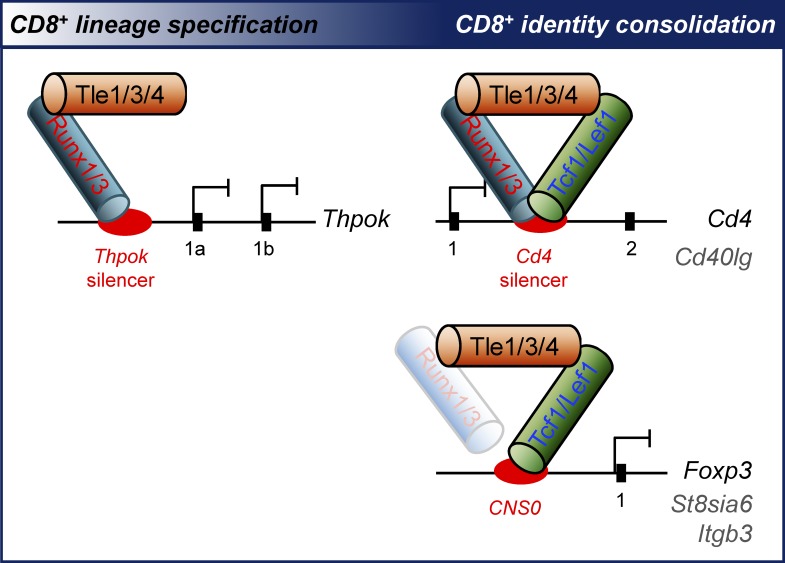
Xing et al demonstrate the requirements for Tle transcriptional corepressors in CD8+ T cell development. Tle proteins are differentially partitioned to the Runx and Tcf/Lef complexes to promote CD8+ lineage choice and establish CD8+ T cell identity, respectively.
Abstract
Tle/Groucho proteins are transcriptional corepressors interacting with Tcf/Lef and Runx transcription factors, but their physiological roles in T cell development remain unknown. Conditional targeting of Tle1, Tle3 and Tle4 revealed gene dose–dependent requirements for Tle proteins in CD8+ lineage cells. Upon ablating all three Tle proteins, generation of CD8+ T cells was greatly diminished, largely owing to redirection of MHC-I–selected thymocytes to CD4+ lineage; the remaining CD8-positive T cells showed aberrant up-regulation of CD4+ lineage-associated genes including Cd4, Thpok, St8sia6, and Foxp3. Mechanistically, Tle3 bound to Runx-occupied Thpok silencer, in post-selection double-positive thymocytes to prevent excessive ThPOK induction and in mature CD8+ T cells to silence Thpok expression. Tle3 also bound to Tcf1-occupied sites in a few CD4+ lineage-associated genes, including Cd4 silencer and St8sia6 introns, to repress their expression in mature CD8+ T cells. These findings indicate that Tle corepressors are differentially partitioned to Runx and Tcf/Lef complexes to instruct CD8+ lineage choice and cooperatively establish CD8+ T cell identity, respectively.
Graphical Abstract
Introduction
The Drosophila melanogaster Groucho and its evolutionarily conserved mammalian Transducin-Like Enhancer of split (Tle) homologues are transcriptional corepressors (Turki-Judeh and Courey, 2012). In mammals, there are four full-length Tle proteins, Tle1–4, while other two partially homologous proteins, Tle5 and Tle6, are expressed in truncated forms (Gasperowicz and Otto, 2005; Buscarlet and Stifani, 2007). Tle proteins do not have the capacity to directly bind DNA, but interact with sequence-specific transcription factors in diverse protein families (Jennings and Ish-Horowicz, 2008; Turki-Judeh and Courey, 2012). As a result, Tle proteins demonstrate critical regulatory roles in a wide range of organogenesis, including neurogenesis, osteogenesis, and hematopoiesis, as well as development of kidney and pancreas (Agarwal et al., 2015). In the blood lineage cells, Tle4 interacts with Pax5 and PU.1 transcription factors, suggesting a role in B cell development and function (Eberhard et al., 2000; Linderson et al., 2004). All Tle proteins interact with Tcf1 and Lef1 downstream of the Wnt signaling pathway (Brantjes et al., 2001; Daniels and Weis, 2005; Staal and Sen, 2008), suggestive of involvement in T cell development and function (Xue and Zhao, 2012; Steinke and Xue, 2014). Tle1 is shown to bind Runx1 (Levanon et al., 1998), which is essential for the generation and maintenance of hematopoietic stem/progenitor cells (HSPCs; Cai et al., 2015). In line with such broadly interacting partners, germline deletion of Tle4 in mice causes a profound reduction in cellularity of hematopoietic cells including HSPCs and B cells (Wheat et al., 2014). Ablation of Tle1, on the other hand, exhibits grossly normal hematopoiesis, but results in excess production of inflammatory cytokines by macrophages (Ramasamy et al., 2016). In addition, Tle1 and Tle4 appear to function as tumor suppressors in the context of myeloid leukemia (Dayyani et al., 2008; Shin et al., 2016). In spite of the advances, the precise roles of these Tle proteins in development and function of immune cells have not been elucidated.
T lymphocytes are essential for cellular immune responses against foreign pathogens. T cell development follows stage-wise maturation stages in the thymus, starting from CD4–CD8– double negative (DN) thymocytes, which then mature into the CD4+CD8+ double positive (DP) stage (Yang et al., 2010). After proper positive and negative selections, the DP thymocytes give rise to CD4+CD8lo intermediate cells, which then differentiate into MHC class II–restricted CD4+ and MHC class I–restricted CD8+ single positive (SP) T cells (Singer et al., 2008; He et al., 2010). The differentiation of bipotent thymic precursors, including post-select TCRβ+ DP and CD4+CD8lo intermediate thymocytes, into SP T cells represents a critical lineage decision, which is influenced by the timing, intensity, and duration of signals derived from TCR and cytokines (Singer et al., 2008). These signals are integrated into a transcriptional network in the nucleus to stipulate the CD4+ versus CD8+ T cell lineage choice (Taniuchi and Ellmeier, 2011; Xiong and Bosselut, 2011; Issuree et al., 2017). At the center of the network are the mutually antagonistic ThPOK and Runx/CBF transcription factors. ThPOK is both necessary and sufficient for instructing the CD4+ lineage specification (He et al., 2005; Sun et al., 2005), while the expression of both Runx1 and Runx3, or their obligatory cofactor CBFβ, is absolutely necessary to ensure generation of CD8+ lineage cells (Egawa and Littman, 2008; Setoguchi et al., 2008). ThPOK expression is induced in the bipotent thymic precursors by TCR (He et al., 2008), and this induction depends on Tox, Gata3, Tcf1, and Lef1 transcription factors (Wang et al., 2008; Aliahmad et al., 2011; Steinke et al., 2014). On the other hand, ThPOK expression is antagonized through a Thpok silencer in the gene locus, which is occupied by Runx factors (He et al., 2008; Setoguchi et al., 2008) and Mazr (Sakaguchi et al., 2010). In addition, Mazr appears to synergize with either Runx1 or Runx3 to promote the thymic precursors to a CD8+ T cell fate (Sakaguchi et al., 2015).
Upon lineage decision, the lineage-committed CD4+ and CD8+ T cells undergo further intrathymic maturation including down-regulation of CD69 and CD24 (Xing et al., 2016b). An important maturation process is to solidify the individual cell identity by silencing lineage-inappropriate genes (Gullicksrud et al., 2017). Whereas mature CD4+ and CD8+ T cells have remarkably similar transcriptomes, a few genes have been identified as lineage signature genes, such as Cd4, Thpok, St8sia6, Cd40lg, Lgmn, and Itgb3 for CD4+ T cells and Cd8a, Cd8b, Runx3d, Nkg7, and Itgae for CD8+ T cells (Mingueneau et al., 2013). In CD8+ lineage T cells, Runx/CBF is essential for silencing the expression of Cd4 and Thpok (Taniuchi et al., 2002; Setoguchi et al., 2008). Our recent study has demonstrated that Tcf1 and Lef1 do not only contribute to Cd4 gene silencing, but also broadly repress the expression of CD4+ signature genes and differentiated CD4+ helper T cell genes including Foxp3 and Rorc in CD8+ T cells (Steinke et al., 2014; Xing et al., 2016a).
Among key factors programming CD8+ lineage choice and cell identity, both Tcf1/Lef1 and Runx factors use Tle proteins as cofactors. The Tle proteins contain five defined domains, with the N-terminal glutamine-rich domain (namely, Q domain) and C-terminal tryptophan-aspartate (WD)–repeat domain responsible for most of protein–protein interactions (Buscarlet and Stifani, 2007; Turki-Judeh and Courey, 2012). The full-length Tcf1 and Lef1 contain an N-terminal β-catenin binding domain, C-terminal high-mobility-group (HMG) DNA binding domain, and a context-dependent regulatory (CRD) domain that harbors histone deacetylase activity (Xing et al., 2016a). Both CRD and HMG domains in Tcf1 and Lef1 contribute to interaction with the Q domain in Tle proteins (Arce et al., 2009; Chodaparambil et al., 2014). On the other hand, both Runx1 and Runx3 use their conserved C-terminus VWRPY motifs to interact with the WD-repeat domain in Tle proteins (Levanon et al., 1998). Genetic deletion of the VWRPY motifs in both Runx1 and Runx3 (Runx1ΔV/ΔVRunx3ΔV/ΔV) result in aberrant up-regulation of CD4 and ThPOK in mature CD8+ T cells (Yarmus et al., 2006; Seo et al., 2012), suggesting a direct involvement of Runx–Tle complex in regulating CD8+ T cell identity. In Runx1ΔV/ΔVRunx3ΔV/ΔV mice, however, the frequency of CD8+ T cells was diminished in mature thymic but not in splenic T cell compartments (Seo et al., 2012). In a fetal thymic organ culture study, forced expression of Runx1, either WT or ΔVWRPY mutant, showed similar capacity to increase the frequency of CD8+ at the expense of CD4+ T cells (Telfer et al., 2004). Therefore, it remains to be unquivocally defined if Runx–Tle interaction contributes to regulation of CD8+ versus CD4+ T cell lineage choice.
As discussed above, genetic targeting of individual Tle factors revealed distinct functions in different cell types. There are several outstanding questions, e.g., whether the Tle factors are functional distinct or redundant in the process of T cell development and if there is a gene dose effect given that several Tle genes coexist in the mammalian genome. Importantly, the known Tle-interacting factors, such as Tcf1/Lef1 and Runx1/3, have quite diverse biological effects in T cells, i.e., Tcf1/Lef1 and Runx1/Runx3 are differentially required for CD4+ and CD8+ T cell lineage choice, respectively, but both cooperatively regulate Cd4 gene silencing in mature CD8+ T cells. It is unknown if Tle proteins are partitioned to different partners to exert their regulatory functions during T cell development. To address these unanswered questions, we conditionally targeted Tle1, Tle3, and Tle4, which were abundantly expressed in T lineage cells. Our detailed analyses revealed that the Runx–Tle complex interacted with a Thpok silencer and repressed Thpok expression in bipotent thymic precursors as well as committed CD8+ T cells, indicating that Tle corepressors coopt Runx factors, but not Tcf1/Lef1, for CD8+ T cell lineage decision and specific repression of Thpok in CD8+ T cells. On the other hand, Tcf1/Lef1–Tle complex was broadly required for suppression of CD4+ signature genes (except for Thpok) and differentiated helper lineage-associated genes in CD8+ T cells, demonstrating that Tle corepressors predominantly partner with Tcf1/Lef1 to establish CD8+ T cell identity. These findings uncover the physiological requirements of Tle corepressors for T cell development in stage-specific and gene context-dependent manner.
