The IFT-B complex powered by kinesin-II is believed to be responsible for anterograde ciliary protein trafficking. Funabashi et al. identify composite interactions of the kinesin-II heterotrimer KIF3A–KIF3B–KAP3 with the IFT-B–connecting tetramer IFT38–IFT52–IFT57–IFT88 and show that their interaction is crucial for ciliogenesis.
Abstract
Intraflagellar transport (IFT) is crucial for the assembly and maintenance of cilia and is mediated by IFT particles containing IFT-A and IFT-B complexes. IFT-B powered by heterotrimeric kinesin-II and IFT-A powered by the dynein-2 complex are responsible for anterograde and retrograde protein trafficking, respectively. However, little is known about the molecular basis of the trafficking of these IFT particles regulated by kinesin and dynein motors. Using the visible immunoprecipitation assay, we identified in this study a three-to-four protein interaction involving the kinesin-II trimer KIF3A–KIF3B–KAP3 and the IFT-B–connecting tetramer IFT38–IFT52–IFT57–IFT88; among the kinesin-II subunits, KIF3B contributed mainly to IFT-B binding. Furthermore, we showed that the ciliogenesis defect of KIF3B-knockout cells can be rescued by the exogenous expression of wild-type KIF3B but not by that of its mutant compromised with respect to IFT-B binding. Thus, interaction of heterotrimeric kinesin-II with the IFT-B–connecting tetramer is crucial for ciliogenesis via the powering of IFT particles to move in the anterograde direction.
Introduction
Cilia are axonemal microtubule-based protrusions that are found on the surface of most eukaryotic cells and serve as cellular antennae by mechanosensation of extracellular stimuli, such as fluid flow, and chemosensation of developmental signals, such as the Hedgehog signal (Mukhopadhyay and Rohatgi, 2014; Bangs and Anderson, 2017). The crucial roles of cilia as cellular antennae have been highlighted by the fact that defects in the assembly and functions of cilia cause a wide spectrum of congenital disorders, collectively called the ciliopathies (Braun and Hildebrandt, 2017).
Specific proteins including various G protein–coupled receptors and ion channels exist on the ciliary membrane (Mukhopadhyay and Rohatgi, 2014). However, for the assembly and maintenance of cilia, tubulins must be continuously trafficked toward the distal tip (Bhogaraju et al., 2014). Although the ciliary membrane is continuous with the plasma membrane, the ciliary interior and ciliary membrane are segregated from the cytoplasm and plasma membrane by the presence of the transition zone, which serves as a diffusion and permeability barrier (Wei et al., 2015; Verhey and Yang, 2016). Therefore, the biogenesis and functions of cilia are dependent not only on the proper bidirectional trafficking of ciliary proteins but also on their proper entry and exit across the transition zone.
Bidirectional trafficking of ciliary proteins along the axonemal microtubules, and probably their ciliary entry and exit, are mediated by intraflagellar transport (IFT) particles containing two large multisubunit complexes, IFT-A and IFT-B, composed of 6 and 16 subunits, respectively (Taschner and Lorentzen, 2016; Prevo et al., 2017; Nakayama and Katoh, 2018). We and others recently demonstrated that the IFT-B complex can be divided into the core (B1) and peripheral (B2) subcomplexes, which are connected by composite interactions involving IFT38–IFT52–IFT57–IFT88 (Boldt et al., 2016; Katoh et al., 2016; Taschner et al., 2016). The IFT-B complex is responsible for anterograde protein trafficking from the ciliary base to the tip powered by kinesin-2 motor proteins, whereas the IFT-A complex mediates retrograde trafficking powered by dynein-2 (Prevo et al., 2017). Deficiency in any of the IFT-B subunits often leads to extremely short or no cilia, indicating a crucial role of the trafficking of ciliary proteins, particularly tubulins, in ciliary assembly (Lechtreck, 2015; Taschner and Lorentzen, 2016; Nakayama and Katoh, 2018).
Two different types of kinesin-2 motors have been implicated in ciliary anterograde protein trafficking: heterotrimeric kinesin-II composed of the motor subunits KIF3A and KIF3B and the accessory protein KAP3 (also known as KIFAP3), and homodimeric KIF17 (Scholey, 2013). In vertebrates, heterotrimeric kinesin-II appears to contribute mainly to anterograde trafficking. Furthermore, in a fla10/kif3a mutant strain of Chlamydomonas reinhardtii, at the nonpermissive temperature, movement of IFT particles ceases and flagella are shortened (Kozminski et al., 1995), and levels of the IFT proteins within flagella are decreased (Cole et al., 1998). In contrast, the role of KIF17 is unclear, at least in vertebrates, as KIF17-knockout (KIF17-KO) mice and zebrafish are viable with a marginal defect in ciliogenesis (Yin et al., 2011; Pooranachandran and Malicki, 2016). Furthermore, we have recently shown that ciliogenesis and the trafficking of most ciliary proteins appear normal in KIF17-KO cells and that KIF17 is a cargo of, rather than a motor for, the IFT-B complex; namely, it is imported into cilia by binding to the IFT-B complex via the IFT46–IFT56 dimer (Funabashi et al., 2017). However, the molecular basis for the interaction between the IFT-B complex and heterotrimeric kinesin-II has been poorly understood to date, although Kif3-KO mice were reported ∼20 yr ago to show ciliary morphogenesis defects (Nonaka et al., 1998; Marszalek et al., 1999). We therefore aimed to determine how heterotrimeric kinesin-II regulates IFT-B trafficking. In this study, we identified a composite interaction involving the kinesin-II trimer KIF3A–KIF3B–KAP3 and the IFT-B–connecting tetramer IFT38–IFT52–IFT57–IFT88 and show that exogenous expression of a KIF3B mutant compromised in IFT-B binding cannot restore the ciliogenesis defect observed in KIF3B-KO cells.
Results
KIF3B is an essential component of the kinesin-II trimer
We first aimed to clarify how the subunits of heterotrimeric kinesin-II are bound to each other using the visible immunoprecipitation (IP; VIP) assay, which we recently developed as a simple and versatile assay for protein–protein interactions, in which not only binary but also one-to-many and many-to-many protein interactions can be visually detected without the need for electrophoresis and immunoblotting (Katoh et al., 2015, 2016).
