Abstract
Amplicons—large, highly identical segmental duplications—are a prominent feature of mammalian Y chromosomes. Although they encode genes essential for fertility, these amplicons differ vastly between species, and little is known about the selective constraints acting on them. Here, we develop computational tools to detect amplicon copy number with unprecedented accuracy from high-throughput sequencing data. We find that one-sixth (16.9%) of 1,216 males from the 1000 Genomes Project have at least one deleted or duplicated amplicon. However, each amplicon’s reference copy number is scrupulously maintained among divergent branches of the Y chromosome phylogeny, including the ancient branch A00, indicating that the reference copy number is ancestral to all modern human Y chromosomes. Using phylogenetic analyses and simulations, we demonstrate that this pattern of variation is incompatible with neutral evolution and instead displays hallmarks of mutation-selection balance. We also observe cases of amplicon rescue, in which deleted amplicons are restored through subsequent duplications. These results indicate that, contrary to the lack of constraint suggested by the differences between species, natural selection has suppressed amplicon copy number variation in diverse human lineages.
Keywords: Y chromosome, sex chromosome, haplogroup, amplicon, CNV, AZFc, spermatogenesis, evolution, infertility, selection
Introduction
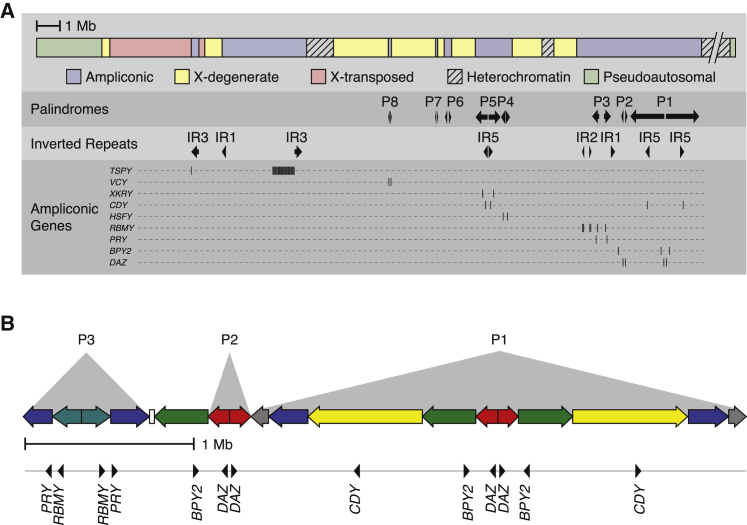
The human Y chromosome does not recombine with a homologous chromosome along the vast majority of its length.1 As a result, it developed a unique and complex genomic structure compared to the other chromosomes.2 In particular, the Y chromosome contains distinct classes of DNA sequence, most strikingly the ampliconic sequence: large segments ranging from tens of kilobases to megabases, comprising 10.2 Mb in total, that are present in two or more copies on the Y chromosome (Figure 1A). The amplicons are typically arranged in palindromes and have extremely high sequence identity—amplicon copies differ by as few as 3 in 100,000 base pairs—that is maintained by gene conversion.3 Remarkably, the genes within the amplicons are functionally coherent: they are expressed predominantly or exclusively in the testis.4 The azoospermia factor c (AZFc) region, composed of ampliconic units interleaved in an intricate pattern, is of particular functional importance: it contains five protein-coding gene families (PRY [MIM: 400019, 400041], RBMY [MIM: 400006], BPY2 [MIM: 400013], DAZ [MIM: 400003], and CDY [MIM: 400016]), each of which is found in a different ampliconic unit (Figure 1B). Large deletions within the region can remove all copies of multiple gene families and cause spermatogenic failure (MIM: 400042, 415000).5, 6 Other amplicon variants cause or increase the risk of spermatogenic failure, sex reversal (MIM: 400045), Turner syndrome, and testis cancer (MIM: 273300).7, 8, 9, 10, 11, 12
Figure 1.
Genomic Structure of the Human Y Chromosome
(A) The human Y chromosome contains five major sequence classes (see Skaletsky et al.2 for details). Ampliconic sequence includes palindromes with a small spacer sequence between copies and widely spaced inverted repeats. Arrows: palindrome or inverted repeat copies; arrow direction indicates copy orientation. Locations of ampliconic protein-coding genes are also shown.
(B) AZFc region encompassing palindromes 1–3, containing multiple copies of six amplicon units with ≥99.94% nucleotide identity. Arrows: blue, teal, green, red, gray, and yellow amplicon units (teal and red arrows include single-copy spacer sequence); arrow direction indicates copy orientation. Small white rectangle: single copy of IR1. Locations of protein-coding genes are shown below AZFc architecture.
Little is known about the evolutionary forces that govern the formation, maintenance, and diversification of Y chromosome amplicons. Although amplicons are present on other mammalian Y chromosomes, the genetic content and genomic structure of those amplicons differ wildly between species. For example, the rhesus macaque Y chromosome contains only 500 kb of ampliconic sequence, although it does share some genes with human amplicons.13 The mouse Y chromosome contains 88 Mb of ampliconic sequence comprised primarily of around 170 copies of a 0.5 Mb unit, and its ampliconic gene content is almost completely different than that of the primate Y chromosome.14 Even in chimpanzees, which diverged from humans only 7 million years ago and whose autosomal euchromatic sequences are 99% identical to humans,15 only around half of the 14.7 Mb of ampliconic sequence is shared with the human Y chromosome, and even the shared portion is drastically rearranged.16 This high level of divergence suggests that the ampliconic regions are evolving in the absence of selective pressures acting to maintain their genetic content and architecture, or even that diversifying selection is actively driving the amplicons to differentiate. However, because of this extreme divergence, the evolutionary history of the amplicons cannot be reconstructed by comparing Y chromosomes of different species.
Amplicon variation within species, on the other hand, occurs over a timescale that is conducive to the study of amplicon evolution. Copy number variants (CNVs)—deletions and duplications of ampliconic sequence—have been studied on the human Y chromosome for decades. These CNVs are often caused by non-allelic homologous recombination (NAHR), as the large, nearly identical amplicon copies represent prime targets for this mechanism.9, 17 Early studies of amplicon CNVs each focused on the detection of a single type of CNV.5, 18, 19 Later studies, bolstered by developing technology, described many types of CNVs in larger numbers of men.11, 20, 21, 22, 23, 24 Amplicon CNVs have recently been discovered on the Y chromosomes of chimpanzees, macaques, gorillas, and mice.25, 26, 27, 28 Some amplicon CNVs have been implicated in spermatogenic failure, sex reversal, Turner syndrome, and testis cancer.5, 6, 7, 8, 9, 10, 11, 12 However, the amplicon CNVs with well-described phenotypes represent only a small part of the spectrum of amplicon variation; the vast majority of amplicon CNVs that have been discovered have no known effect on spermatogenesis or any other trait.20, 21, 22, 23
Even though amplicon CNVs have been the subject of intense investigation, most previous studies made only nominal attempts to reconstruct the evolution of the amplicons, instead focusing on documenting amplicon variation. Here, we present a detailed reconstruction of Y chromosome amplicon evolution in humans. Our study is made possible by the meticulous sequencing of the reference human Y chromosome, advances in technology that made large sequencing datasets available, and phylogenetic studies of the Y chromosome that enabled the construction of a detailed tree of the males in our dataset.29, 30, 31 With these three tools, we obtain accurate amplicon CNV calls in 1,216 males and map those CNV calls onto the phylogenetic tree. We then use that tree to test evolutionary models in a more precise and powerful manner than previous studies, allowing us to better describe the evolutionary pressures acting on the amplicons.
We find that 16.9% of males in our dataset have an amplicon CNV and that such CNVs are caused by both NAHR between amplicon copies and other mechanisms. Although these CNVs are present in almost all major haplogroups (branches of the Y chromosome phylogeny), the reference copy number of each amplicon is maintained among divergent branches, including in haplogroup A00, which diverged from all other human Y chromosomes more than 200 kya.32 Further, the distribution of males with CNVs within the phylogenetic tree of these Y chromosomes is incompatible with a model of neutral evolution and instead is indicative of mutation-selection balance. Finally, we observe cases of amplicon rescue, in which deleted amplicons are restored through subsequent duplication of other nearly identical amplicon copies. These results indicate that although amplicons are susceptible to large-scale rearrangements, selection acts to maintain amplicon copy number among diverse human lineages.
