Abstract
The Trypanosoma theileri group includes several trypanosome species hardly distinguishable due to the lack of discriminating morphological characters. Trypanosomes belonging to this group have been isolated from different bovine, ovine, and cervids in Europe, Africa, Asia, and Americas. The principal vectors of the T. theileri group are considered tabanid flies; however, T. melophagium is transmitted exclusively by sheep keds. In 2016, 128 sand flies out of 2,728 trapped in Valsamoggia municipality, Italy, were individually dissected and an unknown trypanosome strain, named TrPhp1, was isolated from a female of the sand fly Phlebotomus perfiliewi. Sequence analysis placed this trypanosome in the T. theileri group with very high homology to other trypanosomes detected in European cervids. This is the first report of the T. theileri group isolation from a sand fly, and the possible role of this insect group in the trypanosome transmission cycle is discussed. Within the T. theileri group, the phylogenetic analysis distinguished several lineages, which, unfortunately, do not correspond with their host specificity and their taxonomic status remains ambiguous.
1. Introduction
Genus Trypanosoma Gruby, 1843 (Euglenozoa: Kinetoplastea: Trypanosomatidae), infects all classes of vertebrate hosts, but most attention is directed to species that cause human and animal diseases and heavy economic losses [1]. Some trypanosomes can be highly pathogenic for their hosts including humans, such as the causative agents of Chagas disease (T. cruzi) or sleeping sickness (T. brucei); other species are considered to be veterinary important parasites causing serious diseases (e.g., nagana, surra, and dourine) and significant losses in livestock, mainly in Africa and Asia. In Europe, two trypanosome subgenera have been identified in ungulates. While Trypanosoma evansi and T. equiperdum (subgenus Trypanozoon) may result in fatal diseases of their hosts, members of the subgenus Megatrypanum (Trypanosoma theileri, T. cervi, T. melophagium, T. stefanskii, and T. theodori and other species described in the past on morphological basis) are generally considered to be nonpathogenic.
Within the subgenus Megatrypanum, the Trypanosoma theileri group includes trypanosomes of at least three well-defined species with available sequencing data obtained in Europe, Americas, Asia, and Africa: bovine T. theileri, ovine T. melophagium, and deer T. cervi. In general, the trypanosome species determination is not easy for the lack of discriminating morphological characters. In the past, many species have been defined principally on the basis of their epidemiological features, such as host and vector source, while nowadays modern biomolecular methods can help in the species delimitation but are not always conclusive [2]. The current status of the three above-mentioned trypanosome species constituting the T. theileri group is based mainly on their life cycle and host specificity, rather than on available sequencing data. Phylogenetic analyses support the existence of several genotypes and two main lineages (clades TthI and TthII), and some separation of the bovine, ovine, and cervid trypanosomes is accepted [3–6]; however, the clear separation and differentiation of these species have only a very weak support.
Trypanosoma theileri has been first identified in cattle in 1902 by Laveran and Bruce [1]. It is primarily found in cattle (Bos taurus), but other bovids such as bison (Bison bison) and water buffalo (Bubalus bubalis) [6] may be infected as well. In 1975, Kingston and Morton differentiated the trypanosomes found in a wapiti (Cervus elaphus canadensis; Wyoming, USA) from the cattle parasite T. theileri Laveran 1902 and named the new species Trypanosoma cervi Kingston and Morton, 1975 [7]. Since that time, T. cervi has been identified worldwide in mule deer (Odocoileus hemionus, O. virginianus), elk (Alces alces), reindeer (Rangifer tarandus), red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus). Previous records of T. theileri and T. theileri-like infections based only on parasite morphology in various European wild deer populations may, in fact, represent finding of morphologically hardly differentiable Trypanosoma cervi. The last member of the T. theileri trinity, Trypanosoma melophagium described by Flu in 1908, is strictly restricted by its host specificity on sheep. Trypanosomes of the subgenus Megatrypanum are hardly identified in vertebrate host blood, and even isolation in culture is not an easy tool for their identification. In the case of T. melophagium it is peculiar that the trypanosomes were easy to find in huge numbers in sheep keds, the insect host, whereas infection in sheep, the mammal host, is rather cryptic.
