A large number of lysosome-localized proteins control mTORC1 signaling. Rag guanosine triphosphatase (GTPase) heterodimers play a central role in this pathway by recruiting mTORC1 to lysosomes. Meng and Ferguson reveal how folliculin, a tumor suppressor, coordinates nucleotide states within Rag GTPase heterodimers.
Abstract
Folliculin (FLCN) is a tumor suppressor that coordinates cellular responses to changes in amino acid availability via regulation of the Rag guanosine triphosphatases. FLCN is recruited to lysosomes during amino acid starvation, where it interacts with RagA/B as a heterodimeric complex with FLCN-interacting proteins (FNIPs). The FLCN–FNIP heterodimer also has GTPase-activating protein (GAP) activity toward RagC/D. These properties raised two important questions. First, how is amino acid availability sensed to regulate lysosomal abundance of FLCN? Second, what is the relationship between FLCN lysosome localization, RagA/B interactions, and RagC/D GAP activity? In this study, we show that RagA/B nucleotide status determines the FLCN–FNIP1 recruitment to lysosomes. Starvation-induced FLCN–FNIP lysosome localization requires GAP activity toward Rags 1 (GATOR1), the GAP that converts RagA/B to the guanosine diphosphate (GDP)-bound state. This places FLCN–FNIP recruitment to lysosomes under the control of amino acid sensors that act upstream of GATOR1. By binding to RagA/BGDP and acting on RagC/D, FLCN–FNIP can coordinate nucleotide status between Rag heterodimer subunits in response to changes in amino acid availability.
Introduction
Loss-of-function mutations in the folliculin (FLCN) gene cause Birt-Hogg-Dubé syndrome, a disease characterized by benign hair follicle tumors (fibrofolliculomas), pulmonary cysts, spontaneous pneumothorax, and renal carcinoma (Nickerson et al., 2002). Birt-Hogg-Dubé syndrome patients typically inherit one germline FLCN mutation, and somatic second hit mutations result in the complete loss of FLCN function in the cells that give rise to disease pathology (Schmidt and Linehan, 2018). These diverse Birt-Hogg-Dubé syndrome patient phenotypes raise important questions about the cellular functions of the FLCN protein. Although loss-of-function mutations in human FLCN predominantly affect specific tissues, the FLCN gene is ubiquitously expressed and evolutionarily conserved from yeast to humans and thus likely controls fundamental aspects of cell physiology (Schmidt and Linehan, 2018).
The FLCN protein contains a differentially expressed in normal and neoplastic cells (DENN) domain as its major folded domain (Nookala et al., 2012). This places FLCN within a family of proteins that are best characterized as regulators of intracellular membrane traffic through their actions as guanine nucleotide exchange factors (GEFs) for Rab GTPases (Allaire et al., 2010; Yoshimura et al., 2010; Marat et al., 2011; Wu et al., 2011). FLCN forms heterodimeric complexes with either FLCN-interacting protein (FNIP) 1 or 2 (Baba et al., 2006; Hasumi et al., 2008; Takagi et al., 2008). Bioinformatic analyses predicted that FNIP proteins also contain a DENN domain as their major structural feature (Zhang et al., 2012; Levine et al., 2013). These predictions are supported by a crystal structure of a portion of the yeast FNIP1/2 orthologue, Lst4, which contains a longin module, the first element of the full DENN domain, in its N terminus (Pacitto et al., 2015). However, contrary to the expectations raised by other DENN domain proteins that act as Rab GEFs, FLCN–FNIP communicate amino acid abundance to the mTORC1 signaling pathway by serving as a GTPase-activating protein (GAP) for the lysosome-localized RagC/D GTPases (Tsun et al., 2013; Péli-Gulli et al., 2015).
In mammals, the Rag GTPases are obligate heterodimers composed of RagA or RagB isoforms bound to RagC or RagD (Sekiguchi et al., 2001). Rag nucleotide status is tightly regulated by intracellular amino acid availability. When amino acid levels are high, the RagA/BGTP-RagC/DGDP form of the heterodimer recruits mTORC1 to the surface of lysosome by interacting with the Raptor subunit (Kim et al., 2008; Sancak et al., 2008, 2010; Gong et al., 2011). Once at the lysosome, mTORC1 is activated through an interaction with Rheb, a small GTPase that is tightly regulated by growth factor signaling and cellular energy status (Menon et al., 2014; Saxton and Sabatini, 2017). The nucleotide state of individual Rag GTPases in the heterodimer is tightly regulated by distinct GAPs and GEFs whose activities are controlled by amino acid availability. In addition to FLCN–FNIP (RagC/D GAP), these include GAP activity toward Rags 1 (GATOR1; RagA/B GAP) and Ragulator (RagA/B GEF; Bar-Peled et al., 2012, 2013; Panchaud et al., 2013; Tsun et al., 2013; Péli-Gulli et al., 2015).
Identification of Rags as direct binding partners and targets for FLCN–FNIP represented an important step forward in understanding how amino acid availability regulates mTORC1 signaling and by extension suggested that defects in this process contribute to Birt-Hogg-Dubé syndrome pathogenesis (Petit et al., 2013; Tsun et al., 2013). However, these studies also generated new puzzles related to underlying mechanisms. Although the purified FLCN–FNIP complex stimulates GTP hydrolysis by RagC/D in vitro, FLCN was nonetheless also reported to bind to RagA/BGDP (Petit et al., 2013; Tsun et al., 2013). The preferential binding to the GDP-bound form of a small GTPase is a property commonly seen in GEFs (Bos et al., 2007; Cherfils and Zeghouf, 2013). This property combined with the presence of a DENN domain in both FLCN and FNIP proteins suggested the possibility of a RagA/B GEF activity for the FLCN–FNIP heterodimer. However, as GEF activity of FLCN–FNIP toward RagA/B has not been detected, it remains unclear whether and how RagA/B binding by FLCN–FNIP contributes to their GAP activity toward RagC/D. Also, although binding between FLCN and Rags is stimulated by amino acid starvation (Petit et al., 2013; Tsun et al., 2013), the specific amino acid sensors and signal transduction pathways that mediate such regulation remain unknown. This has led to speculation that novel amino acid sensors might function upstream of FLCN–FNIP heterodimers (Wolfson and Sabatini, 2017).
Motivated by the questions outlined above, we sought to elucidate the mechanisms supporting starvation-induced recruitment of FLCN–FNIP to lysosomes and the functional significance of interactions between FLCN–FNIP and RagA/B. Using knockout (KO) and epitope tag knock-in cells generated by CRISPR-Cas9 gene editing combined with biochemical and imaging approaches, we have now established RagA/BGDP as a major determinant of FLCN–FNIP recruitment to lysosomes in amino acid–starved cells and place such regulation downstream of GATOR1, the RagA/B GAP. Our results provide a mechanism to explain the acute recruitment of FLCN–FNIP to lysosomes in amino acid–starved cells and furthermore suggest a role for FLCN–FNIP-dependent coordination of nucleotide states within the Rag heterodimers.
Results
Rag GTPases are required for FLCN recruitment to lysosomes
As a first step toward elucidating the relationship between FLCN subcellular localization, FLCN–Rag interaction, and biochemical function, we sought to validate tools for the immunofluorescent detection of FLCN. Previously, using HeLa cells that were engineered to contain a 3×HA tag within the endogenous FLCN gene locus, we observed that FLCN was dispersed in the cytoplasm under basal growth conditions but was selectively recruited to lysosomes in response to amino acid starvation (Petit et al., 2013). This finding was subsequently confirmed by other laboratories that used an anti-FLCN antibody to detect the endogenous FLCN protein (Tsun et al., 2013; Martina et al., 2014). However, it was also recently reported (based on the same antibody) that FLCN localizes to the nucleus and acts on Rags in this subcellular compartment (Wu et al., 2016). Although intriguing, this finding stands at odds with the lysosome localization data for FLCN. To rigorously evaluate FLCN localization, we revisited this topic using the anti-FLCN antibody on both WT and newly generated FLCN KO cells (Fig. S1 A and Table S1). We observed cytoplasmic staining for FLCN under basal growth conditions and FLCN enrichment on lysosomes in response to starvation in the WT cells (Fig. 1 B). The cytoplasmic and lysosomal signals for FLCN were specific as they were absent in FLCN KO cells (Fig. S1, B and C). In contrast, this anti-FLCN antibody also yielded a strong nuclear signal that was present in both control and FLCN KO cells and thus represents nonspecific staining (Fig. S1 B). Although the strong nonspecific nuclear signal for this anti-FLCN antibody does not rule out the possibility of a small nuclear pool of FLCN, these results clearly establish the specificity of this immunofluorescence assay for assessing lysosome recruitment of the endogenous FLCN protein in amino acid–starved cells. In subsequent experiments, we will combine the use of this antibody with a series of genetically modified cell lines to investigate the mechanisms that dynamically control FLCN recruitment to lysosomes.
