Abstract
Pituitary adenylate cyclase-activating polypeptide (PACAP) has been conserved remarkably during evolution and is widely expressed in the mammalian brain. In Drosophila, mutation of the PACAP homologue results in behavioral defects, including impaired olfaction-associated learning and changes in ethanol sensitivity. Here, we report the generation of mice lacking the PACAP gene (PACAP−/−). PACAP−/− mice were born in the expected Mendelian ratios but had a high early-mortality rate. The surviving adult PACAP−/− mice displayed remarkable behavioral changes; they exhibited hyperactive and explosive jumping behaviors in an open field, increased exploratory behavior, and less anxiety in the elevated plus maze, emergence, and novel-object tests. Analysis of PACAP−/− mice brains revealed that the serotonin metabolite 5-hydroxyindoleacetic acid was slightly decreased in the cortex and striatum compared with wild-type mice. The present study provides evidence that PACAP plays a previously uncharacterized role in the regulation of psychomotor behaviors.
Pituitary adenylate cyclase-activating polypeptide (PACAP) is a member of the vasoactive intestinal peptide (VIP)/secretin/glucagon family of peptides and exists in two amidated forms, PACAP38 and PACAP27, that share an identical 27-aa N terminus and are alternatively processed from a 176-aa precursor called preproPACAP (1, 2). The primary structure of PACAP38 has been conserved significantly during evolution from protochordates to mammals, suggesting that the peptide exerts important activities throughout the vertebrate phylum (1, 2). In Drosophila, recent molecular cloning and transgenic rescue experiments in the memory-mutant amnesiac, which has behavioral defects that include impaired olfaction-associated learning and changes in ethanol sensitivity, demonstrated that the amnesiac gene encodes a neuropeptide homologous to vertebrate PACAP (3, 4). In addition, mammalian PACAP activated both the cAMP and Ras/Raf signal-transduction pathways in Drosophila neurons, suggesting a neuromodulatory role of amnesiac (Drosophila PACAP) in specific neuronal populations (5). In mammals, PACAP occurs in neuronal elements, where it acts as a pleiotropic neuropeptide via three heptahelical G protein-linked receptors—one PACAP-specific (PAC1) receptor and two receptors that it shares with VIP (VPAC1 and VPAC2). PACAP stimulates several different signaling cascades in neurons, leading to the activation of adenylate cyclase, phospholipase C, and mitogen-activated protein kinase and the mobilization of calcium (1, 2, 6). Histochemical studies have shown that PACAP immunoreactivity is observed in several brain regions, including the dopamine (DA) and serotonin (5-HT) systems, with high concentrations found in the nucleus accumbens, hypothalamus, amygdala, substantia nigra, and dorsal raphe (7–9). PAC1 receptor also is expressed throughout the target areas of both the mesocorticolimbic and nigrostriatal DA systems as well as 5-HT system (10). In addition, VPAC1 and VPAC2 receptors also are expressed in these systems (11). These histochemical studies suggest a functional relationship between PACAP neurons and DA and 5-HT neurons. Pharmacological studies show that PACAP has neurotrophic and neuroprotective actions on mesencephalic DA neurons (12), cortical neurons (13), cerebellar granule cells (14), and other neurons (1, 2). PACAP increases tyrosine hydroxylase activity in the nucleus accumbens (15) and stimulates interleukin-6 production in astrocytes (16). PACAP also is implicated in synaptic plasticity in the hippocampus (17). However, the relevance of these pharmacological PACAP responses to the actual physiological activities of endogenous PACAP has not been addressed, because potent and selective low-molecular-weight PACAP antagonists have not yet been developed (2). Previously, we cloned the PAC1 receptor cDNA (6) and the genes for the PAC1 receptor (18), VPAC1 receptor (19), and the PACAP ligand (20) and analyzed their localization in the nervous system (10, 21, 22). Recently, we have generated PAC1-receptor exon 2-deficient mice; however, they showed no obvious behavioral phenotype (23). In addition, Jamen et al. (24, 25) also reported the generation of PAC1-receptor knockout mice, which similarly showed no obvious phenotypic changes in behavior. In the present study, we have generated mice deficient in PACAP (PACAP−/−) to understand the in vivo function of PACAP-dependent signaling. PACAP−/− mice display markedly increased locomotor activity, novelty-related exploration, and explosive jumping behavior. This aberrant behavior is ameliorated by the antipsychotic drug haloperidol. Finally, the 5-HT metabolite 5-hydroxyindoleacetic acid (5-HIAA) is slightly decreased in the cortex and striatum of the PACAP−/− mouse brain. The present study provides evidence that PACAP plays a previously uncharacterized role in the regulation of psychomotor behaviors.
Materials and Methods
All animal care and handling procedures were approved by the Institutional Animal Care and Use Committee of Osaka University.
Generation of PACAP−/− Mice.
