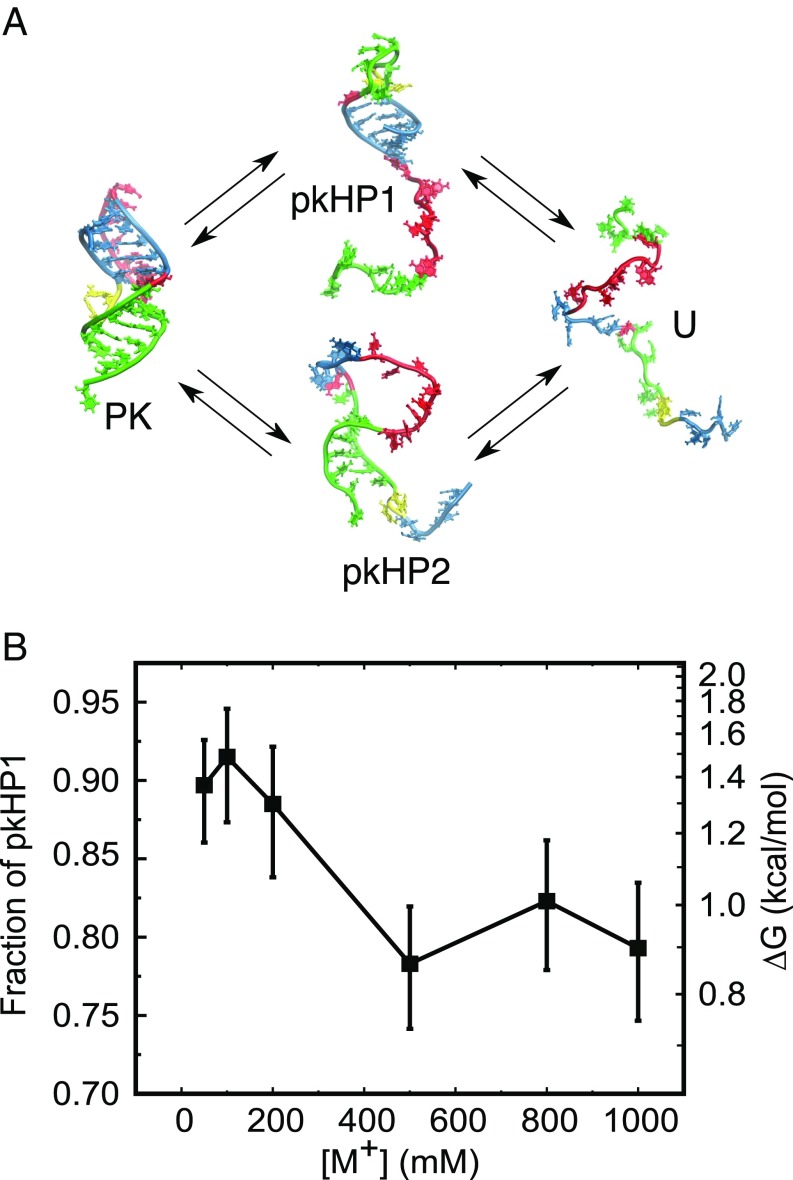
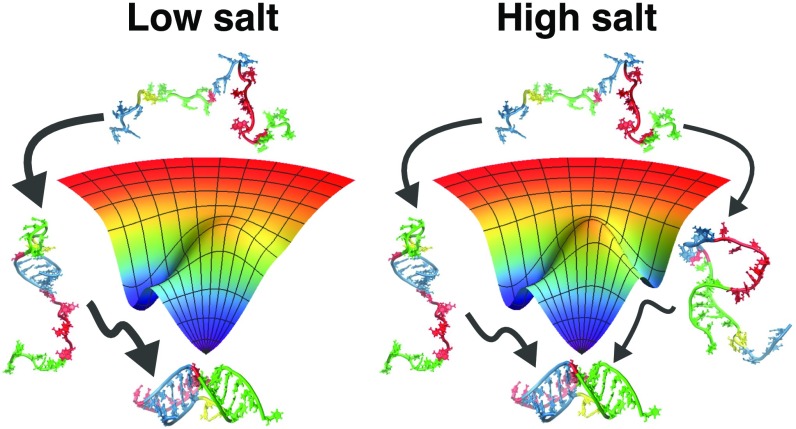
Significance
The assembly mechanism of RNA, vital to describing its functions, depends on both the sequence and the metal ion concentration. How the latter influences the folding trajectories remains an important unsolved problem. Here, we examine the folding pathways of an RNA pseudoknot (PK) with key functional roles in transcription and translation, using a combination of experiments and simulations. We demonstrate that the PK, consisting of two hairpins with differing stabilities, folds by parallel pathways. Surprisingly, the flux between them is modulated by monovalent salt concentration. Our work shows that the order of assembly of PKs is determined by the relative stability of the hairpins, implying that the folding landscape can be controlled by sequence and ion concentration.
Keywords: RNA folding, laser temperature-jump spectroscopy, time-resolved fluorescence, fluorescent nucleotide analogs, coarse-grained simulations
Abstract
The functions of RNA pseudoknots (PKs), which are minimal tertiary structural motifs and an integral part of several ribozymes and ribonucleoprotein complexes, are determined by their structure, stability, and dynamics. Therefore, it is important to elucidate the general principles governing their thermodynamics/folding mechanisms. Here, we combine laser temperature-jump experiments and coarse-grained simulations to determine the folding/unfolding pathways of VPK, a variant of the mouse mammary tumor virus (MMTV) PK involved in ribosomal frameshifting. Fluorescent nucleotide analogs (2-aminopurine and pyrrolocytidine) placed at different stem/loop positions in the PK serve as local probes allowing us to monitor the order of assembly of VPK that has two constituent hairpins with different intrinsic stabilities. We show that at 50 mM KCl, the dominant folding pathway populates only the more stable hairpin intermediate; as the salt concentration is increased, a parallel folding pathway emerges involving the less stable hairpin as an alternate intermediate. Notably, the flux between the pathways is modulated by the ionic strength. Our findings support the principle that the order of PK structure formation is determined by the relative stabilities of the hairpins, which can be altered by sequence variations or salt concentrations. The experimental results of salt effects on the partitioning between the two folding pathways are in remarkable agreement with simulations that were performed with no adjustable parameters. Our study not only unambiguously demonstrates that VPK folds by parallel pathways but also showcases the power of combining experiments and simulations for a more enriched description of RNA self-assembly.
The long-standing problem of how RNA molecules fold is still incompletely understood despite decades of research (1). The functions of RNA molecules are determined by their 3D folded structures, as well as by binding to ligands. The underlying folding landscapes have potentially many minima separated by free-energy barriers, which implies that their dynamics are governed by transitions between them (2). Therefore, elucidating the folding mechanisms of RNA molecules and the time scales in which they interconvert between different states is critical for understanding how structure and dynamics influence their functions. Such studies can be leveraged into the functional characterization of newly discovered RNAs and could lead to designer RNA molecules with desirable functions.
It has been shown that RNA self-assembly, especially for large ribozymes, can occur through multiple pathways, described by the kinetic partitioning mechanism (KPM), according to which a fraction of molecules fold rapidly and another fraction fold slowly as they become trapped in misfolded conformations corresponding to the minima in the rugged folding landscape (3). Evidence for the KPM was first obtained for the Tetrahymena ribozyme (4). Subsequently, it has also been determined that the folding landscapes of even RNA hairpins are rugged (5). The relatively fast folding times of nucleic acid hairpins and the ability to enumerate all possible folded and misfolded conformations have made hairpins particularly useful as model systems to validate various computational studies of the folding mechanisms (6–14). However, hairpins lack tertiary interactions, and they therefore provide only limited insights into the assembly of larger RNA molecules.
The KPM is a general scheme that explains the global folding mechanism of how ribozymes assemble. However, the structural details of the possible intermediates and how the flux between pathways is controlled by counter ions cannot be quantitatively predicted using KPM alone. Here, we have focused on the folding mechanisms of RNA pseudoknots (PKs) that serve as minimal tertiary structural motifs. In contrast to ribozymes that fold on times of the order of 1–1,000 s (15, 16), we previously reported that small H-type PKs that do not have deep misfolded traps can fold in a few milliseconds (17) in accord with predictions from simulations of coarse-grained RNA models (18). Thus, RNA PKs provide a bridge between rapidly folding secondary structure elements and the slower reorganization of ribozymes. They are ideal systems to explore the interplay between competing secondary and tertiary interactions that determine the diverse folding pathways and provide a way to investigate the role ions play in modulating the flux between different assembly pathways.
The H-type PKs form when the nucleotides in a hairpin loop form base pairs with nucleotides downstream of that hairpin, creating a structure with two stems and two loops (19) (Fig. 1). They are involved in several biological functions related to replication, RNA processing, and transcriptional and translational regulation (20–23). They also stabilize the structures of larger RNA assemblies, such as the ribosome, ribozymes, and ribonucleoproteins (24, 25), and are often the rate-limiting step in their folding pathways. Thus, understanding the global principles that determine the thermodynamics and folding pathways of PKs could provide insights into their functions and into the self-assembly of larger RNA complexes.
Fig. 1.
