Abstract
Polyploidy is one of the main forces that drives the evolution of plants and provides great advantages for breeding. Somatic hybridization by protoplast fusion is used in citrus breeding programs. This method allows combining the whole parental genomes in a single genotype, adding complementary dominant characters, regardless of parental heterozygosity. It also contributes to surpass limitations imposed by reproductive biology and quickly generates progenies that combine the required traits. Two allotetraploid somatic hybrids recovered from the citrus rootstocks—Citrus macrophylla (CM) and Carrizo citrange (CC)—were characterized for morphology, genome composition using molecular markers (SNP, SSR, and InDel), and their tolerance to iron chlorosis, salinity, and Citrus tristeza virus (CTV). Both hybrids combine the whole parental genomes even though the loss of parental alleles was detected in most linkage groups. Mitochondrial genome was inherited from CM in both the hybrids, whereas recombination was observed for chloroplastic genome. Thus, somatic hybrids differ from each other in their genome composition, indicating that losses and rearrangements occurred during the fusion process. Both inherited the tolerance to stem pitting caused by CTV from CC, are tolerant to iron chlorosis such as CM, and have a higher tolerance to salinity than the sensitive CC. These hybrids have potential as improved rootstocks to grow citrus in areas with calcareous and saline soils where CTV is present, such as the Mediterranean region. The provided knowledge on the effects of somatic hybridization on the genome composition, anatomy, and physiology of citrus rootstocks will be key for breeding programs that aim to address current and future needs of the citrus industry.
Keywords: rootstock breeding, protoplast fusion, genome instability, tetraploid, polyploid
Introduction
The Mediterranean basin is ranked first among regions in the export of fresh market citrus fruits (FAO, 2016). This region has some adverse biotic and abiotic conditions that affect citrus cultivation. The rootstock is a key element for citrus production because it can confer tolerance to these constraints. The graft-transmissible disease tristeza, caused by the Citrus tristeza virus (CTV), is one of the most important limiting factors and has a strong economic impact that necessitates dramatic changes in citrus production (Cambra et al., 2000; Moreno et al., 2008). The damage brought by CTV is caused by the scion-rootstock combination, the CTV strain, and the environmental conditions (Ballester-Olmos et al., 1993). Most agricultural lands in the Mediterranean basin have two soil limiting conditions: alkalinity and, to a lesser extent, salinity. Soil alkalinity was traditionally managed using sour orange (SO) or Citrus aurantium L. as rootstock. Nevertheless, SO is very sensitive to Quick Decline disease caused by CTV. This limiting condition has forced the use of alternative rootstocks despite the highly desirable agronomic traits that SO induces to citrus trees. Among the main rootstocks, Cleopatra mandarin (C. reshni Hort. ex Tan.) and C. macrophylla W. (CM) are tolerant to calcareous soils, although CM is sensitive to severe CTV strains (Cambra et al., 2000). One of the main rootstocks used worldwide is Carrizo citrange (CC) [C. sinensis (L.) Osb. × Poncirus trifoliata (L.) Raf.] which is tolerant to CTV but sensitive to iron chlorosis in alkaline soils (Castle et al., 2009). Citrus is among the most salt-sensitive perennial crops (Maas, 1993). The tolerance of citrus trees to soil salinity depends greatly on the rootstock ability to restrict ion transport to the scion and this is a heritable trait (Walker, 1986). Cleopatra mandarin and CM are suited for saline soils because they restrict ion transport to the aerial part, whereas CC is sensitive to this condition as it quickly accumulates the ions and reaches toxic concentrations (Gomez-Cadenas et al., 2003). CC is considered a good rootstock for inducing high yield, big fruit size and high fruit quality to the grafted variety. CM induces vigor to citrus trees, early bearing, very high yield, and has an excellent adaptation to calcareous and saline soils. However, this rootstock is sensitive to cold temperatures, moderately sensitive to CTV, and reduces fruit quality. Therefore, CC and CM, used as citrus rootstocks, have complementary characteristics.
Rootstock breeding programs are carried out by sexual or somatic hybridization. The recovery of rootstock hybrids by sexual hybridization is hampered by citrus reproductive biology (apomixis) and the high heterozygosity of the citrus genomes. Most citrus genotypes are apomictic, except for citrons (C. medica L.), pummelos (C. maxima (L.) Osb.), clementines (C. clementina Hort. ex Tan.), and some mandarin hybrids. The seeds of non-apomictic genotypes generally contain only one sexual embryo, whereas the seeds of apomictic genotypes generally contain one sexual embryo and one or more nucellar embryos. The development of nucellar embryos in citrus apomictic genotypes can be initiated before fertilization, and the competition between the zygotic and nucellar embryos often results in the failure of the zygotic embryos (Wakana and Uemoto, 1987; Koltunow, 1993). In addition, the high heterozygosity level of citrus species (Herrero et al., 1996; Ollitrault et al., 2003; Barkley et al., 2006) produces a wide segregation pattern of parental traits in the progenies. The probability of having individuals that combine all the desired traits is usually very low. Therefore, a large number of individuals in these progenies need to be evaluated to find and select those that combine the desirable characteristics of the two parents. In contrast, somatic hybridization by protoplast fusion allows combining the genomes of both parents in only one genotype regardless of their level of heterozygosity, adding their dominant complementary characters (Ollitrault et al., 2000) and to overcome the sexual incompatibility between parents. This methodology is used worldwide for rootstock breeding (Grosser et al., 2000; Ollitrault et al., 2007; Dambier et al., 2011; Grosser and Gmitter, 2011).
Somatic hybridization in citrus is performed by the fusion of protoplasts derived from leaf mesophyll with protoplasts derived from embryogenic callus. In citrus, it has not yet been possible to regenerate plants from leaf protoplasts. Protoplasts isolated from embryogenic callus or leaf protoplasts that incorporate the mitochondrial genome from callus protoplasts are the only ones that have the ability to produce embryos and subsequently, plants (Kobayashi et al., 1991; Grosser and Gmitter, 2005; Guo et al., 2007). Therefore, it is necessary to have different callus lines of rootstock genotypes with favorable traits for the establishment of rootstock breeding programs based on somatic hybridization. Embryogenic callus can be easily obtained in apomictic mandarins and sweet oranges by in vitro ovule culture (Rangan et al., 1969; Ollitrault et al., 1994; Perez et al., 1998). However, it can be very difficult or has never been achieved in other genotypes that are essential for rootstock breeding such as SO, P. trifoliata, and the interspecific hybrids citranges and citrumelos (C. paradisi Macf. × P. trifoliata). Selective agents are not needed to select citrus somatic hybrids after somatic hybridizations. Instead, potential hybrids are identified among all the regenerated plants by ploidy and genetic composition analyses. The genetic analysis is often performed using a small number of molecular markers (Guo et al., 2007; Dambier et al., 2011; Grosser and Gmitter, 2011) that display the complementary allelic configuration of the parents disregarding their homogeneous distribution in the different linkage groups (LGs), hence impeding a detailed study of chromosome stability. Besides molecular analysis, a large number of plants is required for detailed physiological and agronomical evaluations of the somatic hybrids to determine their potential utility as new rootstocks (Dambier et al., 2011). Somatic hybrids go through a long juvenile phase, which often takes more than 6 years, delaying the production of seeds to obtain the plants needed to carry out the experiments (Krajewski and Rabe, 1995). This citrus juvenile phase is one of the main constraints in rootstock breeding programs. However, in vitro micropropagation allows the generation of a large number of clonal plants in a short time, avoiding the delay that juvenility would impose, which is a great advantage for rootstock breeding programs (Bordas et al., 2015). In this study, the genetic composition of two somatic hybrids, obtained by CM and CC protoplast fusion (Pensabene-Bellavia et al., 2015), was analyzed using single sequence repeats (SSR), single nucleotide polymorphism (SNP), and insertion or deletion (InDel) markers. Both somatic hybrids were morphologically described and their behavior was evaluated under salinity, iron deficiency, and CTV inoculation. The main objective of this study was to perform an early and detailed evaluation of the somatic hybrids to determine their potential utility as rootstocks for the Mediterranean citrus industry.
Materials and methods
Plant material and greenhouse conditions
Diploid CC and CM and allotetraploid somatic hybrids SMC-58 and SMC-73 were used for the experiments. Diploid CC and CM seeds were collected from the Citrus Germplasm Bank of pathogen-free plants at the Instituto Valenciano de Investigaciones Agrarias (IVIA) (Navarro et al., 2002; Navarro, 2015) and somatic hybrids SMC-58 and SMC-73 were recovered by protoplast fusion isolated from CM embryogenic callus and CC leaf mesophyll leaves (Pensabene-Bellavia et al., 2015). The somatic hybrids were micropropagated by Agromillora Research S.L. using the methodology described by Bordas et al. (2015). The seeds of CC and CM were germinated in a greenhouse using a sterile substrate composed of peat, coconut fiber, and perlite (50:25:20:5), supplemented with 1.38 g kg−1 of calcium superphosphate, and irrigated twice weekly with the Hoagland and Arnon (1950) nutrient solution modified for citrus (5 mM Ca(NO3)2, 1.4 mM KNO3, 2 mM MgSO4, 0.6 mM H3PO4, 20 μM Fe-EDDHA, 7.6 μM ZnSO4·7H2O, 0.50 μM CuSO4·5H2O, 50 μM H3BO3, 0.50 μMMoO3, and 54 μM MnSO4·H2O). The pH of the nutrient solution was adjusted to 6.0 with 1 M of KOH or H2SO4. After eight weeks, homogeneous seedlings, which were selected based on size uniformity, were transplanted individually to opaque plastic 0.5 L pots filled with a substrate composed of peat, coconut fiber, sand, and perlite (40:25:25:10). Seedlings and micropropagated plants of similar size were then randomized over the experimental area. A row of plants, not included in the experiment, was placed around the perimeter as a border. Plants were grown under greenhouse conditions with supplementary light (250 μmol m−2 s−1, 400–700 nm) to extend the photoperiod to 16 h. The temperature ranges were 16–18°C at night and 26–28°C during the day. Relative humidity (RH) was maintained at around 80%.
Genetic characterization
Nuclear genomes were characterized using 23 SSR and 59 SNP markers selected from the 9 LG of the Clementine genetic map (Ollitrault et al., 2012a; Tables 1–4). Cytoplasmic genomes were characterized with 3 mitochondrial InDel markers, 5/rrn18-1 (Duminil et al., 2002), nad2/4-3, and nad7/1-2 (Froelicher et al., 2011), and 5 chloroplastic SSR markers: NTCP7, NTCP9 CCMP2, CCMP5 (Cheng et al., 2005), and CCMP6 (Bryan et al., 1999; Weising and Gardner, 1999; Table 4). All the analyses were performed in the somatic hybrids, the parents (CM and CC), and the CM embryogenic callus used for protoplast fusion. For these characterizations, genomic DNA was isolated using the methodology described by Dellaporta et al. (1983) with few modifications (0.5 M EDTA, pH 8.0, 1 M Tris-HCl, pH 8.0, 5 M NaCl, 2% MATAB, 1% PEG 6000 and 0.5% Na2SO3) and was performed using different DNA extractions from leaves of different branches of both somatic hybrids.
Table 1.
Molecular markers analyzed indicating the type of marker, locus name, linkage groups (LGs) 1 and 2 and location within LG in centimorgans (cM), bibliographic reference in the literature, and GeneBank accession.
| Type | Locus | Location | References | ||
|---|---|---|---|---|---|
| LG | cM | Bibliography | GeneBank | ||
| SNP | *CiC4827-01 | 1 | 20.5 | Ollitrault et al., 2012b | ET072918 |
| SNP | CiC2110-01 | 1 | 28.8 | Ollitrault et al., 2012b | ET099643 |
| SSR | CiBE5720 | 1 | 58.5 | Ollitrault et al., 2010b | ET082224 |
| SNP | *CiC4581-01 | 1 | 63.7 | Ollitrault et al., 2012b | ET109034 |
| SNP | *ACO-P353 | 1 | 80.4 | Ollitrault et al., 2012a | JX630066 |
| SNP | ACO-C601 | 1 | 83.4 | Ollitrault et al., 2012a | JX630065 |
| SNP | CiC0599-01 | 1 | 102.4 | Ollitrault et al., 2012b | ET093125 |
| SNP | TSC-C80 | 1 | 111.6 | García-Lor et al., 2013a | JX630084 |
| SSR | JK-taa15 | 1 | 119.7 | Kijas et al., 1997 | none |
| SNP | *F3H-M309 | 2 | 19.6 | García-Lor et al., 2013a | JX630066 |
| SNP | *F3H-C341 | 2 | 20.0 | García-Lor et al., 2013a | JX630067 |
| SNP | *F3H-P30 | 2 | 20.0 | García-Lor et al., 2013a | JX630066 |
| SNP | *PEPC-M316 | 2 | 32.6 | García-Lor et al., 2013a | JX630067 |
| SNP | PEPC-C328 | 2 | 32.6 | García-Lor et al., 2013a | JX630067 |
| SSR | mCrCIR07D05 | 2 | 75.6 | Cuenca et al., 2011 | FR677574 |
| SNP | SOS1-M50 | 2 | 78.5 | García-Lor et al., 2013a | JX630068 |
| SNP | *CiC3712-01 | 2 | 93.9 | Ollitrault et al., 2012b | ET079481 |
| SNP | CCC1-P727 | 2 | 110.9 | García-Lor et al., 2013a | JX630069 |
| SNP | *CCC1-M85 | 2 | 110.9 | García-Lor et al., 2013a | JX630069 |
| SSR | JK-TAA41 | 2 | 131.8 | Kijas et al., 1997 | none |
| SNP | *PKF-C64 | 2 | 131.2 | García-Lor et al., 2013a | JX630076 |
| SNP | TRPA-M593 | 2 | 132.3 | García-Lor et al., 2013a | JX630070 |
| SNP | PKF-M186 | 2 | 133.5 | García-Lor et al., 2013a | JX630076 |
Non-polymorphic markers.
Table 4.
