Abstract
Although the link between the inhalation of nanoparticles and cardiovascular disease is well established, the causal pathway between nanoparticle exposure and increased activity of blood coagulation factors remains unexplained. To initiate coagulation tissue factor bearing epithelial cell membranes should be exposed to blood, on the other side of the less than a micrometre thin air-blood barrier. For the inhaled nanoparticles to promote coagulation, they need to bind lung epithelial-cell membrane parts and relocate them into the blood. To assess this hypothesis, we use advanced microscopy and spectroscopy techniques to show that the nanoparticles wrap themselves with epithelial-cell membranes, leading to the membrane’s disruption. The membrane-wrapped nanoparticles are then observed to freely diffuse across the damaged epithelial cell layer relocating epithelial cell membrane parts over the epithelial layer. Proteomic analysis of the protein content in the nanoparticles wraps/corona finally reveals the presence of the coagulation-initiating factors, supporting the proposed causal link between the inhalation of nanoparticles and cardiovascular disease.
Keywords: TiO2 nanoparticles, membrane disruption, lipid wrapping, STED microscopy, tissue factor relocation, coagulation cascade interference
Nanoparticles (NPs) are small (<100 nm in diameter) organic and/or inorganic particles that have attracted great scientific interest because their size positions them between bulk materials and molecular structures, making it possible to produce sophisticated products such as solar panels, specialty alloys, and plastics. Unfortunately, because of their increasingly widespread use in such products, NPs are being dispersed into the air and are a major contributing factor to air pollution. The OECD has suggested that air pollutants, including NPs, might be responsible for the deaths of as many as 3.6 million people every year,1 and as early as 2004 the Expert Panel on Population and Prevention Science of the American Heart Association (AHA) highlighted the strong nanotoxicological effects of NPs and the links between inhaled NPs and cardiovascular disease,2 which have later on been confirmed in animal models.3 Recently, Miller et al. have located NPs in blood clots, indicating their ability to penetrate the alveolar epithelium, that is, lung air–blood barrier, even in human.4 In addition, several studies proved that exposure to some NPs can lead to the systemic inflammation5 and increased blood coagulation connected with cardiovascular diseases.6 However, molecular mechanisms of activation of clotting factors in blood remain unknown. Not surprisingly, the research community prioritized the need to elucidate the unknown molecular mechanisms involved in the sequence of events leading to cardiovascular diseases, including atherosclerosis and stroke.7
Blood coagulation is initiated in vivo, when tissue factor, a transmembrane protein, abundant in lung epithelium,8 is exposed to blood, such as at a site of injury.9 The intriguing question we need to answer is how the lung epithelial cell membranes with tissue factor, located on the other side of the air-blood barrier, can cross this barrier after having been exposed to NPs,and potentially activate the coagulation cascade.
The mechanism we are proposing relies on the affinity of TiO2 nanoparticles for lipids and proteins in cellular membranes, a property that on model membranes10 as well as in the lung surfactant layer11 was proven to build-up a lipid wrap, that is, a corona, around this and other metal-oxide NPs. If the NP surface binds to the membrane strongly enough, it is reasonable to expect that such a NP can act as a carrier of membrane parts together with the aforementioned coagulation-activating factors.
Some of the most ubiquitous NPs are the dioxides of titanium and silicon (TiO2 and SiO2). These are abundant materials in nature as well as common additives in food, cosmetics, paints, solar panels, catalysts, etc. As a result, they have often been included in nanotoxicological studies. For example, strong affinities for lipids have been demonstrated in the cases of TiO2 and SiO2,12 and for SiO2-NPs such an affinity has been recognized as the cause of the disruption of lipid vesicles with subsequent wrapping of the NP into a lipid bilayer.13 TiO2 has been characterized as having a specifically high affinity for phospholipids.14 However, TiO2–NPs have so far been found incapable of disrupting lipid bilayers and lipid wrapping.15 On the other hand, several in vivo toxicological studies have revealed that TiO2 NPs accumulate in and cause dysfunctions of various lipid-rich environments, such as pulmonary surfactants,16 endothelial cell junctions in lung blood vessels,17 the placenta, fetal livers, and the brain.18 Similarly, while studying the photo- and cytotoxicity of TiO2 nanotubes19 we also observed that these NPs were able to disintegrate the membrane of breast-cancer cells, generating a dispersed membrane haze that was not resolvable by conventional optical microscopy (see Figure S1 in the Supporting Information). We thus hypothesize that the strong interactions between TiO2 NPs and lipids can relocate the parts of endothelial cell membranes, which include the coagulation initiating tissue factor.
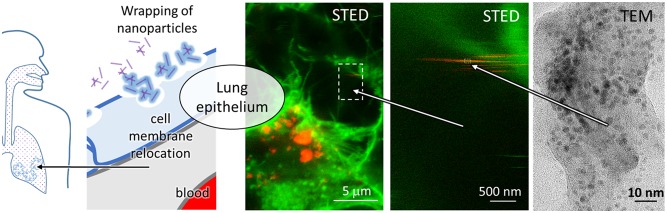
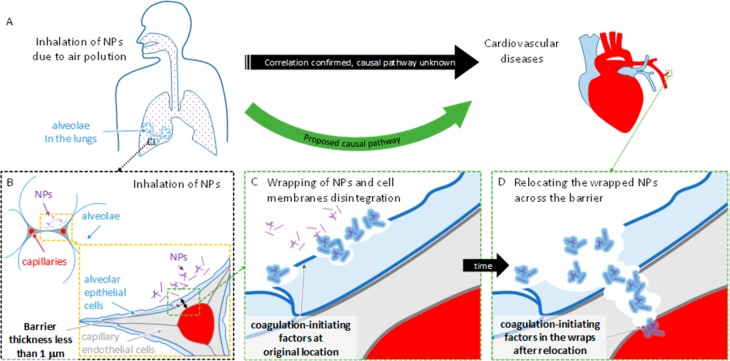
To test this hypothesis, we here employ various advanced observation technologies, such as super-resolution STED fluorescence microscopy and microspectroscopy, fluorescence fluctuation techniques, and electron microscopy, to provide clear evidence (i) that these NPs generally impair the integrity of the lipid membranes, (ii) that upon exposure to TiO2 NPs the membranes of the living lung epithelial cells disintegrate and wrap around the native surface of the TiO2, and (iii) to the best of our knowledge for the first time we observe the wrapping of the cellular membranes around the NPs, which generates freely diffusible carriers that are able to relocate patches of epithelial membrane away from their original position on the surface of the epithelial cells. Finally, (iv) the proteomic analysis of the NP-bound membranes and of the secreted proteins confirmed the presence of many membrane proteins including the coagulation-associated factors. Taking into account the 500 nm thin layer of lung epithelial and capillary endothelial cells (see the model in Figure 1), this formation of mobile membrane-wrapped NPs may be responsible for the relocation of the membrane-anchored, coagulation-activating factors into the blood, possibly leading to a systemic inflammation and the progression of cardiovascular disease.
Figure 1.
Hypothesis concerning NP relocation across the air-to-blood barrier. (A) Inhalation of NPs because of air pollution is known to be correlated with cardiovascular diseases, but the causal pathway is unknown. (B) The barrier separates the aerial space of lung alveoli from the blood (red) through a thin layer of alveolar epithelial cells (light blue) and capillary endothelial cells (light gray). Through inhalation, the NPs come into contact with alveolar epithelial cells. (C) NPs wrap themselves with membranes and cause their disruption. (D) Wrapped NPs diffuse across the damaged layer, relocating the coagulation-initiating factors to the blood.
