Abstract
Clostridium perfringens, a rapid-growing pathogen known to secrete an arsenal of >20 virulent toxins, has been associated with intestinal diseases in both animals and humans throughout the past century. Recent advances in genomic analysis and experimental systems make it timely to re-visit this clinically and veterinary important pathogen. This Review will summarise our understanding of the genomics and virulence-linked factors, including antimicrobial potentials and secreted toxins of this gut pathogen, and then its up-to-date clinical epidemiology and biological role in the pathogenesis of several important human and animal-associated intestinal diseases, including pre-term necrotising enterocolitis. Finally, we highlight some of the important unresolved questions in relation to C. perfringens-mediated infections, and implications for future research directions.
Introduction
Clostridium perfringens (formerly known as Bacillus aerogenes capsulatus, Bacillus perfringens, Bacillus welchii or Clostridium welchii) is a Gram positive, spore-forming, anaerobic, rod-shaped bacterium1. It was first isolated and identified as a novel bacterium in 1891 by William H. Welch from the autopsy of a 38-year-old man, where gas bubbles were observed within infected blood vessels. This gas-forming trait (the original name Bacillus aerogenes capsulatus, ‘aerogenes’ literally means ‘air-producing’ in Latin) was later linked to gas gangrene symptoms seen in British and French soldiers during World War I2.
C. perfringens is associated with diverse environments including soils, food, sewage, and as a member of the gastrointestinal (GI) tract microbial community (i.e., microbiota) of both diseased, and non-diseased humans and animals. Notably it has been associated with humans for thousands of years as evidenced by the recent identification of C. perfringens using next generation sequencing (NGS) technology in the mummified GI tract of a >5000-year-old Neolithic ‘Tyrolean Iceman’ (also known as Ötzi), found in an Alpine glacier in 19913.
Clinically, C. perfringens has been constantly associated with various significant systemic and enteric diseases, in both humans and animals, including gas gangrene (Clostridial myonecrosis), food poisoning and non-foodborne diarrhoea, and enterocolitis4,5. Importantly, C. perfringens strains are known to secrete >20 identified toxins or enzymes that could potentially be the principal virulence factors involved in pathophysiology6.
In this Review, we explore phenotypic and genomic features of this important and re-emerging pathogen, including virulence factors, and antimicrobial resistance (AMR) profiles, and the clinical impact of this bacterium in relation to numerous medically important intestinal diseases, across several host species. Finally, we highlight some of the important unresolved questions in relation to C. perfringens-associated infections, and implications for future research directions.
Isolation, identification and typing methods
There are numerous methods for isolation of C. perfringens, and biochemical and molecular methods for identification as detailed in Table 1.
Table 1.
Common isolation, identification and typing methods used in C. perfringens research
| Method | Method details in brief | Refs |
|---|---|---|
| Isolation | ||
| Direct plating | Direct plating on TSC-EYA + 18–24 h anaerobic incubation at 37 °C. Pitch black colonies with opaque halos are presumptively C. perfringens. | 146 |
| Ethanol pre-treatment | Ethanol pre-treatment (50% ethanol) for 30 mins + plating on Fastidious Anaerobe Agar (sometimes supplemented with 0.1% taurocholate). Colonies that exhibit beta-haemolysis are preliminarily identified as C. perfringens. | 147 |
| Biochemical identification | ||
| Nagler’s reaction | Known as lecithinase (alpha-toxin) test. Egg-yolk agar plates are split into two halves, where one half contains anti-alpha-toxin, and following anaerobic incubation, positivity is defined as turbidity around colonies on the anti-alpha-toxin-free side which confirms C. perfringens. | 148 |
| Reverse CAMP test | Streptococcus agalactiae is streaked on blood agar, and C. perfringens streaked perpendicular to S. agalactiae. After 24–48 h of anaerobic incubation, a ‘bow-tie’ zone will form due to the synergistic haemolysis, this confirms C. perfringens. | 149 |
| Molecular identification | ||
| 16S ribosomal RNA PCR | Appropriating full-length 16S rRNA universal primers/smaller region of 16S rRNA gene in PCR to amplify 16S rRNA gene + sequencing, and identify using informatics approach (based on sequence similarity >97% to assign taxonomy). | 30 |
| MALDI-TOF Mass Spectrometry | Rapid and inexpensive identification method based upon the mass spectrum analysis of highly-conserved ribosomal proteins from whole bacterial cells—apply bacterial colonies straight onto the MALDI-TOF metal target and followed by appropriate treatments and analysis. | 4 |
| Typing | ||
| Multiplex PCR | Multiplex PCR approach is commonly used to amplify key toxins genes to classify C. perfringens into 7 (A to G) different toxinotypes according to the toxin genes combination. | 150 |
| Genome-wide sequence search | Genome-wide search on relevant toxin genes using sequence similarity search program e.g. BLAST for toxinotyping on C. perfringens genomes. | 10 |
As C. perfringens is a pathogen, typing methods are currently used to differentiate between strains that may be associated with serious infection (Table 1). C. perfringens strains, clinically well-known for toxin production, are typed (recently updated for seven toxinotypes: A–G; previously only types A–E7) according to the combination of typing toxins (Fig. 1) they produce, i.e., α-toxin, β-toxin, ε-toxin and ι-toxin, enterotoxin (CPE) and NetB8. Historically this method indicated that certain toxins were associated with certain hosts and diseases e.g., type B (particularly the β-toxin it harbours), is exclusively linked to dysentery in sheep, and rarely seen in other hosts9. Food-poisoning associated CPE was genotyped typically in type F strains (previously named as CPE-positive type A; not to be confused with heat-resistant type C strains) although CPE can also be produced by certain type C, D and E strains, whereas β2-toxin and θ-toxin (perfringolysin O; PFO) could be found in any toxinotypes1,10,11. However, it is important to note that no single strain is known to produce the entire panoply of toxins10.
Fig. 1. C. perfringens current toxinotyping system.
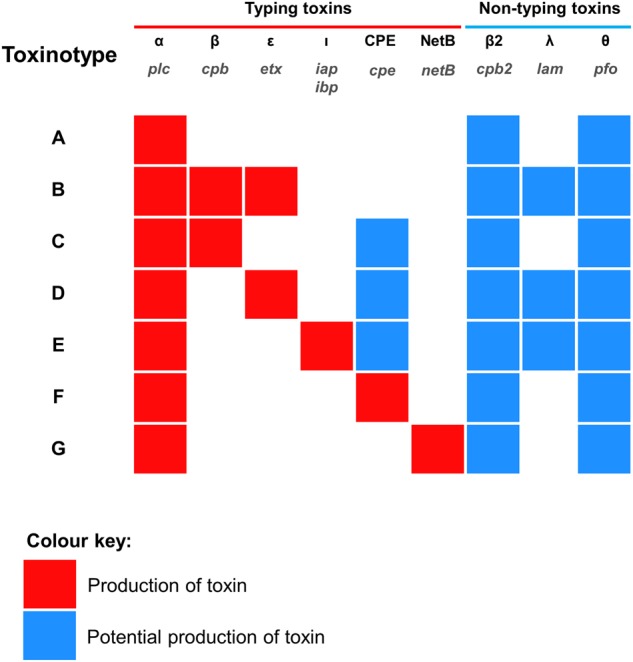
The names of toxin genes are printed in grey
Genomic insights
C. perfringens has a relatively low GC content, between 27–28%, and genome sizes range from 3.0–4.1 Mb, with 2500–3600 predicted genes present in each individual genome10. The first ever complete genome sequence of C. perfringens (strain 13) was published in early 2002; a historical gas gangrene-associated strain from 1939 (this also represents the first Gram-positive anaerobic pathogen to be sequenced)12. Virulence-linked genes within the sequence data include phospholipase C (α-toxin), hyaluronidase (µ-toxin), collagenase (κ-toxin), PFO (θ-toxin), and β2 toxin. Moreover, 61 sporulation-related and germination-related genes were encoded, which supports the fact that C. perfringens is a spore-former. Notably, most virulence genes in this type A strain were not found on genomic islands, or near insertion/transposase sequences, and GC content of those genes were similar to the average GC percentage, implying that horizontal gene transfer (HGT) of these toxin genes was an ancient evolutionary event. This study also highlighted differences in environmental adaption genes, including glycolytic enzymes, and sporulation-linked genes, whereas recent studies indicated that phylogenetic relationships between C. perfringens (based on core genome analysis) does not relate to clone origin or toxinotypes1,10
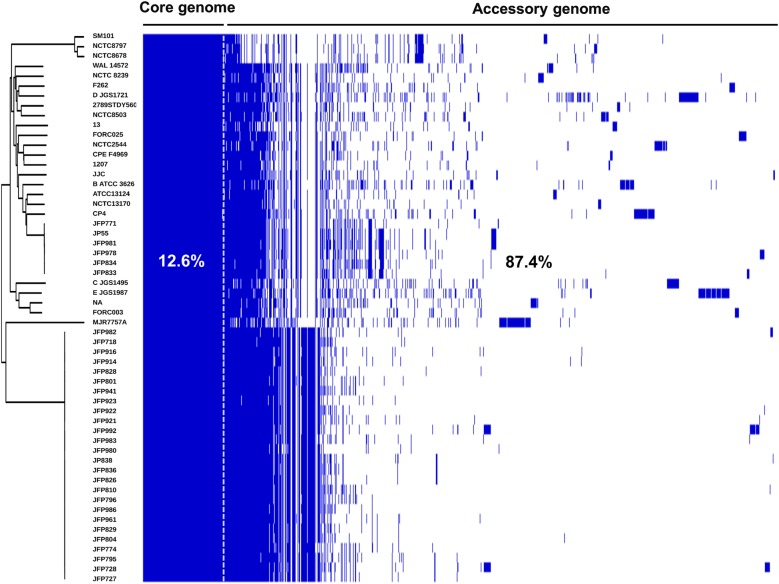
A recent large-scale genomic study that investigated 56 strains C. perfringens strains, representing the largest C. perfringens genomic study to date, revealed a diverse pangenome (a repertoire of genes in a defined number of genomes), with only 12.6% core genes (common genes that are present in each genome) (Fig. 2)10. A wide array of toxin genes was profiled in this study: α-toxin (plc), α-clostripain (ccp) and microbial collagenase (colA) were conserved in all examined genomes, the pore-forming toxin PFO gene pfo was found to be encoded in most genomes (>90%), whereas the food-poisoning causative toxin CPE gene cpe was consistently detected in historical food poisoning isolates (isolated before 1950s). Importantly, prevalence of aminoglycoside-resistant/anti-defensin gene mprF (100% detection), and tetracycline-resistant efflux protein genes tetA (P) (>75% detection) encoded within the genomes underlined the potential antimicrobial resistant threat of C. perfringens-associated infection. This study also suggested that these diverse genetic variations may have been driven by HGT, especially prophage insertion, within the ‘CRISPR-free’ genomes (no single CRISPR prophage defence system detected in >70% of the genomes).
