Abstract
Single cell mass spectrometry (MS) is uniquely positioned for the sequencing and identification of peptides in rare cells. Small peptides can take on different roles in subcellular compartments. Whereas some peptides serve as neurotransmitters in the cytoplasm, they can also function as transcription factors in the nucleus. Thus, there is a need to analyze the subcellular peptide compositions in identified single cells. Here, we apply capillary microsampling MS with ion mobility separation for the sequencing of peptides in single neurons of the mollusk Lymnaea stagnalis, and the analysis of peptide distributions between the cytoplasm and nucleus of identified single neurons that are known to express cardioactive Phe-Met-Arg-Phe amide-like (FMRFamide-like) neuropeptides. Nuclei and cytoplasm of Type 1 and Type 2 F group (Fgp) neurons were analyzed for neuropeptides cleaved from the protein precursors encoded by alternative splicing products of the FMRFamide gene. Relative abundances of nine neuropeptides were determined in the cytoplasm. The nuclei contained six of these peptides at different abundances. Enabled by its relative enrichment in Fgp neurons, a new 28-residue neuropeptide was sequenced by tandem MS.
Introduction
Within a eukaryotic cell, enzymes, metabolites, and signaling molecules are highly compartmentalized, which is due to their localized production and organelle functionality1,2. Some small peptides can play distinct roles in cells and organisms depending on their subcellular locations. For example, a well-known neurotransmitter in mammalian central nervous system (CNS), neuropeptide Y (NPY), packaged in dense core vesicles in the cytoplasm, can be secreted via regulated exocytosis to transduce signals3, whereas the NPY in the nucleus might regulate specific gene expression4. In the CNS of most animal species, neurons are highly heterogeneous cell populations that produce, store and release specific types of neuropeptides5,6. To understand the localized regulatory functions and the related neurodegenerative diseases, subcellular analysis of neuropeptide distributions in individual neurons preferably of known function is necessary7,8. Animal models with a relatively simpler nervous system and low number of neurons include the pond snail (Lymnaea stagnalis) with several easily distinguishable neurons of known function. For example, the 15–18 F-group (Fgp) neurons are known to participate in the physiological functions of cardioexcitatory effects, penial control, and withdrawal in molluscan species9,10. Whereas Phe-Met-Arg-Phe amide-like (FMRFamide-like) neuropeptides have been detected in these cells, their subcellular localization is not fully explored. One of the major challenges for subcellular analysis is to produce organelles with high purity and minimum chemical and mechanical perturbations. Facing these challenges, development of new approaches for analyzing subcellular compartments is necessary.
Cell fractionation by centrifugation has been widely used for isolating organelles in bulk samples followed by molecular analysis11,12. However, using this method, cell-to-cell heterogeneity is obscured, and extensive sample preparation steps are required to yield subcellular fractions with high purity. Flow cytometry has been adapted for localization and quantification of specific proteins at a rate of thousands of cells per second13,14. With the development of advanced fluorescent probes and optical techniques, the subcellular locations, movements, and amounts of pre-labelled peptides and proteins can be determined and quantified by fluorescence microscopy imaging15–17. Immunofluorescence, which employs antibodies with fluorescent labels to target specific epitopes on the molecules of interest, has been used for the visualization of subcellular peptide and protein distributions in fixed cells or tissues4,18,19. However, some antibodies are limited in their specificity due to alterations of biochemical composition in live cells induced by sample fixation20, false detection of other molecular species containing the targeted epitope, i.e., cross-immunoreactivity21, and failing to recognize modified forms of peptides or proteins present in cells22,23.
With its high specificity and sensitivity, mass spectrometry (MS) is a reliable technique for identification and quantitation of a broad range of molecules, including metabolites, lipids, peptides and proteins in single cells24–26. However, sampling and detection of these chemicals within subcellular structures by MS pose significant challenges, such as the fast molecular turnover rates for some compounds, low sample volumes, limited amounts of analytes and the difficulties of isolating individual compartments with high purity. Only a few MS methods have been used for molecular analysis on the subcellular level. With high spatial resolution, secondary ion MS27,28 and matrix-assisted laser desorption ionization (MALDI)-MS29–32 have been applied for subcellular imaging of metabolites, lipids, peptides, pharmaceuticals and antibiotics in individual cells under vacuum conditions. A recent study demonstrated the extraction of lipid droplets from single cells followed by matrix deposition for MALDI-MS analysis33. The peptide content of dense core vesicles has been studied by capillary microsampling combined with MALDI-MS analysis34. A neurotransmitter, D-aspartate, was quantitatively determined in subcellular regions of individual Aplysia californica neurons by capillary electrophoresis laser induced fluorescence detection35. With the capability to analyze subcellular compartments under ambient conditions, laser ablation electrospray ionization (LAESI) MS36, live single cell MS37–39, and nanospray MS40 have been developed for analyzing metabolites and lipids in cell cytoplasm, nucleus, granules and lipid droplets. Recently, mass cytometry has been adapted for imaging of the pre-labelled proteins by metal tagged antibodies in tumor tissues with subcellular spatial resolution41.
The capillary microsampling technique utilizes a pulled capillary with a sharp tip to extract the contents of single cells followed by electrospray ionization (ESI) MS with ion mobility separation (IMS). It has been applied for metabolic and lipidomic analysis of single Arabidopsis thaliana (A. thaliana) epidermal cells42 and human hepatocellular carcinoma cells43. Using this technique, the presence of distinct metabolic pathways in different A. thaliana cell types was studied42. Cellular heterogeneity among individual hepatocytes in response to xenobiotic treatment and due to passing through specific mitotic stages were also revealed43,44. These studies were conducted on the cellular contents without a distinction of particular subcellular regions, and they obscured the compositional variations within a cell.
Here we apply capillary microsampling ESI-IMS-MS for subcellular analysis of single identified Fgp neurons from the L. stagnalis CNS. Distinct sets of neuropeptide species are detected from the cytoplasm of different Fgp neurons with alternative mRNA splicing of the FMRFamide gene. The neuropeptide levels in the cytoplasm and nucleus of Type 2 Fgp neurons show discernible differences. A 28-residue neuropeptide was found for the first time and sequenced by tandem MS in a single Type 2 Fgp neuron.
