Abstract
Antibiotic treatments often fail to completely eradicate a bacterial infection, leaving behind an antibiotic-tolerant subpopulation of intact bacterial cells called persisters. Persisters are considered a major cause for treatment failure and are thought to greatly contribute to the recalcitrance of chronic infections. Pseudomonas aeruginosa infections are commonly associated with elevated levels of drug-tolerant persister cells, posing a serious threat to human health. This study represents the first time a novel large molecule polycationic glycopolymer, poly (acetyl, arginyl) glucosamine (PAAG), has been evaluated against antibiotic and carbonyl cyanide m-chlorophenylhydrazone induced P. aeruginosa persisters. PAAG eliminated eliminated persisters at concentrations that show no significant cytotoxicity on human lung epithelial cells. PAAG demonstrated rapid bactericidal activity against both forms of induced P. aeruginosa persister cells resulting in complete eradication of the in vitro persister cells within 24 h of treatment. PAAG demonstrated greater efficacy against persisters in vitro than antibiotics currently being used to treat persistent chronic infections such as tobramycin, colistin, azithromycin, aztreonam, and clarithromycin. PAAG caused rapid permeabilization of the cell membrane and caused significant membrane depolarization in persister cells. PAAG efficacy against these bacterial subpopulations suggests it may have substantial therapeutic potential for eliminating recurrent P. aeruginosa infections.
Keywords: persister cells, chronic infection, glycopolymer, Pseudomonas aeruginosa, PAAG
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that often causes nosocomial infections in immunocompromised patients and is one of the primary agents responsible for pulmonary decline and early mortality in patients with cystic fibrosis (CF; Mendelson et al., 1994; Dunn and Wunderink, 1995; Govan and Deretic, 1996; Gibson et al., 2003). P. aeruginosa reaches relatively high densities in the CF lung, and a substantial fraction of the cells present are in a low metabolic activity state correlated with persister cell status (Yang et al., 2008). The frequent use of high doses of bactericidal antibiotics during chronic infections may lead to selective mutations that produce heightened levels of persisters (Keren et al., 2004). Multiple lines of evidence suggest that the recalcitrant nature of P. aeruginosa infections in CF lungs is caused by a drug-tolerant subpopulation of persister cells (Burns et al., 1999; Gilligan, 2006; Yang et al., 2008; Mulcahy et al., 2010).
Persisters are a small fraction of non-replicating, metabolically quiescent bacteria tolerant to antibiotic killing (Keren et al., 2004; LaFleur et al., 2010; Mulcahy et al., 2010). These antibiotic-tolerant bacterial cells have a growth-arrested phenotype and are capable of recommencing growth after a stress event (Lewis, 2007, 2008; Kim and Wood, 2016). Due to their state of metabolic dormancy, persisters have a high tolerance against traditional classes of antibiotics such as fluoroquinolones, aminoglycosides, and beta-lactams, which are only effective against metabolically active cells. Antibiotics that are bactericidal against planktonic cells are typically ineffective against persister cells (Hoyle et al., 1990). Once the local antibiotic concentration drops and the nutrients are available (Kim et al., 2018), persisters can become metabolically active again and reestablish the infection (Lewis, 2007, 2008) causing the relapsing chronic infections often observed in CF patients (Lewis, 2008).
The ineffectiveness of conventional systemic antibiotics for treating chronic pulmonary infections have led to treatment with high doses of inhaled antibiotics including azithromycin, aztreonam, and tobramycin (Mearns, 1972; Geller et al., 2002; Zindani et al., 2006). During such treatments, aerosolized tobramycin can reach peak concentrations of 1,237 μg/g of sputum, which is ≥25 times higher than the minimum inhibitory concentration (MIC) of most tested clinical isolates of P. aeruginosa (Geller et al., 2002). Inhalation treatments with levofloxacin achieve up to 1,760 μg/g of sputum, a concentration that is >50 times higher than MIC of clinical isolates of P. aeruginosa (King et al., 2010). Tobramycin and levofloxacin at these concentrations effectively kill actively growing resistant bacteria but induce a stress event that supports persister cell phenotype development (King et al., 2010; Lewis, 2010). Inhaled tobramycin has long been identified to control but not eradicate P. aeruginosa infections in patients with chronic lung infections (Ramsey et al., 1999; Gibson et al., 2003). The decrease in efficacy of tobramycin over treatment time can be attributed to and is consistent with an increase in the numbers of persisters (Koeva et al., 2017).
The limited activity of traditional antibiotics against persisters is due to attenuation of active bacterial transport mechanisms along with low metabolic rates (Davis, 1987; Allison et al., 2011). Metabolite stimulation of the proton motive force (PMF) has been shown to awaken the cells (Kim et al., 2018) therefore improve the uptake of aminoglycosides and increase effectiveness of bacterial persister killing, helping to clear the infection (Allison et al., 2011; Koeva et al., 2017). Fructose in combination with gentamicin was observed to be effective against Escherichia coli as well as Staphylococcus aureus persisters (Lebeaux et al., 2014). Metabolite enabled killing of P. aeruginosa persisters has been observed with aminoglycosides and fructose as well as mannitol and tobramycin (Barraud et al., 2013; Orman and Brynildsen, 2013). Arginine and nitrate were also described as useful additives in improving the uptake of aminoglycosides (Borriello et al., 2006). A recently published study suggests a fumarate antibacterial potentiator used in combination with tobramycin enhanced killing of persister cells compared to tobramycin alone (Koeva et al., 2017). Together these studies show the influence of bacterial transport mechanisms and metabolism on antibiotic tolerance exhibited by persister cells.
Another potential class of antibacterial used in persister treatment and research are antimicrobial peptides (AMPs). AMPs are known for their ability to disrupt bacterial membranes and/or translocate across membranes to bind intracellular targets. A major drawback to this approach is the potential toxicity to mammalian cells (Zhao et al., 2013; Kang et al., 2014). AMPs potential to disrupt bacterial membranes is less dependent on the metabolic status of the bacterial cell (Hurdle et al., 2011). Colistin disrupts the outer membrane of Gram-negative species and is bactericidal against persisters (Cui et al., 2016), but the concentration required to eradicate persisters is above clinically acceptable concentrations, leading to toxicity, including renal and neurological side effects (Cui et al., 2016). Despite major progress in understanding the metabolic changes and mechanisms of bacterial persister cell formation, several impediments to clinical applications remain (Barraud et al., 2013; Chowdhury et al., 2016; Wood, 2017). The observation that membrane-disrupting antibiotics can kill persisters is an important discovery that suggests a therapeutic way forward.
Poly (acetyl, arginyl) glucosamine (PAAG), is among a novel class of polysaccharide therapeutics with antimicrobial activity against a wide range of pathogenic Gram-negative and Gram-positive bacterial species including methicillin-resistant Staphylococcus aureus (MRSA) (Narayanaswamy et al., 2018), multiple species of Burkholderia (Narayanaswamy et al., 2017), E. coli (Tang et al., 2010), and non-tuberculous Mycobacteria (NTM; Unpublished data). The antimicrobial efficacy of PAAG has been attributed to its polycationic nature (Tang et al., 2010; Narayanaswamy et al., 2017). PAAG rapidly permeabilizes Gram negative bacteria (Tang et al., 2010; Narayanaswamy et al., 2017) at the doses that we report in this study, which have minimal cytotoxic effects in vitro when tested against the human epithelial lung cell line, A549.
Aims of the present study were to (i) evaluate the ability of PAAG to eradicate antibiotic-induced P. aeruginosa persisters, commonly implicated in relapsing and chronic lung infections, (ii) evaluate the susceptibility of carbonyl cyanide m-chlorophenylhydrazone (CCCP)-induced persisters to membrane active agents like colistin and polycationic glycopolymer PAAG, (iii) characterize the membrane activity of PAAG and other antibiotics on P. aeruginosa persister-like cells in the presence and absence of CCCP, and (iv) test the cytotoxic effects of PAAG on A549 lung epithelial cells.