Results and Discussion
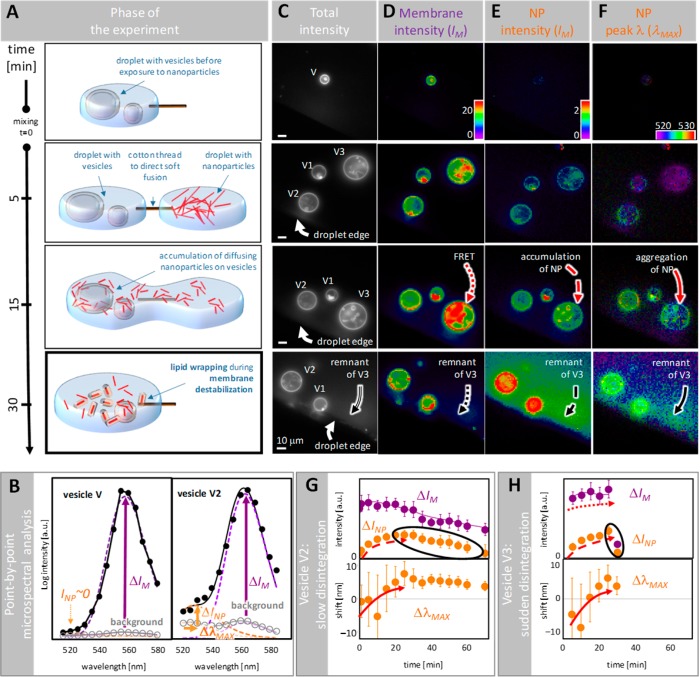
To test our hypothesis about membrane disruption and relocation by NPs we use TiO2 NPs in the form of 10 nm diameter nanotubes (length of few hundreds of nanometers, Materials and Methods). First, we investigate the effects of the affinity of the NPs for lipids in model membrane systems, specifically giant unilamellar lipid vesicles (GUVs) with various compositions (Supplementary Table S1). For the fluorescent imaging we label the NPs with the organic dye Alexa 488 (NP488; Figure S2), and the vesicles with a PC-BODIPY 530/550 fluorescent lipid analogue (phosphocholine (PC)-BODIPY 530/550). We induce the interaction between the vesicles and the NPs by gently fusing a droplet of labeled vesicles with a droplet of labeled NPs along a cotton thread on a microscope coverslip, resulting in a slow increase in the concentration of NPs around the vesicles (Figure 2a). We use fluorescence microspectroscopy (FMS)20,21 (i.e., fluorescence imaging with recording of the fluorescence emission spectra (Figure 2b) for each image pixel) to determine the total fluorescence intensities as well as the spectral maxima of the NP-bound and membrane probes at and near the surface of the vesicles (Figure 2c–f).
Figure 2.
Destabilization of lipid bilayers as seen by fluorescence microspectroscopy (FMS). (A) A droplet of lipid vesicles is gently fused with a droplet of NPs and monitored by spectral-sensitive FMS every 5 min for 1 h. (B) The acquired fluorescence emission spectra before (left) and after mixing (right) is decomposed (unmixed) in every pixel located on the vesicles, denoted V, V1, V2, and V3, into the NP component (INP) and membrane component (IM). Fluorescence signal of the background, that is, a micron away from the membrane, is subtracted from both of the components INP and IM, resulting in ΔINP and ΔIM, respectively. In the case of the NP component, the wavelength shift with respect to the background signal is also derived and denoted as ΔλMAX. (C–F) Besides total fluorescence images (C), resulting spatially resolved intensities IM (D) and INP (E) are presented as false color maps together with the NPs’ spectral peak position (F). (G,H) As an example, average values of ΔIM, ΔINP, ΔλMAX are presented for the vesicle V2 (G) and vesicle V3 (H). Error bars represent standard deviation of parameter values taken from approximately 50–70 pixels along the perimeter of the vesicle. Dashed, solid, and dotted red arrows indicate the accumulation of NPs on the membrane, aggregation of NPs, and energy transfer from NPs to membrane probes, respectively. The black arrows point to the debris of the disrupted vesicle (V3). The encircled data in panel (G) shows the outflow of a lipid-wrapped NP from the vesicle, suggesting the membrane’s gradual disruption.
After the induction of the NP–vesicle interaction, the intensity, and thus concentration of the NP-bound probes increases markedly at the surface of the vesicles (compare left and right spectrum at the membrane of vesicles, obtained before and after mixing, respectively; also see dashed red arrows in Figure 2e,g,h), clearly indicating the accumulation of NPs on the membrane due to the affinity of the TiO2 for lipids. In addition, the emission spectra of the NP-bound probes reveals a red-shift once bound to the membrane (solid red arrows in Figure 2f–h). Such a red shift is characteristic of the situation when Alexa 488 molecules are close to each other and therefore indicate the aggregation of the NPs.22 Further, membrane-binding and the accumulation of the NPs bring NP-bound probes close to the membrane-bound probes, so fostering the so-called Förster resonance energy transfer (FRET), which results in the observed increase in the signal of the membrane probes (dotted red arrows in Figure 2d,h). It is important to note that the NPs accumulate on the vesicles, despite a mild electrostatic repulsion. However, the accumulation and aggregation of the TiO2–NPs on the membranes is impaired and the FRET efficiency only diminishes when the electrostatic repulsion is substantially amplified (Figure S9).
Following the accumulation of the TiO2–NPs on the membranes, the latter become increasingly wobbly, and in some cases completely disintegrate, leaving behind a haze of signals from both the membrane and the NPs (black arrows in Figure 2d,e and the last data points in Figure 2h). Considering the known interactions of TiO2 with lipid bilayers23 and the similar membrane-wrapping characteristics of silica nanoparticles,13 we interpret this disruption and the occurrence of a hazy background as the creation of membrane-wrapped NPs (Figure 2a, bottom panel), which, though spatially unresolvable with a conventional microscope, can be identified via their spectral fingerprint obtained by FMS. The persisting red shift in the emission spectrum from the NPs (black arrow in Figure 2f) suggests that lipid wrapping also occurs for the aggregated NPs.
For other vesicles that maintain their shapes, that is, that do not disintegrate, we observe a slow but continuous decrease in the signals from both the NPs and the membranes (vesicle V2, encircled and purple dots in Figure 2g), even after correction for photobleaching is applied. The remaining decay of the signal therefore highlights the slow and steady outflow of both the NPs and the lipids from the GUVs membrane, which we see as a quasi-continuous disruption of the membrane due to the membrane-wrapping of the NPs. Consistent with this interpretation, the time courses of the signals from both NPs and membranes correlate perfectly (the purple line in Figure 2g; for details see the Supporting Information).
