Abstract
An infectious retroviral particle contains 1000–1500 molecules of the nucleocapsid protein (NC) that cover the diploid RNA genome. NC is a small zinc finger protein that possesses nucleic acid chaperone activity that enables NC to rearrange DNA and RNA molecules into the most thermodynamically stable structures usually those containing the maximum number of base pairs. Thanks to the chaperone activity, NC plays an essential role in reverse transcription of the retroviral genome by facilitating the strand transfer reactions of this process. In addition, these reactions are involved in recombination events that can generate multiple drug resistance mutations in the presence of anti-HIV-1 drugs. The strand transfer reactions rely on base pairing of folded DNA/RNA structures. The molecular mechanisms responsible for NC-mediated strand transfer reactions are presented and discussed in this review. Antiretroviral strategies targeting the NC-mediated strand transfer events are also discussed.
Keywords: Nucleocapsid protein, Strand transfer, Reverse transcription, Retrovirus, HIV
Highlights
-
•
DNA strand transfers are essential steps of reverse transcription in retroviruses.
-
•
DNA strand transfers rely on base pairing of structured nucleic acids.
-
•
In vitro studies identified NC as an actor who facilitates DNA strand transfers.
-
•
NC increases the rate of transfers, thanks to its nucleic acid chaperone activity.
-
•
The nucleic acid-NC complexes are potential targets for antiretroviral therapy.
Abbreviations list
- AIDS
acquired immunodeficiency syndrome
- CA
capsid protein
- HIV-1
human immunodeficiency virus type 1
- HIV-2
human immunodeficiency virus type 2
- HTLV-1
human T-cell leukemia virus type 1
- HTLV-2
human T-cell leukemia virus type 2
- MA
matrix protein
- MLV
murine leukemia virus
- MMTV
mouse mammary tumor virus
- MPMV
Mason-Pfizer monkey virus
- NC
nucleocapsid protein
- PBS
primer binding site
- poly(A)
polyadenylation signal
- cpoly(A)
DNA sequence complementary to poly(A)
- PPT
polypurine tract
- R
repeat
- r
DNA sequence complementary to R
- RSV
Rous sarcoma virus
- RT
reverse transcriptase
- SIV
simian immunodeficiency virus
- (−) ssDNA
minus-strand strong-stop DNA
- TAR
transactivator response element
- cTAR
DNA sequence complementary to TAR
- U5
unique sequence at the 5′-end
- u5
DNA sequence complementary to U5
- U3
unique sequence at the 3′-end
- u3
DNA sequence complementary to U3
1. Introduction
The family Retroviridae is divided into two subfamilies (Orthoretrovirinae and Spumaretrovirinae). This review only deals with retroviruses belonging to the subfamily Orthoretrovirinae. Retroviruses are RNA viruses that infect vertebrates and invertebrates and cause a wide variety of diseases, such as the acquired immunodeficiency syndrome (AIDS), cancers, leukemias and lymphomas. Retrovirus particles contain two copies of the single-stranded RNA genome that are converted into a single double-stranded DNA by reverse transcription in infected cells [1]. Reverse transcription is a complex process requiring at least two virus-encoded proteins, reverse transcriptase (RT) and nucleocapsid protein (NC) [2]. Although all retroviral RTs possess both the required DNA polymerase and RNase H activities, their structures and subunit composition are significantly different [3]. For example, RTs of gammaretroviruses such as murine leukemia virus (MLV) are monomeric enzymes, whereas RTs of lentiviruses such as human immunodeficiency virus type 1 (HIV-1) are heterodimeric enzymes. The combinations of drugs targeting crucial steps of the virus life cycle (entry, reverse transcription, integration and maturation) are widely used in the treatment of HIV-1 infection [4]. However, the development of new drugs from various classes is required because, in rare cases, treatment failures occur, mostly due to the emergence of drug resistance. Since reverse transcription is only blocked by RT inhibitors to date, it would be interesting to have drugs targeting other actors of this essential step. HIV-1 NC is a potential target because it plays an important role in reverse transcription [2].
2. General features of retroviral nucleocapsid proteins
In retroviruses, the gag gene encodes the Gag precursor polyprotein which is cleaved by the retroviral protease (PR) to yield the mature proteins, MA, CA and NC. Mature nucleocapsid proteins (NCs) are small proteins (<100 amino acids) that cover the RNA genome in infectious retroviral particles. Thus, Rous sarcoma virus (RSV) and HIV-1 particles contain about 1000–1500 NC molecules [5], [6], [7], i.e. one NC molecule for 12–18 nucleotides.
2.1. Sequences and structures of NCs
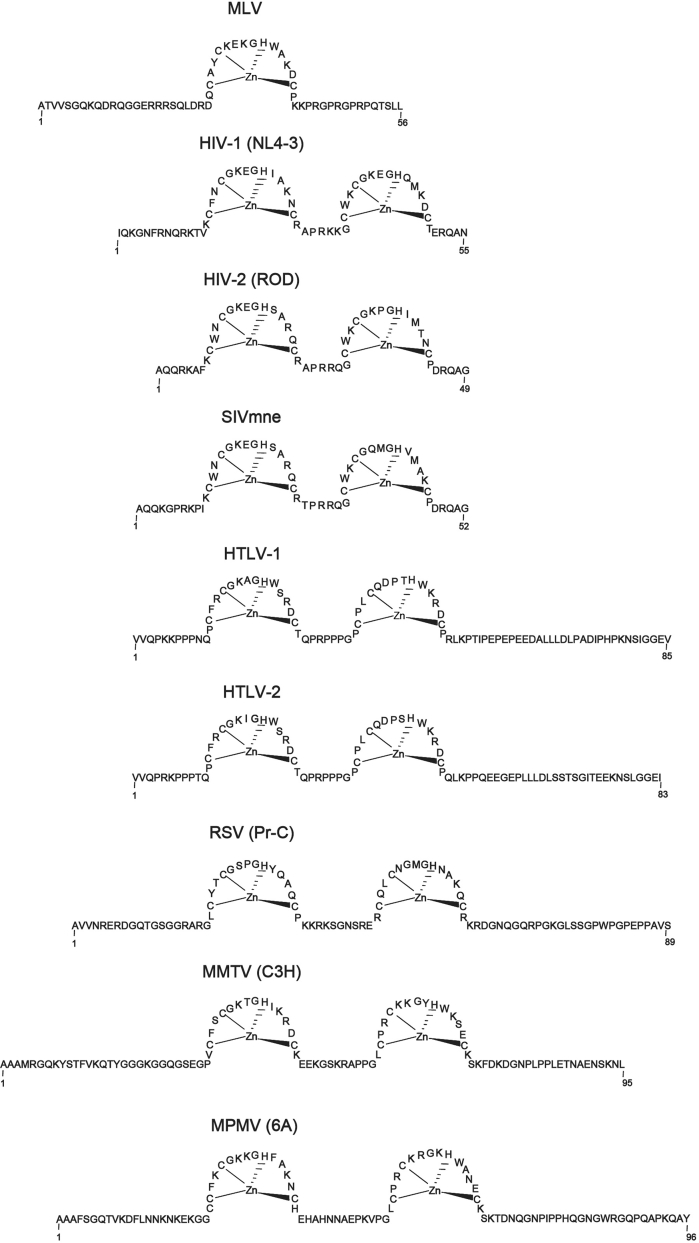
NC of retroviruses contains one or two highly conserved zinc-finger domains formed by the CX2CX4HX4C sequence (referred to as the CCHC motif) [8] (Fig. 1).
Fig. 1.
Sequence and structural features of retroviral NCs from MLV (Genbank #: J02255), HIV-1 (Genbank #: AF324493), HIV-2 (Genbank #: M15390), SIVmne (Genbank #: M32741), HTLV-1 (Genbank #: D13784), HTLV-2 (Genbank #: AF139382), RSV (Genbank #: J02342), MMTV (Genbank #: AF228552), MPMV (Genbank #: M12349).
With the exception of HTLV-1 NC (pI∼7) [9], NCs containing one or two CCHC motifs are basic proteins. The three-dimensional (3D) structures of NCs of six different retroviruses have been investigated by nuclear magnetic resonance spectroscopy (NMR). The 3D structures of MPMV and SIV NCs have been determined using truncated proteins [10], [11], whereas the full-length proteins were used for HIV-1 [12], [13], [14], [15], HIV-2 [16], MLV [17] and MMTV [18]. These structural studies confirmed the folding of the CCHC motif into the zinc finger structure. In contrast, they showed that the N- and C-terminal regions and the linker between the two zinc fingers are flexible and poorly structured. The two zinc fingers behave as independently folded domains on a flexible polypeptide chain [14], [15]. However, a transient interaction between the two zinc fingers of HIV-1 NC has been deduced from NMR data [13].
2.2. Nucleic acid binding properties of NCs
NCs bind to both DNA and RNA with a preference for single-stranded molecules and their binding constant depends on the nature, sequence, and folding of nucleic acids, and can thus vary by several orders of magnitude (see Refs. [8], [19], [20], [21], [22] and references therein). It should be emphasized that HIV-1 NC at high concentrations binds the double-stranded regions of DNA and RNA nonspecifically through electrostatic interactions [23], [24], [25]. The binding site of NCs encompasses 5 to 8 nucleotides (see Ref. [21] and references therein). An in vitro selection method called SELEX (Systematic Evolution of Ligands by EXponential enrichment) allowed to identify high-affinity RNA binding sites for extended forms of HIV-1 NC [26], [27]. Interestingly, most of these sites can form stem-loop structures exhibiting the GU motif in the apical loop. All NCs, except HTLV-1 NC, bind nucleic acids with a preference for sequences containing unpaired guanine residues [19], [28], [29], [30], [31], [32], [33]. This could result from the type-specific nucleotide bias in retroviral genomes, e.g. HIV-1 RNA is A-rich and C-poor, whereas HTLV-1 RNA is C-rich and G-poor [34].
