ABSTRACT
Lipid mediators, derived from arachidonic acid metabolism, play an important role in immune regulation. The functions of bioactive eicosanoids range from modulating cytokine signaling and inflammasome formation to anti-inflammatory and pro-resolving activities. Human pathogenic fungi such as Candida albicans, Candida parapsilosis, Cryptococcus neoformans and Aspergillus fumigatus have been shown to produce such lipid mediators, associated with their virulence. To date, investigations into the molecular mechanisms of fungal eicosanoid biosynthesis in different species have revealed that several genes are associated with prostaglandin production. However, these routes remain uncharacterized in C. parapsilosis with early results suggesting it uses pathways distinct from those found in C. albicans. Therefore, we aimed to identify and characterize C. parapsilosis genes involved in eicosanoid biosynthesis. Following arachidonic acid treatment of C. parapsilosis cells, we identified several genes interfering with prostaglandin production. Out of the identified genes, homologues of a multi copper oxidase (FET3), an Acyl-CoA thiolase (POT1) and an Acyl-CoA oxidase (POX1-3) were found to play a significant role in prostaglandin synthesis. Furthermore, all three genes were confirmed to enhance C. parapsilosis pathogenicity, as the corresponding deletion mutants were cleared more efficiently by human macrophages and induced higher levels of pro-inflammatory cytokines. In addition, the mutants were less virulent than the wild-type strain in a mouse model of systemic infection. Taken together, we identified three genes that regulate eicosanoid biosynthesis in C. parapsilosis and impact the fungus’ virulence.
KEYWORDS: Candida parapsilosis, fungal eicosanoids, immunomodulation, host-pathogen interaction, virulence
Introduction
Eicosanoids, are bioactive lipid molecules derived from the 20-carbon poly-unsaturated fatty acid, arachidonic acid. Prostaglandins and leukotrienes are two major groups of eicosanoids produced in almost all mammalian cells. Their production is initiated by phospholipases that mediate the release of arachidonic acid from the cell membrane and their activity is generally facilitated by binding to G-protein coupled receptors (GPCRs) and peroxisomal proliferator-activated receptors (PPARs) in different cells. Lipid mediators play a major role in inflammasome activation and the maintenance of cellular homoeostasis and in some cases, they act as pro-resolving/anti-inflammatory immune modulators (e.g. lipoxins). Three major enzymatic pathways are known to regulate eicosanoid synthesis, namely cyclooxygenase (COX), lipoxygenase (LOX) and cytochrome P450 proteins. Several drugs can block these enzymes and inhibit the generation of bioactive lipid mediators. For example, nonsteroidal anti-inflammatory drugs (NSAIDs), such as indomethacin or ibuprofen, target mainly the COX enzymes, thus prostaglandin biosynthesis, to alleviate inflammation [1–3]. Interestingly, the recognition of the C. albicans cell wall β-glucan by dectin-1 on the surface of macrophages enhances macrophage cytosolic phospholipase A2 activity and COX expression, thus promoting prostaglandin biosynthesis in the host [4,5]. Prostaglandin E2 (PGE2) is known to inhibit the Th1 response and promote Th2 immune responses [6,7]. Following C. albicans recognition, the induced PGE2 production also enhances Th17 response [8].
Human pathogenic fungi can also produce eicosanoids, mainly PGE2, themselves [9]. However, they lack COX and LOX required for their production, which suggests the presence of alternative routes for prostaglandin biosynthesis. Such routes have been identified in C. albicans (FET3 and OLE2 mediated pathways) [9], C. neoformans (LAC1) [10] and Aspergillus (PPO gene family) species [11]. These fungal eicosanoids play a role in fungal pathogenesis, as deletion of LAC1 in C. neoformans resulted in attenuated virulence while disruption of PPO genes drastically reduced asexual spore formation in Aspergillus nidulans and leads to a hyper-virulent phenotype in A. fumigatus [11]. Additionally, the yeast to hyphal transition in C. albicans is regulated in part by PGE2 and mature biofilms also produce this prostaglandin [12,13].
C. parapsilosis, is an important opportunistic human fungal pathogen, which belongs to the CTG clade of the Ascomycetes family. After C. albicans, it is the second or third leading cause of invasive candidiasis [14]. In particular, low birth weight infants are at increased risk for C. parapsilosis infection [15,16]. Previously, we demonstrated that C. parapsilosis is also able to produce immunomodulatory prostaglandins from exogenous arachidonic acid. Although, in contrast with C. albicans, the fatty acid desaturase OLE2 is not involved in the process; hence, the molecular mechanisms behind eicosanoid biosynthesis remain to be elucidated [17].
In the current study, our aim was to identify and characterize genes involved in C. parapsilosis eicosanoid biosynthesis. Following arachidonic acid induction, we performed RNA sequencing and rigorously investigated gene function through characterizing the generated corresponding deletion mutant strains. We determined that homologues of a multi copper oxidase (FET3), an Acyl-CoA thiolase (POT1) and an Acyl-CoA oxidase (POX1–3) significantly impacted eicosanoid synthesis. Furthermore, according to our results, all three genes influence the virulence of C. parapsilosis in vitro and also regulate pathogenicity in vivo.
Results
Identification of differentially regulated genes following arachidonic acid induction
To identify genes involved in the biosynthesis of lipid mediators, we performed global transcriptomic analysis on C. parapsilosis GA1 [18] yeast cells following growth in the presence of arachidonic acid (AA). As a control [zero amount versus background exposure increase (OH)], equal amounts of cells were grown without the addition of arachidonic acid. Altogether, 151 genes showed significantly altered expression (fold change > 1.5) in the presence of arachidonic acid when compared to control. Out of the 151 genes, 68 genes were upregulated, while 83 genes showed decreased expression levels (Table S1). Hierarchical clustering and principal component analysis (PCA) (Figure S1) showed that the three replicates from each group clustered together by condition, confirming the reliability of the obtained results.
Functional categorization of the genes by Gene Ontology (GO) term analysis using the candida genome database [19] showed, that 14% of the genes involved in lipid metabolic processes (GO:0006629) are upregulated (Figure 1(a)). The heat map indicates all of the upregulated ORFs in the induced condition (Figure 1(b)). Detailed GO term annotations of the upregulated genes can be found in supplementary Table S1. For further analyzes, we selected six upregulated genes with known lipid metabolic process regulatory homologous in C. albicans, (Table 1). Using these genes, we further validated the RNA sequencing data results by performing qRT-PCR analysis (Table 2). qRT-PCR confirmed that the six genes were upregulated following arachidonic acid pretreatment in both C. parapsilosis GA1 as well as in a second strain, CLIB 214. As five out of six genes showed higher expression levels in the CLIB 214 type strain, we subsequently used this strain for further analyzes.
Figure 1.
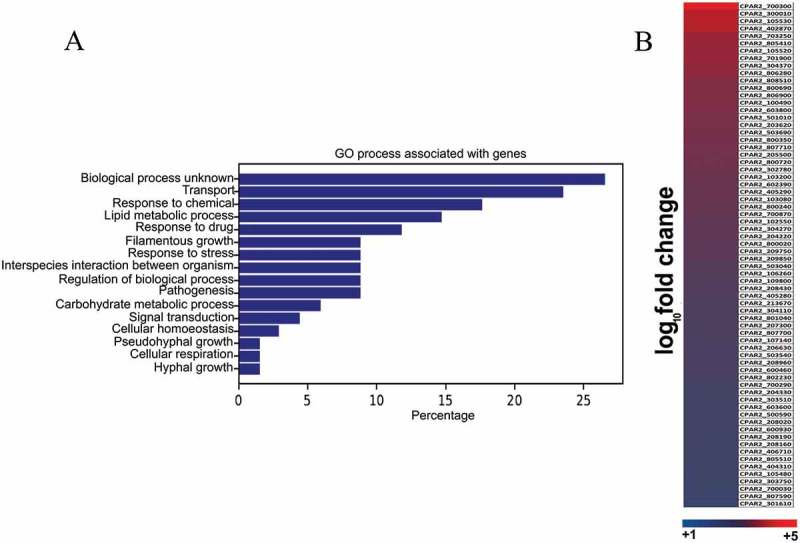
RNA sequencing and data analysis. (a) Global gene expression analysis was performed on the wild type C. parapsilosis strain after 3 hours of growth in presence of arachidonic acid. Functional analysis of genome wide expression data suggests that lipid metabolism and transport related pathways are significantly altered in the presence of arachidonic acid. (b) Heat map shows all the C. parapsilosis genes upregulated in presence of arachidonic acid. For further information see supplementary table S1.
Table 1.
Six up-regulated genes from the RNA sequencing data analysis and their homologues in C. albicans and their fold change expression values in C. parapsilosis.
| CPAR2 GeneID | Candida albicans homologue | Fold change |
|---|---|---|
| CPAR2_807710 | POX1-3(2) | 2.48 |
| CPAR2_205500 | ECI1 | 2.48 |
| CPAR2_102550 | FAA21 | 2.19 |
| CPAR2_800020 | POT1 | 2.10 |
| CPAR2_807700 | POX1-3(1) | 1.80 |
| CPAR2_603600 | FET3 | 1.63 |
Table 2.
