Cryo-EM structures of the epithelial calcium channel TRPV6-calmodulin complex reveal a mechanism of Ca2+-induced inactivation.
Abstract
Calcium (Ca2+) plays a major role in numerous physiological processes. Ca2+ homeostasis is tightly controlled by ion channels, the aberrant regulation of which results in various diseases including cancers. Calmodulin (CaM)–mediated Ca2+-induced inactivation is an ion channel regulatory mechanism that protects cells against the toxic effects of Ca2+ overload. We used cryo-electron microscopy to capture the epithelial calcium channel TRPV6 (transient receptor potential vanilloid subfamily member 6) inactivated by CaM. The TRPV6-CaM complex exhibits 1:1 stoichiometry; one TRPV6 tetramer binds both CaM lobes, which adopt a distinct head-to-tail arrangement. The CaM carboxyl-terminal lobe plugs the channel through a unique cation-π interaction by inserting the side chain of lysine K115 into a tetra-tryptophan cage at the pore’s intracellular entrance. We propose a mechanism of CaM-mediated Ca2+-induced inactivation that can be explored for therapeutic design.
INTRODUCTION
Calcium (Ca2+) acts as a cellular second messenger and regulates numerous physiological processes including neuronal excitability, muscle contraction, bone formation, enzymatic activity, and cell proliferation and apoptosis (1, 2). Ion channels mediate many of these processes that, if misregulated, can result in altered Ca2+ homeostasis and a variety of diseases, including cancers (1). Ca2+-induced inactivation is a common regulatory mechanism for different ion channel families, including voltage-gated potassium (2), sodium (3), calcium (4, 5), cyclic nucleotide–gated (6), ryanodine (7), and ionotropic glutamate receptor channels (8). The influx of calcium into a cell often initiates a negative feedback mechanism that inactivates Ca2+-permeable ion channels to protect cells from calcium overload. In many cases, Ca2+-induced inactivation directly uses the universal calcium sensor calmodulin (CaM) (9), but the structural mechanisms of CaM-dependent Ca2+-induced inactivation remain largely unknown. Here, we studied such a mechanism using the transient receptor potential (TRP) channel TRPV6 (TRP vanilloid subfamily member 6).
TRPV6 and its close relative TRPV5 (~75% sequence identity) are highly Ca2+-selective (10), function as calcium uptake channels in epithelial tissues, and undergo Ca2+-induced inactivation through direct interaction with CaM (11, 12). We determined the structures of human TRPV6 (hTRPV6) and rat TRPV6 (rTRPV6) in complex with CaM using cryo-electron microscopy (cryo-EM). On the basis of our structures, we propose a mechanism of CaM-dependent Ca2+-induced inactivation that can be explored for therapeutic design.
RESULTS AND DISCUSSION
Cryo-EM structure of TRPV6-CAM inactivation complex
We have previously solved the structure of full-length hTRPV6 in the open, conducting state (13). To study the structural basis of CaM-dependent Ca2+-induced inactivation of TRPV6, we purified full-length hTRPV6 in the same manner as was done previously, but in the presence of human CaM that was expressed separately in bacteria and purified using affinity and size exclusion chromatography (see Materials and Methods). Subjected to single-particle cryo-EM, this mixture yielded diverse particle orientations and resolved secondary structure elements in two-dimensional (2D) class averages (fig. S1, A and B). Three-dimensional classification of particles resulted in a single predominant 3D class that was refined to a resolution of 4.0 Å, with no symmetry applied (fig. S1, C to H). Compared to the previous hTRPV6 reconstruction (13), the presence of CaM in our cryo-EM density was obvious (fig. S2, A and B), and the quality of the map (fig. S3) allowed the building of the entire hTRPV6-CaM complex guided by available structures of hTRPV6 (13) and CaM (14).
Our hTRPV6-CaM reconstruction and resulting structure demonstrate that one hTRPV6 tetramer binds one two-lobe CaM molecule (Fig. 1, A to D). CaM docks into a ~50 Å × 50 Å cavity underneath the channel that is enclosed by an intracellular skirt ~110 Å wide, which is composed mostly of ankyrin repeat domains (ARDs). The cavity is large enough for CaM to fit almost entirely into and be hardly visible from the side (Fig. 1, A and B). CaM adopts an unusual head-to-tail arrangement for its N- and C-terminal lobes (Fig. 1E and fig. S4). The highly flexible region, which connects the N- and C-terminal lobes (residues 75 to 83) (15) and forms a linker helix (LH)in the canonical dumbbell conformation of CaM (16), folds into two helices (h4 and h5) connected by a loop. The N- and C-terminal lobes of CaM bind to the proximal and distal portions of the hTRPV6 C terminus, respectively.
