Summary
Since the discovery of SUMO twenty years ago, SUMO conjugation has become a widely-recognized post-translational modification that targets a myriad of proteins in many processes. Great progress has been made in understanding the SUMO pathway enzymes, substrate sumoylation, and the interplay between sumoylation and other regulatory mechanisms in a variety of contexts. As these research directions continue to generate insights into SUMO-based regulation, several mechanisms by which sumoylation and desumoylation can orchestrate large biological effects are emerging. These include the ability to target multiple proteins within the same cellular structure or process, respond dynamically to external and internal stimuli, and modulate signaling pathways involving other post-translational modifications. Focusing on nuclear function and intracellular signaling, this review highlights a broad spectrum of historical data and recent advances with the aim of providing an overview of mechanisms underlying SUMO-mediated global effects to stimulate further inquiry into intriguing roles of SUMO.
eTOC
Dynamic SUMO conjugation and deconjugation of proteins exert a wide range of effects in biology. This review summarizes current understanding of several mechanisms that underlie SUMO-mediated global effects on nuclear function and intracellular signaling to stimulate further inquiry into intriguing roles of SUMO.
Brief overview of sumoylation and desumoylation processes
SUMO proteins are a family of conserved eukaryotic protein modifiers of approximately 100 amino acids. SUMO conjugation to the lysine(s) of substrates is carried out by SUMO E1, E2, and E3 enzymes (Johnson, 2004). Organisms examined so far contain only a single SUMO E1 and E2 enzyme but multiple SUMO E3 enzymes. The SUMO E1 uses ATP hydrolysis to covalently link SUMO to its active site cysteine and subsequently transfer SUMO to the active site on the E2. With the help of a SUMO E3 (or ligase), the E2 further transfers SUMO onto substrates (Fig 1A). SUMO is often conjugated at the sumoylation consensus sequence, ψKxE/D (ψ: hydrophobic residues; x: any amino acid; K: sumoylation site), which is recognized by the E2, or its reverse sequence (Rodriguez et al., 2001; Sampson et al., 2001). SUMO E3s support productive configurations for SUMO transfer by simultaneously binding the SUMO-charged E2 and the substrate (Streich and Lima, 2016; Werner et al., 2012). The multiple SUMO E3s within a cell have both distinct and overlapping substrates (Pichler et al., 2017). While lower eukaryotes contain only one SUMO, higher eukaryotes possess at least three SUMO isoforms, namely SUMO1–3. These isoforms differ in several respects, such as in their SUMO E3 preferences or ability to form poly-SUMO chains by conjugation of one SUMO to another SUMO molecule via different lysine residues (Pichler et al., 2017). Cell line studies further suggest that while the majority of SUMO1 is conjugated to substrates, SUMO2/3 mostly becomes conjugated under stress conditions. These differences suggest that SUMO isoforms can have distinct functions and regulation.
Figure 1: Dynamic SUMO conjugation cycle and its multiple effects on nuclear structure and functions.
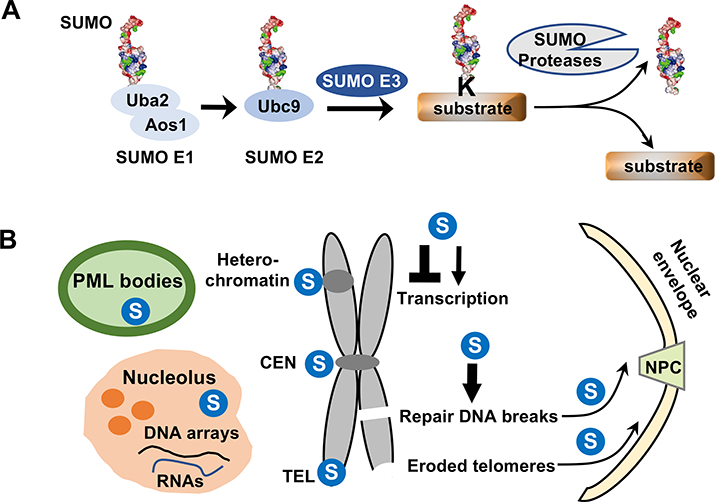
A. Conjugation and deconjugation of SUMO (shown in 3D structure rendering) are outlined. SUMO forms a thioester bond with the heterodimeric E1 (Aos1/Uba2) in an ATP-dependent manner. SUMO is then transferred to the E2 (Ubc9), again forming a thioester bond. SUMO is conjugated to the lysine residue (K) on the substrate with the help of SUMO E3. Only a single SUMO conjugation is shown, but multiple SUMOs or SUMO chains can also be found on substrates. SUMO proteases cleave SUMO from the substrate.
B. A brief summary of major effects of sumoylation on nuclear structure and functions. Nuclear domains and chromosomal regions enriched with SUMO are indicated by  . Sumoylation also regulates transcription and DNA lesion repair as indicated. Arrow: positive effects; lines: negative effects. Arrows pointing to the nuclear pore complex (NPC) and nuclear envelope indicate SUMO-mediated DNA movement toward these locations.
. Sumoylation also regulates transcription and DNA lesion repair as indicated. Arrow: positive effects; lines: negative effects. Arrows pointing to the nuclear pore complex (NPC) and nuclear envelope indicate SUMO-mediated DNA movement toward these locations.
Once conjugated to substrates, SUMO can exert a variety of effects. These include changing substrate interactions with DNA, RNA or other proteins, altering conformation or enzymatic activities, and modulating other modifications (Flotho and Melchior, 2013). Among these effects, the most frequently described has been interactions between SUMO and SUMO interaction motifs (SIMs). Canonical SIMs contain a core of hydrophobic residues preceded or followed by negatively charged amino acids; they contact a hydrophobic pocket on SUMO with neighboring basic residues (Hecker et al., 2006; Song et al., 2004). The SUMO:SIM association is generally weak, but can be enhanced by the binding of multiple SIMs to SUMO chains. Effects of sumoylation can be reversed when the modification is removed by SUMO-specific proteases, or desumoylases, which are also functionally important (Hickey et al., 2012) (Fig 1A).
