Summary
How genes shape diverse plant and animal body forms is a key question in biology. Unlike animal cells, plant cells are confined by rigid cell walls, and cell division plane orientation and growth rather than cell movement determine overall body form. The emergence of plants on land coincided with a new capacity to rotate stem cell divisions through multiple planes, and this enabled three-dimensional (3D) forms to arise from ancestral forms constrained to 2D growth. The genes involved in this evolutionary innovation are largely unknown. The evolution of 3D growth is recapitulated during the development of modern mosses when leafy shoots arise from a filamentous (2D) precursor tissue. Here, we show that a conserved, CLAVATA peptide and receptor-like kinase pathway originated with land plants and orients stem cell division planes during the transition from 2D to 3D growth in a moss, Physcomitrella. We find that this newly identified role for CLAVATA in regulating cell division plane orientation is shared between Physcomitrella and Arabidopsis. We report that roles for CLAVATA in regulating cell proliferation and cell fate are also shared and that CLAVATA-like peptides act via conserved receptor components in Physcomitrella. Our results suggest that CLAVATA was a genetic novelty enabling the morphological innovation of 3D growth in land plants.
Keywords: land plant evolution, plant evo-devo, plant cell division plane, 3D growth, CLAVATA
Highlights
-
•
CLAVATA originated in the last common ancestor of land plants
-
•
CLAVATA regulates cell proliferation, fate, and growth in Physcomitrella
-
•
CLAVATA orients cell division planes in Physcomitrella and Arabidopsis
-
•
CLEs act via receptors that are conserved between Physcomitrella and Arabidopsis
Whitewoods, Cammarata, et al. show that a conserved CLAVATA (CLV) pathway arose in the last common ancestor of land plants. CLV regulates cell division plane orientation during the 2D to 3D growth transition in a moss, and roles for CLV are shared between mosses and flowering plants, suggesting that CLV enabled 3D growth to arise in land plants.
Introduction
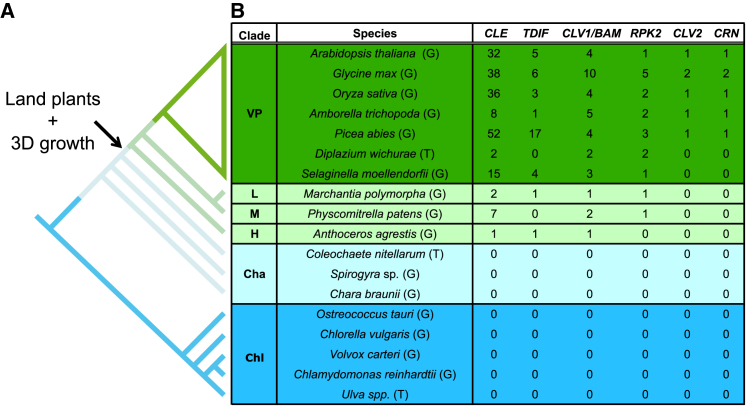
The conquest of land was enabled by a series of innovations that allowed plant forms to radiate and occupy new volumes of space in the sub-aerial environment [1]. Among these, the innovation of shooting systems with organs positioned radially around an upright stem stands out as a primer for massively increased plant productivity and diversity [1]. Such three-dimensional (3D) growth forms first arose as a consequence of a novel stem cell function gained by land plants, namely the capacity to rotate stem cell divisions through multiple plane orientations [1, 2, 3]. The algal sister lineages of land plants are unable to rotate stem cell divisions through multiple planes and are therefore generally constrained to smaller filamentous or mat-like (two-dimensional [2D]) growth forms (Figure 1A) [1, 3]. The evolutionary transition from 2D to 3D growth is recapitulated during the development of modern mosses when a branching, filamentous (protonemal) precursor tissue (2D) gives rise to 3D gamete-producing leafy shoots (gametophores) [6]. Previous studies have shown that gametophores and filament branches initiate similarly as hemispherical outgrowths from parent filaments and that their divergent 2D or 3D fates are specified stochastically by APETALA2-type (APB) transcription factor activity [7]. During a single-celled stage of outgrowth development, persistent APB activity and cell swelling mark a switch to gametophore fate (3D), whereas loss of APB activity marks filament fate (2D) [6, 7]. A strongly oblique cell division is the first reliable morphological marker of gametophore development [6, 7]. This is followed by a second oblique apical cell division, which is approximately perpendicular to the first, after which division planes rotate during two successive rounds of division to establish a tetrahedral apical stem cell [6]. The tetrahedral apical cell divides in spiraling planes to replace itself and produce daughter cells that generate the 3D gametophore axis and leaves [6]. The mechanisms regulating such novel and rotating stem cell division plane orientations during evolutionary and developmental transitions to 3D growth are unknown.
Figure 1.
The CLV Pathway Originated in the Last Common Ancestor of Land Plants, Concomitantly with 3D Growth
(A) Phylogenetic relationships among land plants and their freshwater algal sister lineages redrawn from [4] and [5], respectively. Although chlorophytes and charophytes undergo stem cell divisions in a single orientation (2D growth), land plants undergo stem cell divisions in multiple orientations to generate elaborate three-dimensional forms (3D growth).
(B) The number of CLV pathway homologs was determined by BLAST against genome or draft genome (G) and transcriptome (T) databases as described in STAR Methods. Cha, charophytes; Chl, chlorophytes; H, hornworts; L, liverworts; M, mosses; VP, vascular plants.
See also Figures S1, S2, and S3 and Table S1.
In Arabidopsis, the CLAVATA (CLV) and WUSCHEL (WUS) pathways act in a feedback loop to regulate many aspects of stem cell function, including cell fate [8, 9], proliferation [9, 10, 11], and growth [12]. CLV3 encodes a small, secreted peptide that is expressed in the upper cell layers of the central zone and can move throughout the meristem [13, 14, 15]. CLV1 is expressed in the underlying cell layers of the central zone and encodes a receptor-like kinase that acts as a receptor for CLV3 [11, 16] in conjunction with CLV2, CORYNE (CRN), RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2), and BARELY ANY MERISTEM (BAM) [17, 18]. WUS activity promotes meristem cell proliferation [19], and CLV signaling restricts the size of the WUS expression domain [13]. WUS acts non-cell autonomously, moving from the organizing center to the uppermost meristem cell layers, where it promotes CLV3 expression [20], thereby closing the feedback loop that maintains meristem size.
Results
The CLAVATA Pathway Originated in the Last Common Ancestor of Land Plants
To determine how the CLV pathway evolved and identify potential roles for CLV in Physcomitrella stem cell function, we first queried publicly accessible genome and transcriptome databases from a wide range of green algae and land plants for CLV3-like (CLE), CLV1/BAM, RPK2, CLV2, and CRN homologs (Figure 1B; Table S1). We found no CLV pathway homologs in the chlorophyte or charophyte algae sampled but found at least one CLE homolog and one CLV1/BAM homolog in each early-diverging bryophyte lineage and all other land plants, suggesting that the core CLV signaling module comprises at least one CLE peptide and a CLV/BAM receptor-like kinase. RPK2 homologs were present in all land plants sampled except the hornwort, Anthoceros agrestis. In Physcomitrella, we identified seven genes with a conserved CLE domain encoding a 12-amino-acid peptide motif similar to CLV3, but sequences outside the conserved CLE domain were divergent (Figure 1; Table S1). The genome encodes four CLV3-like peptides: PpCLEs 1, 2, and 3 encode the peptide motif RMVPTGPNPLHN; PpCLE4 encodes the motif RMVPSGPNPLHN; PpCLEs 5 and 6 encode the motif RLVPTGPNPLHN; and PpCLE7 encodes the motif RVVPTGPNPLHN. Neighbor-joining phylogenetic reconstructions showed that, although hornworts and liverworts have CLEs resembling the tracheary element differentiation inhibitory factor (TDIF)-like CLEs that regulate vascular development in Arabidopsis, Physcomitrella does not, consistent with an evolutionary loss in mosses (Figure S1; Data S1). Receptor-like kinase phylogenies were reconstructed by maximum likelihood analysis using amino acids from the conserved kinase domain (Figures S2 and S3; Data S2 and S3). Clades encompassing CLV1/BAM-like sequences from each land plant lineage or containing RPK2-like sequences from each lineage except hornworts were resolved. Both CLV1/BAM and RPK2 phylogenies were broadly congruent with current hypotheses of land plant evolution [4, 21], thereby indicating orthology. Two Physcomitrella genes were incorporated in the CLV1/BAM clade, and these were named Physcomitrella CLAVATA1a and 1b (PpCLV1a and PpCLV1b). One RPK2 homolog was found and named PpRPK2, but no CLV2 or CRN homologs were found. These sequence data indicate that the core components of the CLV pathway first arose in the last common ancestor of land plants, alongside the evolutionary innovation of 3D growth [22].