Fig. 1. Structure of the hTRPV6-CaM complex.
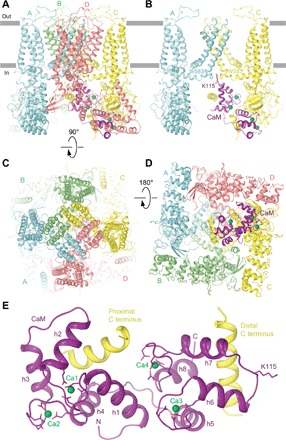
(A to D) Side (A and B), top (C), and bottom (D) views of hTRPV6-CaM with hTRPV6 subunits (A to D) colored cyan, green, yellow, and pink, and CaM colored purple. Calcium ions are shown as green spheres. In (B), only two of four hTRPV6 subunits are shown, with the front and back subunits removed for clarity. Side chains of TRPV6 residue W583 and CaM residue K115 are shown as sticks. (E) Expanded view of CaM bound to the proximal and distal portions of the TRPV6 C terminus. Side chains of CaM residues coordinating calcium ions in the EF hand motifs and K115 are shown as sticks.
We also expressed, purified, and solved the structure of full-length rTRPV6 using cryo-EM (fig. S5). Although we did not add CaM to the purified rTRPV6 sample, we observed strong CaM density in the rTRPV6 3D reconstruction, which was very similar to the CaM density in the hTRPV6-CaM reconstruction (fig. S2, B and C) and which was not observed in a C-terminally truncated rTRPV6 reconstruction or electron density (13, 17). We therefore concluded that our full-length rTRPV6 reconstruction revealed rTRPV6 in complex with endogenous CaM. At an overall resolution of 3.6 Å, the aforementioned cryo-EM map exhibited excellent density for the majority of amino acid side chains (fig. S6) and allowed for the building of a high-quality rTRPV6-CaM model (table S1). The rTRPV6-CaM model was nearly identical to the hTRPV6-CaM model (root mean square deviation = 0.964 Å2; fig. S7), suggesting that both structures represent the same functional state. Given its higher overall resolution, we used the rTRPV6-CaM structure to characterize the binding interfaces between TRPV6 and CaM (Fig. 2).
Fig. 2. Contact interfaces between CaM and TRPV6.
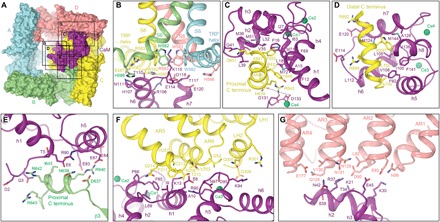
(A) Surface representation of rTRPV6-CaM viewed intracellularly with TRPV6 subunits and CaM colored similarly to Fig. 1. Boxes indicate rTRPV6-CaM interfaces expanded in (B) to (G). (B to G) Interfaces between CaM and different regions of rTRPV6, with residues involved in rTRPV6-CaM binding shown as sticks and calcium ions as green spheres.
TRPV6-CAM inactivation complex stabilized by multiple surfaces
Previous studies identified multiple CaM-binding sites on TRPV5/6, including regions of the ankyrin repeat, transmembrane, and C-terminal domains (11, 12, 18–22), and have suggested a stoichiometry of one CaM molecule per TRPV6 tetramer (18, 23) and two CaM molecules per TRPV5 tetramer (24). These studies have also proposed that CaM binds TRPV6 via two binding motifs, a 1-5-10 motif in the ARD (residues 88-92-97) and a 1-8-14 motif in the distal C terminus (residues 643-650-656). In contrast, our TRPV6-CaM structures reveal six surface regions of TRPV6 that interact with the two lobes of CaM (Fig. 2A). In the center of the first region is lysine K115, which protrudes from a loop between helices h6 and h7 of the C-terminal lobe of CaM and makes contact with all four TRPV6 subunits by sticking its side chain into the intracellular orifice of the ion channel pore formed by four tryptophans (W582 in rTRPV6 and W583 in hTRPV6), one from each TRPV6 subunit at the S6 bundle crossing (Fig. 2B). These tryptophans are highly conserved in TRPV6 and TRPV5 (fig. S8) and trap the side chain of K115 by forming a tight square cage with 4.2 Å between the plane of each indole ring and the ammonium group of K115. This tryptophan cage provides a unique environment for an atypically strong cation-π interaction between the π system of four tryptophan indole rings and the positively charged ammonium group of lysine and has not been observed in any other protein structure. Earlier studies that analyzed interactions of CaM with isolated peptides from the distal C terminus of either TRPV6 or TRPV5 suggested that K115 is involved in CaM binding to the channel (12). Although the peptides used in these studies did not include W583, their results support the role of K115 in CaM-protein interactions. The importance of W583 for CaM-dependent Ca2+-induced inactivation was demonstrated in experiments with TRPV5, where mutation of the tryptophan homologous to W583 to alanine induced cell death due to increased calcium influx (25). Several other charged and polar residues in the h5-h6 loop of CaM, including T117, which was previously implicated in CaM-TRPV6 binding (12), and in the TRP helices of TRPV6 further strengthen the TRPV6-CaM interface in close proximity to K115 (Fig. 2B).
