Abstract
The genus Carboxydocella forms a deeply branching family in the class Clostridia and is currently represented by three physiologically diverse species of thermophilic prokaryotes. The type strain of the type species, Carboxydocella thermautotrophica 41T, is an obligate chemolithoautotroph growing exclusively by hydrogenogenic CO oxidation. Another strain, isolated from a hot spring at Uzon caldera, Kamchatka in the course of this work, is capable of coupling carboxydotrophy and dissimilatory reduction of Fe(III) from oxic and phyllosilicate minerals. The processes of carboxydotrophy and Fe(III) reduction appeared to be interdependent in this strain. The genomes of both isolates were sequenced, assembled into single chromosome sequences (for strain 41T a plasmid sequence was also assembled) and analyzed. Genome analysis revealed that each of the two strains possessed six genes encoding diverse Ni,Fe-containing CO dehydrogenases (maximum reported in complete prokaryotic genomes), indicating crucial role of carbon monoxide in C. thermautotrophica metabolism. Both strains possessed a set of 30 multiheme c-type cytochromes, but only the newly isolated Fe-reducing strain 019 had one extra gene of a 17-heme cytochrome, which is proposed to represent a novel determinant of dissimilatory iron reduction in prokaryotes. Mössbauer studies revealed that strain 019 induced reductive transformation of the abundant ferric/ferrous-mica mineral glauconite to siderite during carboxydotrophic growth. Reconstruction of the C. thermautotrophica strains energy metabolism is the first comprehensive genome analysis of a representative of the deep phylogenetic branch Clostridia Incertae Sedis, family V. Our data provide insights into energy metabolism of C. thermautotrophica with an emphasis on its ecological implications.
Keywords: Carboxydocella, thermophile, Kamchatka hot springs, genomics, hydrogenogenic carboxydotrophy, Fe(III) reduction, Fe(III) silicate minerals, Firmicutes
Introduction
Carbon monoxide (CO) is a common although minor component of gasses in terrestrial hot springs. In the hot springs of Yellowstone National Park (United States) and Uzon Caldera (Kamchatka, Russia), its concentration was reported to be about 1–6 10-7 mole fraction in the gas bubbling through the water (Shock et al., 2005, 2010) or 20–30 nM of dissolved gas (Kochetkova et al., 2011). The main sources of CO in hydrothermal vents are volcanic gasses (Menyailov and Nikitina, 1980; Symonds et al., 1994; Allard et al., 2004) and thermochemical or photochemical decomposition of organic matter (Hellebrand and Schade, 2008). Carbon monoxide is a strong reductant [E0′(CO/CO2) is ca. -520 mV at pH 7.0], and can be involved in a number of microbial metabolic redox reactions (Shock et al., 2005, 2010; Diender et al., 2015) coupled with the reduction of various inorganic electron acceptors.
Key enzymes of CO oxidation pathways are carbon monoxide dehydrogenases (CODHs), which catalyze the oxidation of CO with water to CO2. CODHs from aerobes, encoded by cox genes, contain Cu and Mo as cofactors, while anaerobic CODHs, encoded by the distantly homologous cooS or cdh genes, contain Ni and Fe in active centers, where Ni activates CO and Fe provides the nucleophilic water. As distinct from the water-gas shift reaction, where the electrons and protons generated by CO oxidation are directly released as H2, CODHs keep the protons and electrons separated, and the electrons are transferred to the terminal acceptors via electron-carrier proteins. In case of hydrogenogenic carboxydotrophs and their [Ni,Fe]-CODHs, protons may serve as these terminal acceptors, and H2 is eventually released. [Ni,Fe]-CODHs are frequently associated with acetyl-CoA synthases (ACSs), where the [Ni,Fe]-CODH component reduces CO2 to CO, condensed with a methyl group and CoA by ACS to produce acetyl-CoA, or oxidizes CO formed upon acetyl-CoA cleavage (Jeoung et al., 2014).
A number of prokaryotes are capable of using CO as an electron donor, and some of them can also use CO as a carbon source (King and Weber, 2007; Oelgeschläger and Rother, 2008; Sokolova and Lebedinsky, 2013; Tiquia-Arashiro, 2014; Diender et al., 2015). In various terrestrial hydrotherms, potential activity or the presence of CO-oxidizing anaerobes has been revealed (Kochetkova et al., 2011; Brady et al., 2015; Yoneda et al., 2015), and the number of newly isolated CO-oxidizers is increasing permanently (Balk et al., 2009; Yoneda et al., 2012, 2013; Sokolova and Lebedinsky, 2013; Tiquia-Arashiro, 2014). Among cultivated thermophilic anaerobic CO-oxidizing species, hydrogenogenic carboxydotrophs are in majority, moreover, in certain hot springs, they comprise a significant portion of the microbial population (Yoneda et al., 2015). These bacteria oxidize carbon monoxide via the following reaction: CO + H2O → CO2 + H2 (ΔG0′ = -20 kJ). A representative of thermophilic hydrogenogenic CO-trophic microorganisms, Carboxydocella thermautotrophica, is the type species of the genus Carboxydocella, which forms a deep phylogenetic branch of the order Clostridiales, Incertae Sedis, family V (Sokolova et al., 2009; Sokolova, 2015). Carboxydocella species were found to be widely distributed in neutral hot springs with moderately thermophilic conditions (Slepova et al., 2006, 2007; Kochetkova et al., 2011; Slobodkina et al., 2012; Brady et al., 2015; Sokolova, 2015). So far, three Carboxydocella species have been validly described: C. thermautotrophica (Sokolova et al., 2002), C. sporoproducens (Slepova et al., 2006), and C. manganica (Slobodkina et al., 2012). While C. thermautotrophica and C. sporoproducens are carboxydotrophs, C. manganica is unable to oxidize CO.
Some thermophilic anaerobes isolated from various sedimentary environments of volcanic origin are capable of both hydrogenogenic CO-oxidation and dissimilatory ferric iron reduction from Fe(III) oxides (Sokolova et al., 2004; Slobodkin et al., 2006; Zavarzina et al., 2007; Slepova et al., 2009; Yoneda et al., 2012, 2013). At the moment, the interconnections between the oxidative and reductive branches of energy metabolism in these organisms remain unclear. Fe-bearing silicates (clay and mica minerals) comprising the most abundant Fe(III) source in volcanic areas (Eroschev-Shak et al., 1998, 2005) have not been tested as electron acceptors for growth of carboxydotrophs. Microbial redox transformation of these minerals is poorly understood so far, especially at elevated temperatures. Dissimilatory reduction of structural Fe from clays has been documented for a few thermophilic species (Pentráková et al., 2013). Less is known about bioreduction of Fe-rich micas, which are widely distributed in igneous and sedimentary rocks. The ability to reduce structural Fe in the mica mineral biotite was only demonstrated for resting cell suspensions of two mesophilic Fe(III)-reducing bacteria Geobacter sulfurreducens and Shewanella oneidensis (Brookshaw et al., 2014a,b). Bacteria of these species are the main models used for the investigation of microbial redox interactions with Fe(III) minerals, and the current concept of extracellular electron transfer in prokaryotes is mostly based on the data obtained for these microorganisms. Recent reviews of this concept (Shi et al., 2016; White et al., 2016) highlight the key role of multiheme c-type cytochromes in this process and the existence of two major pathways for direct electron transfer from respiratory chain to electron acceptor outside the cell. The first pathway is mediated by porin-cytochrome complexes linking intracellular electron shuttles and Fe(III)-reducing cytochromes on the cell surface. The second pathway is based on electrically conductive cell appendages (pili or “nanowires”). Genomic studies revealed that the determinants of both pathways are widespread among prokaryotes, although in vivo activity of porin-cytochrome complexes or nanowires has been documented for a restricted number of Fe(III)-reducers (Shi et al., 2012, 2014, 2016).
Here we report physiological and genomic characterization of two strains of Carboxydocella thermautotrophica. The type strain C. thermautotrophica 41T is an obligate chemolithoautotroph growing exclusively by hydrogenogenic CO oxidation (Sokolova et al., 2002), while a new isolate of the same species, obtained during the current work (strain 019), differs significantly in its physiology, namely, by its capacity for Fe(III) reduction from oxic and mica minerals, which depends on CO availability. This physiological versatility of C. thermautotrophica strains has lead us to the proposal of an important ecological role for this taxon, such as coupling the transformation of carbon oxides and Fe minerals in terrestrial sedimentary environments of Kamchatka hot springs.
Materials and Methods
Strains
The type strain C. thermautotrophica 41T (Sokolova et al., 2002) was obtained from DSMZ collection (DSMZ 1236). The strain 019 has been isolated from a core sample of the ground taken at East Thermal Field at Uzon Caldera (Kamchatka Peninsula, Russia).
Sampling
A core was taken at East Thermal Field at Uzon Caldera (Kamchatka Peninsula) at an edge of Zavarzin Pool. Zavarzin Pool, 2.5 m in diameter, is formed by several hot springs discharging inside it. In the pool, thin brown layers of microbial mats were developed covered with a thick layer of sulfur. Temperature and pH of the water varied in different spots at the pool from 46 to 57°C and from 6.10 to 6.75, respectively. The core was taken at a 0.5-m distance from the pool and to 40-cm depth. The upper 10 cm of the core were black, next 15 cm in the middle had vertical black and white stripes, and the bottom 15 cm were almost white. The pH of the water filling the core was 5.8, its temperature was 55°C, and Eh was -225 mV (hereafter all the Eh values are presented vs. the potential of standard hydrogen electrode [SHE] at pH 6.5). Subsamples of top, middle and bottom layers of the core were taken anaerobically in tightly sealed bottles and designated as DC03-018, DC03-019, and DC03-020, respectively.
Enrichment and Isolation
For enrichment and isolation of hydrogenogenic carboxydotrophic prokaryotes and their subsequent cultivation Media 1 and 2 were used. Medium 1 contained (g l-1): NH4Cl, 1; MgCl2.6H2O, 0.33; CaCl2.6H2O, 0.1; KCl, 0.33; KH2PO4, 0.5; resazurin, 0.001; 1 ml l-1 of trace mineral solution (Kevbrin and Zavarzin, 1992); 1 ml l-1 of vitamin solution (Wolin et al., 1963), NaHCO3 (0.5 g l-1). Medium 1 was reduced with Na2S. 9H2O (1.0 g l-1), and pH was adjusted to 6.8 with 6N HCl. Medium 2 had the composition similar to Medium 1 except the presence of hydromorphic ferric oxide (ferrihydrite), while sodium sulfide was omitted. Medium 1 had a redox potential characteristic of strictly anaerobic conditions (Eh value of -430 mV), and Medium 2 had a redox potential of -90 mV. Hereafter, strictly anaerobic conditions of Medium 1 are referred to as “low Eh” and anaerobic conditions of Medium 2 are referred to as “high Eh”. Microaerobic or aerobic growth conditions are specified where necessary. Yeast extract (0.5 g l-1) or sodium acetate (0.2 g l-1) or lactate (0.3 g l-1) were added as additional carbon sources when indicated. Glass vessels (60 ml total volume) sealed with butyl rubber stoppers (Bellco Glass Inc.) and containing 10 ml of liquid Medium 2 and 50 ml gas phase (100% CO) were inoculated with suspended core samples and incubated at 60°C. In chemical controls with non-inoculated media 1 or 2, equilibrium concentration of gaseous CO2, released due to thermal decomposition of bicarbonate at 60°C, was under the detection level at the beginning of incubation and did not exceed 0.03% (v/v) of the gas phase within further 6 days of incubation. Growth was determined using light microscopy and by monitoring CO utilization and gaseous products formation as described previously (Sokolova et al., 2002). After a number of serial end point dilution transfers in Medium 2 pure cultures were isolated from colonies obtained in roll-tubes prepared in 15-ml Hungate tubes with Medium 1 solidified by 5% agar, under 100% CO in the gas phase. Well-separated colonies were transferred to the liquid Medium 1.
Physiological Studies
Utilization of molecular hydrogen, CO2, acetate, lactate (2 g/l) or glycerol (0.2%, v/v) by the new isolate was tested in liquid Medium 1 with 100% N2 or 80:20% H2/CO2 as the gas phase in presence or absence of potential electron acceptors. The acceptors tested were sodium nitrate, nitrite, sulfate, thiosulfate, AQDS, as well as mineral acceptors ferrihydrite (prepared as described by Gavrilov et al., 2012), glauconite (from Severo-Stavropol’skoye underground gas storage reservoir, Russia), nontronite (from the collection of clay minerals of the Faculty of Soil Science, Lomonosov Moscow State University) and diatomite (from Inzenskoye deposit, Russia). Soluble acceptors were added from pre-sterilized stock solutions to a concentration of 10 mM (5 mM in case of nitrite); Fe(III) minerals were added to Medium 2 before sterilization to achieve initial Fe(III) content of 90 mM in case of ferrihydrite and 20 mM in case of glauconite, nontronite, or diatomite.
CO, CO2, and H2 were analyzed using a ‘3700’ custom-modified gas chromatograph (ZIOC RAS Special Design Tech. Dept., Russia) supplied with Phoenix v.3.6.0 analytical software (BSoft, Russia) and equipped with zeolite NaX 80–100 mesh and Chromosorb-102 60-80 mesh 3 m columns and TCD detector, which were all conditioned at 30°C. Extra pure grade Ar was used as the carrier gas. Volatile fatty acids were determined by high performance liquid chromatography (HPLC) on a Stayer chromatograph (Aquilon, Russia) equipped with an Aminex HPX_87H column (Bio-Rad) and a Smartline 2300 refractometric detector (Knauer, Germany), with 5 mM H2SO4 as the eluent. Growth on iron-containing minerals was traced by the increase of the content of HCl-extractable Fe(II) in the mineral phase of Medium 2. The cell density was determined by direct cell counting under a CX41 phase-contrast microscope (Olympus). Temperature and pH growth optima were deduced from growth rates determined on Medium 1.