2.3. Nucleic acid chaperone activity of NCs
NCs possess nucleic acid chaperone activity that facilitates the rearrangement of DNA and RNA into the most thermodynamically stable structures usually those containing the maximum number of base pairs [22], [35]. This activity relies on three properties [2]: 1) nucleic acid aggregation activity, which is mainly due to the basic residues; 2) moderate duplex destabilization activity by the zinc fingers; 3) rapid on-off binding kinetics. Thanks to these properties, NCs are able to promote base pairing of complementary DNA/RNA sequences that form secondary structures such as hairpins. However, the length of base paired nucleic acid helices limits the NC-mediated hybridization process. Indeed, NCs can destabilize only short base paired regions (typically less than twelve consecutive base pairs) flanked by duplex ends, loops, bulges or mismatches [21], [24], [36], [37]. This suggests a co-evolution between the secondary structures of retroviral genomes and the NC destabilizing activity [36]. However, there is no report showing that retroviral RNAs are more or less structured than cellular mRNAs.
Three different forms (NCp15, NCp9 and NCp7) of the HIV-1 NC domain appear sequentially during the cleavage of the Gag precursor polyprotein. Although NCp15 and NCp9 are not supposed to be present during the reverse transcription process, their chaperone properties have been investigated to get a better understanding of the nucleic acid chaperone activity of NCp7 [38], [39], [40]. The chaperone activity of NCp7 and NCp9 is high, whereas that of NCp15 is weak [39], [40]. Single-molecule stretching experiments showed that the residence time of the protein on DNA decreases from NCp15 to NCp7, suggesting that the high chaperone activity of NCp7 is related to its 'fast' binding and unbinding properties [38]. Dynamic light scattering studies showed that NCp15 induces much smaller nucleic acid aggregates relative to those induced by NCp9 and NCp7 [39]. This is probably due to the numerous glutamic acid residues contained in the p6 domain of NCp15 [39]. Recent studies suggest that the p1 and p1-p6 domains in NCp9 and NCp15, respectively, modulate the nucleic acid binding properties of the NC domain [39], [41]. NCp15 and NCp9 are not as efficient as NCp7 in the reverse transcription process [40]. Thanks to their chaperone activity, NCs play essential roles in reverse transcription.
3. NC is an essential actor of reverse transcription
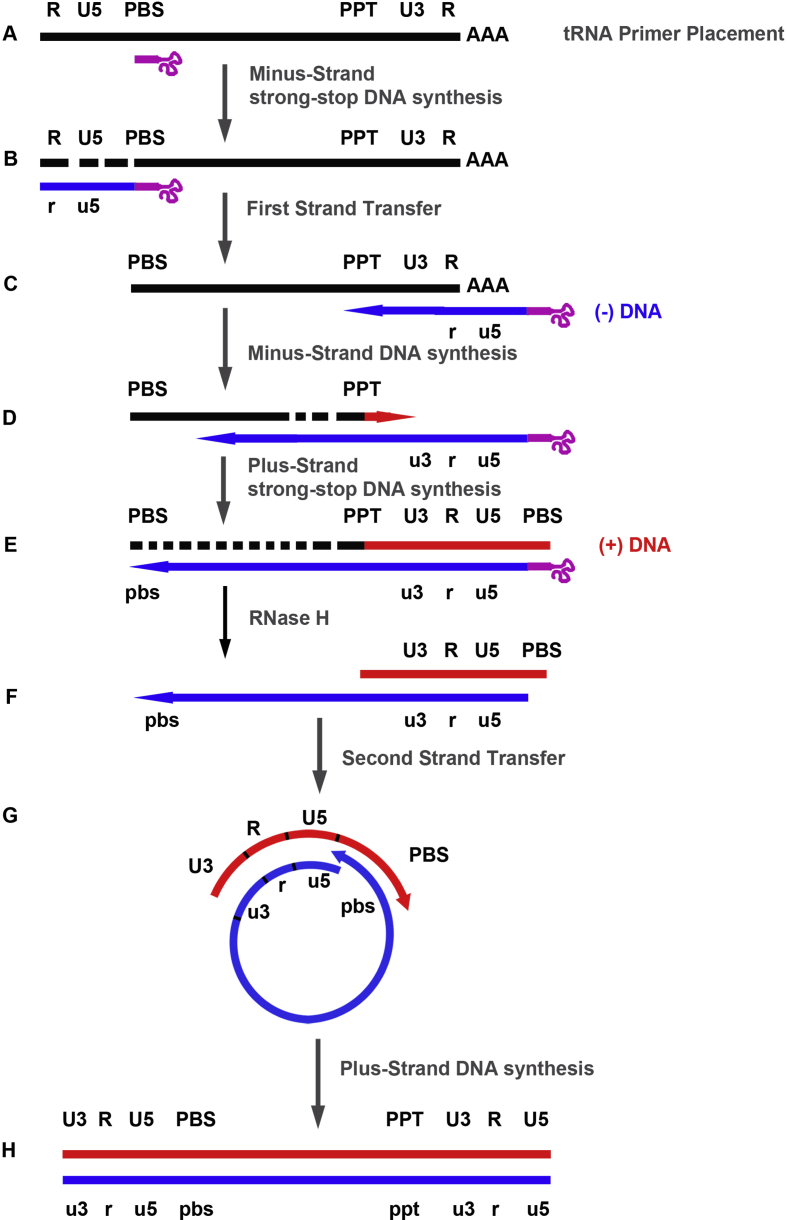
NC is present mainly in its mature form during reverse transcription. As a domain of the Gag precursor polyprotein, NC is also a key element in the assembly step of retroviral particle [42], [43], [44], [45]. Thus, it can be difficult to identify by site-directed mutagenesis the roles performed by NC in its mature and precursor forms during the retroviral life cycle. In vitro models have therefore been developed to determine the mechanisms of action of mature NC in reverse transcription ([35] and references therein). Reverse transcription is a multistep process allowing conversion of the single-stranded retroviral RNA into a double-stranded DNA (Fig. 2) [46]. This process requires at least three steps of base pairing interactions: 1) tRNA primer placement; 2) first strand transfer; 3) second strand transfer. Numerous in vitro studies are consistent with an important role of mature NC in these three essential steps (see Refs. [2], [22], [35] and references therein).
Fig. 2.
Reverse transcription of the retroviral RNA genome. A) The 3′-end of a cellular tRNA (pink line) is annealed to the PBS region of the retroviral RNA (black line). B) Minus-strand DNA (blue line) synthesis is initiated from the tRNA primer to the 5′-end of genome; this product is named minus-strand strong-stop DNA ((−) ssDNA); the genomic RNA is degraded by the RNase H activity of RT. C) The (−) ssDNA is transferred to the 3′-end of the genomic RNA. D) After the first strand transfer, elongation of minus-strand DNA and RNase H degradation continue; plus-strand DNA (red line) synthesis is initiated from a polypurine tract (PPT) sequence that is resistant to RNase H cleavage. E) Elongation of the two DNA strands until the PBS sequences. F) Retroviral RNA, the tRNA and PPT primers are degraded by the RNase H activity of RT. G) The second strand transfer is mediated by base pairing of the complementary PBS sequences at the 3′-ends of minus-strand DNA and plus-strand strong-stop DNA. H) Elongation of minus- and plus-strand DNAs, resulting in a linear double-stranded DNA with a long terminal repeat (U3 R U5) at each end.
During the tRNA primer placement involving two folded RNA structures, the 3′-terminal 18 nucleotides of the tRNA primer are base-paired to the primer binding site (PBS). Interestingly, additional base pairing interactions between the tRNA primer and the retroviral genome have been reported by different groups ([47] and references therein). Because destabilization of both the tRNA and the PBS region is required for duplex formation, the optimal temperature is about 70 °C in the absence of NC [48], [49]. In contrast, NC enhances duplex formation by several orders of magnitude at 37 °C, suggesting the in vivo role of this retroviral protein in the tRNA placement [50], [51], [52]. Interestingly, in virio analysis of tRNALys3 placement supports the notion that mature NC is responsible for complete annealing of tRNALys3 to HIV-1 genomic RNA [53].
As shown in Fig. 2 (steps C and G), there are two obligatory strand transfers that occur at defined positions during reverse transcription. In addition, internal strand transfer events occur from any position in the retroviral genome to the same position in the second genomic RNA copy during minus-strand DNA synthesis, thus accounting for the high rate of retroviral recombination (see Ref. [54] and references therein). Because it is difficult to determine by ex vivo cell assays the molecular mechanisms of strand transfer reactions, in vitro systems have been developed. It is likely that the in vitro systems partially mimic the in vivo context of the reverse transcription process. However, many studies using in vitro models of HIV-1 reverse transcription suggest that NC could enhance the two obligatory transfers and the internal transfers in the in vivo context.
4. NC increases the rate of the first strand transfer
The first strand transfer has been extensively studied. This process can occur in an intra- or intermolecular manner because a retroviral particle contains two copies of its genomic RNA [55], [56]. This transfer requires base pairing of the repeat (R) region at the 3′-end of the retroviral RNA with the complementary r region at the 3′-end of minus-strand strong-stop DNA ((−) ssDNA) (Fig. 2, step C). The R regions of various retroviruses display different lengths, ranging from 16 (mouse mammary tumor virus) to 247 (human T-cell leukemia virus type 2) nucleotides [57]. Long R regions do not lead to increased efficiency of reverse transcription. A long R region contains elements that play other roles in the retroviral replication cycle (e.g. the TAR hairpin for the activation of transcription in HIV-1). During (−) ssDNA synthesis, the R-U5 region of the RNA template needs to be degraded by the RNase H activity of RT in order to allow subsequent (−) ssDNA transfer (Fig. 2, step B). Partial (−) ssDNAs can be transferred prior to complete reverse transcription of the R region [57], [58], [59]. However, the great majority of first strand transfers occur after the synthesis of the full-length (−) ssDNA [60], [61].
In vitro reverse transcription assays using RT, a donor RNA template corresponding to the 5′-end of the retroviral genome and an acceptor RNA template corresponding to the 3′-end of the retroviral genome, have been developed to investigate the molecular mechanisms involved in the first strand transfer [62], [63], [64], [65], [66], [67], [68]. Using this type of reconstituted system, it has been shown that RNase H degradation of the donor RNA template is required for transfer of (−) ssDNA to the acceptor RNA template [62], [68], [69]. These in vitro results are consistent with results obtained from ex vivo replication studies performed with mutant retroviruses lacking RNase H activity [70], [71]. The first strand transfer can be efficient in the absence of NC when the in vitro assay system relies on short RNA templates containing truncated R regions [62], [67]. This is probably due to the inability of the acceptor template and the DNA copy of the donor template to form extensive secondary structures. Consistent with this notion, an in vitro study confirmed that in the absence of NC, efficient strand transfer occurs with weakly structured templates, but not with highly structured templates [72]. Interestingly, the first strand transfer is strongly enhanced by NC when the in vitro assay system involves relatively long RNA templates containing the entire R region [63], [64], [65], [73], [74]. In vitro quantifications with RNAs containing the full-length R region showed that NC increases transfer efficiency by five-to tenfold [65], [75]. In contrast, NC stimulates transfer efficiency by two-to fivefold in the presence of truncated R regions [67], [76]. Most of in vitro studies have been carried out with HIV-1 NC and suggested that it enhances the efficiency of the first strand transfer by different mechanisms as described below.