SUMO affects nuclear structures and functions
In the early 2000s, examination of SUMO and sumoylation machinery mutants captured some striking nuclear structural defects, such as fragmented nucleoli, declustered telomeres, and heterochromatin breakdown (Hari et al., 2001; Nacerddine et al., 2005; Shin et al., 2005; Xhemalce et al., 2004; Zhao and Blobel, 2005). Meanwhile, SUMO enzymes and sumoylated proteins were found to be enriched at nuclear structures, such as PML and Polycomb bodies (Kagey et al., 2003; Muller et al., 1998; Sternsdorf et al., 1997). These early findings hinted at the possibility that SUMO may globally affect biological processes via modulation of nuclear structures. Indeed, insights into cellular membraneless structures suggest that SUMO’s ability to facilitate protein-protein interactions can contribute to their formation. In general, proteins capable of intra- or inter-molecular multivalent interactions can form large oligomers and phase separate from the surrounding solution (Banani et al., 2017; Hyman et al., 2014). These proteins can then use their modular interaction domains or intrinsically disordered regions to recruit additional macro-molecules, expanding liquid droplets. These droplets and membraneless structures can undergo fusion, fission, and rapid molecular exchange with the surrounding solution, yet high concentrations of macro-molecules within the structures may promote certain biological processes (Banani et al., 2017).
The role of SUMO in phase separation and PML body formation
PML bodies provide an example of how SUMO:SIM interactions could contribute to the phase separation-mediated formation of nuclear structures. PML bodies host more than 150 proteins with a wide range of functions, such as DNA repair, stress response, senescence, antiviral immunity, and tumor suppression (Sahin et al., 2014). These proteins appear to carry out some of their roles within PML bodies, though a unifying model for PML body functions remain to be established (Lallemand-Breitenbach and de The, 2018). It was noted early on that PML proteins and many other PML body constituents are sumoylated and contain SIMs; importantly, mutations affecting their sumoylation or SIMs were shown to impair PML body formation or constituent recruitment (Sahin et al., 2014; Shen et al., 2006). These findings and additional data suggested a model for PML body formation, wherein sumoylation of self-associated PML proteins recruits SIM-containing partner proteins, and sumoylation of the latter leads to additional SUMO:SIM interactions and PML body expansion (Banani et al., 2017; Sahin et al., 2014; Wang et al., 2018). Modeling SUMO:SIM interactions in engineered proteins shows that they are sufficient for driving phase separation in vitro, providing strong support for this model (Banani et al., 2016).
The effects of SUMO on nucleolar and proteinaceous structures
The nucleolus is the site of ribosome assembly, RNA processing, and cell cycle regulation, amongst other functions. Its domains also exhibit liquid droplet-like behaviors, such as diffusion and fusion (Brangwynne et al., 2011; Feric et al., 2016). How SUMO affects these behaviors has not been directly interrogated, but sumoylation and desumoylation both influence nucleolar structure and function. On one hand, nucleoli contain a proportion of sumoylation enzymes and substrates, and some of these enzymes and sumoylation events promote nucleolar integrity and functions (Ayaydin and Dasso, 2004; Heun, 2007; Matafora et al., 2009; Srikumar et al., 2013; Takahashi et al., 2008; Westman et al., 2010; Zhao and Blobel, 2005). On the other hand, desumoylation enzymes, such as SENP3 and 5 that target SUMO2/3-conjugates, are also enriched in nucleoli and are required for nucleolar function (Di Bacco et al., 2006; Finkbeiner et al., 2011; Liang et al., 2017; Yun et al., 2008). These data may suggest that specific sumoylation events and balanced sumoylation levels are required for nucleolar biology. Nucleoli additionally contain abundant ribosomal DNA units and RNA species capable of macromolecular association. A productive line of future inquiry may be exploring the potential collaborations between these DNA/RNA molecules and sumo in sculpting nucleoli.
While SUMO helps to “build up” membraneless organelles, it can also “break down” inactive proteinaceous structures, some of which underlie neurodegenerative diseases. A few examples include the ability of sumoylation to prevent protein inclusions formed by the transcriptional corepressor subunit Cyc8, to solubilize DNA end resection protein Sae2, and to reduce aggregation of translation factor CPEB3 or transcription factor androgen receptor (Drisaldi et al., 2015; Mukherjee et al., 2009; Oeser et al., 2016; Sarangi et al., 2015). Future study on how SUMO contributes to the dynamic assembly and disassembly of other proteinaceous and membraneless structures will expand our understanding of the structural roles of SUMO.
SUMO modulates chromosome structures and functions
SUMO and SUMO pathway enzymes associate with chromatin and SUMO substrates are enriched among DNA-binding proteins (Chymkowitch et al., 2015; Flotho and Melchior, 2013; Hendriks and Vertegaal, 2016; Neyret-Kahn et al., 2013; Niskanen et al., 2015). These observations corroborate with early genetic data that SUMO deficiency drastically changes chromosome integrity and segregation (Hari et al., 2001; Nacerddine et al., 2005; Shin et al., 2005; Tanaka et al., 1999; Xhemalce et al., 2004). Recent studies further elucidated how dynamic sumoylation regulates specific chromosome structures, such as centromeres, telomeres, heterochromatin, and broken regions, as summarized below (Fig 1B).
Sumoylation and desumoylation are critical for centromeric structures and functions
Centromeres contain specialized histones and silenced chromatin and support the assembly of kinetochores for microtubule attachment during mitosis. SUMO pathway enzymes are enriched at centromeres and kinetochores in multiple organisms, and sumoylation of numerous proteins concertedly regulate centromeric structure and function as reviewed recently (Cubenas-Potts and Matunis, 2013). One highly conserved SUMO substrate is Topoisomerase II (or Top2) (Clarke and Azuma, 2017). Sumoylation of Top2 C-terminal non-catalytic region leads to centromeric recruitment of Top2 itself to decatenate intertwined DNA before anaphase, as well as other mitotic factors (Claspin, Haspin and Aurora B kinases) to promote centromeric segregation (Azuma et al., 2003; Bachant et al., 2002; Dawlaty et al., 2008; Edgerton et al., 2016; Ryu et al., 2010; Ryu et al., 2015a). The SUMO pathway also affects centromeric histones and other mitotic regulators (Mukhopadhyay and Dasso, 2017). For example, sumoylated Orc2 recruits the KDN5A demethylase to centromeres to convert H3K4me3 into H3K4me2, thus permitting non-coding RNA production from the locus and subsequent heterochromatin maintenance (Huang et al., 2016). In another recent example, SUMO removal was shown to help extract the Aurora B kinase from chromatin and relocate it to the spindle midzone, which is an essential transition during mitosis (Pelisch et al., 2014). How Aurora B is extracted is unclear, but may involve the Cdc48 segregase and cofactors, which use ATP hydrolysis to remove sumoylated proteins such as the centromeric histone CENP-A from DNA (Franz et al., 2016; Merai et al., 2014).