Figure 1.
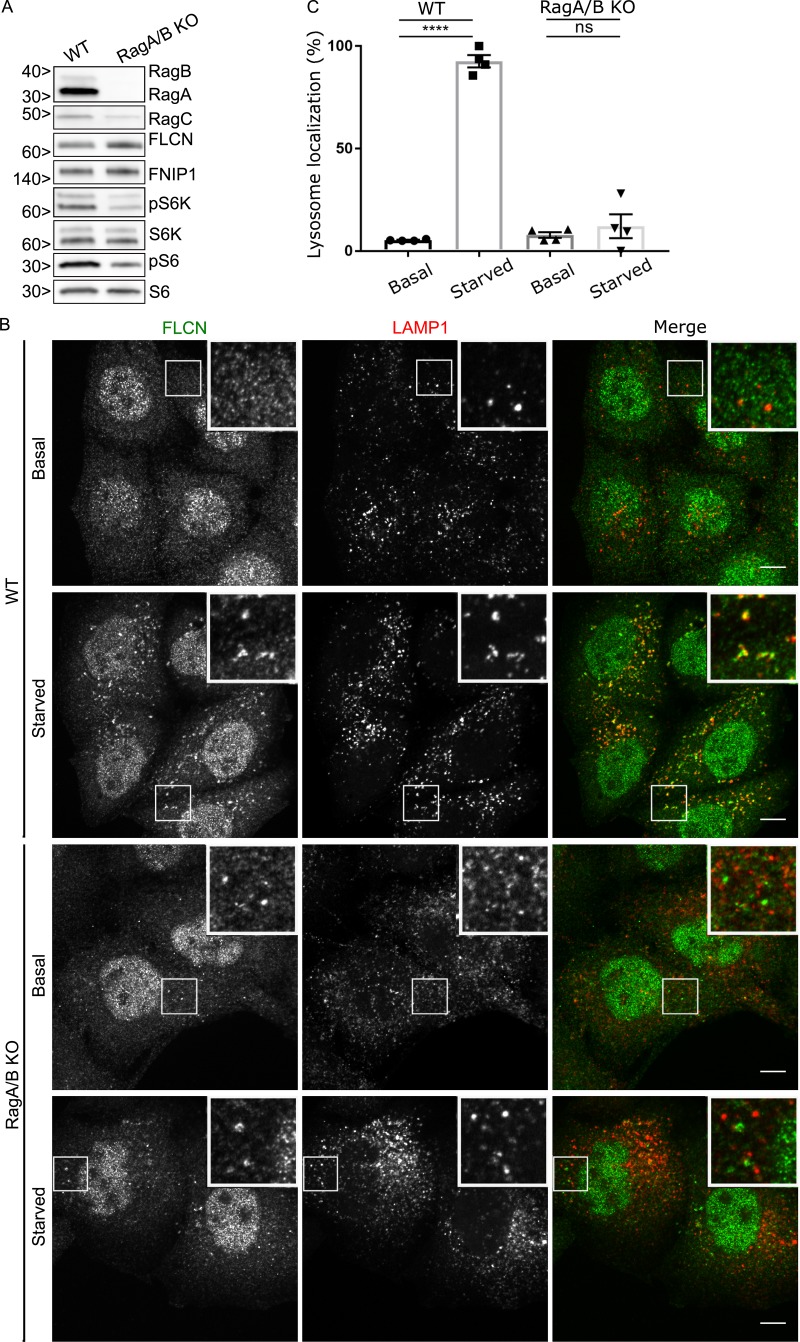
RagA and RagB are essential for starvation-dependent recruitment of FLCN to lysosomes. (A) Western blot analysis of WT and RagA/B KO HeLa cells. Molecular masses are given in kilodaltons. (B) Representative immunofluorescence experiment showing the effect of starvation on FLCN subcellular localization in WT and RagA/B KO HeLa cells. Bars, 10 µm. Insets are 12 µm wide. (C) Quantification of FLCN lysosome localization (percentage of cells with five or more FLCN spots that are also LAMP1 positive; mean ± SEM; n = 4 experiments, 76–80 cells per condition; ****, P < 0.0001; ANOVA with Bonferroni’s post hoc test).
Rag GTPases represent a major node for the integration of signals relating to amino acid availability (Wolfson and Sabatini, 2017), and the interaction between FLCN and Rags is enhanced during starvation (Petit et al., 2013; Tsun et al., 2013). However, it was unclear whether this was a cause or consequence of FLCN at lysosomes. We addressed this problem by testing whether Rag-depleted cells can still support starvation-induced recruitment of FLCN to lysosomes. To this end, we used CRISPR-Cas9 genome editing to generate RagA+B double-KO HeLa cells (Fig. 1 A and Table S1). Consistent with the known role for RagA/B interactions in supporting RagC stability (Sancak et al., 2008), RagC protein levels were diminished in the absence of RagA+B. Meanwhile, FLCN and FNIP1 protein levels were modestly elevated (Fig. 1 A). This change can potentially be explained by increased nuclear localization of transcription factor EB in Rag-depleted cells combined with the established role for transcription factor EB and its closely related transcription factors in regulating FLCN expression (Roczniak-Ferguson et al., 2012; Martina et al., 2014; Di Malta et al., 2017).
The RagA+B KO resulted in a reduction in mTORC1 activity as measured by the phosphorylation states of ribosomal protein S6 kinase (S6K) and ribosomal protein S6 (Fig. 1 A). Furthermore, lysosome localization of mTOR was lost under basal growth conditions in RagA+B KO cells (Fig. S3 A), and starved RagA+B KO cells failed to restore mTOR lysosome localization in response to amino acid refeeding (Fig. S3 B). Having established that the RagA+B KO cells exhibited the expected mTOR localization and signaling defects, we next tested FLCN localization and found that starvation-induced recruitment of FLCN to lysosomes was abolished (Fig. 1, B and C). We also observed that although the RagA+B KO cells did not efficiently recruit FLCN to lysosomes upon starvation, these cells constitutively contained non-lysosomal FLCN puncta (Fig. 1 B). The identity of such FLCN puncta and their relationship to the loss of RagA+B remains to be determined. Nonetheless, their presence does not affect our major conclusions regarding the lack of FLCN at lysosomes in the absence of RagA+B.
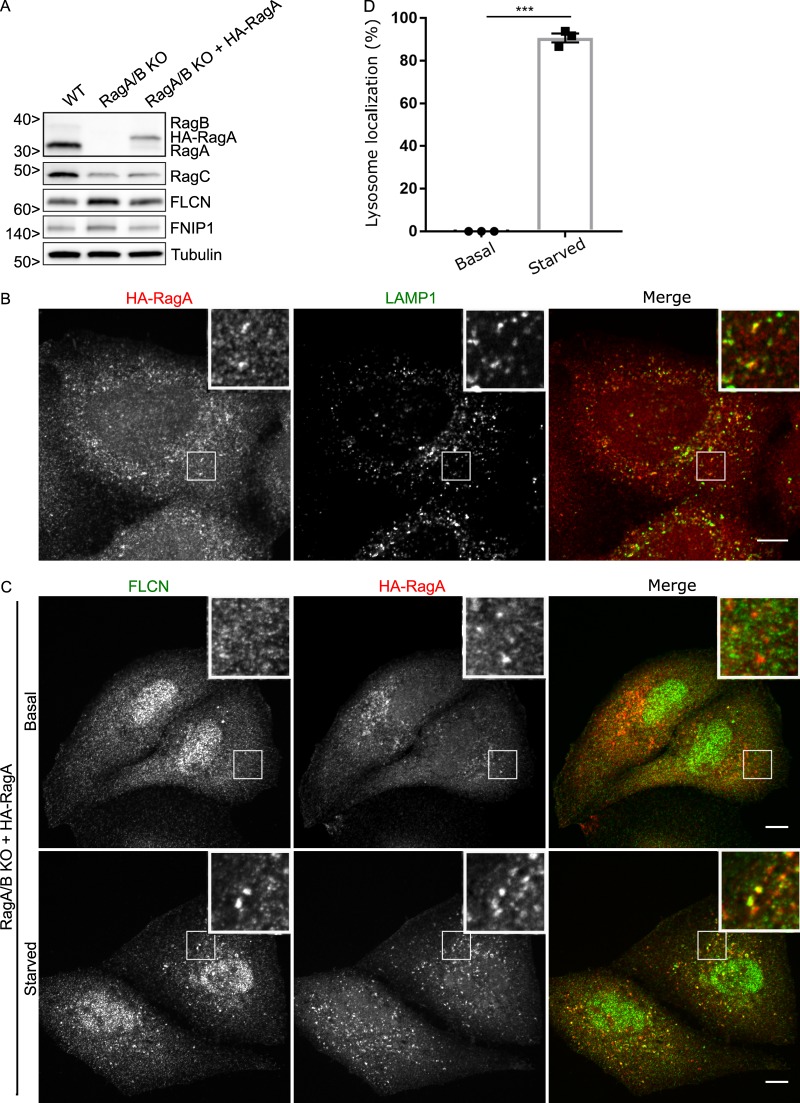
To further test the specificity of the RagA+B KO on FLCN lysosomal localization, we stably reintroduced HA-RagA into RagA+B KO cells (Fig. 2 A). As expected, the HA-RagA protein was found at lysosomes under both basal and starved conditions (Fig. 2 B), and FLCN was selectively recruited to these lysosomes in starved cells (Fig. 2, C and D). Separate from our major focus on the lysosome recruitment of FLCN, we noted that the constitutive non-lysosomal FLCN puncta that were present in the RagA+B KO cells were absent after the reintroduction of RagA. This therefore represents a specific impact of the RagA/B KO on behavior of the FLCN protein that is of unknown physiological relevance.