Confirmation of the RNA sequencing data: fold change values of the 6 selected genes in both C. parapsilosis strains determined by qRT-PCR analysis.
| CPAR GeneID | CLIB | GA1 |
|---|---|---|
| CPAR2_807710 | 20.96 | 4.70 |
| CPAR2_205500 | 7.27 | 2.41 |
| CPAR2_102550 | 11.81 | 3.14 |
| CPAR2_800020 | 3.63 | 2.17 |
| CPAR2_807700 | 7.48 | 1.08 |
| CPAR2_603600 | 0.67 | 3.21 |
In order to determine the role of the identified genes in eicosanoid biosynthesis, we generated homozygous deletion mutant strains for each candidate gene by applying a gene disruption method previously introduced by Holland et al [20].
Homozygous deletion mutants of CPAR2_603600, CPAR2_800020 and CPAR2_807710 genes showed a significant reduction in extracellular lipid mediator production
Using liquid chromatography-mass spectrometry (LC/MS), we analyzed all null mutant strains for their ability to produce eicosanoids. This approach has previously demonstrated that the storage of arachidonic acid results in the production of auto-oxidation products [21,22].
Therefore, we also incubated 100 µM arachidonic acid in PBS at 30°C for 24 hours to measure the amount of spontaneously produced eicosanoids without the presence of fungal cells.
The LC/MS data for the secretory eicosanoid analysis revealed that the deletion mutant strains of CPAR2_603600, CPAR2_800020 and CPAR2_807710 produced less prostaglandin D2 (PGD2), although in case of CPAR2_807710 this reduction was not significant. Further, all three mutant strains showed a significant decrease in PGE2, production. In contrast, a reduction in 15-keto-prostaglandin E2 (15-keto-PGE2) production was measurable only in case of CPAR2_807710 (Figure 2). CPAR2_102550Δ/Δ, CPAR2_205500Δ/Δ and CPAR2_807700Δ/Δ did not show any difference in PGD2, PGE2 and 15-keto-PGE2 production (Figure S2).
Figure 2.
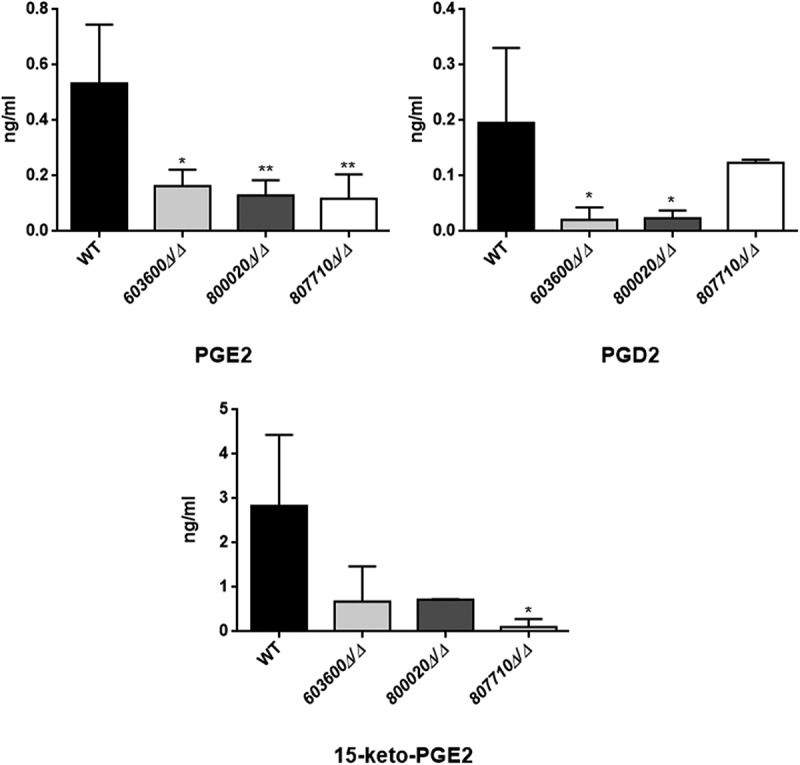
Reduced eicosanoid production by C. parapsilosis mutant strains. C. parapsilosis CLIB 214 wild type strain and three null mutant strains 603600Δ/Δ, 800020Δ/Δ and 807710Δ/Δ were grown for 24 hours at 30°C in the presence of 100 μM arachidonic acid in PBS. PBS with only arachidonic acid served as control. After sterile filtration, 100 µl of cell-free samples were analyzed with LC/MS. Among the examined eicosanoids, the three null mutants showed significant reductions in PGE2 (CPAR2_603600Δ/Δ, CPAR2_800020Δ/Δ and CPAR2_807710Δ/Δ) and one strain in 15-keto-PGE2 (CPAR2_807710∆/∆) production. Strains 603600∆/∆ and 800020∆/∆ had a strong trend toward a lower production of PGD2 compared to the wild type strain. *P < 0.05, **P < 0.01.
Fungal 5-D2-isoprostane identified during LC/MS analysis
The LC/MS data for the secreted eicosanoids also revealed, that C. parapsilosis incubations gave rise to an uncommon isoprostane likely identified to be a 5-D2-IsoProstane which is considered an autoxidative isomer of PGD2 (Figure 3). Investigating the selective ion trace m/z 351 -> 115 and comparing retention times (RT) and tandem mass spectra we observed a signal which overlapped with an authentic standard of lipoxin A4 (LXA4), however, some differences in the fragment intensities during tandem mass spectrometry prompted us to further evaluate the identity of this signal. We used a secondary solvent system for confirmatory analysis (data not shown). This analysis revealed that the signal suspected to be LXA4 was indeed something else as authentic standard and analyte no longer co-eluted. In order to propose a possible structure for the obtained signal we carried out tandem mass spectrometry as well as MS3 analysis of the obtained peak. As can be seen from Figure 3, based on the mass spectrometric data, we propose the observed oxylipin to be a 5-D2-IsoP [23]. The fragment m/z 115 is indicative of a 5-hydroxy group, in combination with the MS/MS fragments m/z 217 and 271 as well as the MS3 fragments m/z 215 and 106 we propose 5-D2-IsoP to be a likely candidate molecule for the observed signal. According to our data, besides the low production of prostaglandins, mutant strains incubations of CPAR2_603600, CPAR2_800020 and CPAR2_807710 also contained less 5-D2-IsoP compared to the wild type (Figure 3). This reduction was significant in all the mutant strains. No significant differences were found in case of CPAR2_102550Δ/Δ, CPAR2_205500Δ/Δ and CPAR2_807700Δ/Δ in terms of 5-D2-IsoP (Figure S2).
Figure 3.
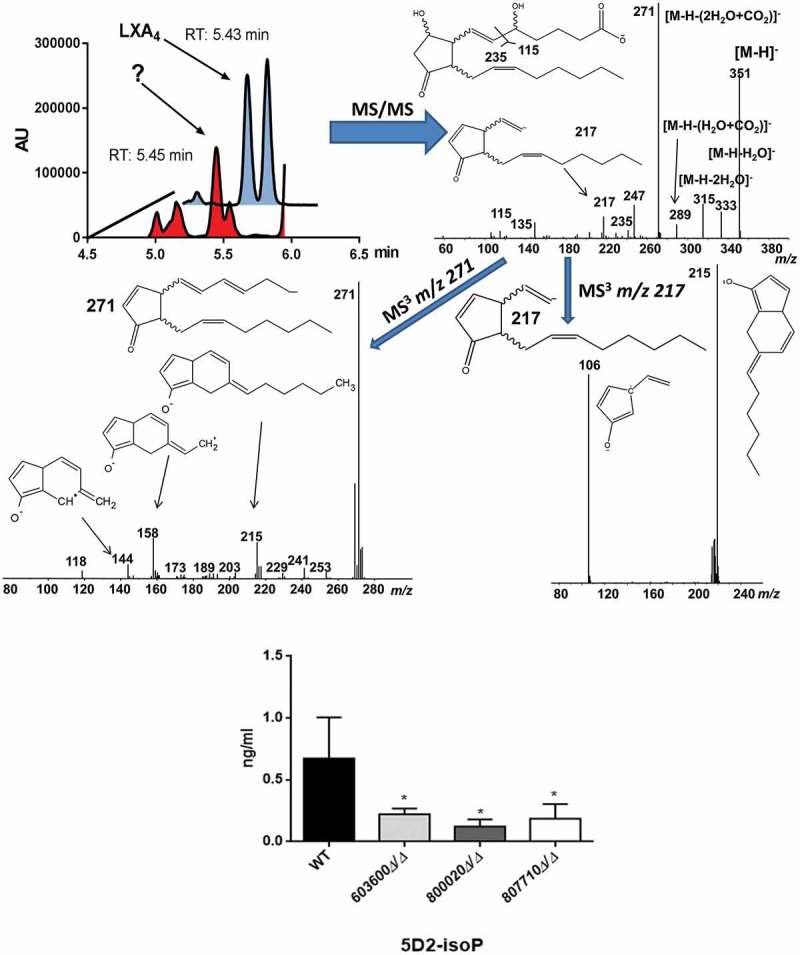
Suggestive identification and measurement of fungal 5D2-IsoP in wild type and eicosanoid mutant strains. Upper left, LC/MS analysis of cell supernatant after growing C. parapsilosis wild type strain in presence of 100 μM arachidonic acid in PBS. Blue trace shows the elution of an authentic LXA4 standard (1 ng/mL) in the transition m/z 351-> 115. Red trace shows the elution of an overlapping signal in a representative C. parapsilosis sample. (Upper right) MS/MS spectrum of the signal co-eluting with LXA4 and chemical structure of the candidate molecule 5D2-IsoP. Middle MS3 spectra of m/z 271 (left) and m/z 217 (right). Lower panel, CPAR2_800020Δ/Δ showed a significant reduction in 5D2-IsoP production, although 5D2-IsoP levels also decreased in CPAR2_603600Δ/Δ and CPAR2_807710Δ/Δ strains.
