Abstract
Infections by carbapenem-resistant Enterobacteriaceae are difficult to manage owing to broad antibiotic resistance profiles and because of the inability of clinically used β-lactamase inhibitors to counter the activity of metallo-β-lactamases often harbored by these pathogens. Of particular importance is New Delhi metallo-β-lactamase (NDM), which requires a di-nuclear zinc ion cluster for catalytic activity. Here, we compare the structures and functions of clinical NDM variants 1–17. The impact of NDM variants on structure is probed by comparing melting temperature and refolding efficiency and also by spectroscopy (UV-visible, 1H NMR, and EPR) of di-cobalt metalloforms. The impact of NDM variants on function is probed by determining the minimum inhibitory concentrations of various antibiotics, pre-steady–state and steady-state kinetics, inhibitor binding, and zinc dependence of resistance and activity. We observed only minor differences among the fully loaded di-zinc enzymes, but most NDM variants had more distinguishable selective advantages in experiments that mimicked zinc scarcity imposed by typical host defenses. Most NDM variants exhibited improved thermostability (up to ∼10 °C increased Tm) and improved zinc affinity (up to ∼10-fold decreased Kd, Zn2). We also provide first evidence that some NDM variants have evolved the ability to function as mono-zinc enzymes with high catalytic efficiency (NDM-15, ampicillin: kcat/Km = 5 × 106 m−1 s−1). These findings reveal the molecular mechanisms that NDM variants have evolved to overcome the combined selective pressures of β-lactam antibiotics and zinc deprivation.
Keywords: antibiotic resistance, antibiotics, metalloenzyme, enzyme kinetics, protein evolution, enzyme mutation, Co(II)-substituted enzyme, metallo-β-lactamase, NDM-1
Introduction
Carbapenem-resistant Enterobacteriaceae continue to be classified as an “urgent threat,” the highest hazard level assigned by the Centers for Disease Control and Prevention (1). The five carbapenemases currently of primary public concern include Klebsiella pneumonia carbapenemase, New Delhi metallo-β-lactamase (NDM),7 Verona integrin-encoded metallo-β-lactamase (VIM), imipenemase (IMP), and oxacillinase-48–like carbapenemase (OXA-48) (2). Three of these carbapenemases (NDM, VIM, and IMP) are metal-dependent β-lactamases that are not susceptible to any of the β-lactamase inhibitors incorporated into combination drugs used in the clinic. Of these three β-lactamases, NDM is the most widespread in patients in the United States, with infections bearing a blaNDM gene reported in 34/50 states (as of December, 2017, https://www.cdc.gov/hai/organisms/cre/trackingcre.html).8
The genes encoding NDM continue to evolve, with discovery of more than 20 variants (NDM-1 through NDM-21 at the time of writing, 16 at the start of this project). Most of these mutations occur at sites distant from the active site, and the functions they confer are not immediately obvious. A comparison of NDM-1 through NDM-8 showed only minor differences in kcat/Km values (≤5-fold) for a panel of diverse β-lactam drugs (3). However, a considerable increase in thermostability was noted for many of the variants, suggesting the functional impact of NDM mutations may be to confer stability. We subsequently found that the selective advantages conferred by the M154L mutation, found in more than half of NDM variants, were only revealed under experimental conditions where zinc availability was limited, mimicking the paucity of free zinc ions at common infection sites (4). Improvements in resistance conferred by enzyme activity correlate with an improved affinity for the more weakly bound zinc ion (Zn2, ligated by Asp-124, Cys-208, and His-250) in the di-nuclear zinc ion cluster required for NDM catalysis. A subsequent study confirmed improved β-lactam resistance at low zinc concentrations for most NDM variants in a comparison of NDM-1 through NDM-16, and the variants that had the most improved resistance at low zinc concentrations were shown to outcompete others when challenged with zinc deprivation and β-lactam treatment (5). Here, we probed the structure and function of NDM-1 through NDM-17 to determine the mechanism underlying improved β-lactam resistance under conditions of zinc scarcity. The mutations found in these NDM variants reflect an evolutionary trend to increase thermostability through optimization of hydrophobic packing in the core of the protein, to increase Zn2 affinity through small conformational adjustments of the zinc-coordinating residues, and to confer on NDM the ability to function as a mono-zinc enzyme with high catalytic efficiency. To our knowledge, this is the first report that some NDM variants have gained the ability to function with only 1 eq of bound zinc ion. Our findings suggest that NDM genes are evolving improved zinc affinity and mono-zinc activity in response to the combined selective pressure of limited zinc availability and β-lactam treatment, likely as a result of nutritional immunity imposed by host defense systems.
Results and discussion
We conducted microbial, biochemical, and biophysical characterization of the clinically-derived NDM variants NDM-1 through NDM-17 to determine whether there were any shared characteristics that might reveal which selective pressures are driving the evolution of the corresponding genes. With the exception of NDM-10, these variants contain 0–3 mutated residues relative to NDM-1 (Fig. 1), often occurring in combinations, notably with the most resistant variants containing the M154L substitution. Each of these substitutions occurs at a site that does not directly interact with zinc or β-lactam substrates (Fig. 2). As described below, each variant was characterized in terms of function, through microbiological tests of resistance, steady-state kinetics, pre-steady–state kinetics, and ligand-binding assays. The structure of each enzyme was probed by using multiple spectroscopic techniques, denaturation and refolding experiments, and metal content analysis. Taken together, these findings prompted additional studies on zinc affinity and the zinc dependence of antibiotic hydrolysis. We found that most of the clinically-derived mutations result in increased resistance only observed under conditions where the supply of zinc is limited, mimicking conditions found at common infection sites. Throughout, we use the term “resistance” in the broad sense of conferring growth in the presence of antibiotics rather than in its more narrow technical application as meeting a defined threshold for clinical resistance. Characteristics found to be common among the most resistant NDM variants include an increase in thermostability and refolding efficiency, a structurally-perturbed Zn2 site, and increased Zn2 affinity, indicating that protein instability and zinc scarcity may be two selective pressures driving the evolution of blaNDM in the clinic. Notably, NDM-1 is only active as a di-zinc protein, but some clinically-derived variants have evolved high catalytic efficiency as mono-zinc enzymes.
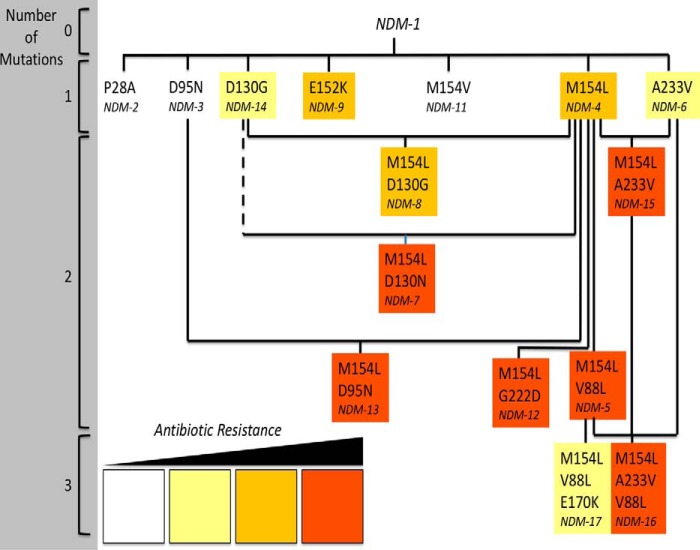
Figure 1.
Mutated residues in clinically derived NDM variants. Variants containing combinations of between 0 and 3 mutations are shown in relation to each other. NDM-10 (Table S1) is omitted. Solid lines indicate shared mutations. The dashed line indicates a shared site of mutation. NDM variants are colored to summarize their relative resistance in zinc-depleted conditions, as determined herein (Table 2): white for 0 to <2 dilution increases of MIC values in two or more antibiotics; yellow for ≥2 dilution increases in MIC values for two or more antibiotics; orange for ≥3 dilution increases in MIC values for two or more antibiotics; and red for ≥4 dilution increases in MIC values for two or more antibiotics.
Figure 2.
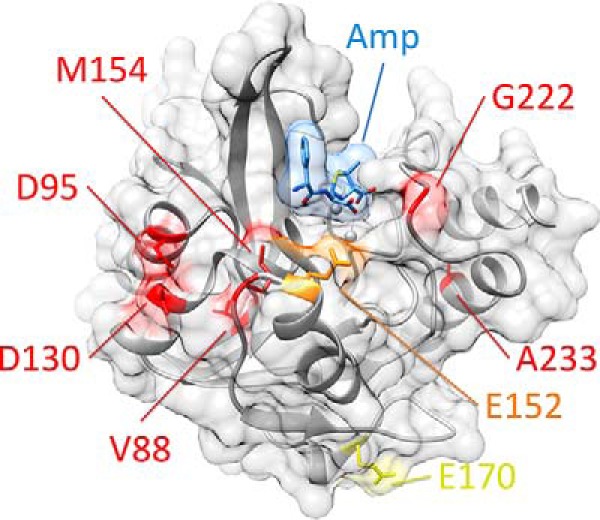
Position of residues mutated in clinically isolated variants of NDM. NDM is shown as a ribbon diagram with zinc atoms as spheres, coated by a transparent surface (all in gray). Hydrolyzed ampicillin is shown as sticks, with a transparent surface (blue). Positions mutated in clinically isolated variants are color-coded by the highest resistance variant in which each occurs (low in yellow, medium in orange, and high in red, see Fig. 1). Figure was prepared using Protein Data Bank code 4HL2 for di-zinc NDM-1 bound to hydrolyzed ampicillin.
Functional characterization of NDM variants
Plasmids encoding NDM-1 through NDM-17 were constructed (Tables S1 and S2) and used to express the enzyme variants in a common genetic background (Escherichia coli DH10B) as their full-length forms, including an N-terminal lipidation signal, with expression controlled by a common native promoter (taken from blaNDM-1). Relative protein concentrations of the NDM variants appeared to be similar to one another in cultured bacteria, with the exceptions of NDM-2, which showed a double band, and NDM-10, which showed decreased protein levels (Fig. S1). Using these strains, resistance was determined against a wide panel of clinically used β-lactam drugs, including a penem (ampicillin), cephems (cephalothin, cefotaxime, ceftazidime, and cefepime), and carbapenems (imipenem and meropenem). Minimum inhibitory concentration (MIC) values determined using standard conditions showed that none of the variants, except for NDM-10, conferred substantially different resistance than NDM-1 (MIC values were ≤2 dilutions different) (Table 1). NDM-10 is unlike the other variants because it has five unique mutations (Table S1), two of which occur in the substrate-binding β-hairpin loop (G69D and A74T), and MIC determinations actually show a decreased resistance to ampicillin, imipenem, and meropenem, likely due, at least in part, to decreased protein levels. It is possible that the mutations in NDM-10 might have evolved to confer increased resistance to an antibiotic not tested here. In summary, none of the clinically-derived NDM variants show increased resistance under these conditions.