Materials and Methods
Bacterial Strains, Reagents, and Growth Conditions
The bacterial strain used in the study was P. aeruginosa PA01 (a kind donation from Paul Orwin, CSUSB). Bacteria were initially streaked from -80°C glycerol stock onto Luria–Bertani agar (LB Agar; Difco, Fisher Scientific) plates and incubated at 37°C overnight. Minimal BM2 medium (Poole et al., 1991; Reffuveille et al., 2014; Mlynarcik and Kolar, 2017) with glucose as the carbon source [phosphate ammonia base (0.07 M (NH4)2SO4 – 9.25 g, 0.4 M K2HPO4 – 69.7 g, 0.22 KH2PO4 – 29.9 g, pH 7.0), 50 mM MgSO4, 10 mM FeSO4, and 40% glucose] and LB broth was used to culture P. aeruginosa cells. Planktonic stationary-phase and exponential cultures were prepared as previously described (40). Planktonic stationary-phase cultures were prepared by inoculating from a fresh LB agar plate into 5 mL LB broth (Difco, Fisher Scientific) and incubating them in a shaker incubator overnight at 37°C. For experiments, 1 mL of the overnight culture was centrifuged at 13,000 rpm for 1 min, then washed and resuspended in BM2 glucose media. Planktonic exponential-phase cultures were prepared by inoculating 107 cells/mL in BM2 glucose media and growing them to an optical density (600 nm) of 0.6–0.8. The bacterial cells were exposed to PAAG (Synedgen, Inc.), azithromycin (TCI), tobramycin (Sigma), clarithromycin (Fluka), aztreonam (TCI), and colistin (TCI) at the concentrations and time periods stated throughout the article. CCCP was purchased from Sigma-Aldrich. Stock solution of 0.2 mg/mL CCCP was diluted in DMSO and stored as aliquots at -20°C.
Antimicrobial Susceptibility Assay
Sterile aqueous stock solutions of PAAG (10–1,000 μg/mL), azithromycin (8–512 μg/mL), ciprofloxacin (0.25–64 μg/mL), ofloxacin (0.25–64 μg/mL), clarithromycin (8–512 μg/mL), tobramycin (2–128 μg/mL), colistin (8–512 μg/mL), aztreonam (8–512 μg/mL), and CCCP (200 μg/mL) were used to perform MIC) assays. Serial dilutions of each antibiotic/PAAG were prepared in the presence or absence of CCCP in 50 μL Mueller Hinton Broth (MHB) in 96-well microtiter plates. Exponential phase broth culture of P. aeruginosa PA01, diluted to a cell density of 1.5 × 108 CFU/mL, were verified by total viable count. Then 50 μL of the inoculum is added to each well of the microtiter plates. Positive and negative growth controls were included in each plate to verify the test method. Plates were incubated at 37°C and MIC values were determined for each antimicrobial as the lowest concentration at which growth was inhibited. The experiment was performed in triplicate with three independent cultures.
Time Kill Assay
Planktonic stationary phase cultures were prepared as described above. Briefly overnight cultures of P. aeruginosa cells were pelleted (13,000 rpm for 1 min), washed and resuspended in BM2 glucose media. An inoculum of 20 μL P. aeruginosa suspension was added into 180 μL of BM2 glucose media containing concentrations of the antimicrobials, four times their MIC [PAAG (100–200 μg/mL), tobramycin (16 μg/mL), clarithromycin (256 μg/mL), colistin (256 μg/mL), aztreonam (256 μg/mL), azithromycin (256 μg/mL)] in 96-well plates. The plates were sealed with Parafilm, loaded onto microplate shakers, and placed in a 37°C incubator for 10, 30 min and 1, 2, 4, 8, 24 h. At each time point, 10-fold serial dilutions were made with BM2 glucose media and the dilutions were spot plated onto LB agar plates and incubated overnight at 37°C to yield visible colonies. The experiments were performed in triplicate with three independent cultures.
CCCP-Induced Persisters
Pretreatment with CCCP at a concentration of 200 μg/mL for 3 h has been observed to induce a 5,000-fold increase in the number of persister-like cells in cultures subsequently exposed to antibiotics (Grassi et al., 2017). The concentration of CCCP and the treatment time required to induce persistence in stationary-phase cultures of P. aeruginosa has been previously described by Grassi et al. (2017). Overnight cultures of P. aeruginosa were incubated with 200 μg/mL of CCCP for 3 h at 37°C with shaking. Following pretreatment with CCCP, bacteria were washed twice in BM2 glucose media, pelleted (13,000 rpm for 1 min), and re-suspended in BM2 glucose media to a final density of 1.5 × 108 CFU/mL. Bacterial suspensions induced with CCCP were exposed to the following antibiotics at concentrations four times the MIC: tobramycin (16 μg/mL), clarithromycin (256 μg/mL), aztreonam (256 μg/mL), azithromycin (256 μg/mL). Untreated and CCCP-pretreated bacteria were used as viability controls. The surviving persister cells were determined by CFU counting at 10, 30 min and 1, 2, 4, 8, 24 h of incubation at 37°C. The experiment was performed in triplicate with three independent cultures.
Effect of PAAG on CCCP-, Antibiotic-Induced Persister Cells
To test the ability of PAAG or colistin to potentiate the effect of antibiotics and thereby eliminate the surviving population of antibiotic-induced persister cells, planktonic stationary phase cultures were exposed to antibiotics [tobramycin (16 μg/mL), clarithromycin (256 μg/mL), or aztreonam (256 μg/mL)] and placed in a 37°C incubator for 3 h prior to treatment with PAAG (200 μg/mL) or colistin (256 μg/mL).
CCCP-induced persisters were generated by pretreating P. aeruginosa cultures to CCCP (200 μg/mL) for 3 h at 37°C as described above. Briefly, the CCCP-induced persister cells were pelleted and resuspended in BM2 glucose media and 20 μL was added to wells containing 180 μL of PAAG or colistin at a concentration of 100–200 or 256 μg/mL respectively.
The plates were sealed with Parafilm, loaded onto microplate shakers in a 37°C incubator for 24 h. At 30 min and 1, 2, 4, 8, 24 h 10-fold dilutions were made with BM2 glucose media and spot plated onto LB plates to determine CFU/mL. The experiment was performed in triplicate with three biological replicates.
Propidium Iodide (PI) Uptake Assay
CCCP-induced cultures of P. aeruginosa were generated as mentioned above and were treated with antibiotics [tobramycin (16 μg/mL), clarithromycin (256 μg/mL), colistin (256 μg/mL), aztreonam (256 μg/mL), azithromycin (256 μg/mL), or PAAG (100–200 μg/mL)] for 3 h, pelleted, washed and resuspended in BM2 glucose media. PI was used at a concentration of 17 μg/mL as described in previous studies (Kwan et al., 2015). All assays were performed at room temperature. PI at a concentration of 17 μg/mL was added to the wells of a 96 well plate containing 1.5 × 108 CFU/mL of the prepared bacterial culture and fluorescence was measured via SpectraMax Gemini XPS (Molecular Devices). PAAG (100 and 200 μg/mL), ciprofloxacin (2 μg/mL), ampicillin (100 μg/mL), tobramycin (16 μg/mL), clarithromycin (256 μg/mL), colistin (256 μg/mL), aztreonam (256 μg/mL), azithromycin (256 μg/mL) were added to the wells containing the mixture. Cells treated with 0.1% Triton X-100 was used as a positive control. PI alone and PI on cells were used as negative controls. The mixture was mixed thoroughly prior to obtaining fluorescence measurements. Fluorescence was measured at excitation and emission wavelengths of 535 and 625 nm, every 10 min up to 4 h. The experiment was performed in triplicate with three independent cultures.