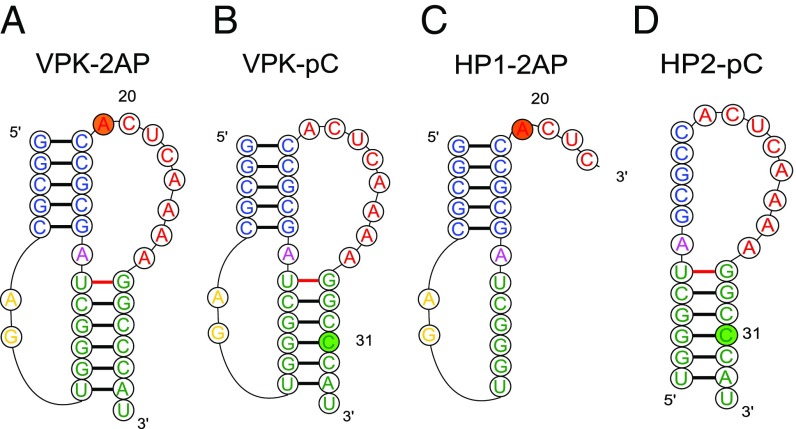
RNA constructs used in this study. PK VPK samples were labeled either with 2AP at position 20 (A, VPK-2AP) or with pC at position 31 (B, VPK-pC). (C) Hairpin HP1-2AP is a truncated sequence consisting of nucleotides 1–23 of VPK-2AP. (D) Hairpin HP2-pC is a truncated sequence consisting of nucleotides 8–34 of VPK-pC. Colors represent different regions of the PK: blue, stem 1; yellow, loop 1; green, stem 2; magenta, junction; and red, loop 2. In the truncated hairpins, nucleotides 8–14 of VPK stem 2 become part of the loop in HP1-2AP; similarly, nucleotides 14–19 of VPK stem 1 become part of the loop in HP2-pC.
Single-molecule pulling experiments and, more recently, nanopore-induced unfolding experiments have shown that PK folding/unfolding can occur through several complex pathways that include the formation of transient hairpins and nonnative structures (26–34). These experiments have also unveiled that the RNA molecules do not fold into a PK conformation in many instances, but fold into alternate structures instead (30). However, the energy landscapes and folding pathways inferred from pulling experiments could differ from those obtained by changing the temperature, as demonstrated for RNA hairpins (10, 35–37). NMR dynamics and single-molecule FRET experiments have also been performed in the absence of force, but these have been restricted to studying the dynamics of interconversion between a PK and its intermediate folds (usually structures with folded hairpins), and thus have not provided a full picture of the folding/unfolding process of PKs (38–40).
Computational studies have predicted that PKs fold through diverse mechanisms, depending on their sequence, by parallel pathways, cooperatively in an all-or-none manner, or by a combination of both mechanisms, exploring several intermediate states, some of which contain nonnative base pairing or alternate structures (18, 41–47). The principle that assembly routes of PKs depend on the stabilities of individual secondary structural elements (18) explains the diversity of the folding mechanisms of PK. However, to our knowledge, experimental demonstration of folding through parallel pathways and the impact of ions in affecting the partitioning between these pathways is lacking. This is due, in part, to experimental limitations that do not allow probing of folding kinetics in the submillisecond region, where hairpins and possible folding intermediates could form.
Here, we take advantage of fluorescent nucleotide analogs (48–51) judiciously placed at various positions in either the stem or loop region of a PK to explore alternative pathways in PK folding, and we use a laser temperature jump (T-jump) approach to study their folding kinetics with microsecond resolution. We apply these fluorescent approaches to study the folding mechanisms of VPK, a variant of the mouse mammary tumor virus (MMTV) PK that promotes efficient frameshifting in MMTV (52–54). To ascertain how ions alter the flux between distinct folding pathways, we use experiments and simulations to determine the thermodynamics and folding kinetics of VPK over a broad range of ionic conditions. The results provide an unparalleled experimental measure, confirmed independently by coarse-grained simulations using a model with no adjustable parameters, of the partitioning of the folding process into different folding pathways. We find that at 50 mM KCl, the dominant folding pathway of VPK populates the more stable constituent hairpin of VPK as an intermediate. However, as the salt concentration is increased, a parallel folding pathway emerges involving the other less stable hairpin as an intermediate, with the flux through the alternative pathway increasing with increasing salt concentrations. The combined experiments and simulations directly demonstrate that PKs can fold via parallel pathways. We further establish that monovalent ions can modulate the flux through distinct folding pathways. As enunciated previously (18), the order of assembly is largely determined by the relative stabilities of the hairpins, which can be changed by altering the ionic strength.
Results
Fluorescent Nucleotide Analogs as Probes of PK Unfolding Thermodynamics.
Previous thermodynamics studies on VPK unfolding were carried out using either UV absorbance measurements (17, 55) or differential scanning calorimetry studies (55). While these studies demonstrated hierarchical unfolding of VPK, they could not discern whether unfolding occurred via parallel pathways. Our previous T-jump studies on VPK (17) also relied on absorbance changes that could not distinguish between the folding of one stem or the other or on fluorescence changes of an extrinsic (fluorescein) label attached at the 5′-end, which could only sense the unfolding of stem 1 and did not provide any insights into possible parallel pathways. Here, we used fluorescent nucleotide analogs at different positions to probe experimentally potential parallel pathways for VPK unfolding. We prepared two PK constructs in which we substituted one nucleotide of the VPK sequence for a fluorescent analog: VPK-2AP, with 2-aminopurine (2AP) instead of adenine at position 20, and VPK-pC, with pyrrolocytidine (pC) instead of cytosine at position 31 (Fig. 1 A and B). In parallel, we also prepared truncated sequences that could form hairpins (HP1 and HP2) that mimicked the constituent hairpins of VPK, with 2AP and pC probes in HP1-2AP and HP2-pC, respectively, located in the same context as in VPK (Fig. 1 C and D).
To assess the degree to which the incorporated fluorescent nucleotides perturb the RNA structures, we performed absorbance melting measurements on unlabeled and labeled VPK (SI Appendix, Fig. S1). At 50 mM KCl, the absorbance melting profiles for all of the VPK constructs show two transitions that were analyzed in terms of a minimal three-state model , where U is defined as the unfolded state (SI Appendix, SI Methods 1.3). The thermodynamic parameters from this analysis are summarized in SI Appendix, Table S1; the uncertainties in these parameters were determined as described in SI Appendix, SI Methods 1.5 and Fig. S2. The melting temperatures (s) for the two transitions in the unlabeled VPK are at ∼47 °C and ∼85 °C (Fig. 2), consistent with previous studies (17, 55). While 2AP has a negligible effect on the stability of the fully folded PK, with for at ∼48 °C, it appears to significantly stabilize the partially unfolded intermediate hairpin states, with for shifted to ∼97 °C (SI Appendix, Fig. S1C). pC slightly stabilizes both the folded PK and the intermediate hairpin states, with for the first transition at ∼53 °C and for the second transition at ∼89 °C (SI Appendix, Fig. S1D).
Fig. 2.
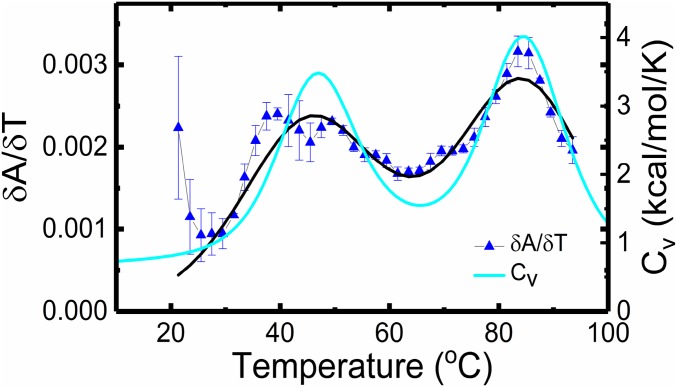
Thermodynamics of unlabeled VPK. The first derivative of the absorbance with respect to temperature (), from measurements on unlabeled VPK (SI Appendix, Fig. S1 A and B), is plotted as a function of temperature (blue). Measurements were done in 10 mM 3-(N-morpholino)propanesulfonic acid buffer (pH 7.0) at 50 mM KCl. The data (▲) are the averages of two to three independent sets of measurements; the errors are the SEM. The continuous black line is a fit to a three-state model, with transitions at 46.6 ± 0.4 °C and 85.5 ± 0.3 °C. The heat capacity of VPK at 50 mM obtained from simulations is shown in cyan, with transitions observed at 47 °C and 84 °C.
Although incorporation of nucleotide analogs does have a detectable effect on the stability of the RNA structures, they nonetheless show promise in reporting on the nature of the transitions and the pathways for PK unfolding, as discussed below. Our choice for the placement of the probes was motivated by the following considerations. We placed 2AP at a position adjacent to the end of stem 1 since VPK does not have a suitably placed adenine in either of its stems (other than at the 3′-end, which could be prone to “fraying” ends). The 2AP in this position is particularly sensitive to the conformational changes (discussed below) that occur along the unfolding pathway that involves the partially folded intermediate state pkHP1 (Fig. 3A, top pathway). On the other hand, the pC location was chosen to directly monitor the loss of stem 2 either when the PK unfolds via pkHP1 (Fig. 3D, top pathway) or when the residual pkHP2 unfolds (Fig. 3D, bottom pathway).
Fig. 3.
