Abstract
Background
The C-terminal 42 kDa region of merozoite surface protein-1 of Plasmodium falciparum (PfMSP-142) is the target of an immune response. It has been recognised as one of the promising candidate antigens for a blood-stage malaria vaccine. Genetic structure of PfMSP-142 has been considered to be largely conserved in the P. falciparum population. However, only limited information is currently available. This study aimed to analyse genetic diversity and the effect of natural selection on PfMSP-142 among the Myanmar P. falciparum population and compare them with publicly available PfMSP-142 from global P. falciparum populations.
Methods
A total of 69 P. falciparum clinical isolates collected from Myanmar malaria patients in Upper Myanmar in 2015 were used. The PfMSP-142 region was amplified by polymerase chain reaction, cloned and sequenced. Genetic structure and natural selection of this region were analysed using MEGA4 and DnaSP programs. Polymorphic nature and natural selection in global PfMSP-142 were also investigated.
Results
All three allele types (MAD20, K1, and RO33) of PfMSP-142 were identified in Myanmar isolates of P. falciparum. Myanmar PfMSP-142 displayed genetic diversity. Most polymorphisms were scattered in blocks 16 and 17. Polymorphisms observed in Myanmar PfMSP-142 showed a similar pattern to those of global PfMSP-142; however, they were not identical to each other. Genetic diversity of Myanmar PfMSP-142 was relatively lower than that of PfMSP-142 from different geographical regions. Evidence of natural selection and recombination were found. Comparative analysis of genetic polymorphism and natural selection in the global PfMSP-142 population suggested that this region was not tightly conserved in global PfMSP-142 as previously thought and is under the complicated influence of natural selection and recombination.
Conclusions
Global PfMSP-142 revealed limited, but non-negligible, genetic diversity by allele types and geographical origins. Complicated natural selection and potential recombination might have occurred in global PfMSP-142. Comprehensive monitoring of genetic diversity for global PfMSP-142 would be needed to better understand the polymorphic nature and evolutionary aspect of PfMSP-142 in the global P. falciparum population. More thought would be necessary for designing a vaccine based on PfMSP-142.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-3027-x) contains supplementary material, which is available to authorized users.
Keywords: Plasmodium falciparum, PfMSP-142, Myanmar, Polymorphism, Natural selection
Background
Malaria is one of the most important health burdens worldwide, especially in tropical and subtropical regions. It results in approximately 216 million clinical cases and about 450,000 deaths annually [1]. Despite many decades of intense study and efforts to control or eliminate malaria, it remains a significant global health problem. Currently, there is no commercially available malaria vaccine. Therefore, developing an effective vaccine is urgently needed to combat this disease. Among human-infecting malaria parasites, Plasmodium falciparum is the most medically important malaria parasite that is responsible for most malaria-related deaths globally [1]. However, due to the complexity of this parasite’s life-cycle and genetic variations of major vaccine candidate antigens, developing an effective malaria vaccine is still a difficult challenge.
Merozoite surface protein-1 (MSP-1) is a 195 kDa major surface glycoprotein of merozoites. This protein binds to human erythrocytes in a sialic acid-dependent manner. It plays a pivotal role in erythrocyte invasion by merozoites [2, 3]. MSP-1 is synthesised as a large precursor form which subsequently undergoes two independent proteolytic cleavage events. First, the large precursor protein is cleaved into four polypeptides (83, 30, 38 and 42 kDa) that form non-covalently associated complex. The 42 kDa fragment (MSP-142) is further cleaved into 33 kDa (MSP-133) and 19 kDa (MSP-119) fragments when merozoites invades erythrocyte [3–5]. Only MSP-119 remains on the merozoite surface and enters into erythrocyte whereas the others are released from the merozoite [6]. MSP-1 is highly immunogenic. Species-specific natural immune responses against this antigen have been reported in patients naturally exposed to P. falciparum and P. vivax [7–10]. In particular, the C-terminal region of MSP-1, MSP-119 or MSP-142, is of particular interest since individuals naturally infected with malaria acquire humoral immune responses against this domain [11–15]. Antibodies that recognise the C-terminal 42 kDa region of P. falciparum MSP-1 (PfMSP-142) can inhibit the growth of the parasite or invasion of the merozoite into host erythrocytes [16–19]. Antibodies that react to PfMSP-142 are associated with protection against P. falciparum infection in endemic settings [20, 21]. Both PfMSP-119 and PfMSP-142 have shown potential as subunit vaccines in animal models by conferring protection against infections [17, 22–25]. These results suggest that PfMSP-142 is a promising candidate antigen for blood stage vaccine development.
Similar to other P. falciparum antigens, PfMSP-1 also shows polymorphism due to different mechanisms, including single nucleotide polymorphisms (SNPs), insertion/deletion of tandem repeats, and meiotic recombination [26, 27]. PfMSP-1 is divided into 17 distinct blocks that encode conserved, semiconserved, or variable regions of the protein [26, 27]. Among the variable regions, block 2 is the most polymorphic region in which repeat length polymorphisms are commonly identified. Size polymorphisms of this region between alleles enable this region as a molecular marker for parasite genotyping [28]. Therefore, the polymorphic nature of PfMSP-1 block 2 in global isolates has been extensively studied [29–37]. Meanwhile, PfMSP-142 has been considered to be broadly conserved in different alleles of P. falciparum [26, 27]. However, only limited information for genetic diversity for this region is currently available. Genetic diversity analysis of MSP-142 in small numbers of P. falciparum and P. vivax isolates suggests natural selection may be acting on the area and natural selection is an important factor in the maintenance and generation of genetic polymorphism observed in MSP-142 [38]. Considering that PfMSP-142 is a leading candidate for vaccine development, in-depth understanding of the polymorphic nature of this region would be beneficial to design optimised vaccine formulation. In this study, genetic diversity and natural selection of PfMSP-142 in the Myanmar P. falciparum population were analysed. Data from this study were also compared with PfMSP-142 sequences available for P. falciparum isolates from other malaria-endemic countries to gain insights into genetic polymorphism and natural selection of PfMSP-142 in the global population(s).
Methods
Study areas and blood samples
A total of 69 blood samples collected from Plasmodium falciparum-infected Myanmar malaria patients were used. The patients enrolled in the study attended the public health centres in towns and villages of Naung Cho, Pyin Oo Lwin and Tha Beik Kyin townships in Upper Myanmar in 2015 with clinical symptoms of malaria. Malaria transmission in these areas was heterogeneous and seasonal with most clinical cases reported during the rainy season. Plasmodium falciparum infection was confirmed by Giemsa-stained thick and thin blood smear examination. Finger-prick blood samples were collected from the P. falciparum-infected patients before drug treatment. Plasmodium falciparum infection was further confirmed by polymerase chain reaction (PCR) targeting 18S ribosomal RNA (rRNA) gene [39, 40]. The mean age of patients who donated the blood samples was 32.7 years-old and ranged between 13–57 years. Written consent was obtained from each patient before blood collection.
Amplification and sequencing analysis of Myanmar PfMSP-142
Parasite genomic DNA was extracted from the blood filters using a QIAamp Blood Kit (Qiagen, Valencia, CA, USA). A nested PCR amplified the full-length PfMSP-142 region flanking nucleotide positions from 3915 to 5163 in 3D7 sequence (XM_001352134) (Fig. 1). The oligonucleotide primers for the first round PCR were 5'-GAA ACT GTA TAA TAT TAA CAT GAG-3' and 5'-GAT AAT GAG GAA TAT TTA GAT CAA-3'. The primers for nested PCR were 5'-TTA CCC ATT TTT GGA GAA TCC GA-3' and 5'-TTA AGG TAA CAT ATT TTA ACT CCT-3'. The cycling condition for both amplification reactions was 95 °C for 10 min, followed by 30 cycles for 95 °C for 1 min, 53 °C for 1 min, and 72 °C for 1.5 min, and one cycle of the final extension at 72 °C for 10 min. Ex Taq DNA polymerase (Takara, Otsu, Japan) was used in all PCR steps to minimize the nucleotide misincorporation during PCR amplification. Each PCR product was analysed on 1.2% agarose gel, purified from the gel, and then ligated into T&A cloning vector (Real Biotech Corporation, Banqiao City, Taiwan). Each ligation mixture was transformed into Escherichia coli DH5α competent cells, and positive clones with the appropriate insert were selected by colony PCR with the nested primers. The cloned gene sequences were analysed by automatic nucleotide sequencing. To verify the sequences, at least two clones from each isolate were sequenced in both directions. Some isolates underwent three or four-fold sequence coverage to confirm the existence of rare polymorphisms. The nucleotide and deduced amino acid sequences of PfMSP-142 were analysed using EditSeq and SeqMan in the DNASTAR package (DNASTAR, Madison, WI, USA). The nucleotide sequences obtained in this study have been deposited in the GenBank database under the accession numbers MF943252-MF943320.
Fig. 1.
