Abstract
High-throughput mapping of cellular differentiation hierarchies from single-cell data promises to empower systematic interrogations of vertebrate development and disease. Here, we applied single-cell RNA sequencing to >92,000 cells from zebrafish embryos during the first day of development. Using a graph-based approach, we mapped a cell state landscape that describes axis patterning, germ layer formation, and organogenesis. We tested how clonally related cells traverse this landscape by developing a transposon-based barcoding approach (“TracerSeq”) for reconstructing single-cell lineage histories. Clonally related cells were often restricted by the state landscape, including a case in which two independent lineages converge on similar fates. Cell fates remained restricted to this landscape in chordin-deficient embryos. We provide web-based resources for further analysis of the single-cell data.
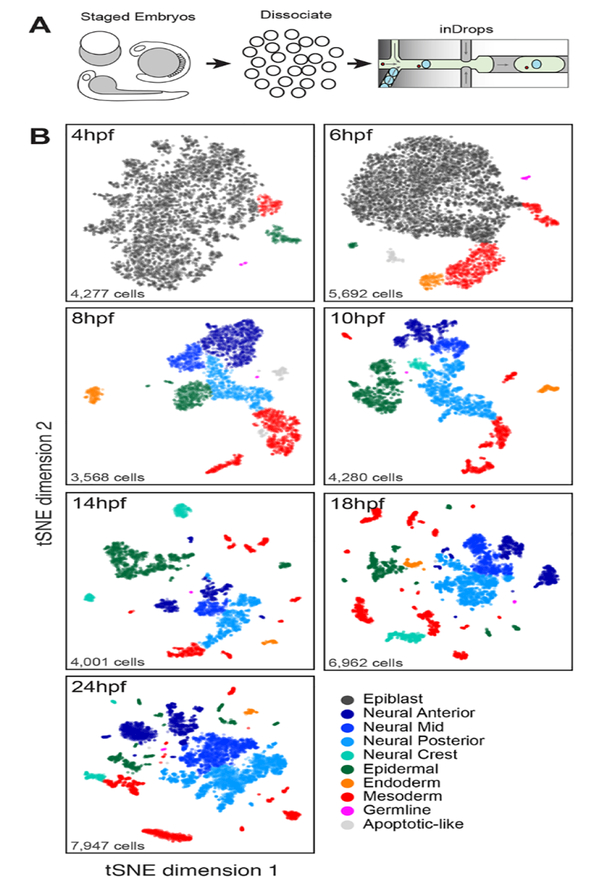
A major goal of developmental biology is to understand the progression of embryonic cell lineages from pluripotency to adulthood (1). Fate mapping, and analysis of mutant pheno-types, have explained much of what we know of development, yet we still lack a systematic atlas of all cell states in a developing embryo. Owing to technical advances in single-cell RNA sequencing (scRNA-seq) (2–6), it is now possible to assemble comprehensive single-cell atlases describing complex and dynamic in vivo biological processes. Here, we utilized inDrops scRNA-seq (4, 7) to collect over 92,000 single-cell transcriptomes from dissociated wild-type and mutant zebrafish embryos during the first 24 hours of embryonic development (Fig. 1, A and B, and fig. S1). For different developmental stages, we sampled 0.17x to 0.97x of the total cells per embryo, sufficient to detect cell states as rare as 0.1–0.5% of all cells (fig. S1C), including germ cells which were detected in all timepoints (Fig. 1B and table S2). From this dataset, clustering of the wild-type transcriptomes revealed an expanding set of epidermal, neural, mesodermal, and endodermal cell states over developmental time, many of which could be specifically annotated based on expression of marker genes (Fig. 1B, fig. S2A, and table S2) (8). We collected seven biological replicates for the final timepoint (24 hours post-fertilization, hpf), which demonstrated consistency of both transcriptional signatures and cell state proportions across independent specimens (fig. S2, B and C).
Fig. 1. A single-cell transcriptional atlas of the zebrafish embryo.
(A) Experimental workflow: Single-cell suspensions were dissociated from staged zebrafish embryos and introduced into the inDrops microfluidic device. Single-cell transcriptome libraries were prepared and sequenced by RNA-seq. (B) tSNE maps for each timepoint, constructed in dimensionality-reduced PCA subspace defined by highly co-variable genes (see methods). Cells are colored by germ layer identities inferred from expressed marker genes (see also fig. S2A and table S2).
A single-cell graph of cell state progression in the developing zebrafish embryo
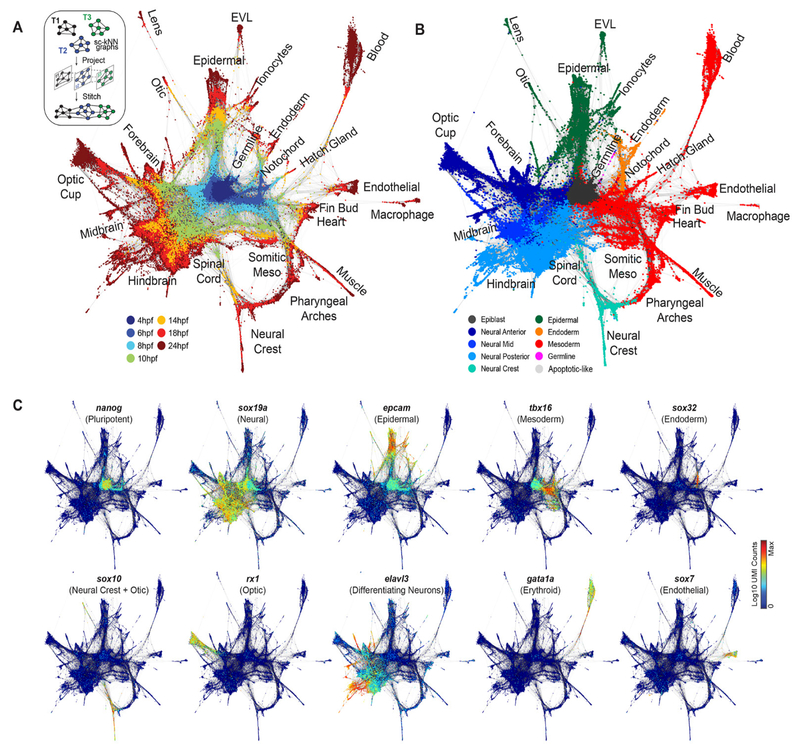
We sought to map trajectories of cell state during development by linking cell states across time. Several computational approaches exist to infer orderings of asynchronous processes from scRNA-seq data (9–11), typically by projecting all cells into a single low-dimensional latent space. Such strategies may be ill-suited to map gene expression in developing embryos, which exhibit dramatically increasing cell state dimensionality and continuous changes in the sets and numbers of cell state-defining genes (fig. S2, D and E). To overcome these obstacles, we developed a graph-based strategy for locally embedding consecutive timepoints on the basis of biological variation that they share, rather than using a global coordinate system for all timepoints. This approach first constructs a single-cell k-nearest-neighbor graph for each timepoint ti, with nodes representing cells and edges linking neighbors in a low-dimensional subspace; it then joins the graphs by identifying neighboring cells in pairs of adjacent time points, using a coordinate system learned from the future (ti+1) timepoint (see methods). The resulting graph spans all time points, and allows application of formal graph-based methods for data analysis. When applied to our zebrafish data, the full graph forms a branching network (Fig. 2A). Inspection of numerous domain and cell-type specific transcriptional markers shows that major initial branches represent neural, epidermal, and mesendodermal states undergoing progressive and spatially restricted differentiation (Fig. 2, B and C, and fig. S3). We also noted distinct and early branching events for germline, notochord, enveloping layer (EVL) epidermis, and the prechordal plate.
