Abstract
Accumulating evidence points to a significant role of the circadian clock in the regulation of splicing in various organisms, including mammals. Both dysregulated circadian rhythms and aberrant pre-mRNA splicing are frequently implicated in human disease, in particular in cancer. To investigate the role of the circadian clock in the regulation of splicing in a cancer progression context at the systems-level, we conducted a genome-wide analysis and compared the rhythmic transcriptional profiles of colon carcinoma cell lines SW480 and SW620, derived from primary and metastatic sites of the same patient, respectively. We identified spliceosome components and splicing factors with cell-specific circadian expression patterns including SRSF1, HNRNPLL, ESRP1, and RBM 8A, as well as altered alternative splicing events and circadian alternative splicing patterns of output genes (e.g., VEGFA, NCAM1, FGFR2, CD44) in our cellular model. Our data reveals a remarkable interplay between the circadian clock and pre-mRNA splicing with putative consequences in tumor progression and metastasis.
Keywords: Circadian clock, Colorectal cancer progression, Spliceosome, Splicing factors, Alternative splicing, Differential rhythmicity
Graphical Abstract
Highlights
-
•
The spliceosome pathway shows a phase-shifted circadian expression in a cellular model of colorectal cancer progression.
-
•
Splicing-related genes exhibit circadian rhythms that differ between primary tumor and metastatic colorectal cancer cells.
-
•
Changes in the clock are associated with alternative splicing and circadian splicing patterns during tumor progression.
In most organisms, a variety of behavioral, physiological and cellular processes undergo time-of-day-dependent oscillations that are coordinated by an endogenous timing system - the circadian clock. Well known examples of circadian regulated cellular processes are the cell cycle, metabolism and, most recently, RNA processing including splicing. Aberrant pre-mRNA splicing is frequently implicated in cancer. Recent studies point to an interplay of the circadian clock and splicing, as well as to a role of the circadian clock in cancer. Yet the putative interplay between the clock and splicing in a cancer context remains elusive. By means of a comprehensive genome-wide analysis of time-course data for a colorectal cancer progression model, we explored the role of the circadian clock in the regulation of mRNA splicing, and its impact in the generation of alternatively spliced products that may influence tumor progression. In particular, we showed that alterations in the core-clock affect the rhythmicity of splicing factors and of spliceosome components during metastasis. These alterations correlate with the observed changes in alternative splicing patterns of well-known tumor progression-related genes. Our findings thus highlight a likely role for the clock as a regulator of alternative splicing events associated with tumor progression.
1. Introduction
Behavior, physiology, and cellular processes of most organisms undergo 24-h rhythms in activity that are generated by an endogenous biological timing system – the circadian clock. On the cellular level, the circadian clock regulates the timing of numerous biological processes such as cell growth and survival, DNA damage response, and metabolism [1, 2]. Disruptions of such processes can lead to aberrant cellular proliferation and are linked to cancer [[3], [4], [5]].
In mammals, a core-clock network (CCN) of genes and proteins forms cell-autonomous transcriptional/translational feedback loops that generate robust 24-h rhythms [6, 7]. A heterodimer complex consisting of the protein clock circadian regulator (CLOCK) and proteins from the aryl hydrocarbon receptor nuclear translocator like (ARNTL, also known as BMAL) family (BMAL1,2) regulates the transcriptional activation of the period circadian regulator (PER1,2,3), cryptochrome circadian regulator (CRY1,2), the nuclear receptor subfamily 1 group D (NR1D1,2, also known as REV-ERBA,B) and the RAR related orphan receptor (RORA,B) clock gene families. PER and CRY proteins inhibit the CLOCK:BMAL-mediated transcription, thereby forming a negative feedback loop. Members of the REV-ERB and ROR families regulate BMAL transcription via negative and positive feedbacks, respectively, and contribute to the fine-tuning of its expression. These interconnected feedback loops further drive the rhythmic expression of clock-controlled genes (CCGs) [8] detectable in 40–80% of all protein-coding genes in a tissue-dependent manner [9, 10].
Additional layers of post-transcriptional regulation account for the subsequent transmission of rhythmic information. These include alternative polyadenylation, mRNA degradation, translation, and alternative splicing (AS) [[11], [12], [13]]. AS of pre-mRNAs allows for the differential processing of multi-exon genes and for a subsequent reprogramming of the output isoform which significantly increases the transcriptome and proteome complexity [14]. The splicing process is catalyzed by the spliceosome [15, 16] and aided by a large number of auxiliary cis-acting regulatory elements and trans-acting factors – splicing factors (SFs) that regulate AS of specific pre-mRNAs. SFs which include members of the serine arginine rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) have crucial roles in both marking the splice site for spliceosome assembly and in fine-tuning of AS events by blocking or promoting access of the spliceosome to a 5′ or 3′ splice site [17]. The correct choice of the splice sites used and the resulting AS decisions are essential during development and cell differentiation, and for tissue-specificity [18].
Links between the circadian clock and splicing have been reported in Arabidopsis [19, 20], Drosophila [21], and mice [[22], [23], [24]]. In mammals, SFs modulate the mRNA expression or stability of the core-clock genes Cry1, Per1, and Per3 and the translation of the core-clock gene Rev-ErbB and the CCG arylalkylamine N-acetyltransferase (AANAT) [[25], [26], [27], [28]], suggesting a reciprocal interplay of the circadian clock and the splicing machinery.
Similarly to the impact caused by dysregulation of the clock, aberrant AS is also associated with various aspects of tumor biology such as control of proliferation and programmed cell death, metabolism, apoptosis, angiogenesis, and metastasis [16, 29]. Isoform switches in different tumor types are linked to losses in functional protein domain families that are frequently mutated in cancer, possibly conferring a selective advantage to tumor cells similar to that caused by somatic mutations in cancer drivers [30]. Moreover, various genes with alternative patterns of splicing that correlate with cancer progression such as the MDM2 proto-oncogene (MDM2) [31], the vascular endothelial growth factor (VEGF) [32], and the glycolytic enzyme pyruvate kinase M (PKM) [33] are also CCGs, as shown by our group and others [34]. As such, it is conceivable that a circadian regulation of splicing may play a key role in cancer progression. A clock-controlled timing of AS events might serve as a complex trans-acting mechanism via which the clock regulates the temporal diversity of proteins. Yet, a comprehensive analysis at the systems-level regarding the circadian regulation of splicing in cancer is still lacking.
In this study, we investigated the interplay between the circadian clock and pre-mRNA splicing regulation in a well-defined model of colorectal cancer (CRC) progression [35]. CRC is an extremely interesting model system for clock-cancer studies given the known promising impact of chronomodulated treatment in patients with metastatic CRC [36], the use of circadian rhythms as a biomarker for CRC patient survival [37], and the altered expression of clock genes and proteins found in cell lines and in tumor tissues of patients with CRC [[38], [39], [40]]. Following a whole-transcriptome approach, we compared the circadian phenotypes of two CRC cell lines which originate from samples resected from the primary tumor and the metastasis of the same patient and which are known to have different clock properties [38].
We identified diverse robustly circadian spliceosome components and SFs that exhibited differing oscillatory patterns in the CRC model and investigated the impact of their differential rhythmicity in terms of alterations in AS events between the cell lines. Interestingly, some of the circadian splicing-regulated genes including serine and arginine rich splicing factor 1 (SRSF1), hnRNP L like (HNRNPLL), epithelial splicing regulatory protein 1 (ESRP1), and RNA binding motif protein 8A (RBM 8A) showed differences in their respective peak phases and amplitudes which are likely to alter the outcome of AS events of their target genes. We further discovered several angiogenesis and cell-cell adhesion-associated genes which exhibited static and circadian changes in splicing, e.g., VEGFA, neural cell adhesion molecule 1 (NCAM1), fibroblast growth factor receptor 2 (FGFR2), and CD44. Aberrant splicing of these genes results in the production of isoforms that are known to confer additional oncogenic properties to tumor cells.
Taken together, our results suggest an important role of the circadian system in the regulation of splicing processes during metastasis and shed light into the complexity and the power of the circadian regulation of splicing switches and the resulting AS products which are likely to impact on tumor progression.
2. Materials and Methods
2.1. Cell Culture
Human colorectal carcinoma cell lines SW480 and SW620 were purchased from the ATCC. The SW480 cell line is derived from a primary Duke's stage B colon carcinoma and the SW620 cell line from a mesenteric lymph node metastasis in the same patient. Cells were maintained in DMEM low glucose (Lonza, Walkersville, MD, USA) culture medium supplemented with 10% FBS (Life technologies, Inc., Wilmington, DE, USA), 1% penicillin-streptomycin (Life technologies, Inc., Wilmington, DE, USA), 1% HEPES (Life technologies, Inc., Wilmington, DE, USA), and 2 mM Ultraglutamine (Lonza, Walkersville, MD, USA) at 37 °C in a humidified atmosphere with 5% CO2.
2.2. Single Live-Cell Microscopy
For single live-cell microscopy, SW480 and SW620 cells were transfected with a REV-ERBα-VNP plasmid using X-tremeGENE HP DNA Transfection reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. REV-ERBα-VNP positive cells were sorted via fluorescence-activated cell sorting and cultured under normal growth conditions until use. For the experiment, cells were plated in μ-Slide 8 Well Glass Bottom dishes (ibidi, Martinsried, DE). Time-course live-cell microscopy was performed using a CSU-X spinning disc confocal microscope (Nikon Corp., Tokio, JP). Pictures were taken and fluorescence intensity was determined every 30 min for 38 h.
2.3. Sample Preparation for 24-h Time-Course Microarrays
Cells were seeded in triplicates in 6-well plates with a density of 1–2 × 106 cells one day prior to the experiment. On the next day, cells were synchronized by medium change using the normal culture medium. Samples were taken every 3 h for a time-course of 24 h and prepared for RNA extraction. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) protocol including 15 min DNase I digestion according to the manufacturer's instructions. Prior to the purification procedure, medium was discarded and cells were washed twice with PBS and lyzed in RLT buffer (Qiagen, Valencia, CA, USA). RNA was eluted in 30–50 μl RNase-free water. The final RNA concentration was determined with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). RNA was then stored at −80 °C until use. Microarray hybridization was carried out by the Labor für funktionelle Genomforschung (LFGC, Charité – Universitätsmedizin Berlin) using GeneChip Human Transcriptome Arrays 2.0 (HTA 2.0) (Affymetrix Inc., Santa Clara, CA, USA).
2.4. Gene Expression Analysis
The microarray gene expression analysis was conducted in R. Expression data was pre-processed for all time points of both cell lines as one batch using the RMA (Robust Multichip Average) methodology [41, 42] as implemented in the R package oligo [43]. Transcript clusters were annotated with NCBI Gene IDs using Affymetrix HTA 2.0 annotation data (hta20transcriptcluster.db, v3.5). For genes annotated by multiple transcript clusters, only the transcript cluster with the highest mean expression over all time points in both cell lines was retained, resulting in a filtered dataset containing data for 24,213 transcripts. The gene expression data has been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5876.
2.5. Rhythmicity Analysis
To detect transcripts exhibiting rhythmic behavior with a 24-h period in their expression, successive filtering steps were applied. For the detection of circadian rhythmicity, the Rhythmicity Analysis Incorporating Nonparametric methods (RAIN) algorithm was used [44]. Acrophases of the 24-h cycling transcripts were either estimated by fitting a robust harmonic regression (p < 0·5) to the time-course data using the R package HarmonicRegression [45] or determined by RAIN. Likewise, amplitudes were either estimated by fitting the robust harmonic regression (p < 0·5) and calculating the fold change (FC) amplitude from the resulting relative amplitude (AmpFC = (1 + Amprel)/(1 − Amprel)) or calculated as the peak-to-trough ratio of the maximum and the minimum expression value. A mean log2 expression threshold of 2·5 and a FC amplitude threshold of 1·15 were used to filter for expressed transcripts with significant rhythms. The p-values of the remaining set of cycling transcripts were Benjamini-Hochberg (BH) adjusted for multiple testing with an adjusted p-value cut-off of 0·08 determining robustly circadian transcripts.
