Abstract
Metformin is currently considered as a promising anticancer agent in addition to its anti-diabetic effect. To better individualize metformin therapy and explore novel molecular mechanisms in cancer treatment, we conducted a pharmacogenomic study using 266 lymphoblastoid cell lines (LCLs). Metformin cytotoxicity assay was performed using the MTS assay. Genome-wide association (GWA) analyses were performed in LCLs using 1.3 million SNPs, 485k DNA methylation probes, 54k mRNA expression probe sets, and metformin cytotoxicity (IC50s). Top candidate genes were functionally validated using siRNA screening, followed by MTS assay in breast cancer cell lines. Further study of one top candidate, STUB1, was performed to elucidate the mechanisms by which STUB1 might contribute to metformin action. GWA analyses in LCLs identified 198 mRNA expression probe sets, 12 SNP loci, and 5 DNA methylation loci associated with metformin IC50 with P-values <10−4 or <10−5. Integrated SNP/methylation loci-expression-IC50 analyses found 3 SNP loci or 5 DNA methylation loci associated with metformin IC50 through trans-regulation of expression of 11 or 26 genes with P-value <10−4. Functional validation of top 61 candidate genes in 4 IPA networks indicated down regulation of 14 genes significantly altered metformin sensitivity in two breast cancer cell lines. Mechanistic studies revealed that the E3 ubiquitin ligase, STUB1, could influence metformin response by facilitating proteasome-mediated degradation of cyclin A. GWAS using a genomic data-enriched LCL model system, together with functional and mechanistic studies using cancer cell lines, help us to identify novel genetic and epigenetic biomarkers involved in metformin anticancer response.
Introduction
Metformin is a widely used antidiabetic drug with very low incidence of side effect. In the last decade, many in vitro, in vivo, and retrospective epidemiological studies have suggested metformin could be a highly promising chemopreventive and chemotherapeutic agent for many types of cancer, particularly in cancer patients with type 2 diabetes (T2D) (1–5). It might be related to the risk factors shared between T2D and cancer (6). Therefore, there are many ongoing clinical trials to assess metformin’s anticancer effect at antidiabetic therapeutic doses (4). There are two well-known mechanisms related to the anticancer action of metformin: one is the indirect inhibition of the PI3K-mTOR signalling pathway through the reduction of insulin level; the other is the direct inhibition of the mitochondrial respiratory complex I, which could cause energetic stress via LKB1/AMPK dependent and/or independent pathway (7,8). In addition, metformin could also cause cell cycle arrest and promote apoptosis (9,10). However, the therapeutic effect of metformin in cancer remains unclear (11,12) and there are no pharmacogenomic biomarkers for selecting responsive patients.
Most pharmacogenomic (PGx) studies of metformin were performed in T2D patients with glycemic response (HbA1c level) as a response endpoint. Considerable inter-individual variability exists with glycemic response, and one-third of T2D patients fail metformin treatment (13,14). A genome-wide complex trait analysis (GCTA) estimated that the heritability for glycemic response to metformin was 20–34%, which might involve many individual variants with minor effects (14). Further genome-wide association (GWA) analysis identified one SNP, rs11212617, located in the ATM gene that was associated with glycemic response (15). This association has been replicated in several additional European populations (16). The variable glycemic response might be partially due to the considerable variation in plasma metformin drug level. One study reported that the mean metformin concentrations range from 0.4 to 1.3 mg/l after 1000 mg twice daily (13). Many pharmacogenetic studies on metformin pharmacokinetic (PK) pathway genes provide substantial evidence that genetic variations in organic cation transporters (OCT1-3) encoded by SLC22 family genes (SLC22A1-3) and multidrug and toxin extrusion proteins (MATE1-2) encoded by SLC47 family genes (SLC47A1-2) are associated with metformin anti-diabetic response (17–25). Recently, Goswami et al reported that genetic polymorphisms in transcription factors (such as SP1) that regulated expression of these transporters were also associated with metformin PK and HbA1c change (13). Moreover, genetic variations in pharmacodynamic (PD) pathway genes have also been found to be associated with metformin response such as LKB1 and AMPK (26). However, metformin is not FDA approved as an anticancer agent. As a result, there are very few clinical studies available to investigate the pharmacogenomics of metformin as an anticancer agent. One study of 44 patients with castration-resistant prostate cancer (CRPC) identified that the SNP rs622342 in OCT1was associated with reduced metformin-related toxicity and an increased risk of tumour progression (27). In addition to germline genetic variation, somatic mutations in cancer cells have also been reported to be related to metformin sensitivity such as genetic mutations in mitochondrial respiratory complex I and LKB1/AMPK deficiency caused by genetic mutations in these genes (28,29).
Here we took advantage of an in vitro EBV-transformed lymphoblastoid cell line model (LCL), ‘Human Variation Panel’, to investigate metformin anticancer response by exploring additional new molecular mechanisms and identifying novel germline genetic and epigenetic biomarkers involved in metformin response. The LCL system contains 287 cell lines with enriched multi-omics data including 1.3 million genotyped and 5.4 million imputed SNPs, 54K mRNA expression probe sets, and 485,577 DNA methylation probes. We have previously used this system for many other PGx biomarker discovery studies as well as for functional studies of clinical GWAS signals (30–32). In this study, we first performed metformin cytotoxicity assay and calculated IC50 as a metformin response phenotype. GWA analyses were then performed using SNP vs IC50, expression vs IC50, and methylation vs IC50, respectively. Since SNP and methylation might influence metformin cytotoxicity through the regulation of gene expression, integrated SNP-expression-IC50 and methylation-expression-IC50 analyses were also performed. After ingenuity pathway analysis (IPA) of the top 65 candidate genes identified through the integrated analysis, 61 genes in 4 IPA networks were selected for functional validation in breast cancer cell lines using siRNA knockdown followed by MTS assay. We found knockdown of 25 genes altered metformin sensitivity in MDA-MB 231 and 14 were replicated in Hs578T cells. Furthermore, one of the top candidates, STUB1, an E3 ubiquitin ligase that is involved in the proteasome dependent degradation of many proteins (33), was found to influence metformin sensitivity through its function as an E3 ubiquitin ligase for cyclin A.
Results
Genome-wide association studies (GWAS) using LCL
Metformin cytotoxicity
Metformin cytotoxicity studies were performed to determine the range of variation in metformin response among all 266 LCLs. IC50 was calculated to represent metformin cytotoxicity. The range of metformin IC50s among all 266 LCLs was 0.440–46.289 mmol/l, and the median value was 2.319 mmol/l. There was no significant difference in metformin IC50s between genders (P-value = 0.566) or among races (P-value = 0.484) (Supplementary Material, Fig. S1).
GWA analysis with expression vs. IC50 values
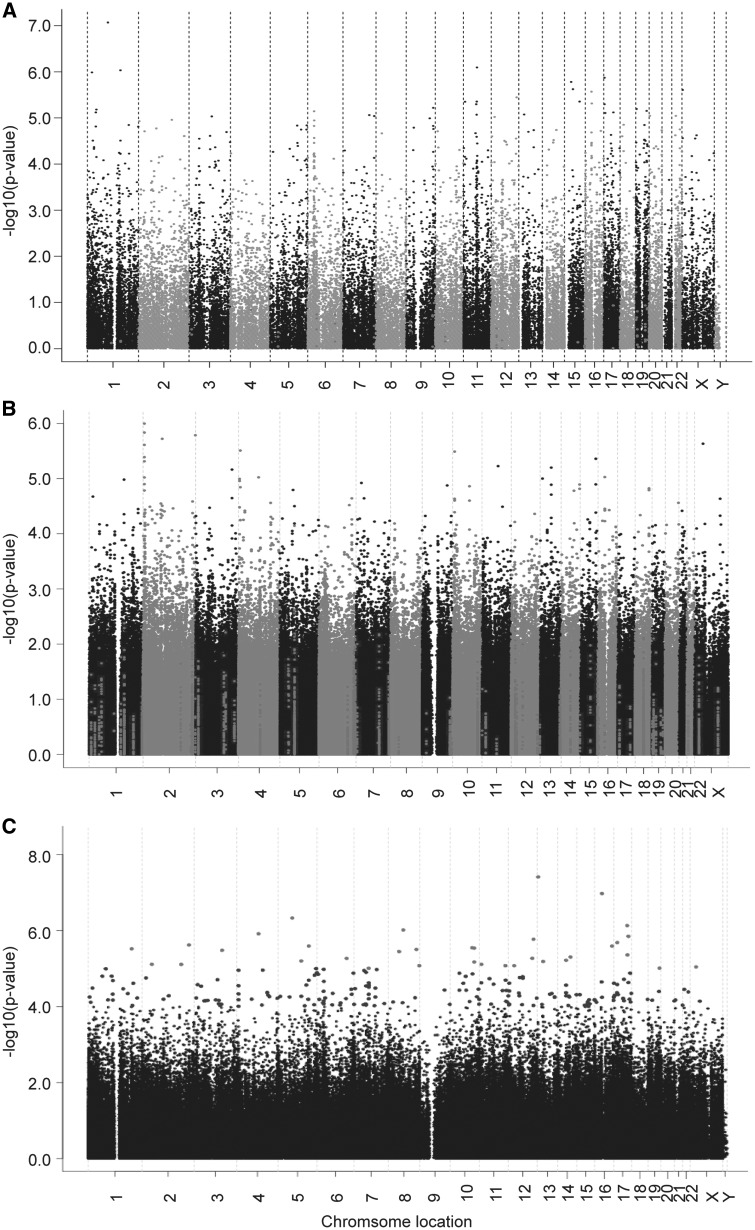
To identify genes whose expression levels might be associated with metformin response, correlation analysis was performed using 54K gene expression probe sets and metformin IC50 values. The association analysis identified that 198 probe sets representing 171 unique annotated genes were correlated with metformin IC50 with P value < 10−4 and q value < 0.02. The top two probe sets representing genes RPL5 and FIBP remained significant after Bonferroni correction (Supplementary Material, Table S1, Fig. 1A). With regard to all known membrane transporters involved in metformin pharmacokinetics (7,34), the expression levels were low in the LCLs (Supplementary Material, Table S2). Further analysis of all possible SLC and ABC transporters showed that the expression of SLC38A10 and SLC25A36 were associated with metformin IC50 with P value < 10−4 (Supplementary Material, Table S1).