Material and Methods
Annotation of Y Chromosome Amplicons
When the human Y chromosome was sequenced, the methods used to annotate the amplicons were sufficient to describe the overall ampliconic structure.2, 6 However, we wanted a more precise description of amplicon coordinates for this study. Therefore, we re-annotated the Y chromosome amplicons as follows. We divided the reference Y chromosome sequence into overlapping 100-base-pair windows and aligned these windows to the entire reference genome (hg38) using bowtie2, with settings to return up to 10 alignments with alignment score −11 or greater. (These alignments were also used for masking repetitive sequence; see below.) We then created a bedGraph file of the Y chromosome, in which the value for each base equals the number of times the window beginning at that base aligned to the genome, and visualized the file using the UCSC Genome Browser.33, 34 Then, using the previous amplicon annotations as a guide, we inspected the regions of the Y chromosome where windows aligned to more than one location on the Y chromosome. We determined the precise start and end coordinates of previously described amplicons, gaps in amplicon copies that have low identity to corresponding copies, and several short and previously undescribed amplicons. Table S1 is a list of coordinates for the amplicons studied in this paper. Table S2 is a complete list of amplicons that we annotated with this method.
1000 Genomes Project Data
We analyzed whole-genome sequencing data of 1,225 males from the 1000 Genomes Project.29 We downloaded FASTQ files from the 1000 Genomes FTP site. To ensure the proper alignment of reads, we used only files with paired-end reads and read length ≥ 50 nucleotides. We then aligned the FASTQ files to the latest build of the human reference genome (hg38) using bowtie2.35
GC Bias Correction and Repeat Masking
The GC content of DNA affects read depth in high-throughput sequencing.36 To correct for this effect, we used custom python scripts to build a GC bias curve for each sequencing library and correct sequencing depth based on those curves. See Supplemental Material and Methods for a full description of our method.
We also masked repetitive sequence from our subsequent analyses. From our re-annotation of the amplicons, we had data on the number of times each 100-bp window of the Y chromosome aligned elsewhere in the genome. We masked all windows that aligned more than the expected number of times. For example, in the green amplicon, which has three copies, we masked all windows that aligned four or more times. Because many genomic repetitive elements are divergent enough that sequencing reads align to them uniquely, this method is significantly less stringent than masking by RepeatMasker (see Web Resources), which was used as part of our GC correction pipeline. As a result, we masked a smaller percentage of the amplicon sequence (18% versus 50%), which increases the accuracy of our subsequent analyses. Table S3 contains the processed depths of each amplicon and control region for each male.
Whole Amplicon CNV Detection
To detect CNVs that delete or duplicate whole copies of amplicons, we calculated the mean depth of 15 amplicons by dividing the total depth in all copies of that amplicon by the combined size of the copies. We also calculated the mean depth of four single-copy regions on the Y chromosome to act as negative controls in the filtering steps below. We then normalized by the mean depth of a 1-Mb single-copy region of the Y. After normalization, the expected depth of an amplicon with the reference copy number is 1. We called copy number of amplicons based on the total amplicon depth. The thresholds for calling an amplicon CNV were the midpoints between the expected depths for each copy number of an amplicon. For example, the blue amplicon has four copies in the reference genome. In a male with four copies, the expected depth is 1; in a male with three copies, the expected depth is 0.75; and in a male with two copies, the expected depth is 0.5. Therefore, a male will be called as having three copies if depth is between 0.625 and 0.875.
Partial Amplicon CNV Detection
To detect CNVs that delete or duplicate only part of an amplicon, we used a modified version of the binary segmentation algorithm. Our input was non-overlapping 100-bp windows of depth for each amplicon copy. We removed windows with ≥25% masked bases. We calculated change points in the mean as described in Sen and Srivastava,37 except we used the Mann-Whitney U statistic rather than the t-statistic. We do so because the t test assumes normality, and sequencing depth is not distributed normally.38 Because of this modification, we cannot use previous methods to determine statistical significance. Instead, for each amplicon, we manually assessed the males with the 64 highest U-statistic values. (Each copy of an amplicon has its own change point; we defined the U-statistic value of an amplicon as the lowest of its copies’ values.) A partial CNV was called if the following three criteria were met. (1) The change points in each amplicon copy were close to each other. (2) The direction of the shift in mean across the change point was the same in each amplicon copy. (3) In each amplicon copy, the predicted copy number based on depth was different on each side of the change point. Several exceptions were made to criterion 1 in cases where an entire duplication or deletion was contained in a single amplicon. In such cases, two change points were present in the single amplicon, and the maximally significant change point differed in different amplicon copies. When such cases were obvious upon visual inspection, we called them as partial CNVs. We also removed four males from the dataset that had extremely noisy depth and an abnormally high number of change points.
It should be noted that this method is less powerful than the more recent circular binary segmentation algorithms that are commonly used to detect genomic CNVs.39 Our choice of a less powerful algorithm was intentional, as we wish to detect only the largest and most obvious partial amplicon CNVs. Detecting small CNVs is its own technical challenge that is beyond the scope of this study. Further, because most small CNVs are unlikely to affect functional regions of the amplicons, they are probably under little selective pressure as a group; adding small CNVs to our analyses could drown out the observed signatures of selection on large CNVs.
Male Specificity of Ampliconic Sequence
We confirmed that no reads from elsewhere in the genome aligned to the amplicons or control regions. We measured amplicon depth as described above from whole-genome sequencing data of 15 females and 5 males from the 1000 Genomes Project, using a 5-Mb region of chromosome 2 (chr2: 80,000,000–85,000,000) as the normalization region. Single-copy sequence on the Y is unfit for this purpose, since its expected depth in females is 0.
Filtering of Copy Number Calls
We performed two filtering steps. First, for each amplicon, we calculated the median depth of the 100-bp windows used in partial amplicon CNV detection. In 40 males, the predicted copy number (using thresholds as described above) was different using this median value and the mean depth. In 38 of these males, mean and median copy number calls differed in a single amplicon, and the copy number state using the mean values was either the reference state or a common and predicted CNV state. Two males that did not fit these criteria were removed from the dataset. Second, we calculated the mean and standard deviation of the depth of each control region, excluding four males with large short-arm deletions that remove control regions 2 and 3. There were 28 males with two or more control regions more than two standard deviations away from their means. 27 of these males had either the reference copy number or a common and predicted CNV state. The one male that had neither was removed from the dataset. Table S4 contains filtered copy number calls of each male.
We estimated call accuracy rate from males with two libraries as follows. Assume a library will contain erroneous CNV calls with a fixed probability x. The probability of both libraries having correct CNV calls is (1 − x)2. We found that 89/92 (96.6%) of males with two libraries had concordant amplicon calls. Further assuming that the chance of both libraries having erroneous calls that are concordant with each other is negligible, we can solve for x = 1.6%. We removed two of the males with discordant copy number calls from the dataset. The third male had concordant CNV calls of other amplicons that matched a common predicted CNV, and we adjusted the discordant call to conform to that state.
See Figure S1 for a schematic of the complete CNV-calling pipeline.
Multi-color FISH
Cell lines of 12 males from the 1000 Genomes Project (HG00142, HG00271, HG01187, HG01890, HG02394, HG02982, HG03445, NA12812, NA18504, NA18960, NA18983, and NA20520) were obtained from the NHGRI Sample Repository for Human Genetic Research at the Coriell Institute for Medical Research. Two-color interphase FISH was performed as previously described.40 We scored at least 200 cells for each set of probes in each cell line. Images were recolored to match the color of amplicon names.
Amplicon Architecture Prediction
We simulated all AZFc architectures formed by one, two, or three NAHR events between amplicon copies, as previously described (Figure S3).20 Males with amplicon CNVs were matched to architectures with the same copy number of each amplicon. When multiple architectures matched, we chose the architecture(s) formed by the fewest NAHR events.