Fig. 2. The linearised pangenome of 56 C. perfringens strains reveals significant genetic diversity.
Each coloured block represents synteny (homolog: identical gene with >95% sequence similarity) in the pangenome. Figure adapted from Kiu et al.10
The substantial genetic divergence in the C. perfringens pangenome suggests there may be additional novel virulence-related genes encoded within the ‘accessory genome’, in addition to the plasmid-borne toxins, known to be primarily responsible for specific disease pathologies. Thus, it is important to analyse plasmid-encoded genes/toxins, in tandem to chromosomally-encoded genes in the accessory genome, to provide a more complete genomic picture of C. perfringens. Potentially, with the aid of bioinformatics tools, more novel toxins or virulence genes will be discovered and identified for investigation in relation to C. perfringens pathogenicity, and disease transmission tracking in hospital and environmental settings, as has been performed for the related gut pathogen C. difficile13. To fully determine and research the pangenome, and discover potentially novel virulence genes, more isolate sequencing will be required, including addition of isolates from a range of hosts and environmental conditions, which may pave the way for translational research into new treatment strategies against C. perfringens infections.
Plasmids
Importantly, C. perfringens strains are known to carry plasmids, which often encode virulence-associated proteins including toxins and antimicrobials14. Disease-associated toxins including; CPE, ε-toxin, ι-toxin, NetB, β2-toxin and binary enterotoxin (BEC) have all been detected on C. perfringens plasmids10,15. HGT of plasmids between strains is reported to be via the tcp conjugation locus that exists in most plasmids16. Important toxin-carrying plasmids will be briefly reviewed in order of toxinotypes. For comprehensive reviews on C. perfringens plasmids, and other related insertion sequences see14,15.
CPE is known to be involved in food poisoning and non-foodborne gastroenteritis. It was reported that up to 70% of food poisoning cases, causative C. perfringens strains (mainly type F, previously known as CPE-positive type A) were shown to encode chromosomal-cpe, rather than plasmid-cpe, while the latter was associated with non-foodborne gastroenteritis (responsible for 5–15% of all cases)17. Some cpe-associated plasmids have also been shown to encode cpb2 genes or iab/ibp genes (e.g., pCPF5603)14. Type B strains carry plasmids which encode one of the deadliest toxins on earth, ε-toxin, and β-toxin. Plasmid pCP8533etx encodes etx and cpb2 genes, but not cpb18. Most plasmids in type B strains also possess other virulence genes encoding λ-toxin and urease19. Similarly, plasmids in type D strains also carry cpe and/or cpb2 genes, and plasmids range in size from 45 to 110 kb20. C. perfringens type C strains possess plasmid-borne β-toxin gene cpb, and other plasmids reported to carry other toxin genes including cpb2, tpeL, or cpe21. In type E strains, two major families of plasmids have been identified: (1) plasmids carry iap/ibp genes, λ-toxin gene and cpe gene, however, cpe contains nonsense and missense mutations in the cpe ORF and is thus non-functional22. (2) These plasmids, including pCPPB-1, encode both iap/ibp genes and cpe gene but not the λ-toxin gene23. NetB is a chicken NE-associated toxin, which is plasmid encoded. These netB-encoded plasmids can potentially carry other virulence genes including tetracycline resistance genes (e.g., pJIR3537 and pCW3) and cpb2 gene (e.g., pJIR3844)24.
Notably, many toxin plasmids of C. perfringens are highly similar, sharing up to 35–40 kb of identical sequences15. In addition, some C. perfringens isolates can carry multiple near-identical plasmids that encode different toxin and AMR genes24. However, no one isolate has ever been found to possess plasmids that encode all key toxins; only certain plasmid combination could can be maintained, which suggests plasmid incompatibilities, due to presence of distinct plasmid segregation systems, and warrants further research14,25,26. Furthermore, the universal existence of tcp locus indicates that conjugative plasmid transfer could be a key HGT event for increasing virulence of C. perfringens strains.
Virulence-related factors
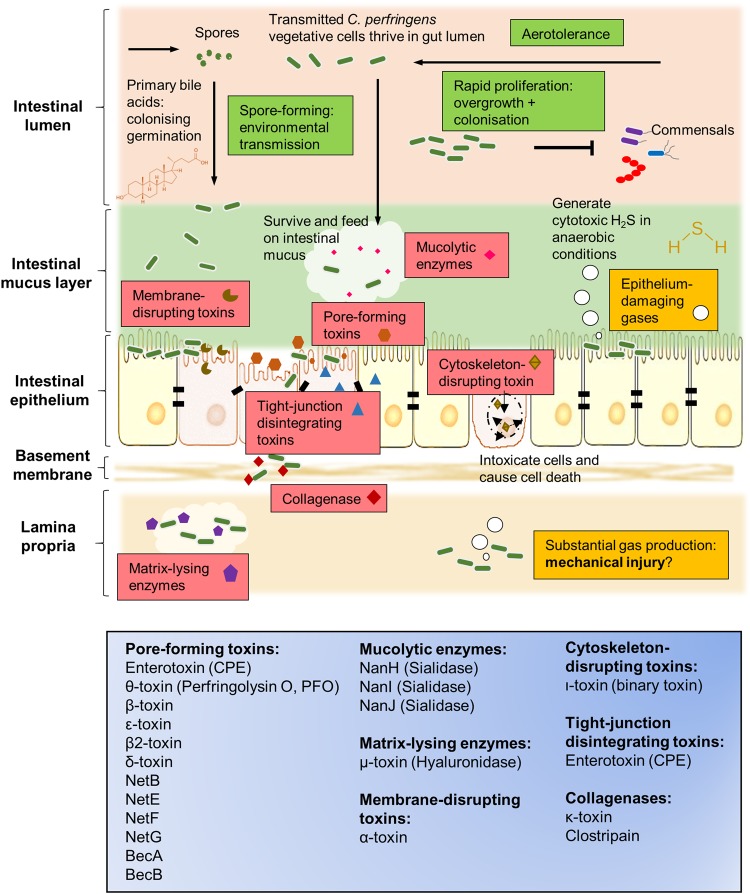
C. perfringens can generate a complement of extracellular toxins and hydrolytic enzymes (>20), survive in aerobic environments (i.e., oxygen tolerance), produce toxic gases, and rapidly grow. Therefore, it possesses the capacity to effect various histotoxic (i.e., toxigenic to tissues) infections in humans, including gas gangrene in contaminated wounds, gastroenteritis (including foodborne and non-foodborne diarrhoea) in human adults, necrotic enteritis (NE) in animals, and recent links to NEC in pre-term infants27. Relevant virulence factors of C. perfringens are represented and summarised in Fig. 3.
Fig. 3. Graphical representation of disease-linked virulence factors of C. perfringens in the context of intestinal infections.
Classification of toxins according to general mechanisms of action are shown in the bottom blue box
Transmission and colonisation
Oxygen sensitivity
C. perfringens is commonly known as a strict anaerobe; however, this bacterium can also survive in the presence of oxygen, and/or low concentrations of superoxide or hydroxyl-radical-generating compounds28. Notably, as an aero-tolerant anaerobe, C. perfringens could potentially survive through aerobic environments (such as on surfaces in hospital wards29) and trigger disease development in aerophillic environments (i.e., adult/pre-term infant gut), and additionally oxygen-exposed tissues (such as gas gangrene), which may facilitate bacterial host-to-host transmission30.
Sporulation
C. perfringens is a spore-former, which enables this bacterium to survive in extreme or nutrient-depleted conditions. This spore-forming characteristic plays a vital role in the transmission of this Gram-positive bacterium from diverse environments to hosts, leading to infection, including food poisoning in human adults31. Some spores of C. perfringens (especially food-poisoning associated strains) had been demonstrated to resist extreme temperature conditions, which may contribute to C. perfringens survival and subsequent disease pathology32,33. Molecular regulation of sporulation in C. perfringens is less well studied compared to the model bacterium Bacillus subtilis. Previous studies have indicated that the transcriptional regulator CcpA (encoded by ccpA) is required for effectual sporulation34 and Spo0A (spo0A), a transcriptional factor known to be present in Clostridium species including C. perfringens, is associated with sporulation initiation35. Sigma factors, including SigF, SigE, SigG and SigK (genes sigF, sigE, sigG and sigK respectively) are known to regulate the sporulation process of C. perfringens36,37. In addition to this, it was also reported that C. perfringens sporulation processes could be regulated by an Agr-like quorum-sensing system, potentially promoted by the codY gene in type A food-poisoning strains38,39. Importantly, CPE is known to be produced by C. perfringens during sporulation, which correlates with the pathogenesis of food poisoning-associated diarrhoea31.
Germination
Germination of C. perfringens can be triggered by small molecules termed germinants—commonly known as sugars, nucleosides, amino acids, salts and purines40. Importantly, primary bile acids (including glycocholate, cholate and taurocholate) in the human GI tract are also known to act as potent germinants in Clostridium species, whereas secondary bile acids, such as deoxycholate (derivatives generated by certain gut microbes), can inhibit the in vitro proliferation of C. perfringens41. This proposed ‘colonising germination’ survival modus operandi of spore-formers, including C. perfringens, potentially acts as a signal to initiate growth, and to persist in the intestinal environment30. This persistence, and long-term gut colonisation may correlate with the ongoing symptoms (up to several weeks) of nonfoodborne diarrhoea in patients42. From a colonisation resistance perspective, inhibition of germination, by secondary bile products produced by resident commensal bacteria, implies a long-standing competition for limited nutrients and niches within the gut.
Rapid proliferation
Notably, C. perfringens represents one of the fastest-growing organisms currently known, and is reported to have a very short 8–12 min generation time when cultured at 43 °C in optimal media, and 12–17 min at 37 °C43. Rapid growth of this bacterium potentially predisposes the host to tissue infection, as in the cases of avian NE, bovine necro-haemorrhagic enteritis (could be <5 h), and gas gangrene. Also, as C. perfringens has a two-fold quicker generation time, when compared to intestinal commensals like wild-type Escherichia coli (typically 20–30 min in Luria-Bertani broth), this could represent a potent mechanism for outgrowing other resident bacteria, leading to efficient gut colonisation.
