Abstract
Fetuin, also known as alpha-2-Heremans Schmid glycoprotein (AHSG), belongs to some of the most abundant glycoproteins secreted into the bloodstream. In blood, fetuins exhibit functions as carriers of metals and small molecules. Bovine fetuin, which harbors 3 N-glycosylation sites and a suggested half dozen O-glycosylation sites, has been used often as a model glycoprotein to test novel analytical workflows in glycoproteomics. Here we characterize and compare fetuin in depth, using protein from three different biological sources: human serum, bovine serum, and recombinant human fetuin expressed in HEK-293 cells, with the aim to elucidate similarities and differences between these proteins and the post-translational modifications they harbor. Combining data from high-resolution native mass spectrometry and glycopeptide centric LC-MS analysis, we qualitatively and quantitatively gather information on fetuin protein maturation, N-glycosylation, O-glycosylation, and phosphorylation. We provide direct experimental evidence that both the human serum and part of the recombinant proteins are processed into two chains (A and B) connected by a single interchain disulfide bridge, whereas bovine fetuin remains a single-chain protein. Although two N-glycosylation sites, one O-glycosylation site, and a phosphorylation site are conserved from bovine to human, the stoichiometry of the modifications and the specific glycoforms they harbor are quite distinct. Comparing serum and recombinant human fetuin, we observe that the serum protein harbors a much simpler proteoform profile, indicating that the recombinant protein is not ideally engineered to mimic human serum fetuin. Comparing the proteoform profile and post-translational modifications of human and bovine serum fetuin, we observe that, although the gene structures of these two proteins are alike, they represent quite distinct proteins when their glycoproteoform profile is also taken into consideration.
Keywords: fetuin, alpha-2-HS glycoprotein, glycoprotein, serum proteins, native mass spectrometry, glycopeptides, proteoforms, hybrid mass spectrometry, N-glycosylation, O-glycosylation
Introduction
The fetuins are a group of related proteins belonging to the cystatin superfamily. These multifunctional proteins were decades ago already identified in various mammals including humans.1,2 Fetuin was discovered in 1944 by Kai Pedersen in fetal calf serum.3 There has been some initial confusion related to the fetuin naming, which led to the mixed use of the name alpha-2-HS glycoprotein in some species and fetuin in others. Since 1990, bovine fetuin and human alpha-2-HS glycoprotein have been considered as species homologues.4 Here, for the naming we follow the recommendation by Brown et al. in 1992 and use the name “fetuin” for both human fetuin (hFet) and bovine fetuin (bFet).5 Fetuins are known for their complicated heterogeneous structure and many reported discrepancies related to their putative biological functions. Despite years of research, the biological function of fetuins and a true understanding of their biological importance is still unclear. Some consensus has been found in the role of hFet in calcium metabolism6,7 and insulin signaling.8 hFet is also extensively studied for its potential relevance as a metabolic biomarker.9,10 Increased levels of hFet have been linked to higher risk of cardiovascular disease (CVD) and incident type 2 diabetes (T2DM).11,12 However, there are several obstacles preventing the use of hFet as a biomarker for those and any other diseases. These include, for example, a lack of reference values, inconsistent values from various commercial enzyme-linked immunosorbent assays (ELISA), and the unknown effect of post-translational modifications (PTMs) on hFet clinical measurements.13 One source of confusion may also originate from some in vitro studies, where recombinant human fetuin was used.14,15 Serum hFet is predominantly synthesized in the liver, where its N-glycosylation pattern originates.16 The glycosylation machinery is species-specific, and thus, proteins produced by different expression systems provide products with distinct glycosylation patterns.17−19 Recombinant human fetuin (rhFet) synthesized in human embryonic kidney cells (HEK-293) is often applied for antibody validation, ELISA assays, immunoprecipitation, or protein functional assays. Therefore, we included this rhFet in our study to investigate potential structural differences between serum-derived hFet and recombinant rhFet.
All described fetuins are glycoproteins, and especially the glycosylation profile of bFet is well-documented in the literature.20,21 For that reason, bFet has also been widely used as a standard glycoprotein for method development in glycoproteomics. Post-translational modifications (PTMs) on hFet have been less described, and even the primary structure of mature hFet is somehow elusive. Amino acid sequence alignment of hFet and bFet shows a relatively high sequence similarity (∼70%), suggesting a high degree of similarity (Figure S1 in the Supporting Information). Also, fetuins from other mammalian species reflect a high sequence conservation showing 60–70% homology at the amino acid level and 80–90% homology at the cDNA level.5 Nevertheless, mature hFet harbors some unique features, as it is present in serum in the form of two chains connected to each other by a single interchain disulfide bridge, while other fetuins, including bFet, are found in serum in a single-chain form.22 PTMs of proteins, in general, play an important role in regulating their structure, function, and interactions.23,24 Regarding hFet and bFet, the published data document the presence of N- and O-glycosylation sites and a few phosphosites.25,26 Although the types of modifications are alike in both proteins, the number, structures, and distribution on their primary structure is distinct. The O-glycosylation sites are less conserved than the N-glycosylation sites.4 Additionally, the number of reported phosphosites on bFet is higher in comparison to hFet.20,25 Differences in the structure of N-linked glycans released from fetuins isolated from various biological sources are well-documented.5 These findings supported the concept of species specificity of N-glycan structure in glycoproteins from different species.27,28 In this work, we follow up these earlier studies and extend them by an in-depth site-specific characterization of fetuins from three different biological sources using state-of-the-art hybrid mass spectrometry (MS) approaches. In our earlier works, we showed the great utility of combining high-resolution MS and peptide-centric MS for comprehensive and unbiased analysis of blood serum protein PTMs.29−31 Here we aim to provide detailed information illustrating the differences between hFet, bFet, and rhFet, with an emphasis on their primary structure and PTMs. Our data provide new evidence for post-translational events occurring on the three fetuins and show how similar gene products synthesized in various species can mature into very different molecules with potentially different functions.
Materials and Methods
Chemicals and Materials
hFet (alpha-2-HS glycoprotein; Uniprot Code: P02765), bFet (bovine fetuin; Uniprot Code: P12763), and rhFet (recombinant alpha-2-HS glycoprotein expressed in HEK293 cells), dithiothreitol (DTT), iodoacetamide (IAA), trifluoroacetic acid (TFA), ammonium bicarbonate (ABC), and ammonium acetate (AMAC) were purchased from Sigma-Aldrich (Steinheim, Germany). Acetonitrile was purchased from Biosolve (Valkenswaard, The Netherlands). Sequencing-grade trypsin was obtained from Promega (Madison, WI). Gluc-C, Lys-C, PNGaseF,32 and Sialidase were obtained from Roche (Indianapolis, IN). Alkaline phosphatase was purchased from New England Biolabs (Ipswich, MA).
