Abstract
The length and density of rumen papillae starts to increase during weaning and growth of ruminants. This significant development increases the intraruminal surface area and the efficiency of VFA (acetate, propionate, butyrate, etc.) uptake. Thus, it is important to investigate the factors controlling the growth and development of rumen papillae during weaning. This study aimed to compare the transcriptomes of rumen papillae in suckling and weaned calves. Total RNA was extracted from the rumen papillae of 10 male Japanese Black calves (5 suckling calves, 5 wk old; 5 weaned calves, 15 wk old) and used in RNA-sequencing. Transcript abundance was estimated and differentially expressed genes were identified and these data were then used in Ingenuity Pathway Analysis (IPA) to predict the major canonical pathways and upstream regulators. Among the 871 differentially expressed genes screened by IPA, 466 genes were upregulated and 405 were downregulated in the weaned group. Canonical pathway analysis showed that “atherosclerosis” was the most significant pathway, and “tretinoin,” a derivative of vitamin A, was predicted as the most active upstream regulator during weaning. Analyses also predicted IgG, lipopolysaccharides, and tumor-necrosis factor-α as regulators of the microbe-epithelium interaction that activates rumen-related immune responses. The functional category and the up-regulators found in this study provide a valuable resource for studying new candidate genes related to the proliferation and development of rumen papillae from suckling to weaning Japanese Black calves.
Keywords: atherosclerosis pathway, Japanese Black calves, RNA sequencing, rumen papillae, transcriptome, tretinoin
INTRODUCTION
The rumen has physiologically essential functions such as nutrient uptake, transportation, and metabolism (Roh et al., 2007; Roh et al., 2016). Therefore, this organ is crucial for ruminant life, body maintenance, and growth (Warner et al., 1956; Tamate et al., 1962). A dramatic morphological development of rumen papillae occurs when feed is converted to roughage or concentrated from milk. Highly concentrated diets induce significant increases in rumen papillae density and length, as VFA (acetate, propionate, butyrate, among others) and lactate derive from intraruminal fermentation (Stobo et al., 1966). In neonatal Holstein calves, diets based on grain and on orchard-grass hay significantly increased the length and density of rumen papillae, whereas a milk-based diet had little effect on the development of these structures (Connor et al., 2013). Feeding VFA, particularly butyrate, significantly induced morphological development and promoted an increase in the length and density of the rumen epithelium (Sakata and Tamate, 1978; Shen et al., 2004; Gorka et al., 2011; Kato et al., 2011). Although many studies have been conducted on rumen development, only a few reported gene expression variations, regulatory factors, and gene networks in the rumen epithelium of calves fed different diets. Transcription factors, such as CREBBP (cAMP response element binding protein-binding protein) and TTF2 (transcription termination factor 2), were found to control several regulatory networks in the rumen epithelium by butyrate infusion using RNA-sequencing (RNA-seq) and Gene Ontology (GO) (Baldwin et al., 2012). Connor et al. (2013) reported that lipid metabolism, cell morphology and death, cellular growth and proliferation, molecular transport, and the cell cycle were predicted by GO analysis using Ingenuity Pathway Analysis (IPA) on microarray data obtained from the rumen epithelium of Holstein calves fed on a milk replacer and solid feed. Using digital differential display (DDD), Kato et al. (2015) identified candidate genes related to rumen development in suckling and weaned Japanese Black cattle (Kato et al., 2015). The present study reports the first application of RNA-seq to comprehensively investigate the transcriptomes of rumen papillae in suckling and weaned Japanese Black calves, aiming to identify differentially expressed genes (DEG) related to the development of rumen papillae.
MATERIALS AND METHODS
This study was conducted in accordance with the “Guidelines for the National Agriculture and Food Research Organization (NARO) Institute of Livestock and Grassland Science” and was approved by the Animal Care Committee of the NARO Institute of Livestock and Grassland Science.
Animals
Ten Japanese Black male calves were used in the experiment. Information on sires, dams, natural dams, and calves used in this study is presented in Supplementary Tables 1 and 2. All calves were bred at the Grassland Management Research Division, NARO Institute of Livestock and Grassland Science. Feed and management practices were as described previously (Kato et al., 2015; Suzuki et al., 2016). Japanese Black male calves born from March 2013 to May 2014 were assigned sequentially to the suckling and weaned groups. Finally, 5 calves in each group were used for this experiment. All calves were raised by natural suckling, and although none were creep fed, all had free access to dams’ feed. Calves in the suckling group were slaughtered at 5 wk of age, and their average body weight at slaughter was 51 ± 2 kg. Calves in the weaned group were separated from their dams at 12 wk of age and then raised by group feeding. All weaned calves were slaughtered at 15 to 16 wk of age, and their average body weight was 126 ± 8 kg. From 12 to 15–16 wk, weaned calves were only fed grower feed and timothy hay at 09:00 and 16:00 h to reach an average daily gain of about 0.6 kg, following the Japanese Feeding Standard for Beef Cattle (Agriculture, Forestry and Fisheries Research Council Secretariat, 2000). Grower feed components (CP 19.1% dry matter basis) were 39% grain (corn, milo, and wheat), 36% chaff and bran (bran, corn gluten meal, corn distiller’s dried grains with solubles, and draff), 11% vegetable oil cake (soybean and rapeseed), and 14% others (alfalfa, alfalfa meal, molasses, and minerals) in raw matter basis. Calves were allowed ad libitum access to water and mineral blocks. The blood hormones and metabolites of suckling and weaned calves were reported in our previous study (Suzuki et al., 2016).
Tissue Collection
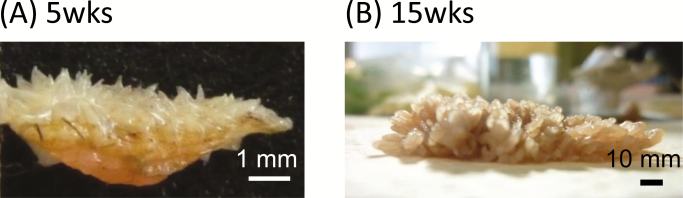
After slaughter, rumen papillae tissues (approximately 1 × 1 cm) were collected from the ventral cranial sac. Excised rumen papillae tissues of suckling and weaned calves are shown in Figure 1. The papillae layer was manually separated from the muscular layer using surgical scissors and rinsed with PBS to remove residual feed particles. All tissue samples were frozen immediately in liquid nitrogen and stored at −80 °C until total RNA extraction was performed.
Figure 1.
Rumen papillae in suckling (A) and weaned (B) calves of Japanese Black cattle. Calves in the suckling group were slaughtered at 5 wk of age. Weaned calves were separated from their dams at 12 wk of age, and, after weaning, they were raised by group feeding. All weaned calves were slaughtered at 15 to 16 wk of age. The bar represents the scale (mm).