The PACAP gene-targeting vector was constructed from genomic DNA clones (λMPL4 and λMPL18; ref. 20) isolated from a 129/SvJ mouse genomic library. A 2.1-kb PvuII fragment of the PACAP gene containing part of exon 5 and the 3′ flanking region was inserted 3′ to the neomycin resistant (neo) gene (derived from pGEM7-PGK-neo-polyA) in pBluescript KS(+). An MC1 promoter-driven diphtheria toxin A-fragment (DT) gene (derived from pMC1DTpA) then was inserted 5′ to the neo gene. Subsequently, a 5.3-kb HindIII genomic-DNA fragment containing exons 1A–4 was inserted between the DT and neo genes to generate the PACAP-targeting vector (Fig. 1A). The linearized vector was electroporated into 129/Ola mouse-derived E14tg2a embryonic stem (ES) cells. Targeted clones were identified by Southern blot analysis with external 0.42-kb and 1.1-kb probes and microinjected into C57BL/6 embryonic day 3.5 blastocysts. Two highly chimeric males showed germ-line transmission and were mated with C57BL/6 wild-type females to produce F1 heterozygous mice. F1 heterozygotes were mated with C57BL/6 mice to produce the F2- and F3-generation mice that were used in this study, unless otherwise specified. The null allele of PACAP also was backcrossed five times onto an Institute of Cancer Research (ICR) mouse background. Wild-type mice and mice homozygous for the mutant PACAP gene were obtained from the intercross of heterozygous animals, and experiments were conducted with adult (3- to 5-months old) mice. Reverse transcription–PCR was performed as described (26) by using the following PACAP gene exon-specific primers: exon 3, 5′-AGA AGA CGA GGC TTA CGA CCA G-3′ (sense); exon 4, 5′-ACG ACC GAC TGC AGG TAC TTC-3′ (antisense); and exon 5, 5′-TTT CTT GAC AGC CAT TTG TTT TCG G-3′ (antisense). The β-actin housekeeping gene was simultaneously reverse-transcribed and amplified as described (27). In situ hybridization analysis was performed on parasagittal brain sections as described (10). Two different cDNA fragments of mouse PACAP (20)—a 431-bp cDNA fragment (−116 to 315, where +1 represents the nucleotide position of the ATG-initiation codon) spanning exons 2–4 and a 198-bp fragment (340) containing part of the exon 5 coding sequence deleted by homologous recombination—were used as templates to synthesize [35S]CTP-labeled cRNA probes. The expression of the biologically active mature PACAP isoform, PACAP38, was studied in brain by an RIA kit (Peninsula Laboratories).
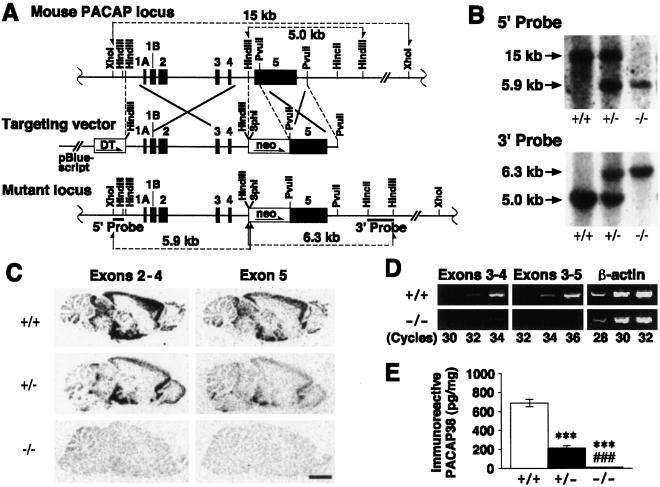
Figure 1.
Targeted disruption of the PACAP gene and characterization of PACAP−/−, PACAP+/−, and wild-type mice. (A) Alignment of the PACAP locus with the targeting vector and the mutant locus. Black boxes 1A–5, exons 1A–5; DT, MC1 promoter with diphtheria toxin A-fragment gene; neo, phosphoglycerate kinase promoter with neomycin-resistance gene. (B) Southern blot analysis of tail DNA digested either with XhoI and SphI and hybridized with a 5′ probe (Upper) or with HindIII and hybridized with a 3′ probe (Lower). (C) In situ hybridization analysis of parasagittal brain sections with two [35S]cRNA probes specific for exons 2–4 and exon 5. (Bar = 2.5 mm.) (D) Reverse transcription–PCR analysis of RNA from midbrain and diencephalon. PCR was performed at the indicated number of cycles with a sense primer derived from PACAP exon 3 and antisense primers derived from PACAP exon 4 (Left) and exon 5 (Center). The β-actin housekeeping gene was simultaneously amplified as an internal standard (Right). (E) Analysis of PACAP38 levels in the midbrain and diencephalon from PACAP+/+ (n = 7), PACAP+/− (n = 6), and PACAP−/− (n = 5) mice by RIA. ***, P < 0.001 vs. PACAP+/+ mice; ###, P < 0.001 vs. PACAP+/− mice, Student's t test.
Open-Field Test.
Motor activity was quantified with an automated video tracking system, the video-image motion analyzer AXIS-90 (Neuroscience, Tokyo, Japan). A computer program was used to overlay grid lines defining 25 separate regions within a circular open field (60 cm in diameter, 30-cm deep, and illuminated with 100 lux of white light; see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.). Paths taken by each mouse were stored permanently as x-y coordinate sequences, and parameters indicative of locomotor activity were assessed. Vertical activity (rearing and jumping) also were scored. In the haloperidol study, the drug was injected intraperitoneally 20 min before monitoring locomotor activity, which was measured for 90 min by another multichannel activity-monitoring system, Supermex (Muromachi Kikai, Tokyo, Japan). Subsequently, the same mice were tested for catalepsy in the bar test as described (28).