Figure 2.
Reintroduction of RagA into RagA+B KO cells results in starvation-dependent recruitment of FLCN to lysosomes. (A) Western blot analysis of WT, RagA+B KO, and RagA+B KO HeLa cells expressing HA-RagA. Molecular masses are given in kilodaltons. (B) Representative immunofluorescence images showing HA-RagA localization to LAMP1-labeled lysosomes. Bars, 10 µm. Insets are 10.2 µm wide. (C) Representative immunofluorescence experiment showing the effect of starvation on FLCN lysosome localization in cells expressing HA-RagA. Bars, 10 µm. Insets are 10.2 µm wide. (D) Quantification of FLCN lysosome localization (percentage of cells with five or more FLCN spots that are also RagA positive; mean ± SEM; n = 3 experiments; 33–43 cells per condition; ***, P < 0.001; Student's t test).
As an additional strategy for investigating the relationship between FLCN and the Rag GTPases, we took advantage of siRNA-mediated knockdown of RagC. RagA/B levels were reduced after siRNA-mediated RagC depletion (Fig. S2 A). This matches expectations based on the known requirement for Rag heterodimerization for optimal stability (Sancak et al., 2008). Meanwhile, as seen in Fig. 1 for the RagA+B KO cells, the levels of FLCN were modestly increased in the absence of RagC (Fig. S2 A). Most importantly, we observed that amino acid starvation failed to stimulate FLCN recruitment to lysosomes in RagC-depleted cells (Fig. S2, B and C). Rag GTPases are thus essential for starvation-induced recruitment of FLCN to lysosomes.
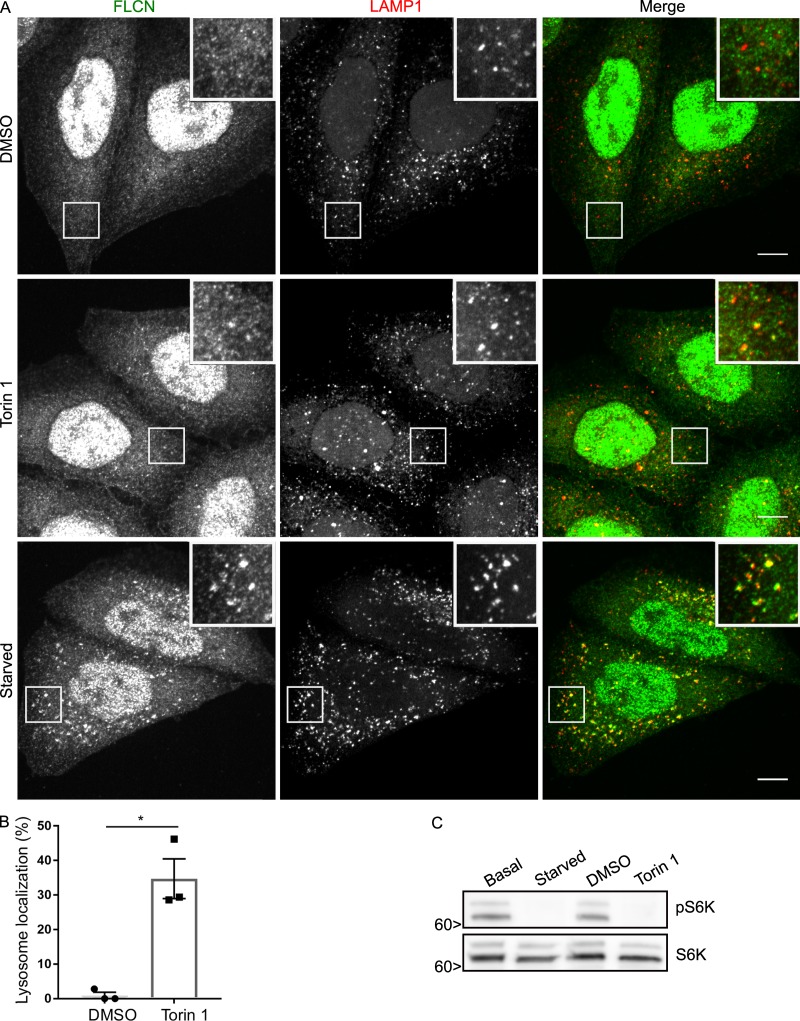
How, then, is starvation sensed and communicated to FLCN? mTORC1 signaling was previously reported to negatively regulate FLCN–FNIP localization to lysosomes (Martina et al., 2014; Péli-Gulli et al., 2017). As mTORC1 is inactivated during starvation, the loss of mTORC1-dependent inhibition could potentially explain the increased localization of FLCN to lysosomes in starved cells. We thus directly compared the impact of starvation versus pharmacological mTOR inhibition on FLCN localization. Although Torin 1 treatment modestly increased FLCN levels at lysosomes (Fig. 3 A), starvation elicited a more robust effect (see quantification in Fig. 3 B vs. Fig. 2 C) even though mTORC1 signaling was suppressed to the same extent as when cells were starved (Fig. 3 C). These results indicate that reduced mTORC1 signaling is not the major explanation for the starvation-induced increase in FLCN abundance at lysosomes.
Figure 3.
Investigation of the effects of mTOR inhibition on FLCN lysosome localization. (A) Immunofluorescence analysis showing the effect of DMSO (vehicle control) versus Torin 1 (250 nM for 1 h) and starvation on FLCN localization in HeLa cells. Bars, 10 µm. Insets are 12 µm wide. (B) Quantification of the impact of Torin 1 treatment on FLCN localization to lysosomes (percentage of cells with five or more FLCN spots that are also LAMP1 positive; mean ± SEM; n = 3 experiments; 76–77 cells per condition; *, P < 0.05; Student’s t test). (C) Western blots showing the similar effects of starvation and Torin 1 on mTORC1 signaling. Molecular masses are given in kilodaltons.
GATOR1 and RagAGDP are critical for starvation-induced recruitment of FLCN to lysosomes
The acute regulation of FLCN levels at lysosomes by changes in amino acid availability raised questions about the underlying amino acid sensing mechanism. Although it has been speculated that a novel amino acid sensor might function upstream of FLCN (Wolfson and Sabatini, 2017), the critical dependence of FLCN on Rags for its lysosome localization as well as the fact that FLCN interacts preferentially with the GDP/nucleotide-free form of RagA/B (Petit et al., 2013; Tsun et al., 2013) suggested to us that FLCN recruitment to lysosomes might simply depend on its interaction with RagA/BGDP. If this were the case, then known amino acid–sensing pathways that act upstream of RagA/B could explain the amino acid sensitivity of FLCN localization to lysosomes.
