Abstract
Antimicrobial peptides (AMPs) are typically thought of as molecular hole punchers that directly kill pathogens by membrane permeation. However, recent work has shown that AMPs are pleiotropic, multifunctional molecules that can strongly modulate immune responses. In this review, we provide a historical overview of the immunomodulatory properties of natural and synthetic antimicrobial peptides, with a special focus on human cathelicidin and defensins. We also summarize the various mechanisms of AMP immune modulation and outline key structural rules underlying the recently-discovered phenomenon of AMP-mediated Toll-like receptor (TLR) signaling. In particular, we describe several complementary studies demonstrating how AMPs self-assemble with nucleic acids to form nanocrystalline complexes that amplify TLR-mediated inflammation. In a broader scope, we discuss how this new conceptual framework allows for the prediction of immunomodulatory behavior in AMPs, how the discovery of hidden antimicrobial activity in known immune signaling proteins can inform these predictions, and how these findings reshape our understanding of AMPs in normal host defense and autoimmune disease.
Keywords: antimicrobial peptides, toll-like receptors, innate immunity, autoimmune diseases, liquid crystals, superselectivity
1. Organization of Review
Antimicrobial peptides (AMPs) have been studied for over 40 years in the context of innate host defense. Initial studies focused on understanding their microbicidal mechanisms and identifying common structural and physicochemical features essential to this activity [1,2]. However, within the last 10 years, dramatic progress has been made in elucidating how AMPs interact with the host innate and adaptive immune systems, and how this synergizes with their direct activity on the pathogen. With the rise of drug-resistant infections, there is a resurgence of interest in the design and development of AMPs with potent, selective activity and favorable immunomodulatory profiles [3–6]. In a complementary direction of inquiry, AMPs have also been repeatedly implicated in the dysregulation of inflammation, such as in chronic autoinflammatory diseases like lupus and psoriasis. Therefore, improving AMP-mediated immune signaling can bolster responses to infections [7], while disruption of this behavior could provide a therapeutic opportunity for autoimmune diseases [8].
In this review, we provide a summary of recent studies on the immunomodulatory activities of AMPs, including both endogenous and synthetically designed AMPs. While some effort has been made to show the history of this conceptual development, we note that reviews of this nature are always incomplete, and involve foregrounding of select work. Moreover, because of the diversity in sequence and structure of AMPs, it is difficult to predict immunomodulatory behavior for all AMPs and identify universal themes. We highlight recent work from our group that suggests a deterministic mechanism for how a surprisingly broad range of AMPs signals to the immune system. Using an unconventional combination of techniques from physics, chemistry, computational biology, and immune activation experiments, we describe a novel physical basis for AMP-induced immunomodulation via Toll-like receptors (TLRs). We find that AMPs can structurally organize and scaffold immune ligands into spatially periodic nanocrystalline and sometimes liquid crystalline complexes, and that the crystallinity of AMP–nucleic acid complexes can determine the degree of immune amplification in well-defined in vitro systems [9,10]. This represents a significant generalization of the central paradigm in immunology. Innate immune receptors can recognize not just pathogen-associated molecular patterns (PAMPs) of single ligand molecules, but also recognize nanocrystalline arrangements of AMPs and ligands, ultimately leading to potent immunomodulation that depends sensitively on crystallinity parameters such as the inter-ligand spacing and the number of repeating ligands. Finally, we review recent literature that focuses on immune signaling molecules with hidden antimicrobial activity (“kinocidins”), and how these molecules may augment and modify our perspectives on AMP immunomodulation.
2. Immunomodulatory Activity of Antimicrobial Peptides
Based on experiments both in vitro and in vivo, the immunomodulatory profiles of AMPs have been extensively studied, with human cathelicidin LL37 and defensins being the best-characterized [11]. Typical experiments that are conducted to assess the immunomodulatory behavior of AMPs include measurements of in vitro cytokine production from immune cells, real-time quantitative PCR monitoring of the expression of genes involved in inflammation, and challenge with TLR-ligands in mouse models of infection or inflammatory disease [12]. AMPs are usually co-administered with canonical immune ligands such as lipopolysaccharide (LPS), CpG oligonucleotides, and Poly(I:C) (viral double-stranded RNA mimic), and resulting changes in cytokine production are measured. From the body of literature, it is clear that AMPs are involved in triggering both pro-inflammatory and anti-inflammatory immune responses, and do so through distinct mechanisms that depend on biological context.
Examples of AMP-induced immunomodulation are legion (Figure 1): An early study in mice demonstrated that LL37 is a potent antisepsis agent that inhibits macrophage stimulation by bacterial endotoxins like LPS and lipoteichoic acid (LTA), and protects mice from endotoxemia [13,14]. LL37 upregulated production of MCP-1, IL-8, but not TNF-α, and led to recruitment of immune cells to the site of infection [15]. More specifically, LL37 chemoattracts leukocytes via the formyl peptide receptor-like 1 (FPRL1) [16]. In a similar manner, human β-defensins are chemotactic for immune cells by binding to the CCR6 receptor, a chemokine receptor found on dendritic cells and memory T cells [17]. In the human airway, defensins induce significant IL-8 production from airway epithelial cells, which lead to neutrophil chemotaxis [18]. In addition, human neutrophil peptide (HNP) defensins demonstrate significant direct chemotactic activity for monocytes [19]. HNP defensins are also involved in modulation of host antibody responses. HNPs directly induced proliferation of and cytokine production from T helper cells, enhanced systemic IgG responses, and fostered B cell-T cell interactions to bridge innate and adaptive immunity [20]. In mice, the murine β-defensin 2 (mDF2β), a small AMP produced in response to bacterial infections of mucosal tissue and skin, has also been shown to directly activate immature dendritic cells as an endogenous ligand for TLR4, resulting in the production of pro-inflammatory chemokines and cytokines [21]. The rhesus macaque θ-defensin RTD-1 inhibits production of pro-inflammatory cytokines induced by a variety of TLR ligands in vitro, including CpG DNA and LPS [22]. In a mouse model of bacterial sepsis, RTD-1 also suppressed pro-inflammatory cytokines and enhanced survival [23]. Granulysin is a human AMP produced by cytotoxic T-cells that forms membrane pores and induces apoptosis [24]. During Propionibacterium acnes infection, a fragment of granulysin (31–50v44w) was reported to inhibit production of IL-12 and chemokines MCP-1, IP-10, and MDC from human monocytes [25]. The porcine cathelicidin PR-39 is a broad-spectrum AMP that was observed to induce IL-8 production from macrophages. Truncation mutants of PR-39 were also found to differentially regulate the production of TNF-α [26]. Dual-functional synthetic host defense peptide mimics have been recently developed that capture both antimicrobial and immunomodulatory properties, while circumventing the issues common to peptide-based antimicrobials, such as hemolysis and short half-lives due to rapid degradation in vivo. These synthetic mimics of AMPs have demonstrated the ability to effectively kill Staphylococcus aureus and inhibit TLR2-mediated inflammation normally associated with the bacterial infection [27,28]. One synthetic design in particular, Clavanin-MO, derived from a marine tunicate AMP, was shown to recruit leukocytes, induce production of GM-CSF, IFN-ɣ, and MCP-1, stimulate secretion of the anti-inflammatory cytokine IL-10, and inhibit production of pro-inflammatory cytokines IL-12 and TNF-α [29].
Figure 1. Antimicrobial peptides can modulate immune responses through multiple mechanisms.
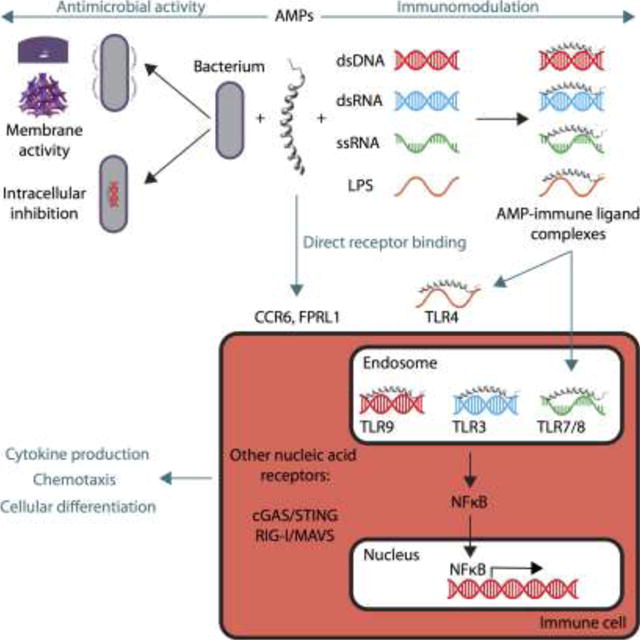
AMPs are multifunctional molecules that can kill microbes by punching holes in their membranes and inhibiting intracellular machinery (upper left). AMPs can also signal to the host immune system (upper right), and exhibit both pro-inflammatory and anti-inflammatory properties. AMPs can interact directly with receptors on immune cells, or form complexes with immune ligands to enable modulation of multiple PRRs like TLRs. (Note that the illustrations of AMP–immune ligand complexes here are purely schematic in nature and do not reflect the actual structures of the nanocomplexes. See text for details). AMP–dsDNA, –dsRNA, and –ssRNA complexes can enter endosomes of immune cells and bind to their respective TLRs, triggering activation of signal transduction pathways that lead to modulation of inflammation (lower right). Downstream consequences of AMP immune signaling include activation of transcription factors, cytokine production, immune cell chemotaxis, and cellular differentiation and proliferation, eventually leading to induction of the adaptive immune response (lower left).
We have summarized only ~10 examples of endogenous and synthetic AMPs that possess immunomodulatory activity above; there are more that exhibit this behavior. To make sense of the vast diversity of evidence documenting the immune signaling activity of AMPs, we suggest a conceptual framework for AMP activity based on recent structural studies, and ask whether we can deduce deterministic rules governing AMP-mediated immunomodulation. To do this, we first analyze the molecular interactions between AMPs, nucleic acid immune ligands, and their respective TLRs.