The important SUMO-based regulatory events exemplified above could explain the drastic chromosome segregation defects arising from acute chemical inhibition of SUMO E1 or depletion of SUMO E2 and specific desumoylases (He et al., 2017; Mukhopadhyay and Dasso, 2017; Nacerddine et al., 2005; Pelisch et al., 2014). Strikingly, aneuploidy-prone SUMO pathway mutants may produce adaptive situations wherein gaining an extra chromosome partially resets cellular homeostasis, as seen in yeast cells lacking the Ulp2 desumoylase (Ryu et al., 2016). How exactly aneuploidy can benefit cells and be maintained in this situation remains to be understood. Another future question to consider is whether centromeric regions and/or kinetochores experience SUMO-facilitated phase separation given the abundant SUMO-mediated interactions at these sites.
The SUMO pathway contributes to heterochromatin formation and maintenance
The SUMO pathway also regulates other heterochromatic loci in addition to centromeric regions. In particular, sumoylation of the heterochromatin assembly factor HP1 promotes its association with RNA transcripts located at these regions to achieve initial HP1 targeting (Maison et al., 2011). Subsequent HP1 propagation along heterochromatin involves its binding to H3K9me3, catalyzed by Suv39h1 (Bannister et al., 2001; Lachner et al., 2001). Enhancement of HP1 sumoylation by Suv39h1 further provides a positive feed-forward mechanism in heterochromatin establishment (Maison et al., 2016). Sumoylation of the fission yeast HP1 homolog has also been implicated in heterochromatin regulation (Shin et al., 2005). Interestingly, interaction between SENP7 desumoylase and HP1 is important for maintaining the latter at heterochromatin, although the underlying mechanism is unclear (Maison et al., 2012). As such, temporal control of the HP1 sumoylation cycle appears to be key for heterochromatic structures. As HP1 phase separation is suggested to promote heterochromatin formation, addressing how its sumoylation and desumoylation are linked to this phenomenon will shed light on the potential effects of SUMO-based phase separation in chromatin domain dynamics (Larson et al., 2017; Strom et al., 2017).
SUMO is required for chromosome movement in multiple contexts
When heterochromatin experiences DNA double strand breaks (DSBs), sumoylation enables the broken DNA ends to relocate outside this region in order to prevent illegitimate repair such as those between repetitive sequences (Ryu et al., 2015b; Torres-Rosell et al., 2007). Analogously, SUMO helps target eroded telomeres and persistent DSBs in other parts of the genome to the nuclear periphery (Churikov et al., 2016; Horigome et al., 2016) (Fig 1B). Such targeting involves the association between sumoylated DNA repair proteins and SUMO-targeted ubiquitin ligases (STUbLs, more below) via SUMO:SIM interactions (Seeber and Gasser, 2017). Mechanisms enabling directional DNA end movement just began to emerge and involve a collaboration between nuclear actin and myosin with the Smc5/6 SUMO E3 complex (Caridi et al., 2018).
Studies highlighted in this section suggest a fundamental role for SUMO in the formation and movement of chromosomal structures (Fig 1B). Such a role could effectively modulate multiple forms of DNA transaction within these structures. In addition, SUMO is well known to regulate individual DNA transaction, such as transcription and DNA repair, by directly targeting proteins involved in these processes; as these topics have been extensively reviewed, we refer readers to some of these articles for details (Rosonina et al., 2017; Sarangi and Zhao, 2015; Schwertman et al., 2016; Wei and Zhao, 2017).
Global changes in sumoylation levels
Sumoylation states are highly dynamic, enabling rapid responses to changing external and internal stimuli. Studies from multiple organisms have begun to delineate the various mechanisms that enable large-scale changes of sumoylation levels and how this leads to alteration in multiple processes at once to achieve coordinated regulation. This section assesses three mechanisms and their implications.
STUbLs and their antagonists regulate sumoylation levels
The STUbL enzymes provide a major means of changing global sumoylation levels. Initially demonstrated in yeasts, it is now well accepted that STUbLs use their SIM arrays to bind poly-SUMO chains on proteins and poly-ubiquitinate these proteins, which often leads to their proteasome-mediated degradation (Mullen and Brill, 2008; Prudden et al., 2007; Sun et al., 2007; Uzunova et al., 2007; Xie et al., 2007). STUbLs target both SUMO pathway enzymes and distinct groups of substrates (Fig 2A). For example, the yeast STUbLs can target Siz SUMO E3s; similarly a human STUbL, RNF4, targets SUMO E2 and multiple PIAS E3s that are heavily sumoylated (Kumar et al., 2017; Nie and Boddy, 2015; Westerbeck et al., 2014). In addition, RNF4 and another STUbL, Arkadia, affects the PML proteins. In particular, STUbL-mediated degradation of the PML-RARA fusion proteins, the causative agent for acute promyelocytic leukemia, underlies the therapeutic effect of arsenic in this disease (Erker et al., 2013; Lallemand-Breitenbach et al., 2008; Poulsen et al., 2013; Tatham et al., 2008). In another context, PML-mediated sumoylation of misfolded proteins located inside PML bodies can facilitate their RNF4-mediated degradation to reduce the protein aggregates associated with neurodegenerative disease (Chu and Yang, 2011; Guo et al., 2014). These findings corroborate the observation that proteasomes can be targeted to PML bodies when their subunits are sumoylated (Lamoliatte et al., 2017), suggesting that multiple SUMO-based mechanisms act in concert to prevent undesired protein aggregation.
Figure 2: Examples of SUMO pathway regulation.
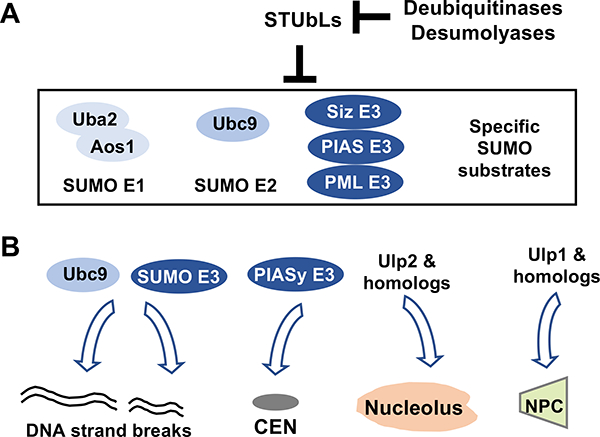
A. SUMO E1, E2, multiple SUMO E3s, and specific SUMO substrates can be targeted by STUbL-mediated protein degradation, an effect that can be counteracted by specific deubiquitinases or desumoylases.