Sample Preparation for Native MS
Unprocessed hFet, bFet, and rhFet in deionized water, containing 25–30 μg of the protein, were buffer-exchanged into 150 mM aqueous ammonium acetate (AMAC) (pH 7.2) by ultrafiltration (vivaspin500, Sartorius Stedim Biotech, Germany) with a 10 kDa cutoff filter. The protein concentration was measured by UV absorbance at 280 nm and adjusted to 2–3 μM prior to native MS analysis. The enzyme PNGase was used to remove the N-glycans of the fetuins, and sialidase was used to cleave sialic acid residues.32 Alkaline phosphatase was used for the removal of phosphate groups. DTT (4 mM) was used to reduce the disulfide bonds between the A chain and the B chain in hFet. All samples for different treatments were buffer-exchanged to 150 mM AMAC (pH 7.2) prior to native MS analysis.
Native MS Analysis of hFet, bFet, and rhFet
Samples were analyzed on a modified Exactive Plus Orbitrap instrument with extended mass range (EMR) (Thermo Fisher Scientific, Bremen) using a standard m/z range of 500–10 000, as described in detail previously.33 The voltage offsets on the transport multipoles and ion lenses were manually tuned to achieve optimal transmission of protein ions at elevated m/z. Nitrogen was used in the higher-energy collision dissociation (HCD) cell at a gas pressure of 6–8 × 10–10 bar. The MS parameters were used typically: spray voltage 1.2–1.3 V, source fragmentation 30 V, source temperature 250 °C, collision energy 30 V, and resolution (at m/z 200) 17 500. The mass spectrometer was calibrated using CsI clusters as described previously.33
Native MS Data Analysis
The accurate masses of observed hFet, bFet, and rhFet proteoforms were extracted by deconvoluting the electrospray ionization (ESI) spectrum to zero-charge spectrum using Intact Mass software by Protein Metrics in ver. 1.5.34 For PTM composition analysis, data was processed manually and glycan structures were deduced on the basis of known biosynthetic pathways. The average masses were used for these calculations, including hexose/mannose/galactose (Hex/Man/Gal, 162.1424 Da), N-acetylhexosamine/N-acetylglucosamine (HexNAc/GlcNAc, 203.1950 Da), deoxyhexose (dHex, 146.1430 Da), N-acetylneuraminic acid (Neu5Ac, 291.2579 Da), and phosphorylation (Pho, 79.9799 Da). All used symbols and text nomenclature are based on the recommendation of the Consortium for Functional Glycomics.35
In-Solution Digestion for Peptide-Centric Proteomics
All proteins (bFet, hFet, and rhFet) were reconstituted in 50 mM ABC at a concentration of 1 mg/mL, reduced with 4 mM DTT at 56 °C for 30 min, and alkylated with 8 mM IAA at room temperature for 30 min in the dark. bFet was digested for 3 h with Glu-C at an enzyme-to-protein-ratio of 1:75 (w/w) at 37 °C, and the resulting peptide mixtures were further treated by using trypsin (1:100; w/w). hFet and rhFet were digested for 3 h with Lys-C at an enzyme-to-protein ratio of 1:75 (w/w) at 37 °C, and the resulting peptide mixtures were further treated by using Glu-C (1:100; w/w). All proteolytic digests containing modified glycopeptides were desalted by using GELoader tips filled with POROS Oligo R3 50 μm particles, dried, and reconstituted in 20 μL of 0.1% FA prior to liquid chromatography (LC)-MS and MS/MS analysis.36
LC-MS and MS/MS Analysis
All peptides generated from fetuin (typically 300 fmol) were separated and analyzed using an Agilent 1290 Infinity HPLC system (Agilent Technologies, Waldbronn, Germany) coupled online to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Reversed-phase separation was accomplished using a 100 μm inner diameter 2 cm trap column (in-house packed with ReproSil-Pur C18-AQ, 3 μm) (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) coupled to a 50 μm inner diameter 50 cm analytical column (in-house packed with Poroshell 120 EC-C18, 2.7 μm) (Agilent Technologies, Amstelveen, The Netherlands). Mobile-phase solvent A consisted of 0.1% formic acid in water, and mobile-phase solvent B consisted of 0.1% formic acid in acetonitrile. The flow rate was set to 300 nL/min. A 45 min gradient was used as follows: 0–5 min, 100% solvent A; 13–44% solvent B within 20 min; 44–100% solvent B within 3 min; 100% solvent B for 1 min; and 100% solvent A for 17 min. For the MS scan, the mass range was set from 375 to 1500 m/z at a resolution of 120 000, and the automatic gain control (AGC) target was set to 4 × 105. For the MS/MS measurements, both higher-energy collision dissociation (HCD) and electron-transfer combined with higher-energy collision dissociation (EThcD) were used and performed with normalized collision energy of 35%. For the MS/MS scan, the mass range was set from 125 to 2000 m/z; the AGC target was set to 5 × 104. The precursor isolation width was 1.6 Da, and the maximum injection time was set to 200 ms.
LC-MS and MS/MS Data Analysis
The raw data files were processed using Proteome Discoverer 2.2 software (Thermo Fisher Scientific) (PD 2.2) equipped with the Byonic software node (Protein Metrics, Inc.).37 The following parameters were used for data searches in Byonic: precursor ion mass tolerance, 10 ppm; product ion mass tolerance, 20 ppm; fixed modification, Cys carbamidomethyl; variable modification, Met oxidation, STY phosphorylation, and both N- and O- glycosylation from mammalian glycan databases. The allowed number of peptide missed cleavages was set to 3. The protein database used contained the hFet (Uniprot Code: P02765) or bFet (Uniprot Code: P12763) amino acid sequences. Site-specific quantification of the fetuin PTMs was performed as follows. Each peptide that contains PTM sites was normalized individually so that the sum of all its proteoform areas was set to 100%. The average peptide ratios from all measurements were taken as a final estimation of the abundance. The extracted ion chromatograms (XICs) were obtained using the software Thermo Proteome Discoverer 2.2.0.388. The glycan structures of each glycoform were manually annotated. Hereby reported glycan structures are depicted without the linkage type of the glycan units because the acquired MS/MS patterns do not provide such information.
Combining Native MS and Peptide-Centric Proteomic Data
Validation of the obtained proteoform profiles of all three fetuins was assessed by an integrative approach combining the native MS data with the peptide-centric proteomics data. This approach has been described in detail previously.29 Briefly, in silico data construction of the “intact protein spectra” was performed based on the masses and relative abundances of all site-specific PTMs derived from the peptide-centric analysis. Subsequently, the constructed spectra were compared to the experimental native MS spectra of the fetuins. The similarity between the two independent data sets (native MS spectra and constructed spectra based on peptide-centric data) was expressed by a Pearson correlation factor. All R scripts used for the spectra simulation are available at github (https://github.com/Yang0014/glycoNativeMS).