Table 1.
MIC values for NDM variants under standard conditions
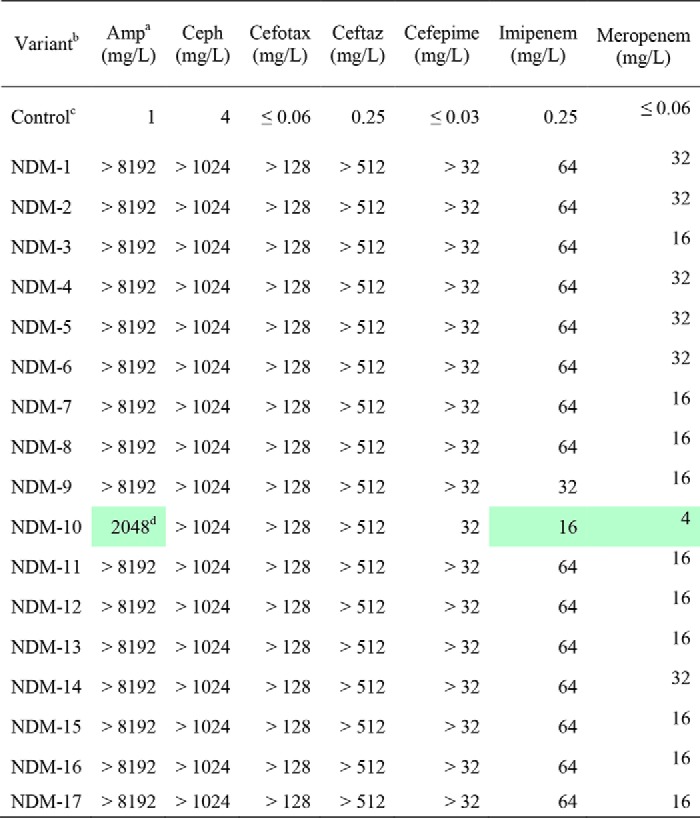
a Antibiotic abbreviations used are as follows: Amp, ampicillin; Ceph, cephalothin; Cefotax, cefotaxime; Ceftaz, ceftazidime.
b Variants are all expressed in the same strain, E. coli DH10B.
c Control is an expression vector with no blaNDM insert (pHSG298).
d Reductions in MIC values (relative to NDM-1) ≥ 2-dilutions are marked in green. Control is not included in the color coding.
Guided by our earlier finding that the M154L mutation increases resistance when zinc is not readily available (4), we measured the resistance conferred by NDM variants against the same panel of antibiotics, but also included EDTA to lower available zinc concentrations. A few of the NDM variants did not reveal any selective advantage under these altered conditions; NDM-10 showed decreased resistance, and the single mutants NDM-2, -3, and -11 show resistance similar to that of NDM-1 (Table 2). However, all of the other NDM variants confer a selective advantage under these conditions. The gains in resistance were significant, with increases of 2, 3, or even ≥4 dilutions in MIC values measured for at least two different antibiotics (we summarize the resistance for each NDM variant throughout this paper with a color code of yellow, orange, or red as described in Fig. 1). The change in resistance to ampicillin (a penam) was most impacted by mutations found in the clinic, and resistance to cephems (especially cephalothin) was the least impacted. This rank order is similar to what we reported earlier for the zinc dependence on in vitro activity of the M154L substitution, a mutation that is shared by the majority of the NDM variants (5). So, we next prepared purified NDM proteins and characterized their structure and function in vitro.
Table 2.
MIC values for NDM variants under zinc-limited conditions (50 μm EDTA)
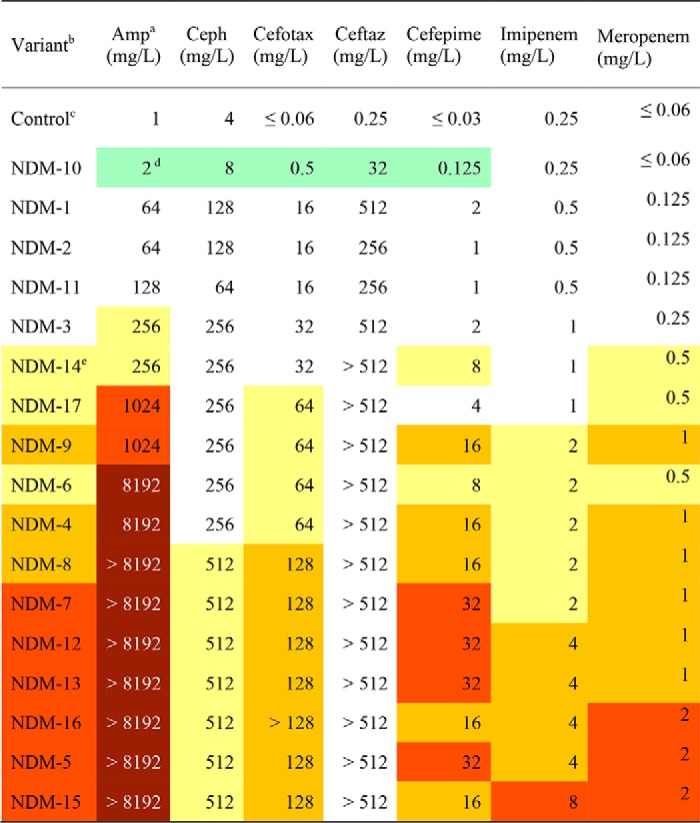
a Antibiotic abbreviations used are as follows: Amp, ampicillin; Ceph, cephalothin; Cefotax, cefotaxime; Ceftaz, ceftazidime.
b Variants are all expressed in the same strain, E. coli DH10B.
c Control is an expression vector with no blaNDM insert (pHSG298).
d Reductions in individual MIC values (relative to NDM-1) are marked as follows: 2 dilution decrease in green, 2 dilution increase in yellow, 3 dilution increase in orange, 4 dilution increase in red, > 4 dilution increase in brown. Control is not included in color-coding.
e Resistance for each variant is summarized and color-coded as described in the legend for Fig. 1.
DNA sequences coding for NDM variants were cloned into overexpression vectors under the control of an inducible promoter to express soluble versions of each protein by omitting the first 35 amino acids (which contains the lipidation signal). NDM-2 was not included in these studies because the P28A mutation resides within the truncated sequence. NDM-10 was also not included because initial trials of expression and purification in this system resulted in low yields (<1 mg/liter) of impure protein as compared with the other variants that typically resulted in high yields (> 30 mg/liter) of highly-purified proteins (Fig. S2). Using these procedures, NDM-1 copurified with 1.8 ± 0.1 eq of zinc ions, and all of the other variants copurified with similar equivalents of zinc (1.4 ± 0.1 to 2.0 ± 0.1) (Table S3), consistent with formation of di-zinc active sites in each purified protein.
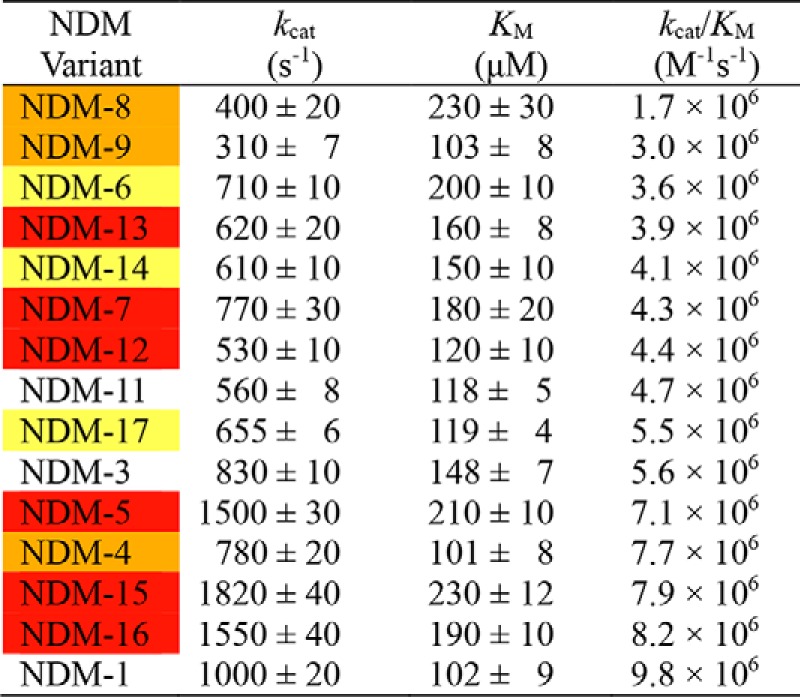
To compare the function of NDM variants, steady-state kinetic parameters were determined for hydrolysis of a penam (ampicillin), a cephem (chromacef), and a carbpenem (meropenem) by each purified NDM in the presence of excess zinc (10 μm) to ensure a fully-loaded di-zinc active site (Table 3 and Table S4). The results with ampicillin (Table 3, sorted by smallest to largest kcat/Km value and color-coded by overall resistance) are representative of the other antibiotics tested (Table S4), with a few minor exceptions. For each antibiotic tested, NDM-1 retains the highest kcat/Km value, with all of the other variants having values at most ∼6-fold lower. Km values for the variants are almost all marginally higher (up to ∼2-fold), and kcat values vary to either slightly higher (∼2-fold) or lower (∼3-fold) values, but only within a limited range. Therefore, when the NDM variants have fully-occupied di-zinc active sites, they all exhibit similar steady-state catalytic parameters, indicating they are all efficient catalysts of β-lactam hydrolysis with kcat/Km values in the range of 106–107 m−1 s−1 for all the substrates tested. These findings are similar to those reported earlier comparing steady-state kinetic analysis of di-zinc NDM-1 and NDM-3 through NDM-8 (3).
Table 3.
Steady-state kinetic parameters of NDM variants for ampicillin hydrolysis in the presence of excess zinc (10 μm)
As a second method for assessing any changes in function due to mutations found in the clinic, we determined the dissociation constants for inhibitor binding to each of the NDM variants. l-Captopril, a commonly used inhibitor of most metallo-β-lactamases, is a competitive inhibitor of NDM-1 and binds by displacing the bridging hydroxide in the di-nuclear zinc active site with a thiolate (6). l-Captopril is not used in the clinic to treat NDM infections, so it likely does not provide a selective pressure driving the evolution of these variants. However, the l-captopril/NDM-1 interaction is well-characterized and can be used to detect any impact that mutations have on ligand binding at the heart of the active site. Isothermal titration calorimetry was used to determine the Kd values for NDM-1 and l-captopril (Kd = 4 ± 1 μm), and the values are similar to other reports of the affinity of this interaction (Fig. 3) (7). In comparison, the dissociation constants for l-captopril with the other NDM variants only occurred within a very narrow range (1.3–5 μm), indicating that the mutations derived from the clinic do not greatly impact affinity of an active-site ligand (Fig. S4 and Table S5). These results are consistent with the steady-state kinetic parameters, which showed only minor changes to Km values of substrates for di-zinc NDM variants. These findings do contrast with a similar analysis of inhibitor binding to variants of the related VIM, which shows a large difference (∼500-fold) in IC50 for one particular inhibitor between VIM-1 and VIM-38, indicating that mutations in this VIM variant impact inhibitor affinity, likely by structural changes in an active-site loop that directly contacts the inhibitor (although affinity for other inhibitors is not as greatly impacted) (8).