Total RNA Purification
Tissues were soaked in 200 μL RNAiso Plus (TAKARA Bio Inc., Shiga, Japan) in liquid nitrogen and homogenized using a Multibeads shocker (YASUIKIKAI Inc., Osaka, Japan), as indicated by the manufacturer. Homogenization was carried out twice at 2,000 rpm for 10 to 15 s, and 300 μL RNAiso was then added at 25 °C. The homogenate was collected in 1.5-mL tubes, 100-μL chloroform was added to the homogenate, and the solution was thoroughly mixed. After settling for 3 min at room temperature, the solution was centrifuged at 12,000 × g for 15 min at 4 °C, and the supernatant was collected. The supernatant was then mixed with 700 μL 70% ethanol. Total RNA was further purified and gDNA was removed using NucleoSpin RNA columns (MACHEREY-NAGEL, Düren, Germany), following the manufacturer’s instructions. Purified total RNA was quantified in a NanoDrop ND-1000 Spectrophotometer V3.7.1 (THERMO FISHER SCIENTIFIC, Waltham, MA, USA) at wavelengths of 230, 260, and 280 nm. The purity of total RNA was determined as the A260/A280 ratio, with expected values over 1.8, and was verified by running samples on 1.0% agarose gels. All samples were stored at −80 °C.
RNA-sequencing
The RNA Integrity Number (RIN) was confirmed in a Bioanalyzer 2100 using RNA nano kit (AGILENT, Palo Alto, CA, USA) to check if the purified total RNA could be used in RNA-seq. The average RIN in suckling and weaned groups was 7.1 (5.9 to 7.9) and 8.0 (7.5 to 8.4), respectively. Quality check of total RNA, library preparation, and RNA-seq were conducted at the NODAI Genome Research Center, Tokyo University of Agriculture. Single 50-bp reads were performed on a HiSeq 2500 platform (ILLUMINA, San Diego, CA, USA) using the TruSeq RNA Sample Preparation Kit v2 (ILLUMINA). Analysis of high-quality sequences was performed as in a previous report (Endo et al., 2013). Sequences were retained as high quality when passing the Illumina quality-filtering pipeline GERALD under default settings with the following conditions: only contain reads that passed filtering based on cluster intensities and noise estimates; demultiplexing; adapter masking; and ignore the first and last bases. These sequences were then aligned to the bovine reference genome (bosTau6) and to the exon–exon splice junction database downloaded from the University of California Santa Cruz (UCSC) sequence and annotation database (http://hgdownload.cse.ucsc.edu/downloads.html#cow). Alignment was performed in CLC Genomics Workbench (Qiagen, Valencia, CA, USA) using the defaults parameters. The EM estimation algorithm was used to iteratively estimate the abundance of transcripts and to assign reads to transcripts according to their abundances (Cappe and Moulines, 2009; Li and Dewey, 2011). Raw and normalized (i.e., Reads Per Kilobase of transcript per Million mapped reads [RPKM]) read counts (Mortazavi et al., 2008) were also calculated in CLC Genomics Workbench. After data normalization and fold change computation based on the estimated relative abundances of reads, the statistical significance of each pairwise comparison (suckling vs. weaned groups) was determined using the digital gene expression (DGE) empirical analysis tool incorporated in the EdgeR Bioconductor package (Robinson et al., 2010) and in the CLC Genomics Workbench. The DGE tool implements an “exact test” for two group comparisons, which is similar to Fisher’s exact test. The test is applicable to count data only and is designed for the analyses of differentially expressed RNA-seq data. In addition to P-values, FDR-corrected P-values were also calculated (Benjamini et al., 2001). Differentially expressed genes (fold change ≥ 2 and FDR-corrected P-value < 0.05) were analyzed regarding their biological processes and functions, and assigned to canonical pathways using IPA with the default parameters in General Settings, Networks, Data sources, Confidence, Species, Tissues & Cell Lines, and Mutation (Qiagen, www.qiagen.com/ingenuity). Fisher’s exact test was used in the analysis of gene set enrichment in the functional categories.
RESULTS
The Sequencing Results and the Dataset of Analyzed Genes
Sequencing of total RNA from rumen papillae in two lanes yielded, on average, 1067.4 and 1011.8 Mb and 21,353,595 and 20,238,186 sequence reads for suckling and weaned groups, respectively. The numbers of raw reads and mapped reads per sample are shown in Supplementary Table 3. Sequence data from RNA-seq were deposited in Sequence Read Archive (DRA) of DNA Data Bank of Japan (DDBJ). The accession number is DRA005801. After sequence alignment, 82% to 86% of the reads within each sample were mapped to the reference genome (Supplementary Table 3). Gene expression levels were then compared between groups based on these data, using the CLC Genomic Workbench. Overall, 17,001 of the 24,616 genes in the UCSC annotation were expressed, and 871 of these genes were differentially expressed, with fold change ≥ 2 and FDR < 0.05. Compared with the suckling group, weaning calves presented 466 upregulated and 405 downregulated genes (Supplementary Table 4). However, it should be noted that 43 of the 871 DEGs showed low expression level (RPKM < 1), which might have affected the large fold change detected.
Canonical Pathway Analysis
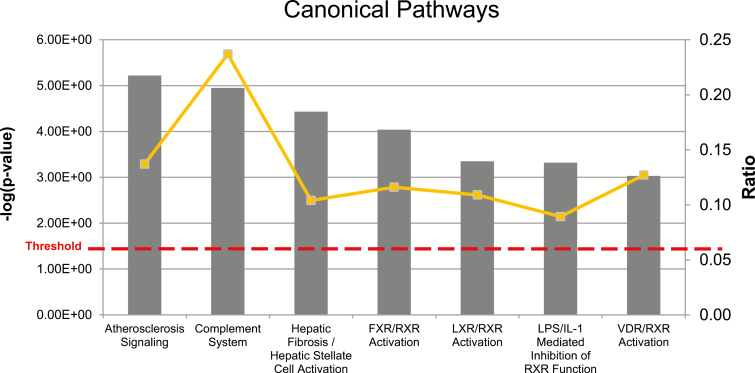
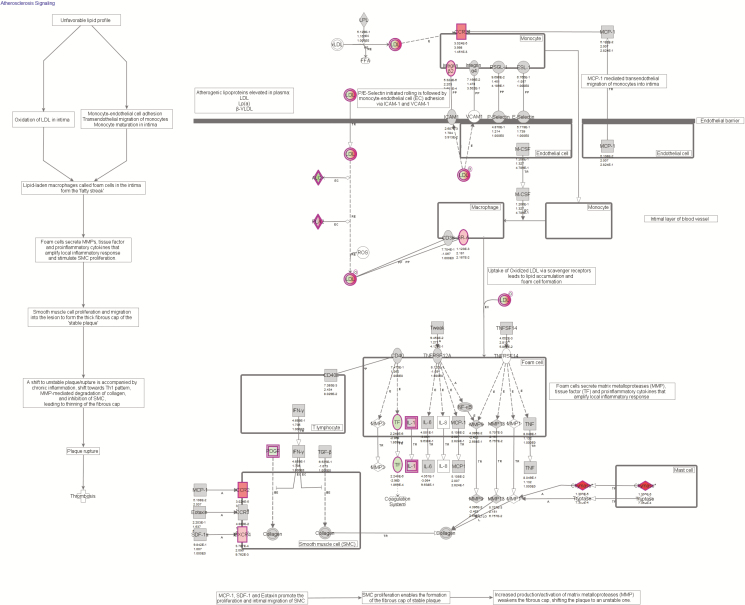
The seven major canonical pathways (P < 0.05) of DEGs found through IPA are displayed in Figure 2. Seventeen DEGs were found in the top canonical pathway “Atherosclerosis Signaling” (Figure 3, Table 1).
Figure 2.
