Hansen et al. review recent structural data that have provided insight into the function and allosteric modulation of NMDA receptors.
Abstract
NMDA-type glutamate receptors are ligand-gated ion channels that mediate a Ca2+-permeable component of excitatory neurotransmission in the central nervous system (CNS). They are expressed throughout the CNS and play key physiological roles in synaptic function, such as synaptic plasticity, learning, and memory. NMDA receptors are also implicated in the pathophysiology of several CNS disorders and more recently have been identified as a locus for disease-associated genomic variation. NMDA receptors exist as a diverse array of subtypes formed by variation in assembly of seven subunits (GluN1, GluN2A-D, and GluN3A-B) into tetrameric receptor complexes. These NMDA receptor subtypes show unique structural features that account for their distinct functional and pharmacological properties allowing precise tuning of their physiological roles. Here, we review the relationship between NMDA receptor structure and function with an emphasis on emerging atomic resolution structures, which begin to explain unique features of this receptor.
Introduction
The vast majority of the excitatory neurotransmission in the central nervous system (CNS) is mediated by vesicular release of glutamate, which activates both pre and postsynaptic G-protein–coupled metabotropic glutamate receptors and ionotropic glutamate receptors (iGluRs). iGluRs are ligand-gated cation channels that are divided into three major structurally distinct functional classes: the α-amino-3-hydroxy-5-methyl-4-isoxasolepropionic acid (AMPA) receptors, kainate receptors, and NMDA receptors (Traynelis et al., 2010; Fig. 1 A). The nomenclature for these functional classes was initially based on the activating agonist, and subsequent molecular cloning revealed cDNAs encoding multiple subunits within the three classes of iGluRs. An intriguing fourth class of iGluRs (GluD1-2) have structural resemblance to AMPA and kainate receptors but do not function as ion channels under normal circumstances (Yuzaki and Aricescu, 2017). Several unique properties distinguish NMDA receptors from other glutamate receptors, including voltage-dependent block by extracellular Mg2+, high permeability to Ca2+, and the requirement for binding of two coagonists, glutamate and glycine (or d-serine), for channel activation (Traynelis et al., 2010). These features have a profound impact on the physiological roles of NMDA receptors and have therefore been the topic of intense investigation.
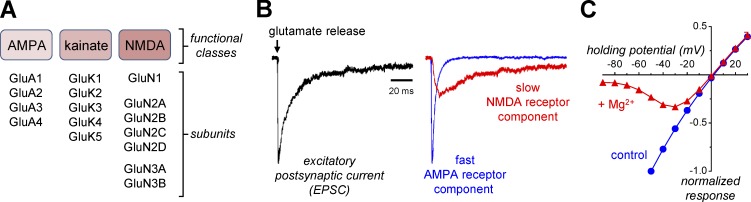
Figure 1.
Functional classes of iGluRs. (A) iGluRs are divided into AMPA, kainate, and NMDA receptors with multiple subunits cloned in each of these functional classes. (B) EPSCs from central synapses can be divided into fast AMPA or slow NMDA receptor–mediated components in the absence of Mg2+ using the AMPA receptor antagonist CNQX or the NMDA receptor antagonist AP5. The figure is adapted from Traynelis et al. (2010). (C) The relationships between NMDA receptor current response and membrane potential (i.e., holding potential) in the presence and absence of 100 µM extracellular Mg2+ reveal the voltage-dependent Mg2+ block, which is relieved as the membrane potential approaches 0 mV (i.e., with depolarization). Data are from Yi et al. (2018).
At central synapses, glutamate release activates iGluRs that mediate an inward current and thereby depolarize the postsynaptic neurons. These excitatory postsynaptic currents (EPSCs) can be described primarily by two temporally distinct components corresponding to activation of AMPA and NMDA receptors. AMPA receptors mediate a synaptic current with rapid rise time and decay, whereas NMDA receptor activation mediates a current that activates more slowly with a time course that endures for tens to hundreds of milliseconds (Hestrin et al., 1990; Sah et al., 1990; Trussell et al., 1993; Geiger et al., 1997; Fig. 1 B). At rest, the NMDA receptor pore is strongly blocked in a voltage-dependent manner by extracellular Mg2+, but this block can be released by the depolarization that accompanies rapid activation of AMPA receptors, particularly when there is a series of closely spaced synaptic events (Fig. 1 C). Thus, the current mediated by NMDA receptors is dependent on both the membrane potential and frequency of synaptic release, rendering these receptors coincidence detectors that respond uniquely to simultaneous presynaptic release of glutamate and postsynaptic depolarization with a slow synaptic current that allows substantial influx of external Ca2+ into the dendritic spine (Bourne and Nicoll, 1993; Seeburg et al., 1995; Nevian and Sakmann, 2004). This increase in intracellular Ca2+ serves as a signal that leads to multiple changes in the postsynaptic neuron, including changes that ultimately produce either short-term or long-term changes in synaptic strength (Lau and Zukin, 2007; Holtmaat and Svoboda, 2009; Traynelis et al., 2010; Higley and Sabatini, 2012; Zorumski and Izumi, 2012; Paoletti et al., 2013; Volianskis et al., 2015). The nature of these changes (e.g., increased or decreased synaptic strength) depends on the frequency and duration of synaptic NMDA receptor activation (Citri and Malenka, 2008; Granger and Nicoll, 2013), thereby providing the brain with a mechanism for encoding information (Hunt and Castillo, 2012; Morris, 2013).
NMDA receptors are unique among synaptic receptors in their requirement for the binding of two agonists, glutamate and glycine (or d-serine; Johnson and Ascher, 1987; Kleckner and Dingledine, 1988; Benveniste and Mayer, 1991; Clements and Westbrook, 1991, 1994). Synaptic NMDA receptors are temporally controlled by the synaptic release of glutamate for activation, because extracellular glycine (or d-serine) is thought to be continuously present at fairly constant concentration. The distinction of glycine or d-serine appears to depend on brain region in addition to the subcellular localization of the receptor (Wolosker, 2007; Oliet and Mothet, 2009; Mothet et al., 2015). For example, some data suggested that d-serine is the dominant coagonist at synapses, with glycine being more important at extrasynaptic sites (Papouin et al., 2012). Although this is an intriguing subdivision, more work will be required to confirm this idea as a general principle. Furthermore, glycine and d-serine are unlikely to be present at concentrations that saturate the coagonist binding site (Berger et al., 1998; Bergeron et al., 1998; Billups and Attwell, 2003). Thus, the requirement for a coagonist enables an additional layer of regulation of NMDA receptor function, in which synaptic activation can be modulated by changes in the ambient levels of glycine/d-serine (Ahmadi et al., 2003; Sullivan and Miller, 2012; Meunier et al., 2017).
There is a rapidly increasing body of data from crystal or cryo-EM structures of intact NMDA receptors or individual domains, which provides a structural framework in which to consider biophysical properties of the receptors and allosteric modulation. In this review, we will focus on how emerging structural understanding has provided functional insight into key properties of the NMDA receptor that are relevant to its roles in the CNS.
Subunit composition of NMDA receptors
Seven genes encode the NMDA receptor subunits: a single GRIN1 gene encodes GluN1, four GRIN2 genes encode GluN2A-D, and two GRIN3 genes encode GluN3A-B (Traynelis et al., 2010). All known NMDA receptors are heterotetrameric assemblies of subunits, which together form a central ion channel pore with striking similarity to an inverted potassium channel. The stoichiometry of the NMDA receptor has been definitively shown to be two glycine-binding GluN1 and two glutamate-binding GluN2 subunits (i.e., GluN1/2 receptors; Ulbrich and Isacoff, 2007; Karakas and Furukawa, 2014; Lee et al., 2014; Fig. 2). However, subunit assembly and physiological roles of the glycine-binding GluN3 subunits remain elusive, and the GluN3 subunits will not be considered in this review (Cavara and Hollmann, 2008; Henson et al., 2010; Low and Wee, 2010; Pachernegg et al., 2012; Kehoe et al., 2013; Pérez-Otaño et al., 2016).
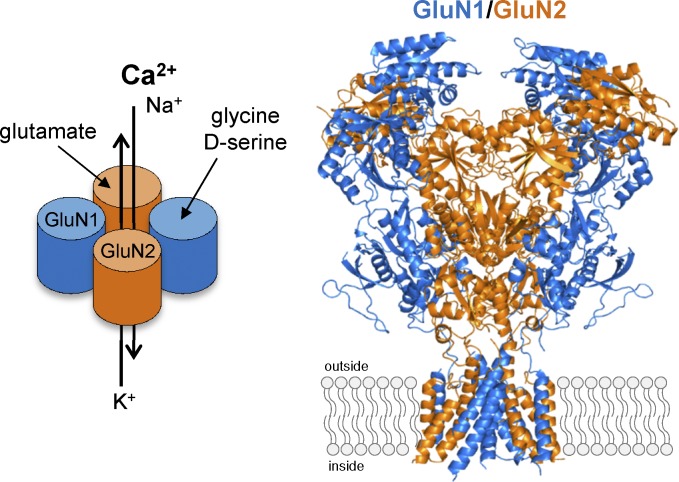
Figure 2.
Subunit stoichiometry and subunit arrangement of GluN1/2 NMDA receptors. The crystal structure of the intact GluN1/2B NMDA receptor (the intracellular CTD omitted from structure; Protein Data Bank accession no. 4PE5; Karakas and Furukawa, 2014) definitively demonstrated that GluN1 and GluN2 subunits assemble as heterotetramers with an alternating pattern (i.e., 1-2-1-2). The NMDA receptor is therefore comprised of two glycine-binding GluN1 and two glutamate-binding GluN2 subunits (i.e., GluN1/2 receptors) that form a central cation-permeable channel pore.
When glutamate is released into the synaptic cleft, it reaches a high concentration (∼1.1 mM) for a brief duration of time, decaying with a time constant of ∼1.2 ms (Clements et al., 1992) as a result of diffusion and active removal of glutamate from the synaptic cleft by excitatory amino acid transporters (i.e., glutamate transporter; Divito and Underhill, 2014). In the synaptic cleft, glutamate will bind to AMPA (and/or kainate) and NMDA receptors, inducing the necessary conformational changes that trigger opening of the ion channel pore, a process referred to as gating. The NMDA receptor–mediated component of the EPSC continues to pass current for tens to hundreds of milliseconds after synaptic glutamate is removed (Lester et al., 1990), which is in part a reflection of agonist binding affinity but also because the receptor activation mechanism involves pregating steps as well as repeated transitions between glutamate-bound open and closed conformational states until glutamate eventually unbinds and the EPSC is terminated (Lester et al., 1990; Lester and Jahr, 1992; Erreger et al., 2005a; Zhang et al., 2008). The functional consequences of the gating reaction mechanism are strongly dependent on the identity of the GluN2 subunit (Monyer et al., 1992, 1994; Vicini et al., 1998; Wyllie et al., 1998). The four different GluN2 subunits thus create substantial diversity among NMDA receptors, and assembly of receptors that contain different GluN2 subunits with distinct properties allows tuning of the synaptic response time course and variation in parameters that control synaptic strength and plasticity. This diversity exerts many effects on neuronal function, circuit properties, and nervous system development.
The glycine-binding GluN1 subunit
The GluN1 subunit, which binds glycine and d-serine, is an obligatory subunit in all functional NMDA receptors and is therefore widely expressed in virtually all central neurons. Three exons in the GluN1 subunit can be alternatively spliced to produce eight different isoforms (Durand et al., 1992; Nakanishi et al., 1992; Sugihara et al., 1992; Hollmann et al., 1993). Exon 5 encodes 21 amino acids in the GluN1 amino-terminal domain (ATD), exon 21 encodes 37 amino acids in the carboxyl-terminal domain (CTD), and exon 22 encodes 38 amino acids in the CTD. Deletion of exon 22 eliminates a stop codon and produces a frameshift, which results in the inclusion of 22 alternative amino acids in the mature polypeptide chain. The GluN1 splice variants show variation in regional and developmental profiles (Laurie and Seeburg, 1994; Zhong et al., 1995; Paupard et al., 1997) and endow the receptor with unique function and pharmacology (see below).
One important property of NMDA receptors containing GluN1 with residues encoded by exon 5 (e.g., GluN1-1b) is reduced agonist potency (i.e., increased EC50, the concentration that produces a half-maximal response; Traynelis et al., 1995, 1998). Consistent with the effect on agonist potency, the GluN1-1b splice variant accelerates deactivation of the NMDA receptor response after removal of glutamate, resulting in EPSCs with a shorter duration (Rumbaugh et al., 2000; Vance et al., 2012; Swanger et al., 2015; Yi et al., 2018). These actions may reflect interactions between the ATD and both the GluN1 and GluN2 agonist-binding domains (ABDs) created by residues encoded by exon 5 (Regan et al., 2018). In addition, GluN1-1b alleviates inhibition of NMDA receptor function by GluN2B-selective antagonists, such as ifenprodil, reduces inhibition by extracellular Zn2+ and protons, and virtually eliminates potentiation by extracellular polyamines (Durand et al., 1992, 1993; Zhang et al., 1994; Traynelis et al., 1995, 1998; Pahk and Williams, 1997; Mott et al., 1998; Rumbaugh et al., 2000; Yi et al., 2018).
Alternative splicing of exons 21 and 22 changes the amino acid composition of the intracellular GluN1 CTD, which interacts with PSD-95, calmodulin, and the neurofilament subunit NF-L (Traynelis et al., 2010). These proteins are involved in surface trafficking and anchoring of receptors at synaptic sites, and alternative splicing of exons 21 and 22 influences cell surface distribution of NMDA receptors (Scott et al., 2001, 2003; Mu et al., 2003; Wenthold et al., 2003). The CTD of GluN1 is a binding site for calmodulin (Ehlers et al., 1996; Iacobucci and Popescu, 2017b), as well as a target of kinases and phosphatases (Tingley et al., 1993, 1997). The relationships between functional roles and structural features of the residues encoded by GluN1 exon 21 and 22 are not yet fully understood.
The glutamate-binding GluN2 subunits
The four glutamate-binding GluN2A-D subunits provide the CNS with a means of controlling NMDA receptor properties as a function of developmental period and brain region (Fig. 3 A). Many studies have described the variation in expression profiles of these subunits, which ultimately control important features of the synaptic NMDA receptor component (Monyer et al., 1992, 1994; Watanabe et al., 1992; Ishii et al., 1993; Akazawa et al., 1994; Zhong et al., 1995). Attempts to pharmacologically control specific NMDA receptor subtypes have, not surprisingly, focused on the development of small molecules that can distinguish between the GluN2 subunits (Ogden and Traynelis, 2011; Strong et al., 2014; Vyklicky et al., 2014; Zhu and Paoletti, 2015; Hackos and Hanson, 2017; Burnell et al., 2018). These efforts are driven in part by the hope that GluN2 subunit–selective pharmacological probes will allow targeting of unique circuits at specific developmental periods to bring about a desired therapeutically beneficial effect.
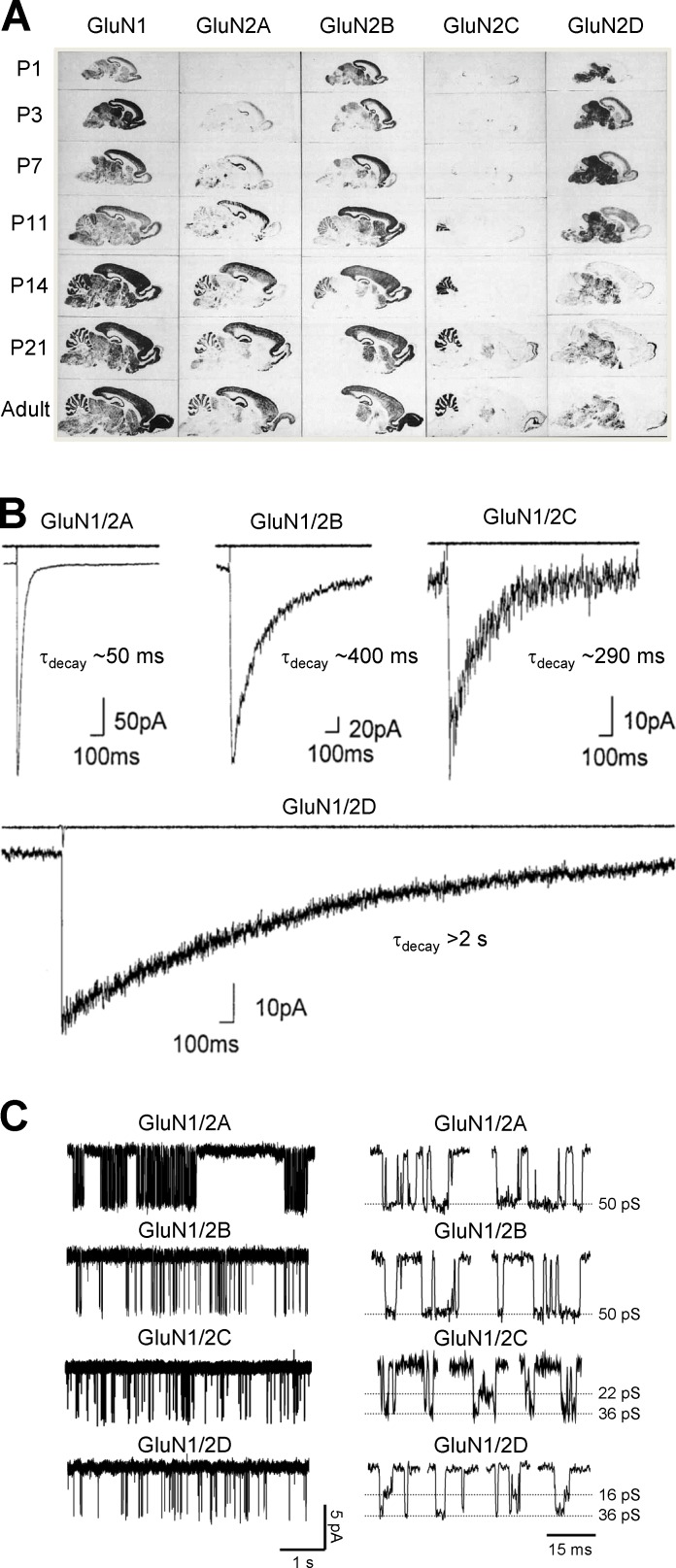
Figure 3.
Expression and functional properties of NMDA receptor subtypes determined by the GluN2 subunit. (A) Autoradiograms obtained by in situ hybridizations of oligonucleotide probes to parasagittal sections of rat brain at indicated postnatal (P) days reveal distinct regional and developmental expression of GluN2 subunits. Fig. 3 A is modified from Akazawa et al., 1994 with permission from the Journal of Comparative Neurology. (B) Whole-cell patch-clamp recordings of responses from recombinant diheteromeric NMDA receptor subtypes expressed in HEK293 cells. The receptors are activated by a brief application of glutamate (1 ms of 1 mM glutamate) indicated by the open tip current in the upper trace. Fig. 3 B is adapted from Vicini et al. (1998) with permission from the Journal of Neurophysiology. (C) Single-channel recordings of currents from outside-out membrane patches obtained from HEK293 cells expressing recombinant NMDA receptor subtypes. The single-channel recordings demonstrate distinct open probabilities and channel conductances depending on the GluN2 subunit in the diheteromeric NMDA receptor. Highlights of individual openings are shown on the left. Adapted from Yuan et al. (2008).
Among the many differences in functional properties governed by the GluN2 subunit, several are particularly noteworthy (Erreger et al., 2004; Traynelis et al., 2010; Paoletti et al., 2013; Wyllie et al., 2013; Glasgow et al., 2015). The potency of glutamate is influenced by the GluN2 subunits. For example, the EC50 for glutamate-activating NMDA receptors containing two GluN1 and two GluN2D subunits is more than fivefold lower (i.e., more potent) than that for GluN1/2A, whereas GluN1/2B and GluN1/2C receptors show intermediate EC50 values (Erreger et al., 2007; Chen et al., 2008; Hansen et al., 2008). The time course of deactivation after removal of glutamate, which controls the duration of the synaptic EPSC (Lester et al., 1990), varies over 100-fold for the different GluN2 subunits (Fig. 3 B). The time constants describing the exponential deactivation time course (τdecay) are ∼40–50 ms for GluN1/2A, ∼300–400 ms for GluN1/2B and GluN1/2C, and ∼4 s for GluN1/2D (Monyer et al., 1992; Vicini et al., 1998; Wyllie et al., 1998; Yuan et al., 2009). Interestingly, intrareceptor allosteric interactions render the potency of glycine and d-serine at the GluN1 subunit sensitive to the identity of the GluN2 subunit (Sheinin et al., 2001; Chen et al., 2008; Dravid et al., 2010; Jessen et al., 2017; Maolanon et al., 2017). For example, the potency of glycine at GluN1/2A receptors is ∼10-fold less than at GluN1/2D receptors (Chen et al., 2008).
Multiple biophysical properties are also controlled by the GluN2 subunit. GluN1/2A and GluN1/2B have higher single-channel conductance than GluN1/2C and GluN1/2D receptors (Erreger et al., 2004; Traynelis et al., 2010; Paoletti et al., 2013; Wyllie et al., 2013; Glasgow et al., 2015; Fig. 3 C). GluN1/2A and GluN1/2B also show higher Ca2+ permeability and are more sensitive to Mg2+ block than GluN1/2C and GluN1/2D (Monyer et al., 1992, 1994; Burnashev et al., 1995; Kuner and Schoepfer, 1996; Qian et al., 2005; Siegler Retchless et al., 2012). These biophysical differences are important, as the sensitivity to voltage-dependent Mg2+ block can influence the temporal window for spike timing–dependent plasticity (Nevian and Sakmann, 2004, 2006; Carter and Jahr, 2016). Furthermore, the probability that the channel will be open when all agonist-binding sites are occupied by agonists (i.e., the open probability) is strongly dependent on GluN2 identity (Fig. 3 C). The open probability is ∼0.5 for recombinant GluN1/2A, ∼0.1 for GluN1/2B, and <0.02 for GluN1/2C and GluN1/2D (Erreger et al., 2004; Traynelis et al., 2010; Paoletti et al., 2013; Wyllie et al., 2013; Glasgow et al., 2015). The GluN2 subunits also control inhibition of NMDA receptors by endogenous modulators, such as protons and extracellular Zn2+ (Traynelis et al., 1995, 1998; Paoletti et al., 1997).