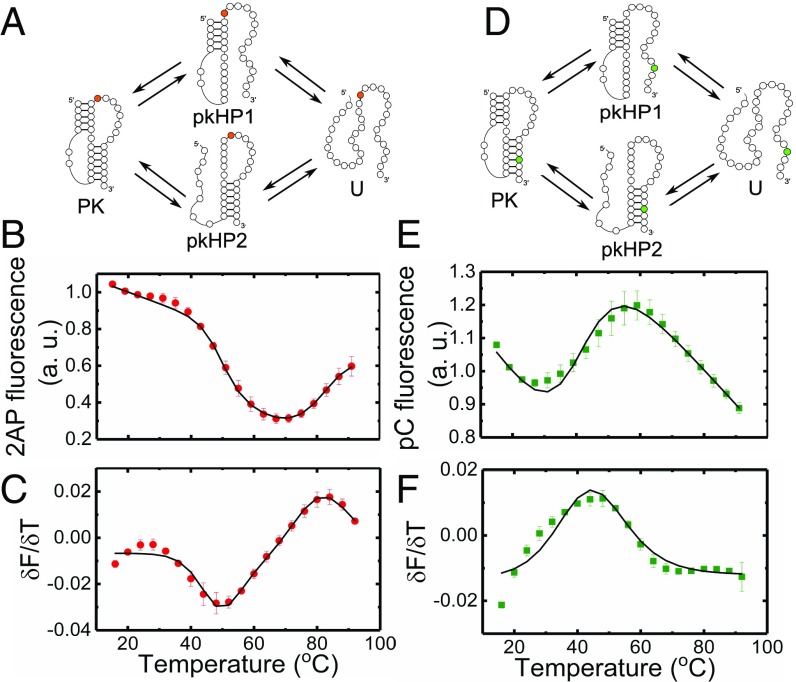
Thermodynamics of VPK melting from fluorescence experiments. Folding/unfolding schemes for VPK-2AP (A) and VPK-pC (D) are shown. The 2AP fluorescence of VPK-2AP (B) and pC fluorescence of VPK-pC (E) are plotted as a function of temperature. Measurements were done in 10 mM 3-(N-morpholino)propanesulfonic acid buffer (pH 7.0) at 50 mM KCl. The corresponding first derivatives of the fluorescence with respect to temperature () are plotted as a function of temperature for VPK-2AP (C) and VPK-pC (F). The data (● and ■) are the averages of two to three independent sets of measurements. The errors are the SEM; error bars not visible are smaller than the symbols. For clarity, only every other data point is presented. The continuous lines are fits to a three-state model.
Melting Profiles at 50 mM KCl Reveal That VPK Unfolds Predominantly via Unfolding of Stem 2.
We first examined the melting profiles of VPK-2AP and VPK-pC from fluorescence measurements in 50 mM KCl (Fig. 3). As controls, we also measured the effect of temperature on the intrinsic quantum yield of the 2AP and pC probes in the context of 5-nt-long reference samples, 2AP-ref and pC-ref, respectively (SI Appendix, Fig. S3). While the control measurements revealed a monotonic decrease in 2AP fluorescence, corresponding measurements on VPK-2AP revealed two unfolding transitions, similar to the absorbance studies; the first transition showed a decrease in 2AP fluorescence, followed by an increase in the second transition (Fig. 3B). A decrease in fluorescence in the first transition is attributed to the loss of stem 2 in the PK structure, which likely enables the 2AP label to stack better over stem 1. These results suggest that unfolding via the pkHP1 intermediate (Fig. 3A, top pathway) is the dominant trajectory in VPK unfolding; unfolding via pkHP2 where stem 1 melts first (Fig. 3A, bottom pathway) is not expected to dramatically affect 2AP fluorescence. Consistent with this assertion, an increase in 2AP fluorescence in the second transition is attributed to the unfolding of the residual hairpin pkHP1, with concomitant loss of 2AP stacking interactions. Again, significant change in 2AP fluorescence is not expected in the unfolding of the alternative pkHP2 hairpin, with 2AP located in the loop of that hairpin.
Analysis of the fluorescence melting profile of VPK-2AP using a three-state model revealed a low temperature transition (attributed to ) with 50 °C and a high temperature transition (attributed to ) with 86 °C (SI Appendix, Table S1). The s for the transition in VPK-2AP obtained from absorbance and fluorescence measurements are in good agreement. The for the transition is lower in fluorescence measurements compared with the ∼97 °C obtained from the absorbance measurements. This discrepancy is attributed to partial unstacking of 2AP from stem 1 of the pkHP1 intermediate as a result of “premelting” or “end-fraying” of that stem before complete unfolding. Similar discrepancies between 2AP fluorescence and absorbance melting profiles were observed in previous studies of DNA hairpins (35).
To further examine whether PK pkHP1 U is indeed the dominant unfolding pathway at 50 mM KCl, we measured the fluorescence melting profiles of VPK-pC. Strikingly, this melting profile exhibited only one dominant transition (Fig. 3 E and F), with 46 °C consistent with the unfolding of the PK. The unfolding of any residual hairpins expected in the second melting transition, as seen in absorbance measurements on this construct, is largely undetected by the pC probe located in stem 2. Instead, the pC fluorescence before and after the dominant transition decreases monotonically with increasing temperature, reminiscent of the intrinsic temperature dependence of the control sample pC-ref (SI Appendix, Fig. S3).
We expect the pC fluorescence in VPK-pC to increase upon melting of stem 2, from loss of stacking interactions (56), and to stay largely unperturbed upon melting of stem 1, either in the step (Fig. 3D, bottom pathway) or in the step (Fig. 3D, top pathway). The observation that pC fluorescence increases in a single transition at low, but not high, temperatures further reaffirms that nearly all of stem 2 is lost when the PK unfolds. Altogether, the fluorescence melting profiles of VPK-2AP and VPK-pC strongly support the conclusion that the dominant pathway for VPK unfolding populates pkHP1 as the intermediate state at low (50 mM) monovalent ion concentrations.
We note here that the PK unfolding transition measured by pC fluorescence (at ∼46 °C) is lower than the same transition measured by absorbance (at ∼53 °C) on the same VPK-pC construct (SI Appendix, Table S1). Our explanation for this difference is that the pC probe as positioned in stem 2 could be sensing some local premelting of VPK that could likely distort the assessed for the PK unfolding transition. Indeed, a closer look at the VPK-pC fluorescence melting profiles (Fig. 3 E and F) shows a somewhat broad transition that appears to have contributions from baselines that deviate from linearity, which is assumed in all our fits. The nonlinear baselines are apparent in VPK-pC measurements at higher (100 mM–1 M) salt, as discussed later.
2AP-Probed Thermodynamics and Folding/Unfolding Kinetics Measured on Hairpin HP1.
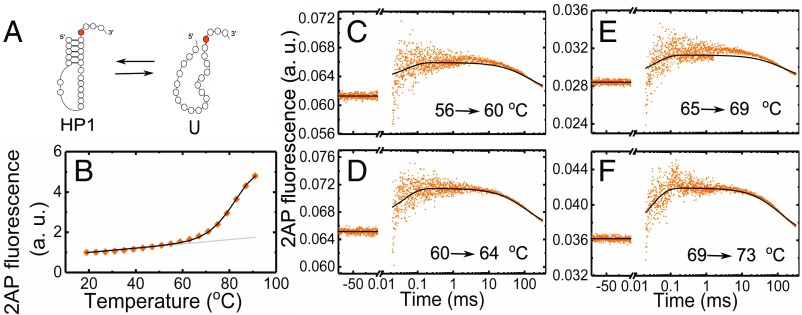
To isolate the thermodynamics and folding/unfolding kinetics of the pkHP1 ⇔ U transition, we studied a shorter sequence with the same nucleotide composition (nucleotides 1–23) as stem 1 and loop 1 of VPK (HP1-2AP in Fig. 1C). Nucleotides 20–23 were added at the 3′-end of stem 1 to approximately mimic the dangling end in the pkHP1 hairpin conformation, and 2AP was incorporated at the same position 20 as in VPK. The fluorescence melting profiles of HP1-2AP, also measured at 50 mM KCl, revealed an increase in 2AP fluorescence in a single (one-step) unfolding transition with 84 °C (Fig. 4B and SI Appendix, Fig. S4 A and D and Table S2), similar to the 2AP-probed of the second transition in VPK-2AP at ∼86 °C (SI Appendix, Table S1). These results further support our conclusion that the second transition in VPK unfolding corresponds to the melting of stem 1 when the intermediate pkHP1 unfolds.
Fig. 4.
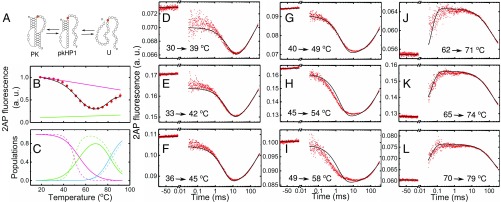
Global analysis of thermodynamics and kinetics measurements on HP1-2AP. (A) Minimal two-state folding/unfolding scheme for HP1-2AP: the fully folded hairpin state (HP1) and the fully unfolded state (U). (B) Fluorescence melting profile of HP1-2AP. The data (♦) are the averages of two independent sets of measurements. The errors are the SEM; error bars not visible are smaller than the symbols. For clarity, only every other data point is presented. The continuous black line is from a global fit of the equilibrium and kinetics data to a two-state kinetic model, as described in SI Appendix, SI Methods 1.9; the gray line represents the temperature dependence of 2AP fluorescence in the hairpin state, as obtained from this global fit. (C–F) Relaxation traces from T-jump measurements on HP1-2AP are shown for four different sets of initial and final temperatures, with an average T-jump 4.2 ± 0.8 °C. Each trace shows the fluorescence intensity level at the initial temperature before the arrival of the IR pulse (denoted by negative times on the x axis); a sharp increase in fluorescence immediately after the IR pulse that heats the sample, reflecting an increase in 2AP fluorescence, possibly from rapid unstacking, followed by relaxation kinetics in a time window of ∼30 μs to ∼1 ms; and then the decay of the temperature of the sample back to the initial temperature on time scales >10 ms. The continuous black lines in each of the panels are from the global fit of the equilibrium and kinetics data.