Elevated Plus Maze.
The movement of mice was recorded for 5 min in an elevated crossbar with two walled and two open arms. The total path length, as well as the number of entries into and time spent on the open arms, was assessed as described (29) with AXIS-90.
Emergence Test.
The emergence test was performed as described (30) with several modifications. Briefly, the open field contained a white plastic cylinder with an open end (11 cm in diameter, 6 cm deep) located centrally. Mice were placed into the cylinder and tested for 15 min. A trained observer blind to genotype scored the following behaviors: the latency of emergence from the cylinder (defined as placement of all four paws into the open field); the total time spent inside the cylinder; and the exploratory behavior (assessed by the frequency of rearing to the wall of the cylinder during the first 5 min).
Novel-Object Test.
The novel-object test was performed as described (30) with several modifications. Briefly, mice were familiarized with the open field by a 2-day preexposure (day 1 for 60 min and day 2 for 30 min). On day 3, mice were placed in the open field for 30 min. Then, a novel white cup was placed into the center of the open field, and mice were tested for an additional 10 min. The number of entries made into the center with a diameter of 12 cm (center access) and time spent in the center were assessed by AXIS-90.
Measurements of Monoamine Neurotransmitters and Metabolites.
Fresh-frozen brain areas were assayed for levels of DA, 3,4-dihydroxyphenylacetic acid, homovanillic acid, 5-HT, and 5-HIAA by using high-performance liquid chromatography with electrochemical detection, as described (31).
Results
Characterization of PACAP−/− Mice.
The mouse PACAP gene (20) was disrupted in ES cells by homologous recombination through inactivation of part of exon 5, from which the mature PACAP (PACAP38 and PACAP27) protein is expressed (Fig. 1A). Four positive ES clones were obtained, and two of them were selected and used to generate mutant mice, whose genotypes were confirmed by Southern hybridization (Fig. 1B). In situ hybridization analysis with a probe corresponding to the disrupted region indicated that the expression of the mature peptide-coding sequence was greatly reduced in heterozygous (PACAP+/−) mice compared with wild-type mice and disappeared in PACAP−/− mice (Fig. 1C). The complete deletion of this sequence also was confirmed by reverse transcription-PCR (Fig. 1D). In addition, expression of the upstream coding sequence was reduced (Fig. 1D) in a gene dose-dependent manner (Fig. 1C). The absence of PACAP38 expression in the brain (midbrain and diencephalon) of PACAP−/− mice was demonstrated by RIA; furthermore, PACAP38 expression was greatly reduced in PACAP+/− mice compared with wild-type mice (Fig. 1E).
Mendelian segregation of pup genotypes from heterozygous breeding (n = 117) was observed at birth, with a genotype distribution of 23.9%, 49.6%, and 26.5% for PACAP−/−, PACAP+/−, and wild-type mice, respectively. However, PACAP−/− pups had a high mortality rate, and the genotype distribution upon weaning (n = 531) was 15.6%, 54.8%, and 29.6% for PACAP−/−, PACAP+/−, and wild-type mice, respectively, suggesting a significant loss of PACAP−/− pups (P < 0.001, χ2 analysis). Homozygous mating of PACAP-deficient mice resulted in fewer successful pregnancies, thereby yielding few pups (data not shown). PACAP−/− female mice could nurse their newborn pups, although PACAP−/− pups had a high mortality rate, irrespective of their parent genotypes.
Hyperactivity in a Novel Environment.
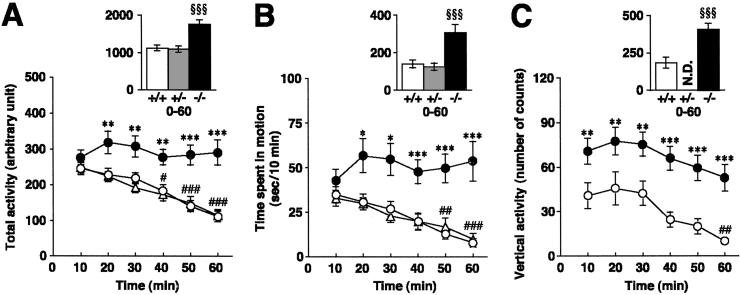
We analyzed the behavior of PACAP−/− mice in an open field to assess their locomotor activity in novel environments. As shown in Fig. 2 A and B, PACAP−/− mice exhibited higher levels of locomotor activity (total activity), spent more time in motion than wild-type mice, and showed minimal habituation to the novel environment for 60 min, at which time wild-type mice were habituated significantly. Tracks of locomotor patterns during the first and last 150 s clearly show that, although the initial levels of locomotion were similar for both genotypes, PACAP−/− mice were more active than wild-type mice at the end of the session (Fig. 3D). Analysis of the mean speed of motion revealed hyperkinetic movement of PACAP−/− mice (PACAP+/+, 20.0 ± 0.59 cm/s; PACAP−/−, 23.7 ± 0.66 cm/s, P < 0.001, Mann–Whitney U test). Furthermore, PACAP−/− mice spent less time engaged in licking/grooming behaviors (PACAP+/+, 19.7 ± 2.5%; PACAP−/−, 10.0 ± 1.9%, P < 0.001, Mann–Whitney U test). There was no difference between PACAP+/− mice and wild-type mice in behavioral measures in the open field (Fig. 2 A and B). Vertical activity, including rearing and jumping (see below), was much higher in PACAP−/− mice than in wild-type mice, indicating increased exploratory activity (Fig. 2C). PACAP−/− mice that have ≈97% of ICR genetic background similarly showed higher levels of locomotor activity than wild-type littermate control mice (P < 0.01) or wild-type inbred ICR mice (P < 0.001) in the open field (total activity for 90 min in arbitrary units, n = 10–11; PACAP−/−, 1896 ± 84; PACAP+/+, 1435 ± 101; inbred ICR, 1322 ± 63).