For the differential rhythmicity analysis, the robust DODR method [46] was used with a period of 24 h on all transcripts that were previously identified as robustly circadian in at least one of the cell lines. DODR p-values were BH adjusted for multiple testing with an adjusted p-value cut-off of 0·05 determining transcripts with differential rhythmicity. Transcripts were further divided in transcripts with higher amplitude in one of the two cell lines when their absolute log2 amplitude change in one cell line compared to the other was >0·5 and in transcripts with a pure phase shift when their FC amplitude was >1·15 in both cell lines, their absolute log2 amplitude change smaller than 0·1 and the phase shift between the cell lines >1 h.
Noise-robust soft clustering of circadian gene expression patterns was performed with the R package Mfuzz using default parameters [96]. Prior to clustering, the log2 gene expression intensities were transformed to the standard normal distribution.
The relationship between expression values of a set of 12 core-clock genes was quantified by calculating the Spearman correlation between each pair of genes using the R package DeltaCCD [47]. For the reference Spearman correlations, the samples from the murine multi-organ circadian dataset GSE54650 were used [9]. A data set of 34 CRC cell lines (GSE97023) originating from a single study using identical culture methods [48] was further used as a comparison for the circadian signature in CRC.
The circadian time (CT) of both CRC cell lines was predicted using the R package ZeitZeiger [49] with the samples from GSE54650 as the training set (sumabsv = 2, nSPC = 2). All possible alignments of time after synchronization to CT (full hour) were tested. The optimal alignment for the prediction alignment was determined by minimizing the mean absolute error of all time points.
2.6. Functional Annotation and Gene Phase Set Enrichment Analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (v6.8) [50] was used to compute enriched Gene Ontology (GO) terms and pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) for circadian gene clusters against a background of the expressed transcripts in each cell line. The resulting annotations were grouped in annotation clusters based on common gene members. The enrichment score of each annotation cluster is defined as the geometric mean (in -log10 scale) of the individual annotations' non-adjusted p-values (p < 0·05). The DAVID resource was also used to compute enriched GO terms for the three sets of differentially rhythmic transcripts and for the candidate genes with differential AS events between the CRC cell lines (p < 0·05).
Circadian pathways were determined by Phase Set Enrichment Analysis (PSEA) [51] based on the sets of circadian transcripts. Gene sets were downloaded from the Molecular Signatures database (MSigDB) C2 (KEGG gene sets) [52]. Sets containing fewer than five circadian transcripts were excluded from the analysis. The Kuiper test was used to identify circadian gene sets by comparing the acrophases of all circadian transcripts (rounded to the full hour) belonging to each gene set to a uniform background distribution and by testing for nonuniformity (q < 0·01).
2.7. Analysis of Alternatively Spliced Exons
Putative alternatively spliced exons were predicted by the Finding Isoforms using Robust Multichip Analysis (FIRMA) method using the R package aroma.affymetrix [53] and a custom chip definition file (CDF) for HTA 2.0 data from Brainarray (v19) [54]. FIRMA can be used to detect alternative splicing of internal cassette exons from exon array data of single samples without replicates. For each cell line and each time point, FIRMA scores were computed for all probesets (which usually coincide with an exon) and annotated with Ensembl exon IDs. Exons with a log2 expression below 2·5 were marked as absent. Exons that were absent in more than half of the samples of a cell line were excluded from the analysis, as were transcripts where at least half of the probesets in more than half of the samples of a cell line were absent. Paired differences in FIRMA scores were computed as the log2 FC of two FIRMA scores for the same exon at the same time point. Exon-level expression and FIRMA scores for individual genes were visualized at the genomic level using the R package GenomeGraphs [55]. Circadian changes in FIRMA scores over time with a period of 24 h were determined by RAIN (p < 0·05).
3. Results
3.1. Transcriptome Analysis of a CRC Model Reveals a Dysregulated Core-Clock and a Phase-Shift of Oscillations in Metastasis
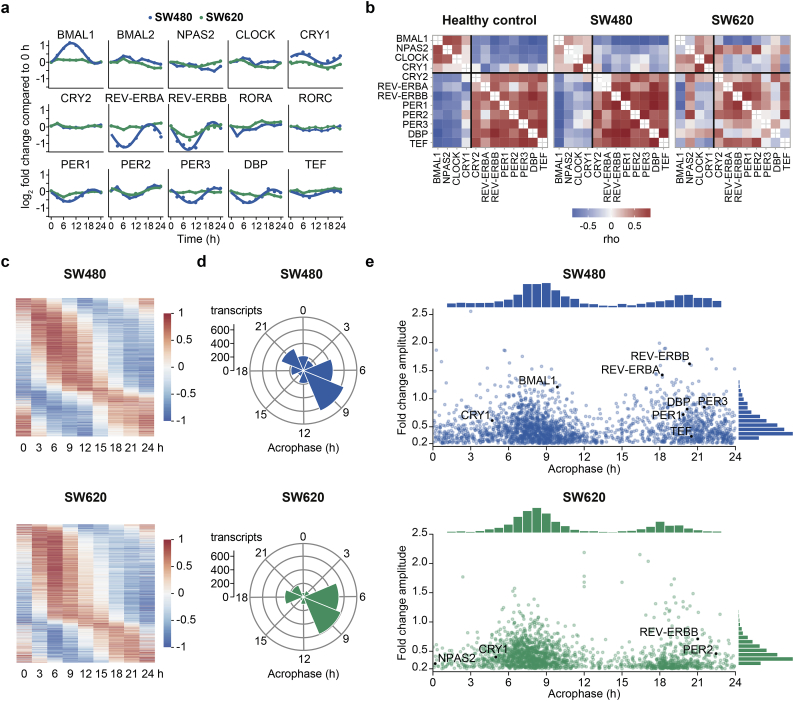
To investigate the possible link between the circadian clock and pre-mRNA splicing in a cancer context, we took the CRC cell lines SW480 (derived from a primary carcinoma) and SW620 (derived from a lymph node metastasis from the same patient) as a model system of tumor progression. We profiled the circadian transcriptome of the cell lines by measuring mRNA expression levels every 3 h for 24 h using whole transcriptome arrays (Fig. 1). First, we analyzed the temporal expression profiles of the 14 genes that form the CCN and two clock-regulated transcription factors, the D-box binding PAR bZIP transcription factor (DBP) and the TEF, PAR bZIP transcription factor (TEF), that are known to confer circadian rhythmicity to downstream CCGs [56] (Fig. 1a). With the exception of REV-ERBB that exhibited low expression levels, all core-clock genes were expressed in both CRC cell lines. However, the oscillations of core-clock genes were severely diminished in the metastatic cell line (SW620) when compared to their expression in the primary tumor-derived cell line (SW480). Several clock genes showing strong rhythms in SW480 cells such as BMAL1, REV-ERBA, PER3, and DBP were not oscillating in SW620 cells while others such as REV-ERBA and CRY1 oscillated in a circadian manner but with lower amplitudes. This observation is in line with previous work from our group where we observed strong and weak oscillations of the promoter activity of BMAL1 for SW480 and SW620 cells, respectively [38]. Time-course measurements of a REV-ERBα-VNP fusion protein also revealed a differential clock phenotype of the cell lines at the single-cell level (Fig. S1a).
Fig. 1.
Transcriptome analysis of the CRC cell lines SW480 and SW620 reveals a dysregulated core-clock in the metastatic cell line and differential profiles of global circadian gene expression.
(a) Transcriptional expression of core-clock genes in primary tumoral SW480 cells (blue) and metastatic SW620 cells (green). Transcripts that were identified as circadian are represented by fitted harmonic regression curves. (b) Heatmaps of Spearman correlation (rho) between each pair of core-clock genes for SW480 and SW620 cells in comparison to a healthy mammalian reference set (GSE54650). (c) Median-normalized, acrophase-ordered expression heatmaps of the 24-h cycling transcripts in the primary tumor cell line SW480 and the metastasis-derived cell line SW620. (d) 3-h acrophase bins of the 24-h cycling transcripts in SW480 cells and SW620 cells. (e) Acrophase and amplitude distributions of the 24-h cycling transcripts in SW480 and SW620. Each transcript is represented by a dot, circadian transcripts from (a) are highlighted in black. Transcripts with 24-h rhythms were determined by RAIN (BH adjusted p < 0·08). Acrophases were either estimated by fitting a harmonic regression (p < 0·5) to the time-course data or determined by RAIN. Amplitudes were either estimated by fitting a harmonic regression (p < 0·5) to the time-course data or calculated as the peak-to-trough ratio of the expression value. The minimal FC amplitude for cycling transcripts was set to 1·15.
We further quantified the extent of circadian perturbation in the CRC cell lines by computing the Spearman correlation between the expression values of each pair of 12 core-clock genes or CCGs and comparing it to a reference signature of clock gene co-expression from healthy mouse organs [49] (Fig. 1b). Remarkably, the correlation pattern of SW480 cells closely resembled that of the reference, exhibiting two groups of genes (BMAL1, neuronal PAS domain protein 2 (NPAS2), CLOCK, and CRY1 vs. CRY2, REV-ERBA, REV-ERBB, PER1, PER2, PER3, DBP, and TEF) that are positively correlated with genes within the group but negatively correlated with genes from the other group. In SW620 cells, however, the separation in two groups was less distinct and the genes NPAS2 and DBP showed a negative correlation with genes within their group, indicating a stronger dysregulation of the clock in the metastatic cell line. Furthermore, we compared the clock gene correlation patterns of both cell lines to that of a single-time point data set of 34 CRC cell lines [48]. Across the 34 cell lines, the correlation is even more diminished than in the metastatic cell line, implying that a dysregulation of the core-clock is a common feature of CRC cell lines (Fig. S2a).
Using the ZeitZeiger method, [49] we tried to predict the CT of all samples of both CRC cell lines. ZeitZeiger is a supervised learning method that uses maximum-likelihood to predict a periodic variable of oscillatory systems based on a high-dimensional training data set. It can also be used to detect phase-shifted or dysregulated oscillators, as indicated by high errors and/or low log-likelihoods of the predictions. We used a multi-organ predictor [49] of healthy mice tissues [9] as the training set and the CRC gene expression data of all time points grouped together as the test sets. For SW480 cells, the time of day could be reasonably well predicted (1·37 h ± 1·48 h, mean absolute error ± SD) with a mean log-likelihood of the predicted CT for all time points of 0·17 (Fig. S2b), assuming that the synchronization of the cells corresponded approximately to the onset of darkness (0 h after synchronization = 13 CT). For SW620 cells, a reliable prediction of the CT was not possible due to consistently high errors of the predictions. The mean log-likelihood of the predicted CT for all time points of SW620 cells was significantly lower than that of SW480 cells (−1·65, p < 0·001 by two-sided Wilcoxon signed-rank test) (Fig. S1b), again indicating a stronger dysregulation of the clock in the metastatic cell line.