5. Mechanisms of NC-mediated first strand transfer
5.1. HIV-1 NC prevents self-priming of (−) ssDNA
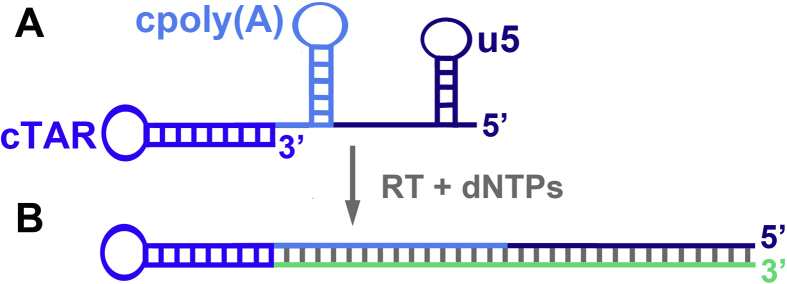
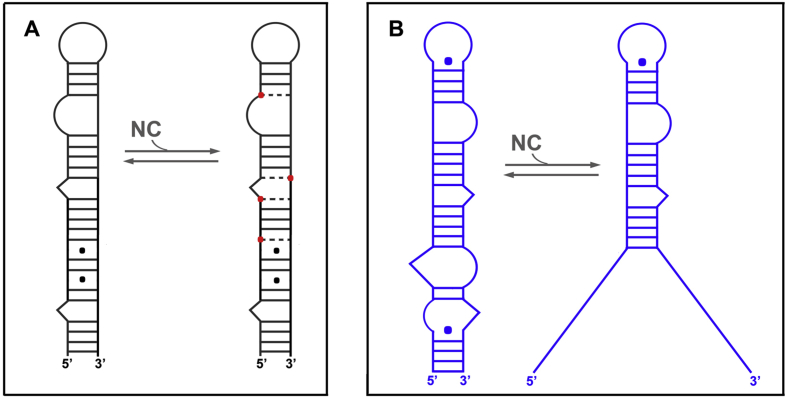
Based on reverse transcription assays performed with a donor RNA template in which the U5 region was partially deleted, Guo et al. [65] suggested that HIV-1 NC increases the efficiency of transfer by preventing self-priming of (−) ssDNA. Indeed, after degradation of the donor RNA template by the RNase H domain of RT, (−) ssDNA can dissociate from the template and its 3′-end can form the cTAR hairpin, which can self-prime and be extended by RT in the absence of NC (Fig. 3).
Fig. 3.
Self-priming of HIV-1 (−) ssDNA. A) Folding of (−) ssDNA into three hairpins [37]; in the 5′ to 3′ direction, the three shades of blue indicate the u5, cpoly(A) and cTAR domains; the r region is composed of cpoly(A) and cTAR domains. B) Elongation of (−) ssDNA in the presence of RT and dNTPs; the green line indicates the extended DNA strand. The figure is not drawn to scale.
DNA products generated by self-priming of (−) ssDNA are dead-end products for the first strand transfer because their 3′-ends are not complementary to the acceptor RNA template. NC significantly reduces self-priming of (−) ssDNA in the absence of an acceptor template when the reverse transcription assay is performed with a donor RNA template containing the entire R-U5-PBS region [77]. This inhibitory effect is likely due to a 5′-terminal RNA fragment (14–18 nucleotides) that remains annealed to the 3′-end of (−) ssDNA, because it forms a blunt-end duplex that is cleaved poorly by RT-RNase H [78]. In other words, NC favors the formation of this RNA-DNA duplex but not hairpin formation by the 3′-end of (−) ssDNA. Consistent with this notion, NC has little or no effect on self-priming of synthetic truncated (−) ssDNAs in the absence of RNA templates [77], [79], [80]. In contrast, NC greatly reduces self-priming of synthetic truncated (−) ssDNAs in the presence of an acceptor RNA template, presumably because NC facilitates the formation of RNA-DNA duplexes containing more base pairs than the hairpin structures of DNAs [79], [80]. Consistent with the role of NC-dependent inhibition of self-priming in first strand transfer, self-primed products are not detected in endogenous reverse transcription reactions with HIV-1 particles that are disrupted by a non-ionic detergent [65]. Note that in the presence of this type of mild detergent, approximately 40% of NC remains associated with the viral nucleoprotein complex and the synthesis of (−) ssDNA is preserved [81].
5.2. HIV-1 NC increases RNase H cleavage of the donor RNA template
In vitro studies also suggested that HIV-1 NC increases the efficiency of the first strand transfer by increasing RNase H degradation of the donor RNA template during synthesis of (−) ssDNA [68], [82]. In addition, Wisniewski et al. [78] found that NC greatly increases secondary RNase H cleavage of blunt-ended duplexes. To explain the effect of NC on enhanced donor cleavage, a model has been proposed in which NC interacts with the RNase H domain of RT [83]. Co-immunoprecipitation, far Western blotting and chemical cross-linking assays suggest that NC and RT interact, possibly through a zinc finger-dependent mechanism [84], [85]. However, it has not been demonstrated that this interaction is directly responsible for the RNase H activity stimulation.
Interestingly, Purohit et al. [86] showed that NC stimulation of RNase H activity is mostly due to the nucleic acid chaperone activity of NC, i.e. NC improves annealing of the primer to the donor template and thus generates more DNA-RNA duplexes that are extended by RT and cleaved by the RT-RNase H domain. It is therefore likely that NC-mediated enhancement of first strand transfer does not result directly from increasing RNase H activity by an interaction between NC and RT.
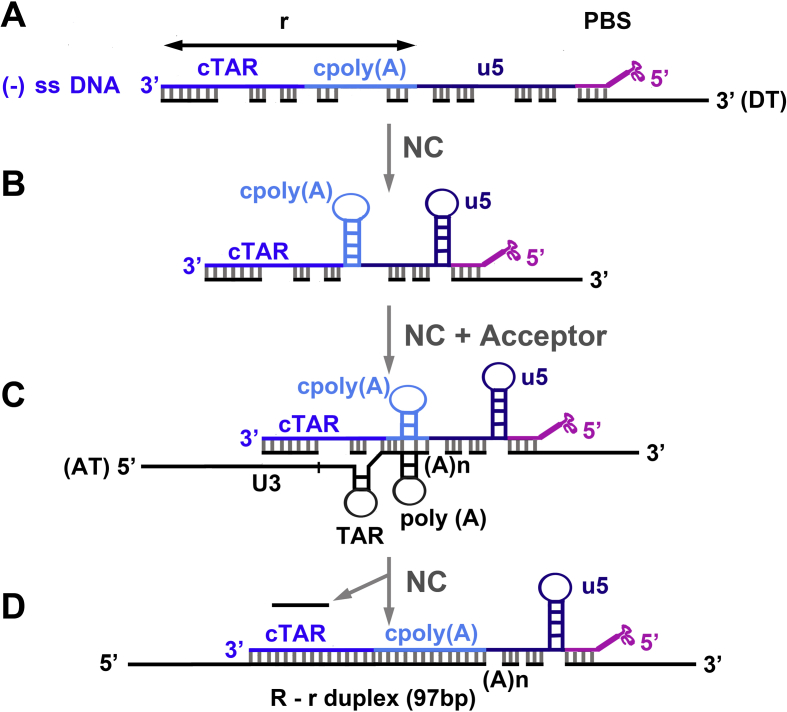
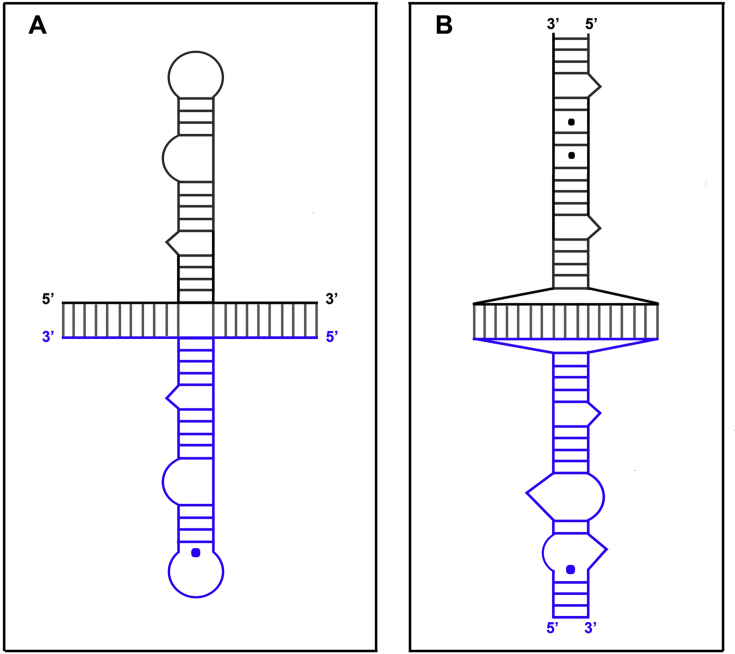
More recently, Hergott et al. [87] showed that NC is required for removal of 5′-terminal RNA fragments (14–18 nucleotides) in the presence of an acceptor RNA template. In fact, NC's destabilizing activity promotes the displacement of short RNAs by the longer acceptor RNA, which forms a thermodynamically more stable duplex with (−) ssDNA (Fig. 4D).
Fig. 4.
A possible mechanism for NC-mediated annealing of (−) ssDNA to the 3′-end of the HIV-1 RNA genome. A) (−) ssDNA has been synthesized from the tRNA primer (pink) to the 5′-end of the donor RNA template (DT); the DT (black line) has been degraded by the RNase H activity of RT, a 5′-terminal RNA fragment (14–18 nt) remains annealed at the 3′-end of (−) ssDNA. In the 5′ to 3′ direction, the three shades of blue indicate the u5, cpoly(A) and cTAR domains of (−) ssDNA. B) Formation of two hairpins by the u5 and cpoly(A) domains; the 5′-terminal RNA fragment prevents the formation of the cTAR hairpin. C) NC promotes partial base pairing of the poly(A) hairpin at the 3′-end of the acceptor RNA template (AT) with the complementary cpoly(A) hairpin at the 3′-end of (−) ssDNA. D) The 5′-terminal RNA fragment and small RNA fragments are released in the presence of NC and AT. The figure is not drawn to scale.