The amino acid sequence of the intracellular CTD is highly variable among GluN2 subunits, thereby producing pronounced differences in interaction sites for phosphatases, kinases, and proteins responsible for anchoring at synaptic sites and surface trafficking (Wenthold et al., 2003; Traynelis et al., 2010; Sanz-Clemente et al., 2013; Aman et al., 2014; Lussier et al., 2015). As a result of this variation, the GluN2 subunits therefore influence cell-surface expression, subcellular localization, and recycling/degradation of NMDA receptor subtypes.
Diheteromeric and triheteromeric NMDA receptors
The NMDA receptor subunits assemble into receptors with varying composition and distinct functional properties and roles in the CNS. At least two different GluN2 subunits are expressed in most neurons, and thus virtually all neurons have the opportunity to signal through triheteromeric NMDA receptors that contain two GluN1 and two different GluN2 subunits (Chazot et al., 1994; Sheng et al., 1994; Chazot and Stephenson, 1997; Luo et al., 1997; Cathala et al., 2000; Piña-Crespo and Gibb, 2002; Brickley et al., 2003; Jones and Gibb, 2005; Al-Hallaq et al., 2007; Rauner and Köhr, 2011; Tovar et al., 2013; Huang and Gibb, 2014; Swanger et al., 2015). Multiple lines of evidence support the expression of triheteromeric NMDA receptors of the GluN1/2A/2B, GluN1/2A/2C, and GluN1/2B/2D subtypes in neurons (Chazot et al., 1994; Sheng et al., 1994; Takahashi et al., 1996; Chazot and Stephenson, 1997; Luo et al., 1997; Sundström et al., 1997; Dunah et al., 1998; Tovar and Westbrook, 1999; Piña-Crespo and Gibb, 2002; Brickley et al., 2003; Dunah and Standaert, 2003; Jones and Gibb, 2005; Lu et al., 2006; Al-Hallaq et al., 2007; Brothwell et al., 2008; Gray et al., 2011; Rauner and Köhr, 2011; Tovar et al., 2013; Huang and Gibb, 2014; Swanger et al., 2015, 2018). Many important properties of triheteromeric NMDA receptors in the CNS are still poorly understood, and the combinations of GluN2 subunits that can form triheteromeric receptors have not been fully established. This knowledge gap persists because triheteromeric NMDA receptors have been difficult to study in isolation (Chazot et al., 1994; Brimecombe et al., 1997; Vicini et al., 1998; Tovar and Westbrook, 1999; Hatton and Paoletti, 2005; Hansen et al., 2014; Stroebel et al., 2014). That is, coexpression of GluN1 with two different GluN2 subunits (e.g., GluN2A and GluN2B) will produce three populations of functional NMDA receptors, including diheteromeric GluN1/2A and GluN1/2B as well as triheteromeric GluN1/2A/2B receptors (Brimecombe et al., 1997; Vicini et al., 1998; Hatton and Paoletti, 2005; Hansen et al., 2014; Stroebel et al., 2014). A wealth of information exists describing the function, pharmacology, and regulation of recombinant diheteromeric NMDA receptors that contain two copies each of GluN1 and a single type of GluN2 (e.g., GluN1/GluN2A). In contrast, relatively little is known about how the coassembly of two different GluN2 subunits affects receptor properties, including the deactivation time course, concentration dependence, and voltage dependence of Mg2+ block and the sensitivity to subunit-selective allosteric modulators. Similarly, phosphorylation sites and trafficking properties of the intracellular GluN2 CTDs have been extensively studied in diheteromeric receptors, whereas the regulation of triheteromeric NMDA receptors that possess two distinct GluN2 CTDs remains elusive (Tang et al., 2010). Knowledge of the key NMDA receptor properties is an essential step to understand the roles of triheteromeric receptors in the brain. One recent advance that has enabled a determination of the functional and pharmacological properties of some triheteromeric NMDA receptors has been to control cell surface expression of receptors with known GluN2 subunit composition (Hansen et al., 2014; Yi et al., 2017, 2018). This method has provided information about the properties of triheteromeric GluN1/2A/2B receptors, which are distinct from the properties of the diheteromeric receptors that contain composite subunits (Hansen et al., 2014; Stroebel et al., 2014; Cheriyan et al., 2016; Hackos et al., 2016; Serraz et al., 2016; Yi et al., 2016, 2018). Importantly, these properties are not simply the average of the respective diheteromeric NMDA receptor properties. This new approach should allow new opportunities to develop therapeutic agents that target disease-relevant triheteromeric NMDA receptors (Khatri et al., 2014; Yuan et al., 2014; Hackos et al., 2016; Serraz et al., 2016; Yi et al., 2016; Swanger et al., 2018).
NMDA receptor structure and function
All glutamate receptor subunits share a similar architecture that comprises four domains: a large extracellular ATD, a bilobed ABD, a pore-forming transmembrane domain (TMD), and an intracellular CTD (Fig. 4 A). The TMD is formed by three transmembrane helices (M1, M3, and M4) and a reentrant loop (M2). In iGluRs, the reentrant loop lines the intracellular portion of the ion channel pore, whereas elements of the third transmembrane segment (M3) form the extracellular region of the pore. Among NMDA receptor subtypes, the residues in the pore region, which influence ion permeation, are highly conserved. A key determinant of ion permeation, which controls divalent ion permeability and Mg2+ block, resides at the apex of the reentrant M2 loop and is often referred to as the Q/R/N site on the basis of amino acids at this position in AMPA, kainate, and NMDA receptors (Wollmuth, 2018).
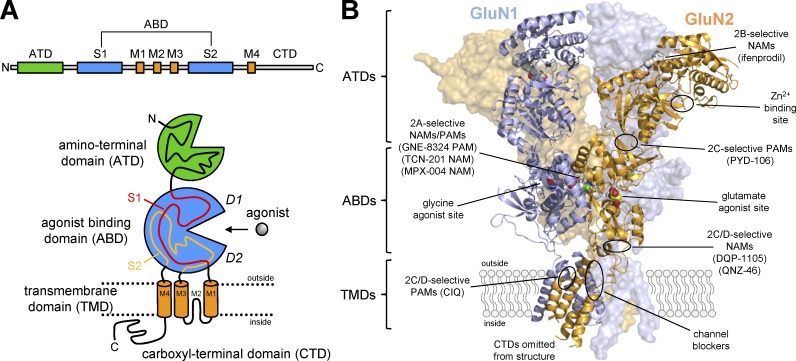
Figure 4.
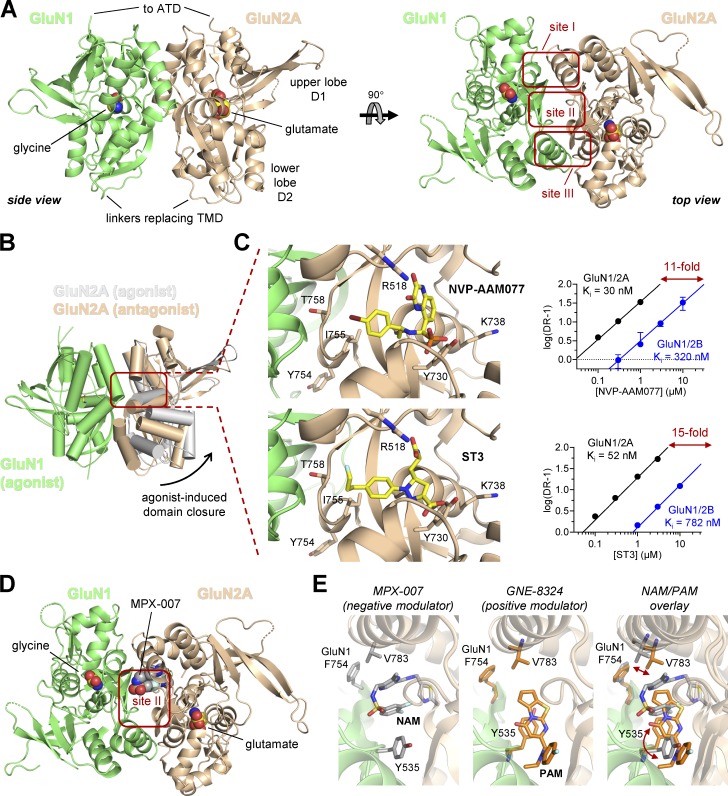
Domain organization and ligand-binding sites in NMDA receptors. (A) The linear representation of the polypeptide chain illustrates the segments that form the four semiautonomous subunit domains shown in the cartoon, which are the extracellular ATD, the ABD, the TMD formed by three transmembrane helices (M1, M2, and M4) and a membrane reentrant loop (M2), and the intracellular CTD. The ABD is formed by two polypeptide segments (S1 and S2) that fold into a bilobed structure with an upper lobe (D1) and lower lobe (D2). The agonist-binding site is located in the cleft between the two lobes. (B) The crystal structure of the GluN1/2B NMDA receptor (Protein Data Bank accession no. 4PE5; Karakas and Furukawa, 2014) shows the subunit arrangement and the layered domain organization. The binding sites for agonists (and competitive antagonists) as well as predicted and known binding sites for PAMs and NAMs are highlighted. The figure is adapted from Hansen et al. (2017).
The ATDs from each subunit adopt bilobed structures formed by the first ∼350 amino acids that associate as back-to-side heterodimers between GluN1 and GluN2. The ATDs play important roles in assembly and strongly modulate NMDA receptor function (Atlason et al., 2007; Gielen et al., 2009; Yuan et al., 2009; Farina et al., 2011). Furthermore, the ATDs create binding sites for allosteric modulators, including extracellular Zn2+ and a diverse series of GluN2B-selective antagonists (exemplified by ifenprodil; Karakas et al., 2009, 2011; Romero-Hernandez et al., 2016; Tajima et al., 2016; Fig. 4 B).
The ABD is formed by the S1 and S2 segments of the polypeptide chain, which are separated by the M1, M2, and M3 segments. The ABDs form kidney-shaped bilobed structures that contain an upper lobe (D1) and a lower lobe (D2) with the agonist-binding site residing in the cleft between these two lobes (Fig. 4 A). The ABD structure, intra- and intersubunit interactions, and its influence on receptor function have been studied for more than two decades. More recently, crystallographic and cryo-EM data have provided the first glimpses of the domain organization of inactive and active GluN1/2B NMDA receptors, providing mechanistic hypotheses by which the different domains and their cognate ligands influence receptor function (Karakas and Furukawa, 2014; Lee et al., 2014; Tajima et al., 2016; Zhu et al., 2016; Regan et al., 2018; Song et al., 2018). We will consider each of these domains in more detail below.
Structure and function of GluN1 and GluN2 ABDs
Keinänen and colleagues were the first to demonstrate that recombinant glutamate receptor ABDs can be generated as soluble proteins by linking the S1 and S2 polypeptide sequences with an artificial peptide linker (Kuusinen et al., 1995; Arvola and Keinänen, 1996). Subsequent work by Gouaux and colleagues resulted in the first crystal structures of glutamate receptor ABDs (Armstrong et al., 1998; Armstrong and Gouaux, 2000). The water-soluble ABD proteins produced by this approach retain ligand-binding activities comparable to those in full-length glutamate receptors, indicating that structural integrity and characteristics of the agonist-binding pocket are retained in isolated ABDs. ABD structures for GluN1, GluN2, and GluN3 subunits have been solved in complex with agonists, antagonists, and allosteric modulators (Furukawa and Gouaux, 2003; Furukawa et al., 2005; Inanobe et al., 2005; Yao and Mayer, 2006; Yao et al., 2008, 2013; Vance et al., 2011; Hansen et al., 2013; Kvist et al., 2013; Jespersen et al., 2014; Hackos et al., 2016; Volgraf et al., 2016; Yi et al., 2016; Lind et al., 2017; Romero-Hernandez and Furukawa, 2017). In addition to NMDA receptor subunits, numerous crystal structures for AMPA and kainate receptor subunits (Pøhlsgaard et al., 2011; Kumar and Mayer, 2013; Karakas et al., 2015) have provided insight into the mechanism underlying full and partial agonism, suggested molecular determinants of subunit selectivity, and demonstrated mechanism and binding pose for competitive antagonists.
The GluN2A ABD in complex with the GluN1 ABD provided the first structural information about a GluN1/GluN2 subunit interface within the NMDA receptor complex, in addition to the binding mode for glutamate and glycine between the two lobes (D1 and D2) of GluN2A and GluN1, respectively (Furukawa et al., 2005; Fig. 5 A). Multiple water molecules reside in close proximity to the agonists, and some form a hydrogen-bonding network that interacts with the ligand. The glycine-binding pocket in GluN1 is considerably smaller and more hydrophobic than the glutamate-binding pocket in GluN2 (Furukawa and Gouaux, 2003; Furukawa et al., 2005; Inanobe et al., 2005; Yao et al., 2008, 2013). Residues within the glutamate-binding pocket that make atomic contacts with agonists or competitive antagonists are mostly conserved in the GluN2 subunits, and it has therefore proven difficult to identify ligands that bind to this site with strong selectivity between the different NMDA receptor subtypes. However, recent crystallographic data have revealed the structural basis for binding of antagonists with modest selectivity (Lind et al., 2017; Romero-Hernandez and Furukawa, 2017). Selectivity in these cases is driven by space outside the conventional binding pocket that competitive antagonists can exploit in a GluN2-dependent manner (Fig. 5 B).
Figure 5.
Crystal structures of NMDA receptor ABDs. (A) Structures of the soluble GluN1/2 ABD heterodimers reveal the subunit interface and back-to-back dimer arrangement of the ABDs. The structure shown here is for the GluN1/2A ABD heterodimer with bound glutamate and glycine shown as spheres (Protein Data Bank accession no. 5I57; Yi et al., 2016). The top view of the structure highlights sites I–III at the subunit interface. (B) Overlay of crystal structures of GluN1/2A ABD heterodimers in complex with glycine and either glutamate agonist (Protein Data Bank accession no. 5I57; Yi et al., 2016) or a competitive glutamate site antagonist (Protein Data Bank accession no. 5U8C; Romero-Hernandez and Furukawa, 2017). Activation of NMDA receptors requires agonist-induced ABD closure. Competitive antagonists bind the ABD without inducing domain closure, thereby preventing receptor activation. (C) Magnified views of the glutamate-binding site with bound GluN2A-preferring antagonists NVP-AAM077 (Protein Data Bank accession no. 5U8C; Romero-Hernandez and Furukawa, 2017) or ST3 (Protein Data Bank accession no. 5VII; Lind et al., 2017). Schild analyses demonstrated that NVP-AAM077 has 11-fold and ST3 has 15-fold preference for GluN1/2A over GluN1/2B receptors (data adapted from Lind et al., 2017). The crystal structures reveal a binding mode in which NVP-AAM077 and ST3 occupy a cavity that extends toward GluN1 at the subunit interface, and mutational analyses show that the GluN2A preference of these antagonists is primarily mediated by four nonconserved residues (Lys738, Tyr754, Ile755, and Thr758) that do not directly contact the ligand but are positioned within 12 Å of the glutamate-binding site. (D) Structure of the agonist-bound GluN1/2A ABD heterodimer with the NAM MPX-007 bound at site II in the subunit interface (Protein Data Bank accession no. 5I59; Yi et al., 2016). (E) Magnified views of site II in GluN1/2A ABD heterodimer with bound MPX-007 (NAM; Protein Data Bank accession no. 5I59; Yi et al., 2016) or PAM GNE-8324 (Protein Data Bank accession no. 5H8Q; Hackos et al., 2016). The overlay illustrates the distinct effects of NAM and PAM binding on Val783 in GluN2A and Tyr535 in GluN1. The GluN2A selectivity of the NAMs and PAMs binding at this modulatory site is mediated by Val783 in GluN2A, which is nonconserved among GluN2 subunits (Phe in GluN2B and Leu in GluN2C/GluN2D).
The residues at the heterodimer interface between the GluN1 and GluN2 ABDs modulate receptor function in several important ways. Three separate areas of contact between GluN1 and GluN2A can be seen in the ABD heterodimer crystal structures (referred to as sites I, II, and III; Furukawa et al., 2005; Fig. 5 A). Sites I and III consist of hydrophobic residues from both GluN1 and GluN2, and nonpolar interactions between these residues mediate ABD heterodimerization (Furukawa et al., 2005). The heterodimeric arrangement of GluN1 and GluN2A ABDs is similar to the homodimeric arrangement found in some AMPA and kainate receptors. In AMPA receptors, allosteric modulators such as cyclothiazide and aniracetam bind to sites equivalent to site I + III and site II, respectively, resulting in block of desensitization and slowing of deactivation speeds (Sun et al., 2002; Jin et al., 2005). The aromatic ring of Tyr535 in GluN1 has a positional overlap with that of aniracetam bound in AMPA receptors, therefore acting like a natural “tethered ligand” incorporated in the primary sequence (Furukawa et al., 2005). Consistently, mutations of Tyr535 in GluN1 alters deactivation time course of NMDA receptors, suggesting that the heterodimer interface can influence factors controlling deactivation, such as agonist dissociation or channel open time (Furukawa et al., 2005; Borschel et al., 2015). Recent crystallographic studies have shown that site II of the GluN1/2A ABD heterodimer contains the binding sites for both positive and negative allosteric modulators with strong selectivity for GluN2A (Hackos et al., 2016; Volgraf et al., 2016; Yi et al., 2016). Together, these results identify the heterodimer ABD interface as an important locus for modulation of NMDA receptor function (Fig. 5 C).
NMDA receptors are sensitive to the redox potential, and reducing conditions can enhance NMDA receptor–mediated current responses (Aizenman et al., 1989; Tang and Aizenman, 1993; Köhr et al., 1994; Choi and Lipton, 2000). This sensitivity appears to be mediated by a pair of conserved cysteine residues (C744 and C798) within the GluN1 subunit (Sullivan et al., 1994; Choi et al., 2001). These two residues interact as a disulfide bond in the GluN1/2A ABD heterodimer structure, and reduction of this conformational constraint in GluN1, but not GluN2, enhances NMDA receptor function (Sullivan et al., 1994; Talukder et al., 2011). Several other disulfide bonds exist in ABD crystal structures of both GluN1 and GluN2 subunits, but functional effects of their reduction or oxidation have not yet been described (Takahashi et al., 2015).
Structures of the GluN1-GluN2A ABD heterodimer in complex with various agonists, partial agonists, and antagonists have suggested a structural basis for their modes of action (Furukawa and Gouaux, 2003; Furukawa et al., 2005; Inanobe et al., 2005; Vance et al., 2011; Hansen et al., 2013; Yao et al., 2013; Jespersen et al., 2014). Binding of glycine and glutamate to GluN1 and GluN2 ABDs, respectively, produces a rapid ABD rearrangement that involves reduction of the angle between the D1 and D2 lobes, producing a clamshell-like closure of the bilobed domain (Fig. 5 B). This agonist-mediated ABD closure triggers formation of hydrogen bonds between residues from the upper and lower lobes, which are hypothesized to stabilize the agonist-bound ABD structure (Kalbaugh et al., 2004; Paganelli et al., 2013). The energy provided by agonist binding and ABD closure triggers the receptor to undergo a series of conformational changes that ultimately open the ion channel pore. Thus, ABD closure that results from agonist binding is the initial conformational change that ultimately triggers the process of ion channel gating. Binding of competitive antagonists, such as the glycine site antagonist DCKA and the glutamate site antagonist D-AP5, stabilizes an open cleft conformation that is incapable of triggering channel gating (Fig. 5 B).
The stabilization of the NMDA receptor ABDs in a closed cleft conformation by agonist binding and in an open cleft conformation by competitive antagonist binding is similar to that found for the AMPA and kainate receptor ABDs (Pøhlsgaard et al., 2011; Kumar and Mayer, 2013; Karakas et al., 2015). However, one notable difference exists. Multiple structures of AMPA receptor ABDs in complex with partial agonists show partial domain closure that correlates with their efficacy (Pøhlsgaard et al., 2011). In contrast, multiple structures of ABD with bound partial agonists, such as d-cycloserine, ACPC, and ACBC in GluN1 and NMDA and Pr-NHP5G in GluN2 show virtually identical degrees of domain closure compared with structures with full agonists (Inanobe et al., 2005; Vance et al., 2011; Hansen et al., 2013). However, these crystal structures capture only one conformation of the isolated ABDs, which may be influenced by the lack of interacting domains (ATD and TMD) and is further stabilized by contacts in the crystal lattice. This caveat to crystal structures of the isolated ABDs is highlighted by recent single-molecule FRET and molecular dynamics studies that provide insight into the dynamic behavior of the NMDA receptor ABDs (Yao et al., 2013; Dai et al., 2015; Dai and Zhou, 2015; Dolino et al., 2015, 2016). These studies suggest that the ABDs fluctuate between open and closed cleft conformations even in the absence of agonist (i.e., the apo state). However, binding of full agonist changes the energy landscape for ABD conformations to strongly favor a fully closed conformation, whereas binding of partial agonists is less efficient in changing this landscape, thereby enabling the ABD to adopt conformations with intermediate domain closure more frequently than full agonists. Hence, a conformational selection mechanism is likely to account for partial agonism in NMDA receptors despite the lack of crystallographic data showing intermediate domain closure for partial agonists.