Canonical pathway analysis generated by Ingenuity Pathway Analysis (IPA). The P-values and ratios of the 7 major pathways in suckling and weaned groups are displayed. Pathways were considered as most important at P < 0.05.
Figure 3.
The atherosclerosis-signaling pathway in rumen papillae. The Ingenuity Pathway Analysis (IPA) indicated that 17 genes were related to this pathway, and 12 of them functioned in the rumen papillae as demonstrated in this figure.
Table 1.
Genes integrating the atherosclerosis pathway
| Symbol | Entrez gene name | Fold change1 | FDR2 | Location | Type(s) |
|---|---|---|---|---|---|
| RBP4 | Retinol binding protein 4, plasma | 6.461 | 1.13E-09 | Extracellular space | Transporter |
| PLA2G4D | Phospholipase A2, group IVD (cytosolic) | 6.146 | 2.98E-03 | Cytoplasm | Enzyme |
| CMA1 | Chymase 1, mast cell | 5.912 | 7.36E-04 | Extracellular space | Peptidase |
| CCR2 | Chemokine (C-C motif) receptor 2 | 3.998 | 1.45E-03 | Plasma membrane | G-protein coupled receptor |
| IL33 | Interleukin 33 | 2.655 | 2.40E-05 | Extracellular space | Cytokine |
| PLA2G4F | Phospholipase A2, group IVF | 2.533 | 3.50E-02 | Cytoplasm | Enzyme |
| APOA2 | Apolipoprotein A-II | 2.389 | 7.94E-06 | Extracellular space | Transporter |
| ITGB2 | Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 2.253 | 3.60E-04 | Plasma membrane | Transmembrane receptor |
| PON1 | Paraoxonase 1 | 2.232 | 1.48E-02 | Extracellular space | Phosphatase |
| PLA2G16 | Phospholipase A2, group XVI | 2.21 | 9.76E-04 | Nucleus | Enzyme |
| MSR1 | Macrophage scavenger receptor 1 | 2.181 | 2.17E-02 | Plasma membrane | Transmembrane receptor |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 2.09 | 9.76E-03 | Plasma membrane | G-protein coupled receptor |
| PDGFA | Platelet-derived growth factor alpha polypeptide | −2.225 | 3.31E-04 | Extracellular space | Growth factor |
| PLA2G3 | Phospholipase A2, group III | −2.448 | 3.23E-03 | Extracellular space | Enzyme |
| APOA1 | Apolipoprotein A-I | −2.681 | 1.01E-02 | Extracellular space | Transporter |
| F3 | Coagulation factor III (thromboplastin, tissue factor) | −2.96 | 1.87E-04 | Plasma membrane | Transmembrane receptor |
| ALOX12B | Arachidonate 12-lipoxygenase, 12R type | −5.499 | 1.67E-02 | Cytoplasm | Enzyme |
1Fold change: Values indicate the relative gene expression. Positive values indicate higher expression in weaned than in suckling calves, and negative values indicate the opposite.
2FDR = false discovery rate.
Upstream Regulators
Based on the differentially increased or decreased expression of genes in the dataset, IPA revealed the upstream regulators that were activated or inhibited within each group. Tables 2 and 3 show the top 5 upstream regulators with P < 0.05 and z-scores, respectively. Tretinoin was predicted as the most significant regulator (Table 2); the P-value of 112 target molecules downstream of tretinoin also changed (Table 4). Seventy target molecules were upregulated during weaning, whereas 42 were downregulated (Table 4). Seven molecules were predicted by IPA as the most activated (z-score > 2.000), whereas “estrogen receptor” was inhibited (z-score < −2.000) (Table 3).
Table 2.
The 5 major upstream regulators generated by ingenuity pathway analysis (IPA)
| Upstream regulator | Fold change1 | Predicted activation state | Activation z-score2 | P-value of overlap3 | Mechanistic network4 |
|---|---|---|---|---|---|
| Tretinoin | 1.918 | 2.19E-16 | 301 (20) | ||
| TGFβ15 | −1.073 | 1.089 | 1.15E-14 | 285 (17) | |
| Phorbol myristate acetate | 0.371 | 1.24E-14 | 253 (17) | ||
| PPARA6 | 1.364 | Activated | 2.155 | 3.17E-14 | 207 (16) |
| Lipopolysaccharide | Activated | 2.516 | 2.35E-13 | 283 (16) |
1Fold change: Values indicate the relative gene expression. Positive values indicate higher expression in weaned than in suckling calves, and negative values indicate the opposite.
2 z-Score: Values indicate a statistically significant pattern match between up- and down-regulation patterns.
3 P-value of overlap: Values indicate a statistically significant enrichment of the network-regulated genes in the database.
4Mechanistic network: The number of molecules that are downstream of the regulators and the number of regulators involved in that particular network in parentheses.
5TGFβ1: tumor growth factor beta-1.
6PPARA: Peroxisome proliferator-activated receptor alpha.
Table 3.
Upstream regulators activated (z-score > 2.00) and inhibited (z-score < -2.00) in the rumen of Japanese Black cattle during weaning
| Upstream regulator | Molecule type | P-value of overlap1 | Predicted activation state | Activation z-score2 |
|---|---|---|---|---|
| IgG | Complex | 1.38E-09 | Activated | 3.039 |
| Lipopolysaccharide | Chemical drug | 2.35E-13 | Activated | 2.516 |
| Alitretinoin | Chemical drug | 7.57E-06 | Activated | 2.241 |
| PPARδ3 | Ligand-dependent nuclear receptor | 3.40E-07 | Activated | 2.232 |
| PPARα4 | Ligand-dependent nuclear receptor | 3.17E-14 | Activated | 2.155 |
| Pirinixic acid | Chemical toxicant | 2.34E-08 | Activated | 2.075 |
| TNF5 | Cytokine | 6.08E-12 | Activated | 2.007 |
| Estrogen receptor | Group | 1.97E-07 | Inhibited | −2.232 |
1P-value of overlap: Values indicate a statistically significant encrichment of the network-regulated genes in the database.
2 z-Score: Values indicate a statistically significant match between up- and down-regulation patterns.
3PPARδ = peroxisome proliferator-activated receptor delta.
4PPARα = peroxisome proliferator-activated receptor alpha.
5TNF = tumor necrosis factor.
Table 4.