To measure the folding/unfolding kinetics of HP1-2AP, we collected relaxation traces in response to an ∼3–10 °C T-jump perturbation by recording 2AP fluorescence as a function of time (Fig. 4 C–F). We also performed T-jump studies on the reference sample 2AP-ref; this 5-nt RNA strand is not expected to exhibit any folding/unfolding kinetics, and hence served as a control (SI Appendix, Fig. S5). T-jump measurements on 2AP-ref showed a rapid initial drop (faster than our earliest observable time of 30 μs), which reflected a drop in 2AP quantum yield as a consequence of the T-jump, followed by a slow relaxation on >10-ms time scales characteristic of the decay (“recovery”) of the heated volume temperature back to its initial pre–T-jump value (SI Appendix, SI Methods 1.8 and Fig. S5B).
In contrast to control measurements, T-jump measurements on HP1-2AP showed a rapid increase in 2AP fluorescence immediately after the T-jump, followed by a further increase in the time window of 30 μs to 1 ms, and then a decrease in 2AP fluorescence back to the pre–T-jump value on a 10- to 300-ms time window (Fig. 4 C–F). The initial increase in the 2AP-probed kinetics immediately after the T-jump is consistent with equilibrium measurements on HP1-2AP, which show a gradual increase in 2AP fluorescence between 20 and 60 °C, before the melting transition, ascribed to increased unstacking of 2AP from the hairpin’s stem as the temperature is raised (Fig. 4B). In T-jump measurements, this premelting is too fast to be resolved and appears as a sudden jump in 2AP fluorescence. The relaxation kinetics observed in the 30-μs to 1-ms time window are attributed to hairpin folding/unfolding kinetics, while the slower relaxation on time scales >10 ms mimics the T-jump recovery decay profile seen in the 2AP-ref control measurements (SI Appendix, Fig. S5B).
Global Analysis of Thermodynamics and Folding/Unfolding Kinetics of HP1-2AP.
We next analyzed the equilibrium and kinetics data on HP1-2AP in terms of a two-state system: HP1 ⇔ U (Fig. 4A), using a global analysis approach as described in SI Appendix, SI Methods 1.9, with the equilibrium populations parameterized as described in SI Appendix, SI Methods 1.3. The relaxation traces were described in terms of the time-dependent change in the populations as a function of time. To this end, the rate coefficients for the hairpin-to-unfolded transition at each temperature () were determined by two parameters: the rate coefficient () at a reference temperature (, chosen to be 85 °C, and the enthalpy barrier () for that transition; the folding rate coefficients at each temperature () were obtained from the unfolding rate coefficients and the equilibrium parameters, as described in SI Appendix, SI Methods 1.9. Additionally, the size of the T-jump and the recovery time constants were varied independently for each kinetics trace. The best fit results from this global analysis are shown in Fig. 4, and the equilibrium and kinetics parameters are summarized in SI Appendix, Table S3.
Global Analysis of Thermodynamics and Folding/Unfolding Kinetics of VPK-2AP.
T-jump kinetics traces on VPK-2AP were obtained in a manner similar to HP1-2AP. Kinetics traces obtained at temperatures below ∼60 °C (Fig. 5 D–I) displayed a sharp drop in 2AP fluorescence immediately after the T-jump, resembling the behavior of the 2AP-ref control (compare Fig. 5 D–I with SI Appendix, Fig. S5B); relaxation kinetics were observed in the time window from 30 μs to 10 ms, followed by T-jump recovery kinetics on time scales >10 ms, as before. In contrast, at temperatures above ∼60 °C (Fig. 5 J–L), the traces are analogous to the T-jump response of HP1-2AP (compare Fig. 5 J–L with Fig. 4 C–F). These results complement thermodynamics experiments and suggest that T-jump measurements in the temperature range of 30–60 °C report on the folding/unfolding kinetics of the PK to the intermediate hairpin conformations, while measurements in the temperature range 60–80 °C report on the folding/unfolding kinetics of the residual hairpins.
Fig. 5.
Global analysis of thermodynamics and kinetics measurements on VPK-2AP. (A) Three-state folding/unfolding scheme for VPK-2AP: the fully folded PK, a partially folded hairpin state with stem 1 formed but not stem 2 (pkHP1), and unfolded RNA (U). VPK-2AP fluorescence equilibrium and kinetics data were simultaneously fitted using this model. (B) Fluorescence melting profile of VPK-2AP; the data (●) are the averages of two independent sets of measurements. The error bars represent the SEM; error bars not visible are smaller than the symbols. For clarity, only every other data point is presented. The continuous black line is from a global fit of the equilibrium and kinetics data to a three-state kinetic model, as described in the main text; the pink (green) line represents the temperature dependence of 2AP fluorescence in the PK (pkHP1) state, as obtained from this global fit. (C) Equilibrium populations of the three states indicated in A are plotted as a function of temperature: PK (pink), pkHP1 (green), U (blue). The populations are calculated from either the parameters from the global kinetic modeling (continuous lines) or the analysis of the equilibrium data alone (dashed lines; SI Appendix, Table S1). (D–L) Relaxation traces from T-jump measurements on VPK-2AP are shown for nine different sets of initial and final temperatures, with an average T-jump 9.1 ± 3.5 °C. The continuous black lines are from a global fit of the equilibrium and kinetics data.
The combined evidence from the equilibrium melting experiments on VPK-2AP, VPK-pC, and HP1-2AP showed that the VPK PK in 50 mM KCl unfolds primarily via a pathway that populates pkHP1 as an intermediate. With this insight, we described the folding/unfolding of VPK in terms of a minimal kinetic model with three states: the fully folded PK, the intermediate hairpin (pkHP1), and the completely unfolded state (U) (Fig. 5A). Using a strategy similar to that described for HP1-2AP, we analyzed the 2AP-probed equilibrium melting profile of VPK-2AP and all of the kinetics traces measured over a range of temperatures in terms of a minimal set of parameters, as described in SI Appendix, SI Methods 1.9.
The results from this global analysis are shown in Fig. 5, and the best-fit parameters are summarized in SI Appendix, Table S3. Reassuringly, the melting parameters obtained from the global analysis are consistent with the parameters obtained from the equilibrium-only data, as indicated by the reasonably good agreement between the equilibrium populations of the three states obtained from the two descriptions (Fig. 5C). The temperature dependence of all of the rate coefficients in this three-state model, obtained from the global analysis, are represented in SI Appendix, Fig. S6.
VPK Fluorescence Melting Profiles at High Salt Concentrations Reveal Evidence of a Second Pathway.
The results at ionic conditions of 50 mM KCl support PK unfolding through one major pathway having pkHP1 as an intermediate. The strongest evidence for this conclusion came from the melting profile of VPK-pC, with the pC probe located in stem 2; in 50 mM KCl, VPK-pC exhibited only a single dominant transition at low temperatures, indicating complete loss of stem 2 in that transition (Fig. 3 E and F). However, our previous computational studies uncovered parallel pathways involving both pkHP1 and pkHP2 at 1 M NaCl (18). To examine this apparent discrepancy between experiments and simulations and to investigate whether a second pathway emerges at higher salt conditions, we measured the effect of varying the salt concentration on the thermodynamics of PK unfolding. To this end, we monitored the fluorescence melting profiles of VPK-2AP at 500 mM and 1 M NaCl (SI Appendix, Fig. S7) and VPK-pC at 100 mM, 200 mM, 500 mM, and 1 M NaCl (Fig. 6). As controls, we also measured the melting profiles of the isolated hairpins over the same salt range: unlabeled HP1 (SI Appendix, Fig. S8), HP1-2AP (SI Appendix, Fig. S4), unlabeled HP2 (SI Appendix, Fig. S9), and labeled HP2-pC (SI Appendix, Figs. S10–S12). The thermodynamic parameters for the hairpins are summarized in SI Appendix, Tables S2 and S4.
Fig. 6.