On the whole-transcriptome level, we identified ~9·5% circadian transcripts in SW480 and ~8·0% circadian transcripts in SW620 cells (Fig. 1c). Interestingly, our data showed that rhythmic genes were expressed during the entire circadian day with two broad phase peaks (Fig. 1d). In SW480 cells, 47·3% of the circadian transcripts peaked between 6 and 12 h and 23·3% between 19 and 24/0 h after synchronization, while in SW620 cells, 57·9% of the transcripts peaked between 4 and 10 h and 17·7% between 16 and 21 h after synchronization. This implies a phase variation of 2–3 h in the overall transcriptome-level circadian rhythmicity from SW480 to SW620 cells, strongly indicating a shift of clock activity in our cellular model system. Furthermore, oscillating transcripts in SW620 cells had lower amplitudes (median FC amplitude of 0·36) than in the primary tumor-derived SW480 cells (median FC amplitude of 0·43) (Fig. 1e). Less than 600 transcripts were identified to be oscillating in both cell lines (Fig. S2c), yet many of the non-intersect transcripts also showed circadian oscillations in the respective other cell line, albeit partly with changes in their time of peak expression (acrophase) (Fig. S2d). Apparently, the dysregulation of the core-clock from the primary tumor to the metastatic CRC cell line leads to a striking reprogramming of the overall circadian rhythmicity that is characterized by phase shifts and loss of amplitude in clock-controlled genes.
3.2. Differential Rhythmicity of Circadian Transcripts Is Linked with Tumor Progression
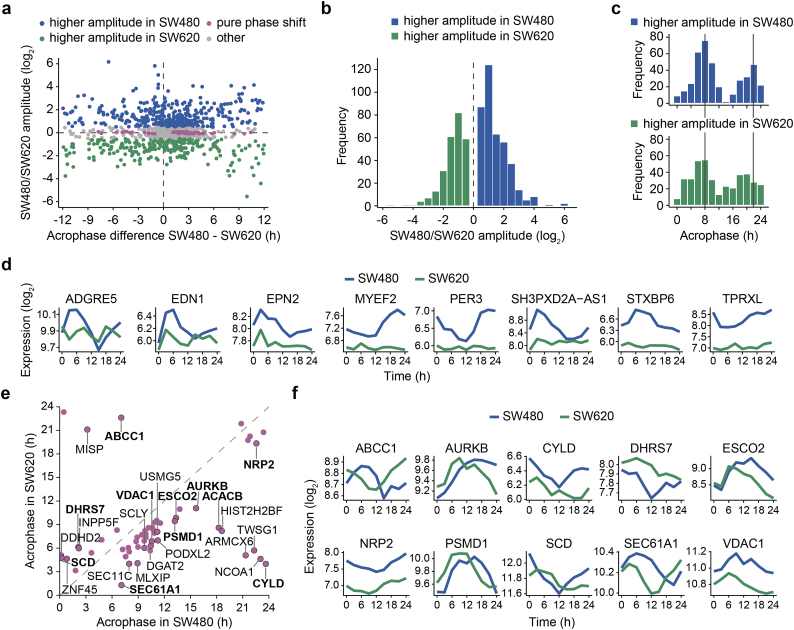
To attain a better understanding regarding the changes in circadian rhythmicity during tumor progression, we compared the amplitude and acrophase of individual transcripts from both cell lines using the robust DODR method [46] (Fig. 2). We computed DODR p-values for all 3606 pairs of circadian transcripts that oscillated in at least one of the CRC cell lines. The resulting 1005 transcripts with significant differential rhythmicity between the cell lines (BH adjusted p < 0.05) were further grouped in three sets of transcripts exhibiting a higher amplitude in SW480 cells (39·5%), a higher amplitude in SW620 cells (25·9%), and a pure phase shift (5·6%) (Fig. 2a) (see Materials and Methods for criteria). In line with the observed weaker oscillations of the circadian system in SW620 cells, there was a tendency for higher amplitudes in the circadian transcripts with a higher amplitude in SW480 cells, as opposed to those with a higher amplitude in SW620 cells (Fig. 2b). Interestingly, we further observed a shift in the acrophase distributions of the circadian transcripts. Transcripts with a higher amplitude in SW620 cells tended to have earlier phases than those with a higher amplitude in SW480 cells (Fig. 2c), again pointing to a temporal reprogramming of clock activity in the metastatic cells.
Fig. 2.
Circadian transcripts exhibit differential rhythmicity in the CRC progression model.
(a) Changes in rhythmicity (amplitudes and/or acrophases) from the primary tumor cell line SW480 to the metastatic cell line SW620 of the transcripts that showed 24-h rhythms in either one of the cell lines were estimated using the robust DODR method. The transcripts with BH adjusted p < 0·05 were further divided in transcripts with a higher amplitude in SW480 cells (blue, log2 amplitude change >0·5), a higher amplitude in SW620 cells (green, log2 amplitude change < −0·5) and transcripts with a pure phase shift (pink, FC amplitude >1·15 in both cell lines, absolute log2 amplitude change <0·1 and phase shift >1 h). (b) log2 amplitude changes from SW480 to SW620 cells for transcripts with DODR adjusted p < 0·05 and absolute log2 amplitude change >0·5. (c) Acrophases for transcripts with higher amplitudes in SW480 cells (upper panel, blue) and SW620 cells (lower panel, green). (d) Time-course for the eight transcripts with the largest loss of oscillations in SW620 cells (absolute log2 amplitude change >4, FC amplitude >1·25 in SW480 cells). (e) Acrophases in SW480 and SW620 cells for transcripts with pure phase shifts. Transcripts with absolute phase shifts >3 h (black border) are labeled with the gene name. Selected transcripts represented in (f) are written in bold. (f) Time-course for selected phase-shifted transcripts.
A set of eight transcripts had an absolute log2 FC > 4 and all exhibited a loss of oscillations in SW620 cells (Fig. 2d). The set includes the core-clock gene PER3 and five other protein-coding genes (adhesion G protein-coupled receptor E5 (ADGRE5), endothelin 1 (EDN1), epsin 2 (EPN2), myelin expression factor 2 (MYEF2), sytaxin binding protein 6 (STXBP6)), as well as two non-coding RNA genes (tetrapeptide repeat homeobox like (TPRXL), SH3PXD2A antisense RNA 1 (SH3PXD2A-AS1)) with interesting functions in tumor development and progression. MYEF2 is a paralog of the SF HNRNPM and acts a suppressor factor in myelinating and erythroid cells [57]. ADGRE5, EDN1, and STXBP6 are all involved in cell-cell adhesion [[58], [59], [60]], while EPN2 is involved in clathrin-mediated endocytosis and acts as a suppressor of VEGF-mediated angiogenesis [61]. The non-coding RNA genes TPRXL and SH3PXD2A-AS1 have both been linked to cancer as well [62, 63].
Moreover, we identified 25 circadian transcripts with estimated pure phase shifts >3 h between the CRC cell lines (Fig. 2d, f). These phase-shifted transcripts encode for several membrane-related proteins including dehydrogenase/reductase 7 (DHRS7), neuropilin2 (NRP2), proteasome 26S subunit, non-ATPase 1 (PSMD1), stearoyl-CoA desaturase (SCD), and Sec61 translocon alpha 1 subunit (SEC61A1). Other phase-shifted circadian transcripts are involved in anion transmembrane transport (ATP binding cassette subfamily C member 1 (ABCC1) and voltage dependent anion channel 1 (VDAC1)) or the cell cycle (aurora kinase B (AURKB), CYLD lysine 63 deubiquitinase (CYLD), and establishment of sister chromatid cohesion N-acetyltransferase 2 (ESCO2)).
To analyze whether the three sets of differentially rhythmic transcripts between the CRC cell lines are involved in biologically related processes or share molecular functions, we conducted a functional annotation analysis against the complete sets of 24-h cycling transcripts in the respective cell line using the DAVID resource (Fig. S3). Transcripts that oscillated in SW480 cells but lost circadian rhythmicity in the metastatic cell line were enriched for GO terms related to i.a. chromatin silencing (p = 0·005) and modification (p = 0·016), homophilic cell adhesion (p = 0·013), and the regulation of mitotic nuclear division (p = 0·031). Conversely, transcripts with higher amplitudes in the metastatic cell line were enriched for GO terms related, i.a., to the regulation of transcription from an RNA polymerase II promoter (p = 0·021) and the regulation of smooth muscle cell proliferation (p = 0·036). Transcripts that oscillated in both cell lines but exhibited phase-shifts were enriched for GO terms related to i.a. DNA binding (p = 0·004), replication fork processing (p = 0·014), and retinoid X receptor binding (p = 0·032). Overall, our results point to an association of differential rhythmicity of cycling transcripts in the CRC model with various hallmarks of tumor progression.
3.3. Circadian Pathway Analysis Identifies Temporally Coordinated Expression of Spliceosome-Related Genes
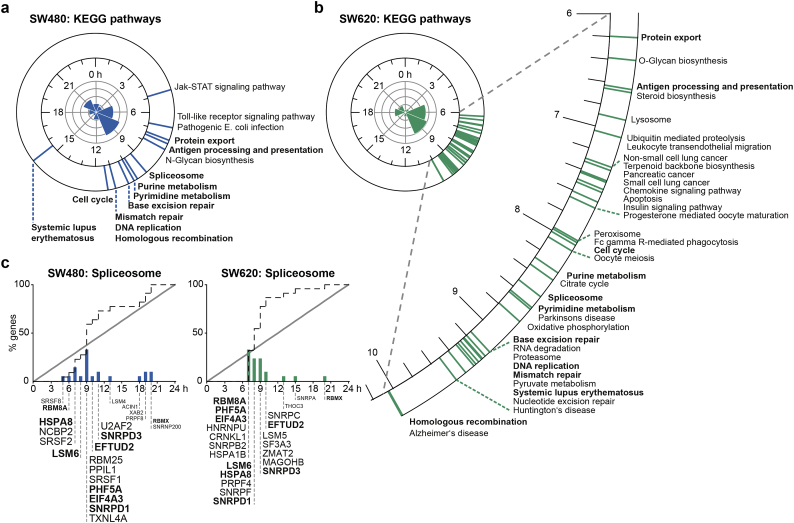
To further explore the functional relevance of the rhythmically transcribed genes, we grouped the complete sets of circadian transcripts into four clusters based on their temporal expression in SW480 (Fig. S4a) and in SW620 cells (Fig. S5a), respectively. For each cluster, we computed enriched GO terms and KEGG pathways against the background of all expressed transcripts in the respective cell line. Both in SW480 and SW620 cells, the circadian transcript clusters were enriched for genes involved in various biological processes, including DNA binding, the cell cycle, mRNA splicing, protein folding, cell-cell adhesion, and metabolism (Fig. S4b and Fig. S5b). Interestingly, for some of the biological processes enriched in both cell lines, the associated transcript clusters showed expression peaks at different times of the circadian day. Strikingly, mRNA splicing was enriched for the cluster of circadian transcripts that peaked at about 9 h after synchronization in SW480 cells (p = 8·1e-5), whereas in SW620 cells, the same process was enriched for the cluster of circadian transcripts that peaked earlier at about 6 h after synchronization (p = 2·5e-5). Other processes such as cell-cell adhesion were enriched for several circadian transcript clusters in the same cell line, indicating that while the genes were rhythmically transcribed, the pathways themselves were not enriched for a particular circadian phase. In order to identify biologically related gene sets with temporally coordinated expression at a certain time of day, we conducted a PSEA for both cell lines (Fig. 3). In contrast to the functional annotation of clustered circadian gene sets, PSEA only identifies biological processes which peak once during the circadian day. Nearly all significantly temporally coordinated pathways (q < 0·01) in the cells took place between 4 and 12 h after synchronization for SW480 cells (Fig. 3a) and between 6 and 10 h for SW620 cells (Fig. 3b). Common temporally coordinated pathways in SW480 and SW620 cells included protein export (7·4/6·2 h after synchronization in SW480/SW620 cells), antigen processing and presentation (7·6/6·6 h), and the spliceosome (9·4/8·5 h), as well as nucleotide metabolic processes (purine metabolism: 9·8/8·3 h; pyrimidine metabolism: 10·0/8·6 h) followed by DNA replication (11·1/9·2 h), DNA repair-related pathways (base excision repair: 10.3/9.2 h; mismatch repair: 11·1/9·4 h, homologous recombination: 11·1/10·0 h), and the cell cycle (11·4/8·0 h). In the primary tumor cell line SW480, the analysis further revealed a significant temporal orchestration for important signaling pathways such as the Jak-STAT signaling pathway (4·9 h) and the Toll-like receptor pathway (6·8 h), whereas in the metastatic SW620 cells, several cancer-associated pathways were temporally coordinated, including small cell lung cancer (7·4 h), non-small cell lung cancer (7·3 h), and pancreatic cancer (7·4 h), as well as metabolic pathways such as oxidative phosphorylation (8·8 h) and pyruvate metabolism (9·3 h).
