Arnold et al. report that eosinophils in the gastrointestinal tract are conditioned by IFN-γ to restrict Th1 responses and promote tissue homeostasis. Eosinophils control Th1 cells in acute and chronic infection and in the steady state and possess bactericidal properties.
Abstract
Eosinophils are predominantly known for their contribution to allergy. Here, we have examined the function and regulation of gastrointestinal eosinophils in the steady-state and during infection with Helicobacter pylori or Citrobacter rodentium. We find that eosinophils are recruited to sites of infection, directly encounter live bacteria, and activate a signature transcriptional program; this applies also to human gastrointestinal eosinophils in humanized mice. The genetic or anti–IL-5–mediated depletion of eosinophils results in improved control of the infection, increased inflammation, and more pronounced Th1 responses. Eosinophils control Th1 responses via the IFN-γ–dependent up-regulation of PD-L1. Furthermore, we find that the conditional loss of IFN-γR in eosinophils phenocopies the effects of eosinophil depletion. Eosinophils further possess bactericidal properties that require their degranulation and the deployment of extracellular traps. Our results highlight two novel functions of this elusive cell type and link it to gastrointestinal homeostasis and anti-bacterial defense.
Introduction
Eosinophils arise in the bone marrow (BM) from granulocyte-monocyte progenitors and are released into the circulation as terminally differentiated cells, which follow an eotaxin gradient to their target tissues. The differentiation and survivalof eosinophils depends on the cytokine IL-5. The tissue distribution of eosinophils at steady-state is asymmetric, with large reservoirs of eosinophils found in the lower gastrointestinal (GI) tract, uterus, ovaries, and thymus and few or no eosinophils residing in the skin, esophagus, and lungs (Throsby et al., 2000; Gouon-Evans and Pollard, 2001; Wu et al., 2011; Chu and Berek, 2013). The lamina propria (LP) of the GI tract hosts by far the most abundant population of resident eosinophils, which comprise 20–30% of the total GI LP leukocyte population (Jung and Rothenberg, 2014). The presence of eosinophils in the skin and lung is generally associated with disease. Eosinophilia is a major hallmark of allergic asthma and atopic dermatitis, but is also typical of chronic inflammatory conditions of the lower GI tract. The tissue-damaging and toxic consequences of eosinophilia at these sites have been attributed to several factors released by activated eosinophils through degranulation, which include eosinophil cationic protein (ECP, which forms pores in the membranes of target cells), eosinophil peroxidase (EPX; which produces reactive oxygen species such as superoxide, hypobromite, and peroxide), and the major basic protein (an enzyme with bactericidal and helminthotoxic properties; Lehrer et al., 1989; Rosenberg and Dyer, 1995; Rosenberg et al., 2013; Soragni et al., 2015). Eosinophils are rapidly recruited to sites of tissue damage in response to injury or exposure to pathogens, which directly promotes the pathogenesis of inflammatory bowel diseases (McGovern et al., 1991; Winterkamp et al., 2000; Aydemir et al., 2004; Griseri et al., 2015). Whereas the pathogenic properties of eosinophils are relatively well understood, less is known about their function in the steady-state. Two recent studies have reported that GI eosinophils promote intestinal homeostasis by inducing IgA class switching in B cells (Chu et al., 2014), and also by inducing the development of Peyer’s patches and enhancing intestinal mucus secretion (Jung et al., 2015).
Here, we set out to examine the role of GI tract eosinophils in the steady-state as well as during acute or chronic bacterial infection. We took advantage of three complementary models of eosinophil deficiency, one constitutive and two inducible, in conjunction with a chronic and an acute infection model using the bacterial pathogens Helicobacter pylori and Citrobacter rodentium, respectively, to dissect the contribution of eosinophils to GI tract homeostasis and to anti-bacterial inflammation and immunity. We found that eosinophils contribute to homeostasis and restrict immunopathology by locally suppressing Th1 responses. We further show that eosinophils directly encounter live bacteria in the GI LP and demonstrate the bactericidal activity of eosinophils against Citrobacter but not Helicobacter. The results provide evidence of two novel functions of this versatile cell type and further reveal IFN-γ as master regulator of eosinophil activity.
Results
Eosinophil evolution in the GI tract in the steady-state and during bacterial infection
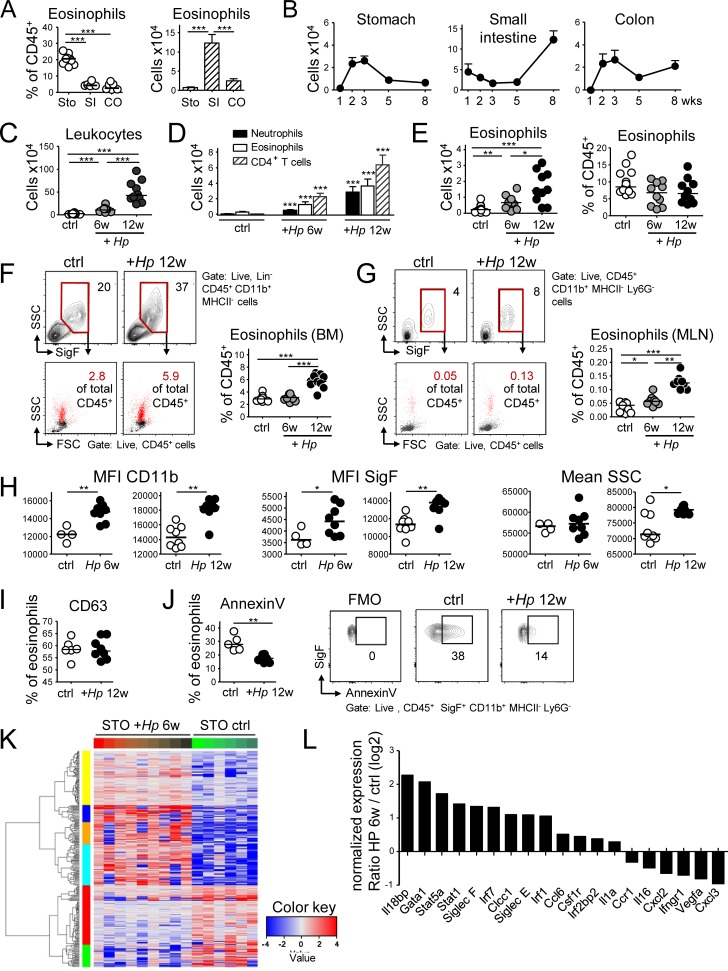
The lower GI tract is populated by a large resident pool of eosinophils (Jung and Rothenberg, 2014); in contrast, little is known about eosinophils and their function in the upper GI tract. We first compared eosinophil numbers and frequencies in the stomach, small intestine, and colon at steady-state and monitored their evolution in these organs during the first 8 wk of life. Eosinophils were identified in GI LP preparations as live CD45+CD11b+MHCII−Ly6G−SiglecF+ cells that constitutively express the chemokine receptor CCR3, the IL-5 receptor, F4/80 and CD44 (Fig. S1, A–D). Eosinophil frequencies, but not absolute numbers per organ, were much higher in the stomach than in the intestines of adult mice (Fig. 1 A); the gradual population of the stomach by eosinophils paralleled that of the colon, but not of the small intestine, peaked at around the time of weaning, and reached an equilibrium in adult animals between 5 and 8 wk of age (Fig. 1 B).
Figure 1.
Comparative analysis of the evolution and composition of the murine GI LP eosinophil population at steady-state and during H. pylori infection. (A) Frequencies (in percent of live CD45+ leukocytes) and absolute counts per organ of CD11b+MHCII−Ly6G−Siglec-F+ eosinophils in the three indicated compartments in 8-wk-old mice. n = 6–8; STO, stomach; SI, small intestine; CO, colon. (B) Absolute eosinophil counts per organ at the indicated age (in weeks). n = 4–5 per time point, shown as mean + SEM. (C–L) Eosinophil recruitment to, and activation in, the H. pylori–infected gastric LP at 6 and 12 wk post infection (p.i.), as assessed by flow cytometry and RNA sequencing. (C) Absolute numbers of CD45+ leukocytes in the gastric LP; n = 10–15, pooled from two independent studies. (D) Absolute numbers of the indicated gastric LP leukocyte populations of the mice shown in C. (E) Gastric LP eosinophil numbers and frequencies of the mice shown in C. (F) Frequencies of eosinophils in the BM of the mice shown in C, shown with representative FACS plots of eosinophils as identified by their Siglec F staining and granularity and back-gated into FSC/SSC plots of all CD45+ live cells. Lineage markers include CD3, CD4, B220, TCRα/β, NK 1.1, Ter119, and GR1. (G) Frequencies of eosinophils in the MLNs of the mice shown in C; representative FACS plots show the same gates as in F. In C–G, four and eight control mice were age-matched to the 6- and 12-wk infection, respectively. (H) Activation state of the gastric LP eosinophils shown in E, as assessed by CD11b and Siglec-F expression and side scatter. MFI, mean fluorescence intensity. (I) Frequencies of degranulated eosinophils as identified by their CD63 expression, of all gastric LP eosinophils at 12 wk p.i. (J) Frequencies of apoptotic eosinophils as identified by Annexin V binding, of all gastric LP eosinophils at 12 wk p.i. relative to controls, shown with representative FACS plots. (K) Heat map of the top 100 most differentially regulated transcripts in FACS-sorted gastric LP eosinophil samples from nine infected relative to six naive mice. (L) Selected log2 expression ratios of transcripts potentially associated with either regulatory or pro-inflammatory functions of eosinophils. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as calculated by Mann-Whitney test. Horizontal lines in scatter plots indicate medians throughout. Each symbol represents one mouse.
We next investigated the impact of experimental infection of the stomach with the gastric pathogen H. pylori on eosinophil numbers, frequencies, and phenotype. The number of eosinophils increased fivefold due to the infection and eosinophils, along with CD4+ T cells and neutrophils, constituted the numerically dominant leukocyte population in the infected stomach (Fig. 1, C–E). Higher eosinophil counts in the infected stomach were paralleled by increased eosinophil development in the BM and their accumulation in the mesenteric lymph nodes (MLNs; Fig. 1, F and G). Eosinophils in the infected stomach were activated as indicated by their elevated expression of CD11b and SiglecF and by their increased granularity relative to eosinophils in the steady-state stomach (Fig. 1 H). In contrast, the expression of CD63, a marker of eosinophil degranulation, did not change (Fig. 1 I). Eosinophils in the infected stomach further showed lower rates of Annexin V positivity, which is indicative of their extended survival (Fig. 1 J). The transcriptional signature as determined by RNA sequencing of sorted, >95% pure eosinophil populations clearly segregated eosinophils from infected and uninfected stomachs (Fig. 1 K), suggesting that gastric eosinophils respond to bacterial infection. Among the most differentially expressed genes were Siglecf and various transcription factors such as Irf1, Irf7, Stat1, Stat5a, and Gata1 (Fig. 1 L). Overall, the results suggest that eosinophils comprise an abundant leukocyte population already in the steady-state stomach, and that bacterial infection further augments their numbers as well as their activation, which in turn is accompanied by increased eosinopoiesis in the BM.