Figure 2.
Open-field measures. Total activity (A) and time spent in motion (B) are shown for PACAP+/+ (white circles), PACAP+/− (white triangles), and PACAP−/− (black circles) mice (n = 20 for all groups). (C) Vertical activity is shown for PACAP+/+ and PACAP−/− mice (n = 14 for both groups). *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. PACAP+/+ mice. #, P < 0.05; ##, P < 0.01; ###, P < 0.001 vs. first 10 min, ANOVA, followed by post hoc Fisher Protected Least Significant Difference (PLSD) test; §§§, P < 0.001 vs. PACAP+/+ mice, Kruskal–Wallis ANOVA, followed by Mann–Whitney U test. N.D., not determined.
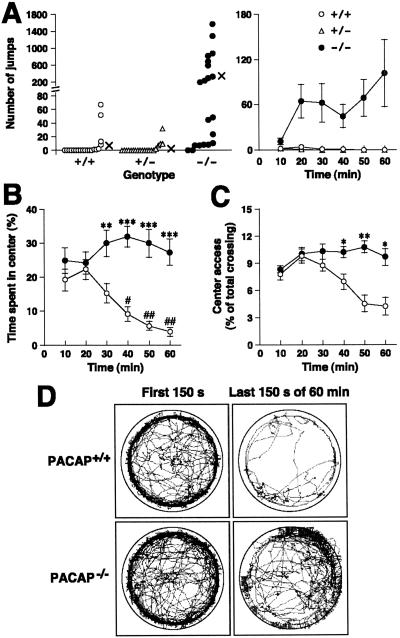
Figure 3.
Jumping behavior, increased central entry, and locomotor pattern of PACAP−/− mice in the open field. (A) Number of jumps in the open field. The actual number of jumps observed in individual mice and mean values (×) for 60 min (Left), and means ± SE at 10-min intervals (Right) are indicated. (n = 20 for all groups.) (B and C) Time spent in the center (B) and center access (C) (percentage of crossings into the center zone out of the total number of crossings) are shown for PACAP+/+ (white circles) and PACAP−/− (black circles) mice (n = 20 for all groups). *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. PACAP+/+ mice. #, P < 0.05 and ##, P < 0.01 vs. first 10 min, ANOVA, followed by post hoc Fisher PLSD test. (D) Examples of locomotor patterns of PACAP+/+ (Upper) and PACAP−/− (Lower) mice during the first 150 s (Left) and last 150 s (Right) of a 60-min recording. Tracks of representative 10 mice in each group are shown. The tracks outward across the circular boundary represent the jumping behavior (see supporting information).
Interestingly, PACAP−/− mice elicited explosive jumping behavior in the open field arena, which was only rarely observed in wild-type or PACAP+/− mice (Fig. 3 A and D). In almost all of the PACAP−/− mice, the frequency of jumping escalated over time, with a maximum of more than 1,500 times during a 60-min observation period. The behavior of a mouse that jumped 1,577 times can be viewed in the video that is published as supporting information on the PNAS web site, www.pnas.org. PACAP−/− mice with ICR background again exhibited explosive jumping behavior in the open field more frequently than wild-type littermate controls (P < 0.001) or wild-type inbred ICR mice (P < 0.01; number of jumps for 90 min, n = 10–11; PACAP−/−, 225 ± 49; PACAP+/+, 21 ± 8; inbred ICR, 30 ± 18).
Increased Exploration and Reduced Anxiety.
Increased exploratory-related and reduced anxiety-related behavior in PACAP−/− mice was demonstrated by zone monitoring in the open-field, on the elevated plus maze, and by the emergence and novel-object tests. In the open field, PACAP−/− mice entered the center region more often and spent more time in the center than wild-type mice (Fig. 3 B–D). When tested on an elevated plus maze, PACAP−/− mice showed a tendency to enter the open arms of the maze more often than wild-type controls. They also spent much more time on the open arms than wild-type controls (Table 1). The emergence test is a free exploration paradigm designed to reduce anxiety in mice confronted with a novel open-field environment with no possibility of escape by providing a safe enclosure to assess approach or exploratory behavior in rodents (30). PACAP−/− mice exhibited shortened latencies for emergence and decreased total time spent in the cylinder, and demonstrated increased exploratory behavior (assessed by the frequency of rearing to the wall of the cylinder) compared with wild-type mice (Table 1). The novel-object test is another free exploration paradigm that provides animals the opportunity to explore a novel object (30). Upon introduction of the novel object (a white cup), PACAP−/− mice increased the frequency of center access and time spent in the center more than wild-type controls (Table 1). Thus, PACAP−/− mice exhibited elevated behavioral responses to novelty with increased exploration and less anxiety compared with wild-type controls.