Results
Loss of Tle3 diminishes CD8+ T cell output
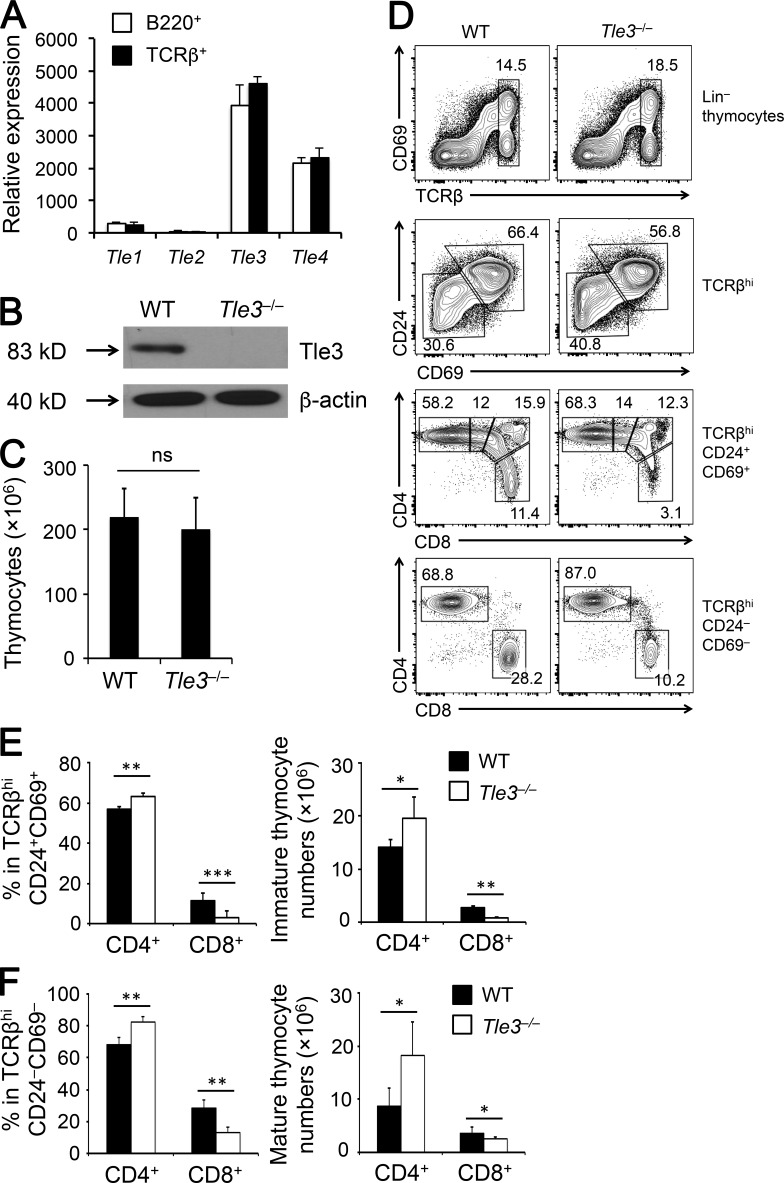
Among Tle genes that encode full-length Tle proteins, Tle3 was more abundantly expressed than Tle4 in splenic T and B cells, whereas Tle1 and Tle2 were only detected at much lower levels (Fig. 1 A). This expression pattern was observed in developing thymocytes, as determined with RNA-Seq by the Immunological Genome Project (Fig. S1). We therefore conditionally targeted the Tle3 gene first by homologous recombination (Fig. S2, A and B), and the resulting Tle3FL/+ mouse strain was crossed to Vav-Cre transgene to inactivate Tle3 in all hematopoietic cells. As expected, Tle3 protein was completely ablated in thymocytes of Vav-Cre+Tle3FL/FL (referred to as Tle3−/−) mice (Fig. 1 B). Tle3−/− and littermate controls had similar thymic cellularity (Fig. 1 C). The thymocyte maturation process in Tle3−/− mice appeared grossly normal, showing a modest increase in the DN and CD4 frequency (Fig. S2, C and D). When focusing on the post-select TCRβhi thymocytes, in both CD69+CD24+ immature and CD69–CD24– mature subsets, Tle3−/− mice showed consistent reduction in the frequency and numbers of CD8+ SP thymocytes, with concomitant increase in CD4+ SP thymocytes (Fig. 1, D–F). Similar increase in CD4+ and decrease in CD8+ T cells were observed in the periphery (Fig. S2 E). These data suggest that Tle3 is important for optimal CD8+ T cell production and may contribute to regulation of CD8+ versus CD4+ T cell lineage choice.
Figure 1.
Loss of Tle3 specifically diminishes generation of CD8+ SP thymocytes. (A) Tle gene expression in splenic T and B cells by qRT-PCR. Data are means ± SD (n = 3 from two experiments). (B) Immunoblot of Tle3 protein in total thymocytes, with β-actin as loading control. Data are representative from two experiments. (C) Total thymic cellularity of WT and Tle3−/− mice. Data are means ± SD (n = 7 from at least three experiments). (D–F) Characterization of post-select thymocytes. TCRβhi post-select thymocytes were detected in WT and Tle3−/− mice (top row in D), and further fractionated into CD69+CD24+ immature and CD69–CD24– mature subsets (second row in D). The immature subset was detected for CD4+, CD4+CD8lo, DP, and CD8+ populations (clockwise from top left in third row in D), and the mature subset was detected for CD4+ and CD8+ populations (last row in D). (E and F) Cumulative data on frequency (left panels) and numbers (right panels) of CD4+ and CD8+ SP thymocytes in the immature subset (E) and mature subset (F) are shown. Data are means ± SD (n ≥ 6 from at least three experiments). ns, not statistically significant; *, P < 0.05; ** P < 0.01; ***, P < 0.001 by Student’s t test.
Loss of Tle3 directs CD8+ to CD4+ T cell lineage by affecting ThPOK-Runx3 balance
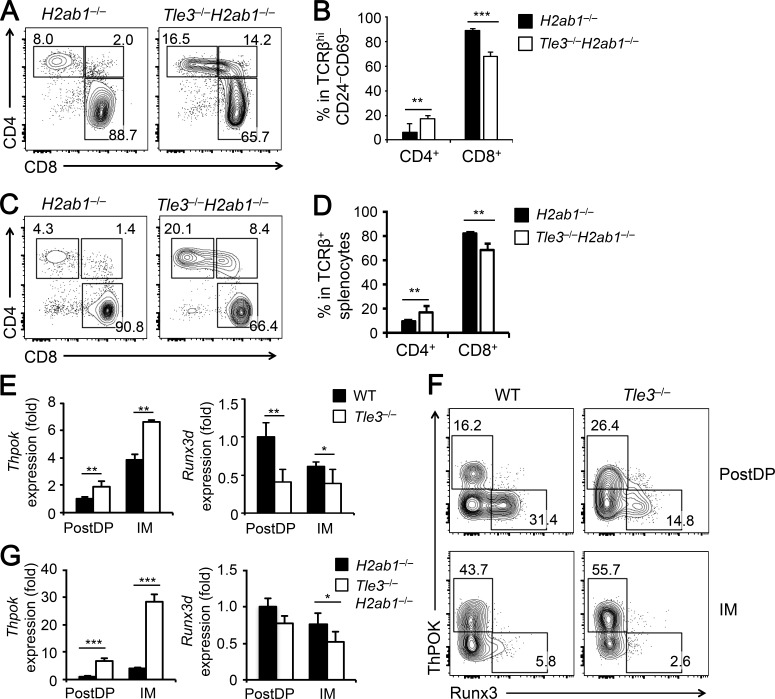
To further test the possibility that Tle3 regulates CD8+ versus CD4+ T cell lineage choice, we crossed Tle3−/− mice to the H2ab1−/− background, where the expression of MHC-II I-A and I-E molecules is defective and generation of CD4+ T cells is greatly impaired (Grusby et al., 1991). TCRβ+CD69–CD24– mature thymocytes in H2ab1−/− mice are predominantly CD8+ SP cells (Fig. 2 A). In Tle3−/−H2ab1−/− mice, however, the frequency of CD8+ SP thymocytes was reduced by ∼25%, and that of CD4+ SP thymocytes was doubled (Fig. 2, A and B). In the periphery, there was a consistent reduction in CD8+ and increase in CD4+ splenic T cells (Fig. 2, C and D). These data suggest that Tle3 deficiency causes redirection of MHC-I–selected cells to a CD4+ T cell fate. We also noted a portion of Tle3−/−H2ab1−/− mature thymic or splenic CD8+ T cells that expressed the CD4 coreceptor (called CD8*4 herein to distinguish from the true DP thymocytes; Fig. 2, A and C). This CD8*4 population may represent an alteration of CD8+ T cell identity in the absence of Tle3 and during lineage redirection (also see below).
Figure 2.
Tle3 instructs CD8+ T cell lineage choice by repressing ThPOK. (A and B) Analysis of mature thymocytes on the H2ab1−/− background. TCRβhiCD69–CD24– mature thymocytes in H2ab1−/− and Tle3−/−H2ab1−/− mice were analyzed for the distribution of CD4+ and CD8+ SP cells. Also marked is a CD8*4 population that is evident in Tle3−/−H2ab1−/− mice (A). Cumulative data on frequency of mature CD4+ and CD8+ SP thymocytes are shown in B. Data are means ± SD (n ≥ 6 from at least three experiments). (C and D) Analysis of splenic T cells on the H2ab1−/− background. TCRβ+ splenocytes from H2ab1−/− and Tle3−/−H2ab1−/− mice were analyzed for the distribution of CD4+ and CD8+ SP cells. Cumulative data on frequency of mature CD4+ and CD8+ T cells are shown in D. Data are means ± SD (n ≥ 6 from at least three experiments). (E) Analysis of Thpok and Runx3d gene expression in Tle3-targeted cells. From the TCRβhi CD69+CD24+ post-select immature thymocytes, DP (PostDP) and CD4+CD8lo intermediate (IM) cells were sorted from WT or Tle3−/− mice and analyzed by qRT-PCR. For relative expression of each gene, its expression was first normalized to the Hprt house-keeping gene in the same cell type; the gene expression in WT PostDP cells were set at 1, and its relative expression in all other cell types were normalized accordingly. Data are means ± SD from two experiments (n = 4). (F) Analysis of ThPOK and Runx3 protein gene expression in Tle3-targeted cells. PostDP and IM thymocytes from mice of indicated genotypes were intracellularly stained for ThPOK and Runx3 expression. Frequency of positive populations is shown, with the gating based on corresponding isotype control staining. Data are representative from three experiments with similar results (n = 4). (G) Analysis of Thpok and Runx3d gene expression on the H2ab1−/− background. PostDP and IM thymocytes were sorted from H2ab1−/− and Tle3−/−H2ab1−/−mice and analyzed for gene expression as in E. Data are means ± SD from two experiments (n = 4). In B, D, E, and G, *, P < 0.05; ** P < 0.01; ***, P < 0.001 by Student’s t test.
To investigate the mechanism by which Tle3 regulates CD8+ T cell fate decision, we analyzed the expression of key transcriptional regulators that promote CD4+ lineage choice, including ThPOK, Gata3, Tox, Myb, Tcf1, and Lef1, and those that promote CD8+ lineage choice, Runx1, Runx3, CBFβ, and Mazr (encoded by Patz1). In both post-select DP and CD4+CD8lo intermediate thymocytes, the expression of Thpok was increased, and that of Runx3d, a CD8+-specific transcript from the Runx3 distal promoter, was decreased in Tle3−/− cells (Fig. 2 E). The other key factors were not significantly affected in Tle3−/− cells (Fig. S3 A). ThPOK induction and Runx3 repression were also observed on the protein level in Tle3−/− post-select DP thymocytes, and this protein expression pattern is further skewed toward stronger induction of ThPOK in Tle3−/− cells during transition to CD4+CD8lo intermediate stage (Fig. 2 F).