Molecular markers analyzed indicating the type of marker, locus name, linkage groups (LGs) 8 and 9 and location within LG in centimorgans (cM), bibliographic reference in the literature, and GeneBank accession.
| Type | Locus | Location | References | ||
|---|---|---|---|---|---|
| LG | cM | Literature | GeneBank | ||
| SSR | mCrCIR07B05 | 8 | 31.7 | Froelicher et al., 2011 | AM489747 |
| SSR | CiBE0214 | 8 | 40.4 | Ollitrault et al., 2010b | ET088913 |
| SNP | CiC5164-02 | 8 | 45.6 | Ollitrault et al., 2012b | ET111943 |
| SNP | CiC1749-05 | 8 | 103 | Ollitrault et al., 2012b | ET097636 |
| SNP | *CiC4876-07 | 9 | 2.7 | Ollitrault et al., 2012b | ET080580 |
| SNP | *CiC5087-01 | 9 | 15.9 | Ollitrault et al., 2012b | ET111514 |
| SSR | mCrCIR07F11 | 9 | 49.6 | Kamiri et al., 2011 | FR677567 |
| SNP | CiC2518-02 | 9 | 53.5 | Ollitrault et al., 2012b | ET101955 |
| SSR | Ci08C05 | 9 | 55.1 | Froelicher et al., 2011 | AJ567415 |
| SNP | *LCYB-P736 | 9 | 78.9 | García-Lor et al., 2013a | JX630084 |
| SNP | LCYB-M480 | 9 | 78.9 | García-Lor et al., 2013a | JX630084 |
| SNP | HYB-M62 | 9 | 102.3 | García-Lor et al., 2013a | AF315289 |
| SNP | HYB-C433 | 9 | 102.3 | García-Lor et al., 2013a | JX630087 |
| Indel | nad2/4-3 | Mitocondrial | Froelicher et al., 2011 | ||
| Indel | * nad7/1-2 | Mitocondrial | Froelicher et al., 2011 | ||
| Indel | * 5/rrn18-1 | Mitocondrial | Duminil et al., 2002 | ||
| cpSSR | CCMP2 | Chloroplastic | Cheng et al., 2003 | ||
| cpSSR | CCMP5 | Chloroplastic | Cheng et al., 2003 | ||
| cpSSR | CCMP6 | Chloroplastic | Cheng et al., 2003 | ||
| cpSSR | NTCP7 | Chloroplastic | Cheng et al., 2003 | ||
| cpSSR | * NTCP9 | Chloroplastic | Cheng et al., 2003 | ||
Non-polymorphic markers.
Table 2.
Molecular markers analyzed indicating the type of marker, locus name, linkage groups (LGs) 3, 4, and 5 and location within LG in centimorgans (cM), bibliographic reference in the literature, and GeneBank accession.
| Type | Locus | Location | References | ||
|---|---|---|---|---|---|
| LG | cM | Literature | GeneBank | ||
| SNP | INVA-M437 | 3 | 30.2 | García-Lor et al., 2013a | JX630071 |
| SNP | MDH-M519 | 3 | 34.8 | García-Lor et al., 2013a | JX630072 |
| SNP | *MDH-MP69 | 3 | 34.8 | García-Lor et al., 2013a | JX630072 |
| SNP | *CiC4681-02 | 3 | 92.8 | Ollitrault et al., 2012b | ET109640 |
| SNP | NCED3-M535 | 3 | 101.3 | García-Lor et al., 2013a | JX630086 |
| SNP | CiC5796-12 | 3 | 109.9 | Ollitrault et al., 2012b | ET0822752 |
| SNP | ATMR-M728 | 3 | 141.9 | García-Lor et al., 2013a | JX630073 |
| SNP | ATMR-C372 | 3 | 141.9 | García-Lor et al., 2013a | JX630073 |
| SSR | Ci08A10 | 3 | 144.9 | Froelicher et al., 2011 | AJ567414 |
| SNP | CHS-M183 | 3 | 167.3 | García-Lor et al., 2013a | JX630074 |
| SNP | *CHS-P57 | 3 | 167.3 | García-Lor et al., 2013a | JX630074 |
| SNP | *CiC4240-04 | 4 | 7.1 | Ollitrault et al., 2012b | ET106812 |
| SNP | CHI-M598 | 4 | 11.0 | García-Lor et al., 2013a | JX630075 |
| SSR | mCrCIR07D06 | 4 | 16.3 | Cuenca et al., 2011 | FR677581 |
| SNP | CiC2840-01 | 4 | 17.0 | Ollitrault et al., 2012b | ET103429 |
| SNP | CiC3740-02 | 4 | 43.9 | Ollitrault et al., 2012b | ET079647 |
| SSR | mCrCIR03G05 | 4 | 75.1 | Cuenca et al., 2011 | FR677578.1 |
| SNP | *CiC6213-07 | 4 | 85.5 | Ollitrault et al., 2012b | ET085253 |
| SNP | CiC1380-05 | 5 | 17.2 | Ollitrault et al., 2012b | ET072553 |
| SNP | CiC5788-16 | 5 | 41.5 | Ollitrault et al., 2012b | ET082679 |
| SNP | *CiC5842-02 | 5 | 77.3 | Ollitrault et al., 2012b | ET083106 |
| SNP | *NADK2-M285 | 5 | 86.0 | García-Lor et al., 2013a | JX630077 |
| SSR | mCrCIR06A12 | 5 | 98.7 | Froelicher et al., 2011 | AM489742 |
| SNP | DFR-M240 | 5 | 105.7 | García-Lor et al., 2013a | JX630074 |
Non-polymorphic markers.
Table 3.
Molecular markers analyzed indicating the type of marker, locus name, linkage groups (LGs) 6 and 7 and location within LG in centimorgans (cM), bibliographic reference in the literature, and GeneBank accession.
| Type | Locus | Location | References | ||
|---|---|---|---|---|---|
| LG | cM | Literature | GeneBank | ||
| SSR | *mCrCIR04H09 | 6 | 0.0 | Ollitrault et al., 2012a | FR692370 |
| SNP | CiC4356-06 | 6 | 6.2 | Ollitrault et al., 2012b | ET107540 |
| SSR | MEST132 | 6 | 26.9 | Aleza et al., 2011 | DY276930 |
| SSR | CiBE4818 | 6 | 28.3 | Ollitrault et al., 2010b | ET110604 |
| SSR | CiBE0733 | 6 | 42.2 | Ollitrault et al., 2010b | ET094202 |
| SNP | *CiC2128-01 | 6 | 61.2 | Ollitrault et al., 2012b | ET111354 |
| SSR | mCrCIR02B11 | 6 | 69.2 | Ollitrault et al., 2012a | FR692358 |
| SNP | PSY-M30 | 6 | 69.7 | García-Lor et al., 2013a | JX630080 |
| SNP | PSY-C461 | 6 | 69.7 | García-Lor et al., 2013a | JX630080 |
| SNP | CiC3056-02 | 6 | 70.5 | Ollitrault et al., 2012b | ET075329 |
| SSR | *CiBE6256 | 6 | 84.6 | Ollitrault et al., 2010b | ET085615 |
| SNP | *AocM290 | 6 | 85.9 | Ollitrault et al., 2012b | JX630079 |
| SNP | AocC593 | 6 | 85.9 | Ollitrault et al., 2012b | DY293375 |
| SSR | MEST123 | 6 | 93.0 | Aleza et al., 2011 | DY276100 |
| SSR | CiBE5866 | 6 | 99.8 | Ollitrault et al., 2010b | ET083232 |
| SSR | mCrCIR07E05 | 7 | 13.1 | Froelicher et al., 2011 | AM489749 |
| SNP | *CiC1444-03 | 7 | 13.6 | Ollitrault et al., 2012b | ET073216 |
| SNP | DXS-M618 | 7 | 40.7 | García-Lor et al., 2013a | JX630082 |
| SNP | DXS-C545 | 7 | 40.7 | García-Lor et al., 2013a | JX630082 |
| SNP | FLS-P129 | 7 | 46.0 | García-Lor et al., 2013a | JX630083 |
| SSR | *mCrCIR03E06 | 7 | 75.1 | Ollitrault et al., 2012a | FR692363 |
| SSR | Ci07C07 | 7 | 98.0 | Froelicher et al., 2011 | AJ567409 |
Non-polymorphic markers.
SSR markers
PCR amplifications were performed using Thermocycler ep gradient S (Eppendorf®, Germany) in 10 μL final volume, containing 0.8 U of Taq DNA polymerase (Fermentas®, Germany), 10 ng of citrus template DNA, 0.2 mM wellRED (Sigma®, Germany) dye-labeled forward primer, 0.2 mM non-dye-labeled reverse primer, 0.2 mM each dNTP, and PCR reaction buffer 10X composed of 750 mM Tris-HCl, pH 9.0, 50 mM KCl, 200 mM (NH4)2SO4, 1.5 mM MgCl2, and 0.0001% BSA. The cycling program was set as follows: denaturation for 5 min at 94°C followed by 40 repeats of 30 s at 94°C, 1 min at the annealing temperature of each primer pair, 45 s at 72°C, and a final elongation step of 4 min at 72°C. Capillary electrophoresis was carried out using a CEQ™ 8,000 Genetic Analysis System (Beckman Coulter Inc., USA). The PCR products were initially denatured for 2 min at 90°C, injected for 30 s at 2 kV, and subsequently separated for 35 min at 6 kV. Alleles were sized based on a DNA size standard (400 bp). The GenomeLab™ GeXP v.10.0 genetic analysis software was used for data collection. Allele dosage was calculated using MAC-PR (microsatellite DNA allele counting-peak ratio) method (Esselink et al., 2004), validated in citrus by Cuenca et al. (2011).
SNP markers
Genetic analysis of SNP markers was performed using KASPar technology by LGCgenomics (http://www.lgcgenomics.com). Primers were designed by LGCgenomic from each SNP locus flanking sequence (approximately 50 nt on each side of the SNP). The KASPar genotyping system is a competitive allele-specific dual Förster Resonance Energy Transfer (FRET)-based assay for SNP genotyping. A detailed description of specific conditions and reagents can be found in Cuppen (2007). Identification of allele dosage in heterozygous somatic hybrids was carried out based on the relative allele signals described by Cuenca et al. (2013) and Aleza et al. (2015).
Identification of the genetic structure of somatic hybrids and their parents
Allelic configurations of the somatic hybrids SMC-58 and SMC-73 and their parents, CM and CC, were determined using the SSR and SNP genotyping data. For markers showing different alleles for the parents (A1A2 + A3A4, and A1A2 + A3A3), the somatic hybrids genotype was directly annotated. In the case of parents sharing alleles for a given marker (A1A2 + A2A2 o A1A2 + A2A3), the allelic configuration of the somatic hybrids was based on the estimated allele dosage.
Morphologic characterization
Plant morphology was evaluated on 9-month-old plants that were cultivated in the greenhouse under the above described conditions. Twelve plants of CC, CM, and the somatic hybrids (SMC-58 and SMC-73) were chosen for performing the evaluation. Measurements were taken on plant height, internodal length, and leaf number. Leaf greenness of 3 mature leaves was measured in each plant using a SPAD device (Minolta®, Japan) and the mean value of 5 readings was taken. The length (l) and width (w) of the main leaflet were also registered in the same leaves. Leaf index, representative of leaf shape, was calculated from l/w relations. Additionally, all the relevant International Plant Genetic Resources Institute (IPGRI) descriptors for each genotype were annotated (IPGRI, 1999).
Iron chlorosis tolerance evaluation
Twelve homogeneous plants of each genotype were trimmed to a single stem and transplanted to 0.5 L pots and then grouped into two groups based on substrate type. Control substrate was composed of peat and sand (2:3) with added 0.4% (w/v) Ca(H2PO4)2, whereas chlorosis-inducing substrate had additional 10% (v/v) of CaCO3 added to the mix. Plants were previously acclimated and were maintained for 4 weeks under the irrigation and climatic conditions previously described. Any new lateral branching shoots were detected and eliminated every 3 days to focus the growth in a single shoot. A plastic ring was placed on top of the stem to differentiate the newly developed biomass prior to the initiation of irrigation treatments. Plants growing on normal substrate were irrigated with the solution previously described, which contained 20 μM Fe-EDDHA, and were chosen as the control treatment (Ct). Plants growing on the chlorosis-inducing substrate were irrigated with a similar solution than Ct treatment but deficient in iron (2 μM Fe-EDDHA) and containing carbonates (10 mM NaHCO3). These conditions were considered the chlorosis-inducing treatment (Ch). Plants were randomized over the experimental area with a guard row and irrigated twice weekly for 10 weeks. After treatments, the new shoot was taken from each plant, rinsed with deionized water, and separated into leaves and stems. They were then fresh-weighed individually and dried in a forced draft oven at 70°C for 48 h until constant dry weight (DW) was obtained. Plant growth was measured using the shoot (leaf and stem) DW and iron content analysis was performed using leaves. The chlorophyll content in leaves was monitored by measuring changes in leaf greenness with a SPAD chlorophyll meter (Minolta, Japan). Two fully expanded leaves per plant were marked with labels and five readings were taken per leaf, avoiding the midrib, at the initial and final days of the trial period. Leaf greenness index was calculated as the ratio of final/initial SPAD readings. Values below 1 indicate greenness descent over the trial period (Castle et al., 2009). The iron concentration was measured from dry tissues (0.5 g) that were burnt in a muffle furnace for 12 h at 550°C. Iron was extracted with 2% nitric acid (Hiperpur, Panreac) in an ultrasonic bath (Fungilab®, Spain) for 30 min at 40°C and the concentration was measured using atomic absorption spectrometry in an ASS Analyst200 (Perkin Elmer®, USA).
Salinity tolerance evaluation
Forty homogeneous plants of each genotype trimmed to a single stem were selected and divided into groups that were irrigated with the basal nutrient solution described above. Either 0 (control, Ct) or 40 mM NaCl (salt-treated, +S) was added to each group. Pots were irrigated with 400 mL of solution per pot every 3 days. Excess solution was drained out of the pot to avoid salt accumulation in the substrate. A plastic ring was placed at the top of the stems to differentiate the newly developed biomass before irrigation treatments were initiated. Leaf gas exchange parameters were registered weekly using a portable infra-red gas analyzer LCpro+ (ADC Bioscientific Ltd., UK). Net CO2 assimilation (ACO2) and transpiration (E) rates were monitored between 10 a.m. and 2 p.m. The measurements were taken in two mid-stem leaves of 9 plants per treatment and genotype by taking 3 consecutive measurements on each leaf. Photosynthetically active radiation (PAR) at the leaf surface was adjusted to 1,000 μmol m−2 s−1, which exceeds the saturating value for citrus, and atmospheric CO2 concentration was not manipulated. Relative humidity and temperature in the greenhouse were recorded during each measurement event and were maintained by the conditions previously described. Dry weight of new leaves, leaf abscission percentage, and leaf burned area percentage were evaluated on leaves after 20 days of salt treatment and 5 mid-stem leaves, roots, and stems were sampled for analysis. Plant organs were rinsed with deionized water and 10% (w/v) Tween 20 (Sigma-Aldrich Co., Germany) and dried in a forced draft oven at 65°C for 48 h until reaching constant DW. Dried samples were crushed separately in a hammer-mill and were stored at room temperature to determine iron concentration in different organs. Chloride (Cl−) was determined by silver ion titration using a Corning 926 chloridometer (Corning®) as described by Gilliam (1971). Sodium (Na+) and potassium (K+) concentration were determined by inductively coupled plasma atomic emission spectroscopy (ICP-AOES iCAP 6000, Thermo Scientific). Samples (0.5 g) were pre-digested overnight with 2% HNO3 and 0.1% (w/v) Triton-X 100 (Sigma-Aldrich Co.) prior to processing on a digestion block at 120°C. The digestion tubes were then removed and cooled at room temperature. 2.0 mL of a 70% ultra-trace-metal-grade HClO4 was then added to the sample and heated at 220°C until white fumes were produced. Digest was diluted to a 25 mL with ultrapure water (Campbell and Plank, 1998) and filtered in n° 1 Whatman paper.