Eosinophil deficiency promotes gastric effector T cell responses and immune control of H. pylori
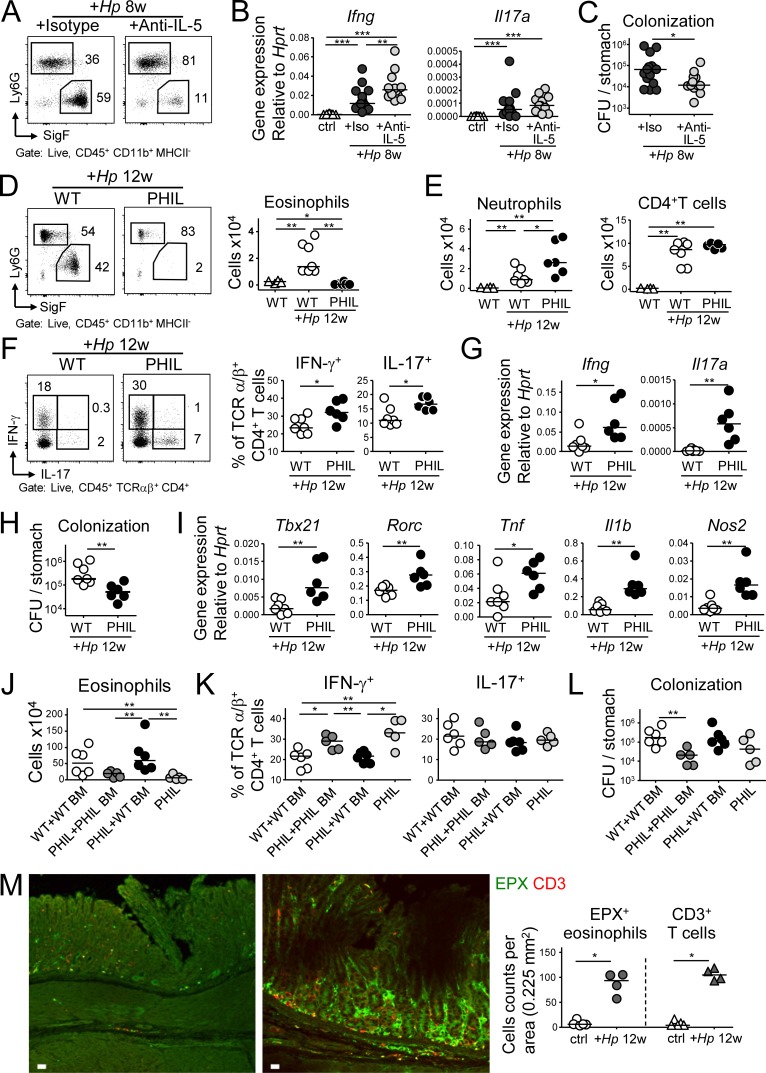
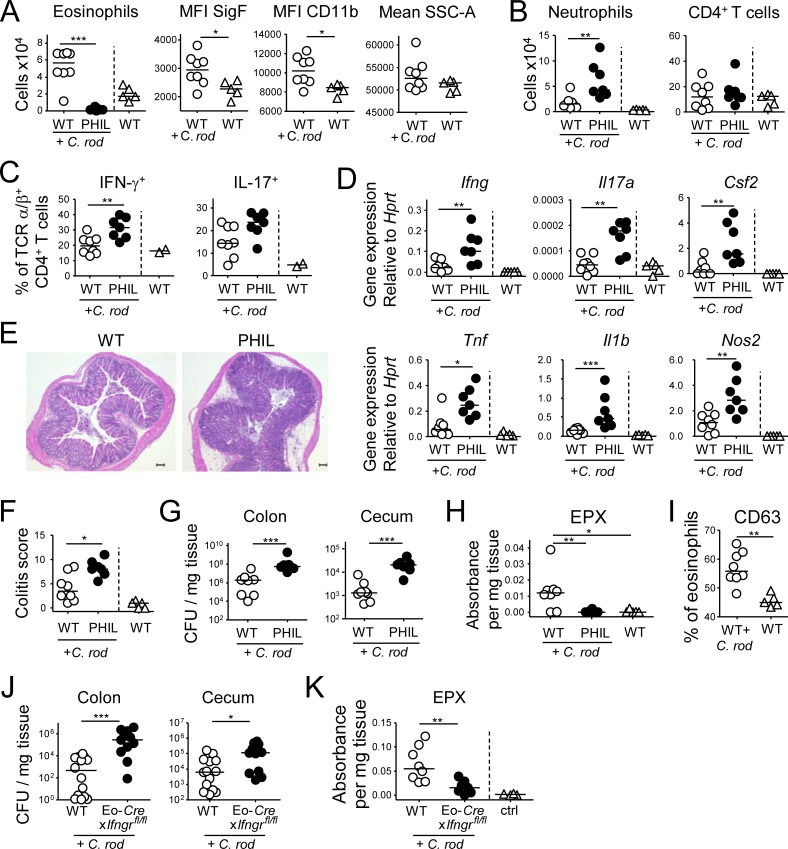
As we had observed sustained accumulation of eosinophils in the H. pylori–infected gastric mucosa, we next sought to investigate the functional role of eosinophils in this pathological condition using two independent approaches to reduce eosinophil numbers. First, we continuously administered a neutralizing antibody targeting the eosinopoietin IL-5 (Griseri et al., 2015) during an 8-wk infection time course. This treatment strongly reduced eosinophil frequencies (and as a consequence increased neutrophil frequencies) and resulted in increased production of gastric Ifng transcripts and in lower bacterial counts, but did not affect Il17 transcript levels (Fig. 2, A–C; and Fig. S2 A). To confirm the effects of eosinophil depletion in a second model, we infected mice that transgenically express diphtheria toxin under the EPX (EPO) promoter, which supports high level expression exclusively in eosinophil lineage-committed cells (PHIL mice; Lee et al., 2004). PHIL mice were devoid of eosinophils, both in the gastric LP (Fig. 2 D and Fig. S2 B) and in all other examined tissues (data not shown), but had normal complements of other major leukocyte populations (Fig. S2 C). Upon H. pylori infection, PHIL mice exhibited higher neutrophil infiltration, showed increased amounts of gastric mucosal Ifng and Il17 transcripts as well as higher Th1 and Th17 cell frequencies, and were colonized at lower levels than their nontransgenic littermates (Fig. 2, E–H). The expression of the Th1- and Th17-specific transcription factors T-bet and RORγt was increased as well in infected PHIL mice (Fig. 2 I). Interestingly, eosinophil deficiency had no effect on the expression of the Th2-specific transcription factor GATA-3 or on the Th2 cytokines IL-4 and IL-5 (Fig. S2 D). Transcript levels of other pro-inflammatory cytokines (TNF-α and IL-1β) and anti-microbial enzymes (NOS2) and the eosinophil recruiting cytokine eotaxin (CCL11), but not of CCL5 or CSF2, were elevated as well (Fig. 2 I and Fig. S2 E). The higher Th1 and Th17 cell frequencies of infected PHIL mice were not attributable to changes in the regulatory T cell compartment, which was similar in PHIL mice and their WT littermates (Fig. S2 F). To prove that the phenotype of PHIL mice was linked to their defect in eosinophil hematopoiesis, we reconstituted neonatal PHIL mice with WT or PHIL BM before infection with H. pylori. As neonatal hematopoiesis takes place in the liver, and hematopoietic stem cells and colony-forming progenitor cells migrate from liver to BM in the first few days of life (Wolber et al., 2002), we chose the intrahepatic route for reconstitution. Interestingly, all aspects of the phenotype of PHIL mice, i.e., eosinophil deficiency, elevated Th1 responses and reduced H. pylori colonization, were reversed by reconstitution of PHIL mice with WT, but not PHIL BM (Fig. 2, J–L).
Figure 2.
The antibody-mediated or genetic depletion of eosinophils enhances Th1 and Th17, as well as innate immune responses to H. pylori and improves clearance of the infection. (A–C) Mice were infected with H. pylori and treated with either anti–IL-5 or isotype control antibody for 8 wk. (A) The eosinophil depletion efficiency was ∼80%, as shown in representative FACS plots of gastric LP leukocyte preparations. (B) Gastric mucosal expression of Ifng and Il17 as determined by qRT-PCR and normalized to Hprt. Data are pooled from two independent experiments. (C) H. pylori colonization of the mice shown in B. (D–I) Eosinophil-deficient PHIL mice were infected with H. pylori for 12 wk. (D) The gastric eosinophil depletion efficiency in PHIL mice was >95%, as shown in representative FACS plots (left) and for all mice of the study relative to uninfected WT controls. (E) Neutrophil and CD4+ T cell numbers in the gastric LP of infected WT and PHIL mice, relative to uninfected WT controls. (F) Th1 and Th17 cell frequencies in the gastric LP of infected WT and PHIL mice. (G) Gastric mucosal expression of Ifng and Il17 as determined by qRT-PCR and normalized to Hprt. (H) H. pylori colonization of the mice shown in D–G. Data in D–H are representative of two independently conducted experiments. (I) Gastric mucosal expression of the indicated transcripts, as determined by qRT-PCR and normalized to Hprt. Note that PHIL mice were infected for 6 or 12 wk, whereas the anti–IL-5 treatment could only be continued for up to 8 wk. (J–L) Neonatal WT or PHIL mice were intrahepatically reconstituted at birth with BM from WT or PHIL donors, infected at 4 wk of age with H. pylori and sacrificed 6 wk later. (J) Eosinophil numbers in the gastric LP of reconstituted, infected mice, relative to nonreconstituted, infected PHIL mice. (K and L) Th1 and Th17 cell frequencies and H. pylori colonization of the mice shown in J. (M) Representative immunofluorescence microscopy images of FFPE sections of a control (left) and a 12 wk-infected (H. pylori PMSS1, right) WT stomach, stained with antibodies specific for EPX (for eosinophils) and CD3 (for T cells), along with the quantification of eosinophil and T cell counts per 0–225 mm2. Bars, 10 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as calculated by Mann-Whitney test.
To investigate whether eosinophils and T cells reside in similar niches in the H. pylori–infected gastric mucosa, we stained sections for both EPX to visualize eosinophils and for CD3 to visualize T cells; both populations were found in close proximity to one another at the base of the glands and throughout the infected mucosa and were much more abundant in the infected than the uninfected gastric mucosa (Fig. 2 M), thereby confirming the results of flow cytometric quantification. In summary, the ablation of eosinophils results in unrestricted Th1 responses and a stronger overall inflammatory reaction to bacterial challenge, which in turn promotes bacterial infection control; these results indicate that eosinophils have a regulatory rather than pro-inflammatory function in this bacterial infection model.