The second TRPV6-CaM interface is formed between the N-terminal lobe of CaM and the proximal α-helical portion of the TRPV6 C terminus of subunit C (residues 639 to 651) via its 1-5 motif (residues 646–650, signature sequence YxxxF) (Fig. 2C). The third interface is formed between the C-terminal lobe of CaM and the distal α-helical portion of the TRPV6 C terminus of subunit C (residues 692 to 704) via its 1-4 motif (residues 697–700, signature sequence WxxL) (Fig. 2B). Previous studies have demonstrated the importance of W697 for TRPV6-CaM binding and CaM-dependent Ca2+-induced inactivation of TRPV6 (18, 26). In addition, R701, the residue that follows the 1-4 motif in TRPV6 and was previously implicated in TRPV6 inactivation (12), is in close proximity to the C-terminal tail of CaM (residues 145 to 148) and might be involved in polar interactions with it. The proximal and distal portions of the TRPV6 C terminus were not visible in TRPV6 structures without CaM (13, 17), emphasizing their unique role in CaM-dependent Ca2+-induced inactivation. It is possible that these regions adopt α-helical conformations only upon CaM binding. Supporting this hypothesis, the C termini of subunits A and D in the TRPV6-CaM structures are also invisible, while the proximal portion of the C terminus of subunit B has a helical conformation (Fig. 2E) that is different from the corresponding region of subunit C (Fig. 2C and fig. S9). Therefore, the proximal portion of the C terminus of subunit B, which forms the fourth binding interface between TRPV6 and CaM, adjusts its conformation, depending on its chemical environment.
The fifth and sixth binding interfaces include the intracellular skirt domains AR5-AR6 and the LHs 1-2 of TRPV6 subunit C (Fig. 2F), and AR1-AR4 of subunit D (Fig. 2G). Together, all six interfaces between TRPV6 and CaM cover a surface area of 3176 Å2. Despite the large size of the TRPV6-CaM interfaces, their interactions seem to be rather weak and allow mobility of CaM with respect to TRPV6. Supporting this conclusion, the density for CaM in both the hTRPV6-CaM and rTRPV6-CaM structures is of lower resolution compared to the channel core (figs. S1 and S5). An exception to the generally weaker density for CaM is the portion from which K115 projects and seems to bind extremely tightly at the intracellular entrance to the TRPV6 pore (figs. S2, S3, and S6).
Distinct pore architecture of the TRPV6-CAM complex
The hTRPV6-CaM and rTRPV6-CaM structures reveal a distinct TRPV6 pore architecture (Fig. 3A). Comparing the radius of the hTRPV6 pore in the hTRPV6-CaM structure to the previously solved TRPV6 structures in the closed and open states (13), it becomes obvious that the pore architecture is different in all three states (Fig. 3B). Remarkably, despite their differences, the selectivity filter of the TRPV6 pore in the inactivated state is similar to that of the closed and open states (fig. S10), strongly supporting its exclusive role in ion permeation (13, 17). On the other hand, the pore profile at the ion channel gate region of the inactivated state is distinct from those of the closed and open states of TRPV6. The narrowest pore constriction of the hTRPV6-CaM inactivated state is similar in diameter to that of the hTRPV6 closed state except that the constriction is formed more extracellularly, by I575, instead of by M578 in the closed state. Because of the aliphatic nature of the isoleucine side chain, this narrow constriction creates a hydrophobic seal impermeable to cations. CaM also directly prevents permeation through the TRPV6-CaM pore, which plugs the pore by inserting the side chain of K115 into the intracellular pore orifice. Accordingly, the tryptophans that form the cage surrounding K115 get pulled toward the center of the pore, making the radius of the pore at its intracellular entrance smaller in the inactivated state than in the closed and open states (Fig. 3B).
