Lee et al. examine the dynamics of membrane proteins within the ciliary membrane using quantum dots and 2P Super FRAP. They show that ciliary membrane proteins diffuse rapidly within highly fluid local membrane domains delimited by actin filaments.
Abstract
Physical properties of primary cilia membranes in living cells were examined using two independent, high-spatiotemporal-resolution approaches: fast tracking of single quantum dot–labeled G protein–coupled receptors and a novel two-photon super-resolution fluorescence recovery after photobleaching of protein ensemble. Both approaches demonstrated the cilium membrane to be partitioned into corralled domains spanning 274 ± 20 nm, within which the receptors are transiently confined for 0.71 ± 0.09 s. The mean membrane diffusion coefficient within the corrals, Dm1 = 2.9 ± 0.41 µm2/s, showed that the ciliary membranes were among the most fluid encountered. At longer times, the apparent membrane diffusion coefficient, Dm2 = 0.23 ± 0.05 µm2/s, showed that corral boundaries impeded receptor diffusion 13-fold. Mathematical simulations predict the probability of G protein–coupled receptors crossing corral boundaries to be 1 in 472. Remarkably, latrunculin A, cytochalasin D, and jasplakinolide treatments altered the corral permeability. Ciliary membranes are thus partitioned into highly fluid membrane nanodomains that are delimited by filamentous actin.
Introduction
Cilia have been described as antennae that probe the physical environment surrounding cells and tissues (Pazour and Witman, 2003). To subserve this function, signal transduction proteins, including seven transmembrane-spanning G protein–coupled receptors (GPCRs) and their downstream effectors, are enriched within ciliary membranes, where they detect light, odorants, and secreted molecules, among other signals (Gerdes et al., 2009; Garcia-Gonzalo and Reiter, 2012; Pearring et al., 2013; Mukhopadhyay and Rohatgi, 2014; Reiter and Leroux, 2017). Signal transduction involves the diffusional encounter between membrane proteins. An activated GPCR sequentially collides with and activates G proteins, which then diffuse to modulate downstream effectors. This arrangement allows response amplification and provides sites for feedback regulation (Pugh and Lamb, 2000). The physical properties of cilia membranes are, therefore, central to understanding cilium signaling and function and cilia-related diseases, yet they remain poorly understood largely because primary cilia are smaller than the spatial resolution limit of conventional live-cell microscopy and because they are difficult to isolate to homogeneity.
Recent studies have taken advantage of the exquisite high spatial resolution afforded by single-molecule tracking to examine the transport of GPCRs and GPCR-like transmembrane proteins within cultured inner medullary collecting duct (IMCD) cell primary cilia and found that they mostly sample the ciliary membrane space via diffusion, with occasional, short-duration coupling to motor proteins (Ye et al., 2013; Milenkovic et al., 2015). It was also shown that ligand binding or receptor activation could result in local binding (Milenkovic et al., 2015) or coupling to intraflagellar transport (IFT) motors via BBsomes (Ye et al., 2018). More recently, a study of single IFT-88 molecules entering the cilium base and moving through the transition zone were found to undergo several changes in transport speed before emerging into the cilium, suggesting that several steps are involved in moving through the diffusion barrier at the cilium base (Yang et al., 2017 Preprint). The relatively slow tracking of proteins in these studies, however, did not allow analysis of the membrane structure or fluidity at a level previously achieved for the plasma membrane (Fujiwara et al., 2002; Kusumi et al., 2005, 2014; Suzuki et al., 2005).
Here we directly examined the physical properties of living IMCD3 cell primary cilia membranes using two independent, high-spatial-resolution and high-temporal-resolution approaches to examine the dynamics of intrinsic membrane proteins, tracking the movements of individual proteins within the ciliary membrane using quantum dots (Qdots) and a novel two-photon (2P) FRAP approach that we call 2P Super FRAP. We chose to examine the transport behavior of two-cilium–enriched GPCRs: the somatostatin receptor type 3 (SSTR3), a rhodopsin-like GPCR found within cilia of developing central nervous system neurons, where it plays a role in neural differentiation (Händel et al., 1999), and a chimera of the rhodopsin apoprotein, opsin (Rho), which is localized to photoreceptor sensory cilia (Palczewski, 2006; Deretic and Wang, 2012; Goldberg et al., 2016). Our results showed that intrinsic ciliary membrane proteins diffused rapidly within highly fluid local membrane domains delimited by actin filaments.
Results
SSTR3-EGFP or a chimera of bovine rhodopsin with the third intracellular loop replaced by that of the SSTR3 receptor (Rhoi3S-EGFP; Geneva et al., 2017), each with an N-terminal myc tag (Fig. 1 A), were expressed in IMCD3 cells, and cilium elaboration was induced by serum starvation. The distribution of EGFP-labeled GPCRs appeared homogeneous in live ciliary membranes (Fig. 1 B), in agreement with previous studies (Geneva and Calvert, 2010; Hu et al., 2010; Trivedi et al., 2012; Ye et al., 2013; Geneva et al., 2017). GPCR dynamics were then probed using two independent approaches.
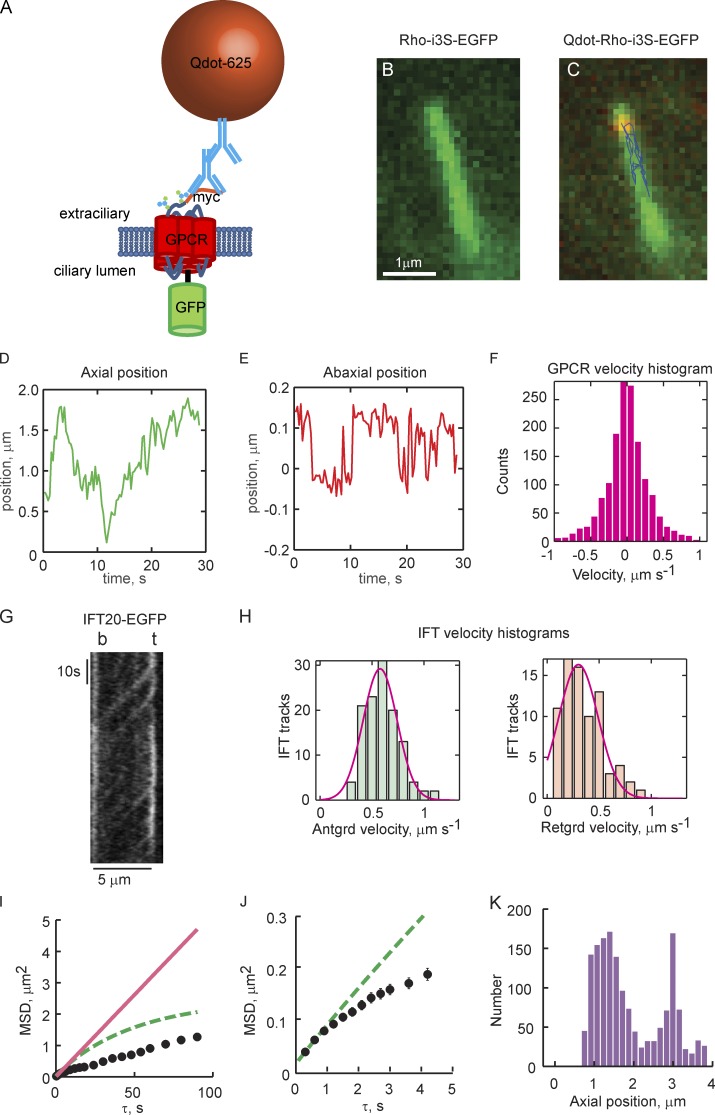
Figure 1.
Superresolution tracking of single Qdot-labeled GPCRs reveals 2D transport on ciliary membranes that is distinct from IFT-based motor transport. (A) Schematic of the GPCR constructs used for single-molecule tracking and ensemble FRAP experiments. (B) Image of a cilium taken with the GFP channel shows uniform distribution of myc-Rhoi3S-EGFP. (C) Overlay of GFP and Qdot channels shown with tracking results from a single Qdot625-labeled myc-Rhoi3S-EGFP. Axial and circumferential movements (abaxial) are apparent (see Videos 1–4). (D) Axial position time course (kymograph) obtained from Qdot tracking. (E) Time course of movements perpendicular to the cilium axis. Zero in the ordinate indicates the center of mass of the cilium. (F) Instantaneous axial velocity histogram. Anterograde velocities are positive; retrograde are negative. Note the symmetry of the histogram, indicating that the distribution of velocities is the same in the anterograde and retrograde directions. (G) Representative IFT20-EGFP kymograph obtained under identical conditions (Video 5). Note smooth contiguous movement along the full length of the cilium. (H) Anterograde and retrograde IFT velocity histograms pooled from the slopes of 122 anterograde and 84 retrograde tracks from 14 cilia obtained from six independent experiments. Red lines, Gaussian fittings of the histograms. Anterograde, mean = 0.594, SD = 0.173 µm/s; retrograde, mean = 0.340, SD = 0.204 µm/s. (I and J) MSD analysis reveals significant subdiffusion of GPCRs within ciliary membranes. Black circles represent time-averaged MSDs of an individual Qdot-labeled GPCR plotted as a function of the time step size (τ). Error bars (SEM) are on the order of the size of the symbols; n > 1,000 for each τ. Red lines, linear fittings to MSD(τ = 0.3 and 0.6 s) from which apparent Dax was estimated; dashed lines, predicted MSD(τ) based on estimated Dax and Eq. 1. (J) MSD(τ) plot on expanded scale. (K) Axial position histogram for the individual GPCR tracking experiment. The length of a given cilium was divided into 20 bins into which axial positions were distributed. Zero on the abscissa represents the base of the cilium. In D–F and I–K, results are representative from one tracking experiment. Total number of Qdot-labeled myc-Rhoi3S-EGFP tracked = 9, on eight individual cilia, taken from four independent experiments (Table 1).
Single-molecule tracking allowed GPCR transport to be evaluated at high spatial resolution
Several basic modes of GPCR transport along the ciliary membrane are possible, including Brownian motion, motor transport, and local binding to stationary objects. We began the study with an evaluation of the relative contributions of these modes to the overall movement of the GPCRs, tracking single molecules labeled with Qdots (Fig. 1, C–F). Tracking on our system allowed 20-nm precision (Fig. S1). Because the major modes of GPCR transport are most apparent in the axial direction, GPCR transport was examined in detail using axial kymographs (Fig. 1 D). SSTR3 and Rhoi3S displayed similar patterns of movement (Fig. S2, C and D; and Videos 3 and 4) that were distinct from motor protein–driven transport typified by components of the IFT system. The GPCRs interspersed Brownian movement with apparent processive or stalled movements of varying duration (Figs. 1 D and S2, C and D). IFT proteins generally traveled as focal puncta from one end of the cilium to the other without interruption (Fig. 1 G and Video 5). Examination of the instantaneous axial velocities of the GPCRs (Fig. 1 F) and IFT (Fig. 1 H) also showed significant differences. Anterograde and retrograde velocities of the GPCRs were symmetrical, peaking near zero and falling exponentially to a maximum rate of ~1–1.5 µm/s. In contrast, velocities of the motor-driven IFT proteins were approximately normally distributed, with mean velocities of 0.60 ± 0.02 µm/s (mean ± SEM) and 0.34 ± 0.02 µm/s, anterograde and retrograde, respectively, in good agreement with previous studies (Follit et al., 2006; Ye et al., 2013; Broekhuis et al., 2014).
Mean square displacement (MSD) shows that GPCR transport along the cilium appears constrained
MSD along the axial dimension of the cilium was performed to assess the major modes of GPCR transport. The analysis had qualitative predictions that gave some insight into the mechanisms of GPCR movement (Saxton and Jacobson, 1997; Kusumi et al., 2005, 2014; Metzler et al., 2014): If GPCR transport is mainly via diffusion, then the relationship between the time-averaged square displacement <x2> and the time step duration τ will be linear, with a slope proportional to the effective diffusion coefficient. If significant motor transport occurred, the relationship would initially arc upward, referred to as superdiffusion. Confinement of the GPCRs to local regions would result in a downward-arcing relation, known as subdiffusion. Invariably, the MSD(τ) plots for the individual GPCRs examined in this study had downward-arching shapes (Fig. 1, I and J). This result showed that the movements of the GPCRs along the ciliary membrane axis were “freer” at shorter τs and constrained at longer τs, and thus demonstrated subdiffusion-like behavior. Some degree of subdiffusion is expected, however, given the small size of primary cilia.
Observed constraint on GPCR motion exceeded that predicted by the boundaries of the ciliary membrane
To examine the impact of boundaries at the base and tip of the cilia on GPCR motion, we calculated the expected MSD(τ) relation for diffusion within a cilium of finite length. The cilium membrane may be approximated as the surface of a right circular cylinder with diameter of ~300 nm. The chief variable in cilium membrane area, then, is the cilium length. The MSD(τ) relation for a membrane protein moving by diffusion along the ciliary axis x of length lx may be described as
| (1) |
where σ = (2Dx)0.5, Dx is the diffusion coefficient in the x direction, and
is the MSD(τ) at which the relation saturates at large τ because of the no-flux boundaries at the cilium base and tip, x = 0, and x = lx (Kusumi et al., 1993). We compared the MSD(τ) predicted from Eq. 1 to the MSD(τ) obtained for each cilium (Fig. 1, I and J, green lines). The MSD(τ) relation of the tracking data followed the MSD(τ) predicted by Eq. 1 only through the first one or two τs (∼0.3–0.6 s), after which it peeled off to lower than predicted values, thus showing impediments to free axial diffusion other than the absolute length of the cilium (Table 1). Long-term GPCR tracking in most cases showed sampling of the entire cilium length (Fig. 1 K, fraction of cilium sampled [fcs]; and Table 1). This indicates that the constraint on GPCR movements could not have resulted from absolute confinement to a subregion of the cilium.