B. SUMO E2, SUMO E3s, and desumoylases can be targeted to specific nuclear and chromosomal structures to induce large scale changes in sumoylation.
As sumoylation is essential, STUbL-mediated degradation of sumoylated proteins must be reined in by counter-regulatory mechanisms to sustain SUMO homeostasis (Fig 2A). Recent studies suggest several means of antagonizing STUbLs. For example, the Ataxin and USP11 deubiquitinases counteract the effects of RNF4 in maintaining PML and DNA repair protein sumoylation levels (Hendriks et al., 2015; Pfeiffer et al., 2017). During DNA replication, the USP7 deubiquitinase can remove ubiquitin conjugated to SUMO, thus preventing degradation of sumoylated proteins at chromatin around replisomes (Lecona et al., 2016). Functional studies suggest that these mechanisms are important for DNA repair and replication (Hendriks et al., 2015; Lecona et al., 2016; Pfeiffer et al., 2017). SUMO removal from substrates by desumoylases provides another way of counteracting the effects of STUbLs. For example, the yeast Ulp2 desumoylase protects rDNA-bound sumoylated proteins from STUbL-mediated degradation, thus promoting ribosomal DNA stability (Gillies et al., 2016; Liang et al., 2017).
SUMO machinery localization is critical for regulating sumoylation levels.
Differential localization of SUMO pathway enzymes provides another means of controlling sumoylation levels (Fig 2B). For example, while the yeast Ulp2 desumoylase is mainly localized inside the nucleus with nucleolar enrichment, the other yeast desumoylase, Ulp1, is tethered to nuclear pore complexes (Panse et al., 2003; Srikumar et al., 2013). Ulp1 sequestration at the nuclear rim is important for sustaining global sumoylation levels and for processes sensitive to SUMO homeostasis, such as DNA repair and transcription (Palancade et al., 2007; Texari et al., 2013). Similarly, anchoring the Ulp1 homolog SENP1 at nuclear pore complexes is required for proper DNA repair (Chow et al., 2012; Duheron et al., 2017). SUMO E3s also show differential localization, such as the presence of the PML SUMO E3 at PML bodies, as described above.
Altering enzymatic localization is an effective mechanism for adjusting cellular sumoylation levels in response to environmental changes. A well-established example is the upregulation of sumoylation levels of many DNA repair and checkpoint proteins in yeast and human cells after genotoxic stress. This is achieved partly by targeting SUMO E2 and specific SUMO E3s to DNA damage sites (Cremona et al., 2012; Galanty et al., 2009; Morris et al., 2009; Psakhye and Jentsch, 2012) via association between sumoylation enzymes and DNA lesion recognition complexes, including the ssDNA-binding complex RPA and DSB-binding MRX complex (Chen et al., 2016; Chung and Zhao, 2015). The SUMO pathway enzymes also change localization during the cell cycle. For example, when cells enter mitosis, the SUMO E3 PIASy and the SENP1 and SENP2 desumolyases are targeted to centromeric regions and kinetochores to orchestrate dynamic modifications required for mitotic progression (Cubenas-Potts et al., 2013; Ryu et al., 2010). Further investigation of enzymatic localization in additional situations and cell types will provide a fuller understanding of SUMO pathway regulation.
Modulation of the SUMO pathway enzymes by other protein modifications
Regulation of the SUMO machinery by other protein modifiers provides yet another strategy for altering sumoylation levels. For example, three other classes of modifications can induce large changes in SUMO E2 activity. In response to hypoxia, SUMO E2 acetylation reduces its interaction with sumoylation consensus sites, thus diminishing sumoylation of many proteins (Hsieh et al., 2013). On the other hand, SUMO E2 phosphorylation by CDK1/cyclin B during mitosis increases its overall activity toward forming thioester bonds with SUMO (Su et al., 2012; Wen et al., 2017). Finally, sumoylation of SUMO E2 favors SUMO chain formation in yeast and target discrimination in mammals (Klug et al., 2013; Knipscheer et al., 2008). Phosphorylation also exerts regulatory effects on other SUMO pathway enzymes. For example, Cdc5 phosphorylation of yeast Ulp2 desumoylase downregulates its function in mitosis, which is thought to preserve appropriate sumoylation levels required for mitosis (Baldwin et al., 2009). In another example, mTOR-mediated phosphorylation of SENP3 favors its interaction with nucleolar scaffold protein NPM1 to promote nucleolar targeting and SUMO2/3 removal at this location (Raman et al., 2014).
As outlined above, adjusting sumoylation levels on a large scale can be achieved by STUbLs and their antagonists, as well as by alteration of SUMO pathway enzyme localization and modifications. Such adjustment provides an economic means of simultaneously modulating multiple pathways, thus facilitating complex processes such as cell cycle progression and environment adaptation. We next review how the requisite tight integration of SUMO-based regulation with other signaling systems operating during the same cellular transitions is achieved.
Crosstalk between SUMO-based and other signaling processes
The above sections described specific situations in which the SUMO machinery is affected by other protein modifications and vice versa. On a larger scale, the physical and genetic interactions between sumoylation and signaling pathways mediated by other post-translational modifications remain to be explored for many contexts. Current progress on crosstalk between SUMO and phosphorylation- and ubiquitin-based signaling is summarized below.
Interactions with the DNA damage checkpoint pathways
Sumoylation and the DNA damage checkpoint pathway interact in multiple ways (Fig 3A). In both yeast and human cells, DNA damage-induced sumoylation and checkpoint-mediated phosphorylation occur in parallel but with significant substrate overlap (Cremona et al., 2012; Munk et al., 2017; Psakhye and Jentsch, 2012). These studies also show that changes in the checkpoint pathway can alter sumoylation events. In particular, reducing the function of the ATR checkpoint kinase or its cofactors leads to increased sumoylation of multiple proteins. Whether this reflects a direct role of checkpoint proteins in down-regulating sumoylation is not well understood, though the ATM checkpoint kinase can positively regulate SENP2 transcription in specific context (Lee et al., 2011). However, the opposite has also been reported, as ATM appears to promote sumoylation in the absence of ATR (Munk et al., 2017), suggesting context-dependent crosstalk.
Figure 3: Examples of crosstalk between SUMO-based regulation and other signaling processes.