Results
Native MS Reveals Remarkable Differences in Structural Heterogeneity among hFet, bFet, and rhFet
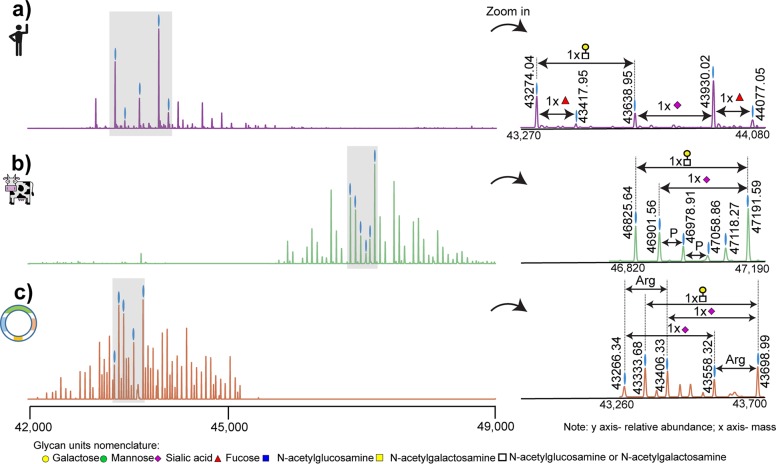
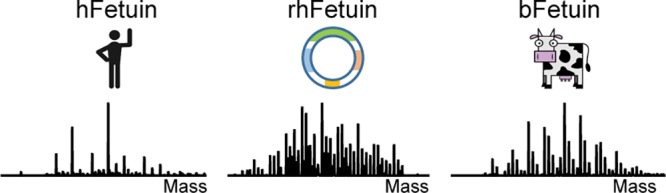
We started our investigation by acquiring high-resolution native ESI-MS spectra of hFet, bFet, and rhFet. Even at first glance, deconvoluted zero-charge spectra show remarkable differences among these three samples (Figure 1). When recording the full proteoform profile of intact hFet by native MS, >30 peaks could be base-line resolved. This number is in pronounced contrast to the number of detected peaks in the native MS spectra of bFet (>40 peaks) and rhFet (>50 peaks). According to the literature, the molecular heterogeneity of fetuins is mainly caused by N-glycosylation and O-glycosylation. Indeed, the observed mass differences among the most abundant peaks in all three native spectra of the fetuins correspond to the presence of glycans. Nevertheless, a closer look at the proteoform profiles reveals some other less-expected structural variants. The heterogeneity of bFet native spectrum is significantly enriched by the presence of many lower-intensity peaks, indicating the attachment of phosphate moieties (+80 Da) (Figure 1b). The most complicated proteoform profile among all three samples can be observed for rhFet (Figure 1c). Interestingly, many proteoform signals in rhFet coexist in pairs differing from each other by a mass of 156 Da. This is likely due to the mass increment of arginine, which will be discussed later.
Figure 1.
Deconvoluted zero-charge mass spectra of (a) hFet, (b) bFet, and (c) rhFet. The zoom-ins on the right depict the most abundant peaks in the spectra to more clearly show the observed mass differences in each spectrum that originated mainly from distinct glycan moieties. The presence of a third N-glycosylation site in bFet increases the molecular weight and glycan heterogeneity of bFet compared to that of hFet. In (c), all proteoforms of rhFet are present in pairs, due to the co-occurrence of proteoforms with and without arginine, making the spectrum twice as complex as that of hFet. The glycan nomenclature used is indicated at the bottom.
Due to the high complexity of the data, we decided to focus on the full annotation of the native MS spectrum of hFet, and here refer to bFet or rhFet only in specific cases, also as the glycoproteome profile of bFet has already been well-characterized.20,38−40 The protein backbone amino acid sequence of hFet represents an average mass of 37 177.01 Da. This mass was calculated based on the gene sequence of hFet lacking the N-terminal signal peptide, including the mass shifts induced by the 6 disulfide bonds and the absence of arginine at position 322.22 Determining the exact backbone mass allowed us to calculate a mass shift of 6751.81 Da induced by the PTMs on the most abundant peak in the hFet native MS spectrum (43 930.02 Da). Next, we enzymatically treated hFet, attempting to remove either all N-glycans, the sialic acid moieties, or the phosphates, which results in specific mass shifts that we subsequently recorded by native MS. For the specific cleavage of N-glycosylations, we used PNGase F, sialidase for the removal of sialic acids, and finally alkaline phosphatase for the release of phosphate residues. Incubation with PNGase F resulted in the removal of only one N-glycan (Figure S2a). The mass difference of 2204 Da between the most abundant intact hFet proteoform with 43 930.02 Da (m/z = 3380.24) and the N-deglycosylated hFet with 41 724.80 Da (m/z = 3210.60) indicated the attachment of a N-glycan with the carbohydrate composition of HexNAc4Hex5Neu5Ac2. It is well-known that hFet contains two N-glycosylation sites. However, even prolonged incubations with PNGase F did not lead to the complete removal of N-glycans under native conditions. This is a well-documented problem attributed to the lower accessibility of the second N-glycosylation site due to steric hindrance. Sialidase treatment of hFet resulted in a pronounced simplification of the structural heterogeneity of the hFet proteoforms (Figure S2b), implying that the heterogeneity of hFet is mainly due to extensive modification with variable amounts of sialic acids. In total, 8 sialic acids were removed from the most abundant hFet proteoform as indicated by a mass shift of 2330 Da (8 × 291 Da). Lastly, we subjected hFet to treatment with alkaline phosphatase, which resulted in the cleavage of one phosphate group from all hFet proteoforms (Figure S2c). Although the composition of the second N-glycan on the most abundant hFet proteoform could not be determined due to the incomplete removal of N-glycans, the presence of this N-glycan is undoubtable based on the calculated PTM mass and information in the literature.16 The mass differences 365 (HexNAc1Hex1) and 656 Da (HexNAc1Hex1Neu5Ac1) between the particular proteoforms correspond either to variability in the number of antennas on the N-glycans and/or the presence of O-glycans. Combining all this information, we can assume that the overall PTM composition of the most abundant hFet proteoform includes two N-glycans, several O-glycans, and one phosphate moiety.
Native MS of hFet Treated with DTT Reveals Its Two-Polypeptide Chain Structure
In addition to the structural variability originating from various PTMs on fetuins, the primary polypeptide architecture is another prominent origin of differences between hFet and bFet. Almost three decades ago, Kellermann et al. isolated hFet from fresh human serum in the presence of proteinase inhibitors and determined that the major circulating form of hFet is likely a two-polypeptide-chain protein with a heavy chain (A chain) of 321 residues and a light chain (B chain) of 27 residues22 (Figure S3). This circulating form of hFet contains a propeptide (also called connecting peptide) with a missing C-terminal arginine residue (position 322) attached to the A chain. The A chain and the B chain are connected to each other by a single interchain disulfide bridge. We treated hFet with DTT to disrupt this linkage and validated the hypothesized arrangement of the primary structure. The subsequent recorded mass spectra, shown in Figure 2, reveal that the B chain was released under reducing conditions from the A chain and confirm the two-polypeptide-chain form of hFet. Notably, the released B chain appeared to be, at least, in two structural variants, unmodified and modified with a glycan (HexNAc1Hex1Neu5Ac1). This confirms not only the existence of hFet in its two-polypeptide form but also the presence of one O-glycan on the B chain.