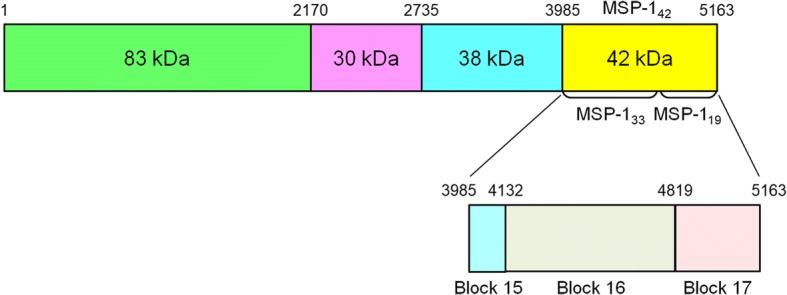
Schematic structure of PfMSP1. PfMSP1 is a large precursor protein that cleaved into four polypeptides (83, 30, 38 and 42 kDa). The 42 kDa fragment (MSP-142) is further cleaved into 33 kDa (MSP-133) and 19 kDa (MSP-119) fragments when merozoite invades erythrocyte. In this study, the full-length PfMSP-142 fragment flanking nucleotide positions from 3915 to 5163 in 3D7 sequence (XM_001352134) was amplified by a nested PCR. The numbers of nucleotide indicate coordinates according to the 3D7 sequence (XM_001352134)
Sequence analysis and neutrality test
Nucleotide sequence polymorphism analysis was performed on the 69 Myanmar PfMSP-142 sequences. The number of segregating sites (S), haplotypes (H), haplotype diversity (Hd), nucleotide diversity (π), and the average number of pairwise nucleotide differences within the population (K) were estimated using the DnaSP ver. 5.10.00 [41]. The π value was calculated to estimate the stepwise diversity throughout the entire PfMSP-142 based on a sliding window of 100 bases with a step size of 25 bp. The rates of synonymous (dS) and non-synonymous (dN) substitutions were estimated and were compared with the Z test (P < 0.05) in MEGA4 program [42] using Nei & Gojobori’s method [43] with the Jukes-Cantor correction. To evaluate the neutral theory of evolution, the Tajima’s D [44] value and Fu and Li’s D and F statistics [45] were analysed using the DnaSP ver. 5.10.00 [41]. Positive values for Nei-Gojobori (dN-dS), Tajima’s D, and the Fu and Li’s D and F tests correspond to positive natural selection, whereas negative values correspond to negative or purifying selection.
Population diversity of PfMSP-142 among global P. falciparum isolates
Population diversity of PfMSP-142 in global P. falciparum isolates was analysed. The PfMSP-142 sequences from Thailand (n = 74, AB276006-AB276018, AB502546-AB502586 and D13343-D13363), Philippines (n = 43, AB502587-AB502628 and AB715434), Papua New Guinea (PNG: n = 77, AB502629-AB502704 and X05624), Solomon Islands (n = 41, AB502705-AB502745), Vanuatu (n = 91, AB715435-AB715519 and AB116596-AB116601), Ghana (n = 33, AB502514-AB502545 and AB276005), and Tanzania (n = 71 , AB502443-AB502513) were included in this study to analyse genetic diversity in global PfMSP-142. Nucleotide diversity and the effect of natural selection for global PfMSP-142 were analysed with the same methods described above. Recombination parameter (R), which included the effective population size and probability of recombination between adjacent nucleotides per generation, and minimum number of recombination events (Rm) were analysed by DnaSP ver. 5.10.00 [41].
Haplotype network analysis
To investigate relationships among PfMSP-142 haplotypes, the haplotype network for a total of 499 global PfMSP-142 sequences including 69 Myanmar sequences and the 430 publicly available sequences of global isolates deposited in the GenBank database listed above was constructed using the program NETWORK version 5.0.0.3 with the Median Joining algorithm [46].
Results
Sequence polymorphism in Myanmar PfMSP-142
Sequence analysis of 69 Myanmar PfMSP-142 showed three different allele types (MAD20, K1, and RO33) in the Myanmar P. falciparum population. The MAD20 allele type was the most prevalent (37/69, 53.6%), followed by K1 (18/69, 26.1%), and RO33 (14/69, 20.3%) allele types. Sequence analysis of MAD20 allele types revealed a total of 21 distinct haplotypes in Myanmar sequences (Additional file 1: Figure S1). When compared to MAD20 (X05624) reference sequence, 19 SNPs were identified in MAD20 allele of Myanmar PfMSP-142, including five synonymous and 14 non-synonymous SNPs. These non-synonymous SNPs caused di-morphic amino acid changes at 14 different positions. Most amino acid changes were found in block 16 and block 17 regions: seven amino acid changes (E105G, F183Y, D220N, F242L, T245H, S269N and Q290P) in block 16 and six amino acid changes (E307Q, T341A, T354K, S363N, R364G and L379F) in block 17. Meanwhile, only one amino acid change (I54F) was identified in block 15. These results suggest that blocks 16 and 17 show polymorphic characters in MAD20 allele types of Myanmar PfMSP-142, which might contribute to inner allele diversity. For K1 allele types, a total of six distinct haplotypes were identified in Myanmar PfMSP-142 sequences. Three synonymous and four non-synonymous SNPs were observed in these sequences when compared to K1 (X03371) reference sequence (Additional file 2: Figure S2). These four non-synonymous SNPs resulted in four di-morphic amino acid changes: E21G in block 15, R116G and H249R in block 16 and Q297E in block 17. For RO33 allele type, six different haplotypes were identified in Myanmar PfMSP-142. When these sequences were compared to RO33 (Z35326) reference sequence, one tri-morphic (P244H/T) and eight di-morphic amino acid changes (F54I, L204I, D219N, Y245N, Q306E, K353T, N362S and G363R) were found in RO33 allele types of 14 Myanmar PfMSP-142 sequences (Additional file 3: Figure S3). All amino acid changes except one (F54I) were observed in blocks 16 and 17. Collectively, sequence analysis of 69 Myanmar PfMSP-142 indicated that most amino acid changes were mainly distributed in blocks 16 and 17 in all three allele types of Myanmar PfMSP-142. All cysteine residues in block 17 were tightly conserved in all allele types of Myanmar PfMSP-142. No insertion or deletion was detected in these sequences. Among 27 amino acid changes identified in these three allele types of Myanmar PfMSP-142, 18 have been previously reported. However, the other nine substitutions (E105G, F242L, S269N, Q290P and T341A in MAD20 allele types; E21G, R116G and H249R in K1 allele types; L204I in RO33 allele types) were novel ones that have not been reported hitherto.
Comparative analysis of amino acid polymorphisms between Myanmar PfMSP-142 and global PfMSP-142
Overall patterns of amino acid polymorphisms identified in each allele type (MAD20, K1 and RO33) of Myanmar PfMSP-142 were similar to those of global PfMSP-142. However, they differed slightly from each other. MAD20 allele types of Myanmar PfMSP-142 showed a different pattern of amino acid polymorphism compared to those from other geographical regions (Fig. 2). Compared to MAD20 (X05624) sequence, a total of 22 amino acid polymorphisms were identified in global PfMSP-142. Their frequencies and distributions differed by country. Most amino acid polymorphisms in Myanmar and global PfMSP-142 were mainly found in blocks 16 and 17. Five amino acid changes (I54F, E307Q, T354K, S363N and R364G) were commonly identified in all global PfMSP-142 analysed in this study, although their frequencies were different by country or geographic area. I54F, the only amino acid change identified in block 15, was more prevalent in African (Ghana and Tanzania) PfMSP-142 than that in other origins. Frequencies of F183Y and T245P were relatively high in African PfMSP-142, but they were low or not detected in Asian (Myanmar, Thailand and Philippines) or the Pacific (PNG, Solomon Islands, and Vanuatu) PfMSP-142. D220N was not identified or very rarely identified in Pacific PfMSP-142.
Fig. 2.
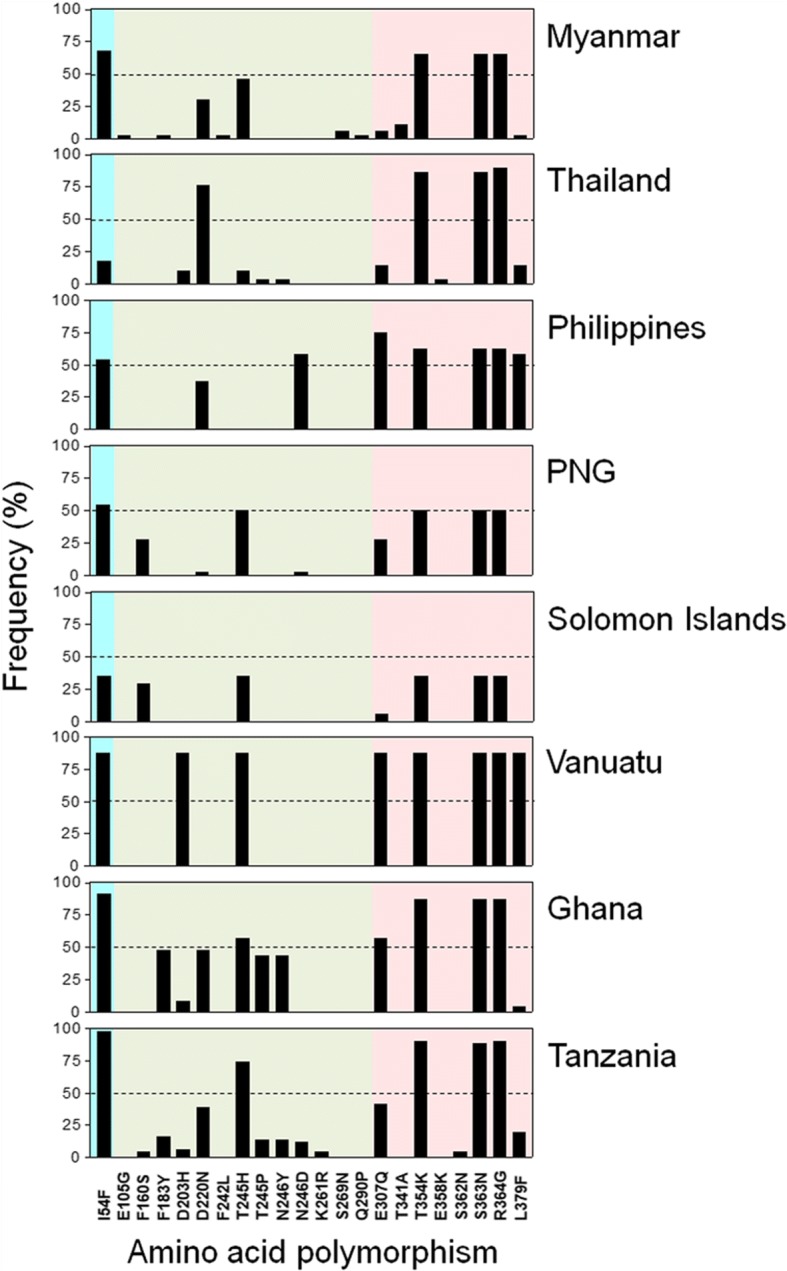
Comparison of amino acid polymorphisms in MAD20 allele types of PfMSP-142 among global Plasmodium falciparum isolates. Positions and frequencies of amino acid changes found in MAD20 allele types of PfMSP-142 in global P. falciparum isolates are compared. Blocks are indicated by different colors: block 15 (sky blue), block 16 (green), block 17 (pink). Abbreviation: PNG, Papua New Guinea
In the case of K1 allele type, only Thailand sequences have been reported so far. Overall amino acid change patterns of K1 allele types of PfMSP-142 from Myanmar and Thailand were not similar (Fig. 3). E21G, R116G and H249R were identified only in Myanmar PfMSP-142, although frequencies of R116G and H249R were low. Meanwhile, D322S was found just in Thailand PfMSP-142. Q297E was identified in both Myanmar and Thailand PfMSP-142, but its frequency was higher in Myanmar (94.4%) than in Thailand (25.0%).