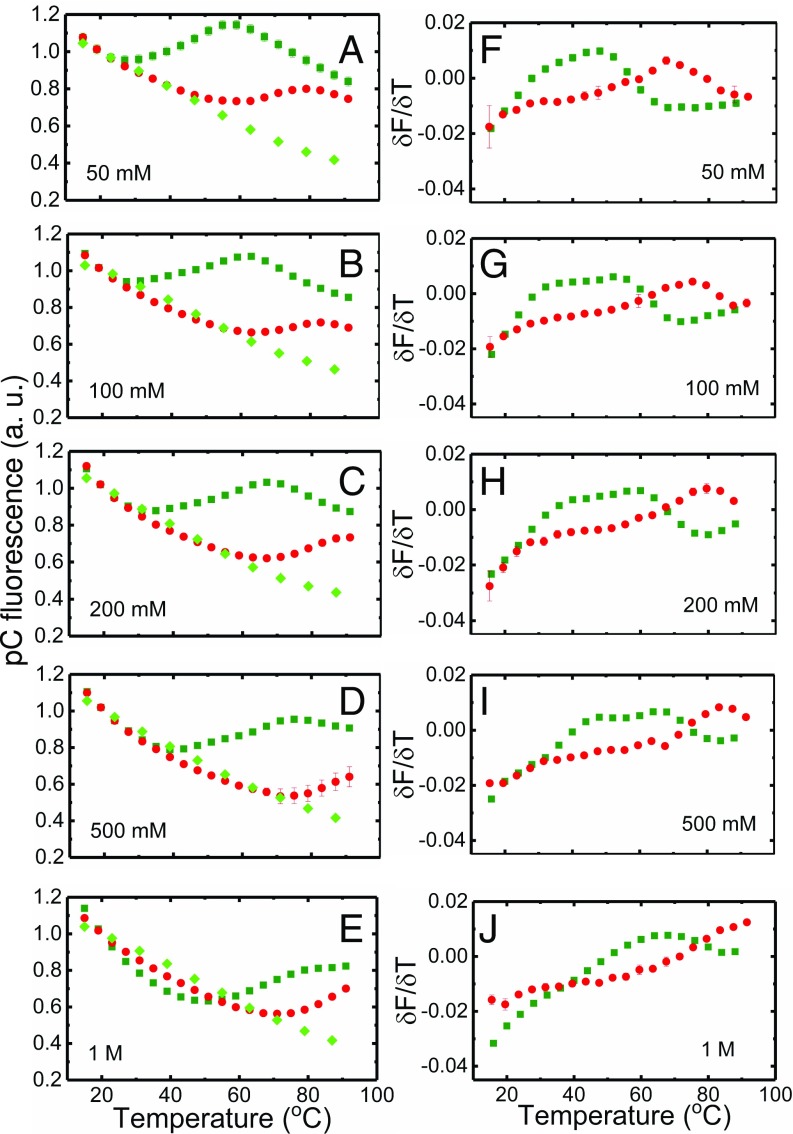
Salt dependence of VPK-pC melting profiles. (A–E) Fluorescence melting profiles of VPK-pC (dark green) and HP2-pC (red) are shown at different monovalent salt concentrations; corresponding measurements on reference sample pC-ref are also shown (light green). All data are normalized to match at 20 °C. (F–J) Corresponding first derivatives δ obtained from the melting profiles are plotted as a function of temperature. The data (■, ●, and ♦) are the averages of two independent sets of measurements; the errors are the SEM; error bars not visible are smaller than the symbols.
We first discuss our results on VPK-2AP. While the 2AP probe proved to be a good sensor of the pathway, it is expected to be less sensitive to the pathway. We therefore anticipated that the emergence of a parallel pathway at the higher salt conditions, via the intermediate pkHP2, should manifest itself as a decrease in the amplitude of the 2AP fluorescence change observed in the two transitions. Indeed, our experiments are consistent with this scenario (SI Appendix, Fig. S7). We further quantified the fraction of molecules that unfolded via the two pathways by fitting the higher salt melting profiles with a four-state parallel pathway model, as described in SI Appendix, SI Methods 1.4. The resulting fits to the VPK-2AP melting profiles reveal the emergence of the second pathway that populates pkHP2 at the higher salt concentrations (SI Appendix, Fig. S13). The parameters from these fits are summarized in SI Appendix, Table S5.
Our thermodynamics data on VPK-pC measured at different NaCl concentrations revealed a similar scenario (Fig. 6). The melting profile of VPK-pC in 100 mM NaCl resembled that in 50 mM KCl, with a predominantly single transition flanked by baselines that reflect the temperature dependence of intrinsic pC fluorescence (as measured by pC-ref, also plotted in Fig. 6). However, at higher salt concentrations, the slope of the high temperature baseline decreased and eventually even changed sign, becoming positive at 1 M NaCl. The change in the high temperature slope, as the salt concentration is increased, is indicative of a structural transition in the RNA involving base unstacking and/or breaking of base pairs as sensed by the pC probe. These observations match the expectation that pC fluorescence will increase upon unfolding of stem 2, pointing to the presence of a parallel unfolding pathway with pkHP2 as intermediate. Indeed, thermodynamic measurements of the HP2-pC hairpin (with pC located in stem 2; Fig. 1D) support these conclusions; these melting profiles are also shown in Fig. 6 for comparison, and the fits to the hairpins in terms of a two-state model are shown in SI Appendix, Fig. S11.
To quantify the populations in the two pathways, we modeled the salt dependence of VPK-pC in terms of a four-state parallel pathway model, as described in SI Appendix, SI Methods 1.4. The results from this analysis, shown in SI Appendix, Fig. S14 (with parameters summarized in SI Appendix, Table S6), again demonstrate the emergence of a second pathway in the folding/unfolding of VPK, similar to what we obtained with VPK-2AP, with an increasing population appearing in the intermediate pkHP2 as the salt concentration is increased.
Interestingly, the derivative data on VPK-pC (Fig. 6, Right) show that at salt concentrations of 100–500 mM NaCl (not so clear at 50 mM or 1 M salt), two peaks are observed in the lower temperature transition that are not well fitted by our model (SI Appendix, Fig. S14). These data indicate additional steps in the unfolding of the PK to the intermediate hairpin states. While the origin of these two peaks is not evident from experiments alone, we cannot rule out premelting of stem 2 or contributions from misfolded conformations that are sensed by the pC probe and are not included in our minimal four-state scheme. The peaks also broaden as the salt is increased, which likely has contributions from a nonlinear baseline for the folded PK state (also evident in the nonlinear baseline of the folded HP2-pC hairpin) and the emergence of a second pathway in the PK unfolding transition with s that are relatively close together (SI Appendix, Table S7).
Simulations Reveal Parallel Pathways Depending on the Salt Concentration.
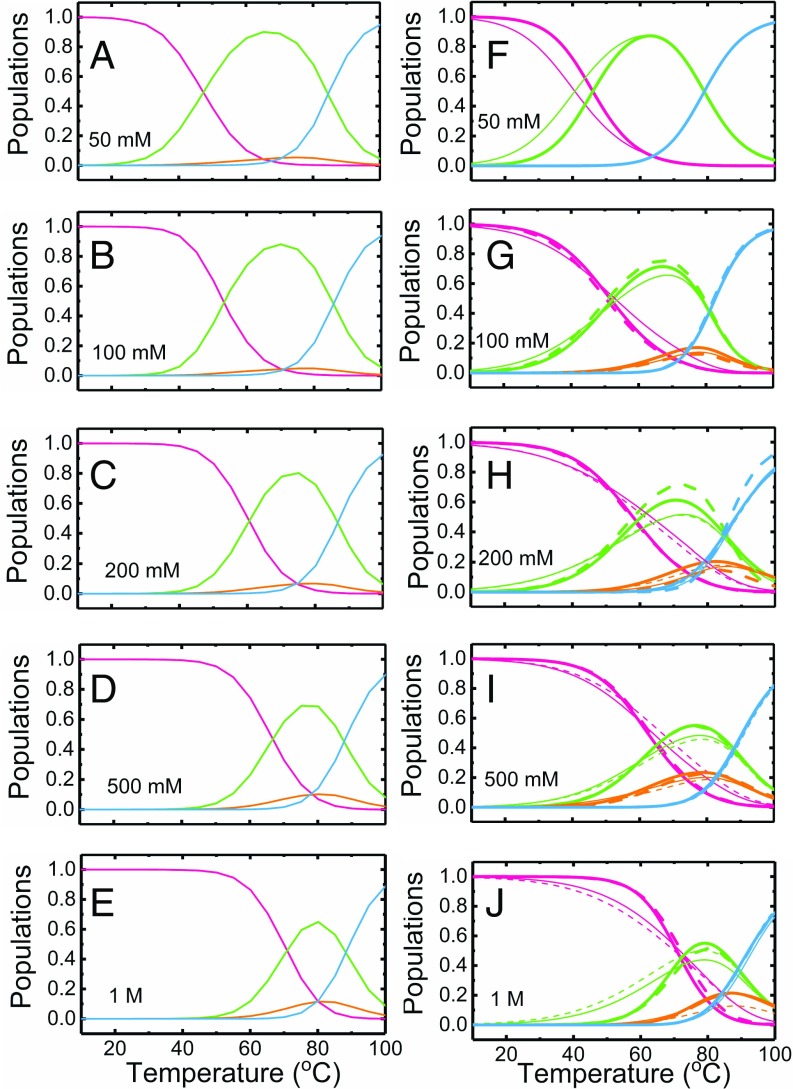
To support the experimental findings and obtain structural insights, we conducted molecular dynamics simulations using a coarse-grained RNA model (57) that is based on the three interaction-site model introduced to simulate nucleic acids (7) (SI Appendix, SI Methods 1.10). It should be stressed that no parameters in the computations were adjusted to reproduce any aspect of the experiments. We note that the simulation results were used to constrain some of the parameters describing the VPK-2AP and VPK-pC melting profiles, as described in SI Appendix, SI Methods 1.4. Before performing kinetics simulations, we did equilibrium simulations at 50 mM, 100 mM, 200 mM, 500 mM, and 1 M salt concentrations, varying the temperature by the replica-exchange method (58). Calculated heat capacities (SI Appendix, Fig. S15) show two distinct transitions at 47 °C and 84 °C (50 mM) and at 70 °C and 88 °C (1 M), respectively. These are in excellent agreement with previous experiments (17, 59), as well as with absorbance melting profiles in this study (Fig. 2). From the equilibrium ensemble, we also obtained temperature-dependent populations of the four states at each salt condition. These population profiles (Fig. 7 A–E) are compared with population profiles obtained from experimental analysis of VPK-pC melting profiles at different salt conditions (Fig. 7 F–J and SI Appendix, Fig. S14 K–O). The results from the simulations closely resemble the results from experiments, with the population of molecules unfolding via pkHP2 increasing as we increase the salt concentration. Furthermore, the s along each pathway (defined as the temperature at which two interconverting states are equally populated) obtained from simulations and experiments are also in remarkably good agreement (Fig. 8 and SI Appendix, Table S7). These heat capacity and population analyses validate the consistency between our experiments and simulations for a range of salt concentrations. The combination of experiments and simulations shows, based on thermodynamic data, that increasing the salt concentration results in folding or unfolding by two pathways.
