Abstract
We challenge the view that our species, Homo sapiens, evolved within a single population and/or region of Africa. The chronology and physical diversity of Pleistocene human fossils suggest that morphologically varied populations pertaining to the H. sapiens clade lived throughout Africa. Similarly, the African archaeological record demonstrates the polycentric origin and persistence of regionally distinct Pleistocene material culture in a variety of paleoecological settings. Genetic studies also indicate that present-day population structure within Africa extends to deep times, paralleling a paleoenvironmental record of shifting and fractured habitable zones. We argue that these fields support an emerging view of a highly structured African prehistory that should be considered in human evolutionary inferences, prompting new interpretations, questions, and interdisciplinary research directions.
Keywords: human evolution, evolutionary genetics, paleoanthropology, paleoecology, Middle Stone Age, African origins
Highlights
The view that Homo sapiens evolved from a single region/population within Africa has been given primacy in studies of human evolution.
However, developments across multiple fields show that relevant data are no longer consistent with this view.
We argue instead that Homo sapiens evolved within a set of interlinked groups living across Africa, whose connectivity changed through time.
Genetic models therefore need to incorporate a more complex view of ancient migration and divergence in Africa.
We summarize this new framework emphasizing population structure, outline how this changes our understanding of human evolution, and identify new research directions.
A Different View of African Origins
The lineage of Homo sapiens probably originated in Africa at least ∼500 thousand years ago (ka) [1], and the earliest observed morphological manifestations of this clade appeared by ∼300 ka [2]. Early H. sapiens fossils do not demonstrate a simple linear progression towards contemporary human morphology. Instead, putative early H. sapiens remains exhibit remarkable morphological diversity and geographical spread. Together with recent archaeological and genetic lines of evidence, these data are consistent with the view that our species originated and diversified within strongly subdivided (i.e., structured) populations, probably living across Africa, that were connected by sporadic gene flow 1, 3, 4, 5, 6, 7, 8. This concept of ‘African multiregionalism’ [1] may also include hybridization between H. sapiens and more divergent hominins (see Glossary) living in different regions 1, 9, 10, 11, 12. Crucially, such population subdivisions may have been shaped and sustained by shifts in ecological boundaries 7, 13, 14, challenging the view that our species was endemic to a single region or habitat, and implying an often underacknowledged complexity to our African origins.
In this paper we examine and synthesize fossil, archaeological, genetic, and paleoenvironmental data to refine our understanding of the time-depth, character, and maintenance of Pleistocene population structure. In doing so, we attempt to separate data from inference to stress that using models of population structure fundamentally changes interpretations of recent human evolution.
The Morphological Diversity and Spread of the Homo sapiens Clade
The constellation of morphological features characterizing H. sapiens is debated. This has strongly impacted on interpretations of recent human origins by variably including or excluding different fossils from interpretative analyses. For example, different morphological criteria and analytical methods have been used to support both a gradual, mosaic-like process of modernization of our species or, conversely, a punctuated speciation (e.g., [1]).
Extant human crania are characterized by a combination of features that distinguish us from our fossil relatives and ancestors, such as a small and gracile face, a chin, and a globular braincase. However, these typical modern human features emerge in a mosaic-like fashion within the H. sapiens clade. The oldest currently recognized members of the H. sapiens clade, from Jebel Irhoud in North Africa, have a facial morphology very similar to extant H. sapiens, as well as endocranial volumes that fall within the contemporary range of variation [2]. However, their braincase shapes are elongated rather than globular, suggesting that distinctive features of brain shape, and possibly brain function, evolved within H. sapiens 2, 5 (Figure 1). Other early H. sapiens fossils from Florisbad in South Africa (∼260 ka), Omo Kibish (∼195 ka) and Herto (∼160 ka), both in Ethiopia, are morphologically diverse 1, 16. This diversity has led some researchers to propose that fossils such as Jebel Irhoud and Florisbad actually represent a more primitive species called ‘H. helmei’, using the binomen given to the Florisbad partial cranium in 1935 17, 18. In a similar vein, the fossil crania from Herto [19], which combine a relatively globular braincase with a robust occipital and large face, were described as the subspecies H. sapiens idaltu because they fall outside the variation of recent humans.
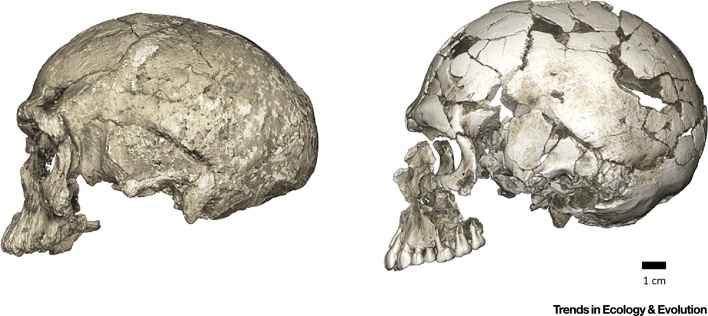
Figure 1.
Evolutionary Changes of Braincase Shape from an Elongated to a Globular Shape. The latter evolves within the H. sapiens lineage via an expansion of the cerebellum and bulging of the parietal. (Left) Micro-computerized tomography scan of Jebel Irhoud 1 (∼300 ka, North Africa). (Right) Qafzeh 9 (∼95 ka, the Levant).
However, we view H. sapiens as an evolving lineage with deep African roots, and therefore prefer to recognize such fossils as part of the diversity shown by early members of the H. sapiens clade. The full suite of cranial features characterizing contemporary humans does not appear until fairly recently, between about ∼100–40 ka [20]. The character and chronology of early H. sapiens fossils, together with their geographic distribution across Africa, suggests that evolution may at times have progressed independently in different regions, in populations that were often semi-isolated for millennia by distance and/or ecological barriers, such as hyperarid regions or tropical forests.
Further insights into the geographic extent and potential habitat diversity of early H. sapiens populations can be gained from more recent forager populations in Africa, which were also strongly structured. For example, Later Stone Age (LSA) human remains highlight both the retention of ‘archaic’ traits and the maintenance of considerable morphological diversity into the terminal Pleistocene 11, 21. In the Holocene, the skeletal record becomes much richer, but there remains considerable spatial variation in morphology. Variation between populations in different regions and environments of Africa may have been shaped by isolation-by-distance and local environmental adaptations 22, 23, 24, 25, 26. For example, challenging environments (e.g., deserts, rainforest) and isolation have likely played a significant role in shaping the population structure of Holocene African foragers and isolated hunter-gatherers across the tropics 25, 27.
Ultimately, the processes underlying the emergence of any ‘package’ of derived features diagnostic of early H. sapiens anatomy remain incompletely understood. However, the data do not seem to be consistent with the long-held view that human ancestry is derived predominantly from a single African region hosting a panmictic population. Instead, H. sapiens likely descended from a shifting structured population (i.e., a set of interlinked groups whose connectivity changed through time), each exhibiting different characteristics of anatomical ‘modernity’. The discovery that the primitive-looking H. naledi dates to between ∼335 ka and 236 ka [28], and that the Broken Hill 1 Homo heidelbergensis skull may date to ∼300–125 ka [29], also shows that other hominin species in Africa coexisted with H. sapiens, raising the possibility of African archaic interbreeding. Future research should attempt to determine which features evolved before the appearance of our species and which primarily developed within the evolutionary history of our species. Another key area concerns understanding the extent to which different processes shaped observed changes. For example, the narrowing of the pelvis may reflect different processes including neutral genetic drift, adaptation to ecological variation, and life-history variation.
