Abstract
Facioscapulohumeral dystrophy (FSHD) is caused by the mis-expression of the double-homeodomain transcription factor DUX4 in skeletal muscle cells. Many different cell culture models have been developed to study the pathophysiology of FSHD, frequently based on endogenous expression of DUX4 in FSHD cells or by mis-expression of DUX4 in control human muscle cells. Although results generated using each model are generally consistent, differences have also been reported, making it unclear which model(s) faithfully recapitulate DUX4 and FSHD biology. In this study, we systematically compared RNA-seq data generated from three different models of FSHD—lentiviral-based DUX4 expression in myoblasts, doxycycline-inducible DUX4 in myoblasts, and differentiated human FSHD myocytes expressing endogenous DUX4—and show that the DUX4-associated gene expression signatures of each dataset are highly correlated (Pearson’s correlation coefficient, r ∼ 0.75-0.85). The few robust differences were attributable to different states of cell differentiation and other differences in experimental design. Our study describes a model system for inducible DUX4 expression that enables reproducible and synchronized experiments and validates the fidelity and FSHD relevance of multiple distinct models of DUX4 expression.
Introduction
Facioscapulohumeral dystrophy (FSHD) is a prevalent form of muscular dystrophy that is currently untreatable and incurable. FSHD is caused by the derepression of the D4Z4 macrosatellite array at chromosome 4q35 (1), resulting in the ectopic expression of the DUX4 gene encoding a germline transcription factor. DUX4 expression is toxic to somatic cells in culture and leads to muscle atrophy in vivo (2–6). These data, as well as strong genetic evidence demonstrating an essential requirement for at least one copy of a polyadenylation-competent DUX4 gene in FSHD, implicate DUX4 as the primary driver of FSHD (7). Consequently, determining whether there is a core set of molecular pathways dysregulated by DUX4 that correlate with transcriptional abnormalities in FSHD cells is of paramount importance to uncover the mechanisms behind DUX4-induced myopathy and develop effective models of the disease for therapeutic development.
The gene expression signature of DUX4 and FSHD has been characterized by several studies, many of which used different model systems of DUX4 expression and/or FSHD disease models and tissues. Such studies have reported diverse genes and biological pathways that are affected in FSHD and by DUX4 expression, including early stem cell and embryonic genes (8,9), cancer testis antigens (8,9), retroelements and repetitive elements (8,10), genes involved in RNA processing and splicing (8,9), inflammatory response (8), myogenesis (3,8,11), Wnt/β-catenin signalling pathway (12,13), oxidative stress response (11), protein homeostasis (14), RNA quality control (15), and PAX targets (3,13), among others. However, not all of these affected genes and/or pathways have been consistently reported by all studies. These differences might reflect the complexity of the disease and raise the possibility that disease models that reproduce all of the major aspects of FSHD might remain out of reach (16).
An alternative perspective is that because DUX4 expression causes FSHD, models based on the expression of DUX4 in skeletal muscle cells should reproduce the major cell-autonomous transcriptional features of FSHD. This perspective is supported by the finding that muscle biopsies from FSHD-affected individuals mis-express the same set of genes that are also upregulated in cultured FSHD muscle cells and in control skeletal muscle cells transduced with a DUX4-expressing vector (17). However, a recent study reported a rather limited overlap between the gene expression signature associated with expression of endogenous DUX4 in FSHD myocytes and the set of DUX4-regulated genes that were previously identified by transduction of non-FSHD human myoblasts with a lentivirus expressing DUX4 (9). Although the authors carefully noted that the differences might arise from different biological and/or technical variables, this finding raised the concern that exogenous expression of DUX4 in non-FSHD skeletal muscle cells might not be a good model for gene expression changes that occur in FSHD muscle cells.
FSHD is a complex disease and it is important to determine whether different model systems yield disparate results or whether they show convergence on similar biological pathways. Differences in DUX4-induced genes in different model systems of FSHD could arise from true biological differences, distinct cell types or contexts, and/or technical differences, such as the use of different data analysis strategies. Here, we used a consistent data analysis pipeline to determine the gene expression profile of three different cell culture models of FSHD: control muscle cells with an inducible DUX4, control muscle cells transduced with a lentiviral vector expressing DUX4, and FSHD muscle cells expressing endogenous DUX4. We report that all three cell culture models exhibited highly similar gene expression patterns and that the few differences, such as viral immune response or myogenic differentiation, were attributable to the differences in experimental design. These results indicate that expressing DUX4 in control skeletal muscle cells accurately recapitulates DUX4-associated differences in gene expression observed in FSHD muscle cells that endogenously express DUX4, and license multiple distinct models of DUX4 expression as relevant tools for studying FSHD biology.
Results
Codon altering DUX4 enables inducible expression
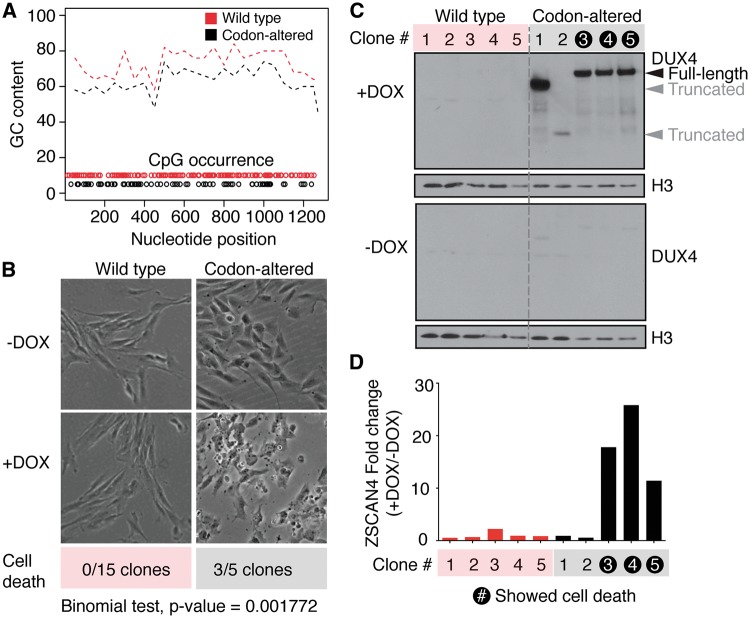
The D4Z4 repeat locus is normally hypermethylated and epigenetically silenced in somatic tissue. The DUX4 open reading frame (ORF) has 130 individual CpG sites that might be subject to DNA methylation, suggesting that the high (73%) GC content of the DUX4 cDNA could contribute to gene silencing. We therefore speculated that reducing the CpG content of the DUX4 cDNA might be required for efficient transgene expression. Hence, we re-coded the DUX4 cDNA to reduce the total number of CpG sites while preserving the protein sequence (Fig. 1A). The codon-altered and wild-type DUX4 ORF sequences were then cloned into the pCW57.1 lentiviral vector (Addgene plasmid #41393) such that a doxycycline-inducible promoter regulates DUX4 and the puromycin resistance gene (puroR) is constitutively driven by the PGK promoter. Clonal cell lines expressing wild-type or codon-altered DUX4 were isolated from immortalized human control myoblasts (MB135) transduced with the corresponding DUX4 constructs using puromycin selection. None of the fifteen tested wild-type DUX4 clones exhibited apoptosis following doxycycline induction, whereas three of five tested codon-altered DUX4 clones exhibited complete cellular detachment/death (Fig. 1B). These data suggest that DUX4 is not efficiently expressed in wild-type DUX4 cells, and that reducing the CpG content of DUX4 cDNA overcomes this barrier to expression (P = 0.001 by the two-sided binomial test for equality of proportions).