Physcomitrella CLAVATA Pathway Components Are Expressed during the 3D Growth Phase
To investigate Physcomitrella CLV activity, we first analyzed gene expression patterns in relation to the transition between 2D filamentous and 3D gametophore growth (Figures 2, S4, and S5). By RT-PCR, we detected PpCLE1, 2, and 7 peptide-encoding gene expression in gametophores (Figure S4). We were unable to detect expression of PpCLEs 3, 4, and 5, but we found PpCLE6 expression in protonemal filaments. Receptor-encoding genes PpRPK2, PpCLV1a, and PpCLV1b were co-expressed in gametophores, although PpRPK2 expression was evident earlier than PpCLV1a and PpCLV1b in day 10 filamentous tissues (Figure S4). These results were broadly consistent with reports from transcriptome data (Figure S5) [23, 24]. We also constructed promoter::NLSGFPGUS (promoter::NGG) fusion lines for PpCLE1, PpCLE2, PpCLE7, PpCLV1a, PpCLV1b, and PpRPK2 as RT-PCR showed that these 6 genes were upregulated at around the time of gametophore initiation (see Strategy for generation of promoter::NLSGUSGFP reporter lines in Methods S1; Figure 2). In 3-week-old spot cultures (Figures 2A–2F), PpCLE1::NGG, PpCLE2::NGG, PpCLE7::NGG, and PpCLV1a::NGG lines accumulated local signal in various protonemal cell types around the buds (Figures 2G–2J and 2M–2P). PpCLV1b::NGG and PpRPK2::NGG lines accumulated signal in buds, and the signal was strongest toward the apex (Figures 2K, 2L, 2Q, and 2R). Whereas all lines accumulated signal in gametophore axes and leaves (Figures 2S–2J’), there was variation in the pattern, timing, and intensity of signal accumulation between lines. Notably, PpCLE1::NGG, PpCLE2::NGG, PpCLE7::NGG, and PpCLV1a::NGG signal accumulation in gametophores was delayed with respect to PpCLV1b::NGG and PpRPK2::NGG lines (Figures 2M–2X). These beta-glucuronidase (GUS) accumulation patterns suggested highly dynamic foci of expression for PpCLEs 1, 2, and 7 and PpCLV1a, PpCLV1b, and PpRPK2 in Physcomitrella, prompting us to investigate roles for CLV pathway components in gametophore initiation and development, i.e., during the transition to 3D growth.
Figure 2.
CLV Pathway Components Are Expressed in Physcomitrella Protonemata and Gametophores
(A– J’) GUS staining of PpCLE1::NGG (A, G, M, S, Y, and E’), PpCLE2::NGG (B, H, N, T, Z, and F’), PpCLE7::NGG (C, I, O, U, A’, and G’), PpCLV1a::NGG (D, J, P, V, B’, and H’), PpCLV1b::NGG (E, K, Q, W, C’, and I’), and PpRPK2::NGG (F, L, R, X, D’, and J’) lines revealed complex expression dynamics.
Although PpCLE::NGG and PpCLV1a::NGG signal accumulated in protonemal tissues close to buds (G–J and M–P; arrows indicate signal in protonemata), PpCLV1b::NGG and PpRPK2::NGG signal accumulated mainly in the apical region of buds (Q and R). At two later stages of gametophore development (S–X and Y–J’), all promoters were active in gametophores, although the patterns and intensity of activity varied between reporters and by developmental stage. PpCLE1::NGG lines stained most strongly in leaves (S, Y, and E’), PpCLE2::NGG lines stained most strongly in leaves and gametophore bases (T, Z, and F’), and PpCLE7::NGG lines accumulated stain in rhizoid tips (arrow in U), leaf bases (arrow in A’), and hairs around the apex and the gametophore axis (G’). PpCLV1a::NGG lines did not stain intensely at early stages of gametophore development (P and V) but accumulated signal in gametophore axes and leaves at later stages (B' and H'). In contrast, PpCLV1b::NGG and PpRPK2::NGG lines accumulated signal in gametophore axes and leaves from early stages of development (W and X), and strong signal was detected in branches initiating at gametophore bases (arrows in X, C’, and D’).
All tissues in (A)–(D’) were stained in a solution containing 0.5 mM FeCN for times specified in (A)–(F), and gametophores in (E’)–(J’) were stained three times longer in a solution containing 2 mM FeCN.
The scale bars in (A)–(F) represent 1 mm, the scale bars in (M)–(R) represent 100 μm, and insets in (G)–(L) indicate position of buds in (M)–(R). The scale bars in (S)–(J’) represent 1 mm. See also Methods S1 and Table S4.
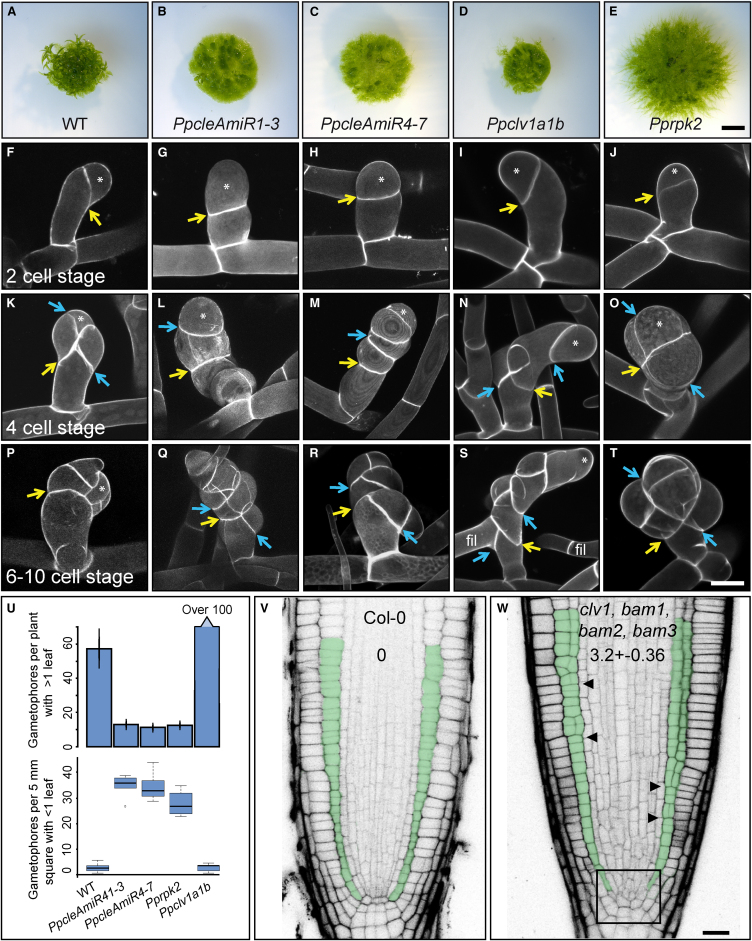
Physcomitrella Mutants Lacking CLAVATA Function Have a Defective 2D to 3D Growth Transition
To identify the functions of CLV pathway components, we used artificial microRNAs (AmiRNAs) to silence expression of PpCLEs 1, 2, and 3 and PpCLEs 4, 5, 6, and 7 (see Strategy for generating PpcleAmiR lines in Methods S1). We used a CRISPR-Cas9 approach to disrupt the function of PpCLV1 paralogs (see CRISPR/Cas9 strategy for generating Ppclv1 mutants in Methods S1), and gene targeting was used to abrogate PpRPK2 function (see Strategy for generating Pprpk2 KO lines in Methods S1). PpcleAmiR1-3, PpcleAmiR4-7, Ppclv1a1b, and Pprpk2 lines were able to form dense protonemal tissues and thus had a relatively normal 2D growth phase (Figures 3A–3E). However, all four mutant classes had defective development during the 3D growth phase, with a reduction in the overall number of mature gametophores and defects in gametophore development (Figures 3A–3E and 3U). Further examination revealed many more gametophore buds with 1 or fewer leaves in PpcleAmiR1-3, PpcleAmiR4-7, and Pprpk2 mutants than in wild-type (WT) plants (Figure 3U), and Ppclv1a1b mutants had many small gametophores arrested at a later stage of development (Figure 3U). These data suggested early defects in gametophore development with potential feedback onto the gametophore initiation process. To determine how WT and mutant phenotypes diverged during development, we imaged gametophore buds at 2-cell, 4-cell, and a later stage of bud development [6] (Figures 3F–3T). Although WT gametophores initiated normally and showed characteristic oblique cell division plane orientations, the plane of the first division was strongly disrupted in PpcleAmiR1-3 and PpcleAmiR4-7 mutants, and it was set at a shallow angle relative to the main growth axis (compare Figure 3F to Figures 3G and 3H). A second round of cell division from the apical cell also had misset division planes that were frequently parallel rather than perpendicular to the first division plane, and a subset of gametophores therefore formed finger-like projections in place of gametophores (compare Figure 3K to Figures 3L and 3M). At developmental stages where the tetrahedral shape of the apical cell is normally established [6], mutants also had defects indicating problems with growth and cell fate specification, appearing to reiterate divisions normally characteristic of the first gametophore initial (compare Figure 3P to Figures 3Q and 3R). Ppclv1a1b mutant phenotypes diverged from WT after the 2-cell stage, subsequently showing a similar pattern of division to PpcleAmiR1-3 and PpcleAmiR4-7 mutants (Figures 3K–3N and 3P–3S), and some cells reverted to filament identity (Figure 3S). Pprpk2 mutant defects were less severe than Ppcle and Ppclv1a1b defects at the earliest developmental stages, and at later stages, swollen cell shapes suggested growth defects as well as division plane defects (Figure 3T). The mutant phenotypes above suggest key roles for the Physcomitrella CLV pathway in modulating cell division planes, cell fate, growth, and proliferation during the 2D-3D developmental transition. The formation of long projections of swollen cells in Ppcle mutants (e.g., Figures 3L and 3M) suggests that gametophore identity is attained normally, as cell swelling is a characteristic of gametophore rather than filament initials. The manifestation of plane orientation defects in the first division suggests that WT and mutant gametophore development diverge at the single-celled stage, after cell fate is specified.
Figure 3.
