Summary
Tissue damage and infection are deemed likewise triggers of innate immune responses. But whereas neutrophil responses to microbes are generally protective, neutrophil recruitment into damaged tissues without infection is deleterious. Why neutrophils respond to tissue damage and not just to microbes is unknown. Is it a flaw of the innate immune system that persists because evolution did not select against it, or does it provide a selective advantage? Here, we dissect the contribution of tissue damage signaling to antimicrobial immune responses in a live vertebrate. By intravital imaging of zebrafish larvae, a powerful model for innate immunity, we show that prevention of tissue damage signaling upon microbial ear infection abrogates leukocyte chemotaxis, and reduces animal survival, at least in part, through suppression of cytosolic phospholipase A2 (cPla2), which integrates tissue damage- and microbe-derived cues. Thus, microbial cues are insufficient, and damage signaling is essential for antimicrobial neutrophil responses in zebrafish.
eTOC
Tissue damage signaling in the absence of pathogens causes harmful neutrophil infiltration into damaged organs with no obvious benefit for the host. By intravital imaging of zebrafish larvae, Huang and Niethammer reveal a beneficial role for tissue damage signaling as prerequisite for rapid neutrophil recruitment to microbial infection sites.
Introduction
The physiological purpose of tissue damage-induced (“sterile”) neutrophilic inflammation is puzzling given its adverse clinical effects, such as delayed healing and regeneration of injured tissue (Dovi et al., 2003, 2004; Schofield et al., 2013; Simpson and Ross, 1972). To test its possible functions in the context of infection, tissue damage-induced, but not microbe-induced signaling must be selectively blocked. Such experiment is difficult to devise: Firstly, bacteria must traverse epithelial barriers to infect a host, and this typically involves barrier damage that leads to damage signaling. Secondly, microbial and tissue damage-derived signals act through a shared set of pattern recognition receptors (PRRs, for example, Toll-like receptors and inflammasomes), which precludes their selective downstream inhibition.
Various studies have shown that sterile injury suffices for local neutrophil recruitment in vivo (Lämmermann et al., 2013; McDonald et al., 2010; Ng et al., 2011). Microbial cues (Pathogen Associated Molecular Patterns, PAMPs) act through similar pattern recognition pathways as damage associated molecular patterns (DAMPs), and some PAMPs are potent neutrophil chemoattractants (Bloes et al., 2015). Thus, microbes, just as tissue damage, are thought to suffice for local neutrophil recruitment. But this assumption has been little tested in vivo, likely for the experimental difficulties discussed above. Per the canonical DAMP paradigm, deciphering the full contribution of host damage signals to microbial detection requires to first elucidate all DAMPs, and then test the effects of eliminating them (Kono and Rock, 2008). Given their number and possible redundancy, such strategy seems problematic.
To this end, zebrafish enable an alternative, more feasible experimental design. The innate immune responses of this freshwater vertebrate resemble those of mammals (Trede et al., 2004), but can be non-invasively imaged in real-time at animal throughputs unattainable with rodent models. At 2–3 days after fertilization, zebrafish larvae do not yet have adaptive immunity (Trede et al., 2004), which reduces mechanistic complexity. When these larvae are injured, environmental fresh water enters the tissue to cause osmotic cell swelling around the injury site. Cell- and organelle swelling (“cytotoxic edema”, “ballooning degeneration”, “hydropic degeneration”) is a pathological hallmark of tissue damage in all animals (Liang et al., 2007; Majno and Joris, 1995; Vanden Berghe et al., 2014). The direct physiological consequences of these dramatic morphological changes have been recently investigated in the zebrafish model. There, cell- and nuclear swelling, mediate neutrophil recruitment through membrane stretch-induced phospholipase (cPla2) activation, which initiates, potentially in conjunction with NADPH oxidase activity, the production of chemotactic lipids (eicosanoids) (Enyedi et al., 2013, 2016; Niethammer et al., 2009). Here, we prevent swelling-induced damage signaling during microbial infection by bathing zebrafish larvae in isotonic solution, which reveals it as prerequisite for rapid, protective leukocyte recruitment to infection sites.
Results
Microbial ear infection triggers heterogeneous neutrophil responses in zebrafish larvae
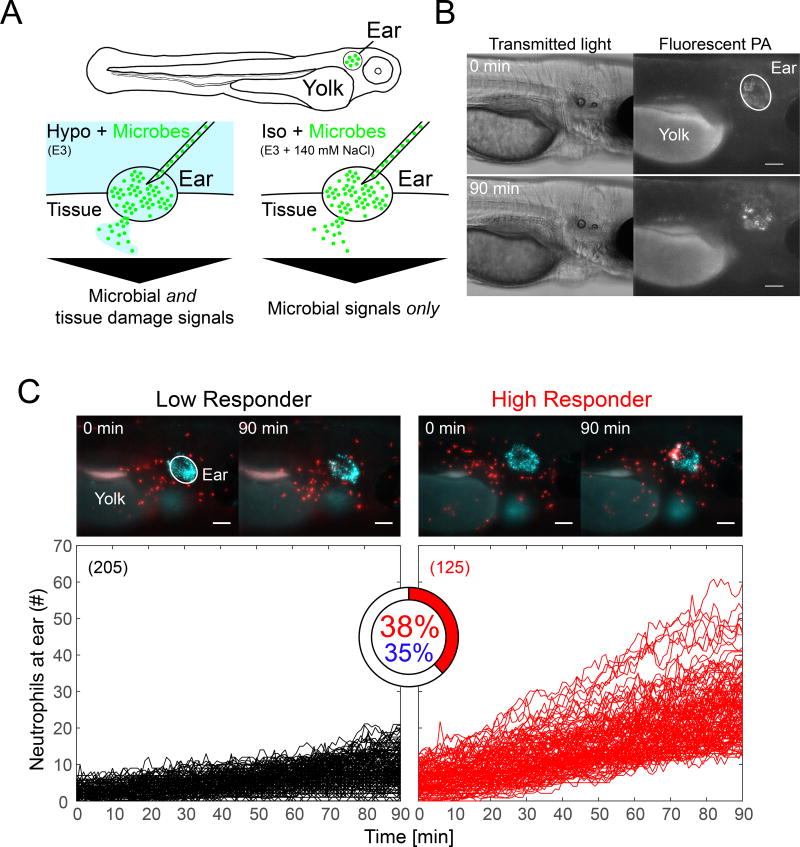
To generate focal infection in zebrafish, we injected ~2·104 colony-forming units (CFU) Pseudomonas aeruginosa (PA) or Escherichia coli (E. coli) suspension into the otic vesicle (ear) of three day old larvae (Figure 1A) (Deng et al., 2012). The suspensions were supplemented with 1 µm fluorescent beads to label the infection site. The beads acted as bright and faithful fluorescent proxy for bacterial dispersion, akin to fluorescent PA (Figure 1B, C; Movie S1). Assuming a spherical shape, we estimated the ear volume to ~1 nL. To rupture the luminal ear integument by overpressure, we injected a ~ two times larger volume of PA suspension. The damage caused by the injection (Figure S1A) gives microbes and bathing solution access to the fish interstitium and allows rapid equilibration of ear fluid and bathing solution, as indicated by the rapid loss of 4 kD fluorescent dextran from the ear lumen (Movie S2).
Figure 1. Ear infection of zebrafish larvae with Pseudomonas aeruginosa (PA) triggers strong neutrophil responses in a subset of animals.
(A) Cartoon scheme of experiment. (B) Ear injection of microbes generates a localized source of infection as visualized with EGFP-tagged PA (shown: hypotonic bathing conditions). Panel represents n=6 injection experiments. (C) Upper panel, time-lapse montage of neutrophil recruitment to infected ears in a Low Responder (left) and a High Responder (right) animal at indicated times. Panels represent n=205 (left) and n=125 (right) injection experiments, respectively. Neutrophils are in red. Fluorescent beads (blue) mark the ear region. Scale bar, 100 µm. Lower panel, neutrophil recruitment kinetics in Hypo + PA fish computationally classified into low (black) and high (red) responders by comparison to user-generated training sets. High responder index is depicted as pie chart. Blue percentage, an unsupervised (that is, no user-defined training sets) Gaussian distribution clustering algorithm determines a similar HR-index. Parentheses, number of injection experiments. Please see also Figure S1.
We imaged neutrophil recruitment by time-lapse microscopy in fluorescent lysC (Hall et al., 2007) neutrophil reporter larvae for 90 min following ear injection of bacteria or buffer. Under standard, hypotonic bathing conditions (Hypo + PA), infection triggered strong leukocyte recruitment in ~ one third of the animals (Figure 1C, “High Responders”, HR). These strong responses were characterized by the synchronized convergence of neutrophils onto the ear infection site (Movie S3). Two thirds of the animals displayed sporadic, unsynchronized neutrophil migration (Figure 1C, “Low Responders”, Movie S3). The relatively high number of observed animals facilitated appreciation and quantification of this response heterogeneity. Although the precise factors that contribute to it remain elusive, we speculate that biological thresholds, (epi-)genetic variability, stochasticity of injection-induced ear rupture, and others, might play a role. For unbiased phenotype classification (Figure 1C), blinded neutrophil recruitment curves were randomly selected from a pool of ~1600 experiments across 41 experimental conditions, and manually classified as high or low responder “training sets”. These training sets were then used to automatically calculate the frequency of high responders in each experimental group (HR-index, HRi). Unsupervised phenotype classification without user-selected reference sets using a Gaussian mixture distribution model (Figure 1C, S1B) yielded similar results, which underlines the robustness of the method.
Selective suppression of tissue damage signals inhibits protective neutrophil recruitment to infections
In cell culture experiments, microbes suffice for attracting neutrophils through formylated peptides, and other emitted chemotactic factors (Nuzzi et al., 2007; Schiffmann et al., 1975). Serum is often present before or during these experiments, possibly mimicking aspects of a wound microenvironment. Likewise, the isolation of primary neutrophils involves injury-inducing procedures. Although these in vitro assays are valuable for dissecting fundamental chemotactic signaling mechanisms, it is uncertain which—if any—physiological state they resemble.
