Abstract
An endogenous molecular clockwork drives various cellular pathways including metabolism and the cell cycle. Its dysregulation is able to prompt pathological phenotypes including cancer. Besides dramatic metabolic alterations, cancer cells display severe changes in the clock phenotype with likely consequences in tumor progression and treatment response. In this study, we use a comprehensive systems-driven approach to investigate the effect of clock disruption on metabolic pathways and its impact on drug response in a cellular model of colon cancer progression. We identified distinctive time-related transcriptomic and metabolic features of a primary tumor and its metastatic counterpart. A mapping of the expression data to a comprehensive genome-scale reconstruction of human metabolism allowed for the in-depth functional characterization of 24 h-oscillating transcripts and pointed to a clock-driven metabolic reprogramming in tumorigenesis. In particular, we identified a set of five clock–regulated glycolysis genes, ALDH3A2, ALDOC, HKDC1, PCK2, and PDHB with differential temporal expression patterns. These findings were validated in organoids and in primary fibroblasts isolated from normal colon and colon adenocarcinoma from the same patient. We further identified a reciprocal connection of HKDC1 to the clock in the primary tumor, which is lost in the metastatic cells. Interestingly, a disruption of the core-clock gene BMAL1 impacts on HKDC1 and leads to a time-dependent rewiring of metabolism, namely an increase in glycolytic activity, as well as changes in treatment response. This work provides novel evidence regarding the complex interplay between the circadian clock and metabolic alterations in carcinogenesis and identifies new connections between both systems with pivotal roles in cancer progression and response to therapy.
Keywords: Circadian clock, Colorectal cancer, Metabolic rewiring, Tumor progression, High-throughput circadian data, Metabolic network reconstruction, Circadian regulation of metabolism, Treatment response, Glycolysis
Highlights
-
•
Primary and metastatic colon cancer cells from the same patient show differential time-dependent metabolic profiles.
-
•
Perturbations of the circadian clock induce differential alterations at the transcriptome level including metabolic pathways.
-
•
Core-clock gene (BMAL1) knockdown affects metabolic activity of cancer cells and impinges on treatment response.
An evolutionary conserved molecular clockwork allows organisms to adapt physiology and biological processes to the geophysical time by driving various cellular pathways including metabolism and the cell cycle. In addition to dramatic metabolic alterations, cancer cells show severe changes in the biological clock likely to affect tumor progression and treatment response. Increasing efforts have been made to elucidate the connection between the circadian clock circuitry, tumor progression and cancer-associated metabolic alterations, yet a more detailed knowledge of this interplay is still missing. In this study, we investigated distinctive time-related transcriptomic and metabolic features in a cellular model of colon cancer progression and patient samples. We show that a disrupted biological clock leads to altered temporal profiles of gene expression, metabolic reprogramming and changes in drug response, and identify the hexokinase HKDC1 as a crucial element in this connection. Our results provide novel evidence regarding the complex interplay between the circadian clock and metabolic alterations in carcinogenesis and pave the way to a possible optimization of cancer treatment.
1. Introduction
The circadian clock is an internal timing system that allows organisms to adapt biological processes and behavior to the geophysical time and it is operated by a set of genes and proteins hardwiring transcriptional and translational regulatory feedback loops. In mammals, these feedback loops drive the oscillatory expression of various target genes and regulate cellular processes involved in tumor development and progression [1], including metabolism and cell cycle [2]. A close link between the circadian clock and metabolism was previously reported [3,4] and increasing evidence points to a role of dysregulated biological clocks in cancer onset [5], as well as to an important role for the clock in the response to anticancer treatments [6,7].
Cancer cells show a vast number of metabolic alterations and specific metabolic pathways are likely to play a role in cell transformation. A well-known example for changes in the cellular metabolism in tumors is the Warburg effect – the predominant use of glycolysis even in the presence of oxygen – which is present in many cancer cells [5]. Another metabolic pathway that came into focus during the last years is the altered glucose metabolism in cancer that is required to achieve the anabolic demands of tumor cells [8]. Although enhanced aerobic glycolysis has been used as a marker to distinguish cancer cells from normal cells for decades, an elevated glucose metabolism was only very recently defined as one of the hallmarks of cancer [9]. Cancer cells maintain high rates of glycolysis and concurrent oxidative phosphorylation to supply other pathways that generate macromolecules to fulfil the metabolic demands of proliferating cells [8,10]. Several metabolic genes such as glucose-6-phosphatase (G6P), phosphoenolpyruvate carboxykinase 2 (PCK2) and solute carrier family 2 member 2 (GLUT2) show a circadian expression pattern [5]. Furthermore, numerous metabolites have been shown to oscillate in a circadian fashion [3,11], further strengthening the role of the circadian clock as a regulator of metabolism. Moreover, a reciprocal interplay between the circadian clock and mitochondrial respiratory activity has recently been shown [12,13]. In addition to the biological clock influence on metabolic pathways, drug response pathways that influence pharmacokinetics and pharmacodynamics are also under circadian control [14]. Consequently, improved treatment response and tolerability has been achieved by chronomodulated chemotherapy [15] and time-dependent radiotherapy scheduling [[16], [17], [18], [19]]. However, despite the recent studies that support the role of the circadian clock in metabolic control, the mechanisms by which the circadian clock might influence cellular metabolism in cancer remain unclear.
In the present study, we investigated the role of a dysregulated biological clock in an in vitro model of colon cancer progression. As a model system, we used established cancer cell lines derived from a primary colorectal adenocarcinoma and a lymph node metastasis from the same patient, in addition to primary fibroblasts isolated from normal colon and colon adenocarcinoma of the same patient and organoids to further explore our findings. At the transcriptome level, we quantified differences in gene expression that impact on metabolic pathways. A genome-scale reconstruction of a human metabolic network allowed for the in-depth functional characterization of the 24 h oscillating genes. Based on different oscillatory profiles of selected metabolic pathways, glycolysis and oxidative phosphorylation, we identified a set of clock-regulated metabolic genes. Among those is the hexokinase HKDC1, which we found to be able to mediate clock-driven metabolic reprogramming in tumorigenesis. A subsequent disruption of the core-clock gene encoding for the Aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL, also known as BMAL1) led to time-dependent rewiring of metabolism in both cell lines, as well as changes in treatment response. Thus, our data demonstrates the existence of a reciprocal interplay between metabolic genes and the circadian clock, and identifies novel connections between both systems that can play a pivotal role in colon cancer progression and in response to therapy.
2. Materials and Methods
2.1. Cell Culture
SW480 (RRID:AB_148468), SW620 (RRID:CVCL_0547) and HepG2 (RRID:CVCL_0027) cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) low glucose (Lonza, Basel, CH) culture medium supplemented with 10% fetal bovine serum (FBS) (Life technologies, Carlsbad, CA, USA), 1% penicillin-streptomycin (Life technologies), 2 mM Ultraglutamine (Lonza) and 1% HEPES (Life technologies). Normal fibroblasts (NFs) and tumor-associated fibroblasts (TAFs) were cultivated in gelatin-coated dishes and maintained in Minimum Essential Medium Eagle (MEM)-α (Gibco, Waltham, MA, USA) medium supplemented with 10% Hyclone FBS (GE healthcare, Little Chalfont, UK), 1× antibiotic-antimycotic (Gibco) and 1× MEM Non-Essential Amino Acids Solution (Gibco). Human fallopian tube organoids were maintained in Advanced DMEM/F12 (Invitrogen, Carlsbad, CA, USA) supplemented with 12 mM HEPES, 1% GlutaMAX, 2% B27, 1% N2, 10 ng/ml human EGF (all from Invitrogen), 100 ng/ml human noggin, 100 ng/ml human FGF-10 (both from Peprotech, Rocky Hill, NJ, USA), 1 mM nicotinamide, 9 μM ROCK inhibitor (Y-27632, both from Sigma-Aldrich, St. Louis, MO, USA) and 0.5 μM TGF-β RI Kinase Inhibitor IV (SB431542, Calbiochem, San Diego, CA, USA), Wnt3a and RSPO1 conditioned medium (housemade stocks) in Matrigel (Corning, NY, USA). Cells were isolated, maintained and cultured as previously described [20]. All cells were incubated at 37 °C in a humidified atmosphere with 5% CO2.
2.2. Primary Fibroblasts
Fibroblasts were derived from colorectal cancer patients. NFs were isolated from normal colon segment with a distance of approximately 20 cm from the tumor and TAFs were isolated from the tumor of the same patient with moderately differentiated adenocarcinoma and lymph node metastasis. Tissue pieces were washed rigorously with phosphate buffered saline (PBS), antibiotic-antimycotic (10×) (Lonza), gentamycin (10×) (Lonza) and cut into pieces for incubation overnight at 37 °C in DMEM (Euroclone, Milano, IT), antibiotic-antimycotic (1×), gentamycin (1×) in the presence of collagenase type II 150 U/ml (Gibco). After incubation, the solution was centrifuged briefly to remove undigested tissue pieces and plated on gelatin (1 g/l) pre-treated dishes in αMEM (Euroclone) supplemented with 10% defined FBS (GE-Healthcare), non-essential amino acids (Euroclone), L-alanine-l-glutamine (Euroclone), penicillin/streptomycin (5×) (Euroclone), antibiotic-antimycotic (5×), and gentamycin (5×).
2.3. Human Fallopian Tube Organoids
Human fallopian tube samples were provided by the department of Gynecology, Charité Medical University of Berlin, Germany. Scientific usage of the samples for experimental purposes was approved by the ethics commission of the Charité (EA1/002/07), and all subjects gave informed consent to their tissues being used in scientific research. Fragments were sourced from standard surgical procedures for benign gynaecological disease. Only anatomically normal fallopian tubes were used. Tubes were transported and dissected within 2–3 h of removal.
2.4. Ethics Statement
Primary fibroblasts: The protocol and the scientific usage of the samples for experimental purposes was approved by IRCCS SDN (Comitato Etico per la Sperimentazione Clinica Progetto N:ro 2013_01_02) and Monaldi Hospital ethical committees (Deliberazione del Direttore Generale n:o 1239). Human fallopian tube organoids: Scientific usage of the samples for experimental purposes was approved by the ethics commission of the Charité Medical University of Berlin (EA1/002/07), and all subjects gave informed consent to their tissues being used in scientific research.
2.5. Lentivirus Production
Lentiviral elements containing a BMAL1-promoter-driven luciferase (BLH) or a PER2-promoter-driven luciferase (PLB) were generated as previously described [21]. For lentivirus production, HEK293T (RRID:CVCL_0063) cells were seeded in 175 cm [2] culture flasks and co-transfected with 12.5 μg packaging plasmid psPAX, 7.5 μg envelope plasmid pMD2G and 17.5 μg expression plasmid using the CalPhos mammalian transfection kit (Clontech, Fremont, CA, USA) according to the manufacturer's instruction. To harvest the lentiviral particles, the supernatant was centrifuged at 4100 x g for 15 min to remove cell debris and passed through a 45 μm filter (Sarstedt, Nümbrecht, DE). The lentiviral particles were stored at −80 °C.
2.6. Transduction with Lentiviral Vectors
For lentiviral transduction, cells were seeded in 6-well plates. On the day of transduction, 1 ml medium and 1 ml of lentiviral particles was added. 8 μg/ml protamine sulfate (Sigma) and 4 μg/ml polybrene was used to enhance transduction efficiency. The next day, the medium was replaced and selection medium was added one day later (complete growth medium containing appropriate antibiotic) to obtain stably transduced cells and incubated at 37 °C with 5% CO2 atmosphere. Untransduced cells treated with the same antibiotic concentration were used as selection controls.
2.7. Bioluminescence Measurements
For live-cell bioluminescence recordings, SW480 and SW620 cells were maintained in phenol red-free DMEM (Gibco) containing 10% FBS, 1% penicillin-streptomycin. NFs and TAFs were maintained in phenol red-free MEM-α (Gibco) medium supplemented with 10% Hyclone FBS (GE healthcare), 1× antibiotic-antimycotic (Gibco) and 1× MEM Non-Essential Amino Acids Solution (Gibco). Human fallopian tube organoids were maintained in normal growth medium. All media were supplemented with 250 μM D-Luciferin (PJK, Kleinblittersdorf, DE). Cells were synchronized by medium change prior to measurement. BMAL1-promoter-(BLH/BLP)-reporter activity and PER2-promoter-(PLB) activity was measured, using a LumiCycle instrument (Actimetrics, Wilmette, IL, USA) for five consecutive days. Raw luminescence data were de-trended by the 24 h running average using the Chronostar software [22]. The first 12 h of measurement were removed from the analysis, since the first data collection is comparatively very noisy due to technical limitations of the device.
2.8. Respirometric Measurements
Cultured cells were synchronized by medium change, gently detached from the dish by trypsinization, washed in PBS, harvested by centrifugation at 500 x g for 5 min and immediately assessed for O2 consumption with a high resolution oxymeter (Oxygraph-2 k, Oroboros Instruments, Innsbruck, AT). About 8–10 × 106 viable cells/ml were assayed in DMEM at 37 °C; after attainment of a stationary endogenous substrate-sustained resting oxygen consumption rate (OCR), 2 μM antimycin A plus 2 μM rotenone were added and the mitochondria-mediated OCR calculated by subtracting the inhibitors-insensitive respiration (<5% of the overall OCR) and normalized to the cell number.
2.9. Cell Synchronization
For all experiments, cells were synchronized by medium change. For the time-dependent analysis of glycolysis, mitochondrial respiration, viability, cytotoxicity and apoptosis, cells were synchronized at three different timepoints prior to the treatment. Treatment was carried out at 18 h, 21 h and 24 h after synchronization. Timepoint 0 h is defined as the timepoint of treatment. Untreated control cells were prepared in the same way, but treated with a vehicle control.