ThPOK is induced in post-select thymocytes, more potently in CD4+CD8lo intermediate cells, by MHC-II–mediated TCR stimulation (He et al., 2008), and consistent with this knowledge, Thpok expression was >10-fold lower in H2ab1−/− post-select DP and CD4+CD8lo intermediate thymocytes compared with MHC-II-sufficient cells; on the other hand, Runx3d expression was elevated by about twofold in H2ab1−/− over WT cells (Fig. S3 B). On the H2ab1−/− background, loss of Tle3 caused markedly elevated Thpok expression, modestly reduced Runx3d expression, while having little impact on other genes in both post-select DP and CD4+CD8lo intermediate thymocytes (Fig. 2 G and Fig. S3 C). These data suggest that Tle3 primarily impinges on the ThPOK-Runx3 balance to regulate CD4+ versus CD8+ T cell lineage choice.
Triple deletion of Tle1, Tle3, and Tle4 impairs generation of lineage-committed CD8+ T cells
The CD8+ to CD4+ T cell lineage redirection did occur in the absence of Tle3, but was not efficient, suggesting functional redundancy among the Tle corepressor proteins. We next crossed Vav-Cre to existing Tle1FL/FL and Tle4FL/FL mouse strains (Wheat et al., 2014; Ramasamy et al., 2016) to generate Vav-Cre+Tle1FL/FL, Vav-Cre+Tle4FL/FL, and Vav-Cre+Tle1FL/FLTle4FL/FL mice (referred to as Tle1−/−, Tle4−/−, and Tle1−/−Tle4−/−, respectively). Immunoblotting confirmed effective ablation of Tle1 and Tle4 proteins in total thymocytes of Tle1−/−Tle4−/− mice (Fig. S4 A). Loss of Tle1 and/or Tle4 did not result in significant differences in thymic cellularity or thymocyte maturation stages (Fig. S4, B and C). Focused analyses of the immature and mature post-select thymocytes showed that neither frequency nor numbers of CD4+ or CD8+ SP cells was detectably altered by ablating Tle1 and/or Tle4 (Fig. S4, D–G). These data suggest that loss of Tle1 and Tle4 was compensated by other homologous Tle factor(s).
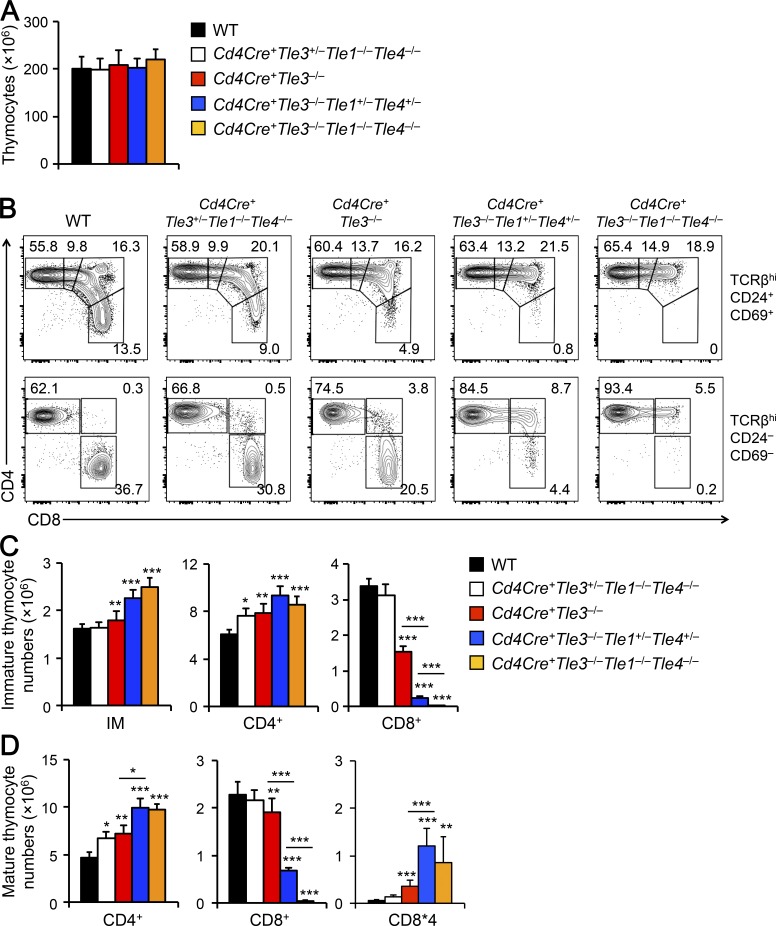
To further address redundancy among Tle proteins, we generated Vav-Cre+Tle1−/−Tle3−/−Tle4−/− mice. Compared with littermate controls, ablating three Tle proteins profoundly reduced thymic cellularity by >90%, which was likely secondary to the impact on hematopoietic stem/progenitor cells and/or early thymocytes (unpublished data). To bypass the requirements for Tle proteins at early developmental stages and to quantitatively measure the impact of Tle1/3/4 on CD8+ T cell lineage choice, we used Cd4Cre to ablate Tle proteins at the DP stage of thymocyte maturation. Late deletion of three Tle proteins in various combinations did not affect thymic cellularity (Fig. 3 A), hence providing an opportunity to directly assess the impact on the size of CD4+ and CD8+ T cell compartments. In the immature post-select thymocytes, CD8+ SP cells were diminished in both frequency and numbers in Cd4Cre+Tle3−/− (Tle1+/+Tle4+/+) mice and were almost absent in Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mice; the reduction in CD8+ SP cells was accompanied by increases in the frequency and numbers of CD4+CD8lo intermediates and CD4+ SP thymocytes (Fig. 3, B and C). Similarly, CD8+ SP cells were reduced upon loss of Tle3 and were absent in Tle1/3/4-deficient mature post-select thymocytes, with corresponding gain in frequency and number of CD4+ SP cells (Fig. 3, B and D). We also observed that the Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mature thymocytes contained a CD8*4 population, due to derepression of the CD4 coreceptor on Tle1/3/4-deficient CD8+ T cells (see below). These data suggest that Tle3, along with Tle1 and Tle4, are essential for generating lineage-committed CD8+ T cells.
Figure 3.
Loss of Tle1, Tle3 and Tle4 abrogates CD8+ T cell development. (A) Total thymic cellularity in mice of indicated genotypes. (B–D) Characterization of post-select thymocytes. TCRβhi post-select thymocytes were detected in mice of indicated genotypes, and further fractionated into CD69+CD24+ immature (top) and CD69–CD24– mature subsets (bottom). The immature subset was detected for CD4+, CD4+CD8lo, DP, and CD8+ populations (clockwise from top left, top panels), and the mature subset was detected for CD4+ and CD8+ populations, as well as CD8*4 cells (bottom panels). (C) Cumulative data on numbers of CD4+, CD4+CD8lo, DP, and CD8+ cells in the immature post-select thymocytes. (D) Cumulative data on numbers of CD4+ SP, CD8+ SP, and CD8*4 cells in the mature post-select thymocytes. Data in A, C, and D are means ± SD (n = 4–9 from at least three experiments). Statistical significance among multiple groups was first assessed with one-way ANOVA followed by Bonferroni correction. *, P < 0.05; ** P < 0.01; ***, P < 0.001 by Student’s t test for comparison with WT (unmarked) or comparison between groups marked with a horizontal line.
In addition to functional redundancy among Tle proteins, we investigated if the Tle gene dosage contributed to CD8+ T cell regulation. In Cd4Cre+Tle3+/−Tle1−/−Tle4−/− mice where only one Tle3 allele was retained with both Tle1 and Tle4 completely ablated, the reduction in CD8+ SP cells was not obvious (Fig. 3 B). In Cd4Cre+Tle3−/−Tle1+/−Tle4+/− mice where Tle3 was ablated with only one allele each of Tle1 and Tle4 was retained, the reduction in CD8+ SP cells was much more severe than that observed in Cd4Cre+Tle3−/− (Tle1+/+Tle4+/+) mice, but was less profound compared with triple Tle gene deletion (Fig. 3, B–D). In addition, Cd4Cre+Tle3−/−Tle1+/−Tle4+/− mature post-select thymocytes contained CD8*4 cells, similar to Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mice (Fig. 3, B and D). These data collectively suggest that Tle3 has a predominant role, and expression of Tle1 and Tle4 from both alleles is necessary for fully supporting development of CD8+ T cells.
Triple deletion of Tle1, Tle3, and Tle4 causes strong CD8+ to CD4+ T cell lineage redirection
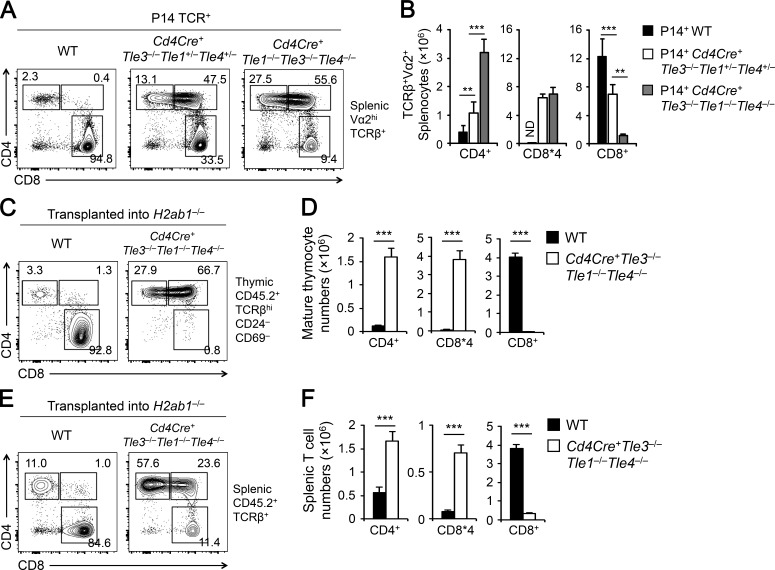
To further demonstrate that the roles of Tle proteins lie with CD8+ T lineage choice, we took two different approaches. The first is to cross the Cd4Cre-mediated triple Tle deletion mouse strain with an MHC-I-restricted P14 TCR transgene on a Rag-sufficient background. In WT P14+ transgenic mice, the vast majority of Vα2+ T cells in the periphery were CD8+ SP T cells (Fig. 4, A and B). In P14+Cd4Cre+Tle3−/−Tle1+/−Tle4+/− and P14+Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mice where complete Tle3 ablation was coupled with hetero- or homozygous inactivation of Tle1/4, respectively, the Vα2+CD8+ SP T cells were progressively lost, giving rise to in either completely directed CD4+ lineage T cells or CD8*4 cells (Fig. 4, A and B).
Figure 4.
Loss of Tle1, Tle3 and Tle4 strongly directs CD8+ to CD4+ T cell lineage. (A and B) Characterization of CD8+ and CD4+ T cell lineage distribution in the presence of an MHC-I–restricted P14 TCR transgene. Splenic Vα2+ T cells from mice of indicated genotypes were analyzed for CD8+ and CD4+ lineage distribution. Cumulative data on numbers of splenic CD4+ SP, CD8+ SP, and CD8*4 T cells are means ± SD (n ≥ 3 from at least two experiments). ** P < 0.01; ***, P < 0.001 by Student’s t test for indicated pairwise comparison. (C–F) Characterization of CD8+ and CD4+ T cell lineage distribution in H2ab1−/− recipients. Total BM cells from CD4Cre+ Tle3−/−Tle1−/−Tle4−/− or WT littermates (CD45.2+) were transplanted into irradiated CD45.1+H2ab1−/− recipient mice. 6 wk later, the recipients were analyzed for CD8+ and CD4+ lineage distribution in CD45.2+TCRβhiCD69–CD24– mature thymocytes (C and D) and in CD45.2+TCRβ+ splenocytes (E and F). Cumulative data on numbers of CD4+ SP, CD8+ SP, and CD8*4 cells are means ± SD for mature thymocytes (D) and for splenocytes (F). n ≥ 5 from two experiments. ***, P < 0.001 by Student’s t test.