Figure 2.
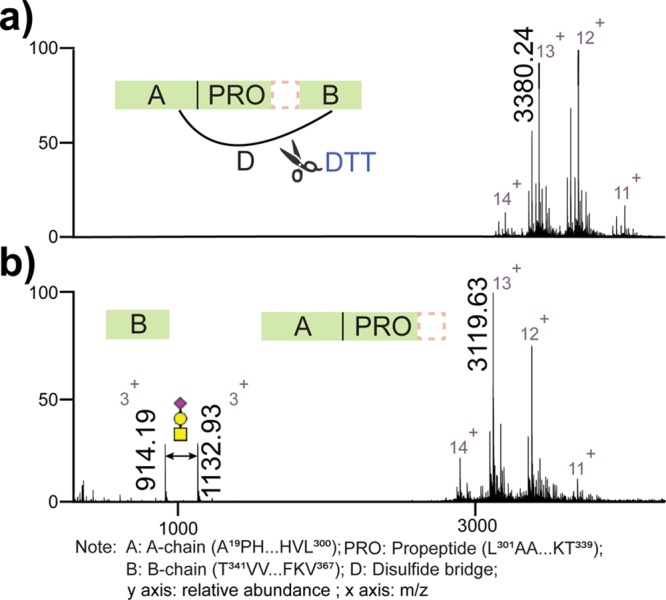
Full native ESI-MS spectra of intact hFet sprayed from aqueous ammonium acetate. (a) Schematic cartoon showing that the B chain is connected to the A chain by a disulfide bridge. (b) Full native ESI-MS spectra of hFet upon treatment by DTT. The released B chain and A chain are observed. The peaks at m/z of 914.19 and 1132.93 correspond to the B chain and B chain with 1 O-glycan (HexNAc1Hex1Neu5Ac1), respectively. Comparing the most abundant peak on the charge state 13+ with m/z of 3380.24 in (a) and 3119.63 in (b), the mass difference indeed originates from the released B chain harboring the O-glycan. The dashed line box represents missing C-terminal A-chain arginine.
Site-Specific Characterization of PTMs on hFet, bFet, and rhFet by Peptide-Centric Proteomics
Because the fetuins harbor at least three different types of PTMs, their analysis at the peptide level is a challenging task. We used two different combinations of proteolytic enzymes for the fetuin digestion. After a careful inspection of the fetuin amino acid sequences, we digested hFet and rhFet with Lys-C and Glu-C and bFet with trypsin and Glu-C. Combining tryptic/Lys-C with Glu-C specificity for the digestion of fetuins resulted in a set of peptides with a suitable length for subsequent sequencing by LC-MS/MS analysis. Lys-C was selected instead of trypsin for the digestion of hFet and rhFet to enable the confirmation of the absence/presence of the C-terminal Arginine on the A-chain. After enzymatic digestion, the peptide mixtures were subjected to EThcD fragmentation to obtain extensive fragment ions of both the glycan and the peptide moieties of glycopeptides. In addition to the PTM identification, we also assessed the relative abundances of the different (glyco/phospho)peptide isoforms. As a result, we identified and relatively quantified peptide isoforms from the putative N- and O-glycosylation and phosphorylation sites on all three investigated fetuins. The list of all modified peptide isoforms on hFet, bFet, and rhFet and their relative quantification, based on XICs, can be found in Table S1. Annotated HCD/EThcD spectra of all glycopeptides observed for hFet are provided in the Supporting Information.
Comparison of the Glycosylation Profile of hFet, bFet, and rhFet
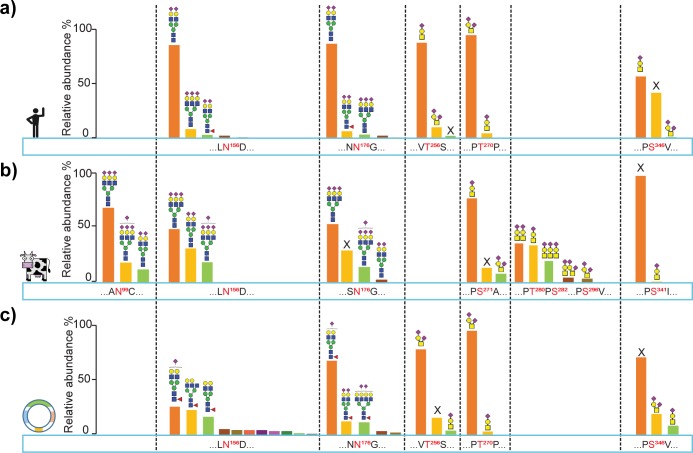
A summary of the site-specific glycosylation patterns in all investigated fetuins is depicted in Figure 3. Focusing first on the N-glycosylation in hFet and bFet (Figure 3a and b), we note that the three N-glycosylation consensus sequences in bFet are well-conserved in fetuins of most mammals. One exception, however, is hFet, which has the site N99 in bFet replaced by an arginine, preventing its N-glycosylation in hFet. If we next compare the N-glycans on the other two conserved sites (N156 and N176), both bFet and hFet contain complex N-glycans but differ in their structural composition and level of microheterogeneity. Sialylated diantennary complex type structures dominate on hFet and are typical for human serum proteins synthesized in the liver. Some less abundant glycoforms were found to be core-fucosylated, which is in sharp contrast to bFet, where no fucosylation of N-glycans was observed at all. The N-glycans present on bFet show a higher degree of branching, and also their quantitative distribution is more equal compared to the relatively more homogeneous hFet. In addition to this, approximately one-third of bFet molecules bear no glycan structure on the N176 site. Comparing next the N-glycosylation patterns on hFet and rhFet (Figure 3a and c), we observe remarkable differences at both N156 and N176 N-glycosylation sites. The most noticeable difference is the observed extensive microheterogeneity on both N-glycosylation sites in rhFet, represented by a repertoire of complex core fucosylated glycan structures with a large variety of branches.
Figure 3.
Overview of the observed qualitative and semiquantitative site-specific glycosylation in (a) hFet, (b) bFet, and (c) rhFet. The conserved glycosylation sites are depicted in the same column. Relative abundances of peptide proteoforms were estimated from their corresponding ion chromatograms (XICs). On a given modification site, the abundance of peptide proteoforms were normalized to 100%. All O-glycan structures and N-glycan structures attached to the 3 most abundant peptide isoforms of each site are depicted; further details of occupancy on each site is provided in the Table S1. (X means unmodified.)