TLRs were first discovered in Drosophila and were found to comprise one arm of the innate immune system [30,31]. Innate immune cells can detect microbial pathogens through pattern-recognition receptors (PRRs), which sense signature PAMPs from microorganisms, signal to the host the presence of infection, and trigger inflammatory responses. The TLRs, a family of PRRs, are transmembrane proteins that have specialized recognition of multiple ligand classes. TLR2 senses a wide range of lipoproteins/lipopeptides, peptidoglycans, and lipoteichoic acids from bacteria. Moreover, it heterodimerizes with either TLR1 or TLR6 to specifically recognize triacylated or diacylated lipopeptides, respectively [32,33]. LPS, a major constituent of the outer wall Gram-negative bacteria, is detected by TLR4, and bacterial flagellin, a structural protein component of flagella, is recognized by TLR5 [34,35]. Several TLRs are also known to detect nucleic acids and are implicated in antiviral responses. Double-stranded RNA (dsRNA), typically generated from viral replication, is recognized by TLR3 [36,37], while TLR7 and TLR8, which are similar in structure, both respond to single-stranded RNA (ssRNA) [38–40]. TLR9 has been identified as the receptor for unmethylated CpG-rich DNA motifs of bacteria [41] and viral double-stranded DNA (dsDNA) [42,43].
The interaction of TLRs with PAMPs activates signal transduction pathways that lead to the production of pro-inflammatory cytokines. In general, TLRs 1, 2, 4, 5, and 6 primarily detect molecules unique to bacteria, and thus, allows for efficient discrimination of self and non-self. On the other hand, TLRs 3, 7, 8, and 9 recognize nucleic acids, which are not exclusive to microbial pathogens (Figure 1). In these cases, discrimination of self and non-self is typically achieved by the accessibility of the ligands to TLRs. More specifically, this subgroup of TLRs are localized to intracellular compartments [44–47], and therefore, are exposed to foreign nucleic acids released into the compartments by endocytosed pathogens but not to the self-nucleic acids.a However, under certain circumstances, such as poor clearance of apoptotic cells or cellular damage, self-nucleic acids may become aberrantly accessible to TLRs and can lead to autoimmunity [34,51–54]. Structural studies of the receptor–ligand complexes for TLR1–TLR2 heterodimer [55,56], TLR2–TLR6 heterodimer [57], TLR3 homodimer [58,59], TLR4 homodimer [55,56], and TLR5 homodimer [60] revealed that they all share an “m”-shaped architecture, composed of two individual horseshoe-shaped TLR ectodomains [61]. These findings together suggest that the activation of TLRs may be governed by a common mechanism.
The structural basis for activation of these TLRs by nucleic acids have been studied using X-ray crystallography [58,61–63]. While activation of TLRs has been shown to modulate and induce the production of AMPs from immune cells [64–67], the direct immunomodulatory effects of AMPs on TLRs are less clear. AMPs have been found within or co-localize and interact with a variety of immune cells, including dendritic cells, monocytes, B cells, and T cells [15,68–70]. AMPs are also produced by a variety of non-canonical immune cells such as keratinocytes [71]. A multitude of studies have demonstrated links between AMP expression, modulation of innate immune receptors like TLRs, and the subsequent upregulation or downregulation of pro-inflammatory and anti-inflammatory cytokines (Figure 1). We describe several recent studies of AMP interactions with TLRs, and how an understanding of the structural rules underlying TLR activation can enable prediction of immunomodulatory abilities of a broad range of molecules.
3. AMP-mediated immunomodulation via Toll-like receptors
3.1 dsDNA binding and TLR9 activation
TLR9 is typically thought of as a sensor for CpG DNA, which is relevant to viral and bacterial infections [63]. However, in a landmark study, Lande et al. demonstrated that human cathelicidin LL37 could potently activate plasmacytoid dendritic cells (pDCs) by forming immune complexes with human genomic dsDNA, accessing endosomal compartments, and binding to TLR9 (Figure 1) [72,73]. In the normal course of infection, LL37 enhances appropriate immune responses by binding to microbial DNA and inducing TLR9 activation. However, binding of LL37 to self-DNA and inappropriate activation of pDCs can break immune tolerance to self-nucleic acids and lead to autoimmunity. Indeed, overexpression of LL37 in autoimmune diseases like lupus [74] and psoriasis [75] has been linked to exacerbation of inflammation via TLR9 hyperactivation in pDCs and keratinocytes [76]. Interestingly, neutrophil extracellular traps (NETs), which are prevalent in autoimmune diseases [77,78], have been shown to release LL37–DNA complexes, which contribute to dysregulation of inflammation in lupus via pDC activation [74]. Intracellular LL37 can also enhance DNA-mediated TLR9 activation in B cells [79], in addition to dendritic cells and macrophages [80].
However, the ability to enable immune recognition of DNA and induce TLR9 activation in immune cells is not unique to LL37. Subsequent studies have shown that the AMPs human β-defensin 2 (hbD2), human β-defensin 3 (hbD3), and lysozyme can also co-assemble with dsDNA to activate pDCs, in a manner analogous to LL37 [81]. In the context of the immune response to malaria, histone–DNA complexes from Plasmodium falciparum were shown to activate dendritic cells via TLR9 [82]. Histones have also been well-characterized as antimicrobial peptides [83,84], in addition to their DNA-binding properties, suggesting that histones play dual roles in microbial defense and immunomodulation. Other DNA-binding proteins have also demonstrated the ability to enable TLR9 recognition of DNA. High-mobility group box-1 protein (HMGB1) is a nuclear DNA-binding protein released from necrotic cells that forms immune complexes with DNA and stimulates cytokine production through a TLR9-dependent pathway [85]. Other members of the HMGB family also facilitate recognition of nucleic acids by PRRs [86–88]. Interestingly, even pathogenic amyloids, some of which have been shown to have hidden antimicrobial activity [89,90], are able to induce chemoattraction [91] and bind to dsDNA and induce type I IFN production via TLR9 [92].
One potentially interesting question to ask is whether TLR9-activation by AMP–DNA immune complexes and the subsequent induction of an adaptive immune response resulting in autoantibody production against dsDNA leads to a self-amplifying pathway that further potentiates TLR9 activation. In a study of immune complexes obtained from lupus patients, anti-dsDNA autoantibodies were found to form complexes with DNA, enter endosomal and lysosomal compartments, and activate dendritic cells through cooperation of CD32 and TLR9 [93]. In a separate study, immune complexes formed between anti-nucleosome autoantibodies and mammalian DNA were shown to activate TLR9 in B cells [51].
From the examples above, it is evident that immune amplification of TLR9-mediated inflammation can be triggered by the formation of various protein–DNA complexes. Furthermore, this phenomenon is general and occurs across a broad range of molecules, including but clearly not limited to AMPs. What is equally clear from these studies is that a large number of natural AMPs and related molecules can induce TLR9 signaling through dsDNA binding, but many other molecules that also bind to dsDNA cannot. To delineate the necessary and sufficient criteria for immune activation by AMP–dsDNA complexes, we utilized high-resolution small-angle synchrotron X-ray scattering (SAXS) to solve the structures of complexes formed between dsDNA and AMPs, and correlated these findings with measurements of pDC IFN-α production [9]. We found that cationic molecules (such as AMPs) can form columnar nanocrystalline complexes with dsDNA, and that the inter-DNA spacing between parallel DNA columns changes with the identity of the cationic molecule. For 15 AMP–DNA and polycation–DNA complexes, measured inter-DNA spacings were correlated with induced IFN-α production from pDCs. Two functional classes of complexes were found: Complexes with inter-DNA spacings in the range of d = 3–4 nm led to strong amplification of IFN production (e.g., LL37–DNA and hbD3–DNA), while those with inter-DNA spacings outside of this range, d < 3 nm or d > 4 nm, resulted in significantly lower levels of IFN production, despite the fact that both classes of complexes have roughly equivalent levels of endosomal access. Interestingly, the optimum spacing of 3–4 nm is strongly suggestive, as it is approximately commensurate with the steric size of TLR9. Electrostatic interactions between the AMP–dsDNA complex and TLR9 are optimized when the inter-DNA spacing is in this range. Accordingly, the presentation of a spatially-periodic “grill” of parallel dsDNA by the nanocrystalline complex allows multivalent intercalative binding of the AMP–dsDNA complex to a cluster of TLR9 embedded in the endosomal membrane [94] (Figure 2A, B).
Figure 2. Nanocrystalline ordering of dsDNA and dsRNA by AMPs modulates TLR-mediated inflammation.
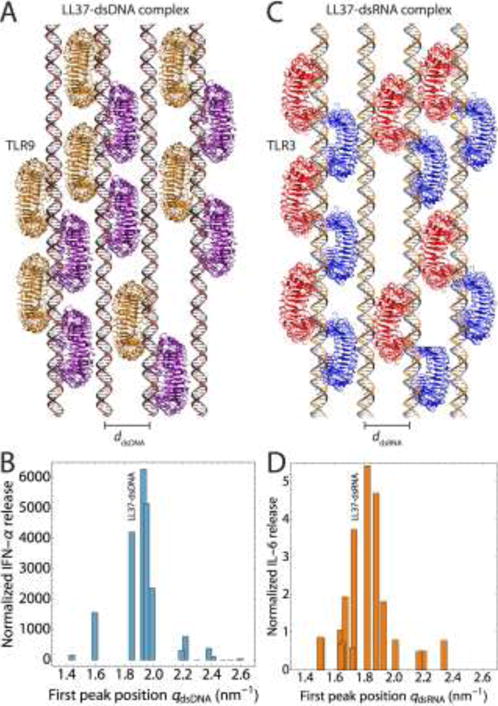
(A) Schematic of nanocrystalline LL37–dsDNA complexes binding to clustered arrays of TLR9 dimers in the endosomal membrane (ddsDNA = 3.40 nm). (B) IFN-ɑ production from human pDCs as a function of the inter-dsDNA spacing of AMP–dsDNA complexes. Complexes with inter-DNA spacings (ddsDNA) well-matched with the steric size of TLR9 leads to drastic amplification of inflammation, due to optimized multivalent intercalative binding of LL37–dsDNA to TLR9. The position of the LL37–dsDNA complex is labeled. Maximal IFN-ɑ production occurs for complexes with spacings in the range of 3.1–3.5 nm. Experimental data is adapted with permission from [9,94]. (C) Schematic of nanocrystalline LL37–dsRNA complexes binding to clustered arrays of TLR3 dimers in the endosomal membrane (ddsRNA = 3.63 nm). (D) IL-6 production from human keratinocytes as a function of the inter-dsRNA spacing of AMP–dsRNA complexes. Complexes with inter-RNA spacings (ddsRNA) well-matched with the steric size of TLR3 leads to strong amplification of inflammation, while those with poorly matched spacings do not. The position of the LL37–dsRNA complex is labeled. Maximal IL-6 production occurs for complexes with spacings in the range of 3.3–3.7 nm. Experimental data is adapted with permission from [10]. The optimal inter-ligand spacings for dsRNA is slightly larger than that for dsDNA, in agreement with the differences in steric size of TLR9 and TLR3, and the diameters of dsDNA and dsRNA. Schematics of TLR binding were generated in VMD (http://www.ks.uiuc.edu/Research/vmd/) using crystal structures from the PDB (3WPC for TLR9 and 3CIY for TLR3).
