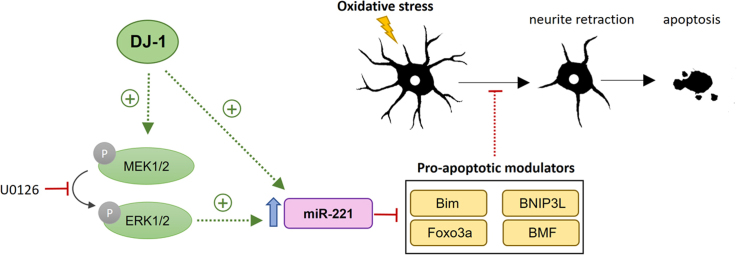
Abstract
DJ-1 is a highly conserved protein that protects neurons against oxidative stress and whose loss of function mutations are linked to recessively inherited Parkinson's disease (PD). While a number of signaling pathways have been shown to be regulated by DJ-1, its role in controlling cell survival through non-coding RNAs remains poorly understood. Here, using a microarray screen, we found that knocking down DJ-1 in human neuroblastoma cells results in down-regulation of microRNA-221 (miR-221). This is one of the most abundant miRNAs in the human brain and promotes neurite outgrowth and neuronal differentiation. Yet the molecular mechanism linking miR-221 to genetic forms of PD has not been studied. Consistent with the microarray data, miR-221 expression is also decreased in DJ-1-/- mouse brains. Re-introduction of wild-type DJ-1, but not its PD-linked pathogenic M26I mutant, restores miR-221 expression. Notably, over-expression of miR-221 is protective against 1-methyl-4-phenylpyridinium (MPP+)-induced cell death, while inhibition of endogenous miR-221 sensitizes cells to this toxin. Additionally, miR-221 down-regulates the expression of several pro-apoptotic proteins at basal conditions and prevents oxidative stress-induced up-regulation of bcl-2-like protein 11 (BIM). Accordingly, miR-221 protects differentiated DJ-1 knock-down ReNcell VM human dopaminergic neuronal cells from MPP+-induced neurite retraction and cell death. DJ-1 is a known activator of the mitogen-activated protein kinase (MAPK)/extracellular-regulated kinase (ERK) pathway and may modulate miR-221 levels in part through this pathway. We found that inhibiting ERK1/2 decreases miR-221 levels, whereas over-expressing ERK1 in DJ-1 knock-down cells increases miR-221 levels. These findings point to a new cytoprotective mechanism by which DJ-1 may increase miR-221 expression through the MAPK/ERK pathway, subsequently leading to repression of apoptotic molecules. The inability of a pathogenic DJ-1 mutant to modulate miR-221 further supports the relevance of this mechanism in neuronal health and its failure in DJ-1-linked PD.
Keywords: microRNA (miRNA), Parkinson's disease, Autosomal recessive, PARK7, miR-221, DJ-1, Oxidative stress
Graphical abstract
1. Introduction
Loss-of-function mutations in DJ-1 (PARK7) are linked to autosomal recessively inherited Parkinson's disease (PD), and this gene is implicated in sporadic PD as well [1], [2], [3]. DJ-1 is a highly conserved homodimeric protein that is directly cytoprotective in neurons [4] and astroglia [5] under oxidative stress conditions [6], [7]. Additionally, DJ-1 influences the transcriptome and proteome of a cell by regulating transcription factors and post-transcriptional processes, often interfering with pro-apoptotic processes [8], [9] or upregulating antioxidant proteins [10], [11].
microRNAs (miRNA) are short non-coding RNAs of about 22 nucleotides long that negatively regulate the post-transcriptional network by recognizing target mRNAs through base-pairing, and catalyzing transcript degradation or inhibiting mRNA translation [12]. Perturbations in miRNA expression have been implicated in a number of nervous system disorders including PD, suggesting that miRNA dysregulation is involved in the pathogenesis of these diseases [13].
DJ-1 can act as a direct transcriptional co-activator [14] and can modulate signal transduction to affect the transcriptome [15], [16]. It may also have a role in direct RNA binding and post-transcriptional regulation [11]. In order to investigate miRNAs regulated by DJ-1, as well as their impact on the transcriptome and downstream processes involved in PD pathogenesis, we profiled miRNA expression in a human neuroblastoma cell model and found miR-221 to be down-regulated in DJ-1 knock down (KD) cells compared with controls. miR-221, one of the most abundant miRNA species in the human brain [17], has a critical role in cell survival [18], apoptosis [19], [20], [21], [22], and neuritogenesis [23], [24], [25]. Interestingly, two separate miRNA profiling studies in PD patients have shown that decreased serum miR-221 may serve as a biomarker for diagnosis and disease stage evaluation [26], [27]. In in vitro PD models, miR-221 was shown to regulate Transferrin receptor type 2 in SH-SY5Y cells challenged with the dopaminergic toxin 1-methyl-4-phenylpyridinium (MPP+)[28], and to be protective by regulating PTEN in PC12 cells challenged with 6-hydroxydopamine [29]. Additionally, miR-221 is reported to be differentially expressed in the cingulate gyrus of PD patients and correlated with the downregulation of the expression of SNCA, PARK2, and LRRK2, genes linked to both autosomal recessive and dominant forms of PD [30], [31].
Here, we report for the first time a mechanistic link between DJ-1 and mir-221. We found that knocking-down DJ-1 in a cellular model and knocking it out in the mouse brain result in down-regulation of miR-221. While re-introducing wild-type DJ-1 restores miR-221 expression, its PD-linked pathogenic M26I mutant fails to do the same. miR-221 mediates the cytoprotective activity of DJ-1 by repressing the expression of several pro-apoptotic proteins and protects differentiated ReNcell VM human dopaminergic neuronal cells from cell death and neurite retraction induced by MPP+. DJ-1 may modulate miR-221 levels, in part, through the mitogen-activated protein kinase (MAPK)/extracellular-regulated kinase (ERK) pathway. These findings suggest that the modulation of miR-221 by DJ-1 is protective in the context of pathogenic mechanisms of PD including cell death [32] and neurite degeneration [33].
2. Results
2.1. miR-221 expression is modulated by wild-type DJ-1 but not its pathogenic mutant
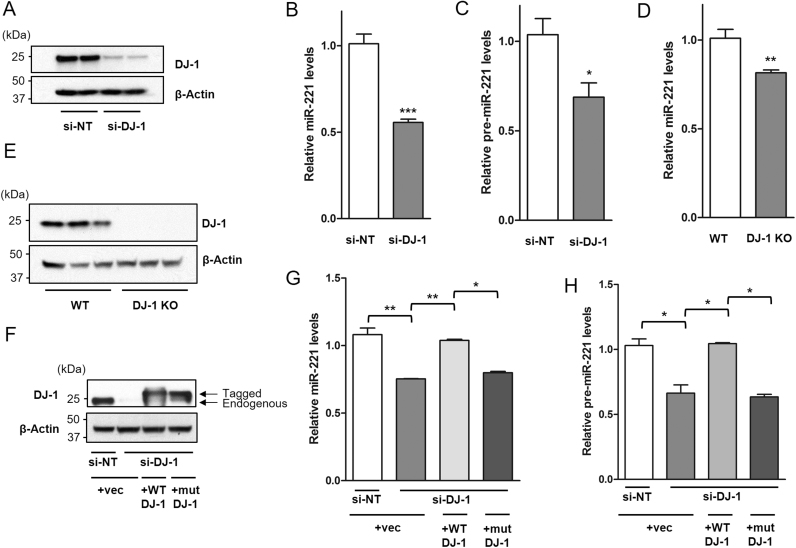
To identify novel RNA transcripts regulated by DJ-1, we performed an Affymetrix Genechip® Human Gene 2.0 ST RNA expression microarray, which covered 40,716 RefSeq transcripts, to identify RNA species that are differentially expressed in a DJ-1 knock-down (KD) model of human dopaminergic neuroblastoma SH-SY5Y cells. A pool of four different small interfering RNAs (siRNAs) directed against DJ-1 was used to achieve potent and specific knock-down (Fig. 1A). Among the down-regulated transcripts, microRNA-221 (miR-221) exhibited the greatest fold reduction, with an expression level of 45% in DJ-1 KD cells compared to that in control cells.
Fig. 1.
miR-221 expression is modulated by wild-type DJ-1 but not its pathogenic mutant. SH-SY5Y cells were transfected with either pooled DJ-1 siRNA (si-DJ-1) or non-targeting control siRNA (si-NT). (A) Immunoblotting shows that DJ-1 is knocked down effectively by pooled DJ-1 siRNA. (B) DJ-1 knock-down (KD) decreases mature miR-221 and (C) precursor pre-miR-221. (D) DJ-1 knockout mouse brains have lower levels of mature miR-221 compared to wild-type mice. (E) DJ-1 knockout mice lack DJ-1 expression. (F) SH-SY5Y cells were transfected with either si-NT or si-DJ-1 for 24 h, followed by transfection with either empty control vector, FLAG-tagged wild-type DJ-1, or FLAG-tagged pathogenic M26I mutant DJ-1. Immunoblotting shows expression of FLAG-tagged wild-type and mutant DJ-1 protein, and the downregulation of endogenous DJ-1 protein in DJ-1 KD cells. (G) Transfection of wild-type DJ-1 but not its pathogenic M26I mutant in DJ-1 KD cells rescued mature miR-221 expression and (H) precursor pre-mir-221 levels. Data are presented as means ± S.E.M. Asterisks denote statistically significant differences (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001) relative to control. (B–D) analyzed using two-tailed student's t-test. (G, H) analyzed using one-way ANOVA with Bonferroni post-hoc multiple comparisons test.