Fig. 3.
Circadian pathway analysis identifies temporally coordinated expression of spliceosome-related genes in CRC cell lines.
Summary of significantly phase-clustered (q < 0·01) circadian pathways in SW480 (a) and SW620 cells (b). The circular axis (blue lines: SW480; green lines: SW620) represents the average phase of all circadian genes in a pathway from the KEGG database. Pathways that are temporally coordinated in both cell lines are written in bold. The circular histograms show the 3-h acrophase bins of the 24-h cycling transcripts. (c) The spliceosome is among the phase-clustered pathways that demonstrated circadian coordination in both SW480 (left panel) and SW620 cells (right panel) cells. The dotted line depicts the empirical cumulative distribution of phases belonging to genes that are part of the spliceosome pathway. The solid line depicts the uniform distribution. The height of the bars corresponds to the percentage of circadian genes in the spliceosome pathway that peak at a certain time after synchronization. Individual gene names are shown with a larger font size corresponding to a greater contribution of the gene to the overall phase clustering of the pathway. Gene acrophases were rounded to the full hour.
To assess temporal shifts from SW480 to SW620 cells on the gene set level, we applied PSEA to the phase differences of the 597 circadian transcripts that cycled in both cell lines. Again, the spliceosome was among the phase-clustered pathways whose associated genes showed significant shifts in their peak expression (0·76 h, q < 0·01) (Table S1). In both cell lines, the sets of the circadian spliceosome-associated genes were unimodally distributed, though the composition of the sets and the peak times of the genes differed between the primary tumor and the metastatic cell line (Fig. 3c). These results imply that the rhythmic orchestration of the spliceosome undergoes subtle but significant changes during CRC metastasis that go in hand with the dysregulation of the circadian system in SW620 cells. Even though other pathways have also been identified as being temporally coordinated in a circadian manner in our data, we decided to focus on the spliceosome and splicing regulation in the further analyses.
3.4. Spliceosome Components and Splicing Regulators Exhibit Differential Circadian Transcriptional Rhythms in Tumor and Metastatic Cells
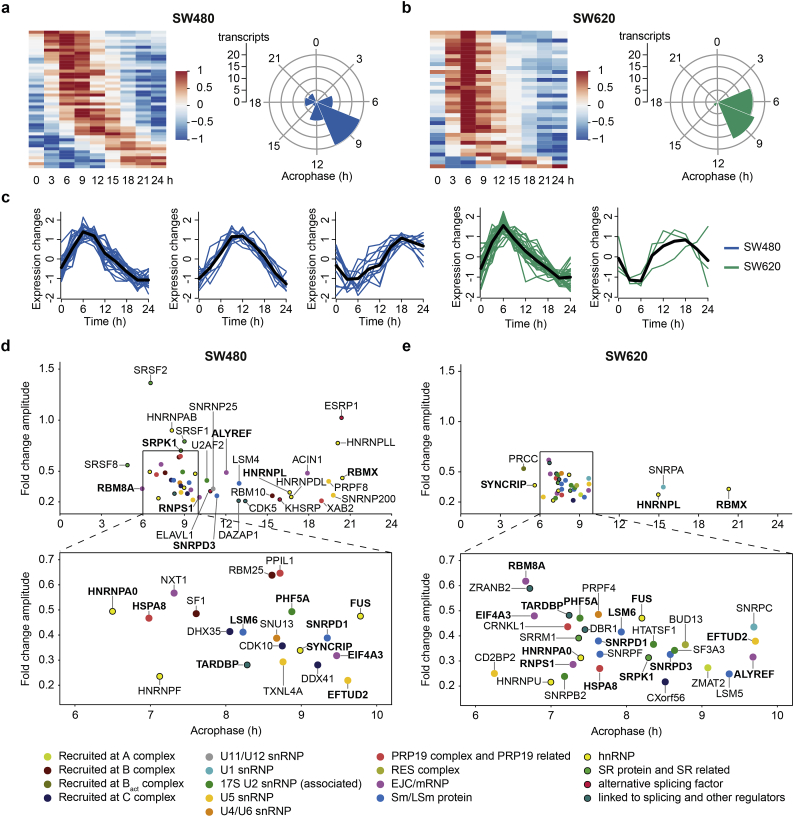
We analyzed the mRNA expression of a curated list of 254 spliceosome components and splicing regulators from the literature [64] and from two public databases for spliceosome components, SpliceosomeDB [65], and for human SFs, SpliceAid-F [66] (Fig. 4). The list consists of components recruited at different complexes of the spliceosome (A, B, Bact, and C complex), different snRNPs (U11/12, U1, 17S U2, U5, and U4/U6 snRNP), other spliceosomal complexes and proteins (pre-mRNA-processing factor 19 (PRP19) and retention and splicing (RES) complex, exon junction complex (EJC)/mRNP, and Sm/like Sm (LSm) proteins), as well as hnRNPs, SR proteins, and other splicing regulators. In SW480 cells, 46 transcripts (~18%) of the splicing-related genes showed 24-h oscillations (Fig. 4a) compared to 35 transcripts (~14%) in SW620 cells (Fig. 4b), which is nearly twice the respective percentages of circadian expression we found in the whole transcriptome for our CRC model system. As expected from the results of the functional annotation and the circadian pathway analysis, the acrophases of the 24-h cycling spliceosome components and splicing regulators showed a phase shift between SW480 and SW620 cells: 30 out of the 35 24-h cycling spliceosome components and splicing regulators in SW620 cells peaked between 6 and 10 h after synchronization whereas in SW480 cells, the phases were distributed throughout the circadian day with a peak of expression around 9 h after synchronization. Clustering of the oscillatory profiles of circadian spliceosome components and splicing regulators yielded three main clusters for SW480 cells and two clusters for SW620 cells (Fig. 4c).
Fig. 4.
Spliceosome components and SFs show circadian transcriptional rhythms in primary and metastatic CRC.
Median-normalized, acrophase-ordered expression heatmap (left panel) and 3-h acrophase bins (right panel) of the circadian spliceosome components and SFs in SW480 (blue) (a) and SW620 cells (green) (b). (c) Expression patterns of clustered circadian spliceosome components and SFs in SW480 and SW620 cells. Acrophases and amplitudes of the circadian spliceosome components and SFs in SW480 (d) and SW620 (e). Transcripts that show circadian oscillations in both cell lines are written in bold. The components are color-coded according to their protein class/family or their recruitment to the spliceosome complex (lime: recruited at A complex, maroon: recruited at B complex, olive: recruited at Bact complex, navy: recruited at C complex, gray: U11/U12 snRNP, cyan: U1 snRNP, green: 17S U2 snRNP or U2 snRNP associated, dark yellow: U5 snRNP, orange: U4/U6 snRNP, light red: PRP19 complex and PRP19 related, light olive: RES complex, purple: EJC/mRNP, blue: Sm/LSm protein, yellow with border: hnRNP, green with border: SR protein and SR related, red with border: alternative SF, teal with border: linked to splicing and other regulators).
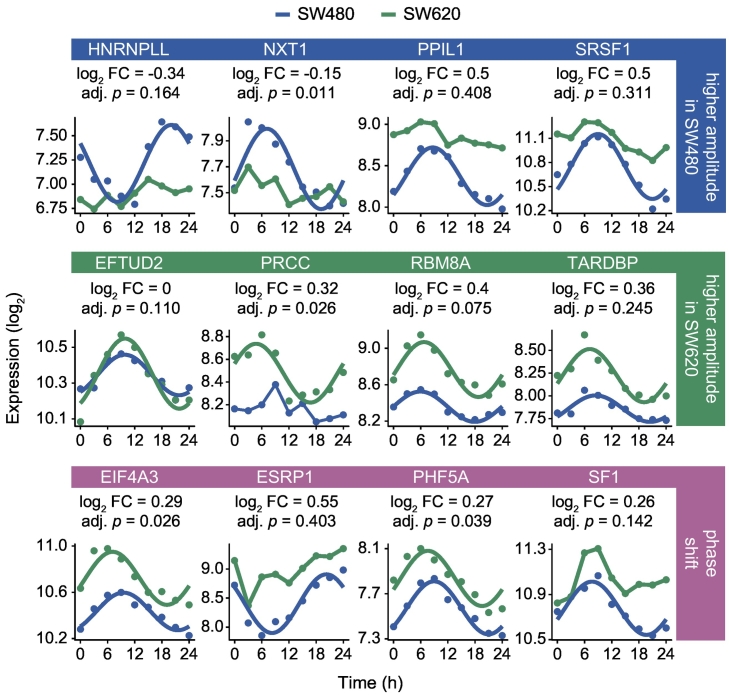
The 24-h cycling transcripts in the CRC cell lines belong to diverse spliceosomal complexes and splicing-related protein classes (Fig. 4d, e). However, neither acrophases nor amplitudes were clustered for specific complexes or genes encoding for proteins from the same class or family, implying a complex temporal regulation of splicing. We further investigated the differentially rhythmic patterns of splicing-related transcripts that cycle in either one of the cell lines (Fig. S6). We selected robustly circadian splicing-related genes that showed differences in their rhythmicity between both cell lines and grouped them according to amplitude changes and phase shifts (Fig. 5). Transcripts with higher amplitudes in SW480 cells include HNRNPLL, nuclear transport factor 2 like export factor 1 (NXT1), peptidylprolyl isomerase like 1 (PPIL1), and SRSF1. The expression of HNRNPLL and NXT1 lost its oscillatory pattern and was decreased in SW620 cells, while the opposite behavior could be observed for PPIL1 and SRSF1 where the mean expression over all time points increased. Despite the observed dysregulation of the clock and the smaller number of 24-h rhythmic splicing-related genes in the metastatic cell line, there were also several transcripts with higher amplitudes in SW620 cells, including elongation factor Tu GTP binding domain containing 2 (EFTUD2), papillary renal cell carcinoma (PRCC), RBM8A, and TAR DNA binding protein (TARDBP). With the exception of EFTUD2, all genes exhibited stronger oscillatory patterns and higher expression levels in the metastatic cell line. A similar change in expression levels could be observed for the phase-shifted transcripts eukaryotic translation initiation factor 4A3 (EIF4A3), ESRP1, PHD finger protein 5A (PHF5A), and splicing factor 1 (SF1).
Fig. 5.
Differentially rhythmic expression patterns of splicing-related genes in CRC progression include amplitude changes and phase shifts.
Selected circadian transcriptional expression patterns in SW480 (blue line) and SW620 cells (green line) of spliceosome components and SFs that showed higher amplitudes in SW480 cells (blue box), higher amplitudes in SW620 cells (green box), as well as phase-shifted oscillations that persisted in both CRC cell lines (pink box). Transcripts that were identified as circadian are represented by fitted harmonic regression curves. The log2 FC was calculated as the change of the mean expression over all time points from SW480 to SW620 cells. The adjusted p-value denotes the results of the DODR analysis and was calculated as previously described.