To understand these effects further, we formulated a mathematical model and conducted Monte-Carlo simulations of TLR9 binding to AMP–dsDNA complexes. As the inter-DNA spacing between parallel DNA ligands was varied, we observed different regimes of binding. At inter-DNA spacings smaller than the steric size of TLR9 (< 3 nm), low numbers of TLR9 were bound to the DNA due to sterically reduced accessibility to binding sites. Similar effects were observed when the inter-DNA spacings were too large (> 4 nm). However, as the inter-DNA spacing approached the optimal range (3–4 nm), we observed a drastic increase in the number of bound TLR9, experimentally correlating with the two orders of magnitude higher levels of immune activation and cytokine production [9,94].
The effect observed in experiments and in simulations is a variation of the phenomenon of “superselectivity”, one amplified by electrostatic effects. Superselectivity is defined as a superlinear relationship between the surface density of receptors and the surface density of bound ligands. The idea of superselectivity driven by ligand clustering is quite general. Nanoparticles and multivalent polymers have been recently engineered to bind superselectively to specific receptors in the context of cancer therapy and tissue targeting [95–97]. This strategy has been extended to the targeting of multiple receptors simultaneously [98]. The quantitative description of superselectivity is cognate to the idea of an “immunological synapse” in immunology, which was proposed in the context of specific ligand–receptor interactions [99], in which a cluster of ligands binding to a cluster of receptors drives a qualitatively different downstream response compared to single ligand–receptor interactions. This effect has been described for T-cell signaling [100], antigen-B cell receptor interactions [101], cell-surface binding of glycosaminoglycan [102], and dectin-glucan interactions [103]. To see how self-assembly of AMPs with dsDNA drives potent superselective binding of clustered DNA to TLR9 in pDCs, thereby driving dramatic upregulation of pro-inflammatory cytokines [9,94], it is helpful to consider the two binding surfaces that ultimately interact with one another. An “m”-shaped TLR9 dimer, which features two highly cationic horseshoe-shaped TLR9 ectodomains that form a groove for binding anionic dsDNA, can spatially organize with one another on the endosomal membrane to optimize binding to DNA. The surface of an LL37–DNA complex effectively presents a “grill” of periodic DNA ligands to these TLR9 receptors. When DNA ligands on the surface of an LL37–DNA complex has a periodicity that matches up with the periodicity of the cationic surfaces of clustered TLR9 dimers, strong intercalative binding can occur in which the TLR9 horseshoes penetrate the grill and effectively crosslink DNA. In fact, simulations show a strong recruitment of TLR9 to the LL37-DNA complex, resulting in superselective binding between the organized DNA and large numbers of TLR9 [9].
In the present context, a brief digression to recent advances in our understanding of electrostatic interactions in aqueous media is helpful, since such interactions govern the organization of DNA (and other nucleic acids) in nanocrystalline complexes, as well as the binding of these complexes to TLR. DNA and RNA are biological polyelectrolytes that carry uncompensated negative charge. Although these charges are strongly screened by the dielectric response of water and by the effects of salt, counterion entropy can result in surprisingly strong interactions between such charged objects. Like-charged objects repel because of the osmotic pressure of “squeezed” counterion distributions, and oppositely charged objects attract because of the entropy gain from the release of counterions into the bulk solution: the free energy gain upon binding between two oppositely charged macro-ions scales as kBT multiplied by the number of counterions released (with an even larger contribution if one includes ion hydration effects). Recent work that combines experiments and computer simulations shows that the osmotic pressure of counterions released during electrostatic binding of oppositely charged macro-ions can enhance the stability of polyelectrolyte complexes [104,105]. Examples of this type of electrostatic self-organization include chromosomes [106–110], non-viral gene delivery systems such as cationic polymers [111,112], dendrimers [113,114], and cationic lipids [115–118].
The degree of multivalent binding between TLR9 receptors and the DNA nanocrystalline complex depends in part on the size of the crystalline domain presented to the TLR9 receptors and thereby on the finite lateral size of the DNA bundle. Experiments show that the electrostatic complexes always appear to grow to a limited size [119,120], which will tend to limit the number of bound TLRs. It has been suggested that kinetics may limit the size of these polyelectrolyte aggregates [121]. Finite-sized polyelectrolyte bundles at equilibrium may also occur if steric effects from the finite size of the compensating ions prevent the bundle from reaching charge neutrality [122]. Likewise, frustration inherent in the bundle structure may cost an energy penalty [116,122]. Recent theoretical work has proposed that finite-sized condensed bundles are a consequence of the chirality of the semiflexible polyelectrolytes, which results in elastic strain from lattice shear increases as the bundle grows [123].
Until recently, we could not predict which molecules can modulate the immune system via TLR9 by binding to dsDNA. The results above provide a molecular recipe that quantitatively correlates the structural parameters of AMP–dsDNA complexes with their immunogenic potential via TLR9 recognition (Figure 2A, B). We highlight one recent example from host–pathogen interactions, where structural characterization of an amyloid–DNA complex enables mechanistic understanding of its immunogenic potential. An emerging field of study in host-pathogen interactions is centered on the biological role of bacterial functional amyloids. Examples include the phenol-soluble modulins from S. aureus [124,125], the FapC amyloid from Pseudomonas aeruginosa [126], and curli from Salmonella typhimurium and Escherichia coli [127]. These functional amyloids are produced in biofilms and self-assemble with extracellular DNA to form core structural components of the biofilm matrix [124]. Recent work has shown that curli forms complexes with extracellular bacterial DNA in biofilms, and leads to hyperactivation of dendritic cells and subsequent production of type I interferons [128]. Furthermore, these complexes triggered the production of autoantibodies in a mouse model of lupus, suggesting a link between infectious disease and autoimmunity [128]. The immune response to curli–DNA occurs in two steps: the curli–DNA nanocrystals gain endosomal access and entry into immune cells by binding to TLR2, and subsequent immune amplification occurs via multivalent binding to TLR9 [129]. Structural characterization using SAXS revealed that curli–DNA complexes form nanocrystals with a well-defined inter-DNA spacing in the range capable of amplifying TLR9-mediated inflammation [129].
The framework of described above is distinct from known mechanisms through which AMPs interact with TLRs. (Some cationic AMPs can block LPS-mediated activation of macrophages via direct binding to LPS, and significantly inhibit TNF-α production [130]. Cathelicidin can also inhibit TLR4-mediated induction of dendritic cell maturation and cytokine production in allergic contact immune responses [131].) In contrast, the mechanism of crystalline recognition by TLRs allows for a physical mode of amplification based on multivalent binding. Moreover, this mechanism of recognition leads to a number of interesting consequences. For example, since the existence of dsDNA and LL37 are both necessary for high levels of TLR9 activation, LL37 indirectly becomes an auto-antigen that TLR9 detects without direct TLR9–LL37 binding. In a more general compass, these results suggest that it is possible for TLR9 to indirectly detect and respond potently to a broad range of molecules in the environment that are not traditional TLR9 agonists. It will be interesting to explore these directions in the context of nanotoxicology.
3.2 dsRNA binding and TLR3 activation
Similar to TLR9 and CpG DNA, TLR3 was initially shown to sense viral dsRNAs in the context of infection [132]. However, recent work has shown that TLR3 also plays an important role in sensing skin injury by binding to non-coding self-dsRNA [133,134]. TLR3 is expressed at especially high levels in human keratinocytes, and plays a role in inducing regeneration after skin injury [135]. Similarly, keratinocytes also produce large amounts of AMPs, and are important cellular mediators of microbial defense in the skin [71,136,137]. Although many studies have shown that AMPs can influence TLR9 activation via DNA binding, AMPs have also been reported to modulate TLR3-mediated immune responses to dsRNA (Figure 1). LL37 and other cationic peptides have been shown to enhance or inhibit TLR3 signaling by viral dsRNAs [138–140]. Other RNA-binding proteins have also been implicated in enhancement of TLR3 activation in immune cells [141,142].
Based on these findings, we hypothesized that the structural ordering of dsRNAs by AMPs can influence TLR3 activation, similar to how scaffolding of dsDNA by AMPs modulates TLR9 activation. To test this hypothesis, we set out to define the structural rules underlying TLR3 activation by AMPs, cognate to our previous work on TLR9. Using SAXS, we measured the crystallinity parameters of 15 AMP–DNA and polycation–dsRNA complexes and correlated them with measurements of keratinocyte IL-6 production. This model system was chosen in the context of innate immune responses to dsRNA in the skin, where defense against microbial infection is critical [137]. In addition, in autoimmune diseases like psoriasis, IL-6 production by keratinocytes play a role in aberrant cytokine production in response to LL37 [134]. We find that, similar to the TLR9–dsDNA system, AMP–dsRNA complexes segregate into two classes: Complexes with inter-dsRNA spacings commensurate with the steric size of TLR3 (~3.3–3.7 nm) lead to a ~5 to 10-fold amplification in IL-6 production from keratinocytes (e.g., LL37–dsRNA and buforin–dsRNA) [10,143]. Conversely, complexes with inter-dsRNA spacings much smaller or larger (< 3 nm or > 4 nm) than the steric size of TLR3 lead to low levels of activation (e.g., HIV TAT–dsRNA, lysozyme–dsRNA). A statistically significant, nonlinear relationship was found between the inter-dsRNA spacing and the quantitative amount of IL-6 induction (Figure 2C, D).
To investigate interactions between the nanocrystalline dsRNA immune complexes and TLR3 in greater detail, we developed an improved statistical mechanical model and computer simulation [9,94]. The specific question we engage is whether the distance between ligands is the only structural characteristic of the nanocrystal that is relevant to immune activation. We expanded the flexibility of our model by taking into account an additional structural characteristic of crystallinity in addition to the inter-ligand spacing: the size of the nanocrystal (“domain size”), which gives the average number of ligand repeats in the crystal. By correlating theoretical predictions to the experimental SAXS measurements of crystallinity, we reveal that immune activation (and hyperactivation) via TLR3 is a function of the inter-dsRNA spacing and the domain size [10]. As the inter-dsRNA spacing is increased from below the steric size of TLR3 to optimal spacings, the number of bound TLR3 drastically increased in a superselective manner. For AMP–dsRNA complexes that exhibit optimal steric matching to TLR3, an increase in the domain size of the crystal led to an increase in the quantitative amount of immune activation. However, those exhibiting poor steric matching to TLR3 did not. Intriguingly, AMP–dsRNA complexes with slightly larger than optimal spacings can compensate for reduced activation with a larger domain size, to a certain extent, suggesting that a broader range of AMPs may have the ability to induce TLR3 signaling. In summary, TLR3 activation can be modulated by the ordered presentation of nanocrystalline AMP–dsRNA immune complexes [10].