Table 1. Individual GPCR and cilia parameters: low frame rate (3–5 fps) tracking.
| Ciliuma | Tracking time | Cilium length | GPCR densityb | GPCRs on ciliumc | Amd | Daxe | Dmf | MIg | fcsh | MSDinti |
|---|---|---|---|---|---|---|---|---|---|---|
| min | μm | μm−2 | % | μm2/s | μm2/s | μm2 | ||||
| R1 | 10.5 | 3.7 | 269.2 | 932 | 0.24 | 0.074 | 0.148 | 0.77 | 1.0 | 0.079 |
| R2 | 3.0 | 4.2 | 1,487.5 | 5,833 | 1.34 | 0.026 | 0.051 | 0.35 | 0.7 | 0.020 |
| R3 | 4.0 | 1.9 | 4,965.2 | 8,735 | 4.47 | 0.035 | 0.069 | 0.78 | 1.0 | 0.020 |
| R4 | 17.1 | 3.7 | 2,790.7 | 9,721 | 2.51 | 0.135 | 0.270 | 0.50 | 1.0 | 0.079 |
| R5 (1) | 12.0 | 2.9 | 1,653.2 | 4,592 | 1.49 | 0.015 | 0.030 | 0.61 | 1.0 | 0.009 |
| R5 (2) | 12.0 | 0.081 | 0.163 | 0.34 | 1.0 | 0.049 | ||||
| R6 | 8.4 | 3.8 | 1,607.6 | 5,795 | 1.45 | 0.030 | 0.059 | 0.42 | 0.9 | 0.018 |
| R7 | 4.5 | 4.3 | 30.5 | 123 | 0.03 | 0.145 | 0.290 | 0.26 | 1.0 | 0.070 |
| R8 | 9.7 | 2.4 | 873.2 | 1,969 | 0.79 | 0.095 | 0.189 | 0.70 | 1.0 | 0.056 |
| Mean R | 3.6 | 1,744.9 | 4,713 | 1.54 | 0.071 | 0.141 | 0.53 | 0.96 | 0.045 | |
| SEM R | 0.3 | 512.5 | 1,172 | 0.47 | 0.016 | 0.032 | 0.07 | 0.03 | 0.009 | |
| S1 | 11.9 | 5.6 | 557.5 | 2,947 | 0.50 | 0.085 | 0.170 | 0.39 | 1.0 | 0.027 |
| S2 (1) | 12.7 | 4.8 | 10,676.0 | 48,368 | 9.61 | 0.167 | 0.334 | 0.62 | 1.0 | 0.099 |
| S2 (2) | 12.7 | 0.180 | 0.361 | 0.55 | 1.0 | 0.099 | ||||
| S3 (1) | 9.2 | 7.4 | 6,534.6 | 45,267 | 5.88 | 0.063 | 0.126 | 0.63 | 1.0 | 0.037 |
| S3 (2) | 9.2 | 0.032 | 0.065 | 0.62 | 1.0 | 0.020 | ||||
| S4 | 28.3 | 7.0 | 7,292.1 | 47,937 | 6.56 | 0.079 | 0.157 | 0.21 | 0.7 | 0.047 |
| S5 | 14.7 | 8.1 | 1,591.5 | 12,077 | 1.43 | 0.141 | 0.282 | 0.28 | 1.0 | 0.079 |
| S6 | 7.5 | 10.0 | 1,761.3 | 16,631 | 1.59 | 0.075 | 0.149 | 0.50 | 0.8 | 0.064 |
| S7 | 14.8 | 4.7 | 2,591.2 | 11,404 | 2.33 | 0.109 | 0.217 | 0.44 | 1.0 | 0.075 |
| S8 | 4.1 | 8.2 | 2,612.2 | 20,191 | 2.35 | 0.121 | 0.242 | 0.39 | 0.9 | 0.096 |
| S9 | 5.2 | 6.4 | 1,461.0 | 8,748 | 1.31 | 0.086 | 0.172 | 0.19 | 0.3 | 0.010 |
| S10 | 6.3 | 7.2 | 2,412.5 | 16,257 | 2.17 | 0.138 | 0.276 | 0.49 | 1.0 | 0.099 |
| S11 (1) | 16.0 | 7.3 | 1,487.1 | 10,205 | 1.34 | 0.129 | 0.258 | 0.46 | 1.0 | 0.056 |
| S11 (2) | 16.0 | 0.111 | 0.222 | 0.56 | 1.0 | 0.056 | ||||
| Mean S | 7.2 | 3,543.4 | 21,821 | 3.19 | 0.108 | 0.216 | 0.45 | 0.91 | 0.062 | |
| SEM S | 0.5 | 1,061.4 | 5,640 | 0.86 | 0.011 | 0.022 | 0.04 | 0.05 | 0.008 |
Individual cilia: R, Rhoi3S; S, SSTR3. Numbers in parentheses identify individual Qdot-GPCR for cilia in which more than one was labeled.
GPCR density was estimated by calibration of the optical system with recombinant EGFP of known concentrations. Confinement of the EGFP label to the ciliary membrane and the sampling by the optical system were accounted for as described previously (Peet et al., 2004).
Total number of GPCRs within the given cilium.
Membrane area fraction occupied by the expressed GPCRs.
Dax is the 1D axial diffusion coefficient estimated from the initial slope of the MSD(τ) plot for an individual Qdot-GPCR.
Effective membrane diffusion coefficient: Dm ≈ 2Dax. Dax underestimates the true 2D membrane diffusion coefficient approximately twofold. Note the similarity of the mean microscopic Rhoi3S diffusion coefficient in ciliary membranes, Dm, reported here to the mean macroscopic lateral diffusion coefficient estimated for rhodopsin within discrete lobules of photoreceptor disc membranes, Dlat = 0.130 ± 0.012 µm2/s (Najafi et al., 2012a).
Mobility index (see text).
Fraction of cilium length sampled by the indicated GPCR.
MSD value at the y-intercept of the linear regression of the first two MSD(τ) points.
2D random walk on ciliary membranes recapitulates single GPCR transport behaviors but fails to reproduce subdiffusion
A simulation of membrane protein transport on the cilium membrane was generated to examine the impact of various modes of GPCR transport on the MSD(τ) relation (Fig. 2 and Theory). The approach was based on a random walk on a cylinder surface into which other modes of transport, including transient local binding, transient motor transport, and transient confinement to corral domains, were incorporated. Results from the pure random walk simulation, based on the parameters of the SSTR3-expressing cilium displayed in Fig. 2 G, showed that the pure random walk contained many of the axial movement behaviors observed in tracking experiments (Fig. 2 B and Video 6), including apparently processive movements (Fig. 2 B, black arrows), some of which traversed the entire cilium length. Periods of low axial movements were also observed in the model output (Fig. 2 B, red arrows). Averaging ∼1,000 MSDs for a given τ produced results that largely agreed with the Eq. 1 prediction (Fig. 2 D), thus validating the random walk simulation. The results show that stretches of unidirectional movement and slow movement are inherent to the random walk and, alone, do not indicate discrete motor transport or local binding events.
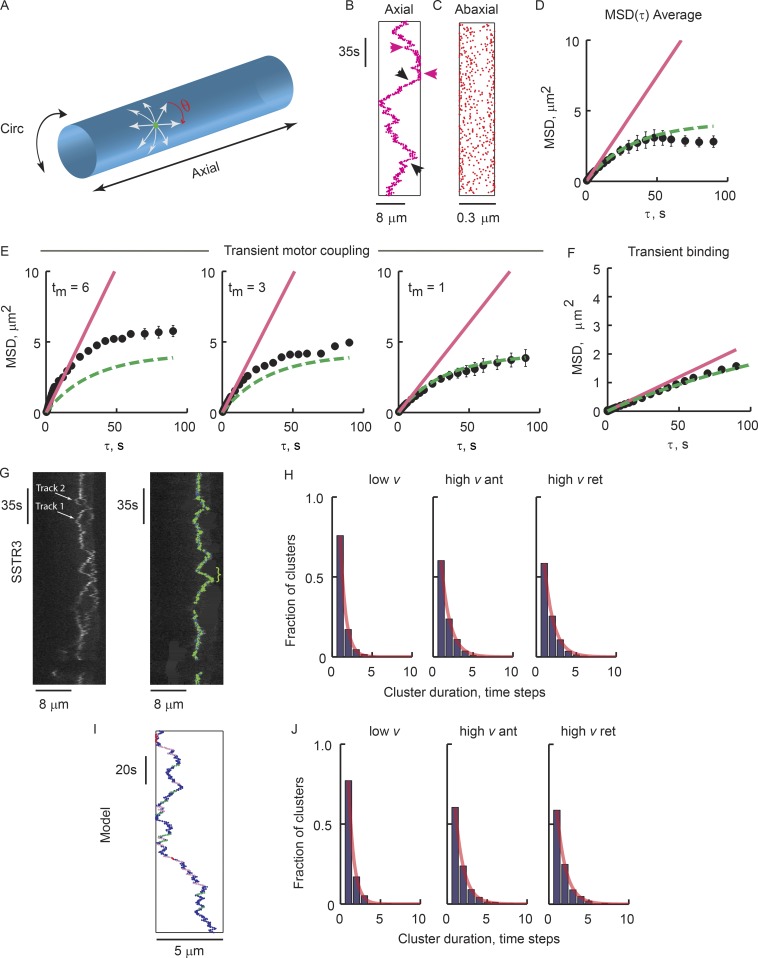
Figure 2.
Simulated random walk on the cilium membrane recapitulates the patterns of axial GPCR movements. (A) Schematic of the cilium membrane. Green dot, a theoretical GPCR; θ, randomly generated angle that determines movement direction; arrows, a subset of possible trajectories; Circ, circumferential. (B–F) Simulation results with parameters found for the SSTR3 tracking shown in G: Dm = 0.15 µm2/s, lax = 8 µm, time step = 0.3 s (Video 6). (B) Axial position kymograph from a single simulation run. Red and black arrowheads indicate regions of low and high axial velocity, respectively. (C) 2D projection of the 3D circumferential positions from run in B show expected denser population of points at the edges. (D) Black symbols represent mean MSD(τ) of five runs; in each run, the MSD represents the mean of >1,000 squared displacements. Error bars = SEM. (E and F) Results from simulations including transient coupling to motors or stationary objects. In each plot, black circles represent the mean from five independent simulations with lax = 5 µm and Dm = 0.15 µm2/s. Straight lines connect the first two MSD(τ) points. Dashed lines are the Eq. 1 predictions. (E) Stochastic, transient coupling to an axial moving motor, where Pm = 0.2, vm = 0.6 µm/s, and coupling duration, tm, are indicated (Video 7). (F) Transient binding to immobile objects, where Pb = 0.2, tb = 10 time steps (Video 6). Note that with transient motor transport, MSD(τ) consistently exceeds the Eq. 1 prediction, whereas transient binding leads to an overall reduced slope. (G–J) Comparison of GPCR motion and pure random walk simulation shows similar frequencies of apparent processive motion and absence of motion. (G) Kymograph segment of an SSTR3 tracking experiment (Video 3). Left, raw kymograph showing two Qdot-labeled SSTR3s. Right, Track 2 was masked for track 1 analysis. Overlaid markers plot detected positions; colors indicate instantaneous velocity categories: blue, v < 0.2 µm/s, green, v ≥ 0.2 µm/s. (H) Frequency of sequential low-velocity clusters (low v), high-velocity clusters, anterograde (high v ant) or retrograde (high v ret), pooled from 14 tracked Qdot-labeled GPCRs obtained in eight independent experiments (Table 1). Lines are exponential fittings. (I) Axial positions from 180 s of a pure random walk simulation. Clusters of sequential displacements are color coded: low v, red; high v anterograde, purple; high v retrograde, green. (J) Velocity cluster histograms from five simulation runs. Lines as in H.
Transient coupling to motor proteins or local binding sites was not supported by simulation
Simulations that include transient GPCR association with motors were examined to understand the potential impact of active transport on the MSD(τ) plots. Motor displacement rate vm = 0.6 µm/s was used, which represents the mean anterograde IFT velocity. Coupling of the GPCR to motors was allowed anywhere on the cilium surface with probability Pm = 0.2, the approximate frequency of motor coupling reported in Ye et al. (2013). The duration of the motor coupling, tm, was varied from 1 to 12 time steps (0.3–3.6 s) and the model output was subjected to the MSD(τ) analysis. With the exception of tm = 1, which produced an MSD(τ) plot that was indistinguishable from the pure Brownian motion prediction, all transient motor-driven simulations resulted in MSD(τ) that exceeded Eq. 1 prediction (Fig. 2 E and Video 7).
We explored the possibility that the subdiffusion observed in GPCR movement could be caused by transient local binding events. A model of stochastic, transient binding to immobile structures located anywhere on the cilium, with probability of binding Pb = 0.2 and binding duration tb = 10 time steps (3 s), resulted in a nearly linear MSD(τ) relation (Fig. 2 F and Video 8). The apparent diffusion coefficient of the simulated GPCRs was uniform over all τ, a result that does not match the MSD(τ) behavior of the GPCRs in the living cilia.
