Louis, MO)). Osteogenic medium was changed every 3 days.
2.3. Characterization of mouse Col2.3GFP ESC and Col2.3GFP iPSC lines
Immunofluorescence staining
Cells on cover glasses were fixed with 4% paraformaldehyde for 15 minutes at room temperature. For Oct4 staining, cells were permeabilized with 1% TritonX-100/PBS for 30 minutes at room temperature. Subsequently, cells were blocked with PBS-BT (1× PBS, 3% BSA, and 0.1% Triton X-100) for 30 minutes at room temperature. Coverslips were then incubated in primary and secondary antibodies diluted in PBS-BT. Oct4 antibody was purchased from Santa Cruz (cat NO: sc-9081, Santa Cruz Biotechnology, Inc.). SSEA1 antibody was obtained from Millipore (cat NO: MAB4301, EMD Millipore Corporation, Billerica, MA). Images were acquired on a fluorescence microscope (EVOS, AMEP-4615).
Karyotyping
Mouse Col2.3GFP ESCs and Col2.3GFP iPSCs were grown in feeder free conditions. Actively dividing cells were subjected to karyotyping analysis by the Cytogenetics Laboratory of Stanford Hospital and Clinics. Chromosomes were analyzed by the Giemsa-trypsin-Wrights binding method. Twenty metaphase cells were analyzed.
Pluripotency analysis after differentiation in vitro and in vivo
Mouse Col2.3GFP ESCs and Col2.3GFP iPSCs were grown in feeder free conditions and then subjected to EB formation for 10 days. Germ layer markers were analyzed by quantitative RT-PCR (qRT-PCR). RNA was extracted using the PureLink® RNA Mini Kit (Invitrogen, Carlsbad, CA) and then used for reverse transcription with the iScript Kit (Bio-Rad, Hercules, CA). qRT-PCR reactions were run using 50 nM forward and reverse primers with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). Real-time PCR was performed using the Bio-Rad iCycler. Relative mRNA levels were normalized to levels of β-Actin. PCR was performed in triplicate for each sample, and 3 independent experiments were carried out. Primer sequences are described in Supplementary Table S4.
In vivo pluripotency of mouse Col2.3GFP ESCs and Col2.3GFP iPSCs was analyzed by teratoma formation assay in severe combined immunodeficient (SCID) mice (Cb17SCID, Charles River Laboratories International, Inc.) with approval of the Administrative Panel on Laboratory Animal Care (APLAC) of Stanford University. Cell colonies were dissociated by trypsin and 1 × 106 cells were suspended in PBS, combined with matrigel, and then subcutaneously injected into 4~5-week-old mice (both female and male). After 3–4 weeks, the tumors were removed, fixed in 4 % formalin, and embedded in paraffin. The histology of in vivo differentiated three germ layers – respiratory (endoderm), cartilage (mesoderm), and neural rosettes (ectoderm) – was analyzed after hematoxylin and eosin staining [46, 47].
2.4. Isolation of calvarial osteoblasts
Calvarial osteoblast cells were prepared by serial collagenase digestion as previously described [48]. Briefly, calvariae were dissected from neonatal pups and plated followed by serial collagenases digestion with total six digests. Typically fractions 1 and 2 were discarded, and fractions 3–6 were collected and plated in a 10 cm dish. The media was changed the following day to remove nonadherent hematopoietic cells, leaving adherent osteoblasts. The calvarial osteoblasts were maintained and matured in αMEM medium containing 10 % FBS and 1× penicillin-streptomycin (all from Invitrogen, Carlsbad, CA) and supplemented with 50 µg/mL ascorbic acid. Cells were harvested and examined after three weeks.
2.5. Alizarin Red S staining
The mouse Col2.3GFP ESC- and iPSC-derived osteoblast cells were rinsed twice with PBS and fixed in 10 % formaldehyde for 10 minutes. After fixation and wash, the cells were stained with 2% alizarin red S solution for 20 minutes. After removal of unincorporated excess dye with distilled water, the mineralized nodules were stained with alizarin red.
2.6. Flow Cytometry
To harvest cells from stem cell differentiation, cells were dissociated with 0.25 % trypsin, filtered through a 70 µm cell strainer, washed once in Staining Buffer (BD Biosciences, San Diego, CA) and suspended in PBS.
For freshly isolated cells from mice long bones, soft tissue and bone marrow were removed and the bone chopped into small pieces. Small pieces of bone were digested in αMEM medium containing 2 mg/mL collagenase type II at 37°C for 45 minutes twice. Cell supernatants were filtered through a 70 µm cell strainer and combined in PBS for staining. Cells were stained with biotin anti-mouse CD45 antibody, biotin anti-mouse Ter-119 antibody and biotin anti-mouse CD31 antibody (cat NO: 103104, 116204, and 102404; BioLegend, San Diego, CA). After washing, cells were stained with Streptavidin APC/Cy7 (cat NO: 405208; BioLegend) and Propidium iodide (PI).
The cells were analyzed by a flow cytometer (FACSCAN, BD Bioscience) in the Stanford Shared FACS Facility. The CD45−/Ter119−/CD31− and GFP+ cells or negative cells were sorted by fluorescence-activated cell sorting (FACS) using a flow cytometer (Verdi, BD Bioscience) fitted with an 85 µM nozzle. Data were analyzed using the Flowjo software (Tree Star, Inc., Ashland, Oregon). Live cell GFP was analyzed by fluorescence microscopy (EVOS, AMEP-4615).
2.7. RNA-sequencing
RNA was extracted with the PureLink® RNA Mini Kit, and purified using the Dynabeads® mRNA Purification Kit (Invitrogen). RNA sequencing libraries were generated from purified mRNA using the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA). For ultra-low amounts of RNA, RNA was extracted using the NucleoSpin® RNA XS kit (Clontech, Mountain View, CA). Libraries were prepared with the SMART-Seq v4 Ultra Low Input RNA Kit followed by the Low Input Library Prep Kit (Clontech, Mountain View, CA). Several libraries were combined and average length 420 bp fragments were sequenced using paired end reads (2 × 100 bp) on the Illumina HiSeq 2500 and HiSeq 4000 platforms (Stanford Personalized Medicine Sequencing Core), with an average of 30 million reads per sample. Paired end sequencing reads (100bp) were generated and aligned to the mouse reference sequence NCBI Build 37/mm9 with the STAR (v2.4.2a) algorithm [49]. Normalization of RefSeq annotated gene expression level and differential expression analysis were performed using Bioconductor package DESeq2 in R (Version 3.2.2) [50]. rlog-transformed values (the regularized-logarithm transformation for count data) was calculated for each gene. Genes with False Discovery Rate (FDR) <0.05 were defined as differentially expressed genes. Gene ontology analysis was performed at Gene Ontology Consortium (http://geneontology.org/). The function enrichment analyses and concept-gene network analysis were implemented using the Bioconductor package GeneAnswers (version 2.14.0) [51]. All transcriptome raw data are publically available in GEO (accession number GSE110434).
2.8. Statistical Analysis
Statistical analysis was performed using Microsoft Excel and GraphPad Prism 5 (GraphPad Software, San Diego, CA). Quantitative data are presented as the mean ± SEM. Statistical Significance was assessed by the Multiple t test followed by correct for multiple comparisons using the Holm-Sidak method.