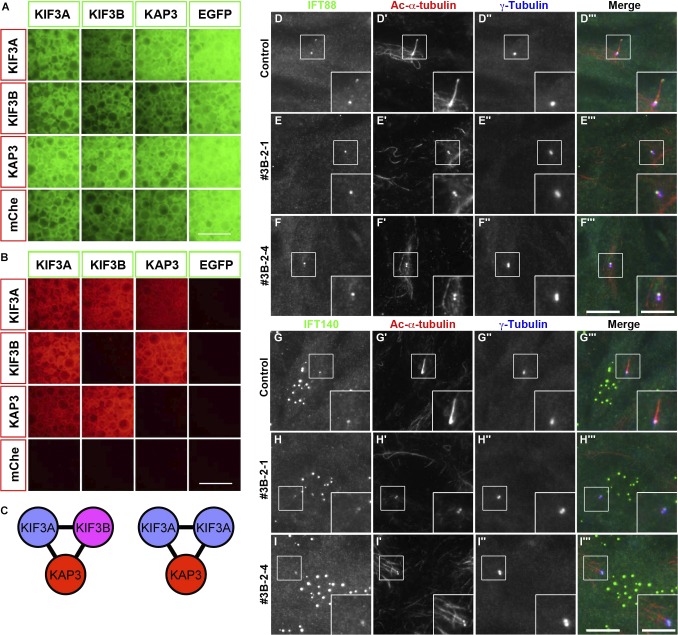
Lysates prepared from cells coexpressing EGFP-fused KIF3A, KIF3B, or KAP3 and its mCherry (mChe)–fused constructs in all possible combinations and processed them for IP with GST-tagged anti–GFP nanobodies (Nbs) prebound to glutathione-Sepharose beads. As depicted by red signals on the precipitated beads under a conventional fluorescence microscope, KIF3A interacted with all proteins (KIF3A, KIF3B, and KAP3; Fig. 1, A and B). However, KIF3B interacted with KIF3A and KAP3, but not with itself. These results demonstrate that in addition to the heterotrimer composed of KIF3A, KIF3B, and KAP3 (Fig. 1 C, left), the kinesin-II trimer can also comprise two KIF3A molecules and one KAP3 molecule without the need for KIF3B (Fig. 1 C, right).
Figure 1.
KIF3B is an essential component of the kinesin-II trimer. (A and B) Lysates prepared from HEK293T cells coexpressing the indicated combination of EGFP-fused and mChe-fused kinesin-II subunits were processed for the VIP assay using GST–anti-GFP Nb prebound to glutathione–Sepharose beads. EGFP (A) and mChe (B) signals on the precipitated beads were observed under a microscope. Bars, 100 µm. (C) Kinesin-II models predicted from the binary interaction data. (D–I) Control RPE1 cells (D and G) and KIF3B-KO cell lines 3B-2-1 (E and H) and 3B-2-4 (F and I) were serum-starved to induce ciliogenesis and triple immunostained for either IFT88 (D–F) or IFT140 (G–I), together with Ac-α-tubulin (D′–I′) and γ-tubulin (D″–I″). Bars: (main images) 10 µm; (insets) 5 µm.
To address whether KIF3B is an essential component of the kinesin-II trimer at the cellular level, we established hTERT-RPE1 cell lines defective in KIF3B, although a previous study showed that Kif3b-KO mice are embryonic lethal (Nonaka et al., 1998). Among the several independent KIF3B-KO cell lines established using a CRISPR/Cas9 system with our original modifications (Katoh et al., 2017; Materials and methods), we used two KO cell lines in the following experiments: the 3B-2-1 cell line has a 1-nt deletion in one KIF3B allele and a forward integration of the donor knock-in vector in the other allele (Fig. S1 A, lanes 5–7; and Fig. S1 B), and 3B-2-4 has an 8-nt deletion in one allele and a forward integration of the donor vector in the other allele (Fig. S1 A, lanes 8–10; and Fig. S1 C).
As expected from the phenotype of KO mice (Nonaka et al., 1998), no cilia were observed in these KIF3B-KO cell lines (Fig. 1, D–I; compare E′, F′, H′, and I′ with D′ and G′). Thus, KIF3A is unable to functionally compensate for the absence of KIF3B, at least with respect to ciliary protein trafficking, although KIF3A constructs engineered to form a homodimer were reported to undergo processive movement along microtubules in vitro (Andreasson et al., 2015; Guzik-Lendrum et al., 2015). The phenotype of KIF3B-KO cells was in striking contrast with that of KIF17-KO cells, in which ciliogenesis did not appear to be impaired (Funabashi et al., 2017).
When the KIF3B-KO cells were immunostained for IFT88 and IFT140, which are subunits of the IFT-B and IFT-A complexes, respectively, both IFT88 (Fig. 1, E and F) and IFT140 (Fig. 1, H and I) were localized on one of two γ-tubulin–positive centrioles; note that as described previously (Hirano et al., 2017; Takahara et al., 2018), the available anti–IFT140 antibody also stained undetermined nuclear structures in RPE1 cells (17460-1-AP; ProteinTech). These observations suggest that both the IFT-A and IFT-B complexes, and thereby the IFT particles, can be assembled around the basal body in the absence of cilia because of the lack of functional kinesin-II. The observations are also in compatible with previous studies on a C. reinhardtii fla10/kif3a mutant strain that flagella are shortened and flagellar levels of the IFT proteins are decreased at the nonpermissive temperature (Kozminski et al., 1995; Cole et al., 1998) and with a recent study indicating that IFT proteins are recruited to the basal body before recruitment of motor proteins in C. reinhardtii flagella (Wingfield et al., 2017).
