Significance
This work demonstrates that exopolysaccharide (EPS) produced by Vibrio cholerae can protect against exogenous type 6 secretion system (T6SS) attacks from different bacterial species. This protection does not affect the ability of the EPS-producing cell to use their own T6SS to attack other bacteria, indicating that EPS does not simply increase the physical distance between cells to afford protection. Furthermore, this protective effect does not depend on other genes involved in biofilm biogenesis, suggesting that EPSs can determine the outcomes of antagonistic bacterial–bacterial interactions independent of their typical function in biofilm formation. Thus, extracellular biopolymers of bacteria may play a role in modulating resistance to exogenous attacks by other bacteria as well as providing potential resistance to host innate immune mechanisms.
Keywords: capsule, secretion, antibacterials, commensal, microbiota
Abstract
The type 6 secretion system (T6SS) is a nanomachine used by many Gram-negative bacteria, including Vibrio cholerae, to deliver toxic effector proteins into adjacent eukaryotic and bacterial cells. Because the activity of the T6SS is dependent on direct contact between cells, its activity is limited to bacteria growing on solid surfaces or in biofilms. V. cholerae can produce an exopolysaccharide (EPS) matrix that plays a role in adhesion and biofilm formation. In this work, we investigated the effect of EPS production on T6SS activity between cells. We found that EPS produced by V. cholerae cells functions as a unidirectional protective armor that blocks exogenous T6SS attacks without interfering with its own T6SS functionality. This EPS armor is effective against both same-species and heterologous attackers. Mutations modulating the level of EPS biosynthesis gene expression result in corresponding modulation in V. cholerae resistance to exogenous T6SS attack. These results provide insight into the potential role of extracellular biopolymers, including polysaccharides, capsules, and S-layers in protecting bacterial cells from attacks involving cell-associated macromolecular protein machines that cannot readily diffuse through these mechanical defenses.
In their natural environment, bacteria engage in a continuous arms race as they compete for nutrients and physical space. The type 6 secretion system (T6SS) is a widely used weapon in the antibacterial arsenal of many Gram-negative species (1–3). The T6SS is a nanomachine capable of delivering toxic effector proteins into adjacent eukaryotic and bacterial cells (4–7). Mechanistically, the apparatus can be thought of as a spear gun that can, in an instant, plunge a poison-tipped spear into an adjacent target cell using mechanical energy captured during the assembly of the loaded gun. Vibrio cholerae T6SS is assembled in stages, starting with the formation of a transmembrane baseplate complex that anchors the apparatus to the cell envelope (8). This baseplate recruits a spearhead consisting of a trimer of VgrG subunits (9) and a PAAR-domain–containing tip protein (10). The baseplate and spearhead complex serve as the nucleation point for the assembly of the spear shaft, consisting of polymerized Hcp protein subunits (11). The toxic effectors of the T6SS associate with different components of this spear through interactions with PAAR proteins (10), VgrG (12, 13), Hcp proteins (14), or adapter proteins (15, 16). Finally, a sheath comprised of VipA and VipB subunits assembles around the Hcp tube in an extended conformation that produces a high-energy state (17–19). The sheath subunits can undergo a rapid conformational change resulting in contraction of the extended structure to propel the Hcp/VgrG/PAAR spear complex and their associated effectors out of the predatory T6SS+ cell and into adjacent target cells (20). Thus, the T6SS is considered a contact-dependent antibacterial machine although its absolute range of action has not been fully explored (3).
Many bacteria use the T6SS to gain a competitive advantage when growing in mixed-species biofilms. When a T6SS+ strain is grown in competition with a T6SS-sensitive strain, the growth of the sensitive strain is massively reduced in vitro (5, 7, 21) and in vivo (22–24). Unlike many other contact-dependent systems, most T6SSs characterized to date do not require a specific receptor in target cells to deliver toxic cargo or recognize prey cells. This property allows a bacterium using a single T6SS to attack a wide variety of target species. The T6SS of V. cholerae can target most Gram-negative cells and phagocytic eukaryotic cells (7, 25), but lacks potency against Gram-positive bacterial species, suggesting that a thick peptidoglycan layer can provide a barrier to T6SS effector delivery. The range of prey sensitivities to T6SS attack is not understood in molecular terms and there is little work that addresses the role of mechanical barriers in defense against T6SS attack. Along with versatility in target range within certain bacterial groups, such as Gram-negative organisms, the T6SS of most predatory species is associated with multiple effectors, each with distinct enzymatic activities that attack periplasmic as well as cytoplasmic target macromolecules (10, 12, 26–28). This multivalent character of the T6SS effector activity repertoire likely presents a difficult challenge for prey bacteria to evolve resistance against the individual effectors themselves.
Despite the robustness of the T6SS, some bacteria have evolved mechanisms to combat it. For example, Pseudomonas aeruginosa has evolved a unique regulatory mechanism of sensing exogenous T6SS attacks and assembling its own T6SS apparatus to launch a targeted retaliatory attack (29). In contrast, Bacteroides species in the human gut have acquired a large repertoire of resistance genes against various effectors, including those that they themselves do not express (30).
Because the T6SS is a contact-dependent system with a submicrometer range, another way for sensitive bacteria to evade killing is to avoid direct contact with T6SS-carrying bacteria. For example, planktonic bacteria floating in liquid culture do not engage in T6SS-dependent killing (6, 7). This physical separation of the bacteria prevents cells with a functional T6SS from reaching their susceptible target. Along similar lines, physical barriers between cells could provide an alternative form of resistance to T6SS killing, though no such barriers have been previously reported. In this study, we explored whether secreted exopolysaccharide (EPS) could serve as such a barrier. In V. cholerae, EPS is produced by the products of vps genes contained in two operons, VpsI and VpsII, encoding VpsA-K and VpsL-Q, respectively (31). When bacteria encounter a solid surface, they activate the expression of these Vps operons and begin secreting EPS, which helps cells adhere to surfaces and lays the foundation for additional matrix proteins to stabilize adhesion, resulting in biofilm formation (32–34).
