Abstract
Developmental toxicity of compounds, which women of reproductive age are exposed to, should be assessed to minimize the incidence of miscarriage and birth defects. The present study examined the potential developmental toxicity of resveratrol, a dietary supplement widely marketed with various health claims, using the P19C5 embryoid body (EB) morphogenesis assay, which evaluates adverse effects of chemical exposures on tissue growth and axial elongation. Resveratrol (trans isoform) impaired morphogenesis at 4 μM and higher, creating smaller and rounder EBs, whereas cis isoform, and glucuronated and sulfonated metabolites did not. Trans-resveratrol also altered expression levels of developmental regulator genes involved in embryonic patterning, such as Wnt3a, Tbx6, and Cyp26a1. To investigate the mechanisms of trans-resveratrol action, the roles of estrogen receptor, sirtuin 1 (SIRT1), and DNA replication in EB morphogenesis were examined. Neither activators of estrogen receptors (diethylstilbestrol [18 μM] and raloxifene [8 μM]) nor activator of SIRT1 (SRT1720 [2.4 – 3.2 μM]) caused morphological and molecular alterations that are comparable to trans-resveratrol (10 μM). By contrast, a reduction in the DNA replication rate with aphidicolin (0.4 μM) or hydroxyurea (40 μM) created smaller and rounder EBs and altered the expression levels of Wnt3a, Tbx6, and Cyp26a1 in a manner similar to trans-resveratrol. Consistently, trans-resveratrol significantly reduced the rate of EdU incorporation in P19C5 cells. These results suggest that a reduction in the DNA replication rate is one of the mechanisms by which trans-resveratrol impacts EB development. This study provides mechanistic insight for further investigations on the developmental toxicity of trans-resveratrol.
Keywords: Resveratrol, Dietary supplement, Developmental toxicity, Morphogenesis, In vitro assay, Stem cell
1. Introduction
Numerous chemical compounds exhibit developmental toxicity, causing embryonic death or malformation in the reproductive tract of the pregnant mother. Whether a given compound adversely affects embryo development depends on its chemical nature as well as concentration, duration, and timing of exposure in utero (Jelinek, 2005; Schardein and Macina, 2006; Daston et al., 2010; Friedman, 2010). Pharmaceutical compounds are usually investigated for their developmental toxicity through experimentations using pregnant model animals and through human case and cohort studies. By contrast, most non-pharmaceutical compounds, such as herbicides, pesticides, cosmetics, industrial byproducts, excipients, and dietary supplements, are much less studied for developmental toxicity. Because the amount and frequency of exposure are not strictly regulated or monitored for non-pharmaceutical compounds, it is difficult to assess their developmental toxicity through human studies. Although experimentations with pregnant animals are routinely conducted in developmental toxicity research, testing of individual non-pharmaceutical compounds would sacrifice enormous numbers of animals, which is not only costly but also unethical from an animal welfare standpoint. To reduce such burden of animal-based research, non-animal alternatives, namely in vitro tests, are highly desired to evaluate the developmental toxicity of compounds. While in vitro tests alone may not be sufficient to fully predict potential harm to embryos in utero, they can yield valuable information on concentration-response relations between specific adverse outcomes and compound exposures. In vitro tests can also provide insight into the mechanisms of developmental toxicity, because they are more amenable to molecular interrogations than in vivo tests. The information obtained from in vitro tests can serve as the foundation for designing animal- and human-based studies in an effective manner.
One of the in vitro tests to evaluate the developmental toxicity of compounds utilizes embryoid body morphogenesis of the mouse P19C5 stem cell line (reviewed in Marikawa, 2018). P19C5 cells possess developmental characteristics similar to the epiblast, the pluripotent embryonic precursor of the entire fetal body. P19C5 cells can be induced to differentiate in vitro as embryoid bodies (EBs) by aggregation culture in hanging drops. During the first two days of culture, P19C5 EBs grow as spherical cell aggregates. By the fourth day of culture, EBs have transformed into an elongated shape with a distinct morphological polarity (Lau and Marikawa, 2014). Spatial and temporal gene expression profiles suggest that the in vitro development of EBs represents gastrulation, the morphogenetic process of body patterning and elongation along the cranial-caudal embryonic axis. Morphological and molecular changes in EBs are controlled by key morphogenetic signals, such as Wnt, Nodal, Fgf, and retinoic acid, in a manner consistent with their regulatory roles in gastrulation (Li and Marikawa, 2015). Importantly, in vitro development of P19C5 EBs is impaired by chemical exposures that are known to cause developmental toxicity in vivo (Warkus et al., 2016; Warkus and Marikawa, 2017). As a reference list for developmental toxicity validation, Daston et al. (2014) compiled 39 chemical exposures, i.e., in vivo concentrations of specific compounds that exhibit adverse effects on embryos or lack thereof. EB growth and morphogenesis, which are quantitatively measured using morphometric parameters of EBs at the end of 4-day culture, are significantly altered by the adverse exposures of the Daston reference list, but not by the non-adverse exposures, with a total concordance of 71.4 to 82.9% (Warkus and Marikawa, 2017). P19C5 EBs also provide insight into the molecular mechanisms of developmental toxicity through the examination of how gene expression profiles are altered by chemical exposures (Li and Marikawa, 2016; Warkus and Marikawa, 2018). Thus, P19C5 EBs can be effectively used as an in vitro model to investigate the developmental toxicity of compounds.
The objective of the present study is to examine the developmental toxicity of a dietary supplement, resveratrol, using the P19C5 EB model. Resveratrol (3,5,4’-trihydroxy-trans-stilbene) is a natural compound produced by several plants, including grapes and blueberries (Signorelli and Ghidoni, 2005). Numerous in vitro and animal studies have implicated the beneficial effects of resveratrol against various diseases, such as cancers, cardiovascular diseases, inflammatory diseases, and diabetes (Baur and Sinclair, 2006; Park and Pezzuto, 2015). Several molecules have been suggested as targets of resveratrol, including the estrogen receptor (Gehm et al., 1997; Bowers et al., 2000), sirtuin 1 (SIRT1; Howits et al., 2003), phosphodiesterase (PDE; Park et al., 2012), AMP-activated protein kinase (AMPK; Baur et al., 2006), DNA polymerases (Stivala et al., 2001), and ribonucleotide reductase (Fontecave et al., 1998). Nonetheless, the molecular mechanisms underlying the therapeutic properties of resveratrol are still elusive (Kulkarni and Cantó, 2015). While most studies on resveratrol focus on its beneficial effects, the information on its potential developmental toxicity is scarce. Most anti-cancer drugs that are approved by the Food and Drug Administration (FDA) are placed under the Pregnancy Risk Category X, i.e., contraindicated for use during pregnancy due to their developmental toxicity. Because resveratrol suppresses proliferation and survival of various types of cancer cells (Jang et al., 1997; Park and Pezzuto, 2015; Singh et al., 2015), it is possible that developing embryos may also be susceptible to resveratrol exposure. Resveratrol is sold as a dietary supplement with health benefit claims that are not approved by the FDA, and is consumed by people, including women of reproductive age, without a physician’s prescription or monitoring. As a result, human studies are essentially impossible to establish an association between resveratrol intake and adverse reproductive outcomes. Intensive investigations are warranted through well-controlled experiments to examine the developmental toxicity of resveratrol. Specifically, the elucidation of the lowest-observed-adverse-effect level (LOAEL) that impairs embryo development and the molecular mechanisms underlying the adverse effects should help assess a potential risk of resveratrol for developing embryos.
Here, we first determined the concentration-response relationship of resveratrol, based on the morphogenetic impact on P19C5 EBs. Next, we examined gene expression profiles of resveratrol-treated EBs to evaluate which developmental regulators were affected by exposure to resveratrol. Lastly, we compared the morphological and molecular impact of resveratrol with other compounds, namely modulators of the estrogen receptor, SIRT1 activity, and DNA replication, to gain insight into the molecular mechanisms underlying the adverse effects of resveratrol. The data provided by the present study serve as a stepping-stone for future studies on the developmental toxicity of resveratrol, which should help determine whether this dietary supplement is to be consumed, and if so how much, by women of reproductive age.
2. Materials and methods
2.1. Chemicals
All chemical compounds used in the present study were commercially obtained, namely from Selleck Chemicals (Houston, TX), Santa Cruz Biotechnology (Dallas, TX), Sigma-Aldrich (St. Louis, MO), and Cayman Chemical (Ann Arbor, MI), and their details are described in Table 1. Chemical structures of some of the compounds are shown in Fig. 1, A. Note that trans-resveratrol was obtained from three different suppliers to confirm the consistency of the morphogenetic and molecular effects. One of them was a part of the Anti-Diabetic Compound Library (Selleck Chemicals), which consists of 33 chemicals that are widely used for diabetes research and treatment, including trans-resveratrol. The preparation of cis-resveratrol may contain 1–5% of trans-resveratrol, according to the product information of the supplier (www.caymanchem.com/product/10004235).
Table 1.
Chemicals used for the present study
| Compound Name | CASRN | Vendor (catalog number) | Stock concentration*1 | Figures*2 |
|---|---|---|---|---|
| trans-Resveratrol | 501-36-0 | Selleck (L2900; Anti-diabetes Compound Library) | 10 mM | 2A |
| Santa Cruz (200808) | 50 mM | 2BC, 3AB, 5DEFG | ||
| Cayman (70675) | 10 mM | 2DE, 3C | ||
| cis-Resveratrol | 61434-67-1 | Cayman (10004235) | 10 mM | 2DE, 3C |
| Resveratrol-3-O-sulfate | 858127-11-4 | Cayman (14942) | 10 mM | 2DE, 3C |
| Resve ratrol-3-O-D-glucuronide | 387372-17-0 | Cayman (13832) | 10 mM | 2DE, 3C |
| Resveratrol-4’-O-D-glucuronide | 387372-20-5 | Cayman (13833) | 10 mM | 2DE, 3C |
| Diethylstilbestrol | 56-53-1 | Santa Cruz (204720) | 50 mM | 4AC |
| Raloxifene | 82640-04-8 | Santa Cruz (204230) | 50 mM | 4BD |
| SRT1720 | 925434-55-5 | Sigma-Aldrich (567860) | 10 mM | 5ABCD |
| EX527 | 49843-98-3 | Sigma-Aldrich (E7034) | 50 mM | 5EFG |
| Aphidicolin | 38966-21-1 | Cayman (14007) | 1 mM | 6AC, 7ABC |
| Hydroxyurea | 127-07-1 | Sigma-Aldrich (H8627) | 100 mM | 6BD, 7ABC |
CASRN, Chemical Abstracts Service Registry Number
All compounds are dissolved in dimethyl sulfoxide (DMSO), except for hydroxyurea (dissolved in H2O)
Figure numbers that show experimental data using the corresponding chemical stocks
Fig. 1.