CTV tolerance evaluation
Six plants of CC, CM, SMC-58, and SMC-73 were inoculated by bark grafting with CTV T388 strain (+CTV), which is a very aggressive strain to CM (Moreno et al., 1990; Ballester-Olmos et al., 1993). The inoculum was obtained from the IVIA citrus virus and virus-like collection. After 25 days, plants were pruned leaving 5 cm above the inoculum to induce a new shooting. Control treatment (Ct) was applied to 3 plants that were not inoculated with the virus. The plants were cultivated under the above-described greenhouse conditions for 12 months. Plant size and the weight of roots and aerial parts were registered, and CTV symptoms were evaluated in leaves and stem wood.
Statistical analysis
Data were subjected to analysis of variance (ANOVA). Means were separated using Duncan's multiple range test at P < 0.05 with the Statgraphics Plus, version 5.1 (Statistical Graphics, Englewood Cliffs) software.
Results
Genetic characterization
Genetic analysis of the CM callus used for protoplast fusion did not show any differences when compared to the tree of the IVIA Citrus Germplasm Bank for the 34 SSR markers analyzed (Pensabene-Bellavia, 2009). Somatic hybrids SMC-58 and SMC-73 and their parents (CC and CM) were analyzed using 90 markers (82 from the nuclear genome and 8 from the cytoplasmic genome). Fifty-nine of the markers analyzed in nuclear genome were SNPs, whereas 23 were SSRs. All of them were distributed on the 9 LGs of the reference genetic citrus map (Ollitrault et al., 2012a) with a coverage between 5 and 15 markers per LG. Thirty six of the fifty-nine SNPs analyzed were polymorphic between parents, whereas polymorphism was found in 20 of the 23 SSRs analyzed (Tables 1–4).
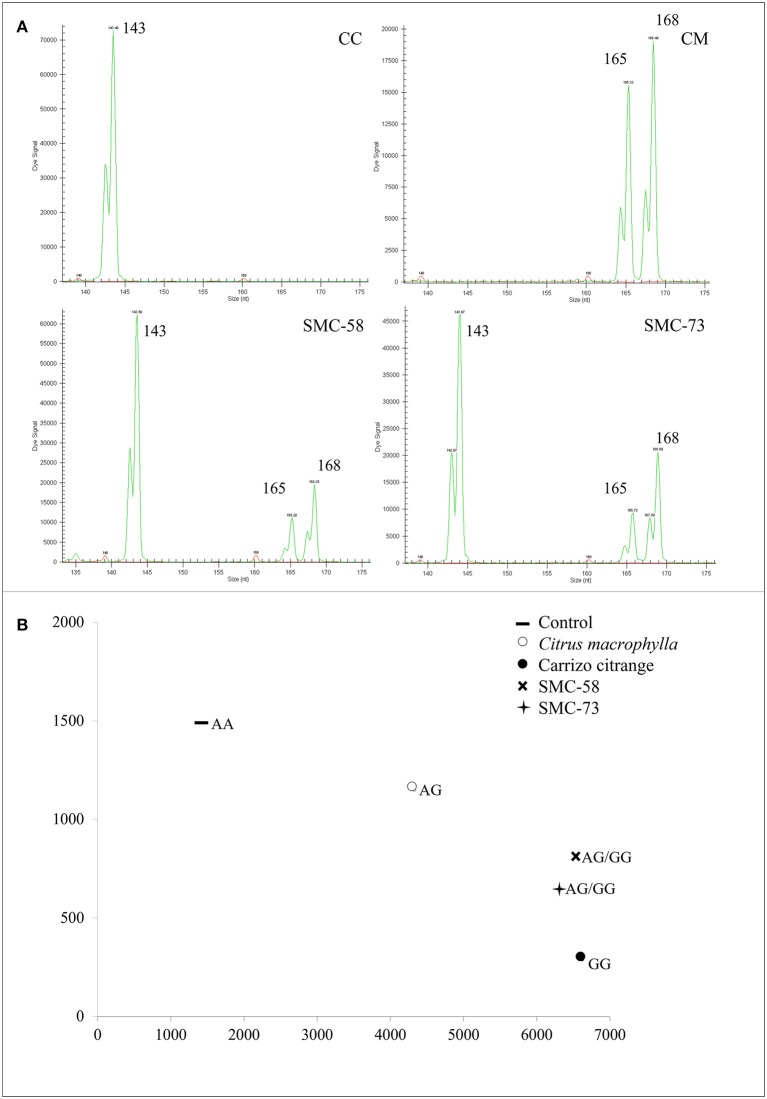
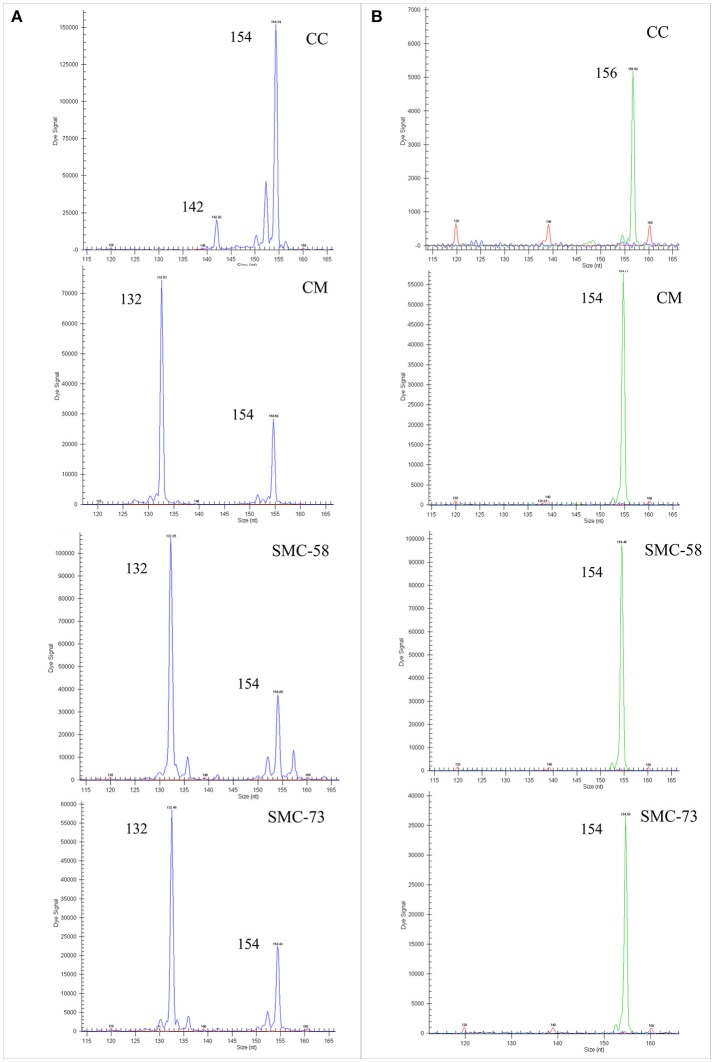
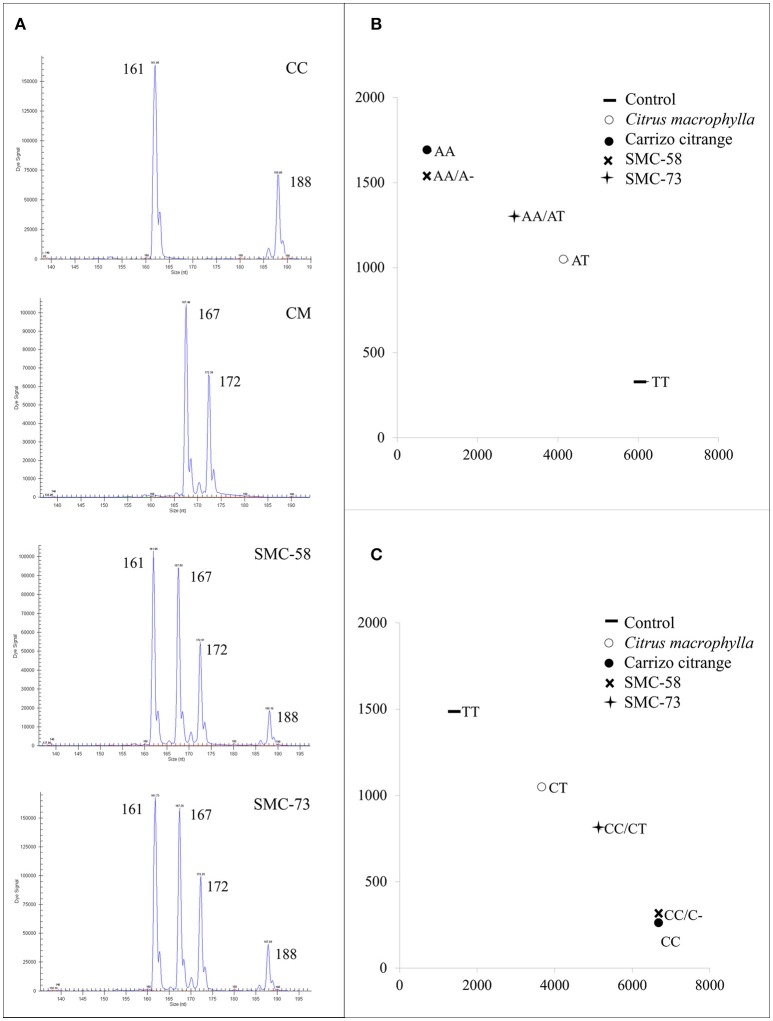
The LG 1 was analyzed with 7 polymorphic markers, consisting of 5 SNPs and 2 SSRs (Table 5). Somatic hybrids SMC-58 and SMC-73 displayed allelic configurations that correspond with the addition of both genome parents as seen with the JK-TAA15 SSR marker (Figure 1A) or the ACO-C601 SNP marker (Figure 1B). The LG 2 was analyzed using 5 SNP markers and 2 SSRs that were polymorphic between parents. The addition of the alleles from both parents was observed in the two somatic hybrids for all the analyzed markers, except for the JK-TAA41 SSR marker (Figure 2A) that showed the loss of the 142 nt allele from CC in both somatic hybrids. The LG 3 was analyzed using 6 SNPs and 1 SSR marker that were polymorphic between parents. Somatic hybrids combined all the alleles from each parent, except for Ci08A10 SSR marker (Figure 2B) that lost the 156 nt allele from CC in both somatic hybrids. The LG 4 was analyzed using 3 SNPs and 2 SSR markers (Table 6) and both somatic hybrids combined all the parental alleles. Figure 3A shows results obtained for Ci07D06 SSR marker as an example. The LG 5 was analyzed using 3 SNPs and 1 SSR marker and results did not show allelic losses in these loci. The LG 6 was analyzed using 5 SNPs and 6 SSR markers. Somatic hybrids showed allelic losses in 5 of the 6 SSR markers analyzed (CiBE4818, CiBE0733, mCrCIR02B11, MEST123, and CiBE5866). The origin of lost alleles was CM, except for the locus CiBE0733 that lost the CC allele. Three of these losses were shared between the somatic hybrids, whereas 1 and 2 of them affected SMC-58 and SMC-73, respectively. Besides, on 3 of the 5 SNP markers (CiC4356-06, PSY-C461, and AocC593), the SMC-58 somatic hybrid lost the T allele from CM, whereas these differences were not observed in SMC-73 (Table 6). Figures 3B and C are examples of the results obtained for the PSY-C461 and AocC593 SNP markers, displaying the T allele lost in the SMC-58 somatic hybrid. The LG 7 was analyzed with 3 SNPs and 2 SSR markers. The 239 nt allele from CM was lost on Ci07C07 SSR locus in both somatic hybrids. On the LG 8, 2 SNPs and 2 SSR markers were analyzed, whereas on the LG 9, 4 SNPs and 2 SSR markers were used. In both LGs, the hybrids displayed allelic configurations that correspond with the addition of both genome parents (Table 7).
Table 5.
Genetic analysis using SNP and SSR nuclear markers located on LGs 1, 2, and 3 performed on SMC-58 and SMC-73 somatic hybrids that were obtained by protoplast fusion between C. macrophylla (CM) and Carrizo citrange (CC).
| Locus | LG | CC | CM | SMC-58 | SMC-73 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CiC2110-01 | 1 | yC | A | A | A | C | A | A | A | C | A | A | A |
| CiBE5720 | z308 | 329 | 308 | 320 | 308 | 329 | 308 | 320 | 308 | 329 | 308 | 320 | |
| ACO-C601 | G | G | A | G | G | G | A | G | G | G | A | G | |
| CiC0599-01 | G | A | G | G | G | A | G | G | G | A | G | G | |
| TSC-C80 | G | G | G | T | G | G | G | T | G | G | G | T | |
| JK-TAA15 | 143 | 165 | 168 | 143 | 165 | 168 | 143 | 165 | 168 | ||||
| PEPC-C328 | A | A | A | G | A | A | A | G | A | A | A | G | |
| mCrCIR07D05 | 2 | 189 | 195 | 189 | 195 | 189 | 195 | ||||||
| SOS1-M50 | A | G | A | A | A | G | A | A | A | G | A | A | |
| CCC1-P727 | C | T | C | C | C | T | C | C | C | T | C | C | |
| JK-TAA41 | 142 | 154 | 132 | 154 | 142 | 154 | 132 | 154 | 142 | 154 | 132 | 154 | |
| TRPA-M593 | C | G | C | C | C | G | C | C | C | G | C | C | |
| PKF-M186 | T | T | C | T | T | T | C | T | T | T | C | T | |
| INVA-M437 | C | T | C | C | C | T | C | C | C | T | C | C | |
| MDH-M519 | 3 | C | T | C | C | C | T | C | C | C | T | C | C |
| NCED3-M535 | G | T | T | T | G | T | T | T | G | T | T | T | |
| CiC5796-12 | A | A | C | C | A | A | C | C | A | A | C | C | |
| ATMR-M728 | T | T | G | G | T | T | G | G | T | T | G | G | |
| ATMR-C372 | A | A | A | G | A | A | A | G | A | A | A | G | |
| Ci08A10 | 156 | 154 | 156 | 154 | 156 | 154 | |||||||
| CHS-M183 | C | C | G | G | C | C | G | G | C | C | G | G | |
SNP alleles: A, adenine; C, cytosine; T, thymine; G, guanine.
SSR allele: Numbers are the allele size in nucleotides. Lost alleles are marked in gray.
Figure 1.
Allelic configurations on LG 1 that correspond to the addition of both parents. (A) Electropherograms of the somatic hybrids SMC-58 and SMC-73 displaying four different alleles from their parents C. macrophylla (CM) and Carrizo citrange (CC) using JK-TAA15 SSR marker. Numbers indicate the size of the amplified allele in nucleotides (nt). (B) Plot of allele signals of ACO-C601 SNP marker in somatic hybrids and their parents. CM displayed AG alleles, Carrizo citrange displayed GG alleles and somatic hybrids display AGGG alleles. Letters indicate the allelic configuration for each genotype as: A, adenine; C, cytosine; T, thymine; G, guanine.
Figure 2.
Electropherograms of the somatic hybrids SMC-58 and SMC-73 and their parents C. macrophylla (CM) and Carrizo citrange (CC) displaying allelic losses. (A) JK-TAA41 SSR marker on LG 2. (B) Ci08A10 SSR marker on LG 3. Numbers indicate the size of the amplified allele in nucleotides (nt).
