Summary
Similar to other genera and species of bacteria, whole genomic sequencing has revolutionized how we think about and address questions of basis Vibrio biology. In this review we examined 36 completely sequenced and annotated members of the Vibrionaceae family, encompassing 12 different species of the genera Vibrio, Aliivibrio and Photobacterium. We reconstructed the phylogenetic relationships among representatives of this group of bacteria using three housekeeping genes and 16S rRNA sequences. With an evolutionary framework in place, we first describe the occurrence and distribution of primary and alternative sigma factors; global regulators present in all bacteria. Among Vibrio we show that the number and function of many of these sigmas differs from species to species. We also describe the role of the Vibrio specific regulator ToxRS in Vibrio fitness and survival. Examination of the biochemical capabilities was and still is the foundation of classifying and identifying new species of Vibrio. Using comparative genomics, we examine the distribution of carbon utilization patterns among Vibrio as a possible marker for understanding bacteria-host interactions. Finally, we discuss the significant role horizontal gene transfer has played in Vibrio evolution, specifically, the distribution and structure of integrons.
Within the family Vibrionaceae 12 genera have thus far been proposed; Aliivibrio, Allomonas, Beneckea, Catenococcus, Echinimonas, Enterovibrio, Grimontia, Listonella, Lucibacterium, Photobacterium, Salinivibrio, and Vibrio (1). Nearly 140 species have been described and over 110 species names with standing in nomenclature are found within the genus Vibrio (2–4). Members of the genus Vibrio are abundant in various marine habitats worldwide and are associated with a wide range of living organisms from corals, sea grass and sponges to zooplankton, shellfish and fish as well as humans. Several species are pathogenic to aquatic life and humans, commonly in the form of vibriosis and gastrointestinal illnesses, respectively. One of the most significant human infections is the diarrheal disease cholera. In 2011 the World Health Organization (WHO) reported that global cholera incidents were on the rise causing an estimated 3-5 million cases and 100,000-200,000 deaths per year. Many Vibrio species are commensals or symbionts, while many are also halophiles and can bioluminescence and most are motile. The ecological diversity of Vibrionaceae is propelled in part by phenotypic innovations driven by both adaptive changes and horizontal gene transfer (HGT). The accessibility and rapid acquisition of whole genome sequences now allows us to investigate the evolutionary events that contribute to Vibrio diversity, versatility and abundance.
As of May 2013 the National Center for Biotechnology Information (NCBI) genome data base contained 36 completely sequenced and annotated members of the Vibrionaceae family, encompassing 12 different species of the Vibrio, Aliivibrio and Photobacterium genera. All species examined to date have two chromosomes, which is now an accepted characteristic of this family (5). There are currently nearly 700 genome sequences in progress of members of the Vibrionaceae family. General characteristics of the completely sequenced species such as genome size, genomic composition, isolation source and phenotypic features can be found in Table 1. Here, we examine and describe the phylogenetic relationships among representatives from the completely sequenced genomes as well as additional species whose genomes are in progress using three housekeeping genes and 16S rRNA sequences. We then describe some of the traits that are important in the adaptation of different species to different niches and discuss these traits in terms of their distribution among species, genera and clades.
Table 1.
Genome features of sequenced Vibrio species
| Species | Size | GC | Plasmid | # of Genes | tRNA genes | rRNA genes | Int | Tnp | Isolation source | Location |
|---|---|---|---|---|---|---|---|---|---|---|
| V. harveyi BAA-1116 | 6.1 | 45.4% | 1 | 6252 | 121 | 24 | 23 | 52 | Marine ocean | unknown |
| Vibrio sp. EJY3 | 5.5 | 44.9% | none | 4935 | 121 | 28 | 20 | 22 | West Sea coast | Korea |
| Vibrio sp. Ex25 | 5.1 | 44.9% | none | 4676 | 124 | 34 | 10 | 0 | hydrothermal vent | East Pacific Rise |
| V. parahaemolyticus RIMD2210633 | 5.1 | 45.4% | none | 5032 | 126 | 34 | 6 | 8 | Homo sapiens | Thailand |
| V. splendidus LGP32 | 5.0 | 43.9% | none | 4572 | 114 | 26 | 7 | 27 | Hollow oyster | France |
| V. vulnificus CMCP6 | 5.1 | 46.7% | none | 4665 | 111 | 28 | 7 | 44 | Homo sapiens | South Korea |
| V. vulnificus MO6-24/O | 5.0 | 46.9% | none | 4701 | 111 | 28 | 6 | 19 | Homo sapiens | US |
| V. vulnificus YJ016 | 5.3 | 46.7% | 1 | 5202 | 112 | 28 | 13 | 48 | Homo sapiens | Taiwan |
| V. anguillarum 775 | 4.1 | 44.5% | none | 3836 | 84 | 21 | 4 | 80 | Coho salmon | Pacific Ocean |
| V. furnissii NCTC 11218 | 4.9 | 50.7% | none | 4584 | 100 | 22 | 13 | 7 | Homo sapiens | UK |
| V. cholerae IEC224 | 4.1 | 47.5% | none | 3787 | 98 | 25 | 11 | 28 | Homo sapiens | Brazil |
| V. cholerae LMA3894-4 | 3.7 | 48.0% | none | 3270 | 92 | 25 | 4 | 6 | Tucunduba creek | Brazil |
| V. cholerae M66-2 | 3.9 | 47.6% | none | 3812 | 97 | 22 | 5 | 20 | Homo sapiens | Indonesia |
| V. cholerae MJ-1236 | 4.2 | 47.32% | none | 3894 | 98 | 22 | 13 | 19 | Homo sapiens | Bangladesh |
| V. cholerae N16961 | 4.0 | 47.5% | none | 3998 | 98 | 25 | 6 | 21 | Homo sapiens | Bangladesh |
| V. cholerae O395 | 4.1 | 47.5% | none | 4055 | 96 | 25 | 12 | 28 | Homo sapiens | India |
| V. cholerae 2010EL-1786 | 4.1 | 47.5% | none | 3946 | 79 | 22 | 10 | 26 | Homo sapiens | Haiti |
| V. salmonicida LFI1238 | 4.7 | 39.0% | 4 | 4075 | 105 | 37 | 11 | 438 | Atlantic cod | Norway |
| V. fischeri ES114 | 4.3 | 38.4% | 1 | 3984 | 119 | 37 | 5 | 1 | Squid light organ | Hawaii |
| V. fischeri MJ11 | 4.5 | 38.2% | 1 | 4175 | 124 | 12 | 4 | 1 | Squid light organ | Japan |
| P. profundum SS9 | 6.4 | 41.7% | 1 | 5746 | 169 | 48 | 9 | 212 | 2500 meters | Sulu Trough |
Phylogenetic analysis
A total of 60 strains encompassing 37 different species were used to construct a concatenated phylogenetic tree of three housekeeping (HK) genes and a tree based on 16S rRNA (Fig. 1 and Fig. 2). Genes from chromosomes I and II were used in the generation of the concatenated tree and include RNA polymerase subunit beta (rpoB), malate dehydrogenase (mdh), both found on chromosome I, and dihydrorotase (pyrC) found on chromosome II. All trees were generated using the Neighbor-Joining method in MEGA 5.0 (6, 7). The Jukes-Cantor method was used to compute evolutionary distances (8). The bootstrap test was repeated 1000 times and values over 50% are shown next to branches (9). The concatenated tree grouped the 60 examined strains into 11 distinct clades similar to groupings of Vibrionaceae previously described (Fig. 1). The 16S rRNA tree was in general congruent with the concatenated housekeeping (HK) tree; however, it displayed shorter branch lengths, indicating that this gene is more highly conserved. In the 16S rRNA tree, minor differences were noted within the Campbellii and Parahaemolyticus clades. Vibrio sp. AND4 and Vibrio sp. EJY3 do not group within these clades as they do in the concatenated HK tree. They did however, branch off of a shared node, although distinctly (Fig. 2). Additionally, in the Orientalis clade, V. caribbenthicus and V. nigripulchritudo is grouped together in this clade but not in the HK gene tree (Fig. 1 and Fig. 2).
Fig. 1.
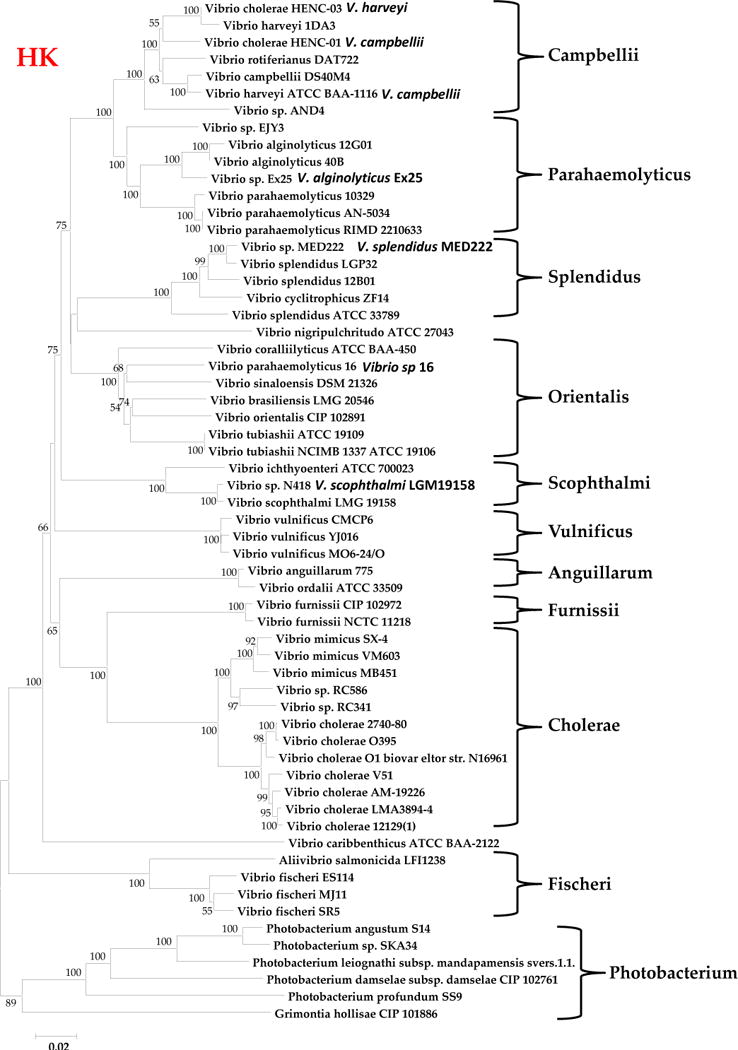
Evolutionary relationships of Vibrio species based the concatenated housekeeping genes rpoB, mdh, and pyrC. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 2.24860938 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. The analysis involved 60 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair. There were a total of 6012 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (120).
Fig. 2.
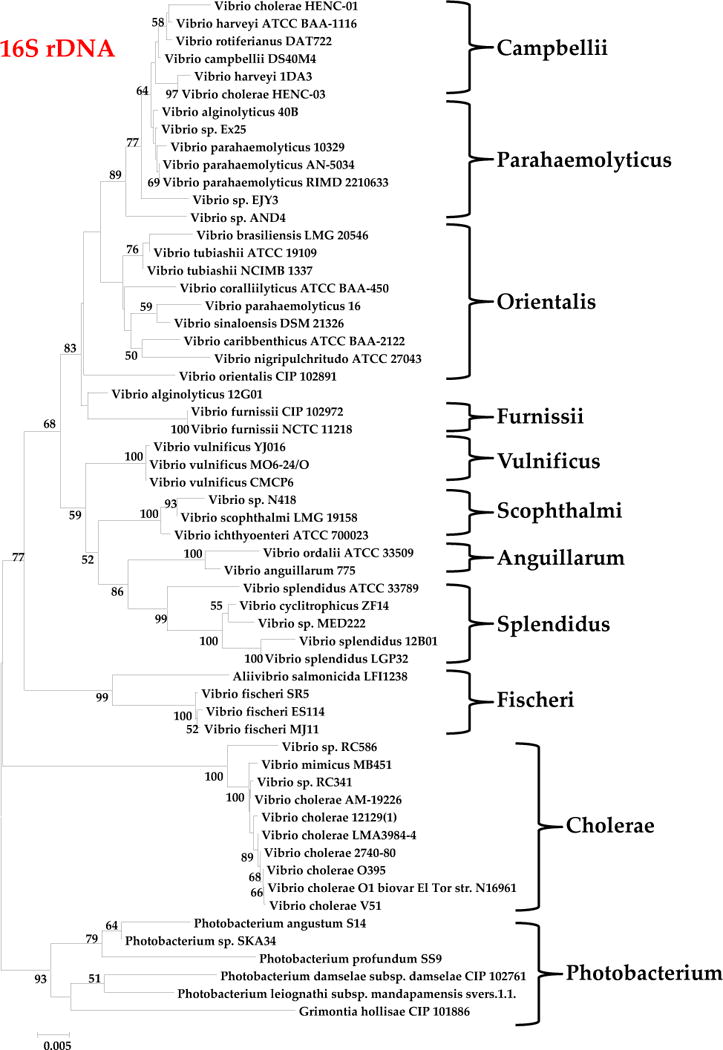
Evolutionary relationships of taxa based on 16S rDNA sequence. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 0.47043464 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (30). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site (52). The analysis involved 58 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1564 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (120).
The Campbellii clade encompassed four species, V. campbellii, V. harveyi, V. rotiferianus, and Vibrio sp AND4 (Fig. 1). Within this clade were two strains named V. cholerae HENC-03 and V. cholerae HENC-01, which are sequenced strains from Haiti (Fig. 1). Our analysis of additional HK loci from these strains and whole genome sequence analysis between HENC-03, HENC-01 and V. harveyi indicated that these strains are bona fide members of the Campbellii clade and should be renamed. Recently, it was shown based on comparative genome hybridization and multilocus locus sequence analyses, that both V. haveryi strains ATCC BAA-116 and HY01 were V. campbellii isolates (10). In our sequence analysis, V. haveryi ATCC BAA-116 clustered closely with V. campbellii DS40M4. Based on HK and 16S rRNA data, HENC-03 appears to be a V. harveyi isolate and HENC-01 a V. campbellii isolate. For both the HK and 16S rRNA trees, the Parahaemolyticus clade branches with the Campbellii clade (Fig. 1 and Fig. 2). This clade contained four species, V. parahaemolyticus (3 strains), V. alginolyticus, which branched with Vibrio sp Ex25, and Vibrio sp EJY3 which clustered within this group but was distinct from the other representatives (Fig. 1).