5.3. HIV-1 NC increases the rate of annealing of the complementary hairpins
The U3R region (551 nucleotides in length) at the 3′-end of the HIV-1 RNA genome and the full-length (−) HIV-1 ssDNA (181 nucleotides in length) are the key actors in the annealing process leading to the first strand transfer (Fig. 2). The U3R region folds into fourteen hairpins in the complete HIV-1 RNA genome extracted from virions [88]. Recently, Chen et al. [37] showed that the full-length (−) HIV-1 ssDNA folds into three hairpins in vitro. Interestingly, the complementary R/r sequences folds into two hairpins named TAR and poly(A) in RNA and cTAR and cpoly(A) in (−) ssDNA [24], [37], [66], [88], [89]. Unfolding of these complementary hairpins is necessary to form the DNA-RNA heteroduplex (97 base pairs). The first annealing study was performed with an RNA molecule (81 nucleotides representing the 3′-part of the R region) and a DNA molecule (81 nucleotides) corresponding the 3′-part of the (−) ssDNA [90], i.e. the poly(A) domain was partially deleted in both molecules. Under these conditions, NC accelerates the annealing reaction roughly 3000-fold and the data are consistent with first-order kinetics: while several models are possible, these authors favor a model in which the rate–limiting step is a step of melting of the nucleic acid species by NC followed by a fast nucleation rate. Using larger DNA (131 nucleotides) and RNA (148 nucleotides) molecules containing the full-length R/r sequences, the annealing reaction follows pseudo-first-order kinetics [91]. This latter study is compatible with an annealing model in which the fast nucleation step corresponds to formation of an unstable short heteroduplex between the R/r sequences while the slow step reflects formation of the stable heteroduplex (97 base pairs). In this model, unfolding and rearrangement of each reactant occur during the slow step.
Ex vivo analysis of the HIV-1 R sequence by site-directed mutagenesis suggests that cTAR DNA-TAR RNA annealing is required for efficient strand transfer [61]. This is in agreement with in vitro studies suggesting that the TAR/cTAR hairpins are far more important in the first strand transfer than the poly(A)/cpoly(A) hairpins [66], [92]. Furthermore, mutations in the TAR apical loop decrease the first strand transfer in vitro, suggesting that the annealing process is initiated via a loop-loop complex formed by the apical loops of the TAR and cTAR hairpins [66]. Consistent with these findings, efficient annealing of cTAR DNA to the 3′-end of the HIV-1 RNA genome relies on sequence complementarities between TAR and cTAR apical loops [24]. Therefore, oligonucleotides forming the TAR/cTAR hairpins (55 or 59 nucleotides) or the mini-TAR/mini cTAR hairpins (26 or 27 nucleotides) were used in many in vitro studies investigating the annealing mechanism [24], [89], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102]. The TAR RNA hairpin can bind a single NC molecule with high affinity (110 ± 2 nM) [103]. More precisely, NC binds the TAR apical loop specifically without changing the folding of the TAR hairpin, and this requires the NC zinc fingers [24]. This loop displays a highly flexible guanine that probably plays an essential role in NC binding [104]. An early study based on absorbance spectroscopy suggests that NC destabilizes only one base pair per TAR RNA molecule [89]. In contrast, NC destabilizes seven to eight base pairs of the cTAR DNA hairpin under similar conditions [89]. Recently, destabilization of the TAR hairpin by NC at high concentrations has been investigated by combining single-molecule optical tweezers measurements with a quantitative mfold-based model [105]. NC does not destabilize the lower part of the hairpin under these conditions, suggesting that the apical and/or the upper internal loops are the best site for initiation of the annealing reaction. Indeed, this study suggests that NC can destabilize the hairpin by targeting four specific sites corresponding to guanines adjacent to unstable stem regions, such as G.U wobble pairs, bulges and loops (Fig. 5A). Among these sites, the guanine located in the apical stem was also identified by NMR as a key site for the initiation of destabilization and the preferential destabilization of the neighboring base pairs has been shown [102]. The apical and internal loops of the cTAR DNA hairpin are weak and strong binding sites for NC, respectively [98]. Taken together, biochemical and biophysical studies are consistent with a dynamic structure of the cTAR DNA hairpin in the absence of NC, involving equilibrium between both the closed conformation and the partially open “Y” conformation (Fig. 5B) [79], [89], [97], [98], [106]. Furthermore, these studies support the notion that NC destabilizes the lower stem and thus shifts the equilibrium toward the “Y” conformation.
Fig. 5.
Destabilization of the complementary hairpins by NC. A) TAR RNA destabilization; the red dots indicate the guanines that are key sites for the initiation of destabilization, the black dots indicate the GU wobble base pairs. B) cTAR DNA destabilization; the blue dots indicate the TG mismatched base pairs.
As mentioned above, the apical loop and the 5′ and 3′-ends of the cTAR hairpin are accessible for initiation of the NC-mediated annealing reaction. Base pairing of the complementary hairpins under high concentrations of NC relies mainly on nucleation through the 3’/5′ termini, resulting in the formation of a zipper intermediate (Fig. 6A) [94], [96]. In contrast, both the apical loops (Fig. 6B) and the 3’/5′ termini are the initiation sites for the base pairing interaction under sub-saturating concentrations of NC (NC:nucleotide ratio equal to or lower than 1:8 [96]). The first step of the reaction is bimolecular and fast relative to the second step consisting in the formation of the long DNA-RNA duplex (55–57 bp). This latter step is monomolecular and requires a sufficient quantity of NC (NC:nucleotide ratio equal to or greater than 1:14 [102]).
Fig. 6.
Proposed initiations sites for NC-mediated annealing of cTAR DNA (blue) to TAR RNA (black). A) Zipper pathway involving initial base pairing of the 3′/5′ termini. B) Loop-loop pathway involving initial formation of an extended loop-loop duplex.
NMR studies help to understand how NC destabilizes and associates the TAR and cTAR hairpins at the molecular scale. The determination of the three-dimensional structures of NC:nucleic acid complexes using the classical NMR strategies and namely the use of nuclear Overhauser effects, has been achieved for three RNA-protein complexes using the full-length NC (NC(1–55)) [107], [108], [109] and for three DNA-protein complexes using a truncated version of NC (NC(12–53) [110], NC(12–55) [111] and NC(11–55) [100]). These complexes, despite some differences, exhibit common features that have been described in detail in several publications [14], [22], [100]. Comparison of the 3D-structures combined with the results of various experiments monitoring the extent of destabilization of the nucleic acid secondary structure allowed to identify the main characteristics associated with stem destabilization. In the structures for which the stem is destabilized, it is possible to observe contacts between hydrophobic amino acids belonging to ZF1 (T/I24, V13) and stem base pairs [14], [111], [112]. These observations are in good agreement with experiments showing that ZF1 is more important than ZF2 in stem destabilization [113], [114], [115], [116].
Another interesting feature emerging from our detailed comparison of the 3D-structures was the fact that, inside a class of nucleic acid (i.e. DNA or RNA), the N-to C-terminal polarity of the peptidic chain is either parallel (DNA) either antiparallel (RNA) to the 5′-3′ direction of the nucleic acid chain [100]. For example, in the DNA:NC complexes, a guanine is always inserted inside the hydrophobic pocket of the ZF2 and makes strong stacking interactions with the W37 aromatic side chain, and subsequently the ZF1 residues (especially the F16 aromatic side chain) contact the nucleotides located at the 5′ side of the guanine mentioned above. The recognition of the single-stranded nucleic acid chain polarity is reminiscent of that observed for other single-stranded DNA binding proteins [117]. However, the reversion of the binding polarity when the nature of the chain is changed from DNA to RNA is quite astonishing. A similar behavior has been observed when considering the inversion of the binding orientation of reverse transcriptase on DNA-DNA duplexes versus RNA-DNA hybrids [118]. Note that NC differential binding to DNA and RNA relies on the analysis of complexes that differ in many aspects: the full-length protein was used with RNA, whereas it was a truncated form with DNA; furthermore, the nucleic acid sequences strongly diverge between the studies. Therefore, it is necessary to perform new experiments specifically designed to confirm or deny the differential binding of NC to DNA and RNA.
It is likely that oligonucleotides forming the TAR/cTAR hairpins partially mimic the base pairing interaction between the r sequence of (−) ssDNA with the R sequence of the genomic RNA. Consistent with this hypothesis, our recent results suggest that the preferential binding of NC to four sites triggers unfolding of the three-dimensional structure of (−) ssDNA, thus facilitating the r-R interaction [37]. In addition, we showed that the cTAR element in ssDNA does not exhibit a secondary structure identical to that in oligonucleotides. More precisely, the cTAR sequence in (−) ssDNA does not form the closed conformation in the absence of NC and its “Y” conformation is more open in the presence of NC. Interestingly, our recent study suggests that the NC-mediated annealing process does not depend on a single pathway (zipper intermediate or loop-loop complex) [37]. Additional in vitro studies with the full-length (−) ssDNA and the U3R region of the RNA genome are necessary to elucidate the NC-mediated annealing process in the natural context.
6. Internal strand transfer events
Two copies of the single-stranded retroviral RNA are co-packaged as a dimer into the same virion [1]. When the two co-packaged RNAs are non-identical, a recombinant can be generated [54]. Recombination results from reverse transcriptase switching between the two co-packaged RNAs during DNA synthesis [54]. There are approximately 5–14 recombination events per genome per reverse transcription cycle in vivo [119]. Most recombination events within internal regions of the dimeric genome of retroviruses result from DNA strand transfer events [54]. These internal strand transfer events occur during minus-strand DNA synthesis (Fig. 2, step C). In vitro reverse transcription assays using RT, a donor RNA template and an acceptor RNA template, have been developed to investigate the molecular mechanisms responsible for internal strand transfer reactions (see Ref. [54] and references therein). Similar mechanisms underlie the first strand transfer and the internal strand transfer events, i.e. the transfer reaction relies on an intermolecular DNA-acceptor RNA interaction that requires RNase H degradation of the donor RNA template.