2.10. Measurement of Glycolytic Activity and Mitochondrial Respiration
Glycolytic activity and mitochondrial respiration were determined using a seahorse machine (Agilent, Santa Clara, CA, USA) and the Seahorse XF Glycolysis Stress Test Kit (Agilent) or the Seahorse XF Cell Mito Stress Test Kit (Agilent). Cells were seeded one day prior to the assay in 96-well seahorse plates. The assays were performed according to the manufacturer's instructions. For the Glycolysis stress test, seahorse XF base medium (Agilent) supplemented with 2 mM l-Glutamine (Thermo Fisher Scientific, Waltham, MA, USA) was used and 10 mM glucose, 1 μM oligomycin and 50 mM 2-DG were used for the injections during the assay. The XF Glycolysis Stress Test presents the measure of glycolysis as the extracellular acidification rate (ECAR) reached by a given cell after the addition of saturating amounts of glucose. The glycolytic capacity is the maximum ECAR reached by a cell following the addition of oligomycin, effectively shutting down oxidative phosphorylation and driving the cell to use glycolysis to its maximum capacity. For the cell mito-stress test, seahorse XF base medium supplemented with 2 mM l-Glutamine, 5.5 mM glucose (Sigma) and 1 mM sodium pyruvate (Sigma) was used and 2 μM oligomycin, 0.5 μM FCCP and 0.5 μM rotenone/antimycin A were used for the injections during the assay. Data was analyzed using the wave software (Agilent). Data was normalized using a CyQuant (Invitrogen) assay according to the manufacturer's manual.
2.11. Measurement of Cell Viability, Cytotoxicity and Apoptosis
To determine cell viability, cytotoxicity and apoptosis, the ApoTox-Glo Triplex Assay (Promega, Madison, WI, USA) was used. The test measures live-cell protease activity using a fluorogenic, cell-permeant peptide substrate (GF-AFC Substrate), dead-cell protease activity using a cell-impermeant, fluorogenic peptide substrate (bis-AAF-R110 Substrate) and caspase-3/7 activation as a key indicator of apoptosis. Cells were seeded in black 96-well half-area plates with clear bottom. The assay was performed according to the manufacturer's instructions.
2.12. RNA Extraction, cDNA Synthesis (Reverse Transcription) and Quantitative Real-Time PCR (qPCR)
Total RNA was isolated using the RNeasy Mini kit (Qiagen, Venlo, NL) according to the manufacturer's manual. Prior to the purification procedure, medium was discarded and cells were washed twice with PBS and lysed in RLT buffer (Qiagen). To digest genomic DNA, an optional on column DNase digestion was performed, using the RNase-free DNase Set (Qiagen). RNA was eluted in 30–50 μl RNase-free water. The final RNA concentration was measured using a Nanodrop 1000 (Thermo Fisher Scientific). RNA was then stored at −80 °C until use. For RT-qPCR analysis, the extracted RNA was reverse transcribed into cDNA using random hexamers (Eurofins MWG Operon, Huntsville, AL, USA) and Reverse Transcriptase (Life technologies). RT-qPCR was performed using human QuantiTect Primer assays (Qiagen) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) in 96-well plates. TBP or GAPDH were used as reference genes. The qPCR reaction and the subsequent melting curve were performed using a CFX Connect Real-Time PCR Detection System (Biorad). A melting curve analysis was performed to detect potential unspecific amplification products. Cq values were determined using the regression method. Relative gene expression was calculated using the 2-ΔΔCt method [23]. Biological and technical replicates were included into the analysis. The mean and the standard error of the mean were calculated. Oscillatory behavior for clock and clock-regulated genes and proteins in SW480 and SW620 cells was determined by the RAIN algorithm (see Microarray Analysis for details). Circadian parameters (acrophase, relative amplitude, mesor) were estimated for both RT-PCR and protein data by fitting a linear sine-cosine function using the R package HarmonicRegression [24]. The analysis is provided in Supplementary Table 1. The harmonic regression procedure fits the model y(t) = m + acos(ωt) + bsin(ωt) in order to estimate absolute amplitudes (A = √(a2 + b2)) and phases (tan φ = b/a) along with confidence intervals and p-values [24]. The relative amplitude is determined by dividing the absolute amplitude A by the mean value m (A/m).
2.13. shRNA-Mediated Knockdown
For the knockdown of BMAL1 and HKDC1, a TRC lentiviral shRNA glycerol set (Dharmacon, Lafayette, CO, USA) specific for each gene was used consisting of five individual shRNAs. The construct that gave best knockdown efficiency was determined by gene expression analysis and used for further experiments.
2.14. Treatment with WZB117 and Oxaliplatin
Treatment concentrations were determined based on the experimentally determined IC50 value (Fig. S7) or based on literature search [25]. For the experimental determination of WZB117 treatment concentration, cells were seeded in 96 well plates one day prior to treatment. Cells were treated with 1.25 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 40 μM, 80 μM, 160 μM, 320 μM or 640 μM WZB117 for 48 h. The cytotoxicity was determined using the CytoTox-Glo™ Cytotoxicity Assay (Promega) following the manufacturer's manual. The following treatment concentrations were used: WZB117: 80 μM, oxaliplatin: 5 μM. Treatment duration was 24 h. The structure of WZB117 (CAS number 1223397–11-2) as well as oxaliplatin (CAS number 61825–94-3) is already published.
2.15. Proliferation Curves
Cells were seeded at the same density and counted for four consecutive days 72 h – 144 h. Cells were counted and cell viability was checked using an automated cell counter (Luna, Biozym, Hessisch Oldendorf, DE) in a 1:1 trypan blue dilution.
2.16. Western Blotting Analysis
Cells were synchronized by medium change as described above, gently detached from the dish, sedimented by low-speed centrifugation and resuspended in lysis buffer. Aliquots containing 40 mg of proteins from each cell lysate were subjected to SDS polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Bio-Rad) using Trans Blot Turbo Transfer System. Membranes were probed with the following primary antibodies: PDH-E1-alpha (1:1000; Abcam, Cambridge, UK); PSER293-PDH-E1-alpha (1:500; Abcam); PCK2 (1:1000; Cell Signaling Technology, Denvers, MA, USA); HKDC1 (1:1000; Sigma-Aldrich); BMAL1 (1:5000; Millipore, Burlington, MA, USA); β-actin (1:10000; Sigma-Aldrich); GAPDH (1:10000; Sigma-Aldrich). After incubation with corresponding suited horseradish peroxidase-conjugated secondary antibody (1:2500; Cell Signaling Technology), signals were developed using the enhanced chemiluminescence kit (ClarityTM Western ECL Substrate, Bio-Rad), acquired by ChemiDoc Imaging System XRS+ (Bio-Rad) and analyzed for densitometry with the ImageJ Lab 4.1 software.
2.17. Sample Preparation for 24 H Time-Course Microarrays
Cells were seeded in triplicates in 6-well plates one day prior to the experiment. On the next day, cells were synchronized by medium change and samples were taken every 3 h for 24 h and prepared for RNA extraction. Microarray hybridization was carried out by the Labor für funktionelle Genomforschung (LFGC, Charité - Universitätsmedizin Berlin) using Affymetrix GeneChip Human Transcriptome Array 2.0. The microarray dataset has been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5876.
2.18. Microarray Analysis
The microarray analysis was conducted in R. Gene expression data was pre-processed for all samples as one batch using the RMA (Robust Multichip Average) methodology [26,27]. To detect genes exhibiting rhythmic behavior with a period of 24 h in their expression intensities, the Rhythmicity Analysis Incorporating Nonparametric methods (RAIN) algorithm was applied to identify robustly 24 h oscillating transcripts with a p-value <.05 [28]. Since the algorithm works optimally with periods that are a multiple of the sampling interval, a period of 24 h was chosen which is the nearest approximation to the observed period of 24.59 h in SW480 cells.
2.19. Clustering of Gene Expression
All genes associated to the three metabolic pathways glycolysis, Warburg effect and oxidative phosphorylation were clustered according to their temporal expression patterns if they showed 24 h oscillations (RAIN analysis) in at least one of the two tested cell lines. Prior to the clustering, the RMA-preprocessed gene expression values were transformed to the standard normal distribution. Clustering of the genes was performed with Mfuzz, an R package for soft clustering of microarray data that uses the fuzzy c-means algorithm [29]. For every gene and cluster, the algorithm generates gradual membership values ranging between 0 and 1, indicating the degree of membership of this gene for the given cluster. The fuzzyfication parameter [30] for all genes sets was set to 1.25. The cluster number was set to 3 in order to obtain clusters with a good agreement between fuzzy and hard clustering, suitable for a broad visualization of the differences in the oscillatory behavior of the gene groups as determined by the partition coefficient for a range of cluster numbers.
2.20. Over-Representation Analysis
For the over-representation analysis of 24 h oscillating genes, genes were grouped into three sets: genes oscillating in both cell lines, genes oscillating only in SW480 cells and genes oscillating only in SW620 cells. Over-representation analysis was performed using ConsensusPathDB (http://cpdb.molgen.mpg.de/). p-values were computed by ConsensusPathDB according to the hypergeometric test based on the number of physical entities present in both the predefined set and user-specified list of physical entities. The p-values were corrected for multiple testing using the false discovery rate method.
All pathways with a p-value <.01 and containing at least two overlapping members of the defined sets (genes oscillating in both cell lines, genes oscillating only in SW480 cells, genes oscillating only in SW620 cells) were taken into consideration. The results are provided as Supplementary Table 2.
2.21. Metabolic Network Analysis
Gene identifiers were mapped on a global reconstruction of human metabolism (RECON 2.2 [31],. The metabolic network reconstruction RECON 2.2 encompasses 5324 metabolites, 7785 reactions and 1673 associated genes and represents one of the most complete and best annotated consensus of a human metabolic network reconstruction available. Of all 25,037 genes considered in the microarray analysis, 1636 genes were mapped on the network reconstruction and thereby linked with detailed reaction annotations for further analysis. The respective files are provided as Supplementary Table 3.
2.22. Statistical Analysis
The experiments were carried out with at least three biological replicates for each condition. All results are represented as mean ± SEM. Statistical analysis of the results was performed using either one-way ANOVA followed by Tukey's multiple comparisons test or by two-tailed unpaired t-test, based on the experimental design. A p-value <.05 was considered as statistical significant (* = p < .05; ** = p < .01; *** = p < .001).
3. Results
3.1. The SW480 Colon Cancer Cell Line and its Metastatic Counterpart SW620 Show Phenotypic Changes in the Expression of Core-Clock Genes and Clock Output Pathways
Results from our previous work showed large phenotypic variations in circadian rhythms for colorectal cancer (CRC) cell lines that are reflected in the dissimilarity of their oscillatory core-clock gene expression patterns [32]. Based on these results, we wished to further explore the consequences of clock alterations in a colon cancer cell model. For the current study, we selected two of the previously analyzed CRC cell lines, namely the colorectal adenocarcinoma cell lines SW480 and SW620, as an experimental model to investigate the interplay of a dysregulated biological clock and the resultant metabolic output in colon cancer progression. These two cell lines have the same genetic background, but different metastatic potential: the SW480 cell line is derived from a primary Duke's stage B colon carcinoma, and the SW620 cell line is derived from a mesenteric lymph node metastasis in the same patient.
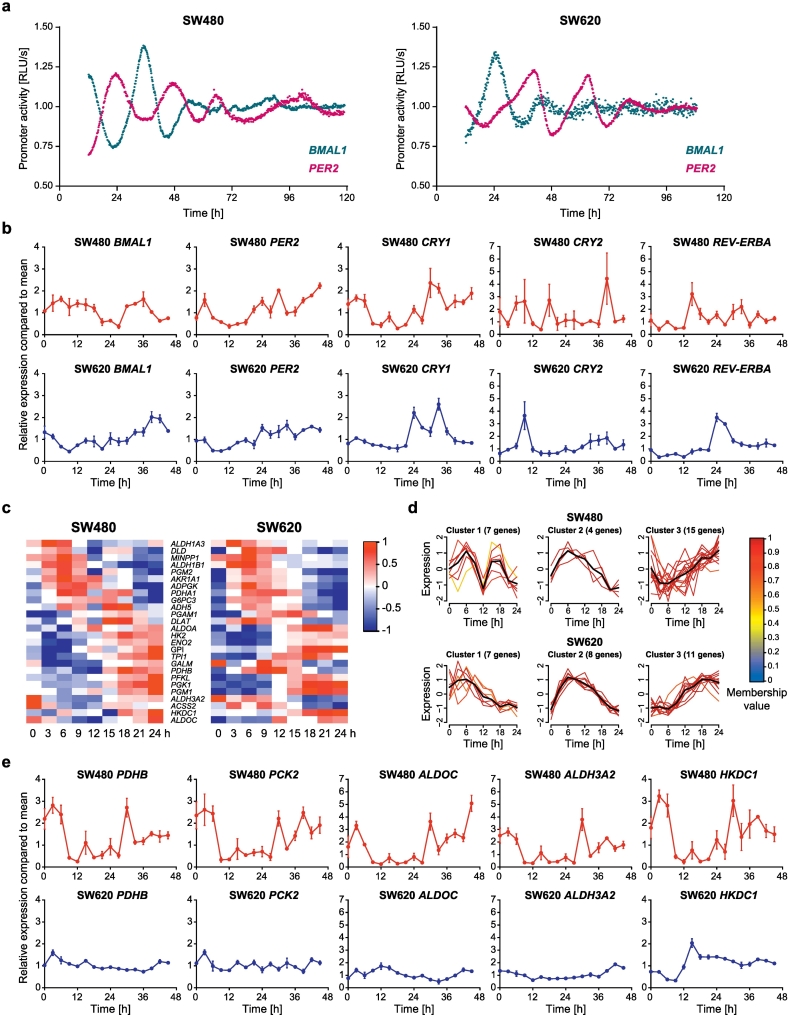
The cell lines exhibit different clock phenotypes: the SW480 cells showed robust oscillations (T = 24.59 h ± 0.34, n = 3) with BMAL1 and period circadian regulator 2 (PER2) oscillating in antiphase, as expected (Fig. 1a). The oscillations in SW620 cells had a delayed phase of around 12 h for both BMAL1 and PER2, as compared to the observations in the SW480 cells (Fig. 1a). To further investigate the observed differences at the transcriptome level, we performed a 24-hour time-course analysis of gene expression using transcriptome microarrays for both CRC cell lines. Out of 14 core-clock genes, 8 were oscillating in SW480 cells with a period of ca. 24 h (p < .05), but only 4 in SW620 cells, and the overall expression patterns were different (Supplementary Fig. 1a).