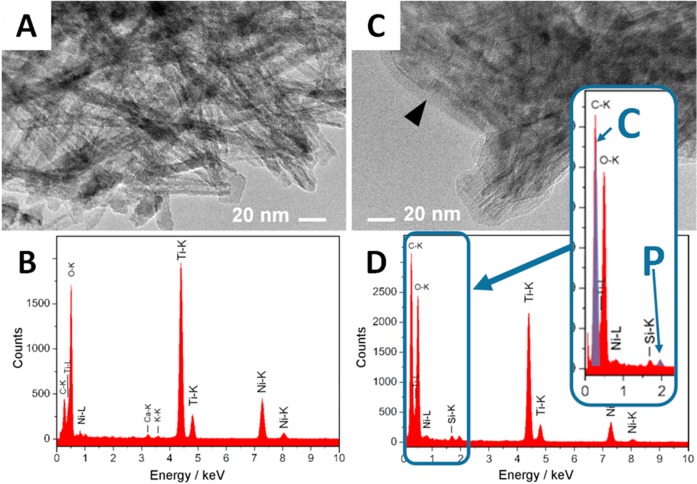
To spatially resolve the haze of the combined NP-membrane signal after disruption of GUV, we employ high-resolution, electron-dispersive transmission electron spectroscopy (EDXS TEM) on dried samples. In the absence of vesicles, the NPs have clear, crystalline-like edges (Figure 3a), and the EDS analysis reveals no elements other than those expected in the TiO2–NP or in the sample support (Figure 3b). In contrast, after exposure of the TiO2–NPs to the membrane vesicles, an amorphous layer appears around the NPs (Figure 3c), giving rise to lipid-associated carbon and phosphorus signals (Figure 3d). The lipid wrapping around the aggregated NPs (Figure 3c) is consistently observed for many TEM images, supporting our interpretation of the creation of membrane-wrapped NPs.
Figure 3.
EDXS TEM analysis of the lipid wraps around the TiO2 nanoparticles. (A,B), TEM image (A) and EDXS spectra (B) of the nanoparticles. (C,D) TEM image (C) and EDXS spectra (D) of the NPs 30 min after mixing with lipid vesicles. Arrow points to the amorphous layer of lipids around the aggregate of NPs. Inset in (D) highlights carbon (denoted as C) and phosphorus (denoted as P), associated with the lipids. Other elements originate mainly from NPs (Ti, O) and partially from support (Ni, Si, C).
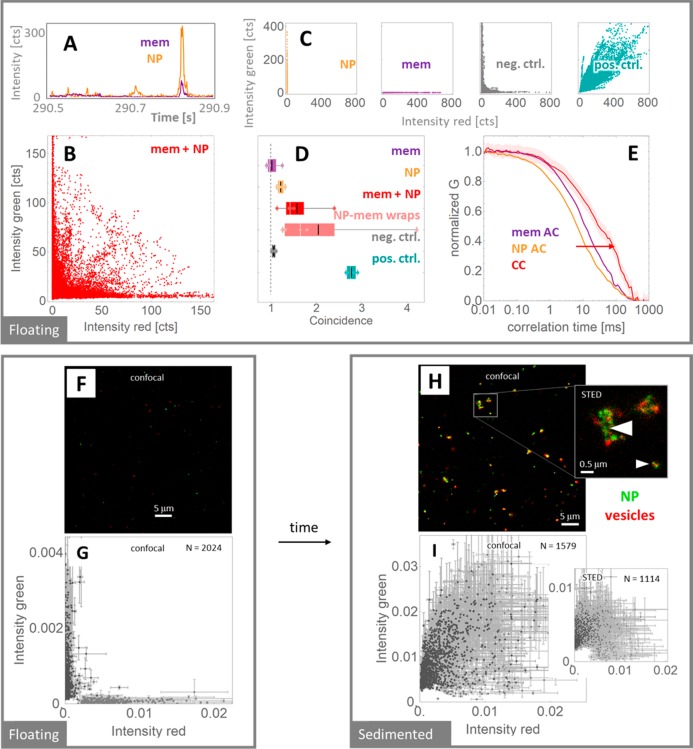
To further confirm our above observations and to exclude artifacts, that is, from the drying protocol involved in the preparation of samples for the TEM, we test the adhesion and aggregation of the NPs at membrane vesicles in dispersion, investigating the diffusion properties of the NPs and smaller vesicles (200 nm in diameter) using single-molecule intensity time traces and dual-color fluorescence (cross-)correlation spectroscopy (F(C)CS).24,25 Here, the time fluctuations of the recorded fluorescence signals are analyzed as the objects diffuse through the excitation spot of two coaligned focused laser beams, allowing a determination of simultaneous transits, that is, codiffusion and thus interaction, and absolute mobility. After incubation of the NPs labeled with Alexa 488 with vesicles labeled with a far-red fluorescent membrane dye Atto 647N DPPE for 90 min, we observed a clear signature of codiffusing in the recorded time traces (Figure 4a). The pairs of fluorescent intensities concomitantly acquired in the two detection channels (Figure 4b) reveal simultaneous transits of both NPs and vesicles, as in the positive control of a codiffusing species (vesicles stained with two membrane probes, Atto 488 DOPE and Atto 647N DPPE; Figure 4c, labeled as pos.ctrl.). In contrast, such codiffusing signatures are absent in negative controls such as NPs or vesicles alone, or a mixture of two dispersions with differently labeled vesicles (Figure 4c, labeled as NP, mem and neg.ctrl.). The coincidence analysis of the fluorescence time traces26 confirms our observations and highlights the fact that the extent of codiffusion for incubated NPs and vesicles is almost as high as in the case of a specifically prepared reference sample of fully membrane-wrapped NPs (NP-mem wraps; Figure 4d). Finally, the calculated FCS and FCCS data reveal that the average transit times of the codiffusing NPs and vesicles are considerably longer than the average transit times for NPs and vesicles alone (Figure 4e). This slowing down is another indication of the formation of mobile aggregates of NPs and membrane vesicles.
Figure 4.
Detection of mobile aggregates of nanoparticles and vesicles via fluorescence fluctuations and fluorescence intensity distribution from a two-laser excitation spot. (A) After incubation of vesicles with NPs for 90 min the time traces of the fluorescence intensities of the probes on the NPs (NP) and vesicles (mem) are recorded, featuring simultaneous spikes when membranes and NPs transit the beams at the same time. (B) Such simultaneous transits are analyzed by plotting these concomitantly detected intensity pairs occurring during 5 min. (C) Control experiments for (B) are samples of NPs (NP) and vesicles (mem) alone, mixture of noninteracting vesicles with different dyes (neg.ctrl.), and perfectly codiffusing, dual-labeled vesicles (pos.ctrl.). (D) The coincidence analysis of the time traces of all the samples at 1 min intervals are shown together with the membrane wraps sample (NP-mem wraps). (E) Associated normalized fluorescence auto- and cross-correlation curves (AC and CC, respectively) showing the codiffusion of the mixed NPs and vesicles and the formation of NP-vesicle aggregates. (F,H) The interaction between nanoparticles and vesicles is imaged with two-channel confocal imaging immediately after mixing NP and vesicles (F) and 1 h later (H). (G,I) The associated scatterplots of the signal-intensity pairs from the sites, where a significant signal can be detected in at least one channel (see Figure S11 for further explanation), reveal that the colocalization of the membrane and the NPs increase with time after mixing. Super-resolution STED images (inset in (H)) and the associated scatterplot (inset in (I)) indicate NP-mediated aggregation of vesicles (large arrow) as well as supposedly single membrane-wrapped NPs (small arrow).