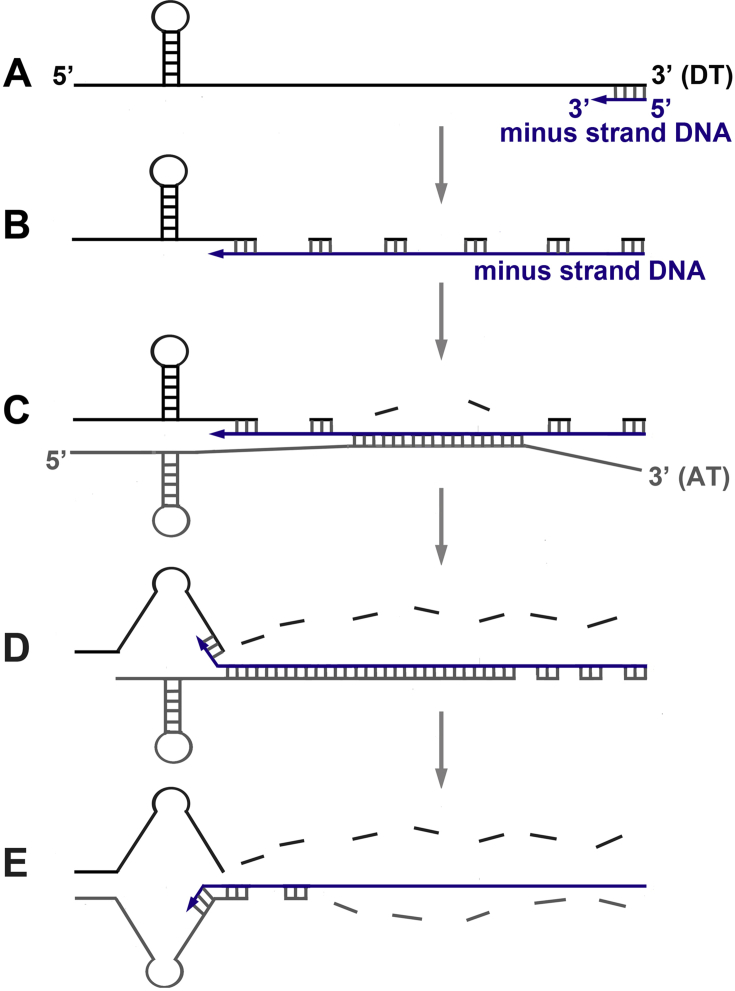
The DeStefano group determined the roles of NC and RNA structures in the internal strand transfer process [72], [114], [120]. In the absence of NC, transfer efficiency is significantly greater with weakly structured RNA templates than with highly structured RNA templates. NC slightly increases transfer in weakly structured RNA templates, whereas it strongly increases this reaction in highly structured RNA templates. The duplex destabilizing activity of the N-terminal zinc finger of NC plays an important role in internal strand transfer that occurs in a highly structured template. In vitro studies support the notion that the acceptor RNA structure plays an essential role in the internal transfer process [121], [122], [123]. An early study suggested that binding of NC to the acceptor RNA is mainly responsible for the enhancement of internal strand transfer events [121]. However, this hypothesis has not been confirmed by other studies. The Negroni group showed that a hairpin on the donor RNA is dispensable, whereas it is required on the acceptor RNA [122]. Depending on RNA templates, the crossover points of transfer are within or outside the hairpin [122], [124], [125].
During reverse transcription of the donor RNA template, RT pauses at the level of hairpins that induce internal strand transfer [120], [122], [124]. Efficient strand transfer can be triggered by RT pausing, but this is not an obligatory step in the strand transfer process (see Ref. [54] and references therein). Furthermore, NC-mediated enhancement of internal strand transfer does not result from an increase of RT pausing; indeed, NC increases the ability of RT to synthesize DNA through pause sites [54] and it does not modify the RT pausing pattern [121]. Depending on RNA templates, NC may [121], [123] or may not change [124], [125] the distribution of the crossover points of transfer. NC can increase internal strand transfer by promoting a donor-acceptor interaction through a loop-loop interaction or a G-quartet structure [125], [126], i.e. it creates a proximity effect between the two RNA templates. Nevertheless, the main role of NC is to increase the efficiency of acceptor invasion by promoting annealing of minus-strand DNA to the acceptor RNA. The acceptor invasion model has been proposed by Negroni and Buc [121] and is consistent with the results of many studies (see Ref. [54] and references therein). In this model (Fig. 7), RT-RNase H cleavage of the donor RNA provides the “invasion” site, i.e. a region of single-stranded DNA that interacts with the acceptor RNA. The “invasion” step (also termed “docking”) corresponds to the first base pairing interaction between DNA and the acceptor RNA that occurs upstream of the crossover points of transfer. This initial interaction propagates by branch migration, displacing the cleaved fragments of the donor template. Then, an intermolecular template exchange occurs, resulting in the capture of the DNA terminus by the acceptor template (Fig. 7, step E). The last step corresponds to DNA synthesis from the acceptor template. Thanks to its chaperone activity, NC can accelerate the invasion, branch migration and exchange steps described above; it is therefore not surprising that this viral protein increases the internal strand transfer.
Fig. 7.
The acceptor invasion model. A) Minus-strand DNA (blue line) is annealed to the donor RNA template (DT, black line). B) DNA synthesis and RNase H cleavage of the donor RNA template. C) Acceptor docking facilitated by NC; this step allows the acceptor RNA template (AT, gray line) to anneal to the nascent DNA. D) Branch migration facilitated by NC; this step brings AT in proximity to the 3′-end of DNA and allows RNase H-directed cleavages of AT. E) NC-facilitated annealing of AT to the 3′-end of DNA.
7. Second strand transfer
The second strand transfer depends on base pairing of the complementary PBS sequences at the 3′-ends of minus-strand DNA and plus-strand strong-stop DNA (Fig. 2, step G). In vitro reconstituted systems have been developed to investigate the molecular mechanisms involved in the second strand transfer [127], [128], [129], [130]. Using these systems, it was shown that HIV-1 NC increases the rate of the second strand transfer by two-to threefold [129], [130]. The transfer reaction requires that the tRNA primer is removed from the 5′-end of minus-strand DNA (Fig. 2, step F). Removal of the primer involves the RNase H activity of RT and is probably facilitated by the NC's duplex destabilization activity [127], [128], [129], [131].
The main role of NC is to facilitate base pairing of the complementary PBS sequences. An NMR study showed that the 18-nucleotide PBS sequence at the 3′-end of minus-strand DNA, termed (−) PBS, folds into a stable stem-loop, containing a five-residue loop and a bulged thymine in a guanosine-cytosine-rich stem [112]. This study also suggests that NC facilitates the base pairing interaction between (−) PBS and (+) PBS by destabilizing the stem-loop of (−) PBS. Another NMR study performed with truncated forms of NC and (−) PBS showed that NC binding to (−) PBS opens its apical loop; this may favor the formation of a “kissing complex” with (+) PBS. Finally, NC destabilizes the base pair adjacent to the apical loop by direct contacts involving several hydrophobic amino acids of its N-terminal zinc finger [111]. Fluorescence and UV–visible absorption spectroscopy studies suggest that the (−) and (+) PBS sequences fold into a stable stem-loop that binds three NC molecules [132]. Furthermore, fluorescent studies showed that a truncated form of NC weakly destabilizes the PBS hairpins and increases the (−)/(+) PBS annealing kinetics by 6-fold, mainly by promoting the base pairing interaction between the apical loops [133].
8. Strand transfer inhibitors
Antiretroviral therapy, comprising at least two reverse transcriptase inhibitors (RTIs) plus another inhibitor, is the recognized standard of care. RTIs, which prevent the conversion of the retroviral RNA genome into double-stranded DNA, target the DNA polymerase activity of RT. The response to antiretroviral therapy can be limited by the emergence of resistance to RTIs. As expected, drugs targeting the RNase H activity of RT are also inhibitors of the strand transfer reactions [134]. To date, although there are several types of RT-RNase H inhibitors, these are not enough selective to treat HIV-1 infected individuals [135]. It would therefore be interesting to possess a new class of drugs that would specifically inhibit the strand transfer reactions, without targeting both activities of RT. Moreover, strand transfer inhibitors could prevent the recombination events that are responsible for genetic diversity which generates multidrug-resistant retroviruses and complicates vaccine development.
Given the important roles of NC in its mature form and as a domain of the Gag precursor polyprotein in strand transfer and in genomic RNA packaging, respectively, it is considered as a potential antiviral target. NC is inhibited by compounds that promote zinc ejection from the fingers and display antiviral activity [136], [137]. These compounds disrupt NC zinc coordination by zinc chelation or covalent modifications of the target [137]. Most of zinc ejectors are toxic due to nonspecific interactions with other cellular targets. Because stable thioesters such as S-acyl-2-mercaptobenzamide thioesters (SAMTs) exhibit preferential selectivity for NC, over that of cellular zinc fingers, the development of new zinc ejectors is pursued. More precisely, it has been shown that the C-terminal zinc finger is a preferential target of SAMTs relative to the N-terminal zinc finger [138], [139], [140]. Others classes of NC inhibitors have been identified from high-throughput screening [139], [141]. These inhibitors that display moderate anti-HIV-1 activity in ex vivo cell assays are small molecules that bind to the NC without inducing zinc ejection and covalent modifications of the protein. Visible absorption spectroscopy and NMR studies performed with this type of inhibitor showed that two inhibitor molecules bind specifically and reversibly to the two zinc fingers of NC through stacking interactions involving Phe16 and Trp37 [142].
Inhibition of the NC-mediated strand transfer reactions by compounds targeting nucleic acids represents an alternative strategy. Early studies showed that actinomycin D, an antibiotic and anti-cancer drug which binds to single- and double-stranded DNA, inhibits the first strand transfer by interacting with the (−) ssDNA [91], [143]. However, actinomycin D is not a potent anti-HIV-1 agent in ex vivo cell assays [144]. More recently, 2,6-disubstituted-anthraquinones targeting the TAR RNA and cTAR DNA hairpins have been developed [145], [146]. These compounds are threading intercalators bearing a polyaromatic nucleus di-substituted at opposite positions with cationic side-chains that stabilize the interaction with the phosphodiester backbone. By stabilizing the secondary structures of TAR and cTAR, the anthraquinone derivatives inhibit the NC-mediated TAR-cTAR annealing process. In addition, anthraquinone derivatives with long cationic side-chains compete with NC for the same binding sites on TAR [146]. The antiviral activity of anthraquinone derivatives in ex vivo cell assays has not been reported. Compounds recognizing a NC-nucleic acid complex involved in a strand transfer reaction have not been developed to date.