Genes regulated by tretinoin in rumen papillae tissue
| Symbol | Entrez gene name | Fold change1 | FDR2 | Location | Type(s) |
|---|---|---|---|---|---|
| SFTPC | Surfactant protein C | 15.731 | 6.95E-05 | Extracellular space | Other |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A | 14.833 | 3.44E-04 | Nucleus | Transcription regulator |
| MSLN | Mesothelin | 14.337 | 5.37E-03 | Extracellular space | Other |
| CYP4F2 | Cytochrome P450, family 4, subfamily F, polypeptide 2 | 14.255 | 5.63E-17 | Cytoplasm | Enzyme |
| NEU4 | Sialidase 4 | 7.214 | 1.56E-03 | Cytoplasm | Enzyme |
| SLC27A2 | Solute carrier family 27 (fatty acid transporter), member 2 | 7.161 | 2.84E-15 | Cytoplasm | Transporter |
| SIX1 | SIX homeobox 1 | 6.675 | 1.69E-04 | Nucleus | Transcription regulator |
| TNC | Tenascin C | 6.034 | 8.28E-04 | Extracellular space | Other |
| CES1 | Carboxylesterase 1 | 5.812 | 3.44E-04 | Cytoplasm | Enzyme |
| CYP1A1 | Cytochrome P450, family 1, subfamily A, polypeptide 1 | 5.111 | 4.60E-03 | Cytoplasm | Enzyme |
| SLA | Src-like-adaptor | 3.589 | 9.81E-05 | Plasma membrane | Other |
| AREG | Amphiregulin | 3.566 | 1.95E-03 | Extracellular space | Growth factor |
| A2M | α-2-macroglobulin | 3.543 | 5.08E-03 | Extracellular space | Transporter |
| DUSP4 | Dual specificity phosphatase 4 | 3.494 | 4.82E-03 | Nucleus | Phosphatase |
| EBI3 | Epstein-Barr virus induced 3 | 3.48 | 6.85E-04 | Extracellular space | Cytokine |
| BMP6 | Bone morphogenetic protein 6 | 3.453 | 6.37E-05 | Extracellular space | Growth factor |
| ALDH1A2 | Aldehyde dehydrogenase 1 family, member A2 | 3.445 | 7.74E-03 | Cytoplasm | Enzyme |
| CCR1 | Chemokine (C-C motif) receptor 1 | 3.425 | 1.12E-03 | Plasma membrane | G-protein coupled receptor |
| ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 3.274 | 2.77E-03 | Cytoplasm | Enzyme |
| PLEK | Pleckstrin | 3.273 | 1.37E-02 | Cytoplasm | Other |
| ALPL | Alkaline phosphatase, liver/bone/kidney | 3.185 | 8.35E-05 | Plasma membrane | Phosphatase |
| CCR5 | Chemokine (C-C motif) receptor 5 (gene/pseudogene) | 3.121 | 7.27E-03 | Plasma membrane | G-protein coupled receptor |
| CTGF | Connective tissue growth factor | 3.116 | 1.03E-02 | Extracellular space | Growth factor |
| TLR5 | Toll-like receptor 5 | 3.1 | 2.96E-03 | Plasma membrane | Transmembrane receptor |
| PTPN22 | Protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | 2.883 | 1.53E-02 | Cytoplasm | Phosphatase |
| THBS1 | Thrombospondin 1 | 2.851 | 1.03E-02 | Extracellular space | Other |
| NOV | Nephroblastoma overexpressed | 2.818 | 4.67E-02 | Extracellular space | Growth factor |
| ANPEP | Alanyl (membrane) aminopeptidase | 2.817 | 2.10E-02 | Plasma membrane | Peptidase |
| SCML1 | Sex comb on midleg-like 1 (Drosophila) | 2.814 | 5.60E-03 | Nucleus | Transcription regulator |
| SOAT1 | Sterol O-acyltransferase 1 | 2.774 | 1.47E-07 | Cytoplasm | Enzyme |
| MMP28 | Matrix metallopeptidase 28 | 2.76 | 1.13E-09 | Extracellular space | Peptidase |
| CTSL | Cathepsin L | 2.678 | 2.61E-02 | Cytoplasm | Peptidase |
| C1QA | Complement component 1, q subcomponent, A chain | 2.625 | 7.58E-03 | Extracellular space | Other |
| VSIG4 | V-set and immunoglobulin domain containing 4 | 2.623 | 2.56E-02 | Plasma membrane | Other |
| AMICA1 | Adhesion molecule, interacts with CXADR antigen 1 | 2.523 | 7.94E-06 | Plasma membrane | Other |
| CD52 | CD52 molecule | 2.522 | 5.02E-03 | Plasma membrane | Other |
| CD101 | CD101 molecule | 2.504 | 4.32E-03 | Plasma membrane | Other |
| FABP4 | Fatty acid binding protein 4, adipocyte | 2.496 | 3.53E-03 | Cytoplasm | Transporter |
| HMGCS2 | 3-Hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) | 2.491 | 4.20E-05 | Cytoplasm | Enzyme |
| IGFBP5 | Insulin-like growth factor binding protein 5 | 2.452 | 9.78E-04 | Extracellular space | Other |
| CD86 | CD86 molecule | 2.404 | 5.74E-03 | Plasma membrane | Transmembrane receptor |
| RBP2 | Retinol binding protein 2, cellular | 2.401 | 2.18E-03 | Cytoplasm | Transporter |
| APOA2 | Apolipoprotein A-II | 2.389 | 7.94E-06 | Extracellular space | Transporter |
| SEMA7A | Semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) | 2.374 | 1.44E-02 | Plasma membrane | Transmembrane receptor |
| BAMBI | BMP and activin membrane-bound inhibitor | 2.374 | 2.89E-03 | Plasma membrane | Other |
| GGT1 | γ-Glutamyltransferase 1 | 2.365 | 2.67E-02 | Plasma membrane | Enzyme |
| POMC | Proopiomelanocortin | 2.343 | 1.19E-04 | Extracellular space | Other |
| CD68 | CD68 molecule | 2.337 | 4.21E-03 | Plasma membrane | Other |
| CD38 | CD38 molecule | 2.332 | 3.81E-02 | Plasma membrane | Enzyme |
| SLAMF7 | SLAM family member 7 | 2.316 | 2.30E-02 | Plasma membrane | Other |
| CYP4F3 | Cytochrome P450, family 4, subfamily F, polypeptide 3 | 2.302 | 1.12E-03 | Cytoplasm | Enzyme |
| ECI2 | Enoyl-CoA delta isomerase 2 | 2.289 | 2.31E-05 | Cytoplasm | Enzyme |
| TYROBP | TYRO protein tyrosine kinase binding protein | 2.288 | 2.73E-03 | Plasma membrane | Transmembrane receptor |
| RGS5 | Regulator of G-protein signaling 5 | 2.276 | 4.61E-02 | Plasma membrane | Other |
| ITGB2 | Integrin, β 2 (complement component 3 receptor 3 and 4 subunit) | 2.253 | 3.60E-04 | Plasma membrane | Transmembrane receptor |
| ITGAM | Integrin, α M (complement component 3 receptor 3 subunit) | 2.245 | 2.53E-02 | Plasma membrane | Transmembrane receptor |
| MGST3 | Microsomal glutathione S-transferase 3 | 2.237 | 2.79E-05 | Cytoplasm | Enzyme |
| BNIP3L | BCL2/adenovirus E1B 19kDa interacting protein 3-like | 2.231 | 2.39E-06 | Cytoplasm | Other |
| HLA-DMB | Major histocompatibility complex, class II, DM beta | 2.207 | 1.58E-04 | Plasma membrane | Transmembrane receptor |
| FOLR2 | Folate receptor 2 (fetal) | 2.206 | 1.12E-02 | Plasma membrane | Transporter |
| MFNG | MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | 2.177 | 1.38E-02 | Cytoplasm | Enzyme |
| SPI1 | Spi-1 proto-oncogene | 2.172 | 4.23E-03 | Nucleus | Transcription regulator |