Fig. 1.
SW480 and SW620 cells serve as a model for tumor progression with changes in the core-clock and metabolic output pathways.
(a) To investigate circadian clock activity, SW480 (left panel) and SW620 (right panel) cells were lentivirally transduced with a BMAL1-promoter (blue) or PER2-promoter (pink) driven luciferase construct. Bioluminescence was measured for five consecutive days. Shown is one representative replicate for each condition. (b) 45 h time-course RT-qPCR measurements of selected core-clock genes (BMAL1, CRY1, CRY2, PER2 and REV-ERBα) in SW480 (dark red) and SW620 (dark blue) cells. Data are expressed as mean ± SEM, n = 3. (c) Phase-ordered, median-normalized heatmap of glycolytic genes oscillating with a 24 h period in at least one of the two tested cell lines. (d) Clustering of metabolic genes based on their time-dependent expression profile. The colors of the gene expression patterns indicate gradual membership values, reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black lines indicate the cluster centres. (e) 45 h time-course RT-qPCR measurements of selected core-clock genes (ALDH3A2, ALDOC, HKDC1, PCK2 and PDHB) in SW480 (dark red) and SW620 (dark blue) cells. Data are expressed as mean ± SEM, n = 3. See also Supplementary Fig 1 and 2 and Supplementary Tables 1 and 2.
We confirmed these observations by 45 h qPCR data of selected core-clock genes and carried out a detailed statistical analysis of the circadian parameters (period, amplitude and phase of the oscillations) by fitting a harmonic regression to the expression data as described in Materials and Methods (Fig. 1b, Supplementary Table 1). Minor discrepancies regarding the circadian parameters between results displayed in Fig. 1a and b occur from different methods used and different sampling intervals. In Fig. 1A, the promoter activity of BMAL1 and PER2 was measured, whereas in Fig. 1B, mRNA levels of BMAL1 and PER2 (as well as other clock genes) were determined. While the first method measures the activity of the promoter of the clock gene, the second quantifies directly the RNA concentration.
Furthermore, we observed a global shift in the set of 24 h oscillating genes between both cell lines. In total, we found similar percentages of oscillating genes when analyzing the oscillations independently in each cell line, namely 16.0% (3998) of all genes were oscillating in SW480 cells and 14.8% (3693) were oscillating in SW620 cells. However, we found that only 5.5% (1385) of all genes oscillate in both cell lines, which points to a switch in the 24 h oscillating genetic landscape for these cells. A subsequent functional annotation analysis for genes with 24 h oscillations in both cell lines revealed several metabolic pathways to be overrepresented, namely glucose metabolism (p = 1.06E-05) and glycolysis (p = 3E-03). Interestingly, the functional analysis of genes oscillating only in SW620 cells showed enriched pathways involved in mitochondrial respiration, including the tricarboxylic acid (TCA) cycle (p = 5.85E-04), mitochondrial electron transport chain (p = 7.66E-05), and the Warburg effect (p = 1.04E-06) (Supplementary Table 2).
To obtain more insights into the metabolic differences between both cell lines, we mapped the expression data to a comprehensive genome-scale reconstruction of human metabolism (RECON 2.2). This metabolic reconstruction specifies all known metabolic reactions occurring in a human cell and provides manually curated relationships between genes, proteins and reactions [31]. This allowed us to annotate 1636 metabolic genes (out of 25,037 genes identified in the microarray analysis) with detailed reaction annotations (Supplementary Table 3). Out of this metabolic gene set, 374 genes (22.7%) were identified as 24 h oscillating in SW480 cells and 367 (22.4%) were identified as 24 h oscillating in SW620 cells. For both cell lines, the percentage of 24 h oscillating genes associated with cellular metabolism was therefore higher than the percentage of 24 h oscillating genes in the whole transcriptome. In addition, the genome-scale reconstruction of human metabolism allowed us to evaluate the functional roles of 24 h oscillating transcripts in greater detail. We distinguished between metabolic genes that are oscillating in SW480 only, oscillating in SW620 only, and oscillating in both cell lines. Interestingly, the pathway annotations oxidative phosphorylation and glycolysis were among the most frequent in all three sets, but with a different ranking as shown in Supplementary Table 3. In particular, the pathways glycolysis (position 5) and oxidative phosphorylation (position 2) ranked the highest in gene sets that oscillate only in the SW620 cells. These results indicate a shift in oscillating genes, whereas general metabolic categories are preserved. Members of distinct metabolic pathways oscillate with different significant weights (ranking) in the metastatic cell line SW620 as compared to the primary tumor cell line SW480.
A following clustering of the gene sets involved in the glycolysis pathway, oxidative phosphorylation or the Warburg effect revealed different mRNA expression patterns of these genes in both cell lines (Fig. 1c-d and Supplementary Fig. 1b-c). We further evaluated the expression data for these gene sets and selected five putatively clock-controlled metabolism-related genes with differential mRNA expression profiles in both cell lines as candidate genes, based on the 24 h microarray data. The expression profiles of the five candidate genes ALDH3A2, ALDOC, HKDC1, PCK2 and PDHB were verified via qPCR (Fig. 1e, Supplementary Table 1). All candidate genes belong to the KEGG pathway glycolysis and gluconeogenesis. In particular, ALDH3A2 is involved in oxidative stress response [33], ALDOC catalyzes the conversion of fructose-1,6-bisphosphate to glyceraldehyde-3-phosphate and dihydroxyacetone phosphate [34], HKDC1 catalyzes the phosphorylation of glucose [35], PCK2 catalyzes the conversion of oxaloacetate to phosphoenolpyruvate [36]. PDHB catalyzes the overall conversion of pyruvate to acetyl-CoA and carbon dioxide, and provides the primary link between glycolysis and the TCA cycle [37]. All five genes had higher fold changes over time in the SW480 cells. In particular, HKDC1 showed a circa 13 h phase-shift in its peak expression between both cell lines. This is consistent with the observed phase shift in the oscillation profile of BMAL1 in SW620 as compared to SW480 cells (Fig. 1a). This might be a hint for a circadian regulation of HKDC1 expression via BMAL1, though further targeted studies will be needed to test this hypothesis in future work. Among the top candidates, HKDC1 oscillated with the best p-value (p = 5.7E-04, SW480; p = 1.6E-05, SW620) and the highest relative amplitude (0.562 in SW480; 0.353 in SW620) for both cell lines, as estimated by the harmonic regression analysis (Supplementary Table 1) and was thus chosen for further investigation.
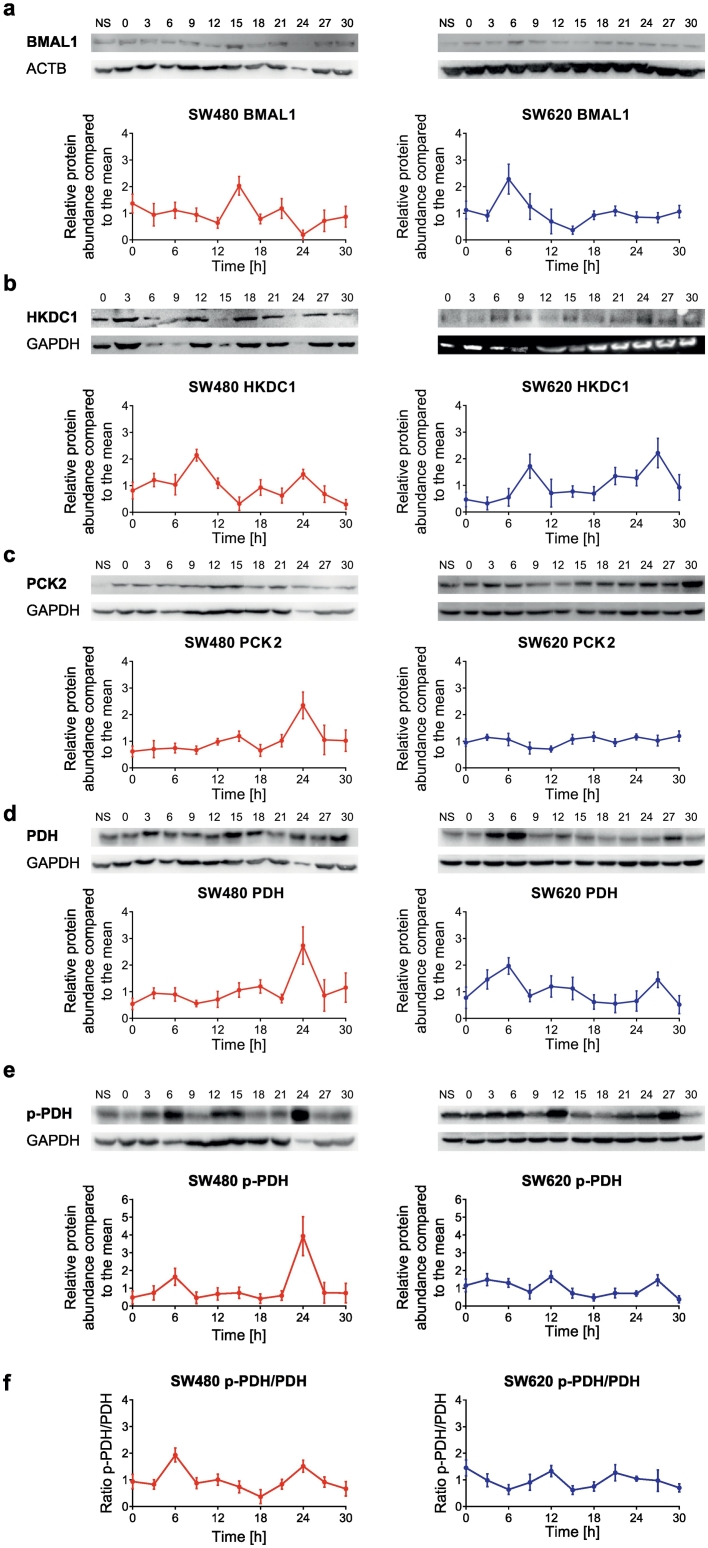
In accordance with the differential mRNA expression of metabolism-related genes, SW480 and SW620 cells displayed distinct out-of-phase profiles of mitochondrial respiration following in vitro synchronization (Supplementary Fig. 1d). As previously reported for other cell lines [12], the oscillatory oxygen consumption rates (OCRs) of both CRC cell lines showed ultradian patterns. To evaluate the possible consequences of rhythmic mRNA expression changes at the protein level, we performed time-course western blot (WB) analysis for our candidate metabolism-related genes. The time-course profiles of BMAL1, HKDC1, pyruvate dehyrogenase (PDH) and phosphoenolpyruvate carboxykinase 2 (PCK2) differed also at the protein level between both cell lines (Fig. 2a-f). In particular, the peaks in protein abundance changed between both cell lines for all tested proteins with phase shifts up to 18 h (Supplementary Table 1). BMAL1 (phase = 16.3 h in SW480) and HKDC1 (phase = 8.7 h in SW480) showed antiphasic peaks in protein abundance (Fig. 2a-b). For PDH and PCK2, the expression changes over time were higher in SW480 compared to SW620 cells, as also observed at the mRNA level.
Fig. 2.
Time-course western blot data of SW480 and SW620 cells.
(a-f) 30 h time-course western blots of BMAL1 (a), HKDC1 (b), PCK2 (c), PDH (d), p-PDH (e) and p-PDH/PDH (f) in SW480 (left panel) and SW620 (right panel) cells. One representative blot for each condition is shown. Data in the graph is presented as mean ± SEM, n = 2.
We further investigated the clock gene-mediated expression of the PDH-subunit E1-β (PDHB). The activity of the PDH complex is known to be regulated by the reversible phosphorylation state of the PDH-subunit E1-α at Ser293 [38]. This post-translational modification is controlled by the relative activities of PDH kinases (PDK) and PDH phosphatases (PDP) [39]. To analyze this mechanism in our cellular model system, we performed WB of the PDH-subunit E1-α with antibodies specific for either one of the two forms. Both the expression of PDH and its phosphorylated form (P-PDH) displayed an oscillatory profile in synchronized SW480 and SW620 cells, albeit with different amplitudes and phases (Fig. 2d-e). Interestingly, the ratio between phosphorylated PDH and unphosphorylated PDH revealed a shift in activity between both cell lines (Fig. 2f). Most notably, the P-PDH/PDH ratios resulted in changes that were in anti-phase with the mitochondrial OCRs which is consistent with the fact that P-PDH has lower pyruvate oxidation activity, as previously reported [40]. In order to validate the general property of synchronized cells to undergo autonomous oscillatory mitochondria-related respiratory activity, we tested these findings in an independent cell line (i.e. human hepatocarcinoma-derived HepG2). Consistently, the changing phosphorylation state of PDH as a function of the synchronization time appears to be a common feature irrespective of the cell type. (Supplementary Fig. 1e).