9. Conclusion
Because it is difficult to determine by ex vivo cell assays the molecular mechanisms of strand transfer events that occur during reverse transcription of retroviral genomic RNA, in vitro systems have been developed. The in vitro systems partially mimic the in vivo context of the reverse transcription process. However, they showed that: 1) the strand transfer reactions require base pairing of complementary DNA/RNA sequences that form secondary structures such as hairpins; 2) NC promotes the base pairing interaction between two hairpins, thanks to its nucleic acid chaperone activity. NC differential binding to DNA and RNA is an attractive hypothesis proposed by our group [100] that needs to be confirmed by additional experiments. Furthermore, most of the annealing reactions have been investigated with short hairpins that partially mimic the nucleic acids involved in the strand transfer events. Therefore, additional in vitro studies with longer DNA and RNA molecules will be necessary to elucidate the NC-mediated annealing process in the natural context. Because a nucleic acid-NC complex is better structured than either molecule alone, it would be interesting to design inhibitors targeting the interface formed by the two macromolecules. Advancements in understanding of mechanisms and structures responsible for strand transfer should contribute to the design of new anti-HIV-1 agents.
Conflict of interest
The authors report no conflict of interest.
Funding
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and Sidaction the grant number is : 15-2-AEQ-04-01.
References
- 1.Dubois N., Marquet R., Paillart J.C., Bernacchi S. Retroviral RNA dimerization: from structure to functions. Front. Microbiol. 2018;9:527. doi: 10.3389/fmicb.2018.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin J.G., Mitra M., Mascarenhas A., Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herschhorn A., Hizi A. Retroviral reverse transcriptases. Cell. Mol. Life Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laskey S.B., Siliciano R.F. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat. Rev. Microbiol. 2014;12:772–780. doi: 10.1038/nrmicro3351. [DOI] [PubMed] [Google Scholar]
- 5.Vogt V.M., Simon M.N. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs J.A., Simon M.N., Gross I., Krausslich H.G., Fuller S.D., Vogt V.M., Johnson M.C. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 7.Chertova E., Chertov O., Coren L.V., Roser J.D., Trubey C.M., Bess J.W., Jr., Sowder R.C., Barsov E., Hood B.L., Fisher R.J., Nagashima K., Conrads T.P., Veenstra T.D., Lifson J.D., Ott D.E. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darlix J.L., Lapadat-Tapolsky M., de Rocquigny H., Roques B.P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 9.Stewart-Maynard K.M., Cruceanu M., Wang F., Vo M.N., Gorelick R.J., Williams M.C., Rouzina I., Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Kaluarachchi K., Giedroc D.P. Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein Sci. 1998;7:2265–2280. doi: 10.1002/pro.5560071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morellet N., Meudal H., Bouaziz S., Roques B.P. Structure of the zinc finger domain encompassing residues 13-51 of the nucleocapsid protein from simian immunodeficiency virus. Biochem. J. 2006;393:725–732. doi: 10.1042/BJ20051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers M.F., Henderson L.E., Chance M.R., Bess J.W., Jr., South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C., III, Hare D.R. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee B.M., De Guzman R.N., Turner B.G., Tjandra N., Summers M.F. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 14.Zargarian L., Tisné C., Barraud P., Xu X., Morellet N., René B., Mély Y., Fossé P., Mauffret O. Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein zinc fingers. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102150. e102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshmukh L., Schwieters C.D., Grishaev A., Clore G.M. Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR, x-ray scattering and protein engineering. Chemphyschem. 2016;17:1548–1552. doi: 10.1002/cphc.201600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsui T., Tanaka T., Endoh H., Sato K., Tanaka H., Miyauchi E., Kawashima Y., Nagai-Makabe M., Komatsu H., Kohno T., Maeda T., Kodera Y. The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7. Biochemistry. 2009;48:4314–4323. doi: 10.1021/bi802364b. [DOI] [PubMed] [Google Scholar]
- 17.Demene H., Jullian N., Morellet N., de Rocquigny H., Cornille F., Maigret B., Roques B.P. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of Moloney murine leukemia virus. J. Biomol. NMR. 1994;4:153–170. doi: 10.1007/BF00175244. [DOI] [PubMed] [Google Scholar]
- 18.Klein D.J., Johnson P.E., Zollars E.S., De Guzman R.N., Summers M.F. The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry. 2000;39:1604–1612. doi: 10.1021/bi9922493. [DOI] [PubMed] [Google Scholar]
- 19.Fisher R.J., Rein A., Fivash M., Urbaneja M.A., Casas-Finet J.R., Medaglia M., Henderson L.E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams M.C., Rouzina I., Wenner J.R., Gorelick R.J., Musier-Forsyth K., Bloomfield V.A. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 22.Darlix J.L., Godet J., Ivanyi-Nagy R., Fossé P., Mauffret O., Mély Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 23.Lapadat-Tapolsky M., de Rocquigny H., Van G.D., Roques B., Plasterk R., Darlix J.L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanevsky I., Chaminade F., Ficheux D., Moumen A., Gorelick R., Negroni M., Darlix J.L., Fossé P. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J. Mol. Biol. 2005;348:1059–1077. doi: 10.1016/j.jmb.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Yeh Y.S., Barbara P.F. HIV-1 nucleocapsid protein bends double-stranded nucleic acids. J. Am. Chem. Soc. 2009;131:15534–15543. doi: 10.1021/ja9070046. [DOI] [PubMed] [Google Scholar]
- 26.Berglund J.A., Charpentier B., Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S.J., Kim M.Y., Lee J.H., You J.C., Jeong S. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2002;291:925–931. doi: 10.1006/bbrc.2002.6521. [DOI] [PubMed] [Google Scholar]
- 28.Vuilleumier C., Bombarda E., Morellet N., Gerard D., Roques B.P., Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 29.Urbaneja M.A., McGrath C.F., Kane B.P., Henderson L.E., Casas-Finet J.R. Nucleic acid binding properties of the simian immunodeficiency virus nucleocapsid protein NCp8. J. Biol. Chem. 2000;275:10394–10404. doi: 10.1074/jbc.275.14.10394. [DOI] [PubMed] [Google Scholar]
- 30.Morcock D.R., Katakam S., Kane B.P., Casas-Finet J.R. Fluorescence and nucleic acid binding properties of bovine leukemia virus nucleocapsid protein. Biophys. Chem. 2002;97:203–212. doi: 10.1016/s0301-4622(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 31.Dey A., York D., Smalls-Mantey A., Summers M.F. Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus. Biochemistry. 2005;44:3735–3744. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 32.Morcock D.R., Kane B.P., Casas-Finet J.R. Fluorescence and nucleic acid binding properties of the human T-cell leukemia virus-type 1 nucleocapsid protein. Biochim. Biophys. Acta. 2000;1481:381–394. doi: 10.1016/s0167-4838(00)00181-3. [DOI] [PubMed] [Google Scholar]
- 33.Grohman J.K., Gorelick R.J., Lickwar C.R., Lieb J.D., Bower B.D., Znosko B.M., Weeks K.M. A guanosine-centric mechanism for RNA chaperone function. Science. 2013;340:190–195. doi: 10.1126/science.1230715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van H.F., van der Kuyl A.C., Berkhout B. On the nucleotide composition and structure of retroviral RNA genomes. Virus Res. 2014;193:16–23. doi: 10.1016/j.virusres.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Thomas J.A., Gorelick R.J. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beltz H., Piemont E., Schaub E., Ficheux D., Roques B., Darlix J.L., Mély Y. Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2004;338:711–723. doi: 10.1016/j.jmb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Maskri O., Chaminade F., René B., Benkaroun J., Godet J., Mély Y., Mauffret O., Fossé P. Structural insights into the HIV-1 minus-strand strong-stop DNA. J. Biol. Chem. 2016;291:3468–3482. doi: 10.1074/jbc.M115.708099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruceanu M., Urbaneja M.A., Hixson C.V., Johnson D.G., Datta S.A., Fivash M.J., Stephen A.G., Fisher R.J., Gorelick R.J., Casas-Finet J.R., Rein A., Rouzina I., Williams M.C. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W., Naiyer N., Mitra M., Li J., Williams M.C., Rouzina I., Gorelick R.J., Wu Z., Musier-Forsyth K. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Res. 2014;42:7145–7159. doi: 10.1093/nar/gku335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu T., Gorelick R.J., Levin J.G. Selection of fully processed HIV-1 nucleocapsid protein is required for optimal nucleic acid chaperone activity in reverse transcription. Virus Res. 2014;193:52–64. doi: 10.1016/j.virusres.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanwar H.S., Khoo K.K., Garvey M., Waddington L., Leis A., Hijnen M., Velkov T., Dumsday G.J., McKinstry W.J., Mak J. The thermodynamics of Pr55Gag-RNA interaction regulate the assembly of HIV. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006221. e1006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu K., Heng X., Summers M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutluay S.B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., Bieniasz P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Meshri S.E., Dujardin D., Godet J., Richert L., Boudier C., Darlix J.L., Didier P., Mély Y., de Rocquigny R.H. Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: a fluorescence lifetime imaging microscopy investigation. J. Mol. Biol. 2015;427:1480–1494. doi: 10.1016/j.jmb.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Racine P.J., Chamontin C., de Rocquigny H., Bernacchi S., Paillart J.C., Mougel M. Requirements for nucleocapsid-mediated regulation of reverse transcription during the late steps of HIV-1 assembly. Sci. Rep. 2016;6:27536. doi: 10.1038/srep27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilboa E., Mitra S.W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 47.Sleiman D., Goldschmidt V., Barraud P., Marquet R., Paillart J.C., Tisné C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012;169:324–339. doi: 10.1016/j.virusres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Isel C., Marquet R., Keith G., Ehresmann C., Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 49.Fossé P., Mougel M., Keith G., Westhof E., Ehresmann B., Ehresmann C. Modified nucleotides of tRNAPro restrict interactions in the binary primer/template complex of M-MuLV. J. Mol. Biol. 1998;275:731–746. doi: 10.1006/jmbi.1997.1487. [DOI] [PubMed] [Google Scholar]