Fig. 2. Single-cell graph reveals a continuous developmental landscape of cell states.
(A) Overview of graph construction strategy, and a force-directed layout of the resulting single-cell graph (nodes colored by collection timepoint). For each cell, up to 20 within- or between-timepoint mutual nearest neighbor edges are retained. (B) Single-cell graph, colored by germ layer identities inferred from differentially expressed marker genes (see table S2). (C) Single-cell graphs, colored by log10 expression counts for indicated cell type-specific marker genes.
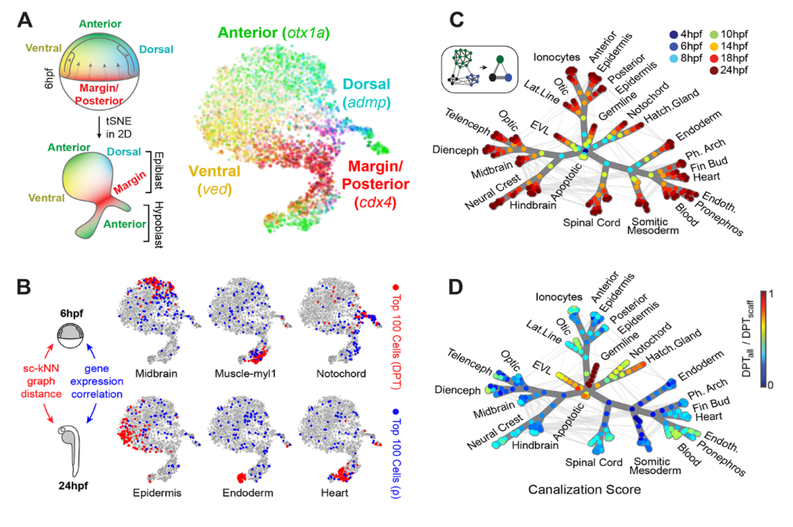
To test whether this graph recapitulates known lineage relationships, we used a measure of graph distance (Diffusion Pseudotime or “DPT”) (12) to explore long-range temporal connections between cell states. Cell states of the early gas-trula (shield stage, 6hpf) are defined largely by positional marker genes (Fig. 3A), yet these cells are connected – through the single-cell graph – to tissue-specific states that emerge later (e.g., pharyngula stage, 24hpf). We found that the shield stage cells with the shortest mean graph distance to each particular 24hpf tissue were clustered, and expressed spatial marker genes predicted from previous in vivo fate mapping studies (13–16), e.g., 24hpf neural tissues mapped to the 6hpf dorsal anterior epiblast (Fig. 3B and fig. S4). Conversely, direct comparison of 6hpf and 24hpf gene expression states failed to capture lineage relationships (blue points, Fig. 3B and fig. S4).
Fig. 3. Single-cell and coarse-grained graphs encode progenitor-fate relationships.
(A) tSNE map of 6hpf epiblast and hypoblast states, colored by normalized transcript counts for select positional marker genes. Overlapping color gradients demonstrate continuous expression domains defined by position. Diagram relates positions of cells in the tSNE map to theoretical positions in the embryo. (B) In silico fate predictions for 6hpf embryo cells. The top 100 cells with predicted 24hpf fate outcomes are indicated for shortest graph diffusion distances (red) or direct single-cell gene expression correlation distances (blue) between 6hpf cells and 24hpf cluster centroids. (C) Construction and overview of the coarse-grained graph (see also fig. S5). Nodes indicate states (groups of transcriptionally similar cells), colored by timepoint. Weighted edges connect similar states within or between timepoints. Spanning tree edges connecting each node to the 4hpf root state through the top weighted edges are highlighted in dark grey. (D) Coarse-grained graph nodes are colored by a “canalization” score, defined as the ratio of diffusion distances between each node and the 4hpf root node through state tree edges only vs. through all graph edges. Highly canalized regions of the graph correspond to branches with the fewest off-tree edges.