Another candidate clock-regulated gene revealed by our study was the glycolytic gene HKDC1. HKDC1 is known to catalyze the phosphorylation of glucose in the glycolytic pathway, but its role in cancer remains largely unknown [35]. In our cellular model system of colon cancer progression, HKDC1 showed time-dependent changes in its mRNA expression levels that differed between SW480 and SW620 cells (13 h phase shift, Supplementary Table 1). In order to compare the gene expression of the core-clock gene BMAL1 and HKDC1 in a normal scenario, we used human organoids derived from normal fallopian tubes. Organoids mimic the physiology and anatomy of the donor-tissue very closely [20,41]. Hence, this cellular model system allows for the analysis of the circadian clock in healthy cells in an organ-like context. The comparison of SW480 and SW620 cells with organoids derived from non-cancer tissues was used to evaluate the relative similarity of both cell lines to a normal tissue. The BMAL1-promoter activity of the organoids showed robust circadian oscillations over a time-course of 330 h (T = 24.64 h ± 0.1, n = 3) (Supplementary Fig. 2a) similar to the period measured in SW480 cells (T = 24.59 h ± 0.34, n = 3). We further carried out a 24 h time-course gene expression analysis in the organoids and compared the results with those of the CRC cellular model system. BMAL1 showed circadian oscillations with a phase at 8–12 h (Supplementary Fig. 2b), as also observed in the SW480 cells (Fig. 1b). Thus, the time-dependent expression pattern of BMAL1 seems to be a characteristic of normal cells, as well as of non-metastatic cancer cells.
Moreover, the changes of HKDC1 mRNA expression over time in human fallopian tube organoids followed a very similar pattern to that observed in SW480 cells with an expression minimum at around 12 h, in antiphase to BMAL1. (Supplementary Fig. 2c). Thus, it seems that the putative interplay between BMAL1 and HKDC1 expression in the primary CRC cell line SW480 is different from that of the metastatic cell line SW620, despite their common genetic origin, and instead resembles more closely the phenotype observed in the organoids.
3.2. The Knockdown of the Core-Clock Gene BMAL1 Induces Differential Metabolic Changes in SW480 and SW620 Cells
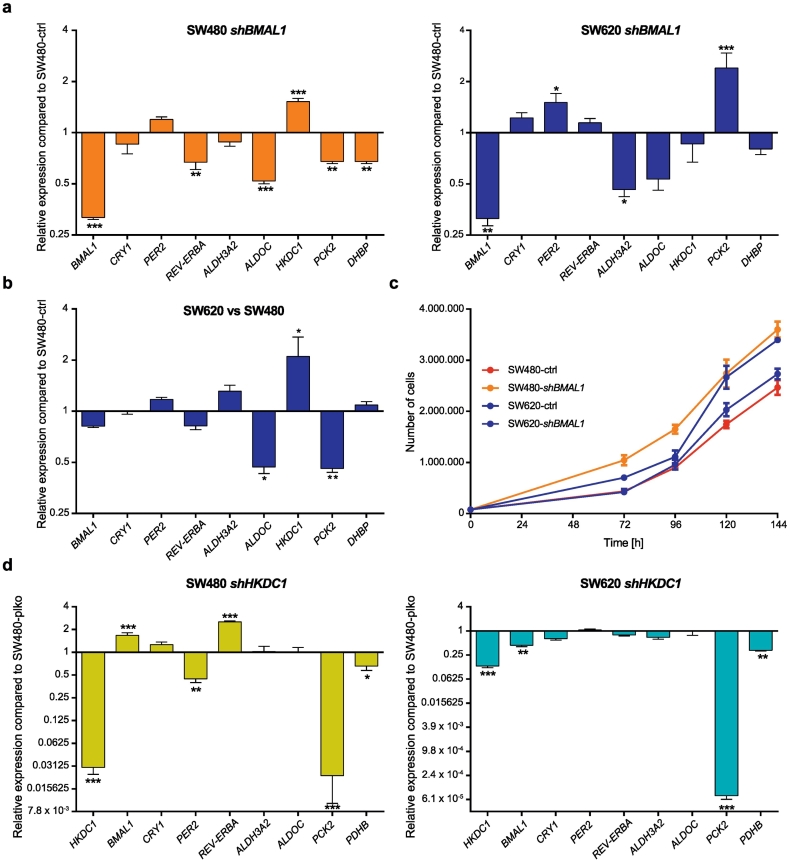
The global analysis of different clock phenotypes revealed metabolic changes between SW480 and SW620 cells. To further investigate the connection between altered circadian regulation and the temporal expression patterns of metabolic genes, we knocked down the core-clock gene BMAL1 in SW480 and SW620 cells and evaluated the resulting output in gene expression and metabolic activity. The knockdown (KD) is carried out a specific time point, but the selected cells contain a stable gene KD (Materials and Methods) so we no longer have a single timepoint effect, but the gene is constantly lower expressed than in the wild type condition. As expected, the KD of BMAL1 in SW480 (KD-efficiency: 68.3%) and in SW620 (KD-efficiency: 68.7%) cells led to an almost complete loss of oscillations in BMAL1-promoter activity (Supplementary Fig. 3a-b, Supplementary Fig. 4). We also tested the effect of BMAL1-KD on the expression of other core-clock genes, as well as on the previously selected clock-regulated metabolic candidate genes via qPCR. HKDC1 expression was significantly upregulated after BMAL1-KD in SW480 cells (p < .001), whereas nuclear receptor subfamily 1 group D member 1 (REV-ERBα) (p < .01), ALDOC (p < .001), PCK2 (p < .01) and PDHB (p < .01) were significantly downregulated (Fig. 3a, left panel). Interestingly, the KD of BMAL1 in SW620 cells led to different effects. While PER2 (p < .05) and PCK2 (p < .001) were significantly upregulated, ALDH3A2 (p < .05) was significantly downregulated (Fig. 3a, right panel). Thus, the KD of the core-clock gene BMAL1 does not only affect core-clock genes, but also impacts on the expression levels of metabolic genes in both cell lines. Remarkably, the expression profile of SW620 control (SW620-ctrl) cells resembles the expression profile of SW480-shBMAL1 cells (Fig. 3a-b).
Fig. 3.
Consequences of BMAL1- and HKDC1-KD on gene expression and proliferation in SW480 and SW620 cells.
(a) Gene expression analysis of selected core-clock (BMAL1, CRY1, REV-ERBα, PER2) and metabolic genes (ALDOC, ALDH3A2, HKDC1, PCK2 and PDHB) in SW480 (left panel) and SW620 (right panel) cells after BMAL1-KD. Gene expression is shown compared to the corresponding control-gene. Mean ± SEM, n = 3. (b) Gene expression analysis of selected core-clock (BMAL1, CRY1, REV-ERBα, PER2) and metabolic genes (ALDOC, ALDH3A2, HKDC1, PCK2 and PDHB) in SW620 cells compared to SW480 cells. Gene expression is shown compared to SW480 cells. Mean ± SEM, n = 3. (c) Proliferation curves of SW480-ctrl, SW480-shBMAL1, SW620-ctrl and SW620-shBMAL1 cells. (d) Gene expression analysis of selected core-clock (BMAL1, CRY1, REV-ERBα, PER2) and metabolic genes (ALDOC, ALDH3A2, HKDC1, PCK2 and PDHB) in SW480 (left panel) and SW620 (right panel) cells after HKDC1-KD. Gene expression is shown compared to the corresponding control-gene. Mean ± SEM, n = 3. Significant changes (p < .05) between different synchronization timepoints of the same condition are marked with *. * = p < .05, ** = p < .01, *** = p < .001. See also Supplementary Figs. 3 and 4.
Furthermore, SW480-shBMAL1 cells proliferate faster than SW480-ctrl cells (p < .05 for all timepoints), and their proliferation profile resembles that of the SW620 metastasis-derived cells (Fig. 3c) (p < .05 for the last two timepoints, but no significant differences for the other timepoints, (Fig. 3c, Supplementary Table 4). These findings corroborate published data regarding the role of the biological clock as a tumor suppressor [[42], [43], [44], [45], [46], [47], [48], [49]] and lead to the assumption that a dysregulated clock promotes a more metastatic phenotype.
We further evaluated the impact of BMAL1-KD on cell viability, cytotoxicity and apoptosis at three different time points after cell synchronization (18 h, 21 h and 24 h). Viability and cytotoxicity exhibited time-dependent profiles that decreased with later synchronization time points in both cell lines, with the highest effect at 18 h and lowest at 24 h (Supplementary Fig. 5a-b). After BMAL1-KD, the time-dependent profile was inverted compared to the normal scenario in both cell lines and viability and cytotoxicity increased with time, with stronger changes in SW480-shBMAL1 cells. The apoptotic profile of the cells did not show a time-dependency, but BMAL1-KD led to significantly lower apoptosis rates in both SW480 (p < .001) and SW620 (p < .001) cells (Supplementary Fig. 5c). Thus, the KD of BMAL1 may confer an advantage in terms of cell progression towards malignant phenotypes, in agreement with our recent findings in a model system of mouse embryonic fibroblasts [50].
In addition, our data shows an upregulation of HKDC1 expression in SW480-shBMAL1 cells (Fig. 3a) which points to a putative interplay between both genes. To attain further insights into this interplay, we knocked down HKDC1 in SW480 and SW620 cells and evaluated the output phenotypes in gene expression and the subsequent effect on metabolic activity. The KD influenced the expression of several core-clock and metabolic candidate genes. In particular, for SW480 cells, HKDC1-KD led to a significant upregulation of BMAL1 (p < .001) and REV-ERBα (p < .001), whereas PER2 (p < .01), PCK2 (p < .001) and PDHB (p < .05) were downregulated (Fig. 3d, left panel). In SW620 cells, BMAL1 (p < .01), PCK2 (p < .001) and PDHB (p < .01) were significantly downregulated upon HKDC1-KD (Fig. 3d, right panel). In the control conditions, HKDC1 expression was higher in SW620 cells compared to SW480 cells, whereas BMAL1 expression was lower (Fig. 3b).
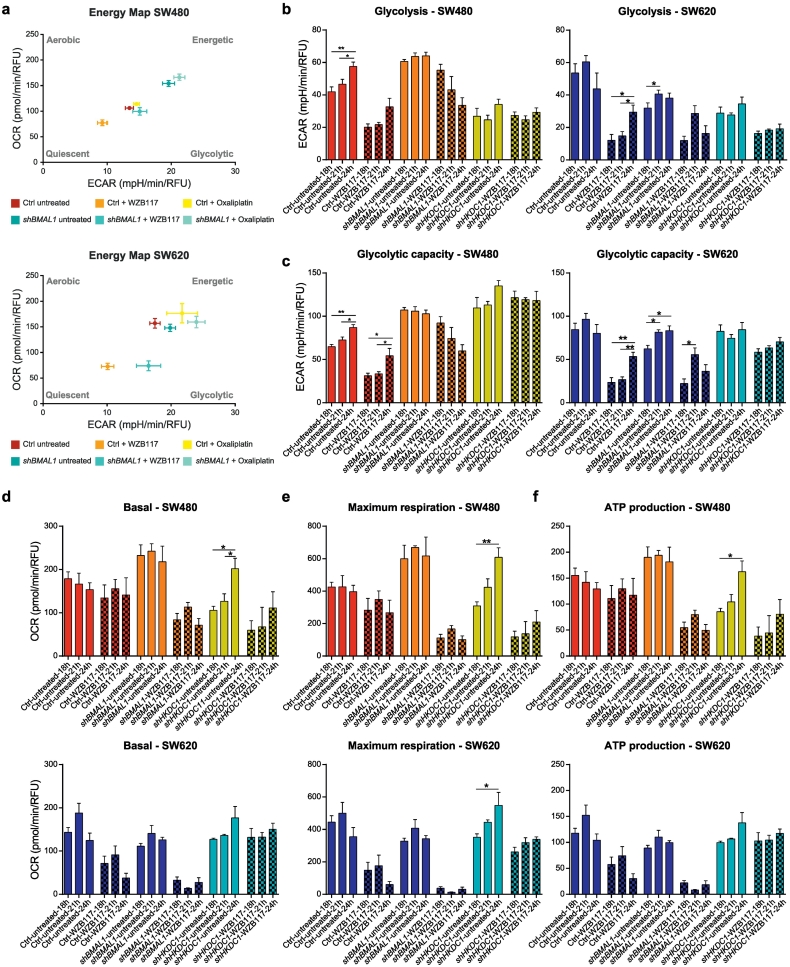
Additionally, we carried out a 24 h time-course qPCR analysis to examine the effect of BMAL1-KD on HKDC1 expression (Supplementary Fig. 3c). In both cell lines, the HKDC1 expression profile changed upon BMAL1-KD (Fig. 1e and Supplementary Fig. 3c). These results further reinforce a connection between the core-clock, clock-controlled metabolism-related genes and metabolic activity. We also investigated the metabolic profiles upon BMAL1-KD in both cell lines: we evaluated the energy phenotype based on the extracellular acidification rate (ECAR) and the OCR of the cells, and determined the metabolic phenotype of SW480 and SW620 cells (Fig. 4a). In SW480 cells, the KD of BMAL1 led to a change towards a more energetic phenotype, while for SW620 cells, no change upon BMAL1-KD was observed. Remarkably, the alterations of the energy phenotype of SW480-shBMAL1 cells led to a phenotype similar to the one observed in SW620 cells, further supporting our hypothesis that clock gene disruption leads to a more metastatic phenotype, and reinforcing the role of the circadian clock as a tumor suppressor.
Fig. 4.
BMAL1- and HKDC1-KD leads to metabolic changes in SW480 and SW620 cells and altered drug response.
(a) Energy map of SW480 (upper panel) and SW620 (lower panel) control and shBMAL1 cells untreated or after treatment with WZB117 or oxaliplatin. The mean of three treatment timepoints after synchronization is shown (18 h, 21 h, 24 h). Mean ± SEM, n = 16. (b) Glycolysis of SW480 (left panel) and SW620 (right panel) control, shBMAL1 and shHKDC1 cells at different timepoints after synchronization. Cells were either untreated or treated with WZB117. Mean ± SEM, n = 5. (c) Glycolytic capacity of SW480 (left panel) and SW620 (right panel) control, shBMAL1 and shHKDC1 cells treated at three different timepoints after synchronization (18 h, 21 h, 24 h). Cells were either untreated or treated with WZB117. Mean ± SEM, n = 5. (d) Basal respiration of SW480 (upper panel) and SW620 (lower panel) control, shBMAL1 and shHKDC1 cells treated at three different timepoints after synchronization (18 h, 21 h, 24 h). Cells were either untreated or treated with WZB117. Mean ± SEM, n = 5. (e) Maximum respiration of SW480 (upper panel) and SW620 (lower panel) control, shBMAL1 and shHKDC1 cells synchronized at different timepoints. Cells were either untreated or treated with WZB117. Mean ± SEM, n = 5. (f) ATP production of SW480 (upper panel) and SW620 (lower panel) control, shBMAL1 and shHKDC1 cells synchronized at different timepoints. Cells were either untreated or treated with WZB117. Mean ± SEM, n = 5. * = p < .05, ** = p < .01, *** = p < .001. For more data, see also Supplementary Fig. 5.