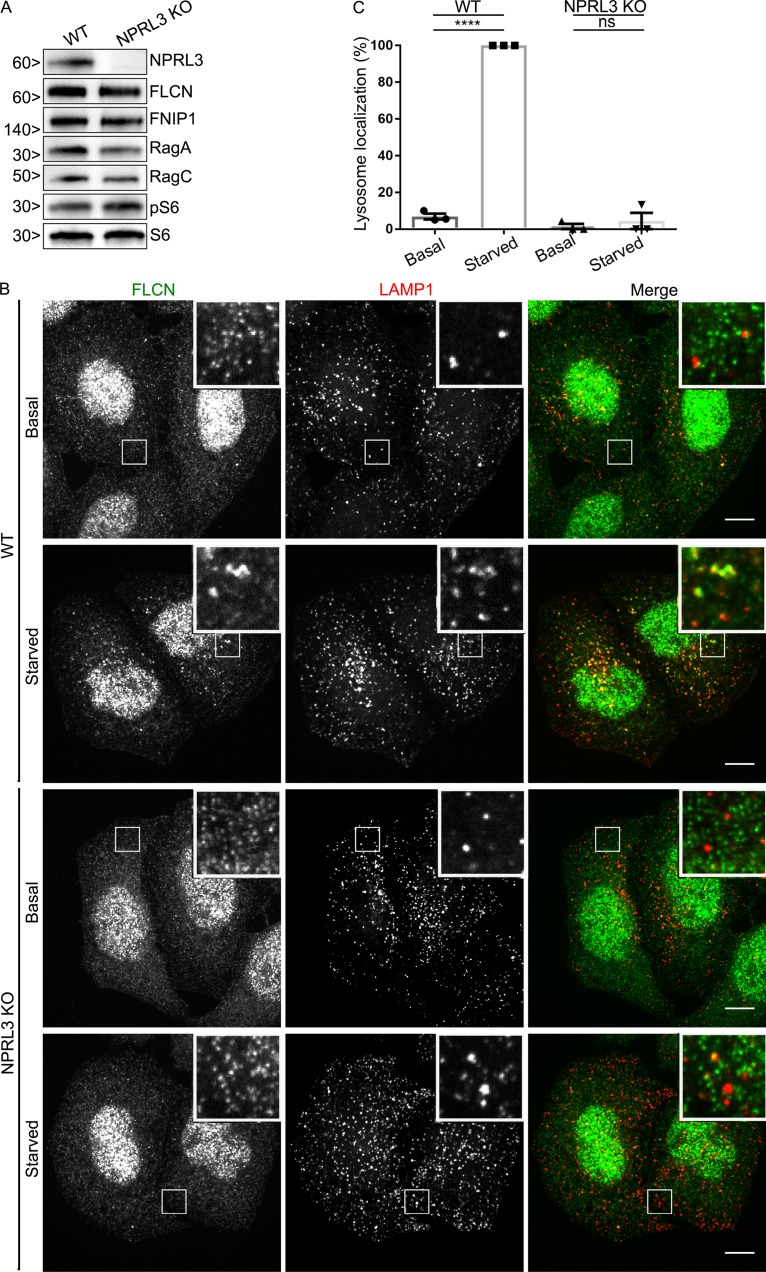
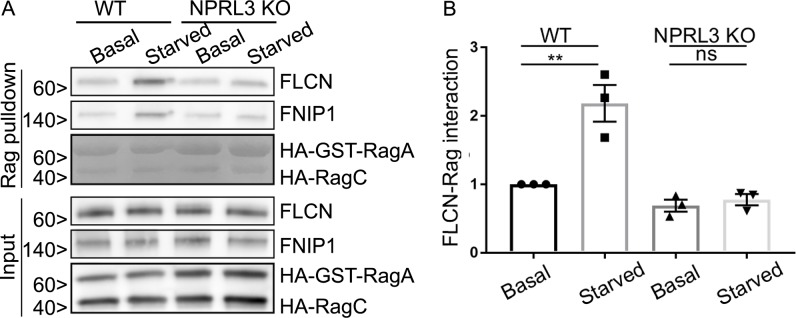
The major regulator of RagA/B GTPase activity is GATOR1 (Bar-Peled et al., 2013; Panchaud et al., 2013; Shen et al., 2018). GATOR1 is a heterotrimeric protein complex made up of NPRL2, NPRL3, and DEPDC5, whose GAP activity toward RagA/B is tightly controlled by amino acid availability via signals received from upstream amino acid sensors (Bar-Peled et al., 2013; Panchaud et al., 2013; Chantranupong et al., 2014, 2016; Parmigiani et al., 2014; Kim et al., 2015; Wolfson et al., 2016). Each of the three GATOR1 subunits is required for normal function of the complex, and human mutations in each of these genes cause a form of epilepsy that is thought to arise from mTORC1 hyperactivation (Bar-Peled et al., 2013; Baldassari et al., 2016). To determine the role of GATOR1-dependent control of RagA/B nucleotide status in the recruitment of FLCN to lysosomes during amino acid starvation, we used CRISPR-Cas9 gene editing to knock out the NPRL3 subunit. In addition to DNA sequencing (Table S1), immunoblotting confirmed the loss of NPRL3 protein from the KO cells and furthermore showed that RagA/B, RagC, FLCN, and FNIP1 proteins are all still present (Fig. 4 A). Consistent with the established amino acid starvation–induced GAP activity of GATOR1 toward RagA/B that suppresses mTORC1 activity, mTORC1 signaling was resistant to amino acid starvation in NPRL3 KO cells (Fig. S4, A and B). Having documented the expected consequences of NPRL3 KO on mTOR, we turned our attention to FLCN and observed that both the starvation-induced recruitment of FLCN to lysosomes (Fig. 4, B and C) and FLCN–Rag interactions were eliminated in NPRL3 KO cells (Fig. 5, A and B). These new results support a model wherein the GATOR1-mediated conversion of RagA/B to the GDP-bound state under starved conditions controls both FLCN–Rag interactions and FLCN recruitment to lysosomes.
Figure 4.
GATOR1 is important for FLCN recruitment to lysosomes. (A) Western blots of WT and NPRL3 KO cells. Molecular masses are given in kilodaltons. (B) Spinning-disk confocal images of FLCN localization in WT and NPRL3 KO cells (HeLa) under basal and starved conditions. Bars, 10 µm. Insets are 7.4 µm wide. (C) Quantification of FLCN lysosome localization (percentage of cells with five or more FLCN spots that are also LAMP1 positive; mean ± SEM; n = 3 experiments; 50–60 cells per condition; ****, P < 0.0001; ANOVA with Bonferroni’s post hoc test).
Figure 5.
FLCN selectively interacts with inactive RagA. (A) Representative Western blots of GST pulldowns in WT and NPRL3 KO cells transfected with HA-GST-RagA + HA-RagC in basal and starved conditions. Because of their high abundance, HA-GST-RagA and HA-RagC in the pulldown condition were detected by Ponceau S staining. Molecular masses are given in kilodaltons. (B) Quantification of FLCN abundance in Rag pulldown (**, P < 0.01; ANOVA with Bonferroni’s post hoc test).
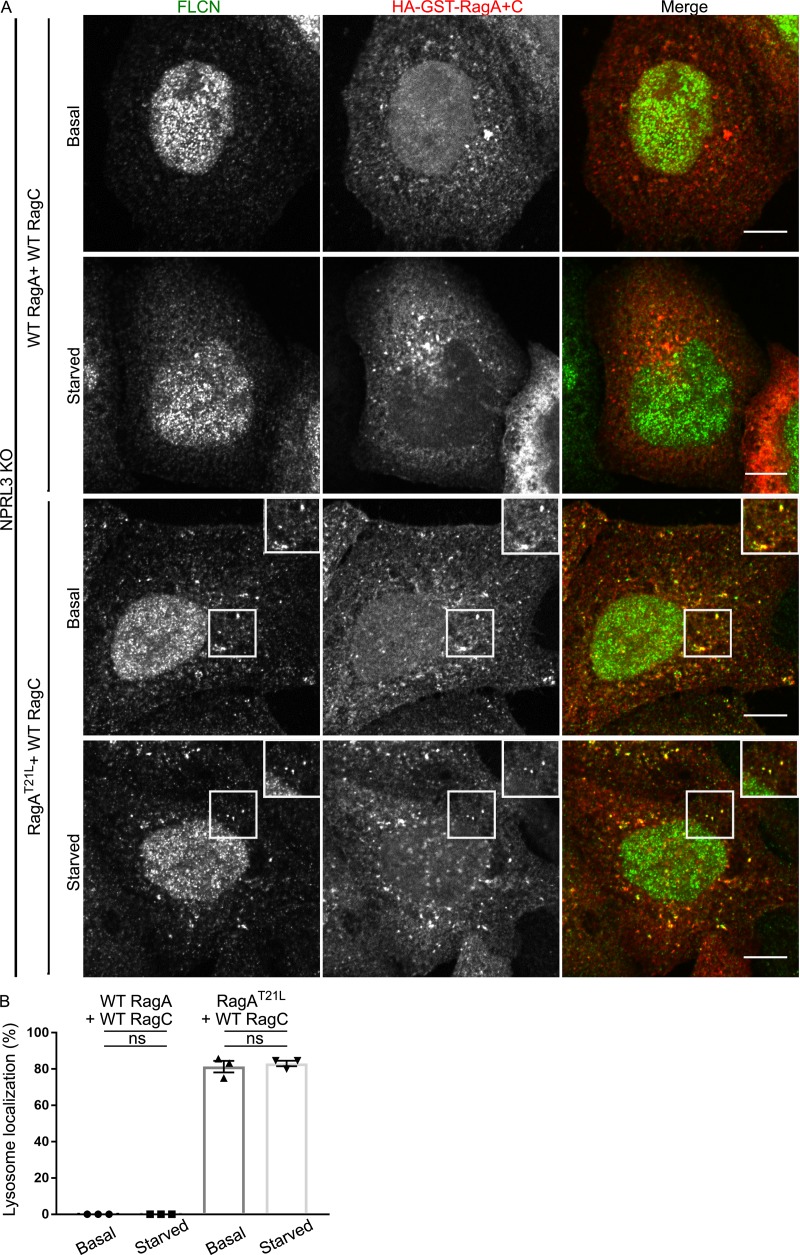
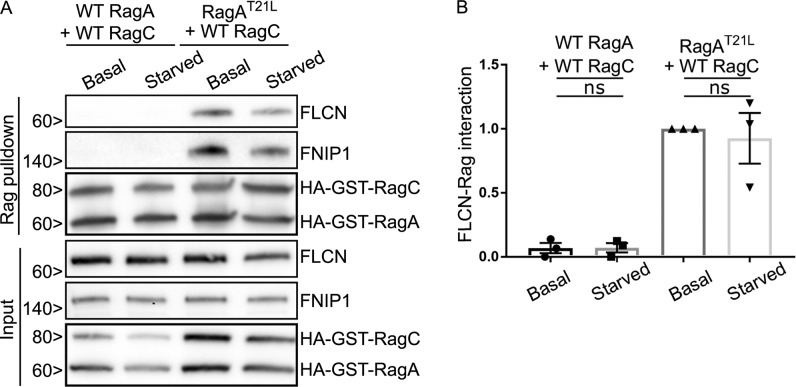
To further test the hypothesis that RagA/B nucleotide status is the key determinant of FLCN recruitment to lysosomes, we investigated FLCN localization in NPRL3 KO cells that were transfected with WT versus mutant forms of RagA + RagC. Although WT RagA and RagC did not support FLCN localization to lysosomes in NPRL3 KO cells, expression of a RagA mutant (RagAT21L) that favors the GDP-bound/nucleotide-free state resulted in constitutive localization of FLCN at lysosomes in NPRL3 KO cells that was no longer regulated by starvation (Fig. 6, A and B). We similarly observed that FLCN coimmunoprecipitated strongly with RagAT21L-RagCWT but minimally with RagAWT-RagCWT in the NPRL3 KO cells and that this interaction was also insensitive to starvation (Fig. 7, A and B). These results further support the conclusion that GATOR1-dependent control of the RagA nucleotide state drives FLCN recruitment to lysosomes when amino acids are scarce. Although we did not directly test RagB in these experiments, previous observations of RagB interactions with FLCN combined with the extensive sequence conservation between RagA and RagB (>99% identical in the GTPase domain) strongly suggest that a common mechanism supports RagA and RagB interactions with FLCN (Petit et al., 2013; Tsun et al., 2013).