Heterotrimeric kinesin-II interacts with the IFT-B complex via the IFT-B–connecting tetramer
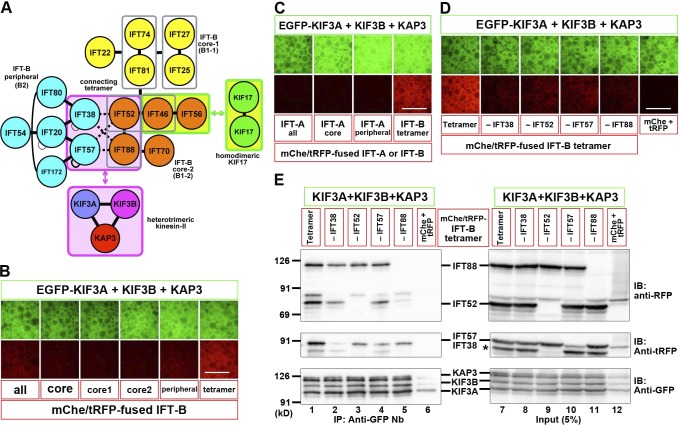
We then addressed whether heterotrimeric kinesin-II can interact with the IFT-B complex as there are virtually no direct data regarding their mode of interaction, although C. reinhardtii kinesin-II was reported to coimmunoprecipitate with components of the IFT-B complex (Liang et al., 2014). Furthermore, a yeast two-hybrid study performed more than 10 yr ago suggested an interaction between KIF3B and IFT20 (Baker et al., 2003), but our preliminary VIP-based analysis did not detect this interaction. We therefore took advantage of the VIP assay for many-to-many protein interactions (Katoh et al., 2016) to reveal the interaction mode between the kinesin-II heterotrimer and the IFT-B complex. When lysates of HEK293T cells coexpressing all kinesin-II subunits fused to EGFP and all IFT-B subunits (16 subunits) fused to mChe or TagRFP (tRFP) were immunoprecipitated with GST–anti-GFP Nb, weak but substantial red signals were observed on the precipitated beads (Fig. 2 B, leftmost row), indicating that kinesin-II interacts with IFT-B. When the IFT-B subunits were grouped into those of the core (B1) subcomplex, peripheral (B2) subcomplex, and the connecting tetramer (IFT38–IFT52–IFT57–IFT88) that constitutes the interface between the core and peripheral subcomplexes (Katoh et al., 2016) and expressed as mChe or tRFP fusions, relatively strong red signals were detected when subunits of the connecting tetramer were coexpressed with EGFP-fused kinesin-II subunits (Fig. 2 B, rightmost row).
Figure 2.
Interaction of heterotrimeric kinesin-II with the IFT-B–connecting tetramer. (A) A model for the interaction of IFT-B with homodimeric KIF17 and heterotrimeric kinesin-II. (B) Identification of an interaction between heterotrimeric kinesin-II and the IFT-B–connecting tetramer. Lysates from cells coexpressing EGFP-fused kinesin-II subunits and all IFT-B subunits, subunits of all core, core 1, core 2, or peripheral subcomplexes, or connecting tetramer fused to mChe/tRFP were processed for the VIP assay. (C) Lack of an interaction between the IFT-A complex and heterotrimeric kinesin-II. Lysates from cells coexpressing EGFP-fused kinesin-II subunits and mChe-fused IFT-A subunits (all, core, or peripheral) were subjected to the VIP assay. (D and E) Subtractive VIP assay and immunoblotting (IB) analysis to determine subunits of the IFT-B–connecting tetramer required for its interaction with kinesin-II. Lysates from cells coexpressing EGFP-fused kinesin-II subunits and all but one (as indicated) subunits of the IFT-B tetramer fused to mChe/tRFP were processed for the VIP assay (D) or immunoblotting analysis (E) using the following antibodies: an anti-RFP antibody (top), which reacts with mChe; an anti-tRFP antibody (middle), which reacts with tRFP and cross-reacts with the mChe portion of mChe-IFT52 (indicated by an asterisk); or an anti-GFP antibody (bottom). Note that the bands for tRFP-fused IFT38 and IFT57 were overlapped with each other. As a negative control, a mixture of mChe-fused and tRFP-fused IFT56 was used in place of mChe/tRFP-fused tetrameric subunits (labeled as mChe+tRFP). Bars, 100 µm.
We also analyzed whether kinesin-II can interact with the IFT-A complex as it was proposed that in the sensory cilia of Caenorhabditis elegans the IFT-A complex moves along the axonemal middle segment together with kinesin-II (Ou et al., 2005), although there was no direct evidence for the IFT-A–kinesin-II interaction. However, we could not detect an interaction of the kinesin-II trimer with all, core, or peripheral subunits of the IFT-A complex (Fig. 2 C).
We then used the subtractive VIP assay to determine which subunits in the IFT-B tetramer are important for its interaction with kinesin-II. As shown in Fig. 2 D, omitting any one subunit of the tetramer reduced the red signals, indicating that all the tetramer components make substantial contributions to the kinesin-II interaction. The VIP results were confirmed by conventional immunoblotting analysis (Fig. 2 E). The band intensities were moderately reduced by omitting any one of the tetrameric subunits (top and middle; compare lanes 2–5 with lane 1), most prominently by omitting IFT88 (lane 5); note that the bands for tRFP-IFT38 and tRFP-IFT57 overlapped with each other as described previously (Katoh et al., 2016).
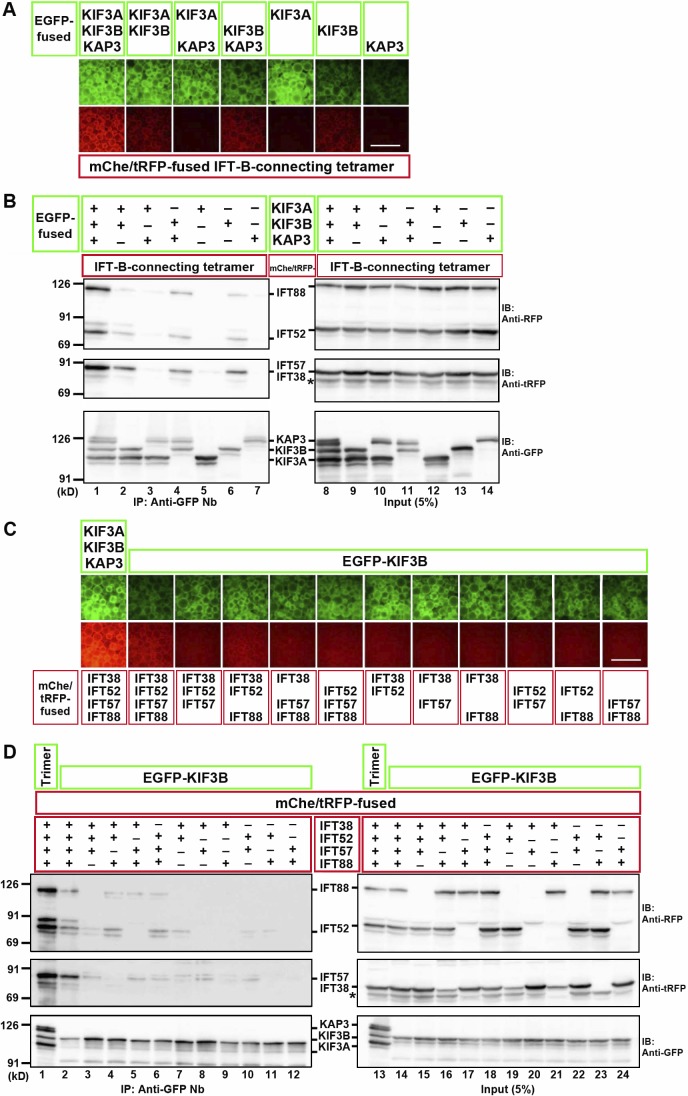
We then aimed to identify which subunits of the kinesin-II trimer are important for its interaction with the IFT-B–connecting tetramer. As shown in Fig. 3 A, although the interaction was strongest when all the kinesin-II subunits were coexpressed, a substantial interaction was detected when KIF3B was expressed on its own or in combination with KIF3A or KAP3. Immunoblotting analysis confirmed the VIP results (Fig. 3 B); intense bands for the IFT-B tetrameric subunits were detected when all kinesin-II subunits were present (top and middle, lane 1), and relatively weak bands were detected when KIF3B was included (lanes 2, 4, and 6).