The Splendidus and Orientalis clades branched together on the HK tree. Three different species were found within the Splendidus clade including V. splendidus, represented by 3 strains, with strain ATCC33789 divergent from the other two strains LPG32 and 12B01 examined. Vibrio sp. MED222 clustered closely with V. splendidus LGP32 suggesting that it is likely a V. splendidus isolate. V. cyclitrophicus also branched within this clade (Fig. 1). The Orientalis clade contained six highly divergent species that were not closely related to one another: V. coralliilyticus, V. parahaemolyticus strain 16, V. sinaloensis, V. brasiliensis, V. orientalis, and V. tubiashii. These species were also present in the same clade in the 16S rRNA tree. The Orientalis clade represents at least six different groupings. V. parahaemolyticus strain 16 is misidentified and should be renamed Vibrio sp. 16 as it is unrelated to V. parahaemolyticus on both the HK and 16S rRNA trees.
The Scophthalmi clade contains three species based on both HK and 16S rRNA trees, V. ichthyoenteri which is distantly related to V. scophthalmi and Vibrio sp N418, which are highly related to one another and are likely the same species (Fig. 1). The Anguillarum/Ordalii group contained V. anguillarum and V. ordalii that were both closely related to one another and also may represent a single species. In the 1985 paper by MacDonell and Colwell, based on 5S rRNA sequence V. anguillarum was placed in a new genus Listonella along with V. damselae and V. pelagia. Since then L. damseale is now placed in the genus Photobacterium based on 16S rRNA gene sequence. We retain the name V. anguillarum in agreement with Thompson and colleagues since this species is nested within Vibrio clades and is highly related to V. ordalii (2–4, 11). It was also demonstrated that L. pelagia is highly related to V. splendidus and clearly belongs in the genus Vibrio (2–4, 11). The Vulnificus clade consists of three V. vulnificus strains that are all highly related to one another suggesting that there is limited genetic diversity within this species. This clade does not cluster closely with any other clade examined in the HK gene tree. The Cholerae clade consisted of four species: V. cholerae and V. mimicus, which are distinct species, and Vibrio sp RC341 and Vibrio sp RC586, which clustered more closely with V. mimicus than V. cholerae. The Fischerii and Photobacterium clades formed divergent groups from the other Vibrio species clades in both HK and 16S rRNA trees (Fig. 1 and Fig. 2).
Overall, among the housekeeping genes examined from these strains, rpoB has the highest amount of conserved sites and pyrC (chromosome II) has the least (Table 2), congruent with the high variability among genes found on chromosome II. As expected, the 16S rRNA gene has the highest overall percentage of conserved sites (83.8%). The most variability was found in the Photobacterium clade for all 4 genes examined demonstrating that Photobacterium species are highly divergent from one another and may represent more than one genus (Table 2). The next most highly variable clade was Orientalis, which showed a high number of polymorphic sites among all four genes examined (Table 2). The most highly conserved clades include Anguillarum, Cholerae, Furnissii and Vulnificus. When all clades are examined, the majority of the variability is represented by parsimony-informative sites. When each clade is examined separately, the majority of the variability is due to singletons particularly in the pyrC gene in the following clades: Photobacterium, Orientalis, Fischerii, Scopthalmi and Splendidus (Table 2).
Table 2.
Total and clade-specific variable site parsimony-informative sites and singleton percentages in the HK genes.
| Gene mdh | Variable sites % | Parsimonious sites % | Singletons % | |||
|---|---|---|---|---|---|---|
| TOTAL | 480 | 51.1 | 440 | 46.8 | 40 | 4.3 |
| Cholerae (C) | 26 | 2.8 | 8 | 0.9 | 18 | 1.9 |
| Mimicus (M) | 125 | 13.3 | 30 | 3.2 | 95 | 10.1 |
| C+M | 163 | 17.3 | 108 | 11.5 | 55 | 5.9 |
| Furnissii | 9 | 1.0 | 0 | 0.0 | 0 | 0.0 |
| Photobacterium | 290 | 30.9 | 110 | 11.7 | 180 | 19.1 |
| Fischeri | 178 | 18.9 | 37 | 3.9 | 141 | 15.0 |
| Campbellii | 187 | 19.9 | 122 | 13.0 | 65 | 6.9 |
| Parahaemolyticus | 197 | 21.0 | 136 | 14.5 | 61 | 6.5 |
| Orientalis | 264 | 28.1 | 170 | 18.1 | 92 | 9.8 |
| Splendidus | 122 | 13.0 | 36 | 3.8 | 86 | 9.1 |
| Scopthalmi | 120 | 12.8 | 0 | 0.0 | 120 | 12.8 |
| Vulnificus | 16 | 1.7 | 0 | 0.0 | 16 | 1.7 |
| Anguillarum | 13 | 1.4 | 0 | 0.0 | 0 | 0.0 |
| rpoB | ||||||
| TOTAL | 1595 | 39.5 | 1429 | 35.4 | 166 | 4.1 |
| Cholerae | 43 | 1.1 | 16 | 0.4 | 27 | 0.7 |
| Mimicus | 146 | 3.6 | 80 | 2.0 | 66 | 1.6 |
| C+M | 248 | 6.1 | 185 | 4.6 | 63 | 1.6 |
| Furnissii | 12 | 0.3 | 0 | 0.0 | 0 | 0.0 |
| Photobacterium | 748 | 18.5 | 221 | 5.5 | 527 | 13.1 |
| Fischeri | 290 | 7.2 | 26 | 0.6 | 264 | 6.5 |
| Campbellii | 296 | 7.3 | 79 | 2.0 | 217 | 5.4 |
| Parahaemolyticus | 229 | 5.7 | 113 | 2.8 | 116 | 2.9 |
| Orientalis | 348 | 8.6 | 163 | 4.0 | 185 | 4.6 |
| Splendidus | 225 | 5.6 | 48 | 1.2 | 177 | 4.4 |
| Scopthalmi | 43 | 1.1 | 0 | 0.0 | 43 | 1.1 |
| Vulnificus | 27 | 0.7 | 0 | 0.0 | 27 | 0.7 |
| Anguillarum | 35 | 0.9 | 0 | 0.0 | 0 | 0.0 |
| pyrC | ||||||
| TOTAL | 585 | 56.1 | 547 | 52.5 | 38 | 3.6 |
| Cholerae | 116 | 11.1 | 72 | 6.9 | 44 | 4.2 |
| Mimicus | 164 | 15.7 | 138 | 13.2 | 26 | 2.5 |
| C+M | 108 | 10.4 | 39 | 3.7 | 69 | 6.6 |
| Furnissii | 23 | 2.2 | 0 | 0.0 | 0 | 0.0 |
| Photobacterium | 388 | 37.2 | 158 | 15.2 | 229 | 22.0 |
| Fischeri | 203 | 19.5 | 15 | 1.4 | 188 | 18.0 |
| Campbellii | 180 | 17.3 | 84 | 8.1 | 96 | 9.2 |
| Parahaemolyticus | 229 | 22.0 | 135 | 13.0 | 94 | 9.0 |
| Orientalis | 340 | 32.6 | 222 | 21.3 | 118 | 11.3 |
| Splendidus | 183 | 17.6 | 62 | 6.0 | 121 | 11.6 |
| Scopthalmi | 181 | 17.4 | 0 | 0.0 | 181 | 17.4 |
| Vulnificus | 37 | 3.6 | 0 | 0.0 | 37 | 3.6 |
| Anguillarum | 16 | 1.5 | 0 | 0.0 | 0 | 0.0 |
In the following sections, we will use the concatenated HK tree to describe and discuss the distribution of different factors and traits within Vibrio species that has allowed them to occupy diverse niches. First, we describe the distribution of sigma factors and their role in global gene regulation in response to different environmental and host signals. Then, we discuss the Vibrio specific two component regulator ToxRS and its role in gene regulation among different species. The presence of Vibrio in diverse niches and their interactions with eukaryotic hosts is driven by the need to exploit new food sources. Thus, we will describe a subset of nutritional traits that specific species have adopted that enable and enhance host-vibrio interactions. Lastly, we will describe the role of horizontal gene transfer in the evolution of Vibrio with specific attention to superintegron distribution and structure.
Distribution and Functionality of Sigma Factors throughout the Vibrionaceae Family
Sigma factors are essential dissociable subunits of the bacterial RNA polymerase (RNAP). In bacteria, RNAP is not capable of initiating transcription or recognizing specific promoters without a sigma factor forming a holoenzyme. The sigma factor subunit has domains, which enables it to recognize and direct the RNAP to specific promoters. Once the holoenzyme is bound to the promoter, transcription initiation can occur.
There are two major families of sigma factors that will be discussed in this section; the sigma-70 family and the sigma-54 family. The sigma-70 family encompasses the largest group of sigma factors, and includes a primary sigma-70, responsible for transcription of housekeeping genes, and several alternative sigma factors, which regulate transcription in response to various stimuli (12). The sigma-54 family encompasses one sigma factor designated RpoN in Gram-negative bacteria. RpoN is functionally similar but structurally divergent from the sigma-70 family.
Alternative sigma factors enable the cell to globally alter transcription in response to various stimuli, including stress conditions that may occur within the bacterium’s natural environment. The repertoire of alternative sigma factors available varies widely between different species of Vibrio, including closely related species, and may reflect the lifestyle of a particular species (Table 3). For example, some Vibrio species possess as few as 8 sigma factors while others possess as many as 12 sigmas. All Vibrio species contain a single copy of the housekeeping sigma-70 factor, known as RpoD, and a single copy of RpoH, which regulates the cytosolic heat shock response. A phylogeny of Vibrio species based on these proteins has an identical topology to that for the HK tree demonstrating that these are highly conserved protein and are ancestral to the group (data not shown). All Vibrio species analyzed also contained a copy of the Escherichia coli RpoS homologue, which in this species is known as the stress response sigma factor. We identified a number of Vibrio species that also encoded an additional divergent copy of RpoS, two of which RpoX and RpoQ are described in the literature (Fig. 3) (13, 14). FliA, which regulates polar flagellum synthesis or FliAP in Vibrio, is present in all Vibrio species. A number of Vibrio species also possess a second flagella sigma factor known as FliAL or LafS, which regulates the synthesis of the lateral flagella in those organisms (Table 3 and Fig. 3). The polar flagellar system is required for swimming motility in a liquid media whereas the lateral system is required for swarming motility in viscous or solid media. The two flagellar systems have been studied extensively in V. parahaemolyticus and were shown to be distinct systems. Knockout deletions of the polar flagellum system do not affect the lateral flagella system and vice a versa (15–18).
Table 3.
Number of primary and alternative sigma factors present among Vibrio species
| Species | Total No. Sigmas | Primary | Alternative | Flagellar | ECF-type | Sigma-54 |
|---|---|---|---|---|---|---|
| V. alginolyticus | 12 | 1 | 3 | 2 | 5 | 1 |
| V. parahaemolyticus | 11 | 1 | 2 | 2 | 5 | 1 |
| V. harveyi | 12 | 1 | 3 | 2 | 5 | 1 |
| V. vulnificus | 9 | 1 | 2 | 1 | 4 | 1 |
| V. anguillarum | 9 | 1 | 2 | 1 | 4 | 1 |
| V. cholerae | 8 | 1 | 2 | 1 | 3 | 1 |
| V. furnissii | 10 | 1 | 2 | 2 | 4 | 1 |
| V. splendidus | 10 | 1 | 3 | 1 | 4 | 1 |
| V. salmonicida | 8 | 1 | 3 | 1 | 2 | 1 |
| V. fischeri | 10 | 1 | 3 | 1 | 4 | 1 |
Fig. 3.
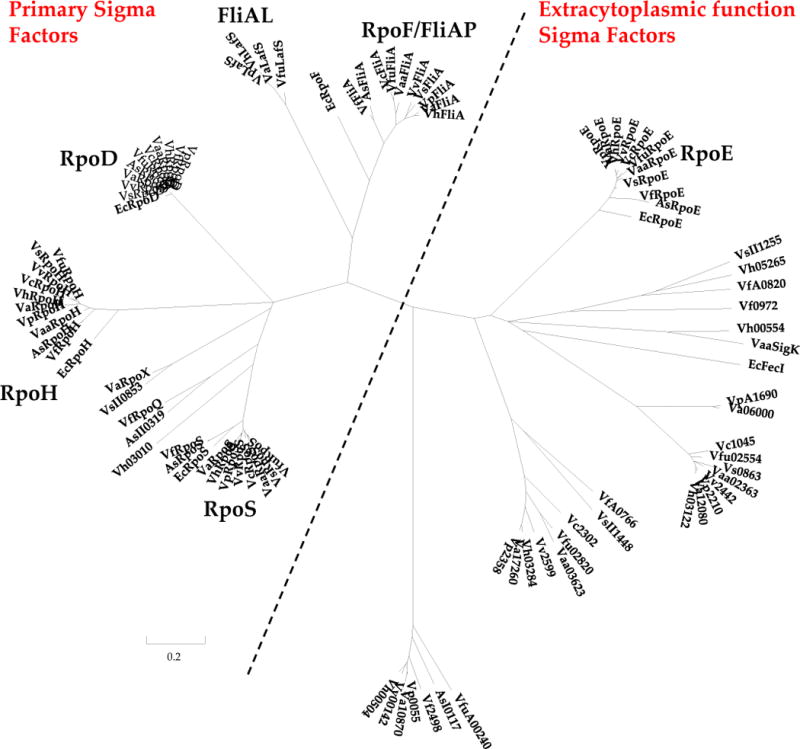
Phylogenetic tree constructed from the alignment of sigma-70 family sigma factors using the amino acid sequences of the highly conserved domains 2 and 4. The program MEGA was used to construct a neighbor-joining tree using the Poisson model, complete deletion and a bootstrap value of 1000 (120). Abbreviations are: Ec, E.coli; Vp, V. parahaemolyticus; Vc, V. cholerae; Vv; V. vulnificus; Vh, V. harveyi; Vf, V. fischeri; Va, V. alginolyticus; Vs, V. splendidus; Vaa, Vibrio anguilarium; Vfu, Vibrio furnissii. Phylogenetic analysis shows that there are two main subfamilies of sigma factors, primary and extracytoplasmic function (ECF)- type. Phylogenetic analysis also shows that predicted RpoD, RpoF, RpoS, RpoH and RpoE sigma factors encoded by each Vibrio species analyzed do cluster with and are homologs to those found in E. coli. The analysis also shows additional primary-like alternative sigma factors found in five of the species that are closely related to RpoS. The analysis also demonstrates that there are multiple putative ECF factors found within the Vibrio species analyzed, forming distinct clades.