Not only do these novel findings in L. stagnalis demonstrate the feasibility of detecting peptide localization in single identified neurons with sub-cellular resolution but they also open new avenues for the analysis of sub-cellular level changes underpinning memory function and dysfunction in both invertebrate and vertebrate model organisms.
Results
Peptides resulting from alternative mRNA splicing in cytoplasm
A microscope image of a freshly prepared L. stagnalis CNS with the individual ganglia labelled is shown in Supplementary Fig. 1a. Some of the neuron types are identifiable based on their size, color, location, and electrophysiological characterisctics10,45–47. In the left lateral region of the visceral ganglion, a cluster of ~15–18 cells with diameters of ~45–105 µm can be identified as the Fgp neurons48. Individual Fgp neurons were isolated by a fire-polished capillary with an inner tip diameter of ~150–200 µm and transferred into a drop of saline in a Petri dish (see Supplementary Fig. 1b).
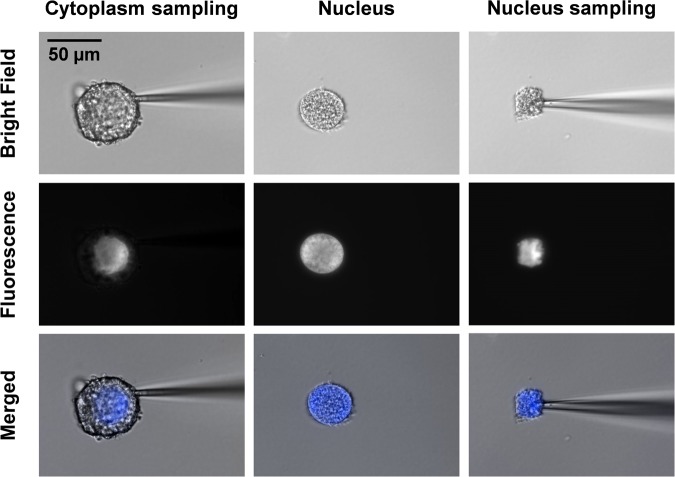
To visualize the nucleus, the double stranded DNA (dsDNA) was stained with Hoechst 33342 before the isolation of the single neuron from the ganglion. Figure 1 shows the bright field (top row), fluorescence (middle row), and merged (bottom row) microscope images for subcellular microsampling of a single Fgp neuron. The observed fluorescence signal was associated with the position of the nucleus. A capillary tip with an opening of ~10 µm was used to extract the cytoplasm. After cytoplasm sampling, the nucleus remained intact in shape and morphology. A second capillary was inserted to extract the contents of the nucleus. The sampled volumes and masses were estimated to be ~1.5 pL (1.5 ng) and ~0.4 pL (0.4 ng) from the cytoplasm (total volume ~75 pL) and nucleus (total volume ~25 pL), respectively. On average, ~2% of the total cytoplasm or nucleus volume was extracted and analyzed by capillary microsampling ESI-IMS-MS. As a result, a three-dimensional dataset comprised of ion abundances as a function of drift time (DT) and mass-to-charge ratio (m/z) was generated for each subcellular measurement. The corresponding mass spectrum integrated over all DT values was exported for further data processing.
Figure 1.
Bright field (top row), fluorescence (middle row) and merged (bottom row) microscope images for subcellular sampling of cytoplasm and nucleus from a single L. stagnalis Fgp neuron. Scale bar is 50 µm.
Previous studies have shown the presence of alternative mRNA splicing of the FMRFamide gene in L. stagnalis Fgp neurons49,50. The spliced mRNA encode two types of protein precursors to produce distinct sets of neuropeptides by post-translational processing, which are mutually exclusively expressed in different Fgp neurons51. The two protein precursor sequences, Type 1 and Type 2, predicted by DNA sequencing in previous studies are shown in the top panels of Fig. 2a,b, respectively, with the corresponding signal peptides marked by orange color52,53. The positive ion mode mass spectra obtained from the cytoplasm of Type 1 and Type 2 Fgp neurons are shown in the bottom panels of Fig. 2a,b, respectively. In the cytoplasm spectrum of Type 1 Fgp neurons, two tetrapeptides, FLRFamide and FMRFamide are identified (see Supplementary Table 1). In the cytoplasm spectrum of Type 2 Fgp neurons, nine neuropeptides, including five heptapeptides, are detected and identified (see Supplementary Table 2). The limited volume of subcellular compartments, and the viscosity of the nucleus samples present significant challenges for neuropeptide detection and identification. Most of the identified neuropeptide sequences are consistent with those predicted from previous cDNA analysis, marked by green in the top panels of Fig. 2a and b 52,53. Collision cross section (CCS) values for these neuropeptides are measured by ion mobility separation and reported for the first time (see Supplementary Tables 1 and 2). These CCS values provide additional information for enhanced molecular identification.
Figure 2.
Protein precursor sequences predicted from cDNA analysis for (a) Type 1 and (b) Type 2 Fgp neurons are shown in top panels. Mass spectra from corresponding cytoplasms are shown in bottom panels of Fig. 2a and b. Signal peptide sequences are highlighted in orange, whereas neuropeptides are marked by green. In Fig. 2b, newly discovered 28-residue neuropeptide is shown in third row from the bottom in the protein precursor 2 sequence, with the cysteine residue marked by red oval conflicting with the glycine residue in our de novo peptide sequence (see Fig. 3).
Single cell tandem MS for neuropeptide identification
The neuropeptide identifications are based on the fragmentation patterns of the peptide ions in single cell tandem ESI-IMS-MS, from MALDI-MS/MS of a visceral ganglion, or nano-liquid chromatography (LC)-MS/MS of peptide extractions from five L. stagnalis CNS. A previous study showed that only ~340 neurons expressed these FMRFamide-like neuropeptides, which represented 1.5% of all the neurons in the snail CNS54. For peptide identification, extracts from five snail CNS are studied by nanoLC-MS/MS, and three of these peptides, GDPFLRFamide, SDPFLRFamide, SDPYLRFamide, are detected and identified. The corresponding nanoLC-MS/MS spectra are shown in Supplementary Fig. 2. MALDI-MS/MS spectra of eight neuropeptides obtained from a visceral ganglion are shown in Supplementary Fig. 3. Two other neuropeptide species, a 35- and a 28-amino acid residue peptide, show low abundances in the MALDI mass spectra.