Figure 3.
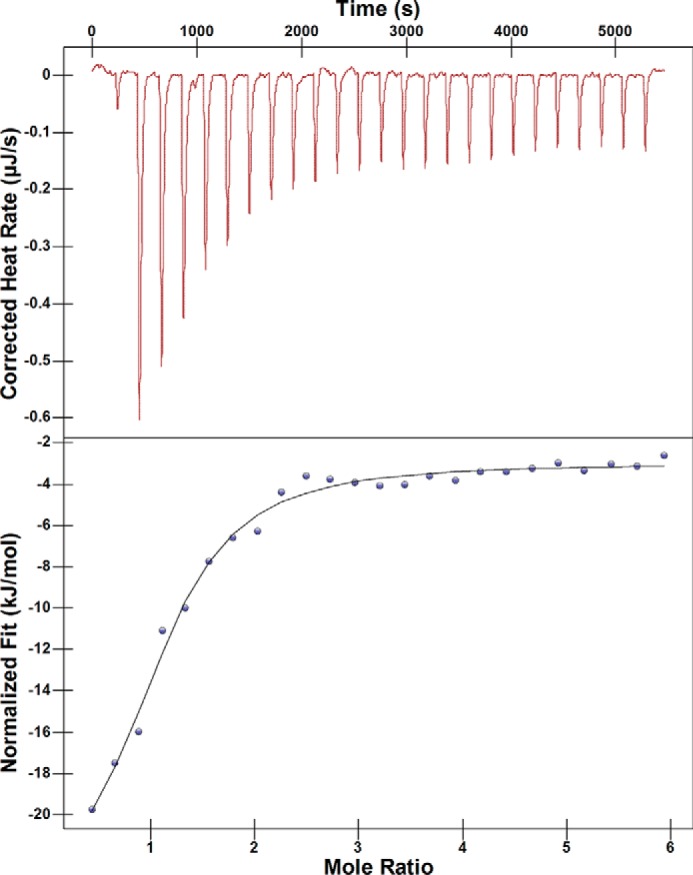
ITC determination of l-captopril/di-zinc NDM-1 dissociation constant. A representative experiment is shown for ITC of NDM-1 and l-captopril, with the thermograms and fits for the other NDM variants included in Fig. S4. Values for Kd of all variants are given in Table S5. Fits above give Kd and stoichiometry for di-zinc NDM1 and l-captopril binding as 4 ± 1 μm and 1.0 ± 0.1, respectively.
As a third assay of function to compare NDM variants, we used stopped-flow spectroscopy to monitor pre-steady–state kinetics of the formation and decay of a distinctive reaction intermediate. NDM-1 uses a mechanism shared by many metallo-β-lactamases in the B1 superfamily in which a substrate β-lactam carbonyl carbon atom is attacked by the hydroxide ion bridging the di-nuclear zinc site, with Zn1 coordinating and polarizing the β-lactam carbonyl oxygen and Zn2 coordinating and stabilizing the developing negative charge on the β-lactam nitrogen leaving group (9). For a review depicting this intermediate and several possible mechanisms of B1 metallo-β-lactamases, please see Ref. 9. With some substrates, especially those with conjugated electron withdrawing groups, this leaving group is stabilized as an anionic nitrogen directly coordinated to Zn2, and the subsequent slow step in turnover is protonation of this nitrogen, followed by facile release of product (10). The anionic intermediate found in NDM-1–catalyzed hydrolysis of the chromogeneic cephem chromacef absorbs strongly at 575 nm and is monitored throughout turnover (11). Unlike the other functional assays described above, this assay does reveal clear differences between NDM variants, which can be qualitatively separated into three groups (Fig. 4 and Fig. S3). The first group (NDM-1, -6, and -9) shows rapid formation of the intermediate followed by a slow decay. The second group (NDM-3–5, -12, and -14) shows an initial rapid formation of intermediate followed by a longer, slower phase of intermediate accumulation of lower amplitude, followed by a slow decay. The third group (NDM-7, -8, -11, -13, and -15–17) shows an initial rapid formation of intermediate, followed by a lag phase, and then a second rapid phase of intermediate accumulation before the final slow decay. The variants within groups do have some commonalities. Group 1 consists of variants with single amino acid changes only, none of which occur at the Met-154 site. Group 3 contains variants that all have a mutation at the Met-154 site, with group 2 containing a mixture. Additional studies outside the scope of this report will be required to quantify the specific kinetic differences and underlying mechanistic differences between these variants. They are not addressed here because it is unlikely that the differences have a large impact on resistance; none of the three observed groups correlate with increased resistance. However, this assay does provide a sensitive probe that reveals changes to the kinetic mechanism caused by these mutations at the active site, particularly changes that arise from altering the intermediate/Zn2 interaction, which is notable because prior crystal structures of NDM variants only reveal minor differences neighboring the Zn2 site (4).
Figure 4.
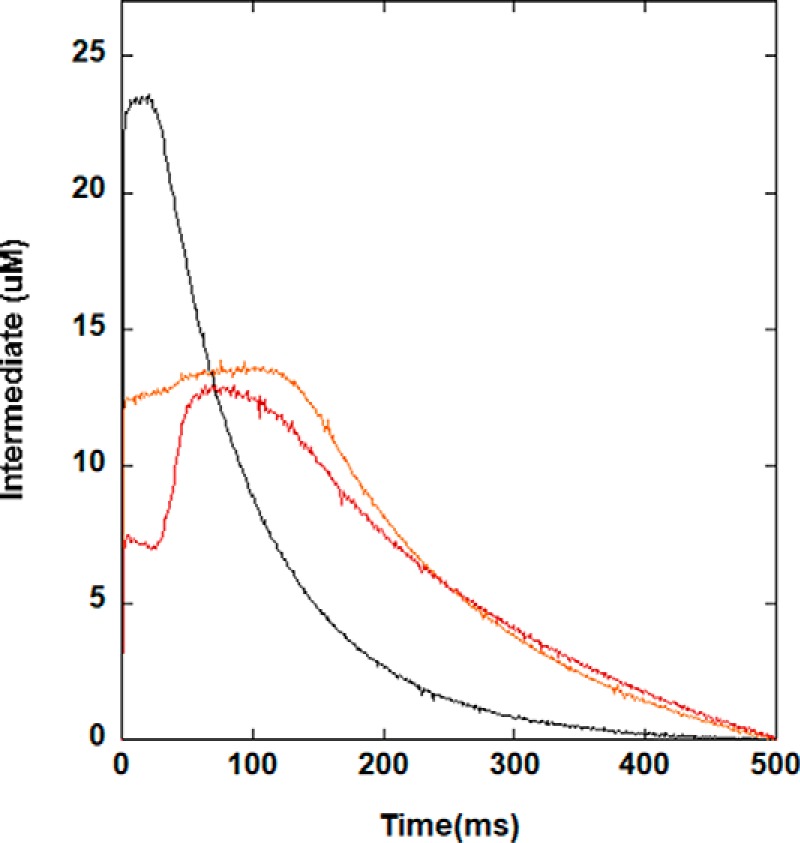
Stopped-flow kinetics of chromacef turnover by di-zinc(II) NDM variants. Using conditions described under “Experimental procedures,” the amount of anionic intermediate that absorbs at 575 nm was monitored. Representative variants NDM-1 (black), NDM-4 (orange), and NDM-15 (red) are shown above, and kinetic traces for the remaining NDM variants are given in Fig. S3.
Structural characterization of NDM variants
Using the same soluble NDM variants described above, we probed the structure of these proteins through a variety of methods. Two techniques did not reveal any changes that could be correlated with resistance; far-UV–CD spectroscopy of the purified variants showed essentially no changes resulting from any mutation, indicating that any structural change did not impact the major secondary structural elements assessable by this technique (Fig. S5). The predicted α-helical (39%) and β-sheet (12%) content for all variants is consistent with prior reports and the X-ray structure of NDM-1 (6). Intrinsic fluorescence of Trp residues was also compared. The NDM variants all contain four Trp residues, with one Trp residue (Trp-93) contributing a hydrophobic surface to the active-site cavity. Intrinsic Trp fluorescence yield and spectra are impacted by quenching due to either solvent or neighboring residues and can be used to detect conformational changes in proteins (12). Using λex, 280 nm the most intense emission peaks for the NDM variants occurred at ∼345 nm and varied in intensity, indicating that the environment of one or more of the Trp residues is changed by these mutations (Fig. S6). Although the variants with lower fluorescence intensities tended to contain a mutation at the Met-154 site, these changes did not correlate with the observed increases in resistance.
In contrast to the experiments above, the thermostability and refolding propensity of NDM variants do correlate with increases in resistance. Differential scanning fluorimetry (DSF) with SYPRO Orange was used to monitor thermal unfolding of the purified variants and to obtain a melting temperature (Tm) for each (Fig. 5). All of the NDM variants derived from the clinic lead to higher Tm values than NDM-1, indicating that each combination of mutants results in a net increase in thermostability, with NDM-16 being the most stable variant, ∼10 °C more stable than NDM-1. NDM variants with only one mutation generally show no increase or modest increases in resistance, and these variants also show the lowest increase in Tm values. The M154L mutation in NDM-4 appears to be a turning point for NDM variants as every variant harboring Leu-154 exhibits higher thermostability when combining the M154L mutation with one or two additional mutations. A previous report noted similar increases in thermostability for the variants NDM-3 to NDM-8 over that of NDM-1, although a direct comparison of Tm values with those herein is problematic because zinc content was not specified in the previous study (3). We also assessed the ability of these proteins to refold by mimicking their refolding after transport into the periplasmic space. Because NDM does not contain a twin-arginine leader sequence, it is likely translocated into the periplasm in unfolded form and requires subsequent refolding and metallation. Conditions were chosen to mimic an environment with low zinc availability, and both high and low protein concentrations were used. Notably, all of the mutations derived from the clinic lead to greater folding efficiencies than NDM-1 when assayed at low zinc and low protein concentrations (Fig. 6). In general, NDM variants with more mutations tended to show a greater ability to refold, with the most efficient variants all combining the M154L mutation with others. Taken together, results from these two assays indicate that clinically-derived NDM mutations help to favor the folded form of the protein, even in conditions where there is insufficient zinc available to form a di-zinc active site.