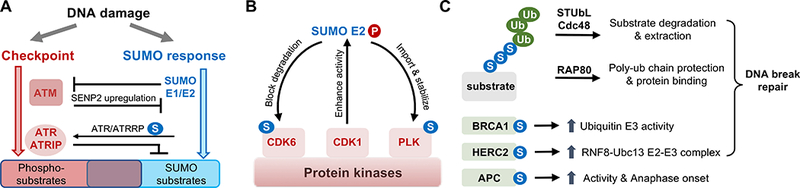
A. Interactions with the DNA damage checkpoint pathways. As described in the text, genome stress such as those caused by genotoxin treatment can induce a SUMO-based response and the ATM/ATR checkpoint kinase-mediated response. The two responses are independent but overlapping, and exhibit context-dependent and multi-layered genetic interactions. The enzymes in the two pathways show both positive (arrows) and negative (lines) genetic interactions as summarized in the text. In addition, they can target a group of common substrates, which provide another layer of crosstalk.
B. SUMO E2 interactions with protein kinases. As described in the text, CDK1-mediated phosphorylation of SUMO E2 can increases E2 activity. Phosphorylated SUMO E2 can promote the sumoylation of the polo-kinase (PLK) and CDK6 in different cellular contexts. These sumoylation events can positively influence substrate kinase functions.
C. Crosstalk with ubiquitination pathways. As detailed in the text, the hybrid SUMO-ubiquitin chain generated by STUbLs can lead to protein degradation or Cdc48 segregase-mediated protein extraction, but may also result in recruitment of proteins like RAP80, which recognize hybrid chains. RAP80 recruits additional proteins to DNA breaks and favors end-joining repair. SUMO can also modify ubiquitin ligases, such BRCA1, HERC2, and APC. Their sumoylation promotes specific activities or scaffolding roles, as depicted. Several types of this regulation occur during DSB repair.
Changing sumoylation levels also influences checkpoint functions. For example, sumoylation of ATR and its cofactor ATRIP in human cells supports genotoxin survival by boosting ATRIP interactions with other checkpoint factors and thus enhancing the checkpoint response (Wu et al., 2014; Wu and Zou, 2016). In contrast, ATM-mediated DNA damage checkpoint and cellular survival are promoted by transient inactivation of SUMO E1 and E2 via oxidative stress-induced disulfide bond formation between their catalytic cysteines (Stankovic-Valentin et al., 2016). These rather complex interactions between sumoylation and DNA damage checkpoint proteins under various conditions require further investigation to deconvolute the specific outcomes linked to distinct molecular mechanisms.
Interactions with other protein kinase-based signaling pathways
Recent proteomic data has helped to paint a more comprehensive picture of SUMO’s functional interactions with protein kinases. Microarray-based assays found that a large number of kinases are sumoylated in vitro (Uzoma et al., 2018). Consistently, another proteomic study reported co-regulation of sumoylation and phosphorylation (Hendriks et al., 2017). In particular, sumoylated lysines are abundantly found near CDK1 consensus sites, and sumoylation of these sequences showed dependence on CDK1. This finding is in line with previously reported phosphorylation-dependent sumoylation (Hietakangas et al., 2006) and the aforementioned ability of CdK1 to enhance SUMO E2 activity (Su et al., 2012; Wen et al., 2017). One outcome of CDK1-mediated SUMO E2 regulation is that phosphorylated E2 interacts with the Polo-kinase, leading to the latter’s SUMO1 conjugation. This in turn serves to promote Polo-kinase nuclear import and stabilization, driving mitotic progression (Wen et al., 2017). Such a kinase-SUMO E2-kinase regulatory relay has also been observed in other contexts (Fig 3B). In glioblastoma cells, CDKI-mediated phosphorylation of SUMO E2 promotes SUMO1 conjugation of CDK6, saving the latter from degradation by blocking its ubiqutination and consequently favoring G1-S phase transition and cancer development (Bellail et al., 2014). With the identification of a large number of substrates subject both to sumoylation and phosphorylation, additional modes of crosstalk between these modifications, as well as elucidation of their functional importance, are anticipated in the future.
Interactions with ubiquitination pathways
Hybrid SUMO-ubiquitin chains generated by STUbLs often lead to protein degradation, but they can also recruit proteins that recognize both ubiquitin and SUMO (Fig 3C). At DSB sites, such chains recruit the RAP80 scaffold protein that can preserve polyubiquitin chains as landing pads for recruitment of DNA repair proteins, such as 53BP1, that favor end joining DNA repair, and sequent recombination repair proteins, such as BRCA1 (Guzzo et al., 2012; Lombardi et al., 2017). Additionally, SUMO plays important roles in modulating multiple ubiquitin E3 functions in DSB repair (Figure 3C). For example, SUMO1 modification of BRCA1 can increase its ubiquitin E3 activity, while similar modification of HERC2 favors the formation of ubiquitin E2-E3 complexes composed of RNF8 and UBC13 (Danielsen et al., 2012; Morris et al., 2009). The combined effects of sumoylation and ubiquitination in these situations generally help recruit DNA repair factors and enhance their functions, but may also enable disassembly of repair complexes through STUbL or the Cdc48 segregase (Nie and Boddy, 2016; Schwertman et al., 2016). Sumoylation also regulates ubiquitination enzymes in other cellular contexts. For example, sumoylation of a subunit of the APC ubiquitin E3 in mitosis was recently suggested to help activate the latter for timely anaphase onset (Eifler et al., 2018; Lee et al., 2018). Of note, SUMO and ubiquitin can also antagonize each other’s effects, sometimes via modification of the same site(s), as has been shown for DNA replication factor PCNA (Zilio et al., 2017).
Conclusion and outlook
A large body of work has established critical roles for sumoylation and desumoylation in many aspects of biology. While SUMO-mediated effects in specific processes have been reviewed, it is also useful to bring together findings from diverse arenas to derive common underlying regulatory mechanisms and identify the most significant roles of SUMO. By stepping back to take a broader perspective, the current body of knowledge offers a few take-home messages regarding the global effects of SUMO on nuclear function. First, as outlined above, dynamic sumoylation of multiple substrates can contribute to membraneless nuclear structures and functions therein. Similarly, the SUMO pathway and its many substrates serve necessary structural and regulatory functions at chromosome regions such as centromeres and heterochromatin. Moreover, sumoylation promotes DNA end movement and can directly affect transcription and DNA repair. Some of these events are a part of cellular response system entailing sumoylation induction of numerous proteins. The dynamic machinery localization. Finally, the SUMO pathway shows intricate interactions with kinase- and ubiquitin-mediated regulation and signaling processes such as during the DNA damage response and cell cycle progression.