Table 6.
Genetic analysis using SNP and SSR nuclear markers located on LGs 4, 5, and 6 performed on SMC-58 and SMC-73 somatic hybrids that were obtained by protoplast fusion between C. macrophylla (CM) and Carrizo citrange (CC).
| Locus | LG | CC | CM | SMC-58 | SMC-73 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHI-M598 | 4 | yC | C | G | C | C | C | G | C | C | C | G | C |
| mCrCIR07D06 | z162 | 188 | 167 | 172 | 162 | 188 | 167 | 172 | 162 | 188 | 167 | 172 | |
| CiC2840-01 | T | T | C | C | T | T | C | C | T | T | C | C | |
| CiC3740-02 | G | C | G | G | G | C | G | G | G | C | G | G | |
| mCrCIR03G05 | 213 | 219 | 199 | 213 | 219 | 199 | 213 | 219 | 199 | ||||
| CiC1380-05 | 5 | T | T | C | C | T | T | C | C | T | T | C | C |
| CiC5788-16 | G | A | A | A | G | A | A | A | G | A | A | A | |
| mCrCIR06A12 | 92 | 103 | 86 | 92 | 103 | 86 | 92 | 103 | 86 | ||||
| DFR-M240 | C | G | C | C | C | G | C | C | C | G | C | C | |
| CiC4356-06 | 6 | C | C | C | T | C | C | C | T | C | C | C | T |
| MEST132 | 231 | 244 | 244 | 231 | 244 | 244 | 231 | 244 | 244 | ||||
| CiBE4818 | 151 | 162 | 154 | 151 | 162 | 154 | 151 | 162 | 154 | ||||
| CiBE0733 | 240 | 245 | 235 | 240 | 245 | 235 | 240 | 245 | 235 | ||||
| mCrCIR02B11 | 232 | 232 | 248 | 232 | 232 | 248 | 232 | 232 | 248 | ||||
| PSY-M30 | C | G | G | G | C | G | G | G | C | G | G | G | |
| PSY-C461 | A | A | A | T | A | A | A | T | A | A | A | T | |
| CiC3056-02 | G | A | A | A | G | A | A | A | G | A | A | A | |
| AocC593 | C | C | C | T | C | C | C | T | C | C | C | T | |
| MEST123 | 239 | 246 | 250 | 239 | 246 | 250 | 239 | 246 | 250 | ||||
| CiBE5866 | 214 | 222 | 214 | 222 | 214 | 222 | |||||||
SNP alleles: A, adenine; C, cytosine; T, thymine; G, guanine.
SSR allele: Numbers are the allele size in nucleotides. Lost alleles are marked in gray.
Figure 3.
Allelic configurations of the somatic hybrids SMC-58 and SMC-73 and their parents C. macrophylla (CM) and Carrizo citrange (CC). (A) Ci07D06 SSR marker on LG 4 where somatic hybrids display four different alleles from their parents. Numbers indicate the size of the amplified allele in nucleotides (nt). Plot of allele signals of (B) PSY-C461 and (C) AocC593 SNP markers on LG 6. For PSY-C461 SNP marker, CM displayed AT alleles, Carrizo citrange displayed AA alleles, SMC-58 AAA alleles with the loss of T allele from CM and SMC-73 with the alleles of both parents, CM, and CC. For AocC593, CM displayed CT alleles, Carrizo citrange displayed CC alleles, SMC-58 CCC alleles with the loss of T allele from CM and SMC-73 with the alleles of both parents, CM and CC. Letters indicate the allelic configuration for each genotype as: A, adenine; C, cytosine; T, thymine; G, guanine.
Table 7.
Genetic analysis using SNP and SSR nuclear markers located on LGs 7, 8, and 9 and mitochondrial (mt) and chloroplastic (cp) markers performed on SMC-58 and SMC-73 somatic hybrids that were obtained by protoplast fusion between C. macrophylla (CM) and Carrizo citrange (CC).
| Locus | LG | CC | CM | SMC-58 | SMC-73 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mCrCIR07E05 | 7 | z119 | 128 | 116 | 119 | 128 | 116 | 119 | 128 | 116 | |||
| DXS-M618 | yG | G | A | A | G | G | A | A | G | G | A | A | |
| DXS-C545 | G | G | C | G | G | G | C | G | G | G | C | G | |
| FLS-P129 | C | T | T | T | C | T | T | T | C | T | T | T | |
| Ci07C07 | 212 | 239 | 212 | 239 | 212 | 239 | |||||||
| mCrCIR07B05 | 8 | 196 | 203 | 210 | 196 | 203 | 210 | 196 | 203 | 210 | |||
| CiBE0214 | 312 | 309 | 312 | 309 | 312 | 309 | |||||||
| CiC5164-02 | C | C | T | T | C | C | T | T | C | C | T | T | |
| CiC1749-05 | G | T | T | T | G | T | T | T | G | T | T | T | |
| mCrCIR07F11 | 9 | 160 | 162 | 164 | 160 | 162 | 164 | 160 | 162 | 164 | |||
| CiC2518-02 | T | A | T | T | T | A | T | T | T | A | T | T | |
| Ci08C05 | 153 | 153 | 156 | 153 | 153 | 156 | 153 | 153 | 156 | ||||
| LCYB-M480 | T | C | T | T | T | C | T | T | T | C | T | T | |
| HYB-M62 | A | A | C | C | A | A | C | C | A | A | C | C | |
| HYB-C433 | G | G | A | G | G | G | A | G | G | G | A | G | |
| nad2/4-3 | mt | 261 | 251 | 251 | 251 | ||||||||
| CCMP2 | cp | 197 | 203 | 197 | 203 | 197 | |||||||
| CCMP5 | cp | 93 | 95 | 93 | 95 | 93 | |||||||
| CCMP6 | cp | 133 | 135 | 133 | 135 | 133 | |||||||
| NTCP7 | cp | 182 | 188 | 182 | 188 | 182 | |||||||
SNP alleles: A, adenine; C, cytosine; T, thymine; G, guanine.
SSR allele: numbers are the allele size in nucleotides. Lost SSR alleles and modified SNP alleles are marked in gray.
In summary, somatic hybrids SMC-58 and SMC-73 combine the parental alleles from CC and CM in 45 of the 56 nuclear markers analyzed (80%). However, allelic losses were found in 11 of the loci analyzed. The origin of lost alleles was CM in 8 loci and CC in 3 loci. Most of the lost alleles, 8 of the 11, were located on the LG 6 (Table 6) and 2 of them have a CC origin, whereas 6 come from CM. The rest of lost alleles were located on LGs 2, 3 (Table 5), and 7 (Table 7). On LGs 2 and 3, the origin of lost alleles was CC, whereas on LG 7, the origin of lost alleles was CM. We found alleles that are lost only in one or the other when comparing the genetic configuration of both somatic hybrids. Therefore, they are genetically different. SMC-58 lost 3 SNP alleles from CM (Figures 3B and C) that were identified in SMC-73 and SMC-73 lost 2 SSR alleles (mCrCIR02B11 and MEST 123 loci), one from CM and the other one from CC, although these alleles were present in the SMC-58 somatic hybrid. We investigated the parental origin of these 11 lost alleles. CM is a hybrid of C. micrantha W. and C. medica (Curk et al., 2016), two of the Citrus ancestral species. CC has C. sinensis and P. trifoliata in its pedigree and C. sinensis is a secondary species that originated from crosses between C. maxima and C. reticulata (García-Lor et al., 2013b). In Ci08A10, Cibe4818, and Cibe5866 SSR markers, we cannot decipher the parental origin of the lost alleles because Ci08A10 is homozygous for CC and for the last 2 markers, the lost allele is shared between C. micrantha and citron. Regarding Ci07C07 SSR marker and CiC4356-06, PSY-C461 and AoC-C593 SNP markers, the lost allele comes from citron, whereas for mCrCIR02B11, JK-TAA41, and Cibe0733 SSR markers lost alleles come from C. micrantha, P. trifoliata, and C. sinensis, respectively. The later lost allele is shared between C. maxima and C. reticulata parental species of sweet orange.
The cytoplasmic genome was analyzed using 1 mitochondrial marker (nad2/4-3) and 4 chloroplastic markers (CCMP2, CCMP5, CCMP6, and NTCP7) (Table 8) that were polymorphic between CC and CM. Both somatic hybrids had the 251 nt allele of the mitochondrial marker nad2/4-3 that belongs to the embryogenic parental CM. However, chloroplastic genome analysis showed that the origin of SMC-73 chloroplasts was CC for the 4 markers analyzed, whereas SMC-58 combined both CM and CC alleles for these markers (Table 7). As an example, in Figure 4, we display the addition of both parental alleles in the SMC-58 somatic hybrid for the NTCP7 SSR marker.
Table 8.
Plant morphology of 9-month-old plants of the somatic hybrids SMC-58 and SMC-73 obtained by protoplast fusion between Carrizo citrange (CC) and C. macrophylla (CM).
| Genotype | Plant height (cm) | Leaf number | Internodal length (cm) | Leaf greenness (SPAD) | Leaf morphological index (l/w) |
|---|---|---|---|---|---|
| CC | 143.6a | 41.9a | 3.43a | 73.82a | 2.51a |
| CM | 111.9b | 36.8b | 3.05b | 60.93b | 2.14b |
| SMC-58 | 59.1d | 19.2d | 3.08bc | 72.78a | 1.74c |
| SMC-73 | 81.5c | 26.6c | 3.14c | 75.99a | 1.79c |
Values are the mean of 12 plants (n = 12). Different letters within each column indicate significant differences for P ≤ 0.05 on multiple range Duncan's test.
Figure 4.
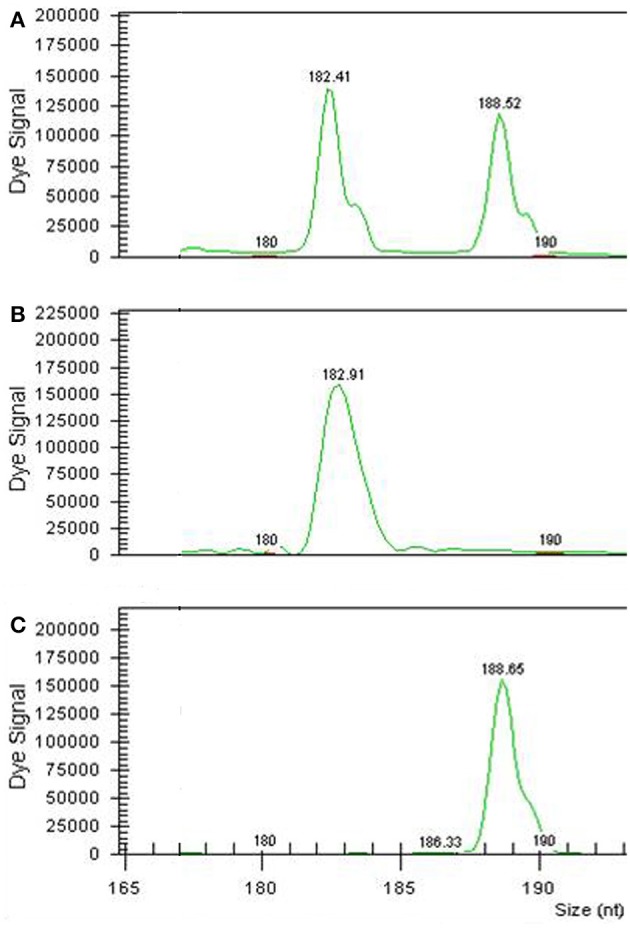
Electropherogram of the chloroplastic SSR marker NTCP7 analyzed in the SMC-58 somatic hybrid (A) and his parents CC (B) and CM (C) Numbers mean the size in nucleotides of each amplified allele.
Plant morphology
Plant morphology was evaluated in 9-month-old plants of CC, CM, SMC-58, and SMC-73. Somatic hybrids had a slower growth than both the parents and were prone to lateral branching (Figures 5A–D). The height of both the somatic hybrids was shorter than CC or CM (Table 8). Differences in growth were also found between the hybrids as SMC-58 grew 20% less than SMC-73. Internodal length was longer in CC than in CM, whereas SMC-58 was similar to CM, and SMC-73 had an intermediate length between parents. The leaf morphological index obtained from the length/width ratio of the main leaflet was lower in somatic hybrids than in both the parents and was similar between SMC-58 and SMC-73. This indicates that the morphology of the measured leaves is round shaped, which is a character that is typical of tetraploid citrus plants (Barrett and Hutchison, 1978). Leaf greenness of somatic hybrids was similar between them and resembled CC, whereas CM had 12% lower leaf greenness than these genotypes. Somatic hybrid plants, SMC-58 and SMC-73, have a spiral phyllotaxis pattern, where leaves and straight thorns of intermediate length (16–40 mm) appear together. These characteristics are similar to those of both the parents. The leaves, showing brevipetiolate attachment to the lamina, are odd-pinnate, and the number of leaflets within the same plant varies between one, as seen with CM, and three, as seen with CC (Figures 5C and D). The somatic hybrid SMC-58 shows mainly one or two leaflets per leaf and trifoliate leaves are also present. The somatic hybrid SMC-73 shows mainly trifoliate leaves, even though simple and bifoliate leaves also appear. The leaf size is small (10–20 cm2) and heterogeneous. The petiole is shorter than the lamina and has narrow obdeltate wings with articulate junction to the lamina. The main leaflet has a length/width ratio between 1.5 and 1.8 and shape varies from elliptic, like in CC, to obovate, as in CM. The leaf margins are crenate, and the apex is obtuse in both somatic hybrids.
Figure 5.
Plants of (A) Carrizo citrange (CC), (B) C. macrophylla (CM) and somatic hybrids (C) SMC-58 and (D) SMC-73 cultivated for 5 months in greenhouse conditions.
Tolerance to iron deficiency
Leaf greenness decreased in all the genotypes under the chlorosis-inducing treatment (Ch). Carrizo citrange had a greater greenness decline than CM and the somatic hybrids showed intermediate values between parents (Table 9). In terms of growth, the shoot developed under the Ch treatment in somatic hybrids had similar leaf biomass than in CM, whereas these values were higher than in CC. In control conditions, SMC-73 had similar growth to that of CC and SMC-58 grew less than both parents. Iron concentration in the leaves developed under the Ch treatment was higher in CM than in CC and somatic hybrids had intermediate concentrations between them (Table 9).
Table 9.
Leaf greenness (f:i), increase in shoot biomass (DW g) and iron concentration in shoot leaves (DW ppm) of Carrizo citrange (CC), C. macrophylla (CM) and the somatic hybrids SMC-58 and SMC-73 cultivated in greenhouse conditions for 10 weeks, either in control conditions (20 μM Fe-EDDHA) or in iron-deficient conditions (10% (v/v) CaCO3 10 mM NaHCO3, 2 μM Fe-EDDHA).
| Treatment | Genotype | Leaf greenness (f/i)X | Increase in shoot biomass (DW g) | Iron concentration (ppm) |
|---|---|---|---|---|
| Control | CC | 0.84 a | 1.36 b | 48.3 a |
| CM | 1.11 b | 1.76 c | 46.7 a | |
| SMC-58 | 1.02 b | 0.89 a | 38.7 a | |
| SMC-73 | 0.89 ab | 1.21 b | 43.5 a | |
| Iron-deficient | CC | 0.30 a | 0.42 a | 16.5 a |
| CM | 0.70 c | 0.56 b | 33.1 b | |
| SMC-58 | 0.56 b | 0.52 b | 23.9 ab | |
| SMC-73 | 0.52 b | 0.61 b | 21.7 ab |
SPAD final/initial, values below 1 indicate greenness decrease. Values are the mean of six plants (n = 6). Different letters in each column indicate significant differences for P ≤ 0.05 on multiple range Duncan's test.