The qualitative concept of clustering of immune ligands to drive immune responses is not a new concept [144]. However, the observation that ordered, crystalline arrangements of dsRNA ligands within AMP–dsRNA complexes can enhance TLR3 signaling, and that such enhancement can be quantitatively correlated with structural parameters describing the degree of crystallinity are significant. Indeed, it is surprising that the innate immune system can recognize crystallinity in this way, rather than just PAMPs. In light of recent work on TLR9-mediated immune amplification by AMP–dsDNA complexes in pDCs [9], these results suggest general mechanisms through which innate immunity can be amplified: The observation of cognate phenomena across multiple cell types, TLRs, and cytokines is consistent with a general conceptual framework in which the ordered presentation of immune ligands by AMPs (and other molecules) drive immune responses, and likely can be extended to other TLRs and immune ligands (e.g., TLR4 and LPS).
3.3 ssRNA binding and TLR7/TLR8 activation
As with nucleic-acid-sensing TLR9 and TLR3, the recognition of ssRNA by TLR7 and TLR8 [38–40] is also crucial in the defense against invading pathogens. Studies have demonstrated that TLR7 and TLR8 recognize guanosine (G)- and uridine (U)-rich viral ssRNA [38], ssRNA viruses such as vesicular stomatitis virus (VSV) and influenza [40], and more recently, bacterial and mitochondrial RNA [145]. TLR7/8–ssRNA binding subsequently recruits MyD88 to activate the NF-κB and IRF7 pathways, which lead to the production of pro-inflammatory cytokines and type I IFN. In addition to these natural ssRNA agonists, synthetic imidazoquinoline-like molecules, including imiquimod (R-837), resiquimod (R-848), and S-27609, as well as guanosine analogues, such as loxoribine, have been shown to activate TLR7. Resiquimod has also been identified to activate TLR8 [146].
TLR7 and TLR8 are closely related phylogenetically and structurally to each other and to TLR9 [147,148], and are both found in intracellular endosomal compartments [45,46]. However, several key differences exist among the two receptors [146], such as distinct expression patterns among specific cell types. TLR7 expression primarily occurs in B cells and pDCs, and is associated with the production of IFN-α and IFN-α-regulated cytokines. In contrast, TLR8 is predominantly expressed in monocytes/macrophages and myeloid dendritic cells (mDCs), and is involved in the production of pro-inflammatory cytokines such as TNF-α [146,149–152]. Furthermore, TLR7 and TLR8 have been shown to detect different sequence-specific RNA motifs [149,153]. Taken together, the distinction in cell-type expression among the two receptors to result in divergent cytokine induction profiles points to their specialized and complementary functionalities [146]. Their concerted action would therefore enable efficient and specific immune responses to different types of pathogens [151,154]. In fact, this specialization of TLRs is not unique to TLR7 and TLR8, and occurs among other TLRs. Substantial data has been collected on dendritic cell functional heterogeneity, such as dendritic cell subset-specific TLR expression and production of cytokines involved in inducing distinct effector cells [155–157].
The aberrant transportation of self-nucleic acids to endosomes enables the activation of TLRs, which can promote autoimmunity. In fact, self-RNA has been shown to act as ligands for TLR7 and TLR8 [158] and their internalization into endosomal compartments exacerbates autoimmune diseases such as systemic lupus erythematosus (SLE) [159], psoriasis [160], and antiphospholipid syndrome (APS) [161]. Interestingly, expression of TLR8, but not TLR7, is correlated to disease activity in patients with systemic arthritis, which suggests that independent signaling through TLR8 contributes to the pathogenesis of the disease [162,163]. Furthermore, oligodeoxynucleotides have been shown to antagonize nucleic-acid-sensing TLRs to alleviate disease conditions for psoriasis [164] and SLE [165] mouse models. Like we have observed for TLR9 and TLR3, these findings also implicate a role of TLR7 and TLR8 in the development of autoimmunity and excessive inflammation.
Normally, self-nucleic acids are rapidly degraded in the extracellular environment and are unable to reach the endosomal compartments [44]. However, self-nucleic acids that are aberrantly transported into endosomes can access nucleic-acid-sensing TLRs and trigger pDC activation, promoting autoimmunity (Figure 1). In the case of psoriasis, self-DNA forms complexes with LL37 and gains access to endosomal TLR9, which triggers pDCs to produce IFN-α and initiate the autoimmune process [72]. LL37–self-RNA complexes have been observed in psoriatic skin in vivo [160] and high levels of LL37 have also been reported in a variety of chronic inflammatory diseases, such as rosacea [166], rheumatoid arthritis [167], ulcerative colitis [168], chronic nasal inflammatory disease [169], sarcoidosis [170], and cystic fibrosis [171]. To examine the possible involvement of TLR7 and TLR8 in the pathogenesis of psoriasis, Ganguly et al. investigated whether LL37 is also capable of interacting with self-RNA released by dying cells to activate immune cells. Similarly, LL37 was found to complex with self-RNA via electrostatic interactions and reach the endosomal compartments of dendritic cells, inducing TLR7 activation in pDCs and secretion of IFN-α. Moreover, LL37–self-RNA complexes also triggered the activation of TLR8 in mDCs to produce TNF-α and IL-6. Interestingly, TLR7 in pDCs and TLR8 in mDCs were activated by self-RNA mixed with LL37, but not when self-RNA was administered alone or mixed with scrambled peptide GL37. Furthermore, complexation of self-RNA with LL37 was shown to protect self-RNA against degradation from extracellular RNases, which normally prevent RNA from accessing intracellular compartments [160]. From these findings, it will be interesting to see if the complexation of AMPs with RNA ligands activate TLR7 and TLR8 in a cognate, structural manner similar to that identified for AMP–dsDNA complexes with TLR9 [9] and AMP–dsRNA complexes with TLR3 [10]. Present efforts are under way to do such studies.
4. Conclusions and Outlook
In this review, we discussed the immunomodulatory properties of natural and synthetic AMPs and their associated mechanisms. AMPs are known to kill microbes through direct activity, including membrane permeation, disruption of electrochemical gradients, and inhibition of metabolic machinery. However, recent evidence shows that AMPs can also orchestrate host immune responses by communicating with the innate and adaptive immune systems through receptor modulation. AMPs can directly bind to various cell-surface or intracellular receptors on immune cells, triggering chemotaxis, differentiation/maturation, and cytokine production (Figure 1).
We also highlighted an underappreciated facet of AMP immunomodulation: AMPs can also organize immune ligands such as dsDNA and dsRNA into nanocrystalline complexes that amplify TLR9 and TLR3 activation (Figure 2). Furthermore, the degree of immune amplification can be quantitatively correlated from the crystallinity parameters of AMP–nucleic acid complexes. Indeed, the same physicochemical features of AMPs that enable their antimicrobial and membrane activity (e.g., cationic charge, amphipathicity) also enable strong electrostatic binding to nucleic acids like dsDNA and dsRNA., it will be interesting to see whether these general mechanisms extend beyond TLRs to various cytosolic nucleic acid sensors. AMP involvement in these corollary pathways have been implicated in earlier studies. For example, LL37 can induce IFN-α production from keratinocytes by binding to cytosolic DNA [172]. LL37 drives IFN-β production in epidermal keratinocytes during skin injury via the mitochondrial antiviral-signaling protein (MAVS). LL37 enables recognition of self-dsRNA through MAVS in addition to TLR3 [134]. In pDCs, LL37 transports dsDNA into endosomal compartments, triggering TLR9 activation [72]. However, in non-pDC dendritic cells, LL37 enables cytosolic DNA sensing via STING and TBK1 kinase [173].
In parallel direction of inquiry, it will be informative to critically dissect the optimal structural and physicochemical requirements for AMP antimicrobial and immunomodulatory activity via TLRs, and compare and contrast these competing requirements. Perhaps it is worthwhile to revisit old questions regarding the structural requirements of AMPs in the context of immunomodulatory effects reviewed here. We have known for some time that cationic charge and hydrophobicity are required for AMP activity, and have interpreted that in the perspective of membrane activity. We can ask broader questions now. How does immunomodulatory behavior change with cationic charge and hydrophobicity? Can we predict which AMPs will be more strongly antimicrobial or immunomodulatory and which innate immune receptors will be involved? Can we rationally design AMPs and AMP-like molecules, and tune both antimicrobial and immunomodulatory properties independently? The majority of immunomodulatory AMPs were first characterized as antimicrobial peptides, and then were later found to possess immunomodulatory activity. However, recent work shows that many traditional immune-signaling molecules that exert their functions through specific membrane-bound receptors (such as cytokines) also possess hidden direct antimicrobial activity. In fact, an increasing body of evidence shows that this categorical delineation between antimicrobial molecules and immunomodulatory molecules is becoming progressively blurred [174]. Perhaps the emerging “kinocidin” family of proteins can provide insight into how nature multiplexes antimicrobial and immunomodulatory activity. Kinocidins are cytokines with intrinsic antimicrobial activity [175], and many share core physicochemical and structural similarities with AMPs. For example, various members of the chemokine family of cytokines contain a β-sheet rich “ɣ-core” motif that is also characteristic of classical defensins [175,176], and exhibit facial amphiphilicity and net cationic charge [174,177]. Many of these molecules contain modular components that share structural homology with α-helical and β-sheet AMPs [178,179]. For example, in a pioneering screen of over 30 human chemokines from these families, 21 were found to possess potent broad-spectrum antimicrobial activity comparable to human defensins, suggesting that chemokines may also directly kill microbes in addition to their chemotactic functions [180]. In another series of studies, the platelet chemokine (C-X-C motif) ligand 4 (CXCL4)/platelet factor 4 (PF-4), its rabbit analogue thrombin-induced platelet microbicidal protein 1 (tPMP-1), and a synthetic congener peptide derived from CXCL4 C-terminal domain was found to have potent antimicrobial properties [179,181–184]. Other studies have shown that IL-8 and MCP-1, two important mediators of chemotaxis, also have antimicrobial activity [185]. Strikingly, IL-8, CXCL4/PF4, and MCP-1 were all found to share strong structural homology with human β-defensins, namely the ɣ-core motif, indicating that a common scaffold has been optimized by nature for dual antimicrobial and immunomodulatory activity. Recently, the cytokines IL-26 and IFN-β were also discovered to have hidden antimicrobial activity [186,187] [188] [189]. Other molecules in innate immunity such as complement C3a and the S100 class of endogenous calcium-binding proteins have also exhibited dual antimicrobial and immunomodulatory activities [190–193]. The development of tools to detect hidden antimicrobial activity in proteins has enabled the possibility of uniting disparate peptide taxonomies under a common umbrella of multifunctional proteins that exhibit different degrees of antimicrobial activity, receptor-binding activity, and immunomodulatory ability [194–196].