Mössbauer Studies
Mössbauer spectra of 57Fe nuclei were recorded at room temperature on a custom-modified MS-1104Em spectrometer (Research Institute of Physics, Southern Federal University, Russia), which was equipped with a 57Co-source in a Rh matrix and operated in a constant acceleration mode. The spectrometer was calibrated using standard α-Fe absorbent. The SpectrRelax software (Matsnev and Rusakov, 2014) was used to analyze the Mössbauer spectra. Iron species were determined colorimetrically in HCl extracts of culture subsamples with ferrozine (Fe2+) or potassium thiocyanate (Fe3+), as previously described (Gavrilov et al., 2012). Insoluble parts of silicate minerals were separated from HCl extracts by centrifugation at 12 kG for 10 min on an Eppendorf table top centrifuge. Non-extractable iron content of glauconite was estimated from Mössbauer spectral data under the assumption that the recoil-free fraction (probability of Mössbauer effect) is equal for iron atoms located in different positions in various mineral phases, and accordingly, relative intensities of subspectra (area of subspectra) are equal to the relative content of iron atoms in these positions.
DNA Isolation
For DNA isolation cells were cultivated on Medium 1 in the absence of minerals under optimal conditions, collected by centrifugation at 9 kG for 15 min and disrupted with glass beads using Minilys homogenizer (Bertin Technologies). DNA was extracted using QIAamp DNA mini kit (Qiagen).
Phylogenetic Reconstructions
Phylogenetic trees were constructed using the Maximum Likelihood method based on the Jones–Taylor–Thornton model (Jones et al., 1992) in MEGA6 (Tamura et al., 2013).
For the putative iron reducing cytochrome, separate reconstructions were performed for C-terminus (first 500 aa residues) and the remaining part of the sequence (501–1486 aa residues), containing all the conservative multiheme cytochrome domains (i.e., putative “catalytic” domain of the protein). Blastp analysis was performed against UniProtKB prokaryotic database on April, 2018. For the catalytic domain of the protein, only the hits with Ev < 0.001, query coverage ≥50% and identity >20% (which is relevant for multiheme cytochromes, Sharma et al., 2010) from cultivated strains were considered. The analysis retrieved 57 sequences, which were then filtered through 0.9 filter using CD-hit utility1 and the resulting set of 50 sequences was aligned with built-in Muscle at default parameters in MEGA6.
Genome Sequencing and Assembly
Sequencing projects were started in May 2013 and finished in August 2014. For sequencing of the genomes of C. thermautotrophica strains both paired-end and mate-paired DNA libraries were used. Paired end libraries were prepared from 1 μg of genomic DNA with NEBNextTM Ultra DNA library preparation kit (New England Biolabs, United States) according to manufacturer’s instructions to obtain mean library size of 500 bp. Mate-paired libraries were prepared with NexteraTM Mate Pair Library Prep Kit (Illumina Inc., United States) using bead-based size selection protocol. Both paired-end mate-paired libraries were sequenced using 2 × 250 bp reads with MiSeqTM Personal Sequencing System (Illumina Inc., United States). After sequencing all reads were subjected to stringent quality filtering with CLC Genomics Workbench 8.5 (Qiagen, Germany). After filtering, overlapping paired-end library reads were merged with SeqPrep tool2, resulting in 215,048 and 227,940 single merged reads; and 895,488 and 892,408 read pairs for strains 41T and 019, respectively. Fragment size of both paired-end libraries was 500–700 bp. Mate-paired reads were treated with NextClip tool (Leggett et al., 2014), resulting in 2,382,016 and 2,123,106 read pairs with mean insert sizes of 2,439 and 2,382 bp for strains 41T and 019, respectively. Reads were assembled with both ALLPATHS-LG (Butler et al., 2008) and SPADES 3.8.0 (Nurk et al., 2013) assemblers and refined manually using CLC Genomics Workbench 8.5 software (Qiagen, Germany). Orientation of contigs and final filling of sequence gaps was made by PCR with outward-oriented primers to contigs termini and subsequent Sanger sequencing of resulting amplicons. Final average genome coverage was 247× and 218× for strains 41T and 019, respectively. During assembly of strain 41T genome an additional circular contig, corresponding to plasmid, was also assembled.
The complete genome sequences of the C. thermautotrophica type strain 41T and strain 019 have been deposited in DDBJ/EMBL/GenBank under accession numbers CP028491 (strain 019), CP028514 (strain 41T) and CP028515 (plasmid of strain 41T). Related projects information and sample details have been deposited in NCBI database under accession numbers PRJEB11520, PRJEB11521, and SAMN07757920, SAMN07757921, respectively.
Genome Annotation
Gene prediction and primary annotation was performed with IMG/M ER System (Chen et al., 2017). Refining of the automated annotations and other predictions were done manually according to genome annotation protocol (Toshchakov et al., 2015).
Subcellular localization of multiheme cytochromes was predicted based on the results of six different on-line prediction services – SignalP 4.1, TatP 1.0, SecretomeP 2.0a and TMHMM 2.0 (all at CBS Prediction Servers3), as well as PSORTb 3.0.24 and Phobius5.
Conserved domains were predicted with HMMSCAN6 considering Pfam, TIGRFAM, Gene3D, Superfamily, PIRSF and TreeFam protein families databases.
Results
Enrichment and Isolation of Strain 019, Cell Morphology of the New Isolate
The C. thermautotrophica Fe(III)-reducing strain 019 was obtained from a core sample taken near Zavarzin thermal pool at Uzon Caldera (Kamchatka). The bottles with anaerobic Medium 2 (high Eh) containing sodium acetate and ferrihydrite under a 100% CO atmosphere and inoculated with the middle (black-to-white) part of the core sample (subsample DC03-019) showed an increase in pressure from 140 to 170 kPa after 2 days of incubation at 60°C. Growth of small rod-shaped cells was observed. The CO content in the gas phase decreased to 40% and about 30% H2 and 30% CO2 were formed. After several transfers in the same medium the enrichments were serially tenfold diluted and transferred to the Medium 1 (low Eh) with sodium acetate and without Fe(III) under 100% CO, and then to roll-tubes with the same solidified Medium 1. After 2 days of incubation at 60°C, round white colonies about 0.5 mm in diameter were observed, which were transferred to a similar liquid medium. As a result, an isolate designated strain 019 was obtained.
Cells of the isolate 019 were straight rods with a length of 1–1.5 μm and a width of about 0.4–0.5 μm, arranged singly or in pairs. Cells were motile due to lateral flagella but singular cell aggregates on magnetite were also observed (Supplementary Figure S1).
Growth Characteristics of the New Isolate
Physico-Chemical Parameters
Growth of strain 019 was observed in the temperature range of 45–68°C, with an optimum at 58°C, and in the pH range 6.5–7.6, with an optimum at pH 7.0. Strain 019 grew anaerobically in a wide range of culture conditions: autotrophically or heterotrophically, by respiration or by fermentation. Aerobic or microaerobic growth was not observed with any of the tested concentrations of oxygen in the gas phase, i.e., under 100% air, a mixture of CO and air (4:1 v/v), or CO with 0.5, 1, or 2% O2.
Carbon Substrates
Autotrophic growth of strain 019 under CO was observed in the absence of any organic carbon sources. H2 as the electron donor with Fe(III), thiosulfate or nitrate as electron acceptors did not support autotrophic growth under a H2/CO2 gas phase. Organic substrates either stimulated carboxydotrophic growth or supported the growth in the absence of CO. Acetate and lactate but not pyruvate were utilized concomitantly with complete CO oxidation and hydrogen formation, the presence of the organic acid salt raising the cell yield up to 10 times compared to autotrophic growth conditions. Fermentative growth of strain 019 was observed on yeast extract, sucrose, glucose, maltose, or pyruvate under pure N2 or CO atmosphere. Galactose, cellobiose, cellulose, lactose, glycerol, peptone were not utilized. Glucose was fermented to lactate, acetate and H2.
Electron Donors and Acceptors
During autotrophic growth on CO coupled to hydrogenogenic carboxydotrophy, strain 019 utilizes protons as electron acceptors in its energy metabolism. Among electron acceptors other than protons, only Fe(III) stimulated growth of the strain, but it was exclusively utilized in the presence of CO (Figure 1). Thiosulfate and nitrate did not affect growth of the organism with organic acids under CO and inhibited autotrophic growth under CO (data not presented). Sulfate, nitrite or AQDS inhibited growth of strain 019 under any of the conditions tested.
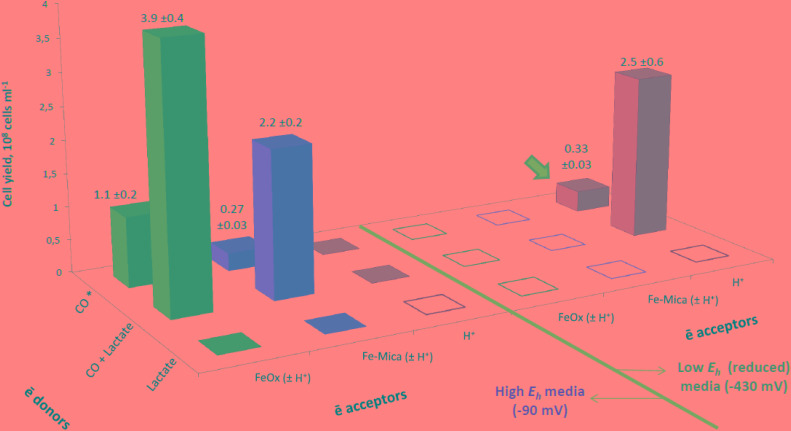
FIGURE 1.
Cell yield of strain 019 cultures as dependent on availability of Fe(III), organic carbon and a reductant in the medium. Indicated above the bars are cell yields and corresponding error limits (×108 cells ml-1). Note that two types of anaerobic culture media (high Eh and low Eh) were used to test each couple of electron donor and acceptor with both C. thermautotrophica strains. The initial Eh values were identical in the culture media of a particular type and are indicated on the diagram. Low Eh medium was pre-reduced with sodium sulfide (refer to “Materials and Methods” section for details). Electron donor/acceptor couples which have been tested but did not support the growth of a C. thermautotrophica strain are shown as flat filled squares. Empty squares indicate donor/acceptor couples which are biochemically senseless (lactate and H+) or physico-chemically restricted [rapid chemical reduction of Fe(III) minerals by sulfides]. Asterisk indicates autotrophic growth conditions when CO was the only carbon source and electron donor available in the culture medium. Thick arrow indicates the conditions supporting the growth of the type strain 41T; data on its cell yields are not presented.
Utilization of protons and Fe(III) as electron acceptors by strain 019 during autotrophic or heterotrophic (with lactate) growth appeared to depend on redox potential of the culture medium (Figure 1). In high Eh anaerobic medium, the growth was only possible in the presence of both CO and an Fe(III) mineral. In contrast, in low Eh culture medium, strain 019 grew either by carboxydotrophy in the absence of electron acceptors other than protons or by fermentation, i.e., with organic electron donors in the absence of both CO and Fe(III). Checking the Fe(III) reducing ability of strain 019 in low Eh medium appeared to be impossible due to rapid chemical redox interactions at 60°C between insoluble Fe(III) and sulfides, added to decrease the redox potential.
In parallel experiments, the type strain 41T grew only at low Eh by autotrophic hydrogenogenic CO oxidation without additional electron acceptors (Figure 1), as described previously by Sokolova et al., 2002.
Fe(III) Reduction From Minerals by Strain 019
The growth of strain 019 in the presence of both Fe(III) and CO was accompanied by Fe(III) reduction, complete CO oxidation and the formation of apparently equimolar quantities of CO2 and H2. In our experiments we used insoluble Fe(III) forms, which are typical for physico-chemical conditions in natural habitats of C. thermautotrophica. No soluble Fe(II) or Fe(III) was detected in cultures during their growth with various Fe(III) minerals.
Fe(III) Reduction From Ferrihydrite
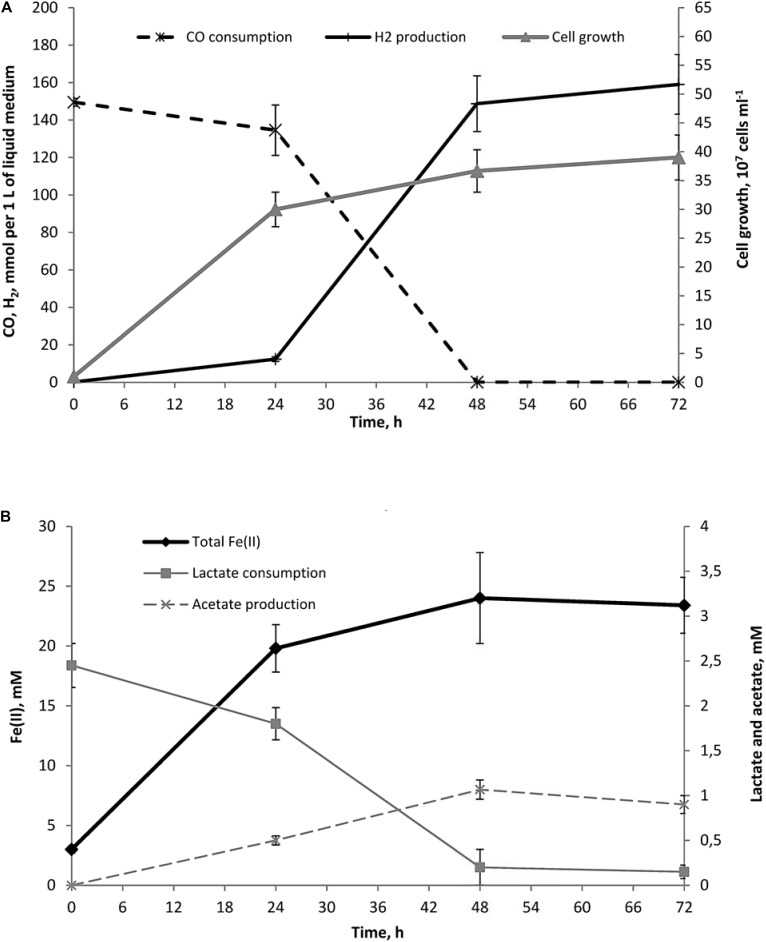
Fe(III) supplied as an oxic mineral ferrihydrite provided maximal cell yield of strain 019 (Figure 1). During the growth, about 23% of insoluble Fe(III) from ferrihydrite was reduced to Fe(II) in the form of a black magnetic mineral (presumably, magnetite) with concomitant decrease of the culture Eh down to -360 mV. The cell yield of autotrophic cultures grown on CO increased three times in the presence of ferrihydrite, in spite of the high initial Eh of the medium. Lactate stimulated the growth on CO with ferrihydrite (Figure 1). Consumption of 2.3 mM lactate was accompanied by the formation of 1.1 mM acetate (Figure 2B), indicating that about half of the oxidized lactate was utilized as an electron donor in catabolic reactions, while the rest of lactate was likely consumed as a carbon source. In contrast, only 0.6 mM lactate was consumed by cultures grown on CO and lactate in the absence of Fe(III) at low Eh, and no acetate production was detected in these growth conditions. Kinetic experiments revealed that maximal growth rate in high Eh medium containing ferrihydrite, CO and lactate was observed within the first 24 h of incubation simultaneously with the highest rates of Fe(III) reduction and lactate conversion to acetate. About 77% of the lactate consumed by the end of this growth phase was converted to acetate (Figure 2B), while only 10% of CO was oxidized with equimolar H2 formation. Maximal rate of hydrogenogenic carboxydotrophy was achieved later, when cell growth and Fe(III) reduction slowed down (Figure 2).