This result from microarray analysis was confirmed by real time quantitative PCR (RT-qPCR) in both DJ-1 KD SH-SY5Y cells (Fig. 1B) and in the cerebral cortex of DJ-1 knock-out mice (Fig. 1D–E). In addition to down-regulation of miR-221 expression, DJ-1 KD in SH-SY5Y cells also decreased the precursor form of miR-221 (pre-miR-221), indicating that DJ-1 may regulate miR-221 prior to its processing to the mature form (Fig. 1C).
To examine the pathogenic consequence of DJ-1 in modulating miR-221 expression, the effect of wild-type and a PD-linked mutant DJ-1 were tested next. Over-expression of FLAG-tagged wild-type DJ-1 but not the empty control vector could re-constitute both mature miR-221 and pre-miR-221 levels in DJ-1 KD cells. However, FLAG-tagged pathogenic M26I mutant DJ-1 [34] could not rescue the down-regulation of miR-221 or pre-miR-221 caused by DJ-1 knock-down (Fig. 1F–H). This provides further evidence that DJ-1 may regulate miR-221 expression whereas PD-causing loss-of-function mutations in DJ-1 nullify its ability to regulate miR-221. This finding raises the possibility that miR-221 could be involved in a pathway that mediates a neuroprotective function of DJ-1.
2.2. miR-221 promotes cell survival under MPP+ stress
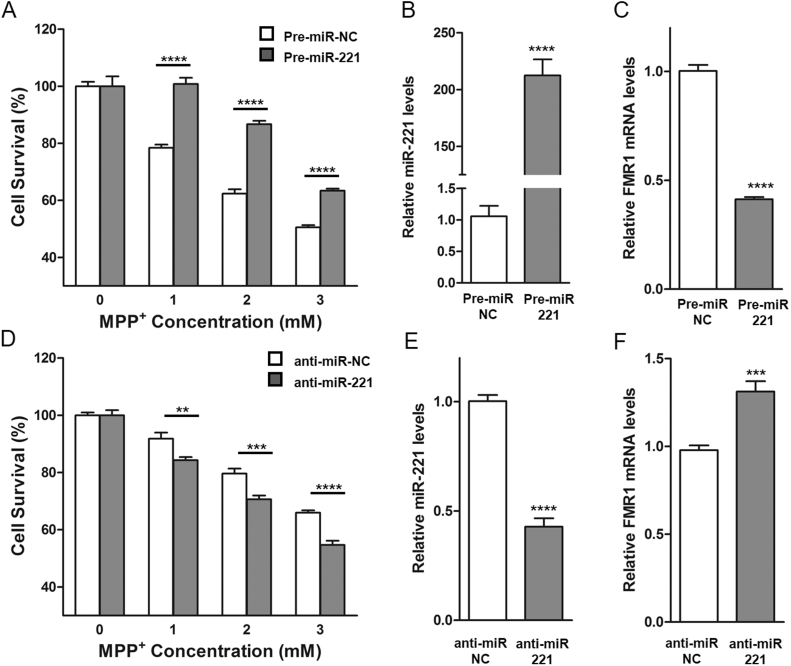
DJ-1 is known to confer protection and increase cell survival against the dopaminergic neurotoxin MPP+ [4], [6], [35], [36]. To investigate whether miR-221 mirrors the cytoprotective effect of DJ-1, cells were made to over-express miR-221 by transfection with the precursor form of miR-221, pre-miR-221. They were then challenged with MPP+ for 24 h, and survival was determined by MTS assay. Exogenous over-expression of miR-221 significantly increased cell survival under MPP+ stress compared to cells transfected with scrambled pre-miR control (Fig. 2A). RT-qPCR confirmed increased levels of mature miR-221 in transfected cells (Fig. 2B). The functionality of miR-221 over-expression in these cells was verified by down-regulation of a known miR-221 target, fragile X mental retardation 1 (FMR1) mRNA [37] (Fig. 2C).
Fig. 2.
miR-221 is cytoprotective under MPP+insult. (A) SH-SY5Y cells were transfected with either pre-miR negative control (pre-miR-NC) or pre-miR-221. Transfected cells were treated with indicated concentrations of MPP+ for 24 h before assessing cell survival using MTS assay. Over-expression of miR-221 resulted in significantly higher cell viability following MPP+ treatment. (B) Transfected cells showed robust increase in mature miR-221 levels and (C) down-regulation of known miR-221 mRNA target, fragile X mental retardation 1 transcript (FMR1). (D) SH-SY5Y cells were transfected with either anti-miR negative control (anti-miR-NC) or anti-miR-221 that binds to endogenous mature miR-221. Cell survival of transfected cells was assessed after MPP+ treatment at the indicated concentrations for 24 h, and showed that inhibition of endogenous miR-221 resulted in a significant decrease of cell viability. (E) Anti-miR-221 transfected cells showed a decrease in mature miR-221 levels and (F) up-regulation of FMR1. Data are presented as means ± S.E.M. Asterisks denote statistically significant differences (**p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001) relative to control. (B, C, E, F) analyzed using two-tailed student's t-test. (A, D) analyzed using two-way ANOVA with Bonferroni post-hoc multiple comparisons test.
Conversely, when endogenous miR-221 was inhibited by anti-miR-221 (a single-stranded oligonucleotide that inhibits mature miR-221 by complementary binding) [22], cell survival decreased significantly in response to MPP+(Fig. 2D). The effects of anti-miR-221 in reducing miR-221 levels (Fig. 2E) and up-regulating FMR1 mRNA levels (Fig. 2F) were confirmed. These findings show that miR-221 indeed mirrors the cytoprotective effect of DJ-1 and that a decrease of endogenous miR-221 renders cells more sensitive to toxic stress. That is, miR-221 at even endogenous levels is sufficient to be cytoprotective, suggesting that the modulation of miR-221 by DJ-1 may have physiological significance.
2.3. miR-221 targets transcripts of apoptotic proteins and rescues DJ-1 knock-down cells from cell death
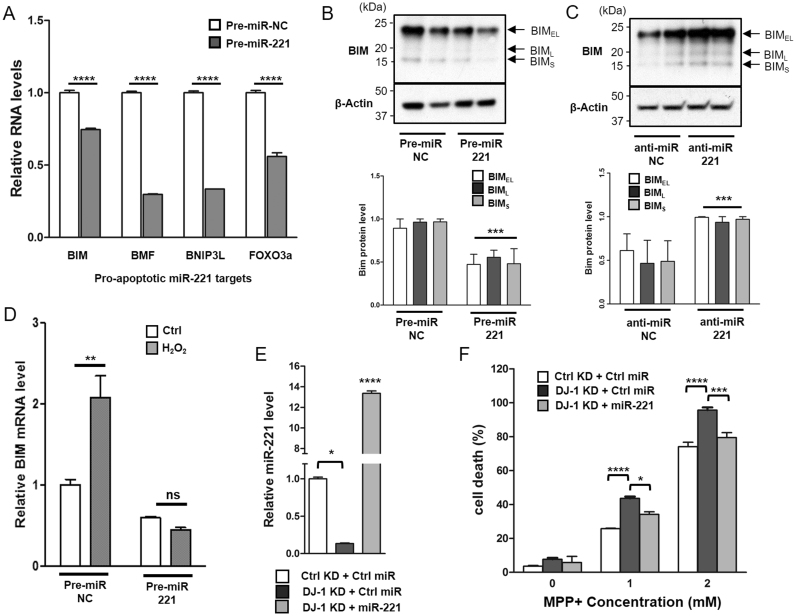
One way by which DJ-1 confers cytoprotection is through interfering with apoptotic cascades and cell death processes [36], [38], [39], [40]. To study whether miR-221 may be acting similarly, we used miR target prediction and database tools, TargetScanHuman [12], [41], miRBase [42], and miRTarBase [43], to identify pro-apoptotic proteins that are predicted to be regulated by miR-221. Among targets with conserved sites predicted to pair with miR-221 seed sequence are apoptotic activators from the BCL2 family that have only the BH3 domain [44]: bcl-2-like protein 11 (BIM), bcl2 modifying factor (BMF), and bcl2 interacting protein 3-like (BNIP3L). Also predicted is forkhead box O3 (FOXO3a), a transcription factor implicated in the expression of genes necessary for cell death. Over-expression of miR-221 in SH-SY5Y cells significantly down-regulated the mRNA levels of all four targets implicated in promoting apoptosis, demonstrating that miR-221 may be acting at multiple levels to inhibit cell death (Fig. 3A). These apoptotic transcripts have been shown to be down-regulated in multiple cell types as well, such as BIM in PC12 cells [25], BMF in hepatocellular carcinoma cells [20], BNIP3L in 293T cells [45], and FOXO3a in human breast cancer cell lines [46].
Fig. 3.
miR-221 Down-Regulates transcripts involved in apoptosis, including Pro-Apoptotic protein BIM, and protects DJ-1 knock-down cells from cell death. (A) Compared to cells transfected with negative control pre-miR (pre-miR-NC), SH-SY5Y cells transfected with pre-miR-221 showed a decrease of mRNA transcripts implicated in apoptosis, including BIM, BMF, BNIP3L, and FOXO3A. (B) All three isoforms of BIM protein (BIMEL, BIML, BIMS) were decreased by over-expression of miR-221 and (C) increased by the inhibition of miR-221 using anti-miR-221. (D) Pre-miR-221 transfected cells were treated with 250 uM H2O2 for 24 h. Compared to control, over-expression of miR-221 prevented the oxidative stress mediated induction of pro-apoptotic BIM transcript. (E) Cells were made to stably express non-targeting control (Ctrl) short hairpin RNA (shRNA) or pooled DJ-1 shRNA. Cells were then transduced with either lentiviral control miR (Ctrl miR) or miR-221 for 24 h. Stable DJ-1 KD decreases the levels of mature miR-221, while transduction with lentiviral miR-221 robustly increases its levels. (F) Cells were then treated with the indicated concentrations of MPP+ for 24 h, and cell death was assessed using LDH assay. Compared to control KD cells, stable DJ-1 KD leads to increased cell death, which is rescued when miR-221 is over-expressed. Data are presented as means ± S.E.M. Asterisks denote statistically significant differences (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001) relative to control. ns = not significant. (A, D, F) analyzed using two-way ANOVA with Bonferroni post-hoc multiple comparisons test. (B, C) analyzed using two-tailed student's t-test. (E) Analyzed using one-way ANOVA with Bonferroni post-hoc multiple comparisons test.