| IKZF1 | IKAROS family zinc finger 1 (Ikaros) | 2.165 | 8.46E-03 | Nucleus | Transcription regulator |
| CFP | Complement factor properdin | 2.165 | 7.82E-03 | Extracellular space | Other |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 2.09 | 9.76E-03 | Plasma membrane | G-protein coupled receptor |
| CIDEA | Cell death-inducing DFFA-like effector a | 2.085 | 3.59E-02 | Cytoplasm | Other |
| RPL6 | Ribosomal protein L6 | 2.055 | 1.27E-02 | Cytoplasm | Other |
| COL4A1 | Collagen, type IV, alpha 1 | 2.024 | 1.40E-03 | Extracellular space | Other |
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B | 2.009 | 3.27E-04 | Plasma membrane | Transmembrane receptor |
| MAOB | Monoamine oxidase B | 2.002 | 7.89E-03 | Cytoplasm | Enzyme |
| KLF9 | Kruppel-like factor 9 | −2.011 | 1.35E-04 | Nucleus | Transcription regulator |
| BTC | betacellulin | −2.042 | 2.68E-02 | Extracellular space | Growth factor |
| MYBL2 | v-myb avian myeloblastosis viral oncogene homolog- like 2 | −2.109 | 2.85E-02 | Nucleus | Transcription regulator |
| TSC22D3 | TSC22 domain family, member 3 | −2.116 | 1.31E-04 | Nucleus | Transcription regulator |
| SOX6 | SRY (sex determining region Y)-box 6 | −2.129 | 2.14E-02 | Nucleus | Transcription regulator |
| DLL1 | δ-Like 1 (Drosophila) | −2.154 | 1.79E-03 | Plasma membrane | Enzyme |
| KRT15 | Keratin 15 | −2.166 | 1.11E-03 | Cytoplasm | Other |
| HES1 | Hes family bHLH transcription factor 1 | −2.169 | 6.79E-03 | Nucleus | Transcription regulator |
| CEACAM1 | Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | −2.172 | 1.12E-02 | Plasma membrane | Other |
| KIF23 | Kinesin family member 23 | −2.181 | 2.57E-03 | Cytoplasm | Other |
| HOXD4 | Homeobox D4 | −2.212 | 1.81E-02 | Nucleus | Transcription regulator |
| HEY1 | Hes-related family bHLH transcription factor with YRPW motif 1 | −2.22 | 1.12E-02 | Nucleus | Transcription regulator |
| DUSP1 | Dual specificity phosphatase 1 | −2.259 | 3.10E-02 | Nucleus | Phosphatase |
| MYB | v-myb avian myeloblastosis viral oncogene homolog | −2.285 | 1.84E-02 | Nucleus | Transcription regulator |
| PTCH1 | Patched 1 | −2.319 | 3.84E-03 | Plasma membrane | Transmembrane receptor |
| IGFBP3 | Insulin-like growth factor binding protein 3 | −2.437 | 7.34E-03 | Extracellular space | Other |
| IGF2 | Insulin-like growth factor 2 | −2.472 | 3.57E-05 | Extracellular space | Growth factor |
| FGF2 | Fibroblast growth factor 2 (basic) | −2.476 | 2.82E-02 | Extracellular space | Growth factor |
| COL4A5 | Collagen, type IV, alpha 5 | −2.481 | 4.70E-03 | Extracellular space | Other |
| ITGA2 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | −2.507 | 6.26E-05 | Plasma membrane | Transmembrane receptor |
| GJB5 | Gap junction protein, beta 5, 31.1kDa | −2.615 | 7.49E-03 | Plasma membrane | Transporter |
| IGFBP2 | Insulin-like growth factor binding protein 2, 36kDa | −2.711 | 1.65E-03 | Extracellular space | Other |
| IGFBP6 | Insulin-like growth factor binding protein 6 | −2.797 | 1.94E-03 | Extracellular space | Other |
| STC1 | Stanniocalcin 1 | −2.923 | 5.90E-03 | Extracellular space | Kinase |
| F3 | Coagulation factor III (thromboplastin, tissue factor) | −2.96 | 1.87E-04 | Plasma membrane | Transmembrane receptor |
| HS3ST1 | Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | −3.044 | 6.26E-05 | Cytoplasm | Enzyme |
| RNASE1 | Ribonuclease, RNase A family, 1 (pancreatic) | −3.056 | 2.31E-02 | Extracellular space | Enzyme |
| GATA6 | GATA binding protein 6 | −3.173 | 1.12E-02 | Nucleus | Transcription regulator |
| TERT | Telomerase reverse transcriptase | −3.239 | 3.14E-03 | Nucleus | Enzyme |
| GHR | Growth hormone receptor | −3.268 | 2.20E-05 | Plasma membrane | Transmembrane receptor |
| CHL1 | Cell adhesion molecule L1-like | −3.721 | 1.88E-02 | Plasma membrane | Other |
| SYCP3 | Synaptonemal complex protein 3 | −4.417 | 2.38E-05 | Nucleus | Other |
| SLC12A2 | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | −4.847 | 3.85E-07 | Plasma membrane | Transporter |
| CFTR | Cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | −4.953 | 1.01E-06 | Plasma membrane | Ion channel |
| OAS2 | 2ʹ-5ʹ-Oligoadenylate synthetase 2, 69/71kDa | −5.376 | 7.18E-05 | Cytoplasm | Enzyme |
| BMPR1B | Bone morphogenetic protein receptor, type IB | −6.669 | 2.95E-03 | Plasma membrane | Kinase |
| PITX2 | Paired-like homeodomain 2 | −7.073 | 3.43E-02 | Nucleus | Transcription regulator |
| RXRG | Retinoid X receptor, gamma | −7.723 | 2.15E-02 | Nucleus | Ligand-dependent nuclear receptor |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 | −8.077 | 1.09E-09 | Plasma membrane | Kinase |
| HOXB9 | Homeobox B9 | −8.378 | 4.76E-02 | Nucleus | Transcription regulator |
| DSG1 | Desmoglein 1 | −9.067 | 3.88E-09 | Plasma membrane | Other |
| EYA2 | EYA transcriptional coactivator and phosphatase 2 | −19.365 | 2.07E-05 | Nucleus | Phosphatase |
1Fold change: Values indicate the relative gene expression. Positive values indicate higher expression in weaned than in suckling calves, and negative values indicate the opposite.
2FDR = false discovery rate.
DISCUSSION
This study comprises the first application of RNA-seq to investigate the transcriptional changes and to predict the genes underlying the physiological and morphological modifications accompanying the transition from suckling to weaning in the rumen papillae of Japanese Black calves. In our previous study, HMGCS2, AKR1C1, and FABP3 were identified as candidate up-regulated genes in the rumen epithelium after weaning and growth (Kato et al., 2015). However, based on RNA-seq analysis, only the expression of HMGCS2 increased in weaned calves compared with suckling calves. These results reflected the limited dataset in the EST library in silico. However, the present results from RNA-seq analysis revealed several possible pathways and upregulators by analyzing global expression profiles during the weaning, growth, and development of rumen papillae.
