Abstract.
Cerebral malaria (CM) remains an important cause of morbidity and mortality. Risk for developing CM partially depends on host genetic factors, including variants encoded in the type I interferon (IFN) receptor 1 (IFNAR1). Type I IFNs bind to IFNAR1 resulting in increased expression of IFN responsive genes, which modulate innate and adaptive immune responses. To comprehensively study IFNAR1 genetic variant associations in Malawians with CM or uncomplicated malaria, we used a tag single nucleotide polymorphism approach, based on the HapMap Yoruba in Ibadan, Nigeria, population database. We identified three novel (rs914142, rs12626750, and rs1041867) and one previously published (Chr21:34696785 [C > G]) IFNAR1 variants to be associated with CM. Some of these variants are in gene regulatory regions. Chr21:34696785 (C > G) is in a region encoding histone modifications and transcription factor–binding sites, which suggests gene regulatory activity. Rs12626750 is predicted to bind embryonic lethal abnormal vision system-like RNA-binding protein 1, a RNA-binding protein which can increase the type I IFN response. Furthermore, we examined these variants in an expression quantitative trait loci database and found that a protective variant, rs914142, is associated with lower expression of IFNAR1, whereas the CM-associated variant rs12626750 was associated with increased IFNAR1 expression, suggesting that activation of the type I IFN pathway may contribute to pathogenesis of CM. Future functional studies of IFNAR1 variants are now needed to clarify the role of this pathway in severe malarial diseases.
INTRODUCTION
Plasmodium falciparum infection continues to cause high rates of morbidity and mortality, with 216 million cases and an estimated 445,000 deaths in 2016, largely in African children.1,2 Malaria has affected human populations for millennia, resulting in evolutionary pressure and selection for host mutations that confer protection against malaria-related death. Some of these variants alter the human red cell niche of the parasite, such as sickle cell trait, G6PD deficiency, and β-thalassemia, whereas other protective variants are found in immune response genes, including CD36, CD40 ligand, TNF-α, IFN-γ, IL-4, and IL-12.3 Type I interferon (IFN) receptor 1 (IFNAR1) genetic variants have also been associated with protection to severe malaria.4–6 IFNAR1 encodes a membrane protein that forms one of the two chains of a receptor for type I IFN cytokines. Type I IFNs modulate host response to viral, mycobacterial, and other infections by binding to IFNAR1 which in turn induces expression of IFN responsive genes (IRGs), which have broad immunomodulatory properties.7–10 It is unknown how variants in IFNAR1 provide protection from severe malaria.
The expression of type I IFN cytokines and IRGs is increased during natural malaria infection, though their roles in immunity is unclear.11–13 Animal models of malaria infection demonstrate both protective and harmful effects of the type I IFN pathway depending on the model studied.12,14,15–19 These data reflect the balancing act that type I IFN pathway plays on modulating host responses to infection over a range of clinical presentations from severe to mild disease.20
To explore the contribution of specific IFNAR1 variants to malaria disease severity, we examined IFNAR1 polymorphisms with a tag SNP approach, through a case–control study of Malawian children with cerebral malaria (CM) and uncomplicated malaria (UM). To explore the function of the disease-associated variants, we determined if they resided in regulatory sequences and mined an expression quantitative trait loci (eQTL) database.
MATERIALS AND METHODS
Study subjects.
The CM subjects were selected from an ongoing observational study of the pathogenesis of CM in Blantyre, Malawi. The inclusion criteria included the World Health Organization definition for CM (Blantyre coma score < 3, peripheral asexual P. falciparum, and no other obvious cause of coma) and the presence of malarial retinopathy.21–23 Study subjects were between 6 months and 12 years of age. Subjects who were human immunodeficiency virus (HIV) infected or had a positive blood or cerebrospinal fluid bacterial culture were excluded from the analysis. Samples analyzed for this study were randomly chosen from enrolled subjects in the 2009, 2011, and 2013–2014 rainy seasons. The UM subjects were selected from the Mfera cohort study, a longitudinal study of UM in the Chikwawa District of Malawi, a wet, low-lying rural district in the Shire Valley with high malaria transmission, which is 22 miles from the CM study site.24 Enrolled subjects were aged between 1 and 31 years, were HIV negative, and had microscopy-confirmed P. falciparum infection on peripheral blood smear analysis. Uncomplicated malaria subjects were excluded from the study if they had acute illness requiring hospitalization, had signs or symptoms of severe malaria, or were receiving chronic medication with any drug that has antimalarial activity (e.g., HIV exposed infants).22 We analyzed all UM samples collected between June 2014 and September 2014 and all subjects aged < 12 years who were enrolled between October 2014 and January 2015. For both groups, parental consent was obtained before study enrollment, a clinical examination was performed, and data were extracted from the study record.