Fig. 3.
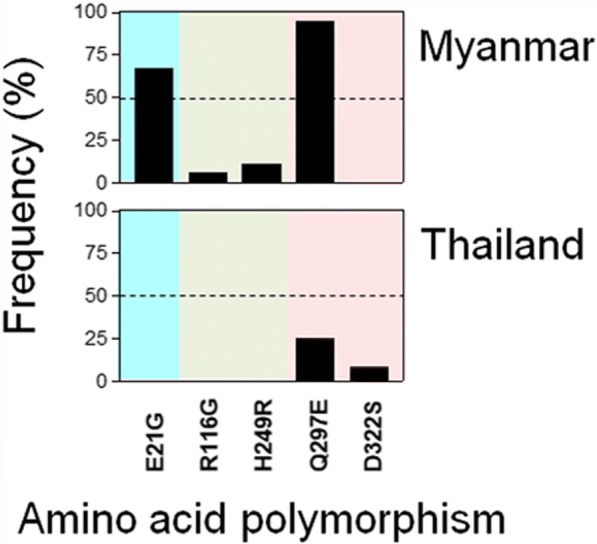
Comparison of amino acid polymorphisms in K1 allele types of PfMSP-142 among global Plasmodium falciparum isolates. Positions and frequencies of amino acid changes found in K1 allele types of PfMSP-142 in Myanmar and Thailand P. falciparum isolates are compared. Blocks are indicated by different colors: block 15 (sky blue), block 16 (green), block 17 (pink)
The pattern of amino acid polymorphisms in RO33 allele types of global PfMSP-142 was also analysed (Fig. 4). A total of 18 amino acid polymorphisms were identified in global PfMSP-142. The most notable amino acid changes that differed among global RO33 allele types of PfMSP-142 were F54I, P244T, Y245N and Q306E. These amino acid changes were detected in all Asian and Pacific PfMSP-142 analysed in this study with high frequencies up to 78%. However, they were less frequent in African PfMSP-142. The three amino acid changes, K353T, N362S and G363R, were also detected with high frequencies in Asian and Pacific PfMSP-142, but not in African PfMSP-142. Unlike other Asian or Pacific PfMSP-142, Myanmar PfMSP-142 showed a lower level of polymorphisms for these three amino acids. Meanwhile, African PfMSP-142 had a higher frequency of amino acid polymorphisms at P244H compared to Asian or Pacific PfMSP-142. D202H and S361N were only identified in African PfMSP-142, although their frequencies were not high.
Fig. 4.
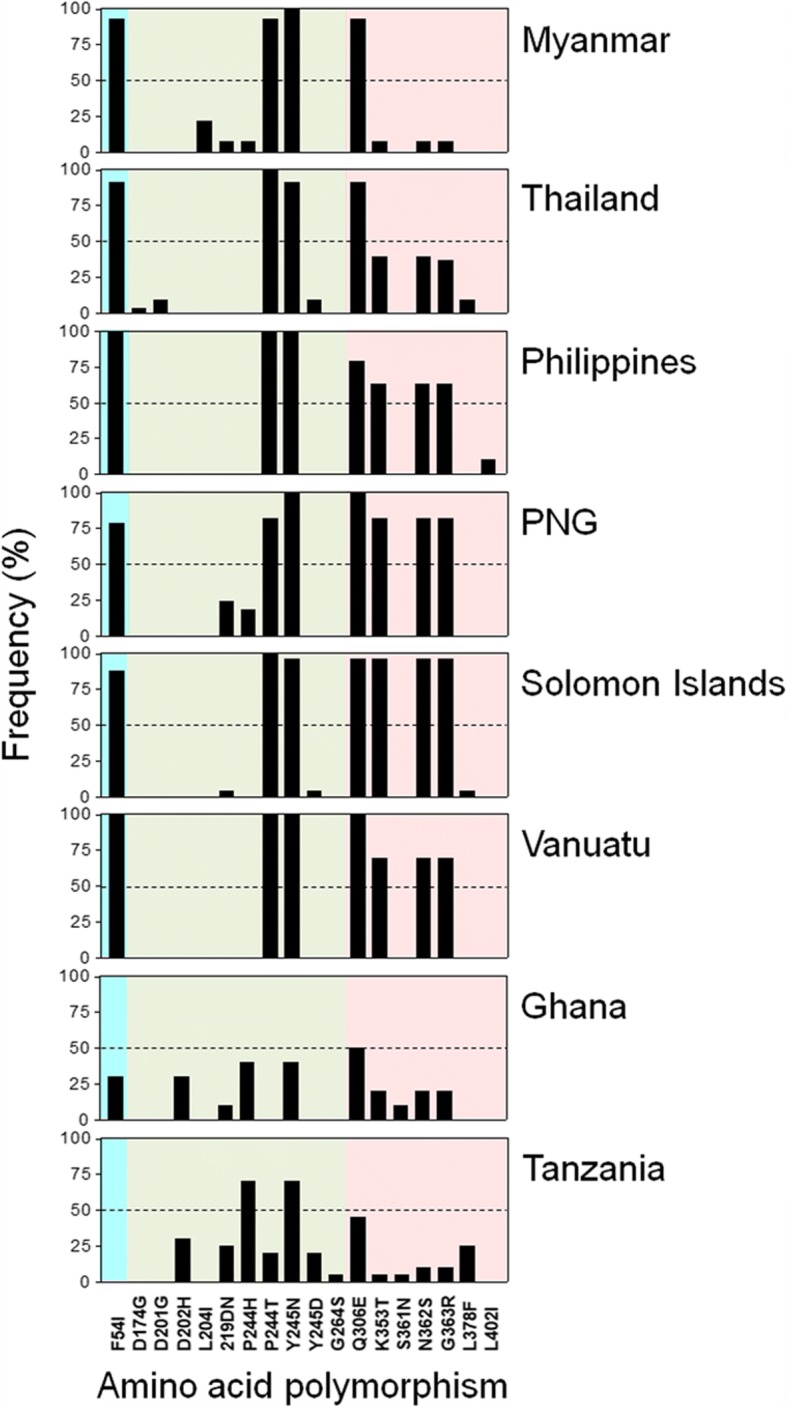
Comparison of amino acid polymorphisms in RO33 allele types of PfMSP-142 among global Plasmodium falciparum isolates. Positions and frequencies of amino acid changes found in RO33 allele types of PfMSP-142 in global P. falciparum isolates are compared. Blocks are indicated by different colors: block 15 (sky blue), block 16 (green), block 17 (pink). Abbreviation: PNG, Papua New Guinea
Nucleotide diversity and natural selection of Myanmar PfMSP-142
Nucleotide sequence analysis was performed to determine nucleotide diversity and genetic differentiation of Myanmar PfMSP-142. The K-values for MAD20, K1 and RO33 allele types of Myanmar PfMSP-142 were 4.300, 1.431 and 1.506, respectively, with the highest K-value at block 16 for all three allele types (Table 1). In MAD20 allele types, overall haplotype diversity (Hd) and nucleotide diversity (π) for 37 Myanmar PfMSP-142 sequences were 0.902 ± 0.040 and 0.00351 ± 0.00033, respectively. The π value for each block was 0.00313 ± 0.00049 for block 15, 0.00272 ± 0.00035 for block 16 and 0.00533 ± 0.00062 for block 17. The Hd for 18 K1 allele types of Myanmar PfMSP-142 was 0.765 ± 0.080, and the π was 0.00120 ± 0.00026. When analysing the π value for each block, the highest value was predicted at block 15 (0.00214 ± 0.00037).
Table 1.