In contrast, detection and identification of these two neuropeptides is achieved by single cell ESI-IMS-MS/MS. This is due to the lack of signal dilution by the 98.5% of cells devoid of these peptides. Single cell tandem mass spectrum of the ion with m/z 990.468 and charge state of +4 assigned as acidic peptide based on fragmentation pattern and predicted neuropeptide sequences from previous cDNA analysis is shown in Supplementary Fig. 4.
A new neuropeptide with 28-amino acid residues is detected and characterized for the first time from the cytoplasm of a single Type 2 Fgp neuron. For the identification of the 28-residue peptide, the DT vs. m/z plot based on the tandem MS of the monoisotopic m/z 1111.183 in charge state +3 is shown in Fig. 3a. The precursor ion with a charge state of +3, and the fragment ions with charge states of +2, and +1, are separated in the DT vs. m/z plot, and highlighted by distinct ellipses. Figure 3b shows the mass spectra corresponding to the three different charge states with the y- and b-type fragment ions labeled in the top and middle panels. Based on this data, the peptide sequence is identified as NNLNNYALEDEDGKLTSDIIDDQFQRYQ with only a single fragmentation missed at the amino terminus. Surprisingly, the thirteenth amino acid residue in this peptide is identified by tandem MS as glycine instead of the cysteine predicted from cDNA data (see the residue underlined in the sequence above vs. the circled residue in Fig. 2b)53. As tandem MS is a direct structural characterization method for identifying the primary structure, we assume that the difference between the codon in the previous literature corresponding to cysteine, and the glycine identified in our study is due to either single nucleotide polymorphism, or a misidentification in the original cDNA data. This discrepancy requires further analysis.
Figure 3.
Single cell tandem (a) DT vs. m/z plot and (b) spectra for identification of a peptide with monoisotopic m/z 1111.183 and a charge state of +3. This ion is identified as a 28-residue peptide with a de novo sequence of NNLNNYALEDEDGKLTSDIIDDQFQRYQ based on the fragments labelled in the spectra. Fragment and precursor ions in different charge states are separated in distinct mobility regions.
Neuropeptide distributions between cytoplasm and nucleus
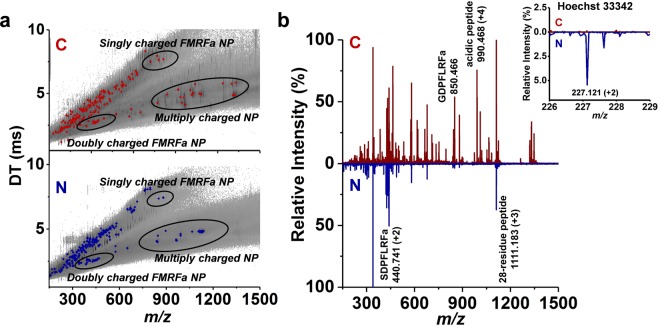
To investigate the subcellular distributions of these neuropeptides, the cytoplasm (n = 8) and nucleus (n = 5) of Type 2 neurons were separately sampled and analyzed by capillary microsampling ESI-IMS-MS. In Fig. 4a, representative DT vs. m/z plots for cytoplasm (top) and nucleus (bottom) are shown with the singly and multiply charged neuropeptides separated and highlighted in distinct mobility regions. The corresponding mass spectrum for the cytoplasm (top) exhibited peaks corresponding to nine peptides, whereas the spectrum for the nucleus (bottom) only contained peaks from six peptides (see Fig. 4b). Differences in the relative ion abundances between the two subcellular compartments are also observed. The inset in Fig. 4b shows that the ion signal for Hoechst 33342 (m/z 227.121) is observed in the nucleus but it is absent in the cytoplasm. This confirms the selectivity of the subcellular sampling process. Due to the lower volumes and higher viscosity of the samples from the nucleus, the absolute abundances and variety of the related ions are relatively low compared to the data from the cytoplasm. It is very challenging to control the extracted volume from cytoplasm and especially the nucleus. Based on all single cell data, ~200 and ~50 molecular features are detected in the cytoplasm and nucleus, respectively. Many of these peaks are related to metabolites and some of them possibly to other, yet unidentified peptides.
Figure 4.
(a) Representative DT vs. m/z plots, and (b) mass spectra for the cytoplasm (C) and the nucleus (N). Inset shows that the m/z 227.121 peak for Hoechst 33342 can be detected in the nucleus but not in the cytoplasm indicating successful selective sampling.
The normalized abundances of the neuropeptides identified in the cytoplasm (IC) and nucleus (IN) are shown in Supplementary Table 2. The normalization is based on the sum of sample related peak abundances in the spectra. All charge states and adduct forms are accounted for calculating the abundances of specific neuropeptides. Some of the neuropeptides in the cytoplasm, i.e., SDPYLRFamide, SDPFFRFamide, and GPSRSSFPRYamide, are not detected in the nucleus. To find the neuropeptide species with the most variance between the cytoplasm and nucleus, multivariate statistical analysis, in particular orthogonal partial least squares-discriminant analysis (OPLS-DA), was performed. The generated S-plot in Supplementary Fig. 5 highlights the neuropeptides with high correlation and covariance. Our results show that some neuropeptides, i.e., GPSRSSFPRYamide, SKPYMRFamide, FLRFamide, and 35-residue acidic peptide, are the distinguishing features of the cytoplasm spectra, whereas others, i.e., GDPFLRFamide, SDPFLRFamide, PFLRFamide, and Hoechst 33342 dye, are more associated with the spectra from the nucleus. A major difference is found (p < 0.006) in the [28-residue peptide]/[acidic peptide] relative abundance ratio that rises from 1.6 ± 0.5 in the cytoplasm to 9.3 ± 3.3 in the nucleus (see Fig. 5a). This is mainly due to the very low abundance of the acidic peptide in the nucleus (see the insets in Fig. 5b).