Eosinophils restrict GI tract T cell responses in the steady-state to promote mucosal homeostasis
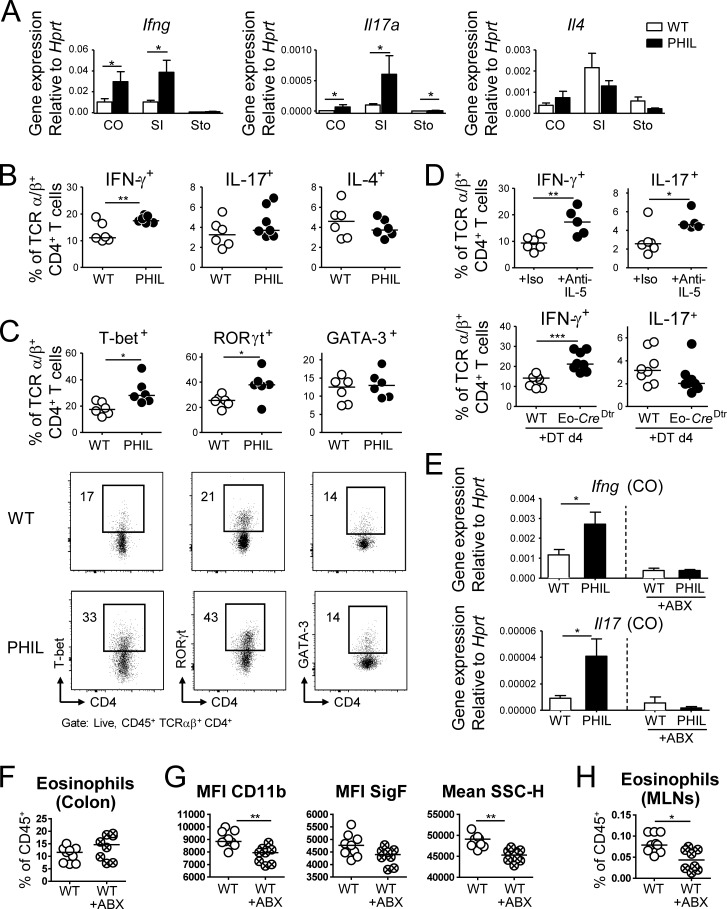
We next sought to elucidate eosinophil functions in the steady-state GI tract, focusing specifically on T cell responses to the normal GI tract microbiota. We quantified transcripts for T cell cytokines in the gastric, small intestinal and colonic mucosa of PHIL and WT mice. Ifng and Il17, but not Il4 transcripts were strongly overexpressed in the colonic and small intestinal mucosa of PHIL mice when compared with WT mice (Fig. 3 A), indicating a deregulated Th1 and Th17, but not Th2 response in the absence of eosinophils. The excessive colonic Th1 and, less conclusively, Th17 response of PHIL mice was confirmed by intracellular cytokine staining for IFN-γ and IL-17 and by staining for the transcription factors T-bet and RORγt; GATA-3+ or IL-4+ Th2 cells on the other hand did not differ between the two strains (Fig. 3, B and C; and Fig. S2 G). We also assessed cytokine production by innate lymphoid cells (ILCs, which we identified as CD45+ Lin− TCRα/β− CD4− Th1.2+ and IL-7R+). Although a clearly detectable population of these cells was positive for either IFN-γ or IL-17, their frequencies did not change under conditions of eosinophil deficiency (Fig. S2 H). The depletion of eosinophils with anti–IL-5, or by diphtheria toxin administration to Eo-CreDtr mice, confirmed the increased presence of colonic Th1 cells observed in PHIL mice (Fig. 3 D). The excessive expression of Ifng and Il17, but not of Il4 transcripts in the colonic mucosa of PHIL mice, was dependent on their gut microbiota, as treatment with a cocktail of antibiotics reversed the increased expression of these cytokines (Fig. 3 E and Fig. S2 I). Interestingly, while the frequency of colonic eosinophils was not affected by the antibiotic treatment (Fig. 3 F and Fig. S2 J), the depletion of commensal bacteria reduced the activation state of eosinophils in WT mice as assessed by their CD11b and Siglec-F expression and granularity (Fig. 3 G) and reduced their frequency in the MLNs (Fig. 3 H). The combined results suggest that constituents of the gut microbiota provide a tonic signal to GI-associated eosinophils that presumably is necessary for their homeostatic function, with homeostasis defined here as the absence of inappropriate, potentially tissue-damaging T cell responses.
Figure 3.
Eosinophil depletion enhances colonic microbiota-dependent Th1 responses. (A) GI tissues from six WT and five PHIL mice were analyzed by qRT-PCR for the expression of Ifng, Il17, and Il4, normalized to Hprt. STO, stomach; SI, small intestine; CO, colon. (B and C) Th1, Th17 and Th2 frequencies in the colonic LP of 6-wk-old WT and PHIL mice at steady-state, as assessed by intracellular cytokine staining (B) and staining for the lineage-specific transcription factors T-bet, RORγt, and GATA-3, shown with representative FACS plots (C). (D) Th1 and Th17 cell frequencies in the colonic LP of WT mice treated with anti–IL-5 or isotype control antibody for 10 d and of WT and Eo-CreDtr mice treated with diphtheria toxin (DT) for four days. Data are representative of two to three independent experiments. (E) Colonic mucosal expression of the indicated cytokines, of mice that were subjected to a 7-wk regimen of bacterial eradication by quadruple antibiotic therapy relative to untreated controls. (F) Eosinophil frequencies in the colons of the mice shown in E. (G) Activation state of the colonic LP eosinophils shown in F. (H) Eosinophil frequencies in the MLNs of the mice shown in E–G. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as calculated by Mann-Whitney test.
Eosinophils suppress T cell proliferation and cytokine production upon bacterial contact
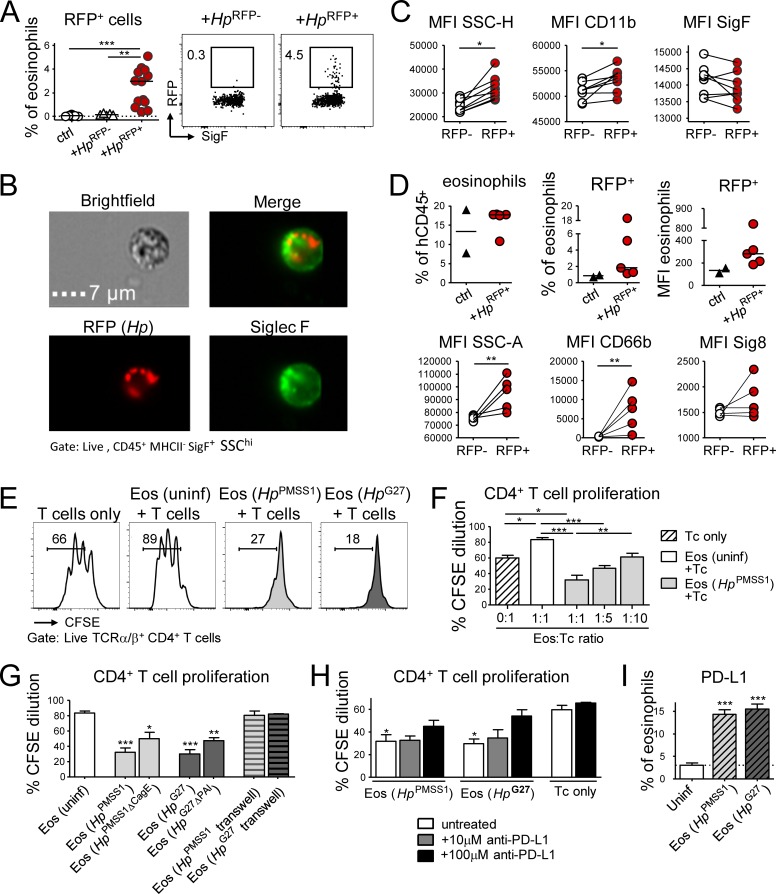
Eosinophil regulatory activity is strongly dependent on GI microbes; we therefore asked whether GI tract eosinophils directly encounter live bacteria in their tissues of residence. We took advantage of RFP+ H. pylori that we can readily detect inside professional phagocytes (Arnold et al., 2017). In chronically infected mice, 1–4% of gastric LP eosinophils were RFP+ (Fig. 4 A); some cells were found by imaging flow cytometry analysis to harbor more than one bacterium (Fig. 4 B and Fig. S3 A). Gastric RFP+ eosinophils were more activated than RFP− eosinophils within the same stomach, as defined by their increased CD11b expression and higher granularity (Fig. 4 C). As H. pylori is a human pathogen and eosinophils are known to infiltrate the gastric mucosa in H. pylori–infected patients presenting with gastritis (McGovern et al., 1991), we asked whether human eosinophils share the ability of their murine counterparts to interact with live H. pylori in its gastric niche. To this end, we reconstituted NSG mice with CD34+ human cord blood–derived hematopoietic stem and progenitor cells, infected them with RFP+ H. pylori, and examined their gastric LP leukocyte populations. We indeed detected human CD33+ HLA-DR− Siglec-8+ eosinophils in the gastric LP of infected as well as control mice; in fact, eosinophils made up ∼15% of all hCD45+ cells in the gastric LP, and a fraction of them were RFP+ in mice infected with fluorescent H. pylori (Fig. 4 D and Fig. S3 B). Human RFP+ eosinophils were more activated than RFP− eosinophils in the same stomach, as judged by their granularity as well as their expression of CD66b (a human myeloid-specific activation marker) and Siglec-8 (Fig. 4 D).
Figure 4.
Eosinophils encounter RFP+H. pylori in the gastric LP and suppress T cell proliferation in a PD-1/PD-L1–dependent manner. (A–C) Mice were infected at 1 wk of age with RFP+ or WT (RFP−) H. pylori for 3 mo. (A) Frequency of RFP+ eosinophils among all gastric LP eosinophils. Representative FACS plots are shown along with the quantification of two pooled experiments. (B) Representative ImageStream-derived image of a SiglecF+ eosinophil that colocalizes with multiple RFP+ bacteria. Bar, 7 µm. (C) Activation state of RFP+ relative to RFP− eosinophils of the same stomach of the mice shown in A. (D) Mice were reconstituted at birth with human cord blood HSCs and infected at 4 wk of age with RFP+ H. pylori for 6 wk; frequencies of human eosinophils among all human CD45+ leukocytes, and of RFP+ human eosinophils among all human gastric LP eosinophils are shown in the top panel. The MFI of the RFP channel is also shown. Bottom panel: activation state of RFP+ relative to RFP− human eosinophils of the same stomach, as judged by CD66b and Siglec-8 expression and granularity. (E–H) Naive or H. pylori–infected, in vitro differentiated eosinophils were co-cultured at 1:1 or the indicated ratios with CSFE-labeled immunomagnetically isolated splenic CD4+ T cells in the presence of anti-CD3/CD28-coated beads, IL-2, and IL-5. Two different strains of H. pylori (PMSS1 and G27) were used in parallel to infect eosinophil cultures for 18 h. (E) Representative FACS plots of the CFSE dilution of anti-CD3/CD28 bead-activated T cells relative to T cells co-cultured with naive or H. pylori–educated eosinophils. (F) Quantification of the CFSE dilution of T cells co-cultured with the indicated eosinophil:T cell ratios. (G) Quantification of the CFSE dilution of T cells co-cultured with eosinophils that had previously been exposed to the indicated WT and mutant H. pylori strains, with and without separation by a trans-well filter. (H) Quantification of the CFSE dilution of T cells in eosinophil co-cultures, in the presence or absence of the indicated concentrations of anti–PD-L1 blocking antibody. Means +SEM of three technical replicates of one representative experiment of two or three are shown in F–H. (I) PD-L1 expression of eosinophils infected with the indicated H. pylori strains for 18 h. Statistics were calculated by Mann-Whitney test in A–D and by one-way ANOVA with Bonferroni’s correction in F–I and are relative to the control condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next addressed in vitro whether bacterial encounter stimulates regulatory activities in murine eosinophils. We differentiated eosinophils in vitro from murine WT BM, which yielded eosinophil cultures of >90% purity as judged by flow cytometric analysis using SiglecF as lineage marker (Fig. S3 C), and exposed them to live H. pylori before co-culturing with T cells. H. pylori–educated, but not naive eosinophils, suppressed anti-CD3/CD28–induced T cell proliferation; this effect was observed with two different patient isolates of H. pylori (Fig. 4 E) and was strongest at high eosinophil:T cell ratios (Fig. 4 F). The suppressive activity of eosinophils required their direct contact with H. pylori, as the separation of bacteria and eosinophils by a trans-well filter abrogated the effect (Fig. 4 G). The interaction of H. pylori with its target cells is facilitated by the Cag-pathogenicity island (PAI)–encoded type IV secretion system (T4SS; Kwok et al., 2007). Our observation that CagE-deficient or Cag-PAI–deficient H. pylori, which both fail to assemble a functional T4SS, are less potent at inducing T cell–suppressive activities in eosinophils than their isogenic WT strains (Fig. 4 G) further underscores the contact dependency of the effect. Eosinophil-driven T cell suppression was independent of the enzyme indoleamine-pyrrole 2,3-dioxygenase (IDO; data not shown), but was partly abrogated by PD-L1 neutralization (Fig. 4 H and Fig. S3 D). Along the same line, we found that in vitro H. pylori–educated eosinophils displayed elevated expression of PD-L1 (Fig. 4 I). To assess the T cell suppressive activity of eosinophils directly ex vivo, we cultured sorted splenic eosinophils with T cells and measured T cell proliferation upon CD3/CD28 activation. Similar to BM-derived eosinophils, splenic eosinophils inhibited T cell proliferation only if they had previously been exposed to live H. pylori (Fig. S3 E). To test whether H. pylori–educated eosinophils suppress cognate antigen-induced T cell activation, we co-cultured eosinophils with ovalbumin-pulsed BM-derived APCs plus ovalbumin-specific CD4+ T cells (OT-II). H. pylori–educated, but not control eosinophils, suppressed the production of IFN-γ by OT-II cells (Fig. S3 F).