- 50.Prats A.C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J.L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y., Khorchid A., Gabor J., Wang J., Li X., Darlix J.L., Wainberg M.A., Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA(3Lys) genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J. Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hargittai M.R., Gorelick R.J., Rouzina I., Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 53.Seif E., Niu M., Kleiman L. In virio SHAPE analysis of tRNA(Lys3) annealing to HIV-1 genomic RNA in wild type and protease-deficient virus. Retrovirology. 2015;12:40. doi: 10.1186/s12977-015-0171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basu V.P., Song M., Gao L., Rigby S.T., Hanson M.N., Bambara R.A. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 55.van Wamel J.L., Berkhout B. The first strand transfer during HIV-1 reverse transcription can occur either intramolecularly or intermolecularly. Virology. 1998;244:245–251. doi: 10.1006/viro.1998.9096. [DOI] [PubMed] [Google Scholar]
- 56.Yu H., Jetzt A.E., Ron Y., Preston B.D., Dougherty J.P. The nature of human immunodeficiency virus type 1 strand transfers. J. Biol. Chem. 1998;273:28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]
- 57.Klaver B., Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lobel L.I., Goff S.P. Reverse transcription of retroviral genomes: mutations in the terminal repeat sequences. J. Virol. 1985;53:447–455. doi: 10.1128/jvi.53.2.447-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramsey C.A., Panganiban A.T. Replication of the retroviral terminal repeat sequence during in vivo reverse transcription. J. Virol. 1993;67:4114–4121. doi: 10.1128/jvi.67.7.4114-4121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kulpa D., Topping R., Telesnitsky A. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 1997;16:856–865. doi: 10.1093/emboj/16.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohi Y., Clever J.L. Sequences in the 5' and 3' R elements of human immunodeficiency virus type 1 critical for efficient reverse transcription. J. Virol. 2000;74:8324–8334. doi: 10.1128/jvi.74.18.8324-8334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peliska J.A., Benkovic S.J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 63.Darlix J.L., Vincent A., Gabus C., de Rocquigny H., Roques B. Trans-activation of the 5' to 3' viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. III. 1993;316:763–771. [PubMed] [Google Scholar]
- 64.Allain B., Lapadat-Tapolsky M., Berlioz C., Darlix J.L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo J., Henderson L.E., Bess J., Kane B., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berkhout B., Vastenhouw N.L., Klasens B.I., Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7:1097–1114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Werner S., Vogel-Bachmayr K., Hollinderbaumer B., Wohrl B.M. Requirements for minus-strand transfer catalyzed by Rous sarcoma virus reverse transcriptase. J. Virol. 2001;75:10132–10138. doi: 10.1128/JVI.75.21.10132-10138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y., Balakrishnan M., Roques B.P., Fay P.J., Bambara R.A. Mechanism of minus strand strong stop transfer in HIV-1 reverse transcription. J. Biol. Chem. 2003;278:8006–8017. doi: 10.1074/jbc.M210959200. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y., Balakrishnan M., Roques B.P., Bambara R.A. Acceptor RNA cleavage profile supports an invasion mechanism for HIV-1 minus strand transfer. J. Biol. Chem. 2005;280:14443–14452. doi: 10.1074/jbc.M412190200. [DOI] [PubMed] [Google Scholar]
- 70.Tanese N., Telesnitsky A., Goff S.P. Abortive reverse transcription by mutants of Moloney murine leukemia virus deficient in the reverse transcriptase-associated RNase H function. J. Virol. 1991;65:4387–4397. doi: 10.1128/jvi.65.8.4387-4397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blain S.W., Goff S.P. Effects on DNA synthesis and translocation caused by mutations in the RNase H domain of Moloney murine leukemia virus reverse transcriptase. J. Virol. 1995;69:4440–4452. doi: 10.1128/jvi.69.7.4440-4452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Derebail S.S., Heath M.J., DeStefano J.J. Evidence for the differential effects of nucleocapsid protein on strand transfer in various regions of the HIV genome. J. Biol. Chem. 2003;278:15702–15712. doi: 10.1074/jbc.M211701200. [DOI] [PubMed] [Google Scholar]
- 73.Henriet S., Sinck L., Bec G., Gorelick R.J., Marquet R., Paillart J.C. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Post K., Olson E.D., Naufer M.N., Gorelick R.J., Rouzina I., Williams M.C., Musier-Forsyth K., Levin J.G. Mechanistic differences between HIV-1 and SIV nucleocapsid proteins and cross-species HIV-1 genomic RNA recognition. Retrovirology. 2016;13:89. doi: 10.1186/s12977-016-0322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y., Balakrishnan M., Roques B.P., Bambara R.A. Steps of the acceptor invasion mechanism for HIV-1 minus strand strong stop transfer. J. Biol. Chem. 2003;278:38368–38375. doi: 10.1074/jbc.M305700200. [DOI] [PubMed] [Google Scholar]
- 76.Wu T., Heilman-Miller S.L., Levin J.G. Effects of nucleic acid local structure and magnesium ions on minus-strand transfer mediated by the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2007;35:3974–3987. doi: 10.1093/nar/gkm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Driscoll M.D., Hughes S.H. Human immunodeficiency virus type 1 nucleocapsid protein can prevent self-priming of minus-strand strong stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J. Virol. 2000;74:8785–8792. doi: 10.1128/jvi.74.19.8785-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wisniewski M., Chen Y., Balakrishnan M., Palaniappan C., Roques B.P., Fay P.J., Bambara R.A. Substrate requirements for secondary cleavage by HIV-1 reverse transcriptase RNase H. J. Biol. Chem. 2002;277:28400–28410. doi: 10.1074/jbc.M201645200. [DOI] [PubMed] [Google Scholar]
- 79.Hong M.K., Harbron E.J., O'Connor D.B., Guo J., Barbara P.F., Levin J.G., Musier-Forsyth K. Nucleic acid conformational changes essential for HIV-1 nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer. J. Mol. Biol. 2003;325:1–10. doi: 10.1016/s0022-2836(02)01177-4. [DOI] [PubMed] [Google Scholar]
- 80.Heilman-Miller S.L., Wu T., Levin J.G. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J. Biol. Chem. 2004;279:44154–44165. doi: 10.1074/jbc.M401646200. [DOI] [PubMed] [Google Scholar]
- 81.Khan M.A., Aberham C., Kao S., Akari H., Gorelick R., Bour S., Strebel K. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 2001;75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peliska J.A., Balasubramanian S., Giedroc D.P., Benkovic S.J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 83.Cameron C.E., Ghosh M., Le Grice S.F., Benkovic S.J. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lener D., Tanchou V., Roques B.P., Le Grice S.F., Darlix J.L. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- 85.Druillennec S., Caneparo A., de Rocquigny H., Roques B.P. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- 86.Purohit V., Roques B.P., Kim B., Bambara R.A. Mechanisms that prevent template inactivation by HIV-1 reverse transcriptase RNase H cleavages. J. Biol. Chem. 2007;282:12598–12609. doi: 10.1074/jbc.M700043200. [DOI] [PubMed] [Google Scholar]
- 87.Hergott C.B., Mitra M., Guo J., Wu T., Miller J.T., Iwatani Y., Gorelick R.J., Levin J.G. Zinc finger function of HIV-1 nucleocapsid protein is required for removal of 5'-terminal genomic RNA fragments: a paradigm for RNA removal reactions in HIV-1 reverse transcription. Virus Res. 2013;171:346–355. doi: 10.1016/j.virusres.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess J.W., Jr., Swanstrom R., Burch C.L., Weeks K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bernacchi S., Stoylov S., Piemont E., Ficheux D., Roques B.P., Darlix J.L., Mély Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 90.You J.C., McHenry C.S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 91.Guo J., Wu T., Bess J., Henderson L.E., Levin J.G. Actinomycin D inhibits human immunodeficiency virus type 1 minus-strand transfer in in vitro and endogenous reverse transcriptase assays. J. Virol. 1998;72:6716–6724. doi: 10.1128/jvi.72.8.6716-6724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moumen A., Polomack L., Roques B., Buc H., Negroni M. The HIV-1 repeated sequence R as a robust hot-spot for copy-choice recombination. Nucleic Acids Res. 2001;29:3814–3821. doi: 10.1093/nar/29.18.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu H.W., Cosa G., Landes C.F., Zeng Y., Kovaleski B.J., Mullen D.G., Barany G., Musier-Forsyth K., Barbara P.F. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys. J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Godet J., de Rocquigny H., Raja C., Glasser N., Ficheux D., Darlix J.L., Mély Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 95.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 96.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J. Mol. Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu H.W., Zeng Y., Landes C.F., Kim Y.J., Zhu Y., Ma X., Vo M.N., Musier-Forsyth K., Barbara P.F. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5261–5267. doi: 10.1073/pnas.0700166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanevsky I., Chaminade F., Chen Y., Godet J., René B., Darlix J.L., Mély Y., Mauffret O., Fossé P. Structural determinants of TAR RNA-DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2011;39:8148–8162. doi: 10.1093/nar/gkr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zargarian L., Kanevsky I., Bazzi A., Boynard J., Chaminade F., Fossé P., Mauffret O. Structural and dynamic characterization of the upper part of the HIV-1 cTAR DNA hairpin. Nucleic Acids Res. 2009;37:4043–4054. doi: 10.1093/nar/gkp297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bazzi A., Zargarian L., Chaminade F., Boudier C., de Rocquigny H., René B., Mély Y., Fossé P., Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39:3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bazzi A., Zargarian L., Chaminade F., de Rocquigny H., René B., Mély Y., Fossé P., Mauffret O. Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets. PLoS One. 2012;7:e38905. doi: 10.1371/journal.pone.0038905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Belfetmi A., Zargarian L., Tisné C., Sleiman D., Morellet N., Lescop E., Maskri O., René B., Mély Y., Fossé P., Mauffret O. Insights into the Mechanisms of RNA Secondary Structure Destabilization by the HIV-1 Nucleocapsid Protein. RNA. 2016;22:506–517. doi: 10.1261/rna.054445.