3.5. Circadian Regulation of AS in CRC Progression
To investigate whether the observed differences in the oscillatory profiles of the splicing machinery lead to actual differences in the AS patterns of the CRC cell lines, we applied the FIRMA method [68]. FIRMA is a widely used algorithm for the detection of AS from exon microarray data (58) that formulates the detection of AS as an outlier detection problem: For each exon, a score is computed based on the residuals of the observed probe-level expression within that exon from the estimated expression produced by fitting the standard RMA model to the array. A log2 FIRMA score of 0 indicates no departure from the model while high or low scores can be indicative of exon inclusion or skipping, respectively.
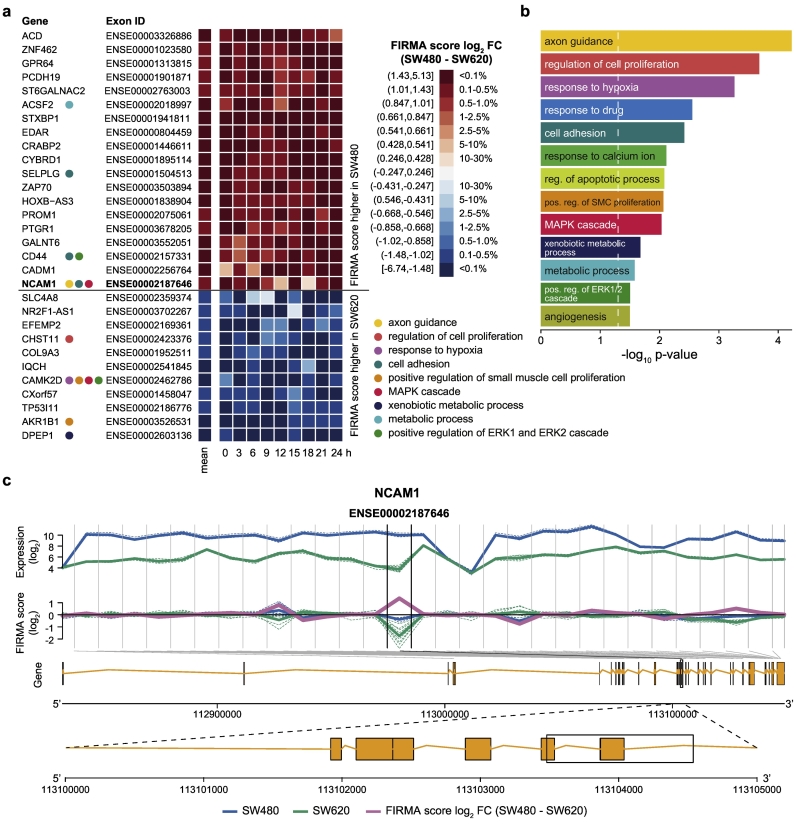
We compared the AS patterns between the two cell lines by computing the paired differences in FIRMA scores from SW480 to SW620 cells for identical time points. Of the 401,839 analyzed exons left after filtering, 299 exons (0·0007%) (256 genes) had significantly different (BH adjusted p < .001) and high (mean absolute FIRMA score log2 FC ≥ 1) FIRMA scores and were taken as candidate exons for differential AS events in CRC progression (Fig. 6a, Table S2). Functional annotation of all 256 candidate genes with differential AS events between SW480 and SW620 cells yielded various enriched biological processes (Table S3) of which many are well-known to be associated with tumor progression and metastasis, such as the regulation of cell proliferation (p = 0·0002) and apoptotic processes (p = 0·0083), response to drug (p = 0·0028), cell adhesion (p = 0·0038), and angiogenesis (p = 0·0316) (Fig. 6b). The most enriched term, however, was the process of axon guidance (p = 5·9e-05) that has recently been reported as a potential mediator of colon cancer metastasis [69]. Among the top 30 candidate exons (mean absolute FIRMA score log2 FC ≥ 1·35), there were several genes reported to have AS variants that play a role in invasion and metastasis such as NCAM1 (Fig. 6c) and CD44 (Fig. S7a). CD44 encodes for a transmembrane protein involved in epithelial-to-mesenchymal transition (EMT). Another EMT-associated candidate gene is the fibroblast growth factor receptor 2 (FGFR2) (Fig. S7b).
Fig. 6.
Differential AS events in SW480 and SW620 cells are associated with proliferation, angiogenesis, and cell adhesion.
(a) Paired differences in FIRMA scores of the top 30 candidate exons with differential AS events in SW480 and SW620 cells (BH adjusted limma p < 0·001 and absolute FIRMA score log2 FC ≥ 1·35). The color scale is not evenly spaced but based on the percentiles of all FIRMA scores for all probesets and samples after filtering. The respective genes are annotated with GO terms that were found to be enriched in (b). (b) Selected enriched GO terms (biological processes) for the 256 candidate genes with differential AS events (BH adjusted limma p < .001 and absolute FIRMA score log2 FC ≥ 1). (c) Exon-level expression (upper panel), FIRMA scores (middle panel), and genomic representation (lower panel) of the candidate gene NCAM1. Vertical lines separate the individual probesets covering the gene. For each probeset, the exon-level expression and the FIRMA scores of the individual time points are depicted by a dotted line and the respective mean values are depicted by a solid line for SW480 (blue) and SW620 (green), respectively. The mean FIRMA score log2 FC (SW480-SW620) is shown in pink. Gray diagonal lines indicate the localization of the probesets within the genome.
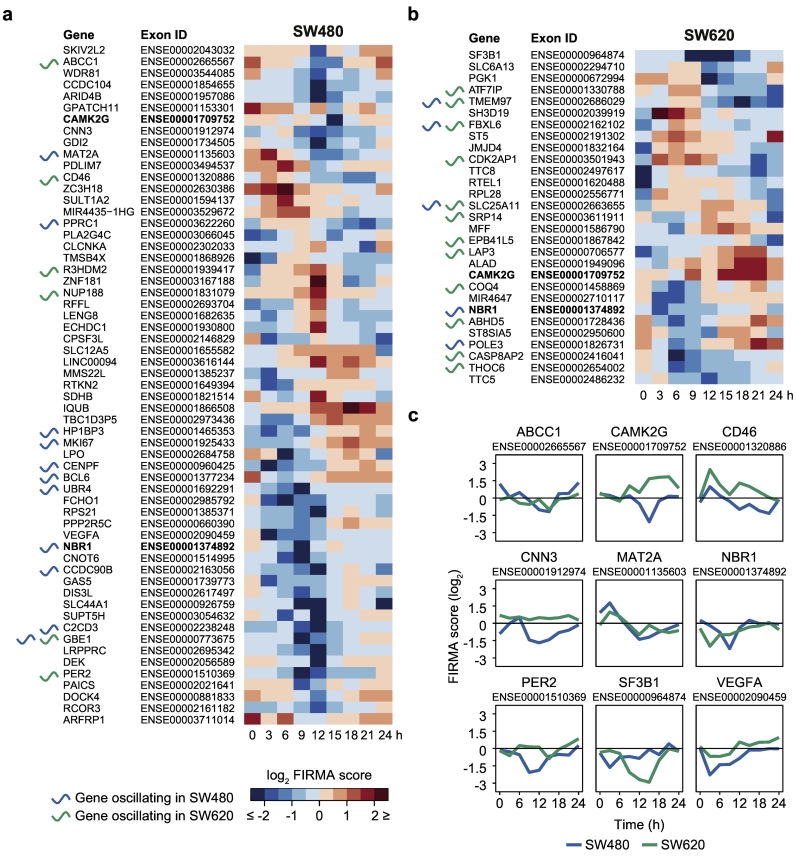
We further tested the array data for circadian rhythmicity in the changes of FIRMA scores over time and identified 59 candidate circadian alternative exons in SW480 cells (59 genes) (Fig. 7a) and half as many in SW620 cells (29 exons, 29 genes) (Fig. 7b). Interestingly, many of the candidate genes also oscillated at the transcriptional level in one or both of the CRC cell lines. They include the core-clock gene PER2 (Fig. 1a) and the multi-drug transporter protein-encoding gene ABCC1 that also exhibited phase-shifted circadian expression patterns between the cell lines at the transcriptional level (Fig. 2f). Only two of the candidate genes, calcium/calmodulin dependent protein kinase II gamma (CAMK2G) and NBR1, autophagy cargo receptor (NBR1), showed circadian AS patterns in both cell lines, pointing to a global reprogramming of circadian AS events during CRC progression. Strikingly, the circadian FIRMA score profiles of CAMK2G and NBR1 also exhibited phase-shifts in their rhythmic AS patterns (Fig. 7c). The DNA damage gene NBR1 was also cyclically expressed at the transcriptome-level in SW480 cells and has been reported as a putative CLOCK target gene in CRC [70]. Circadian AS events were also predicted for CD46, a gene encoding for the complement inhibitor membrane cofactor protein that is also involved in T cell regulation and infection processes, and methionine adenosyltransferase 2A (MAT2A) that are both overexpressed in CRC [71, 72] (Fig. 7c). Further interesting candidates are calponin 3 (CNN3), a proposed marker for lymph node metastasis in CRC [73], VEGFA, and splicing factor 3b subunit 1 (SF3B1) (Fig. 7c). In most cases, the oscillating FIRMA scores of the candidate genes were concordant with the temporal patterns of the respective exons (Fig. S8).
Fig. 7.
Circadian AS event in CRC cell lines include PER2, SF3B1, and VEGFA.
Acrophase-ordered exons with 24-h cycling FIRMA scores (RAIN p < 0·05 and peak-through-ratio of FIRMA score ≥ 2) in SW480 (a) and SW620 cells (b). Exons that showed circadian AS in both cell lines are written in bold. The respective genes are marked with an oscillatory curve if they showed transcriptional rhythms in SW480 (blue) and/or SW620 (green) cells. (c) Circadian FIRMA scores for selected exons with circadian AS events in either SW480 (blue) and/or SW620 cells (green).
Altogether, we identified distinct oscillatory phenotypes for spliceosome components and splicing regulators in our CRC model system. This indicates a strong cross-regulation between the circadian system and splicing with noteworthy implications in tumor development. Interestingly, these phenotypes correlate with changes in AS patterns – both static and circadian – in well-known tumor progression-related genes which points to a role of the circadian clock in the regulation of splicing that can be hijacked in metastasis.
4. Discussion
Even though systemic dysregulations of the circadian clock in different cancer types have been recently reported [47, 74], a circadian systems analysis in a cancer model that allows for the in depth identification of time-specific processes which impact tumor progression has been missing. In this work, we used a comprehensive systems biology approach to couple circadian molecular processes with mRNA splicing, its subsequent impact on AS, and its possible consequences in tumorigenesis. We used a CRC cellular model to analyze tumor progression-specific differences in circadian transcriptional rhythmicity and mRNA splicing patterns. The cellular model chosen includes two cell lines with the same genetic background but different metastatic potential and has been commonly used to investigate primary and metastatic behavior in CRC [[75], [76], [77]]. We performed a time-course genome-wide screening of the two CRC cell lines using whole transcriptome arrays which allowed for the identification of cell-specific 24-h cycles in the expression of gene transcripts, as well as for the prediction of circadian-regulated AS events at the exon level.