Figure 1.
Metformin genome-wide association analyses with (A) mRNA expression/(B) SNP/(C) DNA methylation in the LCLs. The y-axis represents the –log10 (P-value) for the association of individual expression array probe set/SNP/DNA methylation probe. All of the data are plotted on the x-axis based on the chromosomal locations of their genes, SNPs or DNA methylation probes.
GWA analysis with SNP vs. IC50 values
To identify SNPs associated with metformin response, GWA analysis was performed using metformin IC50 values and 1.3 million SNPs. As shown in Figure 1B and Supplementary Material, Table S3, 18 SNPs were found to be associated with metformin IC50 with P-value < 10−5 and 1772 SNPs with P-value < 10−3. The SNP with the lowest P-value of 1.01 × 10−6 (r = -0.300, MAF = 0.209) was the rs7368844 SNP located on chromosome 2. Another five adjacent SNPs were also found to be associated with metformin IC50 with P-value < 10−5 (Fig. 1B, Supplementary Material, Table S6). Linkage disequilibrium (LD) analysis of these 6 SNPs in each of the 3 ethnic groups showed that all the SNPs with MAF >0.05 were in strong LD in CA and HCA (D’ ≥ 0.962, r2 ≥ 0.737), while in a slightly less significant LD in AA (D’ ≥ 0.835, r2 ≥ 0.230) (Supplementary Material, Table S4). Further imputation analysis was performed in the region of 200 kb up-/downstream of the most significant SNP, rs7368844 using 1000 genome as a reference. Two adjacent imputed SNPs (rs2950163 and rs7559267) were more significant than the genotyped SNP rs7368844 (P-value = 2.49 × 10−7 and 9.38 × 10−7, respectively, (Supplementary Material, Table S5). According to the UCSC database, there were two long noncoding RNAs (LINC01105, LOC400940) and one miRNA (MIR7158) located in this SNP locus region on chromosome 2 (200 kb up-/downstream of the most significant SNP, rs7368844). The expression of these two non-coding RNAs in our LCLs was not available. We looked at the GTEX and found that LINC01105 and MIR7158 were not expressed in EBV-transformed lymphocyte and no information was available for LOC400940. Therefore, we could not determine the cis-eQTL relationships for these SNPs. Moreover, the trans-eQTL analysis was conducted using all 54k mRNA expression probe sets, and none of the expression probe sets was found to be associated with the most significant 6 observed SNPs on chromosome 2 with P-value < 10−4 (Supplementary Material, Table S7).
Integrated analysis with SNP, mRNA and IC50 values
Since SNPs might affect metformin response through the influence on gene expression level, similarly like a previous study (31), we performed integrated SNP Locus-mRNA-IC50 analysis. We focused on 83 SNPs from 12 SNP loci and 198 top mRNA expression probe sets that were associated with metformin IC50 with P-value <10−3 or <10−4, respectively (Supplementary Material, Table S1, Supplementary Material, Table S6, Supplementary Material, Fig. S2A). Each SNP locus was defined as a 50 kb region containing at least 1 SNP with P-value < 10−5 and 1 SNP with P-value < 10−3. Seven SNPs located in three SNP loci were found to be associated with the mRNA expression of 11 genes with P-value < 10−4, and these 11 gene expression levels were also associated with metformin IC50 with P-value < 10−4 (Supplementary Material, Table S7). All 7 SNPs were trans-correlated with the expression of 11 genes. Five SNPs in strong linkage disequilibrium (LD) with each other (r2 > 0.9) located on chromosome 16 were tran-associated with 6 out of these 11 gene expression (P-value < 10−4), including genes, THAP7, BAG1, STARD3, and SH3BGRL3, etc (Supplementary Material, Table S7).
Imputation analysis of 200 kb up/downstream of the SNP (rs4889609) that were most significantly associated with metformin IC50 values in this chromosome 16 SNP locus identified additional SNPs. There were 70 SNPs including genotyped and imputed SNPs associated with metformin IC50 with P-value < 10−2 in this region. None of the imputed SNPs was more significantly associated with metformin IC50 values than the observed SNP, rs4889609 (Supplementary Material, Table S8). SNP rs4889609 was located at approximately 18 kb upstream of STX4 gene, and ENCODE data suggested that it might lie in an enhancer element. The association analysis in LCLs showed a trend of association of this rs4889609 SNP with the expression of STX4 gene (P-value = 0.107), which was also positively correlated with the expression of STARD3 and SH3BGRL3 genes (P = 2.95 ×10−6 and 2.04 × 10−3), respectively. The other three SNPs (rs35468353, rs11862744, rs17839567) in this chromosome 16 SNP locus, in strong LD with the SNP rs4889609, were found to be trans-associated with the expression of STARD3 and SH3BGRL3 genes with P-value < 10−4, respectively (Supplementary Material, Table S7). These three SNPs were located at approximately 5–6.5 kb downstream of STX4 gene, and also showed a trend of association with the expression of STX4 gene (P-value < 0.10). The expression of STARD3 and SH3BGRL3 genes were negatively correlated with metformin IC50 with P-value < 10−4, respectively (Supplementary Material, Table S7).
Integrated analysis with DNA methylation, mRNA and IC50 values
DNA methylation array data, including 485,577 methylation probes, were obtained for all the LCLs by the Infinium HumanMethylation450 BeadChip. Quality control (QC) was performed for all of these probes prior to conducting statistical analysis. Specifically, we removed 39,790 probes either located at/near SNP or not annotated as ‘cg’ probes, and 990 probes with a call rate < 0.98. Therefore, 444,797 probes were used in the genome-wide association analysis and 210 DNA methylation probes were found to be associated with metformin IC50 with P value < 10−4 (Fig. 1C, Supplementary Material, Table S9). The top two probes, cg11072570 and cg05390473, were associated with metformin IC50 with P-value of 4.00 × 10−8 and 1.10 × 10−7 respectively, and remained significant after Bonferroni correction (Fig. 1C, Supplementary Material, Table S9).
Since SNP might affect DNA methylation status and in turn, gene expression level, we looked at the top 210 DNA methylation probes (P-value <10−4) and the top 1772 SNPs (P-value <10−3), that were associated with metformin IC50 respectively, and we found that none of the SNPs were located in the region of the DNA methylation probes (Supplementary Material, Table S3 and S9). This was likely due to the removal of 36659 methylation probes located at/near 38688 SNPs during our QC process for methylation probes. To make sure that we did not miss any SNPs that might be functional, we specifically examined the top 1772 SNPs and found 3 SNPs (rs11126017, rs12763379, and rs7934876) were in the region of DNA methylation probes were associated with metformin IC50 with P-value <10−3. In addition, we identified additional 2266 SNPs that were strongly correlated with these top 1772 SNPs with r2 > 0.9. Among these 2266 SNPs, two SNPs (rs2274245 and rs4809373) were also in the region of DNA methylation probes. Since we removed any methylation probes harbouring or near an SNP due to previous finding suggesting that the detected DNA methylation level could be affected by SNPs in the region of methylation probes (35), the cis SNP-methylation relationship for these five SNPs could not be determined . We further conducted the integrated correlation analysis of these five SNPs with all 54k mRNA expression probe sets and metformin IC50 in LCLs. SNP rs12763379 in intron 3 of the PYROXD2 gene was trans-correlated with expression of GIMAP2 gene with P-value of 1.96 × 10−5, which was also associated with metformin IC50 with P-value of 3.22 × 10−3. SNP rs12763379 was also found to be positively cis-correlated with expression of PYROXD2 gene in multiple tissues (including breast tissue) from GTEX data, although no significant correlation was found in our LCLs and GTEX EBV-transformed lymphocyte, which might be related to the low expression of PYROXD2 gene in these cells.
Next we focused on the integrated methylation-mRNA-IC50 analysis (Supplementary Material, Fig S2B). Like the SNP analysis, we focused on the DNA methylation loci (Supplementary Material, Table S10). Each DNA methylation locus was defined as a region (based on Illumina annotation) containing at least two methylation probes located in the region containing a known gene that was associated with metformin IC50 with P value < 10−4, the gene was expressed in the LCLs, and the DNA methylation probes were cis-correlated with the gene expression with P-value < 10−4. Based on the criteria, we identified 5 methylation loci, in which 5 genes were located nearby. In addition to the cis correlation, we also wanted to determine the tans-correlation for these 5 methylation loci. We then correlated the 15 methylation probes in these 5 DNA methylation loci with the top 198 mRNA expression probe sets that were associated with metformin IC50 (P-value < 10−4) and identified 11 methylation probes in these 5 DNA methylation loci were trans-correlated with mRNA expression of 26 genes (Supplementary Material, Table S11, Supplementary Material, Fig. S2B). The expression of these 26 genes was also correlated with metformin IC50 as well as the expression of those 5 genes within the methylation loci with P-value < 10−4, respectively (Supplementary Material, Table S11). The mRNA expression for 3 of these 5 genes (DUSP5, HOXB2, and MAP2K6) were found to be associated with metformin IC50 with moderate P-value <10−2. Finally, methylation-mRNA-IC50 analysis of all top 210 DNA methylation probes and 54K mRNA expression probe sets identified another gene, CDC42BPA, which contained only one methylation probe (cg03890680) that was cis-associated with its own gene expression and metformin IC50 with P-value < 10−4. The expression of CDC42BPA gene was also correlated with metformin IC50 with moderate P-value < 10−2.