Notably, miR-221 over-expression represses the protein expression of BIM, a pro-apoptotic protein shown to be crucial in neuronal apoptosis, particularly in the death of dopaminergic neurons in an N-methyl-4-phenyl 1,2,3,6-tetrahydropyridine (MPTP) model of PD [47], [48]. miR-221 decreases the levels of all three pro-apoptotic BIM protein isoforms, BIM-short (BIMS), BIM-long (BIML), and BIM-extra long (BIMEL) (Fig. 3B), while miR-221 inhibition increases BIM protein levels (Fig. 3C). Remarkably, accumulation of BIM transcript following hydrogen peroxide (H2O2) treatment is completely prevented by miR-221 transfection (Fig. 3D).
As DJ-1 KD cells exhibit increased susceptibility to MPP+ neurotoxicity compared to control cells [36], whether miR-221 could rescue this susceptibility was investigated next. First, cells were made to stably express non-targeting control (Ctrl) short hairpin RNA (shRNA) or pooled DJ-1 shRNA. Cells were then transduced with either lentiviral control miR (Ctrl miR) or miR-221 for 24 h. Stable DJ-1 KD led to decreased levels of mature miR-221, while transduction with lentiviral miR-221 robustly increased its levels (Fig. 3E). Next, cells were challenged with MPP+ for 24 h, and cell death was assessed using lactate dehydrogenase (LDH) release. Stable DJ-1 KD cells, which exhibit a marked decrease in miR-221 levels, showed increased cell death in response to MPP+ treatment. However, transduction with lenti-miR-221 completely rescued DJ-1 KD cells to levels seen with control KD conditions, indicating that miR-221 may play a significant role in DJ-1 mediated cytoprotection (Fig. 3F).
2.4. miR-221 protects human neurons from MPP+-induced toxicity
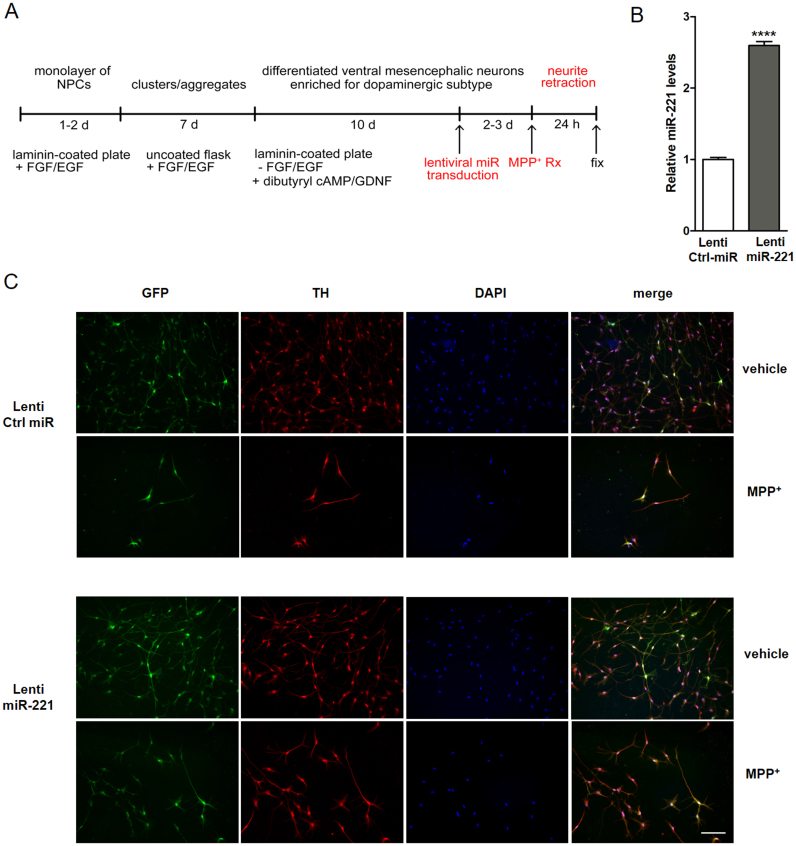
To extend the findings obtained with SH-SY5Y cells, we also examined the impact of miR-221 in ReNcell VM cells, which are human ventral mesencephalic neural progenitor cells (NPCs) that can be terminally differentiated to tyrosine hydroxylase positive dopaminergic neuronal subtype with clearly defined cell bodies and extensive neurite outgrowths through an established pre-aggregation method [49] (Fig. 4A). First, ReNcell VM NPCs were terminally differentiated to non-proliferative dopaminergic neurons and transduced with either lentiviral vector over-expressing miR-221 (lenti-miR-221) or its scrambled control vector (lenti-Ctrl miR) at a multiplicity of infection (MOI) of 10 to achieve high transduction efficiency (Fig. 4B). These lentiviral constructs contain Green Fluorescent Protein (GFP) in a bicistronic transcript so that transduction efficiency can be easily verified. Upon exposure of Ctrl miR-transduced cells to MPP+, loss of neurite morphology and resultant retraction were observed, which are some of the earliest signs of neurotoxin-induced degeneration in cultured neuronal cells [50], [51]. This toxic effect was mitigated in cells over-expressing lentiviral miR-221 (Fig. 4C).
Fig. 4.
miR-221 mitigates the loss of neuron morphology in terminally differentiated, human ReNcell VM dopaminergic neurons treated with MPP+. To investigate the role of miR-221 in terminally differentiated human neuronal cells, (A) neural progenitor cells (NPCs) derived from the ventral mesencephalon (ReNcell VM) were terminally differentiated to dopaminergic neurons using an established pre-aggregation protocol. (B) Differentiated ReNcell VM cells were transduced with lentivirus containing control (lenti-Ctrl miR) or miR-221 (lenti-miR-221). Cells transduced with lenti-miR-221 showed robust up-regulation of mature miR-221. (C) These cells were treated with 0.5 mM MPP+ or vehicle (DMEM) for 24 h. Differentiated ReNcell VM cells exhibited GFP expression from viral transduction, stained positively for the dopaminergic neuron marker tyrosine hydroxylase (TH). Following MPP+ exposure, cells exhibited loss of neurite morphology, which was mitigated by miR-221 over-expression (Scale bar: 100 µm). Data are presented as means ± S.E.M. Asterisks denote statistically significant differences (****p ≤ 0.0001) relative to control. (B) analyzed using two-tailed student's t-test.
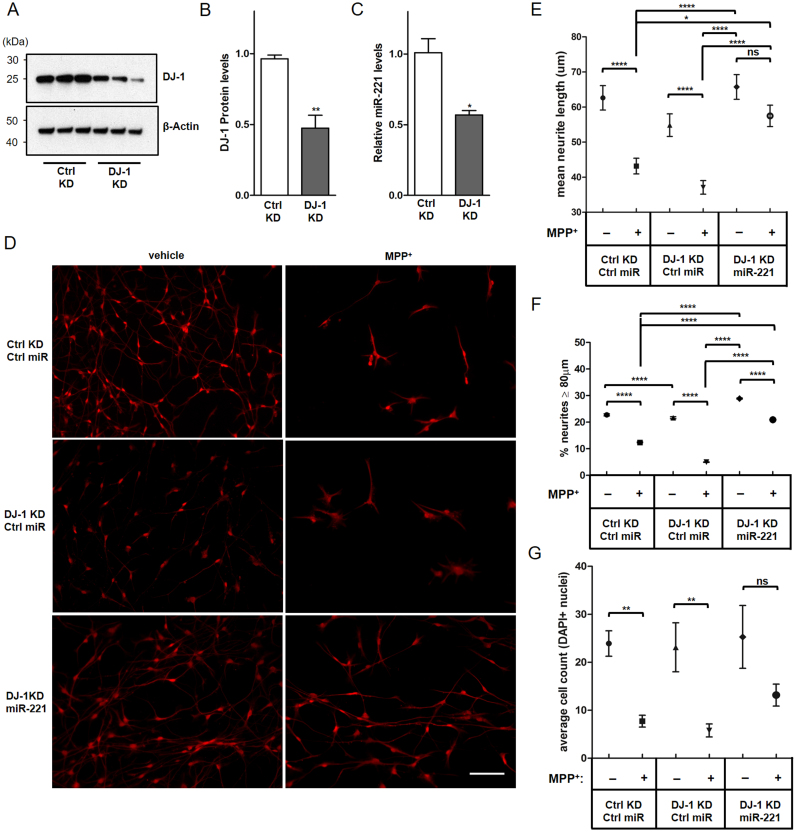
Next, to assess the effect of miR-221 on DJ-1 KD cells, NPCs were made to stably express DJ-1 shRNA or non-targeting control shRNA prior to differentiation (Fig. 5A, B). A pool of three different shRNAs targeting DJ-1 were used to achieve specific and potent knock-down. When terminally differentiated, DJ-1 KD post-mitotic ReNcell VM neurons exhibited decreased miR-221 expression compared to control KD cells (Fig. 5C), replicating the finding in SH-SY5Y cells above (Fig. 1B). DJ-1 KD or control differentiated neurons were then transduced with either lenti-miR-221 or lenti-Ctrl miR, and subjected to MPP+ challenge for 24 h. Following fixation and imaging, neurotoxicity was assessed by measuring neurite length and cell count. While MPP+ caused significant shortening of neurites in all cells that were transduced with Ctrl miR, DJ-1 KD cells transduced with lenti-miR-221 were protected from neuritic retraction (Fig. 5D, E).