Disease initiations
Histotoxic gas production
Clostridial myonecrosis (better known as gas gangrene), is accompanied by profuse gas production, which could in theory be the mechanical determinant of tissue injury, alongside rapid cell proliferation of C. perfringens. Importantly, C. perfringens is known to generate hydrogen sulphide44 (using ubiquitous sulphuric sources from the environment), a readily-permeable toxic substance to human cells, which when produced in excess is associated with other intestinal inflammatory diseases (e.g., Ulcerative Colitis)45. Thus, C. perfringens-associated gas production represents an understudied virulence trait in the pathogenesis of gut infection, such as infant NEC (discussed in more detail below), which symptoms significantly mimic those of gas gangrene; necrosis of tissue accompanied by abundant gas production46,47.
Toxins and virulent enzymes
Presently, 23 virulence genes that encode toxins and virulent enzymes have been identified in C. perfringens (summarised in Table S1); the most ‘prolific’ toxin-producing pathogen currently known. Some key toxins will be briefly discussed and summarised in this sub-section.
Alpha-toxin
Alpha-toxin (encoded by gene plc or cpa), which is produced by all strains of C. perfringens, has been shown to hydrolyse cell membrane phospholipids, which eventually leads to cell necrosis; a key characteristic in gas gangrene. Indeed, this toxin is essential for gas gangrene pathology, as shown in an in vivo myonecrosis model48. Mechanistically, α-toxin may play three major roles in gas gangrene pathology; firstly, it is able to impact the transferring of immune cells such as neutrophils to infected tissues (mechanism currently unknown), hence potentially reducing pathogen clearance at infected sites. Secondly, it can cause constriction of blood vessels, which may reduce the blood supply to tissues, thus creating a micro-aerophillic environment conducive to C. perfringens overgrowth. Thirdly, this toxin can activate inflammation cascades in host cell metabolism (arachidonic acid and protein kinase C), which may lead to direct immune-mediated pathology of tissues49.
Beta-toxin
Beta-toxin, encoded by gene cpb, is a plasmid-encoded pore-forming toxin that is thought to be intestinal-necrotic, and also important for systemic enterotoxaemia, in humans and neonatal animals, as shown in in vivo studies50,51. C. perfringens type C strains that possess the cpb gene were associated with historical Clostridial gut infections; Darmbrand (post-world war II in Germany), and Pig Bel (Papua New Guinea)52. Previous intestinal loop studies have indicated a synergistic effect with CPE (encoded by cpe)53. Furthermore, this toxin shares sequence similarity with several pore-forming toxins produced by Staphylococcus aureus – ɣ toxin (A: 22 and B:28%), α-toxin (28%) and leucocidin (S:17% and F:28%)54.
Perfringolysin O
PFO (also θ-toxin, encoded by gene pfoA or pfo), is also a pore-forming toxin that acts on cholesterol-comprising cell membranes. This toxin has been shown to be involved in the pathogenesis of gas gangrene, and haemorrhagic enteritis in calves55,56. Notably, PFO shares structural homology with similar pore-forming toxins identified in Streptococcus, Bacillus, Listeria and many other genera57. θ-toxin is also known for its capacity to induce tumour necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) expression in the host, and it could activate apoptosis through p38 MAPK (mitogen-activated protein kinase) pathway as demonstrated in in vitro models58. Interestingly, it has synergistic cytotoxic effects with α-toxin on bovine epithelial cells, highlighting the significant role of this toxin in disease development56,59.
Beta2-toxin
Beta2-toxin (encoded by cpb2 plasmid gene), a pore-forming cytolytic toxin shares <15% sequence homology with β-toxin, distinguished itself as a novel toxin produced by C. perfringens54. β2-toxin-producing C. perfringens strains have been associated with gut diseases such as NE in piglets, and enterocolitis in foals60. Importantly, this pore-forming toxin has been suggested as an accessory toxin in C. perfringens-associated non-foodborne diarrhoea61, and proposed to play an important role in pre-term NEC in potential synergistic effects with antibiotic gentamicin62.
Epsilon-toxin
This deadly toxin, is plasmid-encoded by etx gene C. perfringens type B and D strains, and is essential for pathogenesis. Epsilon toxin is involved in animal (goat and sheep) enterotoxaemia63. ε-toxin is currently thought to be the most potent of all toxins produced by C. perfringens, evidenced by the LD50 of 70 ng/kg body weight, ranked only behind C. botulinum and C. tetani neurotoxins. It has been demonstrated to affect various organs, such as kidneys and liver, through unknown mechanisms, that allow this toxin to enter systemic circulation64. Due to its potential use as a biological weapon, ε-toxin-producing C. perfringens strains are on the export control list in a number of countries including USA and the UK.
Iota-toxin
This cytoskeleton-damaging toxin (also considered as a major toxin produced by C. perfringens), consists of two cell-binding monomers Ia (enzymatic component) and Ib (binding component; encoded on plasmids by iap and ibp gene respectively), which act synergistically to first translocate the toxin into host cells, then exert enzymatic activity on ADP-ribosylating actin to disassemble the cytoskeleton, which eventually leads to apoptosis and cell death65.
Microbial collagenase
Microbial collagenase (also known as κ-toxin), encoded by chromosomal gene colA, is a key toxin produced by C. perfringens that degrades collagen. This enzyme might contribute to intestinal infection, as collagen, the substrate for collagenase, is a primary component of intestinal connective tissues/basal membrane of human and animal hosts, thus disruption may damage basal integrity, which may lead to eventual tissue necrosis56,66. However, Awad et al.67 indicated that κ-toxin is not a major determinant in a Clostridial myonecrosis mouse model (gas gangrene), despite its capacity to hydrolyse collagen.
Enterotoxin (CPE)
CPE (encoding gene cpe) is recognised as the key toxin to cause food-poisoning and non-foodborne diarrhoea, it has also been demonstrated to disrupt the intercellular claudin tight junctions in gut epithelial cells68,69. Importantly, this pore-forming toxin was demonstrated to bind and necrotise human ileal and colonic epithelium in vitro, and can induce cell death i.e., apoptosis, via the caspase-3 pathway70. Hence, the potential pathogenesis mechanisms underlying food poisoning could result from CPE-induced tight junction rearrangements or pore-formation.
Sialidase
Three sialidases, encoded by genes nanH, nanI and nanJ in C. perfringens, are also named as neuraminidases or exo-sialidases (NanI and NanJ). This group of enzymes represent important virulence factors during C. perfringens-mediated tissue infection; they catalyse hydrolysis of terminal sialic acids from glycoprotein, glycolipids and polysaccharides of cell membranes that aids in bacterial attachment to host cells71. This mucolytic potential suggests that C. perfringens may utilise intestinal mucus as a nutrient source, and thereby potentiates intestinal colonisation, which has been modelled using the in vitro Caco-2 cell line72. Importantly, in vitro studies have also demonstrated that α-toxin associated with NanI (exo-alpha-sialidase) increased the virulence of C. perfringens73. Furthermore, NanI was shown to potentiate the virulence of ε-toxin, β-toxin and CPE, via binding-enhancing (ε-toxin) and proteolytic activation (β-toxin and CPE) mechanisms, potentially enhancing C. perfringens pathogenesis74. Notably, in a gas gangrene mouse model, NanI and NanJ are not essential for virulence75.
NetB
NetB, a pore-forming toxin (encoded by netB), shares limited amino acid sequence similarity with beta-toxin (38% sequence identity), α-toxin from S. aureus (31% sequence identity) and δ-toxin (40% sequence identity) from C. perfringens76. Less than a decade ago, this chicken-NE associated toxin (plasmid-borne), was discovered and shown to be essential (instead of α-toxin) for lesion formation both in cell line models, and avian in vivo models via genetic mutant strains, and thus fulfilled molecular Koch’s postulates as a disease determinant77. This toxin is important in avian agriculture as netB-positive C. perfringens strains recovered from broiler chickens (healthy birds) can be as high as 61%, however expression of NetB was shown to be lower in healthy birds, when compared with NE-associated chickens (92% vs. 29%)78. In addition, netB was later identified as part of a plasmid-encoded pathogenicity locus named NELoc-1 capable of being transferred via conjugation, indicating the potential spread in relation to chicken NE epidemiology24,79. Please refer to Review by Rood et al.80 for a comprehensive description on NetB and poultry NE.
These extracellular toxins represent potent virulence factors central to intestinal disease development. However, not all toxins are secreted by all strains of C. perfringens (excluding α-toxin), hence, in silico identification of virulence genes using NGS techniques and bioinformatics tools are essential for rapid and comprehensive geno-toxinotyping of C. perfringens, compared with conventional molecular tools.
Antimicrobial resistance (AMR)
AMR traits in C. perfringens pose a serious clinical treatment concern, due to its capacity to generate an array of lethal toxins. Presently there are several phenotyping studies published (based on in vitro susceptibility testing of minimal inhibitory concentration; MIC, Table S2) on the AMR profile of C. perfringens, however, there is currently limited genetic/WGS-based AMR genotyping studies.
Tetracycline resistance in C. perfringens was first described in 1968, when 11 strains of C. perfringens were tested, and found to possess ‘some degree’ of resistance to tetracycline, thus penicillin G (or, benzylpenicillin) was recommended as the drug of choice for treating Clostridial infection81. Plasmid-carrying tetracycline resistance (tet components) in C. perfringens was genetically shown for the first time in porcine samples (three strains)82. Tetracycline resistance elements tetA(P), was then detected in 81 tetracycline-resistance C. perfringens strains (100%), with 93% of these strains carrying a secondary resistance gene, either tetB(P) or tet(M), notably, no single strain possesses all tet genes83. Multidrug-resistant C. perfringens strains were first reported back in 1977, multiple strains (n = 7) isolated from porcine faeces were demonstrated to be resistant against tetracycline (MIC > 32 μg/ml), erythromycin (MIC > 128 μg/ml), clindamycin (MIC > 64 μg/ml), and lincomycin (MIC > 128 μg/ml)84. A PCR-based AMR study on 160 environmental strains (water, soil and sewage) revealed encoded macrolide resistance genes erm(B)(26%), erm(Q)(1%), and mef(A)(18%), in addition to the commonly found tetA(P)(53%), tetB(P)(22%), and tet(M)(8%)85.
Macrolide and Lincosamide resistance (mainly erythromycin and lincomycin) appears widespread86, and therefore is considered ineffective in treating C. perfringens infections. This is supported by a recent multidrug-resistance study of 260 strains of C. perfringens isolated from diarrheal neonatal piglets in Thailand, where higher resistance was observed for erythromycin, lincomycin and tylosin87.