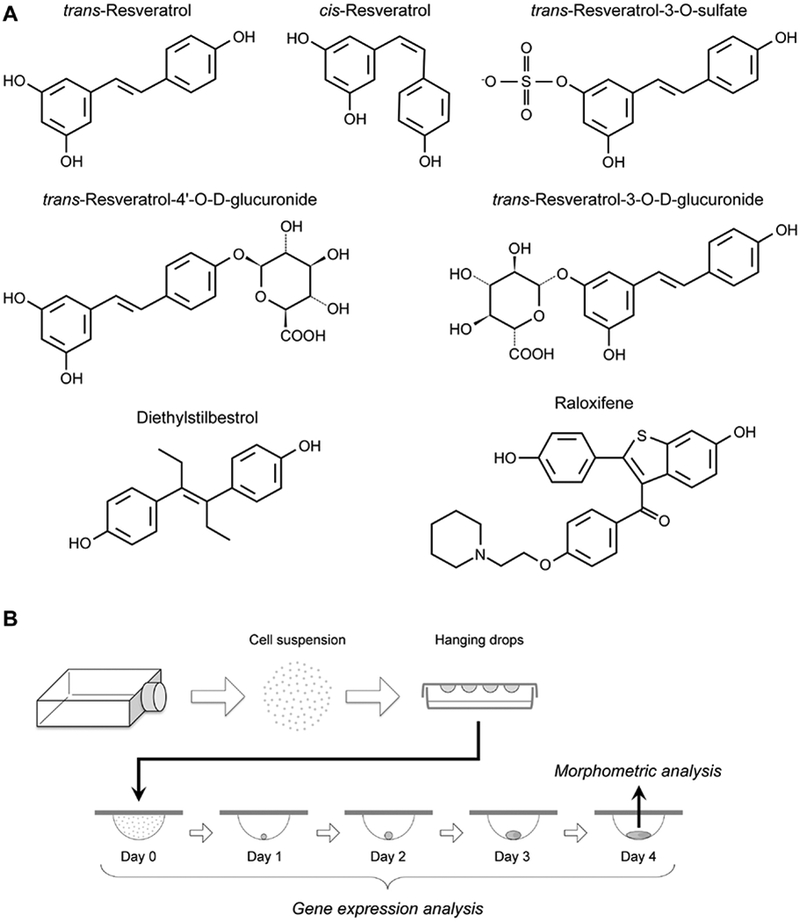
The chemical structures of the compounds evaluated in the present study (A), and the experimental scheme to assess the morphogenetic and molecular impact of compound exposures using P19C5 embryoid bodies (B).
2.2. Cell culture
P19C5 cells were propagated and used to generate embryoid bodies (EBs), according to the method previously described (Lau and Marikawa, 2014). Briefly, cells were dissociated with Trypsin-EDTA, and suspended in the culture medium at the density of 10 cells/μL with the specified amount of a test chemical, containing the final concentration of 1% dimethyl sulfoxide (DMSO). Drops (20 μL each) of cell suspension were spotted on the inner surface of Petri dish lids for hanging drop culture. EBs were removed from hanging drops for morphometric analyses at Day 4 of culture and gene expression analyses at Days 0, 1,2, 3, and 4 (Fig. 1, B). Experiments to assess the morphological impact on EBs by chemical treatments were conducted in three biological replicates using different collections of cell suspensions. For each replicate, 16 hanging drops were generated per treatment in parallel with 16 control (i.e., 1% DMSO only) hanging drops.
2.3. Morphometric analyses
EBs were placed together in a dish filled with phosphate-buffered saline for photography, using an AxioCam MRm digital camera connected to an Axiovert 200 microscope with Hoffman modulation-contrast optics (Carl Zeiss, Thornwood, NY). Image files were converted to JPEG format and opened in the ImageJ program (http://rsb.infonihgov/ij) for morphometric analyses. Morphological parameters, specifically area and circularity (= 4 × π × area / perimeter2), were measured on ImageJ by manually tracing the circumference of individual EBs using the polygon selection tool. Circularity was converted to Elongation Distortion Index (EDI = 1 / circularity – 1), which is more reflective of the extent of EB elongation: the more an EB elongates, the higher its EDI (Marikawa et al., 2009). Area was used as a proxy for the size of EB, whereas EDI was used to gauge the extent of EB axial elongation. Because the average area and EDI of control EBs were slightly different among experimental replicates, normalization was performed against the average values of control EBs in each replicate to calculate relative area and relative EDI, expressed as a percentage (i.e., control = 100%). Relative area and relative EDI data from all replicates were compiled, and their averages are shown with standard deviation. In the present study, a chemical treatment was defined as having an adverse morphogenetic effect when Day 4 EBs exhibited average relative area and/or average relative EDI that was lower than that of control by more than 30%.
2.4. Gene expression analyses
Quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed to determine the relative expression levels of developmental regulator genes (Table 2). Total RNA was extracted from EBs using TRI reagent (Sigma-Aldrich) and Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA), and processed for cDNA synthesis using M-MLV Reverse Transcriptase (Promega, Madison, WI) and oligo-dT (18) primer. Quantitative PCR was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) as follows: initial denaturation at 94°C for 5 min, followed by up to 45 cycles of 94°C for 15 sec, 60°C for 20 sec, and 72°C for 40 sec. Data files were opened in CFX Manager software (Bio-Rad) and Ct values were transferred to the Excel program for further analyses. Actb, which encodes β-Actin, was used as a housekeeping gene to normalize the expression levels of other genes. Actb has been effectively used as a housekeeping gene in the previous studies to evaluate gene expression levels in P19C5 EBs under various experimental conditions (Lau and Marikawa, 2014; Li and Marikawa, 2015, 2016; Yuan and Marikawa, 2017; Warkus and Marikawa, 2018). In addition, microarray analyses of EBs at Days 0 (dissociated cells immediately before aggregation), 1, 2, and 4 showed that the Actb transcript was expressed at a similar level during the course of development (Supplementary material). Gene expression analyses were conducted using three independent sets of samples as biological replicates using different collections of cell suspensions. Each set consisted of 9 samples: Day 0, control EBs at Days 1 to 4, and compound-treated EBs at Days 1 to 4, all of which were originated from the same cell suspension. Relative expression levels were calculated for each set of experiment, as previously described (Warkus and Marikawa, 2018), and the averages of the three replicates are shown with standard deviations.
Table 2.
Developmental regulator genes examined in the present study
| Gene Name | Characteristics* | Primer Sequences (5’ → 3’) | References |
|---|---|---|---|
| Actb | a. Cytoskeletal actin b. Ubiquitous c. House-keeping |
F: GAGAGGGAAATCGTGCGTGACATC R: CAGCTCAGTAACAGTCCGCCTAGA |
|
| Aldh1a2 | a. Aldehyde dehydrogenase b. Trunk region c. Retinoicacidsynthesis |
F: CTTGCCTCACAACAAGTGAGCTTC R: TCACCCAGGTTAGAGACTGGCTTC |
Haselbeck etal., 1999 |
| Brachyury | a. T-box transcription factor b. Early primitive streak c. Mesendoderm specification |
F: CCTCGGATTCACATCGTGAGAGTT R: AGTAGGTGGGCGGGCGTTATGACT |
Herrmann, 1991 |
| Cdx1 | a. Homeodomain transcription factor b. Early primitive streak c. Axial patterning |
F: TCAGGACTGGACATGAGGTAGAGG R: TGGGAAGGTGGGCATGAGCAGGTA |
Meyer and Gruss, 1993 |
| Cyp26a1 | a. Cytochrome P450 oxidase b. Posterior end c. Retinoicacid catabolism |
F: CGGAGCTGTGTAGGCAAAGAGTTT R: CCTGGAAGTGGGTAAATCTTGCAG |
Sakai et al., 2001 |
| Fgf8 | a. Fgf signaling ligand b. Posterior end c. Mesendoderm specification |
F: GTTGCACTTGCTG GTTCTCTGCCT R: AGTCCTTGCCTTTGCCGTTGCTCT |
Ohuchi et al., 1994 |
| Hoxal | a. HOX transcription factor b. Anterior class c. Axial patterning |
F: CCCTTTCCTTCCACACTGTCTTGT R: AAGACCCGTAAACTCTGCTCTGGA |
Wellik, 2009 |
| Hoxb9 | a. HOX transcription factor b. Posterior class c. Axial patterning |
F: AAGCAGGGAGIGGI I I IA IGAAGG R: GGGATAGGAATGTATGAATGGGGA |
Wellik, 2009 |
| Hoxc6 | a. HOX transcription factor b. Central class c. Axial patterning |
F: TTCGCCACAGGAGAATGTCGTGTT R: CGAGTTAGGTAGCGGTTGAAGTGA |
Wellik, 2009 |
| Pou5f1 | a. POU domain transcription factor b. Epiblast c. Pluripotencymaintenance |
F: AGGCAGGAGCACGAGTGGAAAGCA R: GGAGGGCTTCGGGCACTTCAGAAA |
Nichols et al., 1998 |
| Tbx6 | a. T-box transcription factor b. Posteriorend c. Axial stem cell differentiation |
F: GGCCTCTCTTCCACCCTTTAGTTC R: CACTAGTAACAAGGCCCCCAGGAG |
Chapman etal., 1996 |
| Wnt3 | a. Wnt signaling ligand b. Early primitive streak c. Initiation of gastrulation |
F: CAGATGCCCGCTCAGCTATGAACA R: AGCAGCACCAGTGGAAGACGCAAT |
Liu et al., 1999 |
| Wnt3a | a. Wnt signaling ligand b. Posteriorend c. Axial stem cell differentiation |
F: GCCACAAGAGCTTCCTGATTGGTA R: CCAGGCAGAAGACAGTCAGTCACC |
Takada et al., 1994 |
Characteristics are described based on: a. Molecularfunction, b. Major expression domains around the gastrulation stage (mouse embryonic stages from E5.5to E8.5), and c. Functional significance in earlyembryo development
2.5. Cell viability assay
Impact on cell proliferation and viability was assessed with the CellTiter-Glo Luminescent Cell Viability Assay System (Promega), as described previously (Warkus et al., 2016). Briefly, cells were seeded in 96-well plates at a density of 100 cells/well in 100 μL of culture medium containing 1% DMSO with or without a test compound. After 4 days of culture, cells were treated with CellTiter-Glo Reagent. The resulting luminescence was measured as a readout of ATP amount, which serves as a quantitative proxy for the number of metabolically active cells. The intensity of the luminescence was normalized to the control level (1% DMSO without a test compound) in each set of experiments and reported as relative light units. All experiments were repeated independently three times, and the results are shown as mean ± standard deviation.