Here we report that the EPS produced by V. cholerae can act as a type of “armor” to protect against T6SS attacks from different bacterial species. This armor does not affect the ability of the EPS-producing cell to use T6SS to attack other bacteria, indicating that EPS works to repel attacks rather than creating additional physical distance between cells. The effectiveness of the armor is tunable, as mutations that lead to up-regulation of EPS biosynthesis confer increased resistance to T6SS attacks. EPS-mediated resistance to T6SS attacks suggests a role for other EPSs in determining the outcomes of antagonistic bacterial–bacterial interactions that may have a profound influence on ecological and pathogenic success in microbial communities.
Results
To explore the impact of the physical barrier created by EPS on the T6SS, we constructed an in-frame deletion of the vpsA gene from the chromosome of V. cholerae V52, a strain that is constitutive for T6SS expression. VpsA encodes an essential component of the biosynthetic pathway involved in EPS production (31). Deletion of vpsA resulted in a marked reduction in biofilm formation for both wild-type and a T6SS knockout mutant carrying an in-frame deletion in vasK (SI Appendix, Fig. S1A). However, visually under the microscope, the vpsA mutant was indistinguishable from the wild-type parent, with no measurable differences in cell-to-cell distance between the two strains. Additionally, deletion of vpsA did not significantly alter expression of T6SS genes in both the main and auxiliary gene clusters. We observed less than a twofold difference between the wild-type and vpsA knockout strains in T6SS transcripts from the main T6SS cluster compared with the nearly 20-fold reduced expression of the same genes when the T6SS structural gene vasK is knocked out (SI Appendix, Fig. S1B). Furthermore, the vpsA mutant was clearly able to secrete Hcp in a T6SS-dependent manner at levels compared with a wild-type strain (SI Appendix, Fig. S1C). These results indicate that the vpsA deletion does not significantly affect T6SS expression and functional secretion of its substrates.
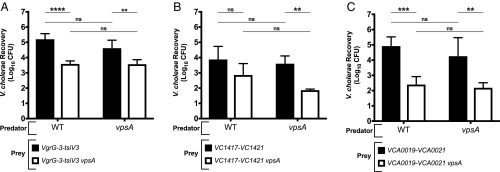
The T6SS of V. cholerae is associated with three different effector–immunity pairs. When any of the immunity proteins are absent, the cells become susceptible to intoxication by the cognate effector protein delivered by the parental wild-type strain (35). We exploited this property to determine whether EPS could affect T6SS functionality in bacterial competition assays. By using isogenic predator and prey bacteria that were altered in T6SS effector and EPS genes, we were able to directly measure the effect of EPS production on the sensitivities of strains to T6SS attack. To make the prey strains sensitive to V. cholerae T6SS, we used knockouts of the operons encoding each of the T6SS effectors and their cognate immunity proteins: vgrG3-tsiV3 (∆VCA0123-4) (Fig. 1A), the tseL operon (∆VC1417-21) (Fig. 1B), or the vasX operon (∆VCA0019-21) (Fig. 1C). The vpsA knockouts of each of these prey strains were cocultured with wild-type or vpsA knockout predators. Prey lacking vpsA were at least 100-fold more susceptible to killing than prey with functional EPS production (black vs. white columns, Fig. 1). In contrast, deletion of vpsA in the predator strain had no effect on T6SS-dependent killing for all three of the effectors tested (Fig. 1). Taken together, these results suggest that while the EPS produced by V. cholerae protects against exogenous T6SS attack, it does not prevent its own T6SS from efficiently attacking other bacterial cells.
Fig. 1.
Deletion of vpsA in prey but not predator V. cholerae strains results in reduced T6SS-dependent killing. Wild-type and vpsA predator V52 were competed against prey V52 with and without deletion of vpsA. Prey stains were sensitive to (A) VgrG-3 (∆vgrG-3∆tsiV3), (B) TseL (∆VC1417-21), or (C) VasX (∆VCA0019-21). Data represent the mean ± SD of at least three biological replicates. Student’s t test was performed to compare each condition (ns, P > 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P < 0.0001).
