Zinc finger protein 521 (ZFP521), a DNA-binding protein containing 30 Krüppel-like zinc fingers, has been implicated in the differentiation of multiple cell types, including hematopoietic stem and progenitor cells (HSPC) and B lymphocytes. Here, we report a novel role for ZFP521 in regulating the earliest stages of hematopoiesis and lymphoid cell development via a cell-extrinsic mechanism.
KEYWORDS: hematopoietic stem cell, ZFP521/ZNF521, lymphopoiesis, bone marrow microenvironment, hematopoietic progenitors
ABSTRACT
Zinc finger protein 521 (ZFP521), a DNA-binding protein containing 30 Krüppel-like zinc fingers, has been implicated in the differentiation of multiple cell types, including hematopoietic stem and progenitor cells (HSPC) and B lymphocytes. Here, we report a novel role for ZFP521 in regulating the earliest stages of hematopoiesis and lymphoid cell development via a cell-extrinsic mechanism. Mice with inactivated Zfp521 genes (Zfp521−/−) possess reduced frequencies and numbers of hematopoietic stem and progenitor cells, common lymphoid progenitors, and B and T cell precursors. Notably, ZFP521 deficiency changes bone marrow microenvironment cytokine levels and gene expression within resident HSPC, consistent with a skewing of hematopoiesis away from lymphopoiesis. These results advance our understanding of ZFP521's role in normal hematopoiesis, justifying further research to assess its potential as a target for cancer therapies.
INTRODUCTION
Considerable attention has been dedicated to elucidating the identities of dysregulated genes in cancer initiation and progression. Dissecting their roles under normal homeostatic conditions is integral to understanding how these proteins function and contribute to malignant transformation. The DNA binding protein zinc finger protein 521 (ZFP521) was first identified as ecotropic viral integration site 3 (Evi3) in B cell leukemias in the AKXD mouse strain (1–3). ZFP521 comprises 30 Krüppel-like zinc finger domains. Its paralog, encoded by Zfp423 (Ebfaz), is also a frequent site of viral integration in murine B cell lymphomas (4). These retroviral insertions are localized to the 5′ regulatory sequences of the genes and are associated with their dysregulation and increased expression of wild-type (WT) transcripts (1–4). In addition, fusion proteins of the human homologue, ZNF521 (EHZF), with PAX5 (PAX5:ZNF521) have been observed in human pre-B-cell acute lymphoblastic leukemia (5, 6).
Zfp521 and Zfp423 are broadly expressed, with highest expression in the brain and heart (3). Their proteins have been implicated in the function and differentiation of early progenitor cells in neural (7–9) and adipose (10, 11) tissues, the erythroid lineage (12), and bone development (13–17). Zfp521/ZNF521 is expressed in early hematopoietic stem cells and is absent in peripheral cell populations, including mature B cells (3–5, 18, 19). In contrast, Zfp423 was not detected in bone marrow (BM) or splenic B cell populations. Importantly, knockdown of Zfp521 reduces repopulation by hematopoietic stem and progenitor cells (HSPC) and promotes their differentiation to become B cells in vitro (20–22), implicating ZFP521 as a regulator of early hematopoietic stem and progenitor cell function.
A speculated role for ZFP521 in regulating B lymphopoiesis, specifically, has been based largely on observations implicating ZFP521/ZNF521 in the posttranslational regulation of early B cell factor (EBF) family members, including EBF1, a master regulator of B cell development (23–27). Overexpression of Zfp521 in AKXD mouse leukemias is associated with the increased expression of Ebf1 as well as many of its target genes, including Cd79a (mb-1) and Cd79b (B29) (2). However, exogenous overexpression of Zfp521/ZNF521 in HEK293 cells abrogates EBF1-driven transcription in a dose-dependent fashion (2, 19). Furthermore, overexpression of ZNF521 in human B lymphoid cell lines or of Zfp521 in murine pre-B cell lymphomas reduced EBF1-dependent transcription (5, 22).
Zinc finger domains 24 to 30 are sufficient for ZFP521 interaction with EBF1, although this region alone is insufficient to inhibit EBF1 function (22). In addition to the zinc finger motifs, the product of Zfp521 includes 12 residues at its N terminus that bind nucleosome remodeling and deacetylase (NuRD) complexes (19, 22, 28, 29). This N-terminal motif was dispensable for ZNF521 inhibition of EBF1 in B lymphoid cell lines (22). In contrast, a separate report indicated that deletion of the N-terminal motif reduced, but did not ablate, ZFP521 repression of EBF1 transactivation of a B29 (Cd79b product) plasmid reporter but blocked EBF1 activation of the bone-relevant target, Ccl9 (14). The disparity in the literature regarding how ZFP521 regulates EBF1 highlights the need for evaluating the normal, endogenous role of ZFP521 in B cells in vivo.
Here, we further define the expression patterns of Zfp521 in hematopoietic progenitor compartments and characterize the consequences of Zfp521 deficiency on lymphopoiesis in vivo. Mice deficient in Zfp521 exhibit reduced BM and thymic cellularity. These mice have altered frequencies and phenotypes of early hematopoietic progenitor cells, as well as B and T cell progenitors. Cytokine expression analysis revealed increased levels of myeloid cell-associated soluble factors in the BM microenvironment of Zfp521-deficient mice. Correlating with the cytokine array, transcriptome sequencing (RNA-seq) analysis of Zfp521−/− hematopoietic stem and progenitor cells revealed transcriptome changes consistent with cytokine signaling changes and myeloid cell-biased differentiation programs. Collectively, these data support a previously undescribed role for ZFP521 in the regulation of hematopoiesis indirectly through soluble factors in the microenvironment.
RESULTS
Zfp521 deficiency alters the frequency of hematopoietic stem and progenitor cells.
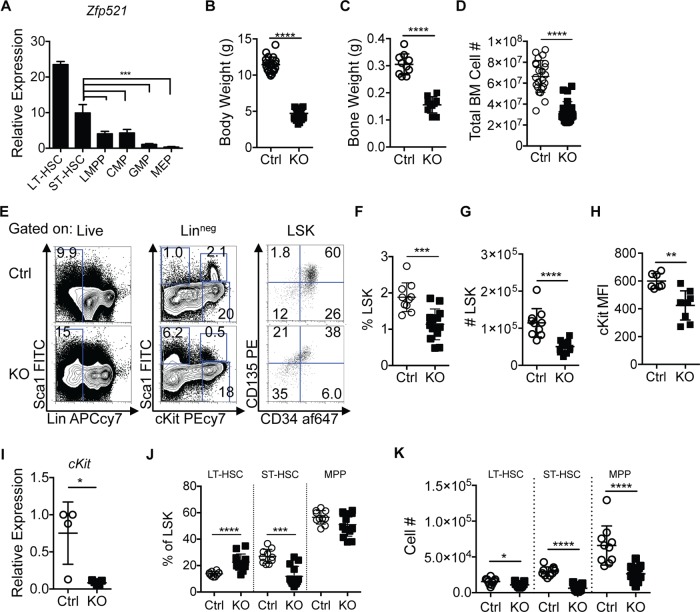
Previous studies indicated that Zfp521 is expressed abundantly in hematopoietic stem cells (HSC) and decreases with differentiation (3, 19). However, these experiments were not performed using highly purified populations of hematopoietic cells. To assess the expression of Zfp521 throughout hematopoiesis, we purified hematopoietic stem and progenitor populations from wild-type C57BL/6 mice and measured Zfp521 transcripts using reverse transcription-quantitative PCR (qRT-PCR) (Fig. 1A). We confirmed that the highest expression level of Zfp521 transcripts was observed within the long-term HSC (LT-HSC) compartment (Linneg Sca1+ cKit+ CD34neg CD135neg). Zfp521 transcript levels decreased progressively with differentiation to short-term HSC (ST-HSC; Linneg Sca1+ cKit+ CD34+ CD135neg), lymphoid-primed multipotent progenitor cells (LMPP; Linneg Sca1+ cKit+ CD34+ CD135hi), common myeloid progenitors (CMP; Linneg Sca1neg cKit+ CD34+ CD16/32neg), granulocyte-macrophage progenitors (GMP; Linneg Sca1neg cKit+ CD34+ CD16/32+), and megakaryocyte-erythrocyte progenitors (MEP; Linneg Sca1neg cKit+ CD34neg CD16/32neg).
FIG 1.