Cytoplasmic Membrane Depolarization Assay
The membrane depolarization activity of PAAG and colistin was determined using the membrane potential-sensitive fluorescent dye 3,3′-Dipropylthiadicarbocyanine iodide (DiSC3-5) (Suzuki et al., 2003). CCCP-induced persister cells of P. aeruginosa PA01 were grown as described above and subsequently treated with tobramycin (16 μg/mL) for 3 h at 37°C. The cells were collected by centrifugation and washed three times with 5 mM HEPES (pH 7.8) buffer. The cells were then resuspended in 5 mM HEPES (pH 7.8) buffer with 0.2 mM EDTA to an OD600 of 0.05 (45). DiSC3(5) and KCl was added at the final concentration of 0.4 uM and 0.1 M, respectively, and incubated at 37°C shaker (150 rpm) incubator for 20 min until the fluorescence quenching was achieved. PAAG and colistin at a final concentration of 50–200 and 256 μg/mL, respectively, was added, and the fluorescence was monitored under shaking conditions at 37°C at an excitation wavelength of 622 nm and an emission wavelength of 670 nm subsequently after 15 min of incubation. Wells with cells and the dye were used as background. Triton X-100 (0.1% v/v), a commonly used membrane disruptor, was used as the positive control (Saar et al., 2005). The experiment was performed in triplicate with three independent cultures.
Cytotoxicity Assay
The cytotoxicity of PAAG was investigated on A549 cells (ATCC CRM-CCL-185) using the Pierce lactate dehydrogenase (LDH) Cytotoxicity Kit (Promega, Madison, WI,United States). Cells were seeded in a 96-well plate at a concentration of 2.0 × 104 cells/well. Then, 100 μL of F12K media was added to each well-containing varying concentrations of PAAG (65–1,000 μg/mL) and incubated for 24 h. Briefly, 50 μL of media was transferred from each treatment well to a new 96-well plate, then mixed with 50 μL of LDH reaction mixture and incubated at room temperature for 30 min. After incubation, 50 μL of stop solution was added and the plate was read using a wavelength of 490 and 680 nm on a microplate reader (SpectraMax Gemini XPS; Molecular Devices). The untreated cell population represents the baseline levels of LDH from A549 cells and a lysis buffer control (provided in the kit) was used to normalize the data. 10× lysis buffer (provided by the manufacturer) was used as the positive control. The % cytotoxicity was calculated according to the manufacturer’s instructions. Briefly, % cytotoxicity equals the difference between the PAAG-treaded LDH activity and the Spontaneous LDH activity (water-treated control), divided by the difference of the maximum LDH activity (lysis buffer) and the spontaneous LDH activity, multiplied by 100. Data represented as % viability. The experiment was performed in triplicate.
Statistical Analysis
All the in vitro experiments were performed in triplicate. The statistical analysis was conducted using GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA, United States). Results are expressed as means ± standard error of the mean (SEM).
Results
Antibacterial Activity of the Antimicrobials Against Planktonic Cells
The MIC’s of PAAG, tobramycin (TOB), azithromycin (AZI), clarithromycin (CLA), aztreonam (ATM), ofloxacin (OFX), ciprofloxacin (CIP), and colistin (CST) were determined to be 30, 4, 64, 64, 64, 0.5, 0.5, and 64 μg/mL, respectively, as shown in Table 1. The CSLI standard clinical breakpoints are also noted (CLSI, 2017). As shown in Table 1, presence of CCCP resulted in substantial reduction in the MIC of colistin. Though the MIC’s of other antibiotics showed a slight increase in MIC in the presence of CCCP (Table 1).
Table 1.
Antibiotic susceptibility of planktonic P. aeruginosa PA01 to antimicrobials.
| P. aeruginosa PA01 | MIC (μg/mL) |
|||||||
|---|---|---|---|---|---|---|---|---|
| PAAG | CST | TOB | ATM | AZI | CLA | OFX | CIP | |
| 30 [30] | 64 (R) [4 (S)] | 4 (S) [8 (I)] | 64 (R) [128 (R)] | 64 [128] | 64 [64] | 0.5 (S) [0.5 (S)] | 0.5 (S) [0.25 (S)] | |
Clinical breakpoints for antibiotics according to CSLI standards were ≤2 μg/mL (S), ≥4 μg/mL (R) for colistin (CST); ≥16 μg/mL (R), 8 μg/mL (I), ≤4 μg/mL (S) for tobramycin (TOB); ≥32 μg/mL (R), 16 μg/mL (I), ≤8 μg/mL (S) for aztreonam (ATM); ≥8 μg/mL (R), 4 μg/mL (I), ≤2 μg/mL (S) for ofloxacin (OFX); ≥4 μg/mL (R), 2 μg/mL (I), ≤1 μg/mL (S) for ciprofloxacin (CIP). R, resistant strains; S, Sensitive strains; I, Intermediate. MIC’s in [] represent were taken in the presence of CCCP.
Bactericidal Effects of Antimicrobials
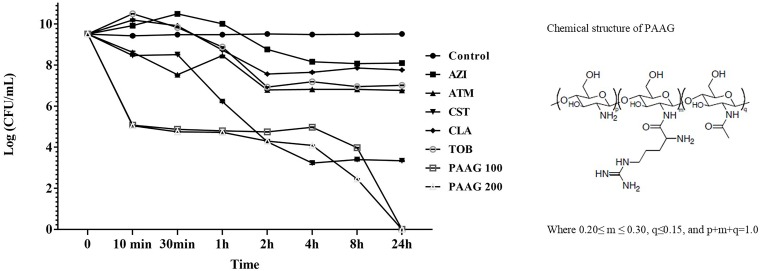
Kill curves for each antibiotic were assessed to identify the antibiotic concentrations enabling survival of a small drug-tolerant subpopulation (Figure 1). Antibiotics like tobramycin, and aztreonam exhibited a 3-log reduction in CFU/mL. Azithromycin and clarithromycin displayed 2-log reduction in CFU/mL. The afore mentioned antibiotics demonstrated limited bactericidal activity even at concentrations four times its MIC indicative of limited bactericidal activity against stationary phase cultures of P. aeruginosa. Colistin, as a cell wall disruptor, demonstrated potent bactericidal activity against stationary phase cultures, resulting in a 5-log reduction in CFU/mL after 3 h of treatment (P < 0.001; Figure 1). Treatment with PAAG at concentrations of 100 and 200 μg/mL, resulted in a 6-log reduction in CFU/mL compared to the untreated control within 10 min (P < 0.001) and total eradication by 24 h.
FIGURE 1.
PAAG kills stationary phase Pseudomonas aeruginosa cultures without forming persisters. Stationary phase P. aeruginosa cultures were challenged with tobramycin (16 μg/mL), clarithromycin (256 μg/mL), colistin (256 μg/mL), aztreonam (256 μg/mL), azithromycin (256 μg/mL), or PAAG (100–200 μg/mL) for 24 h. Samples were aliquoted and plated for viable plate count (CFU/mL) at multiple timepoints over 24 h. Unchallenged control samples were aliquoted directly prior to antibiotic treatment. Data represents average of three independent experiments, and the error bars indicate the standard error of the mean.
At concentrations four times their MIC, all the antibiotics except PAAG tested demonstrated a biphasic time-kill curve with a slow decrease in CFU/mL of stationary phase cultures over the first 3 h of treatment, followed by stabilization of the viable population at 24 h (Figure 1).
PAAG Potentiates Killing of Antibiotic-Induced Persisters
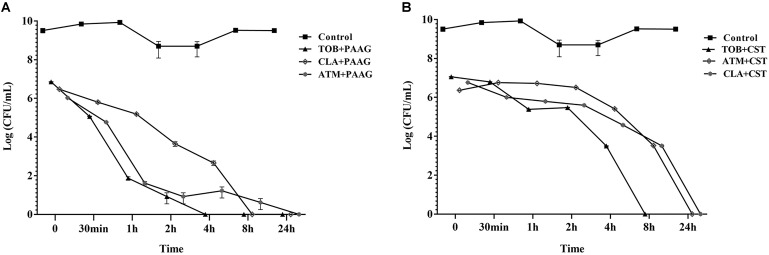
Antibiotic tolerant cells were isolated by treating stationary phase cultures of P. aeruginosa with tobramycin, aztreonam or clarithromycin as described in Section “Materials and Methods.” The surviving antibiotic tolerant bacteria were further challenged with PAAG or colistin at a concentration of 200 and 256 μg/mL respectively (Figure 2). Treatment with PAAG demonstrated a 6- to 7-log reduction (CFU/mL) within 2–4 h of treatment and complete eradication within 24 h (Figure 2A). Colistin at concentrations 4× MIC was observed to be bactericidal (3- to 4-log reduction) against the antibiotic induced persister population in 2 h of treatment and resulted in complete eradication of the antibiotic tolerant population of the P. aeruginosa cells in 8–24 h of treatment (Figure 2B).