A Pan-African Cultural Patchwork
Across Africa, the virtual abandonment of handheld large cutting tools such as handaxes, and an increased emphasis on prepared core technologies and hafting, marked a profound technological reconfiguration of hominin material culture. These technological changes, which define the transition to the Middle Stone Age (MSA), seem to have occurred across Africa at a broadly similar time; for example at ∼300 ka both at Jebel Irhoud, where they are found with early H. sapiens fossils [16], and at Olorgesailie in East Africa [30], and at ∼280 ka in southern Africa at Florisbad [31]. Currently, the earliest dates in West Africa are younger, at ∼180 ka, but the region remains very poorly characterized [32]. The MSA is associated with H. sapiens fossils, but both H. naledi and H. heidelbergensis probably persisted into the late Middle Pleistocene.
Clear regionally distinctive material culture styles, typically involving complex stone tools, first emerged within the MSA. For example, the Central African MSA includes heavy-duty axes, bifacial lanceolates, backed flakes and blades, picks and segments, probably from at least the late Middle Pleistocene [33]. In the Late Pleistocene, grassland and savannah expansion in North Africa led to dense human occupation associated with specific regional technological features such as tanged implements (Figure 2) [34]. At approximately the same time there was an emergence of comparably distinctive industries in parts of southern Africa. As in North Africa, some of these industries are also associated with other aspects of complex material culture such as ochre, bone tools, shell beads, and abstract engravings (Figure 2) [35].
Figure 2.
Middle Stone Age Cultural Artefacts. (A–D) Bifacial foliates from northern Africa (A, Mugharet el Aliya; B–D, Adrar Bous). (E–G) Bifacial foliates from southern Africa (Blombos Cave). (H,I) Tanged tools from northern Africa. (J) Segmented piece bearing mastic residue from southern Africa (Sibudu). (K) Engraved ochre fragment (Blombos Cave). (L–N) Engraved ostrich eggshell fragments from southern Africa (Diepkloof). (O,P) Bone points from southern Africa (Sibudu and Blombos Cave, respectively). (Q) Bone point from northern Africa (El Mnasra). (R–V) Perforated Trivia gibbosula shells from northern Africa (R,S, Grotte de Pigeons; T–V, Rhafas, Ifri n’Ammar, and Oued Djebbana, respectively). (W–Aa) Perforated Nassarius kraussianus shells from Blombos Cave. (Ab) Conus ebraeus shell bead (Conus 2) from southern Africa (Border Cave). (Ac) Ochre fragment shaped by grinding from southern Africa (Blombos Cave). All scales are 1 cm. Boxed items indicate rescaled artefacts. Images reproduced, with permission, from (A–D, H, I) The Stone Age Institute; (E–G, J–P, Ac) from [35]; (Q) from [47]; and (R–Ab) from 35, 47, 48.
Such regionalization is typically linked with the emergence of ‘modern’ cognition. However, it arguably also reflects the interaction between demographic variables (e.g., increased population density) 36, 37, 38 and the learned traditions of long-lived regional subpopulations or demes (Figure 2). For example, northern and southern Africa, apart from being geographically distant, were also separated by environmental factors as a consequence of the expansion and contraction of forests in equatorial Africa, synchronous with amelioration in northern Africa. Other factors, such as habitat variability and adaptation to local environmental conditions, are also likely to play some role in material culture diversification.
Although geographical differences are clear at the continental scale, localized spatial patterning is harder to discern. Similarities between regions may have been produced by occasional contact or by convergent adaptation to common environmental conditions. In East Africa, for example, although there is certainly some variation, there appears to be underlying continuity in material culture throughout much of the MSA (e.g., [39]). In many regions, ‘generic’ MSA assemblages that do not carry an obvious signal of regionalization are common [40]. In a cognitive model, these differences suggest that not all these early populations manifested a ‘modern mind’. However, such assemblages are augmented by shifting frequencies of tool types that appear to be spatially or temporally indicative, and likely reflect demographic factors. In some parts of Africa, the full suite of generalized MSA characteristics continues largely unchanged until the Pleistocene/Holocene boundary [41], matching the morphological patterns, and suggesting that the end of the MSA may have been as structured and mosaic-like as its beginnings. This view has support from LSA material culture. Despite superficial similarities in LSA lithic miniaturization, the cultural record shows continued differentiation and derivation into the Holocene, supporting the biological evidence for variable population dynamics that did not result in wide-scale homogenization [42].
The reasons for, and therefore implications of, the geographic and temporal structuring of MSA cultural diversity are still poorly characterized and likely reflect several processes. These include adaptations to different environments [43]. Long-term, large-scale population separation may also have been the norm for much of Pleistocene Africa (Box 1; i.e., isolation by distance and isolation by habitat, representing null models to be rejected). Rare and spatially explicit models exploring Pleistocene technological innovations have also linked cultural complexity with variation in regional patterns of population growth, mobility, and connectedness (e.g., 36, 44, 45), supported by evidence of long-distance transfer of stone raw material (e.g., [46]).
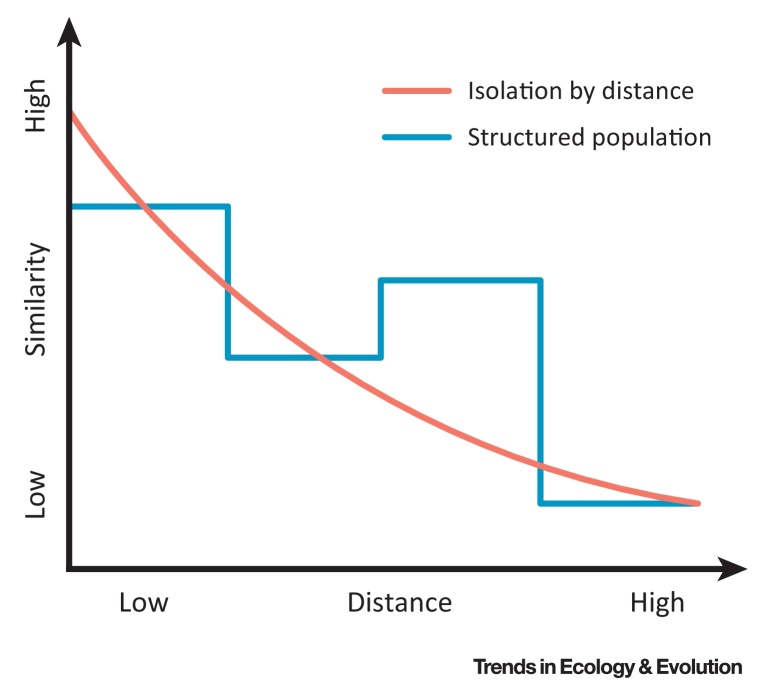
Box 1. Isolation by Distance (IBD).