Figure 6.
RagAGDP determines FLCN recruitment to lysosomes. (A) Representative immunofluorescence images showing the localization of FLCN and Rags in NPRL3 KO cells transfected with WT RagA or RagAT21L + WT RagC under basal growth and starved conditions. Bars, 10 µm. Insets are 10.8 µm wide. (B) Quantification of FLCN lysosome localization (percentage of cells with five or more FLCN spots that are Rag positive; mean ± SEM; n = 3 experiments; 27–31 cells per condition; ANOVA with Bonferroni’s post hoc test).
Figure 7.
FLCN selectively interacts with RagAGDP. (A and B) Representative Western blots (A) and quantification (B) of GST-pulldowns in NPRL3 KO cells transfected with WT RagA or RagAT21L + WT RagC, basal and starved conditions (ANOVA with Bonferroni’s post hoc test). Molecular masses are given in kilodaltons.
FNIP1 is recruited to the surface of lysosomes by Rag GTPases
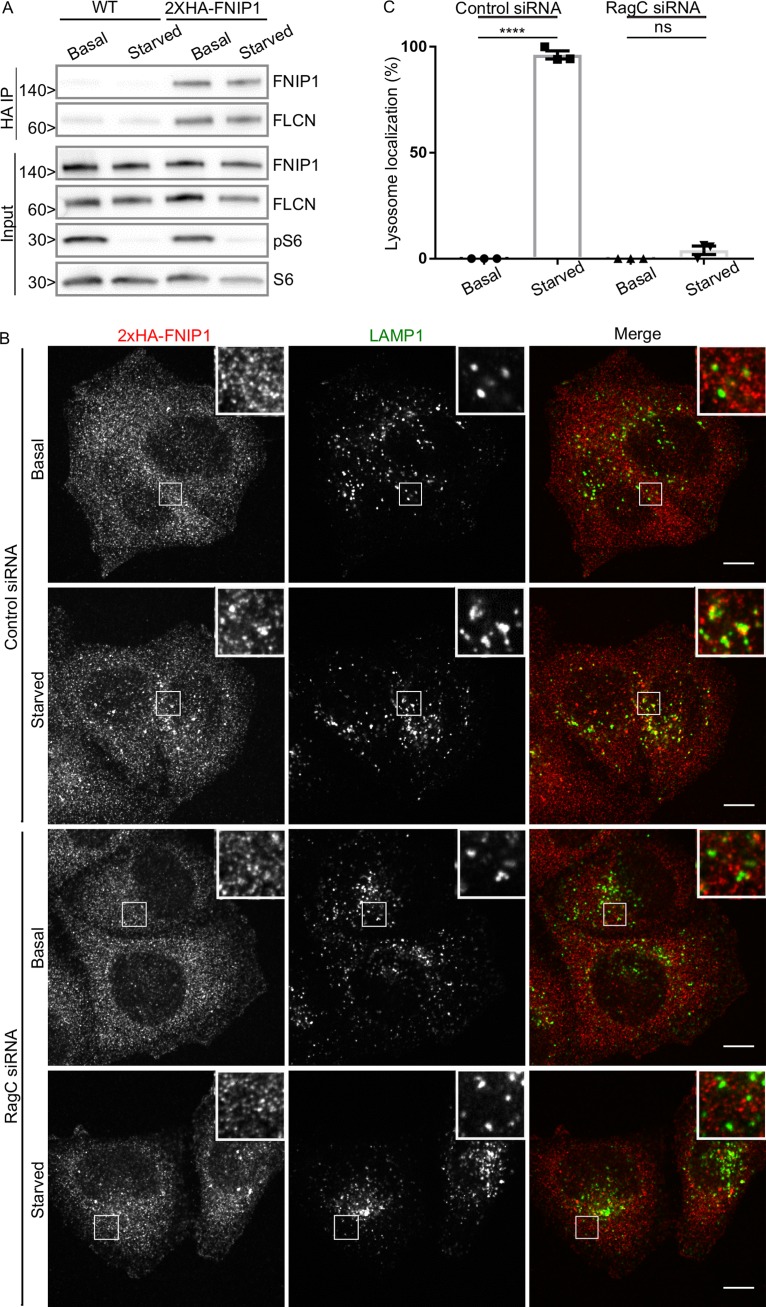
Previous studies showed that FNIP1 and FNIP2 interact with lysosome-localized Rag GTPases as part of a complex with FLCN (Petit et al., 2013; Tsun et al., 2013). However, overexpressed GFP-tagged FNIP1 was constitutively localized to lysosomes (Petit et al., 2013). As the subcellular localization of the endogenous FNIP proteins has not been established, it was unclear whether FNIP proteins cycle on and off lysosomes along with FLCN. To directly establish the subcellular localization of endogenous FNIP1, we used CRISPR-Cas9 genome engineering to generate an N-terminally epitope-tagged 2×HA-FNIP1 protein expressed from its endogenous locus in HeLa cells. Analysis of genomic PCR amplicons from the edited region of the FNIP1 gene revealed that the 2×HA tag was inserted into two FNIP1 alleles by homologous recombination, whereas a frameshift mutation causing deletion of 2 bp inactivated the remaining FNIP1 allele in this near triploid cell line (Table S1). Thus, the 2×HA-FNIP1 protein completely replaced the untagged form of FNIP1 in these cells. As predicted based on its calculated molecular weight, the 2×HA-FNIP1 protein migrated as a band of ∼150 kD after immunoblotting with an anti-HA antibody (Fig. 8 A). Copurification of FLCN with 2×HA-FNIP1 via anti-HA immunoprecipitation revealed that the FLCN–FNIP1 interaction was insensitive to changes in amino acid availability (Fig. 8 A). The insensitivity of the FLCN–FNIP1 interaction to amino acid starvation suggests that the FLCN–FNIP1 complex stays intact while cycling on and off of lysosomes in a Rag-dependent manner.
Figure 8.