2.6. SIRT1 activity assay
SIRT1 activity was measured using the SIRT-Glo Assay System (Promega), which utilizes an acetylated, cell-permeable, luminogenic peptide as an SIRT substrate, according to the manufacturer’s instruction. Briefly, purified SIRT1 enzyme (Cayman Chemical) was incubated with EX527 at various concentrations for 30 minutes at room temperature, and then mixed with the substrate, followed by luminescence measurement. The intensity of the luminescence was normalized to the control level (no EX527; set as 100%) in each experiment and reported as SIRT1 activity. Experiments were repeated independently three times, and the results are shown as mean ± standard deviation.
2.7. EdU assay
The efficiency of DNA replication was determined based on incorporation of EdU (5-ethynyl-2’-deoxyuridine) using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Molecular Probes, Eugene, OR), according to the manufacturer’s instruction. Cells were plated in 24-well plates at the density of 45,000 cells/well, and cultured in 450 μL of medium containing a test compound or vehicle only (1% DMSO). 24 hours later, 50 μL of culture medium containing 1 mM EdU was added into each well, and cultured for one more hour. Cells were then fixed in 3.7% formaldehyde, and stained for EdU (with Alexa Fluor 488 Azide) and nucleus (with Hoechst 33342). Images of the EdU and nuclear staining were captured with a fluorescence microscope using the same exposure length for all specimens of the same set of experiment (i.e., control and compound-treated cells), and opened in ImageJ for further analyses. The mean signal intensity of the EdU staining was normalized by the total nuclear area (measured after binary conversion of the Hoechst 33342 signal), and was reported as relative EdU intensity. Experiments were repeated independently three times, and the results are shown as mean ± standard deviation.
2.8. Statistical analyses
All adverse morphogenetic effects shown in the present study were statistically significant (P < 0.01), based on two-sample t-test that was performed between compound-treated group and the control group. For temporal gene expression analyses, two-sample t-test was performed between control and compound-treated groups for each time point to determine significant changes in relative expression levels (P < 0.05). For comparisons at Day 2, relative expression levels were normalized against the control in each set of experiment, and the averages of the three replicates were compared between the two treatment groups by two-sample t-test to determine significant differences in relative expression levels (P < 0.05). For cell viability assay, SIRT1 activity assay, and EdU assay, two-sample t-test was performed between control and compound-treated groups to determine significant differences (P < 0.01).
3. Results
3.1. Embryoid body morphogenesis is impacted by trans-resveratrol
P19C5 EBs were exposed to individual chemicals of the Anti-Diabetic Compound Library, including trans-resveratrol (Fig. 1, A; hereafter simply referred to as “resveratrol” unless otherwise stated) at the concentrations of 0.1, 1, 10, and 100 μM, and analyzed for morphometric parameters (area and EDI) at Day 4. For resveratrol, adverse morphogenetic effects (see Materials and methods) were observed when EBs were exposed to 10 μM or higher, but not to 1 μM or lower concentrations (Fig. 2, A). At exposure to 10 μM, EBs were smaller (i.e., lower relative area) and rounder (i.e., lower relative EDI) than the control. Cells did not survive to form EBs when treated at 100 μM. To further refine the concentration-response relationship, EBs were exposed to 2, 4, 6, 8, and 10 μM of resveratrol (Fig. 2, B, C). Here, we used another lot of resveratrol from a different supplier to verify the morphogenetic effects (Table 1). At 10 μM, EBs were smaller and rounder (Fig. 2, C), similar to the effects observed with the compound from the library. Resveratrol also yielded adverse morphogenetic effects at 4 μM and higher, showing concentration-dependent gradual reduction in both area and EDI (Fig. 2, B). The smaller EB size raises the possibility that cell proliferation was reduced by resveratrol during hanging drop culture. Consistently, resveratrol also diminished proliferation of undifferentiated P19C5 cells, as the numbers of viable cells were significantly lower in monolayer culture after 4 days of treatment at 4 μM or higher (Fig. 2D).
Fig. 2.
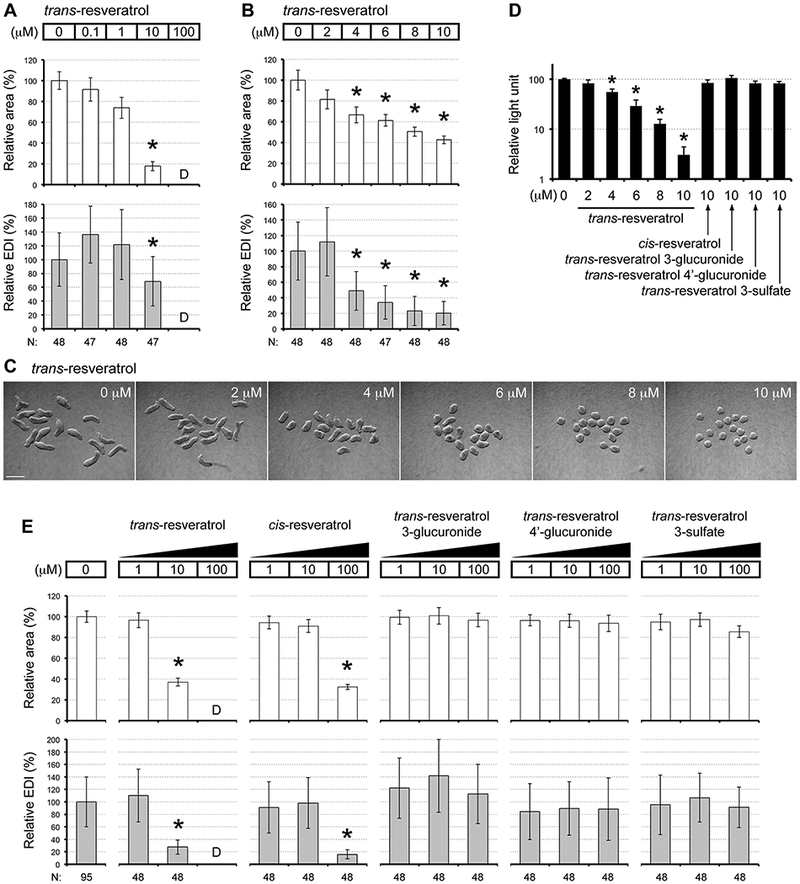
Embryoid body morphogenesis is impacted by trans-resveratrol. (A, B) Morphometric parameters of Day 4 embryoid bodies (EBs) treated with resveratrol at different concentrations. Graphs show averages of relative area (white columns) and relative EDI (gray columns) with error bars of standard deviation. Numbers at the bottom (N) are numbers of EBs scored. No area or EDI value is available when EBs were dead (noted as “D”). Asterisks indicate adverse impacts, which are defined as reduction in average area or average EDI by more than 30% relative to controls. All adverse impacts are statistically significant (P < 0.01; two-sample t-test). (C) Representative images of EBs from one set of experiment, showing control EBs (0 μM) and those treated with different concentrations of resveratrol. Scale bar = 500 μm. (D) Impact of resveratrol on cell proliferation, evaluated by the cell viability assay (mean + standard deviation; n=3). Cells in monolayer culture were exposed to the compounds for 4 days. Asterisks indicate significant difference (P < 0.01; two-sample t-test) in mean relative light unit between control and compound treatment. (E) Morphometric parameters of Day 4 EBs treated with resveratrol-related compounds at different concentrations, presented in the same form at as described for (A, B) above.
To examine specificity of the resveratrol effects, we evaluated the morphogenetic impact of other related chemicals, namely a geometric isomer (cis-resveratrol) and major metabolites of trans-resveratrol (resveratrol 3-glucuronide, resveratrol 4’-glucuronide, and resveratrol 3-sulfate) (Fig. 1, A). cis-Resveratrol is found in various plants along with trans-resveratrol, whereas the metabolites are generated in the liver by glucuronidation and sulfation after ingestion of trans-resveratrol (Baur and Sinclair, 2006). We examined the morphogenetic effects of these chemicals at 1, 10, and 100 μM, in parallel with another lot of trans-resveratrol that was obtained from the same supplier for the cis-isomer and the metabolites (Table 1). Consistent with the earlier observations, trans-resveratrol caused adverse morphogenetic effects at 10 μM (smaller and rounder EBs) and at 100 μM (cell death), but not at 1 μM. By contrast, cis-resveratrol adversely affected EB morphogenesis only at 100 μM (smaller and rounder) (Fig. 2, E). Note that according to the product information from the supplier, the preparation of cis-resveratrol may contain 1–5% of trans-resveratrol (see Materials and methods). Thus, it is unclear whether the adverse effects caused by 100 μM of cis-resveratrol were partly due to the trans-resveratrol contaminant, which was possibly present at 1–5 μM. Regardless, the result clearly indicates that cis-resveratrol was much less potent than trans-resveratrol to cause adverse morphogenetic effects. No adverse effect was observed with any of the three metabolites at all the concentrations tested (Fig. 2, E), suggesting that they are much less potent than unmetabolized trans-resveratrol in causing developmental toxicity. The cytostatic effect to reduce proliferation of undifferentiated cells was also specific to trans-resveratrol, as shown by the cell viability assay in monolayer culture (Fig. 2, D)
3.2. Resveratrol alters gene expression profiles in embryoid bodies
To gain mechanistic insight into how resveratrol exerts the adverse morphogenetic effects, we compared gene expression profiles between control and resveratrol-treated EBs. Resveratrol was evaluated at 10 μM for this study, because this concentration robustly affected both relative area and relative EDI. Twelve developmental regulator genes were examined, whose functions and expression patterns during mouse gastrulation are summarized in Table 2.
In control EBs, a pluripotency maintenance factor (Pou5f1) was markedly down-regulated by Day 1, and the genes critical for the initial stage of gastrulation (Wnt3, Brachyury, Cdx1, and Fgf8) were up-regulated with the highest peak at Day 1 (Fig. 3, A). The expression patterns of these genes were essentially unaffected by resveratrol treatment, although the peak level of Fgf8 was slightly reduced with statistical significance. By contrast, drastic alterations by resveratrol were observed for the genes that control the differentiation of axial stem cells at the posterior end during the later stages of gastrulation (Wnt3a and Tbx6). The Day 2 peak expression of these genes was markedly diminished by resveratrol. For the HOX genes, which are the key regulators of anterior-posterior axial patterning, the anterior class (Hoxa1) was mostly unaffected, whereas the central class (Hoxc6) and the posterior class (Hoxb9) were diminished on Days 3 and 4 by resveratrol. During embryogenesis, expression of HOX genes and other axial patterning genes are regulated by retinoic acid signaling, whose activity and localization are strictly controlled by synthesis (involving Aldh1a2) and degradation (involving Cyp26a1) of retinoic acid (Piersma et al., 2017). In EBs, the expression pattern of Aldh1a2 was mostly unaffected by resveratrol. By contrast, the expression level of Cyp26a1 was strikingly elevated on Days 1 and 2 (Fig. 3, A). Thus, the expression patterns of developmental regulators were differentially affected by resveratrol. Specifically, genes crucial for the early stage of gastrulation were largely unaffected, whereas those functioning in the posterior end at the later stages were significantly altered.