The extra-cytoplasmic function (ECF) family of sigma factors is highly divergent from the primary and primary alternative sigma factors just described. This can be seen visually in Figure 3, with the ECF sigma factors forming distinct divergent clades. The number and type of ECFs varies even among closely related species with some species possessing 3 ECF factors and others possessing as many as 5. ECF factors typically respond to cell wall/cell envelope stress, iron levels and the oxidation state of the cell (19). The most widely studied ECF factor is RpoE from E. coli. All of the Vibrio species analyzed have a copy of the E. coli RpoE homologue. Many ECF factors have an associated anti-sigma factor, which generally suggests that tight regulation of the ECF factor is required.
The sigma-54 family of sigma factors, which is represented by just RpoN, is less divergent than the sigma-70 family of sigma factors and all of the Vibrio species analyzed contain a single copy of the E. coli RpoN homologue. RpoN was initially discovered for its role in nitrogen metabolism, however it has been found to have a much more complex role and is important in a large number of cellular processes. RpoN is a global regulator, which requires an activator protein known as bacterial enhancing binding proteins (bEBPs) (20). In V. parahaemolyticus RpoN is also the master regulator of both polar and lateral flagellar synthesis (18).
Phylogeny and Function of the Alternative Sigma Factor RpoS
The role of RpoS has been characterized in a number of Vibrio species where it has been found to function in response to starvation stress conditions. In V. cholerae, it was shown that RpoS played a role in in hydrogen peroxide, hyperosmolarity, and carbon starvation stress response as well as in the production of HA/protease that processes the cholera toxin (21). It was also suggested that RpoS was not required for mouse intestinal survival (21). Another study examining in vivo colonization of V. cholerae, showed that RpoS was required for colonization in a suckling CD1 mouse model, as well as playing a role in enhanced resistance to hydrogen peroxide but not acid stress (22). It is important to note that these two in vivo studies utilized different strains as well as different growth conditions which may account for the differences in the role of RpoS in colonization. More recently it was shown that RpoS is important for the mucosa escape response, as it was demonstrated that expression of motility and chemotaxis genes was up regulated by RpoS during this response in vivo (23). In the rpoS deletion mutant, levels of cholera toxin were 10-100-fold higher. Taken together these data suggested that RpoS represses virulence and stimulates motility to facilitate transmission (23).
In V. vulnificus, it has been shown that RpoS protects cells from acid stress, oxidative stress and nutrient starvation and also that RpoS positively regulates extracellular enzymes such as albuminase, caseinase and elastase, which may be required for survival under certain environmental conditions as well as for host adaptations (24, 25). In V. anguillarum, it has been shown that RpoS positively regulated expression of metalloproteases and an rpoS deletion mutant showed reduced virulence in zebra fish (26).
In V. parahaemolyticus, it was demonstrated that RpoS had a limited involvement in acid stress and did not play a role in osmotic stress (27). However, RpoS plays a role in the viable nonculturable state for both V. vulnificus and V. parahaemolyticus as it was found to be constitutively expressed even at below 15°C (28, 29). The effect on colonization of an rpoS deletion mutation in V. parahaemolyticus was examined in both a mouse model of colonization and in oyster colonization and in both animal models RpoS was not required (30–32). Analysis of an rpoS deletion mutant in V. harveyi, found that this mutant was more sensitive to stationary phase stress and high concentrations of ethanol in comparison to the wild-type, but was unaffected by high osmolarity or hydrogen peroxide stresses (10). In V. alginolyticus an rpoS mutant was more sensitive than wild-type to ethanol, hyperosmolarity, and hydrogen peroxide (33).
Taken together, these studies demonstrated that RpoS can play a role in starvation, osmolarity, ethanol, hydrogen peroxide and acid stress responses depending on the species. These data suggest that RpoS has evolved different roles among different Vibrio species and the specific role of RpoS in each potentially has evolved to allow changes in the lifestyles of the species. Consistent with this, are the several studies that have shown variation in the RpoS regulon amongst these species (21–23, 34).
As shown in the phylogenetic analysis in figure 3 and table 3, a number of the Vibrio species possess additional RpoS-like sigma factors. This divergence from the canonical RpoS suggests that it may also have diverged in function. In our analysis of the available Vibrio sequenced genomes, we identified five RpoS-like copies that were divergent from each other and RpoS (Fig. 3). RpoX was originally identified in V. alginolyticus and was shown to be involved in biofilm formation and stress response (13). We found that the distribution of RpoX is confined to a handful of species; all V. alginolyticus and V. splendidus (Fig.3). We identified a homologue of RpoX in Vibrio sp MED222, Vibrio sp HENC-01, HENC-02, and HENC-03, which are not shown in figure 3. RpoQ was characterized in V. fischeri, and has 45% amino acid identity to the RpoS protein in this species (14). Overexpression of RpoQ in V. fischeri led to increased chitinase activity but decreased motility and luminescence (14). It was shown that RpoQ was controlled by LuxO via LitR, a LuxR homologue (14). We found that RpoQ is confined to V. fischeri, V. salmonicida and V. shilonii isolates. We identified a third divergent RpoS-like sequence that was present in V. harveyi 1DA3 (locus tag Vh03010) and has not been characterized to date. We identified closely related homologues of Vh03010 in V. alginolyticus, V. splendidus, Vibrio sp MED222, Vibrio sp HENC-01, HENC-02, and HENC-03, V. cyclitropicus, and V. shilonii isolates. As stated earlier all Vibrio species contain a copy of the canonical RpoS. We found three species that had three RpoS-like proteins, V. shilonii, P. damsela, Photobacterium sp SKA3 and one species that had 4 RpoS-like proteins, V. angustum. These RpoS-like sigma factors have diverged significantly from RpoS suggesting that they may have evolved functions distinct from that of RpoS.
Distribution and function of RpoH, FliAP and FliAL
Although all Vibrio species contain a single E. coli RpoH homologue, this sigma factor has remained relatively uncharacterized across the Vibrio species. The V. cholerae RpoH can functionally complement an E. coli rpoH mutant suggesting it has evolved a similar role in both species (35). A global gene expression and phenotypic analysis of a V. cholerae rpoH deletion mutant by Slamti and colleagues suggested that in this species, rpoH promotes growth at temperatures ranging from 15-42°C (36). The rpoH deletion mutant could only be made in the presence of a plasmid-borne inducible copy of rpoH suggesting this is an essential gene. Not surprisingly, the rpoH mutant was severely attenuated in a suckling mouse model of colonization (36).
The flagellar sigma factors FliAP and FliAL have been best characterized in V. parahaemolyticus (15–17). A study in 1993 by McCarter and co-workers identified the genes involved in the swarming flagellar motor of V. parahaemolyticus using transposon mutagenesis (16). They identified sigma-28 (FliAL) which is required for the synthesis of lateral flagella (16). In V. parahaemolyticus, FliAP has been shown to be important in the development of the single polar flagellum required for swimming motility and 3 out of 4 flagellins are dependent on FliAP. The other flagellin expression is dependent on RpoN (15). Both fliAP and fliAL are regulated by RpoN, which is discussed below, and an rpoN mutant is non-flagellated and defective in swimming and swarming motility in V. parahaemolyticus (18). Both the FliAP and FliAL sigma factors are present in all members of the Campbelli and Parahaemolyticus clades indicating that the retention of two flagella systems is conserved in these clades. The presence of both flagella regulators has an unusual distribution. The two regulators are present in all members of the Campbelli, Parahaemolyticus, and Furnissii clades and in all V. mimicus isolates, as well as in V. coralliilyticus, V. brasilllensis, V. caribbenthicus, and several Photobacterium species. These data suggest that the two flagellar systems are ancestral to the Campbellii, Parahaemolyticus and Cholerae clades and lost from V. cholerae isolates.
Distribution and function of the sigma-54 family sigma factor RpoN
RpoN, the only member of the sigma-54 family, has been characterized in V. alginolyticus, V. anguillarum, V. cholerae, V. harveyi, V. fischeri, and V. parahaemolyticus (18, 37–43). In V. cholerae RpoN has been shown to be required for motility, nitrogen metabolism, biofilm formation, quorum sensing, type 6 secretion system (T6SS) synthesis, and virulence (41, 42, 44–48). RpoN is required for colonization in an infant mouse model of cholera and the defect was not entirely related to the lack of motility or glutamine synthetase expression (44). Klose and colleagues recently determined that the V. cholerae RpoN regulon includes more than 500 genes in serogroup O1 pathogenic isolates (48). They showed that RpoN played a role in the regulation of flagella synthesis, ammonium assimilation, virulence factor synthesis, and dicarboxylic acid metabolism (48). A study published in 2012 showed that RpoN differentially regulates the type IV secretion system and flagella operons in V. cholerae serogroup O37 strain V52 (42). They identified 68 RpoN binding sites and 82 operons that were positively controlled by RpoN. The 82 operons identified included genes for motility, all T6SS hcp 1 and hcp 2 operons, nitrogen metabolism, quorum sensing regulator HapR (LuxR homologue), formate dehydrogenase, phage shock, and over 40 genes of unknown function indicating the significant contributions of RpoN to cellular processes in V. cholerae (42). In V. fischeri, RpoN was shown to play a role in motility, biofilm formation, luminescence and colonization of the squid host (40, 49). The rpoN mutant examined in these studies was non motile, produced less biofilm, grew poorly on minimal media and could not colonize its symbiotic squid host. The rpoN mutant also had 3-4 fold higher levels of bioluminescence, suggesting that RpoN normally represses bioluminescence (40, 49). In V. anguillarum, it was shown that RpoN is essential for flagellum production as well as virulence in fish. An rpoN mutant exhibited loss of motility, but no loss of virulence when fish were intraperitoneally injected, however virulence was significantly reduced when fish were orally infected (38). In V. anguillarum, RpoN is also required for protease secretion, exopolysaccharide production, and biofilm formation (43). RpoN is required for formation of the polar flagellum synthesis and motility in V. alginolyticus (37).
RpoN was shown to play a role in quorum sensing in V. harveyi, via the sigma-54 activator LuxO. When LuxO activates RpoN to allow formation of a functional holoenzyme, this leads to expression of LuxR, the regulator of luminescence and other phenotypes (39). This was also shown to be the case in V. cholerae, V. alginolyticus and V. parahaemolyticus. In V. alginolyticus RpoN regulate hcp1, a hallmark of T6SS as well as other T6SS genes and motility via VasH a sigma-54 activator protein (42, 50). As mentioned previously, in V. cholerae T6SS gene clusters are also under the control of RpoN via VasH, and in V. parahaemolyticus RpoN is also required for hcp1 expression, which is repressed by OpaR (LuxR), the quorum sensing output regulator in V. parahaemolyticus that also represses the T3SS1 cluster (41, 42, 50–52).
Our recent studies of the function of RpoN in V. parahaemolyticus RIMD2210633, a clinical serogroup O3:K6 isolate, have uncovered a novel role for this sigma (18). We constructed an in-frame deletion of rpoN (VP2670) and examined the effects in vivo using our newly developed streptomycin-treated adult mouse model of colonization (18, 31). We showed that deletion of rpoN rendered V. parahaemolyticus aflagellar and therefore non-motile, and also resulted in reduced biofilm formation and a defect in glutamine synthetase production. We performed in vivo competition assays between the rpoN mutant and a wild-type marked with the beta-galactosidase gene lacZ (strain WBWlacZ) to screen for white and blue colonies. We demonstrated that the rpoN mutant colonized significantly more proficiently than WBWlacZ.
Interestingly, when we constructed mutants defective in the polar flagellum biosynthesis by knocking out the fliAP sigma factor and the lateral flagellar synthesis system by deletion of fliAL sigma, these mutants also out-competed WBWlacZ, but not to the same level as the rpoN mutant. These data suggested that lack of motility is not the sole cause of the fitness effect. Previous work in E. coli also showed that motility mutants were better in vivo colonizes and this work indicated that the motility mutants were metabolically more fit (53). Therefore we determine whether there were differences in growth of the rpoN mutant and the wild-type strain (18). In an in vitro growth competition assay in mouse intestinal mucus, the rpoN mutant also out-competed wild-type and exhibited faster doubling times when grown in mucus and on individual components of mucus. We determine the doubling times of the mutant and wild-type in mucus sugars and showed that the mutant had faster doubling times. We then examined the expression pattern of genes in the pathways for the catabolism of mucus sugars and showed that these genes had significantly higher expression levels in ΔrpoN than in wild-type (18).
In summary, it appears that the RpoN regulon is vast and complex within the Vibrio genus. Common themes are found such as a requirement for RpoN for flagellation, motility, biofilm formation and regulation of T3SS, T6SS and the quorum sensing regulator LuxR. Differences arise in the role of RpoN in the virulence of a particular species, which probably reflects the different requirements for colonization and virulence among the species and the animal models used rather than differences in RpoN regulation per se. Overall, studies indicate that similar to other genera, sigma factors are an important component of global transcriptional regulation in Vibrio with much variability seen in the number of alternative sigma factors. This variation may be reflective of the different lifestyles of each species and may suggest different requirements for gene transcription regulation in response to changing cellular and extracellular conditions.
Phylogeny and function of ECF-type sigma factors
The most well studied ECF-type sigma factor is RpoE and it is required for cell envelope stress response. RpoE has been characterized in studies in V. vulnificus, V. harveyi, V. angustum, and V. parahaemolyticus and multiple studies in V. cholerae (54–59). We identified a close homologue of the E. coli RpoE in all sequenced Vibrio indicating this ECF is conserved (Fig. 3). In V. cholerae, RpoE was shown to play an important role in intestinal survival and virulence in an infant mouse model (55). In this first study, an rpoE mutant was created in O395, a biotype classical V. cholerae strain, and was highly attenuated for virulence and sensitive to ethanol stress. The rpoE mutant, similar to wild-type, was not defective against heat, bile salts, hydrogen peroxide, polymyxin B, osmolarity and pH (5.5 to 10) stresses (55). In contrast, a study by Waldor’s group, examining an rpoE mutant in a biotype El Tor background strain showed that this mutant was sensitive to both a bioactive peptide P2 and polymixin B antimicrobial peptides (56). They determined that the outer membrane protein OmpU was a key determinant of basal rpoE expression and proposed that misfolded OmpU protein in the periplasm may be the signal that allows release of RpoE tethered to the cell membrane (56). Using next generation high-throughput sequencing, they found that most rpoE mutants could be constructed only in the presence of suppressor mutations suggesting rpoE is an essential gene in this V. cholerae El Tor strain (58). The authors found that out of all of the independently constructed rpoE mutants, 75% had a suppressor mutation in the promoter region of ompU (58).
In V. vulnificus, an rpoE mutant was more sensitive to membrane perturbing agents such as ethanol and SDS as well as hydrogen peroxide and there was a slight increase in sensitivity to heat and cold shock but was not attenuated for virulence in mice (57). In V. harveyi it was suggested that rpoE is essential in this species as an rpoE mutant could not be constructed (59). Instead mutants were constructed in the rpoE regulatory region rseABC. It was demonstrated that the overexpression of rpoE resulted in a reduction of hemolytic activity and attenuation for colonization in shrimp (59).
In V. parahaemolyticus, we constructed an in-frame deletion of the E. coli rpoE homologue from V. parahaemolyticus RIMD2210633 (32). We found that rpoE is essential for survival during cell envelope stress conditions such as polymixin B, hydrogen peroxide and ethanol stresses but was not required for acid, bile salt, and SDS stress responses. Our in vivo colonization analysis in a streptomycin-treated adult mouse model of colonization demonstrated that the rpoE mutant was severely defective compared to the wild type strain (32). These data suggest that RpoE is required for in vivo colonization probably through its role in cell envelope stress tolerance. Unlike in V. cholerae, it appears that in V. parahaemolyticus, OmpU may not be a signal for RpoE release from the membrane since our ompU deletion mutant strain was resistant to polymixin B and ethanol stress unlike the rpoE mutant which is sensitive to both these stresses (32).
We identified several divergent ECF type sigmas among Vibrio species and these along with fliAL were variably present among different clades. These divergent ECFs may not be required for extracytoplasmic cell envelope stress but may have evolved for a range of functions. V. parahaemolyticus contained four additional divergent ECF type sigmas, VP0055, VP2210, VP2358 and VPA1690. VP2210 was present in all sequenced Vibrio species with the exception of V. salmonicida and its function remains unknown. In a number of Vibrio species this gene is embedded among fatty acid oxidation and transport genes, which may suggest a possible role for this regulator. VP2358 is also widely distributed, present in most species with the exception of V. fischeri, V. salmonicida and Photobacterium species (Fig. 3). The function of VP2358 is also unknown and the genes that surround this sigma include a putative anti-sigma factor which has a cupin domain and may be involved in sensing reactive oxygen species. Other genes surrounding this sigma are involved in DNA damage repair and amino acid biosynthesis. Homologues of VP0055 are present in all members of the Campbellii and Parahaemolyticus clades as well as in V. vulnificus, V. fischeri, V. salmonicida, Vibrio sp 16, V. sinolensis, V. furnissii, and Photobacterium AK15. The function of VP0055 is unknown, however in all Vibrio species which possess this sigma, the genes surrounding it are genes involved in gluconate metabolism. VPA1690 is only present in V. parahaemolyticus, V. alginolyticus and Vibrio sp Ex25 strains. The genes immediately adjacent to this sigma are conserved hypothetical proteins and transcriptional regulators, as well as genes involved in alkyl hydroperoxide reduction which may suggest a possible function.
Vibrio specific global regulator ToxRS
Apart from sigma factors, bacteria also utilize two-component regulators to help control gene transcription in response to changing environmental conditions. Among Vibrio species the number of two component systems varies from species to species. The ToxRS system is a two-component regulatory system present in all Vibrio and has been characterized extensively in V. cholerae and to a much lesser extend in V. parahaemolyticus (27, 30, 31, 60–73). We reconstructed the evolutionary history of ToxR for the same set of isolates that we examined for the HK and 16S rRNA gene trees (Fig. 4). We obtained a ToxR phylogeny that had a topology similar to HK and 16S rRNA trees for all the major clades described indicating that this regulatory system is ancestral to the family Vibrionaceae (Fig. 4).
Fig. 4.
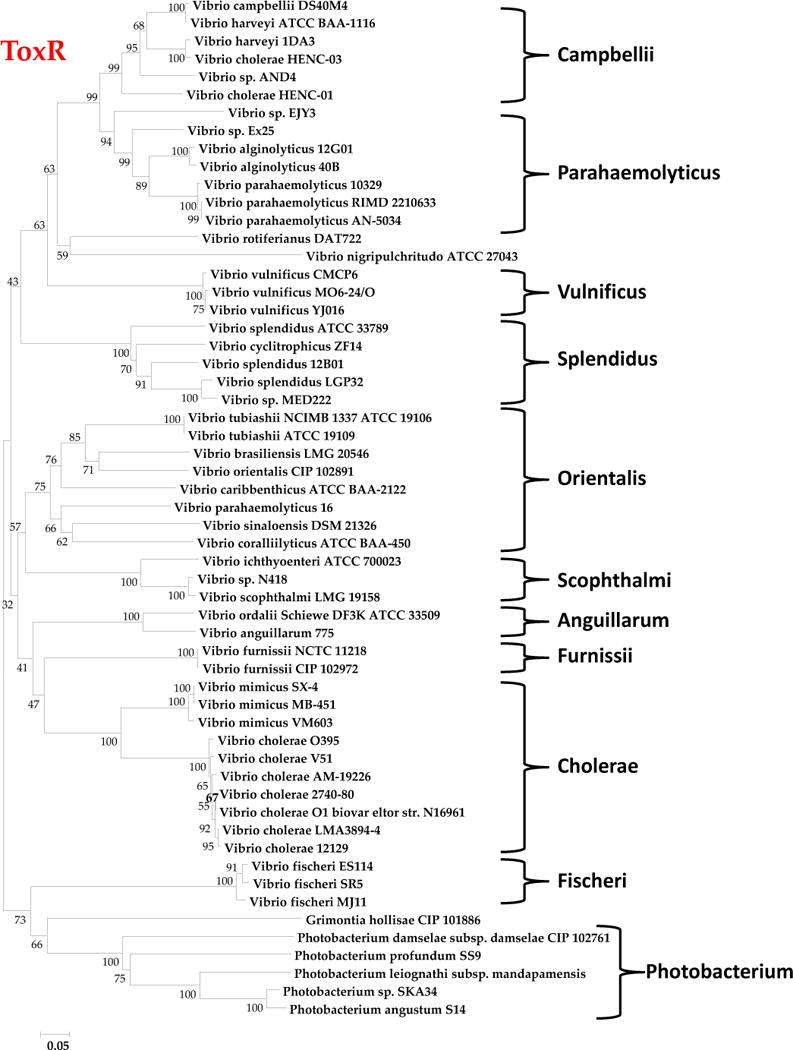
Phylogenetic analysis of toxR. This phylogenetic tree illustrates the relationship among 57 Vibrio species. The tree was constructed in MEGA5 using the Neighbor-Joining method and a bootstrap value of 1000 and complete deletion (30, 52, 120).
In V. cholerae the ToxRS regulon can be separated into two branches, the ToxT dependent and independent pathways (73–76). ToxR has been shown to act as a co-activator of the toxT gene, which encodes a transcription factor that positively regulates the expression of the genes for cholera toxin and the toxin co-regulated pilus (TCP), which are essential virulence and colonization factors, respectively (60, 61, 66, 73). ToxR regulation of the outer membrane porin OmpU has been shown to be important for survival in the face of organic acid stress, bile stress, and antimicrobial peptides in this species (69, 70, 77). As discussed above, there is evidence that indicates OmpU in V. cholerae may act as a signal in the activation of RpoE, by an as yet unknown mechanism (56, 58). The role of OmpU in in vivo survival is not entirely known, as V. cholerae ompU deletion mutants only exhibit a slight decrease in colonization in the infant mouse model of colonization (56, 58). In V. splendidus, OmpU was shown to be required for antimicrobial peptide resistance and to be an essential component for colonization of the oyster host (78, 79).
Unlike V. cholerae, the V. parahaemolyticus ToxRS regulon does not contain the ToxT pathway and its regulon. Klose and co-workers demonstrated that a toxR mutant strain had an altered outer membrane profile, specifically OmpU (70). Our group demonstrated that ToxRS positively regulates the V. parahaemolyticus OmpU homologue and was sensitive to acid and bile salt stresses (27, 31). We showed that under acid conditions toxRS and ompU were highly expressed. In a toxRS mutant strain, ompU expression was severely reduced and cells were sensitive to both acid and bile salt stresses. The toxRS mutant compared to the wild type strain using a pretreated streptomycin adult mouse model exhibited a severe defective in intestinal colonization (27, 31). In addition, we demonstrated that the V. parahaemolyticus ToxRS system was a negative regulator of the T3SS-1 contained in chromosome I. This regulation by ToxRS was indirect and was mediated by the LeuO transcription factor homologue encoded by vp0350. ToxRS was shown to be a positive regulator of leuO expression and LeuO is a negative regulator of T3SS-1 gene expression (31, 80). This ToxRS pathway is thus far unique to V. parahaemolyticus (31).
Carbon utilization: sugars and amino sugars
The adaptability to new nutritional sources is crucial for a species to switch between macro- and micro-environments. Survival in the nutrient limited conditions of the water column and marine sediments requires that an organism efficiently utilizes the available nutrients. Adaptation to host environments also requires the ability to switch from nutrient poor to nutrient rich and from free-living to host associated conditions and also requires that a species can compete with the resident microbiota for nutrients. In the next section, we will discuss some of the major nutrient capabilities of Vibrio species and how generalization and specialization within and between species may have aided niche expansion.
Chitin is a very abundant substance found in the marine environment, with an estimated 10x1011 tons produced annually. Chitin is composed primarily of a polymer of β-1,4 N-acetylglucosamine (GlcNAc) and Vibrio are key to the degradation of chitin, which contributes significantly to nitrogen and carbon cycling in the marine ecosystem (14, 48, 52, 81–90). Chitin provides a readymade source of GlcNAc, which is an important metabolite and source of energy for all Vibrio species. The ability to use chitin as a carbon and nitrogen source is universally found within Vibrio species. It has been shown in V. furnissii, that there is a strong chemotactic response to GlcNAc oligomers, as well as being able to adhere to chitin particles through GlcNAc-lectin association (81–83). Chitin comes in many varying forms in the wild and Vibrio species have been shown to produce several chitinases to cope with this diversity. For example, V. harveyi has six chitinase genes, and among Vibrio in general the number can range from 2 to 8. Extracellular chitinases break down the large polymers into smaller subunits of GlcNAc. The oligiosaccharides are transported by a specific chitin porin (91). Chitin degradation appears to be tightly controlled in the presence of other energy sources, as seen in V. furnissii, where considerable chitinase release was only found in the absence of any other carbon source (86, 88, 91–95). In Vibrio species chitin degradation is controlled by a chitin sensor (ChiS) kinase (87, 96).
Many Vibrio species also interact with a large number of eukaryotic hosts where chitin is absent. Many of these commensal and pathogenic host interactions involve bacterial colonization of the mucus covered epithelium surfaces. For example, the mammalian gastro-intestinal epithelium contains a mucus gel layer that is the site of attachment of some species. The main constituent of mucus is mucin, a glycoprotein composed of oligosaccharides that include ribose, mannose, hexuronates (gluconate and glucuronate), galactose, fucose, arabinose, GlcNAc, N-acetylagalactosamine (GalNAc) and N-acetylneuraminic acid (Neu5Ac or sialic acid). Therefore mucin constitutes a direct carbohydrate source that can offer ecological advantages to Vibrio gastrointestinal species especially if they can utilize a unique carbon source or one more efficiently than the resident commensal species. We examined the genome of sequenced Vibrio species and found that the genes for the metabolism of mucin sugars are widely present. For example, we found that all Vibrio species contain the genes required for the metabolism (and transport) of ribose, mannose, gluconate, galactose, GlcNAc and GalNAc.
In the case of the catabolism of glucuronate, arabinose, fucose, and sialic acid, a wide variation in the presence of these genes was found. While species from 8 of the major clades identified in Figure 1 carry genes for glucuronate catabolism, these genes were missing from all representatives in the Cholerae, Fischeri and Orientalis clades. These data suggest that the ability to utilize glucuronate as a nutrient source is ancestral to the family Vibrionaceae and was likely loss from V. cholerae, V. fischeri and V. orientalis. The genes for arabinose catabolism are only present in two species, V. parahaemolyticus and V. furnissii, which suggests that this is may be a recently acquired trait in the family Vibrionaceae. In V. parahaemolyticus the arabinose catabolism gene cluster is present in a 25 kb region encompassing VPA1652 to VPA1679 that also contains a ferric uptake system (97). This region was identified by genome comparisons with V. cholerae and V. vulnificus which lack this region (97). The ability to utilize arabinose is not present in all strains of this species for example the environmental strain UCM-V493 does not contain the arabinose gene cluster (98).
The genes for fucose catabolism were present in only V. vulnificus, V. mimicus, V. nigripulchritudo, V. shilonii, V. harveryi HENC-01 and Vibrio sp EJY3. Our examination of the fucA gene from this pathway shows that its closest homologue is present in enteric species. In V. vulnificus YJ016, the fucose catabolism gene cluster is present within a 117 kb island. The genomic island is inserted at a tRNA serine locus, contains an integrase and has an overall GC content of 43% (99). The island region encompasses VV2149 to VV2262 in YJ016 and is largely absent in MO6-24/O and CMCP6 but the fucose catabolism cluster is present in MO6-24/O. Taken together, these data strongly suggests that this is a newly acquired trait in these species and not ancestral to the group.
N-acetylneuraminic acid commonly known as sialic acid is an amino sugar utilized in humans and is ubiquitous among metazoans of the deuterostome lineage (100–102). Sialic acid is an integral component of mucin. Genes involved in the catabolism of sialic acid are present in less than 25% of sequenced Vibrio genomes (103, 104). These genes are present in 5 clades of Vibrionaceae; Photobacterium, Fischeri, Cholerae, Vulnificus and Orientalis clades. Within the Cholerae clade the ability to uptake and catabolize sialic acid is broadly distributed, present in many isolates of both V. cholerae and V. mimicus (103–106). In these species, the genes are present within a pathogenicity island (PAI) named Vibrio pathogenicity island-2 (VPI-2), which is found only in pathogenic isolates (105, 106). The VPI-2 region has all the hallmarks of a PAI; a GC content lower than the rest of the genome and encodes an integrase that is adjacent to a tRNA serine locus that (105, 106). In V. cholerae non O1/non O139 isolates and V. mimicus isolates that cause inflammatory diarrhea, a T3SS is present on the VPI-2 island (107–110). Within V. vulnificus, the ability to uptake and catabolize sialic acid is present in clinical type isolates but absent from environmental isolates (111). These genes are present in chromosome II and are not associated with a horizontally acquired region (111). In the Orientalis clade three species, V. orientalis, Vibrio spp 16 and V. sinaloensis, contain the genes for sialic acid catabolism. These genes are also present in V. ichthyoenteri, V. nigripulchritudo, V. shilonii and Vibrio sp. MED222.
The ability to catabolize sialic acid as a sole carbon and energy source has been shown to be a significant in vivo fitness factor (111–114). We demonstrated that deletion of the nanA gene in V. cholerae, which encodes the first enzyme in the sialic acid catabolic pathway, resulted in decreased colonization compared to wild-type in an infant mouse model of cholera (112). It was also demonstrated in V. vulnificus that sialic acid catabolism is important for mouse intestinal colonization (115). These data indicate that the ability to utilize sialic acid, an abundant carbon and nitrogen source in the human gut, is important in in vivo survival. A tripartite ATP independent periplasmic (TRAP) transporter, SiaPQM (vc1777-vc1779) is adjacent to the sialic acid catabolism genes contained within the Vibrio pathogenicity island-2 (VPI-2) in V. cholerae pathogenic isolates (103, 104, 106, 109, 112). Thomas and colleagues have clearly demonstrated that SiaPQM functions biochemically as a sialic acid transporter in vitro (114, 116). We showed a strong correlation between the presence of VPI-2 and the ability of Vibrio to grow on sialic acid as a sole carbon source (113). We demonstrated that a strain of V. cholerae N16961 defective in vc1777, encoding the large membrane protein component of the TRAP transporter, does not growth on sialic acid (113). Interestingly, a report by Sharma and colleagues proposed that an entirely different TRAP transporter, DctPQM encoded by vc1927-vc1929, was the sole sialic acid transporter in V. cholerae (117). However, our bioinformatics, genomic analysis, biochemical and genetic analyses determine that DctPQM is a C4-dicarboxylate-specific TRAP transporter and disruption of vc1929 results in a defect in growth on C4-dicarboxylates but not sialic acid (113, 114). These data demonstrate that SiaPQM and not DctPQM is the sole sialic acid transporter in V. cholerae (113, 114).
Horizontal gene transfer
Horizontal gene transfer has played an important role in the emergence of different Vibrio species by allowing strains and species to expand their niche. In this next section we will provide an overview of the major HGT events that have occurred in the Vibrio species whose complete genome sequence is available. Specifically, we will give a detailed account of the genetic diversity and phylogeny of superintegrons within these sequenced strains.
Vibrio cholerae is the most widely studied Vibrioneceae species due to its current and historical impact on human health. Of the seven completely sequenced strains of V. cholerae, six were isolated from humans, N16961 (Bangladesh), O395 (India), IEC224 (Brazil), M66-2, MJ-1236 (Banglasdesh), 2010EL-1786 (Haiti) and one, V. cholerae LMA3894-4 (Brazil), was acquired from creek water (Table 1). Within the genome of choleragenic isolates are three regions acquired by horizontal gene transfer that are essential for virulence (118). Cholera toxin, the cause of the secretory diarrhea, is encoded within a filamentous phage named CTXphi. The intestinal colonization factor the type IV Toxin coregulated pilus (TCP) is encoded within a pathogenicity island named VPI-1 or the TCP island. Sialidase and sialic acid uptake and catabolism genes are present on a non-homologous island named VPI-2 or the sialic acid metabolism island (62, 105, 106, 112, 119, 120). The first V. cholerae genome to be sequenced was V. cholerae N16961 in 2000, an El Tor biotype strain responsible for the ongoing seventh cholera pandemic (121). Two sixth pandemic biotype classical strains O395 and M66-2 are also available. The most striking difference between the classical and El tor strains is the presence of two putative pathogenicity islands: Vibrio seventh pandemic island I (VSP-I) and VSP-II in N16961. VSP-I is proposed to be important for host adaptation through the production of a regulatory cyclic di-nucleotide (122). The CTXΦ is absent from M66-2 whereas the classical O395 strain contains an extra copy of the phage in chromosome II, identical to that found in chromosome I (123). Additionally, the rstR gene of the CTXΦ in strains N16961 and O395 differ significantly for each other (123). V. cholerae MJ-1236 is described as a hybrid strain. It contains a 33 kb excisable element Kappa phage and a 133 kb ICE-like element, both found in chromosome I and absent from N16961. Additionally, genomic island 14 (GI-14), a 19 kb island, is also a common link between MJ-1236 and the classical strain. On the large chromosome, MJ-1236 contains both VPI-1 and VPI-2 as well as VSP-I and VSP-II, however a second copy of VSP-I can be found on the smaller chromosome (124). The core genomic region of the CTX phage contains ctxAB of the classical strains as well as the El Tor “pre-CTX” region. There is even more variability in the RS2 region of MJ-1236 as it contains a classical rstR gene, an El Tor rstB gene and a rstA gene that differs from both biotypes.
The seventh cholera pandemic hit Latin America in 1991 and in 1994, a sucrose fermenting defective strain, V. cholerae ICE224, which caused outbreaks until 1996 was isolated and eventually sequenced (125). When compared with the N16961 genomes, SNPs were seen in 122 genes and other Latin American strains shared 48 of these SNPs. Additionally, distinguishing it from N16961 is a 49 kb insertion of the WASA1 bacteriophage at an attachment site in the membrane alanine aminopeptidase gene (125). The strain 2010EL-1786 was isolated from Haiti in 2010 and determined to be of the El Tor biotype, however it contained the ctxB gene of the classical biotype. This isolate, as well as all isolates from Haiti contained a tcpA allele of a Bangladeshi El Tor strain (CIRS 101) which carries an El Tor CTXΦ, but produces the classical cholera toxin (126).
Two other important Vibrio human pathogens are V. vulnificus which causes wound infection and septicemia in immune-compromised patients and V. parahaemolyticus, which causes gastroenteritis via consumption of raw infected seafood (127–129). V. vulnificus is an opportunistic pathogen that in susceptible hosts can have a greater than 50% mortality rate (130). Three complete genome sequences are available and all are clinical isolates. V. vulnificus YJ016 contains a plasmid, pYJ016, not found in the other two V. vulnificus strains that are sequenced, CMCP6 and MO6-24/O (131). Chromosome sizes of V. vulnificus are comparable to V. parahaemolyticus RIMD2210633. The V. vulnificus chromosome II is significantly larger than in V. cholerae N16961. Additionally, V. vulnificus YJ016 contains 1134 more genes than N16961. This increase in gene number and genome size may be accounted for by the higher number of duplication events that occurred in V. vulnificus YJ016 compared to V. cholerae N16961 (131). Comparative genome analysis of YJ016 and CMCP6 identified 14 regions ranging in size from 14 to 117 kb, which had the characteristics of recently horizontally acquired DNA (99). These 14 genomic islands each contain an integrase gene, had aberrant GC content and dinucleotide frequency (99). Most of the genomic islands were absent from a collection of V. vulnificus isolates suggesting that in this species each strain may contain its own repertoire of horizontally acquired DNA (99). We identified a genome region among clinical isolates named region XII that contained genes required for the transport and catabolism of glycoaminoglycans, a potential source of carbon and nitrogen in vivo. Two metabolic pathways were encoded within this region, lacto-n-biose and chondroitin sulfate as well as putative transporters for each of these substrates (Fig. 5). Thus, this region may potentially be involved in host interactions that allow it to survive in vivo (132). The complete region XII metabolic island is also present in Vibrio sp RC341, five V. mimicus isolates and V. nigripulchritudo (Fig.5).
Fig. 5.
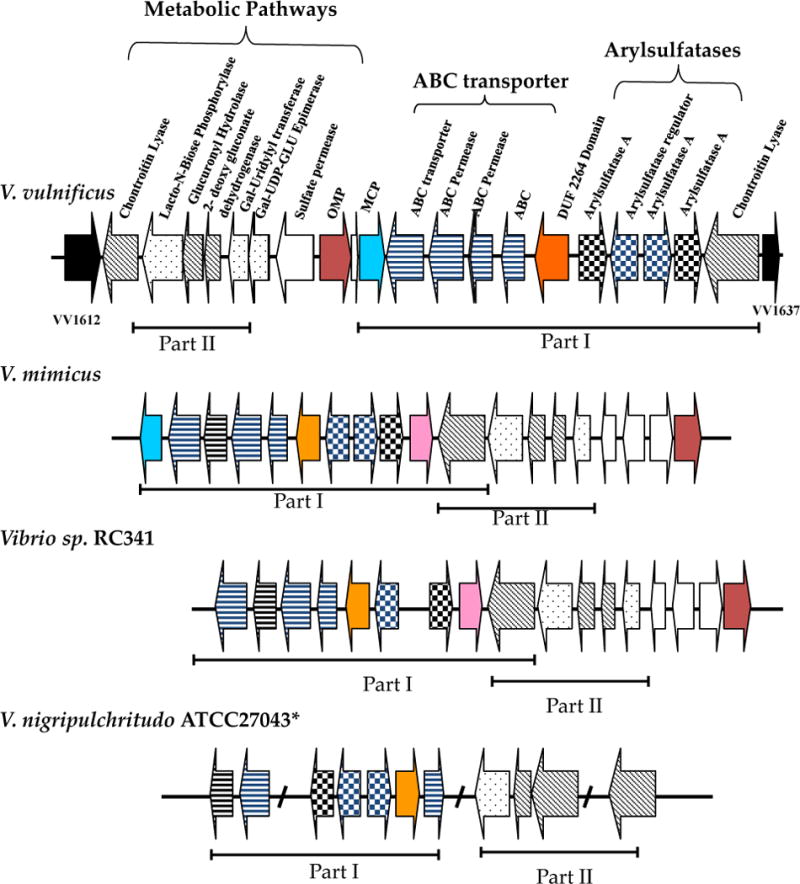
Genome context and arrangement of region XII cluster among Vibrio species. Open reading frames (ORFs) are indicated as arrows, the direction of which shows the direction of transcription, numbers underneath ORFs represent locus tags. ORFs of similar color represent homologous pathway genes among the different species examined. Asterisk represents incomplete genome sequence. The region is present in all V. mimicus strains examined.
We performed systematic BLAST analysis for each of the ORFs of V. parahaemolyticus RIMD2210633 compared with each of the ORFs from the genome sequences of V. cholerae N16961, V. vulnificus YJ016 and CMCP6, and V. fischeri ES114 (97). We identified 24 regions, gaps in the genome atlas, of greater than 10 kb that were unique to RIMD2210633. These regions included an integron, f237 phage, two T3SSs, a T6SS and 7 Vibrio parahaemolyticus genomic islands named VPaI-1 to VPaI-7 (97). Comparative genomic analysis of RIMD2210633 with V. parahaemolyticus AQ3810, an O3:K6 isolate recovered in 1983, identified five regions unique to RIMD2210633, VPaI-1, VPaI-2, VPaI4, VPaI-5 and VPaI-7 (97, 133). RIMD2210633 encoded an additional T6SS as well as an additional T3SS encoded on VPaI-7. Our data showed that there was considerable genomic flux and that the highly virulent clone of RIMD2210633 arose from an O3:K6 isolate that acquired at least seven novel regions (97, 133). We recently sequenced an environmental isolate of V. parahaemolyticus strain UCM_V493. This strain lacked all seven islands including the T3SS on chromosome 2 and the T6SS on chromosome 1 (98).
Vibrio splendidus is a species associated with invertebrate mortalities worldwide. The complete genome sequence for strain LGP32 is available along with partial genome sequences for strain MED222 and 12B01 (78). Genome comparison studies identified 409 genes unique to LGP32 and many of these genes were present in regions that had characteristics of genomic islands (78). An RS-like phage and 9 transposons were are identified. Unusually one of the most divergent acquired genetic elements, the integron, was missing in strain 12B01 and in strain LGP31 it only contained a limited number of cassettes (78).
The genome sequence of V. angullarium strain 775 was compared with draft sequences of strains 96F and rv22 and V. ordalii (4). V. anguillarum is known to cause vibriosis in nearly 50 different fish species as well as mollusks and crustaceans worldwide. Two serogroups O1 and O2 of the 28 recognized serogroups are associated with virulent strains. The analysis of the genome sequence identified 10 genomic islands ranging in size from 4 to 140 kb that encoded integrases and transposases and three regions were adjacent to tRNA loci (4). Although V. anguillarum and V. ordalii are closely related their genome size differed significantly, V. anguillarum has a 4 MB genome compared to V. ordalii 3.4 MB genome. Analysis of V. anguillarum strain 86F identified a T3SS, which was related to the T3SS identified on chromosome II of V. parahaemolyticus RIMD2210933, and to that found in V. cholerae non choleragenic strains and V. mimicus (4). The T3SS was missing for the other strains examined although remnants were present in strain 775 and V. ordalii suggesting it was deleted from these strains (4).
Vibrio salmonicida also known as Aliivibrio salmonicida, is a halophilic psychrophile responsible for cold water vibriosis (CV) in farmed Atlantic salmon and rainbow trout as well as captive Atlantic cod. Similar to all Vibrio, V. salmonicida contains two chromosomes as well as 4 circular plasmids, which encompass 2.7% of the entire genome and 111 protein coding sequences (85). Plasmid pVSAL840 contains the tra locus, associated with plasmid conjugation, which shows high synteny with pYJ016 of V. vulnificus YJ016. pVSAL43 and pVSAL54 are predicted to encode acetyltransferases enabling antigenic variation at the cell surface and thus possible improved protection from the host immune response. Plasmid pVSAL320 contains an iron ABC transporter that may be involved in the non-siderophore uptake of ferrous iron (85). These plasmids are not necessary to cause CV in salmon (134), but based on encoded proteins it appears they may play a role in the pathogenicity of V. salmonicida. Another defining characteristic of V. salmonicida is the extremely large number of insertion sequence (IS) elements found on both chromosomes and plasmids, totaling 288. For comparison, the closest relative of V. salmonicida, V. fischeri, contains only one IS element. A whole genome comparison with V. fischeri reveals breaks in synteny in V. salmonicida in regions flanked by IS elements. V. fischeri contains more than 70% orthologous CDSs to V. salmonicida. V. fischeri is a known symbiont of the squid Euprymna scolopes and is required for bioluminescence in the squid light organ. V. fischeri ES114 is able to colonize E. scolopes whereas V. fischeri MJ11 a Japanese fish species isolate is unable to colonize squid. Comparative genomic studies revealed that RscS, a two-component sensor kinase necessary for colonization of the squid is absent from the MJ11 strain (135). Photobacterium profundum SS9 was isolated from the Sulu Trough at 2500 meters and is a peizophile that can withstand high pressure (136). In addition to the 2 bipartite chromosomes, P. profundum SS9 contains an 80 kb plasmid. P. profundum SS9 is 25% larger than V. parahaemolyticus and V. vulnificus (136). SS9 contains 2 complete operons for F1F0 ATP synthase, 3 cbb3 cytochrome oxidase genes and an unusual di-heme cytochrome c gene, all believed to be necessary for metabolic activity at high pressures (136).
Within all these genomes, signatures of horizontally acquired DNA can be found either in the form of extrachromosomal elements such as plasmids or as chromosomally encoded genetic elements such as IS elements, transposons, genomic islands, prophages and integrons. The number and type of horizontally acquired element differs between species and also within species. Many of these elements contain functions that are critical for a specific lifestyle and many others are of as yet unknown function. One of the most well studied and intriguing mobile genetic element is the integron and in the next section we will describe the intergon genome context and structure that is found within the completed genome sequences of Vibrio.
Superintegrons in Vibrio
Integrons are assembly platforms that incorporate exogenous gene cassettes by site-specific recombination and convert them to functional genes. To date, three classes of integrons (intI1 to intI3) implicated in multi-resistance phenomena have been characterized and are classified based on the homology of their integrase protein which ranges from 43 to 58% (137). These multi-resistant integrons (MRIs) are defective in self-transposition but they are often found in association with insertion sequences (ISs), transposons, and/or conjugative plasmids which can serve as vehicles for their mobility. Therefore, the MRIs are also called mobile integrons. All integrons characterized to date are composed of three key elements necessary for the capture of exogenous genes: a gene (intI) encoding an integrase belonging to the tyrosine-recombinase family; a primary recombination site (attI); and an outward-orientated strong promoter (Pc) that directs transcription of the captured genes (138, 139). The superintegron (SI) is a distinctive class of integrons. Like MRI, the SI is also composed of an integrase (intIA) gene and other key elements. The intIA gene product has 45 to 50% identity with the three known integrases, IntI1 to IntI3 (137). In comparison to MRIs which contain only 5-8 gene cassettes, SI contains a large array of gene cassettes (137, 139). For example, in V. cholerae N16961, the SI is 126 kb in size and contains 179 gene cassettes (215 ORFs) (140, 141). Another striking difference between MRIs and the SI is that the SI does not seem to be mobile as it is located on the chromosome and not associated with mobile DNA elements (139, 142).
The SI is one of the largest genetic elements identified so far in the V. cholerae genome as well as in other vibrios. In comparison to other genetic elements, the SI is dynamic in terms of capturing or releasing gene cassettes (141). The gene cassettes can exist as circular molecules that are unable to replicate or as an integrated element at the attI site in an integron (143). The attI site has a 7 bp core site (CS) with the consensus of GTTAGGC or GTTRRRY (143). The gene cassettes found in integron platforms normally have only two functional components, a gene or non-coding DNA and an imperfect inverted repeat located at the 3′ end of the gene known as 59-base element (59-be) or attC site (141, 143). The attC site is a diverse family of sequences which varies in length (57 to 141 bp) and functions as a recognition site for the site-specific integrase. The most highly conserved feature of the attC site is a 7 bp core site (CS) at the 3′ end with the consensus of GTTAGGC or GTTRRRY (which is identical to that of the attI) and an inverse core site (ICS) at the 5′ end with the consensus of GCCTAAC or RYYYAAC (141, 143). The CS of an attC is perfectly complementary to its ICS.
Like attC sites in MRIs, the gene cassettes in SI are also followed by short (123 to 126 bp) repeat sequences commonly referred as XXR where the first X stands for genus, the second X represents species and R represents repeat. For example, VCR indicates V. cholerae repeat (144, 145). Generally, cassette genesis occurs through recombination between attI and VCR sequences catalyzed by IntIA, the site-specific recombinase (139, 141). MRIs play a leading role in the acquisition and spread of antibiotic resistance genes among Gram-negative bacteria, especially among Enterobacteriaceae and Pseudomonas (141, 146). In contrast, the SI gene cassettes encode proteins for metabolic activities, virulence, DNA-modification enzymes as well as antimicrobial agents, however, most of their functions are still unknown (139, 141).
NCBI microbial genome database shows numerous ongoing projects for whole genome sequencing of diverse Vibrio species and among them are 20 strains of 11 different species whose whole genome sequences have been completed (Table 4). We performed a PSI-BLAST (Position-Specific Iterated BLAST) search using the V. cholerae SI integrase, IntIA (VCA0291) as a seed sequence on these 20 strains of 11 different species. We found that all 20 strains possessed a copy of IntIA, which is the primary indicator of the presence of an SI (Table 4). We constructed a phylogenetic tree based on the intIA gene and found that the sequences clustered the Vibrio strains in a species-specific manner (Fig. 6). The intIA sequence was 100% conserved in all 7 sequenced V. cholerae genomes. Similarly, 3 V. vulnificus strains shared identical intIA sequences and formed a cluster which branched with intIA from V. parahaemolyticus strains. Two undefined Vibrio species strains EX25 and EJY3 clustered with the V. parahaemolyticus clade. Vibrio sp EX25 was very close to V. parahaemolyticus and Vibrio sp. EJY3 was more similar to V. harveyi. Vibrio anguillarum, V. furnissii and V. splendidus intIA genes were more distantly related to each other (Fig. 6). The two V. fischeri and one V. salmonicida intIA sequences clustered together and distinct from the other vibrios examined (Fig. 6). The ancestry and evolutionary relationship between the intIA gene sequences was compared with a concatenated tree of three housekeeping genes for the same set of strains. The topology of intIA for all the Vibrio species analyzed was congruent with the HK tree (Fig. 6).
Table 4.
Superintegron (SI) in Vibrio species
| Species | Strain ID | Size (Kb) | (GC SI/Genome) | No. XXR | No. ORF | 5′-flanking ORF | 3′-flanking ORF |
|---|---|---|---|---|---|---|---|
| V. cholerae | N16961 | 125 | 42/48 | 177 | 216 | VC0290 (L20) | VC0507 (Tnp) |
| IEC224 | 125 | 42/48 | 177 | 218 | O3Y_14818 (L20) | O3Y_15923 (Tnp) | |
| MJ-1236 | 119 | 42/47 | 137 | 163 | VCD_000985 (L20) | VCD_000819 (Tnp) | |
| 2010EL-1786 | 99 | 42/48 | 136 | 160 | Vch1786_II0036 (L20) | Vch1786_II0197 (Tnp) | |
| LMA3894-4 | 37 | 43/48 | 53 | 50 | VCLMA_B0258 (L20) | VCLMA_B0309 (HP) | |
| O395 | 119 | 41/48 | 139 | 188 | VC395_A0327 (L20) | VC395_A0516(Tnp) | |
| M66-2 | 98 | 42/48 | 138 | 169 | VCM66_A0289 (L20) | VCM66_A0466(Tnp) | |
| V. vulnificus | CMCP6 | 151 | 41/47 | 218 | 152 | VV1_2400 (L20) | VV1_2550 (Tnp) |
| YJ016 | 138 | 41/47 | 187 | 203 | VV1942 (L20) | VV1738 (Tnp) | |
| MO6-24/O | 129 | 41/47 | 195 | 179 | VVMO6_01279 (L20) | VVMO6_01459 (HP) | |
| Vibrio sp. | Ex25 | 114 | 41/45 | 95 | 127 | VEA_003168 (CHP) | VEA_003296 (GSH transferase) |
| V. parahaemolyticus | RIMD2210633 | 47 | 40/45 | 68 | 77 | VP1866 (CHP) | VP1788 (Tnp) |
| Vibrio sp. | EJY3 | 190 | 43/45 | 31 | 173 | VEJY3_09680 (acyltransferase) | VEJY3_08800 (HP) |
| V. harveyi (campbellii) | BAA-1116 | 1 | 43/45# | 0& | 1 | VIBHAR_02002 (GPD) | VIBHAR_02004 (HP) |
| V. anguillarum | 775 | 68 | 41/45 | 88 | 120 | VAA_01071 (L20) | VAA_00312 (Tnp) |
| V. furnissii | NCTC 11218 | 39 | 42/51 | 63 | 35 | vfu_B01510 (ATPase ABC tr.) | vfu_B00012 (AAFGH) |
| V. splendidus | LGP32 | 18 | 40/44 | 17 | 24 | VS_1891 (Cytochrome P450) | VS_1866 (HP) |
| V. salmonicida | LFI1238 | 22 | 36/39 | 22 | 27 | VSAL_II0533 (HP) | VSAL_II0561 (Tnp) |
| V. fischeri | ES114 | 29 | 35/38 | 37 | 36 | VFA0664 (HP) | VFA0628 (MOBHMT) |
| MJ11 | 102 | 35/38 | 130 | 126 | VFMJ11_A0760 (HP) | VFMJ11_A0633 (HP) |
XXR: 1st X = Genus, 2nd X = Species, R = Repeat;
Including intIA gene;
Solitary intIA represents SI
Only 2 VHRs were identified, one in plasmid pBIBHAR (CP000791) and the other in chromosome II (CP000790).
C(HP): Conserved (Hypothetical protein); MOBHMT: 3-methyl-2-oxobutanoate hydroxymethyltransferase
GPD = Phage late control gene D protein
ATPase ABC tr. = ATPase of ABC transporter with duplicated ATPase domains
AAFGH = arginase/agmatinase/formimionoglutamate hydrolase; GSH = glutathione
Fig. 6.
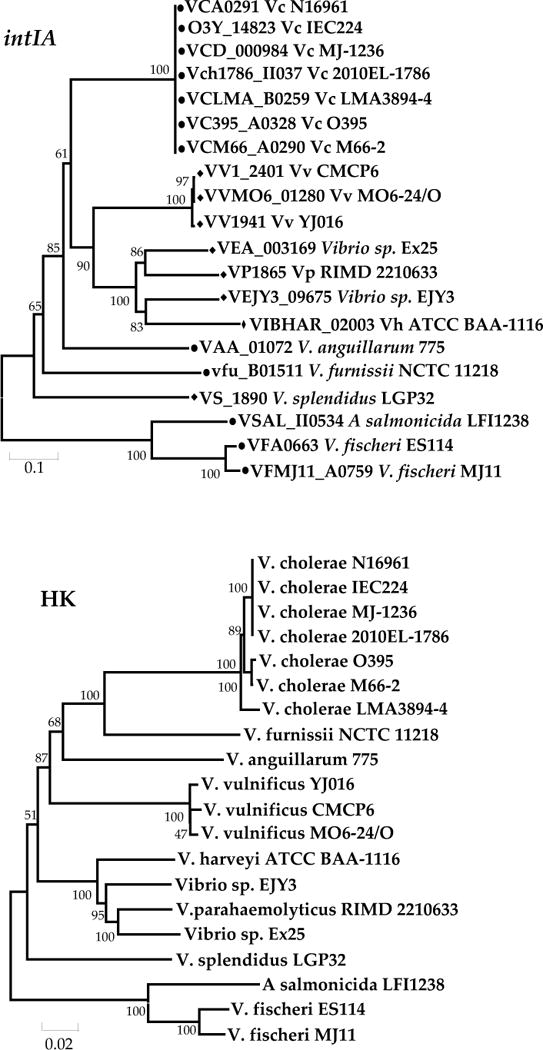
Phylogenetic tree based on Neighbor-Joining method (13) using (A) intIA and (B) concatenated sequences of rpoB, mdh and pyrC genes of 20 Vibrio strains of 11 different species whose whole genome sequences are completed. The locus tag of each gene was used whenever appropriate. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (30). The evolutionary distances were computed using the Jukes-Cantor method (52) and evolutionary analyses were conducted in MEGA5 (120). Here, Black circle and diamond indicate the strains that bear the superintegron in Chr II and Chr I, respectively.
Next, we examined the insertion site of the SI and the SI content among the different Vibrio species. As observed by Rowe-Magnus and colleagues, the intIA gene among V. cholerae strains were all located downstream of the same rplT gene that codes for the ribosomal L20 protein (Table 4) (144, 145). Though the V. vulnificus clade branched with the V. parahaemolyticus clade, their chromosomal locations were different. The SI from the V. vulnificus strains had the same chromosomal location as V. cholerae at the rplT locus. However, within the V. parahaemolyticus clade, the SI was present at a different chromosomal location. V. parahaemolyticus and Vibrio sp. EX25 had their SI inserted downstream of a conserved hypothetical protein VP1866 and VEA_003168, whose homolog is VC1310 in V. cholerae. The SI in V. harveyi strains was located downstream of VIBHAR_02002, which encodes a phage late control gene D protein, GPD and in Vibrio sp. EJY3 the SI was inserted at VEJY3_09680, which encodes an acyltransferase involved in lipid metabolism. The intIA gene in the V. fischeri strains were located downstream of a hypothetical protein with no homolog present in V. cholerae N16961. The intIA gene in V. furnissii was located downstream of vfu_B01510, which encodes an ABC transporter ATPase. In V. cholerae, V. anguillarum, and V. furnissii, the SI is located in chromsome II whereas in all other species examined the SI is present on chromosome I. As suggested previously, the conserved chromosomal location of the SI within the relevant subclades can be helpful in identifying the SI structure from other species within the same subclade (144).
The intIA gene transcription is opposite to the SI gene cassettes and, hence, gene cassettes are always predicted to be present upstream of intIA (139, 141). Therefore, we extracted from each genome 200 kb of DNA upstream of the intIA gene. The presence of VXR (Vibrio species repeat) was determined in this 200 kb sequence by in silico hybridization using VCR/VXR sequence as a probe as well as by the XXR program developed by Mazel and his colleagues (144). As summarized in Table 4, SIs from all 7 V. cholerae strains were highly diverse in terms of size, number of ORFs and VXR. The rearrangement of gene cassettes is an important feature of SI diversity in V. cholerae as well as in other vibrios (139–141, 144). Interestingly, we found 99% identical SI structures in two strains of V. cholerae, N16961 and IEC224, which were isolated from different geographic locations and timepoints, from Bangladesh in 1975 and from Brazil in 1994 respectively. There were anomalies in total ORF numbers such as N16961, which contained 216 and IEC224 contained 218, but this was due to the difference in ORF annotation. The presence of identical SIs as well as their conserved phylogenetic grouping indicates a shared evolutionary origin.
Among the completed genome sequences of Vibrio species, the size of the SI varied from 22 kb in V. salmonicida to 151 kb in V. vulnificus CMCP6. Vibrio harveyi strain ATCC BAA-1116 did not contain any SI gene cassette, only a solitary intIA gene. Repeated attempts to identify a VXR (VHR) sequence in 200 kb± intIA gene were unsuccessful. Two sequences that fulfilled the VHR criteria were found, one in plasmid pBIBHAR (CP000791) and the other in chromosome II. Draft genome sequence of a V. harveyi strain CAIM 1792 showed the presence of SI sequence of 87.2 kb in chromosome I (147). Therefore, the presence of a solitary intIA gene in ATCC BAA-1116 strain may indicate the early stage of SI evolution or loss of an entire gene cassette region. The NCBI database does not contain any completed genome of V. campbellii, we did find in V. campbellii strain DS40M4 an SI element of 68.9 kb in size. The percent GC content of a region can be used as an indicator of evolutionary origin. We examined the %GC content of the SI elements compared to the entire genome and found that the SI %GC content was always lower (Table 4). To date, only a handful of gene cassette functions have been demonstrated such as antibiotic resistance, virulence and metabolic activity (139, 141, 144, 145). The majority of genes within SIs do not have any counterpart in the GenBank database and their function and role in cell fitness and survival remains to be determined.
Acknowledgments
Research on Vibrio species in the Boyd group is supported by a National Science Foundation grant IOS-0918429 and a National Science Foundation CAREER award DEB-0844409. JBL was supported by the Chemistry-Biology Interface graduate program and a University of Delaware graduate student fellowship. WBW was supported in part by a Dissertation fellowship and JJK was supported by a BOYCAST Indian government fellowship.
References
- 1.Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int J Syst Evol Microbiol. 2007;57:2823–2829. doi: 10.1099/ijs.0.65081-0. [DOI] [PubMed] [Google Scholar]
- 2.Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, Alves N, Jr, Ussery DW, Iida T, Thompson FL. Genomic taxonomy of Vibrios. BMC Evol Biol. 2009;9:258. doi: 10.1186/1471-2148-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol. 2005;71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson FL, Thompson CC, Dias GM, Naka H, Dubay C, Crosa JH. The genus Listonella MacDonell and Colwell 1986 is a later heterotypic synonym of the genus Vibrio Pacini 1854 (Approved Lists 1980)–a taxonomic opinion. Int J Syst Evol Microbiol. 2011;61:3023–3027. doi: 10.1099/ijs.0.030015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trucksis M, Michalski J, Deng YK, Kaper JB. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci U S A. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 8.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Lin B, Wang Z, Malanoski AP, O’Grady EA, Wimpee CF, Vuddhakul V, Alves N, Jr, Thompson FL, Gomez-Gil B, Vora GJ. Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep. 2010;2:81–89. doi: 10.1111/j.1758-2229.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Annu Rev Microbiol. 2011;65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 13.Zhao JJ, Chen C, Zhang LP, Hu CQ. Cloning, identification, and characterization of the rpoS-like sigma factor rpoX from Vibrio alginolyticus. J Biomed Biotechnol. 2009;2009:126986. doi: 10.1155/2009/126986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao X, Studer SV, Wassarman K, Zhang Y, Ruby EG, Miyashiro T. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. MBio. 2012;3 doi: 10.1128/mBio.00285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarter LL. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol. 1995;177:1595–1609. doi: 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarter LL, Wright ME. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J Bacteriol. 1993;175:3361–3371. doi: 10.1128/jb.175.11.3361-3371.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart BJ, McCarter LL. Lateral flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 2003;185:4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitaker WB, Richards GP, Boyd EF. Loss of sigma factor RpoN increases intestinal colonization of Vibrio parahaemolyticus in an adult mouse model. Infect Immun. 2014;82:544–556. doi: 10.1128/IAI.01210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho TD, Ellermeier CD. Extra cytoplasmic function sigma factor activation. Curr Opin Microbiol. 2012;15:182–188. doi: 10.1016/j.mib.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappas M, Bose D, Zhang X. Bacterial enhancer-binding proteins: unlocking sigma54-dependent gene transcription. Curr Opin Struct Biol. 2007;17:110–116. doi: 10.1016/j.sbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz FH, Schoolnik GK. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merrell D, Tischler A, Lee S, Camilli A. Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect Immun. 2000;68:6691–6696. doi: 10.1128/iai.68.12.6691-6696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosche TM, Smith DJ, Parker EE, Oliver JD. RpoS involvement and requirement for exogenous nutrient for osmotically induced cross protection in Vibrio vulnificus. FEMS Microbiol Ecol. 2005;53:455–462. doi: 10.1016/j.femsec.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Hulsmann A, Rosche TM, Kong IS, Hassan HM, Beam DM, Oliver JD. RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl Environ Microbiol. 2003;69:6114–6120. doi: 10.1128/AEM.69.10.6114-6120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber B, Croxatto A, Chen C, Milton DL. RpoS induces expression of the Vibrio anguillarum quorum-sensing regulator VanT. Microbiology. 2008;154:767–780. doi: 10.1099/mic.0.2007/014167-0. [DOI] [PubMed] [Google Scholar]
- 27.Whitaker WB, Parent MA, Naughton LM, Richards GP, Blumerman SL, Boyd EF. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl Environ Microbiol. 2010;76:4720–4729. doi: 10.1128/AEM.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith B, Oliver J. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol. 2006;72:1445–1451. doi: 10.1128/AEM.72.2.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coutard F, Lozach S, Pommepuy M, Hervio-Heath D. Real-time reverse transcription-PCR for transcriptional expression analysis of virulence and housekeeping genes in viable but nonculturable Vibrio parahaemolyticus after recovery of culturability. Appl Environ Microbiol. 2007;73:5183–5189. doi: 10.1128/AEM.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF. Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl Environ Microbiol. 2012;78:7455–7466. doi: 10.1128/AEM.01594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect Immun. 2012;80:1834–1845. doi: 10.1128/IAI.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haines-Menges B, Whitaker WB, Boyd EF. Alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect Immun. 2014;82:3667–3677. doi: 10.1128/IAI.01854-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian Y, Wang Q, Liu Q, Ma Y, Cao X, Z Y. Role of RpoS in stress survival, synthesis of extracellular autoinducer 2, and virulence in Vibrio alginolyticus. Arch Microbiol. 2008;190:585–594. doi: 10.1007/s00203-008-0410-6. [DOI] [PubMed] [Google Scholar]
- 34.Dong T, Schellhorn HE. Role of RpoS in virulence of pathogens. Infect Immun. 2009;78:887–897. doi: 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu GK, Chowdhury R, Das J. The rpoH gene encoding sigma 32 homolog of Vibrio cholerae. Gene. 1997;189:203–207. doi: 10.1016/s0378-1119(96)00849-9. [DOI] [PubMed] [Google Scholar]
- 36.Slamti L, Livny J, Waldor MK. Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J Bacteriol. 2007;189:351–362. doi: 10.1128/JB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawagishi I, Nakada M, Nishioka N, Homma M. Cloning of a Vibrio alginolyticus rpoN gene that is required for polar flagellar formation. J Bacteriol. 1997;179:6851–6854. doi: 10.1128/jb.179.21.6851-6854.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Toole R, Milton DL, Horstedt P, Wolf-Watz H. RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology. 1997;143(Pt 12):3849–3859. doi: 10.1099/00221287-143-12-3849. [DOI] [PubMed] [Google Scholar]
- 39.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 40.Wolfe AJ, Millikan DS, Campbell JM, Visick KL. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl Environ Microbiol. 2004;70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao B, Mo ZL, Xiao P, Pan HJ, Lan X, Li GY. Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-012-4372-x. [DOI] [PubMed] [Google Scholar]
- 44.Klose KE, Mekalanos JJ. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 45.Klose KE, Mekalanos JJ. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klose KE, Novik V, Mekalanos JJ. Identification of multiple sigma54-dependent transcriptional activators in Vibrio cholerae. J Bacteriol. 1998;180:5256–5259. doi: 10.1128/jb.180.19.5256-5259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prouty MG, Correa NE, Klose KE. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol. 2001;39:1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 48.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJ, Yildiz F, Klose KE. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol. 2009;191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Millikan DS, Ruby EG. FlrA, a sigma54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol. 2003;185:3547–3557. doi: 10.1128/JB.185.12.3547-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernard CS, Brunet YR, Gavioli M, Lloubes R, Cascales E. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gode-Potratz CJ, McCarter LL. Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol. 2011;193:4224–4237. doi: 10.1128/JB.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabich AJ, Leatham MP, Grissom JE, Wiley G, Lai H, Najar F, Roe BA, Cohen PS, Conway T. Genotype and phenotypes of an intestine-adapted Escherichia coli K-12 mutant selected by animal passage for superior colonization. Infect Immun. 2011;79:2430–2439. doi: 10.1128/IAI.01199-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hild E, Takayama K, Olsson RM, Kjelleberg S. Evidence for a role of rpoE in stressed and unstressed cells of marine Vibrio angustum strain S14. J Bacteriol. 2000;182:6964–6974. doi: 10.1128/jb.182.24.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovacikova G, Skorupski K. The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect Immun. 2002;70:5355–5362. doi: 10.1128/IAI.70.10.5355-5362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathur J, Davis BM, Waldor MK. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol. 2007;63:848–858. doi: 10.1111/j.1365-2958.2006.05544.x. [DOI] [PubMed] [Google Scholar]
- 57.Brown RN, Gulig PA. Roles of RseB, sigmaE, and DegP in virulence and phase variation of colony morphotype of Vibrio vulnificus. Infect Immun. 2009;77:3768–3781. doi: 10.1128/IAI.00205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis BM, Waldor MK. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res. 2009;37:5757–5767. doi: 10.1093/nar/gkp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rattanama P, Thompson JR, Kongkerd N, Srinitiwarawong K, Vuddhakul V, Mekalanos JJ. Sigma E regulators control hemolytic activity and virulence in a shrimp pathogenic Vibrio harveyi. PLoS One. 2012;7:e32523. doi: 10.1371/journal.pone.0032523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 61.Miller VL, Mekalanos JJ. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci U S A. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SH, Hava DL, Waldor MK, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 64.Pfau JD, Taylor RK. Genetic footprint on the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol. 1996;20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 65.Skorupski K, Taylor RK. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 66.DiRita VJ, Engleberg NC, Heath A, Miller A, Crawford JA, Yu R. Virulence gene regulation inside and outside. Philos Trans R Soc Lond B Biol Sci. 2000;355:657–665. doi: 10.1098/rstb.2000.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das S, Chakrabortty A, Banerjee R, Roychoudhury S, Chaudhuri K. Comparison of global transcription responses allows identification of Vibrio cholerae genes differentially expressed following infection. FEMS Microbiol Lett. 2000;190:87–91. doi: 10.1111/j.1574-6968.2000.tb09267.x. [DOI] [PubMed] [Google Scholar]