Figure 5.

(a) Box-and-whisker plot for [28-residue peptide]/[acidic peptide] ratio shows significant difference (p < 0.006) between cytoplasm (n = 8) and nucleus (n = 5). (b) Mass spectra for acidic peptide (+4, m/z 990.468) and 28-residue peptide (+3, m/z 1111.183) from cytoplasm (C) and nucleus (N). Acidic peptide shows significantly lower abundances in nucleus compared to cytoplasm.
To obtain additional information on the subcellular distributions of neuropeptides in Fgp neurons, immunostaining of L. stagnalis CNS by anti-FMRFamide antibody was performed. In Supplementary Fig. 6, confocal microscope images show the presence of FMRFamide (red) only in the cytoplasm (left panel), with the nucleus counterstained by Hoechst 33342 (blue, middle panel). The merged image is shown in the right panel of Supplementary Fig. 6. This observation is consistent with our MS-based results showing FMRFamide neuropeptide present in the cytoplasm of Type 1 Fgp neurons (see Supplementary Table 1).
Discussion
Most of the neuropeptides are produced in the cytoplasm through the cleavage of larger protein precursors by post-translational processing. For example, the immunostaining images in our study show that FMRFamide is only observed in the cytoplasm of L. stagnalis Fgp neurons. Peptides can be present in different forms within cell cytoplasm and nucleus. A previous study showed FMRFamide neuropeptide was found to be packaged in dense core vesicles in A. californica neurons55. The antibodies used in immunostaining only recognize the accessible epitopes of these molecules, whereas peptides in their modified forms (e.g., amidated) may not be detected. Other studies showed that due to peptide degradation by peptidases in cells, free peptides were unstable unless binding with proteins or chromatins56. The level of free peptides is determined by the relative rate of their production and degradation.
Capillary microsampling MS enables the identification of peptides in subcellular compartments without specifically targeting them. Considering the high abundance of the peptide signal in these cells, it is possible that in addition to the free peptides, this method also detects the peptides released from dense core vesicles34 and detached from noncovalent associations with proteins and chromatin during the electrospray process. This untargeted approach opens the door to the detection of localization of multiple peptides (10 in the current study) irrespective of preexisting knowledge on their presence in a cell and the availability of immunoreagents.
Despite the low level of neuropeptides at the tissue level, individual neurons with specific function can exhibit significantly higher concentrations. The high neuropeptide content of these cells enabled the identification of multiple known peptides and the first detection and de novo sequencing of a new 28-residue peptide. Due to the untargeted nature of the MS method, similar findings in other rare cell types can be anticipated even in the presence of posttranslational modifications.
Reports on the detection of peptides in the nucleus are relatively rare in the literature. Immunostaining was used to detect NPY in the nucleus and the nuclear envelope and the authors hypothesized that NPY in the nucleus might play an important role in regulating gene transcription4. Large scale separation of nuclei followed by MS has revealed the presence of arginine vasopressin in the nuclei of lymphocytes57. However, the mechanisms behind these observations are still not well understood. These neuropeptides can be actively transported or transferred through diffusion into the nucleus58.
Finding six peptides in the nucleus of a neuron demonstrates the general utility of the capillary microsampling ESI-IMS-MS approach for subcellular measurements. This is further strengthened by detecting dramatically different peptides in the cytoplasms of Type 1 and Type 2 Fgp neurons. Comparing Supplementary Tables 1 and 2 reveals the presence of two and nine peptides in the cytoplasms of Type1 and Type 2 cells, respectively.
Relative quantitation of subcellular neuropeptide levels is a challenging task. The results demonstrate that capillary microsampling ESI-IMS-MS enables the analysis of subcellular distributions of FMRFamide-like neuropeptides between cytoplasm and nucleus in individual Type 2 Fgp neurons from L. stagnalis CNS. Relative levels of some neuropeptides show distinct differences between the cytoplasm and nucleus indicating the presence of selective active transport, and/or a difference in production and degradation rates between them. For example, higher levels of the heptapeptides, GDPFLRFamide (p < 0.06) and SDPFLRFamide (p < 0.09), are present in the nucleus compared to the other neuropeptides. Likewise, the 35-residue acidic peptide is found with significantly higher abundance in the cytoplasm (p < 0.01).
The capillary microsampling ESI-IMS-MS approach can be potentially applied for the local analysis of subcellular gradients for a broad range of molecules, such as metabolites, lipids and peptides in diverse cell types. The combination of this method with single cell transcriptomics can serve as a foundation for single-cell systems biology and for subcellular analysis of individual neurons after external stimuli or genetic modification to answer fundamental questions in neuroscience.
Methods
Chemicals and materials
HPLC grade methanol and water, and Hoechst 33342 (H1399) were purchased from Thermo Fisher Scientific (Waltham, MA, USA), whereas Instant Ocean sea salt was obtained from Instant Ocean (Blacksburg, VA, USA). Trypsin (T9201), NaCl (S7653), KCl (P9333), CaCl2 (C5670), MgCl2 (M4880), HEPES (H4034), CaSO4 (238988), 2,5-dihydroxybenzoic (DHB, 85707), paraformaldehyde (158127), phosphate buffered saline (PBS, P4417), Dulbecco’s PBS (DPBS, D8537), Triton X-100 (TX, T8787), bovine serum albumin (BSA, A3912), and rabbit anti-FMRFamide primary antibody (AB15348) were purchased from Sigma Aldrich (St. Louis, MO, USA). NorthernLights™ 557-conjugated Anti-Rabbit IgG secondary antibody (red; Cat. No. NL004) was obtained from R&D System (Minneapolis, MN, USA). Snail saline was prepared at final concentrations of 51.3 mM NaCl, 1.7 mM KCl, 4.1 mM CaCl2, and 1.5 mM MgCl2, and 2 mM HEPES in autoclaved water with pH adjusted to 7.9 by 0.1 M NaOH. The DHB matrix solution was prepared in methanol at a final concentration of 10 mg/ml.
Sample preparation
Pond snails, L. stagnalis, were maintained in deionized water (18.2 MΩ·cm) supplemented with 0.26 g/L sea salt and 0.34 g/L CaSO4 at room temperature (~20 °C). Snails of ~3–4 months of age and shell length of ~10–15 mm were dissected in saline. Under observation by a stereomicroscope (E-Zoom6, Edmund Optics, Barrington, NJ, USA), the CNS was extracted from the animal by a pair of fine scissors (50086, World Precision Instruments, Sarasota, FL, USA) and pinned out on the bottom of a Sylgard (Sylgard 184, Dow Corning, Midland, MI, USA) dish containing saline (see Supplementary Fig. 1a). After removing the outer sheath membrane by two pairs of fine forceps (500085, World Precision Instruments, Sarasota, FL, USA), the CNS was treated with 0.2% trypsin solution, prepared in saline, for 10 minutes followed by rinsing three times by saline. To stain the dsDNA contained in the nucleus, the CNS was treated for 5 minutes with 1 µg/ml Hoechst 33342 made in saline. After replacing the staining solution with saline, the inner sheath membrane surrounding the visceral ganglion was teared off by the forceps to expose the neurons of interest. A fire-polished capillary with an inner diameter opening of ~150–200 µm was filled with the saline and attached to a micrometer syringe (GS-1100, Gilmont, Gilmer, TX, USA). The capillary was used to isolate individual Fgp neurons from the visceral ganglion by applying gentle negative pressure (see Supplementary Fig. 1a). The isolated Fgp neurons were transferred to a 10 μL drop of saline on a Petri dish by applying positive pressure.
Subcellular sampling
Initially, glass capillaries (TW100F-3, World Precision Instruments, Sarasota, FL, USA) were pulled by a micropipette puller (P-1000, Sutter Instrument, Novato, CA) installed with a box filament (FB255B, Sutter Instrument, Novato, CA). Sharp capillary tips with long tapering and <1 µm opening were produced. The pulling program was set as Heat = 492, Pull = 60, Velocity = 90, and Delay = 200, at Pressure = 500. Then the capillary tips were cut with a pre-cleaned ceramic scoring wafer (Cat. No. 20116, Restek Corporation, Bellefonte, PA, USA) to generate larger openings with inner diameters of ~10 µm.
A capillary holder (IM-H1, Narishige, Tokyo, Japan) attached with a sampling capillary tip was mounted on a micromanipulator (TransferMan NK2, Eppendorf, Hauppauge, NY, USA) on an inverted microscope (IX71, Olympus, Tokyo, Japan). For sampling the neuron cytoplasm, the capillary tip was gently inserted into the cell without touching the nucleus and a negative pressure was applied through the capillary tip with a syringe attached from the back. After cytoplasm sampling, the nucleus retained its shape and morphology and a second capillary was used to extract its contents. The volumes of the sampled cytoplasm and nucleus were estimated to be ~1.5 pL and ~0.4 pL, respectively. To check for the presence or absence of Hoechst 33342 in the samples, the capillary tips containing the subcellular contents were examined by a fluorescence microscope with a filter cube (49000, Chroma, Bellows Falls, VT, USA).
Single cell mass spectrometry
The capillary tips containing the sampled subcellular contents (cytoplasm or nucleus) were backfilled with 1.0 µL electrospray solution. The electrospray solution was 50% methanol supplemented with 0.1% acetic acid. The capillaries were inserted in a microelectrode holder (MEW-F10A, Warner Instruments, Hamden, CT, USA) attached with a platinum wire of 200 µm in diameter and ~5 cm in length (Alfa Aesar, Ward Hill, MA, USA). The platinum wire was in contact with the solution to provide electrical connection to the capillary tip. A high positive voltage of +2000 V was applied to the microelectrode by a regulated power supply (PS350, Stanford Research Systems Inc., Sunnyvale, CA, USA) to generate electrospray in front of a quadrupole time-of-flight (TOF) mass spectrometer equipped with a traveling wave (T-wave) ion mobility separation system (Synapt G2-S, Waters Co., Milford, MA, USA).
Data analysis
A three-dimensional dataset comprised of ion abundances as a function of drift time (DT) and mass-to-charge ratio (m/z), i.e., a DT vs. m/z plot, was acquired in positive ion mode for each subcellular sample. DriftScope 2.8 (Waters Co., Milford, MA, USA) software was used to process the DT vs. m/z plots and derive the ionic CCS values from the measured DTs using singly charged polyalanine oligomer (n = 4–14) mixture as the calibrant. The CCS calibration curve is shown in Supplementary Fig. 759. The mass spectra integrated over all DTs were visualized by MassLynx 4.1 (Waters Co., Milford, MA, USA) and exported to mMass software60 for further data processing. The sample related peak lists of m/z and ion abundances were generated by mMass through a process of peak picking, deisotoping, and subtraction of the background ions originating from both the electrospray solution and saline, and summing the abundances of all the ionic species for identified molecular features. The processed peak lists for cytoplasm and nucleus spectra were imported into MetaboAnalyst for statistical analysis, in particular, orthogonal partial least squares-discriminant analysis (OPLS-DA) using Pareto scaling61.
Neuropeptide identifications were based on a combination of accurate mass measurements and tandem MS by capillary microsampling MS at the single cell level or MALDI-MS on an L. stagnalis visceral ganglion using DHB as the matrix (see Supplementary Tables 1 and 2). The MALDI tandem MS experiments were performed on a MALDI LTQ Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA, USA). The neuropeptide sequences predicted by cDNA analysis in previous studies were compared to our assignments based on tandem MS52,53.