The IPA data used in the present study were based on human, rat, and mouse and therefore had limitations for bovine pathway analysis. However, analysis revealed that 7 pathways significantly changed (FDR < 0.05) from suckling to weaned calves, especially “Atherosclerosis signaling.” Atherosclerosis is a subsequent, nonadaptive response of blood vessels to damage, induced by chronic inflammation. So far, there are no direct evidences of a relationship between rumen epithelial cells and atherosclerosis-related genes. The main factors regulating atherosclerosis are thought to be high-cholesterol levels (in particular low-density lipoprotein [LDL] cholesterol), hypertension, and blood fat (Levine et al., 1995; Glasser et al., 1996). During the atherogenesis, circulating monocytes are recruited and differentiated, and tissue macrophages capture cholesterol and oxidize LDL in order to form lipid-foam cells (Moreno and Mitjavila, 2003). Intraruminal VFA are thought to be the substrate of cholesterol, and the in vivo synthesis of cholesterol may become active by increasing VFA levels through feeding concentrate or high-grain diets (Sakata and Tamate, 1978). Holstein cattle fed grain-based feed presented changes in the expression level of genes related to cholesterol synthesis and homeostasis, and the synthesis of cholesterol was regulated via intracellular sterol regulatory element-binding protein (SREBP) (Steele et al., 2011). In the present study, 12 of the 17 DEGs in the atherosclerosis pathway were upregulated during weaning, whereas 5 genes were downregulated (Table 1). In monogastric animals, all these genes, except APOA1, may act as regulators or initiators of atherosclerosis (Miller-Kasprzak et al., 2004; Takahashi et al., 2005; Sato et al., 2008; Meisel et al., 2011). Thus, cholesterol synthesis might be promoted by grain-based feeds during weaning, and atherosclerosis might advance. Decreasing the expression of the genes related to cholesterol synthesis may help relieve the inflammation caused by atherosclerosis, at least partly. This suggests that cholesterol homeostasis and inflammation might have been activated in the rumen epithelium during weaning, by feeding starter and roughage.
Among the upstream regulators indicated by IPA, tretinoin was predicted as the most active molecule in the rumen papillae of Japanese Black cattle during weaning (P = 2.19E-16; Table 2). However, tretinoin was not yet reported as an upregulating factor in the rumen epithelium. Other important factors controlling the differentiation of rumen epithelium at weaned are TGFB1 and PPARA (Connor et al., 2014). Tretinoin is also known as “All-trans retinoic acid (ATRA),” which is a derivative of vitamin A. Alitretionin, a form of vitamin A, is predicted to play a role on rumen development (Table 3). There are some reports that vitamin A or its derivatives are related to epithelium functioning. For example, vitamin A was related to the maintenance of lung epithelial cells in rat (Takahashi et al., 1993). In addition, vitamin A and its derivatives, including retinol, retinal, and retinoic acid, function as effective inhibitors of mammary epithelial cell proliferation in cattle, and it is possible that retinoid, an analogue of vitamin A, can regulate mammary growth and development (Purup et al., 2001). The expression of SLC26A3, downregulated in adenoma (Makela et al., 2002), is increased by ATRA in intestinal epithelial cells (Priyamvada et al., 2015). Only a few studies have reported the effects of vitamin A in rumen papillae. Although the effects of vitamin A differ according to gastrointestinal track sites, villus height in the ileum and jejunum was enhanced by the supplementation of vitamin A in newborn calves (Schottstedt et al., 2005). The present study suggests that tretinoin (probably also retinol or retinoic acid) might be an important factor mediating the development of the rumen epithelium during the transition period from suckling to weaned.
The IPA analysis also indicated that IgG, lipopolysaccharides (LPS), and tumor-necrosis factor-α (TNFα) are activated in the rumen papillae of weaned calves. These upstream regulators are involved in immune responses and inflammation. Because IgG is vital to provide adequate immunological protection and resistance to disease (Conneely et al., 2014), in rumen papillae it is needed to induce adequate immunity and protection against pathogenic organisms. Changes in LPS, which are produced from dead bacteria, are caused by the alteration of bacterial diversity during weaning. Grain-induced subacute ruminal acidosis and the increment of a grain-based diet in lactating dairy cows increased the free ruminal LPS (Gozho et al., 2007). The concentration of ruminal LPS increased in steers fed grain-based diets and caused a variety of metabolic and immunologic alterations in the host (Andersen et al., 1994; Gozho et al., 2005). Moreover, the mucosal epithelium becomes more permeable and susceptible to apoptosis by the presence and increment of endotoxins (Chin et al., 2006). In addition, TNFα may be produced in the rumen epithelium by increasing the LPS released from rumen microbes in weaned calves (Pfeffer et al., 1993; Pasparakis et al., 1996; Raabe et al., 1998). Because TNFα is paracrine and endocrine, it may only act on the inflammation of rumen epithelium after weaning. The presence of IgG, LPS, and TNFα in rumen papillae contributes to microbe-epithelium interactions that activate rumen-related immune responses.
The IPA analysis also revealed that PPARα, PPARδ, and pirinixic acid might activate the growth and proliferation of rumen papillae (Table 3). Both PPARα and PPARδ are nuclear-receptor stimulating genes involved in fatty acid intake and metabolism, ketogenesis, and epidermal proliferation (Schoonjans et al., 1996; Burdick et al., 2006; Takahashi et al., 2006; Badman et al., 2007; Laarman et al., 2012; Naeem et al., 2012; Connor et al., 2013; Benesch et al., 2014). These PPARs might be increased to adapt to fatty acid metabolism, ketogenesis, and development of rumen papillae in weaned calves. Both PPARα and PPARδ heterodimerize with retinoid X receptor (RXR) (Klemm et al., 2001). The FXR/RXR-, LXR/RXR-, and LPS/IL-1-mediated inhibition of the RXR function, and the VDR/RXR activation obtained in the canonical pathway analysis suggest the importance of RXR (Figure 1). Tretinoin was identified as the most important upregulator (Table 2), and alitretinoin (9-cis retinoic acid) has been identified as a ligand of RXR (Heyman et al., 1992), whereas pirinixic acid (Wy-14,643) is a ligand of PPARα (Schaefer et al., 2008). Because Wy-14,643 increased the gene expression of monocarboxylate transporters (MCT)1, but not the MCT4 of VFA, in cultured ovine rumen epithelial cells (Benesch et al., 2014), pirinixic acid and PPARα/δ activation of RXR by alitretinoin might be important to adapt to VFA absorption and, therefore, are related to the growth and development of the rumen epithelium during the weaning period. However, estrogen receptors were inhibited in rumen papilla of weaned calves. There are 2 types of estrogen receptors: estrogen receptor 1 (ESR1) and estrogen receptor 2 (ESR2). In the present study, the expression of ESR2, not ESR1, was lower in the rumen papillae of weaned calves, and this molecule is known to have an antiproliferative role in the epithelium of immature uterus (Weihua et al., 2000). Thus, ESR2 might be downregulated in the rumen papillae after weaning to block antiproliferative effects. The upstream regulators predicted in this study need to be further validated.
In conclusion, the RNA-seq analysis of the rumen papillae of suckling and weaned calves provided a comprehensive view of the relative abundance and differential expression of several protein-coding genes. This information provides a valuable resource for studying atherosclerosis signaling, as candidate networks were found, including the activation of PPAR/RXR, which act as upregulators of the proliferation and development of rumen papillae in the transition from suckling to weaning in Japanese Black calves.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
This work was partly supported by JSPS KAKENHI (grant 15K14839) and the Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture. We are grateful to the staff of the Grassland Research Support Center, Institute of Livestock and Grassland Science, NARO, for animal supplies and management.