The heterogeneity of the fetuin glycosylation patterns is further increased by the presence of O-glycosylation. The O-glycopeptides identified and characterized in the present study cover all known hFet O-glycosylation sites and were used to determine the composition and occupancy of the attached O-glycans (Figure 3a). As mentioned earlier, O-glycosylation sites are less conserved among fetuins, which also partly explains the observed major differences in O-glycosylation patterns. hFet/rhFet contain in total three reported O-glycosylation sites (T256, T270, and S346), while bFet harbors five sites (S271, T280, S282, S296, and S341). In all three investigated fetuins, we observed O-glycopeptides bearing simple mucin-type core 1 O-glycans with one or two sialic acids. Regarding the two conserved O-glycosylation sites on hFet and bFet (Figure 3a and b), T270 on hFet harbors mostly disialylated O-glycans while monosialylated structures reside on the analogous site S271 on bFet. Site S271 occurs in a cluster together with additional O-glycosylation sites T280, S282, and S296, which are absent in hFet. In both fetuins the second conserved O-glycosylation site is only partially occupied by glycosylation. S346 on hFet is unmodified in ∼40% of the molecules and bears mostly monosialylated O-glycans. Occupancy of S341 on bFet is negligible.20 The last hFet O-glycosylation site T256 is not present on bFet and is almost fully occupied by monosialylated O-glycans. Differences in the O-glycosylation patterns between hFet and rhFet are rather marginal (Figure 3a and c). Site T256 differs somewhat in the degree of sialylation, T270 is almost identical, and S346 on rhFet has a seemingly lower occupancy when compared to hFet.
Differences in Phosphorylation of hFet, bFet, and rhFet
The third type of PTM occurring on fetuins is phosphorylation. hFet contains two documented phosphorylation sites (S138 and S330) and bFet supposedly four (S138, S320, S323, and S325). Similar to the O-glycosylation sites, the phosphosites are less conserved among fetuins. hFet and bFet have two consensus phosphosites (S138 and S330/S325); however, their occupancy varies. While S138 on hFet was always found to be fully occupied, the analogous site on bFet was found to be occupied in only 10% of the proteoforms. The second phosphosite is located in a much less conserved region. In hFet, this sequence domain corresponds to the C-terminal A-chain propeptide and accommodates the partially phosphorylated site S330. The site S325 on bFet is situated in close proximity to the other two phosphosites, S320 and S323, and their occupancy is also only partial. Because of the low abundance of the phosphorylated peptides and phosphate lability, we were not able to unambiguously localize the neighboring phosphosites on bFet. Finally, in sharp contrast to hFet, we did not find any evidence of phosphorylation on rhFet.
Data Integration and Major Structural Differences among hFet, bFet, and rhFet
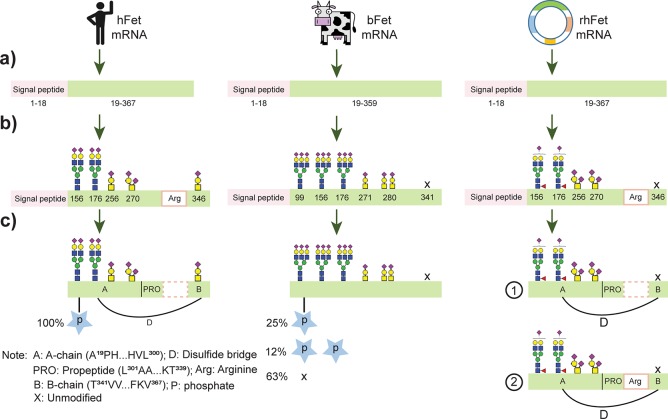
Having both the native MS data and peptide-centric data on all three fetuins available, we cross-validated the data to obtain a comprehensive view of the proteoform profiles of the fetuins. Figure 4 highlights the major differences observed among the most abundant proteoforms of hFet, bFet, and rhFet. In addition to the described structural differences based on various PTMs, hFet differs from bFet by its unique two-polypeptide-chain structure. The native MS data on rhFet suggested incomplete cleavage of the C-terminal arginine at position 322. This observation was further supported by two identified Lys-C peptides with amino acid sequences 321(K)TRTVVQPSVGAAAGPVVPPCPGRIRHFK(V)348 and 323 (R)TVVQPSVGAAAGPVVPPCPGRIRHFK(V)348, respectively (Figure S4). From this data, we conclude that rhFet occurs as a mixture of a one- and two-chain polypeptide forms, differing from each other by the presence/absence of the C-terminal arginine at the A-chain.
Figure 4.
Overall comparison of the protein structure and occurring PTMs on hFet, bFet, and rhFet. After (a) translation from single transcript, single-chain preproteins of hFet and bFet contain 367 and 365 amino acids, respectively. Then (b) N-glycosylation occurs in the endoplasmic reticulum followed by the modification of O-glycosylation sites. As in Figure 3, we depict the most abundant proteoforms at each glycosylation site. Distinctively, a high degree of fucosylation occurs on the N-glycosylation sites of rhFet, not observed for hFet. Also, bFet contains mainly triantennary complex glycans, whereas hFet predominantly biantennary glycan structures. The dominant O-glycans in all fetuins are of the core 1 mucin-type harboring one or two sialic acids. (c) The final step, proteolytic processing and phosphorylation, likely happens after glycosylation. The signal peptides are removed from the preproteins. For hFet, some unknown proteinase cleaves the C-terminal arginine at position 322 and converts hFet into a two-chain polypeptide form as we describe in more detail in Figure S3. Interestingly, we identified both the single-chain and two-chain polypeptide forms in rhFet, suggesting that in the recombinant expression system the cleavage of arginine is incomplete (Figure S4). For the phosphorylation, we found that all proteoforms of the hFet are fully monophosphorylated, whereas the phosphosite occupancy on bFet was about 25% monophosphorylated, 12% doubly phosphorylated, and 63% nonphosphorylated. On rhFet we found no evidence at all for protein phosphorylation.
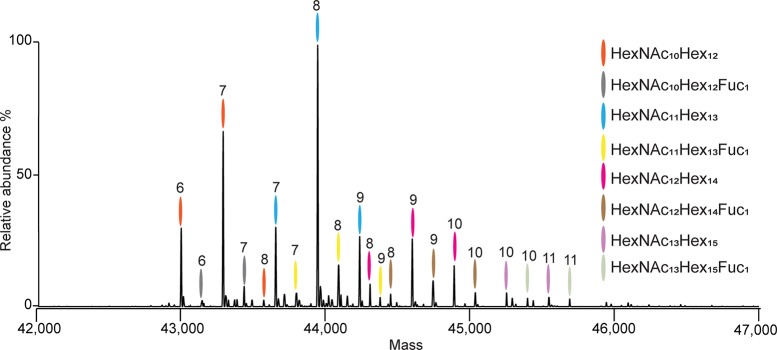
Next, we cross-validated the peptide-centric and native MS data on hFet and bFet by a correlative comparison between the native MS spectra and an in silico constructed MS spectrum based on all the quantitative information we gathered from the LC-MS/MS peptide-centric data. For hFet and bFet, we achieved a high degree of correlation (∼0.9) between our native MS and peptide-centric MS approach (Figure S5). Therefore, all hFet and bFet species predicted from the peptide-centric data were filtered by taking 1% cutoff in relative intensity of the peaks in the experimental native spectrum, and mass deviations were manually checked. Applying these criteria resulted in a list containing 21 hFet and 33 bFet distinct proteoforms (Table S2). As an example of our data, we provide the fully annotated native MS spectrum of hFet demonstrating the (near) completeness of our analysis (Figure 5). Although we could explain most of the ion signals detected in the native MS spectra from hFet and bFet, some unmatched low-abundant ion signals are still present. Those mostly correspond to adducts bearing Na+ and/or K+ ions, which represent frequent artifacts formed during the ESI ionization process.