Structures of intact tetrameric NMDA receptors
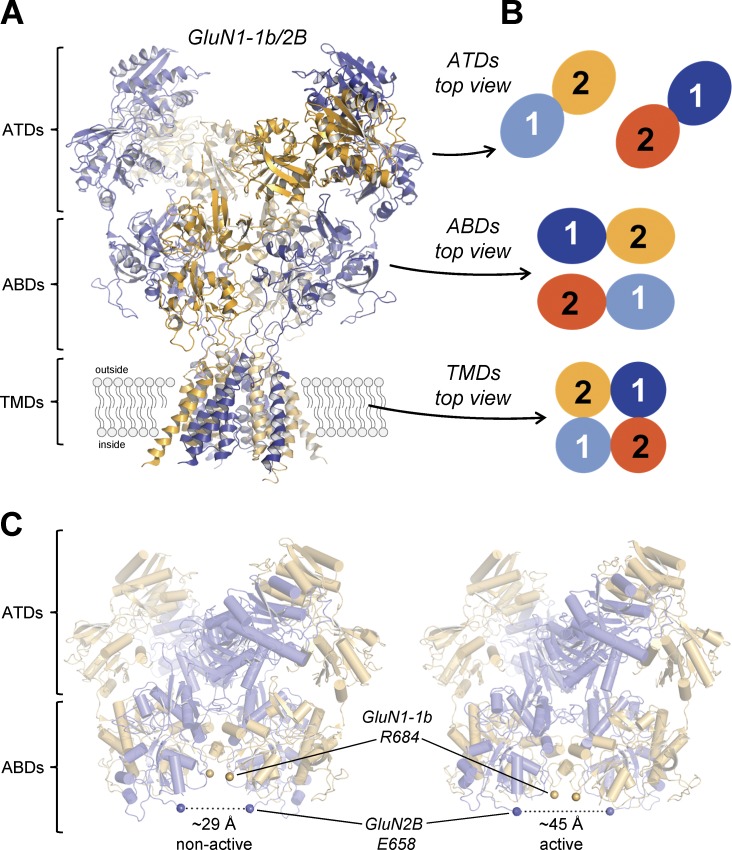
The first structures of intact NMDA receptors (GluN1/2B extracellular domains and TMDs) confirmed the hypothesized domain organization and showed that GluN1 and GluN2B subunits exist in an alternating pattern (i.e., 1-2-1-2) within the tetrameric assembly (Karakas and Furukawa, 2014; Lee et al., 2014; Fig. 6). These studies also confirmed that the NMDA receptor structure shares certain characteristics with AMPA and kainate receptors. First, the receptor subunits adopt a layered structure, with one layer formed by TMDs and two extracellular layers formed by ABD heterodimers and ATD heterodimers. Second, the TMDs have a quasi-fourfold symmetry, whereas the extracellular portion shows twofold symmetry between the two ABD heterodimers and ATD heterodimers in a dimer-of-dimer arrangement. Thus, there is a symmetry mismatch between the TMD layer and the extracellular layers of the receptor. Third, there is a remarkable subunit crossover between the ABD layer and the ATD layer (Fig. 6). In addition, the NMDA receptor has several unique structural features when compared with AMPA and kainate receptors (Karakas and Furukawa, 2014; Lee et al., 2014). For example, there are extensive contacts between the two GluN1/2 ABD heterodimers that are not present in AMPA and kainate receptor structures. These contacts may provide the structural basis of the GluN2 subunit dependence of glycine potency. In addition, the NMDA receptor ATDs show a different arrangement, leading to distinct subunit interfaces compared with AMPA and kainate receptors. Importantly, the ATDs form extensive contacts with the upper lobe of the ABD whereas the ATD–ABD interactions are minimal in AMPA and kainate receptors. These interactions give the NMDA receptor a more compact “hot air balloon–like” appearance, which is distinct from the more Y-shaped AMPA and kainate receptors. The ABD–ATD interactions also create a protein–protein interface at which modulators can bind (Khatri et al., 2014; Kaiser et al., 2018). A recent study also showed that the motif encoded by exon 5, which controls pH sensitivity, deactivation time course, and agonist potency, is also located at the ABD–ATD interface and modulates the ABD–ATD interaction (Regan et al., 2018). The structure of the NMDA receptor thus reveals unique intra- and interdomain contacts that provide a framework for understanding allosteric interactions between subunits, as well as allosteric modulation by small-molecule ligands.
Figure 6.
Structure of the intact NMDA receptors. (A) Structure of the glycine- and glutamate-bound GluN1-1b/2B NMDA receptor without CTDs (Protein Data Bank accession no. 5FXI; Tajima et al., 2016). (B) The GluN1 (1) and GluN2 (2) subunits are arranged as a dimer of heterodimers at the ATD and ABD layers in a 1-2-1-2 fashion. Note that the heterodimer pairs are interchanged between the ATD and ABD layers (i.e., subunit crossover). In the TMD layer, the GluN1 and GluN2 subunits are arranged as a tetramer with pseudo-fourfold symmetry. (C) Comparison of the two major conformational states observed in the presence of glycine and glutamate by cryo-EM/single-particle analysis. Shown in spheres are the Cα of the gating ring residues, GluN1-1b Arg684 and GluN2B Glu658, which are adjacent to the pore-forming M3 transmembrane helices. In the nonactive (Protein Data Bank accession no. 5FXI; Tajima et al., 2016) and active (Protein Data Bank accession no. 5FXG; Tajima et al., 2016) conformations, the distances between the two GluN2B Glu658 Cα atoms are ∼29 Å and ∼45 Å, respectively, indicating that degrees of tension in the ABD–TMD loops are different.
Although the crystallographic structures of intact NMDA receptors advance our understanding of the structure–function relationship, they nevertheless capture only a low energy conformation among the many conformations that the NMDA receptor moves through en route to activation. In these crystal structures, glycine and glutamate were bound to GluN1 and GluN2B, respectively, and the GluN2B-selective negative allosteric modulator ifenprodil was bound to the interface between GluN1 and GluN2B ATDs (Karakas and Furukawa, 2014; Lee et al., 2014). The structures represent the agonist-bound, inhibited receptor with the ion channel closed. However, recent cryo-EM data have described multiple conformations in the extracellular region, providing the first dynamic pictures of NMDA receptor conformational changes and insight into the structural mechanism of receptor activation and allosteric modulation (Tajima et al., 2016; Zhu et al., 2016). Unfortunately, the TMDs for the active and antagonist-bound states are not well resolved in the cryo-EM structures, limiting mechanistic insights into gating and antagonism. However, in the active conformation, distances between the residues that are in proximity of the TMDs increase as much as ∼20 Å in the context of the heterotetramer (Fig. 6 C). This “dilation” of the gating ring likely generates sufficient tensions in the ABD–TMD linker for rearrangement of the helices that form the gate (Kazi et al., 2014; Twomey and Sobolevsky, 2018). This tension can lead to reorientation of the M3 helix in AMPA receptors as well as a kink at an alanine residue that appears to serve as a hinge. Whether or not gating of NMDA receptor ion channels involves similar conformational alterations of the ABD–TMD linker and the TMD demonstrated in the recent AMPA receptor structures (Twomey and Sobolevsky, 2018) remains to be seen, although the gating motifs are highly conserved. Nevertheless, given that the relative orientation of the ABDs and TMDs is distinct in NMDA receptors, structural data are required to evaluate whether these ideas transfer between AMPA and NMDA receptors.
Control of NMDA receptor function by the ATD
The ATD adopts a bilobed structure, which is unrelated to the ABD, with R1 and R2 referring to upper and lower lobes, respectively (Karakas et al., 2009, 2011). Furthermore, there is a unique dimer-of-dimer arrangement of the NMDA receptor ATDs compared with the ATDs in AMPA and kainate receptors (Karakas and Furukawa, 2014; Lee et al., 2014; Meyerson et al., 2014; Sobolevsky, 2015; Tajima et al., 2016; Zhu et al., 2016). This arrangement, which is revealed in crystal and cryo-EM structures of intact iGluRs, is characterized by a protein–protein interface formed by the upper R2 lobes from the GluN1 and GluN2 subunits, whereas the lower R1 lobes, which connect to the ABDs, are almost completely separated.
Many of the GluN2-specific differences between NMDA receptor subtypes are caused by sequence variation in the GluN2 ATDs (Gielen et al., 2009; Yuan et al., 2009). Consistent with this idea, chimeric GluN2 subunits that swap the ATD between GluN2A and GluN2D shift the open probability, deactivation time course, and agonist potency toward that of the subunit contributing the ATD (Gielen et al., 2009; Yuan et al., 2009). Although it remains unclear how the ATD controls NMDA receptor function, the mechanism likely involves intra- and intersubunit allosteric interactions between the ATDs and ABDs that influence the configuration of the GluN1/GluN2 ABD heterodimer and thereby impact channel activation (Gielen et al., 2008; Zhu et al., 2013; Tajima et al., 2016). Thus, some GluN2-specific functional and pharmacological NMDA receptor properties are presumably controlled by distinct conformations adopted by the ATDs in a GluN2-specific manner (Hansen et al., 2014; Zhu et al., 2014; Sirrieh et al., 2015b; Lü et al., 2017; Sun et al., 2017).
Ligand binding to the ATD
Crystal structures have demonstrated that GluN2B-selective negative allosteric modulators (NAMs), such as ifenprodil and Ro 25–6981, bind to a modulatory site located at the subunit interface between GluN1 and GluN2B ATDs (Karakas et al., 2011; Karakas and Furukawa, 2014; Lee et al., 2014; Stroebel et al., 2016). These crystal structures revealed that only one residue in this modulatory site is different between GluN2A and GluN2B subunits, but sensitivity to ifenprodil is not introduced by converting this or other residues in GluN2A to that in GluN2B (Karakas et al., 2011; Burger et al., 2012). This stems from the fact that the intersubunit arrangements in GluN1/2A and GluN1/2B ATD heterodimers are distinct from each other, as demonstrated in the recent crystal structure of the GluN1/2A ATD heterodimer (Romero-Hernandez et al., 2016). Specifically, the “pocket” in the GluN1/GluN2 subunit interface is ideally sized to accommodate ifenprodil analogues in GluN1/2B, whereas such a pocket is absent in GluN1/2A because of the different subunit arrangement characterized by a ∼10° rotation compared with GluN1/2B. Multiple lines of investigation, including cryo-EM structures of intact NMDA receptors, functional studies, and computational analyses, suggest that ifenprodil inhibition involves closure of the GluN2B ATD bilobes with accompanying changes in the arrangement of the GluN1/2B ATD heterodimers (Burger et al., 2012; Tajima et al., 2016), indicating that both clamshell conformation and subunit arrangement are coupled to function of the NMDA receptor ion channel.
Functional and structural studies have converged on a structural model for NMDA receptor modulation by Zn2+ and ifenprodil, where modulator binding regulates receptor function through rearrangement of the ATD layer and GluN2 ATD clamshell opening and closing (Sirrieh et al., 2013, 2015a). In GluN1/2B, opening of the ATD bilobes robustly alters inter-GluN1/GluN2 subunit arrangement within the ATD, which results in a ∼13° rotation between the GluN1/2B ABD dimers and dilation of the gating ring (Tajima et al., 2016). NAMs such as ifenprodil and zinc favor the closure of the bilobed GluN2B ATD thereby “locking” the subunit arrangement in a way that prevents dilation of the gating ring. Interestingly, the zinc-bound GluN2A ATD is ∼13° more open compared with the zinc-bound GluN2B ATD (Karakas et al., 2009; Romero-Hernandez et al., 2016). This may explain in part the observation that the extent of zinc inhibition is smaller in GluN2A than GluN2B (Rachline et al., 2005).
NMDA receptors containing GluN1 with exon 5 (e.g., the GluN1-1b splice variant) have reduced sensitivity to all three allosteric modulators (Zn2+, ifenprodil, and spermine; Traynelis et al., 1995; Mott et al., 1998; Yi et al., 2018). In recent cryo-EM structures, the 21 amino acids encoded by exon 5 are placed just above the GluN1-GluN2 ABD heterodimer interface between the ATD and ABD layers, positioned to influence allosteric interactions between GluN2 ATD clamshell motions and GluN1-GluN2 ABDs (Regan et al., 2018). Furthermore, GluN2C residues from both the ATD and ABD that influenced the activity of PYD-106, a GluN2C-selective positive allosteric modulator (PAM), have been identified and molecular modeling proposed a modulatory binding site located in a pocket at the ATD–ABD interface of GluN2C (Khatri et al., 2014; Kaiser et al., 2018). These studies all point to the ATD as the major structural determinant of GluN2-specific variation among NMDA receptor subtypes. For this reason, allosteric modulation of NMDA receptors by the ATD is intensely investigated, and drug discovery studies are poised to identify novel ATD ligands with therapeutic potential.
Channel gating in NMDA receptors
All three transmembrane helices (M1, M3, and M4) and the membrane-reentrant pore-forming loop (M2) are involved in the process of pore opening (i.e., channel gating; Schneggenburger and Ascher, 1997; Krupp et al., 1998; Villarroel et al., 1998; Ren et al., 2003; Talukder et al., 2010; Kazi et al., 2013; Ogden and Traynelis, 2013; Alsaloum et al., 2016). The transmembrane helix M3 forms a helical bundle crossing that physically occludes the pore, and thus M3 helices must change their position before ions can pass through the channel pore (Jones et al., 2002; Yuan et al., 2005; Chang and Kuo, 2008). The M3 transmembrane helix contains nine amino acids (SYTANLAAF) that are almost fully conserved in iGluRs throughout the animal kingdom. Multiple structural and functional studies suggest that these residues comprise the activation gate and that dilation of the M3 helical bundle crossing is thought to be the key change that allows ion conduction (Beck et al., 1999; Sobolevsky et al., 2002a; Chang and Kuo, 2008).
What sequence of events leads to M3 rearrangement? Agonist binding to the bilobed ABDs involves a clamshell closure around the ligands that must be the first step in a sequence of conformational changes that lead to gating. These are followed by multiple short-lived, intermediate conformations that precede a rapid transition from the closed to the open state of the ion channel, inferred by brief, kinetically distinguishable closed states in the single-channel record and the relatively slow time course for receptor activation by supersaturating agonist (Banke and Traynelis, 2003; Popescu et al., 2004; Auerbach and Zhou, 2005; Erreger et al., 2005a; Schorge et al., 2005; Kussius and Popescu, 2009; Fig. 7). However, there is poor understanding of the protein conformations that represent the rate limiting steps en route to channel opening. Moreover, the lifetimes of some of these intermediate conformations are brief, suggesting they are unlikely to be captured in crystal structures or cryo-EM studies, leaving functional experiments as the most feasible (yet imperfect) way to glean clues as to how these changes might control channel opening. Recent functional studies have built explicit models of channel activation in which specific conformations are hypothesized for each of the four subunits (Gibb et al., 2018). Moreover, work with disease-causing mutations identified in human patients has provided key insights into the elements that comprise the gating control mechanism. Residues in the region connecting the S1 segment of the ABD with the M1 transmembrane helix (i.e., the pre-M1 linker) are invariant in the healthy population, and a locus for disease-associated mutations in various neurological diseases (Ogden et al., 2017). In addition, the region connecting the S2 segment of the ABD with the M4 transmembrane helix (i.e., the pre-M4 linker) also appears to be implicated in patients with NMDA receptor missense mutations, and both the pre-M1 and pre-M4 linkers are close enough to be in contact with the conserved SYTANLAAF motif in the M3 helical bundle crossing. These three elements (pre-M1, SYTANLAAF, and pre-M4) appear positioned to form a gating control mechanism (Chen et al., 2017), and it is possible that kinetically distinct conformational states may be the result of rearrangements of this triad of interacting regions. Moreover, the different amino acid sequences for pre-M1 and pre-M4 that exist for GluN1 and GluN2 as well as different positions of these elements in relation to the gating ring could lead to distinct lifetimes for intermediate conformations that must be traversed before rapid pore dilation (Erreger et al., 2005a; Dravid et al., 2008; Kussius and Popescu, 2009; Amico-Ruvio and Popescu, 2010; Vance et al., 2012).
Figure 7.
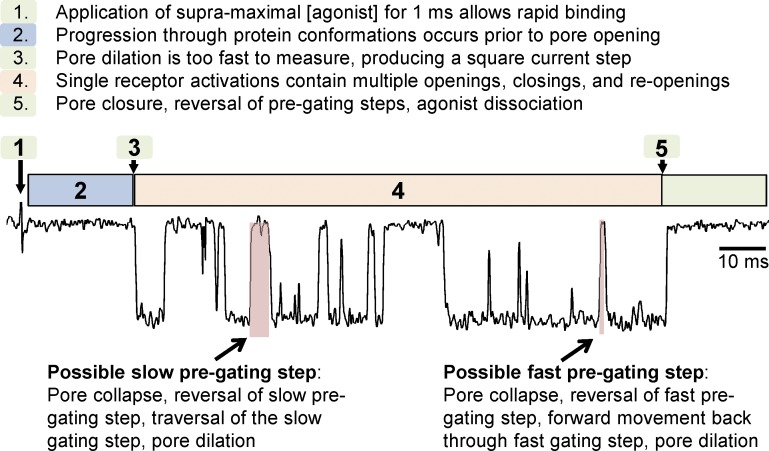
Single-channel recordings of NMDA receptor gating. Recording of receptor activation (i.e., channel gating or pore dilation) in an excised outside-out membrane patch containing a single GluN1/2B receptor exposed to 1 mM glutamate plus 30 µM glycine for 1 ms as indicated. In this example, receptor activation results in a characteristically long burst of channel openings and closings (duration 128 ms). Evaluation of closed periods within the GluN1/2B activation suggests that two kinetically distinct pregating steps exist (i.e., fast and slow steps; see Fig. 8 D for model). Some (but not all) closures within the activation will reflect reversal of pore dilation, reversal of a single pregating step, followed by forward movement back through the pregating step and pore dilation. Two possible closures that might reflect the slow and fast pregating components are highlighted in red. Data are from Banke and Traynelis (2003).
Kinetic models for NMDA receptor activation
The sequence of protein conformational changes that trigger channel gating can be described as reaction schemes (i.e., kinetic models) with agonist binding steps and transitions between different conformational states of the receptor (Fig. 8). The first widely applied kinetic model for NMDA receptor gating was solely designed to account for the time course of the macroscopic current response and consisted of two identical but independent glutamate binding steps, one desensitized state, one closed state, and one open state (Lester and Jahr, 1992). This simple kinetic model appeared to effectively capture key features of macroscopic NMDA receptor responses but was not intended to describe the complexity observed in single-channel recordings (Ascher et al., 1988; Howe et al., 1991; Traynelis and Cull-Candy, 1991; Gibb and Colquhoun, 1992). Furthermore, the usefulness of the Lester and Jahr model was limited by the lack of glycine-binding steps required for receptor activation. Kinetic models that account for both glutamate- and glycine-binding steps as well as intersubunit interactions between the glutamate and glycine ABDs have also been developed (Benveniste et al., 1990), and these models could capture additional features of the time course of NMDA receptors, including glycine-dependent desensitization (see below).
Figure 8.
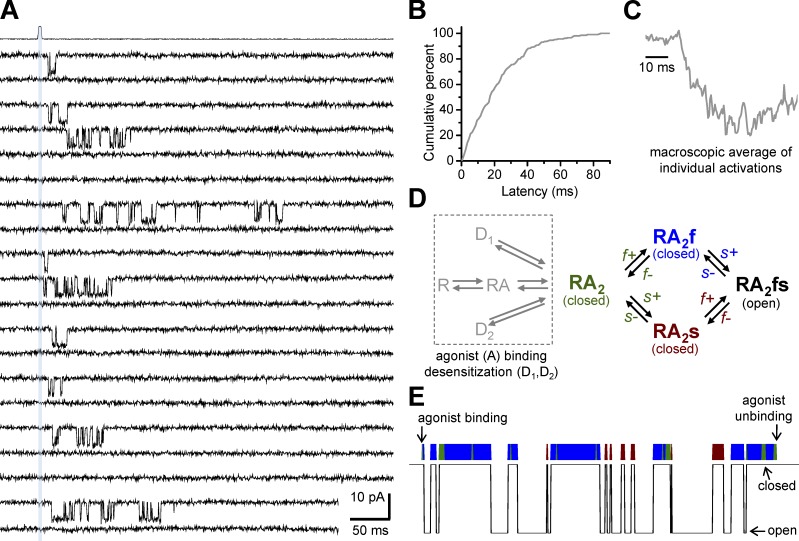
Application of a gating reaction mechanism of NMDA receptors. (A) Individual responses from a recombinant GluN1/2B channel in an excised outside-out patch activated by 1 ms application of maximally effective glutamate and glycine (indicated by the gray vertical bar and the open tip recording above the channel recordings). The patch contained a single active channel, which allowed analysis of the variable delay before channel opening. NMDA receptors bind agonist rapidly and subsequently open after a multimillisecond delay that reflects transition through kinetically distinct protein conformations before pore dilation (i.e., channel gating). Note that although application of maximal glutamate and glycine always produces a binding event, not all binding events lead to channel opening. Reproduced from Erreger et al. (2005a). (B) The cumulative plot of latency to opening after application of 1 mM glutamate for 1 ms. (C) The average of all individual recordings of single activations produced a macroscopic waveform with a characteristic rise time. (D) Evaluation of closed periods within the GluN1/2B activation suggested a model where two pregating steps can occur in any order and explosive opening of the pore, which occurs faster than the resolution of the recordings, is assumed to happen instantaneously once both pregating steps have been traversed. (E) Simulation of a single activation for a GluN1/2B channel (using the model in D) illustrates how brief gaps can contain information about forward rates for the fast kinetically distinct pregating step. The color above the simulation indicates occupancy in the corresponding closed state of the model in D. The slow step often reverses again through the fast state (green) before reopening.
Newer, more complex kinetic models have been proposed that better describe single-channel data by incorporating multiple steps between binding and gating (Banke and Traynelis, 2003; Popescu et al., 2004; Auerbach and Zhou, 2005; Erreger et al., 2005a; Schorge et al., 2005). Investigations of macroscopic and single-channel responses to partial and full agonists suggest that agonist binding to either GluN1 or GluN2 controls distinct steps in the kinetic model (Banke and Traynelis, 2003; Auerbach and Zhou, 2005; Erreger et al., 2005a; Schorge et al., 2005; Fig. 8), although it has also been suggested that partial agonists can impact all pregating steps irrespective of the subunit they bind to (Kussius and Popescu, 2009; Kussius et al., 2010). In some of these models, the actions of allosteric modulators are accounted for by explicitly representing the modulator bound and unbound receptor as independent states (Banke et al., 2005; Erreger and Traynelis, 2008; Amico-Ruvio et al., 2011). Other models for channel blockers and other use-dependent modulators have been described that exclusively allow modulators to bind to the open state (Huettner and Bean, 1988; MacDonald et al., 1991; Blanpied et al., 1997, 2005; Dravid et al., 2007; Kussius et al., 2009; Paganelli and Popescu, 2015; Glasgow et al., 2017).