Fig. 7.
Salt dependence of the thermodynamics of VPK unfolding. The equilibrium populations of the folded and unfolded conformations of VPK, described in terms of a four-state parallel pathway model under different monovalent salt conditions, are plotted as a function of temperature, as obtained from either coarse-grained simulations (A–E) or fits to the VPK-pC fluorescence melting profiles (F–J). Populations are shown for the fully folded VPK (pink), partially folded hairpins pkHP1 and pkHP2 (green and orange, respectively), and fully unfolded U (blue). Four sets of populations are shown: two from fits to the VPK-pC melting profiles (thick lines) and two from fits to the derivatives (thin lines), with either the or the transition temperature constrained from the simulation results (continuous and dashed lines, respectively).
Fig. 8.
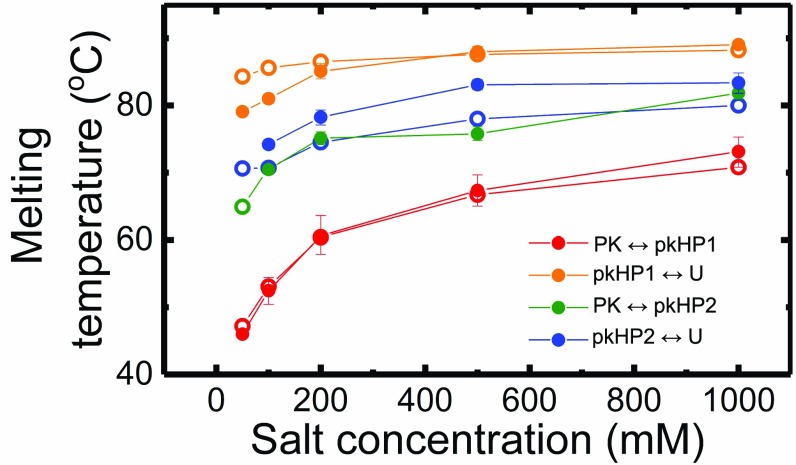
Transition temperatures for the four-state parallel pathway model for VPK PK. The transition (or melting) temperatures for the PK-to-hairpin state and hairpin-to-unfolded state for each of the two pathways are plotted as a function of salt, as obtained from simulations (○) and experiments (●). The experimental values are the average of the s obtained from four sets of fits to the VPK-pC fluorescence melting profiles, as also shown in Fig. 7. The error bars represent the compounded uncertainties in the parameters from all four sets of fits, computed as described in SI Appendix, Table S7. Note that the transition temperature for is undetermined from experiments at 50 mM salt.
Next, we performed a series of kinetics simulations to reveal the details of the folding pathways. We generated more than 200 folding trajectories at various concentrations of monovalent salt. Each simulation was started from an unfolded conformation prepared at 120 °C, and the temperature was quenched to 37 °C so that the folding reactions took place regardless of the salt concentration. After the trajectories reached the folded PK state, we analyzed in each trajectory whether stem 1 folded first (pkHP1 pathway) or stem 2 folded first (pkHP2 pathway). We find that at all salt concentrations examined, the pkHP1 pathway was dominant. However, some fraction of molecules fold via the pkHP2 pathway (Fig. 9). At 50 mM salt, 90% of folding trajectories fold via the pkHP1 pathway. At higher salt concentrations ([M+] > 500 mM), the pathway through pkHP1 becomes less dominant (∼80%). This is consistent with the results from the thermodynamic measurements on VPK-2AP and VPK-pC at higher salt concentrations.
Fig. 9.
Parallel folding pathways of the VPK PK. (A) States of VPK are designated as PK (folded PK), pkHP1 and pkHP2 [partially folded (hairpin) states], and U (unfolded state). In the top pathway, stem 1 folds first to form the pkHP1 hairpin state; in the bottom pathway, stem 2 folds first to form the pkHP2 hairpin state. The structures representing the four states were taken from simulations. (B) Salt dependence of the VPK folding pathways from coarse-grained simulations. The fraction of molecules that fold via the pathway that populates pkHP1 as an intermediate is plotted as a function of the monovalent salt concentrations. The error bars represent the 95% exact confidence intervals. The free-energy difference between the two pathways, calculated based on the fraction of pkHP1, is marked on the right axis.
We note here that while there are differences in the quantitative comparison of experiments and simulations, those differences are not unexpected, given that fluorescent probes in our experimental studies are likely to perturb the thermodynamics and that the choice of baselines in modeling the data also affects the fitted populations, while simulations were done on unlabeled RNA molecules and have no adjustable parameters.
Discussion
We have investigated the thermodynamics and folding/unfolding kinetics of a PK using fluorescent nucleotide analogs and molecular simulations to probe salt-dependent changes in the folding pathways. Altogether, the experiments and simulations provide a consistent description of VPK folding mechanisms and reveal how the ionic conditions modulate the flux through different folding pathways.
Two-Step Assembly of VPK.
At low ionic conditions (50 mM KCl), VPK fold/unfolds predominantly by a single pathway that has two distinct transitions. In the first transition, single-stranded RNA folds to an intermediate conformation, which we identify as hairpin structure pkHP1, in which stem 1 of the PK is formed but stem 2 is not. This intermediate structure takes ∼170–430 μs to fold from a fully unfolded PK at 37 °C (SI Appendix, Table S3). These folding times are consistent with our results on the truncated HP1 sequence (Fig. 1), which forms the same hairpin structure as pkHP1 but without the long dangling end. The folding times for the isolated HP1 were found to be ∼40–280 μs at 37 °C (SI Appendix, Table S3) under identical ionic conditions. The slightly longer times for pkHP1 in comparison to HP1 may be due to the presence of the long dangling end in pkHP1, which could present increased intrachain interactions, and thus slow down the overall folding (60). Nevertheless, the similarity in the folding time of pkHP1 and the truncated HP1 shows that the species sampled at the low KCl concentration is the structure in which the more stable hairpin is formed.
In the second transition, pkHP1 folds to a stable PK, with folding rates faster than the unfolding rates below ∼50 °C. This behavior contrasts with the folding of some riboswitches that exist predominantly in hairpin conformations and only fold to stable H-type PKs upon ligand binding (40), suggesting an important role for stability in the functioning of the VPK PK. At 37 °C, the folding time for the pkHP1-to-PK step is ∼5–13 ms (SI Appendix, Table S3); this time is five- to 10-fold slower than our previously reported value of 1 ms for the unlabeled VPK PK, which was obtained from time-resolved absorbance measurements (17). Comparison of the thermodynamics of unlabeled VPK with VPK-2AP, using absorbance measurements for both, shows that incorporation of the 2AP label significantly increases the stability of the intermediate hairpin structure pkHP1; the for the unfolded-to-hairpin transition is found to be ∼10 °C higher in VPK-2AP than in the unlabeled VPK (SI Appendix, Table S1). This increased stability could arise from stronger stacking interactions between stem 1 and 2AP at position 20 (in VPK-2AP) than the corresponding interactions between stem 1 and adenine (in unlabeled VPK). Therefore, a likely scenario consistent with our thermodynamics and kinetics results is that the transition from pkHP1 to the folded PK conformation requires partial unstacking of the nucleotide at position 20 that prefers to be stacked over stem 1 in the intermediate hairpin structure. This unstacking would then be energetically more costly for the more stable intermediate pkHP1 conformation in VPK-2AP. Hence, the folding of VPK-2AP would be correspondingly slower than the folding of unlabeled VPK.
Our simulations of the folding kinetics at 50 mM monovalent salt also showed predominant folding via a pkHP1 pathway with only 10% folding via pkHP2; this fraction increased to ∼20% at 1 M monovalent salt (Fig. 9B). At 37 °C in 50 mM salt, the simulations predict that the folding from the unfolded state (U) to pkHP1 occurs at ∼70 μs and that the folding from pkHP1 to the folded PK structure occurs at ∼1.5 ms (SI Appendix, Table S8). For the parallel pathway, simulations indicate that the intermediate pkHP2 structure folds in ∼50 μs, followed by folding to the PK in ∼110 μs (SI Appendix, Table S8). Thus, the total folding time through this minor pathway is faster than through the predominant one, suggesting that at the given ionic condition, folding is thermodynamically, and not kinetically, controlled.
For the dominant pathway, both folding times are shorter than the folding times obtained from experiments on VPK-2AP (∼270 μs and ∼8 ms, respectively). It should be emphasized that the predicted folding times are in excellent agreement with the ∼1-ms folding time previously reported for the unlabeled VPK (17). We surmise that the mismatch between our measurements on VPK-2AP and the predictions from the computational studies may be, in part, due to the stabilizing effect of the 2AP probe on the intermediate pkHP1 conformation. The enhanced stability of 2AP-labeled pkHP1 could also favor that pathway over the pkHP2 pathway and account for why experiments implicate predominantly a single folding pathway, while simulations indicate ∼10% folding trajectories via the pkHP2 pathway even at 50 mM salt (Fig. 9). We also cannot rule out the possibility that experiments may potentially miss the small (∼10%) fraction of molecules that fold via the alternative pathway in VPK-2AP at 50 mM KCl.