Figure 5.
Fully annotated zero charge deconvoluted native mass spectrum of hFet. The overall PTM compositions were assigned based on the accurate mass measurements of the intact protein proteoforms. All proteoforms contain 1 phosphate moiety. The number of sialic acids attached is marked at the top of each peak. For example, the most abundant peak is marked in blue and number 8, as it corresponds to the glycan composition HexNAc11Hex13Neu5Ac8 and one phosphate moiety.
Discussion
To disclose that similar genes can lead to a plethora of distinct and different proteoforms, we here meticulously analyzed and compared fetuin originating from three different biological sources, using an integrative MS approach allowing an all-inclusive analysis of protein PTMs. Both hFet and bFet PTMs have been the subject of several structural studies.20,38−43 However, there has been, as far as we know, no study describing data on all three types of modifications (i.e., N-glycosylation, O-glycosylation, and phosphorylation) on hFet and bFet, pinpointing the major structural differences in a qualitative and quantitative site-specific manner. Earlier studies also reported on differences in fetuin DNA sequences, amino acid sequences, and PTMs among a range of species.44,5,39,45 Those studies provided the first evidence for specific structural variabilities among fetuins. Most mammalian fetuins show a high degree of sequence conservation, but their final protein structure can be significantly altered by species-specific PTMs. This intriguing phenomenon complicates structural and functional studies of proteins in general. Our main aim here was to demonstrate how three fetuins (hFet, bFet, and rhFet) exist in very different proteoform populations, as this likely affects their function and should thus be taken into consideration.
Serum-derived hFet and bFet are well-studied glycoproteins and bear both N- and O-linked glycans. bFet is often used as a model glycoprotein in glycoproteomics. Therefore, we did not expect any surprise observations in our analysis, and indeed, our findings are in good agreement with earlier studies. Nonetheless, our hybrid MS approach has the capacity to provide additional information regarding overall structural heterogeneity of the fetuins, which includes not only site-specific characterization of their PTMs but also analysis of their matured primary structure and its possible variants. Although native hFet is known to be present in serum in the form of a two-polypeptide chain linked by disulfide,46 no study provided evidence clarifying whether the processing of the hFet primary structure results in one or more sequence variants. We confirm here the existence of the proposed two-chain form architecture of hFet and further show that the cleavage of arginine at position 322 is complete in native hFet. In our hFet samples, no single-chain proteoforms or proteoforms missing the propeptide were detected. In contrast to the relatively simple proteoform profile of hFet, the native spectrum of rhFet exhibits remarkably more complexity. Major differences between hFet and rhFet originate from more complex glycosylation due to extensive core-fucosylated glycans on a various number of antennas. Furthermore, our data revealed that rhFet exists in two sequence variants differing from each other by the absence/presence of the C-terminal arginine on the A chain. This is likely caused by incomplete processing of the rhFet in the HEK-293 cells. In consequence, rhFet proteoforms coexist as a mixture of the one- and two-chain polypeptide forms, creating another source of structural diversity. Another striking difference between hFet and rhFet is that the former is for 100% a singly phosphorylated protein, whereas phosphorylation is completely missing in rhFet.
These findings seriously question whether the rhFet studied here represents a good model for wild-type serum hFet. Commercially available recombinant fetuin may be produced by various expression systems and is mostly used for scientific purposes. For example, recombinant fetuin produced by insect cells has been used for studies on the inhibitory effect of human fetuin on insulin-induced autophosphorylation of the insulin receptor.15,8,14 Relatively recently, FLAG-tagged human fetuin synthesized in HEK-293T cells has been used to define the mechanism by which fetuin modulates cellular adhesion.47 With respect to our findings on rhFet, we propose that any future functional study performed with fetuin produced in HEK-293 cells (or any other expression system) should be critically evaluated, given the distinct structural differences demonstrated here in between hFet and rhFet.
Acknowledgments
We acknowledge support from The Netherlands Organization for Scientific Research (NWO) funding the large-scale proteomics facility Proteins@Work (Project 184.032.201) embedded in The Netherlands Proteomics Centre. A.J.R.H. acknowledges further support by the NWO TOP-Punt Grant 718.015.003. This project has received additional funding from the European Union’s Horizon 2020 research and innovation programme under Grant agreement 686547 (MSMed).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00318.