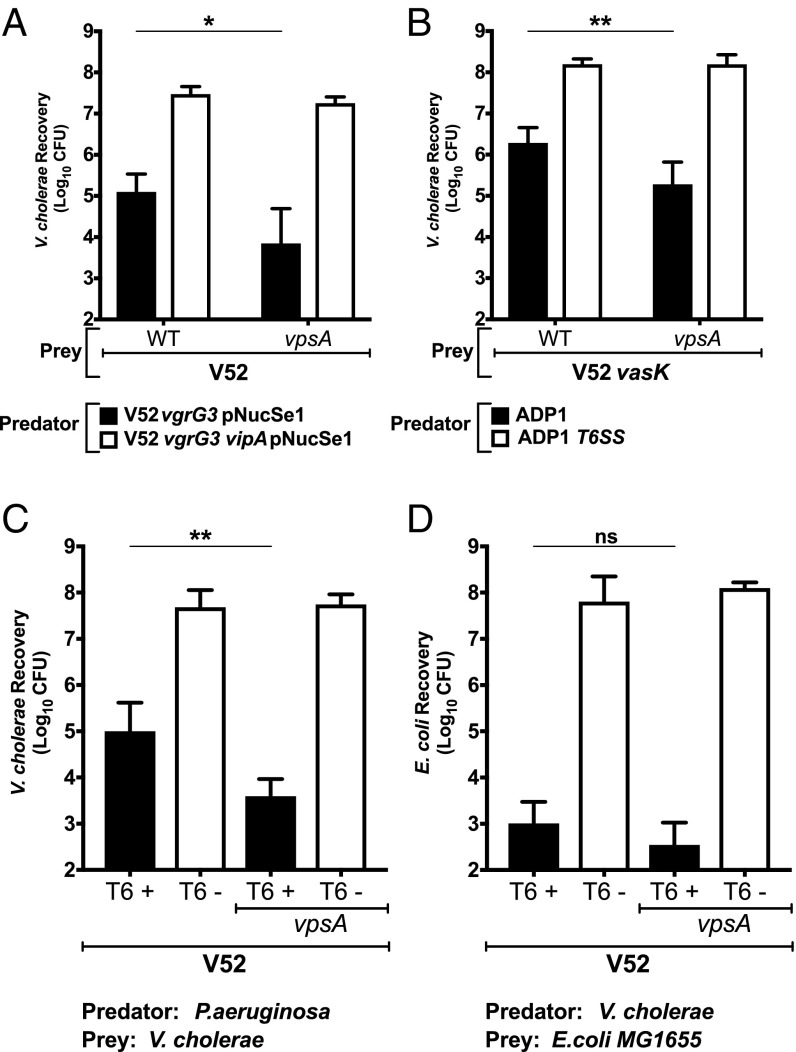
Because the protective property of EPS was not dependent on the specific effector sensitivity of the prey strain, we hypothesized that EPS was blocking the penetration of the exogenous secretion apparatus into the target EPS+ cell rather than providing a targeted defense against a particular effector activity. If this hypothesis is correct, we reasoned that EPS should also protect V. cholerae from T6SS attacks by heterologous organisms. We previously engineered V. cholerae V52 to deliver a chimeric VgrG protein carrying a nuclease domain from Salmonella enterica subsp. arizonae (28), allowing us to evaluate EPS protection against a non-V. cholerae effector. When competed against this isogenic predator, the V52 bacteria lacking vpsA were 10-fold more susceptible to T6SS killing than its wild-type EPS+ parent (Fig. 2A), indicating that EPS-mediated protection is likely not dependent on effector type when scored between homologous cells of the same bacterial species.
Fig. 2.
EPS protects V. cholerae against T6SS attacks from heterologous organisms. (A) Competition between V. cholerae V52 expressing the chimeric VgrG3-NucSe1 nuclease effector from S. enterica subsp. arizonae and T6SS− V52vasK, with or without the vpsA deletion. (B) Competition between A. baylyi ADP1 and T6SS− V52vasK, with and without the vpsA deletion. (C) Competition between P. aeruginosa PAO1 and T6SS+ V52 and T6SS− V52vasK, each with or without the vpsA deletion. (D) Competition between T6SS+ V52 and T6SS− V52vasK, each with or without the vpsA deletion, against E. coli MG1655. Data represent the mean ± SD of three biological replicates. Student’s t test was performed to compare each condition (ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01).
We also evaluated the impact of EPS in cross-species interactions by coculturing the vpsA mutant and its isogenic parent with Acinetobacter baylyi ADP1. We used a T6SS− mutant of V52 (deleted in vasK) as prey to avoid complications arising from killing of the A. baylyi predator by the T6SS of the V. cholerae prey. We found that the vpsA mutant of V52 vasK was 10-fold more susceptible to A. baylyi killing than its EPS+ wild-type parent (Fig. 2B).
P. aeruginosa has a unique posttranslational regulatory mechanism that allows it to assemble and fire its T6SS with precision at cells that attack the P. aeruginosa with their own T6SS (29). As such, killing of V. cholerae by the P. aeruginosa T6SS reflects both V. cholerae T6SS activity and the sensitivity of V. cholerae to the P. aeruginosa T6SS counterattack. We found that V. cholerae carrying the vpsA deletion were 100-fold more sensitive to killing by P. aeruginosa (black bars, Fig. 2C). In contrast, V. cholerae lacking a functional T6SS were not killed by P. aeruginosa, regardless of whether they carried the vpsA mutation or not (white bars, Fig. 2C). In homologous species competitions, EPS production by V. cholerae strains acting as predators had no effect on T6SS activity directed against T6SS-sensitive V. cholerae (Fig. 1). EPS production also did not affect predatory activity directed against a heterologous species, Escherichia coli MG1655, in that this strain was equally susceptible to killing by both wild-type and vpsA mutant V. cholerae (black bars, Fig. 2D). These data suggest that the increased killing by the P. aeruginosa T6SS was indeed due to increased sensitivity of V. cholerae vpsA mutants to P. aeruginosa T6SS counterattacks rather than the ability of VPS mutants to induce more counterattacks through better T6SS function. Stated another way, we saw no evidence that T6SS+ EPS− mutants carry out more efficient attacks on prey and thus induced more lethal counterattacks by P. aeruginosa. Rather, EPS simply blocks counterattacks that occur with similar frequencies when P. aeruginosa encounters either a wild-type or EPS mutant that has a functional T6SS.