FIGURE 2.
(A,B). Effect of antimicrobials against antibiotic- induced P. aeruginosa persister cells. Activity of PAAG and colistin at a concentration of 200 and 256 μg/mL, respectively, was assessed against stationary phase cultures of P. aeruginosa PA01 pretreated with tobramycin (16 μg/mL), aztreonam (256 μg/mL), or clarithromycin (256 μg/mL) for 3 h. After the addition of PAAG or colistin samples were aliquoted and plated for colony count (CFU/mL) at multiple time points over 24 h. Non-challenged control samples were plated immediately prior to PAAG treatment. T = 0 is 3 h post-antibiotic treatment. Data represents average ± standard error of the mean of three independent experiments.
Induction of Persistence in the Presence of CCCP
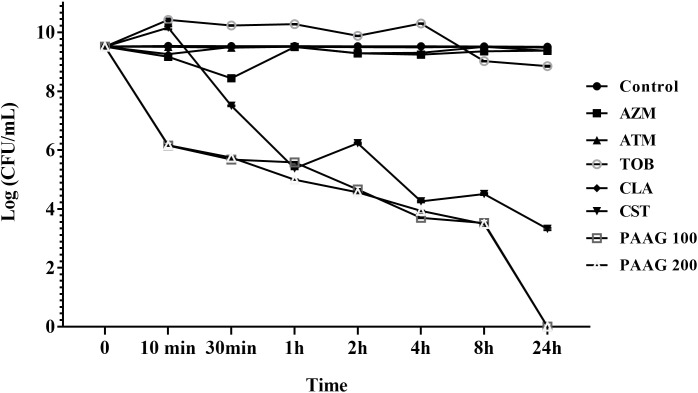
The ability of CCCP to induce persistence in P. aeruginosa cells was evaluated by exposing CCCP-pretreated cultures to different classes of antibiotics with diverse mechanisms of action such as fluoroquinolones, namely the monobactam (aztreonam), aminoglycoside (tobramycin), and macrolide (clarithromycin and azithromycin) as previously described (Grassi et al., 2017). Pretreatment with CCCP (200 μg/mL) for 3 h resulted in significant increase in the survival of P. aeruginosa PA01 cells against the different classes of antibiotics. As shown in Figure 3, stationary-phase cultures pretreated with CCCP for 3 h demonstrated tolerance to treatment with antibiotics namely aztreonam, azithromycin, clarithromycin and tobramycin as compared to the untreated controls. Pretreatment with CCCP increased the surviving cells to the afore mentioned antibiotic treatments by 67–73% compared to the CCCP-untreated where ∼1.7% of the population survived the antibiotic exposure. On the other hand, treatment of P. aeruginosa cultures with CCCP (200 μg/mL) lead to a 7-log decrease in the number of bacteria tolerant to colistin, leaving a small subpopulation of persister cells behind (0.1%). Similarly, treatment with PAAG resulted in 4-log reduction of the bacteria within 1 h of treatment (Figure 3). Unlike colistin, PAAG completely eradicated the CCCP-pretreated P. aeruginosa cells in 24 h of treatment.
FIGURE 3.
Effect of CCCP to enhance persistence in stationary phase cultures of P. aeruginosa. CCCP at a concentration of 200 μg/mL was used to induce persistence was assessed by evaluating the log reduction of cells surviving the treatment with PAAG (100–200 μg/mL) tobramycin (16 μg/mL), clarithromycin (256 μg/mL), colistin (256 μg/mL), aztreonam (256 μg/mL), azithromycin (256 μg/mL) following 3 h exposure to CCCP. Data are reported as mean ± standard error of the mean of the mean of at least three independent experiments.
Effect of PAAG on CCCP-Induced Persisters
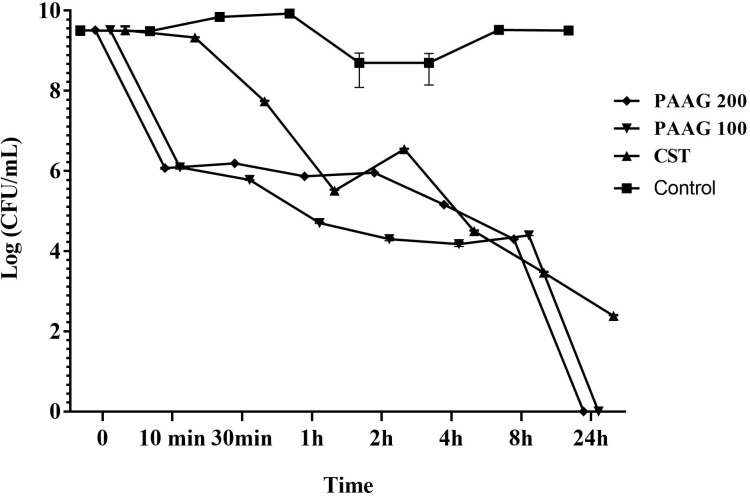
PAAG’s anti-persister activity was also evaluated based on its competence in eliminating persister-like cells formed in CCCP pretreated P. aeruginosa cultures. PAAG displayed bactericidal activity against CCCP-induced persister cells of P. aeruginosa and achieved complete killing of the initial bacterial inoculum at concentrations as low as 100 μg/mL (Figure 4). Colistin on the other hand was able to eliminate most of the CCCP-induced persisters of P. aeruginosa within 24 h of treatment, however, it was observed to leave a sub population of tolerant cells behind even at concentrations 4× its MIC.
FIGURE 4.
Effect of PAAG compared to a membrane active antibiotic against CCCP induced persisters of P. aeruginosa PA01. Activity of PAAG at concentrations of 200 and 100 μg/mL and colistin (256 μg/mL) against CCCP-induced P. aeruginosa PA01 cells pretreated with tobramycin (16 μg/mL). Anti-persister activity of PAAG was assessed by CFU counting at 10, 30 min and 1, 2, 4, 8, 24 h. Data represented as average values from three independent experiments ± standard error of the mean.
Permeabilization of Persisters
Propidium iodide (PI) uptake assay was used to measure the ability of PAAG and the other antimicrobials used in this study to permeabilize CCCP-induced persister cells. PI shifts emission upon entry into bacteria and subsequent intercalation into the bacterial DNA. Cells exposed to PI in the absence of antibiotics were used to form a baseline PI fluorescence (Figure 5). No significant change in fluorescent intensities was observed upon treating the persister cells with ciprofloxacin (1 μg/mL), tobramycin (16 μg/mL), azithromycin (256 μg/mL), aztreonam (256 μg/mL), and clarithromycin (256 μg/mL) (Figure 5). Even after 4 h of incubation, the antibiotics were unable to permeabilize the persister cells (Figure 5). Treatment with colistin (256 μg/mL), a membrane active antibiotic, resulted in permeabilization of the persister cells as indicated by corresponding increase in fluorescence intensity. A steady increase in fluorescence intensity was detected with increasing PAAG concentrations (100–200 μg/mL) within 10 min of treatment when compared to the control (Figure 5). Both concentrations of PAAG tested showed the ability to permeabilize the bacteria to a comparable level to colistin.
FIGURE 5.