Annotated HCD/EThcD spectra of all glycopeptides observed for hFet (PDF)
Amino acid sequence alignment between hFet and bFet; native MS spectra of hFet treated by PNGaseF, sialidase, and alkaline phosphatase; schematic of the two-polypeptide-chain structure of hFet; LC-MS/MS spectra of Lys-C peptides from rhFet; and comparison of the intact protein native MS spectrum with the constructed spectrum based on the peptide-centric proteomics data in hFet and bFet (PDF)
List of all modified peptide isoforms on hFet, bFet, and rhFet and their relative quantification (XLSX)
List containing identified hFet and bFet proteoforms (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Bürgi W.; Schmid K. Preparation and Properties of Zn-Alpha 2-Glycoprotein of Normal Human Plasma. J. Biol. Chem. 1961, 236 (4), 1066–1074. [PubMed] [Google Scholar]
- Heremans J. F.Les Globulines Du Système Gamma Du Plasma Humain; Editions Arscia S.A. Paris: Brussels, 1961; pp 119–138; 10.5169/seals-307477. [DOI] [Google Scholar]
- Pedersen K. O. Fetuin, a New Globulin Isolated from Serum. Nature 1944, 154, 575. 10.1038/154575a0. [DOI] [Google Scholar]
- Dziegielewska K. M.; Brown W. M.; Casey S. J.; Christie D. L.; Foreman R. C.; Hill R. M.; Saunders N. R. The Complete cDNA and Amino Acid Sequence of Bovine Fetuin. Its homology with alpha 2HS glycoprotein and relation to other members of the cystatin superfamily. J. Biol. Chem. 1990, 265 (6), 4354–4357. [PubMed] [Google Scholar]
- Brown W. M.; Dziegielewska K. M.; Saunders N. R.; Christie D. L.; Nawratil P.; Müller-Esterl W. The Nucleotide and Deduced Amino Acid Structures of Sheep and Pig Fetuin: Common Structural Features of the Mammalian Fetuin Family. Eur. J. Biochem. 1992, 205 (1), 321–331. 10.1111/j.1432-1033.1992.tb16783.x. [DOI] [PubMed] [Google Scholar]
- Schinke T.; Amendt C.; Trindl A.; Pöschke O.; Müller-Esterl W.; Jahnen-Dechent W. The Serum Protein Alpha2-HS Glycoprotein/Fetuin Inhibits Apatite Formation in Vitro and in Mineralizing Calvaria Cells. J. Biol. Chem. 1996, 271 (34), 20789–20796. 10.1074/jbc.271.34.20789. [DOI] [PubMed] [Google Scholar]
- Reynolds J. L. Multifunctional Roles for Serum Protein Fetuin-A in Inhibition of Human Vascular Smooth Muscle Cell Calcification. J. Am. Soc. Nephrol. 2005, 16 (10), 2920–2930. 10.1681/ASN.2004100895. [DOI] [PubMed] [Google Scholar]
- Mathews S. T.; Chellam N.; Srinivas P. R.; Cintron V. J.; Leon M. A.; Goustin A. S.; Grunberger G. Alpha 2-HSG, a Specific Inhibitor of Insulin Receptor Autophosphorylation, Interacts with the Insulin Receptor. Mol. Cell. Endocrinol. 2000, 164 (1–2), 87–98. 10.1016/S0303-7207(00)00237-9. [DOI] [PubMed] [Google Scholar]
- Janapatla R. P.; Hsu M-H.; Liao W.-T.; Chien K.-Y.; Lee H.-Y.; Chiu C.-H. Low Serum Fetuin-A as a Biomarker to Predict Pneumococcal Necrotizing Pneumonia and Hemolytic Uremic Syndrome in Children. Medicine (Philadelphia, PA, U. S.) 2016, 95 (13), e3221. 10.1097/MD.0000000000003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin G. A.; McEvoy L. K.; Barrett-Connor E.; Daniels L. B.; Ix J. H. Fetuin-A, a New Vascular Biomarker of Cognitive Decline in Older Adults. Clin. Endocrinol. 2014, 81 (1), 134–140. 10.1111/cen.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K.; Bartz T. M.; Mukamal K. J.; Djoussé L.; Kizer J. R.; Tracy R. P.; Zieman S. J.; Rimm E. B.; Siscovick D. S.; Shlipak M.; et al. Fetuin-A, Type 2 Diabetes, and Risk of Cardiovascular Disease in Older Adults: The Cardiovascular Health Study. Diabetes Care 2013, 36 (5), 1222–1228. 10.2337/dc12-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan N.; Fritsche A.; Weikert C.; Boeing H.; Joost H.-G.; Häring H.-U.; Schulze M. B. Plasma Fetuin-A Levels and the Risk of Type 2 Diabetes. Diabetes 2008, 57 (10), 2762–2767. 10.2337/db08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K. N.; Teran-Garcia M. From Infancy to Aging: Biological and Behavioral Modifiers of Fetuin-A. Biochimie 2016, 124, 141–149. 10.1016/j.biochi.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Srinivas P. R.; Goustin A. S.; Grunberger G. Baculoviral Expression of a Natural Inhibitor of the Human Insulin Receptor Tyrosine Kinase. Biochem. Biophys. Res. Commun. 1995, 208 (2), 879–885. 10.1006/bbrc.1995.1417. [DOI] [PubMed] [Google Scholar]
- Srinivas P. R.; Deutsch D. D.; Mathews S. T.; Goustin A. S.; Leon M. A.; Grunberger G. Recombinant Human Alpha 2-HS Glycoprotein Inhibits Insulin-Stimulated Mitogenic Pathway without Affecting Metabolic Signalling in Chinese Hamster Ovary Cells Overexpressing the Human Insulin Receptor. Cell. Signalling 1996, 8 (8), 567–573. 10.1016/S0898-6568(96)00110-6. [DOI] [PubMed] [Google Scholar]
- Wilson N. L.; Schulz B. L.; Karlsson N. G.; Packer N. H. Sequential Analysis of N- and O-Linked Glycosylation of 2D-PAGE Separated Glycoproteins. J. Proteome Res. 2002, 1 (6), 521–529. 10.1021/pr025538d. [DOI] [PubMed] [Google Scholar]
- Arnold J. N.; Wormald M. R.; Sim R. B.; Rudd P. M.; Dwek R. A. The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annu. Rev. Immunol. 2007, 25 (1), 21–50. 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- Chalabi S.; Panico M.; Sutton-Smith M.; Haslam S. M.; Patankar M. S.; Lattanzio F. A.; Morris H. R.; Clark G. F.; Dell A. Differential O-Glycosylation of a Conserved Domain Expressed in Murine and Human ZP3. Biochemistry 2006, 45 (2), 637–647. 10.1021/bi0512804. [DOI] [PubMed] [Google Scholar]
- Rudd P. M.; Dwek R. A. Glycosylation: Heterogeneity and the 3D Structure of Proteins. Crit. Rev. Biochem. Mol. Biol. 1997, 32 (1), 1–100. 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- Windwarder M.; Altmann F. Site-Specific Analysis of the O-Glycosylation of Bovine Fetuin by Electron-Transfer Dissociation Mass Spectrometry. J. Proteomics 2014, 108, 258–268. 10.1016/j.jprot.2014.05.022. [DOI] [PubMed] [Google Scholar]
- Carr S. A.; Huddleston M. J.; Bean M. F. Selective Identification and Differentiation of N- and O-Linked Oligosaccharides in Glycoproteins by Liquid Chromatography-Mass Spectrometry. Protein Sci. 1993, 2 (2), 183–196. 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann J.; Haupt H.; Auerswald E. A.; Müller-Ester W. The Arrangement of Disulfide Loops in Human Alpha 2-HS Glycoprotein. J. Biol. Chem. 1989, 264 (24), 14121–14128. [PubMed] [Google Scholar]