Trypanosomes are primarily dixenous parasites principally transmitted by insects (from orders Diptera, Heteroptera, or Siphonaptera) and leeches. Members of the T. theileri group circulate between wild or domestic ruminants and their invertebrate vectors. While T. theileri is transmitted by tabanid flies, sheep keds are the only known vectors for T. melophagium. The situation in T. cervi is more complicated and the existence of several trypanosome species among different cervid hosts and/or different species in the same hosts from distant regions cannot be ruled out. It is assumed that the principal vector is the deer ked (Lipotena spp.), but tabanid flies are also mentioned as possible vectors; however, the existence of other vectors is also considered [6]. Only one report suggests that tabanids are also potential vectors due to cross infection among cattle and cervids using the gut content of tabanid flies. Regarding all morphological and poor molecular data from cervid trypanosomes, it seems clear that the evolutionary history of this trypanosomes is very complex and far from being elucidated.
Trypanosome parasites have not been reported from Italian sand flies. In 2016, screening for Leishmania parasites in sand flies collected in Bologna province, we isolated an unknown trypanosomatid, which was named TrPhp1 (Trypanosoma-Phlebotomus perfiliewi-1). The aim of this study was to characterize this trypanosomatid, isolated from a sand fly female of Phlebotomus (Larroussius) perfiliewi Parrot, 1930.
2. Methods
2.1. Collection of Sand Flies and Isolation of Flagellates
In the frame of leishmaniasis surveillance, sand flies were collected in a site located in the Bologna province, Emilia-Romagna region, Italy. Collection was made on the 27-28th of July 2016, with an attractive trap baited with carbon dioxide. From a total of 2,728 sand flies trapped, a subsample of 82 males was morphologically identified at optical microscope [8, 9] after clarification with chlorolactophenol, recording 79 Ph. perfiliewi and 3 Ph. perniciosus. A number of 128 females were anesthetized with cold, dissected in cold sterile saline, and the guts were observed microscopically (× 400 magnification). The hindgut of one specimen was heavily infected with a large agglomerate of flagellate protozoan, with characteristics of the family Trypanosomatidae (Figure 1(a), video (available here)). No blood remnants were observed in the gut. The sand fly specimen was identified morphologically as Ph. perfiliewi (Figure S1); this identification was confirmed by the sequence of the mitochondrial gene cytochrome c oxidase I (COI) obtained according to Hebert et al. [10] utilizing LCO1490 and HCO2198 primer pair (sequence deposited in GenBank as KY646194).
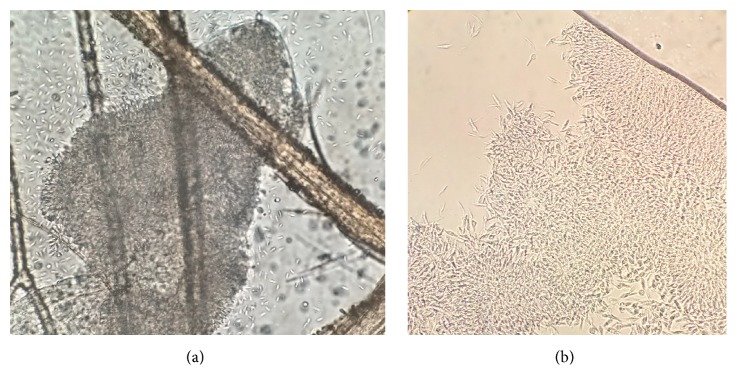
Figure 1.
Photographs of the isolated trypanosome strain (TrPhp1) as was originally found in the sand fly hindgut (a) and in culture (b); light microscope (400×).
The detected flagellates were inoculated into Evans' modified Tobie's medium (EMTM) containing 15% defibrinated rabbit blood and later transferred to Schneider's medium supplemented with 10% fetal calf serum for mass cultivation and further characterization.
2.2. Morphological Characterization of the Isolate
Reared protozoa were firstly observed at light microscopy (400× magnification). To examine their ultrastructural features, aliquots of flagellate cultures were concentrated by centrifugation and prefixed in Karnovsky's fixative in cacodylate buffer (pH 7.2). After postfixation in 2% OsO4 in cacodylate buffer for 1.5 hours at 4°C, samples were washed in corresponding buffer, dehydrated in ethanol series, transferred to propylene oxide, and embedded in Epon 812. Semithin sections were stained with 0.05% toluidine blue in 1% sodium tetraborate and examined by optical microscopy. Thin sections (80nm) were stained with saturated uranyl acetate, followed by Reynolds lead citrate, and examined with Zeiss EM 900 electron microscope at 80 kV (Figure 2).