Rags are required for starvation-induced lysosomal recruitment of FNIP1. (A) Representative Western blots of anti-HA immunoprecipitation (IP) from WT HeLa cells and a HeLa line expressing 2×HA-FNIP1 from the endogenous locus. Molecular masses are given in kilodaltons. (B) Representative immunofluorescence experiment showing 2×HA-FNIP1 localization in fed and starved conditions ± siRNA-mediated RagC depletion. Bars, 10 µm. Insets are 7.3 µm wide. (C) Quantification of FNIP1 lysosome localization as defined by percentage of cells with five or more FNIP1 spots that are also LAMP1 positive (mean ± SEM; n = 3 experiments; 46–54 cells per condition; ****, P < 0.0001; ANOVA with Bonferroni’s post hoc test).
To test this hypothesis, FNIP1 subcellular localization was next investigated. As we observed for FLCN, 2×HA-FNIP1 was not abundant on lysosomes under basal cell growth conditions (fed state), but was enriched on lysosomes when cells were starved (Fig. 8, B and C). As we had previously observed constitutive lysosome localization for FNIP1-GFP after transient transfection (Petit et al., 2013), these results emphasize a potential pitfall that can arise from relying solely on overexpressed GFP-tagged proteins for the study of protein subcellular localization.
Similar to FLCN, siRNA-mediated RagC depletion prevented the starvation-induced enrichment of 2×HA-FNIP1 on lysosomes (Fig. 8, B and C). Based on these observations, we conclude that FNIP1 is corecruited to lysosomes with FLCN in a Rag-dependent manner. This result parallels the behavior of the yeast FNIP orthologue (Lst4) that also localizes to and redistributes from the vacuolar membrane in response to changes in nutrient availability (Péli-Gulli et al., 2015, 2017). However, in contrast with our observations from human cells, the vacuole recruitment of yeast FLCN–FNIP homologues (Lst7-Lst4) is Rag (Gtr1-Gtr2)–independent. Although this could simply reflect species-specific differences in the details of how this pathway is regulated, these observations also raise the possibility of additional, evolutionarily conserved, FLCN–FNIP binding partners at lysosomes.
Discussion
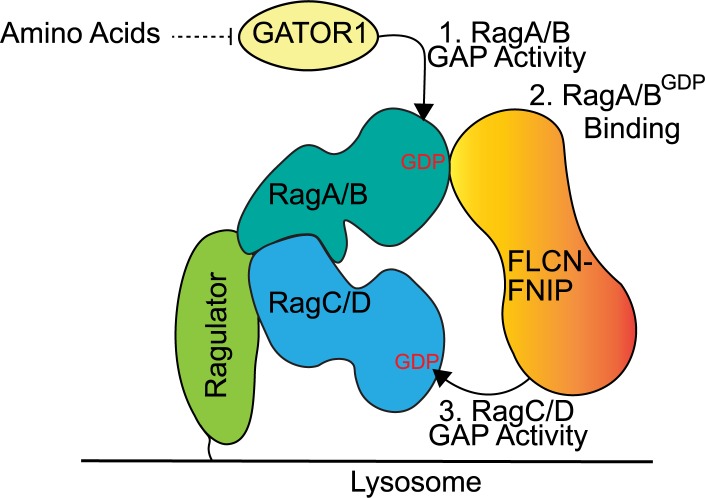
In this study, we identify GATOR1-dependent control of RagA/B nucleotide status as a major determinant of the amino acid–regulated recruitment of FLCN–FNIP to lysosomes and place the RagC/D GAP activity of FLCN–FNIP downstream of RagA/B regulation by the GATOR1 pathway (Fig. 9). These new findings help to resolve the seemingly contradictory previous studies reporting that the FLCN–FNIP heterodimer binds to RagA/B but act as a RagC/D GAP (Petit et al., 2013; Tsun et al., 2013). Coordination of Rag nucleotide status is likely to be critical as experiments that used Rag mutants with different nucleotide binding preferences have found that the most active form of the Rag GTPase heterodimers consists of GTP-bound RagA/B and GDP-bound RagC/D. This combination of Rag mutants renders mTORC1 signaling resistant to amino acid depletion and maximally supports Rag interactions with the MiT-TFE transcription factors (Kim et al., 2008; Sancak et al., 2008; Martina and Puertollano, 2013).
Figure 9.
Schematic diagram summarizing key steps in the coordination of FLCN–FNIP recruitment to and function at lysosomes. 1. When amino acids are scarce, the GAP activity of GATOR1 promotes the GDP-bound state of RagA/B. 2. FLCN–FNIP are recruited to lysosomes by interacting with RagA/BGDP. 3. FLCN–FNIP exhibit GAP activity toward RagC/D.
The recruitment of FLCN–FNIP to lysosomes by RagA/BGDP raises new questions about the molecular details of the interfaces between these proteins. It is interesting that both FLCN and the FNIPs contain DENN domains and are thus part of the larger DENN family that is best characterized for GEF activity toward various Rab GTPases (Allaire et al., 2010; Yoshimura et al., 2010; Marat et al., 2011; Wu et al., 2011). As it is a general property of GEFs to selectively bind to the GDP-bound state of their target GTPases, a behavior that reflects the trapping of an intermediate state in the process of nucleotide exchange (Bos et al., 2007), it is possible that FLCN–FNIP binds to RagA/BGDP in a GEF-like manner that does not lead to nucleotide exchange but rather brings the FLCN–FNIP complex into close proximity to RagC/D and thus facilitates their GAP activity toward RagC/D (Fig. 9). Such a pseudoGEF-GAP function for FLCN–FNIP toward RagA/B and RagC/D, respectively, would thus help to coordinate nucleotide status within Rag heterodimers and would act in addition to recently proposed direct coordination of nucleotide state between RagA/B and RagC/D subunits within the Rag heterodimer (Shen et al., 2017).
Since the initial discovery that the Rags play a critical, lysosome-localized role in coordinating cellular response to changes in amino acid availability, >20 additional proteins have been identified that participate directly in this process (Saxton and Sabatini, 2017). These include amino acid sensors, amino acid transporters, Rag GEF and GAP proteins, multiple scaffold proteins, and many additional proteins of uncertain function that are implicated in this pathway via genetic and proteomic experiments. The complexity of this machinery likely reflects the importance of tightly matching mTORC1 anabolic signaling to ongoing changes in nutrient and growth factor availability. However, as multiple proteins can directly interact with the Rags, questions arise about how the functions of these many proteins are spatially and temporally controlled. With respect to the FLCN–FNIP complex, our data place its interaction with RagA/BGDP downstream of the GAP activity of GATOR1. Conversely, the GEF activity of the pentameric Ragulator complex promotes exchange of GDP for GTP on RagA/B (Bar-Peled et al., 2012) and is thus expected to negatively regulate interactions of RagA/B with FLCN–FNIP. However, the details of how FLCN–FNIP interactions and Ragulator GEF activity change dynamically upon amino acid exposure remain to be established. For example, it is not known whether the FLCN–FNIP and Ragulator complexes compete for access to the GDP-bound form of RagA/B. Furthermore, even though interactions between the C-terminal roadblock domains of the Rags with Ragulator are well established, little is known about how Ragulator stimulates the release of GDP from the RagA/B GTPase domain (Su et al., 2017; Yonehara et al., 2017; Zhang et al., 2017). These points illustrate the need for a better understanding of the mechanism behind Ragulator’s GEF activity toward the Rags. New structural and biochemical insights into these questions will be important for understanding not just the normal functioning of this pathway, but may also be relevant for explaining how loss of FLCN, a tumor suppressor, causes a paradoxical increase in mTOR signaling in Birt-Hogg-Dubé syndrome (Schmidt and Linehan, 2018).
In addition to functioning at lysosomes, FLCN was recently reported to regulate the Rags from within the nucleus (Wu et al., 2016). FLCN was also proposed to control nuclear exit of TDP-43, an RNA binding protein whose cytoplasmic aggregates are a hallmark of several neurodegenerative diseases (Xia et al., 2016). Although our study does not exclude the existence of such intranuclear functions for FLCN, we note that our staining for both FLCN and FNIP1 did not detect a major nuclear pool of either protein. More generally, the strong nonspecific nuclear immunofluorescence signal for FLCN that we observed in FLCN KO cells emphasizes the need for robust controls in immunofluorescence studies (both those that are directly related to FLCN as well as those in the larger field of cell biology).
In summary, our demonstration that RagA/BGDP is a major determinant of both FLCN–FNIP lysosome localization and Rag GTPase interactions that lies downstream of the GATOR1 GAP explains the starvation-dependent regulation of FLCN–FNIP protein recruitment to lysosomes. These findings also raise important new questions about how the structure of the FLCN–FNIP heterodimer supports interactions and functions of this complex toward RagA/B and RagC/D. For example, which parts of the complex are responsible for RagA/B binding versus stimulating GTP hydrolysis by RagC/D? Furthermore, how are such regions either similar to or different from those DENN domains that function as Rab GEFs? It will be additionally important to define how the Rag coordination mediated by FLCN–FNIP is integrated with findings from recent in vitro research concerning intersubunit crosstalk within the Rag heterodimers that pushes the Rag GTPases into two locked states in which one is GDP-bound and the other is GTP-bound (Shen et al., 2017). Efforts to address such mechanistic questions are well justified based on the evolutionarily conserved role played by FLCN–FNIP proteins in supporting the ability of cells to adapt their metabolism to changes in nutrient availability as well as important human health questions arising from the role played by FLCN as a tumor suppressor.