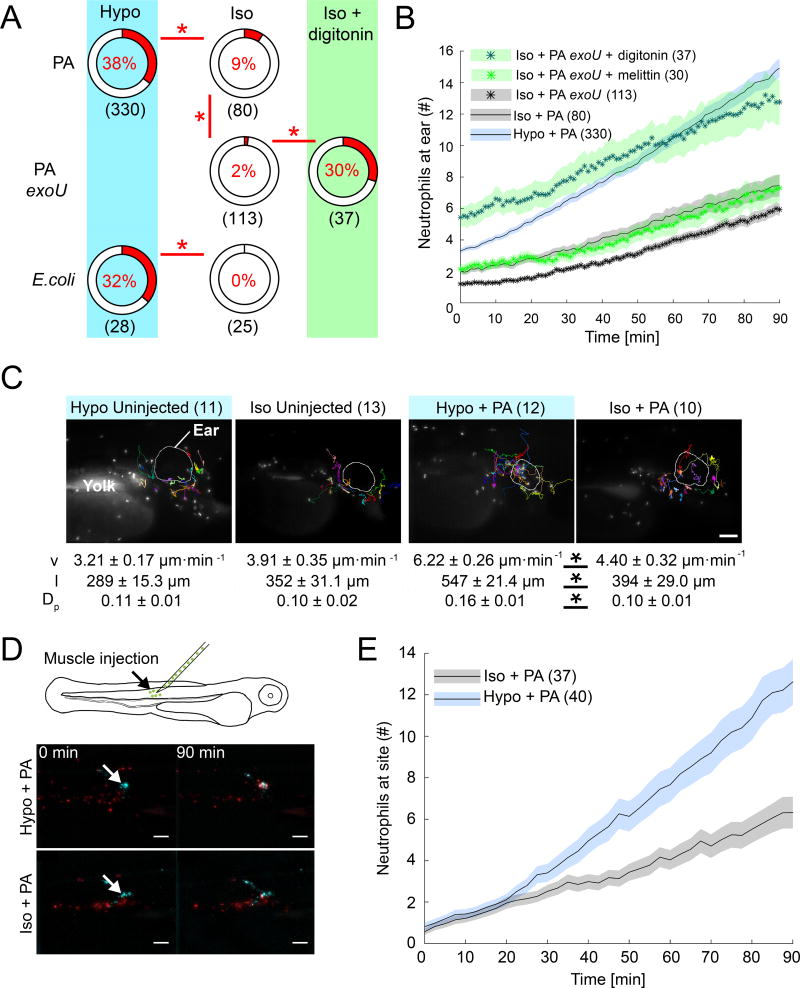
Inhibition of tissue damage signaling by isotonic bathing (Iso + PA) strongly reduced neutrophil recruitment into infected ears, although microbe viability was unaffected (Figure S2A). Bathing tonicity also did not visibly alter injection-induced cell lysis at the ear (Figure S1A), confirming previous findings in the tail fin injury assay (Enyedi et al., 2013; Gault et al., 2014). Under isotonic bathing conditions, strong leukocyte responses, undistinguishable from those under hypotonic conditions, were still sporadically observed in response to PA, but not E. coli (TOP10) infection (Figure 2A, Movie S3, Figure S2B). The basal ability of virulent PA (strain PA14), but not harmless lab E. coli, to induce neutrophil recruitment even in the absence of osmotic tissue damage signals, may be due to PA’s cytotoxicity (Dacheux et al., 2001). As mentioned above, necrotic cell demise involves cell- and organelle swelling even under isotonic, extracellular conditions. The bacterial phospholipase ExoU is a major PA virulence factor that causes necrosis, likely through disrupting cell plasma membranes (Finck-Barbançon et al., 1997; Hauser, 2009). To test whether basal neutrophil recruitment to PA infection under isotonic bathing conditions was caused by PA-mediated cytotoxicity, we conducted experiments with a PA strain mutant for ExoU (PA exoU). Indeed, neutrophil recruitment to injected PA exoU was much reduced as compared to wild type PA. Remaining neutrophil responses may be due to residual, ExoU-independent cytotoxicity. Importantly, co-injecting PA exoU together with the non-microbial membrane disruptors melittin (naturally occurs in venom of Apis mellifera, honeybee) and digitonin (naturally occurs in Digitalis purpurea, foxglove), respectively, restored neutrophil recruitment (Figure 2A, B). Melittin, but not digitonin, moderately decreased bacterial viability, in line with melittin’s known antimicrobial functions (Figure S2C) (Picoli et al., 2017), but control experiments with heat-killed PA suggested that bacterial viability does not affect neutrophil recruitment in our system (Figure S2D, E). Altogether, these experiments argue that necrosis, for instance, induced by bacterial virulence, can functionally replace extracellular hypotonicity to some extent. This makes a broad case for tissue damage signaling—osmotic or otherwise—as essential trigger of antimicrobial neutrophil responses in vivo.
Figure 2. Selective suppression of tissue damage signaling abrogates neutrophil responses to infection sites.
(A) HR indices after PA or E. coli infection in the presence (Hypo) or absence (Iso) of osmotic tissue damage signaling, and as a function of microbial cytotoxicity (ExoU). HR indices are depicted as pie charts. Asterisks, Fisher’s exact test p<0.05. Note, the Hypo + PA reference set is the same as in the other figures. Parentheses, number of injection experiments (B) Average neutrophil recruitment as a function of endogenous, microbial (ExoU) and exogenous (digitonin, melittin) cytotoxicity. Shaded area, SEM of n injection experiments (parentheses). (C) Neutrophil tracking analysis. Top panel, images representative of n (parentheses) injection experiments. Lower panel, table of leukocyte migration parameters. v, migration velocity. l, path length. Dp, directional persistence. Shown are average values ± SEM for indicated number of injection experiments (parentheses). Asterisks, one-way Anova between indicated groups p<0.05. Scale bar, 100 µm. (D) Top, scheme of alternative injection site. Bottom, still images of neutrophil recruitment to muscle infection sites at indicated times in the presence (Hypo) and absence (Iso) of osmotic tissue damage signaling. Image panels represent n=40 (Hypo + PA) and n=37 (Iso + PA) injection experiments. Arrow, injection site. Scale bars, 100 µm. (E) Average neutrophil recruitment to muscle injection sites as a function of osmotic tissue damage signaling. Shaded area, SEM of n injection experiments (parentheses). Please also see Figure S2.
Average neutrophil speed, path length, persistence was decreased when larvae were infected under isotonic (Iso + PA) as compared to hypotonic bathing-conditions (Figure 2C). Isotonicity did not decrease random neutrophil migration in uninfected (that is, uninjected and undamaged) larvae. Thus, integumental damage is required for hypoosmotic stimulation of neutrophil recruitment, likely by enabling entry of hypotonic bathing solution into the organism, which then gives rise to local cell swelling at the infected ear.
The residual high responder occurrence in animals infected under isotonic conditions (Iso + PA) (Figure 2A, Movie S3), the lack of isotonic inhibition of random leukocyte migration (Figure 2C), and our previous findings that isotonic inhibition can be bypassed by externally added eicosanoids (Enyedi et al., 2013), argue against a generalized inhibition of neutrophil motility by bath tonicity. Furthermore, isotonic bathing was not toxic on all experimental time scales, and likewise blocked neutrophil recruitment to an alternative infection site in the trunk muscle (~2·104 CFU per injection; Figure 2D, E, Movie S4). We wondered whether hypotonicity may stimulate rapid neutrophil recruitment through regulating chemokine expression, which is a well-studied effector mechanism of classical PAMP- and DAMP-signaling. Arguing against this possibility, blocking protein synthesis by bathing fish in cycloheximide prior and during infection showed little effect in our assay (Figure S1B, S3).
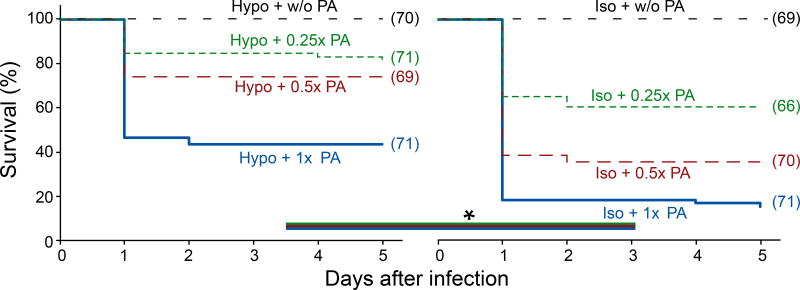
The HR-index turned out to be predictive of post-infection survival: In the presence of tissue damage signals, 38% of the larvae exhibited strong neutrophil responses, with 44% surviving. Without tissue damage signals, only 9% of the animals showed strong neutrophil recruitment, and only 15% survived (Figure 2A, 3). Most animals succumbed to ear infection after a day. The proximity of the infection site to the central nervous system, and the high cytotoxicity of PA14 probably promoted their rapid demise. Isotonic bathing was only performed within the first two hours after infection. Thus, immediate damage signaling upon infection seems critical for post-infection survival, providing a selective advantage.
Figure 3. Selective suppression of osmotic tissue damage signaling decreases post-infection survival.
Meier-Kaplan plots of post-infection survival of PA-infected larvae in the presence or absence of osmotic tissue damage signaling. Different lines in each graph refer to different PA concentrations. Asterisks, log-rank test p<0.05. Parentheses, number of injection experiments.