Having observed that activated eosinophils express high levels of PD-L1 and that PD-L1 neutralization at least partly abrogated eosinophil-driven T cell suppression (Fig. 4, H and I), we asked whether PD-L1 blockade would affect eosinophil activity and T cell responses in the steady-state colon. Mice that had been treated with anti–PD-L1 antibody for 1 wk exhibited normal eosinophil frequencies, but strongly reduced eosinophil-specific expression of PD-L1 itself, as well as of the activation and degranulation markers Siglec-F and CD63 (Fig. S3, G and H). Although CD4+ and CD8+ T cell frequencies were normal, their expression of IFN-γ was increased upon PD-L1 blockade (Fig. S3, I and J). In conclusion, we found that eosinophils in the GI tract robustly suppress proliferation and cytokine production by T cells, with PD-L1 expression emerging as a potentially useful marker for GI tract eosinophils with regulatory properties.
The regulatory function of eosinophils depends on IFN-γ signaling
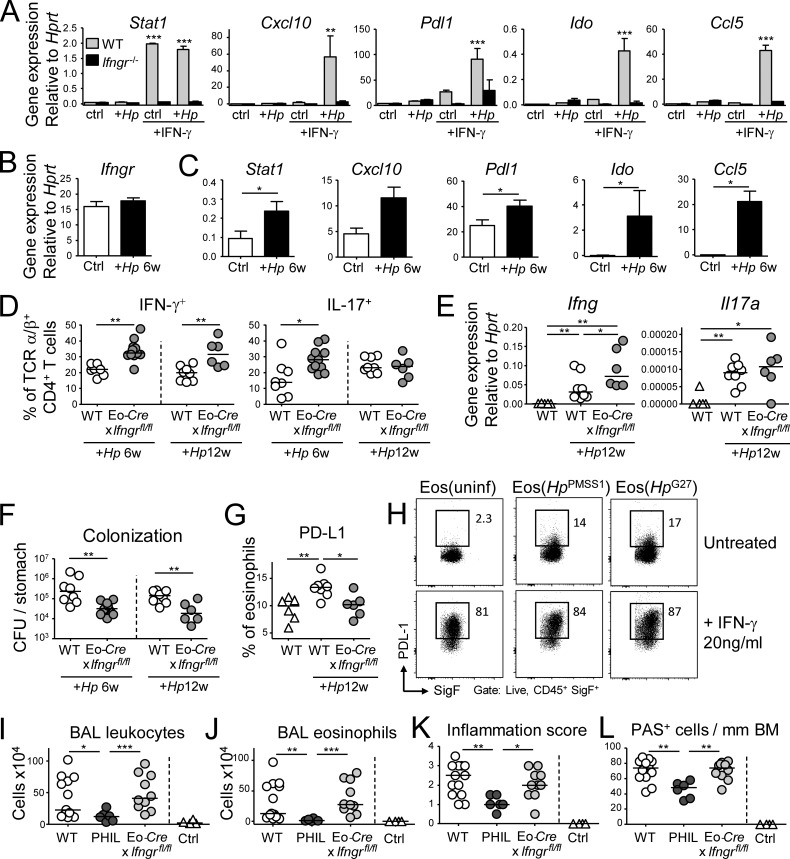
We have previously identified IFN-γ as a major determinant of the outcome of the H. pylori–host interaction (Sayi et al., 2009; Arnold et al., 2011). Furthermore, IFN-γ is a known regulator of PD-L1 expression in various cell types. Because H. pylori–educated eosinophils expressed higher amounts of PD-L1 (Fig. 4 I), we cultured H. pylori–educated eosinophils in the presence or absence of IFN-γ and monitored their gene expression by targeted qRT-PCR. In addition to Pdl1, several additional IFN-γ target genes including Cxcl10 (encoding IP10), Stat1, Ido1, and Ccl5 were up-regulated by H. pylori in the presence of IFN-γ in WT, but not Ifngr1−/− eosinophils (Fig. 5 A). With the exception of Stat1, the up-regulation of all transcripts required the presence of H. pylori in addition to IFN-γ (Fig. 5 A). The same transcripts were up-regulated in eosinophils sorted from the gastric LP of H. pylori–infected mice, whereas the expression of Ifngr1 itself was unaffected by exposure to H. pylori (Fig. 5, B and C). Additional transcripts up-regulated upon H. pylori exposure of eosinophils in vitro, i.e., those encoding TNF-α, IL-1β, and Cox-2 did not show the same strict dependence on IFN-γR signaling and also were not differentially regulated in eosinophils from infected relative to uninfected mice (Fig. S4, A and B).
Figure 5.
IFN-γ signaling conditions eosinophils to suppress Th1 responses, but is not required for the pathogenic functions of eosinophils in allergic asthma. (A) In vitro differentiated WT or Ifngr−/− eosinophils were co-cultured for 18 h with H. pylori PMSS1 (multiplicity of infection of 10) with or without 20 ng/ml rIFN-γ and subjected to qRT-PCR of the indicated transcripts. Means of three technical replicates + SEM are shown for one representative of two experiments. (B and C) Gastric LP eosinophils were FACS-sorted from naive and H. pylori–infected (6 wk) mice and subjected to qRT-PCR of the indicated transcripts. n = 6–8. (D–G) Eo-Cre+ × Ifngrfl/fl mice and their Cre-negative littermates (WT) were infected with H. pylori for 6 or 12 wk. (D) Th1 and Th17 cell frequencies in the gastric LP of infected WT and EoCre+ × Ifngrfl/fl mice. (E) Gastric mucosal expression of Ifng and Il17 as determined by qRT-PCR. (F) H. pylori colonization of the mice shown in D and E. (G) PD-L1 expression on gastric LP eosinophils at 12 wk p.i., as assessed by FACS of leukocyte preparations. 6-wk time point data are pooled from two independent studies; 12-wk time point data are representative of two experiments. (H) In vitro differentiated eosinophils were infected with the indicated strains of H. pylori and/or cultured in the presence of 20 ng/ml rIFN-γ for 18 h. PD-L1 expression as assessed by FACS is shown. (I–L) Eo-Cre+ × Ifngrfl/fl mice, their Cre-negative littermates, and PHIL mice were sensitized and challenged with house dust mite allergen. (I) Total leukocytes in 1 ml of bronchoalveolar lavage fluid (BALF). (J) Total eosinophils in 1 ml of BALF. (K and L) Pulmonary inflammation and goblet cell metaplasia, as assessed on stained lung sections. BM, basal membrane. Results in I–L are pooled from two experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as calculated by Mann-Whitney test (B–K) or by one way ANOVA with Bonferroni’s correction (A).
We next set out to address whether the regulatory function of eosinophils depends on cell-intrinsic IFN-γR signaling. To this end, Eo-Cre mice were crossed with Ifngrfl/fl mice to obtain eosinophil-specific conditional IFN-γR knockout mice. The fidelity of eosinophil-specific Cre expression was addressed using Eo-Cre × Rosa26Rfp reporter mice, which confirmed that all RFP+ cells in the gastric LP of Eo-Cre × Rosa26Rfp mice were eosinophils as defined by their expression of Siglec-F (Fig. S4 C). However, only half of all Siglec-F+ eosinophils were RFP+, both in the steady-state and during H. pylori infection (Fig. S4 D), indicating that the peroxidase promoter driving Cre expression is active in only 50% of all gastric eosinophils. Furthermore, the RFP+ (peroxidase expressing) fraction of eosinophils was more likely to become activated upon H. pylori infection than their RFP− counterparts (Fig. S4 E).
Interestingly, Eo-Cre × Ifngrfl/fl mice mirrored the phenotype of eosinophil-deficient PHIL mice and exhibited higher frequencies of Th1 cells, more abundant mucosal Ifng transcripts, and significantly lower H. pylori colonization at both 6 and 12 wk after infection than control mice (Fig. 5 D-F). The eosinophil-specific loss of IFN-γR abrogated the H. pylori–induced increase of PD-L1 expression (Fig. 5 G). In vitro exposure of eosinophils to both H. pylori and IFN-γ confirmed that the cytokine has a major impact on PD-L1 expression, which is further enhanced by H. pylori (Fig. 5 H).
Finally, to assess whether the eosinophil-specific loss of IFN-γR signaling compromises other eosinophil functions, we subjected Eo-Cre × Ifngrfl/fl, PHIL and WT mice to allergic sensitization and challenge with house dust mite allergen. Whereas PHIL mice were protected against eosinophil-dependent hallmarks of allergic asthma, Eo-Cre × Ifngrfl/fl mice did not differ from their WT littermates with respect to lung inflammation, airway eosinophilia, and goblet cell hyperplasia (Fig. 5, I–L). We conclude that eosinophil activity is regulated by IFN-γ in Th1/Th17-driven, but not Th2-driven conditions and pathologies.
Eosinophils suppress Th1 and Th17 responses in an acute infection model
To address the function of GI-associated eosinophils in a second bacterial infection model, we challenged PHIL and WT mice with the murine attaching/effacing intestinal pathogen C. rodentium, which colonizes the mucosa of cecum and colon and disseminates to the MLNs and spleen in immunocompromised mice. C. rodentium further induces self-limiting colitis that is both mediated and controlled by the immune system. In WT mice, infection with C. rodentium resulted in increased frequencies and activation of eosinophils in the colon (Fig. 6 A). In eosinophil-deficient PHIL mice, we observed increased frequencies of neutrophils, Th1, and Th17 cells, as well as increased expression of transcripts for pro-inflammatory cytokines relative to WT controls (Fig. 6, B–D; and Fig. S5 A). The C. rodentium–induced expression of anti-microbial molecules (e.g., S1009a, CXCL1, and CXCL2) was enhanced in PHIL mice (Fig. S5 A). Thus, the regulatory features of eosinophils seem independent of the type of bacterial infection and may represent a function of general importance. We further observed more severe C. rodentium–induced colitis in PHIL mice (Fig. 6, E and F; and Fig. S5 B), which is in agreement with their stronger immune activation. Unexpectedly, the bacterial load in the cecum and colon of PHIL mice was higher than in WT mice (Fig. 6 G), which suggested an additional role for eosinophils in C. rodentium clearance that was not observed with H. pylori. Indeed, C. rodentium effectively triggered eosinophil degranulation and peroxidase release in vivo as assessed by CD63 expression and the release of EPX into the supernatants of colonic explants cultured ex vivo, respectively (Fig. 6, H and I). Interestingly, the above mentioned conditional strain lacking IFN-γR signaling in the eosinophil compartment shared the defect of PHIL mice with respect to C. rodentium clearance, a feature that could be attributed to its inability to release EPX (Fig. 6, J and K). Overall, the results from the second bacterial challenge model used here confirm the dominant regulatory activity of eosinophils also during an acute infection and further highlight a major role of eosinophils as effector cells contributing to the clearance of some bacterial species, but not others.