IBD is the expectation that genetic differences correlate positively geographic distances as a consequence of the fact that mating is more likely to occur at shorter than longer distances. This concept, although well established in genetics, is rarely applied to Pleistocene archaeological and human fossil material, despite its potential value as a null model for observed cultural or morphological differences between materials from different sites [49].
Because archaeological problems are concerned with identifying the processes that generate observed patterns of cultural variation over time and space, the lack of equivalent null models is particularly problematic. Processes generating cultural variation constitute a complex balance between patterns of inherited knowledge, local innovation, modes of cultural transmission, local adaptation, and shifting population dynamics (e.g., population size, density, or mobility [36]). Without null models of ‘cultural similarity’ as a baseline, it is difficult to escape simplistic, narrative inferences about the past. Similarly, human fossil data can be interpreted in several ways depending on the taxonomy employed, but spatial variation is likely to relate to the same factors that influence genetic similarities between regional populations [50].
In an archeological context, the expectation of an IBD model is that cultural similarity will decrease with distance, with a degree of spatial autocorrelation (e.g., [51]). This expectation represents the simplest explanation of the observed variation. If this null model does not provide an adequate explanation of the data, more complex models can be invoked to explain patterns observed either in the residuals from the null model or in the raw data as a whole. For example, a more complex population structure can be theoretically differentiated from an IBD model if patterns of spatial autocorrelation are discrete rather than continuous (Figure I), pointing towards the formation of distinct biological or cultural clusters which may correlate with other features (e.g., genetic, morphological, or environmental). Many factors could promote the formation of such clusters, including assortativity by cultural similarity, or conformity to local norms of, for example, tool production.
Alt-text: Box 1
Major new archaeological research directions should include: (i) unraveling the relative contributions of different African regions/habitats to recent human evolution; (ii) understanding shifting patterns of population structure through the differential appearance, expansion, contraction, and disappearance of regionally distinct artefact forms (Box 1); and (iii) exploiting the growing interface between archaeology, ecology, morphology, and genetics to explore the extent to which material culture patterning is coupled or decoupled from these associated (but potentially independent) axes.
Why Genetic Models Must Incorporate a More Complex View of Ancient Migration and Divergence in Africa
The starting point for most genetic studies of human origins has been to investigate the depth of present-day diversity between and within African populations. Most studies have used simple ‘tree-like’ demographic models to infer population split times, neglecting or simplifying population structure, even if sometimes considering a degree of gene flow between branches (Box 2). Such studies have produced a variety of split-time estimates, with the KhoeSan populations of southern Africa, who retain the greatest levels of genetic diversity among human populations today, comprising one branch of the deepest divergence inferred, at 150–300 ka 52, 53, 54, 55, 56, 57, 58. Some authors have interpreted this, in conjunction with a gradient of south to north decreasing genetic diversity within Africa, as favoring a single-origin model for modern humans with a locus in southern Africa rather than in eastern Africa 59, 60. Variation in inferred split times reflects a variety of different methodologies, model assumptions, and data sources, with a general trend for more recent analyses to infer older dates. In addition to ancient gene flow and structure, more recent population movements within Africa, such as the expansion of Bantu-speaking peoples from West Africa at 2–1.5 ka 52, 61, will have obscured signatures of older demographic processes, as will have episodes of ‘back-to-Africa’ migration from Europe and southwest Asia into several regions of the continent 60, 62, 63, 64, 65.
Box 2. Modeling Population Structure.
Population genetic modeling for demographic inference often assumes panmixia, wherein all individuals in a population have an equal chance of mating with one another. In such cases there is often an implicit assumption that panmixia occurs at the whole-species level. However, real populations rarely meet this condition, owing to the existence of spatial structure at the species level or other stratification of individuals within the population. Population structure and its consequences for genetic data can be modeled in various ways. For example, Wright’s n-island model assumes that populations are subdivided into n different islands/demes that are connected through gene flow. This simple model assumes that all demes are panmictic, have the same size, and exchange genes with all other islands at the same rate. Other models include tree models in which an ancestral population splits into two or more populations that may themselves later split. The n-island model ignores space (i.e., differing levels of connectivity between populations), whereas in tree models geographic location may be implicit but is not explicitly modeled. For example, two populations that are geographically close may be assumed to share a more recent splitting event than populations that are more distant. Other models (e.g., stepping stone) may incorporate geographic space explicitly by connecting demes via gene flow only with their spatial neighbors.
Another distinction among model types is whether they incorporate change over time. The stepping-stone and n-island models have no temporal aspect. Conversely, tree models may generate very different results depending on the timing of sampling in relation to the splitting events and gene flow between populations. More complex models allow demographic expansions or contractions in space, often based on simulations 71, 72. Finally, the class of ‘meta-population models’ include demes of variable sizes that can be connected by gene flow and colonization events. The population in each deme can also become extinct and be recolonized by individuals coming from one or several other populations.
This diversity in range of structured population demographic models, with varying levels of complexity, leads to some arbitrariness in which models are chosen. Because we know that the human past was complex, it is often assumed that more complex models of that past are more realistic. However, more complexity means more parameters and more ways for a model to differ from reality. This means that unless informed a priori by secure information, or fitted to substantial quantities of conditioning data, more complex models can be more wrong, not more realistic. The past two decades have also seen the development of many different inference methodologies, the results of which are often difficult to compare. This is because they often make different assumptions (tree splitting versus spatial distribution), explain different aspects of the data (allele-frequency spectrum, AFS; versus IICR), and can be computationally demanding – challenging interpretation, explanatory power, and validation, respectively. Models are valuable tools for understanding and interpreting data, but we should not be surprised if a single family of models is unable to explain all patterns of human genetic diversity.
Alt-text: Box 2
Models incorporating more complex population structure can be considerably more parameter-rich and therefore more difficult to test computationally, particularly with limited data. However, they do offer a more generalized and flexible view of past demography – one that can accommodate, but not be limited to, more traditional population tree models. Furthermore, the increased availability of genomic data and developments in analytical methodologies now permit inference under more complex and realistic models. These developments have shown that structure cannot be neglected, and can cause patterns in genetic data that are similar to those generated by other forms of demographic change (e.g., 66, 67). For example, inferred changes in effective population size (Ne) may result from changes in connectivity between ancient populations rather than from, or in addition to, changes in census population size 7, 68. Indeed, the relationship between inferred Ne and census population size is not straightforward, and may even be counterintuitive when structure exists 7, 66, 68. The geographical scale at which population genetic structure may have existed is also difficult to infer. For example, one recent genomic study showed substantial structure between pre-agricultural human populations separated by only tens or hundreds of kilometers [69]. These insights challenge the view that the early prehistory of our species can be well approximated by population growth within a single lineage [70].