- Spiro R. G. Protein Glycosylation: Nature, Distribution, Enzymatic Formation, and Disease Implications of Glycopeptide Bonds. Glycobiology 2002, 12 (4), 43R–56R. 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- Seo J.; Lee K.-J. Post-Translational Modifications and Their Biological Functions: Proteomic Analysis and Systematic Approaches. J. Biochem. Mol. Biol. 2004, 37 (1), 35–44. 10.5483/BMBRep.2004.37.1.035. [DOI] [PubMed] [Google Scholar]
- Huang L. J.; Lin J. H.; Tsai J. H.; Chu Y. Y.; Chen Y. W.; Chen S. L.; Chen S. H. Identification of Protein O-Glycosylation Site and Corresponding Glycans Using Liquid Chromatography-Tandem Mass Spectrometry via Mapping Accurate Mass and Retention Time Shift. J. Chromatogr. A 2014, 1371, 136–145. 10.1016/j.chroma.2014.10.046. [DOI] [PubMed] [Google Scholar]
- Haglund A. C.; Ek B.; Ek P. Phosphorylation of Human Plasma Alpha2-Heremans-Schmid Glycoprotein (Human Fetuin) in Vivo. Biochem. J. 2001, 357 (2), 437–445. 10.1042/bj3570437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M. Biochimica et Biophysica Acta N-Linked Protein Glycosylation in the ER ☆. Biochim. Biophys. Acta, Mol. Cell Res. 2013, 1833 (11), 2430–2437. 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Dell A.; Galadari A.; Sastre F.; Hitchen P. Similarities and Differences in the Glycosylation Mechanisms in Prokaryotes and Eukaryotes. Int. J. Microbiol. 2010, 2010, 148178. 10.1155/2010/148178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Liu F.; Franc V.; Halim L. A.; Schellekens H.; Heck A. J. R. Hybrid Mass Spectrometry Approaches in Glycoprotein Analysis and Their Usage in Scoring Biosimilarity. Nat. Commun. 2016, 7, 13397. 10.1038/ncomms13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc V.; Yang Y.; Heck A. J. R. Proteoform Profile Mapping of the Human Serum Complement Component C9 Revealing Unexpected New Features of N-, O-, and C-Glycosylation. Anal. Chem. 2017, 89 (6), 3483–3491. 10.1021/acs.analchem.6b04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc V.; Zhu J.; Heck A. J. R. Comprehensive Proteoform Characterization of Plasma Complement Component C8αβγ by Hybrid Mass Spectrometry Approaches. J. Am. Soc. Mass Spectrom. 2018, 29 (6), 1099–1110. 10.1007/s13361-018-1901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati S.; Yang Y.; Barendregt A.; Heck A. J. R. Detailed Mass Analysis of Structural Heterogeneity in Monoclonal Antibodies Using Native Mass Spectrometry. Nat. Protoc. 2014, 9 (4), 967–976. 10.1038/nprot.2014.057. [DOI] [PubMed] [Google Scholar]
- Rose R. J.; Damoc E.; Denisov E.; Makarov A.; Heck A. J. R. High-Sensitivity Orbitrap Mass Analysis of Intact Macromolecular Assemblies. Nat. Methods 2012, 9 (11), 1084–1086. 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- Bern M.; Caval T.; Kil Y. J.; Tang W.; Becker C.; Carlson E.; Kletter D.; Sen K. I.; Galy N.; Hagemans D.; et al. Parsimonious Charge Deconvolution for Native Mass Spectrometry. J. Proteome Res. 2018, 17 (3), 1216–1226. 10.1021/acs.jproteome.7b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Richards J. C.. Functional Glycomics and Glycobiology: An Overview. In Functional Glycomics: Methods and Protocols; Li J., Ed.; Humana Press: Totowa, NJ, 2010; pp 1–8. [DOI] [PubMed] [Google Scholar]
- Kussmann M.; Nordhoff E.; Rahbek-Nielsen H.; Haebel S.; Rossel-Larsen M.; Jakobsen L.; Gobom J.; Mirgorodskaya E.; Kroll-Kristensen A.; Palm L.; et al. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Sample Preparation Techniques Designed for Various Peptide and Protein Analytes. J. Mass Spectrom. 1997, 32 (6), 593–601. . [DOI] [Google Scholar]
- Bern M.; Kil Y. J.; Becker C.. Byonic: Advanced Peptide and Protein Identification Software. Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, 2012; Unit 13.20; 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Yuan J.; Wang Z.; Huang L. Separation of One-Pot Procedure Released O-Glycans as 1-Phenyl-3-Methyl-5-Pyrazolone Derivatives by Hydrophilic Interaction and Reversed-Phase Liquid Chromatography followed by Identification Using Electrospray Mass Spectrometry and Tandem Mass Spectrometry. J. Chromatogr. A 2013, 1274, 107–117. 10.1016/j.chroma.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Hayase T.; Rice K. G.; Dziegielewska K. M.; Kuhlenschmidt M.; Reilly T.; Lee Y. C. Comparison of N-Glycosides of Fetuins from Different Species and Human Alpha 2-HS-Glycoprotein. Biochemistry 1992, 31 (20), 4915–4921. 10.1021/bi00135a024. [DOI] [PubMed] [Google Scholar]
- Edge A. S.; Spiro R. G. Presence of an O-Glycosidically Linked Hexasaccharide in Fetuin. J. Biol. Chem. 1987, 262 (33), 16135–16141. [PubMed] [Google Scholar]
- Hoffmann M.; Marx K.; Reichl U.; Wuhrer M.; Rapp E. Site-Specific O -Glycosylation Analysis of Human Blood Plasma Proteins. Mol. Cell. Proteomics 2016, 15 (2), 624–641. 10.1074/mcp.M115.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A.; Rüetschi U.; Larson G.; Nilsson J. LC-MS/MS Characterization of O-Glycosylation Sites and Glycan Structures of Human Cerebrospinal Fluid Glycoproteins. J. Proteome Res. 2013, 12 (2), 573–584. 10.1021/pr300963h. [DOI] [PubMed] [Google Scholar]
- Halim A.; Nilsson J.; Rüetschi U.; Hesse C.; Larson G. Human Urinary Glycoproteomics; Attachment Site Specific Analysis of N - and O -Linked Glycosylations by CID and ECD. Mol. Cell. Proteomics 2012, 11 (4), M111.013649. 10.1074/mcp.M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M.; Dziegielewska K. M.; Saunders N. R.; Møsllgård K. Fetuin–an Old Friend Revisited. BioEssays 1992, 14 (11), 749–755. 10.1002/bies.950141105. [DOI] [PubMed] [Google Scholar]
- Elzanowski A.; Barker W. C.; Hunt L. T.; Seibel-Ross E. Cystatin Domains in Alpha-2-HS-Glycoprotein and Fetuin. FEBS Lett. 1988, 227 (2), 167–170. 10.1016/0014-5793(88)80890-1. [DOI] [PubMed] [Google Scholar]
- Araki T.; Yoshioka Y.; Schmid K. The Position of the Disulfide Bonds in Human Plasma Alpha 2 HS-Glycoprotein and the Repeating Double Disulfide Bonds in the Domain Structure. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1989, 994 (3), 195–199. 10.1016/0167-4838(89)90293-8. [DOI] [PubMed] [Google Scholar]
- Nangami G.; Koumangoye R.; Shawn Goodwin J.; Sakwe A. M.; Marshall D.; Higginbotham J.; Ochieng J. Fetuin-A Associates with Histones Intracellularly and Shuttles Them to Exosomes to Promote Focal Adhesion Assembly Resulting in Rapid Adhesion and Spreading in Breast Carcinoma Cells. Exp. Cell Res. 2014, 328 (2), 388–400. 10.1016/j.yexcr.2014.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.