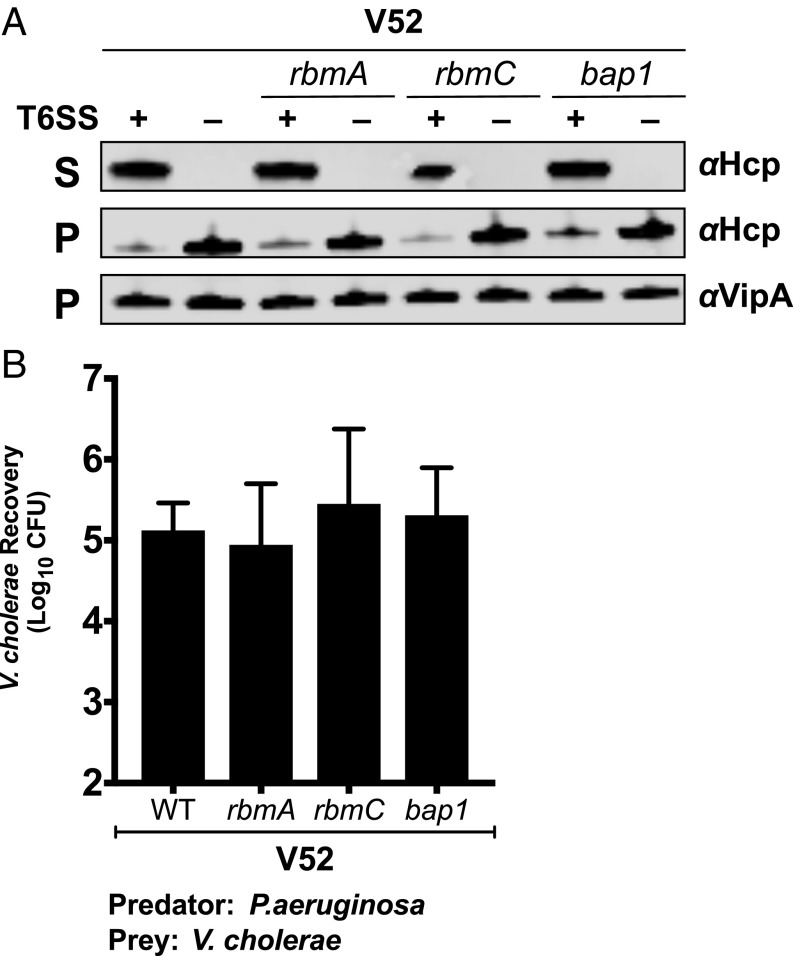
Having observed a protective effect of EPS, we evaluated whether other components of the biofilm matrix could also contribute to T6SS armor. We constructed strains with deletions in the rbmA, rbmC, and bap1 genes, which are essential for mature biofilm formation and integrity (33, 36–38). Deletions of these matrix genes did not alter T6SS-dependent Hcp secretion (Fig. 3A). We measured T6SS killing of these matrix component mutants in competition against P. aeruginosa. The absence of these genes did not result in increased sensitivity to T6SS killing (Fig. 3B), suggesting that production of EPS rather than biofilm formation per se is the critical determining factor in the protective function of this type of T6SS armor.
Fig. 3.
Deletion of biofilm matrix genes does not impact V. cholerae T6SS. (A) Secretion of Hcp into the supernatant (S) was measured in the rbmA, rbmC, and bap1 mutants, and Hcp and VipA expression was measured in the cell pellet (P). (B) Competition between P. aeruginosa PAO1 and T6SS+ V52 and rbmA, rbmC, and bap1 mutants. Data represent the mean ± SD of three biological replicates.
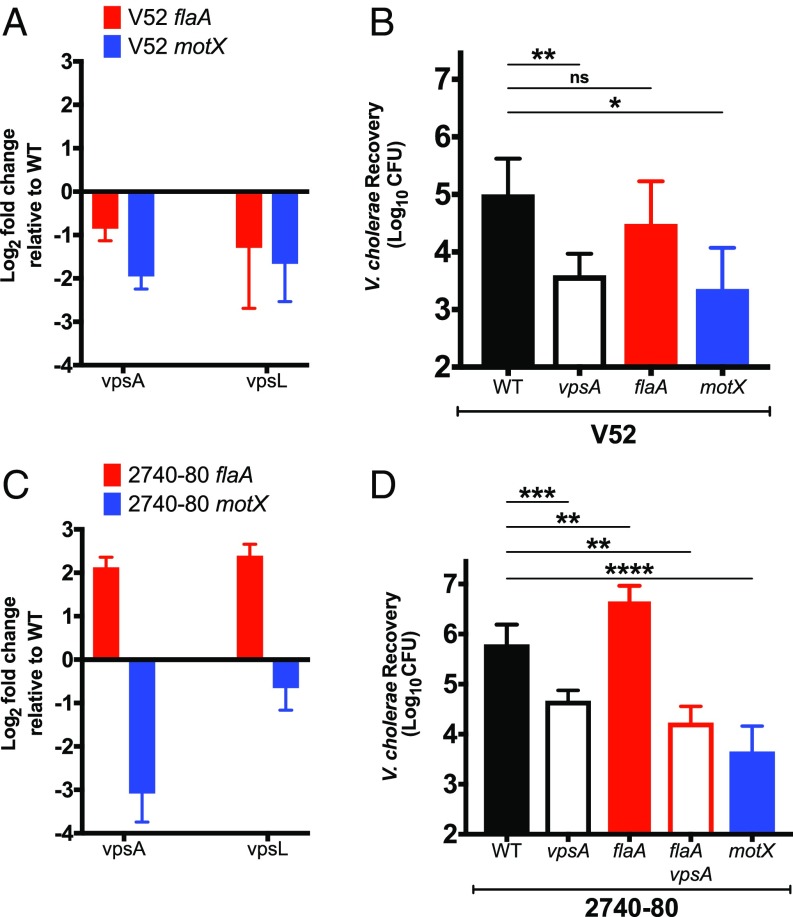
Because deletions of genes responsible for EPS biosynthesis resulted in susceptibility to T6SS-mediated killing, we evaluated whether mutations that modulate EPS gene expression could correspondingly change sensitivity to those attacks. In V. cholerae strain MO10 (O139 serotype), deletion of the core flagellin gene flaA results in increased EPS production, while deletion of sodium motor gene motX suppresses EPS expression (39). However, in V52 (O37 serotype), deletion of flaA had a minimal effect on the expression of EPS biosynthesis genes vpsA and vpsL (Fig. 4A) and, correspondingly, no significant effect on the sensitivity to killing by P. aeruginosa (Fig. 4B). In contrast, deletion of motX reduced EPS gene expression by approximately fourfold (Fig. 4A), which resulted in a corresponding increase in sensitivity to P. aeruginosa T6SS-mediated killing (Fig. 4B).