Figure 1.
Codon altering allows stable, inducible expression of DUX4 in human myoblasts. (A) Graphical depiction of GC percentage and CpG occurrence of the codon-altered (black) and wild-type (red) DUX4 coding regions. GC percentage was calculated over 50 base pair sliding windows. The positions of CpG dinucleotides are indicated by open circles. (B) Phase contrast images of monoclonal cells encoding wild-type or codon-altered DUX4 expression constructs, with or without doxycycline induction. The number of clonal cell lines that exhibited cell death among all that were tested is shown. (Binomial test for equality of proportions, P Value = 0.001). (C) Western blot analysis for DUX4 expression on lysates from 5 clones encoding wild-type or codon-altered DUX4, with or without induction with doxycycline for 8 hours. Histone 3 (H3) serves as a loading control. Black arrowhead indicates full-length DUX4 product. (D) qRT-PCR analysis of a DUX4 transcriptional target, ZSCAN4, shown as fold-change over uninduced cells in the various clones. The clones that exhibited cell death upon doxycycline induction are highlighted.
To confirm that DUX4 was induced by doxycycline, we measured transgene mRNA and DUX4 protein levels, as well as levels of ZSCAN4 mRNA, a direct transcriptional target of DUX4, in five individual codon-altered DUX4 and five wild-type DUX4 myoblast clones upon doxycycline induction. None of the five wild-type DUX4 clones showed induction of DUX4 mRNA, DUX4 protein, or ZSCAN4 mRNA, whereas all of the codon-altered DUX4 clones showed induction of the transgene (Fig. 1C). Three of the five codon-altered DUX4 clones—the same clones that exhibited cell death upon doxycycline addition (Fig. 1B)—showed induction of full-length DUX4 protein and activated ZSCAN4 transcription (Fig. 1C and D). In contrast, the two codon-altered DUX4 clones that did not exhibit cell death upon doxycycline addition expressed truncated forms of the DUX4 protein and did not activate ZSCAN4 expression (Fig. 1C), suggesting that the absence of cell death in these clones was due to a deletion or truncating mutation that prevented DUX4 activity and subsequent cytotoxicity. Together, these results indicate that decreasing the GC content of the DUX4 ORF likely facilitates DUX4 mRNA expression from an integrated DUX4 transgene. Although it remains to be determined, the mechanism might be due to more efficient silencing of the WT-DUX4 transgene because the inducible expression of the WT-DUX4 was initially robust and declined over time following transduction compared to the CA-DUX4, and transduction with WT-DUX4 lentivirus produced fewer puromycin resistant colonies compared to CA-DUX4 lentivirus used at equivalent titers (Supplementary Material, Fig. S1A and B), however, further work will be needed to determine whether this represents epigenetic silencing or another process.
Codon-altered DUX4 faithfully reproduces transcriptional and post-transcriptional dysregulation previously reported for wild-type DUX4
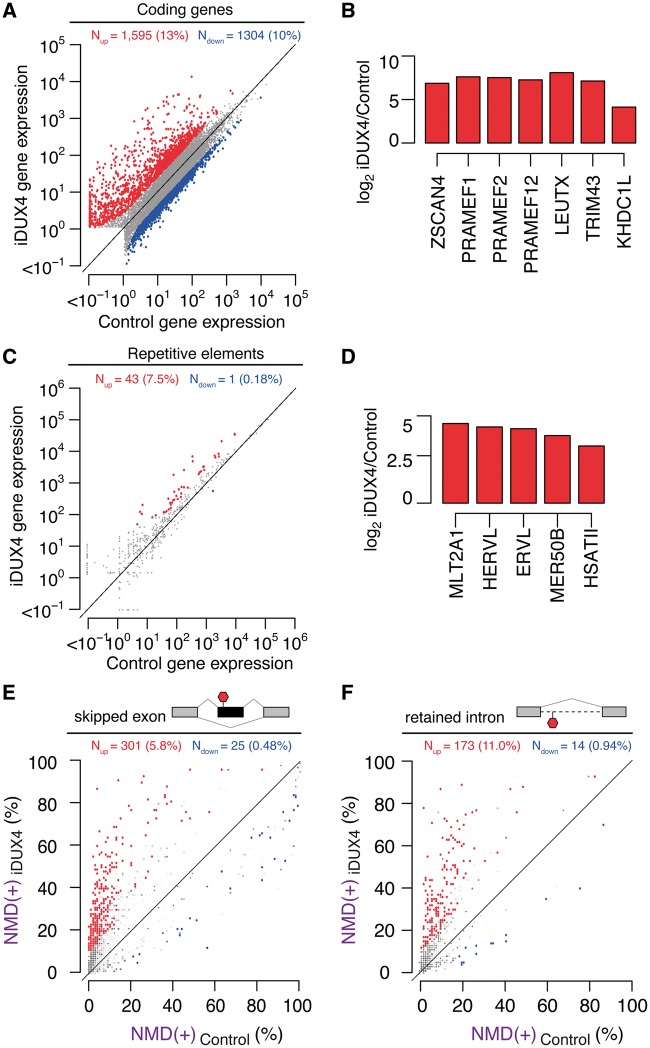
We next confirmed that codon-altered DUX4 induction drove the same transcriptional response and inhibition of RNA quality control that have been previously reported for DUX4 (8,10,15). We conducted RNA-seq on a codon-altered DUX4 myoblast cell line 14 hours after doxycycline induction (referred to as ‘iDUX4’ hereafter) or the same time point without doxycycline. We sequenced each sample to a depth of ∼50 million reads to allow accurate measurement of isoform expression. A scatter plot of gene expression showed robust DUX4-induced expression of hundreds of genes as well as mild, but statistically significant, repression of a smaller set of genes (Fig. 2A). Known targets of DUX4, such as ZSCAN4, several PRAME genes, LEUTX, TRIM43 and KHDC1L, were highly activated (Fig. 2B). Some of the most downregulated genes were involved in the immune response, such as IL7R, and the extracellular matrix, both pathways that were previously shown to be affected by DUX4 expression (by (8) and (9), respectively). Specific classes of endogenous retroviral elements and repetitive genomic sequences, including HSATII, were also upregulated, consistent with previous reports (10) (Fig. 2C and D). Finally, RNA isoforms that contain premature termination codons and are normally degraded via nonsense-mediated RNA decay (NMD) exhibited high expression in cells expressing DUX4 (Fig. 2E and F), as previously reported (15). Together, these data demonstrate that codon-altered DUX4 faithfully recapitulates the transcriptional and post-transcriptional changes that previous studies have reported for expression of wild-type DUX4 in muscle cells.