The ability of AMPA receptor subunits to operate semi-independently (Rosenmund et al., 1998; Jin et al., 2003; Kristensen et al., 2011) and the modular domain architecture of glutamate receptor structures raise the possibility that independent conformational changes in different subunits may progress within the sequence of steps leading to channel opening (Gibb et al., 2018). Some kinetic models suggest that such subunit-specific conformational changes are required in all four NMDA receptor subunits to trigger channel gating and that these structural changes can occur in any order to arrive at an intermediate state that can subsequently transition to the open state of the ion channel (Banke and Traynelis, 2003; Auerbach and Zhou, 2005; Erreger et al., 2005a,b; Schorge et al., 2005). Other models account for macroscopic and single-channel responses by including a few sequential gating steps in a linear kinetic model with an implicit order for slow and fast gating steps (Popescu et al., 2004; Kussius and Popescu, 2009). These models have been used to explore the kinetic aspects of modal gating (Zhang et al., 2008; Iacobucci and Popescu, 2017a), an intriguing phenomenon that is readily apparent in cell-attached recordings from GluN1/GluN2A receptors. Gating modes are defined by different open probabilities and open times and have been described for GluN1/2A and GluN1/2B receptors (Popescu and Auerbach, 2003; Popescu et al., 2004; Amico-Ruvio and Popescu, 2010; Popescu, 2012) but are rarely observed in GluN1/2D (Vance et al., 2013). Although the mechanism remains elusive, mode switching has been proposed to influence the time course of the synaptic current (Zhang et al., 2008).
All these kinetic models for NMDA receptor gating that faithfully describe both macroscopic responses and single-channel data require both multiple pregating steps and multiple open states. The interpretation of this observation is that ion channel opening in NMDA receptors is not directly coupled to agonist-induced ABD closure; instead, the receptor must proceed through a sequence of conformational changes that couple agonist binding to ion channel gating.
Structural determinants of ion permeation and channel block
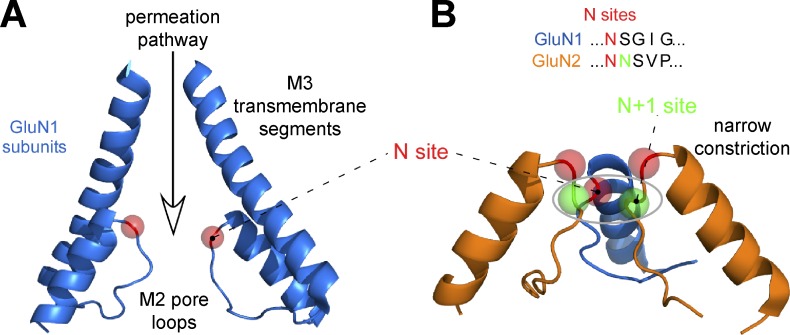
The ion channel pore in NMDA receptors can be divided into the intracellular and extracellular vestibules separated by a narrow constriction (Fig. 9). The narrow restriction resides approximately halfway across the membrane at the apex of the membrane reentrant loop M2 (i.e., the Q/R/N site) and is often referred to as the selectivity filter because of its role as key determinant of Ca2+ permeability, single-channel conductance, and channel block (Wollmuth and Sobolevsky, 2004; Traynelis et al., 2010; Glasgow et al., 2015). The residue at the Q/R/N site is asparagine (N) in both GluN1 and GluN2, but the contribution of this residue to ion permeation is asymmetric between GluN1 and GluN2 subunits (Burnashev et al., 1992; Wollmuth et al., 1996, 1998; Sobolevsky et al., 2002b). This is because the narrow constriction is formed by the Q/R/N site asparagine in GluN1 but by the asparagine residue adjacent to the Q/R/N site (i.e., Q/R/N +1 site) in GluN2. The asymmetric contribution by GluN1 and GluN2 is revealed by substitutions of the Q/R/N site residue in GluN2 that have weak effects on Ca2+ permeability and dramatically reduce Mg2+ block, whereas the same substitutions of the Q/R/N site residue in GluN1 dramatically reduce Ca2+ permeability and have weak effects on Mg2+ block (Burnashev et al., 1992; Wollmuth et al., 1998). However, mutations at the Q/R/N +1 site in GluN2 strongly reduce Mg2+ block (Wollmuth et al., 1998). Functional data therefore suggest a structural asymmetry, where the apexes of M2 in GluN1 and GluN2 are slightly staggered (Sobolevsky et al., 2002b). Diheteromeric GluN1/3 receptors have glycine/arginine residues at the Q/R/N and Q/R/N +1 and show both markedly reduced Ca2+-permeability and Mg2+-block compared with GluN1/2 receptors (Cavara and Hollmann, 2008; Henson et al., 2010; Low and Wee, 2010; Pachernegg et al., 2012; Kehoe et al., 2013). Recent data describing the pore of the AMPA receptor in the open state reinforce the idea that the apex of the reentrant loops form a constriction that impacts ion permeation (Twomey et al., 2017). The structural basis for this functional asymmetry will require high-resolution images of the NMDA receptor in the open state.
Figure 9.
General pore structure of NMDA receptors. (A) Pore-lining elements contributed by the GluN1 subunit (blue; Protein Data Bank accession no. 5UN1; Song et al., 2018). The M3 transmembrane segment lines the extracellular part of the permeation pathway, whereas the M2 pore loop lines the intracellular part with the N site asparagine (red circle) positioned at the tip of the M2 pore loop. The channel is in the closed conformation. (B) The narrow constriction is formed by nonhomologous asparagine residues, the GluN1 N site and the GluN2 N+1 site (Wollmuth et al., 1996; Song et al., 2018). The GluN2B subunit is colored orange. For both GluN1 and GluN2, the N site asparagine residue is positioned at the tip of the M2 loop.
Determinants of ion permeation
NMDA receptor ion channels are permeable to the physiologically relevant Ca2+, Na+, and K+ ions. The different NMDA receptor subtypes display similar permeability to Na+ and K+ ions (PK/PNa = 1.14) but are more permeable to Ca2+ relative to monovalent ions (PCa/PX = 1.8–4.5), with variation in Ca2+ permeability that depends on the GluN2 subunit (Burnashev et al., 1995; Schneggenburger, 1996, 1998; Sharma and Stevens, 1996; Jatzke et al., 2002). However, NMDA receptors also exhibit block by external Ca2+, despite being highly Ca2+ permeable, which can be observed as a reduction in channel conductance in single-channel data (Premkumar and Auerbach, 1996; Wyllie et al., 1996; Premkumar et al., 1997; Dravid et al., 2008). The concurrent block by Ca2+ and the high Ca2+ permeability are not incompatible properties but are expected if multiple Ca2+-binding sites are located in the ion channel pore of NMDA receptors (Premkumar and Auerbach, 1996; Sharma and Stevens, 1996). Studies have suggested that one Ca2+-binding site is located at the Q/R/N site, whereas another, more external Ca2+-binding site could be formed by a cluster of charged DRPEER residues in GluN1 (Watanabe et al., 2002; Karakas and Furukawa, 2014). The external Ca2+-binding site is located at the external entrance to the ion channel above the transmembrane helix M3 of GluN1. Although structural elements of Ca2+ permeation in GluN1/N2 subunits have been identified, the mechanism of Ca2+ permeation remains unknown.
Determinants of channel block
GluN1/2A and GluN1/2B channels are more strongly blocked by extracellular Mg2+ than GluN1/2C and GluN1/2D channels (Monyer et al., 1994; Kuner and Schoepfer, 1996; Qian et al., 2005; Clarke and Johnson, 2006; Siegler Retchless et al., 2012). This channel block is highly dependent on the membrane potential (i.e., voltage dependent), and the IC50 values (the concentrations that produce half-maximal inhibition) for block by external Mg2+ are 2 µM, 2 µM, 14 µM, and 10 µM for GluN1/2A, GluN1/2B, GluN1/2C, and GluN1/2D, respectively, at a holding potential of −100 mV (Kuner and Schoepfer, 1996). The dependency of Mg2+ block on the GluN2 subunit is influenced by multiple structural elements, but a main determinant appears to be a single residue at the S/L site located in the M3 transmembrane helix (Siegler Retchless et al., 2012). The residue at the S/L site, which is a serine in GluN2A/B and a leucine in GluN2C/D, is not lining the ion channel pore but has been suggested to interact with tryptophan residues in the membrane reentrant loop M2 of GluN1 (Siegler Retchless et al., 2012). This interaction between GluN1 and the GluN2 S/L site also appears to be a key determinant of GluN2 subunit–specific variation in channel conductance and Ca2+ permeability (Siegler Retchless et al., 2012). Although important insight into the structural mechanism of GluN2 subunit–dependent control of Mg2+ block is still missing, it is possible that structural elements, including the GluN2 S/L site, govern Mg2+ block by influencing binding sites for permeant ions in the channel pore (Antonov and Johnson, 1999; Zhu and Auerbach, 2001a,b; Qian et al., 2002; Qian and Johnson, 2006).
Channel block by organic cations
The NMDA receptor ion channel pore can be blocked in a voltage-dependent manner by a wide range of organic cations with diverse chemical structures (Huettner and Bean, 1988; Brackley et al., 1993; Parsons et al., 1995). These compounds almost exclusively block open channels in activated NMDA receptors and are positively charged at physiological pH, a mechanism of channel block termed uncompetitive or use dependent. In general, the open channel blockers can be classified into three categories based on their interaction with the channel: (1) “foot-in-the-door” or sequential blockers (e.g., aminoacridine derivatives and tetrapentylammonium) can only bind to the channel when it is open and prevent channel closure when bound (Benveniste and Mayer, 1995; Sobolevsky, 1999; Sobolevsky et al., 1999; Bolshakov et al., 2003; Barygin et al., 2009); (2) partial trapping blockers (e.g., amantadine and memantine) obstruct channel closure but are unable to completely prevent it (Blanpied et al., 1997, 2005; Chen and Lipton, 1997; Mealing et al., 1999; Kotermanski et al., 2009; Johnson et al., 2015); and (3) trapping blockers (e.g., Mg2+, ketamine, phencyclidine [PCP], and MK-801) are trapped inside the channel pore as it closes, and agonists can unbind while the trapping blocker remains bound (Sobolevsky and Yelshansky, 2000; Poulsen et al., 2015). Some channel blockers have also been shown to facilitate channel closure, presumably by interacting with the channel gate (Blanpied et al., 2005; Johnson et al., 2015). Channel blockers proposed to have bifunctionality include nitromemantine derivatives that bind the ion channel pore, facilitating the targeting of a nitro group to a redox-mediated regulatory site on the receptor (Takahashi et al., 2015).
In general, the open channel blockers are considered nonselective among NMDA receptor subtypes (Dravid et al., 2007), but some channel blockers, such as ketamine and memantine, display 5- to 10-fold preference for GluN2C/D-containing receptors over GluN2A/B-containing receptors in the presence of 1 mM extracellular Mg2+ (i.e., under physiological conditions; Kotermanski and Johnson, 2009). NMDA receptor channel blockers have robust neuroprotective effects in animal models of CNS disorders that involve excessive NMDA receptor activation, such as stroke, epilepsy, and traumatic brain injury. However, clinical trials have not been successful because of dose-limiting side effects, patient heterogeneity, and a narrow temporal window for intervention that could have confounded interpretation (Ikonomidou and Turski, 2002; Farin and Marshall, 2004; Muir, 2006; see also Table S2 in Yuan et al., 2015). NMDA receptor channel blockers that bind with high affinity, such as ketamine and PCP, are typically dissociative anesthetics, and their clinical use is limited by psychomimetic side effects. Nonetheless, there is an intense interest in use of ketamine or similar molecules for the treatment of major depressive disorder because of several promising clinical trials in recent years based on the discovery of antidepressant effects for NMDA receptor antagonists (Niciu et al., 2014; Abdallah et al., 2015; Zanos et al., 2018).
Channel blockers such as memantine, which is approved in the treatment of moderate to severe Alzheimer’s disease, have lower affinity than ketamine and PCP and show faster blocking/unblocking kinetics (Parsons et al., 1993). These kinetic properties have been suggested to contribute to an improved therapeutic index with respect to psychomimetic effects, perhaps because of reduced channel block during normal synaptic transmission (Chen and Lipton, 2006), although the mechanism by which memantine may contribute to a symptomatic benefit in Alzheimer’s disease is not well understood. Interestingly, Glasgow et al. (2017) have proposed that memantine stabilizes occupancy of a desensitized state of GluN1/2A receptors, whereas ketamine reduces occupancy of a GluN1/2B desensitized state (Glasgow et al., 2017). Thus, the affinity of these blockers for their binding site in the channel may be allosterically affected by the conformational changes in the receptor protein associated with desensitization. Given the prevalence of triheteromeric GluN1/2A/2B receptors in the brain, it will be important to evaluate memantine and ketamine block at these triheteromeric receptors.
Endogenous mechanisms of functional modulation
NMDA receptors are complex macromolecular membrane-bound protein complexes, and their functional properties and membrane trafficking can be altered by extracellular ions, phosphorylation, and intracellular binding proteins. Additionally, the differences between various diheteromeric and triheteromeric NMDA receptor subtypes create selective actions of many of these types of modulation. Here, we will describe various forms of endogenous regulation of NMDA receptor function.
Modulation by protons
Extracellular protons inhibit NMDA receptor function with an IC50 of ∼50 nM, corresponding to a pH of ∼7.3 (Giffard et al., 1990; Traynelis and Cull-Candy, 1990, 1991; Vyklický et al., 1990). Thus, neuronal NMDA receptors are tonically inhibited by protons at physiological pH and are therefore poised to respond to small changes in extracellular pH that can occur under physiological conditions caused by release of protons from acidic synaptic vesicles or movement of protons across the plasma membrane by pumps (Chesler, 2003). Furthermore, pathological conditions, including seizure and ischemia, produce extracellular acidification, which can decrease pH to levels that strongly inhibit NMDA receptor function (Chesler, 2003).
The sensitivity of the NMDA receptors to inhibition by extracellular protons depends on the GluN2 subunit (Traynelis et al., 1995), with GluN2A-, GluN2B-, and GluN2D-containing NMDA receptors showing proton IC50 values near physiological pH (7.0–7.4). In contrast, GluN2C-containing receptors are less sensitive to changes in pH, with an IC50 value near pH 6.0 (Traynelis et al., 1995; Low et al., 2003). NMDA receptors that include GluN1 subunits containing the alternatively spliced exon 5 in the ATD (i.e., GluN1-1b) are notably less sensitive to protons (Traynelis et al., 1995). Proton inhibition is voltage independent, and without effect on glutamate potency; low pH produces modest shifts in the glycine potency (Tang et al., 1990; Traynelis and Cull-Candy, 1990, 1991; Traynelis et al., 1995). The structural determinants underlying proton inhibition are unknown, although mutations at the ABD interface, linkers to pore-forming elements, and within the M2 reentrant loop can all influence pH sensitivity (Low et al., 2003; Gielen et al., 2008). This suggests that NMDA receptor gating is tightly coupled to proton inhibition of the receptor. This idea is consistent with the observation that channel blockers appear to sense the protonation state of the receptor (Dravid et al., 2007).
The actions of ATD modulators appear to involve a change in the pKa of the proton sensor that leads to enhancement or reduction of tonic proton inhibition at physiological pH. Thus, both Zn2+ and ifenprodil enhance proton sensitivity, which will increase tonic inhibition at resting pH (Pahk and Williams, 1997; Mott et al., 1998; Traynelis et al., 1998; Choi and Lipton, 1999; Erreger and Traynelis, 2008; Bhatt et al., 2013). In contrast, the binding of extracellular polyamines reduces the sensitivity to extracellular pH, resulting in potentiation due to reduced tonic inhibition by physiological levels of protons (Traynelis et al., 1995; Kashiwagi et al., 1996, 1997).
Actions of extracellular Zn2+
Extracellular Zn2+ binds with high affinity to the GluN2A ATD, with an IC50 value in the nanomolar range at GluN1/GluN2A receptors (Williams, 1996; Chen et al., 1997; Paoletti et al., 1997; Traynelis et al., 1998). In contrast, the IC50 for Zn2+ inhibition of GluN1/GluN2B receptors resulting from binding to the GluN2B ATD is in the low micromolar range (Rachline et al., 2005). Crystallographic and functional data show that the Zn2+-binding site is located within the cleft formed by the two lobes R1 and R2 of the ATD (Karakas et al., 2009; Romero-Hernandez et al., 2016). Multiple observations suggest a mechanism of Zn2+ modulation that involves a change in the angle between the two lobes R1 and R2, in addition to twisting motions around the hinge region of the bilobed ATD clamshell (Paoletti et al., 2000; Gielen et al., 2008; Karakas et al., 2009; Romero-Hernandez et al., 2016). Binding of Zn2+ stabilizes a conformation of the GluN2 ATD, which is presumably accompanied by structural changes at the GluN1/GluN2 ABD layers that favor channel closure (Gielen et al., 2008; Romero-Hernandez et al., 2016). Previous studies have suggested that Zn2+ binding can enhance the proton sensitivity (Choi and Lipton, 1999). In support of this idea, there is a strong correlation between mutations that perturb the IC50 of Zn2+ inhibition and their effect on proton IC50 (Traynelis et al., 1998). Furthermore, single-channel analysis can detect changes in the protonation rates for Zn2+-bound receptors, supporting the idea that Zn2+ alters the equilibrium between NMDA receptors and protons at physiological pH (Erreger and Traynelis, 2008). The incomplete inhibition by high-affinity Zn2+ binding is consistent with enhancement of proton sensitivity, because Zn2+ binding produces a leftward shift of the proton inhibition curve, leading to more complete inhibition at acidic pH (Traynelis et al., 1998; Choi and Lipton, 1999; Low et al., 2000; Erreger and Traynelis, 2008). Interestingly, triheteromeric GluN1/2A/2B receptors retain a high-affinity Zn2+ binding, although there is reduced inhibition at maximally effective concentrations of Zn2+ (Hatton and Paoletti, 2005; Hansen et al., 2014; Stroebel et al., 2014). Higher concentrations of Zn2+ can produce a voltage-dependent channel block (Williams, 1996), but it remains unclear whether changes in extracellular Zn2+ in brain tissue are large enough to produce voltage-dependent channel block (Vogt et al., 2000; Anderson et al., 2015).
The affinity for Zn2+ at the GluN2A ATD is high enough such that Zn2+ contamination in physiological levels of salts can produce significant inhibition (Paoletti et al., 1997). The effects of contaminant Zn2+ in functional experiments can be removed by inclusion of even low concentrations of divalent ion chelators, such as 10 µM EDTA. The high affinity of such chelators for Zn2+ means that even low micromolar levels of chelator will bind virtually all of the nanomolar-contaminating Zn2+ ions but exert minimal effects on millimolar concentrations of Ca2+ or Mg2+ (Anderson et al., 2015).
Positive and negative allosteric modulation by neurosteroids
Several endogenous neurosteroids positively and negatively modulate NMDA receptor activity (Traynelis et al., 2010), although the actions of these lipophilic molecules are complex. For instance, pregnenolone sulfate has dual actions on NMDA receptor responses, having both inhibitory and potentiating activity over a wide range of potencies (Horak et al., 2006). The potentiating actions of pregnenolone sulfate are most prominent when applied before receptor activation, whereas inhibitory actions arise when applied continuously (Horak et al., 2006). The dual actions of pregnenolone sulfate lead to divergent effects depending on the GluN2 subunit; when applied during steady-state NMDA receptor responses, GluN1/2A and GluN1/2B are potentiated, whereas GluN1/2C and GluN1/2D are inhibited.
Other neurosteroid analogues, such as pregnanolone sulfate, inhibit all NMDA receptor subtypes (i.e., they are pan-inhibitors) in a use-dependent manner through actions that involve the extracellular portion of the conserved M3 SYTANLAAF motif (Malayev et al., 2002). For GluN2A, pregnanolone sulfate has been proposed to increase the occupancy of a desensitized state (Kussius et al., 2009). In contrast, 24(S)-hydroxycholesterol and related analogues appear to be pan-potentiators, whereas 25(S)-hydroxycholesterol may antagonize actions of endogenous 24(S)-hydroxycholesterol (Paul et al., 2013; Linsenbardt et al., 2014). Some of these neurosteroids and analogues have been shown to exhibit agonist dependency, although this property is difficult to assess, because neurosteroids can have distinct actions on NMDA receptors, dependent on the timing of modulator application (i.e., see pregnenolone sulfate actions above). Additionally, steroid derivatives may partition into the membrane en route to their active site, which will alter the concentration–response relationship of their actions (Borovska et al., 2012; Vyklicky et al., 2015). A recent study reported that cholesterol modulates NMDA receptor function and its removal inhibited receptor activity (Korinek et al., 2015), suggesting that the membrane environment influences NMDA receptor activity and may be an important determinate of neurosteroid action. Although a clear binding site has not been resolved, it seems likely that neurosteroid derivatives interact directly with the receptor rather than simply alter membrane fluidity. A subset of neurosteroid inhibitors also have voltage-dependent actions, suggesting that they may inhibit NMDA receptors through blocking the channel (Vyklicky et al., 2015). These findings highlight the complexity associated with neurosteroid activity, and more work is required to delineate the mechanism of action of these compounds.
Desensitization of NMDA receptors
The process of desensitization is broadly defined as a decrease in a response in the continued presence of a stimulus. NMDA receptors exhibit several forms of desensitization, which can be distinguished on the basis of time course and mechanism, including glycine-, Zn2+-, and Ca2+-dependent desensitization. Most ligand-gated channels can desensitize in the continued presence of agonist by a mechanism thought to involve a conformational change to a stable and long-lived agonist-bound closed state. NMDA receptors can also desensitize in the continued presence of glutamate and glycine, presumably by this same mechanism, in a manner that is independent of glycine, Zn2+, and Ca2+. NMDA receptors exhibit only weak desensitization compared with the relatively strong desensitization of AMPA and kainate receptors. However, this desensitization is sensitive to intracellular dialysis, being more prominent in excised outside-out membrane patches (Sather et al., 1990, 1992), and is perturbed by mutations in a wide range of domains, including the conserved M3 gating motif, the pre-M1 linker region, the ion channel pore, the ABD, and the TMD–ABD interface (Chen et al., 2004; Hu and Zheng, 2005; Alsaloum et al., 2016).