Recent WGS studies on AMR genes have detected mepA (multidrug-resistance gene), using various public AMR databases on C. perfringens strains88, and also tetracycline resistant genes tetA(P), tetB(P) and tet38. The genes, mprF and rpoB (rifampin-resistant) have also been reported to be encoded88. Anti-defensin gene mprF (possibly involved in multidrug-resistant, including resistance against gentamicin) was recently reported in a large-scale genomic study of C. perfringens (n = 56 strains) to be present in 100% of the genomes10. In this latest genomic study, tetA(P) was detected in 75% of the 56 strains, which is more prevalent than tetB(P)(42%). Interestingly, ANT(6)-Ib, an aminoglycoside resistance gene, was also reported to be encoded in a C. perfringens toxinotype C strain. Although mainly anaerobic bacteria like C. perfringens may have reduced transport of aminoglycosides intracellularly, there are also findings that C. perfringens are sensitive to aminoglycosides like Gentamicin at higher concentration, which indicates that C. perfringens might also have acquired additional resistance to aminoglycosides89,90.
WGS will be a key tool in the fight against C. perfringens AMR, particularly in rapid diagnostics, however gold-standard phenotypic MIC tests will also be required to clinically determine the antimicrobial susceptibilities, and facilitate antibiotic management.
Clinical associations in humans and animals
C. perfringens has been constantly associated with gut diseases across both animal and human hosts (summarised in Table S3) as described in this section.
Animal hosts
Poultry NE
Poultry NE was first documented in England in 196191. Since then, NE has been consistently reported in every continent around the globe. Importantly, C. perfringens is unequivocally identified as the key aetiological organism of NE in broiler chickens92. Global financial impact of NE is significant, with an estimated economic loss of 6 billion US$ in 2015, projecting profit loss per bird at >US$0.06293.
Classic NE pathologies are characterised by gaseous lesions and mucosa necrosis in gas-filled distended small intestine, however, these can also involve kidney and liver as secondary infection sites94. Proposed key biological factors that contribute to NE are hydrolytic enzymes (e.g., collagenase)95, toxin production (traditionally understood as α-toxin, and more recently NetB and TpeL, both are pore-forming toxins, were linked to the onset of NE96. Other NE-predisposing factors include: a high-protein diet that favours the growth of C. perfringens, and environmental stress (e.g., high stocking density), which alters gut microbiota/host immunity that eventually increases risk of infection97.
Equine acute necrotising enterocolitis
Acute necrotising enterocolitis (ANEC) in foals/horses (caused by C. perfringens) is a severe intestinal disease that resembles the classic clinical signs of C. perfringens-infections; rapid disease development and necrotic intestines (mainly the colon)98. ANEC symptoms are characterised by bloody diarrhoea, followed by a haemorrhagic and necrotic gut99. Type F C. perfringens strains (previously type A) harbouring both CPE and β2-toxin are often associated with this deadly condition98. More recently, NetE, NetF and NetG toxins (pore-forming toxins) were proposed to underlie the pathogenesis of foal NEC100.
Swine enterocolitis
C. perfringens type A strains (also less frequently, CPE-harbouring type F strains) are widely considered as the invasive agent of non-haemorrhagic EC in piglets, although the actual pathogenesis remains undefined. Similar to other C. perfringens-intestinal infection, this disease commonly affects one-week-old piglets, suspected to inherit (i.e., via vertical transmission during birth) the bacterium from the microbiota of mother sows101. Symptoms involve severe diarrhoea (non-haemorrhagic), accompanied by necrotic mucosa and atrophy of intestinal villi. β2-toxin was initially believed to drive the development of this disease (backed by epidemiological studies in 1990s), and as such was used as a diagnostic biomarker, although this has become controversial in recent years60. Type C strains that carry β-toxin are associated with haemorrhagic EC in piglets and largely affect 1 to 4 days old neonates101. In contrast to type A-infection (to a lesser degree, type F-infection), type C-infection is characterised by haemorrhagic NE, which is proposed to be driven by the presence of type C strains and low trypsin secretion (trypsin can inactivate β-toxin) in the immature host gut102.
Human hosts
Darmbrand
Darmbrand, which literally means ‘burning (fire) bowels’ in German, was used to describe a particular type of necrotic inflammatory gut disease (also known as enteritis necroticans, EN) associated with C. perfringens, that occurred epidemically post-second World War (1944–1949) in north-west Germany103. Darmbrand causative strains were later classified as type C strains that carry β-toxin, and these strains are highly resistant to heat104. Many of these type C strains also harbour CPE, which has been shown to act synergistically with β-toxin in an intestinal loop model and therefore CPE was suggested to play a role in some cases of EN53. Notably, the heat-resistance of this strain might be attributed to the production of a small acid-soluble protein (Ssp4) that could potentially play a central role in dissemination of C. perfringens strains33. Darmbrand was believed to be facilitated by poor post-war sanitary (bacterial contamination), and malnutrition (protein-malnourished) conditions, as this disease disappeared within a few years after its first recognition.
Pigbel
Enteritis necroticans (EN), or commonly known as pig-bel in the highlands of Papua New Guinea (PNG), is a form of inflammatory gut disease that was associated with pork-feasting activities that took place among PNG Highlanders in outbreaks first documented back in 1966105. The classical description of the symptoms is ‘spontaneous gangrene of small intestine, without obvious vascular or mechanical cause’ which resembles Darmbrand, and occurred particularly in children106. The aetiology of this fatal infection was thought to be large co-consumption of C. perfringens type C-contaminated pork (due to poor hygiene), and large amount of sweet potatoes which contain trypsin inhibitor (trypsin secreted in the gut could break down β-toxin secreted by C. perfringens)107. Similar cases of EN were also reported in Uganda108 and Indonesia109. Notably, EN-like cases were also reported (rare cases) in developed countries, including USA, in exclusively diabetic subjects, who also have attenuated trypsin production110.
Acute watery diarrhoea (food poisoning)
C. perfringens has been associated with food poisoning since it was first documented in both the UK and the USA in the 1940s111. C. perfringens-type A food poisoning, is ranked the second most prevalent bacterial food poisoning in the US, estimated at 1 million cases per annum, after Salmonella112. In the European Union (EU) member countries, C. perfringens-linked food-poisoning outbreaks were projected at approximately 5 million cases per year (2011)113. C. perfringens has also been reported to affect elderly communities, especially in care homes (North East of England, 2012–2014; 83% of the outbreaks reported from care homes)114. C. perfringens-linked foodborne cases are suggested to be under-reported due to its self-limiting symptoms, thus the statistics published based on laboratory-confirmed cases may be lower than the actual numbers, suggesting a higher actual epidemiological impact.
Hallmark symptoms (self-limiting, lasting 12–24 h; mortality is uncommon) include, rapid appearance within 8–14 h after food ingestion, intestinal cramp, watery diarrhoea without fever or vomiting115. Currently, food poisoning C. perfringens cases are thought to be caused by CPE (encoded by cpe gene). This secreted pore-forming toxin is also demonstrated to disrupt intestinal tight junction barriers and initiate disease development68.
Non-foodborne diarrhoea
C. perfringens has also been associated with non-foodborne diarrhoea (a distinct disease entity from food poisoning, characterised primarily by more severe symptoms and longer duration), which includes antibiotic-associated diarrhoea (AAD), and sporadic diarrhoea (SD)116,117. AAD typically occurs in 5–25% of patients after administration of broad-spectrum antibiotics118. Non-foodborne diarrhoea typically affects older adults that are >60 years of age (although SD is also associated with younger age groups)119. Clinical symptoms are abdominal pain, prolonged diarrhoea (>3 days to several weeks), and bloody stools120,121. Patients suffering from non-foodborne diarrhoea, particularly AAD, can become seriously ill due to diarrhoea-induced dehydration, and may progress to develop colitis, although full recovery is common122.
Although other pathogenic bacteria including C. difficile (most common AAD pathogen; ~25% of cases123) and S. aureus have also been implicated in AAD, enterotoxigenic C. perfringens type F strains (producing CPE) are estimated to be responsible for up to 15% of all AAD cases116,124,125. CPE, produced by C. perfringens type F strains (encoded by plasmid-borne cpe gene), has been reported to be the aetiological agent for C. perfringens-associated non-foodborne diarrhoea, as evidenced by the high prevalence in diarrhoea patients, but not healthy individuals. Importantly, type F strains have also been linked to SD, although to a lesser extent116. AAD-associated C. perfringens strains are also described to be more adherent to Caco-2 intestinal cells, when compared to other food-poisoning strains, which is attributed to the production of sialidase NanI126. The spore-forming nature of C. perfringens could also potentially contribute to the commonly observed disease persistence and relapse127. Molecular detection of faecal CPE or PCR confirmation of cpe gene represents the current clinical diagnostic method for C. perfringens-AAD128,129.
Pre-term infant NEC
Pre-term infant NEC has been clinically linked with C. perfringens since the 1970s, as type A-C. perfringens were isolated from necrotic tissues in many NEC cases. Notably, it was even called ‘gas gangrene of the bowel’ owing to its reflection of the highly similar diseased histology with the infamous tissue myonecrosis47.
NEC has ~14% prevalence in pre-term infants less than 1000 g birth weight (i.e., extremely low birth weight, ELBW), a high mortality of 30% with surgical NEC, and mortality up to 50%. NEC-related deaths account for 10% of all mortality causes in neonatal intensive care units (NICUs) according to a UK-nationwide study130, and is the most severe and lethal neonatal GI emergency worldwide131. The additional annual cost for treating NEC in pre-term infants is conservatively estimated at £13.5 million in the UK (excluding long-term post-surgical treatment), whilst in the US $1 billion is estimated to be spent annually by neonatal departments on NEC132.
The involvement of bacteria in the pathogenesis of NEC has been strongly implicated since the disease was first described133. Both Klebsiella spp., and C. perfringens, have been linked to clinical NEC in recent years4. Importantly, C. perfringens was isolated from most neonates (70%), as early as the fifth day of life, which supports the theory of pathogen colonisation that then leads to microbial invasion. Classic NEC symptoms such as pneumatosis intestinalis, suggests gas-producing Gram-positive bacteria including Clostridium spp. (also implicated consistently in recent metagenomics studies), as the key causative agent134,135. Most recently, the emergence of two extensive pre-term infant NEC cohort studies that profiled metagenomic faecal samples, have both indicated that C. perfringens is significantly abundant in the infant gut microbiota prior to NEC development4,5. These studies are supported by the substantial number of NEC studies (14 studies to date since 1970s, more than any individual bacterial agents, as summarised in Table S4) that have reported C. perfringens as a potential pathogenic agent, in addition to its established pathogenesis in many other neonatal animal gut diseases, and further supported by in vivo studies136. Thus, C. perfringens appears to be a pathogenic agent of microbial-NEC, i.e., C. perfringens-associated NEC, and may link to specific disease symptoms137.