Tolerance to salinity
The differences in behavior between somatic hybrids and their parents under salinity (+S) were evaluated according to the growth rates, leaf symptoms, ion accumulation, and gas exchange parameters. Carrizo citrange plants subjected to salinity had 25% lower DW than control plants at the end of the experimental period (Figure 6A), indicating their sensitive behavior. In contrast, CM, that is salt-tolerant, showed similar growth in both, +S or Ct treatments. The SMC-73 hybrid had similar behavior to CM regarding growth, given that +S treatment did not affect this parameter. SMC-58 had 16% lower DW under the +S treatment than in Ct conditions, although this growth reduction was lower than in the sensitive CC. Leaf symptoms induced by salt toxicity were intense in CC plants that had 20% of their leaf area burned. Meanwhile, leaves of the tolerant CM were free of burns (Figure 6B) and somatic hybrids showed very mild leaf toxicity symptoms with only 2% (Figure 6C) of their leaf area affected by burns (Figure 6B). Leaf abscission was lower in SMC-73 or SMC-58 than in CC with 6, 3, and 7% of leaves affected, respectively. Nevertheless, the tolerant CM did not suffer this symptom.
Figure 6.
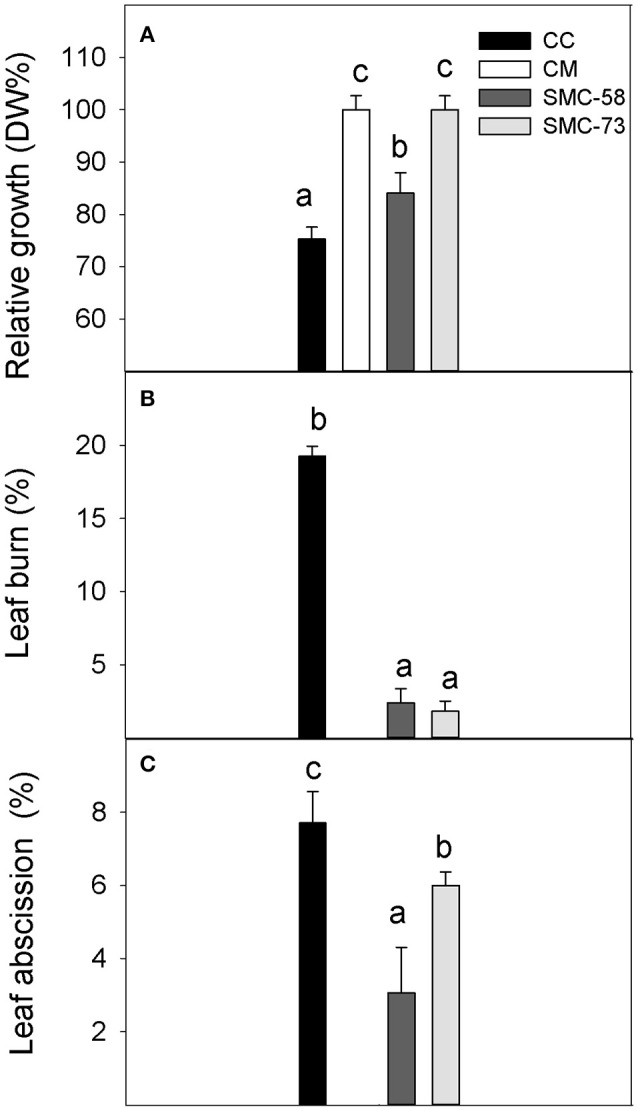
Influence of salinity on Carrizo citrange (CC), C. macrophylla (CM) and somatic hybrids SMC-58 and SMC-73 plants. (A) Relative growth (DW %), (B) Leaf burn damage (%) and (C) Leaf abscission. Plants were cultivated for 20 days under glasshouse conditions and saline treatment (40 mM NaCl). Values are the mean of six plants (n = 6) and standard error. Different letters indicate significant differences for P ≤ 0.05 on Duncan's multiple range test.
Overall, Cl− and Na+ molar concentrations in leaf tissue water were higher in the saline treatment (+S) than in the Ct treatment for all the genotypes (Table 10). The parent CM, which is tolerant to salinity, had the lower Cl− and Na+ concentrations under the +S treatment, and were 2.2 and 1.4-fold, respectively, higher than in control plants. Carrizo citrange, which is considered salt-sensitive, raised Cl− and Na+ leaf concentrations that were 4.1 and 2.6-fold higher in +S treatment than in Ct treatment, respectively. Somatic hybrid SMC-58 subjected to +S treatment had lower Cl− concentration and similar Na+ concentration than CC. Specifically, Cl− and Na+ leaf concentration in SMC-58 were 4.2 and 1.8-fold, respectively, higher in +S than in Ct plants. The SMC-73 plants subjected to +S treatment had leaf Cl− concentrations similar to CC, whereas leaf Na+ concentration was higher than in CC plants. More precisely, leaf Cl− and Na+ concentrations in SMC-73 increased by 3.9 and 2.3-fold, respectively, in salt-treated plants when compared to Ct plants. Therefore, the data show that both somatic hybrids had lower Cl− exclusion capacity than the salt-tolerant parent CM. However, SMC-58 had greater exclusion capacity than the salt-sensitive parent CC, whereas SMC-73 had similar exclusion capacity to CC. Regarding Na+ exclusion, the behavior observed in SMC-58 was similar to CC, whereas SMC-73 plants accumulated less Na+ in their leaves, showing more tolerance than the sensitive parent CC. The concentration of K+ in plants subjected to salinity was not different from Ct plants in the tolerant CM. Leaf concentration of K+ decreased by 21%, 20%, and 33%, respectively, in CC, SMC-58, and SMC-73. Salt-treated CM plants did not differ from Ct plants in their ACO2 and E rates (Table 11). The salt-sensitive parent CC had reduced ACO2 rates by 39% and E rates by 18% when compared to Ct plants. Somatic hybrids SMC-58 and SMC-73 subjected to salinity reduced E rates by 33% and 43%, respectively, when compared to Ct plants. Similarly, these salt-treated plants reduced ACO2 by 32% and 53%, respectively. The data show that gas exchange parameters were more affected by salinity in SMC-73 than in SMC-58. Therefore, the former genotype was similar to CC, whereas the latter had a behavior more similar to CM. In summary, results show that somatic hybrids have an intermediate behavior between the tolerant rootstock CM and the sensitive CC. However, the differences found between SMC-58 and SMC-73 indicate that SMC-58 is better adapted to salinity than SMC-73 because the response was globally more similar to the tolerant parent CM.
Table 10.
Leaf Cl−, Na+ and K+ concentration (mM in tissue water) in Carrizo citrange (CC), C. macrophylla (CM) and SMC-58 and SMC-73 somatic hybrids.
| Treatment | Ion | CC | CM | SMC-58 | SMC-73 |
|---|---|---|---|---|---|
| Control | Cl− | 56.3a | 36.5a | 43.5a | 53.4a |
| Na+ | 140.0b | 85.7a | 217.9c | 202.0c | |
| K+ | 571.1a | 607.0b | 556.2a | 609.0b | |
| Saline | Cl− | 233.2c | 80.9a | 183.0b | 205.4c |
| Na+ | 367.8b | 117.5a | 393.7b | 466.0c | |
| K+ | 462.0a | 621.2b | 446.0a | 468.4a |
Values are the mean of 3 plants (n = 3). Different letters in each line indicate significant differences for P ≤ 0.05 at Duncan's multiple range test. Plants were cultivated in a greenhouse for 20 days, either in saline (40 mM NaCl) or control conditions.
Table 11.
Transpiration (E, mmol H2O·m−2·s−1) and net assimilation (ACO2, μmol CO2·m−2·s−1) rates in Carrizo citrange (CC), C. macrophylla (CM) and SMC-58 and SMC-73 somatic hybrids.
| Treatment | Parameter | CC | CM | SMC-58 | SMC-73 |
|---|---|---|---|---|---|
| Control | E | 0.76 c | 1.37 a | 1.32 a | 1.16 b |
| ACO2 | 6.23 b | 9.99 a | 9.21 a | 8.74 a | |
| Saline | E | 0.62 c | 1.32 a | 0.89 b | 0.66 c |
| ACO2 | 3.82 c | 10.1 a | 6.23 b | 4.14 c |
Values are the mean of six plants (n = 6). Different letters in each line indicate significant differences for P ≤ 0.05 at Duncan's multiple range test. Plants were cultivated in a greenhouse for 20 days, either in saline (40 mM NaCl) or control conditions.
Tolerance to CTV
Plants inoculated with T388 CTV strain were evaluated for growth and symptoms. CM showed reduced growth as evident by the shorter height of the plants (Figure 7A) and the lower aerial and root biomass (Figures 7B and C). Overall, plant biomass decreased by 35% (Figure 7D) in CM. These plants also showed yellow leaves with vein corking (Figure 8A) and stem pitting (Figure 8B). Meanwhile, CC plants and the somatic hybrids SMC-58 and SMC-73 were not different from control plants in their growth (Figure 7D), and neither showed the disease symptoms (Figures 8C–H).
Figure 7.
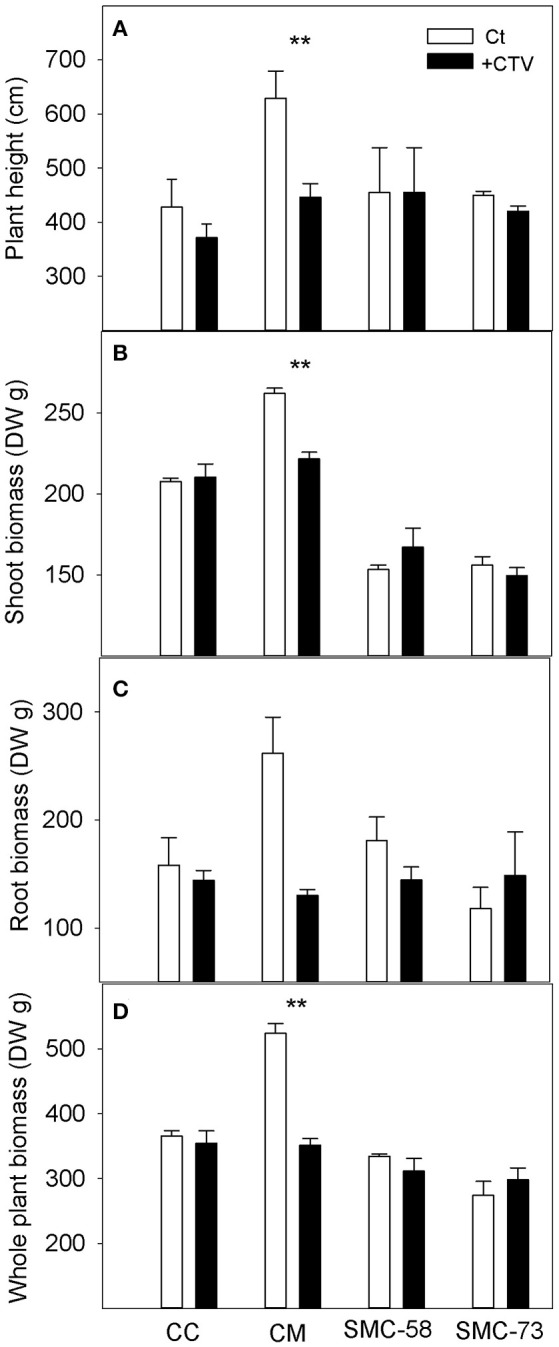
Influence of Citrus tristeza virus (CTV) on Carrizo citrange (CC), C. macrophylla (CM) and somatic hybrids SMC-58 and SMC-73 plants. (A) Plant height (cm), (B) Aerial parts plant biomass (DW g), (C) Root biomass, (D) Whole plant biomass. Plants were either inoculated with the T388 strain of CTV (+CTV) or treated as non-inoculated controls (Ct) and cultivated for 12 months under glasshouse conditions. Values are the mean between 3 and 6 plants and standard error. **Significant differences for P ≤ 0.01 on Duncan's multiple range test.
Figure 8.

Symptoms on plants inoculated with the T388 strain of Citrus tristeza virus: (A) Yellowing and vein corking in leaf of C. macrophylla (CM). (B) Stem pitting in CM. (C,D) Symptomless leaf and stem of Carrizo citrange. (E,F) Symptomless leaf and stem of somatic hybrid SMC-58. (G,H) Symptomless leaf and stem of somatic hybrid SMC-73.
Discussion
Genetic characterization of the somatic hybrids
Two allotetraploid somatic hybrids were previously obtained from protoplasts isolated from callus of CM and from leaves of CC (Pensabene-Bellavia et al., 2015). These somatic hybrids, their parents, and the embryogenic callus of CM were analyzed to verify their origin and genetic structure of the hybrids. Fifty-six nuclear molecular markers distributed uniformly on the 9 LGs of the clementine genetic map (Ollitrault et al., 2012a) and 5 cytoplasmic markers that were polymorphic between the parents were used for the analysis. Nuclear molecular markers confirmed that (i) the embryogenic callus of CM did not show differences compared to leaves of CM for all these markers and (ii) the somatic hybrids added the parental genomes although some parental alleles were lost. Specifically, 11 lost alleles have been identified on different LGs (2, 3, 6, and 7) although most of them are located on LG 6 (8 of the 11 alleles). Differences in 5 markers have also been found between the somatic hybrids. Most of the lost alleles observed in the somatic hybrids have their origin in the embryogenic parent CM (7 of them) even though 4 of them correspond to the leaf parent CC. For some markers, it has been possible to identify the parental origin of the lost allele. Most of the lost alleles come from C. medica, whereas others come from C. micrantha, C. sinensis, and P. trifoliata. These results suggest that the loss of parental alleles occurred during the somatic hybridization process and the differences observed between hybrids seem to be limited to sub-chromosomal level because the flow cytometry analysis did not show differences in the number of chromosomes (2n = 4x = 36) between somatic hybrids or when compared to the tetraploid control. Furthermore, genetic analysis has been performed using leaf samples of the somatic hybrids taken from different branches and the same differences between somatic hybrids and parents were found. However, no differences were identified between analyses of different DNA preparations, discarding the presence of chimeras. Most of the absent alleles that have been identified are located on the same LG and consist of deleted fragments (SSR allele absence) and punctual variations in a small number of nucleotides (non-observed SNP alleles). This indicates that there is chromosome instability in this complex intergeneric combination given that the genomes of the four citrus ancestral species and related genera are present in the somatic hybrids. Previous studies performed in citrus (Xu et al., 2014) and other species (Sundberg and Grimelius, 1991; Sun et al., 2014; Smyda-Dajmund et al., 2016) state that chromosome losses, genomic deletions, and epigenetic alterations are more frequent in somatic hybrids between parents that have a distant genetic relationship than in those from closely related parents. In SMC-58 and SMC-73, most lost alleles do not come from the species that are genetically more distant, C. micrantha and P. trifoliata. This finding suggests that there is no bias against the most dissimilar genomes when somatic hybridization is performed. Therefore, the identified losses might be either random or caused by some other effect. It has also been reported that genomic losses in citrus somatic hybrids are parent-biased toward the callus parent (Xu et al., 2014), which might explain that most of the alleles lost in SMC-58 and SMC-73 come from CM. Overall, the wide diversity of the genomes combined in SMC-58 and SMC-73 and the different origin of the parental protoplasts used to perform the fusions might explain the uneven genomic losses that we observed. Nevertheless, further studies would be required to verify these hypotheses.
Differences between SMC-58 and SMC-73 have also been found in the cytoplasmic genome. Both hybrids have the CM mitochondrial genome. However, SMC-73 has the CC chloroplastic genome, whereas chloroplastic genome recombination was detected in SMC-58. Citrus somatic hybrids predominantly inherit the mitochondrial genome from the embryogenic parent (Kobayashi et al., 1991; Saito et al., 1993; Yamamoto and Kobayashi, 1995; Moriguchi et al., 1997; Moreira et al., 2000; Cabasson et al., 2001; Ollitrault et al., 2001; Guo et al., 2002; Xiao et al., 2014) even though there are some reports of mitochondrial recombination events (Vardi et al., 1987; Moriguchi et al., 1997; Cheng et al., 2003; Dambier et al., 2011). Recently, Cai et al. (2017) have demonstrated that mitochondrial genome of protoplasts isolated from embryogenic callus is essential for plant regeneration after protoplast fusion experiments. However, chloroplastic genome is randomly inherited from one of the parents or shows recombination (Grosser et al., 2000; Dambier et al., 2011; Aleza et al., 2016). It has also been proven that mitochondrial and chloroplastic genomes are involved in differences in aroma and organoleptic fruit properties (Fanciullino et al., 2005; Satpute et al., 2015), disease resistance (Tusa et al., 2000; Omar et al., 2017), floral developmental disturbances, and male sterility (Guo et al., 2004; Zheng et al., 2012). Nevertheless, there is no information available about how these new combinations and rearrangements occurring on the cytoplasmic genome affect the agronomical behavior of citrus rootstocks. Most publications on citrus somatic hybridization report the symmetric addition of nuclear parental genomes (Ollitrault et al., 2010a) although subchromosomal variations have been detected in citrus somatic hybrids (Olivares-Fuster, 1998; Froelicher, 1999; Guo et al., 2007; Xu et al., 2014). However, studies describing them are scarce. Different hypotheses have been suggested for these kind of changes such as extended periods of in vitro culture (Oberwalder et al., 1998; Guo and Deng, 1999), genetic divergence between parents, and increased ploidy level (Sundberg and Grimelius, 1991; Miranda et al., 1997). Genetic analysis of somatic hybrids has been usually performed with a small number of molecular markers, enough to confirm their hybrid origin but not sufficient to identify these variations (Oberwalder et al., 1998; Guo and Deng, 1999). In potato (Solanum spp) somatic hybrids, similar variations have also been recently described using DArT markers (Diversity Array Technology) (Smyda-Dajmund et al., 2016). More than 5,000 markers distributed across the potato genome were analyzed in the somatic hybrids and 2,000 were found to be polymorphic between parents. Among them, between 13.9% and 29.6% of alleles were found to be lost in the somatic hybrids. The identification of genomic changes in somatic hybrids justifies the need for performing a detailed genetic analysis of the plants obtained by somatic hybridization to gather information on their genetic structure. This information is key to optimize and interpret the data on physiological behavior of the somatic hybrids to use them as rootstocks.
Performance of SMC-58 and SMC-73 somatic hybrids as potential citrus rootstocks
Several rootstock breeding programs based on somatic hybridization are currently being carried out across the world. In Florida, a large number of somatic hybrids have been obtained, which stand out for their good adaptation to the local soil, inducing good fruit quality, and high yields (Grosser et al., 2015). Breeding programs focused on somatic hybridization have also been carried out in the Mediterranean basin (Dambier et al., 2011), as well as in China (Guo et al., 2002, 2007), Brazil (Mendes-da-Gloria et al., 2000; Mourao et al., 2008), and Mexico (Medina-Urrutia et al., 2004). These data reveal that somatic hybridization is an efficient approach to produce new citrus rootstock candidates. The morphology of somatic hybrids SMC-58 and SMC-73 shows some intermediate characters between the parents. This type of inheritance has also been described in somatic hybrids between Citrus and related genera such as Citropsis, Severinia, and Microcitrus (Smith et al., 2013) and also between different Citrus species (Olivares-Fuster, 1998). The growth of somatic hybrids when compared with their parents is slower as it has also been described in several citrus allotetraploid somatic hybrids (Grosser et al, 1998; Grosser et al., 2012). This character is related to the increase in ploidy level (Lee, 1988, 1990). Citrus tetraploid hybrids have been used to increase the tree density in orchards to maximize the management efficiency. Furthermore, tetraploid rootstocks do not reduce the yield efficiency of the scion (Ruiz et al., 2016a) and produce fruits with excellent organoleptic qualities (Grosser et al., 2015). Therefore, the profitability of citrus plantations can be increased using tetraploid citrus rootstocks (Grosser et al., 1995, 2015; Grosser and Chandler, 2003).
We have evaluated the tolerance/susceptibility of CM + CC somatic hybrids to the severe T388 CTV strain. This strain causes different symptoms in susceptible citrus genotypes. The symptoms include seedling yellows, vein corking, or stem pitting when used either as varieties or rootstocks and the quick-decline of trees grafted onto SO (Moreno et al., 2008; Lee and Keremane, 2013). CM was found very sensitive to T388, whereas CC and the two CM + CC somatic hybrids were found to be tolerant. In Spain and in other Mediterranean countries, severe strains of CTV have been identified even though the incidence is low (Moreno et al., 2008). However, Toxoptera citricida (Kirkaldy), which is a very efficient vector of severe CTV strains, is already present in northern Portugal and north western Spain. The probable introduction of this aphid into the citrus producing areas would predictably cause a dispersion of severe CTV strains that would affect the trees grafted onto CM (Ilharco et al., 2005). Therefore, it is very important to have alternatives to this rootstock that can be used in alkaline and saline soils, where CC is not a good choice. In other studies, the quick decline was evaluated in somatic hybrids obtained from SO and several tolerant species, but global conclusions could not be reached. While somatic hybrids between SO and Rangpur lime (C. limonia Osb.) or Rough lemon (C. jambhiri Lush) were tolerant, hybrids obtained from SO and trifoliate orange (P. trifoliata) or Cleopatra mandarin were susceptible to this disease (Grosser et al., 1996). The inheritance of some traits such as CTV tolerance in the somatic hybrids is clearly coupled with the dominance or codominance of the trait in relation to the parental combinations (Bassene et al., 2009; Gmitter et al., 2012).
The performance of the somatic hybrids in the presence of soil carbonates, which are abundant in the Mediterranean citrus producing areas, is similar to the tolerant CM and much better than CC. The CM + CC hybrids are also more tolerant to salinity than CC. Enhanced tolerance to these stresses as well as to drought and boron excess has also been described in citrus rootstocks with increased ploidy (Saleh et al., 2008; Grosser et al., 2012; Allario et al., 2013; Tan et al., 2015; Ruiz et al., 2016a,b,c). These somatic hybrids have already started to yield fruits even though fruits are still sporadic and scarce. A large number of apomictic seeds per fruit were found. This characteristic is very important for citrus rootstocks as their clonal propagation and cultivation in nurseries are made easy. Field experiments have already been initiated in collaboration with Agromillora Research S.L. and will allow, within a few years, to confirm the data obtained in the greenhouse experiments and to collect additional information about fruit quality and yield induced by the grafted variety. All this information will be analyzed to determine if any of the studied somatic hybrids can be used commercially, which would be a great advantage for the Mediterranean citriculture.
The importance of performing in-depth molecular and physiological characterization of somatic hybrids
The main goal of citrus rootstock improvement based on somatic hybridization by protoplast fusion is to recover allotetraploid somatic hybrids between parents displaying complementary characteristics as seen in our study. Previous studies on somatic hybridization variability carried out in the past decades reveal that characters expressed by the hybrids can be non-additive. The hybrid phenotype can differ from the addition of parental effects given that allopolyploidization triggers gene expression changes and modifies epigenetics altering the phenotype (Bassene et al., 2009, 2010; Dambier et al., 2011; Xu et al., 2014). Some studies discuss to what extent these changes are caused by de novo interactions established between genomes coming from different species (Hegarty et al., 2009) or to the ploidy gain (Dambier et al., 2011; Tan et al., 2017). Allopolyploidization, generated either by sexual or somatic hybridization, involves the coexistence of parental genomes in a single nucleus. Additionally, in the case of allotetraploid somatic hybrids, changes also take place in cytoplasmic genome composition. The new genomic configuration is associated with diverse reorganizations and modifications affecting the structure and regulation of the new somatic hybrid genome (Comai et al., 2000; Ozkan et al., 2001; Wang et al., 2006a; Soltis and Soltis, 2009; Flagel and Wendel, 2010). This event, coined as genomic shock (Song et al., 1995), has a dynamic and stochastic nature and is composed of diverse processes such as fragment elimination or exchange at sub-chromosomic or chromosomic level, modifications in the methylation pattern, gene repression/expression changes, and activation of transposable elements (Chen, 2007; Xu et al., 2014) among others. These changes modify the gene expression either by altering the sequence or by epigenetic regulation (Comai, 2005). In addition, these changes may confer genome plasticity to improve the adaptation of the hybrids to the environment (Chen, 2007). The neoregulation of parental genomes in allopolyploid plants would greatly explain the obtention of genotypes and phenotypes that were absent in the diploid pool (Osborn et al., 2003) and the non-additive inheritance (He et al., 2003; Albertin et al., 2006; Hegarty et al., 2006; Wang et al., 2006a,b; Chen, 2007, 2010; Flagel et al., 2008; Flagel and Wendel, 2010). The study of genome expression in neopolyploids has recently gained importance, as it has been proposed as a useful approach to understand how genomes work and evolve (Gmitter et al., 2012; Gianinetti, 2013).
Somatic hybridization is more efficient than sexual hybridization as a method for citrus breeding when parents display a complex reproductive biology such as apomixis and high heterozygosity, as in the case of most of the rootstocks used. Nevertheless, it is still necessary to regenerate an adequate number of plants from each fusion to perform further screenings that verify their characteristics and agronomic behavior. This is essential to properly assess their usefulness in breeding programs, yet, the molecular basis of the traits that shape the rootstock agronomical behavior is still unknown. The new genetic (Ollitrault et al., 2012a) and genomic (Wu et al., 2014) tools available nowadays along with the affordable sequencing technologies are paving the way for the availability of numerous molecular markers and genetic information. This knowledge will contribute to the understanding of the molecular processes behind these traits and shorten the time required to perform additional evaluations. It is also essential to have rapid screening methods for early evaluations in greenhouse conditions. This will maximize the efficiency of breeding programs in terms of time, resources, and labor costs. Only those traits that are strictly necessary should be considered for long-term field evaluations.
Conclusion
Somatic hybrids SMC-58 and SMC-73 are promising citrus rootstocks for areas with the presence of CTV and calcareous and saline soils. They have punctual sub-chromosomic losses and show differences in morphology and physiological behavior, both between them and when compared with their parents. This is an evidence of genomic alterations that affect each hybridization event individually and are somehow independent from parental combinations. These identified genetic variations, along with the possible neoregulation events, the new cytoplasmic combinations, and the ploidy gain, might be the underlying phenotypic differences found between the hybrids and the phenotypic deviation from parental additive inheritance. Further investigation on somatic hybrids can add great value to citrus breeding programs as it can reveal information crucial to understand the principles operating in citrus genome expression, regulation, and evolution.
Author contributions
LN, EP-M, PO, and RM conceived the study and were in charge of the direction and planning. MR, AQ, GP-B, EP-M, LN, and RM contributed in the experiment design. MR, AQ, GP-B, and AG-L performed the experiments. MR, AG-L, and PA analyzed the data. MR and PA took the lead in interpreting the results and writing the manuscript with input and review from LN, EP-M, and PO.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financed by the Spanish MINECO Ministry, Project AGL2011-26490 and by agreement between IVIA and Agromillora Research S.L. MR was supported by grant co-funded by the Centre de Coopération International en Recherche Agronomique pour le Développement (CIRAD) and the Valencian Institute for Agricultural Research (IVIA), DOCV [2010/8910]. We are very grateful to Dr. Mireia Bordas from Agromillora Research that made the micropropagation of the rootstocks, Dr. José Guerri, and Dr. Karelia Velázquez for their help in the CTV analysis, the technical assistants Frederique Ollitrault, Mª Carmen Prieto, Enric Alcaide, Carmen Casamayor, Teresa García, Carmen Ortega, and Antonio Navarro, and the greenhouse technical team headed by José Antonio Pina, Rafa Montalt, and Diego Conchilla for their kind support. We would also like to thank Yoko Hiraoka for her useful editing suggestions.