Table 1.
Increased exploratory activity and reduced anxietyrelated behavior of PACAP−/− mice as revealed by the elevated plus maze, emergence, and novel-object tests
| Test | PACAP+/+ | PACAP+/− | PACAP−/− |
|---|---|---|---|
| Elevated plus maze (n = 7–8 mice per genotype) | |||
| Total path length (cm/5 min) | 747 ± 109 | 902 ± 113 | 1158 ± 83* |
| Open arm entries (%) | 24.3 ± 7.1 | 18.8 ± 6.5 | 37.0 ± 4.3# |
| Time spent in open arms (%) | 3.2 ± 1.4 | 9.6 ± 5.7 | 20.1 ± 2.0**# |
| Emergence test (n = 16 mice per genotype) | |||
| Latency of emergence (s) | 888 ± 12 | — | 587 ± 71** |
| Time spent in the cylinder (s) | 888 ± 12 | — | 600 ± 69** |
| Exploratory behavior | 16.0 ± 3.5 | — | 30.8 ± 2.7** |
| Novel object test (n = 8 mice per genotype) | |||
| Number of center access | |||
| No cup | 13.6 ± 4.6 | — | 13.5 ± 4.3 |
| Cup | 6.5 ± 3.4 | — | 30.3 ± 5.2**§ |
| Time spent in the center region (s) | |||
| No cup | 11.7 ± 4.6 | — | 18.2 ± 8.2 |
| Cup | 13.2 ± 7.1 | — | 79.9 ± 24.6**§ |
Values are means ± SE.
, P < 0.05,
, P < 0.01, and
, P < 0.001 vs. PACAP+/+ group;
, P < 0.05 vs. PACAP+/− group;
, P < 0.05 vs. no cup, Kruskal–Wallis ANOVA, followed by Mann–Whitney U test (elevated plus maze and novel-object tests) and Mann–Whitney U test (emergence test).
Effects of Haloperidol on Hyperactive Behavior and Catalepsy.
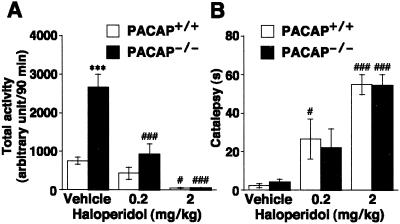
The antipsychotic drug haloperidol was tested for its ability to attenuate the hyperactive behavior of PACAP−/− mice (Fig. 4). Haloperidol (0.2 mg/kg) effectively reduced the hyperactivity in PACAP−/− mice (65 ± 10% reduction) to the control level in wild-type mice, although the same dose of haloperidol resulted in moderate impairment of locomotor activity in wild-type mice (43 ± 21% reduction; Fig. 4A). PACAP−/− mice did not show spontaneous catalepsy, and the cataleptogenic effect of haloperidol (0.2 and 2 mg/kg) in PACAP−/− mice was quite similar to that in wild-type mice, suggesting that the extrapyramidal system (postsynaptic striatal DA receptors) of PACAP−/− mice is functionally normal (Fig. 4B).
Figure 4.
Effects of haloperidol on locomotor activity. (A) PACAP+/+ and PACAP−/− mice (n = 6–7) were injected intraperitoneally with 0.2 and 2 mg/kg haloperidol or vehicle (0.5% carboxymethyl cellulose sodium) and, 20 min later, were placed in the open field for automated measurement of locomotor parameters over a 90-min test period. (B) Subsequently, each mouse was tested for catalepsy in the bar test. Values are means ± SE. ***, P < 0.001 vs. PACAP+/+ mice. #, P < 0.05 and ###, P < 0.001 vs. vehicle; ANOVA, followed by post hoc Fisher PLSD test.
Analyses of Monoamine Turnover in PACAP−/− Mice.
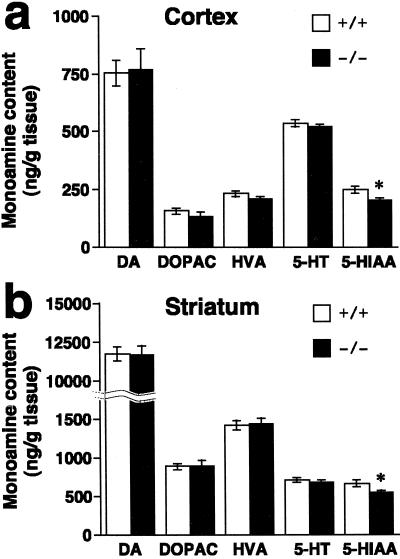
The effect of PACAP deficiency on monoamine turnover was determined by HPLC (Fig. 5). In the cortex and striatum, the 5-HIAA levels were statistically lower (slightly) in PACAP−/− mice compared with wild-type mice (81% and 82%, respectively). The cortical and striatal 5-HIAA/5-HT ratios were reduced by ≈15% in PACAP−/− mice. No significant differences in levels of monoamines and their metabolites have been observed so far in any other brain regions between wild-type and PACAP−/− mice (data not shown).