Suppression of osmotic damage signaling does not abrogate microbe recognition
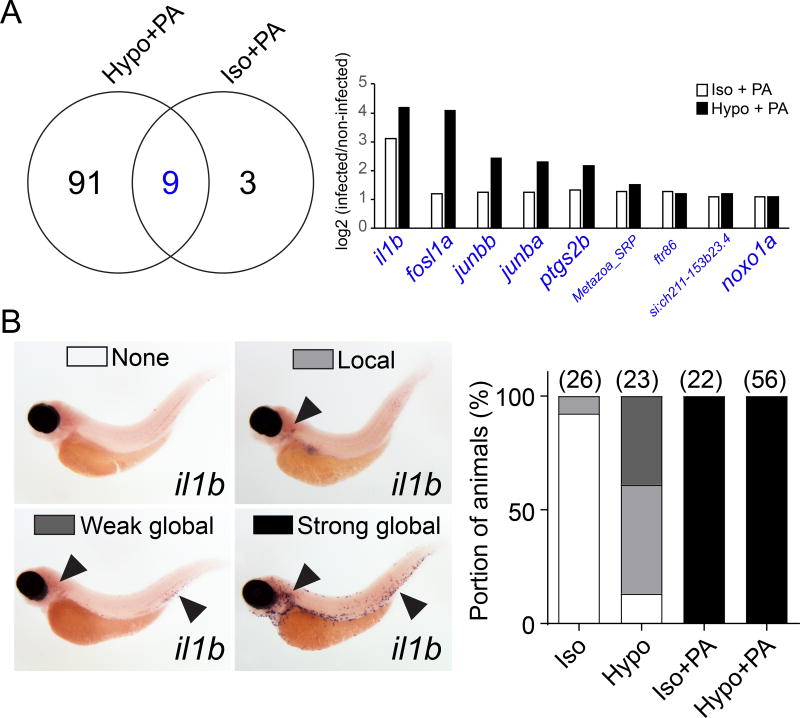
To test whether the suppression of tissue damage signaling interfered with microbial pattern recognition, we employed mRNA sequencing after PA infection. Comparing uninfected larvae, or larvae infected with PA in hypotonic (Hypo + PA), or isotonic (Iso + PA) bathing solution showed that tissue damage signaling was required for full, transcriptional responses to PA-infection (that is, two times up- or down-regulation of 100 genes; Figure 4A, Table S1). Nevertheless, microbial signals alone induced a subset of immune genes. Those included AP-1 subunits (fosl1, junba, junbb), cyclooxygenase 2 (ptgs2b), interleukin 1β (il1b), and NADPH oxidase organizer 1a (noxo1a), which are known host effectors of bacterial lipopolysaccharides (LPS) (Eliopoulos et al., 2002; Forn-Cuní et al., 2017; Kawahara et al., 2005; van der Vaart et al., 2013). Even without microbes, tissue wounding is known to induce expression of the genes coding for AP-1 subunits, IL-1β, and PTGS2 (Chen and Nuñez, 2010; Fitsialos et al., 2007). This may explain why the absence of hypotonic wound cues blunted their induction. Whole mount in situ hybridization revealed strong and systemic il1b mRNA expression (possibly in macrophages) upon infection, irrespective of osmotic bathing conditions (Figure 4B). Hence, without osmotic tissue damage signaling, bacterial cues are still recognized and induce robust upregulation of important inflammatory genes.
Figure 4. Absence of osmotic tissue damage signaling does not block microbial detection.
(A) Left panel, Venn plot of significantly (padj<0.05), at least two times up- or down-regulated mRNAs at 60 min after PA ear infection in the absence (Iso + PA) or presence (Hypo + PA) of osmotic tissue damage signaling. Right panel, log2-fold regulation of intersection gene set (blue). Large font, known LPS-downstream effectors. See Table S1 for more detail. Results are derived from n=3 independent mRNAseq experiments (B) In situ hybridization for il1b mRNA. Left panel, representative images of different classes of staining patterns observed in indicated number of injection experiments (parentheses, right panel). Right panel, quantification of staining pattern frequency.
cPla2 integrates tissue damage- and microbe-induced signaling
Per our mRNAseq results, we speculated that the PA were detected through LPS signaling. Indeed, leukocyte recruitment to an LPS-mutant of PA (Feinbaum et al., 2012) was markedly reduced, whereas a multi-flagellated PA mutant (fleN) (Feinbaum et al., 2012) did not significantly augment responses (Figure 5A). In mammals, LPS is recognized by Toll-like receptors (TLRs), or by noncanonical inflammasomes, which trigger IL-1 signaling (Barton and Medzhitov, 2003; Kayagaki et al., 2013). Myd88, a conserved signaling adaptor for TLRs and the IL-1 receptor, mediates LPS recognition in zebrafish (van der Vaart et al., 2013). Antisense silencing of myd88 with a published morpholino (van der Sar et al., 2006), but not a myd88 mismatch morpholino, attenuated leukocyte recruitment to wild type PA (Figure 5A), consistent with the notion that host detection of PA, at least in part, occurs through LPS signaling. By contrast, morpholino silencing of pycard (ASC), a conserved inflammasome adaptor, had little effect on rapid leukocyte recruitment (Figure S4A).
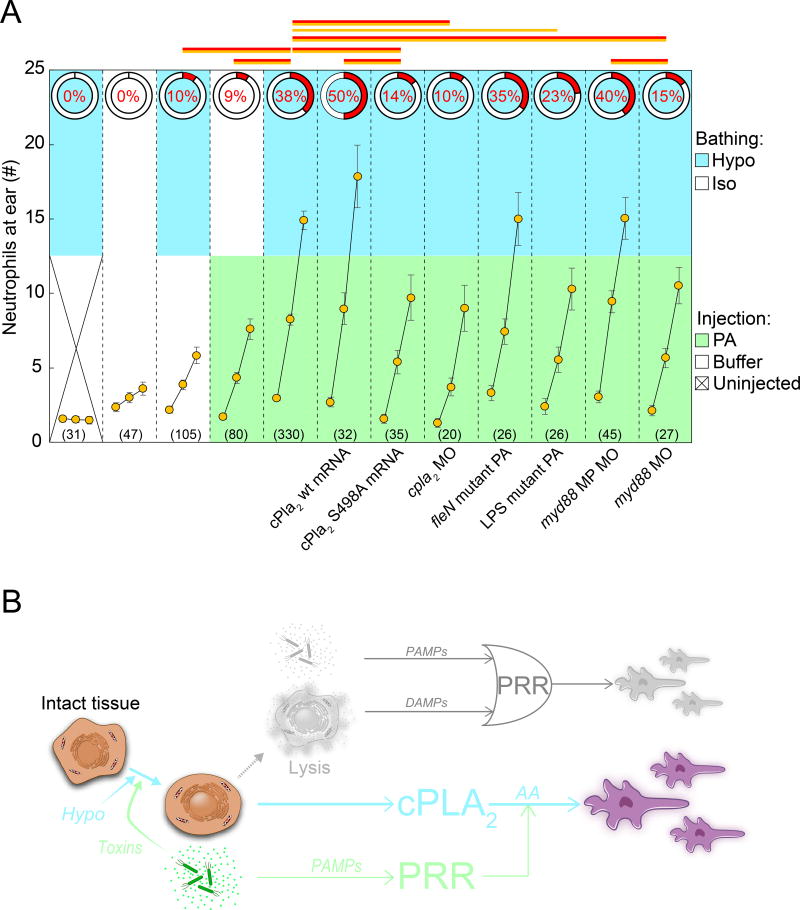
Figure 5. cPla2 integrates tissue damage and microbial cues.
(A) Summary of genetic pathway perturbations (indicated below the graph). Two measurements are given: the HR-index (red, pie charts), and average leukocyte recruitment curves (orange circles, simplified 3-timepoint-plot format: 0’, 45’, 90’, see Figure S3A). Upper graph section, color code indicates respective bathing conditions. Lower graph section, color code indicates injection conditions. Note, the injection buffer tonicity is always the same as bath tonicity. Error bars, SEM of indicated number (parentheses) of injection experiments. Red lines, Fisher’s exact test p<0.05 comparing HR indices. Orange lines, t-test p<0.05 comparing average leukocyte numbers at 90’. cPla2 wt mRNA, cPla2 overexpression by injection of mRNA into one-cell stage embryos. cPla2 S498A mRNA, cPla2 phosphorylation site-mutant overexpression by injection of mRNA into one-cell stage embryos. cpla2 MO, published splice blocking morpholino (Enyedi et al., 2013). fleN mutant PA (multi-flagellated), ear injection of a flagella mutant of PA (Feinbaum et al., 2012). LPS mutant PA, ear injection of O-antigen mutated PA (ORF_11) (Feinbaum et al., 2012). myd88 MP MO, misprime control morpholino. myd88 MO, published translation blocking myd88 morpholino (van der Sar et al., 2006). Note, the Hypo + PA reference set is the same as in other figures. (B) Regulatory diagrams juxtaposing the classical view (upper panel, muted colors) of inflammation initiation with the model supported by this study (lower panel). Classically, either PAMPs released by microbes or DAMPs released by lytic host cells are thought to function as primary triggers for inflammation and leukocyte recruitment. This logical OR relationship is depicted by its standard symbol in the upper diagram. Our study suggests that only tissue damage can function as primary, inflammatory trigger in vivo, and that microbial signals act as amplifiers of tissue damage signaling. Depicted on the left side is the necrotic sequence of cell morphology changes. Arrows mark the proposed regulatory role of each necrotic intermediate. Whereas classic DAMP signaling is triggered by cell lysis, the tissue damage signaling pathway proposed in this study is triggered upstream of cell lysis by cell- and nuclear swelling. Note that necrotic cell lysis causes strong additional nuclear swelling through extranuclear colloid osmotic pressure decrease, which can further activate nuclear cPla2 (Enyedi et al., 2016). For simplicity, this post-lytic nuclear swelling is omitted from the diagram. PAMP, pathogen associated molecular pattern. DAMP, damage associated molecular pattern. PRR, pattern recognition receptor. cPLA2, cytosolic phospholipase A2. AA, arachidonic acid. Please also see Figure S3 and S4.