Functional validation of candidate genes in breast cancer cell lines
Since nongenetic factors (such as EBV transformation) might confound the results of these association studies and gene regulation is tissue specific (31), we performed the functional validation study using siRNA knockdown, followed by MTS assay for selected candidates in breast cancer cell lines to confirm the top association results from the LCLs.
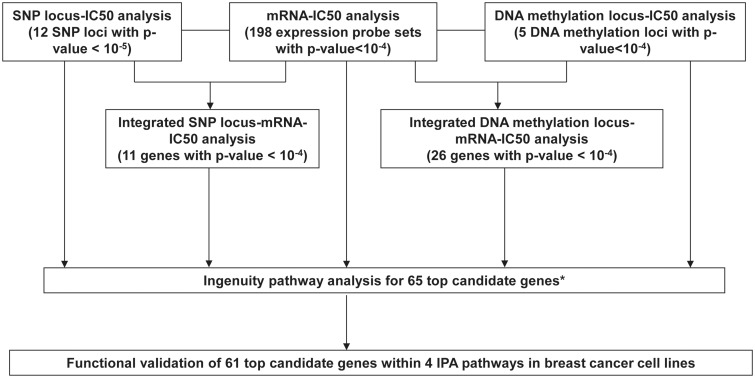
We found that only two mRNA expression probe sets and two DNA methylation probes were associated with metformin IC50 with Bonferroni corrected P-value <0.05 through the GWAS using these 266 LCLs. Therefore, for the purpose of functional validation, we used arbitrary and less stringent P-value cut-offs to select top candidate genes based on the strategy we defined (Fig. 2). We have applied a similar strategy in many of our previous works using the LCLs (30,31,36). The selection criteria were gene expression associated with metformin IC50 with at least one probe set with P-value <10−5 or one probe set with P-value < 10−4 and one probe set with P-value <10−3; genes located in SNP loci or DNA methylation loci; integrated SNP-mRNA-IC50 analysis or integrated Methylation-mRNA-IC50 analysis with P value <10−4; as well as one gene (CDC42BPA) from methylation analysis. After checking the specificity, expression level, and variation in expression level of these probe sets in all the LCLs, 65 top candidate genes were selected for Ingenuity Pathway analysis (IPA). IPA analysis identified four IPA networks including 61 out of the 65 top genes. The four networks included cellular development, cellular assembly and organization, cell death and survival, and cellular function and maintenance (Supplementary Material, Fig. S3).
Figure 2.
Schematic diagram of the strategy used to select genes for functional validation. Genome-wide association studies were performed using metformin IC50 with 1.3 million SNPs, 54K expression probe sets or 485K DNA methylation probes. Integrated SNP/Methylation-Expression-IC50 analyses were performed using SNP loci that contained at least 1 SNP with P-value < 10−5 and 1 SNP with P-value < 10−3, or DNA methylation loci that located in the expressed gene region and contained at least two probes associated with metformin IC50 with P-value <10−4, 54K expression probe sets and metformin IC50 to identify SNP loci/methylation loci associated with metformin IC50 through their influence on gene expression. Ingenuity pathway analysis (IPA) was performed for the top 65 candidate genes. Finally 61 genes within 4 IPA networks were selected for functional validation studies using siRNA screening approach in breast cancer cell lines. * One gene, CDC42BPA, was also included in the top 65 candidates, which contained one methylation probe associated with its own gene expression and metformin IC50 with P-value < 10−4, and which gene expression was associated with metformin IC50 with P-value < 10−2.
We then functionally validated 61 candidate genes within these 4 IPA networks using two breast cancer cell lines, MDA-MB 231 and Hs578T cells (Supplementary Materials, Figs S4 and S5, Table 1). First, siRNA screening followed by MTS assay was performed for all the 61 candidate genes in one breast cancer cell line, MDA-MB 231. We found that knockdown of 25 genes significantly desensitized breast cancer cells to metformin treatment. These 25 genes validated in MDA-MB231 cell included 13 genes from the expression-IC50 association analysis, three genes from the SNP locus-IC50 association analysis, five genes from the integrated SNP locus-mRNA-IC50 analysis and nine genes from the integrated methylation locus-mRNA-IC50 analysis. The reduced sensitivity after knockdown experiments for 22 genes identified from the expression-IC50 or integrated SNP locus/methylation locus-mRNA-IC50 analyses was consistent with the association direction in LCLs. Although it was difficult to validate the direction of the three genes (MID1IP1, POMP, STX4) from the SNP locus-IC50 analysis, the reduced metformin sensitivity after knocking down STX4 gene was consistent with a positive correlation between the expression of STX4 gene and the expression of STARD3 and SH3BGRL3 genes. We further computed the percentage of variation in metformin IC50 that might be explained by all the genomic features associated with these 25 genes. We found that the variation of the 29 expression probe sets, 13 SNPs and 11 DNA methylation probes related to these 25 genes could explain 25% of variation in metformin IC50 values. In addition, we also took the individual profile including top IC50-associated 12 SNP loci, 198 expression probe sets or five methylation loci to determine their contribution to the observed variation. We found that a higher percentage of variation (43%) in the metformin IC50 of LCLs was explained by the 83 SNPs from all 12 SNP loci, which might partially due to the fact that SNPs might influence functions through multiple regulations including expression, epigenomics, protein function and protein–protein interactions. Fourteen out of those 25 genes were confirmed to desensitize cells to metformin after knockdown using another breast cancer cell line, Hs578T (Supplementary Material, Fig. S5).
Table 1.
A total of 61 candidate genes selected for siRNA screening followed by MTS assay
| Basis for selection |
MTS assay |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Gene symbol | IPA Network | Exp-IC50 analysis | SNP-IC50 analysis | Methy-IC50 analysis | SNP-Exp-IC50 analysis | Methy-Exp-IC50 analysis | MDA-MB 231 | Hs578T |
| 1 | ADD1 | 4 | <10−5, <10−27, <10−5, <10−13, <10−4 | ||||||
| 2 | AES | 3 | <10−5 | ||||||
| 3 | AGPAT1 | 3 | <10−4 | resistant | |||||
| 4 | ATP2A3 | 4 | <10−5 | resistant | |||||
| 5 | AUTS2 | 3 | <10−5 | ||||||
| 6 | BAG1 | 1 | <10−5, <10−4, <10−4 | ||||||
| 7 | BAG6 (BAT3) | 1 | <10−4 | ||||||
| 8 | VPS51 (C11orf2) | 2 | <10−5 | resistant | resistant | ||||
| 9 | C14orf105 | 3 | <10−5 | ||||||
| 10 | C16orf58 | 2 | <10−5 | ||||||
| 11 | CCDC88B | 2 | <10−5 | ||||||
| 12 | CD99 | 4 | <10−5 | ||||||
| 13 | CDC25B | 1 | <10−5 | <10−5, <10−27, <10−5, <10−4, <10−5 | resistant | resistant | |||
| 14 | CDC42BPA* | 4 | <10−14, <10−5, <10−2 | resistant | |||||
| 15 | CDK16 (PCTK1) | 1 | <10−4 | <10−5, <10−4, <10−4 | |||||
| 16 | CDK8 | 1 | <10−5 | ||||||
| 17 | CIZ1 | 2 | <10−5 | ||||||
| 18 | CLTB | 1 | <10−4 | ||||||