Figure 3.
KIF3B mainly contributes to the interaction with the IFT-B–connecting tetramer. (A and B) Subtractive VIP assay and immunoblotting (IB) analysis to determine subunits of kinesin-II required for its interaction with the IFT-B tetramer. (C and D) Subtractive VIP assay and immunoblotting analysis to determine subunits of the IFT-B–connecting tetramer required for its interaction with KIF3B. Details are essentially the same as described in the legend for Fig. 2 (C and D), except that exposure time for red fluorescence was 0.2 s. Bars, 100 µm. In B and D, asterisks indicate the position of mChe-IFT52, with which anti–tRFP antibody cross-reacted.
We also attempted to determine which subunits of the IFT-B–connecting tetramer are important for its interaction with KIF3B. As shown in Fig. 3 (C and D), omitting any one subunit of the connecting tetramer moderately reduced the interaction with KIF3B (compare lane 2 with lanes 3–6), and omitting two subunits, particularly omitting combinations including IFT52 (lanes 8, 9, and 12), greatly reduced the interaction. It is noteworthy that the interactions involving KIF3B and any two or three subunits of the IFT-B–connecting tetramer were extremely weak compared with the interaction involving the kinesin-II trimer and all the IFT-B tetrameric subunits (compare lane 1 with the other lanes).
Thus, a maximal interaction between the kinesin-II and IFT-B complexes requires the three-to-four protein interaction involving all the kinesin-II subunits and all the connecting IFT-B subunits (Fig. 2 A); however, among the kinesin-II trimer, KIF3B appears to make a major contribution to IFT-B binding. This is also consistent with the fact that KIF3A cannot compensate for the absence of KIF3B. However, we were unable to determine the most important subunit in the IFT-B–connecting tetramer in the interaction with kinesin-II.
Regions of the KIF3B protein required for its interactions with KIF3A, KAP3, and IFT-B can be dissected
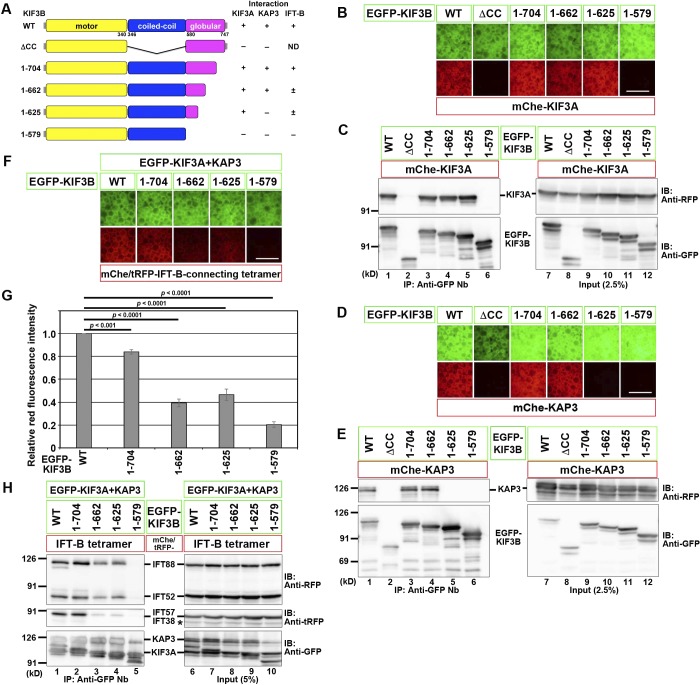
In an attempt to discriminate between the region of KIF3B responsible for the formation of the kinesin-II heterotrimer and that required for the kinesin-II–IFT-B interaction, we constructed various deletion and truncation mutants of KIF3B. When the coiled-coil (CC) region was deleted (ΔCC; Fig. 4 A), KIF3B lost its ability to interact with both KIF3A and KAP3 (Fig. 4, B–E, lane 2), confirming that the CC region is essential for formation of the kinesin-II trimer. When truncated from the C terminus, KIF3B(1–704) and KIF3B(1–662) retained the ability to interact with both KIF3A and KAP3 (lanes 3 and 4). However, KIF3B(1–625) lost the ability to interact with KAP3 but not with KIF3A (lane 5), whereas KIF3B(1–579), which lacks the entire globular domain (Fig. 4 A), lost the ability to interact with both KIF3A and KAP3 (lane 6). Thus, the interaction of KIF3B with both KIF3A and KAP3 requires its CC region and at least the N-terminal part of the globular domain, although overlapping but distinct regions of KIF3B are required for its interaction with KIF3A and KAP3. These results are compatible with previous studies based on single-particle electron microscopic analysis of recombinant KIF3A–KIF3B and KIF3A–KIF3B–KAP3, suggesting that the KIF3A–KIF3B heterodimer appears to have two heads, a middle rod, and a globular tail, and KAP3 appears to wrap around the latter (Yamazaki et al., 1995, 1996).
Figure 4.
Regions of the KIF3B protein responsible for its interactions with KIF3A, KAP3, and the IFT-B–connecting tetramer. (A) Schematic representation of the KIF3B constructs used and their domain organizations. On the right side, interactions of these constructs with KIF3A, KAP3, and the IFT-B–connecting tetramer are summarized. +, strong interaction; ±, weak interaction; −, no interaction; ND, not determined. (B–F and H) VIP assay (B, D, and F) and immunoblotting (IB) analysis (C, E, and H) to determine the regions of KIF3B responsible for its interaction with KIF3A (B and C), KAP3 (D and E), or the IFT-B–connecting tetramer (F and H). Bars, 100 µm. In H, an asterisk indicates the position of mChe-IFT52, with which anti–tRFP antibody cross-reacted. (G) Red fluorescence intensities in the acquired images shown in F were measured using ImageJ, and relative fluorescence intensities are expressed as bar graphs. Values are means ± SD of three independent experiments. P-values were determined by one-way ANOVA followed by Tukey’s post hoc analysis.
We then analyzed whether any of the EGFP-fused KIF3B truncation constructs together with EGFP-fused KIF3A and KAP3 can interact with the IFT-B–connecting tetramer; all the tetrameric subunits were fused to mChe or tRFP (Fig. 4, F–H); relative red fluorescence intensities in the acquired images were quantitatively analyzed (Fig. 4 G). KIF3B(1–704) (lane 2) interacted with the IFT-B tetramer, comparably with KIF3B(WT) (lane 1). In striking contrast, KIF3B(1–579), which cannot interact with KIF3A or KAP3, no longer interacted with the IFT-B tetramer (lane 5). In the case of KIF3B(1–662), which retained its ability to interact with both KIF3A and KAP3, and KIF3B(1–625), which retained its ability to interact with only KIF3A, the interaction with the IFT-B tetramer was moderately attenuated (lanes 3 and 4); in particular, there was a prominent reduction in the amount of coimmunoprecipitated IFT38–IFT57. Thus, even though KIF3B(1–662) appears to retain the ability to form the kinesin-II heterotrimer, it is partially compromised with respect to its interaction with the IFT-B complex.
Phenotype of KIF3B-KO cells exogenously expressing WT or mutant KIF3B
We then analyzed whether the exogenous expression of KIF3B(WT) or its truncation mutants characterized above (Fig. 4) can rescue the no-cilia phenotype of the KIF3B-KO cell lines. As shown in Fig. 5 (A–A′′′), the exogenous expression of EGFP-fused KIF3B(WT) in the KIF3B-KO cell line 3B-2-1 restored ciliogenesis, although the localization of KIF3B(WT)-EGFP around the ciliary base was unclear because of the relatively high background signal throughout the cytoplasm (Fig. 5 A′′′). Similarly, KIF3B(1–704)-EGFP, which retains the ability to form the kinesin-II heterotrimer and interact with the IFT-B complex, also rescued the ciliogenesis defect (Fig. 5 B). In striking contrast, exogenous expression of KIF3B(1–662), which retained the ability to form the kinesin-II heterotrimer (Fig. 4, A–E) but demonstrated a partially compromised IFT-B binding (Fig. 4, F–H), failed to rescue the ciliogenesis defect of the KIF3B-KO cell line (Fig. 5 C). In addition, neither KIF3B(1–625) nor KIF3B(1–579) rescued the KIF3B-KO phenotype (Fig. 5, D and E). Essentially the same results were obtained using the other KO cell line, 3B-2-4 (Fig. 5 G). It is unlikely that the C-terminal truncation of KIF3B affected its motor activity as Albracht et al. (2014) and Guzik-Lendrum et al. (2015) recently reported that an artificial heterodimer of the KIF3A and KIF3B constructs, both of which lacked the entire globular domain and the majority of the CC region, retained the ability to undergo processive movement along microtubules in vitro. Collectively, these results indicate that the binding of kinesin-II to the IFT-B complex via the connecting tetramer is crucial for ciliogenesis.
Figure 5.
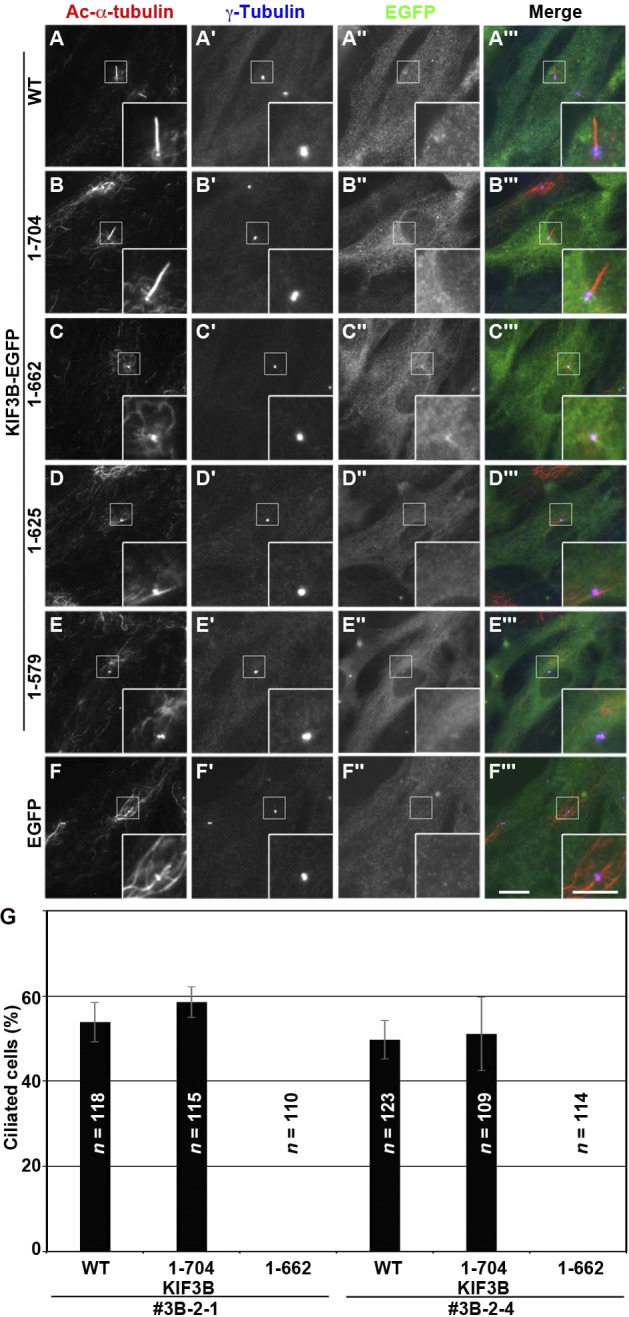
KIF3B–IFT-B interaction is essential for ciliogenesis. (A–F) The KIF3B-KO cell line 3B-2-1 stably expressing EGFP-fused KIF3B(WT) (A), KIF3B(1–704) (B), KIF3B(1–662) (C), KIF3B(1–625) (D), KIF3B(1–579) (E), or EGFP (F) were triple immunostained for Ac-α-tubulin (A–F), γ-tubulin (A′–F′), and GFP (A″–F″). Bars: (main images) 10 µm; (insets) 5 µm. (G) Ciliated cells of the KIF3B-KO cell lines (3B-2-1 and 3B-2-4) stably expressing EGFP-fused KIF3B(WT), KIF3B(1–704), or KIF3B(1–662) were counted, and percentages of ciliated cells are represented as bar graphs. Values are shown as means ± SEM of three independent experiments. In each set of experiments, 31–50 cells with EGFP signals were analyzed, and the total number of cells analyzed (n) for each sample is shown.