We previously found that osmotic tissue damage cues are transduced via biomechanical cPla2 activation upon cell- and nuclear swelling (Enyedi et al., 2013, 2016). Indeed, the cPla2 product arachidonic acid induced leukocyte recruitment in the absence of osmotic tissue damage signals (Figure S1B, S3B). Antisense silencing of cpla2 (pla2g4aa) with different, and published morpholinos attenuated neutrophil recruitment (Figure 5A, S4B, C). cPla2 overexpression by mRNA injection increased neutrophil recruitment. 5-lipoxygenase (5-LOX) and cyclooxygenases (COX) metabolize arachidonic acid downstream of cPLA2 into chemotactic leukotrienes, oxo-eicosanoids, or prostaglandins (Dennis and Norris, 2015; Rådmark et al., 2015). Aspirin, which inhibits the prostaglandin branch of the eicosanoid cascade, did not block neutrophil recruitment—if anything, we noticed stimulation, possibly through AA shunting into the 5-LOX pathway (Figure S1B, S3B). The 5-LOX antagonist Zileuton, and the LOX- and COX dual-antagonist Licofelone attenuated neutrophil recruitment (Figure S1B, S3B). Together with our previous work (Enyedi et al., 2013, 2016), this suggests that the eicosanoid cascade detects osmotic damage cues during microbial infection through its 5-LOX branch, which generates potent neutrophil chemoattractants, such as 5-oxo-ETE and leukotriene B4 (LTB4). The question remained of how microbes may feed into this pathway.
PA induce cPLA2 phosphorylation (Hurley et al., 2006, 2011) through a LPS-TLR-Myd88-MAP kinase signaling axis (Lindner et al., 2009; Qi and Shelhamer, 2005; Ruipérez et al., 2009), and cPLA2 phosphorylation augments the enzyme’s hydrolytic activity, which unfolds after membrane attachment (Tucker et al., 2009). In line with others (Schievella et al., 1995; Tucker et al., 2009), we previously found that phosphorylation does not increase membrane recruitment of cPla2 (Enyedi et al., 2016). Instead, cPla2 membrane interactions are stimulated by Ca2+ and nuclear membrane stretch, which are both induced by tissue damage (Enyedi et al., 2016). Phosphosite-specific antibodies for zebrafish cPla2 are not available, but we could detect stimulation of cPla2 phosphorylation upon ear infection by affinity purification of phosphoproteins from larval extracts (Figure S4D). It is conceivable that these whole-animal phosphorylation changes underestimate local increase of phosphorylation at the ear infection site. To test whether cPla2 phosphorylation controlled antimicrobial neutrophil responses in vivo, we mutated cPla2’s primary, conserved phosphorylation site (S498A), and injection of the S498A mutant mRNA strongly suppressed PA-induced neutrophil recruitment (Figure 5A). Together with published results, these experiments indicate that cPla2 mechanochemically integrates microbe and tissue damage cues (Figure 5B).
Discussion
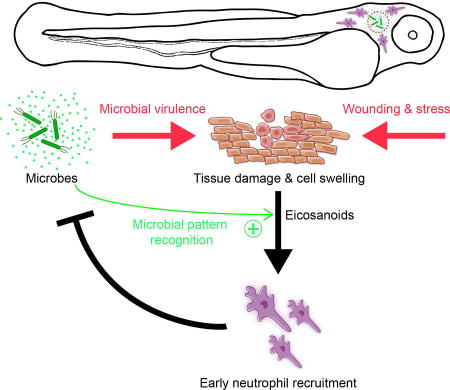
We uncoupled tissue damage- and microbe-induced signaling during bacterial infection in vivo and showed that neutrophils ignore bacteria in the absence of tissue damage signals. These data contest the literature view that either microbial or tissue damage cues suffice for triggering innate immune responses.
The literature view is supported by the notion that PAMPs and DAMPs use a shared set of PRRs, for instance, the DAMP HMGB1 can signal through TLR4, just as the PAMP LPS. Likewise, host-intrinsic, mitochondrial and host-extrinsic, microbial formylated peptides bind to the same formyl peptide receptors. Our results show that tissue damage and microbial signals are not equally capable of activating innate immune responses in vivo. Namely, the PAMPs contained in E. coli, or PA exoU trigger no, or almost no strong neutrophil responses, unlike osmotic damage signals. Thus, in our model, tissue damage cues are primary triggers of rapid, anti-microbial neutrophil recruitment, which is then further amplified by microbial pattern detection. Accordingly, if pathogen-associated factors (such as ExoU) also cause host damage, they can initiate neutrophil responses on their own.
The underlying signaling hierarchy seems hardwired within cPLA2’s molecular activation circuit, which involves primary activation by membrane translocation through damage-induced Ca2+ and nuclear membrane stretch (Enyedi et al., 2013, 2016), and subsequent amplification of hydrolytic activity by phosphorylation.
The limited involvement of transcriptionally-induced chemokines in the immediate neutrophil response to infection is not entirely surprising: Lipid chemoattractants can be synthesized within seconds, whereas peptide chemoattractants that require prior gene expression are expected to function with some delay. Thus, fast enzymatic mechanisms can provide temporary immune protection before more sophisticated transcriptional mechanisms have gained traction.
Although zebrafish do not possess adaptive immunity at an early larval stage, the conceptual parallels to the “danger”, “injury”, or related “two-signal models” of adaptive immunity are undeniable (Land and Messmer, 2012; Matzinger, 2002; Vance, 2000). As an example for co-stimulatory signaling through lipids, dendritic cells were recently shown to stimulate adaptive immune responses upon coincidence of host-derived lipid DAMPs (OxPAPC) and microbe-derived LPS through caspase 11 and inflammasome-dependent IL-1β in mice (Zanoni et al., 2016). In our study, morpholino-mediated silencing of pycard revealed no evidence for inflammasome involvement in rapid neutrophil recruitment. Regardless, to enforce analogy, arachidonic acid, just as OxPAPC, could be considered a lipid DAMP: Polyunsaturated fatty acids, such as arachidonic or linolenic acid, are ancient tissue damage signals. In plants, which typically do not contain arachidonic acid, linolenic acid fuels jasmonic acid synthesis through the octadecanoid pathway, a plant “counterpart” of the eicosanoid pathway in animals. Jasmonic acid is a central regulator of biotic and abiotic stress responses, including responses to wounds (Niethammer, 2016). We note that the biomedical literature often uses the term “DAMP” more narrowly to describe cytoplasmic factors that leak from lysing cells and bind to PRRs.
Could our observations help explain neutrophil tolerance of benign, for example, commensal bacteria (Bäckhed et al., 2005; Nathan, 2006)? In the absence of external wound signals, harmless microbes do not seem to be attacked by neutrophils unlike virulent ones. More research is needed to test the physiological implications of this finding and make a broader case. Meanwhile, our results provide a rationale for why tissue damage signaling to neutrophils might have evolved in the first place: as primary trigger for neutrophil responses to infection.
Experimental models and subject details
General fish procedures
Adult wt and transgenic reporter casper (White et al., 2008) zebrafish, and larvae were maintained as described (Nüsslein-Volhard and Dahm), and subjected to experiments according to institutional animal healthcare guidelines with the approval of the Institutional Animal Care and Use Committee (IACUC). The lines used in each experiment, as well as experimental numbers are detailed in Table S1. For injection assays, 2.5–3 days post-fertilization (2.5–3 dpf) larvae were anaesthetized in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) containing 0.2 mg/mL ethyl 3-aminobenzoate methanesulfonate (Sigma).
Method Details
Plasmid construction
The TG(lysC:Casper1GR) transgenic line was created using the Tol2kit system as previously described (Kwan et al., 2007), by recombining the lysC promoter (Hall et al., 2007), the Casper1GR transgene (that is, a TagGFP and a TagRFP connected by a YVAD motif, originally developed as FRET-sensor for caspase activation (Liu et al., 2014) and here used as fluorescent neutrophil marker), and a SV40 polyadenylation sequence into to the pDestTol2CG2 vector backbone.
pCS2+cPla2mKate2 for cPla2 overexpression by mRNA injection was generated as previously described (Enyedi et al., 2016). Serine-to-Alanine mutation at position 498 (S498A) was achieved using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). Primers used for mutagenesis were: forward, 5’-TCAACCCTGCCCTTCGCTCCCTTCAGCGGCATC – 3’; reverse, 5’ – GATGCCGCTGAAGGGAGCGAAGGGCAGGGTTGA – 3’ (Enyedi et al., 2016). Sequencing confirmed the mutation.
Generation of transgenic lines
A solution containing the lysC:Casper1GR transgene plasmid and transposase mRNA was injected into the cytosol of one-cell stage Casper embryos. Injected larvae with mosaic cardiac EGFP (enhanced green fluorescent protein) expression were raised to sexual maturity and screened by crossing to wt fish to identify founders. Founders were crossed to wt caspers, or subsequent F1 transgenic siblings were crossed together to obtain the 2.5–3 dpf transgenic larvae used in the ear infection experiment.
Bacterial strains and growth conditions
Wild-type, and EGFP-tagged Pseudomonas aeruginosa (strain PA14) were generously provided by Dr. Joao Xavier (MSKCC, New York, NY). PA14 mutant strains ORF_11, fleN, and exoU (Feinbaum et al., 2012) were generously provided by Dr. Cole Haynes (University of Massachusetts Medical School, Worcester, MA). One Shot TOP10 chemically competent E. coli (Thermo Fisher) were used as nonpathogenic bacteria. Wt PA14 and E. coli liquid culture was grown in LB medium. Mutant PA strains were cultured in LB medium containing 15 µg/mL Gentamycin (IBI Scientific). All cultures were incubated at 37 °C in a rotary shaker at 250 r pm. Heat-killed PA14 were prepared by heating the bacteria at 65 °C for 1 h (Matsumoto et al., 1998).
