SUMMARY
Natural killer (NK) cells of the innate immune system are the first line of defense against infectious agents and cancer cells. However, only a few mechanisms that regulate eradication of tumors by NK cells have been identified. In this review, we present an account of epigenetic mechanisms that modulate the ability of NK cells to eradicate cancer cells. To date, several drugs that target epigenetic modifiers have shown clinical efficacy in cancer. Therefore, once a given epigenetic modifier is validated as a regulator of NK cell function, it can be targeted for NK cell-based cancer immunotherapies.
Epigenetics, NK cells and Cancer
The human immune system provides protection against a wide variety of pathogens and diseases, including cancer. The innate immune system comprises several cell types, including natural killer (NK) cells, which are large granular lymphocytes that represent 10 to 15% of the total circulating lymphocytes [1] but can also be tissue resident [2, 3]. NK cells play an important role in the protection against infectious pathogenic agents and serve as the first line of immunological defense against tumor initiation and progression [4, 5]. Unlike other immune cell types that are slow to attain cytolysis activity, NK cells can readily recognize and eradicate pathogen-infected, stressed, and transformed cancer cells [4, 5].
The antitumor effects of NK cells were first shown against implanted mouse tumors in the early 1970s [6, 7]. Conversely, impaired NK cell function was later shown to increase tumor growth and metastasis [8, 9]. A recent long-term epidemiological study revealed that decreased NK cell activity is associated with higher risks of developing various cancers [10], whereas high numbers of tumor-infiltrating NK cells are associated with favorable outcomes in colorectal carcinoma, gastric cancer, and squamous cell lung cancer patients [11]. NK cells control tumor growth by recruiting conventional type 1 dendritic cells to the tumor microenvironment [12]. These observations highlight the potential use of NK cells in cancer immunotherapy.
Immunotherapies have been successful in multiple unrelated types of cancer [13], as immune suppression and immune evasion by cancer cells are known to contribute to tumor development and progression [14, 15]. Tumorigenic and metastatic cellular states are the result of a complex multistep process involving various genetic and epigenetic changes. In this review, we focus on epigenetic mechanisms in cancer and NK cells that affect NK cell-mediated recognition of rogue cancer cells and their eradication (also see, Table 1) [16–42].
Table 1.
Role of NK cells in cancer initiation and progression.
| S. NO | Cancer type | Role of NK cells | Reference |
|---|---|---|---|
| 1 | Lymphoma/Leukemia | NCR1-deleted mice are shown to develop tumors faster than wild-type mice. Soluble MICA and MICB was detected in leukemia patient sera and may prevent NK cell function. CAR-NK cells expressing anti-CD19 show enhanced NK cell killing towards leukemic cells. |
[16] [17] [18] |
| 2 | Neuroblastoma | CD155 levels correlate with NK cell cytotoxicity in neuroblastoma cells isolated from patients. | [19] |
| 3 | Hepatocellular carcinoma | Reduced ULBP1 and MICA/MICB expression correlate with early recurrence and reduced overall survival in HCC patients. Impairment of NK cell function leads to Hepatitis B and C virus induced HCC. |
[20], [21] [22] |
| 4 | Breast cancer | IL15−/− mice show enhanced breast cancer metastasis, in part, in a NK cell dependent manner. Mesenchymal stem cells overexpressing Sirt1 inhibits breast tumor growth via recruiting NK cells. |
[23] [24] |
| 5 | Fibrosarcoma | DNAM1 receptor deficient mice show increased MCA-induced fibrosarcoma development. | [25] |
| 6 | Cervical cancer | MICA/B and ULBP1 independently predict better overall survival in cervical cancer patients. | [26] |
| 7 | Prostate cancer | NKG2D-deficient mice develop prostate tumors that expressed higher amount of NKG2D ligand compared to wild-type mice. | [27] |
| 8 | Glioma | MICA overexpression sensitizes glioma cells to NK cell-mediated eradication. | [28] |
| 9 | Colorectal Cancer | Higher expression of MICA correlates with better colorectal cancer patient survival. | [29] |
| 10 | Melanoma | Ectopic expression of NKG2D ligands in B16 cells leads potent rejection of tumor cells. Blocking of NCR and DNAM1 receptors using specific antibodies reduces NK cell killing of melanoma cells. |
[30] [31] |
| 11 | Multiple myeloma | NKp30 ligand BAT3 promotes tumor lysis in a NK cell dependent manner. | [32] |
| 12 | Head and Neck cancer | Head and neck cancer patients with high NK cell activity have better disease-free survival. | [33] |
| 13 | Renal cell carcinoma | IL-21 activated NK cells inhibit the growth of renal cell carcinoma in mice. | [34] |
| 14 | Lung cancer | Gefitinib enhances NK cell-mediated clearance of EGFR mutant lung cancer cells. | [35] |
| 15 | Ovarian cancer | Depletion of ascorbic acid reduces NK cell cytotoxicity to ovarian cancer cells. IL-2 treatment significantly delays tumor growth in SCID mice engrafted with ovarian cancer. |
[36] [37] |
| 16 | Endometrial cancer | HLA-E expression predicts the prognostic benefit of infiltrating NK cells in endometrial cancer | [38] |
| 17 | Pancreatic Cancer | Inhibition of NK cell checkpoint CD96 prevents relapse of pancreatic cancer. | [39] |
| 18 | Gastric Cancer | NK cells exhibit strong anti-tumor activity against gastric cancer cells. | [40] |
| 19 | Thyroid Cancer | NK cells inhibit the growth and metastasis of anaplastic thyroid cancer. NK cells eradicate anaplastic thyroid cancer in ULBP2/5/6-dependent manner and chemoattract CXCR3-positive NK cells. |
[41] [42] |
Development and regulation of NK cells and mechanisms of cytotoxicity
Throughout life, NK cells develop primarily in bone marrow from hematopoietic stem cells, which differentiate first into common lymphoid progenitors and then into NK/T cell progenitors, undergoing a series of coordinated differentiation steps and acquiring different markers, receptors, and functions [43–45]. Recent studies have also demonstrated that NK cell precursors migrate to and undergo further differentiation in secondary lymphoid tissues, such as those of the uterus and liver [46–48]. We refer readers to some outstanding reviews for more details on the origin and maturation of NK cells [43, 49–52].