As a second approach, we transplanted the bone marrow (BM) cells from CD45.2+ Cd4Cre+Tle3−/−Tle1−/−Tle4−/− or WT mice into irradiated CD45.1+H2ab1−/− recipients. As expected, WT BM-derived post-select mature thymocytes are largely restricted to the CD8+ T cell lineage, because of the defective MHC-II expression in the H2ab1−/− recipients (Fig. 4, C and D). In contrast, mature thymocytes derived from Cd4Cre+Tle3−/−Tle1−/−Tle4−/− BM cells contained few CD8+ SP cells and were mostly CD4+ or CD8*4 cells (Fig. 4, C and D). In the periphery, whereas WT TCRβ+ cells were largely CD8+, most of Cd4Cre+Tle3−/−Tle1−/−Tle4−/− TCRβ+ cells were CD4+ (Fig. 4, E and F). Although CD44hi memory phenotype cells were not excluded from the analyses of BM chimeras, these data from both approaches indicate that MHC-I–selected thymocytes are strongly redirected from CD8+ to CD4+ T cell lineage in the absence of Tle proteins.
Tle proteins repress ThPOK expression by binding to its silencer
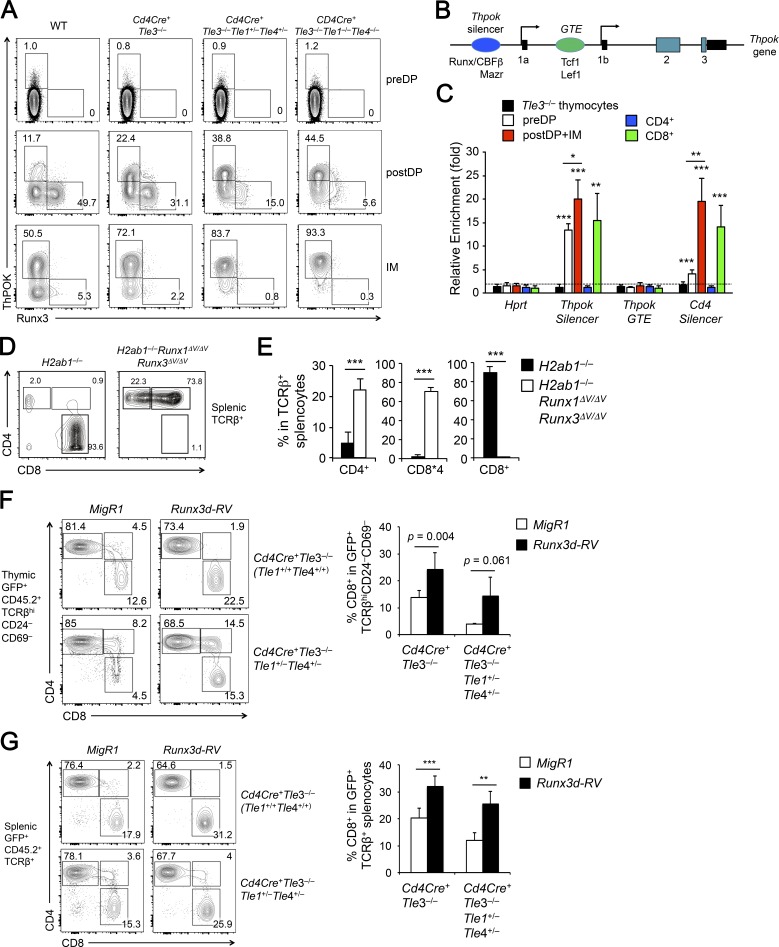
Using VavCre-mediated deletion, we found that Tle3-deficient post-select DP and CD4+CD8lo intermediate thymocytes showed increased ThPOK and decreased Runx3 expression (Fig. 2 F). We validated this finding using Cd4Cre-mediated Tle3 deletion (Fig. 5 A). When Tle3 deficiency was combined with hetero- or homozygous deletion of Tle1 and Tle4, the post-select DP thymocytes showed progressive up-regulation of ThPOK, accompanied by progressively diminished Runx3 expression (Fig. 5 A). Significantly, almost all Tle1/3/4-deficient CD4+CD8lo intermediate thymocytes expressed ThPOK (Fig. 5 A), highlighting the redundancy among Tle proteins in repressing ThPOK. We also analyzed TCRβ+Vα2+CD44med-lo DP thymocytes from P14+ Cd4Cre+Tle3−/−Tle1−/−Tle4−/− and P14+ WT mice and found that whereas ThPOK is minimally detected in WT, it was elevated in Tle1/3/4-deficient MHC-I–restricted DP thymocytes (Fig. S5 A). It is of note that CD69–TCRβlo-med preselect DP thymocytes did not aberrantly up-regulate ThPOK in the absence of Tle proteins (Fig. 5 A), and this finding was consistent with lack of ThPOK up-regulation, as measured by a Thpok locus-driven GFP reporter, in preselect DP thymocytes where Runx–Tle interaction was abrogated (Tanaka et al., 2013). ThPOK expression is induced by TCR signals (He et al., 2008), and the lack of ThPOK expression in preselect DP thymocytes is likely ascribed to absence of activating signals and/or specific chromatin configuration at the Thpok locus that is not sensitive to loss of Tle proteins at the preselect DP stage. These considerations suggest that Tle protein-dependent repression of ThPOK only becomes operative at developmental stages after positive selection.
Figure 5.
Tle proteins ensure balanced expression of ThPOK and Runx3 to instruct lineage choice. (A) Analysis of ThPOK and Runx3 protein gene expression with late deletion of Tle proteins. CD69–TCRβlo-med preselect DP (preDP), PostDP, and IM thymocytes from mice of indicated genotypes were intracellularly stained for ThPOK and Runx3 expression. Data are representative from three experiments with similar results (n ≥ 4). (B) Diagram showing the structure of Thpok gene locus. Black rectangles denote noncoding exons or noncoding sequence in an exon, and cyan rectangles denote coding exons. Ovals mark previously defined cis-regulatory elements, and their known associated transcription factors are also shown. (C) ChIP analysis of enriched Tle3 binding. TCRβlo-med preselect DP thymocytes (PreDP), PostDP + IM thymocytes, and splenic CD4+ and CD8+ T cells were sort-purified from WT mice, and VavCre+Tle3−/− total thymocytes were used as a negative control. The cells were used in ChIP with an anti-Tle3 antibody or control rabbit IgG. Relative enrichment of Tle3 binding was determined by normalizing anti-Tle3 to IgG signals at each indicated genomic location. Data are means ± SD from four experiments. Dotted line marks no enrichment. *, P < 0.05; ** P < 0.01; ***, P < 0.001 by Student’s t test compared with Tle3−/− cells or for indicated pairwise comparison. (D and E) Characterization of CD8+ and CD4+ T cell lineage distribution of Runx1ΔV/ΔV Runx3ΔV/ΔV mice on H2ab1−/− background. TCRβ+ splenocytes from indicated mice were analyzed for CD8+ and CD4+ lineage distribution (D), and cumulative data on the frequency of CD4+ SP, CD8+ SP, and CD8*4 cells are means ± SD (E, n ≥ 3 from two experiments). Note that cell numbers are not dependable readouts in this case, because Runx1ΔV/ΔV Runx3ΔV/ΔV mice showed growth retardation and early onset of inflammatory phenotypes. ***, P < 0.001 by Student’s t test. (F and G) Forced expression of Runx3 “rescues” CD8+ T cell developmental defect due to Tle deficiency. CD45.2+BM cells from Cd4Cre+Tle3−/− (top rows) or Cd4Cre+Tle3−/−Tle1+/–Tle4+/− (bottom rows) mice were infected with empty vector (MigR1) or Runx3d-expressing retrovirus (Runx3d-RV), followed by transplantation into irradiated CD45.1+ recipients. 6 wk later, CD45.2+GFP+ cells were detected in mature thymocytes (F) and splenic T cells (G) and further analyzed for CD4+ and CD8+ lineage distribution. Contour plots are representative from two experiments, and cumulative data on the frequency of CD8+ SP mature thymocytes (F) and splenic T cells (G) are shown as means ± SD (n = 4). ** P < 0.01; ***, P < 0.001 by Student’s t test, unless specified otherwise.
The Thpok gene expression is regulated by several cis-regulatory elements, including a Thpok silencer and a general T-lymphoid element (GTE; Fig. 5 B; He et al., 2010). Whereas Runx–CBFβ complex binds to the Thpok silencer to repress its expression (Egawa and Littman, 2008; He et al., 2008; Setoguchi et al., 2008), Tcf1 and Lef1 promote Thpok induction at least partly through acting on the GTE (Steinke et al., 2014). We performed chromatin immunoprecipitation (ChIP) using an anti-Tle3 antibody on mixed post-select DP and CD4+CD8lo intermediate thymocytes. Tle3 showed strong enriched binding to the Thpok silencer but not GTE (Fig. 5 C), suggesting that Tle proteins predominantly cooperate with Runx3 to directly repress Thpok expression in the bipotent thymic precursors and thus favor CD8+ T cell production. To further determine the dynamic interaction of Tle3 with the Thpok locus, we performed Tle3 ChIP on preselect DP thymocytes and mature CD4+ and CD8+ splenic T cells and found that Tle3 was associated with Thpok silencer in preselect DP thymocytes (Fig. 5 C). It is of note that Runx3 is only weakly expressed in preselect DP thymocytes (Egawa and Littman, 2011), Tle3 might be recruited to the Thpok silencer by Runx1 and poised for Thpok regulation for a later stage. On the other hand, enriched binding of Tle3 to the Thpok silencer was retained in mature CD8+, but lost in CD4+ SP T cells (Fig. 5 C), suggesting a continued involvement of Tle proteins in Thpok repression in mature CD8+ T cells (see below).
Runx1 and Runx3 interact with Tle proteins through their C-terminal VWRPY motifs (Levanon et al., 1998). In mice harboring deletion of the VWRPY sequence in both Runx1 and Runx3 proteins (i.e., Runx1ΔV/ΔVRunx3ΔV/ΔV mice), the post-select mature thymocytes or splenic T cells did not contain CD8+ SP cells, because of complete derepression of the CD4 coreceptor (Seo et al., 2012). When the Runx1ΔV/ΔVRunx3ΔV/ΔV mice were crossed to H2ab1−/− background, a substantial portion of peripheral T cells appeared in the CD4+ lineage, together with CD8*4 cells, whereas T cells in control H2ab1−/− mice were predominantly in the CD8+ lineage (Fig. 5, D and E). Thus, abrogating Runx and Tle interactions resulted in CD8+ to CD4+ lineage direction, albeit not as efficient as ablating Tle1/3/4 proteins (compare with Fig. 4, E and F). Nonetheless, this observation further corroborates a critical requirement for Runx–Tle complex in restraining Thpok expression and instructing the CD8+ T cell lineage choice.
The Runx3d transcription start site (TSS) is bound by both Runx3 and Tcf1 in splenic CD8+ T cells based on previous ChIP-Seq studies (Fig. S5 B; Lotem et al., 2013; Shan et al., 2017). By ChIP analysis, we did find enriched binding of Tle3 at the Runx3d TSS in splenic CD8+ T cells; by comparison, Tle3 binding to Runx3d or Runx3 TSSs was considerably weaker in post-select DP and CD4+CD8lo intermediate thymocytes (Fig. S5 C). These binding events may not be functionally consequential, because Runx3d expression was similar between WT and Tle1/3/4-deficient mature CD8+ SP thymocytes (see below and Fig. 6 B). Because ThPOK and Runx3 are known to have mutually antagonistic effects, we posit that decreased expression of Runx3 in Tle-deficient bipotent thymic precursors is likely secondary to aberrant up-regulation of ThPOK.
Figure 6.