Membrane permeabilization of CCCP- induced P. aeruginosa persister cells using propidium iodide (PI) through spectrophotometry. Permeabilization of persister cells formed in stationary phase cultures of P. aeruginosa cultures were treated with ciprofloxacin (100 μg/mL), tobramycin (16 μg/mL), colistin (256 μg/mL), azithromycin (256 μg/mL), aztreonam (256 μg/mL), and clarithromycin (256 μg/mL) or PAAG (100–200 μg/mL) and measured by spectrophotometry over 4 h. PI fluorescence was measured pre- and post-treatment with antibiotics or PAAG. CCCP-induced cells with PI was used as a baseline control. The experiment was repeated with there independent cultures and the data is represented as the average ±standard error of the mean.
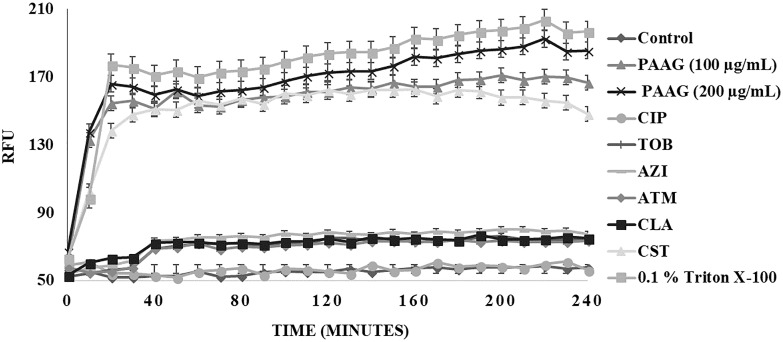
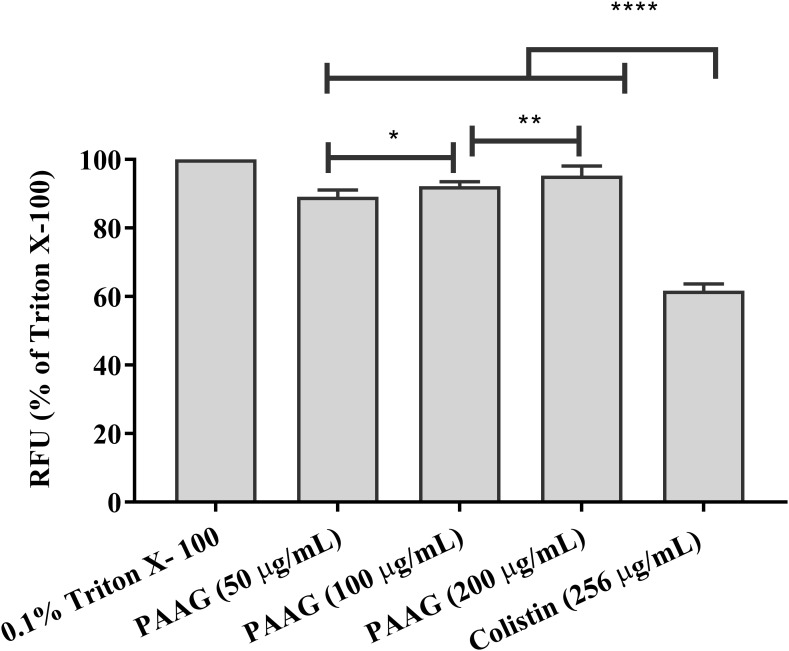
Membrane Depolarization Assay
Persister cells, although metabolically different from planktonic cells, are expected to remain susceptible to membrane active agents (Ouberai et al., 2011). At concentrations of 50–200 μg/mL PAAG was able to depolarize the membrane of P. aeruginosa persister cells within 10 min of treatment (Figure 6). After a 15 min stabilization period, addition of PAAG at concentrations of 50–200 μg/mL resulted in a dose dependent increase in fluorescence intensity corresponding to the change in membrane potential. PAAG at a concentration of 200 μg/mL lead to 91% increase in fluorescent intensity compared to the control, which was statistically more than colistin at 4× its MIC which resulted in a 60% increase in fluorescence intensity.
FIGURE 6.
PAAG upon permeabilization causes depolarization of the cytoplasmic membrane. Effect of PAAG or colistin on the cytoplasmic membrane of CCCP-induced P. aeruginosa persister cells incubated with DiSC3(5). Results expressed in relative fluorescence units (RFU) observed at 670 nm as compared to negative control. 0.1% Triton X-100 was used as positive control. Cells with DiSC3(5) was used as the baseline control. Data represented as mean ± SEM of three determinations. ∗∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05. The lines were used to compare the statistical significance between the different concentration of PAAG treatments.
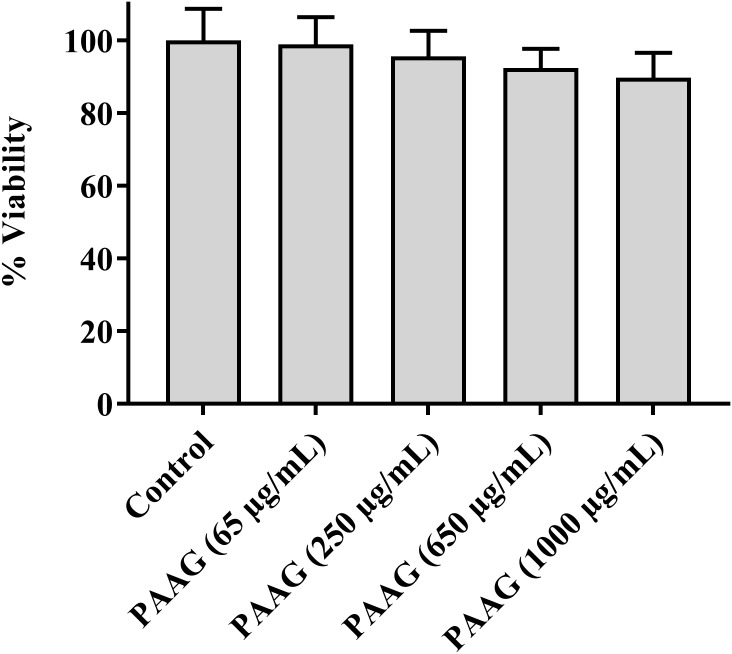
Cytotoxicity Assay
Cytotoxicity of the PAAG glycopolymer was assessed by investigating its effect on A549 cells using an LDH pierce cytotoxicity assay. LDH, a stable cytoplasmic enzyme present in all cells, is rapidly released into the cell culture supernatant when the plasma membrane is damaged. The results indicated that at the concentration range used in this study (65–250 μg/mL), PAAG demonstrated minimal cytotoxic effects (Figure 7).
FIGURE 7.
Cytotoxicity assay showing the percent cell viability after incubating A549 cells with PAAG for 24 h. A549 cells were plated onto 96-well plate at an initial seeding density of 2.0 × 104 cells/well. PAAG was dissolved in serum-free F12K media at concentrations ranging from 65 to 1,000 μg/mL. The plates were incubated for 24 h and compound mediated cytotoxicity was determined through pierce LDH cytotoxicity assay. No significant difference compared to the control (Untreated A549 cells). 10× lysis buffer (provided by the manufacturer) was used as the positive control. Data represented as mean ± standard error of the mean. The experiment was performed in triplicate (n = 5).
Discussion
The importance of persister cells in chronic relapsing infections has been emphasized over recent years and has resulted in an increased body of research focused on elucidating strategies aimed at the eradication of these tolerant bacteria (Crunkhorn, 2018; de Breij et al., 2018). Several novel approaches have been suggested for the elimination of these persistent cells, including reverting the phenotype of the bacteria back to a metabolically active state and therefore making it more susceptible to antibiotics by means of chemical compounds or sugars exposure (Pan et al., 2012; Marques et al., 2014; Zhang, 2014). However, identification of antimicrobial agents with a mechanism of action capable of targeting both actively dividing and persistent cells remains an important unmet need. Undeniably, eradication of entire bacterial populations is necessary to avoid the relapse of an infection and thereby minimize minimizing the chance of developing antibiotic resistance during protracted antibiotic treatments (Shan et al., 2017). The current study evaluates the impact of a novel glycopolymer, PAAG, against P. aeruginosa, a prototypic persister-forming, bacterial pathogen. Herein commonly used conventional classes of antibiotics such as β-lactams, aminoglycosides, macrolides, and polymyxin are compared to a novel large-molecule glycopolymer.
Antibiotic mechanism of action and varied target specificity are important for selecting the right agent for different clinical settings. However, even though antibiotics such as tobramycin (aminoglycoside), azithromycin and clarithromycin (macrolides), and aztreonam (β-lactam) target different bacterial pathways, they all share the requirement for the bacteria to be metabolically active. Bacterial persistence is a survival mechanism bacterium can employ to avoid killing by multiple classes of antibiotics without the need for genetically encoded resistance. The necessity for active growth was displayed in the observation of reduced antibiotic activity of the afore mentioned agents compared to PAAG, only 2–3 log reduction in bacterial counts, against stationary phase, non-replicating, cultures of P. aeruginosa (Figure 1). Bacteria treated with these antibiotics at concentrations four times their MIC resulted in a population of persistent cells (Figure 1). The reduced metabolic activity of stationary phase cells coupled with the stress of the antibiotics is the accepted mechanism for driving the bacteria into a persistent state (Cabral et al., 2018). However, colistin, a polymyxin, disrupts bacterial membranes to kill P. aeruginosa, whether or not actively growing. The increased activity of colistin against dormant cells was shown by the significant reduction of the stationary phase cultures of P. aeruginosa (CFU/mL) compared to other conventional antibiotics tested (Figure 1). However, colistin alone was still unable to completely eradicate the stationary phase cultures of P. aeruginosa. Conversely, both concentrations of PAAG tested were able to completely eradicate stationary phase cultures of P. aeruginosa within 24 h (9-logs), also shown in Figure 1. Due the slow growth of persisters and the need to become active again to grow, PAAG killing was monitored until 72 h post-treatment at which point there was still not growth observed.
Excessive antibiotic treatment can drive persister formation and lead to recurrence of infection once the local antibiotic concentration has dropped. To evaluate PAAG’s activity in potentiating the antibiotics against antibiotic-induced persistent bacteria, P. aeruginosa was first treated with either tobramycin, aztreonam or clarithromycin, all at 4× their tested MICs (Table 1) to eradicate the sensitive bulk of the population. No significant difference in killing was observed beyond concentrations 4× MIC (data not shown). The resultant subpopulation of cells tolerant to the antibiotic treatment (antibiotic-induced persistent bacteria) were then treated with PAAG or colistin at a concentration of 200 or 256 μg/mL, respectively (Figure 2) in addition to the other antibiotics. Treatment with PAAG, like colistin, resulted in eradication (6- to 7-log reduction) of the antibiotic-induced persistent populations of P. aeruginosa. This result suggests that PAAG is not only able kill the bacteria by itself but observed to potentiate the current antibiotics in treating P. aeruginosa similar to previous observations with S. aureus and Burkholderia (Narayanaswamy et al., 2017).
Persister studies are frequently complicated by low levels of naturally forming persisters in antibiotic-based methods and difficulties in separating the surviving persister subpopulation from the substantial proportion of dead bacteria. Consequently, CCCP has been used to enhance generation of persisters (Kwan et al., 2013; Grassi et al., 2017). The development of a persister-like status in CCCP-treated cells has been linked to the inhibition of ATP synthesis and the consequent drop in their metabolic activity (Kwan et al., 2013). The CCCP model system forms persisters through ATP depletion, recently demonstrated in S. aureus, P. aeruginosa, and E. coli (Davis, 1987; Morita et al., 2012; Grassi et al., 2017). CCCP-induced persisters would be expected to show increased tolerance against antibiotics that required metabolic activity to function. Indeed, pretreatment with CCCP was observed to protect P. aeruginosa cells from killing by tobramycin, azithromycin, clarithromycin, and aztreonam (Figure 3). Colistin treatment, on the other hand, lead to a 5-log reduction of the CCCP-pretreated persister cells due to its ability to kill growth-arrested bacteria (Figure 3). Colistin has been previously shown to demonstrate better bactericidal activity in the presence of CCCP than alone, because treatment with CCCP was found to alter the charge of the lipopolysaccharide (LPS) of the bacteria making them more susceptible to colistin (Pamp et al., 2008; Fernández et al., 2013). Treatment with PAAG, like colistin, resulted in significant reduction of the CCCP-induced persister cells. However, unlike colistin that only had a 5-log reduction, both concentrations of PAAG (100 and 200 μg/mL) eradicated 9-logs of persisters within 24 h (Figure 3).
Colistin is a polycationic, amphiphilic peptide that interacts with the negative charges of the lipid A portion of LPS and intercalates into the membrane, causing bacterial permeabilization and cell death (Pamp et al., 2008; Fernández et al., 2013). As a polycationic glycopolymer, PAAG has been shown to permeabilize S. aureus, a Gram-positive bacterium (Narayanaswamy et al., 2017), to synergize antibiotics against Burkholderia strains (Narayanaswamy et al., 2017) and is hypothesized to also target the negative charges of the LPS similar to colistin. The ability of PAAG to also kill metabolically inactive persister cells suggested that PAAG’s mechanism of action also disrupted P. aeruginosa bacterial membranes leading to permeabilization. To investigate PAAG permeabilization of the bacteria, a propidium-iodide (PI) assay was used. PI can only intercalate into the DNA if the bacteria are permeabilized or become permeable overtime as they die. As expected, the control and cytoplasmic active antibiotics (ciprofloxacin, tobramycin, azithromycin, aztreonam, and clarithromycin) did not lead to permeabilization and did not cause significant death (Figure 5). In contrast, the rapid increase in the fluorescence intensity shown in Figure 5 indicated that both colistin and PAAG permeabilized the CCCP-induced P. aeruginosa persister cells rapidly. To determine the extent to which PAAG leads to disruption of the bacterial membranes and to compare its activity with colistin, a membrane depolarization assay was used. Cytoplasmic membrane depolarization was assessed using a membrane potential dependent probe, 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)]. Upon permeabilization and disruption of the membrane, the membrane potential is dissipated, and DiSC3(5) is released into the medium causing an increase in fluorescence. PAAG treatment depolarized the membrane of P. aeruginosa persister cells in a dose dependent manner (Figure 6). All concentrations of PAAG tested showed significantly greater ability to depolarize the cytoplasmic membrane compared to colistin (P < 0.0001), suggestive of PAAG’s mechanism by which it completely eradicated the CCCP-induced persister cells (Figure 4).
Patients with relapsing chronic infections including those with CF, chronic obstructive pulmonary disease (COPD), bronchiectasis and immune compromise are often plagued by infections from both Gram-negative and Gram-positive pathogens. The variety of infecting organisms and antibiotic resistance status have resulted in a wide range of different therapeutic needs (Ventola, 2015). Currently, treatment of infections caused by persisters requires prolonged and repeated exposure to strain-specific antibiotics, with elevated risk of generating genetic antibiotic resistance and relapse (Zindani et al., 2006). Tobramycin and aztreonam, drugs that are currently being used to treat recalcitrant CF infections, must be given only periodically to minimize doses that can ultimately cause harmful side effects (Mearns, 1972; Zindani et al., 2006). A recent 10-year safety study on clarithromycin and azithromycin link their use to increased deaths in immune compromised individuals with heart conditions (Spoering and Lewis, 2001; Krahn et al., 2012). Colistin, a last resort antibiotic to treat chronic infcetions, has been prescribed with increasing frequency, out of necessity, and is often associated with side-effects of nephrotoxicity and neurotoxicity (Falagas et al., 2005; Honoré et al., 2014; Miano et al., 2018). A recent study published sheds further evidence into the cytotoxicity of polymyxins as they were found to induce apoptosis in human lung epithelial cells in a concentration and time dependent manner (Ahmed et al., 2017). To assess if PAAG also displays cytotoxicity the LDH assay was used. LDH release, from the lung epithelial cell line A549, was assayed using a range of concentrations of PAAG (65–1,000 μg/mL). Limited release of LDH was observed at 1,000 μg/mL indicative of minimal cytotoxic effects (Figure 7). Although this result is limited to one cell line the proposed method of drug delivery, inhalation, made the choice of A549 cells particularly relevant. However, studies are still ongoing, currently no significant decrease in activity has been observed against clinical isolates of Pseudomonas in artificial sputum media (data not shown).