References
- Albertin W., Balliau T., Brabant P., Chèvre A. M., Eber F., Malosse C., et al. (2006). Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173, 1101–1113. 10.1534/genetics.106.057554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P., Cuenca J., Hernández M., Juárez J., Navarro L., Ollitrault P. (2015). Genetic mapping of centromeres in the nine Citrus clementina chromosomes using half-tetrad analysis and recombination patterns in unreduced and haploid gametes. BMC Plant Biol. 15:80 10.1186/s12870-015-0464-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P., Froelicher Y., Schwarz S., Agustí M., Hernández M., Juárez J., et al. (2011). Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Ann. Bot. 108, 37–50. 10.1093/aob/mcr099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P., García-Lor A., Juarez J., Navarro L. (2016). Recovery of citrus cybrid plants with diverse mitochondrial and chloroplastic genome combinations by protoplast fusion followed by in vitro shoot, root or embryo micrografting. Plant Cell Tiss. Organ. Cult. 126, 205–217. 10.1007/s11240-016-0991-84109849 [DOI] [Google Scholar]
- Allario T., Brumos J., Colmenero-Flores J. M., Iglesias D. J., Pina J. A., Navarro L., et al. (2013). Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 36, 856–868. 10.1111/pce.12021 [DOI] [PubMed] [Google Scholar]
- Ballester-Olmos J. F., Pina J. A., Carbonell E. A., Moreno P., Hemoso de Mendoza A., Cambra M., et al. (1993). Biological diversity of citrus tristeza virus (Ctv) Isolates in Spain. Plant Pathol. 42, 219–229. 10.1111/j.1365-3059.1993.tb01494.x [DOI] [Google Scholar]
- Barkley N. A., Roose M. L., Krueger R. R., Federici C. T. (2006). Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor. App. Gen. 112, 1519–1531. 10.1007/s00122-006-0255-9 [DOI] [PubMed] [Google Scholar]
- Barrett H. C., Hutchison D. J. (1978). Spontaneous tetraploidy in apomictic seedlings of citrus. Econ. Bot. 32, 27–45. 10.1007/BF02906727 [DOI] [Google Scholar]
- Bassene J. B., Froelicher Y., Dhuique-Mayer C., Mouhaya W., Ferrer R. M., Ancillo G., et al. (2009). Non-additive phenotypic and transcriptomic inheritance in a citrus allotetraploid somatic hybrid between C. reticulata and C. limon: the case of pulp carotenoid biosynthesis pathway. Plant Cell Rep. 28, 1689–1697. 10.1007/s00299-009-0768-1 [DOI] [PubMed] [Google Scholar]
- Bassene J. B., Froelicher Y., Dubois C., Ferrer M. R., Navarro L., Ollitrault P., et al. (2010). Non-additive gene regulation in a citrus allotetraploid somatic hybrid between C. reticulata Blanco and C. limon (L.) Burm. Heredity 105, 299–308. 10.1038/hdy.2009.162 [DOI] [PubMed] [Google Scholar]
- Bordas M., Torrents J., Navarro L. (2015). Micropropagation for evaluation of new citrus somatic hybrid rootstocks. Acta Hortic. 1065, 329–334. 10.17660/ActaHortic.2015.1065.39 [DOI] [Google Scholar]
- Bryan G. J., McNicoll J., Ramsay G., Meyer R. C., De Jong W. S. (1999). Polymorphic simple sequence repeats markers in chloroplast genomes of Solanaceous plants. Theor. Appl. Gen. 99, 859–867. 10.1007/s001220051306 [DOI] [Google Scholar]
- Cabasson C. M., Luro F., Ollitrault P., Grosser J. W. (2001). Non-random inheritance of mitochondrial genomes in Citrus hybrids produced by protoplast fusion. Plant Cell Rep. 20, 604–609. 10.1007/s002990100370 [DOI] [Google Scholar]
- Cai X., Fu J., Guo W. (2017). Mitochondrial genome of callus protoplast has a role in mesophyll protoplast regeneration in citrus: evidence from Transgenic GFP somatic homo-fusion. Hortic. Plant J. 3, 177–182. 10.1016/j.hpj.2017.10.001 [DOI] [Google Scholar]
- Cambra M., Gorris M. T., Marroquín C., Román M. P., Olmos A., Martinez M. C., et al. (2000). Incidence and epidemiology of Citrus tristeza virus in the Valencian Community of Spain. Virus Res. 71, 85–95. 10.1016/S0168-1702(00)00190-8 [DOI] [PubMed] [Google Scholar]
- Campbell C. R., Plank C. O. (1998). Preparation of plant tissue for laboratory analysis, in Handbook of Reference Methods for Plant Analysis, ed Kalra Y. P. (Boca Raton, FL: CRC Press; ), 37–49. [Google Scholar]
- Castle W. S., Nunnallee J., Manthey J. A. (2009). Screening citrus rootstocks and related selections in soil and solution culture for tolerance to low-iron stress. Hortic. Sci. 44, 638–645. [Google Scholar]
- Chen Z. J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann. Rev. Plant Biol. 58, 377–406. 10.1146/annurev.arplant.58.032806.103835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15, 57–71. 10.1016/j.tplants.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., de Vicente M. C., Meng H., Guo W., Tao N., Deng X. (2005). A set of primers for analyzing chloroplast DNA diversity in Citrus and related genera. Tree Phys. 25, 661–672. 10.1093/treephys/25.6.661 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Guo W., Deng X. (2003). Molecular characterization of cytoplasmic and nuclear genomes in phenotypically abnormal Valencia orange (Citrus sinensis) plus Meiwa kumquat (Fortunella crassifolia) intergeneric somatic hybrids. Plant Cell Rep. 21, 445–451. 10.1007/s00299-002-0532-2 [DOI] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. 10.1038/nrg1711 [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A. P., Winter K., Holmes-Davis R., Reynolds S. H., Stevens Y., et al. (2000). Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell 12, 1551–1567. 10.1105/tpc.12.9.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca J., Aleza P., Navarro L., Ollitrault P. (2013). Assignment of SNP allelic configuration in polyploids using competitive allele-specific PCR: application to citrus triploid progeny. Ann. Bot. 111, 731–742. 10.1093/aob/mct032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca J., Froelicher Y., Aleza P., Juárez J., Navarro L., Ollitrault P. (2011). Multilocus half-tetrad analysis and centromere mapping in citrus: evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv 'Fortune'. Heredity (Edinb) 107, 462–470. 10.1038/hdy.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppen E. (2007). Genotyping by allele-specific amplification (KASPar). CSH Protoc. 007:pdb.prot4841. 10.1101/pdb.prot4841 [DOI] [PubMed] [Google Scholar]
- Curk F., Ollitrault F., García-Lor A., Luro F., Navarro L., Ollitrault P. (2016). Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann Bot. 117, 565–583. 10.1093/aob/mcw005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambier D., Benyahia H., Pensabene-Bellavia G., Kaçar Y. A., Froelicher Y., Belfalah Z., et al. (2011). Somatic hybridization for citrus rootstock breeding: an effective tool to solve some important issues of the Mediterranean citrus industry. Plant Cell Rep. 30, 883–900. 10.1007/s00299-010-1000-z [DOI] [PubMed] [Google Scholar]
- Dellaporta S. L., Wood J., Hicks J. B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. 10.1007/BF02712670 [DOI] [Google Scholar]
- Duminil J., Pemonge M. H., Petit R. J. (2002). A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA. Mol. Ecol. Res. 2, 428–430. 10.1046/j.1471-8286.2002.00263.x [DOI] [Google Scholar]
- Esselink G. D., Nybom H., Vosman B. (2004). Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting-peak ratios) method. Theor. Appl. Genet. 109, 402–408. 10.1007/s00122-004-1645-5 [DOI] [PubMed] [Google Scholar]
- Fanciullino A. L., Gancel A. L., Froelicher Y., Luro F., Ollitrault P., Brillouet J. M. (2005). Effects of nucleo-cytoplasmic interactions on leaf volatile compounds from citrus somatic diploid hybrids. J. Agric. Food Chem. 53, 4517–4523. 10.1021/jf.0502855 [DOI] [PubMed] [Google Scholar]
- FAO (2016). Food and Agriculture Organization of the United Nations. FAOSTAT Statistics Database. Available online at: www.fao.org/faostat/en/#home (Accessed Feb 08, 2017).
- Flagel L., Udall J., Nettleton D., Wendel J. (2008). Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6:16. 10.1186/1741-7007-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F. (2010). Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186, 184–193. 10.1111/j.1469-8137.2009.03107.x [DOI] [PubMed] [Google Scholar]
- Froelicher Y. (1999). Organisation de la Diversité dans le Genre Clausena Burm.f . (RUTACEAE) et modalité de son explotation en création variétal. Dissertation, University of Paris-sud. [Google Scholar]
- Froelicher Y., Mouhaya W., Bassene J. B., Costantino G., Kamiri M., Luro F., et al. (2011). New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny. Tree Genet. Genomes 7:49–61. 10.1007/s11295-010-0314-x [DOI] [Google Scholar]
- García-Lor A., Ancillo G., Navarro L., Ollitrault P. (2013a). Citrus (Rutaceae) SNP markers based on competitive allele-specific PCR; transferability across the aurantioideae subfamily. App. Plant Sci. 1:1200406. 10.3732/apps.1200406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Lor A., Curk F., Snoussi-Trifa H., Morillon R., Ancillo G., Luro F., et al. (2013b). A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees' group (Citrinae, Rutaceae) and the origin of cultivated species. Ann Bot. 111, 1–19. 10.1093/aob/mcs227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinetti A. (2013). A criticism of the value of midparent in polyploidization. J. Exp. Bot. 64, 4119–4129. 10.1093/jxb/ert263 [DOI] [PubMed] [Google Scholar]
- Gilliam J. W. (1971). Rapid measurement of chlorine in plant materials. Proc. Soil Sci. Soc. Am. J. 35, 512–513. 10.2136/sssaj1971.03615995003500030051x [DOI] [Google Scholar]
- Gmitter F. G., Chen C., Machado M. A., de Souza A. A., Ollitrault P., Froehlicher Y., et al. (2012). Citrus genomics. Tree Genet. Genomes 8, 611–626. 10.1007/s11295-012-0499-2 [DOI] [Google Scholar]
- Gomez-Cadenas A., Iglesias D. J., Arbona V., Colmenero-Flores J. M., Primo-Millo E., Talon M. (2003). Physiological and molecular responses of Citrus to salinity. Rec. Res. Dev. Plant Mol. Biol. 1, 281–298. [Google Scholar]
- Grosser J. W., Barthe G. A., Castle B., Gmitter F. G., Lee O. (2015). The development of improved tetraploid citrus rootstocks to facilitate advanced production systems and sustainable citriculture in Floridam, in XII International Citrus Congress - International Society of Citriculture (Valencia), Vol. 1065, 319–327. [Google Scholar]
- Grosser J. W., Chandler J. L. (2003). New Citrus rootstocks via protoplast fusion. Acta Hortic. 622, 491–497. 10.17660/ActaHortic.2003.622.54 [DOI] [Google Scholar]
- Grosser J. W., Garnsey S. M., Halliday C. (1996). Assay of sour orange somatic hybrid rootstocks for quick-decline disease caused by citrus tristeza virus. Proc. Int. Soc. Citr. 1, 353–356 [Google Scholar]
- Grosser J. W., Gmitter F. G. Jr. (2005). Applications of somatic hybridization and cybridization in crop improvement, with citrus as a model. In Vitro Cell Dev. Plant 41, 220–225 10.1079/IVP2004634 [DOI] [Google Scholar]
- Grosser J. W., Gmitter F. G. (2011). Protoplast fusion for production of tetraploids and triploids: applications for scion and rootstock breeding in citrus. Plant Cell Tiss. Organ. Cult. 104, 343–357. 10.1007/s11240-010-9823-4 [DOI] [Google Scholar]
- Grosser J. W., Gmitter F. G., Castle W. S., Chandler J. L. (1995). Production and evaluation of citrus somatic hybrid rootstocks: progress report. Proc. Fla. State Hort. Soc. 108, 140–143. [Google Scholar]
- Grosser J. W., Jiang J., Louzada E. S., Chandler J. L., Gmitter F. G. (1998). Somatic hybridization, an integral component of citrus cultivar improvement: II. Rootstock improvement. HortScience 33, 1060–1061. [Google Scholar]
- Grosser J. W., Ollitrault P., Olivares-Fuster O. (2000). Somatic hybridization in citrus: An effective tool to facilitate variety improvement. In Vitro Cell. Dev. Biol. Plant 36, 434–449. 10.1007/s11627-000-0080-9 [DOI] [Google Scholar]
- Grosser J. W., Omar A. A., Gmitter J. A., Syvertsen J. P. (2012). Salinity tolerance of valencia orange trees on allotetraploid rootstocks. Proc. Fla. State Hort. Soc. 125, 50–55. [Google Scholar]
- Guo W., Deng X. (1999). Intertribal hexaploid somatic hybrid plants regeneration from electrofusion between diploids of Citrus sinensis and its sexually incompatible relative, Clausena lansium. Theor. Appl. Genet. 8, 581–585. 10.1007/s001220051107 [DOI] [Google Scholar]
- Guo W., Prasad D., Cheng Y., Serrano P., Deng X., Grosser J. (2004). Targeted cybridization in citrus: transfer of Satsuma cytoplasm to seedy cultivars for potential seedlessness. Plant Cell Rep. 22, 752–758. 10.1007/s00299-003-0747-x [DOI] [PubMed] [Google Scholar]
- Guo W. W., Cheng Y. J., Deng X. X. (2002). Regeneration and molecular characterization of intergeneric somatic hybrids between Citrus reticulata and Poncirus trifoliata. Plant Cell Rep. 20, 829–834. 10.1007/s00299-001-0399-7 [DOI] [Google Scholar]
- Guo W. W., Wu R. C., Cheng Y. J., Deng X. X. (2007). Production and molecular characterization of Citrus intergeneric somatic hybrids between red tangerine and citrange. Plant Breed 126, 72–76. 10.1111/j.1439-0523.2006.01315.x [DOI] [Google Scholar]
- He P., Friebe B. R., Gill B. S., Zhou J. M. (2003). Allopolyploidy alters gene expression in the highly stable hexaploid wheat Plant Mol. Biol. 52, 401–414. 10.1023/A:1023965400532 [DOI] [PubMed] [Google Scholar]
- Hegarty M. J., Barker G. L., Brennan A. C., Edwards K. J., Abbott R. J., Hiscock S. J. (2009). Extreme changes to gene expression associated with homoploid hybrid speciation. Mol. Ecol. 18, 877–889. 10.1111/j.1365-294X.2008.04054.x [DOI] [PubMed] [Google Scholar]
- Hegarty M. J., Barker G. L., Wilson I. D., Abbott R. J., Edwards K. J., Hiscock S. J. (2006). Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Current Biol 16, 1652–1659. 10.1016/j.cub.2006.06.071 [DOI] [PubMed] [Google Scholar]
- Herrero R., Asíns M. J., Pina J. A., Carbonell E. A., Navarro L. (1996). Genetic diversity in the orange subfamily aurantioideae. Genetic relationships among genera and species. Theor. App. Genet. 93, 1327–1334. 10.1007/BF00223466 [DOI] [PubMed] [Google Scholar]
- Hoagland D. I., Arnon D. R. (1950). The water culture method to grow plants without soil. Circular. Calif Agric. Exp. Stat. 347, 1–32. [Google Scholar]
- Ilharco F. A., Sousa-Silva C. R., Alvarez-Alvarez A. (2005). First report on Toxoptera citricidus (Kirkaldy) in Spain and Continental Portugal (Homoptera, Aphidoidea). Agric. Lus 51, 19–21. 10.1111/epp.12181 [DOI] [Google Scholar]
- IPGRI (1999). Descriptors for Citrus. Rome: International Plant Genetic Resources Institute. [Google Scholar]
- Kamiri M., Stift M., Srairi I. Costantino, G., El Moussadik A., Hmyene A., et al. (2011). Evidence for non-disomic inheritance in a Citrus interspecific tetraploid somatic hybrid between C. reticulata and C. limon using SSR markers and cytogenetic analysis. Plant Cell Rep. 30, 1415–1425. [DOI] [PubMed] [Google Scholar]
- Kijas J. M. H., Thomas M. R., Fowler J. C. S., Roose M. L. (1997). Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor. Appl. Genet. 94, 701–706. [Google Scholar]
- Kobayashi S., Ohgawara T., Fujiwara K., Oiyama I. (1991). Analysis of Cytoplasmic Genomes in Somatic Hybrids between Navel Orange (Citrus sinensis Osb) and Murcott Tangor. Theor. Appl. Genet. 82, 6–10. 10.1007/BF00231270 [DOI] [PubMed] [Google Scholar]
- Koltunow A. (1993). Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski A., Rabe E. (1995). Bud age affects sprouting and flowering in clementine mandarin (Citrus reticulata Blanco). HortScience 30, 1366–1368. [Google Scholar]
- Lee L. S. (1988). Citrus Polyploidy: Origins and Potential for Cultivar Improvement. Aust. J. Agric. Res. 39, 735–747. 10.1071/AR9880735 [DOI] [Google Scholar]
- Lee L. S. (1990). Prospects for using citrus tetraploids for rootstocks. Proc. Fla. State Hortic. Soc. 108, 140–143. [Google Scholar]
- Lee R. F., Keremane M. L. (2013). Mild strain cross protection of tristeza: a review of research to protect against decline on sour orange in Florida. Front. Microbiol. 4:259. 10.3389/fmicb.2013.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas E. V. (1993). Salinity and citriculture. Tree Physiol. 12, 195–216. 10.1093/treephys/12.2.195 [DOI] [PubMed] [Google Scholar]
- Medina-Urrutia V., Madera K. F. L., Serrano P., Ananthakrishnan G., Grosser J. W., Guo W. W. (2004). New intergeneric somatic hybrids combining amblycarpa mandarin with six trifoliate/trifoliate hybrid selections for lime rootstock improvement. HortScience 39, 355–360. [Google Scholar]
- Mendes-da-Gloria F. J., Mourao F. D., Camargo L. E. A., Mendes B. M. J. (2000). Caipira sweet orange plus Rangpur lime: a somatic hybrid with potential for use as rootstock in the Brazilian citrus industry. Genet. Mol. Biol. 23, 661–665. 10.1590/S1415-47572000000300026 [DOI] [Google Scholar]
- Miranda M., Motomura T., Ikeda F., Ohgawara T., Saito W., Endo T., et al. (1997). Somatic hybrids obtained by fusion between Poncirus trifoliata (2x) and Fortunella hindsii (4x) protoplasts. Plant Cell Rep. 16:401 10.1007/BF01146782 [DOI] [PubMed] [Google Scholar]
- Moreira C. D., Chase C. D., Gmitter F. G., Grosser J. W. (2000). Inheritance of organelle genomes in citrus somatic cybrids. Mol. Breed. 6, 401–405. 10.1023/A:1009632223708 [DOI] [Google Scholar]
- Moreno P., Ambros S., Albiach-Marti M. R., Guerri J., Pena L. (2008). Plant diseases that changed the world. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 9, 251–268. 10.1111/j.1364-3703.2007.00455.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P., Guerri J., Muñoz N. (1990). Identification of Spanish strains of citrus tristeza virus (CTV) by analysis of double-stranded RNAs (dsRNA). Phytopathology 80 477–482. 10.1094/Phyto-80-477 [DOI] [Google Scholar]
- Moriguchi T., Motomura T., Hidaka T., Akihama T., Omura M. (1997). Analysis of mitochondrial genomes among Citrus plants produced by the interspecific somatic fusion of ‘Seminole' tangelo with rough lemon. Plant Cell Rep. 16, 397–400. 10.1007/s002990050247 [DOI] [PubMed] [Google Scholar]
- Mourao F. D. A., Pio R., Januzzi M., Beatriz M., Azevedo F. A., Schinor E. H., et al. (2008). Evaluation of citrus somatic hybrids for tolerance to Phytophthora nicotianae and citrus tristeza virus. Sci. Hortic. 115, 301–308. 10.1016/j.scienta.2007.10.004 [DOI] [Google Scholar]
- Navarro L. (2015). The Spanish citrus industry. Acta Hortic. 1065, 41–48. 10.17660/ActaHortic.2015.1065.1 [DOI] [Google Scholar]
- Navarro L., Pina J. A., Juárez J., Ballester-Olmos J. F., Arregui J. M., Ortega C., et al. (2002). The citrus variety improvement program in Spain in the period 1975-2001, in Proceedings of the 15th Conference of the International Organization of Citrus Virologists, (Riverside, CA), 306–316. [Google Scholar]
- Oberwalder B., Schilde-Rentschler L., Ruoss B., Wittemann S., Ninnemann H. (1998). Asymmetric protoplast fusions between wild species and breeding lines of potato-effect of recipients and genome stability. Theor. Appl. Genet. 97, 1347–1354. 10.1007/s001220051028 [DOI] [Google Scholar]
- Olivares-Fuster O. (1998). Hibridación Somática de Cítricos. Dissertation, Universidad Politécnica de Valencia. [Google Scholar]
- Ollitrault F., Terol J., Pina J. A., Navarro L., Talon M., Ollitrault P. (2010b). Development of SSR Markers from Citrus Clementina (Rutaceae) bac end sequences and interspecific transferability in citrus. Am. J. Bot. 97, 124–129. 10.3732/ajb.1000280 [DOI] [PubMed] [Google Scholar]
- Ollitrault P., Dambier D., Cabasson C., Allent V., Engelman F. (1994). Optimized management of citrus embryogenic calli for breeding programmes. Fruits 49, 394–397. [Google Scholar]
- Ollitrault P., Dambier D., Froelicher Y., Luro F., Cottin R. (2001). La diversite des agrumes: structuration et exploitation par hybridation somatique. C. R. Acad. Agric. 8, 197–221. [Google Scholar]
- Ollitrault P., Guo W., Grosser J. W. (2007). Recent advances and evolving strategies in Citrus somatic hybridization, in Citrus Genetics, Breed Andbiotechnology, ed Khan I. H. (Wallingford: CABI Publishing; ), 235–260. [Google Scholar]
- Ollitrault P., Guo W., Grosser W. (2010a). Somatic hybridization, in Citrus Genetics, Breeding and Biotechnology, ed Khan I. (Cambridge, MA: CAB International; ), 235–260. [Google Scholar]
- Ollitrault P., Jacquemond C., Dubois C., Luro F. (2003). Citrus, in Genetic Diversity of Cultivated Tropical Plants, eds Hamon P., Seguin M., Perrier X., Glaszmannn J. C. (Enfield: Science Publishers Inc; ), 193–217. [Google Scholar]
- Ollitrault P., Terol J., Chen C., Federici C. T., Lotfy S., Hippolyte I., et al. (2012a). A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genom 13:593 10.1186/1471-2164-13-593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault P., Terol J., Garcia-Lor A., Bérard A., Chauveau A., Froelicher Y., et al. (2012b). SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genom 13:13 10.1186/1471-2164-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault P., Vanel F., Froelicher Y., Dambier D. (2000). Creation of triploid Citrus hybrids by electrofusion of haploid and diploid protoplasts. Acta Hortic. 535, 191–198. 10.17660/ActaHortic.2000.535.23 [DOI] [Google Scholar]
- Omar A. A., Murata M., Yu Q., Gmitter F. G., Jr., Chase C. D., James H., et al. (2017). Production of three new grapefruit cybrids with potential for improved citrus canker resistance. In Vitro Cell. Dev. Biol. Plant 53:256–269 10.1007/s11627-017-9816-7 [DOI] [Google Scholar]
- Osborn T. C., Pires J. C., Birchler J. A., Auger D. L., Chen Z. J., Lee H. S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. 10.1016/S0168-9525(03)00015-5 [DOI] [PubMed] [Google Scholar]
- Ozkan H., Levy A. A., Feldman M. (2001). Allopolyploidy induced rapid genome evolution in the wheat (Aegilops-Triticum) grup. Plant Cell 13, 1735–1747. 10.1105/tpc.13.8.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensabene-Bellavia G. (2009). Aplicación de la hibridación somática a la mejora de la citricultura española, Dissertation, Universidad politécnica de Valencia. [Google Scholar]
- Pensabene-Bellavia G., Ruiz M., Aleza P., Olivares-Fuster O., Ollitrault P., Navarro L. (2015). Chromosome instability in Carrizo citrange + Citrus macrophylla somatic hybrids. Sci. Hortic. 1065, 677–685. 10.17660/ActaHortic.2015.1065.85 [DOI] [Google Scholar]
- Perez R. M., Galiana A. M., Navarro L., Duran-Vila N. (1998). Embryogenesis in vitro of several Citrus species and cultivars. J. Hortic. Sci. Biotech. 73, 419–429. 10.1080/14620316.1998.11511050 [DOI] [Google Scholar]
- Rangan T. S., Murashige T., Bitters W. P. (1969). In vitro studies of zygotic and nucellar embryogenesis in Citrus, in Proceeding of 1st international Citrus Symptoms, Vol. 1, eds Chapman H. D. (Riverside, CA: University of California; ), 225–229. [Google Scholar]
- Ruiz M., Quiñones A., Martínez-Alcántara B., Aleza P., Morillon R., Navarro L., et al. (2016a). Tetraploidy enhances boron-excess tolerance in carrizo citrange (Citrus sinensis L. Osb. x Poncirus trifoliata L. Raf). Front. Plant Sci. 7:701. 10.3389/fpls.2016.00701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M., Quiñones A., Martínez-Alcántara B., Aleza P., Morillon R., Navarro L., et al. (2016b). Effects of salinity on diploid (2x) and doubled diploid (4x) Citrus macrophylla genotypes. Sci. Hortic. 207, 33–40. 10.1016/j.scienta.2016.05.007 [DOI] [Google Scholar]
- Ruiz M., Quiñones A., Martínez-Cuenca M. R., Aleza P., Morillon R., Navarro L., et al. (2016c). Tetraploidy enhances the ability to exclude chloride from leaves in Carrizo citrange seedlings. J. Plant Phys. 205, 1–10. 10.1016/j.jplph.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Saito W., Ohgawara T., Shimitzu J., Ishii S., Kobayashi S. (1993). Citrus cybrid regeneration following cell-fusion between nucellar cells and mesophyll-cells. Plant Sci. 88, 195–201. 10.1016/0168-9452(93)90091-D [DOI] [Google Scholar]
- Saleh B., Allario T., Dambier D., Ollitrault P., Morillon R. (2008). Tetraploid citrus rootstocks are more tolerant to salt stress than diploid. C. R. Biol. 331, 703–710. 10.1016/j.crvi.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Satpute A. D., Chen C., Gmitter F. G. Jr, Ling, P., Yu Q., Grosser M. R., et al. (2015). Cybridization of grapefruit with ‘dancy' mandarin leads to improved fruit characteristics. J. Am. Soc. Hortic. Sci. 140:427–435. [Google Scholar]
- Smith M. W., Gultzow D. L., Newman T. K. (2013). First fruiting intergeneric hybrids between Citrus and Citropsis. J. Am. Soc. Hortic. Sci. 138, 57–63. [Google Scholar]
- Smyda-Dajmund P., Sliwka J., Wasilewicz-Flis I., Jakuczun H., Zimnoch-Guzowska E. (2016). Genetic composition of interspecific potato somatic hybrids and autofused 4x plants evaluated by DArT and cytoplasmic DNA markers. Plant Cell Rep. 35, 1345–1358. 10.1007/s00299-016-1966-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P. S., Soltis D. E. (2009). The role of hybridization in plant speciation. Ann. Rev. Plant Biol. 60, 561–588. 10.1146/annurev.arplant.043008.092039 [DOI] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T. C. (1995). Rapid genome changes in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 7719–7723. 10.1073/pnas.92.17.7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Xu C. H., Wang M. Q., Zhi D. Y., Xia G. M. (2014). Genomic changes at the early stage of somatic hybridization. Genet. Mol. Res. 13, 1938–1948. 10.4238/2014.March.17.21 [DOI] [PubMed] [Google Scholar]
- Sundberg E., Grimelius K. (1991). Effects of parental ploidy level and genetic divergence on chromosome elimination and chloroplast segregation in somatic hybrids within Brassicaceae. Theor. Appl. Genet. 83, 81–88 10.1007/BF00229229 [DOI] [PubMed] [Google Scholar]
- Tan F. Q., Tu H., Liang W. J., Long J. M., Wu X. M., Zhang H. Y., et al. (2015). Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. Junos cv. Ziyang xiangcheng) suggest its potential value for stress resistance improvement. BMC Plant Biol 15:89 10.1186/s12870-015-0450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F. Q., Tu H., Wang R., Wu X. M., Xie K. D., Chen J. J., et al. (2017). Metabolic adaptation following genome doubling in citrus doubled diploids revealed by non-targeted metabolomics. Metabolomics 13:143 10.1007/s11306-017-1276-x [DOI] [Google Scholar]
- Tusa N., Del Bosco S. F., Nigro F., Ippolito A. (2000). Response of cybrids and a somatic hybrid of lemon to Phoma tracheiphila infections. HortScience 35:125–127. [Google Scholar]
- Vardi A., Breiman A., Galun E. (1987). Citrus cybrids -production by donor-recipient protoplast-fusion and verification by mitochondrial-DNA restriction profiles. Theor. Appl. Genet. 75, 51–58. 10.1007/BF00249142 [DOI] [Google Scholar]
- Wakana A., Uemoto S. (1987). Adventive embryogenesis in citrus. I. the occurrence of adventive embryos without pollination or fertilization. Am. J. Bot. 74, 517–530. [Google Scholar]
- Walker R. R. (1986). Sodium exclusion and potassium-sodium selectivity in salt treated Trifoliate orange (Poncirus trifoliata) and Cleopatra mandarin (Citrus reticulata) plants. Aust. J. Plant Phys. 135, 293–303. [Google Scholar]
- Wang J., Tian L., Lee H. S., Chen Z. J. (2006b). Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics 173, 965–974. 10.1534/genetics.106.056580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H., Wei N., Jiang H., Watson B., et al. (2006a). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172, 507–517. 10.1534/genetics.105.047894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weising K., Gardner R. C. (1999). A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42, 9–19. 10.1139/g98-104 [DOI] [PubMed] [Google Scholar]
- Wu G. A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. 10.1038/nbt.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Biswas M. K., Li M., Deng X., Xu Q., Guo W. (2014). Production and molecular characterization of diploid and tetraploid somatic cybrid plants between male sterile Satsuma mandarin and seedy sweet orange cultivars. Plant Cell Tiss. Organ. Cult. 116, 81–88. 10.1007/s11240-013-0384-1 [DOI] [Google Scholar]
- Xu S., Cai D., Tan F., Fang Y., Xie K., Grosser J. W., et al. (2014). Citrus somatic hybrid: an alternative system to study rapid structural and epigenetic reorganization in allotetraploid genomes. Plant Cell Tiss. Organ. Cult. 119, 511–522. 10.1007/s11240-014-0551-z [DOI] [Google Scholar]
- Yamamoto M., Kobayashi S. (1995). A cybrid plant produced by electrofusion between Citrus unshiu (satsuma mandarin) and C.sinensis (sweet orange). Plant Tiss. Cult. Lett. 12, 131–137. 10.5511/plantbiotechnology1984.12.131 [DOI] [Google Scholar]
- Zheng B. B., Wu X. M., Ge X. X., Deng X. X., Grosser J. W., Guo W. W. (2012). Comparative transcript profiling of a male sterile cybrid pummelo and its fertile type revealed altered gene expression related to flower development. PLoS ONE 7:e43758. 10.1371/journal.pone.0043758 [DOI] [PMC free article] [PubMed] [Google Scholar]