Figure 5.
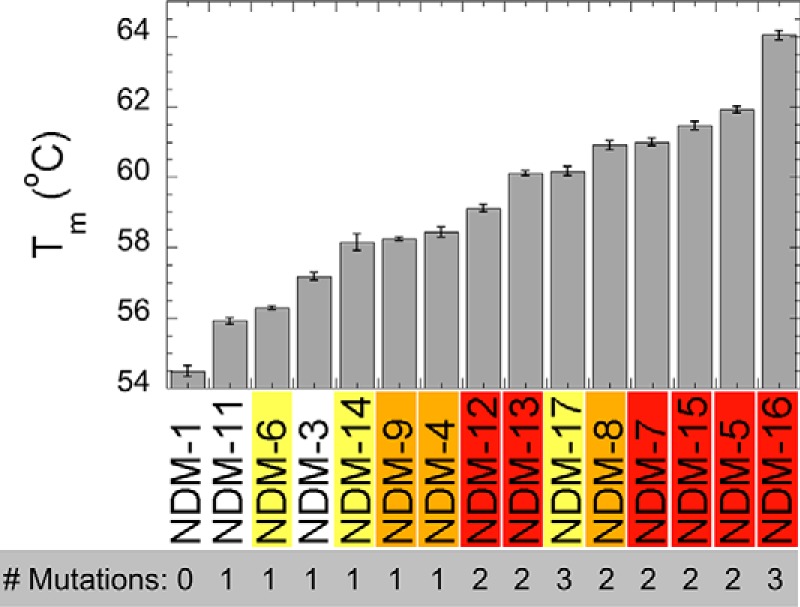
Melting temperatures of NDM variants. SYPRO Orange fluorescent dye was mixed with each variant (5 μm) in HEPES buffer (20 mm) with NaCl (150 mm) at pH 7.5. Unfolding in response to heating was monitored by fluorescence at 570 nm.
Figure 6.
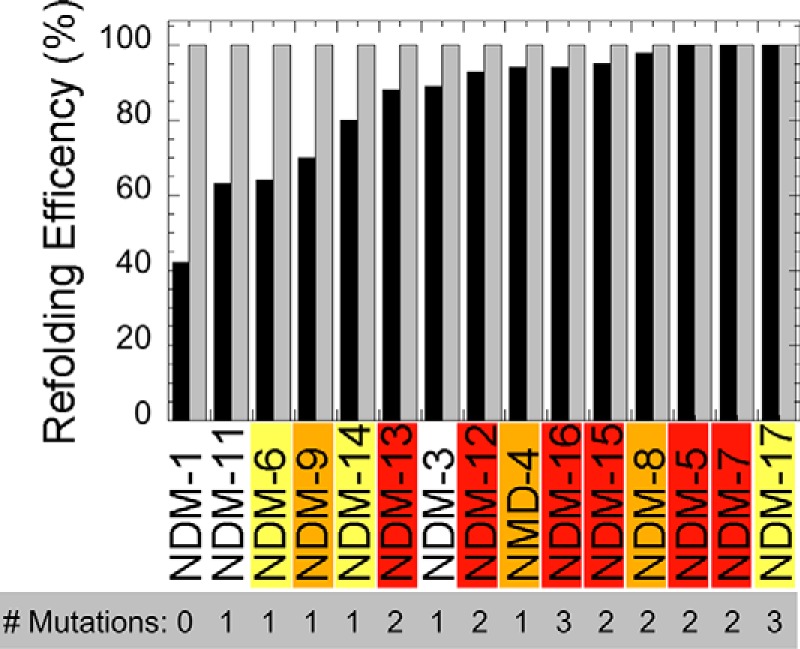
Relative refolding efficiencies of apo-NDM variants. Each purified metal-free variant at low (gray bars, 35 μm) or high (black bars, 350 μm) concentrations was refolded by dialysis and normalized by concentration of the variant before refolding. Each bar is the average of duplicate experiments.
Conversion of purified NDM variants into di-cobalt(II) NDM metalloforms provides a spectroscopic window to probe the protein structure at the active site of these enzymes. We first examined the UV-visible spectra of the mutants. Interaction of Cys-208, which serves as a Zn2 ligand, with Co(II) results in a ligand-to-metal charge transfer transition at 320 nm that shows variable intensity across the NDM variants (Fig. 7). Nearly all of the variants show lower charge transfer intensity than NDM-1, consistent with weaker binding of the Zn2 metal ion to Cys-208. The variants also display a set of ligand field transitions that are characteristic of high-spin cobalt(II) at 510, 549, 615, and 640 nm (Fig. 7, inset). The shape of these bands does change greatly depending on the mutation (with perhaps the exception of NDM-6), reflecting only subtle structural changes occurring in the vicinity of the metal site. The extinction coefficients in the low-energy pair of ligand field bands are not affected significantly, whereas those of the higher energy pair of bands change by up to 50%. Based on our previous work on NDM-1 (13), which showed a more symmetric set of ligand field bands with Co(II) in the Zn1 site (CoCd–NDM-1) and a more asymmetric set of bands for the Zn2 site (ZnCo–NDM-1), the data in Fig. 7 may indicate that the mutations are shifting the spectra toward the Zn2 structure, which is more asymmetric and five-coordinate. However, we believe the observation of more intense ligand field bands is a consequence of weaker Co–S bonding, and the optical spectra are consistent with maintenance of the overall active-site structure, including the ligand sets to both metal ions.
Figure 7.
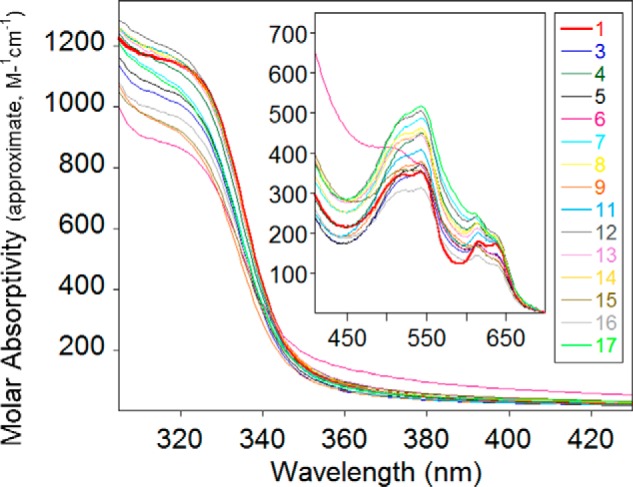
UV-visible spectra of cobalt(II)-loaded NDM variants. The spectrum of NDM-1 (red) is shown in bold for reference. All samples (300 μm) were in HEPES (50 mm), NaCl (150 mm) at pH 6.8.
NMR spectroscopy of di-cobalt(II) NDM variants was used to more closely examine structural changes at or near the metal-binding sites. Resonances at 64, 72, and 79 ppm were previously assigned to solvent-exchangeable protons on His-120, His-122, and His-189 in the Zn1 site, whereas resonances at 48, 107, and 165 ppm have been assigned, respectively, to protons on Asp-124, His-250, and Cys-208 at the Zn2 site (13). The spectrum of di-cobalt(II) NDM-1 shown here (Fig. 8) reproduces what we have reported earlier (4, 14). In comparison, all of the variants differ from NDM-1, yet they are similar to one another. All of the mutations result in a change in the resonances assigned to Cys-208, changing from a minimally-split geminal pair in NDM-1 (162 and 168 ppm) to what appears to be a much less symmetric pair with one line at ∼200 ppm. With the exception of NDM-6, it appears the partner resonance from Cys-208 is centered near ∼75 ppm (in NDM-6, it appears the partners are at 160 and 200 ppm). Similarly, nearly all of the variants display reduced intensity at 48 ppm (Asp-124), with the appearance of a new resonance of near equal intensity at 56 ppm (the only apparent exception being NDM-11), neither of which appear to be solvent-exchangeable (Fig. S7). Both of these observations indicate a change in dihedral angle with respect to the magnetic axes of the Co(II) ion in the Zn2 site, which could reflect a repositioning of the ligands or a reorientation of the magnetic axes. Given the scale of the mutations (1–2 remote amino acids), we do not expect a dramatic rearrangement of the active site, and therefore we interpret the changes as a reflection of a slight repositioning of the Asp and Cys ligands at the Zn2 site. Interestingly, the third protein-derived ligand at this site (His-107) appears unaffected by all of the mutations. The result of these slight alterations appears to be a Zn2 metal ion that is more intimately coupled to its environment, based on an increase in the number of resonances between 40 and −40 ppm, which have been shown to represent Co(II)-second sphere interactions in the MBLs (14). These observations are consistent with the lower Kd, Zn2 values we determined for these variants (see below).
Figure 8.
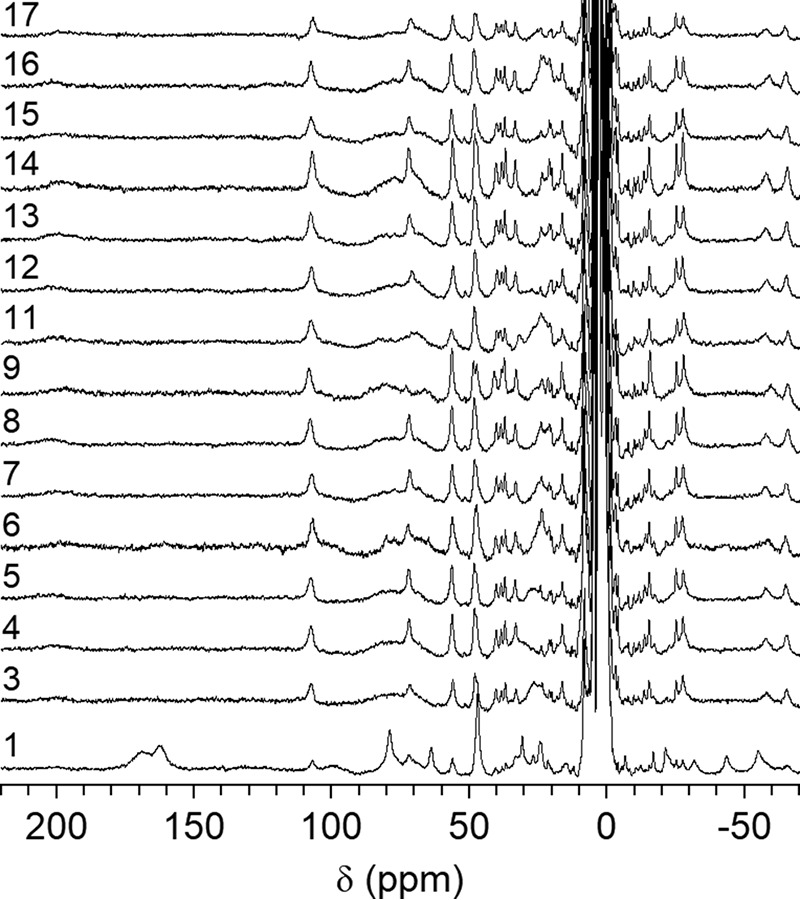
1H NMR spectra (300 MHz) of cobalt(II)-substituted NDM variants.