Being universal and resilient in habitable niches, spore former C. perfringens may readily pass on to in-hospital neonates through either environmental or oral transmission, and then initiate diseases in the intestine138. Proposed underlying mechanisms for C. perfringens-associated NEC include reduced bowel peristalsis in pre-term infants (increased pathogen retention time), universal administration of broad-spectrum antibiotics139 (that leads to reduced microbiota diversity, and the potential rapid overgrowth of resistant spores), reduced gut barrier integrity (non-existent mucus layer), lack of protective bacteria4,5,131 (including Bifidobacterium species, which may antagonise the growth of C. perfringens), and immature/underdeveloped immune system that is ineffective at fighting off pathogenic bacteria46 (Fig. 4). Bile salt factors may also be linked to disease onset. Reduced diversity of the microbiota in pre-term infants, including bacteria that can de-conjugate bile salts to secondary bile acids, may allow germination of C. perfringens, which has also been directly indicated in the pathogenesis of the well-studied C. difficile colitis (both species form spores and secrete colitis-related toxins)140.
Fig. 4. Proposed infection mechanisms that underlie C. perfringens-associated NEC.
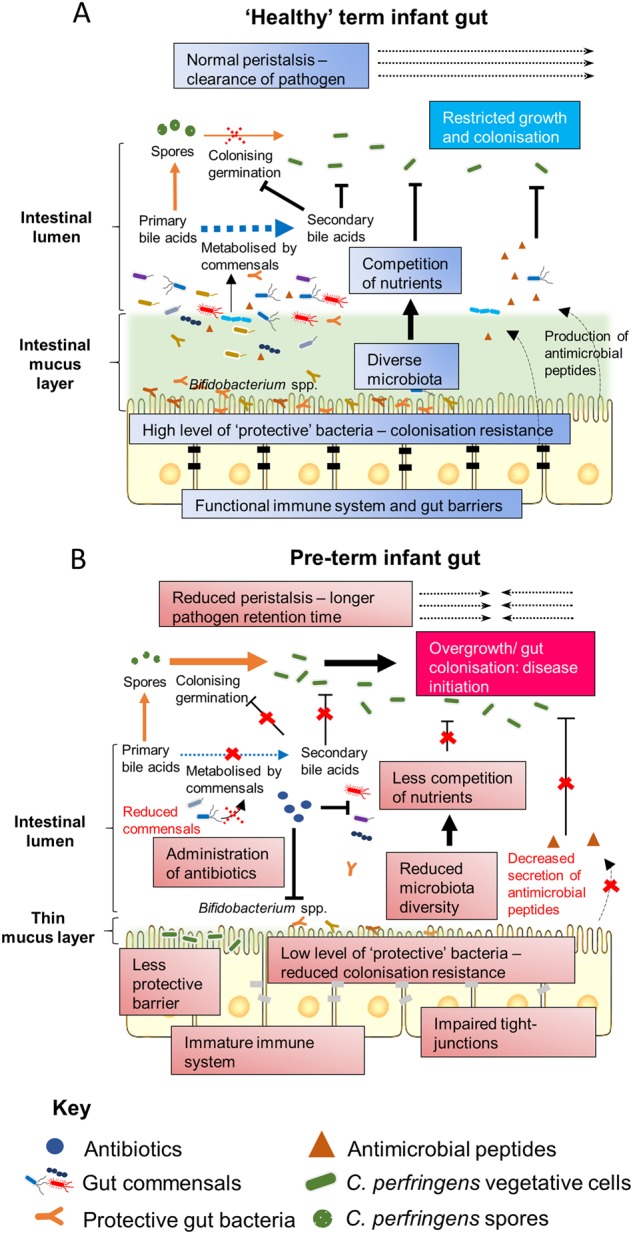
a In non-NEC ‘healthy’ term infant gut. b In pre-term infant gut that leads to C. perfringens-associated NEC
To date, no particular toxin(s) has been specifically linked with pre-term NEC, although β2-toxin has been suggested4. Notably, C. perfringens can also be part of a ‘healthy’ human microbiota (as low as 3.5% in pre-term neonates, nevertheless this is controversial141), thus suggesting other factors play a role. Antibiotic administration in pre-term babies is very high, with up to 97% of pre-term infants residing in NICU exposed to at least one dosage of broad-spectrum antibiotics142. These antibiotics eliminate ‘protective’ members of the gut microbiota, and in tandem with presence of AMR-C. perfringens, may facilitate successful colonisation and infection143. An immature gut barrier in neonatal infants may also expose gut niches for overgrowth, and finally the presence of ‘hyper-virulent’ strains of C. perfringens may be implicated. However, to comprehensibly answer these important questions a more global genomic approach, i.e., WGS and analysis, and validation using suitable in vivo models is required.
Advancement of WGS and genomic analyses, particularly those focussing on other gut pathogens including C. difficile, K. pneumoniae and Salmonella enterica has proved successful in understanding the pathogenic role of these bacteria in disease development and tracking bacterial transmission144,145. These enteric pathogens have all been sequenced extensively with genomic information made public on NCBI database (as of May 2018; bracketed digits indicate number of genomes): C. difficile (1322), E. coli (10541), C. botulinum (225), K. pneumoniae (3927) and S. enterica (8780). Yet, including a batch addition of 30 genomes towards the end of 2016, there are only 59 publicly available genomes of C. perfringens (May 2018), highlighting the lack of in-depth understanding of this pathogen with respect to its genomic information. Importantly, no NEC-associated pre-term infant isolates are known to be sequenced at present, and genomic investigation may allow identification of putative/novel NEC-associated virulence factors, that may facilitate intervention strategies and/or therapy development.
Future research directions
The clinical link between intestinal diseases and C. perfringens is clear and defined, however the underlying factors responsible for specific aspects of pathology remains uncertain (Box 1, Box 2). Deciphering the genes that are involved in the sporulation, germination, enzyme/toxin production, oxygen tolerance, AMR and other novel virulence factors could lead to more targeted clinical preventions in C. perfringens-associated adult and neonatal intestinal diseases, whether it be humans or animals. Important questions may be answered utilising WGS and in silico tools, to delve into the genetic makeup of this notorious, yet under-studied pathogen. In tandem further in vitro and in vivo studies should be carried out to confirm the importance of suspected novel virulence traits, i.e., to fulfil Molecular Koch’s postulates. These approaches may help establish future platforms for disease prevention strategies such as vaccines, phage therapy, microbiota-based therapeutics, or implementation of specific antibiotic administration policies. Furthermore, understanding the genomes will potentially enable epidemiological tracking (as is taking place for other pathogenic bacteria), allowing us to pinpoint the origin and route of spread of isolates in the hospital settings, which is vital within clinical contexts.
Box 1.
Glossary
16S rRNA gene: a housekeeping gene (of ribosomes) that is conserved across all species therefore is widely used as a chronological/biological identity marker in evolutionary genetics and genomics.
Accessory gene: a gene that is absent in one or more strains in an analysed pangenome.
Antimicrobial resistance: a biological characteristic that enable a microorganism to survive and thrive in the presence of antibiotics.
Core gene: a gene that is present in all strains in an analysed pangenome.
Genotyping: a process to differentiate/identify characteristics of genes (genotypes).
Germination: the process which microorganisms grow from a dormant state (usually from endospores).
Necrotising Enterocolitis: severe gut inflammation (often accompanied by necrosis of the gut) which occurs mostly in pre-term infants with high mortality.
Pangenome: The repertoire of collective genes in closely related organisms.
Phenotyping: a process to observe the expression/traits (phenotypes) of an organism’s genotype.
Sporulation: a process certain species/genus of bacteria (e.g., Firmicutes) undergo to produce a tough external structure (spore/endospore) to withstand unfavourable conditions in order to survive through.
Box 2.
What are the key knowledge gaps in understanding C. perfringens as a pathogen, and what are the potential approaches to investigate/mitigate the virulence of C. perfringens in clinical diseases?
Questions
Compared to other pathogens (e.g., C. difficile), there is limited WGS data for C. perfringens, what is the genomic diversity of this important pathogen?
Can we use WGS to ‘type’ C. perfringens isolates, and can WGS be used in diagnostics (including AMR profiling), and to track spread in outbreaks settings?
C. perfringens causes diverse diseases in a wide host range, but what are the microbial virulence factors responsible for pathology, and is this host/strain specific?
What are the host immune cells/signalling cascades involved in C. perfringens-associated clearance and/or pathology?
C. perfringens can reside as a ‘normal’ member of the gut microbiota, what are the external factors that may lead to overgrowth and disease?
Can C. perfringens be directly linked to NEC pathogenesis?
-
What are the therapeutic approaches that could be used to prevent C. perfringens infection in multi-host species?
Approaches
Isolate diverse strains from various environments, i.e., from both case and control samples
Perform comprehensive bioinformatics analyses to understand evolution, e.g., SNP based phylogeny, and investigate variants and spread clinical settings
Carry out pan-GWAS/comparative genomics to identify/predict specific virulence genes related to disease development.
Develop/optimise clinical diagnostics from faecal samples via e.g. ultra-long read real-time sequencing method such as Oxford Nanopore
Develop and characterise in vivo infection models for different disease types (e.g., NEC and gastroenteritis)
Pinpoint specific virulence genes involved in infection via bacterial transcriptomics (RNAseq) and knock-out bacterial strains, to fulfil molecular Koch’s postulates.
Perform microbiota profiling (e.g., 16S rRNA metagenomics or shotgun metagenomics) to investigate the impact of C. perfringens to microbiota members, and the role of microbiome in C. perfringens infections
Understand host defence mechanisms using immunological approaches, and knock out in vivo models
Develop therapeutics for C. perfringens infections including phage therapy, vaccines, microbiota treatment i.e., probiotics.
Electronic supplementary material
Acknowledgements
This review was supported by a Wellcome Trust Investigator Award (100974/C/13/Z), and the Biotechnology and Biological Sciences Research Council (BBSRC); Institute Strategic Programme Gut Microbes and Health BB/R012490/1, and Institute Strategic Programme Gut Health and Food Safety BB/J004529/1 to L.J.H. R.K. is a recipient of University of East Anglia International Bursary (Faculty of Medical and Health Sciences).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0144-8).