Glycine-dependent NMDA receptor desensitization
Glycine-dependent NMDA receptor desensitization is present only in subsaturating glycine concentrations (Mayer et al., 1989) and occurs as a result of a negative allosteric interaction between the glutamate- and glycine-binding sites, such that the binding of glutamate decreases the glycine affinity and vice versa (Benveniste et al., 1990; Lester et al., 1993). Thus, when glutamate binds to GluN2 in the absence of high concentrations of glycine, the current will initially rise to a peak and then decline to a new equilibrium as glycine unbinds from the receptor after the allosteric reduction in glycine affinity. The time course for the desensitization is dictated by glycine unbinding (time constant ∼0.3 s) and is temporally close to the synaptic NMDA receptor time course, raising the possibility that glycine-dependent desensitization could impact synaptic signaling when glycine is subsaturating (Berger et al., 1998). Recent structural data for NMDA receptors provide plausible models for the negative allosteric coupling between glutamate- and glycine-binding sites, given their close proximity (Karakas and Furukawa, 2014; Lee et al., 2014; Tajima et al., 2016; Zhu et al., 2016). However, the structural features that enable glycine-dependent desensitization remain poorly understood.
Zn2+-dependent NMDA receptor desensitization
Extracellular Zn2+ inhibits GluN1/GluN2A and GluN1/GluN2B receptors in a voltage-independent manner through a binding site located in the ATD (Williams, 1996; Traynelis et al., 1998; Choi and Lipton, 1999; Fayyazuddin et al., 2000; Low et al., 2000; Paoletti et al., 2000; Rachline et al., 2005; Karakas et al., 2009). However, NMDA receptors also display a rapid component of desensitization in the presence of extracellular Zn2+ that occurs by a mechanism similar to glycine-dependent desensitization (Chen et al., 1997). This is the result of a positive intrasubunit allosteric interaction between glutamate binding to the GluN2 ABD and Zn2+ binding to the GluN2A ATD (Zheng et al., 2001; Erreger and Traynelis, 2005). As a result, in the presence of subsaturating concentrations of Zn2+, the glutamate-induced increase in Zn2+ affinity will cause a relaxation of the response to a new equilibrium as more Zn2+ ions bind and inhibit the receptor. The time course for Zn2+-dependent desensitization therefore follows the time course for Zn2+ binding.
Ca2+-dependent NMDA receptor inactivation
NMDA receptors also undergo Ca2+-dependent desensitization or inactivation, which requires an increase in intracellular Ca2+ over several seconds (Clark et al., 1990; Legendre et al., 1993; Vyklický, 1993; Rosenmund et al., 1995). The magnitude of this form of desensitization varies with different NMDA receptor subtypes; it is most prominent for GluN2A-containing NMDA receptors and more limited for GluN2B- and GluN2C-containing NMDA receptors (Medina et al., 1995; Krupp et al., 1996). The proposed mechanism involves an increase in the intracellular Ca2+ in the vicinity of the NMDA receptor that triggers uncoupling of the receptor from filamentous actin (Rosenmund and Westbrook, 1993). In addition, calmodulin binding to the GluN1 CTD may play a role in this form of desensitization (Ehlers et al., 1996; Rycroft and Gibb, 2002; Iacobucci and Popescu, 2017b); Ca2+-dependent desensitization is absent in NMDA receptors containing GluN1 splice variants that lack calmodulin-binding sites (Ehlers et al., 1996, 1998) or harbor mutations in calmodulin-binding sites in the GluN1 CTD (Zhang et al., 1998; Krupp et al., 1999).
Conclusion
Recent discoveries from genetic analyses that link NMDA receptors to specific disease conditions, the emerging evidence of the antidepressant effects of NMDA receptor antagonists, and accelerating identification of new subunit-selective modulators have reinvigorated the long-standing interest in NMDA receptors as therapeutic targets. The field now seems poised to achieve new levels of understanding of the functional roles of NMDA receptors in physiology and disease. The rapidly increasing volume of crystallographic and cryo-EM data has created an opportunity to view function through structure, which will inevitably improve our insight to the mechanisms by which agonist binding is linked to channel gating and how different NMDA receptor subunits contribute to the conformational changes that occur during gating and allosteric modulation. These new developments can lead novel perspectives in the NMDA receptor field and create exciting opportunities to study the physiological roles of different diheteromeric and triheteromeric NMDA receptor subtypes in distinct subcellular locations and neuronal populations. These converging advances in NMDA receptor pharmacology, mechanistic understanding of receptor function, and the basis of CNS diseases involving NMDA receptor dysfunction are already catalyzing the development of new therapeutic strategies.
Acknowledgments
This work was supported by grants from National Institutes of Health to S.F. Traynelis (NS036654 and NS065371), K.B. Hansen (GM103546 and NS097536), and L.P. Wollmuth (NS088479).
S.F. Traynelis is a consultant for Janssen Pharmaceuticals, Inc., the principal investigator on a research grant awarded to Emory University from Janssen Pharmaceuticals, a member of the Scientific Advisory Board for Sage Therapeutics, and co-founder of NeurOp, Inc. K.B. Hansen is the principal investigator on a research grant awarded to University of Montana from Janssen Pharmaceuticals. The other authors declare no competing financial interests.
Author contributions: All authors wrote the manuscript.
Lesley C. Anson served as editor.
References
- Abdallah C.G., Sanacora G., Duman R.S., and Krystal J.H.. 2015. Ketamine and rapid-acting antidepressants: a window into a new neurobiology for mood disorder therapeutics. Annu. Rev. Med. 66:509–523. 10.1146/annurev-med-053013-062946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi S., Muth-Selbach U., Lauterbach A., Lipfert P., Neuhuber W.L., and Zeilhofer H.U.. 2003. Facilitation of spinal NMDA receptor currents by spillover of synaptically released glycine. Science. 300:2094–2097. 10.1126/science.1083970 [DOI] [PubMed] [Google Scholar]
- Aizenman E., Lipton S.A., and Loring R.H.. 1989. Selective modulation of NMDA responses by reduction and oxidation. Neuron. 2:1257–1263. 10.1016/0896-6273(89)90310-3 [DOI] [PubMed] [Google Scholar]
- Akazawa C., Shigemoto R., Bessho Y., Nakanishi S., and Mizuno N.. 1994. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J. Comp. Neurol. 347:150–160. 10.1002/cne.903470112 [DOI] [PubMed] [Google Scholar]
- Al-Hallaq R.A., Conrads T.P., Veenstra T.D., and Wenthold R.J.. 2007. NMDA di-heteromeric receptor populations and associated proteins in rat hippocampus. J. Neurosci. 27:8334–8343. 10.1523/JNEUROSCI.2155-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaloum M., Kazi R., Gan Q., Amin J., and Wollmuth L.P.. 2016. A Molecular Determinant of Subtype-Specific Desensitization in Ionotropic Glutamate Receptors. J. Neurosci. 36:2617–2622. 10.1523/JNEUROSCI.2667-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman T.K., Maki B.A., Ruffino T.J., Kasperek E.M., and Popescu G.K.. 2014. Separate intramolecular targets for protein kinase A control N-methyl-D-aspartate receptor gating and Ca2+ permeability. J. Biol. Chem. 289:18805–18817. 10.1074/jbc.M113.537282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amico-Ruvio S.A., and Popescu G.K.. 2010. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys. J. 98:1160–1169. 10.1016/j.bpj.2009.12.4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amico-Ruvio S.A., Murthy S.E., Smith T.P., and Popescu G.K.. 2011. Zinc effects on NMDA receptor gating kinetics. Biophys. J. 100:1910–1918. 10.1016/j.bpj.2011.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.T., Radford R.J., Zastrow M.L., Zhang D.Y., Apfel U.P., Lippard S.J., and Tzounopoulos T.. 2015. Modulation of extrasynaptic NMDA receptors by synaptic and tonic zinc. Proc. Natl. Acad. Sci. USA. 112:E2705–E2714. 10.1073/pnas.1503348112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov S.M., and Johnson J.W.. 1999. Permeant ion regulation of N-methyl-D-aspartate receptor channel block by Mg(2+). Proc. Natl. Acad. Sci. USA. 96:14571–14576. 10.1073/pnas.96.25.14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N., and Gouaux E.. 2000. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 28:165–181. 10.1016/S0896-6273(00)00094-5 [DOI] [PubMed] [Google Scholar]
- Armstrong N., Sun Y., Chen G.Q., and Gouaux E.. 1998. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 395:913–917. 10.1038/27692 [DOI] [PubMed] [Google Scholar]
- Arvola M., and Keinänen K.. 1996. Characterization of the ligand-binding domains of glutamate receptor (GluR)-B and GluR-D subunits expressed in Escherichia coli as periplasmic proteins. J. Biol. Chem. 271:15527–15532. 10.1074/jbc.271.26.15527 [DOI] [PubMed] [Google Scholar]
- Ascher P., Bregestovski P., and Nowak L.. 1988. N-methyl-D-aspartate-activated channels of mouse central neurones in magnesium-free solutions. J. Physiol. 399:207–226. 10.1113/jphysiol.1988.sp017076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlason P.T., Garside M.L., Meddows E., Whiting P., and McIlhinney R.A.. 2007. N-Methyl-D-aspartate (NMDA) receptor subunit NR1 forms the substrate for oligomeric assembly of the NMDA receptor. J. Biol. Chem. 282:25299–25307. 10.1074/jbc.M702778200 [DOI] [PubMed] [Google Scholar]
- Auerbach A., and Zhou Y.. 2005. Gating reaction mechanisms for NMDA receptor channels. J. Neurosci. 25:7914–7923. 10.1523/JNEUROSCI.1471-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke T.G., and Traynelis S.F.. 2003. Activation of NR1/NR2B NMDA receptors. Nat. Neurosci. 6:144–152. 10.1038/nn1000 [DOI] [PubMed] [Google Scholar]
- Banke T.G., Dravid S.M., and Traynelis S.F.. 2005. Protons trap NR1/NR2B NMDA receptors in a nonconducting state. J. Neurosci. 25:42–51. 10.1523/JNEUROSCI.3154-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barygin O.I., Gmiro V.E., Kim K.Kh., Magazanik L.G., and Tikhonov D.B.. 2009. Blockade of NMDA receptor channels by 9-aminoacridine and its derivatives. Neurosci. Lett. 451:29–33. 10.1016/j.neulet.2008.12.036 [DOI] [PubMed] [Google Scholar]
- Beck C., Wollmuth L.P., Seeburg P.H., Sakmann B., and Kuner T.. 1999. NMDAR channel segments forming the extracellular vestibule inferred from the accessibility of substituted cysteines. Neuron. 22:559–570. 10.1016/S0896-6273(00)80710-2 [DOI] [PubMed] [Google Scholar]
- Benveniste M., and Mayer M.L.. 1991. Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys. J. 59:560–573. 10.1016/S0006-3495(91)82272-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M., and Mayer M.L.. 1995. Trapping of glutamate and glycine during open channel block of rat hippocampal neuron NMDA receptors by 9-aminoacridine. J. Physiol. 483:367–384. 10.1113/jphysiol.1995.sp020591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M., Clements J., Vyklický L. Jr., and Mayer M.L.. 1990. A kinetic analysis of the modulation of N-methyl-D-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J. Physiol. 428:333–357. 10.1113/jphysiol.1990.sp018215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A.J., Dieudonné S., and Ascher P.. 1998. Glycine uptake governs glycine site occupancy at NMDA receptors of excitatory synapses. J. Neurophysiol. 80:3336–3340. 10.1152/jn.1998.80.6.3336 [DOI] [PubMed] [Google Scholar]
- Bergeron R., Meyer T.M., Coyle J.T., and Greene R.W.. 1998. Modulation of N-methyl-D-aspartate receptor function by glycine transport. Proc. Natl. Acad. Sci. USA. 95:15730–15734. 10.1073/pnas.95.26.15730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt J.M., Prakash A., Suryavanshi P.S., and Dravid S.M.. 2013. Effect of ifenprodil on GluN1/GluN2B N-methyl-D-aspartate receptor gating. Mol. Pharmacol. 83:9–21. 10.1124/mol.112.080952 [DOI] [PubMed] [Google Scholar]
- Billups D., and Attwell D.. 2003. Active release of glycine or D-serine saturates the glycine site of NMDA receptors at the cerebellar mossy fibre to granule cell synapse. Eur. J. Neurosci. 18:2975–2980. 10.1111/j.1460-9568.2003.02996.x [DOI] [PubMed] [Google Scholar]
- Blanpied T.A., Boeckman F.A., Aizenman E., and Johnson J.W.. 1997. Trapping channel block of NMDA-activated responses by amantadine and memantine. J. Neurophysiol. 77:309–323. 10.1152/jn.1997.77.1.309 [DOI] [PubMed] [Google Scholar]
- Blanpied T.A., Clarke R.J., and Johnson J.W.. 2005. Amantadine inhibits NMDA receptors by accelerating channel closure during channel block. J. Neurosci. 25:3312–3322. 10.1523/JNEUROSCI.4262-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshakov K.V., Gmiro V.E., Tikhonov D.B., and Magazanik L.G.. 2003. Determinants of trapping block of N-methyl-d-aspartate receptor channels. J. Neurochem. 87:56–65. 10.1046/j.1471-4159.2003.01956.x [DOI] [PubMed] [Google Scholar]
- Borovska J., Vyklicky V., Stastna E., Kapras V., Slavikova B., Horak M., Chodounska H., and Vyklicky L. Jr. 2012. Access of inhibitory neurosteroids to the NMDA receptor. Br. J. Pharmacol. 166:1069–1083. 10.1111/j.1476-5381.2011.01816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borschel W.F., Cummings K.A., Tindell L.K., and Popescu G.K.. 2015. Kinetic contributions to gating by interactions unique to N-methyl-D-aspartate (NMDA) receptors. J. Biol. Chem. 290:26846–26855. 10.1074/jbc.M115.678656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H.R., and Nicoll R.. 1993. Molecular machines integrate coincident synaptic signals. Cell. 72(Suppl):65–75. 10.1016/S0092-8674(05)80029-7 [DOI] [PubMed] [Google Scholar]
- Brackley P.T., Bell D.R., Choi S.K., Nakanishi K., and Usherwood P.N.. 1993. Selective antagonism of native and cloned kainate and NMDA receptors by polyamine-containing toxins. J. Pharmacol. Exp. Ther. 266:1573–1580. [PubMed] [Google Scholar]
- Brickley S.G., Misra C., Mok M.H., Mishina M., and Cull-Candy S.G.. 2003. NR2B and NR2D subunits coassemble in cerebellar Golgi cells to form a distinct NMDA receptor subtype restricted to extrasynaptic sites. J. Neurosci. 23:4958–4966. 10.1523/JNEUROSCI.23-12-04958.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimecombe J.C., Boeckman F.A., and Aizenman E.. 1997. Functional consequences of NR2 subunit composition in single recombinant N-methyl-D-aspartate receptors. Proc. Natl. Acad. Sci. USA. 94:11019–11024. 10.1073/pnas.94.20.11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothwell S.L., Barber J.L., Monaghan D.T., Jane D.E., Gibb A.J., and Jones S.. 2008. NR2B- and NR2D-containing synaptic NMDA receptors in developing rat substantia nigra pars compacta dopaminergic neurones. J. Physiol. 586:739–750. 10.1113/jphysiol.2007.144618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger P.B., Yuan H., Karakas E., Geballe M., Furukawa H., Liotta D.C., Snyder J.P., and Traynelis S.F.. 2012. Mapping the binding of GluN2B-selective N-methyl-D-aspartate receptor negative allosteric modulators. Mol. Pharmacol. 82:344–359. 10.1124/mol.112.078568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N., Schoepfer R., Monyer H., Ruppersberg J.P., Günther W., Seeburg P.H., and Sakmann B.. 1992. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science. 257:1415–1419. 10.1126/science.1382314 [DOI] [PubMed] [Google Scholar]
- Burnashev N., Zhou Z., Neher E., and Sakmann B.. 1995. Fractional calcium currents through recombinant GluR channels of the NMDA, AMPA and kainate receptor subtypes. J. Physiol. 485:403–418. 10.1113/jphysiol.1995.sp020738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell E.S., Irvine M., Fang G., Sapkota K., Jane D.E., and Monaghan D.T.. 2018. Positive and Negative Allosteric Modulators of N-Methyl-d-aspartate (NMDA) Receptors: Structure-Activity Relationships and Mechanisms of Action. J. Med. Chem. 10.1021/acs.jmedchem.7b01640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B.C., and Jahr C.E.. 2016. Postsynaptic, not presynaptic NMDA receptors are required for spike-timing-dependent LTD induction. Nat. Neurosci. 19:1218–1224. 10.1038/nn.4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathala L., Misra C., and Cull-Candy S.. 2000. Developmental profile of the changing properties of NMDA receptors at cerebellar mossy fiber-granule cell synapses. J. Neurosci. 20:5899–5905. 10.1523/JNEUROSCI.20-16-05899.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavara N.A., and Hollmann M.. 2008. Shuffling the deck anew: how NR3 tweaks NMDA receptor function. Mol. Neurobiol. 38:16–26. 10.1007/s12035-008-8029-9 [DOI] [PubMed] [Google Scholar]
- Chang H.R., and Kuo C.C.. 2008. The activation gate and gating mechanism of the NMDA receptor. J. Neurosci. 28:1546–1556. 10.1523/JNEUROSCI.3485-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazot P.L., and Stephenson F.A.. 1997. Molecular dissection of native mammalian forebrain NMDA receptors containing the NR1 C2 exon: direct demonstration of NMDA receptors comprising NR1, NR2A, and NR2B subunits within the same complex. J. Neurochem. 69:2138–2144. 10.1046/j.1471-4159.1997.69052138.x [DOI] [PubMed] [Google Scholar]
- Chazot P.L., Coleman S.K., Cik M., and Stephenson F.A.. 1994. Molecular characterization of N-methyl-D-aspartate receptors expressed in mammalian cells yields evidence for the coexistence of three subunit types within a discrete receptor molecule. J. Biol. Chem. 269:24403–24409. [PubMed] [Google Scholar]
- Chen H.S., and Lipton S.A.. 1997. Mechanism of memantine block of NMDA-activated channels in rat retinal ganglion cells: uncompetitive antagonism. J. Physiol. 499:27–46. 10.1113/jphysiol.1997.sp021909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.S., and Lipton S.A.. 2006. The chemical biology of clinically tolerated NMDA receptor antagonists. J. Neurochem. 97:1611–1626. 10.1111/j.1471-4159.2006.03991.x [DOI] [PubMed] [Google Scholar]
- Chen N., Moshaver A., and Raymond L.A.. 1997. Differential sensitivity of recombinant N-methyl-D-aspartate receptor subtypes to zinc inhibition. Mol. Pharmacol. 51:1015–1023. 10.1124/mol.51.6.1015 [DOI] [PubMed] [Google Scholar]
- Chen N., Li B., Murphy T.H., and Raymond L.A.. 2004. Site within N-Methyl-D-aspartate receptor pore modulates channel gating. Mol. Pharmacol. 65:157–164. 10.1124/mol.65.1.157 [DOI] [PubMed] [Google Scholar]
- Chen P.E., Geballe M.T., Katz E., Erreger K., Livesey M.R., O’Toole K.K., Le P., Lee C.J., Snyder J.P., Traynelis S.F., and Wyllie D.J.. 2008. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J. Physiol. 586:227–245. 10.1113/jphysiol.2007.143172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Tankovic A., Burger P.B., Kusumoto H., Traynelis S.F., and Yuan H.. 2017. Functional Evaluation of a De Novo GRIN2A Mutation Identified in a Patient with Profound Global Developmental Delay and Refractory Epilepsy. Mol. Pharmacol. 91:317–330. 10.1124/mol.116.106781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyan J., Balsara R.D., Hansen K.B., and Castellino F.J.. 2016. Pharmacology of triheteromeric N-Methyl-D-Aspartate Receptors. Neurosci. Lett. 617:240–246. 10.1016/j.neulet.2016.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler M. 2003. Regulation and modulation of pH in the brain. Physiol. Rev. 83:1183–1221. 10.1152/physrev.00010.2003 [DOI] [PubMed] [Google Scholar]
- Choi Y.B., and Lipton S.A.. 1999. Identification and mechanism of action of two histidine residues underlying high-affinity Zn2+ inhibition of the NMDA receptor. Neuron. 23:171–180. 10.1016/S0896-6273(00)80763-1 [DOI] [PubMed] [Google Scholar]
- Choi Y.B., and Lipton S.A.. 2000. Redox modulation of the NMDA receptor. Cell. Mol. Life Sci. 57:1535–1541. 10.1007/PL00000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chen H.V., and Lipton S.A.. 2001. Three pairs of cysteine residues mediate both redox and zn2+ modulation of the nmda receptor. J. Neurosci. 21:392–400. 10.1523/JNEUROSCI.21-02-00392.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A., and Malenka R.C.. 2008. Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology. 33:18–41. 10.1038/sj.npp.1301559 [DOI] [PubMed] [Google Scholar]
- Clark G.D., Clifford D.B., and Zorumski C.F.. 1990. The effect of agonist concentration, membrane voltage and calcium on N-methyl-D-aspartate receptor desensitization. Neuroscience. 39:787–797. 10.1016/0306-4522(90)90261-2 [DOI] [PubMed] [Google Scholar]
- Clarke R.J., and Johnson J.W.. 2006. NMDA receptor NR2 subunit dependence of the slow component of magnesium unblock. J. Neurosci. 26:5825–5834. 10.1523/JNEUROSCI.0577-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J.D., and Westbrook G.L.. 1991. Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron. 7:605–613. 10.1016/0896-6273(91)90373-8 [DOI] [PubMed] [Google Scholar]
- Clements J.D., and Westbrook G.L.. 1994. Kinetics of AP5 dissociation from NMDA receptors: evidence for two identical cooperative binding sites. J. Neurophysiol. 71:2566–2569. 10.1152/jn.1994.71.6.2566 [DOI] [PubMed] [Google Scholar]
- Clements J.D., Lester R.A., Tong G., Jahr C.E., and Westbrook G.L.. 1992. The time course of glutamate in the synaptic cleft. Science. 258:1498–1501. 10.1126/science.1359647 [DOI] [PubMed] [Google Scholar]