For subsequent DNA extraction and genetic analysis (Whatman, NJ), whole blood was collected at enrollment in ethylenediaminetetraacetic acid tubes by venipuncture and spotted onto FTA classic cards. Study participants received standard treatment of malaria. Institutional Review Board approvals were obtained from the Albert Einstein College of Medicine, Michigan State University, the University of Maryland, and the University of Malawi College of Medicine Research and Ethics Committee.
IFNAR1 genotyping.
To study IFNAR1 variants, DNA was extracted from the blood spots using DNeasy Blood and Tissue kits (Qiagen, Redwood City, CA). When a blood spot was unavailable for the CM subjects, DNA was extracted according to manufacturer instructions from an enrollment aliquot of whole blood that was stored in Tri-reagent BD at −80°C (Molecular Research Center, Cincinnati, OH).
We used multiple strategies to select IFNAR1 gene SNPs to provide a comprehensive genetic association analysis. For a tag SNP approach, we first identified 16 tagging SNPs from the Hapmap database of all highly linked SNPs (R2 > 0.9) found in the Yoruba in Ibadan, Nigerian (YRI) population within the IFNAR1 coding and noncoding sequences including 2 kb upstream using Haploview (Supplemental Figure 1).25,26 These tagging SNPs included one in the 5′ untranslated region (UTR), 11 in introns, and four in the 3′ UTR. Second, we selected the two IFNAR1 coding variants with a minor allele frequency > 0.01 in the YRI population from the 1000 Genomes and Exome Sequencing Project database.27,28 Finally, we analyzed the five published IFNAR1 SNPs associated with malaria disease severity in other geographical sites (rs1012325, rs2257167, rs2253923, rs2856968, and Chr21:34696785 [C > G]).4–6 After removing two overlapping variants, a total of 21 variants were examined.
We designed polymerase chain reaction (PCR) and iPLEX extension primers for each SNP of interest using Assay Design Suite software to iPLEX (Sequenom, San Diego, CA). Sample DNA was PCR amplified using a multiplex reaction and analyzed using matrix assisted laser desorption ionization-time of flight mass spectrometry analysis (MassArray, San Diego, CA). Mass spectrometry peaks were visualized in the MassARRAY Typer 4.0 software. The iPLEX assay genotyping was performed in the Genomics Shared Facility at Albert Einstein College of Medicine.
Statistical analysis.
The Hardy–Weinberg equilibrium (HWE) test is used as an essential quality control step in the population-based genetic association studies. Deviation from HWE has been associated with problems such as genotyping errors, batch effects, population stratification, and selection bias. Failure to remove variants that are out of HWE may lead to biased study inferences.29 Thus, the HWE test was performed to test the genotype distribution of all the variants. Variants that followed HWE (P > 0.01) were further analyzed. For each variant, three association tests were performed—allelic (major allele A versus minor allele a), genotypic (AA versus Aa versus aa), and logistic regression using an additive model (AA = 0, Aa = 1, and aa = 2) adjusted for age. Statistical differences among groups were assessed by Fisher’s exact and logistic regression tests of association in software package PLINK.30 A two-tailed P value of < 0.05 was considered significant. We also report the false discovery rate (FDR) method of Benjamin and Hochberg MassARRAY.31
Evaluation of regulatory elements associated with identified loci.