Fig. 5.
miR-221 prevents MPP+-induced neurite retraction and cell loss in DJ-1 KD ReNcell VM dopaminergic neurons. ReNcell NPCs were made to stably express either control non-targeting shRNA or pooled DJ-1 shRNA, then differentiated into post-mitotic dopaminergic neurons. (A) Immunoblotting shows that DJ-1 is knocked down effectively by DJ-1 shRNA. (B) Quantification of (A). (C) As seen previously in SH-SY5Y cells, DJ-1 KD decreases miR-221 levels in differentiated ReNcell VM neurons. (D) Control KD (Ctrl KD) or DJ-1 KD cells were transduced with either lenti-Ctrl miR or lenti-miR-221 and exposed to 0.5 mM MPP+ or vehicle (DMEM) for 24 h. MPP+ exposure resulted in cell loss and neuritic retraction (Scale bar: 100 µm). (E) Measurements of neurite length indicated that miR-221 protects DJ-1 KD neurites from MPP+. (F) Compared to control KD cells, long neurites (≥ 80 µm, longer than 75th percentile of all neurites) were less preserved among DJ-1 KD cells treated with MPP+. This effect was reversed when DJ-1 KD cells were made to over-express miR-221. (G) MPP+ treatment significantly decreased the cell count in Ctrl miR transduced neurons, whereas miR-221 transduced neurons showed a smaller, non-significant decrease in cell count. For (E, G), n = 450 neurites per group, representative of length measurements taken from 15 microscopic fields per experimental group across three biological replicates. For (F), mean values of percent neurites ≥ 80 µm long were generated with bootstrap resampling, where each sample run contained n = 100 neurites per group. Resampling runs were repeated 500 times to generate confidence intervals for mean values of percent neurites ≥ 80 µm. For (G), Average cell count was assessed by counting DAPI-positive nuclei in these fields. For (B, C, E, G), data are presented as means ± S.E.M. For (F), data are presented as means ± Confidence Interval. Asterisks denote statistically significant differences (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001) relative to control. ns = not significant. (B, C) analyzed using two-tailed student's t-test. (E, F, G) Analyzed using one-way ANOVA with Bonferroni post-hoc multiple comparisons test.
As expected from the loss of DJ-1's protective effects, DJ-1 KD cells transduced with lenti-Ctrl miR and treated with MPP+ exhibited decreased preservation of its long neurites (defined as being longer than 75th percentile of all neurites, or greater than or equal to 80 µm) compared to control KD cells transduced with lenti-Ctrl-miR and treated with MPP+, with only 5% of total measured neurites exceeding 80 µm in DJ-1 KD cells compared to 12% in control KD cells. Notably, this effect on neuritic retraction brought about with MPP+ in DJ-1 KD cells was mitigated by over-expressing miR-221, with 21% of MPP+ treated neurites exceeding 80 µm in length (Fig. 5F). Finally, while MPP+ treatment significantly decreased the number of neurons in plates transduced with lenti-Ctrl miR, cells over-expressing miR-221 showed a smaller, non-significant decrease in cell count, suggesting that miR-221 provides protection for DJ-1 KD cells from MPP+-induced neurotoxicity (Fig. 5G).
2.5. DJ-1 regulates miR-221 levels through the MAPK/ERK pathway
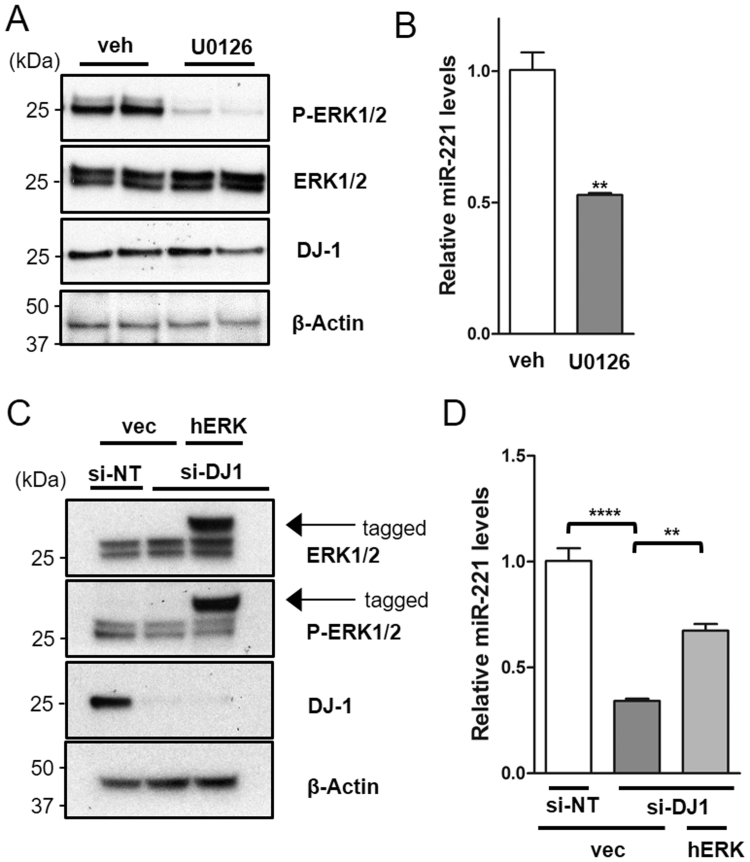
The above findings showing that knocking-down DJ-1 decreases miR-221 levels whereas the re-introduction of DJ-1 rescues miR-221 levels, and that miR-221 mirrors much of DJ-1's cytoprotective effects in both SH-SY5Y and ReNcell VM cells, suggested that miR-221 may be modulated downstream of DJ-1 to mediate some of DJ-1's cytoprotective effects. Accordingly, we set out to investigate the mechanism by which DJ-1 could regulate miR-221 expression. As miR-221 is induced by ERK signaling activated by nerve growth factor (NGF) [25], and DJ-1 positively modulates mitogen-activated protein kinase (MAPK)/extracellular-regulated kinase (ERK) signaling [52], [53], we hypothesized that DJ-1 could regulate miR-221 levels through its effect on the MAPK/ERK pathway. To test this possibility, SH-SY5Y cells were treated with the MEK1/2 inhibitor, U0126 for 24 h, which decreased the levels of phospho-ERK1/2 without impacting the levels of total ERK1/2 or DJ-1 (Fig. 6A). This inhibition of ERK1/2 phosphorylation led to a robust down-regulation of miR-221 (Fig. 6B), confirming that miR-221 levels are indeed affected by the activation status of the MAPK/ERK pathway. Next, to determine whether the down-regulation of miR-221 levels seen in DJ-1 KD cells could be rescued by increasing the activation of the ERK pathway, DJ-1 KD cells were transfected with FLAG-tagged human ERK1 (hERK1) or control plasmid for 48 h. hERK1 over-expressing cells exhibited increased phospho-ERK without impacting the levels of DJ-1 (Fig. 6C). Notably, hERK1 overexpression was able to partially rescue the decreased miR-221 levels in DJ-1 KD cells (Fig. 6D). Together, these results show that DJ-1 may in part regulate miR-221 levels through ERK activation.
Fig. 6.
DJ-1 modulates miR-221 expression through the MAPK/ERK1/2 pathway. To investigate whether DJ-1 could modulate miR-221 levels through its effect on the MAPK/ERK pathway, (A) SH-SY5Y cells were treated with MEK1/2 inhibitor, U0126 (50 µM), or vehicle (DMSO) for 24 h. This led to a decrease in levels of active, phosphorylated ERK1/2 without impacting the levels of total ERK1/2 or DJ-1 protein. (B) The inhibition of ERK1/2 phosphorylation led to down-regulation of miR-221 levels. (C) DJ-1 KD cells were transfected with FLAG-tagged hERK1 or vector-only control for 48 h. hERK1 overexpression increased phosphorylation of ERK1 in DJ-1 KD cells (D) and partially rescued the decrease in miR-221 levels. Data are presented as means ± S.E.M. Asterisks denote statistically significant differences (**p ≤ 0.01, ****p ≤ 0.0001) relative to control. (B) analyzed using two-tailed student's t-test. (D) analyzed using one-way ANOVA with Bonferroni post-hoc multiple comparisons test. Veh = vehicle. Vec = vector-only control.
3. Discussion
Advances in our understanding of genomic architecture have revealed that 60% of the transcriptome consists of transcripts without protein coding capacity, known as non-coding RNA (ncRNA) [54]. One prominent class of ncRNA is miRNAs, which has become increasingly recognized as a critical contributor to gene expression and maintenance in the brain, especially in neural cells, which are highly active transcriptionally [55]. The role of miRNAs is well-established in neurodegenerative disorders, and their therapeutic potential is recognized in PD [13], [56], [57]. For example, miR-7 targets the mRNA encoding the key PD-associated protein α-synuclein, repressing its expression and protecting cells against oxidative stress [58], [59], [60]. In addition, microRNAs have also been shown to modulate other key proteins that are dysregulated in PD, such as LRRK2, and LRRK2 in turn, has been shown to modulate microRNA, indicating that regulation can be bidirectional [61], [62], [63], [64].
Though DJ-1 has been shown to regulate cellular RNA composition through its effect on signal transduction, transcription, and post-transcriptional processes, microRNAs modulated by DJ-1 have not yet been extensively examined. miR-221 is one of the top 20 most abundant microRNA species in the human brain [17] and found to be highly expressed in the brains of rat and mouse models [65], [66]. Identified in our microarray screen as being down-regulated when DJ-1 is knocked down, miR-221 re-capitulates many of the varied functionalities of DJ-1 in cancer [67] and neuroprotection [68], [69], including its ability to facilitate oncogenesis [19], [21] as well as its ability to support neuronal outgrowth and differentiation in PC12 cells [23].