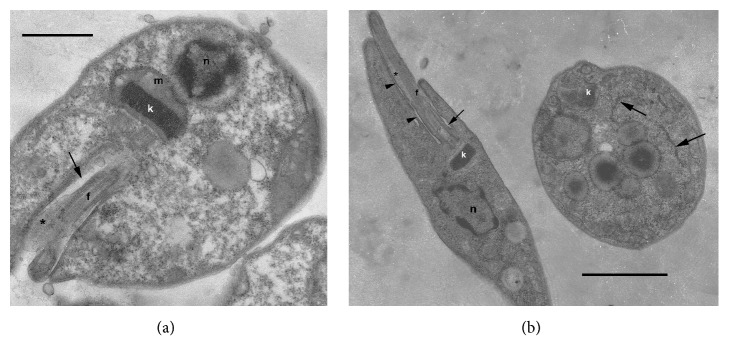
Figure 2.
TEM micrographs of the culture stages of the isolated trypanosome strain (TrPhp1); epimastigotes (a, bb-left) and an amastigote (b-right). Arrow: flagellar pocket; white arrows: endoplasmic reticulum; arrow heads point to the area of adhesion of the flagellum to the cell body; asterisk: paraflagellar rod; f: flagellum; k: kinetoplast; n: nucleus; m: mitochondrion; bar: 1.65 μm (a), 0.9 μm (b).
2.3. Biomolecular Identification and Phylogeny of the Isolate
Two markers were utilized to identify the TrPhp1 trypanosome, i.e., the ribosomal DNA internal transcribed spacer sequence (ITS-1) region of 320 bp and the 18S small subunit ribosomal RNA (SSU rRNA) gene of 927 bp. From the replicating parasite, DNA was extracted according to Rugna et al. [11], and then the two targets were amplified with the primer pair LITSR / L5.8S [12] for ITS-1 and the primer pair TRY927F / TRY927R [13] for the SSU rRNA, according to reported references. The obtained amplicons were purified with the AMPure XP PCR purification kit (Agencourt) and sequenced by CEQ 8000 sequencer (Beckman Coulter), using the GenomeLab DTCS Quick Start Kit (Beckman Coulter). The results were analyzed and assembled using CEQ 8000 version 8.0 software.
Homologue ITS-1 and SSU rRNA sequences of other trypanosomes were retrieved from GenBank with a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi), aligned to the obtained sequences with the ClustalW algorithm, and then refined manually. Phylogenetic analysis was performed by software Beast 2 [14], which uses a Bayesian approach to infer phylogenetic tree. Between the available substitutions models, that with the lowest Bayesian information criterion score, estimated by MEGA6 [15], was selected (Tamura-Nei +G +I for SSU, Jukes-Cantor +G for ITS-1). The coalescent constant population was selected as priors with a chain length of 10 million. Obtained trees were summarized and shown by FigTree software. The same alignments were used to obtain trees with neighbor-joining and minimum evolution methods using MEGA6 [15].
3. Results
3.1. Morphological Characterization
A culture of trypanosomes was obtained from the gut of the specimen of Ph. perfiliewi. On slide smears prepared from the liquid culture, rosettes of flagellates were observed. The parasites showed a low degree of pleomorphism in culture, with abundant epimastigotes and scarce amastigotes. These trypanosomes differed from those observed in the slide directly obtained from the sand fly, which was smaller and rounded, with shorter flagella (Figure 1, Figure S2).
Transmission electron microscopy performed on cultured cells revealed the typical epimastigote morphotypes and rare amastigote forms (Figure 2). Epimastigotes were characterized by a centrally located nucleus, rod-shaped kinetoplast, and paraflagellar rod extending partially into the flagellar pocket.
3.2. Biomolecular Identification and Phylogeny of Trypanosoma sp.
The nucleotide sequences of the ITS-1 and SSU rRNA genes were submitted to GenBank and assigned the accession numbers KY672996 and KY681802, respectively.
BLAST searches of the sequences generated in this study suggested the parasite belongs to the genus Trypanosoma. The obtained ITS-1 sequence was of 223 bp and showed 100% similarity with three sequences belonging to the Trypanosoma theileri-like isolates D30 and TC2 (a.c.: AY773714, HQ664845, JN798601) from deer and sheep, respectively. The obtained SSU rRNA gene sequence of 819 bp showed 100% similarity with four sequences (a.c.: KJ195885, KJ195879, KJ397590 AJ009165), which were related to different Trypanosoma theileri-like isolates including D30, from Cervus sp. and Dama dama.