Fig. 4.
Up- or down-regulation of EPS genes result in corresponding increase or decrease in resistance to T6SS-dependent killing. (A) Expression of EPS production genes vpsA and vpsL in flaA and motX mutants of V. cholerae V52 were measured by qRT-PCR. (B) P. aeruginosa PAO1 was competed against T6SS+ V52 and T6SS− V52 vasK with no additional mutations, deletion of vpsA, deletion of flaA, or deletion of motX. (C) Expression of vpsA and vpsL genes required for EPS production in flaA and motX mutants of V. cholerae 2740-80 were measured by qRT-PCR. (D) P. aeruginosa PAO1 was competed against T6SS+ 2740-80 and T6SS− 2740-80 vipA with no additional mutations, deletion of vpsA, deletion of flaA, deletion of flaA vpsA, or deletion of motX. Data represent the mean ± SD of three biological replicates. Student’s t test was performed to compare each condition (ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001, ****P ≤ 0.0001).
Because EPS gene regulation may be fundamentally different in strain V52 compared with that previously reported for strain MO10, we also analyzed V. cholerae O1 El Tor strain 2740-80. This O1 V. cholerae strain is more closely related to the O139 strain MO10 (40) but still has a constitutively active T6SS (41). In this strain, deletion of flaA resulted in an approximately fourfold up-regulation of vpsA and vpsL genes. Deletion of motX in this strain resulted in an approximately eightfold down-regulation of vpsA and twofold down-regulation of vpsL (Fig. 4C). Like strain V52, deletion of vpsA in 2740-80 increased its sensitivity to P. aeruginosa T6SS (Fig. 4D). However, unlike in V52, deletion of flaA in 2740-80 resulted in a significant increase in resistance to P. aeruginosa T6SS (Fig. 4D). Deletion of vpsA in the flaA deletion background suppressed this effect in 2740-80 (Fig. 4D), confirming that the increased resistance to P. aeruginosa T6SS was indeed due to increased vps gene expression. We conclude that different genetic modules controlling the level of EPS production allow for tuning of the level of resistance to T6SS attacks by heterologous bacterial species.
Discussion
Biofilms are a known mode of defense used by bacteria to protect themselves from attacks by biological predators such as eukaryotic protists and macrophages (42), prokaryotic organisms such as bdellovibrios (43), and myxococci (44, 45) and bacteriophages (46). It is also appreciated that bacterial biofilms are more resistant to noxious small and macromolecules, including antimicrobial substances such as antibiotics, bacteriocins, and host antimicrobial peptides and proteins (e.g., defensins and complement) (47, 48). Although biofilms play a role in allowing bacterial prey cells to resist these exogenous attacks, it is not clear which components of the biofilm provide resistance and how these mechanistically work. Here we show that the EPS that V. cholerae produces to drive biofilm formation also blocks the lethal activity of the dynamic antimicrobial T6SS when expressed by nearby predatory cells. For this reason, we propose that EPS is a type of armor that can repel T6SS attacks even when bacteria are not embedded in fully developed biofilms.
In V. cholerae, production and secretion of EPS is the first step in biofilm formation, as cells switch from a motile planktonic state to being nonmotile and surface-attached. Because T6SS is a contact-dependent system—its activity is only manifested among cells on a solid surface (6, 7)—the timing of EPS production coincides precisely with when T6SS activity becomes relevant within the bacterial population.
It has been previously reported that the physical cell-to-cell distance of adjacent cells changes during biofilm formation (36, 49). However, we found that EPS produced by V. cholerae functions as a barrier against T6SS attack without interfering with the production or function of the T6SS in attacking other bacteria. This unidirectional defense means that the protective capacity of EPS is not mediated by creating physical distance, because that would diminish T6SS activity of both the predator and the prey. Because EPS armor is equally effective in blocking different T6SS effectors with varying enzymatic activities from multiple different species, our data suggest that it acts as a physical barrier, preventing penetration of the outermembrane by exogenous T6SS attacks rather than providing intrinsic resistance to any specific effector activity.