Neuropeptide extraction
Neuropeptide extraction for nanoLC-MS/MS was obtained from five L. stagnalis CNS and prepared following a previous published protocol with slight modifications62. Five L. stagnalis CNS was prepared in saline and rinsed in cold acidified methanol (90% methanol, 9% acetic acid, and 1% water). The CNS was transferred in a 0.5 mL centrifuge tube and quenched with liquid N2. The frozen CNS tissue was grinded by a pestle (Cat. No. 12–141–367, Thermo Fisher Scientific, Waltham, MA, USA) using a pestle motor (Cat. No. 12–141–361, Thermo Fisher Scientific, Waltham, MA, USA). To prevent protein degradation during sample preparation, a protease inhibitor tablet (A32963, Thermo Fisher Scientific, Waltham, MA, USA) dissolved in 80 mL of 20 mM EDTA solution was prepared. The centrifuge tube containing the grinded tissue was added with 50 µL acidified methanol and 50 µL EDTA solution with protease inhibitor. Cell lysis was further performed by sonication using a probe sonicator (Q125, QSONICA, Melville, NY, USA). The sample was centrifuged at 14,000 g at 4 °C for 10 min and the supernatant was transferred in a new tube. To remove larger proteins, the extracted contents were filtered through a 10 kDa MWCO membrane (Cat. No. 88513, Thermo Fisher Scientific (Waltham, MA, USA). The filtrate was dried by a vacuum concentrator (Labconco, Kansas City, MO). The dried sample was reconstituted in 50 µL of 2% acetonitrile solution with 0.1% formic acid for nanoLC-MS/MS injection.
nanoLC-MS/MS
Peptides separation and identification were performed using a reversed-phase nano-liquid chromatography (model Dionex Ultimate 3000 RSLCnano, Thermo Scientific, Waltham, MA) equipped with a 20 µL injection loop coupled to Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, San Jose, CA, USA). Peptides were trapped on a C18 trapping column (Cat No. 160454, Thermo Fisher Scientific, Waltham, MA, USA) followed by chromatographic separation using a C18 analytical column (75 μm ID, 500 mm length, 3 μm beads with 10 nm pore size, Cat No. 164570, Thermo Fisher Scientific, Waltham, MA, USA). The mobile phases were composed of (A) water with 0.1% formic acid and (B) acetonitrile with 0.1% formic acid. An 85 min gradient was used: 0–10 min, 2% B; 10–10.2 min, 2%-5% B; 10.2–65 min, 5%-80% B; 65–70 min, 80% B; 70–80 min, 80–2% B; 80–85 min, 2% B. Tandem MS data was collected under positive ion data dependent acquisition (DDA) mode using HCD fragmentation at 28% normalized collision energy. Full single-stage (MS1) scans were acquired in Orbitrap analyzer at a resolution of 60,000 FWHM, whereas tandem MS scans were collected in the ion trap analyzer.
To document peptide identification, tandem MS raw data and the mzML files from all three instruments (Synapt G2-S, MALDI LTQ Orbitrap XL, and Orbitrap Fusion Tribrid) were uploaded to the PeptideAtlas repository (http://www.peptideatlas.org/). The identifier number for the dataset is PASS01199.
Immunohistochemistry
L. stagnalis CNS was dissected and pinned out on a Sylgard dish, and fixed overnight in 4% paraformaldehyde buffered with 0.1 M phosphate buffer (pH 7.4) at 4 °C. The fixed CNS was washed twice in 1 × PBS solution (pH 7.4) for 10 min, followed by incubation overnight in PBS containing 20% sucrose at 4 °C. Cryomicrotome sections of ~16 µm thickness were made and mounted on slides. To block the non-specific binding sites, the slides were washed with PBS containing 0.25% Triton X-100 and 0.25% bovine serum albumin at room temperature. After washing, the slides were incubated overnight at 4 °C with rabbit anti-FMRFamide primary antibody diluted in 1:1000 in PBS-TX-BSA. The slides were further washed twice with PBS-TX for 10 min, followed by incubation for 6 h at 4 °C with the fluorophore-conjugated donkey anti-rabbit IgG secondary antibody at 1:1000 dilution. After washing with modified DPBS, the nuclei were stained with Hoechst (1 μg/mL in DPBS) for 10 min at room temperature. The slides were washed with DPBS and covered with fluorescent mounting medium (Dako Fluorescence Mounting Medium, S3023). The stained tissue sections were analyzed with a Leica DMi8 confocal microscope.
Electronic supplementary material
Acknowledgements
Research was sponsored by the U.S. Army Research Office and the Defense Advanced Research Projects Agency and was accomplished under cooperative agreement number W911NF-14–2–0020. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office, DARPA, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation hereon. IHC experiments were supported by Hungarian Brain Research Project (2017–1.2.1-NKP-2017–00002). The authors thank John Young in the Tabin Lab at Harvard Medical School, Department of Genetics, for providing the initial cohort of experimental animals.
Author Contributions
A.V. and L.Z. conceived the study, designed the experiments, and conducted the data analysis. L.Z. and N.K. raised the snails and performed the dissection of the animals. L.Z. conducted the mass spectrometry experiments and evaluated the data. I.K., G.K., Z.Z., and Z.P. provided L. stagnalis CNS expertise and instructions. L.Z. and A.V. wrote the manuscript with input from all authors.
Competing Interests
The authors declare that they have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29704-z.