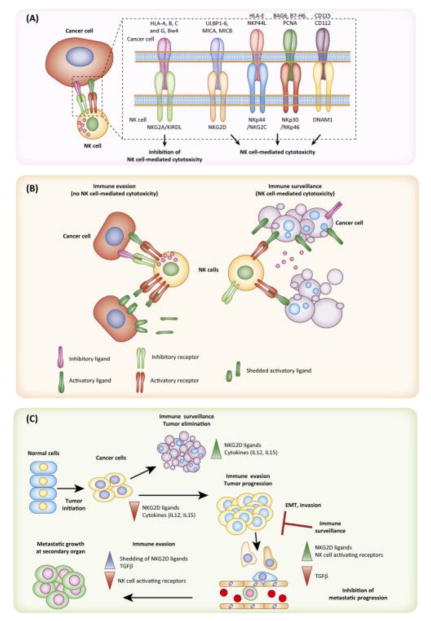
Although cytokines play an important role in the development and activation of NK cells [53], NK cell activities, including cytotoxicity towards target cells (e.g., cancer cells), are largely regulated by the activation or inhibition of their receptors. This is in contrast to B and T cells, primary cell types of the adoptive immune system, which have antigen specificity and a higher degree of flexibility through the arrangement of gene clusters. Nevertheless, NK cell receptors have some flexibility via the recognition of different ligands on target cells, such as the NK group 2D (NKG2D) ligands [54–56] (Figure 1A–B).
Figure 1. Function and regulation of NK cells.
A. NK cell ligands on cancer cells are recognized by NK cell receptors on NK cells, which promotes NK cell-mediated eradication of cancer cells. B. Cancer cells develop a variety of mechanisms to evade eradication by NK cells. Immune evasion mechanisms include shedding of NK cell ligands by cancer cells, downregulation of activating receptors on NK cells, and the overexpression of inhibitory ligands by cancer cells. C. Cancer cells must evade NK cells, which play important roles during the multiple stages of tumor development and progression.
Cancer cells develop multiple strategies to evade NK cell-mediated cytotoxicity (Figure 1C). One mechanism involves the loss or shedding of NKG2D ligands, which in turn downregulates the expression of NKG2D receptor on NK cells [57, 58]. Additionally, cancer cells can also downregulate NK-activating receptors [59]. By contrast, the elevated expression of NKG2A receptors in infiltrating NK cells reduces their cytotoxic activity against renal carcinoma cells [60]. Tumor cells can also avoid eradication by NK cells by expressing the ligands for inhibitory receptors, such as non-classical human leukocyte antigens (HLA-E or HLA-G) [61, 62], and by releasing immunosuppressive cytokines, such as transforming growth factor beta (TGF-β), interleukin-10, prostaglandin, and indoleamine 2,3-dioxygenase [63–65]. These factors not only decrease NK cell activity but also inhibit NK cell maturation [66]. Additional immune inhibitory checkpoint receptors, such as programmed cell death protein 1 (PD-1), are expressed on NK cells and cancer cells expressing programmed death ligand 1 (PD-L1) inhibit the anti-cancer abilities of NK cells [67]. Similarly, the ability of NK cells to control cancer growth is modified by other immunomodulatory surface proteins (TIGIT, TIM-3, and LAG3) that affect a variety of phenotypes, including NK cell effector function and NK cell exhaustion [68–70].
Epigenetic regulation of NK cells in cancer
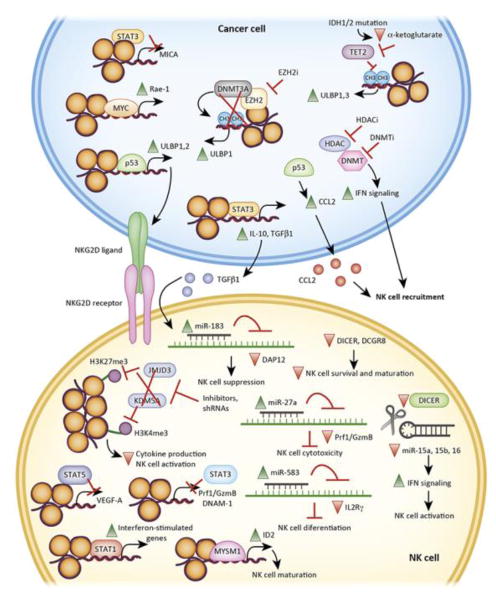
The effect of key immune cells (B and T cells or NK cells) against cancer cells are subject to epigenetic regulation. For example, the epigenetic state of cancer cells either makes them vulnerable to immune clearance or promotes immune evasion [71, 72]. Here, we present evidence for the role of epigenetic modulators and the mechanisms that regulate NK cell-mediated cancer cell eradication (Figure 2. Key Figure).
Figure 2, Key Figure. Epigenetic mechanisms regulate eradication of tumors by NK cells.
Epigenetic regulators (transcription factors, chromatin regulators, DNA modifying enzymes, and miRNAs) regulate NK cell activity and cancer cell eradication. This can occur by cancer cells developing NK cell evasion mechanisms, for example by the downregulation of NK cell ligands (e.g., ULBP1, ULBP2, and MICA) or by the secretion of chemokines that prevent NK cell recruitment (e.g., IL-10, TGFβ1, and CCL2). Similarly, NK cells can undergo changes in the tumor microenvironment to be become less effective, for example by the downregulation of activating receptors (e.g., NKG2D receptor) and the attenuation of their activity due to dampened IFN signaling.
Epigenetic alterations are reversible and heritable changes that do not involve alteration in the DNA sequence itself. These changes are typically associated with DNA modifications, such as CpG DNA methylation or posttranslational modifications of histone proteins. However, for the purpose of this review, we use a broader definition of epigenetic alterations that includes additional non-genetic changes, such as changes in transcription factors and noncoding RNA expression/function.
Transcription factors
Transcription factors are sequence-specific DNA binding proteins that, along with other factors (e.g., epigenetic modulators, transactivators.) can either activate or repress the transcription of target genes. Several transcription factors, including nuclear factor kappa B, Cbl-C, and runt-related transcription factor 3, have been shown to modulate NK cell function and alter the eradication of tumors by NK cells [73–75]. The tumor suppressor protein p53 and the MYC oncogene are two transcription factors with a well-established relevance in both cancer and immunity [76, 77].