Although modern genomic data have been shaped by, and thus contain large amounts of information on, past demography, these data can be explained by many different models of population history (equifinality). Moreover, all such models are necessarily abstractions and simplifications of the true population histories, and the discrepancies involved may be particularly problematic for certain questions about the past (mis-specification). This means that structure can be difficult to unambiguously detect, and even harder to reconstruct. For example, several studies on African populations have identified genes with coalescence times on the order of 1 million years, which could be interpreted as indicating admixture with archaic hominins 9, 10, 73. However, even in a single population some very old coalescence times (>1 My) are expected for humans (Figure 3), and therefore inferences based on the tail of the distribution of coalescence times, which are particularly sensitive to model mis-specification, need to be interpreted with caution. Indeed, several authors have argued that deep coalescence times are compatible with a single human lineage in Africa with deep population structure 7, 68, 74.
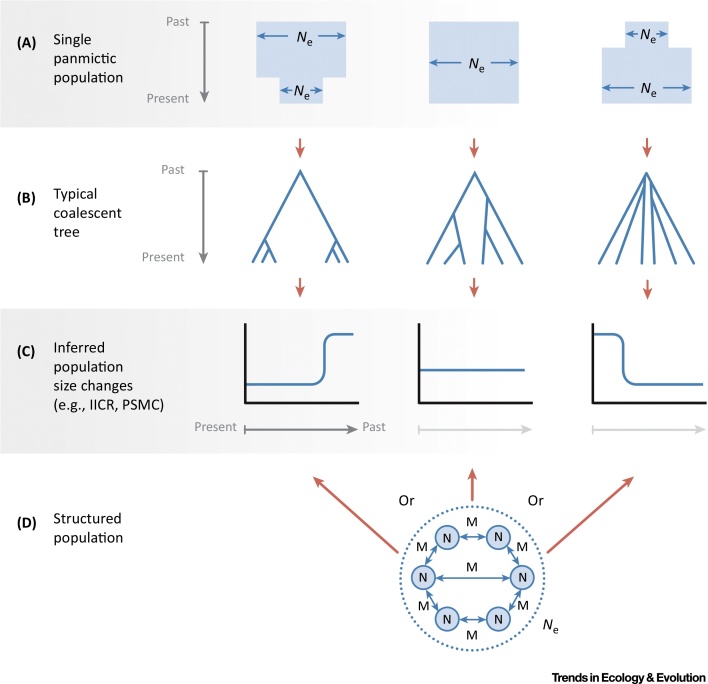
Figure 3.
Inferring Population Size Change in Unstructured and Structured Populations
For a Figure360 author presentation of Figure 3, see the figure legend at https://doi.org/10.1016/j.tree.2018.05.005
The probability, or expected rate, of coalescence of lineages in a single panmictic population is inversely proportional to the population size at the time. Different panmictic population size histories (A) therefore shape the temporal distribution of coalescent events (B). When estimated for many regions of the genome, the temporal distribution of these nodes can be used to estimate the instantaneous inverse coalescence rate (IICR), which in a single panmictic population is a direct proxy for the population size (C). Software such as the pairwise sequentially Markovian coalescent (PSMC) or the multiple sequentially Markovian coalescent (MSMC) can be used to estimate the IICR/population size change in the past. However, when data are sampled from a structured meta-population consisting of subpopulations connected by migration (D), changes in migration through time, and/or in sampling, can generate any IICR-inferred population size history without any actual change in the meta-population size (Ne).
Ancient DNA (aDNA) data can provide additional resolution, and studies on Holocene individuals recently revealed extensive structure and migratory activity during that period 8, 75. Ancient Pleistocene-aged aDNA would be more informative, but is difficult to obtain because tropical environments are mostly unfavorable for DNA preservation. However, a recent study showed that Late Pleistocene aDNA can be retrieved in some African regions [65]. These studies demonstrate that inferences from patterns of human genomic diversity need to consider fluctuating population structure over long periods, in addition to the range of panmictic African population origin models.
Environmental and Ecological Drivers of Population Structure
The genetic, fossil, and archaeological data discussed above indicate that H. sapiens evolved in highly structured populations, probably across many regions of Africa. Elucidating the degree of and mechanisms underlying population structure will require consideration of Middle and Late Pleistocene environmental variability in both space and time (e.g., [13]) (Figure 4). Refugia have been highlighted as key catalysts of evolutionary change [76], and certainly would have generated population structure. Nevertheless, some regions acting as ‘backwaters’ and isolated habitat islands may also have been central in the persistence of relict populations. Research has emphasized broad asynchronous environmental changes in different African regions (e.g., 13, 77). The northern and southern tips of the continent are most strongly affected by winter westerly precipitation, variation in which is largely driven by changes in Atlantic Ocean circulation. However, most of Africa experiences monsoonal rainfall associated with the intertropical convergence zone (ITCZ), the strength and location of which varies according to changes in insolation that are driven primarily by precessional aspects of Milankovitch forcing. Consequently, parts of tropical Africa that are currently humid likely experienced numerous episodes of extreme aridity in the past 78, 79. At the same time that the monsoon migrated northwards, the Sahara contracted, and networks of lakes and rivers expanded across much of north Africa 80, 81, 82, with matching conditions in parts of southwest Asia. Finer-scale shifts in the monsoon are also evident. For example, in West Africa the extent of savannah and forested areas is strongly affected by small changes in patterns of rainfall 83, 84.
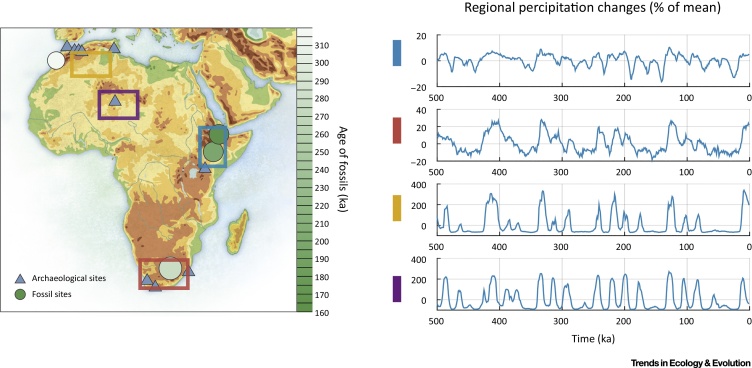
Figure 4.
Middle and Late Pleistocene African Environmental Variability in Space and Time. (Left) Map of Africa with key archaeological and fossil sites discussed in the text. Colored boxes indicate averaged regions for simulated precipitation changes from the transient glacial/interglacial LOVECLIM climate model experiment [81]. (Right) Precipitation changes (%) relative to the long-term 784 thousand year mean in the key regions highlighted in left panel, as simulated by transient 784 thousand year-long LOVECLIM climate model simulation [81]. From top to bottom the regions are eastern equatorial Africa, southern Africa, northwestern Africa, and the central Sahara region.
Climate therefore varied greatly, and periods of relatively increased aridity or humidity were asynchronous across Africa. Crucially, these factors are major drivers of faunal population structure and speciation 85, 86, illustrated by numerous sub-Saharan animals exhibiting similar phylogenetic patterns in their distribution. For example, Bertola and colleagues [87] show that dozens of species exhibit distinct populations in the two major evolutionary realms of west/central Africa and east/southern Africa. Many also show a further subdivision between east Africa and southern Africa, signifying important refuges in these three regions. These species occupy a range of trophic levels, suggesting that climate affected whole ecological communities.