Haplogroups
The haplogroups of 1,210 males in the 1000 Genomes Project are already annotated. For the remaining 15, we determined haplogroups using Ytree. The phylogenetic tree in Figure 5B was built using estimates of divergence time from Poznik et al.31
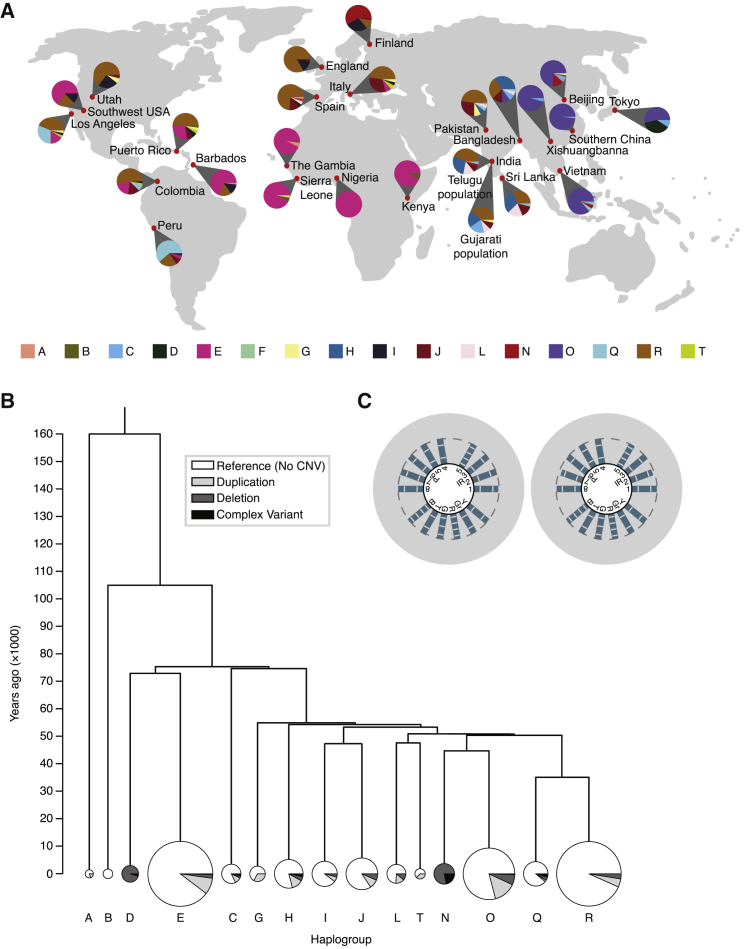
Figure 5.
Amplicon CNVs Are Distributed throughout the Y Chromosome Phylogenetic Tree
(A) Distribution of Y chromosome haplogroups in 1000 Genomes Project populations. Sri Lankan and Indian Telugu samples were collected from a population living in the United Kingdom; Gujarati Indian samples were collected from a population living in Houston, Texas.
(B) Phylogenetic tree of major Y chromosome haplogroups represented in our dataset. Pie charts: proportions of males with different CNV classes in each haplogroup. Pie chart area is proportional to the number of males from that haplogroup in our dataset.
(C) Copy number calls of two males from haplogroup A00.
Modification of Detailed Phylogenetic Tree
In many of our subsequent analyses, we used the detailed phylogenetic tree of the 1000 Genomes Project Y chromosomes (Figure S7, Data S1) built by Poznik et al.31 We modified the tree from that study in two ways. First, we manually identified instances where the tree architecture was inconclusive because no SNPs differed between three or more branches, but two or more of those branches contained the same CNV. We corrected the tree in such instances where its original, arbitrarily determined architecture contradicted the architecture implied by the CNVs.
Second, due to low sequencing coverage in individual males, SNPs may be missing from branches near the tips of the tree, leading to those branches being depicted as shorter than they actually are. This would, in turn, cause CNVs to appear to cluster in the more recent past, even if they were in fact distributed evenly over time. Because such clustering is a key result of this study, it is essential to correct for this effect. First, any branch with length 0 was changed to have length 0.5. Then, we adjusted each branch as follows. As described in Poznik et al., assuming that (1) a SNP is detected if two reads covering the site of the SNP are observed, and (2) the number of reads at a given site can be described with a Poisson distribution with mean equal to the overall sequencing coverage of the Y chromosome, we expect the length of a branch to be reduced by x(p0 + p1), where x = the observed branch length as measured in SNPs and pi = the probability of observing exactly i sequencing reads at a given site.31 Therefore, we divide each branch length by 1 − (p0 + p1) to correct for this reduction, using the combined sequencing depth of each individual descended from that branch to calculate p0 and p1. This method is imperfect: as discussed in Poznik et al., it is intractable to completely model and correct for the effect of missing SNPs. However, our method of correction extends the lengths of the terminal branches of the tree so that each is at least as long as its expected true length. Therefore, our correction is, at worst, incorrectly extending the terminal branches of the tree at the expense of the more ancient branches, so we can be confident that the clustering of CNVs in the more recent branches of the tree is not an artifact caused by this effect.
Calculation of Amplicon Mutation Rate
To calculate a lower bound of the amplicon CNV mutation rate, we divided the number of mutation events in the detailed phylogenetic tree by the total evolutionary time traversed by the tree. The number of mutation events, as determined below by Fitch’s algorithm, was 139.41 The total branch length of the tree after correction for missing SNPs as described above was 69,029 SNPs. We converted SNPs to generations as described below to obtain a total branch length of 363,369 generations. These values yielded a rate of 3.83 × 10−4 mutations per father-to-son Y transmission. We expect that the true mutation rate is higher, as selection is depressing the number of mutations observed by removing Y chromosomes with mutations from the population.
Calculating CNV Distribution over the Phylogenetic Tree
We defined branch age as the mean distance from the child node of the branch to the leaves that descend from that node, plus half the length of the branch (Figure S9). See Supplemental Material and Methods for further discussion of this test.
Estimation of Selection Coefficient
We calculated an overall selection coefficient for amplicon CNVs using the canonical equation for mutation-selection balance in a haploid context s = μ/q, where s = the selection coefficient of CNVs, μ = the CNV mutation rate, and q = the frequency of CNVs in the population.42 With an estimated mutation rate of 3.83 × 10−4 mutations per father-to-son Y transmission and a CNV frequency of 16.9% (206/1,216), s = 0.0023.
Simulation of Neutral Evolution
For simulation of amplicon CNV evolution, we used a simple model based on mutation occurring in the detailed phylogenetic tree. We chose to use this model for several reasons. First, a simple model is easier to interpret and draw qualitative conclusions from, which was the goal of our simulations. Second, a simple model has fewer components that can cause artifactual outcomes. Third, the model is computationally tractable, allowing us to perform millions of simulations and sample a broad range of parameter space. While our simulations do, for example, fail to accurately model cases of amplicon rescue, such events appear to play only a minor role in maintaining amplicon copy number. Therefore, while our simulations do not exactly replicate the process of amplicon evolution, they provide insight into what neutral evolution of amplicon copy number would look like on the empirical phylogenetic tree of Y chromosomes.
Our simulation model works as follows. Nodes of the tree can be in one of two states: reference or mutated. Each simulation began at the root of the tree and traveled along each branch to the leaves. Every generation, there was a fixed probability of mutation from the reference state to a mutated state. We also used a model in which, every generation, there was a fixed probability of reversion from the mutated state to the reference state. Generations are measured by converting branch lengths of the tree, measured in number of SNPs, to years, as described in Poznik et al.31 Briefly, the Y chromosome mutation rate is estimated as 0.76 × 10−9 SNP mutations per bp per year as calculated by Fu et al.43 The total number of bases analyzed to build the tree is approximately 10,000,000. Therefore, (0.76 × 10−9 SNP mutations per bp per year × 107 bp)−1 = 131.6 years per SNP mutation. We then converted years to generations, assuming a generation time of 25 years. Because each SNP corresponds to 5.26 generations, we simulated fractional generations at the end of each branch.
For our simulations of mutation, we used 24 mutation rates ranging from 5 × 10−1 to 1 × 10−6 mutations per generation and performed 10,000 simulations with each mutation rate. For our simulations of mutation with reversion, we used 24 mutation rates and 24 reversion rates, each ranging from 5 × 10−1 to 1 × 10−6 mutations per generation, equaling 576 combinations of mutation and reversion rate, and performed 10,000 simulations with each combination of rates. We implemented Fitch’s algorithm to count the number of mutation events that occurred.41 When calculating mutation events in the real tree, we did not distinguish between different types of CNV events, as our model has only two states: reference and mutated. Therefore, the number of real mutation events counted here is lower than the number used above when calculating the amplicon mutation rate. Simulations were run using custom python scripts and the ete3 python module.
Haplogroup A00 Males
We analyzed two haplogroup A00 males who were sequenced by Karmin et al.44 We downloaded Y chromosome BAM files of these males from the Estonian Biocenter data repository and converted the BAM files to FASTQ files using bedtools.45 We then processed these files using the same pipeline as the 1000 Genomes Project samples. The exception is that we did not perform GC bias correction in these samples. Autosomal data are necessary for GC correction, and only Y chromosome data were available for these males.