Bacterial infection through ear or muscle injection
Bacteria were resuspended in either E3 or isotonic E3 medium, and the optical density was measured at 600 nm with a Ultraspec 10 cell density meter (Amersham Biosciences). To prepare the final inoculum, the bacteria suspension was diluted or pelleted by centrifugation at 1800×g for 10 min, followed by resuspension to achieve the desired bacterial density (OD600=7). Fluorescent beads (Blue-green FluoSpheres, Life Technologies) were added to the bacterial aliquots prior to injection for a final ratio of 1:1. Ear injection was performed as previously described (Deng et al., 2012). Muscle injection was performed by injecting bacteria into the segmented muscle above the yolk extension. Injections were performed using a Nanoject II microinjector (Drummond Scientific). After bacterial infection, a serial-diluted inoculum was plated to quantify colony forming units (CFU). The volume of the inner ear was estimated by approximating the ear vesicle as a sphere and measuring its radius.
Widefield fluorescence imaging
For all imaging experiments, transgenic, transparent (TG(lysC:Casper1GR), casper background) zebrafish larvae expressing TagRFP in neutrophils were embedded in 1% low melting (LM) agarose (Gold Biotechnologies) and maintained in standard, hypotonic E3, or isotonic E3 medium (that is, standard E3 supplemented with 140 mM NaCl), containing the indicated compounds. Every minute, starting at ~10–15 min post injection, images were acquired. TagRFP and FluoSphere fluorescence were excited with an LED light source (Lumencor) using 549/15 and 438/24 bandpass filters, respectively, together with a multispectral dichroic (59022 bs, Chroma). Both TagRFP fluorescence, and the red tail of the FluoSphere emission was acquired with a 632/60 emission filter (Chroma). Fluorescence of EGFP-tagged PA was excited using a 475/28 bandpass filter together with a multispectral dichroic (59022 bs, Chroma), and acquired using a 525/50 emission filter (Chroma). Images were captured at room temperature (~25 °C) using NIS-Elements (Nikon) on an Eclipse Ti microscope (Nikon) equipped with a 10× plan-apochromat NA 0.45 air objective lens, a Clara CCD camera (Andor), and a motorized stage.
Survival assay
2.5–3 dpf zebrafish larvae were injected with serial-diluted PA suspended in either E3 or isotonic E3 medium. Larvae injected with isotonic suspension were kept in isotonic E3 fish bathing solution for 2 h before transferring them back to standard, hypotonic E3 bathing solution. Larval viability was monitored for 5 days after injection. Dead larvae were scored by loss of tissue transparency and heartbeat.
Image processing and data analysis
Leukocyte recruitment to the ear or muscle was computationally measured by counting neutrophil number within the ear or muscle region per frame for 90 min (1 min/frame for ear injections, and 2.5 min/frame for muscle injections). Time-lapse movies were background subtracted using the MOSAIC background subtractor plugin (Sbalzarini and Koumoutsakos, 2005) (http://mosaic.mpi-cbg.de/?q=downloads/imageJ) in ImageJ/FIJI (Schindelin et al., 2012). All images of the background-subtracted time-lapse movies were maximum intensity projected (11 z-planes per frame, 8 µm per z-step) using ImageJ/FIJI. The ear vesicle outlines were detected based on dispersed bead fluorescence. To define the infection-area (that is, region of interest, ROI) after muscle injection, all image filenames were blinded in CellProfiler (Lamprecht et al., 2007), and the approximate needle injection site as marked by bead fluorescence was manually outlined as ROI. We used two different computational methods to count fluorescent neutrophils in the ear, whose accuracy and suitedness we initially confirmed through manual measurements. When neutrophils did not visibly aggregate into large clusters, we used local fluorescence intensity maxima detection (implemented in a Python script, Anaconda distribution) for determining the position and number of leukocytes within an image. Because neutrophil cluster formation posed a challenge for detecting individual leukocytes by local maxima detection, we analyzed time-lapse movies with neutrophil clusters using a custom analysis pipeline in CellProfiler (Lamprecht et al., 2007) and Matlab (MathWorks). Briefly, average neutrophil fluorescent intensity was estimated from the first 10 frames of each time-lapse movie. Neutrophil numbers were then determined by dividing total neutrophil fluorescence intensity of the ear by average neutrophil fluorescence. Visual inspection confirmed that this method more accurately estimated leukocyte numbers in the presence of clustering than local maxima detection. To approximate neutrophil numbers within the muscle ROI, the above described CellProfiler pipeline and manual counting were applied depending on whether neutrophils formed visible aggregates. For clearer data presentation in Figure 5 and S3, a segmented line fit was used to represent average leukocyte recruitment kinetics within the first (0–45 min) and second half (46–90 min) of each time-lapse movie (Figure S3A). The number of high responders in each experimental condition was computationally identified: Neutrophil response curves were blindly selected from a total pool of 1600 experiments across 41 experimental conditions, including those reported in the manuscript, and manually classified into representative high and low responders training sets (with 40 curves per set). The Euclidian distance of leukocyte recruitment curves to the median of the two training sets (Matlab ‘pdist2’ command) was calculated. Curves more similar to the median of the high responder training set than to the median of the low responder training set were computationally classified as “high responders”, and vice versa. User-unsupervised, Gaussian mixture distribution model clustering (McLachlan and Peel, 2000) into two clusters using the Matlab ‘fitgmdist’ command gave largely similar results (Figure S1B). In this fully automated method, all available leukocyte recruitment curves (from 1813 individual injection experiments at that time, including those described in the figures) were used to generate the two-component Gaussian mixture distribution model. For more details, please see the supplemental Matlab scripts (Data File 1).
Leukocyte trajectory analysis
Trajectory analysis was carried out on a subset of the time-lapse movies used for computational quantification of neutrophil recruitment to the ear (see above). Trajectories were manually generated using the MTrackJ plugin of ImageJ/FIJI (Meijering et al., 2012). The ear region was identified by dispersed bead fluorescence, or by transmitted light morphology as appropriate. Leukocytes that were <100 µm away from the ear outline in the first frame were followed by manual tracking. Average migration velocity (v), path length (l), and path directional persistence (Dp) were calculated as previous described (Enyedi et al., 2013).
mRNA preparation and injection
For the cPla2 or S498AcPla2 overexpression experiments (Figure 5), pCS2+cPla2mKate2 or pCS2+cPla2S498AmKate2 were linearized, and in vitro transcribed using the mMessage mMachine SP6 kit (Thermo Fisher). 2.3 nL of 500 ng/µl mRNA was injected into one-cell stage zebrafish embryos.
Pharmacological treatments
Larvae were preincubated for 40–60 min in E3 or isotonic E3 supplemented with the following compounds: 20 µM Zileuton (Cayman), 100 µM Aspirin (Acetylsalicylic acid, ACROS Organics), 50 µM Licofelone (Cayman), 400 µg/mL cycloheximide (CHX, Sigma). 0.5% digitonin (Cayman) and 120 µM melittin (Millipore) were injected into the ear without preincubation. 25 µM arachidonic acid (Sigma) was injected into the ear, and simultaneously applied in the bathing solution. Dimethyl sulfoxide (DMSO, Sigma) was used as a solvent for all water non-soluble compounds with a maximal concentration of 1%. Isotonic E3 was used to dissolve digitonin and melittin. Compounds were present throughout the experiments. To confirm the inhibitory efficiency of CHX, 2.5–3 dpf TG(hsp70:cPla2mKate2) (Enyedi et al., 2016) zebrafish were pretreated with 400 µg/mL CHX or 0.1% DMSO for 1 h at 28°C followed by 1 h heat shock at 37°C. Heat shock-induced expression o f mKate2 fluorescence was measured over 5 hours by fluorescence microscopy (Figure S3C).
Antisense morpholinos
One-cell stage embryos were injected with 2.3 nL morpholino (Gene Tools) diluted in water. With one exception, only previously characterized morpholinos were used (Enyedi et al., 2013; Progatzky et al., 2014; van der Sar et al., 2006). Please see Key Resources Table for information on morpholinos.