DNA sequence polymorphism and test of neutrality for Myanmar PfMSP-142
| Allele type/ Fragment | Segregating sites | Singleton variable sites | Parsimony informative sites | Total no. of mutations | K | H | Hd ± SD | π ± SD | dN-dS | Tajima’s D (P) | Fu & Li’s D (P) | Fu & Li’s F (P) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAD20 | ||||||||||||
| Block 15 | 2 | 0 | 2 | 2 | 0.604 | 4 | 0.536 ± 0.066 | 0.00313 ± 0.00049 | 0.50117 (P>0.10) |
0.78020 (P>0.10) |
0.81005 (P>0.10) |
|
| Block 16 | 10 | 6 | 4 | 10 | 1.880 | 8 | 0.769 ± 0.043 | 0.00272 ± 0.00035 | -0.65200 (P>0.10) |
-2.01692 (P>0.10) |
-1.86022 (P>0.10) |
|
| Block 17 | 7 | 2 | 5 | 7 | 1.817 | 6 | 0.632 ± 0.072 | 0.00533 ± 0.00062 | 0.23510 (P>0.10) |
-0.20232 (P>0.10) |
-0.07845 (P>0.10) |
|
| Full | 19 | 8 | 11 | 19 | 4.300 | 21 | 0.902 ± 0.040 | 0.00351 ± 0.00033 | 0.0028 | -0.18385 (P>0.10) |
-1.6252 (P>0.10) |
-0.99150 (P>0.10) |
| K1 | ||||||||||||
| Block 15 | 1 | 0 | 1 | 1 | 0.471 | 2 | 0.471 ± 0.082 | 0.00214 ± 0.00037 | 1.16615 (P>1.10) |
0.66689 (P>0.10) |
0.91015 (P>0.10) |
|
| Block 16 | 4 | 2 | 2 | 4 | 0.641 | 4 | 0.477 ± 0.134 | 0.00102 ± 0.00037 | -1.34736 (P>0.10) |
-0.70114 (P>0.10) |
-1.00781 (P>0.10) |
|
| Block 17 | 2 | 1 | 1 | 2 | 0.320 | 3 | 0.307 ± 0.132 | 0.00093 ± 0.00042 | -1.09629 (P>0.10) |
-0.55220 (P>0.10) |
-0.79776 (P>0.10) |
|
| Full | 7 | 3 | 4 | 7 | 1.431 | 6 | 0.765 ± 0.080 | 0.00120 ± 0.00026 | −0.0012 | -1.00674 (P>0.10) |
-0.51291 (P>0.10) |
-0.74949 (P>0.10) |
| RO33 | ||||||||||||
| Block 15 | 1 | 1 | 0 | 1 | 0.143 | 2 | 0.143 ± 0.119 | 0.00074 ± 0.00062 | -1.15524 (P>0.10) |
-1.39749 (P>0.10) |
-1.52388 (P>0.10) |
|
| Block 16 | 4 | 3 | 1 | 4 | 0.791 | 4 | 0.571 ± 0.132 | 0.00115 ± 0.00039 | -1.22200 (P>0.10) |
-1.41428 (P>0.10) |
-1.55406 (P>0.10) |
|
| Block 17 | 4 | 4 | 8 | 4 | 0.571 | 3 | 0.275 ± 0.148 | 0.00168 ± 0.00108 | -1.79759 (P>0.10) |
-2.27380 (P>0.10) |
-2.44883 (P>0.10) |
|
| Full | 9 | 8 | 1 | 9 | 1.506 | 6 | 0.736 ± 0.109 | 0.00123 ± 0.00041 | 0.0015 | -1.79616 (P<0.05) |
-2.26934 (P<0.05) |
-2.45049 (P<0.05) |
Abbreviations: K average number of pairwise nucleotide differences, H number of haplotypes, Hd haplotype diversity, π observed average pairwise nucleotide diversity, dN rate of non-synonymous mutations, dS rate of synonymous mutations
The Hd and π values for 14 RO33 allele types of Myanmar PfMSP-142 were 0.736 ± 0.109 and 0.00123 ± 0.00041, respectively. The highest π value for RO33 allele type was identified at block 17 (0.00168 ± 0.00108). The dN-dS value was also estimated to examine whether natural selection contributed to the genetic diversity in Myanmar PfMSP-142. The dN-dS values for MAD20 and RO33 allele types were 0.0028 and 0.0015, respectively, implying that positive natural selection might have occurred in these sequences. Meanwhile, dN-dS value for K1 allele type was -0.0012, suggesting that negative natural selection might have affected the allele type, but this value was not significant (Table 1). To further analyse the occurrence of natural selection in Myanmar PfMSP-142 population, Tajima’s D values were calculated. Estimated Tajima’s D values for MAD20, K1 and RO33 allele types were all negative: -0.18385 (P > 0.10) for MAD20 allele types, -1.00674 (P > 0.10) for K1 allele types and -1.79616 (P < 0.05) for RO33 allele types. These results suggest that all three allele types of Myanmar PfMSP-142 are under population size expansion and/or purifying selection pressure. Fu and Li’s D and F values for all three allele types of Myanmar PfMSP-142 were also negative (Table 1).
Nucleotide diversities in global PfMSP-142
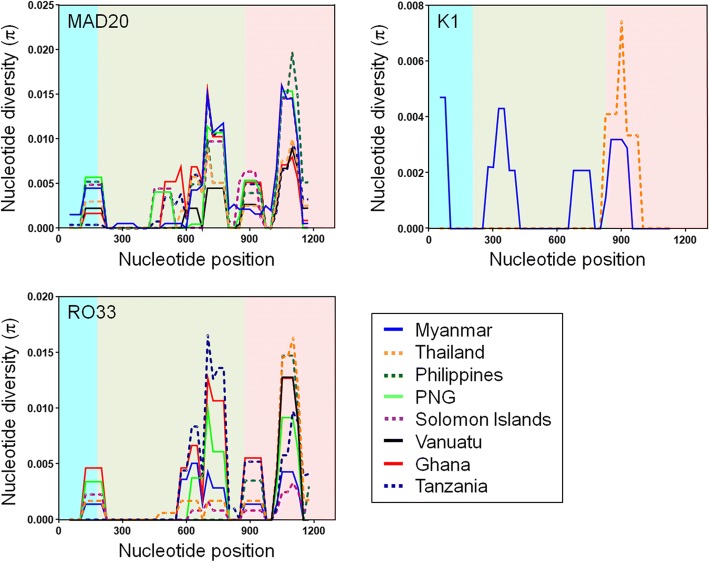
Nucleotide diversity of global PfMSP-142 was analysed to understand the overall pattern of nucleotide diversity in global PfMSP-142. Among PfMSP-142 sequences from global isolates, including Myanmar, Thailand, Philippines, PNG, Solomon Islands, Vanuatu, Ghana and Tanzania, only 18 sequences of Myanmar isolates and 12 sequences of Thailand isolates were grouped to K1 allele types. Other sequences belonged to MAD20 or RO33 allele types. Values for nucleotide diversity (π) differed by allele types and isolates with different geographical origins (Table 2). Among MAD20 allele types of global PfMSP-142, African PfMSP-142 showed higher haplotype diversity (Hd) values than those from other geographical origins. Values of π for full-length PfMSP-142 ranged from 0.00168 to 0.00351 in these isolates. However, all isolates showed similar patterns with the highest π values at block 17. Sliding window plot analysis of π also showed that values observed among global PfMSP-142 sequences peaked at blocks 16 and 17, although the overall pattern was slightly distinct throughout sequences (Fig. 5). In the cases of K1 allele types, Myanmar and Thailand PfMSP-142 had different patterns of π values for each block. Myanmar PfMSP-142 showed nucleotide diversities in blocks 15 to 17, although their π values were not high. Meanwhile, Thailand PfMSP-142 sequences were tightly conserved at blocks 15 and 16 (Table 2; Fig. 5). Similar patterns of dissimilar nucleotide diversity were also identified for RO33 allele types of global PfMSP-142. Unlike PfMSP-142 from Myanmar, Thailand, PNG, Solomon Islands and Ghana, PfMSP-142 from Philippines, Vanuatu and Tanzania did not show nucleotide diversities at block 15 and/or block 16 (Table 2; Fig. 5). The highest value of π was found at block 17 for all isolates, ranging between 0.00122–0.00593. Based on the results of the full-length PfMSP-142 analysis, African PfMSP-142 had higher π values than PfMSP-142 from other geographical isolates.
Table 2.
Genetic polymorphisms and tests of neutrality for global PfMSP-142
| Allele type/ Country (No. of sequences) | K | H | Hd ± SD | π ± SD | dN-dS | Tajima’s D (P-value) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Block15 | Block16 | Block17 | Full | Block15 | Block16 | Block17 | Full | |||||
| MAD20 | ||||||||||||
| Myanmar (n = 37) |
4.300 | 21 | 0.902 ± 0.040 | 0.00313 ± 0.00049 | 0.00272 ± 0.00035 | 0.00533 ± 0.00062 | 0.00351 ± 0.00033 | 0.0208 | 0.50117 (P>0.10) |
-0.65200 (P>0.10) |
0.23510 (P>0.10) |
-0.18385 (P>0.10) |
| Thailand (n = 29) |
2.620 | 6 | 0.473 ± 0.110 | 0.00153 ± 0.00048 | 0.00156 ± 0.00050 | 0.00364 ± 0.00094 | 0.00214 ± 0.00055 | 0.0027 | 0.25334 (P>0.10) |
-0.42016 (P>0.10) |
-0.53026 (P>0.10) |
-0.46786 (P>0.10) |
| Philippines (n = 24) |
3.880 | 5 | 0.652 ± 0.081 | 0.00268 ± 0.00018 | 0.00144 ± 0.00014 | 0.00694 ± 0.00082 | 0.00317 ± 0.00033 | 0.0040 | 1.57336 (P>0.10) |
1.89198 (0.05<P<0.10) |
2.22277 (P<0.05) |
2.60308 (P<0.01) |
| PNG (n = 44) |
4.098 | 7 | 0.753 ± 0.034 | 0.00263 ± 0.00011 | 0.00220 ± 0.00013 | 0.00607 ± 0.00029 | 0.00335 ± 0.00011 | 0.0035 | 1.67820 (P>0.10) |
0.80323 P>0.10 |
1.99671 (0.05<P<0.10) |
1.84977 (0.05<P<0.10) |
| SI (n = 17) |
3.985 | 4 | 0.596 ± 0.099 | 0.00251 ± 0.00041 | 0.00205 ± 0.00036 | 0.00612 ± 0.00097 | 0.00326 ± 0.00052 | 0.0014 | 1.23804 (P>0.10) |
1.67363 (P>0.10) |
1.32924 (P>0.10) |
1.79153 (0.05<P<0.10) |
| Vanuatu (n = 48) |
2.052 | 3 | 0.228 ± 0.075 | 0.00116 ± 0.07200 | 0.00097 ± 0.00031 | 0.00339 ± 0.00110 | 0.00168 ± 0.00054 | 0.0019 | -0.01160 (P>0.10) |
-0.01781 (P>0.10) |
-0.36701 (P>0.10) |
-0.25695 (P>0.10) |
| Ghana (n = 23) |
3.715 | 14 | 0.925 ± 0.041 | 0.00086 ± 0.00051 | 0.00324 ± 0.00019 | 0.00385 ± 0.00097 | 0.00304 ± 0.00029 | 0.0038 | -0.66215 (P>0.10) |
1.91092 (0.05<P<0.10) |
-0.09195 (P>0.10) |
0.84400 (P>0.10) |
| Tanzania (n = 51) |
3.585 | 29 | 0.958 ± 0.014 | 0.00041 ± 0.00027 | 0.00296 ± 0.00028 | 0.00430 ± 0.00060 | 0.00293 ± 0.00023 | 0.0035 | -1.46227 (P>0.10) |
0.05855 (P>0.10) |
0.25010 (P>0.10) |
-0.15863 (P>0.10) |
| K1 | ||||||||||||
| Myanmar (n = 18) |
1.431 | 6 | 0.765 ± 0.080 | 0.00214 ± 0.00037 | 0.00102 ± 0.00037 | 0.00093 ± 0.00042 | 0.00120 ± 0.00026 | -0.0012 | 1.16615 (P>1.10) |
-1.34736 (P>0.10) |
-1.09629 (P>0.10) |
-1.00674 (P>0.10) |
| Thailand (n = 12) |
0.742 | 3 | 0.530 ± 0.136 | 0 | 0 | 0.00215 ± 0.00081 | 0.00062 ± 0.00023 | 0.0008 | 0 | 0 | -0.82879 (P>0.10) |
-0.82879 (P>0.10) |
| RO33 | ||||||||||||
| Myanmar (n = 14) |
1.506 | 6 | 0.736 ± 0.109 | 0.00074 ± 0.00062 | 0.00115 ± 0.00039 | 0.00168 ± 0.00108 | 0.00123 ± 0.00041 | 0.0015 | -1.15524 (P>0.10) |
-1.22200 (P>0.10) |
-1.79759 (P>0.10) |
-1.79616 (P>0.10) |
| Thailand (n = 33) |
2.375 | 6 | 0.680 ± 0.060 | 0.00088 ± 0.00043 | 0.00058 ± 0.00025 | 0.00529 ± 0.00055 | 0.00195 ± 0.00028 | 0.0024 | -0.46603 (P>0.10) |
-1.04700 (P>0.10) |
1.23456 (P>0.10) |
0.21599 (P>0.10) |
| Philippines (n = 19) |
2.023 | 4 | 0.678 ± 0.088 | 0 | 0 | 0.00593 ± 0.00083 | 0.00166 ± 0.00023 | 0.0021 | 0 | 0 | 1.28875 (P>0.10) |
1.28875 (P>0.10) |
| PNG (n = 33) |
2.257 | 4 | 0.447 ± 0.093 | 0.00179 ± 0.00043 | 0.00144 ± 0.00030 | 0.00270 ± 0.00076 | 0.00185 ± 0.00047 | 0.0023 | 0.60324 (P>0.10) |
0.78518 (P>0.10) |
0.56203 (P>0.10) |
0.89252 (P>0.10) |
| SI (n = 24) |
0.811 | 3 | 0.236 ± 0.109 | 0.00118 ± 0.00053 | 0.00024 ± 0.00022 | 0.00122 ± 0.00110 | 0.00066 ± 0.00048 | 0.0008 | -0.24844 (P>0.10) |
-1.51469 (P>0.10) |
-1.99611 (P<0.05) |
-1.99292 (P<0.05) |
| Vanuatu (n = 43) |
1.278 | 2 | 0.432 ± 0.057 | 0 | 0 | 0.00375 ± 0.00051 | 0.00106 ± 0.00014 | 0.0013 | 0 | 0 | 0.90772 (P>0.10) |
0.90772 (P>0.10) |
| Ghana (n = 10) |
4.022 | 6 | 0.844 ± 0.103 | 0.00242 ± 0.00068 | 0.00252 ± 0.00056 | 0.00534 ± 0.00144 | 0.00329 ± 0.00054 | 0.0041 | 0.81980 (P>0.10) |
0.86725 (P>0.10) |
0.12431 (P>0.10) |
0.61111 (P>0.10) |
| Tanzania (n = 20) |
3.694 | 13 | 0.953 ± 0.028 | 0 | 0.00250 ± 0.00030 | 0.00437 ± 0.00086 | 0.00303 ± 0.00032 | 0.0038 | 0 | 0.37812 (P>0.10) |
-0.37024 (P>0.10) |
0.03013 (P>0.10) |
Abbreviations: K average number of pairwise nucleotide differences, Hd haplotype diversity, π observed average pairwise nucleotide diversity, dN rate of non-synonymous mutations, dS rate of synonymous mutations, PNG Papua New Guinea, SI Solomon Islands
Fig. 5.
Nucleotide diversity of PfMSP-142 in global P. falciparum isolates. Sliding window plot analysis illustrates nucleotide diversity (π) values in global PfMSP-142 sequences analysed in this study. A window size of 100 bp and a step size of 25 bp were used. Blocks are indicated by different colors: block 15 (sky blue), block 16 (green), block 17 (pink). Abbreviation: PNG, Papua New Guinea
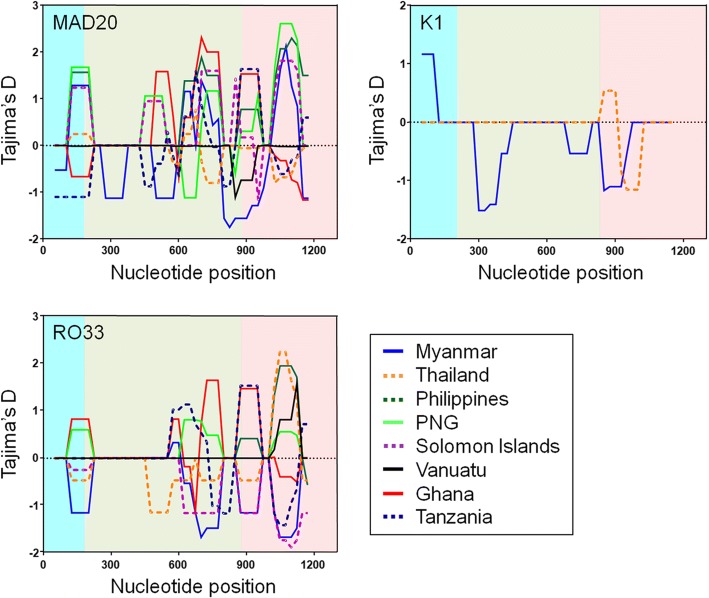
Natural selection and recombinant events in global PfMSP-142
To analyse the effect of natural selection on genetic diversity of global PfMSP-142, the value of dN-dS was estimated. The dN-dS values for full-length PfMSP-142 in global isolates were all positive except for K1 allele types in Myanmar PfMSP-142, suggesting that balancing selection might have occurred in PfMSP-142 of global P. falciparum population (Table 2). Global PfMSP-142 sequences showed positive or negative Tajima’s D values (Table 2). The value for each block in global PfMSP-142 also differed by allele types and geographical origins. A sliding window plot analysis also revealed that PfMSP-142 from different geographical regions showed different patterns for Tajima’s D across the PfMSP-142 gene (Fig. 6). The minimum number of recombination events between adjacent polymorphic sites (Rm) for global PfMSP-142 was analysed. Recombination parameters were different among global PfMSP-142 by allele types and isolates from different countries (Table 3). The higher value of the recombination parameters (Rm and R) indicate that high meiotic recombination is taking place between the sites generating genetic diversity in the gene. The highest R-values were predicted for African PfMSP-142 (Ghana and Tanzania). The Rm values for MAD20 allele types of Ghana and Tanzania were 4 and 4, respectively. The values for RO33 allele types in Ghana and Tanzania were 2 and 3, respectively. The values of R (both between adjacent sites and for the entire domain) were also highest in Tanzania (0.0444 and 54.3 for MAD20 allele types and 0.0439 and 53.5 for RO33 allele types), followed by Ghana (0.0340 and 41.6 for MAD20 allele types and 0.0187 and 22.8 for RO33 allele types).
Fig. 6.
Tajima’s D of PfMSP-142 in global P. falciparum isolates. Sliding window plot analysis of Tajima’s D was performed for global PfMSP-142 sequences. A window size of 100 bp and a step size of 25 bp were used. Blocks are indicated by different colors: block 15 (sky blue), block 16 (green), block 17 (pink). Abbreviation: PNG, Papua New Guinea
Table 3.