Figure 5.
Tissue content of monoamines and their metabolites in cortex and striatum. Levels of the monoamine neurotransmitters and the major metabolites were assayed in cortex (a) and striatum (b) of PACAP+/+ (n = 14) and PACAP−/− (n = 13) mice. Data are expressed as means ± SE. *, P < 0.05 vs. PACAP+/+ mice, Student's t test. DOPAC, 3,4-dihydroxyphenylacetic acid, HVA, homovanillic acid.
Discussion
The results of behavioral experiments with PACAP−/− mice demonstrate that disruption of the PACAP gene in mice lead to perturbations in psychomotor behaviors, especially the exploratory component of locomotor behavior, implicating PACAP in psychotic brain functions. Furthermore, the 5-HIAA level was decreased slightly in the cortex and striatum of the PACAP−/− mouse brain.
It is commonly believed that locomotor hyperactivity is associated with increased DA tone (32). In PACAP−/− mice, DA turnover was unchanged and the incidence of haloperidol-induced catalepsy was quite similar to that in wild-type mice. These findings suggest that the locomotor hyperactivity in PACAP−/− mice probably may not be a result of increased nigrostriatal dopaminergic activity. Alternatively, the importance of 5-HT in controlling the locomotor activity has been demonstrated in a study with 5-HT1B-receptor knockout mice (33). Moreover, a relative balance of the DA and 5-HT systems seems to be important for normal motor activity, and alterations in any of the parameters that control this delicate homeostatic situation might underlie hyperactive states (32). This may explain, at least in part, the locomotor hyperactivity of PACAP−/− mice.
Several lines of evidence suggest that dysfunction of serotonergic pathways, especially those mediated by the 5-HT1A receptor, is associated with anxiety-related traits (34). Although it has not been concluded that PACAP−/− mice are less anxious, if, indeed, they are less anxious, reduced 5-HT turnover (5-HIAA/5-HT ratio) cannot explain it. On the other hand, DA functions, particularly those mediated by the D4 receptor, are involved in novelty-related exploratory behavior, as reported in D4-receptor knockout mice (30, 35). These mutant mice are less active in open-field tests and exhibit reduced exploration of novel stimuli in contrast to the phenotypes observed in PACAP−/− mice.
One of the striking findings of the present study was that PACAP−/− mice showed abnormal jumping behavior in the open field arena. In NIH Swiss mice, an N-methyl-d-aspartate (NMDA)-receptor antagonist MK-801 is known to precipitate explosive episodic jumping behavior that can be attenuated by haloperidol (36). In addition, dysfunction of the NMDA receptor by MK-801 or the targeted disruption of its gene produces psychotic symptoms that closely resemble the positive and negative symptoms of schizophrenia (37, 38). In view of the effects of MK-801, investigation of possible common mechanisms involved in the MK-801-induced abnormal behaviors and those of PACAP−/− mice would be warranted.
Currently, it is accepted that multiple genes of small effect, rather than a single causative gene, act in concert with nongenetic factors to increase the risk of mental disorder (39). The present study shows that PACAP−/− mice display marked behavioral abnormalities without having marked changes in specific neuronal pathways in their brains. To date, we have found no change at the level of gene expression of PACAP-receptor subtypes, tyrosine hydroxylase, and DA D2 receptor in the various brain regions of PACAP−/− mice (data not shown). Of course, there are many other functional molecules for which expression levels should be determined. It is possible that PACAP−/− mice have several small but significant changes in the activity of neuronal networks, and that they collectively cause behavioral abnormalities.
Because specific low-molecular-weight antagonists and agonists to the different PACAP-receptor subtypes are not available, the physiological role of PACAP as well as of each receptor subtype in brain function have not been addressed fully. PAC1 receptor-deficient mice (23–25) did not show any apparent behavioral changes like those observed in PACAP−/− mice. PACAP interacts with three receptors: PACAP-preferring PAC1, VIP-shared VPAC1, and VPAC2 receptors. Lack of signal transmission of PACAP through the VPAC receptors may explain the behavioral changes in PACAP−/− mice.
Several lines of evidence suggest that PACAP acts as a neurotrophic factor (12–14) and plays a role in mammalian neurogenesis (40). For instance, the PAC1 receptor is expressed at very high levels in ventricular zones throughout the embryonic neuraxis, and PACAP likely regulates the development of the general features of the neuronal phenotype (40). Therefore, it is possible also that the lack of PACAP affects developmental processes, resulting in the observed behavioral abnormalities and in a high early mortality rate in PACAP−/− mice.
Because it was demonstrated that the Drosophila mutant amnesiac, which displays behavioral defects, has a mutation in a neuropeptide gene, the in vivo role of the mammalian homolog PACAP has remained an open question (3). In this study, we have shown that disruption of the PACAP gene in mice leads to major alterations in psychomotor activity. Recent genetic linkage studies have suggested that a locus for schizophrenia as well as bipolar affective disorder is located on chromosome 18p11 (41), where the human PACAP gene resides (42). It is now important to determine whether the mutation in the PACAP locus is implicated in disease in these select families.