STAR*METHODS.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Rabbit phospholipase A2 | Abcam | Cat#ab135825 |
| anti-Mouse beta-Actin | Sigma-Aldrich | Cat#A5441; RRID: AB_476744 |
| Goat anti-Rabbit IgG-HRP | Santa Cruz Biotechnology | Cat#sc-2054; RRID: AB_631748 |
| Goat anti-Mouse IgG-HRP | Santa Cruz Biotechnology | Cat#sc-2005; RRID: AB_631736 |
| anti-Digoxigenin-AP, Fab fragments | Roche | Cat#11093274910 |
| Bacterial and Virus Strains | ||
| wt PA14 | Joao Xavier (MSKCC) | N/A |
| eGFP-tagged PA14 | Joao Xavier (MSKCC) | N/A |
| PA14 exoU | Feinbaum et al., 2012 | N/A |
| PA14 fleN | Feinbaum et al., 2012 | N/A |
| PA14 ORF_11 | Feinbaum et al., 2012 | N/A |
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher | Cat#C404010 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Arachidonic acid | Sigma | Cat#A9673 |
| Acetylsalicylic acid (Aspirin) | ACROS Organics | Cat#AC158180025 |
| Cycloheximide | Sigma | Cat#01810 |
| Digitonin | Cayman | Cat#14952 |
| Dimethyl sulfoxide | Sigma | Cat#276855 |
| ethyl 3-aminobenzoate methanesulfonate | Sigma | Cat#10521 |
| Gentamycin | IBI Scientific | Cat#IB02030 |
| Licofelone | Cayman | Cat#10007692 |
| Agarose LM | Gold Biotechnology | Cat#A-204-100 |
| Melittin | Millipore | Cat#444605 |
| Zileuton | Cayman | Cat#10006967 |
| Nitrotetrazolium Blue chloride (NBT) | Sigma-Aldrich | Cat#N6876 |
| 5-Bromo-4-chloro-3-indolyl phosphate p-toluidine salt (BCIP) | Sigma | Cat#B8503 |
| Paraformaldehyde | Aldrich | Cat#441244 |
| Methanol | Fisher | Cat#A452-4 |
| Formamide DI deionized | AmericanBio | Cat#AB00600 |
| Maleic acid | Sigma-Aldrich | Cat#M0375 |
| Sodium chloride | Fisher | Cat#BP358-212 |
| Trizma base | Sigma | Cat#T1503 |
| Glycerol | Sigma | Cat#G2025 |
| Critical Commercial Assays | ||
| mMessage mMachine SP6 Kit | Thermo Fisher | Cat#AM1340 |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Fisher | Cat#K1621 |
| PhosphoProtein Purification Kit | QIAGEN | Cat#37101 |
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | Cat#210518 |
| DIG RNA Labeling Kit | Roche | Cat#11175025910 |
| Deposited Data | ||
| mRNA sequencing data | This paper | GEO: GSE111528 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: wt casper | White et al., 2008 | N/A |
| Zebrafish: TG(lysC:Casper1GR) | This paper | N/A |
| Zebrafish: TG(hsp70:cPla2mKate2) | Enyedi et al., 2016 | N/A |
| Sequence-Based Reagents | ||
| Forward primer for S498A mutagenesis: 5′- TCAACCCTGC CCTTCGCTCCCTTCAGCGGCATC – 3′ | Enyedi et al., 2016 | N/A |
| Reverse primer for S498A mutagenesis: 5′ – GATGCCGCT GAAGGGAGCGAAGGGCAGGGTTGA – 3′ | Enyedi et al., 2016 | N/A |
| cpla2 splice MO1: 5′- AAGCGTCACTTACTATAATGTTGGA-3′ | Enyedi et al., 2013 | N/A |
| cpla2 splice MO2: 5′-TCCCTTTTAGAATTTACCTTGCCGA-3′ | This paper | N/A |
| cpla2 ATG MO: 5′-ATGCTCAACTATAATGTTGGACATT-3′ | Enyedi et al., 2013 | N/A |
| pycard MO: 5′-CAATTGCACTTACATTGCCCTGTGT-3′ | Progatzky et al., 2014 | N/A |
| myd88 MO: 5′-TAGCAAAACCTCTGTTATCCAGCGA-3′ | van der Sar et al., 2006 | N/A |
| myd88 MisPrime MO: 5′-TAcCAtAACCTgTGTTATCgAGgGA-3′ | van der Sar et al., 2006 | N/A |
| Forward primer to confirm the knockdown of pycard MO: 5′ -CGCGTCACAAAGTCTGCAAT-3′ | Progatzky et al., 2014 | N/A |
| Reverse primer to confirm the knockdown of pycard MO: 5′ -CATCAGAGGGAGCACCTTTGC-3′ | Progatzky et al., 2014 | N/A |
| Forward primer to generate il1b antisense probe: 5′- ATGGC ATGCGGGCAATAT-3′ | This paper | N/A |
| Reverse primer to generate il1b antisense probe: 5′- TAATA CGACTCACTATAGGGCTGCAGCTCGAAGTTAATGATG-3′ | This paper | N/A |
| Software and Algorithms | ||
| ImageJ/FIJI | Schindelin et al., 2012 | https://fiji.sc/ |
| MTrackJ plugin of ImageJ/FIJI | Meijering et al., 2012 | https://imagescience.org/meijering/software/mtrackj |
| MOSAIC plugin of ImageJ/FIJI | Sbalzarini and Koumoutsakos, 2005 | http://mosaic.mpi-cbg.de/?q=downloads/imageJ |
| MATLAB | Mathworks Inc., USA | http://mathworks.com |
| Anaconda Python 2.7 | Anaconda Inc., USA | https://www.anaconda.com |
| CellProfiler | Lamprecht et al., 2007 | http://cellprofiler.org |
| Minitab | Minitab Inc., USA | http://www.minitab.com |
| Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism |
| Other | ||
| TRIzol Reagent | Life technologies | Cat#15596026 |
| FluoSpheres polystyrene, 1.0 µm, blue-green (430/465) | Life technologies | Cat#F13080 |
| PhosSTOP EASYpack | Roche | Cat#04906837001 |
| cOmplete Tablets EASY pack | Roche | Cat#04693116001 |
| NuPage 4-12% Bis-Tris Protein Gels | Thermo Fisher | Cat#NP0321BOX |
| Dynabeads Oligo(dT)25 | Invitrogen | Cat#61002 |
| RIPA Buffer | Cell Signaling | Cat#9806S |
| Glass beads, acid-washed 425-600 µm | Sigma-Aldrich | Cat#G8772 |
| Proteinase K | Roche | Cat#03115879001 |
| Saline sodium citrate (SSC) buffer, 20X solution | AmericanBio | Cat#AB13156 |
| Tween-20 | Sigma | Cat#P1379 |
| Bovine Serum Albumin (BSA) | Sigma | Cat#A2153 |
| Fetal Bovine Serum (FBS) | Corning | Cat#35-010-CV |
| Heparin sodium salt from porcine intestinal mucosa | Sigma-Aldrich | Cat#H4784 |
| Ribonucleic acid from torula yeast | Sigma | Cat#R6625 |
| Phosphate buffered saline (PBS) | Sigma | Cat#79382 |
| 2.0 mL MT Graduated Mixed tube | Fisher Scientific | Cat#05-408-146 |
| Amersham ECL Western Blotting Detection Reagents | GE Healthcare | Cat#RPN2106 |
cpla2 silencing was confirmed SDS-PAGE and western blotting of morphant and control lysates using a zebrafish cross-reactive cPLA2 antibody (Abcam; 1:500). To confirm gene silencing with the pycard MO, morphant mRNA was isolated from 2.5–3 dpf larvae with Oligo (dT)25 Dynabeads (Invitrogen), followed by one-step RT-PCR using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Primer sequences were as follows: pycard forward, 5' -CGCGTCACAAAGTCTGCAAT-3'; pycard reverse, 5' -CATCAGAGGGAGCACCTTTGC-3' (Progatzky et al., 2014).
SDS-PAGE and western blotting
2.5–3 dpf larvae were anaesthetized and homogenized in RIPA buffer (Cell Signaling Technology) supplemented with 1× protease inhibitor cocktail (cOmplete, Roche). Homogenization was performed for 45 s in a FastPrep 24 Cell Disrupter (MP Biomedicals) at maximum speed (6.5 m/s) in the cold room. Protein concentration was quantified by standard bicinchoninic acid assay (BCA) assay. Equal amounts of protein were separated by NuPAGE 4%–12% Bis-Tris gel electrophoresis according to the manufacturer’s protocol (Life Technologies), and wet-transferred to a nitrocellulose membrane in Tris-Glycine transfer buffer containing 20% methanol at 120 V for 90 min at 4°C. Western blot membranes were probed with ant i-cPLA2 (Abcam; 1:500), and anti-β-actin (SIGMA-ALDRICH; 1:5000) primary antibodies, followed by incubation with goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology; 1:5000) and goat anti-mouse IgG-HRP (Santa Cruz Biotechnology; 1:5000), respectively. Amersham ECL prime western blotting detection reagent (GE Healthcare) was used for western band detection according to manufacturer`s protocol. Chemiluminescence detection was performed with an ImageQuant LAS 500 device (GE Healthcare).
Measuring cPla2 phosphorylation in larval lysates by affinity purification
Phosphorylation-specific antibodies are not available for zebrafish cPla2. To measure phosphorylated cPla2 in infected zebrafish larvae, we resorted to an affinity purification scheme. At least 30 fish were injected with standard E3 with or without PA (OD600=7) as described above. Phosphorylated proteins were isolated using PhosphoProtein Purification Kit (Qiagen) according to manufacturer’s protocol with three modifications: (i) 1× PhosSTOP phosphotase inhibitor (Roche) was added to PhosphoProtein lysis buffer. (ii) Fish were homogenized with FastPrep 24 Cell Disrupter as described above. (iii) Fish tissue homogenate was diluted in 300 µl lysis buffer. The column eluate was concentrated to <50 µl using Nanosep Ultrafiltration Columns from the kit. Western blotting analysis was performed as described above. To confirm equal lysate-loading onto the phospho-affinity column, the column flow-through (FT, Figure S4D) was analyzed by SDS-PAGE and western blotting using an anti-β-actin antibody.
Embryo collection, RNA extraction and sequencing
PA resuspended in either standard or isotonic E3 were injected into the otic vesicle of 2.5–3 dpf zebrafish larvae. Uninjected zebrafish larvae served as control. ~20 larvae from each group were euthanized 1 h after injection. After homogenization, total RNA was extracted using Trizol reagent (Life technologies) according to manufacturer`s protocol. The following procedures where performed by the MSKCC Genomics and Bioinformatics core facilities. Briefly, after Quant-It quantification and quality control of Agilent BioAnalyzer, 500 ng of total RNA underwent polyA selection and Truseq library preparation according to instruction provided by Illumina (TruSeq Stranded mRNA Library Prep Kit), with 8 cycles of PCR. Samples were barcoded and run on a Hiseq 2500 in a 50bp/50bp Paired end run, using the TruSeq SBS Kit v4 (Illumina). An average of 42 million paired reads was generated per sample. The percent of mRNA bases was close to 53% on average. The output data (FASTQ files) were mapped to the zebrafish genome GCz10 (UCSC) using the rnaStar aligner (Dobin et al., 2013) that mapped reads genomically and resolved reads across splice junctions. A two-pass-mapping method (Engström et al., 2013) was used in which the reads were mapped twice. The first mapping pass used a list of known annotated junctions from Ensemble. Junctions found in the first pass were then added to the known junctions, and a second mapping pass was done (on the second pass the RemoveNoncanoncial flag was used). After mapping, the output SAM files were post processed using the PICARD tools to: add read groups, AddOrReplaceReadGroups which additionally sorted the file and converted it to the compressed BAM format. The expression count matrix was then computed from the mapped reads using HTSeq (www-huber.embl.de/users/anders/HTSeq). The raw count matrix generated by HTSeq was then processed using the R/Bioconductor package DESeq (www-huber.embl.de/users/anders/DESeq), which was used to both normalize the full dataset and analyze differential expression between sample groups.