To examine if there were infrequent motor coupling or local binding events not detected by MSD analysis, each tracking experiment was searched for clusters of sequential movements. We determined instantaneous velocities of the GPCRs and segregated them into two categories: those with velocities lower than the observed retrograde IFT rates (ν ≤ 0.2 µm/s) and those with higher velocities (Fig. 2 G). The durations and frequencies of sequential low-velocity events, and high-velocity events in either the anterograde or retrograde direction, were determined and histograms plotted (Fig. 2 H). The histograms of high-velocity clusters, in either direction, fell exponentially with cluster size, yielding a cluster decay constant, kcl, of ∼1.2 time steps for both SSTR3 and Rhoi3S. Low-velocity cluster durations declined with kcl = 1.3 time steps. Long-duration, low-velocity clusters were found, with 16 clusters of 10,000 having durations longer than 10 time steps and occasional low-velocity stretches of 36 time steps (∼11 s) or longer. This result indicates that GPCRs bind to relatively immobile structures with low frequency.
Remarkably, the temporal cluster analysis of the pure random walk model results did not differ substantially from GPCR tracking results (Fig. 2, I and J). The simulation results, however, did not contain the infrequent longer-duration, low-velocity clusters found in GPCR tracking, indicating that those events were likely genuine, in agreement with similar observations (Milenkovic et al., 2015). The simulations led to the conclusion that significant motor-based GPCR transport did not occur under our experimental conditions. This result stands for cilia expressing vastly different densities of the labeled GPCRs, from ∼100 to tens of thousands per cilium (Table 1), and thus does not appear to be an artifact of overexpression. The result is in agreement with a recent study that unliganded SSTR3 in IMCD3 cilia did not show coupling to motors (Ye et al., 2018).
High-speed GPCR tracking reveals highly fluid, corralled membrane domains
Subdiffusion has been reported to arise from several mechanisms in live cells, including viscoelastic effects of membranes, local membrane curvature (Quemeneur et al., 2014), molecular crowding (Weiss et al., 2004), and corralling of the GPCRs into fluid membrane domains (Kusumi et al., 1993, 2014; Suzuki et al., 2005; Oh et al., 2014). With the exception of local corralling, the MSD(τ) relations for these mechanisms are expected to smoothly curve downward and to be well fitted with a single exponential with time exponent <1. Corralling into subdomains, on the other hand, is expected to result in MSD(τ) relations that exhibit more than one discrete phase, as originally shown using high-speed particle tracking (Kusumi et al., 1993; Oh et al., 2014).
We increased the sampling rate for Qdot tracking to 200 frames per second (fps), thus allowing τs down to 5 ms to be assessed (Fig. 3). At this frame rate, the Qdots appeared to stay within relatively small regions on the ciliary membranes, sampling them intensively, and then moving on to another discrete region (Fig. 3 B). The time spent within a given region varied from less than a second to about a second. Comparison of the apparent sizes of the regions to the positions recorded for the Qdot-labeled GPCR on a glutaraldehyde-fixed cell (Fig. 3 B, fixed) shows that the apparent confinement of the GPCRs was not caused by binding to stationary objects.
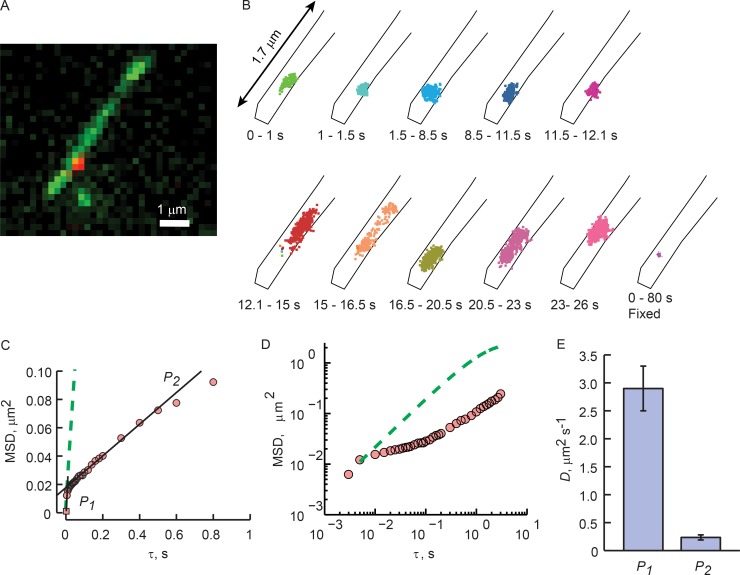
Figure 3.
High-speed imaging reveals corralled GPCR diffusion. (A) A still from a Rhoi3S tracking experiment acquired at 200 fps. (B) Time sequence of the positions of the GPCR tracked in B during the indicated experimental time frame, shown on expanded scale. Black lines represent the boundaries of the cilium. The bottom right plot represents the positions of a Qdot-labeled Rhoi3S fixed with glutaraldehyde (Fig. S1). (C) MSD(τ) analysis (symbols). Black lines represent linear regressions of two plot phases: P1, the first two points; P2, τ = 0.1–0.5 s. The slope of P1 represents diffusion that is least impacted by corral confinement, and that of P2 represents diffusion that is substantially impeded by the corral boundaries. Green dashed line represents Eq. 1 prediction based on the diffusion coefficient estimated from the P1 slope. (D) Log–log plot of the results from E. Note that the Eq. 1 prediction is linear, whereas the tracking results have several phases. In A–D, results are representative from a single tracking experiment. (E) Mean diffusion coefficients computed from the slopes of the two phases. Error bars are SEM. n = 18 cells/18 individual cilia obtained in seven individual experiments (Table 2).
High-speed acquisition revealed two discrete phases of the MSD(τ) plot: a steep rise from 5 to ∼20 ms, followed by a dramatically shallower rise from ∼20 to 500 ms (Fig. 3 C). Linear regressions of the two phases showed that the first phases had, on average, 13-fold steeper slopes than the second phases, corresponding to mean diffusion coefficients of Dm1 = 2.9 ± 0.40 µm2/s and Dm2 = 0.24 ± 0.05 µm2/s (Fig. 3 E, n = 18; and Table 2). For this analysis, the MSD(τ) for Qdot-labeled GPCRs on glutaraldehyde-fixed cells was used to determine the noise floor of our imaging system and thus the lowest measurable MSD(τ) value (Fig. S1 D).
Table 2. Corralled diffusion fitting parameters: high-speed (200-fps) tracking.
| Cilium | Cilium length | Dm1a | Dm2b | Dcorc | Pbcd | lcore | Trf |
|---|---|---|---|---|---|---|---|
| μm | μm2/s | μm2/s | μm2/s | Μm | s | ||
| R9 | 3.8 | 2.2 | 0.24 | 4.6 | 0.0024 | 0.147 | 0.61 |
| R10 | 2.3 | 0.83 | 0.02 | 2.83 | 0.0008 | 0.274 | 1.21 |
| R11 | 5.3 | 3.4 | 0.16 | 6.4 | 0.0012 | 0.367 | 1.20 |
| R12 | 2.8 | 4.7 | 0.34 | 8.4 | 0.0032 | 0.274 | 0.48 |
| R13 | 2.9 | 2.6 | 0.16 | 5.2 | 0.0018 | 0.315 | 0.85 |
| R14 | 2.3 | 5.1 | 0.27 | 8.5 | 0.0016 | 0.367 | 1.39 |
| R14 | 3.8 | 3.2 | 0.12 | 4.3 | 0.0014 | 0.421 | 1.41 |
| R15 | 2.1 | 0.78 | 0.18 | 3.2 | 0.0083 | 0.130 | 0.13 |
| R16 | 4.5 | 1.3 | 0.11 | 3.3 | 0.0033 | 0.183 | 0.39 |
| R17 | 3.0 | 1.7 | 0.25 | 4.8 | 0.0056 | 0.222 | 0.25 |
| R18 | 5.1 | 2.0 | 0.14 | 4.8 | 0.0018 | 0.239 | 0.82 |
| S12 | 7.3 | 2.8 | 0.18 | 4.3 | 0.0017 | 0.245 | 0.94 |
| S13 | 5.9 | 7.5 | 0.11 | 7.5 | 0.0033 | 0.431 | 0.67 |
| S14 | 3.3 | 2.7 | 0.25 | 7.7 | 0.0040 | 0.268 | 0.29 |
| S15 | 3.8 | 5 | 0.14 | 8 | 0.0031 | 0.395 | 0.50 |
| S16 | 7.1 | 1.4 | 0.10 | 7 | 0.0022 | 0.205 | 0.57 |
| S17 | 2.3 | 1.1 | 0.10 | 4.2 | 0.0026 | 0.173 | 0.46 |
| S18 | 3.3 | 3.2 | 0.11 | 3.2 | 0.0026 | 0.329 | 0.53 |
| Mean | 3.9 | 2.9 | 0.235 | 5.46 | 0.0028 | 0.274 | 0.71 |
| SEM | 0.37 | 0.4 | 0.046 | 0.45 | 0.0004 | 0.020 | 0.09 |
GPCR membrane diffusion coefficient computed from slope of first two MSD(τ) points (first phase).
GPCR membrane diffusion coefficient computed from slope of the second phase of the MSD(τ) plot.
GPCR membrane diffusion coefficient found by fitting MSD(τ) with corralled random walk simulation. This represents the Dm that is least affected by corral confinement.
Probability of a GPCR crossing a corral boundary upon encountering the boundary, found by fitting MSD(τ) with corralled random walk simulation.
Mean length of corral in the axial direction, computed from y-intercept of the linear regression of the second phase of the MSD(τ) plot.
Mean residence time for a GPCR within a corral, found by fitting MSD(τ) with corralled random walk simulation.
Estimation of the mean corral size
Previous studies of GPCR and lipid corralling on the plasma membrane estimated the corral size by direct observation of the tracking positions over time (Kusumi et al., 1993; Fujiwara et al., 2002; Suzuki et al., 2005). Taking this approach, examination of the clusters of GPCR positions in Fig. 3 B suggested that corrals may vary in size, with some clusters appearing quite compact and others more dispersed. However, several factors make drawing conclusions about corrals from direct observation of Qdot positions tenuous. The cylindrical geometry of the cilium makes it uncertain where the GPCRs are along the cilium circumference. The narrowness of the cilium reduced detectable corral jumping to one direction. Finally, and perhaps most importantly, as with the appearance of apparent processive motion, pure random walks may appear to have confinement regions, when observed by eye, when in fact they do not (Saxton, 1995).
We thus chose a statistical approach to detecting and estimating the mean corral size. We reasoned that, in the case of finite diffusion space, the MSD(τ) relation would reach a plateau at an MSD value that is proportional to the size of the diffusion space. This can be seen from Eq. 1, where the second term drops out at large τ:
and therefore
This result shows that the amplitude of the MSD(τ) plot at the break in slope, MSDcor, is proportional to the axial length of the corral. On average, MSDcor = 0.015 ± 0.002 µm2, yielding an estimate of the mean size of the corrals of lcor = 274 ± 20 nm (Table 2).
Estimation of the permeability of the corral boundaries and the mean residence time of the GPCRs within the corrals
We next estimated the permeability of the corrals and the mean time that a GPCR remained within a given corral by fitting the high-temporal-resolution MSD(τ) plots with a confined random walk model (Fig. 4, Video 9, and Theory). We introduced a lattice of uniformly spaced corral boundaries along the length of the simulated ciliary membrane (Fig. 4 A), with period corresponding to the mean corral length, lcor (Table 2). The ends of the cilium were treated as reflective no-flux boundaries, and the corral boundaries were treated as semipermeable, with the probability of GPCR crossing upon encounter being Pbc. Pbc and the diffusion coefficient for the GPCR within the corral, Dcor, were varied, and model MSD(τ) was compared with GPCR tracking results (Fig. 4 C). Dcor and Pbc were determined from root mean square error (RMSE) minimization (Fig. 4 D). The mean values from seven independent experiments were Dcor = 5.46 ± 0.44 µm2/s and Pbc = 2.8 × 10−3 ± 0.4 × 10−3 (Table 2). Importantly, the Dcor was slightly larger than Dm1 estimated by the initial slope of the high-temporal-resolution MSD(τ) analysis (Fig. 3 C and Table 2), indicating that even at 200 fps, the diffusion coefficient of the GPCRs within the corral was underestimated by the initial slope of the MSD(τ) plot.
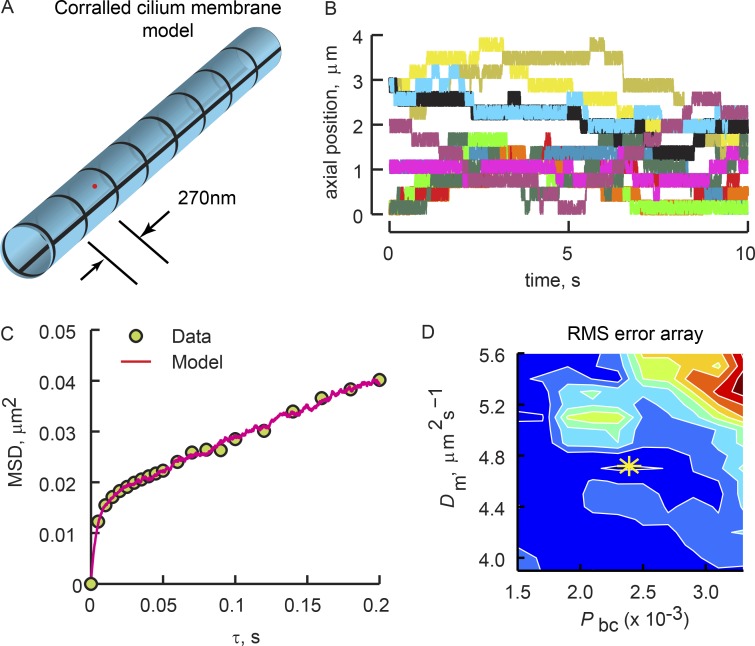
Figure 4.