3. Results
3.1. Generation of mouse Col2.3GFP ESC line and osteoblast differentiation
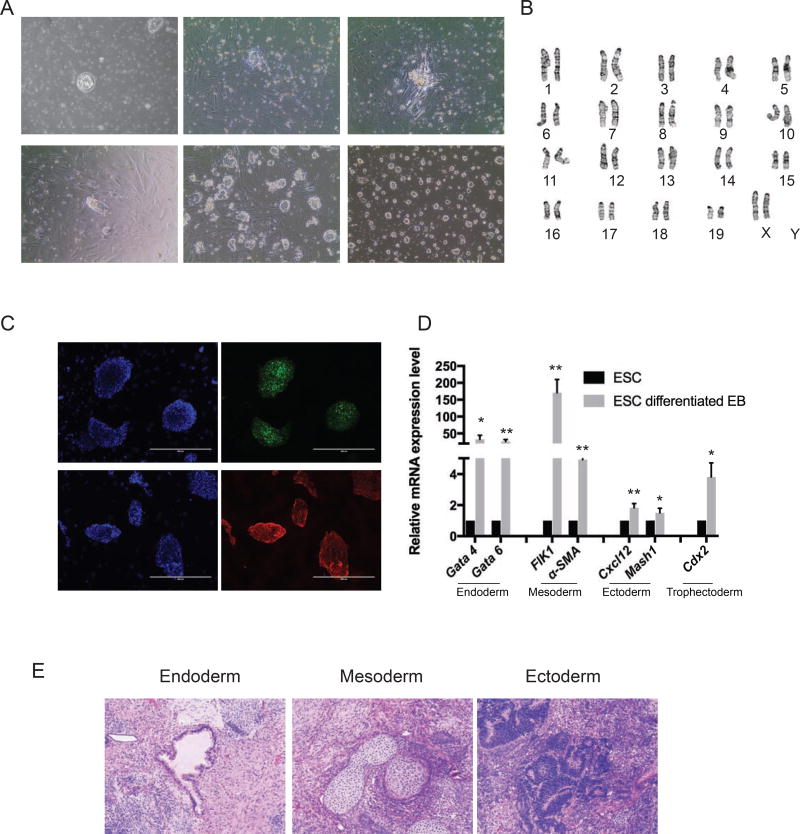
We derived ESC lines from Col2.3GFP transgenic mice. Briefly, blastocysts were flushed out from the uterine horn and plated on the MEF feeder plate in ESC derivation medium (Fig. 1Aa). Outgrowth from blastocysts could be seen by day 2 (Fig. 1Ab). At day 5, the outgrowth was ready to passage by mechanical disaggregation (Fig. 1Ac). ESC lines were generated by several passages and maintained either on feeder cells or in feeder free and serum free conditions (Fig. 1Ad, e and f). Established mouse Col2.3GFP ESC lines exhibited typical ESC cellular morphology featuring compact dome-like colony structure and distinct colony borders. We selected a line showing the best morphology and carrying the Col2.3GFP transgene for further characterization. Karyotype analysis of the Col2.3GFP ESC line revealed a normal diploid female (40, XX) line (Fig. 1B). Col2.3GFP ESCs express the pluripotency markers Oct4 and SSEA-1 (Fig. 1C). We next tested the ability of Col2.3GFP ESCs to form all three germ layers using differentiation in vitro and teratoma formation. In culture medium without anti-differentiation factors LIF and two inhibitors (PD0325901 and CHIR99021), Col2.3GFP ESCs spontaneously differentiated and formed three-dimensional multicellular aggregates termed embryoid bodies (EBs). Markers for multiple lineages, including endoderm (Gata4 and Gate 6), mesoderm (FLK1 and α-SMA), ectoderm (Cxcl12 and Msh1) and trophectoderm (Cdx2) were significantly upregulated in EBs compared to undifferentiated Col2.3GFP ESCs (Fig. 1D). To test whether Col2.3GFP ESCs can form teratomas containing cells of all three lineages, we inoculated Col2.3GFP ESCs subcutaneously in immunocompromised SCID mice and found teratoma formation after 3–4 weeks. Histological analysis of teratomas revealed tissues from all three germ layers: respiratory (endoderm), cartilage (mesoderm), and neural rosettes (ectoderm) (Fig. 1E). Together these results confirmed the pluripotency of Col2.3GFP ESCs.
Fig. 1. Generation of mouse Col2.3GFP ESC line.
(A) Derivation of mouse Col2.3GFP ESC line. (a) The isolated blastocyst on feeder cells. (b) Day 2 at passage 0 with blastocyst outgrowths. (c) Day 5 at passage 0, with formation of a stem cell cluster ready for passage. (d) Day 2 at passage 1. (e) Representative image of mouse Col2.3GFP ESC line maintained on feeder cells. (f) Representative image of mouse Col2.3GFP ESC line maintained in feeder-free and serum-free conditions. Scale bar: 200 µm. (B) A representative chromosome spread of mouse Col2.3GFP ESC line (karyotype 40, XX). (C) Immunofluorescence staining for pluripotency markers Oct4 and SSEA1. Nuclei were stained with DAPI. Scale bars: 400 µm. (D) Mouse Col2.3GFP ESCs differentiated into three germ layers cells in vitro. qRT-PCR analysis with lineage specific markers in undifferentiated mouse Col2.3GFP ESCs and differentiated Col2.3GFP ESC embryonic bodies. *, p<.05; **, p<.005 (relative to ESC). (E) Hematoxylin-eosin stained sections of teratoma formation with endoderm (respiratory), mesoderm (cartilage) and ectoderm (neural rosettes) tissues. Scale bar: 200 µm.
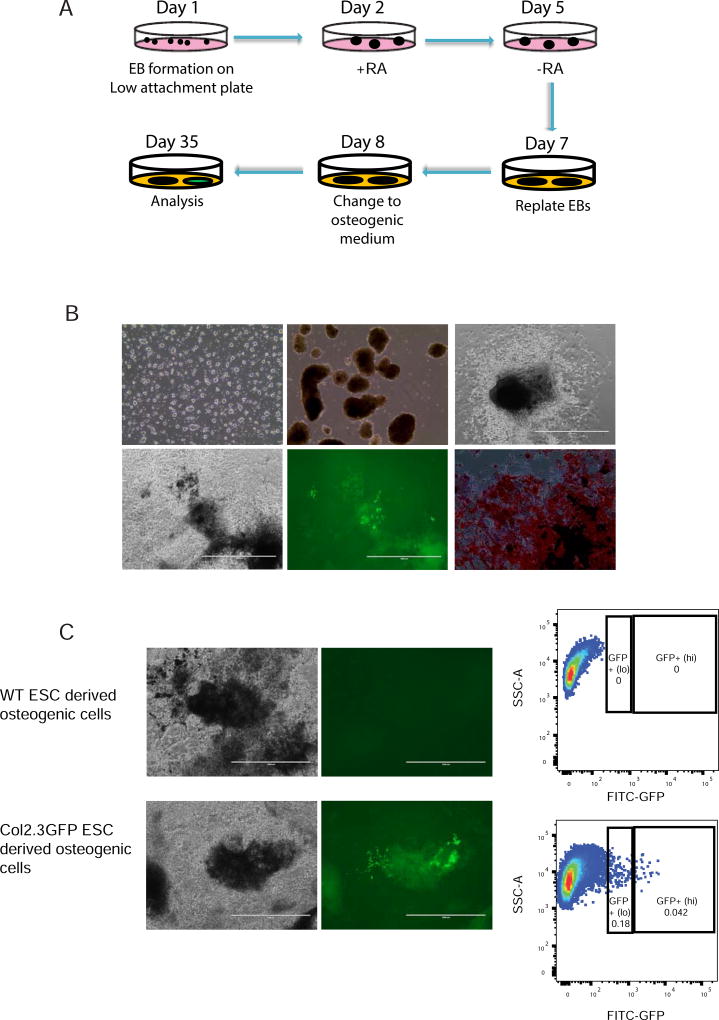
Next, we tested the osteogenic differentiation potential of the Col2.3GFP ESCs (Fig. 2A). Col2.3GFP ESCs were first allowed to spontaneously differentiate as EBs between days 1 to 6 (Fig. 2Bb). EBs were exposed to retinoic acid (RA) from days 2 to 5 to suppress cardiomyocyte formation, then replated on day 7 and subjected to osteogenic differentiation. After 35–40 days in the osteogenic medium, mineralized nodule formation was evident and confirmed by robust alizarin red staining (Fig. 2Bd and f). Osteogenic cells expressing Col2.3GFP appeared after day 20 and gradually increased till after day 35. The expression of Col2.3GFP was observed in differentiated cells (Fig. 2Be) and confirmed by flow cytometry (Fig. 2C). We further separated GFP+(lo) and GFP+(hi) populations, which we have previously reported can distinguish early and late stages of osteoblast differentiation, respectively [42], and selected GFP+(hi) mature osteoblasts for further analysis.
Fig. 2. Differentiation of mouse Col2.3GFP ESCs toward osteoblasts.
(A) Schematic diagram of osteogenic differentiation of ESCs. (B) Representative images of differentiation. (a) Day 0 when differentiation of mouse Col2.3GFP ESCs is initiated. (b) Day 5 with formation of embryonic bodies. (c) Day 11 of differentiation. (d) Day 34 of differentiation (brightfield). (e) Day 34 of differentiation (live cell GFP image). (f) Day 34 of differentiation (Alizarin Red S staining). Scale bars: 200 µm (a,b,f), 1 mm (c-e). (C) Sorting of GFP+ cells at day 35. Cells differentiated from wide type (WT) ESCs were used as a sorting gate negative control for GFP expression. Scale bars: 1 mm.