Figure 6.
Eosinophils suppress C. rodentium–specific Th1 and Th17 responses and colitis. (A–H) PHIL mice and their WT littermates were infected with C. rodentium for 12 d. (A) Colonic LP eosinophil numbers of infected PHIL and WT relative to naive WT mice and their activation state. (B) Colonic neutrophil and CD4+ T cell numbers of the mice shown in A. (C) Th1 and Th17 cell frequencies in the colonic LP of C. rodentium–infected mice. (D) Cytokine and inflammatory gene expression in the colonic mucosa of C. rodentium–infected mice as determined by qRT-PCR. (E and F) C. rodentium–induced colitis as assessed by histopathological examination of H&E-stained sections. Representative sections of C. rodentium–infected WT and PHIL mice are shown in E, along with colitis scores in F. (G) C. rodentium colonization of the colons and cecums of the mice shown in A–E. (H) EPX expression in the colonic mucosa of the mice shown in A–G, as assessed by ELISA. Data in A–H are representative of two independent experiments. (I) CD63 expression as a marker of eosinophil degranulation, as assessed by FACS of colonic leukocyte preparations from the naive and C. rodentium–infected mice shown in A–H. (J and K) Eo-Cre+ × Ifngrfl/fl mice and their Cre-negative (WT) littermates were infected with C. rodentium for 12 d and assessed with respect to their colonization levels (J) and EPX expression in the colon as assessed by ELISA (K). Bar, 100 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as calculated by Mann-Whitney test.
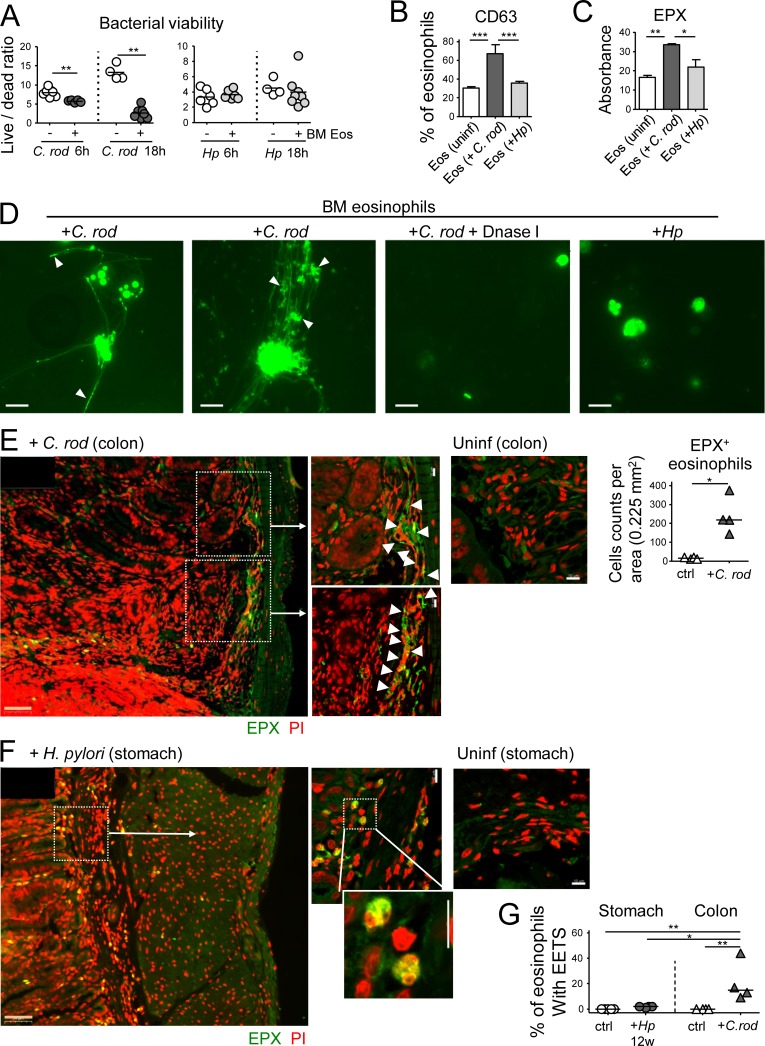
Eosinophils contribute to bacterial clearance through the formation of extracellular traps
To address whether eosinophils are capable of killing C. rodentium and/or H. pylori in vitro, we differentiated eosinophils from murine BM, FACS-sorted them to >99% purity, and co-cultured them with either C. rodentium or H. pylori for 6 or 18 h, after which the bacterial viability was determined by live/dead staining and FACS analysis. As expected based on the in vivo data, C. rodentium, but not H. pylori, was highly susceptible to killing by eosinophils in this in vitro assay (Fig. 7 A). Reduced C. rodentium viability coincided with eosinophil degranulation as judged by CD63 expression, as well as the release of EPX (Fig. 7, B and C). The formation of eosinophil extracellular traps (EETs) was induced by C. rodentium, but not H. pylori (Fig. 7 D). The dissolution of EETs by DNase treatment abrogated the bactericidal activity of eosinophils toward C. rodentium (Fig. 7 D and Fig. S5, C and D). C. rodentium–induced EETs formed independently of the enzyme peptidylarginine deiminase 4 (PAD4), which deiminates histone H3 and H4 and is required for extracellular trap formation by neutrophils (data not shown; Hemmers et al., 2011). To address whether EETs are released in response to either bacterium in vivo, we stained formalin-fixed paraffin-embedded (FFPE) sections for EPX and with propodium iodide (PI) to visualize EETs as described previously (Yousefi et al., 2008). We observed that eosinophils were more abundant in the C. rodentium–infected relative to uninfected colonic LP and showed evidence of degranulation and EET formation. Co-localization of EPX with EETs was also observed (Fig. 7 E). In contrast, we found no evidence of eosinophil degranulation (i.e., non–cell-associated EPX) or EET formation (nonnuclear PI) in the stomachs of H. pylori infected mice (Fig. 7, F and G). The combined results indicate that exposure of eosinophils to C. rodentium or its components, but not to H. pylori, provides a stimulus for eosinophil degranulation and EET formation, resulting in bacterial killing.
Figure 7.
Eosinophils contribute to C. rodentium, but not H. pylori control through degranulation and extracellular trap formation. (A) In vitro differentiated eosinophils were co-cultured with either C. rodentium or H. pylori for 6 or 18 h; bacterial viability was determined by live/dead staining and FACS. (B) CD63 expression as a marker of eosinophil degranulation, as assessed by FACS of in vitro infected and naive eosinophil cultures. (C) EPX expression, as assessed by ELISA of the supernatants of eosinophil cultures. (D) Eosinophil extracellular trap (EET) formation by eosinophils infected with C. rodentium, but not H. pylori; white arrowheads point to bacteria caught in EETs. DNase I treatment dissolves EETs. (E and F) Representative confocal immunofluorescence microscopy images of FFPE sections of a C. rodentium–infected colon (12 d, E) and an H. pylori–infected stomach (12 wk, F), along with uninfected control sections of both tissues (EPX, green; propidium iodide, PI, red). Arrows point to regions of high eosinophil density and of colocalized extracellular EPX and EETs. The quantification of eosinophils in C. rodentium–infected colons is shown in E. Bars, 50 µm in low magnification (orange), 10 µm in high magnification images (white). (G) Quantification of eosinophils with EETs in stomachs and colons of four H. pylori–infected and four C. rodentium–infected mice relative to their uninfected controls. Statistics were calculated by Mann-Whitney test in A–E and by one-way ANOVA with Bonferroni’s correction in G. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
The role of eosinophils in the control of bacterial infections, and of bacterially induced immunopathology, has to date remained largely unexplored. We show here that the frequencies of mucosal Th1 cells are strongly increased in the absence of eosinophils under three very different experimental conditions (i.e., in the steady-state colon, during acute infection of cecum and colon, and during chronic infection of the stomach). Eosinophils appear to rather selectively target Th1 responses and have no or only inconsistent suppressive effects on Th2 and Th17 cells in our hands; in contrast, Sukagawa et al. have observed the preferential down-regulation of Th17 cells by small intestinal eosinophils at steady-state and have linked this phenomenon to the suppression of Th17 differentiation (Sugawara et al., 2016). While we have in some experiments also detected a significant expansion of the Th17 compartment due to eosinophil depletion, the effects on Th1 cells were stronger and generally more consistent. We did not observe clear differences in Th1 or Th17 frequencies in the MLNs due to the absence of eosinophils (data not shown) and therefore conclude that eosinophils control Th1 expansion in the tissue rather than their priming in the draining lymph nodes.
The regulatory capacity of eosinophils toward Th1 cells depends at least in part on their PD-L1 expression, but, as observed in the H. pylori infection model, is not necessarily associated with degranulation. Consistent with their preference for Th1 cells, we found that eosinophils are cell-intrinsically conditioned by IFN-γ levels in their residential tissues; IFN-γ signaling is required for PD-L1 expression and T cell suppression and other regulatory activities, but not for their detrimental functions in an (eosinophil dependent) model of house dust mite–induced allergic asthma, a pathology that is clearly driven by Th2 cells. The cellular sources that provide the first wave of IFN-γ and thus condition eosinophils in their tissues of residence are not known; IFN-γ–producing ILCs, which are abundant in the steady-state colon, may potentially serve as an important source of IFN-γ in that tissue. Direct contact between PD-L1–expressing eosinophils and PD-1+ T cells in the inflamed mucosa likely accounts for some of the suppressive activity; other immunomodulatory determinants expressed by eosinophils (such as prostaglandin E2 synthase/cyclooxygenase 2 and its product PGE2, or transforming growth factor β, TGF-β) may additionally contribute to the overall phenotype. We could not substantiate a critical role for IDO and tryptophan metabolism, at least in in vitro suppression assays. Our results are consistent with a previous study conducted on purified human eosinophils that also documented IFN-γ–dependent effects on eosinophil effector functions such as their degranulation and superoxide production (Yamaguchi et al., 2008). We hypothesize that the relative abundance of eosinophils in the GI tract (their largest reservoir in the body), and their strict regulation by IFN-γ, has specifically evolved to restrict inappropriate Th1 responses to bacterial commensals and pathogens in the context of type 1 immunity.
Interestingly, we found that eosinophils directly encounter bacteria (or at least their protein products) in infected GI tract tissues. A substantial fraction of murine eosinophils, as well as human eosinophils in mice reconstituted with a human immune system, were positive for RFP when exposed to RFP-expressing bacteria. Eosinophils that had come in contact with H. pylori were generally more activated than their naive counterparts in the same stomach, and eosinophils from infected tissues exhibited dramatically different gene expression profiles than steady-state eosinophils as judged by RNA sequencing of sorted populations. Many of the transcripts identified in the RNA sequencing approach could subsequently be validated, and were synergistically regulated by a bacterial (H. pylori) signal in conjunction with IFN-γ. Neither signal alone was sufficient to drive differential gene expression, and only synergistically activated transcripts could be confirmed in vivo. These data indicate that eosinophil activity in tissues is tightly controlled, preventing inappropriate degranulation on the one hand, and Th1-driven immunopathology on the other. The regulation of eosinophil activity by a microbial stimulus, in conjunction with a strongly tissue-damaging cytokine such as IFN-γ, thus is consistent with their critical role in tissue homeostasis within the GI tract.