Materials and methods
Cell culture and transfection
HeLa M cells (provided by P. De Camilli, Yale University, New Haven, CT) and HEK293FT cells (Thermo Fisher Scientific) were grown in DMEM (high glucose, with l-glutamine), 10% FBS, and 1% penicillin/streptomycin supplement (all from Invitrogen). Where indicated, cells were starved by incubation in amino acid–free RPMI (US Biological) for 2 h. Amino acid refeeding was achieved by adding 1× MEM amino acid supplement (Invitrogen) into RPMI. Plasmid transfections were performed with 0.1 µg plasmid DNA, 0.3 µl Fugene 6 transfection reagent (Promega), and 20 µl Opti-MEM (Invitrogen) that was added to 500 µl media containing 20,000 cells per well in 24-well plates. siRNA transfections were performed with 7.5 µl 20 µM siRNA, 5 µl RNAiMAX transfection reagent (Invitrogen), and 500 µl Opti-MEM (Invitrogen) that was added to 2 ml media containing 120,000 cells per well in six-well plates. Experiments were performed 2 d after transfections. For transfection of larger dishes, the volumes of reagents and the number of cells were increased proportionally to surface area of the dish. Control siRNA was purchased from Integrated DNA Technologies (5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′), and RagC siRNA was purchased from GE Healthcare (5′-GCAAUUAUCAAGCUGAAUA-3′).
Lentiviral production and transduction
The lentivirus encoding HA-RagA was produced by transfecting HEK293FT cells with pxPAX2, pCMV-VSV-G, and pLenti-III-PGK using Fugure 6. Cell culture media containing the virus was collected 48 h after transfection and subsequently passed through a 0.45-µm filter before being added to HeLa RagA+B KO cells together with 8 µg/ml polybrene (EMD Millipore). 1 d after transduction, cells were selected with 2 µg/ml puromycin.
Plasmids
pRK5 plasmids encoding HA-GST–tagged human WT RagA, WT RagC, and mutant RagA (T21L) were acquired from D. Sabatini (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (Sancak et al., 2008). pRK5 HA-RagC was also acquired from D. Sabatini via Addgene (Shen et al., 2017). The PX459 V2.0 plasmid was acquired from F. Zhang (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (Ran et al., 2013). Sequences for guide RNAs that were inserted into this plasmid are summarized in Table S2. For packaging of lentivirus to generate cells that stably express HA-RagA, we used pCMV-VSV-G from B. Weinberg (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (Stewart et al., 2003) and PsPAX2 from D. Trono (École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) via Addgene. HA-RagA cDNA was cloned into the pLenti-III-PGK packaging vector (G305; ABM) after its amplification by PCR. Sequences for PCR primers are summarized in Table S4.
Antibodies
Antibodies used in this study are described in Table S5.
Immunoprecipitations and immunoblotting
Cells were lysed in TBS + 1% Triton X-100 + protease and phosphatase inhibitor cocktails (Roche), and insoluble material was removed by centrifugation for 6 min at 20,000 g. For pulldown of HA-GST-Rags, glutathione Sepharose 4B (GE Healthcare) was incubated with the lysates for 2 h while rotating samples at 4°C followed by washes with lysis buffer and elution in 2× Laemmli buffer. For anti-HA immunoprecipitation, the same procedure was performed with anti-HA affinity matrix (Roche) for 3 h. Immunoblotting was performed with 4–15% gradient Mini-PROTEAN TGX precast polyacrylamide gels and nitrocellulose membranes (Bio-Rad Laboratories). Blots were blocked with 5% milk, and antibodies were incubated with 5% milk or bovine serum albumin in TBS with 0.1% Tween 20. Chemiluminescent detection of HRP signals was performed via a VersaDoc imaging station (Bio-Rad Laboratories). ImageJ (National Institutes of Health) was used to quantify band intensities.
Immunofluorescence and microscopy
Immunofluorescent detection and confocal microscopy were performed largely as previously described (Petit et al., 2013). For detection of 2×HA-FNIP1, cells were grown on glass coverslips and fixed with 4% paraformaldehyde/0.1 M sodium phosphate buffer, pH 7.2, for 30 min. Cells were then permeabilized with PBS + 0.1% Triton X-100 for 10 min and then blocked with PBS + 5% normal donkey serum (Jackson ImmunoResearch Laboratories, Inc.) for 1 h. Subsequent antibody incubations were performed in this buffer. For detection of FLCN, antibody incubations were performed in PBS + 0.1% saponin + 10% FBS. Spinning-disk confocal microscopy was performed using the UltraView VoX system (PerkinElmer) including an inverted microscope (Ti-E Eclipse; Nikon; equipped with 60× CFI Plan Apochromat VC, 1.4 NA, oil immersion) and a spinning-disk confocal scan head (CSU-X1; Yokogawa) driven by Volocity (PerkinElmer) software. Colocalization of FLCN and 2×HA-FNIP1 with LAMP1- or HA-tagged Rags was determined by visual identification of FLCN/FNIP1 puncta followed by determination of LAMP1- or HA-tagged Rag colocalization on a spot-by-spot basis. In this strategy, five colocalized spots were set as the threshold for counting a cell as having lysosome-localized FLCN or FNIP1 as was previously described (Starling et al., 2016).
CRISPR-Cas9 genome editing
For insertion of a 2×HA epitope tag into the endogenous FNIP1 locus via CRISPR-Cas9–mediated genome editing, we designed an asymmetric single-stranded DNA donor with 36 bp on the protospacer-adjacent motif–distal side and 91 bp on the protospacer-adjacent motif–proximal side of the break (Richardson et al., 2016). We furthermore used preassembled Cas9 ribonucleoprotein complexes using recombinant protein and RNAs ordered from Integrated DNA Technologies (Lin et al., 2014). In brief, 100 pmol CRISPR RNA (crRNA), 100 pmol transactivating crRNA, 100 pmol single-stranded DNA repair template, and 100 pmol Cas9 nuclease 3×NLS protein were electroporated into one million HeLa M cells (Amaxa kit R, program A-24; Lonza). Clonal cell populations were screened for HA signal by immunoblotting. Genomic DNA surrounding the start codon of FNIP1 was amplified, cloned, and sequenced as previously described (Amick et al., 2016).
Use of the CRISPR-Cas9 genome editing for generating KO cell lines was mostly as described previously (Amick et al., 2016). Annealed guide RNA oligonucleotides (Integrated DNA Technologies) were designed with the help of the CRISPR design tool (http://crispr.mit.edu), cloned into the Bbs1-digested PX459 V2.0 vector, and transformed into Stbl3-competent Escherichia coli cells. After sequencing to confirm successful cloning, 1.0 µg PX459 V2.0 plasmid containing guide RNA was transfected into 100,000 HeLa M cells per well in a six-well plate. Cells were selected with 2 μg/ml puromycin for 2 d to kill nontransfected cells. After subsequent replating at single cell density, KO clonal cell lines were identified by Western blotting and confirmed by sequencing of PCR-amplified genomic DNA. Sequences for oligonucleotide primers that were used to amplify genomic DNA are summarized in Table S3.
Statistical analysis
Data were analyzed using Prism (GraphPad software), and tests are specified in the figure legends. All error bars represent SEM. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 presents validation of FLCN immunofluorescence via analysis of FLCN KO cells. Fig. S2 demonstrates the impact of siRNA-mediated RagC depletion on FLCN localization to lysosomes. Fig. S3 shows the effect of RagA+B KO on mTOR subcellular localization. Fig. S4 documents the regulation of mTOR signaling in NPRL3 KO cells. Table S1 provides genotyping information for KO cell lines. Table S2 defines oligonucleotide sequences relating to gene-editing experiments. Table S3 contains sequences for oligonucleotides for PCR. Table S4 contains oligonucleotide primers for PCR amplification of HA-RagA cDNA. Table S5 summarizes the antibodies that were used in this study.
Supplementary Material
Acknowledgments
We appreciate significant contributions from Agnes Ferguson relating to the establishment of key assays related to the investigation of FLCN subcellular localization and FLCN–Rag interactions as well as outstanding laboratory management.
Grants from the National Institutes of Health (GM105718 and AG047270) and the Ellison Medical Foundation to S.M. Ferguson provided financial support for this research.
The authors declare no competing financial interests.
Author contributions: J. Meng and S.M. Ferguson conceived of and designed all experiments, analyzed and interpreted data, and wrote the manuscript. J. Meng was responsible for performing all experiments.