More striking is the apparent disappearance of two of the three resonances assigned to the His ligands of the Zn1 site in nearly every variant. Specifically, resonances at 64 and 79 ppm are no longer observed, with only the peak at 72 ppm being retained in all but NDM-11. NDM-6 still shows evidence of all three His resonances, and we note here that this was the only variant that did not appear to shift a cysteine β-methylene proton to a position directly under the His resonances, suggesting that they may simply be unresolved in the other variants. Alternatively, the mutations could result in broadening of the His resonances themselves. Comparison of spectra in H2O/D2O offered little insight, showing only a couple of candidates that were of insufficient intensity (Fig. S7).
Finally, we used electron paramagnetic resonance (EPR) spectroscopy of the di-cobalt(II) NDM variants to probe structural changes at the metal center due to mutation. Perpendicular mode EPR spectra show overlapping broad signals, typical of high-spin cobalt(II) ions; the largest difference in the spectra between mutants was the depth of the derivative at 1900 G (Fig. 9 and Fig. S8). Parallel mode spectra show differences in the negative signal at 800 G (Fig. 9 and Fig. S8). The intensity of this signal has been correlated to the coupling strength between cobalt(II) ions in other di-nuclear metallo-β-lactamases. Notably, the changes at 800 G for di-cobalt(II) NDM variants appear to have some correlation with resistance, with variants showing the most intense coupling (NDM-1, -3, -6, and -9) corresponding to less resistance, and those with less coupling (e.g. NDM-15) corresponding, in general, to variants with more resistance at conditions where zinc is scarce. Uncoupling may indicate that the more resistant NDM variants have di-cobalt(II) sites that are better positioned to function in the absence of the Zn2 metal ion. In sum, the di-cobalt(II) metalloforms of NDM variants serve as a sensitive probe of active-site structure, indicating that mutations derived from the clinic do perturb the structure surrounding both active-site metal ions and that uncoupling of the metal sites is found in many of the variants that show increased resistance when exogenous zinc is not readily available.
Figure 9.
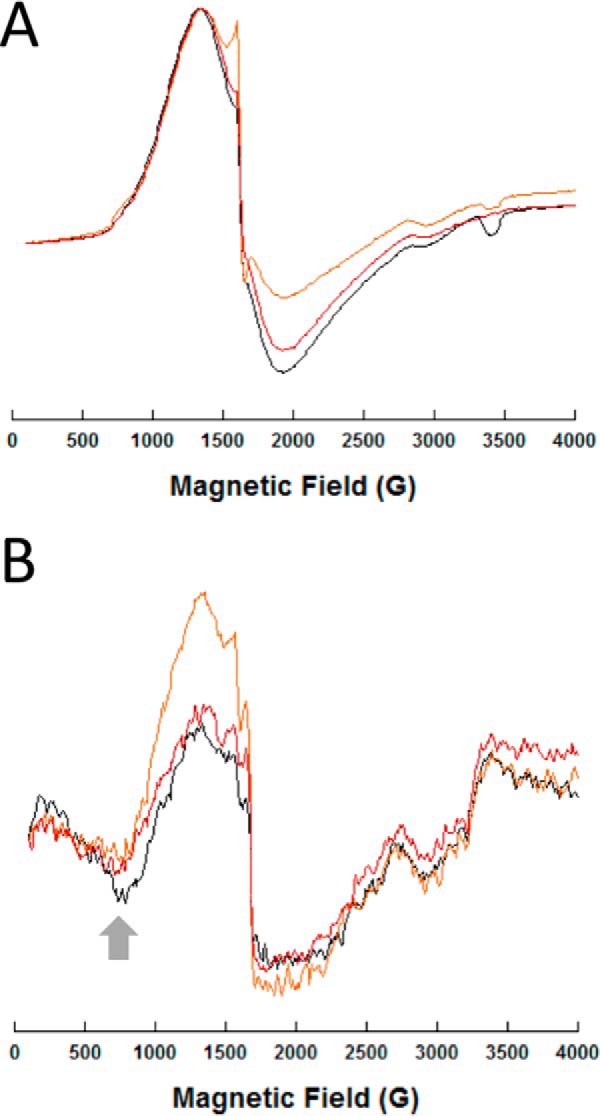
EPR spectra of representative di-cobalt(II) NDM variants. Spectra for all the NDM variants are given in supporting information. Perpendicular (A) and parallel (B) CW-EPR spectra of the metalloforms (0.5 mm), as labeled, are shown. Sharp spikes at 1600 G are due to a minor contamination from iron. The arrow in the right panel (800 G) indicates the feature associated with cobalt(II)–cobalt(II) coupling. NDM-1 is shown in black, NDM-4 in orange, and NDM-15 in red.
Characterization of zinc dependence in NDM variants
A number of the findings described above indicate that clinically-derived mutations of NDM impact structure around the metal-binding sites and that perturbing the metal center (particularly Zn2) impacts function. So, we were led to further characterize the function of NDM variants as a variable of zinc concentration. Previously, we found that ampicillin hydrolysis was quite sensitive to differing zinc concentrations, and fits to this dependence were used to estimate the Kd value of zinc at the Zn2 site (4, 15). We repeated this assay for the NDM variants (Fig. 10, Fig. S9, and Table 4). Because of the low concentrations of enzyme used in the kinetic assays, extensive cuvette washing procedures were introduced here to reduce sources of exogenous zinc ions other than those supplied intentionally NDM-1 showed no activity (within error) at low zinc concentrations (at or lower than 1 nm), indicating that the mono-zinc form (zinc bound at the Zn1 site only) is inactive. Additional zinc raises NDM-1 activity to a maximum value (≈10 μm), with the midpoint of the transition fitted as the Kd value for zinc binding at the Zn2 site (1.0 ± 0.2 μm). NDM-1 also shows a loss of activity at high zinc concentrations (at or lower than 100 μm). This behavior is fitted as described under “Experimental procedures” to give the activity (% of maximum activity for each variant) at low zinc concentrations, which indicates NDM activity in its mono-zinc form, and the midpoint of the first transition, which is assigned as the Kd value for zinc binding to the Zn2 site. The values for mono-zinc activity (%) and Kd, Zn2 were determined for each NDM variant, ranked by zinc affinity, and listed in Table 4. For the NDM variants, affinity for Zn2 appeared to best correlate with resistance. Those variants with no increase or smaller increases in resistance had Kd values close to that of NDM-1, but the variants that show the most resistance have the lowest Kd values, with NDM-12 and NDM-15 binding Zn2 ∼10-fold more tightly than NDM-1. These results are consistent with other reports that NDM variants can confer increased resistance when zinc availability is limited and that bacteria harboring NDM variants with greater zinc affinity can outcompete those carrying NDM with lower affinities when challenged with antibiotic under conditions where zinc is scarce (4, 5).
Figure 10.
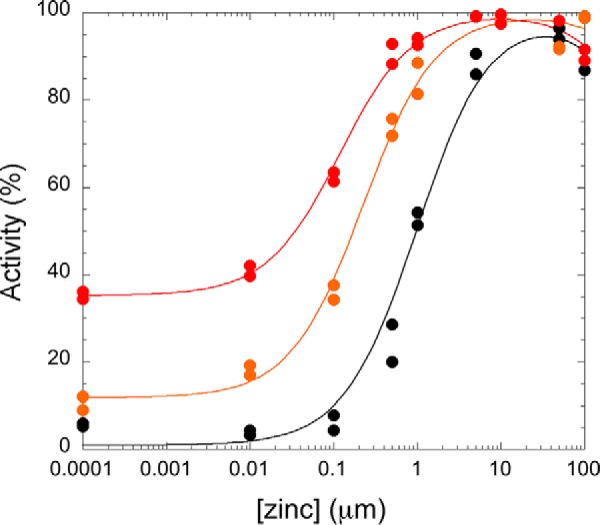
Zinc(II) dependence of representative NDM variants for hydrolysis of ampicillin. Rates of ampicillin hydrolysis were determined with ampicillin (750 μm) as described under “Experimental procedures” and normalized to the largest rate for each variant set to 100%. Representative traces are shown for NDM-1 (black circle), NDM-4 (orange circle), and NDM-15 (red circle), but data for all variants tested are given in Table 4 and Fig. S9. As described under “Experimental procedures,” for each variant the transition is fit as a Kd value for Zn2 and a starting value that describes the relative activity under mono-zinc conditions.
Table 4.
Fits to zinc dependence of NDM variants
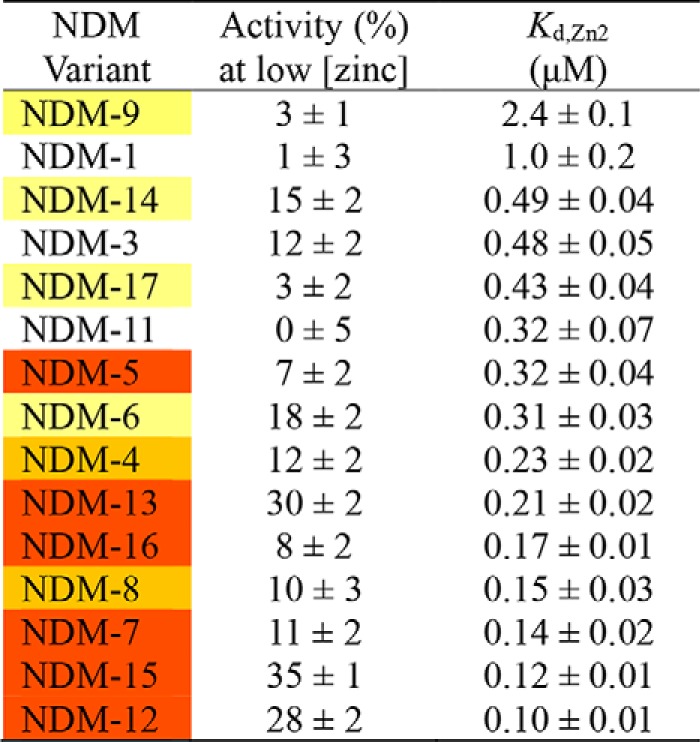
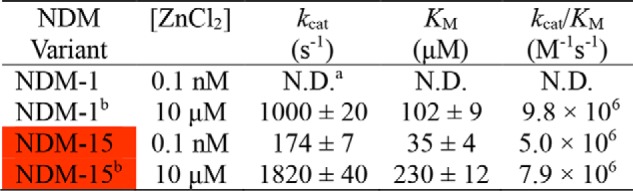
Unexpectedly, we found that some mutations enable NDM to function as a mono-zinc enzyme (Table 4). The steady-state kinetic parameters for NDM-1 and NDM-15 were measured and compared under mono-zinc and di-zinc conditions (Table 5 and Fig. S10). Although NDM-1 does not show appreciable activity under mono-zinc conditions, mono-zinc NDM-15 has high catalytic efficiency, with a kcat/Km value that is only 2-fold less than di-zinc NDM-1. Because Km values are not dependent on enzyme concentration, the observation that the Km values of mono- and di-zinc NDM-15 are significantly different indicates that these are authentically two different forms of the enzyme, rather than just differing amounts of the di-zinc form. Although mono-zinc activity is observed in some of the NDM variants with the highest resistance, the magnitude of activity does not correlate overall to resistance, suggesting that resistance (as measured herein) is conferred more effectively by an increase in Zn2 affinity.