In the present study, we provide evidence that wild-type DJ-1, but not its pathogenic M26I mutant, is able to modulate miR-221 levels, adding relevance to the role of miR-221 in DJ-1 linked PD pathogenesis. Indeed, similarly to DJ-1 [36], a simple decrease in endogenous levels of miR-221 negatively affects cell survival, while over-expression of mir-221 protects DJ-1 KD cells from cell death under MPP+ stress. One way miR-221 protects cells may be through interfering with apoptotic mechanisms triggered under pathophysiological conditions. We show that miR-221 suppresses the transcription of many pro-apoptotic BH-3 only transcripts including bcl-2-like protein 11 (BIM), bcl2 modifying factor (BMF), and bcl2 interacting protein 3-like (BNIP3L). BNIP3L [70], [71] and BMF [72] have both been implicated in apoptotic neuronal death, and BIM in particular has been closely investigated in the context of neurodegenerative diseases. BIM is up-regulated in the MPTP model of PD and affects the degeneration of dopaminergic neurons [48], [73]. It is also reportedly elevated in Alzheimer's disease neurons and is required for β-Amyloid-induced neuronal apoptosis [74]. In addition, BIM is increased in neurons expressing mutant huntingtin protein as well as in Huntington's disease mouse models [75]. miR-221 effectively down-regulates the major BIM protein isoforms BIMS, BIML, BIMEL, which are produced by alternative splicing, and all contain 3’-UTRs that harbor the putative miR-221 seed sequence binding site. Of these isoforms, BIMEL is considered the predominant pro-apoptotic species in neuronal cells [48].
We also found that miR-221 down-regulates FOXO3a, a transcription factor that induces BIM expression [76] and is implicated in the positive regulation of BMF [77] and BNIP3L [78]. Additionally, FOXO3a-mediated induction of BIM is implicated in neurodegeneration brought about by β-Amyloid-treatment [79]. All these observations suggest that miR-221 may function to antagonize apoptosis at many levels to protect cells.
Accordingly, we show that when miR-221 is over-expressed in terminally differentiated ReNcell VM dopaminergic neurons, it protects DJ-1 KD cells from toxin-induced loss of neurite morphology and retraction, a process that precedes apoptosis in cultured neurons [50]. In fact, retrograde axonal degeneration of dopaminergic neurons and loss of striatal terminals is a predominant early feature of PD that precedes cell body loss [80]. Dopaminergic neurons of the substantia nigra pars compacta (SNpc) extend impressively long axonal projections to the striatum, and the soma of these neurons often makes up less than 1% of the total cell volume as one neuron may give rise to over 150,000 presynaptic terminals in the striatum [81], [82]. This fact underscores the importance of the neuritic complexity of SN neurons and may underlie the observation that long, unmyelinated or poorly myelinated dopaminergic neuronal projections are particularly vulnerable to neurodegenerative pathologies due to higher energy demand [83]. Interestingly, neurons lacking DJ-1 have been shown to have reduced microtubule dynamics, and striatal medium spiny neurons (MSNs) from DJ-1-deficient mice have been shown to display impaired dendritic complexity and reduced dendritic spine densities, suggesting that DJ-1 may be necessary to maintain the integrity of the synaptic network [69]. Similarly, DJ-1 may regulate miR-221 to contribute to the maintenance of long neuritic projections, and increase neuronal functionality and target innervation. This may be especially significant in the early stages of PD and provides feasible therapeutic avenues in manipulating miR-221 levels for the neurorestoration of dopaminergic projections.
Finally, DJ-1 may regulate miR-221 in part through its actions on the MAPK/ERK pathway. We show that pharmacologic inhibition of ERK1/2 down-regulates miR-221 levels, whereas ERK1 over-expression in DJ-1 KD cells partially rescues miR-221 levels, suggesting that miR-221 may be modulated by additional pathways that are regulated by DJ-1. For example, c-Jun N-terminal kinase (JNK) activation leading to increased activity of c-Jun (AP1) transcription factor has been reported to regulate miR-221 expression in a non-small cell lung cancer model, and miR-221 has also been reported to be substantially induced by JNK inhibition in mesenchymal cells [84], [85]. DJ-1 negatively regulates JNK activation through its inhibitory effect on apoptosis signal-regulating kinase 1 (ASK1) [36], [38], [86], a mitogen-activated protein kinase kinase kinase 5 (MAP3K5) that activates the MKK4/MKK7-JNK apoptotic axis [87]. Therefore, it would be interesting to study whether DJ-1 regulates miR-221 through multiple alternative signaling pathways, including MAPK/ERK signaling and ASK1/MKK4/MKK7-JNK signaling.
In conclusion, we provide evidence for the first time that DJ-1, but not its pathogenic mutant, modulates miR-221 expression in part through the MAPK/ERK pathway. This further elucidates how DJ-1 may regulate the transcriptome to provide cytoprotection in the context of pathogenic mechanisms of PD. Future investigations into the role of miR-221 and its target genes in in vivo models are needed to further elucidate its potential as a disease biomarker and its therapeutic applicability.
4. Materials and methods
4.1. Materials
Expression vectors encoding FLAG-tagged wild-type or M26I DJ-1 were previously described [10], [36]. DJ-1 null mice were a kind gift from Ted Dawson (Johns Hopkins University), and all mouse brain samples used in this study were collected from 3-month old males. A mixture of four different DJ-1 silencing RNA (SMARTpool siGENOME siRNA; si-DJ1) as well as scrambled control siRNA (siRNA-NT) was purchased from GE Healthcare/Dharmacon. Pre-miR-221 and scrambled miRNA control (miR-SC) was purchased from Ambion. Custom anti-miR-221 [22] was synthesized by Integrated DNA Technologies (IDT). MPP+ was purchased from Sigma. U0126 was purchased from Millipore. pFLAG-CMV-hERK1 was a gift from Melanie Cobb (Addgene, plasmid # 49328).
4.2. Cell culture, ReNcell VM differentiation, and chemicals
HEK293T (ATCC) and human neuroblastoma SH-SY5Y (ATCC) cells were cultured in Dulbecco's Modified Eagle's Medium/Ham's F-12 1:1 Mix (DMEM/F12; GE Healthcare/Hyclone) supplemented with 10% fetal bovine serum (Atlanta Biologicals).
ReNcell VM (ventral mesencephalic/midbrain) human neural progenitor cell line (NPC) was purchased from EMD Millipore. The proliferative ReNcell NPCs were plated onto laminin (Sigma) coated plates and maintained in DMEM/F12 supplemented with 2% B27 neural supplement (Life Technologies/Thermo Fisher Scientific), Glutamax (Thermo Fisher Scientific), 10 U/mL heparin (Sigma), 50 μg/mL gentamycin (Thermo Fisher Scientific), 20 ng/mL basic fibroblast growth factor (bFGF; Peprotech), and 20 ng/mL epidermal growth factor (EGF; Peprotech). To promote terminal differentiation into dopaminergic neurons, pre-aggregation protocol [49] was employed. In short, ReNcell NPCs were propagated in a monolayer on laminin-coated plates in media supplemented with bFGF and EGF. When the plate reached 80% confluence, cells were gently removed from the plate using 1× Accutase cell detachment solution (Sigma), then cultured in non-coated flasks for 7 days until neurosphere formation was observed. These neurospheres were collected, triturated, seeded on laminin-coated plates or slides, and incubated in media without bFGF or EGF, but supplemented instead with 1 mM dibutyryl-cAMP (Santa Cruz Biotechnologies) and 2 ng/mL glial cell derived neurotropic factor (GDNF; Peprotech). Viral transductions were conducted after 10 days of differentiation when neuronal morphology could be observed. MPP+ treatments and other experiments were carried out 2–3 days after transduction. Differentiated cells were verified to stain with the mature neuronal marker, Microtubule Associated Protein 2 (MAP2) and the dopaminergic marker, Tyrosine Hydroxylase (TH). No staining could be observed with the glial cell marker, Glial Fibrillary Acidic Protein (GFAP), indicating that all ReNcell VM neurons derived from the pre-aggregation protocol were indeed dopaminergic neuronal subtypes.
4.3. Transfections
Cells were transfected with siRNA (40 nM), pre-miR (50 nM), and anti-miR (300 nM) using lipofectamine RNAiMAX (Thermo Fisher Scientific/Invitrogen). Reverse transfections with RNAiMAX were performed as follows: siRNA/pre-miR/anti-miR were mixed with RNAiMAX in Opti-MEM (Thermo Fisher) inside the wells, and incubated at room temperature for 10–20 min to allow for the formation of RNA-lipid complexes. Then cells were added to the wells containing the complexes so that they would be 50–60% confluent on the following day. Plasmid DNA transfections using expression vector containing FLAG-tagged DJ-1, or FLAG-tagged M26I mutant DJ-1, or empty expression vector were performed using Lipofectamine 3000 (Thermo Fisher Scientific/Invitrogen). Briefly, cells were plated to be 70–90% confluent in 12-well plates at the time of transfection. One ug of DNA plasmid was mixed with lipofectamine 3000 in Opti-MEM and incubated at room temperature for 10–15 min. The DNA-lipid complexes were then added to cells in a drop-wise fashion. Cells were visually analyzed 4–6 h post-transfection to assess for any reagent toxicity.
4.4. RNA microarray and databases
RNA from DJ-1 knock-down and control knock-down SH-SY5Y cells was harvested for microarray analysis (Affymetrix GeneChip® Human Gene 2.0 ST RNA expression microarray) to identify RNA species that were differentially expressed. TargetScanHuman [12], [41], miRBase [42], and miRTarBase [43] were used to predict potential microRNA targets, and miRTarBase [43] was used to identify experimentally verified microRNA targets.