In our view, it will be worthwhile in future work to work towards a more nuanced understanding of the self-assembly of AMPs and related molecules with traditional immune ligands in the context of inflammation. These studies also question the conventional demarcation between “pro-inflammatory” and “anti-inflammatory” AMPs and cytokines. Context matters especially to pleiotropic molecules, whether it is the tissue-specific expression of immunomodulatory mediators or activation of multiple receptors in specific cells in different tissues. For example, an immune signaling molecule may induce an anti-inflammatory response via binding to its cell-surface receptor, but could simultaneously induce a pro-inflammatory response by binding to nucleic acids and signaling through TLRs. Instead of measuring a binary response, we propose models to quantitatively predict and measure the degree of inflammation on a continuous scale as a function of the biophysical and structural properties of AMPs, immune ligands, and their targets.
Finally, the ability of AMPs to modulate the innate system via mechanisms described here is a double-edged sword: In the normal context of infection, AMPs bolster the immune response by enabling immune cell signaling, recruitment, and proliferation. However, overexpression of AMPs has clearly been associated with exacerbation of many additional chronic inflammatory diseases like systemic sclerosis, vasculitis, atherosclerosis, and others. Further structure–function and mechanistic studies will be required to fully map out the immunomodulation landscapes of AMPs, their pathophysiological role in disease, and strategies to regulate their immunomodulatory abilities.
Acknowledgments
E.Y.L. acknowledges support from the Systems and Integrative Biology Training Program (T32GM008185), the Medical Scientist Training Program (T32GM008042), and the Dermatology Scientist Training Program (T32AR071307) at UCLA. E.Y.L. and G.C.L.W. also acknowledge an Early Career Research Grant and a Discovery Grant from the National Psoriasis Foundation, respectively. G.C.L.W. and M.W.L. acknowledge support from NIH Grant 1R21AI122212. X-ray research was conducted at Stanford Synchrotron Radiation Lightsource, SLAC National Laboratory, supported by the US DOE Office of Basic Energy Sciences under Contract no. DE-AC02-76SF00515.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 3.Jenssen H, Hancock REW. Therapeutic potential of HDPs as immunomodulatory agents. Methods Mol Biol. 2010;618:329–347. doi: 10.1007/978-1-60761-594-1_20. [DOI] [PubMed] [Google Scholar]
- 4.Easton DM, Nijnik A, Mayer ML, Hancock REW. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends in Biotechnology. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nijnik A, Hancock R. Host defence peptides: antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg Health Threats J. 2009;2:e1. doi: 10.3134/ehtj.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haney EF, Hancock REW. Peptide design for antimicrobial and immunomodulatory applications. Peptide Science. 2013;100:572–583. doi: 10.1002/bip.22250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholls EF, Madera L, Hancock REW. Immunomodulators as adjuvants for vaccines and antimicrobial therapy. Annals of the New York Academy of Sciences. 2010;1213:46–61. doi: 10.1111/j.1749-6632.2010.05787.x. [DOI] [PubMed] [Google Scholar]
- 8.Morizane S, Gallo RL. Antimicrobial peptides in the pathogenesis of psoriasis. J Dermatol. 2012;39:225–230. doi: 10.1111/j.1346-8138.2011.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt NW, Jin F, Lande R, Curk T, Xian W, Lee C, et al. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat Mater. 2015;14:696–700. doi: 10.1038/nmat4298. [DOI] [PubMed] [Google Scholar]
- 10.Lee EY, Takahashi T, Curk T, Dobnikar J, Gallo RL, Wong GCL. Crystallinity of Double-Stranded RNA-Antimicrobial Peptide Complexes Modulates Toll-Like Receptor 3-Mediated Inflammation. ACS Nano. 2017 doi: 10.1021/acsnano.7b05234. acsnano.7b05234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowdish DME, Davidson DJ, Hancock REW. Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol. 2006;306:27–66. doi: 10.1007/3-540-29916-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowdish DME, Davidson DJ, Scott MG, Hancock REW. Immunomodulatory activities of small host defense peptides. Antimicrobial Agents and Chemotherapy. 2005;49:1727–1732. doi: 10.1128/AAC.49.5.1727-1732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mookherjee N, Brown KL, Bowdish DME, Doria S, Falsafi R, Hokamp K, et al. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J Immunol. 2006;176:2455–2464. doi: 10.4049/jimmunol.176.4.2455. [DOI] [PubMed] [Google Scholar]
- 14.Ruan Y, Shen T, Wang Y, Hou M, Li J, Sun T. Antimicrobial peptide LL-37 attenuates LTA induced inflammatory effect in macrophages. Int Immunopharmacol. 2013;15:575–580. doi: 10.1016/j.intimp.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Scott MG, Davidson DJ, Gold MR, Bowdish D, Hancock REW. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J Immunol. 2002;169:3883–3891. doi: 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- 16.Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J Immunol. 2005;174:6257–6265. doi: 10.4049/jimmunol.174.10.6257. [DOI] [PubMed] [Google Scholar]
- 17.Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, et al. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 18.Van Wetering S, Mannesse-Lazeroms SP, Van Sterkenburg MA, Daha MR, Dijkman JH, Hiemstra PS. Effect of defensins on interleukin-8 synthesis in airway epithelial cells. Am J Physiol. 1997;272:L888–96. doi: 10.1152/ajplung.1997.272.5.L888. [DOI] [PubMed] [Google Scholar]
- 19.Territo MC, Ganz T, Selsted ME, Lehrer R. Monocyte-chemotactic activity of defensins from human neutrophils. J Clin Invest. 1989;84:2017–2020. doi: 10.1172/JCI114394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lillard JW, Boyaka PN, Chertov O, Oppenheim JJ, McGhee JR. Mechanisms for induction of acquired host immunity by neutrophil peptide defensins. Proc Natl Acad Sci U S a. 1999;96:651–656. doi: 10.1073/pnas.96.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, et al. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science. 2002;298:1025–1029. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- 22.Tongaonkar P, Trinh KK, Schaal JB, Tran D, Gulko PS, Ouellette AJ, et al. Rhesus macaque θ-defensin RTD-1 inhibits proinflammatory cytokine secretion and gene expression by inhibiting the activation of NF-κB and MAPK pathways. Journal of Leukocyte Biology. 2015;98:1061–1070. doi: 10.1189/jlb.3A0315-102R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaal JB, Tran D, Tran P, Ösapay G, Trinh K, Roberts KD, et al. Rhesus macaque theta defensins suppress inflammatory cytokines and enhance survival in mouse models of bacteremic sepsis. PLoS ONE. 2012;7:e51337. doi: 10.1371/journal.pone.0051337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenger S, Hanson DA, Teitelbaum R, Dewan P, Niazi KR, Froelich CJ, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282:121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 25.McInturff JE, Wang SJ, Machleidt T, Lin TR, Oren A, Hertz CJ, et al. Granulysin-derived peptides demonstrate antimicrobial and anti-inflammatory effects against Propionibacterium acnes. Journal of Investigative Dermatology. 2005;125:256–263. doi: 10.1111/j.0022-202X.2005.23805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veldhuizen EJA, Schneider VAF, Agustiandari H, van Dijk A, Tjeerdsma-van Bokhoven JLM, Bikker FJ, et al. Antimicrobial and immunomodulatory activities of PR-39 derived peptides. PLoS ONE. 2014;9:e95939. doi: 10.1371/journal.pone.0095939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Som A, Navasa N, Percher A, Scott RW, Tew GN, Anguita J. Identification of synthetic host defense peptide mimics that exert dual antimicrobial and anti-inflammatory activities. Clin Vaccine Immunol. 2012;19:1784–1791. doi: 10.1128/CVI.00291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaker HD, Som A, Ayaz F, Lui D, Pan W, Scott RW, et al. Synthetic mimics of antimicrobial peptides with immunomodulatory responses. J Am Chem Soc. 2012;134:11088–11091. doi: 10.1021/ja303304j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva ON, de la Fuente-Núñez C, Haney EF, Fensterseifer ICM, Ribeiro SM, Porto WF, et al. An anti-infective synthetic peptide with dual antimicrobial and immunomodulatory activities. Sci Rep. 2016;6:35465. doi: 10.1038/srep35465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uematsu S, Akira S. Toll-Like receptors (TLRs) and their ligands. Handb Exp Pharmacol. 2008;183:1–20. doi: 10.1007/978-3-540-72167-3_1. [DOI] [PubMed] [Google Scholar]
- 31.Blasius AL, Beutler B. Intracellular Toll-like Receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi O, Kawai T, Mühlradt PF, Morr M, Radolf JD, Zychlinsky A, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 34.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 35.O’Neill LAJ, Golenbock DT, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 36.Akira S, Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunology Letters. 2003;85:85–95. doi: 10.1016/S0165-2478(02)00228-6. [DOI] [PubMed] [Google Scholar]
- 37.Carpenter S, O’Neill LAJ. How important are Toll-like receptors for antimicrobial responses? Cellular Microbiology. 2007;9:1891–1901. doi: 10.1111/j.1462-5822.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 38.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 39.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 40.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S a. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 42.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. Journal of Experimental Medicine. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krug A, French AR, Barchet W, Fischer JAA, Dzionek A, Pingel JT, et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 45.Heil F, Ahmad-Nejad P, Hemmi H, Hochrein H, Ampenberger F, Gellert T, et al. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur J Immunol. 2003;33:2987–2997. doi: 10.1002/eji.200324238. [DOI] [PubMed] [Google Scholar]