We next tested the extent to which the single-cell graph represents a simple tree-like hierarchy of discrete states. For this, we ‘coarse-grained’ the graph by collapsing groups of similar cells into state nodes; edges between state nodes were weighted by the number of original single-cell connecting edges. A spanning tree was then traced through the most densely weighted edges to a 4hpf root state (Fig. 3C and fig. S5A). This spanning tree (the ‘state tree’) reflects many specific aspects of early development. In the neural plate, we observe notable branch points for the optic cup, the diencephalon, telencephalon, mesencephalon, and rhombencephalon, with associated states for region-specific post-mitotic neurons (e.g., eomes+ and dlx1+ neurons in distinct forebrain branches). The neural plate also includes neural crest, which branches to include cell states for melanoblasts, iridoblasts and xanthoblasts. In the lateral plate / ventral mesoderm, the state tree encodes extensive branching into hematopoietic cells, endothelial cells, heart, pharyngeal arches, the pronephritic duct, and fin buds. In the endoderm, two branch points give rise to cell states for pancreatic primordium (which includes insulin+ cells) and the pharyngeal pouch. In the epidermal lineage, branch points differentiate the otic placode, lateral line, ionocytes, and several states expressing markers for annotated “mucous-secreting” cells (8). To facilitate data exploration, we developed web-based interfaces for the state tree and the full single-cell graph (www.tinyurl.com/scZfish2018). These tools permit interactive examination of: the inferred state hierarchy; expression for any gene of interest; and differential expression analysis between states, state combinations, or single cells.
Although many major cell state transitions are captured in the state tree, more complex features are evident in the coarse-grained and single-cell graphs. Off-tree interconnections between states, for example, were evident for (1) the neural crest and pharyngeal arches, (2) spinal cord and somitic mesoderm, (3) the neural plate, and others (Fig. 3C and fig. S5A). To formalize the degree to which the developmental landscape can be approximated as a hierarchy with discrete, non-looping branches, we defined a ‘canalization score’ (Fig. 3D, see legend for definition), which reflects the off-tree connectivity of each coarse-grained state node. This analysis revealed widespread regions of ‘low canalization’, particularly in the neural plate and somitic mesoderm. These observations suggest that, in contrast to the classic notion of a cell lineage, the zebrafish cell state landscape cannot be fully represented as a tree.
Cell lineage history does not invariantly reflect cell state graph topology
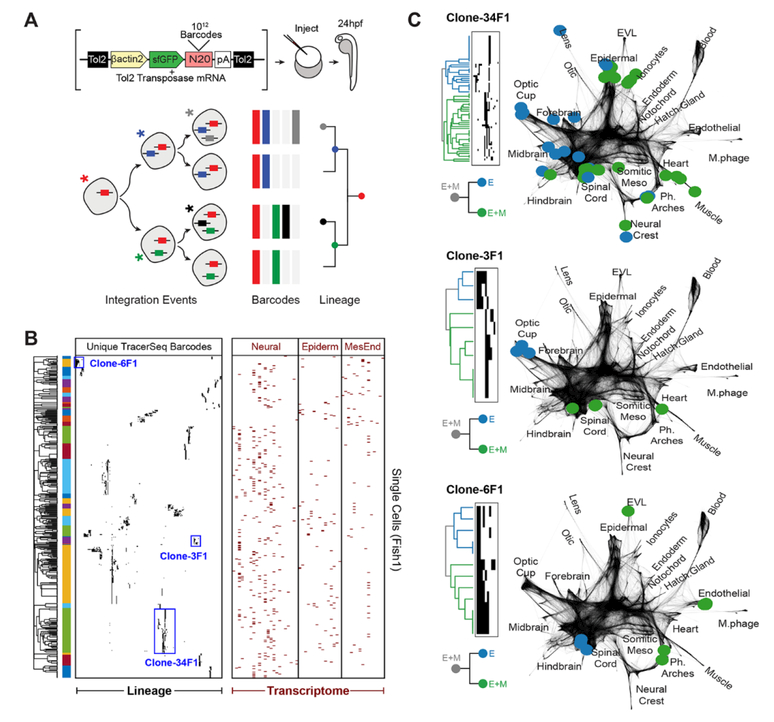
Although the single-cell and coarse-grained graphs represent an inferred landscape of developmental cell states, they do not reveal how individual cells traverse these states. A simple prediction would be that individual cell histories mirror graph topology. We tested this prediction by developing an inDrops-compatible strategy for recording in vivo lineage histories at the single-cell level: Sequencing of Transcribed Clonally Encoded Random Barcodes (“TracerSeq”). TracerSeq utilizes the Tol2 transposase system (17) to randomly integrate GFP reporter cassettes driven by the beta-actin promoter (actb2) into the zebrafish genome. To render each integration event unique and detectable by RNA-seq, we utilized Gibson assembly (18) without subsequent amplification to introduce a random 20mer sequence barcode into the GFP 3′ UTR (Fig. 4A and fig. S6). Because transgenic insertions can occur asynchronously over successive cell divisions, TracerSeq barcodes can facilitate the construction of lineage trees (Fig. 4A). TracerSeq offers an advantage over related Cas9-based approaches (19, 20), which can generate identical edits and/or large barcode deletions in independent lineages at non-trivial frequencies. By contrast, TracerSeq barcodes are uniformly distributed over a large sequence space (e.g., 420 = 1012 unique sequences), facilitating straightforward calling of genetic clones (fig. S7). The small (20bp) locus size also greatly simplifies the construction, sequencing, and analysis of TracerSeq inDrops libraries.
Fig. 4. Single-cell transcriptomic barcoding of cell lineages using TracerSeq.
(A) Method overview. (B) Clustered heatmap for 1/5 TracerSeq embryos (see also fig. S9, A to D) displaying lineage and transcriptome information for each cell. Heatmap rows are single cells for which both transcriptome and >1 TracerSeq barcodes were recovered. Columns denote unique TracerSeq barcodes (left, black squares: ≥1 UMI) and tissue identities (right, red squares) inferred from cluster annotations (table S2). Heatmaps were clustered using Jaccard similarity and average linkage. (C) Examples of TracerSeq founder clones with positions of constituent cells (colored nodes) overlaid on the single-cell graph. Graph edges are shown in dark grey. Colors indicate the first lineage bifurcation within each founder clone. In the three cases shown, the founder clone included cells that differentiated into both ectodermal and mesodermal states, while one of the two first subclones was restricted to ectoderm.
The use of TracerSeq to analyze potentially small clones of cells (each restricted to a single embryo) requires high-efficiency tissue dissociation and transcriptomic barcoding methods. We therefore optimized a high-yield cell dissociation and recovery protocol for individual 24hpf zebrafish embryos (fig. S1D and methods) and leveraged the high cell barcoding efficiency (>80%) of the inDrops platform (7). We then sequenced individual embryos (N=5) at 24hpf (fig. S7) that were injected at the 1-cell stage with the TracerSeq library, generating combined lineage+transcriptome datasets for 1,269 clonal barcodes distributed over 4,342 single cells (fig. S8). 2,361 of these cells (54%) were each marked by ≥2 distinct barcode integrations; 624 cells (14%) were marked by ≥5 integrations (fig. S8). Hierarchical clustering of TracerSeq barcodes organized these cells into over a hundred distinct founder clones with internal nested clone structures (Fig. 4B and fig. S9, A to D). We then compared the lineage history and inferred transcriptional history of each founder clone by embedding its constituent cells onto the single-cell graph (Fig. 4C). We found that the largest clones often marked a wide diversity of cell states. In multiple cases, however, additional barcode integrations in the same founder clone marked cells that were state-restricted. For example, one such clone (34F1), marked cells of the neural plate, epidermal tissues, and muscle, but contained a sub-clone restricted to anterior ectoderm. Similar lineage restriction events could be described for other founder clones (Fig. 4C). These observations suggest that the current timing of TracerSeq integrations encompasses the transition from unrestricted pluripotency to the first fate restriction events appearing in the zebrafish embryo.
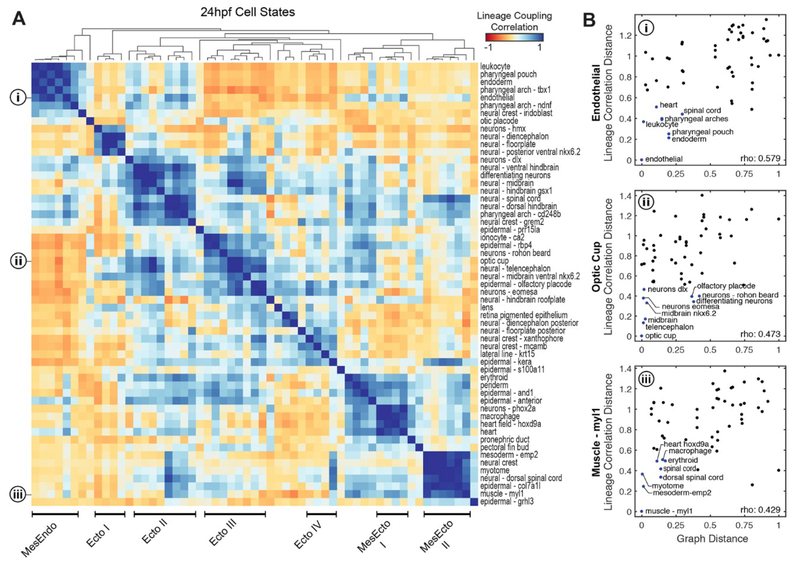
To investigate lineage relationships more systematically, we assessed the likelihood of recovering shared TracerSeq barcodes from all pairs of transcriptional states in the 24hpf zebrafish embryo. We first calculated a lineage coupling score (fig. S9E and methods), defined as the number of shared bar-codes relative to randomized data (z-score standardized), with values ranging from positive (coupled fates) to negative (anti-coupled fates). Hierarchical clustering of the pairwise correlation between coupling scores revealed structured groups of cell states (Fig. 5A), which comprised related tissues and/or inferred germ layer derivatives. These included one distinct group that contained both mesodermal and endodermal derivatives, 4 groups containing ectodermal derivatives, and 2 groups containing mixtures of ectoderm and mesoderm. Several of these lineage groups are corroborated by prior fate mapping studies. We discuss here three examples. The first major lineage group, (‘MesEndo’), includes derivatives of both lateral plate mesoderm and endoderm. These tissues originate from the marginal blastomeres of the early zebrafish gastrula, which involute first during gastrulation to form the hypoblast, and then rapidly migrate toward the animal pole (13, 15, 21). The observed lineage isolation of these tissues is thus consistent with an early spatial partitioning of this region, further reflected in Fig. 5A by negative lineage correlations to most other states. A second group, (Fig. 5A, ‘Ecto III’), captures strong lineage couplings between anterior neural tissues including the optic cup, midbrain, and telencephalon (16), and also to anterior epidermal derivatives such as the olfactory placode (22). These tissues are coupled to a lower degree with another group (‘Ecto II’), which includes couplings between the hindbrain, spinal cord, and neural crest (grem2+). The third example we note is a group coupling ectoderm and mesoderm (Fig. 5A, ‘MesEcto II), including muscle (myl1+), myotome, spinal cord, posterior neural crest, and epidermal states. These correlations mirror development of posterior body regions, which trace their origins to blastomeres proximal to the medial and ventral margin (13). These mesodermal-spinal cord couplings might also be explained by the presence of a later population of transient, multipotent neuromesodermal progenitor cells (NMPs) in the embryonic tailbud, which give rise to both of these populations (23–25). Interestingly, these lineage groups tend to be organized by position (e.g., along the A-P axis) rather than strictly by germ layer/tissue origin (e.g., neural, epidermal, mesodermal).
Fig. 5. TracerSeq reveals systematic relationships between cell lineage and cell state.
(A) Heatmap of TracerSeq lineage coupling scores (see methods) between pairs of 24hpf states, clustered by correlation distance and average linkage. Groups of states with similar lineage coupling signatures are annotated. (B) Quantitative relationships between lineage coupling correlation distances and scaled state tree diffusion distances for (i) endothelial, (ii) optic cup, and (iii) myl+ muscle states (see also fig. S10, A to F).
We next questioned how clonal relationships compared with cell state relationships. A simplistic model of development is that cells progressively diverge in state as they diverge in lineage. Developing embryos, however, could violate this prediction in at least two ways: first, clonally distinct embryonic fields can give rise to similar cell types (i.e., ‘convergent clones’); second, major transcriptional changes might drive related cells into qualitatively dissimilar states, possibly even late in development (i.e., ‘ divergent clones’). Overlaying TracerSeq lineage correlation scores on the cell state graph and comparing these scores to graph-derived state distances (Fig. 5B and fig. S10) revealed that some nearby states on the state graph were indeed clonally correlated, as expected by the simplistic model. However, nearby cell states also frequently displayed weak clonal correlations, suggesting convergent differentiation. These patterns were evident amongst state relationships for endothelial, optic cup, and muscle tissues (Fig. 5B and fig. S10, A to F), and systematically when examining all states (fig. S10G).
We observed considerably fewer cases of divergent clonal behavior (fig. S10G). However, one notable example manifested as apparent looping of the neural crest into the pharyngeal arches, which originate in the graph from both neural plate and lateral plate mesoderm and merge at 18–24hpf (Fig. 2, A and B, and fig. S11A). While the contribution of neural crest to various mesenchymal tissues is well established (26– 28), the transcriptional information reflected by the graph loop alone does not reveal which annotated pharyngeal arch states arise from neural crest. TracerSeq data, however, provides a clear signature of distinct clonal patterns between pharyngeal arch states: one pharyngeal arch state (ph.arch-tbx1) is a member of the “MesEndo” lineage group with mesodermal clonal associations, while the second pharyngeal arch state (ph.arch-cd248b) is clonally related to neural crest and posterior neural states (Fig. 5A and fig. S11, B to F). These data indicate that cells in the ph.arch-cd248b state diverged from a neural plate lineage and subsequently converged with other lateral plate-derived states. The ability of embryonic clones to undergo dramatic converging/diverging behaviors thus underscores a continued need for independent measurements of both cell state and lineage in the mapping of cell fate hierarchies.
Robustness of cell type transcriptional programs following a signaling perturbation
Single-cell maps of vertebrate development can in principle facilitate unbiased, systematic analyses of mutant pheno-types and disease states. We used scRNA-seq to analyze the mutant phenotype for chordin, a well-studied developmental gene encoding a secreted BMP inhibitor expressed in the organizer and required for patterning the early dorsal-ventral axis (29–33). Chordin disruption leads to changes in gross embryo morphology, with an expansion of ventral tissues and a reduction of dorsal tissues (30). scRNA-seq is uniquely suited to address how every tissue in the embryo changes in abundance, and in gene expression, while also allowing detection of qualitatively new states, or combinations of states, if they occur.
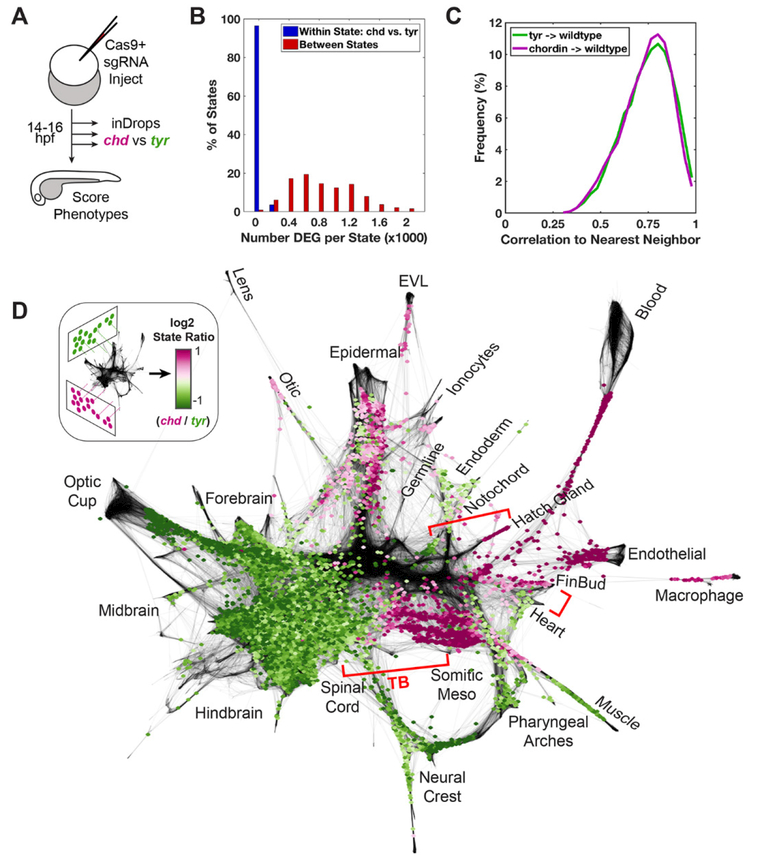
We used CRISPR/Cas9 (34) to disrupt the chordin locus, resulting in highly penetrant clutches of mutant zebrafish embryos (fig. S12). inDrops profiling was performed on chordin-targeted and control embryos (tyrosinase-targeted, see methods) in a narrow time series corresponding to ~14–16hpf (Fig. 6A). After sequencing, we classified each of the chordin-and control-targeted cells to reference cell clusters of the 14hpf wild-type embryo (fig. S13 and methods) and tested for altered gene expression. We reasoned that a qualitatively new cell state, if formed as a result of the aberrant patterning, would manifest as widespread changes in gene expression following mutation, with a magnitude comparable to the differences between wild-type embryonic states. Applying this criterion, we found no evidence of a qualitatively novel cell state following chordin depletion. Rather, the number of genes differentially expressed within states was modest compared to the differences defining the wild-type states of the 14hpf embryo (Fig. 6B and fig. S14A). Moreover, a tSNE mapping of CRISPR-targeted cells (fig. S13, A to C) identified only a single cluster uniquely occupied by chordin-mutant cells (fig. S13D), distinguished primarily by a heat-shock/stress-like transcriptional signature. This same stress signature was elevated in multiple states in chordin targeted embryos (fig. S14A).
Fig. 6. Regulatory features of the developmental landscape identified by genetic perturbation.