- 68.Osorio CR, Klose KE. A region of the transmembrane regulatory protein ToxR that tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among Vibrio species. J Bacteriol. 2000;182:526–528. doi: 10.1128/jb.182.2.526-528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Provenzano D, Klose KE. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci U S A. 2000;97:10220–10224. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun. 2000;68:1491–1497. doi: 10.1128/iai.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Provenzano D, Lauriano CM, Klose KE. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J Bacteriol. 2001;183:3652–3662. doi: 10.1128/JB.183.12.3652-3662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Childers BM, Klose KE. Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol. 2007;2:335–344. doi: 10.2217/17460913.2.3.335. [DOI] [PubMed] [Google Scholar]
- 74.Krukonis ES, Yu RR, Dirita VJ. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol. 2000;38:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 75.Morgan S, Felek S, Gadwal S, Koropatkin NM, Perry JW, Bryson AB, Krukonis E. The two faces of ToxR: activator of ompU, co-regulator of toxT in Vibrio cholerae. Mol Microbiol. 2011;81:113–128. doi: 10.1111/j.1365-2958.2011.07681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goss T, Morgan SJ, French EL, Krukonis E. ToxR recognizes a direct repeat element in the toxT, ompU, ompT, and ctxA promoters of Vibrio cholerae to regulate transcription. Infect Immun. 2013;81:884–895. doi: 10.1128/IAI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Merrell DS, Bailey C, Kaper JB, Camilli A. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J Bacteriol. 2001;183:2746–2754. doi: 10.1128/JB.183.9.2746-2754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duperthuy M, Binesse J, Le Roux F, Romestand B, Caro A, Got P, Givaudan A, Mazel D, Bachere E, Destoumieux-Garzon D. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol. 2010;12:951–963. doi: 10.1111/j.1462-2920.2009.02138.x. [DOI] [PubMed] [Google Scholar]
- 79.Duperthuy M, Schmitt P, Garzon E, Caro A, Rosa RD, Le Roux F, Lautredou-Audouy N, Got P, Romestand B, de Lorgeril J, Kieffer-Jaquinod S, Bachere E, Destoumieux-Garzon D. Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc Natl Acad Sci U S A. 2011;108:2993–2998. doi: 10.1073/pnas.1015326108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gode-Potratz CJ, Chodur DM, McCarter LL. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J Bacteriol. 2010;192:6025–6038. doi: 10.1128/JB.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bassler B, Gibbons P, Roseman S. Chemotaxis to chitin oligosaccharides by Vibrio furnissii, a chitinivorous marine bacterium. Biochem Biophys Res Commun. 1989;161:1172–1176. doi: 10.1016/0006-291x(89)91365-x. [DOI] [PubMed] [Google Scholar]
- 82.Bassler BL, Gibbons PJ, Yu C, Roseman S. Chitin utilization by marine bacteria. Chemotaxis to chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24268–24275. [PubMed] [Google Scholar]
- 83.Bassler BL, Yu C, Lee YC, Roseman S. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24276–24286. [PubMed] [Google Scholar]
- 84.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012;14:1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- 85.Hjerde E, Lorentzen MS, Holden MT, Seeger K, Paulsen S, Bason N, Churcher C, Harris D, Norbertczak H, Quail MA, Sanders S, Thurston S, Parkhill J, Willassen NP, Thomson NR. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics. 2008;9:616. doi: 10.1186/1471-2164-9-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keyhani NO, Wang LX, Lee YC, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Characterization of an N,N′-diacetyl-chitobiose transport system. J Biol Chem. 1996;271:33409–33413. doi: 10.1074/jbc.271.52.33409. [DOI] [PubMed] [Google Scholar]
- 87.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park JK, Keyhani NO, Roseman S. Chitin catabolism in the marine bacterium Vibrio furnissii. Identification, molecular cloning, and characterization of A N, N′-diacetylchitobiose phosphorylase. J Biol Chem. 2000;275:33077–33083. doi: 10.1074/jbc.M001042200. [DOI] [PubMed] [Google Scholar]
- 89.Pruzzo C, Vezzulli L, Colwell RR. Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol. 2008;10:1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 90.Svitil AL, Chadhain S, Moore JA, Kirchman DL. Chitin Degradation Proteins Produced by the Marine Bacterium Vibrio harveyi Growing on Different Forms of Chitin. Appl Environ Microbiol. 1997;63:408–413. doi: 10.1128/aem.63.2.408-413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keyhani NO, Li XB, Roseman S. Chitin catabolism in the marine bacterium Vibrio furnissii. Identification and molecular cloning of a chitoporin. J Biol Chem. 2000;275:33068–33076. doi: 10.1074/jbc.M001041200. [DOI] [PubMed] [Google Scholar]
- 92.Keyhani NO, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic beta-N-acetylglucosaminidase. J Biol Chem. 1996;271:33425–33432. doi: 10.1074/jbc.271.52.33425. [DOI] [PubMed] [Google Scholar]
- 93.Keyhani NO, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J Biol Chem. 1996;271:33414–33424. doi: 10.1074/jbc.271.52.33414. [DOI] [PubMed] [Google Scholar]
- 94.Keyhani NO, Roseman S. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc Natl Acad Sci U S A. 1997;94:14367–14371. doi: 10.1073/pnas.94.26.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keyhani NO, Wang LX, Lee YC, Roseman S. The chitin disaccharide, N,N′-diacetylchitobiose, is catabolized by Escherichia coli and is transported/phosphorylated by the phosphoenolpyruvate:glycose phosphotransferase system. J Biol Chem. 2000;275:33084–33090. doi: 10.1074/jbc.M001043200. [DOI] [PubMed] [Google Scholar]
- 96.Benedek O, Schubert S. Mobility of the Yersinia High-Pathogenicity Island (HPI): transfer mechanisms of pathogenicity islands (PAIS) revisited (a review) Acta Microbiol Immunol Hung. 2007;54(2):89–105. doi: 10.1556/AMicr.54.2007.2.1. [DOI] [PubMed] [Google Scholar]
- 97.Boyd EF, Cohen AL, Naughton LM, Ussery DW, Binnewies TT, Stine OC, Parent MA. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 2008;8:110. doi: 10.1186/1471-2180-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalburge SS, Polson SW, Boyd Crotty K, Katz L, Turnsek M, Tarr CL, Martinez-Urtaza J, Boyd EF. Complete Genome Sequence of Vibrio parahaemolyticus Environmental Strain UCM-V493. Genome Announc. 2014;2 doi: 10.1128/genomeA.00159-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quirke AM, Reen FJ, Claesson MJ, Boyd EF. Genomic island identification in Vibrio vulnificus reveals significant genome plasticity in this human pathogen. Bioinformatics. 2006;22:905–910. doi: 10.1093/bioinformatics/btl015. [DOI] [PubMed] [Google Scholar]
- 100.Varki A. Diversity in the sialic acids. Glycobiology. 1992;2:25–40. doi: 10.1093/glycob/2.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Varki A. Loss of N-glycolylneuraminic acid in humans. Mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthrop. 2001;33:54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Almagro-Moreno S, Boyd EF. Bacterial catabolism of nonulosonic (sialic) acid and fitness in the gut. Gut Microbes. 2010;1:45–50. doi: 10.4161/gmic.1.1.10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jermyn WS, Boyd EF. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology. 2002;148:3681–3693. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- 106.Jermyn WS, Boyd EF. Molecular evolution of Vibrio pathogenicity island-2 (VPI-2): mosaic structure among Vibrio cholerae and Vibrio mimicus natural isolates. Microbiology. 2005;151:311–322. doi: 10.1099/mic.0.27621-0. [DOI] [PubMed] [Google Scholar]
- 107.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci U S A. 2005;102:3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Y, Johnson JA, Pusch GD, Morris JG, Jr, Stine OC. The genome of non-O1 Vibrio cholerae NRT36S demonstrates the presence of pathogenic mechanisms that are distinct from O1 Vibrio cholerae. Infect Immun. 2007 doi: 10.1128/IAI.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol. 2008;190:636–647. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Okada N, Iida T, Park KS, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun. 2009;77:904–913. doi: 10.1128/IAI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lubin JB, Kingston JJ, Chowdhury N, Boyd EF. Sialic acid catabolism and transport gene clusters are lineage specific in Vibrio vulnificus. Appl Environ Microbiol. 2012;78:3407–3415. doi: 10.1128/AEM.07395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Almagro-Moreno S, Boyd EF. Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine. Infect Immun. 2009;77:3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chowdhury N, Norris J, McAlister E, Lau SY, Thomas GH, Boyd EF. The VC1777-VC1779 proteins are members of a sialic acid-specific subfamily of TRAP transporters (SiaPQM) and constitute the sole route of sialic acid uptake in the human pathogen Vibrio cholerae. Microbiology. 2012;158:2158–2167. doi: 10.1099/mic.0.059659-0. [DOI] [PubMed] [Google Scholar]
- 114.Thomas GH, Boyd EF. On sialic acid transport and utilization by Vibrio cholerae. Microbiology. 2011;157:3253–3254. doi: 10.1099/mic.0.054692-0. discussion 3254–3255. [DOI] [PubMed] [Google Scholar]
- 115.Jeong H, Oh MH, Kim BS, Lee MY, Han HJ, Choi S. The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect Immun. 2009;77:3209–3217. doi: 10.1128/IAI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mulligan C, Leech AP, Kelly DJ, Thomas GH. The membrane proteins SiaQ and SiaM form an essential stoichiometric complex in the sialic acid tripartite ATP-independent periplasmic (TRAP) transporter SiaPQM (VC1777-1779) from Vibrio cholerae. J Biol Chem. 2011;287:3598–3608. doi: 10.1074/jbc.M111.281030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma SK, Moe TS, Srivastava R, Chandra D, Srivastava BS. Functional characterization of VC1929 of Vibrio cholerae El Tor: role in mannose-sensitive haemagglutination, virulence and utilization of sialic acid. Microbiology. 2011;157:3180–3186. doi: 10.1099/mic.0.050245-0. [DOI] [PubMed] [Google Scholar]
- 118.Faruque SM, Mekalanos JJ. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 2003;11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 119.Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, Reeves PR. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U S A. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 121.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davies B, Bogard R, Young T, Mekalanos J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Feng L, Reeves PR, Lan R, Ren Y, Gao C, Zhou Z, Cheng J, Wang W, Wang J, Qian W, Li D, Wang L. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS One. 2008;3:e4053. doi: 10.1371/journal.pone.0004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Grim CJ, Hasan NA, Taviani E, Haley B, Chun J, Brettin TS, Bruce DC, Detter JC, Han CS, Chertkov O, Challacombe J, Huq A, Nair GB, Colwell RR. Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J Bacteriol. 2010;192:3524–3533. doi: 10.1128/JB.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Garza DR, Thompson CC, Loureiro EC, Dutilh BE, Inada DT, Junior EC, Cardoso JF, Nunes MR, de Lima CP, Silvestre RV, Nunes KN, Santos EC, Edwards RA, Vicente AC, de Sa Morais LL. Genome-wide study of the defective sucrose fermenter strain of Vibrio cholerae from the Latin American cholera epidemic. PLoS One. 2012;7:e37283. doi: 10.1371/journal.pone.0037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis. 2011;17:2113–2121. doi: 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nair G, Ramamurthy T, Bhattacharya S, Dutta B, Takeda Y, Sack D. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oliver JD. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol Infect. 2005;133:383–391. doi: 10.1017/s0950268805003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oliver JD, Warner RA, Cleland DR. Distribution and ecology of Vibrio vulnificus and other lactose-fermenting marine vibrios in coastal waters of the southeastern United States. Appl Environ Microbiol. 1982;44:1404–1414. doi: 10.1128/aem.44.6.1404-1414.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jones M, Oliver JD. Vibrio vulnificus: disease and pathogenesis. Infect Immun. 2009;77:1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL, Liu YM, Chen HJ, Shen AB, Li JC, Su TL, Shao CP, Lee CT, Hor LI, Tsai SF. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 2003;13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cohen AL, Oliver JD, DePaola A, Feil EJ, Boyd EF. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl Environ Microbiol. 2007;73:5553–5565. doi: 10.1128/AEM.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hurley CC, Quirke A, Reen FJ, Boyd EF. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics. 2006;7:104. doi: 10.1186/1471-2164-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Valla S, Frydenlund K, Coucheron DH, Haugan K, Johansen B, Jorgensen T, Knudsen G, Strom A. Development of a gene transfer system for curing of plasmids in the marine fish pathogen Vibrio salmonicida. Appl Environ Microbiol. 1992;58:1980–1985. doi: 10.1128/aem.58.6.1980-1985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vezzi A, Campanaro S, D’Angelo M, Simonato F, Vitulo N, Lauro FM, Cestaro A, Malacrida G, Simionati B, Cannata N, Romualdi C, Bartlett DH, Valle G. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science. 2005;307:1459–1461. doi: 10.1126/science.1103341. [DOI] [PubMed] [Google Scholar]
- 137.Mazel D, Dychinco B, Webb VA, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 138.Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 139.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 140.Chowdhury N, Asakura M, Neogi SB, Hinenoya A, Haldar S, Ramamurthy T, Sarkar BL, Faruque SM, Yamasaki S. Development of simple and rapid PCR-fingerprinting methods for Vibrio cholerae on the basis of genetic diversity of the superintegron. J Appl Microbiol. 2010;109:304–312. doi: 10.1111/j.1365-2672.2009.04658.x. [DOI] [PubMed] [Google Scholar]
- 141.Rowe-Magnus DA, Guerout AM, Mazel D. Super-integrons. Res Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 142.Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-research0011. RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stokes HW, O’Gorman DB, Recchia GD, Parsekhian M, Hall RM. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 144.Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rowe-Magnus DA, Guerout AM, Ploncard P, Dychinco B, Davies J, Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci U S A. 2001;98:652–657. doi: 10.1073/pnas.98.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hall RM, Collis CM. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updat. 1998;1:109–119. doi: 10.1016/s1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 147.Espinoza-Valles I, Soto-Rodriguez S, Edwards RA, Wang Z, Vora GJ, Gomez-Gil B. Draft genome sequence of the shrimp pathogen Vibrio harveyi CAIM 1792. J Bacteriol. 2012;194:2104. doi: 10.1128/JB.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


