Abstract
Gemcitabine has been considered a first-line chemotherapy agent for the treatment of pancreatic cancer. However, the initial response rate of gemcitabine is low and chemoresistance occurs frequently. Aptamers can be effectively internalized into cancer cells via binding to target molecules with high affinity and specificity. In the current study, we constructed an aptamer-based gemcitabine delivery system, APTA-12, and assessed its therapeutic effects on pancreatic cancer cells in vitro and in vivo. APTA-12 was effective in vitro and in vivo in pancreatic cancer cells with high expression of nucleolin. The results of in vitro cytotoxicity assays indicated that APTA-12 inhibited the growth of pancreatic cancer cell lines. In vivo evaluation showed that APTA-12 effectively inhibited the growth of pancreatic cancer in Capan-1 tumor-bearing mice compared to mice that received gemcitabine alone or vehicle. These results suggest that the gemcitabine-incorporated APTA-12 aptamer may be a promising targeted therapeutic strategy for pancreatic cancer.
Keywords: aptamer, aptamer-drug conjugates, G-quadruplex, gemcitabine, pancreatic cancer
Graphical Abstract
Introduction
Pancreatic cancer is one of the most lethal tumors, and the prognosis of pancreatic cancer has not improved despite recent developments in cancer treatment. There were 48,960 newly diagnosed pancreatic cancer cases and 40,560 deaths recorded in the United States in 2015.1 Curative surgery is difficult in most advanced pancreatic cancers, and radiation therapy, conventional chemotherapy, and even targeted chemotherapy have shown disappointing results when applied to treat inoperable pancreatic cancers. The 5-year survival rate of pancreatic cancer patients remains only 5%.2
Gemcitabine, a deoxycytidine analog, has been used as a standard treatment for pancreatic cancer, but its response rate is very low. Gemcitabine can be internalized into cancer cells via nucleoside transporters, including human equilibrative nucleoside transporter 1 (hENT1) and human concentrative nucleoside transporters 1/3 (hCNT1/3).3 However, low expression levels of hENT1 and hCNT3 contribute to decreased efficacy of gemcitabine and increased chemoresistance.4, 5 In addition, gemcitabine has low molecular weight and high hydrophilicity and is easily degraded by cytidine deaminase, leading to poor cellular penetration.6, 7 To enhance the cellular uptake of gemcitabine, drug delivery systems, including nanoparticle, liposomes, albumin, and chitosan, have been broadly investigated.8
Aptamers are single-stranded oligonucleotides with highly specific target binding affinity that have been exploited in various fields. Aptamers show three-dimensional (3D) folding, which results in high affinity to specific targets, similar to antibodies.9 Aptamers may have the potential to substitute for antibodies because of their low immunogenicity and toxicity, thermal stability, large-scale production, and ease of chemical synthesis and chemical modification.10 Contrary to antibodies, the small molecular weight of aptamers allows rapid renal excretion, which displays a high target-to-background ratio in in vivo imaging.11, 12 Consequently, large numbers of aptamers have shown promise as target-specific imaging agents.13 Despite these initial successful trials, aptamer-based cancer therapy has been challenging. Indeed, few aptamers have been reported to have antiproliferative effects against colon cancer14 and ErbB2-positive breast cancer.15 Rapid distribution through the bloodstream and renal excretion via the urinary tract can be an advantage when aptamers are developed as diagnostic agents; however, this can be a major pitfall with respect to therapy. Degradation by nucleases in serum is another pitfall when aptamers are administered intravenously. However, aptamers are still attractive therapeutic agents because of their high target affinity and unique advantages.
AS1411 is an aptamer with high guanine content that results in a unique structure known as the G-quadruplex, which makes the 3D structure relatively stable relative to other aptamers. AS1411 binds to the cellular protein, nucleolin, which is known to be highly expressed by cancer cells.16 This aptamer has been shown to have anticancer effects against many cancers; therefore, some clinical studies of AS1411 were initiated.17 However, the results of phase II clinical trials of AS1411 were not good enough for it to be adopted as a first-line therapeutic agent.18
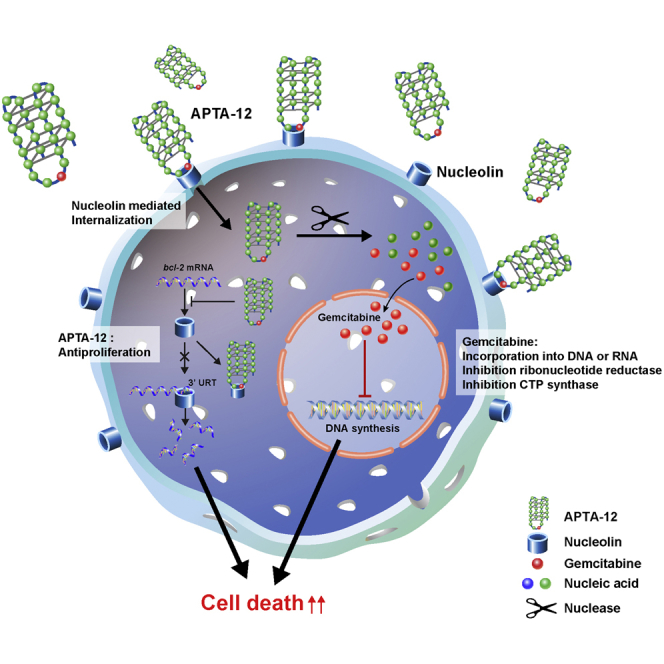
Recently, aptamer-drug conjugates (ApDCs) have been widely investigated in an attempt to overcome their poor cellular penetration, nonspecific toxicity, and immunogenicity.10, 19, 21 When compared with antibody-drug conjugates, development of ApDCs is easier and more convenient because aptamers are synthesized via chemical processes.22 In this study, we developed a gemcitabine-incorporated AS1411, APTA-12, as an ApDC and demonstrated that the developed compound has promising chemotherapeutic effects against pancreatic cancer.
Results
Design and Binding Properties of APTA-12
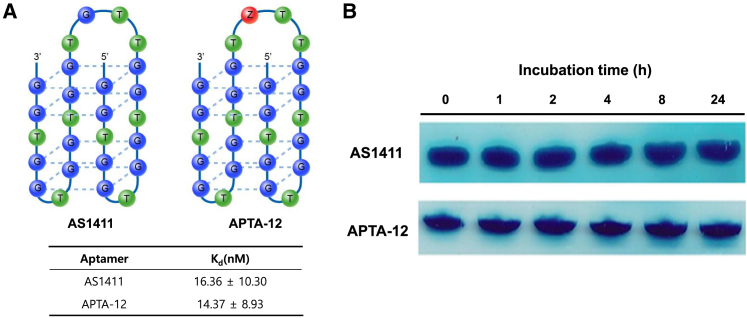
The objective of the project was to deliver the cytotoxic anticancer drug gemcitabine using oligonucleotide aptamers; thus, cell-type-specific internalization of the aptamer AS1411 was chosen as a model. Gemcitabine, one of the most widely used chemotherapeutic drugs, is a nucleoside deoxycytidine analog that can be incorporated into an oligonucleotide.23, 24 It has been reported that AS1411 forms a stable G-quadruplex structure and has the internal stem-loop structure located between 12 and 15 nucleotides.25 APTA-12 aptamers were created by single substitution of a guanine residue at position 14 of the AS1411 sequence with a gemcitabine phosphoramidite (Figure 1A). The sequence of the APTA-12 aptamer was 5′-GGT GGT GGT GGT TZT GGT GGT GGT GG-3′ (Z, gemcitabine). Gemcitabine was intentionally incorporated into the loop region of the AS1411 aptamer to avoid altering the tertiary structure and protect it from fast degradation by nucleases.