Fig. 3.
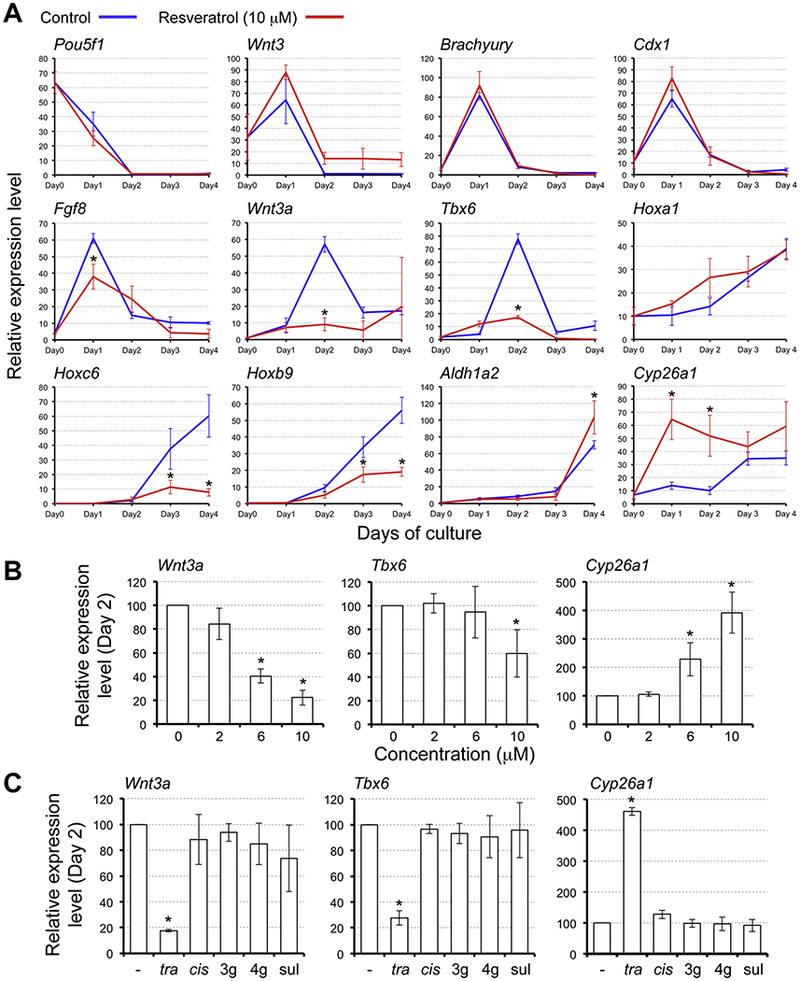
Resveratrol alters gene expression profiles in embryoid bodies. Expression levels of developmental regulator genes were determined by quantitative RT-PCR analyses. (A) Temporal expression profiles in EBs over the 4-day culture period. Horizontal axes represent days of culture whereas vertical axes represent relative expression levels in arbitrary units. Blue and red lines correspond to the relative expression levels (mean ± standard deviation; n=3) in control EBs and EBs treated with 10 μM trans-resveratrol, respectively. Asterisks indicate significant reduction or increase (P < 0.05; two-sample t-test) in mean relative expression levels by resveratrol treatment on a given day of EB culture. (B) Relative expression levels (mean ± standard deviation; n=3) in Day 2 EBs treated with trans-resveratrol. (C) Relative expression levels (mean ± standard deviation; n=3) in Day 2 EBs treated with resveratrol-related compounds at 10 μM. -: compound, tra: trans-resveratrol, cis: cis-resveratrol, 3g: trans-resveratrol 3-glucuronide, 4g: trans-resveratrol 4’-glucuronide, sul: trans-resveratrol 3-sulfate. Asterisks in (B, C) indicate significant reduction or increase (P < 0.05; two-sample t-test) in mean relative expression levels as compared to the control (0 in B, − in C).
We then examined whether gene expression profiles are altered by lower concentrations of resveratrol that affected EB morphogenesis mildly (6 μM) or did not affect significantly (2 μM) (Fig. 2, B). Analyses were focused on the expression levels of Wnt3a, Tbx6, and Cyp26a1 on Day 2, because they were most dramatically affected by 10 μM resveratrol. Neither of Wnt3a, Tbx6 nor Cyp26a1 was significantly altered by 2 μM resveratrol compared to control (0 μM). However, 6 μM resveratrol reduced Wnt3a and increased Cyp26a1 significantly, but by a lesser extent than 10 μM resveratrol (Fig. 3, B). Thus, the concentration-response relationship of resveratrol to alter gene expression levels was largely in line with the morphogenetic effects.
We also examined the impact of the cis isoform and the trans-resveratrol metabolites on the expression of Wnt3a, Tbx6, and Cyp26a1. While trans-resveratrol consistently altered the levels of Wnt3a, Tbx6, and Cyp26a1, none of the other compounds did (Fig. 3, C). This result further corroborates that the molecular changes caused by resveratrol correlate with its morphogenetic effects.
3.3. Estrogen receptor-modulating agents differ from resveratrol in the molecular impact
Resveratrol resembles the structure of an agonist of the estrogen receptor, diethylstilbestrol (DES; Fig. 1, A), and has been shown to activate the estrogen receptor (Gehm et al., 1997; Bowers et al., 2000). As shown in the previous study, DES impairs P19C5 EB morphogenesis (Warkus et al., 2016). EB morphogenesis is also affected by raloxifene (Fig. 1, A; Warkus et al. 2016), which is a selective estrogen receptor modulator (SERM) that potentiates activation of the estrogen receptor. This raises the possibility that the developmental toxicity induced by resveratrol may be mediated through activation of the estrogen receptor. To test this possibility, we evaluated the molecular impacts of DES and raloxifene using the P19C5 EB model.
First, the dose-response relationships were refined for the two agents to determine which concentrations exert adverse morphogenetic effects similarto 10 μM resveratrol (i.e., average relative area = 40–50% and average relative EDI = 20–30%; Fig. 2, B). Exposures to DES at 18 μM and raloxifene at 8 μM yielded smaller and rounder EBs that were comparable to those exposed to resveratrol (Fig. 4, A, B), although raloxifene-treated EBs appeared more opaque compared to resveratrol- or DES-treated EBs.
Fig. 4.
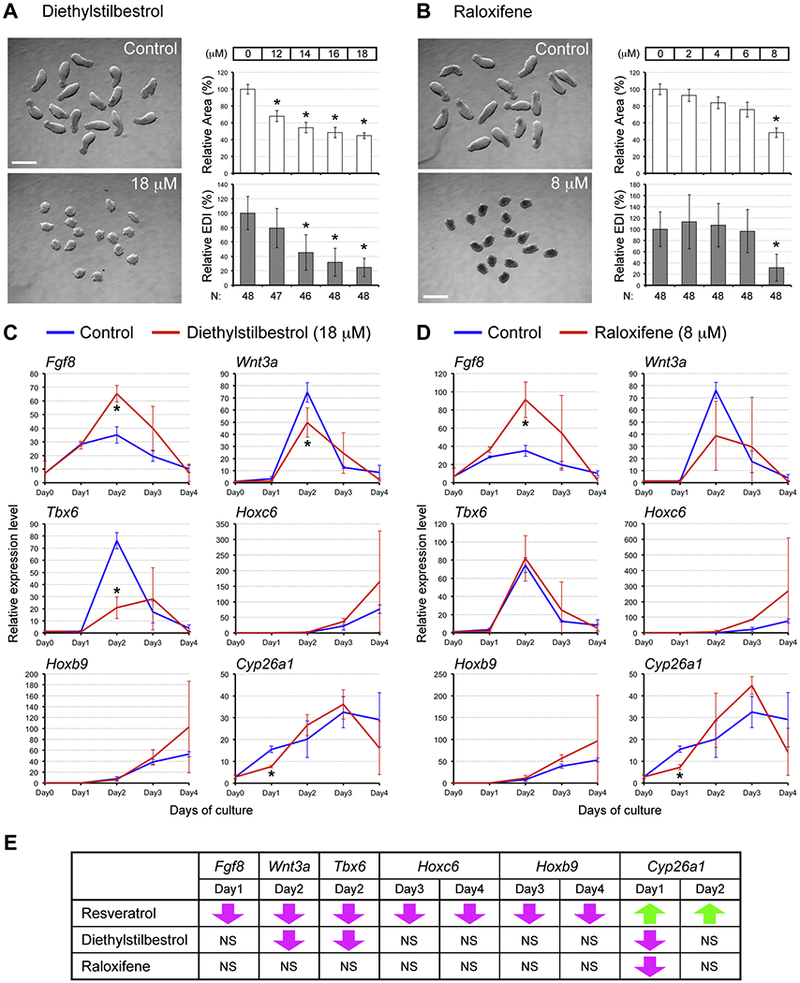
Estrogen receptor-modulating agents differ from resveratrol in the molecular impact. (A, B) Representative images (left) and morphometric parameters (right) of Day 4 EBs treated with diethylstilbestrol (DES; A) and with raloxifene (B). Graphs in (A, B) show averages of relative area (white columns) and relative EDI (gray columns) ± standard deviation. The numbers of EBs scored are shown at the bottom (N). Asterisks indicate adverse impacts. Scale bars = 500 μm. (C, D) Impact of DES (18 μM; C) and raloxifene (8 μM; D) on relative expression levels (mean ± standard deviation; n=3) of the developmental regulator genes over the 4-day culture. Asterisks indicate significant changes (P < 0.05; two-sample t-test) in mean relative expression levels by the estrogen receptor-modulating agent. (E) Comparisons of gene expression changes between resveratrol and the estrogen receptor-modulating agents. Downward magenta arrows indicate significant reduction compared to the control level, whereas upward green arrows indicate significant elevation. NS: No significance.