Fitting of corralled diffusion model to the high-speed tracking results reveals low probability of corral boundary crossing. (A) Schematic of the corralled ciliary membrane simulation (see Theory). (B) Kymographs from 12 separate runs of the corralled diffusion simulation that yielded the best fit of the particle tracking results from the experiment shown in Fig. 3. Note the large variation in corral residence time, Tr, resulting from the stochastic corral boundary crossings. See Table 2, cilium R9 for fitting parameters. (C) MSD(τ) plots of the high-speed tracking experiment from Fig. 3 (symbols) and the simulation that provided the best fit (red line). (D) Array of the RMSE between the model and tracking data. White lines are iso-error lines. Colors are heat maps depicting relative regional error, where red is larger and dark blue is smaller error. The yellow asterisk shows the minimal error, found as the centroid of the smallest iso-error line, indicating the values of Dm and Pbc producing the best fit of the data.
Related to the probability of corral boundary crossing is the mean corral residence time, Tr. Tr was estimated from the simulations used to fit the high-temporal-resolution MSD(τ) plots by summing the time steps from corral entry, ten, to corral exit, tex, and averaging over all corral visits. Tr varied broadly, as expected for stochastic boundary crossing (Fig. 4 B). On average, Tr = 0.71 ± 0.09 s (Table 2).
2P Super FRAP confirms corralled diffusion found by Qdot tracking
The Qdot tracking approach, which involves labeling of the GPCRs with a 20-nm-diameter Qdot coupled via two antibodies (Fig. 1 A), raises the possibility that extraciliary structures, including the glycocalyx, or receptor oligomerization may impact the results. To address these potential caveats, we used a completely independent, high-resolution approach to examining GPCR dynamics.
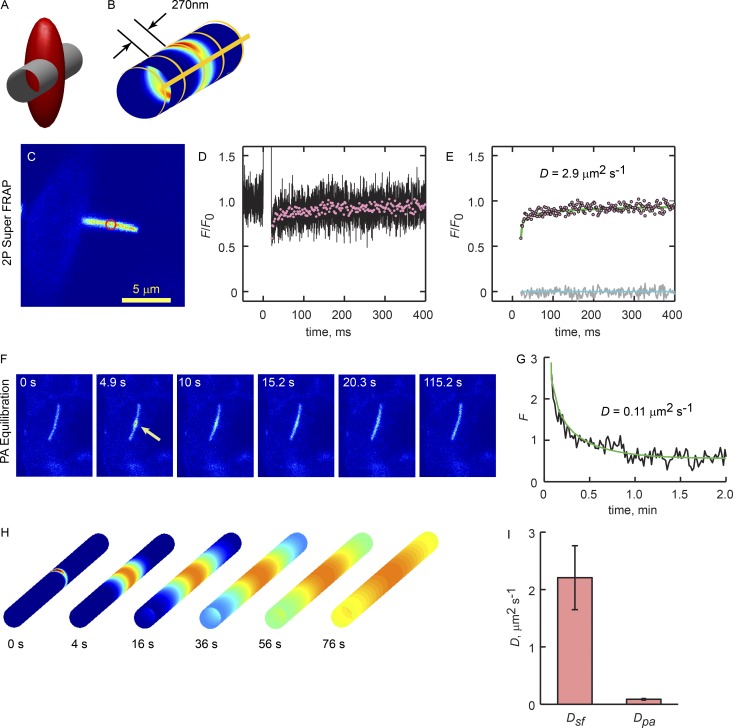
We noted that the mean corral width of ∼270 nm is on the order of the width of the 2P point spread function (psf) formed by focusing a titanium:sapphire (Ti:S) laser to the diffraction limit with a 1.2-NA water-immersion objective (Fig. 5, A and B; Calvert et al., 2007). Thus, exposure of EGFP-labeled GPCRs within the cilium to very brief, intense psf-shaped laser pulses would produce nonuniform, corral-sized bleaching patterns (Fig. 5 B). Thus, with sufficient sampling rates, ensemble GPCR movements within the corral domains could be directly monitored (see Materials and methods). The mean diffusion coefficient obtained with this approach, D Super FRAP or Dsf, was 1.86 ± 0.32 µm2/s (n = 7), in reasonable agreement with the fast phase from the single-molecule tracking results, Dm1 (Table 2).
Figure 5.
2P Super FRAP verifies Qdot tracking results. (A) Schematic showing the geometries of a 300-nm-diameter cilium membrane and the psf of the Ti:S laser focused to the diffraction limit by the 1.2-NA water-immersion objective. The red ellipsoid is an iso-intensity surface representing the psf at 1/e Imax (Imax being at the center of the ellipsoid). (B) Heat map of the relative psf intensity at the ciliary membrane, and thus the pattern of photoconverted GPCRs produced by a brief laser pulse. Yellow lines show the pattern of corral boundaries used in the ensemble diffusion model (see Theory). (C) Confocal image of an SSTR3-EGFP cilium used in a 2P Super FRAP experiment. Red circle indicates the bleach position. (D) Mean of 25 bleach-recovery curves acquired at 200 kHz (black line). A 10-ms bleach pulse began at time 0. Red symbols represent 10-fold downsampled recovery phase (decimation). (E) Fitted ensemble diffusion model (line) and error residuals (gray line). Lower green line is 0. Best fit was determined by varying Dm and finding the minimum RMSE, as described in Calvert et al. (2007), Materials and methods. (F) Time image series of SSTR3-PAGFP equilibration. PAGFP was photoconverted by a 10-ms exposure from the Ti:S laser tuned to 820 nm (Calvert et al., 2007). Equilibration was monitored with serial 1P confocal scans (488 nm, 1.31 s/frame) at a single z plane. (G) Time course of PAGFP relaxation from the photoconversion site (black trace). Smooth green line is the best fit of the ensemble diffusion model where Dm = 0.11 µm2/s. (H) Time series images of the ensemble diffusion model where the source at time 0 represents the distribution of photoconverted molecules after a brief 2P laser exposure. Circumferential corral boundaries were present in the model membrane every 270 nm along the axis, as shown in B. (I) Mean membrane diffusion coefficients obtained with 2P Super FRAP, Dsf, and long-term equilibration by PAGFP FRAPa, Dpa. Error bars are SEM. Mean diffusion coefficients were significantly different, P < 0.05, as determined from pairwise t test. 2P Super FRAP n = 7 cilia obtained from three independent experiments. PAGFP FRAPa n = 8 cilia obtained from five independent experiments.
We next examined the movement of ensembles of GPCRs on the ciliary membrane over longer distances and times. Here, we expressed SSTR3 labeled with the photoactivatable variant of GFP, PAGFP (Patterson and Lippincott-Schwartz, 2002) and the fluorescence recovery after photoactivation (FRAPa) approach we developed previously (Calvert et al., 2007). This high signal-to-noise FRAP approach allowed tracking of the GPCRs along the full length of the cilium, albeit at a much lower temporal resolution of 3 Hz. The time course of relaxation was fitted with the ensemble diffusion model with periodic semipermeable boundaries where the diffusion coefficients were reduced 13-fold (Fig. 5 I). The mean diffusion coefficient for SSTR3-PAGFP in this regimen was DPA = 0.11 ± 0.013 µm2/s (n = 8), again in reasonable agreement with the estimate from phase 2 of the Qdot tracking results, Dm2 (Table 2). Thus, two independent approaches resulted in remarkably similar results at both short and long times and distances.
Actin filament perturbations changed the permeability of corral boundaries
The results thus far are remarkably similar to those reported by Kusumi and colleagues (Kusumi et al., 2005; Ritchie et al., 2005; Suzuki et al., 2005; Hagiwara et al., 2011), who showed in a series of ground-breaking articles that lipids and the μ opioid receptor underwent hop diffusion between plasma membrane domains delimited by F-actin. Thus, we examined the possibility that the ciliary corrals were formed by actin filaments.
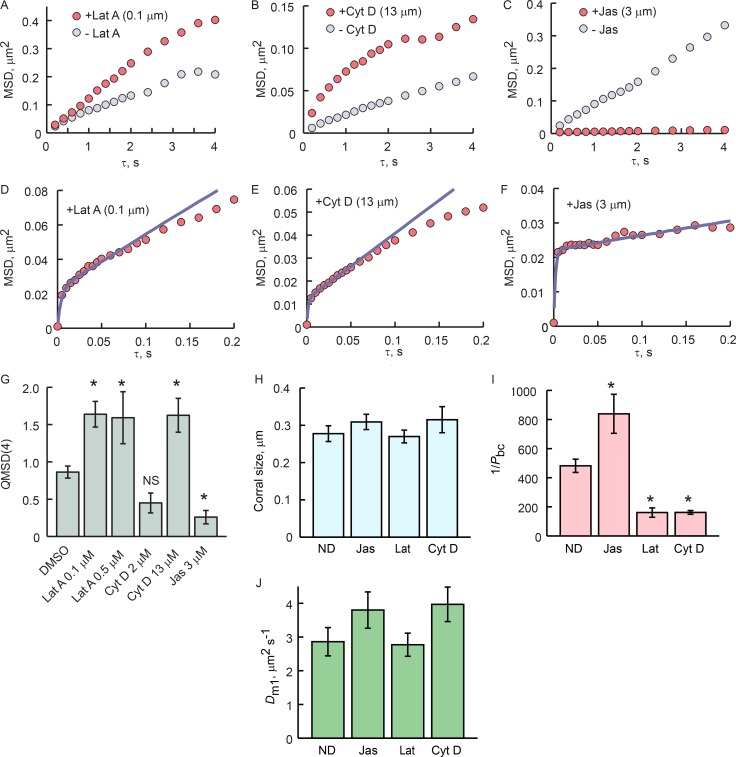
Treatment with 0.1 or 0.5 µM latrunculin A resulted in marked increases in the MSD(τ) slope, with MSD(τ = 4 s) significantly larger than in the pretreatment control in 7 of 11 trials (Fig. 6, A and G). Treatment with cytochalasin D gave a more nuanced result. 2 µM cytochalasin D on average had no effect (Fig. 6 G) where treatment with 13 µM resulted in significantly steeper MSD(τ) slope (Fig. 6, B and G). Treatment with 3 µM jasplakinolide invariably resulted in marked reduction in MSD(τ) slope and MSD(τ = 4 s) (Fig. 6, C and G). All treatments were compared with DMSO alone, which resulted in slight reduction in the MSD(τ) slope in two of six trials with no change in the remainder, for an overall mean of ∼10% reduction in mobility at MSD(τ = 4) (Fig. 6 G).
Figure 6.
Actin filament–disrupting drugs impact cilia membrane corral permeability. (A–C) MSD(τ) plots from 5 Hz GPCR-tracking experiments with and without F-actin–perturbing drugs. GPCRs were tracked for 5 min without drug, incubated with drug for 10 min, and tracked again for 5 min. Similar experiments were performed with DMSO alone. Quotients of MSD(τ = 4) before and after treatment (QMSD(4)) are shown in G. (D–F) 200-Hz tracking and fitting with corralled diffusion model with drug showed that F-actin perturbation altered the permeability of the corral boundaries but had no effect on the size of the corrals or Dm1. Note different scaling of ordinates. (H) Mean sizes of the corrals measured were not significantly different. (I) Inverse mean probability of boundary crossing, 1/Pbc. 1/Pbc is a measure of the number of times a GPCR encounters a boundary before it manages to cross. (J) The mean corral diffusion coefficients in each condition are not statistically different. (G–J) ND, no drug (vehicle control). Error bars are SEM. (G and I) Asterisks denote significant difference from DMSO at P < 0.05 (G) or P < 0.01 (I), as determined by ANOVA and Bonferroni post hoc analysis. In A–C and G, latrunculin A 0.1 µM, n = 7; five independent experiments; latrunculin A 0.5 µM, n = 11; six independent experiments; cytochalasin D 13 µM, n = 14, nine independent experiments; cytochalasin D 2 µM, n = 11; six independent experiments; DMSO controls, n = 6; two independent experiments. In D–F and H–J, ND, n = 18, seven independent experiments; jasplakinolide, n = 7; two independent experiments; latrunculin A n = 11, six independent experiments; cytochalasin D, n = 7, two independent experiments.
Fast tracking experiments showed that F-actin–perturbing drugs did not, however, change the corral size, as evident from the lack of change in the amplitude of the break between the two phases of the MSD(τ) plot (Fig. 6, D–F and H). Fitting of the MSD(τ) relations of the treated and nontreated cilia showed that the main impact of the drugs was to change the permeability of the corral boundaries; Dm1 was not changed (Fig. 6, I and J). Treatment of cilia with F-actin–perturbing drugs in FRAPa experiments resulted in significant changes in the effective diffusion coefficients (Fig. 7), thus independently confirming the tracking results.