Also outlined in this review are many questions that remain to be explored. For example, could SUMO:SIM interactions help to form a wide range of nuclear structures beyond PML bodies, and could these include centromere and heterochromatin domains? How can the SUMO system collaborates with nuclear actin and myosin to drive DNA movement? What are the mechanisms targeting SUMO pathway enzymes to specific sites, and how these can change during the cell cycle and upon changes in internal and external stimuli? What are the specific functional interactions between the SUMO system and the phosphorylation and ubiquitination machinery in different contexts. Addressing these questions can be greatly facilitated by further evolution of techniques for studying sumoylation. For example, improvement of mass spectrometry-based detection of sumoylation sites can help to connect specific sumoylation events with unique biological outcomes. Mapping the complete set of sumoylation sites for low abundance proteins and directly monitoring sumoylated protein forms in tissues are still challenging. Another direction that awaits further development pertains to the understanding of sumoylation in human disease. Besides those mentioned above, such as acute promyelocytic leukemia and neurodegenerative diseases, the SUMO pathway is known to be related to tumorigenesis and host defenses against viral infections (Flotho and Melchior, 2013; Seeler and Dejean, 2017). The mechanism of SUMO-associated pathogenesis in many diseases is just beginning to be fleshed out, and a better understanding will pave the way for new diagnostic and treatment strategies aimed at modulating this dynamic post-translation modification.
Acknowledgement
We thank Michael Matunis, Xiaolu Yang, Richard Gardner, Xiao Peng and other Zhao lab members for their comments. Due to space limitations, we apologize to colleagues whose work is not cited here. We acknowledge the support of NIH grant GM080670.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayaydin F, and Dasso M (2004). Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 15, 5208–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, and Dasso M (2003). SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol 163, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, and Elledge SJ (2002). The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 9, 1169–1182. [DOI] [PubMed] [Google Scholar]
- Baldwin ML, Julius JA, Tang XY, Wang YC, and Bachant J (2009). The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle 8, 3406–3419. [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Bio 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional control of phase-separated cellular bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, and Kouzarides T (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Bellail AC, Olson JJ, and Hao C (2014). SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun 5, 4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, and Hyman AA (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA 108, 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, et al. (2018). Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Chuang YC, Chuang CN, Cheng YH, Chang CR, Leng CH, and Wang TF (2016). S. cerevisiae Mre11 recruits conjugated SUMO moieties to facilitate the assembly and function of the Mre11-Rad50-Xrs2 complex. Nucleic Acids Res 44, 2199–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KH, Elgort S, Dasso M, and Ullman KS (2012). Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus 3, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, and Yang X (2011). SUMO E3 ligase activity of TRIM proteins. Oncogene 30, 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, and Zhao X (2015). DNA break-induced sumoylation is enabled by collaboration between a SUMO ligase and the ssDNA-binding complex RPA. Genes Dev 29, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Charifi F, Eckert-Boulet N, Silva S, Simon MN, Lisby M, and Geli V (2016). SUMO-dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep 15, 1242–1253. [DOI] [PubMed] [Google Scholar]
- Chymkowitch P, Nguea PA, Aanes H, Koehler CJ, Thiede B, Lorenz S, Meza-Zepeda LA, Klungland A, and Enserink JM (2015). Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res 25, 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ, and Azuma Y (2017). Non-Catalytic roles of the topoisomerase II C-terminal domain. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, and Zhao X (2012). Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell 45, 422–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubenas-Potts C, Goeres JD, and Matunis MJ (2013). SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol Biol Cell 24, 3483–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubenas-Potts C, and Matunis MJ (2013). SUMO: A multifaceted modifier of chromatin structure and function. Dev Cell 24, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen JR, Povlsen LK, Villumsen BH, Streicher W, Nilsson J, Wikstrom M, Bekker- Jensen S, and Mailand N (2012). DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol 197, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, and van Deursen JM (2008). Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase II alpha. Cell 133, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, and Gill G (2006). The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol 26, 4489–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisaldi B, Colnaghi L, Fioriti L, Rao N, Myers C, Snyder AM, Metzger DJ, Tarasoff J, Konstantinov E, Fraser PE, et al. (2015). SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep 11, 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duheron V, Nilles N, Pecenko S, Martinelli V, and Fahrenkrog B (2017). Localisation of Nup153 and SENP1 to nuclear pore complexes is required for 53BP1-mediated DNA doublestrand break repair. J Cell Sci 130, 2306–2316. [DOI] [PubMed] [Google Scholar]
- Edgerton H, Johansson M, Keifenheim D, Mukherjee S, Chacon JM, Bachant J, Gardner MK, and Clarke DJ (2016). A noncatalytic function of the topoisomerase II CTD in Aurora B recruitment to inner centromeres during mitosis. J Cell Biol 213, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler K, Cuijpers SAG, Willemstein E, Raaijmakers JA, El Atmioui D, Ovaa H, Medema RH, and Vertegaal ACO (2018). SUMO targets the APC/C to regulate transition from metaphase to anaphase. Nat Commun 9, 1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erker Y, Neyret-Kahn H, Seeler JS, Dejean A, Atfi A, and Levy L (2013). Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 33, 2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Raman N, and Muller S (2011). SUMO routes ribosome maturation. Nucleus 2, 527–532. [DOI] [PubMed] [Google Scholar]
- Flotho A, and Melchior F (2013). Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82, 357–385. [DOI] [PubMed] [Google Scholar]
- Franz A, Ackermann L, and Hoppe T (2016). Ring of change: CDC48/p97 drives protein dynamics at chromatin. Front Genet 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, and Jackson SP (2009). Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies J, Hickey CM, Su D, Wu ZP, Peng JM, and Hochstrasser M (2016). SUMO pathway modulation of regulatory protein binding at the ribosomal DNA Locus in Saccharomyces cerevisiae. Genetics 202, 1377–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LL, Giasson BI, Glavis-Bloom A, Brewer MD, Shorter J, Gitler AD, and Yang XL (2014). A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol Cell 55, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo CM, Berndsen CE, Zhu J, Gupta V, Datta A, Greenberg RA, Wolberger C, and Matunis MJ (2012). RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal 5, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari KL, Cook KR, and Karpen GH (2001). The Drosophila Su(var)2–10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev 15, 1334–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XY, Riceberg J, Soucy T, Koenig E, Minissale J, Gallery M, Bernard H, Yang XF, Liao H, Rabino C, et al. (2017). Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol 13, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, and Dikic I (2006). Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281, 16117–16127. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal ACO, and Nielsen ML (2017). Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol 24, 325–336. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Schimmel J, Eifler K, Olsen JV, and Vertegaal ACO (2015). Ubiquitin-specific protease 11 (USP11) deubiquitinates hybrid small ubiquitin-like modifier (SUMO)-ubiquitin chains to counteract RING finger protein 4 (RNF4). J Biol Chem 290, 15526–15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, and Vertegaal ACO (2016). A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 17, 581–595. [DOI] [PubMed] [Google Scholar]