Fig. 3. Inactivated TRPV6 pore.
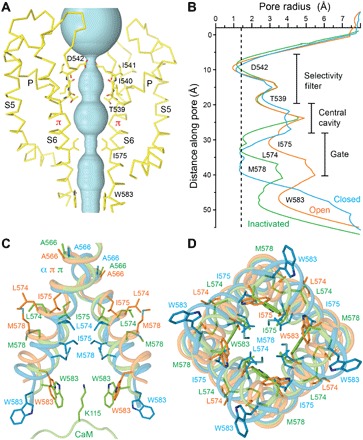
(A) Ion conduction pathway (cyan) in hTRPV6-CaM, with residues lining the selectivity filter and around the gate shown as sticks. Only two of four hTRPV6 subunits are shown, with the front and back subunits removed for clarity. (B) Pore radius calculated using HOLE (41) for hTRPV6-CaM (inactivated state, green), hTRPV6 (PDB ID: 6BO8; open state, orange), and hTRPV6-R470E (PDB ID: 6BOA; closed state, cyan) (13). Dashed line corresponds to 1.4 Å (radius of a water molecule). (C and D) S6 helices in superposed structures of the inactivated (green), open (orange), and closed (cyan) states described in (A), with the residues contributing to gating shown as sticks. The portion of CaM encompassing K115 is shown in (C).
Mechanism of CAM-induced inactivation of TRPV6
The difference in the pore profile of the inactivated state compared to that of the closed and open states is the result of different conformational changes that accompany the gating processes of activation and inactivation (Fig. 3, C and D). While transitioning from the closed to the open state, the middle portion of the α-helical S6 undergoes an α-to-π transition at the alanine A566 gating hinge (13, 17). This energetically unfavorable transition is compensated for by the formation of a salt bridge between Q473 in the S4-S5 elbow and R589 in the TRP helix and a hydrogen bond between D489 in S5 and T581 in S6 (13, 17). As a result of the α-to-π transition, the lower portion of S6 in the open state rotates by ~100°, exposing a different set of residues to the pore, including M578, which creates an activation gate.
In contrast, during inactivation, the S6 helices maintain the α-to-π transition as in the open state. As a result, their lower portions do not rotate, and so they expose the same residues to the pore as they do in the open state. Instead, the S6 helices tilt toward the center of the pore at the alanine A566 gating hinge (Figs. 3C and 4). Because of the pore constriction, I575, which lines the bottom of the central cavity in the open state, hydrophobically seals the pore, creating its inactivation gate. The D489-T581 hydrogen bond, which compensates for the energetically unfavorable α-to-π transition in S6 upon channel opening, is also present in the inactivated state. The tilting of the S6 helices that accompanies inactivation does, however, break the Q473-R589 salt bridge. We hypothesize that the additional energy needed to compensate for the unfavorable α-to-π helical transition in S6 in the inactivated state comes from the cation-π interaction between the tryptophan cage and K115, which functions as an inactivating plug. While many questions about CaM-dependent Ca2+-induced inactivation of ion channels remain to be answered, our TRPV6-CaM structural model (Fig. 4) provides insight into this process and can be exploited to advance the design of novel therapeutics.
Fig. 4. Mechanism of TRPV6 inactivation.
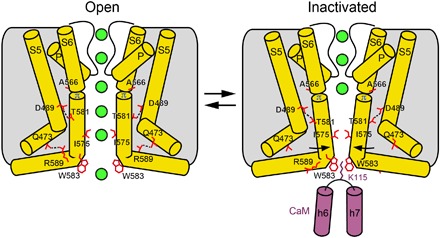
Cartoons represent the structural changes associated with CaM-induced inactivation of TRPV6. Only two of four TRPV6 subunits are shown, with the front and back subunits removed for clarity. The transition from the open to inactivated state involves tilting of the lower portions of the S6 helices toward the center of the pore at the alanine A566 gating hinge and closure of the pore stopping the permeation of ions (green spheres). The loss of the R589-Q473 salt bridges, which stabilize the α-to-π helical transition in S6 in the open state, is compensated by a cation-π interaction between K115 and the π-system of four tryptophans forming a cage at the pore intracellular entrance.
MATERIALS AND METHODS
Constructs
The full-length hTRPV6 (residues 1 to 725) and rTRPV6 (residues 1 to 727) were introduced into a plasmid Eric Gouaux (pEG) BacMam vector (27), with a C-terminal thrombin cleavage site (LVPRG), followed by a streptavidin affinity tag (WSHPQFEK). Human CaM complementary DNA was cloned into the bacterial expression vector pCDFDuet-1 with a C-terminal His tag.