Next, we examined the gene expression profiles in EBs treated with DES at 18 μM and raloxifene at 8 μM. Expression analyses were focused on the genes that were significantly altered by resveratrol, specifically Fgf8, Wnt3a, Tbx6, Hoxc6, Hoxb9, and Cyp26a1 (Fig. 3, A). DES increased the level of Fgf8 on Day 2, reduced Wnt3a at Day 2, reduced Tbx6 on Day2, and reduced Cyp26a1 on Day 1 (Fig. 4, C). No statistically significant alteration was observed for Hoxc6 and Hoxb9. For raloxifene treatment, Fgf8 was increased on Day 2, whereas Cyp26a1 was reduced on Day 1 (Fig. 4, D). The other genes, including Wnt3a and Tbx6, were not significantly affected by raloxifene. When compared with the effects of resveratrol, the molecular impacts of DES and raloxifene were markedly different (Fig. 4, E). Out of 9 types of significant alterations caused by resveratrol, only 2 (i.e., reduction in the Wnt3a and Tbx6 levels at Day 2) were shared by DES and none was shared by raloxifene. Most prominently, Cyp26a1 was markedly elevated by resveratrol on Day 1, whereas it was reduced by DES and raloxifene. Thus, despite the similarity in morphogenetic effects (i.e., creating smaller and rounder EBs), resveratrol altered gene expression profiles differently from DES and raloxifene, suggesting that the developmental toxicity of resveratrol is not mediated by activation of the estrogen receptor.
3.4. Activation of SIRT1 is not sufficient to cause morphogenetic or molecular effects similar to resveratrol
SIRT1, which is a nicotinamide adenine dinucleotide (NAD)-dependent deacetylase, was initially implicated as a direct target of resveratrol (Howitz et al., 2003), although more recent studies suggest that this is not the case (Borra et al., 2005; Kaeberlein et al., 2005; Pacholec et al., 2010). Regardless, activation of SIRT1 appears to be essential for the actions of resveratrol in several experimental settings (Kulkarni and Cantó, 2015). Thus, we investigated whether activation of SIRT1 is involved in the effects of resveratrol in P19C5 EBs.
First, we examined whether SRT1720, a pharmacological activator of SIRT1 (Milne et al., 2007), exhibits morphogenetic and molecular effects comparable to resveratrol. While various concentrations of SRT1720 were evaluated, none of them were able to make a morphological impact similar to resveratrol. At 2.4 μM or lower, SRT1720 did not show adverse morphogenetic impact (Fig. 5, A). By contrast, no cells survived to form EBs when exposed to 3.6 μM of SRT1720. At intermediate concentrations (2.8 and 3.2 μM), mixed responses were observed: cells in some hanging drops did not survive to form EBs (i.e., dead), whereas those in the other drops formed EBs (i.e., live) (Fig. 5, B). Notably, EBs that survived were relatively in good shape. No adverse morphogenetic effects were observed for EBs exposed to 2.8 μM, whereas those exposed to 3.2 μM exhibited reduced relative EDI, although it was not as extensive as resveratrol (Fig. 5, C). Furthermore, unlike resveratrol, SRT1720 treatments (2.4, and 3.2 μM) did not significantly alter the expression levels of Wnt3a, Tbx6, and Cyp26a1 (Fig. 5, D). These results suggest that activation of SIRT1 is not sufficient to account for the adverse effects of resveratrol.
Fig. 5.
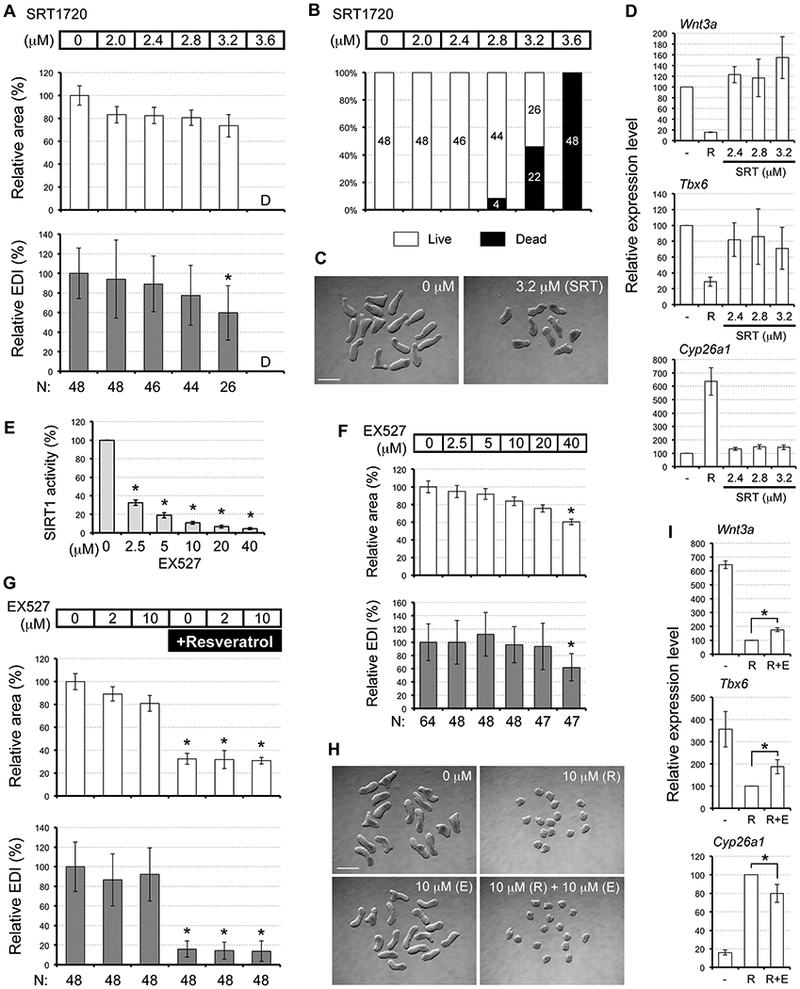
Activation of SIRT1 is not sufficient to cause morphogenetic or molecular effects similar to resveratrol. (A) Morphometric parameters of Day 4 EBs treated with SRT1720. Graphs show averages of relative area and relative EDI ± standard deviation. The numbers of EBs scored are shown at the bottom (N). Area or EDI value is not available for 3.6 μM treatment, as EBs died or did not form (noted as “D”). (B) Percentage of hanging drops with different concentrations of SRT1720 that yielded live EBs. Numbers of hanging drops are indicated in columns. (C) Representative images of Day 4 EBs. In this set of experiment, nine out of 16 hanging drops with 3.2 μM SRT1720 (SRT) yielded live EBs, whereas all 16 control drops yielded live EBs. Scale bar= 500 μm. (D) Relative expression levels (mean ± standard deviation; n=3) of developmental regulator genes in Day 2 EBs treated with SRT1720 (SRT), compared to resveratrol (R; 10 μM) in the same sets of experiments. (E) Inhibition of SIRT1 by EX527. Purified SIRT1 protein is exposed to EX527 for 30 minutes and incubated with a luminogenic substrate to assess the SIRT1 activity. Luminescence intensity of EX527-treated SIRT1 is normalized by that of untreated SIRT1 (0 μM; set as 100%) and presented as SIRT1 activity (mean ± standard deviation; n=3). Asterisks indicate significant reduction (P < 0.01; two-samplet-test). (F) Morphometric parameters of EBs treated with EX527. Asterisks indicate adverse morphogenetic impacts. (G) Morphometric parameters of Day 4 EBs treated with EX527 with or without 10 μM resveratrol. Asterisks indicate adverse morphogenetic impacts. (H) Representative images of Day 4 EBs treated with/without resveratrol (R) and with/without EX527 (E). Scale bar= 500 μm. (I) Relative expression levels (mean ± standard deviation; n=3) of developmental regulator genes in Day 2 EBs treated with resveratrol (R; 10 μM) and with or without EX527 (E; 10 μM). Expression levels are normalized by the resveratrol treatment, which is shown as 100. Asterisks indicate significant difference (P < 0.05; two-sample t-test) in mean relative expression levels between R and R+E.
Next, we tested whether activation of SIRT1 is required for the effects of resveratrol. EX527 is a pharmacological inhibitor of SIRT1 (Napper et al., 2005), and has been shown to alleviate the impact of resveratrol in several experimental systems (Kim et al., 2011; Price et al., 2012; Desquiret-Dumas et al., 2013; Joe et al., 2015; Safaeinejad et al., 2017). For example, the effects of resveratrol (25 μM) to promote mitochondrial function are abrogated by cotreatment with EX527 (10 μM) in skeletal muscle cells (Price et al., 2012). Thus, we investigated whether co-treatment with EX527 can rescue resveratrol-treated EBs from the adverse effects. EX527 that we obtained from a chemical supplier was effective in inhibiting the SIRT1 activity in an in vitro assay at the concentrations between 2.5 μM (by >65%) and 40 μM (by >95%) (Fig. 5, E). However, treatment of EBs with EX527 alone at the high concentration (40 μM) caused adverse morphogenetic effects (Fig. 5, F). Therefore, for the rescue experiment, we used low (2 μM) and medium (10 μM) concentrations of EX527, the latter of which was still able to inhibit the SIRT1 activity by about 90% (Fig. 5, E). Nonetheless, both concentrations of EX527 did not significantly alleviate the morphogenetic effects of resveratrol (Fig. 5, G, H). By contrast, the molecular impact of resveratrol was slightly but significantly alleviated by EX527 at 10 μM. The repressed expression of Wnt3a and Tbx6 by resveratrol was slightly restored by co-treatment with EX527, whereas the heightened expression of Cyp26a1 was slightly diminished (Fig. 5, I). These results suggest that the activity of SIRT 1 is required, to a certain extent, for the molecular impact of resveratrol on EBs.
3.5. Resveratrol reduces the rate of DNA replication to cause morphogenetic and molecular effects
To further explore possible targets linked to the effects of resveratrol, we evaluated the role of DNA replication. Resveratrol has been shown to directly bind to and inhibit the activity of DNA polymerases (Stivala et al., 2001; Locatelli et al., 2005). Resveratrol also inhibits ribonucleotide reductase, which is an enzyme required to generate deoxyribonucleotides for DNA replication (Fontecave et al., 1998). Direct actions of resveratrol on these enzymes on DNA replication have been suggested to account for the anti-cancer activity of resveratrol (Pirola and Fröjdö, 2008).