Discussion
Anterograde ciliary protein trafficking is believed to be mediated by the IFT-B complex powered by the kinesin-2 motor, whereas retrograde trafficking is thought to be mediated by the IFT-A complex powered by dynein-2. We answered in this study the long-standing question since the discovery that Kif3-KO mice demonstrate ciliary morphogenesis defects (Nonaka et al., 1998; Marszalek et al., 1999); namely, the molecular basis for the trafficking of IFT particles driven by kinesin-2. By taking advantage of the flexible VIP-based method, we unraveled the interaction between heterotrimeric kinesin-II and the IFT-B complex. The most robust interaction was observed between the kinesin-II trimer and the IFT-B–connecting tetramer composed of IFT38–IFT52–IFT57–IFT88 (Fig. 2 A). The subtractive VIP assay demonstrated that omitting any one of the kinesin-II subunits or any of the IFT-B–connecting subunits moderately attenuates the interaction (Fig. 2, D and E; and Fig. 3, A–D). We previously showed that KIF17 interacts with the IFT-B complex via the IFT46–IFT56 dimer (Fig. 2 A) and does not act as a motor for IFT-B; rather, it is a cargo of IFT-B (Funabashi et al., 2017). Thus, our previous and present studies unequivocally show that homodimeric KIF17 and heterotrimeric kinesin-II interact with distinct parts of the IFT-B complex (Fig. 2 A) and play distinct roles within cilia.
Because the VIP-based methods are coimmunoprecipitation assays of proteins exogenously coexpressed in cells, it is an important issue to be addressed whether the interaction identified by the VIP-based methods occurs between endogenous proteins. A previous study on C. reinhardtii FLA8/KIF3B showed that HA-tagged FLA8/KIF3B can coimmunoprecipitate the endogenous IFT-B proteins only from a flagellar fraction (Liang et al., 2014), although they did not show the interaction mode between kinesin-II and IFT-B. However, our attempts to detect an interaction of endogenous or exogenously expressed kinesin-II with IFT-B have been unsuccessful so far. Fractions of cellular kinesin-II and IFT-B that participate in their interaction within cilia might be very small, thereby resulting in our failed detection of the interaction of the endogenous proteins.
Analysis of KIF3B truncation mutants expressed in KIF3B-KO cells demonstrated that the exogenous expression of KIF3B(1–704), which retains its ability to form the kinesin-II trimer and interact with the IFT-B tetramer but not the expression of KIF3B(1–662), which can also form a trimer but demonstrates a compromised IFT-B interaction, is able to rescue the ciliogenesis defect caused by the loss of KIF3B. Taking into account that the ciliogenesis defect reflects a lack of the trafficking of tubulins, which are building blocks of axonemal microtubules, and that the IFT-B complex is responsible for tubulin trafficking (Lechtreck, 2015; Taschner and Lorentzen, 2016), a straightforward explanation for the observations in the KIF3B(1–662) rescue experiment would be that the “no-cilia” phenotype was caused by uncoupling of the IFT-B complex from the KIF3B(1–662) –containing kinesin-II trimer. Of course, it remains possible that an unidentified protein that can interact with the C-terminal region of KIF3B is implicated in ciliogenesis because the loss of the IFT-B–binding ability of KIF3B(1–662) was not complete. However, we believe it unlikely that some unknown protein binds to the KIF3B C-terminal region as previous single-particle analyses of recombinant KIF3A–KIF3B and KIF3A–KIF3B–KAP3 by electron microscopy suggested that KAP3 covers the globular tail region of the KIF3A–KIF3B dimer (Yamazaki et al., 1995, 1996).
The IFT-B–connecting tetramer constitutes the interface between the core and peripheral subcomplexes (Taschner and Lorentzen, 2016; Nakayama and Katoh, 2018): IFT52 and IFT88 from the core subcomplex, and IFT38 and IFT57 from the peripheral subcomplex (Fig. 2 A). Given that the kinesin-II trimer interacts with the connecting tetramer, the hypothesis that kinesin-II triggers the complete IFT-B assembly by clamping the core and peripheral subcomplexes and thereby serves as a cue for the anterograde trafficking of IFT particles at the ciliary base is somewhat intriguing. However, our preliminary analyses did not detect an enhancement of the interaction between the IFT-B core and peripheral subcomplexes in the presence of kinesin-II. In view of the recent study on C. reinhardtii flagella indicating that IFT proteins are first recruited to the basal body and in turn kinesin-II is recruited (Wingfield et al., 2017), it is likely that the binding of kinesin-II to preassembled IFT particles triggers entry into cilia and the subsequent anterograde trafficking of IFT particles.
Liang et al. (2014) indicated that calcium-dependent phosphorylation of FLA8 at a serine residue in the globular domain disrupts the interaction between kinesin-II and IFT-B and inhibits IFT entry. However, our preliminary analysis using phosphomimetic mutants of human KIF3B has so far failed to show a decrease in the kinesin-II–IFT-B interaction. Although components involved in IFT are highly conserved between C. reinhardtii and vertebrates, the detailed mechanism underlying regulation of the kinesin-II–IFT-B interaction might be different.
Materials and methods
Plasmids, antibodies, and reagents
cDNAs for human KIF3A (clone ID: IRAK106P17) and KAP3 (clone ID: IRAK034F02) were provided by the RIKEN BioResource Research Center through the National BioResource Project of the MEXT/AMED, Japan, and human KIF3B cDNA (clone ID: ORK00515) was provided by the Kazusa DNA Research Institute, Japan. Expression vectors for kinesin-2 and IFT proteins as well as their deletion constructs used in this study are listed in Table S1; many of them were constructed in our previous studies (Katoh et al., 2016; Funabashi et al., 2017; Hirano et al., 2017). The following antibodies were used in this study: monoclonal mouse anti–Ac-α-tubulin (6-11-B-1) and anti–γ-tubulin (GTU88; Sigma-Aldrich), polyclonal rabbit anti-IFT88 (13967-1-AP) and anti-IFT140 (17460-1-AP; ProteinTech), polyclonal rabbit anti-GFP (A11122; Molecular Probes), monoclonal mouse anti-GFP (JL-8; BD), polyclonal rabbit anti-RFP (PM005; MBL Life Science), polyclonal rabbit anti-tRFP (AB233; Evrogen), and Alexa Fluor–conjugated secondary antibodies (A11034, A21240, and A21147; Molecular Probes). GST-tagged anti–GFP Nb prebound to glutathione–Sepharose 4B beads were prepared as described previously (Katoh et al., 2015).