| 19 | CNNM3 | 4 | <10−5, <10−27, <10−9, <10−18, <10−4 | resistant | resistant | ||||
| 20 | CYB5R2 | 3 | <10−5 | <10−5, <10−27, <10−9, <10−19, <10−5 | |||||
| 21 | CYTH2 (PSCD2) | 4 | <10−5, <10−27, <10−7, <10−11, <10−4 | resistant | |||||
| 22 | DIS3 | 2 | <10−4 | ||||||
| 23 | DUSP5 | 1 | <10−5 | ||||||
| 24 | EPS8 | 4 | <10−5 | <10−5, <10−27, <10−36, <10−44, <10−5 | |||||
| 25 | ESCO1 | 3 | <10−4 | ||||||
| 26 | ESD | 2 | <10−4 | ||||||
| 27 | FAM107B | 3 | <10−5, <10−11, <10−4, <10−7, <10−4 | resistant | resistant | ||||
| 28 | FAM111B | 2 | <10−4 | ||||||
| 29 | FASTK | 3 | <10−5 | ||||||
| 30 | FIBP | 2 | <10−6 | ||||||
| 31 | FXYD5 | 4 | <10−4 | resistant | |||||
| 32 | HOXB2 | 1 | <10−4 | ||||||
| 33 | HOXB7 | 1 | <10−4 | <10−5, <10−27, <10−6, <10−12, <10−4 | resistant | resistant | |||
| 34 | INTS5 | 3 | <10−5 | <10−3, <10−4, <10−5 | resistant | ||||
| 35 | IRF1 | 1 | <10−5, <10−23, <10−7, <10−13, <10−4 | ||||||
| 36 | ITGAL | 1 | <10−5 | ||||||
| 37 | MAP2K6 | 1 | <10−5 | ||||||
| 38 | MED12 | 1 | <10−4 | ||||||
| 39 | MED16 | 1 | <10−4 | ||||||
| 40 | MID1IP1 | 3 | <10−5 | resistant | resistant | ||||
| 41 | NFYC | 3 | <10−5 | ||||||
| 42 | POMP | 3 | <10−5 | resistant | resistant | ||||
| 43 | PPDPF (C20orf149) | 2 | <10−4 | ||||||
| 44 | PPP2R4 | 1 | <10−4 | <10−5, <10−18, <10−4, <10−7, <10−4 | |||||
| 45 | PRDX5 | 3 | <10−4 | ||||||
| 46 | PRKD2 | 4 | <10−5 | resistant | |||||
| 47 | PTP4A2 | 4 | <10−5, <10−4, <10−4, <10−10, <10−4 | ||||||
| 48 | REEP5 | 2 | <10−5, <10−7, <10−5, <10−4, <10−4 | resistant | resistant | ||||
| 49 | SBF1 | 1 | <10−4 | resistant | |||||
| 50 | SCAMP2 | 2 | <10−5 | ||||||
| 51 | SH3BGRL3 | 2 | <10−4, <10−4, <10−4 | resistant | resistant | ||||
| 52 | STARD3 | 3 | <10−4, <10−4, <10−4 | resistant | |||||
| 53 | STUB1 | 1 | <10−4 | resistant | resistant | ||||
| 54 | STX4 | 1 | <10−5 | resistant | resistant | ||||
| 55 | TBCB | 2 | <10−4 | resistant | resistant | ||||
| 56 | TFRC | 1 | <10−5, <10−27, <10−9, <10−17, <10−4 | ||||||
| 57 | THAP7 | 1 | <10−5 | <10−4, <10−4, <10−5 | resistant | ||||
| 58 | TNRC6B | 2 | <10−4 | ||||||
| 59 | TRIO | 4 | <10−5, <10−27, <10−5, <10−9, <10−4 | resistant | |||||
| 60 | USP21 | 4 | <10−6 | <10−5, <10−4, <10−6 | resistant | resistant | |||
| 61 | ZNF496 | 2 | <10−5, <10−27, <10−5, <10−8, <10−4 | resistant | resistant | ||||
Note:P-values under ‘Basis for selection’ indicate the association significance of individual candidate genes with metformin IC50 found by given association analysis. The P-values under ‘SNP-expression-IC50 analysis’ were for SNP-IC50, SNP-expression and expression-IC50 association analyses of individual candidate genes (Supplementary Material, Table S7). The P-values under ‘Methylation-expression-IC50 analysis’ were for Methy1-IC50, Methy1-Exp1, Methy1-Exp2, Exp1-Exp2, and Exp2-IC50 analyses of individual candidate genes (Supplementary Material, Table S11). * CDC42BPA, the P-values were for methylation-expression, methylation-IC50, and expression-IC50 association analyses of CDC42BPA gene. For the MTS assay, ‘resistant’ indicates that the gene knockdown desensitized metformin cytotoxicity when compared with control siRNA. All of the experiments with significant changes were performed in triplicate and were replicated at least three times.
STUB1 influences metformin response through acting as an E3 ligase for cyclin A
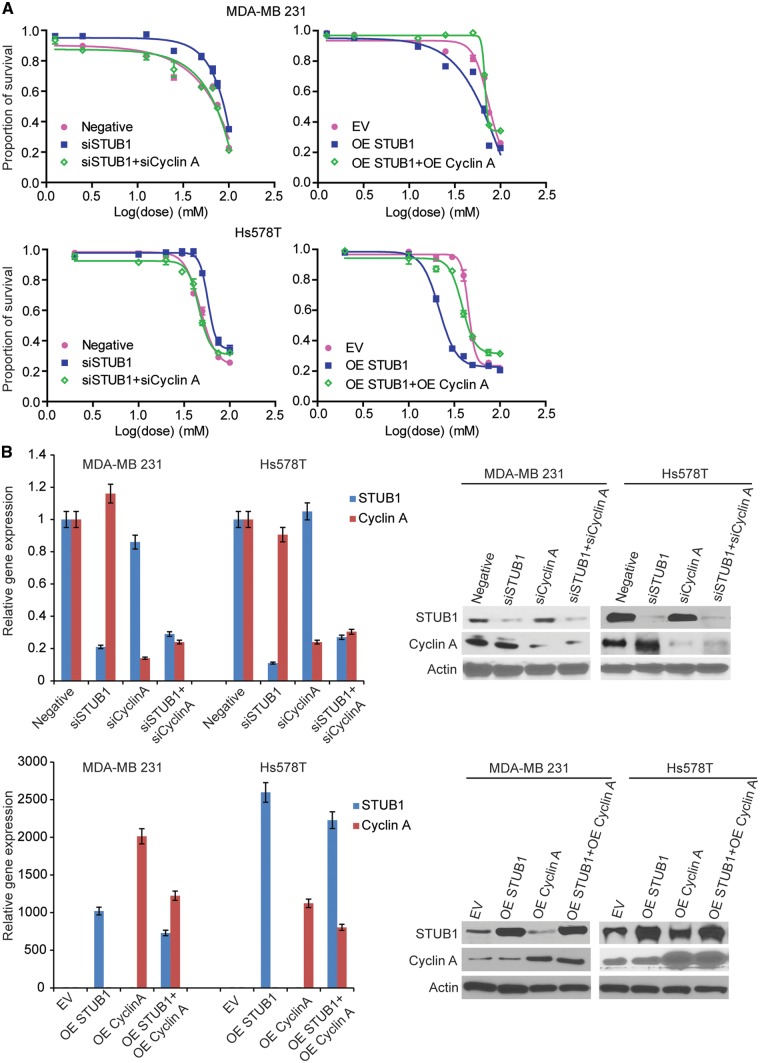
Among the 14 genes that were validated in two breast cancer cells, STUB1 gene is one of the top candidates from the expression vs IC50 analysis. Its expression in LCLs was negatively associated with metformin IC50 (p value = 5.71× 10−5) (Supplementary Material, Table S1). It is well known that STUB1 (STIP1 homology and U-box containing protein 1), also called CHIP (C terminus of Hsc70-interacting protein), is an E3 ubiquitin ligase. It contains a tetratricopeptide repeat (TPR) at the N terminus mediating the interaction with Hsc/Hsp70, and a U-box domain at the C terminus with E3 ubiquitin ligase activity (33). In both breast cancer cell lines, RNA interference experiments showed that knockdown of STUB1 significantly desensitized cells to metformin treatment (Fig. 3A, left panel; and Fig. 3B, upper panel), and overexpression of STUB1 significantly sensitized cells to metformin treatment (Fig. 3A, right panel; Fig. 3B, lower panel). Metformin could induce cell cycle arrest in the G0-G1 phase, resulting in a significant decrease of cells in S phase as well as a decrease of cyclin A and cyclin D protein level (9,37,38). Here we provided evidence that STUB1 could influence cell cycle through promoting the degradation of cyclin A, which might be a novel mechanism involved in the STUB1 regulation of metformin response.
Figure 3.
STUB1 regulated metformin sensitivity through cyclin A. MDA-MB 231 and Hs578T cells were transfected with negative control, STUB1, and cyclin A siRNA; MDA-MB 231 and Hs578T cells were transfected with empty vector, STUB1, and cyclin A plasmids. (A) MTS cytotoxicity assay was performed. (B) Knockdown efficiency and overexpress efficiency were assessed by qRT-PCR and western blot.
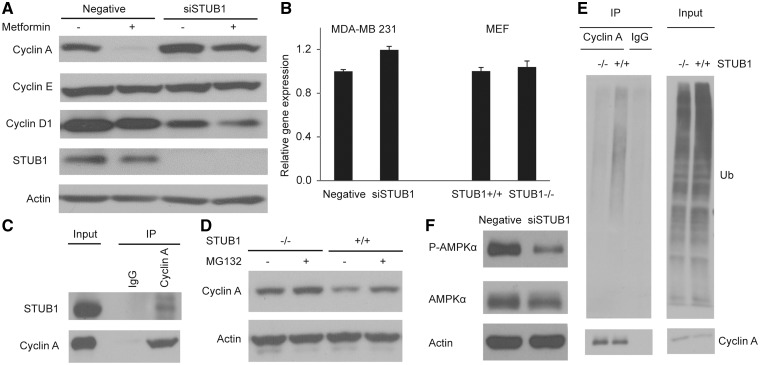
We first determined the effect of STUB1 downregulation on the protein levels of cyclins D, E and A, all of which play a critical role in G1/S transition (39). We found that STUB1 knockdown resulted in a significant increase in cyclin A protein level and a decrease in cyclin D protein level while the cyclin E level was not affected (Fig. 4A). The decrease in cyclin D level might be due to the increase in cell proportion in S phase (40). qRT-PCR experiments showed no significant change in cyclin A mRNA levels (Fig. 4B). In STUB1-/- MEF cells, we also observed an increase in cyclin A protein level without a significant change in mRNA level (Fig. 4B and D). Therefore, given the known function of STUB1 as an E3 ligase, we hypothesized that STUB1 might alter metformin response through promoting proteasome-mediated degradation of cyclin A. The endogenous immunoprecipitation experiments performed in the MDA-MB 231 cells showed an interaction between STUB1 and cyclin A (Fig. 4C). To demonstrate that STUB1 polyubiquitinated cyclin A, cyclin A was immunoprecipitated in wild-type (WT) and STUB1-/- MEF cells. STUB1-/- MEF cells showed a significant reduction in polyubiquitinated cyclin A when compared to WT STUB1 MEF cells (Fig. 4E). Furthermore, treatment of proteasome inhibitor MG132 in WT STUB1 MEFs rescued STUB1-induced degradation of cyclin A (Fig. 4D).