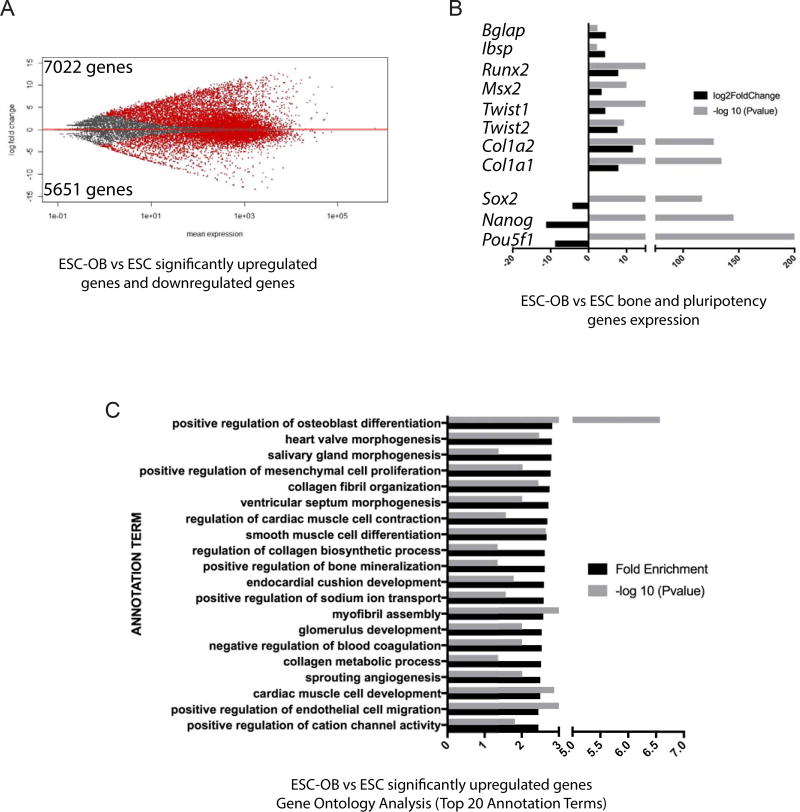
To examine the extent of osteoblast differentiation, we performed RNA-sequencing (RNA-seq) on undifferentiated day 0 Col2.3GFP ESCs (ESC), total differentiated cells at day 35 (ESC-OB), and sorted GFP+(hi) cells at day 35 (ESC-OB GFP+). The gene expression changes of ESC-OB vs ESC are shown in Fig. 3A. A total of 7022 gene and 5651 genes were found to be significantly upregulated and downregulated, respectively. As expected, pluripotency marker genes Pou5f1, Nanog and Sox2 were significantly downregulated, while osteoblast-related genes Runx2, Msx2, Twist1/2, Bglap, Ibsp, Col1a1 and Col1a2 were significantly upregulated (Fig. 3B). We validated the upregulation of these osteoblast genes by qRT-PCR (Supplementary Fig. S1A). Gene Ontology Analysis demonstrated that significantly upregulated genes were highly enriched in positive regulation of osteoblast differentiation, the top annotation term (Fig. 3C). Within the top 20 annotation terms, upregulated genes were also enriched in positive regulation of bone mineralization and other categories related to bone development and maturation including collagen fibril organization, regulation of collagen biosynthetic process, and collagen metabolic process (Fig. 3C). The enrichment of pathways involved in development of other organs of mesoderm origin, such as heart and kidney, indicated that there was still some heterogeneity among differentiated cells.
Fig. 3. Significantly upregulated genes from mouse Col2.3GFP ESC-OB are enriched in osteoblast differentiation and bone mineralization-related categories.
(A) Gene expression changes in mouse ESC-OB versus ESC. The log2-fold change for gene expression level is plotted on the y-axis and the average of the counts normalized by size factor is shown on the x-axis. Each gene is represented with a dot. Differentially expressed genes with an adjusted p (FDR) value below 0.05 are shown in red. Compared to ESC population, 7022 genes were significantly upregulated and 5651 genes were downregulated in the ESC-OB population. (B) Osteoblast and bone related gene expression levels. Log2-fold change of expression level and −log10 (P value) in mouse ESC-OB versus ESC are shown on the X axis. (C) Gene Ontology Analysis of biological process for significantly upregulated genes in mouse ESC-OB versus ESC population. Gene fold enrichment and −log10 (P value) of top 20 annotation terms were shown.
Significantly downregulated genes were mostly enriched in categories relating to DNA replication and RNA processing, confirming that fundamental cellular biological processes such as cell cycle and cell proliferation are changed during osteogenesis (Supplementary Fig. S1B). Genes Signaling Pathway Analysis for upregulated genes revealed that, within the top 50 categories, several bone development and mineralization related pathways such as ECM-receptor interaction, axon guidance, regulation of actin cytoskeleton, osteoclast differentiation, TGF-beta signaling pathway, protein processing in endoplasmic reticulum, calcium signaling pathway, tight junction, Wnt signaling pathway, Hedgehog signaling pathway, adherens junction, etc., are involved in Col2.3GFP ESC osteogenic differentiation (Supplementary Table S1). Taken together, these data confirmed that Col2.3GFP ESCs can be differentiated into osteoblasts.
3.2. Generation of mouse Col2.3GFP iPSC line and osteoblast differentiation
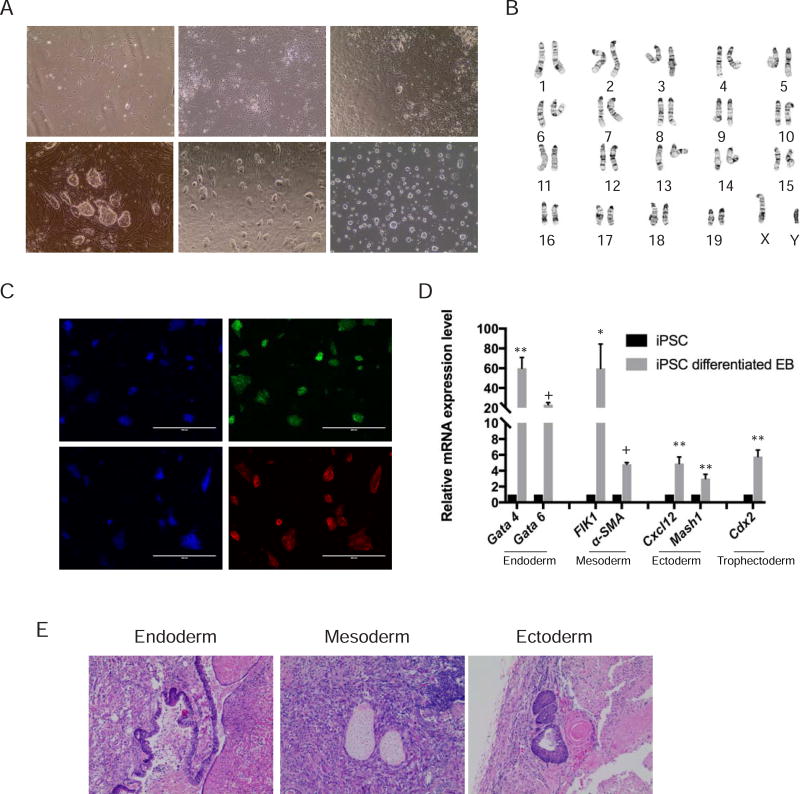
Since iPSCs can be readily derived from somatic cells and represent a potential source of patient-matched pluripotent stem cells, we sought to examine the osteogenic differentiation potential of iPSCs as well. We transduced tail tip fibroblasts from Col2.3GFP transgenic mice (Fig. 4Aa) with lentivirus containing four pluripotency markers Oct4, Klf4, Sox2, and c-Myc. Some cell clusters were visible by day 5, and stem cell like clones appeared by day 7 (Fig. 4Ab–c). Clones of reprogrammed cells were picked based on morphology and replated on MEF feeder cells. The cells expanded rapidly in culture and were trypsinized and propagated every 2–3 days (Fig. 4Ad). Established mouse Col2.3GFP iPSC lines could be maintained on either feeder cells or in feeder-free and serum-free conditions (Fig. 4Ae and f). The line selected for further characterization exhibited a normal karyotype (40, XY) (Fig. 4B) and expressed the pluripotency markers Oct4 and SSEA-1 (Fig. 4C). Col2.3GFP iPSCs differentiated into EBs expressed multiple lineages markers, including endoderm (Gata4 and Gate 6), mesoderm (FLK1 and α-SMA), ectoderm (Cxcl12 and Msh1) and trophectoderm (Cdx2) (Fig. 4D). Similarly, Col2.3GFP iPSCs formed teratomas in vivo containing all three germ layers: respiratory (endoderm), cartilage (mesoderm), and neural rosettes (ectoderm) (Fig. 4E).
Fig. 4. Generation of mouse Col2.3GFP iPSC line.