PAAG has shown activity against both Gram-positive and Gram-negative pathogens (Narayanaswamy et al., 2017; Wood, 2017) despite their structural differences. Though this preliminary study is limited to persisters formed in a single bacterial species, P. aeruginosa, this opportunistic pathogen has served as the main model for persister studies as it produces high levels of drug-tolerant persister cells. PAAG as a membrane-acting glycopolymer eradicates P. aeruginosa persister cells in vitro at doses that show minimal cytotoxicity to human epithelial lung cells even at the highest dose tested (1 mg/mL; Figure 7). Studies are ongoing to better understand the broad spectrum of PAAG’s activity against other important pathogens in chronic lung infections in vitro and how PAAG’s activity translates to an in vivo setting. The findings of this study show strong potential for this new class of glycopolymer drugs in the fight against chronic lung pathogens and antibiotic resistance.
Author Contributions
VN study design, formal analysis, validation, visualization, writing – original draft, writing – revisions, review, and editing. VN, LK, and KD investigation and methodology. ST formal analysis, initial study design, and writing – first draft. WW, SB, and ST conceptualization, funding acquisition, resources, writing – review and editing. AL study design, writing – final revisions, editing, and review.
Conflict of Interest Statement
VN, LK, KD, ST, AL, SB, and WW are paid employees of Synedgen. ST, SB, and WW have ownership and patents affiliated with Synedgen, and SB and WW are board members. The glycopolymer used in this study is protected by US Patent number 8,119,780 B2 and others pending in development of drug products to treat lung infections and related indications. The potential conflicts noted have not impacted or influenced the findings of this manuscript.
Acknowledgments
We thank Dr. Paul Orwin (CSUSB) for his kind donation of the strain PA01 and for reviewing and providing critical comments on the manuscript.
Footnotes
Funding. The funders, through Synedgen, Inc., provided support in the form of salaries for authors VN, LK, KD, ST, AL, SB, and WW, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in Section “Author Contributions.”
References
- Ahmed M. U., Velkov T., Lin Y. W., Yun B., Nowell C. J., Zhou F., et al. (2017). Potential toxicity of polymyxins in human lung epithelial cells. Antimicrob. Agents Chemother. 61:e2690-16. 10.1128/AAC.02690-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison K. R., Brynildsen M. P., Collins J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473 216–220. 10.1038/nature10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N., Buson A., Jarolimek W., Rice S. A. (2013). Mannitol enhances antibiotic sensitivity of persister bacteria in Pseudomonas aeruginosa biofilms. PLoS One 8:e84220. 10.1371/journal.pone.0084220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello G., Richards L., Ehrlich G. D., Stewart P. S. (2006). Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50 382–384. 10.1128/AAC.50.1.382-384.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. L., Van Dalfsen J. M., Shawar R. M., Otto K. L., Garber R. L., Quan J. M., et al. (1999). Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J. Infect. Dis. 179 1190–1196. 10.1086/314727 [DOI] [PubMed] [Google Scholar]
- Cabral D. J., Wurster J. I., Belenky P. (2018). Antibiotic persistence as a metabolic adaptation: stress. metabolism, the host, and new directions. Pharmaceuticals 11:14. 10.3390/ph11010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury N., Wood T. L., Martínez-Vázquez M., García-Contreras R., Wood T. K. (2016). DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 113 1984–1992. 10.1002/bit.25963 [DOI] [PubMed] [Google Scholar]
- CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Crunkhorn S. (2018). Antibacterials: synthetic peptides eradicate resistant infections. Nat. Rev. Drug Discov. 17:166. 10.1038/nrd.2018.31 [DOI] [PubMed] [Google Scholar]
- Cui P., Niu H., Shi W., Zhang S., Zhang H., Margolick J., et al. (2016). Disruption of membrane by colistin kills uropathogenic Escherichia coli persisters and enhances killing of other antibiotics. Antimicrob. Agents Chemother. 60 6867–6871. 10.1128/AAC.01481-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. D. (1987). Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breij A., Riool M., Cordfunke R. A., Malanovic N., de Boer L., Koning R. I., et al. (2018). The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Trans. Med. 10:eaan4044. 10.1126/scitranslmed.aan4044 [DOI] [PubMed] [Google Scholar]
- Dunn M., Wunderink R. G. (1995). Ventilator-associated pneumonia caused by Pseudomonas infection. Clin. Chest Med. 16 95–109. [PubMed] [Google Scholar]
- Falagas M. E., Rizos M., Bliziotis I. A., Rellos K., Kasiakou S. K., Michalopoulos A. (2005). Toxicity after prolonged (more than four weeks) administration of intravenous colistin. BMC Infect. Dis. 5:1. 10.1186/1471-2334-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L., Álvarez-Ortega C., Wiegand I., Olivares J., Kocíncová D., Lam J. S., et al. (2013). Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57 110–119. 10.1128/AAC.01583-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller D. E., Pitlick W. H., Nardella P. A., Tracewell W. G., Ramsey B. W. (2002). Pharmacokinetics and bioavailability of aerosolized tobramycin in cystic fibrosis. Chest 122 219–226. 10.1378/chest.122.1.219 [DOI] [PubMed] [Google Scholar]
- Gibson R. L., Burns J. L., Ramsey B. W. (2003). Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168 918–951. 10.1164/rccm.200304-505SO [DOI] [PubMed] [Google Scholar]
- Gilligan P. H. (2006). Is there value in susceptibility testing of Pseudomonas aeruginosa causing chronic infection in patients with cystic fibrosis? Expert Rev. Anti Infect. Ther. 4 711–715. 10.1586/14787210.4.5.711 [DOI] [PubMed] [Google Scholar]
- Govan J. R., Deretic V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi L., Di Luca M., Maisetta G., Rinaldi A. C., Esin S., Trampuz A., et al. (2017). Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front. Microbiol. 8:1917. 10.3389/fmicb.2017.01917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré P. M., Jacobs R., Joannes-Boyau O., Lochy S., Boer W., De Waele E., et al. (2014). Continuous renal replacement therapy-related strategies to avoid colistin toxicity: a clinically orientated review. Blood Purif. 37 291–295. 10.1159/000363495 [DOI] [PubMed] [Google Scholar]
- Hoyle B. D., Jass J., Costerton J. W. (1990). The biofilm glycocalyx as a resistance factor. J. Antimicrob. Chemother. 26 1–5. 10.1093/jac/26.1.1 [DOI] [PubMed] [Google Scholar]
- Hurdle J. G., O’neill A. J., Chopra I., Lee R. E. (2011). Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9 62–75. 10.1038/nrmicro2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. J., Park S. J., Mishig-Ochir T., Lee B. J. (2014). Antimicrobial peptides: therapeutic potentials. Expert Rev. Anti Infect. Ther. 12 1477–1486. 10.1586/14787210.2014.976613 [DOI] [PubMed] [Google Scholar]