FIGURE 2.
Dynamics of growth, Fe(II) production, lactate consumption, acetate formation and concomitant hydrogenogenic carboxydotrophy of C. thermautotrophica strain 019 growing on CO and lactate with ferrihydrite. (A) Cell growth, CO consumption and H2 production; (B) Fe(II) production, lactate consumption and acetate formation.
Fe(III) Reduction From Phyllosilicates
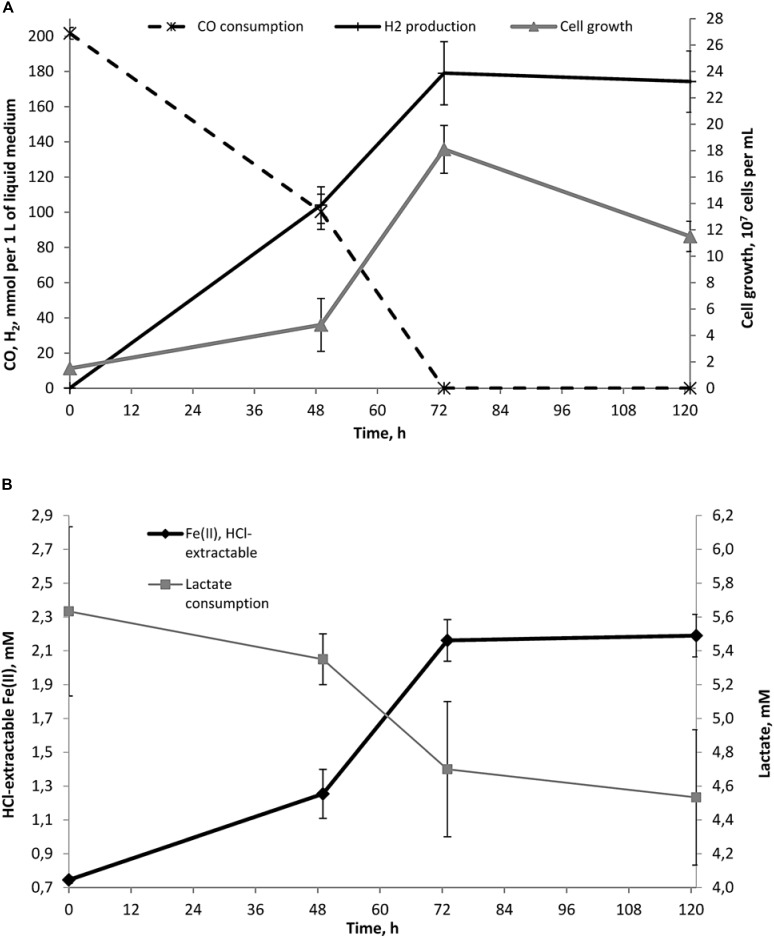
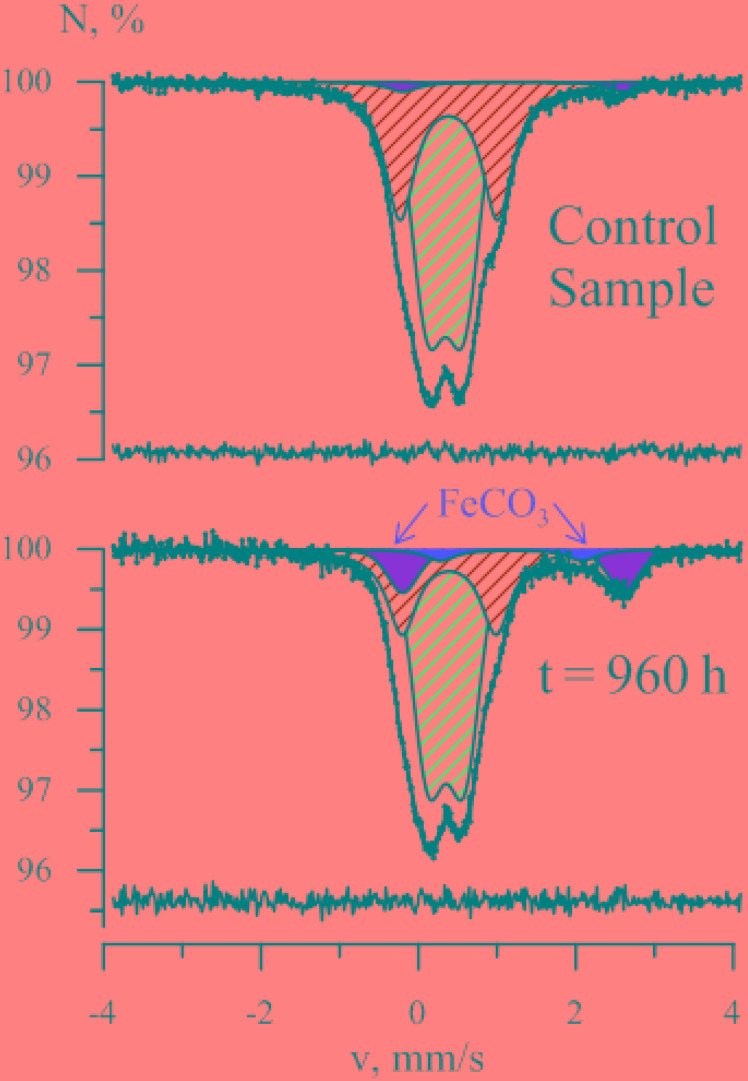
As strain 019 was isolated from a mineral core sample, it was tested for the ability to utilize structural Fe(III) from crystal lattice of phyllosilicates – the group of minerals previously detected in Zavarzin hot pool (Rozanov et al., 2014) right nearby the site where samples for our enrichments were obtained. We tested mixed valence Fe(III/II) minerals: the Fe-mica glauconite, clay mineral nontronite and siliceous sedimentary rock diatomite containing admixtures of natural Fe oxides (Distanov, 1987). Growth of strain 019 was observed in three consecutive transfers with diatomite and glauconite (Figure 3). Growth with diatomite was observed only in the presence of lactate as a potential carbon source, while growth with glauconite was observed both in the presence and in the absence of organic substrates. Growth with glauconite correlated with an increase in HCl-extractable Fe(II) (Figure 4) and non-extractable structural Fe(II) content (Supplementary Figure S2) of the Fe-mica mineral. During autotrophic growth, only a minor increase of glauconite Fe(II) content (by ca. 0.2 mM) was observed and the cell yield correlated with that detected in the absence of Fe(III) minerals at low Eh. Lactate stimulated the growth with glauconite 10-fold (Figure 1) and Fe(II) production from glauconite sevenfold (up to 1.42 mM). About 1 mM of lactate was consumed concomitantly with glauconite reduction but no acetate production was detected, indicating utilization of lactate as the carbon source only (Figure 4B). The growth was accompanied by a pronounced decrease of Eh down to -520 mV. In a control experiment, simulating the initial Fe(III)/Fe(II) ratio of glauconite by using a mixture of ferrihydrite and magnetite, the Eh value of the cultures rapidly decreased from -90 to -320 mV within the first 8 h of incubation. Changes in Eh of uninoculated controls with glauconite or ferrihydrite/magnetite mixture were much lower. The growth of strain 019 in the presence of lactate, CO and glauconite started with active CO oxidation and H2 formation. The rates of lactate consumption and Fe(III) reduction were minimal in this growth phase, but after 49 h of incubation, maximal rates of growth and of all of the mentioned metabolic processes were achieved simultaneously (Figure 4). Fe(III) reduction from the mica mineral by strain 019 rapidly ceased upon exhaustion of CO, followed by a start of cell lysis (Figure 4). However, to trace possible minor structural changes in glauconite, induced by microbial Fe(III) reduction but not leading to an increase in the HCl-extractable Fe(II) content of the mineral, we continued incubation of the cultures further on. Mössbauer investigations revealed that the relative amounts of Fe2+ and Fe3+ atoms in glauconite remained virtually constant within the first 78 h of incubation, until cell lysis started. During subsequent incubation, the relative content of ferrous atoms in glauconite increased almost twice from 2.2 ± 0.5% to 5.7 ± 0.4% by the 166th hour, and this trend was strengthening within further 794 h of incubation, until all the cells have been completely lysed (Supplementary Figure S2). Mössbauer spectra of the studied samples within all the incubation period were of paramagnetic type with a superposition of quadrupole doublets. Final spectra, captured at the 960th hour (Figure 5), were clearly fitted by four quadrupole doublets with equal line widths, depicting the formation of a small relative amount of a new mineral phase containing Fe2 + atoms in the octahedral oxygen environment. This phase was identified as siderite (FeCO3), which indicated the reduction of Fe3+ atoms in glauconite lattice structure. The relative concentration of siderite in the sample, obtained at the end of the entire 960-h incubation period, was I = 3.9 ± 1.3%. No changes in the mineral structure were detected in abiotic controls (Figure 5), as well as no production of H2 or Fe(II), no decrease in lactate concentration, and no interactions of CO or H2 with Fe(III) were observed within the same incubation period.
FIGURE 3.
Growth of C. thermautotrophica strain 019 with Fe(III)-containing silica mineral and rock on lactate under 100% CO. Diamonds – growth with glauconite at high Eh (–90 mV); squares – growth with diatomite at high Eh; circles – control growth without external electron acceptors at low Eh (–430 mV).
FIGURE 4.
Dynamics of growth, Fe(II) production, lactate consumption and concomitant hydrogenogenic carboxydotrophy of C. thermautotrophica strain 019 growing on CO and lactate with glauconite. (A) Cell growth, CO consumption and H2 production; (B) lactate consumption and Fe(II) formation. No acetate production has been recorded during the growth with glauconite.
FIGURE 5.
Mössbauer spectra of the mineral phase formed in glauconite-reducing culture of C. thermautotrophica strain 019 grown with CO and lactate. Characteristic peaks of siderite are marked red. Samples of the mineral phase were collected for analysis at the 960th hour of incubation after complete cell lysis.
No growth or Fe(III) reduction from any of the tested Fe(III) minerals was observed with the type strain 41T under the same cultivation conditions. Neither could the type strain grow by fermentation.
Phylogenetic Position of Strain 019
The average nucleotide identity (ANI) value between the genomes of strains 019 and 41T, calculated using ANI calculator7 with default parameters, was 99.7%; while the species-delimiting value, corresponding to the 70% level of in vitro DNA–DNA hybridization, is 95% (Goris et al., 2007). Thus, in silico hybridization of the genomes shows the affiliation of strains 019 and 41T with the same species, Carboxydocella thermautotrophica.
Genomic Properties of C. thermautotrophica Strains 019 and 41T
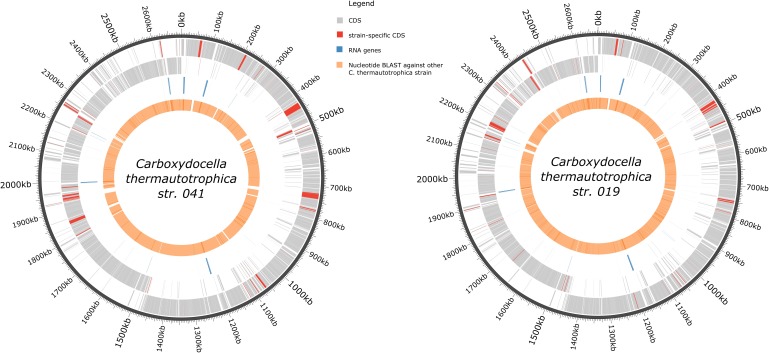
The genome of C. thermautotrophica type strain consists of one circular chromosome of a total length of 2690058 base pairs (49,14% GC content) and one circular plasmid of 53067 nucleotides (41% GC content). Read coverage of plasmid was 3.5 times higher than average chromosome coverage, suggesting 3–5 copies of plasmid per cell. The length of C. thermautotrophica strain 019 circular chromosome is 2676584 base pairs (49,14% GC content), and no plasmids have been identified in this strain.
For the C. thermautotrophica 041T chromosome, 2810 genes were predicted, 2697 of which are protein-coding genes and 113 are RNA genes (Table 1). 65.5% of genes were assigned to at least one COG cluster with IMG annotation pipeline (Huntemann et al., 2015). The distribution of hits to COG functional categories is presented in Supplementary Table S1. Genome has five complete ribosomal operons with 16S rRNA genes showing at least 99.6% identity with each other. General genomic features of C. thermautotrophica strain 019 were similar to those of the type strain chromosome and are presented in Table 1 and Figure 6.
Table 1.
General features of replicons of C. thermautotrophica strains.
| C. thermautotrophica 019 chromosome | C. thermautotrophica 041T chromosome | C. thermautotrophica 041T plasmid | |
|---|---|---|---|
| Replicon length, bp | 2676584 | 2690058 | 53067 |
| GC mol % | 49.14 | 49.14 | 41.3 |
| Number of genes | 2809 | 2810 | 51 |
| RNA genes | 120 | 113 | 0 |
| tRNA | 71 | 73 | 0 |
| rRNA | 15 | 15 | 0 |
| Protein-coding genes | 2689 | 2697 | 51 |
| Assigned to COG | 1775 | 1787 | 14 |
| Number of GIs | 9 | 8 | NA |
| Total length of GIs, bp | 214072 | 211619 | NA |
FIGURE 6.
Graphic representation of the genomes of C. thermautotrophica strains. Rings from outside to inside: genomic coordinates (dark gray); plus-strand common (gray) and strain-specific (red) CDSs; minus-strand common (gray) and strain-specific (red) CDSs; RNA genes (blue); blastn hits with e-value cutoff 10-5 vs. the other C. thermautotrophica strain.
The plasmid of strain 41T encoded 51 proteins, 21 of their ORFs showed dispersed weak to moderate (23–58%) hits to the chromosome-encoded in silico proteomes of both strains, and two of the ORFs revealed weak hits to the proteome of strain 019 only. Little if any of these proteins are of clear metabolic relevance. Presence in the plasmid of the IS1182 mobile element, along with genes of restriction-modification system, suggests that it may be a selfish mobile element (Kobayashi, 2001; Koonin, 2011).
Comparative Genome Analysis of CO and Fe(III) Metabolism in C. thermautotrophica Strains 019 and 41T
Oxidative Phosphorylation
A set of genes of the proton-translocating type I NADH-dehydrogenase (complex I) nuoABCDHIJKLMN is encoded in the same order by CFE_1311–1321 in strain 019 and CTH_1331–1341 in strain 41T. Subunits NuoEFG, essential to provide the catalytic site for NADH oxidation, are encoded separately in both genomes: CFE_2217–2219 in strain 019 and CTH_2319–2321 in strain 41T. NuoEFG proteins in both strains share weak sequence similarity with their homologs from UniProt database. Gene clusters of the respiratory complex II (succinate dehydrogenase) are duplicated in both genomes (CFE_1726–1728 and CFE_2117–2119 in strain 019, and CTH_1740–1742 and CTH_2150–2152 in strain 41T). Oxidative phosphorylation in both strains is performed via F0F1-type bacterial ATP-synthases, encoded by CFE_0343–0352 in strain 019 and CTH_0352–0361 in strain 41T.
Inorganic Carbon Assimilation
Both C. thermautotrophica strains possess full sets of genes for the Wood–Ljungdahl pathway of inorganic carbon assimilation. The genes of the so-called Western (carbonyl) branch of the pathway and genes of the final steps of the Eastern (methyl) branch are encoded in each of the strains in a large gene cluster (Supplementary Table S2) similar to the acs gene cluster of Moorella thermoacetica ATCC 39073 (Pierce et al., 2008), a model organism to study the Wood–Ljungdahl pathway. Interestingly, in C. thermautotrophica strains additional genes of methylenetetrahydrofolate reductase subunits MetVF are also present in a smaller gene cluster (Supplementary Table S2), and here they occur together with hdrCBA and mvhD genes, which in M. thermoacetica adjoin the metVF genes in the large acs cluster. Other Wood–Ljungdahl pathway genes of C. thermautotrophica strains are dispersed over the chromosomes (Supplementary Table S2), as it is in M. thermoacetica. Genes encoding key enzymes of other known autotrophic pathways (Fuchs, 2011; Nunoura et al., 2018; Mall et al., 2018) could not be found.
Both C. thermautotrophica strains also possess genes that might extend the anabolic function of the Wood–Ljungdahl pathway to the catabolic capacity for acetogenesis, including acetogenesis on H2 + CO2. The genomes harbor genes of phosphotransacetylase (two non-homologous genes in each genome) and acetate kinase (Supplementary Table S2), which provide for acetate and ATP production from acetyl-CoA and ADP + Pi, and genes encoding close homologs (49–73% amino acid sequence identity, Supplementary Table S2) of the subunits of enzymatic complexes that promote reductant balance and energy conservation during acetogenesis by M. thermoacetica (Schuchmann and Müller, 2014; Basen and Müller, 2017), including an energy converting hydrogenase additional to the CO-induced one, considered in the next subsection.
However, despite the apparent presence of all required acetogenesis determinants, we failed to obtain growth of strains 41T and 019 on H2 + CO2 mixture or to detect acetate production during autotrophic growth under CO.
Genomic Determinants of Carboxydotrophy
A common metabolic feature of C. thermautotrophica strains is their ability to grow at the expense of hydrogenogenic oxidation of carbon monoxide. In the type strain 41T, this ability appeared to be restricted by the redox potential of the culture medium, manifesting itself only at its low values. In contrast, strain 019 grew by CO oxidation also at relatively high Eh of -90 mV, but only in the presence of Fe(III) minerals, which were utilized as electron acceptors in parallel with protons.
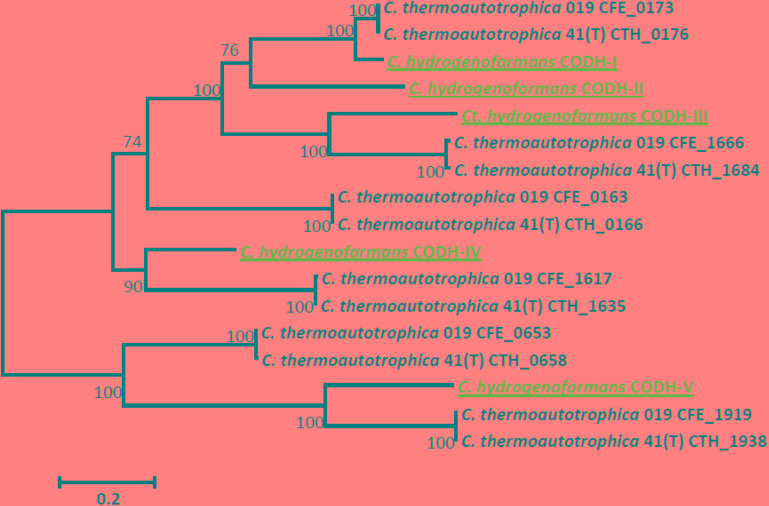
Each of the genomes of C. thermautotrophica strains 41T and 019 harbors six cooS genes, encoding anaerobic (Ni,Fe-containing) CODHs, and no genes encoding aerobic (Mo,Cu-containing) CODHs. Quite recently (see the “Discussion” section for the latest data), the highest number of [Ni,Fe]-CODHs encoded in available completely sequenced genomes was five, one of such few genomes belonging to the well-studied hydrogenogenic carboxydotroph Carboxydothermus hydrogenoformans (see Svetlitchnyi et al., 2004; Wu et al., 2005 for substantiation of the integrity of the cooS3 gene in the original isolate of C. hydrogenoformans). The functional roles of the five [Ni,Fe]-CODHs of C. hydrogenoformans have been studied by various approaches, and for four of them functions have been established (Svetlitchnyi et al., 2001, 2004; Soboh et al., 2002; Hedderich, 2004; Wu et al., 2005). The phylogenetic relations C. thermautotrophica [Ni,Fe]-CODHs with the [Ni,Fe]-CODHs of C. hydrogenoformans are shown in Figure 7.
FIGURE 7.
Phylogenetic tree of Carboxydocella thermautotrophica [Ni,Fe]-CODHs. The tree was constructed by Maximum Likelihood method using MEGA6 (Tamura et al., 2013) at default parameters after aligning sequences with built-in Muscle at default parameters. Underlined blue are the [Ni,Fe]-CODHs of Carboxydothermus hydrogenoformans, which were used as references for function prediction.
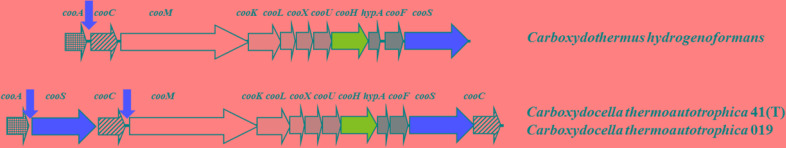
The roles of two [Ni,Fe]-CODHs in each C. thermautotrophica strain can be deduced from high similarity of their amino acid sequences and genomic contexts with [Ni,Fe]-CODH-I and [Ni,Fe]-CODH-III of C. hydrogenoformans. The cooSI-like genes CFE_0173 and CTH_0176 are parts of gene clusters (Figure 8) highly similar to the C. hydrogenoformans gene cluster that includes, along with cooSI, genes of an energy-converting hydrogenase (ECH) and is responsible for hydrogen formation from CO + H2O with transmembrane potential generation (Svetlitchnyi et al., 2001, 2004; Soboh et al., 2002; Hedderich, 2004). However, in the C. thermautotrophica strains this gene cluster has a more complicated pattern, including a second cooC gene in the downstream region and a second cooS gene (CFE_0163 and CTH_0166) in the upstream region, between the cooC gene and the cooA gene of the CO- and redox-sensing transcription regulator protein CooA (Roberts et al., 2001). This additional cooS gene does not have close homologs in C. hydrogenoformans.
FIGURE 8.
Gene clusters determining hydrogenogenic carboxydotrophy in Carboxydothermus hydrogenoformans and Carboxydocella thermautotrophica strains 41T and 019. cooA: gene of redox- and CO-sensitive transcriptional regulator; cooS (red horizontal arrow): [Ni,Fe]-CODH gene; cooC: gene of [Ni,Fe]-CODH accessory nickel-insertion protein; cooMKLXUH: genes of energy-converting hydrogenase (ECH); hypA: gene of hydrogenase maturation protein; cooF: gene of ferredoxin-like protein; cooH (blue horizontal arrow): gene of ECH catalytic subunit. Red vertical arrows show locations of CooA-binding sites. The locus tags for the gene clusters are CHY_1835-1824, CTH_0165-0177, and CFE_0162-0174. Note that the exact role of the upstream ‘enigmatic’ cooSC operon (CTH_0166-167 and CFE_0163-0164) in carboxydocellas remains unclear (refer to the text for details).
The operon formula of the cluster in C. thermautotrophica strains is apparently cooA-cooSC-cooMKLXUHhypAcooFSC-, i.e., the two [Ni,Fe]-CODHs are encoded in distinct operons. However, these operons are evidently co-regulated by the above-mentioned CO- and redox-sensing regulator protein CooA. In both C. thermautotrophica strains, CooA-specific binding sites, conforming to the long-known consensus formula TGTCRNNNNNNYGACR (Fox et al., 1996), are located upstream of the cooSC and cooMKLXUHhypAcooFSC operons (Figure 8). The role of the cooSC operon is obscure, but the fact that it is regulated by CooA, sensing CO at low redox potential conditions (Roberts et al., 2001), provides grounds to speculate that the [Ni,Fe]-CODHs CFE_0163 and CTH_0166 have a role to play in CO oxidation with hydrogen production.
The [Ni,Fe]-CODHs of C. thermautotrophica strains that are similar to C. hydrogenoformans [Ni,Fe]-CODH-III, involved in the Wood–Ljungdahl pathway of acetyl-CoA synthesis from C1 units and low-potential reductants (Svetlitchnyi et al., 2004; Wu et al., 2005), are encoded by CTH_1684 in strain 41T and CFE_1666 in strain 019. The genes of these [Ni,Fe]-CODHs occur in gene clusters (Supplementary Table S2) similar to the acs gene clusters of C. hydrogenoformans (Wu et al., 2005) and M. thermoacetica ATCC 39073 (Pierce et al., 2008). The genes of the Wood–Ljungdahl pathway, which evidently provides for the autotrophic capacity of C. thermautotrophica strains, are considered in more detail above, in the subsection Inorganic carbon assimilation.
The [Ni,Fe]-CODHs of C. thermautotrophica strains CFE_1617 and CTH_1635 are similar to the C. hydrogenoformans [Ni,Fe]-CODH-IV (Figure 7), thought to be involved in defense against oxidative stress (Wu et al., 2005). However, these C. thermautotrophica [Ni,Fe]-CODH genes differ in their genomic context from the C. hydrogenoformans [Ni,Fe]-CODH-IV gene: they are in gene clusters that do not include rubrerythrin gene, which in C. hydrogenoformans is thought to be responsible for the final step in a chain of reactions performing hydrogen peroxide reduction to water at the expense of electrons derived from CO. Thus, currently, we do not have any hypothesis about the role of the [Ni,Fe]-CODHs CFE_1617 and CTH_1635.
The remaining fifth and sixth [Ni,Fe]-CODHs of C. thermautotrophica strains (CFE_0653, CFE_1919, CTH_0658, CTH_1938) are similar to C. hydrogenoformans’ [Ni,Fe]-CODH-V, whose function has not been even hypothetically supposed (Wu et al., 2005). CFE_0653 and CTH_0658 seem to be alone in their operons (judging from the 70-100 bp intergenic spaces upstream and downstream), and CFE_1919 and CTH_1938 are alone judging from the directions of transcription of the neighboring genes. Thus, we are unable to make any predictions about the functions of these [Ni,Fe]-CODHs from purely genomic analysis. Moreover, the genomically lone [Ni,Fe]-CODHs CFE_1919 and CTH_1938 have an altered pattern of the ligands of the active site [Ni-4Fe-5S] C-cluster (Dobbek et al., 2001): they lack the Cys295 residue (C. hydrogenoformans CooSII numbering), thought to be important for Ni-coordination in the C-cluster (Inoue et al., 2013).