Tle proteins are required for suppressing CD4+ lineage-associated genes in mature CD8+ T cells. (A) Tle1/3/4-deficient CD8*4 T cells expressed both CD8α and CD8β at reduced protein levels. CD8*4 cells were identified in TCRβhiCD69–CD24– mature thymocytes in Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mice and analyzed for CD4, CD8α and CD8β expression, with direct comparison to WT CD4+ and CD8+ mature thymocytes. n = 3 from two experiments. (B) Tle1/3/4-deficient CD8*4 T cells expressed CD8+ lineage signature genes. CD8*4 cells were sorted from TCRβhiCD69–CD24– mature thymocytes in Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mice, along with CD4+ and CD8+ SP mature thymocytes from WT mice, and analyzed for the expression of indicated CD8+ signature genes. For each gene, its expression in WT CD8+ T cells was set at 1, and its relative expression in other samples was normalized accordingly. n ≥ 4 from two experiments. (C and D) Comparison of Tle1/3/4 and Tcf1/Lef1 function in suppressing ThPOK, Foxp3, and Rorγt in CD8+ T cells. CD4+ and CD8*4 cells were identified in TCRβhiCD69–CD24– mature thymocytes in Cd4Cre+Tle3−/−Tle1−/−Tle4−/− and Cd4Cre+Tcf7−/−Lef1−/− mice (top panel in C), further gated on CD44med-lo cells and analyzed for Foxp3 and Rorγt expression (bottom panel in C), ThPOK, and Tcf1 (D) by intracellular staining, with direct comparison with WT CD4+ and CD8+ SP mature thymocytes. Values in parentheses in histograms denote geometric mean fluorescence intensity of ThPOK or Tcf1 for the whole population. Data are representative from two experiments with similar results (n ≥ 3). (E and F) Tle1/3/4-deficient CD8*4 T cells showed aberrant up-regulation of CD4+ lineage genes. Mature thymocytes (E) or splenic T cells (F) were sorted and analyzed for the expression of indicated CD4+ signature genes (E) or CD4+ enriched genes (F). For each gene, its expression in WT CD4+ T cells was set at 1, and its relative expression in other samples was normalized accordingly. Data are means ± SD (n ≥ 4 from at least two experiments). *, P < 0.05; ** P < 0.01; ***, P < 0.001 by Student’s t test for indicated pairwise comparison.
Forced expression of Runx3 helps restore CD8+ T cell development in Tle-targeted mice
ThPOK and Runx3 have mutually antagonistic effects. Genetic inactivation of ThPOK abrogates CD4+ T cell development and causes increased expression of Runx3d in post-select DP and CD4+CD8lo intermediate thymocytes (He et al., 2005; Egawa and Littman, 2008). On the other hand, genetic deletion of Runx3 together with its homologue Runx1 or deletion of the Runx cofactor CBFβ abrogates CD8+ T cell development and caused aberrant induction of ThPOK (Egawa et al., 2007; Muroi et al., 2008; Setoguchi et al., 2008). Our data above indicate a critical role of Tle proteins in restraining ThPOK induction in the bipotent thymic precursors. We next sought to determine if restoring the ThPOK–Runx3 balance in the thymic precursor cells would rectify the defect in CD8+ T cell development caused by Tle deficiency. Because Runx3 antagonizes Thpok, probably involving transcriptional as well as functional repression, we chose to overexpress Runx3d. To this end, we cloned Runx3d cDNA into a bicistronic MigR1 retroviral vector (Runx3d-RV), with GFP as an expression indicator. We then used empty vector MigR1 or Runx3d-RV retrovirus to infect BM HSPCs from Cd4Cre+Tle3−/− or Cd4Cre+Tle3−/−Tle1+/−Tle4+/− mice, followed by transplantation into CD45.1+ recipients to generate BM chimeras. These donor mice, rather than Cd4Cre+Tle3−/−Tle1−/−Tle4−/− mice, were selected to at least partly preserved Tle1 and Tle4 expression so that they could interact with ectopically expressed Runx3 protein for Thpok repression. Focused analysis of GFP+ CD45.2+ mature thymocytes showed that reduction of CD8+ SP cells was recapitulated in the recipients of MigR1-infected Cd4Cre+Tle3−/− or Cd4Cre+Tle3−/−Tle1+/−Tle4+/− BM cells (compare Fig. 5 F with Fig. 3 B). In contrast, the recipients of Runx3d-RV-infected Cd4Cre+Tle3−/− or Cd4Cre+Tle3−/−Tle1+/−Tle4+/− BM cells showed increased frequency of CD8+ SP cells in GFP+CD45.2+ mature thymocytes (Fig. 5 F). The “restoration” of CD8+ T cell production by forced expression of Runx3d from Tle3-deficient BM cells appeared to be more effective and consistent in splenic T cell pool (Fig. 5 G). We further confirmed that CD8+ T cells lacking Tle3, alone or in combination with Tle1 and Tle4 heterozygous deletion, were not associated with compensatory up-regulation of Tle2, 5, or 6 (Fig. S5 D), and that the “rescued” CD8+ T cells remained indeed deficient for Tle3, excluding the possibility that these cells arose from those having escaped Cd4Cre-mediated excision (Fig. S5 E). These data demonstrate that forced expression of Runx3d is sufficient to counter-balance the induced ThPOK due to Tle3 deficiency and help direct the bipotent thymic precursor cells to CD8+ T cell lineage.
Tle proteins engage both Runx and Tcf/Lef transcription factors to establish CD8+ T cell identity
In addition to diminishing generation of CD8+ SP T cells, loss of Tle3, in combination with heterozygous or homozygous deletion of Tle1 and Tle4, resulted in appearance of a CD8*4 subset in mature post-select thymocytes (Fig. 3 B). This subset was particularly evident when coupled with a P14 TCR transgene or on an H2ab1−/− background in mature thymocytes and/or splenic T cells (Fig. 4 A, C, and E). The CD8*4 population was previously observed when Runx3 was ablated alone or in combination with Runx1 (Egawa et al., 2007) or when both Tcf1 and Lef1 were targeted (Steinke et al., 2014), highlighting critical roles of Runx1/3 and Tcf1/Lef1 transcription factors in Cd4 gene silencing in mature CD8+ T cells. Consistent with these findings, the CD8*4 population in Cd4Cre+Tle3−/−Tle1−/−Tle4−/− post-select mature thymocytes expressed both CD8α and CD8β chains (Fig. 6 A) and known CD8+ signature genes including Runx3d, Nkg7, and Itgae (Fig. 6 B). These data indicate that the Tle1/3/4-deficient CD8*4 cells are in the CD8+ lineage and further suggest a requirement for Tle proteins in silencing Cd4 in CD8+ T cells. Indeed, Tle3 directly bound to the Cd4 silencer in mature CD8+ T cells (Fig. 5 C), consistent with known direct association of both Runx3 and Tcf1 with the Cd4 silencer (Egawa et al., 2007; Steinke et al., 2014).
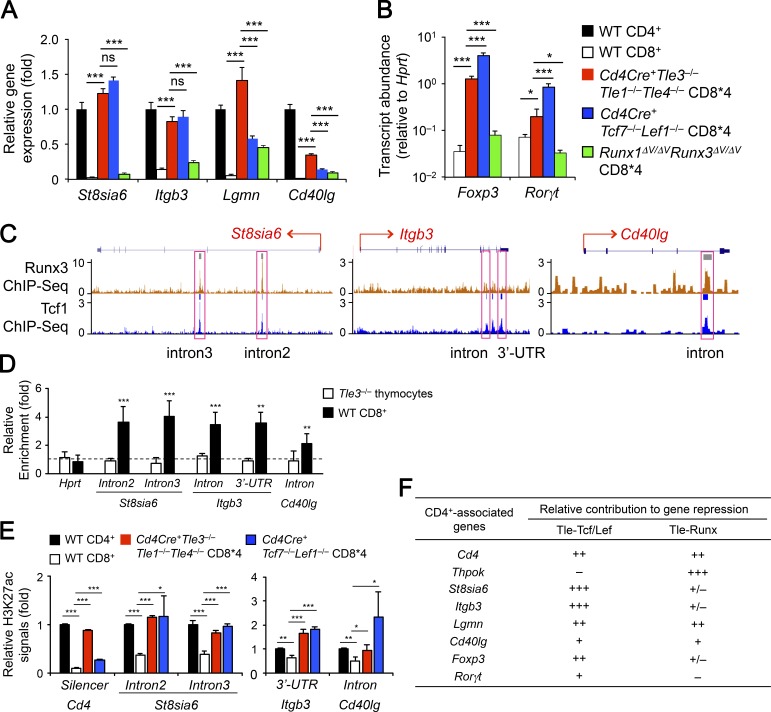
To establish and maintain CD8+ lineage integrity, it is essential to repress or silence CD4+ lineage-associated genes in addition to Cd4, such as Foxp3 and Rorγt, which encode the lineage-defining transcription factors of regulatory T and IL-17–expressing helper T cells, respectively. By intracellular staining, we found that the Tle1/3/4-deficient CD8*4 mature thymocytes showed elevated expression of Foxp3 and Rorγt in at least a portion of the population (Fig. 6 C). A microarray-based comparison of CD4+ and CD8+ T cell transcriptomes identifies six CD4+ signature genes that are expressed at least fivefold higher in CD4+ than in CD8+ T cells (including Thpok, Cd40lg, St8sia6, Lgmn, and Itgb3, in addition to Cd4) and another set of 102 CD4+ enriched genes that are expressed at least twofold, but less than fivefold in CD4+ than in CD8+ T cells (Mingueneau et al., 2013). Among CD4+ signature genes, ThPOK protein showed strong derepression in Tle1/3/4-deficient CD8*4 mature thymocytes (Fig. 6 D). Importantly, Tcf1 expression was not detectably affected by loss of Tle proteins (Fig. 6 D), excluding the possibility of a secondary effect. Tle1/3/4-deficient CD8*4 mature thymocytes also exhibited strong aberrant expression of other CD4+ signature genes such as St8sia6, Itgb3, and Lgmn and modest up-regulation of Cd40lg (Fig. 6 E). In addition, Tle1/3/4-deficient CD8*4 T cells showed increased expression of several CD4+ enriched genes, including Nrp1, Sytl2, St3gal2, Tmem64, Gata3, Tbc1d4, and Plxnd1 (Fig. 6 F), similar to what we found in Tcf1/Lef1-deficient CD8*4 T cells (Steinke et al., 2014). These observations indicate that Tle corepressors are broadly involved in repressing CD4+ lineage-associated genes in CD8+ T cells.
Because both Runx1/3 and Tcf1/Lef1 are involved in regulation of CD8+ T cell identity-related genes (Gullicksrud et al., 2017), we next investigated the relative contribution of Runx–Tle complex and Tcf/Lef–Tle complex, with a focus on the CD4+ signature genes. The CD8*4 T cells in Runx1ΔV/ΔVRunx3ΔV/ΔV mice showed increased Thpok expression, as detected with a Thpok locus-driven GFP reporter (Seo et al., 2012). In contrast, the CD8*4 cells in Cd4Cre+Tcf7fl/flLef1fl/fl (called Cd4Cre+Tcf7−/−Lef1−/−) mice did not show detectable up-regulation of ThPOK protein (Fig. 6 C). Coupled with strong binding of Tle3 to the Thpok silencer in mature CD8+ T cells (Fig. 5 C), we posit that the Runx–Tle complex is solely responsible for Thpok silencing in mature CD8+ T cells, a regulatory circuit inherited from the lineage choice stage.