- Dai J., and Zhou H.X.. 2015. Reduced curvature of ligand-binding domain free-energy surface underlies partial agonism at NMDA receptors. Structure. 23:228–236. 10.1016/j.str.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Wollmuth L.P., and Zhou H.X.. 2015. Mechanism-Based Mathematical Model for Gating of Ionotropic Glutamate Receptors. J. Phys. Chem. B. 119:10934–10940. 10.1021/acs.jpcb.5b00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divito C.B., and Underhill S.M.. 2014. Excitatory amino acid transporters: roles in glutamatergic neurotransmission. Neurochem. Int. 73:172–180. 10.1016/j.neuint.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolino D.M., Cooper D., Ramaswamy S., Jaurich H., Landes C.F., and Jayaraman V.. 2015. Structural dynamics of the glycine-binding domain of the N-methyl-D-aspartate receptor. J. Biol. Chem. 290:797–804. 10.1074/jbc.M114.605436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolino D.M., Rezaei Adariani S., Shaikh S.A., Jayaraman V., and Sanabria H.. 2016. Conformational Selection and Submillisecond Dynamics of the Ligand-binding Domain of the N-Methyl-d-aspartate Receptor. J. Biol. Chem. 291:16175–16185. 10.1074/jbc.M116.721274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid S.M., Erreger K., Yuan H., Nicholson K., Le P., Lyuboslavsky P., Almonte A., Murray E., Mosely C., Barber J., et al. 2007. Subunit-specific mechanisms and proton sensitivity of NMDA receptor channel block. J. Physiol. 581:107–128. 10.1113/jphysiol.2006.124958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid S.M., Prakash A., and Traynelis S.F.. 2008. Activation of recombinant NR1/NR2C NMDA receptors. J. Physiol. 586:4425–4439. 10.1113/jphysiol.2008.158634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid S.M., Burger P.B., Prakash A., Geballe M.T., Yadav R., Le P., Vellano K., Snyder J.P., and Traynelis S.F.. 2010. Structural determinants of D-cycloserine efficacy at the NR1/NR2C NMDA receptors. J. Neurosci. 30:2741–2754. 10.1523/JNEUROSCI.5390-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah A.W., and Standaert D.G.. 2003. Subcellular segregation of distinct heteromeric NMDA glutamate receptors in the striatum. J. Neurochem. 85:935–943. 10.1046/j.1471-4159.2003.01744.x [DOI] [PubMed] [Google Scholar]
- Dunah A.W., Luo J., Wang Y.H., Yasuda R.P., and Wolfe B.B.. 1998. Subunit composition of N-methyl-D-aspartate receptors in the central nervous system that contain the NR2D subunit. Mol. Pharmacol. 53:429–437. 10.1124/mol.53.3.429 [DOI] [PubMed] [Google Scholar]
- Durand G.M., Gregor P., Zheng X., Bennett M.V., Uhl G.R., and Zukin R.S.. 1992. Cloning of an apparent splice variant of the rat N-methyl-D-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc. Natl. Acad. Sci. USA. 89:9359–9363. 10.1073/pnas.89.19.9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand G.M., Bennett M.V., and Zukin R.S.. 1993. Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc. Natl. Acad. Sci. USA. 90:6731–6735. 10.1073/pnas.90.14.6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M.D., Zhang S., Bernhadt J.P., and Huganir R.L.. 1996. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 84:745–755. 10.1016/S0092-8674(00)81052-1 [DOI] [PubMed] [Google Scholar]
- Ehlers M.D., Fung E.T., O’Brien R.J., and Huganir R.L.. 1998. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J. Neurosci. 18:720–730. 10.1523/JNEUROSCI.18-02-00720.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K., and Traynelis S.F.. 2005. Allosteric interaction between zinc and glutamate binding domains on NR2A causes desensitization of NMDA receptors. J. Physiol. 569:381–393. 10.1113/jphysiol.2005.095497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K., and Traynelis S.F.. 2008. Zinc inhibition of rat NR1/NR2A N-methyl-D-aspartate receptors. J. Physiol. 586:763–778. 10.1113/jphysiol.2007.143941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K., Chen P.E., Wyllie D.J., and Traynelis S.F.. 2004. Glutamate receptor gating. Crit. Rev. Neurobiol. 16:187–224. 10.1615/CritRevNeurobiol.v16.i3.10 [DOI] [PubMed] [Google Scholar]
- Erreger K., Dravid S.M., Banke T.G., Wyllie D.J., and Traynelis S.F.. 2005a Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J. Physiol. 563:345–358. 10.1113/jphysiol.2004.080028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K., Geballe M.T., Dravid S.M., Snyder J.P., Wyllie D.J., and Traynelis S.F.. 2005b Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J. Neurosci. 25:7858–7866. 10.1523/JNEUROSCI.1613-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K., Geballe M.T., Kristensen A., Chen P.E., Hansen K.B., Lee C.J., Yuan H., Le P., Lyuboslavsky P.N., Micale N., et al. 2007. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol. Pharmacol. 72:907–920. 10.1124/mol.107.037333 [DOI] [PubMed] [Google Scholar]
- Farin A., and Marshall L.F.. 2004. Lessons from epidemiologic studies in clinical trials of traumatic brain injury. Acta Neurochir. Suppl. (Wien). 89:101–107. [DOI] [PubMed] [Google Scholar]
- Farina A.N., Blain K.Y., Maruo T., Kwiatkowski W., Choe S., and Nakagawa T.. 2011. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J. Neurosci. 31:3565–3579. 10.1523/JNEUROSCI.6041-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyazuddin A., Villarroel A., Le Goff A., Lerma J., and Neyton J.. 2000. Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron. 25:683–694. 10.1016/S0896-6273(00)81070-3 [DOI] [PubMed] [Google Scholar]
- Furukawa H., and Gouaux E.. 2003. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 22:2873–2885. 10.1093/emboj/cdg303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H., Singh S.K., Mancusso R., and Gouaux E.. 2005. Subunit arrangement and function in NMDA receptors. Nature. 438:185–192. 10.1038/nature04089 [DOI] [PubMed] [Google Scholar]
- Geiger J.R.P., Lübke J., Roth A., Frotscher M., and Jonas P.. 1997. Submillisecond AMPA receptor-mediated signaling at a principal neuron-interneuron synapse. Neuron. 18:1009–1023. 10.1016/S0896-6273(00)80339-6 [DOI] [PubMed] [Google Scholar]
- Gibb A.J., and Colquhoun D.. 1992. Activation of N-methyl-D-aspartate receptors by L-glutamate in cells dissociated from adult rat hippocampus. J. Physiol. 456:143–179. 10.1113/jphysiol.1992.sp019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb A., Ogden K.K., McDaniel M.J., Vance K.M., Kell S.A., Butch C., Burger P., Liotta D.C., and Traynelis S.F.. 2018. A structurally-derived model of subunit-dependent NMDA receptor function. J. Physiol. 10.1113/JP276093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M., Le Goff A., Stroebel D., Johnson J.W., Neyton J., and Paoletti P.. 2008. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 57:80–93. 10.1016/j.neuron.2007.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M., Siegler Retchless B., Mony L., Johnson J.W., and Paoletti P.. 2009. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 459:703–707. 10.1038/nature07993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffard R.G., Monyer H., Christine C.W., and Choi D.W.. 1990. Acidosis reduces NMDA receptor activation, glutamate neurotoxicity, and oxygen-glucose deprivation neuronal injury in cortical cultures. Brain Res. 506:339–342. 10.1016/0006-8993(90)91276-M [DOI] [PubMed] [Google Scholar]
- Glasgow N.G., Siegler Retchless B., and Johnson J.W.. 2015. Molecular bases of NMDA receptor subtype-dependent properties. J. Physiol. 593:83–95. 10.1113/jphysiol.2014.273763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow N.G., Povysheva N.V., Azofeifa A.M., and Johnson J.W.. 2017. Memantine and Ketamine Differentially Alter NMDA Receptor Desensitization. J. Neurosci. 37:9686–9704. 10.1523/JNEUROSCI.1173-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger A.J., and Nicoll R.A.. 2013. Expression mechanisms underlying long-term potentiation: a postsynaptic view, 10 years on. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130136 10.1098/rstb.2013.0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.A., Shi Y., Usui H., During M.J., Sakimura K., and Nicoll R.A.. 2011. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 71:1085–1101. 10.1016/j.neuron.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackos D.H., and Hanson J.E.. 2017. Diverse modes of NMDA receptor positive allosteric modulation: Mechanisms and consequences. Neuropharmacology. 112(Pt A):34–45. 10.1016/j.neuropharm.2016.07.037 [DOI] [PubMed] [Google Scholar]
- Hackos D.H., Lupardus P.J., Grand T., Chen Y., Wang T.M., Reynen P., Gustafson A., Wallweber H.J., Volgraf M., Sellers B.D., et al. 2016. Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function. Neuron. 89:983–999. 10.1016/j.neuron.2016.01.016 [DOI] [PubMed] [Google Scholar]
- Hansen K.B., Bräuner-Osborne H., and Egebjerg J.. 2008. Pharmacological characterization of ligands at recombinant NMDA receptor subtypes by electrophysiological recordings and intracellular calcium measurements. Comb. Chem. High Throughput Screen. 11:304–315. 10.2174/138620708784246040 [DOI] [PubMed] [Google Scholar]
- Hansen K.B., Tajima N., Risgaard R., Perszyk R.E., Jørgensen L., Vance K.M., Ogden K.K., Clausen R.P., Furukawa H., and Traynelis S.F.. 2013. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol. Pharmacol. 84:114–127. 10.1124/mol.113.085803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.B., Ogden K.K., Yuan H., and Traynelis S.F.. 2014. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 81:1084–1096. 10.1016/j.neuron.2014.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.B., Yi F., Perszyk R.E., Menniti F.S., and Traynelis S.F.. 2017. NMDA Receptors in the Central Nervous System. Methods Mol. Biol. 1677:1–80. 10.1007/978-1-4939-7321-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton C.J., and Paoletti P.. 2005. Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron. 46:261–274. 10.1016/j.neuron.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Henson M.A., Roberts A.C., Pérez-Otaño I., and Philpot B.D.. 2010. Influence of the NR3A subunit on NMDA receptor functions. Prog. Neurobiol. 91:23–37. 10.1016/j.pneurobio.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hestrin S., Nicoll R.A., Perkel D.J., and Sah P.. 1990. Analysis of excitatory synaptic action in pyramidal cells using whole-cell recording from rat hippocampal slices. J. Physiol. 422:203–225. 10.1113/jphysiol.1990.sp017980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley M.J., and Sabatini B.L.. 2012. Calcium signaling in dendritic spines. Cold Spring Harb. Perspect. Biol. 4:a005686 10.1101/cshperspect.a005686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M., Boulter J., Maron C., Beasley L., Sullivan J., Pecht G., and Heinemann S.. 1993. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 10:943–954. 10.1016/0896-6273(93)90209-A [DOI] [PubMed] [Google Scholar]
- Holtmaat A., and Svoboda K.. 2009. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10:647–658. 10.1038/nrn2699 [DOI] [PubMed] [Google Scholar]
- Horak M., Vlcek K., Chodounska H., and Vyklicky L. Jr. 2006. Subtype-dependence of N-methyl-D-aspartate receptor modulation by pregnenolone sulfate. Neuroscience. 137:93–102. 10.1016/j.neuroscience.2005.08.058 [DOI] [PubMed] [Google Scholar]
- Howe J.R., Cull-Candy S.G., and Colquhoun D.. 1991. Currents through single glutamate receptor channels in outside-out patches from rat cerebellar granule cells. J. Physiol. 432:143–202. 10.1113/jphysiol.1991.sp018381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., and Zheng F.. 2005. Molecular determinants of glycine-independent desensitization of NR1/NR2A receptors. J. Pharmacol. Exp. Ther. 313:563–569. 10.1124/jpet.104.080168 [DOI] [PubMed] [Google Scholar]
- Huang Z., and Gibb A.J.. 2014. Mg2+ block properties of triheteromeric GluN1-GluN2B-GluN2D NMDA receptors on neonatal rat substantia nigra pars compacta dopaminergic neurones. J. Physiol. 592:2059–2078. 10.1113/jphysiol.2013.267864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner J.E., and Bean B.P.. 1988. Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: selective binding to open channels. Proc. Natl. Acad. Sci. USA. 85:1307–1311. 10.1073/pnas.85.4.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D.L., and Castillo P.E.. 2012. Synaptic plasticity of NMDA receptors: mechanisms and functional implications. Curr. Opin. Neurobiol. 22:496–508. 10.1016/j.conb.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci G.J., and Popescu G.K.. 2017a NMDA receptors: linking physiological output to biophysical operation. Nat. Rev. Neurosci. 18:236–249. 10.1038/nrn.2017.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci G.J., and Popescu G.K.. 2017b Resident Calmodulin Primes NMDA Receptors for Ca2+-Dependent Inactivation. Biophys. J. 113:2236–2248. 10.1016/j.bpj.2017.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidou C., and Turski L.. 2002. Why did NMDA receptor antagonists fail clinical trials for stroke and traumatic brain injury? Lancet Neurol. 1:383–386. 10.1016/S1474-4422(02)00164-3 [DOI] [PubMed] [Google Scholar]
- Inanobe A., Furukawa H., and Gouaux E.. 2005. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 47:71–84. 10.1016/j.neuron.2005.05.022 [DOI] [PubMed] [Google Scholar]
- Ishii T., Moriyoshi K., Sugihara H., Sakurada K., Kadotani H., Yokoi M., Akazawa C., Shigemoto R., Mizuno N., Masu M., et al. 1993. Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J. Biol. Chem. 268:2836–2843. [PubMed] [Google Scholar]
- Jatzke C., Watanabe J., and Wollmuth L.P.. 2002. Voltage and concentration dependence of Ca(2+) permeability in recombinant glutamate receptor subtypes. J. Physiol. 538:25–39. 10.1113/jphysiol.2001.012897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen A., Tajima N., Fernandez-Cuervo G., Garnier-Amblard E.C., and Furukawa H.. 2014. Structural insights into competitive antagonism in NMDA receptors. Neuron. 81:366–378. 10.1016/j.neuron.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen M., Frederiksen K., Yi F., Clausen R.P., Hansen K.B., Bräuner-Osborne H., Kilburn P., and Damholt A.. 2017. Identification of AICP as a GluN2C-Selective N-Methyl-d-Aspartate Receptor Superagonist at the GluN1 Glycine Site. Mol. Pharmacol. 92:151–161. 10.1124/mol.117.108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R., Banke T.G., Mayer M.L., Traynelis S.F., and Gouaux E.. 2003. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 6:803–810. 10.1038/nn1091 [DOI] [PubMed] [Google Scholar]
- Jin R., Clark S., Weeks A.M., Dudman J.T., Gouaux E., and Partin K.M.. 2005. Mechanism of positive allosteric modulators acting on AMPA receptors. J. Neurosci. 25:9027–9036. 10.1523/JNEUROSCI.2567-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.W., and Ascher P.. 1987. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 325:529–531. 10.1038/325529a0 [DOI] [PubMed] [Google Scholar]
- Johnson J.W., Glasgow N.G., and Povysheva N.V.. 2015. Recent insights into the mode of action of memantine and ketamine. Curr. Opin. Pharmacol. 20:54–63. 10.1016/j.coph.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., and Gibb A.J.. 2005. Functional NR2B- and NR2D-containing NMDA receptor channels in rat substantia nigra dopaminergic neurones. J. Physiol. 569:209–221. 10.1113/jphysiol.2005.095554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.S., VanDongen H.M.A., and VanDongen A.M.J.. 2002. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J. Neurosci. 22:2044–2053. 10.1523/JNEUROSCI.22-06-02044.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T.M., Kell S.A., Kusumoto H., Shaulsky G., Bhattacharya S., Epplin M.P., Strong K.L., Miller E.J., Cox B.D., Menaldino D.S., et al. 2018. The Bioactive Protein-Ligand Conformation of GluN2C-Selective Positive Allosteric Modulators Bound to the NMDA Receptor. Mol. Pharmacol. 93:141–156. 10.1124/mol.117.110940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbaugh T.L., VanDongen H.M., and VanDongen A.M.. 2004. Ligand-binding residues integrate affinity and efficacy in the NMDA receptor. Mol. Pharmacol. 66:209–219. 10.1124/mol.66.2.209 [DOI] [PubMed] [Google Scholar]
- Karakas E., and Furukawa H.. 2014. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 344:992–997. 10.1126/science.1251915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E., Simorowski N., and Furukawa H.. 2009. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 28:3910–3920. 10.1038/emboj.2009.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E., Simorowski N., and Furukawa H.. 2011. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 475:249–253. 10.1038/nature10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E., Regan M.C., and Furukawa H.. 2015. Emerging structural insights into the function of ionotropic glutamate receptors. Trends Biochem. Sci. 40:328–337. 10.1016/j.tibs.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K., Fukuchi J., Chao J., Igarashi K., and Williams K.. 1996. An aspartate residue in the extracellular loop of the N-methyl-D-aspartate receptor controls sensitivity to spermine and protons. Mol. Pharmacol. 49:1131–1141. [PubMed] [Google Scholar]
- Kashiwagi K., Pahk A.J., Masuko T., Igarashi K., and Williams K.. 1997. Block and modulation of N-methyl-D-aspartate receptors by polyamines and protons: role of amino acid residues in the transmembrane and pore-forming regions of NR1 and NR2 subunits. Mol. Pharmacol. 52:701–713. 10.1124/mol.52.4.701 [DOI] [PubMed] [Google Scholar]
- Kazi R., Gan Q., Talukder I., Markowitz M., Salussolia C.L., and Wollmuth L.P.. 2013. Asynchronous movements prior to pore opening in NMDA receptors. J. Neurosci. 33:12052–12066. 10.1523/JNEUROSCI.5780-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R., Dai J., Sweeney C., Zhou H.X., and Wollmuth L.P.. 2014. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 17:914–922. 10.1038/nn.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe L.A., Bernardinelli Y., and Muller D.. 2013. GluN3A: an NMDA receptor subunit with exquisite properties and functions. Neural Plast. 2013:145387 10.1155/2013/145387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri A., Burger P.B., Swanger S.A., Hansen K.B., Zimmerman S., Karakas E., Liotta D.C., Furukawa H., Snyder J.P., and Traynelis S.F.. 2014. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Mol. Pharmacol. 86:548–560. 10.1124/mol.114.094516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N.W., and Dingledine R.. 1988. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 241:835–837. 10.1126/science.2841759 [DOI] [PubMed] [Google Scholar]
- Köhr G., Eckardt S., Lüddens H., Monyer H., and Seeburg P.H.. 1994. NMDA receptor channels: subunit-specific potentiation by reducing agents. Neuron. 12:1031–1040. 10.1016/0896-6273(94)90311-5 [DOI] [PubMed] [Google Scholar]
- Korinek M., Vyklicky V., Borovska J., Lichnerova K., Kaniakova M., Krausova B., Krusek J., Balik A., Smejkalova T., Horak M., and Vyklicky L.. 2015. Cholesterol modulates open probability and desensitization of NMDA receptors. J. Physiol. 593:2279–2293. 10.1113/jphysiol.2014.288209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotermanski S.E., and Johnson J.W.. 2009. Mg2+ imparts NMDA receptor subtype selectivity to the Alzheimer’s drug memantine. J. Neurosci. 29:2774–2779. 10.1523/JNEUROSCI.3703-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotermanski S.E., Wood J.T., and Johnson J.W.. 2009. Memantine binding to a superficial site on NMDA receptors contributes to partial trapping. J. Physiol. 587:4589–4604. 10.1113/jphysiol.2009.176297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen A.S., Jenkins M.A., Banke T.G., Schousboe A., Makino Y., Johnson R.C., Huganir R., and Traynelis S.F.. 2011. Mechanism of Ca2+/calmodulin-dependent kinase II regulation of AMPA receptor gating. Nat. Neurosci. 14:727–735. 10.1038/nn.2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp J.J., Vissel B., Heinemann S.F., and Westbrook G.L.. 1996. Calcium-dependent inactivation of recombinant N-methyl-D-aspartate receptors is NR2 subunit specific. Mol. Pharmacol. 50:1680–1688. [PubMed] [Google Scholar]
- Krupp J.J., Vissel B., Heinemann S.F., and Westbrook G.L.. 1998. N-terminal domains in the NR2 subunit control desensitization of NMDA receptors. Neuron. 20:317–327. 10.1016/S0896-6273(00)80459-6 [DOI] [PubMed] [Google Scholar]
- Krupp J.J., Vissel B., Thomas C.G., Heinemann S.F., and Westbrook G.L.. 1999. Interactions of calmodulin and alpha-actinin with the NR1 subunit modulate Ca2+-dependent inactivation of NMDA receptors. J. Neurosci. 19:1165–1178. 10.1523/JNEUROSCI.19-04-01165.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., and Mayer M.L.. 2013. Functional insights from glutamate receptor ion channel structures. Annu. Rev. Physiol. 75:313–337. 10.1146/annurev-physiol-030212-183711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T., and Schoepfer R.. 1996. Multiple structural elements determine subunit specificity of Mg2+ block in NMDA receptor channels. J. Neurosci. 16:3549–3558. 10.1523/JNEUROSCI.16-11-03549.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius C.L., and Popescu G.K.. 2009. Kinetic basis of partial agonism at NMDA receptors. Nat. Neurosci. 12:1114–1120. 10.1038/nn.2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius C.L., Kaur N., and Popescu G.K.. 2009. Pregnanolone sulfate promotes desensitization of activated NMDA receptors. J. Neurosci. 29:6819–6827. 10.1523/JNEUROSCI.0281-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius C.L., Popescu A.M., and Popescu G.K.. 2010. Agonist-specific gating of NMDA receptors. Channels (Austin). 4:78–82. 10.4161/chan.4.2.10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusinen A., Arvola M., and Keinänen K.. 1995. Molecular dissection of the agonist binding site of an AMPA receptor. EMBO J. 14:6327–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist T., Steffensen T.B., Greenwood J.R., Mehrzad Tabrizi F., Hansen K.B., Gajhede M., Pickering D.S., Traynelis S.F., Kastrup J.S., and Bräuner-Osborne H.. 2013. Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site. J. Biol. Chem. 288:33124–33135. 10.1074/jbc.M113.480210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.G., and Zukin R.S.. 2007. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 8:413–426. 10.1038/nrn2153 [DOI] [PubMed] [Google Scholar]