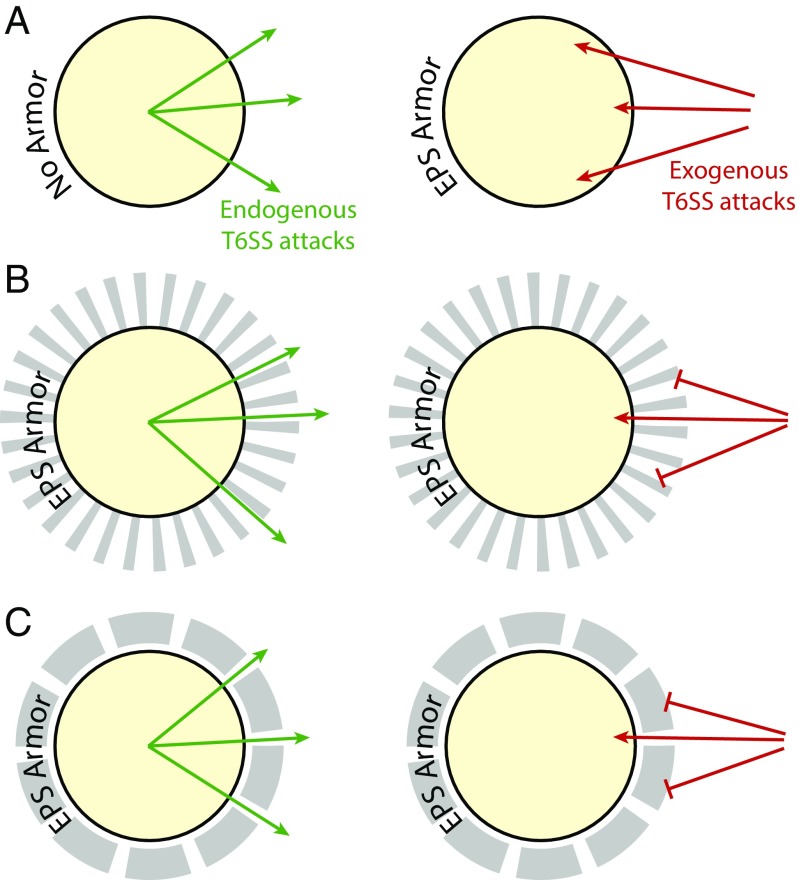
The question thus becomes, how can a physical obstruction act as a unidirectional barrier? Visualization of EPS through fluorescence labeling (36) and electron microscopy (50) suggest that EPS forms a relatively uniform coating around clusters of EPS-producing cells. Thus, the defensive properties of the EPS likely involve the macromolecular structure of the EPS matrix rather than the localization of EPS. At present, the structural organization of EPS is not known, but we can envision a couple of ways in which its geometric structure could confer a unidirectional protection against T6SS. For example, if the polymer structure of EPS consists of open channels perpendicular to the surface of the EPS producer, then the ability to mechanically block exogenous attacks might be dependent on the angle of such attacks (Fig. 5 A and B). Such a structure would allow for a T6SS apparatus perpendicular to the cellular surface to pass out of the predatory cell unobstructed. So, while all T6SS attacks by the EPS-producing predator will be unimpeded, only a small subset of exogenous attacks from a heterologous T6SS+ predatory cell will have a direct path to penetrate the cell envelope of the EPS producer. Alternatively, the EPS could consist of a solid coating of the cellular surface with small gaps in the covering, analogous to arrow slits in a castle wall (Fig. 5 A and C). In this model, the EPS-producing cell would need to localize the construction of its T6SS apparatus to the site of gaps in the EPS covering by coordinating EPS production with secretion machine assembly. Such coordination would have precedents, as other biopolymers, such as pili and flagella, are assembled and secreted in highly localized fashions (51, 52). In V. cholerae, its polar flagellum is even coated by a sheath that appears to be an extension of the outermembrane complete with lipopolysacchride components. Thus, either by chance or design, some T6SS apparatuses of a predatory cell might fire through gaps in the EPS armor that otherwise present a difficult to penetrate, protected surface to random exogenous attacks.
Fig. 5.
Model for EPS-mediated T6SS armor. (A) In the absence of EPS, both endogenous and exogenous T6SS attacks go unimpeded. (B) In this model, openings in the EPS are perpendicular to the cell surface, allowing all endogenous outward attacks to be unobstructed while only allowing a small subset of exogenous inward attacks through. (C) In this model, the EPS armor is a solid covering of the cell surface with small gaps analogous to castle wall arrow slits. Each individual T6SS apparatus is built specifically at these gaps to allow unobstructed endogenous outward T6SS activity, while exposing only a small surface area to exogenous inward attack.
When A. baylyi acts as the predator, EPS– V. cholerae cells are 10 times more susceptible to killing than EPS+ V. cholerae (Fig. 2B). This is in contrast to P. aeruginosa, in which the EPS– V. cholerae cells are 100-fold more susceptible (Fig. 2C). One explanation for this difference could be differences in bacterial cell geometry. For example, A. baylyi cells are smaller than V. cholerae and nearly coccoid in shape, which might allow it to fire its T6SS apparatus more efficiently and overcome the EPS barrier of neighboring prey. Alternatively, this difference in the efficacy of the EPS armor could be due to differences in the T6SS apparatus of the respective predator species (e.g., T6SS structure length, mechanical force generated by the contraction event, “sharpness” of the associated PAAR proteins, and so forth). In this study, we also observed that changes in EPS gene expression result in a commensurate increase and decrease in protection against T6SS attacks, opening the door for fine-tuning the sensitivity of a given bacterium to T6SS depending on the prey and predators it encounters during its natural ecology or while receiving other signals for T6SS expression, DNA uptake, or biofilm formation (53).
Although EPS is a component of biofilm, the T6SS armor effect of EPS does not require formation of a complete biofilm, as deletion of other biofilm matrix components did not affect sensitivity to T6SS attacks (Fig. 3B). Thus, the role of EPS in regulating bacterial interactions may be driven more by the properties of this polysaccaride when surrounding individual bacteria, rather than its role in creating a matrix for a multicellular structurally sound biofilm. In this regard, it is interesting to note that capsular polysaccharides (54), slime layers (55), long O-side chains of some lipopolysaccharides (56), and S-layer protein arrays (57) are extracellular components of bacterial cells that are generally thought to provide individual cells with protection from phagocytic cells and macromolecular complexes, such as the complement membrane attack complex and bacteriophages through primarily mechanical exclusion. The results reported here suggest that some of these surface structures might also impede efficient T6SS-mediated attacks by mechanisms similar to those observed for EPS and V. cholerae. Thus, the virulence-enhancing properties of these myriad surface structures could also reflect a role they might play in providing resistance to T6SS attacks emanating from members of the commensal microbiota. Given the prevalence of the T6SS in gut microbial communities (58, 59), EPS production and other T6SS defense mechanisms are likely to be key factors that regulate the interplay between different microbial species and should be considered when developing antimicrobials or other biological therapies, including vaccines and probiotics.
Materials and Methods
Bacterial Strains and Growth Conditions.
All strains and plasmids used in this study are listed in SI Appendix, Table S1. Strains were grown in LB (10 g/L tryptone, 5 g/L yeast extract, 5 g/L sodium chloride) at 37 °C with shaking at 300 rpm. Antibiotic concentrations that were used include: streptomycin (100 µg mL−1), irgasan (5 µg mL−1), gentamicin (15 µg mL−1), chloramphenicol (E. coli, 20 µg mL−1; V. cholerae, 5 µg mL−1), and kanamycin (50 µg mL−1).
Construction of Deletions in V. cholerae.
In-frame deletions in V. cholerae were constructed using allelic exchange with the suicide vectors pWM91 and pDS132, as previously described (5). All cloning constructs were verified by sequencing.
Biofilm Formation in Microtiter Plates.
Biofilm formation was assayed as previously described with some slight modifications (60). V. cholerae strains were grown overnight in LB with 100 µg mL−1 streptomycin. Following overnight growth, strains were diluted to final OD600 = 0.05 and 100 µL was inoculated into a single well of a polystyrene 96-well microtiter dish. The plates were incubated at 30 °C for 24 h then rinsed three times in Milli-Q water and dried at room temperature for 30 min. Each well was stained by the addition of 125 μL of 0.1% crystal violet for 15 min, followed by four to six rinses of each well in Milli-Q water. Crystal violet in each well was then solubilized in 150 μL 30% acetic acid. Then, 100 μL of the solubilized crystal violet was transferred to a new plate and the absorbance at 550 nm was measured using a plate reader (Bio-Tek).
Bacterial Killing Assay.
Overnight cultures of V. cholerae, E. coli, A. bayli, and P. aeruginosa were diluted 100:1 in LB and grown at 37 °C until mid-log (OD600 = 0.6–0.8). Cultures were centrifuged at 8,000 × g for 5 min and resuspended in LB to final concentration of OD600 = 10. Competition assays were set up at predator-to-prey ratio of 1:1 and incubated on LB agar plates at 37 °C for 2 h (V. cholerae vs. V. cholerae, V. cholerae vs. A. bayli ADP1, and V. cholerae vs. E. coli competitions) or 3 h (V. cholerae vs. P. aeruginosa competitions). For competitions involving the VgrG3-NucSe1 effector, the predator-to-prey ratio was increased to 10:1. Following incubation, mixtures were resuspended in 1 mL LB. Serial dilutions were spotted on plates to select for prey strains and CFUs counted. Three to six biological replicates were performed.
Western Blots on Supernatant and Cell Lysate.
Overnight cultures of V. cholerae were diluted 100:1 in LB and grown shaking at 37 °C until mid-log (OD600 = 0.6–0.8). A 1-mL culture was centrifuged at 8,000 × g for 5 min and resuspended in 125 μL LB. Cells were incubated at room temperature for 15 min and centrifuged again. Next, 75 μL of the supernatant was removed and added to 25 μL 4× SDS buffer and boiled at 95 °C for 10 min. The pellet was resuspended to OD600 = 10 in 1× SDS buffer and boiled at 95 °C for 10 min. Following brief cooling on ice, proteins were separated by SDS/PAGE. Following transfer on a nitrocellulose membrane (iBlot2; Invitrogen), the membranes were blotted for VipA and Hcp, respectively (Rabbit, peptide anti-Hcp and anti-VipA antibody; Genescript). A fluorescent secondary antibody (Li-Cor IRDye 680RD) was used for detection and membranes were imaged using a Li-Cor Odyssey Blot Imager (Li-Cor Biotechnology).
RNA Extraction and qRT-PCR.
Overnight cultures of V. cholerae were diluted 100:1 in LB and grown at 30 °C shaking at 200 rpm for 5 h, as previously described with some modifications (61). Next, 2 mL of each culture was centrifuged and resuspended in 1 mL TRIzol (Thermo Fisher Scientific). RNA was extracted using a kit according to the manufacturer’s instructions (Ambion) and each sample was treated with Turbo DNA-free kit to remove any contaminating DNA (Ambion). RNA was quantified by nanodrop (Thermo-Fisher Scientific), and qRT-PCR reactions were set up using Kapa SYBR Fast One-Step Universal qRT MasterMix and run on a thermocycler (Eppendorf Mastercycler RealPlex2). RNA levels were normalized to that of gyrA for each strain. At least three biological replicates were performed for each experiment.
Statistical Analysis.
Statistical tests were performed using by Student’s t test with Prism (Graphpad Software). All data presented are the mean ± SD.
Supplementary Material
Acknowledgments
We thank Stephen Lory for critical reading of the manuscript and members of the J.J.M. laboratory for helpful discussion and suggestions. This work was supported by National Institutes of Allergy and Infectious Diseases Grant AI-01845 (to J.J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808469115/-/DCSupplemental.