Table 5.
Steady-state kinetic parameters for ampicillin hydrolysis by NDM-1 and NDM-15 under monozinc and dizinc conditions
Although active mono-zinc NDM has not previously been reported, other B1 metallo-β-lactamases are known to function as mono-zinc enzymes, as reviewed previously (9). Our current experiments using NDM variants suggest that the catalytically active zinc ion may reside in the Zn1 site. However, studies of other mono-zinc B1 metallo-β-lactamases suggest migration of the metal ion between Zn1 and Zn2 site is facile, and localization at the Zn2 site may predominate during turnover of some substrates. So further studies will be required to elucidate the specific catalytic mechanism of mono-zinc NDM variants (16, 17).
Conclusions
Our microbial and biochemical characterization of the NDM variants NDM-1 through NDM-17 reveal some shared attributes that suggest common selective pressures are driving the evolution of blaNDM genes in clinical settings. NDM-2 and NDM-10 are excluded from this summary for reasons described earlier. Somewhat counterintuitively, mutations in the NDM variants do not occur at the active site of the enzyme. They do not confer increased resistance to any of the antibiotics tested when zinc is plentiful. They do not improve the ability of di-zinc NDM to bind or to turn over any antibiotic we tested. Nor do they alter inhibitor affinity or induce any large conformational changes in the protein. Their contribution to bacterial resistance, however, is revealed under conditions where zinc availability is limited.
The NDM variants that correlate with increased resistance contain mutations that impact the equilibrium of the NDM protein between its unfolded and folded states. We designed an in vitro experiment to refold denatured NDM in the absence of zinc to mimic the refolding of NDM as it translocates into the periplasm under conditions where zinc is scarce. Notably, all of the NDM variants have an improved refolding efficiency. The variants with higher refolding efficiencies tend to confer more resistance, although there are some exceptions. We also probed the equilibrium between unfolded and folded states by measuring thermostability. Again, all of the NDM variants have improved thermostability and increase the Tm values over an impressive range of 10 °C. Three of the mutations used by NDM variants likely achieve stabilization by either optimizing hydrophobic side-chain packing (M154L), as shown using X-ray crystallography (4), or by burying an additional hydrophobic surface into the core of the protein (A233V and V88L). Other proteins that have evolved increased thermostability have used similar strategies (18). The contribution of these three particular NDM mutations appear to be additive, because the individual contributions of M154L (Tm, NDM-4 − Tm, NDM-1 = 3.9 ± 0.3 °C), A233V (Tm, NDM-6 − Tm, NDM-1 = 1.8 ± 0.3 °C), and V88L (Tm, NDM-5 − Tm, NDM-4 = 3.5 ± 0.2 °C) can account for the thermostability of the triple mutant NDM-16 (Tm, exptl = 9.5 ± 0.3, Tm, calc'd = 9.2 ± 0.8). The other mutations that combine with M154L to improve thermostability of NDM variants occur on the surface of the protein and may improve stability through other mechanisms such as decreasing loop flexibility (G222D) or removing a charge in one of two closely positioned aspartate residues that may have a repulsive interaction with each other (D130N and D95N). The mutations that stabilize the hydrophobic core of NDM, particularly M154L, likely serve as global suppressors that enable the introduction of other mutations, which can add function at the price of structural destabilization. Global suppressing mutations have been previously noted in the evolution of serine β-lactamases (19–21). For example, in the NDM variants, introduction of the E170K mutation decreases Tm by 1.7 ± 0.2 °C (Tm, NDM-17 − Tm, NDM-5), but the stabilizing effect of the other two mutations found in NDM-17 more than compensates to yield an overall thermostability that is still greater than that of NDM-1. The stabilization of the folded form of NDM likely impacts the longevity of these enzymes by resisting thermal denaturation and proteolysis (22), and it may also improve activity by facilitating zinc binding (see below).
We found that the NDM variants that correlate with increased resistance also have altered zinc affinity and perturbed structure at their di-nuclear zinc sites. All but one of the NDM variants has increased Zn2 affinity. These improvements span a 10-fold range, extending Kd, Zn2 into the low nanomolar range. Of all the metrics we tested, the improvement of Kd, Zn2 appears to best correlate with the measured increase in resistance. We note that increased zinc affinity and stabilization of folded NDM are likely not unrelated. Zinc binding stabilizes the folded structure of metallo-β-lactamases in general, and NDM in particular (3, 4, 22). The increased ability of NDM variants to refold in the absence of zinc ions and to maintain their folded structures will increase the concentration of fully-formed zinc-binding sites available to better scavenge zinc ions. However, the effects of the structural changes in these variants are not limited to zinc affinity. NDM variants also contain mutations that impact the kinetic mechanism of the enzyme, that progressively uncouple the two metal-binding sites from one another, and notably that have resulted in some NDM variants that now function effectively as mono-zinc enzymes with catalytic efficiencies similar to that of di-zinc NDM-1. To the best of our knowledge, this is the first report of NDM variants that are active as mono-zinc enzymes. Although the ability to function as a mono-zinc enzyme does not correlate as well as Kd, Zn2 with increased resistance (as measured herein), the ability to function with only 1 eq of zinc will undoubtedly help to maintain resistance in environments with limited zinc availability.
Human pathogens have evolved numerous resistance mechanisms to overcome antibiotic treatment, with the expression of metallo-β-lactamases such as NDM providing just one example of a highly effective resistance determinant. However, antibiotics are not the only existential challenge that pathogens face. The mammalian innate immune system can quickly and effectively respond to a wide range of infectious agents, using as part of this response S100 family proteins to compete with microorganisms by sequestering extracellular zinc in a strategy termed nutritional immunity (23). We reported earlier that two selected NDM variants appeared to have evolved in response to the selective pressure provided by nutritional immunity by increasing the affinity of Zn2 (4). This finding was confirmed independently (5) and was extended to show that additional NDM variants also offered better resistance in conditions of zinc scarcity, and strains expressing some of these variants could outcompete those expressing NDM-1 when challenged with β-lactam antibiotics in media with limited zinc availability. Here, we further extend these and other studies comparing NDM variants (3). Our studies reveal that the mechanisms used to overcome zinc scarcity include optimization of the hydrophobic core of NDM and introduction of small conformational perturbations at the metal center. These mechanisms increase stability, increase Zn2 affinity, and allow NDM to function as a mono-zinc enzyme. Bacteria have evolved a number of different strategies to overcome the challenges of nutritional immunity. Here, the combined selective pressures provided by β-lactam antibiotics and zinc scarcity appear to be driving the evolution of NDM genes in clinical settings.
Experimental procedures
Cloning of blaNDM variants for cell viability assays
The blaNDM-1 gene was cloned from K. pneumoniae 246-61A, a clinical isolate from New Delhi, in 2007 (24). The upstream primer was designed to include the native promoter. Both forward and reverse oligomers included a BamHI restriction site for cloning. The PCR product was first cloned into the PCR XL-TOPO vector (Invitrogen). After verification by sequencing, the XL-TOPO clone was digested with BamHI, and the NDM-1 sequence was inserted into BamHI-digested pHSG-298 vector (Takara, Dalian, China). Coding sequences for NDM variants were generated by site-directed mutagenesis using nonoverlapping primers (IDT, Fig. S2) and the NDM-1/pHSG298 plasmid as template. After PCR, the amplified genes were added directly to a kinase/ligase/DpnI enzyme mix (New England Biolabs) for rapid room temperature circularization and template removal. A 2-μl aliquot of the ligation mixture was used to transform 30 μl of Escherichia coli DH5α chemically competent cells (Lucigen), and the transformation mixtures were spread on lysogeny broth (LB) plates containing 50 μg/ml kanamycin. Plasmid DNA, purified from a single colony, was used to sequence and confirm the insert DNA coding NDM-1 and the native promoter (Eurofins Genomics). Verified NDM variants plasmids were transformed into E. coli DH10B for MIC measurements.
Minimum inhibitory concentration measurements
MIC measurements were performed in triplicate using the Mueller-Hinton (M-H) agar dilution method, according to the Clinical Laboratory and Standards Institute (CLSI) protocol (25). Briefly, bacterial cultures containing blaNDM variants cloned into a uniform vector were grown overnight at 37 °C in M-H broth. The cultures were diluted, and a Steers replicator was used to deliver 10 μl of a diluted overnight culture containing ∼104 CFU. MICs were determined for the following: imipenem, meropenem (Fresnius Kabi), cefepime (WG Critical Care), and ampicillin, cephalothin, ceftazidime, and cefotaxime (Sigma). MICs were also determined (as above) with the addition of 50 μm EDTA (United States Biochemical Corp.) to the M-H agar to limit zinc(II) availability.
Cloning, overexpression, and purification of NDM variants
A codon-optimized NDM-1 sequence (residues 36–270) (15) fused with an N-terminal tobacco etch virus (TEV) cleavage site was synthesized by Genscript Biotech Corp. The synthesized fragment was subcloned into pET15b between NdeI and XhoI. NDM variants were generated by site-directed mutagenesis using nonoverlapping primers (IDT, Fig. S2) and pET15b–His6–TEV–NDM-1 plasmid as template. After PCR, the amplified genes were added directly to a kinase/ligase/DpnI enzyme mix (New England Biolabs) for rapid room temperature circularization and template removal. A 2-μl aliquot of the ligation mixture was used to transform 30 μl of E. coli DH5α chemically competent cells (Lucigen), and the transformation mixtures were spread into lysogeny broth (LB) plates containing 100 μg/ml carbenicillin. Plasmid DNA, purified from a single colony, was used to sequence and confirm the inserted sequence (Eurofins Genomics).