In half of all cancers, p53 is either mutated or deleted, and the loss of p53 has been shown to directly contribute to cancer initiation and progression [78]. A previous study showed that in cancer cells, p53 induces the expression of the NKG2D ligands ULBP1 and ULBP2 and stimulates interferon gamma (IFN-γ) production by co-cultured NK cells [79]. However, a direct role of ULBP1 and ULBP2 in mediating the production of IFN-γ was not tested in this study. In addition, p53 induces the senescence of cancer cells and NK cell-mediated clearance [80], an observation that was confirmed in a more detailed study using an Hras-transformed liver cancer model with doxycycline-inducible p53 shRNA [81]. In this model, p53 increased the secretion of multiple chemokines, particularly C-C motif chemokine ligand 2 (CCL2), which recruited NK cells to senescent tumors, rather than inducing the expression of NK cell ligands [81]. Similar to CCL2, mutations in p53 regulate the levels of other chemokines and result in the inhibition of NK cell recruitment [82]. Additional studies examining the impact of aneuploidy on p53 induction and tumor development and/or control found that DNA replication stress from complex structural aneuploidy resulted in p53 activation and senescence in cancer cells, which also secreted cytokines that promoted eradication by NK cells [83, 84].
MYC represses the expression of class 1 human leukocyte antigen in melanoma cells and potentially enhances NK cell-mediated eradication [85]. Studies using an Eμ-MYC-driven mouse model of lymphoma showed that MYC is necessary for the transcription of NKG2D ligand Rae-1 [86]. Downregulation of NKG2D receptor on NK cells was observed in lymphoma-bearing mice, indicating there may be a compensatory mechanism in vivo by which lymphomagenesis progresses via inactivation of NK cell-mediated cytotoxicity. The possibility that the vast number of lymphoma cells exhausted the NK cells and hampered their ability to regulate lymphoma progression was also not ruled out [86]. Additionally, a recent study using a mouse lung model of KRasG12D-driven adenoma found that MYC cooperated with coactivated oncogenic Ras to cause immune suppression in part by reprogramming the stroma, which was largely driven by C-C motif chemokine ligand 9 and interleukin-23. In this model, the inactivation of MYC after tumor development reversed all the changes in the tumor stroma and caused tumor regression, which was largely dependent on returning NK cells [14]. MYC is also shown to enhance the expression of PD-L1 in cancer cells, which promotes immune evasion [82] and attenuates the ability of NK cells to clear cancer cells [67].
The signal transducer and activator of transcription (STAT) family of transcription factors also regulate NK cell function. STAT proteins play a diverse role in several biological processes and are important regulators of both the innate and the adoptive immune response [87]. For example, STAT5 plays an important role in IL-2 and IL-15-mediated signal transduction and in promoting NK cell survival, cytotoxicity, and maturation [88, 89]. STAT5 regulation of NK cell-mediated angiogenesis can act as a molecular switch to shift from tumor surveillance to tumor promotion. STAT5 normally downregulates VEGFA in NK cells, but the inhibition of STAT5 increases VEGFA production, resulting in NK cell-mediated angiogenesis and tumor growth [90]. This study suggested that STAT5 inhibitors need to be used with caution as anti-cancer agents because they may cause unexpected tumor promoting effects. Similarly, mice with a targeted mutation of STAT1 demonstrate reduced NK cell cytolytic activity in vitro and a failure to reject implanted tumor in vivo [91]. Interestingly, the inability to reject tumors in these mice was dependent upon IFN-α and IFN-γ and was not due to reduced NK cell number [91]. An analysis of mice lacking STAT3 showed normal development and normal NK cell number, but an alteration in the kinetics of IFN-γ production due to a lack of STAT3 binding to IFN-γ promoter was observed [92]. Strikingly, the loss of STAT3 in NK cells enhances tumor surveillance in various in vivo models of hematological diseases. The reduced tumor burden is partially due to increased expression of the activating ligand DNAX accessory molecule 1 as well as the lytic enzymes perforin and granzyme B [92]. Similarly, another study suggested that inhibition of STAT3 increases NK cell cytotoxicity to cancer cells by upregulating the NKG2D ligand major histocompatibility complex class I-related chain A levels by directly binding to its promoter [93]. These findings have therapeutic implications because they suggest that STAT3 inhibitors will enhance NK cell activity towards cancer cells and inhibit tumor growth in a non-autonomous manner. Overall, these studies of STAT proteins and their role in the regulation of NK cell function reveal their surprisingly diverse and non-redundant functions. A thorough evaluation of the cross-talk among different STAT transcription factor family members and their ability to regulate NK cells will require further investigation.
Chromatin regulators and DNA modifiers
Chromatin regulatory proteins are identified as “writers” “erasers,” and “readers” of histone, marks or CpG methylating or demethylating enzymes on the basis of their function [94, 95]. Chromatin regulation is important for NK cell development and function. For example, NK cell maturation requires the activity of the histone H2A deubiquitinase MYSM1 [96]. Epigenetic modulation of NK cell responses was shown is studies testing the effects of inhibitors of histone deacetylases (entinostat and givinostat) and DNA methyltransferases (5-azacytidine [Aza]) in mouse models of epithelial ovarian cancer [97]. Pretreatment of tumor epithelial cells with Aza significantly reduced ascites, alterations in the number and activation state of immune cells, including NK cells, and increase in overall survival. Treatment of ovarian cancer cells with Aza also increased IFN signaling, which was required for the tumor-suppressive effects [97]. Although the relative contribution of NK cells to these effects was not examined, there was an increase in activated NK cells in the tumor microenvironment [97]. The broad-spectrum inhibitors tested in this study have effects that include aspects beyond epigenetic regulation. Therefore, the specific epigenetic mediators responsible, including those for IFN pathway regulation, were not revealed. Another interesting study showed that IDH1 and IDH2 mutant gliomas escape NK cell immune surveillance via DNA methylation-based downregulation of NKG2D ligands (ULBP1 and ULBP3) [98]. These ligands were re-expressed upon cell treatment with Aza, which led to lysis of glioma cells by NK cells. These results are important because in some cancer types, including acute myelogenous leukemia, IDH1 and IDH2 mutations result in global DNA hypermethylation due to reduced α-ketoglutarate levels and TET2 function [98].