References
- 1.Martin W. Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond, Ser B: Biol Sci. 2010;365:847–855. doi: 10.1098/rstb.2009.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diekmann Y, Pereira-Leal JB. Evolution of intracellular compartmentalization. Biochem J. 2013;449:319–331. doi: 10.1042/BJ20120957. [DOI] [PubMed] [Google Scholar]
- 3.Ramamoorthy P, Whim MD. Trafficking and fusion of neuropeptide Y-containing dense-core granules in astrocytes. J Neurosci. 2008;28:13815–13827. doi: 10.1523/JNEUROSCI.5361-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacques D, et al. Presence of neuropeptide Y and the Y-1 receptor in the plasma membrane and nuclear envelope of human endocardial endothelial cells: modulation of intracellular calcium. Can J Physiol Pharmacol. 2003;81:288–300. doi: 10.1139/y02-165. [DOI] [PubMed] [Google Scholar]
- 5.Lagercrantz H, Ringstedt T. Organization of the neuronal circuits in the central nervous system during development. Acta Paediatr. 2001;90:707–715. doi: 10.1111/j.1651-2227.2001.tb02792.x. [DOI] [PubMed] [Google Scholar]
- 6.Hua JYY, Smith SJ. Neural activity and the dynamics of central nervous system development. Nat Neurosci. 2004;7:327–332. doi: 10.1038/nn1218. [DOI] [PubMed] [Google Scholar]
- 7.Buchberger, A., Yu, Q. & Li, L. J. Advances in mass spectrometric tools for probing neuropeptides. In: Annual Review of Analytical Chemistry, Vol 8 (eds Cooks, R. G. & Pemberton, J. E.) (2015). [DOI] [PMC free article] [PubMed]
- 8.Qi M, Philip MC, Yang N, Sweedler JV. Single cell neurometabolomics. ACS Chem Neurosci. 2018;9:40–50. doi: 10.1021/acschemneuro.7b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell GA, Schot LPC, Dockray GJ. Identification and probable role of a single neuron containing the neuropeptide helix FMRFamide. Nature. 1983;304:638–640. doi: 10.1038/304638a0. [DOI] [PubMed] [Google Scholar]
- 10.Santama N, Benjamin PR. Gene expression and function of FMRFamide-related neuropeptides in the snail Lymnaea. Microsc Res Tech. 2000;49:547–556. doi: 10.1002/1097-0029(20000615)49:6<547::AID-JEMT5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Cox B, Emili A. Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat Protoc. 2006;1:1872–1878. doi: 10.1038/nprot.2006.273. [DOI] [PubMed] [Google Scholar]
- 12.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166:1324–1337. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gohar AV, et al. Subcellular localization-dependent changes in EGFP fluorescence lifetime measured by time-resolved flow cytometry. Biomedical Optics Express. 2013;4:1390–1400. doi: 10.1364/BOE.4.001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns TJ, et al. High-throughput precision measurement of subcellular localization in single cells. Cytometry Part A. 2017;91A:180–189. doi: 10.1002/cyto.a.23054. [DOI] [PubMed] [Google Scholar]
- 15.Fehr M, et al. Development and use of fluorescent nanosensors for metabolite imaging in living cells. Biochem Soc Trans. 2005;33:287–290. doi: 10.1042/BST0330287. [DOI] [PubMed] [Google Scholar]
- 16.Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Rizk A, et al. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat Protoc. 2014;9:586–596. doi: 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
- 18.Stadler C, et al. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods. 2013;10:315–+. doi: 10.1038/nmeth.2377. [DOI] [PubMed] [Google Scholar]
- 19.Kiss T, Battonyai I, Pirger Z. Down regulation of sodium channels in the central nervous system of hibernating snails. Physiol Behav. 2014;131:93–98. doi: 10.1016/j.physbeh.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Hobro AJ, Smith NI. An evaluation of fixation methods: Spatial and compositional cellular changes observed by Raman imaging. Vib Spectrosc. 2017;91:31–45. doi: 10.1016/j.vibspec.2016.10.012. [DOI] [Google Scholar]
- 21.Mao, S. P. et al. Verification of the cross immunoreactivity of A60, a mouse monoclonal antibody against neuronal nuclear protein. Front Neuroanat10 (2016). [DOI] [PMC free article] [PubMed]
- 22.Pezzella F, et al. Expression of the bcl-2 oncogene protein is not specific for the 14-18 chromosomal translocation. Am J Pathol. 1990;137:225–232. [PMC free article] [PubMed] [Google Scholar]
- 23.Bordeaux J, et al. Antibody validation. BioTechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342:1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 25.Comi TJ, Do TD, Rubakhin SS, Sweedler JV. Categorizing cells on the basis of their chemical profiles: progress in single-cell mass spectrometry. J Am Chem Soc. 2017;139:3920–3929. doi: 10.1021/jacs.6b12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Vertes A. Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angew Chem Int Ed. 2018;57:4466–4477. doi: 10.1002/anie.201709719. [DOI] [PubMed] [Google Scholar]
- 27.Passarelli MK, et al. Single-cell analysis: visualizing pharmaceutical and metabolite uptake in cells with label-free 3D mass spectrometry imaging. Anal Chem. 2015;87:6696–6702. doi: 10.1021/acs.analchem.5b00842. [DOI] [PubMed] [Google Scholar]
- 28.Tian H, Six DA, Krucker T, Leeds JA, Winograd N. Subcellular chemical imaging of antibiotics in single bacteria using C60-secondary ion mass spectrometry. Anal Chem. 2017;89:5050–5057. doi: 10.1021/acs.analchem.7b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zavalin A, et al. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J Mass Spectrom. 2012;47:1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 30.Korte AR, Yandeau-Nelson MD, Nikolau BJ, Lee YJ. Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal Bioanal Chem. 2015;407:2301–2309. doi: 10.1007/s00216-015-8460-5. [DOI] [PubMed] [Google Scholar]
- 31.Dueñas, M. E., Feenstra, A. D., Korte, A. R., Hinners, P. & Lee, Y. J. Cellular and subcellular level localization of maize lipids and metabolites using high-spatial resolution MALDI mass spectrometry imaging. In: Maize: Methods and Protocols (eds Lagrimini, L. M.). Springer New York (2018). [DOI] [PubMed]
- 32.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 33.Phelps MS, Sturtevant D, Chapman KD, Verbeck GF. Nanomanipulation-coupled matrix-assisted laser desorption/ionization-direct organelle mass spectrometry: a technique for the detailed analysis of single organelles. J Am Soc Mass Spectrom. 2016;27:187–193. doi: 10.1007/s13361-015-1232-9. [DOI] [PubMed] [Google Scholar]
- 34.Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat Biotechnol. 2000;18:172–175. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 35.Miao H, Rubakhin SS, Sweedler JV. Subcellular analysis of D-Aspartate. Anal Chem. 2005;77:7190–7194. doi: 10.1021/ac0511694. [DOI] [PubMed] [Google Scholar]
- 36.Stolee JA, Shrestha B, Mengistu G, Vertes A. Observation of subcellular metabolite gradients in single cells by laser ablation electrospray ionization mass spectrometry. Angew Chem Int Ed. 2012;51:10386–10389. doi: 10.1002/anie.201205436. [DOI] [PubMed] [Google Scholar]
- 37.Mizuno H, Tsuyama N, Harada T, Masujima T. Live single-cell video-mass spectrometry for cellular and subcellular molecular detection and cell classification. J Mass Spectrom. 2008;43:1692–1700. doi: 10.1002/jms.1460. [DOI] [PubMed] [Google Scholar]
- 38.Masujima T. Live single-cell mass spectrometry. Anal Sci. 2009;25:953–960. doi: 10.2116/analsci.25.953. [DOI] [PubMed] [Google Scholar]
- 39.Date S, Mizuno H, Tsuyama N, Harada T, Masujima T. Direct drug metabolism monitoring in a live single hepatic cell by video mass spectrometry. Anal Sci. 2012;28:201–203. doi: 10.2116/analsci.28.201. [DOI] [PubMed] [Google Scholar]
- 40.Horn PJ, et al. Visualization of lipid droplet composition by direct organelle mass spectrometry. J Biol Chem. 2011;286:3298–3306. doi: 10.1074/jbc.M110.186353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, et al. In situ metabolic analysis of single plant cells by capillary microsampling and electrospray ionization mass spectrometry with ion mobility separation. Analyst. 2014;139:5079–5085. doi: 10.1039/C4AN01018C. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Vertes A. Energy charge, redox state, and metabolite turnover in single human hepatocytes revealed by capillary microsampling mass spectrometry. Anal Chem. 2015;87:10397–10405. doi: 10.1021/acs.analchem.5b02502. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Sevinsky CJ, Davis BM, Vertes A. Single-Cell Mass Spectrometry of Subpopulations Selected by Fluorescence Microscopy. Anal Chem. 2018;90:4626–4634. doi: 10.1021/acs.analchem.7b05126. [DOI] [PubMed] [Google Scholar]
- 45.Janes TA, Xu FL, Syed NI. Graded hypoxia acts through a network of distributed peripheral oxygen chemoreceptors to produce changes in respiratory behaviour and plasticity. Eur J Neurosci. 2015;42:1858–1871. doi: 10.1111/ejn.12940. [DOI] [PubMed] [Google Scholar]
- 46.McCrohan CR, Benjamin PR. Synaptic relationships of the cerebral giant-cells with moto-neurons in the feeding system of Lymnaea-stagnalis. J Exp Biol. 1980;85:169–186. doi: 10.1242/jeb.85.1.169. [DOI] [PubMed] [Google Scholar]
- 47.Ferguson GP, Benjamin PR. The whole-body withdrawal response of Lymnaea-stagnalis .1. Identification of central motoneurons and muscles. J Exp Biol. 1991;158:63–95. doi: 10.1242/jeb.158.1.63. [DOI] [PubMed] [Google Scholar]
- 48.Benjamin PR, Winlow W. The distribution of three wide-acting synaptic inputs to identified neurons in the isolated brain of Lymnaea stagnalis (L.) Comparative Biochemistry and Physiology a-Physiology. 1981;70:293–307. doi: 10.1016/0300-9629(81)90182-1. [DOI] [Google Scholar]
- 49.Saunders SE, Kellett E, Bright K, Benjamin PR, Burke JF. Cell-specific alternative RNA splicing of an FMRFamide gene transcript in the brain. J Neurosci. 1992;12:1033–1039. doi: 10.1523/JNEUROSCI.12-03-01033.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamin PR, Burke JF. Alternative mRNA splicing of the FMRFamide gene and its role in neuropeptidergic signalling in a defined neural network. Bioessays. 1994;16:335–342. doi: 10.1002/bies.950160508. [DOI] [PubMed] [Google Scholar]
- 51.Nassel DR. Peptidergic neurohormonal control systems in invertebrates. Curr Opin Neurobiol. 1996;6:842–850. doi: 10.1016/S0959-4388(96)80036-5. [DOI] [PubMed] [Google Scholar]
- 52.Linacre A, et al. Cardioactive neuropeptide Phe-Met-Arg-Phe-NH2 (FMRFamide) and novel related peptides are encoded in multiple copies by a single gene in the snail Lymnaea stagnalis. J Neurosci. 1990;10:412–419. doi: 10.1523/JNEUROSCI.10-02-00412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kellett E, et al. Genomic organization of the FMRFamide gene in Lymnaea: multiple exons encoding novel neuropeptides. J Neurosci. 1994;14:6564–6570. doi: 10.1523/JNEUROSCI.14-11-06564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bright K, et al. Mutually exclusive expression of alternatively spliced FMRFamide transcripts in identified neuronal systems of the snail Lymnaea. J Neurosci. 1993;13:2719–2729. doi: 10.1523/JNEUROSCI.13-06-02719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreiner T, Sossin W, Scheller RH. Localization of Aplysia Neurosecretory Peptides to Multiple Populations of Dense Core Vesicles. J Cell Biol. 1986;102:769–782. doi: 10.1083/jcb.102.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reits E, et al. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/S1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 57.Ekman R, Gobom J, Persson R, Mecocci P, Nilsson CL. Arginine vasopressin in the cytoplasm and nuclear fraction of lymphocytes from healthy donors and patients with depression or schizophrenia. Peptides. 2001;22:67–72. doi: 10.1016/S0196-9781(00)00357-0. [DOI] [PubMed] [Google Scholar]
- 58.Bergquist, J. & Ekman, R. Dynamics of cell nucleus/lymphocyte nuclei-associated peptides studied by mass spectrometry and future peptidomic aspects. In: Mass Spectrometry and Hyphenated Techniques in Neuropeptide Research (eds Silberring, J. & Ekman, R.). John Wiley & Sons (2002).
- 59.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 60.Strohalm M, Hassman M, Kosata B, Kodicek M. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun Mass Spectrom. 2008;22:905–908. doi: 10.1002/rcm.3444. [DOI] [PubMed] [Google Scholar]
- 61.Xia JG, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen RB, Ma MM, Hui LM, Zhang J, Li LJ. Measurement of Neuropeptides in Crustacean Hemolymph via MALDI Mass Spectrometry. J Am Soc Mass Spectrom. 2009;20:708–718. doi: 10.1016/j.jasms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.