To further evaluate the glycolytic and mitochondrial capability of the cells in the BMAL1-KD, we quantified glycolysis and glycolytic capacity, as well as basal respiration, ATP production and maximum respiration at three different time points after synchronization (see Materials and Methods). Cells were measured 18 h, 21 h and 24 h, after synchronization, respectively. Glycolytic capacity and glycolysis showed significant time-dependent alterations in SW480-ctrl cells, but not in SW480-shBMAL1 cells (Fig. 4b-c, left panel). The loss of time dependent variations in glycolytic activity after BMAL1-KD might hint at a circadian control of the timing of glycolytic activity in CRC cells.
SW620 cells also showed changes in glycolytic activity (glycolysis and glycolytic capacity) between control and shBMAL1-cells, but no significant time-dependent effects could be observed (Fig. 4b-c, right panel). Glycolysis was significantly increased (p < .05) after BMAL1-KD in SW480 cells whereas the same perturbation led to a significant decrease (p < .05) in SW620 cells. In addition to the glycolytic activity, the mitochondrial function also plays a role in tumor growth and metastasis [10]. Published data show that highly metastatic tumors increase mitochondrial Nicotinamide adenine dinucleotide phosphate (NADPH) and undergo further metabolic changes to fight stress caused by reactive oxygen species (ROS) [51]. To assay the mitochondrial activity in the tested cells, we measured basal respiration, ATP production and maximal respiration. Our data shows similar time-dependent effects on the mitochondrial activity, although not as strong as for the glycolytic activity (Fig. 4d-f). In general, a KD of BMAL1 led to a slight increase in metabolic activity in SW480 cells, but to the opposite change in SW620 cells. Moreover, upon BMAL1-KD, HKDC1 expression was upregulated in SW480 cells, but not in SW620 cells. This is likely to result from the overall increased metabolic activity of SW480-shBMAL1 cells. Thus, we propose HKDC1 as a putative mediator of time-dependent effects on energy production metabolic pathways.
Overall, our data shows that a dysregulation of the biological clock leads to alterations in expression of metabolic genes, as well as in metabolic activity that could be pivotal in colon cancer progression.
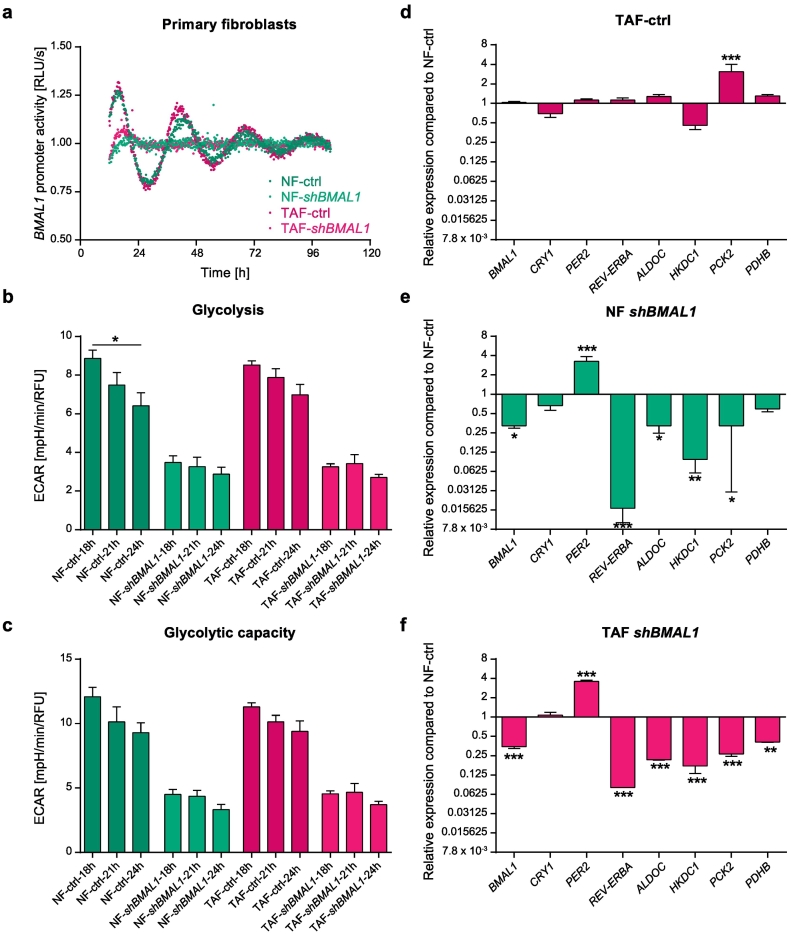
3.3. The Dysregulation of the Circadian Clock Alters Metabolism in Primary Stromal Cells
Cancer cells are under continuous bidirectional communication with tumor stroma that nurtures and supports tumor progression. Stroma modifies proliferation, local migration, metastasis, metabolism and drug resistance responding to the needs of cancer cells [52]. To compare the results gained from the cancer cell lines with patient-derived primary stromal cells, we used normal fibroblasts (NFs) and tumor-associated fibroblasts (TAFs) derived from the same colon cancer patient. Each NF-TAF pair is a singly unique model system reflecting the status of the patient. The NF-TAF analyzed was derived from a patient with lymph node metastasis, with no other distant organ metastasis observed. Therefore, this patient represents best the epithelial cancer cell model system we are using (SW480 vs SW620).
Bioluminescence recordings of NFs and TAFs showed the same BMAL1-expression phase observed in the SW480 cells (Fig. 5a and Fig. 1a). An analysis of clock gene and metabolic gene expression in TAFs compared to NFs revealed a significant upregulation of PCK2 expression (p < .001) and a downregulation of HKDC1 (Fig. 5d). We then perturbed the clock via BMAL1-KD in both NFs and TAFs and measured BMAL1-promoter activity over five consecutive days. As expected, the knockdown led to a complete loss of oscillations for both cell types (Fig. 5a). We further analyzed the effect of BMAL1-KD on different core-clock genes, as well as on our previously selected candidate clock-regulated metabolic genes. In NFs as well as in TAFs, BMAL1-KD led to the upregulation of PER2 (p < .001) and downregulation of REV-ERBα (p < .001). The same tendency was also observed after BMAL1-KD in SW480 cells (Fig. 3a). Furthermore, BMAL1-KD led to the downregulation of ALDOC (NF, p < .05; TAF, p < .001), HKDC1 (NF, p < .05; TAF, p < .001) and PCK2 (NF, p < .01; TAF, p < .001), reinforcing the data for the CRC cells regarding the impact of BMAL1 on the expression of the metabolic genes (Fig. 5e-f). In TAFs, BMAL1-KD additionally led to the downregulation of PDHB (p < .01) (Fig. 5f). An evaluation of the glycolytic activity and glycolytic capacity revealed additional metabolic differences between NFs and TAFs. While a significant time-dependent effect of glycolysis could be observed in NFs, where glycolytic activity decreased at later time points, this effect could not be observed in TAFs. The knockdown of BMAL1 led to a decrease of glycolysis and glycolytic capacity in NFs, as well as in TAFs (Fig. 5b, c). This data corroborates our hypothesis that the circadian clock influences the timing of metabolism and is able to reprogram metabolic activity in general.
Fig. 5.
The circadian clock in primary fibroblasts.
(a) Normal fibroblasts (NFs) and tumor associated fibroblasts (TAFs) were lentivirally transduced either with an empty-vector (ctrl) or with a shBMAL1-construct. Additionally, cells were lentivirally transduced with a BMAL1-promoter driven luciferase construct. Bioluminescence was measured for five consecutive days. Shown is one representative replicate for each condition. Glycolysis (b) and glycolytic capacity (c) of control and shBMAL1 NFs and TAFs at three different timepoints after synchronization (18 h, 21 h, 24 h). Mean ± SEM, n = 5. Significant changes (p < .05) between different synchronization timepoints of the same condition are marked with *. Gene expression analysis of selected core-clock (BMAL1, CRY1, REV-ERBα, and PER2) and metabolic genes (ALDOC, HKDC1, PCK2 and PDHB) in control (d) and shBMAL1 NFs (e) and TAFs (f). Gene expression is shown compared to the corresponding control-gene. Mean ± SEM, n = 3. * = p < .05, ** = p < .01, *** = p < .001.
3.4. A Role for HKDC1 as a Mediator of Metabolic Changes in SW480 and SW620 Cells upon Circadian Disruption
To further evaluate the role of HKDC1 in the regulation of clock-mediated metabolic changes, we analyzed the effect of its downregulation both at the clock and metabolism level in our cellular model system (Supplementary Fig. 4). As expected, given its upstream role in the glycolytic pathway, the KD of HKDC1 led to a decrease of glycolysis in both cell lines (p < .05) (Fig. 4b) whereas only in SW480 cells, the glycolytic capacity increased after HKDC1-KD (Fig. 4c, left panel). In particular, in SW480-shHKDC1 cells, basal respiration, ATP production and maximum respiration showed time-dependent effects and increased in earlier time points after synchronization (Fig. 4e, upper panel), whereas no significant time-dependent changes were observed in SW480-ctrl and SW480-shBMAL1 cells. In SW620-shHKDC1 cells, only maximum respiration showed synchronization time-dependent effects, whereas ATP production and basal respiration remained unaffected in SW620 cells (Fig. 4d-f).
Together, these results point to a negative interplay between HKDC1 and the circadian clock, in particular for SW480 cells, since the KD of BMAL1 leads to a higher HKDC1 expression and consequently to a higher metabolic activity in these cells, and a KD of HKDC1 led to higher BMAL1 expression and slightly lower glycolytic activity.
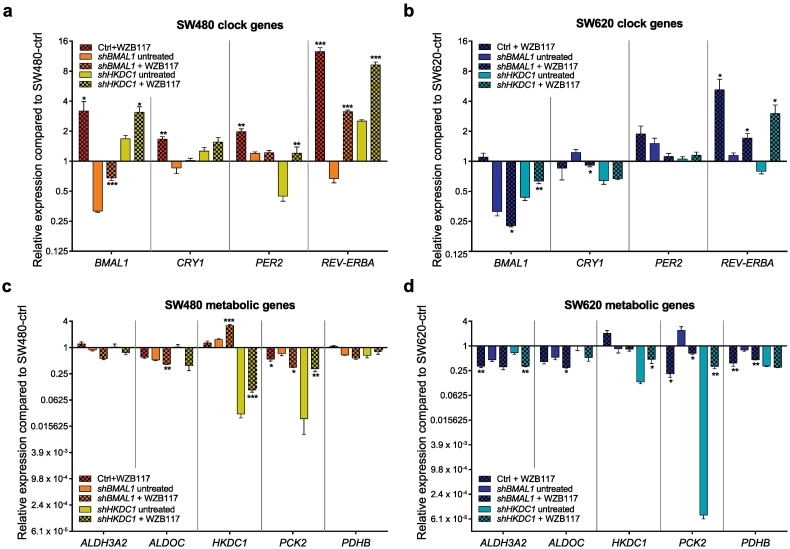
3.5. BMAL1 and HKDC1 Knockdown Affect Treatment Response
Given the metabolic changes induced in SW480 and SW620 cells by BMAL1-KD, we hypothesized that a subsequent effect on treatment response is likely to occur as well. Consequently, we treated ctrl, shBMAL1 and shHKDC1 cells with WZB117, a glucose transporter (GLUT1) inhibitor, and evaluated its effect on the gene expression level, as well as on the metabolic activity (Fig. 6a-d and Fig. 4b-f). In SW480-ctrl cells, WZB117 treatment led to a significant upregulation of all tested core-clock genes (BMAL1 (p < .05), cryptochrome circadian regulator 1 (CRY1) (p < .01), PER2 (p < .01), REV-ERBa (p < .001)) (Fig. 6a). This effect fits with the observed increase of glycolysis upon KD of BMAL1 in the absence of treatment in SW480 cells. Furthermore, these results are in agreement with the reported inhibitory effect of intracellular glucose on the expression of the core-clock genes BMAL1, PER1 and PER2 [53]. Upon KD of BMAL1 in SW480 cells, we measured an upregulation of BMAL1 (p < .001) and REV-ERBα (p < .001) after treatment. This upregulation of core-clock genes was also observed for SW480-shHKDC1 cells after WZB117 treatment where PER2 was additionally upregulated (p < .01). In contrast to these observations, of all tested core-clock genes only REV-ERBα (p < .05) expression was significantly upregulated after treatment in SW620 cells in all conditions. As expected, BMAL1 (p < .05) and CRY1 (p < .05) were downregulated in SW620-shBMAL1 cells, whereas BMAL1 (p < .01) was upregulated in SW620-shHKDC1 cells. Overall, the effect of WZB117 treatment on core-clock gene expression is weaker in SW620 cells as compared to SW480 cells. Additionally, we analyzed the outcome of WZB117 treatment on gene expression of metabolic genes, which showed diverse effects for the different control and KD cell scenarios. Upon WZB117 treatment, PCK2 expression was significantly downregulated in SW480 and SW480-shBMAL1 cells (p < .05), but upregulated in SW480-shHKDC1 cells (p < .01). The same effect could be observed in SW620 cells. While HKDC1 expression was not affected in SW480- and SW620-ctrl cells, it was upregulated in both shHKDC1 cells, as well as in the SW480-shBMAL1 cells (p < .001). Interestingly, we observed no significant effect on PDHB expression in any of the SW480 conditions, but measured a downregulation after treatment in SW620 ctrl and -shBMAL1 cells (p < .01). (Fig. 6c-d).