Whole mount in situ hybridization (WISH)
To generate the il1b antisense riboprobe, probe template was amplified from zebrafish cDNA by PCR with a T7-modified antisense primer (Forward, 5’- ATGGCATGCGGGCAATAT-3’, reverse, 5’-TAATACGACTCACTATAGGGCTGCAGCTCGAAGTTAATGATG-3’) and was transcribed in vitro with T7 polymerase with conjugated Digoxigenin-11-uridine-5’-triphosphate (DIG-11-UTP) from DIG RNA Labeling Kit (T7) (Roche).
WISH was performed as previously described (Gerlach and Wingert, 2014). Briefly, ~ 20 2.5–3 dpf TG(lysC:Casper1GR) larvae were fixed in 4% paraformaldehyde (PFA, Aldrich) overnight at 4 °C. Following removal of PFA, larvae were washed two times with 1× PBST (1× PBS (Sigma), 0.1 % Tween-20 (Sigma)) followed by a wash with 100% methanol (Fisher) at room temperature. Larvae were first immersed in fresh 100% methanol for 20 min at −20 °C, then subsequently wa shed with 50% and 30% methanol (in PBST), and finally with PBST without methanol. Larvae were permeabilized with 10 µg/mL proteinase K (Roche, diluted in PBST) for ~22 min, and rinsed twice with PBST. Larvae were stored in 4% PFA overnight at 4°C. PFA was removed, and larvae were washed three times with PBST, and transferred to flat bottom microcentrifuge tubes (Fisher Scientific). Following removal of PBST, larvae were incubated in 1 mL HYB+ hybridization solution (50% formamide (AmericanBio), 5× SSC (AmericanBio), 0.1% Tween-20, 5 mg/mL yeast torula RNA (Sigma), 50 µg/µL heparin (Sigma-Aldrich)) for 4 h at 70 °C. After HYB+ was removed, 500 µL 1 ng/µL il1b probe diluted in HYB+ was added, and larvae were incubated overnight at 70°C. Larvae were washed at 70 °C with 50% formamide/2× SSCT (2× SSC, 0.1% Tween-20) for 30 min (twice), for 15 min with 2× SSCT, and for 30 min with 0.2× SSCT (twice). Larvae were then washed twice in MABT (Maleic Acid Buffer: 0.1 M maleic acid (Sigma-Aldrich), 0.15 M sodium chloride (Fisher), 0.23 M Trizma base (Sigma), 0.1% Tween-20, pH=7.4) for 5 min at room temperature. Larvae were incubated in blocking solution (2% BSA (Sigma), 10% FBS (Corning), 70% MABT) for 2 h at room temperature. Anti-Digoxigenin (Anti-DIG) secondary antibody (Roche, 1:5000) recognizing DIG conjugated to alkaline phosphatase was applied at 4 °C overnight. To remove extra secondary antibody, larvae were quickly washed three times in MABT, followed by twelve 15 min washes in MABT. Colorimetric substrates of NBT (Sigma) and BCIP (Sigma) were applied to the larvae to generate a colorimetric signal. This reaction was monitored every 20 min to prevent background-overstaining and stopped at ~90 min by washing in PBST and subsequent fixation in 4% PFA. Larvae were mounted in glycerol (Sigma) for colorimetric imaging. Images were acquired with on stereo microscope (Nikon) equipped with a Nikon DS-Fi1 CCD color camera (Nikon).
Quantification and statistical analysis
All error bars indicate standard errors of means (SEM). For most mean value comparisons, p-values were derived by unpaired, two-tailed t-tests assuming unequal variances (heteroscedastic) using Prism (GraphPad), Excel (Microsoft), or Matlab. For mean value comparison of tracking parameters in Figure 2C, a one-way Anova test was applied (Matlab). For comparison of HR-indices, a two-tailed Fisher’s exact test (Minitab) was used. For comparison of Kaplan-Meier survival curves, a log-rank test (Minitab) was used. For statistical analysis, animal experiments from different experimental days were aggregated. Sample sizes were not predetermined. The experiments were not randomized, and the investigators were not blinded to allocation during experiments or outcome assessment. A detailed summary of experimental numbers for all experiments is provided in Table S1.
Data and software availability
The RNA-seq data of ear infected zebrafish larvae under different experimental conditions has been deposited in the Gene Expression Omnibus (GEO, NCBI) under the accession code GSE111528.
Supplementary Material
Table S1. Related to Figure 1–5 and Figure S1–S4. Differential gene expression profiles (mRNAseq) and experimental numbers.
Data File 1. Related to STAR Methods. Matlab scripts used for leukocyte recruitment analysis.
Movie S1. Related to Figure 1. Zebrafish ear injected with fluorescent PA.
Time-lapse movie showing retention of EGFP-labeled P. aeruginosa within the zebrafish ear under hypotonic (top panel), and isotonic (bottom panel) bathing conditions. Scale bar, 100 µm. Time, h:min.
Movie S2. Related to Figure 1. Zebrafish ear injected with fluorescent beads and dextran. Time-lapse movie showing concomitant retention of fluorescent beads within (left panel), and leakage of 4 kD fluorescent dextran from the ear (right panel). Scale bar, 100 µm. Time, h:min.
Movie S3. Related to Figure 1. Leukocyte recruitment to ear infection.
Time-lapse movie montage showing “high responder” and “low responder” neutrophil recruitment to infected zebrafish ears in the absence (Iso + PA) or presence (Hypo + PA) of osmotic tissue damage signaling. Red, neutrophils in TG(lysC:Casper1GR) fish. Blue, fluorescent beads. Scale bar, 100 µm. Time, h:min.
Movie S4. Related to Figure 2. Leukocyte recruitment to trunk infection.
Time-lapse movie montage showing neutrophil recruitment to infected zebrafish trunk muscle in the absence (Iso + PA), or presence (Hypo + PA) of osmotic tissue damage signaling. Red, neutrophils in TG(lysC:Casper1GR) fish. Blue, fluorescent beads. Scale bar, 100 µm. Time, h:min.
Highlights.
Live imaging of zebrafish reveals a beneficial role for tissue damage signaling
Lack of damage signaling abrogates early neutrophil migration to microbial infection
Cytosolic phospholipase A2 (cPla2) integrates microbial and tissue damage cues
Tissue damage cues initiate, and microbial patterns amplify neutrophil migration
Acknowledgments
The authors thank Alexander Rudensky, Michael Glickman, Timothy Mitchison, and Martin Wuehr, as well as Weili Tian, and Xiaojing Mu for comments on the manuscript or study. The authors thank Shan Ba for discussion of data analysis, and Joao Xavier and Cole Haynes for the kind gift of PA strains. Research was supported by the NIH/NIGMS grant R01GM099970, an American Asthma Foundation Scholar award to PN, the MSKCC Functional Genome Initiative, and in part through the NIH/NCI Cancer Center Support grant P30CA008748. Specifically, the authors thank the Integrated Genomics Operation Core and Bioinformatics Core at MSKCC for their assistance. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
PN conceived the study. PN and CH designed the experiments. CH performed all experiments and image analysis. PN wrote image analysis scripts in Matlab and Python and assisted with data analysis in Matlab. PN and CH wrote the paper. All authors read, commented on, and agreed to the final version of the manuscript.
Declaration of Interests
The authors declare no competing interests.