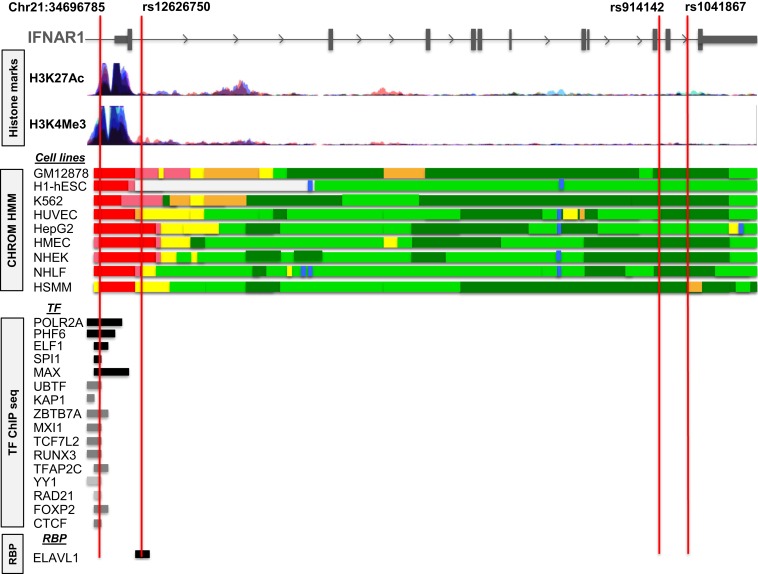
Encyclopedia of DNA Elements at University California, Santa Cruz (UCSC), (ENCODE) and ROADMAP epigenomics project data were assessed using Haploreg, UCSC genome browser, and RegulomeDB to characterize the region where the variants were found.32–34 Histone marks, transcription factor–binding tracks, and RNA-binding protein tracks in Figure 3 were downloaded from the UCSC Genome Browser.
Figure 3.
Loci associated with cerebral malaria are in gene regulatory regions. Representative regulatory motif tracks from the University of California, Santa Cruz, Genome Browser and ENCODE showing histone mark peaks and transcription factor–binding and RNA-binding proteins at the location of Chr21:34696785, rs12626750, rs914142, and rs1041867. Different chromatin state segmentations based on histone marks defined by Hidden Markov Model (HMM) in ENCODE data are represented by different colors. Red, light red, orange, light yellow, green, light green, and blue colors indicate active promoter, weak promoter, strong enhancer, weak enhancer, transcriptional elongation/transition, weak transcription, and insulator, respectively. RBP = RNA-binding protein; TF = transcription factor.
RESULTS
The 21 IFNAR1 SNPs analyzed include previously identified disease-associated SNPs and tagging SNPs to identify novel variants associated with retinopathy-positive CM compared with UM subjects. Of the 21 SNPs examined, one did not meet the quality control criteria of the iPLEX assay and was eliminated from the analysis. Individual sample data were included in the analysis if ≥ 14 of 20 SNPs provided high-quality data. Samples from all 87 retinopathy-positive CM subjects and 54 of the 71 UM subjects passed the quality criteria and were analyzed (Table 1). The CM subjects had higher temperatures and respiratory rates and lower hemoglobin values (Mann–Whitney; P value < 0.001). There was no significant difference in peripheral parasitemia between UM and CM subjects (Mann–Whitney; P value = 0.083). The CM subjects were significantly younger, with lower body weights and heights (Mann–Whitney; P value < 0.001). Because older age is associated with protection from severe disease, we included age as a covariate in the logistic model.35
Table 1.
Characteristics of subjects with UM and CM
| Characteristic | UM (N = 54) | CM (N = 87) | P value |
|---|---|---|---|
| Age (months) | 78 (39–123) | 44 (27–62) | < 0.001 |
| Gender (% Male) | 41 | 43 | 0.834 |
| Weight (kg) | 19 (14–26) | 12.2 (10.2–15.4) | < 0.001 |
| Height (cm) | 113 (93–132) | 94 (81–106) | < 0.001 |
| Temperature (°C) | 37.7 (36.3–38.8) | 39.0 (38.2–39.8) | < 0.001 |
| Hemoglobin (g/dL) | 11.2 (10.1–12.2) | 7.3 (5.9–8.3) | < 0.001 |
| Parasitemia (×103/μL) | 23 (1–68) | 32 (1–186) | 0.083 |
| Respirations (breaths/minutes) | 32 (26–36) | 40 (36–52) | < 0.001 |
CM = cerebral malaria; UM = uncomplicated malaria. Continuous variables are compared using the Mann–Whitney test and dichotomous variables are compared with the χ2 test. Values are reported as median and interquartile range for continuous variables and percent for dichotomous variables.