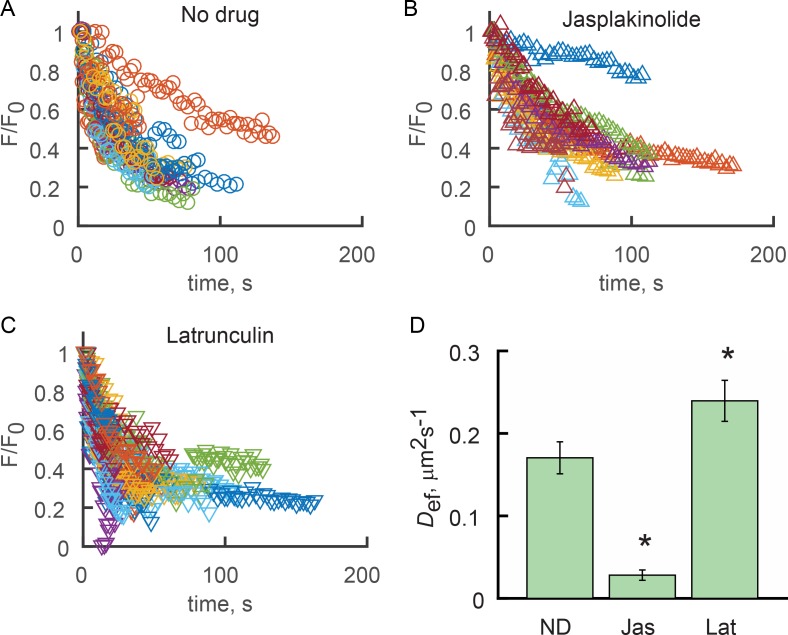
Figure 7.
F-actin–perturbing drugs alter the ensemble longitudinal diffusion of SSTR3 in the absence of Q-dot labeling. (A–C) 2P FRAPa experiments were performed as described in Fig. 5 (F and G), where SSTR3-PAGFP was photoconverted with 100-µs pulses of the Ti:S laser tuned to 850 nm. Groups of fluorescence relaxation curves obtained for cilia treated with vehicle (A), 3 µM jasplakinilode (B), or 0.1 µM latrunculin A (C) show that jasplakinolide treatment slows, whereas latrunculin treatment accelerates, relaxation. Each relaxation curve was fitted with the cilium surface ensemble diffusion model to obtain effective diffusion coefficients (Def). (D) Mean Def for each condition. ND, no drug (vehicle control). Error bars are SEM. Asterisks indicate significant differences from ND (P < 0.01) as determined by ANOVA with Bonferroni post hoc analysis. ND, n = 19 cilia; seven independent experiments; jasplakinolide, n = 17, three independent experiments; latrunculin A, n = 26; three independent experiments.
These results strongly indicate that ciliary membrane corrals are delimited, at least in part, by F-actin and that the dynamics of F-actin may regulate the permeability of the corral boundaries.
F-actin is present within IMCD3 primary cilia
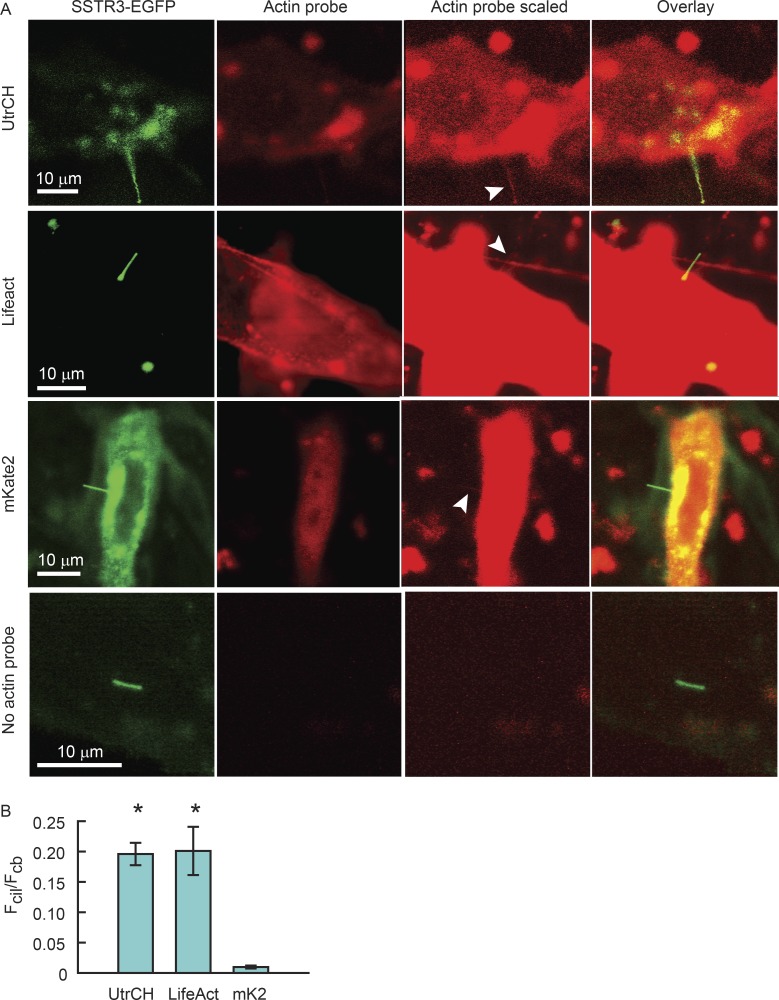
It is not clear if F-actin is present in all cilia. To date, F-actin has been shown to be present in rod photoreceptor-connecting cilia (Chaitin et al., 1984) and within primary cilia under specific conditions where cilia shed ectosomes or their tips (Nager et al., 2017; Phua et al., 2017). F-actin–associated proteins were found to be present in primary cilia by a cilium membrane–specific proteomics approach, and actin itself was not (Kohli et al., 2017), although it was not shown if this membrane-based assay could detect actin associated with other membranes where it is known to reside. Phalloidin did not label cilia on fixed MDCK cells (Francis et al., 2011). However, to our knowledge, there are no studies that directly examine the presence of F-actin in living primary cilia under more physiological conditions. To determine if F-actin was present in the IMCD3 cilia in our study, we individually expressed two F-actin markers, Lifeact-mCherry and RFP-UtrCH, simultaneously with SSTR3-EGFP (Fig. 8 A). Both F-actin probes were detected in the live cilia by confocal microscopy (Fig. 8 A, white arrowheads).
Figure 8.
F-actin is present within IMCD3 cilia at low levels. (A) IMCD3 cells were transfected with plasmids expressing SSTR3-EGFP and UtrCH-RFP, Lifeact-mCherry, or unfused mKate2, and ciliogenesis was initiated by serum starvation. Cells were imaged live by confocal microscopy using identical microscope settings, where green and red channels were not saturated (actin probe). To visualize actin probe signal within cilia, the red channel was scaled arbitrarily after acquisition (actin probe scaled). This scaling did not affect actual pixel counts. White arrowheads indicate red fluorescent signal within the cilia. All three species of red fluorescent proteins were found in cilia, including the unfused mKate2, showing that soluble fluorescent proteins expressed in the cell have access to the ciliary compartment. Actin probe panels displayed at optimal intensity scaling, showing that images were acquired without saturation. Actin probe scaled panels were intensity scaled to reveal cilium signal. (B) Mean ± SEM. Fcil/Fcb ratios. Asterisks indicate significant differences compared with unfused mKate2. UtrCH, n = 9; one independent experiment; Lifeact, n = 22; five independent experiments; unfused mKate2, n = 7; two independent experiments, no probe, n = 5; one independent experiment.
However, because both of these probes are soluble when not bound to F-actin, and thus may simply enter the cilia by diffusion, as we have previously shown in photoreceptors (Najafi et al., 2012b), we confirmed that the ciliary signal indeed reflected F-actin binding. We compared the ratio of fluorescence in the cilia, Fcil, and in the cell body, Fcb, of the Lifeact-mCherry or RFP-UtrCH probes to that of unfused mKate2, which we used as a soluble protein control. Although weak mKate2 signal was also detected in the cilia (Fig. 8 A), the Fcil/Fcb ratio for Lifeact-mCherry or RFP-UtrCH was 20-fold higher than for that of mKate2 (Fig. 8 B), indicating specific F-actin binding.
Discussion
Primary cilia membranes are partitioned into highly fluid corral domains
We have shown that two intrinsic membrane proteins, the GPCRs SSTR3 and Rhoi3S, move within cilium membranes more quickly on short time and distance scales than over longer scales, which fits the broad definition of subdiffusion (Bouchaud and Georges, 1990; Feder et al., 1996; Saxton and Jacobson, 1997). A number of mechanisms may underlie subdiffusion of membrane proteins, including diffusion in an environment crowded by mobile or immobile obstacles (Saxton, 1987; Saxton and Jacobson, 1997; Banks and Fradin, 2005; Cho et al., 2012; Metzler et al., 2014) and viscoelastic effects of interactions with polymers, such as the cytoskeletal components (Weber et al., 2010). These mechanisms manifest MSD(τ) plots that are well described by a power law with respect to time step, τα, where α < 1 (Bouchaud and Georges, 1990; Feder et al., 1996; Saxton and Jacobson, 1997).
Our results with high-speed imaging show a strongly biphasic MSD(τ) that is clearly distinct from power law subdiffusion. This result is similar to previous studies that have reported corralled diffusion of membrane molecules on the plasma membrane. Fujiwara et al. (2002) showed that lipid molecules were transiently confined to 230-nm-diameter membrane regions that were themselves confined to 750-nm-diameter regions within the greater plasma membrane of rat kidney fibroblasts. Suzuki et al. (2005) demonstrated that the μ opioid receptor underwent “hop diffusion” between membrane corrals delimited by actin filaments. Later, Oh et al. (2014) showed that the serine chemoreceptor was transiently confined to a membrane region of 170-nm diameter that resided within a membrane compartment of 290-nm diameter within the polar regions of the Escherichia coli inner membrane.
Treatment of IMCD3 cells with the F-actin–perturbing drugs latrunculin A, cytochalasin D, and jasplakinolide showed that F-actin plays a major role in GPCR corralling in primary cilia. Moreover, the sizes of the corrals we detected, ∼270 nm, were similar to the corrals on the plasma membrane reported by Kusumi and colleagues (Kusumi et al., 2005; Ritchie et al., 2005; Suzuki et al., 2005; Hagiwara et al., 2011). Together with vastly different estimations for the effective diffusion coefficients obtained with high- and low-temporal-resolution 2P FRAP of multiple GPCRs, our results demonstrate that the primary cilium membrane possesses a lattice, much like that of the plasma membrane, that transiently corrals membrane proteins undergoing diffusion and that F-actin plays a significant mechanistic role in the corral structure. Corralled hop diffusion is thus a major mode of GPCR transport within IMCD3 cell primary cilia.
Whether or not F-actin is present within primary cilia is somewhat controversial. Attempts to stain for F-actin in ciliated cultured epithelial cells over the course of decades have failed to definitively show actin within cilia, or even within the periciliary membrane domain (Finck and Holtzer, 1961; Francis et al., 2011). However, a study of crane fly sperm flagella showed F-actin–like structures that bind heavy meromyosin (Forer and Behnke, 1972), and recent cilium proteomics studies showed that actin and actin-associated proteins are found within primary cilia (Konno et al., 2015; Kohli et al., 2017). Recent studies show that primary cilia appear to use actin filaments to shed ectosomes from their distal tips (Nager et al., 2017; Phua et al., 2017). Moreover, it is well established that photoreceptor-connecting cilia contain F-actin, where it appears to play a role in the elaboration of the disc membranes that contain the phototransduction cascade proteins (Chaitin et al., 1984; Chaitin and Bok, 1986; Williams et al., 1988; Chaitin, 1989; Nemet et al., 2014). Our results with mCherry-Lifeact and Tag-RFP-UtrCH showed that F-actin is present in the IMCD3 cilia and support its role as a regulator of corral boundary permeability.
Factors other than F-actin may contribute to corralled GPCR diffusion within cilia
A significant difference between plasma membrane and cilium membrane hop diffusion is the mean residence time, Tr, of the GPCRs within the corrals. In the plasma membrane, Tr for the μ opioid receptor was reported to be a mean of 45 ms for corrals ~210 nm wide and ~1.7 s for 730-nm-wide corrals, in which the smaller corrals were nested (Suzuki et al., 2005). Our estimate of Tr = 0.7 s in ciliary membranes is between these values. It is thus possible that further increasing the temporal resolution of our tracking system will resolve more than one corral structure. Additionally, although the actin destabilizing treatments we used resulted in increases in GPCR mobility at long times and distances, they did not result in increases in mobility that approached the high mobility estimated for the corral domain. The F-actin–destabilizing treatments were rather mild, and higher concentrations of the disrupting chemicals caused general cell morphology changes. Nevertheless, the fact that some mobility constraint remained after F-actin disruption suggests that additional factors may be involved in delimiting the ciliary membrane corrals. Supporting this idea, application of F-actin–perturbing drugs during high-speed tracking experiments showed that their impact was on the permeability of the corral boundaries; the apparent size of the corrals were unchanged (Fig. 6). This result suggests that F-actin may “gate” the corrals, rather than set the absolute coral size.
Interestingly, septin 2, the molecule proposed to form the diffusion barrier at the base of primary cilia (Hu et al., 2010), as well as septins 7 and 9, have recently been shown to be expressed along most of the length of the primary cilium (Ghossoub et al., 2013). Perhaps the septins contribute to corral boundaries. Transient GPCR confinement may also result from formation of ordered, nonfluid lipid raft domains of various sizes interspersed with less ordered, fluid membrane regions. Primary cilia are enriched in gangliosides GM1 and GM3 as well as the cholesterol binding protein prominin 1, suggesting that lipid rafts form in ciliary membranes (Janich and Corbeil, 2007). The observed nonsaturating subdiffusion could arise if GPCRs were excluded from the ordered domains and diffused only in the fluid regions around them, as was recently proposed in a theoretical paper on 2D diffusion in a poly-dispersed medium (Cho et al., 2012). Lipid rafts thus may contribute to corral formation or produce another layer of restricted diffusion beyond that of the F-actin described here. Further study is required to fully understand the ciliary corral boundaries at various time and distance scales.