Zfp521 deficiency alters early HSC and progenitor population phenotypes. (A) BM cells of WT C57BL/6 mice were analyzed for Zfp521 expression by qRT-PCR. Populations analyzed include long-term hematopoietic stem cells (LT-HSC; Linneg Sca1+ cKit+ CD34neg CD135neg), short-term HSC (ST-HSC; Linneg Sca1+ cKit+ CD34+ CD135neg), lymphoid-primed multipotent progenitor cells (LMPP; Linneg Sca1+ cKit+ CD34+ CD135hi), common myeloid progenitors (CMP; Linneg Sca1neg cKit+ CD34+ CD16/32neg), granulocyte-macrophage progenitors (GMP; Linneg Sca1neg cKit+ CD34+ CD16/32+), and megakaryocyte-erythroid progenitors (MEP; Linneg Sca1neg cKit+ CD34neg CD16/32neg). Zfp521 transcript levels were normalized to internal Hprt levels and then to Zfp521 levels in GMP cells. Each biological replicate was run as three technical replicates. Mouse n = 2 LT-HSC, 3 ST-HSC, 4 LMPP, 3 CMP, 3 GMP, 3 MEP. Statistics were generated using one-way analysis of variance (ANOVA). (B) Body weight of 3-week-old Zfp521−/− (KO) and control (Ctrl) littermates. Mouse n = 21 Ctrl, 23 KO. (C) Weight of bones of 3-week-old Zfp521−/− (KO) and control (Ctrl) littermates. Harvested bones include both hind limbs and forelimbs: tibia, femur, pelvis, radius, and ulna. Mouse n = 10 Ctrl, 10 KO. (D) Total BM cell numbers in 3-week-old Zfp521−/− (KO) and littermate control (Ctrl) mice. Mouse n = 23 Ctrl, 25 KO. (E) Representative flow cytometry plots and gating strategy for identification of Linneg cells (left panels) (Lin panel contains B220, CD3, CD11b, Gr1, and Ter119), LSK (middle panels), and LT-HSC, ST-HSC, and MPP populations (right panels). Numbers indicate frequencies of parental population. Data are representative of 10 Ctrl and 13 KO mice. FITC, fluorescein isothiocyanate; PE, phycoerythrin; APC, allophycocyanin. (F) Frequencies of LSK cells within Linneg populations. Mouse n = 10 Ctrl, 13 KO. (G) Total numbers of LSK cells. Mouse n = 10 Ctrl, 13 KO. (H) Mean fluorescence intensity (MFI) of surface cKit expression on LSK cells as assessed by flow cytometry. Mouse n = 7 Ctrl, 9 KO. (I) Transcript expression of cKit in FACS-purified LSK cells was determined by qRT-PCR. Transcript levels were normalized to internal Hprt levels and then to Ctrl cKit levels. Mouse n = 4 Ctrl, 5 KO. (J) Frequencies of LT-HSC, ST-HSC, and MPP populations within LSK populations. Mouse n = 10 Ctrl, 13 KO. (K) Numbers of LT-HSC, ST-HSC, and MPP populations. Mouse n = 10 Ctrl, 13 KO. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To assess the functional role of ZFP521 in early hematopoiesis, hematopoietic stem and progenitor populations of Zfp521 knockout (Zfp521−/−) and Zfp521+/+ littermate control (Ctrl) mice were characterized by flow cytometry. It is important to note that all Zfp521−/− mice die within a few weeks of birth. Thus, our studies were performed using Zfp521−/− and Ctrl mice at 3 weeks of age. Zfp521−/− mice are severely runted, having significantly reduced body mass (Fig. 1B) and bone mass (Fig. 1C). This is likely due to ZFP521's regulation of adipocytes, osteoblasts, and osteoclasts (10, 13, 15, 30). Zfp521−/− mice exhibit reduced BM cellularity (Fig. 1D). However, the numbers of BM cells per gram of bone are the same for Zfp521−/− and Ctrl mice (data not shown), suggesting that reduced occupancy space may be responsible in part for the reduction in BM cellularity.
The frequencies and numbers of HSC-enriched Linneg Sca1+ cKit+ (LSK) cells were reduced in Zfp521−/− mice (Fig. 1E to G). Interestingly, surface expression and transcript levels of cKit, a key regulator of HSC function (31, 32), were significantly decreased on and in Zfp521−/− LSK cells (Fig. 1H and I). The surface expression patterns of CD34 and CD135, used to phenotypically define HSC populations, were altered on Zfp521−/− LSK cells relative to those on the Ctrl (Fig. 1E). Frequencies of LT-HSC were increased, those of ST-HSC were reduced, and multipotent progenitors (MPP; Linneg Sca1+ cKit+ CD34+ CD135+) were unaltered in Zfp521−/− BM (Fig. 1J). The numbers of LT-HSC, ST-HSC, and MPP were all decreased in Zfp521−/− mice (Fig. 1K). These results indicate that ZFP521 plays an important role in early hematopoietic stem and progenitor cell homeostasis.
Zfp521 deficiency impacts early BM myeloid and lymphoid progenitor populations.
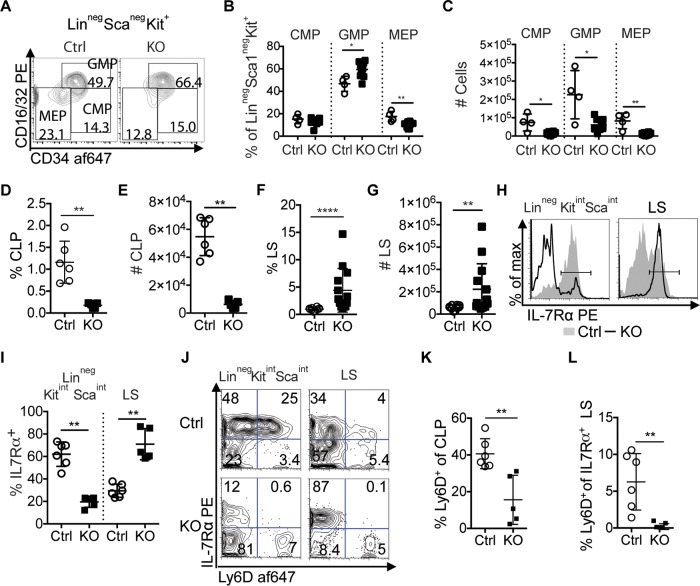
The effects of Zfp521 deficiency on early myeloid and lymphoid progenitor populations were investigated using previously defined stage-specific surface marker expression. Common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP) were analyzed from the bone marrow of 3-week-old Zfp521−/− mice and Ctrl littermates by flow cytometry. While there was no significant difference in the frequency of CMP, GMP were significantly increased and MEP were significantly decreased in Zfp521−/− mice relative to Ctrl littermates (Fig. 2A and B). However, the numbers of CMP, GMP, and MEP were all significantly reduced in the BM of Zfp521-deficient mice (Fig. 2C).
FIG 2.
Frequencies of myeloid and lymphoid progenitors are altered in Zfp521-deficient mice. (A) Representative flow plots of CD16/32 and CD34 expression on Linneg Sca1neg Kit+ cells in the BM of Ctrl and Zfp521−/− (KO) mice. (B) Frequencies of CMP (Linneg Sca1neg cKit+ CD16/32neg CD34+), GMP (Linneg Sca1neg cKit+ CD16/32+ CD34+), and MEP (Linneg Sca1neg cKit+ CD16/32neg CD34neg) within the Linneg Sca1neg Kit+ population. (C) Numbers of CMP, GMP, and MEP cells. (D and E) Frequencies (D) and numbers (E) of CLP (Linneg Sca1int cKitint IL-7-Rα+) within Linneg cells (n = 6 Ctrl, 5 KO). (F and G) Frequencies (F) and numbers (G) of LS (Linneg Sca1+ cKitneg, n = 10 Ctrl, 13 KO) within Linneg populations. (H) Representative histogram of IL-7Rα expression on Linneg Sca1int cKitint (left panel) and LS (right panel) cells. Mouse n = 6 Ctrl, 5 KO. (I) Frequencies of IL-7Rα+ cells within Linneg cKitint Sca1int and LS populations. (J) Representative flow plots of Ly6D expression on Linneg Kitint Sca1int (left column; CLP comprise all IL7Rα+ cells in both top left and top right quadrants of flow plots) and LS cells (right column) in BM of KO (n = 5) and Ctrl (n = 6) mice. (K) Frequencies of Ly6D+ cells within CLP populations. Values were determined as percent Ly6D+ IL-7Rα+ cells of total IL-7Rα+ cells as shown in panel I. (L) Frequencies of Ly6D+ cells within the IL-7Rα+ LS populations. Mouse n = 6 Ctrl, 5 KO. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We next assessed the effects of Zfp521 deficiency on lymphoid progenitor cells. Common lymphoid progenitors (CLP; Linneg cKitint Sca1int interleukin-7 receptor α positive [IL-7Rα+]) give rise to all lymphocyte populations but possess limited myeloid differentiation potential (33). The frequencies and numbers of CLP were significantly reduced in Zfp521−/− mice (Fig. 2D and E). A second phenotypically and functionally distinct subset of early lymphoid progenitors, the Linneg Sca1+ cKitneg (LS) population, has been reported to possess T, B, and NK cell potential but no myeloid potential (34). Zfp521−/− mice possessed increased frequencies and numbers of LS cells (Fig. 2F and G). Notably, the frequencies of IL-7Rα+ cells were reduced in Linneg cKitint Sca1int cell populations but increased in LS cell populations of Zfp521−/− mice (Fig. 2H and I). Previous reports demonstrated that Ly6D marks B-cell-biased CLP (35). Ly6D+ CLP (also known as B-cell-biased lymphoid progenitors or BLP) were significantly reduced in Zfp521−/− mice (Fig. 2J and K). Similarly, frequencies of Ly6D+ cells were reduced within IL-7Rα+ LS populations from Zfp521−/− mice (Fig. 2J and L). These results indicate that germ line Zfp521 deficiency results in reduced CLP populations and increased LS populations.
B and T cell progenitor populations are skewed in Zfp521−/− mice.