In contrast to H. pylori, which activates eosinophils without triggering their degranulation and EET formation and therefore is protected from eosinophil bactericidal activities, the pathogen C. rodentium is readily killed by eosinophils in vitro, and colonizes the murine colon and cecum at much higher levels in their absence. Candidate eosinophil citrobactericidal factors are ECP and MBP, which both were shown to be associated with EETs (von Köckritz-Blickwede and Nizet, 2009) and, in very early work, to have bactericidal activity in vitro (Lehrer et al., 1989; Rosenberg and Dyer, 1995). The deployment of EETs protects mice against sepsis in vivo and has been proposed as an essential mechanism maintaining intestinal barrier function in the face of inflammation-associated epithelial cell damage (Yousefi et al., 2008). Among the few bacterial species with documented susceptibility to eosinophil killing in vitro and in vivo is Pseudomonas aeruginosa (Linch et al., 2009). The fact that the citrobactericidal activities of eosinophils are abrogated by DNase treatment implicates EETs and their associated granule factors.
The concept that eosinophils have roles in homeostasis, tissue protection and the prevention of immunopathology is slowly gaining more attention (Rosenberg et al., 2013; Travers and Rothenberg, 2015; Wen and Rothenberg, 2016). In the lung, a resident eosinophil subset has been identified that possesses regulatory activities, is phenotypically distinct from the inflammatory eosinophils that are recruited in an IL-5–dependent manner during allergen-induced asthma, and in fact suppresses allergen-specific Th2 responses by inhibiting the maturation of allergen-loaded DCs (Mesnil et al., 2016). It is tempting to speculate in this context that the two (EoCre+ and EoCre−) eosinophil populations that we have identified in the GI tract correspond to such specialized subsets, although both subsets are depleted in PHIL mice and by anti–IL-5 treatment. In the GI tract, two recent reports have highlighted homeostatic properties of eosinophils, which could be attributed to their role in promoting the differentiation of IgA-expressing plasma cells and class switching to IgA, as well as the maintenance of intestinal mucus secretion and the development of Peyer's patches (Chu et al., 2014; Jung et al., 2015). Consequently, mice lacking eosinophils had a clearly altered intestinal microbial composition (Chu et al., 2014; Jung et al., 2015). These data are in good agreement with our own results showing that T cell responses directed against the normal colonic and cecal microbiota are under tight eosinophil control at steady-state. Overall, our results add to a growing body of evidence that attributes beneficial functions in homeostasis and immune regulation to this poorly understood cell type.
Materials and methods
Animal experimentation
C57BL/6J (stock 000664), Ifngr1fl/fl (stock 025394), Ifngr1−/− (stock 003288), OT-II (Stock 004194), and NSG (Stock 005557) mice were obtained from the Jackson Laboratory. B6-Gt(ROSA)26Sortm1(Dtr)ThBu (iDTR) mice were described previously (Buch et al., 2005). B6-Gt(ROSA)26Sortm1(Rfp) (Rosa26Rfp) mice were obtained from a local mouse repository. Eosinophil-deficient mice (PHIL; Lee et al., 2004) and mice expressing Cre under the EPX promoter (EoCre; Doyle et al., 2013) were obtained from J.J. Lee (Mayo Clinic, Phoenix, AZ). All strains were bred and maintained under specific pathogen-free conditions in accredited animal facilities at the University of Zürich. Mice were included in experiments at 5–6 wk of age; breeding pairs were set up so that female breeders were phenotypically WT, and each litter was internally controlled. Breeding pairs were renewed every second generation. For the depletion of eosinophils, WT mice were i.p. injected two times per week with 0.25 mg of anti–IL-5 antibody (TREK5; BioXCell) or with anti–horseradish peroxidase isotype control antibody (HRPN; BioXCell) starting from the day of infection. For short-term diphtheria-toxin (DT)–mediated eosinophil depletion, Eo-Cre × iDTRfl/+ mice were treated on four consecutive days with 15 ng/g body weight DT (Sigma). PD-L1 blocking antibody (10F.9G2’ BioXcell) was administered twice with a 3-d interval at 0.5 mg/mouse. For eosinophil reconstitution experiments, newborn PHIL or WT mice were sub-lethally irradiated with 1.3 Gy and injected intra-hepatically with 1.5 × 106 adult BM cells 3 d after birth. Mice were infected with H. pylori 4 wk after reconstitution and analyzed 6 wk later. For the generation of humanized mice, newborn NSG mice were sub-lethally irradiated with 1.3 Gy and injected intra-hepatically with 1.5 × 105 human CD34+ hematopoietic stem cells (HSCs) 3 d after birth. CD34+ cells were isolated from human cord blood with immunomagnetic beads (Miltenyi Biotec) with a yield of 0.5–4 × 106 CD34+ cells per donor (purity >90%) and were stored in liquid nitrogen until use. Mice were infected with H. pylori 3 wk after reconstitution and analyzed 10 wk later. For the depletion of gut commensal microbiota, animals were given ampicillin (1 g l−1; Sigma), vancomycin (500 mg l−1; Applichem), neomycin sulfate (N; 1 g l−1; Applichem), and metronidazole (1 g l−1; Sigma) in their drinking water for 7 wk as described (Diehl et al., 2013) before analysis. For the induction of house dust mite–induced allergic asthma, mice were sensitized on day 0 (1 µg) and challenged on days 8–12 (15 µg) intranasally with house dust mite extract (Greer Laboratorie; XPB70D3A25 Dermatophagoides pteronyssinus). On day 15, mice were sacrificed, and lungs were lavaged with PBS via the trachea. Broncho-alveolar lavage fluid (BAL) cells were counted using trypan blue dye exclusion. Differential cell counts of macrophages, lymphocytes, neutrophils, and eosinophils were performed on cytospins stained with the Microscopy Hemacolor-Set (Merck). For lung histopathology, lungs were fixed by inflation and immersion in buffered 10% formalin and embedded in paraffin. Tissue sections were stained with H&E and periodic acid-Schiff (PAS) and examined in blinded fashion. Peribronchial and perivascular inflammation was scored on a scale from 0 to 4. PAS-positive goblet cells were quantified per 1 mm of basement membrane. All animal experimentation was reviewed and approved by the Zürich Cantonal Veterinary Office (licenses ZH170/2014, ZH24/2013, ZH235/2015, and ZH140/2017 to A.M.).
H. pylori strains, culture conditions and colony counting
Mice were infected orally on two consecutive days with 108 CFU H. pylori PMSS1 at 5 wk of age and analyzed at 6 and 12 wk p.i. For PMSS1 RFP+ infection of C57BL/6 mice, mice were infected on day 7 of age to achieve high levels of bacterial colonization and analyzed after 12 wk p.i. The H. pylori strain used in this study, PMSS1, is a clinical isolate of a patient with duodenal ulcer and the parental strain of the mouse-derivative Sydney strain 1 (SS1) (Arnold et al., 2011). The PMSS1ΔCagE isogenic mutant was described previously (Arnold et al., 2011), as was H. pylori strain G27 and its G27ΔPAI isogenic mutant (Toller et al., 2011). The RFP-expressing PMSS1 was generated by transforming PMSS1 with the plasmid pTM115-RFP used previously in strain SS1 (Keilberg et al., 2016). H. pylori was grown on horse blood agar plates and in liquid culture as described previously (Arnold et al., 2011). Cultures were routinely assessed by light microscopy for contamination, morphology, and motility.
C. rodentium strains, culture conditions, and colony counting
The nalidixic acid (NAL)–resistant C. rodentium strain ICC169 was grown overnight at 37°C in Luria broth (LB) supplemented with NAL (50 µg/ml; Sigma). Mice were infected orally with 5 × 108 bacteria for 12 d. To assess C. rodentium colonization, cecal and colonic tissues were homogenized in PBS, diluted, and plated on LB plates supplemented with NAL. Colonies were counted after 18 h of culture at 37°C. Colonic and cecal bacterial loads were normalized to tissue weight.
Histological assessment of colonic inflammation
Distal colon samples were fixed in buffered 10% formalin solution. Paraffin embedded sections were stained with hematoxylin and eosin and the histopathology of the colon was scored in a blinded fashion using a scoring system considering five categories (each scored on a scale of 0–3): epithelial hyperplasia/damage and goblet cell depletion; leukocyte infiltration in the lamina propria; submucosal inflammation and edema; area of tissue affected; and markers of severe inflammation such as bleeding, crypt abscesses, and necrosis/ulceration. The final score presented (scored 0–15) represents the sums of all categories.
Flow cytometry, cell sorting and counting
For surface staining, cells were stained in PBS with 0.5% BSA with a fixable viability dye and a combination of the following antibodies: anti-mouse B220 (RA3-6B2), CCR3 (J073E5), CD11b (M1/70), CD11c (N418), CD4 (RM4-5), CD44 (IM7), CD45 (30-F11), CD49b (DX5), CD63 (NVG-2), F4/80 (BM8), GR-1 (RB6-8C5), IL-7R (A7R34), Ly6G (1A8), MHC-II (M5/114.15.2), TCR-β (H57-597), and Thy1.2 (53–2.1), all from BioLegend; anti-mouse IL-5R (TRFK5) and Siglec-F (E50-2440) from BD Biosciences; anti-mouse PD-L1 (B7-H11) from eBioscience/Invitrogen; anti-human CD33 (P67.6), CD66b (G10F5), CD45 (HI30), HLA-DR (L243), and Siglec-8 (7C9), all from BioLegend. Fc block (anti-CD16/CD32, Affymetrix) was included to minimize nonspecific antibody binding. Annexin V staining was performed with the Annexin V Apoptosis Detection Set (eBioscience) according to the manufacturer's instructions. For intracellular cytokine staining of T cells, cells were incubated for 3.5 h in complete IMDM containing 0.1 µM phorbol 12-myristate 13-acetate and 1 µM ionomycin with 1:1,000 Brefeldin A (eBioscience) and GolgiStop solutions (BD Biosciences) at 37°C in a humidified incubator with 5% CO2. Following surface staining, cells were fixed and permeabilized with the Cytofix/Cytoperm Fixation/Permeabilization Solution kit (BD Biosciences) according to the manufacturer's instructions. Cells were then stained for 50 min with antibodies to IL-17A (TC11-18H10.1), IFN-γ (XMG1.2), and IL-4 (11B11, all from Biolegend). For the staining of T cell transcription factors, cells were surface stained and fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer's instructions. Cells were then stained for 50 min with antibodies to mouse Foxp3 (MF-14, Biolegend), Gata3 (TWAJ, eBioscience), T-bet (4B10, Biolegend) or Rorγt (Q31-378, BD Bioscience). Total leukocyte counts were determined by adding countBright Absolute Counting Beads (Life Technologies) to each sample before analysis. Samples were analyzed on a LSRII Fortessa (BD Biosciences) or sorted on a FACS Aria III (BD Biosciences) to a purity of >95%. Flow-cytometric analyses were performed using FlowJo software (Tree Star). Where indicated, sorted eosinophils were subjected to cytospin and visualized by microscopy upon staining with the Microscopy Hemacolor-Set (Merck).