A recent study using a chemical screen targeting epigenetic regulators identified the histone methyltransferase enhancer of zeste homolog 2 (EZH2) as a modulator of NK cell function against hepatocellular carcinoma [99]. EZH2 inhibited the expression of ULBP1 and induced the sensitization of hepatocellular carcinoma cells to NK cells in a DNA methylation-dependent manner (via DNA methyltransferase 3A). Furthermore, NK cell differentiation is enhanced in EZH2-null or EZH2 inhibitor (UNC1999 or EPZ005687)-treated hematopoietic stem cells, which were also more effective in eradicating tumor cells [100]. Increase in methylation of lysine 27 of histone 3 in NK cells via inhibition of JMJD3/UTX demethylase was associated with an anti-inflammatory reduction in cytokine production, which confirms the importance of this histone modification in NK cell activation [101]. Similarly, the demethylating enzyme specific for trimethylated lysine 4 of histone 3, Kdm5a, is necessary for NK cell activation, and a deficiency in this enzyme reduces IFN-γ production by activated NK cells, mediated in part by SOCS1 [102].
Collectively, these studies show NK cells undergo multiple histone and DNA methylation-based modifications that affect activation and function, similar to what happens in cancer cells. Of note, some of clinically approved EZH2, IDH1, and IDH2 inhibitors are being tested in immunocompromised preclinical models of cancer, which may impact the evaluation of their anticancer efficacy. In future studies, the use of either allograft mouse tumor systems or mouse models with a humanized immune system should be encouraged, so that the impact of the immune system on the efficacy of these agents can be determined.
microRNAs
MicroRNAs (miRNAs) are small noncoding RNAs that bind to the 3′-untranslated regions of mRNAs, inducing degradation or inhibition of translation [103]. There is sufficient evidence indicating that miRNAs regulate NK cell function [104–106]. For example, knockout of Dicer or Dgcr8, which prevents miRNA maturation [103], results in reduced survival and turnover of mouse NK cells. Survival and maturation of NK cells is also reduced in mice with lymphocyte Dicer1 knockout [60]. Dicer1-deficient NK cells have enhanced activity, showed by increased degranulation and IFN-γ production in response to tumor cells and other stimuli, such as NK cell receptor ligation. The increased IFN-γ production was attributed to the reduced expression of miRNAs (miR-15a, -15b, and -16) that directly repress mouse IFN-γ.
Analysis of miRNA transcriptomes from NK cells derived from peripheral blood, cord blood, and uterine decidua revealed significant differences in miRNA profiles, suggesting different miRNAs effects on different NK cell populations [107]. For example, miR-362-5p is highly expressed in human peripheral blood NK cells and targets cyclin D1 to enhance NK cell function. miRNA-27a* targets Prf1 and GzmB expression to regulate NK cell cytotoxicity [108], and miR-583 is a negative regulator of interleukin 2 receptor gamma expression and blocks NK cell differentiation [109]. Similarly, the expression of miRNA-183 is induced by TGF-β and blocks expression of DNAX-activating protein 12 kDa (DAP12). Inhibition of DAP12 creates an immunosuppressive tumor microenvironment by inhibiting NK cell function [110]. Downregulation of DAP12 is a common feature in all types of lung cancer and its expression is lower in intratumoral NK cells than in peritumoral NK cells [110]. Another TGF-β-induced miRNA, miR-27a-5p, was shown to function by inhibiting perforin and granzyme B expression and by downregulating the expression of C-X-C motif chemokine receptor 1, resulting in a limitation of NK cell migration ability [111].
Epigenetics and NK cell-based immunotherapy
Immune checkpoint therapies with antibodies against cytotoxic T lymphocyte-associated antigen 4 (e.g., ipilimumab) or programmed cell death protein 1 (e.g., pembrolizumab) or its ligand (e.g., avelumab) that engage T-cell-based eradication of cancer cells show significant clinical benefits in Hodgkin’s lymphoma and melanoma [112, 113]. Similarly, success is seen with chimeric antigen receptor T-cell-based therapy [114]. By contrast, NK cell-based therapies are still in the early stages of development. NK cells from multiple myeloma and renal carcinoma patients express PD-1 on their surface and some immune checkpoint blockage therapies that primarily engage T-cells may also engage NK cells [67, 115]. However, the extent to which these therapies involve NK cell activity requires careful analysis.
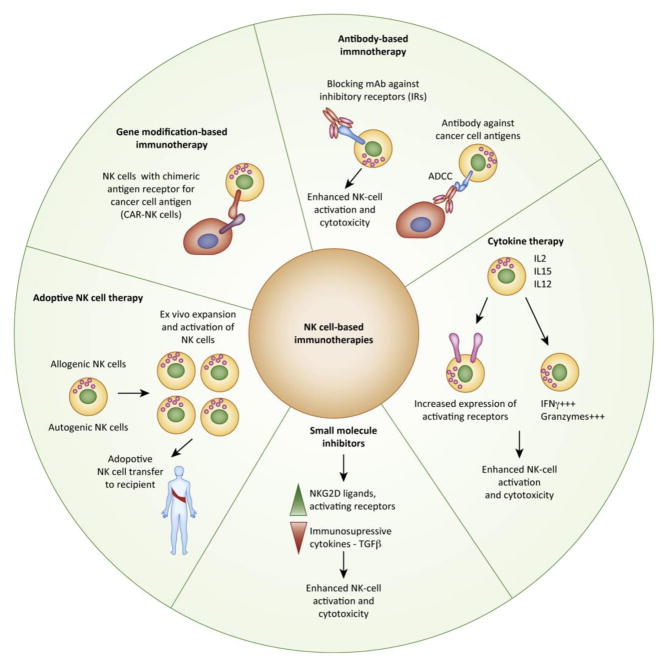
New studies aim to directly enhance NK cells ability to eradicate cancer cells [13, 116]. The clinical utility of NK cell-based therapies is exemplified by the more than 180 ongoing clinical trials using these cells as of April 2018. NK cell-based immunotherapies can be largely categorized into five different types: (i) antibody-based, (ii) cytokine-based, (iii) adoptive NK cells, (iv) gene modification, and (v) small-molecule inhibitors (Figure 3, Key Figure and Table 2).