4.5. RNA extraction, purification, and quantitative RT-PCR
RNA was extracted from cells and male mouse brain tissue using the miRNeasy mini kit (Qiagen). Brain samples were collected and flash frozen on dry ice (− 78.5 °C). Up to 50 mg of tissue was used per sample, and the tissue was disrupted and homogenized using a motorized pellet pestle (Kimble), followed by shearing through a 26 G needle and then vortexing. RNA was quantified using NanoDrop 2000 UV–Vis Spectrophotometer. cDNA was synthesized using miscript II RT kit (Qiagen) and SuperScript III RT kit (Thermo Fisher Scientific/Life Technologies) according to manufacturer's protocols. qPCR was performed using miScript SYBR Green PCR kit (Qiagen) and iTaq Universal SYBR Green Supermix (Bio-Rad) in an Applied Biosystems 7500 real-time PCR system. U6 small nuclear 6 (RNU6-6P) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as endogenous control for the normalization of miRNA qPCRs and mRNA qPCRs, respectively. The relative levels of RNA was compared and expressed as fold of control levels (ΔΔCT value). The following QuantiTect primer assays from Qiagen were used: human GAPDH, PARK7, BIM, FMR1, FOXO3, BNIP3L, and BMF. The following miScript primer assays from Qiagen were used: human RNU6-6P, human mature mir-221-3p, human precursor miR-221, mouse mature miR-221-3p.
4.6. Cell viability and cell death assays
Forty-eight hours after transfection, cells were incubated with the indicated concentrations of toxin (H2O2 or MPP+) in serum-free media for 24 h. Four to six technical replicates were performed for each concentration. CellTiter 96 Aqueous MTS reagent (Promega) was used to measure cell viability. Lactate dehydrogenase (LDH) Cytotoxicity Detection Kit (Takara/Clontech) was used to measure cell death. DMEM without cells was used as a background control, and 2 mM H2O2 was used to induce maximal LDH release conditions and used as a high control.
4.7. Lentiviral production and transduction
DJ-1 short hairpin RNA (shRNA) and control non-silencing shRNA expressing plasmids (pGIPZ) and corresponding lentiviral packaging systems were purchased from GE Healthcare/Dharmacon. These shRNAs were: Clone ID V2LHS_207558 targeting DJ-1 3’-UTR (sequence TATAGACACAATTTGTAGG), Clone ID V3LHS_331989 targeting DJ-1 ORF (sequence TTAAGAACAAGTGGAGCCT), Clone ID V3LHS_331991 targeting DJ-1 ORF (sequence TTCATGAGCCAACAGAGCA).
Hsa-miR-221-3p overexpressing lentiviral vector (lenti-miR-221) and its scrambled control (lenti-miR-SC) were purchased from Applied Biological Materials (ABM) Inc. Lentiviral constructs contain GFP in a bicistronic transcript so that transduction efficiency can be visually verified.
Lentiviral particles were produced using HEK293T cells (ATCC) and purified using Lenti-X Concentrator (Takara/Clontech) according to manufacturer's protocols. Lentiviral particles were incubated with 1 μg/mL polybrene (EMD Millipore) prior to cell infection to increase transduction efficiency, and concentrated virus (1 × 109 TU/mL) was used to transduce differentiated ReNcell VM at a multiplicity of infection (MOI) of 10.
As the pGIPZ system contains puromycin resistance, SH-SY5Y cells with stable expression/integration of DJ-1 shRNA were selected using 4 μg/mL puromycin (Sigma) and stable DJ-1 shRNA expressing ReNcell VM NPCs were selected using 2 μg/mL puromycin.
4.8. Western blotting
Cultured cells and brain tissue were rinsed with ice-cold PBS and lysed in 2% sodium dodecyl sulfate buffer with protease inhibitor cocktail and phosphatase inhibitors (Roche). Cell lysates were sonicated for 10 s three times and quantified using BCA Protein assay reagent (Thermo Fisher Scientific). Depending on the probing antibody used, between 20 μg and 60 μg of total protein were loaded per well at equal amount across all wells for each blot. Western blots were quantified by densitometry using ImageJ software (NIH), and band intensity was normalized to β-actin. Primary antibodies from the following suppliers were used for Western blot analyses: anti-DJ-1 (#A300-744A) from Bethyl; anti-p44/42 MAPK (anti-Erk1/2) (#9102), anti-phospho-p44/42 MAPK (Anti-phospho-Erk1/2) (Thr202/Tyr204) (#9101) from Cell Signaling; anti-Bim (#AB17003) from EMD Millipore; anti-β-Actin (#A2228) from Sigma. The following secondary antibodies from R&D Systems were used: horseradish peroxidase-conjugated anti-rabbit (#HAF008) or anti-mouse antibody (#HAF007).
4.9. Immunocytochemistry
Cells on glass coverslips were washed with PBS, fixed with 4% paraformaldehyde (Sigma), then permeabilized with 0.5% Triton X-100. After blocking with 5% donkey serum, cells were incubated with the dopaminergic marker primary anti-Tyrosine Hydroxylase (TH; #T2928; Sigma) monoclonal antibody diluted in blocking buffer overnight at 4 °C. The following day, the cells were washed with PBS and then incubated with secondary Rhodamine Red Donkey Anti-Mouse (#715-296-150; Jackson Immuno Research) antibody diluted in blocking buffer at room temperature for 1 h. After another PBS wash, cells were incubated with 1 μg/mL 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma) in PBS for 5 min. Subsequently, cells were washed, mounted on glass slides with Permafluor mounting media (Thermo Scientific), and allowed to set overnight at room temperature. Cells were imaged using a Carl Zeiss Axiovert 2000 fluorescence microscope.
4.10. ReNcell VM neurotoxicity assay, neurite length and cell count measurement
ReNcell VM NPCs stably expressing DJ-1 or control shRNA were differentiated using the previously described pre-aggregation method into terminal, TH-positive, dopaminergic neurons [49]. Neurite outgrowth was observed after 10 days of differentiation, and the neurons were transduced with lenti-miR-221 or lenti-miR-SC. After 48 h to allow for the expression of lenti-miR, cells were incubated with 0.5 mM MPP+ in serum-free media for 24 h. Cells were prepared for immunocytochemistry as described above, and imaged on a Zeiss fluorescence microscope at 20×. Five microscopic fields from the center of the slide were imaged from each of the six treatment groups (n = 3 biological replicates). This yielded an average of 150 neurites per experimental sample. Neurite length was measured from the tip of the neurite to the edge of the soma using ImageJ (NIH) software with NeuronJ plug-in [88]. Neurites that extended beyond the microscopic field were not included in measurements. Cell numbers were determined by counting DAPI positive nuclei. In order to reduce bias, both the imaging and measuring neurite length and cell numbers were performed under blind conditions.
4.11. Experimental design and statistical analyses
Statistical analyses were performed with Graph Pad Prism 5.0.4. Data are expressed as the mean ± SEM of at least 3 independent experiments. qPCR data were analyzed with two-tailed student's t-test or one-way ANOVA with Bonferroni multiple comparison test. Neurite length data were analyzed with one-way ANOVA with Bonferroni multiple comparison test. For Fig. 5F, mean values of percent neurites ≥ 80 µm long were generated with bootstrap resampling, where each sample run contained n = 100 neurites per group. Resampling runs were repeated 500 times to generate confidence intervals for mean values of percent neurites ≥ 80 µm. The data from bootstrap resampling runs were analyzed using one-way ANOVA with Bonferroni multiple comparison test. Cell survival and cell death data were analyzed with two-way ANOVA with Bonferroni multiple comparison test. All experiments with cells and animal tissue use at least 3 independent biological samples.
Acknowledgements
E.J. is supported by the National Institutes of Health [NS070898 and NS095003]. M.M.M. is the William Dow Lovett Professor of Neurology and is supported by the Michael J. Fox Foundation for Parkinson's Research, the American Parkinson Disease Association, the New Jersey Health Foundation/Nicholson Foundation, and by the National Institutes of Health [AT006868, NS073994, NS096032, and R01NS101134]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
S.E.O. and M.M.M. jointly designed the study and experiments with conceptual advice from E.J. L.H. performed the initial microarray. S.E.O., H.P., C.S. performed experiments, collected data, and provided technical support. S.E.O. analyzed the data. S.E.O. and M.M.M. interpreted the data. S.E.O. drafted the manuscript with critical revision from E.J. and M.M.M.
Acknowledgments
Conflicts of interest
E.J. and M.M.M. are inventors on US Patent US9255266 “RNA Targeting in alpha-Synucleinopathies.”