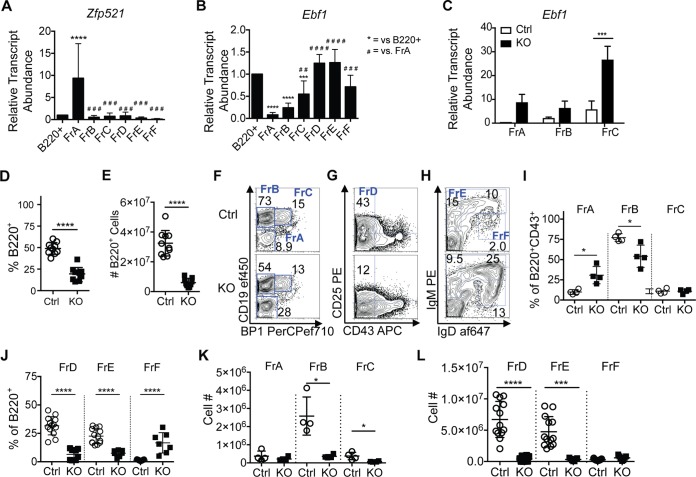
Previous studies have concluded that ZFP521 is a regulator of B lymphopoiesis, largely through its interactions with EBF1 (22). This mechanism was assessed in cells with exogenously coexpressed ZFP521 and EBF1, which resulted in direct interactions between the two proteins. To determine whether Zfp521 and Ebf1 are coexpressed during B lymphopoiesis, we assessed the relative transcripts levels in purified B cell progenitors from wild-type (WT) mice (Fig. 3A and B). The total B220+ cells and pre-pro-B (Hardy fraction A [FrA]; Linneg B220+ CD43+ CD19neg BP1neg), pro-B (FrB; Linneg B220+ CD43+ CD19+ BP1neg/low), early pre-B (FrC; Linneg B220+ CD43+ CD19+ BP1+), late pre-B (FrD; B220+ CD43neg CD25+), immature B (FrE; B220+ IgM+ IgDneg), and recirculating mature B (FrF; B220+ IgMlow IgD+) cells were purified by fluorescence-activated cell sorting (FACS) from the BM, and RNA was extracted for subsequent qRT-PCR analysis. Zfp521 transcripts were highest in pre-pro-B cells, with relatively low expression in all other B progenitor populations (Fig. 3A). In contrast, transcript levels of Ebf1 were low in pre-pro-B cells and increased with differentiation, reciprocal to Zfp521 levels (Fig. 3B). Importantly, Zfp521-deficient early B cell progenitors expressed increased amounts of Ebf1 transcripts (Fig. 3C). These data suggest that ZFP521 negatively regulates the expression of an integral B cell transcription factor gene, Ebf1, at the transcriptional level.
FIG 3.
Zfp521−/− mice possess reduced frequencies of early B cell progenitors. (A and B) qRT-PCR of Zfp521 (A) and Ebf1 (B) in total B220+ BM cells and pre-pro-B (Hardy fraction A [FrA]), pro-B (FrB), early pre-B (FrC), late pre-B (FrD), immature (FrE), and recirculating mature B (FrF) cells in wild-type mice. Transcript amounts were normalized to internal Hprt levels and then to levels in total B220+ cells. Statistics were generated using one-way ANOVA. Each biological replicate includes three technical replicates. Mouse n: B220+, 11 (A) or 10 (B); FrA and FrB, 6; FrC and FrD, 5 (A) or 4 (B); FrE and Fr F, 4 (A) or 3 (B). (C) Transcript levels of Ebf1 were measured in FACS-purified pre-pro-B (FrA), pro-B (FrB), and early pre-B (FrC) cells by qRT-PCR from 3-week-old Zfp521-deficient mice (n = 2) and control littermates (n = 3 to 5). Transcript levels were normalized to internal Hprt levels. Statistics were generated using two-way ANOVA. Each biological replicate includes three technical replicates. (D and E) Frequencies of B220+ cells of singlets (D) and numbers of B220+ cells (E) in BM of Zfp521−/− (KO) and control (Ctrl) mice. Cells were gated on Linneg (NK1.1, Ly6C, CD317, CD3e, CD11b, CD11c, Gr1, Ter119) cells. Mouse n = 10 Ctrl, 9 KO. (F to H) Flow cytometric analysis of B cell progenitor populations in the BM of Ctrl and KO mice. Mouse n: 4 Ctrl and 4 KO for FrA to FrC, 13 Ctrl and 8 KO for FrD to FrF. (F) Pre-pro-B (FrA), pro-B (FrB), and early pre-B (FrC) cells, gated on Linneg B220+ CD43+ BM cells. (G) Late pre-B (FrD) cells were gated on total B220+ BM cells. (H) Immature (FrE) and recirculating mature B (FrF) cells were gated on total B220+ BM cells. (I and J) Percent indicated B cell progenitor population of B220+ CD43+ cells (I) or total B220+ cells (J). (K and L) Numbers of indicated B cell progenitor populations in the BM. *, P < 0.05; **, P < 0.01; *** and ###, P < 0.001;**** and ####, P < 0.0001.
The frequencies and total numbers of B220+ cells were significantly reduced in the BM of Zfp521−/− mice (Fig. 3D and E). Phenotypic analysis of B cell progenitor populations revealed significantly increased frequencies of pre-pro-B cells in Zfp521−/− mice (Fig. 3F and I). Frequencies of pro-B (Fig. 3F and I), late pre-B, and immature B (Fig. 3G, H, and J) cells were all reduced. Interestingly, Zfp521−/− BM contained higher frequencies of recirculating mature B cells (Fig. 3H and J). The numbers of pro-B cells through immature B cells were significantly decreased in Zfp521−/− mice relative to Ctrl littermates, while the numbers of recirculating mature B cells were not affected (Fig. 3K and L). Unlike earlier-stage B cell progenitors in the bone marrow, mature FrF cells migrate to the periphery prior to reentering the bone marrow niche (36). Thus, the lack of ZFP521 has minimal effects on B cells that have matured in the periphery, while ZFP521 deficiency impacts earlier stages of B lymphopoiesis in the bone marrow.
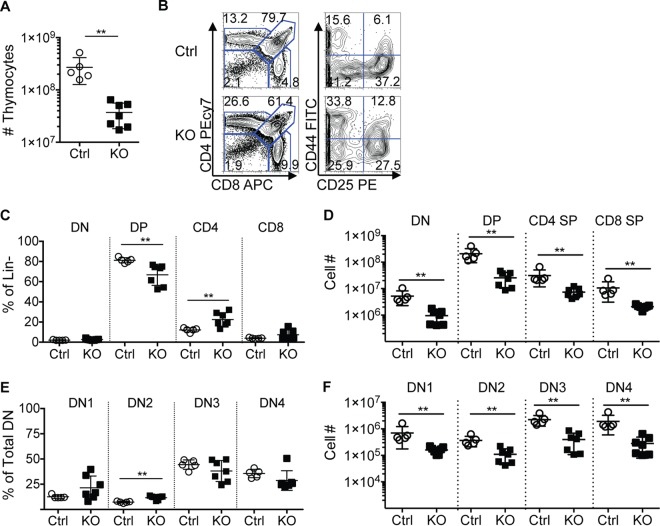
We next assessed the consequences of Zfp521 deficiency on T lymphopoiesis. The thymi of Zfp521−/− mice were smaller than those of Ctrl littermates and exhibited significantly reduced cellularity (Fig. 4A). Analysis of CD4, CD8, CD25, and CD44 surface expression on thymocytes revealed a significant effect of Zfp521 deficiency on T cell progenitor populations (Fig. 4B). Zfp521−/− mice possessed reduced frequencies of double-positive (DP) thymocytes and increased CD4 single-positive (SP) cells (Fig. 4B and C). The total number of CD4neg CD8neg (double negative [DN]), CD4+ CD8+ DP, CD4+ SP, and CD8+ SP cells were all significantly reduced in Zfp521−/− thymi (Fig. 4D). The earliest T cell precursors proceed through phenotypically defined stages: DN1 (CD44+ CD25neg), DN2 (CD44+ CD25+), DN3 (CD44neg CD25+), and DN4 (CD44neg CD25neg) (37, 38). Zfp521−/− mice possessed only slightly higher frequencies of DN2 cells than the Ctrl mice (Fig. 4B and E); however, the numbers of all DN cell populations were significantly reduced in Zfp521−/− thymi (Fig. 4F). These data are the first to implicate ZFP521 in T cell development. Because Zfp521-dependent defects largely occur in the earliest T cell progenitor populations, they may be a consequence of defective precursor BM cells.
FIG 4.
Thymopoiesis is altered in Zfp521−/− mice. (A) Total numbers of thymocytes in Zfp521−/− (KO) and control (Ctrl) littermates at 3 weeks of age. (B) Representative flow cytometry plots of T cell progenitor subsets. Left, CD4 and CD8 expression in Linneg (B220, NK1.1, Gr1, CD11b) thymocytes. Right, CD44 and CD25 expression within double-negative (DN) populations (Linneg CD4neg CD8neg): DN1 (CD44+ CD25neg), DN2 (CD44+ CD25+), DN3 (CD44neg CD25+), and DN4 (CD44neg CD25neg). (C and D) Frequencies (C) and numbers (D) of total DN, CD4+ CD8+ (DP), CD4+, and CD8+ of Linneg thymocytes. (E and F) Frequencies (E) and numbers (F) of DN1 to DN4 populations within total DN populations. Mouse n = 5 Ctrl and 7 KO mice. **, P < 0.01.
Zfp521 interacts with transcriptional regulators of hematopoiesis.