Figure 3, Key Figure. NK cell-based cancer immunotherapies and epigenetic targets.
NK cell-based immunotherapies can be largely categorized into five different types ((i) antibody-based, (ii) cytokine-based, (iii) adoptive NK cells, (iv) gene modification, and (v) small-molecule inhibitors). An improved understanding of epigenetic regulators and their roles in modulating the NK cell functions will open up new opportunities for the use of epigenetic regulator-targeting drugs to enhance NK cell-based immunotherapies. For example, small molecule inhibitors of epigenetic regulators can increase the expression of NKG2D ligands or activating receptors on cancer cells and reduce the production of immunosuppressive cytokines such as TGFβ.
Table 2.
Approved and investigational NK cell-based cancer immunotherapies.
| S.No. | Therapy/Inhibitor/Antibody | Cancer type | Clinical Trial Stage | Clinical trial ID |
|---|---|---|---|---|
| 1 | Autologous NK cells | Advanced Kidney Cancer Metastatic melanoma Digestive cancer |
Phase I/II |
NCT02843607 NCT00631072 UMIN000007527 |
| 2 | Allogenic NK cells | Metastatic Gastrointestinal Carcinoma High-risk AML (HINKL) Lymphoma and Solid tumors |
Phase I/II |
NCT02845999 NCT02229266 NCT01212341 |
| 3 | NK Cell lines (Neukoplast™ (NK-92)) | Hematological Malignancies | Phase I |
NCT00990717 NCT00900809 |
| 4 | CAR-NK cells | Relapsed/Refractory CD33+ AML Metastatic Solid Tumors Acute Lymphoblastic Leukemia |
Pre-clinical/Phase 1/II |
NCT02944162 NCT03415100 NCT01974479 |
| 5 | IL-2 | Lymphoma, lung cancer and melanoma | FDA approved | |
| 6 | IL-15 | Advanced Solid Tumors Acute Myelogenous Leukemia |
Phase I |
NCT01875601 NCT01572493 NCT01385423 |
| 7 | IL-15 Superagonist (ALT-803) | Relapsed/Refractory AML | Phase II | NCT03050216 |
| 8 | Anti-KIR mAB (IPH2101) | Smoldering Multiple Myeloma | Phase I | NCT01248455 |
| 9 | Anti-NKG2A mAB (IPH2201/Monalizumab) | Metastatic squamous cell carcinoma Refractory Lymphoid Malignancies Hematological Malignancies Gynecological Malignancies |
Phase I/II |
NCT02643550 NCT02671435 NCT02921685 NCT02459301 |
| 10 | mAB targeting tumor antigens (Rituximab/Trastuzumab/Cetuximab) | Non-Hodgkin’s Lymphoma Refractory Lymphoid Malignancies Breast and Gastric Cancer Advanced Solid Tumors Recurrent Non-small Cell Lung Cancer |
FDA approved/Phase I/II |
NCT03019640 NCT01181258 NCT02030561 NCT03319459 NCT02845856 |
| 11 | Epigenetic drugs (Decitabine with Donor NK cells and Aldesleukin) | Relapsed/Refractory AML | Phase I | NCT02316964 |
Recent studies with EZH2 inhibitors suggest that the targeting epigenetic regulators may be useful to enhancing NK cell activity [117]. EZH2 inhibitors can also enhance eradication of hepatocellular carcinoma cells by NK cells [99]. Nevertheless, the immunotherapy effects could have arisen from the engagement of other immune cell populations. For example, EZH2 inhibition can promote T-cell-mediated clearance of melanoma cells and cooperate with anti-cytotoxic T-lymphocyte-associated antigen 4-based immunotherapy [117]. Small molecules that target epigenetic regulators have the ability to stimulate anti-cancer immune responses, further highlighting that these drugs represent new enhancers of cancer immunotherapy.
CONCLUDING REMARKS
Despite compelling evidence that NK cells play a role in preventing tumor initiation and progression, a majority of NK cell-based immunotherapies are still in the early stages of development and clinical testing. Additional understanding of the anti-cancer functions of NK cells will aid in the development of more precise and effective NK cell-based immunotherapies (see Outstanding Questions). For example, recent studies have shown important alterations in the super-enhancer functions in immune cells, including NK cells, [118, 119] and a potential future direction is to evaluate the biological impact of super-enhancers on NK cell function and develop drugs to modulate super-enhancers to stimulate NK cell-mediated target cell lysis. Additionally, understanding the epigenetic regulation of NK cell function may have a significant impact on the development of cancer prevention approaches. For example, studies have shown that NK cells are epigenetically reprogrammed and exert stronger activity as a result of exercise [120, 121]. It is still not fully understood why exercise provides cancer prevention benefits, but future studies investigating the direct role of NK cells in mediating those benefits will shed light on this aspect of NK cell function. It is worth noting that several drugs targeting various epigenetic regulators are used clinically for cancer treatment and these drugs can be used either alone or in combination with immunotherapies to enhance NK cell function. The hope is that a combination of drugs targeting epigenetic regulators and NK cell-based therapies will engage both cell intrinsic and extrinsic tumor suppressive pathways to deliver a better clinical outcome for cancer patients.
OUTSTANDING QUESTIONS.
What epigenetic regulators and states modulate NK cell-mediated eradication of cancer cells?
Are paracrine mechanisms important in NK cell-mediated eradication of cancer cells and how are they regulated by epigenetic mechanisms?
How do cancer cells thwart NK cells even when they have heightened stress response pathways that should render them more sensitive to NK cell-mediated eradication?
What aspects of cancer initiation and progression are regulated by NK cells and how does NK cell function evolve with changing epigenetic landscapes in cancer cells?
Trends Box.
NK innate immune cells inhibit tumor initiation and progression. Cancer cells develop several strategies to evade eradication by NK cells.
Epigenetic mechanisms regulate the development and activity of NK cells, and also the ability of cancer cells to evade NK cells.
NK cell-based immunotherapies are in early stages of clinical development. Combination of NK cell-based immunotherapy with investigational or approved epigenetic therapies will likely enhance their efficacy.