Fig. 6.
Consequences of BMAL1- and HKDC1-KD on gene expression after WZB117 treatment.
(a) and (b) Gene expression analysis of selected core-clock genes (BMAL1, CRY1, REV-ERBα and PER2) in SW480 (A) and SW620 (B) control, shBMAL1 and shHKDC1 cells after treatment with WZB117. Data are shown compared to untreated control cells. Mean ± SEM, n = 3. (c) and (d) Gene expression analysis of selected metabolic genes (ALDOC, ALDH3A2, HKDC1, PCK2 and PDHB) in SW480 (C) and SW620 (D) control, shBMAL1 and shHKDC1 cells after treatment with WZB117. Data are shown compared to untreated control cells. Mean ± SEM, n = 3. * = p < .05, ** = p < .01, *** = p < .001 compared to the corresponding untreated sample of the same condition (control, shBMAL1, shHKDC1).
As described above, glycolytic activity showed a different synchronization time-dependent pattern after BMAL1 KD, which was particularly evident for SW480 cells. To investigate the dependence of the time of treatment and the subsequent effects on metabolism in greater depth, we treated cells at three different time points after synchronization. As before, cells were synchronized 18 h, 21 h and 24 h before treatment, respectively. The treatment with WZB117 led to the inhibition of glycolysis in SW480-ctrl, as well as in shBMAL1 cells, but with differential time-dependent effects (Fig. 4b-c). In the SW480-ctrl cells, treatment at all time points led to a significant inhibition of glycolysis, as well as of glycolytic capacity. In contrast to that, upon KD of BMAL1, glycolysis was only inhibited when cells were synchronized 24 h and 21 h before treatment, and glycolytic capacity was only significantly lower when cells were synchronized 24 h before treatment. WZB117 treatment did neither lead to lower glycolysis, nor to lower glycolytic capacity in SW480-shHKDC1 cells, independent of the time of treatment (Fig. 4b-c, left panel). In SW620 cells, the glycolysis rate was lower when treated at time point 21 h or 18 h after synchronization, whereas glycolytic capacity was downregulated for all treatment time points. Comparable results were obtained after BMAL1-KD. In contrast to the observation in SW480 cells, WZB117 treatment led to a slight inhibition of glycolysis and glycolytic activity in SW620-shHKDC1 cells (Fig. 4b-c, right panel). These results lead to the hypothesis that not only the glycolytic activity itself, but also the effect of glycolysis-targeting treatment is time-dependent. Other metabolic indicators, including the basal respiration, maximum respiration and adenosine triphosphate (ATP) production, were not affected upon WZB117 treatment in SW480, but showed an effect for SW480-shBMAL1 cells, independent of the time of treatment. For shHKDC1 cells, while basal respiration and ATP production were not affected, maximum respiration was inhibited independent of the time of treatment (Fig. 4d-f, upper panel). This might result from a weaker input into the glycolytic pathway that subsequently impacts the mitochondrial respiration pathway, leading to an overall inhibition of maximum respiration.
WZB117 treatment in SW620 cells led to differing results when compared to the untreated scenario. Basal respiration, maximum respiration and ATP production were inhibited in SW620-ctrl cells, independent of the treatment time. The results for shBMAL1 and shHKDC1 cells, however, were the same in both cell lines. While basal respiration, ATP production and maximum respiration were affected in shBMAL1 cells, only maximum respiration was inhibited in shHKDC1 cells (Fig. 4d-f, lower panel).
Regarding the energy phenotype, WZB117 treatment led to a shift from an energetic towards a more quiescent phenotype in ctrl as well as in shBMAL1 cells for both cell lines (Fig. 4a). Untreated SW480-shBMAL1 cells displayed a more energetic phenotype than SW480-ctrl cells which was also true after the shift to a more quiescent phenotype upon WZB117 treatment. We further treated the cells with the classical chemotherapeutic oxaliplatin, a platinum complex that is used effectively for CRC treatment [14], to analyze whether resulting effects on cell metabolism are comparable with the ones induced by WZB117. However, the oxaliplatin treatment did neither induce changes regarding the overall energy phenotype in SW480 or in SW620 cells, nor did it lead to glycolysis inhibition (Supplementary Fig. 6a-b). We further observed that viability and cytotoxicity were not affected by WZB117 treatment in SW480-ctrl and -shBMAL1 cells, as they still showed the same synchronization time-dependent pattern as without treatment. Interestingly, after WZB117 treatment, we measured a lower apoptosis rate in SW480-ctrl as well as in -shBMAL1 cells (Supplementary Fig. 5a-c). This data reinforces the role of the circadian clock as a regulator of metabolism which results in time-dependent treatment effects and identifies HKDC1 as a potential mediator involved in this interplay.
Although the selection of a temporal window of six hours is too short to access chrono-pharmacology profiles, we could show that the relative efficacy of the drug on the metabolic fluxes appears to change depending on the post-synchronization time of drug administration. This result provides insights to schedule crono-therapeutic protocols targeting cancer cell metabolism and prompts to further deepen these studies in future work. Our results point to a role of the circadian clock in tumorigenesis, indicating in particular that the disruption of the biological clock leads to a more metastatic phenotype.
4. Discussion
Cancer cell lines show a wide variety of circadian phenotypes, varying from robust oscillations in clock gene expression to their complete disruption [32]. Such alterations in the biological clock influence the cell's metabolism and proliferation which ultimately impact the cancer phenotype [5]. In this study, we conducted a comprehensive analysis of the consequences of clock disruption on cancer metabolism and the resulting influence on the metabolic phenotype and drug response.
We used an experimental cell model of colorectal cancer progression based on the cell lines SW480 (primary tumor) and SW620 (metastatic site from the same patient) to evaluate, in a time-dependent manner, the metabolic consequences of clock disruption at the transcriptome level. The in vitro model we have chosen has been widely used in the scientific literature to investigate primary and metastatic behavior [[54], [55], [56]]. However, other possible explanations for their different behavior must be considered e.g., these two cell lines could represent two clones from the same cancer with different features, not necessarily implying metastatic versus not-metastatic behavior.
In our CRC model, we observed expression level changes for both core-clock genes and downstream clock-regulated genes. Intriguingly, despite their common genetic background and although the total number of 24 h oscillating genes only slightly differed between both cell lines, the resulting intersect of oscillating genes between both cell lines was rather small. This led us to assume a global shift in the expression of circadian-controlled genes in the progression from primary tumor to metastatic cells. Furthermore, as a consequence of changes in the core-clock between both cell lines, we observed differential oscillatory patterns within different metabolic pathways. Thus, changes in the core-clock seem to cause a reprogramming of the circadian system in the metastatic cell lines and appear to contribute to a shift in the metabolic profile which consequently induces major metabolic modifications during carcinogenesis and metastasis.
One of the consequences of this reprogramming is a change of the predominant oscillating metabolic pathways. Indeed, a genome-scale reconstruction of human metabolism and the subsequent functional annotation of the 24 h oscillating gene sets allowed us to identify differential over-represented metabolic pathways between the cell lines. While glycolysis was over-represented in the set of genes oscillating in both cell lines, metabolic pathways such as the TCA cycle and the Warburg effect were specifically over-represented in the set of genes oscillating in SW620 cells. A thorough investigation of glycolytic genes revealed differences in their temporal expression patterns between both cell lines, leading to the hypothesis that the circadian clock differentially regulates members of these pathways in a tumor progression-dependent manner. An in-depth analysis of this set of metabolic genes identified five clock-regulated metabolic candidate genes which exhibited differential expression patterns in our cellular model system. A further time course analysis of the candidate metabolic genes and of the core-clock genes in organoids revealed a genetic profile which rather resembles the profile observed in the primary tumor cell line, SW480. This cell line also seems to have a more normal clock than its counterpart metastatic cell, SW620. One of our top candidate genes is HKDC1, which encodes a recently identified member of the hexokinase family (HKDC1) that plays a role in glucose metabolism [57]. A recently study points to HKDC1 as a potential therapeutic target for lung cancer [58] and a high expression of HKDC1 is associated with poor prognosis and aggressive phenotype in hepatocarcinoma [35]. However, the functions and mechanisms via which HKDC1 influences cell proliferation and cancer progression are still unclear, particularly in the setting of CRC. In our study, metastatic SW620 cells showed higher HKDC1 expression compared to SW480 cells, in agreement with the findings described above. Furthermore, a KD of the core-clock gene BMAL1 led to higher HKDC1 expression and a subsequent increase of glycolysis and cell proliferation, in SW480 cells. Cancer cells are known to have a reprogrammed metabolism [9,51,59], which allows them to fulfil the enhanced energy demands commonly leading to a switch towards hypoxic glycolysis over normoxic glycolysis. Comparably, in our cellular model system, the KD of BMAL1 in SW480 cells led to a significant increase in glycolytic activity (p < .05), supporting our hypothesis that perturbations of the core-clock system lead to a more metastatic phenotype.
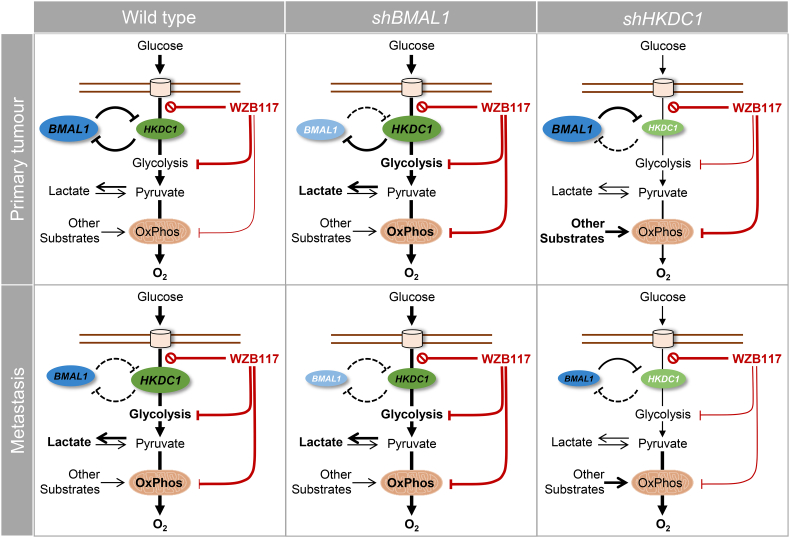
Following the identification of differential expression of metabolic pathways in both cell lines, we further analyzed the impact of clock dysregulation on metabolic activity and drug response. Previous studies point to a role of BMAL1 in mediating tumorigenesis, through influencing the cell cycle [50,60], and to a sensitivity to cancer therapeutics in different cancer types [42,45,61]. Thus, we evaluated the impact of BMAL1-KD and HKDC1-KD on gene expression and drug response in our model system. The KD of BMAL1 led to changes in the expression levels of core-clock genes as well as of metabolic genes, in a cell type-specific manner. Interestingly, while most metabolic genes were downregulated after BMAL1-KD in SW480 cells, HKDC1 expression was upregulated – an effect that was not observed in SW620 cells, likely due to the initial high HKDC1 expression in these cells. On the other hand, HKDC1-KD induced BMAL1 expression in SW480 cells, pointing to a reciprocal control between both genes. In both CRC cell lines, HKDC1-KD led to a strong decrease of PCK2 expression. PCK2 is a mitochondrial enzyme that catalyzes the conversion of oxaloacetate to phosphoenolpyruvate (PEP). The functional role of the mitochondrial isoform PCK2 is likely linked to a cataplerotic role providing a precursor of intermediates for biosynthesis of phospholipids, amino acids and purines thereby fulfilling the increased demand for biomass in rapidly proliferating cells [62]. This is particularly relevant in cancer cells under restricted glucose where glutamine-derived oxaloacetate is converted into PEP by PCK2. However, additional roles of PCK2 have to be considered as well. In particular, PEP can act as an effector metabolite stimulating the activity of the PDH phosphatase. This is consistent with our observation that, at the protein level, a lower expression of PCK2 matches a higher P-PDH/PDH ratio (particularly in SW620 cells). Consequently, the following sequence of events could be tentatively envisioned: PCK2 produces PEP resulting in the activation of the PDH phosphatase, and the subsequent decrease of the ratio between the phosphorylated and the non-phosphorylated forms of PDH. This in turn can lead to the increase in pyruvate oxidation and result in the observed increase of the OCR (Fig. 7). The observed strong effect of shHKDC1 on the expression of clock genes in SW480 led to pronounced variations in the time-dependent metabolic activity. These results show that the interplay between the core-clock and HKDC1 is stronger in SW480 cells than in the metastatic SW620 cells. Consequently, our data hints at a more functional clock in primary cancer cells in respect to their metastatic counterpart, which also correlates with the difference in the clock phenotypes of both cell lines.
Fig. 7.
Schematic representation of the interplay between the biological clock and metabolism in tumorigenesis.
In the primary tumor cell line (SW480), clock gene disruption (shBMAL1) leads to the upregulation of HKDC1 and subsequent activation of metabolic activity (glycolysis and mitochondrial respiration). The downregulation of HKDC1 (shHKDC1) leads to the upregulation of BMAL1. Upon treatment with WZB117, metabolic activity is inhibited in shBMAL1 cells, and glycolytic activity is inhibited to a lesser extent than in SW480-ctrl cells, partially explained by an increase of HKDC1 expression in shBMAL1 cells. In the metastatic cells (SW620) the effect of clock gene disruption (shBmal1) on HKDC1 expression is rather low and consequently Bmal1-KD leads to a slight inhibition of metabolic activity (glycolysis and mitochondrial respiration). After treatment with WZB117, the glycolytic activity is inhibited in shBmal1-cells, and mitochondrial respiration is inhibited to a much larger extent. The effect of WZB117 treatment is weaker in the shHKDC1 cells. The size of the ellipses represents the relative RNA quantity, the width of the connecting lines represents the impact of the interaction.
The differential levels of core-clock gene expression in both cell lines seem to be determinant for the resulting metabolic phenotype upon BMAL1-KD. While the metabolic activity of both cells without BMAL1-KD was similar, glycolysis and mitochondrial respiration were slightly increased in SW480-shBMAL1 cells, whereas the KD of BMAL1 led to a decrease in metabolic activity in SW620 cells. Strikingly, the time-dependent glycolytic activity observed in SW480-ctrl cells is disrupted upon BMAL1-KD. This indicates a circadian regulation of temporal patterns in metabolic activity that are lost upon clock impairment via KD of a core-clock gene. Upon HKDC1-KD, glycolysis decreased in both cell lines, while mitochondrial respiration was not affected which points to a temporal regulation of glycolysis via HKDC1.
We further confirmed the findings from our CRC model system, regarding the circadian regulation of metabolic genes, in primary fibroblasts isolated from normal colon and colon adenocarcinoma of the same patient. In the normal primary fibroblasts, the KD of BMAL1 led to a downregulation of HKDC1 and a subsequent inhibition of glycolytic activity. These results suggest that the disruption of the circadian clock via BMAL1-KD leads to changes in the expression levels of HKDC1 with subsequent effects on glycolysis. In normal cells, the inhibition of HKDC1 and glycolysis is favored, while in primary tumor cells an upregulation of HKDC1 and glycolysis is preferred (Fig. 7). Our study points to a tumor progression-inhibiting role of BMAL1. The suggested tumor-suppressive role of BMAL1 is in line with published data including recent work from our group [50]. Furthermore, after BMAL1-KD, SW480 cells proliferate faster and their proliferation profile resembles that of the metastasis-derived SW620 cells. Thus, we hypothesize that a dysregulated biological clock leads to a more metastatic phenotype. Our hypothesis is further supported by the observation that SW480-shBMAL1 cells develop an active metabolic phenotype that is similar to that of SW620-ctrl cells in order to fulfil the energetic demands of proliferating cells.
In recent years, there have been increasing efforts in cancer therapy to take circadian timing into consideration, aiming at an improved tolerability of anti-cancer drugs and treatment efficacy. Chronotherapy protocols were used in chronomodulated chemotherapy [15] and time-dependent radiotherapy scheduling [[16], [17], [18], [19]]. In line with the idea behind these therapeutic strategies, we treated our CRC model system with a glycolysis inhibitor at three different time points after synchronization to analyze putative time-dependent treatment effects. WZB117, an inhibitor of glucose transport, showed a strong impact on clock gene expression, in line with the described inhibitory effect of intracellular glucose on clock gene expression [53]. In SW480 cells, glucose transport inhibition upregulated the expression of BMAL1 and, upon KD of BMAL1, also of HKDC1. In SW620 cells, the effect of the treatment was more complex: BMAL1 expression was upregulated after shHKDC1 while HKDC1 was upregulated only after WZB117 treatment in HKDC1-KD cells. Among the observed metabolic effects, glucose transport inhibition led to a slight decrease of glycolysis and glycolytic activity in SW620-shHKDC1 cells, while the inhibition of mitochondrial respiration was very strong in SW620 cells after WZB117 treatment, in particular for shBMAL1 cells. This could be due to altered metabolic pathways that might be replaceable in primary tumors, but that are essential for metastasis and could thus lead to the activation of mechanisms that ultimately aid cells to evade treatment.
Metabolic alterations during tumorigenesis are a relevant factor to be considered for drug resistance [10]. While expression changes in some metabolic pathways including glycolysis are frequently observed during tumorigenesis, alterations in other pathways such as oxidative phosphorylation are very heterogeneous [63].
Glycolysis is an attractive anti-cancer target with promising therapeutic potential, since many tumors increase glucose uptake and show elevated glycolysis rates. A possible glycolysis-related target might be the hexokinase HKII, which is upregulated in different tumor types and whose inhibition slows tumor progression [10]. A comparison of normal and tumor cells revealed HK as upregulated, ALDOC as upregulated and PDH as downregulated in most tumors [63]. In our study, the same tendency regarding HKDC1 and PDHB was observed, leading to the assumption that these changes are important for both tumor onset and progression. However, this tendency could not be observed after BMAL1-KD in the metastatic SW620 cells. Although some isoforms of glycolytic enzymes are only present in cancer cells and not in normal cells, such as the hexokinase isoform HK2, the close similarity of different isoforms makes them a difficult target, thus complicating the use of glycolysis as a potential therapeutic target [8]. An alternative candidate for targeting glycolysis might be HKDC1 that we identified here as an important regulator via which the circadian clock influences metabolism in colorectal cancer cells.
Importantly, BMAL1 may impact on metabolic function in different ways: either by influencing the clock machinery, and altering circadian rhythms, and/or by influencing the expression of genes that are direct targets of CLOCK/BMAL1. This could be a result of other potential, non-circadian functions of BMAL1. As such, to differentiate between these two hypotheses and to better characterize the specific output results of BMAL1 KD phenotypes, further studies are necessary.
Altogether, our study shows that the biological clock regulates metabolism in cancer cells with profound implications in the metabolic reprogramming observed in association with tumor progression. Additionally, our data suggests a role for the biological clock in fine-tuning both drug efficiency and timing in cancer treatment, highlighting its crucial function as an elicitor of treatment response.
The following are the supplementary data related to this article.
Changes in the core-clock of SW480 and SW620 cells lead to changes in metabolic pathways. (a) Phase-ordered, median-normalized heatmap of core-clock genes in SW480 and SW620 cells. Phase-ordering was performed based on SW480 expression data and the same ordering of genes was used for SW620 cells. (b) Phase-ordered, median-normalized heatmap of genes included in the oxidative phosphorylation pathway (left panel) and the Warburg effect (right panel) oscillating in at least one of the two tested cell lines. Phase-ordering was performed based on SW480 expression data and the same ordering of genes was used for SW620 cells. (c) Clustering of metabolic genes based on their time-dependent expression profile. All members of the pathways oxidative phosphorylation (left panel) and Warburg effect (right panel) that were oscillating in at least one cell line were grouped into three different clusters. The colors of the gene expression patterns indicate gradual membership values, reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black lines indicate the cluster centers. (d) Resting endogenous respiration in SW480 and SW620 cells. OCR was determined over a period of 30 h. Mean ± SEM, n = 3. (e) PDH and pPDH protein abundance, pPDH/PDH ratio and oxygen consumption rate was determined in HepG2 cells for 24 h. Mean ± SEM, n = 3 (protein abundance). Mean ± SEM, n = 10 (OCR).
The circadian clock in human fallopian tube organoids. (a) Bioluminescence measurements of human fallopian tube organoids transduced with a BMAL1-promoter driven luciferase construct. Bioluminescence was measured for 14 consecutive days. Shown is one representative replicate. 24 h time-course RT-qPCR measurements of BMAL1 (b) and HKDC1 (c) in human fallopian tube organoids. Data are expressed as mean ± SEM, n = 3.
BMAL1 promoter activity in SW480 and SW620 cells and time-dependent gene expression after BMAL1-KD. (a) SW480 and (b) SW620 cells were lentivirally transduced either with an empty-vector (control) or with an shBMAL1-construct. Additionally, cells were lentivirally transduced with a Bmal1-promoter driven luciferase construct. Bioluminescence was measured for five consecutive days. Shown is one representative replicate for each condition. (c) 24 h RT-qPCR measurements of BMAL1 and HKDC1 in SW480-shBMAL1 (upper panel) and SW620-shBMAL1 (lower panel) cells. Data are shown compared to the mean expression. Mean ± SEM, n = 3.
KD efficiency after shRNA-mediated kD of BMAL1 and HKDC1. Gene expression analysis of BMAL1 in SW480 (a) and SW620 (b) cells and HKDC1 in SW480 (c) and SW620 (d) cells after shRNA-mediated BMAL1-KD and HKDC1-KD. Gene expression is shown compared to the corresponding control gene. Mean ± SEM, n = 3. KD efficiency in SW480-shBMAL1 cells: 0.317 ± 0.009 (68.3%). KD efficiency in SW620-shBMAL1 cells: 0.313 ± 0.03 (68.7%). KD efficiency in SW480-shHKDC1: 0.03 ± 0.006 (97%). KD efficiency in SW620-shHKDC1: 0.13 ± 0.013 (87%).
Viability, cytotoxicity and apoptosis in SW480 and SW620 cells. (a) Viability, (b) cytotoxicity and (c) apoptosis in SW480 (upper panel) and SW620 (lower panel) control and shBMAL1 cells synchronized at different timepoints. Cells were either untreated or treated with WZB117 or oxaliplatin. Mean ± SEM, n = 3. Significant changes (p < .05) between different synchronization timepoints of the same condition are marked with *. * = p < .05, ** = p < .01, *** = p < .001.
Metabolic activity of SW480 and SW620 control and shBMAL1 cells after oxaliplatin treatment. (a) Glycolysis of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at three different timepoints after synchronization (18 h, 21 h, 24 h). Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (b) Glycolytic capacity of SW480 (left panel) and SW620 (right panel) control and shBBMAL1 cells at different timepoints. Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (c) Basal respiration of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at different timepoints. Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (d) Maximum respiration of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at three different timepoints (18 h, 21 h, 24 h). Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (e) ATP production of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at three different timepoints (18 h, 21 h, 24 h). Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. Significant changes (p < .05) between different timepoints of the same condition are marked with *. * = p < .05, ** = p < .01, *** = p < .001.
Determination of treatment concentration for WZB117 treatment. To determine the appropriate treatment concentration with WZB117, SW480-ctrl, SW480-shBMAL1, SW620-ctrl and SW620-shBMAL1 cells were treated with different concentrations of WZB117 and the cytotoxicity was determined. Based on the calculated IC50 value, the concentration of treatment was chosen.
Statistical analysis of circadian parameters of genes and proteins. Circadian parameters (p-value for oscillatory behavior, acrophase, amplitude, mesor) for clock and clock-regulated genes and proteins in SW480 and SW620 cells determined using RAIN and harmonic regression analysis.
Over-representation analysis. An over-representation analysis was performed with three sets of genes: genes oscillating in SW480 and SW620 cells, genes oscillating in SW480 cells only and genes oscillating in SW620 cells only.
Metabolic network analysis. Expression data gained from a 24 h microarray analysis of SW480 and SW620 cells was mapped to a reconstruction of human metabolism.
Proliferation curve analysis. Statistical analysis was done for every timepoint of the proliferation curve to test for significant differences between the different conditions.
Acknowledgments
Acknowledgments
We thank Dr. Mirjana Kessler and Susan Jackisch from the group of Prof. Thomas F. Meyer for support with human fallopian tube organoids. We are grateful to Ute Ungethüm from the LFGC (Labor für funktionelle Genomforschung, Charité - Universitätsmedizin Berlin) for technical support regarding the microarrays work.
Financial statement
The work in AR's group was funded by the German Federal Ministry of Education and Research (BMBF)—eBio-CIRSPLICE - FKZ031A316, and by the Dr. Rolf M. Schwiete Stiftung. YL was additionally funded by Jinan Huaiyin Hospital of Shandong Province. LF and YL were additionally funded by the Berlin School of Integrative Oncology (BSIO) of the Charité – Universitätsmedizin Berlin. The work in GM's lab was supported by the “5 × 1000” voluntary contribution, and by a grant (GM) from the Italian Ministry of Health through Department of Medical Sciences, Division of Internal Medicine and Chronobiology Unit (RC1504ME53, RC1603ME43 and RC1703ME43), IRCCS Scientific Institute and Regional General Hospital “Casa Sollievo della Sofferenza”, Opera di Padre Pio da Pietrelcina, San Giovanni Rotondo (FG), Italy.
Conflict of Interest
The authors declare that they have no conflict of interest.
Author Contributions
Conceived and designed the work: AR. Performed the wet lab experiments: LF, YL, RS, OC, AC, KH. Performed the bioinformatics analysis: RE, LF, HK. Analyzed the data: LF, RE, RS, OC, AC, YL, HK, KH, ML, FC, RS, TFM, GM, NC, AR. Contributed with reagents/materials/analysis tools: ML, FC, TFM, GM, NC, AR. Wrote the paper: LF, AR. Critically read and contributed to write the paper: LF, RE, RS, OC, AC, HK, YL, KH, ML, FC, RS, TFM, GM, NC, AR.
References
- 1.Bollinger T., Schibler U. Circadian rhythms - from genes to physiology and disease. Swiss Med Wkly. 2014;144:w13984. doi: 10.4414/smw.2014.13984. [DOI] [PubMed] [Google Scholar]
- 2.Sukumaran S., Almon R.R., Dubois D.C., Jusko W.J. Circadian rhythms in gene expression: relationship to physiology, disease, drug disposition and drug action. Adv Drug Deliv Rev. 2010;62(9–10):904–917. doi: 10.1016/j.addr.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev. 2013;93(1):107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S.A. Circadian metabolism: from mechanisms to metabolomics and medicine. Trends Endocrinol Metab. 2016;27(6):415–426. doi: 10.1016/j.tem.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Sahar S., Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9(12):886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 6.Fu L., Kettner N.M. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. 2013;119:221–282. doi: 10.1016/B978-0-12-396971-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhr L., Abreu M., Pett P., Relogio A. Circadian systems biology: when time matters. Comput Struct Biotechnol J. 2015;13:417–426. doi: 10.1016/j.csbj.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16(10):635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlova N.N., Thompson C.B. The emerging hallmarks of Cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deberardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnaiah S.Y., Wu G., Altman B.J. Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab. 2017;25(5):1206. doi: 10.1016/j.cmet.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Scrima R., Cela O., Merla G. Clock-genes and mitochondrial respiratory activity: evidence of a reciprocal interplay. Biochim Biophys Acta. 2016;1857(8):1344–1351. doi: 10.1016/j.bbabio.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 13.Cela O., Scrima R., Pazienza V. Clock genes-dependent acetylation of complex I sets rhythmic activity of mitochondrial OxPhos. Biochim Biophys Acta. 2016;1863(4):596–606. doi: 10.1016/j.bbamcr.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Ballesta A., Innominato P.F., Dallmann R., Rand D.A., Levi F.A. Systems Chronotherapeutics. Pharmacol Rev. 2017;69(2):161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giacchetti S., Dugue P.A., Innominato P.F. Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: a meta-analysis. Ann Oncol. 2012;23(12):3110–3116. doi: 10.1093/annonc/mds148. [DOI] [PubMed] [Google Scholar]
- 16.Plikus M.V., Vollmers C., de la Cruz D. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc Natl Acad Sci U S A. 2013;110(23):E2106–E2115. doi: 10.1073/pnas.1215935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla P., Gupta D., Bisht S.S. Circadian variation in radiation-induced intestinal mucositis in patients with cervical carcinoma. Cancer. 2010;116(8):2031–2035. doi: 10.1002/cncr.24867. [DOI] [PubMed] [Google Scholar]
- 18.Bjarnason G.A., Mackenzie R.G., Nabid A. Comparison of toxicity associated with early morning versus late afternoon radiotherapy in patients with head-and-neck cancer: a prospective randomized trial of the National Cancer Institute of Canada clinical trials group (HN3) Int J Radiat Oncol Biol Phys. 2009;73(1):166–172. doi: 10.1016/j.ijrobp.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Dallmann R., Okyar A., Levi F. Dosing-time makes the poison: circadian regulation and pharmacotherapy. Trends Mol Med. 2016;22(5):430–445. doi: 10.1016/j.molmed.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Kessler M., Hoffmann K., Brinkmann V. The notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown S.A., Fleury-Olela F., Nagoshi E. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sporl F., Schellenberg K., Blatt T. A circadian clock in HaCaT keratinocytes. J Invest Dermatol. 2011;131(2):338–348. doi: 10.1038/jid.2010.315. [DOI] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Lück S., Thurley K., Thaben P.F., Westermark P.O. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014;9(2):741–751. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Li X.X., Peng J.J., Liang L. RNA-seq identifies determinants of oxaliplatin sensitivity in colorectal cancer cell lines. Int J Clin Exp Pathol. 2014;7(7):3763–3770. [PMC free article] [PubMed] [Google Scholar]
- 26.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaben P.F., Westermark P.O. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29(6):391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar L., Ef M. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futschik M.E., Kasabov N.K. Fuzzy Systems, 2002 FUZZ-IEEE'02 Proceedings of the 2002 IEEE International Conference on; 2002. IEEE; 2002. Fuzzy clustering of gene expression data; pp. 414–419. [Google Scholar]
- 31.Swainston N., Smallbone K., Hefzi H. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Relogio A., Thomas P., Medina-Perez P. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014;10(5) doi: 10.1371/journal.pgen.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Fernandez R., Hernandez J., Martin-Vasallo P., Puopolo M., Palumbo A., Avila J. Expression levels of the oxidative stress response gene ALDH3A2 in granulosa-lutein cells are related to female age and infertility diagnosis. Reprod Sci. 2016;23(5):604–609. doi: 10.1177/1933719115607996. [DOI] [PubMed] [Google Scholar]
- 34.Oppelt S.A., Zhang W., Tolan D.R. Specific regions of the brain are capable of fructose metabolism. Brain Res. 2017;1657:312–322. doi: 10.1016/j.brainres.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z., Huang S., Wang H. High expression of hexokinase domain containing 1 is associated with poor prognosis and aggressive phenotype in hepatocarcinoma. Biochem Biophys Res Commun. 2016;474(4):673–679. doi: 10.1016/j.bbrc.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Montal E.D., Dewi R., Bhalla K. PEPCK coordinates the regulation of central carbon metabolism to promote Cancer cell growth. Mol Cell. 2015;60(4):571–583. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H., Luo X., Li J. Pyruvate dehydrogenase B promoted the growth and migration of the nasopharyngeal carcinoma cells. Tumour Biol. 2016;37(8):10563–10569. doi: 10.1007/s13277-016-4922-4. [DOI] [PubMed] [Google Scholar]
- 38.Rardin M.J., Wiley S.E., Naviaux R.K., Murphy A.N., Dixon J.E. Monitoring phosphorylation of the pyruvate dehydrogenase complex. Anal Biochem. 2009;389(2):157–164. doi: 10.1016/j.ab.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel M.S., Nemeria N.S., Furey W., Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. 2014;289(24):16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holness M.J., Sugden M.C. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 2003;31:1143–1151. doi: 10.1042/bst0311143. Pt 6. [DOI] [PubMed] [Google Scholar]
- 41.Sachs N., Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Kiessling S., Beaulieu-Laroche L., Blum I.D. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017;15(1):13. doi: 10.1186/s12915-017-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu L., Lee C.C. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3(5):350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 44.Savvidis C., Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med. 2012;18:1249–1260. doi: 10.2119/molmed.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng Z.L., Luo H.Y., Yang J. Overexpression of the circadian clock gene Bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin Cancer Res. 2014;20(4):1042–1052. doi: 10.1158/1078-0432.CCR-13-0171. [DOI] [PubMed] [Google Scholar]
- 46.Kuo S.J., Chen S.T., Yeh K.T. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009;454(4):467–474. doi: 10.1007/s00428-009-0761-7. [DOI] [PubMed] [Google Scholar]
- 47.Grechez-Cassiau A., Rayet B., Guillaumond F., Teboul M., Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283(8):4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 48.Geyfman M., Kumar V., Liu Q. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A. 2012;109(29):11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullenders J., Fabius A.W., Madiredjo M., Bernards R., Beijersbergen R.L. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Athman R., Genov N.N., Mazuch J. The Ink4a/Arf locus operates as a regulator of the circadian clock modulating RAS activity. PLoS Biol. 2017;15(12) doi: 10.1371/journal.pbio.2002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vander Heiden M.G., Deberardinis R.J. Understanding the intersections between metabolism and Cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cammarota F., Laukkanen M.O. Mesenchymal stem/stromal cells in stromal evolution and Cancer progression. Stem Cells Int. 2016;2016 doi: 10.1155/2016/4824573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirota T., Okano T., Kokame K., Shirotani-Ikejima H., Miyata T., Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277(46):44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 54.Chen B., Liu Y., Jin X. MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer. 2014;14:443. doi: 10.1186/1471-2407-14-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma Y.S., Yang I.P., Tsai H.L., Huang C.W., Juo S.H., Wang J.Y. High glucose modulates antiproliferative effect and cytotoxicity of 5-fluorouracil in human colon cancer cells. DNA Cell Biol. 2014;33(2):64–72. doi: 10.1089/dna.2013.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh C.S., Wang J.Y., Chung F.Y. Significance of the glycolytic pathway and glycolysis related-genes in tumorigenesis of human colorectal cancers. Oncol Rep. 2008;19(1):81–91. [PubMed] [Google Scholar]
- 57.Irwin D.M., Tan H. Molecular evolution of the vertebrate hexokinase gene family: identification of a conserved fifth vertebrate hexokinase gene. Comp Biochem Physiol Part D Genomics Proteomics. 2008;3(1):96–107. doi: 10.1016/j.cbd.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 58.Li G.H., Huang J.F. Inferring therapeutic targets from heterogeneous data: HKDC1 is a novel potential therapeutic target for cancer. Bioinformatics. 2014;30(6):748–752. doi: 10.1093/bioinformatics/btt606. [DOI] [PubMed] [Google Scholar]
- 59.Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 60.Masri S., Cervantes M., Sassone-Corsi P. The circadian clock and cell cycle: interconnected biological circuits. Curr Opin Cell Biol. 2013;25(6):730–734. doi: 10.1016/j.ceb.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang Q., Cheng B., Xie M. Circadian clock gene Bmal1 inhibits tumorigenesis and increases paclitaxel sensitivity in tongue squamous cell carcinoma. Cancer Res. 2017;77(2):532–544. doi: 10.1158/0008-5472.CAN-16-1322. [DOI] [PubMed] [Google Scholar]
- 62.Vincent E.E., Sergushichev A., Griss T. Mitochondrial phosphoenolpyruvate Carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol Cell. 2015;60(2):195–207. doi: 10.1016/j.molcel.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 63.Hu J., Locasale J.W., Bielas J.H. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31(6):522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Changes in the core-clock of SW480 and SW620 cells lead to changes in metabolic pathways. (a) Phase-ordered, median-normalized heatmap of core-clock genes in SW480 and SW620 cells. Phase-ordering was performed based on SW480 expression data and the same ordering of genes was used for SW620 cells. (b) Phase-ordered, median-normalized heatmap of genes included in the oxidative phosphorylation pathway (left panel) and the Warburg effect (right panel) oscillating in at least one of the two tested cell lines. Phase-ordering was performed based on SW480 expression data and the same ordering of genes was used for SW620 cells. (c) Clustering of metabolic genes based on their time-dependent expression profile. All members of the pathways oxidative phosphorylation (left panel) and Warburg effect (right panel) that were oscillating in at least one cell line were grouped into three different clusters. The colors of the gene expression patterns indicate gradual membership values, reflecting the strength of a gene's association with its respective cluster (red: high membership value, blue: low membership value). The black lines indicate the cluster centers. (d) Resting endogenous respiration in SW480 and SW620 cells. OCR was determined over a period of 30 h. Mean ± SEM, n = 3. (e) PDH and pPDH protein abundance, pPDH/PDH ratio and oxygen consumption rate was determined in HepG2 cells for 24 h. Mean ± SEM, n = 3 (protein abundance). Mean ± SEM, n = 10 (OCR).
The circadian clock in human fallopian tube organoids. (a) Bioluminescence measurements of human fallopian tube organoids transduced with a BMAL1-promoter driven luciferase construct. Bioluminescence was measured for 14 consecutive days. Shown is one representative replicate. 24 h time-course RT-qPCR measurements of BMAL1 (b) and HKDC1 (c) in human fallopian tube organoids. Data are expressed as mean ± SEM, n = 3.
BMAL1 promoter activity in SW480 and SW620 cells and time-dependent gene expression after BMAL1-KD. (a) SW480 and (b) SW620 cells were lentivirally transduced either with an empty-vector (control) or with an shBMAL1-construct. Additionally, cells were lentivirally transduced with a Bmal1-promoter driven luciferase construct. Bioluminescence was measured for five consecutive days. Shown is one representative replicate for each condition. (c) 24 h RT-qPCR measurements of BMAL1 and HKDC1 in SW480-shBMAL1 (upper panel) and SW620-shBMAL1 (lower panel) cells. Data are shown compared to the mean expression. Mean ± SEM, n = 3.
KD efficiency after shRNA-mediated kD of BMAL1 and HKDC1. Gene expression analysis of BMAL1 in SW480 (a) and SW620 (b) cells and HKDC1 in SW480 (c) and SW620 (d) cells after shRNA-mediated BMAL1-KD and HKDC1-KD. Gene expression is shown compared to the corresponding control gene. Mean ± SEM, n = 3. KD efficiency in SW480-shBMAL1 cells: 0.317 ± 0.009 (68.3%). KD efficiency in SW620-shBMAL1 cells: 0.313 ± 0.03 (68.7%). KD efficiency in SW480-shHKDC1: 0.03 ± 0.006 (97%). KD efficiency in SW620-shHKDC1: 0.13 ± 0.013 (87%).
Viability, cytotoxicity and apoptosis in SW480 and SW620 cells. (a) Viability, (b) cytotoxicity and (c) apoptosis in SW480 (upper panel) and SW620 (lower panel) control and shBMAL1 cells synchronized at different timepoints. Cells were either untreated or treated with WZB117 or oxaliplatin. Mean ± SEM, n = 3. Significant changes (p < .05) between different synchronization timepoints of the same condition are marked with *. * = p < .05, ** = p < .01, *** = p < .001.
Metabolic activity of SW480 and SW620 control and shBMAL1 cells after oxaliplatin treatment. (a) Glycolysis of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at three different timepoints after synchronization (18 h, 21 h, 24 h). Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (b) Glycolytic capacity of SW480 (left panel) and SW620 (right panel) control and shBBMAL1 cells at different timepoints. Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (c) Basal respiration of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at different timepoints. Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (d) Maximum respiration of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at three different timepoints (18 h, 21 h, 24 h). Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. (e) ATP production of SW480 (left panel) and SW620 (right panel) control and shBMAL1 cells at three different timepoints (18 h, 21 h, 24 h). Cells were either untreated or treated with oxaliplatin. Mean ± SEM, n = 5. Significant changes (p < .05) between different timepoints of the same condition are marked with *. * = p < .05, ** = p < .01, *** = p < .001.
Determination of treatment concentration for WZB117 treatment. To determine the appropriate treatment concentration with WZB117, SW480-ctrl, SW480-shBMAL1, SW620-ctrl and SW620-shBMAL1 cells were treated with different concentrations of WZB117 and the cytotoxicity was determined. Based on the calculated IC50 value, the concentration of treatment was chosen.
Statistical analysis of circadian parameters of genes and proteins. Circadian parameters (p-value for oscillatory behavior, acrophase, amplitude, mesor) for clock and clock-regulated genes and proteins in SW480 and SW620 cells determined using RAIN and harmonic regression analysis.
Over-representation analysis. An over-representation analysis was performed with three sets of genes: genes oscillating in SW480 and SW620 cells, genes oscillating in SW480 cells only and genes oscillating in SW620 cells only.
Metabolic network analysis. Expression data gained from a 24 h microarray analysis of SW480 and SW620 cells was mapped to a reconstruction of human metabolism.
Proliferation curve analysis. Statistical analysis was done for every timepoint of the proliferation curve to test for significant differences between the different conditions.