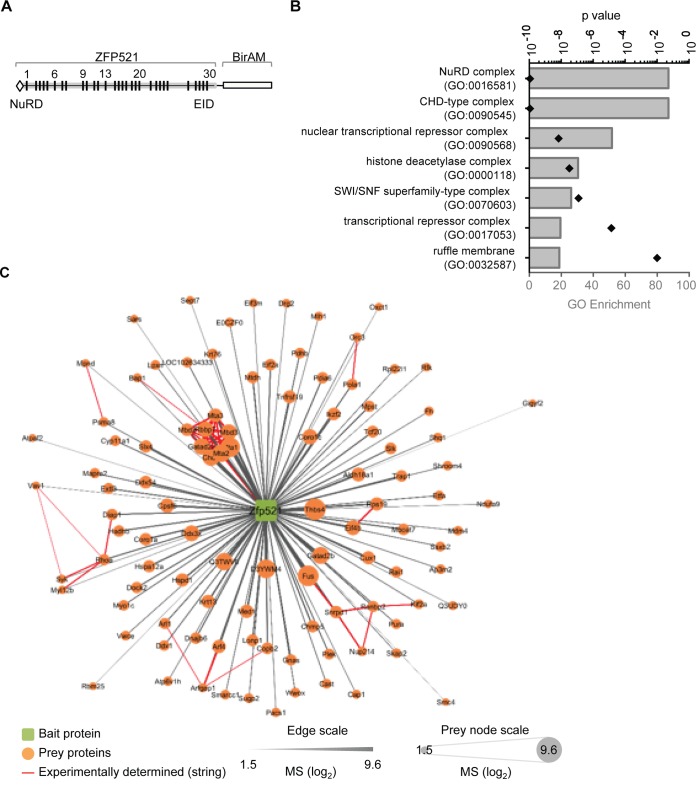
ZFP521 contains numerous potential DNA- and protein-interacting domains, including its 30 zinc finger motifs, yet the list of proteins reported to interact with ZFP521 is quite small. To identify transcriptional regulators that may interact with ZFP521 to regulate lymphopoiesis, we generated a ZFP521:BirAM fusion protein for proximity-dependent biotinylation (BioID) in Ba/F3 cells, a pre-pro-B (FrA)-like cell line that does not express EBF1 (Fig. 5A) (39, 40). A total of 131 proteins were identified as potential ZFP521-interacting partners (Fig. 5B; see Table S1 in the supplemental material), including ZFP521 itself and expected NuRD components (CHD3, Mta1/2/3, and Mbd2/3). Interestingly, ZFP521:BirAM also biotinylated novel interactive candidates, including the lymphopoiesis-associated transcriptional regulators Cut-like homeobox 1 (CUX1) and IKZF2 (Helios), as well as the guanine nucleotide exchange factor VAV1 (Fig. 5C) (41–48). Together, these data define a new regulatory network consisting of ZFP521 and its extensive interactions with epigenetic and transcriptional regulators in hematopoiesis.
FIG 5.
BioID analysis identifies ZFP521-interacting proteins. (A) ZFP521:BirAM fusion protein. The previously identified NuRD binding domain and EBF-interacting domain (EID) are indicated. (B) Network plot of BioID-identified ZFP521-interacting candidates in Ba/F3 cells. The distance and size of individual prey protein nodes denote enrichment in BioID experiments, and red strings indicate interactions among prey proteins identified in previous studies. Each of three independent biological replicates was analyzed with two technical replicates. (C) Gene ontology (GO) analysis of BioID candidate proteins for enrichment of cellular components. The top seven cellular component complexes are shown.
Myeloid cell-associated cytokines and transcriptional signatures are elevated in Zfp521−/− BM environments.
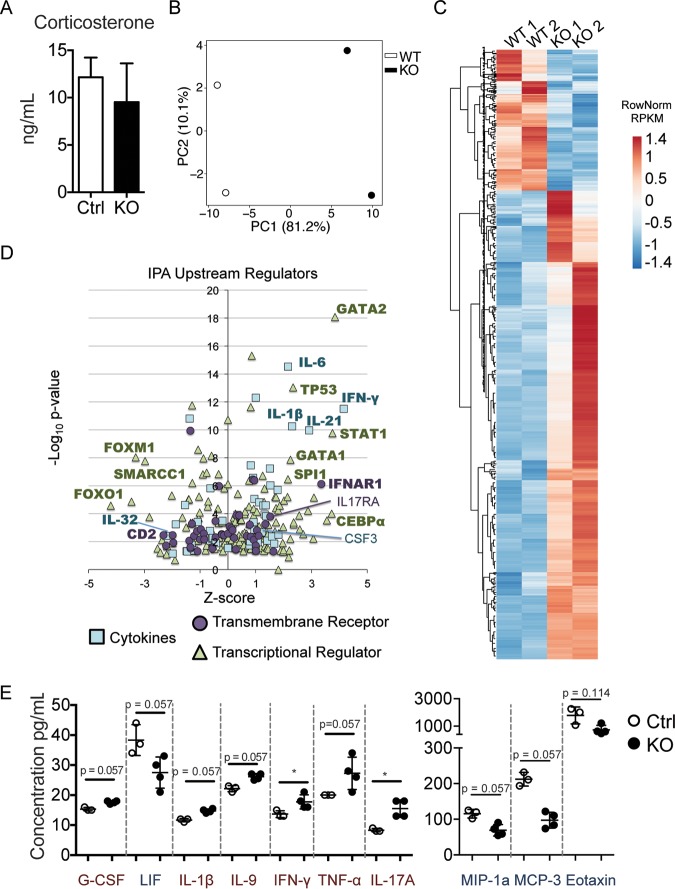
Elevated stress-related hormones, such as the glucocorticoid corticosterone, reduce the frequency of early lymphocyte progenitor populations (49–51), similar to the phenotypes seen in Zfp521−/− mice. Furthermore, Zfp521−/− mice are significantly runted and display abnormal behavior, which may increase the levels of stress hormones (52). However, serum levels of corticosterone were similar between 3-week-old Zfp521−/− mice and Ctrl littermates (Fig. 6A). Thus, stress-induced corticosterone is not the driver of reduced lymphoid progenitors in Zfp52-deficient mice.
FIG 6.
Zfp521 deficiency affects HSC transcriptomes, potentially through circulating factors within the BM microenvironment. (A) The concentration of serum corticosterone was analyzed by ELISA. Mouse n = 3 littermate controls (Ctrl) and 4 Zfp521−/− knockouts (KO). (B and C) LSK cells were sorted from the BM of KO and Ctrl mice, and RNA was purified and processed for RNA-seq analysis (n = 2 mice each). (B) Principal-component analysis was performed using the top 500 most variable genes. (C) Heatmap showing 947 genes significantly altered (fold change of >2 or <0.5, P value adjusted to <0.1; 728 up and 219 down) in Zfp521−/− LSK relative to Ctrl LSK. (D) IPA analysis of upstream regulator candidates. Cytokines, transmembrane receptors, and transcriptional regulators are shown. Candidates of interest are labeled, and regulators called activated or repressed by IPA are in bold. (E) Soluble protein concentrations were analyzed in BM fluid from Ctrl (n = 3) and KO (n = 4) mice by cytokine array. Trending and significantly upregulated (red) and downregulated (blue) proteins are shown. Statistics were generated using the Mann-Whitney test. *, P < 0.05.
To determine candidate pathways and molecules that may instigate the altered phenotype of Zfp521−/− LSK cells, we performed RNA-seq analysis. Principal-component analysis (PCA) was performed to assess relationships between biological replicates (Fig. 6B). A total of 947 genes exhibited significantly changed expression in Zfp521−/− LSK cells relative to the Ctrl (Fig. 6C). Of these, 728 genes were upregulated and 219 were downregulated. Ingenuity pathway analysis (IPA) was performed to identify upstream regulators potentially driving the gene expression changes (Fig. 6D). Expression of multiple cytokines and cytokine receptors was increased (e.g., gamma interferon [IFN-γ], IL-6, IL-21, IL-1β and IFNAR1 [Fig. 6D, in bold]) or decreased (e.g., IL-32). Additionally, transcriptome signatures were enriched for key hematopoietic transcription factors, including CEBPα, SPI1 (PU.1), GATA1, and GATA2. Notably, many of these cytokines and transcription factors induce myelopoiesis and suppress lymphopoiesis (e.g., IL-6, IL-1β, CEBPα, SPI1, and GATA1) (53–56).
To detect microenvironmental mediators that could be potentially responsible for the transcriptional changes seen in Zfp521−/− LSK cells, relative amounts of 36 cytokines and chemokines in Ctrl versus Zfp521−/− BM fluids were measured (Fig. 6E). Of the assayed proteins, five were significantly increased or trending toward increase (granulocyte colony-stimulating factor [G-CSF], IL-1β, IL-9, IFN-γ, IL-17A, tumor necrosis factor alpha [TNF-α]), and four were significantly decreased or trending toward decrease (LIF, MIP-1a, MCP-3, eotaxin) in BM supernatants of Zfp521−/− mice. The cytokine array largely correlated with the RNA-seq IPA analysis, which suggested increased levels of myeloid cell-polarizing cytokines. IPA predicts activation of IL-1β pathways in LSK cells; this conclusion is supported by the increased IL-1β levels that were observed in the BM fluids of Zfp521−/− mice. Additionally, IPA suggests that LIF is inhibited in LSK cells by the lack of ZFP521. Indeed, LIF protein trended toward reduced levels in BM fluids from Zfp521-deficient mice. Most notably, G-CSF increased (nearly significantly) in the BM of Zfp521−/− mice and its downstream pathway is activated (gene name, CSF3) in Zfp521−/− LSK cells. G-CSF has previously been shown to suppress B cell development (57), and G-CSF-mediated reductions in B cell progenitors largely resemble the phenotypes observed in Zfp521−/− BM. Together, these data support a model in which Zfp521 deficiency results in altered cytokine and chemokine levels in the BM. In turn, these changes in the BM microenvironment influence hematopoiesis and the resulting hematopoietic cell functions.