To examine how a reduction in the DNA replication rate may affect the EB development, we employed aphidicolin and hydroxyurea, which are pharmacological inhibitors of DNA polymerases and ribonucleotide reductase, respectively. These inhibitors exhibited morphogenetic effects on EBs in a concentration-dependent manner (Fig. 6, A, B). EBs treated with aphidicolin at 0.4 μM or hydroxyurea at 40 μM were smaller and rounder, similarto resveratrol treatment at 10 μM (i.e., relative area = < 60% and relative EDI = < 30%). Thus, we analyzed gene expression profiles of EBs treated with aphidicolin and hydroxyurea at these concentrations, focusing on Fgf8, Wnt3a, Tbx6, Hoxc6, Hoxb9, and Cyp26a1. The expression patterns of these genes were significantly altered by both aphidicolin and hydroxyurea (Fig. 6, C, D). When com pared with resveratrol, the impacts of aphidicolin and hydroxyurea were markedly similar. Out of 9 types of significant alterations caused by resveratrol, 7 were shared by aphidicolin and 6 were shared by hydroxyurea, including the dramatic up-regulation of Cyp26a1 at Days 1 and 2 (Fig. 6, E). These observations raise the possibility that the major impact of resveratrol is a reduction in the rate of DNA replication, comparable to aphidicolin and hydroxyurea.
Fig. 6.
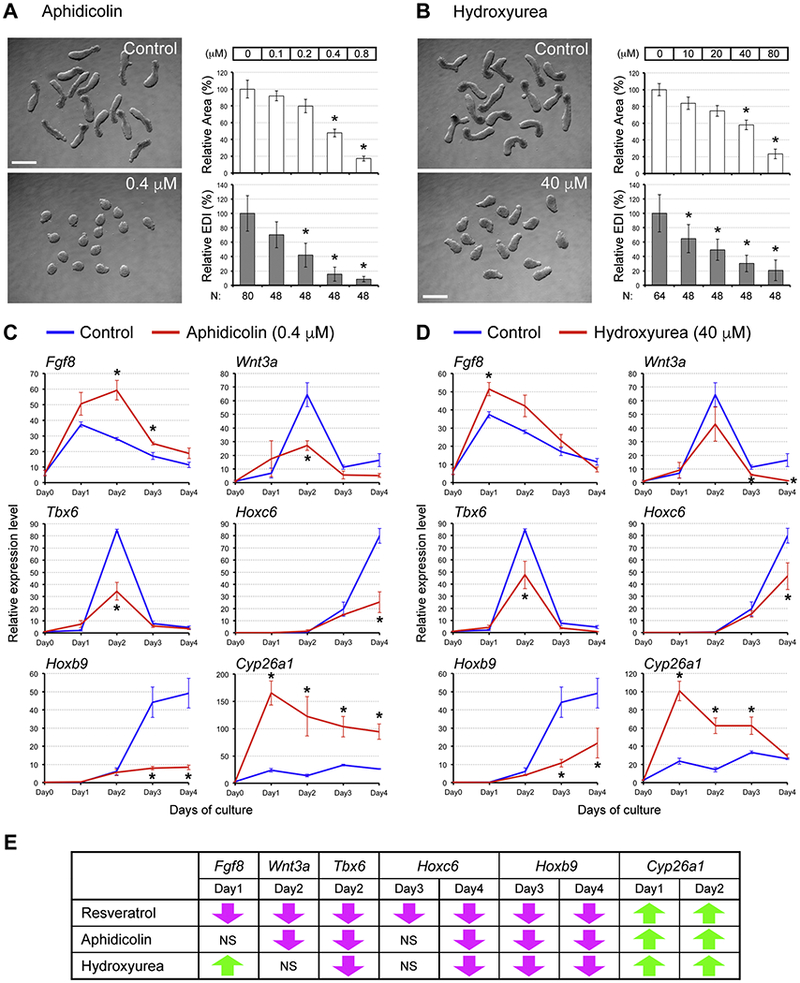
Pharmacological inhibitors of DNA replication cause morphogenetic and molecular effects similar to resveratrol. (A, B) Representative images (left) and morphometric parameters (right) of Day 4 EBs treated with aphidicolin (A) and hydroxyurea (B). Graphs in (A, B) show averages of relative area (white columns) and relative EDI (gray columns) ± standard deviation. The numbers of EBs scored are shown at the bottom (N). Asterisks indicate adverse impacts. Scale bars = 500 μm. (C, D) Impact of aphidicolin (0.4 μM; C) and hydroxyurea (40 μM; D) on gene expression patterns in EBs. Relative expression levels of the developmental regulator genes over the 4 days of culture are shown (mean ± standard deviation; n=3). Asterisks indicate significant change (P < 0.05; two-sample t-test) in mean relative expression levels by the pharmacological inhibitors of DNA replication. (E) Comparisons of gene expression changes between resveratrol, aphidicolin, and hydroxyurea. Downward magenta arrows indicate significant reduction compared to the control level, whereas upward green arrows indicate significant elevation. NS: No significance.
To assess whether resveratrol indeed impairs DNA replication in P19C5 cells, we examined its impact on the incorporation of EdU, a deoxyribonucleotide analog. Efficiency of EdU incorporation was significantly lower than control, when cells were treated with resveratrol (10 μM), aphidicolin (0.4 μM), orhyroxyurea (40 μM) (Fig. 7, A, B). By contrast, neither DES (18 μM) nor raloxifene (8 μM) significantly reduced EdU incorporation, even though both types of exposures created smaller EBs (Fig. 4, A, B) and diminished cell proliferation in monolayer culture (Fig. 7, C). These results suggest that resveratrol reduces the rate of DNA replication, possibly through inhibition of DNA polymerase and/or ribonucleotide reductase, to cause the distinct morphogenetic and molecular effects in EBs.
Fig. 7.
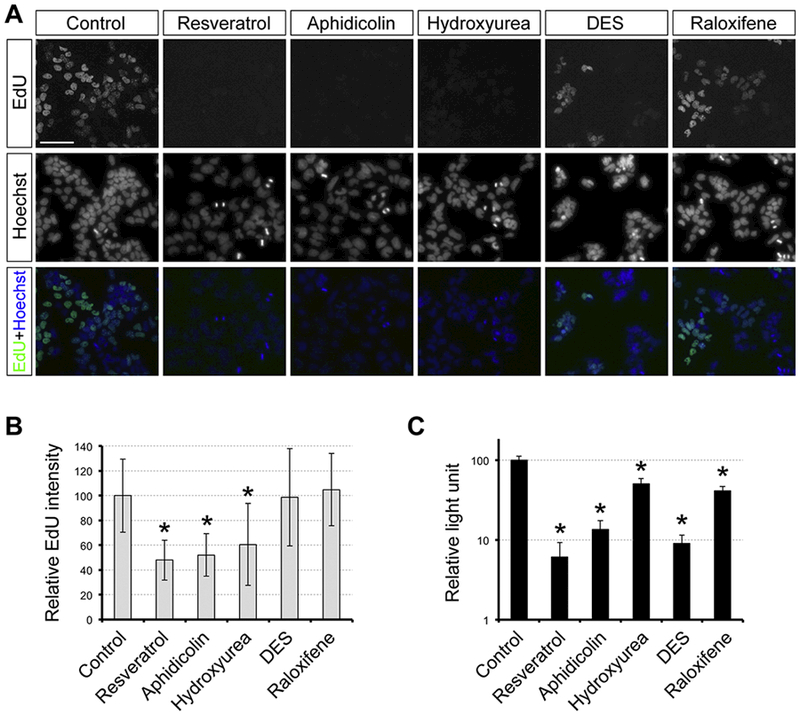
Resveratrol reduces the rate of DNA replication. (A) Evaluation of DNA replication rate by the EdU assay. P19C5 cells in monolayer culture were treated with vehicle control (1% DMSO), trans-resveratrol (10 μM), Aphidicolin (0.4 μM), hydroxyurea (40 μM), diethylstilbestrol (DES; 18 μM), and raloxifene (8 μM) for 24 hours, followed by incubation with EdU (100 μM) for 1 hour. Representative images of EdU labeling and nuclear staining (with Hoechst 33342) are shown. Scale bar = 50 μm. (B) Relative EdU intensity (mean ± standard deviation; n=3) to evaluate the rate of DNA replication under the compound exposures described in (A). Asterisks indicate significant difference (P < 0.01; two-sample t-test) in mean relative intensity between control and compound treatment. (C) Impact of compound exposures on cell proliferation, evaluated by the cell viability assay (mean + standard deviation; n=3). Cells in monolayer culture were exposed to the compounds for 4 days at the same concentrations as described in (A). Asterisks indicate significant difference (P < 0.01; two-sample t-test) in mean relative light unit between control and compound treatment.
4. Discussion
In the present study, the potential developmental toxicity of resveratrol was evaluated using the P19C5 EB morphogenesis model. Resveratrol has been marketed as a dietary supplement with numerous health benefit claims, such as protective effects against cancers, cardiovascular diseases, diabetes, and aging (Baur and Sinclair, 2006; Park and Pezzuto, 2015). Animal studies have also suggested that resveratrol provides protective effects for embryos against teratogenic insults, such as exposures to ethanol, dioxin, and maternal diabetes (Jang et al., 2008; Kumar et al.,2011; Singh et al., 2011, 2012; Vega et al., 2016; Bariani et al., 2017). However, the developmental toxicity of resveratrol by itself has not been thoroughly investigated. Whether a given compound exhibits beneficial or adverse developmental effects is dependent on the dosages or concentrations of exposures (Daston et al., 2010). Thus, it is crucial to determine dose-response relationships for adverse impact of resveratrol as well as other dietary supplements that may be consumed by women of reproductive age. In vitro tests, such as the EB morphogenesis assay, are generally less costly and more humane than animal experimentations, so they may be used as alternatives at the initial stage of investigations to evaluate the LOAEL and molecular mechanisms of potential developmental toxicity, as in the present study. The information acquired from in vitro tests would be valuable in designing adequate and effective in vivo studies with animals and humans.
Adverse morphogenetic effects were observed when P19C5 EBs were treated with trans-resveratrol at 4 μM and higher, raising the possibility that in utero exposure to such concentrations would impair embryo development. The key question is whether such concentrations actually occur in people who take resveratrol as a dietary supplement. Pharmacokinetic studies in humans have shown that orally taken resveratrol is efficiently absorbed, but is rapidly metabolized into glucuronated and sulfated forms (Walle et al., 2004; Sergides et al., 2016), which in the present study did not exhibit adverse impact on EBs. Nonetheless, a significant amount of the unmetabolized form, trans-resveratrol, is still found in the plasma. Based on the studies conducted with healthy volunteers, a single oral dose of 1 and 5 g trans-resveratrol resulted in average peak plasma concentrations (Cmax) of unmetabolized resveratrol at 0.4 μM and 2.4 μM, respectively (Boocock et al., 2007). Repeated oral intakes of 1 and 5 g per day for 29 days resulted in higher Cmax of trans-resveratrol, namely 0.6 μM and 4.2 μM, respectively (Brown et al., 2010), which are not much lower than those which affected EB development. Similar concentrations may occur in developing embryos, as resveratrol has been shown to cross the placenta in rodents and non-human primates (Bourque et al., 2012; O’Tierney-Ginn et al., 2015). It is of note that the plasma concentrations in the above studies were measured in healthy individuals. Some people may have a condition that diminishes metabolism, e.g., liver diseases or concomitant intake of other drugs that interfere with metabolizing enzymes, which in turn may raise plasma concentrations of unmetabolized resveratrol. Thus, further investigations are warranted to determine the plasma concentrations of resveratrol in people with diverse backgrounds, including women of reproductive age. It is also important to evaluate the stability of resveratrol in the culture medium during 4 days of EB development. The concentrations and effectiveness of compounds may be reduced during culture by various factors, such as degradation, metabolic conversion by the cells, and binding to the culture vessel (plastic) and serum proteins. In that case, actual concentrations of resveratrol that affected EB morphogenesis might be lower than 4 μM, which would further raise a concern regarding the developmental toxicity of this dietary supplement.