- 46.Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci U S a. 2003;100:6646–6651. doi: 10.1073/pnas.0631696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, et al. Subcellular Localization of Toll-Like Receptor 3 in Human Dendritic Cells. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 48.Kanno A, Tanimura N, Ishizaki M, Ohko K, Motoi Y, Onji M, et al. Targeting cell surface TLR7 for therapeutic intervention in autoimmune diseases. Nat Commun. 2015;6:6119. doi: 10.1038/ncomms7119. [DOI] [PubMed] [Google Scholar]
- 49.Lindau D, Mussard J, Wagner BJ, Ribon M, Rönnefarth VM, Quettier M, et al. Primary blood neutrophils express a functional cell surface Toll-like receptor 9. Eur J Immunol. 2013;43:2101–2113. doi: 10.1002/eji.201142143. [DOI] [PubMed] [Google Scholar]
- 50.Onji M, Kanno A, Saitoh SI, Fukui R, Motoi Y, Shibata T, et al. An essential role for the N-terminal fragment of Toll-like receptor 9 in DNA sensing. Nat Commun. 2013;4:1949. doi: 10.1038/ncomms2949. [DOI] [PubMed] [Google Scholar]
- 51.Marshak-Rothstein A, Busconi L, Lau CM, Tabor AS, Leadbetter EA, Akira S, et al. Comparison of CpG s-ODNs, chromatin immune complexes, and dsDNA fragment immune complexes in the TLR9-dependent activation of rheumatoid factor B cells. J Endotoxin Res. 2004;10:247–251. doi: 10.1179/096805104225005850. [DOI] [PubMed] [Google Scholar]
- 52.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 53.Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. Intracellular Nucleic Acid Detection in Autoimmunity. Annu Rev Immunol. 2017;35:313–336. doi: 10.1146/annurev-immunol-051116-052331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theofilopoulos AN, Kono DH, Beutler B, Baccala R. Intracellular nucleic acid sensors and autoimmunity. J Interferon Cytokine Res. 2011;31:867–886. doi: 10.1089/jir.2011.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Jin MS, Lee JO. Structures of TLR-ligand complexes. 2008;20:414–419. doi: 10.1016/j.coi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 60.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Botos I, Segal DM, Davies DR. The Structural Biology of Toll-like Receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang JY, Lee JO. Structural biology of the Toll-like receptor family. Annu Rev Biochem. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 63.Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, Uchiyama S, et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 64.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 65.Birchler T, Seibl R, Büchner K, Loeliger S, Seger R, Hossle JP, et al. Human Toll-like receptor 2 mediates induction of the antimicrobial peptide human beta-defensin 2 in response to bacterial lipoprotein. Eur J Immunol. 2001;31:3131–3137. doi: 10.1002/1521-4141(200111)31:11<3131::AID-IMMU3131>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 66.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 67.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S a. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davidson DJ, Currie AJ, Reid GSD, Bowdish DME, MacDonald KL, Ma RC, et al. The cationic antimicrobial peptide LL-37 modulates dendritic cell differentiation and dendritic cell-induced T cell polarization. J Immunol. 2004;172:1146–1156. doi: 10.4049/jimmunol.172.2.1146. [DOI] [PubMed] [Google Scholar]
- 69.Zhang XJ, Wang P, Zhang N, Chen DD, Nie P, Li JL, et al. B Cell Functions Can Be Modulated by Antimicrobial Peptides in Rainbow Trout Oncorhynchus mykiss: Novel Insights into the Innate Nature of B Cells in Fish. Frontiers in Immunology. 2017;8:388. doi: 10.3389/fimmu.2017.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, et al. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood. 2000;96:3086–3093. [PubMed] [Google Scholar]
- 71.Braff MH, Di Nardo A, Gallo RL. Keratinocytes store the antimicrobial peptide cathelicidin in lamellar bodies. J Invest Dermatol. 2005;124:394–400. doi: 10.1111/j.0022-202X.2004.23443.x. [DOI] [PubMed] [Google Scholar]
- 72.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 73.Gilliet M, Lande R. Antimicrobial peptides and self-DNA in autoimmune skin inflammation. Curr Opin Immunol. 2008;20:401–407. doi: 10.1016/j.coi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 74.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Science Translational Medicine. 2011;3:73ra19–73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lande R, Botti E, Jandus C, Dojcinovic D, Fanelli G, Conrad C, et al. The antimicrobial peptide LL37 is a T-cell autoantigen in psoriasis. Nat Commun. 2014;5:5621. doi: 10.1038/ncomms6621. [DOI] [PubMed] [Google Scholar]
- 76.Morizane S, Yamasaki K, Mühleisen B. Cathelicidin antimicrobial peptide LL-37 in psoriasis enables keratinocyte reactivity against TLR9 ligands, Journal of Investigative. Dermatology. 2012;132:135–143. doi: 10.1038/jid.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Radic M, Marion TN. Neutrophil extracellular chromatin traps connect innate immune response to autoimmunity. Semin Immunopathol. 2013;35:465–480. doi: 10.1007/s00281-013-0376-6. [DOI] [PubMed] [Google Scholar]
- 78.Kaplan MJ, Radic M. Neutrophil extracellular traps: double-edged swords of innate immunity. J Immunol. 2012;189:2689–2695. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hurtado P, Peh CA. LL-37 promotes rapid sensing of CpG oligodeoxynucleotides by B lymphocytes and plasmacytoid dendritic cells. J Immunol. 2010;184:1425–1435. doi: 10.4049/jimmunol.0902305. [DOI] [PubMed] [Google Scholar]
- 80.Nakagawa Y, Gallo RL. Endogenous intracellular cathelicidin enhances TLR9 activation in dendritic cells and macrophages. J Immunol. 2015;194:1274–1284. doi: 10.4049/jimmunol.1402388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lande R, Chamilos G, Ganguly D, Demaria O, Frasca L, Durr S, et al. Cationic antimicrobial peptides in psoriatic skin cooperate to break innate tolerance to self-DNA. Eur J Immunol. 2015;45:203–213. doi: 10.1002/eji.201344277. [DOI] [PubMed] [Google Scholar]
- 82.Gowda NM, Wu X, Gowda DC. The Nucleosome (Histone-DNA Complex) Is the TLR9-Specific Immunostimulatory Component of Plasmodium falciparum That Activates DCs. PLoS ONE. 2011;6:e20398–14. doi: 10.1371/journal.pone.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawasaki H, Iwamuro S. Potential roles of histones in host defense as antimicrobial agents. Infect Disord Drug Targets. 2008;8:195–205. doi: 10.2174/1871526510808030195. [DOI] [PubMed] [Google Scholar]
- 84.Lee DY, Huang CM, Nakatsuji T, Thiboutot D, Kang SA, Monestier M, et al. Histone H4 is a major component of the antimicrobial action of human sebocytes. J Invest Dermatol. 2009;129:2489–2496. doi: 10.1038/jid.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 86.Yanai H, Ban T, Taniguchi T. Essential role of high-mobility group box proteins in nucleic acid-mediated innate immune responses. J Intern Med. 2011;270:301–308. doi: 10.1111/j.1365-2796.2011.02433.x. [DOI] [PubMed] [Google Scholar]
- 87.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 88.Urbonaviciute V, Fürnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J Exp Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kagan BL, Jang H, Capone R, Arce FT, Ramachandran S, Lal R, et al. Antimicrobial Properties of Amyloid Peptides. Mol Pharmaceutics. 2011;9:708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS ONE. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stolzenberg E, Berry D, Yang D, Lee EY, Kroemer A, Kaufman S, et al. A Role for Neuronal Alpha-Synuclein in Gastrointestinal Immunity. J Innate Immun. 2017 doi: 10.1159/000477990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Di Domizio J, Dorta-Estremera S, Gagea M, Ganguly D, Meller S, Li P, et al. Nucleic acid-containing amyloid fibrils potently induce type I interferon and stimulate systemic autoimmunity. Proc Natl Acad Sci U S a. 2012;109:14550–14555. doi: 10.1073/pnas.1206923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody–DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee EY, Lee CK, Schmidt NW, Jin F, Lande R, Curk T, et al. A review of immune amplification via ligand clustering by self-assembled liquid-crystalline DNA complexes. Adv Colloid Interface Sci. 2016;232:17–24. doi: 10.1016/j.cis.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Albertazzi L, Martinez-Veracoechea FJ, Leenders CMA, Voets IK, Frenkel D, Meijer EW. Spatiotemporal control and superselectivity in supramolecular polymers using multivalency. Proc Natl Acad Sci U S a. 2013;110:12203–12208. doi: 10.1073/pnas.1303109110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dubacheva GV, Curk T, Mognetti BM, Auzély-Velty R, Frenkel D, Richter RP. Superselective Targeting Using Multivalent Polymers. J Am Chem Soc. 2014;136:1722–1725. doi: 10.1021/ja411138s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dubacheva GV, Curk T, Auzély-Velty R, Frenkel D, Richter RP. Designing multivalent probes for tunable superselective targeting. Proc Natl Acad Sci U S a. 2015;112:5579–5584. doi: 10.1073/pnas.1500622112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Curk T, Dobnikar J, Frenkel D. Optimal multivalent targeting of membranes with many distinct receptors. Proc Natl Acad Sci U S a. 2017;114:7210–7215. doi: 10.1073/pnas.1704226114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 100.Germain RN. T-cell signaling: the importance of receptor clustering. Curr Biol. 1997;7:R640–4. doi: 10.1016/s0960-9822(06)00323-x. [DOI] [PubMed] [Google Scholar]
- 101.Snapper CM, Kehry MR, Castle BE, Mond JJ. Multivalent, but not divalent, antigen receptor cross-linkers synergize with CD40 ligand for induction of Ig synthesis and class switching in normal murine B cells. A redefinition of the TI-2 vs T cell-dependent antigen dichotomy. J Immunol. 1995;154:1177–1187. [PubMed] [Google Scholar]
- 102.Lesley J, Hascall VC, Tammi M, Hyman R. Hyaluronan binding by cell surface CD44. Journal of Biological Chemistry. 2000;275:26967–26975. doi: 10.1074/jbc.M002527200. [DOI] [PubMed] [Google Scholar]