The CLV Pathway Regulates Cell Division Plane Orientations during 3D Growth in Physcomitrella and Arabidopsis
(A–E) Although WT plants (A) developed many normal gametophores, PpcleAmiR1-3 (B), PpcleAmiR4-7 (C), Ppclv1a1b (D), and Pprpk2 (E) mutants had no obvious gametophores. The scale bar represents 0.35 cm.
(F–T) PpcleAmiR1-3, PpcleAmiR4-7, Ppclv1a1b, and Pprpk2 mutants have cell division plane defects at the onset of 3D morphogenesis.
(F–J) The first division of each bud is indicated by a yellow arrow and is set at a strongly oblique angle in WT (F), Ppclv1a1b (I), and Pprpk2 (J) plants, but is weakly oblique in PpcleAmiR1-3 (G) and PpcleAmiR4-7 (H) mutants.
(K–O) Whereas (K) the second division (blue arrow) from the apical cell (asterisk) is normally oblique and roughly perpendicular to the first, in PpcleAmiR1-3 (L), PpcleAmiR4-7 (M) and Ppclv1a1b (N) mutants, it is roughly parallel to the first. Pprpk2 (O) mutants look normal at this stage.
(P–T) The stereotypical divisions that normally generate the tetrahedral shape of the gametophore apical cell at the 6- to 10-celled stage of development (P) are misset in PpcleAmiR1-3 (Q), PpcleAmiR4-7 (R), and Ppclv1a1b (S), and Pprpk2 (T) mutants. The scale bar represents 30 μm.
(U) Bar chart and boxplot showing that gametophore initiation was disrupted in PpcleAmiR1-3, PpcleAmiR4-7, PpCLV1a1b, and Pprpk2 mutants. The number of gametophores with >1 leaf was counted in 5 WT and mutant plants from a single line representing each mutant class. Gametophore buds with <1 leaf were counted from a 5-mm2 area in 3 WT and mutant plants from a single line representing each mutant class. ANOVA, Tukey’s Honest Significant Difference (HSD) test; p < 0.005.
(V and W) Confocal micrographs of WT (Col-0) (V) and clv1/bam1/bam2/bam3 mutant (W) root tips showing disordered cell division plane orientations in the meristem and ground tissue layers. The box in (W) indicates the meristem, and arrowheads indicate the developmental onset of abnormal periclinal division plane orientations in the cortex layer (shaded green). The scale bar represents 20 μm.
See also Figure S6, Methods S1, and Table S4.
Roles for CLAVATA in Regulating Cell Division Plane Orientation Are Conserved between Physcomitrella and Arabidopsis
As roles for CLV in cell division plane orientation were previously unreported, we sought to identify conservation of function with Arabidopsis. To this end, we examined Arabidopsis clv1/bam1/bam2/bam3 quadruple mutant meristems, in which the function of the entire CLV/BAM gene clade is lost [25]. Whereas division plane orientations are normally stereotypic in root meristems, we detected strongly disordered planes in the stem cell niche and ground tissue layers of clv1/bam1/bam2/bam3 mutant roots (Figures 3V, 3W, and S6). Thus, a newly identified role for CLV in cell division plane orientation is conserved between Physcomitrella and Arabidopsis.
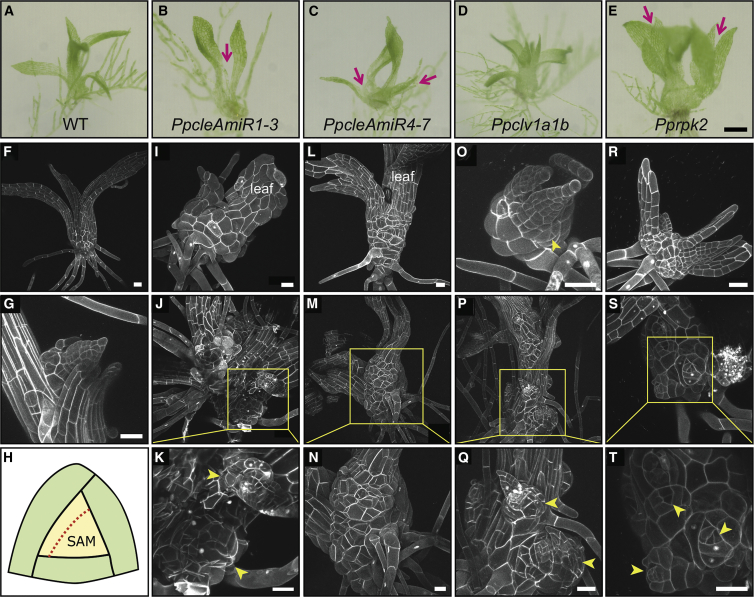
Physcomitrella Mutants with Disrupted CLV Function Have Defective Gametophore Development
In Arabidopsis and other flowering plants, the CLV pathway is known for its role in maintaining the size of the meristematic stem cell pool [26], and increases in the number of stem cells lead to highly enlarged meristems in both clv1 and clv3 (cle) mutants. However, Physcomitrella does not fit the Arabidopsis paradigm of meristem function because the shoot apex comprises a single apical stem cell. The apical cell cleaves merophyte daughter cells in a spiral pattern, and merophytes subsequently divide to generate leaf initials and stem tissues [6]. To investigate whether roles for CLV in regulating stem cell function are conserved between Physcomitrella and Arabidopsis, we imaged one of the largest gametophores from 1-month-old WT and mutant plants using light and confocal microscopy and found that mutant gametophores were reduced in height and had developmental defects (Figure 4). Although PpcleAmiR1-3, PpcleAmiR4-7, and Pprpk2 mutants were most severely reduced in height (Figures 4B, 4C, and 4G), Ppclv1a and Ppclv1b mutants had milder defects (Figures 4D and 4E). PpcleAmiR1-3, PpcleAmiR4-7, Ppclv1a1b, and Pprpk2 mutants had defective leaf development, and Ppclv1b, Ppclv1a1b, and Pprpk2 mutants also had strong cell fate and/or proliferation defects, developing a callus-like mass at the gametophore base (Figures 4L–4N). Closer inspection revealed that these masses arose by the activity of many ectopic apical cells at the gametophore base (Figure 5). These loss-of-function data suggest that CLV has roles in regulating stem cell function that are conserved between Physcomitrella and Arabidopsis.
Figure 4.
Gametophore Phenotypes in PpAmiRcle, Ppclv1, and Pprpk2 Mutants
(A–G) Light micrographs showing height differences between WT (A), PpcleAmiR1-3 (B), PpcleAmiR4-7 (C), Ppclv1a (D), Ppclv1b (E), Ppclv1a1b (F), and Pprpk2 (G) gametophores dissected from 1-month-old plants. The scale bar represents 1 mm.
(H–N) Light micrographs of gametophore bases with arrows showing overproliferation in Ppclv1b (L), Ppclv1a1b (M), and Pprpk2 (N) mutants. WT (H), PpcleAmiR1-3 (I), PpcleAmiR4-7 (J) and Ppclv1a (K) gametophores show no such overproliferation. The scale bar represents 0.5 mm.
Figure 5.
Overproliferation Phenotypes in PpAmiRcle, Ppclv1, and Pprpk2 Mutants
(A–E) Light micrographs of mutant gametophore morphology showing that gametophores (B) arrest, (C and E) develop multiple axes (pink arrows), and (C–E) develop swollen bases relative to (A) WT plants. The scale bar represents 200 μm.
(F and G) Confocal micrographs showing (F) overall gametophore morphology and (G) a branch initiating in a leaf axil in WT plants.
(H) Schematic showing Physcomitrella gametophore apex organization with an apical cell (pale yellow) and rotating division plane orientations.
(I–T) Confocal micrographs showing (I–K) PpcleAmiR1-3 mutant gametophore morphologies, with (I) overproliferation at the gametophore base and (J and K) disorganized growth with ectopic meristems.
(L–N) PpcleAmiR4-7 mutant gametophore morphologies with (L) split leaf phenotypes and (M and N) meristem overproliferation and termination.
(O–Q) Ppclv1a1b mutant gametophore morphology (O), with multiple growth axes and multiple meristems at the gametophore base (P and Q).
(R–T) Pprpk2 mutant gametophore morphology with multiple growth axes (R) and multiple meristems at the gametophore base (S and T).
Yellow arrowheads indicate regions of overproliferation or ectopic meristems. Yellow boxes show regions magnified from (J), (M), (P), and (S) to (K), (N), (Q), and (T). The scale bars represent 50 μm.