We simulated A00 evolution using a model of haploid genetic drift. Males can be in one of two states, reference or mutated. We began with N males, all in the reference state. Each generation, we drew a number x from a binomial distribution B(N, p + m), where p = the fraction of males in the previous generation with the mutated state and m = the mutation rate per generation per individual. We then set the number of males with the mutated state in the next generation to x. We simulated 10,000 generations, corresponding to 250,000 years of history given a generation time of 25 years. Simulations over 8,000 generations yielded similar results (data not shown).
Figure Generation
Plots were generated using Adobe Illustrator and custom python scripts with python modules matplotlib, seaborn, and ete3.46, 47
Results
Sequencing Depth Corresponds to Amplicon Copy Number
To detect amplicon CNVs, we analyzed whole-genome sequencing data from males from the 1000 Genomes Project.29 Detecting copy number variation from sequencing data is a well-studied problem, but widely used tools struggle to accurately call CNVs in the complex ampliconic regions. Therefore, we developed a pipeline to search for amplicon copy number changes (Figure S1). We aligned the data to the entire reference genome, masked genome typical interspersed repeats on the Y chromosome, and computed depth of 15 amplicons and 4 single-copy control regions on the Y chromosome (Table S1). We then adjusted depth to correct for GC content bias and normalize for coverage of the Y chromosome so that, in the absence of a CNV, the expected depth of each control region and amplicon is 1 (Table S3).
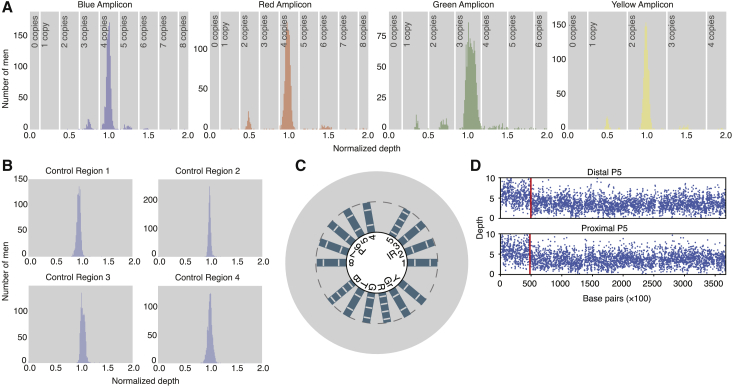
After these steps, the depth of each amplicon provided an estimate of its copy number. When the depth of an amplicon for each individual was plotted as a histogram, we observed extraordinarily clear peaks corresponding to whole-amplicon deletions and duplications (Figure 2A). (The exception is the P7 amplicon, which did not give consistent results because of its small size and was excluded from subsequent analyses.) In contrast, the control regions showed only single peaks centered around a normalized depth of 1 (Figure 2B). The sharp ampliconic peaks imply that most amplicon copy number variation affects whole amplicons at a time, consistent with the idea that NAHR is the dominant mechanism by which Y chromosome amplicon CNVs arise. Because of these peaks, we called amplicon copy number from depth alone and determined the complete amplicon copy number state of each individual (Figures 2C and S2, Table S4).
Figure 2.
Amplicon CNVs Can Be Detected from Sequencing Data
(A) Normalized depth of blue, green, red, and yellow amplicons in 1,216 males. Vertical lines: cutoff at which copy number call changes.
(B) Normalized depth of single-copy control regions in 1,216 males.
(C) Circular plot representing the normalized depth and copy number of each amplicon in a single male. Each bar represents the normalized depth of 1 of the 15 amplicons analyzed (B, blue; T, teal; G, green; R, red; Gr, gray; Y, yellow). Hash marks within each bar represent the cutoffs at which copy number call changes, as shown in (A). Therefore, the number of hashes within a bar equals the copy number of that amplicon. The dotted gray line is drawn at a normalized depth of 1. The background gray circle is drawn at a normalized depth of 2. The male shown in this figure has the reference copy number call for each amplicon; in plots of males with a deletion or duplication, the bar of the affected amplicon is shown in red or green, respectively.
(D) Example of a partial amplicon CNV. Depth in each copy of the amplicon is shown. Blue dots: depth of 100-bp windows. Red lines: predicted change points.
The AZFc region is particularly susceptible to CNVs due to NAHR because, as many of its amplicon copies are in the same orientation (Figure 1B), crossing-over between those copies results in deletion or duplication.6, 20 (In contrast, most other amplicons on the human Y chromosome are in opposite orientations and are susceptible only to inversion through this mechanism.) The AZFc architectures caused by NAHR events can be predicted by enumerating all possible series of one, two, and three NAHR events, and we matched observed copy number states to these predicted architectures (Figure S3). In this way, we used our copy number data to draw conclusions about the arrangement of CNVs and the mechanism by which they arose.
CNVs that cause the deletion or duplication of only part of an amplicon copy can also occur. To detect these, we used a modified version of the binary segmentation algorithm to detect abrupt changes in depth in the middle of amplicons (Figure 2D).37 These change points represent the breakpoints of partial (rather than whole amplicon) CNV events, which are not caused by NAHR between whole amplicon copies. We detected 52 males with partial CNVs, 6 of which had previously been called by our whole amplicon analysis as having the reference amplicon state.
Filtering and Confirmation of Copy Number Calls
To confirm that no reads from elsewhere in the genome align to the amplicons or control regions, we measured amplicon and control region depth of 15 females and 5 males from the 1000 Genomes Project. Normalized ampliconic and control region depth in all females was near 0 (Figure 3A).
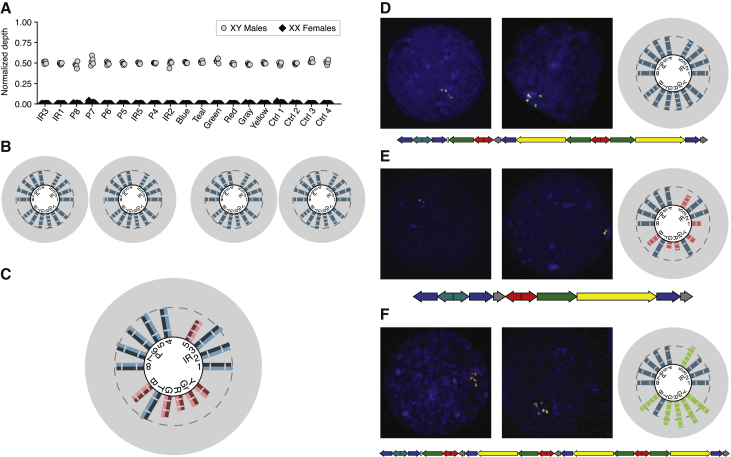
Figure 3.
Validation of Amplicon CNV Calls
(A) Normalized amplicon and control region depth in 15 females and 5 males. Depth in males is ∼0.5 because depth was normalized with an autosomal region.
(B) Amplicon copy number calls in two father-son pairs (left, HG03713 and HG03715; right, HG02371 and HG02372).
(C) Example of a male with two sequencing libraries with concordant amplicon copy number calls. Each half of a bar represents one library.
(D–F) Two-color interphase FISH using probes that hybridize to the green and red amplicons (left) and green and yellow amplicons (middle) in males with (D) the reference AZFc copy number, (E) a b2/b3 deletion, and (F) a gr/gr duplication. Right: copy number calls. Bottom: model of predicted AZFc architecture (these and further AZFc architectures not drawn to exact scale).
To ensure the accuracy of copy number calls, we removed males with either abnormal depth in their control regions or discordance between the mean and median depth of their amplicons. To ensure that noise due to low depth was not introducing artifactual calls to our dataset, we determined that the rate of CNVs did not significantly differ between males with lower depth and males with higher depth (Figure S4). We also compared amplicon copy number in two father-son pairs found in the 1000 Genomes Project. In each pair, the same copy number of each amplicon was present in father and son (Figure 3B). Additionally, 92 males have sequencing data from two independent sequencing libraries that pass the above filtering steps. When amplicon copy number calls were generated independently from both libraries for each male, 89 of the 92 males (96.7%) had concordant copy number calls between libraries, and the three males with discordant calls each differed in only a single amplicon (Figure 3C). From these results, we expect that 98.4% of the males in our dataset have accurate copy number calls for every amplicon.