Functional significance of ciliary membrane corrals
Recent studies suggest that the primary cilium is divided into functional domains, perhaps to segregate signaling cascades, as evident from enrichment of subregions of the ciliary membrane with specific proteins. Inversin (Inv), a poly ankyrin repeat transcription factor involved in Wnt signaling, was shown to be localized to a region of the primary cilium distal to the transition zone and the ciliary necklace and proximal to the body and tip regions (Shiba et al., 2009). This localization pattern was the basis for defining an Inv domain in primary cilia where other proteins are also enriched, including Joubert syndrome–associated protein, ARL-13b/ARL-13, which were proposed to be localized to the Inv domain by a combination of active transport and leaky diffusion barriers (Cevik et al., 2013). These and similar results have led to the proposal that the primary cilium contains at least five distinct subdomains with different functions and different complements of enriched proteins (Blacque and Sanders, 2014). How these domains are established is not currently known; it would be of interest to see if F-actin–perturbing drugs disrupt these domains.
One of the remarkable features of the ciliary corrals is the fluidity of the corralled membrane domains. The mean diffusion coefficient of the GPCRs at short times and distances obtained with high-speed tracking, Dcor = 5.5 µm2/s, is among the largest diffusion coefficients reported for intrinsic membrane proteins. It is more than 10-fold larger than the diffusion coefficient for rhodopsin within photoreceptor disc membranes (Liebman and Entine, 1974; Poo and Cone, 1974; Wey et al., 1981; Gupta and Williams, 1990; Wang et al., 2008; Govardovskii et al., 2009; Najafi et al., 2012a). Similar membrane diffusion coefficients, averaging ∼4.2 µm2/s and ranging up to 30 µm2/s or more, were reported for the μ opioid receptor during high-speed single-molecule tracking in the plasma membranes of normal rat kidney fibroblast and Chinese hamster ovary cells (Suzuki et al., 2005). Given the large diffusion coefficient for the GPCRs in our study, the mean residence time of a GPCR within a corral and the mean corral size, we predicted that the corral membrane area fraction sampled by the GPCRs during a corral visit was ∼0.7 (Fig. S3). This shows that confinement of the GPCRs within corrals will dramatically increase the likelihood of a given GPCR to encounter other proteins, such as a G protein, within the same corral, and would reduce the probability of encountering one outside of the corral. Thus, the corrals may help to generate local signaling nanodomains and could conceivably play regulatory roles in ciliary signaling by limiting the number of interactions between cascade components on the time scale of signal transduction.
Materials and methods
Constructs and plasmids
Protein expression of all constructs was driven by the CMV promoter. The SSTR3-EGFP fusion construct was a gift from K. Mykytyn (Ohio State University, Columbus, OH). The SSTR3-mKate2 fusion protein was created by subcloning mKate2, a gift from E.L. Snapp (Albert Einstein College of Medicine, New York, NY), in place of EGFP in the SSTR3-EGFP construct. The bovine rhodopsin DNA construct with EGFP fused to rhodopsin with the last eight amino acids of rhodopsin appended to the C terminus of EGFP (Trivedi and Williams, 2010) was obtained from D.S. Williams (University of California, Los Angeles, School of Medicine, Los Angeles, CA). A rhodopsin chimera with third intracellular loop of SSTR3 (T236-R258) replacing the third intracellular loop of rhodopsin (E232-K248), Rho-i3S, was constructed using the FastCloning method (Li et al., 2011). Replacing the i3 loop on rhodopsin with the i3 loop pf SSTR3 was necessary to achieve Rho-EGFP enrichment in the IMCD3 cilia. For single-molecule tracking, the SSTR3-EGFP and Rho-i3S chimera were subcloned into PrecisionShuttle mammalian vector with N-terminal Myc tag (PS100012; OriGene). To mark the base of the primary cilia, GFP-xcent (a gift from J. Amack, SUNY Upstate Medical University, Syracuse, NY) and pTagBFP-C (a gift from A. Gross, University of Alabama, Birmingham, AL) was subcloned to make BFP-xcent construct. IFT20-GFP and IFT88-GFP constructs were gifts from G. Pazour (University of Massachusetts Medical School, Worcester, MA). pCS2.0 Tag-RFP-T-UtrCH was obtained from Addgene (plasmid 101279). mCherry-Lifeact-7 was obtained from Addgene (plasmid 54491).
Cell culture
IMCD3 cells were grown in DMEM/F12 culture medium with 10% FBS, penicillin/streptomycin, l-glutamine, sodium pyruvate, and nonessential amino acid solution at 37°C. Transfection was achieved with Lipofectamine 2000 (Life Technologies) or jetPRIME (Polypus Transfection). Before imaging, cells were seeded onto Fisherbrand No. 1, 25-mm cover glass (12-545-86), grown to confluence, and then starved by reducing FBS to 0.2% for 12–48 h to induce cilium elaboration.
Qdot tracking of GPCRs
Cells were incubated for 30 min in blocking buffer (DMEM/F12 1:1, with l-glutamine, Hepes, without Phenol Red, 1% BSA); incubated for 15 min in blocking buffer with 1:20,000 dilution of mouse anti–c-myc antibody (Covance); rinsed three times with blocking buffer; incubated for 10 min with 1:12,000 Qdot 625-nm goat F(ab′)2 anti-mouse IgG (A10195; Thermo Fisher); and washed three times. This protocol achieved sparse GPCR labeling. The bases of cilia were identified just before Qdot tracking by taking images of the cilium in the BFP (BFP-xcent) and EGFP (GPCR) channels. Cilia that lay flat relative to the imaging plane were selected for analysis. Tracking of Qdot 625 was performed at 37°C with an inverted epifluorescence microscope (Olympus IX70) using a 60× 1.45-NA Plan Apo objective. The ∼1-µm depth of focus means that cilia were within ∼10 degrees of the z-plane. Excitation was broadband in a window between 400 and 490 nm. The frame rate was set to 3.333 fps (image acquired every 0.3 s), 5 fps (image acquired every 0.2 s), or 200 fps (image acquired every 5 ms). The low frame rates allowed use of lower excitation intensities: excitation power was <3 mW, because of longer image integration times, thus minimizing photobleaching of the EGFP label that identified cilium position. This allowed tracking over tens of minutes and was an appropriate frame rate for assessing motor-driven movements or local binding events. High-frame-rate (200-fps) acquisition required at least 10-fold higher excitation intensities to offset the lower image integration times. This resulted in more rapid EGFP bleaching and thus prevented tracking for longer than ∼30 s. Images of the red (Qdot) and green (EGFP) channels were acquired simultaneously with an Optosplit III (89North) with a custom set of dichroic mirrors and barrier filters that were arranged to simultaneously project the red and green channels onto different regions of an EMCCD camera chip (iXon DU-897; Andor) running on Nikon Elements software.
MSD analysis
After tracking acquisition, the images were converted to tiff format, and the fluorescent wavelength channels that were recorded simultaneously on the EMCCD chip were separated into individual files using the Fiji Cairn Image Splitter plugin. The Qdot positions in each time series image were determined and linked together to form contiguous trajectories using a particle tracking code written in Matlab (Crocker and Grier, 1996). The Qdot positions were then layered onto the images from the EGFP channel, which shows the position of the cilium, and the displacements along the ciliary axis were determined using a custom Matlab code developed in our laboratory.
The MSD as a function of time steps τ was calculated according to
where τ is the time step interval for the position difference, x is the axial position of the Qdot or the simulated GPCR (in the case of the model), N = n − τ is the number of positional differences, and n is the total number of image time steps. This approach averages all pairs of points with a given time step interval τ (Lee et al., 1991; Zhang et al., 1991; Saxton and Jacobson, 1997).
Confocal and 2P microscopy
Live confocal imaging of mCherry-Lifeact and Tag-RFP-UtrCH was performed on an LSM510 using a Plan-Apochromat 63×/1.4-NA oil-immersion objective (Zeiss) at 30°C. All other confocal and 2P imaging was performed with a custom-built confocal/multiphoton microscope (Peet et al., 2004; Calvert et al., 2007, 2010) consisting of an argon-ion laser (model 163C; Newport Corp.) and a Ti:S laser (Mai Tai HP; Newport Corp.), each of which were focused to the diffraction limit with a 60×, 1.2-NA, water-immersion objective (Plan Apo VC; Nikon). Fluorescence emission was detected with photon-counting avalanche photodiodes (SPCM-AQR-14; PerkinElmer). Data acquisition and instrument control were implemented with computer acquisition and control boards and custom software written in LabView (National Instruments) in collaboration with M. Coleman (Coleman Technologies, Newtown Square, PA). Cells were maintained at 37°C. For FRAP analyses, the 3D coordinates for the photoconversion pulse were manually selected from initial 3D scans of the cells using the 488-nm line of the argon ion laser. 2P laser exposures were controlled by a high-voltage, fast Pockels cell (350-50; Conoptics) capable of modulating the beam power on the microsecond time scale.
2P Super FRAP
The 2P Super FRAP approach is a modification of one we previously described, the point-blast-point-scan FRAP method (Calvert et al., 2007), and is similar to early photobleaching approaches to membrane protein dynamics using rhodopsin optical density (Poo and Cone, 1973, 1974; Liebman and Entine, 1974) or fluorescence photobleaching recovery (Axelrod et al., 1976a,b), with the exception that our approach has subdiffraction-limit spatial resolution and very high temporal resolution, achieving 200-kHz acquisition rate.
Cilia expressing SSTR3-EGFP were scanned in 3D in confocal mode, and a point along the cilium length was selected in x,y,z for 2P Super FRAP (Fig. 5 C). Fluorescence was monitored at that point using the Ti:S laser tuned to 920 nm, ∼3 mW, with an acquisition rate of 200 kHz. A series of 25–50 bleach pulses of 0.1–10-ms duration, ∼30 mW, were applied at time 0, and the recorded traces were normalized to the prebleach fluorescence and averaged (Fig. 5 D, black trace). To reduce oversampling and noise, the data were decimated: i.e., smoothened by 10-window boxcar averaging and then 10-fold downsampled (Fig. 5, D and E, symbols). Minimal diffusion of SSTR3-EGFP occurred during the short bleach exposures, thus allowing fluorescence changes to be monitored as the psf intensity profile bleach pattern relaxed into an approximately homogeneous distribution. The recovery time course was then fitted with the numerical ensemble diffusion model (Fig. 5, E and H; and Theory).
FRAPa of PAGFP-labeled GPCRs
FRAPa was implemented using the point-blast region of interest scan method (Najafi et al., 2012a). PAGFP-labeled SSTR3 was found in cilia by 3D confocal scanning using the 488-nm line of an argon-ion laser. A position along the cilium was selected in 3D where SSTR3-PAGFP was photoconverted from the minimally fluorescent state to the fluorescent state by 0.1–10-ms, 10-mW mean power pulses from the Ti:S laser tuned to 820 nm. The equilibration of the activated molecules was then monitored with serial x–y confocal scans using 488-nm excitation. Rapid focus corrections were made before and after the photoconversion pulse to account for the measured focus difference between 820- and 488-nm illumination caused by objective chromatic aberration (Calvert et al., 2007).
Raw images were processed using custom Matlab (MathWorks) routines to correct for slight field inhomogeneities inherent to the optical system and for nonlinearities in the photon detectors as follows. Full-field scans of solutions of fluorescein were averaged to generate image field-flattening maps. Spatial photon counts in the maps were normalized to the maximum counts, and acquired images were multiplied by the maps (Calvert et al., 2007, 2010; Najafi et al., 2012a). No other image processing was done.
Ensemble diffusion model solutions were computed for various values of D on an appropriate time grid and convolved in 3D with the measured multiphoton psf of the Ti:S laser (2P Super FRAP experiments) or the measured psf of the argon-ion (488-nm) laser (PAGFP FRAPa experiments) to predict the measured fluorescence relaxation. The RMSEs between data and theory were calculated as follows: nine data samples centered on the time at which each theory point was computed were averaged, and the difference between theory and the averaged data point was computed, squared, summed, and divided by the total number of theory points. The optimal fitted trace was found as that which minimized the RMSE (Calvert et al., 2007).
For 2P Super FRAP and PAGFP FRAPa experiments, only SSTR3-EGFP– or SSTR3-PAGFP–expressing cells were analyzed because they more efficiently enrich in the cilia, leaving virtually no signal in the apical membrane and other cellular structures (Geneva et al., 2017).
Determination of the psf intensity profile and the photoconversion pattern at the cilium surface
Direct measurement of the psf profile in our confocal microscope was performed by scanning 0.1-µm-diameter fluorescent microspheres. Fluorescence profiles of the microspheres were fitted with Gaussians, yielding standard deviations (σ) in xy of σxy = 0.14 µm and in z of σz = 0.56 µm. To estimate the EGFP or PAGFP photoconversion profile on the cilium membrane (Fig. 5), the psf intensity profile was approximated as a 3D Gaussian:
where a is the intensity maximum at the 3D center of the profile. For this analysis, we used a normalized psf profile where
The estimate of the profile on the cilium was then obtained from the interpolation of the psf at the surface of a right circular cylinder with diameter of 300 nm whose axis was perpendicular to the z-axis of the psf.
Statistical analysis of significance
Results are presented as mean ± SEM. When pairwise comparisons were made, significance was tested with appropriate t tests. When multiple means were compared, a one-way analysis of variance was performed with α = 0.05, where the null hypothesis was that all mean values were equal. Upon rejection of the null hypothesis with P < 0.05, post hoc comparisons among the mean values were performed by two-sample t test. To account for the variance among the multiple pairs of mean values compared, the Bonferroni correction was applied to α such that αb = α/m, where αb is the corrected α and m is the number of comparisons being made among the means.
Theory
We used two high-spatiotemporal-resolution approaches to explore the dynamics of intrinsic membrane proteins on live primary cilia, single-particle tracking and 2P Super FRAP. In this section, we describe the models of protein transport we used to quantitatively evaluate the results and differentiate between potential modes of transport.
Coordinate system
The primary cilium membrane is essentially the surface of a right circular cylinder and thus is conveniently described in cylindrical coordinates. The spatial coordinate system is defined as where r is the radial distance, z is the axial distance, and θ is the angle about the axis. Owing to the fact that GPCRs are intrinsic membrane proteins and thus confined to the ciliary membrane at r = rm, the radius of the ciliary membrane, modeling of GPCR transport is reduced to two spatial dimensions where z and θ are the only spatial variables.
Random walk simulation
The simulation was based on a 2D, continuous random walk on the surface of a right circular cylinder and implemented in Matlab or the C programming languages (Fig. 2 A). A point on the cylindrical surface, p(rm,θ,z), was randomly chosen as the initial position of a simulated GPCR, and the direction of movement away from the initial and all subsequent positions was determined by a randomly selected angle, φ. The simulated GPCR was then moved a distance d along the φ trajectory on the cylinder surface as determined by d = vts, where ts is the magnitude of the time step and v is the velocity of the GPCR. v was defined by the estimate of the membrane diffusion coefficient Dm1 as:
The cylinder ends, corresponding to the base and tip of the cilium, were treated as reflective, no-flux boundaries. Axial positions of the simulated GPCR (Fig. 2 B) were thus equivalent to the axial kymographs of the GPCR tracking shown in Fig. 1.
To examine the impact of transient motor-driven transport, confinement or local binding on the predicted MSD(τ) relation, stochastic coupling to motors, binding to immobile objects, or transient confinement to axial subdomains were incorporated into the random walk simulation. To accomplish the addition of a motor-driven component, coupling to an anterograde or retrograde motor occurred stochastically with a probability Pm applied at each time step that was arrived at by free diffusion. Once on a motor, the simulated GPCR moved axially a distance dm = ±vm × tm, where vm is the motor displacement velocity. The simulated GPCR moved in a + or – direction for the number of sequential time steps defined by tm, the motor coupling time.
The impact of transient local binding was examined by implementing stochastic coupling of the GPCRs to immobile objects. Here, a freely diffusing molecule executing the random walk bound to an immobile object at arbitrary position within the cilium membrane with a probability Pb. Once bound, the molecule remained stationary for a duration tb after which it resumed the random walk until it again became bound.
We examined the impact of transient corralled diffusion by generating a theoretical ciliary membrane onto which we imposed corral boundaries at arbitrary spacing and with specified permeability to the GPCRs. The boundaries were approximately square, where sides parallel to the axis of the cylinder had a length of L1, and sides perpendicular to the axis had a length of L2 = C/Round(C/L1). Simulated GPCRs executed random walks within the corral domain as described above. When the path of the GPCR movement crossed a boundary, a random decision was made as to whether the GPCR was reflected back into the same corral in which it was diffusing or crossed the boundary and entered the neighboring corral. The boundary reflection versus crossing decision was governed by a probability of corral boundary crossing, Pbc. Matlab code for the random walk simulation is provided in the online supplemental material.
Ensemble diffusion model
The model of ensemble diffusion on the cilium surface was based on molecular diffusion models we previously published (Calvert et al., 2007, 2010; Najafi et al., 2012a), where we described diffusion in 3D in a cylinder. In the present work, diffusion is limited to two spatial dimensions at the cilium surface, and reduced flux corral boundaries were introduced. The general diffusion equation may be written as
| (2) |
where c is the concentration of diffusing substance, t is time, D is the diffusion coefficient, and is the Laplace operator. Eq. 2 is a parabolic partial differential equation that may be applied in any coordinate system. The general diffusion equation in cylindrical coordinates may be written
| (3) |
where Dr, Dθ, and Dz are the diffusion coefficients in the indicated directions, and Qs is a source or sink of the diffusing substance. Because the GPCRs are intrinsic membrane proteins, and thus are confined to the ciliary membrane, Dr = 0 and the GPCRs diffuse only in z and θ at r = rm, the radius of the cylindrical cilium at the membrane. Eq. 3 thus reduces to
| (4) |
Eq. 4 is first order in time and second order in space, requiring one initial condition (IC) and two boundary conditions (BC) in each spatial variable. The IC is
| (5) |
The BCs in θ are
| (6) |
which reflect the symmetry in the source term and assume isotropic diffusion in θ and z. The BCs in z are
| (7) |
which specify no-flux, Neumann boundary conditions at the base and tip of the cilium. The assumption of no-flux at the cilium base appears appropriate on the time frame of the experiments described in light of the presence of a significant diffusional impediment for membrane proteins between the ciliary membrane and the larger apical membrane (Geneva and Calvert, 2010; Hu et al., 2010).
Solutions of Eqs. 4–7 were found using the numerical method of lines, where the partial differential equations specified in Eq. 4 were replaced with ordinary differential equations (ODEs), finite differences approximations of the spatial derivatives (Schiesser, 1991; Schiesser and Griffiths, 2009). For the general grid point p(i,j), where i and j are indices for θ and z, respectively, the spatial derivative approximations were
| (8) |
and
| (9) |
where Δθ and Δz are the spatial increments in θ and z. Note that in the present treatment, the inherent singularities at r = 0 were avoided because of the absence of radial diffusion and because rm ≠ r0. For the complete numerical solution, which includes radial diffusion and the regularization of those singularities, see Calvert et al. (2010).
Approximations of the grid points neighboring the no-flux boundaries at the base and tip of the cilium were
| (10) |
at the cylinder base, where the flux between (i,j – 1), a point that lies outside of the base of the cilium, and (i,j) is taken to be 0, and
| (11) |
at the cylinder tip, where the flux between (i,j + 1), a point that lies outside of the tip of the cilium, and (i,j) is taken to be 0. The inhomogeneous source term was
| (12) |
where is the effective 2P cross section of the fluorophore, γ* is the quantum efficiency of photoconversion, and I(i,j,r = rm) is the photon flux density at the cilium membrane. Substituting Eqs. 8–12 into Eq. 4 gives a system of nθ × nz ODEs in t. The ODEs were programmed in Matlab and integrated forward in t from t = 0 using the Matlab ODE integrators ode15s or ode45.
The model described above is appropriate for diffusion on the ciliary membrane in the absence of corrals. To assess the impact of corrals on ensemble diffusion, we mapped boundaries onto the cylindrical grid that acted to reduce the local flux of theoretical GPCRs. We mapped approximately square corral boundaries onto the cylinder surface where sides parallel to the axis of the cylinder had a length of L1, and sides perpendicular to the axis had a length of L2 = C/Round(C/L1). Transport across the boundaries was diffusive, with diffusion coefficient Dcb distinct from the bulk membrane diffusion constant Dm. Solutions to the diffusion equations with local change in diffusivity at the boundaries were found numerically, as follows. The flux at the grid point neighboring a corral boundary in the positive z direction was approximated as
and at the grid point neighboring the corral boundary in the negative z direction was approximated as
The flux at the grid point neighboring the corral boundary in the positive θ direction was approximated as
| (13) |
and the flux at the grid point neighboring the corral boundary in the negative θ direction was approximated as
| (14) |
Flux between grid points within the corral boundaries was found by solving Eqs. 4, 13, and 14 as described above, where Dz and Dθ were replaced with Dcb. Matlab code for the enesmble diffusion model is provided in the online supplemental material.
Online supplemental material
Fig. S1 shows positional precision of the tracking system. In Fig. S2, axial kymographs of GPCR tracking results reveal a mixture of Brownian motion, apparent processivity, and apparent binding to immobile structures. Fig. S3 shows impact of corrals on sampling of the ciliary membranes by GPCRs. Video 1 is a high-magnification video of the movement of Rhoi3S labeled with a Qdot625. Video 2 is the same as Video 1 with tracking overlaid. Video 3 is a low-magnification video of SSTR3 labeled with Qdot625. Video 4 is a low-magnification video of Rhoi3S labeled with Qdot625. Video 5 shows tracking of IFT20-GFP. Video 6 shows output of a random walk simulation. Video 7 shows output of a random walk simulation with stochastic motor coupling. Video 8 shows output of a random walk simulation with stochastic binding to immobile objects. Video 9 shows output of a random walk simulation with semipermeable corrals. Matlab code for the random walk and ensemble diffusion models are also provided as a ZIP file.
Supplementary Material
Acknowledgments
We thank members of the Center for Vision Research and Mira Krendel for helpful discussions and Zach Fechtner for technical assistance.
This work was supported by National Institutes of Health grant R01EY018421 (P.D. Calvert), a Postdoctoral Award from Fight for Sight (H.Y. Tan), and an unrestricted grant to the State University of New York Upstate Department of Ophthalmology from Research to Prevent Blindness. We are grateful for support from Lions Clubs of Central New York, District 20-Y1.
The authors declare no competing financial interests.
Author contributions: Conceptualization: P.D. Calvert; methodology: S. Lee, H.Y. Tan, P.D. Calvert; software: S. Lee, H.Y. Tan, A. Kruglov, P.D. Calvert; validation: S. Lee, H.Y. Tan, P.D. Calvert; formal analysis: S. Lee, H.Y. Tan, I.I. Geneva, A. Kruglov, P.D. Calvert; investigation: S. Lee, H.Y. Tan, I.I. Geneva, P.D. Calvert; resources: P.D. Calvert; data curation: P.D. Calvert; writing-original draft: S. Lee, P.D. Calvert; writing – review and editing, S. Lee, P.D. Calvert; visualization: S. Lee, P.D. Calvert; supervision: P.D. Calvert; project administration: P.D. Calvert; funding acquisition: P.D. Calvert.