Plasmids containing blaNDM variants as identified by sequencing, were transformed into E. coli EXPRESS BL21(DE3) chemically competent cells (Lucigen). A single colony was transferred into 50 ml of LB containing 100 μg/ml carbenicillin, and the culture was shaken overnight at 37 °C. The overnight culture (10 ml) was transferred into four flasks with each containing 1 liter of LB with 100 μg/ml carbenicillin. The resulting culture was grown at 37 °C with a shaking speed of 220 rpm until an A600 of 0.6 was reached. Protein production was induced by adding ZnCl2 (100 μm) and isopropyl 1-thio-β-d-galactopyranoside (0.5 mm) to each culture. The cell cultures were shaken overnight at 18 °C and then were harvested by centrifugation for 10 min at 8000 × g. The resulting pellets were resuspended in 50 ml of 50 mm HEPES, pH 7.5, containing 500 mm NaCl. Cells were lysed by passing the mixture two times through a French press at a pressure between 15,000–20,000 p.s.i. The insoluble components were removed after pelleting in a centrifuge for 1 h at 32,000 × g. The retained supernatant was dialyzed versus 1 liter of 50 mm HEPES, pH 7.5, containing 500 mm NaCl and 100 μm ZnCl2 overnight and loaded onto a HisTrap HP (5 ml, GE Healthcare) column. The resulting column was washed with 10 volumes of Wash Buffer (50 mm HEPES, pH 7.5, 500 mm NaCl, and 50 mm imidazole). Bound proteins were eluted using Elution Buffer (Wash Buffer with 500 mm imidazole). TEV protease was added at a protease/target protein ratio of 1:100 (w/w), and the mixture was dialyzed overnight against 1 liter of 50 mm HEPES, pH 7.5, containing 500 mm NaCl. Truncated NDM variants were then re-purified by passage through a HisTrap column, which removed the His-TEV and cleaved His-tag peptide, with purified NDM exiting the column in the flow-through fractions.
Purified, recombinant NDM variants (before and after TEV digestion) were analyzed by SDS-PAGE using Fisher EZ-Gel solution (12.5%), a Mini-PROTEAN System (Bio-Rad), and Coomassie Blue stain. Protein concentrations were determined by UV-visible absorbance at 280 nm using the calculated data: ϵ280 nm = 29,450 m−1 cm−1 for variants containing the poly-His tag and ϵ280 nm = 27,960 m−1 cm−1 for the NDM variants without the poly-His tag.
CD
Far-UV CD spectroscopy was performed using a AVIV Model 435 circular dichroism spectrometer (Lakewood, NJ). Spectra were obtained between 185 and 260 nm, with sampling every 1000 nm at 25 °C. Four scans were taken with an average time of 1.0 s per nm with a wait time of 1.0 s between scans. A quartz Suprasil cuvette (Hellma) with a 1-mm path length was used for each sample with a protein concentration of 4 μm in 20 mm potassium phosphate, pH 7.0. The four scans were averaged and exported to Kaleidagraph (Synergy) where the baseline was subtracted. The ratio of α-helices and β-sheets was determined using the on-line program K2D2 (26).
Fluorescence spectroscopy
Fluorescence emission spectra of NDM variants (2 μm protein concentration in 2 ml of 50 mm HEPES, pH 7.5) were obtained on a luminescence spectrometer (model LS-55, PerkinElmer Life Sciences), with an excitation wavelength of 280 nm and an emission spectrum from 300 to 400 nm. The relative tryptophan fluorescence intensity (at 345 nm) was calculated setting the intensity of the buffer to 0% and that of NDM-1 to 100%.
Metal analyses
The zinc content of the NDM variants was determined using a Optima 7300V inductively coupled plasma spectrometer with optical emission spectroscopy (PerkinElmer Life Sciences) detection. Protein samples, used without modification after purification, were diluted to 6 μm with 50 mm HEPES, pH 7.5. Calibration curves were generated using serial dilutions of the Fisher zinc metal standard ranging from 0 to 16 μm, and the emission line at 202.548 nm was chosen for zinc.
Steady-state kinetics
Steady-state kinetic studies were performed using three substrates, chromacef, meropenem, and ampicillin, adjusting substrate concentration to bracket Km values, and enzyme concentration to maintain linearity during the assay time scale. For chromacef, disposable polystyrene cuvettes were used with the final reaction volume of 1 ml. For this substrate, we used concentrations in the range of 0.2–10 μm and enzyme (NDM variants) in the range of 0.75–20 nm. For meropenem and ampicillin, quartz microcuvettes were used for the assay, with a final assay volume of 0.3 ml. We used ampicillin concentrations between 25 and 1000 μm and enzyme concentrations between 5 and 40 nm. Meropenem concentrations were between 10 and 300 μm with enzyme concentrations between 2.5 and 20 nm. Absorbance changes due to either the disappearance of the substrate (meropenem at 300 nm or ampicillin at 235 nm) or the appearance of product (chromacef at 442 nm) were monitored for 0.3 min at room temperature. Assay Buffer contained 50 mm HEPES, pH 7, and 10 μm ZnSO4. The steady-state catalytic parameters Km and kcat values for each substrate/NDM variant pair were derived by fitting the concentration dependence of initial rate measurements to the Michaelis-Menten equation using KaleidaGraph.
Zinc concentration dependence of NDM-catalyzed ampicillin hydrolysis
Two quartz microcuvettes were soaked in diluted detergent overnight and extensively washed and rinsed with deionized water and ethanol before use. After each series of repeated rinses, an aliquot of NDM-1 was assayed for ampicillin hydrolysis (see below for conditions) multiple times in each cuvette without any addition of ZnSO4 until use of the washed cuvette resulted in a constant minimal value. This procedure was repeated before assaying each NDM variant, and for each variant, zinc concentrations were tested from lower to higher concentrations to avoid contamination by excess zinc ion. The final assay solution used to monitor ampicillin hydrolysis (as described above) contained 0.3 ml of 50 mm HEPES, pH 7, 0.6 mm ampicillin, 20 nm NDM variant, with addition of ZnSO4 to the final concentrations of 0.0001, 0.01, 0.1, 0.5, 1, 5, 10, 50, and 100 μm. Observed rates were converted into percentages, using the highest value for each variant as 100%, as described earlier (4, 15). Because most NDM variants showed increasing activity followed by a loss of activity as zinc concentrations were increased, data were fitted using Equation 1,
| (Eq. 1) |
with [zinc] being the concentration of ZnSO4, x being the activity (%) at no added zinc, y being the Kd, Zn2, and z being Kd value for binding an inhibitory zinc at high concentrations. Concentrations of ZnSO4 of >100 μm were not tested, so reliable fits to z were not determined, as they are likely not relevant.
Pre-steady–state kinetics
Stopped-flow UV-visible spectra, using diode array detection of chromacef hydrolyzed by NDM variants, were collected on an Applied Photophysics SX 20 spectrophotometer equipped with a 2-mm path length optical cell. The buffer used in this study was the same buffer used in the steady-state kinetic studies. The temperature was kept constant at 22 °C using a circulating water bath. Absorbance data were converted to concentration data using the following extinction coefficients: substrate Δϵ378 nm = 22,200 m−1 cm−1, product Δϵ44 nm = 18,600 m−1 cm−1, and intermediate Δϵ575 nm = 2200 m−1 cm−1 (27).
DSF
Purified NDM variants were diluted to a final concentration of 5 μm with 20 mm HEPES, pH 7.5, containing 150 mm NaCl. A 5000× SYPRO Orange fluorescent dye stock (Pierce Thermo Inc.) was added to a final concentration of 10×. A Bio-Rad CFX96 RT-PCR instrument was set to the preset channel “HEX” for fluorescent excitation and emission. DSF data were collected in triplicate in a 96-well Frame Star PCR plate, which was covered with a clear thermal-seal film. The temperature was increased in 0.5 °C increments, followed by a 5-s hold at each temperature for equilibration. Fluorescence of SYPRO Orange was monitored at 570 nm. Fluorescence intensity data were fitted to a Boltzmann Sigmoidal curve using Prism 6 (GraphPad software) to determine melting temperatures (Tm) for each NDM variant.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were carried out using a Nano ITC System (TA Instruments, Waters LLC) with a 500-μl cell. All experiments were performed at 25 °C. NDM variants were prepared by diluting the purified stock solutions with 20 mm HEPES, pH 7.5, containing 150 mm NaCl. l-Captopril (Acros Organics) was dissolved in the same buffer. The ITC cell was filled with 50 μm of each NDM variant (300 μl), and the enzyme solutions were titrated with 500 μm l-captopril. The injection volume was 50 μl, and the time between two injections was 210 s, allowing the inhibitor to bind. Kd values were determined by fitting using NanoAnalyze (TA Instruments, Waters LLC).
Preparation of metal-free NDM variants
Metal-free forms of the NDM variants were prepared through a process of metal chelation, enzyme denaturation, and enzyme refolding (4). Each purified protein (diluted to 350 μm in 10 ml) was placed in a 10-kDa MWCO dialysis bag. These samples were dialyzed versus dialysis buffers for 4 h at 4 °C before changing to the subsequent dialysis buffer to include a series of 11 dialysis steps as follows: first, four changes of Buffer A (50 mm HEPES, pH 6.8, containing 150 mm NaCl) with EDTA (2 mm), then urea (6 m, 0.5 liters), and finally six changes of Buffer A. The resulting metal-free enzymes were analyzed by using SDS-PAGE and UV-visible spectroscopy. The refolding efficiency for the variants was estimated by dividing the protein concentration after refolding by the protein concentration before refolding.
UV-visible spectroscopy
Metal-free NDM variants were diluted to 300 μm with 50 mm HEPES, pH 6.8, containing 150 mm NaCl. Two molar equivalents of CoCl2 were added to the protein samples from a 50 mm stock solution. The resulting di-Co(II)-substituted NDM variants were placed in a 500-μl quartz cuvette, and UV-visible spectra were collected on a Lambda 750 UV-visible NIR spectrometer (PerkinElmer Life Sciences) at 25 °C. A blank spectrum of 50 mm HEPES, pH 6.8, containing 150 mm NaCl was used to generate difference spectra. Because of varying levels of precipitation of the various mutants, we were unable to reliably normalize the spectra and quantitatively obtain extinction coefficients. However, as we felt it was important these data be scaled appropriately, we normalized all of the NDM variants to match A280 of NDM-1, which showed the least precipitation. As a consequence, the spectra presented in Fig. 7 can be compared directly to each other but not to extinction coefficients previously reported by us and others for other MBLs.