Acknowledgments
We gratefully acknowledge grants from the National Institutes of Health: R01CA195077-01A1 (NW), R01CA200919-01 (NW) and 1R01 CA218008-01A1 (NW). N.W. is also supported by Research Scholar Grant from American Cancer Society (128347-RSG-15-212-01-TBG. Grant support from Elsa U Pardee Foundation is also acknowledged.
Footnotes
CONFLICT OF INTEREST STATEMENT
Authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bozzano F, Marras F, De Maria A. Natural Killer Cell Development and Maturation Revisited: Possible Implications of a Novel Distinct Lin(−)CD34(+)DNAM-1(bright)CXCR4(+) Cell Progenitor. Front Immunol. 2017;8:268. doi: 10.3389/fimmu.2017.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi FD, et al. Organ-specific features of natural killer cells. Nat Rev Immunol. 2011;11(10):658–71. doi: 10.1038/nri3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 5.Poli A, et al. Revisiting the Functional Impact of NK Cells. Trends Immunol. 2018 doi: 10.1016/j.it.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer. 1975;16(2):216–29. doi: 10.1002/ijc.2910160204. [DOI] [PubMed] [Google Scholar]
- 7.Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5(2):112–7. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 8.Wang B, et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014;74(20):5746–57. doi: 10.1158/0008-5472.CAN-13-2563. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Eliyahu S, et al. Acute alcohol intoxication suppresses natural killer cell activity and promotes tumor metastasis. Nat Med. 1996;2(4):457–60. doi: 10.1038/nm0496-457. [DOI] [PubMed] [Google Scholar]
- 10.Imai K, et al. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. Lancet. 2000;356(9244):1795–9. doi: 10.1016/S0140-6736(00)03231-1. [DOI] [PubMed] [Google Scholar]
- 11.Melero I, et al. T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4(5):522–6. doi: 10.1158/2159-8290.CD-13-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottcher JP, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172(5):1022–1037 e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17(9):1025–36. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 14.Kortlever RM, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 2017;171(6):1301–1315 e14. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14(3):155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 16.Halfteck GG, et al. Enhanced in vivo growth of lymphoma tumors in the absence of the NK-activating receptor NKp46/NCR1. J Immunol. 2009;182(4):2221–30. doi: 10.4049/jimmunol.0801878. [DOI] [PubMed] [Google Scholar]
- 17.Salih HR, et al. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102(4):1389–96. doi: 10.1182/blood-2003-01-0019. [DOI] [PubMed] [Google Scholar]
- 18.Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. 2005;106(1):376–83. doi: 10.1182/blood-2004-12-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castriconi R, et al. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res. 2004;64(24):9180–4. doi: 10.1158/0008-5472.CAN-04-2682. [DOI] [PubMed] [Google Scholar]
- 20.Kamimura H, et al. Reduced NKG2D ligand expression in hepatocellular carcinoma correlates with early recurrence. J Hepatol. 2012;56(2):381–8. doi: 10.1016/j.jhep.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Fang L, et al. MICA/B expression is inhibited by unfolded protein response and associated with poor prognosis in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:76. doi: 10.1186/s13046-014-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun C, et al. Natural killer cell dysfunction in hepatocellular carcinoma and NK cell-based immunotherapy. Acta Pharmacol Sin. 2015;36(10):1191–9. doi: 10.1038/aps.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillgrass A, et al. The absence or overexpression of IL-15 drastically alters breast cancer metastasis via effects on NK cells, CD4 T cells, and macrophages. J Immunol. 2014;193(12):6184–91. doi: 10.4049/jimmunol.1303175. [DOI] [PubMed] [Google Scholar]
- 24.Yu Y, et al. Mesenchymal stem cells with Sirt1 overexpression suppress breast tumor growth via chemokine-dependent natural killer cells recruitment. Sci Rep. 2016;6:35998. doi: 10.1038/srep35998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iguchi-Manaka A, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med. 2008;205(13):2959–64. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H, et al. MICA/B and ULBP1 NKG2D ligands are independent predictors of good prognosis in cervical cancer. BMC Cancer. 2014;14:957. doi: 10.1186/1471-2407-14-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerra N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28(4):571–80. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friese MA, et al. MICA/NKG2D-mediated immunogene therapy of experimental gliomas. Cancer Res. 2003;63(24):8996–9006. [PubMed] [Google Scholar]
- 29.Watson NF, et al. Expression of the stress-related MHC class I chain-related protein MICA is an indicator of good prognosis in colorectal cancer patients. Int J Cancer. 2006;118(6):1445–52. doi: 10.1002/ijc.21510. [DOI] [PubMed] [Google Scholar]
- 30.Diefenbach A, et al. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413(6852):165–71. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakshmikanth T, et al. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest. 2009;119(5):1251–63. doi: 10.1172/JCI36022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogge von Strandmann E, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27(6):965–74. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Schantz SP, et al. Natural killer cell activity and head and neck cancer: a clinical assessment. J Natl Cancer Inst. 1986;77(4):869–75. [PubMed] [Google Scholar]
- 34.Kumano M, et al. Interleukin-21 activates cytotoxic T lymphocytes and natural killer cells to generate antitumor response in mouse renal cell carcinoma. J Urol. 2007;178(4 Pt 1):1504–9. doi: 10.1016/j.juro.2007.05.115. [DOI] [PubMed] [Google Scholar]
- 35.He S, et al. Enhanced interaction between natural killer cells and lung cancer cells: involvement in gefitinib-mediated immunoregulation. J Transl Med. 2013;11:186. doi: 10.1186/1479-5876-11-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JE, et al. Depletion of ascorbic acid impairs NK cell activity against ovarian cancer in a mouse model. Immunobiology. 2012;217(9):873–81. doi: 10.1016/j.imbio.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Silver DF, et al. Effects of IL-12 on human ovarian tumors engrafted into SCID mice. Gynecol Oncol. 1999;72(2):154–60. doi: 10.1006/gyno.1998.5239. [DOI] [PubMed] [Google Scholar]
- 38.Versluis MAC, et al. The prognostic benefit of tumour-infiltrating Natural Killer cells in endometrial cancer is dependent on concurrent overexpression of Human Leucocyte Antigen-E in the tumour microenvironment. Eur J Cancer. 2017;86:285–295. doi: 10.1016/j.ejca.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Brooks J, et al. Perioperative, Spatiotemporally Coordinated Activation of T and NK Cells Prevents Recurrence of Pancreatic Cancer. Cancer Res. 2018;78(2):475–488. doi: 10.1158/0008-5472.CAN-17-2415. [DOI] [PubMed] [Google Scholar]