- Laurie D.J., and Seeburg P.H.. 1994. Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J. Neurosci. 14:3180–3194. 10.1523/JNEUROSCI.14-05-03180.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Lü W., Michel J.C., Goehring A., Du J., Song X., and Gouaux E.. 2014. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 511:191–197. 10.1038/nature13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P., Rosenmund C., and Westbrook G.L.. 1993. Inactivation of NMDA channels in cultured hippocampal neurons by intracellular calcium. J. Neurosci. 13:674–684. 10.1523/JNEUROSCI.13-02-00674.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R.A., and Jahr C.E.. 1992. NMDA channel behavior depends on agonist affinity. J. Neurosci. 12:635–643. 10.1523/JNEUROSCI.12-02-00635.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R.A., Clements J.D., Westbrook G.L., and Jahr C.E.. 1990. Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature. 346:565–567. 10.1038/346565a0 [DOI] [PubMed] [Google Scholar]
- Lester R.A.J., Tong G., and Jahr C.E.. 1993. Interactions between the glycine and glutamate binding sites of the NMDA receptor. J. Neurosci. 13:1088–1096. 10.1523/JNEUROSCI.13-03-01088.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind G.E., Mou T.C., Tamborini L., Pomper M.G., De Micheli C., Conti P., Pinto A., and Hansen K.B.. 2017. Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits. Proc. Natl. Acad. Sci. USA. 114:E6942–E6951. 10.1073/pnas.1707752114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsenbardt A.J., Taylor A., Emnett C.M., Doherty J.J., Krishnan K., Covey D.F., Paul S.M., Zorumski C.F., and Mennerick S.. 2014. Different oxysterols have opposing actions at N-methyl-D-aspartate receptors. Neuropharmacology. 85:232–242. 10.1016/j.neuropharm.2014.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C.M., and Wee K.S.. 2010. New insights into the not-so-new NR3 subunits of N-methyl-D-aspartate receptor: localization, structure, and function. Mol. Pharmacol. 78:1–11. 10.1124/mol.110.064006 [DOI] [PubMed] [Google Scholar]
- Low C.M., Zheng F., Lyuboslavsky P., and Traynelis S.F.. 2000. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proc. Natl. Acad. Sci. USA. 97:11062–11067. 10.1073/pnas.180307497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C.M., Lyuboslavsky P., French A., Le P., Wyatte K., Thiel W.H., Marchan E.M., Igarashi K., Kashiwagi K., Gernert K., et al. 2003. Molecular determinants of proton-sensitive N-methyl-D-aspartate receptor gating. Mol. Pharmacol. 63:1212–1222. 10.1124/mol.63.6.1212 [DOI] [PubMed] [Google Scholar]
- Lu C., Fu Z., Karavanov I., Yasuda R.P., Wolfe B.B., Buonanno A., and Vicini S.. 2006. NMDA receptor subtypes at autaptic synapses of cerebellar granule neurons. J. Neurophysiol. 96:2282–2294. 10.1152/jn.00078.2006 [DOI] [PubMed] [Google Scholar]
- Lü W., Du J., Goehring A., and Gouaux E.. 2017. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science. 355:355 10.1126/science.aal3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Wang Y., Yasuda R.P., Dunah A.W., and Wolfe B.B.. 1997. The majority of N-methyl-D-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol. Pharmacol. 51:79–86. 10.1124/mol.51.1.79 [DOI] [PubMed] [Google Scholar]
- Lussier M.P., Sanz-Clemente A., and Roche K.W.. 2015. Dynamic Regulation of N-Methyl-d-aspartate (NMDA) and α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors by Posttranslational Modifications. J. Biol. Chem. 290:28596–28603. 10.1074/jbc.R115.652750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J.F., Bartlett M.C., Mody I., Pahapill P., Reynolds J.N., Salter M.W., Schneiderman J.H., and Pennefather P.S.. 1991. Actions of ketamine, phencyclidine and MK-801 on NMDA receptor currents in cultured mouse hippocampal neurones. J. Physiol. 432:483–508. 10.1113/jphysiol.1991.sp018396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malayev A., Gibbs T.T., and Farb D.H.. 2002. Inhibition of the NMDA response by pregnenolone sulphate reveals subtype selective modulation of NMDA receptors by sulphated steroids. Br. J. Pharmacol. 135:901–909. 10.1038/sj.bjp.0704543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maolanon A.R., Risgaard R., Wang S.Y., Snoep Y., Papangelis A., Yi F., Holley D., Barslund A.F., Svenstrup N., Hansen K.B., and Clausen R.P.. 2017. Subtype-Specific Agonists for NMDA Receptor Glycine Binding Sites. ACS Chem. Neurosci. 8:1681–1687. 10.1021/acschemneuro.7b00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.L., Vyklicky L. Jr., and Clements J.. 1989. Regulation of NMDA receptor desensitization in mouse hippocampal neurons by glycine. Nature. 338:425–427. 10.1038/338425a0 [DOI] [PubMed] [Google Scholar]
- Mealing G.A., Lanthorn T.H., Murray C.L., Small D.L., and Morley P.. 1999. Differences in degree of trapping of low-affinity uncompetitive N-methyl-D-aspartic acid receptor antagonists with similar kinetics of block. J. Pharmacol. Exp. Ther. 288:204–210. [PubMed] [Google Scholar]
- Medina I., Filippova N., Charton G., Rougeole S., Ben-Ari Y., Khrestchatisky M., and Bregestovski P.. 1995. Calcium-dependent inactivation of heteromeric NMDA receptor-channels expressed in human embryonic kidney cells. J. Physiol. 482:567–573. 10.1113/jphysiol.1995.sp020540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier C., Wang N., Yi C., Dallerac G., Ezan P., Koulakoff A., Leybaert L., and Giaume C.. 2017. Contribution of Astroglial Cx43 Hemichannels to the Modulation of Glutamatergic Currents by D-Serine in the Mouse Prefrontal Cortex. J. Neurosci. 37:9064–9075. 10.1523/JNEUROSCI.2204-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson J.R., Kumar J., Chittori S., Rao P., Pierson J., Bartesaghi A., Mayer M.L., and Subramaniam S.. 2014. Structural mechanism of glutamate receptor activation and desensitization. Nature. 514:328–334. 10.1038/nature13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H., Sprengel R., Schoepfer R., Herb A., Higuchi M., Lomeli H., Burnashev N., Sakmann B., and Seeburg P.H.. 1992. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 256:1217–1221. 10.1126/science.256.5060.1217 [DOI] [PubMed] [Google Scholar]
- Monyer H., Burnashev N., Laurie D.J., Sakmann B., and Seeburg P.H.. 1994. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 12:529–540. 10.1016/0896-6273(94)90210-0 [DOI] [PubMed] [Google Scholar]
- Morris R.G. 2013. NMDA receptors and memory encoding. Neuropharmacology. 74:32–40. 10.1016/j.neuropharm.2013.04.014 [DOI] [PubMed] [Google Scholar]
- Mothet J.P., Le Bail M., and Billard J.M.. 2015. Time and space profiling of NMDA receptor co-agonist functions. J. Neurochem. 135:210–225. 10.1111/jnc.13204 [DOI] [PubMed] [Google Scholar]
- Mott D.D., Doherty J.J., Zhang S., Washburn M.S., Fendley M.J., Lyuboslavsky P., Traynelis S.F., and Dingledine R.. 1998. Phenylethanolamines inhibit NMDA receptors by enhancing proton inhibition. Nat. Neurosci. 1:659–667. 10.1038/3661 [DOI] [PubMed] [Google Scholar]
- Mu Y., Otsuka T., Horton A.C., Scott D.B., and Ehlers M.D.. 2003. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 40:581–594. 10.1016/S0896-6273(03)00676-7 [DOI] [PubMed] [Google Scholar]
- Muir K.W. 2006. Glutamate-based therapeutic approaches: clinical trials with NMDA antagonists. Curr. Opin. Pharmacol. 6:53–60. 10.1016/j.coph.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Nakanishi N., Axel R., and Shneider N.A.. 1992. Alternative splicing generates functionally distinct N-methyl-D-aspartate receptors. Proc. Natl. Acad. Sci. USA. 89:8552–8556. 10.1073/pnas.89.18.8552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevian T., and Sakmann B.. 2004. Single spine Ca2+ signals evoked by coincident EPSPs and backpropagating action potentials in spiny stellate cells of layer 4 in the juvenile rat somatosensory barrel cortex. J. Neurosci. 24:1689–1699. 10.1523/JNEUROSCI.3332-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevian T., and Sakmann B.. 2006. Spine Ca2+ signaling in spike-timing-dependent plasticity. J. Neurosci. 26:11001–11013. 10.1523/JNEUROSCI.1749-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niciu M.J., Henter I.D., Luckenbaugh D.A., Zarate C.A. Jr., and Charney D.S.. 2014. Glutamate receptor antagonists as fast-acting therapeutic alternatives for the treatment of depression: ketamine and other compounds. Annu. Rev. Pharmacol. Toxicol. 54:119–139. 10.1146/annurev-pharmtox-011613-135950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden K.K., and Traynelis S.F.. 2011. New advances in NMDA receptor pharmacology. Trends Pharmacol. Sci. 32:726–733. 10.1016/j.tips.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden K.K., and Traynelis S.F.. 2013. Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Mol. Pharmacol. 83:1045–1056. 10.1124/mol.113.085209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden K.K., Chen W., Swanger S.A., McDaniel M.J., Fan L.Z., Hu C., Tankovic A., Kusumoto H., Kosobucki G.J., Schulien A.J., et al. 2017. Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet. 13:e1006536 10.1371/journal.pgen.1006536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliet S.H., and Mothet J.P.. 2009. Regulation of N-methyl-D-aspartate receptors by astrocytic D-serine. Neuroscience. 158:275–283. 10.1016/j.neuroscience.2008.01.071 [DOI] [PubMed] [Google Scholar]
- Pachernegg S., Strutz-Seebohm N., and Hollmann M.. 2012. GluN3 subunit-containing NMDA receptors: not just one-trick ponies. Trends Neurosci. 35:240–249. 10.1016/j.tins.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Paganelli M.A., and Popescu G.K.. 2015. Actions of bupivacaine, a widely used local anesthetic, on NMDA receptor responses. J. Neurosci. 35:831–842. 10.1523/JNEUROSCI.3578-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganelli M.A., Kussius C.L., and Popescu G.K.. 2013. Role of cross-cleft contacts in NMDA receptor gating. PLoS One. 8:e80953 10.1371/journal.pone.0080953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahk A.J., and Williams K.. 1997. Influence of extracellular pH on inhibition by ifenprodil at N-methyl-D-aspartate receptors in Xenopus oocytes. Neurosci. Lett. 225:29–32. 10.1016/S0304-3940(97)00176-6 [DOI] [PubMed] [Google Scholar]
- Paoletti P., Ascher P., and Neyton J.. 1997. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J. Neurosci. 17:5711–5725. 10.1523/JNEUROSCI.17-15-05711.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P., Perin-Dureau F., Fayyazuddin A., Le Goff A., Callebaut I., and Neyton J.. 2000. Molecular organization of a zinc binding n-terminal modulatory domain in a NMDA receptor subunit. Neuron. 28:911–925. 10.1016/S0896-6273(00)00163-X [DOI] [PubMed] [Google Scholar]
- Paoletti P., Bellone C., and Zhou Q.. 2013. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14:383–400. 10.1038/nrn3504 [DOI] [PubMed] [Google Scholar]
- Papouin T., Ladépêche L., Ruel J., Sacchi S., Labasque M., Hanini M., Groc L., Pollegioni L., Mothet J.P., and Oliet S.H.. 2012. Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell. 150:633–646. 10.1016/j.cell.2012.06.029 [DOI] [PubMed] [Google Scholar]
- Parsons C.G., Gruner R., Rozental J., Millar J., and Lodge D.. 1993. Patch clamp studies on the kinetics and selectivity of N-methyl-D-aspartate receptor antagonism by memantine (1-amino-3,5-dimethyladamantan). Neuropharmacology. 32:1337–1350. 10.1016/0028-3908(93)90029-3 [DOI] [PubMed] [Google Scholar]
- Parsons C.G., Quack G., Bresink I., Baran L., Przegalinski E., Kostowski W., Krzascik P., Hartmann S., and Danysz W.. 1995. Comparison of the potency, kinetics and voltage-dependency of a series of uncompetitive NMDA receptor antagonists in vitro with anticonvulsive and motor impairment activity in vivo. Neuropharmacology. 34:1239–1258. 10.1016/0028-3908(95)00092-K [DOI] [PubMed] [Google Scholar]
- Paul S.M., Doherty J.J., Robichaud A.J., Belfort G.M., Chow B.Y., Hammond R.S., Crawford D.C., Linsenbardt A.J., Shu H.J., Izumi Y., et al. 2013. The major brain cholesterol metabolite 24(S)-hydroxycholesterol is a potent allosteric modulator of N-methyl-D-aspartate receptors. J. Neurosci. 33:17290–17300. 10.1523/JNEUROSCI.2619-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupard M.C., Friedman L.K., and Zukin R.S.. 1997. Developmental regulation and cell-specific expression of N-methyl-D-aspartate receptor splice variants in rat hippocampus. Neuroscience. 79:399–409. 10.1016/S0306-4522(96)00677-X [DOI] [PubMed] [Google Scholar]
- Pérez-Otaño I., Larsen R.S., and Wesseling J.F.. 2016. Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat. Rev. Neurosci. 17:623–635. 10.1038/nrn.2016.92 [DOI] [PubMed] [Google Scholar]
- Piña-Crespo J.C., and Gibb A.J.. 2002. Subtypes of NMDA receptors in new-born rat hippocampal granule cells. J. Physiol. 541:41–64. 10.1113/jphysiol.2001.014001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pøhlsgaard J., Frydenvang K., Madsen U., and Kastrup J.S.. 2011. Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology. 60:135–150. 10.1016/j.neuropharm.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Popescu G.K. 2012. Modes of glutamate receptor gating. J. Physiol. 590:73–91. 10.1113/jphysiol.2011.223750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G., and Auerbach A.. 2003. Modal gating of NMDA receptors and the shape of their synaptic response. Nat. Neurosci. 6:476–483. 10.1038/nn1044 [DOI] [PubMed] [Google Scholar]
- Popescu G., Robert A., Howe J.R., and Auerbach A.. 2004. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 430:790–793. 10.1038/nature02775 [DOI] [PubMed] [Google Scholar]
- Poulsen M.H., Andersen J., Christensen R., Hansen K.B., Traynelis S.F., Strømgaard K., and Kristensen A.S.. 2015. Binding of ArgTX-636 in the NMDA receptor ion channel. J. Mol. Biol. 427:176–189. 10.1016/j.jmb.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L.S., and Auerbach A.. 1996. Identification of a high affinity divalent cation binding site near the entrance of the NMDA receptor channel. Neuron. 16:869–880. 10.1016/S0896-6273(00)80107-5 [DOI] [PubMed] [Google Scholar]
- Premkumar L.S., Qin F., and Auerbach A.. 1997. Subconductance states of a mutant NMDA receptor channel kinetics, calcium, and voltage dependence. J. Gen. Physiol. 109:181–189. 10.1085/jgp.109.2.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian A., and Johnson J.W.. 2006. Permeant ion effects on external Mg2+ block of NR1/2D NMDA receptors. J. Neurosci. 26:10899–10910. 10.1523/JNEUROSCI.3453-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian A., Antonov S.M., and Johnson J.W.. 2002. Modulation by permeant ions of Mg(2+) inhibition of NMDA-activated whole-cell currents in rat cortical neurons. J. Physiol. 538:65–77. 10.1113/jphysiol.2001.012685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian A., Buller A.L., and Johnson J.W.. 2005. NR2 subunit-dependence of NMDA receptor channel block by external Mg2+. J. Physiol. 562:319–331. 10.1113/jphysiol.2004.076737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachline J., Perin-Dureau F., Le Goff A., Neyton J., and Paoletti P.. 2005. The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J. Neurosci. 25:308–317. 10.1523/JNEUROSCI.3967-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauner C., and Köhr G.. 2011. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J. Biol. Chem. 286:7558–7566. 10.1074/jbc.M110.182600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan M.C., Grant T., McDaniel M.J., Karakas E., Zhang J., Traynelis S.F., Grigorieff N., and Furukawa H.. 2018. Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors. Neuron. 98:521–529.e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Honse Y., Karp B.J., Lipsky R.H., and Peoples R.W.. 2003. A site in the fourth membrane-associated domain of the N-methyl-D-aspartate receptor regulates desensitization and ion channel gating. J. Biol. Chem. 278:276–283. 10.1074/jbc.M209486200 [DOI] [PubMed] [Google Scholar]
- Romero-Hernandez A., and Furukawa H.. 2017. Novel Mode of Antagonist Binding in NMDA Receptors Revealed by the Crystal Structure of the GluN1-GluN2A Ligand-Binding Domain Complexed to NVP-AAM077. Mol. Pharmacol. 92:22–29. 10.1124/mol.116.107912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Hernandez A., Simorowski N., Karakas E., and Furukawa H.. 2016. Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain. Neuron. 92:1324–1336. 10.1016/j.neuron.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C., and Westbrook G.L.. 1993. Calcium-induced actin depolymerization reduces NMDA channel activity. Neuron. 10:805–814. 10.1016/0896-6273(93)90197-Y [DOI] [PubMed] [Google Scholar]
- Rosenmund C., Feltz A., and Westbrook G.L.. 1995. Calcium-dependent inactivation of synaptic NMDA receptors in hippocampal neurons. J. Neurophysiol. 73:427–430. 10.1152/jn.1995.73.1.427 [DOI] [PubMed] [Google Scholar]
- Rosenmund C., Stern-Bach Y., and Stevens C.F.. 1998. The tetrameric structure of a glutamate receptor channel. Science. 280:1596–1599. 10.1126/science.280.5369.1596 [DOI] [PubMed] [Google Scholar]
- Rumbaugh G., Prybylowski K., Wang J.F., and Vicini S.. 2000. Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J. Neurophysiol. 83:1300–1306. 10.1152/jn.2000.83.3.1300 [DOI] [PubMed] [Google Scholar]
- Rycroft B.K., and Gibb A.J.. 2002. Direct effects of calmodulin on NMDA receptor single-channel gating in rat hippocampal granule cells. J. Neurosci. 22:8860–8868. 10.1523/JNEUROSCI.22-20-08860.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P., Hestrin S., and Nicoll R.A.. 1990. Properties of excitatory postsynaptic currents recorded in vitro from rat hippocampal interneurones. J. Physiol. 430:605–616. 10.1113/jphysiol.1990.sp018310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Clemente A., Nicoll R.A., and Roche K.W.. 2013. Diversity in NMDA receptor composition: many regulators, many consequences. Neuroscientist. 19:62–75. 10.1177/1073858411435129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather W., Johnson J.W., Henderson G., and Ascher P.. 1990. Glycine-insensitive desensitization of NMDA responses in cultured mouse embryonic neurons. Neuron. 4:725–731. 10.1016/0896-6273(90)90198-O [DOI] [PubMed] [Google Scholar]
- Sather W., Dieudonné S., MacDonald J.F., and Ascher P.. 1992. Activation and desensitization of N-methyl-D-aspartate receptors in nucleated outside-out patches from mouse neurones. J. Physiol. 450:643–672. 10.1113/jphysiol.1992.sp019148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneggenburger R. 1996. Simultaneous measurement of Ca2+ influx and reversal potentials in recombinant N-methyl-D-aspartate receptor channels. Biophys. J. 70:2165–2174. 10.1016/S0006-3495(96)79782-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneggenburger R. 1998. Altered voltage dependence of fractional Ca2+ current in N-methyl-D-aspartate channel pore mutants with a decreased Ca2+ permeability. Biophys. J. 74:1790–1794. 10.1016/S0006-3495(98)77889-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneggenburger R., and Ascher P.. 1997. Coupling of permeation and gating in an NMDA-channel pore mutant. Neuron. 18:167–177. 10.1016/S0896-6273(01)80055-6 [DOI] [PubMed] [Google Scholar]
- Schorge S., Elenes S., and Colquhoun D.. 2005. Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J. Physiol. 569:395–418. 10.1113/jphysiol.2005.095349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.B., Blanpied T.A., Swanson G.T., Zhang C., and Ehlers M.D.. 2001. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21:3063–3072. 10.1523/JNEUROSCI.21-09-03063.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.B., Blanpied T.A., and Ehlers M.D.. 2003. Coordinated PKA and PKC phosphorylation suppresses RXR-mediated ER retention and regulates the surface delivery of NMDA receptors. Neuropharmacology. 45:755–767. 10.1016/S0028-3908(03)00250-8 [DOI] [PubMed] [Google Scholar]
- Seeburg P.H., Burnashev N., Köhr G., Kuner T., Sprengel R., and Monyer H.. 1995. The NMDA receptor channel: molecular design of a coincidence detector. Recent Prog. Horm. Res. 50:19–34. [DOI] [PubMed] [Google Scholar]
- Serraz B., Grand T., and Paoletti P.. 2016. Altered zinc sensitivity of NMDA receptors harboring clinically-relevant mutations. Neuropharmacology. 109:196–204. 10.1016/j.neuropharm.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Sharma G., and Stevens C.F.. 1996. Interactions between two divalent ion binding sites in N-methyl-D-aspartate receptor channels. Proc. Natl. Acad. Sci. USA. 93:14170–14175. 10.1073/pnas.93.24.14170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin A., Shavit S., and Benveniste M.. 2001. Subunit specificity and mechanism of action of NMDA partial agonist D-cycloserine. Neuropharmacology. 41:151–158. 10.1016/S0028-3908(01)00073-9 [DOI] [PubMed] [Google Scholar]
- Sheng M., Cummings J., Roldan L.A., Jan Y.N., and Jan L.Y.. 1994. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 368:144–147. 10.1038/368144a0 [DOI] [PubMed] [Google Scholar]