To validate the copy number calls using an orthogonal and non-computational method, we performed multi-color interphase FISH on lymphoblastoid cell lines of 12 males sequenced in the 1000 Genomes Project. These samples were chosen to represent a range of amplicon structures. We counted the copy number of green, red, and yellow amplicons using probes that hybridize to those amplicons. The FISH analysis confirmed our computational CNV calls (Figures 3D–3F and S5).
Classification and Phenotypic Impact of Observed CNVs
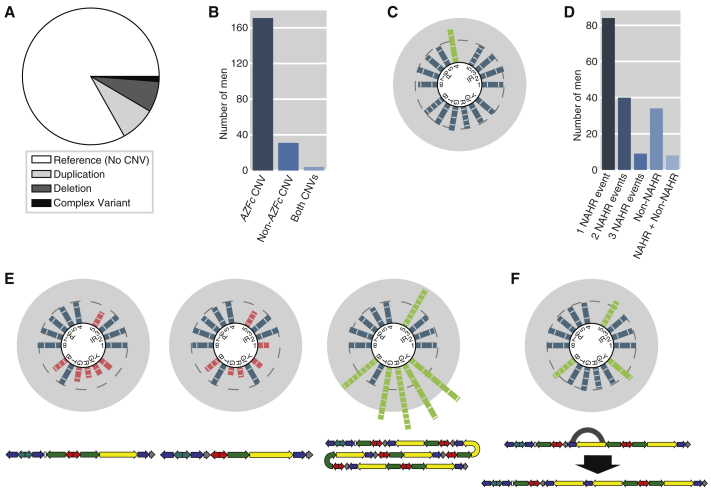
Of the 1,216 males analyzed, 206 (16.9%) had one of 56 distinct CNVs affecting at least one amplicon (Tables S5 and S6). Of these males, 88 (43%) had deletions, 103 (50%) had duplications, and 15 (7%) had complex CNVs—deletions of some amplicons and duplications of others (Figure 4A). These results are in rough concordance with a previous report that found CNVs in 14.7% of Y chromosomes.21 As expected because of the AZFc region’s particular susceptibility to NAHR-mediated CNVs, the majority (171/206) of these males have amplicon CNVs found solely within the AZFc region (Figure 4B). However, we also detected 31 males with non-AZFc CNVs (Figure 4C), as well as 4 males with CNVs both within and outside of the AZFc region.
Figure 4.
Amplicon CNVs
(A) Proportion of males with the reference amplicon state (no CNVs), duplications, deletions, and complex CNVs.
(B) Locations of observed CNVs.
(C) Copy number calls in a male with a non-AZFc CNV.
(D) Predicted mechanism of AZFc CNV formation.
(E) Examples of males with AZFc CNVs predicted to be caused by NAHR. Predicted architectures are shown below. Left: a male with the previously described gr/gr deletion, found in 49 males in our dataset. Middle: a male with the previously described b2/b3 deletion, found in 26 males in our dataset. Right: a male with a duplication found in one male in our dataset.
(F) Example of a male with an AZFc CNV not caused by NAHR between amplicon copies. Bottom: reference AZFc architecture with gray arc showing the predicted breakpoints of the non-NAHR event and predicted CNV architecture.
The majority (133/175, 76%) of males with AZFc CNVs have CNVs that corresponded to the predicted one-, two-, or three-step NAHR events (Figures 4D and S3), supporting our observation that most CNVs affect whole amplicons (Figure 2A). Several of the observed CNVs are well-studied and recurrent, such as gr/gr deletions and b2/b3 deletions, while others are rare (Figure 4E). One-step events (n = 84) were more common than two-step events (n = 40), which were in turn more common than three-step events (n = 9), consistent with our hypothesis that such CNVs are caused by independent and successive NAHR events. However, 34 (19%) of the AZFc CNVs did not correspond to predicted NAHR events, and instead corresponded to the deletion or duplication of a single block of sequence within the AZFc region (Figure 4F). Such CNVs could be caused by non-NAHR mechanisms such as non-homologous end joining, or by NAHR between small targets within amplicon copies, such as genome typical interspersed repeats. An additional eight males had AZFc CNVs that could not be explained by either of the above two mechanisms. Such males likely represent a combination of NAHR and non-NAHR events. Several of these males had evidence of partial amplicon CNVs that, combined with the predicted AZFc architectures, suggested plausible mechanisms for the formation of such CNVs (Figure S6).
The only CNVs in our dataset with demonstrated phenotypic effects are the gr/gr and b1/b3 deletions, which we found in 49 males and 1 male, respectively, and are known risk factors for spermatogenic failure and/or testis cancer.10, 11, 12 We also observed 26 b2/b3 deletions, whose phenotype is less clear, with conflicting reports about whether or not it contributes to spermatogenic failure.11, 48 We observed no males with either of the canonical complete AZFb or AZFc deletions, which both cause spermatogenic failure with high penetrance.6, 18 (The prevalence of these complete AZFb and AZFc deletions in the general population are approximately 1/8,188 and 1/2,320, respectively, so it is unsurprising that we see no such variants in a study of our size.11, 18) The remaining 53 distinct CNVs, which are present in the majority of males with CNVs in our dataset (130/206, 63.1%), have no known strong associations with a phenotype. This reinforces the fact that ampliconic copy number variation is broader than the few well-studied variants that have been brought to the forefront by studies in azoospermic men.
The Reference Amplicon Copy Number State Pervades the Y Chromosome Phylogenetic Tree
We next asked what this variation tells us about the evolution of the amplicons. Because the Y chromosome is inherited from father to son as a single haplotype, an accurate phylogenetic tree of all Y chromosomes can be constructed; major branches of this phylogeny are called haplogroups.30 The 1000 Genomes Project contains samples collected from populations around the globe, so most major haplogroups are represented (Figure 5A). We calculated the proportion of Y chromosomes of each haplogroup that have amplicon CNVs (Figure 5B). Two haplogroups, D and N, have no individuals with the reference copy number, the result of ancestral deletions that are fixed within those haplogroups.19, 49, 50 All other haplogroups (with the exception of B, which has only seven males in our data) contain males with CNVs, but the reference state is present in the majority (62%–93%) of males in each haplogroup. We then mapped the detected CNVs onto a modified version of the detailed phylogenetic tree of the 1000 Genomes Project Y chromosomes built by Poznik et al. (Figure S7, Data S1).31 With this detailed tree, we calculated a lower bound for the amplicon CNV mutation rate of 3.83 × 10−4 mutations per father-to-son Y transmission. Given this high mutation rate, it is surprising that the reference amplicon state is so pervasive. If amplicon CNVs were selectively neutral, we would expect to see a larger number of ancient mutations, which would cause most or all of a haplogroup to have a CNV. This led us to hypothesize that selection was acting to remove amplicon CNVs from the population.
Haplogroup A00 Has Maintained the Reference Copy Number State
The most ancient haplogroup known is A00, which diverged from all other Y chromosomes between 200 and 300 kya.32 We determined amplicon copy number of two A00 males who were sequenced as part of a different study.44 These two males represent an independent experiment of evolution over almost twice as much time as the other males we analyzed. We found that both A00 Y chromosomes have the reference copy number of each amplicon (Figure 5C), implying that the reference amplicon state is the ancestral state. Further, given the amplicon CNV mutation rate calculated above and the time since the divergence of A00 from the reference, it is extremely unlikely that the reference copy number would be maintained in the absence of selection (Figure S8).
Mutations Times Are Skewed toward the Recent Past
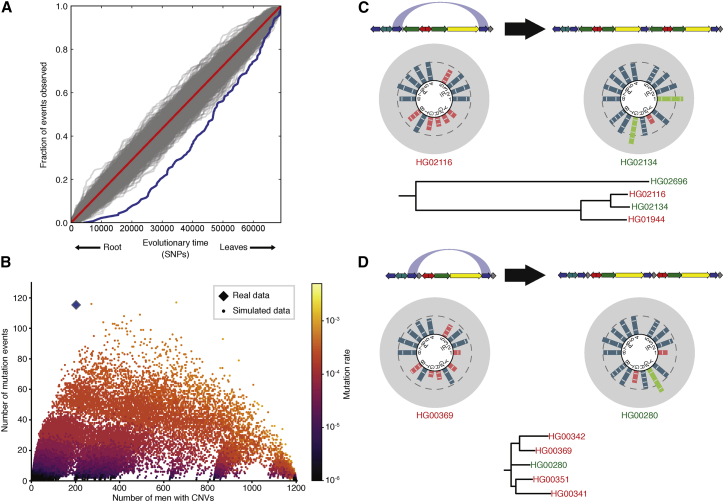
We tested the distribution of CNV mutation events within the detailed phylogenetic tree to determine whether that distribution is compatible with neutral evolution. If amplicon variation were neutral, we would expect mutation events to be distributed evenly between ancient and recent branches of the tree. Instead, we found that mutation events are significantly skewed toward the recent branches of the tree (p = 1.01 × 10−7, KS test, Figures 6A and S9)—71% (99/139) of mutation events took place in the latter 50% of the tree. In contrast, shuffling the tree so that the branches containing mutation events were placed randomly with respect to time resulted in distributions of mutation events that were closer to the uniform distribution that is expected under neutral evolution (Figure 6A). The skew in the real data fits with a history of mutation-selection balance, in which mutations occur at a high rate but are constantly removed from the population due to selection. Recent mutations, which have not yet had enough time to be removed from the population, are therefore more common than ancient mutations.