(A) Left: Overview of the CRISPR experiment. Three pairs of chordin and tyrosinase (control) targeted samples were prepared and processed by inDrops ~14–16hpf. (B) Histogram depicting numbers of differentially expressed genes (DEG) identified in chordin vs. control (tyrosinase) cells for each state (blue bars), compared to DEG numbers when comparing between all state pairs (red bars). DEG were identified by Wilcoxon rank-sum test (adj. p-value < 0.01, absolute log2 fold change >1, average expression >25 transcripts per million). (C) Histogram of Pearson correlation similarities (after PCA-projection) between each chordin/tyrosinase cell and its nearest neighbor from 10hpf, 14hpf, and 18hpf wild-type datasets (see methods). (D) Log2 ratios of cell states with significant differential abundance (FDR < 0.25) in the chordin vs. tyrosinase samples. Purple and green regions correspond to wild-type cell states that are over- or under-represented in the chordin mutant, respectively. Adjacent graph domains with opposing chordin sensitivity are highlighted by brackets. TB: tailbud region (see cdx4 expression in fig. S3).
We next tested whether chordin disruption led to changes in abundance of particular classified cell types. As expected, expansion of states corresponding to ventral tissues (e.g., somitic mesoderm, epidermis, hatching gland, blood and endothelial tissues) at the expense of dorsal tissues (e.g., the neural plate and notochord) was observed (fig. S14, A and B) (30, 35). Additional features could be appreciated by projecting the CRISPR datasets directly onto the wild-type single-cell graph (Fig. 6, C and D). For example, a sharp boundary bisected the lateral plate mesoderm into two compartments of opposing chordin-sensitivity, separating the heart and fin bud progenitor fields. Similar juxtaposed domains of opposing chordin sensitivity were evident in the axial mesoderm, partitioning notochord from hatching gland, and in the tailbud separating spinal cord from somitic mesoderm (Fig. 6D). Strikingly, each of these pairs of phenotypic domains appeared to be organized downstream of an inferred branchpoint in the cell state landscape. These domain pairs, therefore, likely reflect binary fate choices that are tuned by BMP signaling in wild-type embryos.
In a final analysis, we searched for the putative identity of the cells responding to chordin in the tailbud, as this is the site showing the largest expansion (somitic mesoderm) and loss (spinal cord) after perturbation. In zebrafish, chordin is expressed in the embryonic shield, transiently in the neural plate, adaxial cells, and also in the posterior tailbud region (36). All of these expression patterns were confirmed in our single-cell graphs (fig. S15A). Furthermore, in contrast to its earlier expression in the shield, continued expression of chordin in the tailbud was distinct among a large panel of known BMP inhibitor genes (fig. S15A) and was tightly apposed by expression domains for multiple bmp transcripts (fig. S15B). These expression characteristics might explain the elevated chordin sensitivity of posterior body regions. To examine this region in greater detail, we isolated a subgraph of tailbud and descendent cells, Consistent with previous studies, two cell state trajectories branching from a common neuromesodermal-like brachyury+;sox2+ progenitor state were identified, each expressing markers of neural fates (sox3, sox19a, pax6a, neurog1), or somitic fates (tbx16, tbx6, tbx24, msgn1, myod1) (fig. S16, A to C) (25, 37–39). Strikingly, the neural-mesodermal branchpoint coincided with the boundaries of both chordin expression and sensitivity (fig. S16, D and E). The chordin expressing cells in this region of the single-cell graph exhibited a distinct expression profile (fig. S17), including a cadherin (cdh11), early neurogenic markers (her3, her8a, sox19a), and several relatively uncharacterized genes (gig2g, foxb1b, foxb1a). We hypothesize that these cells represent a key transition state at which point tailbud cells initiate a posterior neurogenic program in a chordin-dependent manner.
Discussion
Our study demonstrates a graph-based approach for mapping whole-embryo developmental landscapes, over time, from scRNA-seq data. The graph was constructed with minimal assumptions about development, and describes individual cell states transitioning from pluripotent blastomeres to a large array of cell types and tissues during the first day of zebrafish embryogenesis. This dataset can now be mined to identify temporal and tissue associations for any gene, cell type, or biological process of interest. As with genome annotation efforts over the years, we expect that the annotation of identified cell states may undergo refinement with community input.
As single-cell atlases and landscapes of embryo development become routinely available, one is challenged to reconsider the relationship between a cell lineage (by definition, a tree), and the considerably more topologically complex gene expression landscape through which these cells traverse. Using TracerSeq, we confirmed that differentiating cells of the zebrafish embryo do not invariantly follow tree-like hierarchies. Instead, we observed both widespread convergence in cell states for clonally distant cells and instances in which clonally related cells diverged into distant states. Non-tree like convergence of cell states could be explained by the differentiation of well-separated spatial domains of the embryo into the same basic cell types (e.g., along the A-P axis), whereas divergence could involve mechanisms such as asymmetric cell division or exposure to spatially varying signals (40). We anticipate that the synthesis of single-cell lineage and transcriptome information will continue to be crucial for deciphering how cells traverse state trajectories with complex topologies (e.g., loops or continua).
Single-cell mapping of genetic perturbation data presents a powerful framework for identifying regulatory features of a developmental landscape. Following deletion of the BMP inhibitor, chordin, we showed that the defining transcriptional features of the landscape remained mostly unchanged, yet cell state abundances could be dramatically and reciprocally altered, as if the landscape were “tilted” but cell fates remain canalized. Future systematic mapping of signaling perturbations could be used to reveal the complete signaling logic of the embryo, as cells are specified toward their final fates. Together, these studies demonstrate the power, modularity, and quantitative benefits of unbiased scRNA-seq-based interrogations of embryonic development. We anticipate that similar large-scale datasets will facilitate explorations of additional developmental stages, tissues, and species.
Supplementary Material
ACKNOWLEDGMENTS
We thank A.Ratner for technical support, T.W.Hiscock, V.Savova, S.L.Wolock, S.Mekhoubad and R.M.W. for helpful discussions.
Funding:
D.E.W. acknowledges support from an HHMI-LSRF postdoctoral fellowship and 1K99GM121852. A.M.K was supported by an Edward J Mallinckrodt Foundation Grant and a Burroughs Wellcome Fund CASI Award. S.G.M was supported by R01GM107733 and R01DC015478.