References
- 1.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., van Dongen J.W., Vanacore N., van Swieten J.C., Brice A., Meco G., van Duijn C.M., Oostra B.A., Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 2.Choi J., Sullards M.C., Olzmann J.A., Rees H.D., Weintraub S.T., Bostwick D.E., Gearing M., Levey A.I., Chin L.S., Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 2006;281:10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandopadhyay R., Kingsbury A.E., Cookson M.R., Reid A.R., Evans I.M., Hope A.D., Pittman A.M., Lashley T., Canet-Aviles R., Miller D.W., McLendon C., Strand C., Leonard A.J., Abou-Sleiman P.M., Healy D.G., Ariga H., Wood N.W., de Silva R., Revesz T., Hardy J.A., Lees A.J. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson's disease. Brain: J. Neurol. 2004;127:420–430. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- 4.Taira T., Saito Y., Niki T., Iguchi-Ariga S.M.M., Takahashi K., Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullett S.J., Di Maio R., Greenamyre J.T., Hinkle D.A. DJ-1 expression modulates astrocyte-mediated protection against neuronal oxidative stress. J. Mol. Neurosci. 2013;49:507–511. doi: 10.1007/s12031-012-9904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canet-Aviles R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M.J., Ringe D., Petsko G.A., Cookson M.R. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junn E., Jang W.H., Zhao X., Jeong B.S., Mouradian M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009;87:123–129. doi: 10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang S., Song S., Hong Y.K., Choi G., Suh Y.S., Han S.Y., Lee M., Park S.H., Lee J.H., Lee S., Bang S.M., Jeong Y., Chung W.J., Lee I.S., Jeong G., Chung J., Cho K.S. Drosophila DJ-1 decreases neural sensitivity to stress by negatively regulating Daxx-like protein through dFOXO. PLoS Genet. 2013;9:e1003412. doi: 10.1371/journal.pgen.1003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato I., Maita H., Takahashi-Niki K., Saito Y., Noguchi N., Iguchi-Ariga S.M., Ariga H. Oxidized DJ-1 inhibits p53 by sequestering p53 from promoters in a DNA-binding affinity-dependent manner. Mol. Cell. Biol. 2013;33:340–359. doi: 10.1128/MCB.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Im J.Y., Lee K.W., Woo J.M., Junn E., Mouradian M.M. DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum. Mol. Genet. 2012;21:3013–3024. doi: 10.1093/hmg/dds131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Brug M.P., Blackinton J., Chandran J., Hao L.Y., Lal A., Mazan-Mamczarz K., Martindale J., Xie C., Ahmad R., Thomas K.J., Beilina A., Gibbs J.R., Ding J., Myers A.J., Zhan M., Cai H., Bonini N.M., Gorospe M., Cookson M.R. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. USA. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Mouradian M.M. MicroRNAs in Parkinson's disease. Neurobiol. Dis. 2012;46:279–284. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 14.Xu J., Zhong N., Wang H., Elias J.E., Kim C.Y., Woldman I., Pifl C., Gygi S.P., Geula C., Yankner B.A. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 2005;14:1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 15.Lu L., Sun X., Liu Y., Zhao H., Zhao S., Yang H. DJ-1 upregulates tyrosine hydroxylase gene expression by activating its transcriptional factor Nurr1 via the ERK1/2 pathway. Int. J. Biochem. Cell Biol. 2012;44:65–71. doi: 10.1016/j.biocel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Oh S.E., Mouradian M.M. Regulation of signal transduction by DJ-1. Adv. Exp. Med. Biol. 2017;1037:97–131. doi: 10.1007/978-981-10-6583-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boudreau R.L., Jiang P., Gilmore B.L., Spengler R.M., Tirabassi R., Nelson J.A., Ross C.A., Xing Y., Davidson B.L. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron. 2014;81:294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina R., Zaidi S.K., Liu C.G., Stein J.L., van Wijnen A.J., Croce C.M., Stein G.S. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gramantieri L., Fornari F., Callegari E., Sabbioni S., Lanza G., Croce C.M., Bolondi L., Negrini M. MicroRNA involvement in hepatocellular carcinoma. J. Cell. Mol. Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gramantieri L., Fornari F., Ferracin M., Veronese A., Sabbioni S., Calin G.A., Grazi G.L., Croce C.M., Bolondi L., Negrini M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C.Z., Zhang J.X., Zhang A.L., Shi Z.D., Han L., Jia Z.F., Yang W.D., Wang G.X., Jiang T., You Y.P., Pu P.Y., Cheng J.Q., Kang C.S. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupini L., Bassi C., Ferracin M., Bartonicek N., D'Abundo L., Zagatti B., Callegari E., Musa G., Moshiri F., Gramantieri L., Corrales F.J., Enright A.J., Sabbioni S., Negrini M. miR-221 affects multiple cancer pathways by modulating the level of hundreds messenger RNAs. Front. Genet. 2013;4:64. doi: 10.3389/fgene.2013.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada N., Fujita Y., Kojima T., Kitamoto A., Akao Y., Nozawa Y., Ito M. MicroRNA expression profiling of NGF-treated PC12 cells revealed a critical role for miR-221 in neuronal differentiation. Neurochem. Int. 2012;60:743–750. doi: 10.1016/j.neuint.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Pandey A., Singh P., Jauhari A., Singh T., Khan F., Pant A.B., Parmar D., Yadav S. Critical role of the miR-200 family in regulating differentiation and proliferation of neurons. J. Neurochem. 2015;133:640–652. doi: 10.1111/jnc.13089. [DOI] [PubMed] [Google Scholar]
- 25.Terasawa K., Ichimura A., Sato F., Shimizu K., Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276:3269–3276. doi: 10.1111/j.1742-4658.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 26.Ma W., Li Y., Wang C., Xu F., Wang M., Liu Y. Serum miR-221 serves as a biomarker for Parkinson's disease. Cell Biochem. Funct. 2016;34:511–515. doi: 10.1002/cbf.3224. [DOI] [PubMed] [Google Scholar]
- 27.Ding H., Huang Z., Chen M., Wang C., Chen X., Chen J., Zhang J. Identification of a panel of five serum miRNAs as a biomarker for Parkinson's disease. Park. Relat. Disord. 2016;22:68–73. doi: 10.1016/j.parkreldis.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Asci R., Vallefuoco F., Andolfo I., Bruno M., De Falco L., Iolascon A. Transferrin receptor 2 gene regulation by microRNA 221 in SH-SY5Y cells treated with MPP+ as Parkinson's disease cellular model. Neurosci. Res. 2013;77:121–127. doi: 10.1016/j.neures.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Li L., Xu J., Wu M., Hu J.M. Protective role of microRNA-221 in Parkinson's disease. Bratisl. Lek. Listy. 2018;119:22–27. doi: 10.4149/BLL_2018_005. [DOI] [PubMed] [Google Scholar]
- 30.Tatura R., Kraus T., Giese A., Arzberger T., Buchholz M., Hoglinger G., Muller U. Parkinson's disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Park. Relat. Disord. 2016;33:115–121. doi: 10.1016/j.parkreldis.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Klein C., Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb. Perspect. Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy O.A., Malagelada C., Greene L.A. Cell death pathways in Parkinson's disease: proximal triggers, distal effectors, and final steps. Apoptosis: Int. J. Program. Cell Death. 2009;14:478–500. doi: 10.1007/s10495-008-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke R.E., O'Malley K. Axon degeneration in Parkinson's disease. Exp. Neurol. 2013;246:72–83. doi: 10.1016/j.expneurol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi-Niki K., Niki T., Taira T., Iguchi-Ariga S.M., Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson's disease patients. Biochem. Biophys. Res. Commun. 2004;320:389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- 35.Singer T.P., Ramsay R.R. Mechanism of the neurotoxicity of MPTP. An update. FEBS Lett. 1990;274:1–8. doi: 10.1016/0014-5793(90)81315-f. [DOI] [PubMed] [Google Scholar]
- 36.Junn E., Taniguchi H., Jeong B.S., Zhao X., Ichijo H., Mouradian M.M. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc. Natl. Acad. Sci. USA. 2005;102:9691–9696. doi: 10.1073/pnas.0409635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zongaro S., Hukema R., D'Antoni S., Davidovic L., Barbry P., Catania M.V., Willemsen R., Mari B., Bardoni B. The 3' UTR of FMR1 mRNA is a target of miR-101, miR-129-5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Hum. Mol. Genet. 2013;22:1971–1982. doi: 10.1093/hmg/ddt044. [DOI] [PubMed] [Google Scholar]
- 38.Im J.Y., Lee K.W., Junn E., Mouradian M.M. DJ-1 protects against oxidative damage by regulating the thioredoxin/ASK1 complex. Neurosci. Res. 2010;67:203–208. doi: 10.1016/j.neures.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo J.S., Kim M.Y., Ann E.J., Hong J.A., Park H.S. DJ-1 modulates UV-induced oxidative stress signaling through the suppression of MEKK1 and cell death. Cell Death Differ. 2008;15:1030–1041. doi: 10.1038/cdd.2008.26. [DOI] [PubMed] [Google Scholar]
- 40.Ren H., Fu K., Mu C., Zhen X., Wang G. L166P mutant DJ-1 promotes cell death by dissociating Bax from mitochondrial Bcl-XL. Mol. Neurodegener. 2012;7:40. doi: 10.1186/1750-1326-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell. 2014;53:1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou C.-H., Chang N.-W., Shrestha S., Hsu S.-D., Lin Y.-L., Lee W.-H., Yang C.-D., Hong H.-C., Wei T.-Y., Tu S.-J., Tsai T.-R., Ho S.-Y., Jian T.-Y., Wu H.-Y., Chen P.-R., Lin N.-C., Huang H.-T., Yang T.-L., Pai C.-Y., Tai C.-S., Chen W.-L., Huang C.-Y., Liu C.-C., Weng S.-L., Liao K.-W., Hsu W.-L., Huang H.-D. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puthalakath H., Strasser A. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 2002;9:505–512. doi: 10.1038/sj.cdd.4400998. [DOI] [PubMed] [Google Scholar]