References
- 1.Hassan KA, et al. Genomic analyses of Clostridium perfringens isolates from five toxinotypes. Res. Microbiol. 2015;166:255–263. doi: 10.1016/j.resmic.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Welch WH, Nuttall GHF. A gas-producing bacillus (Bacillus aerogenes capsulatus, Nov, Spec.) capable of rapid development in the body after death. Bull. John Hopkins Hosp. Baltim. 1891;3:81–91. [Google Scholar]
- 3.Lugli GA, et al. Ancient bacteria of the Otzi’s microbiome: a genomic tale from the Copper Age. Microbiome. 2017;5:5. doi: 10.1186/s40168-016-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sim K, et al. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin. Infect. Dis. 2015;60:389–397. doi: 10.1093/cid/ciu822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heida FH, et al. A necrotizing enterocolitis-associated gut microbiota is present in the meconium: results of a prospective study. Clin. Infect. Dis. 2016;62:863–870. doi: 10.1093/cid/ciw016. [DOI] [PubMed] [Google Scholar]
- 6.Revitt-Mills, S. A., Rood, J. I. & Adams, V. Clostridium perfringens extracellular toxins and enzymes: 20 and counting. Microbiology Australia, 114–117 (2015).
- 7.Rood, J. I. et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe (2018) 10.1016/j.anaerobe.2018.04.011. [DOI] [PMC free article] [PubMed]
- 8.Petit L, Gibert M, Popoff M. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999;7:104–110. doi: 10.1016/S0966-842X(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 9.Nagahama M, et al. Recent insights into Clostridium perfringens Beta-toxin. Toxins. 2015;7:396–406. doi: 10.3390/toxins7020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiu R, Caim S, Alexander S, Pachori P, Hall LJ. Probing genomic aspects of the multi-host pathogen Clostridium perfringens reveals significant pangenome diversity, and a diverse array of virulence factors. Front. Microbiol. 2017;8:2485. doi: 10.3389/fmicb.2017.02485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman, J. C., Shrestha, A. & McClane, B. A. Clostridium perfringens Enterotoxin: Action, Genetics, and Translational Applications. Toxins (Basel) 8, 10.3390/toxins8030073 (2016). [DOI] [PMC free article] [PubMed]
- 12.Shimizu T, et al. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl Acad. Sci. USA. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar, N. et al. Genome-based infection tracking reveals dynamics of Clostridium difficile transmission and disease recurrence. Clin. Infect. Dis., civ1031, 10.1093/cid/civ1031 (2015). [DOI] [PMC free article] [PubMed]
- 14.Freedman JC, et al. Clostridium perfringens type A-E toxin plasmids. Res. Microbiol. 2015;166:264–279. doi: 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, et al. Toxin plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 2013;77:208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisniewski JA, Rood JI. The Tcp conjugation system of Clostridium perfringens. Plasmid. 2017;91:28–36. doi: 10.1016/j.plasmid.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Grant KA, et al. The identification and characterization of Clostridium perfringens by real-time PCR, location of enterotoxin gene, and heat resistance. Foodborne Pathog. Dis. 2008;5:629–639. doi: 10.1089/fpd.2007.0066. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto K, et al. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 2006;188:1585–1598. doi: 10.1128/JB.188.4.1585-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayeed S, Li J, McClane BA. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect. Immun. 2010;78:495–504. doi: 10.1128/IAI.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayeed S, Li J, McClane BA. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect. Immun. 2007;75:2391–2398. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurjar A, Li J, McClane BA. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect. Immun. 2010;78:4860–4869. doi: 10.1128/IAI.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Miyamoto K, McClane BA. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect. Immun. 2007;75:1811–1819. doi: 10.1128/IAI.01981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto K, et al. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLoS ONE. 2011;6:e20376. doi: 10.1371/journal.pone.0020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bannam, T. L. et al. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic resistance plasmids. MBio2, 10.1128/mBio.00190-11 (2011). [DOI] [PMC free article] [PubMed]
- 25.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu. Rev. Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 26.Watts TD, Johanesen PA, Lyras D, Rood JI, Adams V. Evidence that compatibility of closely related replicons in Clostridium perfringens depends on linkage to parMRC-like partitioning systems of different subfamilies. Plasmid. 2017;91:68–75. doi: 10.1016/j.plasmid.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 27.McClane, B. A., et al. eds 698–752 (Springer, 2006).
- 28.Briolat V, Reysset G. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J. Bacteriol. 2002;184:2333–2343. doi: 10.1128/JB.184.9.2333-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machida Y, et al. An outbreak of enterocolitis due to Clostridium perfringens in a hospital for the severely disabled. Kansenshogaku Zasshi. 1989;63:410–416. doi: 10.11150/kansenshogakuzasshi1970.63.410. [DOI] [PubMed] [Google Scholar]
- 30.Browne HP, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, J., Paredes-Sabja, D., Sarker, M. R. & McClane, B. A. Clostridium perfringens sporulation and sporulation-associated toxin production. Microbiol. Spectr.4, 10.1128/microbiolspec.TBS-0022-2015 (2016). [DOI] [PMC free article] [PubMed]
- 32.Orsburn B, Melville SB, Popham DL. Factors contributing to heat resistance of Clostridium perfringens endospores. Appl. Environ. Microbiol. 2008;74:3328–3335. doi: 10.1128/AEM.02629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, McClane BA. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 2008;4:e1000056. doi: 10.1371/journal.ppat.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varga J, Stirewalt VL, Melville SB. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 2004;186:5221–5229. doi: 10.1128/JB.186.16.5221-5229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers GS, et al. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16:1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, McClane BA. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect. Immun. 2010;78:4286–4293. doi: 10.1128/IAI.00528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harry KH, Zhou R, Kroos L, Melville SB. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 2009;191:2728–2742. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Chen J, Vidal JE, McClane BA. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect. Immun. 2011;79:2451–2459. doi: 10.1128/IAI.00169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, J., Freedman, J. C., Evans, D. R. & McClane, B. A. CodY promotes sporulation and enterotoxin production by Clostridium perfringens type A strain SM101. Infect. Immun. 85, 10.1128/IAI.00855-16 (2017). [DOI] [PMC free article] [PubMed]
- 40.Setlow P. Spore germination. Curr. Opin. Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Modi N, Wilcox MH. Evidence for antibiotic induced Clostridium perfringens diarrhoea. J. Clin. Pathol. 2001;54:748–751. doi: 10.1136/jcp.54.10.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, McClane BA. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl. Environ. Microbiol. 2006;72:4561–4568. doi: 10.1128/AEM.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuchs A, Bonde GJ. The availability of sulphur for Clostridium perfringens and an examination of hydrogen sulphide production. J. Gen. Microbiol. 1957;16:330–340. doi: 10.1099/00221287-16-2-330. [DOI] [PubMed] [Google Scholar]
- 45.Rowan FE, Docherty NG, Coffey JC, O’Connell PR. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br. J. Surg. 2009;96:151–158. doi: 10.1002/bjs.6454. [DOI] [PubMed] [Google Scholar]
- 46.Neu J, Walker AW. Necrotizing enterocolitis. N. Engl. J. Med. 2011;364:255–264. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen PV, Hansen FH, Halveg AB, Christiansen ED. Necrotising enterocolitis of the newborn--is it gas-gangrene of the bowel? Lancet. 1976;2:715–716. doi: 10.1016/S0140-6736(76)90009-X. [DOI] [PubMed] [Google Scholar]
- 48.Awad MM, Bryant AE, Stevens DL, Rood JI. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 1995;15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. [DOI] [PubMed] [Google Scholar]
- 49.Takehara M, et al. Clostridium perfringens alpha-toxin impairs innate immunity via inhibition of neutrophil differentiation. Sci. Rep. 2016;6:28192. doi: 10.1038/srep28192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timoney, J. F., Gillespie, J. H., Scott, F. W. & Barlough, J. E. Hagan and Bruner’s Microbiology and Infectious Diseases of Domestic Animals. (Comstock Publishing Associates, New York 1988).