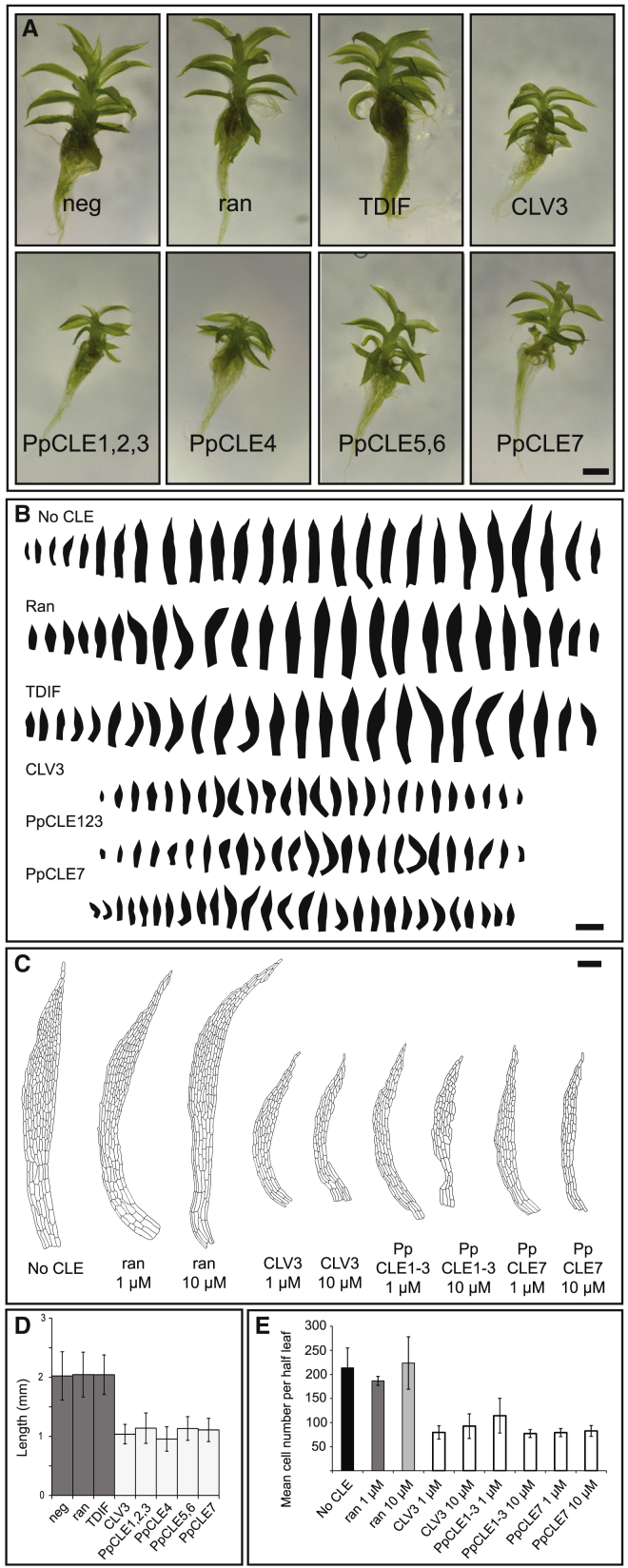
CLE Peptides Can Suppress Cell Proliferation in Physcomitrella Gametophores
To further assay conservation in CLV function, we undertook a gain-of-function approach by applying synthetic CLE peptides to growing plants (Figures 6 and S7). After 4 weeks of growth, we found that treatment with a 1-μM concentration of CLE had no appreciable effect on plant spread or the number of gametophores initiating, indicating that protonemal development is normal (Figure S7). However, although solute controls, a randomized peptide and Arabidopsis CLE41 (a TDIF CLE) have no appreciable effect on gametophore development, Arabidopsis CLV3 and all of the Physcomitrella CLEs cause gametophore dwarfing and a strong reduction in leaf size correlating with a reduction in leaf cell number (Figure 6). Although this phenotype superficially resembles the stunted gametophore phenotypes of PpcleAmiR1-3 and PpcleAmiR4-7 mutants (Figures 4B and 4C), we found no evidence of developmental arrest or meristematic overproliferation following CLE application and no difference in the number of gametophores initiating was detected following CLE treatment (data not shown). These data show that CLEs act through a conserved signaling module to regulate cell proliferation specifically during the 3D growth phase in Physcomitrella.
Figure 6.
Physcomitrella CLEs Suppress Cell Proliferation
(A) Treatment of Physcomitrella plants with 1 μM CLV3-like CLEs from Arabidopsis and Physcomitrella, but not TDIF-like CLEs, causes gametophore and leaf stunting. The scale bar represents 100 μm.
(B) Leaf series from gametophores treated with CLEs expressed during gametophore development. The scale bar represents 1 mm.
(C) Cell outlines of half-leaves in CLE-treated gametophores (leaf 9 was used). The scale bar represents 100 μm.
(D) Height measured from ≥25 gametophores treated with CLEs (n ≥ 25; ANOVA; Tukey’s HSD; p < 0.005).
(E) Leaf 9 cell numbers in CLE-treated half-leaves (n = 3; ANOVA; Tukey’s HSD; p < 0.05).
See also Figure S7.
CLE Peptides Can Act through Receptor Components that Are Conserved between Physcomitrella and Arabidopsis
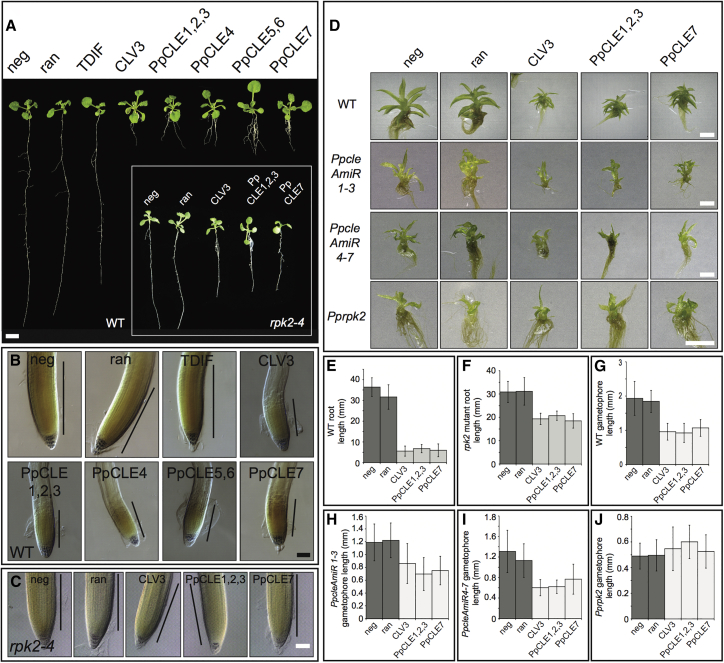
Previous studies in Arabidopsis have shown that application of CLV3-like, but not TDIF-like, CLEs to roots can arrest meristem function [27]. To assay conservation in peptide function, we germinated Arabidopsis seeds on Murashige and Skoog (MS) medium plates containing solute or peptides at a 1 μM concentration. Although solute controls, a randomized peptide, and CLE41 caused no arrest of root development, CLV3 and all of the Physcomitrella CLEs caused a significant reduction in root length in Arabidopsis resulting from collapse of the root meristem (Figures 7A–7C, 7E, and 7F). Physcomitrella CLEs therefore regulate growth and proliferation in a similar manner to CLV3 in Arabidopsis. To confirm that PpCLEs can act through a conserved receptor machinery, we used peptide treatment assays on Arabidopsis and Physcomitrella rpk2 mutants (Figure 7). Whereas treatment of WT Arabidopsis plants with CLV3-like peptides strongly inhibited root growth, rpk2 mutants showed less growth inhibition when treated with Arabidopsis and Physcomitrella peptides (Figures 7A–7C, 7E, and 7F). These data are in line with previously published results showing that RPK2 acts among other receptors to contribute to CLV signaling in Arabidopsis [17] and show that Physcomitrella CLEs can also act via RPK2 in Arabidopsis. To determine whether Physcomitrella CLEs act via PpRPK2, we performed similar experiments in WT, Ppcle, and Pprpk2 mutant backgrounds. Ppcle mutant gametophores are roughly the same size as Pprpk2 mutant gametophores, and we reasoned that, if PpCLEs act via PpRPK2, we should detect a response in Ppcle mutants, but not Pprpk2 mutants. As in previous experiments, we found strong inhibition of gametophore development in WT plants (Figure 7D). Potentially due to lack of positional information, treatment of Ppcle mutants with CLE peptides did not rescue developmental defects but nevertheless induced a gametophore dwarfing response, consistent with an intact receptor machinery (Figures 7D and 7G–7I). In contrast, Pprpk2 mutants showed no morphological response to CLE application, suggesting that PpCLEs act via PpRPK2 in regulating 3D growth (Figures 7D and 7G–7J).
Figure 7.
Physcomitrella CLE Peptides Act via a Conserved Receptor Machinery
(A and B) Treatment of Arabidopsis seedlings with 1 μM CLV3-like CLEs from Arabidopsis and Physcomitrella, but not TDIF-like CLEs, suppresses root meristem proliferation in WT Arabidopsis (n = 30; ANOVA; Tukey’s HSD; p < 0.0005). The scale bar in (A) represents 1 cm and scale bar in (B) represents 100 μm; black bars in (B) represent approximate position and extent of root meristem.
(C and inset in A) Arabidopsis rpk2-4 mutants are resistant to treatment with 1 μM CLE peptides; n ≥ 15; ANOVA; Tukey’s HSD; p < 0.0005. The scale bar represents 100 μm in (C).
(D) Gametophores in plants treated with 10 μM CLEs are stunted in WT, PpcleAmiR1-3, and PpcleAmiR4-7 mutants, but not Pprpk2 mutants (n ≥ 20; ANOVA; Tukey’s HSD; p < 0.0005). The scale bar represents 100 μm.
(E and F) Quantitative data on root length in WT (E) and rpk2 mutant plants (F), supporting inferences from images shown in (A).
(G–J) Quantitative data on gametophore length in WT (G), PpcleAmiR1-3 (H), PpcleAmiR4-7 (I), and Pprpk2 (J), mutant plants supporting inferences from images shown in (D).
Discussion
How Might CLV Pattern Cell Division Plane Orientation?