Quantitative PCR
RNA was isolated from scraped gastric mucosa, homogenized colonic and intestinal tissues, BM-derived eosinophils, or from FACS-sorted cells using the RNeasy Mini kit (QIAGEN) according to the manufacturer’s instructions, including an on-column DNase I digestion step. Complementary DNA synthesis was performed using Superscript III reverse transcription (QIAGEN). Quantitative PCR reactions for the candidate genes were performed using TaqMan Gene Expression Assays (Applied Biosystems by ThermoFisher Scientific): Ccl11 (Mm00441238_m1), Ccl5 (Mm01302427_m1), Csf2 (Mm01290062_m1), Cxcl1 (Mm04207460_m1), Cxcl10 (Mm00445235_m1), Cxcl2 (Mm00436450_m1), Gata3 (Mm00484683_m1), Hprt (Mm03024075_m1), Ido1 (Mm00492590_m1), Ifng (Mm01168134_m1), Ifngr1 (Mm00599890_m1), Il17a (Mm00439618_m1), Il1b (Mm00434228_m1), Il22 (Mm01226722_g1), Il4 (Mm00445259_m1), Il5 (Mm00439646_m1), Il6 (Mm00446190_m1), Nos2 (Mm00440502_m1), Pdl1 (Mm03048248_m1), Ptgs2 (Mm00478374_m1), Rorc (Mm01261022_m1), S100a9 (Mm00656925_m1), Stat1 (Mm01257286_m1), Tbx21 (Mm00450960_m1), and Tnf (Mm00443258_m1). According to the manufacturer’s instructions, all TaqMan Gene Expression Assays have been designed, optimized, and extensively tested to ensure equivalent amplification efficiencies of 100% (±10%) and therefore do not require additional efficacy measurements. Complementary DNA samples were analyzed using a Light Cycler 480 detection system (Roche) with the Second Derivative Maximum method analysis with the high confidence algorithm. Gene expression levels for each sample were normalized to HPRT expression. Mean relative gene expression was determined, and the differences were calculated using the 2ΔC(t) method.
Leukocyte isolation
For lamina propria leukocyte isolation, GI tissues were opened longitudinally, washed and cut into pieces. Peyer’s patches were removed from the small intestine. Pieces were incubated in HBSS with 10% FCS and 5 mM EDTA at 37°C to remove epithelial cells. Gastric tissue was digested at 37°C for 50 min in a shaking incubator with 15 mM Hepes, 500 U/ml of type IV collagenase (Sigma-Aldrich) and 0.05 mg ml−1 DNase I in RPMI-1640 medium supplemented with 10% FBS and 100 U ml−1 penicillin/streptomycin. Intestinal tissues were digested with an equal mixture of 250 U/ml type IV and type VIII (Sigma-Aldrich) collagenase. Cells were then layered onto a 40/80% Percoll gradient, centrifuged, and the interface was washed in PBS with 0.5% BSA. BM cell suspensions were prepared by flushing the marrow out of femur and tibia and were resuspended in PBS with 2% BSA. MLNs were digested at 37°C for 20 min in a shaking incubator with 500 U/ml of type IV collagenase and mashed through a cell strainer. Splenocyte suspensions were prepared by mashing the spleens through a cell strainer using a syringe plunger, followed by red blood cells lysis.
Differentiation of BM-derived eosinophils and APCs
To generate murine BM-derived myeloid cells, BM stem cells from donor mice were flushed from the femur and the tibia and were resuspended in RPMI-1640 medium upon red blood cell lysis. For the generation of BM-derived eosinophils (BM Eos), cells were seeded at a density of 106 cells/ml in RPMI-1640 medium supplemented with 20% heat inactivated FBS, 25 mM Hepes, 100 U ml−1 penicillin/streptomycin, 2 mM glutamine, 1× NEAA, and 1 mM sodium pyruvate, and cultured at 37°C. BM Eos were differentiated with 100 ng/ml mouse stem cell factor (PeproTech) and 100 ng/ml mouse FLT3-Ligand (FLT3-liter; PeproTech) from day 0 to day 4, followed by differentiation with 10 ng/ml mouse IL-5 (PeproTech) only from day 4 onwards, as described (Dyer et al., 2008). Half of the medium was changed every other day and the cell concentration was adjusted to 106 cells/ml until day 14. On day 8, cells were collected and moved to new flasks to remove adherent contaminating cells. On day 13, the nonadherent cells were collected washed, counted, and plated into a 96-well plate. Experiments were performed at day 14. Eosinophil purity was assessed by flow cytometry at day 13 and cell morphology was visualized by microscopy upon cytospin and staining with the Microscopy Hemacolor-Set (Merck). For the generation of BM APCs, cells were seeded at a density of 0.7 × 106 cells/ml and differentiated with 20 ng/ml GM-CSF (PeproTech) for 6 d at 37°C. Cells were replated into 96-well plates prior to being used.
In vitro eosinophil infection, co-culture experiments, EETs analysis, and bacterial viability assay
For in vitro infection experiments, BM eosinophils or FACS-sorted splenic eosinophils were washed and kept in antibiotic-free RPMI-1640 medium supplemented with 10% heat-inactivated FBS and 10 ng/ml IL-5. BM eos were seeded in round-bottom 96-well plates at a density of 125,000 cells/well and infected with H. pylori or C. rodentium at 37°C for 18 h unless specified otherwise. Where indicated, eosinophils were treated with recombinant mouse IFN-γ (20 ng/ml, Peprotech). Where indicated, H. pylori and eosinophils were separated by a transwell during the time of infection (0.4 µm pores; Corning Transwell, Sigma). Upon infection or treatment, cells were washed and either analyzed by flow-cytometry for surface markers or gene expression, or supplemented with 100 U ml−1 penicillin/streptomycin and 25 mg ml−1 Kanamycin to eliminate bacterial contaminants, before T cell co-culture. CD4+ naive T cells were magnetically isolated from splenocyte suspensions with the MagCellect Mouse Naive CD4+ T Cell Isolation kit (R&D Systems). T cells were labeled with CFSE (Life Technologies) and co-cultured with eosinophils for 4 d at a 1:1 ratio in the presence of 30 U/ml recombinant human IL-2 (Proleukin, Prometheus), 10 ng/ml IL-5 and mouse T-Activator CD3/CD28 Dynabeads (Life Technologies). CFSE dilution was assessed by flow cytometry. Where indicated, co-cultures were treated with anti-mouse PD-L1 (B7-H1, BioXcell). For co-culture experiments with splenic eosinophils, 40,000 FACS-sorted eosinophils were co-cultured as described above with 150,000 naive CD4+ T cells. For the antigen-specific setup, 40,000 BM APCs were matured with LPS for 4 h, washed and loaded with OVA peptide 329–339 (5 × 10−5 M, PolyPeptide) 30 min before co-culture with 120,000 naive OTII CD4+ T cells and 40,000 sorted splenic eosinophils, previously infected with PMSS1 for 18 h. IFN-γ production was determined by intracellular cytokine staining. To determine bacterial viability upon infection of eosinophils, cell cultures were resuspended and an aliquot was stained with the LIVE/DEAD BacLight Bacterial Viability and Counting kit (Molecular Probes by Life Technologies) according to the manufacturer’s instructions and analyzed by flow cytometry. For EET formation assay and DNA quantification, 100,000 BM eos were infected with H. pylori and C. rodentium and treated with DNaseI (5 U/per well) where indicated at 37°C for 20 min. Cells further received 5 µM/well Sytox green (Invitrogen) after 10 min and were analyzed for EETs formation and DNA release in the culture supernatants. Cells were then spun, spread onto a glass slide, and visualized for extra cellular trap formation on a Leica DM6 B microscope. To quantify the amount of released DNA, 100 μl of culture supernatant per sample were transferred to a white, clear-bottom 96-well plate (Greiner Bio-One GmbH). The fluorescent emission intensity of DNA-bound Sytox green was measured on a fluorometer (SpectraMax i3, Molecular Devices) at 485 nm (excitation)/527 nm (emission).
Quantitation of secreted EPX
Mid-colon sections (one section per mouse) were weighed and cultured in RPMI 1640 medium with 10% FCS and 100 U/ml penicillin/streptomycin for 24 h at 37°C in a humidified incubator with 5% CO2. Cell-free supernatants were stored at −80°C and normalized to initial tissue weight in subsequent assays. EPX was detected in supernatants of colon explant cultures or BM Eos cultures by Sandwich/Capture ELISA (anti-EPX capture antibody clone MM25-429.1.1; biotinylated anti-EPX detection antibody clone MM25-82.2.1), provided by J.J. Lee, as described previously (Ochkur et al., 2012).
RNA sequencing and data analysis
RNA from FACS-sorted gastric eosinophils was isolated using the Arcturus PicoPure RNA Isolation kit (Applied Biosystems, ThermoFisher Scientific). RNA quality was assessed by the Agilent Tapestation followed by library preparation using the NuGEN Ovation SoLo protocol. Sequencing was subsequently performed on the Illumina HiSeq 2500 instrument. RNA-seq reads were quality-checked with fastqc, which computes various quality metrics for the raw reads. RNA-seq reads were mapped using the STAR aligner to the mouse reference genome build GRCm38. We included the Ensembl transcript annotations in the mapping index. Alignments were counted according using the featureCounts function in the Rsubread Bioconductor package. Statistical analysis of differential expression was conducted with the DESeq2 package. All RNA seq data generated within this study are publicly accessible at the Gene Expression Omnibus (GEO) repository (no. GSE105143).
Imaging flow cytometry
For imaging flow cytometry, cells were prepared and stained as for conventional flow cytometry. Cell acquisition was performed on the Image Stream X Mark II (Merck, Millipore). Analysis was performed with IDEAS software following a standard gating strategy excluding doublets, dead and CD45 negative cells. A g-value of 0.9 was applied to the images while setting the display to pixel data range.
Immunofluorescence and confocal microscopy
Paraffin-embedded tissue sections were deparaffinized and rehydrated with graded ethanol dilutions. After antigen retrieval in pressure cooker using EDTA-Tris buffer (10 mM, pH 9.0), immunofluorescence staining was performed by incubating the paraffin sections with mouse monoclonal anti-EPX antibody (1:300; clone MM25-82 2; obtained from Lee Laboratories, Mayo Clinic) and rabbit monoclonal anti-CD3 antibody (1:20; clone SP7; ThermoFisher Scientific, distributed by LuBioScience GmbH). Thereafter, secondary Alexa Fluor 488–conjugated goat anti-mouse (1:400) and Alexa Fluor 545–conjugated goat anti-rabbit (1:400) antibodies (ThermoFisher Scientific) were applied and tissue samples incubated at RT for 1 h. To detect EETs, tissue sections were incubated with anti-EPX antibody and subsequently stained with PI for additional 10 min. Samples were washed and mounted in Prolong Gold mounting medium and image acquisition was performed using confocal laser scanning microscopy LSM 700 (Carl Zeiss Micro Imaging) with a 20× or 40×/1.40 Oil DIC objective and analyzed with IMARIS software (Bitplane AG). For quantitative analysis, EPX+ and CD3+ infiltrating cells were counted in 10 high-power fields (hpf) of highest activity using an automated slide scanner (3DHISTECH slide scanner, Quant Center software, using Cell Count module). To determine the proportion of eosinophils forming EETs, we analyzed 300 eosinophils in each tissue section.