- 51.Uzal FA, et al. Development and application of new mouse models to study the pathogenesis of Clostridium perfringens type C Enterotoxemias. Infect. Immun. 2009;77:5291–5299. doi: 10.1128/IAI.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murrel TGC. Pigbel in Papua New Guinea: an ancient disease rediscovered. Int. J. Epidemiol. 1983;12:211–214. doi: 10.1093/ije/12.2.211. [DOI] [PubMed] [Google Scholar]
- 53.Ma M, et al. Synergistic effects of Clostridium perfringens enterotoxin and beta toxin in rabbit small intestinal loops. Infect. Immun. 2014;82:2958–2970. doi: 10.1128/IAI.01848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunter SE, Brown JE, Oyston PC, Sakurai J, Titball RW. Molecular genetic analysis of beta-toxin of Clostridium perfringens reveals sequence homology with alpha-toxin, gamma-toxin, and leukocidin of Staphylococcus aureus. Infect. Immun. 1993;61:3958–3965. doi: 10.1128/iai.61.9.3958-3965.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Awad MM, Ellemor DM, Boyd RL, Emmins JJ, Rood JI. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect. Immun. 2001;69:7904–7910. doi: 10.1128/IAI.69.12.7904-7910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goossens E, et al. Rethinking the role of alpha toxin in Clostridium perfringens-associated enteric diseases: a review on bovine necro-haemorrhagic enteritis. Vet. Res. 2017;48:9. doi: 10.1186/s13567-017-0413-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verherstraeten S, et al. Perfringolysin O: the underrated Clostridium perfringens toxin? Toxins. 2015;7:1702–1721. doi: 10.3390/toxins7051702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park JM, Ng VH, Maeda S, Rest RF, Karin M. Anthrolysin O and other gram-positive cytolysins are toll-like receptor 4 agonists. J. Exp. Med. 2004;200:1647–1655. doi: 10.1084/jem.20041215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verherstraeten S, et al. The synergistics necrohemorrhagic action of Clostridium perfringens perfringolysin and alpha toxin in the bovine intestine and against bovine endothelial cells. Vet. Res. 2013;44:45. doi: 10.1186/1297-9716-44-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibert M, Jolivet-Reynaud C, Popoff MR. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene. 1997;203:65–73. doi: 10.1016/S0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 61.Fisher DJ, et al. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 2005;56:747–762. doi: 10.1111/j.1365-2958.2005.04573.x. [DOI] [PubMed] [Google Scholar]
- 62.Vilei EM, et al. Antibiotic-induced expression of a cryptic cpb2 gene in equine beta2-toxigenic Clostridium perfringens. Mol. Microbiol. 2005;57:1570–1581. doi: 10.1111/j.1365-2958.2005.04789.x. [DOI] [PubMed] [Google Scholar]
- 63.Popoff MR. Epsilon toxin: a fascinating pore-forming toxin. FEBS J. 2011;278:4602–4615. doi: 10.1111/j.1742-4658.2011.08145.x. [DOI] [PubMed] [Google Scholar]
- 64.Tamai E, et al. Accumulation of Clostridium perfringens epsilon-toxin in the mouse kidney and its possible biological significance. Infect. Immun. 2003;71:5371–5375. doi: 10.1128/IAI.71.9.5371-5375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hilger H, et al. The long-lived nature of Clostridium perfringens iota toxin in mammalian cells induces delayed apoptosis. Infect. Immun. 2009;77:5593–5601. doi: 10.1128/IAI.00710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Obana N, Nomura N, Nakamura K. Structural requirement in Clostridium perfringens collagenase mRNA 5’ leader sequence for translational induction through small RNA-mRNA base pairing. J. Bacteriol. 2013;195:2937–2946. doi: 10.1128/JB.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Awad MM, et al. Construction and virulence testing of a collagenase mutant of Clostridium perfringens. Microb. Pathog. 2000;28:107–117. doi: 10.1006/mpat.1999.0328. [DOI] [PubMed] [Google Scholar]
- 68.Shinoda T, et al. Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin. Sci. Rep. 2016;6:33632. doi: 10.1038/srep33632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eichner M, et al. In colon epithelia, Clostridium perfringens enterotoxin causes focal leaks by targeting claudins which are apically accessible due to tight junction derangement. J. Infect. Dis. 2017;217:147–157. doi: 10.1093/infdis/jix485. [DOI] [PubMed] [Google Scholar]
- 70.Chakrabarti G, McClane BA. The importance of calcium influx, calpain and calmodulin for the activation of CaCo-2 cell death pathways by Clostridium perfringens enterotoxin. Cell Microbiol. 2005;7:129–146. doi: 10.1111/j.1462-5822.2004.00442.x. [DOI] [PubMed] [Google Scholar]
- 71.Llanco LA, Nakano V, Avila-Campos MJ. Sialidase production and genetic diversity in Clostridium perfringens type A isolated from chicken with necrotic enteritis in Brazil. Curr. Microbiol. 2015;70:330–337. doi: 10.1007/s00284-014-0722-5. [DOI] [PubMed] [Google Scholar]
- 72.Li, J. & McClane, B. A. NanI sialidase can support the growth and survival of Clostridium perfringens strain F4969 using sialyated host macromolecules (Mucin) or Caco-2 cells. Infect. Immun., 10.1128/IAI.00547-17 (2017). [DOI] [PMC free article] [PubMed]
- 73.Flores-Diaz M, et al. A cellular deficiency of gangliosides causes hypersensitivity to Clostridium perfringens phospholipase C. J. Biol. Chem. 2005;280:26680–26689. doi: 10.1074/jbc.M500278200. [DOI] [PubMed] [Google Scholar]
- 74.Theoret, J. R. et al. Native or proteolytically activated NanI sialidase enhances the binding and cytotoxic activity of Clostridium perfringens enterotoxin and beta toxin. Infect. Immun. 86, 10.1128/IAI.00730-17 (2018). [DOI] [PMC free article] [PubMed]
- 75.Chiarezza M, et al. The NanI and NanJ sialidases of Clostridium perfringens are not essential for virulence. Infect. Immun. 2009;77:4421–4428. doi: 10.1128/IAI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keyburn AL, Bannam TL, Moore RJ, Rood JI. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Toxins. 2010;2:1913–1927. doi: 10.3390/toxins2071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keyburn AL, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abildgaard L, Sondergaard TE, Engberg RM, Schramm A, Hojberg O. In vitro production of necrotic enteritis toxin B, netB, by netB-positive and netB-negative Clostridium perfringens originating from healthy and diseased broiler chickens. Vet. Microbiol. 2010;144:231–235. doi: 10.1016/j.vetmic.2009.12.036. [DOI] [PubMed] [Google Scholar]
- 79.Lepp D, et al. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS ONE. 2010;5:e10795. doi: 10.1371/journal.pone.0010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rood JI, Keyburn AL, Moore RJ. NetB and necrotic enteritis: the hole movable story. Avian Pathol. 2016;45:295–301. doi: 10.1080/03079457.2016.1158781. [DOI] [PubMed] [Google Scholar]
- 81.Johnstone FR, Cockcroft WH. Clostridium welchii resistance to tetracycline. Lancet. 1968;1:660–661. doi: 10.1016/S0140-6736(68)92098-9. [DOI] [PubMed] [Google Scholar]
- 82.Rood JI, Buddle JR, Wales AJ, Sidhu R. The occurrence of antibiotic resistance in Clostridium perfringens from pigs. Aust. Vet. J. 1985;62:276–279. doi: 10.1111/j.1751-0813.1985.tb14251.x. [DOI] [PubMed] [Google Scholar]
- 83.Lyras D, Rood JI. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob. Agents Chemother. 1996;40:2500–2504. doi: 10.1128/AAC.40.11.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rood JI, Maher EA, Somers EB, Campos E, Duncan CL. Isolation and characterization of multiply antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob. Agents Chemother. 1978;13:871–880. doi: 10.1128/AAC.13.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soge OO, Tivoli LD, Meschke JS, Roberts MC. A conjugative macrolide resistance gene, mef(A), in environmental Clostridium perfringens carrying multiple macrolide and/or tetracycline resistance genes. J. Appl. Microbiol. 2009;106:34–40. doi: 10.1111/j.1365-2672.2008.03960.x. [DOI] [PubMed] [Google Scholar]
- 86.Slavic D, et al. Antimicrobial susceptibility of Clostridium perfringens isolates of bovine, chicken, porcine, and turkey origin from Ontario. Can. J. Vet. Res. 2011;75:89–97. [PMC free article] [PubMed] [Google Scholar]
- 87.Ngamwongsatit B, et al. Multidrug resistance in Clostridium perfringens isolated from diarrheal neonatal piglets in Thailand. Anaerobe. 2016;38:88–93. doi: 10.1016/j.anaerobe.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 88.Li, C., Yan, X. & Lillehoj, H. S. Complete genome sequence of Clostridium perfringens LLY_N11, a necrotic enteritis-inducing strain isolated from a healthy chicken intestine. Genome Announc. 5, 10.1128/genomeA.01225-17 (2017). [DOI] [PMC free article] [PubMed]
- 89.Bryan, L. E., Kowand, S. K. Van Den Elzen, H. M. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother.15, 7–13 (1979). [DOI] [PMC free article] [PubMed]
- 90.Udhayavel S, Thippichettypalayam Ramasamy G, Gowthaman V, Malmarugan S, Senthilvel K. Occurrence of Clostridium perfringens contamination in poultry feed ingredients: Isolation, identification and its antibiotic sensitivity pattern. Anim. Nutr. 2017;3:309–312. doi: 10.1016/j.aninu.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parish WE. Necrotic enteritis in the fowl (Gallus gallus domesticus). I. Histopathology of the disease and isolation of a strain of Clostridium welchii. J. Comp. Pathol. 1961;71:377–393. doi: 10.1016/S0368-1742(61)80043-X. [DOI] [PubMed] [Google Scholar]
- 92.Zahoor I, Ghayas A, Basheer A. Genetics and genomics of susceptibility and immune response to necrotic enteritis in chicken: a review. Mol. Biol. Rep. 2017;45:31–37. doi: 10.1007/s11033-017-4138-8. [DOI] [PubMed] [Google Scholar]
- 93.Wade, B., and Keybun, A. 2015. The true cost of necrotic enteritis. Poultry World [Online]. Available: https://www.poultryworld.net/Meat/Articles/2015/10/The-true-cost-of-necrotic-enteritis-2699819W/ [Accessed 30 July 2018].
- 94.Timbermont L, Haesebrouck F, Ducatelle R, Van Immerseel F. Necrotic enteritis in broilers: an updated review on the pathogenesis. Avian Pathol. 2011;40:341–347. doi: 10.1080/03079457.2011.590967. [DOI] [PubMed] [Google Scholar]
- 95.Olkowski AA, Wojnarowicz C, Chirino-Trejo M, Laarveld B, Sawicki G. Sub-clinical necrotic enteritis in broiler chickens: novel etiological consideration based on ultra-structural and molecular changes in the intestinal tissue. Res. Vet. Sci. 2008;85:543–553. doi: 10.1016/j.rvsc.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 96.Coursodon CF, Glock RD, Moore KL, Cooper KK, Songer JG. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe. 2012;18:117–121. doi: 10.1016/j.anaerobe.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 97.Drew MD, Syed NA, Goldade BG, Laarveld B, Van Kessel AG. Effects of dietary protein source and level on intestinal populations of Clostridium perfringens in broiler chickens. Poult. Sci. 2004;83:414–420. doi: 10.1093/ps/83.3.414. [DOI] [PubMed] [Google Scholar]
- 98.Gohari IM, et al. Characterization of Clostridium perfringens in the feces of adult horses and foals with acute enterocolitis. Can. J. Vet. Res. 2014;78:1–7. [PMC free article] [PubMed] [Google Scholar]
- 99.Diab SS, et al. Pathology of Clostridium perfringens type C enterotoxemia in horses. Vet. Pathol. 2012;49:255–263. doi: 10.1177/0300985811404710. [DOI] [PubMed] [Google Scholar]
- 100.Gohari, I. M. et al. NetF-producing Clostridium perfringens: clonality and plasmid pathogenicity loci analysis. Infect. Genet. Evol.49, 32–38 (2017). [DOI] [PubMed]
- 101.Songer JG, Uzal FA. Clostridial enteric infections in pigs. J. Vet. Diagn. Invest. 2005;17:528–536. doi: 10.1177/104063870501700602. [DOI] [PubMed] [Google Scholar]
- 102.Niilo L. Clostridium perfringens Type C Enterotoxemia. Can. Vet. J. 1988;29:658–664. [PMC free article] [PubMed] [Google Scholar]
- 103.Hansen, K. et al. Darmbrand - Enteritis Necroticans. (George Thieme, Stuttgart 1949).