- 40.Mimura K, et al. Therapeutic potential of highly cytotoxic natural killer cells for gastric cancer. Int J Cancer. 2014;135(6):1390–8. doi: 10.1002/ijc.28780. [DOI] [PubMed] [Google Scholar]
- 41.Zhu L, et al. Natural Killer Cell (NK-92MI)-Based Therapy for Pulmonary Metastasis of Anaplastic Thyroid Cancer in a Nude Mouse Model. Front Immunol. 2017;8:816. doi: 10.3389/fimmu.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wennerberg E, et al. Human anaplastic thyroid carcinoma cells are sensitive to NK cell-mediated lysis via ULBP2/5/6 and chemoattract NK cells. Clin Cancer Res. 2014;20(22):5733–44. doi: 10.1158/1078-0432.CCR-14-0291. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. 2013;34(12):573–82. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 45.Colucci F, Caligiuri MA, Di Santo JP. What does it take to make a natural killer? Nat Rev Immunol. 2003;3(5):413–25. doi: 10.1038/nri1088. [DOI] [PubMed] [Google Scholar]
- 46.Freud AG, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22(3):295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–86. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 48.Freud AG, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203(4):1033–43. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol. 2011;11(10):645–57. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huntington ND, Vosshenrich CA, Di Santo JP. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat Rev Immunol. 2007;7(9):703–14. doi: 10.1038/nri2154. [DOI] [PubMed] [Google Scholar]
- 51.Pinhas N, et al. Murine peripheral NK-cell populations originate from site-specific immature NK cells more than from BM-derived NK cells. Eur J Immunol. 2016;46(5):1258–70. doi: 10.1002/eji.201545847. [DOI] [PubMed] [Google Scholar]
- 52.Geiger TL, Sun JC. Development and maturation of natural killer cells. Curr Opin Immunol. 2016;39:82–9. doi: 10.1016/j.coi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, Tian Z, Wei H. Developmental and Functional Control of Natural Killer Cells by Cytokines. Front Immunol. 2017;8:930. doi: 10.3389/fimmu.2017.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pegram HJ, et al. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol. 2011;89(2):216–24. doi: 10.1038/icb.2010.78. [DOI] [PubMed] [Google Scholar]
- 55.Long EO, et al. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–58. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–9. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groh V, et al. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419(6908):734–8. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 58.Salih HR, Holdenrieder S, Steinle A. Soluble NKG2D ligands: prevalence, release, and functional impact. Front Biosci. 2008;13:3448–56. doi: 10.2741/2939. [DOI] [PubMed] [Google Scholar]
- 59.Fauriat C, et al. Impaired activating receptor expression pattern in natural killer cells from patients with multiple myeloma. Leukemia. 2006;20(4):732–3. doi: 10.1038/sj.leu.2404096. [DOI] [PubMed] [Google Scholar]
- 60.Schleypen JS, et al. Renal cell carcinoma-infiltrating natural killer cells express differential repertoires of activating and inhibitory receptors and are inhibited by specific HLA class I allotypes. Int J Cancer. 2003;106(6):905–12. doi: 10.1002/ijc.11321. [DOI] [PubMed] [Google Scholar]
- 61.Kohler B, Birkhed D, Olsson S. Acid production by human strains of Streptococcus mutans and Streptococcus sobrinus. Caries Res. 1995;29(5):402–6. doi: 10.1159/000262099. [DOI] [PubMed] [Google Scholar]
- 62.de Kruijf EM, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. 2010;185(12):7452–9. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 63.Chiu J, Ernst DM, Keating A. Acquired Natural Killer Cell Dysfunction in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Front Immunol. 2018;9:267. doi: 10.3389/fimmu.2018.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Z, et al. Notch1 signaling in melanoma cells promoted tumor-induced immunosuppression via upregulation of TGF-beta1. J Exp Clin Cancer Res. 2018;37(1):1. doi: 10.1186/s13046-017-0664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holt D, et al. Prostaglandin E(2) (PGE (2)) suppresses natural killer cell function primarily through the PGE(2) receptor EP4. Cancer Immunol Immunother. 2011;60(11):1577–86. doi: 10.1007/s00262-011-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richards JO, et al. Tumor growth impedes natural-killer-cell maturation in the bone marrow. Blood. 2006;108(1):246–52. doi: 10.1182/blood-2005-11-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benson DM, Jr, et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116(13):2286–94. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity. 2016;44(5):989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He Y, et al. Contribution of inhibitory receptor TIGIT to NK cell education. J Autoimmun. 2017;81:1–12. doi: 10.1016/j.jaut.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Gallois A, et al. Reversal of natural killer cell exhaustion by TIM-3 blockade. Oncoimmunology. 2014;3(12):e946365. doi: 10.4161/21624011.2014.946365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Busslinger M, Tarakhovsky A. Epigenetic control of immunity. Cold Spring Harb Perspect Biol. 2014;6(6) doi: 10.1101/cshperspect.a019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Topper MJ, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 2017;171(6):1284–1300 e21. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon HJ, et al. Stepwise phosphorylation of p65 promotes NF-kappaB activation and NK cell responses during target cell recognition. Nat Commun. 2016;7:11686. doi: 10.1038/ncomms11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liyasova MS, Ma K, Lipkowitz S. Molecular pathways: cbl proteins in tumorigenesis and antitumor immunity-opportunities for cancer treatment. Clin Cancer Res. 2015;21(8):1789–94. doi: 10.1158/1078-0432.CCR-13-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Selvarajan V, et al. RUNX3 is oncogenic in natural killer/T-cell lymphoma and is transcriptionally regulated by MYC. Leukemia. 2017;31(10):2219–2227. doi: 10.1038/leu.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Munoz-Fontela C, et al. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol. 2016;16(12):741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Casey SC, Baylot V, Felsher DW. MYC: Master Regulator of Immune Privilege. Trends Immunol. 2017;38(4):298–305. doi: 10.1016/j.it.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Textor S, et al. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71(18):5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 80.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iannello A, et al. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med. 2013;210(10):2057–69. doi: 10.1084/jem.20130783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18(3):139–147. doi: 10.1038/nrc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soto M, et al. p53 Prohibits Propagation of Chromosome Segregation Errors that Produce Structural Aneuploidies. Cell Rep. 2017;19(12):2423–2431. doi: 10.1016/j.celrep.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 84.Santaguida S, et al. Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Dev Cell. 2017;41(6):638–651 e5. doi: 10.1016/j.devcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Versteeg R, et al. High expression of the c-myc oncogene renders melanoma cells prone to lysis by natural killer cells. J Immunol. 1989;143(12):4331–7. [PubMed] [Google Scholar]