References
- Axelrod D., Koppel D.E., Schlessinger J., Elson E., and Webb W.W.. 1976a Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. 10.1016/S0006-3495(76)85755-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D., Ravdin P., Koppel D.E., Schlessinger J., Webb W.W., Elson E.L., and Podleski T.R.. 1976b Lateral motion of fluorescently labeled acetylcholine receptors in membranes of developing muscle fibers. Proc. Natl. Acad. Sci. USA. 73:4594–4598. 10.1073/pnas.73.12.4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks D.S., and Fradin C.. 2005. Anomalous diffusion of proteins due to molecular crowding. Biophys. J. 89:2960–2971. 10.1529/biophysj.104.051078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., and Sanders A.A.W.M.. 2014. Compartments within a compartment: What C. elegans can tell us about ciliary subdomain composition, biogenesis, function, and disease. Organogenesis. 10:126–137. 10.4161/org.28830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchaud J.-P., and Georges A.. 1990. Anomalous diffusion in disordered media: Statistical mechanisms, models and physical applications. Phys. Rep. 195:127–293. 10.1016/0370-1573(90)90099-N [DOI] [Google Scholar]
- Broekhuis J.R., Verhey K.J., and Jansen G.. 2014. Regulation of cilium length and intraflagellar transport by the RCK-kinases ICK and MOK in renal epithelial cells. PLoS One. 9:e108470 10.1371/journal.pone.0108470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert P.D., Peet J.A., Bragin A., Schiesser W.E., and Pugh E.N. Jr. 2007. Fluorescence relaxation in 3D from diffraction-limited sources of PAGFP or sinks of EGFP created by multiphoton photoconversion. J. Microsc. 225:49–71. 10.1111/j.1365-2818.2007.01715.x [DOI] [PubMed] [Google Scholar]
- Calvert P.D., Schiesser W.E., and Pugh E.N. Jr. 2010. Diffusion of a soluble protein, photoactivatable GFP, through a sensory cilium. J. Gen. Physiol. 135:173–196. 10.1085/jgp.200910322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S., Sanders A.A.W.M., Van Wijk E., Boldt K., Clarke L., van Reeuwijk J., Hori Y., Horn N., Hetterschijt L., Wdowicz A., et al. 2013. Active transport and diffusion barriers restrict Joubert Syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet. 9:e1003977 10.1371/journal.pgen.1003977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitin M.H. 1989. Immunogold localization of actin and opsin in RDS mouse photoreceptors. Prog. Clin. Biol. Res. 314:265–274. [PubMed] [Google Scholar]
- Chaitin M.H., and Bok D.. 1986. Immunoferritin localization of actin in retinal photoreceptors. Invest. Ophthalmol. Vis. Sci. 27:1764–1767. [PubMed] [Google Scholar]
- Chaitin M.H., Schneider B.G., Hall M.O., and Papermaster D.S.. 1984. Actin in the photoreceptor connecting cilium: Immunocytochemical localization to the site of outer segment disk formation. J. Cell Biol. 99:239–247. 10.1083/jcb.99.1.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.W., Kwon G., Sung B.J., and Yethiraj A.. 2012. Effect of polydispersity on diffusion in random obstacle matrices. Phys. Rev. Lett. 109:155901 10.1103/PhysRevLett.109.155901 [DOI] [PubMed] [Google Scholar]
- Crocker J.C., and Grier D.G.. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310. 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Deretic D., and Wang J.. 2012. Molecular assemblies that control rhodopsin transport to the cilia. Vision Res. 75:5–10. 10.1016/j.visres.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder T.J., Brust-Mascher I., Slattery J.P., Baird B., and Webb W.W.. 1996. Constrained diffusion or immobile fraction on cell surfaces: A new interpretation. Biophys. J. 70:2767–2773. 10.1016/S0006-3495(96)79846-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck H., and Holtzer H.. 1961. Attempts to detect myosin and actin in cilia and flagella. Exp. Cell Res. 23:251–257. 10.1016/0014-4827(61)90035-0 [DOI] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., and Pazour G.J.. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792. 10.1091/mbc.e06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forer A., and Behnke O.. 1972. An actin-like component in sperm tails of a crane fly (Nephrotoma suturalis Loew). J. Cell Sci. 11:491–519. [DOI] [PubMed] [Google Scholar]
- Francis S.S., Sfakianos J., Lo B., and Mellman I.. 2011. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J. Cell Biol. 193:219–233. 10.1083/jcb.201009001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., and Kusumi A.. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. 10.1083/jcb.200202050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., and Reiter J.F.. 2012. Scoring a backstage pass: Mechanisms of ciliogenesis and ciliary access. J. Cell Biol. 197:697–709. 10.1083/jcb.201111146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneva I.I., and Calvert P.D.. 2010. High-resolution FRAP of the cilium-localized somatostatin receptor 3 reveals the presence of a lateral diffusion barrier at the cilium base. Biophys. J. 98:376a 10.1016/j.bpj.2009.12.2029 [DOI] [Google Scholar]
- Geneva I.I., Tan H.Y., and Calvert P.D.. 2017. Untangling ciliary access and enrichment of two rhodopsin-like receptors using quantitative fluorescence microscopy reveals cell-specific sorting pathways. Mol. Biol. Cell. 28:554–566. 10.1091/mbc.e16-07-0549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J.M., Davis E.E., and Katsanis N.. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 137:32–45. 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R., Hu Q., Failler M., Rouyez M.C., Spitzbarth B., Mostowy S., Wolfrum U., Saunier S., Cossart P., Jamesnelson W., and Benmerah A.. 2013. Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. J. Cell Sci. 126:2583–2594. 10.1242/jcs.111377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.F.X., Moritz O.L., and Williams D.S.. 2016. Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 55:52–81. 10.1016/j.preteyeres.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govardovskii V.I., Korenyak D.A., Shukolyukov S.A., and Zueva L.V.. 2009. Lateral diffusion of rhodopsin in photoreceptor membrane: A reappraisal. Mol. Vis. 15:1717–1729. [PMC free article] [PubMed] [Google Scholar]
- Gupta B.D., and Williams T.P.. 1990. Lateral diffusion of visual pigments in toad (Bufo marinus) rods and in catfish (Ictalurus punctatus) cones. J. Physiol. 430:483–496. 10.1113/jphysiol.1990.sp018303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A., Tanaka Y., Hikawa R., Morone N., Kusumi A., Kimura H., and Kinoshita M.. 2011. Submembranous septins as relatively stable components of actin-based membrane skeleton. Cytoskeleton (Hoboken). 68:512–525. 10.1002/cm.20528 [DOI] [PubMed] [Google Scholar]
- Händel M., Schulz S., Stanarius A., Schreff M., Erdtmann-Vourliotis M., Schmidt H., Wolf G., and Höllt V.. 1999. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 89:909–926. 10.1016/S0306-4522(98)00354-6 [DOI] [PubMed] [Google Scholar]
- Hu Q., Milenkovic L., Jin H., Scott M.P., Nachury M.V., Spiliotis E.T., and Nelson W.J.. 2010. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 329:436–439. 10.1126/science.1191054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P., and Corbeil D.. 2007. GM1 and GM3 gangliosides highlight distinct lipid microdomains within the apical domain of epithelial cells. FEBS Lett. 581:1783–1787. 10.1016/j.febslet.2007.03.065 [DOI] [PubMed] [Google Scholar]
- Kohli P., Höhne M., Jüngst C., Bertsch S., Ebert L.K., Schauss A.C., Benzing T., Rinschen M.M., and Schermer B.. 2017. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep. 18:1521–1535. 10.15252/embr.201643846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno A., Shiba K., Cai C., and Inaba K.. 2015. Branchial cilia and sperm flagella recruit distinct axonemal components. PLoS One. 10:e0126005 10.1371/journal.pone.0126005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Sako Y., and Yamamoto M.. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65:2021–2040. 10.1016/S0006-3495(93)81253-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., and Fujiwara T.. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Tsunoyama T.A., Hirosawa K.M., Kasai R.S., and Fujiwara T.K.. 2014. Tracking single molecules at work in living cells. Nat. Chem. Biol. 10:524–532. 10.1038/nchembio.1558 [DOI] [PubMed] [Google Scholar]
- Lee G.M., Ishihara A., and Jacobson K.A.. 1991. Direct observation of Brownian motion of lipids in a membrane. Proc. Natl. Acad. Sci. USA. 88:6274–6278. 10.1073/pnas.88.14.6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wen A., Shen B., Lu J., Huang Y., and Chang Y.. 2011. FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11:92 10.1186/1472-6750-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman P.A., and Entine G.. 1974. Lateral diffusion of visual pigment in photorecptor disk membranes. Science. 185:457–459. 10.1126/science.185.4149.457 [DOI] [PubMed] [Google Scholar]
- Metzler R., Jeon J.-H., Cherstvy A.G., and Barkai E.. 2014. Anomalous diffusion models and their properties: Non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys. Chem. Chem. Phys. 16:24128–24164. 10.1039/C4CP03465A [DOI] [PubMed] [Google Scholar]
- Milenkovic L., Weiss L.E., Yoon J., Roth T.L., Su Y.S., Sahl S.J., Scott M.P., and Moerner W.E.. 2015. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc. Natl. Acad. Sci. USA. 112:8320–8325. 10.1073/pnas.1510094112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., and Rohatgi R.. 2014. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin. Cell Dev. Biol. 33:63–72. 10.1016/j.semcdb.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager A.R., Goldstein J.S., Herranz-Perez V., Portran D., Ye F., Garcia-Verdugo J.M., and Nachury M.V.. 2017. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 168:252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi M., Haeri M., Knox B.E., Schiesser W.E., and Calvert P.D.. 2012a Impact of signaling microcompartment geometry on GPCR dynamics in live retinal photoreceptors. J. Gen. Physiol. 140:249–266. 10.1085/jgp.201210818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi M., Maza N.A., and Calvert P.D.. 2012b Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc. Natl. Acad. Sci. USA. 109:203–208. 10.1073/pnas.1115109109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I., Tian G., and Imanishi Y.. 2014. Submembrane assembly and renewal of rod photoreceptor cGMP-gated channel: Insight into the actin-dependent process of outer segment morphogenesis. J. Neurosci. 34:8164–8174. 10.1523/JNEUROSCI.1282-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D., Yu Y., Lee H., Wanner B.L., and Ritchie K.. 2014. Dynamics of the serine chemoreceptor in the Escherichia coli inner membrane: A high-speed single-molecule tracking study. Biophys. J. 106:145–153. 10.1016/j.bpj.2013.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. 2006. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75:743–767. 10.1146/annurev.biochem.75.103004.142743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.H., and Lippincott-Schwartz J.. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., and Witman G.B.. 2003. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15:105–110. 10.1016/S0955-0674(02)00012-1 [DOI] [PubMed] [Google Scholar]
- Pearring J.N., Salinas R.Y., Baker S.A., and Arshavsky V.Y.. 2013. Protein sorting, targeting and trafficking in photoreceptor cells. Prog. Retin. Eye Res. 36:24–51. 10.1016/j.preteyeres.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet J.A., Bragin A., Calvert P.D., Nikonov S.S., Mani S., Zhao X., Besharse J.C., Pierce E.A., Knox B.E., and Pugh E.N. Jr. 2004. Quantification of the cytoplasmic spaces of living cells with EGFP reveals arrestin-EGFP to be in disequilibrium in dark adapted rod photoreceptors. J. Cell Sci. 117:3049–3059. 10.1242/jcs.01167 [DOI] [PubMed] [Google Scholar]
- Phua S.C., Chiba S., Suzuki M., Su E., Roberson E.C., Pusapati G.V., Setou M., Rohatgi R., Reiter J.F., Ikegami K., and Inoue T.. 2017. Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell. 168:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo M., and Cone R.A.. 1973. Lateral diffusion of rhodopsin in the visual receptor membrane. J. Supramol. Struct. 1:354 10.1002/jss.400010411 [DOI] [PubMed] [Google Scholar]
- Poo M., and Cone R.A.. 1974. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 247:438–441. 10.1038/247438a0 [DOI] [PubMed] [Google Scholar]
- Pugh E.N. Jr., and Lamb T.D.. 2000. Phototransduction in Vertebrate Rods and Cones: Molecular Mechanisms of Amplification, Recovery and Light Adaptation. In Handbook of Biological Physics, Vol. 3. Stavenga D.G., de Grip W.J., and Pugh E.N. Jr., editors. New York: Elsevier Science; pp. 183–255. [Google Scholar]
- Quemeneur F., Sigurdsson J.K., Renner M., Atzberger P.J., Bassereau P., and Lacoste D.. 2014. Shape matters in protein mobility within membranes. Proc. Natl. Acad. Sci. USA. 111:5083–5087. 10.1073/pnas.1321054111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K., Shan X.Y., Kondo J., Iwasawa K., Fujiwara T., and Kusumi A.. 2005. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 88:2266–2277. 10.1529/biophysj.104.054106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M.J. 1987. Lateral diffusion in an archipelago. The effect of mobile obstacles. Biophys. J. 52:989–997. 10.1016/S0006-3495(87)83291-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M.J. 1995. Single-particle tracking: Effects of corrals. Biophys. J. 69:389–398. 10.1016/S0006-3495(95)79911-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M.J., and Jacobson K.. 1997. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26:373–399. 10.1146/annurev.biophys.26.1.373 [DOI] [PubMed] [Google Scholar]
- Schiesser W.E. 1991. The Numerical Method of Lines: Integration of Partial Differential Equations. Cambridge, MA: Academic Press. [Google Scholar]
- Schiesser W.E., and Griffiths G.W.. 2009. A compendium of partial differential equation models: Method of lines analysis with MATLAB. New York: Cambridge University Press; 10.1017/CBO9780511576270 [DOI] [Google Scholar]
- Shiba D., Yamaoka Y., Hagiwara H., Takamatsu T., Hamada H., and Yokoyama T.. 2009. Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J. Cell Sci. 122:44–54. 10.1242/jcs.037408 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ritchie K., Kajikawa E., Fujiwara T., and Kusumi A.. 2005. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88:3659–3680. 10.1529/biophysj.104.048538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi D., and Williams D.S.. 2010. Ciliary transport of opsin. Adv. Exp. Med. Biol. 664:185–191. 10.1007/978-1-4419-1399-9_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi D., Colin E., Louie C.M., and Williams D.S.. 2012. Live-cell imaging evidence for the ciliary transport of rod photoreceptor opsin by heterotrimeric kinesin-2. J. Neurosci. 32:10587–10593. 10.1523/JNEUROSCI.0015-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang X., Zhang L., He F., Zhang G., Jamrich M., and Wensel T.G.. 2008. Activation-dependent hindrance of photoreceptor G protein diffusion by lipid microdomains. J. Biol. Chem. 283:30015–30024. 10.1074/jbc.M803953200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., Spakowitz A.J., and Theriot J.A.. 2010. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys. Rev. Lett. 104:238102 10.1103/PhysRevLett.104.238102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M., Elsner M., Kartberg F., and Nilsson T.. 2004. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 87:3518–3524. 10.1529/biophysj.104.044263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey C.L., Cone R.A., and Edidin M.A.. 1981. Lateral diffusion of rhodopsin in photoreceptor cells measured by fluorescence photobleaching and recovery. Biophys. J. 33:225–232. 10.1016/S0006-3495(81)84883-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.S., Linberg K.A., Vaughan D.K., Fariss R.N., and Fisher S.K.. 1988. Disruption of microfilament organization and deregulation of disk membrane morphogenesis by cytochalasin D in rod and cone photoreceptors. J. Comp. Neurol. 272:161–176. 10.1002/cne.902720202 [DOI] [PubMed] [Google Scholar]
- Yang T., Tran M.N.T., Chong W.M., Huang C.-E., and Liao J.-C.. 2017. Intraflagellar transport proteins undergo nonaxonemal staged hindrance between the recruiting distal appendages and the cilium. bioRxiv. (Preprint posted on December 1, 2017). 10.1101/227587 [DOI] [Google Scholar]
- Ye F., Breslow D.K., Koslover E.F., Spakowitz A.J., Nelson W.J., and Nachury M.V.. 2013. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. eLife. 2:e00654 10.7554/eLife.00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Nager A.R., and Nachury M.V.. 2018. BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217:1847–1868. 10.1083/jcb.201709041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Crise B., Su B., Hou Y., Rose J.K., Bothwell A., and Jacobson K.. 1991. Lateral diffusion of membrane-spanning and glycosylphosphatidylinositol-linked proteins: Toward establishing rules governing the lateral mobility of membrane proteins. J. Cell Biol. 115:75–84. 10.1083/jcb.115.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.