We then sort-purified CD8*4 T cells in the spleens of Cd4Cre+Tle3−/−Tle1−/−Tle4−/−, Cd4Cre+Tcf7−/−Lef1−/−, and Runx1ΔV/ΔVRunx3ΔV/ΔV mice to directly compare the impact of ablating Tle proteins, ablating Tcf1/Lef1, and abrogating Runx–Tle interaction on the expression of other CD4+ signature genes. For St8sia6 and Itgb3, the expression was similarly elevated in Tle1/3/4- and Tcf1/Lef1-deficient CD8*4 cells, while the increase in Runx1ΔV/ΔVRunx3ΔV/ΔV CD8*4 cells was negligible (Fig. 7 A), indicating a more predominant role of the Tcf/Lef-Tle complex. For Lgmn and Cd40lg, the up-regulation was the strongest in Tle1/3/4-deficient CD8*4 cells and was modest in Tcf1/Lef1-deficient or Runx1ΔV/ΔVRunx3ΔV/ΔV CD8*4 cells (Fig. 7 A), suggesting the involvement of both Tcf/Lef-Tle and Runx-Tle complexes. For Foxp3 and Rorγt, the up-regulation was clearly detected in Tle1/3/4-deficient CD8*4 cells, but was not as potent as that in Tcf1/Lef1-deficient cells, whereas their expression changes in Runx1ΔV/ΔVRunx3ΔV/ΔV CD8*4 cells were minimal (Fig. 6 C and Fig. 7 B). Collectively, these data suggest that the Tcf/Lef-Tle complex has a broader role in suppressing the lineage-inappropriate genes in mature CD8+ T cells.
Figure 7.
The Tcf/Lef-Tle complex has a broader role in repressing CD4+ lineage-associated genes in mature CD8+ T cells. (A and B) Comparative analysis of CD4+ lineage-associated genes upon loss of Tle1/3/4, Tcf1/Lef1, and Runx-Tle interactions. CD8*4 cells were sorted from TCRβ+ splenocytes in Cd4Cre+Tle3−/−Tle1−/−Tle4−/−, Cd4Cre+Tcf7−/−Lef1−/− or Runx1ΔV/ΔVRunx3ΔV/ΔV mice, along with splenic CD4+ and CD8+ SP T cells from WT mice, and analyzed for the expression of indicated CD4+ lineage-associated genes. For each of CD4+ signature genes in A, its expression in WT CD4+ T cells was set at 1, and its relative expression in other samples was normalized accordingly. Because conventional CD4+ T cells do not express Foxp3, the relative abundance of Foxp3 and Rorγt transcripts in WT CD8+ or various CD8*4 cells were compared directly without normalization to WT CD4+ cells. Data are means ± SD from two experiments (n = 2–3, with each sample measured in duplicates). Statistical significance among multiple groups was first assessed with one-way ANOVA coupled with Bonferroni correction. ns, not statistically significant; *, P < 0.05; ** P < 0.01; ***, P < 0.001 by Student’s t test for indicated pairwise comparison. (C) Tcf1 and Runx3 ChIP-Seq tracks at the St8sia6, Itgb3, and Cd40lg loci in CD8+ T cells. Tcf1 and Runx3 ChIP-Seq data in CD8+ T cells were retrieved from GSE73240 and GSE50131, respectively, and loaded on UCSC genome browser, with horizontal bars on top of each track denoting MACS-called binding peaks. Pink rectangles mark Tcf1 binding peaks that were assessed for Tle3 binding (D) and H3K27ac signals (E). (D) Enriched Tle3 binding at Tcf1-occupied sites in select CD4+ signature genes. Tle3 ChIP was performed on WT splenic CD8+ T cells with VavCre+Tle3−/− total thymocytes as a negative control, and relative enrichment of Tle3 binding was determined at the indicated genomic locations. Data are means ± SD from three to four experiments. Dotted line marks no enrichment. **, P < 0.01; ***, P < 0.001 by Student’s t test. E. Tcf1- and Tle3-cooccupied sites are hyperacetylated in CD8+ T cells lacking Tcf1/Lef1 or Tle1/3/4. Cells were sorted as in A and subjected to H3K27ac ChIP analyses at indicated genomic locations. The H3K27ac signal was first normalized to chromatin input in each cell type, and that in WT CD4+ T cells was set as 1, with relative H3K27ac in other cell types normalized accordingly. Data are means ± SD from two independent experiments with each ChIP sample measured in duplicates or triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t test compared with WT CD8+ T cells. (F) Deduced relative contribution of Tcf/Lef-Tle and Runx-Tle complexes to repression of CD4+ signature genes in CD8+ T cells, based on data in Figs. 6 and 7.
We next determined if Tle proteins is directly involved in repression of CD4+ signature genes. Previously we performed Tcf1 ChIP-Seq in naive CD8+ T cells (Xing et al., 2016a), and found that multiple Tcf1 binding peaks in the intron and/or 3′UTR in St8sia6, Itgb3, and Cd40lg genes (Fig. 7 C). ChIP analysis with an anti-Tle3 antibody revealed enriched binding of Tle3 to all these Tcf1-occupied locations (Fig. 7 D), suggesting a direct recruitment of Tle proteins to the target gene loci by Tcf1. We also retrieved published data of Runx3 ChIP-Seq in naive CD8+ T cells (Lotem et al., 2013) and found cooccupancy by Runx3 at Tcf1 binding peaks in St8sia6 and Cd40lg intron regions (Fig. 7 C). The Runx3 binding to Cd40lg may help recruit Tle proteins to the loci; on the other hand, although Runx–Tle complex did not contribute to St8sia6 repression (Fig. 7 A), the binding of Runx3 to St8sia6 introns may help stabilize Tcf1 binding because of the direct interaction of Runx3 with Tcf1 (Steinke et al., 2014).
For other CD4+ lineage-associated genes, Tcf1 binding peaks were not associated with the Rorγt locus, but were found far upstream of Foxp3 and far downstream of Lgmn, and these two Tcf1 peaks were in intronic regions of neighboring genes (Fig. S5 F). Tle3 showed cooccupancy at the Tcf1 site upstream of Foxp3, but enriched Tle3 binding was not detected at the Lgmn and Rorγt loci (Fig. S5 G), implying regulation through more distal regulatory elements or indirect mechanisms by the Tcf/Lef–Tle complex. Nonetheless, our data indicate that recruitment of Tle proteins by Tcf1 and Lef1 is an important event for directly repressing a few CD4+ signature genes in CD8+ T cells.
We previously reported that Tcf1 and Lef1 have intrinsic histone deacetylase (HDAC) activity, and loss of these factors results in extensive hyperacetylation in mature CD8+ T cells (Xing et al., 2016b). Indeed, the Tcf1 binding peaks at Cd4 silencer and intronic regions of St8sia6, Itgb3, and Cd40lg genes showed increased acetylation of lysine 27 in H3 histone (H3K27ac) in Tcf1/Lef1-deficient CD8*4 cells compared with WT CD8+ T cells (Fig. 7 E). Interestingly, all these locations showed elevated H3K27ac in Tle1/3/4-deficient CD8*4 cells (Fig. 7 E), albeit this trend was less consistent with Foxp3, Rorγt, and Lgmn loci, which did not harbor Tcf1 binding peaks within the gene loci (Fig. S5 H). We posit that Tle proteins may stabilize/enhance the HDAC activity in Tcf1 and Lef1 through direct protein–protein interaction and/or recruit additional HDACs (Yochum and Ayer, 2001; Arce et al., 2009) to enforce the hypo-acetylated status of CD4+ lineage-associated gene loci in mature CD8+ T cells.
Discussion
Due to gene duplication during evolution, four full-length Tle genes are in a mammal’s genome (Jennings and Ish-Horowicz, 2008). Although targeting individual Tle genes showed some specificity in term of organogenesis (Gasperowicz and Otto, 2005; Agarwal et al., 2015), their functional redundancy, especially in immune cells, have not been addressed in vivo. In this study, we ablated three Tle genes that were abundantly expressed in T cells and uncovered physiological requirements for Tle1, Tle3, and Tle4 in CD8+ T cell lineage choice and cell identity. Tcf1 and Lef1 are frequently described as transcriptional suppressors through interactions with Tle proteins in the absence of stimulation by Wnt ligands, and it is only after Wnt stimulation, the stabilized β-catenin enters the nucleus, displaces Tle, and turns Tcf1/Lef1 into transcriptional activators (Staal and Sen, 2008; Xue and Zhao, 2012). In the late stages of T cell development, genetic ablation of Tcf1 and Lef1 greatly diminished the output of CD4+ T cells with concomitant increase in CD8+ T cells (Steinke et al., 2014). However, ablating Tle1/3/4 did not affect CD4+ T cell production, but resembled the effect of targeting Runx/CBF complex in the aspect of CD8+ versus CD4+ T cell lineage choice (Egawa et al., 2007; Egawa and Littman, 2008). Our data indicate that Runx–Tle complex has a predominant role in instructing a CD8+ T cell fate.
One critical target for the Runx–Tle complex was the Thpok silencer. Our data suggest that the Runx–Tle–Thpok silencer interaction is critical for restraining ThPOK induction in the bipotent thymic precursors to CD4+ and CD8+ SP thymocytes, and continues to exert an indispensable role in Thpok gene silencing in the committed CD8+ T cells. The Thpok silencer is bound by Runx factors and Mazr (Setoguchi et al., 2008; Sakaguchi et al., 2010), which seem to work cooperatively in Thpok suppression (Sakaguchi et al., 2015). It has been puzzling that the binding of Runx obligatory cofactor CBFβ to the Thpok silencer is detected not only in DP and CD8+ T cells, but also in CD4+ T cells where Thpok is actively transcribed. Here we showed that Tle3 binding to the Thpok silencer was only detected in DP and CD8+ but not CD4+ T cells, suggesting the presence of Tle corepressors in the Runx–CBF complex is necessary for exerting an repressive role.
Our analyses of Tle1/3/4 deficiency, coupled with MHC-I–restricted TCR or MHC-II deficiency, provided unequivocal experimental support for the role of Tle corepressors in instructing a CD8+ T cell fate. In Runx1ΔV/ΔVRunx3ΔV/ΔV mice where Runx–Tle interaction is abrogated, CD8+ SP cells are absent and replaced by CD8-positive cells showing derepression of the CD4 coreceptor (i.e., CD8*4 cells) within mature thymocyte and splenic T cell compartments (Seo et al., 2012). By further analysis of Runx1ΔV/ΔVRunx3ΔV/Δ mice on MHC-II–deficient background, we were able to clarify that CD8+ to CD4+ lineage redirection did occur when Runx–Tle interaction was interrupted, highlighting an essential requirement for Runx–Tle complex in promoting CD8+ T cell lineage choice. It is noteworthy, however, that the lineage redirection in Runx1ΔV/ΔVRunx3ΔV/ΔV mice was less efficient than that in Tle1/3/4-deficient mice or that in CD4-Cre+Runx1ΔV/ΔVRunx3FL/FL mice (Setoguchi et al., 2008). There might be at least two scenarios that account for the difference. One is that Runx1/3 may have VWPRY motif-independent effects, for example, interacting with non-Tle corepressors or interacting with Tle corepressors through additional contact surface other than the VWPRY motif. The other possibility is that the Tle corepressors might be recruited to the Thpok silencer by other protein factors, and in fact, Mazr, Satb1, and Bcl11b have been shown to bind to the Thpok silencer (Sakaguchi et al., 2015; Kakugawa et al., 2017; Kojo et al., 2017). These possibilities merit further investigation.
Based on the phenotypic characterization of mature CD8+ T cells, Tle1/3/4-deficient mice seemed to be a phenocopy of Runx1ΔV/ΔVRunx3ΔV/ΔV mice, showing complete derepression of both CD4 and ThPOK. However, in-depth analysis revealed that Tle1/3/4-deficient CD8*4 cells were molecularly different from Runx1ΔV/ΔVRunx3ΔV/ΔV CD8*4 cells, with stronger up-regulation of CD4+ signature genes as well as Foxp3 and Rorγt. These features bore strong similarity to Tcf1/Lef1-deficient CD8*4 cells (Xing et al., 2016a). In particular, two CD4+ signature genes, St8sia6 and Itgae, showed similar levels of aberrant expression in Tle1/3/4- and Tcf1/Lef1-deficient CD8*4 cells, suggesting a more dominant role of Tcf/Lef–Tle complex for transcriptional repression at these loci.