- Keren I., Kaldalu N., Spoering A., Wang Y., Lewis K. (2004). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230 13–18. 10.1016/S0378-1097(03)00856-5 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Wood T. K. (2016). Persistent persister misperceptions. Front. Microbiol. 7:2134. 10.3389/fmicb.2016.02134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Yamasaki R., Song S., Zhang W., Wood T. K. (2018). Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol. 10.1111/1462-2920.14093 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- King P., Lomovskaya O., Griffith D. C., Burns J. L., Dudley M. N. (2010). In vitro pharmacodynamics of levofloxacin and other aerosolized antibiotics under multiple conditions relevant to chronic pulmonary infection in cystic fibrosis. Antimicrob. Agents Chemother. 54 143–148. 10.1128/AAC.00248-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeva M., Gutu A. D., Hebert W., Wager J. D., Yonker L. M., O’Toole G. A., et al. (2017). An antipersister strategy for treatment of chronic Pseudomonas aeruginosa infections. Antimicrob. Agents Chemother. 61:e00987-17. 10.1128/AAC.00987-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn T., Gilmour C., Tilak J., Fraud S., Kerr N., Lau C. H. F., et al. (2012). Determinants of intrinsic aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56 5591–5602. 10.1128/AAC.01446-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan B. W., Osbourne D. O., Hu Y., Benedik M. J., Wood T. K. (2015). Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol. Bioeng. 112 588–600. 10.1002/bit.25456 [DOI] [PubMed] [Google Scholar]
- Kwan B. W., Valenta J. A., Benedik M. J., Wood T. K. (2013). Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 57 1468–1473. 10.1128/AAC.02135-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur M. D., Qi Q., Lewis K. (2010). Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 54 39–44. 10.1128/AAC.00860-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D., Chauhan A., Létoffé S., Fischer F., De Reuse H., Beloin C., et al. (2014). pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J. Infect. Dis. 210 1357–1366. 10.1093/infdis/jiu286 [DOI] [PubMed] [Google Scholar]
- Lewis K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5 48–56. 10.1038/nrmicro1557 [DOI] [PubMed] [Google Scholar]
- Lewis K. (2008). Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322 107–131. 10.1007/978-3-540-75418-3_6 [DOI] [PubMed] [Google Scholar]
- Lewis K. (2010). Persister cells and the paradox of chronic infections. Microbe 5 429–437. 10.1038/nature12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques C. N., Morozov A., Planzos P., Zelaya H. M. (2014). The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl. Environ. Microbiol. 80 6976–6991. 10.1128/AEM.01576-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mearns M. B. (1972). Treatment and prevention of pulmonary complications of cystic fibrosis in infancy and early childhood. Arch. Dis. Child. 47 5–11. 10.1136/adc.47.251.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson M. H., Gurtman A., Szabo S., Neibart E., Meyers B. R., Policar M., et al. (1994). Pseudomonas aeruginosa bacteremia in patients with AIDS. Clin. Infect. Dis. 18 886–895. 10.1093/clinids/18.6.886 [DOI] [PubMed] [Google Scholar]
- Miano T. A., Lautenbach E., Wilson F. P., Guo W., Borovskiy Y., Hennessy S. (2018). Attributable risk and time course of colistin-associated acute kidney injury. Clin. J. Am. Soc. Nephrol. 13 542–550. 10.2215/CJN.06980717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarcik P., Kolar M. (2017). Starvation-and antibiotics-induced formation of persister cells in Pseudomonas aeruginosa. Biomed. Papers Med. Faculty Palacky Univ. Olomouc 161 58–67. 10.5507/bp.2016.057 [DOI] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. (2012). Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158 1071–1083. 10.1099/mic.0.054320-0 [DOI] [PubMed] [Google Scholar]
- Mulcahy L. R., Burns J. L., Lory S., Lewis K. (2010). Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192 6191–6199. 10.1128/JB.01651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy V. P., Giatpaiboon S., Baker S. M., Wiesmann W. P., LiPuma J. J., Townsend S. M. (2017). Novel glycopolymer sensitizes Burkholderia cepacia complex isolates from cystic fibrosis patients to tobramycin and meropenem. PLoS One 12:e0179776. 10.1371/journal.pone.0179776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy V. P., Giatpaiboon S. A., Uhrig J., Orwin P., Wiesmann W., Baker S. M., et al. (2018). In vitro activity of novel glycopolymer against clinical isolates of multidrug-resistant Staphylococcus aureus. PLoS One 13:e0191522. 10.1371/journal.pone.0191522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman M. A., Brynildsen M. P. (2013). Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 57 4398–4409. 10.1128/AAC.00372-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouberai M., El Garch F., Bussiere A., Riou M., Alsteens D., Lins L., et al. (2011). The Pseudomonas aeruginosa membranes: a target for a new amphiphilic aminoglycoside derivative? Biochim. Biophys. Acta Biomemb. 1808 1716–1727. 10.1016/j.bbamem.2011.01.014 [DOI] [PubMed] [Google Scholar]
- Pamp S. J., Gjermansen M., Johansen H. K., Tolker-Nielsen T. (2008). Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68 223–240. 10.1111/j.1365-2958.2008.06152.x [DOI] [PubMed] [Google Scholar]
- Pan J., Bahar A. A., Syed H., Ren D. (2012). Reverting antibiotic tolerance of Pseudomonas aeruginosa PAO1 persister cells by (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2 (5H)-one. PLoS One 7:e45778. 10.1371/journal.pone.0045778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K., Neshat S., Heinrichs D. (1991). Pyoverdine-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. FEMS Microbiol. Lett. 62 1–5. [PubMed] [Google Scholar]
- Ramsey B. W., Pepe M. S., Quan J. M., Otto K. L., Montgomery A. B., Williams-Warren J., et al. (1999). Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. New Engl. J. Med. 340 23–30. 10.1056/NEJM199901073400104 [DOI] [PubMed] [Google Scholar]
- Reffuveille F., de la Fuente-Núñez C., Mansour S., Hancock R. E. (2014). A broad-spectrum anti-biofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58 5363–5371. 10.1128/AAC.03163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar K., Lindgren M., Hansen M., Eiríksdóttir E., Jiang Y., Rosenthal-Aizman K., et al. (2005). Cell-penetrating peptides: a comparative membrane toxicity study. Anal. Biochem. 345 55–65. 10.1016/j.ab.2005.07.033 [DOI] [PubMed] [Google Scholar]
- Shan Y., Gandt A. B., Rowe S. E., Deisinger J. P., Conlon B. P., Lewis K. (2017). ATP-dependent persister formation in Escherichia coli. mBio 8:e2267-16. 10.1128/mBio.02267-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering A. L., Lewis K. I. M. (2001). Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183 6746–6751. 10.1128/JB.183.23.6746-6751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Wang Z. Y., Yamakoshi M., Kobayashi M., Nozawa T. (2003). Probing the transmembrane potential of bacterial cells by voltage-sensitive dyes. Anal. Sci. 19 1239–1242. 10.2116/analsci.19.1239 [DOI] [PubMed] [Google Scholar]
- Tang H., Zhang P., Kieft T. L., Ryan S. J., Baker S. M., Wiesmann W. P., et al. (2010). Antibacterial action of a novel functionalized chitosan-arginine against Gram-negative bacteria. Acta Biomater. 6 2562–2571. 10.1016/j.actbio.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. Pharm. Therap. 40 277–283. [PMC free article] [PubMed] [Google Scholar]
- Wood T. K. (2017). Strategies for combating persister cell and biofilm infections. Microb. Biotechnol. 10 1054–1056. 10.1111/1751-7915.12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Haagensen J. A., Jelsbak L., Johansen H. K., Sternberg C., Høiby N., et al. (2008). In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190 2767–2776. 10.1128/JB.01581-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2014). Persisters, persistent infections and the Yin–Yang model. Emerg. Microbes Infect. 3:e3. 10.1038/emi.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao C., Liang G., Zhang M., Zheng J. (2013). Engineering antimicrobial peptides with improved antimicrobial and hemolytic activities. J. Chem. Inform. Model. 53 3280–3296. 10.1021/ci400477e [DOI] [PubMed] [Google Scholar]
- Zindani G. N., Streetman D. D., Streetman D. S., Nasr S. Z. (2006). Adherence to treatment in children and adolescent patients with cystic fibrosis. J. Adolesc. Health 38 13–17. 10.1016/j.jadohealth.2004.09.013 [DOI] [PubMed] [Google Scholar]