Statistical analysis
Statistical analysis was performed with Prism 6.0 (GraphPad Software). The nonparametric Mann-Whitney test was used for all statistical comparisons unless specified otherwise. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
The supplemental material associated with this manuscript consists of five supplemental figures. These show the gating strategy used to identify eosinophils as CD45+CD11b+MHCII−Ly6G−SiglecF+ cells in single-cell preparations of the gastric lamina propria (Fig. S1) and demonstrate that the frequencies of major GI leukocyte populations do not change upon eosinophil depletion in the steady-state and during infection (Fig. S2). The supplemental material further shows that cultured murine eosinophils acquire T cell–suppressive properties upon encountering H. pylori (Fig. S3), a feature that is regulated by IFN-γ (Fig. S4), and that eosinophils suppress C. rodentium–induced colitis and contribute to bacterial clearance by extracellular trap formation (Fig. S5).
Supplementary Material
Acknowledgments
We thank Markus G. Manz and the technical staff of the Department of Hematology for providing access to and preparation of human cord blood HSCs. We thank the personnel of the Laboratory Animal Services Center (LASC) of the University of Zürich for expert animal care. We would also like to thank the staff of the Zurich Functional Genomics Center for RNA sequencing services.
This work was supported by a Swiss National Science Foundation Temporary Backup Schemes Consolidator grant BSCGIO_157841/1 to A. Müller, a Swiss National Science Foundation grant (31003A_152851) to M. van der Broek., an OncoSuisse grant (KFS-3233-08-2013) to M. van der Broek, and the clinical research priority program on Human Hemato-Lymphatic Diseases, University of Zürich.
The authors declare no competing financial interests.
Author contributions: I.C. Arnold designed, performed, and analyzed most of the experiments and co-wrote the manuscript; M. Artola-Borán, P. Tallón de Lara, and A. Kyburz helped with experiments and provided critical advice. C. Taube, K. Ottemann, and M van der Broek provided critical tools and intellectual input. H.-U. Simon and S. Yousefi performed immunofluorescence stainings and their quantification. A. Müller supervised the study and co-wrote the manuscript.
References
- Arnold I.C., Lee J.Y., Amieva M.R., Roers A., Flavell R.A., Sparwasser T., and Müller A.. 2011. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology. 140:199–209. 10.1053/j.gastro.2010.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I.C., Zhang X., Urban S., Artola-Borán M., Manz M.G., Ottemann K.M., and Müller A.. 2017. NLRP3 Controls the Development of Gastrointestinal CD11b+ Dendritic Cells in the Steady State and during Chronic Bacterial Infection. Cell Reports. 21:3860–3872. 10.1016/j.celrep.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Aydemir S.A., Tekin I.O., Numanoglu G., Borazan A., and Ustundag Y.. 2004. Eosinophil infiltration, gastric juice and serum eosinophil cationic protein levels in Helicobacter pylori-associated chronic gastritis and gastric ulcer. Mediators Inflamm. 13:369–372. 10.1155/S0962935104000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T., Heppner F.L., Tertilt C., Heinen T.J., Kremer M., Wunderlich F.T., Jung S., and Waisman A.. 2005. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2:419–426. 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- Chu V.T., and Berek C.. 2013. The establishment of the plasma cell survival niche in the bone marrow. Immunol. Rev. 251:177–188. 10.1111/imr.12011 [DOI] [PubMed] [Google Scholar]
- Chu V.T., Beller A., Rausch S., Strandmark J., Zänker M., Arbach O., Kruglov A., and Berek C.. 2014. Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity. 40:582–593. 10.1016/j.immuni.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Diehl G.E., Longman R.S., Zhang J.X., Breart B., Galan C., Cuesta A., Schwab S.R., and Littman D.R.. 2013. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 494:116–120. 10.1038/nature11809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A.D., Jacobsen E.A., Ochkur S.I., Willetts L., Shim K., Neely J., Kloeber J., Lesuer W.E., Pero R.S., Lacy P., et al. 2013. Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J. Leukoc. Biol. 94:17–24. 10.1189/jlb.0213089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer K.D., Moser J.M., Czapiga M., Siegel S.J., Percopo C.M., and Rosenberg H.F.. 2008. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J. Immunol. 181:4004–4009. 10.4049/jimmunol.181.6.4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V., and Pollard J.W.. 2001. Eotaxin is required for eosinophil homing into the stroma of the pubertal and cycling uterus. Endocrinology. 142:4515–4521. 10.1210/endo.142.10.8459 [DOI] [PubMed] [Google Scholar]
- Griseri T., Arnold I.C., Pearson C., Krausgruber T., Schiering C., Franchini F., Schulthess J., McKenzie B.S., Crocker P.R., and Powrie F.. 2015. Granulocyte Macrophage Colony-Stimulating Factor-Activated Eosinophils Promote Interleukin-23 Driven Chronic Colitis. Immunity. 43:187–199. 10.1016/j.immuni.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmers S., Teijaro J.R., Arandjelovic S., and Mowen K.A.. 2011. PAD4-mediated neutrophil extracellular trap formation is not required for immunity against influenza infection. PLoS One. 6:e22043 10.1371/journal.pone.0022043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., and Rothenberg M.E.. 2014. Roles and regulation of gastrointestinal eosinophils in immunity and disease. J. Immunol. 193:999–1005. 10.4049/jimmunol.1400413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Wen T., Mingler M.K., Caldwell J.M., Wang Y.H., Chaplin D.D., Lee E.H., Jang M.H., Woo S.Y., Seoh J.Y., et al. 2015. IL-1β in eosinophil-mediated small intestinal homeostasis and IgA production. Mucosal Immunol. 8:930–942. 10.1038/mi.2014.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilberg D., Zavros Y., Shepherd B., Salama N.R., and Ottemann K.M.. 2016. Spatial and Temporal Shifts in Bacterial Biogeography and Gland Occupation during the Development of a Chronic Infection. MBio. 7:e01705–16. 10.1128/mBio.01705-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., König W., and Backert S.. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 449:862–866. 10.1038/nature06187 [DOI] [PubMed] [Google Scholar]
- Lee J.J., Dimina D., Macias M.P., Ochkur S.I., McGarry M.P., O’Neill K.R., Protheroe C., Pero R., Nguyen T., Cormier S.A., et al. 2004. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 305:1773–1776. 10.1126/science.1099472 [DOI] [PubMed] [Google Scholar]
- Lehrer R.I., Szklarek D., Barton A., Ganz T., Hamann K.J., and Gleich G.J.. 1989. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 142:4428–4434. [PubMed] [Google Scholar]
- Linch S.N., Kelly A.M., Danielson E.T., Pero R., Lee J.J., and Gold J.A.. 2009. Mouse eosinophils possess potent antibacterial properties in vivo. Infect. Immun. 77:4976–4982. 10.1128/IAI.00306-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern T.W., Talley N.J., Kephart G.M., Carpenter H.A., and Gleich G.J.. 1991. Eosinophil infiltration and degranulation in Helicobacter pylori-associated chronic gastritis. Dig. Dis. Sci. 36:435–440. 10.1007/BF01298871 [DOI] [PubMed] [Google Scholar]
- Mesnil C., Raulier S., Paulissen G., Xiao X., Birrell M.A., Pirottin D., Janss T., Starkl P., Ramery E., Henket M., et al. 2016. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Invest. 126:3279–3295. 10.1172/JCI85664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochkur S.I., Kim J.D., Protheroe C.A., Colbert D., Moqbel R., Lacy P., Lee J.J., and Lee N.A.. 2012. The development of a sensitive and specific ELISA for mouse eosinophil peroxidase: assessment of eosinophil degranulation ex vivo and in models of human disease. J. Immunol. Methods. 375:138–147. 10.1016/j.jim.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg H.F., and Dyer K.D.. 1995. Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem. 270:30234 10.1074/jbc.270.37.21539 [DOI] [PubMed] [Google Scholar]
- Rosenberg H.F., Dyer K.D., and Foster P.S.. 2013. Eosinophils: changing perspectives in health and disease. Nat. Rev. Immunol. 13:9–22. 10.1038/nri3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayi A., Kohler E., Hitzler I., Arnold I., Schwendener R., Rehrauer H., and Müller A.. 2009. The CD4+ T cell-mediated IFN-gamma response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J. Immunol. 182:7085–7101. 10.4049/jimmunol.0803293 [DOI] [PubMed] [Google Scholar]
- Soragni A., Yousefi S., Stoeckle C., Soriaga A.B., Sawaya M.R., Kozlowski E., Schmid I., Radonjic-Hoesli S., Boutet S., Williams G.J., et al. 2015. Toxicity of eosinophil MBP is repressed by intracellular crystallization and promoted by extracellular aggregation. Mol. Cell. 57:1011–1021. 10.1016/j.molcel.2015.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara R., Lee E.J., Jang M.S., Jeun E.J., Hong C.P., Kim J.H., Park A., Yun C.H., Hong S.W., Kim Y.M., et al. 2016. Small intestinal eosinophils regulate Th17 cells by producing IL-1 receptor antagonist. J. Exp. Med. 213:555–567. 10.1084/jem.20141388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M., Herbelin A., Pléau J.M., and Dardenne M.. 2000. CD11c+ eosinophils in the murine thymus: developmental regulation and recruitment upon MHC class I-restricted thymocyte deletion. J. Immunol. 165:1965–1975. 10.4049/jimmunol.165.4.1965 [DOI] [PubMed] [Google Scholar]
- Toller I.M., Neelsen K.J., Steger M., Hartung M.L., Hottiger M.O., Stucki M., Kalali B., Gerhard M., Sartori A.A., Lopes M., and Müller A.. 2011. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA. 108:14944–14949. 10.1073/pnas.1100959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers J., and Rothenberg M.E.. 2015. Eosinophils in mucosal immune responses. Mucosal Immunol. 8:464–475. 10.1038/mi.2015.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Köckritz-Blickwede M., and Nizet V.. 2009. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. (Berl.). 87:775–783. 10.1007/s00109-009-0481-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T., and Rothenberg M.E.. 2016. The Regulatory Function of Eosinophils. Microbiol. Spectr. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterkamp S., Raithel M., and Hahn E.G.. 2000. Secretion and tissue content of eosinophil cationic protein in Crohn’s disease. J. Clin. Gastroenterol. 30:170–175. 10.1097/00004836-200003000-00009 [DOI] [PubMed] [Google Scholar]
- Wolber F.M., Leonard E., Michael S., Orschell-Traycoff C.M., Yoder M.C., and Srour E.F.. 2002. Roles of spleen and liver in development of the murine hematopoietic system. Exp. Hematol. 30:1010–1019. 10.1016/S0301-472X(02)00881-0 [DOI] [PubMed] [Google Scholar]
- Wu D., Molofsky A.B., Liang H.E., Ricardo-Gonzalez R.R., Jouihan H.A., Bando J.K., Chawla A., and Locksley R.M.. 2011. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 332:243–247. 10.1126/science.1201475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Kimura H., Kurabayashi M., Kozawa K., and Kato M.. 2008. Interferon-gamma enhances human eosinophil effector functions induced by granulocyte-macrophage colony-stimulating factor or interleukin-5. Immunol. Lett. 118:88–95. 10.1016/j.imlet.2008.03.005 [DOI] [PubMed] [Google Scholar]
- Yousefi S., Gold J.A., Andina N., Lee J.J., Kelly A.M., Kozlowski E., Schmid I., Straumann A., Reichenbach J., Gleich G.J., and Simon H.U.. 2008. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 14:949–953. 10.1038/nm.1855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.