(A) Generation of mouse Col2.3GFP iPSC line. (a) Fibroblasts derived from Col2.3GFP mice (day 0). (b) Day 5 after transduction of lentivirus containing four pluripotency factors Oct4, Klf4, Sox2, and c-Myc. (c) Day 7 after transduction with formation of stem cell-like clones ready for passage. (d) Day 2 at passage 1. (e) Representative image of mouse Col2.3GFP iPSC line maintained on feeder cells. (f) Representative image of mouse Col2.3GFP iPSC line maintained in feeder-free and serum-free conditions. Scale bar: 200 µm. (B) A representative chromosome spread of mouse Col2.3GFP iPSC line (karyotype 40, XY). (C) Immunofluorescence staining for pluripotency markers Oct4 and SSEA1. Nuclei were stained with DAPI. Scale bars: 400 µm. (D) Mouse Col2.3GFP iPSCs differentiated into three germ layers cells in vitro. qRT-PCR analysis with lineage specific markers in undifferentiated mouse Col2.3GFP iPSCs and differentiated Col2.3GFP iPSCs embryonic bodies. *, p<.05; **, p<.005; +, p<5×10−5 (relative to iPSC). (E) Hematoxylin-eosin stained sections of the teratoma formation with endoderm (respiratory), mesoderm (cartilage) and ectoderm (neural rosettes) tissues. Scale bar: 200 µm.
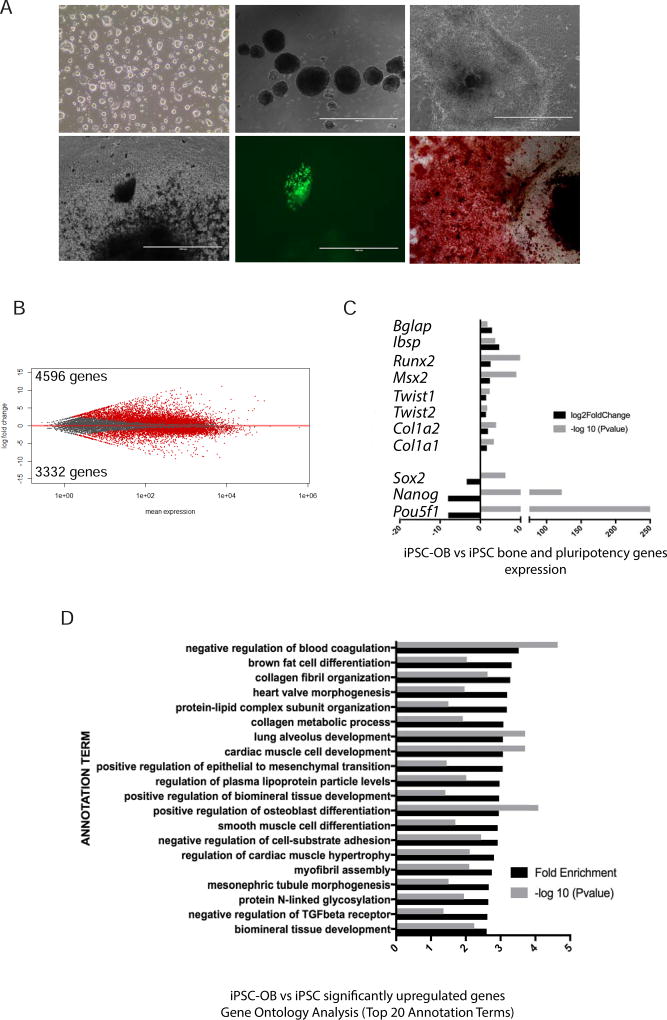
We next examined the ability of the Col2.3GFP iPSCs to differentiate toward the osteoblast lineage. Undifferentiated Col2.3GFP iPSCs (day 0) formed EBs and then were replated and differentiated in osteogenic medium for 35–40 days. Alizarin red staining and the presence of GFP+ cells confirmed the osteogenic potential of Col2.3GFP iPSCs (Fig. 5A). The total differentiated cell population (iPSC-OB) and undifferentiated cells (iPSC) were collected and used for global gene expression profiling analysis. 4596 genes were significantly upregulated including osteoblast-related genes Runx2, Msx2, Twist1/2, Bglap, Ibsp, Col1a1 and Col1a2. 3332 genes were significantly downregulated including pluripotency markers Pou5f1, Nanog and Sox2 (Fig. 5B and C). The upregulated osteogenic related genes were validated by qRT-PCR (Supplementary Fig. S2A). Gene Ontology Analysis revealed that significantly upregulated genes were enriched in osteoblast differentiation and bone development related categories (i.e. positive regulation of osteoblast differentiation, collagen fibril organization, and positive regulation of biomineral tissue development) (Fig. 5D). Of note iPSC-OB were also heterogeneous as shown by enrichment of lineage genes for cardiac and renal development. Gene Ontology Analysis for significantly downregulated genes (Supplementary Fig. S2B) and Genes Signaling Pathway Analysis for significantly upregulated genes (Supplementary Table S2) showed similar categories of enrichment as for ESC-OB. Overall, these data demonstrate that Col2.3GFP iPSCs maintain pluripotency and can differentiate into osteoblast lineage cells.
Fig. 5. Differentiation of mouse Col2.3GFP iPSCs toward osteoblasts.
(A) Representative images of differentiation. (a) Day 0 when differentiation of mouse Col2.3GFP iPSCs is initiated. (b) Day 5 with formation of embryoid bodies. (c) Day 10 of differentiation. (d) Day 35 of differentiation (brightfield). (e) Day 35 of differentiation (live cell GFP image). (f) Day 35 of differentiation (Alizarin Red S staining). Scale bars: 200 µm (a, f), 1 mm (b–e). (B) A plot of gene expression changes in mouse iPSC-OB versus iPSC. The log2-fold change for gene expression level is plotted on the y-axis and the average of the counts normalized by size factor is shown on the x-axis. Each gene is represented with a dot. Differentially expressed genes with an adjusted p (FDR) value below 0.05 are shown in red. Compared to iPSC population, 4596 genes were significantly upregulated and 3332 genes were downregulated in the iPSC-OB population. (C) Osteoblast and bone related gene expression levels. Log2-fold change of expression level and −log10 (P value) in mouse iPSC-OB versus iPSC are shown on the X axis. (D) Gene Ontology Analysis of biological process for significantly upregulated genes in mouse iPSC-OB versus iPSC population. Gene fold enrichment and −log10 (P value) of top 20 annotation terms were shown.
3.3. Mouse ESC-OB and iPSC-OB transcriptomes resemble osteoblasts
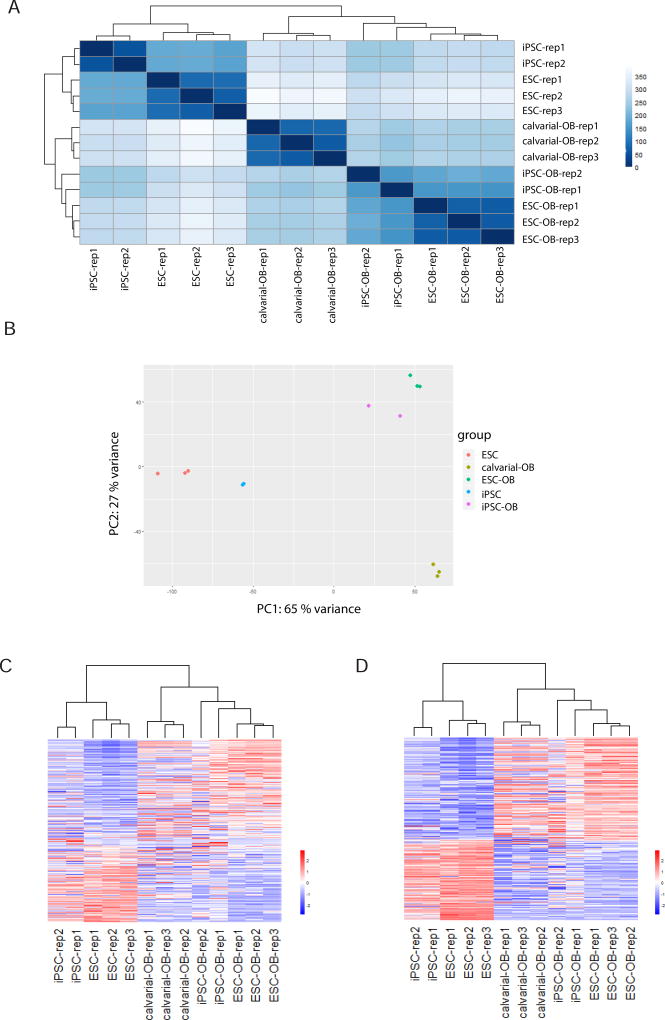
Previous studies have shown that osteoblasts can be differentiated from preosteoblastic cells, mesenchymal stem cells (MSCs), ESCs and iPSCs [1, 8, 11, 52, 53]. The assessment of osteogenic capability is typically based on two criteria: mineralization capacity in vitro (visualized by Alizarin red staining) and the expression of osteoblast-specific markers (i.e. Runx2, BSP, and Osteocalcin). However, the degree of molecular similarity between ESC- or iPSC-derived osteogenic cells and osteoblasts has not been completely elucidated. We therefore compared the global gene expression profiles of ESC-OB and iPSC-OB with primary calvarial cell-derived osteoblasts (calvarial-OB). Based on the global gene expression by RNA-seq analysis, the heatmap of sample-to-sample distance demonstrated that ESC-OB and iPSC-OB were clustered together with calvarial-OB and distinct from ESCs and iPSCs (Fig. 6A), suggesting the similarity of ESC-OB/iPSC-OB to calvarial-OB. The global expression patterns were further analyzed by principal component analysis (PCA), which reduces high-dimensionality data into a limited number of principal components. The first principal component (PC1) captures the largest contributing factor of variation, in this case 65% of variation, and revealed that ESC-OB and iPSC-OB are much closer to calvarial-OB than undifferentiated ESCs or iPSCs. Of note ESC-OB gene expression profile is more similar to calvarial-OB than iPSC-OB, suggesting more effective osteoblast differentiation from ESCs under the conditions studies (Fig. 6B).