Fluxes Through the Dominant Pathways Depend on Salt Concentration.
The major finding in our study is that not only does this PK fold by parallel pathways but the flux between the two pathways also changes as the concentration of monovalent cation is altered. Experiments indicate a predominantly single folding pathway to form the PK structure in 50 mM KCl. In contrast, our previous computational studies, carried out at 1 M NaCl, showed that VPK folds through parallel pathways, with a significant fraction (∼23%; Fig. 9A) of molecules folding through the alternative pathway that populates the less stable pkHP2 (18). To assess the role of ionic strength in modulating the folding pathways partition, we performed equilibrium experiments on VPK-2AP and VPK-pC at various salt concentrations; these measurements indeed uncovered evidence for the appearance of an alternative folding/unfolding pathway as the ionic strength was increased from 100 mM to 1 M NaCl (SI Appendix, Figs. S13 and S14). The measurements on VPK-pC, which directly probe the folding/unfolding of stem 2, were particularly important in unveiling the emergence of the parallel pathway involving pkHP2 at high salt. The populations of the intermediate hairpins pkHP1 and pkHP2 obtained from the experiments showed a trend that was consistent between VPK-2AP and VPK-pC, and provide experimental evidence for parallel folding pathways for PKs that are in remarkable agreement with the predictions from simulations (Figs. 7 and 8).
Assembly Order Is Determined by Stability of Hairpins.
Based on simulations of several PKs, Cho et al. (18) showed that assembly mechanism of PKs is largely determined by stabilities of the constituent secondary structural elements, which are pkHP1 and pkHP2 (Fig. 3). Their stabilities could be changed by sequence as well as ionic conditions. With this stability principle in mind, our experimental and computational results allow us to answer the question: What causes the flux through the minor alternative pathway to increase with increasing salt? To gain insight into how the folding trajectories partition into the parallel pathways, we consider the nature of the intermediate hairpin states formed in each pathway. The pkHP1 state has a stem with five G-C base pairs and a loop of nine nucleotides, while pkHP2 has a stem of six base pairs (four G-C, one A-U, and one G-U) and a loop of 14 nucleotides. Based on the composition of the two stems and the corresponding loop lengths, pkHP1 is more stable than pkHP2. The stem of pkHP1 has a larger G-C content, and is therefore more stable than the stem of pkHP2; additionally, from entropic considerations, the smaller loop of pkHP1 is more stable than the loop of pkHP2 (61). Thus, the stability principle (18) would predict, as observed in experiments and simulations, that the dominant pathway ought to be through pkHP1.
What happens to each of these structures as we increase the salt? Previous studies on RNA hairpin stability at different salt conditions showed that the stability of the RNA hairpin stems is not significantly affected as the monovalent ion concentration is increased but the stability of the loops increases (62). Furthermore, thermodynamic measurements on ssDNA and RNA hairpins of varying loop sizes demonstrated that smaller loops are significantly more stable, compared with larger loops, than expected from entropic considerations, especially in the presence of 100 mM monovalent ions (61, 63). Moreover, computational studies on nucleic acid hairpin folding showed that the difference in stability between hairpins with small and large loops is further amplified as the salt concentration decreases (64). In light of these results, we therefore conclude that while pkHP1 is more stable than pkHP2 at all salt conditions, the difference in the stabilities between the two hairpins decreases as we increase the concentration of monovalent ions (the difference in the s of the two hairpins decreases with increasing concentration as shown in Fig. 8 and SI Appendix, Table S7), thereby increasing the probability that some fraction of VPK molecules will fold via the alternative pkHP2 pathway (Fig. 10). Indeed, this conclusion is supported by our measurements and simulations of the stabilities of hairpins HP1 and HP2 as a function of salt concentrations, which reveal that while HP1 is more stable than HP2 at low salt, the difference in their stabilities decreases with salt (SI Appendix, Fig. S16). Thus, altering the ionic strength or changing the sequence of the hairpins, which changes the relative stabilities of the hairpin stems and/or loops, will result in change in fluxes through the pathways. This demonstration in the present combined experimental and computational study is in complete accord with the stability principle for PK assembly enunciated by Cho et al. (18).
Fig. 10.
Modulation of the folding pathways of the VPK PK by monovalent ions. At a low salt concentration, the PK has a dominant folding pathway, with the hairpin pkHP1 as an intermediate state. As the salt concentration increases, the hairpin pkHP2 becomes stable, causing the emergence of a parallel folding pathway.
Parallel Pathways in VPK.
That there are parallel pathways as the salt concentration increases comes from the following arguments that are consistent between experiments and simulations:
-
i)
The difference (ΔT) between the and s, which is an indication of the relative stabilities of the two helices, is ΔT ≈ 20 °C at 50 mM, as predicted by simulations (first row in SI Appendix, Table S7). Thus, the large difference in the stabilities results in only one detectable pathway (), referred to as I, that dominates at 50 mM.
-
ii)
The only way the alternate pathway, II (), can be populated is if mutations or external conditions are altered such that the stability difference between pkHP1 and pkHP2 is reduced. This difference in ΔT decreases as the salt concentration increases (Fig. 8 and SI Appendix, Table S7), implying that the free-energy difference between the pkHP1 and pkHP2 hairpins decreases. As a consequence, pathway II becomes accessible.
-
iii)
If these results from the experiments and simulations provided above hold, then we expect that the of HP1 should be greater than that of HP2. Furthermore, the gap between the two s should decrease as the salt concentration increases. Both of these predictions, which are consequences of the parallel pathway mechanism, are borne out in experiments and simulations on the isolated hairpins (SI Appendix, Fig. S16).
We note here that the calculated values are in near-quantitative agreement with experiments, which is remarkable because no parameter in the simulations was adjusted to obtain agreement with experiments. We conclude that in silico experiments directly show that, indeed, VPK assembles by parallel pathways, with the dominant one being pathway I at all concentrations, and the experiments are consistent with these results. Taken together, we believe that the inescapable conclusion is that VPK folds by parallel pathways, with salt modulating the flux between the two, as stated in the title of this paper.
It is interesting to contrast the unambiguous demonstration that VPK folds by parallel pathways to the difficulties in showing similar behavior in the folding of small single-domain proteins. The flux through a pathway, say II, relative to a competing route I, is . The magnitude of the quantity (GII − GI), related to the stability difference between pkHP1 and pkHP2 in the intact PK, has to be such that can be measured or inferred, as we have done here for the VPK PK (Fig. 9B). In several RNA PKs, the difference between GII and GI can be altered by changing sequence or ionic strength, as we demonstrate here, while in the folding of small single-domain proteins, it has not been straightforward to tune the folding landscape parameters to modulate the flux through alternate pathways.
Conclusions
Our experiments and modeling provide a remarkably self-consistent picture of RNA PK folding mechanisms and also provide insight into the role of monovalent ion concentrations in partitioning into different folding pathways. It would be most instructive to consider PK folding in the presence of divalent ions, which could have a significant effect on the stabilities of the intermediate states and on the folding pathways, through specific ion–RNA interactions. Further studies that synergistically combine experiments and simulations, as in this study, are needed to elucidate the nuances of folding pathways and mechanisms for these and other RNA PKs, which, in turn, will be invaluable for understanding the folding of larger and more complicated ribozymes.
Methods
RNA Samples.
Unlabeled RNA samples were obtained from Dharmacon, Inc. The 2AP- and pC-labeled samples were obtained from TriLink Biotechnologies. Samples were prepared as described in SI Appendix, SI Methods 1.1. All measurements were made in 10 mM 3-(N-morpholino)propanesulfonic acid buffer (pH 7.0), with either 50 mM KCl or a range of NaCl concentrations from 100 mM to 1 M NaCl.
Equilibrium Melting Experiments.
Absorbance and fluorescence melting profiles were measured and analyzed as described in SI Appendix, SI Methods 1.2–1.5.
T-Jump Experiments.
Kinetics measurements were carried out using a home-built laser T-jump apparatus, which uses 10-ns laser pulses at 1,550 nm, generated by Raman-shifting the 1,064-nm pulses from the output of a neodymium-doped yttrium aluminum garnet (Nd:YAG) laser to rapidly heat a small volume of the sample within ∼10 ns (61, 65). Details of the T-jump apparatus and analysis of the relaxation traces are provided in SI Appendix, SI Methods 1.6–1.8.
Global Analysis of Equilibrium and T-Jump Relaxation Traces.
The equilibrium and kinetics data were simultaneously analyzed in terms of a minimal kinetic scheme, using a master-equation approach, as described in SI Appendix, SI Methods 1.9.
Simulation Model and Protocols.
We performed molecular dynamics simulations using a coarse-grained RNA model. The model and parameters were calibrated and described in detail elsewhere (57). Further details are provided in SI Appendix, SI Methods 1.10.
Supplementary Material
Acknowledgments
We thank Jie Liang and Ke Tang for many helpful discussions and Alex Sarmiento for his extensive help in the implementation of the global modeling algorithms. We acknowledge the Texas Advanced Computing Center at The University of Texas at Austin for providing computational resources. This work was supported, in part, by National Science Foundation Grants MCB-1158217 and MCB-1715649 (to A.A.) and Grant CHE 16-36424 (to D.T.). D.T. also acknowledges additional support by the Collie–Welch Regents Chair (F-0019).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717582115/-/DCSupplemental.