Figure 1.
Binding Affinity and Stability of APTA-12 Aptamer
(A) Schematic representation of gemcitabine-incorporated APTA12 and determination of dissociation constant (KD) values for the AS1411 and APTA-12 aptamers. (B) Serum stability of APTA-12 and AS1411. The electrophoresis bands in the figure represent retained APTA-12 aptamer in 50% serum-containing media after incubation.
A previous report showed that the loop region of the G-quadruplex played a key role in nucleolin binding, and chemical modification of the loop region could alter the protein-binding properties.26 To evaluate the impact of internal incorporation of gemcitabine into the loop region, the equilibrium dissociation constants (KD) of APTA-12 and AS1411 were determined. The APTA-12 aptamer displayed a similar binding affinity with a KD value of 14.37 ± 8.93 nM, and AS1411 showed a KD value of 16.36 ± 10.30 nM (Figure 1A). This result demonstrated that the gemcitabine modification of the loop region of the AS1411 sequence did not alter the binding affinity and that chemically modified APTA-12 retained a slightly higher binding affinity to nucleolin than the AS1411 aptamer.
Stability of APTA-12
To determine the stability of the APTA-12 aptamer, serum stability assays were carried out by incubating the APTA-12 aptamers in human serum. As indicated in Figure 1B, APTA-12 and AS1411 aptamers remained stable for up to 24 hr of incubation. It is known that G-quadruplex-forming oligonucleotides are relatively stable against nuclease degradation.27, 28 The high serum stability of guanine-rich APTA-12 can be explained by its stable G-quadruplex structure.
In Vitro Characterization of APTA-12
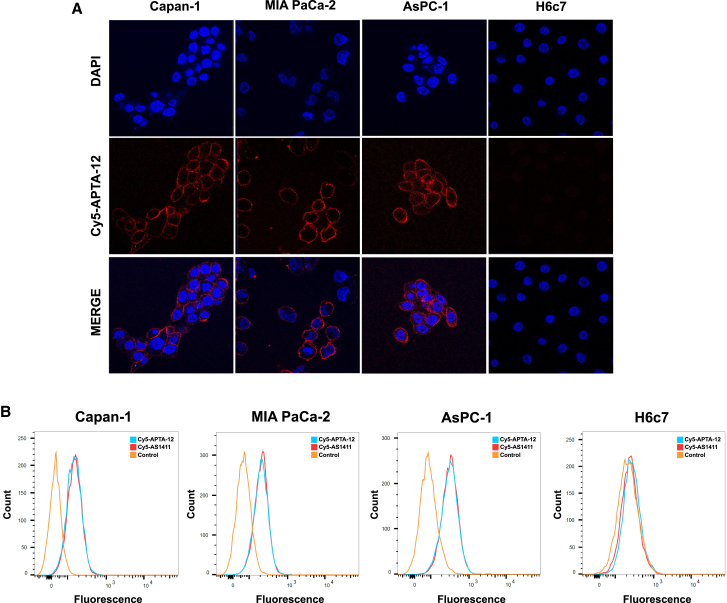
To investigate the binding specificity of the APTA-12 aptamer, pancreatic cancer cells were imaged by confocal microscopy after incubation with Cy5-labeled APTA-12. H6c7 cells were used as a negative control to determine nonspecific binding. As shown in Figure 2A, APTA-12 aptamers bound on the periphery of the Capan-1, AsPC-1, and MIA PaCa-2 cells, while no aptamer binding was observed in control H6c7 cells. Capan-1 cells were treated with Cy5-labeled cytosine-rich oligonucleotide (CRO), as a negative control. Cy5-CRO displayed only a weak fluorescence signals in Capan-1 cells (Figure S1A). To further characterize the binding specificity of the APTA-12 aptamer for pancreatic cancer cell lines, flow cytometric analysis was carried out using Cy5-labeled APTA-12. We found that APTA-12 aptamers were able to bind to all three pancreatic cancer cell lines (Figure 2B). Both APTA-12 and AS1411 aptamers had a relatively high affinity for Capan-1 cells (49.3% and 48.2% shift, respectively) compared to control cells (1.4% and 1.2% shift, respectively). Moreover, APTA-12 was shown to have much higher binding affinity for Capan-1 cells compared to negative control CRO (Figure S1B). These results suggest that binding of APTA-12 aptamer was specific to nucleolin.
Figure 2.
In Vitro Characterization of Gemcitabine-Incorporated APTA-12 Aptamer
(A) Confocal microscopy images of Capan-1, AsPC-1, MIA PaCa-2 cells, and negative control H6c7 cells incubated with Cy5-labeled APTA-12 (red). The nuclei were stained with DAPI (blue). (B) The specific binding capacity of APTA-12 to pancreatic cancer cells was assessed by flow cytometry. Nucleolin-positive Capan-1, MIA PaCa-2, and AsPC-1 cell lines and nucleolin-negative H6c7 cells were stained with Cy5-AS1411 or Cy5-APTA-12.
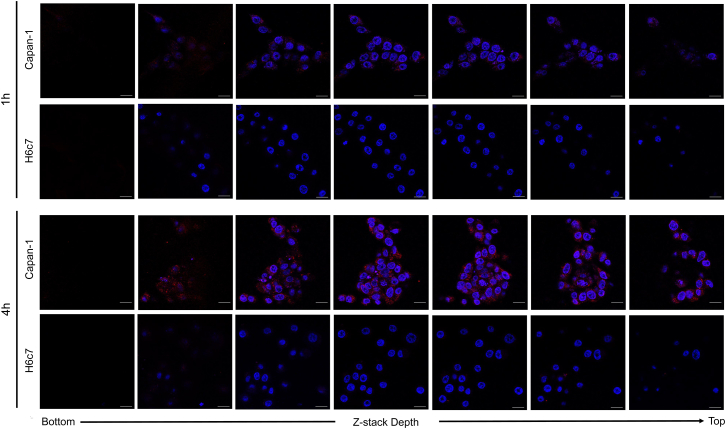
A previous study showed that AS1411 internalized into various cancer cell lines.17 To determine whether APTA-12 aptamers were internalized after binding to the cell-surface target, z stack images of Capan-1 and H6c7 cells were acquired. Confocal z stack images of 5′-Cy5-APTA-12 over a 4-hr period showed that APTA-12 aptamers were efficiently internalized into nucleolin-positive Capan-1 cells; however, the aptamers were not internalized into the H6c7 cells (Figure 3). These results indicated that APTA-12 is able to deliver the anticancer drugs or prodrugs selectively into target cells.
Figure 3.
Representative Confocal Fluorescence Microscopy Z Stack Images of Capan-1 and H6c7 Cells Incubated with Cy5-Labeled APTA-12
Z stack images were acquired from the bottom to the top of the cells after 1 hr and 4 hr incubation of Cy5-APTA-12 (red) at 37°C. Nuclei are stained with DAPI (blue). Scale bars, 20 μm.