Fe(III) Respiration
Screening of both C. thermautotrophica genomes revealed 30 genes in each strain possessing various c-type multiheme cytochrome domains. Almost all the encoded proteins are 99–100% identical in the two organisms (Supplementary Table S3). Four of the cytochrome genes comprise typical nrfAH loci of dissimilatory nitrite reductase complexes and were excluded from further screening of putative Fe(III) reductases. Some of the rest multiheme cytochrome genes could be involved in extracellular electron transfer chain. We found no homologs of putative quinol oxidizing multiheme CymA, regarded to initiate extracellular electron transfer in S. oneidensis (Coursolle and Gralnick, 2010). However, both C. thermautotrophica strains possess homologs of inner membrane cytochromes ImcH and CbcL from G. sulfurreducens, which have been shown to determine high- and low-potential pathways for metal reduction, respectively (Levar et al., 2017). The homologs of ImcH are encoded by CFE_1714 in strain 019 and CTH_1728 in strain 041T, located in identical clusters with two other multiheme cytochrome genes. The homologs of CbcL (Zacharoff et al., 2016) are encoded in two Cbc-like gene clusters in each C. thermautotrophica genome. Those are CFE_2192-2193 and CFE_2225-2226 in strain 019 and CTH_2221-2222 and CTH_2327-2328 in strain 041T. In each of these clusters, one gene encodes a transmembrane b-type diheme domain protein, which could serve for quinol oxidation, and the other encodes a protein with a predicted transmembrane helix and a c-type multiheme domain facing the cell surface, which could accept electrons from the b-type cytochrome and initiate their further transfer to terminal Fe(III) reductases.
As described above, strain 041T does not reduce any forms of Fe(III). Interestingly, among predicted c-type multihemes of C. thermautotrophica strains, only one 17-heme cytochrome was identified exclusively in the Fe(III)-reducing strain 019 (Supplementary Table S3). The protein is encoded by CFE_2239 downstream of a 15-heme and a hexaheme c-type cytochromes CFE_2242-2243 with unknown function and upstream of two cytochrome c maturation proteins and an S-layer homology domain-containing protein CFE_2230. The cytochrome CFE_2239 is predicted to contain signal peptide in its C-terminal part and no transmembrane helixes, and thus is likely to be a secreted protein. Topology prediction with CW-PRED service (Fimereli et al., 2012) indicates probable anchoring of CFE_2239 in the cell wall. However, the protein shares no homology with any of the outer surface cytochromes or proteins related to porin-cytochrome complexes that are suggested to perform the final step of extracellular electron transfer in Shewanella and Geobacter species (Coursolle and Gralnick, 2010; Aklujkar et al., 2013; Shi et al., 2016). The cytochrome CFE_2239 consists of two parts which are likely to have different origin and functions. In the cytochrome CFE_2239, putative catalytic N-terminal part (from 501st to 1486th amino acid residue) harbors all the 17 heme-binding motives organized in several conservative multiheme domains. It has distant homologs among many different multihemes, mainly of proteobacterial origin. Phylogeny reconstruction indicates (Supplementary Figure S3) that this putative catalytic domain could have been acquired by horizontal gene transfer from Geobacter species. The C-terminal part of the protein CFE_2239 was not reliably predicted to contain homologs of any conservative protein domains, however, the search against SwissProt database revealed a few weak homologs among bacterial adhesin proteins and eukaryotic secreted proteins involved in receptor-ligand interactions and adhesion.
Discussion
Bacteria of the genus Carboxydocella represent a distinct metabolic group of thermophilic hydrogenogenic carboxydotrophs (Sokolova et al., 2009). The members of this group are either obligately dependent on CO or capable of gaining energy using other catabolic processes (fermentation or different types of anaerobic respiration). Three species of the genus Carboxydocella have been isolated from various thermal habitats of Kamchatka peninsula (Sokolova et al., 2002; Slepova et al., 2006; Slobodkina et al., 2012). The metabolism of these organisms differs significantly (Supplementary Table S4). Our new isolate 019, obtained from a core sample near Zavarzin thermal pool at Uzon Caldera, shares some features with all validly described Carboxydocella species. However, it belongs phylogenetically to the species with the type strain of which it has the lowest number of common metabolic features: in contrast to strain 019, C. thermautotrophica 41T can only grow autotrophically and cannot reduce Fe(III). Two metabolic processes are manifestly common to both C. thermautotrophica strains – those are inorganic carbon assimilation and hydrogenogenic carboxydotrophy.
Diversity of CO-Dehydrogenases
The genomes of C. thermautotrophica strains 41T and 019 encode six [Ni,Fe]-CODHs each. Until the recent isolation and genomic study of Calderihabitans maritimus (six [Ni,Fe]-CODH genes, including one frameshifted gene (Omae et al., 2017), and the publication of the genome of Clostridium formicaceticum (six [Ni,Fe]-CODH genes, CP020559.1, Karl et al., 2017), the highest number of [Ni,Fe]-CODHs encoded in available completely sequenced genomes was five. Our analysis performed in April 2018 by tblastn in the NCBI Complete Prokaryote Genome Database (20,008 genomes) revealed 379 genomes encoding at least one [Ni,Fe]-CODH; of them, 208 genomes encoded more than one [Ni,Fe]-CODH. Six genomes encoded five [Ni,Fe]-CODHs each, and the genome of Clostridium formicaceticum encoded six [Ni,Fe]-CODHs. Thus, the genomes of C. thermautotrophica strains are top-ranking with respect to the number of [Ni,Fe]-CODHs encoded.
The functional roles of multiple [Ni,Fe]-CODHs have been best studied in Carboxydothermus hydrogenoformans, and to four of them functions have been ascribed (Svetlitchnyi et al., 2001, 2004; Soboh et al., 2002; Hedderich, 2004; Wu et al., 2005). Based on comparison of amino acid sequences and genomic contexts, we managed to ascribe functions to only two of the six [Ni,Fe]-CODHs in each of the C. thermautotrophica strains and to tentatively suppose a possible role for one more [Ni,Fe]-CODH in each strain.
The [Ni,Fe]-CODH encoded immediately adjacent to ECH genes (apparently in a single operon) is evidently responsible in C. thermautotrophica strains for hydrogen formation from CO + H2O with transmembrane potential generation. [Ni,Fe]-CODH–ECH gene clusters occur in all of the sequenced genomes of hydrogenogenic carboxydotrophs, the only exception known so far being Carboxydothermus pertinax Ug1 (Fukuyama et al., 2017). Three types of such gene clusters have been described, composed of homologous genes that, however, differ in their order and phylogeny (genes specific to a particular cluster type are quite few). The gene cluster found in the genomes of C. thermautotrophica strains belongs to the long-known coo-type gene cluster. The first [Ni,Fe]-CODH–ECH gene cluster to be described was the coo gene cluster of Rhodospirillum rubrum S1T (Fox et al., 1996; Kerby et al., 1997). Similar (coo-type) gene clusters were then described in C. hydrogenoformans Z-2901T (Soboh et al., 2002; Wu et al., 2005) (Figure 8), Thermosinus carboxydivorans Nor1T (Techtmann et al., 2012), Desulfotomaculum caboxydivorans CO-1-SRBT (Visser et al., 2014), Calderihabitans maritimus KKC1 (Omae et al., 2017), and mentioned to occur in Carboxydothermus islandicus SET (Fukuyama et al., 2017). Gene clusters of coo-type also occur in the genomes of some other hydrogenogenic carboxydotrophs (‘Thermincola potens’ JR (CP002028.1), Thermincola ferriacetica Z-0001T (LGTE00000000)), as well as in some bacteria for which the capacity has not been tested. A peculiar variant of the coo gene cluster was described in Rubrivivax gelatinosus CBS (Wawrousek et al., 2014). Also known are two other types of [Ni,Fe]-CODH–ECH gene clusters that differ from the coo cluster in the order of homologous genes and their phylogeny. One was found in Caldanaerobacter subterraneus subspecies (Sant’Anna et al., 2015), and the other was revealed in several representatives of the archaeal genus Thermococcus (Lim et al., 2010; Kozhevnikova et al., 2016; Oger et al., 2016). Some of the organisms harboring the gene clusters of the latter two types are known to be capable of hydrogenogenic carboxydotrophy, for others the capacity has not been tested.
Thus, the [Ni,Fe]-CODH–ECH gene cluster peculiar to C. thermautotrophica strains belongs to the long-known, best studied, and most widely occurring coo-type gene clusters. However, in the C. thermautotrophica strains this gene cluster has a more complicated pattern; in particular, it includes a second cooS gene in an adjacent and apparently co-regulated cooSC operon. This co-regulation, performed by the CO- and redox-sensing transcription regulator protein CooA, provides grounds to speculate that this enigmatic [Ni,Fe]-CODH (CFE_0163 in strain 019 or CTH_0166 in strain 41T), which does not have close homologs in C. hydrogenoformans or in finished genomes represented in the NCBI nr database, has a role to play in CO oxidation with hydrogen production. It was shown by Soboh et al. (2002) that the CO-oxidizing:H2-evolving enzyme complex that these authors isolated from C. hydrogenoformans contained both CooS1 and CooS2 at a ratio of about 10:1, although the main function of CooS2 has been proposed to be generation of NADPH for anabolic purposes (Svetlitchnyi et al., 2001). The physiological relevance of the dual composition of the [Ni,Fe]-CODH–ECH enzymatic complex has not been discussed; and we can only mention that CooS1 and CooS2 do not differ significantly in their apparent Km values (Svetlitchnyi et al., 2001). No close homologs of CooS2 are encoded in the genomes of C. thermautotrophica strains, and it is possible that the discussed ‘enigmatic’ [Ni,Fe]-CODH encoded upstream of the [Ni,Fe]-CODH–ECH gene cluster is a minor substitute in the CO-oxidizing:H2-evolving enzyme complex of C. thermautotrophica.
None of the other C. thermautotrophica [Ni,Fe]-CODHs seem to be co-regulated with the CO-oxidizing:H2-evolving enzyme complex, since no CooA-binding sites could be found in proximity of other [Ni,Fe]-CODH genes. Moreover, since the CooA transcription regulator is sensitive both to CO and redox conditions, it may be speculated that the remaining four [Ni,Fe]-CODHs are expressed and active both in the absence and in the presence of electron acceptors other than proton [nitrate for both strains, nitrate or Fe(III) for strain 019]. However, only for one of these [Ni,Fe]-CODHs can we predict function from sequence comparisons with well-studied [Ni,Fe]-CODHs and from genomic context. This [Ni,Fe]-CODH (CTH_1684 and CFE_1666) must be involved in the Wood–Ljungdahl pathway. Interestingly, in some hydrogenogenic autotrophic carboxydotrophs the gene encoding this [Ni,Fe]-CODH becomes frameshifted as a result of cultivation at high CO concentrations. The laboratory-acquired origin of the frameshift and its cause are evident for C. hydrogenoformans (Svetlitchnyi et al., 2004; Wu et al., 2005) and can be supposed for Calderihabitans maritimus. Svetlitchnyi et al. (2004) showed that in a C. hydrogenoformans strain variant with non-frameshifted [Ni,Fe]-CODH, acetyl-CoA synthase existed predominantly as monomer at high CO concentrations and used exogenous CO instead of the CO produced from CO2 by the [Ni,Fe]-CODH. Such one-step assimilation of CO should be more beneficial. In the C. thermautotrophica strains the [Ni,Fe]-CODHs involved in the Wood–Ljungdahl pathway are not impaired despite repeated culture transfers under 100% CO.
In M. thermoacetica the Wood–Ljungdahl pathway plays the roles of both carbon assimilation and energy generation, depending on whether acetyl-CoA is further carboxylated or converted to acetate with ATP formation (Pierce et al., 2008). C. hydrogenoformans has been reported (Henstra and Stams, 2011) to switch to acetogenesis from CO under conditions where hydrogen production from CO becomes thermodynamically unfavorable (low substrate and high product concentrations). Both C. thermautotrophica strains possess genes closely homologous to those thought to be responsible for the acetogenic growth of M. thermoacetica (Schuchmann and Müller, 2014; Basen and Müller, 2017). Especially remarkable is that each of their genomes encodes two methylenetetrahydrofolate reductase homologs and that one of these metVF gene variants is clustered with hdrCBA and mvhD genes, indicative of the formation of the HdrCBA+MetVF+ MvhD methylenetetrahydrofolate-reducing electron-bifurcating complex, which is highly efficient in terms of bioenergetics since it produces low-potential reductant (Mock et al., 2014).
The genomes of each of the C. thermautotrophica strains encoded three [Ni,Fe]-CODHs for which we could not predict or suppose a definite function. We can only make some very general comments about the remaining [Ni,Fe]-CODHs. In both strains, particular [Ni,Fe]-CODH(s) may be involved in autotrophic carboxylation reactions that follow acetyl-CoA synthesis, supplying reducing equivalents to carboxylating enzymes, as it was supposed for one of the [Ni,Fe]-CODHs of Calderihabitans maritimus based on the genomic proximity of its gene with genes of ferredoxin:oxoacid oxidoreductase (Omae et al., 2017). However, in case of the C. thermautotrophica strains, there are no such hints from the genomic contexts.
Anyway, it is evident that CO is an important nutrient in the natural habitat of the C. thermautotrophica strains, and they largely base on it their survival strategy.