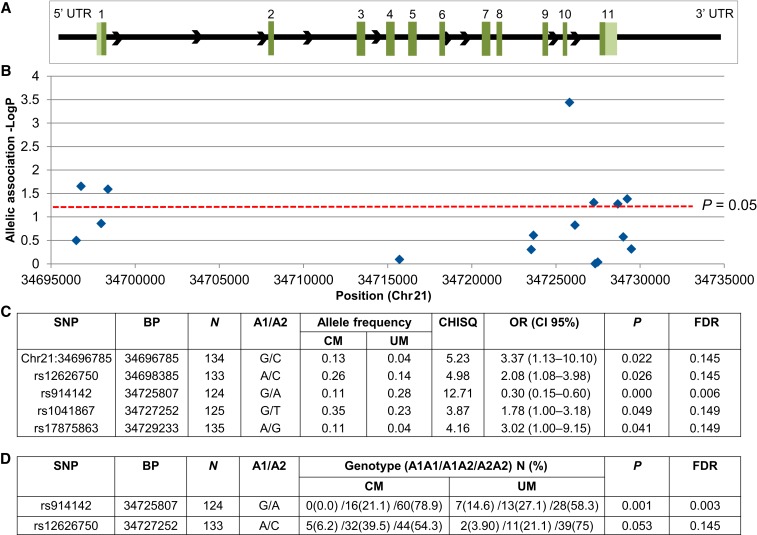
Three (rs2253923, rs7280108, and rs17875834) of the remaining 20 variants deviated from the HWE and were excluded, leaving 17 SNPs for the analysis. To investigate the association of IFNAR1 variants and malaria disease severity, allele frequencies and genotype frequencies were compared between the CM and UM cohorts. We first examined allelic associations between CM and UM and identified five IFNAR1 SNPs with a significant allelic association with CM: rs914142, Chr21:34696785 (C > G), rs12626750, rs17875863, and rs1041867 (χ2; P < 0.05) (Figure 1). These SNPs are in noncoding regions including the 5′UTR, introns, and 3′UTR (Figure 1A and B). The variant rs914142 has the lowest FDR of 0.006 and the other variants have FDRs of less than 0.15. These variants are independent and not in linkage disequilibrium (LD) with each other (Supplemental Figure 1). Distribution of the allelic variants is reported (Figure 1C).
Figure 1.
Allelic and genotypic associations of interferon-alpha receptor 1 (IFNAR1) single nucleotide polymorphisms (SNP) with cerebral malaria. Association study of IFNAR1 variants with cerebral malaria (CM) compared with uncomplicated malaria (UM). Analyzed SNPs were selected using tagging SNP analysis and previously identified disease-associated SNPs. (A) Schematic of IFNAR1 gene structure: exons are marked with their corresponding number. The 5′ untranslated region (UTR) and 3′ UTRs are represented by light green boxes and introns are represented by black lines between exons. Allelic and genotypic association P values were obtained by comparing 87 CM subjects with 54 UM subjects. (B and C) There were five significant alleles (P < 0.05). (D) There were two significant genotypic association results (P < 0.05). A1 = affected/alternate allele; A2 = reference allele; BP = genomic position in base pair according to human GRCh37/hg19 assembly; CI = confidence interval; FDR = false discovery rate; OR = odds ratio.
To then examine differences in genotype, we removed three variants that did not have a homozygous minor allele in our study population (rs17875887, rs1012325, and Chr21:34696785 [C > G]) and examined the remaining 14 variants. Two variants were significantly different between the CM and UM subjects in this analysis: rs914142 and rs12626750. The genotypic frequencies of rs914142 were significantly different (Fisher’s exact; P = 0.001) with 0 (GG), 0.21 (GA), and 0.79 (AA) for the CM group and 0.15 (GG), 0.27 (GA), and 0.58 (AA) for the UM group. The variant rs12626750 also differed between CM and UM (Fisher’s exact; P = 0.053) with 0.06 (AA), 0.40 (AC), and 0.54 (CC) in the CM group and 0.04 (AA), 0.21 (AC), and 0.75 (CC) in the UM group (Figure 1D).