Expression and purification
hTRPV6 and rTRPV6 were expressed and purified as previously described for TRPV6 (13, 17, 28). In brief, hTRPV6 and rTRPV6 in pEG BacMam plasmids were used to generate baculovirus in Sf9 insect cells. This virus was used to transduce human embryonic kidney (HEK) 293 GnTI− cells grown in suspension. Forty-eight to 72 hours after transduction, HEK cells were harvested and washed with phosphate-buffered saline (pH 8.0). Cells were broken by sonication, and cellular membranes were pelleted using ultracentrifugation. rTRPV6 was extracted from cellular membranes using 10 mM lauryl maltose-neopentyl glycol (LMNG) and purified using Strep-Tactin affinity chromatography, followed by size exclusion chromatography in 0.5 mM LMNG. hTRPV6 was extracted from cellular membranes using 20 mM n-dodecyl-β-d-maltopyranoside (DDM) and 0.2 mM cholesteryl hemisuccinate (CHS) supplemented with 20 μM purified CaM and 5 mM Ca2+ for ~2 hours. To maintain the integrity of the hTRPV6-CaM complex, 20 μM CaM was present throughout the entire purification, including affinity and size exclusion chromatography and subsequent amphipol trapping steps.
CaM was subcloned into the pCDEF-duet expression vector that was used to transform BL21(DE3). CaM was purified in two steps, using talon affinity and size exclusion chromatography. The 2L culture of CaM-expressing BL21(DE3) cells was grown in the presence of streptomycin (30 μg/ml) at 37°C for ~2.5 to 3.0 hours. Subsequently, CaM expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C, and cells were grown for another 12 to 14 hours at 200 rpm and 20°C. Cells were harvested by centrifugation, resuspended in 40 ml of lysis buffer [20 mM tris-Cl (pH 8.0) and 150 mM NaCl], and subjected to sonication. The soluble fraction of the resulting slurry containing CaM was recovered by centrifugation. The N-terminally His-tagged CaM was first purified using affinity chromatography. The clarified CaM supernatant was loaded onto a 5-ml TALON metal affinity resin (Clontech) preequilibrated in lysis buffer. After overnight incubation at 20°C, the resin was washed with 30 ml of lysis buffer containing 20 mM imidazole and eluted with lysis buffer containing 250 mM imidazole. The N-terminal His-tag was removed by PreScission Protease at a 1:100 (CaM/PreScission Protease) ratio and again passed through the TALON column to remove the PreScission Protease and the cleaved tag from the CaM-containing solution. Subsequently, concentrated protein was loaded on the Superdex 200 gel filtration column (GE Healthcare) equilibrated with 1.5 column volume of the lysis buffer. CaM fractions were collected and concentrated for structural studies.
Reconstitution of hTRPV6 protein into amphipols
hTRPV6 was incorporated into amphipols as described previously (29). Briefly, for reconstitution in A8-35 amphipols (Anatrace #A835), purified hTRPV6 in the presence of 20 μM CaM and 5 mM Ca2+ was mixed with amphipols in a 1:3 mass ratio (protein/amphipols) and incubated for 3 hours with rotation at 4°C. Subsequently, 7 to 8 mg (per 0.5 ml of protein/amphipol mixture) of Bio-beads SM2 (Bio-Rad) prewet in buffer containing 20 mM tris (pH 8.0), 150 mM NaCl, and 1 mM β-mercaptoethanol was added to the protein/amphipol mixture to facilitate the reconstitution of TRPV6 into amphipols. The mixture was rotated for ~20 hours at 4°C, and the amphipol-solubilized TRPV6 was purified as described above.
Cryo-EM sample preparation and data collection
Au/Au grids were made as described in the literature (30) and used for cryo-EM sample preparation. A Vitrobot Mark IV (FEI) was used to plunge-freeze the grids after the application of 3 μl of protein solution with 100% humidity at 4°C, a blot time of 2 or 3 s, the blot force set to 3, and a wait time of 20 s. A concentration of 0.5 mg/ml was used for the amphipol-solubilized hTRPV6-CaM complex and 2.7 mg/ml for the LMNG-solubilized rTRPV6 protein.