Recombination events in global PfMSP-142. The Ra, Rb and Rm were estimated excluding the sites containing alignment gaps or those segregating for three nucleotides
| Allele type | Country (No. of sequences) | Ra | Rb | Rm |
|---|---|---|---|---|
| MAD20 | Myanmar (n = 37) | 0.0134 | 16.4 | 4 |
| Thailand (n = 29) | 0 | 0.001 | 1 | |
| Philippines (n = 24) | 0.0004 | 0.5 | 0 | |
| PNG (n = 44) | 0.0028 | 3.4 | 2 | |
| SI (n = 17) | 0 | 0.001 | 0 | |
| Vanuatu (n = 48) | 0 | 0.001 | 0 | |
| Ghana (n = 23) | 0.0340 | 41.6 | 4 | |
| Tanzania (n = 51) | 0.0444 | 54.3 | 4 | |
| K1 | Myanmar (n = 18) | 0.0363 | 43.3 | 0 |
| Thailand (n = 12) | 0.0083 | 9.9 | 0 | |
| RO33 | Myanmar (n = 14) | 0.0035 | 4.3 | 0 |
| Thailand (n = 33) | 0.0028 | 3.4 | 1 | |
| Philippines (n = 19) | 0.0028 | 3.4 | 0 | |
| PNG (n = 33) | 0 | 0.001 | 1 | |
| SI (n = 24) | 0 | 0.001 | 0 | |
| Vanuatu (n = 43) | 0 | 0.001 | 0 | |
| Ghana (n = 10) | 0.0187 | 22.8 | 2 | |
| Tanzania (n = 20) | 0.0439 | 53.5 | 3 |
Abbreviations: Ra recombination parameter between adjacent sites, Rb recombination parameter for entire gene, Rm minimum number of recombination events between adjacent sites, PNG Papua New Guinea, SI Solomon Islands
Meanwhile, no recombination event was identified in PfMSP-142 from the Solomon Islands or Vanuatu. Recombination parameters identified in PfMSP-142 were not high, and they differed by geographical population, but a possible recombination event might have occurred in global PfMSP-142.
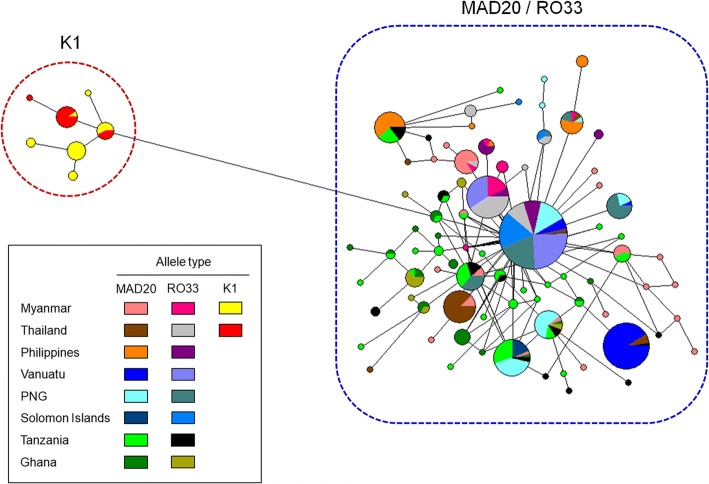
Haplotype network analysis
Haplotype network analysis of PfMSP-142 haplotypes from global P. falciparum populations showed a dense network with extremely complex relationships (Fig. 7). A total of 86 distinct haplotypes were identified in 499 PfMSP-142 sequences analysed, of which 70.9% (61/86) was a singleton. Haplotype prevalence ranged between 0.2–24.2%. K1 allele type formed a separated group from MAD20 and RO33 allele types. Meanwhile, MAD20 and RO33 clustered into one group with mixed patterns probably due to their high sequence similarities. Interestingly, the rate of a singleton was high in African PfMSP-142, MAD20 and RO33 allele types, which support a high level of genetic diversity in the African PfMSP-142 population.
Fig. 7.
Network analysis of global PfMSP-142 haplotypes. Haplotype network was constructed using the program NETWORK version 5.0.0.3 with the Median Joining algorithm. A total of 499 PfMSP-142 sequences were analysed. The size of each circle reflects the richness of the haplotype, which each color showing the geographical origin and allele type of the isolates. The lengths of the lines connecting the circles, measured from their centres, is proportional to the number of base pair substitutions separating the haplotypes. Color of each node indicates different country and allele type
Discussion
The population structure of malaria parasites plays a vital role in maintaining natural immunity against malaria. Several factors including transmission intensity, parasite recombination rate, antimalarial drug resistance status, and effective malaria control measures can affect the population diversity of parasites. In this respect, population structure analysis of malaria parasites is essential to understand the genetic nature and evolutionary aspect of the parasites, especially for vaccine candidate antigens or drug resistance genes. PfMSP-142 is the target of immune protection. Therefore, PfMSP-142 has been recognised as a promising candidate antigen for blood stage vaccine development [17, 20–23]. Unlike other regions of PfMSP-1, PfMSP-142 has been considered as genetically stable among P. falciparum isolates [26, 27]. This is one of the main reasons why it has been recognised as a potential vaccine candidate. However, only limited information is currently available on genetic structure, and natural selection of PfMSP-142 in global P. falciparum isolates. In this study, comparative analysis of population structure and natural selection of PfMSP-142 in Myanmar and global P. falciparum isolates was performed.
Myanmar and global PfMSP-142 displayed limited levels of genetic diversity. However, non-negligible genetic diversity was also found. Thus, it is difficult to regard that PfMSP-142 is genetically well conserved in global P. falciparum population. Genetic diversity of PfMSP-1 has been defined as unique, different allele types: MAD20, K1 and RO33 [26, 27]. Blocks 15, 16 and 17 of PfMSP-1 comprising PfMSP-142 have been considered to be well conserved in P. falciparum population [26, 27, 47]. All three allele types of Myanmar and global PfMSP-142 showed SNPs that were mainly identified in blocks 16 and 17. Overall distribution patterns and frequencies of amino acid changes found in Myanmar and global PfMSP-142 analysed in this study were not identical between or among global PfMSP-142. The implication of these substantial differences in amino acid polymorphisms among global PfMSP-142 by geographical differentiation is currently unclear. Geographical isolation followed by population subdivision may affect genetic differences of PfMSP-142 population in different geographical areas. Major amino acid changes found in global PfMSP-142 were commonly detected among global PfMSP-142, although their frequencies differed among and between populations and only a limited number of PfMSP-142 sequences in each geographical area were analysed in this study. Thus, further examination of PfMSP-142 nucleotide and amino acid variations in a more substantial number of global PfMSP-142 sequences is needed to better understand the polymorphic nature of global PfMSP-142.
Values of haplotype diversity (Hd) and nucleotide diversity (π) were higher in African PfMSP-142 than those in Asian and Pacific PfMSP-142, indicating that African PfMSP-142 possessed higher levels of genetic diversities compared to Asian and Pacific PfMSP-142. For both MAD20 and RO33 allele types, the highest π value was identified at block 17 in global PfMSP-142, suggesting that block 17 had the highest genetic variations. Sliding window plot analysis of π across PfMSP-142 revealed similar patterns of nucleotide diversity in global MAD20 and RO33 allele types of PfMSP-142 analysed here, suggesting that global PfMSP-142 might share similar nucleotide diversity across this gene. In the case of K1 allele types, different patterns of π were identified between Myanmar and Thailand PfMSP-142. Nucleotide diversity in Thailand PfMSP-142 was identified only at block 17 while Myanmar PfMSP-142 showed nucleotide diversity at blocks 15, 16 and 17, albeit π values for these regions were not high. Natural selection analysis of global PfMSP-142 suggests that this region is likely to be under natural selection which may maintain or produce genetic diversity in PfMSP-142 population. The dN-dS value for global PfMSP-142 was positive, implying balancing selection might act in this gene. However, dN-dS value for Myanmar K1 allele types was negative (-0.0012), suggesting negative or purifying natural selection in this region. In the sliding window plot analysis of Tajima’s D, overall patterns of Tajima’s D values across full-length PfMSP-142 showed highly complicated patterns that were distinct between and among global PfMSP-142. These results suggested that global PfMSP-142 was under a complicated influence of natural selection, in which either positive natural selection or purifying selection might have occurred in the population depending on allele types and geographical origins. Possible recombination events in global PfMSP-142 were also predicted, although values were not high. Higher values of recombination events were found in African PfMSP-142 than those in PfMSP-142 from other geographical areas, suggesting that African PfMSP-142 might be under more opportunity for inter- or intra-allelic recombination than other geographical PfMSP-142 probably due to high multiclonal infection rate, subsequent cross-fertilisation and recombination in mosquitoes. These results collectively suggest that PfMSP-142 is not highly conserved in global population as known to date and that complicated natural selection acts on this region. Recombination might also contribute to the genetic diversity of global PfMSP-142, albeit recombination parameters were not high.
Of three blocks of PfMSP-142, block 17 (referred to as PfMSP-119) has attracted considerable attention since this region is functionally critical. PfMSP-119 has two EGF-like motifs that play an important role in the attachment of the parasite to specific receptors on the surface of erythrocyte [6]. Antibodies to one of these two EGF-like motifs in PfMSP-119 are strongly associated with resistance to both clinical malaria and high levels of parasitemia [48]. It has been proposed that PfMSP-119 is conserved mainly between parasite strains with only a few amino acid substitutions [26, 27, 47]. However, results of this study suggested that block 17 also showed greater polymorphic patterns in global isolates than previously known, especially for MAD20 and RO33 allele types. A possible recombination event in this region was also predicted, in concordance with previous reports [26, 27, 47, 49]. SNPs in PfMSP-119 might have evolved to evade the human immune response capable of blocking the parasite from invading erythrocytes [27, 48]. Polymorphic nature, possible natural selection and recombination events observed at block 17 in global isolates raise concern for reduced efficacy of vaccine using PfMSP-119. Therefore, caution is needed when designing polyvalent vaccine constructs targeting PfMSP-119. Comprehensive genetic analysis of global PfMSP-119 is merited to understand the complexity of genetic makeup of this region in the global P. falciparum population.
Conclusions
Comparative analysis of Myanmar PfMSP-142 with global PfMSP-142 revealed that global PfMSP-142 displayed limited levels of genetic diversity in the P. falciparum population. However, results of this study also suggest that global PfMSP-142 is not as genetically conserved as previously thought. Although patterns of major amino acid changes found in global PfMSP-142 were relatively similar, their frequencies differed by allele types and geographical origins. Moreover, some amino acid changes were characteristic only in specific geographical origins. Global PfMSP-142 was under the complicated influence of natural selection, in which either balancing selection or purifying selection might have occurred in the population depending on allelic types and geographical origins. Recombination might have also affected the genetic diversity of the global PfMSP-142 population. These results collectively suggested that more concern would be necessary when designing vaccine based on PfMSP-142 which has been considered to be well conserved in P. falciparum population. This study also warrants continuous monitoring of genetic analysis for PfMSP-142 in global PfMSP-142 to better understand the polymorphic nature and evolutionary aspect of PfMSP-142 in the global P. falciparum population.