We propose that the CLV pathway regulates the 2D to 3D developmental transition in Physcomitrella by orienting gametophore cell division planes and regulating growth and fate. How ligands and receptors act together to do this is not yet clear. One possibility is that CLE ligands diffuse to create a concentration gradient that division planes are patterned against. A similar mechanism involving CLEs patterns cambial meristems in Arabidopsis [28], where CLE41 is synthesized in the phloem and diffuses to bind PXY receptors in neighboring procambial cells, thereby imparting spatial information for periclinal division [28]. Constitutive or ectopic expression of CLE41 disrupts this positional information, resulting in disordered cambial division planes [28]. In Physcomitrella, similar patterning could be achieved by sub-cellular localization of receptors to create a graded CLV response in bud initials, or at later stages of development, patterning could be provided by receptor expression in different portions of buds.
It is also possible that CLV signaling does not directly modulate cell division planes but that CLV influences cell division planes via hormone signaling, cell geometry, and/or cell mechanics. Auxin signaling and the activity of microtubule-interacting proteins, such as CLIP-associated proteins (CLASPs), are known to specify cell division planes in Arabidopsis embryos [29], and auxin signaling modulates the activity of previously identified factors necessary for correct division plane orientation in Physcomitrella buds, including DEK1 and NOG1 [30, 31]. There appears to be a complex interplay between auxin and cytokinin in Physcomitrella [32, 33, 34], and several phenotypes suggest that this interplay is disrupted in Ppcle, Ppclv, and Pprpk2 mutants. For instance, cell fate and proliferation at the gametophore base are perturbed (Figures 4 and 5) and leaf cell proliferation is perturbed in plants treated with CLEs (Figure 6), and these aspects of development are auxin and cytokinin regulated [33, 34]. Linking CLV signaling to the hormone pathways regulating growth and fate will be important in unravelling mechanisms of cell division plane specification during 3D growth.
CLAVATA-Regulated Stem Cell Function Is an Ancestral Feature of Land Plants
The data we present are important in two evolutionary contexts. First, they show that the CLV pathway originated with land plants and that CLV-regulated stem cell proliferation and function is likely to be an ancestral feature of land plants. The acquired capacity of land plants to orient stem cell divisions in multiple planes enabled diversification by permitting plants to develop upright axes with organs arranged in multiple orientations, a crucial step in shoot evolution [1]. Stem cell division plane defects in Ppcle mutants specifically affect the transition to 3D growth and the 3D growth phase, and morphological responses to peptide application are also specific to the 3D growth phase. Thus, in an ancient land plant group, CLV regulates a developmental transition that mirrors an evolutionary transition. The data suggest that CLV was a genetic novelty for a key morphological innovation of land plants.
CLAVATA-Regulated Meristem Functions Originated prior to WOX- and KNOX-Regulated Meristem Functions
Second, the data are important in the context of evolving gene regulatory networks for land plant meristem function. Whereas the first land plant meristems comprised a single gametophytic stem cell, the multicellular sporophyte meristems of vascular plants combine stem cell and more generally proliferative capacities [1]. Class I KNOX genes regulate meristematic proliferation in vascular plants [35, 36], but these roles are not shared between bryophytes and vascular plants. Moss KNOX (MKN) genes are primarily expressed in sporophyte tissues [24, 37], and although loss-of-function mkn2 mutants have elongation defects in sporophytes, they have normal gametophytes [37]. WOX genes are key regulators of stem cell proliferation in Arabidopsis [19]. However, this function was acquired by the recently derived WUS gene clade [38, 39], and the downstream pathways regulated by CLV in Physcomitrella are likely to be distinct from those in Arabidopsis as Ppwox13L mutant gametophores develop normally [40]. Thus, class I KNOX- and WOX-regulated meristem functions were both acquired after the bryophyte-vascular plant divergence. CLV was important in the origin of land plant meristem functions in the gametophyte stage of the life cycle, and we speculate that CLV was recruited to regulate stem cell function in the sporophyte stage of the life cycle prior to the origin of KNOX- and WOX- regulated meristem functions.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli strain DH5α | Widely distributed | N/A |
| E. coli strain DB3.1 | Widely distributed | N/A |
| E. coli strain DH10B | Widely distributed | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Taq polymerase | Widely available | N/A |
| Phusion High-Fidelity DNA polymerase | ThermoFisher | Cat#F530S |
| Novagen KOD Hot Start polymerase | Sigma-Aldrich | Cat#71086 |
| Superscript II reverse transcriptase | ThermoFisher | Cat#18064022 |
| Restriction enzymes for cloning | New England Biolabs | N/A |
| DNase | Fermentas | Cat#EN0525 |
| MS medium | Melford | Cat#M0221 |
| Plant agar | Duchefa | Cat#P1001 |
| Driselase (basidiomycetes sp.) | Sigma-Aldrich | Cat#8037 |
| Polyethylene glycol (PEG) 6000 | Sigma-Aldrich | Cat#81255 |
| G418 disulphate | Melford | Cat#G0175 |
| Hygromycin B | Melford | Cat#H7502 |
| Blasticidin S | Melford | Cat#B1220 |
| α-32P dCTP | GE Healthcare | Cat#PB10205 |
| X-GlcA | Melford | Cat#MB1021 |
| Propidium iodide | Sigma-Aldrich | Cat#P4864 |
| Synthetic CLE peptides (95% purity) | Genecust | N/A |
| Lugol’s stain | Fisher Scientific | Cat#12801823 |
| Critical Commercial Assays | ||
| RNeasy RNA extraction kit | QIAGEN | Cat#74104 |
| Plasmid Plus Midi kit | QIAGEN | Cat#12943 |
| Amersham Rediprime II DNA labeling kit | GE Healthcare | Cat#RPN1633 |
| Dig-High Prime DNA labeling and detection starter kit II | Sigma-Aldrich | Cat#11585614910 |
| Dig Easy Hyb | Sigma-Aldrich | Cat#11585762001 |
| Experimental Models: Organisms/Strains | ||
| Physcomitrella patens Gransden | Widely available | N/A |
| PpCLE1::NGG line | This study | N/A |
| PpCLE2::NGG line | This study | N/A |
| PpCLE7::NGG line | This study | N/A |
| PpCLV1a::NGG line | This study | N/A |
| PpCLV1b::NGG line | This study | N/A |
| PpRPK2::NGG line | This study | N/A |
| PpcleamiR1-3 mutant | This study | N/A |
| PpcleamiR4-7 mutant | This study | N/A |
| Ppclv1a mutant | This study | N/A |
| Ppclv1b mutant | This study | N/A |
| Ppclv1ab double mutant | This study | N/A |
| Pprpk2 mutant | This study | N/A |
| Arabidopsis thaliana Col-0 | Widely available | N/A |
| Arabidopsis thaliana rpk2-4 mutant | [17] | N/A |
| Arabidopsis thaliana clv1,bam1,bam2,bam3 mutant | [25] | N/A |
| Oligonucleotides | ||
| A list of oligonucleotides is given in Table S4 | N/A | N/A |
| Recombinant DNA | ||
| PIG1NGGII construct | [41] | N/A |
| PpCLE1::NGG construct | This study | N/A |
| PpCLE2::NGG construct (NptII) | This study | GenBank: MH310732, MH310732 |
| PpCLE7::NGG construct | This study | N/A |
| PpCLV1a::NGG construct | This study | N/A |
| PpCLV1b::NGG construct | This study | N/A |
| PpRPK2::NGG construct (AphIV) | This study | GenBank: MH310733 |
| pRS300 | [42] | N/A |
| pGREEN (Hyg) | [43] | N/A |
| pGREEN (Kan) | [43] | N/A |
| pBJ36 | [44] | N/A |
| pBRACT211 | [45] | N/A |
| pJH125 | This study | N/A |
| pJH131 | This study | N/A |
| PpcleAmiR1-3 construct | This study | GenBank: MH310734 |
| PpcleamiR4-7 construct | This study | GenBank: MH310735 |
| U3::Ppclv1a sgRNA5 construct | This study | GenBank: MH310736 |
| U3::Ppclv1a sgRNA7 construct | This study | GenBank: MH310737 |
| U6::Ppclv1b sgRNA construct | This study | GenBank: MH310738 |
| pACT::Cas9 construct | [46] | N/A |
| pNRF | [47] | N/A |
| pBHRF108 | [48] | N/A |
| pDONR2.1 | Invitrogen | N/A |
| pGEMT-EASY | Promega | Cat#A1360 |
| Software and Algorithms | ||
| tBLASTn | [49] | N/A |
| SignalP | [50] | v4.0 |
| MEGA | [51] | v7.0.26 |
| Figtree | http://tree.bio.ed.ac.uk/software/figtree/ | v1.4.3 |
| AmiR design software | http://wmd3.weigelworld.org/cgi-bin/webapp.cgi | N/A |
| CRISPR design software | [52] | http://crispor.tefor.net/ |
| ImageJ | http://imagej.net/Welcome | V1.4.8 |
| Adobe Photoshop | Adobe | N/A |
| Adobe Illustrator | Adobe | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jill Harrison (jill.harrison@bristol.ac.uk). Please note that the transfer of transgenic materials will be subject to MTA and any relevant import permits.
Experimental Models and Subject Details
Arabidopsis plant growth
Columbia (Col-0), rpk2-4 (cli) or clv1/bam1/bam2/bam3 mutants [17, 25] were used for Arabidopsis experiments. Homozygous rpk2-4 mutants were confirmed using a BamHI dCAPs screen with a PCR fragment amplified using primers AtRPK2-BamHIF and AtRPK2-BamHIR (see primer list). Seeds were surface sterilized in 5% (v/v) sodium hypochlorite for 10 min and washed three times with sterile de-ionised water. They were then stratified at 4°C in darkness for 48 hr and sown on 0.5 X MS plates containing 0.8% agar [53]. Plants were grown vertically for 7 days at 25°C in a 16 hr light/ 8 hr dark cycle prior to observation (rpk2 experiments) or at 22°C under continuous light (clv1/bam1/bam2/bam3 experiments).