References
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (80-.) 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science (80-.) 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- Bloes DA, Kretschmer D, Peschel A. Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat. Rev. Micro. 2015;13:95–104. doi: 10.1038/nrmicro3390. [DOI] [PubMed] [Google Scholar]
- Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux D, Goure J, Chabert J, Usson Y, Attree I. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 2001;40:76–85. doi: 10.1046/j.1365-2958.2001.02368.x. [DOI] [PubMed] [Google Scholar]
- Deng Q, Harvie EA, Huttenlocher A. Distinct signalling mechanisms mediate neutrophil attraction to bacterial infection and tissue injury. Cell Microbiol. 2012;14:517–528. doi: 10.1111/j.1462-5822.2011.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovi JV, He L-K, DiPietro LA. Accelerated wound closure in neutrophil-depleted mice. J. Leukoc. Biol. 2003;73:448–455. doi: 10.1189/jlb.0802406. [DOI] [PubMed] [Google Scholar]
- Dovi JV, Szpaderska AM, DiPietro LA. Neutrophil function in the healing wound: adding insult to injury? Thromb. Haemost. 2004;92:275–280. doi: 10.1160/TH03-11-0720. [DOI] [PubMed] [Google Scholar]
- Eliopoulos AG, Dumitru CD, Wang C-C, Cho J, Tsichlis PN. Induction of COX-2 by LPS in macrophages is regulated by Tpl2-dependent CREB activation signals. EMBO J. 2002;21:4831–4840. doi: 10.1093/emboj/cdf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Steijger T, Sipos B, Grant GR, Kahles A, Rätsch G, Goldman N, Hubbard TJ, Harrow J, Guigó R, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi B, Kala S, Nikolich-Zugich T, Niethammer P. Tissue damage detection by osmotic surveillance. Nat. Cell Biol. 2013;15:1123–1130. doi: 10.1038/ncb2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi B, Jelcic M, Niethammer P. The Cell Nucleus Serves as a Mechanotransducer of Tissue Damage-Induced Inflammation. Cell. 2016;165:1160–1170. doi: 10.1016/j.cell.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum RL, Urbach JM, Liberati NT, Djonovic S, Adonizio A, Carvunis A-R, Ausubel FM. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 2012;8:e1002813. doi: 10.1371/journal.ppat.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, Wu C, Mende-Mueller L, Frank DW. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- Fitsialos G, Chassot A-A, Turchi L, Dayem MA, LeBrigand K, Moreilhon C, Meneguzzi G, Buscà R, Mari B, Barbry P, et al. Transcriptional signature of epidermal keratinocytes subjected to in vitro scratch wounding reveals selective roles for ERK1/2, p38, and phosphatidylinositol 3-kinase signaling pathways. J. Biol. Chem. 2007;282:15090–15102. doi: 10.1074/jbc.M606094200. [DOI] [PubMed] [Google Scholar]
- Forn-Cuní G, Varela M, Pereiro P, Novoa B, Figueras A. Conserved gene regulation during acute inflammation between zebrafish and mammals. Sci. Rep. 2017;7:41905. doi: 10.1038/srep41905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault WJ, Enyedi B, Niethammer P. Osmotic surveillance mediates rapid wound closure through nucleotide release. J. Cell Biol. 2014;207:767–782. doi: 10.1083/jcb.201408049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach GF, Wingert RA. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota and zeta. Dev. Biol. 2014;396:183–200. doi: 10.1016/j.ydbio.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Micro. 2009;7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley BP, Williams NL, McCormick BA. Involvement of phospholipase A2 in Pseudomonas aeruginosa-mediated PMN transepithelial migration. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290:L703–L709. doi: 10.1152/ajplung.00390.2005. [DOI] [PubMed] [Google Scholar]
- Hurley BP, Pirzai W, Mumy KL, Gronert K, McCormick BA. Selective eicosanoid-generating capacity of cytoplasmic phospholipase A2 in Pseudomonas aeruginosa-infected epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2011;300:L286–94. doi: 10.1152/ajplung.00147.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Kohjima M, Kuwano Y, Mino H, Teshima-Kondo S, Takeya R, Tsunawaki S, Wada A, Sumimoto H, Rokutan K. Helicobacter pylori lipopolysaccharide activates Rac1 and transcription of NADPH oxidase Nox1 and its organizer NOXO1 in guinea pig gastric mucosal cells. Am. J. Physiol. Cell Physiol. 2005;288:C450–7. doi: 10.1152/ajpcell.00319.2004. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszyński A, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science (80-.) 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Kono H, Rock KL. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C-B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, Germain RN. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013;498:371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Bio Techniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Land WG, Messmer K. The danger theory in view of the injury hypothesis: 20 years later. Front. Immunol. 2012;3:349. doi: 10.3389/fimmu.2012.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Bhatta S, Gerzanich V, Simard JM. Cytotoxic edema: mechanisms of pathological cell swelling. Neurosurg. Focus. 2007;22:E2. doi: 10.3171/foc.2007.22.5.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SC, Köhl U, Maier TJ, Steinhilber D, Sorg BL. TLR2 ligands augment cPLA2alpha activity and lead to enhanced leukotriene release in human monocytes. J. Leukoc. Biol. 2009;86:389–399. doi: 10.1189/jlb.1008591. [DOI] [PubMed] [Google Scholar]
- Liu T, Yamaguchi Y, Shirasaki Y, Shikada K, Yamagishi M, Hoshino K, Kaisho T, Takemoto K, Suzuki T, Kuranaga E, et al. Single-cell imaging of caspase-1 dynamics reveals an all-or-none inflammasome signaling response. Cell Rep. 2014;8:974–982. doi: 10.1016/j.celrep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am. J. Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Tateda K, Miyazaki S, Furuya N, Ohno A, Ishii Y, Hirakata Y, Yamaguchi K. Effect of immunisation with Pseudomonas aeruginosa on gut-derived sepsis in mice. J. Med. Microbiol. 1998;47:295–301. doi: 10.1099/00222615-47-4-295. [DOI] [PubMed] [Google Scholar]
- Matzinger P. The danger model: a renewed sense of self. Science (80-.) 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science (80-.) 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- McLachlan G, Peel D. Finite Mixture Models. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2000. [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Meth. Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Nathan C. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- Ng LG, Qin JS, Roediger B, Wang Y, Jain R, Cavanagh LL, Smith AL, Jones CA, de Veer M, Grimbaldeston MA, et al. Visualizing the neutrophil response to sterile tissue injury in mouse dermis reveals a three-phase cascade of events. J. Invest. Dermatol. 2011;131:2058–2068. doi: 10.1038/jid.2011.179. [DOI] [PubMed] [Google Scholar]
- Niethammer P. The early wound signals. Curr. Opin. Genet. Dev. 2016;40:17–22. doi: 10.1016/j.gde.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C (Christiane), Dahm R. Zebrafish: A practical approach. Oxford: Oxford University Press; [Google Scholar]
- Nuzzi PA, Lokuta MA, Huttenlocher A. Analysis of neutrophil chemotaxis. Methods Mol. Biol. 2007;370:23–36. doi: 10.1007/978-1-59745-353-0_3. [DOI] [PubMed] [Google Scholar]
- Picoli T, Peter CM, Zani JL, Waller SB, Lopes MG, Boesche KN, Vargas GDÁ, Hübner S, de O, Fischer G. Melittin and its potential in the destruction and inhibition of the biofilm formation by Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa isolated from bovine milk. Microb. Pathog. 2017;112:57–62. doi: 10.1016/j.micpath.2017.09.046. [DOI] [PubMed] [Google Scholar]
- Progatzky F, Sangha NJ, Yoshida N, McBrien M, Cheung J, Shia A, Scott J, Marchesi JR, Lamb JR, Bugeon L, et al. Dietary cholesterol directly induces acute inflammasome-dependent intestinal inflammation. Nat Commun. 2014;5:5864. doi: 10.1038/ncomms6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H-Y, Shelhamer JH. Toll-like receptor 4 signaling regulates cytosolic phospholipase A2 activation and lipid generation in lipopolysaccharide-stimulated macrophages. J. Biol. Chem. 2005;280:38969–38975. doi: 10.1074/jbc.M509352200. [DOI] [PubMed] [Google Scholar]
- Rådmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim. Biophys. Acta. 2015;1851:331–339. doi: 10.1016/j.bbalip.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Ruipérez V, Astudillo AM, Balboa MA, Balsinde J. Coordinate regulation of TLR-mediated arachidonic acid mobilization in macrophages by group IVA and group V phospholipase A2s. J. Immunol. 2009;182:3877–3883. doi: 10.4049/jimmunol.0804003. [DOI] [PubMed] [Google Scholar]
- Van der Sar AM, Stockhammer OW, van der Laan C, Spaink HP, Bitter W, Meijer AH. MyD88 innate immune function in a zebrafish embryo infection model. Infect. Immun. 2006;74:2436–2441. doi: 10.1128/IAI.74.4.2436-2441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Schievella AR, Regier MK, Smith WL, Lin LL. Calcium-mediated translocation of cytosolic phospholipase A2 to the nuclear envelope and endoplasmic reticulum. J. Biol. Chem. 1995;270:30749–30754. doi: 10.1074/jbc.270.51.30749. [DOI] [PubMed] [Google Scholar]
- Schiffmann E, Corcoran BA, Wahl SM. N-formylmethionyl peptides as chemoattractants for leucocytes. Proc. Natl. Acad. Sci. USA. 1975;72:1059–1062. doi: 10.1073/pnas.72.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield ZV, Woodruff TM, Halai R, Wu MC-L, Cooper MA. Neutrophils--a key component of ischemia-reperfusion injury. Shock. 2013;40:463–470. doi: 10.1097/SHK.0000000000000044. [DOI] [PubMed] [Google Scholar]
- Simpson DM, Ross R. The neutrophilic leukocyte in wound repair a study with antineutrophil serum. J. Clin. Invest. 1972;51:2009–2023. doi: 10.1172/JCI107007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Tucker DE, Ghosh M, Ghomashchi F, Loper R, Suram S, John BS, Girotti M, Bollinger JG, Gelb MH, Leslie CC. Role of phosphorylation and basic residues in the catalytic domain of cytosolic phospholipase A2alpha in regulating interfacial kinetics and binding and cellular function. J. Biol. Chem. 2009;284:9596–9611. doi: 10.1074/jbc.M807299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vaart M, van Soest JJ, Spaink HP, Meijer AH. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis. Model. Mech. 2013;6:841–854. doi: 10.1242/dmm.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE. Cutting edge commentary: A copernican revolution? doubts about the danger theory. The Journal of Immunology. 2000;165:1725–1728. doi: 10.4049/jimmunol.165.4.1725. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR, et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science (80-.) 2016;352:1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 1–5 and Figure S1–S4. Differential gene expression profiles (mRNAseq) and experimental numbers.
Data File 1. Related to STAR Methods. Matlab scripts used for leukocyte recruitment analysis.
Movie S1. Related to Figure 1. Zebrafish ear injected with fluorescent PA.
Time-lapse movie showing retention of EGFP-labeled P. aeruginosa within the zebrafish ear under hypotonic (top panel), and isotonic (bottom panel) bathing conditions. Scale bar, 100 µm. Time, h:min.
Movie S2. Related to Figure 1. Zebrafish ear injected with fluorescent beads and dextran. Time-lapse movie showing concomitant retention of fluorescent beads within (left panel), and leakage of 4 kD fluorescent dextran from the ear (right panel). Scale bar, 100 µm. Time, h:min.
Movie S3. Related to Figure 1. Leukocyte recruitment to ear infection.
Time-lapse movie montage showing “high responder” and “low responder” neutrophil recruitment to infected zebrafish ears in the absence (Iso + PA) or presence (Hypo + PA) of osmotic tissue damage signaling. Red, neutrophils in TG(lysC:Casper1GR) fish. Blue, fluorescent beads. Scale bar, 100 µm. Time, h:min.
Movie S4. Related to Figure 2. Leukocyte recruitment to trunk infection.
Time-lapse movie montage showing neutrophil recruitment to infected zebrafish trunk muscle in the absence (Iso + PA), or presence (Hypo + PA) of osmotic tissue damage signaling. Red, neutrophils in TG(lysC:Casper1GR) fish. Blue, fluorescent beads. Scale bar, 100 µm. Time, h:min.