- 45.Pineau P., Volinia S., McJunkin K., Marchio A., Battiston C., Terris B., Mazzaferro V., Lowe S.W., Croce C.M., Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Leva G., Gasparini P., Piovan C., Ngankeu A., Garofalo M., Taccioli C., Iorio M.V., Li M., Volinia S., Alder H., Nakamura T., Nuovo G., Liu Y., Nephew K.P., Croce C.M. MicroRNA cluster 221-222 and estrogen receptor α interactions in breast cancer. JNCI J. Natl. Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putcha G.V., Moulder K.L., Golden J.P., Bouillet P., Adams J.A., Strasser A., Johnson E.M. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron. 2001;29:615–628. doi: 10.1016/s0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 48.Perier C., Bove J., Wu D.C., Dehay B., Choi D.K., Jackson-Lewis V., Rathke-Hartlieb S., Bouillet P., Strasser A., Schulz J.B., Przedborski S., Vila M. Two molecular pathways initiate mitochondria-dependent dopaminergic neurodegeneration in experimental Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2007;104:8161–8166. doi: 10.1073/pnas.0609874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donato R., Miljan E.A., Hines S.J., Aouabdi S., Pollock K., Patel S., Edwards F.A., Sinden J.D. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007;8:36. doi: 10.1186/1471-2202-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nuydens R., Dispersyn G., Van Den Keiboom G., de Jong M., Connors R., Ramaekers F., Borgers M., Geerts H. Bcl-2 protects against apoptosis-related microtubule alterations in neuronal cells. Apoptosis. 2000;5:43–51. doi: 10.1023/a:1009685609275. [DOI] [PubMed] [Google Scholar]
- 51.Cheng H.-C., Ulane C.M., Burke R.E. Clinical progression in Parkinson's disease and the neurobiology of axons. Ann. Neurol. 2010;67:715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi-Niki K., Kato-Ose I., Murata H., Maita H., Iguchi-Ariga S.M., Ariga H. Epidermal growth factor-dependent activation of the extracellular signal-regulated kinase pathway by DJ-1 protein through Its direct binding to c-Raf protein. J. Biol. Chem. 2015;290:17838–17847. doi: 10.1074/jbc.M115.666271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu L., Cui T., Fan C., Zhao H., Zhao C., Lu L., Yang H. Involvement of ERK1/2 signaling pathway in DJ-1-induced neuroprotection against oxidative stress. Biochem. Biophys. Res. Commun. 2009;383:469–474. doi: 10.1016/j.bbrc.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 54.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G.K., Khatun J., Williams B.A., Zaleski C., Rozowsky J., Roder M., Kokocinski F., Abdelhamid R.F., Alioto T., Antoshechkin I., Baer M.T., Bar N.S., Batut P., Bell K., Bell I., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Falconnet E., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M.J., Gao H., Gonzalez D., Gordon A., Gunawardena H., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Luo O.J., Park E., Persaud K., Preall J.B., Ribeca P., Risk B., Robyr D., Sammeth M., Schaffer L., See L.H., Shahab A., Skancke J., Suzuki A.M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Ruan X., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T., Reymond A., Antonarakis S.E., Hannon G., Giddings M.C., Ruan Y., Wold B., Carninci P., Guigo R., Gingeras T.R. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qureshi I.A., Mehler M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Junn E., Mouradian M.M. MicroRNAs in neurodegenerative disorders. Cell Cycle. 2010;9:1717–1721. doi: 10.4161/cc.9.9.11296. [DOI] [PubMed] [Google Scholar]
- 57.Junn E., Mouradian M.M. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol. Ther. 2012;133:142–150. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Junn E., Lee K.W., Jeong B.S., Chan T.W., Im J.Y., Mouradian M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi D.C., Chae Y.J., Kabaria S., Chaudhuri A.D., Jain M.R., Li H., Mouradian M.M., Junn E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J. Neurosci.: Off. J. Soc. Neurosci. 2014;34:12725–12737. doi: 10.1523/JNEUROSCI.0985-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaudhuri A.D., Kabaria S., Choi D.C., Mouradian M.M., Junn E. MicroRNA-7 promotes glycolysis to protect against 1-methyl-4-phenylpyridinium-induced cell death. J. Biol. Chem. 2015;290:12425–12434. doi: 10.1074/jbc.M114.625962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho H.J., Liu G., Jin S.M., Parisiadou L., Xie C., Yu J., Sun L., Ma B., Ding J., Vancraenenbroeck R., Lobbestael E., Baekelandt V., Taymans J.M., He P., Troncoso J.C., Shen Y., Cai H. MicroRNA-205 regulates the expression of Parkinson's disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2013;22:608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Botta-Orfila T., Morato X., Compta Y., Lozano J.J., Falgas N., Valldeoriola F., Pont-Sunyer C., Vilas D., Mengual L., Fernandez M., Molinuevo J.L., Antonell A., Marti M.J., Fernandez-Santiago R., Ezquerra M. Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson's disease. J. Neurosci. Res. 2014;92:1071–1077. doi: 10.1002/jnr.23377. [DOI] [PubMed] [Google Scholar]
- 63.Dorval V., Mandemakers W., Jolivette F., Coudert L., Mazroui R., De Strooper B., Hebert S.S. Gene and MicroRNA transcriptome analysis of Parkinson's related LRRK2 mouse models. PLoS One. 2014;9:e85510. doi: 10.1371/journal.pone.0085510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gehrke S., Imai Y., Sokol N., Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olsen L., Klausen M., Helboe L., Nielsen F.C., Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS One. 2009;4:e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bak M., Silahtaroglu A., Moller M., Christensen M., Rath M.F., Skryabin B., Tommerup N., Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao J., Lou S., Ying M., Yang B. DJ-1 as a human oncogene and potential therapeutic target. Biochem. Pharmacol. 2015;93:241–250. doi: 10.1016/j.bcp.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Dias V., Junn E., Mouradian M.M. The role of oxidative stress in Parkinson's disease. J. Parkinson's Dis. 2013;3:461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheng C., Heng X., Zhang G., Xiong R., Li H., Zhang S., Chen S. DJ-1 deficiency perturbs microtubule dynamics and impairs striatal neurite outgrowth. Neurobiol. Aging. 2013;34:489–498. doi: 10.1016/j.neurobiolaging.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 70.Gubern C., Camos S., Ballesteros I., Rodriguez R., Romera V.G., Canadas R., Lizasoain I., Moro M.A., Serena J., Mallolas J., Castellanos M. miRNA expression is modulated over time after focal ischaemia: up-regulation of miR-347 promotes neuronal apoptosis. FEBS J. 2013;280:6233–6246. doi: 10.1111/febs.12546. [DOI] [PubMed] [Google Scholar]
- 71.Shi R.Y., Zhu S.H., Li V., Gibson S.B., Xu X.S., Kong J.M. BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci. Ther. 2014;20:1045–1055. doi: 10.1111/cns.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfeiffer S., Anilkumar U., Chen G., Ramirez-Peinado S., Galindo-Moreno J., Munoz-Pinedo C., Prehn J.H. Analysis of BH3-only proteins upregulated in response to oxygen/glucose deprivation in cortical neurons identifies Bmf but not Noxa as potential mediator of neuronal injury. Cell Death Dis. 2014;5:e1456. doi: 10.1038/cddis.2014.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H., Ye Y., Zhu Z., Mo L., Lin C., Wang Q., Wang H., Gong X., He X., Lu G., Lu F., Zhang S. MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson's disease by targeting to bim. Brain Pathol. 2016;26:167–176. doi: 10.1111/bpa.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biswas S.C., Shi Y., Vonsattel J.P., Leung C.L., Troy C.M., Greene L.A. Bim is elevated in Alzheimer's disease neurons and is required for beta-amyloid-induced neuronal apoptosis. J. Neurosci.: Off. J. Soc. Neurosci. 2007;27:893–900. doi: 10.1523/JNEUROSCI.3524-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leon R., Bhagavatula N., Ulukpo O., McCollum M., Wei J. BimEL as a possible molecular link between proteasome dysfunction and cell death induced by mutant huntingtin. Eur. J. Neurosci. 2010;31:1915–1925. doi: 10.1111/j.1460-9568.2010.07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sionov R.V., Vlahopoulos S.A., Granot Z. Regulation of bim in health and disease. Oncotarget. 2015;6:23058–23134. doi: 10.18632/oncotarget.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hornsveld M., Tenhagen M., van de Ven R.A., Smits A.M., van Triest M.H., van Amersfoort M., Kloet D.E., Dansen T.B., Burgering B.M., Derksen P.W. Restraining FOXO3-dependent transcriptional BMF activation underpins tumour growth and metastasis of E-cadherin-negative breast cancer. Cell Death Differ. 2016;23:1483–1492. doi: 10.1038/cdd.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Q., Chen X., Han C., Wang Y., Huang T., Du Y., Dong Z. FGF-2 transcriptionally down-regulates the expression of BNIP3L via PI3K/Akt/FoxO3a signaling and inhibits necrosis and mitochondrial dysfunction induced by high concentrations of hydrogen peroxide in H9c2 cells. Cell. Physiol. Biochem.: Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016;40:1678–1691. doi: 10.1159/000453217. [DOI] [PubMed] [Google Scholar]
- 79.Sanphui P., Biswas S.C. FoxO3a is activated and executes neuron death via Bim in response to beta-amyloid. Cell Death Dis. 2013;4:e625. doi: 10.1038/cddis.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tagliaferro P., Burke R.E. Retrograde axonal degeneration in Parkinson disease. J. Parkinson’s Dis. 2016;6:1–15. doi: 10.3233/JPD-150769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sulzer D. Multiple hit hypotheses for dopamine neuron loss in Parkinson's disease. Trends Neurosci. 2007;30:244–250. doi: 10.1016/j.tins.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 82.Matsuda W., Furuta T., Nakamura K.C., Hioki H., Fujiyama F., Arai R., Kaneko T. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J. Neurosci. 2009;29:444–453. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braak H., Del Tredici K. Poor and protracted myelination as a contributory factor to neurodegenerative disorders. Neurobiol. Aging. 2004;25:19–23. doi: 10.1016/j.neurobiolaging.2003.04.001. [DOI] [PubMed] [Google Scholar]
- 84.Acunzo M., Visone R., Romano G., Veronese A., Lovat F., Palmieri D., Bottoni A., Garofalo M., Gasparini P., Condorelli G., Chiariello M., Croce C.M. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim D., Song J., Jin E.J. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J. Biol. Chem. 2010;285:26900–26907. doi: 10.1074/jbc.M110.115105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Oh S.E., Mouradian M.M. Cytoprotective mechanisms of DJ-1 against oxidative stress through modulating ERK1/2 and ASK1 signal transduction. Redox Biol. 2018;14:211–217. doi: 10.1016/j.redox.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tobiume K., Matsuzawa A., Takahashi T., Nishitoh H., Morita K., Takeda K., Minowa O., Miyazono K., Noda T., Ichijo H. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2:222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meijering E., Jacob M., Sarria J.C., Steiner P., Hirling H., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. Part A: J. Int. Soc. Anal. Cytol. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]