VIP assay and immunoblotting analysis
The VIP assay and subsequent immunoblotting analysis were performed as described previously (Katoh et al., 2015, 2016) with a minor modification (Nishijima et al., 2017); a detailed protocol for the VIP assay has been recently described (Katoh et al., 2018). HEK293T cells on a six-well plate (∼1.6 × 106 cells) were cotransfected with expression vectors for EGFP and mChe or tRFP fusion constructs using 20 µg polyethylenimine max (Polysciences) and cultured for 24 h in DMEM with high glucose (Nacalai Tesque) supplemented with 5% FBS. The transfected cells were lysed in 250 µl HMDEKN cell-lysis buffer (10 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 25 mM KCl, and 0.5% NP-40) containing protease inhibitor cocktail (Nacalai Tesque). After 15 min on ice, lysates were centrifuged at 16,100 g for 15 min. The supernatant (200 µl) was transferred to a 0.2-ml eight-tube strip containing GST-tagged anti–GFP Nb bound to glutathione–Sepharose 4B beads (∼5 µl bed volume of the beads) and incubated for 1 h at 4°C with constant rotation of the tube. After centrifugation of the tube at 2,000 g for 30 s, the precipitated beads were washed three times with 180 µl lysis buffer, transferred to a 96-well plate, and observed using an all-in-one type microscope (BZ-8000; Keyence) with a 20× 0.75 NA objective lens under fixed conditions (sensitivity ISO 400; exposure 1/30 s for green fluorescence; and sensitivity ISO 800, exposure 1/10 s for red fluorescence), unless otherwise noted.
The beads bearing fluorescent fusion proteins were then processed for conventional immunoblotting analysis. Proteins on the beads were separated by SDS-PAGE and electroblotted onto an Immobilon-P membrane (EMD Millipore). The membrane was blocked in 5% skimmed milk and incubated sequentially with primary antibody and peroxidase-conjugated secondary antibody. Protein bands were detected using a Chemi-Lumi One L kit (Nacalai Tesque).
Establishment of KO cell lines using the CRISPR/Cas9 system
The strategy for the KO of genes in hTERT-RPE1 cells (CRL-4000; ATCC) using the CRISPR/Cas9 system using homology-independent DNA repair (version 1 method) was described in detail previously (Katoh et al., 2017). The single gRNA sequence targeting the human KIF3B gene (Table S2) was designed using CRISPRscan (Moreno-Mateos et al., 2015). Double-stranded oligonucleotides for the sequence were inserted into an all-in-one single gRNA expression vector (pSpCas9(BB)-2A-Puro; PX459; 48139; Addgene). hTERT-RPE1 cells were grown on a 12-well plate to ∼3.0 × 105 cells and transfected with 1 µg single gRNA vector and 0.25 µg donor knock-in vector (pDonor-tBFP-NLS-Neo; 80766; Addgene) using X-tremeGENE9 DNA transfection reagent (Roche). After selection in the presence of 600 µg/ml G418, cells with nuclear tBFP signals were isolated. To confirm KO of the KIF3B gene, genomic DNA was extracted from the isolated cells and subjected to PCR using KOD FX Neo DNA polymerase (Toyobo). Three sets of primers (Table S2) were used for PCR to distinguish the following three states of integration of the donor vector: forward integration, reverse integration, and no integration with a small indel (Fig. S1 A). Direct sequencing of the PCR products ensured the KO of both alleles of the KIF3B gene; a small deletion causing a frameshift in one allele and a forward integration of the donor vector in the other allele (Fig. S1, B and C).
Preparation of cells stably expressing EGFP-KIF3B
Lentiviral vectors for the expression of KIF3B constructs were prepared as described previously (Takahashi et al., 2012). In brief, pRRLsinPPT-KIF3B-EGFP or its deletion construct was transfected into HEK293T cells with packaging plasmids (pRSV-REV, pMD2.g, and pMDL/pRRE; provided by P. McPherson, McGill University, Montreal, Quebec; Thomas et al., 2009). Culture media were replaced 8 h after transfection. Culture media containing lentiviral particles were collected at 24, 36, and 48 h after transfection, passed through a 0.45-µm filter (Sartorius), and centrifuged at 32,000 g at 4°C for 4 h using an R15A rotor and Himac CR22G centrifuge (Hitachi Koki). Precipitated viral particles were resuspended in Opti-MEM (Invitrogen). KIF3B-KO cells expressing the EGFP-fused KIF3B construct were prepared by the addition of a lentiviral suspension into the culture medium, followed by a 24-h incubation. These cells were used for immunofluorescence analysis.
Immunofluorescence analysis
hTERT-RPE1 cells were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown on coverslips up to 100% confluence and starved for 24 h in Opti-MEM containing 0.2% BSA.
Immunofluorescence analysis was performed as described previously (Takahashi et al., 2012; Funabashi et al., 2017). Unless otherwise noted, cells on coverslips were fixed and permeabilized with 100% methanol for 5 min at −20°C and washed three times with PBS. The fixed/permeabilized cells were blocked with 10% FBS, incubated sequentially with primary and secondary antibodies diluted in 5% FBS, and observed using an Axiovert 200M microscope (ZEISS).
Online supplemental material
Fig. S1 shows genotypic characterization of the two KIF3B-KO cell lines by genomic PCR (A) and direct DNA sequencing of the PCR products (B and C). Table S1 lists plasmid vectors used in this study, and Table S2 lists oligodeoxyribonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank Peter McPherson for providing plasmids for the production of recombinant lentiviruses and Helena Akiko Popiel for critical reading of the manuscript.
This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas “Cilia and Centrosome” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (15H01211 to K. Nakayama), grants from the Japan Society for the Promotion of Science (15H04370 and 15K14456, to K. Nakayama, and 15K07929 and 18H02403, to Y. Katoh), and grants from the Astellas Foundation for Research on Metabolic Disorders (to K. Nakayama) and the Takeda Science Foundation and Uehara Memorial Foundation (to Y. Katoh).
The authors declare no competing financial interests.
Author contributions: T. Funabashi designed and performed the experiments and prepared the manuscript; M. Okazaki and M. Sugawa performed the experiments; and Y. Katoh and K. Nakayama designed the experiments and prepared the manuscript.