References
- Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., and Ritter B.. 2010. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell. 37:370–382. 10.1016/j.molcel.2009.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amick J., Roczniak-Ferguson A., and Ferguson S.M.. 2016. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol. Biol. Cell. 27:3040–3051. 10.1091/mbc.e16-01-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Hong S.B., Sharma N., Warren M.B., Nickerson M.L., Iwamatsu A., Esposito D., Gillette W.K., Hopkins R.F. III, Hartley J.L., et al. 2006. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA. 103:15552–15557. 10.1073/pnas.0603781103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassari S., Licchetta L., Tinuper P., Bisulli F., and Pippucci T.. 2016. GATOR1 complex: the common genetic actor in focal epilepsies. J. Med. Genet. 53:503–510. 10.1136/jmedgenet-2016-103883 [DOI] [PubMed] [Google Scholar]
- Bar-Peled L., Schweitzer L.D., Zoncu R., and Sabatini D.M.. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150:1196–1208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., Spear E.D., Carter S.L., Meyerson M., and Sabatini D.M.. 2013. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 340:1100–1106. 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L., Rehmann H., and Wittinghofer A.. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 129:865–877. 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Chantranupong L., Wolfson R.L., Orozco J.M., Saxton R.A., Scaria S.M., Bar-Peled L., Spooner E., Isasa M., Gygi S.P., and Sabatini D.M.. 2014. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Reports. 9:1–8. 10.1016/j.celrep.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L., Scaria S.M., Saxton R.A., Gygi M.P., Shen K., Wyant G.A., Wang T., Harper J.W., Gygi S.P., and Sabatini D.M.. 2016. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 165:153–164. 10.1016/j.cell.2016.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., and Zeghouf M.. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93:269–309. 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Di Malta C., Siciliano D., Calcagni A., Monfregola J., Punzi S., Pastore N., Eastes A.N., Davis O., De Cegli R., Zampelli A., et al. 2017. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science. 356:1188–1192. 10.1126/science.aag2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong R., Li L., Liu Y., Wang P., Yang H., Wang L., Cheng J., Guan K.L., and Xu Y.. 2011. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 25:1668–1673. 10.1101/gad.16968011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H., Baba M., Hong S.B., Hasumi Y., Huang Y., Yao M., Valera V.A., Linehan W.M., and Schmidt L.S.. 2008. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 415:60–67. 10.1016/j.gene.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., and Guan K.L.. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10:935–945. 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Ro S.H., Kim M., Park H.W., Semple I.A., Park H., Cho U.S., Wang W., Guan K.L., Karin M., and Lee J.H.. 2015. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 5:9502 10.1038/srep09502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T.P., Daniels R.D., Gatta A.T., Wong L.H., and Hayes M.J.. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 29:499–503. 10.1093/bioinformatics/bts725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B.T., Alla R.K., and Doudna J.A.. 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 3:e04766 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., Dokainish H., and McPherson P.S.. 2011. DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 286:13791–13800. 10.1074/jbc.R110.217067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., and Puertollano R.. 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200:475–491. 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Lishu L., Jeong-A L., Patange S., Raben N., and Puertollano R.. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7:ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S., Dibble C.C., Talbott G., Hoxhaj G., Valvezan A.J., Takahashi H., Cantley L.C., and Manning B.D.. 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 156:771–785. 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson M.L., Warren M.B., Toro J.R., Matrosova V., Glenn G., Turner M.L., Duray P., Merino M., Choyke P., Pavlovich C.P., et al. 2002. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell. 2:157–164. 10.1016/S1535-6108(02)00104-6 [DOI] [PubMed] [Google Scholar]
- Nookala R.K., Langemeyer L., Pacitto A., Ochoa-Montaño B., Donaldson J.C., Blaszczyk B.K., Chirgadze D.Y., Barr F.A., Bazan J.F., and Blundell T.L.. 2012. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2:120071 10.1098/rsob.120071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacitto A., Ascher D.B., Wong L.H., Blaszczyk B.K., Nookala R.K., Zhang N., Dokudovskaya S., Levine T.P., and Blundell T.L.. 2015. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 5:150174 10.1098/rsob.150174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.P., and De Virgilio C.. 2013. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6:ra42 10.1126/scisignal.2004112 [DOI] [PubMed] [Google Scholar]
- Parmigiani A., Nourbakhsh A., Ding B., Wang W., Kim Y.C., Akopiants K., Guan K.L., Karin M., and Budanov A.V.. 2014. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Reports. 9:1281–1291. 10.1016/j.celrep.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péli-Gulli M.P., Sardu A., Panchaud N., Raucci S., and De Virgilio C.. 2015. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Reports. 13:1–7. 10.1016/j.celrep.2015.08.059 [DOI] [PubMed] [Google Scholar]
- Péli-Gulli M.P., Raucci S., Hu Z., Dengjel J., and De Virgilio C.. 2017. Feedback Inhibition of the Rag GTPase GAP Complex Lst4-Lst7 Safeguards TORC1 from Hyperactivation by Amino Acid Signals. Cell Reports. 20:281–288. 10.1016/j.celrep.2017.06.058 [DOI] [PubMed] [Google Scholar]
- Petit C.S., Roczniak-Ferguson A., and Ferguson S.M.. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202:1107–1122. 10.1083/jcb.201307084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., and Corn J.E.. 2016. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34:339–344. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 168:960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt L.S., and Linehan W.M.. 2018. FLCN: The causative gene for Birt-Hogg-Dubé syndrome. Gene. 640:28–42. 10.1016/j.gene.2017.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T., Hirose E., Nakashima N., Ii M., and Nishimoto T.. 2001. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276:7246–7257. 10.1074/jbc.M004389200 [DOI] [PubMed] [Google Scholar]
- Shen K., Choe A., and Sabatini D.M.. 2017. Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Mol. Cell. 68:552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Huang R.K., Brignole E.J., Condon K.J., Valenstein M.L., Chantranupong L., Bomaliyamu A., Choe A., Hong C., Yu Z., and Sabatini D.M.. 2018. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 556:64–69. 10.1038/nature26158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starling G.P., Yip Y.Y., Sanger A., Morton P.E., Eden E.R., and Dodding M.P.. 2016. Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep. 17:823–841. 10.15252/embr.201541382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A., et al. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 9:493–501. 10.1261/rna.2192803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M.Y., Morris K.L., Kim D.J., Fu Y., Lawrence R., Stjepanovic G., Zoncu R., and Hurley J.H.. 2017. Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex. Mol. Cell. 68:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y., Kobayashi T., Shiono M., Wang L., Piao X., Sun G., Zhang D., Abe M., Hagiwara Y., Takahashi K., and Hino O.. 2008. Interaction of folliculin (Birt-Hogg-Dubé gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 27:5339–5347. 10.1038/onc.2008.261 [DOI] [PubMed] [Google Scholar]
- Tsun Z.Y., Bar-Peled L., Chantranupong L., Zoncu R., Wang T., Kim C., Spooner E., and Sabatini D.M.. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell. 52:495–505. 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson R.L., and Sabatini D.M.. 2017. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 26:301–309. 10.1016/j.cmet.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson R.L., Chantranupong L., Saxton R.A., Shen K., Scaria S.M., Cantor J.R., and Sabatini D.M.. 2016. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 351:43–48. 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhou B., Oshiro-Rapley N., Li M., Paulo J.A., Webster C.M., Mou F., Kacergis M.C., Talkowski M.E., Carr C.E., et al. 2016. An Ancient, Unified Mechanism for Metformin Growth Inhibition in C. elegans and Cancer. Cell. 167:1705–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Bradley M.J., Cai Y., Kümmel D., De La Cruz E.M., Barr F.A., and Reinisch K.M.. 2011. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc. Natl. Acad. Sci. USA. 108:18672–18677. 10.1073/pnas.1110415108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q., Wang G., Wang H., Hu Q., and Ying Z.. 2016. Folliculin, a tumor suppressor associated with Birt-Hogg-Dubé (BHD) syndrome, is a novel modifier of TDP-43 cytoplasmic translocation and aggregation. Hum. Mol. Genet. 25:83–96. 10.1093/hmg/ddv450 [DOI] [PubMed] [Google Scholar]
- Yonehara R., Nada S., Nakai T., Nakai M., Kitamura A., Ogawa A., Nakatsumi H., Nakayama K.I., Li S., Standley D.M., et al. 2017. Structural basis for the assembly of the Ragulator-Rag GTPase complex. Nat. Commun. 8:1625 10.1038/s41467-017-01762-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., and Barr F.A.. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381. 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., He F., and Aravind L.. 2012. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front. Genet. 3:283 10.3389/fgene.2012.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wang R., Wang Z., Wang X., Wang F., and Ding J.. 2017. Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat. Commun. 8:1394 10.1038/s41467-017-01567-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.