Phagocytosis and killing of 603600∆/∆, 800020∆/∆ and 807710∆/∆ mutants by human macrophages
Human peripheral blood monocyte derived macrophages (PBMC-DM) were used to characterize the virulence properties of mutant strains with altered eicosanoid producing profiles. We first examined the phagocytic activity of PBMC-DM by fluorescence-activated cell sorting (FACS). Candida yeast cells were labeled with the fluorescent dye Alexa Fluor 488 (a succimidyl ester) and then co-incubated with PBMC-DMs for 2 hours. Our results indicated that PBMC-DMs ingested each of the mutant strains more efficiently compared to the wild type strain (Figure 4). We also examined the yeast cell killing efficiency of PBMC-DMs by comparing the recovered fungal CFUs. Our data showed that each of the mutant strains were killed more effectively by PBMC-DMs in comparison to the wild type strain (Figure 5(a)).
Figure 4.
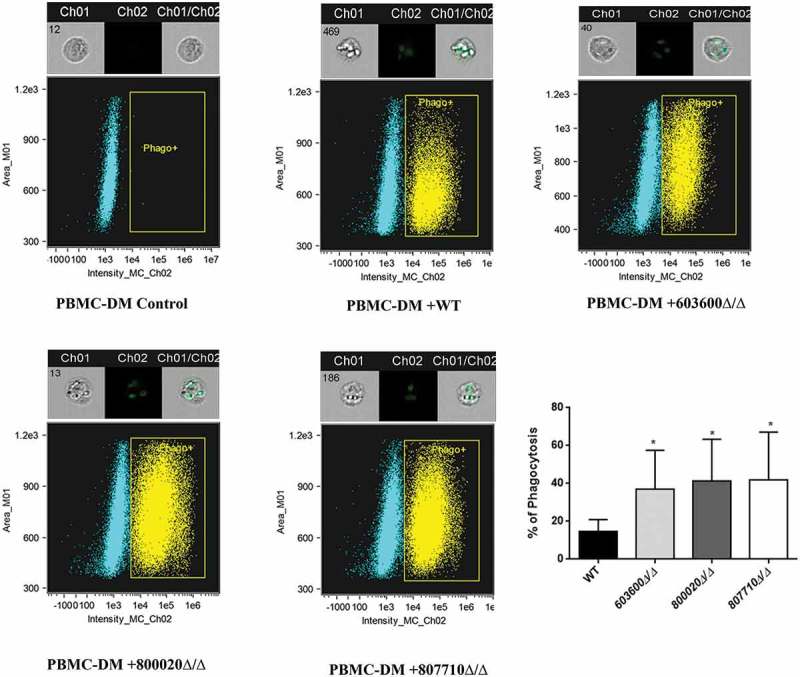
Phagocytosis of wild type and eicosanoid mutant strains by human macrophages by imaging flow cytometry. The phagocytosis of wild type and C. parapsilosis eicosanoid mutant cells by PBMC-DMs was analyzed by an imaging flow cytometer. Yeast cells were labeled with AlexaFluor447 and co-incubated with macrophages for 2 hours at 37°C with 5% CO2. Data were obtained from three independent experiments. Representative dot plots and summarized data of the flow cytometric analysis are shown. *P < 0.05.
Figure 5.
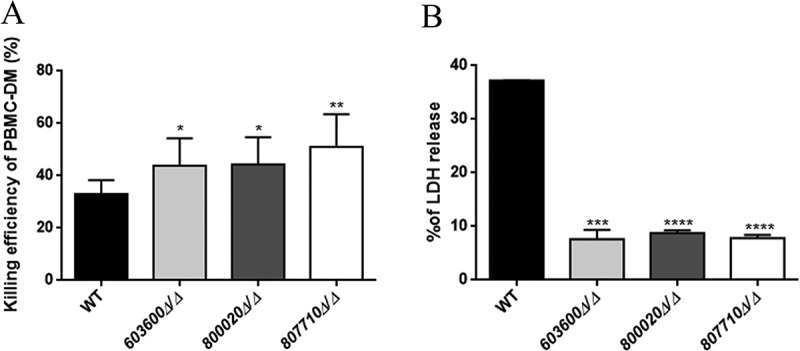
Killing of C. parapsilosis strains by human PBMC-DMs and host cell damage. (a) The efficiency of killing by human macrophages was analyzed by CFU determination. Experiments were performed in triplicates. The obtained data represents the killing efficiency of macrophages gained from 5 healthy donors. (b) Human PBMC-DMs were infected with wild type and C. parapsilosis eicosanoid mutants for 24 hours and LDH release was measured. LDH release is expressed as % of positive control. *P < 0.05, **P < 0.01, ***p < 0.002, ****p < 0.0001.
Host cell damage is decreased by 603600∆/∆, 800020∆/∆ and 807710∆/∆ strains
Next, we examined the mutant strains’ abilities to cause host cell damage by measuring the amount of LDH released by human PBMC-DMs following infection. As shown in Figure 5(b), we found that the PBMC-DMs showed significantly lower LDH release when infected with the mutants compared to the wild type. Our results indicated that 603600∆/∆, 800020∆/∆ and 807710∆/∆ mutant strains show a lower host cell damaging capacity than wild type cells (Figure 5(b)).
Macrophages favor the uptake of 603600∆/∆, 800020∆/∆ and 807710∆/∆ strains over the wild type
We tested the uptake efficiency of the mutants by human PBMC-DMs using a competition assay and compared them to the wild type strain. During the assay, we used differently labeled Candida strains: a GFP tagged wild type strain and mCherry labeled mutant strains. PBMC-DMs were infected with 2 types of fungal cells mixed in a ratio of 1:1 and co-incubated for 2 hours. Interactions were monitored via fluorescent imaging. The percentage of internalized cells was calculated for each strain, and the values for the mutants were compared to those of the wild type’s (Figure 6(a)). We calculated the percentage of phagocytosis by counting the total number of PBMC-DMs (Figure 6(b)). The number of green and red cells inside a single PBMC-DM which phagocytosed both type of cells was also analyzed (Figure 6(c)). Overall our results revealed that, PBMC-DMs significantly preferred the uptake of 603600∆/∆, 800020∆/∆ and 807710∆/∆ cells over the wild type. However, when the PBMC-DMs phagocytosed both type of cells we did not find any significant preference for the mutant cells.
Figure 6.
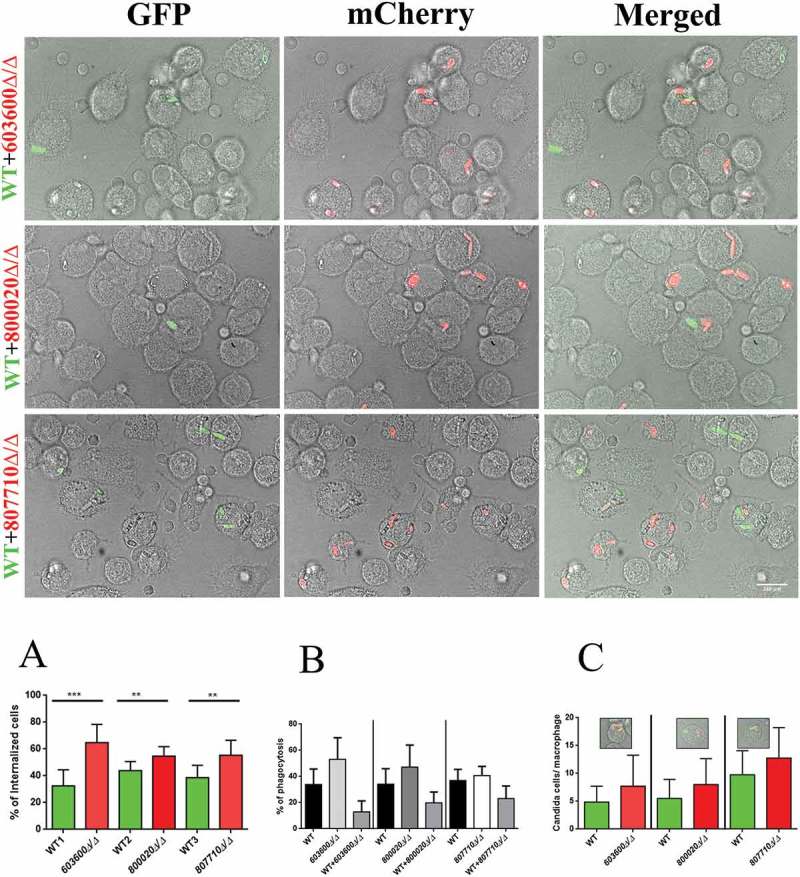
Phagocytic competition assay. The competition assays were performed using combinations of GFP tagged wild type C. parapsilosis and mCherry tagged eicosanoid mutant strains. Equal numbers of yeast cells expressing GFP or mCherry were mixed together and added to human PBMC-DMs at the MOI of 1:2. (a) Significantly higher number of mutant cells (red) were internalized compared to the wild type cells (green). (b) Percentage of phagocytic macrophages in each case (c) Number of macrophages with both wild type and mutant strain in each case did not show any difference. **P < 0.01, ***p < 0.002.
Influence on phagosome-lysosome fusion
Next, we addressed whether the lack of the examined lipid mediators had any effect on phagosome-lysosome fusion in human PBMC-DMs. We analyzed the phagosome-lysosome co-localization after co-incubating pHrodo stained Candida cells with PBMC-DMs for 2 hours. Although, all three mutants are phagocytosed and killed more efficiently by PBMC-DMs, only the 603600∆/∆ strain induced a higher rate of phagosome-lysosome fusion, suggesting the corresponding gene’s influence on phagosome maturation (Figure 7).
Figure 7.
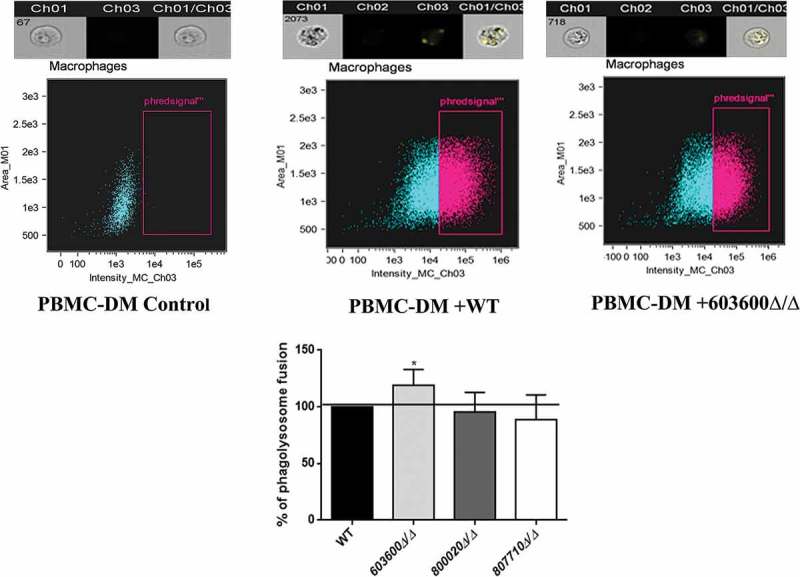
Phagosome-lysosome fusion in response to wild type and eicosanoid mutants. Phago-lysosome co-localization in human PBMC-DMs following the phagocytosis of pHrodo labeled Candida cells. Representative picture of a phagocytosing macrophage during quantitative imaging analysis. Ch1: brightfield image, Ch3: green fluorescence channel. Graph showing the difference between the extent of phago-lysosome fusion in case of the wild type and the mutants. *P < 0.05.
Reduction in prostaglandin production alters the cytokine response
Eicosanoid derived lipid mediators effectively regulate inflammatory responses. Multiple functions, dependent on concentration, location and timing have been described for the prostaglandins (e.g.PGD2 and PGE2) [1-3,24,25]. In order to examine the immunological responses triggered by the 603600∆/∆, 800020∆/∆ and 807710∆/∆, we stimulated human PBMC-DMs for 24 hours with each strain and determined the amount of cytokine and chemokine production. During the experiments we measured pro-IL1β, TNFα, Interleukin-1 receptor antagonist (IL-1ra), interleukin-6 (IL-6), interleukin-8 (IL-8) and interleukin-10 (IL-10) levels.
PBMC-DMs infected with 807710∆/∆ produced significantly higher amounts of the pro-inflammatory cytokine, Il-6, TNFα, chemokine IL-8 and IL-1ra levels compared to the wild type strain. Similarly, PBMC-DMs stimulated with 603600∆/∆ and 800020∆/∆ produced higher amounts of Pro-IL-1β, IL-1ra, Il-6 and TNFα compared to the reference strain, although these differences were not significant. Further, these two mutant strains did not induce IL-8 secretion. In terms of IL-10 production, none of the mutant strains showed a significant difference compared to the wild type (p > 0.05), however, 807710∆/∆ showed a trend towards higher levels of release (Figure 8(a)).
Figure 8.
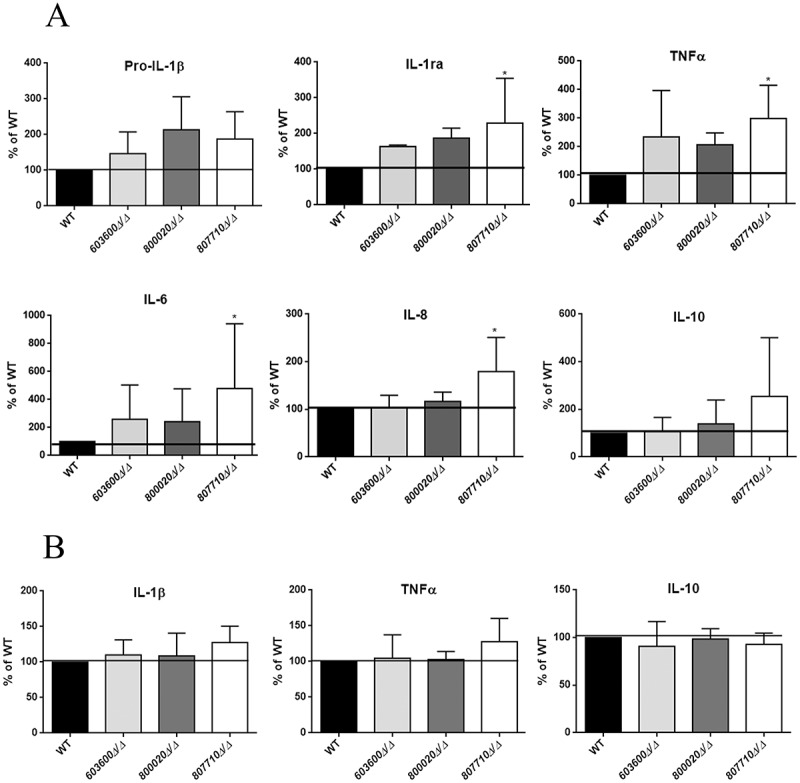
Cytokine secretion of human macrophages in response to wild type and eicosanoid mutants. (a) Pro IL-1β, IL-1ra, TNFα, IL-6, IL-8, and IL-10 levels were measured by ELISA after stimulation of PBMC-DMs with wild type or eicosanoid mutant strains for 24 hours. Data were normalized for each donor to cytokine levels induced by the wild type strain (100%) to minimize donor to-donor variability. (b) IL-1β, TNFα and IL-10 were also measured after infection of human PBMCs with the same strains. Data represent % cytokine production ± SEM for 6 donors. *P < 0.05.
We analyzed the cytokine production by human primary PBMCs following fungal stimuli. PBMCs infected with 807710∆/∆ showed a trend towards a higher IL-1β and TNFα (Figure 8(b)) release, although there was no difference in case of the other mutant strains. These data suggest that fungal eicosanoids may also influence the host cytokine response.
603600∆/∆, 800020∆/∆ and 807710∆/∆ strains show attenuated virulence in vivo
Following in vitro studies, we also aimed to examine the virulence of the 603600∆/∆, 800020∆/∆ and 807710∆/∆ in vivo using a mouse model of disseminated candidiasis. Following the intravenous infection of BALB/c mice, the fungal burdens of different organs were determined three days after the infection. After CFU recovery, we found that mice infected with 603600∆/∆ showed significantly reduced fungal burdens in the liver and kidneys, while 800020∆/∆ inoculated mice also revealed lower fungal burdens in the liver and kidneys. Finally, CFUs recovered following 807710∆/∆ injection were significantly less in the spleen and kidneys compared to those recovered from wild type-infected mice (Figure 9). These results indicate that these eicosanoid biosynthesis genes play role in C. parapsilosis virulence.
Figure 9.
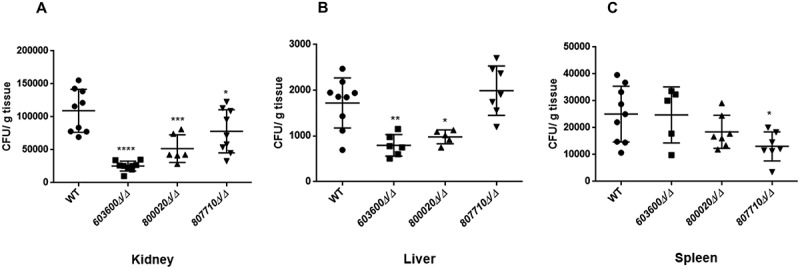
Fungal burden in organs after intravenous infection. Five mice were infected intravenously with 2 × 107/100 µl of C. parapsilosis wild type or eicosanoid mutant cells. CFUs recovered from kidney (a), liver (b), and spleen (c) after 3 days of the infection. The results are pooled data from two independent experiments. *P < 0.05, **P < 0.01, ***p < 0.002, ****p < 0.0001.
Discussion
Eicosanoids, a group of bioactive mediators, are signaling molecules with diverse physiological and pathological functions known to modulate inflammatory responses. Prostaglandins, leukotrienes and lipoxins are groups of eicosanoids involved in pro-, and/or anti-inflammatory responses. During vasodilation prostaglandins and leukotrienes induce increased permeability of post capillary venules and also recruit complement components and leukocytes to the site of inflammation [2]. In contrast, lipoxins act as pro-resolving lipid mediators, as for example, LXA4 inhibits the recruitment of neutrophil and eosinophil granulocytes in post capillary venules [3]. Previously, it has been hypothesized that, during fungal infection the invading fungi induce host prostaglandin biosynthesis that causes a local anti-inflammatory response, which in turn could contribute to fungal invasion [26].
In recent years, several studies have revealed that human pathogenic fungi are able to produce lipid mediators, specifically prostaglandins that might as well contribute to their virulence. This group of pathogens includes species such as A. nidulans, A. fumigatus, C. neoformans, C. albicans and C. parapsilosis.
Investigations have revealed several biosynthetic pathways used by some of these pathogens. As an example, A. nidulans and A. fumigatus have three dioxygenase-encoding genes, namely ppoA, ppoB and ppoC with high sequence similarity to mammalian cyclooxygenases [11]. Besides regulating sexual and asexual sporulation, all three ppo genes contribute to prostaglandin biosynthesis, possibly via oxygenating arachidonic acid, thereby generating the prostaglandin precursor PGH2 [11].
Other studies have revealed that certain pathogenic fungi, such as C. albicans and C. neoformans, do not possess COX-like enzymes, thus they must have evolved prostaglandin biosynthetic pathways different from those of mammals [12,13]. In C. neoformans, a member of the multicopper oxidase family, the Lac1 laccase regulates PGE2 biosynthesis, possibly by converting the prostaglandin precursor PGG2 to PGE2 and15-keto-PGE2 [27].
In C. albicans however, the fatty acid desaturase OLE2 and a multicopper oxidase FET3 play a role in prostaglandin production via novel pathways using exogenous arachidonic acid as precursor [9]. CaFET3, a laccase homolog (and also a member of the Fet family of multicopper oxidases) is suggested to regulate PGE2 biosynthesis through a mechanism similar to that of LAC1 in C. neoformans. On the other hand, Ole2, a putative delta9 desaturase also containing a cytochrome B domain, is hypothesized to regulate PGE2 production via the oxidation of exogenous arachidonic acids [9].
Notably, in these species, the identified genes are pleiotropic regulators as they also participate in mechanisms such as sporulation (ppo genes), cell wall homeostasis (LAC1) or iron uptake (FET3), and thus contribute to virulence in a complex manner. Nevertheless, the role of fungal prostaglandins in virulence is evident as, in all yet examined fungal species, their production has been associated with markedly altered immune responses [11,26]. To expand our knowledge about the role of fungal eicosanoids in pathogenesis, we examined lipid mediator production in another important human fungal pathogen, C. parapsilosis. As the incidence of this species has increased over the past two decades and the patient group at risk includes immunosuppressed children and adults as well as neonates, understanding the pathogenesis of C. parapsilosis has gained increased attention [16,28]. Preliminary studies have started to elucidate the prostaglandin profile of this species [17], however, the involved biosynthetic pathways and the presence or role of other fungal eicosanoids remained elusive. Previously, we have shown that in the presence of exogenous arachidonic acid, C. parapsilosis is capable of producing fungal prostaglandins, although OLE2 is not involved in the synthetic mechanisms, leaving the corresponding biosynthetic processes unexplored [17].
Therefore, in the current study, we aimed to reveal regulators involved in eicosanoid biosynthetic mechanisms and to investigate their roles in the virulence of this species. Following arachidonic acid induction, we identified three genes that significantly influence the biosynthesis of fungal prostaglandins. Our results indicate, that CPAR2_603600, a homologous gene of CaFET3 is involved in PGE2 and PGD2 production, CPAR2_807710, a homologue of the acyl-coenzyme A oxidase, ScPOX1-3, regulates PGE2 and 15-keto-PGE2 synthesis, and CPAR2_800020, a homologue of 3-ketoacyl-CoA thiolase, ScPOT1, influences PGE2 biosynthesis. Using LC/MS analysis, we observed that the disruption of each gene led to a decrease in the corresponding eicosanoids’ production.
While CaFET3 is known to interfere with fungal prostaglandin production [9], no such role has been associated with CaPOX1-3, and CaPOT1 in C. albicans, suggesting a novel function of the corresponding homologues in C. parapsilosis.
The roles of CaFET3 and CaPOT1 in C. albicans’ virulence have been investigated. Deletion of CaFET3 resulted in reduced adhesiveness to fibroblasts, although no significant differences were observed between the virulence of the wild type and the Δ/Δfet3 strain in a mouse model of systemic candidiasis [29]. In contrast with C. albicans, upon the removal of the ortholog of FET3 in C. parapsilosis indeed influenced the fungi’s virulence in vivo, given the lower amount of fungal burden measured. In contrast, CaPOT1, a 3-ketoacyl-CoA thiolase, involved in fatty acid utilization, is not required for virulence in an embryonated chicken egg infection model [30]. However, its ortholog in C parapsilosis is involved in virulence regulation as represented by both our in vitro and in vivo virulence studies. Unfortunately, to date, we lack information about the hypothetical acyl-coenzyme A oxidase’s (CaPOX1-3) role in C. albicans pathogenesis. Interestingly, our results also suggest the involvement of the corresponding gene’s ortholog in the virulence of C. parapsilosis.
These data suggest, that in contrast to C. albicans, the homologous genes of FET3, POT1 and POX1-3 in C. parapsilosis indeed contribute to fungal virulence, although the corresponding mechanisms still need to be elucidated.
Interestingly, CPAR2_603600 might be involved in delaying phagosome-lysosome fusion, a phenomenon not yet investigated in C. albicans, although additional studies are needed to confirm this mechanism hypothesis.
Fungal prostaglandins produced by C. albicans and C. neoformans alter host cytokine responses by down-regulating chemokine (IL-8) and pro-inflammatory cytokine (e.g. TNFα) production while concomitantly up-regulating anti-inflammatory responses via promoting IL-10 release [26]. Our results suggest a similar effect with C. parapsilosis eicosanoids, as mutant strains defective in prostaglandin production induced higher pro-inflammatory cytokine responses, as shown by the increased levels of Pro-IL-1β, IL-6 and TNFα released by human PBMC-DMs. Although, stimulation of human PBMCs did not result in a significant increase of the examined cytokines in case of any of the mutant strains. These data suggest that CPAR2_807710, CPAR2_800020 and CPAR2_603600 contribute unequally to the alteration of host immune responses.
This is the first study reporting the presence of 5D2-IsoP in fungal incubations that might actively contribute to fungal virulence. We have identified three C. parapsilosis eicosanoid biosynthesis regulatory genes, namely CPAR2_807710, CPAR2_800020 and CPAR2_603600, that are involved in the production of fungal prostaglandins. Virulence studies performed with the corresponding null mutant strains suggests that these regulatory genes also influence the fungi’s virulence. Although, further investigation is needed to thoroughly understand the importance of fungal eicosanoids, our results can contribute to a better understanding of host pathogen interactions during candidiasis.
Materials and methods
Strains and growth conditions
All C. parapsilosis strains used in this study are listed in Table S2. Strains were grown in YPD (1% dextrose, 1% peptone and 0.5% yeast extract) at 30°C. For colony selection 2% agar was added in the media. Nourseothricin (NAT) resistant transformants were selected on YPD plates supplemented with 100 μg/ml NAT. Cells transformed with LEU2 and HIS1 markers were selected on synthetic complete media (SC; 2% Dextrose, 0.95% yeast nitrogen base, mixture of amino acid, 2% agar) without leucine and histidine.
RNA extraction
For RNA sequencing, the C. parapsilosis GA1 strain was grown overnight in 2 ml of YPD media at 30°C with continuous shaking applied at 180 rpm. The next day, cells were washed 3 times with PBS and then counted using a Burker’s chamber. Cell concentration was adjusted to 2 × 107 cells per 10 ml PBS supplemented with 500 μM of arachidonic acid in triplicates. As a control, cells were also grown in 10 ml PBS supplemented with ethanol only, as it is the dissolving agent used for arachidonic acid solubilization. After 3 hours of growth at 30°C, RNA was isolated using the Ribopure Yeast RNA isolation Kit (Ambion) following the manufacturer’s instructions. For validating the RNA sequencing data, C. parapsilosis CLIB and GA1 cells were grown as described above and RNA extraction was performed using the same kit.
RNA sequencing and data analysis
RNA-seq library preparation and sequencing, paired-end reads (Illumina Truseq V2 PolyA, not -stranded, V3-150, 2 × 75bp, 25M read) were generated from three biological replicates of induced and non-induced C. parapsilosis cells.
For initial quality assessment and data preprocessing we used FastQC 0.10.1 [http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/] quality assessment and Trimmomatic v0.32 [31] where preset conditions were applied for quality cutoff to remove low quality regions from the raw data [parameters used LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.]. Mapping was carried out against the reference genome obtained from the Candida Genome Database [19]. We used the splice junction sensitive mapper Tophat 2.01.13 [32] with default settings, and mapped by applying the bowtie 2.2.4 [33] short read mapper. The counts per gene were estimated by using flux-capacitor [34]. To estimate differential expression, we used the R package Deseq2 [35] with a cutoff of log2fold change of 1.5. Gene Onthology enrichment was performed using the CGD database [19].
Reverse transcription PCR
A total of 500 ng RNA was used for cDNA synthesis. The cDNA was synthesized using the Revert Aid first Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions.
Real time PCR
Real time PCR was performed in a final volume of 20 μl using Maxima SYBR Green/Fluorescein qPCR Master Mix (2X) (Thermo Scientific). The reaction was performed in C1000 Thermal Cycler (Bio-Rad) using the following protocol: 95°C for 3mins, 95°C for 10 s, 60°C for 30 s, 65°C for 5 s for 50 cycles. Fold change in mRNA expression was calculated by ΔΔCt method (Real–Time PCR applications guide BIO-Rad). TUB4 gene was used as a housekeeping gene for an internal control.
Preparation and transformation of competent cells
All the deletion mutants and fluorescently labeled strains were constructed by chemical transformation. The parental CLIB 214 double auxotrophic strain (CPL2H1) [20] and the mutants were inoculated in 2 ml YPD and grown overnight at 30°C with shaking applied at 180 rpm. Then the culture was diluted to an OD600 of 0.2 in 30 ml YPD broth and grown at 30°C to an OD600 of 0.8–1. The culture was centrifuged at 4000 rpm for 5 min and the pellet was suspended in 3 ml of ice-cold water. The cells were again centrifuged at 4000 rpm for 5 min and washed with 1 ml ice cold TE-LiOAC (0.1M lithium acetate, 10 mM TRIS and 1mM EDTA). After centrifugation the final pellet was dissolved in 300 μl of ice cold TE-LiOAC. A transformation mix was prepared with 100 μl competent cells, 10 μl single stranded salmon sperm DNA (10 mg/ml) and 25–30 μl fusion PCR product and the mixture was kept at 30°C for 30 min. Finally, 700 μl PLATE (0.1M lithium acetate, 10 mM tris, 1mM EDTA and 40% PEG 3350) was added to the transformation mix and cells were kept overnight at 30°C. The next day, cells were heat shocked at 44°C for 15 min, centrifuged and washed with 1 ml of YPD media. The cells were then re-suspended in 1 ml of YPD and incubated at 30°C at 200 rpm for 2–3 hours. After the incubation, the cells were centrifuged and dissolved in 100 μl of YPD, then plated on the respective agar plates.
Generation of C. parapsilosis mutant strains
For gene deletion, the fusion PCR technique was used, as described previously by Holland et al [20]. Target genes are listed in Table 2 (Figure 1) and the primers are used listed in TabS3. Approximately 500bp upstream and 500bp downstream sequences of the target genes were amplified with primer pairs 1,3 and 4,6 using DreamTaq polymerase (Thermo Scientific). Selection markers LEU2 and HIS1 were amplified with primer pairs 2 and 6 from the plasmids pSN40 (LEU2) and pSN52 (HIS1). Finally, all the PCR products were purified using the PCR Purification kit (Quiagen) and fused with Phusion Taq polymerase (Thermo Scientific) using primers 1and 6. The resulting disruption cassette was transformed into the double auxotrophic CLIB 214 strain.
Construction of fluorescent tagged strains
All the C. parapsilosis fluorescently labeled strains were generated using the Gateway® cloning technology (Invitrogen). The fluorescent constructs were expressed under an overexpression promoter targeted in the NEUT5 locus. The fluorescence expression was confirmed after checking the strains under fluorescent microscope.
Sample preparation for secreted eicosanoid measurement
For eicosanoid measurement, 2 × 107 Candida cells were inoculated in 10 ml of PBS+ 100 μM arachidonic acid in triplicates and incubated at 30°C for 24 hours. Three tubes with only PBS and 100 μM arachidonic acid were also incubated and served as control. For secreted eicosanoids, the cell supernatant was collected using sterile filtration after centrifugation. 100 µl sample was complemented with 96 µl MeOH and 4 µl internal standard solution (final concentration: 1 ng/ml PGE2-d4, LTB4-d4 and 15-HETE-d8 and 10 ng/ml DHA-d5) and analyzed without further processing.
Analysis of lipid mediators by LC/MS
Eicosanoids were analyzed using a targeted LC-MS/MS method according to published protocols with a minor modification [36]. The drying temperature was set to 450°C instead of 400°C. Table S4 shows the MRM characteristics of the monitored eicosanoids. When concentrations were determined external calibration was carried out with linear regression using a weighing factor of 1/x2. For calculating the actual concentration of the secreted eicosanoids, the amount of spontaneously generated eicosanoids (PBS+ AA control sample) were subtracted from the total amount of eicosanoids produced by each strain (data not shown). MS3 analysis were carried out under identical conditions using an excitation energy (AF2 value) of either 0.5 or 0.8 (V) as collisional energy in the MS3 experiments, investigating the MS/MS fragments m/z 217 and 271.
Isolation and differentiation of PBMCs
Human PBMCs were isolated from buffy coats of healthy individual by density gradient centrifugation [37]. Briefly, blood samples were first diluted with PBS (1:1 dilution). The diluted blood was overlaid above the Ficoll solution (Ficoll Paque PLUS-GE Healthcare) and then centrifuged at 900 rpm for 30 mins. After centrifugation, the mononuclear cell layer was aspirated with a pipette, collected in a separate 50 ml centrifuge tube and then washed 3 times with PBS. Finally, cells were suspended in RPMI 1640 medium (Lonza) supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were counted and the concentration was adjusted to 5 × 106 cells/ml. For later experiments, 100 μl of 5 × 105 cells in RPMI were added to U-bottom 96-well plates. For macrophage differentiation, cells were added to flat bottom 24-well plates with the concentration of 1ml of 1 × 107 cells per well and incubated for 90 mins at 37°C in the presence of 5% CO2 and 100% humidity. After the incubation, RPMI was removed and cells were washed with 1 ml of PBS. Finally, the cells were re-suspended in X-VIVO 15 media (Lonza) supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin and 10ng/ml recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF, Sigma-Aldrich). The media was changed every 2nd day for 7 days. During the study, PBMCs were used for cytokine measurement only.
Infection of PBMCs and PBMC-DMs
For the infection experiments, C. parapsilosis strains were grown overnight in 2 ml YPD media, washed 3 times with PBS and adjusted to the proper concentrations used for each experiment. For the infection of both human PBMCs (cytokine measurement only) and PBMC-derived macrophages, the multiplicity of infection (MOI) was 1:5. Candida cells were dissolved in 100 μl of either RPMI/PS (PBMC) or GM-CSF-free X-VIVO 15 (PBMC-DM) medium and were added to host cell containing wells.
Killing assay
Human PBMC-DMs (5 × 105/well) were co-incubated with C. parapsilosis cells in 24-well plastic culture plates at a MOI of 1:5. As a control, equal amounts of Candida cells were incubated in the GM-CSF free X-VIVO 15 medium only (500 μl/well). After 3 hours of incubation, macrophages were lysed with PBS + 4% Triton X-100 solution. Control wells with yeast cells only were treated similarly. Finally, the lysates and the cells were serially diluted, plated on YPD plates and incubated for 2 days at 30°C. After incubation, the number of CFUs was determined and the recovered CFUs from each strain were compared. The killing efficiency was calculated as follows: (number of live Candida cells in control wells – number of live Candida cells in co-cultures)/number of live Candida cells in control wells × 100. The experiments were performed with PBMC-DMs derived from five independent donors with triplicates.
Phagocytosis assay using flow cytometry
For the phagocytosis assays, Candida cells were labeled with the fluorescent dye Alexa Fluor 447 carboxylic acid succimidyl ester (Invitrogen) and measured using the FlowSight Imaging flow cytometer (Amnis). For labeling, the yeast cell suspension (100 µl containing 109 cells) was treated with 11 μl of Na2CO3 (1 M, pH 10) and 2 μl Alexa Fluor 447 (1 mg/ml in DMSO) was added followed by incubation for 1 hour at room temperature, carefully protected from light. After the incubation, the cells were washed four times with PBS and adjusted to the proper concentration of 2.5 × 106 (MOI of 1:5). Human PBMC-DMs (5 × 105/well) were infected with labeled Candida cells and kept for 2 hours at 37°C with 5% CO2. Then, the non-phagocytosed Candida cells were washed with PBS and the yeast containing macrophages were detached from the cell culture plates with TrypLETM Express solution (Gibco). Finally, macrophages were collected in FACS buffer (0.5% FBS in PBS), centrifuged and dissolved in 200 μl PBS and measurement using the flow cytometer. The obtained raw data was analyzed by the IDEAS Software (Amnis). The experiments were performed with PBMC-DMs derived from five independent donors.
Phagocytosis competition assay
GFP (wild type) and mCherry (mutant strains) expressing C. parapsilosis strains were grown overnight in YPD at 30°C at 200 rpm. Next day, the cells were harvested by centrifugation at 3000 rpm for 5 mins and washed with PBS. PBMC-DMs were infected with a mixture of fungal cells (wild type and a mutant strain, mixed in a ratio of 1:1), and were co-incubated at 37°C for 2 hours. Then the co-culture was incubated and then washed 2–3 times with PBS before visualized under the fluorescent microscope [38]. At least 300 macrophages and 150 yeast cells were counted for each experiment and the experiments were performed with macrophages derived from three independent donors.
LDH measurement
We determined the release of lactate dehydrogenases from the supernatant, as an indicator of host cell death, by the LDH cytotoxicity detection kit (Takara) according to the manufacturer’s instructions. Cytotoxicity was calculated after subtracting the absorbance value of the infected control from all the test samples: cytotoxicity (%) is calculated as follows: (ODExperimental value/ODPositive control) × 100. Supernatant collected from the cells treated with 1% TritonX-100 was used as positive control. The experiments were performed with PBMC-DMs generated from five independent donors.
Analysis of phagosome lysosome fusion by FACS
For phagosome-lysosome fusion analysis by quantitative imaging (Amnis Flow Sight), the Candida cells were labeled with the fluorescent dye pHrodo® (Invitrogen). First, the yeast cell suspension (100 µl containing 109 cells) was treated with 11 μl of Na2CO3 (1 M, pH 10) and then 2 μl pHrodo (1 mg/ml in DMSO) was added followed by incubation for 1 hr at room temperature in the dark. After the incubation, cells were washed 4 times with HBSS buffer (Hank’s Balanced Salt Solution) (LONZA) and adjusted to the proper concentration (see phagocytosis assay protocol). Human PBMC-derived macrophages were infected with labeled Candida cells at a 1:5 ratio and kept for 2 hrs to allow phagocytosis. Then, the non-phagocytosed Candida cells were washed with 1xPBS and macrophages were detached from cell culture plates by TrypLETM Express solution (Gibco). Finally, macrophages were collected in FACS buffer (0.5% FBS in 1xPBS), centrifuged and dissolved in 1xPBS. Phago-lysosome fusion was then measured by Amnis FlowSight. Data were analyzed by the IDEAS Software (Amnis).
Cytokine measurement by ELISA
The concentration of different cytokines was analyzed by commercial ELISA kits according to the manufacturer’s instructions. The IL-1β, IL-1ra, IL-6, TNFα, IL-10 and IL-8 ELISA kits were obtained from R&D Systems (Abingdon, United Kingdom). For the pro-IL1β measurement, macrophages were lysed by repeated freeze thaw cycles and then the supernatant was collected [39]. The experiments were performed with both PBMCs and macrophages derived from peripheral blood mononuclear cells (PBMC-DMs) were used from at least five independent donors.
Mouse model of systemic Candida infection and fungal burden
For the experiments, a non-lethal experimental mouse model of disseminated candidiasis was used [40]. Briefly, groups of five 8–12-weeks-old female Balb/c mice (22 – 27 g. of weight) were infected with 2 × 107/100 μl C. parapsilosis cells via the lateral tail vein using a syringe with a 32-gauge needle. A group of control mice was injected with 100 µl of sterile PBS. Two independent experiments were performed. The mice were maintained with sterile water and food ad libitum. Three days following infection, animals were euthanized, and their livers, kidneys and spleens were aseptically removed. The organs were weighed and homogenized in sterile PBS with a tissue grinder. CFUs were determined after 48 hours of incubation at 30°C, and CFU/g tissue was calculated.
Ethics statement
The in vivo mouse infection experiments were performed according to National (1998. XXVIII; 40/2013) and European (2010/63/EU) animal ethics guidelines. The experimental protocols were approved by the Animal Experimentation and Ethics Committee of the Biological Research Centre of the Hungarian Academy of Sciences and the Hungarian National Animal Experimentation and Ethics Board (clearance number: XVI./03521/2011.). The University of Szeged granted permissions XII./00455/2011 and XVI./3652/2016 to work with mice.
For PBMC isolation, blood was collected from healthy individuals. The Institutional Human Medical Biological Research Ethics Committee of the University of Szeged gave approval for the procedure and the respective consent documents. Healthy individuals provided written informed consent. The experiments were performed in accordance with guidelines and regulations of the Ethics Committee of University of Szeged and the experimental protocols were approved by the same institutional committee.
Statistical analysis
Unpaired t-tests were used to determine differences between groups in case of phagocytosis assay, killing assay, LDH assay and phagocytic competition assay. Differences were considered statistically significant at P ≤ 0.05. One-way ANOVA with Dunnett’s Post-Hoc test was used to determine the differences between groups in case of eicosanoid analysis, cytokine analysis and in vivo experiment. Graph Pad Prism 6 software was used to perform the statistical analysis.
Funding Statement
TC was supported the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreements FP7-PEOPLE-2013-ITN-606786 ‘ImResFun’ and from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No H2020-MSCAITN-2014-642095. AG was funded by NKFIH NN 113153, by GINOP-2.3.2-15-2016-00035, by GINOP-2.3.3-15-2016-00006 and by OTKA-NKFIH K123952. RT was supported by TÁMOP 4.2.4. A/2-11-1-2012-0001 National Excellence Program - Elaborating and operating an inland student and researcher personal support system convergence program. MH and MG lab was supported by the Leiden University Medical Centre, Leiden, The Netherlands. ET was supported the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreements FP7-PEOPLE-2013-ITN-606786 ‘ImResFun’ and from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No H2020-MSCAITN-2014-642095. TG group acknowledges support of the Spanish Ministry of Economy and Competitiveness grants, ‘Centro de Excelencia Severo Ochoa 2013-2017ʹ SEV-2012-0208, and BFU2015-67107 cofounded by European Regional Development Fund (ERDF); from the CERCA Programme/Generalitat de Catalunya; from the European Union and ERC Seventh Framework Programme (FP7/2007-2013) under grant agreement FP7-PEOPLE-2013-ITN-606786, and a grant from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No H2020-MSCA-ITN-2014-642095.
Acknowledgments
We thank Christophe D’Enfert for sharing the plasmids for the Gateway system and Judith Berman for providing us with the GFP and mCherry plasmids. We also thank Tibor Németh for giving us the GFP expressing wild type strain for the phagocytic competition assay. We are thankful to Ferenc Jankovics from Biological Research Centre, Szeged for helping with the confocal microscopy. We would like to thank Csaba Papp for assisting with the confocal microscopy for the phagocytic competition assay; Katalin Csonka, Erik Zajta, Dhirendra N. Singh for helping with the in vivo mouse experiment. We thank Prof. Dr. Wilfried Niessen (hyphen Mass Spec) for his help with interpreting the MS/MS and MS/MS/MS spectra of the candidate molecule 5D2-IsoP. We are also thankful to Joshua D. Nosanchuk for critically reading the manuscript and give valuable suggestions.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental data
Supplemental data for this article can be accessed here.