Analyses of gene expression profiles in EBs revealed that resveratrol affected several developmental regulators in a differential manner. Gene expression associated with the initial step of gastrulation was largely unaffected, whereas gene expression relevant for the later step was markedly diminished by resveratrol. Interestingly, Cyp26a1 was dramatically elevated by resveratrol on Day 1 by about 5-fold higher than control. Cyp26a1 encodes the cytochrome P450 enzyme responsible for oxidative inactivation of retinoic acid (RA), and Cyp26a1 knockout mouse embryos exhibit morphological and molecular abnormalities similar to those caused by excess RA (Abu-Abed et al., 2001; Sakai et al., 2001). Heightened expression of Cyp26a1 in resveratrol-treated EBs may reduce the endogenous level of RA and diminish expression of Hoxc6 and Hoxb9. However, a reduction in RA signaling alone cannot fully account for the effects of resveratrol on other genes. For example, the expression of Cdx1 and Hoxa1, both of which are known transcriptional targets of RA receptors (Langston and Gudas, 1992; Houle et al., 2003; Li and Marikawa, 2016), was not significantly altered by resveratrol. Also, the Day 2 peak expression of Wnt3a and Tbx6 is not abolished by BMS493, a pharmacological inhibitor of RA signaling (Li and Marikawa, 2015). Thus, resveratrol is likely to impact other molecular pathways in addition to RA signaling.
The molecular targets of resveratrol that are responsible for its various beneficial effects are still elusive, although several candidate molecules have been proposed (Kulkarni and Cantó, 2015). In the present study, we investigated whether such candidates, specifically the estrogen receptor, SIRT1, and DNA polymerase, are involved in the adverse effects of resveratrol on EBs. Despite having a similar morphological impact, DES and raloxifene did not alter the gene expression profiles in a manner comparable to resveratrol, suggesting that the activation of the estrogen receptor is not responsible for the effects of resveratrol. Activation of SIRT1 by SRT1720 did not yield morphological or molecular effects that are comparable to resveratrol either, although inhibition of SIRT1 by EX527 slightly alleviated the molecular impact of resveratrol. By contrast, a reduction in the DNA replication rate by aphidicolin or hydroxyurea caused morphogenetic and molecular impacts strikingly similar to resveratrol. Note that DES and raloxifene diminished cell proliferation but did not cause similar gene expression changes. Thus, the distinct alteration of gene expression profiles, such as up-regulation of Cyp26a1 and down-regulation of Wnt3a and Tbx6, are not simply due to delayed cell cycle or general cytostatic effects. Rather, the alterations are caused more specifically by a reduced rate of DNA replication, although the molecular mechanisms are presently unclear. Also, note that the molecular impact of aphidicolin and hydroxyurea was not entirely identical to resveratrol: Fgf8 at Day 1 was increased by the former two but lowered by the latter. This suggests that resveratrol has additional targets linked to the adverse effects on EBs. Among the other potential targets, of particular interest for future studies, are AMPK (Baur et al., 2006; Dasgupta and Milbrandt, 2007), PDE (Park et al., 2012), mechanistic target of rapamycin (mTOR; Liu et al., 2010), and mitogen-activated protein kinase (MAPK; Miloso et al., 1999), as they are key regulators of signaling pathways essential for proper embryo development.
One of the health benefit claims of resveratrol is amelioration of diabetes (Szkudelski and Szkudelska, 2015; Öztürk et al., 2017). Diabetes is characterized by hyperglycemia and impairment in insulin action, and affects about 8.5% of adults aged 18 and older in the entire world (WHO, 2017). Diabetes in pregnant women during the pre-gestational or the first trim ester stage significantly increases the chance of congenital malformations in the offspring, collectively known as diabetic embryopathy (Allen and Armson, 2007). To minimize diabetic embryopathy, it is important to control diabetes before and during pregnancy. However, treatment of diabetes must be conducted in a careful manner because medications by themselves could be developmentally toxic. The present study was initiated as a part of the screening of the Anti-Diabetic Compound Library to assess the developmental toxicity of various medications for diabetes. While the outcome of the screening is to be reported elsewhere in the future, the adverse effects of resveratrol on EBs raises a concern for this dietary supplement, which is widely available without prescription. Animal studies suggest that resveratrol ameliorates some of the adverse impact of maternal diabetes on embryos (Singh et al., 2011, 2012, 2013). Nonetheless, in these studies, the plasma concentrations of resveratrol, to which embryos may be exposed, are not shown. Further investigations are warranted to determine the proper dosages of resveratrol that yield beneficial effects without reaching the LOAEL to impair embryo development. Importantly, in spite of many studies of the beneficial impact of resveratrol, its therapeutic application has not been approved by the FDA for any indication, and its pregnancy risk has not been thoroughly investigated. Thus, women of reproductive age should be strongly warned about the use of resveratrol.
Supplementary Material
Highlights.
Developmental toxicity of resveratrol is examined using morphogenetic embryoid body
trans-resveratrol, but not cis isoform or metabolites, impairs morphogenesis
Resveratrol alters gene expression patterns of gastrulation regulators
Activation of the estrogen receptor or SIRT1 is not the major action of resveratrol
Reduction in DNA replication rate accounts for the resveratrol action
Acknowledgements
This work was supported by grants from the Johns Hopkins Center for Alternatives to Animal Testing (CAAT), the Alternatives Research & Development Foundation (ARDF), and the National Institutes of Health (NIH; P20GM103457 and R03HD088970). The authors are grateful to Dr. Vernadeth B. Alarcon for reading the article and providing valuable comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Abu-Abed S, Dollé P, Metzger D, Beckett B, Chambon P, Petkovich M 2001. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 15, 226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen VM, Armson BA; GENETICS COMMITTEE; MATERNAL FETAL MEDICINE COMMITTEE. 2007. Teratogenicity associated with pre-existing and gestational diabetes. J. Obstet. Gynaecol. Can 29, 927–934. [DOI] [PubMed] [Google Scholar]
- Bariani MV, Correa F, Leishman E, Domínguez-Rubio AP, Arias A, Stern A, Bradshaw HB, Franchi AM 2017. Resveratrol protects from lipopolysaccharide-induced inflammation in the uterus and prevents experimental preterm birth. Mol. Hum. Reprod 23, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA 2006. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA 2006. Therapeutic potential of resveratrol: the in vivo evidence. Nat. Rev. Drug Discov 5, 493–506. [DOI] [PubMed] [Google Scholar]
- Boocock DJ, Faust GE, Patel KR, Schinas AM, Brown VA, Ducharme MP, Booth TD, Crowell JA, Perloff M, Gescher AJ, Steward WP, Brenner DE 2007. Phase I dose escalation pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive agent. Cancer Epidemiol. Biomarkers Prev 16, 1246–1252. [DOI] [PubMed] [Google Scholar]
- Borra MT, Smith BC, Denu JM 2005. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem 280, 17187–17195. [DOI] [PubMed] [Google Scholar]
- Bourque SL, Dolinsky VW, Dyck JR, Davidge ST 2012. Maternal resveratrol treatment during pregnancy improves adverse fetal outcomes in a rat model of severe hypoxia. Placenta 33, 449–452. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Tyulmenkov VV, Jernigan SC, Klinge CM 2000. Resveratrol acts as a mixed agonist/antagonist for estrogen receptors alpha and beta. Endocrinology 141, 3657–3667. [DOI] [PubMed] [Google Scholar]
- Brown VA, Patel KR, Viskaduraki M, Crowell JA, Perloff M, Booth TD, Vasilinin G, Sen A, Schinas AM, Piccirilli G, Brown K, Steward WP, Gescher AJ, Brenner DE 2010. Repeat dose study of the cancer chemopreventive agent resveratrol in healthy volunteers: safety, pharmacokinetics, and effect on the insulin-like growth factor axis. Cancer Res 70, 9003–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Agulnik I, Hancock S, Silver LM, Papaioannou VE 1996. Tbx6, a mouse T-Box gene implicated in paraxial mesoderm formation at gastrulation. Dev. Biol 180, 534–542. [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Milbrandt J 2007. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. USA 104, 7217–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daston GP, Beyer BK, Carney EW, Chapin RE, Friedman JM, Piersma AH, Rogers JM, Scialli AR 2014. Exposure-based validation list for developmental toxicity screening assays. Birth Defects Res. B Dev. Reprod. Toxicol 101, 423–428. [DOI] [PubMed] [Google Scholar]
- Daston GP, Chapin RE, Scialli AR, Piersma AH, Carney EW, Rogers JM, Friedman JM 2010. A different approach to validating screening assays for developmental toxicity. Birth Defects Res. B Dev. Reprod. Toxicol 89, 526–530. [DOI] [PubMed] [Google Scholar]
- Desquiret-Dumas V, Gueguen N, Leman G, Baron S, Nivet-Antoine V, Chupin S, Chevrollier A, Vessières E, Ayer A, Ferré M, Bonneau D, Henrion D, Reynier P, Procaccio V 2013. Resveratrol induces a mitochondrial complex I-dependent increase in NADH oxidation responsible for sirtuin activation in liver cells. J. Biol. Chem 288, 36662–36675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M, Lepoivre M, Elleingand E, Gerez C, Guittet O 1998. Resveratrol, a remarkable inhibitor of ribonucleotide reductase. FEBS Letters. 421, 277–279. [DOI] [PubMed] [Google Scholar]
- Friedman JM, 2010. The principles of teratology: are they still true? Birth Defects Res. A Clin. Mol. Teratol 88, 766–768. [DOI] [PubMed] [Google Scholar]
- Gehm BD, McAndrews JM, Chien PY, Jameson JL 1997. Resveratrol, a polyphenolic compound found in grapes and wine, is an agonist for the estrogen receptor. Proc. Natl. Acad. Sci. USA 94, 14138–14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbeck RJ, Hoffmann I, Duester G 1999. Distinct functions for Aldh1 and Raldh2 in the control of ligand production for embryonic retinoid signaling pathways. Dev. Genet 25, 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG 1991. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development 113, 913–917. [DOI] [PubMed] [Google Scholar]