References
- 1.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand E, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 9.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazarov S, et al. Cryo-EM reconstruction of type VI secretion system baseplate and sheath distal end. EMBO J. 2018;37:e97103. doi: 10.15252/embj.201797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unterweger D, et al. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 2015;34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silverman JM, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burkinshaw BJ, et al. A type VI secretion system effector delivery mechanism dependent on PAAR and a chaperone-co-chaperone complex. Nat Microbiol. 2018;3:632–640. doi: 10.1038/s41564-018-0144-4. [DOI] [PubMed] [Google Scholar]
- 16.Unterweger D, Kostiuk B, Pukatzki S. Adaptor proteins of type VI secretion system effectors. Trends Microbiol. 2017;25:8–10. doi: 10.1016/j.tim.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Kudryashev M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoued A, et al. Priming and polymerization of a bacterial contractile tail structure. Nature. 2016;531:59–63. doi: 10.1038/nature17182. [DOI] [PubMed] [Google Scholar]
- 19.Kapitein N, et al. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol. 2013;87:1013–1028. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- 20.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murdoch SL, et al. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol. 2011;193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sana TG, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci USA. 2016;113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao W, Caro F, Robins W, Mekalanos JJ. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science. 2018;359:210–213. doi: 10.1126/science.aap8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci USA. 2010;107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray A, et al. Type VI secretion system MIX-effectors carry both antibacterial and anti-eukaryotic activities. EMBO Rep. 2017;18:1978–1990. doi: 10.15252/embr.201744226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell AB, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho BT, Fu Y, Dong TG, Mekalanos JJ. Vibrio cholerae type 6 secretion system effector trafficking in target bacterial cells. Proc Natl Acad Sci USA. 2017;114:9427–9432. doi: 10.1073/pnas.1711219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wexler AG, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong JC, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yildiz FH, Visick KL. Vibrio biofilms: So much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong JN, Yildiz FH. Biofilm matrix proteins. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teschler JK, et al. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol. 2015;13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fong JC, et al. Structural dynamics of RbmA governs plasticity of Vibrio cholerae biofilms. eLife. 2017;6:e26163. doi: 10.7554/eLife.26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan J, Sharo AG, Stone HA, Wingreen NS, Bassler BL. Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc Natl Acad Sci USA. 2016;113:E5337–E5343. doi: 10.1073/pnas.1611494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giglio KM, Fong JC, Yildiz FH, Sondermann H. Structural basis for biofilm formation via the Vibrio cholerae matrix protein RbmA. J Bacteriol. 2013;195:3277–3286. doi: 10.1128/JB.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauriano CM, Ghosh C, Correa NE, Klose KE. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J Bacteriol. 2004;186:4864–4874. doi: 10.1128/JB.186.15.4864-4874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calia KE, Murtagh M, Ferraro MJ, Calderwood SB. Comparison of Vibrio cholerae O139 with V. cholerae O1 classical and El Tor biotypes. Infect Immun. 1994;62:1504–1506. doi: 10.1128/iai.62.4.1504-1506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leid JG, et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 43.Kadouri D, O’Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71:4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berleman JE, et al. Exopolysaccharide microchannels direct bacterial motility and organize multicellular behavior. ISME J. 2016;10:2620–2632. doi: 10.1038/ismej.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan H, He X, Lux R, Luan J, Shi W. Killing of Escherichia coli by Myxococcus xanthus in aqueous environments requires exopolysaccharide-dependent physical contact. Microb Ecol. 2013;66:630–638. doi: 10.1007/s00248-013-0252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutherland IW, Hughes KA, Skillman LC, Tait K. The interaction of phage and biofilms. FEMS Microbiol Lett. 2004;232:1–6. doi: 10.1016/S0378-1097(04)00041-2. [DOI] [PubMed] [Google Scholar]
- 47.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 48.Domenech M, Ramos-Sevillano E, García E, Moscoso M, Yuste J. Biofilm formation avoids complement immunity and phagocytosis of Streptococcus pneumoniae. Infect Immun. 2013;81:2606–2615. doi: 10.1128/IAI.00491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drescher K, et al. Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc Natl Acad Sci USA. 2016;113:E2066–E2072. doi: 10.1073/pnas.1601702113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuhmacher JS, Thormann KM, Bange G. How bacteria maintain location and number of flagella? FEMS Microbiol Rev. 2015;39:812–822. doi: 10.1093/femsre/fuv034. [DOI] [PubMed] [Google Scholar]
- 52.Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol. 2017;15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 53.Veening JW, Blokesch M. Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat Rev Microbiol. 2017;15:621–629, and erratum (2017) 15:629. doi: 10.1038/nrmicro.2017.66. [DOI] [PubMed] [Google Scholar]
- 54.Cooper CA, Mainprize IL, Nickerson NN. Genetic, biochemical, and structural analyses of bacterial surface polysaccharides. Adv Exp Med Biol. 2015;883:295–315. doi: 10.1007/978-3-319-23603-2_16. [DOI] [PubMed] [Google Scholar]
- 55.Costerton JW, Irvin RT, Cheng KJ. The bacterial glycocalyx in nature and disease. Annu Rev Microbiol. 1981;35:299–324. doi: 10.1146/annurev.mi.35.100181.001503. [DOI] [PubMed] [Google Scholar]
- 56.Lerouge I, Vanderleyden J. O-antigen structural variation: Mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 57.Beveridge TJ, et al. Functions of S-layers. FEMS Microbiol Rev. 1997;20:99–149. doi: 10.1111/j.1574-6976.1997.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 58.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verster AJ, et al. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe. 2017;22:411–419.e4. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stutzmann S, Blokesch M. Circulation of a quorum-sensing-impaired variant of Vibrio cholerae strain C6706 masks important phenotypes. MSphere. 2016;1:e00098-16. doi: 10.1128/mSphere.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.