To directly visualize the NPs interacting with the membranes we image the very same samples (NP labeled with Alexa 488 mixed with large unilamellar vesicles labeled with a far-red fluorescent membrane dye Atto 647N DPPE) with confocal and super-resolution STED microscopy, offering better contrast and higher spatial resolution than the wide-field FMS images of Figure 2. Immediately after mixing vesicles and NPs on a microscope coverslip, both entities are clearly separated in the two color channels (Figure 4f), confirmed by the corresponding scatterplots of the fluorescence intensity from the objects recognized in either of the two channels (Figure 4g; see also Figure S11). One hour after mixing, many more objects sediment to the cover glass, mainly micrometer-sized aggregates (Figure 4h) with a strong colocalization of the green signal of the NP and the red signal of the membrane vesicles (Figure 4i). Super-resolved STED microscopy images confirm the NP mediating the aggregation of vesicles (large arrow in the inset of Figure 4h, and increased off-diagonal fraction of data-points in the STED-associated inset of Figure 4i compared to the confocal data), as suggested by our previous experiments. However, several smaller aggregates or individual NPs are still surrounded by the membrane signal (small arrow in the inset of Figure 4h and remaining population of diagonal data-points in Figure 4i), highlighting the possibility of NP-wrapping by membranes despite the larger curvature of LUVs compared to GUVs used before. This explains also the expected27,28 increased overall disorder of the membranes (Figure S13). Images of the control samples for the negative (mixture of two dispersions of differently labeled vesicles) and positive colocalization (vesicles labeled with both probes), as expected, show hardly any changes over time, that is, between initial mixing and full sedimentation (Figure S12).
The results shown above clearly indicate that TiO2–NPs bind to lipid bilayers (for additional chemical and geometrical consideration of the binding properties see Additional Comments in Supporting Information) and further lead to freely diffusible membrane-wrapped NPs and disintegrating vesicles. The most important questions, however, remain unanswered: Can the membrane disruption and wrapping around the NPs be observed in live lung epithelial cells with their far more complex membrane composition? Can the membrane-wrapped NPs freely diffuse around or do they simply adsorb on other cellular structures? And finally, can the cells disintegrate to an extent that allows the relocation of the membrane wrapped NPs across the epithelial layer?
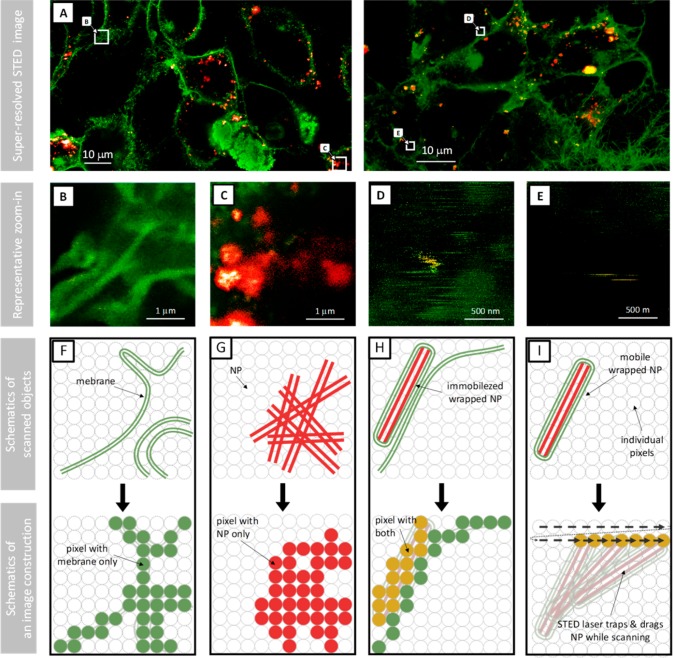
We designed an in vitro experiment on the live lung epithelial cell layer (LA-4 cells) to answer these questions. In detail, to monitor the wrapping of NPs with cell membranes, the free diffusion of the wrapped NPs, and their relocation over the cell layer we expose epithelial cells to TiO2–NPs (fluorescently tagged with the organic dye Alexa 647, NP647) for 2 days (with a cell-surface-to-NP-surface ratio of approximately 1:1) and then label the cell membranes with the membrane dye CellMask Orange and image them with super-resolution STED microscopy (Figure 5a). We observe intact cell membranes including micrometre-long microtubular structures (named microvilli) without bound NPs (Figure 5b; green) in addition to individual or (mostly internalized) aggregated NPs (Figure 5c; red). Moreover, we identify also membrane-wrapped NPs, immobilized either at the cell membrane or at other cellular structures (Figure 5d), via overlapping membrane (green) and NP (red) signals (see zoom-ins in Figure S15 with raw red- and green-channel data). Co-localization of the NP and membrane signal resolved on the scale of 30 nm confirms that the wrapping of NPs with cell membrane indeed occurs under in vitro conditions on and in the living epithelial cells. A more detailed analysis reveals also the occurrence of horizontal stripes, aligned along the line of the scanning direction of our recordings (Figure 5e). These stripes indicate mobile NPs, which are dragged along by the scanning STED laser light (optical trapping is possible due to the high STED laser powers and the high dielectric constant of NPs). After having been dragged for a while, their signal disappears within the time the STED laser comes to the same location within the following line of scan, which can be explained by diffusion of NPs within the line scan time (few milliseconds), propulsion along the optical axis by the photon flux, or complete photobleaching due to prolonged exposure to STED light while dragging. Whatever the mechanism, we argue that the single-pixel-wide stripes are clearly associated with NPs, even when they appear in the green (membrane) channel only (e.g., in Figure 5d). If the NP were labeled homogeneously and STED efficiency of the CellMask and Alexa probes were the same, the red and yellow stripes would directly correspond to bare or wrapped NPs, respectively. However, because many NPs are labeled with fewer Alexa probes and CellMask is depleted less efficiently than Alexa fluorophore (which acquires the signal of the later from smaller space), mobile wrapped NPs can be detected in almost or completely green color as well. The comparison to the nonexposed cells in control samples (Figure S14) clearly indicates that STED under the same conditions produces almost none of the single-pixel wide stripes of pure membrane objects, which allows us to conclude that single-pixel stripes can be exclusively associated with NP. This could be of particular importance because it would allow high-throughput identification of wrapping for large number of nanomaterials without the need for their labeling. Finally, note that the stripes appear on various locations of the images (Figure S15) not only close to cell membranes but also far away from them, further confirming that the (membrane-wrapped) NPs are able to diffuse around and even away from the cells.
Figure 5.
Super-resolution STED imaging of membrane-wrapped mobile NPs following exposure of a live lung epithelial cell layer (LA-4 cells). (A) Super-resolution STED composite images of 2 independent experiments of the cellular plasma membrane (green) after 2 days of incubation with the fluorescently labeled NPs (red) in a complete cell-culturing medium, revealing the accumulation of NPs on and within the membrane structures. (B,C,D,E) Zoom-ins (top) and schematic representation (middle, scanned objects; bottom, image pixel signals; yellow indicates co-occurrence of NP an membrane signal, that is, membrane-wrapped NPs) of the marked regions from (A) represent an intact cell membrane shown in green (B), bare NPs or their aggregates immobilized shown in red (C), membrane-wrapped nanoparticles immobilized on the membrane or other cellular structures shown in yellow (D), and mobile membrane-wrapped NPs, appearing as yellow stripes (indicated by arrows) (E). The gray circles in the schematic representations represent the STED laser positions.
The observed NP-induced relocation of the epithelial cell membranes opens up new important questions: Are the wraps around NP composed of the lipids only or they include the proteins as well? Can the coagulation factors be detected in the wraps as a consequence of the NP-exposure of the epithelial cell layer and the resulting lipid wrapping? How biologically relevant is the epithelial cell disruption and relocation?
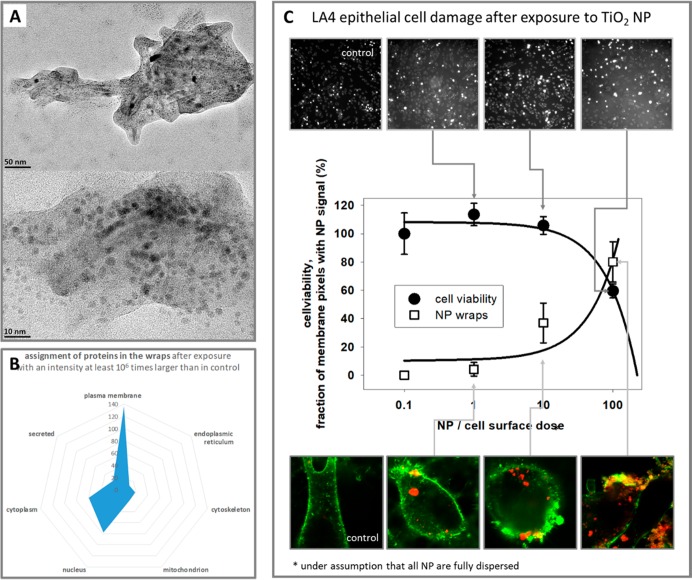
To address this issue, we analyzed the membrane wrapped NPs in the lavage taken from atop of the epithelial layer after a 2 day exposure to NPs. Part of the samples has been used to image membrane-wrapped NPs with TEM while another part has been analyzed by high throughput mass-spectrometry-based quatitative proteomics.. In the TEM images of the wrapped aggregates in the lavage (Figure 6a), NPs, lipids, and proteins can be identified as crystalline tube structures, surrounding amorphous layer, and the dark almost-round objects, respectively. Being much less likely the artifact of drying, which could have potentially affected the model sample in Figure 3, these data confirm that wrapping around NPs can be observed after exposing cell layers to NPs with proteins included. Furthermore, label free quantitative proteomics (LFQ) of the NPs wraps (corona) identified around 550 proteins with LFQ intensity more than 1,000,000 times higher than control (see also Table S2). Among 140 proteins associated with plasma membrane (using UniProt database) tissue factor (F3) with 18,000,000 intensity is clearly identified. Additionally, coagulation factor X (F10) is also identified within secreted proteins with the third largest LFQ intensity of all 550 identified proteins. Taking into account that coagulation factor X (F10) is a substrate for the tissue factor (F3) triggering both coagulation and even systemic inflammation response,29,30 our results clearly indicate that NP-induced membrane disruption and NP-wrapped epithelial cell membrane relocation can interfere with coagulation cascades. The detection of larger populations of proteins found in the lavage after exposure to NP and associated with the nucleus, mitochondrion, and cytoskeleton should be considered in the future as other potentially interfering factors.
Figure 6.
TEM and proteomics analysis of the lavage taken from the NP-exposed LA-4 cell layer with the dose dependence of the LA-4 membrane disruption. (A) TEM images of a typical aggregate of LA-4 cell membrane and NPs after 2 days incubation of LA-4 cell layer with NPs in a complete cell-culturing medium. NPs, lipids, and proteins can be identified with crystalline tube structures, surrounding amorphous layer, and the almost-round dark objects, respectively. (B) Number of proteins and their assignment (using UniProt database) with respect to cellular location as identified in proteomics analysis of the same lavage. The assignment has been done for more than 550 proteins with LFQ intensity more than 1,000,000-times higher than control (see also Table S2). (C) Dose-dependent damage of the NP-exposed LA-4 cell layer characterized via cell viability test of Hoechst to count the number of cell remaining after exposure (top images, black circles, representing median ± standard deviation of 5 measurements) as well as with the fraction of membrane used in wrapping of NP (bottom images, empty squares). The solid lines represent linear response in logarithmic plot as a low-dose part of an expected sigmoidal dose response. See Supporting Information for details.
Finally, the wrapping effect was compared to the cell survival depending on the NP dose (Figure 6c). Because of strong interference of the LDH release test and propidium iodide test with the surface of TiO2 NP (see Supporting Information for more details), the cell number could only be assessed by the Hoechst test (staining the nucleic content of the cells). Despite the simplicity of the test, the images revealed that the number of cells after exposure to NP decreases with the dose (NP-to-cell-surface ratio) (Figure 6c top) in a very similar dependence as the probability of wrapping increases with the dose (Figure 6c bottom).
Conclusions
We have provided ample evidence that TiO2–nanoparticles strongly adsorb onto the membranes and alter the molecular properties of the latter, leading to the formation of a membrane corona (wrap) around the nanoparticles, which includes important membrane proteins associated with the coagulation cascade and can move away from the original location of the cell membranes.
Since numerous epidemiological studies linked the exposure to particulate matter, especially nanoparticles, to pulmonary as well as cardiovascular disease,31 our findings are especially relevant to studies involving the inhalation of nanoparticles. Upon inhalation, the nanoparticles directly interact with various membrane structures in the lungs and could relocate them by diffusing into systemic circulation. It is important to note the thickness of the air–blood barrier, that is, the average distance between the pulmonary surfactant structure and the capillary interior, is only about half a micrometer. We are confident that the ability of the nanoparticles to relocate the cell membrane with phosphatidylserine lipids and tissue factor proteins might be the key missing link in the etiology of nanoparticle-initiated cardiovascular disease, which we demonstrate by detecting a very strongly increased abundance of coagulation factor X on the nanoparticles exposed to lung epithelial cells for 2 days. Future in vivo exposure studies should aim at confirming the presence of the two activating factors in the blood that are delivered by membrane-wrapped nanoparticles.
Furthermore, the observed strong destructive interaction between the nanoparticles and the membranes and the associated formation of lipid wraps can resolve another intriguing mystery, the observed destabilization of the lysosomal membrane. Namely, as the cells try to dispose of the intruding internalized material, the corona is enzymatically degraded in the lysosomes,32 thus exposing the native surface of the nanoparticles to the lysosomal membrane, which can degrade in the same way as discussed in this paper. Such a destabilization of the lysosomal membrane due to exposure to TiO2 nanoparticles33,34 is known to trigger cell death35,36 but has been according to our results insufficiently associated only with the electrostatic interactions between lipids and the cationic nanoparticles.32 Considering that only a few basic studies of nanoparticles interacting with lipids mention a potential role of such an affinity in nanotoxicity, let alone any systemic effects, it is not at all surprising that this notion has not yet appeared in the context of nanoparticle-induced cardiovascular disease. We are therefore convinced that the formation of mobile nanoparticles wrapped with cell membranes should be of broader interest in nanotoxicology.
Material and Methods
Synthesis of Nanoparticles
Sodium titanate nanotubes (NP) were first synthesized under hydrothermal conditions, then transformed into hydrogen titanate nanotubes by ion exchange, and finally thermally treated to transform them into TiO2 nanotubes, as described previously.37
Functionalization and Fluorescent Labeling of the NPs
The 3-(2-aminoethylamino)propyl-trimethoxysilane (AEAPMS) linker was first attached to the NPs in toluene, washed after 16 h, dispersed in ethanol, and dried. The functionalized NPs were then dispersed in a bicarbonate buffer at a pH of 8.4 and tip-sonicated. Alexa 488 SDP, or Alexa 647 NHS, ester in DMSO was added to the nanotubes, sonicated for 2 h, and left stirring overnight at room temperature. To remove the unbound fluorescent probe, the mixture was finally dialyzed 4 times in ethanol and exchanged with distilled water or bicarbonate buffer. The stabilities of the binding of the AEAPMS to the NPs, and of the Alexa to the AEAPMS, were checked by measuring the zeta-potential (Figure S2 in Supporting Information).
Preparation of Liposomes
Giant unilamellar vesicles (GUV) were prepared by the gentle hydration method38 from DOPC and DPPG in 90:10 or 50:50 mol % (GUV 10PG and GUV 50PG, respectively) to achieve different negative charge on the lipid bilayers. For the FMS experiments, the Bodipy 530/550 lipid analogue (PC-BODIPY; GUV530) was added to the lipid mixture, which in combination with Alexa 488 on the NPs also acted as the FRET acceptor.
Large unilamellar vesicles (LUVs) were prepared by the hydration of a dry lipid film of the desired lipid composition (DOPC, or a mixture of DOPC and DPPG in 90:10 mol %) with a fluorescent membrane probe (Atto 647N DPPE (LUV647), Atto 488 DOPE (LUV488), both (LUV488&647), or in-house synthesized polarity-sensitive dye SP23-B (LUVsp)) in the chosen solvent (100 times diluted bicarbonate buffer or water) while vigorous vortexing or rotating, followed by freeze–thaw cycling of the dispersion. The LUVs used for the FCS and imaging were further extruded through 200 nm pores. As another positive codiffusing control, membrane-wrapped NP (LUV640-NP488 wraps) were prepared by the reverse phase evaporation approach; after having added NP488 in a bicarbonate buffer to the mixture of lipids and lipid probes in chloroform and diisopropyl ether, the organic solvents were slowly evaporated.
Please refer to Table S1 for an overview of all the samples of vesicles and further details.
FRET FMS Experiments and References
FMS experiments were performed using a home-built setup described previously:20 Xe–Hg arc lamp (Sutter Lambda LS) with excitation filters (Semrock Brightline) on an inverted microscope (Nikon Ti-2000e), 60× water immersion objective, a tunable narrow-band filter (CRi Varispec VIS-10–20), and an electron-multiplying charge-coupled device camera (Andor iXon3 897). GUV was labeled with PC-BODIPY (final lipid concentration of 3 μM) added to a droplet of Alexa-labeled-TiO2 nanoparticle (NP488) (final concentration of 3 μg/mL). Excitation band of 430 to 490 nm was used, images were taken every 5 min for 1 h over the spectral range 515–580 nm in 5 nm steps.
FMS Analysis
Emission spectra were extracted from a λ-stack, linear unmixing was used to decompose the spectra into two spectral components corresponding to the NP488 (TiO2 nanoparticle) and PC-BODIPY (membrane) contributions (Figure S3c), to increase the accuracy of the spectral analysis at the voxels located on the edge of vesicles, averaging along the vesicular membrane was implemented (Figure S3d). Reference experiments were used to determine the line-shape parameters within the log-normal spectral model,20 except for the peak position (λMAX) of the NP488. The photobleaching was negligible for both probes within each λ-scan but significant for the PC-BODIPY on the time scale of the whole experiment.
TEM
Wrapping of TiO2 nanotubes with lipids and membranes was investigated with a transmission electron microscope (TEM, Jeol 2100, 200 keV) equipped with an energy-dispersive X-ray spectrometer (EDXS) for the chemical microanalysis. Specimens for TEM and EDXS investigation were prepared by mixing giant unilamellar vesicles (GUV530 10PG; composed of DOPC and DOPG (90:10)) with TiO2 nanotubes at room temperature and left to stir for 1 h and then a drop of the solution was deposited on a lacey carbon film supported by a nickel grid (300 mesh).
Fluorescence Fluctuation-Based Experiments
NP488 and LUV647 were incubated for 30 min prior to the experiments. Just before the measurements, the dispersion was diluted with a bicarbonate buffer to the concentrations around 0.1 and 0.01 mg/mL, respectively. Using PicoQuant MicroTime 200 equipped with a 60× water immersion objective and two avalanche photodiodes, time -correlated single-photon counting streams were acquired from a small droplet of the dispersion on a coverslip in the green and red detection channels after excitation with alternating laser pulses with wavelengths of 485 and 640 nm, respectively, for a total duration of at least 5 min. The recorded time-traces were used to generate the scatterplots, to calculate the FCS and FCCS curves, and to determine the coincidence values. For comparison, membrane-wrapped NP488 (LUV640-NP488 wraps) were also measured. As the true negative and positive references for codiffusion, a mixed dispersion of LUV647 and LUV488, or dual-labeled LUV488&647 were used, respectively. The scatterplots of the negative controls confirm that there was no significant crosstalk between the two detection channels.
Confocal and STED Imaging of LUV and NP
Experiments were performed by a Leica SP8 STED instrument equipped with a 100×/1.4 oil immersion STED objective. A dispersion of NP488 and LUV647, sealed between a microscopy slide and a coverslip, was excited by 488 and 633 nm lines from a white light laser, and their emissions were recorded with two hybrid detectors in the wavelength ranges of 495–585 and 640–730 nm, respectively. Both the confocal images were acquired simultaneously, whereas the STED images using STED lasers at 592 and 775 nm were recorded sequentially to avoid severe photobleaching of the red dye by the 592 nm STED laser. For negatively and positively colocalizing controls, the same samples as for FCS were used (a mixture of LUV488 and LUV647, and LUV488&647, respectively). The data of the negative control (Figure S12) confirmed that the crosstalk between the two channels was negligible.
Image Analysis
In every channel of each two-color image, features with intensity above a predefined threshold were recognized as NP/LUV. Within the masked area of each recognized feature, the mean fluorescence intensity in both color channels was read out. The 2D scatterplots were generated by analyzing 3–9 different images of each experiment.
STED Imaging of LA-4 Cells with NP
The LA-4 murine lung epithelial cells were seeded into a 35 mm Ibidi μ-Dish and incubated in the complete culturing medium for a day (F12K medium, 15% FCS, 1% P/S (antibiotics), 1% NEAA (nonessential amino acids)). TiO2 nanotubes, labeled with Alexa 647 (NP647), were resuspended in PBS and diluted in the cell medium to the final concentration of 10 μg/mL. After 1 day of incubation, the samples were washed and the plasma membranes were labeled with CellMask Orange. For super-resolution imaging, an Abberior Instruments STED microscope equipped with a 60× water immersion objective was used. The STED image was acquired close to the top surface of the cells at 10 nm pixel size. CellMask and NP647 were excited with the two pulsed lasers at 561 and 640 nm, respectively, overlaid with a doughnut-shaped STED beam at 775 nm, and their fluorescence recorded with two avalanche photodiodes within 580–625 and 655–720 nm (filters by Semrock), respectively.
Viability Tests
Viability of the LA4 murine lung epithelial cells after exposure to the TiO2 nanotubes was determined by counting the Hoechst reagent 33342 labeled cells in Live Cell Imaging Solution (LCIS). Positive control was done using 0.25% tryton X-100.
TEM of the Lavage
After incubation of the cells with the nanoparticles, the cell supernatant was washed off. The morphology of the plasma membrane-wrapped nanoparticle was investigated with a transmission electron microscope (TEM, Jeol 2100, 200 keV).
Proteomics on Wrapped NP
The same samples used for TEM (lavage) were centrifuged to remove any remaining cellular debris. The supernatant was then centrifuged once more with the pellet containing the so-called NP-Hard Corona complexes. This sample was washed, centrifuged three times, chemically modified (see Supporting Information for more details), resuspended in diluted TFA and stored at 4 °C until MS analysis, which was done on a Thermo Scientific Q Exactive mass spectrometer operated in positive ion mode and connected to a Dionex Ultimate 3000 (RSLCnano) chromatography system. All data was acquired while operating in automatic data dependent switching mode. A high-resolution (70,000) MS scan (300–1600 m/z) was performed to select the 12 most intense ions prior to MS/MS analysis using high-energy collision dissociation (HCD). Proteins were identified and quantified by MaxLFQ39 by searching with the MaxQuant version 1.5 against the Mus musculus reference proteome database (Uniprot). Modifications included C carbamlylation (fixed) and M oxidation (variable). Excel was employed to finally analyze the MaxQuant data, using fold change cut off values of 1,000,000. Pantherdb, String DB, and cytoscape were utilized in the analysis.
All experiments were performed at room temperature.
For further details, see the Supporting Information.
Acknowledgments
This work was supported by Slovenian Research Agency (program P1-0060), EU Horizon 2020 programme (project SmartNanoTox), ESRR (Centre of Excellence NAMASTE), and Marie Skłodowska-Curie Action (FNS-4-NAMOSAT, I.U.) the MRC (Grant MC_UU_12010/unit programs G0902418 and MC_UU_12025), the Wellcome Trust (Grant 104924/14/Z/14), MRC/BBSRC/EPSRC (Grant MR/K01577X/1), MRC/BBSRC/EPSRC (Grant MR/K01577X/1), the Wolfson Foundation (for initial funding of the Wolfson Imaging Centre Oxford), and Oxford-internal funding (John Fell Fund and EPA Cephalosporin Fund). The authors thank Anže Testen, Anja Dobravec, Katarina van Midden, and David Dolhar for their help with the preparation of the samples and measurements, Prof. Dr. Alenka Mertelj (“Jožef Stefan” Institute, Ljubljana, Slovenia) for access to their DLS instrument and help with experiments, the Wolfson Imaging Centre Oxford (Dominic Waithe and Chris Lagerholm), members of the Laboratory of Biophysics (especially Stane Pajk for providing the environment-sensitive probe) and Nano-Immunology lab (especially Erdinc Sezgin for his help with FCS) as well as the Micron Advanced Imaging Unit (Strategic Award 091911) for support.
Glossary
ABBREVIATIONS
- NPs
nanoparticles
- FMS
fluorescence microspectroscopy
- STED
stimulated emission depletion
Supporting Information Available
Supporting Information is available free of charge via the Internet at The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.8b02291.
Motivation, material, equipment, preparation of the samples, measurements of zeta-potential, fluorescence microspectrosopy (FMS), dynamic light scattering (DLS), experiments based on fluorescence fluctuations, confocal and STED fluorescence imaging of LUV and NP, fluorescence spectroscopy of NP and LUV, STED imaging of LA-4 cells and NP, lactate dehidrogenase (LDH) assay, cell viability test (hoechst and propidium iodide), determination of plasma membrane fraction used in wrapping of TiO2 nanotubes, proteomic analysis of lipid-protein corona on TiO2 nanotubes, additional comments, references (PDF)
Author Contributions
M.G., I.U., H.M., B.K., and H.C. prepared the biological samples, P.U. synthesized the nanoparticles, I.U., M.G., B.K., H.M., P.U., M.S., F.S., D.G., H.C., M.C., and S.G. performed the measurements, I.U., M.G., H.M., B.K., D.G., S.H., and F.S. analyzed the data, I.U., T.K., D.G., C.E., and J.S. codesigned the experiments, I.U., Z.A., T.K., I.M., C.E., and J.S. cowrote the manuscript, J.S. directed the project.
The authors declare no competing financial interest.
Supplementary Material
References
- OECD. OECD Environmental Outlook to 2050; Organisation for Economic Co-operation and Development: Paris, 2012. [Google Scholar]
- Brook R. D.; Franklin B.; Cascio W.; Hong Y.; Howard G.; Lipsett M.; Luepker R.; Mittleman M.; Samet J.; Smith S. C.; et al. Air Pollution and Cardiovascular Disease: A Statement for Healthcare Professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation 2004, 109 (21), 2655–2671. 10.1161/01.CIR.0000128587.30041.C8. [DOI] [PubMed] [Google Scholar]
- Choi H. S.; Ashitate Y.; Lee J. H.; Kim S. H.; Matsui A.; Insin N.; Bawendi M. G.; Semmler-Behnke M.; Frangioni J. V.; Tsuda A. Rapid Translocation of Nanoparticles from the Lung Airspaces to the Body. Nat. Biotechnol. 2010, 28 (12), 1300–1303. 10.1038/nbt.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. R.; Raftis J. B.; Langrish J. P.; McLean S. G.; Samutrtai P.; Connell S. P.; Wilson S.; Vesey A. T.; Fokkens P. H. B.; Boere A. J. F.; et al. Inhaled Nanoparticles Accumulate at Sites of Vascular Disease. ACS Nano 2017, 11 (5), 4542–4552. 10.1021/acsnano.6b08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani T.; Nakagawa S.; Kurosawa M.; Mizuashi M.; Ozawa M.; Aiba S. Cellular Basis of the Role of Diesel Exhaust Particles in Inducing Th2-Dominant Response. J. Immunol. 2005, 174 (4), 2412–2419. 10.4049/jimmunol.174.4.2412. [DOI] [PubMed] [Google Scholar]
- Donaldson K.; Stone V.; Seaton A.; MacNee W. Ambient Particle Inhalation and the Cardiovascular System: Potential Mechanisms. Environ. Health Perspect. 2001, 109, 523–527. 10.2307/3454663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA National Center for Environmental Assessment, R. T. P. N.; Sacks J. Provisional Assessment of Recent Studies on Health Effects of Particulate Matter Exposure https://cfpub.epa.gov/ncea/isa/recordisplay.cfm?deid=247132 (accessed Nov 5, 2017).
- Drake T. A.; Morrissey J. H.; Edgington T. S. Selective Cellular Expression of Tissue Factor in Human Tissues. Implications for Disorders of Hemostasis and Thrombosis. Am. J. Pathol. 1989, 134 (5), 1087–1097. [PMC free article] [PubMed] [Google Scholar]
- Hoffman M.; Monroe D. M. A Cell-Based Model of Hemostasis. Thromb. Haemostasis 2001, 85 (6), 958–965. 10.1055/s-0037-1615947. [DOI] [PubMed] [Google Scholar]
- Liu J. Interfacing Zwitterionic Liposomes with Inorganic Nanomaterials: Surface Forces, Membrane Integrity, and Applications. Langmuir 2016, 32 (18), 4393–4404. 10.1021/acs.langmuir.6b00493. [DOI] [PubMed] [Google Scholar]
- Hu Q.; Bai X.; Hu G.; Zuo Y. Y. Unveiling the Molecular Structure of Pulmonary Surfactant Corona on Nanoparticles. ACS Nano 2017, 11, 6832. 10.1021/acsnano.7b01873. [DOI] [PubMed] [Google Scholar]
- Pera H.; Nolte T. M.; Leermakers F. A. M.; Kleijn J. M. Coverage and Disruption of Phospholipid Membranes by Oxide Nanoparticles. Langmuir 2014, 30 (48), 14581–14590. 10.1021/la503413w. [DOI] [PubMed] [Google Scholar]
- Alkhammash H. I.; Li N.; Berthier R.; de Planque M. R. R. Native Silica Nanoparticles Are Powerful Membrane Disruptors. Phys. Chem. Chem. Phys. 2015, 17 (24), 15547–15560. 10.1039/C4CP05882H. [DOI] [PubMed] [Google Scholar]
- Fortunelli A.; Monti S. Simulations of Lipid Adsorption on TiO2 Surfaces in Solution. Langmuir 2008, 24 (18), 10145–10154. 10.1021/la801787s. [DOI] [PubMed] [Google Scholar]
- Jackman J. A.; Zan G. H.; Zhao Z.; Cho N.-J. Contribution of the Hydration Force to Vesicle Adhesion on Titanium Oxide. Langmuir 2014, 30 (19), 5368–5372. 10.1021/la404581d. [DOI] [PubMed] [Google Scholar]
- Schleh C.; Mühlfeld C.; Pulskamp K.; Schmiedl A.; Nassimi M.; Lauenstein H. D.; Braun A.; Krug N.; Erpenbeck V. J.; Hohlfeld J. M. The Effect of Titanium Dioxide Nanoparticles on Pulmonary Surfactant Function and Ultrastructure. Respir. Res. 2009, 10, 90. 10.1186/1465-9921-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setyawati M. I.; Tay C. Y.; Chia S. L.; Goh S. L.; Fang W.; Neo M. J.; Chong H. C.; Tan S. M.; Loo S. C. J.; Ng K. W.; et al. Titanium Dioxide Nanomaterials Cause Endothelial Cell Leakiness by Disrupting the Homophilic Interaction of VE-Cadherin. Nat. Commun. 2013, 4, 1673. 10.1038/ncomms2655. [DOI] [PubMed] [Google Scholar]
- Yamashita K.; Yoshioka Y.; Higashisaka K.; Mimura K.; Morishita Y.; Nozaki M.; Yoshida T.; Ogura T.; Nabeshi H.; Nagano K.; et al. Silica and Titanium Dioxide Nanoparticles Cause Pregnancy Complications in Mice. Nat. Nanotechnol. 2011, 6 (5), 321–328. 10.1038/nnano.2011.41. [DOI] [PubMed] [Google Scholar]
- Garvas M.; Testen A.; Umek P.; Gloter A.; Koklic T.; Strancar J. Protein Corona Prevents TiO2 Phototoxicity. PLoS One 2015, 10 (6), e0129577. 10.1371/journal.pone.0129577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbančič I.; Arsov Z.; Ljubetič A.; Biglino D.; Strancar J. Bleaching-Corrected Fluorescence Microspectroscopy with Nanometer Peak Position Resolution. Opt. Express 2013, 21 (21), 25291–25306. 10.1364/OE.21.025291. [DOI] [PubMed] [Google Scholar]
- Urbančič I.; Ljubetič A.; Arsov Z.; Štrancar J. Coexistence of Probe Conformations in Lipid Phases—a Polarized Fluorescence Microspectroscopy Study. Biophys. J. 2013, 105 (4), 919–927. 10.1016/j.bpj.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsov Z.; Urbančič I.; Štrancar J. Aggregation-Induced Emission Spectral Shift as a Measure of Local Concentration of a pH-Activatable Rhodamine-Based Smart Probe. Spectrochim. Acta, Part A 2018, 190, 486. 10.1016/j.saa.2017.09.067. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu J. A Stable Lipid/TiO2 Interface with Headgroup-Inversed Phosphocholine and a Comparison with SiO2. J. Am. Chem. Soc. 2015, 137 (36), 11736–11742. 10.1021/jacs.5b06642. [DOI] [PubMed] [Google Scholar]
- Haustein E.; Schwille P. Ultrasensitive Investigations of Biological Systems by Fluorescence Correlation Spectroscopy. Methods 2003, 29 (2), 153–166. 10.1016/S1046-2023(02)00306-7. [DOI] [PubMed] [Google Scholar]
- Bacia K.; Kim S. A.; Schwille P. Fluorescence Cross-Correlation Spectroscopy in Living Cells. Nat. Methods 2006, 3 (2), 83. 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- Winkler T.; Kettling U.; Koltermann A.; Eigen M. Confocal Fluorescence Coincidence Analysis: An Approach to Ultra High-Throughput Screening. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (4), 1375–1378. 10.1073/pnas.96.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Gu N. Thermodynamics of Charged Nanoparticle Adsorption on Charge-Neutral Membranes: A Simulation Study. J. Phys. Chem. B 2010, 114 (8), 2749–2754. 10.1021/jp904550b. [DOI] [PubMed] [Google Scholar]
- Wang B.; Zhang L.; Bae S. C.; Granick S. Nanoparticle-Induced Surface Reconstruction of Phospholipid Membranes. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (47), 18171–18175. 10.1073/pnas.0807296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf W. FXa Takes Center Stage in Vascular Inflammation. Blood 2014, 123 (11), 1630–1631. 10.1182/blood-2014-02-553081. [DOI] [PubMed] [Google Scholar]
- Cirino G.; Cicala C.; Bucci M.; Sorrentino L.; Ambrosini G.; DeDominicis G.; Altieri D. C. Factor Xa as an Interface between Coagulation and Inflammation. Molecular Mimicry of Factor Xa Association with Effector Cell Protease Receptor-1 Induces Acute Inflammation in Vivo. J. Clin. Invest. 1997, 99 (10), 2446–2451. 10.1172/JCI119428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook R. D.; Franklin B.; Cascio W.; Hong Y.; Howard G.; Lipsett M.; Luepker R.; Mittleman M.; Samet J.; Smith S. C.; et al. Air Pollution and Cardiovascular Disease: A Statement for Healthcare Professionals From the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation 2004, 109 (21), 2655–2671. 10.1161/01.CIR.0000128587.30041.C8. [DOI] [PubMed] [Google Scholar]
- Wang F.; Yu L.; Monopoli M. P.; Sandin P.; Mahon E.; Salvati A.; Dawson K. A. The Biomolecular Corona Is Retained during Nanoparticle Uptake and Protects the Cells from the Damage Induced by Cationic Nanoparticles until Degraded in the Lysosomes. Nanomedicine 2013, 9 (8), 1159–1168. 10.1016/j.nano.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Jimeno-Romero A.; Oron M.; Cajaraville M. P.; Soto M.; Marigómez I. Nanoparticle Size and Combined Toxicity of TiO2 and DSLS (Surfactant) Contribute to Lysosomal Responses in Digestive Cells of Mussels Exposed to TiO2 Nanoparticles. Nanotoxicology 2016, 10 (8), 1168–1176. 10.1080/17435390.2016.1196250. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Eaton J. W.; Li C. Titanium Dioxide (TiO2) Nanoparticles Preferentially Induce Cell Death in Transformed Cells in a Bak/Bax-Independent Fashion. PLoS One 2012, 7 (11), e50607. 10.1371/journal.pone.0050607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguissola S.; Garry D.; Salvati A.; O’Brien P. J.; Dawson K. A. High Content Analysis Provides Mechanistic Insights on the Pathways of Toxicity Induced by Amine-Modified Polystyrene Nanoparticles. PLoS One 2014, 9 (9), e108025. 10.1371/journal.pone.0108025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S. T.; Adiseshaiah P. P.; Crist R. M. Autophagy and Lysosomal Dysfunction as Emerging Mechanisms of Nanomaterial Toxicity. Part. Fibre Toxicol. 2012, 9, 20. 10.1186/1743-8977-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek P.; Korošec R. C.; Jančar B.; Dominko R.; Arčon D. The Influence of the Reaction Temperature on the Morphology of Sodium Titanate 1D Nanostructures and Their Thermal Stability. J. Nanosci. Nanotechnol. 2007, 7 (10), 3502–3508. 10.1166/jnn.2007.838. [DOI] [PubMed] [Google Scholar]
- Akashi K.; Miyata H.; Itoh H.; Kinosita K. Preparation of Giant Liposomes in Physiological Conditions and Their Characterization under an Optical Microscope. Biophys. J. 1996, 71 (6), 3242–3250. 10.1016/S0006-3495(96)79517-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Hein M. Y.; Luber C. A.; Paron I.; Nagaraj N.; Mann M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 2014, 13 (9), 2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.