It has been well established that Runx factors act on an intronic Cd4 silencer to repress Cd4 gene expression in CD8+ T cells (Taniuchi and Ellmeier, 2011; Issuree et al., 2017). We showed that Tcf1 and Lef1 also bind to the Cd4 silencer and cooperate with Runx factors in Cd4 gene silencing (Steinke et al., 2014). Tle proteins use distinct domains for protein–protein interactions, i.e., binding Tcf1 and Lef1 through its N-terminal Q domain and binding Runx factors through its C-terminal WD-repeat domain (Levanon et al., 1998; Canon and Banerjee, 2003; Chodaparambil et al., 2014). Given that Tcf1 and Runx factors can physically interact with each other (Steinke et al., 2014), it is thus likely that Tcf/Lef, Runx, and Tle proteins form an obligatory tripartite complex to achieve stable Cd4 gene silencing. In fact, disrupting components in any of the three family factors causes CD4 derepression in CD8+ T cells. This mode of action applies to Cd40lg, another CD4+ signature gene, where Tcf/Lef-Tle and Runx-Tle act in concert to protect CD8+ T cell identity.
For all the CD4+ signature genes that harbor direct Tcf1 binding within the gene body, including Cd4, St8sia6, Itgb3 and Cd40lg, Tle proteins (at least Tle3) cooccupied with Tcf1 at these loci. We previously demonstrated that Tcf1 has intrinsic HDAC activity, which is critically required for establishing CD8+ T cell identity by preventing hyperacetylation at key CD4+-associated gene loci (Xing et al., 2016a). All these Tcf1-Tle–cooccupied sites exhibited increased H3K27ac not only in Tcf1/Lef1-deficient but also in Tle1/3/4-deficient CD8+ T cells. This observation suggests a critical involvement of Tle corepressors in histone deacetylation, presumably through stabilizing/enhancing Tcf1 HDAC activity, or independently recruiting other HDAC enzymes (Yochum and Ayer, 2001; Arce et al., 2009). For repressing aberrant up-regulation of genes associated with differentiated CD4+ helper T cells such as Foxp3 and Rorγt, whereas the Runx–Tle complex had little contribution, the Tcf1/Lef1 and Tle protein may act through more complex means. These observations suggest that establishing CD8+ T cell identity is a highly demanding task, involving multiple mechanisms tailored to specific gene context.
In summary, this study reveals in vivo requirements for Tle proteins in CD8+ T cell lineage choice and cell identity. Our findings demonstrate that Tle corepressors are differentially partitioned into Runx and Tcf/Lef complexes to exert their developmental stage-specific effects. This work represents an important step to further decipher the physiological functions of Tle proteins in immune cell development and response to pathogens.
Materials and methods
Mice
Runx1ΔV/ΔVRunx3ΔV/ΔV mice, Tle1- and Tle4-floxed mice were previously described (Seo et al., 2012; Wheat et al., 2014; Ramasamy et al., 2016), and Tle3-floxed mouse strain was generated in-house (detailed below). H2ab1−/− mice were from Taconic (model ABBN12) and crossed to B6.SJL (The Jackson Laboratory) to produce congenic CD45.1+H2ab1−/− mice, which were used as recipients for generation of BM chimeras. All animals were analyzed at 5–10 wk of age and both genders included without randomization or “blinding”. All the BM chimeras were analyzed within 6–10 wk after the BM transplantation. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa.
Conditional targeting of the Tle3 allele
The Tle3 targeting construct was designed to insert two LoxP sites to flank exons 3 and 4 (Fig. S2 A), and deletion of these two exons was predicted to cause frame-shift and ablation of the Tle3 protein. The targeting arms were PCR-amplified from C57BL/6 mouse genomic DNA, sequence-verified, and then cloned into the pNTK LoxpFrtII vector to assemble the targeting construct. The construct was then electroporated into 129 × C57BL/6 F1 hybrid embryonic stem (ES) cells, and ES clones with expected homologous recombination were screened by Southern blotting (Fig. S2 B). Blastocyst injection of the ES cells from the positive clone 160 was performed at the Transgenic Animal Model Core facility, University of Michigan (headed by T.L. Saunders). Germline-transmitted animals were then crossed with the FLP recombinase transgenic mice to remove Frt site-flanked NEO cassette. The resulting allele was designated floxed Tle3 allele (Tle3FL/+), and mice harboring this allele were crossed with the Vav-Cre transgene to inactivate Tle3 in hematopoietic cells. These mice were then crossed with Tle1FL/FL and Tle4FL/FL strains to generate triply targeted mice.
Flow cytometry and cell sorting
Single cell suspension was prepared from thymus and spleen and surface-stained. All fluorochrome-conjugated antibodies were from eBiosciences. The antibodies and their clone numbers are CD4 (RM4-5), CD8α (53-6.7), CD8β (H35-17.2); TCRβ (H57-597), CD24 (M1/69), CD69 (H1.2F3), CD25 (PC61.5), CD44 (IM7), CD45.2 (104), B220 (RA3-6B2), Gr-1 (RB6-C5), NK1.1 (PK136), TER-119 (TER-119), γδTCR (GL3), and Streptavidin (eBiosciences Cat. No. 48-4317-82). For focused analysis of αβ T cell lineage in the thymocytes, we excluded cells expressing the following lineage markers: NK1.1, Gr-1, TER-119, B220, and γδTCR. For intranuclear detection of transcription factors, anti-mouse ThPOK (T43-94, Alexa Fluor 647) with corresponding isotype control rat IgG2b-κ (A95-1, Alexa Fluor 647) and anti-mouse Runx3 (R3-5G4, PE) and corresponding isotype control mouse IgG1-κ (MOPC-21, PE) were obtained from BD Biosciences; anti-Tcf1 rabbit mAb (C63D9, Alexa Fluor 647) and corresponding isotype control rabbit mAb IgG (DA1E, Alexa Fluor 647) were from Cell Signaling Technologies. Data were collected on FACSVerse (BD Biosciences) and analyzed with FlowJo software (Version X, TreeStar). For cell sorting, surface-stained cells were sorted on BD FACSAria II or FACSAria Fusion cell sorter.
Immunoblotting
Cell lysates were prepared from total thymocytes, resolved on SDS-PAGE, followed by immunoblotting with anti-Tle1 (clone C-7, sc137097, mouse monoclonal), anti-Tle3 (clone M-201, sc-9124, rabbit polyclonal), or anti-Tle4 (clone E-10, sc365406, mouse monoclonal) antibodies, with anti–β-actin (clone I-19) detecting equal loading. All antibodies are from Santa Cruz Biotechnology.
Gene expression analysis
For quantification of Tle gene expression, splenic B220+ and TCRβ+ cells were sorted, and the total RNA was extracted from sorted cells and reverse-transcribed as previously described (Xing et al., 2016a). Plasmids containing Tle1, Tle2, Tle3, Tle4, or Hprt coding sequence were used to generate standard curves for each transcript, and each transcript in the sorted cells was determined by quantitative PCR (qPCR). The copy numbers of Tle genes were then calculated assuming that Hprt is expressed at 10,000 copies.
For identification of CD4+ signature genes, published microarray data were retrieved and analyzed (GSE15907; Mingueneau et al., 2013). For comparison of gene expression in gene-targeted and control mice, target cell populations were sorted from thymocytes or splenocytes, RNA extraction, reverse-transcription, and qPCR were performed as described (Xing et al., 2016a). Relative gene expression was calculated as specified in the figure legends. The primer sequences are either previously published (Xing et al., 2016a) or in Table S1.
Generation of BM chimeras
For detection of CD8+ to CD4+ T cell lineage redirection, whole BM cells were isolated from various donors, and 2 × 106 cells were transplanted into CD45.1+H2ab1−/− mice that were irradiated at 1,050 rad. 6–10 wk later, the recipient mice were analyzed.
For detecting the impact of forced expression of Runx3d, BM cells from donor mice were lineage depleted, and then retrovirally infected with empty-vector or Runx3d-expressing MigR1 retrovirus as previously described (Xing et al., 2016a). The infected BM cells, containing 2,000–5,000 GFP+Lin–Sca1+c-Kit+ cells (enriched in HSPCs) were then transplanted into irradiated CD45.1+ B6.SJL mice and analyzed as above.
ChIP
Pre-select DP, post-select DP + CD4+CD8lo intermediate thymocytes, splenic CD4+, and CD8+ T cells were sorted from WT C57BL/6 mice. Total thymocytes from VavCre+Tle3−/− mice were used as negative control. The cells were incubated with 2 mM disuccinimidyl glutarate (Sigma-Aldrich) for 25 min at room temperature and then cross-linked with 1% formaldehyde in medium for 10 min. The fixed cells were processed using truChIP Chromatin Shearing Reagent kit (Covaris) and sonicated for 6 min on Covaris S2 ultrasonicator. The sheared chromatin was in the range of 300–800 bp and was immunoprecipitated with 3 µg anti-Tle3 antibody (Cat. No. 11372-1-AP, rabbit polyclonal antibody, Proteintech Group) or control rabbit IgG, followed by steps of washing as previously described (Xing et al., 2016a). Genomic locations of interest were detected by qPCR, and the relative enrichment of Tle3 binding was determined by normalizing Tle3 ChIP signals to IgG ChIP signals. The primers for assessing enriched Tle3 binding are listed in Table S1.
To assess H3K27ac levels at key CD4+ lineage-associated gene loci, CD8*4 cells were sorted from TCRβ+ splenocytes in Cd4Cre+Tle3−/−Tle1−/−Tle4−/− or Cd4Cre+Tcf7−/−Lef1−/− mice, along with splenic CD4+ CD25– and CD8+ TCRβ+ cells from WT mice. The cells were subjected to ChIP analysis with anti-H3K27ac as previously described (Xing et al., 2016a).
Statistical analysis
For pairwise comparison between gene-targeted animals with control littermates, the Student’s t test was used, with a two-tailed distribution assuming equal sample variance. P values of no more than 0.05 are considered statistically significant; the following asterisk marks are used to indicate the level of significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values ≥0.05 are considered not statistically significant (marked as “ns” or in actual P values). For multiple group comparisons, Bonferroni correction was used to assess statistical significance.
Online supplemental materials
Fig. S1 shows expression changes in Tle, Tcf/Lef, and Runx/CBF factors during T cell development. Fig. S2 documents generation and characterization of Tle3 conditionally targeted mice. Fig. S3 demonstrates Tle3 deficiency does not strongly affect the expression of lineage-specifying transcriptional regulators other than the ThPOK–Runx3 axis. Fig. S4 shows that double deletion of Tle1 and Tle4 did not detectably perturb T cell development. Fig. S5 demonstrates connection of Tle proteins with target genes. Table S1 shows primers for qRT-PCR.
Supplementary Material
Acknowledgments
We thank the University of Iowa Flow Cytometry Core facility (J. Fishbaugh, H. Vignes, and M. Shey) for cell sorting, Radiation Core facility (A. Kalen) for mouse irradiation, Yoram Groner (Weizmann Institute of Science, Israel) for sharing the Runx3ΔV/ΔV mice with I.T. laboratory, B. Zhou for generating the Tle3-targeting construct, and University of Michigan Transgenic Animal Model Core (headed by T.L. Saunders) for blastocyst injection.
The Flow Cytometry Core Facility is supported by the Carver College of Medicine/Holden Comprehensive Cancer Center (the University of Iowa), the Iowa City Veteran’s Administration Medical Center, and the National Center for Research Resources of the National Institutes of Health (NIH; 1S10 OD016199). This study is supported in part by grants from the NIH (AI112579, AI119160, and AI121080 to H.-H. Xue, and CA115772 to D.A. Sweetser), and the Veteran Affairs BLR&D Merit Review Program (BX002903A to H.-H. Xue). C. Liu is supported by the intramural research program of the National Heart, Lung, and Blood Institute, NIH.
The authors declare no competing financial interests.