- 103.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, et al. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature. 2011;472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sanders LK, Guáqueta C, Angelini TE, Lee JW, Slimmer SC, Luijten E, et al. Structure and stability of self-assembled actin-lysozyme complexes in salty water. Phys Rev Lett. 2005;95:108302. doi: 10.1103/PhysRevLett.95.108302. [DOI] [PubMed] [Google Scholar]
- 105.Sanders LK, Xian W, Guáqueta C, Strohman MJ, Vrasich CR, Luijten E, et al. Control of electrostatic interactions between F-actin and genetically modified lysozyme in aqueous media. Proc Natl Acad Sci U S a. 2007;104:15994–15999. doi: 10.1073/pnas.0705898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mohammad-Rafiee F, Golestanian R. Elastic Correlations in Nucleosomal DNA Structure. Phys Rev Lett. 2005;94:238102. doi: 10.1103/PhysRevLett.94.238102. [DOI] [PubMed] [Google Scholar]
- 107.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 Å resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 108.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 109.Schiessel H. The nucleosome: A transparent, slippery, sticky and yet stable DNA-protein complex. Eur Phys J E. 2006;19:251–262. doi: 10.1140/epje/i2005-10049-y. [DOI] [PubMed] [Google Scholar]
- 110.Chen Y, Tokuda JM, Topping T, Sutton JL, Meisburger SP, Pabit SA, et al. Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Research. 2014;42:8767–8776. doi: 10.1093/nar/gku562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.DeRouchey J, Netz RR, Radler JO. Structural investigations of DNA-polycation complexes. Eur Phys J E. 2005;16:17–28. doi: 10.1140/epje/e2005-00003-4. [DOI] [PubMed] [Google Scholar]
- 112.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A Versatile Vector for Gene and Oligonucleotide Transfer Into Cells in Culture and in-Vivo - Polyethylenimine. Proc Natl Acad Sci U S a. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR. Efficient transfer of genetic material into mammalian cells using Starburst polyamidoamine dendrimers. Proc Natl Acad Sci U S a. 1996;93:4897–4902. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Evans HM, Ahmad A, Ewert K, Pfohl T, Martin-Herranz A, Bruinsma RF, et al. Structural polymorphism of DNA-dendrimer complexes. Phys Rev Lett. 2003;91:075501. doi: 10.1103/PhysRevLett.91.075501. [DOI] [PubMed] [Google Scholar]
- 115.Kanduč M, Dobnikar J, Podgornik R. Counterion-mediated electrostatic interactions between helical molecules. Soft Matter. 2009;5:868–877. doi: 10.1039/B811795K. [DOI] [Google Scholar]
- 116.Angelini TE, Liang H, Wriggers W, Wong GCL. Like-charge attraction between polyelectrolytes induced by counterion charge density waves. Proc Natl Acad Sci U S a. 2003;100:8634–8637. doi: 10.1073/pnas.1533355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, et al. Lipofection: a highly efficient. lipid-mediated DNA-transfection procedure. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Radler JO, Koltover I, Salditt T, Safinya CR. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 119.Lai GH, Coridan R, Zribi OV, Golestanian R, Wong GCL. Evolution of growth modes for polyelectrolyte bundles. Phys Rev Lett. 2007;98:187802. doi: 10.1103/PhysRevLett.98.187802. [DOI] [PubMed] [Google Scholar]
- 120.Sayar M, Holm C. Finite-size polyelectrolyte bundles at thermodynamic equilibrium. Europhys Lett. 2007;77:16001–6. doi: 10.1209/0295-5075/77/16001. [DOI] [Google Scholar]
- 121.Ha BY, Liu AJ. Kinetics of bundle growth in DNA condensation. Europhys Lett. 1999;46:624–8. doi: 10.1209/epl/i1999-00311-6. [DOI] [Google Scholar]
- 122.Henle M, Pincus P. Equilibrium bundle size of rodlike polyelectrolytes with counterion-induced attractive interactions. Phys Rev E. 2005;71:060801–4. doi: 10.1103/PhysRevE.71.060801. [DOI] [PubMed] [Google Scholar]
- 123.Grason GM, Bruinsma RF. Chirality and equilibrium biopolymer bundles. Phys Rev Lett. 2007;99:098101. doi: 10.1103/PhysRevLett.99.098101. [DOI] [PubMed] [Google Scholar]
- 124.Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Molecular Microbiology. 2016;99:123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nature Reviews Microbiology. 2013;11:667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dueholm MS, Petersen SV, Sønderkær M, Larsen P, Christiansen G, Hein KL, et al. Functional amyloid in Pseudomonas. Molecular Microbiology. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- 127.C MR, Barnhart Michelle M. Curli Biogenesis and Function. Annual Review of Microbiology. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gallo PM, Rapsinski GJ, Wilson RP, Oppong GO, Sriram U, Goulian M, et al. Amyloid-DNA Composites of Bacterial Biofilms Stimulate Autoimmunity. Immunity. 2015;42:1171–1184. doi: 10.1016/j.immuni.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tursi SA, Lee EY, Medeiros NJ, Lee MH, Nicastro LK, Buttaro B, et al. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog. 2017;13:e1006315. doi: 10.1371/journal.ppat.1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scott MG, Vreugdenhil AC, Buurman WA, Hancock RE, Gold MR. Cutting edge: cationic antimicrobial peptides block the binding of lipopolysaccharide (LPS) to LPS binding protein. J Immunol. 2000;164:549–553. doi: 10.4049/jimmunol.164.2.549. [DOI] [PubMed] [Google Scholar]
- 131.Di Nardo A, Braff MH, Taylor KR, Na C, Granstein RD, McInturff JE, et al. Cathelicidin Antimicrobial Peptides Block Dendritic Cell TLR4 Activation and Allergic Contact Sensitization. J Immunol. 2007;178:1829–1834. doi: 10.4049/jimmunol.178.3.1829. [DOI] [PubMed] [Google Scholar]
- 132.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 133.Adase CA, Borkowski AW, Zhang LJ, Williams M, Sato E, Sanford JA, et al. Non-coding double-stranded RNA and LL-37 induce growth factor expression from keratinocytes and endothelial cells. Journal of Biological Chemistry. 2016;291 doi: 10.1074/jbc.M116.725317. jbc.M116.725317–11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang LJ, Sen GL, Ward NL, Johnston A, Chun K, Chen Y, et al. Antimicrobial Peptide LL37 and MAVS Signaling Drive Interferon-β Production by Epidermal Keratinocytes during Skin Injury. Immunity. 2016;45:119–130. doi: 10.1016/j.immuni.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nelson AM, Reddy SK, Ratliff TS, Hossain MZ, Katseff AS, Zhu AS, et al. dsRNA Released by Tissue Damage Activates TLR3 to Drive Skin Regeneration. Cell Stem Cell. 2015;17:139–151. doi: 10.1016/j.stem.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang LJ, Gallo RL. Antimicrobial peptides. Curr Biol. 2016;26:R14–9. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 137.Büchau AS, Gallo RL. Innate immunity and antimicrobial defense systems in psoriasis. Clin Dermatol. 2007;25:616–624. doi: 10.1016/j.clindermatol.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lai Y, Adhikarakunnathu S, Bhardwaj K, Ranjith-Kumar CT, Wen Y, Jordan JL, et al. LL37 and Cationic Peptides Enhance TLR3 Signaling by Viral Double-stranded RNAs. PLoS ONE. 2011;6:e26632. doi: 10.1371/journal.pone.0026632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen X, Takai T, Xie Y, Niyonsaba F, Okumura K, Ogawa H. Human antimicrobial peptide LL-37 modulates proinflammatory responses induced by cytokine milieus and double-stranded RNA in human keratinocytes. Biochem Biophys Res Commun. 2013;433:532–537. doi: 10.1016/j.bbrc.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 140.Chen X, Takai T, Xie Y, Okumura K, Ikeda S, Ogawa H. Modulation of double-stranded RNA- and cytokine- induced responses of human keratinocytes by LL-37. Journal of Dermatological Science. 2013;69:e14. doi: 10.1016/j.jdermsci.2012.11.339. [DOI] [Google Scholar]
- 141.Yang Y, Wang SY, Huang ZF, Zou HM, Yan BR, Luo WW, et al. The RNA-binding protein Mex3B is a coreceptor of Toll-like receptor 3 in innate antiviral response. Cell Res. 2016;26:288–303. doi: 10.1038/cr.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Valverde Rodrigo A, et al. Viral double-strand RNA-binding proteins can enhance innate immune signaling by toll-like Receptor 3. PLoS ONE. 2011;6:e25837. doi: 10.1371/journal.pone.0025837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee EY, Takahashi T, Curk T, Dobnikar J, Gallo RL, Wong GCL. 070 Liquid crystalline ordering of antimicrobial peptide-RNA complexes controls TLR3 activation. Journal of Investigative Dermatology. 2017;137:S12. doi: 10.1016/j.jid.2017.02.083. [DOI] [Google Scholar]
- 144.Luo J, Obmolova G, Malia TJ, Wu SJ, Duffy KE, Marion JD, et al. Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure. J Mol Biol. 2012;421:112–124. doi: 10.1016/j.jmb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Krüger A, Oldenburg M, Chebrolu C, Beisser D, Kolter J, Sigmund AM, et al. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO Rep. 2015;16:1656–1663. doi: 10.15252/embr.201540861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gorden KB, Gorski KS, Gibson SJ, Kedl RM, Kieper WC, Qiu X, et al. Synthetic TLR Agonists Reveal Functional Differences between Human TLR7 and TLR8. J Immunol. 2005;174:1259–1268. doi: 10.4049/jimmunol.174.3.1259. [DOI] [PubMed] [Google Scholar]
- 147.Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11:362–371. doi: 10.1007/SpringerReference_39030. [DOI] [PubMed] [Google Scholar]
- 148.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11:372–378. doi: 10.2174/978160805362911201010003. [DOI] [PubMed] [Google Scholar]