In Vivo Characterization of APTA-12
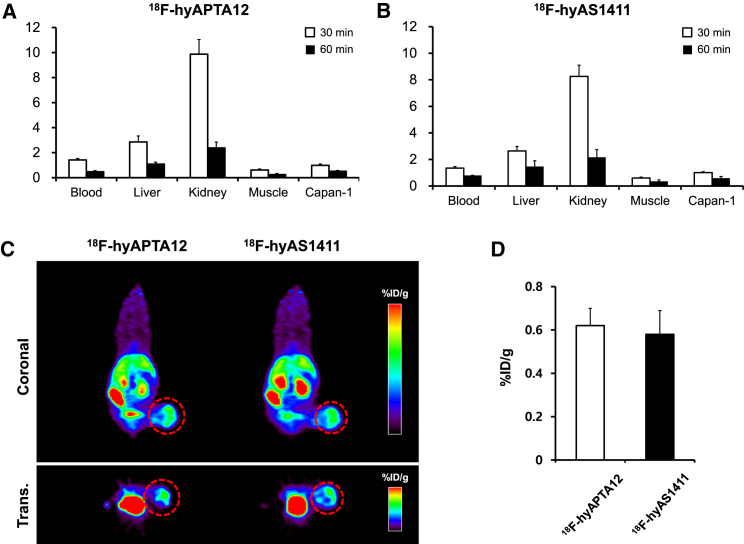
The ex vivo biodistribution of 18F-labeled APTA-12 was investigated in Capan-1 tumor-bearing mice. The blood-pool uptake of 18F-hyAPTA-12 decreased from 1.42 ± 0.12 percent injected dose per gram (%ID/g) at 30 min to 0.47 ± 0.09 %ID/g at 60 min after injection (Figure 4A). Kidney uptake of 18F-hyAPTA-12 also drastically decreased from 9.87 %ID/g at 30 min to 0.51 %ID/g at 60 min after injection. The retention values for 18F-hyAPTA-12 in the blood and tissues were quite similar to those for 18F-hyAS1411 (Figure 4B). This result agreed with previous reports that aptamers were quickly distributed through the bloodstream and rapidly eliminated via the urinary tract.11, 12 The tumor uptake of 18F-hyAPTA-12 averaged 0.98 ± 0.11 %ID/g at 30 min after injection, which decreased to 0.51 ± 0.07 %ID/g at 60 min. Positron emission tomography (PET) images were acquired at 60 min after injection of 18F-hyAPTA-12 and compared to 18F-hyAS1411. MicroPET imaging showed high uptake of 18F-hyAPTA-12 in the liver, kidneys, and intestines, which correlated with the biodistribution studies (Figure 4C). The tumor uptake of 18F-hyAPTA-12 (0.62 ± 0.08 %ID/g, n = 3) was similar to that of 18F-hyAS1411 (0.58 ± 0.11 %ID/g, n = 3) (Figure 4D).
Figure 4.
In Vivo Characterization of F-18-Labeled APTA-12 in Capan-1 Tumor-Bearing Mice
Quantitative analysis of biodistribution of (A) 18F-hyAPTA-12 aptamer and (B) 18F-hyAS1411 aptamer. Mice were sacrificed at 30 min and 60 min after injection with 300 pmol of 18F-hyAPTA-12 or 18F-hyAS1411 (n = 4, at each concentration). The radioactivity was expressed as %ID/g. (C) Representative PET images at 60 min after injection of 18F-hyAPTA-12 or 18F-hyAS1411. The red-dotted circles indicate Capan-1 tumors. (D) Quantification analysis of 18F-labeled APTA-12 and AS1411 accumulation in Capan-1 tumors derived from the PET images at 60 min after injection. The radioactivity was expressed as %ID/g.
Cell Cytotoxicity in Pancreatic Cancer Cell Lines
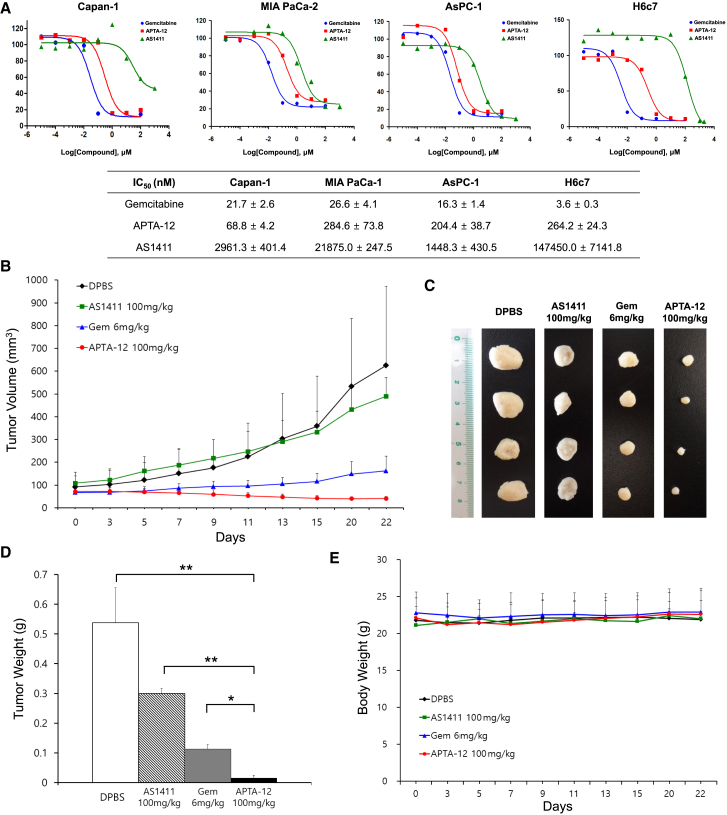
Previous reports showed that AS1411 aptamers have antiproliferative activity against various cancer cell lines.29, 30 And gemcitabine has a broad spectrum of antitumor activity, including pancreatic cancer.31, 32 To assess the potential cytotoxic effects of gemcitabine-incorporated APTA-12 aptamers on pancreatic cancer cells in vitro, IC50 values were determined. As indicated in Figure 5A, APTA-12 inhibited the growth of Capan-1, MIA PaCa-2, and AsPC-1 cells, with IC50 values of 68.8 ± 4.2, 284.6 ± 73.8, and 204.4 ± 38.7 nM, respectively. The lowest IC50 value of APTA-12 was found in Capan-1 cells. The half-maximal inhibitory concentration (IC50) values of APTA-12-treated Capan-1 cells were 43-fold lower than AS1411-treated Capan-1 cells (IC50 = 2,961.3 ± 401.4 nM). Gemcitabine showed similar growth inhibition against pancreatic cancer cells. The IC50 values for the normal cell H6c7 were 3.6 ± 0.3 nM for gemcitabine and 147,450.0 ± 7,141.8 nM for AS1411. In this study, Capan-1 cells were chosen for further in vivo studies because of their high receptor-binding affinity and high sensitivity to the cytotoxic effects of APTA-12.
Figure 5.
Therapeutic Efficacy of Gemcitabine-Incorporated APTA-12 Aptamer against Pancreatic Cancer Cells
(A) IC50 values of gemcitabine-, AS1411-, and APTA-12-treated pancreatic cell lines (Capan-1, MIA PaCa-1, AsPC-1) and normal cell (H6c7) after 6 days treatment at various concentrations (0.00001–1,000 μM). (B) The growth curves of Capan-1 tumor-bearing mice treated with intravenous injections of gemcitabine (6 mg/kg), AS1411 (100 mg/kg), APTA-12 (100 mg/kg), or vehicle (DPBS) alone. (C) Representative image of tumors excised on day 22. (D) Changes in mean tumor weight and (E) the body weights of mice treated with gemcitabine, AS1411, APTA-12, and vehicle. Data are presented as the mean ± SD. *p < 0.05; **p < 0.01.