- Heun P (2007). SUMOrganization of the nucleus. Curr Opin Cell Biol 19, 350–355. [DOI] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, and Hochstrasser M (2012). Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, and Sistonen L (2006). PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A 103, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-Pflugfelder M, Cobb JA, and Gasser SM (2016). PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev 30, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YL, Kuo HY, Chang CC, Naik MT, Liao PH, Ho CC, Huang TC, Jeng JC, Hsu PH, Tsai MD, et al. (2013). Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J 32, 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Cheng JK, Bawa-Khalfe T, Yao XB, Chin YE, and Yeh ETH (2016). SUMOylated ORC2 recruits a histone demethylase to regulate centromeric histone modification and genomic stability. Cell Rep 15, 147–157. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, and Juelicher F (2014). Liquid-liquid phase separation in biology. Annu Rev Cell Dev Bi 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004). Protein modification by SUMO. Annu Rev Biochem 73, 355–382. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, and Wotton D (2003). The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137. [DOI] [PubMed] [Google Scholar]
- Klug H, Xaver M, Chaugule VK, Koid S, Mittler G, Klein F, and Pichler A (2013). Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae. Mol Cell 50, 625–636. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, and Pichler A (2008). Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 31, 371–382. [DOI] [PubMed] [Google Scholar]
- Kumar R, Gonzalez-Prieto R, Xiao ZY, Verlaan-de Vries M, and Vertegaal ACO (2017). The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat Commun 8, 1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll N, Rea S, Mechtler K, and Jenuwein T (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, and de The H (2018). PML nuclear bodies: from architecture to function. Curr Opin Cell Biol 52, 154–161. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, and de The H (2008). Arsenic degrades PML or PML - RAR alpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10, 547–555. [DOI] [PubMed] [Google Scholar]
- Lamoliatte F, McManus FP, Maarifi G, Chelbi-Alix MK, and Thibault P (2017). Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat Commun 8, 14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecona E, Rodriguez-Acebes S, Specks J, Lopez-Contreras AJ, Ruppen I, Murga M, Munoz J, Mendez J, and Fernandez-Capetillo O (2016). USP7 is a SUMO deubiquitinase essential for DNA replication. Nat Struct Mol Biol 23, 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Li B, Yu H, and Matunis MJ (2018). Sumoylation promotes optimal APC/C activation and timely anaphase. Elife 7, e29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Mabb AM, Gill GB, Yeh ETH, and Miyamoto S (2011). NF-kappa B induction of the SUMO protease SENP2: a negative feedback loop to attenuate cell survival response to genotoxic stress. Mol Cell 43, 180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Singh N, Carlson CR, Albuquerque CP, Corbett KD, and Zhou HL (2017). Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity. Genes Dev 31, 802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi PM, Matunis MJ, and Wolberger C (2017). RAP80, ubiquitin and SUMO in the DNA damage response. J Mol Med 95, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Quivy JP, and Almouzni G (2016). The methyltransferase Suv39h1 links the SUMO pathway to HP1 alpha marking at pericentric heterochromatin. Nat Commun 7, 12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, de Oca RM, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. (2011). SUMOylation promotes de novo targeting of HP1 alpha to pericentric heterochromatin. Nat Genet 43, 220–U265. [DOI] [PubMed] [Google Scholar]
- Maison C, Romeo K, Bailly D, Dubarry M, Quivy J-P, and Almouzni G (2012). The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat Struct Mol Biol 19, 458–460. [DOI] [PubMed] [Google Scholar]
- Matafora V, D’Amato A, Mori S, Blasi F, and Bachi A (2009). Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics 8, 2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merai Z, Chumak N, Garcia-Aguilar M, Hsieh TF, Nishimura T, Schoft VK, Bindics J, Slusarz L, Arnoux S, Opravil S, et al. (2014). The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc Natl Acad Sci USA 111, 16166–16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. (2009). The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Thomas M, Dadgar N, Lieberman AP, and Iniguez-Lluhi JA (2009). Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem 284, 21296–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, and Dasso M (2017). The SUMO pathway in mitosis. Adv Exp Med Biol 963, 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, and Brill SJ (2008). Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem 283, 19912–19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, and Dejean A (1998). Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk S, Sigurdosson JO, Xiao ZY, Batth TS, Franciosa G, Von Stechow L, Lopez- Contreras AJ, Vertegaal ACO, and Olsen JV (2017). Proteomics reveals global regulation of protein SUMOylation by ATM and ATR kinases during replication stress. Cell Rep 21, 546–558. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, and Dejean A (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9, 769–779. [DOI] [PubMed] [Google Scholar]
- Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, et al. (2013). Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res 23, 1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M, and Boddy MN (2016). Cooperativity of the SUMO and Ubiquitin pathways in genome stability. Biomolecules 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie MH, and Boddy MN (2015). Pli1PIAS1 SUMO Ligase protected by the nuclear pore-associated SUMO protease Ulp1SENP1/2. J Biol Chem 290, 22678–22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niskanen EA, Malinen M, Sutinen P, Toropainen S, Paakinaho V, Vihervaara A, Joutsen J, Kaikkonen MU, Sistonen L, and Palvimo JJ (2015). Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol 16, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeser ML, Amen T, Nadel CM, Bradley AI, Reed BJ, Jones RD, Gopalan J, Kaganovich D, and Gardner RG (2016). Dynamic sumoylation of a conserved transcription corepressor prevents persistent inclusion formation during hyperosmotic stress. PloS Genet 12, e1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, and Doye V (2007). Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell 18, 2912–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kuster B, Gerstberger T, and Hurt E (2003). Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol 5, 21–27. [DOI] [PubMed] [Google Scholar]
- Pelisch F, Sonneville R, Pourkarimi E, Agostinho A, Blow JJ, Gartner A, and Hay RT (2014). Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nat Commun 5, 5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Luijsterburg MS, Acs K, Wiegant WW, Helfricht A, Herzog LK, Minoia M, Bottcher C, Salomons FA, van Attikum H, et al. (2017). Ataxin-3 consolidates the MDC1-dependent DNA double-strand break response by counteracting the SUMO-targeted ubiquitin ligase RNF4. EMBO J 36, 1066–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Fatouros C, Lee H, and Eisenhardt N (2017). SUMO conjugation - a mechanistic view. Biomol Concepts 8, 13–36. [DOI] [PubMed] [Google Scholar]
- Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA, et al. (2013). RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJP, Tainer JA, McGowan CH, and Boddy MN (2007). SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26, 4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, and Jentsch S (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820. [DOI] [PubMed] [Google Scholar]
- Raman N, Nayak A, and Muller S (2014). mTOR signaling regulates nucleolar targeting of the SUMO-specific isopeptidase SENP3. Mol Cell Biol 34, 4474–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, and Hay RT (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276, 12654–12659. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Akhter A, Dou Y, Babu J, and Theivakadadcham VSS (2017). Regulation of transcription factors by sumoylation. Transcription 8, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Furuta M, Kirkpatrick D, Gygi SP, and Azuma Y (2010). PIASy-dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J Cell Biol 191, 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Yoshida MM, Sridharan V, Kumagai A, Dunphy WG, Dasso M, and Azuma Y (2015a). SUMOylation of the C-terminal domain of DNA topoisomerase II alpha regulates the centromeric localization of Claspin. Cell Cycle 14, 2777–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HY, Wilson NR, Mehta S, Hwang SS, and Hochstrasser M (2016). Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Gene Dev 30, 1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, and Chiolo I (2015b). Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol 17, 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, de The H, and Lallemand-Breitenbach V (2014). PML nuclear bodies: Assembly and oxidative stress-sensitive sumoylation. Nucleus 5, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson DA, Wang M, and Matunis MJ (2001). The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 276, 21664–21669. [DOI] [PubMed] [Google Scholar]
- Sarangi P, Steinacher R, Altmannova V, Fu Q, Paull TT, Krejci L, Whitby MC, and Zhao X (2015). Sumoylation influences DNA break repair partly by increasing the solubility of a conserved end resection protein. PloS Genet 11, e1004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi P, and Zhao X (2015). SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci 40, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman P, Bekker-Jensen S, and Mailand N (2016). Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Bio 17, 379–394. [DOI] [PubMed] [Google Scholar]
- Seeber A, and Gasser SM (2017). Chromatin organization and dynamics in double-strand break repair. Curr Opin Genet Dev 43, 9–16. [DOI] [PubMed] [Google Scholar]
- Seeler JS, and Dejean A (2017). SUMO and the robustness of cancer. Nat Rev Cancer 17, 184–197. [DOI] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, and Pandolfi PP (2006). The mechanisms of PML-nuclear body formation. Mol Cell 24, 805. [DOI] [PubMed] [Google Scholar]
- Shin JA, Choi ES, Kim HS, Ho JC, Watts FZ, Park SD, and Jang YK (2005).SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol Cell 19, 817–828. [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, and Chen Y (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A 101, 14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar T, Lewicki MC, and Raught B (2013). A global S. cerevisiae small ubiquitin-related modifier (SUMO) system interactome. Mol Syst Biol 9, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic-Valentin N, Drzewicka K, Konig C, Schiebel E, and Melchior F (2016). Redox regulation of SUMO enzymes is required for ATM activity and survival in oxidative stress. EMBO J 35, 1312–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T, Jensen K, and Will H (1997). Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol 139, 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC, and Lima CD (2016). Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 536, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, and Karpen GH (2017). Phase separation drives heterochromatin domain formation. Nature 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YF, Yang TH, Huang HT, Liu LF, and Hwang JL (2012). Phosphorylation of Ubc9 by Cdk1 enhances sumoylation activity. PloS One 7, e34250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Leverson JD, and Hunter T (2007). Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26, 4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, and Strunnikov A (2008). Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PloS Genet 4, e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, and Murakami Y (1999). Characterization of a fission yeast SUMO-1 homologue, Pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19, 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, and Hay RT (2008). RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10, 538–546. [DOI] [PubMed] [Google Scholar]
- Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, and Stutz F (2013). The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell 51, 807–818. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, and Lisby M (2007). The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9, 923–931. [DOI] [PubMed] [Google Scholar]
- Uzoma I, Hu J, Cox E, Xia S, Zhou J, Rho H-S, Guzzo C, Paul C, Ajala O, Goodwin CR, et al. (2018). Global identification of SUMO substrates reveals crosstalk between sumoylation and phosphorylation promotes cell migration. Mol Cell Proteomics 17:871–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. (2007). Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282, 34167–34175. [DOI] [PubMed] [Google Scholar]
- Wang P, Benhenda S, Wu H, Lallemand-Breitenbach V, Zhen T, Jollivet F, Peres L, Li Y, Chen S-J, Chen Z, et al. (2018). RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat Commun 9, 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, and Zhao X (2017). Roles of SUMO in replication initiation, progression, and termination. Adv Exp Med Biol 1042, 371–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen DH, Wu JG, Wang L, and Fu Z (2017). SUMOylation promotes nuclear import and stabilization of Polo-like kinase 1 to support its mitotic function. Cell Rep 21, 2147–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Flotho A, and Melchior F (2012). The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell 46, 287–298. [DOI] [PubMed] [Google Scholar]
- Westerbeck JW, Pasupala N, Guillotte M, Szymanski E, Matson BC, Esteban C, and Kerscher O (2014). A SUMO-targeted ubiquitin ligase is involved in the degradation of the nuclear pool of the SUMO E3 ligase Siz1. Mol Biol Cell 25, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman BJ, Verheggen C, Hutten S, Lam YW, Bertrand E, and Lamond AI (2010). A proteomic screen for nucleolar SUMO targets shows sumoylation modulates the function of Nop5/Nop58. Mol Cell 39, 618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Ouyang J, Mori E, Nguyen HD, Marechal A, Hallet A, Chen DJ, and Zou L (2014). SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes Dev 28, 1472–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, and Zou L (2016). The SUMO (Small Ubiquitin-like Modifier) ligase PIAS3 primes ATR for checkpoint activation. J Biol Chem 291, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce B, Seeler JS, Thon G, Dejean A, and Arcangioli B (2004). Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J 23, 3844–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, and Hochstrasser M (2007). The yeast Hex3/Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282, 34176–34184. [DOI] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, and Dasso M (2008). Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol 183, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, and Blobel G (2005). A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102, 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilio N, Eifler-Olivi K, and Ulrich HD (2017). Functions of SUMO in the maintenance of genome stability. Adv Exp Med Biol 963, 51–87. [DOI] [PubMed] [Google Scholar]