In conclusion, the present study proposes a role of PACAP-ergic neurons in regulating psychomotor behaviors acutely or developmentally. The PACAP−/− mouse should be a valuable tool to investigate both normal and pathological processes in which PACAP has been proposed to play a role.
Supplementary Material
Acknowledgments
We thank Mayu Yamamoto, Rie Hatanaka, Ayako Ichibori, and Tatsuya Ojika for expert technical assistance. This research was supported, in part, by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan, and by grants from Uehara Memorial Foundation and Takeda Science Foundation.
Abbreviations
- PACAP
pituitary adenylate cyclase-activating polypeptide
- VIP
vasoactive intestinal peptide
- PAC1 receptor
PACAP receptor
- VPAC receptor
VIP/PACAP receptor
- DA
dopamine
- 5-HT
serotonin
- 5-HIAA
5-hydroxyindoleacetic acid
- ICR mouse
Institute of Cancer Research mouse
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Arimura A. Jpn J Physiol. 1998;48:301–331. doi: 10.2170/jjphysiol.48.301. [DOI] [PubMed] [Google Scholar]
- 2.Vaudry D, Gonzalez B J, Basille M, Yon L, Fournier A, Vaudry H. Pharmacol Rev. 2000;52:269–324. [PubMed] [Google Scholar]
- 3.Feany M B, Quinn W G. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 4.Moore M S, DeZazzo J, Luk A Y, Tully T, Singh C M, Heberlein U. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Y. Nature (London) 1995;375:588–592. doi: 10.1038/375588a0. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto H, Ishihara T, Shigemoto R, Mori K, Nagata S. Neuron. 1993;11:333–342. doi: 10.1016/0896-6273(93)90188-w. [DOI] [PubMed] [Google Scholar]
- 7.Ghatei M A, Takahashi K, Suzuki Y, Gardiner J, Jones P M, Bloom S R. J Endocrinol. 1993;136:159–166. doi: 10.1677/joe.0.1360159. [DOI] [PubMed] [Google Scholar]
- 8.Masuo Y, Suzuki N, Matsumoto H, Tokito F, Matsumoto Y, Tsuda M, Fujino M. Brain Res. 1993;602:57–63. doi: 10.1016/0006-8993(93)90241-e. [DOI] [PubMed] [Google Scholar]
- 9.Piggins H D, Stamp J A, Burns J, Rusak B, Semba K. J Comp Neurol. 1996;376:278–294. doi: 10.1002/(SICI)1096-9861(19961209)376:2<278::AID-CNE9>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto H, Nogi H, Mori K, Ohishi H, Shigemoto R, Yamamoto K, Matsuda T, Mizuno N, Nagata S, Baba A. J Comp Neurol. 1996;371:567–577. doi: 10.1002/(SICI)1096-9861(19960805)371:4<567::AID-CNE6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Usdin T B, Bonner T I, Mezey E. Endocrinology. 1994;135:2662–2680. doi: 10.1210/endo.135.6.7988457. [DOI] [PubMed] [Google Scholar]
- 12.Takei N, Skoglosa Y, Lindholm D. J Neurosci Res. 1998;54:698–706. doi: 10.1002/(SICI)1097-4547(19981201)54:5<698::AID-JNR15>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Morio H, Tatsuno I, Hirai A, Tamura Y, Saito Y. Brain Res. 1996;741:82–88. doi: 10.1016/s0006-8993(96)00920-1. [DOI] [PubMed] [Google Scholar]
- 14.Villalba M, Bockaert J, Journot L. J Neurosci. 1997;17:83–90. doi: 10.1523/JNEUROSCI.17-01-00083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser A, Scholz J, Gansle A. Neuropeptides. 1999;33:492–497. doi: 10.1054/npep.1999.0768. [DOI] [PubMed] [Google Scholar]
- 16.Gottschall P E, Tatsuno I, Arimura A. Brain Res. 1994;637:197–203. doi: 10.1016/0006-8993(94)91233-5. [DOI] [PubMed] [Google Scholar]
- 17.Kondo T, Tominaga T, Ichikawa M, Iijima T. Neurosci Lett. 1997;221:189–192. doi: 10.1016/s0304-3940(96)13323-1. [DOI] [PubMed] [Google Scholar]
- 18.Aino H, Hashimoto H, Ogawa N, Nishino A, Yamamoto K, Nogi H, Nagata S, Baba A. Gene. 1995;164:301–304. doi: 10.1016/0378-1119(95)00391-i. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto H, Nishino A, Shintani N, Hagihara N, Copeland N G, Jenkins N A, Yamamoto K, Matsuda T, Ishihara T, Nagata S, Baba A. Genomics. 1999;58:90–93. doi: 10.1006/geno.1999.5805. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto K, Hashimoto H, Hagihara N, Nishino A, Fujita T, Matsuda T, Baba A. Gene. 1998;211:63–69. doi: 10.1016/s0378-1119(98)00110-3. [DOI] [PubMed] [Google Scholar]