LITERATURE CITED
- Andersen P. H., Hesselholt M., and Jarløv N.. 1994. Endotoxin and arachidonic acid metabolites in portal, hepatic and arterial blood of cattle with acute ruminal acidosis. Acta Vet. Scand. 35:223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badman M. K., Pissios P., Kennedy A. R., Koukos G., Flier J. S., and Maratos-Flier E.. 2007. Hepatic fibroblast growth factor 21 is regulated by pparalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 5:426–437. doi:10.1016/j.cmet.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Baldwin R. L. 6th, Wu S., Li W., Li C., Bequette B. J., and Li R. W.. 2012. Quantification of transcriptome responses of the rumen epithelium to butyrate infusion using RNA-seq technology. Gene Regul. Syst. Bio. 6:67–80. doi:10.4137/GRSB.S9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch F., Dengler F., Masur F., Pfannkuche H., and Gäbel G.. 2014. Monocarboxylate transporters 1 and 4: expression and regulation by PPARα in ovine ruminal epithelial cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 307:R1428–R1437. doi:10.1152/ajpregu.00408.2013 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., and Golani I.. 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125:279–284. [DOI] [PubMed] [Google Scholar]
- Burdick A. D., Kim D. J., Peraza M. A., Gonzalez F. J., and Peters J. M.. 2006. The role of peroxisome proliferator-activated receptor-beta/delta in epithelial cell growth and differentiation. Cell. Signal. 18:9–20. doi:10.1016/j.cellsig.2005.07.009 [DOI] [PubMed] [Google Scholar]
- Cappe O., and Moulines E.. 2009. On-line expectation-maximization algorithm for latent data models. J. R. Stat. Soc. B. 71:593–613. [Google Scholar]
- Chin A. C., Flynn A. N., Fedwick J. P., and Buret A. G.. 2006. The role of caspase-3 in lipopolysaccharide-mediated disruption of intestinal epithelial tight junctions. Can. J. Physiol. Pharmacol. 84:1043–1050. doi:10.1139/y06-056 [DOI] [PubMed] [Google Scholar]
- Conneely M., Berry D. P., Murphy J. P., Lorenz I., Doherty M. L., and Kennedy E.. 2014. Effect of feeding colostrum at different volumes and subsequent number of transition milk feeds on the serum immunoglobulin G concentration and health status of dairy calves. J. Dairy Sci. 97:6991–7000. doi:10.3168/jds.2013-7494 [DOI] [PubMed] [Google Scholar]
- Connor E. E., Baldwin R. L. 6th, Li C. J., Li R. W., and Chung H.. 2013. Gene expression in bovine rumen epithelium during weaning identifies molecular regulators of rumen development and growth. Funct. Integr. Genomics 13:133–142. doi:10.1007/s10142-012-0308-x [DOI] [PubMed] [Google Scholar]
- Connor E. E., Baldwin R. L. 6th, Walker M. P., Ellis S. E., Li C., Kahl S., Chung H., and Li R. W.. 2014. Transcriptional regulators transforming growth factor-β1 and estrogen-related receptor-α identified as putative mediators of calf rumen epithelial tissue development and function during weaning. J. Dairy Sci. 97:4193–4207. doi:10.3168/jds.2013-7471 [DOI] [PubMed] [Google Scholar]
- Endo M., Kawahara-Miki R., Cao F., Kimura K., Kuwayama T., Monji Y., and Iwata H.. 2013. Estradiol supports in vitro development of bovine early antral follicles. Reproduction. 145:85–96. doi:10.1530/REP-12-0319 [DOI] [PubMed] [Google Scholar]
- Glasser S. P., Selwyn A. P., and Ganz P.. 1996. Atherosclerosis: risk factors and the vascular endothelium. Am. Heart J. 131:379–384. [DOI] [PubMed] [Google Scholar]
- Górka P., Kowalski Z. M., Pietrzak P., Kotunia A., Jagusiak W., Holst J. J., Guilloteau P., and Zabielski R.. 2011. Effect of method of delivery of sodium butyrate on rumen development in newborn calves. J. Dairy Sci. 94:5578–5588. doi:10.3168/jds.2011-4166 [DOI] [PubMed] [Google Scholar]
- Gozho G. N., Krause D. O., and Plaizier J. C.. 2007. Ruminal lipopolysaccharide concentration and inflammatory response during grain-induced subacute ruminal acidosis in dairy cows. J. Dairy Sci. 90:856–866. doi:10.3168/jds.S0022-0302(07)71569-2 [DOI] [PubMed] [Google Scholar]
- Gozho G. N., Plaizier J. C., Krause D. O., Kennedy A. D., and Wittenberg K. M.. 2005. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J. Dairy Sci. 88:1399–1403. doi:10.3168/jds.S0022-0302(05)72807-1 [DOI] [PubMed] [Google Scholar]
- Heyman R. A., Mangelsdorf D. J., Dyck J. A., Stein R. B., Eichele G., Evans R. M., and Thaller C.. 1992. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 68:397–406. [DOI] [PubMed] [Google Scholar]
- Kato S., Sato K., Chida H., Roh S. G., Ohwada S., Sato S., Guilloteau P., and Katoh K.. 2011. Effects of na-butyrate supplementation in milk formula on plasma concentrations of GH and insulin, and on rumen papilla development in calves. J. Endocrinol. 211:241–248. doi:10.1530/JOE-11-0299 [DOI] [PubMed] [Google Scholar]
- Kato D., Suzuki Y., Haga S., So K., Yamauchi E., Nakano M., Ishizaki H., Choi K., Katoh K., and Roh S. G.. 2015. Utilization of digital differential display to identify differentially expressed genes related to rumen development. Anim. Sci. j. 87:584–590. doi:10.1111/asj.12448 [DOI] [PubMed] [Google Scholar]
- Klemm D. J., Leitner J. W., Watson P., Nesterova A., Reusch J. E., Goalstone M. L., and Draznin B.. 2001. Insulin-induced adipocyte differentiation. Activation of CREB rescues adipogenesis from the arrest caused by inhibition of prenylation. J. Biol. Chem. 276:28430–28435. doi:10.1074/jbc.M103382200 [DOI] [PubMed] [Google Scholar]
- Laarman A. H., Ruiz-Sanchez A. L., Sugino T., Guan L. L., and Oba M.. 2012. Effects of feeding a calf starter on molecular adaptations in the ruminal epithelium and liver of Holstein dairy calves. J. Dairy Sci. 95:2585–2594. doi:10.3168/jds.2011-4788 [DOI] [PubMed] [Google Scholar]
- Levine G. N., Keaney J. F. Jr, and Vita J. A.. 1995. Cholesterol reduction in cardiovascular disease. Clinical benefits and possible mechanisms. N. Engl. J. Med. 332:512–521. doi:10.1056/NEJM199502233320807 [DOI] [PubMed] [Google Scholar]
- Li B., and Dewey C. N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323. doi:10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä S., Kere J., Holmberg C., and Höglund P.. 2002. slc26a3 mutations in congenital chloride diarrhea. Hum. Mutat. 20:425–438. doi:10.1002/humu.10139 [DOI] [PubMed] [Google Scholar]
- Meisel S. R., Xu X. P., Edgington T. S., Cercek B., Ong J., Kaul S., and Shah P. K.. 2011. Dose-dependent modulation of tissue factor protein and procoagulant activity in human monocyte-derived macrophages by oxidized low density lipoprotein. J. Atheroscler. Thromb. 18:596–603. [DOI] [PubMed] [Google Scholar]
- Miller-Kasprzak E., Niemir Z. I., and Czekalski S.. 2004. [The role of platelet-derived growth factor a (pdgf-a) in hypertension and renal diseases. Part 1: structure and regulation of the pdgf-a gene expression and its role in hypertension]. Pol. Merkur. Lekarski 16:398–401. [PubMed] [Google Scholar]