Author contributions S. Xing performed all the experiments with help from P. Shao, F. Li, X. Zhao, and S. Yu; C. Liu supervised generation of Tle3-targeted mice, W. Seo, J.C. Wheat, S. Ramasamy, J. Wang, I. Taniuchi, and D.A. Sweetser provided essential reagents; S. Ramasamy and I. Taniuchi shared unpublished data on related studies; X. Li and W. Peng analyzed ChIP-seq data in public domain. C. Liu, I. Taniuchi, and D.A. Sweetser provided scientific insights. S. Xing and H.-H. Xue analyzed the data. H.-H. Xue conceived the project, supervised the overall study, and wrote the paper.
References
- Agarwal M., Kumar P., and Mathew S.J.. 2015. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life. 67:472–481. 10.1002/iub.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliahmad P., Kadavallore A., de la Torre B., Kappes D., and Kaye J.. 2011. TOX is required for development of the CD4 T cell lineage gene program. J. Immunol. 187:5931–5940. 10.4049/jimmunol.1101474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L., Pate K.T., and Waterman M.L.. 2009. Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer. 9:159 10.1186/1471-2407-9-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantjes H., Roose J., van De Wetering M., and Clevers H.. 2001. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29:1410–1419. 10.1093/nar/29.7.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M., and Stifani S.. 2007. The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 17:353–361. 10.1016/j.tcb.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Cai X., Gao L., Teng L., Ge J., Oo Z.M., Kumar A.R., Gilliland D.G., Mason P.J., Tan K., and Speck N.A.. 2015. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 17:165–177. 10.1016/j.stem.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J., and Banerjee U.. 2003. In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes Dev. 17:838–843. 10.1101/gad.1064803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodaparambil J.V., Pate K.T., Hepler M.R., Tsai B.P., Muthurajan U.M., Luger K., Waterman M.L., and Weis W.I.. 2014. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 33:719–731. 10.1002/embj.201387188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D.L., and Weis W.I.. 2005. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12:364–371. 10.1038/nsmb912 [DOI] [PubMed] [Google Scholar]
- Dayyani F., Wang J., Yeh J.R., Ahn E.Y., Tobey E., Zhang D.E., Bernstein I.D., Peterson R.T., and Sweetser D.A.. 2008. Loss of TLE1 and TLE4 from the del(9q) commonly deleted region in AML cooperates with AML1-ETO to affect myeloid cell proliferation and survival. Blood. 111:4338–4347. 10.1182/blood-2007-07-103291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D., Jiménez G., Heavey B., and Busslinger M.. 2000. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19:2292–2303. 10.1093/emboj/19.10.2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., and Littman D.R.. 2008. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9:1131–1139. 10.1038/ni.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., and Littman D.R.. 2011. Transcription factor AP4 modulates reversible and epigenetic silencing of the Cd4 gene. Proc. Natl. Acad. Sci. USA. 108:14873–14878. 10.1073/pnas.1112293108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Tillman R.E., Naoe Y., Taniuchi I., and Littman D.R.. 2007. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204:1945–1957. 10.1084/jem.20070133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperowicz M., and Otto F.. 2005. Mammalian Groucho homologs: redundancy or specificity? J. Cell. Biochem. 95:670–687. 10.1002/jcb.20476 [DOI] [PubMed] [Google Scholar]
- Grusby M.J., Johnson R.S., Papaioannou V.E., and Glimcher L.H.. 1991. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 253:1417–1420. 10.1126/science.1910207 [DOI] [PubMed] [Google Scholar]
- Gullicksrud G.A., Shan Q., and Xue H.H.. 2017. Tcf1 at the crossroads of CD4+ and CD8+ T cell identity. Front. Biol. 12:83–93. 10.1007/s11515-017-1445-3 [DOI] [Google Scholar]
- He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., and Kappes D.J.. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833. 10.1038/nature03338 [DOI] [PubMed] [Google Scholar]
- He X., Park K., Wang H., He X., Zhang Y., Hua X., Li Y., and Kappes D.J.. 2008. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 28:346–358. 10.1016/j.immuni.2008.02.006 [DOI] [PubMed] [Google Scholar]
- He X., Park K., and Kappes D.J.. 2010. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu. Rev. Immunol. 28:295–320. 10.1146/annurev.immunol.25.022106.141715 [DOI] [PubMed] [Google Scholar]
- Issuree P.D., Ng C.P., and Littman D.R.. 2017. Heritable Gene Regulation in the CD4:CD8 T Cell Lineage Choice. Front. Immunol. 8:291 10.3389/fimmu.2017.00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings B.H., and Ish-Horowicz D.. 2008. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9:205 10.1186/gb-2008-9-1-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakugawa K., Kojo S., Tanaka H., Seo W., Endo T.A., Kitagawa Y., Muroi S., Tenno M., Yasmin N., Kohwi Y., et al. 2017. Essential Roles of SATB1 in Specifying T Lymphocyte Subsets. Cell Reports. 19:1176–1188. 10.1016/j.celrep.2017.04.038 [DOI] [PubMed] [Google Scholar]
- Kojo S., Tanaka H., Endo T.A., Muroi S., Liu Y., Seo W., Tenno M., Kakugawa K., Naoe Y., Nair K., et al. 2017. Priming of lineage-specifying genes by Bcl11b is required for lineage choice in post-selection thymocytes. Nat. Commun. 8:702 10.1038/s41467-017-00768-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Goldstein R.E., Bernstein Y., Tang H., Goldenberg D., Stifani S., Paroush Z., and Groner Y.. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA. 95:11590–11595. 10.1073/pnas.95.20.11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderson Y., Eberhard D., Malin S., Johansson A., Busslinger M., and Pettersson S.. 2004. Corecruitment of the Grg4 repressor by PU.1 is critical for Pax5-mediated repression of B-cell-specific genes. EMBO Rep. 5:291–296. 10.1038/sj.embor.7400089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotem J., Levanon D., Negreanu V., Leshkowitz D., Friedlander G., and Groner Y.. 2013. Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS One. 8:e80467 10.1371/journal.pone.0080467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M., Kreslavsky T., Gray D., Heng T., Cruse R., Ericson J., Bendall S., Spitzer M.H., Nolan G.P., Kobayashi K., et al. Immunological Genome Consortium . 2013. The transcriptional landscape of αβ T cell differentiation. Nat. Immunol. 14:619–632. 10.1038/ni.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi S., Naoe Y., Miyamoto C., Akiyama K., Ikawa T., Masuda K., Kawamoto H., and Taniuchi I.. 2008. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat. Immunol. 9:1113–1121. 10.1038/ni.1650 [DOI] [PubMed] [Google Scholar]
- Ramasamy S., Saez B., Mukhopadhyay S., Ding D., Ahmed A.M., Chen X., Pucci F., Yamin R., Wang J., Pittet M.J., et al. 2016. Tle1 tumor suppressor negatively regulates inflammation in vivo and modulates NF-κB inflammatory pathway. Proc. Natl. Acad. Sci. USA. 113:1871–1876. 10.1073/pnas.1511380113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Hombauer M., Bilic I., Naoe Y., Schebesta A., Taniuchi I., and Ellmeier W.. 2010. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat. Immunol. 11:442–448. 10.1038/ni.1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Hainberger D., Tizian C., Tanaka H., Okuda T., Taniuchi I., and Ellmeier W.. 2015. MAZR and Runx Factors Synergistically Repress ThPOK during CD8+ T Cell Lineage Development. J. Immunol. 195:2879–2887. 10.4049/jimmunol.1500387 [DOI] [PubMed] [Google Scholar]
- Seo W., Tanaka H., Miyamoto C., Levanon D., Groner Y., and Taniuchi I.. 2012. Roles of VWRPY motif-mediated gene repression by Runx proteins during T-cell development. Immunol. Cell Biol. 90:827–830. 10.1038/icb.2012.6 [DOI] [PubMed] [Google Scholar]
- Setoguchi R., Tachibana M., Naoe Y., Muroi S., Akiyama K., Tezuka C., Okuda T., and Taniuchi I.. 2008. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 319:822–825. 10.1126/science.1151844 [DOI] [PubMed] [Google Scholar]
- Shan Q., Zeng Z., Xing S., Li F., Hartwig S.M., Gullicksrud J.A., Kurup S.P., Van Braeckel-Budimir N., Su Y., Martin M.D., et al. 2017. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18:931–939. 10.1038/ni.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin T.H., Brynczka C., Dayyani F., Rivera M.N., and Sweetser D.A.. 2016. TLE4 regulation of wnt-mediated inflammation underlies its role as a tumor suppressor in myeloid leukemia. Leuk. Res. 48:46–56. 10.1016/j.leukres.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A., Adoro S., and Park J.H.. 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8:788–801. 10.1038/nri2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal F.J.T., and Sen J.M.. 2008. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur. J. Immunol. 38:1788–1794. 10.1002/eji.200738118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke F.C., and Xue H.H.. 2014. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol. Res. 59:45–55. 10.1007/s12026-014-8545-9 [DOI] [PubMed] [Google Scholar]
- Steinke F.C., Yu S., Zhou X., He B., Yang W., Zhou B., Kawamoto H., Zhu J., Tan K., and Xue H.H.. 2014. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 15:646–656. 10.1038/ni.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Liu X., Mercado P., Jenkinson S.R., Kypriotou M., Feigenbaum L., Galéra P., and Bosselut R.. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6:373–381. 10.1038/ni1183 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Naito T., Muroi S., Seo W., Chihara R., Miyamoto C., Kominami R., and Taniuchi I.. 2013. Epigenetic Thpok silencing limits the time window to choose CD4(+) helper-lineage fate in the thymus. EMBO J. 32:1183–1194. 10.1038/emboj.2013.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I., and Ellmeier W.. 2011. Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Adv. Immunol. 110:71–110. 10.1016/B978-0-12-387663-8.00003-X [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., and Littman D.R.. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. 10.1016/S0092-8674(02)01111-X [DOI] [PubMed] [Google Scholar]
- Telfer J.C., Hedblom E.E., Anderson M.K., Laurent M.N., and Rothenberg E.V.. 2004. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J. Immunol. 172:4359–4370. 10.4049/jimmunol.172.7.4359 [DOI] [PubMed] [Google Scholar]
- Turki-Judeh W., and Courey A.J.. 2012. Groucho: a corepressor with instructive roles in development. Curr. Top. Dev. Biol. 98:65–96. 10.1016/B978-0-12-386499-4.00003-3 [DOI] [PubMed] [Google Scholar]
- Wang L., Wildt K.F., Zhu J., Zhang X., Feigenbaum L., Tessarollo L., Paul W.E., Fowlkes B.J., and Bosselut R.. 2008. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat. Immunol. 9:1122–1130. 10.1038/ni.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat J.C., Krause D.S., Shin T.H., Chen X., Wang J., Ding D., Yamin R., and Sweetser D.A.. 2014. The corepressor Tle4 is a novel regulator of murine hematopoiesis and bone development. PLoS One. 9:e105557 10.1371/journal.pone.0105557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Li F., Zeng Z., Zhao Y., Yu S., Shan Q., Li Y., Phillips F.C., Maina P.K., Qi H.H., et al. 2016a Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 17:695–703. 10.1038/ni.3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Wang X., Jameson S.C., and Hogquist K.A.. 2016b Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17:565–573. 10.1038/ni.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., and Bosselut R.. 2011. The enigma of CD4-lineage specification. Eur. J. Immunol. 41:568–574. 10.1002/eji.201041098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H.H., and Zhao D.M.. 2012. Regulation of mature T cell responses by the Wnt signaling pathway. Ann. N. Y. Acad. Sci. 1247:16–33. 10.1111/j.1749-6632.2011.06302.x [DOI] [PubMed] [Google Scholar]
- Yang Q., Jeremiah Bell J., and Bhandoola A.. 2010. T-cell lineage determination. Immunol. Rev. 238:12–22. 10.1111/j.1600-065X.2010.00956.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmus M., Woolf E., Bernstein Y., Fainaru O., Negreanu V., Levanon D., and Groner Y.. 2006. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc. Natl. Acad. Sci. USA. 103:7384–7389. 10.1073/pnas.0602470103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochum G.S., and Ayer D.E.. 2001. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 21:4110–4118. 10.1128/MCB.21.13.4110-4118.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.