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heng X., Kharytonchyk S., Garcia E.L., Lu K., Divakaruni S.S., LaCotti C., Edme K., Telesnitsky A., Summers M.F. Identification of a minimal region of the HIV-1 5'-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dethoff E.A., Hansen A.L., Musselman C., Watt E.D., Andricioaei I., Al-Hashimi H.M. Characterizing complex dynamics in the transactivation response element apical loop and motional correlations with the bulge by NMR, molecular dynamics, and mutagenesis. Biophys. J. 2008;95:3906–3915. doi: 10.1529/biophysj.108.140285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCauley M.J., Rouzina I., Manthei K.A., Gorelick R.J., Musier-Forsyth K., Williams M.C. Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13555–13560. doi: 10.1073/pnas.1510100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Azoulay J., Clamme J.P., Darlix J.L., Roques B.P., Mély Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- 107.De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 108.Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K.J., Wu Z.R., Summers M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 109.Spriggs S., Garyu L., Connor R., Summers M.F. Potential intra- and intermolecular interactions involving the unique-5' region of the HIV-1 5'-UTR. Biochemistry. 2008;47:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morellet N., Demene H., Teilleux V., Huynh-Dinh T., de Rocquigny H., Fournie-Zaluski M.C., Roques B.P. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 111.Bourbigot S., Ramalanjaona N., Boudier C., Salgado G.F., Roques B.P., Mély Y., Bouaziz S., Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription. J. Mol. Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 112.Johnson P.E., Turner R.B., Wu Z.R., Hairston L., Guo J., Levin J.G., Summers M.F. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 113.Guo J., Wu T., Kane B.F., Johnson D.G., Henderson L.E., Gorelick R.J., Levin J.G. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 2002;76:4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heath M.J., Derebail S.S., Gorelick R.J., DeStefano J.J. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J. Biol. Chem. 2003;278:30755–30763. doi: 10.1074/jbc.M303819200. [DOI] [PubMed] [Google Scholar]
- 115.Williams M.C., Gorelick R.J., Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mitra M., Wang W., Vo M.N., Rouzina I., Barany G., Musier-Forsyth K. The N-terminal zinc finger and flanking basic domains represent the minimal region of the human immunodeficiency virus type-1 nucleocapsid protein for targeting chaperone function. Biochemistry. 2013;52:8226–8236. doi: 10.1021/bi401250a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gamsjaeger R., Kariawasam R., Gimenez A.X., Touma C., McIlwain E., Bernardo R.E., Shepherd N.E., Ataide S.F., Dong Q., Richard D.J., White M.F., Cubeddu L. The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus. Biochem. J. 2015;465:337–346. doi: 10.1042/BJ20141140. [DOI] [PubMed] [Google Scholar]
- 118.Abbondanzieri E.A., Bokinsky G., Rausch J.W., Zhang J.X., Le Grice S.F., Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cromer D., Grimm A.J., Schlub T.E., Mak J., Davenport M.P. Estimating the in-vivo HIV template switching and recombination rate. AIDS. 2016;30:185–192. doi: 10.1097/QAD.0000000000000936. [DOI] [PubMed] [Google Scholar]
- 120.Derebail S.S., DeStefano J.J. Mechanistic analysis of pause site-dependent and -independent recombinogenic strand transfer from structurally diverse regions of the HIV genome. J. Biol. Chem. 2004;279:47446–47454. doi: 10.1074/jbc.M408927200. [DOI] [PubMed] [Google Scholar]
- 121.Negroni M., Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6385–6390. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moumen A., Polomack L., Unge T., Veron M., Buc H., Negroni M. Evidence for a mechanism of recombination during reverse transcription dependent on the structure of the acceptor RNA. J. Biol. Chem. 2003;278:15973–15982. doi: 10.1074/jbc.M212306200. [DOI] [PubMed] [Google Scholar]
- 123.Hanson M.N., Balakrishnan M., Roques B.P., Bambara R.A. Effects of donor and acceptor RNA structures on the mechanism of strand transfer by HIV-1 reverse transcriptase. J. Mol. Biol. 2005;353:772–787. doi: 10.1016/j.jmb.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 124.Roda R.H., Balakrishnan M., Kim J.K., Roques B.P., Fay P.J., Bambara R.A. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 2002;277:46900–46911. doi: 10.1074/jbc.M208638200. [DOI] [PubMed] [Google Scholar]
- 125.Balakrishnan M., Roques B.P., Fay P.J., Bambara R.A. Template dimerization promotes an acceptor invasion-induced transfer mechanism during human immunodeficiency virus type 1 minus-strand synthesis. J. Virol. 2003;77:4710–4721. doi: 10.1128/JVI.77.8.4710-4721.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen W., Gorelick R.J., Bambara R.A. HIV-1 nucleocapsid protein increases strand transfer recombination by promoting dimeric G-quartet formation. J. Biol. Chem. 2011;286:29838–29847. doi: 10.1074/jbc.M111.262352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Auxilien S., Keith G., Le Grice S.F., Darlix J.L. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999;274:4412–4420. doi: 10.1074/jbc.274.7.4412. [DOI] [PubMed] [Google Scholar]
- 128.Smith C.M., Smith J.S., Roth M.J. RNase H requirements for the second strand transfer reaction of human immunodeficiency virus type 1 reverse transcription. J. Virol. 1999;73:6573–6581. doi: 10.1128/jvi.73.8.6573-6581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu T., Guo J., Bess J., Henderson L.E., Levin J.G. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 1999;73:4794–4805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Muthuswami R., Chen J., Burnett B.P., Thimmig R.L., Janjic N., McHenry C.S. The HIV plus-strand transfer reaction: determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence. J. Mol. Biol. 2002;315:311–323. doi: 10.1006/jmbi.2001.5205. [DOI] [PubMed] [Google Scholar]
- 131.Guo J., Wu T., Anderson J., Kane B.F., Johnson D.G., Gorelick R.J., Henderson L.E., Levin J.G. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Egele C., Schaub E., Ramalanjaona N., Piemont E., Ficheux D., Roques B., Darlix J.L., Mély Y. HIV-1 nucleocapsid protein binds to the viral DNA initiation sequences and chaperones their kissing interactions. J. Mol. Biol. 2004;342:453–466. doi: 10.1016/j.jmb.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 133.Ramalanjaona N., de Rocquigny H., Millet A., Ficheux D., Darlix J.L., Mély Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J. Mol. Biol. 2007;374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 134.Gabbara S., Davis W.R., Hupe L., Hupe D., Peliska J.A. Inhibitors of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1999;38:13070–13076. doi: 10.1021/bi991085n. [DOI] [PubMed] [Google Scholar]
- 135.Kankanala J., Kirby K.A., Huber A.D., Casey M.C., Wilson D.J., Sarafianos S.G., Wang Z. Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H. Eur. J. Med. Chem. 2017;141:149–161. doi: 10.1016/j.ejmech.2017.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rice W.G., Schaeffer C.A., Harten B., Villinger F., South T.L., Summers M.F., Henderson L.E., Bess J.W., Jr., Arthur L.O., McDougal J.S. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 137.Mori M., Kovalenko L., Lyonnais S., Antaki D., Torbett B.E., Botta M., Mirambeau G., Mély Y. Nucleocapsid protein: a desirable target for future therapies against HIV-1. Curr. Top. Microbiol. Immunol. 2015;389:53–92. doi: 10.1007/82_2015_433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jenkins L.M., Byrd J.C., Hara T., Srivastava P., Mazur S.J., Stahl S.J., Inman J.K., Appella E., Omichinski J.G., Legault P. Studies on the mechanism of inactivation of the HIV-1 nucleocapsid protein NCp7 with 2-mercaptobenzamide thioesters. J. Med. Chem. 2005;48:2847–2858. doi: 10.1021/jm0492195. [DOI] [PubMed] [Google Scholar]
- 139.Garg D., Torbett B.E. Advances in targeting nucleocapsid-nucleic acid interactions in HIV-1 therapy. Virus Res. 2014;193:135–143. doi: 10.1016/j.virusres.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sancineto L., Iraci N., Tabarrini O., Santi C. NCp7: targeting a multitasking protein for next-generation anti-HIV drug development part 1: covalent inhibitors. Drug Discov. Today. 2018;23:260–271. doi: 10.1016/j.drudis.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 141.Shvadchak V., Sanglier S., Rocle S., Villa P., Haiech J., Hibert M., Van D.A., Mély Y., de Rocquigny H. Identification by high throughput screening of small compounds inhibiting the nucleic acid destabilization activity of the HIV-1 nucleocapsid protein. Biochimie. 2009;91:916–923. doi: 10.1016/j.biochi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 142.Goudreau N., Hucke O., Faucher A.M., Grand-Maitre C., Lepage O., Bonneau P.R., Mason S.W., Titolo S. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2:1 inhibitor/NC stoichiometry. J. Mol. Biol. 2013;425:1982–1998. doi: 10.1016/j.jmb.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 143.Jeeninga R.E., Huthoff H.T., Gultyaev A.P., Berkhout B. The mechanism of actinomycin D-mediated inhibition of HIV-1 reverse transcription. Nucleic Acids Res. 1998;26:5472–5479. doi: 10.1093/nar/26.23.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Imamichi T., Murphy M.A., Adelsberger J.W., Yang J., Watkins C.M., Berg S.C., Baseler M.W., Lempicki R.A., Guo J., Levin J.G., Lane H.C. Actinomycin D induces high-level resistance to thymidine analogs in replication of human immunodeficiency virus type 1 by interfering with host cell thymidine kinase expression. J. Virol. 2003;77:1011–1020. doi: 10.1128/JVI.77.2.1011-1020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Frecentese F., Sosic A., Saccone I., Gamba E., Link K., Miola A., Cappellini M., Cattelan M.G., Severino B., Fiorino F., Magli E., Corvino A., Perissutti E., Fabris D., Gatto B., Caliendo G., Santagada V. Synthesis and in vitro screening of new series of 2,6-Dipeptidyl-anthraquinones: influence of side chain length on HIV-1 nucleocapsid inhibitors. J. Med. Chem. 2016;59:1914–1924. doi: 10.1021/acs.jmedchem.5b01494. [DOI] [PubMed] [Google Scholar]
- 146.Sosic A., Sinigaglia L., Cappellini M., Carli I., Parolin C., Zagotto G., Sabatino G., Rovero P., Fabris D., Gatto B. Mechanisms of HIV-1 nucleocapsid protein inhibition by Lysyl-Peptidyl-anthraquinone conjugates. Bioconjug. Chem. 2016;27:247–256. doi: 10.1021/acs.bioconjchem.5b00627. [DOI] [PubMed] [Google Scholar]