Figure 4.
STUB1 functions as an E3 ligase for cyclin A. MDA-MB 231 cells were transfected with negative control or STUB1 siRNA, and treated with 20 mM metformin for 24 h. STUB1+/+ and STUB1-/- MEFs were gifts from Dr. Patterson. (A) The protein levels of Cyclin A, E, and D1 in cell lysates were detected by western blot. (B) The mRNA level of Cyclin A was examined by qRT-PCR. (C) Endogenous interaction between STUB1 and Cyclin A was detected in MDA-MB 231 cells. (D) Cyclin A protein level was determined in MEFs after MG132 treatment. (E) Cyclin A Ub assay was performed in wild-type and STUB1-/-MEFs. (F) The level of phospho-AMPK T172 was detected by western blot.
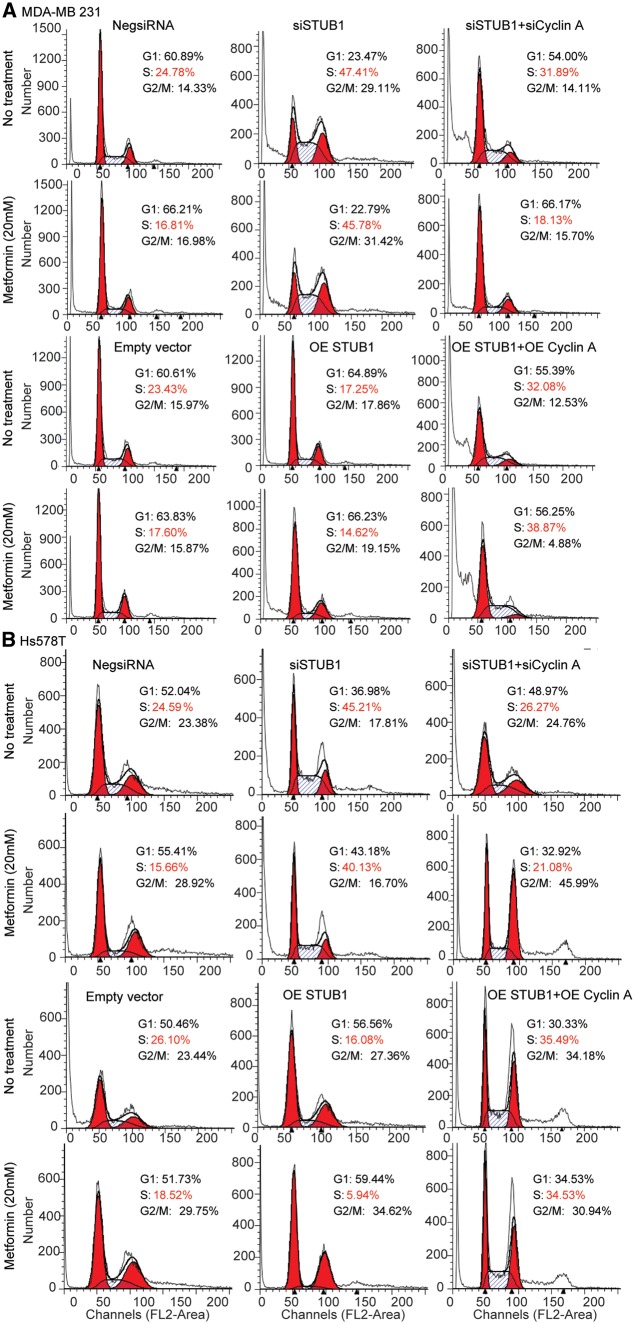
The degradation of cyclin A mediated by STUB1 suggests that the effect of STUB1 on metformin response may depend on cyclin A. To test this hypothesis, we first examined whether STUB1 would have any effect on cell cycle. We found that knockdown of STUB1 in MDA-MB 231 and Hs578T breast cancer cells significantly increased the proportion of cells in S phase while metformin treatment resulted in a reduction of cells in S phase (Fig. 5A and B). STUB1 knockdown could reverse the effect of metformin on cells in S phase compared with control (Fig. 5A). To further investigate whether the effect of STUB1 on metformin response was mediated through cyclin A, cell cycle distribution was analysed following cyclin A knockdown in STUB1 knockdown cells treated with or without metformin. Knockdown of both cyclin A and STUB1 significantly decreased the proportion of cells in S phase comparing with knockdown STUB1 alone in both metformin treated and nontreated two breast cancer cells (Fig. 5A and B). Additionally, overexpression of STUB1 significantly decreased the proportion of cells in S phase (Fig. 5A and B), while overexpression of both cyclin A and STUB1 significantly increased the proportion of cells in S phase, a reversed phenotype when compared to overexpression of STUB1 alone (Fig. 5A and B). Since STUB1 regulated cell cycle through cyclin A, we tested whether cyclin A might be the key factor mediating the effects of STUB1 on metformin response. We silenced cyclin A in STUB1 knockdown cell, and found that knocking down both cyclin A and STUB1 re-sensitized cells to metformin treatment compared with STUB1 knockdown alone in both MDA-MB 231 and Hs578T cells (Fig. 3A and B). Conversely, overexpressing both cyclin A and STUB1 desensitized cells to metformin treatment compared with STUB1 overexpression alone (Fig. 3A and B). Together, these results indicate that STUB1 influences metformin response through its functioning as an E3 ligase for cyclin A.
Figure 5.
STUB1 regulated cell cycle through clyclin A. MDA-MB 231 and Hs578T cells were transfected with negative control, STUB1, and cyclin A siRNA; MDA-MB 231 and Hs578T cells were transfected with empty vector, STUB1, and cyclin A plasmids. (A) Cell cycle profile in MDA-MB 231 cell line was analysed by flow cytometry. (B) Cell cycle profile in Hs578T cell line was analysed by flow cytometry.
Discussion
Many preclinical studies suggest that the antidiabetic drug, metformin, could be used as an anticancer agent (5). Large inter-individual variation in glycemic response to metformin treatment has been observed in the clinic, which might be partly due to the genetic variation in the metformin PK and PD pathway genes (14,18–27,41–45). However, the anti-cancer activity of metformin as well as the pharmacogenomic aspect of metformin in anti-cancer treatment remains unclear. In this study, we took advantage of the in vitro EBV-transformed LCL model to study the pharmacogenomics of metformin as an anticancer agent. This model contains 266 LCLs with enriched multiple omics data and has been successfully used in many pharmacogenomic studies to identify and understand the contribution of germline SNPs or genes to variation in metformin response, particularly genes involved in the pharmacodynamics side. Many previous studies suggested metformin could cause G1/G0 cell cycle arrest (9,37,38), therefore, in our study the metformin response phenotype (IC50 values) could be potentially affected by the cell cycle status of LCLs at baseline. However, this might not be a disadvantage since the variation among cells might allow us to identify genes involved in metformin induced cell cycle changes. It should be noted that some substantial transcriptional differences have been reported between in vitro cell line model and primary tissue, and it may also be affected by long-time cell culture (46,47). In our previous study, we have described the validity of our study design by comparing methylation profiles between our LCLs and other tissue origins like the breast, lungs and colon using the TCGA data. We found no significant difference in these analyses between LCLs and other tissues (48). Obviously, all the results from in vitro cell line model have to be warranted by future studies using additional in vitro or in vivo models. Finally, it should be also noted that LCLs are not cancer cells per se and somatic genomic alterations that strongly influence response to metformin will be missed in screens like this.
Based on the genome wide analysis of SNPs, methylation, basal gene expression, and metformin IC50 data, our study identified 198 top mRNA expression probe sets, 12 SNP loci, and 5 DNA methylation loci associated with metformin IC50 with P-value <10−4 or <10−5, respectively (Fig. 1, Supplementary Materials, Tables S1, S6, S10). Integrated SNP loci/methylation loci-mRNA expression-IC50 analysis indicated that 3 SNP loci was associated with metformin IC50 through trans-regulation of 11 gene expression levels with P-value <10−4 (Supplementary Material, Table S7), and 5 DNA methylation loci through trans-regulation of expression for 26 genes with P-value <10−4 (Supplementary Material, Table S11). Ingenuity pathway analysis of top 65 candidate genes identified 61 genes clustered within 4 networks (Fig. 2, Supplementary Material, Fig. S3), which provided us with new insights into additional mechanisms that might be involved in metformin response. It should be noted that in order to have a reasonable power, we performed GWAS using 266 LCLs from all three different ethnic groups and adjust for the ethnicity. We understand that we might have missed ethnic specific signals. However, considering the power, we choose the strategy described here. Like all model systems, the LCL system used in our study has limitations. EBV transformation might cause chromosomal instability and cellular changes in the LCLs. Variation in drug response in LCLs could be affected by nongenetic factors such as cell growth rate or baseline ATP levels (31). In addition, the regulation of gene expression is tissue specific. Previous studies indicated metformin might be a potential treatment for triple negative breast cancer (49). Therefore, to functionally validate the initial association results obtained using LCLs, we conducted knockdown for the top 61 candidate genes selected based on the IPA using siRNA screening, followed by MTS assay in two different breast cancer cell lines, MDA-MB 231 and Hs578T cells. Knockdown experiments showed that downregulation of 25 genes significantly altered metformin sensitivity in MDA-MB 231 cells (Table 1, Supplementary Material, Fig. S4). 14 out of 25 genes also showed the same effect on metformin response in Hs578T cells (Table 1, Supplementary Material, Fig. S5).