Additional files
Figure S1. Sequence analysis for MAD20 allele types of PfMSP-142 in Myanmar Plasmodium falciparum isolates. Regions corresponding to blocks 15, 16 and 17 are marked with different colors. Dots represent identical residues compared to reference sequences. Underlines indicate regions of epidermal growth factor-like (EGF1 and EGF2) domains. Amino acid changes identified in at least one sequence of the corresponding haplotype are marked in red. Cysteine residues in block 17 are shown in blue with an underline. Total numbers of isolates for each haplotype are listed in the right panel. (TIF 3611 kb)
Figure S2. Sequence analysis for K1 allele types of PfMSP-142 in Myanmar Plasmodium falciparum isolates. Regions corresponding blocks 15, 16 and 17 are marked with different colors. Dots represent identical residues compared to reference sequences. Regions of epidermal growth factor-like (EGF1 and EGF2) domains are indicated by underlines. Amino acid changes identified in at least one sequence of the corresponding haplotype are marked in red. Cysteine residues in block 17 are shown in blue with an underline. Total numbers of isolates for each haplotype are listed in the right panel. (TIF 1834 kb)
Figure S3. Sequence analysis for RO33 allele types of PfMSP-142 in Myanmar Plasmodium falciparum isolates. Regions corresponding blocks 15, 16 and 17 are marked with different colors. Dots represent identical residues compared to reference sequences. Regions of epidermal growth factor-like (EGF1 and EGF2) domains are indicated by underlines. Amino acid changes identified in at least one sequence of the corresponding haplotype are marked in red. Cysteine residues in block 17 are shown in blue with an underline. Total numbers of isolates for each haplotype are listed in the right panel. (TIF 1791 kb)
Acknowledgements
We thank the staffs in Department of Medical Research Pyin Oo Lwin Branch and the health professionals in Naung Cho, Pyin Oo Lwin and Tha Beik Kyin townships for their contribution and technical support in field study. This work was supported by the National Research Foundation (NRF) of Korea grants funded by the Korea government (MSIP) (NRF-2015R1A2A2A01004310) and the Korea government (MEST) (NRF-2015K1A3A9A01034893).
Funding
This work was supported by the National Research Foundation (NRF) of Korea grants funded by the Korea government (MSIP) (NRF-2015R1A2A2A01004310) and the Korea government (MEST) (NRF-2015K1A3A9A01034893).
Availability of data and materials
The data supporting the conclusions of this article are provided within the article and its additional files. The original datasets analysed in the current study are available from the corresponding author upon request. The newly generated sequences were deposited in the GenBank database under the accession numbers MF943252-MF943320.
Abbreviations
- dN
Rate of non-synonymous substitutions
- dS
Rates of synonymous substitutions
- H
Haplotypes
- Hd
Haplotype diversity
- K
Average number of pairwise nucleotide differences within the population
- MSP-1
Merozoite surface protein-1
- PCR
Polymerase chain reaction
- PfMSP-142
C-terminal 42 kDa region of merozoite surface protein-1 of Plasmodium falciparum
- PNG
Papua New Guinea
- Rm
Minimum number of recombination events between adjacent polymorphic sites
- S
Segregating sites
- π
Nucleotide diversity
Authors’ contributions
TLT, JMK and JL performed genetic analysis of PfMSP-142. MM, HJ, JL, KL and KZT contributed the blood sample collection. TLT, JMK, HGL, WMS, TSK and BKN analysed and interpreted the data. TSK and BKN designed and supervised the experiments. TLT and BKN wrote the draft of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval for this study was obtained from the Ethics Review Committee, Department of Medical Research, Myanmar (97/Ethics 2015) and the Ethical Review Committee of Inha University School of Medicine, Korea (INHA 15-013). Informed written consent and permission were obtained from each individual enrolled in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Thị Lam Thái, Email: thailamcnshe.hua@gmail.com.
Hojong Jun, Email: hojongi2@inha.ac.kr.
Jinyoung Lee, Email: jylee5492@gmail.com.
Jung-Mi Kang, Email: gjm9951001@hanmail.net.
Hương Giang Lê, Email: gianglee291994@gmail.com.
Khin Lin, Email: dr.khinlin.dir@gmail.com.
Kyaw Zin Thant, Email: drkz.thant@gmail.com.
Woon-Mok Sohn, Email: wmsohn@gnu.ac.kr.
Tong-Soo Kim, Email: tongsookim@inha.ac.kr.
Byoung-Kuk Na, Email: bkna@gnu.ac.kr.
References
- 1.WHO . World Malaria Report 2017. Geneva: World Health Organization; 2017. [Google Scholar]
- 2.Holder AA, Blackman MJ, Burghaus PA, Chappel JA, Ling IT, McCallum-Deighton N, et al. A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz. 1992;87(Suppl. 3):37–42. doi: 10.1590/S0074-02761992000700004. [DOI] [PubMed] [Google Scholar]
- 3.Holder AA. The carboxy-terminus of merozoite surface protein 1: structure, specific antibodies and immunity to malaria. Parasitology. 2009;136:1455–1456. doi: 10.1017/S0031182009990515. [DOI] [PubMed] [Google Scholar]
- 4.Blackman MJ, Whilev H, Holder AA. Processing of the Plasmodium falciparum major merozoite surface protein-1: identification of a 33-kilodalton secondary processing product which is shed prior to erythrocyte invasion. Mol Biochem Parasitol. 1991;49:35–44. doi: 10.1016/0166-6851(91)90128-S. [DOI] [PubMed] [Google Scholar]
- 5.Blackman MJ, Holder AA. Secondary processing of the Plasmodium falciparum merozoite surface protein-1 (MSP1) by a calcium-dependent membrane-bound serine protease: shedding of MSP-133 as a noncovalently associated complex with other fragments of the MSP1. Mol Biochem Parasitol. 1992;50:307–315. doi: 10.1016/0166-6851(92)90228-C. [DOI] [PubMed] [Google Scholar]
- 6.Blackman MJ, Heidrich HG, Donachie S, McBridge JS, Holder AA. A single fragment of a malaria merozoite surface protein remains on the parasite during red blood cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terrientes ZI, Kramer K, Herrera MA, Chang SP. Naturally acquired antibodies against the major merozoite surface coat protein (MSP-1) of Plasmodium falciparum acquired by residents in an endemic area of Colombia. Mem Inst Oswaldo Cruz. 1994;89(Suppl. 2):55–61. doi: 10.1590/S0074-02761994000600014. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh DR, McBride JS. Antigenicity of recombinant proteins derived from Plasmodium falciparum merozoite surface protein 1. Mol Biochem Parasitol. 1997;85:197–211. doi: 10.1016/S0166-6851(96)02826-5. [DOI] [PubMed] [Google Scholar]
- 9.Mawili-Mboumba DP, Borrmann S, Cavanagh DR, McBride JS, Matsiegui PB, Missinou MA, et al. Antibody responses to Plasmodium falciparum merozoite surface protein-1 and efficacy of amodiaquine in Gabonese children with P. falciparum malaria. J Infect Dis. 2003;187:1137–1141. doi: 10.1086/368414. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Becerra C, Sanz S, Brucet M, Stanisic DI, Alves FP, Camargo EP, et al. Naturally-acquired humoral immune responses against the N- and C-termini of the Plasmodium vivax MSP1 protein in endemic regions of Brazil and Papua New Guinea using a multiplex assay. Malar J. 2010;9:29. doi: 10.1186/1475-2875-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeyrek FY, Babaoglu A, Demirel S, Erdogan DD, Ak M, Korkmaz M, et al. Analysis of naturally acquired antibody responses to the 19-kD C-terminal region of merozoite surface protein-1 of Plasmodium vivax from individuals in Sanliurfa, Turkey. Am J Trop Med Hyg. 2008;78:729–732. [PubMed] [Google Scholar]
- 12.Mehrizi AA, Asgharpour S, Salmanian AH, Djadid ND, Zakeri S. IgG subclass antibodies to three variants of Plasmodium falciparum merozoite surface protein-1 (PfMSP-1(19)) in an area with unstable malaria transmission in Iran. Acta Trop. 2011;119:84–90. doi: 10.1016/j.actatropica.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Moormann AM, Sumba PO, Chelimo K, Fang H, Tisch DJ, Dent AE, et al. Humoral and cellular immunity to Plasmodium falciparum merozoite surface protein 1 and protection from infection with blood-stage parasites. J Infect Dis. 2013;208:149–158. doi: 10.1093/infdis/jit134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Zhao Z, Zhang X, Li X, Zhu M, Li P, et al. Naturally acquired antibody responses to Plasmodium vivax and Plasmodium falciparum merozoite surface protein 1 (MSP1) C-terminal 19 kDa domains in an area of unstable malaria transmission in Southeast Asia. PLoS One. 2016;11:e0151900. doi: 10.1371/journal.pone.0151900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenhouse B, Ho B, Hubbard A, Njama-Meya D, Narum DL, Lanar DE, et al. Antibodies to Plasmodium falciparum antigens predict a higher risk of malaria but protection from symptoms once parasitemic. J Infect Dis. 2011;204:19–26. doi: 10.1093/infdis/jir223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egan AF, Burghaus P, Druilhe P, Holder AA, Riley EM. Human antibodies to the 19kDa C-terminal fragment of Plasmodium falciparum merozoite surface protein 1 inhibit parasite growth in vitro. Parasite Immunol. 1999;21:133–9. [DOI] [PubMed]
- 17.Singh S, Miura K, Zhou H, Muratova O, Keegan B, Miles A, et al. Immunity to recombinant Plasmodium falciparum merozoite surface protein 1 (MSP1): protection in Aotus nancymai monkeys strongly correlates with anti-MSP1 antibody titer and in vitro parasite-inhibitory activity. Infect Immun. 2006;74:4573–80. [DOI] [PMC free article] [PubMed]