Figure 6.
The Reference Amplicon State Has Been Selected For throughout Human Evolution
(A) Distribution of mutation events over the phylogenetic tree. Blue curve: branches of the phylogenetic tree of males in our dataset, sorted by branch age. Red diagonal line: expected distribution if CNVs were selectively neutral. Gray lines: branches of the phylogenetic tree shuffled at random. 1,000 shuffles were performed. See Figure S9 for an in-depth description of this method.
(B) Mutation events versus number of males with CNVs. Each point represents one simulation over the phylogenetic tree of males in our dataset.
(C and D) Amplicon rescue in gr/gr (C) and b2/b3 (D) deletions. Top: a chromosome with the deletion undergoes a blue-to-blue duplication that restores most of the amplicons to the reference copy number. The blue arc shows the targets of NAHR on a single copy of the AZFc region. Middle: males with the pre-rescue and post-rescue AZFc structures. Bottom: phylogenetic trees containing the rescued males. Males in red have the gr/gr (C) or b2/b3 (D) deletion; males in green have the respective rescue amplicon copy number states.
Under a model of mutation-selection balance, we can estimate an overall selection coefficient for amplicon CNVs of s = 0.0023. This value indicates that amplicon CNVs as a whole are weakly deleterious. However, our calculation of the mutation rate assumes that selection is absent; selection removes ancient variants from the tree and therefore causes us to underestimate the mutation rate. A higher mutation rate would, in turn, denote a higher selection coefficient. In support of the assertion that our calculated mutation rate of 3.83 × 10−4 mutations/generation is a considerable underestimate of the true mutation rate, the combined prevalence of complete AZFb and AZFc deletions is 5.53 × 10−4.11, 18 Because these deletions cause spermatogenic failure with almost complete penetrance, their prevalence should equal their mutation rate.11 It is implausible that the mutation rate of just the complete AZFb and AZFc deletions is higher than the combined mutation rate of all amplicon CNVs (which includes the AZFb and AZFc deletions), indicating that the true amplicon CNV mutation rate is much higher than our calculated rate of 3.83 × 10−4 mutations/generation.
The Distribution of CNVs Is Incompatible with a Model of Neutral Evolution
We then simulated the formation of amplicon CNVs using a range of amplicon mutation rates to gain a qualitative understanding of how neutral evolution would behave over the detailed phylogenetic tree and how the real data differs from this neutral behavior. For each simulation, we counted the number of mutation events that occurred during simulation and the total number of males with an amplicon CNV after the simulation was complete (Figure 6B). At high mutation rates, there were many males with CNVs but few observed mutation events, as most mutation events occurred near the root of the tree, and mutated branches could not re-mutate in our model. At intermediate mutation rates, we observed more mutation events but fewer males with CNVs. Finally, at low mutation rates, there were few mutation events and few males with CNVs. However, in our real data, we observed many mutation events and a middling number of males with CNVs, a combination that is incompatible with the neutral model (Figure 6B). A more complex model that allowed for reversion to the ancestral state and subsequent re-mutation matched our real data only when the reversion rate was five to ten times higher than the mutation rate (Figure S10). Such extreme discrepancy between mutation and reversion likely represents selection acting to remove CNVs from the population, rather than true chromosomal reversion.
An alternative interpretation of the pattern of amplicon variation we observe in our real data is a recent change in the amplicon mutation rate, possibly due to environmental factors. However, such a change would have had to occur independently in each haplogroup, many of which are geographically isolated. Our observations are also not explained by bursts of Y chromosome population expansion described in Poznik et al.31 The detailed phylogenetic tree of Y chromosomes used in our analyses is built from SNPs, and its branch lengths are dependent solely on SNPs; as a result, any historical dynamics that affect the tree’s structure and branch length are accounted for when we compare the amplicon CNVs to the tree. Therefore, selection against amplicon CNVs is the most plausible interpretation of our findings.
The above analyses are particularly sensitive to false positive CNV calls, as false calls would appear as recent, isolated mutations. Males that differ from the reference state by a single copy of a single amplicon are the most likely to be false positives. Therefore, we generated a high-confidence CNV callset by excluding 22 such males that we could not validate with partial-amplicon CNV detection or FISH. We then performed the above two analyses using this high-confidence callset; in both cases, the results changed only minimally (Figure S11).
Amplicon Deletions Are Rescued by Subsequent Duplication
In addition to selection acting to eliminate CNVs, variant Y chromosomes can undergo subsequent mutations that restore most or all amplicons to the reference copy number. We used our phylogenetic tree to directly observe this process, which we call amplicon rescue. While we observed no cases of rescued duplications, which—in the absence of phylogenetic evidence—would be indistinguishable from other males with the reference copy number, we did observe several cases of rescued deletions (Figures 6C, 6D, and S12). There may be more instances of rescued deletions or duplications in our dataset that would be revealed with a more densely populated phylogenetic tree but that appear as singleton complex mutations or even the reference copy number state in our tree. Even so, amplicon rescue occurred at a significantly higher rate than analogous duplications on the reference background: we observed nine blue-to-blue duplications on a deletion background (75 males with gr/gr or b2/b3 deletions) versus six blue-to-blue duplications on a reference background (1,010 reference state males; p = 2.00 × 10−7, Fisher exact test). Since both events are mechanistically analogous, we suggest that this difference represents selection favoring rescued Y chromosomes over chromosomes with deletions, rather than a difference in the rate of incidence of such mutation events. Still, the total number of rescued Y chromosomes is low, indicating that the primary effect that maintains amplicon copy number is selection acting to remove any CNVs from the population, not amplicon rescue.
Discussion
Natural selection has played a foundational role in shaping autosomal and X-linked copy number variation.51 However, it has been stated that selection is ineffective on the mammalian Y chromosome, because the Y chromosome does not undergo recombination with a homologous chromosome.52 (This lack of recombination actually makes our study possible, enabling a deep reconstruction of the Y chromosome’s history that is impossible for any autosome or X chromosome.) Demographic studies emphasize the role of neutral processes rather than selection to explain the history of the Y chromosome.31 Some have even speculated that, in the absence of effective selection, the mammalian Y chromosome will eventually decay and be lost.53 On the contrary, we have shown that Y chromosome amplicons have been subject to purifying selection to maintain the ancestral copy number for more than 200,000 years of human history. Previous studies had hypothesized about the effects of selection on amplicons, without providing direct evidence of ampliconic selection.20, 54, 55 Selection also acts to maintain the non-ampliconic regions of the Y chromosome.54, 56, 57 In conjunction, these results demonstrate that the human Y chromosome’s current form is the result of selective forces acting on both its single-copy and multi-copy sequence classes.
Most of the early research into Y chromosome structural variation focused on azoospermic men, leading to the discovery of ampliconic deletions that cause spermatogenic failure.6, 10, 11, 12, 18 This initial focus on variants that affect spermatogenesis was compounded by the fact that infertility clinics were a major resource for such research, resulting in significant ascertainment bias in the set of variants that became well studied and well known. Additionally, almost all of these well-known amplicon CNVs are deletions, the legacy of the early years of Y chromosome research in which the primary method of detecting CNVs was the use of sequence-tagged sites, which can detect deletions but not duplications or inversions.58
The true breadth of amplicon copy number variation has been revealed by recent surveys.20, 21, 22, 23 In accordance with these studies, we found that most amplicon CNVs in the general population do not fall within the small set of CNVs with confirmed phenotypes, and that duplications are more common than deletions. Our results suggest that most or all amplicon CNVs have phenotypic effects that cause selection to remove them from the population. The obvious candidate for this phenotype is a negative effect on spermatogenesis, as ampliconic genes are expressed exclusively in the testis,4 and recent evidence suggests that both deletions and duplications in the AZFc region can increase the risk of spermatogenic failure in certain populations.59 However, the mechanism through which any of these mutations affect spermatogenesis is still a mystery. Because the large CNVs within the AZFc region that cause spermatogenic failure delete multiple genes at once, the gene or genes responsible for the resulting phenotype cannot be determined. Further, studies in model systems are thwarted because most human ampliconic genes are not present on, for example, the mouse Y chromosome.14 For these reasons, the functions of individual human ampliconic genes are unknown, with the only information being that they are expressed in the testis and at least one is crucial for spermatogenesis. It is even possible that noncoding elements within the amplicons play a major phenotypic role. Further complicating this question is the fact that different CNVs change the copy number of different genes, which may cause different molecular phenotypes altogether.