Figure 2.
Inducible expression of codon-altered DUX4 activates germline antigens, endogenous retrotransposons and repetitive elements and inhibits RNA quality control. (A) Scatter plot of gene expression (in transcripts per million) in control (uninduced) versus iDUX4 (doxycycline-induced) myoblasts. Red/blue, genes exhibiting increases/decreases of > 2.5 fold. (N) Numbers of genes with increased/decreased expression; (percentages) fraction of genes that are affected by DUX4 expression. (B) Relative mRNA levels of known DUX4 transcriptional targets in iDUX4 versus control myoblasts expressed as log2 fold-change. (C) Scatter plot of repetitive element expression (in transcripts per million) in control (uninduced) versus iDUX4 (doxycycline-induced) myoblasts. Red/blue, repeat elements exhibiting increases/decreases of > 2.5 fold. (N) Numbers of repetitive elements with increased/decreased expression; (percentages) fraction of repetitive elements that are affected by DUX4 expression. (D) Relative levels of known DUX4-activated repetitive elements in iDUX4 versus control myoblasts expressed as log2 fold-change. (E, F) Isoform ratios of predicted NMD substrates generated by cassette exon alternative splicing (E) or intron retention (F), comparing iDUX4 versus control myoblasts. Red/blue, cassette exons (E) or retained introns (F) exhibiting increases/decreases of ≥10% in isoform ratios for the isoforms that are predicted NMD substrates. Events that do not change significantly are rendered transparent.
Comparison of inducible, viral and endogenous DUX4 expression systems
A recent study of the transcriptome regulated by endogenous DUX4 in differentiated FSHD muscle cells (9) reproduced many of the changes in RNA abundance previously shown to be mediated by DUX4 in myoblasts transduced with a DUX4-expressing lentivirus (8,10,15). However, this study also reported that a large component of the gene expression signature in these FSHD myocytes only occurred in the endogenous expression system. These results raised the possibility that the delivery of exogenous DUX4 to control muscle cells might not faithfully recapitulate the changes in RNA expression mediated by endogenous DUX4 in FSHD muscle cells. However, the different datasets were acquired using different gene expression profiling platforms and analyzed using different statistical methodologies, preventing direct assessment of similarities and differences in the consequences of exogenous versus endogenous DUX4 expression.
We therefore performed a systematic comparison of DUX4-regulated changes in the transcriptome in our inducible codon-altered DUX4 expression system (iDUX4), the endogenous DUX4 expression system (enDUX4), and cells transduced with lentivirus constitutively expressing DUX4 (vDUX4). The specific datasets used in this comparison are as follows: iDUX4 represents a new dataset generated from the MB135 immortalized human myoblasts with the doxycycline inducible codon-altered DUX4 (iDUX4), performed in biological triplicate fourteen hours after DUX4 induction in growth media, with uninduced cells as a control; enDUX4 represents the published dataset of differentiated FSHD myocytes that do or do not express endogenous DUX4, as determined using a DUX4-responsive fluorescent reporter and flow sorting (9); vDUX4 represents a published dataset wherein two different myoblast cell lines (MB135 and 54-1) were transduced with a lentiviral construct that drives constitutive DUX4 expression via the PGK promoter and maintained in growth media for 24 hours (MB135) or 36 hours (54-1) prior to harvesting RNA (15,17). More information about the datasets, including the RNA-seq methodology and the number of mapped reads in each sample, is presented in Table 1.
Table 1.
Description of samples and datasets used in this study
| Sample names | Sample description | RNA-seq details | Number of replicates | RNA-seq read count (control, dux4) |
|---|---|---|---|---|
|
Stable DUX4 expression induced with doxycycline; uninduced sample serves as Control | 50bp SR | 1 |
|
| iDUX4, control | Stable DUX4 expression induced with doxycycline; uninduced sample serves as Control | 100bp SR | 3 |
|
| enDUX4, Control | FSHD patient-derived cell line expressing DUX4 spontaneously during differentiation. DUX4-positive cells isolated by sorting for DUX4-induced BFP. | 50bp PE, treated as 50bp SR | 6 |
|
| vDUX4, Control | DUX4 expression via lentiviral delivery; samples expressing GFP via lentiviral infection serve as control. | 100bp SR | 2 |
|
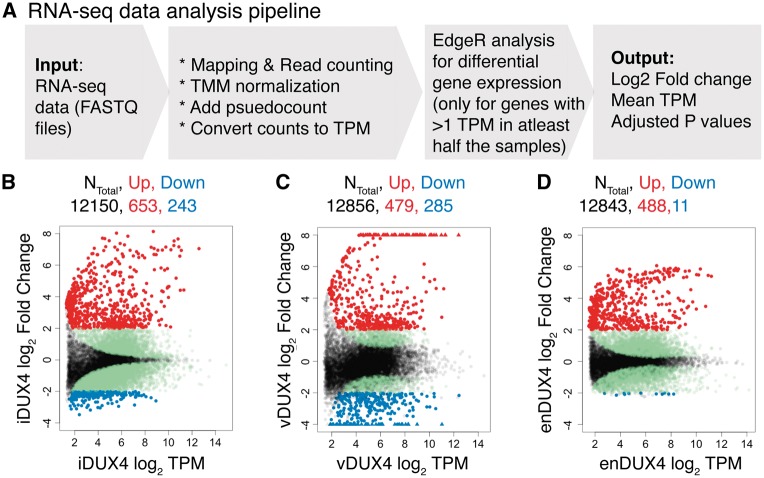
Distinct RNA-seq analysis strategies and statistical methodologies can produce very different results, including the identification of only partially overlapping sets of differentially expressed genes in a given dataset (18). To avoid this confounding factor, we quantified differential gene expression in the enDUX4, iDUX4, and vDUX4 datasets using a common read mapping and analysis pipeline (Fig. 3A; described in Materials and Methods).
Figure 3.
Transcriptional response of endogenous and exogenous DUX4 expression in human myoblasts. (A) Schematic representation of the RNA-seq data analysis pipeline. (B–D) MA plots for inducible, viral and endogenous DUX4-induced transcriptomes. Genes upregulated by more than 4-fold in red; genes downregulated by more than 4-fold in blue; genes with a significant adjusted P-value (< 0.05) that do not meet 4-fold cutoff for differential expression in green and the genes with adjusted P-value > 0.05 in black. For the vDUX4 sample, genes with log2 fold-change >8 or < -4 are plotted as ‘triangles’ at the top and bottom edges of the plot, respectively.
We first compared the global extent of gene induction versus repression associated with DUX4. Plotting average gene expression versus DUX4-associated change in gene expression (“MA” plots) revealed that DUX4 preferentially caused gene upregulation, as expected, in all three datasets. However, the global extent of gene induction or repression varied between the different models of DUX4 expression (Fig. 3B and D, Supplementary Material, Table S1). Endogenous DUX4 expression was associated with more modest changes in gene expression relative to ectopic DUX4 expression, particularly for the few genes exhibiting downregulation in DUX4-expressing cells. In contrast, a substantial number of genes were downregulated in the vDUX4 dataset, likely due to suppression of the innate immune response to the lentiviral transduction by DUX4 (described in detail below).
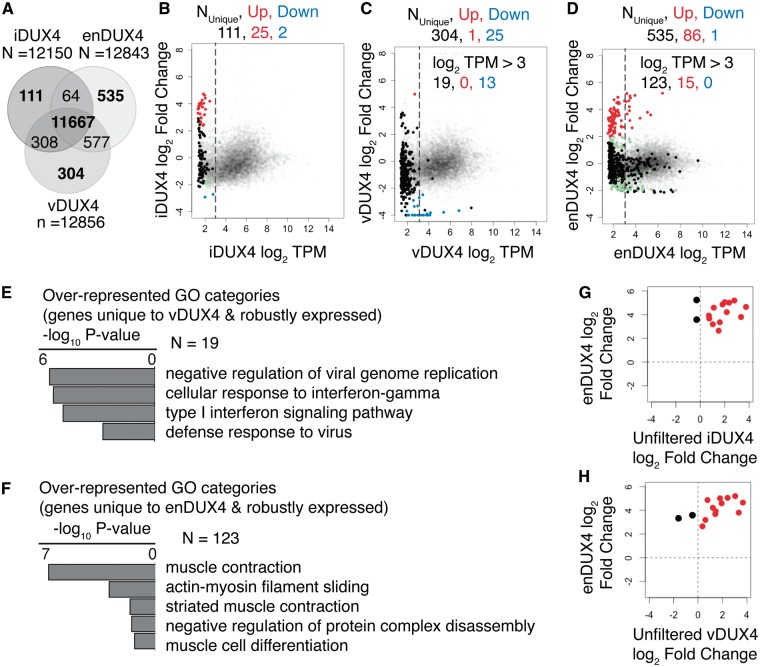
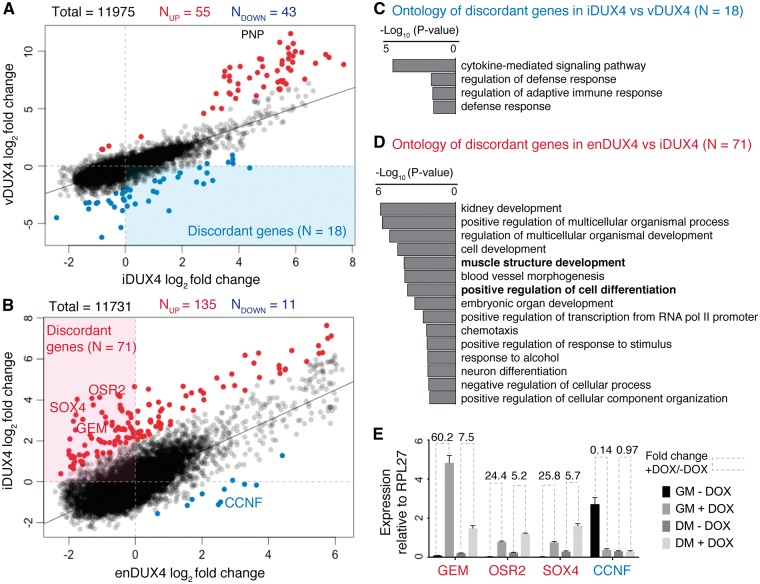
We next tested whether identical sets of genes were expressed in each dataset (irrespective of DUX4 expression). We noted that each dataset contained a set of genes that were not identified as expressed in the other two samples (Supplementary Material, Table S2). (We defined expressed genes as those with a mean expression of at least one transcript per million (TPM), the standard filter recommended by the edgeR package for RNA-seq data analysis (19)). A Venn diagram of the overlap of detected genes among the three datasets (Fig. 4A) showed that the number of dataset-specific genes was particularly large for enDUX4 (n = 535). In comparison, vDUX4 had fewer data-set-specific genes (n = 304), while iDUX4 elicited even less data-set-specific gene expression (n = 111). There are two possible reasons for a gene to be identified as differentially expressed in a given sample: a gene might be robustly expressed only in one dataset, or a gene might be expressed near the detection threshold in all datasets and therefore not reproducibly detectable across all samples. An MA plot highlighting only the dataset-specific genes present in the iDUX4, vDUX4 and enDUX4 datasets showed that most, but not all, such genes were expressed at very low levels in all datasets (Fig. 4B–D). Therefore, to eliminate the noise generated by genes expressed at low levels (based on these MA plots), we limited our analysis to genes expressed above a minimal threshold, defined as an average of 8 TPM (log2 TPM of 3; dotted lines in Fig. 4B–D).
Figure 4.
Genes unique to endogenous DUX4 expression are most relevant to muscle differentiation. (A) Venn diagram showing the overlap between the detected genes in iDUX4, enDUX4 and vDUX4 samples. (B–D) MA plot for inducible, viral and endogenous DUX4-induced transcriptome, highlighting the genes that were uniquely detected in each of the samples. Dotted line represents log2 TPM of 3. Color scheme is same as that of Figure 3B–D. (E–F) Gene Ontology (GO) analysis for the genes unique to vDUX4 and enDUX4, respectively, and expressed at a level above 8 TPM (log2 TPM of 3). (G-H) Scatter plot of log2 fold-change of genes robustly and uniquely upregulated by enDUX4 versus log2 fold-change for the corresponding genes by iDUX4 (G) and vDUX4 (H) calculated without applying a filter for low expressing genes. Black dots represent the discordant genes.
We next sought to understand the biological functions of the minorities of genes that were uniquely expressed in particular datasets. We identified 19 and 123 such genes, respectively, in the vDUX4 and enDUX4 datasets. Gene Ontology (GO) analysis of the 19 genes uniquely expressed in vDUX4 showed that they were mostly genes involved in the viral immune response, consistent with the viral mode of transgene delivery used for vDUX4 (Fig. 4E). The genes unique to enDUX4, on the other hand, were preferentially involved in muscle differentiation or muscle function (Fig. 4F). This enrichment for myogenic processes is likely due to the fact that the enDUX4 dataset assayed differentiated myocytes, whereas the iDUX4 and vDUX4 datasets were derived from undifferentiated, replicating myoblasts.
Of the 123 genes uniquely and robustly detected in the enDUX4 dataset, 15 were upregulated by enDUX4 (log2 fold-change > 2) Fig. 4B–D), indicating that they might be DUX4 targets activated only in the context of myogenesis. If so, then despite their low number, such genes could be very relevant to the FSHD disease process since DUX4 is presumed to be expressed during myogenic differentiation in vivo. Hence, we asked how iDUX4 and vDUX4 affected the expression of these 15 genes, which are poorly expressed in myoblasts and so did not pass the filter for robust expression. We repeated the edgeR analysis for these samples without applying a filter for low expression. We found that 12 of these 15 genes were activated by iDUX4 and vDUX4 (Fig. 4G and H). The three discordant genes—MBD3L4 and FAM151A in iDUX4 and AIRE and FAM151A in vDUX4—showed uncertain RNA-seq read assignment in the enDUX4 samples (data not shown). Accurate read mapping to members of multigene families, such as MBD3L4 and FAM151A, and subsequently quantifying gene expression is a known challenge in bioinformatics (20). For subsequent analyses, we restricted comparisons to only those genes reliably detected and quantified with an average expression greater than 8 TPM in one of the samples being compared, as appropriate.
In summary, while some genes do exhibit dataset-specific expression patterns, our analyses indicate that these differences are primarily due to the distinct basal transcriptomes of myoblasts (iDUX4) versus differentiated myocytes (enDUX4), as well as gene expression changes in response to viral transduction of myoblasts (vDUX4).
Endogenous and exogenous DUX4 produce highly similar transcriptional responses
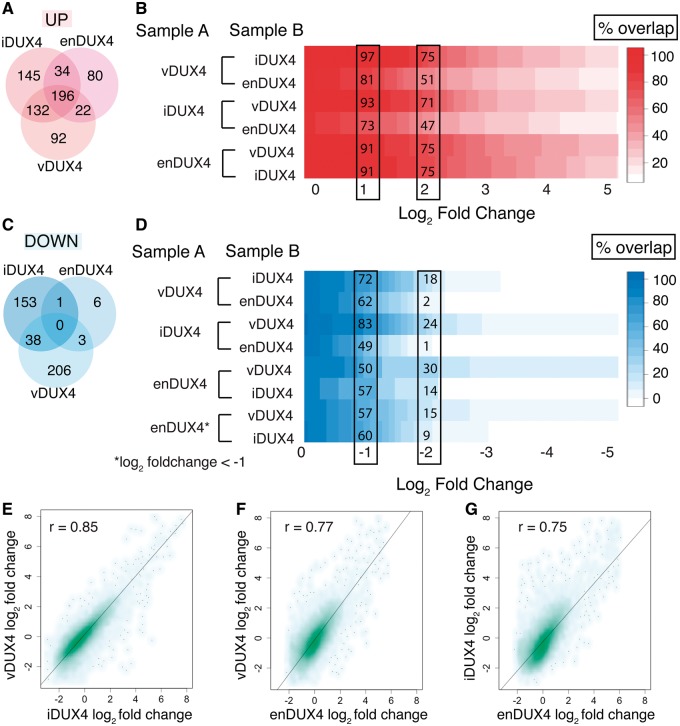
We next measured the similarity in differential gene expression caused by expression of iDUX4, vDUX4, and enDUX4. Venn diagrams are commonly used to assess similarities and differences between datasets and to represent the overlap between lists of significantly up or downregulated genes. For this purpose, a log2 fold-change cutoff of two is often chosen (9). However, it is clear from the MA plots (Fig. 3B–D) that DUX4 activates genes to different extents in the different datasets, such that a particular gene might be induced two-fold in one dataset but three-fold in another. Therefore, using a single, arbitrarily defined fold-change cutoff to select up- or downregulated genes might make the datasets seem artificially dissimilar. We therefore used a more statistically rigorous approach to further explore the differences between the DUX4 expression systems. We measured the percentage overlap between the sets of significantly up- or downregulated genes across a wide range of fold-change cutoffs (Fig. 5A–D). For instance, of all of the genes that were upregulated by more than four-fold (adjusted P-value < 0.05) by vDUX4, 75% (310 genes) and 51% (215 genes) were also upregulated by more than four-fold in iDUX4 and enDUX4, respectively (Fig. 5A–B). As the (arbitrarily defined) fold-change cutoff is relaxed for the comparator group to two-fold, the sets of upregulated genes become increasingly concordant (97% and 81% overlap). This dependence on the fold-change threshold is even more striking for the sets of downregulated genes. There is poor overlap at a cutoff of four-fold, but reasonable concordance at a comparator group cutoff of two-fold (Fig. 5C and D). Moreover, pairwise scatter plots of the fold-changes of the three datasets showed that transcriptional changes caused by DUX4 were indeed highly similar (Pearson’s correlation coefficient, r ∼ 0.75–0.85; P-values < 2.2e-16), despite major differences in cell type (myoblasts versus differentiated muscle cells), timing, expression levels, culture conditions, and the mechanism of expressing the DUX4 protein (Fig. 5E–G). Together, these statistical analyses indicate that DUX4-responsive genes are consistently induced or repressed across the three datasets, although the magnitude of induction or repression may differ.
Figure 5.
Regulated gene sets show significant overlap between samples. (A, C) Venn diagram of upregulated genes showing the overlap between genes with > 2 log2 fold-change (A) or < -2 log2 fold-change (B) and an adjusted P-value < 0.05 in the three datasets. (B) Percent overlap plot shows the overlap of gene sets that are > 2 log2 fold upregulated in Sample A with a significant adjusted P value over a sliding scale of 0 to 5 log2 fold upregulation in Sample B. (D) Percent overlap plot shows the overlap of gene sets that are < -2 log2 fold downregulated in Sample A with a significant adjusted P value over a sliding scale of 0 to -5 log2 fold downregulation in Sample B. (E–G) Scatter plot of log2 fold-change of quantifiable genes in inducible versus viral DUX4 expression (E), endogenous versus viral DUX4 expression (F), and endogenous versus inducible DUX4 expression (G). r - Pearson’s correlation coefficient.
Biological differences between DUX4 expression systems arise from the distinct cellular contexts used
Our comparisons of DUX4-regulated genes revealed highly similar patterns of induction and repression across all datasets, but there were small subsets of genes that responded to DUX4 expression only in particular datasets. In order to understand the origins of these differences, it is important to consider the different cellular contexts in which DUX4 was expressed in each dataset. For example, in the vDUX4 dataset, the lentiviral delivery of DUX4 induced an antiviral innate immune response. In the enDUX4 dataset, the muscle cells were differentiated in the presence of the calcium chelator EGTA to prevent fusion and facilitate FAC sorting, whereas vDUX4 and iDUX4 were expressed in undifferentiated myoblasts. When we plotted normalized log2 fold-changes of all of the genes expressed in the iDUX4 dataset versus the vDUX4 or enDUX4 datasets, we identified a small subset of genes that were comparatively over- or under-expressed in each sample (Fig. 6A and B). For the genes that were comparatively under-induced in the vDUX4 dataset compared to the iDUX4 dataset, the top five enriched GO categories all corresponded to the cellular defense response (Fig. 6C). This enrichment is consistent with our expectation that a lentiviral vector will induce an immune response, as well as the fact that lentiviral GFP induces a stronger response relative to lentiviral DUX4, as DUX4 suppresses the innate immune response (8). Genes that were comparatively more induced in the iDUX4 dataset compared to the enDUX4 dataset were enriched for processes including cell development and differentiation (Fig. 6D), suggesting that the degree of fold-change might be affected by the state of muscle differentiation.
Figure 6.
Differentially regulated genes appear most relevant to the gene expression programs underway during DUX4 expression. (A) Scatter plot of scaled and centered log2 fold-change values of iDUX4 and vDUX4. Line represents a linear model and the genes marked in red are more over-expressed in vDUX4 compared to iDUX4 (residual > 2) and those in blue are more under-expressed in vDUX4 compared to iDUX4 (residual < -2). (B) Scatter plot of scaled and centered log2 fold-change values of enDUX4 and iDUX4. Line represents a linear model and the genes marked in red are more over-expressed in iDUX4 compared to enDUX4 (residual > 2) and those in blue are more under-expressed in iDUX4 compared to enDUX4 (residual < -2). (C–D) GO category analysis of significantly under-expressed genes in vDUX4 compared to iDUX4 (C) and significantly over-expressed genes in iDUX4 compared to enDUX4 (D). (E) qPCR data for a few candidate discordant genes in control and iDUX4 cells in growth media (GM) versus differentiation media (DM).
To confirm that myogenic differentiation is the major contributor to the small differences between the DUX4-induced genes in the iDUX4 and enDUX4 datasets, we induced codon-altered DUX4 with doxycycline (DOX) in the MB135 myoblasts in growth media (GM) and the same cells differentiated into myotubes in differentiation media (DM). RT-qPCR analysis of a set of genes that were relatively repressed in the enDUX4 dataset compared to the iDUX4 dataset showed a similar trend of relative repression in differentiated iDUX4 myotubes compared to undifferentiated iDUX4 myoblasts (Fig. 6E). Together, these results suggest that the differences in gene expression between the enDUX4 and vDUX4 or iDUX4 datasets were primarily due to differences in baseline gene expression in undifferentiated versus differentiated cells, rather than differences in the intrinsic activities of endogenous or exogenous DUX4.
The expression pattern common across the different modes of DUX4 expression recapitulates context-independent functions of DUX4
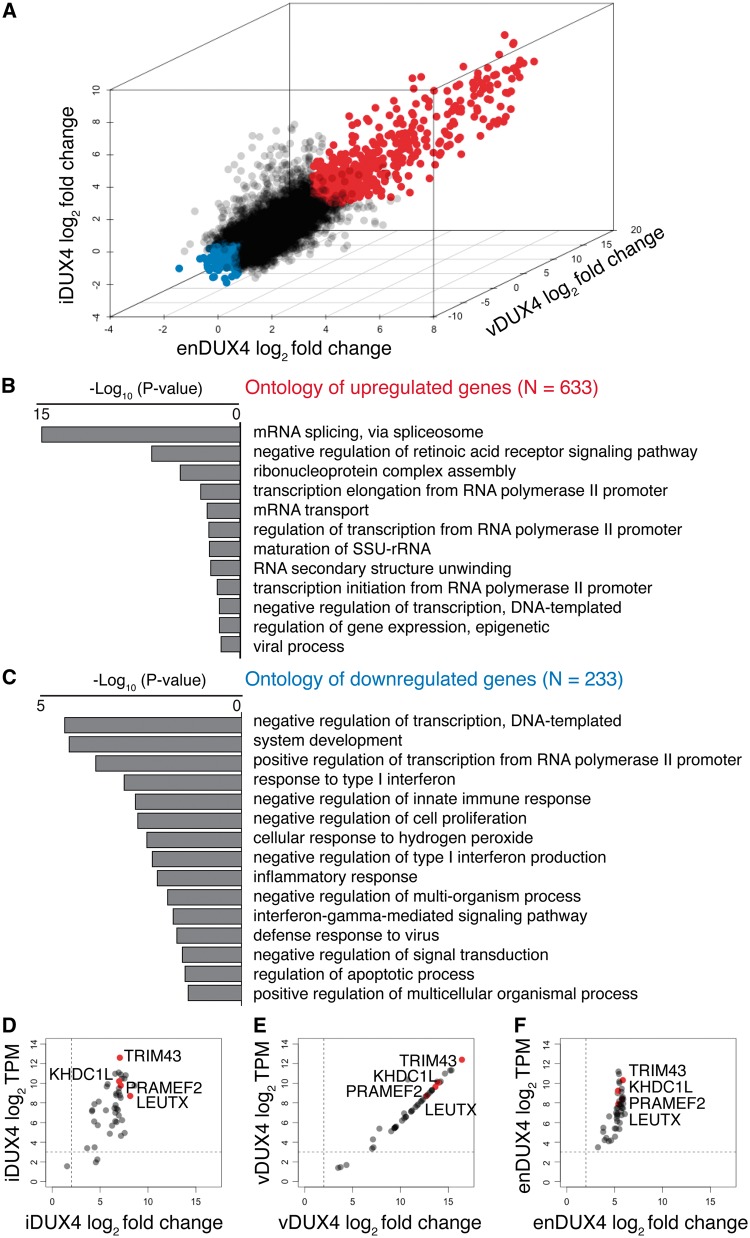
Having explored the differences between the endogenous and exogenous DUX4 expression systems, we next sought to characterize the gene expression program that is common to the different systems. To this end, we performed Gene Ontology analyses of the genes that were induced or repressed by more than two-fold by iDUX4, vDUX4 and enDUX4 (Fig. 7A and Supplementary Material, Table S4). The genes that were upregulated by DUX4 in all three expression systems were enriched for proteins involved in transcription, RNA processing, splicing and transport (Fig. 7B), as has been observed before (8,9). The downregulated genes were involved in viral defense, cell proliferation, and apoptosis, among other gene classes (Fig. 7C). Next, we asked if the FSHD biomarkers previously identified via transcriptome analysis of FSHD muscle biopsies (17) showed activation across all three datasets. Out of the 67 biomarker genes identified by Yao et al. (17), 47 were annotated and/or detectably expressed in the iDUX4, vDUX4 and enDUX4 datasets and all showed high upregulation by both exogenous and endogenous DUX4 as shown by the MA plot (Fig. 7D–F). In conclusion, the core activity of DUX4 is highly similar across different modes of DUX4 expression and the three model systems compared in this study each recapitulate key transcriptome changes that can be found in FSHD muscle.
Figure 7.
Gene sets common to endogenous and exogenous DUX4 expression highlight the core functions of DUX4. (A) 3D scatter plot for the three datasets (iDUX4, enDUX4 and vDUX4) highlighting the genes upregulated by more than 2 log2 fold-change in all samples in red and those downregulated by more than 2 log2 fold-change in all samples in blue. (B) GO analysis of the upregulated genes (marked ‘red' in 6A). (C) GO analysis of downregulated genes (marked ‘blue' in 6A). (D–F) MA plot for 47 biomarkers identified by Yao et al. (17) for the iDUX4 (D), vDUX4 (E) and enDUX4 (F) datasets; The four high-confidence biomarkers (LEUTX, PRAMEF2, TRIM43, KHDC1L) are marked in ‘red'. The horizontal dotted line represents TPM of 8; the vertical dotted line represents fold change of 4.
Discussion
It is important to compare different cellular models of DUX4 expression to determine whether particular models are more or less appropriate for studying FSHD biology. In this study, we found that quite different models of DUX4 expression yielded very similar patterns of DUX4-induced transcriptional changes. While there were differences between systems, such differences were largely explained by the differentiation state of the cells or culture conditions used, which gave rise to distinct patterns of basal gene expression. Overall, the high degree of overlap between DUX4-regulated genes identified across distinct cellular contexts strongly indicates that all three models recapitulate important aspects of FSHD biology.
The current study was motivated, in part, by a prior publication that identified differences in gene expression among different models of FSHD (9). While single fold-change cutoffs are useful for defining gene sets of interest within a dataset, they are not statistically robust when comparing multiple different datasets, as arbitrary cutoffs generate “edge effects”. This statistical effect might explain why this previous study identified relatively little overlap between the differentially expressed genes among different models of FSHD. Correlation analyses conducted here provide strong assurance that studies using different model systems of FSHD can be compared with confidence, provided that the biological state of the cells or variables induced by the experimental design are taken into account.
Consistent with the above conclusions, it is further reassuring that our study, using a newly developed doxycycline-inducible codon-altered DUX4, identified the same DUX4-induced gene expression changes and RNA processing abnormalities reported by prior studies (8,10,15). Our earlier attempts to create an inducible DUX4 were hampered by the difficulty of establishing a stable integrant that was efficiently induced, which was true for multiple different viral preparations and transduced cell types. Decreasing the CG content and the number of CpG dinucleotides substantially increased the recovery of clones with an inducible DUX4, suggesting that the high CG content of the DUX4 coding sequence might trigger silencing, however, the precise mechanism for the more efficient induction of the stably integrated CA-DUX4 needs further study. The similarity of the genes regulated by the codon-altered DUX4 to genes identified in studies using the wild-type DUX4 indicates that the gene expression changes are due to the DUX4 protein and not the RNA, consistent with our prior studies (21). Finally, we expect that the human myoblast cell line expressing doxycycline-inducible DUX4 described here will be a useful tool for FSHD research. This cell line allows the isolation of a large number of clonal cells with synchronized induction of DUX4 expression, which we expect will improve reproducibility and allow for accurate temporal dissection of the molecular events following DUX4 expression.
Materials and Methods
Accession codes
The raw sequence reads for the enDUX4 expression experiments were downloaded from the NCBI sequence read archive (SRA) database under accession number SRP058319 (9). Data generated in this study are available through the NCBI SRA database under accession number GSE85461.
Cell culture
Proliferating human myoblasts were cultured in F10 medium (Gibco/Life Technologies) supplemented with 20% fetal bovine serum (Thermo Scientific), 10ng bFGF (Life Technologies), 1µM dexamethasone (Sigma) and 50U/50µg penicillin/streptomycin (Life Technologies). Differentiation into myotubes was initiated by switching the fully confluent cell monolayer into a low-serum media such as DMEM with 1% horse serum (Life Technologies), supplemented with 10 µg/ml each of insulin and transferrin for 48 hours. To induce DUX4 expression in differentiated iDUX4 cells, doxycycline was added in the last 14 hours of differentiation. 293T cells were cultured in DMEM (Gibco/Life Technologies) supplemented with 10% fetal bovine serum and 50U/50µg penicillin/streptomycin as above.
Codon-altered and wild-type inducible constructs
Wild-type DUX4 was subcloned into the pCW57.1 vector, a gift from David Root (Addgene plasmid #41393) by restriction enzyme digest, using the NheI and SalI sites of the pCW57.1 vector. The codon-altered DUX4, which has ∼73% identity to wild-type DUX4, was synthesized by IDT custom gene synthesis and subcloned into pCW57.1 such that the only discrepancies between the codon-altered and wild-type constructs are within the coding region itself. Sequence is in Supplementary Material, Fig. S2.
Generation of clonal cell lines expressing DUX4
Lentivirus with the inducible wild-type and codon-altered DUX4 transgenes were generated by transfection of the appropriate pCW57.1 vector into 293T cells, along with the packaging and envelope plasmids pMD2.G and psPAX2 using lipofectamine 2000 reagent (ThermoFisher). To generate clonal lines, control human myoblasts, MB135, immortalized with hTERT and CDK4, were plated at low density and transduced with lentivirus at a low multiplicity of infection (MOI < 1) in the presence of polybrene. Cells were selected and maintained in puromycin. After the selection was complete, remaining cells were allowed to grow and form colonies. Individual clones that were well isolated were picked using cloning cylinders, about 10 days after transfection, and expanded in the presence of puromycin. Five individual codon-altered DUX4 clones and 15 wild-type DUX4 clones were picked and used in this study.
RNA extraction and RT-qPCR
Total RNA was extracted from whole cells using either TRIzol reagent (Ambion) or NucleoSpin RNA kit (Macherey-Nagel) following the manufacturer's instructions. Purified RNA was Dnase I treated (ThermoFisher) and heat inactivated prior to cDNA synthesis. First strand cDNA synthesis was performed using SuperScript III reverse transcriptase (ThermoFisher) and following the manufacturer's instructions. Samples were split into duplicate reactions where one underwent a mock, no enzyme treatment as a control. Quantitative PCR was carried out on cDNA using the standard curve method and SYBR green as the detector. The primers used in this study are listed below:
GEM_1F: GAAAAGAACCCCTGGAACGTG
GEM_1R: TGTACTGGTGGGGCTCTTTC
OSR2_1F: TGCCCAGGTTGACCTTTCTG
OSR2_1R: CTGAGGGGACCAACCCTTTC
SOX4_1F: ATCGCTGTTTGGATTTCCTG
SOX4_1R: ACACTGGTGGCAGGTTAAGG
CCNF_1F: GACCATCTTGAGTCTCCCCG
CCNF_1R: AAGAGCTTCAGGTTCCCTGG
RPL27-1L: GCAAGAAGAAGATCGCCAAG
RPL27-1R: TCCAAGGGGATATCCACAGA
ZSCAN4_F: FTGGAAATCAAGTGGCAAAAA
ZSCAN4_R: RCTGCATGTGGACGTGGAC
KHDC1L_F: CACCAATGGCAAAGCAGTGG
KHDC1L_R: TCAGTCTCCGGTGTACGGTG
Protein extraction and immunoblotting
Cells were directly lysed in 2X gel loading buffer with 4% BME, sonicated, and boiled for 10 minutes. Samples were run on a 4–12% polyacrylamide gel and transferred to a PVDF membrane. Membranes were blocked in 5% milk for one hour before overnight incubation with primary antibody at 4°C. Membranes were incubated with secondary antibody for one hour, and the chemiluminescent signal was detected on film. The antibodies used in this study are rabbit anti-DUX4 (E14-3) and rabbit anti-H3 (Abcam; ab1791).