- Siegler Retchless B., Gao W., and Johnson J.W.. 2012. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat. Neurosci. 15:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh R.E., MacLean D.M., and Jayaraman V.. 2013. Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-D-aspartate (NMDA) receptor. J. Biol. Chem. 288:22555–22564. 10.1074/jbc.M113.482356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh R.E., MacLean D.M., and Jayaraman V.. 2015a A conserved structural mechanism of NMDA receptor inhibition: A comparison of ifenprodil and zinc. J. Gen. Physiol. 146:173–181. 10.1085/jgp.201511422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh R.E., MacLean D.M., and Jayaraman V.. 2015b Subtype-dependent N-methyl-D-aspartate receptor amino-terminal domain conformations and modulation by spermine. J. Biol. Chem. 290:12812–12820. 10.1074/jbc.M115.649723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I. 1999. Two-component blocking kinetics of open NMDA channels by organic cations. Biochim. Biophys. Acta. 1416:69–91. 10.1016/S0005-2736(98)00211-9 [DOI] [PubMed] [Google Scholar]
- Sobolevsky A.I. 2015. Structure and gating of tetrameric glutamate receptors. J. Physiol. 593:29–38. 10.1113/jphysiol.2013.264911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I., and Yelshansky M.V.. 2000. The trapping block of NMDA receptor channels in acutely isolated rat hippocampal neurones. J. Physiol. 526:493–506. 10.1111/j.1469-7793.2000.t01-2-00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I., Koshelev S.G., and Khodorov B.I.. 1999. Probing of NMDA channels with fast blockers. J. Neurosci. 19:10611–10626. 10.1523/JNEUROSCI.19-24-10611.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I., Beck C., and Wollmuth L.P.. 2002a Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 33:75–85. 10.1016/S0896-6273(01)00560-8 [DOI] [PubMed] [Google Scholar]
- Sobolevsky A.I., Rooney L., and Wollmuth L.P.. 2002b Staggering of subunits in NMDAR channels. Biophys. J. 83:3304–3314. 10.1016/S0006-3495(02)75331-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Jensen M.O., Jogini V., Stein R.A., Lee C.H., Mchaourab H.S., Shaw D.E., and Gouaux E.. 2018. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature. 556:515–519. 10.1038/s41586-018-0039-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D., Carvalho S., Grand T., Zhu S., and Paoletti P.. 2014. Controlling NMDA receptor subunit composition using ectopic retention signals. J. Neurosci. 34:16630–16636. 10.1523/JNEUROSCI.2736-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D., Buhl D.L., Knafels J.D., Chanda P.K., Green M., Sciabola S., Mony L., Paoletti P., and Pandit J.. 2016. A Novel Binding Mode Reveals Two Distinct Classes of NMDA Receptor GluN2B-selective Antagonists. Mol. Pharmacol. 89:541–551. 10.1124/mol.115.103036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong K.L., Jing Y., Prosser A.R., Traynelis S.F., and Liotta D.C.. 2014. NMDA receptor modulators: an updated patent review (2013-2014). Expert Opin. Ther. Pat. 24:1349–1366. 10.1517/13543776.2014.972938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara H., Moriyoshi K., Ishii T., Masu M., and Nakanishi S.. 1992. Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem. Biophys. Res. Commun. 185:826–832. 10.1016/0006-291X(92)91701-Q [DOI] [PubMed] [Google Scholar]
- Sullivan S.J., and Miller R.F.. 2012. AMPA receptor-dependent, light-evoked D-serine release acts on retinal ganglion cell NMDA receptors. J. Neurophysiol. 108:1044–1051. 10.1152/jn.00264.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.M., Traynelis S.F., Chen H.S., Escobar W., Heinemann S.F., and Lipton S.A.. 1994. Identification of two cysteine residues that are required for redox modulation of the NMDA subtype of glutamate receptor. Neuron. 13:929–936. 10.1016/0896-6273(94)90258-5 [DOI] [PubMed] [Google Scholar]
- Sun W., Hansen K.B., and Jahr C.E.. 2017. Allosteric Interactions between NMDA Receptor Subunits Shape the Developmental Shift in Channel Properties. Neuron. 94:58–64.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Olson R., Horning M., Armstrong N., Mayer M., and Gouaux E.. 2002. Mechanism of glutamate receptor desensitization. Nature. 417:245–253. 10.1038/417245a [DOI] [PubMed] [Google Scholar]
- Sundström E., Whittemore S., Mo L.L., and Seiger A.. 1997. Analysis of NMDA receptors in the human spinal cord. Exp. Neurol. 148:407–413. 10.1006/exnr.1997.6691 [DOI] [PubMed] [Google Scholar]
- Swanger S.A., Vance K.M., Pare J.F., Sotty F., Fog K., Smith Y., and Traynelis S.F.. 2015. NMDA Receptors Containing the GluN2D Subunit Control Neuronal Function in the Subthalamic Nucleus. J. Neurosci. 35:15971–15983. 10.1523/JNEUROSCI.1702-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger S.A., Vance K.M., Acker T.M., Zimmerman S.S., DiRaddo J.O., Myers S.J., Bundgaard C., Mosley C.A., Summer S.L., Menaldino D.S., et al. 2018. A Novel Negative Allosteric Modulator Selective for GluN2C/2D-Containing NMDA Receptors Inhibits Synaptic Transmission in Hippocampal Interneurons. ACS Chem. Neurosci. 9:306–319. 10.1021/acschemneuro.7b00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N., Karakas E., Grant T., Simorowski N., Diaz-Avalos R., Grigorieff N., and Furukawa H.. 2016. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 534:63–68. 10.1038/nature17679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Xia P., Cui J., Talantova M., Bodhinathan K., Li W., Saleem S., Holland E.A., Tong G., Piña-Crespo J., et al. 2015. Pharmacologically targeted NMDA receptor antagonism by NitroMemantine for cerebrovascular disease. Sci. Rep. 5:14781 10.1038/srep14781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Feldmeyer D., Suzuki N., Onodera K., Cull-Candy S.G., Sakimura K., and Mishina M.. 1996. Functional correlation of NMDA receptor epsilon subunits expression with the properties of single-channel and synaptic currents in the developing cerebellum. J. Neurosci. 16:4376–4382. 10.1523/JNEUROSCI.16-14-04376.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder I., Borker P., and Wollmuth L.P.. 2010. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J. Neurosci. 30:11792–11804. 10.1523/JNEUROSCI.5382-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder I., Kazi R., and Wollmuth L.P.. 2011. GluN1-specific redox effects on the kinetic mechanism of NMDA receptor activation. Biophys. J. 101:2389–2398. 10.1016/j.bpj.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L.H., and Aizenman E.. 1993. The modulation of N-methyl-D-aspartate receptors by redox and alkylating reagents in rat cortical neurones in vitro. J. Physiol. 465:303–323. 10.1113/jphysiol.1993.sp019678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.M., Dichter M., and Morad M.. 1990. Modulation of the N-methyl-D-aspartate channel by extracellular H+. Proc. Natl. Acad. Sci. USA. 87:6445–6449. 10.1073/pnas.87.16.6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.T., Badger J.D. II, Roche P.A., and Roche K.W.. 2010. Novel approach to probe subunit-specific contributions to N-methyl-D-aspartate (NMDA) receptor trafficking reveals a dominant role for NR2B in receptor recycling. J. Biol. Chem. 285:20975–20981. 10.1074/jbc.M110.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley W.G., Roche K.W., Thompson A.K., and Huganir R.L.. 1993. Regulation of NMDA receptor phosphorylation by alternative splicing of the C-terminal domain. Nature. 364:70–73. 10.1038/364070a0 [DOI] [PubMed] [Google Scholar]
- Tingley W.G., Ehlers M.D., Kameyama K., Doherty C., Ptak J.B., Riley C.T., and Huganir R.L.. 1997. Characterization of protein kinase A and protein kinase C phosphorylation of the N-methyl-D-aspartate receptor NR1 subunit using phosphorylation site-specific antibodies. J. Biol. Chem. 272:5157–5166. 10.1074/jbc.272.8.5157 [DOI] [PubMed] [Google Scholar]
- Tovar K.R., and Westbrook G.L.. 1999. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. J. Neurosci. 19:4180–4188. 10.1523/JNEUROSCI.19-10-04180.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar K.R., McGinley M.J., and Westbrook G.L.. 2013. Triheteromeric NMDA receptors at hippocampal synapses. J. Neurosci. 33:9150–9160. 10.1523/JNEUROSCI.0829-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis S.F., and Cull-Candy S.G.. 1990. Proton inhibition of N-methyl-D-aspartate receptors in cerebellar neurons. Nature. 345:347–350. 10.1038/345347a0 [DOI] [PubMed] [Google Scholar]
- Traynelis S.F., and Cull-Candy S.G.. 1991. Pharmacological properties and H+ sensitivity of excitatory amino acid receptor channels in rat cerebellar granule neurones. J. Physiol. 433:727–763. 10.1113/jphysiol.1991.sp018453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis S.F., Hartley M., and Heinemann S.F.. 1995. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 268:873–876. 10.1126/science.7754371 [DOI] [PubMed] [Google Scholar]
- Traynelis S.F., Burgess M.F., Zheng F., Lyuboslavsky P., and Powers J.L.. 1998. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J. Neurosci. 18:6163–6175. 10.1523/JNEUROSCI.18-16-06163.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., and Dingledine R.. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62:405–496. 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trussell L.O., Zhang S., and Raman I.M.. 1993. Desensitization of AMPA receptors upon multiquantal neurotransmitter release. Neuron. 10:1185–1196. 10.1016/0896-6273(93)90066-Z [DOI] [PubMed] [Google Scholar]
- Twomey E.C., and Sobolevsky A.I.. 2018. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry. 57:267–276. 10.1021/acs.biochem.7b00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Yelshanskaya M.V., Grassucci R.A., Frank J., and Sobolevsky A.I.. 2017. Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 549:60–65. 10.1038/nature23479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich M.H., and Isacoff E.Y.. 2007. Subunit counting in membrane-bound proteins. Nat. Methods. 4:319–321. 10.1038/nmeth1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K.M., Simorowski N., Traynelis S.F., and Furukawa H.. 2011. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat. Commun. 2:294 10.1038/ncomms1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K.M., Hansen K.B., and Traynelis S.F.. 2012. GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J. Physiol. 590:3857–3875. 10.1113/jphysiol.2012.234062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K.M., Hansen K.B., and Traynelis S.F.. 2013. Modal gating of GluN1/GluN2D NMDA receptors. Neuropharmacology. 71:184–190. 10.1016/j.neuropharm.2013.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini S., Wang J.F., Li J.H., Zhu W.J., Wang Y.H., Luo J.H., Wolfe B.B., and Grayson D.R.. 1998. Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J. Neurophysiol. 79:555–566. 10.1152/jn.1998.79.2.555 [DOI] [PubMed] [Google Scholar]
- Villarroel A., Regalado M.P., and Lerma J.. 1998. Glycine-independent NMDA receptor desensitization: localization of structural determinants. Neuron. 20:329–339. 10.1016/S0896-6273(00)80460-2 [DOI] [PubMed] [Google Scholar]
- Vogt K., Mellor J., Tong G., and Nicoll R.. 2000. The actions of synaptically released zinc at hippocampal mossy fiber synapses. Neuron. 26:187–196. 10.1016/S0896-6273(00)81149-6 [DOI] [PubMed] [Google Scholar]
- Volgraf M., Sellers B.D., Jiang Y., Wu G., Ly C.Q., Villemure E., Pastor R.M., Yuen P.W., Lu A., Luo X., et al. 2016. Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design. J. Med. Chem. 59:2760–2779. 10.1021/acs.jmedchem.5b02010 [DOI] [PubMed] [Google Scholar]
- Volianskis A., France G., Jensen M.S., Bortolotto Z.A., Jane D.E., and Collingridge G.L.. 2015. Long-term potentiation and the role of N-methyl-D-aspartate receptors. Brain Res. 1621:5–16. 10.1016/j.brainres.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklicky V., Korinek M., Smejkalova T., Balik A., Krausova B., Kaniakova M., Lichnerova K., Cerny J., Krusek J., Dittert I., et al. 2014. Structure, function, and pharmacology of NMDA receptor channels. Physiol. Res. 63(Suppl 1):S191–S203. [DOI] [PubMed] [Google Scholar]
- Vyklicky V., Krausova B., Cerny J., Balik A., Zapotocky M., Novotny M., Lichnerova K., Smejkalova T., Kaniakova M., Korinek M., et al. 2015. Block of NMDA receptor channels by endogenous neurosteroids: implications for the agonist induced conformational states of the channel vestibule. Sci. Rep. 5:10935 10.1038/srep10935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklický L., Jr 1993. Calcium-mediated modulation of N-methyl-D-aspartate (NMDA) responses in cultured rat hippocampal neurones. J. Physiol. 470:575–600. 10.1113/jphysiol.1993.sp019876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklický L. Jr., Vlachová V., and Krůsek J.. 1990. The effect of external pH changes on responses to excitatory amino acids in mouse hippocampal neurones. J. Physiol. 430:497–517. 10.1113/jphysiol.1990.sp018304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J., Beck C., Kuner T., Premkumar L.S., and Wollmuth L.P.. 2002. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J. Neurosci. 22:10209–10216. 10.1523/JNEUROSCI.22-23-10209.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Inoue Y., Sakimura K., and Mishina M.. 1992. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 3:1138–1140. 10.1097/00001756-199212000-00027 [DOI] [PubMed] [Google Scholar]
- Wenthold R.J., Prybylowski K., Standley S., Sans N., and Petralia R.S.. 2003. Trafficking of NMDA receptors. Annu. Rev. Pharmacol. Toxicol. 43:335–358. 10.1146/annurev.pharmtox.43.100901.135803 [DOI] [PubMed] [Google Scholar]
- Williams K. 1996. Separating dual effects of zinc at recombinant N-methyl-D-aspartate receptors. Neurosci. Lett. 215:9–12. 10.1016/S0304-3940(96)12924-4 [DOI] [PubMed] [Google Scholar]
- Wollmuth L.P. 2018. Ion permeation in ionotropic glutamate receptors: Still dynamic after all these years. Curr Opin Physiol. 2:36–41. 10.1016/j.cophys.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth L.P., and Sobolevsky A.I.. 2004. Structure and gating of the glutamate receptor ion channel. Trends Neurosci. 27:321–328. 10.1016/j.tins.2004.04.005 [DOI] [PubMed] [Google Scholar]
- Wollmuth L.P., Kuner T., Seeburg P.H., and Sakmann B.. 1996. Differential contribution of the NR1- and NR2A-subunits to the selectivity filter of recombinant NMDA receptor channels. J. Physiol. 491:779–797. 10.1113/jphysiol.1996.sp021257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth L.P., Kuner T., and Sakmann B.. 1998. Adjacent asparagines in the NR2-subunit of the NMDA receptor channel control the voltage-dependent block by extracellular Mg2+. J. Physiol. 506:13–32. 10.1111/j.1469-7793.1998.013bx.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosker H. 2007. NMDA receptor regulation by D-serine: new findings and perspectives. Mol. Neurobiol. 36:152–164. 10.1007/s12035-007-0038-6 [DOI] [PubMed] [Google Scholar]
- Wyllie D.J., Béhé P., Nassar M., Schoepfer R., and Colquhoun D.. 1996. Single-channel currents from recombinant NMDA NR1a/NR2D receptors expressed in Xenopus oocytes. Proc. Biol. Sci. 263:1079–1086. 10.1098/rspb.1996.0159 [DOI] [PubMed] [Google Scholar]
- Wyllie D.J., Béhé P., and Colquhoun D.. 1998. Single-channel activations and concentration jumps: comparison of recombinant NR1a/NR2A and NR1a/NR2D NMDA receptors. J. Physiol. 510:1–18. 10.1111/j.1469-7793.1998.001bz.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie D.J., Livesey M.R., and Hardingham G.E.. 2013. Influence of GluN2 subunit identity on NMDA receptor function. Neuropharmacology. 74:4–17. 10.1016/j.neuropharm.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., and Mayer M.L.. 2006. Characterization of a soluble ligand binding domain of the NMDA receptor regulatory subunit NR3A. J. Neurosci. 26:4559–4566. 10.1523/JNEUROSCI.0560-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Harrison C.B., Freddolino P.L., Schulten K., and Mayer M.L.. 2008. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 27:2158–2170. 10.1038/emboj.2008.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Belcher J., Berger A.J., Mayer M.L., and Lau A.Y.. 2013. Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure. 21:1788–1799. 10.1016/j.str.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Mou T.C., Dorsett K.N., Volkmann R.A., Menniti F.S., Sprang S.R., and Hansen K.B.. 2016. Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors. Neuron. 91:1316–1329. 10.1016/j.neuron.2016.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Traynelis S.F., and Hansen K.B.. 2017. Selective Cell-Surface Expression of Triheteromeric NMDA Receptors. Methods Mol. Biol. 1677:145–162. 10.1007/978-1-4939-7321-7_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Zachariassen L.G., Dorsett K.N., and Hansen K.B.. 2018. Properties of Triheteromeric N-Methyl-d-Aspartate Receptors Containing Two Distinct GluN1 Isoforms. Mol. Pharmacol. 93:453–467. 10.1124/mol.117.111427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Erreger K., Dravid S.M., and Traynelis S.F.. 2005. Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. J. Biol. Chem. 280:29708–29716. 10.1074/jbc.M414215200 [DOI] [PubMed] [Google Scholar]
- Yuan H., Geballe M.T., Hansen K.B., and Traynelis S.F.. 2008. Structure and function of the NMDA receptor. In Structural and Functional Organization of the Synapse. Hell J.W., and Ehlers M.D., editors. Springer, New York: 289–316. 10.1007/978-0-387-77232-5_11 [DOI] [Google Scholar]
- Yuan H., Hansen K.B., Vance K.M., Ogden K.K., and Traynelis S.F.. 2009. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J. Neurosci. 29:12045–12058. 10.1523/JNEUROSCI.1365-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Hansen K.B., Zhang J., Pierson T.M., Markello T.C., Fajardo K.V., Holloman C.M., Golas G., Adams D.R., Boerkoel C.F., et al. 2014. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat. Commun. 5:3251 10.1038/ncomms4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Myers S.J., Wells G., Nicholson K.L., Swanger S.A., Lyuboslavsky P., Tahirovic Y.A., Menaldino D.S., Ganesh T., Wilson L.J., et al. 2015. Context-dependent GluN2B-selective inhibitors of NMDA receptor function are neuroprotective with minimal side effects. Neuron. 85:1305–1318. 10.1016/j.neuron.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki M., and Aricescu A.R.. 2017. A GluD Coming-Of-Age Story. Trends Neurosci. 40:138–150. 10.1016/j.tins.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanos P., Thompson S.M., Duman R.S., Zarate C.A. Jr., and Gould T.D.. 2018. Convergent Mechanisms Underlying Rapid Antidepressant Action. CNS Drugs. 32:197–227. 10.1007/s40263-018-0492-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zheng X., Paupard M.C., Wang A.P., Santchi L., Friedman L.K., Zukin R.S., and Bennett M.V.. 1994. Spermine potentiation of recombinant N-methyl-D-aspartate receptors is affected by subunit composition. Proc. Natl. Acad. Sci. USA. 91:10883–10887. 10.1073/pnas.91.23.10883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ehlers M.D., Bernhardt J.P., Su C.T., and Huganir R.L.. 1998. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron. 21:443–453. 10.1016/S0896-6273(00)80553-X [DOI] [PubMed] [Google Scholar]
- Zhang W., Howe J.R., and Popescu G.K.. 2008. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat. Neurosci. 11:1373–1375. 10.1038/nn.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Erreger K., Low C.M., Banke T., Lee C.J., Conn P.J., and Traynelis S.F.. 2001. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat. Neurosci. 4:894–901. 10.1038/nn0901-894 [DOI] [PubMed] [Google Scholar]
- Zhong J., Carrozza D.P., Williams K., Pritchett D.B., and Molinoff P.B.. 1995. Expression of mRNAs encoding subunits of the NMDA receptor in developing rat brain. J. Neurochem. 64:531–539. 10.1046/j.1471-4159.1995.64020531.x [DOI] [PubMed] [Google Scholar]
- Zhu S., and Paoletti P.. 2015. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr. Opin. Pharmacol. 20:14–23. 10.1016/j.coph.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Zhu Y., and Auerbach A.. 2001a K(+) occupancy of the N-methyl-d-aspartate receptor channel probed by Mg(2+) block. J. Gen. Physiol. 117:287–298. 10.1085/jgp.117.3.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., and Auerbach A.. 2001b Na(+) occupancy and Mg(2+) block of the N-methyl-d-aspartate receptor channel. J. Gen. Physiol. 117:275–286. 10.1085/jgp.117.3.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Stroebel D., Yao C.A., Taly A., and Paoletti P.. 2013. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat. Struct. Mol. Biol. 20:477–485. 10.1038/nsmb.2522 [DOI] [PubMed] [Google Scholar]
- Zhu S., Riou M., Yao C.A., Carvalho S., Rodriguez P.C., Bensaude O., Paoletti P., and Ye S.. 2014. Genetically encoding a light switch in an ionotropic glutamate receptor reveals subunit-specific interfaces. Proc. Natl. Acad. Sci. USA. 111:6081–6086. 10.1073/pnas.1318808111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Stein R.A., Yoshioka C., Lee C.H., Goehring A., Mchaourab H.S., and Gouaux E.. 2016. Mechanism of NMDA Receptor Inhibition and Activation. Cell. 165:704–714. 10.1016/j.cell.2016.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorumski C.F., and Izumi Y.. 2012. NMDA receptors and metaplasticity: mechanisms and possible roles in neuropsychiatric disorders. Neurosci. Biobehav. Rev. 36:989–1000. 10.1016/j.neubiorev.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]