Therefore, faunal speciation largely appears to have been catalyzed by climate-driven habitat fragmentation and interaction between different biomes over time. This provides insights into how human population structure could have been maintained over significant timescales and geographic areas of Africa. In Africa, the concept of ‘refugium networks’ has been specifically implicated in Pleistocene human population subdivisions and expansions [18], and as a result such regions are of major evolutionary interest. Although fragmentation of suitable habitat has been highlighted as a major driver of population structure over and above isolation-by-distance (Box 1), isolation-by-habitat can also play an important role in animal [84] and probably also human population structure 14, 88.
Major challenges remain in integrating fossil, archaeological, and genetic lines of evidence into paleoenvironmental and paleoecological contexts (but see [3] and [72] for diverse attempts to do so). Currently, the chronology of much of the paleoanthropological record remains too coarse to allow any firm conclusions to be drawn about the role of environmental changes. Promising new avenues of research include genomic analyses of fauna, including the identification of commensal species and reconstructions of human habitats through stable-isotope analyses.
Concluding Remarks: Moving Forwards
Available morphological, archaeological, genetic, and paleoenvironmental data indicate that the subdivision of Middle and Late Pleistocene African human populations drove the mosaic-like emergence and evolution of derived H. sapiens morphology. Reproductively semi-isolated populations adapted to local ecologies alongside drift. Such population isolation was likely facilitated by small population sizes. Thus, as with other fauna, gene flow should not be assumed to have been constant through time, or to have occurred at the same rate within and between different regions. Across the large timescales of the Middle and Late Pleistocene, with their strong climatic fluctuations, the number of intermediate populations connecting different regions is also likely to have fluctuated considerably.
Several major unanswered questions flow from this reorientation of recent human origins (see Outstanding Questions). Did key diagnostic morphological characteristics emerge in one region and become elaborated with subsequent dispersals? Or did the transition from ‘archaic’ to ‘modern’ – whether indicated by morphology or material culture – occur gradually, and in a mosaic-like fashion across the continent? If this was the case, did African archaic hybridization also play a role? How does the existing evidence for structure affect our understanding of the history of population size changes and dispersals? Similarly, we have no firm grasp on the concordance that might exist between morphological and cultural structuring. Regional cultural signatures are apparent, raising the possibility that spatially distinct forms of material culture reflect similar patterns of population isolation and aggregation. Filling in these knowledge gaps requires us to reconsider paleoanthropological species concepts which are challenged by the view of deep population structure with sporadic gene flow/admixture.
Ultimately, reconstructing the demographic history of human populations in its full complexity is beyond the power of population genomics alone, necessitating an interdisciplinary approach. In the past this has been achieved by geneticists working with archaeologists and paleoanthropologists to define a narrow set of simplified hypotheses whose genetic outcomes can be compared to identify the models that best explain the data. While such an approach has met with considerable success, a more complete picture will require integrating different data types (genetic, fossil, material culture, paleoclimate and paleoecological data) using the same or analogous models of population structure, size change, and dispersal. This represents a major challenge for ancestral demographic inference over the coming years.
Fully characterizing the nature of this apparent ‘African multiregionalism’ also requires rejecting numerous longstanding, if implicit, assumptions, and formulating new questions. For example, the chronological lag between genetic estimates of population divergence times and morphological changes in the fossil record is not well understood, and should not be assumed to be short – particularly because inferences from genetic data are profoundly influenced by the models or families of models used. For instance, the estimates of population split times that are sometimes published may become less appropriate or relevant in our understanding of human evolution if models of spatial structure are to be used.
Similarly, while a globular braincase does seem to represent a synapomorphy of extant H. sapiens, can it be effectively characterized for application to the fossil record? We emphasize that H. sapiens is a lineage with deep and likely diverse African roots that challenge our use of terms such as ‘archaic H. sapiens’ and ‘anatomically modern humans’. Unless they can be operationalized with more clearly defined traits, such categories will have declining value. Diagnostics of H. sapiens must reflect trajectories of evolution rather than static views of our species – which has changed, and continues to change, at various scales.
The next decade of research will be crucial to resolving these emerging research themes. Contemporary human genomes are now available from across the African continent, together with an increasing number of ancient genomes. Our understanding of paleoecology is also improving thanks to biogeographic reconstructions premised on the genomes of African fauna. Paleoclimate reconstructions are increasingly precise, with rapidly growing proxy data and better models covering key periods. Finally, the expansion of paleoanthropological investigations into neglected areas of Africa will undoubtedly reveal new data that will significantly refine the parameters of recent human evolution.
Outstanding Questions.
In the conventional view, H. sapiens emerged in one region and/or population of Africa. Instead, new data suggest that a variety of transitional human groups, with a mosaic of primitive and derived features, may have lived over an extensive area from Morocco to South Africa between >300 ka and 12 ka.
Three outstanding questions emerge from this view. First, within the African ‘multiregional’ paradigm, which species best fits as the ancestor(s) of H. sapiens? Many aspects of the delicate H. sapiens facial shape may not be derived but instead be primitive retentions from an ancestor with a generalized facial shape. It therefore seems possible that H. sapiens did not evolve from the African forms of H. heidelbergensis (as represented, e.g., by the Bodo skull from Ethiopia, and Broken Hill from Zambia), but from a more primitive H. antecessor or H. erectus-like ancestor, beginning at ∼0.5 Ma 1, 2. However, hybridization during the inception of this process is also a possibility. Resolving the speciation of H. sapiens and the character of ancestral populations represents a crucial first step in understanding the emergence of the morphological features that diagnose our species during the later Middle Pleistocene.
Second, how many populations, environments, and geographical areas of Africa played a role in the origins of H. sapiens? Did adjoining areas of western Asia also play a part? It seems possible that early humans followed the same ecological partitioning and subspeciation patterns that are seen among continentally distributed African mammals, many of which emerged at the same time as the genus Homo. The Sahara may have played a particularly important role in this respect. Other areas, such as regions of forest, may also have supported populations who remained semi-isolated from those in grasslands and savannahs. Addressing the challenges of research in deserts and rainforests will be difficult, but is likely to be rewarding.
Finally, were some of our anatomical traits inherited from transitional African forms before they became extinct? The range of dates for H. naledi and H. heidelbergensis confirms the late survival of at least two archaic species in Africa. The size and environmental diversity of Africa, particularly the poorly investigated forested regions, may have permitted the late survival of more archaic species as well as of early forms of H. sapiens. These discoveries have fuelled speculations that H. sapiens may have interbred with archaic species in Africa itself. Distinguishing admixture between species from the reintegration of diverse H. sapiens lineages represents a major challenge, with significant taxonomic implications.
Acknowledgments
E.M.L.S. and H.S.G. wish to thank the British Academy of Humanities and Social Sciences for funding this research. E.M.L.S. thanks the Wellcome Trust, the Galton Institute and Jesus College Oxford for funding the workshop ‘Human Evolution in Structured Populations’ at the University of Oxford that provided the platform for this Opinion piece. M.G.T. was supported by Wellcome Trust Senior Investigator Award Grant 100719/Z/12/Z. A.M. was supported by the European Research Council Consolidator grant 647787 – LocalAdaptation. C.S. thanks the Calleva Foundation and the Human Origins Research Fund. G.P.R. and C.A.T. thank the American School of Prehistoric Research (Harvard University). J.S.T. was supported by the European Research Council Consolidator grant no. 617627. F.D. thanks the Research Council of Norway and its Centres of Excellence funding scheme, the SFF Centre for Early Sapiens Behaviour (project number 262618), and the LaScArBx research programme (ANR-10-LABX-52). R.W.D. is grateful to the Leverhulme Trust for grant EM-2016-050. R.D. thanks the Wellcome Trust for funding under grants WT207492 and WT206194. L.C. was funded by the LIA BEEG-B (Laboratoire International Associé - Bioinformatics, Ecology, Evolution, Genomics, and Behaviour) (CNRS), the Laboratoire d’Excellence (LabEx) project TULIP (ANR-10-LABX-41; ANR-11-IDEX-0002-02), an Investissement d’Avenir grant (CEBA; ANR-10-LABX-25-01) and the Instituto Gulbenkian de Ciência. For the computed tomography data in Figure 1, we thank the curators of the original fossils in Morocco and Israel, and J-J. Hublin. Finally, we thank Michelle O’Reilly at the Max Planck Institute for the Science of Human History for the design of Figure I in Box 1 and Figure 3, Figure 4.
Figure I.
Simple IBD Model with Cultural Data. Note that similarity can increase with distance under some circumstances, for example when similar habitats are separated by considerable distances, with areas of different habitat types being located between them.
Glossary
- Allele-frequency spectrum (AFS)
also known as or site-frequency spectrum (SFS), AFS is a histogram representing the frequencies of the alleles from multiple loci (e.g., SNPs). Assumes that loci are biallelic, and is used as a summary of genomic data for demographic inference.
- Archaic hominin
umbrella term for any of a broad group of non-Homo sapiens humans, such as the Neanderthals or Homo heidelbergensis (see below). Archaic features within our species refers to the retention of particular traits typically associated with archaic hominins, such as an elongated (versus a modern globular) braincase.
- Core-and-flake technology
stone tool technology focused on the removal of flakes as desired products from a block of raw material, which is ultimately discarded as waste. This stands in contrast to technology involving the shaping of raw material into a product (e.g., a handaxe), where the shaping flakes are discarded as waste.
- Effective population size (Ne)
a measure of genetic drift, and indirectly of some measure of population genetic diversity. Ne represents the size of the ideal population that would have the same drift properties as the real population. Depending on the properties of the genetic data in which we are interested, different effective sizes may be defined. In an ideal population, the different Ne values should be the same, but in real populations this may not be the case. Ne can be seen as the number of individuals actually contributing to the next generation, as opposed to the total number of individuals in a population (which is usually much larger).
- Hafting
the process of attaching a stone or a bone tool to a haft, typically but not always constructed from wood, either through adhesives (gum, resin), bindings, or both.
- Holocene
the recent geological epoch which began at around ∼11.7 ka and which is associated with the current warm period.
- Homo heidelbergensis
originally named for the Mauer mandible in 1908. Some researchers (e.g., [15]) have argued that this species was widespread in Eurasia and Africa during the Middle Pleistocene. Others prefer to distinguish the African samples as H. rhodesiensis, based on the Broken Hill cranium found in 1921.
- Instantaneous inverse coalescence rate (IICR)
the IICR is a time- and sample-dependent parameter. In a panmictic population, the IICR is proportional to Ne. In structured populations, the IICR can be strongly disconnected from changes in Ne.
- Intertropical convergence zone (ITCZ)
a zone near the equator where the northern and southern air masses converge, typically producing low atmospheric pressure and considerable convective rainfall.
- Large cutting tools
blocks of stone that were shaped into large handheld tools for cutting functions, such as stone axes. The flakes chipped off the stone block are referred to as thinning flakes and were typically waste products.
- Last Interglacial
the previous interglacial to the current one; this began at ∼125 ka and ended at ∼109 ka. Interglacials are warm periods characterized by receding ice sheets in the higher latitudes and increased humidity in the mid-latitude arid belt.
- Late Pleistocene
the final part of the Pleistocene geological epoch, beginning with the Last Interglacial at ∼125 ka and lasting until the beginning of the Holocene at ∼11.7 ka.
- Later Stone Age (LSA)
a cultural-technological phase in Africa dating broadly from ∼60 ka to 5 ka. While highly variable and poorly defined, the LSA is characterized by a focus on the production of small tools, such as blades and bladelets, and geometric microliths.
- Middle Pleistocene
the middle part of the Pleistocene geological epoch, beginning at ∼781 ka and ending with the beginning of the Last Interglacial at ∼125 ka ago. The Middle Pleistocene is associated with the emergence of Homo sapiens and Homo neanderthalensis at >300 ka.
- Middle Stone Age (MSA)
a cultural-technological period in Africa characterized by the widespread use of core-and-flake technologies, which were often hafted, at around ∼300 ka. The MSA gave way to the LSA over a protracted period broadly between ∼60 ka and 20 ka which featured the replacement of classic core-and-flake technologies with small and often geometric microlithic tools. The MSA is in general similar to the contemporaneous Middle Paleolithic of Eurasia.
- Milankovitch forcing
the effect on climate of slow changes in the tilt of the Earth’s axis and shape of its orbit that change the amount and distribution of sunlight reaching the Earth.
- Pairwise sequentially Markovian coalescent (PSMC)
a method (and associated program) producing a plot showing changes in Ne from a single diploid (or two haploid) genome(s) [16]. The PSMC actually estimates the IICR and has been used on many species.
- Panmixia
random patterns of mating within a population.
- Precession
climatically relevant change of the Earth’s rotation with a period of 26 000 years. In combination with the slow rotation of Earth’s elliptical orbit around the Sun, the main solar incoming radiation changes vary with a periodicity of 21 000 years. The effect of precession on the climate system occurs preferentially for large elliptic orbits.
- Prepared core technology
a form of core-and-flake technology where the core is shaped in such as a way as to predetermine the shape of subsequent flake removals. It was the dominant technology of the MSA. The most iconic form of prepared core technology is Levallois technology.
- Refugia
regions or areas that remain consistently habitable by a species or association of species through an entire glacial and interglacial climatic cycle.
- Tanged implements
stone tools characterized by a stem or a tang at the base of the tool, which is thought to be inserted into the socket of a wooden haft. This would allow stone spear points to be attached to wooden shafts, for example.
- Upper Paleolithic
a cultural-technological period in Eurasia between ∼45 ka and 10 ka that is generally characterized by an emphasis on blade and bone technology, cave art, carvings, varied personal ornaments, and elaborate burials. In Europe it is typically regarded as a marker of Homo sapiens.
References
- 1.Stringer C.B. The Origin and Evolution of Homo sapiens. Philos. Trans. R. Soc. 2016;B 371 doi: 10.1098/rstb.2015.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hublin J.-J. New fossils from Jebel Irhoud (Morocco) and the pan-African origin of Homo sapiens. Nature. 2017;546:289–292. doi: 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 3.Mirazón Lahr M., Foley R.A. Towards a theory of modern human origins: geography, demography, and diversity in recent human evolution. Am. J. Phys. Anthropol. 1998;107:137–176. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 4.Howell F.C. Species clades and extinctions in the Pleistocene hominin record. J. Anthropol. Res. 1999;55:191–243. [Google Scholar]
- 5.Gunz P. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. PNAS. 2009;106:6094–6098. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scerri E.M.L. Earliest evidence for the structure of Homo sapiens populations in Africa. Quat. Sci. Rev. 2014;101:207–216. [Google Scholar]
- 7.Mazet O. On the importance of being structured: instantaneous coalescence rates and human evolution – lessons for ancestral population size inference? Heredity. 2016;116:362–371. doi: 10.1038/hdy.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skoglund P. Reconstructing prehistoric African population structure. Cell. 2017;171:59–71. doi: 10.1016/j.cell.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wall J.D. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol. Biol. Evol. 2009;26:1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer M.F. Genetic evidence for archaic admixture in Africa. PNAS. 2011;108:15123–15128. doi: 10.1073/pnas.1109300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvati K. The Later Stone Age Calvaria from Iwo Eleru, Nigeria: morphology and chronology. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez F.L. An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am. J. Hum. Genet. 2013;92:454–459. doi: 10.1016/j.ajhg.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blome M.W. The environmental context for the origins of modern human diversity: a synthesis of regional variability in African climate 150,000–30,000 years ago. J. Hum. Evol. 2012;62:563–592. doi: 10.1016/j.jhevol.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Uren C. Fine-scale human population structure in southern Africa reflects ecogeographic boundaries. Genetics. 2016;204:303–314. doi: 10.1534/genetics.116.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stringer C.B. The status of Homo heidelbergensis (Shoetensack 1980) Evol. Anthropol. 2012;21:101–107. doi: 10.1002/evan.21311. [DOI] [PubMed] [Google Scholar]
- 16.Richter D. The age of the hominin fossils from Jebel Irhoud, Morocco, and the origins of the Middle Stone Age. Nature. 2017;546:293–296. doi: 10.1038/nature22335. [DOI] [PubMed] [Google Scholar]
- 17.Foley R., Lahr M. Mode 3 technologies and the evolution of modern humans. Camb. Archaeol. J. 1997;7:3–36. [Google Scholar]
- 18.Mirazón Lahr M., Foley R.A. Human evolution in late Quaternary Eastern Africa. In: Jones S.C., Stewart B.A., editors. Africa from MIS 6 to 2: Population Dynamics and Palaeoenvironments. Springer; 2016. pp. 215–232. [Google Scholar]
- 19.White T.D. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 20.Neubauer S. The evolution of modern human brain shape. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aao5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crevecoeur I. Late Stone Age human remains from Ishango (Democratic Republic of Congo): new insights on Late Pleistocene modern human diversity in Africa. J. Hum. Evol. 2016;96:35–57. doi: 10.1016/j.jhevol.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Grine F.E. Late Pleistocene human skull from Hofmeyr, South Africa, and modern human origins. Science. 2007;315:226–229. doi: 10.1126/science.1136294. [DOI] [PubMed] [Google Scholar]
- 23.Stynder D.D. Craniofacial variation and population continuity during the South African Holocene. Am. J. Phys. Anthropol. 2007;134:489–500. doi: 10.1002/ajpa.20696. [DOI] [PubMed] [Google Scholar]
- 24.Crevecoeur I. Modern human cranial diversity in the Late Pleistocene of Africa and Eurasia: Evidence from Nazlet Khater, Peştera cu Oase, and Hofmeyr. Am. J. Phys. Anthropol. 2009;140:347–358. doi: 10.1002/ajpa.21080. [DOI] [PubMed] [Google Scholar]
- 25.Stock J.T. The skeletal phenotype of ‘Negritos’ from the Andaman Islands and Philippines relative to global variation among hunter-gatherers. Hum. Biol. 2013;85:67–94. doi: 10.3378/027.085.0304. [DOI] [PubMed] [Google Scholar]
- 26.Migliano A.B. Life history trade-offs explain the evolution of human pygmies. PNAS. 2007;104:20216–20219. doi: 10.1073/pnas.0708024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sereno P.C. Lakeside cemeteries in the Sahara: 5000 years of Holocene population and environmental change. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dirks P.H. The age of Homo naledi and associated sediments in the Rising Star Cave, South Africa. eLife. 2017;6 doi: 10.7554/eLife.24231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balzeau A. The internal cranial anatomy of the Middle Pleistocene Broken Hill 1 cranium. Paleoanthropology. 2017;2017:107–138. [Google Scholar]
- 30.Brooks A. Long-distance stone transport and pigment use in the earliest Middle Stone Age. Science. 2018;360:90–94. doi: 10.1126/science.aao2646. [DOI] [PubMed] [Google Scholar]
- 31.Grün R. Direct dating of the Florisbad hominid. Nature. 1996;382:500–501. doi: 10.1038/382500a0. [DOI] [PubMed] [Google Scholar]
- 32.Scerri E.M.L. Oxford Research Encyclopaedia of African History. 2018. The West African Stone Age. Published online 24 January 2018. [Google Scholar]
- 33.Taylor N. Across rainforests and woodlands: a systematic reappraisal of the Lupemban Middle Stone Age in Central Africa. In: Jones S.C., Stewart B.A., editors. Africa from MIS 6 to 2: Population Dynamics and Palaeoenvironments. Springer; 2016. pp. 273–300. [Google Scholar]
- 34.Scerri E.M.L. The North African Middle Stone Age and its place in recent human evolution. Evol. Anthropol. 2017;26:119–135. doi: 10.1002/evan.21527. [DOI] [PubMed] [Google Scholar]
- 35.d’Errico F. Identifying early modern human ecological niche expansions and associated cultural dynamics in the South African Middle Stone Age. PNAS. 2017;114:7869–7876. doi: 10.1073/pnas.1620752114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell A. Late Pleistocene demography and the appearance of modern human behavior. Science. 2009;324:1298–1310. doi: 10.1126/science.1170165. [DOI] [PubMed] [Google Scholar]
- 37.Henshilwood C.S. The Still Bay and Howiesons Poort: ‘Palaeolithic’ techno-traditions in southern Africa. J. World Prehist. 2012;25:205–237. [Google Scholar]
- 38.Mackay A. Coalescence and fragmentation in the late Pleistocene archaeology of southernmost Africa. J. Hum. Evol. 2014;72:26–51. doi: 10.1016/j.jhevol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Groucutt H.S. Stone tool assemblages and models for the dispersal of Homo sapiens out of Africa. Quat. Int. 2015;382:8–30. [Google Scholar]
- 40.Thompson J. Ecological risk, demography, and technological complexity in the Late Pleistocene of Northern Malawi: implications for geographical patterning in the Middle Stone Age. J. Quat. Sci. 2017;33:261–284. [Google Scholar]
- 41.Scerri E.M.L. Persistence of Middle Stone Age technology to the Pleistocene/Holocene transition supports a complex hominin evolutionary scenario in West Africa. J. Archaeol. Sci. Rep. 2017;11:639–646. [Google Scholar]
- 42.Tryon C.A., Faith J.T. A demographic perspective on the Middle to Later Stone Age transition from Nasera rockshelter, Tanzania. Philos. Trans. R. Soc. 2016;B 371 doi: 10.1098/rstb.2015.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foley R.A., Mirazón-Lahr M. The evolution of the diversity of cultures. Philos. Trans. R. Soc. 2011;366:1080–1089. doi: 10.1098/rstb.2010.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Premo L.S., Hublin J.-J. Culture, population structure, and low genetic diversity in Pleistocene hominins. PNAS. 2009;106:33–37. doi: 10.1073/pnas.0809194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grove M. Population density, mobility, and cultural transmission. J. Archaeol. Sci. 2016;74:75–84. [Google Scholar]
- 46.Blegen N. The earliest long-distance obsidian transport: evidence from the ∼200 ka Middle Stone Age Sibilo School Road Site, Baringo, Kenya. J. Hum. Evol. 2016;103:1–19. doi: 10.1016/j.jhevol.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Backwell L., d’Errico F. Osseous projectile weaponry from early to late Middle Stone Age Africa. In: Langley M., editor. Osseous Projectile Weaponry. Springer; 2016. pp. 15–29. [Google Scholar]
- 48.d’Errico F. Additional evidence on the use of personal ornaments in the Middle Paleolithic of North Africa. PNAS. 2009;106:16051–16056. doi: 10.1073/pnas.0903532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loog L. Estimating mobility using sparse data: application to human genetic variation. PNAS. 2017;114:12213–12218. doi: 10.1073/pnas.1703642114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betti L. Distance from Africa, not climate, explains within-population phenotypic diversity in humans. Proc. R. Soc. 2009;B 276:809–814. doi: 10.1098/rspb.2008.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shennan S.J. Isolation-by-distance, homophily and ‘core’ vs. ‘package’ cultural evolution models in Neolithic Europe. Evol. Hum. Behav. 2015;36:103–109. [Google Scholar]
- 52.Tishkoff S.A. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gronau I. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veeramah K.R. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing Data. Mol. Biol. Evol. 2011;29:617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lachance J. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlebusch C.M. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mallick S. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song S. Modeling human population separation history using physically phased genomes. Genetics. 2016;205:385–395. doi: 10.1534/genetics.116.192963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henn B.M. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. PNAS. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pagani L. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am. J. Hum. Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patin E. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science. 2017;356:543–546. doi: 10.1126/science.aal1988. [DOI] [PubMed] [Google Scholar]
- 62.Henn B.M. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012 doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pickrell J.K. Ancient west Eurasian ancestry in southern and eastern Africa. PNAS. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lazaridis I. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van de Loosdrecht M. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science. 2018;360:548–552. doi: 10.1126/science.aar8380. [DOI] [PubMed] [Google Scholar]
- 66.Wakeley J. Nonequilibrium migration in human history. Genetics. 1999;153:1863–1871. doi: 10.1093/genetics/153.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Storz J.F., Beaumont M.A. Testing for genetic evidence of population expansion and contraction: an empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution. 2002;56:154–166. doi: 10.1111/j.0014-3820.2002.tb00857.x. [DOI] [PubMed] [Google Scholar]
- 68.Chikhi L. The IICR (inverse instantaneous coalescence rate) as a summary of genomic diversity: insights into demographic inference and model choice. Heredity. 2018;120:13–24. doi: 10.1038/s41437-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Broushaki F. Early Neolithic genomes from the eastern Fertile Crescent. Science. 2016;353:499–503. doi: 10.1126/science.aaf7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldstein D.B., Chikhi L. Human migrations and population structure: what we know and why it matters. Annu. Rev. Genom. Hum. Genet. 2002;3:129–152. doi: 10.1146/annurev.genom.3.022502.103200. [DOI] [PubMed] [Google Scholar]
- 71.Itan Y. The origins of lactase persistence in Europe. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eriksson A. Late Pleistocene climate change and the global expansion of anatomically modern humans. PNAS. 2012;109:16089–16094. doi: 10.1073/pnas.1209494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu D. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol. Biol. Evol. 2017;34:2704–2715. doi: 10.1093/molbev/msx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eriksson A., Manica A. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. PNAS. 2012;109:13956–13960. doi: 10.1073/pnas.1200567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schlebusch C.M. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science. 2017;358:652–655. doi: 10.1126/science.aao6266. [DOI] [PubMed] [Google Scholar]
- 76.Stewart J.R., Stringer C.B. Human evolution out of Africa: the role of refugia and climate change. Science. 2012;335:1317–1321. doi: 10.1126/science.1215627. [DOI] [PubMed] [Google Scholar]
- 77.Potts R. Environmental dynamics during the onset of the Middle Stone Age in eastern Africa. Science. 2018;360:86–90. doi: 10.1126/science.aao2200. [DOI] [PubMed] [Google Scholar]
- 78.Cohen A.S. Ecological consequences of early Late Pleistocene megadroughts in tropical Africa. PNAS. 2007;104:16422–16427. doi: 10.1073/pnas.0703873104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivory S.J. Environmental change explains cichlid adaptive radiation at Lake Malawi over the past 1.2 million years. PNAS. 2016;113:1895–11900. doi: 10.1073/pnas.1611028113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drake N.A. Ancient watercourses and biogeography of the Sahara explain the peopling of the desert. PNAS. 2011;108:358–462. doi: 10.1073/pnas.1012231108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Timmermann A., Friedrich T. Late Pleistocene climate drivers of early human migration. Nature. 2016;538:92–95. doi: 10.1038/nature19365. [DOI] [PubMed] [Google Scholar]
- 82.Tierney J.E. A climatic context for the out-of-Africa migration. Geology. 2017;45:1023–1026. [Google Scholar]
- 83.deMenocal P. African climate change and faunal evolution during the Pliocene–Pleistocene. Earth Planet. Sci. Lett. 2004;220:3–24. [Google Scholar]
- 84.Miller C.S., Gosling W.D. Quaternary forest associations in lowland tropical West Africa. Quat. Sci. Rev. 2014;84:7–25. [Google Scholar]
- 85.Moodley Y., Bruford M.W. Molecular biogeography: towards and integrated framework for conserving Pan-African biodiversity. PLoS One. 2007;2:e454. doi: 10.1371/journal.pone.0000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Linder H.P. The partitioning of Africa: statistically defined biogeographical regions in sub-Saharan Africa. J. Biogeogr. 2012;39:1189–1205. [Google Scholar]
- 87.Bertola L.D. Phylogeographic patterns in Africa and high resolution delineation of genetic clades in the Lion (Panthera leo) Sci. Rep. 2016;6 doi: 10.1038/srep30807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Montinaro F. Complex ancient genetic structure and cultural transitions in southern African populations. Genetics. 2017;205:303–316. doi: 10.1534/genetics.116.189209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure360: an author presentation of Figure 3