The hTRPV6-CaM and rTRPV6 data were both collected on a Titan Krios (FEI) equipped with a post-column GIF Quantum energy filter, operating at 300 kV, and using a Gatan K2 Summit direct electron detection (DED) camera. For hTRPV6-CaM, a total of 5953 micrographs were collected in counting mode with a pixel size of 1.08 Å across a defocus range of −1.5 to −3.5 μm. The total dose, ~55 e− Å−2, was attained using a dose rate of ~8.0 e− pixel−1 s−1 across 40 frames for 8-s total exposure time. For rTRPV6, a total of 1485 micrographs were collected in counting mode with a pixel size of 1.06 Å across a defocus range of −1.0 to −2.5 μm. For these data, a total dose of ~47 e− Å−2 was attained using a dose rate of ~4.4 e− pixel−1 s−1 across 40 frames for 12-s total exposure time.
Image processing
Frame alignment was performed using MotionCor2 (31), and CTF (contrast transfer function) correction, using Gctf (32) for both data sets, was performed on non–dose-weighted micrographs, while subsequent data processing was performed on dose-weighted micrographs. For both data sets, manual picking for automatic picking template generation, automatic picking, and particle extraction were performed using Relion 2.0 (33), and all subsequent data processing was performed using cryoSPARC (34). For each data set, ~2000 particles were manually selected to generate 2D classes for use in automatic picking.
In processing the hTRPV6-CaM data set, six 2D classes were used as templates for automatically picking 429,167 particles from 4515 micrographs. An ab initio map generated by cryoSPARC from a random selection from the entire pool of particles served as a reference for 3D classification (heterogeneous refinement) into three classes with no symmetry imposed. Ab initio map generation and 3D classification were performed iteratively a total of three times, each time selecting the best of three classes from the previous step as an initial pool for ab initio model generation. Eventually, a pool of 180,600 particles making up the best class from the third round of classification was subjected to homogeneous refinement, which resulted in a 3.98 Å resolution reconstruction. The rTRPV6-CaM data set was processed using a workflow similar to that described above. A total of 274,507 particles were picked and extracted from 1447 micrographs. Ab initio map generation for rTRPV6-CaM was not successful, most likely because of poor angular distribution of particle views in the initial random pool compared to hTRPV6-CaM. Instead, we used the density for hTRPV6-CaM as a reference for rTRPV6-CaM 3D classifications. After three consecutive rounds of 3D classification, a pool of 111,593 particles was subjected to homogeneous refinement, which generated a 3.60 Å resolution reconstruction. No symmetry was ever applied throughout the processing of either the hTRPV6-CaM or the rTRPV6-CaM data sets. The Fourier shell correlation (FSC) and local resolutions were calculated using cryoSPARC, and EM density maps were visualized using UCSF Chimera (35).
Model building
To build models of the hTRPV6-CaM and rTRPV6-CaM complexes in COOT (36), we docked the structures of hTRPV6 [Protein Data Bank (PDB) ID: 6BO8], TRPV6* (PBD ID: 5WO7) (28), and CaM (PDB ID: 2L7L) (37) into the cryo-EM densities and used them as a guide. The resulting models were refined against unfiltered half maps in real space with constraints using PHENIX (38). The models were tested for overfitting (figs. S1 and S5) by shifting their coordinates by 0.5 Å (using shake) in PHENIX and generating densities in Chimera (35) from the shaken models. FSC was calculated between the densities from the shaken models, the half maps used in PHENIX refinement, the other half maps, and the unfiltered sum maps using EMAN2 (39). Structures were visualized, and figures were prepared in PyMOL (40).
Supplementary Material
Acknowledgments
We thank H. Kao for computational support, R. Grassucci and I. S. Fernandez for assistance with microscope operation, and U. Baxa and T. Edwards for help with data collection. We thank H. Colecraft for providing the CaM construct and O. B. Clarke for advice with cryoSPARC. Funding: A.I.S. was supported by the NIH (R01 CA206573 and R01 NS083660), the Amgen Young Investigator Award, and the Irma T. Hirschl Career Scientist Award. L.L.M. and E.C.T. were supported by the NIH (T32 GM008224 and F31 NS093838, respectively). Author contributions: A.K.S., L.L.M., and A.I.S. designed the project. A.K.S. and L.L.M. carried out protein expression, purification, cryo-EM sample preparation, and data collection. A.K.S., L.L.M., E.C.T., and A.I.S. processed and analyzed the cryo-EM data. E.C.T. designed the image processing workflow. A.K.S., L.L.M., and A.I.S. built models and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Data were collected at the Frederick National Laboratory for Cancer Research National Cryo-EM Facility (NIH) and the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy (New York Structural Biology Center) supported by grants from the Simons Foundation (349247), New York Office of Science Technology and Academic Research (NYSTAR), and the NIH (GM103310). Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-8961 (hTRPV6-CaM) and EMD-8962 (rTRPV6-CaM). Model coordinates have been deposited in the PDB under accession numbers 6E2F (hTRPV6-CaM) and 6E2G (rTRPV6-CaM). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaau6088/DC1
Fig. S1. Overview of single-particle cryo-EM for hTRPV6-CaM.