References
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thirumalai D, Woodson SA. Kinetics of folding of proteins and RNA. Acc Chem Res. 1996;29:433–439. [Google Scholar]
- 4.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 5.Narayanan R, et al. Exploring the energy landscape of nucleic acid hairpins using laser temperature-jump and microfluidic mixing. J Am Chem Soc. 2012;134:18952–18963. doi: 10.1021/ja301218e. [DOI] [PubMed] [Google Scholar]
- 6.Sorin EJ, Rhee YM, Nakatani BJ, Pande VS. Insights into nucleic acid conformational dynamics from massively parallel stochastic simulations. Biophys J. 2003;85:790–803. doi: 10.1016/S0006-3495(03)74520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nivón LG, Shakhnovich EI. All-atom Monte Carlo simulation of GCAA RNA folding. J Mol Biol. 2004;344:29–45. doi: 10.1016/j.jmb.2004.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Hyeon C, Thirumalai D. Mechanical unfolding of RNA hairpins. Proc Natl Acad Sci USA. 2005;102:6789–6794. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kannan S, Zacharias M. Folding of a DNA hairpin loop structure in explicit solvent using replica-exchange molecular dynamics simulations. Biophys J. 2007;93:3218–3228. doi: 10.1529/biophysj.107.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyeon C, Thirumalai D. Multiple probes are required to explore and control the rugged energy landscape of RNA hairpins. J Am Chem Soc. 2008;130:1538–1539. doi: 10.1021/ja0771641. [DOI] [PubMed] [Google Scholar]
- 11.Bowman GR, et al. Structural insight into RNA hairpin folding intermediates. J Am Chem Soc. 2008;130:9676–9678. doi: 10.1021/ja8032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia AE, Paschek D. Simulation of the pressure and temperature folding/unfolding equilibrium of a small RNA hairpin. J Am Chem Soc. 2008;130:815–817. doi: 10.1021/ja074191i. [DOI] [PubMed] [Google Scholar]
- 13.Huang X, et al. Constructing multi-resolution Markov state models (MSMs) to elucidate RNA hairpin folding mechanisms. Pac Symp Biocomput. 2010;15:228–239. doi: 10.1142/9789814295291_0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng NJ, Cieplak P. Free energy profile of RNA hairpins: A molecular dynamics simulation study. Biophys J. 2010;98:627–636. doi: 10.1016/j.bpj.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 16.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. Productive folding to the native state by a group II intron ribozyme. J Mol Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 17.Narayanan R, Velmurugu Y, Kuznetsov SV, Ansari A. Fast folding of RNA pseudoknots initiated by laser temperature-jump. J Am Chem Soc. 2011;133:18767–18774. doi: 10.1021/ja205737v. [DOI] [PubMed] [Google Scholar]
- 18.Cho SS, Pincus DL, Thirumalai D. Assembly mechanisms of RNA pseudoknots are determined by the stabilities of constituent secondary structures. Proc Natl Acad Sci USA. 2009;106:17349–17354. doi: 10.1073/pnas.0906625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brierley I, Pennell S, Gilbert RJC. Viral RNA pseudoknots: Versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brierley I, Gilbert RJC, Pennell S. RNA pseudoknots and the regulation of protein synthesis. Biochem Soc Trans. 2008;36:684–689. doi: 10.1042/BST0360684. [DOI] [PubMed] [Google Scholar]
- 22.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peselis A, Serganov A. Structure and function of pseudoknots involved in gene expression control. Wiley Interdiscip Rev RNA. 2014;5:803–822. doi: 10.1002/wrna.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dam E, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- 25.Gilley D, Blackburn EH. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Wen J-D, Tinoco I., Jr Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–2188. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen TM, Reihani SNS, Oddershede LB, Sørensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J Mol Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White KH, Orzechowski M, Fourmy D, Visscher K. Mechanical unfolding of the beet western yellow virus -1 frameshift signal. J Am Chem Soc. 2011;133:9775–9782. doi: 10.1021/ja111281f. [DOI] [PubMed] [Google Scholar]
- 30.Ritchie DB, Foster DAN, Woodside MT. Programmed -1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc Natl Acad Sci USA. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Messieres M, et al. Single-molecule measurements of the CCR5 mRNA unfolding pathways. Biophys J. 2014;106:244–252. doi: 10.1016/j.bpj.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y-J, Wu C-H, Yeh AY-C, Wen J-D. Folding a stable RNA pseudoknot through rearrangement of two hairpin structures. Nucleic Acids Res. 2014;42:4505–4515. doi: 10.1093/nar/gkt1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Mimicking ribosomal unfolding of RNA pseudoknot in a protein channel. J Am Chem Soc. 2015;137:15742–15752. doi: 10.1021/jacs.5b07910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong Z, et al. Mechanical unfolding kinetics of the SRV-1 gag-pro mRNA pseudoknot: Possible implications for -1 ribosomal frameshifting stimulation. Sci Rep. 2016;6:39549. doi: 10.1038/srep39549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansari A, Kuznetsov SV, Shen Y. Configurational diffusion down a folding funnel describes the dynamics of DNA hairpins. Proc Natl Acad Sci USA. 2001;98:7771–7776. doi: 10.1073/pnas.131477798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 37.Cocco S, Marko JF, Monasson R. Slow nucleic acid unzipping kinetics from sequence-defined barriers. Eur Phys J E Soft Matter. 2003;10:153–161. doi: 10.1140/epje/e2003-00019-8. [DOI] [PubMed] [Google Scholar]
- 38.Hengesbach M, Kim N-K, Feigon J, Stone MD. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew Chem Int Ed Engl. 2012;51:5876–5879. doi: 10.1002/anie.201200526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmstrom ED, Nesbitt DJ. Single-molecule fluorescence resonance energy transfer studies of the human telomerase RNA pseudoknot: Temperature-/urea-dependent folding kinetics and thermodynamics. J Phys Chem B. 2014;118:3853–3863. doi: 10.1021/jp501893c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen B, LeBlanc R, Dayie TK. SAM-II riboswitch samples at least two conformations in solution in the absence of ligand: Implications for recognition. Angew Chem Int Ed Engl. 2016;55:2724–2727. doi: 10.1002/anie.201509997. [DOI] [PubMed] [Google Scholar]
- 41.Chen S-J. RNA folding: Conformational statistics, folding kinetics, and ion electrostatics. Annu Rev Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan ZJ, Chen S-J. Salt contribution to RNA tertiary structure folding stability. Biophys J. 2011;101:176–187. doi: 10.1016/j.bpj.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hori N, Denesyuk NA, Thirumalai D. Salt effects on the thermodynamics of a frameshifting RNA pseudoknot under tension. J Mol Biol. 2016;428:2847–2859. doi: 10.1016/j.jmb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biyun S, Cho SS, Thirumalai D. Folding of human telomerase RNA pseudoknot using ion-jump and temperature-quench simulations. J Am Chem Soc. 2011;133:20634–20643. doi: 10.1021/ja2092823. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Zhang J, Wang W. Atomistic analysis of pseudoknotted RNA unfolding. J Am Chem Soc. 2011;133:6882–6885. doi: 10.1021/ja1109425. [DOI] [PubMed] [Google Scholar]
- 46.Bian Y, Zhang J, Wang J, Wang J, Wang W. Free energy landscape and multiple folding pathways of an H-type RNA pseudoknot. PLoS One. 2015;10:e0129089. doi: 10.1371/journal.pone.0129089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Q Nguyen KK, et al. Ensemble simulations: Folding, unfolding and misfolding of a high-efficiency frameshifting RNA pseudoknot. Nucleic Acids Res. 2017;45:4893–4904. doi: 10.1093/nar/gkx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 50.Sarkar K, Meister K, Sethi A, Gruebele M. Fast folding of an RNA tetraloop on a rugged energy landscape detected by a stacking-sensitive probe. Biophys J. 2009;97:1418–1427. doi: 10.1016/j.bpj.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarkar K, Nguyen DA, Gruebele M. Loop and stem dynamics during RNA hairpin folding and unfolding. RNA. 2010;16:2427–2434. doi: 10.1261/rna.2253310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, et al. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: Nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen LX, Tinoco I., Jr The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez RL, Jr, Tinoco I., Jr Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J Mol Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- 55.Theimer CA, Giedroc DP. Contribution of the intercalated adenosine at the helical junction to the stability of the gag-pro frameshifting pseudoknot from mouse mammary tumor virus. RNA. 2000;6:409–421. doi: 10.1017/s1355838200992057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denesyuk NA, Thirumalai D. Coarse-grained model for predicting RNA folding thermodynamics. J Phys Chem B. 2013;117:4901–4911. doi: 10.1021/jp401087x. [DOI] [PubMed] [Google Scholar]
- 58.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 59.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J Mol Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuznetsov SV, Ansari A. A kinetic zipper model with intrachain interactions applied to nucleic acid hairpin folding kinetics. Biophys J. 2012;102:101–111. doi: 10.1016/j.bpj.2011.11.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuznetsov SV, Ren C-C, Woodson SA, Ansari A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008;36:1098–1112. doi: 10.1093/nar/gkm1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vieregg J, Cheng W, Bustamante C, Tinoco I., Jr Measurement of the effect of monovalent cations on RNA hairpin stability. J Am Chem Soc. 2007;129:14966–14973. doi: 10.1021/ja074809o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuznetsov SV, Shen Y, Benight AS, Ansari A. A semiflexible polymer model applied to loop formation in DNA hairpins. Biophys J. 2001;81:2864–2875. doi: 10.1016/S0006-3495(01)75927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan Z-J, Chen S-J. Salt dependence of nucleic acid hairpin stability. Biophys J. 2008;95:738–752. doi: 10.1529/biophysj.108.131524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.