Footnotes
Competing interests: S.G.M. D.E.W, C.W., Z.M.C., J.A.B.: none declared. AMK is a founder of 1Cell-Bio, Inc.
Data and materials accessibility: A web portal providing access to the single-cell and coarse-grained graphs is available at www.tinyurl.com/scZfish2018. Single-cell counts matrices and FASTQ files have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO), accession GSE112294.
REFERENCES AND NOTES
- 1.Schier AF, Talbot WS, Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet 39, 561–613 (2005). doi:10.1146/annurev.genet.37.110801.143752 10.1146/annurev.genet.37.110801.143752MedlineMedline [DOI] [PubMed] [Google Scholar]
- 2.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I, CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016). doi:10.1186/s13059-016-0938-8 10.1186/s13059-016-0938-8MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S, Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014). doi:10.1038/nmeth.2772 10.1038/nmeth.2772MedlineMedline [DOI] [PubMed] [Google Scholar]
- 4.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). doi:10.1016/j.cell.2015.04.044 10.1016/j.cell.2015.04.044MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). doi:10.1016/j.cell.2015.05.002 10.1016/j.cell.2015.05.002MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA, Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc 8, 870–891 (2013). doi:10.1038/nprot.2013.046 10.1038/nprot.2013.046MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L, Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc 12, 44–73 (2017). doi:10.1038/nprot.2016.154 10.1038/nprot.2016.154MedlineMedline [DOI] [PubMed] [Google Scholar]
- 8.Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C, Expression of the zebrafish genome during embryogenesis, ZFIN Direct Data Submission, (2001); http://zfin.org.
- 9.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 (2014). doi:10.1038/nbt.2859 10.1038/nbt.2859MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bendall SC, Davis KL, Amir AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D, Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014). doi:10.1016/j.cell.2014.04.005 10.1016/j.cell.2014.04.005MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, Song H, Single-cell RNA-seq with Waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015). doi:10.1016/j.stem.2015.07.013 10.1016/j.stem.2015.07.013MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016). doi:10.1038/nmeth.3971 10.1038/nmeth.3971MedlineMedline [DOI] [PubMed] [Google Scholar]
- 13.Kimmel CB, Warga RM, Schilling TF, Origin and organization of the zebrafish fate map. Development 108, 581–594 (1990). Medline [DOI] [PubMed] [Google Scholar]
- 14.Melby AE, Warga RM, Kimmel CB, Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development 122, 2225–2237 (1996). Medline [DOI] [PubMed] [Google Scholar]
- 15.Warga RM, Nüsslein-Volhard C, Origin and development of the zebrafish endoderm. Development 126, 827–838 (1999). Medline [DOI] [PubMed] [Google Scholar]
- 16.Woo K, Fraser SE, Order and coherence in the fate map of the zebrafish nervous system. Development 121, 2595–2609 (1995). Medline [DOI] [PubMed] [Google Scholar]
- 17.Kawakami K, Tol2: A versatile gene transfer vector in vertebrates. Genome Biol. 8 (Suppl 1), S7 (2007). doi:10.1186/gb-2007-8-s1-s7 10.1186/gb-2007-8-s1-s7MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA 3rd, Smith HO, Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). doi:10.1038/nmeth.1318 10.1038/nmeth.1318MedlineMedline [DOI] [PubMed] [Google Scholar]
- 19.McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). doi:10.1126/science.aaf7907 10.1126/science.aaf7907MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junker JP, Spanjaard B, Peterson-Maduro J, Alemany A, Hu B, Florescu M, van Oudenaarden A, Massively parallel clonal analysis using CRISPR/Cas9 induced genetic scars. BioRxiv 056499 [Preprint]. 4 January 2017. 10.1101/056499. [DOI] [Google Scholar]
- 21.Warga RM, Kane DA, Ho RK, Fate mapping embryonic blood in zebrafish: Multi- and unipotential lineages are segregated at gastrulation. Dev. Cell 16, 744–755 (2009). doi:10.1016/j.devcel.2009.04.007 10.1016/j.devcel.2009.04.007MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitlock KE, Westerfield M, The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development 127, 3645–3653 (2000). Medline [DOI] [PubMed] [Google Scholar]
- 23.Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas J-F, Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365–376 (2009). doi:10.1016/j.devcel.2009.08.002 10.1016/j.devcel.2009.08.002MedlineMedline [DOI] [PubMed] [Google Scholar]
- 24.Davis RL, Kirschner MW, The fate of cells in the tailbud of Xenopus laevis. Development 127, 255–267 (2000). Medline [DOI] [PubMed] [Google Scholar]
- 25.Kanki JP, Ho RK, The development of the posterior body in zebrafish. Development 124, 881–893 (1997). Medline [DOI] [PubMed] [Google Scholar]
- 26.Le Douarin NM, Dupin E, Multipotentiality of the neural crest. Curr. Opin. Genet. Dev 13, 529–536 (2003). doi:10.1016/j.gde.2003.08.002 10.1016/j.gde.2003.08.002MedlineMedline [DOI] [PubMed] [Google Scholar]
- 27.Le Douarin NM, Creuzet S, Couly G, Dupin E, Neural crest cell plasticity and its limits. Development 131, 4637–4650 (2004). doi:10.1242/dev.01350 10.1242/dev.01350MedlineMedline [DOI] [PubMed] [Google Scholar]
- 28.Le Lièvre CS, Le Douarin NM, Mesenchymal derivatives of the neural crest: Analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol 34, 125–154 (1975). Medline [PubMed] [Google Scholar]
- 29.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM, Xenopus chordin: A novel dorsalizing factor activated by organizer-specific homeobox genes. Cell 79, 779–790 (1994). doi:10.1016/0092-8674(94)90068-X 10.1016/0092-8674(94)90068-XMedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ, Granato M, Brand M, Furutani-Seiki M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nüsslein-Volhard C, dino and mercedes, two genes regulating dorsal development in the zebrafish embryo. Development 123, 95–102 (1996). Medline [DOI] [PubMed] [Google Scholar]