- 18.Moss DK, Remarque EJ, Faber BW, Cavanagh DR, Arnot DE, Thomas AW, et al. Plasmodium falciparum 19-kilodalton merozoite surface protein 1 (MSP1)-specific antibodies that interfere with parasite growth in vitro can inhibit MSP1 processing, merozoite invasion, and intracellular parasite development. Infect Immun. 2012;80:1280–7. [DOI] [PMC free article] [PubMed]
- 19.Chen Q, Liang W, Qian F, Qian B, Cao J, Zhang D, et al. Rice-produced MSP142 of Plasmodium falciparum elicits antibodies that inhibit parasite growth in vitro. Parasite Immunol. 2016;38:635–641. doi: 10.1111/pim.12352. [DOI] [PubMed] [Google Scholar]
- 20.Riley EM, Allen SJ, Wheeler JG, Blackman MJ, Bennett S, Takacs B, et al. Naturally acquired cellular and humoral immune responses to the major merozoite surface antigen (PfMSP1) of Plasmodium falciparum are associated with reduced malaria morbidity. Parasite Immunol. 1992;14:321–337. doi: 10.1111/j.1365-3024.1992.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 21.John CC, O’Donnell RA, Sumba PO, Moormann AM, de Koning-Ward TF, King CL, et al. Evidence that invasion-inhibitory antibodies specific for the 19-kDa fragment of merozoite surface protein-1 (MSP-119) can play a protective role against blood-stage Plasmodium falciparum infection in individuals in a malaria endemic area of Africa. J Immunol. 2004;173:666–672. doi: 10.4049/jimmunol.173.1.666. [DOI] [PubMed] [Google Scholar]
- 22.Chang SP, Case SE, Gosnell WL, Hashimoto A, Kramer KJ, et al. A recombinant baculovirus 42-kilodalton C-terminal fragment of Plasmodium falciparum merozoite surface protein 1 protects Aotus monkeys against malaria. Infect Immun. 1996;64:253–261. doi: 10.1128/iai.64.1.253-261.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Collins W, Egan A, Yadava A, Garraud O, Blackman MJ, et al. Immunogenicity and efficacy in Aotus monkeys of four recombinant Plasmodium falciparum vaccines in multiple adjuvant formulations based on the 19-kilodalton C terminus of merozoite surface protein 1. Infect Immun. 2000;68:2215–2223. doi: 10.1128/IAI.68.4.2215-2223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazumdar S, Mukherjee P, Yazdani SS, Jain SK, Mohmmed A, Chauhan VS. Plasmodium falciparum merozoite surface protein 1 (MSP-1)-MSP-3 chimeric protein: immunogenicity determined with human-compatible adjuvants and induction of protective immune response. Infect Immun. 2010;78:872–883. doi: 10.1128/IAI.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alaro JR, Partridge A, Miura K, Diouf A, Lopez AM, Angov E, et al. A chimeric Plasmodium falciparum merozoite surface protein vaccine induces high titers of parasite growth inhibitory antibodies. Infect Immun. 2013;81:3843–3854. doi: 10.1128/IAI.00522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 27.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1) Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-F. [DOI] [PubMed] [Google Scholar]
- 28.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, et al. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/S0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 29.Kang JM, Moon SU, Kim JY, Cho SH, Lin K, Sohn WM, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131. doi: 10.1186/1475-2875-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bharti PK, Shukla MM, Sharma YD, Singh N. Genetic diversity in the block 2 region of the merozoite surface protein-1 of Plasmodium falciparum in central India. Malar J. 2012;11:78. doi: 10.1186/1475-2875-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmedou Salem MS, Ndiaye M, OuldAbdallahi M, Lekweiry KM, Bogreau H, Konaté L, et al. Polymorphism of the merozoite surface protein-1 block 2 region in Plasmodium falciparum isolates from Mauritania. Malar J. 2014;13:26. doi: 10.1186/1475-2875-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammed H, Mindaye T, Belayneh M, Kassa M, Assefa A, Tadesse M, et al. Genetic diversity of Plasmodium falciparum isolates based on MSP-1 and MSP-2 genes from Kolla-Shele area, Arbaminch Zuria District, southwest Ethiopia. Malar J. 2015;14:73. doi: 10.1186/s12936-015-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohd Abd Razak MR, Sastu UR, Norahmad NA, Abdul-Karim A, Muhammad A, Muniandy PK, et al. Genetic diversity of Plasmodium falciparum populations in malaria declining areas of Sabah, East Malaysia. PLoS One. 2016;11:e0152415. doi: 10.1371/journal.pone.0152415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha P, Gangguly S, Maji AK. Genetic diversity and multiplicity of infection of Plasmodium falciparum isolates from Kolkata, West Bengal, India. Infect Genet Evol. 2016;43:239–44. [DOI] [PubMed]
- 35.Soe TN, Wu Y, Tun MW, Xu X, Hu Y, Ruan Y, et al. Genetic diversity of Plasmodium falciparum populations in southeast and western Myanmar. Parasit Vectors. 2017;10:322. doi: 10.1186/s13071-017-2254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur H, Sehgal R, Goyal K, Makkar N, Yadav R, Bharti PK, et al. Genetic diversity of Plasmodium falciparum merozoite surface protein-1 (block 2), glutamate-rich protein and sexual stage antigen Pfs25 from Chandigarh, North India. Trop Med Int Health. 2017;22:1590–1598. doi: 10.1111/tmi.12990. [DOI] [PubMed] [Google Scholar]
- 37.Gueye NSG, Ntoumi F, Vouvoungui C, Kobawila SC, NKombo M, Mouanga AM, et al. Plasmodium falciparum merozoite protein-1 genetic diversity and multiplicity of infection in isolates from Congolese children consulting in a pediatric hospital in Brazzaville. Acta Trop. 2018;183:78–83. doi: 10.1016/j.actatropica.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Pacheco MA, Poe AC, Collins WE, Lal AA, Tanabe K, Kariuki SK. A comparative study of the genetic diversity of the 42kDa fragment of the merozoite surface protein 1 in Plasmodium falciparum and P. vivax. Infect Genet Evol. 2007;7:180–187. doi: 10.1016/j.meegid.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203. doi: 10.1385/1-59259-271-6:189. [DOI] [PubMed] [Google Scholar]
- 40.Kang JM, Cho PY, Moe M, Lee J, Jun H, Lee HW, et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar J. 2017;16:119. doi: 10.1186/s12936-017-1765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;23:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 43.Nei M, Gojobory T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 44.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandelt HJ, Forst P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 47.Mehrizi AA, Zakeri S, Salmanian AH, Sanati MH, Djadid ND. Plasmodium falciparum: sequence analysis of the gene encoding the C-terminus region of the merozoite surface protein-1, a potential malaria vaccine antigen, in Iranian clinical isolates. Exp Parasitol. 2008;118:378–385. doi: 10.1016/j.exppara.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Egan AF, Morris J, Barnish G, Allen S, Greenwood BM, Kaslow DC, et al. Clinical immunity to Plasmodium falciparum malaria is associated with serum antibodies to the 19-kDa C-terminal fragment of the merozoite surface antigen, PfMSP-1. J Infect Dis. 1996;173:765–769. doi: 10.1093/infdis/173.3.765. [DOI] [PubMed] [Google Scholar]
- 49.Lalitha PV, Malhotra P, Chattopadhyay R, Chauhan VS. Plasmodium falciparum: variations in the C-terminal cysteine-rich region of the merozoite surface protein-1 in field samples among Indian isolates. Exp Parasitol. 1999;92:12–18. doi: 10.1006/expr.1999.4401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence analysis for MAD20 allele types of PfMSP-142 in Myanmar Plasmodium falciparum isolates. Regions corresponding to blocks 15, 16 and 17 are marked with different colors. Dots represent identical residues compared to reference sequences. Underlines indicate regions of epidermal growth factor-like (EGF1 and EGF2) domains. Amino acid changes identified in at least one sequence of the corresponding haplotype are marked in red. Cysteine residues in block 17 are shown in blue with an underline. Total numbers of isolates for each haplotype are listed in the right panel. (TIF 3611 kb)
Figure S2. Sequence analysis for K1 allele types of PfMSP-142 in Myanmar Plasmodium falciparum isolates. Regions corresponding blocks 15, 16 and 17 are marked with different colors. Dots represent identical residues compared to reference sequences. Regions of epidermal growth factor-like (EGF1 and EGF2) domains are indicated by underlines. Amino acid changes identified in at least one sequence of the corresponding haplotype are marked in red. Cysteine residues in block 17 are shown in blue with an underline. Total numbers of isolates for each haplotype are listed in the right panel. (TIF 1834 kb)
Figure S3. Sequence analysis for RO33 allele types of PfMSP-142 in Myanmar Plasmodium falciparum isolates. Regions corresponding blocks 15, 16 and 17 are marked with different colors. Dots represent identical residues compared to reference sequences. Regions of epidermal growth factor-like (EGF1 and EGF2) domains are indicated by underlines. Amino acid changes identified in at least one sequence of the corresponding haplotype are marked in red. Cysteine residues in block 17 are shown in blue with an underline. Total numbers of isolates for each haplotype are listed in the right panel. (TIF 1791 kb)
Data Availability Statement
The data supporting the conclusions of this article are provided within the article and its additional files. The original datasets analysed in the current study are available from the corresponding author upon request. The newly generated sequences were deposited in the GenBank database under the accession numbers MF943252-MF943320.