Many previous studies suggest that in preclinical models, high concentrations of metformin are required to achieve an anticancer effect (4,8,50). In our study, high concentrations of metformin were also required to kill LCLs and breast cancer cells. Therefore, many concerns have been raised with regard to how effective metformin is as an anti-cancer agent at antidiabetic therapeutic concentrations. Even though the treatment concentration is low, the concentration in the target cancer tissues might vary based on the expression of metformin transporters (4,8). Furthermore, because of the low incidence of metformin related side effects, it would be possible to increase the metformin dose as an anticancer therapy or as part of a combination treatment that would be evaluated in future clinical trials (4,8). In addition, the required high metformin concentration might be partially due to the high glucose level in the cell culture medium. It has been suggested that under high glucose condition, metformin-induced energetic stress could cause a cytostatic effect, and reduce energy consumption and improve cell survival, whereas under low glucose condition, metformin treatment leads to an energetic crisis and causes a cytotoxic effect (51). In this study, all of our experiments were performed in a regular cell culture medium that has a high glucose compared with physiological glucose concentration. Therefore, the effect we observed would be warranted after future confirmation at physiological concentration of glucose.
As one of the top candidates, STUB1, showed resistance to metformin treatment in breast cancer cells when knockdown (Fig. 3A and B). STUB1 is known as a U-Box E3 ubiquitin ligase. STUB1 together with HSP70 and HSP90 play a critical role in ubiquitin-mediated protein degradation (33). Previous studies indicated that STUB1 promotes LKB1-mediated activation of AMPK through inducing conformational change of AMPKα subunit (52). However, it also acts as an E3 ligase involved in proteasome-mediated LKB1 degradation (53). We also found that depletion of STUB1 by siRNA in MDA-MB 231 cells could decrease the phosphorylation of AMPK (Fig. 4F), suggesting that this effect was not through STUB1’s effect on the protein degradation of LKB1. In this study, we showed that STUB1 interacted with cyclin A and provided evidence that STUB1 influenced metformin response through its function as an E3 ligase for cyclin A. Cyclin A-CDK2 plays a critical role in G1 to S transition, in which high expression of cyclin A promotes an increase of cells in S phase (39). Our experiments showed that STUB1 knockdown in MDA-MB 231 cells resulted in an increased proportion of cells in S phase by flow cytometry, which could be reversed by knocking down cyclin A (Fig. 5). We observed the same phenomena on metfomin cytotoxicity, indicating the effect of STUB1 on metformin response is through the regulation of cyclin A degradation (Fig. 3). Down regulation of STUBl also resulted in a significant increase of cyclin A at the protein level, but not at the mRNA level (Fig. 4A and B). We also found that STUB1 physically interacted with cyclin A endogenously, and it could promote ubiquitination of cyclin A (Fig. 4C–E). Taken together, our data indicated that STUB1 could impact metformin response via its function as E3 ubiquitin ligase of cyclin A. It suggests a potential novel mechanism with regard to STUB1 in the regulation of cyclin A, and in turn metformin response. In addition, in our GWAS using LCLs, the mRNA expression level of CCNA2 gene, cyclin A, was not associated with metformin IC50, neither did any SNPs in the CCNA2 gene, which suggested that cyclin A protein level, instead of mRNA level, might function as a biomarker predicting metformin response. Certainly, STUB1 could regulate many downstream proteins, there could be other mechanisms in additional to cyclin A may play a role in metformin response.
In summary, our GWAS in LCLs identified 198 mRNA expression probe sets, 12 SNP loci, and five DNA methylation loci associated with metformin IC50 with P-value <10−4 or <10−5. Our functional validation studies in breast cancer cell lines validated 14 top candidate genes that significantly altered metformin cytotoxicity in both MDA-MB 231 and Hs578T breast cancer cell lines. A further mechanistic study of one top candidate gene, STUB1, indicated that it affected metformin sensitivity through its function as an E3 ligase for cyclin A. Currently we are also conducting the follow-up mechanistic study for a few other candidates. These results would provide additional insight into novel mechanisms by which these genes might contribute to variation in response to metformin as an anticancer agent.
Methods
Cell culture and antibodies
As described in our previous publication (31), EBV-transformed LCLs from 96 African-American (AA), 96 Caucasian-American (CA), and 96 Han Chinese-American (HCA) unrelated subjects (sample sets HD100AA, HD100CAU, HD100CHI) were purchased from the Coriell Cell Repository with very low passage number (Camden, NJ). These samples had been anonymized by NIGMS, and all subjects had provided written consent for their experimental use. This study was reviewed and approved by Mayo Clinic Institutional Review Board. Human breast cancer cell lines, MDA-MB 231 and Hs578T were newly obtained from the American Type Culture Collection (Manassas, VA). Wild-type (STUB1+/+) and STUB1-/- mouse embryonic fibroblast (MEF) cells were gifts from Dr. Cam Patterson in the University of North Carolina at Chapel Hill (54). LCLs were cultured in RPMI 1640 medium (Mediatech, Manassas, VA) supplemented with 15% foetal bovine serum (FBS) (Mediatech). MDA-MB 231, Hs 578T, and MEF cell lines were cultured in DMEM medium containing 10% FBS.
Antibodies against cyclin E, cyclin A, ubiquitin were purchased from Santa Cruz Biotechnology (Dallas, TX). Antibody against cyclin D was obtained from Abcam (Cambridge, MA). Anti-STUB1 antibody was purchased from Bethyl Laboratories (Montgomery, TX). Anti β-actin antibody was purchased from Sigma-Aldrich (St. Louis, MO).
Cell proliferation assay
Metformin was purchased from Sigma-Aldrich (St. Louis, MO). The drug was dissolved in PBS and aliquots of stock solutions were frozen at −80°C. As described previously, cell proliferation assays were performed in triplicate at each drug concentration. Specifically, 90 µl of cells (5 × 105 cells/ml) were plated into each well of 96-well plates (Corning, Lowell, MA) (30) and were treated with 10 µl of metformin at the final concentrations of 0, 0.001, 0.01, 0.25, 1, 2.5, 10, 100, 200 mmol/l. Seventy-2 h after metformin treatment, 20 µl of CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay solution (Promega Corporation, Madison, WI) was added to each well and incubated for an additional 3 h. Plates were then read in a Safire2 microplate reader (Tecan AG, Switzerland). Experiments were successfully performed for 266 LCLs (89 AA, 85 CA and 92 HCA). The cell proliferation assays for breast cancer cell lines were conducted in a similar fashion except metformin was added after the cells were seeded overnight. The final concentrations of metformin were 0, 1.25, 2.5, 12.5, 25, 50, 66.7, 75, 100 mmol/l for MDA-MB 231 cells, and 0, 2, 10, 20, 30, 40, 50, 75, 100 mmol/l for Hs578T cells.
Genome-wide SNPs in LCLs
As described previously (31), we genotyped DNA samples from the LCLs in the Genotype Shared Resource (GSR) at Mayo Clinic using Illumina HumanHap 550K and 510S BeadArrays, which contained 561,298 and 493,750 SNPs, respectively. Publicly available Affymetrix SNP Array 6.0 Chip data for additional 643,600 SNPs were also obtained for the same cell lines. All the genotyping data are publicly available from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under SuperSeries accession No. GSE24277. SNPs that deviated from Hardy-Weinberg Equilibrium (HWE) based on the minimum P-value from an exact test for HWE and the stratified test for HWE (P-values <0.001); SNPs with call rates <95%; or SNPs with minor allele frequencies (MAFs) <5% were removed from the analysis.
Expression array assay in LCLs
Total RNA was extracted from each of the cell lines using Qiagen RNeasy Mini kits (QIAGEN, Inc.). RNA quality was tested using an Agilent 2100 Bioanalyzer, followed by hybridization to Affymetrix U133 Plus 2.0 Gene-Chips. The expression array data for all 54k probe sets was used in our previous studies (30,31) and is publicly available from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under SuperSeries accession no. GSE24277 and accession No.GSE23120.
DNA methylation array assay in LCLs
As described previously (48), quality of genomic DNA was tested by electrophoresis in a 1.3% agarose gel, picogreen quantification, and nanodrop measurements. Bisulphate conversion of genomic DNA was conducted using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) followed by hybridization on the HumanMethylation450 BeadChip (Illumina). Raw methylation data was normalized by three steps using the lumi package in Bioconductor and R package (R Development Core Team 2009), including colour bias adjustment, background level adjustment, and quantile normalization across arrays. The methylation level (b-value) for each of the 485,577 CpG sites was calculated as the ratio of methylated signal divided to the sum of methylated and unmethylated signals plus 100. Quality control (QC) was performed to remove probes not annotated as ‘cg’ probes, probes located at or near an SNP, and probes with low call rate (<0.98). Subjects with a call rate <0.98 and outlier probes/samples identified via principal component analysis (PCA) were also removed from future analyses.