- 31.Schulte-Merker S, Lee KJ, McMahon AP, Hammerschmidt M, The zebrafish organizer requires chordino. Nature 387, 862–863 (1997). doi:10.1038/43092 10.1038/43092MedlineMedline [DOI] [PubMed] [Google Scholar]
- 32.Sasal Y, Lu B, Steinbelsser H, De Robertis EM, Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature 378, 419 (1995). doi:10.1038/378419d0 10.1038/378419d0MedlineMedline [DOI] [PubMed] [Google Scholar]
- 33.Piccolo S, Sasai Y, Lu B, De Robertis EM, Dorsoventral patterning in Xenopus: Inhibition of ventral signals by direct binding of chordin to BMP-4. Cell 86, 589–598 (1996). doi:10.1016/S0092-8674(00)80132-4 10.1016/S0092-8674(00)80132-4MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF, Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLOS ONE 9, e98186 (2014). doi:10.1371/journal.pone.0098186 10.1371/journal.pone.0098186MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung AY, Mendenhall EM, Kwan TTF, Liang R, Eckfeldt C, Chen E, Hammerschmidt M, Grindley S, Ekker SC, Verfaillie CM, Characterization of expanded intermediate cell mass in zebrafish chordin morphant embryos. Dev. Biol 277, 235–254 (2005). doi:10.1016/j.ydbio.2004.09.032 10.1016/j.ydbio.2004.09.032MedlineMedline [DOI] [PubMed] [Google Scholar]
- 36.Miller-Bertoglio VE, Fisher S, Sánchez A, Mullins MC, Halpern ME, Differential regulation of chordin expression domains in mutant zebrafish. Dev. Biol 192, 537–550 (1997). doi:10.1006/dbio.1997.8788 10.1006/dbio.1997.8788MedlineMedline [DOI] [PubMed] [Google Scholar]
- 37.Row RH, Kimelman D, Bmp inhibition is necessary for post-gastrulation patterning and morphogenesis of the zebrafish tailbud. Dev. Biol 329, 55–63 (2009). doi:10.1016/j.ydbio.2009.02.016 10.1016/j.ydbio.2009.02.016MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Row RH, Tsotras SR, Goto H, Martin BL, The zebrafish tailbud contains two independent populations of midline progenitor cells that maintain long-term germ layer plasticity and differentiate in response to local signaling cues. Development 143, 244–254 (2016). doi:10.1242/dev.129015 10.1242/dev.129015MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouti M, Delile J, Stamataki D, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, Briscoe J, A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243–261.e7 (2017). doi:10.1016/j.devcel.2017.04.002 10.1016/j.devcel.2017.04.002MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gönczy P, Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol 9, 355–366 (2008). doi:10.1038/nrm2388 10.1038/nrm2388MedlineMedline [DOI] [PubMed] [Google Scholar]
- 41.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF, Stages of embryonic development of the zebrafish. Dev. Dyn 203, 253–310 (1995). doi:10.1002/aja.1002030302 10.1002/aja.1002030302MedlineMedline [DOI] [PubMed] [Google Scholar]
- 42.Manoli M, Driever W, Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae for RNA isolation. Cold Spring Harb. Protoc 2012, pdb.prot069633 (2012). doi:10.1101/pdb.prot069633 10.1101/pdb.prot069633MedlineMedline [DOI] [PubMed] [Google Scholar]
- 43.Van der Maaten L, Hinton G, Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605 (2008). [Google Scholar]
- 44.Van der Maaten L, Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res 15, 3221–3245 (2014). [Google Scholar]
- 45.Rodriguez A, Laio A, Clustering by fast search and find of density peaks. Science 344, 1492–1496 (2014). doi:10.1126/science.1242072 10.1126/science.1242072MedlineMedline [DOI] [PubMed] [Google Scholar]
- 46.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). doi:10.1186/s13059-015-0844-5 10.1186/s13059-015-0844-5MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey JD, A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol 64, 479–498 (2002). doi: 10.1111/1467-9868.00346 [DOI] [Google Scholar]
- 48.Weinreb C, Wolock S, Klein AM, SPRING: A kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248 (2018). doi:10.1093/bioinformatics/btx792 10.1093/bioinformatics/btx792MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Q, Feng J, Huang J, Weighted natural neighborhood graph: An adaptive structure for clustering and outlier detection with no neighborhood parameter. Cluster Comput. 19, 1385–1397 (2016). doi: 10.1007/s10586-016-0598-1 [DOI] [Google Scholar]
- 50.Ting D, Huang L, Jordan M, An analysis of the convergence of graph Laplacians. arXiv:1101.5435 [stat.ML] (28 January 2011). [Google Scholar]
- 51.Jacomy M, Venturini T, Heymann S, Bastian M, ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLOS ONE 9, e98679 (2014). doi:10.1371/journal.pone.0098679 10.1371/journal.pone.0098679MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bastian M, Heymann S, Jacomy M, “Gephi: an open source software for exploring and manipulating networks,” in Third International AAAI Conference on Weblogs and Social Media (Association for the Advancement of Artificial Intelligence, AAAI, 2009). [Google Scholar]
- 53.Hu Y, Efficient, high-quality force-directed graph drawing. Mathematica J. 10, 37–71 (2006). [Google Scholar]
- 54.Koren Y, Drawing graphs by eigenvectors: Theory and practice. Comput. Math. Appl 49, 1867–1888 (2005). doi: 10.1016/j.camwa.2004.08.015 [DOI] [Google Scholar]
- 55.Jao LE, Wente SR, Chen W, Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A 110, 13904–13909 (2013). doi:10.1073/pnas.1308335110 10.1073/pnas.1308335110MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobitski AY, Otte JC, Takamiya M, Schäfer B, Mertes J, Stegmaier J, Rastegar S, Rindone F, Hartmann V, Stotzka R, García A, van Wezel J, Mikut R, Strähle U, Nienhaus GU, An ensemble-averaged, cell density-based digital model of zebrafish embryo development derived from light-sheet microscopy data with single-cell resolution. Sci. Rep 5, 8601 (2015). doi:10.1038/srep08601 10.1038/srep08601MedlineMedline [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.