Physiology of Fe(III) Reduction
The most intriguing physiological feature of C. thermautotrophica strain 019 appeared to be dissimilatory reduction of Fe(III) from minerals, which is obviously coupled to hydrogenogenic CO oxidation. Minerals of Fe(III) are the only form of electron acceptors sustaining C. thermautotrophica growth at elevated Eh of -90 mV (Figure 1). Hydrogenogenic carboxydotrophy with protons as electron acceptors was exclusively performed by the organism under strongly reduced conditions (-430 mV). Notably, strain 019 appeared to reduce structural Fe(III) from silica mineral glauconite. This process has not been previously reported in Fe(III) reducers. In our recent report, we have demonstrated oxidation of structural Fe(II) but not the reduction of Fe(III) from glauconite by the alkaliphilic dissimilatory iron-reducer Geoalkalibacter ferrihydriticus during acetogenic growth (Zavarzina et al., 2016). In contrast to that finding, C. thermautotrophica performed the reduction of structural Fe(III) from the same mineral, as clearly indicated by the formation of siderite (Figure 5) and the increase of extractable Fe(II) concentration (Figure 4) during strain 019 cultivation with glauconite. Achievement of the highest possible Fe(II) content of glauconite during its reduction is clearly marked by decreased line width of Mössbauer spectra (Supplementary Figure S2, Inlay), which is an indicator of the decrease in the ordering of glauconite crystal lattice. Such disordering results from the attack of Fe3+ atoms at the mineral surface, e.g., by bacterial redox systems. Further increase in Mössbauer spectral line width and Fe(II) content of the mineral after the start of cell lysis could be explained by the formation of new solid phase siderite (Supplementary Figure S2). No such changes were observed in abiotic controls, indicating biologically induced process of siderite formation, which, however, is not directly controlled by the redox systems of live cells and proceeds after their lysis.
The reduction of mixed valence Fe(III/II) silica minerals and the oxic Fe(III) mineral ferrihydrite have different impacts on the metabolism of strain 019. When Fe(III) was provided as an electron acceptor in the form of ferrihydrite, cultures of strain 019 revealed two clearly distinguishable growth phases. During the first 24-h phase with high initial Eh and low initial Fe(II) content, maximal growth rate correlated with maximal rates of Fe(III) reduction, lactate conversion to acetate and low rates of carboxydotrophy and hydrogen formation. The second phase started upon accumulation of Fe(II) and concomitant Eh decrease. This ‘high Fe(II)’ phase was characterized by pronounced slowdown of growth and Fe(II) formation, but at the same time, the rate of hydrogenogenic carboxydotrophy increased to its maximum.
We may assume that during the first stage of growth both CO and lactate could be simultaneously utilized as electron donors for Fe(III) reduction in parallel with hydrogenogenic carboxydotrophy. Accordingly, three reactions are likely to sustain the growth of strain 019 at this stage:
| (1) |
| (2) |
| (3) |
Data on metabolite concentrations (Figure 2) allow us to propose the following brutto-equation for energy metabolism of strain 019 at the first stage of its growth on CO and lactate with ferrihydrite:
| (4) |
Given that each lactate molecule donates 4 electrons to Fe3+ and each CO molecule donates 2 electrons to either Fe3+ or H+, reaction (4) implies that the ratio of carbon monoxide spent for iron(III) reduction and for hydrogenogenesis was 13:17, i.e., about 43% of the consumed carbon monoxide was utilized for Fe(III) reduction during the first phase of growth of strain 019 with ferrihydrite and CO. Protons produced in this reaction could be translocated by type I NADH-dehydrogenase or ECH complexes. During further growth of the culture, when Fe(II) accumulates and Eh decreases, hydrogenogenic carboxydotrophy becomes much more active and seems to play the major role in the energy metabolism of strain 019 (Figure 2). Indeed, redox-control of the [Ni,Fe]-CODH–ECH cluster transcription in C. thermautotrophica via CooA regulator could restrict employment of this energy generating complex under high Eh growth conditions. On the other hand, both C. thermautotrophica strains possess four more [Ni,Fe]-CODH clusters which do not depend on CooA and thus on low redox potential. At least three of those could be active at elevated Eh being coupled to Fe(III) reduction in yet unknown way. Formation of magnetite during cell growth coupled to ferrihydrite reduction decreases Eh down to -360 mV and switches on the [Ni,Fe]-CODH–ECH energy generating complex at the second, ‘high Fe(II),’ growth phase of strain 019. As the [Ni,Fe]-CODH–ECH complex deals with soluble electron donor and acceptor inside the cell and includes a single step proton translocation via ECH, it outcompetes the more complicated Fe(III)-reducing extracellular electron transfer pathway for electrons derived from CO oxidation. Metabolite concentrations monitoring (Figure 2) allows us to propose the following brutto-equation for the ‘high Fe(II)’ phase of strain 019 growth with ferrihydrite, lactate and CO:
| (5) |
This reaction, in contrast to reaction (4), implies that the main portion of carbon monoxide consumed during the second growth phase of strain 019 was spent for hydrogenogenesis, and only 1% of it (3 moles of 270) was utilized for ferrihydrite reduction. This switch of the electron acceptor for CO oxidation from Fe(III) to more accessible but less energetically favorable intracellular protons, together with magnetite accumulation, which restricts the access of the cells to Fe(III) atoms on the mineral surface, decreases the growth rate of the organism at high Fe(II) content, in spite of intensified consumption of CO and lactate.
Key Role of CO in Fe(III) Reduction
The inability of strain 019 to reduce any forms of Fe(III) with lactate in the absence of CO indicates the key metabolic role of carbon monoxide utilization pathways in the organism. In particular, even lactate utilization is likely linked to [Ni,Fe]-CODH activity. Lactate enters the metabolic network of C. thermautotrophica via pyruvate-forming lactate dehydrogenase. Pyruvate can be converted to formate and acetyl-CoA by pyruvate-formate lyase (formate C-acetyltransferase, CFE_0574-575). Formate in its turn could further enter Wood–Ljungdahl pathway via formate-tetrahydrofolate ligase (CFE_0088) and finally be condensed with a CO molecule by the ACS complex (Supplementary Table S2) to form acetyl-CoA, which is then converted to acetate with substrate-level phosphorylation (Supplementary Table S2). Previously, acetogenesis from formate and CO via the Wood–Ljungdahl pathway has been proposed for the acetogen Acetobacterium woodii (Bertsch and Müller, 2015). C. thermautotrophica strains possess all the necessary genes for acetogenesis (Supplementary Table S2), including those of the HdrCBA+MetVF+MvhD electron-bifurcating complex, which does not seem to be crucial when the energy requirements are covered by hydrogenogenesis from CO, but is highly efficient in case of acetogenesis. In our experiments, conditions favoring acetogenesis by C. thermautotrophica strain 019 seem to have existed in the high Eh culture medium, when the activity of [Ni,Fe]-CODH–ECH was restricted by redox potential. However, from purely genomic analysis, it is difficult to predict whether or not the acetyl-CoA synthase of C. thermautotrophica strains can directly use exogenous CO under certain conditions. One should also remember that Fe(III) is indispensable electron acceptor for C. thermautotrophica at elevated Eh, and that metabolic link between acetogenesis and Fe(III) reduction in this organism remains to be understood.
Of note is the fact that during organotrophic growth with the Fe(III)-mica mineral glauconite there was no acetate production, and maximal rates of growth, carboxydotrophy, lactate consumption and Fe(III) reduction were observed simultaneously. The most probable cause is the ferrous iron content of the mixed valence mineral glauconite. This Fe(II) could act as an immobilized reductant rapidly decreasing redox potential of the culture medium in the close vicinity of mineral particles and thus stimulating redox-controlled carboxydotrophic growth of the organism. Such a decrease of the Eh value in the medium containing both ferric and ferrous iron mineral forms is enhanced by growing cultures, as revealed in our control experiment with a ferrihydrite/magnetite mixture. In contrast to ferrihydrite, glauconite did not stimulate autotrophic growth of strain 019 on CO (Figure 1). This fact indicates that glauconite reduction is not as energetically favorable as ferrihydrite reduction, and Fe(II) production from glauconite is rather caused by secondary activity of the extracellular electron transfer chain of strain 019. The absence of acetate production during growth on lactate with glauconite and CO, as well as the cessation of Fe(III) reduction from glauconite upon exhaustion of CO, indicate that carbon monoxide serves as the only electron donor for the reduction of this mica mineral.
Genomic Determinants of Extracellular Electron Transfer to Fe(III) Minerals
Utilization of Fe(III) minerals as electron acceptors is challenging and requires extracellular electron transfer chain to be established between enzymatic respiratory system of an organism and Fe atoms on the mineral surface or in the silicate lattice. Crystalline Fe-silicate minerals are less energetically favorable electron acceptors than amorphous or poorly crystalline Fe(III) oxyhydroxides. Nonetheless, the reduction of structural Fe(III) within phyllosilicate clay minerals has been documented for mesophilic, thermophilic and hyperthermophilic microorganisms (Kashefi et al., 2008; Pentráková et al., 2013). However, molecular mechanisms of this process remain poorly understood, as well as no information is still available on microbial reduction of Fe(III) within mica minerals, which are widespread in the Earth’s crust, being precursors of clay minerals in weathering processes. Comprehensively studied are the mechanisms of Fe(III) reduction from oxyhydroxides, such as ferrihydrite. Key genes determining the extracellular electron transfer to these minerals have been revealed and their products have been extensively characterized in model mesophilic proteobacteria (Shi et al., 2016). Extensive data on biochemical mechanisms for Fe(III) oxide reduction have also been obtained for two thermophilic Firmicutes (Carlson et al., 2012; Gavrilov et al., 2012) and three hyperthermophilic archaea of the family Archaeoglobaceae (Manzella et al., 2015; Mardanov et al., 2015; Smith et al., 2015), although their pathways for extracellular electron transfer remain less studied than in mesophilic models. In all the mentioned organisms three major groups of multiheme c-type cytochromes are considered to drive dissimilatory Fe(III) reduction (Shi et al., 2016), those are: cytochromes associated with the cytoplasmic membrane that accept electrons from the quinone pool of the electron transfer chain; intermediate electron-shuttling cytochromes; and a group of cytochromes associated with the outer membrane (or the S-layer), which accept electrons from the shuttles and transfer them to an Fe(III) oxide contacting the cell surface (Xiong et al., 2006; Zhang et al., 2008). Terminal step of electron transfer to Fe(III) oxides is regarded to be provided by porin-cytochrome complexes of pcc-type, first described in Geobacter species, or Mtr-type, first described in Fe-reducing Shewanella spp. Such complexes, albeit not related phylogenetically to each other, have been recently shown to be widespread among various Fe(III)-reducing and Fe(II)-oxidizing prokaryotes (see White et al., 2016 for review), although it is still unclear whether these complexes are universal for all the prokaryotes reducing Fe(III) from insoluble forms. In our work, comparative genome analysis of two strains of a single species, which differ from each other by the ability to reduce Fe(III), revealed the only gene of a cytochrome which exclusively exists in the genome of the iron-reducing strain 019. Thus, this cytochrome is supposed to determine the ability to reduce Fe(III) from both high-potential iron oxides and low-potential Fe-mica minerals. This gene appeared to encode a secreted multiheme c-type cytochrome which shares no homology with any of the components of porin-cytochrome complexes, although its heme-containing part does share homology with some cytochromes of Geobacter species. The C-terminus of the putative terminal Fe(III) reductase gene of strain 019 is homologous to secreted adhesins but not to porins or any other beta-barrel structures inherent in porin-cytochrome complexes involved in redox cycling of iron. We assume the putative Fe(III) reductase of strain 019 (CFE_2239) to appear by a fusion of genes encoding an adhesion protein and a cytochrome acquired from deltaproteobacteria by horizontal gene transfer. Most probable source organisms of this cytochrome gene belong to Geobacteraceae family (Supplementary Figure S3), well known for their Fe(III) reducing activity. This correlates with ecological data indicating proteobacteria to comprise 3–6% of prokaryotic diversity in Zavarzin hot spring (Gumerov et al., 2011; Rozanov et al., 2014), adjoining the natural habitat of strain 019. However, the reduction of structural Fe(III) from mica minerals has not been previously reported either in geobacters or in any other Fe(III) reducers.
Acquiring an Fe(III) reductase that is able to interact with Fe(III)-mica minerals by C. thermautotrophica strain 019 significantly impacts the fitness of this species in an unstable hydrothermal environment. Fe(III) is the only electron acceptor that allows the organism to proliferate at elevated redox values and utilize various organic and inorganic carbon sources and electron donors, depending on their availability. The presence of genes which are supposed to determine high- and low-potential pathways for extracellular electron transfer (ImcH-like and CbcL-like cytochromes) in strain 019 makes the organism less dependent on the form of Fe(III) available. So, upon oxygenation of the environment, enhancing the formation of Fe(III) oxides, the organism can utilize these high-potential electron acceptors both for energy conservation and redox control of its ecological microniche via Fe(II) production. A decrease of redox potential would not inhibit iron reduction in the organism, as its catabolism can be switched to another pathway for extracellular electron transfer – to a low-potential form of the acceptor, namely, to the structural Fe(III) of mica minerals, which are abundant in sedimentary environments. Both pathways for the reduction of Fe(III) minerals could be linked to the electron transfer chain in the cell membrane by CbcL-like quinol-oxidizing multiheme cytochrome complexes. Details of these pathways for Fe(III) reduction are to be studied further.
Conclusion
Our results improve current knowledge on metabolic features of deeply branching Clostridia, and indicate possible ecological importance of C. thermautotrophica in its sedimentary thermal habitats. For the first time dissimilatory reduction of structural Fe(III) from ubiquitous silicate mineral glauconite is described.
Genome analysis provided insights in such ecologically relevant properties of C. thermautotrophica as hydrogenogenic CO-trophy and Fe(III) reduction. An outstanding number of CO dehydrogenases and a novel type of putative terminal reductase of insoluble Fe(III) compounds have been identified in the organism.
The variety of [Ni,Fe]-CODHs in both C. thermautotrophica genomes is supposed to reflect high affinity of the species to this substrate, which is common gaseous component of sedimentary environments of volcanic origin. Fe(III) reducing ability of strain 019 allows it to couple carboxydotrophy with utilization of this high-potential electron acceptor in anoxic sediments. In a more global scale, coupling of Fe(III) reduction with CO oxidation by C. thermautotrophica hardwires the electron flow from carbon monoxide to the mineral constituent of the environment that could be further used as an electrical conductor, facilitating direct interspecies electron transfer (“DIET”), or as an electron-storage material, which supports microbial metabolism of other community members (Shi et al., 2016) and could be called a “biogeobattery.”