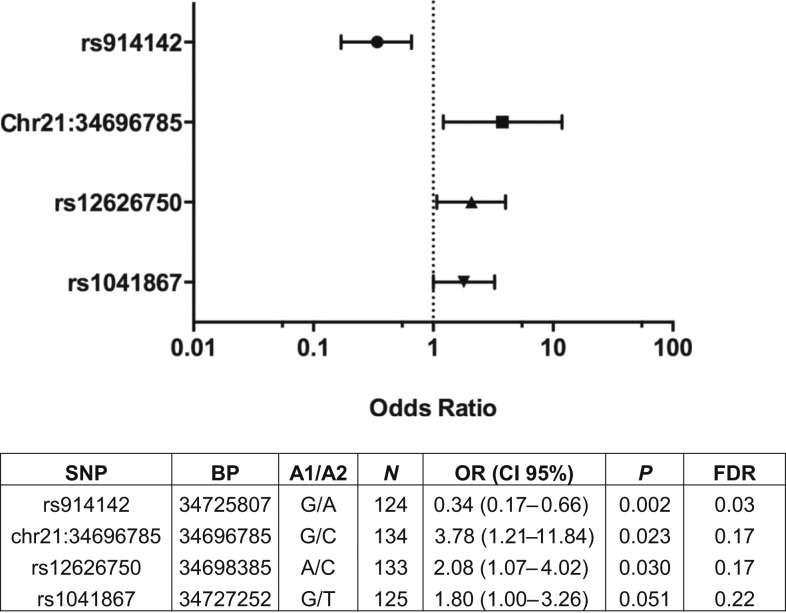
Age is associated with development of clinical immunity; thus, we carried out logistic regression using an additive genetic model with age as a covariate (Figure 2). After age correction, three variants (rs914142, Chr21:34696785 (C > G), and rs12626750) remained significantly associated with disease outcome (P < 0.05) and one variant (rs1041867) approached significance (P = 0.051). For rs12626750, relative to the UM group, subjects carrying the A allele had increased risk of developing CM (odds ratio [OR] = 2.08 [95% confidence interval (CI) = 1.07–4.02], P = 0.030). The G allele of Chr21:34696785(C > G), and rs1041867 showed a significant association with increased risk of CM (OR = 3.78 [95% CI = 1.21–11.84], P = 0.023 and OR = 1.80 [95% CI = 1.00–3.26], P = 0.051), respectively, as compared with UM. Relative to the UM group, subjects carrying the G allele of rs914142 had reduced risk of developing CM (OR = 0.34 [95% CI = 0.17–0.66], P = 0.0016).
Figure 2.
Relationship between interferon-alpha receptor 1 genotypes and malaria disease severity. Odds ratios (OR) and confidence intervals (CI) were determined using logistic regression analysis with an additive model, controlling for age. Dotted line represents null. A1 = affected/alternate allele; A2 = reference allele; BP = genomic position in base pair on chromosome 21 according to human GRCh37/hg19 assembly; FDR = false discovery rate.
To explore a potential functional role of the SNPs identified in the logistic regression model, we examined whether the disease-associated loci are found in functional noncoding sequences involved in gene regulation and mined the epigenetic ENCODE and ROADMAP data for associated SNPs using Haploreg. Functional noncoding regions are defined by the presence of histone marks and predicted and experimentally validated transcription factor–binding and RNA-binding protein data. The gene regulatory and epigenetic context surrounding CM-associated SNPs is shown in Figure 3.
Chr21:34696785(C > G) SNP lies within peaks of histone marks associated with strong promoter activity in nine cell lines including lymphocytes, hepatocytes, and endothelial cells. Moreover, this SNP is located within ChIP-identified binding regions for several transcription factors including POLR2A, PHF8, ELF1, SPI1, and MAX. The variant rs12626750 also falls in a region with predicted promoter and enhancer activity. In addition, rs12626750 localizes in the region predicted to bind the RNA-binding protein embryonic lethal, abnormal vision-like 1 (ELAV1).
To further determine the contribution of these variants on gene expression, we examined a published eQTL meta-analysis of peripheral blood of 5,311 individuals.36 Three of our variants (rs914142, rs12626750, and rs1041867) had significant associations within the eQTL data (FDR = 0) (Supplemental Table 1) by affecting the expression of IFNAR1 and/or interleukin 10 receptor, beta subunit (IL10RB). The protective rs914142 G allele is linked to decreased IFNAR1 expression (P value 3.01E-49) and the susceptible rs12625750 A allele is linked to increased IFNAR1 expression (P value 3.80E-28). All three variants (rs914142, rs12626750, and rs1041867) are associated with alterations in the expression of IL10RB. Two CM-associated variants (rs12626750 and rs1041867) had increased IL10RB (P value 1.06E-06 and 1.17E-10). Conversely, the protective variant (rs914142) produces a decrease in IL10RB expression (P value 5.63E-07). The variant Chr21:34696785 does not have an associated RefSNP (rs) designation and thus we could not evaluate its association with the eQTL data.