DISCUSSION
Much research has been devoted to exploring ZFP521 as a potential regulator of B cell development through its putative inhibition of EBF1. Previous experimental studies employed overexpression of wild-type or mutant ZFP521 protein or short hairpin RNA (shRNA)-mediated knockdown of endogenous Zfp521 transcripts to modulate ZFP521 in B cells. Other studies have linked overexpression of Zfp521 with murine and human B cell leukemogenesis. These studies have advocated a role for ZFP521 in the negative regulation of B cell development via inhibition of EBF1, potentially through direct protein-protein interactions.
Here, we report that mice with germ line disruption of Zfp521 exhibit reductions in the frequency and number of early hematopoietic stem and progenitor cells, including lymphoid progenitor populations. Specifically, Zfp521−/− HSC populations are reduced, express reduced levels of cKit, and exhibit altered expression of CD34 and CD135, which are used to phenotypically classify LT-HSC, ST-HSC, and MPP populations. Zfp521−/− mice possess increased frequencies of GMP and reduced frequencies of MEP. Additionally, LS lymphoid progenitors are increased, while CLP are reduced.
Zfp521−/− mice contain increased frequencies of the earliest B cell progenitor population: pre-pro-B cells. Interestingly, the frequencies of successive B cell progenitor populations (pro-B through immature B) in the BM of Zfp521−/− mice are reduced up until the recirculating mature B cell stage; at this stage, the populations have expanded in the periphery and returned to the BM (36). T cell progenitor populations were also altered in Zfp521−/− mice. Notably, the CD4+ CD8+ DP population of thymocytes was significantly reduced in frequency and number in Zfp521−/− mice. Whether the reductions in lymphoid progenitor populations are due to cell death, altered expression of surface markers used to define these populations, selective developmental arrest, accelerated development, or a combination of cellular responses or reflect real defects present in their precursor progenitor populations remains to be determined.
Importantly, the Zfp521 expression pattern in hematopoiesis is reciprocal to that of Ebf1 expression. Therefore, simultaneous expression of Zfp521 and Ebf1 is limited and quite disproportional, restricting the lymphocyte progenitor populations in which these two transcriptional regulators potentially interact. Although bone marrow reconstitution assays indicated that the competitive or repopulation potential of Zfp521-deficient BM was slightly reduced (data not shown), we did not detect a significant reduction in the ability of Zfp521−/− donor BM to generate B or T cells in wild-type recipients. Furthermore, in vitro B and T cell differentiation assays did not reveal cell-intrinsic defects in the ability of Zfp521−/− LSK cells to differentiate into either of these lymphoid lineages (data not shown). These data are in contrast with our observations in primary Zfp521−/− mice, which exhibit significantly altered frequencies and numbers of B and T cell progenitors. Thus, while we cannot rule out cell-intrinsic regulation of early hematopoiesis and lymphopoiesis by ZFP521, our data suggest the importance of ZFP521 for extrinsic mechanisms acting upon blood development in the BM niche.
Mechanisms for the regulation of hematopoiesis by ZFP521 likely include context-specific interactions with other transcription factors and regulatory proteins. Previous studies revealed very little in this regard, with the notable exceptions of EBF1 and NuRD complexes (14). In this study, we used proximity biotinylation to identify novel interactions between ZFP521 and factors, including the hematopoiesis-associated proteins CUX1, Helios, and VAV1. Notably, transcripts encoding each of these factors are expressed in hematopoietic stem cells, which also express high levels of Zfp521 transcripts (58). ZFP521 is overexpressed in B cell leukemias (1–3, 5, 6), and thus further studies will be required to evaluate interactions between ZFP521 and these transcriptional regulators in the context of leukemia or lymphomagenesis.
Cell-extrinsic regulation of hematopoiesis is mediated through cell-cell interactions between hematopoietic cells and local niche cells, as well as through soluble factors, such as cytokines, chemokines, and hormones. It is notable that increased levels of stress hormones, including glucocorticoids, have been reported to reduce the numbers of intermediate-stage B and T cell progenitors (49–51). Although this phenotype strikingly resembles that of our Zfp521−/− mice, corticosterone levels were not elevated in the serum of Zfp521-deficient mice.
Transcriptomes of the HSC-enriched LSK population in Zfp521-deficient mice displayed significant changes relative to Ctrl LSK. Notably, many pathways downstream of cytokines and cytokine receptors were identified as activated or inhibited by IPA. Indeed, the levels of many of these soluble protein candidates were significantly changed in Zfp521−/− BM microenvironments. Although many of the changes in cytokine levels appear small, it is important to note that these assays measured the mean protein expression across the entire BM compartment. Many of these soluble mediators function in gradients and/or a paracrine fashion. Thus, small changes in mean protein levels may represent large changes within distinct microenvironments. Strong correlations between the RNA-seq data and protein arrays strongly support elevated levels of the myeloid cell-promoting cytokines IL-1β, G-CSF, IFN-γ, and IL-17 and increased signaling through corresponding receptors in the HSC-enriched LSK compartment. Furthermore, IPA identified transcriptome signatures of key myeloid cell-associated transcription factors, including GATA1, CEBPα, and PU.1. However, whether these cytokines are directly regulated by ZFP521 or whether they are causative of the hematopoietic phenotypes observed remains to be determined. For example, the increased amounts of G-CSF may be a consequence of increased IL-1 signaling (58). It is notable that two proinflammatory cytokines, IFN-γ and IL-17, were significantly elevated in the absence of ZFP521. Upregulation of IFN-γ and that of IL-17 have each been linked with reduced hematopoietic stem cell function and increased myelopoiesis (59, 60). IFN-γ is associated with reduced erythropoiesis (61), which is supported by our detection of increased and decreased frequencies of GMP and MEP, respectively, in Zfp521-deficient mice.
Previous reports utilizing competitive BM transplants (BMT) of whole fetal liver cells from Zfp521−/− or Ctrl mice showed a slight reduction in donor chimerism and reduced myeloid reconstitution of Zfp521−/− donors in primary BMT recipients (20). We similarly report that myeloid progenitor populations are altered in 3-week-old Zfp521−/− mice. However, our BMT assays indicated no effects of Zfp521 deficiency on lineage skewing (data not shown). We demonstrate that ZFP521 functions in both cell-intrinsic and -extrinsic capacities. With regard to cell-extrinsic influences, the niche compartments differ between those of fetal liver hematopoietic progenitors and BM-resident hematopoietic progenitors of 3-week-old mice, which may account for the differences observed between these studies. Importantly, results demonstrating that long-term lymphoid reconstitution is unaffected by Zfp521 deficiency (20) support our conclusion that ZFP521 regulation of lymphopoiesis is largely cell extrinsic.
Finally, many of the cytokines identified in the cytokine array are produced and secreted by BM niche cells, including osteoclasts and osteoblasts (60–69). The developmental progression and function of these niche cells are known to be regulated by ZFP521, and their frequencies and function are altered in Zfp521−/− mice (14–17, 30). Collectively, our data support a model in which ZFP521 regulates functions of BM niche cells, including osteoblasts and osteoclasts, resulting in altered cytokine and chemokine production by these cells. In turn, these cytokines influence the fates and functions of the niche cells themselves, as well as surrounding hematopoietic stem and progenitor populations. To address mechanisms contributing to these observations, future studies will utilize high-throughput mRNA sequencing to interrogate ZFP521-dependent changes in transcriptomes of various BM niche cells, as well as chromatin immunoprecipitation sequencing (ChIP-seq) to identify direct targets of ZFP521 in vivo.
MATERIALS AND METHODS
Mice.
Zfp521tm2NgC (Zfp521−/−) mice and PCR screening for wild-type and deleted (knockout) alleles were described previously (13). Zfp521+/− mice were maintained on a mixed 129S1/SvImJ.C57BL/6NHsd background or fully crossed onto the 129S1/SvImJ.C57BL/6NHsd background. Because genetic background-specific effects on the Zfp521−/− phenotype were noted, only Zfp521−/− agouti and Zfp521+/+ WT (Ctrl) agouti littermates were used in our assays. All mice were bred and housed in the pathogen-free Biological Resources Center of National Jewish Health. All experiments were performed with prior approval of the National Jewish Health Institutional Animal Care and Use Committee.
Flow cytometry.
Thymus and BM (tibia, femur, hipbone, humerus, radius, and ulna from both front and back legs) were harvested from 3-week-old Zfp521−/− mice or WT littermates. Single-cell suspensions were hemolyzed and used for flow cytometry using the LSR Fortessa and FACS using MoFlo XDP (Beckman Coulter) and SY3200 (Sony) cell sorters. Data analysis was performed using FlowJo software (TreeStar). Information regarding antibodies is located in Table S2 in the supplemental material. DAPI (4′,6-diamidino-2-phenylindole) was used for dead cell exclusion.
RNA isolation, cDNA synthesis, and qRT-PCR.