References
- 1.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001. November 30;294(5548):1871–1875. PubMed PMID: 11729303. [DOI] [PubMed] [Google Scholar]
- 2.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015. August;15(8):511–523. PubMed PMID: 26139350; PubMed Central PMCID: PMC4606863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence T, Willoughby DA, Gilroy DW. Anti-inflammatory lipid mediators and insights into the resolution of inflammation. Nat Rev Immunol. 2002. October;2(10):787–795. PubMed PMID: 12360216. [DOI] [PubMed] [Google Scholar]
- 4.Suram S, Brown GD, Ghosh M, et al. Regulation of cytosolic phospholipase A2 activation and cyclooxygenase 2 expression in macrophages by the beta-glucan receptor. J Biol Chem. 2006. March 3;281(9):5506–5514. PubMed PMID: 16407295. [DOI] [PubMed] [Google Scholar]
- 5.Suram S, Gangelhoff TA, Taylor PR, et al. Pathways regulating cytosolic phospholipase A2 activation and eicosanoid production in macrophages by Candida albicans. J Biol Chem. 2010. October 1;285(40):30676–30685. PubMed PMID: 20643646; PubMed Central PMCID: PMC2945562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibata Y, Henriksen RA, Honda I, et al. Splenic PGE2-releasing macrophages regulate Th1 and Th2 immune responses in mice treated with heat-killed BCG. J Leukoc Biol. 2005. December;78(6):1281–1290. PubMed PMID: 16204627. [DOI] [PubMed] [Google Scholar]
- 7.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011. April;11(4):275–288. PubMed PMID: 21394104. [DOI] [PubMed] [Google Scholar]
- 8.Smeekens SP, Van De Veerdonk FL, van der Meer JW, et al. The Candida Th17 response is dependent on mannan- and beta-glucan-induced prostaglandin E2. Int Immunol. 2010. November;22(11):889–895. PubMed PMID: 21059767. [DOI] [PubMed] [Google Scholar]
- 9.Erb-Downward JR, Noverr MC. Characterization of prostaglandin E2 production by Candida albicans. Infect Immun. 2007. July;75(7):3498–3505. PubMed PMID: 17470538; PubMed Central PMCID: PMC1932954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erb-Downward JR, Noggle RM, Williamson PR, et al. The role of laccase in prostaglandin production by Cryptococcus neoformans. Mol Microbiol. 2008. June;68(6):1428–1437. PubMed PMID: 18410494; PubMed Central PMCID: PMC3973538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsitsigiannis DI, Bok JW, Andes D, et al. Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect Immun. 2005. August;73(8):4548–4559. PubMed PMID: 16040966; PubMed Central PMCID: PMC1201276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erb-Downward JR, Huffnagle GB. Role of oxylipins and other lipid mediators in fungal pathogenesis. Future Microbiol. 2006. August;1(2):219–227. PubMed PMID: 17661667. [DOI] [PubMed] [Google Scholar]
- 13.Fischer GJ, Keller NP. Production of cross-kingdom oxylipins by pathogenic fungi: an update on their role in development and pathogenicity. J Microbiol. 2016. March;54(3):254–264. PubMed PMID: 26920885; PubMed Central PMCID: PMC5107414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012. December 19;4(165):165rv13 PubMed PMID: 23253612. [DOI] [PubMed] [Google Scholar]
- 15.Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008. October;21(4):606–625. PubMed PMID: 18854483; PubMed Central PMCID: PMC2570155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Asbeck EC, Clemons KV, Stevens DA. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit Rev Microbiol. 2009;35(4):283–309. PubMed PMID: 19821642. [DOI] [PubMed] [Google Scholar]
- 17.Grozer Z, Toth A, Toth R, et al. Candida parapsilosis produces prostaglandins from exogenous arachidonic acid and OLE2 is not required for their synthesis. Virulence. 2015;6(1):85–92. PubMed PMID: 25654274; PubMed Central PMCID: PMC4603437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gacser A, Trofa D, Schafer W, et al. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest. 2007. October;117(10):3049–3058. PubMed PMID: 17853941; PubMed Central PMCID: PMC1974868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrzypek MS, Binkley J, Binkley G, et al. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017. January 4;45(D1):D592–D596. PubMed PMID: 27738138; PubMed Central PMCID: PMC5210628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland LM, Schroder MS, Turner SA, et al. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014. September;10(9):e1004365 PubMed PMID: 25233198; PubMed Central PMCID: PMC4169492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brose SA, Baker AG, Golovko MY. A fast one-step extraction and UPLC-MS/MS analysis for E2/D2 series prostaglandins and isoprostanes. Lipids. 2013. April;48(4):411–419. PubMed PMID: 23400687; PubMed Central PMCID: PMC3608832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galano JM, Lee YY, Oger C, et al. Isoprostanes, neuroprostanes and phytoprostanes: an overview of 25 years of research in chemistry and biology. Prog Lipid Res. 2017. October;68:83–108. PubMed PMID: 28923590. [DOI] [PubMed] [Google Scholar]
- 23.Korner A, Schlegel M, Theurer J, et al. Resolution of inflammation and sepsis survival are improved by dietary Omega-3 fatty acids. Cell Death Differ. 2018. February;25(2):421–431. PubMed PMID: 29053142; PubMed Central PMCID: PMC5762854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.FitzGerald GA. BIOMEDICINE. Bringing PGE(2) in from the cold. Science. 2015. June 12;348(6240):1208–1209. PubMed PMID: 26068834. [DOI] [PubMed] [Google Scholar]
- 25.Laan LC, Williams AR, Stavenhagen K, et al. The whipworm (Trichuris suis) secretes prostaglandin E2 to suppress proinflammatory properties in human dendritic cells. FASEB J. 2017. February;31(2):719–731. PubMed PMID: 27806992; PubMed Central PMCID: PMC5240662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noverr MC, Phare SM, Toews GB, et al. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun. 2001. May;69(5):2957–2963. PubMed PMID: 11292712; PubMed Central PMCID: PMC98248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erb-Downward JR, Huffnagle GB. Cryptococcus neoformans produces authentic prostaglandin E2 without a cyclooxygenase. Eukaryot Cell. 2007. February;6(2):346–350. PubMed PMID: 17158733; PubMed Central PMCID: PMC1797952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow BD, Linden JR, Bliss JM. Candida parapsilosis and the neonate: epidemiology, virulence and host defense in a unique patient setting. Expert Rev Anti Infect Ther. 2012. August;10(8):935–946. PubMed PMID: 23030332; PubMed Central PMCID: PMC3498909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eck R, Hundt S, Hartl A, et al. A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology. 1999. September;145(Pt 9):2415–2422. PubMed PMID: 10517594. [DOI] [PubMed] [Google Scholar]
- 30.Otzen C, Bardl B, Jacobsen ID, et al. Candida albicans utilizes a modified beta-oxidation pathway for the degradation of toxic propionyl-CoA. J Biol Chem. 2014. March 21;289(12):8151–8169. PubMed PMID: 24497638; PubMed Central PMCID: PMC3961645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014. August 1;30(15):2114–2120. PubMed PMID: 24695404; PubMed Central PMCID: PMC4103590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013. April 25;14(4):R36 PubMed PMID: 23618408; PubMed Central PMCID: PMC4053844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012. March 4;9(4):357–359. PubMed PMID: 22388286; PubMed Central PMCID: PMC3322381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010. April 1;464(7289):773–777. PubMed PMID: 20220756; PubMed Central PMCID: PMC3836232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 PubMed PMID: 25516281; PubMed Central PMCID: PMC4302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonasdottir HS, Brouwers H, Kwekkeboom JC, et al. Targeted lipidomics reveals activation of resolution pathways in knee osteoarthritis in humans. Osteoarthritis Cartilage. 2017. July;25(7):1150–1160. PubMed PMID: 28189826. [DOI] [PubMed] [Google Scholar]
- 37.Schlenke P, Kluter H, Muller-Steinhardt M, et al. Evaluation of a novel mononuclear cell isolation procedure for serological HLA typing. Clin Diagn Lab Immunol. 1998. November;5(6):808–813. PubMed PMID: 9801339; PubMed Central PMCID: PMC96206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keppler-Ross S, Douglas L, Konopka JB, et al. Recognition of yeast by murine macrophages requires mannan but not glucan. Eukaryot Cell. 2010. November;9(11):1776–1787. PubMed PMID: 20833894; PubMed Central PMCID: PMC2976302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross O, Poeck H, Bscheider M, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009. May 21;459(7245):433–436. PubMed PMID: 19339971. [DOI] [PubMed] [Google Scholar]
- 40.Ifrim DC, Bain JM, Reid DM, et al. Role of Dectin-2 for host defense against systemic infection with Candida glabrata. Infect Immun. 2014. March;82(3):1064–1073. PubMed PMID: 24343653; PubMed Central PMCID: PMC3957982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