1H NMR spectroscopy
Metal-free NDM variants were concentrated to ∼1 mm with an Amicon Ultra-4 centrifugal unit and an Ultracel-10 membrane. To the concentrated protein, 2 eq of CoCl2 were added. Each NMR sample was buffered with 50 mm HEPES, pH 6.8, containing 150 mm NaCl and 10% D2O. Spectra were collected at 292 K on a Bruker ASX300 (BBI probe) NMR spectrometer operating at a frequency of 300.16 MHz. Spectra were collected using a frequency switching method, applying a long-low power (270 ms) pulse centered at the water frequency, followed by a high power 3-μs pulse centered at 90 ppm (28). This method allowed for suppression of the water signal with enhancement of severely hyperfine-shifted resonances. Spectra consisted of 30,000 transients of 10,000 data points over a 333 ppm spectral window (taq ∼51 ms). Signal averaging took ∼3 h per spectrum.
EPR spectroscopy
EPR samples containing NDM variants included ∼10% (v/v) glycerol as glassing agent. Samples were loaded into EPR tubes and degassed by repeated evacuation/purgation with N2 prior to data collection. Spectra were collected on a Bruker EMX EPR spectrometer, equipped with an ER4116-DM dual mode resonator (9.37 GHz, parallel; 9.62 GHz perpendicular). The data in EPR figures were scaled so that the x axes matched (perpendicular mode field values were scaled by 9.37/9.62). Temperature control was accomplished using an Oxford ESR900 cryostat and temperature controller (4.5 K). Other spectral conditions included the following: microwave power = 0.2 milliwatt; field modulation = 10 G (100 kHz); receiver gain = 104; time constant/conversion time = 41 ms.
Author contributions
Z. C., D. L. T., R. C. P., W. F., and M. W. C. conceptualization; Z. C., P. W. T., L. J., A. B., K. M., D. C., C. M., C. R. B., and J. V. formal analysis; Z. C., P. W. T., L. J., A. B., K. M., D. C., C. M., C. R. B., J. V., and W. F. investigation; Z. C. methodology; Z. C., L. J., A. B., D. L. T., R. C. P., W. F., and M. W. C. writing-original draft; Z. C., A. B., D. L. T., R. C. P., R. A. B., W. F., and M. W. C. writing-review and editing; D. L. T., R. C. P., W. F., and M. W. C. supervision; D. L. T., R. C. P., R. A. B., W. F., and M. W. C. funding acquisition; D. L. T., R. C. P., R. A. B., W. F., and M. W. C. project administration.
Supplementary Material
This work was supported in part by National Institutes of Health Grants GM111926 (to W. F., R. A. B., M. W. C., D. L. T., and R. P.), R01AI100560 (to R. A. B.), R01AI063517 (to R. A. B.), and R01AI072219 (to R. A. B.) from NIGMS and NIAID, National Science Foundation Grant CHE-1509285 (to M. W. C. and D. L. T.), Robert A. Welch Foundation Award F-1572 (to W. F.), Miami University through the Robert H. and Nancy J. Blayney Professorship (to R. C. P.), funds and/or facilities provided by the Cleveland Department of Veterans Affairs Award 1I01BX001974 (to R. A. B.), the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10 (to R. A. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S10, Tables S1–S5, and supporting Refs. 1–6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- NDM
- New Delhi metallo-β-lactamase
- ITC
- isothermal titration calorimetry
- MIC
- minimum inhibitory concentration
- TEV
- tobacco etch virus
- VIM
- Verona integrin-borne metallo-β-lactamase
- IMP
- imipenemase
- DSF
- differential scanning fluorimetry
- MBL
- metallo-β-lactamase.
References
- 1. Solomon S. L., and Oliver K. B. (2014) Antibiotic resistance threats in the United States: stepping back from the brink. Am. Fam. Physician 89, 938–941 [PubMed] [Google Scholar]
- 2. Woodworth K. R., Walters M. S., Weiner L. M., Edwards J., Brown A. C., Huang J. Y., Malik S., Slayton R. B., Paul P., Capers C., Kainer M. A., Wilde N., Shugart A., Mahon G., Kallen A. J., Patel J., McDonald L. C., Srinivasan A., Craig M., and Cardo D. M. (2018) Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms–United States, 2006–2017. MMWR Morb. Mortal. Wkly Rep. 67, 396–401 10.15585/mmwr.mm6713e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Makena A., Brem J., Pfeffer I., Geffen R. E., Wilkins S. E., Tarhonskaya H., Flashman E., Phee L. M., Wareham D. W., and Schofield C. J. (2015) Biochemical characterization of New Delhi metallo-β-lactamase variants reveals differences in protein stability. J. Antimicrob. Chemother. 70, 463–469 10.1093/jac/dku403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stewart A. C., Bethel C. R., VanPelt J., Bergstrom A., Cheng Z., Miller C. G., Williams C., Poth R., Morris M., Lahey O., Nix J. C., Tierney D. L., Page R. C., Crowder M. W., Bonomo R. A., and Fast W. (2017) Clinical variants of New Delhi metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect. Dis. 3, 927–940 10.1021/acsinfecdis.7b00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bahr G., Vitor-Horen L., Bethel C. R., Bonomo R. A., Gonzalez L. J., and Vila A. J. (2018) Clinical evolution of New Delhi metallo-β-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob. Agents Chemother. e01849–17 10.1128/AAC.01849-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King D. T., Worrall L. J., Gruninger R., and Strynadka N. C. (2012) New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J. Am. Chem. Soc. 134, 11362–11365 10.1021/ja303579d [DOI] [PubMed] [Google Scholar]
- 7. Chen X., Li L., Chen S., Xu Y., Xia Q., Guo Y., Liu X., Tang Y., Zhang T., Chen Y., Yang C., and Shui W. (2013) Identification of inhibitors of the antibiotic-resistance target New Delhi metallo-β-lactamase 1 by both nanoelectrospray ionization mass spectrometry and ultrafiltration liquid chromatography/mass spectrometry approaches. Anal. Chem. 85, 7957–7965 10.1021/ac401732d [DOI] [PubMed] [Google Scholar]
- 8. Makena A., Düzgün A. Ö., Brem J., McDonough M. A., Rydzik A. M., Abboud M. I., Saral A., Čiček A. Č., Sandalli C., and Schofield C. J. (2015) Comparison of Verona Integron-Borne metallo-β-lactamase (VIM) variants reveals differences in stability and inhibition profiles. Antimicrob. Agents Chemother. 60, 1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crowder M. W., Spencer J., and Vila A. J. (2006) Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chemical Res. 39, 721–728 10.1021/ar0400241 [DOI] [PubMed] [Google Scholar]
- 10. Wang Z., Fast W., and Benkovic S. J. (1998) Direct observation of an enzyme-bound intermediate in the catalytic cycle of the metallo-β-lactamase from Bacteroides fragilis. J. Am. Chem. Soc. 120, 10788–10789 10.1021/ja982621m [DOI] [Google Scholar]
- 11. Yang H., Young H., Yu S., Sutton L., and Crowder M. W. (2014) Targeting metallo-carbapenemases via modulation of electronic properties of cephalosporins. Biochem. J. 464, 271–279 10.1042/BJ20140364 [DOI] [PubMed] [Google Scholar]
- 12. Teale F. W. (1960) The ultraviolet fluorescence of proteins in neutral solution. Biochem. J. 76, 381–388 10.1042/bj0760381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang H., Aitha M., Marts A. R., Hetrick A., Bennett B., Crowder M. W., and Tierney D. L. (2014) Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-β-lactamase NDM-1. J. Am. Chem. Soc. 136, 7273–7285 10.1021/ja410376s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Orellano E. G., Girardini J. E., Cricco J. A., Ceccarelli E. A., and Vila A. J. (1998) Spectroscopic characterization of a binuclear metal site in Bacillus cereus β-lactamase II. Biochemistry 37, 10173–10180 10.1021/bi980309j [DOI] [PubMed] [Google Scholar]
- 15. Thomas P. W., Zheng M., Wu S., Guo H., Liu D., Xu D., and Fast W. (2011) Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry 50, 10102–10113 10.1021/bi201449r [DOI] [PubMed] [Google Scholar]
- 16. Hemmingsen L., Damblon C., Antony J., Jensen M., Adolph H. W., Wommer S., Roberts G. C., and Bauer R. (2001) Dynamics of mononuclear cadmium β-lactamase revealed by the combination of NMR and PAC spectroscopy. J. Am. Chem. Soc. 123, 10329–10335 10.1021/ja0112240 [DOI] [PubMed] [Google Scholar]
- 17. Llarrull L. I., Tioni M. F., and Vila A. J. (2008) Metal content and localization during turnover in B. cereus metallo-β-lactamase. J. Am. Chem. Soc. 130, 15842–15851 10.1021/ja801168r [DOI] [PubMed] [Google Scholar]
- 18. Goldenzweig A., and Fleishman S. (2018) Principles of protein stability and their application in computational design. Annu. Rev. Biochem. 87, 105–129 10.1146/annurev-biochem-062917-012102 [DOI] [PubMed] [Google Scholar]
- 19. Huang W., and Palzkill T. (1997) A natural polymorphism in beta-lactamase is a global suppressor. Proc. Natl. Acad. Sci. U.S.A. 94, 8801–8806 10.1073/pnas.94.16.8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel M. P., Fryszczyn B. G., and Palzkill T. (2015) Characterization of the global stabilizing substitution A77V and its role in the evolution of CTX-M β-lactamases. Antimicrob. Agents Chemother. 59, 6741–6748 10.1128/AAC.00618-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown N. G., Pennington J. M., Huang W., Ayvaz T., and Palzkill T. (2010) Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases. J. Mol. Biol. 404, 832–846 10.1016/j.jmb.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. González L. J., Bahr G., Nakashige T. G., Nolan E. M., Bonomo R. A., and Vila A. J. (2016) Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat. Chem. Biol. 12, 516–522 10.1038/nchembio.2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cunden L. S., and Nolan E. M. (2018) Bioinorganic explorations of Zn(II) sequestration by human S100 host-defense proteins. Biochemistry 57, 1673–1680 10.1021/acs.biochem.7b01305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castanheira M., Deshpande L. M., Mathai D., Bell J. M., Jones R. N., and Mendes R. E. (2011) Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother. 55, 1274–1278 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. CLSI (2015) Performance Standards for Antimicrobial Susceptibility Testing. 24th Informational Supplement Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 26. Perez-Iratxeta C., and Andrade-Navarro M. A. (2008) K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 8, 25 10.1186/1472-6807-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang H., Aitha M., Hetrick A. M., Richmond T. K., Tierney D. L., and Crowder M. W. (2012) Mechanistic and spectroscopic studies of metallo-β-lactamase NDM-1. Biochemistry 51, 3839–3847 10.1021/bi300056y [DOI] [PubMed] [Google Scholar]
- 28. Riley E. A., Petros A. K., Smith K. A., Gibney B. R., and Tierney D. L. (2006) Frequency-switching inversion-recovery for severely hyperfine-shifted NMR: evidence of asymmetric electron relaxation in high-spin Co(II). Inorg. Chem. 45, 10016–10018 10.1021/ic061207h [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.