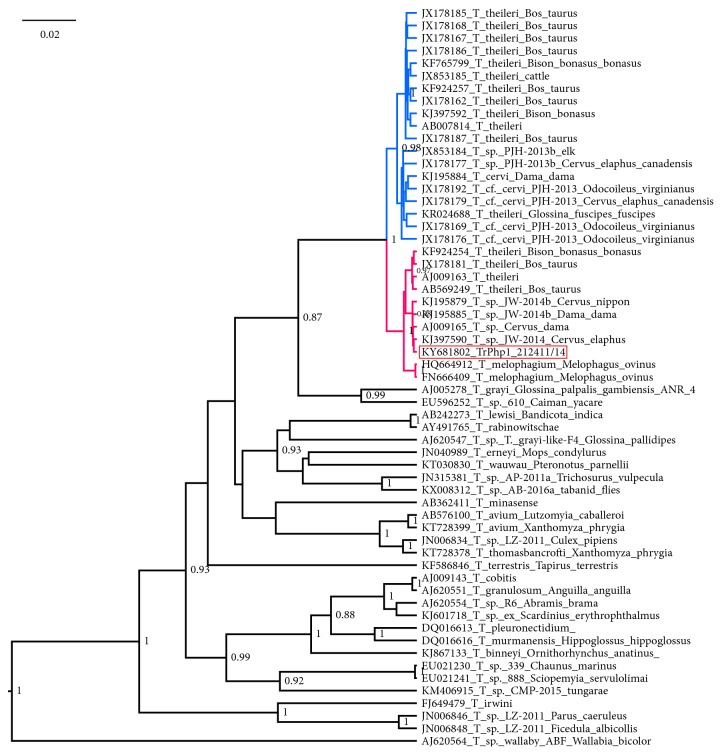
In the SSU rRNA tree (Figure 3), the T. theileri group is separated into two well-supported clades, previously named lineages TthI and TthII [5]; the sequence obtained in this study cluster is the lineage TthII along with sequences classified as T. melophagium and T. theileri. The only species consistent in the topology of this tree is T. melophagium (maximum posterior probability), while—as already suggested [2]—to define other systematic units, additional epidemiological data are needed, as host and vector specificity. This result confirms that SSU rRNA lacks sufficient resolution to discriminate among closely related species [16]. The obtained Bayesian tree is consistent with trees obtained by other methods employed (Figure S3).
Figure 3.
An SSU rRNA-based Bayesian phylogenetic tree demonstrating the position of newly obtained trypanosome (strain TrPhp1) of the Trypanosoma theileri group isolated from a sand fly female captured in Italy. For the other sequences in the tree the GB number is reported along with the Trypanosoma species and source of isolations, if available. Only posterior probabilities over 0.8 are shown at the node. Scale bar in units of substitutions per site. TthI clade in blue, TthII clade in red, font of sequence of isolated trypanosome with red background.
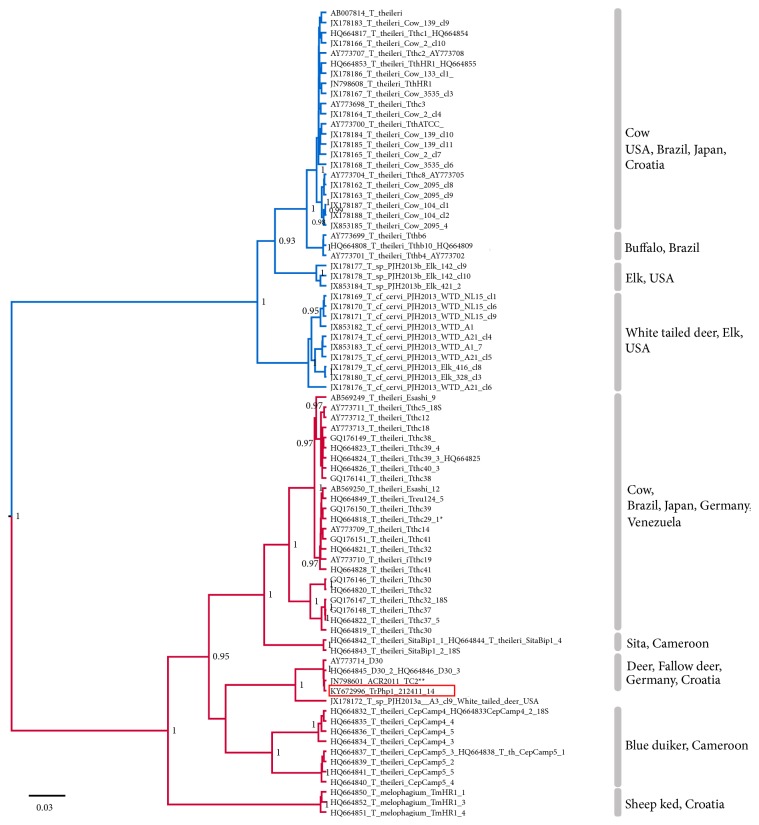
A more detailed view inside the T. theileri group is provided by the ITS-1 tree (Figure 4). This tree bears several clades with high posterior probability, which are quite consistent with the species reported, while sequences classified as T. theileri are spread in different clades. In the ITS1-based phylogenetic tree, our sequences are included in a clade (with the maximum posterior probability) containing other European sequences (from Germany and Croatia), not yet defined at species level but detected in various cervids. Also this obtained Bayesian tree is consistent with trees obtained by other methods (Figure S3).
Figure 4.
Phylogenetic tree of the ITS-1 sequence of isolated trypanosome (strain TrPhp1) and homologue sequences deposed in GenBank (GB numbers reported near the taxon name). Source and countries of isolation of the strains are reported near the square brackets. The tree was reconstructed using the Bayesian method. Only posterior probabilities over 0.8 are shown at the node. Scale bar in units of substitutions per site. TthI clade in blue, TthII clade in red, sequence of isolated trypanosome with red background. Identical sequences available in GenBank: ∗HQ664829 TthcV2-7; HQ664847 Treu124-2; HQ664848 Treu124-3; ∗∗JN798602 TC3; JN798603 TC4; JN798604 TC9; JN798605, TC10; JN798606, TC339; JN798607, TC603.
4. Discussion
To the best of our knowledge, no reports of the Trypanosoma genus in sand flies have been published in Italy; thus this is the first report of the genus Trypanosoma in a naturally infected female Phlebotomus perfiliewi in the country. Despite the fact that the phylogenetic analysis based on SSU rRNA and ITS-1 did not allow us to unambiguously identify the studied parasites at species level, the trypanosome isolated from sand fly definitely belongs to the Trypanosoma (Megatrypanum) theileri group and thus represents the first report of this trypanosome species in sand flies in general.
This group comprises trypanosomes of ruminants with bovine T. theileri assumed as a type-species [17]. As suggested by Votýpka et al. (2015), without additional information about host specificity, the isolated Trypanosoma should be considered an operational taxonomic unit (OTU) within the T. theileri group. Phylogenies inferred in this study placed the newly obtained isolate TrPhp1 in the T. theileri lineage II, which comprised T. melophagium and trypanosome genotypes detected in cervids and cattle [6]. Interestingly, analysis of ITS-1 showed high sequence homology with trypanosomes detected in fallow deer (Dama dama) and red deer (Cervus elaphus) in Germany and Croatia, respectively [5, 17]. According to Garcia et al. [18], genotypes within the T. theileri group appear to be host specific. Based on the checklist of the Monteveglio Abbey Regional Park (Bologna province) and of the adjacent area, the only cervid species present at the sampled site is the European roe deer (Capreolus capreolus) and this information is consistent with the previous findings of Megatrypanum trypanosomes in this host species [19].
In the present study, the trypanosome of the T. theileri group was isolated from Ph. perfiliewi, a sand fly species broadly distributed in the Emilia-Romagna region, northeastern Italy [11, 20]. Natural infection of sand flies by trypanosomes was already reported; for example, trypanosomes of lizards and amphibians were detected in sand flies in Brazil [21, 22] and Ghana [23] and isolated from this insect in California [24] and Pakistan [25], incriminating phlebotomine sand flies as vectors of these protozoa. In addition, trypanosomes of birds and rodents were detected in sand flies in Peru [26] and Thailand [27], respectively, and trypanosomes of marsupials were isolated from sand flies in Brazil [28]. However, the commonly reported vectors of species belonging to the T. theileri group are tabanids and sheep or deer keds, and parasites are transmitted to vertebrate hosts via ingestion of vectors or via defecation. Therefore, the isolation of this trypanosome from a sand fly was unexpected. This finding could be explained by an underestimation of the vectorial competence of sand flies for T. theileri group trypanosomes and in line with the claim that also other vectors than tabanids and keds must be involved in the transmission of the T. theileri group [6]. Indirect support for the possible involvement of sand flies in the T. theileri group transmission is a massive occurrence of parasites in sand fly female gut after the defecation and the parasite localization in the posterior part of the digestive system, suggesting the possible transmission by defecation, the regular way of transmission occurring in the T. theileri group. Moreover, morphological forms present in the sand fly and in culture are consistent with the forms described in cycle of trypanosomes of this group, which pass from epimastigote to metacyclic stages in arthropods vectors [29, 30]. In such a case, the role of sand flies in the trypanosome transmission must be confirmed with an experimental infection of flies and transmission of parasites to vertebrate hosts. However, it is also possible that we recorded only a nonspecific development of trypanosome in the Phlebotomus perfiliewi sand fly female, which may represent a dead-end street for the parasite. This explanation could be supported, for example, by the regular occurrence of the T. theileri group in mosquitoes collected in several localities in Austria [31].
Although many trypanosome species have been known for years, complete life cycles and effect on vertebrate hosts have been elucidated only in a minority of the species. Insufficient knowledge of vectors and possible pathogenicity also concern trypanosomes from the T. theileri group. T. theileri belongs to a group of trypanosomes which are commonly considered nonpathogenic and may persist in cattle for many years without any evidence of clinical disease [32]. On the other hand, disease associated with T. theileri in cattle was sporadically reported [32, 33]. Even though the T. theileri group infections in roe deer and European elk and T. theileri-like infections in deer and chevrotains are documented, little is known about wild ruminants harboring these trypanosomes and being reservoirs for infection of livestock. For these reasons, the potential pathogenicity of the isolated trypanosome, its vertebrate host specificity, and development in possible vectors needs more investigation.
5. Conclusions
In this work we reported the unusual isolation of a trypanosome (the strain named TrPhp1), related to the Trypanosoma theileri group, from the sand fly Phlebotomus perfiliewi. Interestingly, in the dissected sand fly female the developmental stages of the parasite resembled the infective forms characteristic of this trypanosome group. Phylogenetic analysis highlighted a close relation between TrPhp1 and trypanosomes isolated from deer and fallow deer in Europe. These findings could indicate that life cycle of this trypanosome may involve sand flies as vectors and cervids as vertebrate hosts.
Acknowledgments
This work was supported by the Regional Project on Insect-Borne Diseases [COMM07001], funded by Regione Emilia-Romagna. The authors would like to thank Dr. Simone Magnino for the help in revising the paper.
Data Availability
The nucleotide sequences of the isolated trypanosome TrPhp1 were submitted to GenBank: ITS-1 with the accession number KY672996, SSU rRNA with the accession number and KY681802.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Figure S1. The spermatheca of the Phlebotomus perfiliewi female from which the trypanosome strain (TrPhp1) was isolated; tip of the abdomen (above) and detail of the spermatheca (below). Figure S2. Cultured trypanosomes (strain named TrPhp1) stained (Hemacolor®) under optical microscope (1000×). Figure S3. Phylogenetic trees of the SSU and ITS-1 sequence alignments obtained by neighbor-joining and minimum evolution methods. Video. Video of the isolated trypanosome strain (TrPhp1) as was originally found in the sand fly hindgut.
References
- 1.Hoare C. A. The Trypanosomes of Mammals. Vol. 768. Oxford, UK: BlackwellScientific Publications; 1972. [Google Scholar]
- 2.Votýpka J., d'Avila-Levy C. M., Grellier P., Maslov D. A., Lukeš J., Yurchenko V. New Approaches to Systematics of Trypanosomatidae: Criteria for Taxonomic (Re)description. Trends in Parasitology. 2015;31(10):460–469. doi: 10.1016/j.pt.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues A. C., Garcia H. A., Batista J. S., et al. Characterization of spliced leader genes of Trypanosoma (Megatrypanum) theileri: Phylogeographical analysis of Brazilian isolates from cattle supports spatial clustering of genotypes and parity with ribosomal markers. Parasitology. 2010;137(1):111–122. doi: 10.1017/S0031182009991053. [DOI] [PubMed] [Google Scholar]
- 4.Rodrigues A. C., Garcia H. A., Ortiz P. A., et al. Cysteine proteases of Trypanosoma (Megatrypanum) theileri: Cathepsin L-like gene sequences as targets for phylogenetic analysis, genotyping diagnosis. Parasitology International. 2010;59(3):318–325. doi: 10.1016/j.parint.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Martinković F., Matanović K., Rodrigues A. C., Garcia H. A., Teixeira M. M. G. Trypanosoma (Megatrypanum) melophagium in the sheep ked Melophagus ovinus from organic farms in Croatia: Phylogenetic inferences support restriction to sheep and sheep keds and close relationship with trypanosomes from other ruminant species. Journal of Eukaryotic Microbiology. 2012;59(2):134–144. doi: 10.1111/j.1550-7408.2011.00599.x. [DOI] [PubMed] [Google Scholar]
- 6.Fisher A. C., Schuster G., Cobb W. J., et al. Molecular characterization of Trypanosoma (Megatrypanum) spp. infecting cattle (Bos taurus), white-tailed deer (Odocoileus virginianus), and elk (Cervus elaphus canadensis) in the United States. Veterinary Parasitology. 2013;197(1-2):29–42. doi: 10.1016/j.vetpar.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 7.Kingston N., Morton J. K. Trypanosoma cervi sp. n. from elk (Cervus canadensis) in Wyoming. Journal of Parasitology. 1975;61(1):17–23. doi: 10.2307/3279099. [DOI] [PubMed] [Google Scholar]
- 8.Romi R., Khoury C., Bigliocchi F., Maroli M. Fact Sheet on Mites and Insects of Medical Importance [Schede Guida Su Acari E Insetti Di Interesse Sanitario] Rome, Italy: Istituto Superiore di Sanità; 1994. [Google Scholar]
- 9.Dantas-Torres F., Tarallo V. D., Otranto D. Morphological keys for the identification of Italian phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae) Parasites & Vectors. 2014;7(1, article 479) doi: 10.1186/s13071-014-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert P. D. N., Cywinska A., Ball S. L., DeWaard J. R. Biological identifications through DNA barcodes. Proceedings of the Royal Society B Biological Science. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rugna G., Carra E., Corpus F., et al. Distinct leishmania infantum strains circulate in humans and dogs in the Emilia-Romagna region, northeastern Italy. Vector-Borne and Zoonotic Diseases. 2017;17(6):409–415. doi: 10.1089/vbz.2016.2052. [DOI] [PubMed] [Google Scholar]
- 12.Kuhls K., Mauricio I. L., Pratlong F., Presber W., Schönian G. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes and Infection. 2005;7(11-12):1224–1234. doi: 10.1016/j.micinf.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Noyes H. A., Stevens J. R., Teixeira M., Phelan J., Holz P. Erratum: A nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia (International Journal for Parasitology (1999) 29:2 (331-339) PII: S0020751998001672) International Journal for Parasitology. 2000;30(2):p. 228. doi: 10.1016/S0020-7519(00)00026-6. [DOI] [PubMed] [Google Scholar]
- 14.Bouckaert R., Heled J., Kühnert D., et al. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Computational Biology. 2014;10(4) doi: 10.1371/journal.pcbi.1003537.e1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maslov D. A., Votýpka J., Yurchenko V., Lukeš J. Diversity and phylogeny of insect trypanosomatids: All that is hidden shall be revealed. Trends in Parasitology. 2013;29(1):43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues A. C., Paiva F., Campaner M., Stevens J. R., Noyes H. A., Teixeira M. M. G. Phylogeny of Trypanosoma (Megatrypanum) theileri and related trypanosomes reveals lineages of isolates associated with artiodactyl hosts diverging on SSU and ITS ribosomal sequences. Parasitology. 2006;132(2):215–224. doi: 10.1017/S0031182005008929. [DOI] [PubMed] [Google Scholar]
- 18.Garcia H. A., Kamyingkird K., Rodrigues A. C., Jittapalapong S., Teixeira M. M. G., Desquesnes M. High genetic diversity in field isolates of Trypanosoma theileri assessed by analysis of cathepsin L-like sequences disclosed multiple and new genotypes infecting cattle in Thailand. Veterinary Parasitology. 2011;180(3-4):363–367. doi: 10.1016/j.vetpar.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Bose R., Petersen K., Pospichal H., Buchanan N., Tait A. Characterization of Megatrypanum trypanosomes from European Cervidae. Parasitology. 1993;107(1):55–61. doi: 10.1017/S0031182000079403. [DOI] [PubMed] [Google Scholar]
- 20.Calzolari M., Angelini P., Finarelli A. C., et al. Human and entomological surveillance of Toscana virus in the Emilia-Romagna region, Italy, 2010 to 2012. Eurosurveillance. 2014;19(48) doi: 10.2807/1560-7917.ES2014.19.48.20978. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira R. C., De Souza A. A., Freitas R. A., et al. A phylogenetic lineage of closely related trypanosomes (Trypanosomatidae, Kinetoplastida) of anurans and sand flies (Psychodidae, Diptera) sharing the same ecotopes in Brazilian Amazonia. Journal of Eukaryotic Microbiology. 2008;55(5):427–435. doi: 10.1111/j.1550-7408.2008.00342.x. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira T. D. S., Minuzzi-Souza T. T. C., de Andrade A. J., et al. Molecular detection of Trypanosoma sp. and Blastocrithidia sp. (Trypanosomatidae) in phlebotomine sand flies (Psychodidae) in the Federal District of Brazil. Journal of the Brazilian Society of Tropical Medicine. 2015;48(6):776–779. doi: 10.1590/0037-8682-0076-2015. [DOI] [PubMed] [Google Scholar]
- 23.Nzelu C. O., Kato H., Puplampu N., et al. First detection of Leishmania tropica DNA and trypanosoma species in Sergentomyia sand flies (diptera: psychodidae) from an outbreak area of cutaneous leishmaniasis in Ghana. PLOS Neglected Tropical Diseases. 2014;8(2, article e2630) doi: 10.1371/journal.pntd.0002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayala S. C. Trypanosomes in wild California sandflies, and extrinsic stages of Trypanosoma bufophlebotomi. The Journal of Protozoology. 1971;18(3):433–436. doi: 10.1111/j.1550-7408.1971.tb03349.x. [DOI] [PubMed] [Google Scholar]
- 25.Kato H., Uezato H., Sato H., et al. Natural infection of the sand fly Phlebotomus kazeruni by Trypanosoma species in Pakistan. Parasites & Vectors. 2010;3(1) doi: 10.1186/1756-3305-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato H., Gomez E. A., Cáceres A. G., et al. Natural infections of man-biting sand flies by leishmania and trypanosoma species in the northern peruvian andes. Vector-Borne and Zoonotic Diseases. 2011;11(5):515–521. doi: 10.1089/vbz.2010.0138. [DOI] [PubMed] [Google Scholar]
- 27.Phumee A., Tawatsin A., Thavara U., et al. Detection of an Unknown Trypanosoma DNA in a Phlebotomus stantoni (Diptera: Psychodidae) Collected From Southern Thailand and Records of New Sand Flies With Reinstatement of Sergentomyia hivernus Raynal & Gaschen, 1935 (Diptera: Psychodidae) Journal of Medical Entomology. 2017;54(2):429–434. doi: 10.1093/jme/tjw161. [DOI] [PubMed] [Google Scholar]
- 28.Naiff R. D., Barrett T. V., Freitas R. A. Isolation of Trypanosoma freitasi (Kinetoplastida: Trypanosomatidae) from Psychodopygus claustrei (Diptera: Psychodidae). Memórias do Instituto Oswaldo Cruz. 1989;84(2):273–275. doi: 10.1590/S0074-02761989000200020. [DOI] [PubMed] [Google Scholar]
- 29.Hoare C. A. An experimental study of the sheep-trypanosome (t. melophagium flu, 1908), and its transmission by the sheep-ked (melophagus ovinus L.) Parasitology. 1923;15(4):365–424. doi: 10.1017/S0031182000014888. [DOI] [Google Scholar]
- 30.Böse R., Heister N. C. Development of Trypanosoma (M.) theileri in Tabanids. Journal of Eukaryotic Microbiology. 1993;40(6):788–792. doi: 10.1111/j.1550-7408.1993.tb04475.x. [DOI] [PubMed] [Google Scholar]
- 31.Schoener E., Uebleis S. S., Cuk C., et al. Trypanosomatid parasites in Austrian mosquitoes. PLoS ONE. 2018;13(4):p. e0196052. doi: 10.1371/journal.pone.0196052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villa A., Gutierrez C., Gracia E., et al. Presence of Trypanosoma theileri in Spanish cattle. Annals of the New York Academy of Sciences. 2008;1149:352–354. doi: 10.1196/annals.1428.016. [DOI] [PubMed] [Google Scholar]
- 33.Greco A., Loria G. R., Dara S., Luckins T., Sparagano O. First isolation of Trypanosoma theileri in Sicilian cattle. Veterinary Research Communications. 2000;24(7):471–475. doi: 10.1023/A:1006403706224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The spermatheca of the Phlebotomus perfiliewi female from which the trypanosome strain (TrPhp1) was isolated; tip of the abdomen (above) and detail of the spermatheca (below). Figure S2. Cultured trypanosomes (strain named TrPhp1) stained (Hemacolor®) under optical microscope (1000×). Figure S3. Phylogenetic trees of the SSU and ITS-1 sequence alignments obtained by neighbor-joining and minimum evolution methods. Video. Video of the isolated trypanosome strain (TrPhp1) as was originally found in the sand fly hindgut.
Data Availability Statement
The nucleotide sequences of the isolated trypanosome TrPhp1 were submitted to GenBank: ITS-1 with the accession number KY672996, SSU rRNA with the accession number and KY681802.