Our results demonstrate that despite diminished oscillations of core-clock genes and dysregulated correlation patterns in the metastatic cell line in comparison to the primary tumor and a healthy control pool of murine tissues, oscillations of CCGs largely persist on the transcriptome-level. Furthermore, the identity of circadian transcripts and their overall acrophases distribution differed between the cell lines, with a 2–3 h shift in clock activity which may account for the global reprogramming of circadian activity observed in the metastatic cell line. Following a detailed analysis of circadian differential rhythmicity at the single transcript-level, we grouped the oscillating genes in three distinct sets exhibiting amplitude changes (either a higher amplitude in SW480 or in SW620 cells) or pure phase shifts. Interestingly, genes with diminished oscillations in the metastatic cell line are associated with important functional processes relevant to tumor progression such as cell-cell adhesion and angiogenesis. Conversely, genes that showed an increase of oscillations from the primary tumor to the metastatic cell line were associated with the regulation of smooth muscle cell proliferation. The smooth muscle layer is often lacking in tumor vessels and it has been shown that CRC cells induce apoptosis of vascular smooth muscle cells, thereby facilitating their intravasation into the bloodstream and promoting metastasis [78]. The gain and loss of oscillations in specific circadian transcripts might indicate a possible mechanism which enables CRC cells to alter the temporal regulation of smooth muscle cell proliferation and cell-cell adhesion and thus to develop properties that are favorable to the formation of metastases. Genes with pure phase-shifts in transcriptional rhythmicity were less frequent than those showing changes in the oscillatory amplitude, but could also be linked to tumor onset and progression. Thus, our results show that alterations of circadian rhythmicity are wide-spread during CRC tumor progression and that they may have far-reaching consequences on diverse biological functions associated with metastasis. However, other cancer models need to be analyzed in future work to allow for more generalized conclusions regarding the circadian regulation of splicing and its putative role in metastatic behavior.
We further investigated the temporal phenotype of the circadian enriched pathways in our CRC model. Remarkably, the spliceosome was among the biological pathways whose associated genes showed a significantly phase-shifted, temporally coordinated expression in both CRC cell lines. This effect seems to be the outcome of an altered clock. We identified circadian oscillations on the transcriptional level of diverse spliceosome components and SFs, including hnRNPs, SR proteins, and RBM proteins. Many of the circadian splicing-related genes showed changes in amplitudes and phase-shifted expression patterns in CRC progression that correlate with the altered outcome of AS events of their target genes as shown by our data.
The high flexibility of alternatively spliced gene products can provide tumor cells with the ability to produce protein isoforms that are normally expressed only in specific developmental stages, but that are downregulated in adult cells [29]. In cancer, they can promote uncontrolled growth and survival, thereby conferring tumor cells with a selective advantage. A recent study on the functional impact of AS in cancer further revealed that recurrent transcript isoform switches across various cancer types affect protein domain families frequently mutated in cancer and may disrupt protein-protein networks in cancer-related pathways [30]. Interestingly, these AS changes have been found to be negatively correlated to somatic mutations [30]. One possible explanation for their origin might be trans-acting alterations such as the temporal expression changes in spliceosome components and SFs as observed in our CRC progression model.
Even though most alterations in AS during CRC development have been reported to occur during the transition from normal to tumor cells [79], we identified nearly 300 candidate differential AS events between the primary tumor and the metastatic cell line. Many of the genes with predicted AS events were involved in processes relevant to tumor progression, migration, and invasion, such as NCAM1 which encodes for a neural adhesion molecule capable of heterotypic and homotypic binding. Loss of expression of the NCAM-180 isoform due to truncated transcripts was previously reported for SW620 cells and is linked to CRC [80]. Other examples for differential AS events in CRC progression identified in our study are the EMT-associated splicing switches of the genes CD44 and FGFR2. CD44 encodes for a family of cell adhesion molecules, involved in both homotypic and heterotypic interactions with extracellular matrix components [81]. The genomic structure of human CD44 consists of 20 exons, ten of which (v1 to v10) are included or skipped in a variable fashion, defining the attachment properties of the molecule and its subsequent impact on cancer metastasis and EMT [29, 67, 82]. FGFR2 encodes for two isoforms due to mutually exclusive splicing of alternative cassette exons III3b and IIIc: The IIIb isoform is characteristic to epithelial and the IIIc isoforms to mesenchymal cells [83]. A choice between mutually exclusive exons in FGFR2 alters its specificity to bind growth-stimulating proteins in prostate cancer [84]. The splicing of both FGFR2 and CD44 is regulated by two isoforms of the SF ESRP (ESRP1/2) [83]. Both ESRP isoforms are known to be important regulators of splicing switches during EMT [67]. Interestingly, ESRP1 also showed phase-shifted circadian transcriptional rhythms in our CRC model. Thus, it is likely that the observed temporal shift in the circadian expression of the SF ESRP1 during tumor progression leads to switches in the splicing of FGFR2 and CD44 which may result in the production of isoforms that promote malignant transformation by enabling EMT and favoring metastasis.
In addition to the above described static changes in splicing due to temporal changes in the expression of SFs, we also identified exons whose AS patterns themselves changed over time in a circadian manner. Most exons that exhibited circadian oscillations in their splicing patterns in the primary tumor cells, such as ABCC1, CNN3, PER2, and VEGFA, lost them during metastasis, whereas others, such as the SF SF3B1, exhibited a gain of cyclic splicing behavior. According to the position of the candidate exon on the gene, the AS of SF3B1 affects the HEAT repeat domain 8 which encodes for part of the canonical isoform of the gene. Though not much is known regarding the effects of AS of SF3B1, its knockdown in human myeloid cell lines resulted in over 500 significant differentially regulated splicing variants of nearly 400 genes, including TP53 [85]. A similar effect could result from the observed circadian AS of SF3B1 in our system, however, further experiments are necessary to elucidate a possible functional outcome of this mechanism.
The candidate circadian alternatively spliced exon of PER2 is part of the C-terminal region of a previously reported splicing variant of PER2 known as PER2S, which is not conserved and non-homologous to the PER2 sequence [86]. The PER2S isoform was found to be preferentially localized in the nucleolus, suggesting a possible cross-talk of the clock and the nucleolus via PER2S. Interestingly, the disturbance of nucleolar function in response to cellular stress is linked to cancer via regulation of the p53 pathway: The disruption of the nucleolar structure triggers the release of the tumor suppressor ARF and several nucleolar riboproteins into the nucleoplasm where they bind to MDM2, thereby preventing MDM2-mediated p53 degradation, leading to cell cycle arrest or apoptosis (Suzuki et al., 2012). The loss of PER2S circadian AS could potentially lead to alterations in the nucleolar function due to disrupted circadian timing of its components, which may impact p53 activation and promote carcinogenesis via the pathways described above.
Depending on the splice site choice, VEGF can act either as an anti-angiogenic protein or as a pro-angiogenic protein [32]. These alternative splice forms of VEGF are regulated by SR proteins, e.g., SRSF1, which was also reported to act as an oncogene and for which we observed an altered circadian phenotype in the metastatic cell line. SRSF1 itself is phosphorylated by an SR protein kinase (SRPK1/2), which exhibited higher expression levels and an altered oscillation in the SW620 cells. The inhibition of the kinase was shown to reduce angiogenesis, a critical process for cancer progression and metastasis [87].
ABCC1 showed phase-shifted circadian oscillations for both cell lines on the transcriptome-level, as well as circadian patterns of AS of exon 22 that are not observed in the metastatic cell line. ABCC1 encodes for the multidrug resistance-associated protein 1 (MRP1) that plays an important role in conferring resistance to chemotherapeutic drugs in cancer cells [88]. Circadian transcriptional oscillations of ABCC1 have also been observed in Caco-2 cells [89] and diverse tissues in mouse, rat, and monkey [90]. Different splice variants of ABCC1 have been characterized [91], some of which have been found to confer a drug resistance phenotype in ovarian cancer [92]. Even though further studies are necessary to explore the observed phenotype, we hypothesize that the dysregulation of the clock may lead to a reprogramming of the circadian behavior of ABCC1, which might play a relevant role in the known drug resistance of colon cancer patients [93]. Altogether, our findings regarding the circadian regulation of ABCC1 further strengthen the relevance of this gene as a promising drug target in cancer and highlight the role of the clock in optimizing therapy [74, 94, 95]. Thus, the coupling of the circadian clock and splicing regulation may offer novel angles to tackle and elucidate the mechanisms of cancer development and metastasis.
We are still in the beginning of investigating how time may play a role in the splicing process and its subsequent effects on cancer progression. The subtle, yet significant phase shifts observed in the oscillatory patterns of splicing-related genes suggest that a higher resolution of time points and/or a wider temporal window might help to reveal even smaller alterations in the circadian rhythmicity of spliceosome components and SFs in subsequent studies. Additionally, an RNA-seq analysis of the candidate differentially spliced internal cassette exons may help to validate the results of the whole transcriptome array data and to reveal other types of AS events in CRC progression and will be addressed in the future. The data produced so far is intriguing and the usage of further experimental validation regarding the AS events and their elicitors will allow us to fully understand and eventually unravel this process.
The results of our study revealed differential circadian oscillations of spliceosome components and SFs which correlate with AS decisions and add a temporal layer to the regulation of mRNA splicing. Hence, our work sheds light on the role of the circadian clock in the fine-tuning of AS and on how this putative reprogramming of cellular time might enable cells to acquire oncogenic properties which favor tumorigenesis.
The following are the supplementary data related to this article.
Single-cell time-course measurements of REV-ERBα protein abundance in CRC cell lines. The fluorescence intensity of a REV-ERBα-VNP fusion protein was measured for 38 h in single live-cells from SW480 (a) and SW620 (b) (n = 4).
Alterations of the circadian clock in CRC primary tumors and metastasis. (a) Heatmaps of Spearman correlation (rho) between each pair of core-clock genes in a set of 34 CRC cell lines (GSE97023) in comparison to a mammalian reference set (GSE54650). (b) Box-plot of the log-likelihoods of the ZeitZeiger predictions of CT for SW480 (blue) and SW620 (green) samples. (c) Venn diagram of the 24-h cycling transcripts in SW480 and SW620 cells. (d) Median-normalized, SW620-acrophase-ordered expression heatmaps of the 24-h cycling transcripts of SW620 showing their expression in SW480 (left panel) and SW480-acrophase-ordered expression heatmaps of the 24-h cycling transcripts of SW480 showing their expression in SW620 (right panel).
Functional annotation for circadian transcripts with differential rhythmicity. Enriched GO terms for the three sets of circadian transcripts with differential rhythmicity in SW480 and SW620 cells (blue: higher amplitude in SW480 cells; green: higher amplitude in SW620 cells; pink: pure-phase shift).
Functional annotation of SW480 circadian transcript clusters reveals splicing processes among enriched GO and KEGG terms. (a) The circadian transcripts of SW480 were grouped in four clusters. Each colored line represents the RMA-preprocessed expression level of a transcript transformed to the standard normal distribution. The colors of the gene expression patterns indicate gradual membership values reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black line indicates the cluster center for which the acrophase is given as the data point with the highest expression value. (b) Annotation clusters of enriched GO terms and KEGG pathways for the four circadian transcript clusters of SW480. Shown are the first four terms with p < 0·05 for all annotation clusters with an enrichment score ≥ 1·3.
Functional annotation of SW620 circadian transcript clusters reveals splicing processes among enriched GO and KEGG terms. (a) The circadian transcripts of SW620 were grouped in four clusters. Each colored line represents the RMA-preprocessed expression level of a transcript transformed to the standard normal distribution. The colors of the gene expression patterns indicate gradual membership values reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black line indicates the cluster center for which the acrophase is given as the data point with the highest expression value. (b) Annotation clusters of enriched GO terms and KEGG pathways for the four circadian transcript clusters of SW620. Shown are the first four terms with p < 0·05 for all annotation clusters with an enrichment score ≥ 1·3.
Differentially rhythmic expression patterns of splicing-related genes in CRC progression. Circadian transcriptional expression patterns in SW480 (blue line) and SW620 cells (green line) of all spliceosome components and SFs that showed 24-h oscillations in at least one of the cell lines. The adjusted p-value denotes the results of the DODR analysis and was calculated as previously described.