DISCUSSION
The type I IFN pathway is involved in host response to malaria infection and variants within the IFNAR1 have been associated with severe illness in African populations.4–6 To provide a more comprehensive analysis of IFNAR1 variants and malaria disease outcomes, we carried out an association study of the previously identified IFNAR1 variants and novel variants selected through a tagging SNP approach in Malawian children with CM and UM. We identified four disease-associated variants in the logistic regression analysis. All variants are in noncoding regions that have predicted gene regulatory roles, with one CM-associated variant linked to higher IFNAR1 transcript levels and one protective variant associated with lower IFNAR1 transcript level identified from previously published eQTL data. These data generate novel testable hypotheses regarding the role of IFNAR1 in CM.
We examined previously identified disease association IFNAR1 SNPs and tagging SNPs identified in the YRI population from the International HapMap dataset to also provide a comprehensive disease association analysis in a Malawian cohort. We confirmed the association of Chr21:34696785 (C > G), also known as IFNAR1 272354c-g, a variant located at position -576 relative to the transcription start site of IFNAR1, with CM in the Malawian cohort.5 This variant was also reported in association studies of severe malaria from geographically diverse study sites including Gambia, Kenya, and Vietnam, suggesting that this variant is strongly selected for and thus an important candidate for functional studies.5 Our logistic regression analysis also identified three novel IFNAR1 variants associated with disease outcomes.
There are no functional studies on the role of these variants, but these data suggest that they are involved in the regulation of gene expression. Chr21:34696785 (C > G) is located in a transcription factor–binding region which can bind POLR2A, PHF8, ELF1, SPI1, and MAX and this region also contains histone marks.37–40 The variant rs12626750 lies in a region predicted to bind ELAVL1, which stabilizes adenylate uridylate-rich elements–containing mRNAs.41 ELAV-like RNA-binding protein 1 expression has been shown to increase the type I IFN response as it is required for the stabilization of IFN-β and ELAV1 inhibition hampers the cellular type I IFN response.42,43 Both rs914142 and rs1041867 are found in regions with transcriptional elongation activity.
To further analyze how these variants may impact host physiology, we examined previously published eQTL data to identify a role in gene expression and found consistent disease association changes in IFNAR1 expression. The UM-associated variant rs914142 was associated with a decrease in IFNAR1 expression, whereas the CM-associated variants rs12626750 and rs1041867 were associated with increased IFNAR1 expression. This suggests a model where higher IFNAR1 expression drives increased IRG expression and cytokine production to increase the risk for CM. The effect of these variants on the IFNAR1 pathway can now be functionally tested in genetic systems.
The UM-associated variant rs914142 was associated with a decrease in IL10RB expression, whereas CM-associated variants rs12626750 and rs1041867 were associated with increased IL10RB expression. IL10RB encodes part of the interleukin 10 receptor complex and is found in a cytokine receptor gene cluster with IFNAR1 on chromosome 21. IL10RB variants have been found to be associated with severe malaria in prior studies.5,44,45 How IL10RB variants impact severe disease outcomes will require further study.
One potential limitation of this study is that the CM-associated tagging SNPs studied are in intronic or noncoding regions and the true genetic effect on disease outcome could be because of one of the linked variants within the LD block. Second, there are regional genetic differences between Malawians and the YRI population, though studies show good generalizability of HapMap tag SNPs used within a continent of nonisolated populations.46,47 Alternative approaches include designing tagging SNPs from the 1000 Genomes Project using the data from East African populations or sequencing of IFNAR1 directly from Malawian CM and UM samples.27 Our data confirmed the association of Chr21:34696785 (C > G) with severe disease, which has been reported in other African and Southeast Asian countries and provides confidence to the novel associations presented here.
In conclusion, we identified three novel IFNAR1 variants and confirmed one previously described IFNAR1 variant as being associated with CM in Malawi. The in silico and eQTL analyses suggest that these variants may have a regulatory role in IFNAR1 expression. Future studies will determine the function of these variants in the type I IFN pathway and further define the role of these disease-associated mutations in CM.