References
- Albracht C.D., Rank K.C., Obrzut S., Rayment I., and Gilbert S.P.. 2014. Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 289:27836–27848. 10.1074/jbc.M114.583914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson J.O.L., Shastry S., Hancock W.O., and Block S.M.. 2015. The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 25:1166–1175. 10.1016/j.cub.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.A., Freeman K., Luby-Phelps K., Pazour G.J., and Besharse J.C.. 2003. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J. Biol. Chem. 278:34211–34218. 10.1074/jbc.M300156200 [DOI] [PubMed] [Google Scholar]
- Bangs F., and Anderson K.V.. 2017. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9:a028175 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S., Weber K., Engel B.D., Lechtreck K.-F., and Lorentzen E.. 2014. Getting tubulin to the tip of the cilium: one IFT train, many different tubulin cargo-binding sites? BioEssays. 36:463–467. 10.1002/bies.201400007 [DOI] [PubMed] [Google Scholar]
- Boldt K., van Reeuwijk J., Lu Q., Koutroumpas K., Nguyen T.M., Texier Y., van Beersum S.E.C., Horn N., Willer J.R., Mans D.A., et al. UK10K Rare Diseases Group . 2016. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 7:11491 10.1038/ncomms11491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D.A., and Hildebrandt F.. 2017. Ciliopathies. Cold Spring Harb. Perspect. Biol. 9:a028191 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., and Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T., Katoh Y., Michisaka S., Terada M., Sugawa M., and Nakayama K.. 2017. Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol. Biol. Cell. 28:624–633. 10.1091/mbc.e16-09-0648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik-Lendrum S., Rank K.C., Bensel B.M., Taylor K.C., Rayment I., and Gilbert S.P.. 2015. Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules. Biophys. J. 109:1472–1482. 10.1016/j.bpj.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Katoh Y., and Nakayama K.. 2017. Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein-coupled receptors. Mol. Biol. Cell. 28:429–439. 10.1091/mbc.e16-11-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Nozaki S., Hartanto D., Miyano R., and Nakayama K.. 2015. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128:2351–2362. 10.1242/jcs.168740 [DOI] [PubMed] [Google Scholar]
- Katoh Y., Terada M., Nishijima Y., Takei R., Nozaki S., Hamada H., and Nakayama K.. 2016. Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J. Biol. Chem. 291:10962–10975. 10.1074/jbc.M116.713883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Michisaka S., Nozaki S., Funabashi T., Hirano T., Takei R., and Nakayama K.. 2017. Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated, homology-independent knock-in system. Mol. Biol. Cell. 28:898–906. 10.1091/mbc.e17-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Nakamura K., and Nakayama K.. 2018. Visible immunoprecipitation (VIP) assay: a simple and versatile method for visual detection of protein-protein interactions. Bio Protoc. 8:e2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Beech P.L., and Rosenbaum J.L.. 1995. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131:1517–1527. 10.1083/jcb.131.6.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F. 2015. IFT-cargo interactions and protein transport in cilia. Trends Biochem. Sci. 40:765–778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Pang Y., Wu Q., Hu Z., Han X., Xu Y., Deng H., and Pan J.. 2014. FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev. Cell. 30:585–597. 10.1016/j.devcel.2014.07.019 [DOI] [PubMed] [Google Scholar]
- Marszalek J.R., Ruiz-Lozano P., Roberts E., Chien K.R., and Goldstein L.S.B.. 1999. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. USA. 96:5043–5048. 10.1073/pnas.96.9.5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.-D., Fernandez J.P., Mis E.K., Khokha M.K., and Giraldez A.J.. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 12:982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., and Rohatgi R.. 2014. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin. Cell Dev. Biol. 33:63–72. 10.1016/j.semcdb.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K., and Katoh Y.. 2018. Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 163:155–164. 10.1093/jb/mvx087 [DOI] [PubMed] [Google Scholar]
- Nishijima Y., Hagiya Y., Kubo T., Takei R., Katoh Y., and Nakayama K.. 2017. RABL2 interacts with the intraflagellar transport-B complex and CEP19 and participates in ciliary assembly. Mol. Biol. Cell. 28:1652–1666. 10.1091/mbc.e17-01-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., and Hirokawa N.. 1998. Randamization of Left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid. Cell. 95:829–837. 10.1016/S0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., and Scholey J.M.. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587. 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Pooranachandran N., and Malicki J.J.. 2016. Unexpected roles for ciliary kinesins and intraflagellar transport proteins. Genetics. 203:771–785. 10.1534/genetics.115.180943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo B., Scholey J.M., and Peterman E.J.G.. 2017. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 284:2905–2931. 10.1111/febs.14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M. 2013. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu. Rev. Cell Dev. Biol. 29:443–469. 10.1146/annurev-cellbio-101512-122335 [DOI] [PubMed] [Google Scholar]
- Takahara M., Katoh Y., Nakamura K., Hirano T., Sugawa M., Tsurumi Y., and Nakayama K.. 2018. Ciliopathy-associated mutations of IFT122 impair ciliary protein trafficking but not ciliogenesis. Hum. Mol. Genet. 27:516–528. 10.1093/hmg/ddx421 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Kubo K., Waguri S., Yabashi A., Shin H.-W., Katoh Y., and Nakayama K.. 2012. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125:4049–4057. 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- Taschner M., and Lorentzen E.. 2016. The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. 8:a028092 10.1101/cshperspect.a028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M., Weber K., Mourão A., Vetter M., Awasthi M., Stiegler M., Bhogaraju S., and Lorentzen E.. 2016. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 35:773–790. 10.15252/embj.201593164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R.A., and McPherson P.S.. 2009. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J. Biol. Chem. 284:12410–12419. 10.1074/jbc.M809746200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., and Yang W.. 2016. Permeability barriers for generating a unique ciliary protein and lipid composition. Curr. Opin. Cell Biol. 41:109–116. 10.1016/j.ceb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Ling K., and Hu J.. 2015. The essential roles of transition fibers in the context of cilia. Curr. Opin. Cell Biol. 35:98–105. 10.1016/j.ceb.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield J.L., Mengoni I., Bomberger H., Jiang Y.Y., Walsh J.D., Brown J.M., Picariello T., Cochran D.A., Zhu B., Pan J., et al. 2017. IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. eLife. 6:e26609 10.7554/eLife.26609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Nakata T., Okada Y., and Hirokawa N.. 1995. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130:1387–1399. 10.1083/jcb.130.6.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Nakata T., Okada Y., and Hirokawa N.. 1996. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc. Natl. Acad. Sci. USA. 93:8443–8448. 10.1073/pnas.93.16.8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Takei Y., Kido M.A., and Hirokawa N.. 2011. Molecular motor KIF17 is fundamental for memory and learning via differential support of synaptic NR2A/2B levels. Neuron. 70:310–325. 10.1016/j.neuron.2011.02.049 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.