Hematopoietic progenitor cell cDNA was generated from cells as described previously (62). Briefly, LT-HSC, ST-HSC, MPP, GMP, and MEP were FACS purified using a BD FACSAria (BD Biosciences). LSK and B cell progenitors were purified using a SY3200 (Sony) cell sorter. RNA was extracted using the Qiagen RNeasy micro kit, and single-strand DNA was generated using SuperScript reverse transcriptase II (Invitrogen). qRT-PCR was performed using SYBR green (Applied Biosystems) reagent and analyzed using an Applied Biosystems 7300 real-time PCR system. Primer sequences were as follows: Hprt fwd, 5′-GGGGGCTATAAGTTCTTTGCTGACC-3′; Hprt rev, 5′-CCTGTATCCAACACTTCGAGAGGTCC-3′; Zfp521 fwd, 5′-GGTGTTTGAGTCACTGAGCGACATC-3′; Zfp521 rev, 5′-CCTCTCCGAAATCACACCCTTCTC-3′; Ebf1 fwd, 5′-GCCTTCTAACCTGCGGAAATCCAA-3′; Ebf1 rev, 5′-GGAGCTGGAGCCGGTAGTGGAT-3′; cKit fwd, 5′-AAGGGGACACATTTACGGTGG-3′; and cKit rev, 5′-TCCAGAATCGTCAACTCTTGCC-3′.
Proximity-dependent biotinylation (BioID).
Full-length cDNA sequences, including the Zfp521 open reading frame (ORF), were PCR amplified using primer sequences 5′-TTAGGGGATCCACCATGTCTCGCCGCAAGCAAGCGAAACC-3′ and 5′-GAAATGTCGACACTGCTGTGCTGAGTCATCGTATGATTCTG-3′, the product was cut with BamHI and SalI, and the fragment was cloned into a retroviral MigR1 vector between BglII and XhoI sites, resulting in the fusion of BirAM (62) to the C terminus of ZFP521. For a control protein, we employed BirAM harboring the nuclear localization signal (NLS) from simian virus 40 (SV40) on its N terminus. Vectors were cotransfected with pCMV-VSVG into Phoenix cells using X-tremeGene HP DNA transfection reagent (Roche) according to the manufacturer's protocol. Conditioned medium was harvested 2 days after the transfection and used as virus supernatant. Infection of virus into Ba/F3 cells was performed by spin infection (1,800 × g, for 90 min, at 32°C). Three days after the transduction, green fluorescent protein-positive (GFP+) cells were sorted by using the FACSAria III (BD Biosciences) and grown in RPMI medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 10 mM pyruvate, 50 μg/ml gentamicin, and 50 μM β-mercaptoethanol (β-ME). As described previously (63), when 70% confluence was reached in 150-mm culture flasks, 50 μM biotin (Sigma-Aldrich B4501-1G) was spiked into the medium. Cells were incubated for a maximum of 24 h at 37°C. Approximately 100 million cells were pooled, washed twice with ice-cold phosphate-buffered saline (PBS), flash frozen in liquid nitrogen, and stored at −80°C until analysis. Cells were lysed in 10 ml lysis buffer (25 mM Tris HCl [pH 7.6], 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, protease, and phosphatase inhibitor cocktail tablets [Thermo Scientific; 88669]), benzonase nuclease (Sigma; E1014-5KU) was added to a final concentration of 25 U/ml, and the mixture was sonicated and incubated for an hour on an end-over-end rotator at 4°C. Samples were then transferred into a centrifugation tube and spun at 4°C and 16,000 rpm for 30 min to remove cell debris. Streptavidin-Sepharose beads (GE Health Care; 17-5113-01) (30 μl of packed bead volume) were washed and equilibrated in the lysis buffer before being added to cell lysates and incubated at 4°C and 2,000 rpm for 3 h to induce effective conjugation of biotin and streptavidin. The beads were then washed three times with 50 mM ammonium bicarbonate to reduce nonspecific binding, followed by two washes to remove any traces of detergents. The beads were resuspended in 50 mM ammonium bicarbonate with 2 μl of 1 μg/μl trypsin (∼20 U) (Thermo Scientific; reference no. V5113), the tubes were sealed, and samples were incubated overnight at 37°C on an end-over-end rotator. One microliter of 1 μg/μl trypsin was added again the next day to achieve effective trypsinization. The supernatant was saved in a fresh tube, and the beads were washed with fresh 50 mM ammonium bicarbonate and later pooled. The digests were then dried in a vacuum dryer (Savant SPD 1010).
BioID data analysis.
Proteome Discoverer (Thermo Scientific; version 1.3) was used for protein identification and quantitation with the SEQUEST algorithm (Thermo Fisher Scientific, San Jose, CA; version 1.4.0.288) and X! Tandem (CYCLONE; version 2010.12.01.1). Trypsin was chosen as the enzyme, allowing up to two missed cleavages; phosphorylation of serine, threonine, and tyrosine residues, oxidation of methionine residue, and acetylation of protein N terminus were selected. The searches were performed with a precursor ion mass tolerance of up to 10 ppm and a fragment ion mass tolerance of 0.6 Da. The database search was performed against the complete mouse database from UniProt (85,828 entries). All searches were done against a decoy database with a false discovery rate (FDR) of less than 0.01. The minimum peptide length considered was 6, and the FDR was set to 0.01 for both proteins and peptides. Data analysis was achieved using Trans-Proteomic Pipeline (TPP) software (64) from the ProHits software suite (65). Vendor-specific, Thermo “.raw” files were converted into open-format “.mzXML” files using ProteoWizard (66), and database searches were performed by Comet (67). Proteins identified with a ProteinProphet cutoff of 0.85 (corresponding to ≤1% FDR) and with ≥2 unique peptides were analyzed with SAINT Express v.3.3. Each biological replicate was analyzed using two technical replicates. Data were compared to six controls (three biological and three technical replicates) with BirAM conjugated to NLS collapsed to the two highest spectral counts for each prey. A Bayesian FDR of 0.02 (corresponding to a SAINT score of ∼0.80) was used as a cutoff to define high-confidence interactors. ProHits Lite (version 3.0.3) was run on Linux Fedora virtual machine in Oracle VMware (version 5.0.2 r 102096). Output from TPP for each sample served as input to the ProHits Analyst stand-alone application. The sample files were uploaded according to their respective baits, and experiments were analyzed. The search results were further analyzed using a statistical tool, SAINT, with 5,000 iterations, a 1-min fold, and low mode off, along with normalization to calculate interaction confidence scores, which were also integrated into ProHits.
MS analysis.
Each sample with three biological replicates was run in duplicate. The dried samples were reconstituted in 0.1% formic acid, and the peptide concentration was measured using a Nanodrop instrument (ND 2000; Thermo Scientific). Mass spectrometric (MS) analysis was performed on a reverse-phase nano-liquid chromatograph coupled online to an LTQ Orbitrap Velos Pro MS (Thermo Fisher Scientific, Inc.). Each of the samples was separated using an Agilent 1200 Easy-nLC system (Agilent Technologies) with a nano-electrospray ion source (Proxeon). The peptides were trapped using a precolumn (NS-MP-10 Biosphere C18; 5-μm particle size, 120 Å, 100 μm by 20 cm), and separated on a column (NS-AC-10 Biosphere C18; 5-μm particle size, 120 Å, 75 μm by 10.2 cm). A linear gradient from 2% to 35% buffer B (0.1% formic acid in acetonitrile) against buffer A (0.1% formic acid in water) was carried out with a constant flow rate of 300 nl/min and then eluted using a 120-min gradient. Full-scan MS spectra were acquired in positive-mode electrospray ionization with an ion spray voltage of 2.4 kV, a radio frequency lens voltage of 69, and a capillary temperature of 235°C. This was acquired over an m/z of 390 to 2,000 Da at a resolution of 60,000, and the top 20 intense ions were selected for tandem MS (MS/MS) under an isolation width of 1 m/z unit with a minimum ion count of 100 for activation. A collision energy of 35 was used to fragment the ions in the collision-induced dissociation (CID) mode. The selected masses were included in a dynamic exclusion list for 30 s with a repeat duration of 30 s, and an abundance threshold of more than 500 counts was considered.
Corticosterone assay.
Blood was obtained from 3-week-old Zfp521−/− and Ctrl littermates via submandibular bleed. Microtainer tubes (BD Biosciences) were used for serum separation. Serum corticosterone levels were analyzed using the DetectX corticosterone enzyme immunoassay kit (Arbor Assays).
RNA-seq analysis.
BM was harvested from Zfp521−/− mice and Ctrl littermates, and Linneg cKit+ Sca1+ (LSK) cells were FACS purified. Total RNA was isolated using an RNeasy Micro kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Libraries were constructed using NuGEN′s Ovation Ultralow Library systems (NuGEN Technologies, San Carlos, CA) and subsequently subjected to 76 cycles of NextSeq500 sequencing (Illumina, San Diego, CA). Adapters were trimmed, low-quality bases and reads of <30 bp were removed using ea-utils v1.05. FastQC (v0.11.5) analysis was performed to ensure the quality of data. Reads were mapped to the genome using Tophat2 v2.1.1 using reference genome UCSC mm10. Mapped reads with a mapping quality of <10 were removed using samtools v1.5, and coordinates were sorted using PICARD v2.9.4. Genes were quantified using HTSeq v0.8.0, and reads per kilobase of transcript per million mapped reads (RPKMS) were normalized to exonic gene length using a custom R script. A PCA plot was generated using DESeq2 to get variance-stabilized counts, and the top 500 most variable genes were used for the PCA plot. Differential gene expression analysis was performed using DESeq2 (fold change of >2 or <0.5; Benjamini-Hochberg procedure-corrected adjusted P value [Padj] of <0.1). Heatmaps were generated using pheatmap package v1.0.8 (68) in R v3.3.2 (69). Values were normalized for each gene by centering the value on the mean of the RPKMS and scaling by the standard deviation. Candidate pathways activated or inhibited in Zfp521−/− LSK cells were analyzed using Ingenuity Pathway Analysis v01-08.