Transient transfection and RNA interference
siRNA pools for candidate genes and negative control were purchased from Dharmacon (Chicago, IL). Reverse transfection of siRNA was performed in 96-well plates with a seeding density of 3000-5000 breast cancer cells, 0.3 µL of lipofectamineTM RNAiMAX reagent (Life technology, Grand Island, NY), and 30 nmol/l siRNA pools for cell proliferation assay. For immunoblotting and cell cycle assays, the amount of transfection mixture was scaled up proportionally based on the different cell numbers required.
The STUB1 plasmid was purchased from Dharmacon (Chicago, IL). Cyclin A plasmid, CycA-Venus-Flag (1305), was purchased from Addgene (Cambridge, MA). Plasmids were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according the manufacture’s protocol.
Real-time quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was isolated from cultured cells using the Quick-RNATM MiniPrep kit (Zymo Research), followed by qRT-PCR with the Power SYBR® Green RNA-to- 1-Step Kit (AB Foster CA). Specifically, primers purchased from QIAGEN or IDT were used to perform qRT-PCR using the Stratagene Mx3005P Real-Time PCR detection system (Stratagene). All experiments were performed with human β-actin as an internal control. For MEF cell lines, mouse β-actin was used as an internal control.
Immunoprecipitation and immunoblotting
As described previously (55), cells were lysed in NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet p-40) containing protease and phosphatase inhibitors (Roche) on ice for 30 min. After centrifugation at 14,000 g for 10 min at 4ºC, cell lysate were incubated with 4 µg of antibody or normal IgG (Cell Signalling Technology) and protein A Sepharose beads (Amersham Biosciences) overnight at 4ºC. The immunocomplexes were washed with NETN buffer for three times. The precipitates were then eluted in SDS sample buffer and separated by SDS-PAGE. Immunoblotting was performed following standard procedures.
Cell cycle analysis
After treatment of control or 20 mM metformin for 24 h, cells were trypsinized and washed by phosphate-buffered saline (PBS). After fixation in ice-cold 70% ethanol overnight at 4°C, cells were washed with PBS containing 1% BSA, and then resuspended in PBS containing 1% BSA, 20 µg/ml propidium iodine, and 100 µg/ml RNase A. 50000 cells were collected for the Flow cytometry assay per experiment using a FACSCalibur cytometer (BD Biosciences). The percentage of cells in each phase of the cell cycle was estimated with ModFit.
In vivo ubiquitination assay
As described previously (56), MEF cells were treated with MG132 (10 nM) for 12 h and then lysed in NETN buffer containing protease inhibitor, 20 mM Nethylmaleimide (NEM), and 1 mM iodoacetamide. After centrifugation, the lysates were immunoprecipitated with cyclin A antibody overnight at 4ºC. The immunocomplexes were washed with NETN buffer for three times. The precipitates were eluted in SDS sample buffer and then separated by SDS-PAGE.
Statistical Methods
The metformin cytotoxicity phenotype IC50, indicating the drug concentration that inhibits half of maximal cell growth, was calculated based on a logistic model. Three different logistic functions (a four parameter logistic model, a three parameter logistic model with a fixed asymptote at 0%, and a three parameter logistic model with a fixed asymptote at 100%) were used to fit the data with the R package ‘‘drc’’ (http://cran.r-project.org/web/packages/drc/drc.pdf). The best model fit (i.e., lowest mean square error) from the three logistic models was used to determine the cytotoxicity IC50 phenotype. As previously described (31), prior to association analyses, the Van der Waerden (rank) transformed IC50 and SNPs were adjusted for gender, race and population stratification. Similarly, GCRMA normalized and log2 transformed mRNA expression array data were adjusted for gender, race, population stratification, as well as batch effect. For DNA methylation data, the logit of beta values were adjusted for age, gender and race using linear regression model, followed by adding overall mean probe values back followed by back transformation of the values using inverse logit function. The final adjusted beta values on the [0,1] scale were used for analysis.
The pair wised association among adjusted SNP, expression, methylation and IC50 values were completed using Pearson correlations. False Discovery Q-values were also computed for each test. Genes and SNPs were annotated using NCBI Build 37. To integrate all the analysis data together, we used a similar approach as described previously. Specifically, we first identified top SNP loci or methylation probes/loci associated with IC50, then determined which expression probe sets were associated with these SNPs/methylation probes, and finally determined whether the expression probe sets associated with these SNPs/methylation probes were also associated with metformin IC50 values. For the most significant locus on chromosome 2 and SNP locus on chromosome 16, SNPs were imputed for a region including 200 kb on either side of the most significant SNPs. Imputation was performed using Beagle v3.3.1 with 1000 genome data (release of 11/23/2010) as the reference panel with build37 annotation. Chromosomes were divided into 40MB regions and BEAGLE v3.3.1 was ran on these 40MB regions of the genome, plus a 1MB buffer region to the right and left of the main region to provide buffer at the ends of regions as the ends are generally imputed with poor quality. Imputation was run with markers obtained from combining all populations. There were a total of 1094 subjects in the reference group from different populations. In addition, pairwise LD (D’ and R2 values) was calculated using PLINK software (57). To identify SNPs that were correlated with the top 1772 SNPs associated with metformin IC50, Pearson correlation was calculated between each SNP and the rest of SNPs on the same chromosome and r2 >0.9 was used as the cut-off. The pathway analysis for the top genes was performed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems). The percentage of variation in the drug response phenotype explained by the combination of gene expression probe sets, SNPs, and DNA methylation probes was estimated using Gradient Boosting Algorithm. Lastly, the significance of the AUC values between negative control siRNA and gene-specific siRNA was determined by a two-tailed unpaired t-test.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The Mayo Clinic Genotyping Shared Resource is supported by CA15083 (Mayo Comprehensive Cancer Center).
Conflict of Interest statement. None declared.
Funding
This work was supported by NIH grants R01CA196648, R01 CA138461, U19 GM61388 (The Pharmacogenomics Research Network); R01 CA80127, R01 CA84354 and R01 CA105857.
References
- 1. Chlebowski R.T., McTiernan A., Wactawski-Wende J., Manson J.E., Aragaki A.K., Rohan T., Ipp E., Kaklamani V.G., Vitolins M., Wallace R., et al. (2012) Diabetes, metformin, and breast cancer in postmenopausal women. J. Clin. Oncol., 30, 2844–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu B., Fan Z., Edgerton S.M., Deng X.S., Alimova I.N., Lind S.E., Thor A.D. (2009) Metformin induces unique biological and molecular responses in triple negative breast cancer cells. Cell Cycle, 8, 2031–2040. [DOI] [PubMed] [Google Scholar]
- 3. Williams C.C., Singleton B.A., Llopis S.D., Skripnikova E.V. (2013) Metformin induces a senescence-associated gene signature in breast cancer cells. J. Health Care Poor Underserved, 24, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pollak M. (2014) Overcoming Drug Development Bottlenecks With Repurposing: Repurposing biguanides to target energy metabolism for cancer treatment. Nat. Med., 20, 591–593. [DOI] [PubMed] [Google Scholar]
- 5. Pollak M.N. (2012) Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discov., 2, 778–790. [DOI] [PubMed] [Google Scholar]
- 6. Giovannucci E., Harlan D.M., Archer M.C., Bergenstal R.M., Gapstur S.M., Habel L.A., Pollak M., Regensteiner J.G., Yee D. (2010) Diabetes and cancer: a consensus report. CA. Cancer. J. Clin., 60, 207–221. [DOI] [PubMed] [Google Scholar]
- 7. Pernicova I., Korbonits M. (2014) Metformin–mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol., 10, 143–156. [DOI] [PubMed] [Google Scholar]
- 8. Pollak M. (2013) Potential applications for biguanides in oncology. J. Clin. Invest., 123, 3693–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai X., Hu X., Tan X., Cheng W., Wang Q., Chen X., Guan Y., Chen C., Jing X. (2015) Metformin Induced AMPK Activation, G0/G1 Phase Cell Cycle Arrest and the Inhibition of Growth of Esophageal Squamous Cell Carcinomas In Vitro and In Vivo. PLoS One, 10, e0133349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng X.S., Wang S., Deng A., Liu B., Edgerton S.M., Lind S.E., Wahdan-Alaswad R., Thor A.D. (2012) Metformin targets Stat3 to inhibit cell growth and induce apoptosis in triple-negative breast cancers. Cell Cycle, 11, 367–376. [DOI] [PubMed] [Google Scholar]
- 11. Kordes S., Pollak M.N., Zwinderman A.H., Mathot R.A., Weterman M.J., Beeker A., Punt C.J., Richel D.J., Wilmink J.W. (2015) Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. The Lancet. Oncology, 16, 839–847. [DOI] [PubMed] [Google Scholar]