Differential AS events in the EMT-associated genes CD44 and FGFR2. Exon-level expression (upper panel), FIRMA scores (middle panel), and genomic representation (lower panel) of the candidate genes CD44 (a) and FGFR2 (b). Vertical lines separate the individual probesets covering the genes. For each probeset, the exon-level expression and the FIRMA scores of the individual time points are depicted by a dotted line and the respective mean values are depicted by a solid line for SW480 (blue) and SW620 (green), respectively. The mean FIRMA score log2 FC (SW480-SW620) is shown in pink. Gray diagonal lines indicate the localization of the probesets within the genome.
Temporal exon expression patterns of selected exons with circadian AS events. Exon expression profiles for selected exons with circadian AS events in either SW480 (blue) and/or SW620 cells (green).
Phase-clustered pathways whose associated genes showed significant shifts in their peak expression in the CRC progression model.
Candidate exons with differential AS events between SW480 and SW620 cells.
Enriched GO terms (biological processes) for the candidate genes with differential AS events.
Acknowledgments
Acknowledgements
We are grateful to Ute Ungethüm from the LFGC (Charité – Universitätsmedizin Berlin) for technical support regarding the microarrays work, to Pål Westermark for technical support regarding the usage of the R packages RAIN, HarmonicRegression, and DODR, and to Caterina Coli from the group of Juan Valcárcel for providing a list of spliceosome components and splicing regulators.
Funding Sources
The work in A.R.'s group was funded by the German Federal Ministry of Education and Research (BMBF) (eBio-CIRSPLICE - FKZ031A316) and by the Dr. Rolf M. Schwiete Stiftung. R.E. was additionally funded by the Joachim Herz Stiftung and L.F. was additionally funded by the Berlin School of Integrative Oncology (BSIO) of the Charité – Universitätsmedizin Berlin.
Declaration of Interest
The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest concerning this manuscript.
Author Contributions
Conceptualization, A.R.; Methodology, R.E. and A.R.; Investigation, R.E., L.F. and A.R.; Formal Analysis, R.E.; Visualization, R.E.; Writing – Original Draft, R.E. and A.R; Writing – Review and Editing, R.E., L.F., and A.R.; Funding Acquisition, A.R.; Resources, A.R.; Supervision, A.R.
References
- 1.Sahar S., Sassone-Corsi P. Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab. 2012;23(1):1–8. doi: 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamia K.A. F1000Research. 2017. Ticking time bombs: connections between circadian clocks and cancer; p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shostak A. Circadian clock, cell division, and Cancer: from molecules to organism. Int J Mol Sci. 2017;18(4):873. doi: 10.3390/ijms18040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Athman R., Genov N.N., Mazuch J., Zhang K., Yu Y., Fuhr L. The Ink4a/Arf locus operates as a regulator of the circadian clock modulating RAS activity. PLoS Biol. 2017;15(12) doi: 10.1371/journal.pbio.2002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrero-Vargas N.N., Navarro-Espíndola R., Guzmán-Ruíz M.A., del Carmen Basualdo M., Espitia-Bautista E., López-Bago A. Circadian disruption promotes tumor growth by anabolic host metabolism; experimental evidence in a rat model. BMC Cancer. 2017;17(1):625. doi: 10.1186/s12885-017-3636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhr L., Abreu M., Pett P., Relógio A. Circadian systems biology: when time matters. Comput Struct Biotechnol J. 2015;13:417–426. doi: 10.1016/j.csbj.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann R., Childs L., Thomas P., Abreu M., Fuhr L., Herzel H. Assembly of a comprehensive regulatory network for the mammalian circadian clock: a bioinformatics approach. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mure L.S., Le H.D., Benegiamo G., Chang M.W., Rios L., Jillani N. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018:eaao0318. doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim C., Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16(11):1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 12.Kojima S., Green C.B. Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry. 2014;54(2):124–133. doi: 10.1021/bi500707c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mermet J., Yeung J., Naef F. Systems chronobiology: global analysis of gene regulation in a 24-hour periodic world. Cold Spring Harb Perspect Biol. 2017;9(3):a028720. doi: 10.1101/cshperspect.a028720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallego-Paez L., Bordone M., Leote A., Saraiva-Agostinho N., Ascensão-Ferreira M., Barbosa-Morais N. Alternative splicing: the pledge, the turn, and the prestige. Hum Genet. 2017:1–28. doi: 10.1007/s00439-017-1790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Bonnal S., Vigevani L., Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11(11):847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 17.Singh R., Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol. 2005;12(8):645. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 18.Cooper T.A., Wan L., Dreyfuss G. RNA and disease. Cell. 2009;136(4):777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Syed N.H., Prince S.J., Mutava R.N., Patil G., Li S., Chen W. Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J Exp Bot. 2015;66(22):7129–7149. doi: 10.1093/jxb/erv407. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez S.E., Petrillo E., Beckwith E.J., Zhang X., Rugnone M.L., Hernando C.E. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468(7320):112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 21.Hughes M.E., Grant G.R., Paquin C., Qian J., Nitabach M.N. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22(7):1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bélanger V., Picard N., Cermakian N. The circadian regulation of Presenilin-2 gene expression. Chronobiol Int. 2006;23(4):747–766. doi: 10.1080/07420520600827087. [DOI] [PubMed] [Google Scholar]
- 23.Preußner M., Wilhelmi I., Schultz A.-S., Finkernagel F., Michel M., Möröy T. Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice. Mol Cell. 2014;54(4):651–662. doi: 10.1016/j.molcel.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 24.McGlincy N.J., Valomon A., Chesham J.E., Maywood E.S., Hastings M.H., Ule J. Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 2012;13(6):1. doi: 10.1186/gb-2012-13-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preußner M., Heyd F. Post-transcriptional control of the mammalian circadian clock: implications for health and disease. Pflügers Archiv-European Journal of Physiology. 2016:1–9. doi: 10.1007/s00424-016-1820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D., Liang X., Chen X., Guo J. Ribonucleoprotein complexes that control circadian clocks. Int J Mol Sci. 2013;14(5):9018–9036. doi: 10.3390/ijms14059018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo K.-C., Ha D.-C., Lee K.-H., Kim D.-Y., Kim T.-D., Kim K.-T. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol. 2010;30(1):197–205. doi: 10.1128/MCB.01154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim T.-D., Woo K.-C., Cho S., Ha D.-C., Jang S.K., Kim K.-T. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21(7):797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David C.J., Manley J.L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24(21):2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Climente-Gonzalez H., Porta-Pardo E., Godzik A., Eyras E. The functional impact of alternative splicing in Cancer. Cell Rep. 2017;20(9):2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Jeyaraj S., O'Brien D.M., Chandler D.S. MDM2 and MDM4 splicing: an integral part of the cancer spliceome. Frontiers in Bioscience (Landmark edition) 2008;14:2647–2656. doi: 10.2741/3402. [DOI] [PubMed] [Google Scholar]
- 32.Kaida D., Schneider-Poetsch T., Yoshida M. Splicing in oncogenesis and tumor suppression. Cancer Sci. 2012;103(9):1611–1616. doi: 10.1111/j.1349-7006.2012.02356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 34.Bozek K., Relogio A., Kielbasa S.M., Heine M., Dame C., Kramer A. Regulation of clock-controlled genes in mammals. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hewitt R.E., Mcmarlin A., Kleiner D., Wersto R., Martin P., Tsoskas M. Validation of a model of colon cancer progression. J Pathol. 2000;192(4):446–454. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 36.Giacchetti S., Bjarnason G., Garufi C., Genet D., Iacobelli S., Tampellini M. Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: the European Organisation for Research and Treatment of Cancer chronotherapy group. J Clin Oncol. 2006;24(22):3562–3569. doi: 10.1200/JCO.2006.06.1440. [DOI] [PubMed] [Google Scholar]
- 37.Lévi F., Dugué P.-A., Innominato P., Karaboué A., Dispersyn G., Parganiha A. Wrist actimetry circadian rhythm as a robust predictor of colorectal cancer patients survival. Chronobiol Int. 2014;31(8):891–900. doi: 10.3109/07420528.2014.924523. [DOI] [PubMed] [Google Scholar]
- 38.Relógio A., Thomas P., Medina-Pérez P., Reischl S., Bervoets S., Gloc E. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014;10(5) doi: 10.1371/journal.pgen.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazzoccoli G., Vinciguerra M., Papa G., Piepoli A. Circadian clock circuitry in colorectal cancer. World J Gastroenterol: WJG. 2014;20(15):4197. doi: 10.3748/wjg.v20.i15.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazzoccoli G., Pazienza V., Panza A., Valvano M.R., Benegiamo G., Vinciguerra M. ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J Cancer Res Clin Oncol. 2012;138(3):501–511. doi: 10.1007/s00432-011-1126-6. [DOI] [PubMed] [Google Scholar]
- 41.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolstad B.M., Irizarry R.A., Åstrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 43.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thaben P.F., Westermark P.O. Detecting rhythms in time series with RAIN. J Biol Rhythm. 2014;29(6):391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lück S., Thurley K., Thaben P.F., Westermark P.O. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014;9(2):741–751. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 46.Thaben P.F., Westermark P.O. Differential rhythmicity: detecting altered rhythmicity in biological data. Bioinformatics. 2016;32(18):2800–2808. doi: 10.1093/bioinformatics/btw309. [DOI] [PubMed] [Google Scholar]
- 47.Shilts J., Chen G., Hughey J.J. Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ. 2018;6 doi: 10.7717/peerj.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg K.C., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A. Multi-omics of 34 colorectal cancer cell lines-a resource for biomedical studies. Mol Cancer. 2017;16(1):116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughey J.J., Hastie T., Butte A.J. ZeitZeiger: supervised learning for high-dimensional data from an oscillatory system. Nucleic Acids Res. 2016;44(8):e80. doi: 10.1093/nar/gkw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang R., Podtelezhnikov A.A., Hogenesch J.B., Anafi R.C. Discovering biology in periodic data through phase set enrichment analysis (PSEA) J Biol Rhythm. 2016;31(3):244–257. doi: 10.1177/0748730416631895. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bengtsson H., Simpson K., Bullard J., Hansen K. Tech report. 2008. Aroma. affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. [Google Scholar]