Cytokine analysis.
To obtain BM fluid, tibias and femurs were collected from 3-week-old Zfp521−/− mice and Ctrl littermates. The bones were crushed in 300 μl 0.5% bovine serum albumin (BSA)-PBS, and the supernatant was collected into tubes. All samples were normalized to a total volume of 350 μl, centrifuged at 1,200 rpm for 5 min to pellet the cells, and then stored at −80°C. For cytokine measurement, BM fluid was thawed and clarified by spinning at 12,000 × g for 10 min. Twenty-five microliters of BM fluid was subsequently diluted 1:1 with assay diluent and incubated on a Mouse ProcartaPlex 36-plex panel 1A array (Thermo Fisher) according to the manufacturer's instructions. The array was analyzed on a MAGPIX (Luminex Corporation) instrument, and cytokine concentrations were determined based on standard curves run on the same plate for each analyte.
Statistical analysis.
Statistical significance was determined using the Mann-Whitney test, unless otherwise indicated. A minimum of three replicates was used to calculate P values. Data are reported as means ± standard deviations (SD) (GraphPad Prism).
Accession number(s).
RNA sequencing data have been deposited in the Gene Expression Omnibus database under the accession code GSE113543.
Supplementary Material
ACKNOWLEDGMENTS
We thank Greg Kirchenbaum for advice concerning B cell progenitor stains, Rachel Blumhagen for assistance with statistical analysis, and Raul Torres and Mair Churchill for helpful discussions. We are indebted to Seth Welsh, Kara Lukin, and Carissa Dege for their technical support and helpful comments.
We declare no competing financial or other conflicts of interest.
This research was supported by generous grants from the National Institutes of Health, i.e., R01AI081878 (J.R.H.), R01AI098417 (J.R.H.), R21AI115696 (J.R.H.), R01CA117907 (J.M.E.), and K01DK098315 (E.M.P.), by The Wendy Siegel Fund for Leukemia and Cancer Research (J.R.H.), and by the Mary Miller and Charlotte Fonfara-Larose Leukemia and Down Syndrome Research Fund (J.M.E.). C.J.F. and H.L. were supported by NIH Institutional National Service Award 2T32AI074491. J.L.R. was the recipient of NIH F31HL138754. T.A. received an award from The Victor W. Bolie and Earleen D. Bolie Graduate Scholarship Fund. M.S. thanks the Swedish Cancer Foundation and the Swedish Medical Research Council for their support.
C.J.F. designed and conducted experiments, analyzed the results, and wrote the manuscript. T.A. performed flow cytometry on BM myeloid progenitors and endpoint analysis for the BMT assay. T.A. and H.L. performed B cell progenitor flow cytometry and conducted RT-PCR experiments. J.A., K.O., and M.S. executed and analyzed proximity biotinylation experiments; J.K. and S.C. performed the mass spectrometry. K.O. and M.S. generated the RNA-seq data. D.S. provided technical support. A.P., J.M.E., and T.D. performed bioinformatics analysis of RNA-seq data. J.L.R. and E.M.P. performed and analyzed the cytokine array. S.W. generated the Zfp521−/− mice and helped develop the project. J.R.H. developed the project and wrote the paper with C.J.F. All authors reviewed the data and approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00603-17.
REFERENCES
- 1.Justice MJ, Morse HC III, Jenkins NA, Copeland NG. 1994. Identification of Evi-3, a novel common site of retroviral integration in mouse AKXD B-cell lymphomas. J Virol 68:1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentges KE, Weiser KC, Schountz T, Woodward LS, Morse HC, Justice MJ. 2005. Evi3, a zinc-finger protein related to EBFAZ, regulates EBF activity in B-cell leukemia. Oncogene 24:1220–1230. doi: 10.1038/sj.onc.1208243. [DOI] [PubMed] [Google Scholar]
- 3.Warming S, Liu P, Suzuki T, Akagi K, Lindtner S, Pavlakis GN, Jenkins NA, Copeland NG. 2003. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Kruppel-like zinc finger protein. Blood 101:1934–1940. doi: 10.1182/blood-2002-08-2652. [DOI] [PubMed] [Google Scholar]
- 4.Warming S, Suzuki T, Yamaguchi TP, Jenkins NA, Copeland NG. 2004. Early B-cell factor-associated zinc-finger gene is a frequent target of retroviral integration in murine B-cell lymphomas. Oncogene 23:2727–2731. doi: 10.1038/sj.onc.1207452. [DOI] [PubMed] [Google Scholar]
- 5.Hiratsuka T, Takei Y, Ohmori R, Imai Y, Ozeki M, Tamaki K, Haga H, Nakamura T, Tsuruyama T. 2016. ZFP521 contributes to pre-B-cell lymphomagenesis through modulation of the pre-B-cell receptor signaling pathway. Oncogene 35:3227–3238. doi: 10.1038/onc.2015.385. [DOI] [PubMed] [Google Scholar]
- 6.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 7.Alcaraz WA, Gold DA, Raponi E, Gent PM, Concepcion D, Hamilton BA. 2006. Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci U S A 103:19424–19429. doi: 10.1073/pnas.0609184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warming S, Rachel RA, Jenkins NA, Copeland NG. 2006. Zfp423 is required for normal cerebellar development. Mol Cell Biol 26:6913–6922. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han R, Kan Q, Sun Y, Wang S, Zhang G, Peng T, Jia Y. 2012. MiR-9 promotes the neural differentiation of mouse bone marrow mesenchymal stem cells via targeting zinc finger protein 521. Neurosci Lett 515:147–152. doi: 10.1016/j.neulet.2012.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Kang S, Akerblad P, Kiviranta R, Gupta RK, Kajimura S, Griffin MJ, Min J, Baron R, Rosen ED. 2012. Regulation of early adipose commitment by Zfp521. PLoS Biol 10:e1001433. doi: 10.1371/journal.pbio.1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. 2010. Transcriptional control of preadipocyte determination by Zfp423. Nature 464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsubara E, Sakai I, Yamanouchi J, Fujiwara H, Yakushijin Y, Hato T, Shigemoto K, Yasukawa M. 2009. The role of zinc finger protein 521/early hematopoietic zinc finger protein in erythroid cell differentiation. J Biol Chem 284:3480–3487. doi: 10.1074/jbc.M805874200. [DOI] [PubMed] [Google Scholar]
- 13.Hesse E, Saito H, Kiviranta R, Correa D, Yamana K, Neff L, Toben D, Duda G, Atfi A, Geoffroy V, Horne WC, Baron R. 2010. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol 191:1271–1283. doi: 10.1083/jcb.201009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiviranta R, Yamana K, Saito H, Ho DK, Laine J, Tarkkonen K, Nieminen-Pihala V, Hesse E, Correa D, Maatta J, Tessarollo L, Rosen ED, Horne WC, Jenkins NA, Copeland NG, Warming S, Baron R. 2013. Coordinated transcriptional regulation of bone homeostasis by Ebf1 and Zfp521 in both mesenchymal and hematopoietic lineages. J Exp Med 210:969–985. doi: 10.1084/jem.20121187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M, Hesse E, Morvan F, Zhang JP, Correa D, Rowe GC, Kiviranta R, Neff L, Philbrick WM, Horne WC, Baron R. 2009. Zfp521 antagonizes Runx2, delays osteoblast differentiation in vitro, and promotes bone formation in vivo. Bone 44:528–536. doi: 10.1016/j.bone.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hesse E, Kiviranta R, Wu M, Saito H, Yamana K, Correa D, Atfi A, Baron R. 2010. Zinc finger protein 521, a new player in bone formation. Ann N Y Acad Sci 1192:32–37. doi: 10.1111/j.1749-6632.2009.05347.x. [DOI] [PubMed] [Google Scholar]
- 17.Correa D, Hesse E, Seriwatanachai D, Kiviranta R, Saito H, Yamana K, Neff L, Atfi A, Coillard L, Sitara D, Maeda Y, Warming S, Jenkins NA, Copeland NG, Horne WC, Lanske B, Baron R. 2010. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev Cell 19:533–546. doi: 10.1016/j.devcel.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SS, Tsai RY, Reed RR. 1997. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci 17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bond HM, Mesuraca M, Carbone E, Bonelli P, Agosti V, Amodio N, De Rosa G, Di Nicola M, Gianni AM, Moore MA, Hata A, Grieco M, Morrone G, Venuta S. 2004. Early hematopoietic zinc finger protein (EHZF), the human homolog to mouse Evi3, is highly expressed in primitive human hematopoietic cells. Blood 103:2062–2070. doi: 10.1182/blood-2003-07-2388. [DOI] [PubMed] [Google Scholar]
- 20.Garrison BS, Rybak AP, Beerman I, Heesters B, Mercier FE, Scadden DT, Bryder D, Baron R, Rossi DJ. 2017. ZFP521 regulates murine hematopoietic stem cell function and facilitates MLL-AF9 leukemogenesis in mouse and human cells. Blood 130:619–624. doi: 10.1182/blood-2016-09-738591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmfeldt P, Ganuza M, Marathe H, He B, Hall T, Kang G, Moen J, Pardieck J, Saulsberry AC, Cico A, Gaut L, McGoldrick D, Finkelstein D, Tan K, McKinney-Freeman S. 2016. Functional screen identifies regulators of murine hematopoietic stem cell repopulation. J Exp Med 213:433–449. doi: 10.1084/jem.20150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mega T, Lupia M, Amodio N, Horton SJ, Mesuraca M, Pelaggi D, Agosti V, Grieco M, Chiarella E, Spina R, Moore MA, Schuringa JJ, Bond HM, Morrone G. 2011. Zinc finger protein 521 antagonizes early B-cell factor 1 and modulates the B-lymphoid differentiation of primary hematopoietic progenitors. Cell Cycle 10:2129–2139. doi: 10.4161/cc.10.13.16045. [DOI] [PubMed] [Google Scholar]
- 23.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansson R, Welinder E, Ahsberg J, Lin YC, Benner C, Glass CK, Lucas JS, Sigvardsson M, Murre C. 2012. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci U S A 109:21028–21033. doi: 10.1073/pnas.1211427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. 2013. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol 14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 26.Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 27.Seo W, Ikawa T, Kawamoto H, Taniuchi I. 2012. Runx1-Cbfbeta facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med 209:1255–1262. doi: 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin AC, Roche AE, Wilk J, Svensson EC. 2004. The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J Biol Chem 279:55017–55023. doi: 10.1074/jbc.M411240200. [DOI] [PubMed] [Google Scholar]
- 29.Lauberth SM, Rauchman M. 2006. A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J Biol Chem 281:23922–23931. doi: 10.1074/jbc.M513461200. [DOI] [PubMed] [Google Scholar]
- 30.Addison WN, Fu MM, Yang HX, Lin Z, Nagano K, Gori F, Baron R. 2014. Direct transcriptional repression of Zfp423 by Zfp521 mediates a bone morphogenic protein-dependent osteoblast versus adipocyte lineage commitment switch. Mol Cell Biol 34:3076–3085. doi: 10.1128/MCB.00185-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czechowicz A, Kraft D, Weissman IL, Bhattacharya D. 2007. Efficient transplantation via antibody-based clearance of hematopoietic stem cell niches. Science 318:1296–1299. doi: 10.1126/science.1149726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiohara M, Koike K, Kubo T, Amano Y, Takagi M, Muraoka K, Nakao J, Nakahata T, Komiyama A. 1993. Possible role of stem cell factor as a serum factor: monoclonal anti-c-kit antibody abrogates interleukin-6-dependent colony growth in serum-containing culture. Exp Hematol 21:907–912. [PubMed] [Google Scholar]
- 33.Kondo M, Weissman IL, Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91:661–672. doi: 10.1016/S0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 34.Kumar R, Fossati V, Israel M, Snoeck HW. 2008. Lin−Sca1+Kit− bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J Immunol 181:7507–7513. doi: 10.4049/jimmunol.181.11.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loder F, Mutschler B, Ray RJ, Paige CJ, Sideras P, Torres R, Lamers MC, Carsetti R. 1999. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp Med 190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yui MA, Feng N, Rothenberg EV. 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol 185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godfrey DI, Kennedy J, Suda T, Zlotnik A. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3− CD4− CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol 150:4244–4252. [PubMed] [Google Scholar]
- 39.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigvardsson M, O'Riordan M, Grosschedl R. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25–36. doi: 10.1016/S1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 41.Cobb BS, Smale ST. 2005. Ikaros-family proteins: in search of molecular functions during lymphocyte development. Curr Top Microbiol Immunol 290:29–47. [DOI] [PubMed] [Google Scholar]
- 42.Cortes M, Wong E, Koipally J, Georgopoulos K. 1999. Control of lymphocyte development by the Ikaros gene family. Curr Opin Immunol 11:167–171. doi: 10.1016/S0952-7915(99)80028-4. [DOI] [PubMed] [Google Scholar]
- 43.Fujikawa K, Miletic AV, Alt FW, Faccio R, Brown T, Hoog J, Fredericks J, Nishi S, Mildiner S, Moores SL, Brugge J, Rosen FS, Swat W. 2003. Vav1/2/3-null mice define an essential role for Vav family proteins in lymphocyte development and activation but a differential requirement in MAPK signaling in T and B cells. J Exp Med 198:1595–1608. doi: 10.1084/jem.20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, Akashi K, Weissman IL, Fisher AG, Smale ST. 1998. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev 12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, Morgan BA. 1998. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol 8:508–515. doi: 10.1016/S0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 46.Sansregret L, Nepveu A. 2008. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene 412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Serre K, Benezech C, Desanti G, Bobat S, Toellner KM, Bird R, Chan S, Kastner P, Cunningham AF, Maclennan IC, Mohr E. 2011. Helios is associated with CD4 T cells differentiating to T helper 2 and follicular helper T cells in vivo independently of Foxp3 expression. PLoS One 6:e20731. doi: 10.1371/journal.pone.0020731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vadnais C, Davoudi S, Afshin M, Harada R, Dudley R, Clermont PL, Drobetsky E, Nepveu A. 2012. CUX1 transcription factor is required for optimal ATM/ATR-mediated responses to DNA damage. Nucleic Acids Res 40:4483–4495. doi: 10.1093/nar/gks041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laakko T, Fraker P. 2002. Rapid changes in the lymphopoietic and granulopoietic compartments of the marrow caused by stress levels of corticosterone. Immunology 105:111–119. doi: 10.1046/j.1365-2567.2002.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garvy BA, King LE, Telford WG, Morford LA, Fraker PJ. 1993. Chronic elevation of plasma corticosterone causes reductions in the number of cycling cells of the B lineage in murine bone marrow and induces apoptosis. Immunology 80:587–592. [PMC free article] [PubMed] [Google Scholar]
- 51.Sato Y, Suzuki H, Sato T, Suda T, Yoda T, Iwakura Y, Chida D. 2010. The role of endogenous glucocorticoids in lymphocyte development in melanocortin receptor 2-deficient mice. Biochem Biophys Res Commun 403:253–257. doi: 10.1016/j.bbrc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Ohkubo N, Matsubara E, Yamanouchi J, Akazawa R, Aoto M, Suzuki Y, Sakai I, Abe T, Kiyonari H, Matsuda S, Yasukawa M, Mitsuda N. 2014. Abnormal behaviors and developmental disorder of hippocampus in zinc finger protein 521 (ZFP521) mutant mice. PLoS One 9:e92848. doi: 10.1371/journal.pone.0092848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avellino R, Delwel R. 2017. Expression and regulation of C/EBPalpha in normal myelopoiesis and in malignant transformation. Blood 129:2083–2091. doi: 10.1182/blood-2016-09-687822. [DOI] [PubMed] [Google Scholar]
- 54.Rosenbauer F, Tenen DG. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 55.Hauser SP, Kajkenova O, Lipschitz DA. 1997. The pivotal role of interleukin 6 in formation and function of hematopoietically active murine long-term bone marrow cultures. Stem Cells 15:125–132. doi: 10.1002/stem.150125. [DOI] [PubMed] [Google Scholar]
- 56.Dorshkind K. 1988. IL-1 inhibits B cell differentiation in long term bone marrow cultures. J Immunol 141:531–538. [PubMed] [Google Scholar]
- 57.Day RB, Bhattacharya D, Nagasawa T, Link DC. 2015. Granulocyte colony-stimulating factor reprograms bone marrow stromal cells to actively suppress B lymphopoiesis in mice. Blood 125:3114–3117. doi: 10.1182/blood-2015-02-629444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cain DW, Snowden PB, Sempowski GD, Kelsoe G. 2011. Inflammation triggers emergency granulopoiesis through a density-dependent feedback mechanism. PLoS One 6:e19957. doi: 10.1371/journal.pone.0019957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Bruin AM, Voermans C, Nolte MA. 2014. Impact of interferon-gamma on hematopoiesis. Blood 124:2479–2486. doi: 10.1182/blood-2014-04-568451. [DOI] [PubMed] [Google Scholar]
- 60.Mojsilovic S, Jaukovic A, Santibanez JF, Bugarski D. 2015. Interleukin-17 and its implication in the regulation of differentiation and function of hematopoietic and mesenchymal stem cells. Mediators Inflamm 2015:470458. doi: 10.1155/2015/470458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang CQ, Udupa KB, Lipschitz DA. 1995. Interferon-gamma exerts its negative regulatory effect primarily on the earliest stages of murine erythroid progenitor cell development. J Cell Physiol 162:134–138. doi: 10.1002/jcp.1041620116. [DOI] [PubMed] [Google Scholar]
- 62.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Dingar D, Kalkat M, Chan P-K, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, Huang A, Lupien M, Penn LZ, Raught B. 2015. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics 118:95–111. doi: 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 64.Pedrioli PG. 2010. Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol Biol 604:213–238. doi: 10.1007/978-1-60761-444-9_15. [DOI] [PubMed] [Google Scholar]
- 65.Liu G, Zhang J, Larsen B, Stark C, Breitkreutz A, Lin ZY, Breitkreutz BJ, Ding Y, Colwill K, Pasculescu A, Pawson T, Wrana JL, Nesvizhskii AI, Raught B, Tyers M, Gingras AC. 2010. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol 28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kessner D, Chambers M, Burke R, Agus D, Mallick P. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eng JK, Jahan TA, Hoopmann MR. 2013. Comet: an open-source MS/MS sequence database search tool. Proteomics 13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 68.Kolde R. 2015. pheatmap: pretty heatmaps. R package version 1.0.8 https://CRAN.R-project.org/package=pheatmap.
- 69.R Development Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.