Due to the tremendous differences in amplicon structure and content between species, we might expect amplicon structure within species to also be highly variable. Such diversity would not be entirely unexpected—male reproductive genes evolve rapidly.60 Instead, although the amplicons are quite mutable, we found that the ancestral amplicon copy number state has been maintained for 200,000 years of human history. Reconciling the differences in amplicon structure between species with the maintenance of amplicon structure in humans represents the next major challenge in the study of amplicon evolution. We propose two models that can explain this apparent contradiction: amplicon structure can either evolve through a steady progression of intermediate states or undergo times of rapid turnover separated by long periods of stability. The first model could be caused by rare amplicon CNVs with large positive effects, in contrast to the common deleterious CNVs that are subject to the mutation-selection balance that we have observed. The second model could result from external factors changing the optimal amplicon structure; because the amplicons are so mutable, the race to a new optimum would shuffle the amplicon architecture, leaving the new structure unrecognizable compared to the previous one. Two possible historical drivers of such change are the different levels of sperm competition present in primate mating systems and the Y chromosome’s acquisition of the gene DAZ in primates.61, 62, 63 While our present results cannot distinguish between these two models, future studies that observe amplicon evolution over a timescale between the 200,000 years of human history studied here and the 7 million years since the human-chimpanzee split can test them.
Our results also provide insight into the biological role of ampliconic sequence. The ubiquity of ampliconic sequence on mammalian Y chromosomes suggests that amplification itself confers a functional benefit. Theories about this benefit include (1) gene conversion between amplicon copies allows for the rescue of deleterious mutations, (2) multiple copies provide the proper dosage of ampliconic genes, and (3) palindromes allow ampliconic genes to escape meiotic sex chromosome inactivation by pairing with themselves.3, 14, 64, 65, 66 Our finding that fitness is negatively affected by both duplications and deletions of amplicons supports the gene dosage theory, as extra amplicon copies should have no deleterious effect on either gene conversion or escape from inactivation. However, these theories are not mutually exclusive; it is even possible that the initial driver of amplicon formation was gene conversion or escape from inactivation, and only afterward was gene expression tuned to the number of amplicon copies.
This study provides a foundation for deeper investigation of the evolutionary questions presented here, as sequencing technologies grow increasingly powerful and large datasets—some orders of magnitude larger than the one we analyzed—become available.67, 68, 69 For example, these datasets might contain the rare beneficial amplicon CNVs predicted by our first model, which, in a study of our size, would either be absent or occur at such low frequency as to be indistinguishable from the other, deleterious, CNVs. These datasets can also be used to tease out the magnitude of each amplicon’s contribution to reproductive fitness.
Additionally, data from other species can determine whether the maintenance of an ancestral amplicon copy number state is common or restricted to humans. Of course, amplicons are subject to a variety of evolutionary forces that differ between species. For example, the mouse Y chromosome underwent runaway expansion as part of a genetic arms race with the X chromosome.14 Even if some selective pressures favored the ancestral amplicon state in mouse, the opposing pressure to amplify could have overridden them. Studying other species, particularly those with population divergence times greater than the 200,000 years of humans, can also help choose between our two proposed models of amplicon evolution. A promising possibility is the chimpanzee: its Y chromosome is highly ampliconic and has high-quality reference sequence, the most recent common ancestor of the chimpanzee Y chromosomes is more than one million years old, and chimpanzee genomes are beginning to be sequenced in high numbers.70, 71 Future studies, using data from both human and non-human species, will continue to shed light on the evolutionary history of the Y chromosome amplicons and their roles in fitness and reproduction.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank D. Bellott and A. Godfrey for advice on analyses and figures, R. George for assistance with Ytree, D. Poznik for providing a formatted version of the Y chromosome phylogenetic tree, and D. Bellott, J. Hughes, A. Godfrey, and E. Jackson for critical reading of the manuscript. This work was supported by the National Institutes of Health (R01-HG007852) and the Howard Hughes Medical Institute.
Published: August 2, 2018
Footnotes
Supplemental Data include 12 figures, 6 tables, and Supplemental Material and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.07.007.
Web Resources
1000 Genomes Project, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/
Custom code, https://github.com/lsteitz/y-amplicon-evolution
Estonian Biocenter Data Repository, http://www.ebc.ee/free_data/chrY
NHGRI Sample Repository for Human Genetic Research at the Coriell Institute for Medical Research, https://www.coriell.org/1/NHGRI
RepeatMasker, http://www.repeatmasker.org
seaborn, https://doi.org/10.5281/zenodo.54844
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Burgoyne P.S. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- 2.Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 3.Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 4.Lahn B.T., Page D.C. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 5.Vogt P.H., Edelmann A., Kirsch S., Henegariu O., Hirschmann P., Kiesewetter F., Köhn F.M., Schill W.B., Farah S., Ramos C. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum. Mol. Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 6.Kuroda-Kawaguchi T., Skaletsky H., Brown L.G., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Page D.C. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 7.Giachini C., Nuti F., Turner D.J., Laface I., Xue Y., Daguin F., Forti G., Tyler-Smith C., Krausz C. TSPY1 copy number variation influences spermatogenesis and shows differences among Y lineages. J. Clin. Endocrinol. Metab. 2009;94:4016–4022. doi: 10.1210/jc.2009-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson M., Page D.C., de la Chapelle A. Chromosome Y-specific DNA is transferred to the short arm of X chromosome in human XX males. Science. 1986;233:786–788. doi: 10.1126/science.3738510. [DOI] [PubMed] [Google Scholar]
- 9.Lange J., Skaletsky H., van Daalen S.K., Embry S.L., Korver C.M., Brown L.G., Oates R.D., Silber S., Repping S., Page D.C. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathanson K.L., Kanetsky P.A., Hawes R., Vaughn D.J., Letrero R., Tucker K., Friedlander M., Phillips K.A., Hogg D., Jewett M.A. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am. J. Hum. Genet. 2005;77:1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozen S.G., Marszalek J.D., Irenze K., Skaletsky H., Brown L.G., Oates R.D., Silber S.J., Ardlie K., Page D.C. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am. J. Hum. Genet. 2012;91:890–896. doi: 10.1016/j.ajhg.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch M., Cram D.S., Reilly A., O’Bryan M.K., Baker H.W., de Kretser D.M., McLachlan R.I. The Y chromosome gr/gr subdeletion is associated with male infertility. Mol. Hum. Reprod. 2005;11:507–512. doi: 10.1093/molehr/gah191. [DOI] [PubMed] [Google Scholar]