The difference in the ability of two C. thermautotrophica strains to reduce Fe(III) correlates with peculiar ecological factors encouraging strain 019 to evolve or acquire the determinants of extracellular electron transfer to Fe(III) minerals. While strain 41T was isolated from the surface layer of hot spring sediments (Sokolova et al., 2002), strain 019 was obtained from a transition (anaerobic to aerobic) zone of a core rich in Fe-bearing silicates (Rozanov et al., 2014). Further on, a biofilm-like lifestyle in a sedimentary environment with restricted free volume of the mineral phase and tight cell-to-cell contacts favor horizontal gene transfer (Madsen et al., 2012).
Further ecological studies are needed to assess the distribution of C. thermautotrophica and prokaryotes with similar phenotypes in various terrestrial hydrothermal vents, widely represented on the Eurasian continent. This would help to estimate the global role of microbial processes coupling the transformation of carbon monoxide and one of the key components of the Earth’s crust, mica minerals.
Author Contributions
SG, TS, and EB-O convened the research. ST, TS, DZ, AL, and SG designed the research. TS, DZ, and SG performed the cultivation studies. NC and VR performed the Mössbauer studies. ST, AK, and AT performed the genome sequencing and assembling. AL, ST, SG, and IK performed the genome analysis. ST, TS, DZ, NC, EB-O, IK, AL, and SG wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The work of ST, TS, DZ, EB-O, IK, AL, and SG was supported by the RSF project # 17-74-30025.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01759/full#supplementary-material
References
- Aklujkar M., Coppi M. V., Leang C., Kim B. C., Chavan M. A., Perpetua L. A., et al. (2013). Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159 515–535. 10.1099/mic.0.064089-0 [DOI] [PubMed] [Google Scholar]
- Allard P., Burton M., Muré F. (2004). High resolution FTIR sensing of magmatic gas composition during explosive eruption of primitive Etna basalt. Geophys. Res. Abstr. 6:6493. [Google Scholar]
- Balk M., Heilig H. G. H. J., van Eekert M. H. A., van Stams A. J. M., van Rijpstra I. C., van Sinninghe-Damsté J. S., et al. (2009). Isolation and characterization of a new CO-utilizing strain, Thermoanaerobacter thermohydrosulfuricus subsp. carboxydovorans, isolated from a geothermal spring in Turkey. Extremophiles 13 885–894. 10.1007/s00792-009-0276-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basen M., Müller V. (2017). “Hot” acetogenesis. Extremophiles 21 15–26. 10.1007/s00792-016-0873-3 [DOI] [PubMed] [Google Scholar]
- Bertsch J., Müller V. (2015). CO metabolism in the acetogen Acetobacterium woodii. Appl. Environ. Microbiol. 81 5949–5956. 10.1128/AEM.01772-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady A. L., Sharp C. E., Grasby S. E., Dunfield P. F. (2015). Anaerobic carboxydotrophic bacteria in geothermal springs identified using stable isotope probing. Front. Microbiol. 6:897. 10.3389/fmicb.2015.00897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookshaw D. R., Coker V. S., Lloyd J. R., Vaughan D. J., Pattrick R. A. D. (2014a). Redox interactions between Cr(VI) and Fe(II) in bioreduced biotite and chlorite. Environ. Sci. Technol. 48 11337–11342. 10.1021/es5031849 [DOI] [PubMed] [Google Scholar]
- Brookshaw D. R., Lloyd J. R., Vaughan D. J., Pattrick R. A. D. (2014b). Bioreduction of biotite and chlorite by a Shewanella species. Am. Mineral. 99 1746–1754. 10.2138/am.2014.4774ccby [DOI] [Google Scholar]
- Butler J., MacCallum I., Kleber M., Shlyakhter I. A., Belmonte M. K., Lander E. S., et al. (2008). ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18 810–820. 10.1101/gr.7337908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H. K., Iavarone A. T., Gorur A., Yeo B. S., Tran R., Melnyk R. A., et al. (2012). Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 109 1702–1707. 10.1073/pnas.1112905109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. M. A., Markowitz V. M., Chu K., Palaniappan K., Szeto E., Pillay M., et al. (2017). IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45 D507–D516. 10.1093/nar/gkw929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursolle D., Gralnick J. A. (2010). Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 77 995–1008. 10.1111/j.1365-2958.2010.07266.x [DOI] [PubMed] [Google Scholar]
- Diender M., Stams A. J. M., Sousa D. Z. (2015). Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol. 6:1275 10.3389/fmicb.2015.01275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distanov U. G. (1987). “Geological-industrial types of deposits of sedimentary siliceous rocks of the USSR. The criteria of prognosis and searches,” in The Origin and Practical Use of Siliceous Rocks. [In Russian], eds Kholodov V. N., Sadnicki V. I. (Moscow: Nauka; ), 157–167. [Google Scholar]
- Dobbek H., Svetlitchnyi V., Gremer L., Huber R., Meyer O. (2001). Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293 1281–1285. 10.1126/science.1061500 [DOI] [PubMed] [Google Scholar]
- Eroschev-Shak V. A., Zolotarev B. P., Karpov G. A. (2005). Clay minerals in present-day volcano- hydrothermal systems. Volcanol. Seismol. 4 11–24. [Google Scholar]
- Eroschev-Shak V. A., Zolotarev B. P., Karpov G. A., Grigoriev V. S., Pokrovsky B. G., Artamonov A. V. (1998). Secondary alterations of basalts and dacites in the Uzon Caldera, Kamchatka. Lithol. Miner. Resour. 2 195–206. [Google Scholar]
- Fimereli D. K., Tsirigos K. D., Litou Z. I., Liakopoulos T. D., Bagos P. G., Hamodrakas S. J. (2012). “CW-PRED: a HMM-based method for the classification of cell wall-anchored proteins of Gram-positive bacteria,” in Proceedings of the 7th Hellenic Conference on AI, SETN 2012 Artificial Intelligence: Theories and Applications, Lamia, 285–290. 10.1007/978-3-642-30448-4_36 [DOI] [Google Scholar]
- Fox J. D., Yiping H. E., Shelver D., Roberts G. P., Ludden P. W. (1996). Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J. Bacteriol. 178 6200–6208. 10.1128/jb.178.21.6200-6208.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G. (2011). Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol. 65 631–658. 10.1146/annurev-micro-090110-102801 [DOI] [PubMed] [Google Scholar]
- Fukuyama Y., Omae K., Yoneda Y., Yoshida T., Sako Y. (2017). Draft Genome Sequences of Carboxydothermus pertinax and C. islandicus, hydrogenogenic carboxydotrophic bacteria. Genome Announc. 5:e01648-16. 10.1128/genomeA.01648-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov S. N., Lloyd J. R., Kostrikina N. A., Slobodkin A. I. (2012). Fe(III) oxide reduction by a gram-positive thermophile: physiological mechanisms for dissimilatory reduction of poorly crystalline Fe(III) oxide by a thermophilic gram-positive bacterium Carboxydothermus ferrireducens. Geomicrobiol. J. 29 804–819. 10.1080/01490451.2011.635755 [DOI] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Gumerov V. M., Mardanov A. V., Beletsky A. V., Bonch-Osmolovskaya E. A., Ravin N. V. (2011). Molecular analysis of microbial diversity in the Zavarzin Spring, Uzon Caldera, Kamchatka. Microbiology 80 244–251. 10.1134/S002626171102007X [DOI] [PubMed] [Google Scholar]
- Hedderich R. (2004). Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36 65–75. 10.1023/B:JOBB.0000019599.43969.33 [DOI] [PubMed] [Google Scholar]
- Hellebrand H. J., Schade G. W. (2008). Carbon monoxide from composting due to thermal oxidation of biomass. J. Environ. Qual. 37 592–598. 10.2134/jeq2006.0429 [DOI] [PubMed] [Google Scholar]
- Henstra A. M., Stams A. J. M. (2011). Deep conversion of carbon monoxide to hydrogen and formation of acetate by the anaerobic thermophile Carboxydothermus hydrogenoformans. Int. J. Microbiol. 2011:641582. 10.1155/2011/641582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntemann M., Ivanova N. N., Mavromatis K., James Tripp H., Paez-Espino D., Palaniappan K., et al. (2015). The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v.4). Stand. Genomic Sci. 10 4–9. 10.1186/s40793-015-0077-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Takao K., Yoshida T., Wada K., Daifuku T., Yoneda Y., et al. (2013). Cysteine 295 indirectly affects Ni coordination of carbon monoxide dehydrogenase-II C-cluster. Biochem. Biophys. Res. Commun. 441 13–17. 10.1016/j.bbrc.2013.09.143 [DOI] [PubMed] [Google Scholar]
- Jeoung J., Fesseler J., Goetzl S., Dobbek H. (2014). Carbon monoxide. Toxic gas and fuel for anaerobes and aerobes: carbon monoxide dehydrogenases. Met. Ions Life Sci. 14 37–69. 10.1007/978-94-017-9269-1_3 [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Karl M. M., Poehlein A., Bengelsdorf F. R., Daniel R., Dürre P. (2017). Complete genome sequence of the autotrophic acetogen Clostridium formicaceticum DSM 92T using nanopore and illumina sequencing data. Genome Announc. 5:e00423-17. 10.1128/genomeA.00423-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi K., Shelobolina E. S., Elliott W. C., Lovley D. R. (2008). Growth of thermophilic and hyperthermophilic Fe(III)-reducing microorganisms on a ferruginous smectite as the sole electron acceptor. Appl. Environ. Microbiol. 74 251–258. 10.1128/AEM.01580-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerby R. L., Ludden P. W., Roberts G. P. (1997). In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J. Bacteriol. 179 2259–2266. 10.1128/jb.179.7.2259-2266.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevbrin V. V., Zavarzin G. A. (1992). The effect of sulfur compounds on growth of halophilic homoacetic bacterium Acetohalobium arabaticum. Microbiology 61 812–817. [Google Scholar]
- King G. M., Weber C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5 107–118. 10.1038/nrmicro1595 [DOI] [PubMed] [Google Scholar]
- Kobayashi I. (2001). Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29 3742–3756. 10.1093/nar/29.18.3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetkova T. V., Rusanov I. I., Pimenov N. V., Kolganova T. V., Lebedinsky A. V., Bonch-Osmolovskaya E. A., et al. (2011). Anaerobic transformation of carbon monoxide by microbial communities of Kamchatka hot springs. Extremophiles 15 319–325. 10.1007/s00792-011-0362-7 [DOI] [PubMed] [Google Scholar]
- Koonin E. V. (2011). The Logic of Chance: The Nature and Origin of Biological Evolution. Upper Saddle River, NJ: FT Press Science, 295. [Google Scholar]
- Kozhevnikova D. A., Taranov E. A., Lebedinsky A. V., Bonch-Osmolovskaya E. A., Sokolova T. G. (2016). Hydrogenogenic and sulfidogenic growth of thermococcus archaea on carbon monoxide and formate. Microbiology 85 400–410. 10.1134/S0026261716040135 [DOI] [PubMed] [Google Scholar]
- Leggett R. M., Clavijo B. J., Clissold L., Clark M. D., Caccamo M. (2014). Next clip: an analysis and read preparation tool for nextera long mate pair libraries. Bioinformatics 30 566–568. 10.1093/bioinformatics/btt702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levar C. E., Hoffman C. L., Dunshee A. J., Toner B. M., Bond D. R. (2017). Redox potential as a master variable controlling pathways of metal reduction by Geobacter sulfurreducens. ISME J. 11 741–752. 10.1038/ismej.2016.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. K., Kang S. G., Lebedinsky A. V., Lee J. H., Lee H. S. (2010). Identification of a novel class of membrane-bound [NiFe]-Hydrogenases in Thermococcus onnurineus NA1 by in silico analysis. Appl. Environ. Microbiol. 76 6286–6289. 10.1128/AEM.00123-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen J. S., Burmølle M., Hansen L. H., Sørensen S. J. (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65 183–195. 10.1111/j.1574-695X.2012.00960.x [DOI] [PubMed] [Google Scholar]
- Mall A., Sobotta J., Huber C., Tschirner C., Kowarschik S., Bačnik K., et al. (2018). Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359 563–567. 10.1126/science.aao2410 [DOI] [PubMed] [Google Scholar]
- Manzella M. P., Holmes D. E., Rocheleau J. M., Chung A., Reguera G., Kashefi K. (2015). The complete genome sequence and emendation of the hyperthermophilic, obligate iron-reducing archaeon “Geoglobus ahangari” strain 234T. Stand. Genomic Sci. 10:77. 10.1186/s40793-015-0035-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov A. V., Slododkina G. B., Slobodkin A. I., Beletsky A. V., Gavrilov S. N., Kublanov I. V., et al. (2015). The Geoglobus acetivorans genome: Fe(III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 81 1003–1012. 10.1128/AEM.02705-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsnev M. E., Rusakov V. S. (2014). Study of spatial spin-modulated structures by Mössbauer spectroscopy using SpectrRelax. AIP Conf. Proc. 1622 40–49. 10.1063/1.4898609 [DOI] [Google Scholar]
- Menyailov I. A., Nikitina L. P. (1980). Chemistry and metal contents of magmatic gases: the new Tolbachik volcanoes case (Kamchatka). Bull. Volcanol. 43 195–205. 10.1007/BF02597621 [DOI] [Google Scholar]
- Mock J., Wang S., Huang H., Kahnt J., Thauer R. K. (2014). Evidence for a hexaheteromeric methylenetetrahydrofolate reductase in Moorella thermoacetica. J. Bacteriol. 196 3303–3314. 10.1128/JB.01839-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoura T., Chikaraishi Y., Izaki R., Suwa T., Sato T., Harada T., et al. (2018). A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 359 559–563. 10.1126/science.aao3407 [DOI] [PubMed] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A. A., Korobeynikov A., Lapidus A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20 714–737. 10.1089/cmb.2013.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschläger E., Rother M. (2008). Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch. Microbiol. 190 257–269. 10.1007/s00203-008-0382-6 [DOI] [PubMed] [Google Scholar]
- Oger P., Sokolova T. G., Kozhevnikova D. A., Taranov E. A., Vannier P., Lee H. S., et al. (2016). Complete genome sequence of the hyperthermophilic and piezophilic archaeon Thermococcus barophilus Ch 5, capable of growth at the expense of hydrogenogenesis from carbon monoxide and formate. Genome Announc. 4:e01534-15 10.1128/genomeA.01534-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omae K., Yoneda Y., Fukuyama Y., Yoshida T., Sako Y. (2017). Genomic analysis of Calderihabitans maritimus KKC 1, a thermophilic, hydrogenogenic, carboxydotrophic bacterium isolated from marine sediment. Appl. Environ. Microbiol. 83:e00832-17. 10.1128/AEM.00832-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentráková L., Su K., Pentrák M., Stucki J. W. (2013). A review of microbial redox interactions with structural Fe in clay minerals. Clay Miner. 48 543–560. 10.1180/claymin.2013.048.3.10 [DOI] [Google Scholar]
- Pierce E., Xie G., Barabote R. D., Saunders E., Han C. S., Detter J. C., et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10 2550–2573. 10.1111/j.1462-2920.2008.01679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G. P., Thorsteinsson M. V., Kerby R. L., Lanzilotta W. N., Poulos T. (2001). CooA: a heme-containing regulatory protein that serves as a specific sensor of both carbon monoxide and redox state. Prog. Nucleic Acid Res. Mol. Biol. 67 35–63. 10.1016/S0079-6603(01)67024-7 [DOI] [PubMed] [Google Scholar]
- Rozanov A. S., Bryanskaya A. V., Malup T. K., Meshcheryakova I. A., Lazareva E. V., Taran O. P., et al. (2014). Molecular analysis of the benthos microbial community in Zavarzin thermal spring (Uzon Caldera, Kamchatka, Russia). BMC Genomics 15:S12. 10.1186/1471-2164-15-S12-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna F. H., Lebedinsky A. V., Sokolova T. G., Robb F. T., Gonzalez J. M. (2015). Analysis of three genomes within the thermophilic bacterial species Caldanaerobacter subterraneus with a focus on carbon monoxide dehydrogenase evolution and hydrolase diversity. BMC Genomics 16:757. 10.1186/s12864-015-1955-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchmann K., Müller V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12 809–821. 10.1038/nrmicro3365 [DOI] [PubMed] [Google Scholar]
- Sharma S., Cavallaro G., Rosato A. (2010). A systematic investigation of multiheme c-type cytochromes in prokaryotes. J. Biol. Inorg. Chem. 15 559–571. 10.1007/s00775-010-0623-4 [DOI] [PubMed] [Google Scholar]
- Shi L., Dong H., Reguera G., Beyenal H., Lu A., Liu J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14 651–662. 10.1038/nrmicro.2016.93 [DOI] [PubMed] [Google Scholar]
- Shi L., Fredrickson J. K., Zachara J. M. (2014). Genomic analyses of bacterial porin-cytochrome gene clusters. Front. Microbiol. 5:657. 10.3389/fmicb.2014.00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Rosso K. M., Zachara J. M., Fredrickson J. K. (2012). Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem. Soc. Trans. 40 1261–1267. 10.1042/BST20120098 [DOI] [PubMed] [Google Scholar]
- Shock E. L., Holland M., Meyer-Dombard D., Amend J. P., Osburn G. R., Fischer T. P. (2010). Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, USA. Geochim. Cosmochim. Acta 74 4005–4043. 10.1016/j.gca.2009.08.036 [DOI] [Google Scholar]
- Shock E. L., Holland M., Meyer-Dombard D. R., Amend J. P. (2005). “Geochemical sources of energy for microbial metabolism in hydrothermal ecosystems: Obsidian Pool, Yellowstone National Park,” in Geothermal Biology and Geochemistry in Yellowstone National Park Thermal Biology Institute, eds Inskeep W. P., McDermott T. R. (Bozeman, MT: Montana State University; ), 95–112 [Google Scholar]
- Slepova T. V., Rusanov I. I., Sokolova T. G., Bonch-Osmolovskaia E. A., Pimenov N. V. (2007). Radioisotopic assays of rates of carbon monoxide conversion by anaerobic thermophilic prokaryotes. Microbiology 76 523–529. 10.1134/S0026261707050025 [DOI] [PubMed] [Google Scholar]
- Slepova T. V., Sokolova T. G., Kolganova T. V., Tourova T. P., Bonch-Osmolovskaya E. A. (2009). Carboxydothermus siderophilus sp. nov., a thermophilic, hydrogenogenic, carboxydotrophic, dissimilatory Fe(III)-reducing bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 59 213–217. 10.1099/ijs.0.000620-0 [DOI] [PubMed] [Google Scholar]
- Slepova T. V., Sokolova T. G., Lysenko A. M., Tourova T. P., Kolganova T. V., Kamzolkina O. V., et al. (2006). Carboxydocella sporoproducens sp. nov., a novel anaerobic CO-utilizing/H2-producing thermophilic bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 56 797–800. 10.1099/ijs.0.63961-0 [DOI] [PubMed] [Google Scholar]
- Slobodkin A. I., Sokolova T. G., Lysenko A. M., Wiegel J. (2006). Reclassification of Thermoterrabacterium ferrireducens as Carboxydothermus ferrireducens comb. nov., and emended description of the genus Carboxydothermus. Int. J. Syst. Evol. Microbiol. 56 2349–2351. 10.1099/ijs.0.64503-0 [DOI] [PubMed] [Google Scholar]
- Slobodkina G. B., Panteleeva A. N., Sokolova T. G., Bonch-Osmolovskaya E. A., Slobodkin A. I. (2012). Carboxydocella manganica sp. nov., a thermophilic, dissimilatory Mn(IV)- and Fe(III)-reducing bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 62 890–894. 10.1099/ijs.0.027623-0 [DOI] [PubMed] [Google Scholar]
- Smith J. A., Aklujkar M., Risso C., Leang C., Giloteaux L., Holmes D. E. (2015). Mechanisms involved in Fe(III) respiration by the hyperthermophilic archaeon Ferroglobus placidus. Appl. Environ. Microbiol. 81 2735–2744. 10.1128/AEM.04038-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboh B., Linder D., Hedderich R. (2002). Purification and catalytic properties of a CO-oxidizing:H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur. J. Biochem. 269 5712–5721. 10.1046/j.1432-1033.2002.03282.x [DOI] [PubMed] [Google Scholar]
- Sokolova T., Lebedinsky A. (2013). “CO-Oxidizing anaerobic thermophilic prokaryotes,” in Thermophilic Microbes in Environmental and Industrial Biotechnology. Biotechnology of Thermophiles, eds Satyanarayana T., Littlechild J., Kawarabayasi Y. (Dordrecht: Springer; ), 203–231. 10.1007/978-94-007-5899-5_7 [DOI] [Google Scholar]
- Sokolova T. G. (2015). “Carboxydocella,” in Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons, Inc, 1–4. [Google Scholar]
- Sokolova T. G., González J. M., Kostrikina N. A., Chernyh N. A., Slepova T. V., Bonch-Osmolovskaya E. A., et al. (2004). Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 54 2353–2359. 10.1099/ijs.0.63186-0 [DOI] [PubMed] [Google Scholar]
- Sokolova T. G., Henstra A. M., Sipma J., Parshina S. N., Stams A. J. M., Lebedinsky A. V. (2009). Diversity and ecophysiological features of thermophilic carboxydotrophic anaerobes. FEMS Microbiol. Ecol. 68 131–141. 10.1111/j.1574-6941.2009.00663.x [DOI] [PubMed] [Google Scholar]
- Sokolova T. G., Kostrikina N. A., Chernyh N. A., Tourova T. P., Kolganova T. V., Bonch-Osmolovskaya E. A. (2002). Carboxydocella thermoautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring. Int. J. Syst. Evol. Microbiol. 52 1961–1967. 10.1099/ijs.0.02173-0 [DOI] [PubMed] [Google Scholar]
- Svetlitchnyi V., Dobbek H., Meyer-Klaucke W., Meins T., Thiele B., Römer P., et al. (2004). A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans. Proc. Natl. Acad. Sci. U.S.A. 101 446–451. 10.1073/pnas.0304262101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlitchnyi V., Peschel C., Acker G., Meyer O. (2001). Two membrane-associated NiFeS-carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 183 5134–5144. 10.1128/JB.183.17.5134-5144.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds R. B., Rose W. I., Bluth G. J. S., Gerlach T. M. (1994). Volcanic gas studies- methods, results, and applications. Rev. Mineral. 30 1–66. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techtmann S. M., Lebedinsky A. V., Colman A. S., Sokolova T. G., Woyke T., Goodwin L., et al. (2012). Evidence for horizontal gene transfer of anaerobic carbon monoxide dehydrogenases. Front. Microbiol. 3:132. 10.3389/fmicb.2012.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiquia-Arashiro S. M. (2014). Thermophilic Carboxydotrophs and their Applications in Biotechnology (Extremophilic Bacteria). Berlin: Springer, 131. [Google Scholar]
- Toshchakov S. V., Kublanov I. V., Messina E., Yakimov M. M., Golyshin P. N. (2015). “Genomic analysis of pure cultures and communities,” in Hydrocarbon and Lipid Microbiology Protocols. Springer Protocols Handbooks, eds McGenity T., Timmis K., Nogales B. (Berlin: Springer; ), 5–27. 10.1007/8623_2015_126 [DOI] [Google Scholar]
- Visser M., Parshina S. N., Alves J. I., Sousa D. Z., Pereira A. C., Muyzer G., et al. (2014). Genome analyses of the carboxydotrophic sulfate-reducers Desulfotomaculum nigrificans and Desulfotomaculum carboxydivorans and reclassification of Desulfotomaculum caboxydivorans as a later synonym of Desulfotomaculum nigrificans. Stand. Genomic Sci. 9 655–675. 10.4056/sig [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrousek K., Noble S., Korlach J., Chen J., Eckert C., Yu J., et al. (2014). Genome annotation provides insight into carbon monoxide and hydrogen metabolism in Rubrivivax gelatinosus. PLoS One 9:e114551. 10.1371/journal.pone.0114551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White G. F., Edwards M. J., Gomez-Perez L., Richardson D. J., Butt J. N., Clarke T. A. (2016). Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 68 87–138. 10.1016/bs.ampbs.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Wolin E. A., Wolin M. J., Wolfe R. S. (1963). Formation of methane by bacterial extracts. J. Biol. Chem. 238 2882–2886. 10.1016/S0016-0032(13)90081-8 [DOI] [PubMed] [Google Scholar]
- Wu M., Ren Q., Durkin A. S., Daugherty S. C., Brinkac L. M., Dodson R. J., et al. (2005). Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1:e65. 10.1371/journal.pgen.0010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Shi L., Chen B., Mayer M. U., Lower B. H., Londer Y., et al. (2006). High-affinity binding and direct electron transfer to solid metals by purified metal reducing protein OmcA Decaheme cytochrome. J. Am. Chem. Soc. 128 13978–13979. 10.1021/ja063526d [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Kano S. I., Yoshida T., Ikeda E., Fukuyama Y., Omae K., et al. (2015). Detection of anaerobic carbon monoxide-oxidizing thermophiles in hydrothermal environments. FEMS Microbiol. Ecol. 91 1–9. 10.1093/femsec/fiv093 [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Yoshida T., Kawaichi S., Daifuku T., Takabe K., Sako Y. (2012). Carboxydothermus pertinax sp. nov., a thermophilic, hydrogenogenic, Fe(III)-reducing, sulfur-reducing carboxydotrophic bacterium from an acidic hot spring. Int. J. Syst. Evol. Microbiol. 62 1692–1697. 10.1099/ijs.0.031583-0 [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Yoshida T., Yasuda H., Imada C., Sako Y. (2013). A thermophilic, hydrogenogenic and carboxydotrophic bacterium, Calderihabitans maritimus gen. nov., sp. nov., from a marine sediment core of an undersea caldera. Int. J. Syst. Evol. Microbiol. 63 3602–3608. 10.1099/ijs.0.050468-0 [DOI] [PubMed] [Google Scholar]
- Zacharoff L., Chan C. H., Bond D. R. (2016). Reduction of low potential electron acceptors requires the CbcL inner membrane cytochrome of Geobacter sulfurreducens. Bioelectrochemistry 107 7–13. 10.1016/j.bioelechem.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Zavarzina D. G., Chistyakova N. I., Shapkin A. V., Savenko A. V., Zhilina T. N., Kevbrin V. V., et al. (2016). Oxidative biotransformation of biotite and glauconite by alkaliphilic anaerobes: the effect of Fe oxidation on the weathering of phyllosilicates. Chem. Geol. 439 98–109. 10.1016/j.chemgeo.2016.06.015 [DOI] [Google Scholar]
- Zavarzina D. G., Sokolova T. G., Tourova T. P., Chernyh N. A., Kostrikina N. A., Bonch-Osmolovskaya E. A. (2007). Thermincola ferriacetica sp. nov., a new anaerobic, thermophilic, facultatively chemolithoautotrophic bacterium capable of dissimilatory Fe(III) reduction. Extremophiles 11 1–7. 10.1007/s00792-006-0004-7 [DOI] [PubMed] [Google Scholar]
- Zhang H., Tang X., Munske G. R., Zakharova N., Zheng C., Wolff M. A., et al. (2008). In vivo identification of outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel chemical cross-linkers. J. Proteome Res. 7 1712–1720. 10.1021/pr7007658 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.