- Houle M, Sylvestre JR, Lohnes D 2003. Retinoic acid regulates a subset of Cdx1 function in vivo. Development 130, 6555–6567. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196. [DOI] [PubMed] [Google Scholar]
- Jang JY, Park D, Shin S, Jeon JH, Choi BI, Joo SS, Hwang SY, Nahm SS, Kim YB 2008. Antiteratogenic effect of resveratrol in mice exposed in utero to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Eur. J. Pharmacol 591, 280–283. [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM 1997. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 275, 218–220. [DOI] [PubMed] [Google Scholar]
- Jelínek R, 2005. The contribution of new findings and ideas to the old principles of teratology. Reprod. Toxicol 20, 295–300. [DOI] [PubMed] [Google Scholar]
- Joe IS, Jeong SG, Cho GW 2015. Resveratrol-induced SIRT1 activation promotes neuronal differentiation of human bone marrow mesenchymal stem cells. Neurosci. Lett 584, 97–102. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK 2005. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem 280, 17038–17045. [DOI] [PubMed] [Google Scholar]
- Kim DH, Jung YJ, Lee JE, Lee AS, Kang KP, Lee S, Park SK, Han MK, Lee SY, Ramkumar KM, Sung MJ, Kim W 2011. SIRT1 activation by resveratrol ameliorates cisplatin-induced renal injury through deacetylation of p53. Am. J. Physiol. Renal Physiol 301, F427–435. [DOI] [PubMed] [Google Scholar]
- Kulkarni SS, Cantó C 2015. The molecular targets of resveratrol. Biochim. Biophys. Acta 1852, 1114–1123. [DOI] [PubMed] [Google Scholar]
- Kumar A, Singh CK, Lavoie HA, Dipette DJ, Singh US 2011. Resveratrol restores Nrf2 level and prevents ethanol-induced toxic effects in the cerebellum of a rodent model of fetal alcohol spectrum disorders. Mol. Pharmacol 80, 446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston AW, Gudas LJ 1992. Identification of a retinoic acid responsive enhancer 3’ of the murine homeobox gene Hox-1.6. Mech. Dev 38, 217–227. [DOI] [PubMed] [Google Scholar]
- Lau CG, Marikawa Y 2014. Morphology-based mammalian stem cell tests reveal potential developmental toxicity of donepezil. Mol. Reprod. Dev 81, 994–1008. [DOI] [PubMed] [Google Scholar]
- Li AS, Marikawa Y 2015. An in vitro gastrulation model recapitulates the morphogenetic impact of pharmacological inhibitors of developmental signaling pathways. Mol. Reprod. Dev 82, 1015–1036. [DOI] [PubMed] [Google Scholar]
- Li AS, Marikawa Y 2016. Adverse effect of valproic acid on an in vitro gastrulation model entails activation of retinoic acid signaling. Reprod. Toxicol 66, 68–83. [DOI] [PubMed] [Google Scholar]
- Liu M, Wilk SA, Wang A, Zhou L, Wang RH, Ogawa W, Deng C, Dong LQ, Liu F 2010. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. J. Biol. Chem 285, 36387–36394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A 1999. Requirement for Wnt3 in vertebrate axis formation. Nature Genet 22, 361–365. [DOI] [PubMed] [Google Scholar]
- Locatelli GA, Savio M, Forti L, Shevelev I, Ramadan K, Stivala LA, Vannini V, Hübscher U, Spadari S, Maga G 2005. Inhibition of mammalian DNA polymerases by resveratrol: mechanism and structural determinants. Biochem. J 389, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa Y (2018) Stem-cell-based in vitro morphogenesis models to investigate developmental toxicity of chemical exposures Stem Cells in Birth Defects Research and Developmental Toxicology (edited by Rasmussen TP), pp71–89, Wiley, New York, NY. [Google Scholar]
- Marikawa Y, Tamashiro DA, Fujita TC, Alarcón VB 2009. Aggregated P19 mouse embryonal carcinoma cells as a simple in vitro model to study the molecular regulations of mesoderm formation and axial elongation morphogenesis. Genesis 47, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BI, Gruss P 1993. Mouse Cdx-1 expression during gastrulation. Development 117, 191–203. [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH 2007. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miloso M, Bertelli AA, Nicolini G, Tredici G 1999. Resveratrol-induced activation of the mitogen-activated protein kinases, ERK1 and ERK2, in human neuroblastoma SH-SY5Y cells. Neurosci. Lett 264, 141–144. [DOI] [PubMed] [Google Scholar]
- Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, Thomas RJ, Kates M, Jones S, Navia MA, Saunders JO, DiStefano PS, Curtis R 2005. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem 48, 8045–8054. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 95, 379–391. [DOI] [PubMed] [Google Scholar]
- O’Tierney-Ginn P, Roberts V, Gillingham M, Walker J, Glazebrook PA, Thornburg KL, Grove K, Frias AE 2015. Influence of high fat diet and resveratrol supplementation on placental fatty acid uptake in the Japanese macaque. Placenta 36, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Yoshioka H, Tanaka A, Kawakami Y, Nohno T, Noji S. 1994. Involvement of androgen-induced growth factor (FGF-8) gene in mouse embryogenesis and morphogenesis. Biochem Biophys Res Commun 204:882–888. [DOI] [PubMed] [Google Scholar]
- Öztürk E, Arslan AKK, Yerer MB, Bishayee A 2017. Resveratrol and diabetes: A critical review of clinical studies. Biomed. Pharmacother 95, 230–234. [DOI] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K 2010. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem 285, 8340–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Pezzuto JM 2015. The pharmacology of resveratrol in animals and humans. Biochim. Biophys. Acta 1852, 1071–1113. [DOI] [PubMed] [Google Scholar]
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH 2012. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma AH, Hessel EV, Staal YC 2017. Retinoic acid in developmental toxicology: Teratogen, morphogen and biomarker. Reprod. Toxicol 72, 53–61. [DOI] [PubMed] [Google Scholar]
- Pirola L, Fröjdö S 2008. Resveratrol: one molecule, many targets. IUBMB Life. 60, 323–332. [DOI] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA 2012. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 15, 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaeinejad Z, Nabiuni M, Peymani M, Ghaedi K, Nasr-Esfahani MH, Baharvand H 2017. Resveratrol promotes human embryonic stem cells self-renewal by targeting SIRT1-ERK signaling pathway. Eur. J. Cell Biol 96, 665–672. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H 2001. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 15, 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardein JL, Macina OT, 2006. Human Developmental Toxicants: Aspects of Toxicology and Chemistry. CRC Press, Boca Raton, FL. [Google Scholar]
- Sergides C, Chirilă M, Silvestro L, Pitta D, Pittas A 2016. Bioavailability and safety study of resveratrol 500 mg tablets in healthy male and female volunteers. Exp. Ther. Med 11, 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorelli P, Ghidoni R 2005. Resveratrol as an anticancer nutrient: molecular basis, open questions and promises. J. Nutr. Biochem 16, 449–466. [DOI] [PubMed] [Google Scholar]
- Singh CK, Kumar A, Hitchcock DB, Fan D, Goodwin R, LaVoie HA, Nagarkatti P, DiPette DJ, Singh US 2011. Resveratrol prevents embryonic oxidative stress and apoptosis associated with diabetic embryopathy and improves glucose and lipid profile of diabetic dam. Mol. Nutr. Food Res 55, 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CK, Kumar A, LaVoie HA, DiPette DJ, Singh US 2012. Resveratrol prevents impairment in activation of retinoic acid receptors and MAP kinases in the embryos of a rodent model of diabetic embryopathy. Reprod. Sci 19, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CK, Kumar A, Lavoie HA, Dipette DJ, Singh US 2013. Diabetic complications in pregnancy: is resveratrol a solution? Exp. Biol. Med. (Maywood) 238, 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CK, Ndiaye MA, Ahmad N 2015. Resveratrol and cancer: Challenges for clinical translation. Biochim. Biophys. Acta 1852, 1178–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivala LA, Savio M, Carafoli F, Perucca P, Bianchi L, Maga G, Forti L, Pagnoni UM, Albini A, Prosperi E, Vannini V 2001. Specific structural determinants are responsible for the antioxidant activity and the cell cycle effects of resveratrol. J. Biol. Chem 276, 22586–22594. [DOI] [PubMed] [Google Scholar]
- Szkudelski T, Szkudelska K 2015. Resveratrol and diabetes: from animal to human studies. Biochim. Biophys. Acta 1852, 1145–1154. [DOI] [PubMed] [Google Scholar]
- Takada S, Stark KL, Shea MJ, Vassilieva G, McMahon JA, McMahon AP 1994. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev 8, 174–189. [DOI] [PubMed] [Google Scholar]
- Vega CC, Reyes-Castro LA, Rodríguez-González GL, Bautista CJ, Vázquez-Martínez M, Larrea F, Chamorro-Cevallos GA, Nathanielsz PW, Zambrano E 2016. Resveratrol partially prevents oxidative stress and metabolic dysfunction in pregnant rats fed a low protein diet and their offspring. J. Physiol 594, 1483–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walle T, Hsieh F, DeLegge MH, Oatis JE Jr., Walle UK 2004. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab. Dispos 32, 1377–1382. [DOI] [PubMed] [Google Scholar]
- Warkus ELL, Marikawa Y 2018. Fluoxetine inhibits canonical Wnt signaling to impair embryoid body morphogenesis: potential teratogenic mechanisms of a commonly used antidepressant. Toxicol. Sci (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkus ELL, Marikawa Y 2017. Exposure-based validation of an in vitro gastrulation model for developmental toxicity assays. Toxicol. Sci 157, 235–245. [DOI] [PubMed] [Google Scholar]
- Warkus EL, Yuen AA, Lau CG, Marikawa Y 2016. Use of in vitro morphogenesis of mouse embryoid bodies to assess developmental toxicity of therapeutic drugs contraindicated in pregnancy. Toxicol. Sci 149, 15–30. [DOI] [PubMed] [Google Scholar]
- Wellik DM 2009. Hox genes and vertebrate axial pattern. Curr. Top. Dev. Biol 88, 257–278. [DOI] [PubMed] [Google Scholar]
- WHO, 2017. World Health Organization, Diabetes Fact Sheet (Updated November 2017) http://www.who.int/mediacentre/factsheets/fs312/en/ (Accessed 8 July 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