- 13.Hughes J.F., Skaletsky H., Brown L.G., Pyntikova T., Graves T., Fulton R.S., Dugan S., Ding Y., Buhay C.J., Kremitzki C. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soh Y.Q., Alföldi J., Pyntikova T., Brown L.G., Graves T., Minx P.J., Fulton R.S., Kremitzki C., Koutseva N., Mueller J.L. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 16.Hughes J.F., Skaletsky H., Pyntikova T., Graves T.A., van Daalen S.K., Minx P.J., Fulton R.S., McGrath S.D., Locke D.P., Friedman C. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange J., Noordam M.J., van Daalen S.K., Skaletsky H., Clark B.A., Macville M.V., Page D.C., Repping S. Intrachromosomal homologous recombination between inverted amplicons on opposing Y-chromosome arms. Genomics. 2013;102:257–264. doi: 10.1016/j.ygeno.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repping S., Skaletsky H., Lange J., Silber S., Van Der Veen F., Oates R.D., Page D.C., Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Repping S., van Daalen S.K., Korver C.M., Brown L.G., Marszalek J.D., Gianotten J., Oates R.D., Silber S., van der Veen F., Page D.C., Rozen S. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Repping S., van Daalen S.K., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 21.Johansson M.M., Van Geystelen A., Larmuseau M.H., Djurovic S., Andreassen O.A., Agartz I., Jazin E. Microarray analysis of copy number variants on the human Y chromosome reveals novel and frequent duplications overrepresented in specific haplogroups. PLoS ONE. 2015;10:e0137223. doi: 10.1371/journal.pone.0137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espinosa J.R., Ayub Q., Chen Y., Xue Y., Tyler-Smith C. Structural variation on the human Y chromosome from population-scale resequencing. Croat. Med. J. 2015;56:194–207. doi: 10.3325/cmj.2015.56.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei W., Fitzgerald T.W., Ayub Q., Massaia A., Smith B.H., Dominiczak A.F., Morris A.D., Porteous D.J., Hurles M.E., Tyler-Smith C., Xue Y. Copy number variation in the human Y chromosome in the UK population. Hum. Genet. 2015;134:789–800. doi: 10.1007/s00439-015-1562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massaia A., Xue Y. Human Y chromosome copy number variation in the next generation sequencing era and beyond. Hum. Genet. 2017;136:591–603. doi: 10.1007/s00439-017-1788-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghenu A.H., Bolker B.M., Melnick D.J., Evans B.J. Multicopy gene family evolution on primate Y chromosomes. BMC Genomics. 2016;17:157. doi: 10.1186/s12864-015-2187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oetjens M.T., Shen F., Emery S.B., Zou Z., Kidd J.M. Y-chromosome structural diversity in the bonobo and chimpanzee lineages. Genome Biol. Evol. 2016;8:2231–2240. doi: 10.1093/gbe/evw150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomaszkiewicz M., Rangavittal S., Cechova M., Campos Sanchez R., Fescemyer H.W., Harris R., Ye D., O’Brien P.C., Chikhi R., Ryder O.A. A time- and cost-effective strategy to sequence mammalian Y Chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 2016;26:530–540. doi: 10.1101/gr.199448.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan A.P., Pardo-Manuel de Villena F. Sequence and structural diversity of mouse Y chromosomes. Mol. Biol. Evol. 2017;34:3186–3204. doi: 10.1093/molbev/msx250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poznik G.D., Xue Y., Mendez F.L., Willems T.F., Massaia A., Wilson Sayres M.A., Ayub Q., McCarthy S.A., Narechania A., Kashin S., 1000 Genomes Project Consortium Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat. Genet. 2016;48:593–599. doi: 10.1038/ng.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez F.L., Krahn T., Schrack B., Krahn A.M., Veeramah K.R., Woerner A.E., Fomine F.L., Bradman N., Thomas M.G., Karafet T.M., Hammer M.F. An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am. J. Hum. Genet. 2013;92:454–459. doi: 10.1016/j.ajhg.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dohm J.C., Lottaz C., Borodina T., Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen A., Srivastava M.S. On tests for detecting a change in mean. Ann. Stat. 1975;3:98–108. [Google Scholar]
- 38.Sampson J., Jacobs K., Yeager M., Chanock S., Chatterjee N. Efficient study design for next generation sequencing. Genet. Epidemiol. 2011;35:269–277. doi: 10.1002/gepi.20575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 40.Saxena R., de Vries J.W., Repping S., Alagappan R.K., Skaletsky H., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–267. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- 41.Fitch W.M. Toward defining the course of evolution: minimum change for a specified tree topology. Syst. Zool. 1971;20:406–416. [Google Scholar]
- 42.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Q., Li H., Moorjani P., Jay F., Slepchenko S.M., Bondarev A.A., Johnson P.L., Aximu-Petri A., Prüfer K., de Filippo C. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karmin M., Saag L., Vicente M., Wilson Sayres M.A., Järve M., Talas U.G., Rootsi S., Ilumäe A.M., Mägi R., Mitt M. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25:459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 47.Huerta-Cepas J., Serra F., Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu C., Zhang J., Li Y., Xia Y., Zhang F., Wu B., Wu W., Ji G., Gu A., Wang S. The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum. Mol. Genet. 2009;18:1122–1130. doi: 10.1093/hmg/ddn427. [DOI] [PubMed] [Google Scholar]
- 49.Repping S., Skaletsky H., Brown L., van Daalen S.K., Korver C.M., Pyntikova T., Kuroda-Kawaguchi T., de Vries J.W., Oates R.D., Silber S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat. Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 50.Fernandes S., Paracchini S., Meyer L.H., Floridia G., Tyler-Smith C., Vogt P.H. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. Am. J. Hum. Genet. 2004;74:180–187. doi: 10.1086/381132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudmant P.H., Mallick S., Nelson B.J., Hormozdiari F., Krumm N., Huddleston J., Coe B.P., Baker C., Nordenfelt S., Bamshad M. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charlesworth B., Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graves J.A. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 54.Wilson Sayres M.A., Lohmueller K.E., Nielsen R. Natural selection reduced diversity on human y chromosomes. PLoS Genet. 2014;10:e1004064. doi: 10.1371/journal.pgen.1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lucotte E.A., Skov L., Jensen J.M., Macià M.C., Munch K., Schierup M.H. Dynamic copy number evolution of X- and Y-linked ampliconic genes in human populations. Genetics. 2018;209:907–920. doi: 10.1534/genetics.118.300826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rozen S., Marszalek J.D., Alagappan R.K., Skaletsky H., Page D.C. Remarkably little variation in proteins encoded by the Y chromosome’s single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 2009;85:923–928. doi: 10.1016/j.ajhg.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellott D.W., Hughes J.F., Skaletsky H., Brown L.G., Pyntikova T., Cho T.J., Koutseva N., Zaghlul S., Graves T., Rock S. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vollrath D., Foote S., Hilton A., Brown L.G., Beer-Romero P., Bogan J.S., Page D.C. The human Y chromosome: a 43-interval map based on naturally occurring deletions. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 59.Yang B., Ma Y.Y., Liu Y.Q., Li L., Yang D., Tu W.L., Shen Y., Dong Q., Yang Y. Common AZFc structure may possess the optimal spermatogenesis efficiency relative to the rearranged structures mediated by non-allele homologous recombination. Sci. Rep. 2015;5:10551. doi: 10.1038/srep10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyckoff G.J., Wang W., Wu C.I. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 61.Hughes J.F., Skaletsky H., Pyntikova T., Minx P.J., Graves T., Rozen S., Wilson R.K., Page D.C. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 62.Dixson A.F. Univ. Chicago Press; 1998. Primate Sexuality: Comparative Studies of the Prosimians, Monkeys, Apes and Human Beings. [Google Scholar]
- 63.Saxena R., Brown L.G., Hawkins T., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Page D.C. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 64.Connallon T., Clark A.G. Gene duplication, gene conversion and the evolution of the Y chromosome. Genetics. 2010;186:277–286. doi: 10.1534/genetics.110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hallast P., Balaresque P., Bowden G.R., Ballereau S., Jobling M.A. Recombination dynamics of a human Y-chromosomal palindrome: rapid GC-biased gene conversion, multi-kilobase conversion tracts, and rare inversions. PLoS Genet. 2013;9:e1003666. doi: 10.1371/journal.pgen.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warburton P.E., Giordano J., Cheung F., Gelfand Y., Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14(10A):1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gudbjartsson D.F., Helgason H., Gudjonsson S.A., Zink F., Oddson A., Gylfason A., Besenbacher S., Magnusson G., Halldorsson B.V., Hjartarson E. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 68.Nagasaki M., Yasuda J., Katsuoka F., Nariai N., Kojima K., Kawai Y., Yamaguchi-Kabata Y., Yokozawa J., Danjoh I., Saito S., ToMMo Japanese Reference Panel Project Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat. Commun. 2015;6:8018. doi: 10.1038/ncomms9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Telenti A., Pierce L.C., Biggs W.H., di Iulio J., Wong E.H., Fabani M.M., Kirkness E.F., Moustafa A., Shah N., Xie C. Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. USA. 2016;113:11901–11906. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hallast P., Maisano Delser P., Batini C., Zadik D., Rocchi M., Schempp W., Tyler-Smith C., Jobling M.A. Great ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal. Genome Res. 2016;26:427–439. doi: 10.1101/gr.198754.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prado-Martinez J., Sudmant P.H., Kidd J.M., Li H., Kelley J.L., Lorente-Galdos B., Veeramah K.R., Woerner A.E., O’Connor T.D., Santpere G. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.