RNA-seq library preparation and sequencing
The RNA-seq libraries were prepared with polyA-selected RNA (starting with 1 µg of total RNA) using TruSeq RNA Sample Prep Kit (Illumina) either manually (for the pilot dataset in Fig. 2) or using a Perkin Elmer Sciclone NGSx Automated Library Prep Workstation (for the triplicate dataset in Fig. 3). Library size distributions were validated using an Agilent 2100 Bioanalyzer (Agilent Technologies). Indexed libraries were quantified using Qubit® 2.0 Fluorometer (Life Technologies) and pooled for optimal clustering. Sequencing on an Illumina HiSeq 2500 was carried out by the FHCRC Genomics Shared Resource to generate 50 bp single-end reads for the pilot singleton iDUX4 dataset (Fig. 2) and 100 bp single-end reads for the iDUX4 and vDUX4 datasets with replicates (Fig. 3). Image analysis and base-calling were performed using Real time Analysis (RTA) version v1.18 (Illumina), followed by demultiplexing of indexed reads to generate FASTQ files using bcl2fastq Conversion Software v1.8.4 (Illumina).
RNA-seq data analysis
The general pipeline for RNA-seq data analysis is presented in Fig. 3A. Briefly, RNA-seq reads were mapped to the UCSC hg19 (NCBI GRCh37) human genome assembly using bowtie (22), RSEM (23) and TopHat (24), as described by Dvinge et al. (25). Two mismatches were allowed for 50bp reads and three mismatches allowed for 100bp reads. Differential gene expression analysis was performed using the edgeR package (19) as follows: RNA transcript levels normalized using trimmed mean of M value (TMM; (26)) were input to the edgeR program in transcripts per million (TPM). DUX4-expressing samples were compared to the corresponding control samples in biological triplicates (iDUX4), duplicates (vDUX4) or sextuplicates (enDUX4). Only genes expressed with at least 1 TPM in 50% of the samples in a dataset (i.e. in either the controls or the DUX4 samples) were considered for differential expression to avoid noise from poorly expressed genes. edgeR output of log2 fold-change, average TPM and false discovery rate (corrected for multiple hypothesis testing by Benjamini-Hochberg approach) were used in all subsequent analysis (processed data is provided in Supplementary Material, Table S1). All plots were generated using R plotting functions and/or the ggplot2 package (27). All statistical tests were also performed using R functions.
GO category analysis
GO category analysis was conducted using the PANTHER classification system (http://pantherdb.org/geneListAnalysis.do; (28)) using the statistical overrepresentation test against all human genes, using the complete GO biological process annotation. P-values were corrected for multiple hypothesis testing using the Bonferroni correction.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Bradley and Tapscott laboratories for helpful discussions.
Conflict of Interest statement. None declared.
Funding
This work was supported by NIH/NINDS P01 NS069539 (RKB, SJT), PHS NRSA T32 GM007270 NIGMS (SCS), NIH/NHGRI T32 HG00035 (SCS), the Ellison Medical Foundation AG-NS-1030-13 (RKB), Friends of FSH Research (SJT) and the FSH Society FSHS-22014-01 (SJ).
References
- 1. Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet., 2, 26–30. [DOI] [PubMed] [Google Scholar]
- 2. Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O., et al. (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord., 17, 611–623. [DOI] [PubMed] [Google Scholar]
- 3. Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F., et al. (2008) An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. Embo J., 27, 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppee F., Belayew A. (2011) The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One, 6, e26820.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. (2011) DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol., 69, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dandapat A., Bosnakovski D., Hartweck L.M., Arpke R.W., Baltgalvis K.A., Vang D., Baik J., Darabi R., Perlingeiro R.C., Hamra F.K., et al. (2014) Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep., 8, 1484–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. (2010) A unifying genetic model for facioscapulohumeral muscular dystrophy. Science, 329, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., et al. (2012) DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell, 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rickard A.M., Petek L.M., Miller D.G. (2015) Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet., 24, 5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Young J.M., Whiddon J.L., Yao Z., Kasinathan B., Snider L., Geng L.N., Balog J., Tawil R., van der Maarel S.M., Tapscott S.J. (2013) DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet., 9, e1003947.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bosnakovski D., Choi S.H., Strasser J.M., Toso E.A., Walters M.A., Kyba M. (2014) High-throughput screening identifies inhibitors of DUX4-induced myoblast toxicity. Skelet. Muscle, 4, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G. (2013) Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet., 22, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banerji C.R., Knopp P., Moyle L.A., Severini S., Orrell R.W., Teschendorff A.E., Zammit P.S. (2015) beta-Catenin is central to DUX4-driven network rewiring in facioscapulohumeral muscular dystrophy. J. R. Soc. Interface, 12, 20140797.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Homma S., Beermann M.L., Boyce F.M., Miller J.B. (2015) Expression of FSHD-related DUX4-FL alters proteostasis and induces TDP-43 aggregation. Ann. Clin. Transl Neurol., 2, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng Q., Snider L., Jagannathan S., Tawil R., van der Maarel S.M., Tapscott S.J., Bradley R.K. (2015) A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. Elife, 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lek A., Rahimov F., Jones P.L., Kunkel L.M. (2015) Emerging preclinical animal models for FSHD. Trends Mol. Med., 21, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet., 23, 5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soneson C., Delorenzi M. (2013) A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics, 14, 91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y., McCarthy D., Robinson M., Smyth G.K. (2011) edgeR: differential expression analysis of digital gene expression data User's Guide.http://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf; Date Last accessed, September 16, 2016.
- 20. Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szczesniak M.W., Gaffney D.J., Elo L.L., Zhang X., et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol., 17, 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R., et al. (2009) RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet., 18, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li B., Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dvinge H., Ries R.E., Ilagan J.O., Stirewalt D.L., Meshinchi S., Bradley R.K. (2014) Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc. Natl Acad. Sci. U S A, 111, 16802–16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson M.D., Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol., 11, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wickham H. (2009) ggplot2: elegant graphics for data analysis. Springer, New York. [Google Scholar]
- 28. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res., 44, D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.