- Moreno J. J., and Mitjavila M. T.. 2003. The degree of unsaturation of dietary fatty acids and the development of atherosclerosis (review). J. Nutr. Biochem. 14:182–195. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., and Wold B.. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods. 5:621–628. doi:10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Naeem A., Drackley J. K., Stamey J., and Loor J. J.. 2012. Role of metabolic and cellular proliferation genes in ruminal development in response to enhanced plane of nutrition in neonatal Holstein calves. J. Dairy Sci. 95:1807–1820. doi:10.3168/jds.2011-4709 [DOI] [PubMed] [Google Scholar]
- Pasparakis M., Alexopoulou L., Episkopou V., and Kollias G.. 1996. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med. 184:1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer K., Matsuyama T., Kündig T. M., Wakeham A., Kishihara K., Shahinian A., Wiegmann K., Ohashi P. S., Krönke M., and Mak T. W.. 1993. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. Monocytogenes infection. Cell 73:457–467. [DOI] [PubMed] [Google Scholar]
- Priyamvada S., Anbazhagan A. N., Gujral T., Borthakur A., Saksena S., Gill R. K., Alrefai W. A., and Dudeja P. K.. 2015. All-trans-retinoic acid increases SLC26A3 DRA (down-regulated in adenoma) expression in intestinal epithelial cells via HNF-1β. J. Biol. Chem. 290:15066–15077. doi:10.1074/jbc.M114.566356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purup S., Jensen S. K., and Sejrsen K.. 2001. Differential effects of retinoids on proliferation of bovine mammary epithelial cells in collagen gel culture. J. Dairy Res. 68:157–164. [DOI] [PubMed] [Google Scholar]
- Raabe T., Bukrinsky M., and Currie R. A.. 1998. Relative contribution of transcription and translation to the induction of tumor necrosis factor-alpha by lipopolysaccharide. J. Biol. Chem. 273:974–980. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K.. 2010. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. doi:10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh S. G., Kuno M., Hishikawa D., Hong Y. H., Katoh K., Obara Y., Hidari H., and Sasaki S.. 2007. Identification of differentially expressed transcripts in bovine rumen and abomasum using a differential display method. J. Anim. Sci. 85:395–403. doi:10.2527/jas.2006-234 [DOI] [PubMed] [Google Scholar]
- Roh S. G., Suzuki Y., Gotoh T., Tatsumi R., and Katoh K.. 2016. Physiological roles of adipokines, hepatokines, and myokines in ruminants. Asian-Australas. J. Anim. Sci. 29:1–15. doi:10.5713/ajas.16.0001R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T., and Tamate H.. 1978. Rumen epithelial cell proliferation accelerated by rapid increase in intraruminal butyrate. J. Dairy Sci. 61:1109–1113. doi:10.3168/jds.S0022-0302(78)83694-7 [DOI] [PubMed] [Google Scholar]
- Sato H., Kato R., Isogai Y., Saka G., Ohtsuki M., Taketomi Y., Yamamoto K., Tsutsumi K., Yamada J., Masuda S., et al. 2008. Analyses of group III secreted phospholipase A2 transgenic mice reveal potential participation of this enzyme in plasma lipoprotein modification, macrophage foam cell formation, and atherosclerosis. J. Biol. Chem. 283:33483–33497. doi:10.1074/jbc.M804628200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M. B., Pose A., Ott J., Hecker M., Behnk A., Schulz R., Weissmann N., Günther A., Seeger W., and Mayer K.. 2008. Peroxisome proliferator-activated receptor-alpha reduces inflammation and vascular leakage in a murine model of acute lung injury. Eur. Respir. J. 32:1344–1353. doi:10.1183/09031936.00035808 [DOI] [PubMed] [Google Scholar]
- Schoonjans K., Staels B., and Auwerx J.. 1996. The peroxisome proliferator activated receptors (ppars) and their effects on lipid metabolism and adipocyte differentiation. Biochim. Biophys. Acta. 1302:93–109. [DOI] [PubMed] [Google Scholar]
- Schottstedt T., Muri C., Morel C., Philipona C., Hammon H. M., and Blum J. W.. 2005. Effects of feeding vitamin a and lactoferrin on epithelium of lymphoid tissues of intestine of neonatal calves. j. Dairy Sci. 88:1050–1061. doi:10.3168/jds.S0022-0302(05)72773-9 [DOI] [PubMed] [Google Scholar]
- Shen Z., Seyfert H. M., Löhrke B., Schneider F., Zitnan R., Chudy A., Kuhla S., Hammon H. M., Blum J. W., Martens H., et al. 2004. An energy-rich diet causes rumen papillae proliferation associated with more igf type 1 receptors and increased plasma igf-1 concentrations in young goats. J. Nutr. 134:11–17. doi:10.1093/jn/134.1.11 [DOI] [PubMed] [Google Scholar]
- Steele M. A., Vandervoort G., AlZahal O., Hook S. E., Matthews J. C., and McBride B. W.. 2011. Rumen epithelial adaptation to high-grain diets involves the coordinated regulation of genes involved in cholesterol homeostasis. Physiol. Genomics. 43:308–316. doi:10.1152/physiolgenomics.00117.2010 [DOI] [PubMed] [Google Scholar]
- Stobo I. J., Roy J. H., and Gaston H. J.. 1966. Rumen development in the calf. 1. The effect of diets containing different proportions of concentrates to hay on rumen development. Br. j. Nutr. 20:171–188. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Haga S., Nakano M., Ishizaki H., Nakano M., Song S., Katoh K., and Roh S.. 2016. Postweaning changes in the expression of chemerin and its receptors in calves are associated with the modification of glucose metabolism. j. Anim. Sci. 94:4600–4610. doi:10.2527/jas.2016-0677 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Miura T., and Takahashi K.. 1993. Vitamin a is involved in maintenance of epithelial cells on the bronchioles and cells in the alveoli of rats. J. Nutr. 123:634–641. doi:10.1093/jn/123.4.634 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Tanaka T., Kodama T., and Sakai J.. 2006. Peroxisome proliferator-activated receptor delta (PPARdelta), A novel target site for drug discovery in metabolic syndrome. Pharmacol. Res. 53:501–507. doi:10.1016/j.phrs.2006.03.019 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Zhu H., and Yoshimoto T.. 2005. Essential roles of lipoxygenases in LDL oxidation and development of atherosclerosis. Antioxid. Redox Signal. 7:425–431. doi:10.1089/ars.2005.7.425 [DOI] [PubMed] [Google Scholar]
- Tamate H., McGilliard A. D., Jacobson N. L., and Getty R.. 1962. Effect of various dietaries on the anatomical development of the stomach in the calf. J. Dairy Sci. 45:408–420. [Google Scholar]
- Warner R. G., Flatt W. P., and Loosli J. K.. 1956. Dietary factors influencing the development of the ruminant stomach. J. Agric. Food Chem. 4:788–792. [Google Scholar]
- Weihua Z., Saji S., Mäkinen S., Cheng G., Jensen E. V., Warner M., and Gustafsson J. A.. 2000. Estrogen receptor (ER) beta, a modulator of eralpha in the uterus. Proc. Natl. Acad. Sci. USA. 97:5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.