- 12. Hatoum D., McGowan E.M. (2015) Recent advances in the use of metformin: can treating diabetes prevent breast cancer?. Biomed. Res. Int., 2015, 548436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goswami S., Yee S.W., Stocker S., Mosley J.D., Kubo M., Castro R., Mefford J.A., Wen C., Liang X., Witte J., et al. (2014) Genetic variants in transcription factors are associated with the pharmacokinetics and pharmacodynamics of metformin. Clin. Pharmacol. Ther., 96, 370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou K., Donnelly L., Yang J., Li M., Deshmukh H., Van Zuydam N., Ahlqvist E., Spencer C.C., Groop L., Morris A.D., et al. (2014) Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol., 2, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GoDarts, Group U.D.P.S., Wellcome Trust Case Control C., Zhou K., Bellenguez C., Spencer C.C., Bennett A.J., Coleman R.L., Tavendale R., Hawley S.A., et al. (2011) Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet., 43, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Leeuwen N., Nijpels G., Becker M.L., Deshmukh H., Zhou K., Stricker B.H., Uitterlinden A.G., Hofman A., van't Riet E., Palmer C.N., et al. (2012) A gene variant near ATM is significantly associated with metformin treatment response in type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia, 55, 1971–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahlin G., Chen L., Lazorova L., Chen Y., Ianculescu A.G., Davis R.L., Giacomini K.M., Artursson P. (2011) Genotype-dependent effects of inhibitors of the organic cation transporter, OCT1: predictions of metformin interactions. Pharmacogenomics J., 11, 400–411. [DOI] [PubMed] [Google Scholar]
- 18. Becker M.L., Visser L.E., van Schaik R.H., Hofman A., Uitterlinden A.G., Stricker B.H. (2009) Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J., 9, 242–247. [DOI] [PubMed] [Google Scholar]
- 19. Becker M.L., Visser L.E., van Schaik R.H., Hofman A., Uitterlinden A.G., Stricker B.H. (2009) Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes, 58, 745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Pawlikowski B., Schlessinger A., More S.S., Stryke D., Johns S.J., Portman M.A., Chen E., Ferrin T.E., Sali A., et al. (2010) Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet. Genomics, 20, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., Teranishi K., Li S., Yee S.W., Hesselson S., Stryke D., Johns S.J., Ferrin T.E., Kwok P., Giacomini K.M. (2009) Genetic variants in multidrug and toxic compound extrusion-1, hMATE1, alter transport function. Pharmacogenomics J., 9, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Motohashi H., Inui K. (2013) Organic cation transporter OCTs (SLC22) and MATEs (SLC47) in the human kidney. AAPS. J., 15, 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song I.S., Shin H.J., Shim E.J., Jung I.S., Kim W.Y., Shon J.H., Shin J.G. (2008) Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin. Pharmacol. Ther., 84, 559–562. [DOI] [PubMed] [Google Scholar]
- 24. Song I.S., Shin H.J., Shin J.G. (2008) Genetic variants of organic cation transporter 2 (OCT2) significantly reduce metformin uptake in oocytes. Xenobiotica, 38, 1252–1262. [DOI] [PubMed] [Google Scholar]
- 25. Reitman M.L., Schadt E.E. (2007) Pharmacogenetics of metformin response: a step in the path toward personalized medicine. J. Clin. Invest., 117, 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jablonski K.A., McAteer J.B., de Bakker P.I., Franks P.W., Pollin T.I., Hanson R.L., Saxena R., Fowler S., Shuldiner A.R., Knowler W.C., et al. (2010) Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes, 59, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joerger M., van Schaik R.H., Becker M.L., Hayoz S., Pollak M., Cathomas R., Winterhalder R., Gillessen S., Rothermundt C. (2015) Multidrug and toxin extrusion 1 and human organic cation transporter 1 polymorphisms in patients with castration-resistant prostate cancer receiving metformin (SAKK 08/09). Prostate Cancer Prostatic Dis., 18, 167–172. [DOI] [PubMed] [Google Scholar]
- 28. Birsoy K., Possemato R., Lorbeer F.K., Bayraktar E.C., Thiru P., Yucel B., Wang T., Chen W.W., Clish C.B., Sabatini D.M. (2014) Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature, 508, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aldea M., Craciun L., Tomuleasa C., Berindan-Neagoe I., Kacso G., Florian I.S., Crivii C. (2014) Repositioning metformin in cancer: genetics, drug targets, and new ways of delivery. Tumour Biol., 35, 5101–5110. [DOI] [PubMed] [Google Scholar]
- 30. Li L., Fridley B., Kalari K., Jenkins G., Batzler A., Safgren S., Hildebrandt M., Ames M., Schaid D., Wang L. (2008) Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res., 68, 7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niu N., Qin Y., Fridley B.L., Hou J., Kalari K.R., Zhu M., Wu T.Y., Jenkins G.D., Batzler A., Wang L. (2010) Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res., 20, 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ingle J.N., Schaid D.J., Goss P.E., Liu M., Mushiroda T., Chapman J.A., Kubo M., Jenkins G.D., Batzler A., Shepherd L., et al. (2010) Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J. Clin. Oncol., 28, 4674–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonough H., Patterson C. (2003) CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones, 8, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang X., Chien H.C., Yee S.W., Giacomini M.M., Chen E.C., Piao M., Hao J., Twelves J., Lepist E.I., Ray A.S., et al. (2015) Metformin Is a Substrate and Inhibitor of the Human Thiamine Transporter, THTR-2 (SLC19A3). Mol. Pharm., 12, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. (2013) Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics, 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang J., Fridley B.L., Feng Q., Abo R.P., Brisbin A., Batzler A., Jenkins G., Long P.A., Wang L. (2013) Genome-wide association study for biomarker identification of Rapamycin and Everolimus using a lymphoblastoid cell line system. Front. Genet., 4, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Lirio A., Perez-Yarza G., Fernandez-Suarez M.R., Alonso-Tejerina E., Boyano M.D., Asumendi A. (2015) Metformin induces cell cycle arrest and apoptosis in drug-resistant leukemia cells. Leuk. Res. Treatment, 2015, 516460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ben Sahra I., Laurent K., Giuliano S., Larbret F., Ponzio G., Gounon P., Le Marchand-Brustel Y., Giorgetti-Peraldi S., Cormont M., Bertolotto C., et al. (2010) Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer. Res., 70, 2465–2475. [DOI] [PubMed] [Google Scholar]
- 39. Suryadinata R., Sadowski M., Sarcevic B. (2010) Control of cell cycle progression by phosphorylation of cyclin-dependent kinase (CDK) substrates. Biosci. Rep., 30, 243–255. [DOI] [PubMed] [Google Scholar]
- 40. Yang K., Hitomi M., Stacey D.W. (2006) Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Division, 1, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Christensen M.M., Brasch-Andersen C., Green H., Nielsen F., Damkier P., Beck-Nielsen H., Brosen K. (2011) The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenet. Genomics, 21, 837–850. [DOI] [PubMed] [Google Scholar]
- 42. Becker M.L., Pearson E.R., Tkac I. (2013) Pharmacogenetics of oral antidiabetic drugs. Int. J. Endocrinol., 2013, 686315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gong L., Goswami S., Giacomini K.M., Altman R.B., Klein T.E. (2012) Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet. Genomics, 22, 820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Semiz S., Dujic T., Causevic A. (2013) Pharmacogenetics and personalized treatment of type 2 diabetes. Biochem. Med. (Zagreb), 23, 154–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L., Weinshilboum R. (2014) Metformin pharmacogenomics: biomarkers to mechanisms. Diabetes, 63, 2609–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nestor C.E., Ottaviano R., Reinhardt D., Cruickshanks H.A., Mjoseng H.K., McPherson R.C., Lentini A., Thomson J.P., Dunican D.S., Pennings S., et al. (2015) Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol., 16, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bork S., Pfister S., Witt H., Horn P., Korn B., Ho A.D., Wagner W. (2010) DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell, 9, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heyn H., Moran S., Hernando-Herraez I., Sayols S., Gomez A., Sandoval J., Monk D., Hata K., Marques-Bonet T., Wang L., et al. (2013) DNA methylation contributes to natural human variation. Genome Res., 23, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hadad S.M., Coates P., Jordan L.B., Dowling R.J., Chang M.C., Done S.J., Purdie C.A., Goodwin P.J., Stambolic V., Moulder-Thompson S., et al. (2015) Evidence for biological effects of metformin in operable breast cancer: biomarker analysis in a pre-operative window of opportunity randomized trial. Breast Cancer Res. Treat., 150, 149–155. [DOI] [PubMed] [Google Scholar]
- 50. Smith M.A., Houghton P. (2013) A proposal regarding reporting of in vitro testing results. Clin. Cancer. Res., 19, 2828–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Menendez J.A., Oliveras-Ferraros C., Cufi S., Corominas-Faja B., Joven J., Martin-Castillo B., Vazquez-Martin A. (2012) Metformin is synthetically lethal with glucose withdrawal in cancer cells. Cell Cycle, 11, 2782–2792. [DOI] [PubMed] [Google Scholar]
- 52. Schisler J.C., Rubel C.E., Zhang C., Lockyer P., Cyr D.M., Patterson C. (2013) CHIP protects against cardiac pressure overload through regulation of AMPK. J. Clin. Invest., 123, 3588–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gaude H., Aznar N., Delay A., Bres A., Buchet-Poyau K., Caillat C., Vigouroux A., Rogon C., Woods A., Vanacker J.M., et al. (2012) Molecular chaperone complexes with antagonizing activities regulate stability and activity of the tumor suppressor LKB1. Oncogene, 31, 1582–1591. [DOI] [PubMed] [Google Scholar]
- 54. McDonough H., Charles P.C., Hilliard E.G., Qian S.B., Min J.N., Portbury A., Cyr D.M., Patterson C. (2009) Stress-dependent Daxx-CHIP interaction suppresses the p53 apoptotic program. J. Biol. Chem., 284, 20649–20659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu T., Lin Y.H., Leng W., Jung S.Y., Zhang H., Deng M., Evans D., Li Y., Luo K., Qin B., et al. (2014) A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint. Molecular Cell, 56, 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luo K., Zhang H., Wang L., Yuan J., Lou Z. (2012) Sumoylation of MDC1 is important for proper DNA damage response. EMBO J., 31, 3008–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.