- 86.Unni AM, Bondar T, Medzhitov R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc Natl Acad Sci U S A. 2008;105(5):1686–91. doi: 10.1073/pnas.0701675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25(10):496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 88.Eckelhart E, et al. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117(5):1565–73. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- 89.Gotthardt D, Sexl V. STATs in NK-Cells: The Good, the Bad, and the Ugly. Front Immunol. 2016;7:694. doi: 10.3389/fimmu.2016.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gotthardt D, et al. STAT5 Is a Key Regulator in NK Cells and Acts as a Molecular Switch from Tumor Surveillance to Tumor Promotion. Cancer Discov. 2016;6(4):414–29. doi: 10.1158/2159-8290.CD-15-0732. [DOI] [PubMed] [Google Scholar]
- 91.Lee CK, et al. Distinct requirements for IFNs and STAT1 in NK cell function. J Immunol. 2000;165(7):3571–7. doi: 10.4049/jimmunol.165.7.3571. [DOI] [PubMed] [Google Scholar]
- 92.Gotthardt D, et al. Loss of STAT3 in murine NK cells enhances NK cell-dependent tumor surveillance. Blood. 2014;124(15):2370–9. doi: 10.1182/blood-2014-03-564450. [DOI] [PubMed] [Google Scholar]
- 93.Bedel R, et al. Novel role for STAT3 in transcriptional regulation of NK immune cell targeting receptor MICA on cancer cells. Cancer Res. 2011;71(5):1615–26. doi: 10.1158/0008-5472.CAN-09-4540. [DOI] [PubMed] [Google Scholar]
- 94.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gates LA, Foulds CE, O’Malley BW. Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem Sci. 2017;42(12):977–989. doi: 10.1016/j.tibs.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nandakumar V, et al. Epigenetic control of natural killer cell maturation by histone H2A deubiquitinase, MYSM1. Proc Natl Acad Sci U S A. 2013;110(41):E3927–36. doi: 10.1073/pnas.1308888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stone ML, et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc Natl Acad Sci U S A. 2017;114(51):E10981–E10990. doi: 10.1073/pnas.1712514114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bugide S, Green MR, Wajapeyee N. Inhibition of Enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc Natl Acad Sci U S A. 2018 doi: 10.1073/pnas.1802691115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin J, et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci U S A. 2015;112(52):15988–93. doi: 10.1073/pnas.1521740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cribbs A, et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. J Biol Chem. 2018;293(7):2422–2437. doi: 10.1074/jbc.RA117.000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao D, et al. H3K4me3 Demethylase Kdm5a Is Required for NK Cell Activation by Associating with p50 to Suppress SOCS1. Cell Rep. 2016;15(2):288–99. doi: 10.1016/j.celrep.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 103.Ha M, V, Kim N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 104.Shen J, et al. Silencing NKG2D ligand-targeting miRNAs enhances natural killer cell-mediated cytotoxicity in breast cancer. Cell Death Dis. 2017;8(4):e2740. doi: 10.1038/cddis.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Breunig C, et al. MicroRNA-519a-3p mediates apoptosis resistance in breast cancer cells and their escape from recognition by natural killer cells. Cell Death Dis. 2017;8(8):e2973. doi: 10.1038/cddis.2017.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie J, et al. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. 2014;11(5):495–502. doi: 10.1038/cmi.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ni F, et al. MicroRNA transcriptomes of distinct human NK cell populations identify miR-362-5p as an essential regulator of NK cell function. Sci Rep. 2015;5:9993. doi: 10.1038/srep09993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim TD, et al. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 2011;118(20):5476–86. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yun S, et al. Integrated mRNA-microRNA profiling of human NK cell differentiation identifies MiR-583 as a negative regulator of IL2Rgamma expression. PLoS One. 2014;9(10):e108913. doi: 10.1371/journal.pone.0108913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Donatelli SS, et al. TGF-beta-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014;111(11):4203–8. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Regis S, et al. TGF-beta1 Downregulates the Expression of CX3CR1 by Inducing miR-27a-5p in Primary Human NK Cells. Front Immunol. 2017;8:868. doi: 10.3389/fimmu.2017.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ansell SM, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neelapu SS, et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N Engl J Med. 2017;377(26):2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Campbell KS, Hasegawa J. Natural killer cell biology: an update and future directions. J Allergy Clin Immunol. 2013;132(3):536–544. doi: 10.1016/j.jaci.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chiossone L, et al. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol. 2017;31:55–63. doi: 10.1016/j.smim.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 117.Zingg D, et al. The Histone Methyltransferase Ezh2 Controls Mechanisms of Adaptive Resistance to Tumor Immunotherapy. Cell Rep. 2017;20(4):854–867. doi: 10.1016/j.celrep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 118.Koues OI, et al. Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell. 2016;165(5):1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peeters JG, et al. Inhibition of Super-Enhancer Activity in Autoinflammatory Site-Derived T Cells Reduces Disease-Associated Gene Expression. Cell Rep. 2015;12(12):1986–96. doi: 10.1016/j.celrep.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 120.Zimmer P, et al. Exercise-induced Natural Killer Cell Activation is Driven by Epigenetic Modifications. Int J Sports Med. 2015;36(6):510–5. doi: 10.1055/s-0034-1398531. [DOI] [PubMed] [Google Scholar]
- 121.Zimmer P, et al. Impact of exercise on pro inflammatory cytokine levels and epigenetic modulations of tumor-competitive lymphocytes in Non-Hodgkin-Lymphoma patients-randomized controlled trial. Eur J Haematol. 2014;93(6):527–32. doi: 10.1111/ejh.12395. [DOI] [PubMed] [Google Scholar]