- 149.Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, et al. Identification of RNA Sequence Motifs Stimulating Sequence-Specific TLR8-Dependent Immune Responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 150.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, et al. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. Journal of Experimental Medicine. 2001;194:863–869. doi: 10.3410/f.1001021.10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jarrossay D, Napolitani G, Colonna M, Sallusto F, Lanzavecchia A. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur J Immunol. 2001;31:3388–3393. doi: 10.1002/1521-4141(200111)31:11<3388::AID-IMMU3388>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 152.Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, et al. Quantitative Expression of Toll-Like Receptor 1–10 mRNA in Cellular Subsets of Human Peripheral Blood Mononuclear Cells and Sensitivity to CpG Oligodeoxynucleotides. J Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 153.Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, et al. TLR7 Is Involved in Sequence-Specific Sensing of Single-Stranded RNAs in Human Macrophages. J Immunol. 2008;180:2117–2124. doi: 10.4049/jimmunol.180.4.2117. [DOI] [PubMed] [Google Scholar]
- 154.Gringhuis SI, van der Vlist M, van den Berg LM, den Dunnen J, Litjens M, Geijtenbeek TBH. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat Immunol. 2010;11:419–426. doi: 10.1038/ni.1858. [DOI] [PubMed] [Google Scholar]
- 155.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vollmer J, Tluk S, Schmitz C, Hamm S, Jurk M, Forsbach A, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. Journal of Experimental Medicine. 2005;202:1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu Rev Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 160.Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206:1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Prinz N, Clemens N, Strand D, Pütz I, Lorenz M, Daiber A, et al. Antiphospholipid antibodies induce translocation of TLR7 and TLR8 to the endosome in human monocytes and plasmacytoid dendritic cells. Blood. 2011;118:2322–2332. doi: 10.1182/blood-2011-01-330639. [DOI] [PubMed] [Google Scholar]
- 162.Sacre SM, Lo A, Gregory B, Simmonds RE, Williams L, Feldmann M, et al. Inhibitors of TLR8 Reduce TNF Production from Human Rheumatoid Synovial Membrane Cultures. J Immunol. 2008;181:8002–8009. doi: 10.4049/jimmunol.181.11.8002. [DOI] [PubMed] [Google Scholar]
- 163.Guiducci C, Gong M, Cepika AM, Xu Z, Tripodo C, Bennett L, et al. RNA recognition by human TLR8 can lead to autoimmune inflammation. J Exp Med. 2013;210:2903–2919. doi: 10.1084/jem.20131044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jiang W, Zhu FG, Bhagat L, Yu D, Tang JX, Kandimalla ER, et al. A Toll-like receptor 7, 8, and 9 antagonist inhibits Th1 and Th17 responses and inflammasome activation in a model of IL-23-induced psoriasis. J Invest Dermatol. 2013;133:1777–1784. doi: 10.1038/jid.2013.57. [DOI] [PubMed] [Google Scholar]
- 165.Zhu FG, Jiang W, Bhagat L, Wang D, Yu D, Tang JX, et al. A novel antagonist of Toll-like receptors 7, 8 and 9 suppresses lupus disease-associated parameters in NZBW/F1 mice. Autoimmunity. 2013;46:419–428. doi: 10.3109/08916934.2013.798651. [DOI] [PubMed] [Google Scholar]
- 166.Yamasaki K, Di Nardo A, Bardan A, Murakami M, Ohtake T, Coda A, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 167.Paulsen F, Pufe T, Conradi L, Varoga D, Tsokos M, Papendieck J, et al. Antimicrobial peptides are expressed and produced in healthy and inflamed human synovial membranes. J Pathol. 2002;198:369–377. doi: 10.1002/path.1224. [DOI] [PubMed] [Google Scholar]
- 168.Schauber JR, Rieger D, Weiler F, Wehkamp J, Eck M, Fellermann K, et al. Heterogeneous expression of human cathelicidin hCAP18/LL-37 in inflammatory bowel diseases. European Journal of Gastroenterology & Hepatology. 2006;18:615–621. doi: 10.1097/00042737-200606000-00007. [DOI] [PubMed] [Google Scholar]
- 169.Kim ST, Cha HE, Kim DY, Han GC, Chung YS, Lee YJ, et al. Antimicrobial peptide LL-37 is upregulated in chronic nasal inflammatory disease. Acta Otolaryngol. 2003;123:81–85. doi: 10.1080/0036554021000028089. [DOI] [PubMed] [Google Scholar]
- 170.Agerberth B, Grunewald J, Castaños-Velez E, Olsson B, Jörnvall H, Wigzell H, et al. Antibacterial components in bronchoalveolar lavage fluid from healthy individuals and sarcoidosis patients. Am J Respir Crit Care Med. 1999;160:283–290. doi: 10.1164/ajrccm.160.1.9807041. [DOI] [PubMed] [Google Scholar]
- 171.Bucki R, Byfield FJ, Janmey PA. Release of the antimicrobial peptide LL-37 from DNA/F-actin bundles in cystic fibrosis sputum. Eur Respir J. 2007;29:624–632. doi: 10.1183/09031936.00080806. [DOI] [PubMed] [Google Scholar]
- 172.Chi Z, Wang Z, Wang K, Zhu Y, Qin S. Cathelicidin Antimicrobial Peptide LL-37 in Cholesteatoma Enables Keratinocyte Reactivity with Cytosolic DNA. Scandinavian Journal of Immunology. 2014;79:214–221. doi: 10.1111/sji.12149. [DOI] [PubMed] [Google Scholar]
- 173.Chamilos G, Gregorio J, Meller S, Lande R, Kontoyiannis DP, Modlin RL, et al. Cytosolic sensing of extracellular self-DNA transported into monocytes by the antimicrobial peptide LL37. Blood. 2012;120:3699–3707. doi: 10.1182/blood-2012-01-401364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yount NY, Yeaman MR. Structural congruence among membrane-active host defense polypeptides of diverse phylogeny, Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2006;1758:1373–1386. doi: 10.1016/j.bbamem.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 175.Yount NY, Yeaman MR. Multidimensional signatures in antimicrobial peptides. Proc Natl Acad Sci U S a. 2004;101:7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yount NY, Kupferwasser D, Spisni A, Dutz SM, Ramjan ZH, Sharma S, et al. Selective reciprocity in antimicrobial activity versus cytotoxicity of hBD-2 and crotamine. Proc Natl Acad Sci U S a. 2009;106:14972–14977. doi: 10.1073/pnas.0904465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yeaman MR, Yount NY. Unifying themes in host defence effector polypeptides. Nature Reviews Microbiology. 2007;5:727–740. doi: 10.1038/nrmicro1744. [DOI] [PubMed] [Google Scholar]
- 178.Yeaman MR, Yount NY. Code among chaos: immunorelativity and the AEGIS model of antimicrobial peptides. 2005:21–27. [Google Scholar]
- 179.Yeaman MR, Yount NY, Waring AJ, Gank KD, Kupferwasser D, Wiese R, et al. Modular determinants of antimicrobial activity in platelet factor-4 family kinocidins. Biochim Biophys Acta. 2007;1768:609–619. doi: 10.1016/j.bbamem.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, et al. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. Journal of Leukocyte Biology. 2003;74:448–455. doi: 10.1189/jlb.0103024. [DOI] [PubMed] [Google Scholar]
- 181.Yount NY, Cohen SE, Kupferwasser D, Waring AJ, Ruchala P, Sharma S, et al. Context Mediates Antimicrobial Efficacy of Kinocidin Congener Peptide RP-1. PLoS ONE. 2011;6:e26727. doi: 10.1371/journal.pone.0026727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xiong YQ, Bayer AS, Elazegui L, Yeaman MR. A synthetic congener modeled on a microbicidal domain of thrombin- induced platelet microbicidal protein 1 recapitulates staphylocidal mechanisms of the native molecule. Antimicrobial Agents and Chemotherapy. 2006;50:3786–3792. doi: 10.1128/AAC.00038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yeaman MR. Platelets: at the nexus of antimicrobial defence. Nature Reviews Microbiology. 2014;12:426–437. doi: 10.1038/nrmicro3269. [DOI] [PubMed] [Google Scholar]
- 184.Yount NY, Gank KD, Xiong YQ, Bayer AS, Pender T, Welch WH, et al. Platelet microbicidal protein 1: structural themes of a multifunctional antimicrobial peptide. Antimicrobial Agents and Chemotherapy. 2004;48:4395–4404. doi: 10.1128/AAC.48.11.4395-4404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yount NY, Waring AJ, Gank KD, Welch WH, Kupferwasser D, Yeaman MR. Structural correlates of antimicrobial efficacy in IL-8 and related human kinocidins, Biochim. Biophys Acta. 2007;1768:598–608. doi: 10.1016/j.bbamem.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 186.Meller S, Di Domizio J, Voo KS, Friedrich HC, Chamilos G, Ganguly D, et al. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat Immunol. 2015;16:970–979. doi: 10.1038/ni.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee JS, Cua DJ. IL-26 AMPs up the TH17 arsenal. Nat Immunol. 2015;16:897–898. doi: 10.1038/ni.3256. [DOI] [PubMed] [Google Scholar]
- 188.Poli C, Augusto JF, Dauvé J, Adam C, Preisser L, Larochette V, et al. IL-26 Confers Proinflammatory Properties to Extracellular DNA. J Immunol. 2017;198:1600594–3661. doi: 10.4049/jimmunol.1600594. [DOI] [PubMed] [Google Scholar]
- 189.Kaplan A, Lee MW, Wolf AJ, Limon JJ, Becker CA, Ding M, et al. Direct Antimicrobial Activity of IFN-β. J Immunol. 2017;198:4036–4045. doi: 10.4049/jimmunol.1601226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nordahl EA, Rydengård V, Nyberg P, Nitsche DP, Mörgelin M, Malmsten M, et al. Activation of the complement system generates antibacterial peptides. Proc Natl Acad Sci U S a. 2004;101:16879–16884. doi: 10.1073/pnas.0406678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ruan BH, Li X, Winkler AR, Cunningham KM, Kuai J, Greco RM, et al. Complement C3a, CpG oligos, and DNA/C3a complex stimulate IFN-α production in a receptor for advanced glycation end product-dependent manner. J Immunol. 2010;185:4213–4222. doi: 10.4049/jimmunol.1000863. [DOI] [PubMed] [Google Scholar]
- 192.Realegeno S, Kelly-Scumpia KM, Dang AT, Lu J, Teles R, Liu PT, et al. S100A12 Is Part of the Antimicrobial Network against Mycobacterium leprae in Human Macrophages. PLoS Pathog. 2016;12:e1005705. doi: 10.1371/journal.ppat.1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yang Z, Tao T, Raftery MJ, Youssef P, Di Girolamo N, Geczy CL. Proinflammatory properties of the human S100 protein S100A12. Journal of Leukocyte Biology. 2001;69:986–994. [PubMed] [Google Scholar]
- 194.Lee EY, Fulan BM, Wong GCL, Ferguson AL. Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc Natl Acad Sci U S a. 2016;113:13588–13593. doi: 10.1073/pnas.1609893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lee EY, Wong GCL, Ferguson AL. Machine learning-enabled discovery and design of membrane-active peptides. Bioorg Med Chem. 2017 doi: 10.1016/j.bmc.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lee EY, Lee MW, Fulan BM, Ferguson AL, Wong GCL. What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning? Interface Focus. 2017;7:20160153. doi: 10.1098/rsfs.2016.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]