Antitumor Efficacy of APTA-12 In Vivo
The antitumor efficacy of APTA-12 was investigated using Capan-1 tumor xenograft models. Tumors from vehicle-treated mice showed a 6.9-fold increase in size during the treatment schedule. Tumors in mice treated with gemcitabine alone at 6 mg/kg grew slowly to about 2.4 times their initial size (Figure 5B). However, mice injected with 100 mg/kg APTA-12 exhibited approximately 93.4% tumor growth inhibition when compared to control mice injected with Dulbecco’s PBS (DPBS) alone and a 42.5% tumor shrinkage compared to the tumor size at the beginning of treatment (Figure 5C). The average tumor weight of the APTA-12-treated group was significantly reduced compared to tumors of the AS1411- or vehicle-treated groups (unpaired t test; p < 0.01). In addition, 100 mg/kg APTA-12 had a stronger antitumor effect compared with 6 mg/kg gemcitabine (p < 0.05) (Figure 5D).
The body weight of mice was monitored during the treatment. The weight loss of the mice treated with gemcitabine or APTA-12 did not exceed 10% of their initial body weight (Figure 5E). The mean weight in the 100 mg/kg APTA-12-treated group was 22.5 ± 0.5 g and was not different from that of the 100 mg/kg AS1411-treated group (22.7 ± 0.7 g) or 6 mg/kg gemcitabine-treated group (22.9 ± 0.6 g). To evaluate potential toxicity of APTA-12, peripheral red blood cell (RBC) and white blood cell (WBC) counts were measured. There was no significant difference in the levels of RBC and WBC between the gemcitabine-, AS1411-, APTA-12-, and vehicle (DPBS)-treated groups (Table 1). To investigate the effects of APTA-12 on liver and kidney function, blood levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), blood urea nitrogen (BUN), and creatinine were determined. There were no significant differences in serum biochemical values between the APTA-12-treated group and the vehicle-treated group (p ≥ 0.05) (Table 2). This result suggested that APTA-12 treatment caused no major toxicity or side effects.
Table 1.
Comparison of Hematological Values between Gemcitabine-, AS1411-, and APTA-12-Treated Mice (Mean ± SD)
| Parameters | Control | Gemcitabine 6 mg/kg | AS1411 100 mg/kg | APTA-12 100 mg/kg |
|---|---|---|---|---|
| Red blood cell (M/μL) | 10.5 ± 0.3 | 10.2 ± 0.5 | 9.8 ± 0.5 | 9.8 ± 0.6 |
| Hematocrit (%) | 59.6 ± 2.0 | 57.4 ± 2.5 | 54.1 ± 3.6 | 58.8 ± 4.6 |
| Mean corpuscular volume (fL) | 54.4 ± 1.2 | 56.2 ± 0.6 | 55.3 ± 0.8 | 60.2 ± 2.0 |
| Mean cell hemoglobin (pg) | 13.7 ± 0.5 | 12.2 ± 2.9 | 7.7 ± 8.8 | 14.5 ± 0.5 |
| Mean cell hemoglobin concentration (g/dL) | 25.1 ± 0.6 | 21.8 ± 5.0 | 26.0 ± 1.5 | 24.1 ± 0.4 |
| White blood cell (k/μL) | 0.9 ± 0.2 | 0.8 ± 0.2 | 0.7 ± 0.5 | 1.7 ± 0.4 |
| Neutrophil (k/μL) | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.2 | 0.6 ± 0.2 |
| Lymphocyte (k/μL) | 0.6 ± 0.2 | 0.4 ± 0.1 | 0.4 ± 0.1 | 0.9 ± 0.2 |
| Monocyte (k/μL) | 0.1 ± 0.0 | 0.1 ± 0.1 | 0.0 ± 0.0 | 0.2 ± 0.1 |
| Eosinophil (k/μL) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.1 | 0.0 ± 0.0 |
| Basophil (k/μL) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
Table 2.
Serum Biochemical Values of Mice Treated with Gemcitabine, AS1411, and APTA-12 (Mean ± SD)
| Parameters | Control | Gemcitabine 6 mg/kg | AS1411 100 mg/kg | APTA-12 100 mg/kg |
|---|---|---|---|---|
| Alanine aminotransferase (U/L) | 25.5 ± 2.1 | 33.0 ± 1.4 | 34.5 ± 3.5 | 28.5 ± 7.8 |
| Aspartate aminotransferase (U/L) | 128.5 ± 27.6 | 233.0 ± 7.1 | 194.0 ± 63.6 | 174.0 ± 89.1 |
| Alkaline phosphatase (U/L) | 215.0 ± 9.9 | 177.0 ± 15.6 | 131.0 ± 9.9 | 230.0 ± 12.7 |
| Blood urea nitrogen (mg/dL) | 22.2 ± 2.3 | 16.3 ± 0.0 | 10.2 ± 1.2 | 16.4 ± 1.0 |
| Creatinine (mg/dL) | 0.2 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.1 | 0.1 ± 0.0 |
To evaluate the therapeutic effect of APTA-12, 18F-fluorodeoxyglucose (18F-FDG) PET was performed at the end of treatment. The microPET quantification in APTA-12-treated mice showed a clear reduction in the mean standardized uptake value (SUVmean) compared to the vehicle-treated mice (Figure S2A). The SUVmean of tumors was 0.29 ± 0.03 for the APTA-12 (100 mg/kg)-treated group and 1.32 ± 0.06 for vehicle-treated group, which were significantly different (unpaired t test, p < 0.001) (Figure S2B). 18F-FDG PET imaging confirmed that tumor growth in APTA-12-treated mice was markedly inhibited compared to the control group.
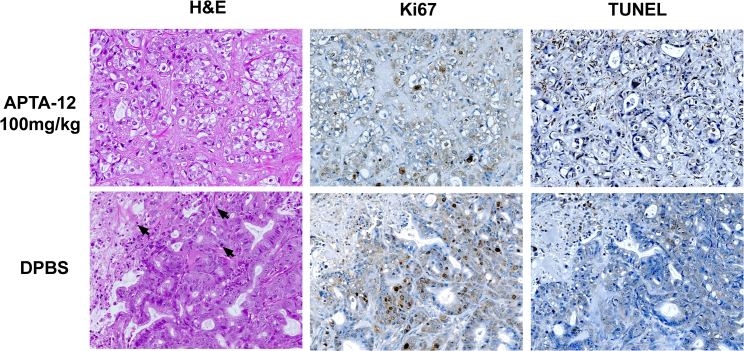
Cell apoptosis and proliferation in tumors were analyzed by H&E, TUNEL, and Ki67 assay (Figure 6). H&E staining of tumor sections from the vehicle-treated group showed a greater number of mitotic cells than the APTA-12-treated group. APTA-12 induced at least a 4-fold increase in the number of TUNEL-positive apoptotic cells compared to vehicle (p < 0.05). Cell proliferation was evaluated by Ki67 immunostaining. The numbers of Ki67-positive cells in the tumors of the APTA-12 (100 mg/kg)-treated group were 3.7-fold lower than the vehicle-treated group (p < 0.05). These results indicated that APTA-12 at a therapeutic dosage efficiently inhibited in vivo tumor growth in Capan-1 tumor-bearing mice.
Figure 6.
H&E, Ki67, and TUNEL Analysis of Tumor Tissues of Capan-1 Tumor-Bearing Mice after Treatment with APTA-12
H&E stain showed that more mitotic figures and necrosis were noted in DPBS-treated group. Black arrows indicate mitotic figures. Ki67-positive cells were quantified by counting five fields from each tumor. The results indicated significant reduction of Ki67 expression in the APTA-12-treated mice (p < 0.05). APTA-12 treatment-induced apoptosis shown by TUNEL staining of Capan-1 tumors. Quantification of TUNEL-positive cells shows that inhibition of APTA-12 led to a four-fold increase in apoptotic cells (p < 0.05). Magnification, ×200.
Discussion
Here, we report the characterization of the gemcitabine-incorporated aptamer, APTA-12, which exhibited potent antitumor activity in pancreatic cancer cells. APTA-12, which binds to nucleolin with high affinity, showed antiproliferative activity in pancreatic cancer cells in vitro. In addition, APTA-12 significantly inhibited tumor growth in Capan-1 tumor-bearing mice in vivo.
The clinical use of anticancer drugs is limited by their general toxicity to proliferating cells and low selectivity.33 Therefore, there has been an effort to develop new anticancer agents that are highly effective and less toxic. Since the discovery that a short antisense oligodeoxynucleotide could inhibit RNA translation and replication of the Rous sarcoma virus in 1978, therapeutic oligonucleotides including antisense oligonucleotides, RNA interference, decoy oligonucleotides, and aptamers have been extensively investigated and applied to the treatment of various cancers.34, 35, 36, 37 Previous studies have shown that guanine-rich oligonucleotides (GROs) bind to specific cellular proteins and inhibit the proliferation of various cancer cells.38 GROs can form stable G-quadruplex structures because three to four guanine bases form a square planar structure called a G-quartet through Hoogsteen base pairing, and these G-quartets spontaneously assemble into four-stranded helical structures termed G-quadruplexes.39, 40 The major disadvantage in the use of oligonucleotides in clinical applications is metabolic lability, rapid systemic clearance, and low cell permeability.41, 42 However, oligonucleotides that form G-quadruplexes display relatively higher cell permeability, thermostability, and nuclease resistance than non-G-quartet-forming oligonucleotides.18, 38, 39 One of the most widely studied G-quadruplex forming oligonucleotides is DNA aptamer AS1411. Aptamers are relatively short (20–60 nucleotides) single-stranded oligonucleotides capable of binding to their target molecules through structural recognition-like antibodies.43, 44 AS1411 can bind to nucleolin that is overexpressed on the surface of cancer cells and then be internalized into the cytoplasm and nucleus, where it exerts antiproliferative effects through inhibition of nuclear factor-κB (NF-κB) signaling or destabilization of bcl-2 mRNA.29, 45 AS1411 has also been investigated for its potential usefulness in the delivery of anticancer drugs.46, 47, 48 Trinh et al.48 assessed the antitumor efficacy of doxorubicin-conjugated AS1411t and found that the AS1411-doxorubicin adduct could efficiently deliver doxorubicin into target cells and inhibit tumor growth without inducing apoptosis.
The main objective of the present study was to develop a method for delivery of the anticancer drug gemcitabine using an oligonucleotide aptamer and evaluate its in vivo antitumor efficacy against pancreatic tumors. Gemcitabine is a deoxycytidine analog that inhibits DNA synthesis via incorporation of its diphosphate metabolite into the DNA strand. Recent studies showed that chemically modified AS1411 aptamer significantly improved targeting affinity and inhibition of cancer cell proliferation.25, 49 We hypothesized that gemcitabine could be incorporated into nucleolin-targeting aptamers and internalized into tumor cells after binding to cell-surface nucleolin. The internalized gemcitabine-incorporated AS1411 then exerted therapeutic effects through destabilization of bcl-2 mRNA and incorporation of gemcitabine into DNA after degradation by nuclease. With this in mind, a gemcitabine-modified G-quadruplex-forming oligonucleotide, APTA-12, was designed and synthesized based on the AS1411 aptamer. To verify the hypothesis, the nucleolin-binding affinity and serum stability of APTA-12 were evaluated. The measured KD values and in vitro serum stability of APTA-12 were comparable to those of the AS1411 aptamer. We also demonstrated that APTA-12 could efficiently deliver gemcitabine into pancreatic cancer cells. In the confocal image of Capan-1, AsPC-1, and MIA PaCa-2 cells, APTA-12 aptamers were selectively bound to the cell surface. z stack imaging showed that internalized APTA-12 aptamers accumulated only in nucleolin-positive cells, while flow cytometric analysis demonstrated that the APTA-12 aptamer had similar or higher affinity to nucleolin protein than the AS1411 aptamer. Furthermore, APTA-12 treatment of pancreatic cancer cells significantly reduced cell proliferation and the IC50 value of APTA-12-treated cells was much lower than that of the AS1411-treated cells. In vivo evaluation showed that APTA-12 effectively inhibited the growth of pancreatic cancer cells in nude mice. [18F]FDG SUVmean in the tumor region of the APTA-12-treated mice decreased by 78% when compared to the vehicle-treated mice. No significant differences in body weights were noted among treatment groups. TUNEL and Ki67 assay showed that APTA-12 induced more apoptotic cell death and reduced the numbers of proliferating tumor cells compared to the control group. Taken together, these results demonstrated the antitumor efficacy of gemcitabine-incorporated APTA-12.
Various types of ApDCs have been developed to efficiently deliver anticancer drugs to target sites.21, 50 Typically, chemotherapy drugs can be conjugated to aptamers via functional linkers or physical intercalation.21 In this study, single gemcitabine was chemically incorporated into the loop region of the DNA aptamer AS1411 without using linkers or physical intercalation. The concept of intrinsic incorporation of gemcitabine into oligonucleotide was first reported by Ray et al.51 In their study, gemcitabine was incorporated into oligonucleotides by mutant RNA polymerases, and gemcitabine-containing oligonucleotides were annealed to the aptamer for targeted drug delivery. Recently, Yoon et al.52 synthesized gemcitabine or 5-fluorouracil-incorporated RNA aptamer and showed that it was internalized and inhibited cell proliferation of pancreatic cancer cells in vitro. Intrinsic incorporation of oncolytic nucleoside analog into aptamers might be a good strategy for the development of ApDCs because of easy chemical modification without loss of affinity and large-scale chemical synthesis. In the present study, we demonstrate that gemcitabine-incorporated APTA-12 exerted growth-inhibitory properties against different pancreatic cancer cell lines and possessed a higher therapeutic effect than gemcitabine alone in in vivo. These results suggest that aptamer-based gemcitabine delivery of APTA-12 is a promising anticancer strategy for targeted pancreatic cancer therapy.
Materials and Methods
Cell Lines and Reagents
The human pancreatic cancer cell lines Capan-1, AsPC-1, and MIA PaCa-2 were purchased from the American Type Culture Collection (Manassas, VA, USA). Noncancerous human pancreatic duct epithelial cell line HPDE6-C7 (H6c7) was purchased from Kerafast (Boston, MA, USA). Cells were grown in RPMI medium (Capan-1 and AsPC-1) or DMEM medium (MIA PaCa-2) supplemented with penicillin G (100 U/mL), streptomycin (100 μg/mL), and 10% fetal bovine serum (FBS). H6c7 cells were maintained in keratinocyte serum-free media supplemented with epidermal growth factor and bovine pituitary extract (Life Technologies, Grand Island, NY). All cells were incubated at 37°C in a humidified incubator with 5% CO2/95% air. Cell culture media, supplements, and serum products were purchased from Invitrogen (Carlsbad, CA, USA). AS1411, APTA-12, CRO, 5′-Cy5-AS1411 and 5′-Cy5-APTA-12 aptamers were synthesized in house. All reagents were commercially available and used without modification.
Synthesis of Aptamers
All aptamers were synthesized using a commercial DNA synthesizer (PolyGen, Langen, Germany) following the standard phosphoramidite method.53 The AS1411 sequence was 5′-GGT GGT GGT GGT TGT GGT GGT GGT GG-3′, and the APTA-12 sequence was 5′-GGT GGT GGT GGT TZT GGT GGT GGT GG-3′ (Z, gemcitabine). CRO (5′-CCT CCT CCT CCT TCT CCT CCT CCT CC-3′) was synthesized as a negative control. A 0.1 M solution of phosphoramidites in anhydrous acetonitrile was used for the synthesis. Gemcitabine phosphoramidite was prepared by a modification of the previously reported procedure.51 Cy5 was coupled to the 5′ termini of AS1411, APTA-12, and CRO. Crude oligonucleotides were purified using a 1260 Infinity II reversed-phase high-performance liquid chromatograph (Agilent Technologies, Santa Clara, CA, USA) on an XTerra RP18 column (10 μm, 7.8 × 300 mm, Waters, Milford, MA, USA). The eluent was acetonitrile containing 0.1 M triethylammonium acetate buffer. The resulting product was characterized by LCMS-2020 (Shimadzu, Kyoto, Japan), and the purity was assessed using the 1260 Infinity II LC system.
Nucleolin-Binding Assay
Nucleolin protein was purchased from Vaxron (Rockaway, NJ, USA). The affinity of nucleolin for AS1411 and APTA-12 was assessed according to the modified method of Carballo et al.54 The 5′ termini of AS1411 and APTA-12 were labeled with phosphorus-32 (32P) using [γ-32P]-ATP (cat. #NEG502A; PerkinElmer, Waltham, MA, USA) and T4 polynucleotide kinase (cat. #M0201L; New England Biolabs, Ipswitch, MA, USA) at 37°C for 30 min. Unincorporated [γ-32P]-ATP was removed using MicroSpin G-25 Columns (GE Healthcare, Buckinghamshire, UK). For the affinity assay, 32P-labeled AS1411 and APTA-12 were incubated with nucleolin protein at various concentrations at 37°C for 15 min. Bound complexes were mixed with Zorbax PSM-300 resin (Agilent Technologies) and captured on a Durapore filter (Millipore, Bedford, MA). Filter plates were exposed overnight and quantitated on a FLA-5100 (Fuji, Düsseldorf, Germany). Dissociation constant (KD) and maximal binding (Bmax) were calculated using SigmaPlot10 (Systat Software, San Jose CA, USA).
In Vitro Stability
AS1411 and APTA-12 were incubated with H4522 serum (Human AB serum, Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 50% and incubated at 37°C for 0, 1, 2, 4, 8, and 24 hr. At each time point, a sample was taken and quick-frozen by placement in a deep freezer. Prior to analysis, samples were quickly thawed at 37°C and diluted with water and PCI (phenol-chloroform-isoamyl alcohol) solution. The mixtures were centrifuged at 10,000 × g for 15 min, and supernatants were used for PAGE analysis. Gels were stained with 0.5% methylene blue solution to reveal the oligonucleotide aptamers.
Confocal Fluorescence Microscopy
Capan-1, AsPC-1, MIA PaCa-2, and H6c7 cells were grown on 3.5-cm glass-bottom dishes (MatTek Corporation, Ashland, MA, USA). To stain the plasma membrane and cytoplasmic nucleolin, the cells were fixed for 20 min at room temperature in PBS containing 4% paraformaldehyde. The cells were incubated with 200 nM of 5′-Cy5-APTA-12 in binding buffer (DPBS supplemented with 4.5 g/L glucose, 5 mM MgCl2, 0.1 mg/mL yeast tRNA, and 1 mg/mL BSA) at 4°C with gentle rocking, and then washed three times with ice-cold PBS. Confocal images were obtained using a Zeiss LSM-700 confocal laser-scanning microscope (Carl Zeiss, Oberkochen, Germany).
For confocal z stack imaging, Capan-1 and H6c7 cells were treated with 200 nM of 5′-Cy5-APTA-12 and then incubated at 37°C in 5% CO2 for 30 min, 1 hr, 2 hr, and 4 hr. At the indicated time points, the cells were washed with binding buffer and fixed with 4% paraformaldehyde. Confocal z stack images were acquired using a Zeiss LSM 770 and processed with ZEN 2010 image software.
Flow Cytometry
Capan-1, AsPC-1, MIA PaCa-2, and H6c7 cells were incubated with 200 pmol of 5′-Cy5-AS1411 or 5′-Cy5-APTA-12 at 4°C for 30 min in binding buffer. Cells were then washed twice with PBS, and flow cytometric analyses were performed on an LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Cell Viability Assays
Cytotoxicity of gemcitabine, AS1411, and APTA-12 was evaluated using a cell-proliferation reagent WST-1 (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s specifications. In brief, Capan-1, AsPC-1, MIA PaCa-2, and H6c7 cells were seeded into 96-well plates at a density of 1 × 104 cells per well and incubated at 37°C in 5% CO2. After 24 hr, the media was removed and replaced with fresh medium containing 5% FBS. The cells were treated with different concentrations (0.00001–1,000 μM) of gemcitabine (Sigma-Aldrich, cat. #G6423), AS1411, or APTA-12 and then incubated at 37°C for 6 days. All experiments were performed in triplicate. Cell viability was measured, and the IC50 was obtained from the dose-response curve using GraphPad PRISM (GraphPad Software, San Diego, CA, USA).
Animal Model
Athymic female nude mice were purchased from Orient Bio (Gyeonggido, South Korea). The animal experiments were performed with the approval of the Institutional Animal Care and Ethics Committee of Yonsei Laboratory Animal Research Center. For the tumor model, a suspension of 1 × 106 Capan-1 cells in PBS was injected subcutaneously in the right thigh of 7-week-old BALB/c nude mice.
Biodistribution Study
For the biodistribution study, 18F-hyAS1411 or 18F-hyAPTA-12 was synthesized as previously described.12 Capan-1 tumor-bearing mice were anesthetized with 2% isoflurane and injected intravenously with radiotracer 18F-hyAPTA-12 (3.7 ± 0.2 megabecquerel [MBq], 300 pmol) via the tail vein. Mice were sacrificed at 30 and 60 min (n = 4 for each group) after injection. Whole blood, tumor, and other major organs and tissues were excised and wet-weighed. Radioactivity in each sample was measured using a Wizard2 2480 gamma counter (PerkinElmer, Woodbridge, Ontario, Canada), and radioactivity uptake was expressed as percent injected dose per gram (%ID/g).
PET Imaging
For PET imaging, Capan-1 tumor-bearing mice were injected intravenously with 18F-hyAPTA-12 (7.4 ± 0.5 MBq, 300 pmol) or 18F-FDG (5.6 ± 0.5 MBq) via tail vein. PET imaging was performed using an Inveon microPET scanner (Siemens, Knoxville, TN, USA). PET images were acquired for 10 min using the static acquisition mode at 60 min after injection, and images were reconstructed by a 3-dimensional ordered-subsets expectation maximum (OSEM) algorithm. Quantification analysis was performed as previously reported.55
In Vivo Tumor Growth Inhibition
The in vivo therapeutic effect of APTA-12 was evaluated in mice bearing Capan-1 xenografts. When the tumor volumes reached approximately 100 mm3, Capan-1 tumor-bearing mice were randomly separated into four groups (n = 8 per group) as follows: group I, tumor control (DPBS); group II, gemcitabine 6 mg/kg; group III, APTA-12 100 mg/kg; group IV, AS1411 100 mg/kg. The absolute amount of gemcitabine in 100 mg/kg APTA-12 was equivalent to 6 mg/kg gemcitabine. Gemcitabine, APTA-12, and AS1411 were dissolved in DPBS and then injected intravenously into mice every other day eight times. Body weight and tumor volume were measured every other day. Tumor volume was determined according to the following formula: tumor volume (mm3) = a × b2 × 0.52, where a was the longest diameter and b was the shortest diameter. The mice were sacrificed on day 22, and the tumors were excised, weighed, and photographed.
Hematological and Biochemical Parameters
Prior to tumor dissection, blood samples were collected through cardiac puncture under the isoflurane anesthesia on day 22. Whole blood was collected in an EDTA-coated Microtainer tube (BD Biosciences, Franklin Lakes, NJ) for complete blood counts (CBC), and centrifuged at 2,500 × g for 10 min. Hematological parameters were analyzed using HEMAVET 950 hematology system (Drew Scientific, Waterbury, CT).
Blood was collected in Microtainer Serum Separator tubes (BD Biosciences), and serum was obtained after centrifugation at 2,000 × g for 20 min. The biochemical parameter testing was performed using an automated biochemistry analyzer (FUJI DRI-CHEM 4000i, Fuji, Tokyo, Japan). Liver function was evaluated based on the serum levels of AST, ALT, and ALP. Renal function was analyzed by serum creatinine and BUN.
Histological and Immunohistochemical Analysis
The tumors were fixed in 4% formalin, embedded in paraffin, and sectioned at 4- to 5-μm thickness. The sections were deparaffinized, dehydrated, and stained with H&E for histological analysis. Tumor cell proliferation was measured by Ki67 staining with anti-Ki67 antibody (Abcam, Cambridge, MA) as described previously.56 The detection of apoptotic cells in tumor tissue sections was performed using the TACS 2TdT-DAB In Situ Apoptosis Detection Kit (Trevigen, Gaithersburg, MD) following the manufacturer’s instructions. Cell nuclei were counterstained with methyl green solution. The number of terminal deoxynucleotidyl TUNEL and Ki67-positive cells was counted by double-blind evaluations. TUNEL and Ki67-positive cells were quantified manually using ImageJ software according to a method described previously.57
Statistical Analysis
Quantitative data were expressed as mean ± SD. Data were compared using one-way ANOVA and Student’s t test. p values < 0.05 were considered statistically significant.
Author Contributions
W.J.K. and J.Y.P. designed and coordinated the research. J.Y.P. wrote the manuscript. J.Y.P., Y.L.C., J.R.C., and S.J.L. conducted all experiments and analyzed the data. S.J.L. and S.H.M. provided essential reagents. W.J.K., W.G.C., Y.J.C., and S.H.M. were involved in final paper editing.
Conflicts of Interest
S.H.M. and S.J.L. are co-inventors of APTA-12. The patent for APTA-12 is held by the Aptabio Therapeutics Inc. The other co-authors declare no conflicts of interest.
Acknowledgments
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI17C1491), and the National R&D Program for Cancer Control, Ministry of Health and Welfare (1320210). The authors would like to thank Dong-Su Jang, (Medical Illustrator, Medical Research Support Section, Yonsei University College of Medicine, Seoul, Korea) for his help with the illustrations and Ji Hae Nahm (Department of Pathology, Yonsei University College of Medicine, Seoul, Korea) for her assistance in immunohistochemical analysis.
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.06.003.
Contributor Information
Soo Jin Lee, Email: novagene2@hanmail.net.
Won Jun Kang, Email: mdkwj@yuhs.ac.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Wolfgang C.L., Herman J.M., Laheru D.A., Klein A.P., Erdek M.A., Fishman E.K., Hruban R.H. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueno H., Kiyosawa K., Kaniwa N. Pharmacogenomics of gemcitabine: can genetic studies lead to tailor-made therapy? Br. J. Cancer. 2007;97:145–151. doi: 10.1038/sj.bjc.6603860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maréchal R., Mackey J.R., Lai R., Demetter P., Peeters M., Polus M., Cass C.E., Young J., Salmon I., Devière J., Van Laethem J.L. Human equilibrative nucleoside transporter 1 and human concentrative nucleoside transporter 3 predict survival after adjuvant gemcitabine therapy in resected pancreatic adenocarcinoma. Clin. Cancer Res. 2009;15:2913–2919. doi: 10.1158/1078-0432.CCR-08-2080. [DOI] [PubMed] [Google Scholar]
- 5.Mackey J.R., Mani R.S., Selner M., Mowles D., Young J.D., Belt J.A., Crawford C.R., Cass C.E. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58:4349–4357. [PubMed] [Google Scholar]
- 6.Vrignaud S., Benoit J.P., Saulnier P. Strategies for the nanoencapsulation of hydrophilic molecules in polymer-based nanoparticles. Biomaterials. 2011;32:8593–8604. doi: 10.1016/j.biomaterials.2011.07.057. [DOI] [PubMed] [Google Scholar]
- 7.Amrutkar M., Gladhaug I.P. Pancreatic cancer chemoresistance to gemcitabine. Cancers (Basel) 2017;9:157. doi: 10.3390/cancers9110157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birhanu G., Javar H.A., Seyedjafari E., Zandi-Karimi A. Nanotechnology for delivery of gemcitabine to treat pancreatic cancer. Biomed. Pharmacother. 2017;88:635–643. doi: 10.1016/j.biopha.2017.01.071. [DOI] [PubMed] [Google Scholar]
- 9.Ruscito A., DeRosa M.C. Small-molecule binding aptamers: selection strategies, characterization, and applications. Front Chem. 2016;4:14. doi: 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H., Zhu X., Lu P.Y., Rosato R.R., Tan W., Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson O., Yan X., Niu G., Weiss I.D., Ma Y., Szajek L.P., Shen B., Kiesewetter D.O., Chen X. PET imaging of tenascin-C with a radiolabeled single-stranded DNA aptamer. J. Nucl. Med. 2015;56:616–621. doi: 10.2967/jnumed.114.149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J.Y., Lee T.S., Song I.H., Cho Y.L., Chae J.R., Yun M., Kang H., Lee J.H., Lim J.H., Cho W.G., Kang W.J. Hybridization-based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials. 2016;100:143–151. doi: 10.1016/j.biomaterials.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 13.Hong H., Goel S., Zhang Y., Cai W. Molecular imaging with nucleic acid aptamers. Curr. Med. Chem. 2011;18:4195–4205. doi: 10.2174/092986711797189691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y.J., Han S.R., Kim N.Y., Lee S.H., Jeong J.S., Lee S.W. An RNA aptamer that binds carcinoembryonic antigen inhibits hepatic metastasis of colon cancer cells in mice. Gastroenterology. 2012;143 doi: 10.1053/j.gastro.2012.03.039. 155–65.e8. [DOI] [PubMed] [Google Scholar]
- 15.Mahlknecht G., Maron R., Mancini M., Schechter B., Sela M., Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc. Natl. Acad. Sci. USA. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bates P.J., Reyes-Reyes E.M., Malik M.T., Murphy E.M., O’Toole M.G., Trent J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta. 2017;1861(5 Pt B):1414–1428. doi: 10.1016/j.bbagen.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg J.E., Bambury R.M., Van Allen E.M., Drabkin H.A., Lara P.N., Jr., Harzstark A.L., Wagle N., Figlin R.A., Smith G.W., Garraway L.A. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs. 2014;32:178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagalkot V., Farokhzad O.C., Langer R., Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 21.Zhu G., Niu G., Chen X. Aptamer-Drug Conjugates. Bioconjug. Chem. 2015;26:2186–2197. doi: 10.1021/acs.bioconjchem.5b00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y.F., Shangguan D., Liu H., Phillips J.A., Zhang X., Chen Y., Tan W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson F.C., Richardson K.K., Kroin J.S., Hertel L.W. Synthesis and restriction enzyme analysis of oligodeoxyribonucleotides containing the anti-cancer drug 2′,2′-difluoro-2′-deoxycytidine. Nucleic Acids Res. 1992;20:1763–1768. doi: 10.1093/nar/20.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toschi L., Finocchiaro G., Bartolini S., Gioia V., Cappuzzo F. Role of gemcitabine in cancer therapy. Future Oncol. 2005;1:7–17. doi: 10.1517/14796694.1.1.7. [DOI] [PubMed] [Google Scholar]
- 25.Fan X., Sun L., Wu Y., Zhang L., Yang Z. Bioactivity of 2′-deoxyinosine-incorporated aptamer AS1411. Sci. Rep. 2016;6:25799. doi: 10.1038/srep25799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotula J.W., Pratico E.D., Ming X., Nakagawa O., Juliano R.L., Sullenger B.A. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Ther. 2012;22:187–195. doi: 10.1089/nat.2012.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop J.S., Guy-Caffey J.K., Ojwang J.O., Smith S.R., Hogan M.E., Cossum P.A., Rando R.F., Chaudhary N. Intramolecular G-quartet motifs confer nuclease resistance to a potent anti-HIV oligonucleotide. J. Biol. Chem. 1996;271:5698–5703. doi: 10.1074/jbc.271.10.5698. [DOI] [PubMed] [Google Scholar]
- 28.Dapić V., Bates P.J., Trent J.O., Rodger A., Thomas S.D., Miller D.M. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry. 2002;41:3676–3685. doi: 10.1021/bi0119520. [DOI] [PubMed] [Google Scholar]
- 29.Soundararajan S., Chen W., Spicer E.K., Courtenay-Luck N., Fernandes D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 30.Mongelard F., Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Ther. 2010;12:107–114. [PubMed] [Google Scholar]
- 31.Burris H.A., 3rd, Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R., Cripps M.C., Portenoy R.K., Storniolo A.M., Tarassoff P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 32.Heinemann V. Gemcitabine: progress in the treatment of pancreatic cancer. Oncology. 2001;60:8–18. doi: 10.1159/000055290. [DOI] [PubMed] [Google Scholar]
- 33.Chari R.V. Targeted cancer therapy: conferring specificity to cytotoxic drugs. Acc. Chem. Res. 2008;41:98–107. doi: 10.1021/ar700108g. [DOI] [PubMed] [Google Scholar]
- 34.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephenson M.L., Zamecnik P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shigdar S., Ward A.C., De A., Yang C.J., Wei M., Duan W. Clinical applications of aptamers and nucleic acid therapeutics in haematological malignancies. Br. J. Haematol. 2011;155:3–13. doi: 10.1111/j.1365-2141.2011.08807.x. [DOI] [PubMed] [Google Scholar]
- 37.Gleave M.E., Monia B.P. Antisense therapy for cancer. Nat. Rev. Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 38.Bates P.J., Kahlon J.B., Thomas S.D., Trent J.O., Miller D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 39.Sha F., Mu R., Henderson D., Chen F.M. Self-aggregation of DNA oligomers with XGG trinucleotide repeats: kinetic and atomic force microscopy measurements. Biophys. J. 1999;77:410–423. doi: 10.1016/S0006-3495(99)76899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B., Zhao X., Lee L.J., Lee R.J. Targeted delivery systems for oligonucleotide therapeutics. AAPS J. 2009;11:195–203. doi: 10.1208/s12248-009-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hicke B.J., Stephens A.W., Gould T., Chang Y.F., Lynott C.K., Heil J., Borkowski S., Hilger C.S., Cook G., Warren S., Schmidt P.G. Tumor targeting by an aptamer. J. Nucl. Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 43.Lin C.H., Patel D.J. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 1997;4:817–832. doi: 10.1016/s1074-5521(97)90115-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girvan A.C., Teng Y., Casson L.K., Thomas S.D., Jüliger S., Ball M.W., Klein J.B., Pierce W.M., Jr., Barve S.S., Bates P.J. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 46.Wu J., Song C., Jiang C., Shen X., Qiao Q., Hu Y. Nucleolin targeting AS1411 modified protein nanoparticle for antitumor drugs delivery. Mol. Pharm. 2013;10:3555–3563. doi: 10.1021/mp300686g. [DOI] [PubMed] [Google Scholar]
- 47.Li X., Yu Y., Ji Q., Qiu L. Targeted delivery of anticancer drugs by aptamer AS1411 mediated Pluronic F127/cyclodextrin-linked polymer composite micelles. Nanomedicine (Lond.) 2015;11:175–184. doi: 10.1016/j.nano.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 48.Trinh T.L., Zhu G., Xiao X., Puszyk W., Sefah K., Wu Q., Tan W., Liu C. A synthetic aptamer-drug adduct for targeted liver cancer therapy. PLoS ONE. 2015;10:e0136673. doi: 10.1371/journal.pone.0136673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho Y., Lee Y.B., Lee J.-H., Lee D.H., Cho E.J., Yu S.J., Kim Y.J., Kim J.I., Im J.H., Lee J.H. Modified AS1411 aptamer suppresses hepatocellular carcinoma by up-regulating galectin-14. PLoS ONE. 2016;11:e0160822. doi: 10.1371/journal.pone.0160822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catuogno S., Esposito C.L., de Franciscis V. Aptamer-mediated targeted delivery of therapeutics: an update. Pharmaceuticals (Basel) 2016;9:69. doi: 10.3390/ph9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray P., Cheek M.A., Sharaf M.L., Li N., Ellington A.D., Sullenger B.A., Shaw B.R., White R.R. Aptamer-mediated delivery of chemotherapy to pancreatic cancer cells. Nucleic Acid Ther. 2012;22:295–305. doi: 10.1089/nat.2012.0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon S., Huang K.-W., Reebye V., Spalding D., Przytycka T.M., Wang Y., Swiderski P., Li L., Armstrong B., Reccia I. Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth. Mol. Ther. Nucleic Acids. 2017;6:80–88. doi: 10.1016/j.omtn.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McBride L.J., Caruthers M.H. An investigation of several deoxynucleosidephosphoramidites useful for synthesizing deoxyoligonucleotides. Tetrahedron Lett. 1983;24:245–248. [Google Scholar]
- 54.Carballo M., Puigdomènech P., Palau J. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 1983;2:1759–1764. doi: 10.1002/j.1460-2075.1983.tb01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J., Lee T.S., Ryu J., Hong S., Kang M., Im K., Kang J.H., Lim S.M., Park S., Song R. RGD peptide-conjugated multimodal NaGdF4:Yb3+/Er3+ nanophosphors for upconversion luminescence, MR, and PET imaging of tumor angiogenesis. J. Nucl. Med. 2013;54:96–103. doi: 10.2967/jnumed.112.108043. [DOI] [PubMed] [Google Scholar]
- 56.Blanco F.F., Sanduja S., Deane N.G., Blackshear P.J., Dixon D.A. Transforming growth factor β regulates P-body formation through induction of the mRNA decay factor tristetraprolin. Mol. Cell. Biol. 2014;34:180–195. doi: 10.1128/MCB.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klauschen F., Wienert S., Schmitt W.D., Loibl S., Gerber B., Blohmer J.U., Huober J., Rüdiger T., Erbstößer E., Mehta K. Standardized Ki67 Diagnostics Using Automated Scoring—Clinical Validation in the GeparTrio Breast Cancer Study. Clin. Cancer Res. 2015;21:3651–3657. doi: 10.1158/1078-0432.CCR-14-1283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.