Fig. S2. Comparison of cryo-EM densities for hTRPV6, hTRPV6-CaM, and rTRPV6-CaM.
Fig. S3. Cryo-EM density for hTRPV6-CaM.
Fig. S4. Structures of CaM bound to ion channel fragments.
Fig. S5. Overview of single-particle cryo-EM for rTRPV6-CaM.
Fig. S6. Cryo-EM density for rTRPV6-CaM.
Fig. S7. Comparison of hTRPV6-CaM and rTRPV6-CaM.
Fig. S8. Sequence alignment of TRPV subunits.
Fig. S9. Superposition of the C termini in different TRPV6 subunits.
Fig. S10. hTRPV6 selectivity filter from different gating states.
Table S1. Cryo-EM data collection, refinement, and validation statistics.
Reference (42)
REFERENCES AND NOTES
- 1.Marchi S., Pinton P., Alterations of calcium homeostasis in cancer cells. Curr. Opin. Pharmacol. 29, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Jow F., Zhang Z.-H., Kopsco D. C., Carroll K. C., Wang K., Functional coupling of intracellular calcium and inactivation of voltage-gated Kv1.1/Kvβ1.1 A-type K+ channels. Proc. Natl. Acad. Sci. U.S.A. 101, 15535–15540 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herzog R. I., Liu C., Waxman S. G., Cummins T. R., Calmodulin binds to the C terminus of sodium channels Nav1.4 and Nav1.6 and differentially modulates their functional properties. J. Neurosci. 23, 8261–8270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin N., Olcese R., Bransby M., Lin T., Birnbaumer L., Ca2+-induced inhibition of the cardiac Ca2+ channel depends on calmodulin. Proc. Natl. Acad. Sci. U.S.A. 96, 2435–2438 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee A., Wong S. T., Gallagher D., Li B., Storm D. R., Scheuer T., Catterall W. A., Ca2+/calmodulin binds to and modulates P/Q-type calcium channels. Nature 399, 155–159 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Chen T.-Y., Yau K.-W., Direct modulation by Ca2+–calmodulin of cyclic nucleotide-activated channel of rat olfactory receptor neurons. Nature 368, 545–548 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Gomez A. C., Yamaguchi N., Two regions of the ryanodine receptor calcium channel are involved in Ca2+-dependent inactivation. Biochemistry 53, 1373–1379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medina I., Filippova N., Bakhramov A., Bregestovski P., Calcium-induced inactivation of NMDA receptor-channels evolves independently of run-down in cultured rat brain neurones. J. Physiol. 495 (Pt. 2), 411–427 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalevskaya N. V., van de Waterbeemd M., Bokhovchuk F. M., Bate N., Bindels R. J., Hoenderop J. G., Vuister G. W., Structural analysis of calmodulin binding to ion channels demonstrates the role of its plasticity in regulation. Pflugers Arch. 465, 1507–1519 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Owsianik G., Talavera K., Voets T., Nilius B., Permeation and selectivity of TRP channels. Annu. Rev. Physiol. 68, 685–717 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Lambers T. T., Weidema A. F., Nilius B., Hoenderop J. G. J., Bindels R. J. M., Regulation of the mouse epithelial Ca2+ channel TRPV6 by the Ca2+-sensor calmodulin. J. Biol. Chem. 279, 28855–28861 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Kovalevskaya N. V., Bokhovchuk F. M., Vuister G. W., The TRPV5/6 calcium channels contain multiple calmodulin binding sites with differential binding properties. J. Struct. Funct. Genomics 13, 91–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGoldrick L. L., Singh A. K., Saotome K., Yelshanskaya M. V., Twomey E. C., Grassucci R. A., Sobolevsky A. I., Opening of the human epithelial calcium channel TRPV6. Nature 553, 233–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau S.-Y., Procko E., Gaudet R., Distinct properties of Ca2+–calmodulin binding to N- and C-terminal regulatory regions of the TRPV1 channel. J. Gen. Physiol. 