- 54.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durinck S., Bullard J., Spellman P.T., Dudoit S. GenomeGraphs: integrated genomic data visualization with R. BMC Bioinformatics. 2009;10(1):2. doi: 10.1186/1471-2105-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gachon F., Fonjallaz P., Damiola F., Gos P., Kodama T., Zakany J. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18(12):1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Riel B., Pakozdi T., Brouwer R., Monteiro R., Tuladhar K., Franke V. A novel complex, RUNX1-MYEF2, represses hematopoietic genes in erythroid cells. Mol Cell Biol. 2012;32(19):3814–3822. doi: 10.1128/MCB.05938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sood S., Szkop K.J., Nakhuda A., Gallagher I.J., Murie C., Brogan R.J. iGEMS: an integrated model for identification of alternative exon usage events. Nucleic Acids Res. 2016;44(11):e109. doi: 10.1093/nar/gkw263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenka G., Tsai M.-H., Lin H.-C., Hsiao J.-H., Lee Y.-C., Lu T.-P. Identification of methylation-driven, differentially expressed STXBP6 as a novel biomarker in lung adenocarcinoma. Sci Rep. 2017;7:42573. doi: 10.1038/srep42573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davenport A.P., Hyndman K.A., Dhaun N., Southan C., Kohan D.E., Pollock J.S. Endothelin. Pharmacol Rev. 2016;68(2):357–418. doi: 10.1124/pr.115.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahman H.A., Wu H., Dong Y., Pasula S., Wen A., Sun Y. Selective targeting of a novel Epsin–VEGFR2 interaction promotes VEGF-mediated AngiogenesisNovelty and significance. Circ Res. 2016;118(6):957–969. doi: 10.1161/CIRCRESAHA.115.307679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torres-Martín M., Kusak M.E., Isla A., Burbano R.R., Pinto G.R., Melendez B. Whole exome sequencing in a case of sporadic multiple meningioma reveals shared NF2, FAM109B, and TPRXL mutations, together with unique SMARCB1 alterations in a subset of tumor nodules. Cancer Gene Ther. 2015;208(6):327–332. doi: 10.1016/j.cancergen.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 63.Hao S., Yao L., Huang J., He H., Yang F., Di Y. Genome-wide analysis identified a number of dysregulated long noncoding RNA (lncRNA) in human pancreatic ductal adenocarcinoma. Technol Cancer Res Treat. 2018;17 doi: 10.1177/1533034617748429. [1533034617748429] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Relógio A., Ben-Dov C., Baum M., Ruggiu M., Gemund C., Benes V. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J Biol Chem. 2005;280(6):4779–4784. doi: 10.1074/jbc.M411976200. [DOI] [PubMed] [Google Scholar]
- 65.Cvitkovic I., Jurica M.S. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res. 2012;41(D1):D41–D132. doi: 10.1093/nar/gks999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giulietti M., Piva F., D'Antonio M., D'onorio De Meo P., Paoletti D., Castrignano T. SpliceAid-F: a database of human splicing factors and their RNA-binding sites. Nucleic Acids Res. 2012;41(D1):D31–D125. doi: 10.1093/nar/gks997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warzecha C.C., Carstens R.P., editors. Seminars in cancer biology. Elsevier; 2012. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Purdom E., Simpson K.M., Robinson M.D., Conboy J., Lapuk A., Speed T.P. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24(15):1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rokavec M., Horst D., Hermeking H. Cellular model of colon cancer progression reveals signatures of mRNAs, miRNA, lncRNAs, and epigenetic modifications associated with metastasis. Cancer Res. 2017;77(8):1854–1867. doi: 10.1158/0008-5472.CAN-16-3236. [DOI] [PubMed] [Google Scholar]
- 70.Alhopuro P., Björklund M., Sammalkorpi H., Turunen M., Tuupanen S., Biström M. Mutations in the circadian gene CLOCK in colorectal cancer. Mol Cancer Res. 2010;8(7):952–960. doi: 10.1158/1541-7786.MCR-10-0086. [DOI] [PubMed] [Google Scholar]
- 71.Chen H., Xia M., Lin M., Yang H., Kuhlenkamp J., Li T. Role of methionine adenosyltransferase 2A and S-adenosylmethionine in mitogen-induced growth of human colon cancer cells. Gastroenterology. 2007;133(1):207–218. doi: 10.1053/j.gastro.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 72.Cho Y.-S., Do M.-H., Kwon S.-Y., Moon C., Kim K., Lee K. Efficacy of CD46-targeting chimeric Ad5/35 adenoviral gene therapy for colorectal cancers. Oncotarget. 2016;7(25):38210. doi: 10.18632/oncotarget.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakarai C., Osawa K., Akiyama M., Matsubara N., Ikeuchi H., Yamano T. Expression of AKR1C3 and CNN3 as markers for detection of lymph node metastases in colorectal cancer. Clin Exp Med. 2015;15(3):333–341. doi: 10.1007/s10238-014-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye Y., Xiang Y., Ozguc F.M., Kim Y., Liu C.-J., Park P.K. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Systems. 2018;6(3):314–328.e2. doi: 10.1016/j.cels.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen B., Liu Y., Jin X., Lu W., Liu J., Xia Z. MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer. 2014;14:443. doi: 10.1186/1471-2407-14-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma Y.S., Yang I.P., Tsai H.L., Huang C.W., Juo S.H., Wang J.Y. High glucose modulates antiproliferative effect and cytotoxicity of 5-fluorouracil in human colon cancer cells. DNA Cell Biol. 2014;33(2):64–72. doi: 10.1089/dna.2013.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh C.S., Wang J.Y., Chung F.Y., Lee S.C., Huang M.Y., Kuo C.W. Significance of the glycolytic pathway and glycolysis related-genes in tumorigenesis of human colorectal cancers. Oncol Rep. 2008;19(1):81–91. [PubMed] [Google Scholar]
- 78.Li W.-W., Wang H.-Y., Nie X., Liu Y.-B., Han M., Li B.-H. Human colorectal cancer cells induce vascular smooth muscle cell apoptosis in an exocrine manner. Oncotarget. 2017;8(37):62049. doi: 10.18632/oncotarget.18893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bisognin A., Pizzini S., Perilli L., Esposito G., Mocellin S., Nitti D. An integrative framework identifies alternative splicing events in colorectal cancer development. Mol Oncol. 2014;8(1):129–141. doi: 10.1016/j.molonc.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huerta S., Srivatsan E.S., Venkatesan N., Peters J., Moatamed F., Renner S. Alternative mRNA splicing in colon cancer causes loss of expression of neural cell adhesion molecule. Surgery. 2001;130(5):834–843. doi: 10.1067/msy.2001.116415. [DOI] [PubMed] [Google Scholar]
- 81.Isacke C.M., Yarwood H. The hyaluronan receptor, CD44. Int J Biochem Cell Biol. 2002;34(7):718–721. doi: 10.1016/s1357-2725(01)00166-2. [DOI] [PubMed] [Google Scholar]
- 82.Cooper D.L., Dougherty G.J. To metastasize or not? Selection of CD44 splice sites. Nat Med. 1995;1(7):635–637. doi: 10.1038/nm0795-635. [DOI] [PubMed] [Google Scholar]
- 83.Warzecha C.C., Sato T.K., Nabet B., Hogenesch J.B., Carstens R.P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33(5):591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carstens R.P., Eaton J.V., Krigman H.R., Walther P.J., Garcia-Blanco M.A. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene. 1997;15(25):3059. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 85.Dolatshad H., Pellagatti A., Fernandez-Mercado M., Yip B.H., Malcovati L., Attwood M. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29(5):1092. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Avitabile D., Genovese L., Ponti D., Ranieri D., Raffa S., Calogero A. Nucleolar localization and circadian regulation of Per2S, a novel splicing variant of the period 2 gene. Cell Mol Life Sci. 2014;71(13):2547–2559. doi: 10.1007/s00018-013-1503-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nowak D.G., Amin E.M., Rennel E.S., Hoareau-Aveilla C., Gammons M., Damodoran G. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms a novel therapeutic strategy for angiogenesis. J Biol Chem. 2010;285(8):5532–5540. doi: 10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kunická T., Souček P. Importance of ABCC1 for cancer therapy and prognosis. Drug Metab Rev. 2014;46(3):325–342. doi: 10.3109/03602532.2014.901348. [DOI] [PubMed] [Google Scholar]
- 89.Ballesta A., Dulong S., Abbara C., Cohen B., Okyar A., Clairambault J. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput Biol. 2011;7(9) doi: 10.1371/journal.pcbi.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ozturk N., Ozturk D., Kavakli I.H., Okyar A. Molecular aspects of circadian pharmacology and relevance for cancer chronotherapy. Int J Mol Sci. 2017;18(10):2168. doi: 10.3390/ijms18102168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grant C.E., Kurz E.U., Cole S.P., Deeley R.G. Analysis of the intron–exon organization of the human multidrug-resistance protein gene (MRP) and alternative splicing of its mRNA. Genomics. 1997;45(2):368–378. doi: 10.1006/geno.1997.4950. [DOI] [PubMed] [Google Scholar]
- 92.He X., Ee P.R., Coon J.S., Beck W.T. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res. 2004;10(14):4652–4660. doi: 10.1158/1078-0432.CCR-03-0439. [DOI] [PubMed] [Google Scholar]
- 93.Hu T., Li Z., Gao C.-Y., Cho C.H. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol. 2016;22(30):6876. doi: 10.3748/wjg.v22.i30.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lévi F., Okyar A., Dulong S., Innominato P.F., Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 95.El-Athman R., Relógio A. Escaping circadian regulation: an emerging Hallmark of Cancer? Cell Systems. 2018;6(3):266–267. doi: 10.1016/j.cels.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 96.Kumar L., Futschik M.E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Single-cell time-course measurements of REV-ERBα protein abundance in CRC cell lines. The fluorescence intensity of a REV-ERBα-VNP fusion protein was measured for 38 h in single live-cells from SW480 (a) and SW620 (b) (n = 4).
Alterations of the circadian clock in CRC primary tumors and metastasis. (a) Heatmaps of Spearman correlation (rho) between each pair of core-clock genes in a set of 34 CRC cell lines (GSE97023) in comparison to a mammalian reference set (GSE54650). (b) Box-plot of the log-likelihoods of the ZeitZeiger predictions of CT for SW480 (blue) and SW620 (green) samples. (c) Venn diagram of the 24-h cycling transcripts in SW480 and SW620 cells. (d) Median-normalized, SW620-acrophase-ordered expression heatmaps of the 24-h cycling transcripts of SW620 showing their expression in SW480 (left panel) and SW480-acrophase-ordered expression heatmaps of the 24-h cycling transcripts of SW480 showing their expression in SW620 (right panel).
Functional annotation for circadian transcripts with differential rhythmicity. Enriched GO terms for the three sets of circadian transcripts with differential rhythmicity in SW480 and SW620 cells (blue: higher amplitude in SW480 cells; green: higher amplitude in SW620 cells; pink: pure-phase shift).
Functional annotation of SW480 circadian transcript clusters reveals splicing processes among enriched GO and KEGG terms. (a) The circadian transcripts of SW480 were grouped in four clusters. Each colored line represents the RMA-preprocessed expression level of a transcript transformed to the standard normal distribution. The colors of the gene expression patterns indicate gradual membership values reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black line indicates the cluster center for which the acrophase is given as the data point with the highest expression value. (b) Annotation clusters of enriched GO terms and KEGG pathways for the four circadian transcript clusters of SW480. Shown are the first four terms with p < 0·05 for all annotation clusters with an enrichment score ≥ 1·3.
Functional annotation of SW620 circadian transcript clusters reveals splicing processes among enriched GO and KEGG terms. (a) The circadian transcripts of SW620 were grouped in four clusters. Each colored line represents the RMA-preprocessed expression level of a transcript transformed to the standard normal distribution. The colors of the gene expression patterns indicate gradual membership values reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black line indicates the cluster center for which the acrophase is given as the data point with the highest expression value. (b) Annotation clusters of enriched GO terms and KEGG pathways for the four circadian transcript clusters of SW620. Shown are the first four terms with p < 0·05 for all annotation clusters with an enrichment score ≥ 1·3.
Differentially rhythmic expression patterns of splicing-related genes in CRC progression. Circadian transcriptional expression patterns in SW480 (blue line) and SW620 cells (green line) of all spliceosome components and SFs that showed 24-h oscillations in at least one of the cell lines. The adjusted p-value denotes the results of the DODR analysis and was calculated as previously described.
Differential AS events in the EMT-associated genes CD44 and FGFR2. Exon-level expression (upper panel), FIRMA scores (middle panel), and genomic representation (lower panel) of the candidate genes CD44 (a) and FGFR2 (b). Vertical lines separate the individual probesets covering the genes. For each probeset, the exon-level expression and the FIRMA scores of the individual time points are depicted by a dotted line and the respective mean values are depicted by a solid line for SW480 (blue) and SW620 (green), respectively. The mean FIRMA score log2 FC (SW480-SW620) is shown in pink. Gray diagonal lines indicate the localization of the probesets within the genome.
Temporal exon expression patterns of selected exons with circadian AS events. Exon expression profiles for selected exons with circadian AS events in either SW480 (blue) and/or SW620 cells (green).
Phase-clustered pathways whose associated genes showed significant shifts in their peak expression in the CRC progression model.
Candidate exons with differential AS events between SW480 and SW620 cells.
Enriched GO terms (biological processes) for the candidate genes with differential AS events.