- 104.Sterne M, Warrack GH. The Types of Clostridium perfringens. J. Pathol. Bacteriol. 1964;88:279–283. doi: 10.1002/path.1700880135. [DOI] [PubMed] [Google Scholar]
- 105.Murrell TG, Roth L, Egerton J, Samels J, Walker PD. Pig-bel: enteritis necroticans. A study in diagnosis and management. Lancet. 1966;1:217–222. doi: 10.1016/S0140-6736(66)90048-1. [DOI] [PubMed] [Google Scholar]
- 106.Murrell TG, Egerton JR, Rampling A, Samels J, Walker PD. The ecology and epidemiology of the pig-bel syndrome in man in New Guinea. J. Hyg. 1966;64:375–396. doi: 10.1017/S0022172400040663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sakurai J, Duncan CL. Some properties of beta-toxin produced by Clostridium perfringens type C. Infect. Immun. 1978;21:678–680. doi: 10.1128/iai.21.2.678-680.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foster WD. The bacteriology of necrotizing jejunitis in Uganda. East Afr. Med. J. 1966;43:550–553. [PubMed] [Google Scholar]
- 109.Gan KH, Sukono D, Satari S, Sujudi RW, Njooo SN. First outbreak of necrotising enteritis caused by Clostridium welchii in Indonesia. Nadj Kedkt. 1967;1:802–806. [Google Scholar]
- 110.Gui L, Subramony C, Fratkin J, Hughson MD. Fatal enteritis necroticans (pigbel) in a diabetic adult. Mod. Pathol. 2002;15:66–70. doi: 10.1038/modpathol.3880491. [DOI] [PubMed] [Google Scholar]
- 111.Know R, MacDonald E. Outbreaks of foodpoisoning in certain Leicester institutions. Med. Offr. 1943;69:21–22. [Google Scholar]
- 112.Scallan E, et al. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Felicio DaSilva, et al. Risk ranking of pathogens in ready-to-eat unprocessed foods of non-animal origin (FoNAO) in the EU: initial evaluation using outbreak data (2007-2011) Int. J. Food Microbiol. 2015;195:9–19. doi: 10.1016/j.ijfoodmicro.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 114.Dolan GP, et al. An epidemiological review of gastrointestinal outbreaks associated with Clostridium perfringens, North East of England, 2012-2014. Epidemiol. Infect. 2016;144:1386–1393. doi: 10.1017/S0950268815002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.DuPont HL. Clinical practice. Bacterial diarrhea. N. Engl. J. Med. 2009;361:1560–1569. doi: 10.1056/NEJMcp0904162. [DOI] [PubMed] [Google Scholar]
- 116.Collie RE, McClane BA. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 1998;36:30–36. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Borriello SP, et al. Enterotoxigenic Clostridium perfringens: a possible cause of antibiotic-associated diarrhoea. Lancet. 1984;1:305–307. doi: 10.1016/S0140-6736(84)90359-3. [DOI] [PubMed] [Google Scholar]
- 118.Hogenauer C, Hammer HF, Krejs GJ, Reisinger EC. Mechanisms and management of antibiotic-associated diarrhea. Clin. Infect. Dis. 1998;27:702–710. doi: 10.1086/514958. [DOI] [PubMed] [Google Scholar]
- 119.Brett MM, Rodhouse JC, Donovan TJ, Tebbutt GM, Hutchinson DN. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhoea. J. Clin. Pathol. 1992;45:609–611. doi: 10.1136/jcp.45.7.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Larson HE, Borriello SP. Infectious diarrhea due to Clostridium perfringens. J. Infect. Dis. 1988;157:390–391. doi: 10.1093/infdis/157.2.390. [DOI] [PubMed] [Google Scholar]
- 121.Mpamugo O, Donovan T, Brett MM. Enterotoxigenic Clostridium perfringens as a cause of sporadic cases of diarrhoea. J. Med. Microbiol. 1995;43:442–445. doi: 10.1099/00222615-43-6-442. [DOI] [PubMed] [Google Scholar]
- 122.Wong S, et al. Use of antibiotic and prevalence of antibiotic-associated diarrhoea in-patients with spinal cord injuries: a UK national spinal injury centre experience. Spinal Cord. 2017;55:583–587. doi: 10.1038/sc.2016.193. [DOI] [PubMed] [Google Scholar]
- 123.Ayyagari A, Agarwal J, Garg A. Antibiotic associated diarrhoea: infectious causes. Indian. J. Med. Microbiol. 2003;21:6–11. [PubMed] [Google Scholar]
- 124.Asha NJ, Wilcox MH. Laboratory diagnosis of Clostridium perfringens antibiotic-associated diarrhoea. J. Med. Microbiol. 2002;51:891–894. doi: 10.1099/0022-1317-51-10-891. [DOI] [PubMed] [Google Scholar]
- 125.Asha NJ, Tompkins D, Wilcox MH. Comparative analysis of prevalence, risk factors, and molecular epidemiology of antibiotic-associated diarrhea due to Clostridium difficile, Clostridium perfringens, and Staphylococcus aureus. J. Clin. Microbiol. 2006;44:2785–2791. doi: 10.1128/JCM.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li J, McClane BA. Contributions of NanI sialidase to Caco-2 cell adherence by Clostridium perfringens type A and C strains causing human intestinal disease. Infect. Immun. 2014;82:4620–4630. doi: 10.1128/IAI.02322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Larcombe S, Hutton ML, Lyras D. Involvement of bacteria other than Clostridium difficile in antibiotic-associated diarrhoea. Trends Microbiol. 2016;24:463–476. doi: 10.1016/j.tim.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 128.Pituch H, et al. Laboratory diagnosis of antibiotic-associated diarrhea: a Polish pilot study into the clinical relevance of Clostridium difficile and Clostridium perfringens toxins. Diagn. Microbiol. Infect. Dis. 2007;58:71–75. doi: 10.1016/j.diagmicrobio.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 129.Kim YJ, et al. Prevalence of Clostridium perfringens toxin in patients suspected of having antibiotic-associated diarrhea. Anaerobe. 2017;48:34–36. doi: 10.1016/j.anaerobe.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 130.Rees CM, Eaton S, Pierro A. National prospective surveillance study of necrotizing enterocolitis in neonatal intensive care units. J. Pediatr. Surg. 2010;45:1391–1397. doi: 10.1016/j.jpedsurg.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 131.Lim JC, Golden JM, Ford HR. Pathogenesis of neonatal necrotizing enterocolitis. Pediatr. Surg. Int. 2015;31:509–518. doi: 10.1007/s00383-015-3697-9. [DOI] [PubMed] [Google Scholar]
- 132.Bisquera JA, Cooper TR, Berseth CL. Impact of necrotizing enterocolitis on length of stay and hospital charges in very low birth weight infants. Pediatrics. 2002;109:423–428. doi: 10.1542/peds.109.3.423. [DOI] [PubMed] [Google Scholar]
- 133.Berdon WE, et al. Necrotizing enterocolitis in the premature infant. Radiology. 1964;83:879–887. doi: 10.1148/83.5.879. [DOI] [PubMed] [Google Scholar]
- 134.Zhou Y, et al. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: a case-control study. PLoS ONE. 2015;10:e0118632. doi: 10.1371/journal.pone.0118632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cassir N, et al. Clostridium butyricum strains and dysbiosis linked to necrotizing enterocolitis in preterm neonates. Clin. Infect. Dis. 2015;61:1107–1115. doi: 10.1093/cid/civ468. [DOI] [PubMed] [Google Scholar]
- 136.Butel MJ, et al. Clostridial pathogenicity in experimental necrotising enterocolitis in gnotobiotic quails and protective role of bifidobacteria. J. Med. Microbiol. 1998;47:391–399. doi: 10.1099/00222615-47-5-391. [DOI] [PubMed] [Google Scholar]
- 137.Hanke CA, et al. Clostridium perfringens intestinal gas gangrene in a preterm newborn. Eur. J. Pediatr. Surg. 2009;19:257–259. doi: 10.1055/s-2008-1038958. [DOI] [PubMed] [Google Scholar]
- 138.Kindley AD, Rboerts PJ, Tulloch WH. Neonatal necrotising enterocolitis. Lancet. 1977;1:649. doi: 10.1016/S0140-6736(77)92082-7. [DOI] [PubMed] [Google Scholar]
- 139.Silverman MA, Konnikova L, Gerber JS. Impact of antibiotics on necrotizing enterocolitis and antibiotic-associated diarrhea. Gastroenterol. Clin. North Am. 2017;46:61–76. doi: 10.1016/j.gtc.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 2016;14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Blakey JL, et al. Development of gut colonisation in pre-term neonates. J. Med. Microbiol. 1982;15:519–529. doi: 10.1099/00222615-15-4-519. [DOI] [PubMed] [Google Scholar]
- 142.Soll RF, Edwards WH. Antibiotic use in neonatal intensive care. Pediatrics. 2015;135:928–929. doi: 10.1542/peds.2015-0707. [DOI] [PubMed] [Google Scholar]
- 143.Alexander VN, Northrup V, Bizzarro MJ. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J. Pediatr. 2011;159:392–397. doi: 10.1016/j.jpeds.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Izumiya H, et al. Whole-genome analysis of Salmonella enterica serovar Typhimurium T000240 reveals the acquisition of a genomic island involved in multidrug resistance via IS1 derivatives on the chromosome. Antimicrob. Agents Chemother. 2011;55:623–630. doi: 10.1128/AAC.01215-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ramos PI, et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genom. 2014;15:54. doi: 10.1186/1471-2164-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kotsanas D, et al. Novel use of tryptose sulfite cycloserine egg yolk agar for isolation of Clostridium perfringens during an outbreak of necrotizing enterocolitis in a neonatal unit. J. Clin. Microbiol. 2010;48:4263–4265. doi: 10.1128/JCM.01724-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sim, K. Defining the gastrointestinal microbiota in premature neonates: Its development and relation to necrotizing enterocolitis Doctor of Philosophy (PhD) thesis, Imperial College London, (2015).
- 148.Nagler FPO. Observations on a reaction between the lethal toxin of Cl. welchii (Type A) and human serum. Br. J. Exp. Pathol. 1939;20:473–485. [Google Scholar]
- 149.Hansen M, Elliott L. New presumptive identification test for Clostridium perfringens: reverese CAMP test. J. Clin. Microbiol. 1980;12:617–619. doi: 10.1128/jcm.12.4.617-619.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.van Asten AJ, van der Wiel CW, Nikolaou G, Houwers DJ, Grone A. A multiplex PCR for toxin typing of Clostridium perfringens isolates. Vet. Microbiol. 2009;136:411–412. doi: 10.1016/j.vetmic.2008.11.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.