- 21.Nogi H, Hashimoto H, Fujita T, Hagihara N, Matsuda T, Baba A. Jpn J Pharmacol. 1997;75:203–207. doi: 10.1254/jjp.75.203. [DOI] [PubMed] [Google Scholar]
- 22.Nogi H, Hashimoto H, Hagihara N, Shimada S, Yamamoto K, Matsuda T, Tohyama M, Baba A. Neurosci Lett. 1997;227:37–40. doi: 10.1016/s0304-3940(97)00295-4. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto H, Shintani N, Nishino A, Okabe M, Ikawa M, Matsuyama S, Itoh K, Yamamoto K, Tomimoto S, Fujita T, et al. J Neurochem. 2000;75:1810–1817. doi: 10.1046/j.1471-4159.2000.0751810.x. [DOI] [PubMed] [Google Scholar]
- 24.Jamen F, Persson K, Bertrand G, Rodriguez Henche N, Puech R, Bockaert J, Ahren B, Brabet P. J Clin Invest. 2000;105:1307–1315. doi: 10.1172/JCI9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauvage M, Brabet P, Holsboer F, Bockaert J, Steckler T. Brain Res Mol Brain Res. 2000;84:79–89. doi: 10.1016/s0169-328x(00)00219-9. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto H, Hagihara N, Koga K, Yamamoto K, Shintani N, Tomimoto S, Mori W, Koyama Y, Matsuda T, Baba A. J Neurochem. 2000;74:501–507. doi: 10.1046/j.1471-4159.2000.740501.x. [DOI] [PubMed] [Google Scholar]
- 27.Kitanaka J, Hashimoto H, Sugimoto Y, Sawada M, Negishi M, Suzumura A, Marunouchi T, Ichikawa A, Baba A. Biochim Biophys Acta. 1995;1265:220–223. doi: 10.1016/0167-4889(94)00225-4. [DOI] [PubMed] [Google Scholar]
- 28.Boulay D, Depoortere R, Oblin A, Sanger D J, Schoemaker H, Perrault G. Eur J Pharmacol. 2000;391:63–73. doi: 10.1016/s0014-2999(99)00916-4. [DOI] [PubMed] [Google Scholar]
- 29.Lister R G. Psychopharmacology (Berl) 1987;92:180–185. doi: 10.1007/BF00177912. [DOI] [PubMed] [Google Scholar]
- 30.Dulawa S C, Grandy D K, Low M J, Paulus M P, Geyer M A. J Neurosci. 1999;19:9550–9556. doi: 10.1523/JNEUROSCI.19-21-09550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda T, Seong Y H, Aono H, Kanda T, Baba A, Saito K, Tobe A, Iwata H. Eur J Pharmacol. 1989;170:75–82. doi: 10.1016/0014-2999(89)90136-2. [DOI] [PubMed] [Google Scholar]
- 32.Gainetdinov R R, Wetsel W C, Jones S R, Levin E D, Jaber M, Caron M G. Science. 1999;283:397–401. doi: 10.1126/science.283.5400.397. [DOI] [PubMed] [Google Scholar]
- 33.Saudou F, Amara D A, Dierich A, LeMeur M, Ramboz S, Segu L, Buhot M C, Hen R. Science. 1994;265:1875–1878. doi: 10.1126/science.8091214. [DOI] [PubMed] [Google Scholar]
- 34.Hoyer D, Clarke D E, Fozard J R, Hartig P R, Martin G R, Mylecharane E J, Saxena P R, Humphrey P P. Pharmacol Rev. 1994;46:157–203. [PubMed] [Google Scholar]
- 35.Rubinstein M, Phillips T J, Bunzow J R, Falzone T L, Dziewczapolski G, Zhang G, Fang Y, Larson J L, McDougall J A, Chester J A, et al. Cell. 1997;90:991–1001. doi: 10.1016/s0092-8674(00)80365-7. [DOI] [PubMed] [Google Scholar]
- 36.Rosse R B, Mastropaolo J, Sussman D M, Koetzner L, Morn C B, Deutsch S I. Clin Neuropharmacol. 1995;18:448–457. doi: 10.1097/00002826-199510000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Mohn A R, Gainetdinov R R, Caron M G, Koller B H. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- 38.Svensson T H. Brain Res Brain Res Rev. 2000;31:320–329. doi: 10.1016/s0165-0173(99)00048-x. [DOI] [PubMed] [Google Scholar]
- 39.Hyman S E. Bull W H O. 2000;78:455–463. [PMC free article] [PubMed] [Google Scholar]
- 40.Jaworski D M, Proctor M D. Brain Res Dev Brain Res. 2000;120:27–39. doi: 10.1016/s0165-3806(99)00192-3. [DOI] [PubMed] [Google Scholar]
- 41.Rojas K, Liang L, Johnson E I, Berrettini W H, Overhauser J. Mol Psychiatry. 2000;5:389–395. doi: 10.1038/sj.mp.4000737. [DOI] [PubMed] [Google Scholar]
- 42.Hosoya M, Kimura C, Ogi K, Ohkubo S, Miyamoto Y, Kugoh H, Shimizu M, Onda H, Oshimura M, Arimura A, Fujino M. Biochim Biophys Acta. 1992;1129:199–206. doi: 10.1016/0167-4781(92)90488-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.