Physcomitrella plant growth
The Gransden strain of Physcomitrella patens [54] was used for all experiments. Plants were grown in sterile culture on BCDAT plates at 23°C in continuous light at 30-50 μmols-1 in Sanyo MLR-351 growth cabinets. BCDAT medium comprises 250mg/L MgSO4.7H2O, 250mg/L KH2PO4 (pH6.5), 1010mg/L KNO3, 12.5mg/L, FeSO4.7H2O, 0.001% Trace Element Solution (0.614mg/L H3BO3, 0.055mg/L AlK(SO4)2.12H2O, 0.055mg/L CuSO4.5H2O, 0.028mg/L KBr, 0.028mg/L LiCl, 0.389mg/L MnCl2.4H2O, 0.055mg/L CoCl2.6H2O, 0.055mg/L ZnSO4.7H2O, 0.028mg/L KI and 0.028mg/L SnCl2.2H2O), 0.92 g/L C4H12N2O6 and 8g/L agar with CaCl2 added to a 1mM concentration after autoclaving. Protonemal cultures for transformation were grown on BCDAT plates overlaid with autoclaved cellophane disks and molecular and phenotypic analyses were undertaken using 1 mm spot cultures unless otherwise stated.
Method Details
Sequence retrieval
CLE genes
Previously described Arabidopsis thaliana and Oryza sativa CLE sequences were respectively retrieved from TAIR and RAP-DB [55]. Selaginella moelendorffii [56], Glycine max [57, 58] and Picea abies [59] CLEs were retrieved from NCBI. To extend taxon sampling within land plants and identify previously unknown CLEs, the CLE domains of Arabidopsis thaliana CLV3 and CLE41 were used as tBLASTn queries with an e-value cutoff of e-100 to screen transcriptome or draft genome assemblies of a basal angiosperm (Amborella trichopoda), a fern (Diplazium wichurae), a hornwort (Anthoceros agrestis), a moss (Physcomitrella patens v1.6 [60]) and a liverwort (Marchantia polymorpha). Positive hits were used in reciprocal BLASTs until no new sequences were retrieved. All sequences retrieved were checked for the presence of a signal peptide [61] using SignalP [50, 62]. Newly identified CLE sequences were named with a two-letter prefix denoting the genus and species and numbered (Table S1). A recent cluster analysis [63] succeeded our analyses with slight variation in CLE numbers between species for reasons explained in [63]. An updated version of the Physcomitrella genome (v 3.1 [64]) also succeeded our analyses, and this includes two further PpCLEs that encode the same CLE motif as PpCLEs 1, 2 and 3. While these were not included in this study, V3 gene IDs including PpCLE8 and PpCLE9 are listed in Table S3. Transcriptomes and draft or complete genomes of charophyte (Coleochaete nitellarum, Spirogyra sp., Chara braunii) and chlorophyte algae (Ulva linza, Chlamydomonas reinhardtii, Volvox carteri, Ostreoscoccus tauri and Chlorella vulgaris) were also searched but no CLEs were found. A previously annotated Chlamydomonas reinhardtii CLE [65] was re-analyzed and discarded due to lack of similarity to other CLEs and a premature in-frame stop codon. A full list of taxa and databases searched is given in Table S2.
CLV1/RPK2 genes
Arabidopsis CLV1 and RPK2 sequences were used to query the databases listed above using tBLASTn searches with an e-value cut-off of e-1000. As the LRR-Receptor kinase family is large, only sequences that retrieved CLV1 or RPK2 as a top hit in reciprocal BLASTs to Arabidopsis were used in further analyses. Newly identified CLV1-like and RPK2-like genes were named with a two-letter prefix denoting the genus and species and given an alphabetical epithet (Table S1). A list of taxa searched is given in Table S2.
Phylogenetic reconstruction
To infer CLE relationships, the conserved 12 amino acid CLE motif from 193 CLEs was used in neighbor joining reconstructions compiled with the JTT model in MEGA7.0.26 [51] (Figure S1; Data S1). This approach was taken because there is little conservation in CLE structure outside the CLE motif and so few characters can only yield limited phylogenetic signal (see [63]). To infer CLV/BAM relationships, 525 conserved amino acid residues from 36 genes were used in maximum likelihood reconstructions with the JTT model in MEGA7.0.26 [51] (Figure S2; Data S2). To infer RPK2 relationships, 782 conserved amino acid residues from 18 genes were used in maximum likelihood reconstructions with the JTT model in MEGA7.0.26 [51] (Figure S3; Data S3). For all analyses 100 bootstrap replicates were performed and support values over 50% (CLE tree) or 70% (CLV1/BAM and RPK2 trees) are represented above the branches.
Molecular biology
RT-PCR
Total RNA was isolated from 4 day-old protonemal cultures and 10, 21 or 28 day old spot cultures using the QIAGEN RNeasy method. RNA was DNase treated prior to reverse transcription with SuperScript II following manufacturer’s guidelines. Semiquantitative RT-PCR was undertaken using UBIQUITIN (Pp1s56_52V6.1) as a loading control. Where possible, primers were designed to span introns to detect genomic contamination, and sequences are listed in Table S4.
Genomic DNA extraction
Genomic DNA was extracted from protonemal cultures using a CTAB (Hexadecyltrimethylammonium bromide) protocol. Snap-frozen tissue was ground in liquid nitrogen and transferred to tubes containing prewarmed extraction buffer (2% CTAB, 1.4 M NaCl, 100 mM Tris pH8.0, 20 mM EDTA pH8.0, 2% PVP and 1 mg/mL RNaseA), with no more than 100 mg of tissue per mL of buffer. Samples were incubated for 10 min at 65°C and an equal volume of 24:1 chloroform:isoamyl alcohol was added and mixed with each sample to form an emulsion. The tubes were centrifuged at high speed (> 10,000 rpm) for 10 min, and the aqueous phase was transferred to a fresh tube prior to DNA precipitation with an equal volume of isopropanol and repeated centrifugation. DNA was washed with 70% ethanol and dissolved in water, 10 mM Tris pH 8.0 or 10 mM Tris pH 8.0 with 1 mM Na2EDTA.
Generation of promoter::NGG constructs
Promoter sequences from PpCLE1 (2.1 kbp), PpCLE2 (2.1 kbp), PpCLE7 (2 kbp), PpCLV1a (2 kbp), PpCLV1b (2.8 kbp) and PpRPK2 (1.4 kbp) were PCR amplified using a proof-reading Taq polymerase and primers listed in Table S4 and cloned directly or via pGEMT Easy into the SmaI site of PIG1NGGII [41] or derivatives with alternative selection cassettes and sequenced prior to linearization and transformation as illustrated in Methods S1.
Generation of AmiR constructs
To generate PplcleAmiR1-3 and PplcleAmiR4-7 constructs, resistance cassettes from pGREEN [45] were first inserted into a blunt-ended HindII site of pBJ36 [44]. A soybean UBIQUITIN promoter from pBRACT211 [45] was inserted into the SmaI site to drive AmiRNA expression and the resultant plasmids were named pJH125 (KanR) and pJH131 (HygR). AmiRNAs were designed according to [42], generated by degenerate PCR using a proof-reading Taq polymerase and the pRS300 plasmid as a template, cloned into pGEMT-EASY and transferred as XmaI/BamHI fragments into pJH125 or pJH131. Silencing constructs were checked by sequencing and digested with SacI for transformation as illustrated in Methods S1.
Generation of CRISPR constructs
Small cassettes containing two BsaI restriction sites and sgRNAs [66] driven by the Physcomitrella U3 or U6 promoter and flanked by attB sites were synthesized and cloned into pDONR201. sgRNA sequences were selected and screened for off target hits in the Physcomitrella V3 genome using http://crispor.tefor.net/. To clone guide RNAs into expression cassettes, two primers consisting of guide sequences with overhangs for U3 and U6 promoters were annealed and ligated into U3 or U6 expression vectors pre-digested with BsaI. Constructs were checked by sequencing and co-transformed with pACT::Cas9 [46] to engineer mutants as illustrated in Methods S1.
Generation of RPK2 KO construct
5′ and 3′ flanking regions were PCR amplified with a proof-reading Taq polymerase and cloned sequentially into pGEMT-EASY using primers listed in Table S4. The resultant plasmid was digested with PmeI and AscI, and the AphIV cassette from pBHRF-108 [48] was ligated between PpRPK2 flanking regions. This plasmid was checked by sequencing and linearized for transformation as illustrated in Methods S1.
Transgenic line generation and phenotype analyses
Moss transformation and line authentication
For gene targeting and AmiR approaches, 10-20 μg of plasmid DNA was isolated using the QIAGEN Plasmid Plus Midi system and linearized as illustrated in Methods S1. For CRISPR approaches, 5-7 μg of Cas9 and pNRF, and 2-3 μg of each gRNA-expressing construct were purified and pooled for transformation [46] at a concentration of at least 1 μg per μL. All solutions for the transformation procedure were prepared prior to commencing transformation [67]. First, a polyethylene glycol (PEG) solution was prepared by adding 10 mL of mannitol/CaNO3 solution (8% mannitol, 0.1 M Ca(NO3), 10 mM Tris pH7.2) to 2 g of molten PEG 6000, and the tube containing the solution was left in a water bath at 45°C. To isolate protoplasts, homogoenous protonemal cultures were grown for 5 days to a week post passage. A 1% driselase solution was prepared in 25 mL 8% mannitol, and the supernatant was removed and filter sterilized into to a clean 50 mL falcon tube following centrifugation. Tissue from 4-6 plates was transferred into the driselase solution and the tissue suspension was left for 30-40 min with intermittent mixing to allow cell wall digestion. The mixture was then transferred into a fresh tube through a 50 μm filter to remove cell and cell wall debris. Protoplasts were sedimented by centrifugation for 3 min at 120 g, resuspended and washed three times in 10 mL of 8.0% mannitol prior to counting with a hematocytometer. Protoplasts were then sedimented and resuspended to a density of 1.2 × 106 per mL in MMM solution (0.5 M mannitol, 0.15 M MgCl2 and 0.1% MES pH5.6). 300 μL aliquots of protoplasts were dispensed into falcon tubes prior to addition of DNA and 300 μL PEG solution, and cells were then heat shocked for 5 min at 45°C. Transformation mixtures were progressively diluted with 1 mL of 8% mannitol solution and washed. Protoplasts were then sedimented by centrifugation as above and washed four more times. After the final wash and spin, protoplasts were resuspended in 5 mL liquid BCD medium (constituents as specified above but without ammonium tartrate or agar) with 8% mannitol, 10 mM CaCl2 and 0.5% glucose, wrapped in aluminum foil and left at 23°C overnight. The next day, the protoplast suspension was plated onto BCDAT plates overlain with cellophane and containing 8% mannitol and 5 g/L glucose, using c.1 mL per plate. Plants were grown under standard conditions until regenerants comprised 10-20 cells. Cellophane discs were then transferred onto BCDAT plates containing antibiotics for selection (25 μg/mL Hyg, 50 μg/mL G418, 100 μg/mL BSD). Plants were grown for 2 weeks on selection plates prior to transfer onto BCDAT plates lacking antibiotic for 2 weeks and then back on to selection plates for a further 2 weeks. All lines were screened by PCR, RT-PCR, Southern analysis or sequencing as illustrated in Methods S1. PCR conditions were standard and primer sequences are listed in Table S4.
Southern hybridization
For PpcleAmiR Southerns, 10-15 μg genomic DNA was digested with EcoRV and fractionated in 0.8% agarose by gel electrophoresis. DNA in each gel was depurinated with 0.2 M HCl for 20 min and denatured with 0.4 M NaOH for 20 min prior to neutralization for 20 min in a solution containing 3 M NaCl and 1 M Tris pH 7.5. Gels were inverted onto a Whatman paper wick inserted into a bath of 20 X SSC solution, and DNA was transferred onto a nitrocellulose membrane by overnight Southern blotting. DNA was UV crosslinked to the membrane and the membrane was rinsed in water prior to immersion in pre-hybridization solution (3 X SSC, 1% SDS, 0.1% sodium pyrophosphate, 5 X Denhardt’s and 200 μg per mL sheared salmon sperm DNA). The probe template was excised with EcoRV and BamHI from the PpcleAmiR1-3 construct and the probe was synthesized using an Amersham Rediprime II DNA labeling kit as per manufacturer’s instructions. Hybridization was undertaken in a 3 X SSC buffer at 58°C and this was followed by two 20 min washes at 58°C in 3 X SSC and 2 X SSC buffers respectively. Membranes were wrapped in Saran Wrap and used to expose X-ray film, and film was then developed using a film processor. For promoter::NGG and Pprpk2 Southerns, 2.5-3 μg genomic DNA was digested as illustrated in Methods S1. Probe templates comprising PIG1 flanking sequence, PpRPK2 coding sequence or a hygromycin resistance cassette were PCR amplified and labeled using the Roche DIG High Prime system. Hybridization was undertaken overnight at 42°C using the Roche DIG Easy Hyb system. Washing and detection were performed using the manufacturer’s protocol from the Roche DIG High Prime DNA labeling and Detection Starter kit II.
Physcomitrella plant imaging
To assess whole plant and gametophore phenotypes, 4 to 5 week-old spot cultures were imaged using a Keyence VHX-1000E digital microscope with a 20-50 X or 50-200 X objective. To analyze bud phenotypes, confocal imaging was undertaken on tissue stained with 0.5 mg/ml propidium iodide using a Leica TCS SP5 microscope with excitation from the 488 or 514 laser line and emission collected at 600-650 nm or using a Zeiss 710 LSM with excitation from a 514 laser line and emission collected at 566-650 nm. To analyze leaf phenotypes, leaves were removed form gametophores, arranged in heteroblastic series, cleared in 1% chloral hydrate overnight, washed in deionised water and treated with 2M NaOH for 2 hr. They were then washed with water and stained with 0.05% toluidine blue for 2 min before destaining for 10 min in water. The stained leaves were then mounted on a slide under a coverslip and imaged to visualize cell outlines. Adobe Illustrator was used to trace leaf outlines to produce silhouettes for illustration purposes (Figure 6). Quantitative analyses of leaf size were performed using ImageJ, and cell numbers were evaluated using the ‘analyze particles’ option [68]. Leaf size comparisons were undertaken using leaves from the same point in the heteroblastic leaf series [33] as stipulated in figure legends.
Arabidopsis plant imaging
Root length was scored from scanned images of plants grown on ½ X MS plates using ImageJ [68]. To visualize rpk2 meristems, roots were stained with Lugol’s stain, cleared, and imaged using a 20 X objective on a Leica DMRXA microscope with DIC [69]. clv1/bam1/bam2/bam3 roots were stained with 15 mM propidium iodide and imaged using a C-Apochromat 40 X/1.20 W Korr objective on a Zeiss LSM710 microscope. Excitation and emission windows for propidium iodide were 560 nm and 566-719 nm respectively. Confocal images were analyzed and processed using ImageJ and Adobe Photoshop.
GUS staining and imaging
Physcomitrella plants grown on BCDAT were cut out of plates with agar and incubated at 37°C in a 100 mM phosphate buffer with 10 mM Tris pH8.0, 1 mM EDTA pH8.0, 0.05% Triton X-100, 1 mg/mL X-GlcA (5-Bromo-4-chloro-3-indolyl-β-D-glucuronic acid) and potassium ferri/ferrocyanide using concentrations and times indicated in Figure 2 and legend. Plants were bleached in 70% ethanol and dissected and mounted in 0.3% low melting point agarose prior to imaging with a Keyence VHX-1000 digital microscope with a 0-50 X or a 50-200 X objective.
CLE peptide application
Synthetic CLE peptides (Genecust, >95% purity) were dissolved in phosphate buffer (50 μm, pH6.8) to stock concentrations of 1 mM and 10 mM. Plants were grown on BCDAT plates containing peptides diluted to concentrations specified in the main text.
Quantification and Statistical Analysis
Quantification and statistical analyses were undertaken as stipulated in main text and SI figures and figure legends.
Data and Software Availability
Genome and transcriptome data were searched as described in Method Details and details of data repositories are listed in Table S2.
Acknowledgments
C.J.H. thanks the Gatsby Charitable Foundation (GAT2962), BBSRC (BB/L02248/1), and Royal Society (UF130563) for funding. J.C. thanks Santander and the Cambridge Overseas Trust for funding his Master’s research. Z.N.V. thanks the Gatsby Charitable Foundation for funding. Z.L.N. and A.D.C. thank the NIH (R35GM119614-01) for funding. P.S. and M.W. thank the Swiss National Science Foundation (160004 and 131726 to P.S.), URPP Evolution in Action (to P.S. and M.W.), and Georges und Antoine Claraz-Schenkung (to P.S. and M.W.) for funding. We thank Takashi Ishida for rpk2 mutant seed. We thank Christophe Dunand, Liam Dolan, and Takayuchi Kohchi for access to Spirogyra and Marchantia genome data. We thank Tomoaki Nishiyama and Stefan Rensing for searching Chara genome data on our behalf. We thank Jon Hughes and Maureen Liu for preliminary expression analyses.
Author Contributions
Database Searches, C.D.W., M.W., P.S., and C.J.H.; Phylogenetic Analyses, C.D.W. and C.J.H.; RT-PCR, C.D.W. and C.J.H.; promoter::NGG Line Generation and Characterization, Z.N.V., C.D.W., X.Y.W., T.A., Y.K., A.C.C., S.S., and C.J.H.; AmiRcle1-3 and AmiRcle4-7 Line Generation and Characterization, C.D.W., J.C., Z.N.V., and C.J.H.; Ppclv1a1b Mutant Generation and Characterization, J.C., Z.N.V., A.H.K.R., M.J.S., and C.J.H.; Pprpk2 Mutant Generation and Characterization, J.C., C.D.W., Z.N.V., X.Y.W., T.A., A.H.K.R., M.J.S., and C.J.H.; Peptide Application Experiments, C.D.W. and C.J.H.; Arabidopsis clv1/bam1/bam2/bam3 Mutant Analysis, A.D.C. and Z.L.N.; Writing the Manuscript, C.J.H.; Editing the Manuscript, C.J.H., C.D.W., J.C., Z.N.V., T.A., P.S., M.J.S., and A.H.K.R.
Declaration of Interests
The authors declare no competing interests.
Published: July 19, 2018
Footnotes
Supplemental Information includes seven figures, four tables, three data files, and supplemental text and is available online at https://doi.org/10.1016/j.cub.2018.05.068.
Supplemental Information
References
- 1.Harrison C.J. Development and genetics in the evolution of land plant body plans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:e20150490. doi: 10.1098/rstb.2015.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmermann W. Main results of the “Telome theory”. Palaeobotanist. 1952;1:456–470. [Google Scholar]
- 3.Graham L.E., Cook M.E., Busse J.S. The origin of plants: body plan changes contributing to a major evolutionary radiation. Proc. Natl. Acad. Sci. USA. 2000;97:4535–4540. doi: 10.1073/pnas.97.9.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickett N.J., Mirarab S., Nguyen N., Warnow T., Carpenter E., Matasci N., Ayyampalayam S., Barker M.S., Burleigh J.G., Gitzendanner M.A. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leliaert F., Smith D.R., Moreau H., Herron M.D., Verbruggen H., Delwiche C.F., De Clerck O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012;31:1–46. [Google Scholar]
- 6.Harrison C.J., Roeder A.H.K., Meyerowitz E.M., Langdale J.A. Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 2009;19:461–471. doi: 10.1016/j.cub.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 7.Aoyama T., Hiwatashi Y., Shigyo M., Kofuji R., Kubo M., Ito M., Hasebe M. AP2-type transcription factors determine stem cell identity in the moss Physcomitrella patens. Development. 2012;139:3120–3129. doi: 10.1242/dev.076091. [DOI] [PubMed] [Google Scholar]
- 8.Brand U., Grünewald M., Hobe M., Simon R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002;129:565–575. doi: 10.1104/pp.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy G.V., Meyerowitz E.M. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 10.Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 11.Clark S.E., Williams R.W., Meyerowitz E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 12.Milani P., Mirabet V., Cellier C., Rozier F., Hamant O., Das P., Boudaoud A. Matching patterns of gene expression to mechanical stiffness at cell resolution through quantitative tandem epifluorescence and nanoindentation. Plant Physiol. 2014;165:1399–1408. doi: 10.1104/pp.114.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 15.Nimchuk Z.L., Tarr P.T., Ohno C., Qu X., Meyerowitz E.M. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 2011;21:345–352. doi: 10.1016/j.cub.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- 18.Nimchuk Z.L. CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS Genet. 2017;13:e1006681. doi: 10.1371/journal.pgen.1006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laux T., Mayer K.F., Berger J., Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 20.Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox C.J., Li B., Foster P.G., Embley T.M., Civán P. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst. Biol. 2014;63:272–279. doi: 10.1093/sysbio/syt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowman J.L., Kohchi T., Yamato K.T., Jenkins J., Shu S., Ishizaki K., Yamaoka S., Nishihama R., Nakamura Y., Berger F. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304.e15. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz-Ramírez C., Hernandez-Coronado M., Thamm A., Catarino B., Wang M., Dolan L., Feijó J.A., Becker J.D. A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant. 2016;9:205–220. doi: 10.1016/j.molp.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Frank M.H., Scanlon M.J. Cell-specific transcriptomic analyses of three-dimensional shoot development in the moss Physcomitrella patens. Plant J. 2015;83:743–751. doi: 10.1111/tpj.12928. [DOI] [PubMed] [Google Scholar]
- 25.Nimchuk Z.L., Zhou Y., Tarr P.T., Peterson B.A., Meyerowitz E.M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development. 2015;142:1043–1049. doi: 10.1242/dev.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somssich M., Je B.I., Simon R., Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 27.Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C.M. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etchells J.P., Turner S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development. 2010;137:767–774. doi: 10.1242/dev.044941. [DOI] [PubMed] [Google Scholar]
- 29.Chakrabortty B., Willemsen V., de Zeeuw T., Liao C.-Y., Weijers D., Mulder B., Scheres B. A microtubule-based mechanism predicts cell division orientation in plant embryogenesis. bioRxiv. 2018 doi: 10.1016/j.cub.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Perroud P.F., Demko V., Johansen W., Wilson R.C., Olsen O.A., Quatrano R.S. Defective Kernel 1 (DEK1) is required for three-dimensional growth in Physcomitrella patens. New Phytol. 2014;203:794–804. doi: 10.1111/nph.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moody L.A., Kelly S., Rabbinowitsch E., Langdale J.A. Genetic regulation of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr. Biol. 2018;28:473–478.e5. doi: 10.1016/j.cub.2017.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashton N.W., Grimsley N.H., Cove D.J. Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta. 1979;144:427–435. doi: 10.1007/BF00380118. [DOI] [PubMed] [Google Scholar]
- 33.Barker E.I., Ashton N.W. Heteroblasty in the moss, Aphanoregma patens (Physcomitrella patens), results from progressive modulation of a single fundamental leaf developmental programme. J. Bryol. 2013;35:185–196. [Google Scholar]
- 34.Coudert Y., Palubicki W., Ljung K., Novak O., Leyser O., Harrison C.J. Three ancient hormonal cues co-ordinate shoot branching in a moss. eLife. 2015;4:e06808. doi: 10.7554/eLife.06808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long J.A., Moan E.I., Medford J.I., Barton M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 36.Harrison C.J., Corley S.B., Moylan E.C., Alexander D.L., Scotland R.W., Langdale J.A. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]
- 37.Sakakibara K., Nishiyama T., Deguchi H., Hasebe M. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol. Dev. 2008;10:555–566. doi: 10.1111/j.1525-142X.2008.00271.x. [DOI] [PubMed] [Google Scholar]
- 38.Nardmann J., Werr W. The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Mol. Biol. 2012;78:123–134. doi: 10.1007/s11103-011-9851-4. [DOI] [PubMed] [Google Scholar]
- 39.Dolzblasz A., Nardmann J., Clerici E., Causier B., van der Graaff E., Chen J., Davies B., Werr W., Laux T. Stem cell regulation by Arabidopsis WOX genes. Mol. Plant. 2016;9:1028–1039. doi: 10.1016/j.molp.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Sakakibara K., Reisewitz P., Aoyama T., Friedrich T., Ando S., Sato Y., Tamada Y., Nishiyama T., Hiwatashi Y., Kurata T. WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development. 2014;141:1660–1670. doi: 10.1242/dev.097444. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa M., Murata T., Sato Y., Nishiyama T., Hiwatashi Y., Imai A., Kimura M., Sugimoto N., Akita A., Oguri Y. Physcomitrella cyclin-dependent kinase A links cell cycle reactivation to other cellular changes during reprogramming of leaf cells. Plant Cell. 2011;23:2924–2938. doi: 10.1105/tpc.111.088005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ossowski S., Schwab R., Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 43.Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- 44.Eshed Y., Baum S.F., Perea J.V., Bowman J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- 45.Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Obando M., Hoffmann B., Géry C., Guyon-Debast A., Téoulé E., Rameau C., Bonhomme S., Nogué F. Simple and efficient targeting of multiple genes through CRISPR-Cas9 in Physcomitrella patens. G3 (Bethesda) 2016;6:3647–3653. doi: 10.1534/g3.116.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer D.G., Delacote F., Charlot F., Vrielynck N., Guyon-Debast A., Le Guin S., Neuhaus J.M., Doutriaux M.P., Nogué F. RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair (Amst.) 2010;9:526–533. doi: 10.1016/j.dnarep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Moody L.A., Kelly S., Coudert Y., Nimchuk Z.L., Harrison C.J., Langdale J.A. Somatic hybridization provides segregating populations for the identification of causative mutations in sterile mutants of the moss Physcomitrella patens. New Phytol. 2018;218:1270–1277. doi: 10.1111/nph.15069. [DOI] [PubMed] [Google Scholar]
- 49.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murashige T., Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 54.Ashton N.W., Cove D.J. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol. Gen. Genet. 1977;154:87–95. [Google Scholar]
- 55.Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- 56.Miwa H., Tamaki T., Fukuda H., Sawa S. Evolution of CLE signaling: origins of the CLV1 and SOL2/CRN receptor diversity. Plant Signaling and Behaviour. 2009;4:477–481. doi: 10.4161/psb.4.6.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortier V., Den Herder G., Whitford R., Van de Velde W., Rombauts S., D’Haeseleer K., Holsters M., Goormachtig S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D.L., Song Q., Thelen J.J., Cheng J. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 59.Strabala T.J., Phillips L., West M., Stanbra L. Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biol. 2014;14:47. doi: 10.1186/1471-2229-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P.F., Lindquist E.A., Kamisugi Y. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 61.Rojo E., Sharma V.K., Kovaleva V., Raikhel N.V., Fletcher J.C. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;14:969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 63.Goad D.M., Zhu C., Kellogg E.A. Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol. 2017;216:605–616. doi: 10.1111/nph.14348. [DOI] [PubMed] [Google Scholar]
- 64.Lang D., Ullrich K.K., Murat F., Fuchs J., Jenkins J., Haas F.B., Piednoel M., Gundlach H., Van Bel M., Meyberg R. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2018;93:515–533. doi: 10.1111/tpj.13801. [DOI] [PubMed] [Google Scholar]
- 65.Oelkers K., Goffard N., Weiller G.F., Gresshoff P.M., Mathesius U., Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 2008;8:1. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaefer D., Zryd J.P., Knight C.D., Cove D.J. Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 1991;226:418–424. doi: 10.1007/BF00260654. [DOI] [PubMed] [Google Scholar]
- 68.Abràmoff M.D., Magalhães P.J., Ram S.J. Image processing with ImageJ. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- 69.Willemsen V., Wolkenfelt H., de Vrieze G., Weisbeek P., Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development. 1998;125:521–531. doi: 10.1242/dev.125.3.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.