Supplementary Material
Supplemental figure and tables
Acknowledgments:
Sample collection was performed by technicians with the Mfera cohort study: Syze Gama, Alex Saidi, Andrew Nyambalo, and Enock Jumbe. Microscopy was performed by John Changalala and John Matiki.
Note: Supplemental figure and table appear at www.ajtmh.org.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.World Health Organization , 2017. World Malaria Report 2017, Geneva, Switzerland: World Health Organization. [Google Scholar]
- 2.World Health Organization , 2015. Guidelines for the Treatment of Malaria. 3rd edition. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 3.Kwiatkowski DP, 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet 77: 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aucan C, Walley AJ, Hennig BJ, Fitness J, Frodsham A, Zhang L, Kwiatkowski D, Hill AV, 2003. Interferon-alpha receptor-1 (IFNAR1) variants are associated with protection against cerebral malaria in the Gambia. Genes Immun 4: 275–282. [DOI] [PubMed] [Google Scholar]
- 5.Khor CC, et al. 2007. Positive replication and linkage disequilibrium mapping of the chromosome 21q22.1 malaria susceptibility locus. Genes Immun 8: 570–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball EA, Sambo MR, Martins M, Trovoada MJ, Benchimol C, Costa J, Antunes Goncalves L, Coutinho A, Penha-Goncalves C, 2013. IFNAR1 controls progression to cerebral malaria in children and CD8+ T cell brain pathology in Plasmodium berghei-infected mice. J Immunol 190: 5118–5127. [DOI] [PubMed] [Google Scholar]
- 7.Stryker GA, Nickell SP, 1995. Trypanosoma cruzi: exposure of murine cells to live parasites in vitro leads to enhanced surface class I MHC expression which is type I interferon-dependent. Exp Parasitol 81: 564–573. [DOI] [PubMed] [Google Scholar]
- 8.Stetson DB, Medzhitov R, 2006. Type I interferons in host defense. Immunity 25: 373–381. [DOI] [PubMed] [Google Scholar]
- 9.Berry MP, et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stifter SA, Feng CG, 2015. Interfering with immunity: detrimental role of type I IFNs during infection. J Immunol 194: 2455–2465. [DOI] [PubMed] [Google Scholar]
- 11.Pichyangkul S, et al. 2004. Malaria blood stage parasites activate human plasmacytoid dendritic cells and murine dendritic cells through a toll-like receptor 9-dependent pathway. J Immunol 172: 4926–4933. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, et al. 2011. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity 35: 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krupka M, Seydel K, Feintuch CM, Yee K, Kim R, Lin CY, Calder RB, Petersen C, Taylor T, Daily J, 2012. Mild Plasmodium falciparum malaria following an episode of severe malaria is associated with induction of the interferon pathway in Malawian children. Infect Immun 80: 1150–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vigario AM, et al. 2007. Recombinant human IFN-alpha inhibits cerebral malaria and reduces parasite burden in mice. J Immunol 178: 6416–6425. [DOI] [PubMed] [Google Scholar]
- 15.Morrell CN, Srivastava K, Swaim A, Lee MT, Chen J, Nagineni C, Hooks JJ, Detrick B, 2011. Beta interferon suppresses the development of experimental cerebral malaria. Infect Immun 79: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palomo J, et al. 2013. Type I interferons contribute to experimental cerebral malaria development in response to sporozoite or blood-stage Plasmodium berghei ANKA. Eur J Immunol 43: 2683–2695. [DOI] [PubMed] [Google Scholar]
- 17.Haque A, et al. 2011. Type I interferons suppress CD4+ T-cell-dependent parasite control during blood-stage Plasmodium infection. Eur J Immunol 41: 2688–2698. [DOI] [PubMed] [Google Scholar]
- 18.Spaulding E, Fooksman D, Moore JM, Saidi A, Feintuch CM, Reizis B, Chorro L, Daily J, Lauvau G, 2016. STING-licensed macrophages prime type I IFN production by plasmacytoid dendritic cells in the bone marrow during severe Plasmodium yoelii malaria. PLoS Pathog 12: e1005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zander RA, Guthmiller JJ, Graham AC, Pope RL, Burke BE, Carr DJ, Butler NS, 2016. Type I interferons induce T regulatory 1 responses and restrict humoral immunity during experimental malaria. PLoS Pathog 12: e1005945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mooney JP, Wassmer SC, Hafalla JC, 2017. Type I interferon in malaria: a balancing act. Trends Parasitol 33: 257–260. [DOI] [PubMed] [Google Scholar]