140, 541–555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A., Backbone dynamics of calmodulin studied by nitrogen-15 relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 31, 5269–5278 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Tidow H., Nissen P., Structural diversity of calmodulin binding to its target sites. FEBS J. 280, 5551–5565 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Saotome K., Singh A. K., Yelshanskaya M. V., Sobolevsky A. I., Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506–511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bate N., Caves R. E., Skinner S. P., Goult B. T., Basran J., Mitcheson J. S., Vuister G. W., A novel mechanism for calmodulin dependent inactivation of transient receptor potential vanilloid 6. Biochemistry 57, 2611–2622 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Bokhovchuk F. M., Bate N., Kovalevskaya N. V., Goult B. T., Spronk C. A. E. M., Vuister G. W., The structural basis of calcium dependent inactivation of the transient receptor potential vanilloid 5 channel. Biochemistry 57, 2623–2635 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Niemeyer B. A., Bergs C., Wissenbach U., Flockerzi V., Trost C., Competitive regulation of CaT-like-mediated Ca2+ entry by protein kinase C and calmodulin. Proc. Natl. Acad. Sci. U.S.A. 98, 3600–3605 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derler I., Hofbauer M., Kahr H., Fritsch R., Muik M., Kepplinger K., Hack M. E., Moritz S., Schindl R., Groschner K., Romanin C., Dynamic but not constitutive association of calmodulin with rat TRPV6 channels enables fine tuning of Ca2+-dependent inactivation. J. Physiol. 577 (Pt. 1), 31–44 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holakovska B., Grycova L., Bily J., Teisinger J., Characterization of calmodulin binding domains in TRPV2 and TRPV5 C-tails. Amino Acids 40, 741–748 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Hirnet D., Olausson J., Fecher-Trost C., Bödding M., Nastainczyk W., Wissenbach U., Flockerzi V., Freichel M., The TRPV6 gene, cDNA and protein. Cell Calcium 33, 509–518 (2003). [DOI] [PubMed] [Google Scholar]
- 24.de Groot T., Kovalevskaya N. V., Verkaart S., Schilderink N., Felici M., van der Hagen E. A. E., Bindels R. J. M., Vuister G. W., Hoenderop J. G., Molecular mechanisms of calmodulin action on TRPV5 and modulation by parathyroid hormone. Mol. Cell Biol. 31, 2845–2853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Wijst J., Leunissen E. H., Blanchard M. G., Venselaar H., Verkaart S., Paulsen C. E., Bindels R. J., Hoenderop J. G., A gate hinge controls the epithelial calcium channel TRPV5. Sci. Rep. 7, 45489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao C. K., Zakharian E., Borbiro I., Rohacs T., Interplay between calmodulin and phosphatidylinositol 4,5-bisphosphate in Ca2+-induced inactivation of transient receptor potential vanilloid 6 channels. J. Biol. Chem. 288, 5278–5290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goehring A., Lee C.-H., Wang K. H., Michel J. C., Claxton D. P., Baconguis I., Althoff T., Fischer S., Garcia K. C., Gouaux E., Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A. K., Saotome K., Sobolevsky A. I., Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci. Rep. 7, 10669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao M., Cao E., Julius D., Cheng Y., Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo C. J., Passmore L. A., Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimanius D., Forsberg B. O., Scheres S. H. W., Lindahl E., Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gifford J. L., Ishida H., Vogel H. J., Fast methionine-based solution structure determination of calcium-calmodulin complexes. J. Biomol. NMR 50, 71–81 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68 (Pt. 4), 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 40.W. L. DeLano, The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA, USA, 2002). [Google Scholar]
- 41.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354– 360, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Strotmann R., Schultz G., Plant T. D., Ca2+-dependent potentiation of the nonselective cation channel TRPV4 is mediated by a C-terminal calmodulin binding site. J. Biol. Chem. 278, 26541–26549 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaau6088/DC1
Fig. S1. Overview of single-particle cryo-EM for hTRPV6-CaM.
Fig. S2. Comparison of cryo-EM densities for hTRPV6, hTRPV6-CaM, and rTRPV6-CaM.
Fig. S3. Cryo-EM density for hTRPV6-CaM.
Fig. S4. Structures of CaM bound to ion channel fragments.
Fig. S5. Overview of single-particle cryo-EM for rTRPV6-CaM.
Fig. S6. Cryo-EM density for rTRPV6-CaM.
Fig. S7. Comparison of hTRPV6-CaM and rTRPV6-CaM.
Fig. S8. Sequence alignment of TRPV subunits.
Fig. S9. Superposition of the C termini in different TRPV6 subunits.
Fig. S10. hTRPV6 selectivity filter from different gating states.
Table S1. Cryo-EM data collection, refinement, and validation statistics.
Reference (42)