Fig. 6. ESC-OB and iPSC-OB resemble calvarial-OB.
(A) Heatmap of sample-to-sample distances using rlog-transformed values of RNA-seq data. For each sample, the distance calculation is based on the contribution of rlog from all the genes. (B) PCA plot using the rlog-transformed values of RNA-seq data. Each color represents a sample type. Each sample has three or two replicates. (C) Heatmap of relative rlog-transformed values of total genes across samples. Each row represents a gene. Each column represents a cell type. (D) Heatmap of relative rlog-transformed values of differentially expressed genes in ESC-OB versus ESC across samples. Each row represents a gene. Each column represents a cell type.
Unsupervised hierarchical clustering of genome-wide gene expression data across ESC, iPSC, ESC-OB, iPSC-OB and calvarial-OB highlighted gene expression patterns between these cell types (Fig. 6C). In agreement with sample distance and PCA analysis, ESC-OB and iPSC-OB were clustered together with calvarial-OB. Second, there were more gene expression differences between the two replicates of iPSC-OB than within the three replicates in ESC-OB, suggesting greater heterogeneity of iPSCs. Third, iPSCs may not fully recapitulate the pluripotent state of bona fide embryonic cells as molecular differences between iPSCs and ESCs were evident in their undifferentiated state. Next we selected differentially expressed genes between ESC-OB and ESC and illustrated the heatmap of these gene expression patterns across all the 13 samples. Compared to whole genome-wide gene expression pattern, the differentially expressed gene patterns in ESC-OB and iPSC-OB were very similar to calvarial-OB, demonstrating that the transcriptional programs of both mouse ESC and iPSC were globally programmed toward that of the osteoblast lineage (Fig. 6D).
3.4. Mouse Col2.3GFP ESC-derived GFP+ cells resemble GFP+ osteoblasts from Col2.3GFP mouse bone
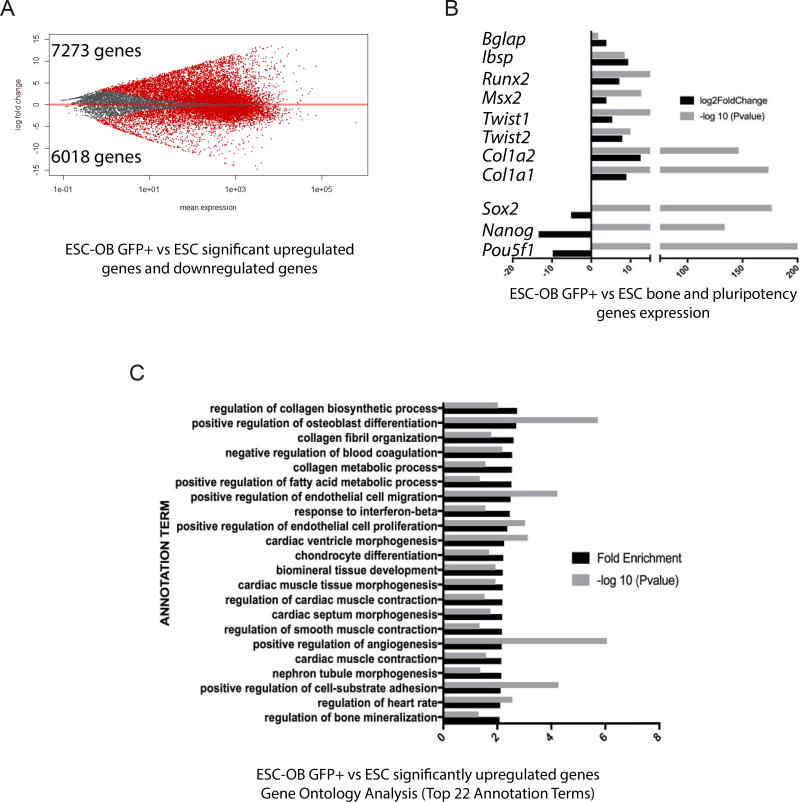
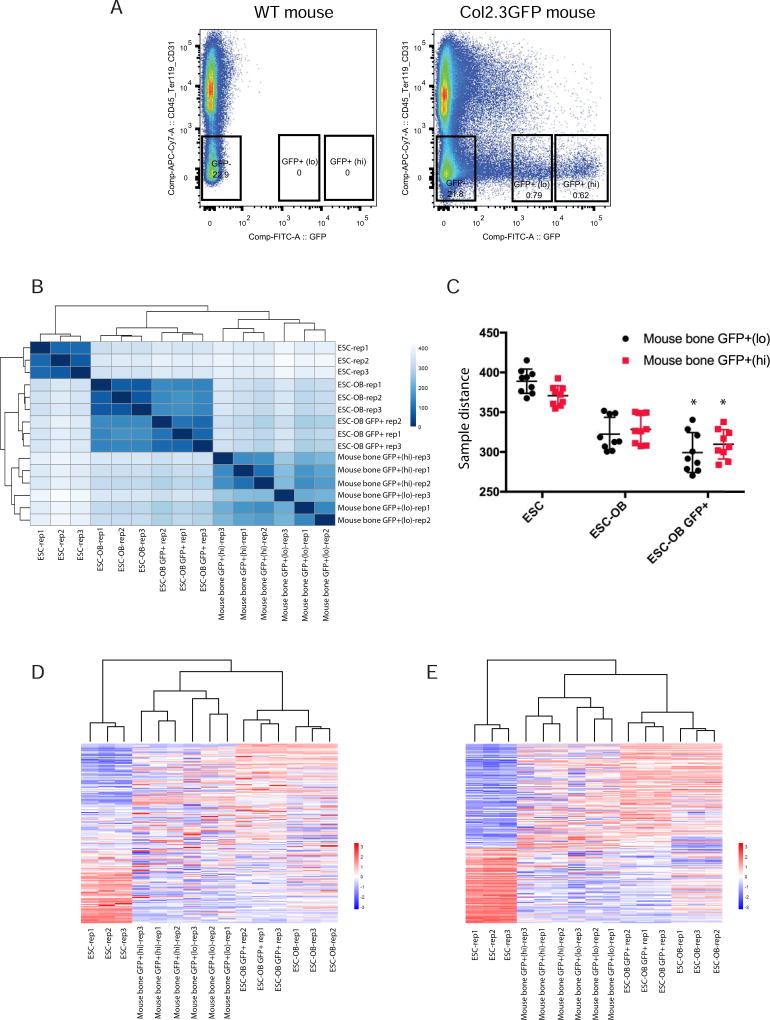
Although our data show that mouse ESC and iPSC derived osteogenic cells resemble osteoblasts in global gene expression, the population of differentiated cells is mixed due to the inherent heterogeneity of differentiating pluripotent stem cells and the low efficiency of osteogenic differentiation. A more enriched osteoblast cell population would be useful for osteoblast cell identification and regenerative medicine. We therefore sorted GFP+ cells from mouse Col2.3GFP ESC-derived osteogenic cells and analyzed the genome-wide gene expression pattern by RNA-seq. As shown in Fig. 7A, a total of 7273 genes were significantly upregulated and 6018 genes were significantly downregulated in ESC-OB GFP+ versus ESC. The osteogenic markers Runx2, Msx2, Twist1/2, Bglap, Ibsp, Col1a1 and Col1a2 were significantly upregulated while pluripotency marker genes Pou5f1, Nanog and Sox2 were significantly downregulated (Fig. 7B). Similarly, significantly upregulated genes were enriched in positive regulation of osteoblast differentiation, collagen fibril organization, collagen metabolic process, and regulation of bone mineralization, etc., which were associated with bone development and maturation (Fig. 7C). Gene Ontology Analysis for significantly downregulated genes (Supplementary Fig. 3) and Genes Signaling Pathway Analysis for significantly upregulated genes (Supplementary Table S3) showed similar categories of enrichment as ESC-OB. Despite the enrichment for GFP+ ESC-OB, we still found some lineage gene enrichment for other mesoderm tissues, which prompted us to rigorously compare ESC-OB GFP+ with freshly isolated Col2.3GFP+ cells from long bones of Col2.3GFP transgenic mice (Fig. 8A). We have previously reported that the intensity of Col2.3GFP expression correlates with osteoblast maturation stage. We therefore sorted freshly isolated mouse bone GFP+(lo) and GFP+(hi) populations from long bones of Col2.3GFP transgenic mice, which represent early and late stage (mature) osteoblasts, respectively [42]. We gated on CD45−/Ter119−/CD31− cells to exclude hematopoietic, erythroid and endothelial lineage cells. A heatmap of sample-to-sample distance showed that ESC-OB and ESC-OB GFP+ were clustered together with mouse bone GFP+(lo) and GFP+(hi) cells and distinct from undifferentiated ESCs (Fig. 8B). The ESC-OB GFP+ population was more similar to ESC-OB than mouse bone GFP+ populations, perhaps due to the low percentage of GFP+ cells in ESC derived osteoblasts or less maturation of the ESC-OB GFP+ population under in vitro differentiation conditions. Direct comparison of the distance between these cell types revealed that the ESC-OB GFP+ population was closer to mouse bone GFP+ populations compared with ESC-OB (Fig. 8C). These data demonstrated that Col2.3GFP+ cells isolated from mouse ESC subjected to osteogenic differentiation are a more enriched osteoblast population. Interestingly, ESC-OB GFP+ was more similar to mouse bone GFP+(lo) than to mouse bone GFP+(hi) populations, suggesting that the ESC derived GFP+ osteoblasts may be less mature.
Fig. 7. Genes from ESC-OB GFP+ cells are enriched in osteoblast differentiation and bone mineralization related categories.
(A) A plot of gene expression changes in mouse ESC-OB GFP+ versus ESC. The log2-fold change for gene expression level is plotted on the y-axis and the average of the counts normalized by size factor is shown on the x-axis. Each gene is represented with a dot. Differentially expressed genes with an adjusted p (FDR) value below 0.05 are shown in red. Compared to the ESC population, 7273 genes were significantly upregulated and 6018 genes were downregulated in the ESC-OB GFP+ population. (B) Osteoblast and bone related gene expression levels. Log2-fold change of expression level and −log10 (P value) in mouse ESC-OB GFP+ versus ESC are shown on the X axis. (C) Gene Ontology Analysis of biological process for significantly upregulated genes in mouse ESC-OB GFP+ versus ESC population. Gene fold enrichment and −log10 (P value) of top 22 annotation terms are shown.
Fig. 8. ESC-OB GFP+ population resembles mouse bone GFP+ population.
(A) FACS sorting of CD45−/Ter119−/CD31−/GFP+(lo), CD45−/Ter119−/CD31−/GFP+(hi), and CD45−/Ter119−/CD31−/GFP− cells from mouse long bone. Three biological replicates were processed. Long bone cells from WT mice were used as sorting gate negative control. (B) Heatmap of sample-to-sample distances using rlog-transformed values of RNA-seq data. (C) The sample distance between ESC, ESC-OB, ESC-OB GFP+ with mouse bone GFP+(lo) and GFP+(hi) populations. *, p<.05 (relative to ESC-OB). (D) Heatmap of relative rlog-transformed values of total genes across samples. Each row represents a gene. Each column represents a cell type. (E) Heatmap of relative rlog-transformed values of differentially expressed genes in ESC-OB GFP+ versus ESC across samples. Each row represents a gene. Each column represents a cell type.
Unsupervised hierarchical clustering of the genome-wide gene expression data across ESC, ESC-OB, ESC-OB GFP+ and mouse bone GFP+(lo)/(hi) populations revealed that ESC-OB and ESC-OB GFP+ were clustered together with mouse bone GFP (lo)/(hi) populations (Fig. 8D). As for the expression pattern of differentially expressed genes in ESC-OB GFP+ versus ESC, the heatmap of gene profiling cross all the 15 samples showed much greater similarity of ESC-OB GFP+ to mouse bone GFP+ (Fig. 8E). Taken together, our data suggest that ESC-differentiated GFP+ cells are a more enriched and differentiated osteoblast population. Isolation of enriched mature osteoblasts from differentiated ESCs is possible and reliable.
4. Discussion
Pluripotent stem cells have the ability to self-renewal indefinitely in vitro while maintaining their differentiation potential. Patient-specific iPSCs could potentially be generated from patients, expanded in vitro, modified, and differentiated for autologous transplantation [54]. Therefore, pluripotent stem cells are promising sources for the cellular regenerative therapies in bone, particularly if combined with encapsulation in biomaterials for cell-based tissue engineering. Both mouse and human ESCs and iPSCs have been tested for osteogenic potential in vitro and in vivo [8, 11, 19, 29, 55–58]. At present the identification of mature osteoblasts derived from differentiated pluripotent stem cells by nondestructive methods has been a significant challenge in the field.
Osteogenic differentiation is typically assessed by expression of osteoblast markers at the mRNA level by qRT-PCR [12, 29, 35, 56, 58] or protein level by immunostaining [30]. For example, Runx2, Ostrix/Sp7, Osteocalcin/Bglap, and BSP/Ibsp are osteoblast-related genes often used as marker for osteoblast differentiation [8]. However, the presence of even a small fraction of osteoblasts within a mixed population is sufficient to significantly increase mRNA levels of selected osteoblast-related genes over baseline. Alizarin Red S staining [15, 30, 57] and mineralized nodule formation [27] have also been used to measure osteogenic potential, but since mineral is deposited in extracellular matrix this does not allow for cell-autonomous identification of osteoblasts. Therefore, a more rigorous method to identify pluripotent stem cell-derived mature osteoblasts is needed.
In this study, we derived ESC and iPSC lines from transgenic mice carrying the Col2.3GFP fluorescent reporter for maturing osteoblasts. Other groups have also generated Col2.3GFP mESCs and hESCs and demonstrated Col2.3GFP expression in mineralized areas in vivo followed by teratoma formation or cranial defect repair. However, reliable generation of GFP+ osteoblasts differentiated from PSCs in vitro has not been shown [36, 56]. In this study, we differentiated mouse Col2.3GFP PSCs to mature GFP+ osteoblasts via EB formation. To our knowledge, this is the first demonstration that GFP+(hi) mature osteoblasts can be successfully purified from differentiation of ESCs in vitro. While the efficiency of deriving mature osteoblasts remains low, our findings reinforce those of other groups that have similarly demonstrated the difficulty in differentiating pluripotent stem cells to late stage osteoblasts in vitro [30, 36, 56, 59]. We have not been able generate GFP+ cells from Col2.3GFP mESCs under monolayer culture, which is agreement with a previous study [36], although we did detect the dramatic upregulation of most of osteogenic markers (e.g. Runx2, Ostrix/Sp7, Osteocalcin/Bglap, Bsp/Ibsp, etc.) and matrix mineralization (data not shown). Our findings confirm that osteoblast gene expression and mineralization are not sufficient to assays of osteogenic maturation of PSCs [36]. A RUNX2-YFP reporter system engineered in human ESCs was used to monitor early stage hESC osteogenic differentiation but did not recapitulate mature osteoblast differentiation as well as bone marrow mesenchymal stem cell-derived osteoblasts [59].
Here we extend these studies by analyzing the global gene profiles of ESCs, iPSCs and their osteogenic progeny by deep RNA-sequencing. We found that osteogenic differentiation significantly upregulated genes enriched in osteoblast and bone development categories; these genes were also involved in bone related signaling pathways. Hierarchical clustering and principle component analyses revealed that ESC- and iPSC-derived osteoblasts were similar to primary calvarial osteoblasts. It should be noted that expression of most osteogenic marker genes in iPSC-OB was lower than in ESC-OB; ESC-OBs were more similar to calvarial-OBs in principal component analysis (PC1); and iPSC-OBs were more heterogeneous than ESC-OBs. Together these findings imply that under the conditions we tested the efficiency of iPSC osteogenic differentiation was less than for ESCs. One possible reason is that iPSCs were not identical to ESCs. Our data revealed gene expression differences between iPSCs and ESCs in their undifferentiated state; iPSCs are closer to calvarial-OB in principle component analysis, perhaps due to the epigenetic “cell memory” as iPSCs were derived from fibroblasts and both fibroblasts and osteoblasts are of mesenchymal origin. Whether the “cell memory” of iPSC affects osteoblast differentiation remains to be fully elucidated [60, 61]. However, the overall cell identity and functionality of iPSC-derived osteoblasts were comparable with ESC-derived osteoblast, indicating that iPSC-derived osteoblasts may be indeed suitable for in vivo bone regeneration. Overall, our data validate the cell identity of mouse pluripotent stem cell-differentiated mature osteoblasts, which resemble calvarial osteoblasts, and confirm that pluripotent stem cells prove useful as an unlimited source of osteoblast cells for bone tissue regeneration.
5. Conclusion
In this study, we derived mouse ESCs and iPSCs from Col2.3GFP transgenic mice. We differentiated mouse Col2.3GFP ESCs and iPSCs to osteogenic cells through EB formation and demonstrated that mature osteoblasts can be enriched by harvesting the GFP+(hi) population. We extensively compared global gene expression profiles with calvarial osteoblasts and validated the mature osteoblast identity of pluripotent stem cell-derived osteoblasts. Furthermore, we show that GFP+ sorted cells are more similar to mouse bone GFP+ population than to unsorted cells based on whole genome expression profiling. Our findings highlight the potential for ESCs and iPSCs as a source of mature osteoblasts for bone regeneration.
Supplementary Material
Acknowledgments
We thank Dr. Axel Schambach (Hannover Medical School, Hannover, Germany) for the 4-in-1 lentiviral reprogramming vector. This worked was funded by National Institute of Health grant DP2OD008466 to J.Y.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The raw data required to reproduce these findings are available to download from GEO (accession number GSE110434).
References
- 1.Panaroni C, Tzeng YS, Saeed H, Wu JY. Mesenchymal progenitors and the osteoblast lineage in bone marrow hematopoietic niches. Curr Osteoporos Rep. 2014;12(1):22–32. doi: 10.1007/s11914-014-0190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Dimitriou R, Jones E, McGonagle D, Giannoudis PV. Bone regeneration: current concepts and future directions. BMC Med. 2011;9:66. doi: 10.1186/1741-7015-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddappa R, Licht R, van Blitterswijk C, de Boer J. Donor variation and loss of multipotency during in vitro expansion of human mesenchymal stem cells for bone tissue engineering. J Orthop Res. 2007;25(8):1029–41. doi: 10.1002/jor.20402. [DOI] [PubMed] [Google Scholar]
- 5.Meijer GJ, de Bruijn JD, Koole R, van Blitterswijk CA. Cell-based bone tissue engineering. PLoS Med. 2007;4(2):e9. doi: 10.1371/journal.pmed.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Chubb R, Oh J, Riley AK, Kimura T, Wu SM, Wu JY. In Vivo Rescue of the Hematopoietic Niche by Pluripotent Stem Cell Complementation of Defective Osteoblast Compartments. Stem Cells. 2017 doi: 10.1002/stem.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu JY. Pluripotent Stem Cells and Skeletal Regeneration--Promise and Potential. Curr Osteoporos Rep. 2015;13(5):342–50. doi: 10.1007/s11914-015-0285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou X. Induced Pluripotent Stem Cells as a new Strategy for Osteogenesis and Bone Regeneration. Stem Cell Rev. 2015 doi: 10.1007/s12015-015-9594-8. [DOI] [PubMed] [Google Scholar]
- 10.Duplomb L, Dagouassat M, Jourdon P, Heymann D. Concise review: embryonic stem cells: a new tool to study osteoblast and osteoclast differentiation. Stem Cells. 2007;25(3):544–52. doi: 10.1634/stemcells.2006-0395. [DOI] [PubMed] [Google Scholar]
- 11.Ma MS, Kannan V, de Vries AE, Czepiel M, Wesseling EM, Balasubramaniyan V, Kuijer R, Vissink A, Copray S, Raghoebar GM. Characterization and comparison of osteoblasts derived from mouse embryonic stem cells and induced pluripotent stem cells. J Bone Miner Metab. 2017;35(1):21–30. doi: 10.1007/s00774-015-0730-y. [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Al-Mansoori L, Opas M. Optimized osteogenic differentiation protocol from R1 mouse embryonic stem cells in vitro. Differentiation. 2015;89(1–2):1–10. doi: 10.1016/j.diff.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Ehnes DD, Price FD, Shrive NG, Hart DA, Rancourt DE, zur Nieden NI. Embryonic stem cellderived osteocytes are capable of responding to mechanical oscillatory hydrostatic pressure. J Biomech. 2015;48(10):1915–21. doi: 10.1016/j.jbiomech.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Camargos BM, Tavares RL, Del Puerto HL, Andrade LO, Camargos AF, Reis FM. BMP-4 increases activin A gene expression during osteogenic differentiation of mouse embryonic stem cells. Growth Factors. 2015;33(2):133–8. doi: 10.3109/08977194.2014.984805. [DOI] [PubMed] [Google Scholar]
- 15.Sidney LE, Kirkham GR, Buttery LD. Comparison of osteogenic differentiation of embryonic stem cells and primary osteoblasts revealed by responses to IL-1beta, TNF-alpha, and IFN-gamma. Stem Cells Dev. 2014;23(6):605–17. doi: 10.1089/scd.2013.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochiai-Shino H, Kato H, Sawada T, Onodera S, Saito A, Takato T, Shibahara T, Muramatsu T, Azuma T. A novel strategy for enrichment and isolation of osteoprogenitor cells from induced pluripotent stem cells based on surface marker combination. PLoS One. 2014;9(6):e99534. doi: 10.1371/journal.pone.0099534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans ND, Swain RJ, Gentleman E, Gentleman MM, Stevens MM. Gene-expression analysis reveals that embryonic stem cells cultured under osteogenic conditions produce mineral non-specifically compared to marrow stromal cells or osteoblasts. Eur Cell Mater. 2012;24:211–23. doi: 10.22203/ecm.v024a15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MJ, Park JS, Kim S, Moon SH, Yang HN, Park KH, Chung HM. Encapsulation of bone morphogenic protein-2 with Cbfa1-overexpressing osteogenic cells derived from human embryonic stem cells in hydrogel accelerates bone tissue regeneration. Stem Cells Dev. 2011;20(8):1349–58. doi: 10.1089/scd.2010.0311. [DOI] [PubMed] [Google Scholar]
- 19.Bilousova G, Jun du H, King KB, De Langhe S, Chick WS, Torchia EC, Chow KS, Klemm DJ, Roop DR, Majka SM. Osteoblasts derived from induced pluripotent stem cells form calcified structures in scaffolds both in vitro and in vivo. Stem Cells. 2011;29(2):206–16. doi: 10.1002/stem.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Bronson S, Niyibizi C. Derivation of murine induced pluripotent stem cells (iPS) and assessment of their differentiation toward osteogenic lineage. J Cell Biochem. 2010;109(4):643–52. doi: 10.1002/jcb.22440. [DOI] [PubMed] [Google Scholar]
- 21.Evans ND, Gentleman E, Chen X, Roberts CJ, Polak JM, Stevens MM. Extracellular matrixmediated osteogenic differentiation of murine embryonic stem cells. Biomaterials. 2010;31(12):3244–52. doi: 10.1016/j.biomaterials.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 22.Tashiro K, Inamura M, Kawabata K, Sakurai F, Yamanishi K, Hayakawa T, Mizuguchi H. Efficient adipocyte and osteoblast differentiation from mouse induced pluripotent stem cells by adenoviral transduction. Stem Cells. 2009;27(8):1802–11. doi: 10.1002/stem.108. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi J. Generation of osteoblasts and chondrocytes from embryonic stem cells. Methods Mol Biol. 2006;330:135–48. doi: 10.1385/1-59745-036-7:135. [DOI] [PubMed] [Google Scholar]
- 24.Bielby RC, Boccaccini AR, Polak JM, Buttery LD. In vitro differentiation and in vivo mineralization of osteogenic cells derived from human embryonic stem cells. Tissue Eng. 2004;10(9–10):1518–25. doi: 10.1089/ten.2004.10.1518. [DOI] [PubMed] [Google Scholar]
- 25.zur Nieden NI, Kempka G, Ahr HJ. In vitro differentiation of embryonic stem cells into mineralized osteoblasts. Differentiation. 2003;71(1):18–27. doi: 10.1046/j.1432-0436.2003.700602.x. [DOI] [PubMed] [Google Scholar]
- 26.Sottile V, Thomson A, McWhir J. In vitro osteogenic differentiation of human ES cells. Cloning Stem Cells. 2003;5(2):149–55. doi: 10.1089/153623003322234759. [DOI] [PubMed] [Google Scholar]
- 27.Buttery LD, Bourne S, Xynos JD, Wood H, Hughes FJ, Hughes SP, Episkopou V, Polak JM. Differentiation of osteoblasts and in vitro bone formation from murine embryonic stem cells. Tissue Eng. 2001;7(1):89–99. doi: 10.1089/107632700300003323. [DOI] [PubMed] [Google Scholar]
- 28.Rutledge K, Cheng Q, Pryzhkova M, Harris GM, Jabbarzadeh E. Enhanced differentiation of human embryonic stem cells on extracellular matrix-containing osteomimetic scaffolds for bone tissue engineering. Tissue Eng Part C Methods. 2014;20(11):865–74. doi: 10.1089/ten.tec.2013.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips MD, Kuznetsov SA, Cherman N, Park K, Chen KG, McClendon BN, Hamilton RS, McKay RD, Chenoweth JG, Mallon BS, Robey PG. Directed differentiation of human induced pluripotent stem cells toward bone and cartilage: in vitro versus in vivo assays. Stem Cells Transl Med. 2014;3(7):867–78. doi: 10.5966/sctm.2013-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanke K, Masaki H, Saito T, Komiyama Y, Hojo H, Nakauchi H, Lichtler AC, Takato T, Chung UI, Ohba S. Stepwise differentiation of pluripotent stem cells into osteoblasts using four small molecules under serum-free and feeder-free conditions. Stem Cell Reports. 2014;2(6):751–60. doi: 10.1016/j.stemcr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, Li X, Meikle MC, Islam I, Cao T. Short periods of cyclic mechanical strain enhance triple-supplement directed osteogenesis and bone nodule formation by human embryonic stem cells in vitro. Tissue Eng Part A. 2013;19(19–20):2130–7. doi: 10.1089/ten.tea.2012.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Peppo GM, Marcos-Campos I, Kahler DJ, Alsalman D, Shang L, Vunjak-Novakovic G, Marolt D. Engineering bone tissue substitutes from human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110(21):8680–5. doi: 10.1073/pnas.1301190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marolt D, Campos IM, Bhumiratana S, Koren A, Petridis P, Zhang G, Spitalnik PF, Grayson WL, Vunjak-Novakovic G. Engineering bone tissue from human embryonic stem cells. Proc Natl Acad Sci U S A. 2012;109(22):8705–9. doi: 10.1073/pnas.1201830109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuznetsov SA, Cherman N, Robey PG. In vivo bone formation by progeny of human embryonic stem cells. Stem Cells Dev. 2011;20(2):269–87. doi: 10.1089/scd.2009.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfred R, Taiani JT, Krawetz RJ, Yamashita A, Rancourt DE, Kallos MS. Large-scale production of murine embryonic stem cell-derived osteoblasts and chondrocytes on microcarriers in serum-free media. Biomaterials. 2011;32(26):6006–16. doi: 10.1016/j.biomaterials.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 36.Repic D, Torreggiani E, Franceschetti T, Matthews BG, Ivcevic S, Lichtler AC, Grcevic D, Kalajzic I. Utilization of transgenic models in the evaluation of osteogenic differentiation of embryonic stem cells. Connect Tissue Res. 2013;54(4–5):296–304. doi: 10.3109/03008207.2013.814646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourne S, Polak JM, Hughes SP, Buttery LD. Osteogenic differentiation of mouse embryonic stem cells: differential gene expression analysis by cDNA microarray and purification of osteoblasts by cadherin-11 magnetically activated cell sorting. Tissue Eng. 2004;10(5–6):796–806. doi: 10.1089/1076327041348293. [DOI] [PubMed] [Google Scholar]
- 38.Walsh S, Jefferiss C, Stewart K, Jordan GR, Screen J, Beresford JN. Expression of the developmental markers STRO-1 and alkaline phosphatase in cultures of human marrow stromal cells: regulation by fibroblast growth factor (FGF)-2 and relationship to the expression of FGF receptors 1–4. Bone. 2000;27(2):185–95. doi: 10.1016/s8756-3282(00)00319-7. [DOI] [PubMed] [Google Scholar]
- 39.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–44. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 40.Bilic-Curcic I, Kronenberg M, Jiang X, Bellizzi J, Mina M, Marijanovic I, Gardiner EM, Rowe DW. Visualizing levels of osteoblast differentiation by a two-color promoter-GFP strategy: Type I collagen-GFPcyan and osteocalcin-GFPtpz. Genesis. 2005;43(2):87–98. doi: 10.1002/gene.20156. [DOI] [PubMed] [Google Scholar]
- 41.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17(1):15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 42.Wu JY, Aarnisalo P, Bastepe M, Sinha P, Fulzele K, Selig MK, Chen M, Poulton IJ, Purton LE, Sims NA, Weinstein LS, Kronenberg HM. Gsalpha enhances commitment of mesenchymal progenitors to the osteoblast lineage but restrains osteoblast differentiation in mice. J Clin Invest. 2011;121(9):3492–504. doi: 10.1172/JCI46406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czechanski A, Byers C, Greenstein I, Schrode N, Donahue LR, Hadjantonakis AK, Reinholdt LG. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat Protoc. 2014;9(3):559–74. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, Filipczyk AA, Halle S, Klump H, Scholer HR, Baum C, Schroeder T, Schambach A. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19(4):782–9. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelakanti RV, Kooreman NG, Wu JC. Teratoma formation: a tool for monitoring pluripotency in stem cell research. Curr Protoc Stem Cell Biol. 2015;32:4A 8 1–17. doi: 10.1002/9780470151808.sc04a08s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh SK, Chua P, Foon KL, Ng E, Chin A, Choo AB, Srinivasan R. Quantitative identification of teratoma tissues formed by human embryonic stem cells with TeratomEye. Biotechnol Lett. 2009;31(5):653–8. doi: 10.1007/s10529-009-9928-1. [DOI] [PubMed] [Google Scholar]
- 48.Wu JY, Purton LE, Rodda SJ, Chen M, Weinstein LS, McMahon AP, Scadden DT, Kronenberg HM. Osteoblastic regulation of B lymphopoiesis is mediated by Gs{alpha}-dependent signaling pathways. Proc Natl Acad Sci U S A. 2008;105(44):16976–81. doi: 10.1073/pnas.0802898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng G, Du P, Krett NL, Tessel M, Rosen S, Kibbe WA, Lin SM. A collection of bioconductor methods to visualize gene-list annotations. BMC Res Notes. 2010;3:10. doi: 10.1186/1756-0500-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Garrett R, Jung Y, Zhang Y, Kim N, Wang J, Joe GJ, Hexner E, Choi Y, Taichman RS, Emerson SG. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood. 2007;109(9):3706–12. doi: 10.1182/blood-2006-08-041384. [DOI] [PubMed] [Google Scholar]
- 53.Kalajzic I, Staal A, Yang WP, Wu Y, Johnson SE, Feyen JH, Krueger W, Maye P, Yu F, Zhao Y, Kuo L, Gupta RR, Achenie LE, Wang HW, Shin DG, Rowe DW. Expression profile of osteoblast lineage at defined stages of differentiation. J Biol Chem. 2005;280(26):24618–26. doi: 10.1074/jbc.M413834200. [DOI] [PubMed] [Google Scholar]
- 54.Fox IJ, Daley GQ, Goldman SA, Huard J, Kamp TJ, Trucco M. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy for treating disease. Science. 2014;345(6199):1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang Y, Georgiou AI, MacFarlane RJ, Klontzas ME, Heliotis M, Tsiridis E, Mantalaris A. Fibronectin stimulates the osteogenic differentiation of murine embryonic stem cells. J Tissue Eng Regen Med. 2017;11(7):1929–1940. doi: 10.1002/term.2090. [DOI] [PubMed] [Google Scholar]
- 56.Xin X, Jiang X, Wang L, Stover ML, Zhan S, Huang J, Goldberg AJ, Liu Y, Kuhn L, Reichenberger EJ, Rowe DW, Lichtler AC. A Site-Specific Integrated Col2.3GFP Reporter Identifies Osteoblasts Within Mineralized Tissue Formed In Vivo by Human Embryonic Stem Cells. Stem Cells Transl Med. 2014;3(10):1125–37. doi: 10.5966/sctm.2013-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levi B, Hyun JS, Montoro DT, Lo DD, Chan CK, Hu S, Sun N, Lee M, Grova M, Connolly AJ, Wu JC, Gurtner GC, Weissman IL, Wan DC, Longaker MT. In vivo directed differentiation of pluripotent stem cells for skeletal regeneration. Proc Natl Acad Sci U S A. 2012;109(50):20379–84. doi: 10.1073/pnas.1218052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S, Kim SS, Lee SH, Eun Ahn S, Gwak SJ, Song JH, Kim BS, Chung HM. In vivo bone formation from human embryonic stem cell-derived osteogenic cells in poly(d,l-lactic-co-glycolic acid)/hydroxyapatite composite scaffolds. Biomaterials. 2008;29(8):1043–53. doi: 10.1016/j.biomaterials.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Zou L, Kidwai FK, Kopher RA, Motl J, Kellum CA, Westendorf JJ, Kaufman DS. Use of RUNX2 expression to identify osteogenic progenitor cells derived from human embryonic stem cells. Stem Cell Reports. 2015;4(2):190–8. doi: 10.1016/j.stemcr.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10(6):678–84. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(12):1117–9. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.