- 21.Barrera V, et al. 2015. Severity of retinopathy parallels the degree of parasite sequestration in the eyes and brains of Malawian children with fatal cerebral malaria. J Infect Dis 211: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization , 2015. Guidelines for the Treatment of Malaria, 3rd edition. Geneva, Switzerland: WHO. [Google Scholar]
- 23.Taylor TE, Fu WJ, Carr RA, Whitten RO, Mueller JS, Fosiko NG, Lewallen S, Liomba NG, Molyneux ME, 2004. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med 10: 143–145. [DOI] [PubMed] [Google Scholar]
- 24.Mathanga DP, Walker ED, Wilson ML, Ali D, Taylor TE, Laufer MK, 2012. Malaria control in Malawi: current status and directions for the future. Acta Trop 121: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.International HapMap Consortium , 2003. The international HapMap project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 27.The 1000 Genomes Project Consortium ; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Exome Variant Server , 2011. NHLBI Exome Sequencing Project (ESP) Available at: http://evs.gs.washington.edu/EVS/. Accessed December 15, 2011.
- 29.Namipashaki A, Razaghi-Moghadam Z, Ansari-Pour N, 2015. The essentiality of reporting Hardy-Weinberg equilibrium calculations in population-based genetic association studies. Cell J 17: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300. [Google Scholar]
- 32.Ward LD, Kellis M, 2012. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle AP, et al. 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aponte JJ, Menendez C, Schellenberg D, Kahigwa E, Mshinda H, Vountasou P, Tanner M, Alonso PL, 2007. Age interactions in the development of naturally acquired immunity to Plasmodium falciparum and its clinical presentation. PLoS Med 4: e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westra HJ, et al. 2013. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45: 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowell J, Koitabashi N, Kass DA, Barth AS, 2014. Dynamic gene expression patterns in animal models of early and late heart failure reveal biphasic-bidirectional transcriptional activation of signaling pathways. Physiol Genomics 46: 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Reilly D, Quinn CM, El-Shanawany T, Gordon S, Greaves DR, 2003. Multiple Ets factors and interferon regulatory factor-4 modulate CD68 expression in a cell type-specific manner. J Biol Chem 278: 21909–21919. [DOI] [PubMed] [Google Scholar]
- 39.Hikami K, et al. 2011. Association of a functional polymorphism in the 3′-untranslated region of SPI1 with systemic lupus erythematosus. Arthritis Rheum 63: 755–763. [DOI] [PubMed] [Google Scholar]
- 40.Kubosaki A, et al. 2010. The combination of gene perturbation assay and ChIP-chip reveals functional direct target genes for IRF8 in THP-1 cells. Mol Immunol 47: 2295–2302. [DOI] [PubMed] [Google Scholar]
- 41.Pruitt KD, et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42: D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herdy B, et al. 2015. The RNA-binding protein HuR/ELAVL1 regulates IFN-β mRNA abundance and the type I IFN response. Eur J Immunol 45: 1500–1511. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi O, 2015. HuR keeps interferon-β mRNA stable. Eur J Immunol 45: 1296–1299. [DOI] [PubMed] [Google Scholar]
- 44.de Weerd NA, Samarajiwa SA, Hertzog PJ, 2007. Type I interferon receptors: biochemistry and biological functions. J Biol Chem 282: 20053–20057. [DOI] [PubMed] [Google Scholar]
- 45.Idro R, Jenkins NE, Newton CR, 2005. Pathogenesis, clinical features, and neurological outcome of cerebral malaria. Lancet Neurol 4: 827–840. [DOI] [PubMed] [Google Scholar]
- 46.Tishkoff SA, et al. 2009. The genetic structure and history of Africans and African Americans. Science 324: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing J, Witherspoon DJ, Watkins WS, Zhang Y, Tolpinrud W, Jorde LB, 2008. HapMap tagSNP transferability in multiple populations: general guidelines. Genomics 92: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables