Abstract.
A quantifiable, stool-based, Mycobacterium tuberculosis (Mtb) test has potential complementary value to respiratory specimens. Limit of detection (LOD) was determined by spiking control stool. Clinical test performance was evaluated in a cohort with pulmonary tuberculosis (TB) (N = 166) and asymptomatic household TB child contacts (N = 105). Stool-quantitative polymerase chain reaction (qPCR) results were compared with sputum acid-fast bacilli (AFB) microscopy, GeneXpert MTB/RIF (Xpert MTB/RIF), and cultures. In Mtb stool-spiking studies, the LOD was 96 colony-forming units/50 mg of stool (95% confidence interval [CI]: 84.8–105.6). Among specimens collected within 72 hours of antituberculosis treatment (ATT) initiation, stool qPCR detected 22 of 23 (95%) of culture-positive cases. Among clinically diagnosed cases that were Xpert MTB/RIF and culture negative, stool qPCR detected an additional 8% (3/37). Among asymptomatic, recently TB-exposed participants, stool PCR detected Mtb in two of 105 (1.9%) patients. Two months after ATT, the Mtb quantitative burden in femtogram per microliters decreased (Wilcoxon signed-rank P < 0.001) and persistent positive stool PCR was associated with treatment failure or drug resistance (relative risk 2.8, CI: 1.2–6.5; P = 0.012). Stool-based qPCR is a promising complementary technique to sputum-based diagnosis. It detects and quantifies low levels of stool Mtb DNA, thereby supporting adjunct diagnosis and treatment monitoring in pulmonary TB.
INTRODUCTION
Despite recent advances, the diagnosis of pulmonary tuberculosis (TB) in certain populations, particularly HIV-positive individuals and children, remains challenging. In 2016, 39% of the estimated 10.4 million new cases of TB went undiagnosed.1 Diagnosis of HIV-associated or childhood TB is hindered by presentation with nonspecific clinical signs and symptoms, the paucibacillary nature of the disease, and difficulty in obtaining induced or expectorated sputum. Misdiagnosis and delays in treatment contribute significantly to increased morbidity and mortality among these groups.2–5 A quarter of HIV-related deaths in adults are secondary to TB, rising to 40% in TB high-burden settings.6,7 The culture remains the reference standard despite its suboptimal sensitivity and lengthy wait time of 2–6 weeks for results.8
Although polymerase chain reaction (PCR) techniques, such as Gene Xpert® MTB/RIF (Xpert; Cepheid, Sunnyvale, CA), commercial PCR tests, and in-house PCR tests, have improved sensitivity for HIV-associated and childhood TB diagnosis compared with smear microscopy, diagnostic yield in paucibacillary and smear-negative TB is still suboptimal.8–12 For example, among children with culture-confirmed TB, the sensitivity of Xpert MTB/RIF to detect Mtb in respiratory and gastric samples ranges from 62% to 66%.9 However, of the 50–80% of children with negative cultures who are clinically diagnosed with TB, Xpert MTB/RIF positivity decreases to 2%.9 Similarly, the sensitivity of Xpert MTB/RIF drops from 89% to 76% in HIV-associated TB in adults compared with non-HIV-associated TB.13 There remains a dire need for adjunct means to bacteriologically confirm HIV-associated, childhood, and other paucibacillary forms of TB.
Respiratory secretions are swallowed and enter the gastrointestinal (GI) track as part of the normal mucociliary escalator function.14,15 Thus, the use of gastric aspirates and stool to detect Mycobacterium tuberculosis (Mtb) is biologically plausible but has varying degrees of success with diagnostic yields of stool Xpert MTB/RIF ranging widely from 38% to 100%.16–18 Thus far, performance of stool Xpert MTB/RIF in children is less sensitive than sputum Xpert MTB/RIF,19–21 which may be related to Xpert MTB/RIF’s limit of detection (LOD) in sputum being (131 colony-forming units [CFU]/mL) compared with its stool LOD (6,800 CFU/mL).22 Improved stool processing has been shown to decrease the LOD for stool to ∼1,000 CFU/gm stool.23 Further improvement in stool DNA extraction could increase test sensitivity and consistency, making stool PCR a more feasible, noninvasive, highly sensitive technique for diagnosing pulmonary TB.
Currently, there are no accurate or precise means to monitor TB treatment. About 80% of individuals with acid-fast bacilli (AFB) sputum smear-positive TB will become smear negative by 2 months of effective treatment, making AFB sputum smear status the current surrogate measure of treatment effectiveness.24 Hence, for the 40% of HIV-associated TB and 76% of pediatric TB that are AFB sputum smear negative, only symptomatic and radiographic treatment monitoring are useful, although imprecise.1,13,25 Although the World Health Organization (WHO) recommends against using Xpert MTB/RIF for treatment monitoring, recent reports show that the semiquantitative measure provided by Xpert MTB/RIF is able to predict treatment failure and offers promise in predicting therapeutic response.26,27 Accurate and precise longitudinal quantification of the Mtb burden during antituberculosis treatment (ATT) could support earlier detection of treatment failures and thereby improve patient outcomes.
By applying a soil DNA isolation kit to stool and using the Mtb insertion sequence 6110 (IS6110),28 we report an improved LOD of a PCR diagnostic test for TB. We tested this novel method using clinical samples from adults and children with culture-confirmed and clinically diagnosed TB and asymptomatic healthy child household contacts (age < 15 years) who were enrolled in a concurrent immunology study.
METHODS
Stool-spiking study.
Stool from a healthy, asymptomatic, QuantiFERON TB Gold–negative, noninfected control subject was spiked with serial 10-log dilutions of strain H37Rv, HN3465, and HN3522 Mtb in a certified BSL-3 facility. Mycobacterium tuberculosis strains were grown at 37°C with 5% CO2 in Middlebrook 7H9 liquid media that had reached log-phase growth. The quantification of Mycobacteria was determined by spectrometry (Thermo Scientific Genesys 10S; Thermo Scientific, Waltham, MA) with an optical density of 1 equating to 3 × 108 CFU/mL as established by an in-house reference standard. Each dilution of H37Rv Mtb was spiked into six separate stool samples. Mycobacterium tuberculosis strains HN3465 and HN3522 were spiked into three separate stool samples per dilution. DNA was isolated and quantitative PCR (qPCR) was performed with quantification of DNA.
Study setting and population.
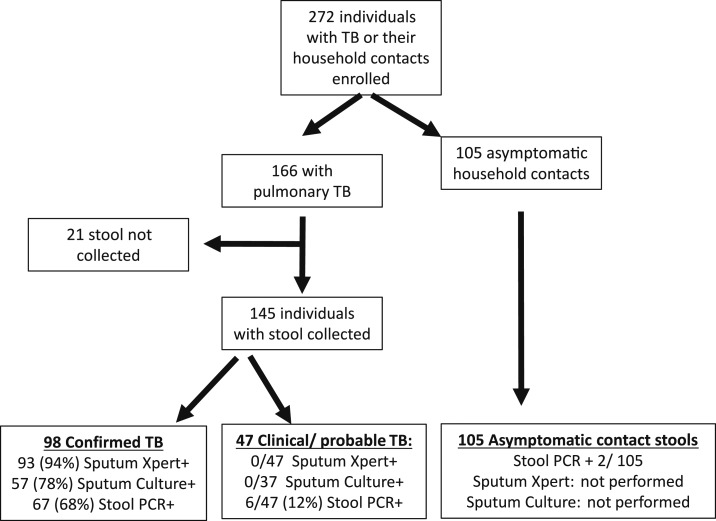
Between October 2014 and May 2017, participants were consecutively enrolled from the Baylor-Swaziland Children’s Foundation TB Clinical Center of Excellence in Mbabane, Swaziland (Figure 1). As part of an ongoing immunology study, participants were enrolled up to 6 weeks after initiating ATT. Individuals with clinical symptoms (e.g., fever, lethargy, and cough) and signs (e.g., wheezing, rales, failure to gain weight, and radiographic findings) consistent with TB were evaluated by a clinician and categorized in accordance with internationally accepted case definitions for research.29 Depending on age, pulmonary TB evaluation included expectorated or induced sputum for adults and older children and induced sputum with nasopharyngeal aspiration or gastric lavage for young children. Radiographic findings were documented at baseline and repeated on follow-up if clinically warranted. Based on a combination of clinical history, exposure to a known TB contact, and radiographic findings, a clinician diagnosed clinical TB and recommended ATT. Diagnosis and treatment decisions were made irrespective of a negative Xpert result and before culture results were available. Individuals were excluded from participation if they had received ATT in the past year or if unable to provide consent. The participants were followed prospectively for a minimum of 6 months after ATT initiation with repeat stool collected at 2 months. Asymptomatic, household contacts of microbiologically confirmed pulmonary TB patients were enrolled to test specificity.
Figure 1.
Cohort enrollment. TB = tuberculosis; PCR = polymerase chain reaction.
Sample collection and processing.
Participants provided both stool and respiratory samples. Fecal samples were collected and within 12 hours, stored without preservatives in a −80°C freezer until batch DNA isolation was performed in Mbabane, Swaziland, as previously described.30 In brief, 50 mg of stool was processed using the MP Fast DNA kit for soil (MP Biochemicals, Solon, OH) with a 6-minute homogenization via bead-beating disruption on the SI-D238 Disruptor Genie (Scientific Industries, Inc., Bohemia, NY) and with 100 μL final volume used to elute DNA. Based on Swaziland National Guidelines, the first respiratory sample was analyzed by Xpert and a second respiratory sample was sent for culture at a later date if the Xpert was positive or if a clinician made a clinical diagnosis of TB despite a negative Xpert. In children, because of difficulty collecting samples, often the respiratory culture was obtained at a later date. Sputum and gastric lavage samples were processed for Xpert as previously described.31 Sputum was collected at time of diagnosis and before ATT initiation, whereas stool was collected at time of study enrollment.
Quantitative PCR (qPCR).
Mycobacterium tuberculosis–specific primers and black hole quencher Fluorescein amidite–labeled minor groove binder probes were selected from previously described Mtb-specific sequences (Table 1) (Integrated DNA Technologies, Coralville, IA). Prior research has demonstrated that this sequence has no cross-reactivity to 35 different nontuberculous mycobacterial strains.28 In this study, spiked, Mycobacterium avium, and Mycobacterium marinum (a kind gift from Dr. Jeffrey Cirillo) DNA were used to confirm the Mtb specificity. Primers were designed using Primer Express Software (Applied Biosytems, Foster City, CA) using IS6110 and purchased from Integrated DNA Technologies. Quantitative PCR was performed using a QuantStudio3 Real-time PCR System (Thermo Fischer, Waltham, MA) using fast chemistry, 40 cycles of 95°C for 3 seconds and 60°C for 30 seconds. Each assay was performed using a total volume of 7 μL consisting of 2 μL of isolated DNA, 900 nM of each primer, 250 nM probe, and 3.5 μL of TaqMan Advanced Fast MasterMix 2X (Applied Biosystems, Foster City, CA). DNA isolated from H37Rv Mtb was logarithmically serially diluted to create a 7-point standard curve performed in triplicate for each plate. The quantitative value, in femtogram per microliters, was determined by comparing the cycle threshold (Ct) value of unknown samples with this 7-point standard curve. A Ct value of 38 was set as the cutoff point for positivity based on the known LOD from our H37Rv DNA standards. Quantitative PCR performed on isolated DNA from clinical samples were run in adjacent duplicate wells. If the two wells were discordant, the samples were again tested in duplicate with the result considered negative if repeat testing remained discordant. The samples were also repeatedly tested if discordant or duplicate results had a Ct difference more than 30%. All plates contained a “No Template Control” consisting of master mix and PCR-grade water as a negative control. Individuals implementing and analyzing qPCR results were blind to Xpert and culture results.
Table 1.
Sequence of the primers and probe
| FWR primer | CCTGAAAGACGTTATCCACCATAC |
| RVS primer | CGGCTAGTGCATTGTCATAGGAG |
| FAM/ZEN probe | TCTCAGTAC/ZEN/ACATCGATCCGGTTCAGC |
FAM = fluorescein amidite; FWR = forward; RVS = reverse.
Statistical analysis.
Statistical analysis was performed using Prism version 6.0 (GraphPad, LaJolla, CA). Correlation of Mtb-spiked stool with quantified DNA was evaluated using Pearson correlation coefficient. Spearman’s rank correlation was used to evaluate the relationship between time on ATT and quantity of isolated DNA. Wilcoxon matched signed-rank test was used to evaluate an individual’s pair-matched Mtb DNA levels before and after ATT initiation. Fisher’s exact test (two tailed) was performed to evaluate detection of Mtb from stool PCR compared with culture and Gene Xpert.
Ethics Statement.
The Baylor-Swaziland Children’s Foundation, Baylor College of Medicine Institutional Review Board, and the Swaziland Scientific and Ethics Council approved this study. All participants underwent written informed consent in compliance with the guidelines for the Protection of Human Subjects and the Declaration of Helsinki.
RESULTS
Limit of detection in stool.
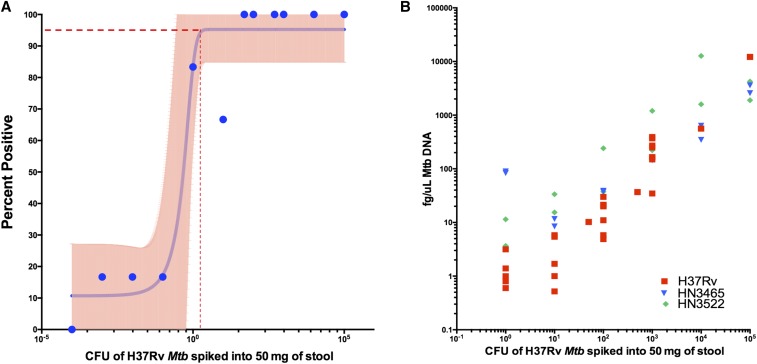
To evaluate the LOD of the qPCR assay in stool, H37Rv Mtb was spiked into 50 mg of healthy control stool and the DNA subsequently isolated using the MP Fast kits.30 The quantity of Mtb, as determined by spectrometer, obtained from spiking 106–10−4 CFU of Mtb into 50 mg of healthy stool had a high correlation with known femtogram per microliters quantities of control H37Rv DNA (Figure 2A; Pearson’s r = 0.998; 95% confidence interval [CI]: 0.9961–0.9992; P < 0.0001). When 1 CFU of H37Rv Mtb was spiked into 50 mg of stool, the assay detected Mtb in five of six occurrences. Ten colony-forming units spiked into 50 mg of stool were detected four of six times, whereas 100–100,000 CFU of H37Rv were detected six of six times (Figure 2B). When 0.1 and 0.01 CFU was spiked into 50 mg of stool, the assay detected Mtb one of six times, whereas zero of six was detected when 0.0001 CFU was spiked into 50 mg of stool. By logistic regression, there was a 95% probability of detecting H37Rv Mtb in spiked stool in samples containing at least 96 CFU/50 mg of stool (Figure 2B; 95% CI: 84.87–105.6).
Figure 2.
Quantity of Mycobacterium tuberculosis (Mtb) DNA detected by polymerase chain reaction (PCR) correlates with known colony-forming units (CFUs) of Mtb. Stool from a healthy control without tuberculosis was spiked with 10-fold dilutions of H37Rv Mtb (N = 6 per concentration, N = 60 total). (A) Stool DNA was isolated and Mtb qPCR quantified (Pearson’s r = 0.998, P value < 0.0001). (B) The percent of assays that detected Mtb was plotted for each concentration. Using logistic regression, there was a 95% probability of detecting Mtb in samples containing at least 95.24 CFU/50 mg of stool (95% confidence interval [CI]: 84.87–105.6) (95% CI shown in pink). This figure appears in color at www.ajtmh.org.
IS6110, although specific to the Mtb complex, can be found in up to 25 copies per strain with H37Rv having 16 copies.28 We, therefore, tested two clinical strains (HN3465 and HN3522) of Mtb that contain seven and 11 copies, respectively, with similar correlation (Figure 2A). There was no cross-reactivity when the assay was performed with M. avium or M. marinum DNA. In previous experiments, there was no amplification of the IS6110 sequence when evaluated against 84 samples of 35 non-tuberculosis mycobacteria strains.28
Clinical validation.
To evaluate the assay’s performance in clinical samples, we compared results of qPCR of Mtb from stool against results from the Xpert and culture of sputum or gastric lavage samples. Within 6 weeks of ATT initiation, 107 adults and 38 children (less than 15 years of age) were consecutively enrolled and provided stool (Figure 1). Forty-six individuals (19 adults and 27 children) had probable TB (Table 2), as defined by consensus definitions and clinical guidelines,1,29 and initiated ATT without Xpert or culture confirmation. Sixty-six percent (96/145) were HIV-infected with 67% (98/145) having TB microbiologically confirmed either by Xpert and/or culture.
Table 2.
Cohort characteristics and test performance
| Micro-confirmed TB (N = 98) | Probable TB (N = 47) | Asymptomatic household contacts (N = 105) | |
|---|---|---|---|
| Age (median and IQR) | 30 (21–37) | 9 (4–31) | 7 (4–11) |
| Gender (%F) | 53/98 (54%) | 26/47 (55%) | 65% |
| HIV status (%+) | 65/98 (66%) | 31/47 (65%) | 22% |
| Median CD4 count (IQR) | 114 (31–324) | 152 (31–378) | 1,030 (603–1,195) |
| Sputum Xpert results | 93/98 (94%) | 0/33 (0%) | NA |
| Sputum culture results* | 57/73 (78%) | 0/21 (0%) | NA |
| Median time stool collected after ATT initiation in days (IQR) | 6 (1–14) | 1 (0–13) | NA |
| Stool PCR results | 67/98 (68%) | 6/47 (12%) | 2/105 (1.9%) |
| Adults | |||
|---|---|---|---|
| Micro-confirmed TB (N = 87) | Probable TB (N = 19) | ||
| Age (median) | 32 (25–41) | 32 (25–41) | |
| HIV status (%+) | 60/87 (68%) | 18/19 (94%) | |
| Median CD4 count (IQR) | 116 (30–323) | 115 (29–320) | |
| Sputum Xpert results | 82/87 (94%) | 0/19 (0%) | |
| Sputum culture results* | 50/65 (77%) | 0/14 (0%) | |
| Median time stool collected after ATT initiation in days | 5 (0–14) | 6 (1–14) | |
| Stool PCR results | 61/87 (70%) | 4/19 (21%) | |
| Children | |||
|---|---|---|---|
| Micro-confirmed TB (N = 10) | Probable TB (N = 28) | ||
| Age (median and IQR) | 7 (3–10) | 6 (2–9) | |
| HIV status (%+) | 5/10 (50%) | 15/27 (55%) | |
| Median CD4 count (IQR) | 181 (28–543) | 214 (27–598) | |
| Sputum Xpert results | 10/10 (100%) | 0/27 (0%) | |
| Sputum culture results* | 6/7 (87%) | 0/23 (0%) | |
| Median time stool collected after ATT initiation in days | 1 (0–14) | 1 (1–13) | |
| Stool PCR results | 5/10 (50%) | 2/27 (7%) | |
ATT = antituberculosis treatment; PCR = polymerase chain reaction; TB = tuberculosis.
According to National Guidelines, respiratory samples for culture are sent only if Xpert is positive or an individual is started on ATT. Therefore, the samples are not sent at the same time.
To evaluate the method’s diagnostic utility, in adults and children, the stool qPCR result was compared with the diagnostic reference standard of sputum culture. Stool was collected as part of an immunology study and was, therefore, collected a median of 4 days (interquartile range: 0–14 days) after ATT initiation. Among individuals with a positive culture who provided stool within 72 hours of ATT initiation, the diagnostic yield from stool qPCR was 22/23 comparable to 19/23 for sputum Xpert (Fisher’s exact P = 0.34). In culture-positive individuals in whom the stool was collected greater than 72 hours since ATT initiation (obtained at a median of 4 days; interquartile 1–interquartile 3 0–14 days after ATT initiation), stool qPCR yield detected 47/57 compared with 52/57 for sputum Xpert (Fisher’s exact P = 0.26). Of note, sputum for Xpert was collected before ATT. In comparison, smear AFB microscopy was positive in 34 of 92 culture-positive samples. Supplemental Tables 1 and 2 and Supplemental Figure 1 depict the performance of sputum AFB smear with stool Mtb microscopy.
There were 47 individuals (19 adults and 28 children) with clinically diagnosed TB. Sputum Xpert was performed on all 47, and in 37 individuals, a second specimen was able to be collected for culture. Stool qPCR was positive in 6/47 (12%) individuals with negative sputum Xpert (Fisher’s exact, P = 0.02) and in 3/37 (8%) individuals with negative sputum Xpert and negative culture (Fisher’s exact, P = 0.23).
Stool qPCR was also performed for 105 asymptomatic, non-diseased individuals with recent (< 6 months) household TB contact. Of these, two contacts were positive for Mtb, indicating a specificity of 98.2 (95% CI: 93.8–99.7%).
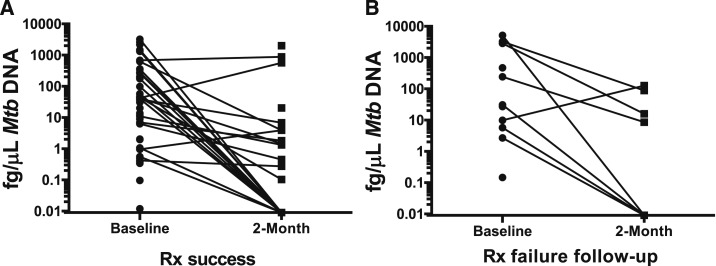
Participants were longitudinally evaluated with comparison of baseline and 2-month follow-up stool qPCR results. Of 73 participants with stool-positive qPCR at baseline, 83% (60/73) provided stool at the 2-month study follow-up (four deceased, one too sick to provide stool, and eight lost to follow-up or transferred to another facility). Individual levels of Mtb DNA significantly declined following initiation of ATT (Figure 3; median difference: 319 fg/μL of DNA, P < 0.0001 Wilcoxon signed-rank test). Among individuals with treatment failure or drug resistance, four of nine (44%) had persistent stool qPCR at 2 months compared with 22 of 119 (18%) with cure or treatment completion (relative risk [RR] 2.8 CI: 1.2–6.5, P = 0.012; Figure 3). In comparison, AFB smear status at 2 months was positive in three of 12 (25%) individuals with treatment failure or drug resistance, compared with one of 78 with cure or treatment completion (RR 15.8; CI: 1.7–141.8; P = 0.013). Sputum AFB smear and stool Mtb PCR had complementary value with AFB smear detecting two positive follow-up samples not detected by stool PCR, whereas stool PCR detected five follow-up–positive cases that were not detected by AFB smear (Supplemental Table 2). In 52 individuals with microbiologically confirmed TB and baseline-negative AFB smear, stool PCR was detected in 31, a 59% increased yield of individuals who would have lacked a test of treatment monitoring (Supplemental Figure 1).
Figure 3.
Mycobacterium tuberculosis (Mtb) burden decreases during antituberculosis treatment (ATT). Individuals with tuberculosis provided stool samples at time of enrollment and again at the 2-month follow-up visit. (A) In most participants with treatment success, quantified Mtb polymerase chain reaction (PCR) on paired samples demonstrated a decrease in time as ATT increases (inverse correlation). (B) Individuals with treatment failure or drug resistance had an increased risk of persistent stool Mtb PCR at 2 months (relative risk 2.8 confidence interval: 1.2–6.5 P = 0.012). Rx = treatment.
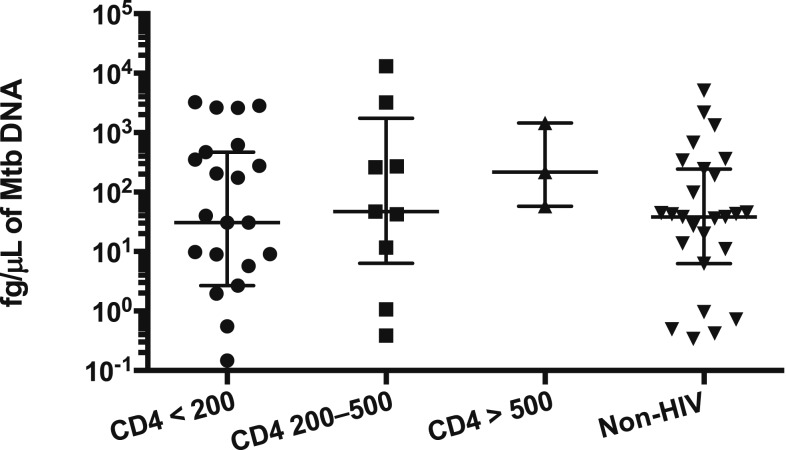
Stool Mtb DNA detection levels did not vary by HIV infection status (Kruskal–Wallis test, P value = 0.493; Figure 4) and were not correlated with CD4 counts (Spearman r = −0.08 P = 0.65). After controlling for time on ATT, the quantity of Mtb DNA detected in stool was similar for individuals with and without HIV infection (Mann–Whitney test, P value 0.527, two tailed).
Figure 4.
Quantitative Mycobacterium tuberculosis (Mtb) levels are similar despite HIV status. Individuals with tuberculosis completed stool Mtb quantitative polymerase chain reaction. Results were compared between individuals with and without HIV infection, and quantitative Mtb femtogram per microliters were similar among HIV-infected and HIV-infected individuals and did not vary by CD4 count (Kruskal–Wallis test, P value = 0.493). Error bars depict the median and interquartile range.
DISCUSSION
The success of Xpert has driven WHO’s rapid rollout of the technology to replace AFB smear microscopy as the initial diagnostic test of choice.32 Despite these significant advances, aspects of TB diagnosis remain challenging as Xpert fails to detect a third of smear-negative, culture-positive adult TB and is positive in only 2% of culture-negative, clinically diagnosed childhood TB.9 Furthermore, sputum is challenging to obtain in young children in resource-limited settings and often inadequate in children and HIV-infected adults with paucibacillary disease, limiting the use of traditional sputum-based diagnostics.
By adapting a DNA isolation method originally designed for soil and modified to increase the yield of GI parasite detection,30 we have refined a qPCR method that affords a stool Mtb LOD of 105 CFU/50 mg of stool. These spiking study results were validated in our clinical cohort showing similar and complementary diagnostic yield among stool samples and respiratory samples collected at similar time points. Addition of this method improved bacteriologic confirmation by 8% in Xpert-negative, culture-negative individuals with a clinical diagnosis of TB. The mycobacteriology marker, IS6110, has demonstrated good specificity with regard to NTM species in our data and others’ data.28,33 This is confirmed in our cohort, with a specificity of 98% among asymptomatic household contacts of TB cases. Although HIV-associated TB is often described as paucibacillary, neither the CD4+ count nor HIV status impacted the burden of Mtb found in stool (Figure 4). This preliminarily suggests that despite HIV-associated TB being characterized by paucibacillary sputum, the Mtb burden, as quantified by stool, may not be lower in HIV-infected individuals compared with HIV-uninfected individuals. Larger studies should, therefore, evaluate if quantitative stool Mtb DNA monitoring has improved treatment monitoring capacity compared with sputum-based techniques.
Currently, TB treatment monitoring is evaluated by AFB sputum smear microscopy, precluding the ability to monitor treatment among smear-negative individuals. Here, we show a quantitative measure of Mtb burden that, at 2 months, correlated with treatment outcome or drug resistance status. Recent evidence suggests that a decrease in sputum Mtb DNA burden at 1 week reliably predicts conversion to culture negativity.27 Despite the modest size of this pilot study and relatively late follow-up measure at 2 months, our method shows promise to detect individuals at increased risk of treatment failure. Earlier application of this method could afford a powerful prognostic tool to detect treatment failure.
Collection of GI tract samples to diagnose TB is a long-standing practice with the first report of gastric lavage described in 1898.34 As physiologic mucociliary escalator function empties respiratory secretions into the GI system, multiple studies have evaluated the ability of stool to diagnose TB. Using manual mixing,35 centrifugation,19 or a sugar flotation method,16,20,22 diagnostic yield of stool using in-house PCR or Xpert have yielded variable results. In contrast, our method uses mechanical homogenization, DNA isolation and purification, and adapted Mtb-specific primers and probes with a resultant improved LOD compared with previous stool methods.22 Except for the BSL-3 spiking studies, all Xpert, DNA extraction, and qPCR were performed in Mbabane, Swaziland, in a peripheral laboratory adjoining a HIV primary care clinic, demonstrating feasibility in a resource-constrained environment. Consistent with WHO guidelines for processing sputum, special care was given in our BSL-2 laboratory to avoid aerosolization.
Although these preliminary results are promising, there are several inherent limitations of this evaluation. Despite the improvement in the LOD, these methods are laborious, and transition to a test with widespread clinical feasibility will require a conversion to point-of-care methods. Furthermore, the clinical cohort was from an immunology study and stool samples were collected a median of 6 days after ATT initiation. Similarly, the stool was not collected at the same time as the respiratory sample evaluated by Xpert. In addition, there are some rare Mtb strains that lack the IS6110 region and, therefore, would test negative by this method.36 Finally, multiple studies have shown increased diagnostic yield of culture with additional sputum samples sent. This study had a real world approach sending a single stool and single sputum for Xpert and culture. Future studies should evaluate the additional yield of multiple stools in comparison to multiple sputum Xperts and culture tests.
In summary, we show that coupled with improved DNA extraction, stool Mtb qPCR has a LOD comparable to culture and increased diagnostic yield when used as an add-on test to existing sputum examinations. In addition to diagnostic potential, the quantitative nature of this method may afford detection of treatment failure earlier than sputum smear microscopy. As highlighted by the widespread rollout of Gene Xpert, PCR capacity is becoming more available to peripheral health-care systems. This method’s diagnostic advantages, use of an easily attained specimen, and ease of application in a peripheral laboratory lacking specialized equipment collectively suggest promise to improve TB diagnosis and treatment among at risk populations in TB high-burden settings.
Supplementary Material
Supplemental figure and tables
Acknowledgments:
We would like to thank all the patients in Swaziland for participating in the study and Dr. Jeffrey Cirillo for gifting us the non-tuberculosis mycobacteria used in the spiking studies.
Note: Supplemental figure and tables appear at www.ajtmh.org.
REFERENCES
- 1.WHO , 2016. Global Tuberculosis Report 2016. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 2.Harries AD, Hargreaves NJ, Kemp J, Jindani A, Enarson DA, Maher D, Salaniponi FM, 2001. Deaths from tuberculosis in sub-Saharan African countries with a high prevalence of HIV-1. Lancet 357: 1519–1523. [DOI] [PubMed] [Google Scholar]
- 3.Harries AD, Nyirenda TE, Banerjee A, Boeree MJ, Salaniponi FM, 1999. Treatment outcome of patients with smear-negative and smear-positive pulmonary tuberculosis in the National Tuberculosis Control Programme, Malawi. Trans R Soc Trop Med Hyg 93: 443–446. [DOI] [PubMed] [Google Scholar]
- 4.Macpherson P, et al. 2011. Risk factors for mortality in smear-negative tuberculosis suspects: a cohort study in Harare, Zimbabwe. Int J Tuberc Lung Dis 15: 1390–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Velez CM, Marais BJ, 2012. Tuberculosis in children. N Engl J Med 367: 348–361. [DOI] [PubMed] [Google Scholar]
- 6.Gupta RK, Lucas SB, Fielding KL, Lawn SD, 2015. Prevalence of tuberculosis in post-mortem studies of HIV-infected adults and children in resource-limited settings: a systematic review and meta-analysis. AIDS 29: 1987–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO , 2015. Global Tuberculosis Report. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 8.Small PM, Pai M, 2010. Tuberculosis diagnosis–time for a game change. N Engl J Med 363: 1070–1071. [DOI] [PubMed] [Google Scholar]
- 9.Detjen AK, DiNardo AR, Leyden J, Steingart KR, Menzies D, Schiller I, Dendukuri N, Mandalakas AM, 2015. Xpert MTB/RIF assay for the diagnosis of pulmonary tuberculosis in children: a systematic review and meta-analysis. Lancet Respir Med 3: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aryan E, Makvandi M, Farajzadeh A, Huygen K, Alvandi AH, Gouya MM, Sadrizadeh A, Romano M, 2013. Clinical value of IS6110-based loop-mediated isothermal amplification for detection of Mycobacterium tuberculosis complex in respiratory specimens. J Infect 66: 487–493. [DOI] [PubMed] [Google Scholar]
- 11.Jeanes C, O’Grady J, 2016. Diagnosing tuberculosis in the 21st century—dawn of a genomics revolution? Int J Mycobacteriol 5: 384–391. [DOI] [PubMed] [Google Scholar]
- 12.Pholwat S, Stroup S, Foongladda S, Houpt E, 2013. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One 8: e57238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steingart KR, Sohn H, Schiller I, Kloda LA, Boehme CC, Pai M, Dendukuri N, 2013. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev 1: CD009593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulmar D, Ornstein OG, 1933. Gastric examination in pulmonary tuberculosis with negative sputum: diagnostic importance. JAMA 101: 835–836. [Google Scholar]
- 15.Alonso JM, 2008. Immunity and pathophysiology of respiratory tract infections. Med Mal Infect 38: 433–437. [DOI] [PubMed] [Google Scholar]
- 16.DiNardo AR, Hahn A, Leyden J, Stager C, Baron EJ, Graviss EA, Mandalakas AM, Guy E, 2015. Use of string test and stool specimens to diagnose pulmonary tuberculosis. Int J Infect Dis 41: 50–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf H, et al. 2008. Diagnosis of pediatric pulmonary tuberculosis by stool PCR. Am J Trop Med Hyg 79: 893–898. [PMC free article] [PubMed] [Google Scholar]
- 18.Kokuto H, Sasaki Y, Yoshimatsu S, Mizuno K, Yi L, Mitarai S, 2015. Detection of Mycobacterium tuberculosis (MTB) in fecal specimens from adults diagnosed with pulmonary tuberculosis using the Xpert MTB/rifampicin test. Open Forum Infect Dis 2: ofv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicol MP, Spiers K, Workman L, Isaacs W, Munro J, Black F, Zemanay W, Zar HJ, 2013. Xpert MTB/RIF testing of stool samples for the diagnosis of pulmonary tuberculosis in children. Clin Infect Dis 57: e18–e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcy O, et al. 2016. Performance of Xpert MTB/RIF and alternative specimen collection methods for the diagnosis of tuberculosis in HIV-infected children. Clin Infect Dis 62: 1161–1168. [DOI] [PubMed] [Google Scholar]
- 21.Walters E, et al. 2017. Xpert MTB/RIF on stool is useful for the rapid diagnosis of tuberculosis in young children with severe pulmonary disease. Pediatr Infect Dis J 36: 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor N, Gaur RL, Baron EJ, Banaei N, 2012. Can a simple flotation method lower the limit of detection of Mycobacterium tuberculosis in extrapulmonary samples analyzed by the GeneXpert MTB/RIF assay? J Clin Microbiol 50: 2272–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banada PP, Naidoo U, Deshpande S, Karim F, Flynn JL, O’Malley M, Jones M, Nanassy O, Jeena P, Alland D, 2016. A novel sample processing method for rapid detection of tuberculosis in the stool of pediatric patients using the Xpert MTB/RIF assay. PLoS One 11: e0151980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nahid P, et al. 2016. Executive summary: Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Clinical Practice guidelines: treatment of drug-susceptible tuberculosis. Clin Infect Dis 63: 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Detjen AKDA, Leyden J, Steingart KR, Menzies D, Schiller I, Dendukuri N, Mandalakas AM, 2013. Systematic Review and Meta-Analysis: Xpert MTB/RIF for the Diagnosis of Pulmonary Tuberculosis, Peripheral Lymph Node TB and TB meningitis in Children. WHO Expert Group Meeting on Use of Xpert MTB/RIF in TB control in low and middle-income settings. WHO Expert Meeting, May 20–21, 2013, Vevrier Du-Lac, France. [Google Scholar]
- 26.Malherbe ST, et al. 2016. Persisting positron emission tomography lesion activity and Mycobacterium tuberculosis mRNA after tuberculosis cure. Nat Med 22: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenai S, et al. 2016. Bacterial loads measured by the Xpert MTB/RIF assay as markers of culture conversion and bacteriological cure in pulmonary TB. PLoS One 11: e0160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barletta F, Vandelannoote K, Collantes J, Evans CA, Arevalo J, Rigouts L, 2014. Standardization of a TaqMan-based real-time PCR for the detection of Mycobacterium tuberculosis-complex in human sputum. Am J Trop Med Hyg 91: 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham SM, et al. 2012. Evaluation of tuberculosis diagnostics in children: 1. Proposed clinical case definitions for classification of intrathoracic tuberculosis disease. Consensus from an expert panel. J Infect Dis 205 (Suppl 2): S199–S208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mejia R, Vicuna Y, Broncano N, Sandoval C, Vaca M, Chico M, Cooper PJ, Nutman TB, 2013. A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am J Trop Med Hyg 88: 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cepheid , 2015. Xpert MTB/RIF Package Insert Available at: http://www.cepheid.com/us/mtbrif-pi. Accessed April 27, 2018.
- 32.Van Rie A, 2013. Xpert MTB/RIF: a game changer for the diagnosis of pulmonary tuberculosis in children? Lancet Glob Health 1: e60–e61. [DOI] [PubMed] [Google Scholar]
- 33.Desjardin LE, et al. 1999. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Respir Crit Care Med 160: 203–210. [DOI] [PubMed] [Google Scholar]
- 34.Foley JA, Andosca JB, 1943. The value of the examination of gastric contents for tubercle bacilli. Ann Intern Med 19: 629–633. [Google Scholar]
- 35.Walters E, Gie RP, Hesseling AC, Friedrich SO, Diacon AH, Gie RP, 2012. Rapid diagnosis of pediatric intrathoracic tuberculosis from stool samples using the Xpert MTB/RIF Assay: a pilot study. Pediatr Infect Dis J 31: 1316. [DOI] [PubMed] [Google Scholar]
- 36.Lok KH, Benjamin WH, Jr., Kimerling ME, Pruitt V, Lathan M, Razeq J, Hooper N, Cronin W, Dunlap NE, 2002. Molecular differentiation of Mycobacterium tuberculosis strains without IS6110 insertions. Emerg Infect Dis 8: 1310–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables